- 1 Laboratory of Zoonotic Pathogens, Rocky Mountain Laboratories, National Institute of Allergy and Infectious Diseases, National Institute of Health, Hamilton, MT, USA
- 2 Center for Predictive Medicine, Department of Microbiology and Immunology, University of Louisville, Louisville, KY, USA
- 3 Rocky Mountain Veterinary Branch, Rocky Mountain Laboratories, National Institute of Allergy and Infectious Diseases, National Institute of Health, Hamilton, MT, USA
Pneumonia is a common manifestation of the potentially fatal disease melioidosis, caused by the select agent bacteria Burkholderia pseudomallei. In this study we describe a new model system to investigate pulmonary melioidosis in vivo using bioluminescent-engineered bacteria in a murine respiratory disease model. Studies were performed to validate that the stable, light producing B. pseudomallei strain JW280 constitutively produced light in biologically relevant host–pathogen interactions. Hairless outbred SKH1 mice were used to enhance the ability to monitor B. pseudomallei respiratory disease, and were found to be similarly susceptible to respiratory melioidosis as BALB/c mice. This represents the first demonstration of in vivo diagnostic imaging of pulmonary melioidosis permitting the detection of B. pseudomallei less than 24 h post-infection. Diagnostic imaging of pulmonary melioidosis revealed distinct temporal patterns of bacterial colonization unique to both BALB/c and SKH1 mice. Validation of these model systems included the use of the previously characterized capsule mutant, which was found to colonize the upper respiratory tract at significantly higher levels than the wild type strain. These model systems allow for high resolution detection of bacterial pulmonary disease which will facilitate studies of therapeutics and basic science evaluation of melioidosis.
Introduction
Burkholderia pseudomallei is a Gram-negative bacterial pathogen responsible for the potentially fatal disease melioidosis. B. pseudomallei has naturally developed the resistance to numerous classes of antibiotics, and presently there is no licensed vaccine for immunoprotection. Susceptible hosts acquire melioidosis through several possible routes of infection including ingestion, percutaneous inoculation, and inhalation (Currie et al., 2000; Inglis et al., 2000). The potential for respiratory acquisition of fatal melioidosis from a bioweapon has contributed to the identification of B. pseudomallei as a category B Select Agent organism (Rotz et al., 2002).
The lung is the most commonly affected organ in human melioidosis (Wiersinga et al., 2006), and radiological imaging approaches have identified the liver and spleen as additional key sites of infection (Muttarak et al., 2008; Lim and Chong, 2010). Mice have been used as an excellent surrogate model which allow for studies of bacterial colonization at all major sites of infection, including the lung, liver, and spleen, approximating numerous aspects of clinical melioidosis (Warawa, 2010). Dissemination and progression of disease in the lung are poorly characterized phenomena in both clinical melioidosis and the mouse model due to the lack of diagnostic tools to monitor disease progression. Bioluminescently engineered B. pseudomallei were recently used to study respiratory melioidosis in mice, in which in vivo detection of bacteria was limited to the upper respiratory tract (URT) with no bioluminescent detection of pulmonary disease (Owen et al., 2009). In the same study, ex vivo bioluminescence was detected additionally in the liver, spleen, and NALT in euthanized animals. A goal of this current study was to establish the ability to detect bacterial colonization of the lung in vivo, early in the infectious process, in order to facilitate both therapeutic and basic science investigations of mechanisms of virulence employed by B. pseudomallei.
Several virulence determinants have been identified as critical to the virulence of B. pseudomallei in animal models including Type 3 Secretion, Type 6 Secretion, LPS, and capsular polysaccharide (CPS I; Deshazer et al., 1998; Reckseidler et al., 2001; Atkins et al., 2002; Stevens et al., 2004; Warawa and Woods, 2005; Burtnick et al., 2011). Of these, all but LPS have been additionally demonstrated to be important for murine respiratory melioidosis (Pilatz et al., 2006; Warawa et al., 2009). We have previously characterized the role of the capsular polysaccharide in respiratory disease and found that a capsule operon mutant is attenuated 101.8 fold and facilitates efficient initial pulmonary colonization, but that capsule is not critically required for dissemination of B. pseudomallei to the liver and spleen (Warawa et al., 2009). In developing the ability to perform diagnostic imaging of respiratory melioidosis, we included use of the capsular polysaccharide mutant to attempt to further characterize the role for this virulence determinant in pulmonary disease.
Materials and Methods
Bacterial Strains and Media
All strains, plasmids, and oligonucleotides are summarized in Table 1. Luria broth (LB; Lennox, 1955) or trypticase soy broth (dialyzed and chelated) (TSBDC; Brett et al., 1997) were used to culture B. pseudomallei strains at 37°C with shaking. E. coli strain TOP10 was used for cloning purposes. Antibiotics were used at the following concentrations: kanamycin (Km: 25 μg/ml), streptomycin (Sm: 100 μg/ml), gentamicin (Gm: 20 μg/ml), and polymyxin B (Pm: 50 μg/ml).
Chromosomal Introduction of Lux Operon
The cloning site of pGSV3-lux was modified to allow for directional cloning of DNA fragments upstream of the luxCDABE operon. A 850-bp fragment was amplified by PCR from pGSV3-lux [pGSV4 ApaI(+)/luxCAatII(−)] and cloned into pCR4. This fragment was excised from pCR4 using EcoRI/AatII, and cloned into the same restriction sites of pGSV3-lux to yield pGSV4. Subsequently, two PCR fragments from the B. pseudomallei genome [5′ara EcoRI(+)/5′ara ApaI(−) and 3′ara NotI(+)/3′ara SpeI(−)] were cloned into pGSV4 as EcoRI/ApaI (812 bp) and NotI/SpeI (867 bp) fragments to yield pGSV7. The tolC promoter was PCR amplified from B. pseudomallei genomic DNA [PtolC(+)/PtolC(−)] and cloned into pGSV7 as an NheI/BamHI fragment. A 7.6-kb EcoRI/KpnI fragment was excised from pGSV7-PtolC and cloned into the same sites of pKAS46 to yield pKAS46-araPtolC-lux, which was transformed into the E. coli strain S17-1. S17-1/ pKAS46-araPtolC-lux was conjugated with B. pseudomallei strains for the insertion of the PtolC-lux construct into the ancestrally deleted arabinose utilization operon in an allelic exchange protocol described elsewhere (Warawa et al., 2009). The DD503::PtolC-lux strain was named JW280, and the capsule mutant JW270::PtolC-lux strain was named JW280Δwcb. Strains JW280 and JW280Δwcb were identified by their ability to produce light, their resistance to streptomycin, and sensitivity to kanamycin.
Infection of Cultured Cells
J774A.1 cells were maintained in DMEM supplemented with 10% heat inactivated fetal bovine serum (Invitrogen) and grown at 37°C with 5% CO2. For infection studies, J774A.1 cells were seeded at 7.5 × 104 cells per well (100 μl) in a white 96-well plate (Greiner Bio-One) 1 day before infection. B. pseudomallei strains were subcultured 1:25 (v/v) from an LB overnight culture into TSBDC and grown at 37°C with shaking for 3 h. The bacterial concentration was estimated from OD600 measurements, bacterial suspensions were appropriately diluted into PBS, and a 5-μl aliquot of B. pseudomallei suspension was used to inoculate each well of J774A.1 cells. Four replicate plates were infected with B. pseudomallei strains JW280 and JW280Δwcb and treated with gentamicin (20 μg/ml final) 1 h after infection. Lux measurements were taken (Victor, Perkin Elmer) from one of the replicate plates at either 3, 5, 7, or 9 h post-infection, then immediately samples were processed for plate enumerations by removing the media, lysing the samples in 0.1% Triton X-100 for 5 min, serially diluting samples in PBS, and enumerating on LB plates.
Animal Infections – Survival Challenge
Murine infection studies were conducted at BSL-3 confinement and were approved by the Rocky Mountain Laboratories Biosafety and Animal Care and Use Committees in accordance with National Institutes of Health guidelines. Intranasal (i.n.) infection of groups of six 8-week-old female BALB/c mice (Charles River Laboratories) was carried out as previously described (Warawa et al., 2009), using lux+ B. pseudomallei strains JW280 and JW280Δwcb. Animals were euthanized at the onset of terminal disease symptoms, thus 50% infectious dose (ID50) values were calculated as opposed to death endpoint based lethal dose (LD50) values. Statistical analysis of survival data was conducted using GraphPad Prism 5 to evaluate significant differences in survival curves by log-rank (Mantel–Cox) test. StatPlus 2008 was used to perform Probit Analysis to estimate ID50 values for JW280 and JW280Δwcb in the SKH1 model.
Histological Analysis of Infected Tissue
Tissues were harvested from animals involved in the survival studies. Tissues were fixed in formalin, processed and stained with hematoxylin and eosin (H&E), and the histopathology of lung, liver, and spleen was scored as previously described (Warawa et al., 2009).
Time Course Lux Imaging
Groups of six BALB/c and SKH1 female 8-week-old mice (Charles River Laboratories) were infected i.n. with 30 μl of PBS bacterial suspension containing either 1 × 104 CFU JW280 or 1 × 106 CFU JW280Δwcb. BALB/c mice were shaved to reduce the amount of fur covering their dorsal thoracic cavities. Mice were anesthetized with 2.5% isoflurane and imaged at ∼12 h intervals, with 5 min exposures from the dorsal perspective using an IVIS Lumina (Caliper Life Sciences). Measurements of lung-specific lux activity were taken by selecting regions of interest (ROI; Living Image 3.0, Caliper Life Sciences) of the dorsally viewed chest cavity and area-normalized backgrounds were subtracted. Animals were euthanized at first presentation of terminal disease symptoms, and lungs were harvested for bacterial enumeration, as described previously (Warawa et al., 2009).
Data Presentation and Statistical Analysis
Unless otherwise stated, Graph Pad Prism 5 was used to plot data and conduct statistical analysis by unpaired Student’s t-test.
Results
Generating Lux+ B. pseudomallei Strains
We sought to engineer a strain of B. pseudomallei which constitutively and stably produced light from the Photorhabdus luminescens luxCDABE operon. To ensure stability of the lux operon in the absence of antibiotic selection, the operon was introduced into the chromosome by allelic exchange. The insertion site chosen for insertion of the lux operon was the site of an ancestrally deleted arabinose utilization operon, known to be present in B. thailandensis while only gene fragments are present in the B. pseudomallei genome (Moore et al., 2004). Several promoters were evaluated for their ability to constitutively produce light under a variety of growth conditions. One such promoter, PtolC, was previously demonstrated to constitutively produce high level photon emission in Pseudomonas aeruginosa (Kadurugamuwa et al., 2003). Therefore, the PtolC-lux construct was introduced into the genome of wild type B. pseudomallei strain DD503 to generate a stable Lux-expressing strain JW280. Similarly, the previously characterized capsular polysaccharide operon mutant, JW270, was engineered to produce light resulting in strain JW280Δwcb.
Intracellular Survival of Lux-Expressing B. pseudomallei
To investigate whether the light production from JW280 and JW280Δwcb correlates with bacterial numbers, B. pseudomallei strains were examined in a cell culture model system. It has been reported that B. pseudomallei survive within cultured macrophages, escape from the phagosome in a type III secretion-dependent mechanism, and replicate in the cytoplasm (Stevens et al., 2002). The capsular polysaccharide was previously found to not be critical for intracellular survival of B. pseudomallei at early time points (Warawa et al., 2009), however capsule mutants may be required for prolonged survival within macrophages (Wikraiphat et al., 2009).In a gentamicin protection assay, light producing B. pseudomallei strains JW280 and JW280Δwcb were found to infect J774A.1 macrophages and proliferate as previously reported (Figure 1A). No significant difference in growth was observed between wild type and capsule mutant at 3, 5, or 7 h post-infection. However, at 9 h post-infection the capsule mutant had fewer organisms in infected J774A.1 macrophages than wild type. This was consistent with previous reports which suggested that the capsule mutant was not as fit as wild type bacteria during late stage infection of macrophages (Wikraiphat et al., 2009). Bioluminescence was measured by plate reader immediately before harvesting samples for bacterial enumeration (counts per second, cps). Changes in both bacterial number and emitted photons were plotted as a function of time for both wild type JW280 (Figure 1B) and the capsule mutant (Figure 1C). Both JW280 and JW280Δwcb produced light consistent with bacterial number from 3 to 7 h post-infection with an average cps/CFU ratio of 2.79 × 10−4 and 3.02 × 10−4, respectively. At 9 h post-infection, relative light production increased with JW280 and JW280Δwcb ratios calculated to be 3.07 × 10−4 and 4.49 × 10−4 cps/CFU, respectively. Thus, both JW280 and JW280Δwcb produce light constitutively during host–pathogen interaction studies, with the potential for variations in the cps/CFU ratio based on bacterial fitness during these interactions.
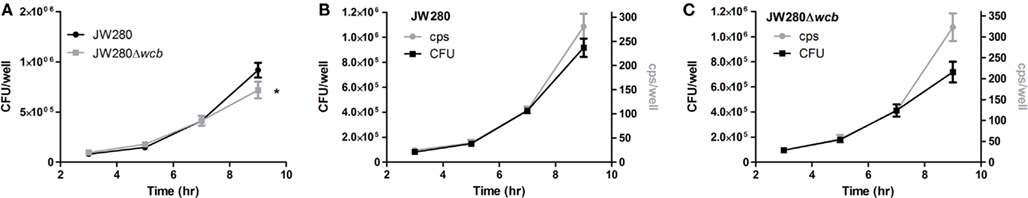
Figure 1. Intracellular survival of B. pseudomallei in J774A.1 cell line. B. pseudomallei strains JW280 and JW280Δwcb were used to infect 7.5 × 104 J774A.1 murine macrophages in triplicate at an MOI of 0.5. After 1 h, extracellular bacteria were killed with gentamicin. At time points of 3, 5, 7, and 9 h post-inoculation, the light production of triplicate set of samples was measured, then samples were lysed with 0.1% Triton X-100 and enumerated by plate counting. The CFU/well was plotted as a function of time for both JW280 and JW280Δwcb(A). Similarly both CFU/well (left y-axis) and cps/well (right y-axis) were jointly plotted as a function of time for JW280 (B) and JW280Δwcb (C). The mean and standard deviation were plotted for each strain/time point. Asterisks indicate significant difference in bacterial colonization of J774A.1 cells at specific time points (*p < 0.05).
SKH1 Mouse Susceptibility to Melioidosis
We have previously demonstrated that a B. pseudomallei capsule mutant is attenuated 101.8 fold when introduced by the i.n. route in the BALB/c mouse model (Warawa et al., 2009). Using both wild type and capsule mutant strains of B. pseudomallei, we examined whether the SKH1 mouse strain represents a viable model for the study of respiratory melioidosis, and whether Lux-producing strains of B. pseudomallei are attenuated. SKH1 mice are an immunocompetent hairless strain that potentially permit enhanced bioluminescent detection due to their lack of fur. A pairwise comparison of lux− and lux+ versions of both wild type (DD503 and JW280) and capsule mutant (JW270 and JW280Δwcb) strains were not significantly different in their virulence in BALB/c mice (Figure 2A, DD503 and JW270 survival curves reproduced as per Warawa et al., 2009). This finding indicates that introduction of the luxCDABE operon into the B. pseudomallei genome did not significantly impact its virulence in a characterized disease model. The Lux-producing wild type and capsule mutant strains were next used to compare the susceptibility of the SKH1 mouse line to melioidosis relative to the previously characterized BALB/c mouse line. Both SKH1 and BALB/c mice were similarly susceptible to respiratory melioidosis when infected with the wild type strain (Figure 2B, left panel). Interestingly, SKH1 mice were more resistant to infection with the capsule mutant than BALB/c mice (Figure 2B, right panel), suggesting that greater resolution may be observed in the SKH1 model when evaluating attenuation of bacterial strains or potentially when studying therapeutic interventions. A dose response evaluation of wild type and capsule mutant strains was next conducted in SKH1 mice to facilitate estimation of the 50% ID50 values. Probit analysis of the survival curve data (Figure 2C) was used to estimate an ID50 of 2.47 × 103 CFU (95% CI: 3.5 × 102 to 1.72 × 104 CFU) for JW280 and 5.23 × 105 CFU (95% CI: 2.47 × 105 to 1.11 × 106 CFU) for JW280Δwcb. It is noteworthy that infection with 107 CFU of JW280Δwcb produced rapid morbidity suggestive of endotoxicity rather than disease, where the ID50 of JW280Δwcb may be higher than estimated. Thus, the capsule mutant is attenuated greater than 200-fold in the SKH1 model.
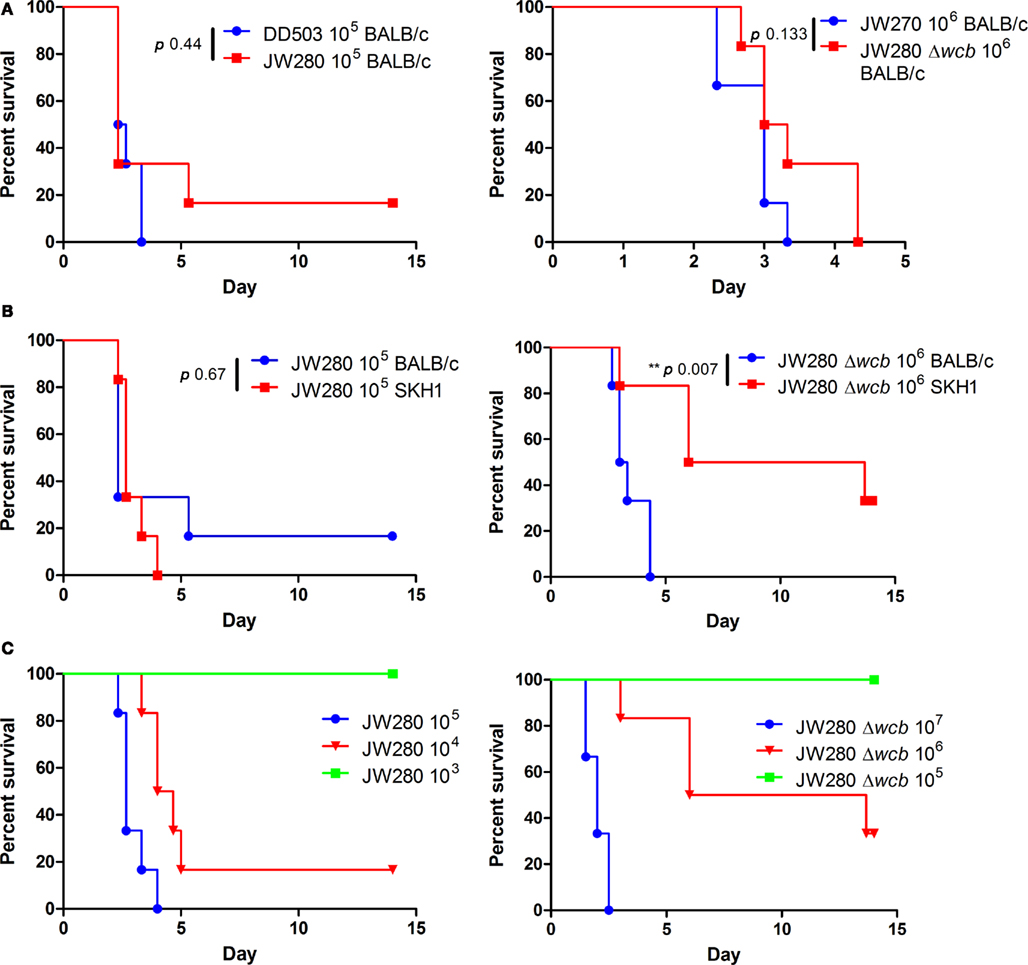
Figure 2. Virulence of Lux-producing B. pseudomallei strains. Groups of six mice were infected by the i.n. route with B. pseudomallei and monitored for 14 days. (A) Groups of BALB/c mice were infected with 105 CFU of wild type strains DD503 (lux−) or JW280 (lux+, left panel) or with 106 CFU of capsule mutant strains JW270 (lux−) or JW280Δwcb (lux+, right panel). (B) Groups of SKH1 or BALB/c mice were infected with 105 CFU of JW280 (left panel) or 106 CFU JW280Δwcb (right panel) to compare the susceptibility of each mouse strain to melioidosis. (C) SKH1 mice were infected with 3- to 10-fold serial dilutions of JW280 (103–105 CFU, left panel) or JW280Δwcb (105–107 CFU, right panel) to facilitate estimation of the ID50 for each strain. Mice were euthanized at the onset of moribund disease. Asterisks indicate significant differences between survival curves using the log-rank (Mantel–Cox) test.
Infected SKH1 tissues were harvested for evaluation of histopathological damage. As previously reported in the BALB/c model (Warawa et al., 2009), both the wild type and capsule mutant strains produce significant pathology in the lung and liver at the moribund stage of disease (Figure 3). Extensive lesions are present throughout the lung associated with inflammatory cell recruitment, fibrin deposition, and cellular necrosis. The liver pathology was characterized by multifocal lesions, and as previously reported, the capsule mutant was found to induce fewer and smaller lesions. Pathology was also detected in the nasal cavity (rhinitis), middle ear (otitis media), and brain (meningitis; data not shown). Blind scoring of the tissue histopathology was conducted to confirm that the capsule mutant indeed produced significantly reduced hepatic damage (Figure 4). It was noted, however, that neither the wild type nor capsule mutant strains induced significant pathology in the spleen. This is in contrast to our previous finding of wild type, but not capsule mutant, pathology being detected in the spleen of BALB/c mice. This suggests that B. pseudomallei may not colonize the spleen of SKH1 to the same extent as in BALB/c mice, or that the response to B. pseudomallei in the spleen of SKH1 is altered from the response in BALB/c mice.
Figure 3. Histological analysis of B. pseudomallei-infected tissues. Photomicrographs of representative tissue samples from SKH1 mice infected i.n. with JW280 (104 CFU), JW280Δwcb (106 CFU), or PBS mock-infected control animals. Samples were formalin-fixed, stained with hematoxylin and eosin (H&E), and imaged by bright field microscopy using either a ×10 (lung) or ×20 (liver) objectives.
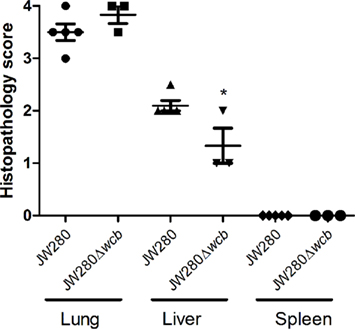
Figure 4. Histopathological scoring of SKH1 infected tissue. Lung, liver, and spleen from SKH1 mice infected i.n. with JW280 (104 CFU) or JW280Δwcb (106 CFU) were fixed, stained with H&E, and scored in a blind study for varying degrees of pathology. The scoring system was defined as: 0 = within normal limits, 1 = minimal, 2 = mild, 3 = moderate, 4 = severe. Individual data points, the mean, and standard deviation are plotted, and statistically significant variation from JW280-infected tissue scoring is indicated by asterisks (*p < 0.05).
In vivo Detection of Respiratory Tract B. pseudomallei Infection
In order to investigate the potential for in vivo diagnostic imaging using bioluminescent B. pseudomallei, SKH1 and BALB/c mice were infected i.n. with 104 CFU of JW280 or 106 CFU of JW280Δwcb, and time course in vivo imaging was conducted. Mice were euthanized at the onset of moribund disease and lung, liver, and spleen tissues were collected to enumerate bacterial colonization at key sites of infection. Both the SKH1 and BALB/c mice in the moribund stage of melioidosis were found to be colonized at similar levels by both wild type and capsule mutant strains of B. pseudomallei, with the exception that there were significantly fewer JW280 detected in the late stage infected lung of BALB/c mice relative to SKH1 mice with a 2.5-fold difference in average bacterial colonization (Figure 5). It was noted that the spleen of SKH1 mice was colonized at a similar level as the spleen of BALB/c mice, suggesting that bacterial colonization does not account for the lack of splenic pathology in SKH1 mice (Figure 4). It was also determined that the lung is the most significantly colonized organ in SKH1 infected mice with an average 104.2 fold more B. pseudomallei in the lung than the liver, and 105.4 fold more B. pseudomallei in the lung than the spleen. In BALB/c mice, the average colonization ratios are 102.8 fold lung to liver and 104.7 fold lung to spleen.
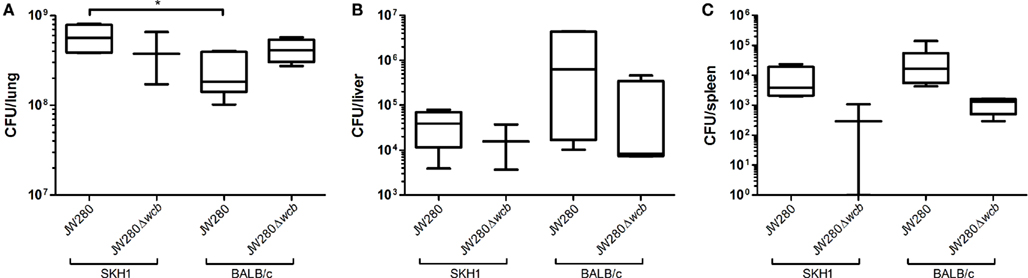
Figure 5. B. pseudomallei colonization of key sites of infection. SKH1 and BALB/c mice infected i.n. with JW280 (104 CFU) or JW280Δwcb (106 CFU) were euthanized at the onset of morbid disease symptoms, organs were harvested, homogenized in PBS and bacteria were enumerated. Data indicates the total number of bacteria detected per organ for lung (A), liver (B), and spleen (C). Significant difference between data sets indicated by asterisk (*p < 0.05).
Images of bioluminescence detection from in vivo diagnostic imaging were collected using an IVIS Lumina system. Figure 6 presents a representative collection of images demonstrating the potential to track developing pulmonary infection in mice over numerous time points in individual mice, where the last image of each panel represents moribund disease. In both mouse models and using both B. pseudomallei strains, in vivo detection of pulmonary infection was successfully achieved. Consistent with the bacterial enumeration data, less light emission is detected in the lung of JW280-infected BALB/c mice at moribund disease relative to JW280 infection of SKH1 mice.
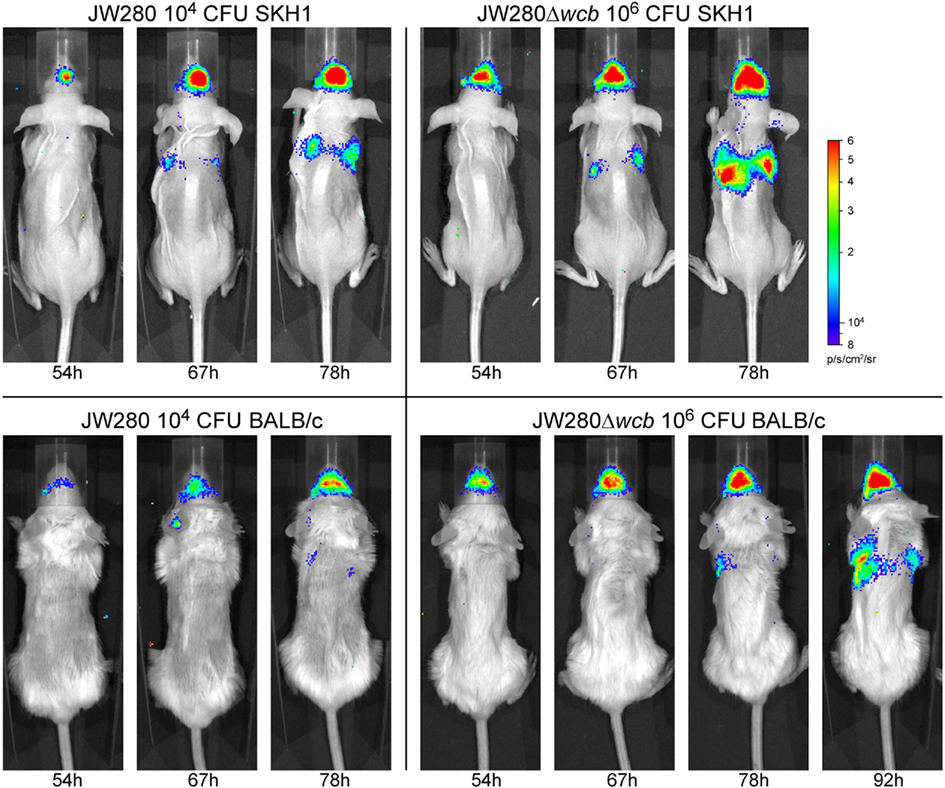
Figure 6. In vivo detection of a developing B. pseudomallei infection. Overlay images captured with an IVIS Lumina system of representative SKH1 and BALB/c mice infected i.n. with JW280 (104 CFU) or JW280Δwcb (106 CFU). Infected mice were imaged twice daily and images from late infection (54–92 h) are presented until the time point at which mice were euthanized due to terminal disease. All images were normalized to the same photon saturation scale.
Photon emission was quantified from both the URT and dorsal thoracic cavity in all imaged mice beginning 24 h post-infection. An analysis of the correlation between total emitted light (total flux) from the thoracic cavity and bacterial colonization of the lung was performed. Trend lines were constrained through the y-axis at the level of noise estimated from mock-infected animals. It was determined that BALB/c mice have a much higher level of noise (2.6 ± 0.7 × 104 p/s) than SKH1 mice (3.1 ± 2.1 × 103 p/s; **p = 0.0064), which was subsequently found to relate to the amount of residual fur present on BALB/c mice (data not shown). In both the BALB/c and SKH1 mouse models, the capsule mutant produced greater light per CFU than the wild type strain in the lung (Figure 7A) as previously observed in cell culture assays (Figure 1), suggesting that the capsule mutant may be experiencing a similar environment in vivo as that modeled in cultured murine macrophages. Using the calculated equations of the trend lines, it was estimated that the level of detection of wild type and capsule mutant bacteria in the lung of SKH1 mice is 5.7 × 106 and 2.1 × 106 CFU, respectively, at one standard deviation of noise above the average level of noise. Similarly, the BALB/c detection levels were estimated to be 2.6 × 107 and 4.4 × 106 CFU for the wild type and capsule mutant strains, respectively.
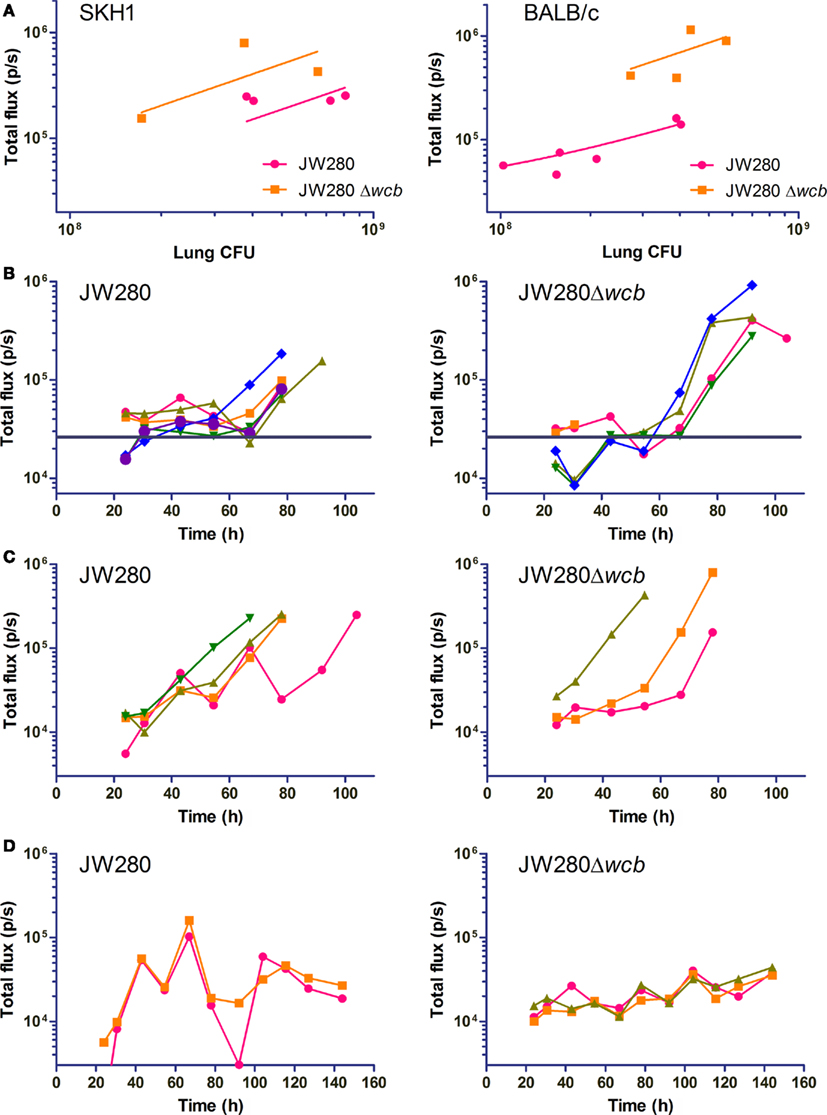
Figure 7. Dynamic assessment of lung colonization by B. pseudomallei. SKH1 and BALB/c mice were infected i.n. with JW280 (104 CFU), JW280Δwcb (106 CFU) or PBS mock-infected, and mice were imaged dorsally with an IVIS Lumina system beginning 24 h post-infection. Living Image 3.0 software was used to enumerate light emission from the lung. PBS mock-infected mice were used to establish baseline noise. At onset of morbid disease, mice were euthanized and bacteria enumerated from the lung. (A) Plot of emitted light immediately before euthanization of mice in morbid disease stage as a function of bacterial burden in the lung. The y-intercept of the trend line was constrained through the estimated noise determined from mock-infected mice. (B,C) Total flux plotted as a function of time for mice which developed morbid disease in BALB/c (B) and SKH1 (C) mice. The average noise level estimated from mock-infected mice is indicated with a horizontal bar (B) or the x-axis (C). (D) Total flux plotted as a function of time for mice which survived the pneumonia phase of disease in SKH1 mice. The average noise level estimated from mock-infected mice is set at the x-axis.
The total flux was also plotted as a function of time for wild type and capsule mutant infections of BALB/c (Figure 7B) and SKH1 (Figure 7C) mice, and the noise level indicated in each plot. The ability to sensitively detect photon emission from BALB/c mice was reduced relative to SKH1 mice, with temporal plots of total flux often overlapping the estimated noise level (Figure 7B, horizontal bar). In the BALB/c model, both the wild type and capsule mutants exhibited a biphasic expansion in the lung characterized by an initial increase in pulmonary bacterial burden until approximately 40 h post-infection, followed by a decrease in lung-associated bacteria and subsequent second phase expansion beginning at approximately 60 h post-infection (Figure 7B).
In the hairless SKH1 model, the low noise (Figure 7C, x-axis) allows for sensitive detection of both wild type and capsule mutant bacteria at 24 h post-infection, with the potential for detection at much earlier time points. Unlike the BALB/c model, a biphasic expansion is not observed, and instead both wild type and capsule mutant strains B. pseudomallei exhibit an initial lag phase prior to logarithmic expansion in the lung (Figure 7C). Of the four wild type-infected SKH1 mice which became morbidly infected, one mouse had two drops in bacterial colonization before the onset of late stage disease, indicating that individual-to-individual alterations in disease kinetics can be detected using this system. The wild type and capsule mutant strains are associated with an initial low rate expansion in the lung until a titer of approximately 3.2 × 104 organisms is achieved, at which point rapid exponential increase is observed. Statistical analysis was conducted on parsed data sets representing early and late stage expansion for both strains, and slopes from logarithmically transformed data were significantly different when comparing early to late stage colonization rates for each strain, but no intra-strain differences were statistically significant (data not shown).
Mice that survived the pneumonia phase of infection (first 5 days) were also evaluated for fluctuations in total flux as a function of time (Figure 7D). These mice were characterized as having oscillatory changes in total flux throughout the imaging conducted to day 6. Wild type-infected total flux plots trended downward from 100 h post-infection, which was consistent with the inability to culture organisms from the lung at the termination of the experiment at 14 days post-infection. In contrast, the oscillating total flux of the capsule mutant survivors (Figure 7D, right panel) trended upward, and at 14 days post-infection, a bacterial burden of 9.5 × 103 (±1.52 × 103) CFU/lung was observed. This data suggests that the capsule mutant potentially establishes a chronic infection in SKH1 mice while survivors of wild type infection effectively clear B. pseudomallei.
A striking feature of the bioluminescence overlay images is the observation of significant colonization of the URT as a result of i.n. infection (Figure 6), as observed previously (Owen et al., 2009). An evaluation of the ratio of nasal cavity total flux was conducted as a function of the level of thoracic cavity total flux in order to estimate the fold increase in number of B. pseudomallei associated preferentially with the nasal cavity. This analysis made use of mice that develop lethal pneumonia and therefore achieve high titers of B. pseudomallei in the lung. At all measured time points, the ratio of nasal cavity flux to thoracic cavity flux was included to account for temporal fluctuations. In both the SKH1 and BALB/c models, the capsule mutant had a significantly higher proportion of bacteria associated with the URT with an average of 4.5-fold higher flux in SKH1 mice and 3.1-fold higher flux in BALB/c mice (Figure 8). These data indicate that the capsule mutant colonizes the nasal cavity at a significantly higher level than wild type B. pseudomallei.
Figure 8. Preferential colonization of the nasal cavity by B. pseudomallei. SKH1 and BALB/c mice were infected i.n. with JW280 (104 CFU) or JW280Δwcb (106 CFU) and mice were imaged dorsally with an IVIS Lumina system beginning 24 h post-infection. The background corrected nasal cavity to thoracic cavity ratio was evaluated at all time points for mice that developed lethal pneumonia. The collective of all experimental ratios were plotted for each group. Significant differences between groups are indicated by asterisks (**p < 0.01, ***p < 0.001).
Discussion
This study focused on characterizing and validating a diagnostic imaging model for investigating respiratory melioidosis in mouse models. The approach we selected was use of optical imaging strategies using B. pseudomallei strains engineered to constitutively produce bioluminescence. Bioluminescence offers the advantage over fluorescent for detection of weak signals due to the enzymatic amplification of signal, and bioluminescence offers the added benefit of detecting only metabolically active bacteria as the production of light requires energy from the cell. The luxCDABE operon from P. luminescens allows for light production in the absence of addition of foreign substrates. This is in contrast to the eukaryotic luciferases which require exogenous addition of D-luciferin. We preferentially selected use of the lux operon as B. pseudomallei are known to traffic in intracellular environments in vivo, and the lux operon allows for stable access to endogenous luciferase substrate using the accessory luxCDE gene products. The same luxCDABE operon was previously introduced by transposon into B. pseudomallei strain 08, where i.n. infections of mice resulted in vivo detection bacteria only in the URT, with ex vivo analysis demonstrating bioluminescence of the liver, spleen, and NALT (Owen et al., 2009). The authors do not comment on the absence of morbid disease when infecting with 3.6 × 105 organisms, however we hypothesize that the small delivery volume (12 μl) may have contributed to a minimal pulmonary infection, and similarly the low bacterial burden reported in the lung (Owen et al., 2009). Thus, B. pseudomallei strain 08 may represent a low virulence strain, or may indicate that infections limited to the URT do not lead to morbid disease due to the importance of the lung in B. pseudomallei virulence. However, our current model has been shown to facilitate sensitive detection of virulent B. pseudomallei in cell culture and mouse disease models, allowing for ratiometric detection of changes in bacterial burdens in vivo.
The bioluminescent strain of B. pseudomallei engineered in this study, JW280, was found to maintain an excellent correlation between bacterial number and photon production in a biologically relevant macrophage cell culture model. Furthermore, stable, chromosomal introduction of the lux operon was not found to significantly attenuate B. pseudomallei virulence. Both wild type and capsule mutant B. pseudomallei strains maintained a close relationship between measured cps and CFU for the majority of time points investigated in macrophage survival assays. At a late time point, the capsule mutant was found to proliferate less efficiently than wild type in J774A.1 cells, and this reduction in fitness was associated with an increase in relative light production. Interestingly, the relative light production of the capsule mutant in the mouse lung was similarly higher than the wild type strain in both BALB/c and SKH1 mice. Prolonged incubation of a capsule mutant in cultured macrophages was previously reported to be associated with a decrease in capsule mutant viability (Wikraiphat et al., 2009), and the data from this current study suggests that the same mechanism affecting capsule mutant viability in cell culture models may be exerted in vivo during pulmonary disease.
This study represents the first characterization of SKH1 mice to investigate melioidosis. SKH1 mice are a hairless mouse model useful for studying a variety of dermatologically important events such as burn models, UV sensitivity, skin aging, and also offer the ability to enhance optical diagnostic imaging for both bioluminescent and fluorescence detection strategies (Benavides et al., 2009). The effect of fur on masking photon emission has been previously estimated to account for up to a 10-fold loss in signal in fluorescence imaging approaches (Nishijo et al., 2009). In this present study, we found that fur also contributes to the noise level which also affects sensitivity of detection, where shaved BALB/c mice have a greater than eight-fold higher noise level than SKH1 mice. Shaving was performed in preference to use of depilatory creams as it has been reported that depilatory creams allow for complete hair regrowth over a 14-day period while shaving results in much slower hair regrowth (Faia et al., 2008).
Unlike nude mice, hairless SKH1 are considered to be an immunocompetent mouse strain which we hypothesized would represent an excellent new model for bioluminescent-based diagnostic imaging of melioidosis. In this study, outbred SKH1 were found to be similarly sensitive to respiratory melioidosis as inbred BALB/c mice. Interestingly, SKH1 mice were found to be more resistant to the virulence of the capsule mutant, such that the capsule mutant attenuation was higher in the SKH1 mouse model than we previously reported in the BALB/c model. Thus, SKH1 mice may allow for higher resolution determination of attenuation when investigating roles of bacterial virulence determinants. We have also demonstrated that SKH1 mice provide a benefit over BALB/c mice in that they: (i) do not have to be shaved, (ii) have no residual fur to mask photon emission or create noise, and (iii) allow for higher levels of lung colonization of wild type B. pseudomallei. The most significant caveat we observed in the SKH1 model is the lack of splenic pathology when infected with wild type B. pseudomallei even though the bacterial burden in SKH1 mice is similar to that of BALB/c mice. We previously characterized the role of the capsular polysaccharide in stimulating a proinflammatory response in the liver and spleen of BALB/c mice associated with wild type-specific elevations in histopathologic damage in these tissues (Warawa et al., 2009). Splenic pathology is a hallmark of clinical melioidosis (Laopaiboon et al., 2009), and it is therefore a concern that SKH1 mice do not allow for a full study of the host response to melioidosis in the spleen. SKH1 mice possess a retroviral leukemia virus which interrupts the hr gene resulting in the autosomal recessive hairless phenotype, and have been reported to possess altered subpopulations of naïve and memory T cells relative to other common mouse strains (Schaffer et al., 2010). While symptoms of leukemia do not exhibit until mice reach adulthood, it is possible that T cell sub-population alterations in 8-week-old SKH1 mice lead to the lack of the host response triggering splenic pathology. Thus, SKH1 mice represent an excellent model for the study of pulmonary disease kinetics during B. pseudomallei infection, but caution should be taken if this strain is used to study the host response in disseminated tissues.
Optical imaging strategies for the detection of B. pseudomallei in mice appear to be limited to detection of pathogen specifically in the lung. We estimated the level of detection of B. pseudomallei in the lung to be 106.3–106.8 CFU in SKH1 mice, and 106.6–107.4 CFU in BALB/c mice. At terminal disease, B. pseudomallei burdens do not exceed 105 CFU in the liver or spleen of BALB/c or SKH1 mice with the exception of the livers of BALB/c-infected mice which may exceed 106 CFU. It is possible that some punctate visualization of hepatic colonization may be detected during terminal disease, but the ability to monitor disease kinetics in the liver or spleen during murine melioidosis may not be achievable with optical diagnostic imaging. Engineering of new B. pseudomallei strains that produce higher levels of light may allow for limited detection in the spleen, but it is likely that the close proximity of the liver to the lung, the significant pigmentation of the liver, and the relatively higher pulmonary colonization rates may mask the hepatic signal. Some strains of B. pseudomallei also exhibit altered organ colonization patterns, such as the NCTC 13178 strain which does not colonize the lung at high titer, even when delivered intranasally (Barnes and Ketheesan, 2005). Similarly, the lux+ B. pseudomallei 08 strain did not colonize the lung at high titer when introduced intranasally (Owen et al., 2009), but it is unclear whether this is due to the low delivery volume or low strain virulence, as discussed above. Therefore not all B. pseudomallei strains may be amenable to bioluminescent engineering for the in vivo detection of pulmonary disease.
Diagnostic imaging was used in this study to identify unique patterns of bacterial expansion in the lungs of BALB/c and SKH1 mice. BALB/c mice were found to be associated with a biphasic expansion while SKH1 mice were more typically associated with a low initial rate of expansion prior to rapid exponential expansion. Net bacterial expansion in the lung can be affected by several variables including: (i) variation in bacterial growth rates in different host niches, (ii) host immune-mediated clearance of bacteria, and (iii) reduction of lung colonization due to dissemination to other key sites of infection. Additional studies will be required to characterize the contribution of each of these variables on the biphasic expansion in BALB/c mice and lag expansion in SKH1 mice. Importantly, this study highlights how diagnostic imaging can critically impact our understanding of how melioidosis progresses in the lung in a mouse strain specific manner. Respiratory disease in the mouse model is typically very acute with symptoms of lethal pneumonia presenting within 3–4 days post-infection. The findings of this study suggest that use of arbitrary time designations to identify stages of disease (e.g., 24 h post-infection designation of early disease) may not allow for meaningful study of host–pathogen interaction in vivo. Diagnostic imaging is therefore a critically important tool to aid in high resolution definition of host–pathogen interactions in the study of both host and pathogen responses during infection.
A phenotype identified for the capsular polysaccharide B. pseudomallei mutant in this study was the elevated colonization of the URT during infection. The high degree of nasal cavity colonization for both the wild type and capsule mutant strains had been an unexpected observation, with the nasal cavity bacterial burden exceeding that of the lung in the majority of instances of diagnostic imaging. This finding suggests that the murine nasal cavity is a primary colonization niche for B. pseudomallei delivered by the i.n. route, while we have not found evidence to suggest that clinical melioidosis is associated with similar URT carriage. The reason for a potential difference between human and murine URT carriage may be due to opportunistic colonization in mice due to the high relative surface area, where mice have been reported to have over 100-fold greater relative surface area than the human nasal cavity (Warawa, 2010). Alternatively, normal resident flora specific to the human URT may outcompete B. pseudomallei in interspecies competition, a reported phenomenon for other respiratory pathogens (Lysenko et al., 2005). Additional studies are required to characterize the murine-specific tolerance for URT-related melioidosis and the role of capsular polysaccharide in directing elevated URT colonization, which may shed light on our general understanding of B. pseudomallei colonization of mucosal surfaces.
In conclusion, we have characterized the first in vivo pulmonary diagnostic imaging model of melioidosis using a fully virulent, stable, bioluminescent strain of B. pseudomallei. We validated the ability to detect B. pseudomallei in the lung of infected mice in both BALB/c and SKH1 mice, with the higher level of sensitivity being achieved in SKH1 mice. Different patterns of bacterial expansion in the lung were observed in the two mouse strains suggesting that altered kinetics of host response and/or dissemination profiles may affect disease in these strains. We also identified the high propensity for URT colonization by B. pseudomallei with the intact capsular polysaccharide modulating the level of colonization of the nasal cavity. These findings highlight the importance of using diagnostic imaging to characterize in vivo models of melioidosis.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by the Intramural Research Program of the NIH, NIAID. We thank Dr. Matthew Lawrenz for his critical evaluation of this manuscript.
References
Atkins, T., Prior, R., Mack, K., Russell, P., Nelson, M., Prior, J., Ellis, J., Oyston, P. C., Dougan, G., and Titball, R. W. (2002). Characterisation of an acapsular mutant of Burkholderia pseudomallei identified by signature tagged mutagenesis. J. Med. Microbiol. 51, 539–547.
Barnes, J. L., and Ketheesan, N. (2005). Route of infection in melioidosis. Emerging Infect. Dis. 11, 638–639.
Benavides, F., Oberyszyn, T. M., Vanbuskirk, A. M., Reeve, V. E., and Kusewitt, D. F. (2009). The hairless mouse in skin research. J. Dermatol. Sci. 53, 10–18.
Brett, P. J., Deshazer, D., and Woods, D. E. (1997). Characterization of Burkholderia pseudomallei and Burkholderia pseudomallei-like strains. Epidemiol. Infect. 118, 137–148.
Burtnick, M. N., Brett, P. J., Harding, S. V., Ngugi, S. A., Ribot, W. J., Chantratita, N., Scorpio, A., Milne, T. S., Dean, R. E., Fritz, D. L., Peacock, S. J., Prior, J. L., Atkins, T. P., and Deshazer, D. (2011). The cluster 1 type VI secretion system is a major virulence determinant in Burkholderia pseudomallei. Infect. Immun. 79, 1512–1525.
Currie, B. J., Fisher, D. A., Howard, D. M., Burrow, J. N., Selvanayagam, S., Snelling, P. L., Anstey, N. M., and Mayo, M. J. (2000). The epidemiology of melioidosis in Australia and Papua New Guinea. Acta Trop. 74, 121–127.
Deshazer, D., Brett, P. J., and Woods, D. E. (1998). The type II O-antigenic polysaccharide moiety of Burkholderia pseudomallei lipopolysaccharide is required for serum resistance and virulence. Mol. Microbiol. 30, 1081–1100.
Faia, K., Mandley, E., Dembski, S., Pien, C., Macdougall, J., Hoyt, J., Campbell, M., Lin, G., Lo, P., Read, M., and Mcgovern, K. (2008). “Abstract #2827: depilation induced anagen as a model to study hedgehog pathway antagonist IPI-926 implications for biomarker development,” in Annual Meeting of the American Association of Cancer Research, San Diego, CA.
Inglis, T. J., Garrow, S. C., Henderson, M., Clair, A., Sampson, J., O’Reilly, L., and Cameron, B. (2000). Burkholderia pseudomallei traced to water treatment plant in Australia. Emerging Infect. Dis. 6, 56–59.
Kadurugamuwa, J. L., Sin, L., Albert, E., Yu, J., Francis, K., Deboer, M., Rubin, M., Bellinger-Kawahara, C., Parr, T. R. Jr., and Contag, P. R. (2003). Direct continuous method for monitoring biofilm infection in a mouse model. Infect. Immun. 71, 882–890.
Laopaiboon, V., Chamadol, N., Buttham, H., and Sukeepaisarnjareon, W. (2009). CT findings of liver and splenic abscesses in melioidosis: comparison with those in non-melioidosis. J. Med. Assoc. Thai. 92, 1476–1484.
Lennox, E. S. (1955). Transduction of linked genetic characters of the host by bacteriophage P1. Virology 1, 190–206.
Lim, K. S., and Chong, V. H. (2010). Radiological manifestations of melioidosis. Clin. Radiol. 65, 66–72.
Lysenko, E. S., Ratner, A. J., Nelson, A. L., and Weiser, J. N. (2005). The role of innate immune responses in the outcome of interspecies competition for colonization of mucosal surfaces. PLoS Pathog. 1, e1. doi: 10.1371/journal.ppat.0010001
Moore, R. A., Deshazer, D., Reckseidler, S., Weissman, A., and Woods, D. E. (1999). Efflux-mediated aminoglycoside and macrolide resistance in Burkholderia pseudomallei. Antimicrob. Agents Chemother. 43, 465–470.
Moore, R. A., Reckseidler-Zenteno, S., Kim, H., Nierman, W., Yu, Y., Tuanyok, A., Warawa, J., Deshazer, D., and Woods, D. E. (2004). Contribution of gene loss to the pathogenic evolution of Burkholderia pseudomallei and Burkholderia mallei. Infect. Immun. 72, 4172–4187.
Muttarak, M., Peh, W. C., Euathrongchit, J., Lin, S. E., Tan, A. G., Lerttumnongtum, P., and Sivasomboon, C. (2008). Spectrum of imaging findings in melioidosis. Br. J. Radiol. 82, 514–521.
Nishijo, K., Hosoyama, T., Bjornson, C. R., Schaffer, B. S., Prajapati, S. I., Bahadur, A. N., Hansen, M. S., Blandford, M. C., Mccleish, A. T., Rubin, B. P., Epstein, J. A., Rando, T. A., Capecchi, M. R., and Keller, C. (2009). Biomarker system for studying muscle, stem cells, and cancer in vivo. FASEB J. 23, 2681–2690.
Owen, S. J., Batzloff, M., Chehrehasa, F., Meedeniya, A., Casart, Y., Logue, C. A., Hirst, R. G., Peak, I. R., Mackay-Sim, A., and Beacham, I. R. (2009). Nasal-associated lymphoid tissue and olfactory epithelium as portals of entry for Burkholderia pseudomallei in murine melioidosis. J. Infect. Dis. 199, 1761–1770.
Pilatz, S., Breitbach, K., Hein, N., Fehlhaber, B., Schulze, J., Brenneke, B., Eberl, L., and Steinmetz, I. (2006). Identification of Burkholderia pseudomallei genes required for the intracellular life cycle and in vivo virulence. Infect. Immun. 74, 3576–3586.
Reckseidler, S. L., Deshazer, D., Sokol, P. A., and Woods, D. E. (2001). Detection of bacterial virulence genes by subtractive hybridization: identification of capsular polysaccharide of Burkholderia pseudomallei as a major virulence determinant. Infect. Immun. 69, 34–44.
Rotz, L. D., Khan, A. S., Lillibridge, S. R., Ostroff, S. M., and Hughes, J. M. (2002). Public health assessment of potential biological terrorism agents. Emerging Infect. Dis. 8, 225–230.
Schaffer, B. S., Grayson, M. H., Wortham, J. M., Kubicek, C. B., Mccleish, A. T., Prajapati, S. I., Nelon, L. D., Brady, M. M., Jung, I., Hosoyama, T., Sarro, L. M., Hanes, M. A., Rubin, B. P., Michalek, J. E., Clifford, C. B., Infante, A. J., and Keller, C. (2010). Immune competency of a hairless mouse strain for improved preclinical studies in genetically engineered mice. Mol. Cancer Ther. 9, 2354–2364.
Simon, R., Priefer, U., and Pühler, A. (1983). A broad range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram-negative bacteria. Biotechnology 1, 784–791.
Skorupski, K., and Taylor, R. K. (1996). Positive selection vectors for allelic exchange. Gene 169, 47–52.
Stevens, M. P., Haque, A., Atkins, T., Hill, J., Wood, M. W., Easton, A., Nelson, M., Underwood-Fowler, C., Titball, R. W., Bancroft, G. J., and Galyov, E. E. (2004). Attenuated virulence and protective efficacy of a Burkholderia pseudomallei bsa type III secretion mutant in murine models of melioidosis. Microbiology 150, 2669–2676.
Stevens, M. P., Wood, M. W., Taylor, L. A., Monaghan, P., Hawes, P., Jones, P. W., Wallis, T. S., and Galyov, E. E. (2002). An Inv/Mxi-Spa-like type III protein secretion system in Burkholderia pseudomallei modulates intracellular behaviour of the pathogen. Mol. Microbiol. 46, 649–659.
Warawa, J., and Woods, D. E. (2005). Type III secretion system cluster 3 is required for maximal virulence of Burkholderia pseudomallei in a hamster infection model. FEMS Microbiol. Lett. 242, 101–108.
Warawa, J. M. (2010). Evaluation of surrogate animal models of melioidosis. Front. Microbio. 1:141. doi: 10.3389/fmicb.2010.00141
Warawa, J. M., Long, D., Rosenke, R., Gardner, D., and Gherardini, F. C. (2009). Role for the Burkholderia pseudomallei capsular polysaccharide encoded by the wcb operon in acute disseminated melioidosis. Infect. Immun. 77, 5252–5261.
Wiersinga, W. J., Van Der Poll, T., White, N. J., Day, N. P., and Peacock, S. J. (2006). Melioidosis: insights into the pathogenicity of Burkholderia pseudomallei. Nat. Rev. Microbiol. 4, 272–282.
Wikraiphat, C., Charoensap, J., Utaisincharoen, P., Wongratanacheewin, S., Taweechaisupapong, S., Woods, D. E., Bolscher, J. G., and Sirisinha, S. (2009). Comparative in vivo and in vitro analyses of putative virulence factors of Burkholderia pseudomallei using lipopolysaccharide, capsule and flagellin mutants. FEMS Immunol. Med. Microbiol. 56, 253–259.
Keywords: bioluminescent diagnostic imaging, hairless mouse model, upper respiratory tract infection, pulmonary disease, Burkholderia pseudomallei, melioidosis, capsular polysaccharide, intranasal infection
Citation: Warawa JM, Long D, Rosenke R, Gardner D and Gherardini FC (2011) Bioluminescent diagnostic imaging to characterize altered respiratory tract colonization by the Burkholderia pseudomallei capsule mutant. Front. Microbio. 2:133. doi: 10.3389/fmicb.2011.00133
Received: 01 May 2011;
Paper pending published: 13 May 2011;
Accepted: 06 June 2011;
Published online: 16 June 2011.
Edited by:
Alfredo G. Torres, University of Texas Medical Branch, USAReviewed by:
Miguel A. Valvano, University of Western Ontario, CanadaIfor Beacham, Griffith University, Australia
Copyright: © 2011 Warawa, Long, Rosenke, Gardner and Gherardini. This is an open-access article subject to a non-exclusive license between the authors and Frontiers Media SA, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and other Frontiers conditions are complied with.
*Correspondence: Jonathan M. Warawa, Center for Predictive Medicine, Department of Microbiology and Immunology, University of Louisville, 505 South Hancock Street, CTRB 619, Louisville, KY 40202, USA. e-mail: jonathan.warawa@ louisville.edu

 Dan Long3
Dan Long3