- Unité de Recherche sur les Maladies Infectieuses et Tropicales Émergentes, UM63, CNRS 7278, IRD 198, INSERM 1095, Institut Hospitalo-Universitaire Méditerranée Infection, Aix-Marseille Université, Marseille, France
The increase and spread of antibiotic resistance (AR) over the past decade in human pathogens has become a worldwide health concern. Recent genomic and metagenomic studies in humans, animals, in food and in the environment have led to the discovery of a huge reservoir of AR genes called the resistome that could be mobilized and transferred from these sources to human pathogens. AR is a natural phenomenon developed by bacteria to protect antibiotic-producing bacteria from their own products and also to increase their survival in highly competitive microbial environments. Although antibiotics are used extensively in humans and animals, there is also considerable usage of antibiotics in agriculture, especially in animal feeds and aquaculture. The aim of this review is to give an overview of the sources of AR and the use of antibiotics in these reservoirs as selectors for emergence of AR bacteria in humans via the food chain.
Introduction
The recent increase and spread of antibiotic resistance (AR) in human pathogens over the past decade has become a global problem, and is mainly due to the overuse and/or misuse of antibiotics in humans. However, there are many other sources of selective pressure such as use of antibiotics in agriculture that are known to be drivers of AR. Moreover, recent genomic and metagenomic studies in humans, animals, and the environment have led to the discovery of a large reservoir of new and/or unknown AR genes in these reservoirs that represent the so-called “ intrinsic resistome” that represent a large subset of non-acquired AR genes whose functions are multiple and complex in nature (Rolain et al., 2012; Baquero et al., 2013). Antibiotics are natural, semi-synthetic, or synthetic compounds with antimicrobial activity and are administered to both humans and animals for several purposes, including treatment, prevention, and growth promotion (animals). The mechanisms of intrinsic or “ natural AR” itself are complex and are of ancient origin and are marked by recent evolutionary adaptations. Intrinsic AR is a natural phenomenon developed by bacteria to protect antibiotic-producing bacteria from their own products and also to increase their chances of survival in highly competitive environments (D’Costa et al., 2011). Moreover, antibiotics produced by natural organisms have many other functions in nature mainly for cell to cell signaling networks and homeostasis of bacterial communities (Martinez and Rojo, 2011; Baquero et al., 2013). Thus production of antibiotics in environment is counterbalanced by natural resistant organisms in a socially cohesive ecological population (Cordero et al., 2012). Emergence of AR genes in pathogenic bacteria in humans is a recent phenomenon that occurred via mobilization from such AR gene pool (D’Costa et al., 2006; Forsberg et al., 2012) either through chromosomal mutations or mainly by lateral transfer of AR determinants from other bacteria. Although most of the AR genes found in human pathogens have a recent origin, there are pieces of evidence that AR genes could be also found in samples from pre-antibiotic era (D’Costa et al., 2011; Bhullar et al., 2012). Finally, because most of AR genes are located in nature in the “ intrinsic resistome” with many different functions, emergence of AR genes in clinical isolates represent only a small part of the total of these genes. Industrial use and release of antibiotics in microbial environments is a recent phenomenon with many consequences on bacterial population and evolution of bacteria in natural environments. It has been well-demonstrated that the use of antibiotics as growth promoters in food animals selects for AR bacteria that may spread from food to humans leading to human infection (Phillips et al., 2004). While there is extensive use of antibiotics in humans and food animals, there is also significant usage of antibiotics in agriculture, especially in animal feeds and in aquaculture. Finally, recent metagenomic studies have demonstrated that native human gut microbiota contain many bacteria that possess several AR-encoding genes that could be laterally transferred in the gut to potentially pathogenic bacteria (Scott, 2002; Salyers et al., 2004; Kazimierczak and Scott, 2007). Consequently, AR genes exist not only in nature and in humans but also in food animals, fish, plants, and vegetables as an enormous reservoir of genes that can be transferred to human pathogens either by direct contact or indirectly through the consumption of contaminated foods. The aim of this review is to give an overview of the reservoirs of these contemporary AR genes that were recently discovered in normal human gut microbiota, food animals, and agriculturally produced foods; to summarize the scientific evidence of the transfer of AR genes of bacteria from these pools; and to explore the emergence of AR bacteria in humans via the food chain.
The Use of Antibiotics in Food Animals and Aquaculture and the Subsequent Emergence of Antibiotic-Resistant Bacteria
Antibiotics are used in food animals and aquaculture as therapeutic agents, prophylactics, and growth promoters (Phillips et al., 2004). For more than 50 years antibiotics used as growth promoters were thought to have only the positive effect of promoting growth in food animals, but the efficacy of growth promotion of these antibiotics were never rigorously tested (Graham et al., 2007). A recent large-scale economic analysis conducted in the United States has demonstrated that the use of antibiotics as growth promoters in food animal production is associated with economic losses for producers (Graham et al., 2007). Moreover, the overuse of antibiotics in food-source animal production has been shown to increase the risk of AR bacterial transfer to humans, and the use of most antibiotics for growth promotion has been banned by the European Union (EU) in 1999 (Casewell et al., 2003) following numerous studies that have linked the decrease of AR prevalence to the ban of antibiotic use in food animal production (Marshall and Levy, 2011). However, in some countries, such as China and United States, antibiotics as growth promoters and therapeutics in animals are not monitored despite a high occurrence of AR bacteria (Zhu et al., 2013). The main problem with the use of antibiotics in food animal production and aquaculture is that many of the antibiotics that are used are from antibiotic classes that are also used in the treatment of human bacterial infections. Thus, the use of one specific antibiotic as a growth promoter may also lead to cross-resistance with antibiotics used in medicine. Even more alarming is the possibility that the use of one antibiotic could select for multiple ARs to functionally unrelated antibiotics because AR genes could be associated with transferable plasmids and transposons (Marshall and Levy, 2011). Although several antibiotics have been withdrawn from use as growth promoters in animal feed production, many others still continue to be used for growth promotion and for the treatment of animals. An emerging field where antibiotics are given for prophylaxis is industrial fish aquaculture, especially shrimp, trout, and salmon (Cabello, 2006). This has resulted in an increase of both AR bacteria in fish pathogens and in AR in the environment (Cabello, 2006). The exchange of AR genes between aquatic environments and terrestrial environments by lateral gene transfer (LGT) occurs via transformation (uptake of free DNA), conjugation (DNA transfer between two bacteria via conjugative plasmids and transposons), conjugative transposition, and transduction (DNA transfer via a bacteriophage) events (Cabello, 2006). Thus, resistant bacteria from foods (animals, fish, plants, and vegetables) that are consumed by humans, as well as direct contact between humans and animals, water, fish, or vegetables, can be a source of AR gene transfer (Schjorring and Krogfelt, 2011).
Sources of Antibiotic Resistance in Human Pathogens
Antibiotic Resistance in Food Animals
Antimicrobial agents administered to animals either for prophylaxis (growth promoters) or for therapy have been known to select for AR bacteria that could be transferred to human pathogens since 1975 with the first demonstration of the use of oxytetracycline as a growth promoter in chickens, which demonstrated an increased selection of tetracycline-resistant Escherichia coli colonization in the chickens, as well as the acquisition of such resistance in E. coli in the gut of the farm family (Levy et al., 1976a,b). This was studied during the previous decades to show the relationships between antibiotic usage as growth promoters in animals and the emergence of AR bacteria in animal and human infections (Marshall and Levy, 2011). Bacterial cross-resistance between antibiotics used in animal feed production and those used in humans has been clearly demonstrated by the use of avoparcin and the subsequent emergence and spread of vancomycin-resistant enterococci (VRE) as a dramatically life-threatening human pathogen (Hammerum et al., 2010). Avoparcin was banned in the EU in 1997 for prophylactic use, which led to a subsequent decrease in the carriage of glycopeptide-resistant Enterococcus faecium (Kazimierczak and Scott, 2007). This decrease was not observed in Denmark where the persistence of vanA genes in pigs was likely due to the use of tylosin (macrolide antibiotic) because the ermB and vanA genes were found to be encoded on the same mobile genetic element (Aarestrup, 2000). A short list of antibiotics that are used in animal food production and their structurally related antibiotics in human medicine is provided in Table 1. Thus, it is well established that AR bacteria that are selected in chickens, pigs, and cattle may be transmitted to the human intestine via the food chain (Salyers et al., 2004). The role of pets and wild animals as reservoirs of AR genes has been also documented (Costa et al., 2008; Poeta et al., 2009). Moreover, other compounds such as metals and biocides widely used in husbandry and veterinary can co-select AR genes such as glycopeptide resistance and macrolide resistance located in a transposon in Enterococcus faecium isolates from pigs in Denmark (Hasman and Aarestrup, 2005).
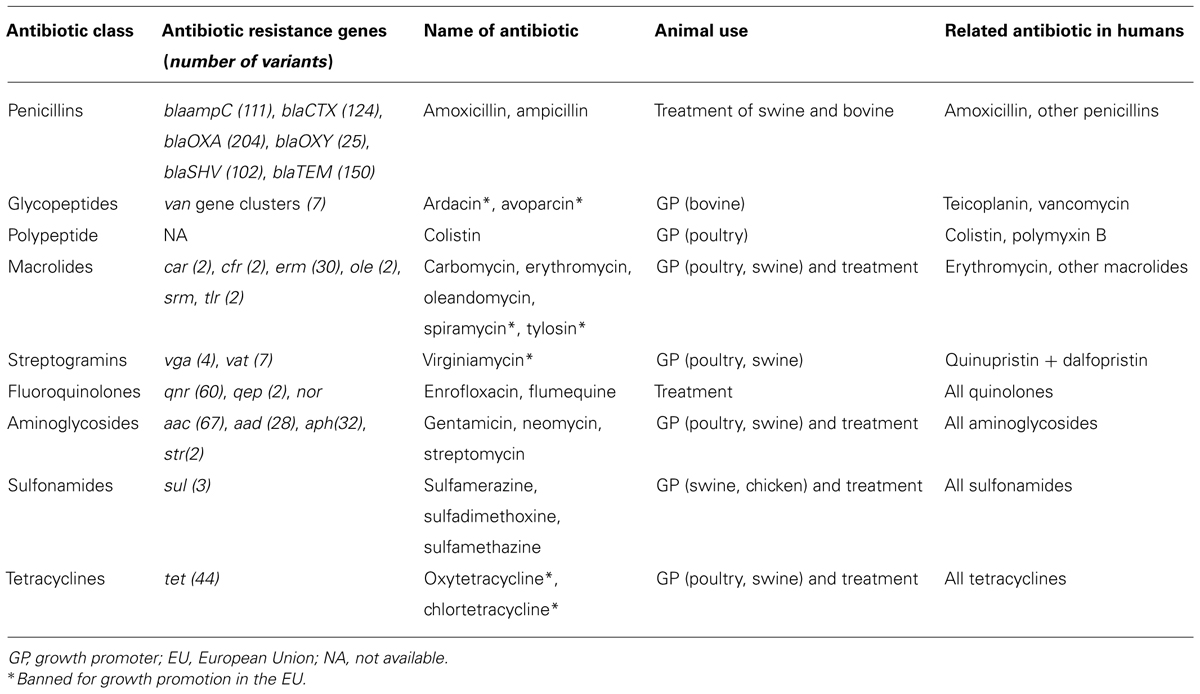
TABLE 1. Antibiotics used in food animal production, antibiotic resistance genes families, and related antibiotics used in humans.
Recent metagenomic studies have revealed that AR genes are numerous in wild animals and food animals, and some recent examples are provided below. The effects of antibiotic exposure (amoxicillin, oxytetracycline and enrofloxacin) and the development of AR in E. coli from the chicken gut has revealed that even short-term exposure can lead to resistant strains (van der Horst et al., 2013). AR genes also increased in abundance and diversity in the intestinal microbiota of swine that received chlortetracycline, sulfamethazine, and penicillin, including resistance to antibiotics that were not administered (i.e., aminoglycoside O-phosphotransferases), which demonstrated the potential collateral effects of indirect selection of resistance to other classes of antibiotics (Looft et al., 2012).
The effects of penicillin supplementation in the feed of broiler chickens was recently assessed using the 454 pyrosequencing method and revealed an elevated percentage of Firmicutes and a decreased proportion of Bacteroidetes, which was similar to what was reported in human obese versus lean individuals (Singh et al., 2013). Similarly, it has been recently shown using functional metagenomic screening that long-term treatment of adult honeybees with oxytetracycline for control of larval pathogens is associated with a high incidence of tetracycline resistance (Tian et al., 2012). Recently, using high-capacity quantitative PCR targeting 244 AR genes, Zhu et al. (2013) showed that the reservoir of AR genes in large-scale commercial swine farms in China was enormous by detecting 149 unique AR genes from different classes of antibiotics of human medical interest. Moreover, along with the detection of numerous AR genes, they found a large enrichment in transposase sequences with some of them involved in mobilization of AR genes (Aziz et al., 2010), suggesting an increased risk of LGT of AR genes from livestock animals to human pathogens (Zhu et al., 2013). Similar findings regarding swine as reservoirs of AR genes have also been reported in the United States (Whitehead and Cotta, 2013). Finally, other foods may contain AR genes that are present in lactic acid bacteria and bifidobacteria, including milk and dairy products, which have been extensively studied and reviewed elsewhere (Ammor et al., 2007, 2008). Foodborne urinary tract infections (FUTIs) has been recently revisited as a new paradigm for antimicrobial-resistant foodborne illness (Nordstrom et al., 2013). One of the most impressive examples of food as a source of antibiotic-resistant bacteria in humans is –the multidrug resistant extraintestinal pathogenic E. coli (ExPEC) that is responsible for community-acquired urinary tract infections (UTIs; Vincent et al., 2010) including the outbreak of trimethoprim–sulfamethoxazole-resistant E. coli UTIs in women from the United States (Manges et al., 2001), a pyelonephritis outbreak (Johnson et al., 2002), and the community outbreak of clonally related extended-spectrum beta-lactamases (ESBLs) type CTX -M infections worldwide (Pitout et al., 2005; Pitout and Laupland, 2008). The source of the strains responsible for these outbreaks has been linked to contaminated meat and other foods suggesting that the use of antimicrobials in food animal production may select for antibiotic-resistant strains of ExPEC. Moreover, a high prevalence of contamination by antimicrobial-resistant and ExPEC in retail foods has been reported, especially in turkey products purchased in grocery stores in retail markets from the United States (Johnson et al., 2005a). There are other examples of foods as reservoirs for AR bacteria that could be transmitted to humans via the food chain. Johnson et al. (2005b) have reported that retail foods may be a source for community transmission of AR ExPEC, which are now recognized as clinically significant foodborne pathogens. This has also been reported in Canada in retail chicken, meat, and pork (Manges et al., 2007; Vincent et al., 2010). AR clinical isolates of E. coli that were indistinguishable from chicken isolates have also been reported in Barcelona, Spain and in Minnesota and Wisconsin, the United States, suggesting that foodborne transmission of AR ExPEC is a very common phenomenon and is a major source of AR genes transmissible to humans via the food chain (Johnson et al., 2006, 2007). A novel mosaic tetracycline resistance-encoding gene, tet(S/M), in a transposon likely acquired from Lactococcus lactis, has been described in foodborne strains of Streptococcus bovis, a human opportunistic pathogen (Barile et al., 2012).
Antibiotic Resistance in Aquaculture
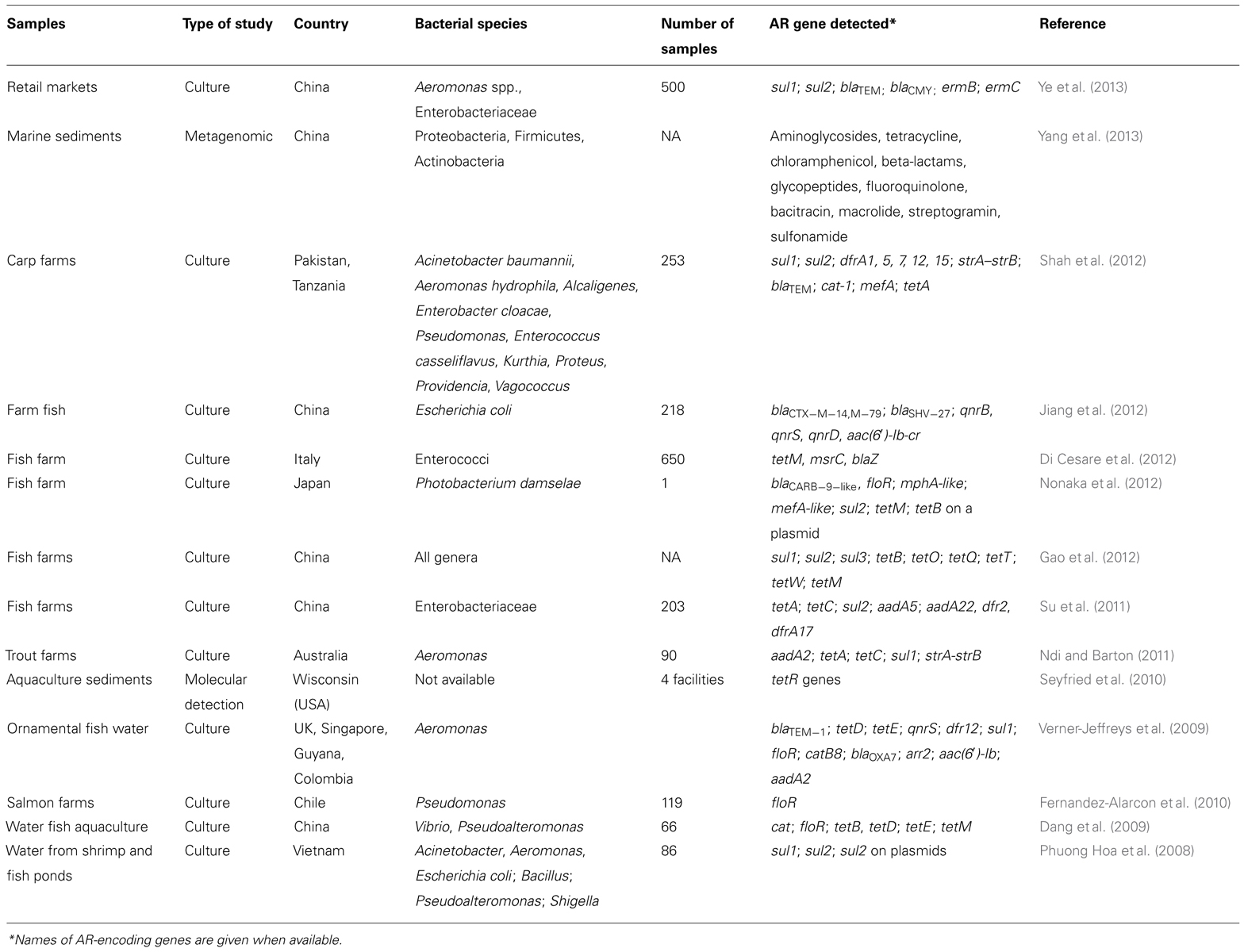
Aquaculture is a relatively recent and growing field where antibiotics are heavily applied, either directly to the water or in fish food, as prophylactics to control infectious diseases, which has resulted in the emergence of AR gene reservoirs in fish and aquatic animals as well as in the aquatic environment (Heuer et al., 2009). Different classes of antibiotics are used, including some that are critically important in human medicine, such as aminopenicillins, macrolides, aminoglycosides, fluoroquinolones, and tetracyclines (Heuer et al., 2009). The indirect spread of AR genes by LGT from antibiotic-resistant bacteria in fish to human pathogens has been widely documented on Aeromonas strains of fish origin that could easily transfer their AR plasmids to human pathogens, such as Aeromonas hydrophila, E. coli, and Salmonella (Cabello, 2006; Heuer et al., 2009). Conversely, transferable multidrug resistant plasmid from Salmonella enterica to Aeromonas salmonicida subsp. salmonicida has been also reported (McIntosh et al., 2008). Yet another example is the emergence of multidrug resistant strains of Vibrio cholerae that were responsible for the Latin American outbreak in 1992, which was likely transmitted by water and seafood (Weber et al., 1994). This was also demonstrated in multidrug resistant Vibrio parahaemolyticus and Vibrio alginolyticus isolates from farmed fishes in Korea (Oh et al., 2011). Similarly, AR genes found in the emerging human pathogen Salmonella enterica serotype Typhimurium DT104, responsible for outbreaks of salmonellosis in humans in Europe and in the United States, likely originated from an aquaculture setting (Cabello, 2006). An AR gene cluster of Salmonella Typhimurium DT104 containing a florfenicol resistance-encoding gene, which is an antibiotic widely used in aquaculture, was detected in these human isolates (Briggs and Fratamico, 1999). A transferable multiple antibiotic and mercury resistance plasmid was also reported in Atlantic Canadian isolates of Aeromonas salmonicida subsp. salmonicida (McIntosh et al., 2008). Thus, there is ample evidence that many AR determinants found in pathogenic human bacteria have a fish origin because these determinants could be detected in fish pathogens (Cabello, 2006). Direct transmission of AR bacteria from aquatic environments, resulting in human infection, has also been reported in cases involving direct contact with water and aquatic environments, drinking water or the consumption of contaminated fish products (Heuer et al., 2009). Although fish and aquaculture environments could be a reservoir of AR genes, marine sediment and oceans also have a wide pool of AR genes (ocean resistome; Yang et al., 2013). A recent metagenomic study using a high-throughput sequencing approach reveals that marine sediments in China contain numerous putative AR genes to 11 different classes of antibiotics of medical importance (especially genes conferring resistance to aminoglycosides, glycopeptides, and bacitracin), including sequences that were highly similar to transposons and plasmids found in human pathogens, suggesting that marine sediment bacteria play a major role in LGT of AR genes (Yang et al., 2013). Moreover, in the latter study, authors were able to identify sequences of a transposon similar to a 5.5-kb mega element that contains macrolide resistance genes. Although these macrolide resistance genes could be located in other genetic platforms, this mega element is the most prevalent molecular mechanism of macrolide resistance in Streptococcus pneumoniae human infections in the United States, Canada, and the UK (Zhanel et al., 2006). Examples of AR genes detected in aquaculture over the last 5 years are summarized in Table 2. Besides the use of antibiotics, there is also a risk of AR transmission by lactic acid bacteria used as probiotics in aquaculture (Muñoz-Atienza et al., 2013).
Antibiotic Resistance in Raw Fruits and Fresh Vegetables
Vegetables could also be a source of AR genes via contaminated raw fruits and fresh vegetables that are not sufficiently treated with water before consumption (Ruimy et al., 2010a). For example, Pseudomonas aeruginosa strains resistant to various classes of antibiotics, including ampicillin, chloramphenicol, and sulfamethoxazole/trimethoprim, have been isolated from the following foods/locations: fresh lettuce, carrots, tomatoes, and cucumbers from markets in Jamaica (Allydice-Francis and Brown, 2012), raw salads contaminated with tetracycline and trimethoprim/sulfamethoxazole-resistant Shigella species in Tunisia (Mokhtari et al., 2012) and fresh lettuce from markets in two cities of Mexico contaminated with AR Salmonella species (Castaneda-Ramirez et al., 2011). Interestingly, in the study from Tunisia, the authors reported a seasonal isolation of Shigella species from human and food samples during summer, suggesting that some human invasive infections could be linked to seasonal vegetable consumption. Seasonal bacterial infections have recently been reported in Klebsiella pneumoniae bloodstream infections on four continents (Anderson et al., 2008). ESBLs of the CTX-M-15 family, an ESBL encoding gene endemic worldwide in E. coli human infections, was detected in a Pseudomonas teessidea strain cultured from a prepackaged retail spinach as well as a novel ESBL of the bla-RAHN-2 family from Rahnella aquatilis, suggesting that saprophytes in common fresh vegetables may be an important source of AR genes that could spread laterally to human pathogens (Raphael et al., 2011). The ESBLs of the RAHN family have been described in R. aquatilis strains that were isolated from raw fruits and vegetables (Ruimy et al., 2010a,b). Traditionally fermented foods of animal and vegetable origins may also contain AR bacteria, such as Enterococcus faecium and Enterococcus faecalis, both of which exhibit a high incidence of resistance to rifampicin, ciprofloxacin, and quinupristin/dalfopristin, and surprisingly, vancomycin (Sánchez et al., 2013). Finally, the recent significant outbreak of hemolytic uremic syndrome in Germany in 2011 was found to be due to a CTX-M-15 ESBL E. coli clone likely originating from fenugreek seeds imported from Egypt (Buchholz et al., 2011; Rasko et al., 2011; Weiser et al., 2013).
Normal Human Gut Microbiota as a Reservoir for AR Genes
Thanks to the use of new technologies, such as high-throughput pyrosequencing, metagenomic studies and exhaustive culture techniques, a description of human microbiota has begun to revolutionize our current understanding of the relationship between gut microbiota composition, several diseases and obesity (Lagier et al., 2012). It is now accepted that numerous factors, including age, geographical provenance and environment, dietary habits, antibiotics and probiotics, can modify the composition of the human gut microbiota (Lagier et al., 2012). Thus, it is not surprising that the human gut microbiota represents an important reservoir of AR genes present in the bacterial community and that these AR genes may be transferred from one bacterium to another bacterium in the gut, especially human pathogens (Salyers et al., 2004). Although AR in commensal E. coli from healthy people was reported more than 40 years ago (Smith and Halls, 1966), recent studies have highlighted an increase and high incidence of AR in commensal E. coli from both children and from adults (Bailey et al., 2010). However, there are few reports dealing with the identity of the AR genes carried by healthy humans. Several functional metagenomic studies have been successfully applied to human gut microbiota, including healthy adults or infant individuals, to discover the presence of a vast diversity of new AR-encoding genes that could be a source for the emergence of new AR genes in human pathogens in the future (Gueimonde et al., 2006; Sommer et al., 2009; Cheng et al., 2012). One of the most recent and surprising findings in humans was that approximately 2/3 of individuals living in a remote community in South America without any antibiotic selection pressure carried bacteria resistant to at least one antibiotic (Bartoloni et al., 2009). Another research area, especially during the past years, is studying the development of gut microbiota in infants to show that gut microbiota is an important reservoir of AR genes (Valles et al., 2012). Antibiotic-resistant bacteria responsible for invasive infections in children are increasing worldwide, and it has been shown using metagenomics that such AR bacteria and AR gene transfer (tetracycline resistance determinants) could be transmitted from mother to child and persist for weeks after birth (de Vries et al., 2011). Thus AR genes and bacteria could be transmitted vertically from the mother’s gastrointestinal tract, birth canal, or breast milk to the infant at a very early age. Moreover, the human gut is known to be a favorable environment for LGT (Palmer et al., 2007), and the spread/transfer of AR genes from non-pathogenic to pathogenic bacteria have been documented even in the case of young breast-fed children (de Vries et al., 2011). Tetracycline resistance-encoding genes (tetA and tetB) have also been detected in 12% of 309 commensal intestinal strains of E. coli from 128 Swedish infants who did not receive tetracyclines during their first year of life (Karami et al., 2006). A recent study in India revealed a high prevalence of multidrug resistant (MDR) and carbapenem-resistant Acinetobacter baumannii colonization from the gut of neonates, including one particular baby in which the same clone was found in both its blood and stool samples (Roy et al., 2010). Infant gut microbiota in Greece at days 4, 30, and 90 after delivery were found to contain as many as 34% of vancomycin-resistant Lactobacillus species (Kirtzalidou et al., 2011). Zhang et al. (2011) reported the presence of several AR genes including tetM, sul2, and blaTEM in Enterococcus, Staphylococcus, Klebsiella, Streptococcus, and Escherichia/Shigella species from the gut microbiota of 16 American infants without antibiotic exposure who had fed on breast milk or infant formula from birth to 1-year-old. A recent study in Australian healthy adults has shown that commensal E. coli represent a huge reservoir of AR-encoding genes including genes conferring resistance to trimethoprim (dfrA1, dfrA5, drfA7, dfrA12, or dfrA17), sulfamethoxazole (sul1, sul2, or sul3), tetracycline (tetA or tetB), and ampicillin (bla TEM; Bailey et al., 2010). Similarly, bla NDM-1 human carriage cases have been reported in several countries in India, China, and Pakistan as well as in the environment (Nordmann et al., 2011).
Although antibiotic treatment in humans directly affects the pathogen responsible for the infectious disease, antibiotic use has been shown to lead to a collateral effect in commensal bacteria in the gut microbiota. In fact when AR genes are selected in the commensal gut microbiota, they could be further transferred to human pathogen. Karami et al. (2007) have shown that a plasmid-encoded beta-lactamase encoding gene of the TEM family from an ampicillin-resistant E. coli was transferable to a susceptible strain in the gut of a children treated with ampicillin for UTIs. It has been shown that the administration of antibiotics could alter the bacterial community structure of the gastrointestinal microbiota in mice and that these modifications are different according to the antibiotic classes administered (Robinson and Young, 2010). Moreover, these modifications are not transient only (Jakobsson et al., 2010) in human gut microbiota; recent data have been accumulated demonstrating that some classes of antibiotics may affect the global composition of the gut microbiota and the persistence of AR bacteria in the human intestine for years (Jernberg et al., 2010). It has been shown that the diversity of gut microbiota is reduced after antibiotic therapy in children (Penders et al., 2006) and in elderly patients (Bartosch et al., 2004, 2005). Deep 16S rRNA pyrosequencing of the human gut microbiota from three healthy humans before and after treatment with ciprofloxacin has also shown a dramatic shift in bacterial abundance (Dethlefsen et al., 2008). Interestingly, it has been demonstrated that antibiotic treatment has a dramatic effect on bacterial diversity even after only 1 week of treatment and that AR, in this case the macrolide resistance gene ermB, can persist for long periods (4 years; Jakobsson et al., 2010). Broad-range culture techniques and metagenomic analyses have confirmed that antibiotic treatment, especially broad-spectrum antibiotic treatment, dramatically changes the gut microbiota in humans, which could lead to the emergence of invasive infections (Raum et al., 2008; Cotter et al., 2012; Perez-Cobas et al., 2012; Dubourg et al., 2013a,b). Löfmark et al. (2006) and Jernberg et al. (2007) have shown that 1 week of clindamycin administration could select for resistant Bacteroides spp., which could persist for as long as 2 years following clindamycin exposure. Antibiotic use could also lead to a collateral effect through the selection of AR bacteria among the normal microbiota, as has been demonstrated with the long-term persistence of clarithromycin-resistant Enterococcus species harboring the ermB gene immediately after treatment to eradicate Helicobacter pylori, which may persist for 1–3 years after antibiotic therapy (Sjolund et al., 2003). Similarly, Jakobsson et al. (2010) have shown that clarithromycin and metronidazole administration in patients suffering from peptic ulcer disease may select for the presence of the ermB gene, which may persist in some cases for up to 4 years post-treatment, demonstrating that the AR gene may be stabilized in the human gut microbiota following selection. Thus, the increased prevalence of AR and possibility LGT between bacteria are important consequences of the long-term persistence of AR genes in gut microbiota (Jernberg et al., 2010).
Conclusion
While the overuse and misuse of antibiotics in humans is undoubtedly associated with the emergence and spread of AR bacteria and AR genes, at least in human infections, recent studies have demonstrated that the use of antibiotics in animals, aquaculture and most importantly, food production, is also a major factor contributing to the emergence of AR bacteria and the spread of AR genes. Moreover, because the potential pool of AR genes in these environments remains largely unknown, with thousands of AR genes yet to be discovered, future observation of AR in these ecosystems is warranted from an ecological perspective. This phenomenon is compounded by the immense number of methods that bacteria have developed for the exchange of AR genes via LGT, recombination, and transposition and also for the purpose of creating new AR genes, as recently exemplified with the discovery of the new chimeric carbapenemase encoding gene NDM-1 (Yong et al., 2009; Toleman et al., 2012). Further studies are warranted and should utilize a combination of culture and non-culture-based techniques to better characterize the resistome in both the human gut microbiota, in response to various conditions and diseases, and in non-human environments, including animals, aquaculture, and food products, to better understand the role of antibiotics in the evolution of these ecosystems.
Conflict of Interest Statement
The author declares that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Aarestrup, F. M. (2000). Characterization of glycopeptide-resistant Enterococcus faecium (GRE) from broilers and pigs in Denmark: genetic evidence that persistence of GRE in pig herds is associated with coselection by resistance to macrolides. J. Clin. Microbiol. 38, 2774–2777.
Allydice-Francis, K., and Brown, P. D. (2012). Diversity of antimicrobial resistance and virulence determinants in Pseudomonas aeruginosa associated with fresh vegetables. Int. J. Microbiol. 2012, 426241. doi: 10.1155/2012/426241
Ammor, M. S., Florez, A. B., and Mayo, B. (2007). Antibiotic resistance in non-enterococcal lactic acid bacteria and bifidobacteria. Food Microbiol. 24, 559–570. doi: 10.1016/j.fm.2006.11.001
Ammor, M. S., Florez, A. B., van Hoek, A. H., de Los Reyes-Gavilan, C. G., Aarts, H. J., Margolles, A., et al. (2008). Molecular characterization of intrinsic and acquired antibiotic resistance in lactic acid bacteria and bifidobacteria. J. Mol. Microbiol. Biotechnol. 14, 6–15. doi: 10.1159/000106077
Anderson, D. J., Richet, H., Chen, L. F., Spelman, D. W., Hung, Y. J., Huang, A. T., et al. (2008). Seasonal variation in Klebsiella pneumoniae bloodstream infection on 4 continents. J. Infect. Dis. 197, 752–756. doi: 10.1086/527486
Aziz, R. K., Breitbart, M., and Edwards, R. A. (2010). Transposases are the most abundant, most ubiquitous genes in nature. Nucleic Acids Res. 38, 4207–4217. doi: 10.1093/nar/gkq140
Bailey, J. K., Pinyon, J. L., Anantham, S., and Hall, R. M. (2010). Commensal Escherichia coli of healthy humans: a reservoir for antibiotic-resistance determinants. J. Med. Microbiol. 59, 1331–1339. doi: 10.1099/jmm.0.022475-0
Baquero, F., Tedim, A. P., and Coque, T. M. (2013). Antibiotic resistance shaping multi-level population biology of bacteria. Front. Microbiol. 4:15. doi: 10.3389/fmicb.2013.00015
Barile, S., Devirgiliis, C., and Perozzi, G. (2012). Molecular characterization of a novel mosaic tet(S/M) gene encoding tetracycline resistance in foodborne strains of Streptococcus bovis. Microbiology 158, 2353–2362.doi: 10.1099/mic.0.058206-0
Bartoloni, A., Pallecchi, L., Rodriguez, H., Fernandez, C., Mantella, A., Bartalesi, F., et al. (2009). Antibiotic resistance in a very remote Amazonas community. Int. J. Antimicrob. Agents 33, 125–129. doi: 10.1016/j.ijantimicag.2008.07.029
Bartosch, S., Fite, A., Macfarlane, G. T., and McMurdo, M. E. (2004). Characterization of bacterial communities in feces from healthy elderly volunteers and hospitalized elderly patients by using real-time PCR and effects of antibiotic treatment on the fecal microbiota. Appl. Environ. Microbiol. 70, 3575–3581. doi: 10.1128/AEM.70.6.3575-3581.2004
Bartosch, S., Woodmansey, E. J., Paterson, J. C., McMurdo, M. E., and Macfarlane, G. T. (2005). Microbiological effects of consuming a synbiotic containing Bifidobacterium bifidum, Bifidobacterium lactis, and oligofructose in elderly persons, determined by real-time polymerase chain reaction and counting of viable bacteria. Clin. Infect. Dis. 40, 28–37. doi: 10.1086/426027
Bhullar, K., Waglechner, N., Pawlowski, A., Koteva, K., Banks, E. D., Johnston, M. D., et al. (2012). Antibiotic resistance is prevalent in an isolated cave microbiome. PLoS ONE 7:e34953. doi: 10.1371/journal.pone.0034953
Briggs, C. E., and Fratamico, P. M. (1999). Molecular characterization of an antibiotic resistance gene cluster of Salmonella typhimurium DT104. Antimicrob. Agents Chemother. 43, 846–849.
Buchholz, U., Bernard, H., Werber, D., Bohmer, M. M., Remschmidt, C., Wilking, H., et al. (2011). German outbreak of Escherichia coli O104:H4 associated with sprouts. N. Engl. J. Med. 365, 1763–1770. doi: 10.1056/NEJMoa1106482
Cabello, F. C. (2006). Heavy use of prophylactic antibiotics in aquaculture: a growing problem for human and animal health and for the environment. Environ. Microbiol. 8, 1137–1144. doi: 10.1111/j.1462-2920.2006.01054.x.
Casewell, M., Friis, C., Marco, E., McMullin, P., and Phillips, I. (2003). The European ban on growth-promoting antibiotics and emerging consequences for human and animal health. J. Antimicrob. Chemother. 52, 159–161. doi: 10.1093/jac/dkg313
Castaneda-Ramirez, C., Cortes-Rodriguez, V., Fuente-Salcido, N., Bideshi, D. K., del Rincon-Castro, M. C., et al. (2011). Isolation of Salmonella spp. from lettuce and evaluation of its susceptibility to novel bacteriocins of Bacillus thuringiensis and antibiotics. J. Food Prot. 74, 274–278. doi: 10.4315/0362-028X.JFP-10-324
Cheng, G., Hu, Y., Yin, Y., Yang, X., Xiang, C., Wang, B., et al. (2012). Functional screening of antibiotic resistance genes from human gut microbiota reveals a novel gene fusion. FEMS Microbiol. Lett. 336, 11–16. doi: 10.1111/j.1574-6968.2012.02647.x
Cordero, O. X., Wildschutte, H., Kirkup, B., Proehl, S., Ngo, L., Hussain, F., et al. (2012). Ecological populations of bacteria act as socially cohesive units of antibiotic production and resistance. Science 337, 1228–1231. doi: 10.1126/science.1219385
Costa, D., Poeta, P., Saenz, Y., Coelho, A. C., Matos, M., Vinue, L., et al. (2008). Prevalence of antimicrobial resistance and resistance genes in faecal Escherichia coli isolates recovered from healthy pets. Vet. Microbiol. 127, 97–105. doi: 10.1016/j.vetmic.2007.08.004
Cotter, P. D., Stanton, C., Ross, R. P., and Hill, C. (2012). The impact of antibiotics on the gut microbiota as revealed by high throughput DNA sequencing. Discov. Med. 13, 193–199.
D’Costa, V. M., King, C. E., Kalan, L., Morar, M., Sung, W. W., Schwarz, C., et al. (2011). Antibiotic resistance is ancient. Nature 477, 457–461. doi: 10.1038/nature10388.
D’Costa, V. M., McGrann, K. M., Hughes, D. W., and Wright, G. D. (2006). Sampling the antibiotic resistome. Science 311, 374–377. doi: 10.1126/science.1120800.
Dang, H., Zhao, J., Song, L., Chen, M., and Chang, Y. (2009). Molecular characterizations of chloramphenicol- and oxytetracycline-resistant bacteria and resistance genes in mariculture waters of China. Mar. Pollut. Bull. 58, 987–994. doi: 10.1016/j.marpolbul.2009.02.016.
Dethlefsen, L., Huse, S., Sogin, M. L., and Relman, D. A. (2008). The pervasive effects of an antibiotic on the human gut microbiota, as revealed by deep 16S rRNA sequencing. PLoS Biol. 6:e280. doi: 10.1371/journal.pbio.0060280.
de Vries, L. E., Valles, Y., Agerso, Y., Vaishampayan, P. A., Garcia-Montaner, A., Kuehl, J. V., et al. (2011). The gut as reservoir of antibiotic resistance: microbial diversity of tetracycline resistance in mother and infant. PLoS ONE 6:e21644. doi: 10.1371/journal.pone.0021644
Di Cesare, A., Vignaroli, C., Luna, G. M., Pasquaroli, S., and Biavasco, F. (2012). Antibiotic-resistant enterococci in seawater and sediments from a coastal fish farm. Microb. Drug Resist. 18, 502–509. doi: 10.1089/mdr.2011.0204
Dubourg, G., Lagier, J. C., Armougom, F., Robert, C., Audoly, G., Papazian, L., et al. (2013a). High-level colonisation of the human gut by Verrucomicrobia following broad-spectrum antibiotic treatment. Int. J. Antimicrob. Agents 41, 149–155. doi: 10.1016/j.ijantimicag.2012.10.012
Dubourg, G., Lagier, J. C., Armougom, F., Robert, C., Hamad, I., Brouqui, P., et al. (2013b). The gut microbiota of a patient with resistant tuberculosis is more comprehensively studied by culturomics than by metagenomics. Eur. J. Clin. Microbiol. Infect. Dis. 32, 637–645. doi: 10.1007/s10096-012-1787-3
Fernandez-Alarcon, C., Miranda, C. D., Singer, R. S., Lopez, Y., Rojas, R., Bello, H., et al. (2010). Detection of the floR gene in a diversity of florfenicol resistant Gram-negative bacilli from freshwater salmon farms in Chile. Zoonoses Public Health 57, 181–188. doi: 10.1111/j.1863-2378.2009.01243.x
Forsberg, K. J., Reyes, A., Wang, B., Selleck, E. M., Sommer, M. O., and Dantas, G. (2012). The shared antibiotic resistome of soil bacteria and human pathogens. Science 337, 1107–1111. doi: 10.1126/science.1220761
Gao, P., Mao, D., Luo, Y., Wang, L., Xu, B., and Xu, L. (2012). Occurrence of sulfonamide and tetracycline-resistant bacteria and resistance genes in aquaculture environment. Water Res. 46, 2355–2364. doi: 10.1016/j.watres.2012.02.004
Graham, J. P., Boland, J. J., and Silbergeld, E. (2007). Growth promoting antibiotics in food animal production: an economic analysis. Public Health Rep. 122, 79–87.
Gueimonde, M., Salminen, S., and Isolauri, E. (2006). Presence of specific antibiotic (tet) resistance genes in infant faecal microbiota. FEMS Immunol. Med. Microbiol. 48, 21–25. doi: 10.1111/j.1574-695X.2006.00112.x
Hammerum, A. M., Lester, C. H., and Heuer, O. E. (2010). Antimicrobial-resistant enterococci in animals and meat: a human health hazard? Foodborne Pathog. Dis. 7, 1137–1146.doi: 10.1089/fpd.2010.0552
Hasman, H., and Aarestrup, F. M. (2005). Relationship between copper, glycopeptide, and macrolide resistance among Enterococcus faecium strains isolated from pigs in Denmark between 1997 and 2003. Antimicrob. Agents Chemother. 49, 454–456. doi: 10.1128/AAC.49.1.454-456.2005
Heuer, O. E., Kruse, H., Grave, K., Collignon, P., Karunasagar, I., and Angulo, F. J. (2009). Human health consequences of use of antimicrobial agents in aquaculture. Clin. Infect. Dis. 49, 1248–1253. doi: 10.1086/605667
Jakobsson, H. E., Jernberg, C., Andersson, A. F., Sjolund-Karlsson, M., Jansson, J. K., and Engstrand, L. (2010). Short-term antibiotic treatment has differing long-term impacts on the human throat and gut microbiome. PLoS ONE 5:e9836. doi: 10.1371/journal.pone.0009836
Jernberg, C., Lofmark, S., Edlund, C., and Jansson, J. K. (2007). Long-term ecological impacts of antibiotic administration on the human intestinal microbiota. ISME J. 1, 56–66. doi: 10.1038/ismej.2007.3
Jernberg, C., Lofmark, S., Edlund, C., and Jansson, J. K. (2010). Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiology 156, 3216–3223. doi: 10.1099/mic.0.040618-0
Jiang, H. X., Tang, D., Liu, Y. H., Zhang, X. H., Zeng, Z. L., Xu, L., et al. (2012). Prevalence and characteristics of beta-lactamase and plasmid-mediated quinolone resistance genes in Escherichia coli isolated from farmed fish in China. J. Antimicrob. Chemother. 67, 2350–2353. doi: 10.1093/jac/dks250
Johnson, J. R., Delavari, P., O’Bryan, T. T., Smith, K. E., and Tatini, S. (2005a). Contamination of retail foods, particularly turkey, from community markets (Minnesota, 1999–2000) with antimicrobial-resistant and extraintestinal pathogenic Escherichia coli. Foodborne Pathog. Dis. 2, 38–49. doi: 10.1089/fpd.2005.2.38
Johnson, J. R., Kuskowski, M. A., Smith, K., O’Bryan, T. T., and Tatini, S. (2005b). Antimicrobial-resistant and extraintestinal pathogenic Escherichia coli in retail foods. J. Infect. Dis. 191, 1040–1049. doi: 10.1086/428451
Johnson, J. R., Kuskowski, M. A., Menard, M., Gajewski, A., Xercavins, M., and Garau, J. (2006). Similarity between human and chicken Escherichia coli isolates in relation to ciprofloxacin resistance status. J. Infect. Dis. 194, 71–78. doi: 10.1086/504921
Johnson, J. R., Manges, A. R., O’Bryan, T. T., and Riley, L. W. (2002). A disseminated multidrug-resistant clonal group of uropathogenic Escherichia coli in pyelonephritis. Lancet 359, 2249–2251. doi: 10.1016/S0140-6736(02)09264-4
Johnson, J. R., Sannes, M. R., Croy, C., Johnston, B., Clabots, C., Kuskowski, M. A., et al. (2007). Antimicrobial drug-resistant Escherichia coli from humans and poultry products, Minnesota and Wisconsin, 2002–2004. Emerg. Infect. Dis. 13, 838–846. doi: 10.3201/eid1306.061576
Karami, N., Martner, A., Enne, V. I., Swerkersson, S., Adlerberth, I., and Wold, A. E. (2007). Transfer of an ampicillin resistance gene between two Escherichia coli strains in the bowel microbiota of an infant treated with antibiotics. J. Antimicrob. Chemother. 60, 1142–1145. doi: 10.1093/jac/dkm327
Karami, N., Nowrouzian, F., Adlerberth, I., and Wold, A. E. (2006). Tetracycline resistance in Escherichia coli and persistence in the infantile colonic microbiota. Antimicrob. Agents Chemother. 50, 156–161. doi: 10.1128/AAC.50.1.156-161.2006
Kazimierczak, K. A., and Scott, K. P. (2007). Antibiotics and resistance genes: influencing the microbial ecosystem in the gut. Adv. Appl. Microbiol. 62, 269–292. doi: 10.1016/S0065-2164(07)62009-7
Kirtzalidou, E., Pramateftaki, P., Kotsou, M., and Kyriacou, A. (2011). Screening for lactobacilli with probiotic properties in the infant gut microbiota. Anaerobe 17, 440–443. doi: 10.1016/j.anaerobe.2011.05.007
Lagier, J. C., Million, M., Hugon, P., Armougom, F., and Raoult, D. (2012). Human gut microbiota: repertoire and variations. Front. Cell. Infect. Microbiol. 2:136. doi: 10.3389/fcimb.2012.00136
Levy, S. B., FitzGerald, G. B., and Macone, A. B. (1976a). Changes in intestinal flora of farm personnel after introduction of a tetracycline-supplemented feed on a farm. N. Engl. J. Med. 295, 583–588. doi: 10.1056/NEJM197609092951103
Levy, S. B., FitzGerald, G. B., and Macone, A. B. (1976b). Spread of antibiotic-resistant plasmids from chicken to chicken and from chicken to man. Nature 260, 40–42.
Löfmark, S., Jernberg, C., Jansson, J. K., and Edlund, C. (2006). Clindamycin-induced enrichment and long-term persistence of resistant Bacteroides spp. and resistance genes. J. Antimicrob. Chemother. 58, 1160–1167. doi: 10.1093/jac/dkl420
Looft, T., Johnson, T. A., Allen, H. K., Bayles, D. O., Alt, D. P., Stedtfeld, R. D., et al. (2012). In-feed antibiotic effects on the swine intestinal microbiome. Proc. Natl. Acad. Sci. U.S.A. 109, 1691–1696. doi: 10.1073/pnas.1120238109
Manges, A. R., Johnson, J. R., Foxman, B., O’Bryan, T. T., Fullerton, K. E., and Riley, L. W. (2001). Widespread distribution of urinary tract infections caused by a multidrug-resistant Escherichia coli clonal group. N. Engl. J. Med. 345, 1007–1013. doi: 10.1056/NEJMoa011265
Manges, A. R., Smith, S. P., Lau, B. J., Nuval, C. J., Eisenberg, J. N., Dietrich, P. S., et al. (2007). Retail meat consumption and the acquisition of antimicrobial resistant Escherichia coli causing urinary tract infections: a case–control study. Foodborne Pathog. Dis. 4, 419–431. doi: 10.1089/fpd.2007.0026
Marshall, B. M., and Levy, S. B. (2011). Food animals and antimicrobials: impacts on human health. Clin. Microbiol. Rev. 24, 718–733. doi: 10.1128/CMR.00002-11
Martinez, J. L., and Rojo, F. (2011). Metabolic regulation of antibiotic resistance. FEMS Microbiol. Rev. 35, 768–789. doi: 10.1111/j.1574-6976.2011.00282.x
McIntosh, D., Cunningham, M., Ji, B., Fekete, F. A., Parry, E. M., Clark, S. E., et al. (2008). Transferable, multiple antibiotic and mercury resistance in Atlantic Canadian isolates of Aeromonas salmonicida subsp. salmonicida is associated with carriage of an IncA/C plasmid similar to the Salmonella enterica plasmid pSN254. J. Antimicrob. Chemother. 61, 1221–1228. doi: 10.1093/jac/dkn123
Mokhtari, W., Nsaibia, S., Majouri, D., Ben, H. A., Gharbi, A., and Aouni, M. (2012). Detection and characterization of Shigella species isolated from food and human stool samples in Nabeul, Tunisia, by molecular methods and culture techniques. J. Appl. Microbiol. 113, 209–222. doi: 10.1111/j.1365-2672.2012.05324.x
Muñoz-Atienza, E., Gomez-Sala, B., Araujo, C., Campanero, C., del Campo, R., Hernandez, P. E., et al. (2013). Antimicrobial activity, antibiotic susceptibility and virulence factors of Lactic Acid Bacteria of aquatic origin intended for use as probiotics in aquaculture. BMC Microbiol. 13:15. doi: 10.1186/1471-2180-13-15
Ndi, O. L., and Barton, M. D. (2011). Incidence of class 1 integron and other antibiotic resistance determinants in Aeromonas spp. from rainbow trout farms in Australia. J. Fish Dis. 34, 589–599. doi: 10.1111/j.1365-2761.2011.01272.x
Nonaka, L., Maruyama, F., Miyamoto, M., Miyakoshi, M., Kurokawa, K., and Masuda, M. (2012). Novel conjugative transferable multiple drug resistance plasmid pAQU1 from Photobacterium damselae subsp. damselae isolated from marine aquaculture environment. Microbes Environ. 27, 263–272. doi: 10.1264/jsme2.ME11338
Nordmann, P., Poirel, L., Walsh, T. R., and Livermore, D. M. (2011). The emerging NDM carbapenemases. Trends Microbiol. 19, 588–595. doi: 10.1016/j.tim.2011.09.005
Nordstrom, L., Liu, C. M., and Price, L. B. (2013). Foodborne urinary tract infections: a new paradigm for antimicrobial-resistant foodborne illness. Front. Microbiol. 4:29. doi: 10.3389/fmicb.2013.00029
Oh, E. G., Son, K. T., Yu, H., Lee, T. S., Lee, H. J., Shin, S., et al. (2011). Antimicrobial resistance of Vibrio parahaemolyticus and Vibrio alginolyticus strains isolated from farmed fish in Korea from 2005 through 2007. J. Food Prot. 74, 380–386. doi: 10.4315/0362-028X.JFP-10-307
Palmer, C., Bik, E. M., DiGiulio, D. B., Relman, D. A., and Brown, P. O. (2007). Development of the human infant intestinal microbiota. PLoS Biol. 5:e177. doi: 10.1371/journal.pbio.0050177
Penders, J., Thijs, C., Vink, C., Stelma, F. F., Snijders, B., Kummeling, I., et al. (2006). Factors influencing the composition of the intestinal microbiota in early infancy. Pediatrics 118, 511–521. doi: 10.1542/peds.2005-2824
Perez-Cobas, A. E., Gosalbes, M. J., Friedrichs, A., Knecht, H., Artacho, A., Eismann, K., et al. (2012). Gut microbiota disturbance during antibiotic therapy: a multi-omic approach. Gut. doi: 10.1136/gutjnl-2012-303184 [Epub ahead of print].
Phillips, I., Casewell, M., Cox, T., De Groot, B., Friis, C., Jones, R., et al. (2004). Antibiotic use in animals. J. Antimicrob. Chemother. 53, 885. doi: 10.1093/jac/dkh149
Phuong Hoa, P. T., Nonaka, L., Hung, V. P., and Suzuki, S. (2008). Detection of the sul1, sul2, and sul3 genes in sulfonamide-resistant bacteria from wastewater and shrimp ponds of north Vietnam. Sci. Total Environ. 405, 377–384. doi: 10.1016/j.scitotenv.2008.06.023
Pitout, J. D., Gregson, D. B., Church, D. L., Elsayed, S., and Laupland, K. B. (2005). Community-wide outbreaks of clonally related CTX-M-14 beta-lactamase-producing Escherichia coli strains in the Calgary health region. J. Clin. Microbiol. 43, 2844–2849. doi: 10.1128/JCM.43.6.2844-2849.2005
Pitout, J. D., and Laupland, K. B. (2008). Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect. Dis. 8, 159–166. doi: 10.1016/S1473-3099(08)70041-0
Poeta, P., Radhouani, H., Pinto, L., Martinho, A., Rego, V., Rodrigues, R., et al. (2009). Wild boars as reservoirs of extended-spectrum beta-lactamase (ESBL) producing Escherichia coli of different phylogenetic groups. J. Basic Microbiol. 49, 584–588. doi: 10.1002/jobm.200900066
Raphael, E., Wong, L. K., and Riley, L. W. (2011). Extended-spectrum Beta-lactamase gene sequences in gram-negative saprophytes on retail organic and nonorganic spinach. Appl. Environ. Microbiol. 77, 1601–1607. doi: 10.1128/AEM.02506-10
Rasko, D. A., Webster, D. R., Sahl, J. W., Bashir, A., Boisen, N., Scheutz, F., et al. (2011). Origins of the E. coli strain causing an outbreak of hemolytic–uremic syndrome in Germany. N. Engl. J. Med. 365, 709–717. doi: 10.1056/NEJMoa1106920
Raum, E., Lietzau, S., von Baum, H., Marre, R., and Brenner, H. (2008). Changes in Escherichia coli resistance patterns during and after antibiotic therapy: a longitudinal study among outpatients in Germany. Clin. Microbiol. Infect. 14, 41–48. doi: 10.1111/j.1469-0691.2007.01841.x
Robinson, C. J., and Young, V. B. (2010). Antibiotic administration alters the community structure of the gastrointestinal micobiota. Gut Microbes 1, 279–284. doi: 10.4161/gmic.1.4.12614
Rolain, J. M., Canton, R., and Cornaglia, G. (2012). Emergence of antibiotic resistance: need for a new paradigm. Clin. Microbiol. Infect. 18, 615–616.
Roy, S., Viswanathan, R., Singh, A., Das, P., and Basu, S. (2010). Gut colonization by multidrug-resistant and carbapenem-resistant Acinetobacter baumannii in neonates. Eur. J. Clin. Microbiol. Infect. Dis. 29, 1495–1500. doi: 10.1007/s10096-010-1030-z
Ruimy, R., Brisabois, A., Bernede, C., Skurnik, D., Barnat, S., et al. (2010a). Organic and conventional fruits and vegetables contain equivalent counts of Gram-negative bacteria expressing resistance to antibacterial agents. Environ. Microbiol. 12, 608–615. doi: 10.1111/j.1462-2920.2009.02100.x
Ruimy, R., Meziane-Cherif, D., Momcilovic, S., Arlet, G., Andremont, A., and Courvalin, P. (2010b). RAHN-2, a chromosomal extended-spectrum class A beta-lactamase from Rahnella aquatilis. J. Antimicrob. Chemother. 65, 1619–1623. doi: 10.1093/jac/dkq178
Salyers, A. A., Gupta, A., and Wang, Y. (2004). Human intestinal bacteria as reservoirs for antibiotic resistance genes. Trends Microbiol. 12, 412–416. doi: 10.1016/j.tim.2004.07.004
Sánchez, V. A., Lavilla, L. L., Benomar, N., Galvez, A., Perez, P. R., and Abriouel, H. (2013). Phenotypic and molecular antibiotic resistance profile of Enterococcus faecalis and Enterococcus faecium isolated from different traditional fermented foods. Foodborne Pathog. Dis. 10, 143–149. doi: 10.1089/fpd.2012.1279
Schjorring, S., and Krogfelt, K. A. (2011). Assessment of bacterial antibiotic resistance transfer in the gut. Int. J. Microbiol. 2011, 312956. doi: 10.1155/2011/312956
Scott, K. P. (2002). The role of conjugative transposons in spreading antibiotic resistance between bacteria that inhabit the gastrointestinal tract. Cell. Mol. Life Sci. 59, 2071–2082.
Seyfried, E. E., Newton, R. J., Rubert, K. F., Pedersen, J. A., and McMahon, K. D. (2010). Occurrence of tetracycline resistance genes in aquaculture facilities with varying use of oxytetracycline. Microb. Ecol. 59, 799–807. doi: 10.1007/s00248-009-9624-7
Shah, S. Q., Colquhoun, D. J., Nikuli, H. L., and Sorum, H. (2012). Prevalence of antibiotic resistance genes in the bacterial flora of integrated fish farming environments of Pakistan and Tanzania. Environ. Sci. Technol. 46, 8672–8679. doi: 10.1021/es3018607.
Singh, P., Karimi, A., Devendra, K., Waldroup, P. W., Cho, K. K., and Kwon, Y. M. (2013). Influence of penicillin on microbial diversity of the cecal microbiota in broiler chickens. Poult. Sci. 92, 272–276. doi: 10.3382/ps.2012-02603
Sjolund, M., Wreiber, K., Andersson, D. I., Blaser, M. J., and Engstrand, L. (2003). Long-term persistence of resistant Enterococcus species after antibiotics to eradicate Helicobacter pylori. Ann. Intern. Med. 139, 483–487.
Smith, H. W., and Halls, S. (1966). Observations on infective drug resistance in Britain. Br. Med. J. 1, 266–269.
Sommer, M. O., Dantas, G., and Church, G. M. (2009). Functional characterization of the antibiotic resistance reservoir in the human microflora. Science 325, 1128–1131. doi: 10.1126/science.1176950
Su, H. C., Ying, G. G., Tao, R., Zhang, R. Q., Fogarty, L. R., and Kolpin, D. W. (2011). Occurrence of antibiotic resistance and characterization of resistance genes and integrons in Enterobacteriaceae isolated from integrated fish farms in South China. J. Environ. Monit. 13, 3229–3236. doi: 10.1039/c1em10634a
Tian, B., Fadhil, N. H., Powell, J. E., Kwong, W. K., and Moran, N. A. (2012). Long-term exposure to antibiotics has caused accumulation of resistance determinants in the gut microbiota of honeybees. MBio 3, e00377–e00412. doi: 10.1128/mBio.00377-12
Toleman, M. A., Spencer, J., Jones, L., and Walsh, T. R. (2012). BlaNDM-1 is a chimera likely constructed in Acinetobacter baumannii. Antimicrob. Agents Chemother. 56, 2773–2776. doi: 10.1128/AAC.06297-11
Valles, Y., Gosalbes, M. J., de Vries, L. E., Abellan, J. J., and Francino, M. P. (2012). Metagenomics and development of the gut microbiota in infants. Clin. Microbiol. Infect. 18(Suppl. 4), 21–26. doi: 10.1111/j.1469-0691.2012.03876.x
van der Horst, M. A., Fabri, T. H., Schuurmans, J. M., Koenders, B. B., Brul, S., and Ter Kuile, B. H. (2013). Effects of therapeutical and reduced levels of antibiotics on the fraction of antibiotic-resistant strains of Escherichia coli in the chicken gut. Foodborne Pathog. Dis. 10, 55–61. doi: 10.1089/fpd.2012.1217
Verner-Jeffreys, D. W., Welch, T. J., Schwarz, T., Pond, M. J., Woodward, M. J., Haig, S. J., et al. (2009). High prevalence of multidrug-tolerant bacteria and associated antimicrobial resistance genes isolated from ornamental fish and their carriage water. PLoS ONE 4:e8388. doi: 10.1371/journal.pone.0008388
Vincent, C., Boerlin, P., Daignault, D., Dozois, C. M., Dutil, L., Galanakis, C., et al. (2010). Food reservoir for Escherichia coli causing urinary tract infections. Emerg. Infect. Dis. 16, 88–95. doi: 10.3201/eid1601.091118
Weber, J. T., Mintz, E. D., Canizares, R., Semiglia, A., Gomez, I., Sempertegui, R., et al. (1994). Epidemic cholera in Ecuador: multidrug-resistance and transmission by water and seafood. Epidemiol. Infect. 112, 1–11.
Weiser, A. A., Gross, S., Schielke, A., Wigger, J. F., Ernert, A., Adolphs, J., et al. (2013). Trace-back and trace-forward tools developed ad hoc and used during the STEC O104:H4 outbreak 2011 in Germany and generic concepts for future outbreak situations. Foodborne Pathog. Dis. 10, 263–269. doi: 10.1089/fpd.2012.1296
Whitehead, T. R., and Cotta, M. A. (2013). Stored swine manure and swine faeces as reservoirs of antibiotic resistance genes. Lett. Appl. Microbiol. 56, 264–267. doi: 10.1111/lam.12043
Yang, J., Wang, C., Shu, C., Liu, L., Geng, J., Hu, S., et al. (2013). Marine sediment bacteria harbor antibiotic resistance genes highly similar to those found in human pathogens. Microb. Ecol. doi: 10.1007/s00248-013-0187-2
Ye, L., Lu, Z., Li, X., Shi, L., Huang, Y., and Wang, H. H. (2013). Antibiotic-resistant bacteria associated with retail aquaculture products from Guangzhou, China. J. Food Prot. 76, 295–301. doi: 10.4315/0362-028X.JFP-12-288
Yong, D., Toleman, M. A., Giske, C. G., Cho, H. S., Sundman, K., Lee, K., et al. (2009). Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob. Agents Chemother. 53, 5046–5054. doi: 10.1128/AAC.00774-09
Zhanel, G. G., Wang, X., Nichol, K., Nikulin, A., Wierzbowski, A. K., Mulvey, M., et al. (2006). Molecular characterisation of Canadian paediatric multidrug-resistant Streptococcus pneumoniae from 1998–2004. Int. J. Antimicrob. Agents 28, 465–471. doi: 10.1016/j.ijantimicag.2006.08.005
Zhang, L., Kinkelaar, D., Huang, Y., Li, Y., Li, X., and Wang, H. H. (2011). Acquired antibiotic resistance: are we born with it? Appl. Environ. Microbiol. 77, 7134–7141. doi: 10.1128/AEM.05087-11
Keywords: antibiotic resistance, multidrug resistant bacteria, resistome
Citation: Rolain J-M (2013) Food and human gut as reservoirs of transferable antibiotic resistance encoding genes.Front. Microbiol. 4:173. doi: 10.3389/fmicb.2013.00173
Received: 29 March 2013; Accepted: 10 June 2013;
Published online: 24 June 2013.
Edited by:
Abelardo Margolles, Consejo Superior de Investigaciones Cientïficas, SpainReviewed by:
Teresa M. Coque, Hospital Universitario Ramón y Cajal, SpainCarlos F. Amábile-Cuevas, Fundación Lusara, Mexico
Copyright: © 2013 Rolain. This is an open-access article distributed under the terms of the Creative Commons Attribution License, which permits use, distribution and reproduction in other forums, provided the original authors and source are credited and subject to any copyright notices concerning any third-party graphics etc.
*Correspondence: Jean-Marc Rolain, Unité de Recherche sur les Maladies Infectieuses et Tropicales Émergentes, UM63, CNRS 7278, IRD 198, INSERM 1095, Institut Hospitalo-Universitaire Méditerranée Infection, Aix-Marseille Université, 27 Boulevard Jean Moulin, 13385 Marseille Cedex 05, France e-mail:amVhbi1tYXJjLnJvbGFpbkB1bml2bWVkLmZy