- 1Department of Microbiology, National Taiwan University College of Medicine, Taipei, Taiwan
- 2Department of Internal Medicine, National Taiwan University Hospital, Taipei, Taiwan
- 3The Institute of Biological Chemistry, Academia Sinica, Taipei, Taiwan
In the O1 strain of Klebsiella, the lipopolysaccharide (LPS) O-antigen is composed of D-galactan I and D-galactan II. Although the composition of the O1 antigen of Klebsiella was resolved more than two decades, the genetic locus involved in the biosynthesis of D-galactan II and the role of D-galactan II in bacterial pathogenesis remain unclear. Here, we report the identification of the D-galactan II-synthesizing genes by screening a transposon mutant library of an acapsulated Klebsiella pneumoniae O1 strain with bacteriophage. K. pneumoniae strain deleted for wbbY exhibited abrogated D-galactan II production; altered serum resistance and attenuation of virulence. Serologic analysis of K. pneumoniae clinical isolates demonstrated that D-galactan II was more prevalent in community-acquired pyogenic liver abscess (PLA)—causing strains than in non-tissue-invasive strains. WbbY homologs, WbbZ homologs, and lipopolysaccharide structures based on D-galactan II also were present in several Gram-negative bacteria. Immunization of mice with the magA-mutant (K−1 O1) (that is, with a LPS D-galactan II-producing strain) provided protection against infection with an O1:K2 PLA strain. Our findings indicate that both WbbY and WbbZ homologs are sufficient for the synthesis of D-galactan II. D-galactan II represents an immunodominant antigen; is conserved among multiple species of Gram-negative bacteria and could be a useful vaccine candidate.
Introduction
Several Gram-negative bacterial pathogens decorate their surfaces with exopolysaccharide (EPS), including capsular polysaccharide (CPS) and lipopolysaccharide (LPS). LPS is composed of lipid A, core oligosaccharide, and O antigenic polysaccharide. The O antigen contributes to serum resistance of several Gram-negative bacteria and also is a determinant of bacterial virulence (Krishnapillai, 1971; Taylor, 1976; Schneider et al., 1982; Tomas et al., 1986; Hong and Payne, 1997; Hsieh et al., 2012).
K. pneumoniae is an opportunistic pathogen that causes several kinds of infections, including pneumonia, bacteremia, urinary tract infection, and community-acquired pyogenic liver abscess (PLA) (Podschun and Ullmann, 1998; Ko et al., 2002; Ramphal and Ambrose, 2006; Tsai et al., 2008). There are 9 O antigens found in Klebsiella species, of which O1 is the most common serotype among clinical K. pneumoniae isolates (Alberti et al., 1993; Hansen et al., 1999). Our previous study showed that among K. pneumoniae causing PLA, the O1 serotype is highly predominant, with a prevalence rate of 90% (Hsieh et al., 2012). The O1 antigen contains two domains: D-Gal I and D-Gal II. The low-molecular-weight (LMW) D-Gal I polymers, which are directly linked to the core oligosaccharide, are composed of repeat units of the structure →3)-β-D-Galf-(1→3)-α-D-Galp-(1→. The wb (rfb) cluster, which is composed of six genes (wzm, wzt, glf, wbbM, wbbN, and wbbO), is required for the production of D-Gal I (Whitfield et al., 1991; Clarke and Whitfield, 1992). The high-molecular-weight (HMW) D-Gal II polymers, which are linked to the distal ends of D-Gal I polymers, consist of repeat units of the structure →3)-α-D-Galp-(1→3)- β-D-Galp-(1→, and are required for the resistance of K. pneumoniae to complement-mediated serum killing (McCallum et al., 1989). However, the location and identity of the genes required for D-Gal II biosynthesis are still unknown.
Bacteriophages target different capsule- or O- types of K. pneumoniae by recognizing surface molecules. Klebsiella bacteriophage O1-A, which infects the non-capsulated K. pneumoniae KD2 strain, has been used to screen and identify LPS mutants with spontaneous chromosomal mutations (McCallum et al., 1989). O1-A phage-resistant KD2 mutants have been classified into 2 basic types: those lacking high-molecular-weight (HMW) LPS and those lacking both LMW- and HMW-LPS. However, phage screening did not identify the genetic determinants of D-Gal II biosynthesis in the K. pneumoniae O1 strain (McCallum et al., 1989).
In the present study, we performed a bacteriophage screen of LPS-deficient mutants of an acapsulated K. pneumoniae O1 strain; these mutants had been derived from the parent strain by transposon mutagenesis. Using this screen, we identified and characterized the genetic locus involved in the biosynthesis of D-Gal II of the O1 antigen; examined the distribution of the D-Gal II antigen in PLA and non-tissue-invasive K. pneumoniae clinical isolates; investigated the distribution of the D-Gal II-synthesizing genes in E. coli O19 and other Gram-negative bacteria; explored the role of D-Gal II of K. pneumoniae in serum-killing and the pathogenesis of bacteremia; and investigated whether antiserum raised against LPS D-Gal II protected against D-Gal II-producing bacterial infection.
Materials and Methods
Ethics Statement
The study was approved by the Research Ethics Committee (REC) of the National Taiwan University Hospital (NTUH) (IRB approval #: 9561701018). All animal procedures were approved under application number 20140062 of the Institutional Animal Care and Use Committee (IACUC) of the National Taiwan University College of Medicine (NTUCM). Procedures were consistent with the recommendations of the Guide for the Care and Use of Laboratory Animals of the National Institutes of Health and Taiwan's Animal Protection Act.
Bacterial Strains and Culture Conditions
The bacterial strains used in this study are listed in Table S1. The K. pneumoniae NTUH-K2044 was isolated from a patient with liver abscess-complicated meningitis and endophthalmitis (Fang et al., 2004). The K-antigen Klebsiella reference strains were obtained from the Statens Serum Institute (Copenhagen, Denmark). A total of 74 clinical isolates of K. pneumoniae were collected from 1997 to 2003 in the National Taiwan University Hospital, as described previously (Hsieh et al., 2008). K. pneumoniae and E. coli strains were cultured in Luria-Bertani (LB) medium or LB medium supplemented with appropriate antibiotics, including 100 μg/mL ampicillin, 50 μg/mL kanamycin, or 100 μg/mL chloramphenicol.
Isolation of K. pneumoniae Bacteriophage O1-1
The magA deletion mutant was co-incubated with water from rivers of Taiwan in LB broth overnight. After centrifugation, the supernatant was filtered using a 0.45-μm filter and spotted onto LB plates overlaid with the magA mutant to detect phage plaques. An agar overlay method was used for isolation of a pure phage preparation and to determine phage titers as described elsewhere (Pieroni et al., 1994).
Construction of Mutant Library of the magA Mutant
A mutant library of the magA-deletion strain was constructed using random mutagenesis as previously described (Fang et al., 2004).
Screening for Phage-Resistant Mutants from the magA-Deletion Transposon Mutant Library
Lytic activity of O1-1 bacteriophage against the magA-deletion transposon mutants was measured with a spot test. A volume of 1 μL of 103 plaque forming units (PFU) of the O1-1 bacteriophage was used; the sensitivity of the magA-deletion transposon mutants against the O1-1 was observed by formation of a clear circular zone.
Semi-Random PCR and DNA Sequencing
Transposon-insertion sites were determined by semi-random PCR and DNA sequencing as described elsewhere (Chun et al., 1997). The primer pairs for semi-random PCR are listed in Table S1.
Determination of the Transcription Initiation Sites of wbbY and wbbZ Genes
5′-Rapid Amplification of cDNA Ends (RACE) -PCR was performed using the SMARTer RACE cDNA Amplication Kit (Clontech Laboratories) following the manufacturer's instructions.
Gene Deletion and Complementation
K. pneumoniae mutated in wbbY, wbbZ [polar and non-polar (NP)], magA wbbY (double mutant), or wza wzb (double mutant) were constructed using the previously described unmarked deletion method (Hsieh et al., 2008), which employs electroporation and selection with a temperature-sensitive vector (pKO3-Km) containing flanking regions for each target gene (Simon et al., 1983). For trans-complementation, wbbY, wbbZ with the intergenic region, wbbY with its intact promoter, or the entire wbbY-wbbZ region from K. pneumoniae NTUH-K2044 and E. coli F8188-41 were amplified by PCR and cloned into the pACYC184 vector (Fermentas). The recombinant pACYC184-derived plasmids were transformed into the wbbY, wbbZ, wbbZ-NP, and wbbY magA (double-mutant) strains by electroporation. The primer pairs for the deletion and complementation constructs are listed in Table S1. All sequence data for the wbbY-wbbZ regions from NTUH-K2044 and F8188-41 have been deposited (GenBank accession numbers KJ451390 and KJ451391).
Bacterial Growth Assays
An 18-h culture of each strain was used to inoculate each 5-ml LB broth aliquot at a ratio of 1:100. Each culture was grown at 37°C and growth was monitored hourly by serial dilution and plating to LB agar with next-day quantitation of colony-forming units (CFU).
Extraction and Quantification of Capsular Polysaccharides (CPS)
The amount of K1 CPS from each strain was determined by assaying uronic acid content as described by Domenico (Domenico et al., 1989).
LPS Analysis
The exopolysaccharide (EPS) extracts (containing both CPS and LPS) were purified by a modified hot water–phenol extraction method, as described previously (Chuang et al., 2006; Hsieh et al., 2012). Samples were separated by 14% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) containing 4 M urea (Tsai and Frasch, 1982) and visualized by silver staining (Tsai and Frasch, 1982). For immunoblots, samples were separated by SDS-PAGE and blotted to a Hybond-C membrane (Amersham, Little Chalfont, UK). Anti-D-Gal II antiserum was obtained from a rabbit immunized with a magA-mutant K. pneumoniae as described elsewhere (LTK BioLaboratories, TW); this serum was diluted 1/50,000 for LPS D-Gal II analysis.
NMR Spectroscopy
Proton spectra were carried out at 500 MHz at 25°C on solutions formulated in D2O by using a spectral width of 8.5 KHz, a 32 K data block, and a 90° pulse in a Bruker AVII-500 LC-NMR (Billerica, Massachusetts, USA) spectrometer with standard pulsed sequences and Bruker software.
wbbY and wbbZ Detection
In order to detect wbbY and wbbZ homologs in various species, PCR was performed using primer pairs specific for wbbY and wbbZ. Primers used in this study are listed in Table S1. PCR was performed as described elsewhere (Chuang et al., 2006).
Serum Killing Assays
The survival of exponential-phase bacteria in non-immune human serum was measured as previously described (Hsieh et al., 2012). Briefly, a log-phase inoculum of 2.5 × 104 CFU was mixed at a 1:3 or 3:1 vol/vol ratio with mixed non-immune human serum donated by 5 healthy volunteers. The final mixture, comprising 75 or 25% non-immune serum by volume, was incubated at 37°C for 3 h. For time-course studies, the bacteria/serum mixture, comprising 75 or 25% non-immune serum by volume, was incubated at 37°C for 1, 2, and 3 h. The colony count was determined by plating of serial dilutions on LB agar, and the mean survival ratio was plotted. A mean survival ratio ≥1 corresponds to serum resistance.
Mouse Inoculation Experiments
Virulence was evaluated by mortality in a murine model of septicemia generated by intraperitoneal (IP) injection. Groups of five-week-old female BALB/c mice were infected IP with the K. pneumoniae NTUH-K2044 (or the isogenic mutants) in 0.1 mL of 0.95% saline (1 × 102 CFU/mouse; 8 mice for each group). The exact inoculation dose was confirmed by serial dilution and plating to LB agar and survival rate of mice was monitored for 4 weeks. For in vivo competition, the wbbY mutant strain and the fully virulent placZ deletion mutant were grown (separately) to log-phase in LB broth, mixed at a 1:1 ratio (the initial dose was 1 × 103 CFU each) in 100 μL sterile saline, and inoculated IP into 5-week-old BALB/c mice (8 for each group). The mice were sacrificed at 24 h post-inoculation, and the spleen and liver were removed and homogenized in 1x PBS. The concentrations of bacteria in each sample were determined as described previously (Hsieh et al., 2010). The output/input ratio of the test strain to the virulent strain, the competitive index (CI), was interpreted as the in vivo survival capacity. For active immunization, five-week-old BALB/cByl mice were immunized three times by once-weekly IP injection with the indicated dose of the live magA single mutant or magA wbbY double mutant. Age-matched, unimmunized control mice (10 mice per group) were primed with an equivalent volume of saline on the same schedule. After 4 weeks, immunized and unimmunized control mice were challenged with the lethal dose 1 × 103 CFU of NTUH-A4528 (O1:K2). The challenged mice were observed for 28 days for mortality and clinical signs. Survival was analyzed by Kaplan-Meier analysis with a log-rank test; a P value <0.05 was considered to be statistically significant. For passive protection, 100 μL of serum derived from immunized and unimmunized control mice were administered to five-week-old BALB/cByl mice by IP injection. One hour after serum injection, mice were infected IP with 1 × 103 CFU of NTUH-A4528 (O1:K2) or 1 × 106 CFU of E. coli F8188-41 per mouse. Three hour after infection, the mice were euthanized and bacterial counts from liver and spleen were determined.
Statistical Analyses
Data are presented as means ± standard deviations (SDs). Statistical significance was assessed by a two-tailed Student's t test using Prism 5 (Graphpad) software. Prevalence was analyzed by chi-square test using SPSS version 12.0 software. Survival was analyzed by Kaplan-Meier analysis with a log-rank test. P values of <0.05 were considered significant.
Results
Isolation of K. pneumoniae Bacteriophage O1-1
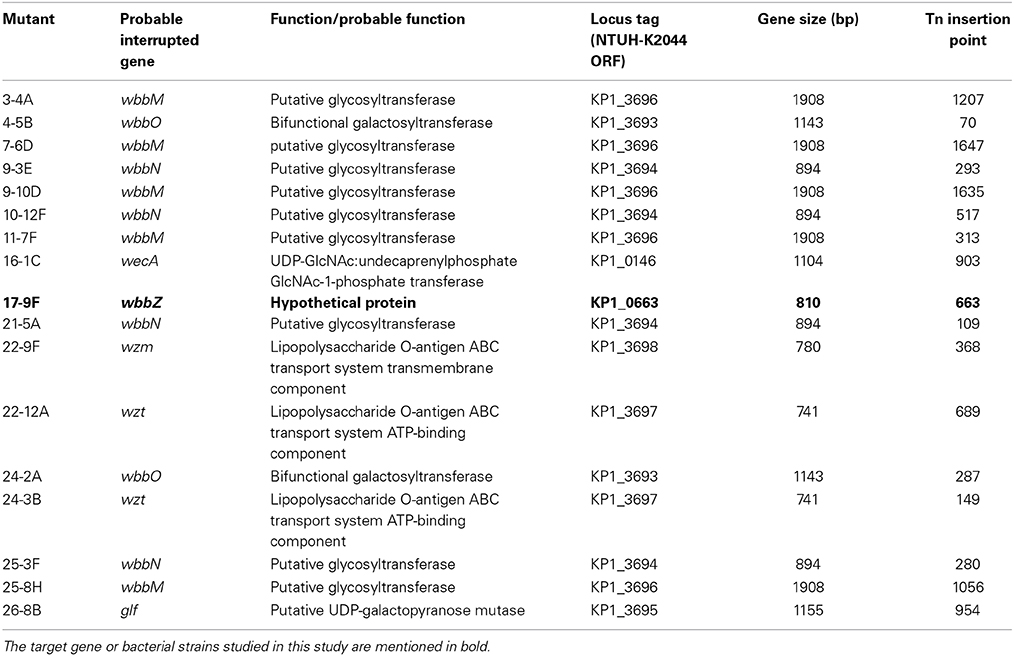
The magA deletion mutant, which was derived from the NTUH-K2044 (K1:O1), lacks the outermost capsule, thereby exposing the O1 antigen (Hsieh et al., 2012). In the present study, we isolated a K. pneumoniae O1-1-specific bacteriophage using the magA mutant. A magA mutant lawn was successfully lysed by bacteriophage O1-1, yielding visible plaques from phage titers as low as 10 PFU (Figure 1A). In contrast, the wild-type, the wbbO mutant (LPS mutant), and the magA wbbO double-mutant (CPS- and LPS-lacking) strains were all resistant to lysis by bacteriophage O1-1, even when the viral titer was elevated to 105 PFU. This result demonstrates that bacteriophage O1-1 did not infect the encapsulated K. pneumoniae bacteria, presumably due to masking of the receptor by CPS.
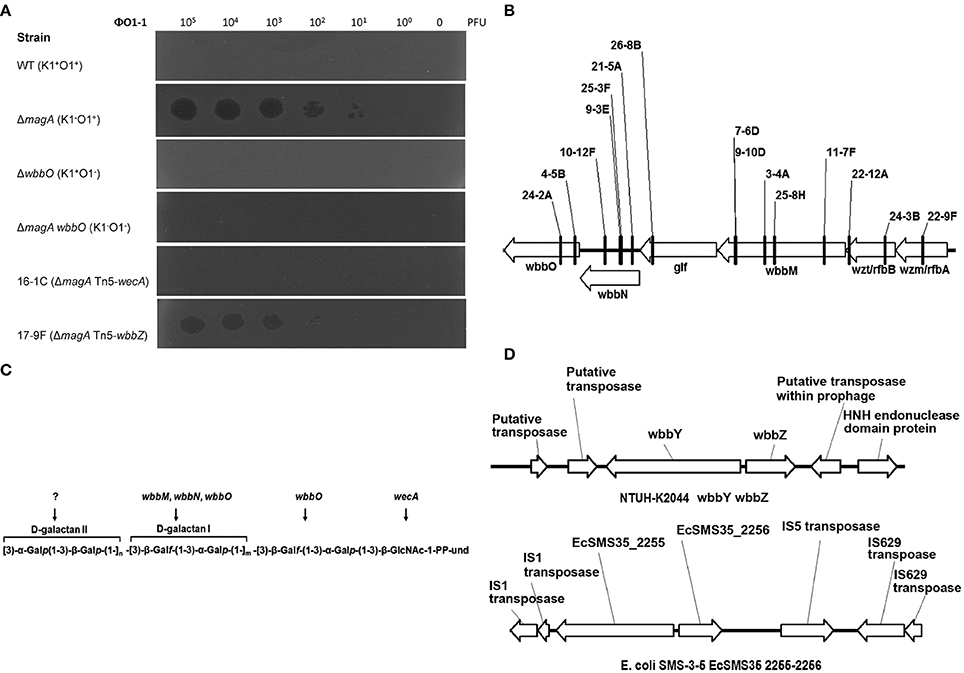
Figure 1. The K. pneumoniae O1 mutants. (A) The K. pneumoniae NTUH-K2044 wild-type, magA deletion, wbbO deletion, magA wbbO double-deletion mutants and magA deletion transposon mutant strains, 16-1C and 17-9F, were grown to mid-log phase at 37°C and spread on LB plates. Serial dilutions of SM buffer containing phage O1-1 were dotted onto the bacterial lawn and grown at 37°C for 16–18 h. PFU, plaque forming unit. (B) Transposon insertion sites within the wb cluster in the NTUH-K2044 magA mutants. Arrows denote the orientations of the ORFs. (C) The composition of K. pneumoniae O1 LPS and the gene responsible for each step. (D) Genetic organization of wbbY, wbbZ (or their homologs), and their flanking regions in K. pneumoniae NTUH-K2044 (upper) and E. coli SMS-3-5 (lower). Arrows denote the orientations of the ORFs.
Screening and Identification of Bacteriophage O1-1-Resistant Mutants
We constructed a magA deletion strain transposon library containing 2500 mutants to screen for genes associated with LPS biosynthesis. Phage O1-1-resistant mutants derived from the magA mutant were classified into two types, distinguished by their sensitivity to the O1-1 phage on spot tests. Among 17 phage O1-1-resistant mutants, 16 isolates (94%) were fully resistant to lysis by phage O1-1, even when the viral titer was elevated to 105 PFU. In Figure 1A, this class is represented by the 16-1C mutant. In contrast, 1 of the 17 phage O1-1-resistant mutants, designated 17-9F, showed visible plaques when the viral titer was raised to 102 PFU (Figure 1A). The transposon-insertion sites of these 17 phage O1-1-resistant mutants were determined by semi-random PCR and DNA sequencing. The resulting sequences were analyzed by comparison to the genome sequence of strain NTUH-K2044 (accession number AP006725). The analyses showed that 15 mutants harbored insertions within the wb cluster: five in wbbM, four in wbbN, two in wbbO, two in wzt, one in wzm, and one in glf (Table 1, and Figure 1B). Mutant 16-1C had an insertion in wecA, which initiates LPS biosynthesis by catalyzing the transfer of GlcNAc-1-phosphate onto undecaprenyl phosphate, generating undecaprenyl-pyrophosphoryl-GlcNAc (Figure 1C) (Lehrer et al., 2007). Mutant 17-9F was disrupted in a hypothetical protein-encoding gene; consistent with The Bacterial Polysaccharide Gene Nomenclature scheme (BPGN) (Reeves et al., 1996), this ORF is designated as wbbZ (Figure 1D).
Genetic Analysis of wbbY and wbbZ
Analysis using the BlastX program showed that the predicted WbbZ protein shares a conserved domain with CsaB of Bacillus subtilis, which is a pyruvyl transferase required for polysaccharide biosynthesis (Mesnage et al., 2000). The gene adjacent to wbbZ, wbbY, encodes a putative glycosyltransferase that might be involved in the synthesis of oligosaccharides, polysaccharides, and glycoconjugates by transferring the sugar moiety from a sugar donor onto an acceptor (Figure 1D). The nearby flanking genes, kp0660, kp0661, and kp0664, are all putative transposase-encoding genes that are likely not associated with the biosynthesis of LPS. Moreover, WbbY and WbbZ share 99% amino acid sequence identity with EcSMS35_2255 and EcSMS35_2256 of E. coli SMS-3-5; these E. coli gene products have been conjectured to contribute to LPS biosynthesis, though the proteins were not further characterized (Fricke et al., 2008). This genetic evidence encouraged us to investigate the association between the wbbY-wbbZ region and LPS biosynthesis of K. pneumoniae.
Glycosyltransferase WbbY is essential for the Production of D-Gal II
The sequence features of the wbbY-wbbZ region suggested that wbbY and wbbZ are transcribed as a divergent gene pair; however, these two genes are only separated by a very short intergenic region consisting of 97 base-pairs between the postulated ATGs of these two ORFs. To avoid the polar effects resulting from the single mutations, that is, that deletion of wbbY affected the function of wbbZ and vice versa. Therefore, we determined the transcription initiation sites of both the wbbY and wbbZ genes by 5′-RACE (5′-Rapid Amplification of cDNA Ends) method. The transcription start site and promoter region of wbbZ were located within the short intergenic region between the wbbY and wbbZ genes. In contrast, the transcription start site and promoter region of the wbbY gene were located in the coding sequences of wbbZ (Figure 2A).
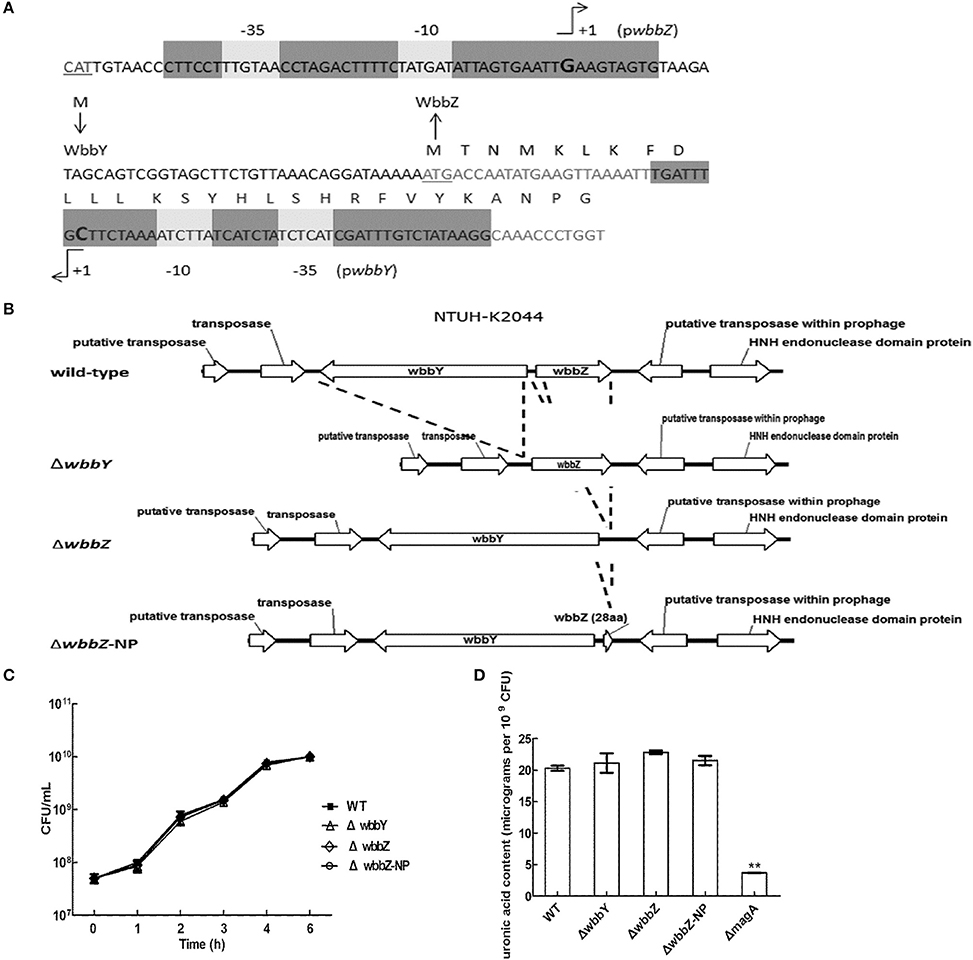
Figure 2. Genetic construction, growth, and CPS production of K. pneumoniae NTUH-K2044 and its isogenic mutants. (A) Mapping of transcription start sites of the wbbY and wbbZ genes. The promoter regions and the proposed −10 and −35 regions are indicated by gray shading. The transcription start sites are shown by +1 with arrowheads and in larger font. Deduced amino acid sequences (in single-letter code) are indicated beginning with an ATG start codon. (B) Schematic diagram representing the construction of deletion mutations in the NTUH-K2044 background. The locations and orientations of the ORFs described in this study are indicated by arrows. The dashed lines refer to joining of the regions flanking the deleted target. (C) Mutation of wbbY or wbbZ does not impair bacterial growth. Overnight cultures of the K. pneumoniae NTUH-K2044 wild-type strain, wbbY, wbbZ, and wbbZ-NP (non-polar) mutant strains were individually inoculated into fresh LB medium and grown at 37°C. The growth of bacteria was monitored periodically every hour by plating of serial dilutions on LB agar and counting CFU. The data represent the means of three independent trials; the error bars represent the standard deviations. (D) The amount of K1 CPS production of the K. pneumoniae NTUH-K2044 wild-type, and of the wbbY, wbbZ, wbbZ-NP, and magA single-mutant strains, was determined by assaying uronic acid content. The data represent the means of three independent trials; the error bars represent the standard deviations. **P < 0.01 by Student's t-test (compared to the wild-type strain); other comparisons were not statistically significant (P ≥ 0.05).
To confirm our phage screening results, we constructed in-frame deletion mutant of wbbY, wbbZ, a non-polar deletion mutant of wbbZ that retained the upstream end of the wbbZ ORF (encoding 28 N-terminal amino acids of WbbZ, and corresponding to the DNA sequences of the presumed minimal wbbY promoter) and examined the production of CPS and LPS in these three mutants (Figure 2B). Growth rate and CPS production by in-frame deletion of wbbY, wbbZ or the non-polar wbbZ mutant strain was similar to that of the wild-type strain (Figures 2C,D). In the case of LPS expression, the wild-type and the magA mutant strains displayed similar patterns in both silver staining and immunoblotting, producing both HMW-LPS and LMW-LPS (Figure 3A). In contrast, the wbbO mutant strain completely lost its ability to produce O-antigen, displaying only the core and lipid-A in silver staining. The wbbY mutant was completely depleted in HMW-LPS. The in-frame wbbZ deletion mutant was impaired in the production of HMW-LPS, such that HMW-LPS was detected only at low levels. Unexpectedly, the non-polar wbbZ mutant expressed HMW-LPS and the molecule was detected by anti-D-Gal II antiserum (Figure 3B).
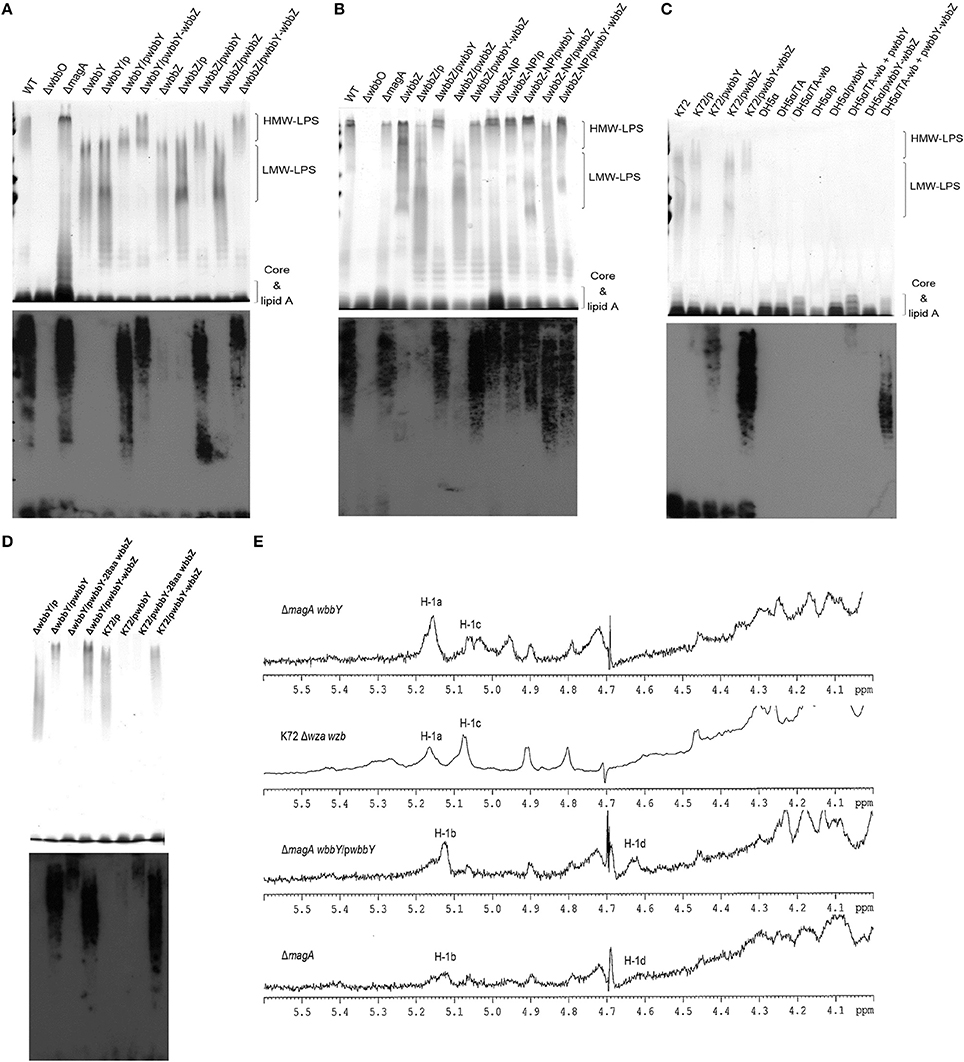
Figure 3. The LPS profiles of K. pneumoniae NTUH-K2044 and its isogenic mutants. (A) EPS specimens were prepared from K. pneumoniae NTUH-K2044, from wbbY and wbbZ single mutants, and from the same mutants harboring complementation plasmids. Extracts from normalized bacterial suspensions (108 CFU) were separated by SDS-PAGE and visualized by silver staining (upper) or immunoblotting with anti-D-Gal II antiserum (lower). HMW-LPS, high-molecular-weight LPS. LMW-LPS, low-molecular-weight LPS. (B) EPS specimens were prepared from K. pneumoniae NTUH-K2044, from wbbZ and wbbZ-NP single mutants, and from mutants harboring complementation plasmids. Extracts from normalized bacterial suspensions (108 CFU) were separated by SDS-PAGE and visualized by silver staining (upper) or immunoblotting with anti-D-Gal II antiserum (lower). (C,D) EPS extracted from Klebsiella K72:O2, E. coli DH5α, and K. pneumoniae wbbY mutant strains harboring the indicated plasmids were separated by SDS-PAGE and the LPS patterns were visualized by silver staining (upper) or immunoblotting with anti-D-Gal II antiserum (lower). (E) The 1H-NMR spectra of the LPS of the wbbY mutant. LPS specimens were prepared from the Klebsiella K72:O2 capsule-deficient mutant (K72Δwza wzb), the K. pneumoniae NTUH-K2044 magA deletion strain, the magA wbbY double-deletion mutants, and the magA wbbY double-deletion strain containing the wbbY-expressing plasmid. The four D-galactosyl residues are indicated (a to d) according to decreasing anomeric 1H shifts.
To compensate for the deficiency in D-Gal II synthesis, we transformed the wbbY and wbbZ single-mutant strains with plasmids containing either (i) wbbY, (ii) wbbZ with the intergenic region between these two genes, or (iii) the wbbY-wbbZ region, and then examined HMW-LPS production in the transformants. Compared to the vector control, the LPS of wbbY mutant harboring the wbbY-expressing plasmid shifted to a region of higher molecular weight, and this molecule was detected by anti-D-Gal II antiserum (Figure 3A). An even higher molecular weight LPS was obtained by introducing the wbbY-wbbZ-expressing plasmid into the wbbY mutant; the size of the LPS produced by this strain resembled that of the wild-type strain. Additionally, introduction of the wbbY-expressing plasmid resulted in a shift to a higher molecular weight of LPS in the wbbZ mutant. The wbbZ-expressing plasmid did not compensate for the lack of production of HMW-LPS in the wbbZ mutant. As expected, the wbbY-wbbZ-expressing plasmid fully restored the production of LPS in the wbbZ mutant. These results indicated that the deletion of the wbbZ gene have additional polar effects on wbbY, presumably by removing the promoter of wbbY. Thus, these results implied that only wbbY was essential for the production of D-Gal II in the K. pneumoniae O1 strain.
wbbY and wbbZ are Sufficient for the Biosynthesis of D-Gal II
To investigate whether the wbbY-wbbZ region was sufficient for the expression of D-Gal II, we individually introduced plasmids containing (i) wbbY, (ii) wbbZ with the intergenic region between these two genes, (iii) wbbY with its intact promoter sequence, or (iv) the entire wbbY-wbbZ region into the Klebsiella K72 reference and the wbbY single-mutant strains. The Klebsiella K72 reference strain is an isolate that produces O2 antigen, which is composed only of D-Gal I and runs as LMW-LPS and is not recognized by anti-D-Gal II antiserum (Figure 3C). Introduction of a plasmid carrying wbbY with its intact promoter sequence enabled the K72 and the wbbY single-mutant strains to produce partial D-Gal II antigen of increased molecular weight that was weakly detected by anti-D-Gal II antiserum (Figure 3D). Interestingly, the introduction of the entire wbbY-wbbZ region into the K72 strain enabled the bacterium to produce D-Gal II. This results parallels the full restoration of D-Gal II antigen production to the wbbY mutant strain upon complementation with the plasmid containing the entire wbbY-wbbZ region.
Additionally, we compared the 1H-NMR spectra of LPS obtained from the wbbY-deletion mutant, from the wbbY mutant harboring the wbbY-expressing plasmid, and from the K72 strain. To rule out the possibility of CPS contamination during LPS analysis, we constructed these mutants in the K-antigen-deficient background. Consistent with the previous NMR assignment (Whitfield et al., 1991), we found that the LPS of the magA wbbY double-deletion mutant strain contained primarily D-Gal I [H-1a: -3)-β-D-Galf-(1- at 5.18 ppm) and H-1c: -3)-α-D-Galp-(1- at 5.06 ppm], resembling the LPS derived from the K72 wza wzb double-deletion mutant strain (D-Gal I). In contrast, the LPS of the magA wbbY double-deletion mutant harboring the wbbY-expressing plasmid and that of the magA mutant strain contained both D-Gal II [H-1b: -3]-α-D-Galp-(1- at 5.15 ppm and H-1d: -3)-β-D-Galp-(1- at 4.66 ppm] and D-Gal I (Figure 3E). In addition, we tried to express the K. pneumoniae O1 antigen in E. coli. Co-transformation of E. coli DH5α with two plasmids containing the wb cluster and the entire wbbY-wbbZ region enabled the bacterium to express a typical smooth lipopolysaccharide that could be detected by anti-D-Gal II antiserum, though the molecular weight was slightly lower than that produced in K. pneumoniae (Figure 3C). No HMW-LPS or smooth LPS was detected in those bacteria containing plasmids carrying the wb cluster only or with vector alone. Taken together, these genetic and chemical analyses indicate that both wbbY and wbbZ were sufficient for the expression of D-Gal II.
Prevalence of the D-Gal II Antigen in K. pneumoniae Clinical Isolates
In order to detect the production of D-Gal II, we generated and evaluated antiserum raised against LPS D-Gal II by immunization of a rabbit with the unencapsulated magA mutant (K−1 O1). Antiserum raised against the magA mutant reacted with all (20 of 20) of randomly selected Klebsiella reference strains, which are known to belong to the O1 serotype and produce D-Gal II. In contrast, this antiserum did not react with the Klebsiella K72 strain, an isolate that is known to belong to the O2 type and produce D-Gal I only (Figure 3C). Therefore, the antiserum isolated from the magA-mutant-immunized rabbit is specific to D-Gal II. Using this tool, we compared the distribution of D-Gal II by immunoblotting of samples from 74 clinical isolates of K. pneumoniae, including 42 PLA strains and 32 non-tissue-invasive strains. The D-Gal II-containing strains accounted for 63.5% of all clinical isolates, with significant differences in representation among the strains that caused PLA (38/42; 90.5%) and those that were not tissue-invasive (9/32; 28.1%) (P < 0.0001, chi-squared test).
Occurrence of wbbY and wbbZ Homologs in the E. coli O19 Strain and other Gram-Negative Bacteria
A previous study indicated that the E. coli O19 antigen is recognized by an anti-Klebsiella O1 monoclonal antibody (McCallum et al., 1989; Fricke et al., 2008). We therefore investigated the link between the wbbY and wbbZ genes and structures based on D-Gal II synthesis in the E. coli O19-type bacterium. Specifically, we carried out PCR analysis using primer pairs located within the wbbY and wbbZ genes. Positive signals were obtained from the acapsulated E. coli O19ab K−:H7 strain, F8188-41; the amplicons were of the same sizes as those obtained from K. pneumoniae NTUH-K2044 and A5054, which are known O1-type strains (Figure 4A). We then carried out immunoblotting analysis using the anti-D-Gal II antiserum. All wbbY-, wbbZ-positive strains, including F8188-41, showed positive signals. In addition, D-Gal II was immunologically detected in all of the wbbY and wbbZ single-mutant and the K72 harboring the F8188-41 wbbY-wbbZ-expressing plasmid strains (Figures 4B,C). These results suggested that D-Gal II is present in the LPS of the E. coli O19 group. Moreover, we also determined the DNA sequences of the wbbY and wbbZ genes from F8188-41, which were clustered on the chromosome and showed 95% DNA sequence identity compared with those from the NTUH-K2044 and another O19-type E. coli strain, SMS-3-5. The WbbY and WbbZ homologs from K. pneumoniae NTUH-K2044 and E. coli SMS-3-5 showed 99% protein sequence identity and belong to the same branch by phylogenetic tree analysis (Figure 4D); the E. coli F8188-41 homologs showed lower protein sequence similarity and sorted as a separate phylogenetic branch. To investigate the presence of WbbY and WbbZ homologs in other bacteria, we searched the NCBI database of encoded sequences from other bacterial genomes by BLASTp. WbbY and WbbZ homologs were detected in another member of the Klebsiella genus (K. oxytoca) and in four other more distantly related Gram-negative bacteria (E. coli, Erwinia piriflorinigrans, Serratia plymuthica, and Commensalibacter intestine). In each of these species, the wbbY and wbbZ gene were adjacently located on the respective chromosome and also as divergent transcription units (Table 2). These observations suggested that the D-Gal II structure described in the present study also may be present in Gram-negative bacteria of multiple genera.
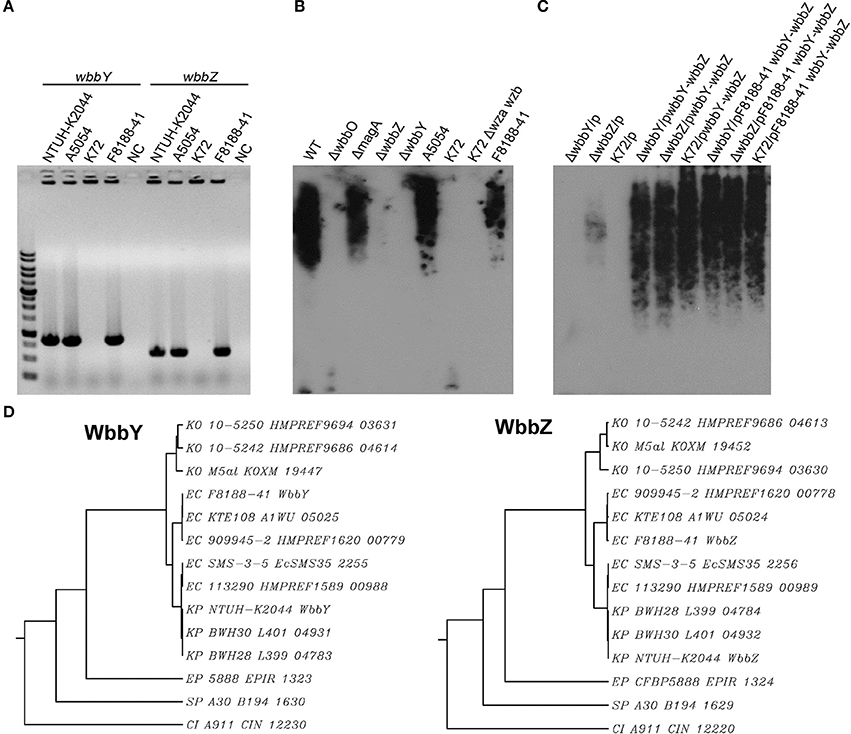
Figure 4. Analysis of WbbY and WbbZ homologs in K. pneumoniae and other Gram-negative bacteria. (A) PCR detection of wbbY (830 bp) and wbbZ (583 bp)—like genes. lane M, 1 kb DNA ladder; NC, no-template control. (B) EPS specimens were prepared from K. pneumoniae NTUH-K2044; isogenic mutants of K2044; strain A5054; the Klebsiella K72 wild type; K72 wza wzb double mutant; and E. coli O19 ab K−:H7 F8188-41. Extracts from normalized bacterial suspensions (108 CFU) were separated by SDS-PAGE and visualized by immunoblotting with anti-D-Gal II antiserum. (C) EPS extracted from the wbbY and wbbZ single-mutant and Klebsiella K72:O2 strains harboring the indicated plasmid was separated by SDS-PAGE and the LPS patterns were analyzed by immunoblotting with anti-D-Gal II antiserum. (D) Phylogenetic analysis of WbbY and WbbZ homologs from different Gram-negative bacteria using the SMART (Simple Modular Architecture Research Tool) database.
D-Gal II is Required for K. pneumoniae's Serum Resistance and Virulence In Vivo
Serum killing assays were performed on the K. pneumoniae wbbY and non-polar wbbZ single-deletion mutants to determine the mutants' sensitivities to serum bactericidality. Exposure to human serum (75% human serum for 3 h) resulted in higher levels of killing in the wbbY mutant than in the wild-type and non-polar wbbZ mutant strains (Figure 5A); the wild-type and non-polar wbbZ mutant strains exhibited similar levels of resistance to serum killing. Moreover, introduction of plasmid carrying either wbbY alone or the entire wbbY-wbbZ region successfully restored serum resistance levels to the wbbY mutant strain. This result confirmed that resistance to serum killing correlated with the production of D-Gal II.
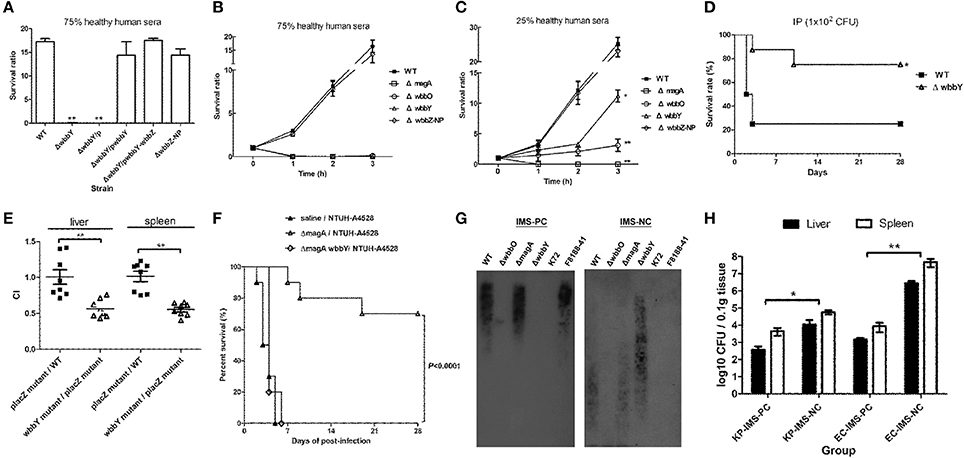
Figure 5. Serum sensitivity and virulence of NTUH-K2044 and its isogenic mutants. (A–C) Serum sensitivity assays of resistance to killing by non-immune healthy human serum of the NTUH-K2044 wild-type; wbbY, wbbZ, wbbZ-NP, magA, and wbbO single-deletion mutants; and the respective complemented strains. The data represent the means of three independent trials; the error bars represent the standard deviations. A mean survival ratio ≥1 corresponds to serum resistance. **P < 0.01 or *P < 0.05 by Student's t test (compared to the wild-type strain). (D) Eight mice per group were infected with the NTUH-K2044 wild-type and the wbbY mutant strains at an intraperitoneal (IP) dose of 1 × 102 CFU/animal. Survival of mice was monitored for 4 weeks. ■, NTUH-K2044; △, wbbY mutant (wbbY vs. parent, P = 0.024; log-rank test). (E) Survival of the isogenic K. pneumoniae wbbY (open triangles) mutant in the in vivo bacterial competition model, using competition against the fully virulent placZ mutant in groups of eight mice. The ratio of lacZ+ or lacZ− K. pneumoniae (close squares) colonies in the contents of the spleen and liver was determined from a single mouse at sacrifice (24 h post-infection). Each symbol represents the competitive index (CI) for each inoculum; and the medians and SDs of the values are shown (for the wild-type strain group vs. placZ mutant group, the CI of spleen and liver were 1.016 ± 0.206 and 1.009 ± 0.286, respectively; wbbY mutant group vs. placZ mutant group, the CI of spleen and liver were 0.553 ± 0.089 and 0.564 ± 0.123, respectively; P = 0.001, Wilcoxon's signed rank test). (F) Survival of mice immunized with magA mutant and then challenged with NTUH-A4528 (O1:K2). Mice (10 per group) were inoculated three times by once-weekly IP injections with 1 × 106 CFU of magA mutant or magA wbbY double-mutant strain. Age-matched, unimmunized control mice were inoculated with saline. On the fourth week, immunized or unimmunized groups were challenged with NTUH-A4528 (1 × 103 CFU per animal, IP). Survival was assessed for 28 days following infection. ▲, non-immunized, NTUH-A4528 challenged; △, magA mutant immunized, NTUH-A4528 challenged; ◊, magA wbbY double mutant immunized, NTUH-A4528 challenged. By log-rank test: P < 0.0001, magA/NTUH-A4528 vs. non-immune/NTUH-A4528 or magA wbbY/NTUH-A4528. (G) Immune response of anti-D-Gal II IgG in magA mutant-immunized mice. Immunoblots developed with magA mutant immune mouse serum (1:500) or magA wbbY double-mutant immune mouse serum (1:500). (H) Anti-D-Gal II specific antibodies reduce growth of NTUH-A4528 (O1:K2) K. pneumoniae and E. coli F8188-41 (O19) in mice. Groups of 4 BALB/c mice were treated with magA mutant immune mouse serum (IMS–positive control, IMS-PC) or with magA wbbY double-mutant immune mouse serum (IMS–negative control, IMS-NC). One hour after injection, mice were infected with 1 × 103 CFU of the NTUH-A4528 (KP-IMS-PC and KP-IMS-NC) or 1 × 106 CFU of E. coli F8188-41 (EC-IMS-PC and EC-IMS-NC) by IP injection. Three hours after infection, the numbers of bacteria in the liver and spleen were determined. Log10 CFU was standardized per 0.1 gram wet organ weight. The black and white bars represent the means for each group for liver and spleen, respectively. Data are presented as means ± SD. **P < 0.01 or *P < 0.05 by Student's t test (KP-IMS-PC vs. KP-IMS-NC and EC-IMS-PC vs. EC-IMS-NC).
To delineate the contribution of CPS, D-Gal I, and D-Gal II to K. pneumoniae's serum resistance, the survival ratio of the wild-type, magA, wbbO, wbbY and non-polar wbbZ single-deletion mutant strains after killing by 75 and 25% human serum was assessed at one-hour intervals through 3 h of exposure (Figures 5B,C). Mutation of magA dramatically reduced the serum resistance of the NTUH-K2044, presumably because the mutant lacked K1 antigen. As expected, wbbO mutation resulted in the loss of O1 antigen; the mutant exhibited elevated susceptibility to serum killing compared to the wild-type, wbbY, and non-polar wbbZ mutant strains. The wbbY mutant, which lacked D-Gal II, showed more susceptible to serum killing than the wild-type and non-polar wbbZ mutant strains. These results demonstrated that the sensitivity to serum killing of the K1:O1 K. pneumoniae strain depended on the integrity of bacterial surface polysaccharides.
To evaluate the contribution of the LPS D-Gal II of K. pneumoniae in bacteremia, we challenged mice with the same dose (1 × 102 CFU) of the wild-type and wbbY deletion-mutant strains. Upon IP infection of mice, the wbbY mutant showed significantly reduced virulence compared to the parent strain (P = 0.024, Figure 5D). Previously, we generated an isogenic lacZ mutant with its promoter deletion and used it as the wild-type strain in the competition assay (Hsieh et al., 2010). In those experiments, no competitive advantage was observed for either the wild type or placZ mutant in the spleen and liver of BALB/c mice. Using the same assay in the present study, the wbbY mutant strain showed lower competitive indices than the wild type or placZ mutant (P < 0.001, Figure 5E). These results indicated that D-Gal II was required for K. pneumoniae's serum resistance and contributed to in vivo virulence.
Protective Effect of Anti-D-Gal II Antibodies During K. pneumoniae Infection
To evaluate the protective efficacy of immunization against D-Gal II, mice were pretreated by IP injection with either magA or magA wbbY (1 × 106 CFU per dose), administered once weekly for 3 weeks. On the fourth week, the animals (in groups of 10) were challenged IP with a lethal dose of highly virulent K. pneumoniae NTUH-A4528 (O1:K2). Within 6 days of NTUH-A4528 infection, 100% of the unimmunized control mice or the mice pre-immunized with the magA wbbY double-mutant strain died. Meanwhile, 70% of the mice pre-immunized with the magA-mutant (K−1 O1) survived without any symptoms of disease through 28 days after challenge (P < 0.0001, Figure 5F). Mice immunized with the magA mutant (K−1 O1) showed anti-D-Gal II immunoglobulin G (IgG) production by immunoblot; mice immunized with the magA wbbY double mutant or unimmunized control mice did not (Figure 5G).
The results of the immunization study were consistent with a separate test in which the passive protective efficacy of anti-D-Gal II antiserum was tested. In the passive immunization study, mice were pretreated with immune mouse sera (negative and positive controls, respectively). The in vivo protective capacity of the sera was tested using a K. pneumoniae or an E. coli septicemia infection model, whereby the mice were inoculated IP with 1 × 103 CFU of the encapsulated K. pneumoniae NTUH-A4528 (O1:K2) or 1 × 106 CFU of the acapsulated E. coli F8188-41 (O19) strain. The bacterial load was determined in both the liver and spleen at 3 h post-infection. Compared to animals pre-treated with the magA wbbY double-mutant immune mouse serum (IMS-NC), mice pretreated with the magA mutant immune mouse serum (IMS-PC) had significantly reduced bacterial loads (Figure 5H). These results indicated that in both active and passive immunization studies, D-Gal II-specific antiserum was able to reduce bacterial dissemination and protect against infection by encapsulated O1:K2 PLA-associated K. pneumoniae and by D-Gal II-containing E. coli in mouse models of septicemia.
Discussion
In this study, we identified 17 K. pneumoniae magA-deletion transposon mutants resistant to the bacteriophage O1-1. Among these mutants, 16 mutants were disrupted in well-characterized LPS-biosynthesis genes: 15 were in the wb-cluster genes, and 1 was in wecA, indicating that a bacteriophage screening method was very useful. The last mutant represented a transposon insertion into wbbZ (formerly designated kp0663), a locus located well away from the wb cluster [located in a region formerly designated kp3698 (wzm) to kp3693 (wbbO)]. This result was consistent with the previous finding that genes involved in the expression of D-Gal II were not closely linked to the wb cluster (Whitfield et al., 1991).
The gene adjacent to wbbZ, wbbY, has apparent carbohydrate transferase functions and conserved among D-Gal II synthesizing organisms. The sequence features of this region suggested that these two genes are adjacently located on the respective chromosome and also as divergent transcription units; however, they are only separated by a very short intergenic region. Indeed, mapping of the transcription initiation sites of the two genes of the NTUH-K2044 revealed that the transcription start site of the wbbY gene was located within the coding region of wbbZ, apparent overlap of regulatory sequences so that wbbZ insertion has polar effects on wbbY. More detailed studies will be needed to clarify the regulation and interaction of these overlapping promoters.
Silver staining revealed that the wbbY mutant had enriched LMW-LPS, but lacked HMW-LPS. Trans-complementation of the wbbY mutation by the wbbY-expressing plasmid resulted in the production of LPS with a molecular weight between those of the LPS of the wild-type and mutant strains; this intermediate molecular weight product was detected by anti-D-Gal II antiserum. These findings suggest that wbbY encodes a glycosyltransferase that has a central role in the elongation of D-Gal II. The D-Gal II structure is the difference between the LPS composition of K. pneumoniae serotypes O1 and O2. Expression of the entire wbbY-wbbZ region in a Klebsiella O2 strain enabled the bacterium to express HMW-LPS that was recognized by anti-D-Gal II antiserum. Furthermore, co-transformation of E. coli DH5α with two plasmids carrying the K. pneumoniae wb cluster and the wbbY-wbbZ regions permitted the E. coli K-12 recipient (which normally produces rough LPS consisting of only lipid A-core) to produce a typical smooth LPS ladder. These pieces of evidence support the contention that both the wbbY and wbbZ genes are sufficient for the biosynthesis of D-Gal II. Determination of their precise functions in D-Gal II assembly awaits further study. The current data suggests that WbbZ participates in some other way, perhaps involving termination of the elongation process, following the step catalyzed by WbbY.
Sequence analysis also showed that WbbY and WbbZ shared a high degree of sequence similarity with two putative LPS biosynthesis genes of SMS-3-5, an E. coli O19:H34 strain (McCallum et al., 1989; Fricke et al., 2008). Interestingly, an early study indicated that the E. coli O19 antigen could be recognized by a Klebsiella O1 antigen-specific monoclonal antibody (McCallum et al., 1989; Fricke et al., 2008). This connection led us to the reasonable assumption that the E. coli O19 group has an O-antigen structure similar to that of Klebsiella O1, as well as the genetic apparatus required for synthesis of O antigen. That is exactly what we demonstrate in this study: a genetic apparatus for D-Gal II biosynthesis is present in the E. coli O19 group. Therefore, it is conceivable that WbbY and WbbZ participate in the assembly of D-Gal II-containing LPS in the K. pneumoniae serotype O1 and the E. coli serotype O19 strains.
Previous studies reported that D-Gal I and structures based on D-Gal I are found in a variety of Klebsiella O serotypes, as well as in Serratia plymuthica, S. marcescens O16 and O20, and Pasteurella hemolytica serotypes 4 and T10 (Oxley and Wilkinson, 1989; Richards and Leitch, 1989; Whitfield et al., 1992; Aucken et al., 1993; Kelly et al., 1993). However, D-Gal II is only found in O-antigen from Klebsiella serotypes O1 and O8, and in S. plymuthica (Aucken et al., 1993; Kelly et al., 1993). In fact, database searches performed as part of the present study indicate WbbY and WbbZ homologs are present in several bacterial genera, especially within the Enterobacteriaceae. Taken together, it is conceivable that WbbY and WbbZ homologs participate in the assembly of D-Gal II-containing LPS in a wider and previously unappreciated range of Gram-negative bacteria.
Deletion of the wbbY gene impaired the survival of the NTUH-K2044 in human serum, presumably because the mutant lacked D-Gal II. This result is consistent with an earlier finding that the presence of D-Gal II is required for the resistance of K. pneumoniae to serum killing (McCallum et al., 1989). The importance of the bacterial surface EPS to serum resistance of the O1:K1 K. pneumoniae strain is ranked in decreasing order from K1 antigen, O1 antigen, D-Gal I, and D-Gal II. Upon in vivo infection, the D-Gal II-deficient K. pneumoniae mutant is less virulent than the parental strain. Loss of D-Gal II production in K. pneumoniae restricts the spread of the bacteria to distant organs, reducing mice mortality. Thus, these findings indicate that mutation of the K. pneumoniae gene coding for D-Gal II biosynthetic machinery results in changes in serum resistance; reducing bacterial dissemination and colonization into deep organs.
The O antigen is the most external component of the LPS. In the present study, D-Gal II-specific antiserum was obtained from a rabbit immunized with a magA mutant (K−1 O1) strain. This result indicates that D-Gal II is the immunodominant antigen of the LPS O1 antigen of K. pneumoniae. The high prevalence (90.5%) of D-Gal II in PLA strains indicates that the D-Gal II-producing strains are predominant among K. pneumoniae isolates causing PLA. Earlier results showed that K antigen and LPS are exposed together at the cell surface in K. pneumoniae O1 strains harboring CPS serotype K2 (Merino et al., 1992). Clements et al. used K. pneumoniae B5055 (O1:K2), a pneumonia strain, to show that immunization with purified LPS can protect mice against lethal challenge with either O1:K2 or O2:K1 K. pneumoniae infection (Clements et al., 2008). In the present study, mice immunized with the magA mutant (K−1 O1) induced the production of antiserum against D-Gal II, and this antiserum had a significant protective efficacy against the PLA-associated O1:K2 strain. In passive immunization studies, D-Gal II-specific antiserum was able to limit the growth and bacterial dissemination of encapsulated O1:K2 PLA-associated K. pneumoniae or acapsulated O19-type E. coli in mouse models of septicemia. Therefore, LPS D-Gal II antigen could be a useful vaccine target conferring broad cross-protection against D-Gal II-containing Gram-negative bacterial pathogens.
In summary, bacteriophage is a powerful screening tool for identification of bacterial surface components. The gene encoding glycosyltransferase WbbY is essential for the production of D-Gal II in K. pneumoniae. Both WbbY and WbbZ homologs are sufficient for the biosynthesis of D-Gal II. LPS D-Gal II is more prevalent in PLA strains than in non-tissue-invasive strains. Besides the Klebsiella O1 strain, the E. coli O19 group and several other species of Gram-negative bacteria also harbor WbbY homologs, WbbZ homologs, and LPS structures based on D-Gal II. Mutation of the gene coding for biosynthesis of the D-Gal II antigen in K. pneumoniae changes serum resistance; reduces bacterial dissemination and colonization into deep organs; and decreases mouse mortality upon infection with K. pneumoniae. D-Gal II is the immunodominant antigen of LPS and immunization of mice against D-Gal II provides protection against infection with an O1:K2 PLA-associated K. pneumoniae strain.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Ministry of Science and Technology, the Excellent Research Projects of National Taiwan University, and the Liver Disease Prevention and Treatment Research Foundation of Taiwan.
Supplementary Material
The Supplementary Material for this article can be found online at: http://www.frontiersin.org/journal/10.3389/fmicb.2014.00608/abstract
Abbreviations
LPS, lipopolysaccharide; D-Gal I, D-galactan I; D-Gal II, D-galactan II; EPS, exopolysaccharide; CPS, capsular polysaccharide; PLA, community-acquired pyogenic liver abscess; LMW, low-molecular-weight; HMW, high-molecular-weight; 5′-RACE, 5′-Rapid Amplification of cDNA Ends; IP, intraperitoneal; PFU, plaque-forming units; CFU, colony-forming units; CI, competitive index.
References
Alberti, S., Hernandez-Alles, S., Gil, J., Reina, J., Martinez-Beltran, J., Camprubi, S., et al. (1993). Development of an enzyme-linked immunosorbent assay method for typing and quantitation of Klebsiella pneumoniae lipopolysaccharide: application to serotype O1. J. Clin. Microbiol. 31, 1379–1381.
Aucken, H. M., Oxley, D., and Wilkinson, S. G. (1993). Structural and serological characterisation of an O-specific polysaccharide from Serratia plymuthica. FEMS Microbiol. Lett. 111, 295–300. doi: 10.1111/j.1574-6968.1993.tb06401.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chuang, Y. P., Fang, C. T., Lai, S. Y., Chang, S. C., and Wang, J. T. (2006). Genetic determinants of capsular serotype K1 of Klebsiella pneumoniae causing primary pyogenic liver abscess. J. Infect. Dis. 193, 645–654. doi: 10.1086/499968
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chun, K. T., Edenberg, H. J., Kelley, M. R., and Goebl, M. G. (1997). Rapid amplification of uncharacterized transposon-tagged DNA sequences from genomic DNA. Yeast 13, 233–240.
Clarke, B. R., and Whitfield, C. (1992). Molecular cloning of the rfb region of Klebsiella pneumoniae serotype O1:K20: the rfb gene cluster is responsible for synthesis of the D-galactan I O polysaccharide. J. Bacteriol. 174, 4614–4621.
Clements, A., Jenney, A. W., Farn, J. L., Brown, L. E., Deliyannis, G., Hartland, E. L., et al. (2008). Targeting subcapsular antigens for prevention of Klebsiella pneumoniae infections. Vaccine 26, 5649–5653. doi: 10.1016/j.vaccine.2008.07.100
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Domenico, P., Schwartz, S., and Cunha, B. A. (1989). Reduction of capsular polysaccharide production in Klebsiella pneumoniae by sodium salicylate. Infect. Immun. 57, 3778–3782.
Fang, C. T., Chuang, Y. P., Shun, C. T., Chang, S. C., and Wang, J. T. (2004). A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J. Exp. Med. 199, 697–705. doi: 10.1084/jem.20030857
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fricke, W. F., Wright, M. S., Lindell, A. H., Harkins, D. M., Baker-Austin, C., Ravel, J., et al. (2008). Insights into the environmental resistance gene pool from the genome sequence of the multidrug-resistant environmental isolate Escherichia coli SMS-3-5. J. Bacteriol. 190, 6779–6794. doi: 10.1128/JB.00661-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hansen, D. S., Mestre, F., Alberti, S., Hernandez-Alles, S., Alvarez, D., Domenech-Sanchez, A., et al. (1999). Klebsiella pneumoniae lipopolysaccharide O typing: revision of prototype strains and O-group distribution among clinical isolates from different sources and countries. J. Clin. Microbiol. 37, 56–62.
Hong, M., and Payne, S. M. (1997). Effect of mutations in Shigella flexneri chromosomal and plasmid-encoded lipopolysaccharide genes on invasion and serum resistance. Mol. Microbiol. 24, 779–791. doi: 10.1046/j.1365-2958.1997.3731744.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hsieh, P. F., Lin, H. H., Lin, T. L., and Wang, J. T. (2010). CadC regulates cad and tdc operons in response to gastrointestinal stresses and enhances intestinal colonization of Klebsiella pneumoniae. J. Infect. Dis. 202, 52–64. doi: 10.1086/653079
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hsieh, P. F., Lin, T. L., Lee, C. Z., Tsai, S. F., and Wang, J. T. (2008). Serum-induced iron-acquisition systems and TonB contribute to virulence in Klebsiella pneumoniae causing primary pyogenic liver abscess. J. Infect. Dis. 197, 1717–1727. doi: 10.1086/588383
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hsieh, P. F., Lin, T. L., Yang, F. L., Wu, M. C., Pan, Y. J., Wu, S. H., et al. (2012). Lipopolysaccharide O1 antigen contributes to the virulence in Klebsiella pneumoniae causing pyogenic liver abscess. PLoS ONE 7:e33155. doi: 10.1371/journal.pone.0033155
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kelly, R. F., Severn, W. B., Richards, J. C., Perry, M. B., Maclean, L. L., Tomas, J. M., et al. (1993). Structural variation in the O-specific polysaccharides of Klebsiella pneumoniae serotype O1 and O8 lipopolysaccharide: evidence for clonal diversity in rfb genes. Mol. Microbiol. 10, 615–625. doi: 10.1111/j.1365-2958.1993.tb00933.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ko, W. C., Paterson, D. L., Sagnimeni, A. J., Hansen, D. S., Von Gottberg, A., Mohapatra, S., et al. (2002). Community-acquired Klebsiella pneumoniae bacteremia: global differences in clinical patterns. Emerg. Infect. Dis. 8, 160–166. doi: 10.3201/eid0802.010025
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Krishnapillai, V. (1971). Uridinediphosphogalactose-4-epimerase deficiency in Salmonella typhimurium and its correction by plasmoid-borne galactose genes of Escherichia coli K-12: effects on mouse virulence, phagocytosis, and serum sensitivity. Infect. Immun. 4, 177–188.
Lehrer, J., Vigeant, K. A., Tatar, L. D., and Valvano, M. A. (2007). Functional characterization and membrane topology of Escherichia coli WecA, a sugar-phosphate transferase initiating the biosynthesis of enterobacterial common antigen and O-antigen lipopolysaccharide. J. Bacteriol. 189, 2618–2628. doi: 10.1128/JB.01905-06
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
McCallum, K. L., Schoenhals, G., Laakso, D., Clarke, B., and Whitfield, C. (1989). A high-molecular-weight fraction of smooth lipopolysaccharide in Klebsiella serotype O1:K20 contains a unique O-antigen epitope and determines resistance to nonspecific serum killing. Infect. Immun. 57, 3816–3822.
Merino, S., Camprubi, S., Alberti, S., Benedi, V. J., and Tomas, J. M. (1992). Mechanisms of Klebsiella pneumoniae resistance to complement-mediated killing. Infect. Immun. 60, 2529–2535.
Mesnage, S., Fontaine, T., Mignot, T., Delepierre, M., Mock, M., and Fouet, A. (2000). Bacterial SLH domain proteins are non-covalently anchored to the cell surface via a conserved mechanism involving wall polysaccharide pyruvylation. EMBO J. 19, 4473–4484. doi: 10.1093/emboj/19.17.4473
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Oxley, D., and Wilkinson, S. G. (1989). Structures of neutral glycans isolated from the lipopolysaccharides of reference strains for Serratia marcescens serogroups O16 and O20. Carbohydr. Res. 193, 241–248. doi: 10.1016/0008-6215(89)85122-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Pieroni, P., Rennie, R. P., Ziola, B., and Deneer, H. G. (1994). The use of bacteriophages to differentiate serologically cross-reactive isolates of Klebsiella pneumoniae. J. Med. Microbiol. 41, 423–429. doi: 10.1099/00222615-41-6-423
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Podschun, R., and Ullmann, U. (1998). Klebsiella spp. as nosocomial pathogens: epidemiology, taxonomy, typing methods, and pathogenicity factors. Clin. Microbiol. Rev. 11, 589–603.
Ramphal, R., and Ambrose, P. G. (2006). Extended-spectrum beta-lactamases and clinical outcomes: current data. Clin. Infect. Dis. 42(Suppl. 4), S164–S172. doi: 10.1086/500663
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Reeves, P. R., Hobbs, M., Valvano, M. A., Skurnik, M., Whitfield, C., Coplin, D., et al. (1996). Bacterial polysaccharide synthesis and gene nomenclature. Trends Microbiol. 4, 495–503. doi: 10.1016/S0966-842X(97)82912-5
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Richards, J. C., and Leitch, R. A. (1989). Elucidation of the structure of the Pasteurella haemolytica serotype T10 lipopolysaccharide O-antigen by n.m.r. spectroscopy. Carbohydr. Res. 186, 275–286. doi: 10.1016/0008-6215(89)84041-8
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schneider, H., Griffiss, J. M., Williams, G. D., and Pier, G. B. (1982). Immunological basis of serum resistance of Neisseria gonorrhoeae. J. Gen. Microbiol. 128, 13–22.
Simon, R., Priefer, U., and Puhler, A. (1983). A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Nat. Biotechnol. 1, 784–791. doi: 10.1038/nbt1183-784
Taylor, P. W. (1976). Immunochemical investigations on lipopolysaccharides and acidic polysaccharides from serum-sensitive and serum-resistant strains of Escherichia coli isolated from urinary-tract infections. J. Med. Microbiol. 9, 405–421. doi: 10.1099/00222615-9-4-405
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tomas, J. M., Benedi, V. J., Ciurana, B., and Jofre, J. (1986). Role of capsule and O antigen in resistance of Klebsiella pneumoniae to serum bactericidal activity. Infect. Immun. 54, 85–89.
Tsai, C. M., and Frasch, C. E. (1982). A sensitive silver stain for detecting lipopolysaccharides in polyacrylamide gels. Anal. Biochem. 119, 115–119. doi: 10.1016/0003-2697(82)90673-X
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Tsai, F. C., Huang, Y. T., Chang, L. Y., and Wang, J. T. (2008). Pyogenic liver abscess as endemic disease, Taiwan. Emerg. Infect. Dis. 14, 1592–1600. doi: 10.3201/eid1410.071254
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Whitfield, C., Perry, M. B., Maclean, L. L., and Yu, S. H. (1992). Structural analysis of the O-antigen side chain polysaccharides in the lipopolysaccharides of Klebsiella serotypes O2(2a), O2(2a,2b), and O2(2a,2c). J. Bacteriol. 174, 4913–4919.
Whitfield, C., Richards, J. C., Perry, M. B., Clarke, B. R., and Maclean, L. L. (1991). Expression of two structurally distinct D-galactan O antigens in the lipopolysaccharide of Klebsiella pneumoniae serotype O1. J. Bacteriol. 173, 1420–1431.
Wu, M. C., Lin, T. L., Hsieh, P. F., Yang, H. C., and Wang, J. T. (2011). Isolation of genes involved in biofilm formation of a Klebsiella pneumoniae strain causing pyogenic liver abscess. PLoS ONE 6:e23500. doi: 10.1371/journal.pone.0023500
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: D-galactan II, Klebsiella pneumoniae, lipopolysaccharide, immunodominant antigen, vaccine
Citation: Hsieh P-F, Wu M-C, Yang F-L, Chen C-T, Lou T-C, Chen Y-Y, Wu S-H, Sheu J-C and Wang J-T (2014) D-galactan II is an immunodominant antigen in O1 lipopolysaccharide and affects virulence in Klebsiella pneumoniae: implication in vaccine design. Front. Microbiol. 5:608. doi: 10.3389/fmicb.2014.00608
Received: 17 September 2014; Paper pending published: 12 October 2014;
Accepted: 24 October 2014; Published online: 19 November 2014.
Edited by:
Evangelos Giamarellos-Bourboulis, University of Athens Medical School, GreeceReviewed by:
Magdalena Chirilă, Iuliu Haţieganu University of Medicine and Pharmacy, RomaniaThomas Tsaganos, Athens Medical School, Greece
Copyright © 2014 Hsieh, Wu, Yang, Chen, Lou, Chen, Wu, Sheu and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jin-Town Wang, Department of Microbiology, National Taiwan University College of Medicine, 1, Sec. 1, Jen-Ai Rd., Taipei 100, Taiwan e-mail:d2FuZ2p0QG50dS5lZHUudHc=
 Pei-Fang Hsieh1
Pei-Fang Hsieh1 Feng-Ling Yang
Feng-Ling Yang Jin-Town Wang
Jin-Town Wang