- 1Unit of Microbiology, Department of Life and Environmental Sciences, Polytechnic University of Marche, Ancona, Italy
- 2Unit of Microbiology, Department of Biomedical Sciences and Public Health, Polytechnic University of Marche Medical School, Ancona, Italy
Φm46.1 – Streptococcus pyogenes bacteriophage carrying mef(A) and tet(O), respectively, encoding resistance to macrolides (M phenotype) and tetracycline – is widespread in S. pyogenes but has not been reported outside this species. Φm46.1 is transferable in vitro among S. pyogenes isolates, but no information is available about its transferability to other Streptococcus species. We thus investigated Φm46.1 for its ability to be transduced in vitro to recipients of different Streptococcus species. Transductants were obtained from recipients of Streptococcus agalactiae, Streptococcus gordonii, and Streptococcus suis. Retransfer was always achieved, and from S. suis to S. pyogenes occurred at a much greater frequency than in the opposite direction. In transductants Φm46.1 retained its functional properties, such as inducibility with mitomycin C, presence both as a prophage and as a free circular form, and transferability. The transductants shared the same Φm46.1 chromosomal integration site as the donor, at the 3′ end of a conserved RNA uracil methyltransferase (rum) gene, which is an integration hotspot for a variety of genetic elements. No transfer occurred to recipients of Streptococcus pneumoniae, Streptococcus oralis, and Streptococcus salivarius, even though rum-like genes were also detected in the sequenced genomes of these species. A largely overlapping 18-bp critical sequence, where the site-specific recombination process presumably takes place, was identified in the rum genes of all recipients, including those of the species yielding no transductants. Growth assays to evaluate the fitness cost of Φm46.1 acquisition disclosed a negligible impact on S. pyogenes, S. agalactiae, and S. gordonii transductants and a noticeable fitness advantage in S. suis. The S. suis transductant also displayed marked overexpression of the autolysin-encoding gene atl.
Introduction
Many bacterial genomes deposited in public databases contain phage DNA integrated into the bacterial chromosome, at times as multiple prophages. Prophages are not passive genetic cargo of the bacterial chromosome but are likely to be active players in cell physiology, since phage DNA is a vector, as other mobile genetic elements, for lateral gene transfer between bacteria (Canchaya et al., 2003).
A neat test case for the role of prophages is Streptococcus pyogenes (Banks et al., 2002). About 90% of the isolates of this species are lysogenic, due to complete or partial prophages integrated into the host chromosome that sometimes contribute up to 12% of the total genome (Canchaya et al., 2003; Ferretti et al., 2004). Transformation appears to play no or only a minor role in lateral DNA transfer in S. pyogenes, conferring on phages a special role in this process. It has been suggested that in this species competence and transformation may have been lost due to the growing role assumed by bacteriophages in population diversity (Ferretti et al., 2004). These S. pyogenes phages or phage-like elements have long been known to encode many virulence factors, but more recently they have also been shown to carry antibiotic resistance genes. In particular, this applies to the macrolide efflux resistance gene mef (A) (Clancy et al., 1996), which is typically associated with a low-level resistance pattern involving, among macrolide-lincosamide-streptogramin B antibiotics, only 14- and 15-membered macrolides (M phenotype; Sutcliffe et al., 1996b). mef (A) is carried by Tn1207.1, a defective transposon originally detected in Streptococcus pneumoniae (Santagati et al., 2000). In S. pyogenes, Tn1207.1 is not found as such, but as part of larger composite elements that have all been shown to be chimeric, i.e., resulting from insertion of a transposon (identical or related to Tn1207.1) into a prophage (Banks et al., 2003; Giovanetti et al., 2005).
The mef (A)-carrying phage varies depending on whether the strain is resistant only to macrolides or also to tetracycline. When M-phenotype isolates of S. pyogenes are tetracycline susceptible, the bacteriophages involved are Φ1207.3 (formerly Tn1207.3, 52,491 bp, accession no. AY657002) (Santagati et al., 2003; Giovanetti et al., 2005; Iannelli et al., 2014) or Φ10394.4 (58,761 bp, accession no. AY445042; Banks et al., 2003, 2004), which are closely related and are integrated into the same chromosomal gene (comEC, encoding a putative competence protein; Santagati et al., 2003; Banks et al., 2003; Brenciani et al., 2004). The only difference is that, in Φ1207.3, Tn1207.1 is the left end of the element, whereas Φ10394.4 presents an additional left-hand region of ~6 kb. When M-phenotype isolates of S. pyogenes are coresistant to tetracycline – a condition that in Italy is more common than macrolide resistance alone (Giovanetti et al., 1999; Brenciani et al., 2004; D’Ercole et al., 2005) – tetracycline resistance is mediated by the tet(O) determinant (Giovanetti et al., 2003), linked to mef (A) in a phage variety whose extensively investigated representative is Φm46.1 (55,172 bp, accession no. FM864213; Giovanetti et al., 2005; Varaldo et al., 2009; Brenciani et al., 2010). Compared to Φ1207.3/Φ10394.4, Φm46.1 has a different integration site, at the 3′ end of a chromosomal gene (rum) encoding an RNA uracil methyltransferase (Brenciani et al., 2010). Electron microscopic analysis following induction with mitomycin C has revealed phage particles with the typical icosahedral head and tail morphology of Siphoviridae in both Φ10394.4 (Banks et al., 2003) and Φm46.1 (Brenciani et al., 2010).
Φ1207.3 and Φ10394.4 have also been detected in Streptococcus species other than S. pyogenes: the former in Streptococcus agalactiae (Marimón et al., 2005) and the latter in viridans group isolates of Streptococcus gordonii and Streptococcus salivarius (Brenciani et al., 2014). Conversely, Φm46.1 has never been reported outside S. pyogenes. Moreover, in in vitro transfer experiments Φ1207.3 was transferred to other Streptococcus species (Santagati et al., 2003), whereas such experiments have never been performed with Φm46.1. In early conjugation assays using S. pyogenes donors whose tet(O)–mef (A) elements had not yet been realized to be phages, mef (A) and tet(O) were co-transferred to a S. pyogenes but not to an Enterococcus faecalis recipient (Giovanetti et al., 2003). More recently lysogenic transfer of Φm46.1 has been reported among S. pyogenes isolates (Di Luca et al., 2010).
In this study, we investigated the ability of Φm46.1 to be transduced to recipients of Streptococcus species other than S. pyogenes. Φm46.1 was transferred to some species but not to others. The chromosomal integration site of Φm46.1 in the transductants corresponded to the one originally detected in S. pyogenes. Investigation of the fitness cost associated with Φm46.1 acquisition disclosed that it varied with the species and that a significant fitness advantage was conferred on the Streptococcus suis transductant.
Materials and Methods
Bacterial Strain
The strain harboring Φm46.1 was the same (S. pyogenes m46, an ST39, emm type 4 throat clinical isolate) where the mef (A)–tet(O) combination and linkage were initially detected (Giovanetti et al., 2003), and from which Φm46.1 was subsequently characterized and sequenced (Brenciani et al., 2010). Phenotypically, the strain is coresistant to erythromycin (MIC, 16 μg/ml; M phenotype) and tetracycline (MIC, 64 μg/ml).
Antibiotics and Susceptibility Tests
Erythromycin and tetracycline were purchased from Sigma Chemical Co. (St. Louis, MO, USA). MICs were determined by a standard broth microdilution method, using S. pneumoniae ATCC 49619 for quality control.
Lysogenic Transfer and Analysis of Transductants
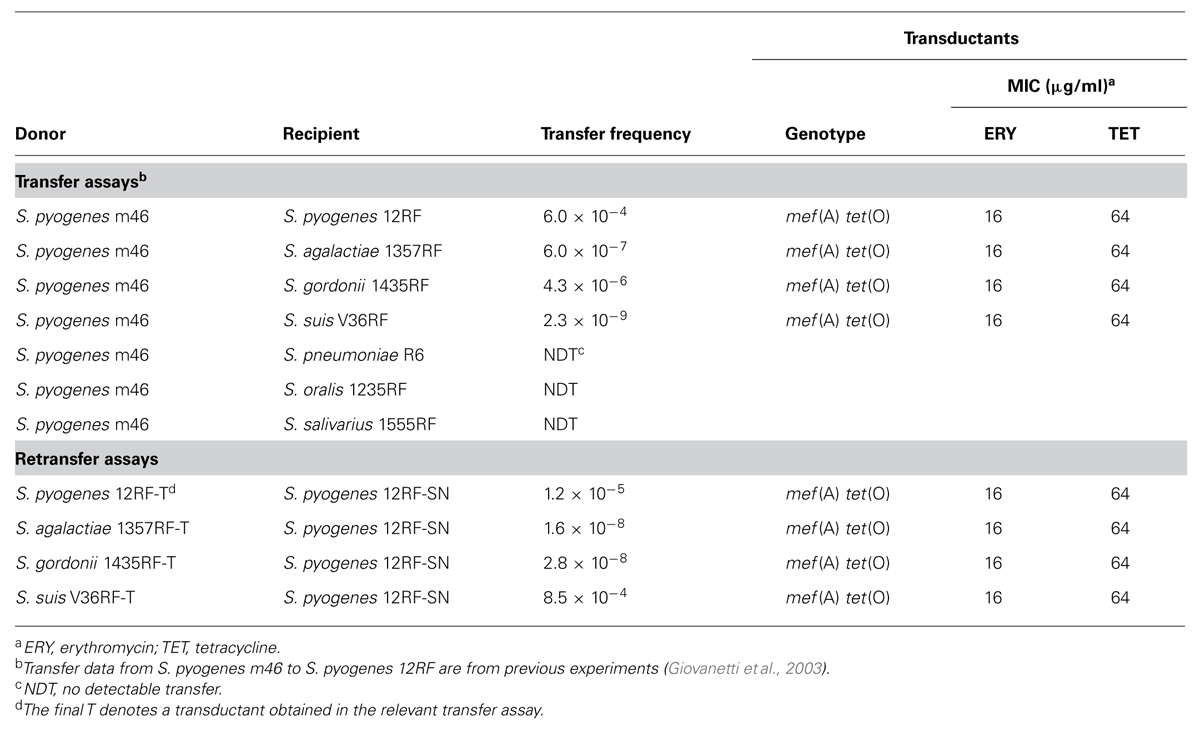
Transfer experiments were performed as described elsewhere (Giovanetti et al., 2002). S. pyogenes m46 was used as the donor. Rifampin- and fusidic acid-resistant (RF) derivatives of erythromycin- and tetracycline-susceptible strains of different Streptococcus species were used as recipients: S. pneumoniae R6RF (Cochetti et al., 2005); S. agalactiae 1357RF (Palmieri et al., 2012); S. gordonii 1435RF (Mingoia et al., 2014); Streptococcus oralis 1235RF (Mingoia et al., 2014); Streptococcus suis v36RF (Palmieri et al., 2012); and S. salivarius 1555RF, an RF derivative obtained for this study from a recently investigated strain (Brenciani et al., 2014). Retransfer experiments were performed using S. pyogenes 12RF-SN, a streptomycin- and nalidixic acid-resistant derivative of our recipient 12RF (Giovanetti et al., 2003), as the recipient. Transductants were selected on plates containing erythromycin (1 μg/ml) plus rifampin (10 μg/ml) and fusidic acid (10 μg/ml), or plus streptomycin (500 μg/ml) and nalidixic acid (10 μg/ml) in retransfer assays. Putative transductants were tested for mef (A) and tet(O) by polymerase chain reaction (PCR) and for erythromycin and tetracycline MICs; the presence of Φm46.1 was checked by PCR mapping in five randomly selected transductants of each species. Transduction frequency was expressed as the number of transductants per recipient. Mating experiments were done at least three times.
PCR Experiments
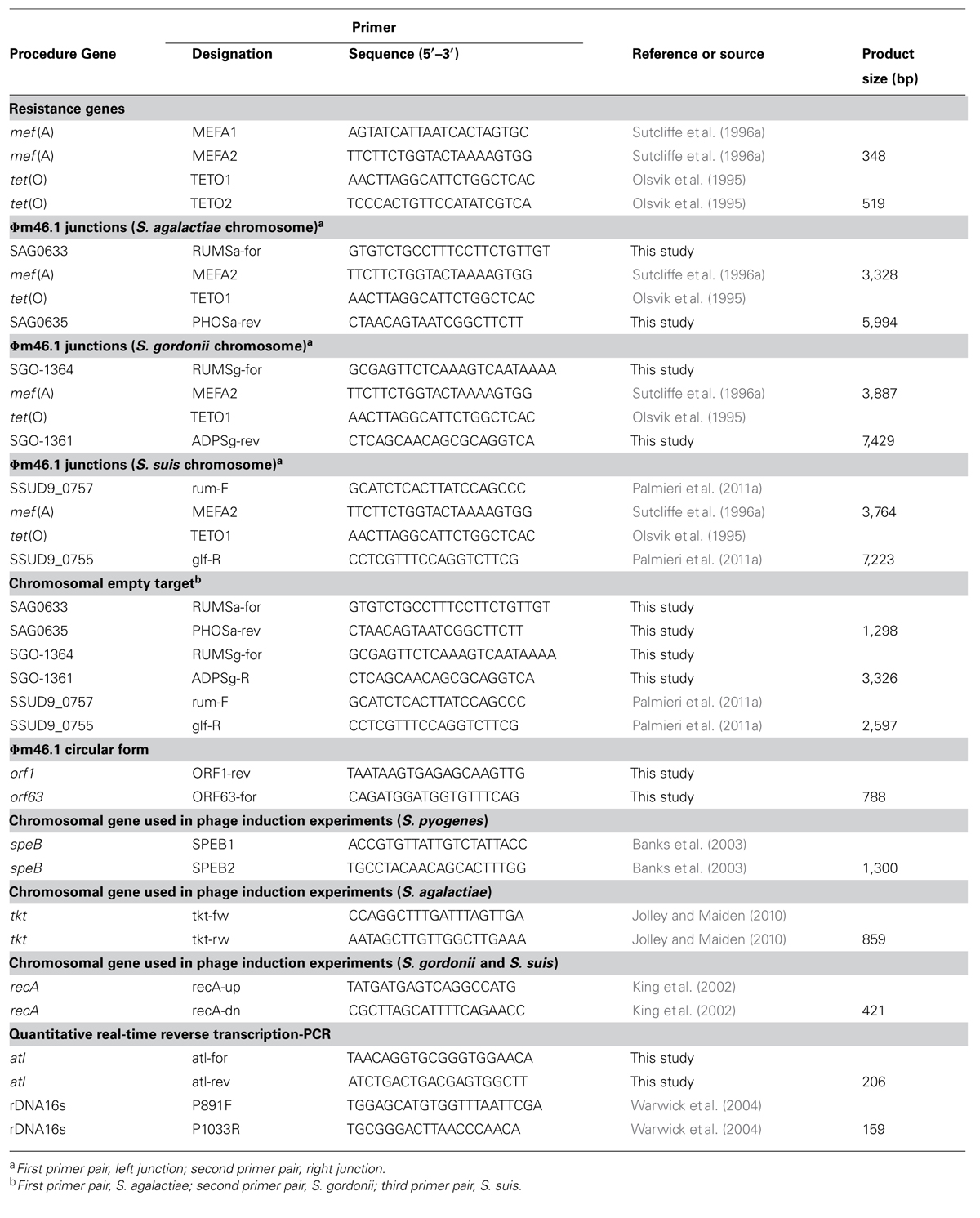
The primer pairs used in PCR experiments are listed in Table 1. DNA preparation and amplification and electrophoresis of PCR products were carried out by established procedures and following recommended conditions for the use of individual primer pairs. The Ex Taq system (TaKaRa Bio, Shiga, Japan) was used when expected PCR products exceeded 3 kb in size.
DNA Sequencing and Sequence Analysis
All PCR products used for sequence analysis were purified using Montage PCR filter units (Millipore Corporation, Bedford, MA, USA). Amplicons were sequenced (bidirectionally or by primer walking) using ABI Prism (Perkin-Elmer Applied Biosystems, Foster City, CA, USA) with dye-labeled terminators. Sequences were analyzed using the Sequence Navigator software package (Perkin-Elmer Applied Biosystems). Sequence similarity and conserved domain searches were carried out using tools (BLAST and CDART) available online at the National Center for Biotechnology Information of the National Library of Medicine (Bethesda, MD, USA; http://www.ncbi.nlm.nih.gov/).
Induction of Φm46.1 with Mitomycin C
For phage induction, one transductant of each species was treated with 0.2 μg/ml mitomycin C (Sigma) for 4 h at 37°C. S. pyogenes m46 was used as a control; phage DNA was extracted and purified as described previously (Brenciani et al., 2010). Induction was monitored by PCR using primer pairs targeting mef (A) and tet(O). Chromosomal genes (speB for S. pyogenes, tkt for S. agalactiae, and recA for S. gordonii and S. suis) were monitored as negative controls using specific PCR primers (Table 1) to confirm that there was no contaminating chromosomal DNA in the phage DNA preparations.
Fitness Assessment
The biological cost of Φm46.1 acquisition was investigated by growth assays, fitness differences being disclosed by exponential growth rates measured in resistant transductants and susceptible recipients. Competitive growth assays could not be used due to transduction events occurring during co-culture of recipients and transductants. One transductant and the recipient of each species [from previous experiments in the case of S. pyogenes (Giovanetti et al., 2003)] were grown overnight in brain heart infusion broth (BHI; Difco Laboratories, Detroit, MI, USA) at 37°C in 5% CO2. Cultures were diluted to an optical density (OD) of 0.1 ± 0.05 at 690 nm and then diluted 1:100 in BHI. From each dilution, a 150-μl aliquot was transferred to a well of a microtiter plate. Growth was monitored at 37°C for 24 h using Multiscan Ascent (Thermo Scientific, Waltham, MA, USA); OD690 measurements were taken every hour. Experiments were repeated three times.
RNA Isolation and Quantitative Real-Time Reverse Transcription-PCR (qRT-PCR)
Cultures were grown overnight in BHI at 37°C and then diluted 100-fold in fresh BHI. Subcultures were collected at the logarithmic phase (OD690 value of 0.8), and total RNA was isolated with the GenElute total RNA purification kit (Sigma) according to the manufacturer’s instructions. Total cDNA was obtained by the QuantiTect Reserve Transcription kit (Qiagen, Hilden, Germany) according to the manufacturer’s instructions. The copy number of a specific cDNA was calculated using the Rotor-Gene Q MDx instrument and the Rotor-Gene SYBR Green PCR kit (Qiagen). The 16S rDNA housekeeping gene was analyzed as an internal control. The primers used for qRT-PCR assays are reported in Table 1.
Results
Lysogenic Transfer and Analysis of Transductants
Transfer of macrolide and tetracycline coresistance from S. pyogenes m46, formerly described to a S. pyogenes recipient (Giovanetti et al., 2003), was obtained in the present study to recipients of S. agalactiae, S. gordonii, and S. suis, but not of S. pneumoniae, S. oralis, and S. salivarius (Table 2). In retransfer experiments using transductants as donors, including a previous S. pyogenes transductant (Giovanetti et al., 2003), erythromycin and tetracycline coresistance was consistently retransferred to S. pyogenes (Table 2). Whereas transduction frequencies were comparable (or slightly lower) in retransfer vs. transfer assays with S. pyogenes, S. agalactiae, and S. gordonii, retransfer from S. suis to S. pyogenes occurred at a much greater frequency (over 104 times) than transfer from S. pyogenes to S. suis.
Transductants exhibited a mef (A) tet(O) genotype and proved macrolide and tetracycline resistant, with MICs identical to those for the donors. The presence of a regular Φm46.1 was confirmed by PCR mapping in the transductants of all species. A transductant from each species was induced with mitomycin C: in all cases Φm46.1 was detected in culture supernatants in a DNAse-resistant form, such as a phage capsid.
Chromosomal Integration Site of Φm46.1 in Different Streptococcal Species
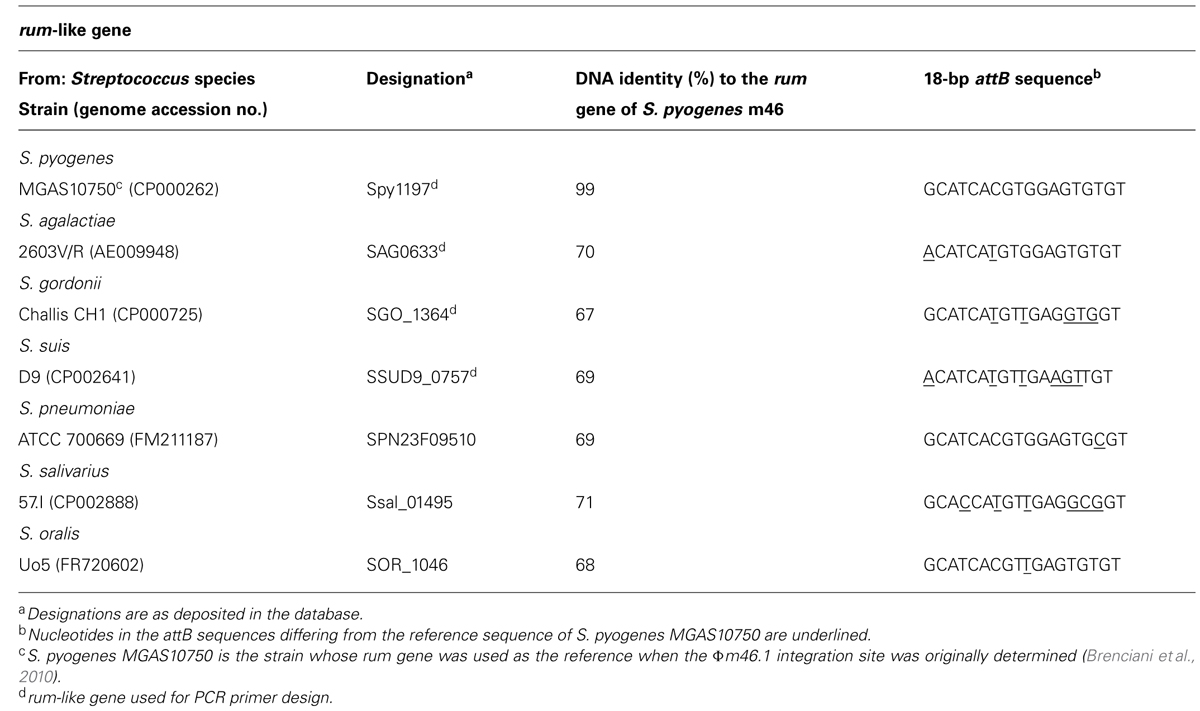
Since in S. pyogenes m46 Φm46.1 is integrated into the chromosome at the 3′ end of the rum gene (Brenciani et al., 2010), we first checked whether it had a similar integration site in the transductants obtained in the above experiments. This possibility was explored in one transductant of each species using two primer pairs, one for the left (attL) and one for the right (attR) junction. The primers of each pair that fell on the phage targeted mef (A) (left junction) or tet(O) (right junction), thanks to the close proximity of the two genes to the respective end of Φm46.1 (Brenciani et al., 2010). To design the primers that fell on the chromosome, homologs of the rum gene of S. pyogenes m46 in the relevant Streptococcus species (S. agalactiae, S. gordonii, and S. suis) were sought by BLASTN assays. A rum-like gene (DNA identity, around 70%) was detected in the genomes of such three species, and the one yielding the closest database match was considered. For S. pyogenes, the rum gene considered was Spy1197 from the genome of MGAS10750, 99% identical to the rum gene of S. pyogenes m46 and previously used as the reference rum gene when the Φm46.1 integration site was originally determined (Brenciani et al., 2010). The rum-like genes investigated are reported in Table 3, where the particular genes chosen to design the primers are also indicated. For attL, the primers to be paired to MEFA2, internal to mef (A), were designed from the respective rum-like gene in the portion upstream of the integration site; for attR, the primers to be paired to TETO1, internal to tet(O), were designed from open reading frames located downstream of the rum-like genes in the respective genomes (Table 1; Figure 1).
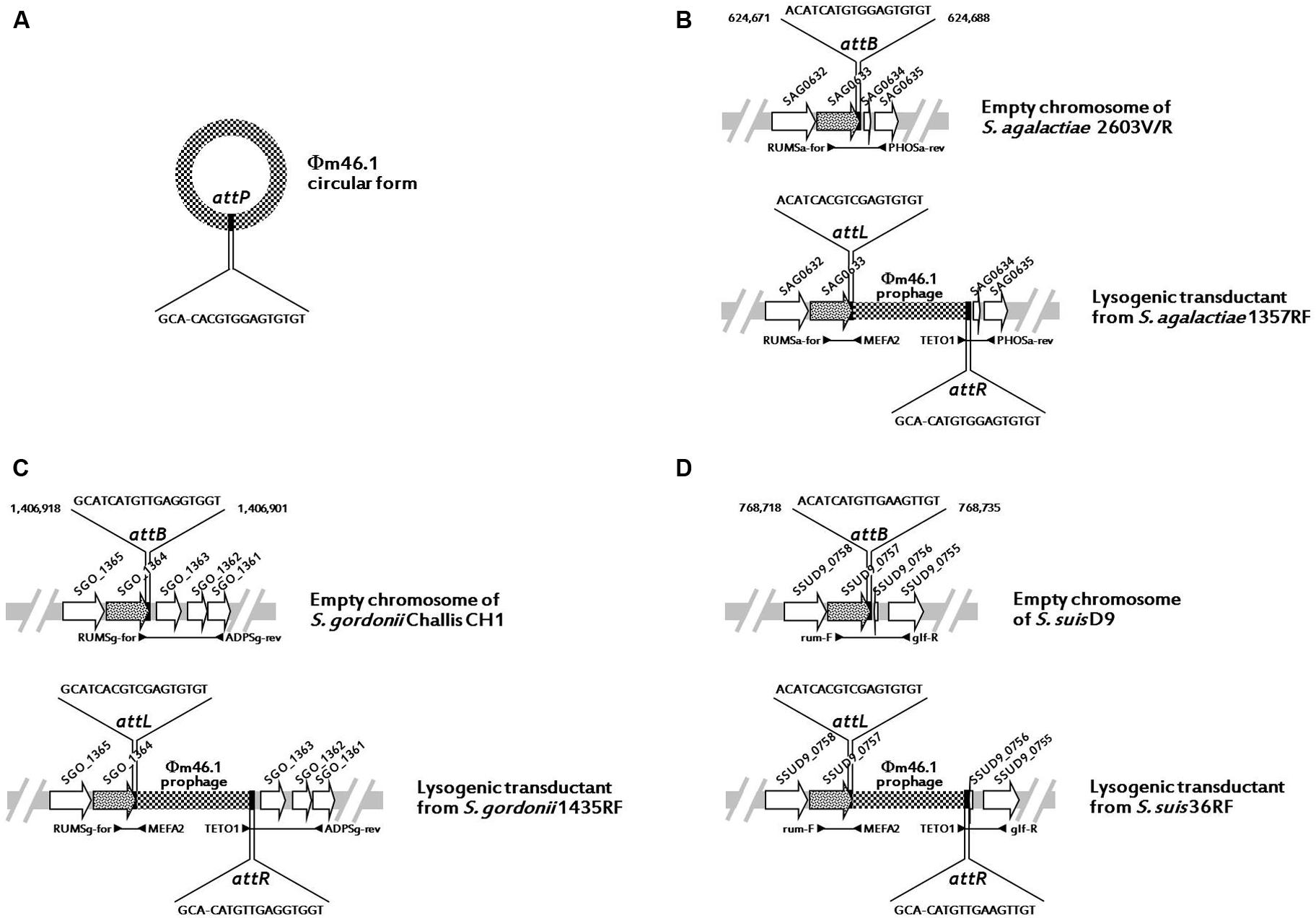
FIGURE 1. Schematic representation (not drawn to scale) of Φm46.1 (checkered) in circular form with the attP sequence (A) and its integration into the chromosome of S. agalactiae (B), S. gordonii (C), and S. suis (D). The chromosome is represented as a light gray bar. ORFs are represented as arrows pointing in the direction of transcription (the rum-like genes are spotted, the other genes are white). The top half of panels (B–D; the chromosome empty target) shows the relevant primer pair and the relevant attB sequence with the corresponding bases of the reference genome. The bottom half of panels (B–D; the lysogenic transductant) shows the relevant primer pairs for attL and attR and the respective sequences with the corresponding bases of the relevant transductant.
All PCR assays performed using these primer pairs yielded positive reactions, and all amplicons were sequenced and analyzed. The results indicated that in all transductants, irrespective of the species, the integration site of Φm46.1 corresponded to the one originally detected in S. pyogenes m46, i.e., at the 3′ end of the respective, species-specific rum gene. These data are schematically illustrated in Figure 1.
Search for Φm46.1 Circular Forms in the Transductants and Core Site Analysis
PCR experiments using an appropriate pair of outward-directed primers (reverse primer targeting orf1 and forward primer targeting orf63 of Φm46.1; Table 1) were performed using one transductant per species. The circular form of Φm46.1 was consistently detected.
Sequence analysis of amplicons from the circular forms, the empty chromosomes, and the attL and attR regions of Φm46.1 from the S. agalactiae, S. gordonii, and S. suis transductants allowed identification of an 18-bp putative core site – i.e., the critical sequence where the site-specific recombination process presumably takes place – largely overlapping with the attachment sequence of Φm46.1 (attP) and the chromosomal attachment sequence (attB; Table 3). The attB site corresponded to bases 624,671 to 624,688 of the genome of S. agalactiae 2603V/R; to bases 1,406,918 to 1,406,901 of the genome of S. gordonii Challis substr. CH1; and to bases 768,718 to 768,735 of the genome of S. suis D9 (Figure 1).
rum-Like Genes and Putative Core Sites of Φm46.1 in the Genomes of the Species Yielding no Transductants
rum-like genes ~70% identical to the rum gene of S. pyogenes m46 were detected by BLASTN assays also in the genomes of the Streptococcus species yielding no transductants (S. pneumoniae, S. oralis, and S. salivarius). An 18-bp sequence largely overlapping with the above-mentioned attB sequences from S. pyogenes, S. agalactiae, S. gordonii, and S. suis was also detected in the rum-like genes from the genomes of these species (Table 3).
Fitness Cost of the Acquisition of Φm46.1
The in vitro growth curves of the recipient and the transductant of S. pyogenes, S. agalactiae, S. gordonii, and S. suis are shown in Figure 2. While the recipient and the transductant of three species (S. pyogenes, S. agalactiae, and S. gordonii) displayed similar growth rates, denoting a negligible impact of Φm46.1 acquisition on fitness, a distinctly greater fitness was observed in S. suis. In this species, the log phase started at 5 h in the transductant compared to 7 h in the recipient; moreover, the transductant displayed a greater growth rate compared to the recipient, with an earlier (at less than 9 h vs. 12 h) and higher (OD690, ~0.60 vs. ~0.35) peak.
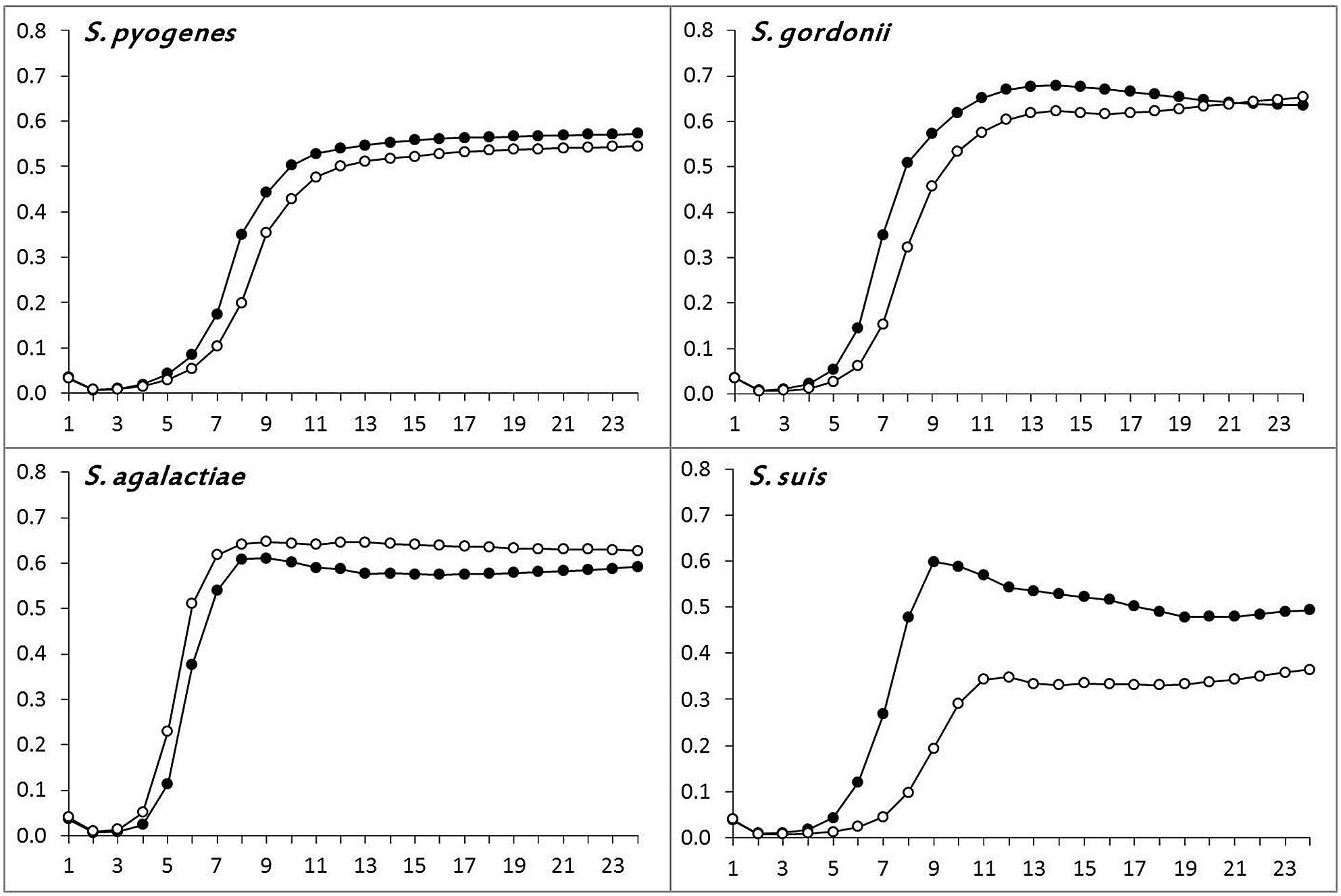
FIGURE 2. Determination of bacterial fitness by growth assays. Comparison of the growth rates of the recipient (∘–∘) and a randomly chosen transductant (∙–∙) of S. pyogenes, S. agalactiae, S. gordonii, and S. suis.
Further insights into the behavior of S. suis appeared to be needed. Since a phage has recently been shown to alter fitness in S. pneumoniae by interfering with autolytic activity (DeBardeleben et al., 2014), and a novel autolysin-encoding gene (designated atl) has recently been described in S. suis (Ju et al., 2012), we investigated atl expression in the S. suis transductant and recipient. qRT-PCR experiments disclosed significant atl overexpression by the transductant compared to the recipient, the mRNA level in the former (300 copies) being at least 60-fold higher than in the latter (<5 copies).
Discussion
In bacteria bacteriophages are less common vehicles of antibiotic resistance genes than other mobile genetic elements such as plasmids or integrative and conjugative elements (ICEs). Φm46.1, the S. pyogenes phage carrying mef (A) and tet(O; Brenciani et al., 2010), is however, closely associated to a major erythromycin-resistant subpopulation of this species, which in Italy is predominant among M phenotype isolates (Spinaci et al., 2004; D’Ercole et al., 2005; Giovanetti et al., 2005). Φm46.1, common in S. pyogenes but unreported outside this species, is known to be transferable in vitro among S. pyogenes isolates (Giovanetti et al., 2003; Di Luca et al., 2010). In contrast, no information is available about its transferability to other Streptococcus species. The primary goal of our study was to explore this point. In vitro transfer assays showed that the recipients of some species (S. agalactiae, S. gordonii, and S. suis) yielded transductants harboring Φm46.1, whereas those of other species (S. pneumoniae, S. oralis, and S. salivarius) did not. In the transductants Φm46.1 retained its functional properties, including inducibility with mitomycin C, presence in the host cell both as a prophage and as free circular DNA, and transferability. In the species to which Φm46.1 was not transduced, failure to transfer did not seem to depend on the lack of the chromosomal integration site. Indeed, this site was consistently found to be at the 3′ end of a species-specific homolog of the rum gene, the chromosome integration site of Φm46.1 that was originally described in S. pyogenes (Brenciani et al., 2010); comparable rum-like genes were detected in the genomes of all Streptococcus species used as recipients, regardless of whether they yielded transductants. Failure of Φm46.1 to be transduced could perhaps reflect a flaw in an earlier step, e.g., during phage adsorption to host surface receptors or viral DNA injection.
A conserved rum gene is commonly found in streptococcal genomes, and its 3′ end is an integration hotspot for a vast array of genetic elements, typically carrying cargo genes encoding antibiotic resistances. In S. pyogenes, besides Φm46.1 [carrying mef (A) and tet(O)], such elements include ICE2096-RD.2 [tet(O)] (Beres and Musser, 2007); ICESp1108 [erm(TR)] (Brenciani et al., 2011); ICESp2905 [erm(TR) and tet(O)] (Brenciani et al., 2011); and ICESp2906 [tet(O)] (Giovanetti et al., 2012). Insertion into the same conserved rum location is also shared by S. pyogenes ICE6180-RD.1 (Beres and Musser, 2007) and the S. agalactiae prophage λSa04 (Brenciani et al., 2010), neither carrying resistance genes. In S. suis, it is important to mention ΦSsUD.1, a bacteriophage with a scaffold closely related to that of Φm46.1, which carries tet(W), a MAS (macrolide–aminoglycoside–streptothricin)-like fragment (Cochetti et al., 2005), and a cadC/cadA cadmium efflux cassette (Palmieri et al., 2011a). The same integration site is shared by a S. suis chimeric element (Hu et al., 2011; Palmieri et al., 2011b), constituted of an ICE and a phage, which carries tet(O) in tandem with tet(40), a mef(E)-containing mega-like structure (Gay and Stephens, 2001), and a MAS-like fragment. The rum 3′ region has very recently been reported to be the chromosomal integration site of genetic elements from other Streptococcus species: in particular of two related vanG-carrying elements conferring vancomycin resistance on S. agalactiae and S. anginosus (Srinivasan et al., 2014), previously unreported in these species. All these data actually suggested a strategy for routine localized screening of these insertions for the acquisition of new resistances (Srinivasan et al., 2014).
Of special interest was the marked difference between transfer and retransfer assays involving S. suis. While Φm46.1 was transferred from S. pyogenes to S. suis at a very low frequency (around 10-9), retransfer from S. suis to S. pyogenes occurred at a considerably greater (>104 times) frequency. This may reflect the fact that S. pyogenes is the usual, natural host of Φm46.1, even though such a large difference between transfer and retransfer frequencies was not seen in the other species. On the other hand, the above-mentioned S. suis bacteriophage ΦSsUD.1 has proved to be transferable to S. pyogenes, but not to S. suis (Palmieri et al., 2011a).
Other intriguing findings, again concerning S. suis but not the transductants of the other species, were observed in fitness-related experiments. In S. suis, Φm46.1 acquisition was associated with a markedly greater growth rate and with overexpression of atl, an autolysin-encoding gene. These phenomena are experimentally very clear, but are not easy to elucidate; the reason why they occurred in S. suis but not in the other species is similarly difficult to explain. It is worth noting that the atl-encoded autolysin is believed to take part, besides cell autolysis, in separation of daughter cells, biofilm formation, fibronectin-binding activity, cell adhesion, and pathogenesis (Ju et al., 2012). In S. pneumoniae, the autolysin LytA has been shown to be activated after prophage induction and to contribute to efficient bacteriophage progeny release (Frias et al., 2009). Still in S. pneumoniae, autolysis-mediated fitness changes dependent on the presence of a prophage have recently been regarded as a new insight into how bacteria and prophages interact and affect bacterial fitness (DeBardeleben et al., 2014). The atl overexpression we observed in the S. suis transductant may suggests that the autolysin target, the cell wall, is more resistant to its enzymatic activity; however, unlike DeBardeleben et al. (2014) who observed increased penicillin resistance in a phage-harboring isolate, we found similar penicillin MICs for the S. suis transductant and recipient (data not shown). Moreover, it is worth noting that Φm46.1 contains a toxin–antitoxin system (Brenciani et al., 2010); such systems, possibly working as selfish entities favoring their own maintenance (Gerdes, 2000), may be involved in interactions with host regulatory networks and in fitness variations (Van Melderen and Saavedra De Bast, 2009).
A final consideration concerns the subpopulation naturally harboring Φm46.1, which in Italy is the majority of M phenotype erythromycin-resistant isolates of S. pyogenes (Spinaci et al., 2004; Giovanetti et al., 2005). The lack of a fitness cost associated with Φm46.1 acquisition by pyogenes may account for the wide circulation of such erythromycin- and tetracycline-coresistant organisms.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Banks, D. J., Beres, S. B., and Musser, J. M. (2002). The fundamental contribution of phages to GAS evolution, genome diversification and strain emergence. Trends Microbiol. 10, 515–521. doi: 10.1016/S0966-842X(02)02461-7
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Banks, D. J., Porcella, S. F., Barbian, K. D., Beres, S. B., Philips, L. E., Voyich, J. M.,et al. (2004). Progress toward characterization of the group A Streptococcus metagenome: complete genome sequence of a macrolide-resistant serotype M6 strain. J. Infect. Dis. 190, 727–738. doi: 10.1086/422697
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Banks, D. J., Porcella, S. F., Barbian, K. D., Martin, J. M., and Musser, J. M. (2003). Structure and distribution of an unusual chimeric genetic element encoding macrolide resistance in phylogenetically diverse clones of group A Streptococcus. J. Infect. Dis. 188, 1898–1908. doi: 10.1086/379897
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Beres, S. B., and Musser, J. M. (2007). Contribution of exogenous genetic elements to the group A Streptococcus metagenome. PLoS ONE 2:e800. doi: 10.1371/journal.pone.0000800
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brenciani, A., Bacciaglia, A., Vignaroli, C., Pugnaloni, A., Varaldo, P. E., and Giovanetti, E. (2010). Φm46.1, the main Streptococcus pyogenes element carrying mef(A) and tet(O) genes. Antimicrob. Agents Chemother. 54, 221–229. doi: 10.1128/AAC.00499-09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brenciani, A., Ojo, K. K., Monachetti, A., Menzo, S., Roberts, M. C., Varaldo, P. E.,et al. (2004). Distribution and molecular analysis of mef(A)-containing elements in tetracycline-susceptible and resistant Streptococcus pyogenes clinical isolates with efflux-mediated erythromycin resistance. J. Antimicrob. Chemother. 54, 991–998. doi: 10.1093/jac/dkh481
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brenciani, A., Tiberi, E., Bacciaglia, A., Petrelli, D., Varaldo, P. E., and Giovanetti, E. (2011). Two distinct genetic elements are responsible for erm(TR)-mediated erythromycin resistance in tetracycline-susceptible and tetracycline-resistant strains of Streptococcus pyogenes. Antimicrob. Agents Chemother. 55, 2106–2112. doi: 10.1128/AAC.01378-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Brenciani, A., Tiberi, E., Tili, E., Mingoia, M., Palmieri, C., Varaldo, P. E.,et al. (2014). Genetic determinants and elements associated with antibiotic resistance in viridans group streptococci. J. Antimicrob. Chemother. 69, 1197–1204. doi: 10.1093/jac/dkt495
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Canchaya, C., Proux, C., Fournous, G., Bruttin, A., and Brüssow, H. (2003). Prophage genomics. Microbiol. Mol. Biol. Rev. 67, 238–276. doi: 10.1128/MMBR.67.2.238-276.2003
Clancy, J., Petitpas, J., Dib-Hajj, F., Yuan, W., Cronan, M., Kamath, A. V., et al. (1996). Molecular cloning and functional analysis of a novel macrolide resistance determinant, mefA, from Streptococcus pyogenes. Mol. Microbiol. 22, 867–879. doi: 10.1046/j.1365-2958.1996.01521.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Cochetti, I., Vecchi, M., Mingoia, M., Tili, E., Catania, M. R., Manzin, A., et al. (2005). Molecular characterization of pneumococci with efflux-mediated erythromycin resistance and identification of a novel mef gene subclass, mef(I). Antimicrob. Agents Chemother. 49, 4999–5006. doi: 10.1128/AAC.49.12.4999-5006.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
DeBardeleben, H. K., Lysenko, E. S., Dalia, A. B., and Weiser, J. N. (2014). Tolerance of a phage element by Streptococcus pneumoniae leads to a fitness defect during colonization. J. Bacteriol. 196, 2670–2680. doi: 10.1128/JB.01556-14
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
D’Ercole, S., Petrelli, D., Prenna, M., Zampaloni, C., Catania, M. R., Ripa, S., et al. (2005). Distribution of mef(A)-containing genetic elements in erythromycin-resistant isolates of Streptococcus pyogenes from Italy. Clin. Microbiol. Infect. 11, 927–930. doi: 10.1111/j.1469-0691.2005.01250.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Di Luca, M. C., D’Ercole, S., Petrelli, D., Prenna, M., Ripa, S., and Vitali, L. A. (2010). Lysogenic transfer of mef(A) and tet(O) genes carried by Φm46.1 among group A streptococci. Antimicrob. Agents Chemother. 54, 4464–4466. doi: 10.1128/AAC.01318-09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ferretti, J. J., Ajdic, D., and McShan, W. M. (2004). Comparative genomics of streptococcal species. Indian J. Med. Res. 119(Suppl.), 1–6.
Frias, M. J., Melo-Cristino, J., and Ramirez, M. (2009). The autolysin LytA contributes to efficient bacteriophage progeny release in Streptococcus pneumoniae. J. Bacteriol. 191, 5428–5440. doi: 10.1128/JB.00477-09
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gay, K., and Stephens, D. S. (2001). Structure and dissemination of a chromosomal insertion element encoding macrolide efflux in Streptococcus pneumoniae. J. Infect. Dis. 184, 56–65. doi: 10.1086/321001
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Gerdes, K. (2000). Toxin-antitoxin modules may regulate synthesis of macromolecules during nutritional stress. J. Bacteriol. 182, 561–572. doi: 10.1128/JB.182.3.561-572.2000
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Giovanetti, E., Brenciani, A., Lupidi, R., Roberts, M. C., and Varaldo, P. E. (2003). Presence of the tet(O) gene in erythromycin- and tetracycline-resistant strains of Streptococcus pyogenes and linkage with either the mef(A) or the erm(A) gene. Antimicrob. Agents Chemother. 47, 2844–2849. doi: 10.1128/AAC.47.9.2844-2849.2003
Giovanetti, E., Brenciani, A., Tiberi, E., Bacciaglia, A., and Varaldo, P. E. (2012). ICESp2905, the erm(TR)-tet(O) element of Streptococcus pyogenes, is formed by two independent integrative and conjugative elements. Antimicrob. Agents Chemother. 56, 591–594. doi: 10.1128/AAC.05352-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Giovanetti, E., Brenciani, A., Vecchi, M., Manzin, A., and Varaldo, P. E. (2005). Prophage association of mef(A) elements encoding efflux-mediated erythromycin resistance in Streptococcus pyogenes. J. Antimicrob. Chemother. 55, 445–451. doi: 10.1093/jac/dki049
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Giovanetti, E., Magi, G., Brenciani, A., Spinaci, C., Lupidi, R., Facinelli, B., et al. (2002). Conjugative transfer of the erm(A) gene from erythromycin-resistant Streptococcus pyogenes to macrolide-susceptible S. pyogenes, Enterococcus faecalis, and Listeria innocua. J. Antimicrob. Chemother. 50, 249–252. doi: 10.1093/jac/dkf122
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Giovanetti, E., Montanari, M. P., Mingoia, M., and Varaldo, P. E. (1999). Phenotypes and genotypes of erythromycin-resistant Streptococcus pyogenes strains in Italy and heterogeneity of inducibly resistant strains. Antimicrob. Agents Chemother. 43, 1935–1940.
Hu, P., Yang, M., Zhang, A., Wu, J., Chen, B., Hua, Y.,et al. (2011). Complete genome sequence of Streptococcus suis serotype 14 strain JS14. J. Bacteriol. 193, 2375–2376. doi: 10.1128/JB.00083-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Iannelli, F., Santagati, M., Santoro, F., Oggioni, M. R., Stefani, S., and Pozzi, G. (2014). Nucleotide sequence of conjugative prophage Φ1207.3 (formerly Tn1207.3) carrying the mef(A)/msr(D) genes for efflux resistance to macrolides in Streptococcus pyogenes. Front. Microbiol. 5:687. doi: 10.3389/fmicb.2014.00687
Jolley, K. A., and Maiden, M. C. J. (2010). BIGSdb: Scalable analysis of bacterial genome variation at the population level. BMC Bioinformatics 11:595. doi: 10.1186/1471-2105-11-595
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Ju, C. X., Gu, H. W., and Lu, C. P. (2012). Characterization and functional analysis of atl, a novel gene encoding autolysin in Streptococcus suis. J. Bacteriol. 194, 1464–1473. doi: 10.1128/JB.06231-11
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
King, S. J., Leigh, J. A., Heath, P. J., Luque, I., Tarradas, C., Dowson, C. G.,et al. (2002). Development of a multilocus sequence typing scheme for the pig pathogen Streptococcus suis: identification of virulent clones and potential capsular serotype exchange. J. Clin. Microbiol. 40, 3671–3680. doi: 10.1128/JCM.40.10.3671-3680.2002
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Marimón, J. M., Valiente, A., Ercibengoa, M., García-Arenzana, J. M., and Pérez-Trallero, E. (2005). Erythromycin resistance and genetic elements carrying macrolide efflux genes in Streptococcus agalactiae. Antimicrob. Agents Chemother. 49, 5069–5074. doi: 10.1128/AAC.49.12.5069-5074.2005
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Mingoia, M., Morici, E., Morroni, G., Giovanetti, E., Del Grosso, M., Pantosti, A., et al. (2014). Tn5253 family integrative and conjugative elements carrying mef(I) and catQ determinants in Streptococcus pneumoniae and Streptococcus pyogenes. Antimicrob. Agents Chemother. 58, 5886–5893. doi: 10.1128/AAC.03638-14
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Olsvik, B., Olsen, I., and Tenover, F. C. (1995). Detection of tet(M) and tet(O) using the polymerase chain reaction in bacteria isolated from patients with periodontal disease. Oral Microbiol. Immunol. 10, 87–92. doi: 10.1111/j.1399-302X.1995.tb00124.x
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Palmieri, C., Magi, G., Mingoia, M., Bagnarelli, P., Ripa, S., Varaldo, P. E.,et al. (2012). Characterization of a Streptococcus suis tet(O/W/32/O)-carrying element transferable to major Streptococcal pathogens. Antimicrob. Agents Chemother. 57, 4697–4702. doi: 10.1128/AAC.00629-12
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Palmieri, C., Princivalli, M. S., Brenciani, A., Varaldo, P. E., and Facinelli, B. (2011a). Different genetic elements carrying the tet(W) gene in two human clinical isolates of Streptococcus suis. Antimicrob. Agents Chemother. 55, 631–636. doi: 10.1128/AAC.00965-10
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Palmieri, C., Varaldo, P. E., and Facinelli, B. (2011b). Streptococcus suis, an emerging drug-resistant animal and human pathogen. Front. Microbiol. 2:235. doi: 10.3389/fmicb.2011.00235
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Santagati, M., Iannelli, F., Cascone, C., Campanile, F., Oggioni, M. R., Stefani, S., et al. (2003). The novel conjugative transposon Tn1207.3 carries the macrolide efflux gene mef(A) in Streptococcus pyogenes. Microb. Drug Resist. 9, 243–247. doi: 10.1089/107662903322286445
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Santagati, M., Iannelli, F., Oggioni, M. R., Stefani, S., and Pozzi, G. (2000). Characterization of a genetic element carrying the macrolide efflux gene mef(A) in Streptococcus pneumoniae. Antimicrob. Agents Chemother. 44, 2585–2587. doi: 10.1128/AAC.44.9.2585-2587.2000
Spinaci, C., Magi, G., Zampaloni, C., Vitali, L. A., Paoletti, C., Catania, M. R.,et al. (2004). Genetic diversity of cell-invasive erythromycin-resistant and susceptible group A streptococci determined by analysis of the RD2 region of the prtF1 gene. J. Clin. Microbiol. 42, 639–644. doi: 10.1128/JCM.42.2.639-644.2004
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Srinivasan, V., Metcalf, B. J., Knipe, K. M., Ouattara, M., McGee, L., Shewmaker, P. L., et al. (2014). vanG element insertions within a conserved chromosomal site conferring vancomycin resistance to Streptococcus agalactiae and Streptococcus anginosus. MBio 5, e01386–14. doi: 10.1128/mBio.01386-14
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Sutcliffe, J., Grebe, T., Tait-Kamradt, A., and Wondrack, L. (1996a). Detection of erythromycin-resistant determinants by PCR. Antimicrob. Agents Chemother. 40, 2562–2566.
Sutcliffe, J., Tait-Kamradt, A., and Wondrack, L. (1996b). Streptococcus pneumoniae and Streptococcus pyogenes resistant to macrolides but sensitive to clindamycin: a common resistance pattern mediated by an efflux system. Antimicrob. Agents Chemother. 40, 1817–1824.
Van Melderen, L., and Saavedra De Bast, M. (2009). Bacterial toxin–antitoxin systems: more than selfish entities? PLoS Genet. 5:e1000437. doi: 10.1371/journal.pgen.1000437
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Varaldo, P. E., Montanari, M. P., and Giovanetti, E. (2009). Genetic elements responsible for erythromycin resistance in streptococci. Antimicrob. Agents Chemother. 53, 343–353. doi: 10.1128/AAC.00781-08
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Warwick, S., Wilks, M., Hennessy, E., Powell-Tuck, J., Small, M., Sharp, J.,et al. (2004). Use of quantitative 16S ribosomal DNA detection for diagnosis of central vascular catheter-associated bacterial infection. J. Clin. Microbiol. 42, 1402–1408. doi: 10.1128/JCM.42.4
Pubmed Abstract | Pubmed Full Text | CrossRef Full Text | Google Scholar
Keywords: Streptococcus speciesm46.1bacteriophagesmef(A)tet(O)transductionchromosomal integrationfitness cost
Citation: Giovanetti E, Brenciani A, Morroni G, Tiberi E, Pasquaroli S, Mingoia M and Varaldo PE (2015) Transduction of the Streptococcus pyogenes bacteriophage Φm46.1, carrying resistance genes mef (A) and tet(O), to other Streptococcus species. Front. Microbiol. 5:746. doi: 10.3389/fmicb.2014.00746
Received: 25 September 2014; Paper pending published: 18 October 2014;
Accepted: 09 December 2014; Published online: 08 January 2015.
Edited by:
Adam Paul Roberts, University College London, UKReviewed by:
Giovanni Gherardi, University Campus Biomedico, ItalyOrietta Massidda, Università di Cagliari, Italy
Copyright © 2015 Giovanetti, Brenciani, Morroni, Tiberi, Pasquaroli, Mingoia and Varaldo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Andrea Brenciani, Unit of Microbiology, Department of Biomedical Sciences and Public Health, Polytechnic University of Marche Medical School, Via Tronto 10/A, 60126 Ancona, Italy e-mail:YW5kcmVhYnJlbmNpYW5pQHlhaG9vLml0
 Eleonora Giovanetti
Eleonora Giovanetti  Andrea Brenciani
Andrea Brenciani Gianluca Morroni
Gianluca Morroni Erika Tiberi
Erika Tiberi Sonia Pasquaroli
Sonia Pasquaroli Marina Mingoia
Marina Mingoia Pietro E.Varaldo
Pietro E.Varaldo