- 1Institute of Hygiene, Microbiology and Environmental Medicine, Medical University of Graz, Graz, Austria
- 2Institute for Medical Informatics, Statistics and Documentation, Medical University of Graz, Graz, Austria
The aim of the study was to analyze the antimicrobial susceptibility of Austrian clinical Klebsiella sp. and Enterobacter sp. isolates linked to patient-related data over a time period from 1998 to 2014. The main findings of this study were (i) a marked difference of antibiotic susceptibility rates between different infection sites for both Klebsiella sp. and Enterobacter sp., (ii) significantly greater percentages of resistant isolates among both Klebsiella sp. and Enterobacter sp. in male patients compared to female patients and (iii) significantly greater percentages of resistant isolates among both Klebsiella sp. and Enterobacter sp. from hospital-derived samples compared to samples from the community. In conclusion, our statistical data analysis clearly indicated a strong association of patient-related data and Klebsiella sp. and Enterobacter sp. susceptibility profiles.
Introduction
Whereas the antibiotic resistance crisis in Gram-positive pathogens largely seems to be kept under control, the emergence and dissemination of antibiotic resistance in Gram-negative pathogens risk getting out of control (Arya et al., 2008; Slama, 2008; Bush et al., 2011; Livermore, 2012). To gain useful information on overall trends in a specific region and alert for more effective prevention and control measures, surveillance studies based on large data sets are of particular importance in this field (World Health Organization [WHO], 2014). Multivariate statistical analysis further allows studying possible relationships between antimicrobial resistance data and patient-related factors; however, there are only a limited number of studies up to now on that issue mainly dealing with Escherichia coli. The main findings of these studies are that susceptibilities to several antimicrobial agents are strongly depending on infection site as well as patient location (hospital/community), gender and age (Livermore et al., 2003; Miguel Sahuquillo-Arce et al., 2011; Badura et al., 2015). Under the hypothesis that the same is true for Klebsiella sp. and Enterobacter sp., the aim of this study is to analyze a large amount of antibiotic resistance data (almost 39.000 isolates) from our geographical region according to patient-related factors. The application of multivariate statistical analysis enables us to detect significant associations between patient’s characteristics and the percentages of resistant isolates what is of key importance to further identify possible risk factors related to antibiotic-resistant Klebsiella and Enterobacter infections.
Materials and Methods
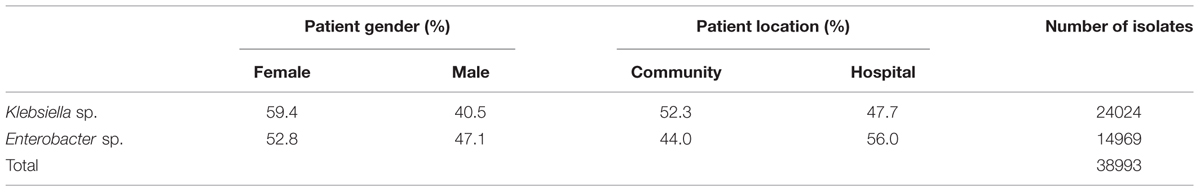
A total of 24.024 Klebsiella sp. and 14.969 Enterobacter sp. isolates were included in the present analysis. The study design of this retrospective observational study (data origin, laboratory methods, and statistical analysis) is largely similar to our previous report on E. coli (Badura et al., 2015). Data were retrieved from the Laboratory of Medical Bacteriology and Mycology of the Medical University of Graz, Austria from January 1998 to March 2014. In the event of multiple isolates from one person in a year, only the first one was considered. For each isolate, patient–related data (age, gender), patient location (community/hospital) and culture site were obtained. Culture sites were subdivided into the following categories: urinary tract (UT), genital tract (GT), wounds (WOU), respiratory tract (RES), and blood (BLO); patient age was categorized as follows: <1, 1–9, 10–19, 20–29, 30–39, 40–49, 50–59, 60–69, 70–79, > = 80 years.
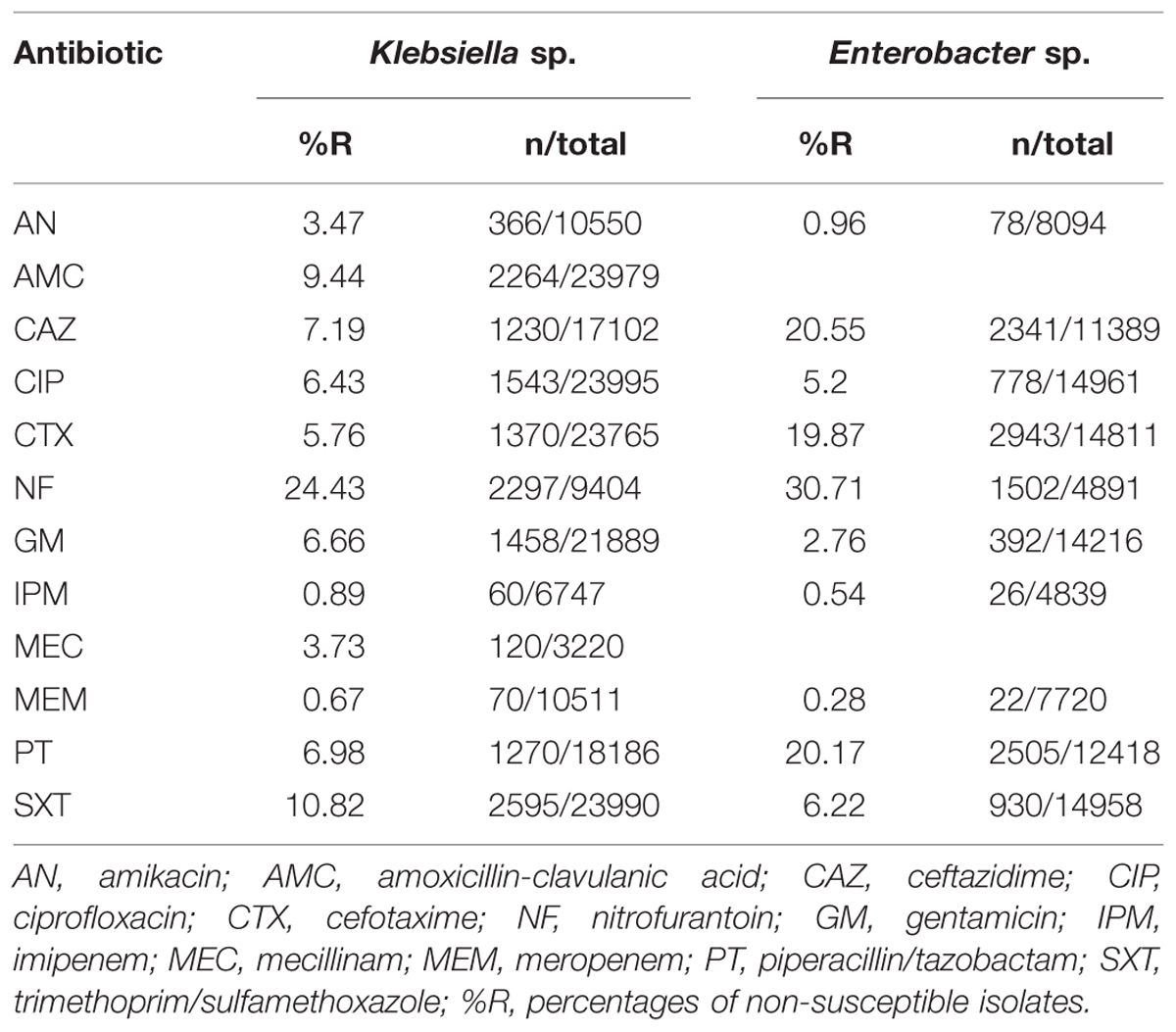
All isolates had been tested for antibiotic susceptibilities in the routine microbiology laboratory in the same way as reported in a previous study using the disk diffusion method or a VITEK 2 system (bioMérieux, Marcy l’Etoile France) (Badura et al., 2015). From January 1st, 1998 to May 31st, 2011 results were interpreted using the criteria recommended by the Clinical and Laboratory Standards Institute (CLSI), formerly known as National Committee for Clinical Laboratory Standards (NCCLS) (CLSI, 2012). From June 1st, 2011 results were interpreted using the criteria recommended by the European Committee on Antimicrobial Susceptibility Testing (EUCAST breakpoint tables v1.2, 2011) in its respective current version. For this study, resistance to the following 12 antibiotic agents was analyzed: amikacin (AN), amoxicillin-clavulanic acid (AMC; analyzed for Klebsiella sp. only), ceftazidime (CAZ), ciprofloxacin (CIP), cefotaxime (CTX), nitrofurantoin (NF; analyzed for urinary tract isolates only), gentamicin (GM), imipenem (IPM), mecillinam (MEC; analyzed for Klebsiella sp. urinary tract isolates only); meropenem (MEM), piperacillin/tazobactam (PT), and trimethoprim/sulfamethoxazole (SXT). Resistant and intermediate resistant isolates were combined.
Statistical Analysis
For each of the two microbes and for each antibiotic under consideration, the absolute and relative frequencies of resistant samples were determined in total, per culture site, patient location, age group, gender and year of acquisition. In order to assess the influence of the covariates on antibiotic resistance, multivariable logistic regression analysis was performed. For binary covariates, odds ratios and 95% confidence intervals are presented and are adjusted for year of acquisition, patient gender, patient age, patient location and resistance phenotype; for culture site a model containing the aforementioned covariates as well as cultures site has been tested against a model without culture site and p values from the respective Chi-square test are presented. Statistical analysis was performed using the R package, version 3.1.1 and SPSS, version 21.0 (R Development Core Team, 2008; IBM Corp, 2012).
Ethics Statement
The study was approved by the Ethics Committee of the Medical University of Graz (26-502 ex 13/14); patient records were anonymized prior to analysis.
Results
The underlying data basis for the present analysis is shown in Table 1. In total, 38.993 clinical isolates of Klebsiella sp. and Enterobacter sp. were included in the statistical analysis.
Species Distribution
Within the Klebsiella samples, K. pneumoniae (61%) and K. oxytoca (27.1%) were the most frequently recovered species; within the Enterobacter samples, E. cloacae complex (59.2%) and E. aerogenes (15.4%) were the most frequently recovered species.
Overall and Annual Resistance Percentages
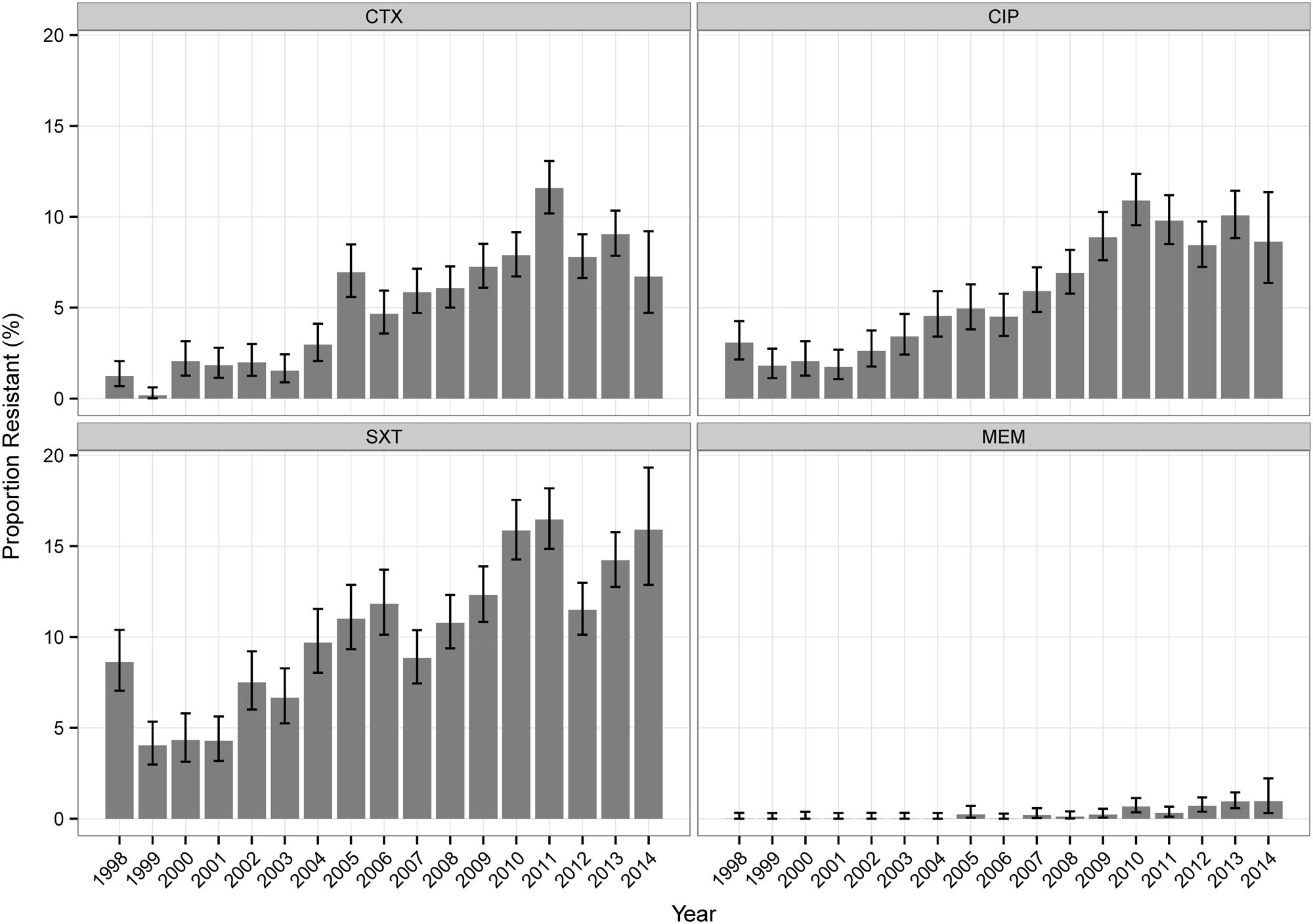
Table 2 shows the overall resistance percentages of all tested isolates. For Klebsiella sp. isolates, NF (analyzed for UT isolates only) and SXT showed the highest values, for Enterobacter sp. the highest values were found for NF, CAZ, PT, and CTX. For Klebsiella sp. isolates, most antibiotics (CAZ, CTX, NF, CIP, SXT, IPM, and MEM) showed increasing percentages of resistant isolates over time (Figure 1, Supplemental Table S1). The percentages of resistant Enterobacter sp. isolates did not change considerably over the investigational period for the majority of antibiotic agents (Supplemental Table S1).
Resistance Percentages According to Patient-Related Data
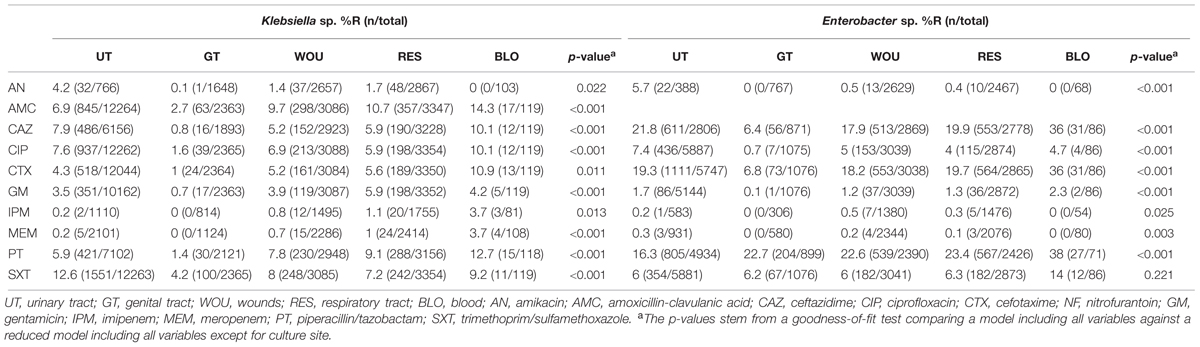
Results of the multivariate analysis of the antibiotic resistance patterns according to culture sites for both pathogens are presented in Table 3. The findings reveal the lowest resistance percentages from samples originating from the male or female genital tract (GT). For Klebsiella sp. isolates, the highest resistance percentages for almost all antibiotics analyzed occurred in blood infections. For Enterobacter, the highest resistance percentages were found in isolates originating from blood (CAZ, CTX, GM, PT, SXT), and urinary tract infections (AN, CIP) (Table 3).
Antibiotic resistance patterns in relation to patient gender are shown in Table 4. Resistance percentages for almost all antibiotic agents analyzed are higher in male patients. A significant difference can be shown for all antibiotics except for AN for Klebsiella sp. isolates and for AN, GM, IPM, MEM, and PT for Enterobacter sp., respectively. Antibiotic resistance patterns in relation to patient location are shown in Table 5. Resistance percentages for almost all antibiotic agents analyzed are significantly higher in hospital treated patients. This is true for both Klebsiella sp. and Enterobacter sp. isolates.
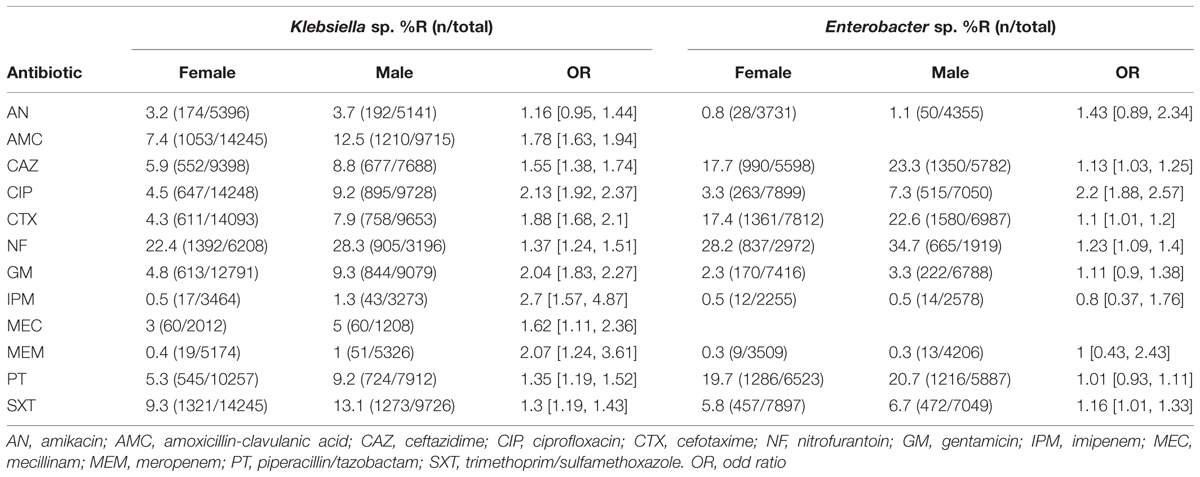
TABLE 4. Antibiotic resistance percentages of Klebsiella sp. and Enterobacter sp. according to patient gender.
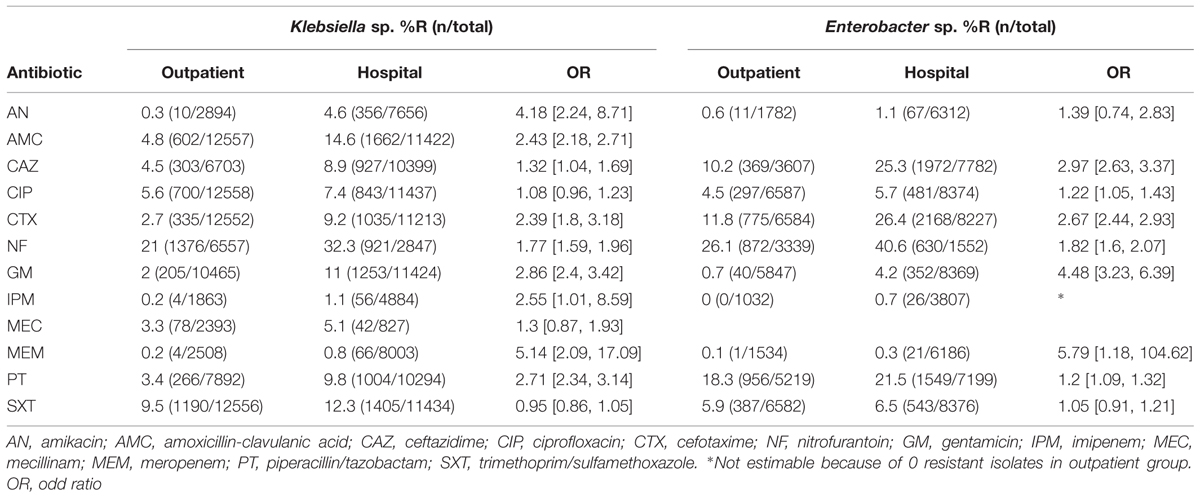
TABLE 5. Antibiotic resistance percentages of Klebsiella sp. and Enterobacter sp. according to patient location.
Regarding the association between antibiotic resistance percentages of Klebsiella sp. and Enterobacter sp. isolates and patient age, the majority of antibiotics show a balanced distribution except for CIP where a constant increase with age can be shown for both pathogen groups (Figure 2).
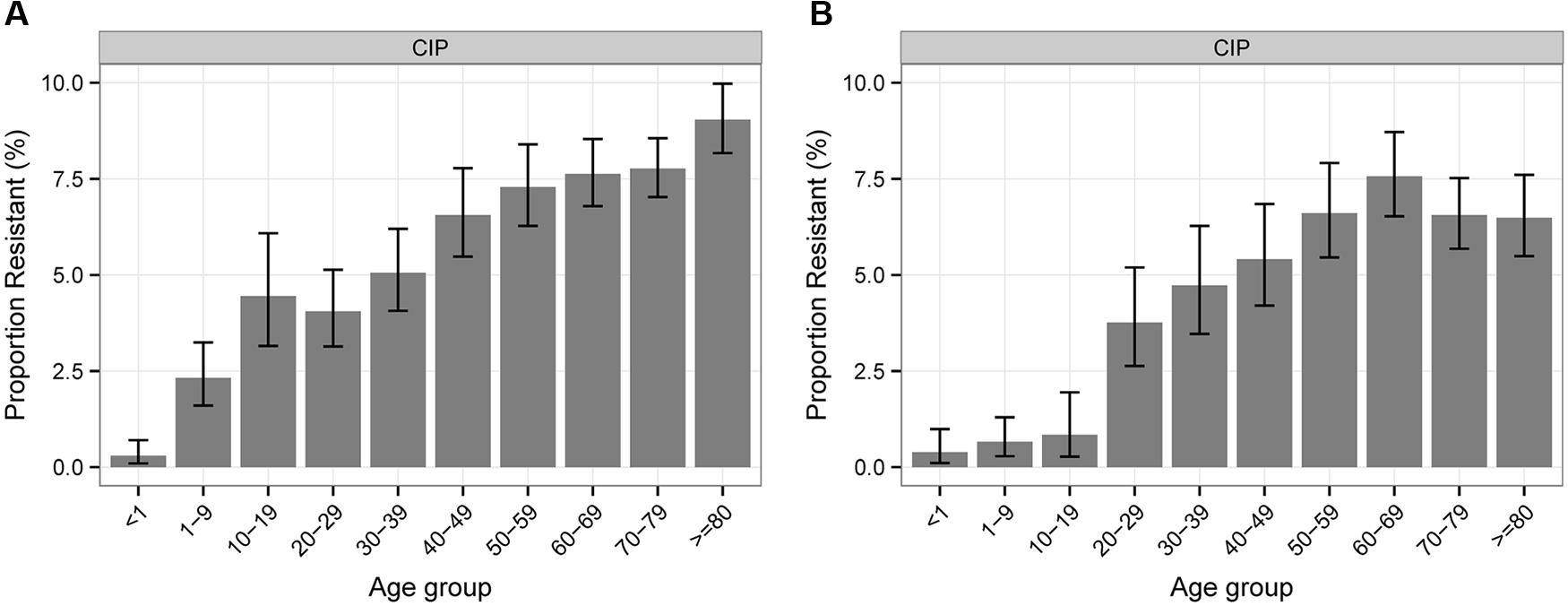
FIGURE 2. Percentages of antibiotic resistance of Klebsiella sp. (A) and Enterobacter sp. (B) to CIP according to patient age.
Discussion
We here report an observational study whose aim is to analyze the susceptibility of Austrian clinical Klebsiella sp. and Enterobacter sp. isolates linked to patient-related data. The underlying database for this statistical analysis covers the period from 1998 to 2014. A major limitation of the present study is that it was not possible to link the data to severity of disease and patient outcome as these data were not available. Additionally, information about the patient’s length of the hospital stay was not included in the analysis which would have been useful for the classification as hospital or community acquired infections. In the present work, infections were categorized depending on the patient’s location at the moment the clinical sample was obtained. The passage from CLSI to EUCAST guidelines in 2011 in Austria may further bias data comparability. Several authors have reported an increase of antibiotic resistance rates for certain Enterobacteriaceae species/drug combinations following EUCAST breakpoints implementation (Hombach et al., 2012; Wolfensberger et al., 2013). However, we did not observe any artificial rise of antibiotic resistance at that time indicating that the change of interpretation criteria does not apparently impact the results of the large data set analyzed.
In general, surveillance is a substantial basis in the combat against antimicrobial resistance in both community and hospital settings; a large number of resistance surveillance reports thus exist about Klebsiella sp. and Enterobacter sp. isolates from various parts of the world describing the current resistance situation (Jones et al., 2004; Bedenic et al., 2010; Gales et al., 2012; Badal et al., 2013; Kaye and Pogue, 2015). Furthermore, studies dealing with the association of antimicrobial resistance data and specific factors depending on the host are of major importance with regard to the identification of possible risk factors related to antibiotic-resistant infections. Several studies aimed to find various patient demographics and clinical factors associated with the presence of antibiotic-resistant Enterobacteriaceae isolates, however, most of them involve particular infections, patient cohorts, anatomic sites or specific resistance problems. These studies thus mostly cover a small amount of isolates consequently limiting statistical data processing (O’Fallon et al., 2010; Metan et al., 2013; Ivady et al., 2015; Moini et al., 2015; Oliveira et al., 2015). Here we present a comprehensive statistical analysis on associations between patient-related data and the percentages of antibiotic resistant Klebsiella sp. and Enterobacter sp. isolates referring on a large database with almost 39.000 isolates from Austria. The main findings of this study are (i) a marked difference of antibiotic susceptibility rates between different infection sites for both Klebsiella sp. and Enterobacter sp., (ii) significantly greater percentages of resistant isolates among both Klebsiella sp. and Enterobacter sp. in male patients compared to female patients and (iii) significantly greater percentages of resistant isolates among both Klebsiella sp. and Enterobacter sp. isolates from hospital-derived samples compared to samples from the community.
To our knowledge, this is the first report to compare percentages of antibiotic resistant Klebsiella sp. and Enterobacter sp. isolates from different infection sites within a specific region. The highest resistance percentages for the majority of antibiotics analyzed can be shown for both pathogen groups originating from blood infections. This is in accordance with previous statistical analyses on the association of the infection site and Escherichia coli susceptibility profiles (Miguel Sahuquillo-Arce et al., 2011; Badura et al., 2015). As reported for Escherichia coli, Klebsiella sp. and Enterobacter sp. isolates originating from the GT generally show the lowest resistance percentages indicating that detection from this culture site often represents the normal patient flora. Interestingly, antibiotic-resistant Klebsiella sp. and Enterobacter sp. infections within our study collection derive mostly from men. The same association between patient sex and antibiotic resistance was previously shown for Escherichia coli, however, the underlying cause remains as yet largely unclear (Livermore et al., 2003; Miguel Sahuquillo-Arce et al., 2011; Badura et al., 2015). Similar to the results from several other studies dealing with antibiotic resistance percentages of Enterobacteriaceae in community and hospital settings we found significantly greater resistance rates for both pathogen groups amongst the hospital-derived isolates suggesting that the problem of antibiotic resistance amongst Gram-negative pathogens is still focused on hospital settings in our region (Boyd et al., 2008; Ferjani et al., 2015; McKay and Bamford, 2015).
Conclusion
Our statistical data analysis clearly indicates a strong association between patient characteristics (gender, localization, infection site) and Klebsiella sp. and Enterobacter sp. susceptibility profiles. To gain further insight in the correlation of bacterial antibiotic resistance and its host, additional studies analyzing large databases of microbiological information are of crucial importance ultimately influencing local antimicrobial therapy guidelines.
Author Contributions
AB: general conception, study design, data acquisition, data analysis, data interpretation, writing of the manuscript; GF: data interpretation, revising of the manuscript; JH and GP: statistical analysis, data interpretation, revising of the manuscript; AG: data interpretation, revising of the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.00034
TABLE S1 | Annual resistance percentages of Klebsiella sp. and Enterobacter sp.
References
Arya, S. C., Agarwal, N., and Agarwal, S. (2008). Emerging Gram-negative antibiotic resistance: daunting challenges, declining sensitivities, and dire consequences. Respir. Care 53, 1749–1750; author reply 1750.
Badal, R. E., Bouchillon, S. K., Lob, S. H., Hackel, M. A., Hawser, S. P., and Hoban, D. J. (2013). Etiology, extended-spectrum beta-lactamase rates and antimicrobial susceptibility of gram-negative bacilli causing intra-abdominal infections in patients in general pediatric and pediatric intensive care units-global data from the study for monitoring antimicrobial resistance trends 2008 to 2010. Pediatr. Infect. Dis. J. 32, 636–640. doi: 10.1097/INF.0b013e3182886377
Badura, A., Feierl, G., Pregartner, G., Krause, R., and Grisold, A. J. (2015). Antibiotic resistance patterns of more than 120 000 clinical Escherichia coli isolates in Southeast Austria, 1998-2013. Clin. Microbiol. Infect. 21:569.e1-7. doi: 10.1016/j.cmi.2015.02.012
Bedenic, B., Goic-Barisic, I., Budimir, A., Tonkic, M., Mihajkevic, L. J., Novak, A., et al. (2010). Antimicrobial susceptibility and beta-lactamase production of selected gram-negative bacilli from two Croatian hospitals: MYSTIC study results. J. Chemother. 22, 147–152. doi: 10.1179/joc.2010.22.3.147
Boyd, L. B., Atmar, R. L., Randall, G. L., Hamill, R. J., Steffen, D., and Zechiedrich, L. (2008). Increased fluoroquinolone resistance with time in Escherichia coli from > 17,000 patients at a large county hospital as a function of culture site, age, sex, and location. BMC Infect. Dis. 8:4. doi: 10.1186/1471-2334-8-4.
Bush, K., Courvalin, P., Dantas, G., Davies, J., Eisenstein, B., Huovinen, P., et al. (2011). Tackling antibiotic resistance. Nat. Rev. Microbiol. 9, 894–896. doi: 10.1038/nrmicro2693
CLSI (2012) Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals; Approved StandardCLSI Document M31-A3, 3rd Edn. Wayne, PA: CLSI.
Ferjani, S., Saidani, M., Amine, F. S., and Boutiba Ben Boubaker, I. (2015). A comparative study of antimicrobial resistance rates and phylogenetic groups of community-acquired versus hospital-acquired invasive Escherichia coli. Med. Mal. Infect. 45, 133–138. doi: 10.1016/j.medmal.2015.01.012
Gales, A. C., Castanheira, M., Jones, R. N., and Sader, H. S. (2012). Antimicrobial resistance among Gram-negative bacilli isolated from Latin America: results from SENTRY Antimicrobial Surveillance Program (Latin America, 2008-2010). Diagn. Microbiol. Infect. Dis. 73, 354–360. doi: 10.1016/j.diagmicrobio.2012.04.007
Hombach, M., Bloemberg, G. V., and Böttger, E. C. (2012). Effects of clinical breakpoint changes in CLSI guidelines 2010/2011 and EUCAST guidelines 2011 on antibiotic susceptibility test reporting of Gram-negative bacilli. J. Antimicrob. Chemother. 67, 622-632 doi: 10.1093/jac/dkr524
Ivady, B., Kenesei, E., Toth-Heyn, P., Kertesz, G., Tarkanyi, K., Kassa, C., et al. (2015). Factors influencing antimicrobial resistance and outcome of Gram-negative bloodstream infections in children. Infection [Epub ahead of print]. doi: 10.1007/s15010-015-0857-8
Jones, M. E., Karlowsky, J. A., Draghi, D. C., Thornsberry, C, Sahm, D. F., and Bradley, J. S. (2004). Rates of antimicrobial resistance among common bacterial pathogens causing respiratory, blood, urine, and skin and soft tissue infections in pediatric patients. Eur. J. Clin. Microbiol. Infect. Dis. 23, 445–455. doi: 10.1007/s10096-004-1133-5
Kaye, K. S., and Pogue, J. M. (2015). Infections caused by resistant gram-negative bacteria: epidemiology and management. Pharmacotherapy 35, 949–962. doi: 10.1002/phar.1636
Livermore, D. M. (2012). Current epidemiology and growing resistance of gram-negative pathogens. Korean J. Intern. Med. 27, 128–142. doi: 10.3904/kjim.2012.27.2.128
Livermore, D. M., Nichols, T., Lamagni, T. L., Potz, N., Reynolds, R., and Duckworth, G. (2003). Ciprofloxacin-resistant Escherichia coli from bacteraemias in England; increasingly prevalent and mostly from men. J. Antimicrob. Chemother. 52, 1040–1042. doi: 10.1093/jac/dkg479
McKay, R., and Bamford, C. (2015). Community- versus healthcare-acquired bloodstream infections at Groote Schuur Hospital, Cape Town, South Africa. S. Afr. Med. J. 105, 363–369. doi: 10.7196/samj.8183
Metan, G., Demiraslan, H., Kaynar, L. G., Zararsiz, G., Alp, E., and Eser, B. (2013). Factors influencing the early mortality in haematological malignancy patients with nosocomial Gram negative bacilli bacteraemia: a retrospective analysis of 154 cases. Braz. J. Infect. Dis. 17, 143–149. doi: 10.1016/j.bjid.2012.09.010
Miguel Sahuquillo-Arce, J., Selva, M., Perpinan, H., Gobernado, M., Armero, C., Lopez-Quilez, A., et al. (2011). Antimicrobial resistance in more than 100,000 Escherichia coli isolates according to culture site and patient age, gender, and location. Antimicrob. Agents Chemother. 55, 1222–1228. doi: 10.1128/AAC.00765-10
Moini, A. S., Soltani, B., Taghavi Ardakani, A., Moravveji, A., Erami, M., Haji Rezaei, M., et al. (2015). Multidrug-resistant Escherichia coli and Klebsiella pneumonia isolated from patients in Kashan, Iran. Jundishapur J. Microbiol. 8:e27517.
O’Fallon, E., Kandel, R., Schreiber, R., and D’Agata, E. M. (2010). Acquisition of multidrug-resistant gram-negative bacteria: incidence and risk factors within a long-term care population. Infect. Control Hosp. Epidemiol. 31, 1148–1153. doi: 10.1086/656590
Oliveira, M. C., Oliveira, C. R., Goncalves, K. V., Santos, M. S., Tardelli, A. C., and Nobre, V. A. (2015). Enterobacteriaceae resistant to third generation cephalosporins upon hospital admission: risk factors and clinical outcomes. Braz. J. Infect. Dis. 19, 239–245. doi: 10.1016/j.bjid.2015.01.006
R Development Core Team (2008). R: A Language and Environment Forstatistical Computing. Vienna: R Foundation for Statistical Computing.
Slama, T. G. (2008). Gram-negative antibiotic resistance: there is a price to pay. Crit. Care 12(Suppl 4), S4. doi: 10.1186/cc6820
Wolfensberger, A., Sax, H., Weber, R, Zbinden, R., Kuster, S. P., and Hombach, M. (2013). Change of antibiotic susceptibility testing guidelines from CLSI to EUCAST: influence on cumulative hospital antibiograms. PLoS ONE 8: e79130. doi: 10.1371/journal.pone.0079130.eCollection2013.
World Health Organization [WHO] (2014). Antimicrobial Resistance: Global Report on Surveillance. Available at: http://apps.who.int/iris/bitstream/10665/112642/1/9789241564748_eng.pdf
Keywords: antibiotic resistance, Klebsiella, Enterobacter, Austria, statistical analysis
Citation: Badura A, Pregartner G, Holzer JC, Feierl G and Grisold AJ (2016) Susceptibility of Austrian Clinical Klebsiella and Enterobacter Isolates Linked to Patient-Related Data. Front. Microbiol. 7:34. doi: 10.3389/fmicb.2016.00034
Received: 28 October 2015; Accepted: 11 January 2016;
Published: 05 February 2016.
Edited by:
Gilberto Igrejas, University of Trás-os-Montes and Alto Douro, PortugalReviewed by:
Jose Luis Balcazar, Catalan Institute for Water Research, SpainAnnalisa Pantosti, Istituto Superiore di Sanità, Italy
Copyright © 2016 Badura, Pregartner, Holzer, Feierl and Grisold. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Alexandra Badura, YWxleGFuZHJhLmJhZHVyYUBtZWR1bmlncmF6LmF0
 Alexandra Badura
Alexandra Badura Gudrun Pregartner2
Gudrun Pregartner2 Judith C. Holzer
Judith C. Holzer Andrea J. Grisold
Andrea J. Grisold