- 1Research Center of Bioenergy and Bioremediation, Southwest University, Chongqing, China
- 2College of Plant Protection, Southwest University, Chongqing, China
- 3Laboratory of Plant Pathology and Biotechnology, Kochi University, Kochi, Japan
- 4Research Institute of Molecular Genetics, Kochi University, Kochi, Japan
The MarR-family of transcriptional regulators are involved in various cellular processes, including resistance to multiple antibiotics and other toxic chemicals, adaptation to different environments and pathogenesis in many plant and animal pathogens. Here, we reported a new MarR regulator PrhN, which was involved in the pathogenesis of Ralstonia solanacearum. prhN mutant exhibited significantly reduced virulence and stem colonization compared to that of wild type in tomato plants. prhN mutant caused identical hypersensitive response (HR) on resistant plants to the wild type. Deletion of prhN gene substantially reduced the expression of type III secretion system (T3SS) in vitro and in planta (mainly in tomato plants), which is essential for pathogenicity of R. solanacearum, and the complemented PrhN could restore its virulence and T3SS expression to that of wild type. T3SS is directly controlled by AraC-type transcriptional regulator HrpB, and the transcription of hrpB is activated by HrpG and PrhG. HrpG and PrhG are homologs but are regulated by the PhcA positively and negatively, respectively. Deletion of prhN gene also abolished the expression of hrpB and prhG, but didn't change the expression of hrpG and phcA. Together, these results indicated that PrhN positively regulates T3SS expression through PrhG and HrpB. PrhN and PhcA should regulate prhG expression in a parallel way. This is the first report on the pathogenesis of MarR regulator in R. solanacearum, and this new finding will improve our understanding on the various biological functions of MarR regulator and the complex regulatory network on hrp regulon in R. solanacearum.
Introduction
The MarR (multiple antibiotic resistance regulator) protein was originally identified as repressor of the multiple antibiotic resistance operon marRAB in Escherichia coli. It is a prototypical member of the MarR-family of transcriptional regulators (Seoane and Levy, 1995; Wilkinson and Grove, 2006). MarR members are widely distributed in many bacteria and archaea and are involved in various cellular processes, including resistance to multiple antibiotics and other toxic chemicals, adaptation to different environments and the pathogenesis of many plant and animal pathogens (Alekshun et al., 2001; Ellison and Miller, 2006). A majority of MarR members are characterized as transcriptional repressors, several of which are activators, i.e., SlyA is suggested to repress and activate expression of target genes (Stapleton et al., 2002; Haque et al., 2009; Zou et al., 2012). The role of MarR members in pathogenesis is to control the expression of some virulence-related genes or virulence-associated traits (Ellison et al., 2004; Wilkinson and Grove, 2004). It has been recently reported that the MarR regulator SlyA regulates Type III secretion system (T3SS) genes in parallel with the T3SS master regulator HrpL and is important for the virulence of Dickeya dadantii 3937 (Haque et al., 2009; Zou et al., 2012). In Xanthomonas campestris pathovar campestris (taxonomically close to Ralstonia solanacearum), the MarR regulator HpaR is positively controlled by HrpG and HrpX and is involved in pathogenesis (Wei et al., 2007). MarR regulators are conserved in many pathogens, but whether some regulators share MarR-like activities in R. solanacearum remains to be characterized.
R. solanacearum is one of the most destructive bacterial plant pathogens and currently, there are no generally effective controls for the lethal bacterial wilt diseases caused by R. solanacearum (Mansfield et al., 2012; Peeters et al., 2013). R. solanacearum is a Gram-negative, soil-borne vascular bacterium, and like in many plant and animal bacterial pathogens, the syringe-like T3SS plays a crucial role in the pathogenicity of R. solanacearum (Angot et al., 2006; Galán and Wolf-Watz, 2006). Bacteria use T3SS to inject virulence effectors, so-called type III effectors, into host cells to interfere with cellular defenses (Gohre and Robatzek, 2008; Poueymiro and Genin, 2009). In R. solanacearum, the T3SS is encoded by a gene cluster of more than 20 hypersensitive response and pathogenicity (hrp) genes; these genes form a cluster known as the hrp regulon (Van Gijsegem et al., 1995; Cornelis and Van Gijsegem, 2000). Transcription of hrp regulon is repressed in nutrient-rich medium and induced in nutrient-poor medium, which may mimic conditions in the intercellular spaces of plants (Arlat et al., 1992; Coll and Valls, 2013). Plant signals or some mimic signals can induce hrp expression at levels to 10 to 20-fold greater than that in nutrient-poor conditions (Jacobs et al., 2012; Monteiro et al., 2012). The T3SS and type III effectors are directly controlled by the AraC-type transcriptional regulator HrpB, which binds directly to the plant-inducible promoter (PIP) motif in the promoter regions of its target genes (Cunnac et al., 2004; Mukaihara et al., 2010). The transcription of hrpB is activated by both HrpG and PrhG, which are close paralogs (72% global identity) and belong to the OmpR/PhoB family of two-component response regulators (Plener et al., 2010; Zhang et al., 2013). The regulation of HrpG on hrpB expression is activated by some plant-related signals which are perceived by the outer membrane receptor PrhA or some unknown receptors and transduced to HrpG through the PrhA-PrhR/PrhI-PrhJ cascade or some unknown pathway (Valls et al., 2006; Yoshimochi et al., 2009b). PrhG is dispensable for this signaling cascade (Zhang et al., 2013; Zuluaga et al., 2013). HrpG has been well illustrated as master regulator since hrpG mutants are impaired in growth in planta and complete lost the pathogenicity in host plants (Vasse et al., 2000; Valls et al., 2006; Yoshimochi et al., 2009b). PrhG mutant can grow the same as do wild-type bacteria in planta and show slightly reduced virulence compared to that of wild-type (Zhang et al., 2013). The global virulence regulator PhcA, which is quorum sensing-dependent, negatively regulates the expression of hrpG through PrhIR in an indirect manner (Genin et al., 2005; Yoshimochi et al., 2009a). PhcA positively regulates prhG expression (Zhang et al., 2013). In this process, PhcA regulates hrpB expression in opposite ways. R. solanacearum may switch from using HrpG to PrhG for hrpB activation in a cell density-dependent manner (Zhang et al., 2013).
In a previous study using transposon mutagenesis, we isolated some prh (positive regulation of hrp regulon) genes from the Japanese R. solanacearum strain OE1-1 (Zhang et al., 2013). OE1-1 causes disease in tomato and tobacco plants (Kanda et al., 2003). We constructed a popA-lacZYA fusion in OE1-1 to monitor the expression profile of the hrp regulon, in which the promoterless lacZYA operon was integrated downstream of the popA gene and shared a promoter with popA (a schematic is available as Figure S1 in Zhang et al., 2013). The popA gene exists as part of an operon with popB and popC, which maps to the left-side of the hrp regulon. The popA operon is directly controlled by HrpB and popA-lacZYA exhibits an expression profile that is identical to that of the hrp regulon under different conditions. The generated reporter strain RK5050 (OE1-1 popA-lacZYA) exhibits bacterial growth and virulence that are identical to OE1-1 in tomato and tobacco plants (Yoshimochi et al., 2009b; Zhang et al., 2011). To better elucidate the regulation mechanism of the hrp regulon in R. solanacearum, we focused on Rsc1721 (prh18 in Zhang et al., 2013 hereafter designated as PrhN). PrhN contains 188 amino acids and is a putative MarR-type transcriptional regulator (https://iant.toulouse.inra.fr/bacteria/annotation/cgi/ralso.cgi). We analyzed the contribution of PrhN to hrp expression and pathogenicity in R. solanacearum, and demonstrated genetically that the newly characterized MarR regulator PrhN positively regulates the hrp regulon indirectly and is required for full virulence of R. solanacearum in host plants.
Materials and Methods
Bacterial Strains and Culture Conditions
Bacterial strains used in this study are listed in Table 1. R. solanacearum strains are derivatives of the strain OE1-1 (phylotype I, race 1, biovar 3) (Kanda et al., 2003) and RS1002 (phylotype I, race 1, biovar 4) (Mukaihara et al., 2004). The OE1-1 strain is pathogenic on tomato and tobacco plants, and the strain RS1002 is pathogenic on tomato plants and elicits a hypersensitive response (HR) in tobacco leaves. E. coli strains DH12S and S17-1 (Simon et al., 1983) were used for plasmid construction and conjugational transfer, respectively. E. coli was grown in Luria-Bertani (LB) medium at 37°C. R. solanacearum was grown at 28°C in rich B medium or in hydroponic plant culture medium containing 2% sucrose (sucrose medium, in which the hrp expression can be well induced) (Yoshimochi et al., 2009b; Zhang et al., 2011). Antibiotics were added into the medium at the following concentrations: ampicillin (Ap), 100 μg ml−1; gentamycin (Gm), 20 μg ml−1; kanamycin (Km), 50 μg ml−1; polymyxin B (PB), 50 μg ml−1.
Construction of prhN Deletion Mutants
Plasmids designed to create deletion mutants were based on pK18mobsacB (Scḧafer et al., 1994). To construct a complete in-frame deletion of prhN, two DNA fragments flanking the prhN gene were amplified from OE1-1 genomic DNA using PrimeSTAR HS DNA polymerase (Takara Japan). The left flanking fragment was amplified using the primers rsc1721A1BamHI (5′-CATGGATCCGATGTCGAGTGCGTAGGC-3′) and rsc1721B1C (5′-CCGGCCCGAACGGGCGTTGCGTGACTCGCGGGCAGA-3′). The right flanking fragment was amplified using the primers rsc1721A2C (5′-TCTGCCCGCGAGTCACGCAACGCCCGTTCGGGCCGG-3′) and rsc1721B2HindIII (5′-CATAAGCTTGCGGTCGTGCAGGTGCGC-3′). rsc1721B1C and rsc1721A2C are full complements that contain 18 bases of nucleic aligning to the left and right flanking fragments, respectively. Two DNA fragments (~600-bp) were purified from an agarose gel and subjected to a second round of PCR amplification using the primers rsc1721A1BamHI and rsc1721B2HindIII. Next, a 1.2-kb fragment was agarose gel-purified and cloned using the TA-blunt ligation kit (Nippon gene) into pBluescript II KS (+) vector predigested with EcoR-V. The resulting plasmid was named pKSdprhN and the sequence was validated. Next, a BamH-I-Hind-III digested DNA fragment from pKSdprhN was sub-cloned into a BamH-I-Hind-III digested pK18mobsacB vector and pK18dprhN was generated.
After the sequence was validated, the plasmid pK18dprhN was transferred from E. coli strain S17-1 into the OE1-1 derivative strains RK5050 (popA-lacZYA), RK5046 (hrpB-lacZYA), RK5120 (hrpG-lacZYA), and RK5212 (prhG-lacZYA), and the RS1002 derivative strain RK10001 (RS1002, popA-lacZYA). The deletion of prhN at its original location was confirmed by colony PCR using primers rsc1721A1BamHI and rsc1721B2HindIII (the expected PCR band size for wild type strains was 1.7-kb; PCR band size for prhN deletion was 1.2-kb). The prhN deletion strains RK5601 (popA-lacZYA, Δ prhN), RK5604 (hrpB-lacZYA, Δ prhN), RK5607 (prhG-lacZYA, Δ prhN), RK5610 (hrpG-lacZYA, Δ prhN), and RK10010 (RK0001, ΔprhN) were obtained.
Complementation Analyses
In the present study, complementation analyses were performed using a Tn7-based site-specific integration system. The gene for complementation was cloned into pUC18-mini-Tn7T-Gm vector (Choi et al., 2005; Zhang et al., 2011). The prhN gene (containing a 613-bp region upstream of prhN, which possibly includes its native promoter) was amplified using primers rsc1721A1BamHI and rsc1721B3HindIII (5′-CATAAGCTTTTACAGCGAGGTGGCCGAAC-3′). The purified ~1.2-kb DNA fragment was cloned using the TA-blunt ligation kit (Nippon gene) into pBluescript II KS (+) predigested with EcoR-V and pKSprhNC was constructed. After the sequence was validated, a BamH-I-Hind-III digested DNA fragment from pKSprhNC was sub-cloned into BamH-I-Hind-III digested pUC18-mini-Tn7T-Gm and pUCprhN was generated. After the sequence was validated, the transposase-containing helping plasmid pTNS2 was used to electroporate pUCprhN into prhN mutants. The prhN gene (containing its possible native promoter) was specifically integrated into a single attTn7 site (25-bp downstream of the glmS gene). Insertion at the attTn7 site was confirmed by colony PCR using primers glmS-down and Tn7R (Zhang et al., 2011).
Construction of prhN-lacZYA Reporter Strain and Deletion with phcA
The promoterless lacZYA fragment from pUClacZYA (Yoshimochi et al., 2009b) was inserted 24-bp downstream of the start codon of prhN, where six nucleotides (GAAAGT) were replaced with BamH I sequence (GGATCC). Two DNA fragments flanking insertion site were amplified using PCR. The primers rsc1721A4EcoRI (5′-CATGAATTCGATGTCGAGTGCGTAGGC-3′) and rsc1721B4BamHI (5′-TCGTCGTCCTCTTCGGATCCCGATTTG-3′) were used to amplify left flanking fragment. rsc1721A4BamHI (5′-CAAATCGGGATCCGAAGAGGACGACGA-3′) and rsc1721B2HindIII were used to amplify the right flanking fragment. The primers rsc1721B4BamHI and rsc1721A4BamHI are full complements. Two DNA fragments were purified from agarose gel and subjected for a second round of PCR amplification using primers rsc1721A4EcoRI and rsc1721B4HindIII. The agarose gel-purified 1.2-kb fragment was cloned using the TA-blunt ligation kit (Nippon gene) into pBluescript II KS (+) predigested with EcoR-V and pKSprhNB was constructed. After the sequence was validated, a EcoR-I-Hind-III digested DNA fragment from pKSprhNB was sub-cloned into EcoR-I-Hind-III digested pK18mobsacB vector and pK18prhNB was generated. Subsequently, a BamH-I digested promoterless lacZYA fragment from pUClacZYA was inserted into BamH-I digested pK18prhNB with the same direction as prhN and pK18prhN-lacZYA was generated.
After the sequence was validated, plasmid pK18prhN-lacZYA was transferred from E. coli S17-1 into R. solanacearum OE1-1 and RK5613 (OE1-1, prhN-lacZYA) was obtained. When the phcA deletion was introduced into RK5613, the plasmid phcA22 (used for the phcA deletion based on pK18mobsacB, Yoshimochi et al., 2009b) was transferred from E. coli strain S17-1 into RK5613, and RK5619 (prhN-lacZYA, Δ phcA) was obtained.
β-Galactosidase Assay
The β-galactosidase assay was performed as described previously (Zhang et al., 2011). Bacterial cells (40 μ l) were used to inoculate 2 ml of fresh medium and incubated with shaking at 28°C for 5 h in B medium or 8 h in sucrose medium (cells where grown to an approximate OD600 of 0.1). When cells were collected at a low density, i.e., at an approximate OD600 of 0.01, 30 μ l of bacterial cells was used to inoculate 40 ml of fresh medium and cells where grown to an approximate OD600 of 0.01 after 5 or 8 h cultivation. Next, 40 ml of cell culture was centrifuged and resuspended into 2 ml of fresh medium. When cells were collected at a high density, i.e., at an approximate OD600 of 1.0, 200 μ l of bacterial cells was used to inoculate 2 ml of fresh medium and cells will grow to an approximate OD600 of 1.0 after 5 or 8 h cultivation. An aliquot (100 or 200 μ l) of cells was used for β-galactosidase assay. The enzyme assay was repeated in at least three independent times and mean values were averaged with standard deviation (SD). Statistical significance was determined by the two-tailed Student's t-test.
Virulence Assays and HR Tests
Virulence assays were performed on wilt-susceptible plants (tomato, Solanum lycopersicum cv. Moneymaker and tobacco, Nicotiana tabacum cv. Bright Yellow) using the soil-soak and petiole inoculation procedures as described previously (Yao and Allen, 2007; Zhang et al., 2013). For the soil-soak inoculation, 2–3-week-old tomato plants or 3–4-week-old tobacco plants were inoculated with bacteria at a final concentration of 107 cfu g−1 of soil and incubated at 25°C under 10000 lx. For petiole inoculation in tomato plants, 2 μ l of bacteria at 107 cfu ml−1 was dropped onto the freshly cut surface of petioles. Plants were inspected for wilt symptoms daily over approximately 20 days post inoculation (dpi). Each test for one strain included at least 12 plants per treatment and each assay was repeated in at least four independent trials. The mean values were averaged with SD. For the HR test in tobacco leaves, N. tabacum BY leaves were infiltrated with approximately 50 μ l of bacterial suspension at 108 cfu ml−1 with a blunt-end syringe. Plants were incubated at 25°C, and HR symptom development was recorded periodically. Each test was repeated at least three times and a representative result is presented.
Bacterial Growth In Planta
Bacteria in planta were collected from stem tissues and quantified by dilution plating as previously described (Zhang et al., 2013). Two-to-three-week-old tomato plants were inoculated with bacteria using the soil-soak procedure and stem pieces (5 cm above the soil and 1 cm in length) were removed at 4 dpi (wilt symptoms reached 1–2) and 7 dpi (wilt symptoms reached 3–4). Each test included at least three samples from different plants and mean values of four independent trials were averaged with SD. Statistical significance was determined by the two-tailed Student's t-test.
β-Galactosidase Assay in Planta
The β-galactosidase assay in planta was performed as described previously (Zhang et al., 2013). Plants (tomato and tobacco) were inoculated with bacteria using petiole inoculation and leaf infiltration procedures, respectively. For petiole inoculation, 2 μ l of bacterial cells (108 cfu ml−1) was placed on the freshly cut surface of tomato petioles. For leaf infiltration, approximately 100 μ l of bacterial cells (104 cfu ml−1) was infiltrated into tobacco and tomato leaves. After incubation for various times, leaf discs (0.38 cm2) and tomato stem pieces (1 cm in length) were removed from plants and bacteria were collected for β-galactosidase assay. Enzymatic activity in planta was determined using a Galacto-Light Plus kit (Applied Biosystems) and the luminescence was evaluated using GloMax 20/20 luminometer (Promega). Enzymatic activity was normalized with luminescence divided by cell number. Each assay included at least three samples from different plants and mean values of at least four independent trials were averaged with SD. Statistical significance was determined by the two-tailed Student's t-test.
Nucleotide Sequence Accession Number
The nucleotide sequences of prhN in strain OE1-1 have been deposited into the DDBJ database under accession number AB981315.
Results
PrhN is Important for popA and hrpB Expression In Vitro
Generated by transposon mutagenesis, a prhN mutant exhibited substantially reduced popA expression, indicating that PrhN is a positive regulator of the popA operon in R. solanacearum OE1-1 (Zhang et al., 2013). PrhN is well conserved in many bacterial strains in addition to R. solanacearum. PrhN in R. solanacearum OE1-1 is more than 96% identical to those in R. solanacearum strains, approximately 80% identical to those in Ralstonia sp. and Cupriavidus sp., approximately 60% identical to those in Chromobacterium sp. and others. Searching the genome databank of R. solanacearum strains, only seldom provides gene products that are presumed to be MarR-type regulators. A genome BLAST search revealed that Rsc1721 is a unique putative MarR-type regulator in GMI1000, whose genome sequence has been completed and well studied (Salanoubat et al., 2002).
In order to confirm this positive regulation of PrhN on popA expression, we constructed a prhN in-frame deletion mutant RK5601 (popA-lacZYA, Δ prhN) from RK5050 (OE1-1, popA-lacZYA) and evaluated popA expression. popA expression in RK5050 is seriously repressed in rich medium and well induced in sucrose medium (approximately 328 Miller Units). In sucrose medium, popA expression in the prhN mutant (RK5601) was significantly reduced to 15 Miller Units (Figure 1A, p < 0.01). The prhN mutant (RK5601) showed identically reduced popA expression as that of a transposon mutant. Strain RS1002 differs from OE1-1 for virulence in tobacco plants, and the prhN deletion in RS1002 also significantly diminished popA expression in sucrose medium (12 Miller Units) (Figure 1A, p < 0.01).
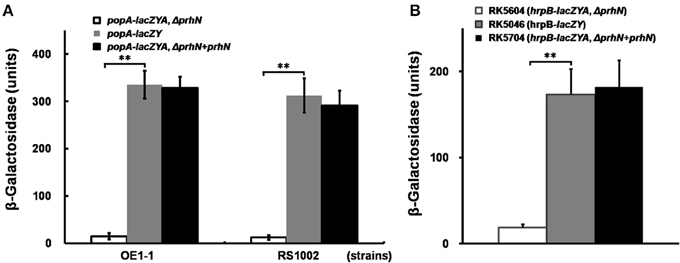
Figure 1. popA and hrpB expression in prhN mutants. (A) popA, (B) hrpB expression in sucrose medium. (A) prhN mutants (popA-lacZAY, ΔprhN), white bars; wild type (popA-lacZAY), gray bars; and strains with complementation (popA-lacZAY, ΔprhN+prhN), dark bars. Derivatives of strain OE1-1: RK5601 (prhN mutant), RK5050 (wild type), and RK5701 (complementation). Derivatives of strain RS1002: RK10010 (prhN mutant), RK10001 (wild type), and RK10013 (complementation). (B) RK5604 (hrpB-lacZAY, ΔprhN), white bars; RK5046 (hrpB-lacZAY), gray bars and RK5704 (complementation, hrpB-lacZAY, ΔprhN+prhN), dark bars. Cells were grown in sucrose medium to an OD600 of about 0.1, which corresponds to 1.8 × 108 cfu ml−1, and treated with SDS–chloroform for β-galactosidase assay. Mean values of at least four independent trials were presented in Miller units with SD (error bars). **P < 0.01.
As popA expression is directly controlled by HrpB, we evaluated the effect of PrhN on hrpB expression. We constructed a prhN in-frame deletion mutant RK5604 (hrpB-lacZYA, Δ prhN) from RK5046 (OE1-1, hrpB-lacZYA) and evaluated hrpB expression in sucrose medium. hrpB expression in the prhN mutant (RK5604) was significantly reduced compared to that of RK5046 (19 vs. 173 Miller Units) (Figure 1B, p < 0.01). The complemented prhN gene (under the control of its native promoter) could completely restore popA and hrpB expression to that of wild-type (Figures 1A,B). As with the control, the empty vector (pUC18-mini-Tn7T-Gm) did not affect popA and hrpB expression (data not shown). All of these findings demonstrate that PrhN is important for the expression of popA and hrpB in R. solanacearum in vitro, and this control is not strain-specific.
PrhN Positively Regulates prhG Expression But is Dispensable for hrpG Expression
As the transcription of hrpB is activated by both HrpG and PrhG, we evaluated the effect of PrhN on expression of hrpG and prhG. We constructed prhN in-frame deletion mutants RK5607 (prhG-lacZYA, Δ prhN) and RK5610 (hrpG-lacZYA, Δ prhN) from RK5120 (hrpG-lacZYA) and RK5212 (prhG-lacZYA), respectively. In sucrose medium, prhG expression in RK5212 (prhG-lacZYA) was approximately 2533 Miller units, and it was significantly reduced to 256 Miller units in RK5607 (prhG-lacZYA, Δ prhN) (Figure 2A, p < 0.01). Complemented PrhN could completely recover prhG expression to that of wild type (Figure 2A). Empty vector did not affect the prhG expression (data not shown). The phcA deletion substantially reduced prhG expression in both B and sucrose media (Zhang et al., 2013). In sucrose medium, prhG expression in the phcA mutant (RK5234) was even slightly lower than that in the prhN mutant (Figure 2A). In rich medium (B medium), RK5212 (prhG-lacZYA) and RK5607 (prhG-lacZYA, Δ prhN) exhibited almost equal prhG expression levels (Figure 2A). These results suggest that prhG expression is positively regulated by PrhN but only in sucrose medium. In both B and sucrose media, almost equal hrpG expression was observed in RK5120 (hrpG-lacZYA) and RK5610 (hrpG-lacZYA, Δ prhN) (Figure 2B), suggesting that PrhN is not involved in the regulation of hrpG expression in R. solanacearum.
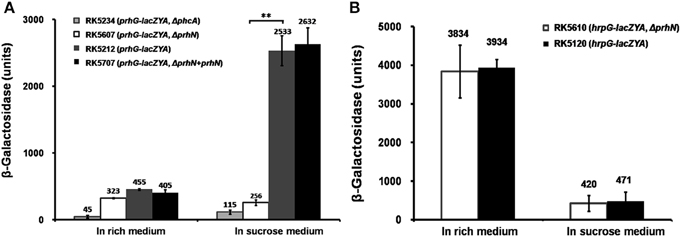
Figure 2. prhG and hrpG expression in prhN mutants. (A) prhG, (B) hrpG expression in rich and sucrose media. (A), RK5234 (prhG-lacZYA, ΔphcA), grayish bars; RK5607 (prhG-lacZYA, ΔprhN), white bars; RK5212 (prhG-lacZYA), gray bars and RK5707 (complementation, prhG-lacZAY, ΔprhN+prhN), dark bars. (B) RK5610 (hrpG-lacZYA, ΔprhN), white bars and RK5120 (hrpG-lacZYA), dark bars. Cells were grown to an OD600 of about 0.1 and enzyme activity was measured. Mean values of at least four independent trials were presented in Miller units with SD (error bars). **P < 0.01.
The Expression of prhN Increases with Cell Density and Regulates prhG Expression in Parallel with PhcA
As PhcA also positively regulates prhG expression (Zhang et al., 2013), we evaluated the effect of PhcA on prhN expression. We constructed a prhN-lacZYA reporter strain RK5613 (OE1-1 prhN-lacZYA) and a phcA deletion mutant RK5619 (prhN-lacZYA, Δ phcA) from RK5613. In rich medium, sucrose medium, and co-cultivation with A. thaliana seedlings, almost equal prhN expression levels were observed in RK5613 (prhN-lacZYA) and RK5619 (prhN-lacZYA, Δ phcA) (Figure 3A), suggesting that PhcA is not involved in the regulation of prhN expression in R. solanacearum. phcA mutants are well known to be deficient in extracellular polysaccharides (EPS) and show stick and matte cellular morphology. The prhN mutant remains mucoid and demonstrates abundant EPS production (data not shown), indicating that PrhN is dispensable for phcA expression in R. solanacearum. PhcA and PrhN should positively regulate prhG expression in a parallel way.
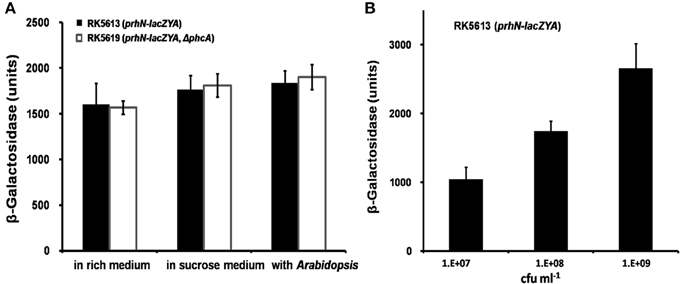
Figure 3. prhN expression under different conditions. (A) prhN expression in phcA mutant in rich medium, sucrose medium and co-cultivation with A. thaliana seedlings. RK5613 (prhN-lacZYA), dark bars and RK5619 (prhN-lacZYA, ΔphcA), white bars. Cells were grown to an OD600 of about 0.1 and enzyme activity was measured. (B) prhN expression at different cell density in sucrose medium. Cells were grown to an OD600 of about 0.01, 0.1, and 1.0 (1 OD600 corresponds to 1.8 × 109 cfu ml−1), and enzyme activity was measured. Mean values of at least four independent trials were presented in Miller units with SD (error bars).
As prhG expression increases constantly with cell densities (Zhang et al., 2013), we evaluated prhN expression levels at different cell densities. In sucrose medium, prhN expression at approximate OD600s of 0.01, 0.1 and 1.0 reached 1045, 1743, and 2654 Miller units, respectively (Figure 3B). The expression profile of prhN with cell density was consistent with that of prhG.
PrhN is Important for Full Virulence of R. solanacearum in Tomato Plants but is not Involved in HR Elicitation
Since PrhN is important for hrpB and prhG expression in R. solanacearum, and hrpB and prhG are important for pathogenicity of R. solanacearum in host plants, we tested the effect of PrhN on virulence in host plants. When RK5050 (OE1-1, popA-lacZYA) was inoculated on host plants (tomato and tobacco plants), it showed virulence and bacterial growth in planta which were identical to those of the parental strain OE1-1 (Yoshimochi et al., 2009b; Zhang et al., 2011). Using soil-soak inoculation in tomato plants, which mimics natural infection from roots, RK5050 wilted tomato plants at 4 dpi and killed plants at 10 dpi (Figure 4A). Several tomato plants inoculated with the prhN mutant (RK5601) began to wilt at 6 dpi and died at 14 dpi. However, most plants remained alive up to 20 dpi (wilt symptoms reached approximately 1–2, Figure 4A). These results suggest that the prhN mutant exhibited less virulence than that of RK5050 in tomato plants. As reported previously (Zhang et al., 2013), the prhG mutant (RK5185) exhibited slightly less virulence than that of RK5050. The prhN mutant (RK5601) exhibited obviously less virulence that of the prhG mutant (RK5185) (Figure 4A). Complementation with PrhN (under the control of its native promoter) could completely restore the virulence of the prhN mutant (RK5601) to that of RK5050 (Figure 4A). As with the control, the empty vector (pUC18-mini-Tn7T-Gm) did not affect virulence (data not shown).
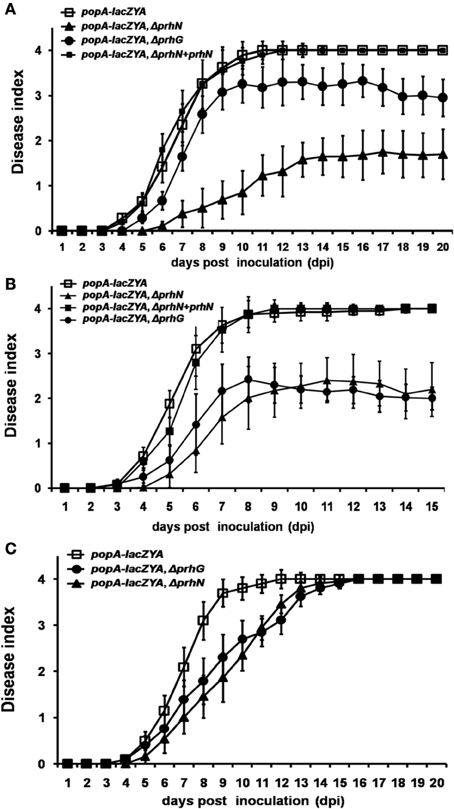
Figure 4. Pathogenicity test of OE1-1 derived strains. A bacterial suspension (108 cfu ml−1) was poured onto the soil supporting growth of plants (A) tomato and (C) tobacco plants at a final concentration of 107 cfu g−1 of soil. (B) 2μ l of bacterial suspension (108 cfu ml−1) was dropped onto freshly cut surfaces of tomato petioles. RK5050 (popA-lacZYA), squares; RK5601 (popA-lacZAY, ΔprhN), closed triangles; RK5185 (popA-lacZAY, ΔprhG), closed circles; and RK5701 (complementation, popA-lacZAY, ΔprhN+prhN), closed squares. Plants were inspected daily for wilt symptoms, and scored on a disease index scale from 0 to 4 (0, no wilting; 1, 1–25% wilting; 2, 26–50% wilting; 3, 51–75% wilting; and 4, 76–100% wilted or dead). Each trial included at least 12 plants for inoculation with one strain and mean values of at least four independent trials were presented in Miller units with SD (error bars).
When tomato plants were challenged using petiole inoculation, which directly introduces cells into the xylem, reduced virulence was also observed with prhN mutant (RK5601), and it exhibited almost identical virulence to that of the prhG mutant (RK5185) (Figure 4B). Complementation with PrhN could completely recover the virulence of the prhN mutant to that of the wild-type strain (RK5050) (Figure 4B). The empty vector did not affect virulence (data not shown). We also evaluated the virulence of the prhN mutant (RK5601) in tobacco plants using soil-soaking inoculation. In contrast to the deleterious effect of the prhN deletion on bacterial virulence in tomato plants, prhN disruption only caused delayed symptom development in tobacco plants (Figure 4C).
We also tested the virulence of the prhN mutant (RK10010), (a derivative of RS1002, which causes disease in tomato plants and HR in tobacco leaves) in tomato plants. Using petiole inoculation, reduced virulence was also observed with the prhN mutant (RK10010) in tomato plants, and complementation with PrhN could completely restore its virulence to that of wild-type (Figure 5A). The empty vector did not affect virulence (data not shown). All of these results demonstrate that PrhN is important for full virulence of R. solanacearum in host plants, and it is not strain-specific.
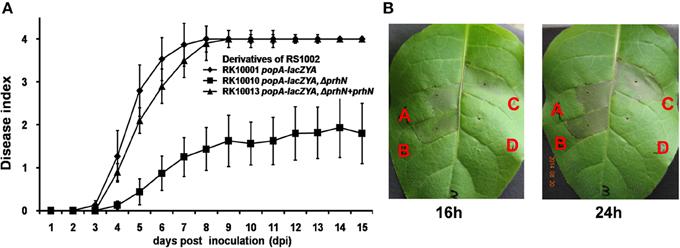
Figure 5. Pathogenicity and HR test of RS1002 derived strains. (A) Pathogenicity test in tomato plants. 2 μ l of bacterial suspension (108 cfu ml−1) was dropped onto freshly cut surfaces of tomato petioles. RK10001(popA-lacZYA), closed rhombus; RK10010 (popA-lacZYA, ΔprhN), closed squares; and RK10013 (complementation, popA-lacZAY, ΔprhN+prhN), closed triangles. Each trial included at least 12 plants for inoculation with one strain and mean values of at least four independent trials were presented in Miller units with SD (error bars). (B) HR test in tobacco leaves. Approximately 50 μ l of bacterial suspension at 108 cfu ml−1 was infiltrated into leaf mesophyll tissue with a blunt-end syringe. (A) RS1002; (B) RK10001 (popA-lacZYA); (C) RK10010 (popA-lacZYA, ΔprhN); and (D) distilled water. Pictures of tobacco leaves were taken at 16 and 24 h post infiltration (hpi). Each trial included four replications and three independent trials were performed. Results presented were from a representative experiment. Similar results were obtained in all other independent experiments.
Since RS1002 elicits a HR in tobacco leaves, we evaluated the effect of PrhN on HR elicitation of RS1002. We infiltrated N. tabacum BY leaves with RS1002, RK10001 (RS10002, popA-lacZYA) and RK10010 (RS1002, popA-lacZYA, Δ phcA), and evaluated the development of necrotic lesions. All three strains were able to cause necrotic lesions (Figure 5B). Lesions appeared at about 16 h post infiltration (hpi) and were maximal at about 24 hpi (Figure 5B). No obvious differences in the development of necrotic lesions were observed between these three strains, suggesting that PrhN is not involved in the HR elicitation by RS1002 in tobacco leaves.
PrhN is Involved in Bacterial Growth In Planta
The prhN mutant (RK5601) is less virulent in tomato plants and it exhibited similar bacterial morphology and growth profiles as did the wild-type strain in B and sucrose media (data not shown). Therefore, we evaluated the bacterial growth of the prhN mutant (RK5601) in planta. We inoculated tomato plants with RK5050 (popA-lacZYA), RK5185 (popA-lacZYA, Δ prhG), RK5601 (popA-lacZYA, Δ prhN), and RK5701 (popA-lacZYA, Δ prhN+prhN) by soil-soak and quantified bacterial cells in stem tissue. As reported previously, the prhG mutant (RK5185) and the wild-type strain (RK5050) can grow well in tomato stems and they proliferated in stem tissues to approximately 108 cfu g−1 at 4 dpi (wilt symptoms reached ~1), and approximately 1010 cfu g−1 at 7 dpi (wilt symptoms reached ~3–4) (Figure 6). The prhN mutant (RK5601) proliferated in the stem to approximately 107 cfu g−1 at 4 dpi and approximately 5 × 108 cfu g−1 at 7 dpi. The prhN mutant (RK5601) proliferated in planta significantly less than did the wild-type bacteria at a difference of one to two orders of magnitude (p < 0.01). Complementation with PrhN could completely restore the bacterial growth of the prhN mutant (RK5601) to that of wild-type bacteria in planta (tomato plants). These results suggest that PrhN is a pathogenicity regulator and the growth deficiency of the prhN mutant in planta might be a cause of its reduced virulence.
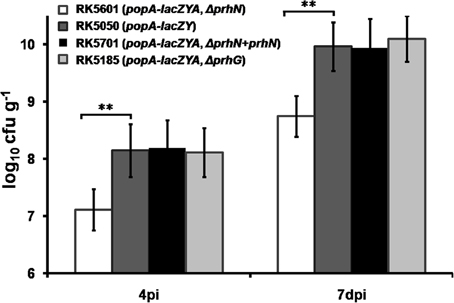
Figure 6. Bacterial growth of prhN mutant in tomato stems. Tomato plants were inoculated with bacteria using a soil-soak method at a final concentration of 107 cfu g−1 of soil. Stem species (approximately 5 cm above the soil and 1 cm in length) were removed at 4 dpi (wilt symptoms reached 1–2) and 7 dpi (wilt symptoms reached 3–4). The cell number was quantified by dilution plating. RK5601 (popA-lacZYA, ΔprhN), white bars; RK5050 (wild type), grayish bars; RK5701 (complementation), dark bars; and RK5185 (popA-lacZYA, ΔprhG), gray bars. Each trial included at least six plants for inoculation with one strain and mean values of at least four independent trials were presented in RLU cell−1 with SD (error bars). **P < 0.01.
PrhN is Important for popA Expression in Tomato Plants but Not in Tobacco Plants
Since PrhG is involved in popA expression of OE1-1 in tomato plants (Zhang et al., 2013), we evaluated the effect of PrhN on popA expression in planta (tomato and tobacco). Strains RK5050 (wild-type), RK5185 (ΔprhG), RK5601 (ΔprhN), and RK5701 (ΔprhN+prhN) were used to inoculate tomato plants using petiole inoculation and bacterial cells were collected from stems periodically for β-galactosidase assay. Since hrpB and hrpG mutants completely lost virulence and bacterial growth in planta, we didn't use them for comparison with prhG and prhN mutants. At 1 dpi, cell numbers in the stem was low, and popA expression in the stem was undetectable. PopA expression in RK5050 increased at 2 and 3 dpi, and then decreased to the basal level at 4 dpi. The prhN mutant showed significantly reduced popA expression at 2 and 3 dpi compared to that of RK5050 (wild-type) in stem xylem vessels (Figure 7A, p < 0.05 at 2 dpi, and p < 0.01 at 3 dpi). As reported previously, the prhG mutant (RK5185) also showed significantly reduced popA expression at 2 and 3 dpi, and is slightly lower than that of the prhN mutant (RK5601) (Figure 7A).
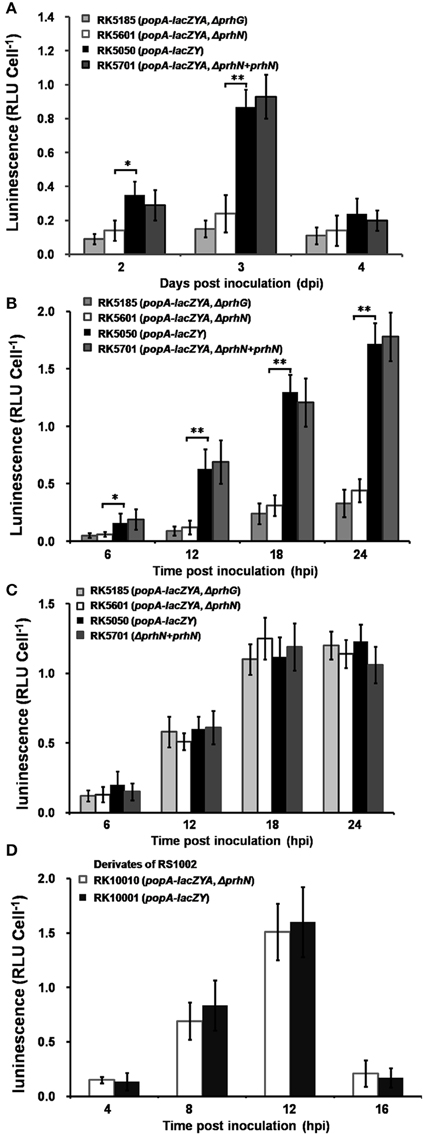
Figure 7. popA expression in planta. Strains RK5185, RK5601, RK5050, and RK5701 were inoculated into tomato stems using petiole inoculation method (A) and infiltrated into tomato leaves (B) and tobacco leaves (C). Stem pieces were removed every day and leaf discs were removed every 6 hpi, and bacterial cells was measured using Galacto-Light Plus. Strains RK10001 and RK10010 were infiltrated into tobacco leaves (D). Tobacco leaf discs were removed every 4 hpi, and bacterial cells was collected for β-galactosidase assay. Each assay included at least three samples from different plants and mean values of at least four independent trials were presented in RLU cell−1 with SD (error bars). *P < 0.05; **P < 0.01.
We also evaluated popA expression in leaf intercellular spaces. Strains RK5050 (wild type), RK5185 (ΔprhG), RK5601 (ΔprhN), and RK5701 (ΔprhN+prhN) were infiltrated into tomato and tobacco leaves and cells were recovered from infiltrated leaves for β-galactosidase assay. PopA expression in RK5050 was undetectable at time 0, and it increased at 12, 18, and 24 hpi. In tomato leaves, the prhN mutant (RK5601) showed significantly reduced popA expression compared to that of RK5050 (Figure 7B, p < 0.05 at 6 hpi and p < 0.01 at 12, 18, 24 hpi). The prhG mutant (RK5185) also showed reduced popA expression, but it is slightly lower than that of the prhN mutant (RK5601) in tomato leaves (Figure 7B). As reported previously (Zhang et al., 2013), the prhG mutant (RK5185) did not show a significant difference in popA expression in tobacco leaves from that in wild-type (Figure 7C). In contrast to tomato leaves, no significant difference was observed between the prhN mutant (RK5601) and wild-type bacteria in tobacco leaves (Figure 7C).
Since RK10001 and RK10010 (ΔprhN) showed almost equal HR elicitation in tobacco leaves, we evaluated popA expression in a prhN mutant (RK10010) in tobacco leaves. RK10001 (wild-type) or the prhN mutant (RK10010) were infiltrated into tobacco leaves and popA expression was evaluated. Necrotic lesions appeared at 16 hpi in tobacco leaves, indicating bacterial cells began to die at this time, and therefore we collected bacterial cells from infiltrated tobacco leaves every 4 h from 4 hpi. PopA expression in RK10001 increased at 4, 8, and 12 hpi, and then decreased quickly at 16 hpi. The prhN mutant (RK10010) showed almost identical popA expression levels as that in wild-type bacteria (Figure 7D), reflecting that the prhN mutant is equivalent in HR elicitation in tobacco leaves to that in wild-type bacteria. These results suggest that PrhN is a regulator of popA expression in tomato plants, and that the reduced popA expression of prhN mutants in planta might be another cause of its reduced virulence in tomato plants, but this control is host plant-specific.
Discussion
The MarR regulator was originally identified as repressor of multiple antibiotic resistance in E coli, and variants are widely distributed in bacteria and involved in the regulation of various cellular processes, including pathogenesis by both plant and human pathogens (Alekshun et al., 2001; Ellison and Miller, 2006). In the present study, we have genetically demonstrated that prhN (rsc1721 in GMI1000 that encodes a putative MarR-type regulator) is involved in the pathogenesis of R. solanacearum in tomato plants. MarR members control expression of some virulence-related genes or virulence associated traits; however, their regulatory targets vary in different pathogens (Ellison et al., 2004; Wilkinson and Grove, 2004).
In R. solanacearum, the hrp regulon and its encoding T3SS are essential for pathogenicity in host plants and for HR elicitation in resistant or non-host plants (Van Gijsegem et al., 1995; Coll and Valls, 2013). The expression of the hrp regulon and type III effectors are directly controlled by HrpB. HrpB controls multiple virulence pathways, including chemotaxis, biosynthesis or catabolism of various low-molecular-weight chemical compounds, and siderophore production and uptake (Jacobs et al., 2012; Coll and Valls, 2013). The expression of hrpB is activated by both HrpG and PrhG in a cell density-dependent manner both in vitro and in planta (Plener et al., 2010; Zhang et al., 2013). HrpB and HrpG are well known to be master regulators since hrpB and hrpG mutants completely lose virulence and bacterial growth in host plants (Vasse et al., 2000; Yoshimochi et al., 2009b). Despite the high similarity (72% global identity), PrhG displays a quite distinct role from HrpG. The prhG mutant exhibits equivalent bacterial growth in planta as do wild-type bacteria and is just slightly less virulent compared to that of wild-type bacteria in tomato plants (Plener et al., 2010; Zhang et al., 2013). Another remarkable difference between PrhG and HrpG is that their expression is positively and negatively regulated by PhcA, respectively. In this study, we genetically demonstrated that PrhN positively regulates prhG expression (only in hrp-inducing conditions), but it is dispensable for hrpG expression. This, in turn, results in the positive regulation of the expression of hrpB and popA (representative of a T3SS and the hrp regulon). PrhG expression increases constantly with cell density and is substantially reduced in phcA deletion mutants in both rich medium and hrp-inducing medium (Zhang et al., 2013). Although, PrhN positively regulates prhG expression only in hrp-inducing conditions, the expression manner of prhG at different cell densities is consistent with that of prhG. It was previously reported that PhcA positively regulates prhG expression (Zhang et al., 2013), while PhcA does not control prhN expression. The phcA deletion caused substantially reduced prhG expression (45 vs. 455 Miller units in rich medium and 115 vs. 2533 Miller units in hrp-inducing condition). The prhN deletion reduced prhG expression to 256 Miller units only in hrp-inducing conditions (Figure 2A). PrhG expression in prhN mutants was slightly higher than that in phcA mutants in hrp-inducing conditions (Figure 2A). Moreover, prhN mutants retain the same cellular morphology as wild-type bacteria (typical abundant mucoid EPS), and are totally different from that of phcA mutants, which usually results in a non-mucoid colony morphology because of deficient EPS production. PhcA and PrhN positively regulate prhG expression in a parallel way. HrpG is well known to be involved in the pathway triggered by plant signals or some mimic plant signals. PrhG is mainly involved in pathways triggered by metabolites and cell density (Plener et al., 2010). prhN expression maintained an almost equivalent level in rich medium, hrp-inducing medium, and co-cultivation with A. thaliana seedlings (Figure 3A), suggesting that PrhN may be not involved in the response to different environmental conditions.
It was previously reported that the expression of popA and hrpB was also reduced in prhG mutants in planta (Zhang et al., 2013). Here we report that PrhN is also important for the expression of popA and hrpB in vitro and in planta (only in tomato plants). Moreover, PrhG was dispensable for bacterial growth in planta, while the prhN mutant proliferated in tomato stems significantly less efficiently (one or two orders of magnitude) than wild-type bacteria (Figure 6). These results suggest that PrhN might control not only prhG expression (and in turn the hrp regulon) but also some unknown pathogenicity determinants. As reported previously, the prhG mutant was slightly less virulent than were wild-type bacteria when challenged tomato plants using soil-soaking inoculation, and it is obviously less virulent than wild-type bacteria on challenged tomato plants using petiole inoculation (Figures 4A,B). The prhN mutant always exhibited less virulence than wild-type bacteria when tomato plants were challenged using both methods. Although, the prhN mutant exhibited almost equivalent virulence as did the prhG mutant when tomato plants were challenged using petiole inoculation (Figure 4B), the prhN mutant exhibited much less virulence than did the prhG mutant when tomato plants were challenged using soil-soaking inoculation (Figure 4A). PrhG expression is positively controlled by PhcA, which becomes active in a cell density-dependent manner (Clough et al., 1997). PrhG mainly functions at high cell density (Zhang et al., 2013). PrhN expression increased constantly with cell density, and it was not regulated by PhcA, suggesting that PrhN might function during the whole infection process. Taken together, these data may explain why the prhN mutant exhibited less virulence than do wild-type bacteria in tomato plants, while the prhG mutant just exhibited less virulence on tomato plants using petiole inoculation.
In tobacco plants, PrhN contributes little to popA expression and disease development in strain OE1-1, suggesting that PrhN might play different roles in disease development in different plants. PrhN positively regulates prhG expression and PrhG was also reported to play different roles in disease development in different plants (Zhang et al., 2013). The contribution of PrhG and PrhN to virulence is consistent in tobacco plants. In strain OE1-1, it was also reported that prhK, prhL, and prhM mutant strains completely lost pathogenicity toward tomato plants, while they were slightly less virulent toward tobacco plants (Zhang et al., 2011). It seems common that some virulence regulators in OE1-1 play different roles in disease development on different plants.
Another R. solanacearum strain RS1002 causes disease in tomato and HR in tobacco leaves. Here, we provide evidence that PrhN does play an important role in disease development of RS1002 in tomato plants. Similar to strain OE1-1, prhN mutants in RS1002 exhibited significantly reduced popA expression under hrp-inducing conditions and obviously reduced virulence in tomato plants. In tobacco leaves, RS1002 and the prhN mutant showed almost equivalent popA expression level. This result might be able to explain why RS1002 and its prhN mutant showed almost identical HR development in tobacco leaves.
MarR family members display various biological functions in many bacteria and archaea. Some MarR family members are involved in the detoxification of aromatic compounds and the regulation of the expression of ferulic catabolic genes (Fiorentino et al., 2007; Calisti et al., 2008). In D. dadantii 3937, the MarR member MfbR is involved in the modulation of virulence gene expression in response to an acidic pH (Reverchon et al., 2009). In X. campestris pv. campestris, the MarR regulator HpaR is important for the production of an extracellular protease in hrp-inducing medium. HpaR is dispensable for the production of extracellular endoglucanase and amylase (Zou et al., 2012). Regulatory targets of MarR members usually vary in different pathogens (Perera and Grove, 2010). The regulatory mechanism of the MarR-type regulator PrhN remains unclear. We plan to elucidate further various biological functions of PrhN in R. solanacearum. We have genetically demonstrated that the newly characterized MarR-type regulator PrhN is important for hrp expression and full virulence of R. solanacearum in tomato plants. PrhN positively regulates the expression of prhG, hrpB and popA, but it is dispensable for the expression of hrpG and phcA. This finding will improve our understanding of complex regulatory pathway of the hrp regulon in R. solanacearum and the various biological functions of MarR regulators.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported in part by grants from the National Natural Science Foundation of China (31200067 to YZ) and the Fundamental Research Foundation for the Central Universities (XDJK2013C156 to YZ), in part by a key project of the China National Tobacco Corporation (110201202002 to WD), and in part by KAKENHI (Grant-in-Aid for Scientific Research) from the Japan Society for the Promotion of Science (16658020 to YH and 17380031 to KO).
References
Alekshun, M. N., Levy, S. B., Mealy, T. R., Seaton, B. A., and Head, J. F. (2001). The crystal structure of MarR, a regulator of multiple antibiotic resistance at 2.3 Å resolution. Nat. Struct. Biol. 8, 710–714. doi: 10.1038/90429
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Angot, A., Peeters, N., Lechner, E., Vailleau, F., Baud, C., Gentzbittel, L., et al. (2006). Ralstonia solanacearum requires F-box-like domain-containing type III effectors to promote disease on several host plants. Proc. Natl. Acad. Sci. U.S.A. 103, 14620–14625. doi: 10.1073/pnas.0509393103
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Arlat, M., Gough, C. L., Zischek, C., Barberis, P. A., Trigalet, A., and Boucher, C. A. (1992). Transcriptional organization and expression of the large hrp gene cluster of Pseudomonas solanacearum. Mol. Plant. Microbe Interact. 5, 187–193. doi: 10.1094/MPMI-5-187
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Calisti, C., Ficca, A. G., Barghini, P., and Ruzzi, M. (2008). Regulation of ferulic catabolic genes in Pseudomonas fluorescens BF13: involvement of a MarR family regulator. Appl. Microbiol. Biotechnol. 80, 475–483. doi: 10.1007/s00253-008-1557-4
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Choi, K. H., Gaynor, J. B., White, K. G., Lopez, C., Bosio, C. M., Karkhoff- Chweizer, R. R., et al. (2005). A Tn7-based broad range bacterial cloning and expression system. Nat. Methods 2, 443–448. doi: 10.1038/nmeth765
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Clough, S., Lee, K., Schell, M., and Denny, T. (1997). A two-component system in Ralstonia (Pseudomonas) solanacearum modulates the production of PhcA-regulated virulence factors in response to 3-hydroxypalmitic acid methyl ester. J. Bacteriol. 179, 3639–3648.
Coll, N. S., and Valls, M. (2013). Current knowledge on the Ralstonia solanacearum type III secretion system. Microb. Biotechnol. 6, 614–620. doi: 10.1111/1751-7915.12056
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Cornelis, G. R., and Van Gijsegem, F. (2000). Assembly and function of type III secretory systems. Annu. Rev. Microbiol. 54, 735–774. doi: 10.1146/annurev.micro.54.1.735
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Cunnac, S., Boucher, C. A., and Genin, S. (2004). Characterization of the cis-acting regulatory element controlling HrpB-mediated activation of the type III secretion system and effector genes in Ralstonia solanacearum. J. Bacteriol. 186, 2309–2318. doi: 10.1128/JB.186.8.2309-2318.2004
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ellison, D. W., Lawrenz, M. B., and Miller, V. L. (2004). Invasin and beyond: regulation of Yersinia virulence by RovA. Trends Microbiol. 12, 296–300. doi: 10.1016/j.tim.2004.04.006
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Ellison, D. W., and Miller, V. L. (2006). Regulation of virulence by members of the MarR/SlyA family. Curr. Opin. Microbiol. 9, 153–159. doi: 10.1016/j.mib.2006.02.003
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Fiorentino, G., Ronca, R., Cannio, R., Rossi, M., and Bartolucci, S. (2007). MarR-like transcriptional regulator involved in detoxification of aromatic compounds in Sulfolobus solfataricus. J. Bacteriol. 189, 7351–7360. doi: 10.1128/JB.00885-07
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Galán, J. E., and Wolf-Watz, H. (2006). Protein delivery into eukaryotic cells by type III secretion machines. Nature 444, 567–573. doi: 10.1038/nature05272
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Genin, S., Brito, B., Denny, T. P., and Boucher, C. (2005). Control of the Ralstonia solanacearum Type III secretion system (Hrp) genes by the global virulence regulator PhcA. FEBS Lett. 579, 2077–2081. doi: 10.1016/j.febslet.2005.02.058
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Gohre, V., and Robatzek, S. (2008). Breaking the barriers: microbial effector molecules subvert plant immunity. Annu. Rev. Phytopathol. 46, 189–215. doi: 10.1146/annurev.phyto.46.120407.110050
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Haque, M. M., Kabir, M. S., Aini, L. Q., Hirata, H., and Tsuyumu, S. (2009). SlyA, a MarR family transcriptional regulator, is important for virulence in Dickeya dadantii 3937. J. Bacteriol. 191, 5409–5418. doi: 10.1128/JB.00240-09
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Jacobs, J. M., Babujee, L., Meng, F., Milling, A., and Allen, C. (2012). The in planta transcriptome of Ralstonia solanacearum: conserved physiological and virulence strategies during bacterial wilt of tomato. MBio. 3:e00114–e00112. doi: 10.1128/mBio.00114-12
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Kanda, A., Ohnishi, S., Tomiyama, H., Hasegawa, H., Yasukohchi, M., Kiba, A., et al. (2003). Type III secretion machinery-deficient mutants of Ralstonia solanacearum lose their ability to colonize resulting in loss of pathogenicity. J. Gen. Plant Pathol. 69, 250–257. doi: 10.1007/s10327-003-0041-3
Mansfield, J., Genin, S., Magori, S., Citovsky, V., Sriariyanum, M., and Ronald, P. (2012). Top 10 plant pathogenic bacteria in molecular plant pathology. Mol. Plant Pathol. 13, 614–629. doi: 10.1111/j.1364-3703.2012.00804.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Monteiro, F., Genin, S., Van Dijk, I., and Valls, M. (2012). A luminescent reporter evidences active expression of Ralstonia solanacearum type III secretion system genes throughout plant infection. Microbiology 158, 2107–2116. doi: 10.1099/mic.0.058610-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mukaihara, T., Tamura, N., and Iwabuchi, M. (2010). Genome-wide identification of a large repertoire of Ralstonia solanacearum type III effector proteins by a new functional screen. Mol. Plant Microbe Interact. 23, 251–262. doi: 10.1094/MPMI-23-3-0251
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Mukaihara, T., Tamura, N., Murata, Y., and Iwabuchi, M. (2004). Genetic screening of Hrp type III-related pathogenicity genes controlled by the HrpB transcriptional activator in Ralstonia solanacearum. Mol. Microbiol. 54, 863–875. doi: 10.1111/j.1365-2958.2004.04328.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Peeters, N., Guidot, A., Vailleau, F., and Valls, M. (2013). Ralstonia solanacearum, a widespread bacterial plant pathogen in the post-genomic era. Mol. Plant Pathol. 14, 651–662. doi: 10.1111/mpp.12038
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Perera, I. C., and Grove, A. (2010). Molecular mechanisms of ligand-mediated attenuation of DNA binding by MarR family transcriptional regulators. J. Mol.Cell Biol. 5, 243–254. doi: 10.1093/jmcb/mjq021
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Plener, L., Manfredi, P., Valls, M., and Genin, S. (2010). PrhG, a transcriptional regulator responding to growth conditions, is involved in the control of the type III secretion system regulon in Ralstonia solanacearum. J. Bacteriol. 192, 1011–1019. doi: 10.1128/JB.01189-09
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Poueymiro, M., and Genin, S. (2009). Secreted proteins from Ralstonia solanacearum: a hundred tricks to kill a plant. Curr. Opin. Microbiol. 12, 44–52. doi: 10.1016/j.mib.2008.11.008
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Reverchon, S., Van-Gijsegem, F., Effantin, G., Zghidi-Abouzid, O., and Nasser, W. (2009). Systematic targeted mutagenesis of the MarR/SlyA family members of Dickeya dadantii 3937 reveals a role for MfbR in the modulation of virulence gene expression in response to acidic pH. Mol. Microbiol. 78, 1018–1037. doi: 10.1111/j.1365-2958.2010.07388.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Salanoubat, M., Genin, S., Artiguenave, F., Gouzy, J., Mangenot, S., Arlat, M., et al. (2002). Genome sequence of the plant pathogen Ralstonia solanacearum. Nature 415, 497–502. doi: 10.1038/415497a
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Scḧafer, A., Tauch, A., Jager, W., Kalinowski, J., Thierbach, G., and Pühler, A. (1994). Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145, 69–73. doi: 10.1016/0378-1119(94)90324-7
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Seoane, A. S., and Levy, S. B. (1995). Characterization of MarR, the repressor of the multiple antibiotic resistance (mar) operon in Escherichia coli. J Bacteriol. 177, 3414–3419.
Simon, R., Priefer, U., and Pühler, A. (1983). A broad host range mobilization system for in vivo genetic engineering: transpo son mutagenesis in gram negative bacteria. Nat. Biotechnol. 1, 784–791. doi: 10.1038/nbt1183-784
Stapleton, M. R., Norte, V. A., Read, R. C., and Green, J. (2002). Interaction of the Salmonella typhimurium transcription and virulence factor SlyA with target DNA and identification of members of the SlyA regulon. J. Biol. Chem. 277, 17630–17637. doi: 10.1074/jbc.M110178200
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Valls, M., Genin, S., and Boucher, C. (2006). Integrated regulation of the type III secretion system and other virulence determinants in Ralstonia solanacearum. PLoS Pathog. 2:e82. doi: 10.1371/journal.ppat.0020082
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Van Gijsegem, F., Gough, C., Zischek, C., Niqueux, E., Arlat, M. F., and Boucher, C. (1995). The hrp locus of Pseudomonas solanacearum, which controls the production of a type III secretion system, encodes eight proteins related to components of the bacterial flagellar biogenesis complex. Mol. Microbiol. 15, 1095–1114. doi: 10.1111/j.1365-2958.1995.tb02284.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Vasse, J., Genin, S., Frey, P., Boucher, C., and Brito, B. (2000). The hrpB and hrpG regulatory genes of Ralstonia solanacearum are required for different stages of the tomato root infection process. Mol. Plant Microbe Interact. 13, 259–267. doi: 10.1094/MPMI.2000.13.3.259
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wei, K., Tang, D. J., He, Y. Q., Feng, J. X., Jiang, B. L., Lu, G. T., et al. (2007). hpaR, a putative MarR family transcriptional regulator, is positively controlled by HrpG and HrpX and involved in the pathogenesis, hypersensitive response, and extracellular protease production of Xanthomonas campestris pathovar campestris. J. Bacteriol. 189, 2055–2062. doi: 10.1128/JB.01331-06
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wilkinson, S. P., and Grove, A. (2004). HucR, a novel uric acid-responsive member of the MarR family of transcriptional regulators from Deinococcus radiodurans. J. Biol. Chem. 279, 51442–51450. doi: 10.1074/jbc.M405586200
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Wilkinson, S. P., and Grove, A. (2006). Ligand-responsive transcriptional regulation by members of the MarR family of winged helix proteins. Curr. Issues Mol. Biol. 8, 51.
Yao, J., and Allen, C. (2007). The plant pathogen Ralstonia solanacearum needs aerotaxis for normal biofilm formation and interactions with its tomato host. J. Bacteriol. 189, 6415–6424. doi: 10.1128/JB.00398-07
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yoshimochi, T., Hikichi, Y., Kiba, A., and Ohnishi, K. (2009a). The global virulence regulator PhcA negatively controls the Ralstonia solanacearum hrp regulatory cascade by repressing expression of the PrhIR signaling proteins. J. Bacteriol. 191, 3424–3428. doi: 10.1128/JB.01113-08
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Yoshimochi, T., Zhang, Y., Kiba, A., Hikichi, Y., and Ohnishi, K. (2009b). Expression of hrpG and activation of response regulator HrpG are controlled by distinct signal cascades in Ralstonia solanacearum. J. Gen. Plant. Pathol. 75, 196–204. doi: 10.1007/s10327-009-0157-1
Zhang, Y., Chen, L., Takehi, Y., Kiba, A., Hikichi, Y., and Ohnishi, K. (2013). Functional analysis of Ralstonia solanacearum PrhG regulating the hrp regulon in host plants. Microbiology 159, 1695–1704. doi: 10.1099/mic.0.067819-0
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zhang, Y., Kiba, A., hikichi, Y., and Ohnishi, K. (2011). prhKLM genes of Ralstonia solanacearum encode novel activators of hrp regulon and are required for pathogenesis in tomato. FEMS Microbiol. Lett. 317, 75–82. doi: 10.1111/j.1574-6968.2011.02213.x
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zou, L., Zeng, Q., Lin, H., Gyaneshwar, P., Chen, G., and Yang, C. H. (2012). SlyA Regulates Type III Secretion System (T3SS) Genes in Parallel with the T3SS Master Regulator HrpL in Dickeya dadantii 3937. Appl. Environ. Microbiol. 78, 2888–2895. doi: 10.1128/AEM.07021-11
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Zuluaga, A. P., Puigvert, M., and Valls, M. (2013). Novel plant inputs influencing Ralstonia solanacearum during infection. Front. Microbiol. 4:349. doi: 10.3389/fmicb.2013.00349
PubMed Abstract | Full Text | CrossRef Full Text | Google Scholar
Keywords: R. solanacearum, type III secretion system, Hrp regulon, pathogenesis, PrhN, MarR type regulator
Citation: Zhang Y, Luo F, Wu D, Hikichi Y, Kiba A, Igarashi Y, Ding W and Ohnishi K (2015) PrhN, a putative marR family transcriptional regulator, is involved in positive regulation of type III secretion system and full virulence of Ralstonia solanacearum. Front. Microbiol. 6:357. doi: 10.3389/fmicb.2015.00357
Received: 10 December 2014; Accepted: 09 April 2015;
Published: 28 April 2015.
Edited by:
Stanton B. Gelvin, Purdue University, USAReviewed by:
Stephane Genin, Institut National de la Recherche Agronomique- Centre National de la Recherche Scientifique, FranceChiu-Ping Cheng, National Taiwan University, Taiwan
Copyright © 2015 Zhang, Luo, Wu, Hikichi, Kiba, Igarashi, Ding and Ohnishi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Ding, College of Plant Protection, Southwest University, BeiBei, Chongqing 400715, China,ZHdpbmc4MThAMTYzLmNvbQ==;
Kouhei Ohnishi, Research Institute of Molecular Genetics, Kochi University, Nankoku, Kochi 783-8502, Japan,a291aGVpb0Brb2NoaS11LmFjLmpw
†These authors have contributed equally to this work.
 Yong Zhang
Yong Zhang Feng Luo1†
Feng Luo1† Kouhei Ohnishi
Kouhei Ohnishi