- 1 Department of Biology, East Carolina University, Greenville, NC, USA
- 2 School of Oceanography, University of Washington, Seattle, WA, USA
Ultramafic rocks in the Earth’s mantle represent a tremendous reservoir of carbon and reducing power. Upon tectonic uplift and exposure to fluid flow, serpentinization of these materials generates copious energy, sustains abiogenic synthesis of organic molecules, and releases hydrogen gas (H2). In order to assess the potential for microbial H2 utilization fueled by serpentinization, we conducted metagenomic surveys of a marine serpentinite-hosted hydrothermal chimney (at the Lost City hydrothermal field) and two continental serpentinite-hosted alkaline seeps (at the Tablelands Ophiolite, Newfoundland). Novel [NiFe]-hydrogenase sequences were identified at both the marine and continental sites, and in both cases, phylogenetic analyses indicated aerobic, potentially autotrophic Betaproteobacteria belonging to order Burkholderiales as the most likely H2-oxidizers. Both sites also yielded metagenomic evidence for microbial H2 production catalyzed by [FeFe]-hydrogenases in anaerobic Gram-positive bacteria belonging to order Clostridiales. In addition, we present metagenomic evidence at both sites for aerobic carbon monoxide utilization and anaerobic carbon fixation via the Wood–Ljungdahl pathway. In general, our results point to H2-oxidizing Betaproteobacteria thriving in shallow, oxic–anoxic transition zones and the anaerobic Clostridia thriving in anoxic, deep subsurface habitats. These data demonstrate the feasibility of metagenomic investigations into novel subsurface habitats via surface-exposed seeps and indicate the potential for H2-powered primary production in serpentinite-hosted subsurface habitats.
Introduction
The potentially vast microbial diversity and biomass of the subsurface biosphere (Whitman et al., 1998) has been frequently noted (Biddle et al., 2006; Huber et al., 2007; Santelli et al., 2008; Schrenk et al., 2010), but there is very little evidence to indicate how much of it is supported by new primary production or by recycling of buried organic carbon. Earth’s mantle is primarily composed of ultramafic rocks that undergo a geochemical process known as serpentinization when they are tectonically uplifted into the crust and exposed to water. Serpentinization is highly exothermic and can release large quantities of hydrogen gas (H2) and variable amounts of methane and low-molecular weight organic compounds (McCollom and Seewald, 2007; Proskurowski et al., 2008). Therefore, serpentinization is a potential source of reducing power and organic carbon for organisms inhabiting the ultramafic subsurface. Actively serpentinizing rocks are present on all of the world’s continents and comprise significant portions of the deep seafloor, and yet they are some of the most poorly understood portions of the biosphere.
The most dramatic example of an ecosystem supported by serpentinization is the Lost City hydrothermal field, which is situated on a serpentinite-rich massif 15 km from the Mid-Atlantic Ridge. Carbonate chimneys at Lost City vent warm (up to 90°C), pH 9–11 fluids rich in calcium, H2 (up to 14 mmol/kg), and methane (1–2 mmol/kg; Kelley et al., 2005). Methane and larger hydrocarbon chains with up to four carbon atoms in Lost City fluids show evidence of an abiogenic origin in the deep subsurface (Proskurowski et al., 2008), but the amount of microbial activity supported by this abiotic source of organics has not been quantified. The anoxic interiors of Lost City carbonate chimneys are dominated by Methanosarcinales-related archaea potentially involved in methane production and oxidation (Schrenk et al., 2004; Brazelton et al., 2011). The oxic chimney exteriors are dominated by aerobic methane- and sulfur-oxidizing bacteria (Brazelton et al., 2006, 2010). The role of H2-metabolizing bacteria in Lost City chimneys, though, has not been explicitly investigated.
In addition to marine hydrothermal systems such as Lost City, Ophiolites abducted onto continents also provide a potential window into subsurface habitats supported by serpentinization. The Tablelands Ophiolite in western Newfoundland, Canada features extensive ultramafic exposures including serpentinites associated with the seepage of highly reducing, pH 12 fluids, and extensive calcium carbonate (travertine) deposits. The Tablelands fluids are enriched in calcium, H2 (up to ∼500 μM), and methane (Szponar et al., submitted). The geochemical similarities between the alkaline springs at the Tablelands and the vent fluids at Lost City strongly suggest that they are the surface expressions of ongoing serpentinization-associated reactions occurring in the underlying ultramafic subsurface.
In order to identify potential inhabitants of the ultramafic subsurface in both marine and continental settings, we conducted a metagenomic survey of two Tablelands alkaline springs as a comparison to a previously published metagenome from a Lost City chimney (Brazelton and Baross, 2009, 2010). In this report, we focus on the potential for H2-fueled microbial activity by investigating the incidence and diversity of sequences encoding hydrogenase enzymes. Hydrogenases catalyze the reversible conversion between molecular hydrogen and its component protons and electrons: H2↔2H+ + 2e−. This reaction is catalyzed by two main classes of hydrogenase: [NiFe]-hydrogenases are required for uptake and oxidation of H2, while [FeFe]-hydrogenases are typically involved in microbial H2 production. Although the two classes share some sequence similarity, they do not appear to be monophyletic (Vignais et al., 2001; Vignais and Billoud, 2007). Therefore, phylogenetic analyses of possible hydrogenase-encoding sequences should reliably indicate a genetic potential for H2 oxidation or H2 production. The results presented here indicate that both types of hydrogenase are abundant in the Tablelands and Lost City metagenomes and that the identity of the H2-metabolizing organisms at both sites are intriguingly similar. Additional metagenomic evidence also indicates the potential for carbon fixation pathways involving carbon monoxide utilization or acetogenesis by H2-metabolizing organisms. In general, this initial metagenomic survey highlights the potential for H2-fueled primary production in the ultramafic subsurface.
Materials and Methods
Lost City Metagenome
The Lost City metagenomic dataset included in this analysis was previously described in Brazelton and Baross(2009, 2010). In summary, a ∼1 kg carbonate chimney sample (H03_072705_R0424) was collected from the central “Poseidon” edifice (Figure 1A) of the Lost City hydrothermal field on 27 July 2005 by the DSV Hercules during the 2005 Lost City expedition aboard the R/V Ronald H. Brown. The fluids venting from this chimney can reach 88°C, pH 10.7, and are highly enriched in H2 (13 mmol/kg) and methane (1.5 mmol/kg; Proskurowski et al., 2008). DNA extraction and purification is described in Brazelton and Baross (2010). Shotgun library construction and Sanger end-sequencing of pUC18 inserts were conducted according to standard protocols at the DOE Joint Genome Institute in 2007. Two libraries were constructed from two subsamples of the same carbonate chimney sample. Reads from both libraries were combined for assembly and for analyses described here. Mean read length for the 46,360 reads from both libraries was 755 bp, and the mean length of all 6324 contigs was 1583 bp, with a maximum of 24 kb. All sequencing reads and assembled contigs are publicly available on the MG-RAST (Meyer et al., 2008) server under project name “Lost City hydrothermal field” and MG-RAST IDs 4461585.3 (reads) and 4470602.3 (contigs).
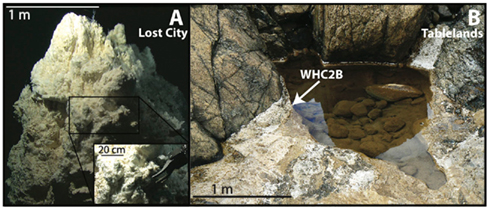
Figure 1. Photographs of the Lost City (A) and Tablelands (B) sites where metagenome samples were collected. The Lost City metagenome sample was grabbed by the manipulator on the ROV Hercules, comprising approximately 1 kg of carbonate minerals and associated biofilm material (A inset) from the Poseidon pinnacle (A) at the Lost City hydrothermal field. The WHC2b metagenome sample was collected by filtering 10 L of fluid from a pH 12 pool approximately 2.5 m long, 1 m wide, and less than 1 m deep (B). Fluid was collected by gentle suction from a depression in the bottom of the pool underneath a ledge [arrow in (B)], which appeared to be a source of the spring fluid. Travertine (carbonate) deposits are visible around the edge of the pool. Lost City photo credit: Deborah Kelley, U. of Washington, IFE, URI–IAO, NOAA.
Collection of Tablelands Samples
Two springs ∼2 km apart were sampled for this study. A fluid sample from the WHC2b spring was collected from the bottom of a pH 12 pool (∼1 m deep) surrounded by travertine (carbonate) deposits in Winter House Canyon on 25 August 2010. The sampling tubing inlet was placed at the highest pH (12.06) and lowest Eh (−733 mV) location detected at the time of sampling, which was a depression at the bottom edge of the pool (Figure 1B). The pool depth is less than 1 m, so the pool represents a very strong redox gradient from the surface (which is in contact with the atmosphere) to the anoxic bottom. At the time of sampling, the measured H2 concentration was ∼0.24 mmol/kg, and a maximum of 0.52 mmol/kg was measured at the same location one year later (Szponar et al., submitted). These concentrations are much lower than that of the astonishingly H2-rich Lost City fluids, but are nevertheless indicative of significant H2 enrichment over background surface waters. The surface temperature of the pool is determined by the air temperature, and the bottom of the pool is typically 1–4°C cooler than the surface. The pH and Eh of fluid flowing through the tube was monitored during sampling to verify that ambient fluid with more neutral pH or more oxidizing Eh did not dilute the sample. Approximately 10 L of fluid was collected from WHC2b by repeated pulls with a 60 mL sterile syringe and stored in sterile bottles until filtration through four replicate 0.2 μm Sterivex (Millipore) filter cartridges. Sterivex filters were immediately frozen in liquid N2, transported on dry ice, and stored at –80°C until DNA extraction.
A fluid sample was collected from a spring (called TLE) associated with a large travertine deposit on the northeastern face of Table Mountain on 17 June 2010. The spring fluid was visibly mixing with surface runoff, as evidenced by the lower pH and higher Eh compared to WHC2b: pH 10.5 and Eh + 25 mV. No H2 was detected at the time of sampling, but concentrations of 65 and 180 μM have been measured at this location by prior sampling efforts (Szponar et al., submitted). The temperature of the spring was not measured at the time of sampling, but subsequent measurements have shown that it is typically much cooler than air temperature (∼9°C with air temperature of ∼20°C) due to its subsurface source and/or its mixing with runoff from snowmelt. The spring was sampled by repeated pulls with a 60 mL sterile syringe for a total of 1.2 L stored in sterile bottles. Filtration, transportation, and storage of the TLE sample was conducted as described above for the WHC2b sample.
DNA Extraction and Sequencing of Tablelands Samples
DNA extraction from Sterivex filters was conducted according to the protocol described by Huber et al. (2002) and Sogin et al. (2006). The DNA yield from the TLE sample was insufficient for metagenomic sequencing, so a minimal amount of whole genome amplification (WGA) was conducted according to the following protocol. RepliG (Qiagen) WGA reactions were incubated for only 3 h according to the manufacturer’s instructions, followed by inactivation of the enzyme at 65°C for 3 min. Four replicate RepliG WGA reactions were pooled, and amplification products were purified with QiAamp (Qiagen) columns, treated with S1 Nuclease at 37°C for 1hr, purified with phenol/chloroform/isoamyl alcohol, and finally precipitated with ethanol and sodium acetate. TRFLP profiles of 16S rRNA amplicons from TLE pre- and post-WGA were nearly identical (86% Sorensen similarity; 75% Jaccard similarity, calculated with EstimateS1 after clustering peaks according to the method of (Abdo et al., 2006). Differences between pre- and post-WGA TRFLP profiles involved the loss of a few minor peaks and moderate increase in the size of major peaks in the post-WGA profiles. Therefore, WGA may have limited the diversity of rare sequences in the TLE metagenomic dataset, but we do not expect any taxonomic bias in the most abundant sequences. Approximately 1 μg of DNA from each sample was submitted to the Engencore facility (University of South Carolina) for shotgun metagenomic pyrosequencing on a Roche Titanium sequencer.
Metagenomic Analysis and Assembly
Low-quality ends of sequencing reads were trimmed in Geneious (Biomatters Ltd.) with default parameters, and reads were dereplicated with the server2 described by Gomez-Alvarez et al. (2009). Dereplicated reads (80,830 reads from 118,348 original reads for WHC2b; 56,965 reads from 135,538 original reads for TLE) were used for de novo metagenomic assembly in Geneious with default parameters for “medium sensitivity.” Taxonomic classification of WHC2b contigs > 2 kb was conducted with the TaxSom server (Weber et al., 2010) with a pre-calculated model for all Archaea and Bacteria. Automated annotation of open reading frames in contig WHC2b.C1 was performed by the RAST server (Aziz et al., 2008). All sequencing reads and assembled contigs for both samples are available on the MG-RAST (Meyer et al., 2008) server under project name “Serpentinite springs” and MG-RAST IDs 4460689.3, 4460690.3, 4461618.3, and 4461619.3. Furthermore, MIMS-compliant metadata are provided in Table A1 in Appendix.
Phylogenetic Analyses
Hydrogenase sequences were detected in metagenomic datasets by TBLASTN (Altschul et al., 1997) using as query a file of PFAM (Finn et al., 2008) seed sequences for the large subunits of [FeFe]-hydrogenase (PF02906) or [NiFe]-hydrogenase (PF00374). For comparative purposes, we included in this tblastn search additional metagenomic datasets that are publicly available in the MG-RAST (Meyer et al., 2008) and DOE Joint Genome Institute IMG/M (Markowitz et al., 2006) databases. For metagenomic reads with tblastn E-values better than 10−5, the aligned regions were translated and included in further phylogenetic analyses. All phylogenetic trees involving metagenomic sequences (Figures 3 and 5–7; Figures A1 and A2 in Appendix) were constructed with a multi-step approach utilizing reference alignments and trees in order to minimize errors and biases introduced by the fragmentary and non-overlapping nature of the metagenomic sequences. For each tree, a multiple sequence alignment was constructed with ClustalX (Larkin et al., 2007) using sequences from the studies by (Vignais et al., 2001; Boyd et al., 2009, 2010) and additional close relatives found in GenBank by blastx. Unaligned metagenomic fragments were added to the reference alignment profile with ClustalX in order to avoid alteration of the relative positions of residues in the reference alignment. Next, a bootstrapped maximum-likelihood tree was constructed from the reference-only alignment using the “-f a” algorithm in RAxML version 7.0.3 (Stamatakis, 2006). The reference-only tree with the highest-likelihood was used as a constraint tree (“-r” flag in RAxML) for 100 inferences from the full alignment (including metagenomic fragments) by RAxML, and bootstrap support values were drawn on the highest-likelihood tree. CODH and ACS trees were constructed using the same method; CODH reference sequences were obtained from (Cunliffe et al., 2008), and ACS reference sequences were obtained from (Gagen et al., 2010). The closest relatives to the 16S rRNA sequences were identified with the aid of the SILVA aligner and database (Pruesse et al., 2007) visualized in ARB (Ludwig et al., 2004). The alignment was exported from ARB, re-aligned with MUSCLE version 3.6 (Edgar, 2004), manually corrected, and built into a bootstrapped maximum-likelihood tree with RaxML.
Results
Taxonomic Analysis of Metagenomic Assemblies
A previous study (Brazelton and Baross, 2010) demonstrated that almost all of the large contigs assembled from Lost City chimney shotgun metagenomic sequences contain open reading frames with significant sequence similarity to at least one protein encoded by the genome of Thiomicrospira crunogena XCL-2, indicating that Thiomicrospira-related organisms dominated the metagenomic assembly.
Figure 2 displays the %GC and length of the 907 contigs with length > 2 kb obtained from the assembly of WHC2b shotgun metagenomic sequencing reads. Despite relatively low sequencing effort, de novo assembly yielded moderately large contigs and revealed a few dominant populations, highlighting the low diversity of the spring fluid. Most of the largest contigs were taxonomically classified by the TaxSOM server (Weber et al., 2010) as members of the Comamonadaceae family within order Burkholderiales (Figure 2). A secondary assemblage of low %GC Firmicutes contigs (orders Bacillales and Clostridiales) is also evident in WHC2b (Figure 2).
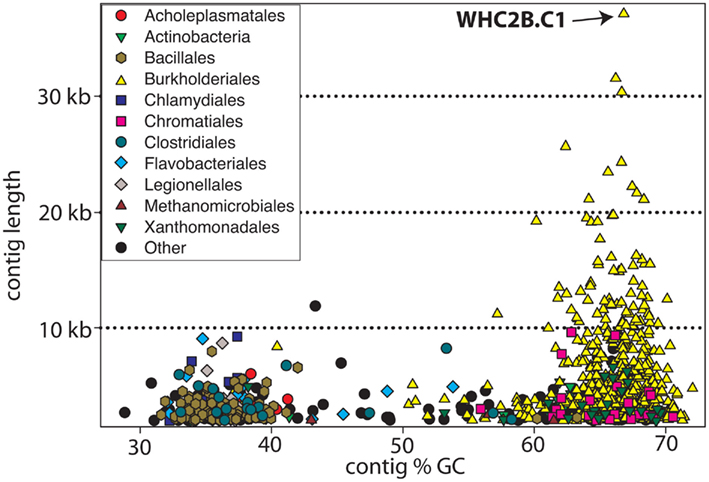
Figure 2. Taxonomic assignment of contigs (plotted with respect to length and %GC) assembled from WHC2b metagenomic sequences. Large Burkholderiales contigs with high %GC (yellow triangles) dominate the assembly. Most of the smaller contigs have low %GC, and many of these are assigned to Gram-positive bacteria including Bacillales and Clostridiales. Only contigs > 2 kb are shown.
The de novo metagenomic assembly from TLE yielded only 118 contigs > 2 kb, and the longest was just 8.5 kb (data not shown). Most of the same taxonomic groups in WHC2b are also present in TLE, including Burkholderiales, Bacillales, and Clostridiales, but the Burkholderiales do not dominate the assembly as they do in WHC2b.
A partial 16S rRNA sequence was identified in one of the Burkholderiales contigs in each Tablelands fluid sample (WHC2b.C991: 1.6 kb, 56% GC, 3.6 mean coverage; TLE.C712: 1.5 kb, 54% GC, 1.3 mean coverage). Maximum-likelihood phylogenetic analysis placed both sequences in genus Hydrogenophaga (Figure 3), which is consistent with the phylogeny of the hydrogenases discussed below. Two Burkholderiales-related 16S rRNA gene clones were also recovered from the same Lost City carbonate structure that is the source of the metagenome discussed here; these clones were previously described in Brazelton et al. (2006). They are 99% similar over 1471 bp (calculated with MatGat; Campanella et al., 2003) to the 16S rRNA sequence of Ralstonia pickettii, an oligotroph known to grow in moist biofilms (Stelzmueller et al., 2006). The 16S rRNA sequence of R. eutropha (the hydrogenases of which are discussed below) is 96% similar to the Lost City clones (Figure 3).
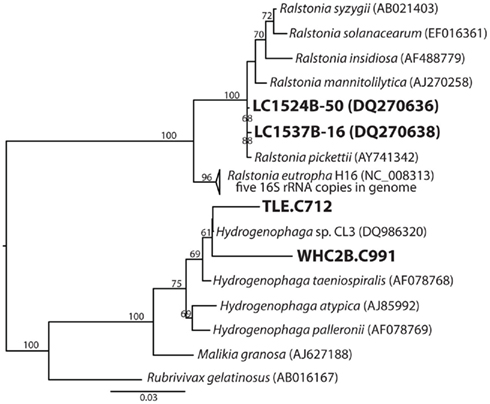
Figure 3. Phylogenetic tree of Burkholderiales 16S rRNA sequences including two 16S rDNA clones from Lost City carbonate chimneys and a partial 16S rRNA coding sequence identified in a metagenomic contig from the Tablelands fluid sample WHC2b.
The largest WHC2b contig (WHC2b.C1: 37 kb, 67% GC, comprising 615 shotgun reads at 6.6 mean coverage) contains putative operons for a [NiFe]-hydrogenase, carbon monoxide dehydrogenase (CODH), and Rubisco (Figure 4). The Rubisco cluster includes RAST annotations for the small and large subunits of Rubisco, six carboxysome shell proteins, and two Rubisco activation proteins. The hydrogenase cluster includes RAST annotations for the small and large subunits of uptake [NiFe]-hydrogenase, a signal transduction histidine kinase (HoxJ), and a transcriptional regulatory protein (HoxA). The CODH cluster is at the edge of the contig and is most likely incomplete. It includes RAST annotations for the three subunits of CODH (CoxMSL) with an upstream CoxC and a downstream CoxD. This arrangement is consistent with that found in type I CODH clusters in Marine Roseobacter Clades (Cunliffe, 2011). The phylogenetic relationships of the [NiFe]-hydrogenase large subunit and CODH large subunit from this contig are described below.
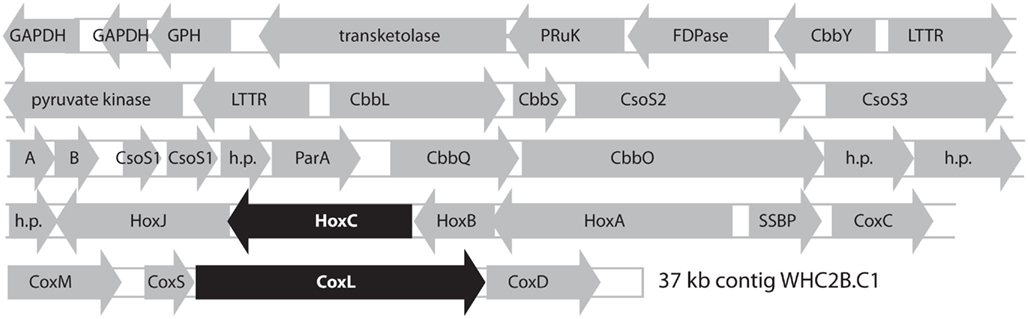
Figure 4. Predicted products of open reading frames identified in the longest contig from Tablelands fluid sample WHC2b (37 kb, 67% GC, comprising 615 shotgun reads at 6.6 mean coverage). The putative large subunits of [NiFe]-hydrogenase (HoxC) and carbon monoxide dehydrogenase (CoxL) are highlighted in black to indicate that their phylogenetic relationships are shown in Figures 5 and Figure A1 in Appendix. GAPDH: NAD-dependent glyceraldehyde-3-phosphate dehydrogenase. GPH, phosphoglycolate phosphatase; PRuK, phosphoribulokinase; FDPase, type I fructose-1,6-bisphosphatase; CbbY, rubisco-associated hydrolase; LTTR, LysR-type transcriptional regulator; CbbL, CbbS, ribulose bisphosphate carboxylase, large and small chains; CbbS, ribulose bisphosphate carboxylase small chain; CsoS2, CsoS3, CsoS1, carboxysome shell proteins; A, B, putative carboxysome peptides; h. p., hypothetical protein; ParA, chromosome (plasmid) partitioning protein; CbbQ, CbbO, rubisco activation proteins; HoxJ, HoxA, hydrogenase regulation proteins; HoxC, HoxB, uptake [NiFe]-hydrogenase, large and small subunits; SSBP, single-stranded DNA binding protein; CoxC, carbon monoxide dehydrogenase signaling protein; CoxM, CoxS, CoxL, carbon monoxide dehydrogenase, medium, small, and large subunits; CoxD, carbon monoxide dehydrogenase accessory protein.
The relatively high coverage and presence of a plasmid partitioning protein in WHC2b.C1 (Figure 4) suggests the possibility that this contig represents a plasmid. Indeed, the H2 oxidation and carbon fixation capabilities of Ralstonia eutropha are encoded in a “megaplasmid,” and loss of this plasmid would result in a strictly heterotrophic lifestyle (Schwartz et al., 2009). The plasmid partitioning protein in WHC2b.C1 does not contain any homologs in the R. eutropha megaplasmid, however, and the contig is not a complete plasmid sequence, so we cannot conclude with certainty whether it is a plasmid. Nevertheless, all sequenced members of the Burkholderiaceae family contain multiple replicons; the additional genetic molecules other than the main chromosome are sometimes called “chromosomes” and sometimes “megaplasmids” (Fricke et al., 2009). No genomes from Hydrogenophaga species (Comamonadaceae family) have been sequenced yet, and we are not aware of any data pertaining to whether their hydrogenases are plasmid-encoded.
Abundance of Sequences Encoding Hydrogenases
We investigated the genetic potential for communities in these samples to metabolize H2 by conducting a tblastn search for potential homologs of the catalytic (large) subunits of [FeFe]-hydrogenase and [NiFe]-hydrogenase. For each metagenome, the number of unique sequencing reads that match at least one hydrogenase is reported in Table 1. For comparative purposes, only unassembled sequences were included in the quantification of hydrogenase abundance because quantitative comparisons of gene abundance among assembled metagenomes is problematic due to their varying assembly strategies and sequencing coverages.
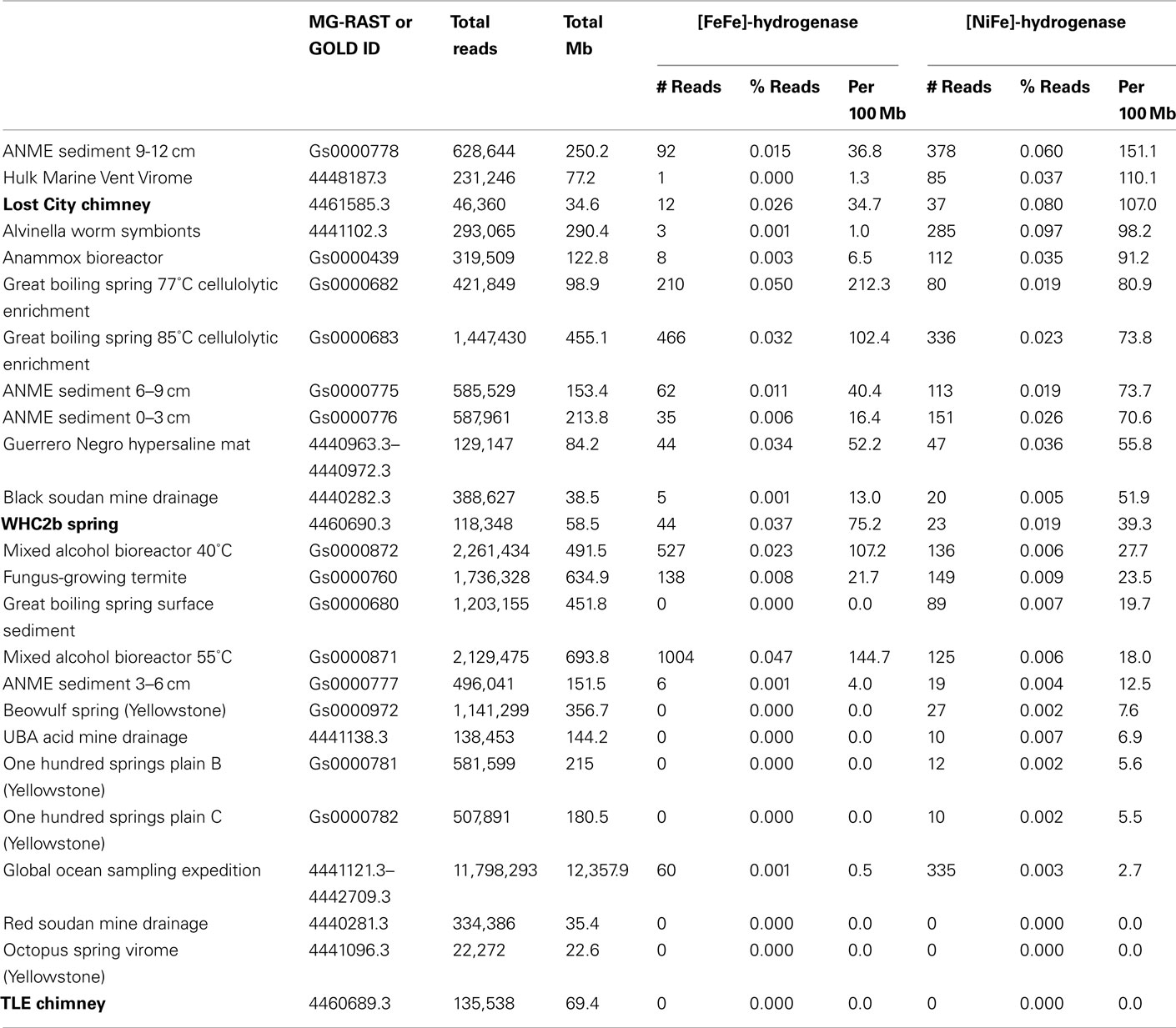
Table 1. [FeFe]-hydrogenase and [NiFe]-hydrogenase abundance in selected metagenomes, as measured by the number of unique shotgun sequencing reads with tblastn matches to Pfam families PF02906 and PF00374, respectively, with E-values better than 10− 5.
The Lost City chimney metagenome and the Tablelands metagenome WHC2b both contained a similar proportion of putative hydrogenase sequences as those found in other metagenomes from environments where H2 metabolism is expected to occur (Table 1). These environments (including methanotrophic sediments, hot springs, marine hydrothermal vents, organic degradation bioreactors, acid mine drainage, and a hypersaline microbial mat) contained >10× greater density of hydrogenases than the combined dataset from the Global Ocean Sampling expedition. Remarkably, zero hydrogenase sequences were detected in the metagenomic data from the TLE spring, which exhibited more moderate pH and Eh values compared to WHC2b.
The Lost City metagenome was exceptionally rich in sequences matching [NiFe]-hydrogenases; only two metagenomes in the MG-RAST and IMG/M databases contained a higher proportion (Table 1). The WHC2b metagenome, in contrast, was exceptionally rich in sequences matching [FeFe]-hydrogenases, which are typically involved in H2 production during fermentation. Indeed, the only metagenomes with a higher proportion of [FeFe]-hydrogenases than WHC2b are derived from cellulose-degrading fermentation bioreactors.
Inferring the abundance of H2-oxidizing or H2-producing organisms from the abundance of hydrogenases genes is problematic because multiple hydrogenases are frequently present in a given genome (Vignais and Billoud, 2007). Furthermore, our tblastn search could have detected hydrogenases found in all three domains of life. Phylogenetic analyses, however, can constrain the taxonomic possibilities for each hydrogenase sequence and therefore enable one to test hypotheses about which hydrogenases are encoded by which organisms. The phylogenetic analyses described below indicate that both the Tablelands and Lost City springs feature [NiFe]-hydrogenases primarily encoded by Betaproteobacteria related to known H2-oxidizers and [FeFe]-hydrogenases encoded by Clostridia-related to known H2-producers. No archaeal or eukaryotic hydrogenases were detected in this study.
[NiFe]-Hydrogenase Diversity
In order to further characterize the putative hydrogenase sequences identified in the tblastn search, we constructed maximum-likelihood phylogenetic trees (Figures 5–7) in which the putative hydrogenase sequences were incorporated into a constraint tree comprised of previously characterized hydrogenases. The resulting [NiFe]-hydrogenase phylogeny (Figure 5) is congruent with that reported by Vignais et al. (2001) and Vignais and Billoud (2007), who identified four monophyletic groups consistent with the original classification by Wu and Mandrand (1993). Each of the four groups includes archaeal and bacterial representatives. None of the sequences in this study, however, exhibited significant phylogenetic affinity with archaeal hydrogenases. The phylogenetic relationships of the putative bacterial hydrogenases are described in detail below.
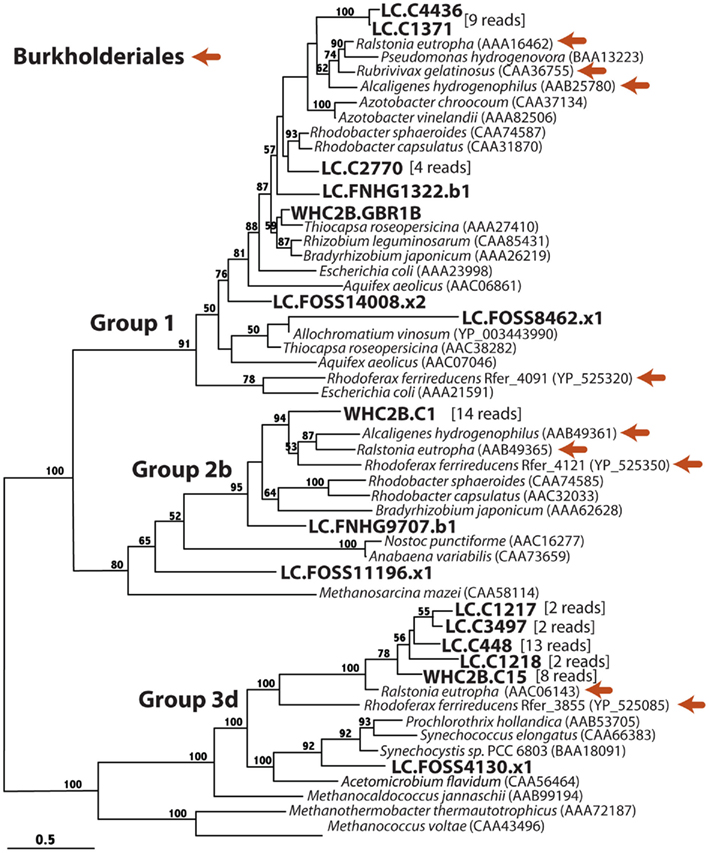
Figure 5. Phylogenetic diversity of putative [NiFe]-hydrogenase detected in metagenomes from a Lost City chimney and Tablelands spring fluid. Burkholderiales references sequences are indicated with orange arrows, and Lost City (LC), and Tablelands (WHC2b) sequences are in large, bold font. If additional metagenomic sequencing reads are associated with the representative on the tree, the number of sequencing reads is indicated in brackets. Group numbers are consistent with the phylogeny reported by Vignais et al. (2001) and Vignais and Billoud (2007). Maximum-likelihood bootstrap support values > 50 are shown.
Group 1 includes the membrane-bound respiratory uptake hydrogenases, which couple H2 oxidation to a cytochrome, resulting in proton pumping across the membrane. Nitrogen-fixing organisms also use Group 1 [NiFe]-hydrogenases to recapture H2 produced by nitrogenase. The [NiFe]-hydrogenase phylogeny (Figure 5) shows that many Lost City sequences and a single WHC2b shotgun sequencing read belong to Group 1. Two Lost City contigs representing nine reads are most similar to hydrogenases from Burkholderiales members Ralstonia eutropha, Rubrivivax gelatinosus, and Alcaligenes hydrogenophilus. A third Lost City contig contained a hydrogenase closely related to that from two Rhodobacter species. Three additional Lost City reads belong to Group 1, but their phylogenetic affiliations are unresolved. Only one read from WHC2b contained a Group 1 [NiFe]-hydrogenase, and its closest relative was from Thiocapsa roseopersicina, a nitrogen-fixing member of order Chromatiales.
Group 2b includes H2-sensing proteins that are involved in H2-regulated gene expression and do not directly provide energy from H2 to the cell (Vignais et al., 2001). They include Alpha- and Beta-proteobacteria. A cluster of 14 reads representing the contig WHC2b.C1 are most similar to Burkholderiales Group 2b hydrogenases. Two Lost City reads with unresolved phylogenetic affiliations are also in the Group 2b clade. Group 2a (not show) is specific to cyanobacterial hydrogenases; none of these were detected by this study.
Group 3 includes cytoplasmic hydrogenases capable of reversible H2 oxidation with a soluble cofactor. Groups 3a and 3c are specific to archaea, and a Group 3c representative (from Methanocaldococcus jannaschii) is shown in Figure 5 as an outgroup to Group 3d, which is specific to bacteria. The contig WHC2b.C15 (21 kb, 64% GC) representing eight shotgun reads clustered with four Lost City contigs and two Burkholderiales Group 3d hydrogenases. An additional Lost City read is most similar to cyanobacterial Group 3d hydrogenases. No sequences in this study shared significant similarity with archaeal hydrogenases.
Group 4 includes H2-producing [NiFe]-hydrogenases that are typically involved in the disposal of excess reducing equivalents in archaea (Vignais and Billoud, 2007). No Group 4 homologs were identified in this study.
[FeFe]-Hydrogenase Diversity
Whereas [NiFe]-hydrogenases are typically involved in H2 uptake and oxidation, almost all [FeFe]-hydrogenases catalyze the production of H2 (Vignais and Billoud, 2007). (The one known exception does not have any phylogenetic affiliation with any sequences in this study.) The enzyme can be monomeric, dimeric, trimeric, or tetrameric in different species, and the variable number and diversity of paralogs in closely related species obscures phylogenetic relationships (Vignais et al., 2001). Therefore, [FeFe]-hydrogenase phylogeny has limited utility for inferring taxonomic affiliations of metagenomic sequences. Nevertheless, the phylogeny illustrated in Figure 6, which is consistent with that reported by Boyd et al. (2009), shows that all of the putative [FeFe]-hydrogenase sequences detected in Lost City and Tablelands samples have close phylogenetic relationships with Clostridia. The largest cluster of putative [FeFe]-hydrogenase sequences represents nine shotgun sequencing reads from WHC2b and shares high sequence similarity with Desulfotomaculum reducens (60% identity, 76% similarity over 140 amino acid residues). Members of genus Desulfotomaculum are typically sulfate-reducers; some species oxidize H2, and others require organic carbon compounds as electron donors. Some species are incapable of sulfate reduction, however, and produce H2 during fermentation as part of a syntrophic relationship with methanogens (Imachi et al., 2006).
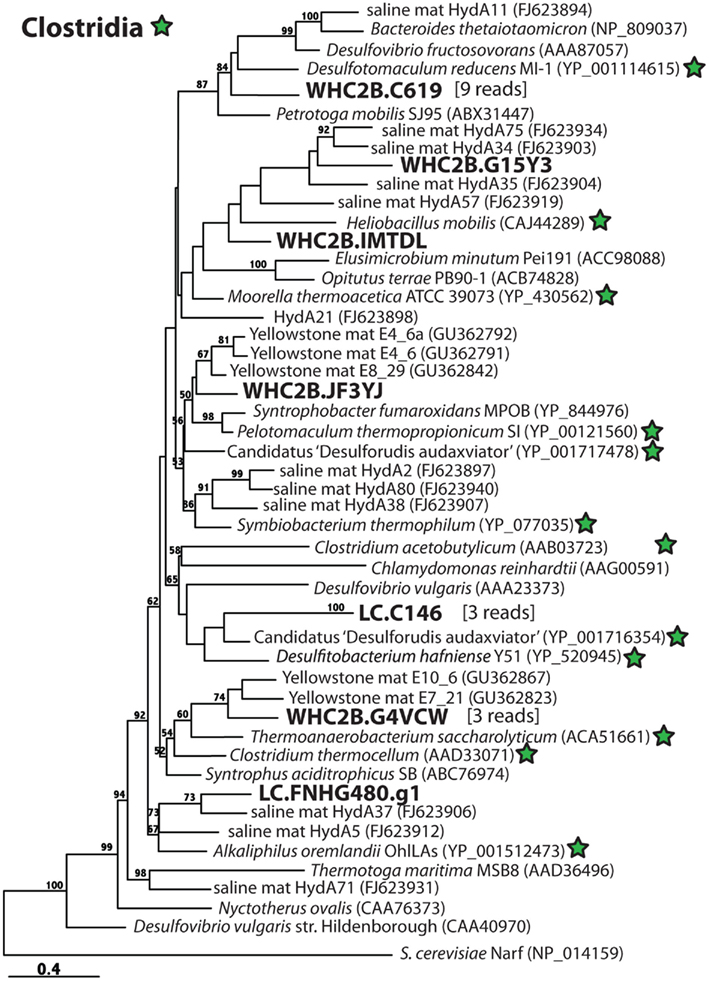
Figure 6. Phylogenetic diversity of putative [FeFe]-hydrogenases detected in metagenomes from a Lost City chimney and Tablelands spring fluid. Clostridia reference sequences are indicated with green stars, and Lost City (LC), and Tablelands (WHC2b) sequences are in large, bold font. The number of metagenomic shotgun sequencing reads associated with each representative on the tree is indicated in brackets. Maximum-likelihood bootstrap support values > 50 are shown.
A 1.9 kb contig from Lost City (LC.C146) contains a predicted [FeFe]-hydrogenase that is most closely related to that of Candidatus “Desulforudis audaxviator” (Figure 6), a deep subsurface organism that is very closely related to the Desulfotomaculum. Its genome sequence encodes multiple hydrogenases, including uptake [NiFe]-hydrogenases that indicate the potential for uptake and oxidation of H2 (Chivian et al., 2008). Only [FeFe]-hydrogenases (Figure 6) and no [NiFe]-hydrogenases related to Clostridia, however, were detected in the Lost City or Tablelands metagenomes.
Figure 7 illustrates phylogenetic relationships among proteins predicted to be [FeFe]-hydrogenases by homology but are clearly divergent from and may not be monophyletic with the well-characterized [FeFe]-hydrogenases in Figure 6. Almost all of these hypothetical [FeFe]-hydrogenases show high similarity to predicted proteins in Clostridia genomes, the one exception being a cluster of 11 reads from WHC2b that is most similar to a predicted protein from the genome of an unidentified Erysipelotrichaceae bacterium. To our knowledge, the only sequence in this tree for which there is any published experimental characterization is the HydII of Thermoanaerobacterium saccharolyticum, which groups with a cluster of eight WHC2b reads. This sequence forms a gene cluster required for H2 production during fermentation by T. saccharolyticum, but deletion of the hydII gene alone had no effect on H2 production (Shaw et al., 2009). Therefore, the sequences in this tree are potentially associated with H2 production but are not likely to encode the catalytic subunits of H2-evolving hydrogenases. It is important to note that these hypothetical hydrogenases are included in the tblastn results for [FeFe]-hydrogenases reported in Table 1, and our phylogenetic analyses were required to distinguish them from homologs of characterized H2-evolving hydrogenases (Figure 6).
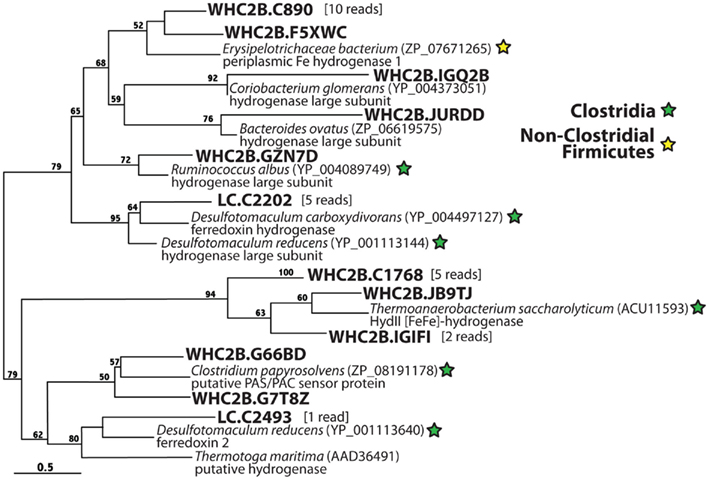
Figure 7. Phylogenetic diversity of hypothetical [FeFe]-hydrogenases detected in metagenomes from a Lost City chimney and Tablelands spring fluid. These sequences show weak similarity to the characterized hydrogenases in Figure 6 but are highly similar to hypothetical hydrogenases.
Carbon Monoxide Dehydrogenase Diversity
The oxidation of carbon monoxide by aerobic carboxydotrophs (organisms that use CO as their sole electron donor and carbon source) is catalyzed by carbon monoxide dehydrogenase (CODH; Ragsdale, 2004; King and Weber, 2007). (Note that this enzyme is distinct from the CODH/ACS enzyme complex that is required for carbon fixation in the Wood–Ljungdahl pathway discussed below.) CODH is encoded by the cox, also called cut, genes, which can be used as marker genes for aerobic carboxydotrophs (King, 2003; Cunliffe et al., 2008; Cunliffe, 2011). The largest WHC2b contig in our dataset (WHC2b.C1, 37 kb) contains putative homologs for the three structural subunits of CODH, CoxMSL. The CoxL homolog shows a close phylogenetic affinity to other Betaprotebacteria CoxL genes (Figure A1 in Appendix) including that of Hydrogenophaga pseudoflava, a facultative anaerobe that can grow autotrophically on H2 or CO (Willems et al., 1989). The only other potential CoxL homolog detected in our datasets by tblastn search was a Lost City shotgun read (Figure A1 in Appendix) that is most similar to Labrenzia aggregata (formerly Stappia aggregata), a marine aerobic carboxydotroph belonging to the alphaproteobacterial order Rhodobacterales (Weber and King, 2007).
Acetyl-CoA Synthase Diversity
Many Clostridia use H2 to generate acetate, so we explored the metagenomic data to test whether the clostridial hydrogenases (Figure 6) are potentially encoded by acetogens. Acetogens and methanogens utilize the Wood–Ljungdahl (also known as reductive acetyl-CoA) pathway for carbon fixation, and the key enzyme acetyl-CoA synthase (ACS) is unique to that pathway (Ragsdale and Pierce, 2008). To investigate the potential for anaerobic carbon fixation via the Wood–Ljungdahl pathway in the Tablelands springs and Lost City chimneys, we conducted a tblastn search for ACS sequences in our metagenomes. The phylogenetic relationships among the three WHC2b shotgun sequencing reads and two Lost City shotgun sequencing reads with potential ACS homologs are displayed in Figure A2 in Appendix. All three WHC2b sequences cluster with the ACS of Dethiobacter alkaliphilus, a H2-oxidizing autotrophic Clostridia that can use acetate as a carbon source but not as an energy source (Sorokin et al., 2008). To our knowledge, there is no published evidence that D. alkaliphilus can carry out acetogenesis. The KEGG annotation of its genome available at the DOE Joint Genome Institute’s IMG database (Markowitz et al., 2008) indicates the presence of at least three additional homologs required for the Wood–Ljungdahl pathway, but it is unclear whether a complete pathway for acetogenesis is present. The ACS of Moorella thermoacetica, a known thermophilic acetogen (Pierce et al., 2008), has 60% identity and 78% similarity over 161 amino acid residues with shotgun read WHC2b.JXG7G. The Lost City sequences, by contrast, group with methanogen ACS sequences (Figure A2 in Appendix). Methanosarcinales-related archaea are known to dominate the anoxic interiors of Lost City chimney biofilm communities (Schrenk et al., 2004), but methanogen sequences are rare in this Lost City metagenomic dataset, presumably because it is derived from more oxidized, exterior portions of the chimney (Brazelton et al., 2006; Brazelton and Baross, 2010).
Discussion
Potential for H2-Fueled Carbon Fixation by Burkholderiales
The metagenomic and phylogenetic data presented above indicate that Betaproteobacteria belonging to order Burkholderiales are potentially important primary producers adapted to the extreme conditions of the Tablelands springs. Their potential for H2 oxidation is indicated by the diversity of uptake [NiFe]-hydrogenase sequences (Figure 5) in the WHC2b spring, which are absent in the TLE spring. Their potential for carbon fixation is indicated by the presence of gene clusters encoding carbon monoxide dehydrogenase (CODH) and Rubisco in the largest WHC2b contig (WHC2b.C1; Figure 4). The phylogenies of the [NiFe]-hydrogenases, CODH, and 16S rRNA sequences in WHC2b contigs indicate close relationships with Hydrogenophaga species and Ralstonia eutropha (now Cupriavidus necator). These organisms are facultatively autotrophic; i.e., they only utilize H2 or fix carbon when organic carbon is unavailable (Willems et al., 1989; Schwartz et al., 2009). Therefore, further characterization of the physiology of these organisms and their access to organic matter in the Tablelands springs is required to estimate their contribution to primary production. Furthermore, all Hydrogenophaga and Ralstonia species are aerobic or facultatively anaerobic, so the corresponding organisms at the Tablelands and Lost City are likely to inhabit oxic–anoxic transition zones where they have access to both H2 and oxygen. Both of these systems feature strong oxygen gradients between the atmosphere and spring water (at the Tablelands) and between oxygenated seawater and hydrothermal fluid (at Lost City), so there is potential in each system for organisms to utilize both H2 and oxygen.
It is unclear whether the TLE spring also hosts H2-fueled carbon fixation. Although a Burkholderiales 16S rRNA sequence was identified in a metagenomic contig from TLE and automated taxonomy classifiers identify many Burkholderiales-related sequences in the TLE metagenome (MG-RAST and TaxSOM, data not shown), no hydrogenases were detected in TLE. The absence of hydrogenases could be due to a combination of lower abundance of Burkholderiales in TLE and lower sequencing depth of the TLE metagenome compared to WHC2b. It is also possible that the Burkholderiales species in TLE have lost their hydrogenase genes, which may have resulted from the loss of the plasmid potentially represented by contig WHC2b.C1, as discussed above in the description of Figure 4.
Most H2-oxidizing autotrophs utilize both membrane-bound (Group 1) and cytoplasmic (Group 3) [NiFe]-hydrogenases. The [NiFe]-hydrogenase in the WHC2b.C1 contig belongs to the Group 2 H2 sensor proteins, which are involved in the regulation of carbon fixation by H2 but do not directly couple H2 oxidation with energy conservation (Vignais et al., 2001). Therefore, the evidence from this one contig indicates only that carbon fixation in the corresponding organism is regulated by the presence of H2 and not necessarily fueled by H2 oxidation. The phylogeny of the Group 2 [NiFe]-hydrogenase in WHC2b.C1, however, is congruent with the phylogeny of the Group 3 [NiFe]-hydrogenase in a 21 kb contig (WHC2b.C15; Figure 5). It seems highly likely that both contigs are derived from the same species, and both hydrogenases are highly similar to putative homologs in Ralstonia eutropha. The Group 1, 2, and 3 hydrogenases in R. eutropha are all encoded in a 452 kb megaplasmid, and it is possible that contigs WHC2b.C1 and WHC2b.C15 are partial sequences of the same plasmid (as described above).
The lack of a Ralstonia-related Group 1 [NiFe]-hydrogenase in the WHC2b metagenome is puzzling, however (Figure 5). Nitrosospira multiformis is one of the few examples listed in the exhaustive survey by Vignais and Billoud (2007) of an organism that has only a Group 3d [NiFe]-hydrogenase and no representative from Group 1. The function of the N. multiformis Group 3d [NiFe]-hydrogenase is unknown but suspected to be the catalysis of NAD reduction by H2 in order to “increase the overall energetic yield from ammonia oxidation” (Norton et al., 2008). Therefore, it is possible that the Ralstonia-like organisms in the Tablelands only utilize H2 to supplementary their primary electron donor (e.g., organic carbon). The current metagenomic dataset from WHC2b is relatively low coverage, however, and additional sequencing at higher coverage may eventually recover a Group 1 homolog.
Both Group 1 and Group 3 [NiFe]-hydrogenases related to R. eutropha are present in several Lost City contigs (Figure 5), indicating that a Ralstonia-related organism with the genetic potential for H2 oxidation also inhabits Lost City chimneys. The Lost City metagenomic dataset is dominated by sequences with high similarity to that of Thiomicrospira crunogena, a cosmopolitan sulfur-oxidizing autotroph in marine hydrothermal vents. Previous studies have noted the inability of T. crunogena to utilize H2 as a sole electron donor despite the presence of a Group 1 [NiFe]-hydrogenase in its genome (Scott et al., 2006). None of the hydrogenases detected in this study have high sequence similarity to the T. crunogena hydrogenase, nor do any of the large Lost City contigs expected to correspond to Thiomicrospira-like organisms contain predicted hydrogenases. Therefore, H2-oxidizing organisms in young, hot Lost City chimneys are most likely aerobic or facultatively anaerobic Betaproteobacteria belonging to order Burkholderiales and appear to be less abundant than the dominant sulfur-oxidizing Thiomicrospira-like population.
Potential for CO Utilization by Burkholderiales
Carbon dioxide is extremely scarce in the highly reducing, high pH fluids of the Tablelands and Lost City, so alternative carbon species may be more favorable substrates for carbon fixation. The largest Tablelands contig (WHC2b.C1; Figure 4) includes the CoxMSL gene cluster which encodes all three subunits of the carbon monoxide dehydrogenase (CODH) used by aerobic carboxydotrophs (Ragsdale, 2004; King and Weber, 2007). This enzyme is frequently plasmid-encoded (Hugendieek and Meyer, 1992), providing additional but not conclusive evidence that the WHC2b.C1 contig represents a plasmid. The phylogeny of the large subunit of CODH from WHC2b.C1 indicates a close phylogenetic relationship with Hydrogenophaga pseudoflava, an aerobic autotrophic member of Burkholderiales that can grow on either H2 or CO (Willems et al., 1989; Kang and Kim, 1999). Therefore, the phylogeny of the CODH in the WHC2b.C1 contig is consistent with that of the [NiFe]-hydrogenases discussed above. CODH is typically involved in aerobic oxidation of CO, but some studies indicate that oxidation of low levels of CO can be coupled to nitrate rather than oxygen (King, 2006). Therefore, CO utilization could be advantageous in Tablelands springs when concentrations of H2, oxygen, and organic compounds are too low to support growth, but any conclusions about the importance of CO in these systems will require further investigations. At Lost City, CO utilization seems unlikely because of the abundance and ubiquity of H2 and because CODH appears to be very rare (identified in only a single shotgun sequencing read).
Potential for H2 Production by Clostridia
Nearly all of the [FeFe]-hydrogenases detected in the WHC2b spring at the Tablelands and in the Lost City chimney have close phylogenetic relationships with putative homologs in Clostridia. [FeFe]-hydrogenases catalyze H2 production by anaerobic bacteria, typically during fermentation, so one would expect them to be prevalent in anoxic environments where H2 production is favorable. Potential subsurface sources of fermentable organic material are indicated by elevated levels of dissolved organic carbon in Lost City fluids (Lang et al., 2010) and the presence of low molecular weight hydrocarbons with potentially abiogenic origins in both Lost City and Tablelands fluids (Proskurowski et al., 2008; Szponar et al., submitted). No hydrogenases were detected in the Tablelands spring (TLE) that was collected from a more dilute and oxidizing seep (pH 10.5, Eh + 25 mV) only ∼2 km from WHC2b (pH 12.06, Eh − 733 mV). Therefore, the presence of [FeFe]-hydrogenases in the WHC2b metagenome supports the notion that the spring is supplied by fluid from an anoxic environment. Their presence in the Lost City chimney could be indicative of anoxic niches within chimney biofilms and/or the contribution of subsurface fluid to the chimney sample. The greater abundance of Clostridia-related 16S rRNA gene sequences in younger, hotter Lost City chimneys is consistent with both of these possibilities (Brazelton et al., 2010).
The community structure of Tablelands springs and Lost City chimneys, as described here, resembles that of deep boreholes in South Africa (Moser et al., 2005; Lin et al., 2006). The subsurface fluids sampled by these boreholes are also basic (pH ∼9) and enriched in H2 (up to 3.7 mM). The shallow fluids described by Moser et al. (2005) are dominated by Betaproteobacteria belonging to the Comamonadaceae family, and deeper fluids are comprised almost exclusively of Clostridia affiliated with genus Desulfotomaculum. The deep subsurface Desulfotomaculum-related organisms are predicted to be sulfate-reducers in these environments, which is consistent with metagenomic data representing the dominant organism, Candidatus “Desulforudis audaxviator” (Chivian et al., 2008). Some closely related species, however, are known to lack the genes required for sulfate reduction and instead subsist on fermentation, producing H2 as part of a syntrophic relationship with methanogens (Imachi et al., 2006). Indeed, no sequences encoding dissimilatory sulfite reductase were identified in the Tablelands metagenomes, a striking result compared to the abundance of hydrogenases. The Lost City metagenome encodes a dissimilatory sulfite reductase with high sequence similarity to multiple Desulfotomaculum species (Brazelton, 2010), but it was detected in only a single sequencing read, indicating that it is far less abundant than the hydrogenases. Therefore, the Clostridia in Tablelands springs and Lost City chimneys are potential sulfate-reducers, but the abundance of [FeFe]-hydrogenases in metagenomic data from both environments indicates that they are more likely to be involved in H2-generating fermentation. It is unclear whether this putative fermentation is syntrophic with H2-utilizing methanogens. Automated annotation predicted very few methanogen sequences in the Tablelands and Lost City metagenomes (data available on the MG-RAST server), but they were present and may be more abundant in deeper habitats that were not well-represented in the samples described in this study.
It is also possible that the Clostridia detected in this study are acetogens that are adapted to the elevated H2 concentrations in the Tablelands and Lost City fluids. No sequences encoding acetyl-CoA synthase were detected at Lost City, but the phylogeny of ACS sequences from WHC2b is consistent with the presence of clostridial acetogens in very low abundance. Acetogens are known to be capable of producing H2 and harboring a wide diversity of [FeFe]-hydrogenases (Kellum and Drake, 1984; Schmidt et al., 2010, 2011), so determining the role of these Clostridia in the H2 budget of these systems will require physiological and biogeochemical investigations. Only three ACS sequences were recovered from WHC2b, however, and none of these were assembled into contigs. Therefore, the current dataset indicates that acetogenesis may occur but does not appear to be prevalent in the Tablelands springs. It is possible that a more representative sample of the subsurface habitat underlying the spring could reveal more abundant evidence of acetogenesis, as well as other anaerobic metabolic pathways.
Conclusion
Both the marine and continental serpentinite springs investigated in this study show evidence of aerobic organisms capable of H2-fueled (or at least H2-regulated) primary production (i.e., Burkholderiales) and anaerobic organisms capable of H2 production from fermentation of organic carbon (i.e., Clostridia). This community structure resembles that of the deep subsurface habitat sampled by a ∼3 km deep borehole in South Africa (Moser et al., 2005), indicating that the surface-exposed springs described in this study provide access to organisms flushed from the subsurface. Furthermore, the remarkably high density of hydrogenases in both the marine and continental springs (Lost City and WHC2b) in this study and their complete absence in a spring showing evidence of extensive mixture with surface runoff (TLE) indicate that the H2-associated metabolic activities discussed here are specific to subsurface processes.
The predicted metabolic characteristics of the dominant organisms in the Tablelands springs are consistent with the known abiogenic products of subsurface serpentinization-associated processes: H2 and low molecular-weight organic compounds. A major unanswered question, however, is whether the H2-oxidizing Burkholderiales subsist on abiogenic H2 generated by serpentinization in the subsurface or if they depend on biogenic H2 produced by Clostridia. In either case, it seems likely that the Burkholderiales in the Tablelands springs and Lost City chimneys inhabit oxic–anoxic interfaces where they have access to both H2 and oxygen. Our metagenomic evidence also suggests that these organisms may be able to survive on carbon monoxide if H2 is unavailable. The Clostridia are likely inhabitants of anoxic, subsurface habitats where they ferment organic compounds into H2, but the source of these organic compounds is unknown. If they are ultimately derived from reduction of carbon by serpentinization-associated reactions (as evidenced in (Proskurowski et al., 2008) and predicted by experiments reviewed in (McCollom and Seewald, 2007), then fermentation of these compounds could be considered a kind of primary production as it would be the generation of new biomass from non-biological carbon and energy.
Therefore, both the H2-oxidizing Burkholderiales and H2-producing Clostridia may be important mediators of carbon and energy exchange between the deep Earth and the surface biosphere. Further research should investigate whether these organisms are bona fide denizens of the anoxic subsurface by probing deeper to obtain more representative samples of deep subsurface habitats.
In particular, the Clostridia and methanogens should be better represented in deeper samples. Nevertheless, the datasets presented here represent a proof-of-concept metagenomic study that demonstrates the potential of surface-exposed springs to yield insights into the microbial diversity of the subsurface biosphere.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We gratefully acknowledge logistical support and scientific inspiration from P. Morrill, N. Szponar, and many other faculty, staff, and students at Memorial University of Newfoundland. This work was supported by ECU startup funds to MOS and a NASA Astrobiology Institute grant through Cooperative Agreement NNA04CC09A to the Geophysical Laboratory at the Carnegie Institution for Science. William J. Brazelton was supported by a NASA Astrobiology Institute postdoctoral fellowship.
Footnotes
References
Abdo, Z., Schüette, U. M., Bent, S. J., Williams, C. J., Forney, L. J., and Joyce, P. (2006). Brief report statistical methods for characterizing diversity of microbial communities by analysis of terminal restriction fragment length polymorphisms of 16S rRNA genes. Environ. Microbiol. 8, 929–938.
Altschul, S. F., Madden, T. L., Schaffer, A. A., Zhang, J. H., Zhang, Z., Miller, W., and Lipman, D. J. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25, 3389–3402.
Aziz, R. K., Bartels, D., Best, A. A., DeJongh, M., Disz, T., Edwards, R. A., Formsma, K., Gerdes, S., Glass, E. M., Kubal, M., Meyer, F., Olsen, G. J., Olson, R., Osterman, A. L., Overbeek, R., McNeil, L. K., Paarmann, D., Paczian, T., Parrello, B., Pusch, G. D., Reich, C., Stevens, R., Vassieva, O., Vonstein, V., Wilke, A., and Zagnitko, O. (2008). The RAST Server: rapid annotations using subsystems technology. BMC Genomics 9, 75. doi: 10.1186/1471-2164-9-75
Biddle, J. F., Lipp, J. S., Lever, M. A., Lloyd, K. G., Sørensen, K. B., Anderson, R., Fredricks, H. F., Elvert, M., Kelly, T. J., Schrag, D. P., Sogin, M. L., Brenchley, J. E., Teske, A., House, C. H., and Hinrichs, K. (2006). Heterotrophic Archaea dominate sedimentary subsurface ecosystems off Peru. Proc. Natl. Acad. Sci. U.S.A. 103, 3846–3851.
Boyd, E. S., Hamilton, T. L., Spear, J. R., Lavin, M., and Peters, J. W. (2010). [FeFe]-Hydrogenase in Yellowstone National Park: evidence for dispersal limitation and phylogenetic niche conservatism. ISME J. 4, 1485–1495.
Boyd, E. S., Spear, J. R., and Peters, J. W. (2009). [FeFe] Hydrogenase genetic diversity provides insight into molecular adaptation in a saline microbial mat community. Appl. Environ. Microbiol. 75, 4620–4623.
Brazelton, W. J. (2010). Ecology of Archaeal and Bacterial Biofilm Communities at the Lost City Hydrothermal Field. Ph.D. dissertation, University of Washington, Seattle, WA.
Brazelton, W. J., and Baross, J. A. (2009). Abundant transposases encoded by the metagenome of a hydrothermal chimney biofilm. ISME J. 3, 1420–1424.
Brazelton, W. J., and Baross, J. A. (2010). Metagenomic comparison of two thiomicrospira lineages inhabiting contrasting deep-sea hydrothermal environments. PLoS ONE 5, e13530. doi: 10.1371/journal.pone.0013530
Brazelton, W. J., Ludwig, K. A., Sogin, M. L., Andreishcheva, E. N., Kelley, D. S., Shen, C.-C., Edwards, R. L., and Baross, J. A. (2010). Archaea and bacteria with surprising microdiversity show shifts in dominance over 1000-year time scales in hydrothermal chimneys. Proc. Natl. Acad. Sci. U.S.A. 107, 1612–1617.
Brazelton, W. J., Mehta, M. P., Kelley, D. S., and Baross, J. A. (2011). Physiological differentiation within a single-species biofilm fueled by serpentinization. MBio. 2, e00127–11.
Brazelton, W. J., Schrenk, M. O., Kelley, D. S., and Baross, J. A. (2006). Methane- and sulfur-metabolizing microbial communities dominate the Lost City hydrothermal field ecosystem. Appl. Environ. Microbiol. 72, 6257–6270.
Campanella, J. J., Bitincka, L., and Smalley, J. (2003). MatGAT: an application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinformatics 4, 29. doi: 10.1186/1471-2105-4-29
Chivian, D., Brodie, E. L., Alm, E. J., Culley, D. E., Dehal, P. S., DeSantis, T. Z., Gihring, T. M., Lapidus, A., Lin, L.-H., Lowry, S. R., Moser, D. P., Richardson, P. M., Southam, G., Wanger, G., Pratt, L. M., Andersen, G. L., Hazen, T. C., Brockman, F. J., Arkin, A. P., and Onstott, T. C. (2008). Environmental genomics reveals a single-species ecosystem deep within Earth. Science 322, 275–278.
Cunliffe, M. (2011). Correlating carbon monoxide oxidation with cox genes in the abundant Marine Roseobacter Clade. ISME J. 5, 685–691.
Cunliffe, M., Schäfer, H., Harrison, E., Cleave, S., Upstill-Goddard, R., and Murrell, J. C. (2008). Phylogenetic and functional gene analysis of the bacterial and archaeal communities associated with the surface microlayer of an estuary. ISME J. 2, 776–789.
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797.
Finn, R. D., Tate, J., Mistry, J., Coggill, P. C., Sammut, S. J., Hotz, H.-R., Ceric, G., Forslund, K., Eddy, S. R., Sonnhammer, E. L., and Bateman, A. (2008). The Pfam protein families database. Nucleic Acids Res. 36, D281–D288.
Fricke, W. F., Kusian, B., and Bowien, B. (2009). The genome organization of Ralstonia eutropha strain H16 and related species of the Burkholderiaceae. J. Mol. Microbiol. Biotechnol. 16, 124–135.
Gagen, E. J., Denman, S. E., Padmanabha, J., Zadbuke, S., Al Jassim, R., Morrison, M., and McSweeney, C. S. (2010). Functional gene analysis suggests different acetogen populations in the bovine rumen and tammar wallaby forestomach. Appl. Environ. Microbiol. 76, 7785–7795.
Gomez-Alvarez, V., Teal, T. K., and Schmidt, T. M. (2009). Systematic artifacts in metagenomes from complex microbial communities. ISME J. 3, 1314–1317.
Huber, J. A., Butterfield, D. A., and Baross, J. A. (2002). Temporal changes in archaeal diversity and chemistry in a mid-ocean ridge subseafloor habitat. Appl. Environ. Microbiol. 68, 1585–1594.
Huber, J. A., Mark Welch, D. B., Morrison, H. G., Huse, S. M., Neal, P. R., Butterfield, D. A., and Sogin, M. L. (2007). Microbial population structures in the deep marine biosphere. Science 318, 97–100.
Hugendieek, I., and Meyer, O. (1992). The structural genes encoding CO dehydrogenase subunits (cox L, M and S) in Pseudomonas carboxydovorans OM5 reside on plasmid pHCG3 and are, with the exception of Streptomyces thermoautotrophicus, conserved in carboxydotrophic bacteria. Arch. Microbiol. 157, 301–304.
Imachi, H., Sekiguchi, Y., Kamagata, Y., Loy, A., Qiu, Y.-L., Hugenholtz, P., Kimura, N., Wagner, M., Ohashi, A., and Harada, H. (2006). Non-sulfate-reducing, syntrophic bacteria affiliated with Desulfotomaculum cluster I are widely distributed in methanogenic environments. Appl. Environ. Microbiol. 72, 2080–2091.
Kang, B. S., and Kim, Y. M. (1999). Cloning and molecular characterization of the genes for carbon monoxide dehydrogenase and localization of molybdopterin, flavin adenine dinucleotide, and iron-sulfur centers in the enzyme of Hydrogenophaga pseudoflava. J. Bacteriol. 181, 5581–5590.
Kelley, D. S., Karson, J. A., Fruh-Green, G. L., Yoerger, D. R., Shank, T. M., Butterfield, D. A., Hayes, J. M., Schrenk, M. O., Olson, E. J., Proskurowski, G., Jakuba, M., Bradley, A., Larson, B., Ludwig, K., Glickson, D., Buckman, K., Bradley, A. S., Brazelton, W. J., Roe, K., Elend, M. J., Delacour, A., Bernasconi, S. M., Lilley, M. D., Baross, J. A., Summons, R. E., and Sylva, S. P. (2005). A serpentinite-hosted ecosystem: the Lost City hydrothermal field. Science 307, 1428–1434.
Kellum, R., and Drake, H. L. (1984). Effects of cultivation gas phase on hydrogenase of the acetogen Clostridium thermoaceticum. J. Bacteriol. 160, 466–469.
King, G. M. (2003). Molecular and culture-based analyses of aerobic carbon monoxide oxidizer diversity. Appl. Environ. Microbiol. 69, 7257–7265.
King, G. M. (2006). Nitrate-dependent anaerobic carbon monoxide oxidation by aerobic CO-oxidizing bacteria. FEMS Microbiol. Ecol. 56, 1–7.
King, G. M., and Weber, C. F. (2007). Distribution, diversity and ecology of aerobic CO-oxidizing bacteria. Nat. Rev. Microbiol. 5, 107–118.
Lang, S. Q., Butterfield, D. A., Schulte, M., Kelley, D. S., and Lilley, M. D. (2010). Elevated concentrations of formate, acetate and dissolved organic carbon found at the Lost City hydrothermal field. Geochim. Cosmochim. Acta 74, 941–952.
Larkin, M. A., Blackshields, G., Brown, N. P., Chenna, R., McGettigan, P. A., McWilliam, H., Valentin, F., Wallace, I. M., Wilm, A., Lopez, R., Thompson, J. D., Gibson, T. J., and Higgins, D. G. (2007). Clustal W and Clustal X version 2.0 . Bioinformatics 23, 2947–2948.
Lin, L.-H., Wang, P.-L., Rumble, D., Lippmann-Pipke, J., Boice, E., Pratt, L. M., Sherwood Lollar, B., Brodie, E. L., Hazen, T. C., Andersen, G. L., DeSantis, T. Z., Moser, D. P., Kershaw, D., and Onstott, T. C. (2006). Long-term sustainability of a high-energy, low-diversity crustal biome. Science 314, 479–482.
Ludwig, W., Strunk, O., Westram, R., Richter, L., Meier, H., Yadhukumar, Buchner, A., Lai, T., Steppi, S., Jobb, G., Förster, W., Brettske, I., Gerber, S., Ginhart, A. W., Gross, O., Grumann, S., Hermann, S., Jost, R., König, A., Liss, T., Lüssmann, R., May, M., Nonhoff, B., Reichel, B., Strehlow, R., Stamatakis, A., Stuckmann, N., Vilbig, A., Lenke, M., Ludwig, T., Bode, A., and Schleifer, K.-H. (2004). ARB: a software environment for sequence data. Nucleic Acids Res. 32, 1363–1371.
Markowitz, V. M., Ivanova, N., Palaniappan, K., Szeto, E., Korzeniewski, F., Lykidis, A., Anderson, I., Mavromatis, K., Mavrommatis, K., Kunin, V., Garcia Martin, H., Dubchak, I., Hugenholtz, P., and Kyrpides, N. C. (2006). An experimental metagenome data management and analysis system. Bioinformatics 22, e359–e367.
Markowitz, V. M., Szeto, E., Palaniappan, K., Grechkin, Y., Chu, K., Chen, I.-M., Dubchak, I., Anderson, I., Lykidis, A., Mavromatis, K., Ivanova, N. N., and Kyrpides, N. C. (2008). The integrated microbial genomes (IMG) system in 2007: data content and analysis tool extensions. Nucleic Acids Res. 36, D528–D533.
McCollom, T. M., and Seewald, J. S. (2007). Abiotic synthesis of organic compounds in deep-sea hydrothermal environments. Chem. Rev. 107, 382–401.
Meyer, F., Paarmann, D., D’Souza, M., Olson, R., Glass, E. M., Kubal, M., Paczian, T., Rodriguez, A., Stevens, R., Wilke, A., Wilkening, J., and Edwards, R. A. (2008). The metagenomics RAST server – a public resource for the automatic phylogenetic and functional analysis of metagenomes. BMC Bioinformatics 9, 386. doi: 10.1186/1471-2105-9-386
Moser, D. P., Gihring, T. M., Brockman, F. J., Fredrickson, J. K., Balkwill, D. L., Dollhopf, M. E., Lollar, B. S., Pratt, L. M., Boice, E., Southam, G., Wanger, G., Baker, B. J., Pfiffner, S. M., Lin, L.-H., and Onstott, T. C. (2005). Desulfotomaculum and Methanobacterium spp. Dominate 4- to 5-kilometer-deep fault. Appl. Environ. Microbiol. 71, 8773–8783.
Norton, J. M., Klotz, M. G., Stein, L. Y., Arp, D. J., Bottomley, P. J., Chain, P. S., Hauser, L. J., Land, M. L., Larimer, F. W., Shin, M. W., and Starkenburg, S. R. (2008). Complete genome sequence of Nitrosospira multiformis, an ammonia-oxidizing bacterium from the soil environment. Appl. Environ. Microbiol. 74, 3559–3572.
Pierce, E., Xie, G., Barabote, R. D., Saunders, E., Han, C. S., Detter, J. C., Richardson, P., Brettin, T. S., Das, A., Ljungdahl, L. G., and Ragsdale, S. W. (2008). The complete genome sequence of Moorella thermoacetica (f. Clostridium thermoaceticum). Environ. Microbiol. 10, 2550–2573.
Proskurowski, G., Lilley, M. D., Seewald, J. S., Fruh-Green, G. L., Olson, E. J., Lupton, J. E., Sylva, S. P., and Kelley, D. S. (2008). Abiogenic hydrocarbon production at lost city hydrothermal field. Science 319, 604–607.
Pruesse, E., Quast, C., Knittel, K., Fuchs, B. M., Ludwig, W., Peplies, J., and Glöckner, F. O. (2007). SILVA: a comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 35, 7188–7196.
Ragsdale, S. W., and Pierce, E. (2008). Acetogenesis and the Wood-Ljungdahl pathway of CO2 fixation. Biochim. Biophys. Acta 1784, 1873–1898.
Santelli, C. M., Orcutt, B. N., Banning, E., Bach, W., Moyer, C. L., Sogin, M. L., Staudigel, H., and Edwards, K. J. (2008). Abundance and diversity of microbial life in ocean crust. Nature 453, 653–656.
Schmidt, O., Drake, H. L., and Horn, M. A. (2010). Hitherto unknown [Fe-Fe]-hydrogenase gene diversity in anaerobes and anoxic enrichments from a moderately acidic fen. Appl. Environ. Microbiol. 76, 2027–2031.
Schmidt, O., Wüst, P. K., Hellmuth, S., Borst, K., Horn, M. A., and Drake, H. L. (2011). Novel [NiFe]- and [FeFe]-hydrogenase gene transcripts indicative of active facultative aerobes and obligate anaerobes in earthworm gut contents. Appl. Environ. Microbiol. 77, 5842–5850.
Schrenk, M. O., Huber, J. A., and Edwards, K. J. (2010). Microbial provinces in the subseafloor. Annu. Rev. Mar. Sci. 2, 279–306.
Schrenk, M. O., Kelley, D. S., Bolton, S. A., and Baross, J. A. (2004). Low archaeal diversity linked to subseafloor geochemical processes at the Lost City Hydrothermal Field, Mid-Atlantic Ridge. Environ. Microbiol. 6, 1086–1095.
Schwartz, E., Voigt, B., Zühlke, D., Pohlmann, A., Lenz, O., Albrecht, D., Schwarze, A., Kohlmann, Y., Krause, C., Hecker, M., and Friedrich, B. (2009). A proteomic view of the facultatively chemolithoautotrophic lifestyle of Ralstonia eutropha H16. Proteomics 9, 5132–5142.
Scott, K. M., Sievert, S. M., Abril, F. N., Ball, L. A., Barrett, C. J., Blake, R. A., Boller, A. J., Chain, P. S., Clark, J. A., Davis, C. R., Detter, C., Do, K. F., Dobrinski, K. P., Faza, B. I., Fitzpatrick, K. A., Freyermuth, S. K., Harmer, T. L., Hauser, L. J., Hugler, M., Kerfeld, C. A., Klotz, M. G., Kong, W. W., Land, M., Lapidus, A., Larimer, F. W., Longo, D. L., Lucas, S., Malfatti, S. A., Massey, S. E., Martin, D. D., McCuddin, Z., Meyer, F., Moore, J. L., Ocampo, L. H. Jr., Paul, J. H., Paulsen, I. T., Reep, D. K., Ren, Q., Ross, R. L., Sato, P. Y., Thomas, P., Tinkham, L. E., and Zeruth, G. T. (2006). The genome of deep-sea vent chemolithoautotroph Thiomicrospira crunogena XCL-2. PLoS Biol. 4, e383. doi: 10.1371/journal.pbio.0040383
Shaw, A. J., Hogsett, D. A., and Lynd, L. R. (2009). Identification of the [FeFe]-hydrogenase responsible for hydrogen generation in Thermoanaerobacterium saccharolyticum and demonstration of increased ethanol yield via hydrogenase knockout. J. Bacteriol. 191, 6457–6464.
Sogin, M. L., Morrison, H. G., Huber, J. A., Mark Welch, D., Huse, S. M., Neal, P. R., Arrieta, J. M., and Herndl, G. J. (2006). Microbial diversity in the deep sea and the underexplored “rare biosphere.” Proc. Natl. Acad. Sci. U.S.A. 103, 12115–12120.
Sorokin, D. Y., Tourova, T. P., Mussmann, M., and Muyzer, G. (2008). Dethiobacter alkaliphilus gen. nov. sp. nov., and Desulfurivibrio alkaliphilus gen. nov. sp. nov.: two novel representatives of reductive sulfur cycle from soda lakes. Extremophiles 12, 431–439.
Stamatakis, A. (2006). RAxML-VI-HPC: maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690.
Stelzmueller, I., Biebl, M., Wiesmayr, S., Eller, M., Hoeller, E., Fille, M., Weiss, G., Lass-Floerl, C., and Bonatti, H. (2006). Ralstonia pickettii-innocent bystander or a potential threat? Clin. Microbiol. Infect. 12, 99–101.
Vignais, P. M., and Billoud, B. (2007). Occurrence, classification, and biological function of hydrogenases: an overview. Chem. Rev. 107, 4206–4272.
Vignais, P. M., Billoud, B., and Meyer, J. (2001). Classification and phylogeny of hydrogenases. FEMS Microbiol. Rev. 25, 455–501.
Weber, C. F., and King, G. M. (2007). Physiological, ecological, and phylogenetic characterization of Stappia, a marine CO-oxidizing bacterial genus. Appl. Environ. Microbiol. 73, 1266–1276.
Weber, M., Teeling, H., Huang, S., Waldmann, J., Kassabgy, M., Fuchs, B. M., Klindworth, A., Klockow, C., Wichels, A., Gerdts, G., Amann, R., and Glöckner, F. O. (2010). Practical application of self-organizing maps to interrelate biodiversity and functional data in NGS-based metagenomics. ISME J. 918–928.
Whitman, W. B., Coleman, D. C., and Wiebe, W. J. (1998). Prokaryotes: the unseen majority. Proc. Natl. Acad. Sci. U.S.A. 95, 6578–6583.
Willems, A., Busse, J., Goor, M., Pot, B., Falsen, E., Jantzen, E., Hoste, B., Gillis, M., Kersters, K., Auling, G., and De Ley, J. (1989). Hydrogenophaga, a new genus of hydrogen-oxidizing bacteria that includes Hydrogenophaga flava comb. nov. (Formerly Pseudomonas flava), Hydrogenophaga palleronii (Formerly Pseudomonas palleronii), Hydrogenophaga pseudoflava (Formerly Pseudomonas pseudoflava and “Pseudomonas carboxydoflava”), and Hydrogenophaga taeniospiralis (Formerly Pseudomonas taeniospiralis). Int. J. Syst. Bacteriol. 39, 319–333.
Wu, L. F., and Mandrand, M. A. (1993). Microbial hydrogenases: primary structure, classification, signatures and phylogeny. FEMS Microbiol. Rev. 10, 243–269.
Appendix
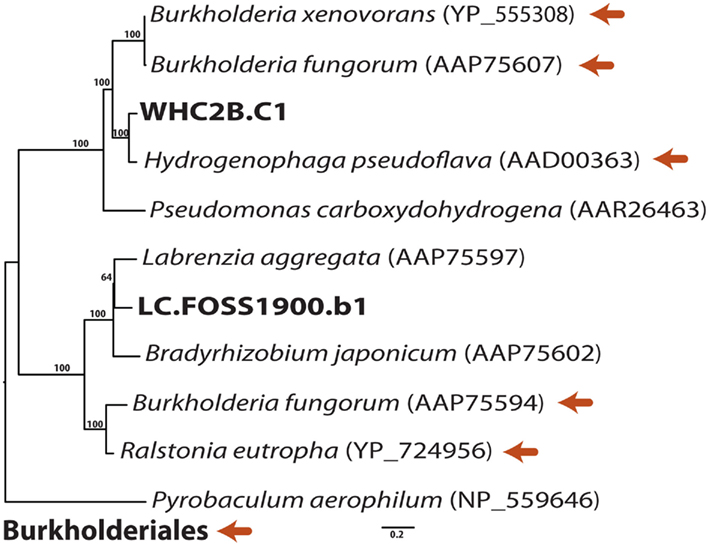
Figure A1. Phylogenetic relationships of putative carbon monoxide dehydrogenase (CODH) sequences in the longest WHC2b contig and in a Lost City shotgun sequencing read. Burkholderiales reference sequences are indicated with orange arrows. Maximum-likelihood bootstrap support values > 50 are shown.
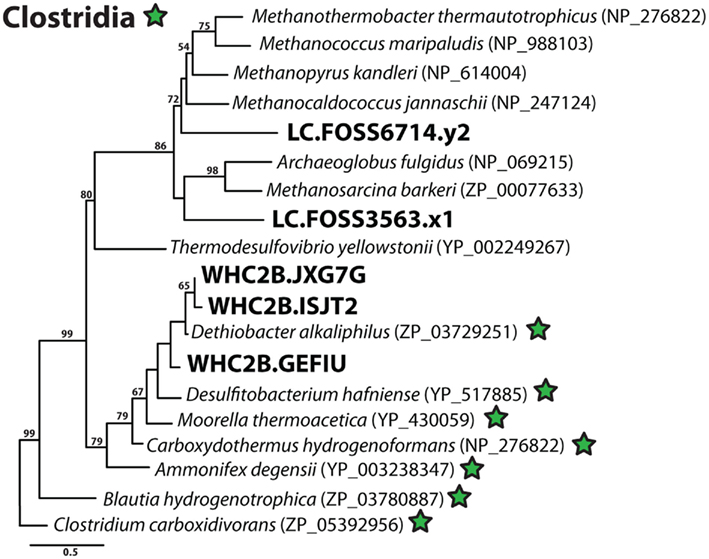
Figure A2. Phylogenetic diversity of putative acetyl-CoA synthase (ACS) sequences in Lost City and Tablelands metagenomic shotgun sequencing reads. Clostridia reference sequences are indicated with green stars, and Lost City (LC), and Tablelands (WHC2b) sequences are in large, bold font. Maximum-likelihood bootstrap support values > 50 are shown.
Keywords: hydrogenase, serpentinization, alkaline spring, metagenome, subsurface
Citation: Brazelton WJ, Nelson B and Schrenk MO (2012) Metagenomic evidence for H2 oxidation and H2 production by serpentinite-hosted subsurface microbial communities. Front. Microbio. 2:268. doi: 10.3389/fmicb.2011.00268
Received: 13 September 2011; Accepted: 16 December 2011;
Published online: 06 January 2012.
Edited by:
Jennifer F. Biddle, University of Delaware, USAReviewed by:
Barbara J. Campbell, University of Delaware, USABrent Craig Christner, Louisiana State University, USA
Copyright: © 2012 Brazelton, Nelson and Schrenk. This is an open-access article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: William J. Brazelton, School of Oceanography, University of Washington, Box 357940, Seattle, WA 98195, USA. e-mail:d2JyYXplbHRvbkBnbWFpbC5jb20=