- Department of Biological Sciences, University of Cincinnati, Cincinnati, OH, USA
Hyperthermophilic archaea exhibit certain molecular-genetic features not seen in bacteria or eukaryotes, and their systems of homologous recombination (HR) remain largely unexplored in vivo. We transformed a Sulfolobus acidocaldarius pyrE mutant with short DNAs that contained multiple non-selected genetic markers within the pyrE gene. From 20 to 40% of the resulting colonies were found to contain two Pyr+ clones with distinct sets of the non-selected markers. The dual-genotype colonies could not be attributed to multiple DNAs entering the cells, or to conjugation between transformed and non-transformed cells. These colonies thus appear to represent genetic sectoring in which regions of heteroduplex DNA formed and then segregated after partial resolution of inter-strand differences. Surprisingly, sectoring was also frequent in cells transformed with single-stranded DNAs. Oligonucleotides produced more sectored transformants when electroporated as single strands than as a duplex, although all forms of donor DNA (positive-strand, negative-strand, and duplex) produced a diversity of genotypes, despite the limited number of markers. The marker patterns in the recombinants indicate that S. acidocaldarius resolves individual mismatches through un-coordinated short-patch excision followed by re-filling of the resulting gap. The conversion events that occur during transformation by single-stranded DNA do not show the strand bias necessary for a system that corrects replication errors effectively; similar events also occur in pre-formed heteroduplex electroporated into the cells. Although numerous mechanistic details remain obscure, the results demonstrate that the HR system of S. acidocaldarius can generate remarkable genetic diversity from short intervals of moderately diverged DNAs.
Introduction
All cellular organisms can transfer strands between two DNA molecules of nearly identical sequences in an active process termed homologous (or generalized) recombination. In addition to promoting genome diversification, homologous recombination (HR) plays various roles that support genome replication and partitioning, and these functions seem to account for its ubiquity (Michel et al., 2001; Burgess, 2004; Kreuzer, 2005). Multiple pathways of HR have been identified in model organisms, and HR proteins have diverged across major phylogenetic groups. Thus, bacteria and eukaryotic cells use distinct sub-families of ssDNA-binding proteins (the RecA and Rad51 families, respectively) to promote strand exchange, and differ further with respect to the helicases, nucleases, and accessory proteins that interact to catalyze and regulate the steps before and after this exchange (Kowalczykowski et al., 1994; Aylon and Kupiec, 2004; Krogh and Symington, 2004). Consistent with their extremely early divergence from the bacterial and eukaryotic lineages, archaea encode a third, structurally distinct sub-family of strand-exchange proteins called the RadA family (Sandler et al., 1999). Although archaea resemble bacteria with respect to certain features of cell and genome structure, archaeal RadA proteins most closely resemble the Rad51 and Dmc1 proteins of eukaryotes (Sandler et al., 1999).
Homologous recombination affects the structure and diversification of all microbial genomes, yet its functional properties in archaea have been investigated for only a few species, primarily extreme halophiles and methanogens. For hyperthermophilic archaea, HR has become increasingly important as the means of targeted gene mutation and other forms of genetic engineering, but it has not been analyzed extensively as a genetic process. Most quantitative analyses of HR in hyperthermophilic archaea have been done in Sulfolobus acidocaldarius, due in part to the availability of natural genetic exchange and various mutants that allow it to be quantified. In Sulfolobus spp., 5-fluoro-orotic acid selects spontaneous pyrE mutants that are uracil auxotrophs, and these mutants provide a convenient assay of recombination events that replace the defective sequence in the recipient genome with the homologous functional sequence.
Detailed analyses of recombinants generated by conjugation and transformation suggest that HR in S. acidocaldarius transfers multiple, short intervals of input DNA unidirectionally to the recipient genome (Hansen et al., 2005; Grogan and Rockwood, 2010). This short-patch, gene-conversion mode of HR mimics bacterial and eukaryotic HR in the absence of functional DNA mismatch repair proteins (Coic et al., 2000; Barnes and McCulloch, 2007; Lin et al., 2009), and thus remains consistent with the natural lack of MutSL homologs in hyperthermophilic archaea (White and Grogan, 2008). A defining feature of this form of HR is that it occurs within a region of heteroduplex formed between complementary strands of the two parental DNAs (Aylon and Kupiec, 2004). However, the genetic analyses performed to date in S. acidocaldarius do not distinguish among three possible alternatives for a heteroduplex formed between a strand of donor DNA and the opposite strand of the recipient chromosome (Figure 1).
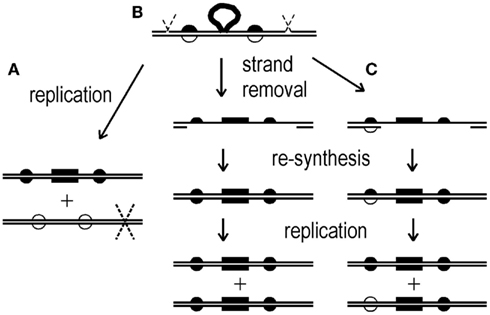
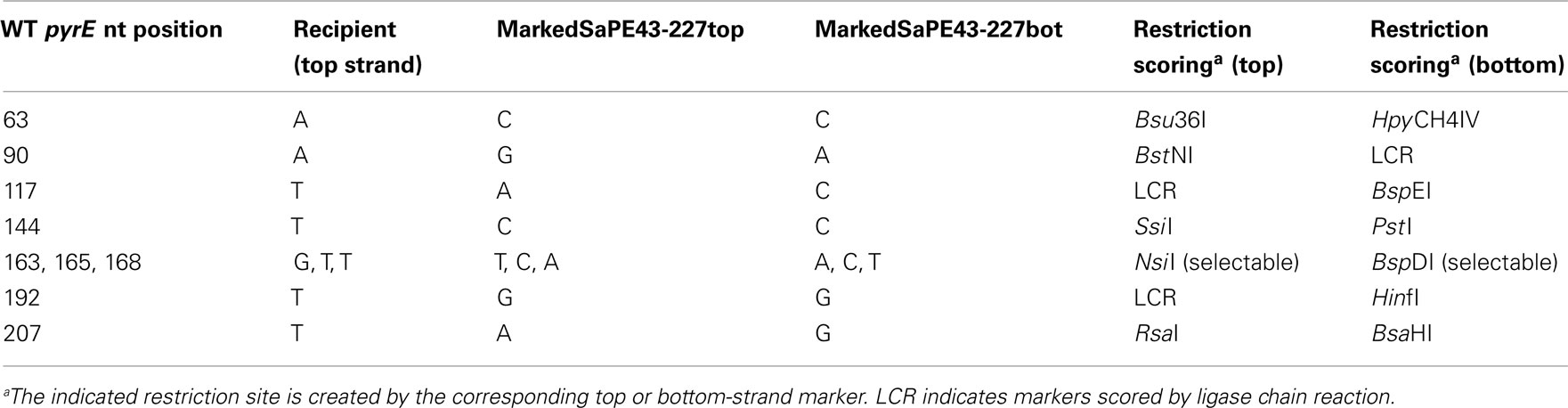
Figure 1. Routes of genetic transformation by non-reciprocal events. All schemes begin with a region of heteroduplex that has formed between a donor (input) DNA and the recipient genome. The donor DNA bears a selectable marker (bar in center) and multiple non-selected markers (dark semi-circles). (A) Incorporation of the donor strand by trimming, gap-filling, and ligation leads to one transformed cell and a non-transformed daughter cell which is lost. (B) Similar incorporation of the donor strand is followed by a large gap on the opposite strand and re-filling, thereby copying all donor markers to the opposite strand (complete conversion) yielding two identical daughter cells. (C) A smaller gap opposite the donor strand results in partial conversion and two distinct daughter cells (genetic sectoring).
For example, if the structure shown in Figure 1 were stabilized by ligation, subsequent DNA replication and cell division would yield one recombinant cell acquiring donor markers and an unaltered recipient cell which, under selective conditions, would not be recovered (Figure 1A). Alternatively, after ligation, a large region of the recipient-strand opposite the donor markers may be removed and the resulting gap re-filled by DNA polymerase, converting all the recipient-strand markers to the donor allele (Figure 1B). In bacteria and eukaryotes, this long-patch excision is a characteristic of the DNA mismatch repair system and promotes the co-repair of markers in heteroduplex DNA (Coic et al., 2000). Replication of the resulting duplex would generate two recombinant daughter cells with the same set of donor markers (Figure 1B). The third alternative is intermediate between these two extremes and would generate a unique, detectable result. In this case, the selected marker, and possibly other markers, are copied to the opposite strand by conversion, as in Figure 1B, but one or more additional markers escape this transfer (Figure 1C). Replication of this modified heteroduplex would segregate two different genotypes, and because both retain the selected marker, the transformant colony retains both genotypes. Such genetically sectored colonies thus imply (i) that a heteroduplex formed during HR and led to transfer of the selected marker by conversion, and (ii) that the conversion did not include one or more additional markers.
In the present study, we used transformation by multiply marked DNA to identify and analyze genetically sectored transformants of S. acidocaldarius. The observed patterns of marker transfer in these transformants indicate sporadic, localized resolution of individual mismatches in a heteroduplex, thereby generating extremely diverse recombinants from short intervals of moderately diverged DNAs.
Materials and Methods
Strains and Growth Conditions
The Sulfolobus strain used for most experiments was S. acidocaldarius strain MR31, which was derived from his-2 (histidine-requiring) mutant DG55 and has an internal deletion of the pyrE gene removing nt 154–171 (Reilly and Grogan, 2001). The pyrE gene encodes orotate phosphoribosyltransferase, which is essential for de novo pyrimidine biosynthesis; a loss-of-function mutation in pyrE thus generates uracil auxotrophs which are also resistant to 5-F orotic acid (Grogan and Gunsalus, 1993; Grogan et al., 2001). For experiments testing the effects of blocking conjugation, mutant SA1 was used; this strain is isogenic to MR31, and contains, in addition to the 18-bp internal pyrE deletion, a deletion of most of the upsE gene encoding an ATP-dependent pilus-assembly protein (Ajon et al., 2011). Cells were cultured at 76–78°C; liquid growth and plating conditions were those described previously (Grogan and Rockwood, 2010).
Donor DNAs
The multicopy plasmid pSaPyrEv3 carries the wild-type pyrE sequence (Saci_1597; Chen et al., 2005) modified with synonymous base-pair substitutions (BPS) regularly spaced along the entire length of the coding sequence (Grogan and Rockwood, 2010). The plasmid does not have a Sulfolobus replication origin and thus cannot replicate in Sulfolobus. Unless otherwise noted, full-length marked pyrE DNA for transformation was amplified from the pyrEv3 cassette by PCR using primers sspyrEfwd and sspyrErev (Table 1). The thermocycler program comprised of an initial denaturation at 95°C for 3 min followed by 39 cycles of denaturation at 95°C for 30 s, annealing at 50°C, and extension at 72°C. This was followed by a final extension at 72°C for 3 min. After PCR and digestions (if any, see below), the amplicons were purified using SureClean (Bioline).
The PCR primers used to amplify the pyrEv3 cassette also provided for the specific digestion of one strand to generate the single-stranded form. The forward primer (sspyrEfwd) was designed to include a 5′-tail with KpnI site (highlighted in red in Table 1) and a 3′-end that anneals to the start of the pyrE (start codon underlined in Table 1). The reverse primer (sspyrErev) has a corresponding 5′-tail with HinP1I site (highlighted in blue in Table 1) and a 3′-end that anneals to end of the pyrE coding sequence (stop codon underlined). Digesting the resulting amplicon with KpnI and HinP1I creates 3′- and 5′-overhangs respectively. The amplicon was then treated with ExoIII, which specifically degrades the 5′-overhang strand, because the strand with 3′-overhang is not a substrate for the exonuclease (Henikoff, 1984). Any residual double-stranded amplicons were eliminated by digestion with SalI, which cuts 5 nt away from the 5′-end of the selectable marker on the sense strand and 1 nt away from the 3′-end of the selectable marker on the anti-sense strand. The double-stranded amplicon cut with SalI did not yield transformants, confirming that this treatment was effective in eliminating ds DNA.
Oligonucleotides were synthesized and de-salted by Integrated DNA technologies (IDT), IA, USA and used without further purification, as described for our previous studies (Grogan and Stengel, 2008; Grogan and Rockwood, 2010). Synthetic donor DNAs “markedSaPE43-227top” and “markedSaPE43-227bot” (Table 2) were designed with regularly spaced non-selected markers as for the pyrEv3 cassette (Grogan and Rockwood, 2010), except that only a 185-bp interval (pyrE nt 43 to 227) was represented. These two ssDNAs were synthesized as high-quality “Ultramers”, and mass spectra provided by the manufacturer indicate that full-length product represented the bulk of both preparations. As in previous studies, no extraneous mutations (i.e., not designed in the oligonucleotides) were detected in any recombinant.
Transformation
S. acidocaldarius was electroporated in a 1-mm cuvette with a single pulse of 1.25 kV at 2 μF and 1 kΩ. Pre- and post-electroporation treatments were those described previously (Grogan and Stengel, 2008). The amount of pyrEv3 DNA was 60–100 ng, and the amount of 185-nt oligo was 10 and 20 pmol. Electroporations were considered saturating with respect to input DNA, as indicated by a slight decrease in transformation efficiency at 20 pmol compared to 10 pmol.
Analysis of Transformants
Up to 24 transformant colonies were picked from independent electroporations of the DNA preparation, drawn mostly from 10-pmol transformations. (A few colonies from 20-pmol DNA transformations were included to determine whether frequency of sectoring varied with amount of DNA, and no such effect was seen.) In initial studies, isolated S. acidocaldarius Pyr+ transformant colonies were suspended in sterile buffer and serially diluted. Dilutions (10−4, 10−5, and 10−6) were plated on selective plates and incubated for 5 days. Eight isolated colonies of sibling cells from each transformant colony were picked and grown in selective liquid medium for about 2 days. Genomic DNA was extracted and a 995-bp fragment containing the pyrE loci was amplified using primers JRset1fwd and JRset1rev (Table 1). Most of the BPSs introduced into the donor DNAs either create or destroy restriction enzyme sites relative to the wild-type sequence, enabling scoring by restriction enzyme digestions and fragment analysis on agarose gels (Grogan and Rockwood, 2010). Restriction-digestion screening using four to five markers equally spaced within the pyrE gene were carried out to identify sibling strains that differed in digestion patterns. Potential cases of sectoring identified by restriction screening were then scored for all polymorphic sites by Sanger sequencing of the PCR amplicons.
In a later streamlined procedure, the original (non-purified) transformant colonies were picked and grown in 3 ml selective medium. Genomic DNA was extracted from 2 ml of culture and pyrE was amplified as described above, while the remaining cells were stored at room temperature until needed for plating (see below). A restriction-digestion screen was carried out on the amplicons to identify samples that showed only partial cleavage, indicating both endonuclease-sensitive and -resistant amplicons in the sample. Apparently dual-genotype cultures were then serially diluted and plated to obtain isolated colonies as described above. The markers in these “sibling colonies” were then scored as described above to confirm distinct clones, and representatives of these sibling clones were then scored at all remaining sites by Sanger sequencing. The two approaches of identifying sectored colonies (i) isolating and analyzing sibling colonies initially from each transformant, vs. (ii) screening non-purified transformant colonies followed by confirmation of purified sibling colonies, yielded comparable efficiencies in detecting sectored colonies from cells transformed with the pyrEv3 cassette. Method (ii) was therefore employed for most of the study, owing to its greater efficiency, to simplify and increase our capacity to screen more samples; only potentially sectored colonies were subjected to confirmatory tests in purified sibling colonies.
Scoring Non-Selected Markers
Most of the BPSs introduced into the native pyrE sequence in the various donor DNAs either create or destroy restriction sites, and were scored in screening procedures by restriction enzyme digestions of PCR amplicons and fragment analysis on agarose gels, as described above. For markers derived from pSaPyrEv3, final scoring was by Sanger sequencing of the amplicons. For oligonucleotide transformants, most markers were scored by restriction. The endonuclease TsoI proved to be unreliable in scoring the marker incorporated at pyrE nt 192, however, so ligase chain reaction (LCR; Wiedmann et al., 1994) was used to score this marker and two other donor alleles that did not alter restriction sites (Table 1). The LCR mixture consisted of approximately 10 ng pyrE DNA, 2 μM of the four probes (Table 1), 1× SYBR Green, 1 × 9°N™ ligase buffer and 5U of 9°N™ (Thermococcus) DNA ligase (New England Biolabs), in a total volume of 20 μl. After an initial denaturation at 95°C for 3 min, the following two-step cycle was used: denaturation at 95°C for 30 s and annealing and ligation at 59.3°C, for 39 cycles. This was followed by a final annealing and ligation at 59.3°C for 3 min. A melting curve of the reaction mixture was obtained from 60 to 95°C by plotting decrease in fluorescence per increment of 0.5°C, and ligation product was identified as a peak at intermediate Tm in the first-derivative curve.
Results
Genetic Evidence of Heteroduplex Segregation
To investigate whether heteroduplex DNA formed by HR can be detected in Sulfolobus spp., we electroporated strain MR31 with a pyrE DNA marked by a set of phenotypically silent mutations (Grogan and Rockwood, 2010). Each of the resulting Pyr+ transformant colonies was then re-suspended in liquid medium, diluted, and re-plated on selective medium to obtain multiple isolated colonies. Four to eight such sub-clones were then analyzed with respect to non-selected (silent) pyrE markers, and apparent cases of two genotypes, indicated by differences found at one or more sites, were confirmed by analysis of all the markers in each of the two distinct daughter lineages (see Materials and Methods). About one-fourth (11/42) of the transformant colonies examined revealed different alleles (base pairs) at one or more marked sites. Similar to previous observations on clonally purified transformants (Grogan and Rockwood, 2010), replacement tracts ranged in length from the entire donor segment down to four markers (Figure 2). Discontinuity was also observed, as typified by two replacement tracts separated by a block of recipient markers (Figure 2).

Figure 2. Dual-genotype transformants. Diagrams show the marker (donor vs. recipient allele) at each of 28 sites distinguishing the input (donor) pyrE sequence from the chromosomal gene (marker positions are not drawn to scale). For details on the marked sites, see Grogan and Rockwood (2010). The donor allele is represented by a filled dot, the recipient allele is an open circle, and the selected marker is represented by a filled square. Each pair of genotypes was recovered from a colony of cells on selective medium that were transformed to the Pyr+ phenotype by double-stranded pyrEv3 DNA (see Materials and Methods).
The recombinants defined several categories of transfer patterns. For example, 9 out of 12 dual-genotype colonies analyzed were pairs in which each sibling incorporated one tract of donor markers, and the resulting tracts shared one endpoint (Figure 2). In four of the colonies, one of the siblings had two separate tracts of donor markers, and in three of these, the second donor tract consisted of a single marker (dsPE1, 4, 6). Other configurations included a single tract in each sibling that differed at both ends (dsPE12).
To confirm that the two-genotype colonies derived from heteroduplex DNA, we first considered whether they could result from two independent recombination events within the same cell. In principle, two DNAs could enter the same recipient cell and recombine separately with two completed copies of the chromosome, for example (Bernander and Poplawski, 1997), or with different arms of a replicating chromosome. In one set of experiments, two distinctly marked 60-nt DNAs which represent the same strand and interval of the pyrE gene (PE AseI+ and PE AseI−; Table 1), were combined to yield an equimolar mixture which was electroporated into recipient cells. A total of 55 Pyr+ transformants were analyzed as unpurified colonies. The selected marker containing an AseI site was recovered in 25 transformants and the alternative allele, without the AseI site, was found in 30. No transformant colony was observed to retain both of these alleles, indicating that most transformants under our conditions reflect incorporation of a single DNA molecule. This implied either that only one DNA enters most cells under these conditions, or that most cells have only one site competent for incorporating exogenous DNA at any given time. In addition, we noted that most of the dual-genotype transformant colonies exhibited a common donor-tract endpoint in pyrE (Figure 2), despite the eight possible left-hand endpoints and 21 possible right-hand endpoints available in this assay. The over-representation of shared endpoints thus argued that the two genotypes originated from a common recombination intermediate.
We also considered whether two-genotype colonies could have resulted from diversification of an initial recombinant cell through a second round of recombination by conjugation with a non-transformed cell on the selective plate. The efficiency of the DNA transfer step in S. acidocaldarius has not been measured experimentally and may not be especially high, since recombinants rarely form at frequencies above 10−4 (Grogan, 1996). However, in contrast to the conjugation system of S. solfataricus, the S. acidocaldarius system functions constitutively (Ajon et al., 2011) and thus cannot be discounted on the basis that the recipient cells were not treated with DNA-damaging agents under our conditions. We therefore tested the dependence of genetic sectoring on conjugation, by performing the same experiments with the conjugation-defective Pil− strain SA1 (Ajon et al., 2011). Dual-genotype colonies represented three of 19 colonies examined, and their patterns of markers resembled those found in the isogenic Pil+ recipient MR31, i.e., uninterrupted tracts of donor markers differing at one end but not the other (Figure 3A). This result demonstrated that the process leading to the dual-genotype colonies was fully functional in the ΔupsE strain, and, therefore, that conjugation is not necessary to produce this class of transformants.
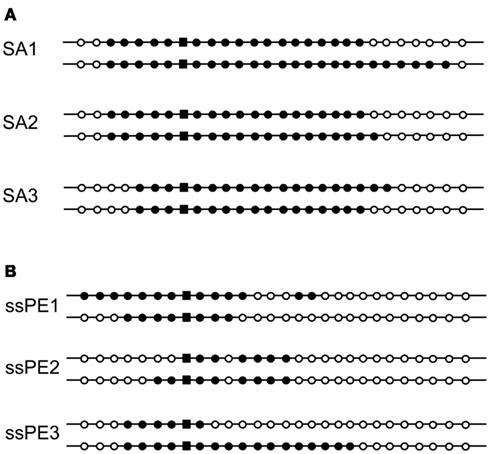
Figure 3. Effects of blocking conjugation or transforming with single-stranded DNA. The genotypes of dual-genotype transformants are shown as for Figure 2. (A) Strain SA1 (conjugation-deficient) transformed with the double-stranded pyrEv3 cassette. (B) Strain MR31 transformed with single-stranded pyrEv3 DNA (see Materials and Methods).
In addition, the hypothesis of conjugation after recombination predicts that, on average, the genotype with the fewer donor markers should be the one generated by conjugation. This reflects the fact that the second recombination event (the one proposed to be initiated by conjugation) would sometimes occur after an additional round of genome replication and thus affect one-fourth or fewer of the clones representing the original HR event. We therefore examined the relative abundance of the genotypes in sectored colonies. Out of six colonies identified using method (i) (see Materials and Methods), five showed greater abundance of genotypes with fewer donor markers and one colony had equal numbers of both, contrary to the prediction of diversification through conjugation. Thus, in all the various control experiments, properties of the dual-genotype colonies remained consistent with segregation of two distinct products of a single HR event.
Genetic Sectoring from ssDNA
In the absence of mutants lacking specific, or even putative, HR functions, we probed mechanistic features of the HR process by altering the donor DNA. We first tested the ability of ssDNA to generate genetic sectoring. Multiply marked pyrE amplicons were digested with exonuclease to remove the top strand; any residual duplex DNA in the preparation was then inactivated by restriction endonuclease before the DNA was electroporated into cells (see Materials and Methods). At least 7% of the resulting transformants were sectored (3/43), and the differences defined by the sibling pairs appeared to be more extensive than those produced by dsDNA (Figure 3B, compare to Figure 2).
Based on this result from enzymatically produced ssDNA, we designed a new assay that used long synthetic oligonucleotides as multiply marked but chemically defined donor. In this scheme, two oligonucleotides represent the top and bottom strands, respectively, of the same 185-bp interval of the functional S. acidocaldarius pyrE gene and incorporated silent BPSs within the selected region and at regular intervals on both sides (Table 2). When electroporated separately into MR31 cells, the two oligonucleotides showed no obvious strand bias (averages of 2.6 vs. 1.8 Pyr+ transformants per pmol for top and bottom strand, respectively). In addition, both top- (T-) and bottom- (B-) strand transformants included sectored colonies, indicating that these short ssDNAs form extensive heteroduplex structures with the S. acidocaldarius chromosome which can segregate two distinct genotypes (Figure 4). The six non-selected, phenotypically silent, markers in each oligonucleotide (Table 2), define 26 = 64 possible genotypes for the transformants in each case. We found 13 of these genotypes among 48 T-strand transformants, and 15 among 48 B-strand transformants. Figure 5 compares the two or three most common of the genotypes recovered under these and other conditions.
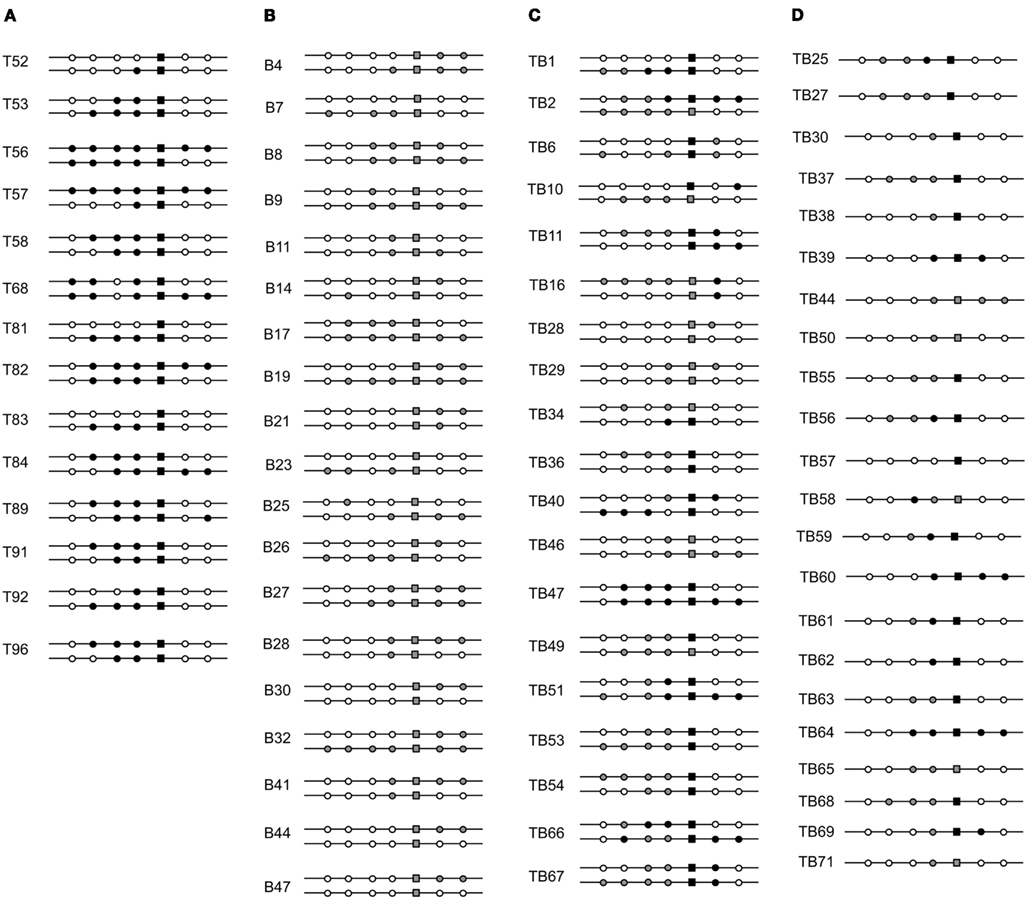
Figure 4. Transformation by differentially marked oligonucleotides. Two synthetic DNAs (Table 2), representing the top strand (T) and bottom strand (B) of base pairs 43–227 of pyrE, were electroporated individually (A,B) or as a duplex (C,D). Markers are given in Table 2; the transformants were selected and analyzed as for Figure 2. (C) Shows pairs of genotypes from sectored colonies, whereas (D) shows the single genotypes of non-sectored colonies recovered in the same experiment. Symbols: top-strand donor allele, black; bottom-strand donor allele, gray; recipient allele, white.
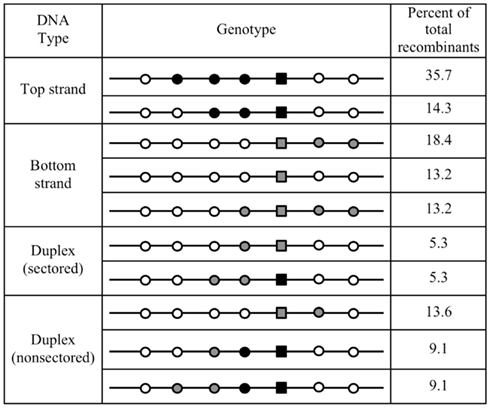
Figure 5. Common genotypes formed by differentially marked oligonucleotides. The two or three most frequent genotypes (data of Figure 4) are indicated with their frequency among all recombinants. It should be noted that the number of potential genotypes is much higher for the duplex than for the individual strands of donor DNA, resulting in lower absolute frequencies of the most frequent genotypes. Symbols follow the scheme of Figure 4.
In the pyrE gene of the transformants, the frequency of the donor allele was 100% at the site of selected marker, and decreased with increasing distance in both directions (Figure 6A). The resulting gradients differed somewhat between T-strand and B-strand transformants however, and the pattern suggested preferential loss of the 3′ portion of both oligonucleotides (Figure 6A). Each transformant contained a continuous block of donor alleles that includes, at a minimum, the selected marker, and often additional, silent markers from the donor; we term this feature the selected donor tract (Grogan and Rockwood, 2010). In most cases, the selected tracts of the two siblings shared one end in common; they differed at both ends in only 8% (1/12) of sectored T-strand colonies, and 16% (3/19) of sectored B-strand colonies.
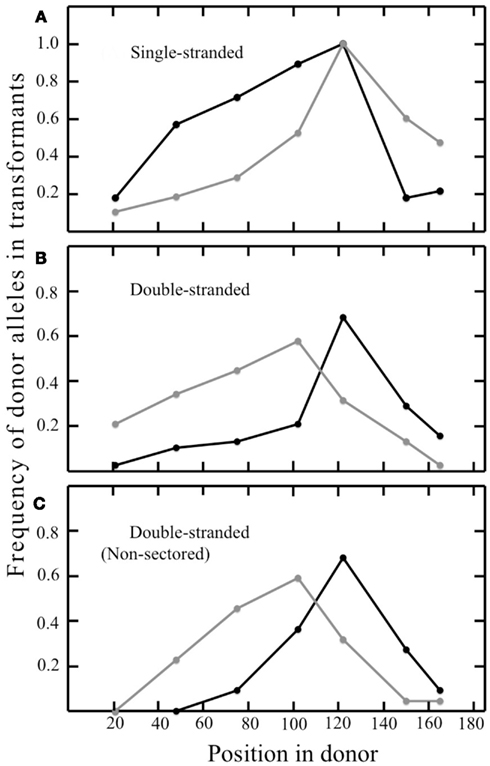
Figure 6. Frequency of strand-specific donor marker incorporation. Graphs show the proportion of recombinants retaining the indicated donor marker at the corresponding position in the synthetic pyrE DNA described in Table 2. Panels and symbols correspond to Figure 4. (A) Sectored transformants from ssDNAs; (B) sectored transformants from dsDNA; (C) non-sectored transformants from dsDNA.
Processing of Pre-Formed Heteroduplex
Both the frequent discontinuity of the recombination tracts and the genetic sectoring implied a form of HR that often separates markers that are very close together, i.e., within 20–30 nt of each other. This pattern would seem difficult to achieve by reciprocal strand exchanges but relatively straightforward by nucleolytic removal of short intervals around mismatches, in a manner resembling excision of the damaged strand during nucleotide excision repair (NER). In this latter scheme, the resulting small gaps would be re-filled by DNA synthesis, thereby copying any allele in the gap to the opposite strand.
To investigate removal of mismatches from a defined heteroduplex, we took advantage of the fact that the two ssDNAs used in the previous study were differentially marked, so that the marker contributed by the top strand could be distinguished from its bottom-strand counterpart at all positions, including the selected region. Equimolar amounts of the two 185-nt strands were therefore annealed in vitro to produce the corresponding duplex with seven regularly spaced mismatches. MR31 cells were electroporated with the resulting duplex, and the pyr+ transformants were analyzed by PCR of the pyrE locus and scoring of the seven markers. The efficiency of transformation by the linear duplex (average of 1 Pyr+ per pmol DNA) was slightly below the efficiencies for each strand separately (see above), while the proportion of sectored transformants exhibiting sectoring was similar [26% (19 of 72 screened) vs. 29 and 31% for T and B strands, respectively].
The sectored transformants were unexpectedly complex. Four of the 19 had acquired markers from only one donor, whereas another four differed with respect to the selected marker, i.e., one sibling had the T-strand allele the other had the B-strand allele (see Figure 4, colonies TB2, TB10, TB34, TB49). These latter recombinants preserve the original duplex configuration at the selected site, but their frequency was not high: 36% (4/11) among all sectored colonies retaining markers from both donor strands, compared to 22% predicted by random association. This suggests that most pre-formed heteroduplex DNAs experienced considerable processing during HR that discarded and resynthesized much of the original duplex.
The sibling pairs that retained both T- and B-strand markers showed a bias in positioning, with B-strand tracts generally extending farther in the left-hand portion, and T-strand tracts extending farther in the right-hand portion. Examining the frequency of T- vs. B-strand markers across all the sectored recombinants confirmed a similar pattern, i.e., lower frequencies of T-strand markers at the left end of the 185-bp interval and lower frequencies of B-strand markers at the right end (Figures 6B,C). This suggested preferential loss of 5′ ends, which is the reverse of the donor vs. recipient bias seen when the same oligonucleotides were electroporated individually (Figure 6A). Thus, although the HR system of transfers markers from both single-stranded and double-stranded DNAs to the S. acidocaldarius genome, processing of the two forms differs in basic respects. We also observed a number of individual clones that incorporated both T- and B-strand donor markers; in each of these cases the transition from one donor source to the other was always immediate, i.e., never interrupted by a recipient marker (Figures 4C,D).
Finally, we investigated whether these patterns of marker transfer from the dually marked duplex to the S. acidocaldarius chromosome were a general feature of HR under these conditions, or alternatively, associated specifically with genetic sectoring. To test this, we analyzed non-sectored Pyr+ transformants from the experiments described above with respect to the T- and B-strand markers. The recombinants appeared qualitatively similar to those recovered from sectored colonies (Figures 4C,D), which was also supported by quantitative comparisons. For example, sectored and non-sectored transformants incorporated statistically indistinguishable numbers of top- and bottom-strand markers (P = 0.778 for sectored and 0.299 for non-sectored, by two-tailed T-test). They also had similar proportions of genotypes incorporating both donor markers (55.3 vs. 54.6%), and similar positions where the transition between the two occurred (pyrE co-ordinate 149 vs. 146). The T-strand allele provided the selected marker in 68.4% of sectored transformants and 68.2% of non-sectored ones, and the two sets of recombinants showed similar profiles of marker incorporation as a function of position within the marked interval (Figures 6B,C). Thus, we found no quantitative evidence that genetic sectoring involves processes distinct from those that generate a single recombinant genotype in cells transformed under these conditions.
Discussion
In the present study, genetic assays applied to a thermoacidophilic crenarchaeote demonstrate for the first time that an archaeon can assimilate three forms of linear DNA (positive-strand, negative-strand, and duplex) into pairs of related recombinant genotypes that represent alternative products of recombination events. The specific features of these sibling pairs, and of the other, non-sectored, recombinants, further indicate that sporadic strand-removal events, many of them highly localized, resolve mismatches in heteroduplex DNA. The apparently stochastic, combinatorial nature of these events distinguish HR in S. acidocaldarius from the dominant modes of HR in eukaryotes and bacteria, and has significant implications for genome stability in Sulfolobus species and other hyperthermophilic archaea.
Incorporation of Linear dsDNA into the S. acidocaldarius Chromosome
Transformation by linear DNA involves recombination in an “ends-out” orientation, and at least two pathways have been proposed for “ends-out” HR in eukaryotes and bacteria, which differ with respect to the predicted segregation of markers flanking the selected marker or gene. The first pathway, sometimes termed “double-crossover,” involves strand exchange, and represents the initial steps of classical DSB/gap repair oriented outward instead of inward: both 5′ ends of the linear duplex are resected to expose 3′ tails, which find their complements in the intact recipient DNA (Figure 7A), and invade the duplex at those sites to generate two displacement (D-) loops (Langston and Symington, 2004). The D-loop intermediate has hDNA flanking the selected marker; sequence divergence of these end regions creates a distinctive configuration of mismatches, defined by the original 3′ tails. This configuration puts input (donor) DNA markers to the right-hand side in one strand and to the left in the other (Figure 7A). If any of these mismatches segregate without being repaired, one daughter cell inherits the donor markers only on the right side of the selected central segment, while the other daughter cell inherits donor markers only on the left side; this arrangement is called the “trans” configuration. The alternative, “strand-assimilation,” pathway (Figure 7B) has been proposed for E. coli expressing the λ red genes (Maresca et al., 2010) and for yeast (Leung et al., 1997), and involves one of the two input strands forming uninterrupted hDNA across the entire region (i.e., the selected region and both flanks), which keeps the flanking markers in a “cis” configuration (i.e., on both sides of the selected gene or marker; Figure 7B).
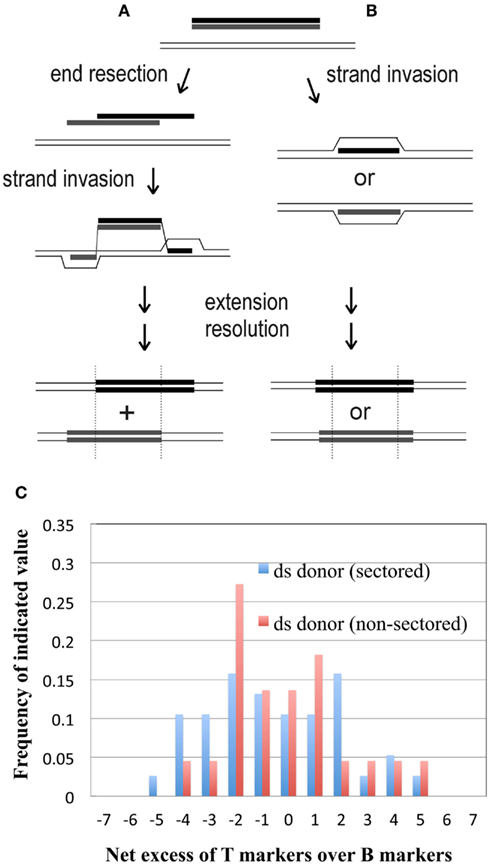
Figure 7. Alternative models of “ ends-out” recombination. Essential features of two alternatives proposed to explain integration of a linear DNA sequence into a cellular genome by HR. (A) “Double-crossover” (strand exchange) model (see Langston and Symington, 2004); (B) “strand-assimilation” model (see Leung et al., 1997); (C) histogram showing the distribution of the net excess of T over B markers among sectored and non-sectored (simple) recombinants from the differentially marked dsDNA.
By genotyping sectored Pyr+ transformant colonies, we could determine the configuration of flanking markers in hDNA that had segregated to yield two distinct selectable Sulfolobus recombinants. The most common configuration lacked the basic, bilateral distribution predicted for ends-out (gene-replacement) HR by both the double-crossover and strand-assimilation models, in that markers segregated on only one side of the central selected region.
Another test of conformity to existing models of “ends-out” HR was provided by the distinct genetic markers in the top and bottom strands of the 185-bp duplex. This short dsDNA documented the original source of the donor markers in all recombinants, including those that did not exhibit sectoring. Both published models of “ends-out” HR predict recombinants that, when considered individually, retain donor markers from either the top or bottom strands of the input duplex, not both (Figures 7A,B). As a test of symmetry, we calculated the net excess of T over B alleles at each marked site for each of the 38 recombinants from sectored colonies and 22 from non-sectored. In both groups (Figure 7C), this measure of marker bias did not give the clearly bimodal distribution predicted by early strand separation (Figure 7A) or complete discarding of one strand before annealing (Figure 7B), even if the choice of strand varied among transformants.
Resolution of Heteroduplex Formed by ssDNA
The genetic sectoring we observed in transformation by ssDNA implies that (i) in the course of HR, the selected marker was retained in the donor strand and also copied to the recipient strand, and (ii) at least one non-selected marker escaped the copying process (depicted schematically in Figure 1C). In our assay, all the non-selected markers are BPS; thus, each sectored transformant generated by ssDNA represents at least one mismatch that was segregated, presumably by replication, without being repaired. However, in only seven of 33 sectored ssDNA transformants was this unrepaired mismatch not accompanied by conversion of at least one other non-selected marker. It seems likely, therefore, that many of the non-sectored ssDNA transformants represent cases in which blocks of markers were copied from donor to chromosomal strand of a heteroduplex before it was replicated, corresponding in Figure 1 to outcome B rather than outcome A.
Conversely, other features of ssDNA transformants argue that mismatches of a heteroduplex intermediate can be repaired individually, not as part of a large block. Specifically, isolated recipient markers (donor-tract interruptions) were generated by both full-length and 185 nt single-stranded forms of pyrE (Figures 2 and 4), which cannot be explained without invoking resolution of mismatches. Our data do not address whether the mismatches triggered the postulated excision events or otherwise influenced their position, but they do provide a test for directionality of the resolution, i.e., whether the donor or the recipient marker was preferentially copied to the opposite strand. This test is provided by the relative abundance of isolated donor markers vs. isolated recipient markers in the transformants, representing resolution of a single mismatch that preserves the donor allele vs. the recipient allele. Among the events in T-strand transformants, one retained the donor allele and three retained the recipient allele. Among B-strand transformants, five retained the donor allele and six retained the recipient allele (the selected marker was excluded from this analysis). Since these ratios are not statistically different from 1, they indicate no directional bias with respect to the discontinuous (donor) strand.
The donor strand in these experiments was an ssDNA 185 nt or shorter annealed to the chromosome; it can thus be considered a plausible mimic of a Sulfolobus Okazaki fragment (Beattie and Bell, 2012), and accordingly, a potential substrate of any hypothetical MutSL-independent post-replication error-correction system that S. acidocaldarius may have. Such a system operating on this type of hDNA would be expected to transfer the recipient allele to the annealed oligonucleotide nearly exclusively. The fact that we observed frequent mismatch resolution but no strand discrimination thus fails to support the hypothesis that S. acidocaldarius has a generalized system of post-replicational error correction. Alternatively, if the mispair resolution we observed does involve components of such a system, the strand discrimination function of that system must require a specific context that was not provided by our experimental conditions.
We also note that patterns of marker transfer similar to those we observe in S. acidocaldarius have been reported for MMR-deficient strains of eukaryotic micro-organisms. Numerous short recombination tracts, including those transferring a single base-pair change, were observed following intragenic HR between diverged sequences in yeast (Coic et al., 2000) and trypanosomes (Barnes and McCulloch, 2007). The behavior appeared only in msh2 mutants, however, indicating that it did not involve MutSL homologs, and in fact was normally masked by their activity. In the yeast assay, the short-patch events were frequently accompanied by segregation of hDNA (Coic et al., 2000), whereas no such segregation was seen in the trypanosomes, for unknown reasons (Barnes and McCulloch, 2007). The similarities between these recombination results and our own thus reinforce the hypothesis that the observed short-patch mode of recombination observed in S. acidocaldarius involves localized resolution of mispairs in heteroduplex DNA without the influence of a post-replicational error-correcting system.
In addition to revealing apparently erratic resolution of mismatches in S. acidocaldarius during HR, both the top- and bottom-strand oligonucleotides exhibited preferential loss of the 3′-end (i.e., right-side markers for T-strand transformants and left-side markers for B-strand transformants; Figure 6A). However, when the same two strands were introduced as a duplex, the 5′-end markers of each strand were lost preferentially (Figures 6B,C), demonstrating that dsDNAs and ssDNAs are processed differently during HR. Resection of 5′ ends of duplex DNA is a critical feature of HR in eukaryotes and bacteria, and may play an important role S. acidocaldarius HR, as well. However, the extent of resection implied by donor markers in the sectored S. acidocaldarius recombinants varied erratically at the two ends of the marked interval. In none of the 19 sectored colonies analyzed did apparent 5′-end resection remove the same number of markers from each end, and in only three cases did it remove at least one marker from each end.
Implications for HR in S. acidocaldarius
These various functional properties are not typical for HR in model micro-organisms, and may reflect a set of recombination mechanisms peculiar to S. acidocaldarius, Sulfolobus species, or hyperthermophilic Archaea as a group. As a basis for designing future studies that could clarify these questions, we offer four functional interpretations that seem mechanistically consistent with our results.
1. S. acidocaldarius processes heteroduplex intermediates of HR by removing single-stranded intervals encompassing individual mispairs or short tracts of mispairs and re-filling the resulting gaps. The intervals are neither especially uniform in size, nor directed with respect to the discontinuous strand of the heteroduplex.
2. S. acidocaldarius incorporates homologous ssDNA into the chromosome to form an extensive heteroduplex. The region of hDNA can be hundreds of bp long, as indicated by the number of markers that can be transferred by one ssDNA. The process that incorporates the ssDNA involves preferential loss of the 3′ end, but remains otherwise undefined.
3. Short-patch conversion can resolve a mispair in any hDNA present in S. acidocaldarius cells at any stage of the HR process. When conversion copies the recipient allele onto the donor side of the heteroduplex, it will often interrupt a string of donor markers, and when it copies the donor allele to the other strand, it will often generate a lone donor marker. Such isolated recipient and donor markers represent the most common form of non-selected marker tract in S. acidocaldarius recombinants (Grogan and Rockwood, 2010). When the transforming DNA has two differentially marked strands, we observe that most of the recombinants inherit donor markers from both strands, and these disparate donor alleles are never separated by recipient alleles. Among sectored transformants, only a minority indicates segregation of the two original donor alleles (positive- vs. negative-strand, or in our terminology, T vs. B) at any position.
Taken together, these properties suggest that localized conversion events copy most of the surviving donor markers from one donor strand to the other before the original strands segregate. These conversion events in pre-formed hDNA are analogous to those proposed to generate discontinuous recombination tracts from ssDNA, but occur within the context of a T:B donor heteroduplex, rather than a donor:recipient heteroduplex.
4. Un-coordinated excision events near the ends of a linear duplex contribute to net resection of 5′ends. Although the pattern of end loss in duplex DNA was biased in favor of 5′ ends, very few sectored transformants indicate the bilateral 5′-end resection of classical HR pathways. In most cases, recombination tracts differed between the two siblings only at one end, and these segregating intervals varied considerably in length. When the two donor strands were distinguished from each other by markers, sectored colonies similarly showed little bilateral resection; a few indicated 5′ resection at each end, but one was mixed (5′ and 3′), one suggested bilateral 3′-end resection, and none had equal numbers of markers removed from the two ends, despite the similar spacing of the markers (Figure 4C).
This lack of uniformity or correlation between the two ends could reflect erratic processing by the HerA helicase and NurA nuclease of S. acidocaldarius, which have been implicated in double-strand break end resection (Constantinesco et al., 2004; Hopkins and Paull, 2008; Quaiser et al., 2008). We note that it also could result, in principle, from the same intrinsically stochastic short-patch excision events proposed to resolve mispairs in hDNA. These events should form extensions (overhangs) of both polarities when they occur very close to one end of the duplex. The 5′ overhangs would be prone to strand re-synthesis primed by the recessed 3′ ends, but no such priming is available for a 3′ extension created by loss of the complementary 5′ end. As a result, 3′ extensions would tend to persist and accumulate. Regardless of how they are formed, 3′-extensions would be expected to promote integration of the donor duplex into the recipient chromosome. However, the “random-excision” pathway we hypothesize predicts that most dsDNAs will have experienced conversion events by the time they integrate into the recipient chromosome. This seems consistent with the high density of T/B conversion events evident in transformants from the dually marked donor duplex (Figure 4C).
Although the four features proposed above leave a number of important steps undefined, including those that form and stabilize heteroduplex DNA, they provide a context for formulating and testing models of HR involving linear DNA in S. acidocaldarius and other hyperthermophilic archaea. The schemes of Figure 8 illustrate a few of the possibilities, but various alternatives also cannot be excluded. Short-patch excision events play central roles in these schemes, and the molecular details of those events also remain unclear, although we favor the idea of a process analogous to NER, in which each mispair promotes formation of a small bubble, one strand of which is excised by structure-specific endonucleases cutting at the boundaries.
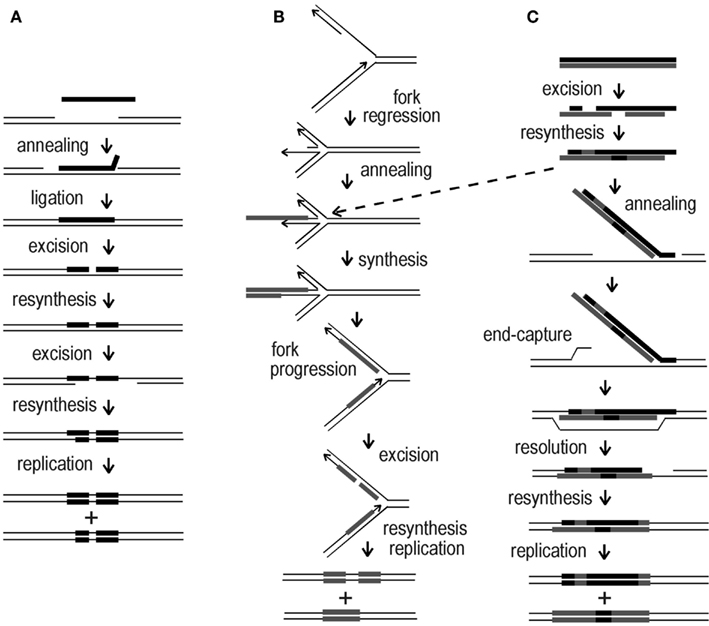
Figure 8. Mechanistic alternatives for “ends-out” HR in Sulfolobus. (A) ssDNA annealing at a transient gap in the chromosome; (B) ssDNA annealing to a transiently regressed replication fork; (C) dsDNA incorporated at a gap in the chromosome, or alternatively, at a regressed replication fork (dotted arrow). Arrowheads indicate 3′ ends, black bars correspond to the top (sense) strand of the pyrE gene, gray bars indicate the bottom (anti-sense) strand.
Figure 8A depicts annealing of a ssDNA (with exclusion of its 3′ end) to a pre-existing gap, incorporation of the base-paired segment by ligation, conversion of one donor marker by a short-patch event, and transfer of a block of markers by a larger conversion event before replication. Figure 8B illustrates an alternative process in which ssDNA anneals to an exposed nascent-strand terminus at a regressed replication fork, followed by second-strand synthesis primed by the terminus, and resumption of replication; this creates two heteroduplex structures of differing length, which can diverge further through short-patch conversions. Figure 8C depicts dsDNA initially undergoing multiple short-patch excision events, which rearrange the markers and expose a 3′ extension. This single-strand extension then anneals to a chromosomal gap and anchors the remaining double-stranded portion until the opposite end can be captured by the recipient genome via some form of strand exchange. Another route to initial capture of the duplex is offered by a regressed replication fork, as indicated by the dotted arrow from scheme Figures 8B,C; in this case, subsequent formation and divergence of two heteroduplexes would follow, as shown in Figure 8B. The schemes, which have deliberately emphasized relatively simple transactions, seem to accommodate nearly all of the marker arrangements observed in recombinants, although we note that ssDNA transformants T11, B11, B25, and B26, and dsDNA transformants TB66 (all shown in Figure 4) imply additional, reciprocal forms of strand exchange not represented in Figure 8.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank J. Rockwood for technical assistance and unpublished data. This work was supported by grant MCB0543910 from the National Science Foundation.
References
Ajon, M., Frols, S., Van Wolferen, M., Stoecker, K., Teichmann, D., Driessen, A. J., Grogan, D. W., Albers, S. V., and Schleper, C. (2011). UV-inducible DNA exchange in hyperthermophilic archaea mediated by type IV pili. Mol. Microbiol. 82, 807–817.
Barnes, R. L., and McCulloch, R. (2007). Trypanosoma brucei homologous recombination is dependent on substrate length and homology, though displays a differential dependence on mismatch repair as substrate length decreases. Nucleic Acids Res. 35, 3478–3493.
Beattie, T. R., and Bell, S. D. (2012). Coordination of multiple enzyme activities by a single PCNA in archaeal Okazaki fragment maturation. EMBO J. 31, 1556–1567.
Bernander, R., and Poplawski, A. (1997). Cell cycle characteristics of thermophilic archaea. J. Bacteriol. 179, 4963–4969.
Chen, L., Brugger, K., Skovgaard, M., Redder, P., She, Q., Torarinsson, E., Greve, B., Awayez, M., Zibat, A., Klenk, H. P., and Garrett, R. A. (2005). The genome of Sulfolobus acidocaldarius, a model organism of the Crenarchaeota. J. Bacteriol. 187, 4992–4999.
Coic, E., Gluck, L., and Fabre, F. (2000). Evidence for short-patch mismatch repair in Saccharomyces cerevisiae. EMBO J. 19, 3408–3417.
Constantinesco, F., Forterre, P., Koonin, E. V., Aravind, L., and Elie, C. (2004). A bipolar DNA helicase gene, herA, clusters with rad50, mre11 and nurA genes in thermophilic archaea. Nucleic Acids Res. 32, 1439–1447.
Grogan, D. W. (1996). Exchange of genetic markers at extremely high temperatures in the archaeon Sulfolobus acidocaldarius. J. Bacteriol. 178, 3207–3211.
Grogan, D. W., Carver, G. T., and Drake, J. W. (2001). Genetic fidelity under harsh conditions: analysis of spontaneous mutation in the thermoacidophilic archaeon Sulfolobus acidocaldarius. Proc. Natl. Acad. Sci. U.S.A. 98, 7928–7933.
Grogan, D. W., and Gunsalus, R. P. (1993). Sulfolobus acidocaldarius synthesizes UMP via a standard de novo pathway: results of a biochemical-genetic study. J. Bacteriol. 175, 1500–1507.
Grogan, D. W., and Rockwood, J. (2010). Discontinuity and limited linkage in the homologous recombination system of a hyperthermophilic archaeon. J. Bacteriol. 192, 4660–4668.
Grogan, D. W., and Stengel, K. R. (2008). Recombination of synthetic oligonucleotides with prokaryotic chromosomes: substrate requirements of the Escherichia coli/lambdaRed and Sulfolobus acidocaldarius recombination systems. Mol. Microbiol. 69, 1255–1265.
Hansen, J. E., Dill, A. C., and Grogan, D. W. (2005). conjugational genetic exchange in the hyperthermophilic archaeon Sulfolobus acidocaldarius: intragenic recombination with minimal dependence on marker separation. J. Bacteriol. 187, 805–809.
Henikoff, S. (1984). Unidirectional digestion with exonuclease III creates targeted breakpoints for DNA sequencing. Gene 28, 351–359.
Hopkins, B. B., and Paull, T. T. (2008). The P. furiosus mre11/rad50 complex promotes 5′ strand resection at a DNA double-strand break. Cell 135, 250–260.
Kowalczykowski, S. C., Dixon, D. A., Eggleston, A. K., Lauder, S. D., and Rehrauer, W. M. (1994). Biochemistry of homologous recombination in Escherichia coli. Microbiol. Rev. 58, 401–465.
Kreuzer, K. N. (2005). Interplay between DNA replication and recombination in prokaryotes. Annu. Rev. Microbiol. 59, 43–67.
Krogh, B. O., and Symington, L. S. (2004). Recombination proteins in yeast. Annu. Rev. Genet. 38, 233–271.
Langston, L. D., and Symington, L. S. (2004). Gene targeting in yeast is initiated by two independent strand invasions. Proc. Natl. Acad. Sci. U.S.A. 101, 15392–15397.
Leung, W., Malkova, A., and Haber, J. E. (1997). Gene targeting by linear duplex DNA frequently occurs by assimilation of a single strand that is subject to preferential mismatch correction. Proc. Natl. Acad. Sci. U.S.A. 94, 6851–6856.
Lin, E. A., Zhang, X. S., Levine, S. M., Gill, S. R., Falush, D., and Blaser, M. J. (2009). Natural transformation of helicobacter pylori involves the integration of short DNA fragments interrupted by gaps of variable size. PLoS Pathog. 5, e1000337. doi:10.1371/journal.ppat.1000337
Maresca, M., Erler, A., Fu, J., Friedrich, A., Zhang, Y., and Stewart, A. F. (2010). Single-stranded heteroduplex intermediates in lambda Red homologous recombination. BMC Mol. Biol. 11, 54. doi:10.1186/1471-2199-11-54
Michel, B., Flores, M. J., Viguera, E., Grompone, G., Seigneur, M., and Bidnenko, V. (2001). Rescue of arrested replication forks by homologous recombination. Proc. Natl. Acad. Sci. U.S.A. 98, 8181–8188.
Quaiser, A., Constantinesco, F., White, M. F., Forterre, P., and Elie, C. (2008). The Mre11 protein interacts with both Rad50 and the HerA bipolar helicase and is recruited to DNA following gamma irradiation in the archaeon Sulfolobus acidocaldarius. BMC Mol. Biol. 9, 25. doi:10.1186/1471-2199-9-25
Reilly, M. S., and Grogan, D. W. (2001). Characterization of intragenic recombination in a hyperthermophilic archaeon via conjugational DNA exchange. J. Bacteriol. 183, 2943–2946.
Sandler, S. J., Hugenholtz, P., Schleper, C., Delong, E. F., Pace, N. R., and Clark, A. J. (1999). Diversity of radA genes from cultured and uncultured archaea: comparative analysis of putative RadA proteins and their use as a phylogenetic marker. J. Bacteriol. 181, 907–915.
White, M. F., and Grogan, D. W. (2008). “DNA stability and repair,” in Thermophiles: Biology and Technology at High Temperatures, eds F. T. Robb, G. Antranikian, D. W. Grogan, and A. J. Driessen (Boca Raton, FL: CRC Press), 179–188.
Keywords: linear DNA, genetic transformation, mismatch repair, gene conversion
Citation: Mao D and Grogan DW (2012) Heteroduplex formation, mismatch resolution, and genetic sectoring during homologous recombination in the hyperthermophilic archaeon Sulfolobus acidocaldarius. Front. Microbio. 3:192. doi: 10.3389/fmicb.2012.00192
Received: 06 March 2012; Accepted: 11 May 2012;
Published online: 05 June 2012.
Edited by:
Frank T. Robb, University of California at Riverside, USAReviewed by:
Barbara Methé, The J. Craig Venter Institute, USAImke Schroeder, University of California Los Angeles, USA
Marla Tuffin, University of the Western Cape, South Africa
Copyright: © 2012 Mao and Grogan. This is an openaccess article distributed under the terms of the Creative Commons Attribution Non Commercial License, which permits non-commercial use, distribution, and reproduction in other forums, provided the original authors and source are credited.
*Correspondence: Dennis W. Grogan, Department of Biological Sciences, University of Cincinnati, 614 Rieveschl Hall, ML0006, Clifton Court, Cincinnati, OH 45221-0006, USA. e-mail:Z3JvZ2FuZHdAdWNtYWlsLnVjLmVkdQ==