- 1Department of Pharmacology and Pharmaceutical Sciences, School of Pharmacy, University of Southern California, Los Angeles, CA, USA
- 2Department of Chemistry, College of Letters, Arts, and Sciences, University of Southern California, Los Angeles, CA, USA
Filamentous fungi are rich resources of secondary metabolites (SMs) with a variety of interesting biological activities. Recent advances in genome sequencing and techniques in genetic manipulation have enabled researchers to study the biosynthetic genes of these SMs. Aspergillus terreus is the well-known producer of lovastatin, a cholesterol-lowering drug. This fungus also produces other SMs, including acetylaranotin, butyrolactones, and territram, with interesting bioactivities. This review will cover recent progress in genome mining of SMs identified in this fungus. The identification and characterization of the gene cluster for these SMs, as well as the proposed biosynthetic pathways, will be discussed in depth.
Introduction
Filamentous fungi, such as species found within the genus Aspergillus, are known to produce a wide variety of natural products displaying a broad spectrum of biological activities. Genome sequencing of members in the genus Aspergillus revealed that the number of secondary metabolite (SM) genes or gene clusters greatly exceeds the number of SMs identified so far. This suggests that more types of SMs still remain to be discovered. Efficient and high-throughput genome sequencing techniques have now greatly facilitated the genome mining of SMs identified in Aspergillus species. More and more studies have shown that most fungal SM genes are clustered, often separated from each other by less than 2 kb within the chromosome (Keller et al., 2005). Striking examples of fungal SM clusters include those responsible for the biosynthesis of fumonisin (Proctor et al., 2003) and sterigmatocystin (Brown et al., 1996). The clustering of these SM genes is fortuitous for researchers since upon discovering one responsible gene within a cluster, other genes that are within close proximity of the identified genes may be involved in the biosynthesis of the same natural product.
Analysis of the fungal genome makes it feasible to characterize these SM gene clusters via genetic manipulations. Verification of function of the relevant genes can be achieved via several approaches. These approaches include targeted deletion or over-expression of the relevant genes in the native organism, heterologous expression in an alternative host, or in vitro biochemical characterization. Recent advances in the genome editing of Aspergillus nidulans have greatly expedited the SM genome mining of this fungal species (Sanchez et al., 2012). One advance is the development of a fusion PCR technique that allows quick synthesis of linear PCR fragments that are used in the transformation of filamentous fungi (Yu et al., 2004; Szewczyk et al., 2006). Another advance concerns the establishment of an efficient gene targeting system in the fungus A. nidulans. This is accomplished via targeted deletion of the A. nidulans homolog (nkuA) of the human KU70 gene which is essential for non-homologous end joining of DNA in DNA double-strand breaks (Nayak et al., 2006; Szewczyk et al., 2006). Later, this ku70 deletion toolbox was expanded to other Aspergillus species and boosted the gene targeting efficiency in these species (Kuck and Hoff, 2010).
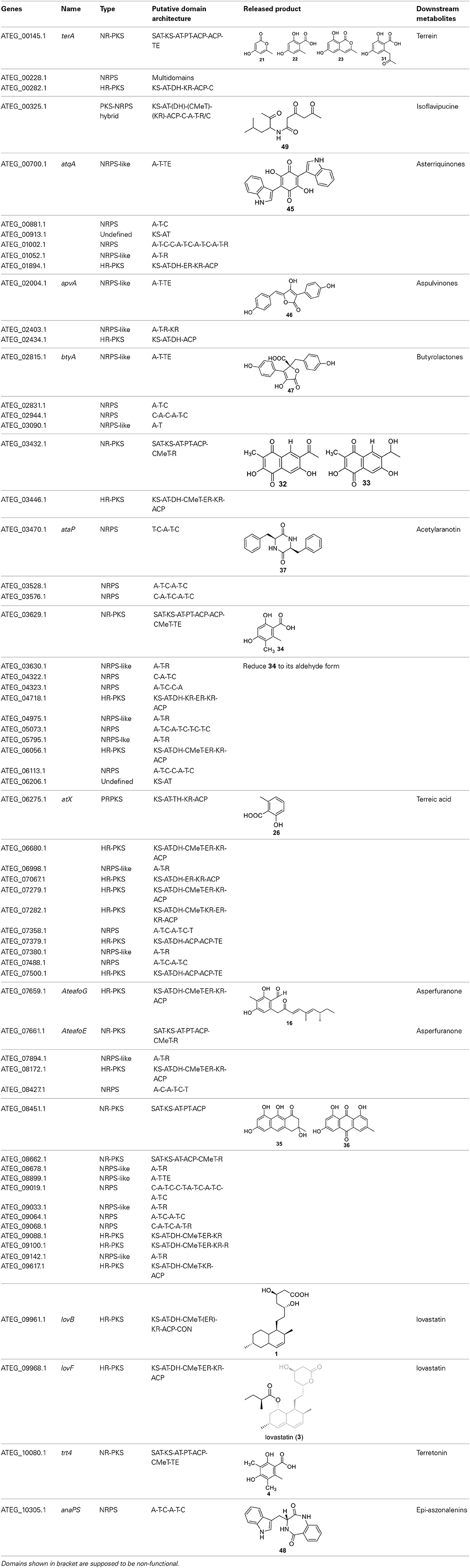
The fungus Aspergillus terreus is known to produce lovastatin, which became the first cholesterol-lowering drug of its class approved for human use in the United States. Besides lovastatin, A. terreus produces a number of biologically relevant compounds such as sulochrin, terretonin, asterriquinone, and butyrolactone. The strain A. terreus NIH 2624, a patient isolate, was sequenced by the Broad Institute as part of the Broad Fungal Genome Initiative. A 10 × coverage genome sequence has been completed, and the data are publicly available through the Broad Institute website. Analysis by Secondary Metabolite Unique Regions Finder (SMURF) showed that A. terreus NIH2624 contains 28 polyketide synthase (PKS) genes, 22 non-ribosomal peptide synthetase (NRPS) genes, one hybrid PKS/NRPS gene, two PKS-like genes, and 15 NRPS-like genes (Khaldi et al., 2010). This review will cover the most current knowledge of the genome mining of SMs produced by A. terreus. The review consists of three major sections: genome mining of PKS-derived natural products, genome mining of NRPS-derived natural products, and genome mining of hybrid PKS-NRPS derived natural products by A. terreus.
Genome Mining of PKS-Derived Natural Products in A. terreus
Biosynthesis of Lovastatin in A. terreus
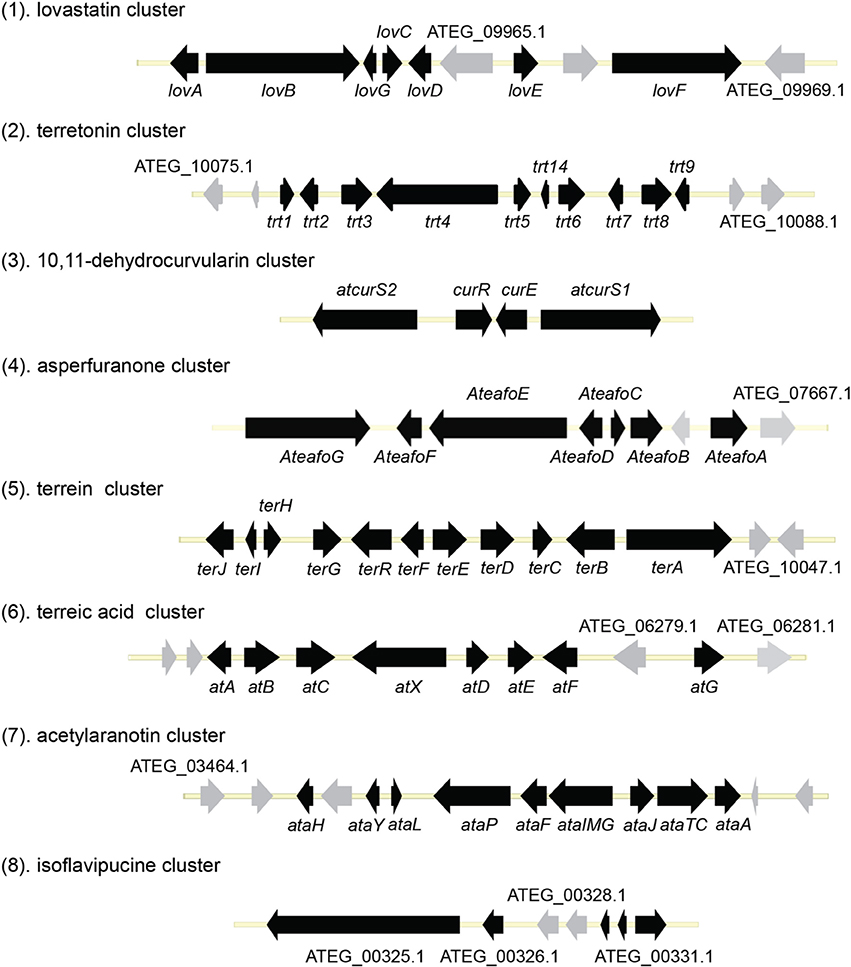
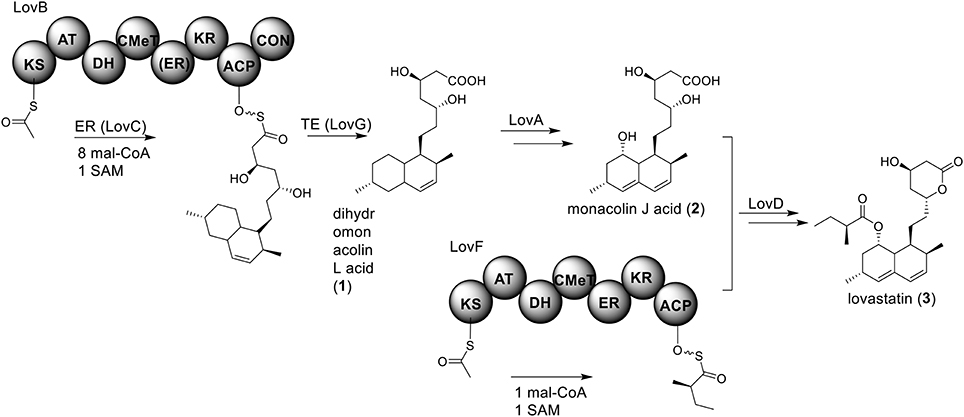
Perhaps the most well-known molecule produced by A. terreus is lovastatin. Lovastatin is an inhibitor of the enzyme (3S)-hydroxymethylglutaryl-coenzyme A (HMG-CoA) reductase which reduces HMG-CoA to mevalonate. The biosynthesis of lovastain has been extensively reviewed in literature reports (Hill, 2006; Cox, 2007; Campbell and Vederas, 2010; Chiang et al., 2010; Chooi and Tang, 2012). Two highly reducing PKSs (HR-PKSs), LovB (lovastatin nonaketide synthase) and LovF, play critical roles in the formation of the lovastatin core structure (Figure 1). In the pathway, the HR-PKS LovB, together with the trans-acting enoyl reductase (ER) protein LovC, are responsible for the production of dihydromonacolin L acid (1). The release of 1 from LovB is catalyzed by a thioesterase LovG (Xu et al., 2013a). The oxidative conversion of 1 to monacolin J acid (2) involves LovA. The other HR-PKS LovF synthesizes the 2-methylbutyrate moiety of lovastatin (3) and the covalent attachment of this moiety to 2 is catalyzed by a transesterase, LovD (Figure 2). Recent efforts in engineering LovD using a directed evolution strategy generated a mutant LovD containing 29 point mutations that greatly improves (~1000 fold) the protein's efficiency in the synthesis of the drug simvastatin (Jimenez-Oses et al., 2014), representing an excellent example of how the combination of synthetic biology and protein chemistry may facilitate drug discovery and production.
Biosynthesis of Terretonin in A. terreus
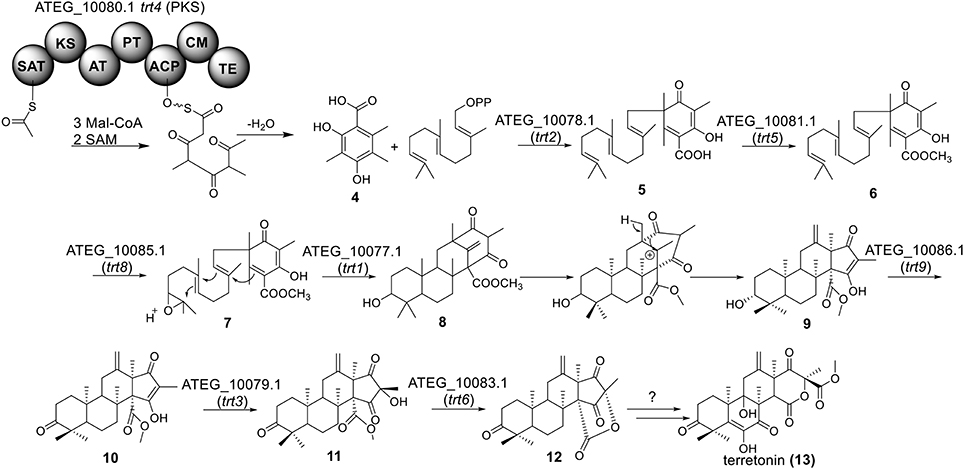
Terretonin is one type of meroterpenoids with mixed origins (Figure 3). Previous studies by Simpson and Vederas in the 1980's using isotopic labeled precursors have shown that terretonin originates from both polyketide and terpenoid pathways (McIntyre et al., 1982, 1989). Recent genome mining studies revealed that the terretonin cluster contains a total of 10 genes (Guo et al., 2012). The cluster includes a non-reducing PKS (NR-PKS) gene, trt4 (ATEG_10080.1), and a prenyltransferase gene, trt2 (Figure 1). Trt4 is responsible for the biosynthesis of 3,5-dimethylorsellinic acid (4) and Trt2 catalyzes the alkylation of compound 4 to give 5 (Figures 1, 3) (Guo et al., 2012; Itoh et al., 2012). The following methylation (5 to 6), epoxidation (6 to 7) and cyclization (7 to 8) are proposed to be catalyzed by proteins encoded by trt5, trt8, and trt1, respectively (Guo et al., 2012; Matsuda et al., 2012). Interestingly, the Abe group discovered that the methylation step catalyzed by Trt5 is an essential step for cyclization (Matsuda et al., 2012). Upon formation of the tetracyclic precursor, Trt9 is proposed to oxidize compound 9 to 10 and Trt3 hydroxylases 10 to give 11 (Figure 3). Next, the putative protein Trt6 catalyzes the intra-lactonization reaction to give 12, while the conversion of 12 to terretonin (13) might involve other enzymes that are not identified by Guo et al. (2012).
Biosynthesis of 10,11-Dehydrocurvularin in A. terreus
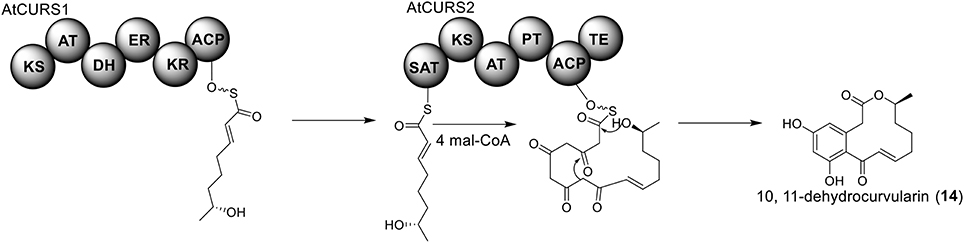
The polyketide origin of the compound 10,11-dehydrocurvularin (14) was established by incorporation of stable isotope labeled precursors (Arai et al., 1989). The biosynthetic cluster for 10,11-dehydrocurvularin was identified in A. terreus AH-02-30-F7. The cluster is proposed to contain four genes, an HR-PKS gene (Atcurs1), an NR-PKS gene (Atcurs2), a GAL4-like transcription regulatory gene (AtcursR) and a major facilitator superfamily gene (AtcursE) (Figure 1) (Xu et al., 2013b). The biosynthesis of 10,11-dehydrocurvularin in A. terreus was characterized by heterologous expressing the HR-PKS and NR-PKS pair in Saccharomyces cerevisiae (Xu et al., 2013b). In the pathway, the HR-PKS AtCURS1 is proposed to synthesize the tetraketide 7(S)-hydroxyotc-2(E)-enoic acid (Figure 4). This tetraketide is then loaded onto the starter unit ACP transacylase domain (SAT) of AtCURS2, followed by four chain extension cycles to release the final product 14. In the cluster, the gene AtcursR is predicted to encode for a fungal transcription regulator and AtcursE may code for an exporter involved in the transporting of 10,11-dehydrocurvularin (14) (Xu et al., 2013b). Further investigation of the product template (PT) domain of AtCURS2, which catalyzes the ring cyclization and aromatization, revealed that the cyclization mode of this domain could be reshaped from the bacterial folding mode, as shown in the biosynthesis of compound 14, to the fungal mode by three selected point mutations (Xu et al., 2013c). Such rational control of fungal polyketide ring cyclization should facilitate the engineering of natural products with novel chemical scaffolds.
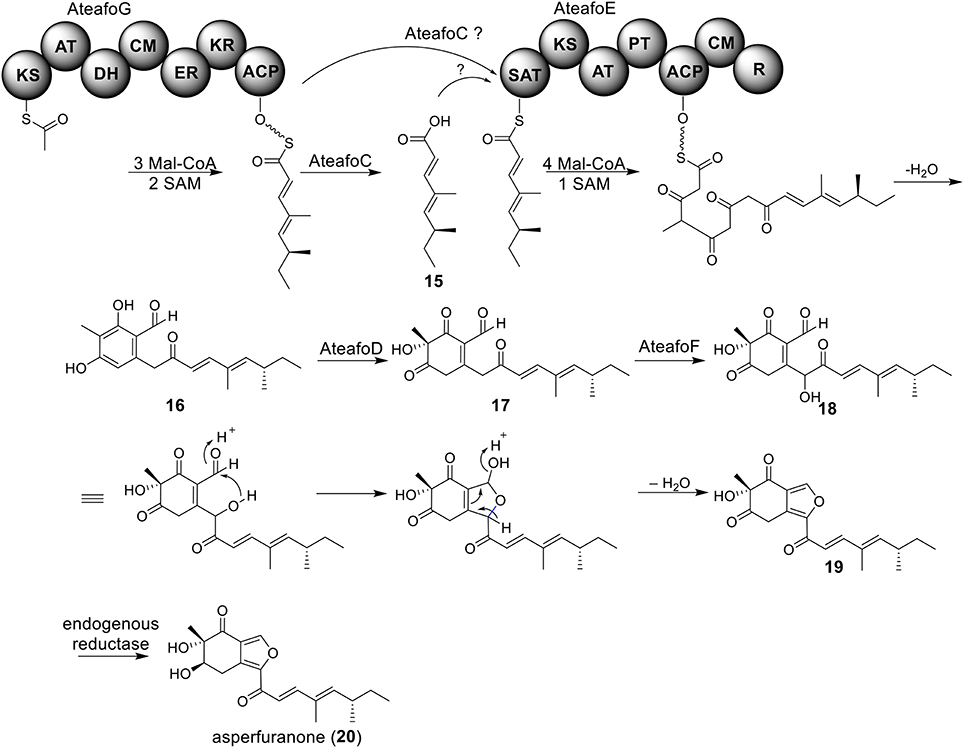
Biosynthesis of Asperfuranone in A. terreus
The azaphilone asperfuranone (20) was first isolated from A. nidulans after activating two PKSs encoded by afoE and afoG (Chiang et al., 2009). Later a highly homologous cluster was identified in the A. terreus genome (Figure 1) (Chiang et al., 2013). In this case, the individual genes from A. terreus were heterologously expressed in A. nidulans and the whole asperfuranone pathway was reconstituted. The asperfuranone cluster in A. terreus contains a total of seven genes including two PKS genes ateafoE and ateafoG (Figure 1). In the pathway, the gene AteafoG encodes the HR-PKS that is responsible for the production of the PK intermediate 15. The enzyme AteafoC might facilitate the transfer of the side chain 15 from AteafoG to AteafoE. Next, AteafoD catalyzes the dearomatization of the precursor 16 to give intermediate 17, followed by hydroxylation by AteafoF to 18. Last, the authors proposed the existence of an endogenous reductase in A. nidulans that catalyzes the conversion of 19 to asperfuranone (20) (Figure 5).
Biosynthesis of Terrein in A. terreus
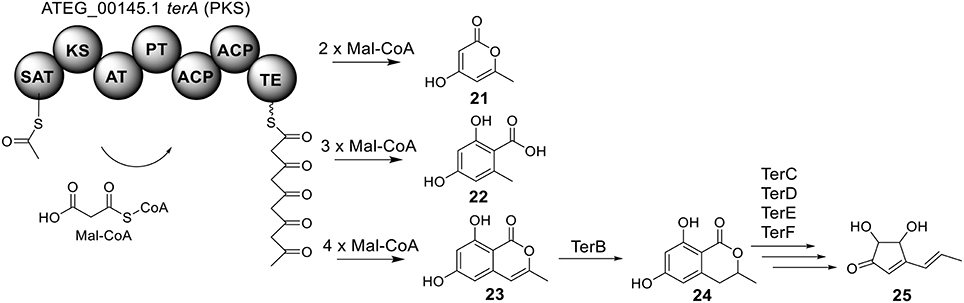
The terrein (25) cluster was serendipitously identified while the Brock group were initially looking for secondary metabolites that are involved in the biosynthesis of conidia pigment in A. terreus (Zaehle et al., 2014). Previous isotope-labeling studies revealed that the biosynthesis of terrein originates from a polyketide-based pathway (Birch et al., 1965) and involves the contraction of a six-membered ring precursor to give the five-membered ring terrein (25) (Hill et al., 1981). The terrein cluster contains a total of 11 genes, including an NR-PKS gene terA (ATEG_00145.1) and a transcription factor gene, terR (Figure 1). In the terrein biosynthetic pathway as shown in Figure 6, the NR-PKS encoded by terA condensates one acetyl-CoA and four malonyl-CoA to synthesize the precursor 2,3-dehydro-6-hydroxymellein (23). Heterologous expression of terA in A. niger revealed that the NR-PKS TerA is also capable of producing compounds 21 and 22 by incorporating different number of malonyl-CoAs as substrates. The reduction of compound 23 to 6-hydroxymellein (6-HM) (24) is probably catalyzed by TerB, but it is also possible that other unspecific ketoreductases may perform this reduction. The authors proposed that the conversion of 6-HM (24) to the final product terrein (25) might involve four genes, terC, terD, terE, and terF while the specific roles of these genes await further verification. Interestingly, the study also revealed that terrein (25) has phytotoxic activity. In the two experiments, terrein (25), rather than the intermediates or shunt products, inhibited root elongation and caused lesions on fruit surface (Zaehle et al., 2014), representing a prime example of how fungal secondary metabolites might play a role in fungus-plant interactions.
Biosynthesis of Terreic Acid in A. terreus
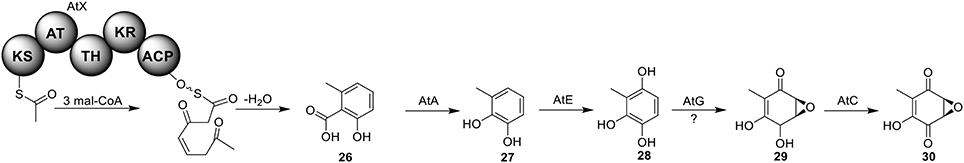
Terreic acid (30) is a natural product isolated from A. terreus with anti-bacterial activity (Yamamoto et al., 1980). A later study recognized its inhibitory effect against Bruton's tyrosine kinase (Btk), and this compound has been used as a chemical probe to examine the function of Btk (Kawakami et al., 1999). Previous research using isotope-labeled precursors has shown that the biosynthesis of terreic acid originated from one polyketide, 6-methylsalicylic acid (6-MSA, 26) (Read and Vining, 1968; Read et al., 1969). The PKS gene atX was cloned and identified as a 6-MSA (26) synthase (Fujii et al., 1996). A recent bioinformatic analysis study predicts that the PKS gene atX and its surrounding genes encodes the biosynthetic cluster which is responsible for terreic acid biosynthesis (Boruta and Bizukojc, 2014). The cluster was experimentally characterized using a targeted gene deletion method (Guo et al., 2014). The cluster contains a total of eight genes, including a partially reducing PKS (PRPKS) gene, atX, and a transcription factor gene atF (Figure 1). The pathway starts with the formation of 6-MSA (26) by AtX using one acetyl-CoA and three malonyl-CoA units as its substrates, followed by a decarboxylative hydroxylation catalyzed by AtA to give 27. Compound 27 could be degraded to a shunt product, (2Z,4E)-2-methyl-2,4-hexadienedioic acid. The degradation of 27 may be catalyzed by a putative catechol 1,2-dioxygenase in A. terreus, but the coding gene for this enzyme is not identified in the terreic acid cluster (Guo et al., 2014). The hydroxylation of 27 to give 28 is possibly catalyzed by AtE. The oxidative conversion of 28 to 29 might involve AtG, but this proposal is not confirmed in the study. Last, AtC catalyzes the oxidation of compound 29 to give the final product terreic acid (30) (Figure 7).
Identification of the Products of Six NR-PKS Genes in A. terreus
The previous studies successfully linked several PKS-derived SMs with their biosynthetic clusters in A. terreus. Using a heterologous expression strategy, researchers also identified the products of six NR-PKS genes in A. terreus (Chiang et al., 2013). The products of these NR-PKSs are shown in Table 1. The NR-PKS encoded by ATEG_00145.1 synthesizes a mixture of compounds, 21, 22, 23, and 31. The plasticity of ATEG_00145.1 is also demonstrated in the study of terrein biosynthesis (Zaehle et al., 2014). The products of the NR-PKS, encoded by ATEG_03432.1, are compounds 32 and 33, and the product of the ATEG_03629.1 encoded NR-PKS is 34. The NR-PKS atrochrysone carboxylic acid synthase (ACAS) encoded by ATEG_08451.1 produces atrochrysone 35 and its derivative emodin 36 with the aid of a β-lactamase encoded by ATEG_08450.1. This result matches the previous finding reported by Awakawa et al. (2009) As expected, ATEG_10080.1 (trt4) synthesizes and releases the PKS product 3,5-dimethylorsellinic acid (4), which is presumed to be the first precursor in the biosynthesis of terretonin (Figure 3) (Guo et al., 2012; Itoh et al., 2012). As aforementioned, the products of the NR-PKS AteafoE and its adjacent HR-PKS AteafoG incorporate into the asperfuranone pathway as shown in Figure 5 (Table 1).
Biosynthesis of NRPS-Derived Natural Products in A. terreus
Biosynthesis of Acetylaranotin in A. terreus
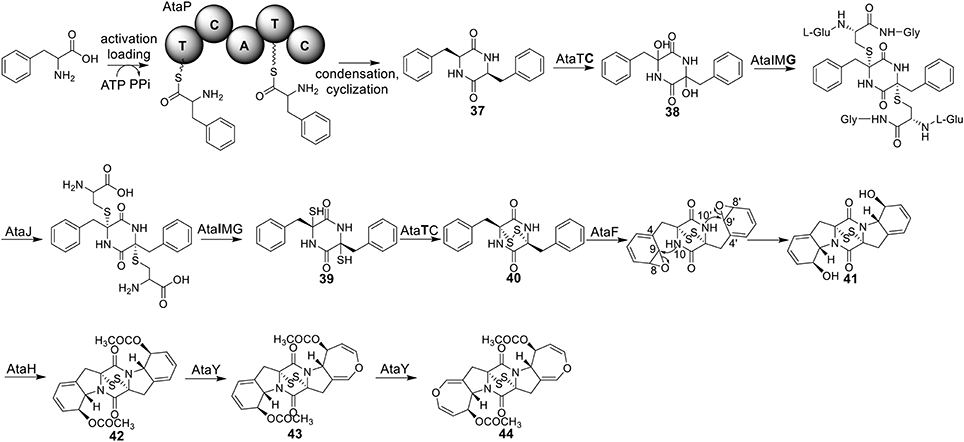
Acetylaranotin belongs to one group of SM toxins named epipolythiodioxopiperizine (ETP). This type of natural products is usually featured by the presence of unique di- or poly-sulfide bridges. The di- or poly-sulfide bridges are presumed to mediate the molecule's cytotoxicity, either by cross-linking vital proteins via cysteine bonds, or by generating reactive oxygen species via redox cycling (Scharf et al., 2011a). An early study using stable isotope-labeled precursors showed that the diketopiperazine (DKP) precursor, compound 37, could be incorporated intact into acetylaranotin (44) (Boente et al., 1981). This indicates that the acetylaranotin pathway may include an NRPS that catalyzes the condensation of two L-phenylalanines to give 37. Bioinformatic analysis of the ETP clusters dispersed in filamentous ascomycetes uncovered two putative clusters in A. terreus that might encode the biosynthetic pathway for acetylaranotin (Patron et al., 2007).
Using the gene targeted deletion strategy, one of the two clusters, named ata cluster, was found to be responsible for acetylaranotin production in A. terreus (Guo et al., 2013b). The ata cluster contains a total of nine genes, including one NRPS gene, ataP (Figure 1). In the pathway, the NRPS AtaP catalyzes the condensation of two phenylalanines to give compound 37. Next, the C domain of the protein AtaTC is proposed to catalyze the dual hydroxylation of compound 37 to 38. A series of studies have fully elucidated the functions of the enzymes that are involved in the installation of the disulfide moiety in gliotoxin (Scharf et al., 2010, 2011b, 2012, 2013; Schrettl et al., 2010; Davis et al., 2011; Gallagher et al., 2012). Similar to their homologs in the gliotoxin cluster, AtaG, AtaJ, AtaI are involved in the conversion of the dual hydroxyl groups in 38 to thiol groups in 39, followed by the AtaT-mediated oxidation to form the transannular disulfide bridge as shown in intermediate 40. Next, AtaF catalyzes the dual epoxidation of 40 followed by the spontaneous nucleophilic attack of the amide nitrogens to yield intermediate 41. The acetylation of 41 to 42 is catalyzed by AtaH, and the oxepine ring formation involves the protein AtaY to yield acetylaranotin 44 (Figure 8). The proteins AtaH and AtaY can function independently, but the detailed catalytic mechanism of AtaY still awaits further investigation. Two recent studies unveiled that the S-alkylation of gliotoxin was catalyzed by a SAM dependent methyltransferase encoded by a gene tmtA/gtmA outside the gliotoxin cluster (Dolan et al., 2014; Scharf et al., 2014). These studies suggest that the S-methylation of acetylaranotin to give bisdethiobis(methylthio)acetylaranotin is likely to be catalyzed by a methyltransferase encoded by a gene outside the acetylaranotin cluster.
Identification of the Products of Four NRPS or NRPS-Like Genes in A. terreus
Screening of the genome-sequenced strain A. terreus NIH 2624 showed that this fungus is able to biosynthesize four main types of secondary metabolites including aspulvinones, butyrolactones, asterriquinones, and aszonalenins (Guo et al., 2013a). The NRPS-origin of these four types of natural products was revealed in previous studies (Kiriyama et al., 1977; Nitta et al., 1983; Balibar et al., 2007; Yin et al., 2009). To link these molecules to their core synthesis NRPS genes, a mutant library was created by individually deleting a total of 21 NRPS or NRPS-like genes in A. terreus. Screening of the mutants' SM profiles, in comparison with that of the wild type, showed that three NRPS-like genes atqA (ATEG_00700.1), apvA (ATEG_02004.1), and btyA (ATEG_02815.1), are responsible for the synthesis of the core structures of asterriquinones, aspulvinones, and butyrolactones, respectively (Table 1) (Balibar et al., 2007; Guo et al., 2013a). The NRPS gene anaPS encoded by ATEG_10305.1 is proposed to biosynthesize the (R)-benzodiazepinedione core (Table 1) (Yin et al., 2009; Guo et al., 2013a). Serendipitously, deletion of one NRPS-like gene atmelA (ATEG_03563.1) generates an albino mutant that abolishes the brown conidia melanin in A. terreus. It is possible that AtmelA synthesizes the first precursor that incorporates into the conidia pigment produced by A. terreus. Since all the identified conidial melanins in other Aspergillus species are derived from a PKS pathway, another possibility is that this NRPS-like gene might be involved, but not directly related in the melanin biosynthesis of A. terreus (Guo et al., 2013a).
An NRPS-Like Gene is Involved in Activating and Reducing of an Aryl Acid Released from an NR-PKS
Zhao group recently reported that an NRPS-like protein encoded by ATEG_03630.1 could catalyze the reduction of compound 34 to an aryl aldehyde (Wang et al., 2014). Compound 34 is released by its adjacent NR-PKS ATEG_03629.1, The putative domain architecture of the NRPS-like protein is A-T-R. The aryl acid product 34, upon its release from the TE domain of the NRPKS, is then loaded onto the A domain. And the R domain is responsible for the reduction of the aryl acid 34 to its aldehyde form (Wang et al., 2014). Interestingly, a subsequent study revealed that the substrate of the A domain of this NRPS-like enzyme could be engineered to anthranilate via bioinformatic analysis and mutagenesis (Wang and Zhao, 2014).
Biosynthesis of Hybrid PKS-NRPS Natural Products in A. terreus
Biosynthesis of Isoflavipucine in A. terreus
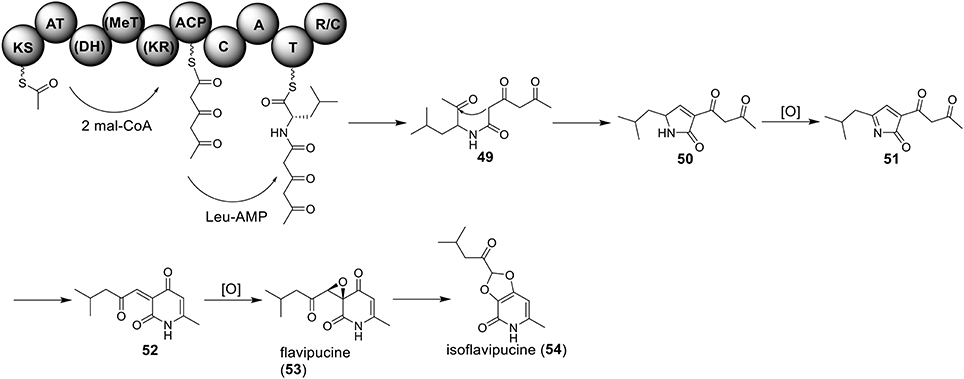
Only one hybrid PKS-NRPS gene, ATEG_00325.1, can be identified in the genome of A. terreus NIH 2624. The Brock group identified that the PKS-NRPS encoded by this hybrid gene is responsible for the core synthesis of isoflavipucine and dihydroisoflavipucine (Gressler et al., 2011). RT–PCR analysis of expression of the hybrid gene and its surrounding genes, under isoflavipucine producing condition, suggested that the cluster for isoflavipucine biosynthesis might contain five genes, ATEG_00325.1, ATEG_00326.1 (transcription factor gene), ATEG_00329.1, ATEG_00330.1, and ATEG_00331.1 (Figure 1) (Gressler et al., 2011). In the pathway, the hybrid PKS-NRS synthesizes and releases the linear precursor (49). The conversion of the precursor 49 to final product includes heterocyclization (49 to 50), oxidation (50 to 51), transamination and ring arrangement (51 to 52), and epoxidation to give flavipucine (53) (Figure 9). However, the enzymes that are responsible for the conversion of the linear precursor 45 to isoflavipucine (54) are not identified in this study and needs further investigation.
Conclusion
In the fungus A. terreus, most of the genome mining experiments are carried out in the strain NIH 2624 because it is the only A. terreus strain in which the genome sequence is publicly available. A combination of bioinformatic analysis and experimental verification have enabled researchers to elucidate the functions of genes and proteins that are involved in the biosynthesis of several SMs produced in A. terreus. However, as shown in Table 1, there are still many other SM genes or gene clusters in A. terreus whose products still remain elusive. With the advance of genome sequencing and manipulation techniques, an increasing amount of effort will focus on deciphering the products of these silent or cryptic genes or gene clusters, as well as engineering the characterized pathways to produce second generation molecules with improved bioactivities.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Chun-Jun Guo was supported by the National Science Foundation—Emerging Frontiers in Research and Innovation-MIKS (Grant 1136903). Research conducted in the lab of Clay C. C. Wang is supported by the National Institute of General Medical Sciences GM084077, the National Science Foundation—Emerging Frontiers in Research and Innovation-MIKS (Grant 1136903), the Department of Energy, Bioenergy Technologies office, the Department of Defense Strategic Environmental Research and Development program (SERDP), and the National Aeronautics and Space Administration Space Biology program.
References
Arai, K., Rawlings, B. J., Yoshizawa, Y., and Vederas, J. C. (1989). Biosyntheses of antibiotic A26771B by Penicillium turbatum and dehydrocurvularin by Alternaria cinerariae: comparison of stereochemistry of polyketide and fatty acid enoyl thiol ester reductases. J. Am. Chem. Soc. 111, 3391–3399. doi: 10.1021/ja00191a042
Awakawa, T., Yokota, K., Funa, N., Doi, F., Mori, N., Watanabe, H., et al. (2009). Physically discrete beta-lactamase-type thioesterase catalyzes product release in atrochrysone synthesis by iterative type I polyketide synthase. Chem. Biol. 16, 613–623. doi: 10.1016/j.chembiol.2009.04.004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Balibar, C. J., Howard-Jones, A. R., and Walsh, C. T. (2007). Terrequinone A biosynthesis through L-tryptophan oxidation, dimerization and bisprenylation. Nat. Chem. Biol. 3, 584–592. doi: 10.1038/nchembio.2007.20
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Birch, A. J., Cassera, A., and Jones, A. R. (1965). The biosynthesis of terrain. Chem. Commun. 167–168. doi: 10.1039/c19650000167
Boente, M. I. P., Kirby, G. W., and Robins, D. J. (1981). Biosynthetic incorporation of cyclo-(L-phenylalanyl-L-phenylalanyl) into bisdethiobis(methylthio)acetylaranotin in Aspergillus terreus. Chem. Commun. 619. doi: 10.1039/c39810000619
Boruta, T., and Bizukojc, M. (2014). Culture-based and sequence-based insights into biosynthesis of secondary metabolites by Aspergillus terreus ATCC 20542. J. Biotechnol. 175, 53–62. doi: 10.1016/j.jbiotec.2014.01.038
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brown, D. W., Yu, J. H., Kelkar, H. S., Fernandes, M., Nesbitt, T. C., Keller, N. P., et al. (1996). Twenty-five coregulated transcripts define a sterigmatocystin gene cluster in Aspergillus nidulans. Proc. Natl. Acad. Sci. U.S.A. 93, 1418–1422. doi: 10.1073/pnas.93.4.1418
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Campbell, C. D., and Vederas, J. C. (2010). Biosynthesis of lovastatin and related metabolites formed by fungal iterative PKS enzymes. Biopolymers 93, 755–763. doi: 10.1002/bip.21428
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chiang, Y. M., Oakley, B. R., Keller, N. P., and Wang, C. C. (2010). Unraveling polyketide synthesis in members of the genus Aspergillus. Appl. Microbiol. Biotechnol. 86, 1719–1736. doi: 10.1007/s00253-010-2525-3
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chiang, Y. M., Oakley, C. E., Ahuja, M., Entwistle, R., Schultz, A., Chang, S. L., et al. (2013). An efficient system for heterologous expression of secondary metabolite genes in Aspergillus nidulans. J. Am. Chem. Soc. 135, 7720–7731. doi: 10.1021/ja401945a
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chiang, Y. M., Szewczyk, E., Davidson, A. D., Keller, N., Oakley, B. R., and Wang, C. C. (2009). A gene cluster containing two fungal polyketide synthases encodes the biosynthetic pathway for a polyketide, asperfuranone, in Aspergillus nidulans. J. Am. Chem. Soc. 131, 2965–2970. doi: 10.1021/ja8088185
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Chooi, Y. H., and Tang, Y. (2012). Navigating the fungal polyketide chemical space: from genes to molecules. J. Org. Chem. 77, 9933–9953. doi: 10.1021/jo301592k
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cox, R. J. (2007). Polyketides, proteins and genes in fungi: programmed nano-machines begin to reveal their secrets. Org. Biomol. Chem. 5, 2010–2026. doi: 10.1039/b704420h
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Davis, C., Carberry, S., Schrettl, M., Singh, I., Stephens, J. C., Barry, S. M., et al. (2011). The role of glutathione S-transferase GliG in gliotoxin biosynthesis in Aspergillus fumigatus. Chem. Biol. 18, 542–552. doi: 10.1016/j.chembiol.2010.12.022
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Dolan, S. K., Owens, R. A., O'keeffe, G., Hammel, S., Fitzpatrick, D. A., Jones, G. W., et al. (2014). Regulation of nonribosomal peptide synthesis: bis-thiomethylation attenuates gliotoxin biosynthesis in Aspergillus fumigatus. Chem. Biol. 21, 999–1012. doi: 10.1016/j.chembiol.2014.07.006
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Fujii, I., Ono, Y., Tada, H., Gomi, K., Ebizuka, Y., and Sankawa, U. (1996). Cloning of the polyketide synthase gene atX from Aspergillus terreus and its identification as the 6-methylsalicylic acid synthase gene by heterologous expression. Mol. Gen. Genet. 253, 1–10. doi: 10.1007/s004380050289
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gallagher, L., Owens, R. A., Dolan, S. K., O'keeffe, G., Schrettl, M., Kavanagh, K., et al. (2012). The Aspergillus fumigatus protein GliK protects against oxidative stress and is essential for gliotoxin biosynthesis. Eukaryot. Cell 11, 1226–1238. doi: 10.1128/EC.00113-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gressler, M., Zaehle, C., Scherlach, K., Hertweck, C., and Brock, M. (2011). Multifactorial induction of an orphan PKS-NRPS gene cluster in Aspergillus terreus. Chem. Biol. 18, 198–209. doi: 10.1016/j.chembiol.2010.12.011
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Guo, C.-J., Knox, B. P., Chiang, Y.-M., Lo, H.-C., Sanchez, J. F., Lee, K.-H., et al. (2012). Molecular genetic characterization of a cluster in A. terreus for biosynthesis of the meroterpenoid terretonin. Org. Lett. 14, 5684–5687. doi: 10.1021/ol302682z
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Guo, C. J., Knox, B. P., Sanchez, J. F., Chiang, Y. M., Bruno, K. S., and Wang, C. C. (2013a). Application of an efficient gene targeting system linking secondary metabolites to their biosynthetic genes in Aspergillus terreus. Org. Lett. 15, 3562–3565. doi: 10.1021/ol401384v
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Guo, C.-J., Sun, W.-W., Bruno, K. S., and Wang, C. C. C. (2014). Molecular genetic characterization of terreic acid pathway in Aspergillus terreus. Org. Lett. 16, 5250–5253. doi: 10.1021/ol502242a
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Guo, C. J., Yeh, H. H., Chiang, Y. M., Sanchez, J. F., Chang, S. L., Bruno, K. S., et al. (2013b). Biosynthetic pathway for the epipolythiodioxopiperazine acetylaranotin in Aspergillus terreus revealed by genome-based deletion analysis. J. Am. Chem. Soc. 135, 7205–7213. doi: 10.1021/ja3123653
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hill, A. M. (2006). The biosynthesis, molecular genetics and enzymology of the polyketide-derived metabolites. Nat. Prod. Rep. 23, 256–320. doi: 10.1039/b301028g
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Hill, R. A., Carter, R. H., and Staunton, J. (1981). Biosynthesis of fungal metabolites. Terrein, a metabolite of Aspergillus terreus Thom. J. Chem. Soc. Perkin Trans. 1, 2570–2576. doi: 10.1039/p19810002570
Itoh, T., Tokunaga, K., Radhakrishnan, E. K., Fujii, I., Abe, I., Ebizuka, Y., et al. (2012). Identification of a key prenyltransferase involved in biosynthesis of the most abundant fungal meroterpenoids derived from 3,5-dimethylorsellinic acid. Chembiochem 13, 1132–1135. doi: 10.1002/cbic.201200124
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Jimenez-Oses, G., Osuna, S., Gao, X., Sawaya, M. R., Gilson, L., Collier, S. J., et al. (2014). The role of distant mutations and allosteric regulation on LovD active site dynamics. Nat. Chem. Biol. 10, 431–436. doi: 10.1038/nchembio.1503
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kawakami, Y., Hartman, S. E., Kinoshita, E., Suzuki, H., Kitaura, J., Yao, L., et al. (1999). Terreic acid, a quinone epoxide inhibitor of Bruton's tyrosine kinase. Proc. Natl. Acad. Sci. U.S.A. 96, 2227–2232. doi: 10.1073/pnas.96.5.2227
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keller, N. P., Turner, G., and Bennett, J. W. (2005). Fungal secondary metabolism – from biochemistry to genomics. Nat. Rev. Microbiol. 3, 937–947. doi: 10.1038/nrmicro1286
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Khaldi, N., Seifuddin, F. T., Turner, G., Haft, D., Nierman, W. C., Wolfe, K. H., et al. (2010). SMURF: genomic mapping of fungal secondary metabolite clusters. Fungal Genet. Biol. 47, 736–741. doi: 10.1016/j.fgb.2010.06.003
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Kiriyama, N., Nitta, K., Sakaguchi, Y., Taguchi, Y., and Yamamoto, Y. (1977). Studies on the metabolic products of Aspergillus terreus. III. Metabolites of the strain IFO 8835. Chem. Pharm. Bull. 25, 2593–2601. doi: 10.1248/cpb.25.2593
Kuck, U., and Hoff, B. (2010). New tools for the genetic manipulation of filamentous fungi. Appl. Microbiol. Biotechnol. 86, 51–62. doi: 10.1007/s00253-009-2416-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Matsuda, Y., Awakawa, T., Itoh, T., Wakimoto, T., Kushiro, T., Fujii, I., et al. (2012). Terretonin biosynthesis requires methylation as essential step for cyclization. Chembiochem 13, 1738–1741. doi: 10.1002/cbic.201200369
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
McIntyre, C. R., Scott, F. E., Simpson, T. J., Trimble, L. A., and Vederas, J. C. (1989). Application of stable isotope labelling methodology to the biosynthesis of the mycotoxin, terretonin, by Aspergillus terreus: incorporation of 13C-labelled acetates and methionine, 2H- and 13C, 18O-labelled ethyl 3,5-dimethylorsellinate and oxygen-18 gas. Tetrahedron 45, 2307–2321. doi: 10.1016/S0040-4020(01)83433-5
McIntyre, C. R., Simpson, T. J., Stenzel, D. J., Bartlett, A. J., Q'brien, E., and Holker, J. S. E. (1982). Biosynthesis of the meroterpenoid metabolites, austin and terretonin: incorporation of 3,5-dimethylorsellinate. Chem. Commun. 781. doi: 10.1039/c39820000781
Nayak, T., Szewczyk, E., Oakley, C. E., Osmani, A., Ukil, L., Murray, S. L., et al. (2006). A versatile and efficient gene-targeting system for Aspergillus nidulans. Genetics 172, 1557–1566. doi: 10.1534/genetics.105.052563
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Nitta, K., Fujita, N., Yoshimura, T., Arai, K., and Yamamoto, Y. (1983). Metabolic products of Aspergillus terreus. IX. Biosynthesis of butyrolactone derivatives isolated from strains IFO 8835 and 4100. Chem. Pharm. Bull. 31, 1528–1533. doi: 10.1248/cpb.31.1528
Patron, N. J., Waller, R. F., Cozijnsen, A. J., Straney, D. C., Gardiner, D. M., Nierman, W. C., et al. (2007). Origin and distribution of epipolythiodioxopiperazine (ETP) gene clusters in filamentous ascomycetes. BMC Evol. Biol. 7:174. doi: 10.1186/1471-2148-7-174
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Proctor, R. H., Brown, D. W., Plattner, R. D., and Desjardins, A. E. (2003). Co-expression of 15 contiguous genes delineates a fumonisin biosynthetic gene cluster in Gibberella moniliformis. Fungal Genet. Biol. 38, 237–249. doi: 10.1016/S1087-1845(02)00525-X
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Read, G., and Vining, L. C. (1968). The biosynthesis of terreic acid. Chem. Commun. 935. doi: 10.1039/c19680000935
Read, G., Westlake, D. W., and Vining, L. C. (1969). Quinone epoxides. V. The biosynthesis of terreic acid. Can. J. Biochem. 47, 1071–1079. doi: 10.1139/o69-171
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sanchez, J. F., Somoza, A. D., Keller, N. P., and Wang, C. C. C. (2012). Advances in Aspergillus secondary metabolite research in the post-genomic era. Nat. Prod. Rep. 29, 351. doi: 10.1039/c2np00084a
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Scharf, D. H., Chankhamjon, P., Scherlach, K., Heinekamp, T., Roth, M., Brakhage, A. A., et al. (2012). Epidithiol formation by an unprecedented twin carbon-sulfur lyase in the gliotoxin pathway. Angew. Chem. Int. Ed. Engl. 51, 10064–10068. doi: 10.1002/anie.201205041
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Scharf, D. H., Chankhamjon, P., Scherlach, K., Heinekamp, T., Willing, K., Brakhage, A. A., et al. (2013). Epidithiodiketopiperazine biosynthesis: a four-enzyme cascade converts glutathione conjugates into transannular disulfide bridges. Angew. Chem. Int. Ed. Engl. 52, 11092–11095. doi: 10.1002/anie.201305059
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Scharf, D. H., Habel, A., Heinekamp, T., Brakhage, A. A., and Hertweck, C. (2014). Opposed effects of enzymatic gliotoxin N- and S-methylations. J. Am. Chem. Soc. 136, 11674–11679. doi: 10.1021/ja5033106
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Scharf, D. H., Heinekamp, T., Remme, N., Hortschansky, P., Brakhage, A. A., and Hertweck, C. (2011a). Biosynthesis and function of gliotoxin in Aspergillus fumigatus. Appl. Microbiol. Biotechnol. 93, 467–472. doi: 10.1007/s00253-011-3689-1
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Scharf, D. H., Remme, N., Habel, A., Chankhamjon, P., Scherlach, K., Heinekamp, T., et al. (2011b). A dedicated glutathione S-transferase mediates carbon-sulfur bond formation in gliotoxin biosynthesis. J. Am. Chem. Soc. 133, 12322–12325. doi: 10.1021/ja201311d
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Scharf, D. H., Remme, N., Heinekamp, T., Hortschansky, P., Brakhage, A. A., and Hertweck, C. (2010). Transannular disulfide formation in gliotoxin biosynthesis and its role in self-resistance of the human pathogen Aspergillus fumigatus. J. Am. Chem. Soc. 132, 10136–10141. doi: 10.1021/ja103262m
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Schrettl, M., Carberry, S., Kavanagh, K., Haas, H., Jones, G. W., O'brien, J., et al. (2010). Self-protection against gliotoxin–a component of the gliotoxin biosynthetic cluster, GliT, completely protects Aspergillus fumigatus against exogenous gliotoxin. PLoS Pathog. 6:e1000952. doi: 10.1371/journal.ppat.1000952
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Szewczyk, E., Nayak, T., Oakley, C. E., Edgerton, H., Xiong, Y., Taheri-Talesh, N., et al. (2006). Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 1, 3111–3120. doi: 10.1038/nprot.2006.405
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, M., Beissner, M., and Zhao, H. (2014). Aryl-aldehyde formation in fungal polyketides: discovery and characterization of a distinct biosynthetic mechanism. Chem. Biol. 21, 257–263. doi: 10.1016/j.chembiol.2013.12.005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Wang, M., and Zhao, H. (2014). Characterization and engineering of the adenylation domain of a NRPS-like protein: a potential biocatalyst for aldehyde generation. ACS Catal. 4, 1219–1225. doi: 10.1021/cs500039v
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Xu, W., Chooi, Y.-H., Choi, J. W., Li, S., Vederas, J. C., Da Silva, N. A., et al. (2013a). LovG: The thioesterase required for dihydromonacolin L release and lovastatin nonaketide synthase turnover in lovastatin biosynthesis. Angew. Chem. Int. Ed. Engl. 52, 6472–6475. doi: 10.1002/anie.201302406
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Xu, Y., Espinosa-Artiles, P., Schubert, V., Xu, Y. M., Zhang, W., Lin, M., et al. (2013b). Characterization of the biosynthetic genes for 10,11-dehydrocurvularin, a heat shock response-modulating anticancer fungal polyketide from Aspergillus terreus. Appl. Environ. Microbiol. 79, 2038–2047. doi: 10.1128/AEM.03334-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Xu, Y., Zhou, T., Zhou, Z., Su, S., Roberts, S. A., Montfort, W. R., et al. (2013c). Rational reprogramming of fungal polyketide first-ring cyclization. Proc. Natl. Acad. Sci. U.S.A. 110, 5398–5403. doi: 10.1073/pnas.1301201110
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yamamoto, H., Moriyama, K., Jinnouchi, H., and Yagishita, K. (1980). Studies on terreic acid. Jpn. J. Antibiot. 33, 320–328. doi: 10.11553/antibiotics1968b.33.320
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yin, W. B., Grundmann, A., Cheng, J., and Li, S. M. (2009). Acetylaszonalenin biosynthesis in Neosartorya fischeri. Identification of the biosynthetic gene cluster by genomic mining and functional proof of the genes by biochemical investigation. J. Biol. Chem. 284, 100–109. doi: 10.1074/jbc.M807606200
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Yu, J.-H., Hamari, Z., Han, K.-H., Seo, J.-A., Reyes-Domínguez, Y., and Scazzocchio, C. (2004). Double-joint PCR: a PCR-based molecular tool for gene manipulations in filamentous fungi. Fungal Genet. Biol. 41, 973–981. doi: 10.1016/j.fgb.2004.08.001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Zaehle, C., Gressler, M., Shelest, E., Geib, E., Hertweck, C., and Brock, M. (2014). Terrein biosynthesis in Aspergillus terreus and its impact on phytotoxicity. Chem. Biol. 21, 719–731. doi: 10.1016/j.chembiol.2014.03.010
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: genome mining, Aspergillus terreus, secondary metabolites, natural products
Citation: Guo C-J and Wang CCC (2014) Recent advances in genome mining of secondary metabolites in Aspergillus terreus. Front. Microbiol. 5:717. doi: 10.3389/fmicb.2014.00717
Received: 29 October 2014; Paper pending published: 15 November 2014;
Accepted: 01 December 2014; Published online: 23 December 2014.
Edited by:
Ozgur Bayram, National University of Ireland Maynooth, IrelandReviewed by:
Axel A. Brakhage, Friedrich Schiller University Jena, GermanySean Doyle, National University of Ireland Maynooth, Ireland
Copyright © 2014 Guo and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Clay C. C. Wang, Department of Pharmacology and Pharmaceutical Sciences, School of Pharmacy, University of Southern California, 1985 Zonal Ave PSC 406A, Los Angeles, CA, USA e-mail:Y2xheXdAdXNjLmVkdQ==
 Chun-Jun Guo
Chun-Jun Guo Clay C. C. Wang
Clay C. C. Wang