- 1Division of Microbiology and Genetics, Institute of Resource Development and Analysis, Kumamoto University, Kumamoto, Japan
- 2Centre for Cellular and Molecular Biology, Hyderabad, India
Noroviruses cause most cases of acute viral gastroenteritis worldwide. The lack of a cell culture infection model for human norovirus necessitates the use of molecular methods and/or viral surrogate models amenable to cell culture to predict norovirus inactivation. Murine norovirus (MNV) may be used to construct a small animal model for studying the biology and pathogenesis of noroviruses because MNV is the only norovirus that replicates in cell culture and a small animal model. However, recent studies have shown that natural MNV infection is widespread in laboratory mouse colonies. We investigated MNV infection in both conventional and specific pathogen-free (SPF) genetically modified mice from Japan and the US, and commercial mice from several animal breeders in Japan, using serological and molecular techniques. MNV antibodies were detected in 67.3% of conventional mice and 39.1% of SPF mice from Japan and 62.5% of conventional mice from the US. MNV antibodies were also found in 20% of commercial SPF C57BL/6 mice from one of three breeders. Partial gene amplification of fecal isolates from infected animals showed that the isolates were homologous to reported MNV sequences. These results suggest that both conventional and SPF laboratory mice, including commercial mice, are widely infected with MNV, which might require considerable attention as an animal model of human disease.
Introduction
Human noroviruses are the major cause of non-bacterial epidemic gastroenteritis worldwide (Patel et al., 2009). The study of human norovirus has been hampered by the lack of a cell culture system. Murine norovirus (MNV) was first isolated and characterized as a sporadic and lethal pathogen in immunocompromised knockout mice (Karst et al., 2003). MNV is the only norovirus that replicates in cell culture and in a small animal model, though several studies have reported norovirus infection in humans, cattle, swine, dogs, and mice (Wobus et al., 2006; Patel et al., 2009). Thus, MNV is expected to be a surrogate for evaluating the resistance of human norovirus to disinfectants and can be used in animal model studies of human norovirus infection.
Natural MNV infection is prevalent in animal facilities around the world (Hsu et al., 2005; Perdue et al., 2007; Pritchett-Corning et al., 2009). These infections might influence not only the results of a mouse model for studying the biology and pathogenesis of noroviruses, but also those of other biological studies (Lencioni et al., 2008; Cadwell et al., 2010). Until recently, no MNV infection had been reported in laboratory mice in Japan. However, the first reports of MNV detected in conventional mouse colonies in Japan were published in 2009 (Goto et al., 2009b; Kitajima et al., 2009). Recent serological analysis of MNV found that MNV infection is also prevalent in conventional animal colonies in Japan (Kitagawa et al., 2010). However, information on the prevalence of MNV infection in specific pathogen-free (SPF) mice colonies and commercial SPF mice in Japan is not currently available. Most researchers in Japan think that MNV infection is absent among SPF mice facilities.
The Institute of Resource Development and Analysis at Kumamoto University collects, preserves, and distributes experimental animals; thus, numerous mice are transferred to the institute from across Japan and from the other countries for embryo freezing or microbiological cleaning (Nakagata and Yamamura, 2009). Our division in the institute checks the microbiological status of genetically modified mice before or after embryo transfer. In the present study, we examined the prevalence of MNV infection in conventional and SPF genetically modified mice and commercial SPF mice using both serological and molecular biological methods.
Materials and Methods
Mice
Ninety-six genetically modified mice derived from 29 facilities in 2011, including 18 mice derived from five different animal facilities in the US which were to be converted to SPF grade by in vitro fertilization, were used in this study. Thirty commercial SPF C57BL/6 mice derived from three breeders in Japan were tested for MNV infection. After arriving at our facility, the mice were kept in the laminar flow racks (negative air pressure) in our quarantine animal rooms, which are separate from the SPF animal rooms, until the collection of embryos and sperm. The quarantine period was less than 1 week, except for five mice used in an experiment for the frequency of detection of MNV. All procedures involving animals and their care were approved by the Animal Care Committee of Kumamoto University in accordance with the Regulations for Animal Experiments at Kumamoto University.
Microbiological Monitoring
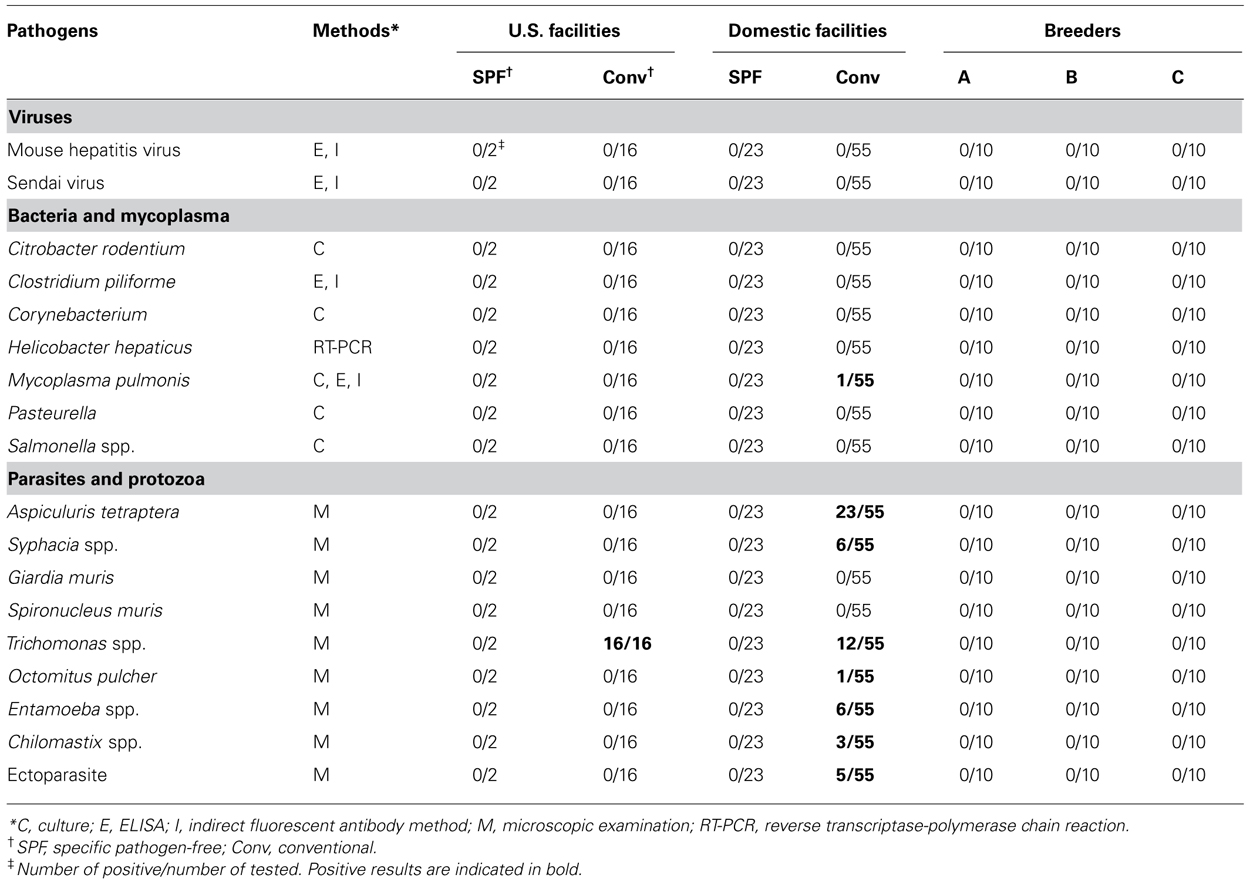
The animals were checked for the presence of specific pathogens using routine methods described previously (Goto et al., 2009a). Briefly, the analysis included a range of viruses, bacteria, mycoplasmas, and protozoans, which are listed in full in Table 1. The absence of all tested pathogens indicated that an animal was SPF grade.
Serological Test for MNV
Serological analysis of MNV infection was performed using an enzyme-linked immunosorbent assay (ELISA) kit (Mouse Norovirus; Biotech Trading Partners, Encinitas, CA, USA) according to the manufacturer’s protocol.
RNA Isolation and Reverse Transcriptase-Polymerase Chain Reaction
Total RNA was extracted from fresh feces as described previously (Nakamura et al., 2005). Primers and reverse transcriptase-polymerase chain reaction (RT-PCR) conditions were described elsewhere (Kitajima et al., 2009). PCR products were separated by 2% agarose gel electrophoresis.
Sequence Analysis
Polymerase chain reaction products were sequenced as described previously (Ohsugi et al., 2004). Nucleotide sequences were aligned using ClustalW. A phylogenetic tree was generated from a bootstrap analysis of 1,000 replicates using the neighbor joining method. Evolutionary distances were computed by the p-distance method using MEGA 5.05 software (Tamura et al., 2011).
Results
Identification of Microbiological Status in Mice Reared Under SPF or Conventional Conditions
Microbiological analysis was performed in 96 mice from 29 animal facilities, including 18 mice from five US facilities. In addition, 10 of each SPF female C57BL/6 mouse were purchased from breeders A, B, and C (Figure 1). Most of the tested mice were negative for viruses and bacteria (Table 1). Mycoplasma pulmonis was isolated from a single mouse from a Japanese conventional facility. Trichomonas spp. contamination was common in US facilities, despite their disparate geographic locations across the country. Almost all of the mice reared under conventional conditions were positive for parasites and protozoa.
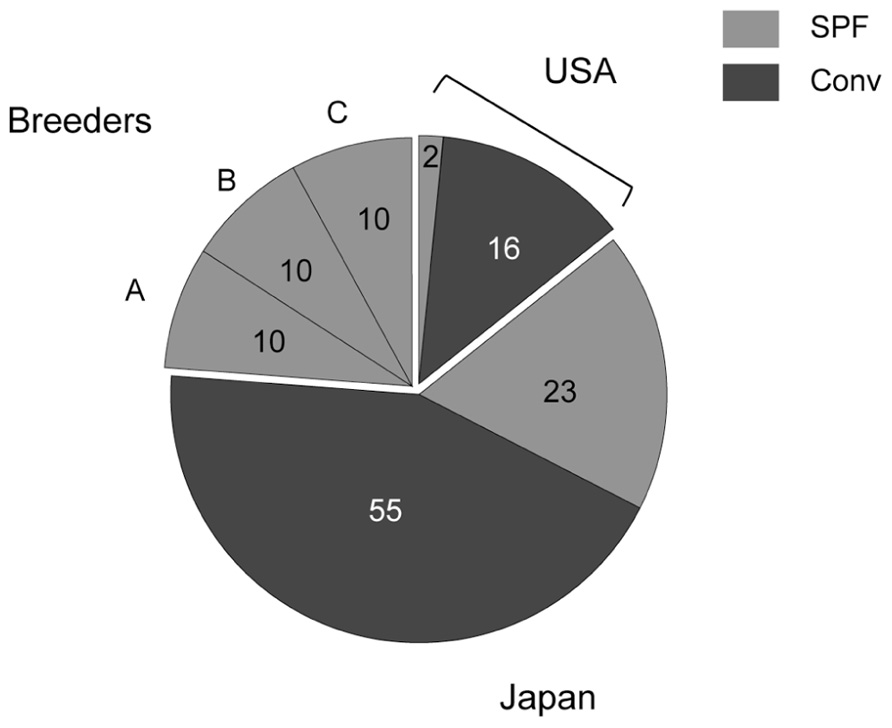
FIGURE 1. Numbers of tested mice. Two SPF and 16 conventional mice derived from US animal facilities, 23 SPF and 55 conventional mice derived from domestic facilities, and 10 of each SPF female C57BL/6 mouse obtained from breeders A, B, and C were tested in this study.
Antibodies to MNV in Animal Facilities in Japan and the US and Commercial Mice in Japan
Specific pathogen-free mice from the US were negative for MNV, whereas 62.5% (10/16) of conventional mice from US animal facilities were positive for MNV (Figure 2A). MNV infection was found in 39.1% (9/23) of SPF and 67.3% (37/55) of conventional mice in Japan. Surprisingly, antibodies were found in 20% (2/10) of commercial SPF mice derived from breeder C.
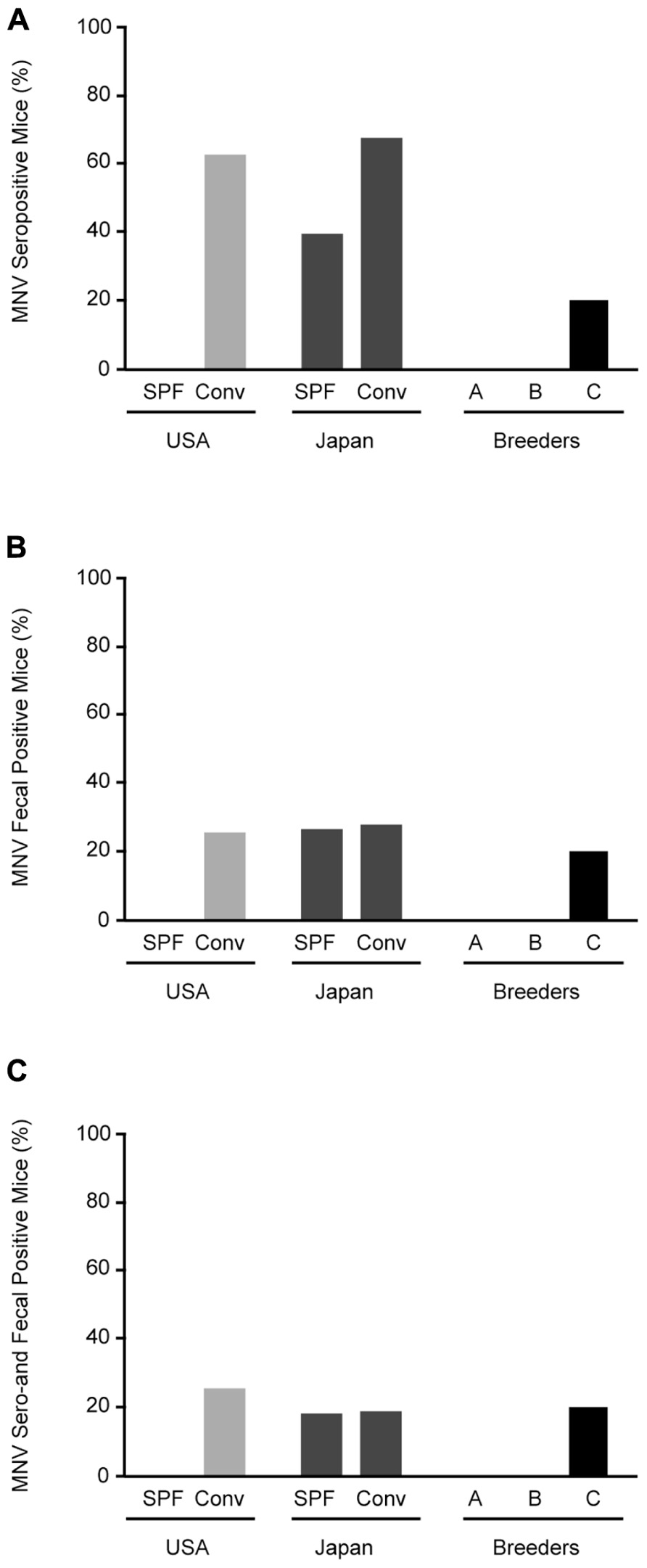
FIGURE 2. Prevalence of MNV infection in conventional and SPF mice. (A) MNV seropositive mice from animal facilities in Japan and the US and breeders in Japan. (B) RT-PCR detection of MNV sequences in feces from tested mice. (C) Mice positive for both MNV antibodies by ELISA and MNV-specific sequences in feces by RT-PCR. SPF, specific pathogen-free; Conv, conventional.
Detection of MNV in FECES from SPF or Conventional Mice
Stool specimens were collected from 126 animals from 32 separate laboratory colonies in Japan and the US and screened for MNV (Figure 2B). No MNV genome segments were isolated from SPF mice from the US, similar to the serological study, whereas MNV was isolated from 25% (4/16) of conventional mice. MNV was isolated from 26% (6/23) of SPF mice and 27.3% (15/55) of conventional mice from Japan. The frequency of MNV isolation was similar between SPF and conventional mice from Japan and the US. In commercial SPF mice, MNV was isolated from 20% (2/10) of the mice from breeder C. The mice positive by both serological and molecular biological methods are shown in Figure 2C. These results suggest that MNV infection is widespread in both conventional and SPF mice, including commercial mice in Japan.
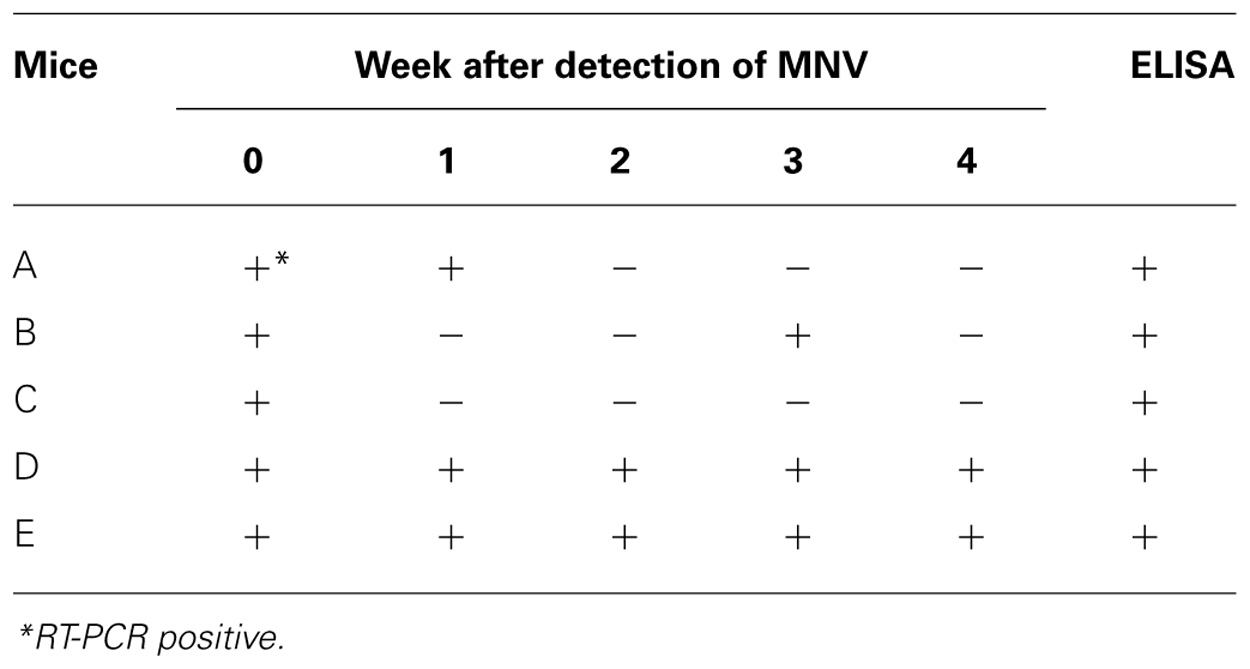
Frequency of Detection of MNV
Five mice were autopsied 4 weeks after MNV-specific sequence detection and tested for antibodies against MNV (Table 2). All tested mice had antibodies for MNV. The MNV-specific sequence was not always detected in the feces of the mice except mice D and E during tested 4 weeks. The sensitivity of RT-PCR did not improve with different PCR conditions, PCR reagents, or primers. Thus, we speculated that the amount of MNV particles in the feces might vary in each mouse due to, for example, host defenses or intestinal circumstances.
Phylogenetic Analysis
The partial nucleotide sequence was determined for the 5′ terminus of the capsid gene (Figure 3). Phylogenetic analysis of the partial capsid sequences of MNV showed widespread expression in various facilities throughout Japan and the US, and that these virus strains comprise reported MNV subgroups. Reference strains MNV-1 and Ku29 derived from an SPF facility in Japan were divergent from the other strain group including reference stains MNV-2, -3, and -4. Ku02 and Ku04 derived from the same facilities in Japan exhibited moderate similarity, but the sequences of Ku22 and Ku23 derived from different conventional US facilities were closely related despite their disparate geographic locations across the country.
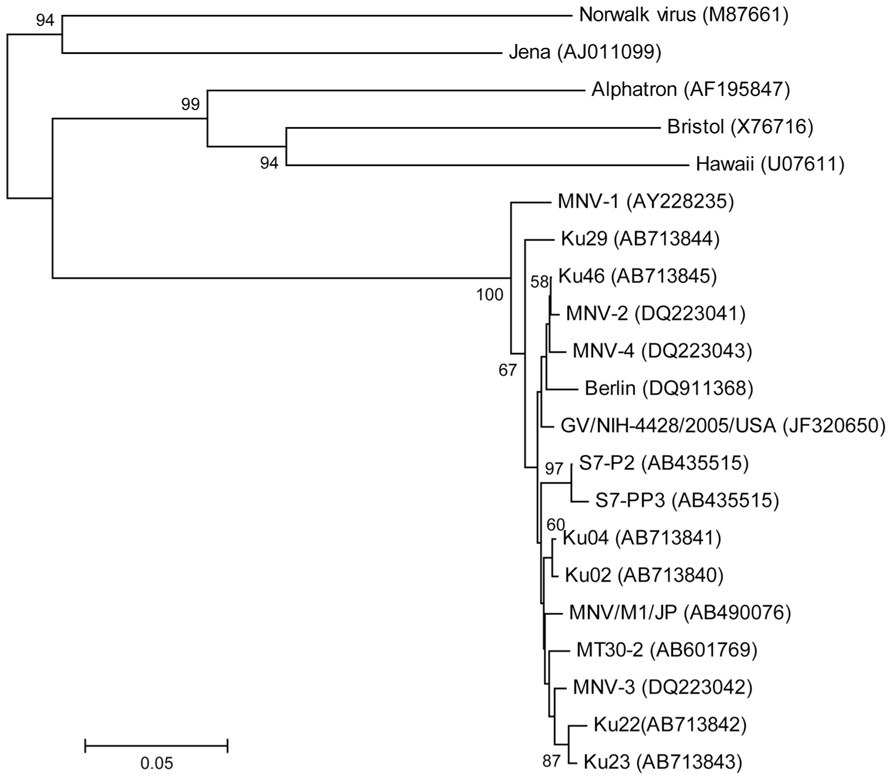
FIGURE 3. Phylogenetic analysis of the partial capsid gene of norovirus (approximately 350 nucleotides). Phylogenetic relationships were inferred using the neighbor joining method, with the evolutionary distances computed using the p-distance model. The statistical support for tree nodes was evaluated by bootstrap analysis (1,000 replicates). Bootstrap values (>50%) are indicated at branch nodes. Scale bar indicates nucleotide substitutions per site. Numbers in parentheses indicate GenBank accession numbers. Human isolates, Norwalk, Alphatron, Bristol, and Hawaii; bovine isolate, Jena; North American isolates, MNV-1, -2, -3, and -4, GV/NIH-4428/2005/USA, Ku22, and Ku 23; European isolate, Berlin; Japanese isolates, S7-P2, S7-PP3, MNV/M1/JP, MT30-2, Ku02, Ku04, Ku46, and Ku29 (SPF mice).
Discussion
In this study, we analyzed MNV infection in mice derived from animal facilities in Japan, the US, and three breeders (Japan) using serological and molecular techniques. We show that MNV infection is widespread among both conventional and SPF mice in Japan, as well as commercial SPF mice. In a previous study, serological analysis revealed that 53.4% of conventional mice are positive for MNV, 48% positive for mouse hepatitis virus, and 31.2% positive for M. pulmonis (Kitagawa et al., 2010). Consistent with this report, we show that MNV is the most prevalent infection in mouse laboratory colonies, and the mice we tested, with the exception of one case, were negative for the antibodies for hepatitis virus and M. pulmonis (Table 1). This report is the first on MNV infection in SPF mice in Japan and commercial mice.
The frequency of MNV infection as detected by RT-PCR was lower than that detected by serological study using ELISA. We detected MNV antibodies in all five mice tested 4 weeks after the first MNV sequences were detected by RT-PCR Using RT-PCR, MNV sequences were detected consistently in the feces of only two mice. Thus, a survey for MNV infection might require the use of serological methods, such as ELISA. The first norovirus to infect mice, MNV-1, caused death in severely immunocompromised mice lacking recombination-activating gene 2 (RAG2) and signal transducer and activator of transcription 1 (STAT-1; RAG2/STAT1-/-) and mice lacking both the alpha/beta interferon (IFN-α/β) and the IFN-γ receptors (IFN-αβγR-/-; Karst et al., 2003; Mumphrey et al., 2007). In this study, the mice infected with MNV exhibited no clinical signs, though the tested mice, with the exception of the mice from breeders, were genetically modified, including compromised immune systems. Recently, MNVs (except MNV-1) were associated with asymptomatic infection and shedding in both normal and genetically modified mice (Hsu et al., 2006; Muller et al., 2007). The origin of MNV in laboratory mouse strains is unknown. Phylogenetic analysis of the partial capsid sequences of MNV in this study showed that the isolated virus strains comprised the reported MNV subgroups, though MNV-1 and Ku29 isolated from the SPF facility were divergent form the main group. However, bootstrap values for the position of Ku29 were not high (<70%). Thus, further study is needed to address whether the Ku29 isolate belongs to a new MNV genotype. These results indicate that attenuated (persistent) strains of MNV, such as MNV-2, -3, and -4, are spread around the world, making the viruses difficult to detect in animals without active screening (Muller et al., 2007; Barron et al., 2011). Using ELISA, we did not detect MNV antibodies in stocked serum from mice transported from domestic animal facilities before 2009 (data not shown). These results suggest that MNV has been spreading through 50% of animal facilities in Japan in only 2 years.
Finally, our results suggest that MNV infections have spread widely throughout the animal facilities in Japan, not only conventional mice colonies, but also SPF mice colonies. The findings warrant further studies to elucidate the spread of MNV infection in SPF colonies, including breeders, in Japan. These results also suggest that SPF laboratory mice, including commercial mice, which might require considerable attention as an animal model for human norovirus infection.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of anycommercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Yoshiteru Tanaka and Keiko Honda for their excellent technical assistance. This work was supported in part by a Grant-in-Aid for Scientific Research from the Japan Society for the Promotion of Science, the Japan Leukaemia Research Fund, and Kumamoto University’s Centers of Excellence (COE) Programs.
References
Barron, E. L., Sosnovtsev, S. V., Bok, K., Prikhodko, V., Sandoval-Jaime, C., Rhodes, C. R., et al. (2011). Diversity of murine norovirus strains isolated from asymptomatic mice of different genetic backgrounds within a single U.S. research institute. PLoS ONE 6:e21435. doi: 10.1371/journal.pone.0021435
Cadwell, K., Patel, K. K., Maloney, N. S., Liu, T. C., Ng, A. C., Storer, C. E., et al. (2010). Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell 141, 1135–1145.
Goto, K., Hayashimoto, N., Ishida, T., Takakura, A., and Kagiyama, N. (2009a). First trial in the developmental phase of the “performance evaluation program” based on the ICLAS animal quality network program: self-assessment of microbiological monitoring methods using test samples supplied by ICLAS. Exp. Anim. 58, 47–52.
Goto, K., Hayashimoto, N., Yasuda, M., Ishida, T., Kameda, S., Takakura, A., et al. (2009b). Molecular detection of murine norovirus from experimentally and spontaneously infected mice. Exp. Anim. 58, 135–140.
Hsu, C. C., Riley, L. K., Wills, H. M., and Livingston, R. S. (2006). Persistent infection with and serologic cross-reactivity of three novel murine noroviruses. Comp. Med. 56, 247–251.
Hsu, C. C., Wobus, C. E., Steffen, E. K., Riley, L. K., and Livingston, R. S. (2005). Development of a microsphere-based serologic multiplexed fluorescent immunoassay and a reverse transcriptase PCR assay to detect murine norovirus 1 infection in mice. Clin. Diagn. Lab. Immunol. 12, 1145–1151.
Karst, S. M., Wobus, C. E., Lay, M., Davidson, J., and Virgin, H. W. T. (2003). STAT1-dependent innate immunity to a Norwalk-like virus. Science 299, 1575–1578.
Kitagawa, Y., Tohya, Y., Ike, F., Kajita, A., Park, S. J., Ishii, Y., et al. (2010). Indirect ELISA and indirect immunofluorescent antibody assay for detecting the antibody against murine norovirus S7 in mice. Exp. Anim. 59, 47–55.
Kitajima, M., Oka, T., Tohya, Y., Katayama, H., Takeda, N., and Katayama, K. (2009). Development of a broadly reactive nested reverse transcription-PCR assay to detect murine noroviruses, and investigation of the prevalence of murine noroviruses in laboratory mice in Japan. Microbiol. Immunol. 53, 531–534.
Lencioni, K. C., Seamons, A., Treuting, P. M., Maggio-Price, L., and Brabb, T. (2008). Murine norovirus: an intercurrent variable in a mouse model of bacteria-induced inflammatory bowel disease. Comp. Med. 58, 522–533.
Muller, B., Klemm, U., Mas Marques, A., and Schreier, E. (2007). Genetic diversity and recombination of murine noroviruses in immunocompromised mice. Arch. Virol. 152, 1709–1719.
Mumphrey, S. M., Changotra, H., Moore, T. N., Heimann-Nichols, E. R., Wobus, C. E., Reilly, M. J., et al. (2007). Murine norovirus 1 infection is associated with histopathological changes in immunocompetent hosts, but clinical disease is prevented by STAT1-dependent interferon responses. J. Virol. 81, 3251–3263.
Nakagata, N., and Yamamura, K. (2009). Current activities of CARD as an international core center for mouse resources. Exp. Anim. 58, 343–350.
Nakamura, N., Yoshizumi, S., Urano, T., and Ohsugi, T. (2005). Differentiation of mouse hepatitis virus genotypes by restriction fragment length polymorphism analysis. Lab. Anim. 39, 107–110.
Ohsugi, T., Kumasaka, T., and Urano, T. (2004). Construction of a full-length human T cell leukemia virus type I genome from MT-2 cells containing multiple defective proviruses using overlapping polymerase chain reaction. Anal. Biochem. 329, 281–288.
Patel, M. M., Hall, A. J., Vinje, J., and Parashar, U. D. (2009). Noroviruses: a comprehensive review. J. Clin. Virol. 44, 1–8.
Perdue, K. A., Green, K. Y., Copeland, M., Barron, E., Mandel, M., Faucette, L. J., et al. (2007). Naturally occurring murine norovirus infection in a large research institution. J. Am. Assoc. Lab. Anim. Sci. 46, 39–45.
Pritchett-Corning, K. R., Cosentino, J., and Clifford, C. B. (2009). Contemporary prevalence of infectious agents in laboratory mice and rats. Lab. Anim. 43, 165–173.
Tamura, K., Peterson, D., Peterson, N., Stecher, G., Nei, M., and Kumar, S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol. Biol. Evol. 28, 2731–2739.
Keywords: genetically modified mice, mice, microbiological monitoring, MNV, mouse norovirus, specific pathogen-free
Citation: Ohsugi T, Matsuura K, Kawabe S, Nakamura N, Kumar JM, Wakamiya M, Morikawa S and Urano T (2013) Natural infection of murine norovirus in conventional and specific pathogen-free laboratory mice. Front. Microbio. 4:12. doi: 10.3389/fmicb.2013.00012
Received: 27 December 2012; Accepted: 14 January 2013;
Published online: 30 January 2013.
Edited by:
Akio Adachi, The University of Tokushima Graduate School, JapanReviewed by:
Akio Adachi, The University of Tokushima Graduate School, JapanHironori Sato, National Institute of Infectious Diseases, Japan
Copyright: © 2013 Ohsugi, Matsuura, Kawabe, Nakamura, Kumar, Wakamiya, Morikawa and Urano. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Takeo Ohsugi, Division of Microbiology and Genetics, Institute of Resource Development and Analysis, Kumamoto University, 2-2-1 Honjo, Kumamoto 860-0811, Japan. e-mail:b2hzdWdpQGdwby5rdW1hbW90by11LmFjLmpw

 Kumi Matsuura1
Kumi Matsuura1