- 1Biosciences Division, Oak Ridge National Laboratory, Oak Ridge, TN, USA
- 2Department of Chemical and Biomolecular Engineering, University of Notre Dame, Notre Dame, IN, USA
- 3Department of Microbiology and Plant Pathology, Forestry and Agricultural Biotechnology Institute, University of Pretoria, Pretoria, South Africa
The complex interactions between plants and their microbiome can have a profound effect on the health and productivity of the plant host. A better understanding of the microbial mechanisms that promote plant health and stress tolerance will enable strategies for improving the productivity of economically important plants. Pantoea sp. YR343 is a motile, rod-shaped bacterium isolated from the roots of Populus deltoides that possesses the ability to solubilize phosphate and produce the phytohormone indole-3-acetic acid (IAA). Pantoea sp. YR343 readily colonizes plant roots and does not appear to be pathogenic when applied to the leaves or roots of selected plant hosts. To better understand the molecular mechanisms involved in plant association and rhizosphere survival by Pantoea sp. YR343, we constructed a mutant in which the crtB gene encoding phytoene synthase was deleted. Phytoene synthase is responsible for converting geranylgeranyl pyrophosphate to phytoene, an important precursor to the production of carotenoids. As predicted, the ΔcrtB mutant is defective in carotenoid production, and shows increased sensitivity to oxidative stress. Moreover, we find that the ΔcrtB mutant is impaired in biofilm formation and production of IAA. Finally we demonstrate that the ΔcrtB mutant shows reduced colonization of plant roots. Taken together, these data suggest that carotenoids are important for plant association and/or rhizosphere survival in Pantoea sp. YR343.
Introduction
The rhizosphere is the site of a complex network of plant–microbe and microbe–microbe interactions which ultimately influence plant health and productivity. Microbes can be considered pathogenic, neutral, or beneficial depending on the plant host with which they associate (Raaijmakers et al., 2009). Among these bacteria is a group of plant growth-promoting bacteria (PGPB) that can colonize within the plant, on leaf or root surfaces, or in the surrounding rhizosphere (Hayat et al., 2010; Barret et al., 2011; Beneduzi et al., 2012). These bacteria can promote plant growth via phytohormone production, nitrogen fixation, and/or enhancement of water and mineral uptake (Kloos et al., 2001; Perrig et al., 2007; Bashan and de-Bashan, 2010; del Amor and Cuadra-Crespo, 2011). For example, the ability of microbes to influence root development via biosynthesis of the auxin indole-3-acetic acid (IAA) is well documented and multiple pathways for microbial IAA production have been described (Spaepen et al., 2007). Although well-studied genera like Azospirillum and Rhizobium are included in the PGPB group, the mechanisms by which these bacteria interact with and influence plant growth are not yet fully understood.
Recently the bacterial community associated with mature Populus deltoides roots was analyzed and shown to be dominated by Proteobacteria, Acidobacteria, and Verrucomicrobia (Gottel et al., 2011; Shakya et al., 2013). Among the organisms isolated from the Populus rhizosphere was the γ-proteobacterium Pantoea sp. YR343. Different species within the Pantoea genera have been shown to be either beneficial or harmful in association with plants (Walterson and Stavrinides, 2015). For example, P. stewartii and P. ananatis are responsible for Stewart’s bacterial wilt disease on corn, leaf blotch disease in Sudangrass, and center rot in onion (Azad et al., 2000; Walcott et al., 2002). P. rwandensis, P. rodasii, P. vagans, and P. eucalypti have been implicated as the causal agents of bacterial blight, leaf lesions, and dieback in eucalyptus (Coutinho et al., 2002; Brady et al., 2009, 2012). In contrast, P. vagans C9-1 is used as a biocontrol agent to protect against fire blight (Stockwell et al., 2010) and P. agglomerans can protect against plant pathogens like Pseudomonas syringae pv. Syringae (Braun-Kiewnick et al., 2000). Strains of P. agglomerans have also been shown to promote plant growth in wheat, rice, and cotton (Ruppel et al., 1992; Amellal et al., 1998; Egamberdiyeva and Höflich, 2001; Verma et al., 2001; Feng et al., 2006). It is thought that P. agglomerans promotes plant growth by enhancing root architecture, which increases the amount of minerals and water that can be taken up by the plant (Sergeeva et al., 2007). P. dispersa and P. agglomerans have been shown to produce IAA and evidence supports the hypothesis that the production of IAA by P. agglomerans is responsible for its plant growth-promoting abilities (Sergeeva et al., 2007; Kulkarni et al., 2013). Additional studies have shown that production of exopolysaccharides by P. agglomerans contributes to soil aggregation and moisture control, which also enhances plant growth (Amellal et al., 1998).
Plants produce reactive oxygen species as a by-product of their metabolism, and these molecules have been implicated in plant defense mechanisms (Apel and Hirt, 2004), stress responses (Gechev et al., 2006), and the establishment of symbiotic relationships (Gechev et al., 2006). Many soil-dwelling and plant-associated bacteria, including Pantoea spp., produce carotenoids, which are pigment molecules found in the cellular membrane (Dussault et al., 2008; Mohammadi et al., 2012). Carotenoids have been studied for their photoprotective and antioxidant activities in some photosynthetic and plant growth-promoting microbes, where carotenoids serve an essential role in protection against singlet oxygen species, protection from oxygen during nitrogen fixation, and in energy transfer during photosynthesis (Foote and Denny, 1968; Cogdell and Frank, 1987; Hartmann and Hurek, 1988; Britton, 1989; Krinsky, 1989, 1993; Hirayama et al., 1994; Edge et al., 1997; Cogdell et al., 2000; Fraser et al., 2001). In addition, carotenoids can modulate membrane fluidity and may play a role in the formation of membrane domains (Huang and Haug, 1974; Chamberlain et al., 1991; Gruszecki, 1999; Gruszecki and Strzalka, 2005; Landrum, 2009; Lopez and Kolter, 2010). Zeaxanthin (both mono- and diglucoside forms) was found to be the predominant carotenoid present in Erwinia herbicola and P. stewartii (Hundle et al., 1991; Mohammadi et al., 2012). The carotenoid biosynthesis pathway has been well characterized in Pantoea and consists of six enzymes: geranylgeranyl diphosphate (GGPP) synthase CrtE, phytoene synthase CrtB, phytoene desaturase CrtI, lycopene cyclase CrtY, β-carotene hydroxylase CrtZ, and the zeaxanthin glucosyltransferase CrtX (To et al., 1994; Sedkova et al., 2005). In P. ananatis, it was found that deletion of the phytoene synthase gene, crtB, resulted in the loss of yellow pigment and increased sensitivity to environmental stress factors (Mohammadi et al., 2012).
Here we describe Pantoea sp. YR343, a non-pathogenic bacterial strain isolated from the rhizosphere of poplar, which was found to be a robust colonizer of plant roots. This motile, rod-shaped bacterium is able to solubilize phosphate and produce IAA. To better understand the molecular mechanisms involved in rhizosphere survival and plant association in Pantoea sp. YR343, we constructed a mutant in which the phytoene synthase gene, crtB, was deleted. As predicted, the crtB mutant is defective in carotenoid production and showed increased sensitivity to reactive oxygen species. In addition, we find that the ΔcrtB mutant is impaired in other behaviors, including biofilm formation, production of IAA, and root colonization.
Materials and Methods
Bacterial Isolation and Growth Conditions
Fine roots and associated rhizophere samples were harvested from native Populus deltoides trees at the Yadkin River in North Carolina in May 2011 and bacteria were isolated from both the endosphere and rhizosphere. A rhizosphere isolate designated YR343 was restreaked three times to R2A agar plates (R2A Agar, VWR) to obtain a purified strain and a draft genome sequence was obtained (Brown et al., 2012). This bacterial strain is referred to as Pantoea sp. YR343 in this paper. Pantoea sp. YR343 was cultured at 28°C in R2A medium (R2A Broth Premix, TEKnova, Inc.) or on R2A agar plates. Cultures were also grown in standard LB, M9, TY (per 1 l, 10 g tryptone, 5 g yeast extract), or SOBG medium (per 1 l, 20 g tryptone, 5 g yeast extract, 0.5 g NaCl, 2.4 g MgSO4, 0.186 g KCl, 50 ml of 40% v/v glycerol). Fluorescent strains of Pantoea sp. YR343 expressing EGFP were engineered either by expressing EGFP from a Gateway-modified pBBR1-MCS5 plasmid (Pelletier et al., 2008) maintained with 10 μg gentamycin ml-1 (referred to as YR343-pGFP) or by chromosomal insertion of EGFP using pBT270 (pUC18-miniTn7T2-PA1/04/03-GFP, a gift from B. S. Tseng, University of Washington) (referred to as YR343-GFP).
Phylogenetic Analysis
Approximately 500 base pairs from the 5′ end of the 16S rRNA gene were amplified using universal primers (Weston et al., 2012). Initial BLAST (16S and gyrB) and Ribosomal Database Project 16S sequence comparisons suggested a strong affiliation of strainYR343 with Pantoea spp. Multi-locus sequence analysis (MLSA) of partial nucleotide sequences of genes rpoB, infB, atpD and gyrB was employed as described (Brady et al., 2008, 2010, 2012). Sequences from Populus rhizosphere isolates GM01, YR343 (Brown et al., 2012) and three other unidentified isolates for which data was available from genome sequencing efforts on IMG were aligned using the Translation alignment tool within GeneiousTM (v6.0.4-www.geneious.com) with 94 total representatives of Pantoea and related taxa of Erwinia, Tatumella and other Enterobacteriaceae for further phylogenetic analysis with Chronobacter sakazakii as an outgroup. Maximum likelihood analysis was conducted using PhyML v3.0, (Guindon et al., 2010) as implemented within GeneiousTM. A general time reversible model was used with each rate category and allowed to vary according to a gamma distribution. An initial best tree was obtained via comparison of a neighbor-joining tree using NNI/SPR analysis, and 100 bootstrap resampling trees were conducted.
Pathogenicity Assay with Pantoea sp. YR343 in Populus deltoides
Populus deltoides WV94 plants were grown from shoot tips that were surface-sterilized by washing for 5 min in a 1% Tween-20 solution, followed by a 1 min wash in 70% ethanol, then a 12 min wash in 10% bleach. Shoot tips were then washed three times in sterile water before inoculation into MS medium (per 1 l of medium: 4.43 g MS salts, 0.5 g MES hydrate, 30 g sucrose, 5 g activated charcoal, 1.5 g Gelrite and 1 ml plant preservative mixture (PPM). The pH was adjusted to 5.7 using KOH prior to addition of activated charcoal and Gelrite). Once the shoot tips had rooted, the Poplar plants were transferred to Magenta boxes containing sterile clay soil mixed with 1X Hoagland’s solution (1.63 g Hoagland Modified Basal Salt Mixture per 1 l water, Phytotechnology Laboratories) to provide plant nutrition. For this assay, there were two treatments using three plants per experiment, along with a set of three control plants. In the first experiment, the clay soil was mixed with approximately 1 × 109 cells of Pantoea sp. YR343 grown in R2A medium prior to planting the newly rooted Poplar plant. For the second experiment, we used a culture with 1 × 108 cells per ml of Pantoea sp. YR343. The control plants were treated by mixing R2A medium in the clay soil (at a similar volume to those treated with Pantoea) and also with R2A medium swabbed onto leaves. Plants were grown for 21 days at 24°C with a 12 h light and 12 h dark photoperiod. All plant samples were measured at the beginning of the study and again at the time of harvest in order to assess the effects of Pantoea sp. YR343 on such characteristics as stem height, leaf number and size, and root area.
Phenotypic Analysis
To compare growth rates of Pantoea sp. YR343 in LB, R2A, or M9 media, overnight cultures were diluted 1:100 into fresh medium and 200 μl was loaded into 12 wells on a honeycomb plate and placed into a Bioscreen C Reader System at room temperature with shaking overnight. Phosphate solubilization was examined using Pikovskaya’s agar medium plates (per 1 l, 0.5 g yeast extract, 10 g dextrose, 5 g Ca3(PO4)2, 0.5 g (NH4)2SO4, 0.2 g KCl, 0.1 g MgSO4⋅7H2O, 0.0001 MnSO4⋅H2O, FeSO4⋅7H2O, 15 g agar) and incubated at 28°C for approximately 1 week prior to imaging. The presence of cellulose in Pantoea sp. YR343 exopolysaccharides was detected by inoculating 200 μl of an overnight culture into a glass-bottom dish containing 5 ml of R2A and grown statically at 25°C for 72 h. After incubation, the culture was stained with a solution of Calcofluor White (5 μg ml-1) (Sigma–Aldrich), and 5 μM SYTO61 (Life Technologies). Cultures were imaged using a Zeiss LSM 710 laser scanning confocal microscope and images were processed using Zen software (Zeiss). Swimming and swarming motility was examined on LB containing 0.3% w/v agar or 0.6% w/v agar supplemented with 0.4% w/v glucose or 0.4% v/v glycerol, based on previous studies (Herrera et al., 2008).
Root Colonization Assays
Arabidopsis thaliana ecotype Col-0 seeds were germinated and grown, as previously described (Wang et al., 2005). Seeds were surface-sterilized by washing in a 15% bleach solution containing 0.01% Tween-20 for 15–20 min, then rinsed in sterile water. Afterward, seeds were washed briefly in 70% ethanol and rinsed several times in sterile water. Sterilized seeds were soaked in double distilled H2O containing 0.1% w/v agar at 4°C for 4 days and then germinated on Murashige and Skoog (MS) agar containing 0.25% w/v sucrose (Murashige and Skoog, 1962). Seedlings were incubated in a growth chamber at 24°C with a 12 h light and 12 h dark photoperiod. After 7 days, 4 – 6 A. thaliana seedlings of equivalent root lengths were transferred to new MS agar plates containing 0.25% w/v sucrose and grown for an additional 7 days. An overnight culture of Pantoea sp. YR343 was then pipetted in a line across the MS plate approximately 1 cm from the bottom edge, while sterile R2A media was used for the untreated control. After 10 days, plants were imaged for differences in root architecture. For analysis of root colonization, we utilized the YR343-pGFP strain. Co-cultured seedlings were harvested and mounted on slides prior to imaging with a Zeiss LSM710 confocal laser scanning microscope.
Populus deltoides WV94 shoot tips were surface-sterilized and grown as described for the pathogenicity studies. Rooted cuttings were planted in sterile clay soil inoculated with Pantoea sp. YR343-pGFP at 2.7 × 107 CFU g-1 soil and incubated at 24°C with a 12 h light and 12 h dark photoperiod for 7 days. Roots and soil were collected for re-isolation of Panteoa sp. YR343 and measurement of colonization. Plants that were inoculated with medium alone were used as controls for bacterial contamination, which was not observed in these assays. Briefly, roots were weighed, washed by vortexing at least 30 s with 3 ml of PBS and a few small glass beads and dilutions of the wash solution was plated on R2A to measure colony forming units (CFU). For soil samples, 1 g of soil was mixed with 4 ml of PBS, then vortexed and plated similarly.
Triticum aestivum (wheat) seeds were sterilized as described for Arabidopsis thaliana, then incubated on wet filter paper in the dark for 3 days. Overnight cultures of wild type Pantoea sp. YR343-GFP and the ΔcrtB mutant were normalized to the same optical density (OD600). Cultures were then inoculated into sterile, molten Fahraeus medium (per 1 l, 100 mg CaCl2⋅H2O, 120 mg MgSO4⋅7H2O, 100 mg KH2PO4, 150 mg Na2HPO4, 5 mg Ferric citrate, and a trace amount of Na2MoO4⋅2H2O, then adjusted to pH 7.5 and added 4 g agar) that was cooled, but not solidified, and then the medium was poured into sterile glass test tubes or magenta boxes and allowed to solidify. Plants treated with sterile R2A medium were used as controls for background contamination. Afterward, germinated seedlings were added to each tube. Three plants were measured for colonization per treatment: uninoculated control, wild type only, ΔcrtB only, and a 1:1 mix of wild type and ΔcrtB. Each plant was inoculated with approximately 1 × 108 cells per plant and grown for 1 week prior to harvesting. Roots were harvested as described above for Populus and analyzed for CFUs per gram of root material. Mixtures of wild type and the ΔcrtB mutant were distinguished by colony color (wild type colonies were yellow, while the mutant colonies were white). Imaging of root colonization was performed by staining root tissue with 5 μM Syto61 (Life Technologies) to stain all bacterial cells that were attached to the root. Upon rinsing, the roots were visualized with a Zeiss LSM710 confocal laser scanning microscope. Wild type cells were distinguished by fluorescence in both the red (Syto61) and green (EGFP) channels, whereas the ΔcrtB mutant fluoresced only in the red channel (Styo61).
Construction of crtB Mutant
A marker-less mutant lacking the phytoene synthetase gene, CrtB (PMI39_03408), was generated by PCR amplification of the DNA regions 500 base pairs upstream (using primers CrtB_up For (XbaI): 5′-GCTCTAGATCCGCGTCCACCTTT-3′ and CrtB up Rev (XhoI): 5′-CCGCTCGAGTCTTACGTCCGTGGC-3′) and 500 base pairs downstream (using primers CrtB_dn For (XhoI): 5′-CCGCTCGAGTCGGCGCGATCCTCC-3′ and CrtB_dn Rev (XbaI): 5′-GCTCTAGATGTTTCGGTCCGCGC-3′) of the crtB gene from Pantoea sp. YR343. PCR products were ligated into pK18mobsacB (Schafer et al., 1994) and the resulting plasmid was verified by restriction digests and transformed into Pantoea sp. YR343 by electroporation and selected on R2A plates containing 50 μg kanamycin ml-1. Colonies lacking yellow pigmentation were screened by PCR to verify that the crtB gene was deleted.
Carotenoid Extraction and Analysis
Carotenoids were extracted from the wild type Pantoea sp. YR343 and ΔcrtB mutant as described (Mohammadi et al., 2012). Briefly, 2 ml of cells grown in LB medium were rinsed in water, resuspended in 1 ml of 100% methanol, and heated at 85°C for 20 min. After centrifugation to remove the cell debris, spectral analysis of the extracted carotenoids was measured in triplicate in the range of 400–500 nm using a BioTek Synergy 2 microplate reader.
Cell Viability Assays
Wild type Pantoea sp. YR343 and ΔcrtB were grown in LB medium at 28°C overnight. Cells were diluted 1:20 into fresh LB and grown 2–3 h at 28°C with shaking. In a 96 well plate, 100 μl of log phase or stationary phase cells were treated with final concentrations of 0.5, 1, 2, 5, 10, and 100 mM hydrogen peroxide and incubated for 1 h with shaking at 28°C. Afterward, each well was treated with 100 μl of Bac-Titer Glo reagent (Promega) according to the manufacturer’s instructions and luminescence was measured on a BioTek Synergy 2 microplate reader.
Raman Spectroscopy
Single bacterial colonies grown on R2A agar plates were transferred to 2 ml of sterile PBS, vortexed, and adjusted to OD600 = 0.2 using a Cary UV-Vis spectrophotometer. 10 μl of the cell suspension was pipetted onto a clean gold-coated silicon wafer and dried for Raman analysis. Raman measurements were performed at room temperature using a confocal Raman microscope (Alpha 300R, WITec GmbH, Germany) equipped with a focused Nd:YAG operating at (aaa = 532 nm), a 40×, Nikon air objective (NA = 0.6), and a coverslip-corrected Nikon water immersion 60× objective (NA = 1). The laser radiation was delivered via a single mode optical fiber through a dichroic beam splitter into the microscope objective and focused to a diffraction limited spot size on the surface of the sample. The scattered Raman radiation was collected by the same objective and focused into a 50 μm diameter multi-mode fiber connected to a UHTS 300 spectrometer equipped with a 600 groove mm-1 grating and a back-illuminated CCD camera (Newton DU970 N-BV, Andor Inc., cooled to -65°C). Each Raman spectrum recorded was an accumulation of 100 spectra acquired with integration time of 0.5 s each at 5 mW incident laser power. For each sample, individual Raman spectra were collected at 15 randomly selected spatial locations and averaged to give the representative spectrum for that sample. Data analysis was performed using commercial graphic software Igor Pro 6.32A (Lake Oswego, OR, USA).
Indole-3-Acetic Acid (IAA) Production Assay
Indole-3-acetic acid production was measured by a colorimetric assay as previously described (Tang and Bonner, 1947). Briefly, 500 μl of overnight cultures was diluted into 50 ml of M9 minimal medium plus L-Tryptophan (200 μg ml-1 final concentration), and incubated overnight at 28°C. Cells were harvested at the same OD and IAA was detected in the supernatant using Salkowski’s reagent (500 ml dH2O, 300 ml concentrated H2SO4, 2.03g FeCl3⋅6H2O) and absorbance was measured at 535 nm using a BioTekSynergy 2 microplate reader. All measurements were performed in triplicate and compared to a standard curve generated from IAA (Sigma–Aldrich). Gas chromatography-mass spectrometry was performed to confirm the presence of IAA using supernatants from wild type and mutant cultures grown in M9 minimal medium with or without L-Tryptophan as described (Weston et al., 2012).
Biofilm Formation Assay
Biofilm formation was measured using the protocol described (O’Toole and Kolter, 1998), with the following modifications. An overnight culture was diluted 1:100 into either LB, R2A, or M9 medium supplemented with 0.4% w/v glucose and grown statically in a 96-well plate covered with breatheable tape in place of the lid (Breathe-EASIER, Diversified Biotech) at 28°C for 72 h. After 72 h, adherent cells were stained with 0.1% w/v crystal violet stain, then the crystal violet associated with biofilms was dissolved using a modified solution which contained 10% w/v SDS dissolved in 80% v/v ethanol (Tram et al., 2013). Absorbance was measured at 550 nm using a BioTekSynergy 2 microplate reader.
Results
Phylogenetic Analysis of Pantoea sp. YR343
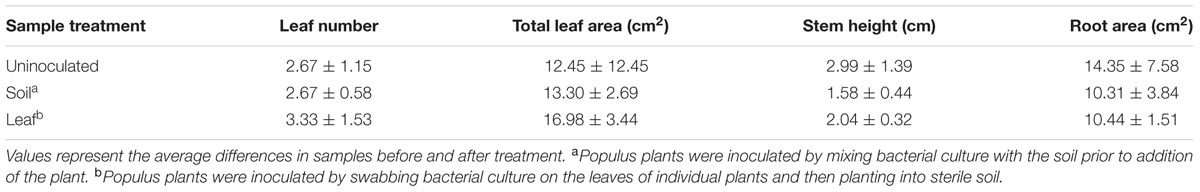
Pantoea sp. YR343 was isolated from the rhizosphere of a native Populus deltoides tree growing in the Yadkin River region of North Carolina (Shakya et al., 2013) and a draft genome sequence was obtained (Brown et al., 2012). Phylogenetic analysis was conducted to compare the Pantoea isolate with known species within the Enterobacteriaceae. The results of multi locus sequence analysis (MLSA) using partial nucleotide sequences from atpD, infB, gyrB, and rpoB (Brady et al., 2012) revealed that Pantoea sp. YR343 clusters (100% of bootstrap replicates) with another Pantoea strain isolated from the Poplar rhizosphere (Pantoea sp. GM01; Brown et al., 2012) within a monophyletic Pantoea group (90%) and forms a unique, well supported (100%) group, basal to the recently described P. rwandensis and P. rodasii, that are known to form lesions on leaves of plantation-grown Eucalyptus trees (Brady et al., 2012) (Figure 1).
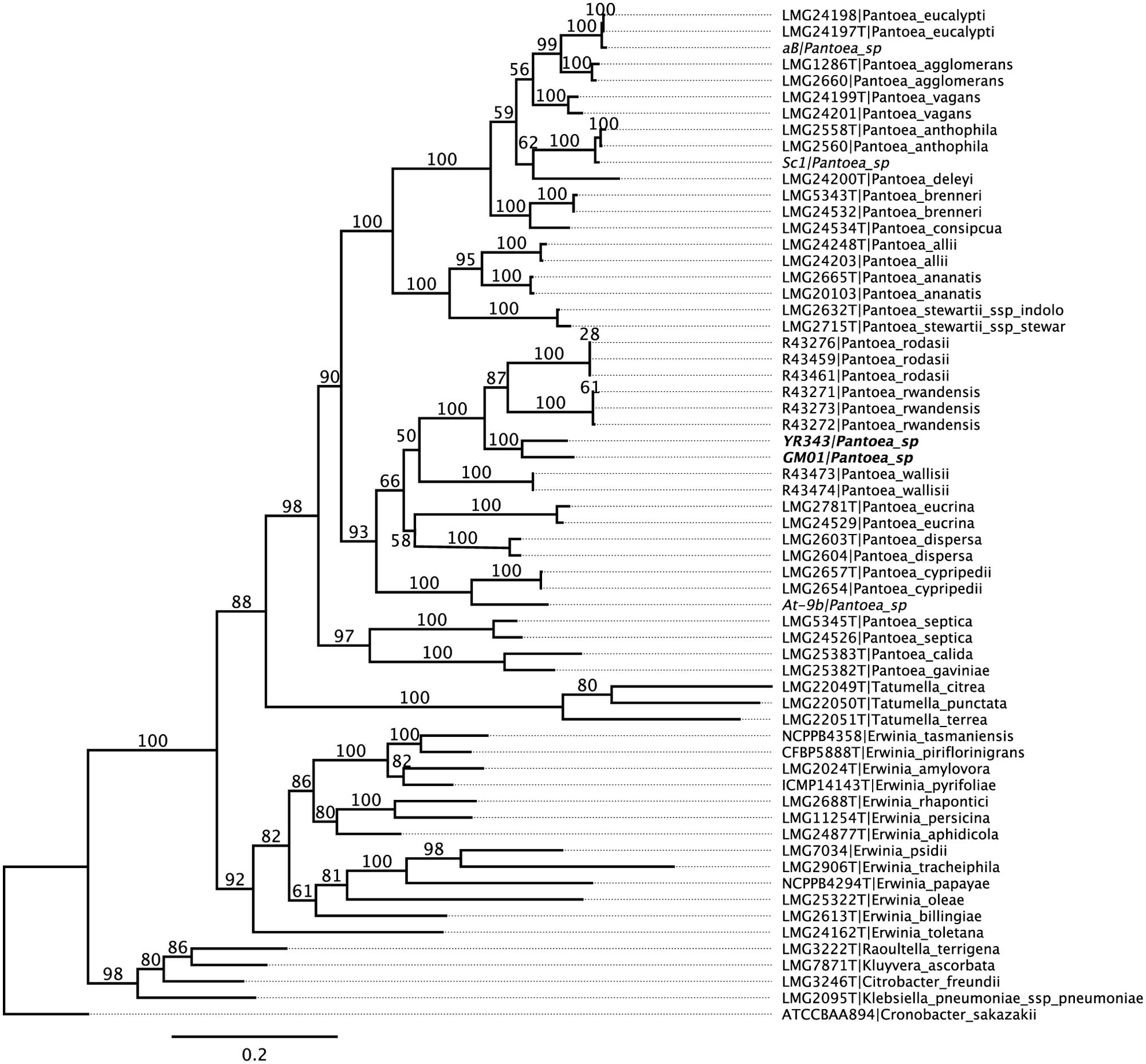
FIGURE 1. Phylogenetic tree of Pantoea sp. YR343 and related species within Enterobacteriaceae. Partial nucleotide sequences from atpD, infB, gyrB, and rpoB were used for multi locus sequence analysis.
Although Pantoea sp. YR343 was isolated from a healthy poplar tree, its close phylogenetic relationship to Pantoea strains found to be the causal agents of leaf blight in Eucalyptus prompted us to test the effect of Pantoea sp. YR343 on cultured poplar cuttings. Populus deltoides WV94 cuttings were grown under sterile conditions in magenta boxes and exposed to Pantoea sp. YR343 at the roots by inoculation into the soil, or on leaves by direct foliar application. No evidence of lesions was observed on the leaves for 21 days following exposure to Pantoea sp. YR343 (data not shown). After 3 weeks, plants were harvested and measured for number of leaves, total leaf area (cm2), stem height (cm), and root area (cm2) (Table 1). Statistical analysis using a t-test indicated no significant differences (p-values >0.05) between control plants, foliar treated plants, or soil inoculated plants, suggesting that Pantoea sp. YR343 is not pathogenic to P. deltoides WV94.
Phenotypic Properties of Pantoea sp. YR343
As determined by phylogenetic analysis, Pantoea sp. YR343 is a member of the Enterobacteriaceae family and falls into the class of γ-proteobacteria. Pantoea sp. YR343 is a Gram-negative bacterium that grows in liquid LB, R2A, or M9 minimal medium using glucose as a carbon source (Figure 2A). When grown on medium containing an insoluble form of calcium phosphate, a zone of clearing formed around colonies of Pantoea sp. YR343, indicating that this species is capable of phosphate solubilization (Figure 2B). Pantoea sp. YR343 growing on LB agar plates produced colonies that were round, smooth, and produced a yellow pigment (Figure 2C). In contrast, Pantoea sp. YR343 produced colonies that were irregularly shaped, wrinkly, and light yellow in color when grown on R2A agar plates (Figure 2C). Pantoea sp. YR343 also appeared to be highly mucoid, particularly when grown on R2A medium. Microscopic observation showed that Pantoea sp. YR343 displays a rod-shaped morphology with cells averaging approximately 2 μm in length (Figure 2D). In addition, Pantoea sp. YR343 was able to form biofilms on abiotic surfaces and the production of cellulose was detected in these biofilms by staining with Calcofluor White (Figure 2D). We also analyzed motility behavior in this organism using LB medium containing either 0.3% (swimming) or 0.6% (swarming) agar (Figure 2E). After 18 h, the cells had moved from the center to the edges of the plate, consistent with swimming motility behavior. Swarming motility was also observed, particularly when Pantoea sp. YR343 was grown on LB supplemented with 0.4% glycerol (Figure 2E). Finally, we used GC-MS analyses to examine whether Pantoea sp. YR343 could produce IAA, which is synthesized from the amino acid tryptophan (Patten and Glick, 1996). The tryptophan-dependent production of IAA was confirmed by GC-MS analyses and measured at approximately 0.5 μg/ml.
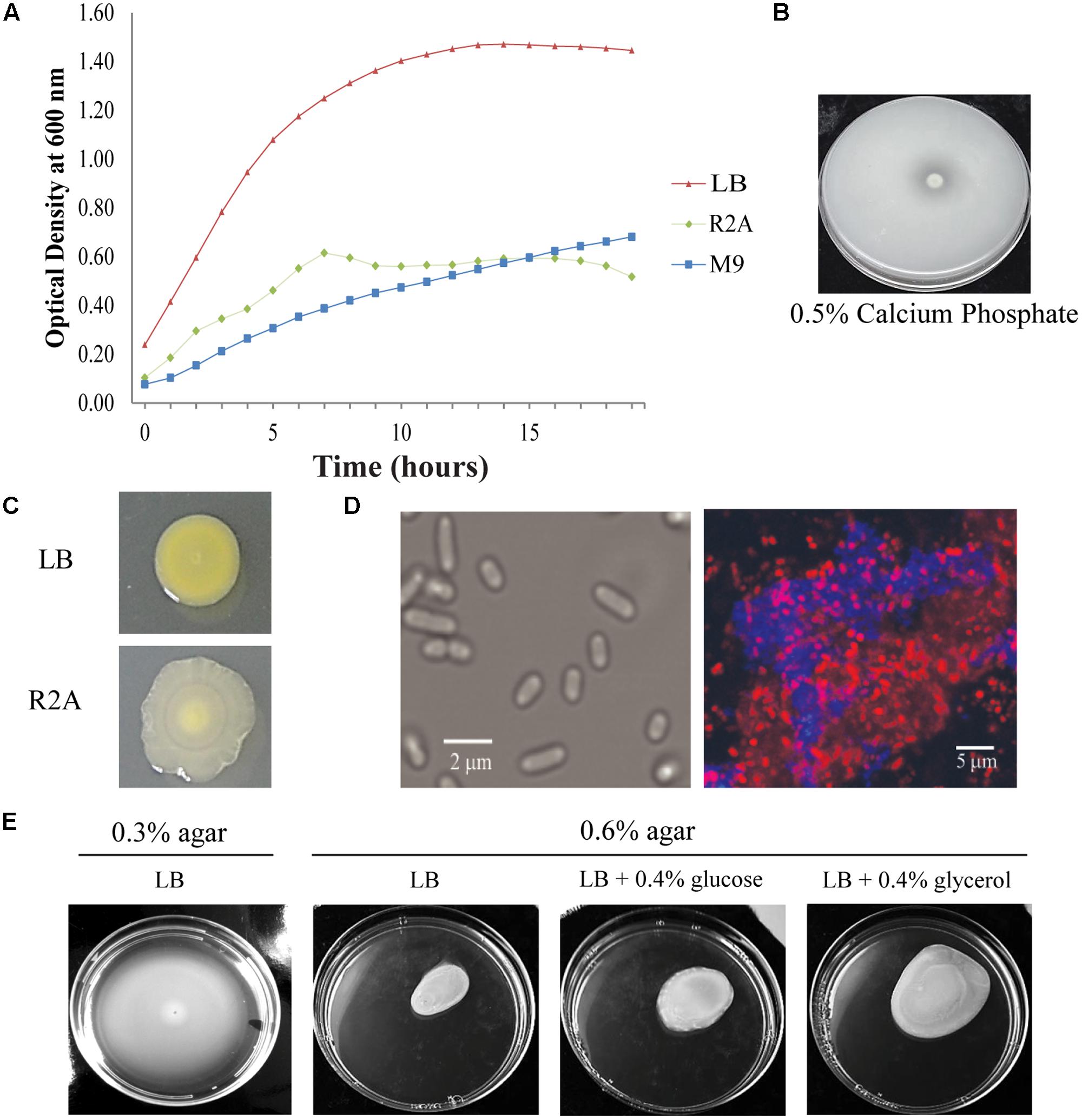
FIGURE 2. Phenotypic characteristics of Pantoea sp. YR343. (A) Growth of wild type Pantoea sp. YR343 grown in liquid LB, R2A, and M9 media. (B) Phosphate solubilization by Pantoea sp. YR343 is shown by a zone of clearing surrounding the colony. (C) Colony morphology of Pantoea sp. YR343 on LB (top) versus R2A (bottom) agar plates. (D) Bright field image of Pantoea sp. YR343 grown in R2A medium (left). (Right) Cellulose production in Pantoea sp. YR343 biofilm as detected by Calcofluor White staining (blue). The distribution of cells is indicated by staining with Syto61 (red). (E) Swimming and swarming motility of Pantoea sp. YR343. Swimming motility was characterized using LB medium with 0.3% agar (left) and swarming motility was characterized on LB medium with 0.6% agar, or on LB medium with 0.6% agar supplemented with 0.4% glucose or glycerol (right).
Pantoea sp. YR343 Colonizes Plant Roots
We next analyzed the ability of Pantoea sp. YR343 to colonize plant surfaces. Co-cultures of Arabidopsis seedlings and Pantoea sp. YR343 showed that seedlings grown in the presence of the microbe had enhanced lateral root density compared to control seedlings (Figure 3A). To enable detection of Panteoa sp. YR343 on plant roots, we constructed a fluorescent strain (YR343-pGFP) in which GFP was expressed from a Gateway-modified pBBR1 plasmid (Pelletier et al., 2008). Using this fluorescent strain, we found that YR343-pGFP readily attached to the surface of Arabidopsis roots (Figure 3B). The same fluorescent strain was then used to determine whether Pantoea associated directly with Populus roots. Sterile P. deltoides WV94 cuttings were exposed to YR343-pGFP at the roots by inoculation into the soil at a concentration of 2.7 × 107 CFU g-1 of soil. YR343-pGFP was re-isolated from the roots and soil at day 7 to determine its distribution and confirm viability on plants and in the soil. Pantoea sp. YR343 was preferentially associated with the plant as indicated by 3.5 × 106 CFU g-1 of root, compared to 3.0 × 102 CFU g-1 of soil. Consistent with this data, we observed YR343-pGFP cells attached to P. deltoides roots using confocal microscopy, whereas no bacteria were detected in the un-inoculated control plants (Figure 3C). Indeed, numerous locations along the roots were covered in bacteria. The bacteria were aggregated into what appeared to be biofilms on the surface of the roots and along the root hairs. Because the roots underwent several washes when they were harvested, the bacteria colonizing the Populus roots were firmly attached, as the transiently attached cells were presumably washed away.
FIGURE 3. Colonization of plant roots by Pantoea sp. YR343. (A) Arabidopsis thaliana plants were grown on MS plates in the absence or presence of Pantoea sp. YR343 for 10 days prior to imaging. (B) Corresponding to the pictures in (A) are images of roots taken with confocal microscopy. The image on top shows roots from an uninoculated plant, while the bottom image shows roots colonized by YR343-pGFP. (C) Populus deltoides WV94 cuttings were grown in the presence or absence of Panteoa sp. YR343 expressing GFP. (Top) untreated P. deltoides WV94 plant; (bottom) P. deltoides WV94 cultured with YR343-pGFP for 7 days. In all images, Pantoea sp. YR343 is detected by GFP fluorescence (green) and plant roots are detected using autofluorescence (red).
Pantoea sp. YR343 Produces Zeaxanthin Which Plays a Role in Withstanding Oxidative Stress
Pantoea sp. YR343 produced a yellow pigment under all growth conditions tested; however, it was the most apparent when cells were grown to stationary phase in LB medium. Genomic comparisons of the carotenoid biosynthesis operon in Pantoea sp. YR343 and P. ananatis LMG 20103 (De Maayer et al., 2010) indicated that the amino acid sequences of each gene from Pantoea sp. YR343 was more than 50% identical to those from P. ananatis LMG20103. Moreover, the carotenoid biosynthesis genes in Pantoea sp. YR343 were also arranged with an operon structure similar to that of P. ananatis and P. stewartii (Sedkova et al., 2005) (Figure 4A).
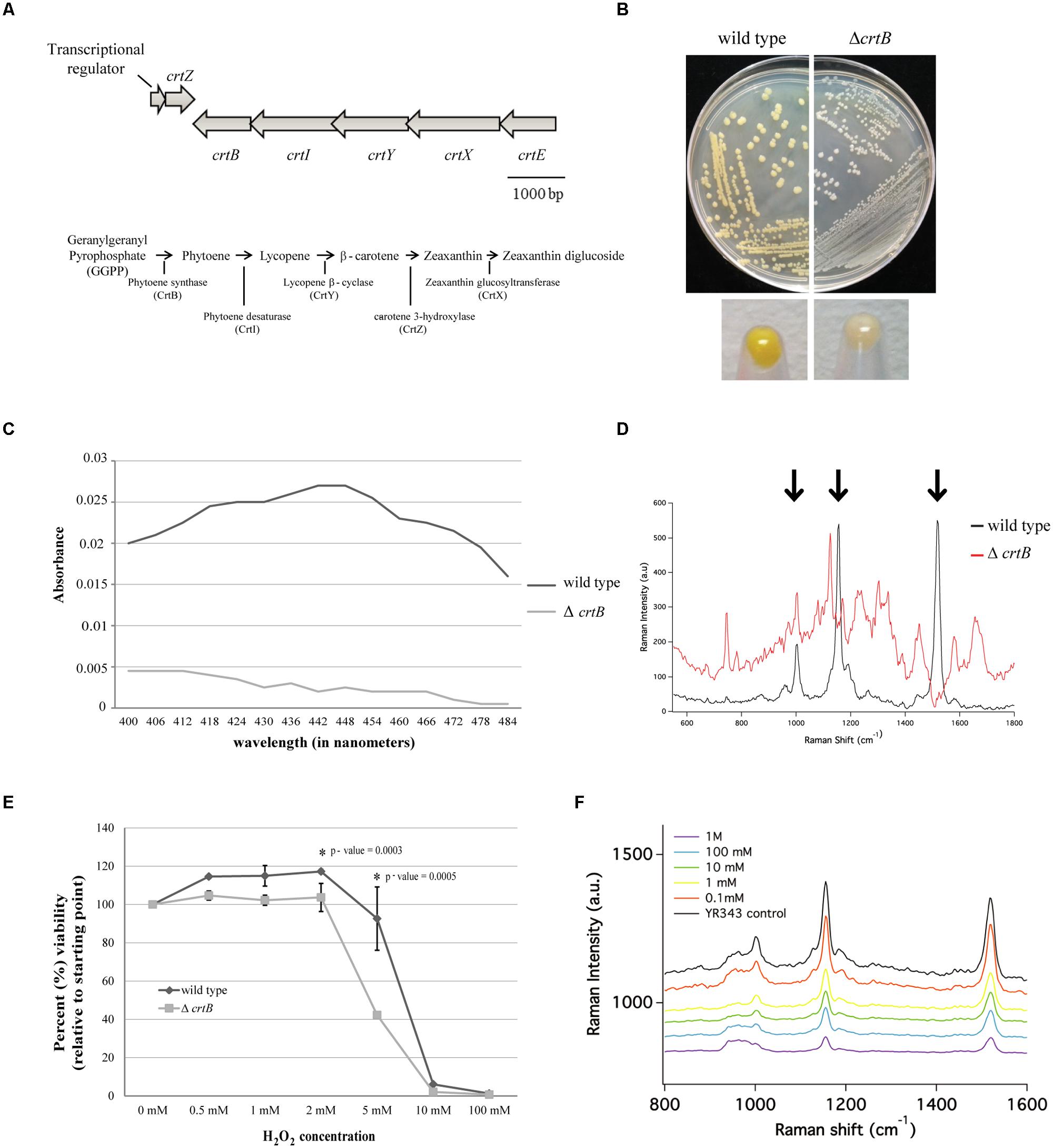
FIGURE 4. Identification and characterization of a carotenoid mutant in Pantoea sp. YR343. (A) Top, genomic structure of operon regulating carotenoid production. Bottom, predicted carotenoid biosynthesis pathway in Pantoea sp. YR343 based on genomic comparisons. PMI39_03412 encodes a product with 60% amino acid identity and 74% amino acid similarity to P. ananatis CrtE; PMI39_03408 encodes a product with 66% amino acid identity and 76% amino acid similarity to P. ananatis CrtB; PMI39_03409 encodes a product with 81% amino acid identity and 87% amino acid similarity to P. ananatis CrtI; PMI39_03410 encodes a product with 58% amino acid identity and 74% amino acid similarity to P. ananatis CrtY; PMI39_0340 encodes a product with 84% amino acid identity and 92% amino acid similarity to P. ananatis CrtZ, and PMI39_03411 encodes a product with 53% amino acid identity and 61% amino acid similarity to P. ananatis CrtX. (B) LB plates streaked with wild type Pantoea sp. YR343 (left) and ΔcrtB (right) and grown for 48 h showing loss of pigmentation in the mutant strain. (C) Methanol extraction of carotenoids from wild type Pantoea sp. YR343 and ΔcrtB. Graph represents the range of absorbances between 400 and 500 nm measured from one of two replicates. (D) Raman spectroscopy of wild type Pantoea sp. YR343 and ΔcrtB. The wild type strain shows a spectra dominated by peaks (highlighted by arrows) corresponding to zeaxanthin. These peaks are reduced in the ΔcrtB mutant. (E) Sensitivity of wild type and ΔcrtB mutant cells to increasing concentrations of hydrogen peroxide. Cell viability was measured using the Bac-Titer Glo assay and plotted as a percentage relative to the untreated control, measured as 100%. (F) Raman spectroscopy of wild type Pantoea sp. YR343 treated with different concentrations of hydrogen peroxide shows a decrease in peak intensity at 1500 cm-1 upon treatment with hydrogen peroxide.
Since carotenoids play a role in resistance to reactive oxygen species and, thus, may influence plant association, we constructed a mutant defective in carotenoid production by deleting the crtB gene that encodes for phytoene synthase, the enzyme responsible for the conversion of geranylgeranyl pyrophosphate into phytoene which serves as a precursor for the synthesis of carotenoids (Sandmann and Misawa, 1992). As predicted, the ΔcrtB mutant no longer produced a yellow pigment and the colonies appeared white (Figure 4B). This was further confirmed by analyzing the spectroscopic profiles of pigments extracted from wild type and ΔcrtB mutant cells. The pigments extracted from wild type cells showed a spectra consistent with zeaxanthin as has been described for other Pantoea strains (Hundle et al., 1991; Mohammadi et al., 2012). Conversely, little to no pigment was extracted from the ΔcrtB mutant (Figure 4C). We chose to further characterize this carotenoid using Raman spectroscopy. A representative Raman spectrum of wild type Pantoea sp. YR343 and the ΔcrtB mutant from 550 cm-1 to 1800 cm-1 is shown in Figure 4D. The spectrum for the wild type exhibits three prominent bands at 1520 cm-1, 1155 cm-1, and 1002 cm-1 which are characteristic of carotenoid compounds, and arise from in-phase C = C and C–C stretching and in-plane CH3 rocking vibrations, designated ν1 = 1520 cm-1, ν2 = 1155 cm-1, and ν3 = 1002 cm-1 (Merlin, 1985; Withnall et al., 2003; Schulz et al., 2005; Goodwin et al., 2006; de Oliveira et al., 2010). In contrast, these bands are less prominent in the ΔcrtB mutant.
We then tested the protective role of this carotenoid under oxidative stress created by exposure to hydrogen peroxide. Compared to the wild type strain, the ΔcrtB mutant was more sensitive to the effects of increasing concentrations of hydrogen peroxide as determined by a viability assay (Figure 4E). To demonstrate that hydrogen peroxide has a specific effect on carotenoids, we examined the effect of hydrogen peroxide on wild type cells using Raman spectroscopy. By this method, we observed a steady decrease in the spectral peak intensity corresponding to zeaxanthin (Figure 4F) which correlates to the loss of cell viability. Furthermore, in contrast to the viability assays which show a precipitous decrease in viability only at higher concentrations of 5–10 mM H2O2, the Raman spectra in Figure 4F clearly show progressive loss of carotenoid-related peaks beginning at H2O2 concentrations as low as 1 mM.
The crtB Mutant is Defective in Biofilm Formation, Indole-3-Acetic Acid Production, and Plant Colonization
Since carotenoids have also been implicated in modulating membrane properties (Huang and Haug, 1974; Chamberlain et al., 1991; Gruszecki, 1999; Gruszecki and Strzalka, 2005; Landrum, 2009; Lopez and Kolter, 2010), we next examined whether this carotenoid affected other biological behaviors, such as growth, IAA production, biofilm formation, and root colonization. Comparisons of growth in liquid cultures between wild type and the ΔcrtB mutant showed that the growth rates were very similar when cells were grown in LB or R2A medium (data not shown), but that the ΔcrtB mutant reached stationary phase at a lower cell density compared to wild type cells when grown in M9 minimal medium (Figure 5A).
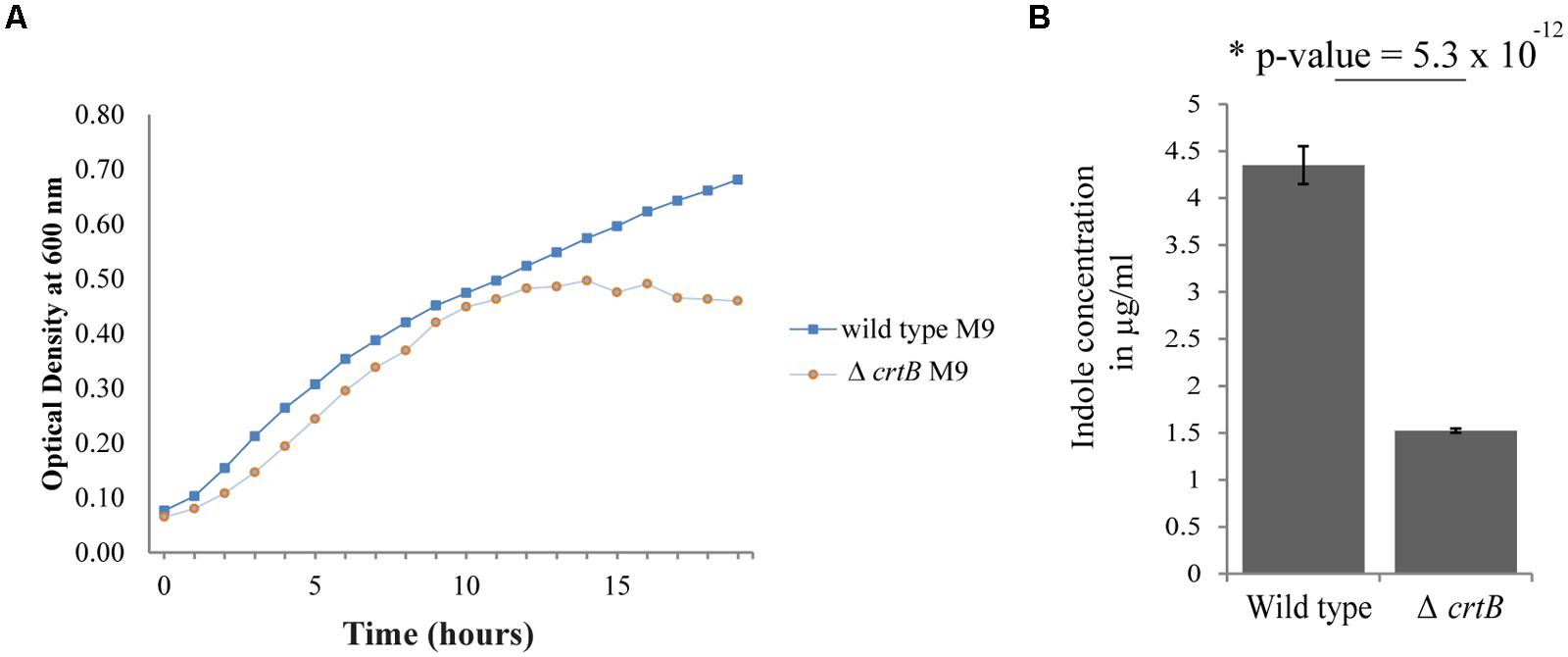
FIGURE 5. Loss of carotenoids affects growth and phytohormone production. (A) Growth curves of wild type Pantoea sp. YR343 and ΔcrtB in M9 minimal medium. (B) Comparison of indole-3-acetic acid production in the wild type Pantoea sp. YR343 and the ΔcrtB mutant. The production of indoles was measured using a colorimetric assay.
We then compared production of indolic compounds, including IAA, in Pantoea sp. YR343 and the ΔcrtB mutant when grown in the presence of tryptophan. Wild type and mutant cells were grown to the same optical density and the supernatants were measured for the presence of IAA. Somewhat unexpectedly, we found that the ΔcrtB mutant was defective in IAA production as determined using a colorimetric assay (Figure 5B). Indeed, we observed a nearly threefold decrease in indole production from 4.35 ± 0.20 μg/ml indoles in wild type cells compared to 1.52 ± 0.02 μg/ml indoles in ΔcrtB.
We next examined the ability of wild type and ΔcrtB cells to form biofilms using two different formats: biofilm formation on abiotic surfaces using 96-well plates with breathable tape, and biofilm formation at the air-liquid interface (pellicles) using glass test tubes. Using the 96-well plate assay, we found that the ΔcrtB mutant was impaired in biofilm formation, with the defect more apparent when cells were grown in LB medium (Figure 6A). Likewise, ΔcrtB mutant cells were also impaired in pellicle formation (Figure 6B). In this experiment, the mutant cells tended to settle at the bottom of the tube rather than form a biofilm at the air-liquid interface like wild type cells.
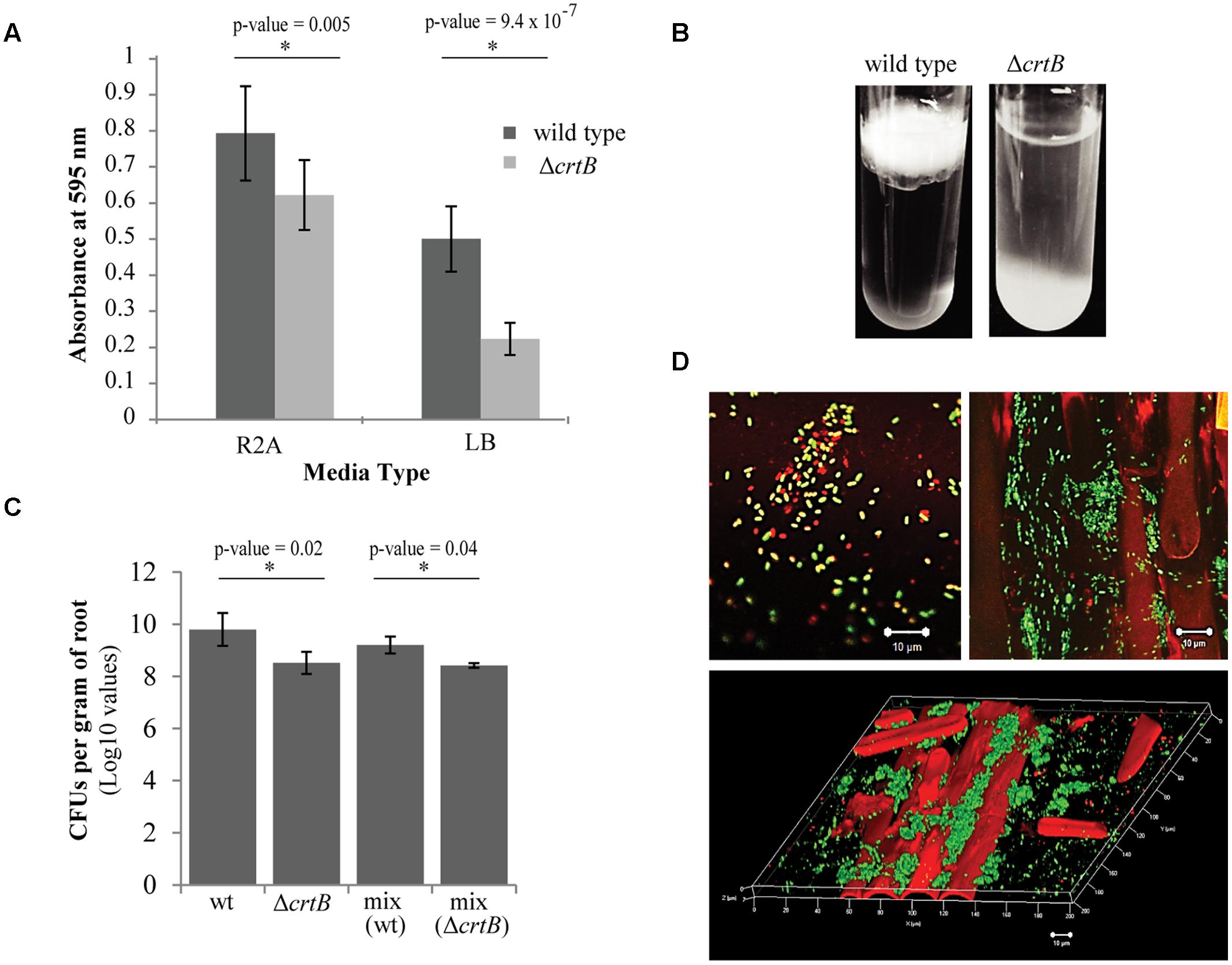
FIGURE 6. Loss of carotenoids affects biofilm formation and root colonization. (A) Comparison of biofilm formation between wild type Pantoea sp. YR343 and the ΔcrtB mutant in plastic 96-well plates measured by the crystal violet assay (B) Pellicle formation assay performed in SOBG medium in glass test tubes. Wild type Pantoea sp. YR343 forms pellicles on top of the liquid medium, while the ΔcrtB mutant settles to the bottom of the tube. (C) Wheat root colonization assay by wild type Pantoea sp. YR343 and ΔcrtB is described as the Log10 value of colony forming units (CFUs) per gram of root material. (D) Images of wheat roots treated with YR343-GFP and ΔcrtB for 1 week and stained with Syto61. Top left image shows a group of motile cells outside of the plant. Green represents the wild type population, while red represents the ΔcrtB mutant population. Top right image shows a wheat root (visualized using autofluorescence in the red channel) colonized predominantly by wild typeYR343-GFP (green). Bottom image shows a three-dimensional view of a colonized wheat root.
Due to the observation that the ΔcrtB mutant is impaired in biofilm formation and IAA production, we speculated that this mutant may also be affected in root colonization. For these studies, wheat was used as the host plant due to its hardiness and ease of growth. Three plants were tested for each of four conditions, including uninoculated control, wild type only, ΔcrtB only, and a 1:1 mix of wild type and ΔcrtB. After 1 week, the roots were harvested and washed prior to plating in order to compare the amount of CFU per treatment. We found that the wild type cells colonized the roots significantly better than the ΔcrtB mutant and this behavior is also observed in the 1:1 competition condition (Figure 6C).
Root colonization by Pantoea sp. YR343 wild type and ΔcrtB mutants was also examined using microscopy. In this case, Pantoea sp. YR343 with a chromosomal insertion of EGFP was used as the wild type strain (YR343-GFP) and the plants were inoculated as described above. After 1 week, the roots were harvested, washed and stained with Syto61, which is a cell-permeable dye that fluoresces red. Cells that only fluoresced red were assumed to be ΔcrtB mutants, and cells that fluoresced red and green were assumed to be wild type cells expressing GFP. Examination of the plant roots indicated that there were significantly more wild type cells (green) than mutant cells (red) attached to the roots (Figure 6D). These cells appeared to colonize in crevices on the plant root (Figure 6D) as shown in the 3D reconstruction from a z-stack using confocal microscopy (Figure 6D). Because of the predominance of wild type cells along the roots, we also examined the composition of cells in the wash based on the assumption that this might represent loosely attached cells. In this case, we found a mixture of wild type and ΔcrtB mutant cells (Figure 6D), most of which appeared to be motile (data not shown). Similar results were obtained using two fluorescent strains, wild type YR343-GFP and the ΔcrtB mutant strain with a chromosomal insertion of mCherry (data not shown).
Discussion
Pantoea sp. YR343 was isolated from the rhizosphere of a healthy Populus deltoides tree in North Carolina and is a robust colonizer of plant roots. Based on phylogeny, Pantoea sp. YR343 is most closely related to Pantoea sp. GM01 which was isolated from the rhizosphere of a P. deltoides tree in Tennessee. Like Pantoea sp. YR343, the colonies of Pantoea sp. GM01 are yellowish in color and this strain is predicted to produce carotenoids based on its genomic sequence. Our phylogenetic analyses also indicated that both Pantoea sp. YR343 and GM01 are closely related to P. rwandensis, P. rodasii, P. vagans, and P. eucalypti, all of which were isolated from eucalyptus trees (Coutinho et al., 2002; Brady et al., 2009, 2012). Importantly, however, P. rwandensis, P. rodasii, P. vagans, and P. eucalypti were isolated from diseased eucalyptus trees and have been implicated as the causal agents of bacterial blight, leaf lesions, and dieback. Despite the close phylogenetic relationship, we have not observed any pathogenicity associated with Pantoea sp. YR343 using P. deltoides, T. aestivum (wheat), or A. thaliana as plant hosts. Since only two isolates are available from Populus in the Eastern USA at present, it is unclear at this point whether these isolates represent a single or multiple new species. It will be interesting to perform comparative genomic analyses on new and related Pantoea strains as they become available to resolve these systematics issues and to determine whether any host-specificity or pathogenicity-related factors can be identified.
Pantoea sp. YR343 possesses a number of characteristics that may promote its ability to survive in the rhizosphere and associate with plant hosts, including both swimming and swarming motility, the ability to solubilize phosphate, and the production of IAA. Motility, directed by chemotaxis, is an important means by which bacteria may avoid adverse conditions, while detecting and colonizing roots within the soil environment (Bashan, 1986; de Weert et al., 2002; Somers et al., 2004). Swarming motililty has been associated with virulence in the plant pathogen, P. stewartii, which colonizes the plant xylem, blocking flow and causing wilting in its plant hosts, including corn (Herrera et al., 2008). We observed enhanced swarming motility in Pantoea sp. YR343 in the presence of 0.4% glycerol, as compared to medium containing 0.4% glucose. Interestingly, this phenotype was the direct opposite of that shown for P. stewartii (Herrera et al., 2008), suggesting a possible difference in metabolism and preferred growth environment for Pantoea sp. YR343. Pantoea sp. YR343 can also solubilize phosphate, which has been implicated in plant-growth promotion due to increasing the availability of usable phosphate to the plants (Rodriguez and Fraga, 1999).
We also observed that Pantoea sp. YR343 produces cellulose during biofilm growth as a component of the exopolysaccharide matrix. Examination of Enterobacteriaceae genomes shows the existence of two distinct cellulose synthesis gene clusters, represented by the operon structure found in Gluconacetobacter xylinus (Wong et al., 1990) compared to the operon structure found in Escherichia coli (Zogaj et al., 2001). While the catalytic proteins, encoded by bscA and bscB, are conserved in both types of cellulose synthesis gene clusters, there are genes unique to each operon for which the functions are not well known (Romling, 2002). The majority of bacteria that produce cellulose possess a single cellulose synthesis gene cluster; however, Pantoea sp. YR343 possesses two gene clusters with distinct organizations representing both classes of cellulose synthesis gene clusters. Genomic comparisons indicate the presence of two cellulose synthase operons in other Pantoea spp. and some related Klebsiella spp. as well. Whether these cellulose gene clusters are differentially regulated and/or produce distinct cellulose synthase complexes has yet to be determined.
Our analyses indicate that Pantoea sp. YR343 readily forms biofilms on abiotic and biotic surfaces. Interestingly, our initial attempts at measuring biofilm formation using a 96-well assay were inconsistent until we replaced the lid of the 96-well plate with breathable tape. This observation suggests that oxygen-sensing may play a role in biofilm formation in Pantoea sp. YR343, although we have not yet explored this hypothesis in depth. Using microscopy and engineered strains of Pantoea sp. YR343 that express fluorescent proteins, we have observed the attachment of Pantoea sp. YR343 on plant root surfaces. On rare occasions, we have also observed Pantoea sp. YR343 within plants (data not shown), suggesting that it can survive as a plant endophyte, at least transiently.
The transient production of reactive oxygen species (oxidative bursting) is a common plant defense mechanism during the early stages of plant–microbe interactions (Doke et al., 1996; Lamb and Dixon, 1997; Wojtaszek, 1997; Torres et al., 2006; Nanda et al., 2010). Many beneficial microbes have developed a number of strategies for overcoming such plant defense mechanisms (Zamioudis and Pieterse, 2012). For example, oxidative stress has been overcome by up-regulation of antioxidant pathways in Gluconaacetobacter diazotrophicus (Alqueres et al., 2010), production of superoxide dismutase in Rhizobium species (Santos et al., 2000), and synthesis of alkyl hydroperoxide reductase in Azospirillum brasilense (Wasim et al., 2009). Similarly, many microbes produce carotenoids, which play a vital role in the survival of cells under harsh conditions, such as oxidative stress, extremes in pH, and resistance to toxins (Liaaen-Jensen and Andrewes, 1972; Krinsky, 1989; Kulkarni et al., 2015). Thus, we hypothesized that carotenoids may play a role in rhizosphere survival and/or plant association. Genomic analyses of Pantoea sp. YR343 identified candidate genes encoding all of the biosynthetic enzymes involved in carotenoid biosynthesis (To et al., 1994; Sedkova et al., 2005). As predicted from these analyses, loss of phytoene synthase activity by deletion of the crtB gene, resulted in a strain that was unable to produce any detectable carotenoids and showed increased sensitivity to reactive oxygen species.
The production of carotenoids by Pantoea sp. YR343 proved to be a distinguishing feature for Raman spectroscopic analysis, in that the signals produced from the presence of carotenoids, particularly zeaxanthin, dominated the spectra, as seen in Figure 4D. It is likely that the Raman intensities of the carotenoid bands are pre-resonantly enhanced, which is a Raman scattering process that occurs when the frequency of the excitation laser beam lies just below the frequency of an electronic transition of the chromophore in the irradiated molecule. This process results in the selective enhancement, by factors of up to 106, of the Raman intensities of bands coupled to the electronic transition of the chromophore, which can be advantageous in the analysis of complex systems (Robert, 1990). Due to the selective enhancement afforded by resonance Raman scattering, molecular vibrations arising from other cellular components, such as DNA and proteins, are represented by relatively weak bands compared to the carotenoid bands in the Pantoea sp. YR343 Raman spectrum. The strong Raman signal in wild type Pantoea sp. YR343 also allowed us to follow the effect of H2O2 on carotenoids. In these analyses, we see a steady decrease in the Raman spectra associated with carotenoids as the cells are exposed to increasing concentrations of H2O2. The decrease in Raman signal intensity was apparent even at H2O2 concentrations that did not result in decreased cell viability, consistent with the protective role of carotenoids in the presence of free oxygen radicals (Krinsky, 1989, 1993; Hirayama et al., 1994; Edge et al., 1997).
Consistent with other Pantoea strains (Sergeeva et al., 2007; Kulkarni et al., 2013), we observed that Panteoa sp. YR343 produces IAA, which is synthesized from the amino acid tryptophan (Patten and Glick, 1996), an amino acid commonly found in plant root exudates (Kamilova et al., 2006). The genome of Pantoea sp. YR343 encodes a single, conserved ipdC gene which encodes indole pyruvate decarboxylase, suggesting that this strain synthesizes IAA using the indole pyruvate pathway (Patten and Glick, 1996). Other potential pathways to IAA production include the indole-acetamide pathway, although it is difficult to determine by genome comparisons whether this pathway is complete in Pantoea sp. YR343. Interestingly, IAA is produced in some pathogenic species of Pantoea that induce tumor or gall formation via the tryptamine pathway (Manulis et al., 1998). Our genome comparisons, however, suggest that Pantoea sp. YR343 does not have a complete tryptamine pathway. Surprisingly, we observed a decrease in IAA production by the ΔcrtB mutant. This was unexpected since there was no obvious connection (e.g., common enzymes or substrates) between the pathways involved in carotenoid production and the pathways involved in IAA production. In addition to their protective role, however, carotenoids have been implicated in modulation of membrane fluidity and may play a role in the formation of membrane domains (Huang and Haug, 1974; Chamberlain et al., 1991; Gruszecki, 1999; Gruszecki and Strzalka, 2005; Landrum, 2009; Lopez and Kolter, 2010). From this perspective, the decrease in IAA production by the ΔcrtB mutant may be a consequence of changes in membrane fluidity or organization. For example, uptake of tryptophan, which is a precursor to IAA production, may be defective in the ΔcrtB mutant.
Likewise, we also observed changes in biofilm formation in the ΔcrtB mutant. Indeed, the mutant was defective in both surface attachment and pellicle formation. In other organisms, such as Bacillus subtilis, it was shown that zaragozic acid, an inhibitor of squalene synthesis, could reduce the ability of B. subtilis to form biofilms (Lopez and Kolter, 2010). Squalene is a precursor in the production of hopanoids which, like carotenoids, have been show to modulate membrane fluidity and organization in bacteria (Kannenberg and Poralla, 1999; Kulkarni et al., 2015). Thus, we hypothesize that changes in membrane organization as a consequence of the loss of carotenoids could impact the function of membrane proteins involved in signaling and transport, and result in the observed defects in IAA production and biofilm formation in Pantoea sp. YR343.
Ultimately, the phenotypes associated with the loss of carotenoids in Pantoea sp. YR343 resulted in a mutant that was defective in plant association and/or rhizosphere survival. We observed a significant decrease in the number of ΔcrtB mutant cells associated with plant roots compared to wild type based on both CFU counts and microscopy. There are several possible explanations for the role carotenoids may have in plant association. For example, carotenoids may be needed for survival of Pantoea sp. YR343 in the soil, which would therefore influence its ability to associate with a plant root. Alternatively, carotenoids may play a role in overcoming plant defense mechanisms, including oxidative bursting, during colonization of the root surface. That the ability to overcome oxidative stress is important for root colonization was shown in Gluconoacetobacter diazotrophicus PAL5 by demonstrating that production of superoxide dismutase and glutathione reductase were essential for colonization of rice roots (Alqueres et al., 2013). Another possibility is that the loss of carotenoids affects membrane organization and impacts the signaling pathways involved in plant recognition. Finally, the loss of carotenoids may affect the ability of Pantoea sp. YR343 to establish a strong interaction with the plant root, perhaps due to defects in IAA production or in biofilm production. Future studies are aimed at elucidating the mechanisms by which carotenoids impact membrane organization in Pantoea sp. YR343.
Author Contributions
All authors contributed intellectual input and data analyses and assisted in manuscript preparation. JM-F, AB, and MD developed the experimental plan. AB, SF, DP, and JM-F performed phenotypic characterization and mutant analyses; CS and TC performed phylogenetic analyses, NE and TT performed GC-MS analyses, RM, SP, and PB performed Raman spectroscopy analyses, and AB, SJ, and DW conducted pathogenicity analyses. JM-F and AB wrote the paper.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to acknowledge Dr. Gladys Alexandre (University of Tennessee, Knoxville) for experimental advice and supplying the pK18mob-sacB vector as well as supplying wheat seeds obtained from the University of Tennessee, Knoxville. We would also like to acknowledge Justin Jagodinski for his preliminary work characterizing the ΔcrtB mutant. This research was sponsored by the Genomic Science Program, U.S. Department of Energy, Office of Science, Biological and Environmental Research, as part of the Plant Microbe Interfaces Scientific Focus Area (http://pmi.ornl.gov). Work at the University of Notre Dame was supported by DOE grant SC0006642 (RM) and by a subcontract from Oak Ridge National Laboratory (SP). This manuscript has been authored by UT-Battelle, LLC under Contract No. DE-AC05-00OR22725 with the U.S. Department of Energy. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a non-exclusive, paid-up, irrevocable, world-wide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes. The Department of Energy will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan (http://energy.gov/downloads/doe-public-access-plan).
References
Alqueres, S., Meneses, C., Rouws, L., Rothballer, M., Baldani, I., Schmid, M., et al. (2013). The bacterial superoxide dismutase and glutathione reductase are crucial for endophytic colonization of rice roots by Gluconacetobacter diazotrophicus PAL5. Mol. Plant Microbe Interact. 26, 937–945. doi: 10.1094/mpmi-12-12-0286-r
Alqueres, S. M., Oliveira, J. H., Nogueira, E. M., Guedes, H. V., Oliveira, P. L., Camara, F., et al. (2010). Antioxidant pathways are up-regulated during biological nitrogen fixation to prevent ROS-induced nitrogenase inhibition in Gluconacetobacter diazotrophicus. Arch. Microbiol. 192, 835–841. doi: 10.1007/s00203-010-0609–601
Amellal, N., Burtin, G., Bartoli, F., and Heulin, T. (1998). Colonization of wheat roots by an exopolysaccharide-producing Pantoea agglomerans strain and its effect on rhizosphere soil aggregation. Appl. Environ. Microbiol. 64, 3740–3747.
Apel, K., and Hirt, H. (2004). Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399. doi: 10.1146/annurev.arplant.55.031903.141701
Azad, H. R., Holmes, G. J., and Cooksey, D. A. (2000). A new leaf blotch disease of sudangrass caused by Pantoea ananatis and Pantoea stewartii. Plant Dis. 84, 973–979. doi: 10.1094/Pdis.2000.84.9.973
Barret, M., Morrissey, J. P., and O’Gara, F. (2011). Functional genomics analysis of plant growth-promoting rhizobacterial traits involved in rhizosphere competence. Biol. Fertil. Soils 47, 729–743. doi: 10.1007/s00374-011-0605-x
Bashan, Y. (1986). Migration of the rhizosphere bacteria Azospirillum brasilense and Pseudomonas fluorescens towards wheat roots in the soil. J. General Microbiol. 132, 3407–3414.
Bashan, Y., and de-Bashan, L. E. (2010). How the plant growth-promoting bacterium Azospirillum promotes plant growth - a critical assessment. Adv. Agron. 108, 77–136. doi: 10.1016/S0065-2113(10)08002-8
Beneduzi, A., Ambrosini, A., and Passaglia, L. M. P. (2012). Plant growth-promoting rhizobacteria (PGPR): their potential as antagonists and biocontrol agents. Genet. Mol. Biol. 35, 1044–1051. doi: 10.1590/S1415-47572012000600020
Brady, C. L., Cleenwerck, I., van der Westhuizen, L., Venter, S. N., Coutinho, T. A., and De Vos, P. (2012). Pantoea rodasii sp. nov., Pantoea rwandensis sp. nov., and Pantoea wallisii sp nov., isolated from Eucalyptus. Int. J. Syst. Evol. Microbiol. 62, 1457–1464. doi: 10.1099/Ijs.0.032615–32610
Brady, C., Cleenwerck, I., Venter, S., Vancanneyt, M., Swings, J., and Coutinho, T. (2008). Phylogeny and identification of Pantoea species associated with plants, humans and the natural environment based on multilocus sequence analysis (MLSA). Syst. Appl. Microbiol. 31, 447–460. doi: 10.1016/j.syapm.2008.09.004
Brady, C. L., Cleenwerck, I., Venter, S. N., Engelbeen, K., De Vos, P., and Coutinho, T. A. (2010). Emended description of the genus Pantoea, description of four species from human clinical samples, Pantoea septica sp. nov., Pantoea eucrina sp. nov., Pantoea brenneri sp. nov. and Pantoea conspicua sp. nov., and transfer of Pectobacterium cypripedii (Hori 1911) Brenner et al. 1973 emend. Hauben et al. 1998 to the genus as Pantoea cypripedii comb. nov. Int. J. Syst. Evol. Microbiol. 60, 2430–2440. doi: 10.1099/Ijs.0.017301–17300
Brady, C. L., Venter, S. N., Cleenwerck, I., Engelbeen, K., Vancanneyt, M., Swings, J., et al. (2009). Pantoea vagans sp. nov., Pantoea eucalypti sp. nov., Pantoea deleyi sp. nov. and Pantoea anthophila sp. nov. Int. J. Syst. Evol. Microbiol. 59, 2339–2345. doi: 10.1099/Ijs.0.009241–9240
Braun-Kiewnick, A., Jacobsen, B. J., and Sands, D. C. (2000). Biological Control of Pseudomonas syringae pv. syringae, the causal agent of basal kernel blight of barley, by antagonistic Pantoea agglomerans. Phytopathology 90, 368–375. doi: 10.1094/PHYTO.2000.90.4.368
Britton, G. (1989). “Carotenoid biosynthesis — an overview,” in Carotenoids, eds N. Krinsky, M. Mathews-Roth and R. Taylor (New York, NY: Springer), 167–184.
Brown, S. D., Utturkar, S. M., Klingeman, D. M., Johnson, C. M., Martin, S. L., Land, M. L., et al. (2012). Twenty-one genome sequences from Pseudomonas species and 19 genome sequences from diverse bacteria isolated from the rhizosphere and endosphere of Populus deltoides. J. Bacteriol. 194, 5991–5993. doi: 10.1128/Jb.01243–1212
Chamberlain, N. R., Mehrtens, B. G., Xiong, Z., Kapral, F. A., Boardman, J. L., and Rearick, J. I. (1991). Correlation of carotenoid production, decreased membrane fluidity, and resistance to oleic acid killing in Staphylococcus aureus 18Z. Infect. Immun. 59, 4332–4337.
Cogdell, R. J., and Frank, H. A. (1987). How carotenoids function in photosynthetic bacteria. Biochim. Biophys. Acta 895, 63–79. doi: 10.1016/s0304-4173(87)80008–80003
Cogdell, R. J., Howard, T. D., Bittl, R., Schlodder, E., Geisenheimer, I., and Lubitz, W. (2000). How carotenoids protect bacterial photosynthesis. Philos. Trans. R. Soc. B Biol. Sci. 355, 1345–1349. doi: 10.1098/rstb.2000.0696
Coutinho, T. A., Preisig, O., Mergaert, J., Cnockaert, M. C., Riedel, K. H., Swings, J., et al. (2002). Bacterial blight and dieback of Eucalyptus species, hybrids, and clones in South Africa. Plant Dis. 86, 20–25. doi: 10.1094/Pdis.2002.86.1.20
De Maayer, P., Chan, W. Y., Venter, S. N., Toth, I. K., Birch, P. R. J., Joubert, F., et al. (2010). Genome sequence of Pantoea ananatis LMG20103, the causative agent of Eucalyptus blight and dieback. J. Bacteriol. 192, 2936–2937. doi: 10.1128/Jb.00060–10
de Oliveira, V. E., Castro, H. V., Edwards, H. G. M., and de Oliveira, L. F. C. (2010). Carotenes and carotenoids in natural biological samples: a Raman spectroscopic analysis. J. Raman Spectrosc. 41, 642–650. doi: 10.1002/jrs.2493
de Weert, S., Vermeiren, H., Mulders, I. H., Kuiper, I., Hendrickx, N., Bloemberg, G. V., et al. (2002). Flagella-driven chemotaxis towards exudate components is an important trait for tomato root colonization by Pseudomonas fluorescens. Mol. Plant Microbe Interact. 15, 1173–1180. doi: 10.1094/mpmi.2002.15.11.1173
del Amor, F. M., and Cuadra-Crespo, P. (2011). Plant growth-promoting bacteria as a tool to improve salinity tolerance in sweet pepper. Funct. Plant Biol. 39, 82–90. doi: 10.1071/FP11173
Doke, N., Miura, Y., Sanchez, L. M., Park, H. J., Noritake, T., Yoshioka, H., et al. (1996). The oxidative burst protects plants against pathogen attack: mechanism and role as an emergency signal for plant bio-defence–a review. Gene 179, 45–51. doi: 10.1016/S0378-1119(96)00423-4
Dussault, D., Caillet, S., Le Tien, C., and Lacroix, M. (2008). Carotenoids’ influence on radiotolerance of Pantoea agglomerans, a plant pathogen. Lett. Appl. Microbiol. 47, 208–213. doi: 10.1111/j.1472-765X.2008.02410.x
Edge, R., McGarvey, D. J., and Truscott, T. G. (1997). The carotenoids as anti-oxidants–a review. J. Photochem. Photobiol. B 41, 189–200. doi: 10.1016/S1011-1344(97)00092-4
Egamberdiyeva, D., and Höflich, G. (2001). “Influence of growth promoting bacteria from Uzbekistan and Germany on the growth and nutrient uptake of cotton and wheat on different soils,” in Plant Nutrition: Food Security and Sustainability of Agro-Ecosystems Through Basic and Applied Research Developments in plant and soil sciences, eds W. Horst, M. K. Schenk, A. Bürkert, N. Claassen, H. Flessa, W. B. Frommer, et al. (Berlin: Springer), 674–675.
Feng, Y., Shen, D., and Song, W. (2006). Rice endophyte Pantoea agglomerans YS19 promotes host plant growth and affects allocations of host photosynthates. J. Appl. Microbiol. 100, 938–945. doi: 10.1111/j.1365-2672.2006.02843.x
Foote, C. S., and Denny, R. W. (1968). Chemistry of singlet oxygen. VII. Quenching by beta-carotene. J. Am. Chem. Soc. 90, 6233–6235. doi: 10.1021/ja01024a061
Fraser, N. J., Hashimoto, H., and Cogdell, R. J. (2001). Carotenoids and bacterial photosynthesis: the story so far. Photosynth. Res. 70, 249–256. doi: 10.1023/A:1014715114520
Gechev, T. S., Van Breusegem, F., Stone, J. M., Denev, I., and Laloi, C. (2006). Reactive oxygen species as signals that modulate plant stress responses and programmed cell death. Bioessays 28, 1091–1101. doi: 10.1002/bies.20493
Goodwin, J. R., Hafner, L. M., and Fredericks, P. M. (2006). Raman spectroscopic study of the heterogeneity of microcolonies of a pigmented bacterium. J. Raman Spectrosc. 37, 932–936. doi: 10.1002/jrs.1523
Gottel, N. R., Castro, H. F., Kerley, M., Yang, Z., Pelletier, D. A., Podar, M., et al. (2011). Distinct microbial communities within the endosphere and rhizosphere of Populus deltoides roots across contrasting soil types. Appl. Environ. Microbiol. 77, 5934–5944. doi: 10.1128/AEM.05255-11
Gruszecki, W. I. (1999). “Carotenoids in Membranes,” in The Photochemistry of Carotenoids, eds H. A. Frank, A. J. Young, G. Britton and R. J. Cogdell (Berlin: Springer).
Gruszecki, W. I., and Strzalka, K. (2005). Carotenoids as modulators of lipid membrane physical properties. Biochim. Biophys. Acta 1740, 108–115. doi: 10.1016/j.bbadis.2004.11.015
Guindon, S., Dufayard, J. F., Lefort, V., Anisimova, M., Hordijk, W., and Gascuel, O. (2010). New algorithms and methods to estimate maximum-likelihood phylogenies: assessing the performance of PhyML 3.0. Syst. Biol. 59, 307–321. doi: 10.1093/sysbio/syq010
Hartmann, A., and Hurek, T. (1988). Effect of carotenoid overproduction on oxygen tolerance of nitrogen fixation in Azospirillum brasilense Sp7. J. Gen. Microbiol. 134, 2449–2455. doi: 10.1099/00221287-134-9-2449
Hayat, R., Ali, S., Amara, U., Khalid, R., and Ahmed, I. (2010). Soil beneficial bacteria and their role in plant growth promotion: a review. Ann. Microbiol. 60, 579–598. doi: 10.1007/s13213-010-0117–111
Herrera, C. M., Koutsoudis, M. D., Wang, X., and von Bodman, S. B. (2008). Pantoea stewartii subsp. stewartii exhibits surface motility, which is a critical aspect of Stewart’s wilt disease development on maize. Mol. Plant Microbe Interact. 21, 1359–1370. doi: 10.1094/MPMI-21-10-1359
Hirayama, O., Nakamura, K., Hamada, S., and Kobayasi, Y. (1994). Singlet oxygen quenching ability of naturally occurring carotenoids. Lipids 29, 149–150. doi: 10.1007/BF02537155
Huang, L., and Haug, A. (1974). Regulation of membrane lipid fluidity in Acholeplasma laidlawii: effect of carotenoid pigment content. Biochim. Biophys. Acta 352, 361–370. doi: 10.1016/0005-2736(74)90228-4
Hundle, B. S., Beyer, P., Kleinig, H., Englert, G., and Hearst, J. E. (1991). Carotenoids of Erwinia herbicola and an Escherichia coli HB101 strain carrying the Erwinia herbicola carotenoid gene cluster. Photochem. Photobiol. 54, 89–93. doi: 10.1111/j.1751-1097.1991.tb01989.x
Kamilova, F., Kravchenko, L. V., Shaposhnikov, A. I., Azarova, T., Makarova, N., and Lugtenberg, B. (2006). Organic acids, sugars, and L-tryptophane in exudates of vegetables growing on stonewool and their effects on activities of rhizosphere bacteria. Mol. Plant Microbe Interact. 19, 250–256. doi: 10.1094/MPMI-19-0250
Kannenberg, E. L., and Poralla, K. (1999). Hopanoid biosynthesis and function in bacteria. Naturwissenschaften 86, 168–176. doi: 10.1007/s001140050592
Kloos, K., Mergel, A., Rösch, C., and Bothe, H. (2001). Denitrification within the genus Azospirillum and other associative bacteria. Austral. J. Plant Physiol. 28, 991–998.
Krinsky, N. I. (1989). Antioxidant functions of carotenoids. Free Radic. Biol. Med. 7, 617–635. doi: 10.1016/0891-5849(89)90143-3
Krinsky, N. I. (1993). Actions of carotenoids in biological systems. Annu. Rev. Nutr. 13, 561–587. doi: 10.1146/annurev.nu.13.070193.003021
Kulkarni, G., Busset, N., Molinaro, A., Gargani, D., Chaintreuil, C., Silipo, A., et al. (2015). Specific hopanoid classes differentially affect free-living and symbiotic states of Bradyrhizobium diazoefficiens. MBio 6:e01251. doi: 10.1128/mBio.01251-1215
Kulkarni, G. B., Nayak, A. S., Sajjan, S. S., Oblesha, A., and Karegoudar, T. B. (2013). Indole-3-acetic acid biosynthetic pathway and aromatic amino acid aminotransferase activities in Pantoea dispersa strain GPK. Lett. Appl. Microbiol. 56, 340–347. doi: 10.1111/lam.12053
Lamb, C., and Dixon, R. A. (1997). The oxidative burst in plant disease resistance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 251–275. doi: 10.1146/annurev.arplant.48.1.251
Landrum, J. T. (2009). Carotenoids: Physical, Chemical, and Biological Functions and Properties. Boca Raton: CRC Press.
Liaaen-Jensen, S., and Andrewes, A. G. (1972). Microbial carotenoids. Annu. Rev. Microbiol. 26, 225–248. doi: 10.1146/annurev.mi.26.100172.001301
Lopez, D., and Kolter, R. (2010). Functional microdomains in bacterial membranes. Genes Dev. 24, 1893–1902. doi: 10.1101/gad.1945010
Manulis, S., Haviv-Chesner, A., Brandl, M. T., Lindow, S. E., and Barash, I. (1998). Differential involvement of indole-3-acetic acid biosynthetic pathways in pathogenicity and epiphytic fitness of Erwinia herbicola pv. gypsophilae. Mol. Plant Microbe Interact. 11, 634–642. doi: 10.1094/Mpmi.1998.11.7.634
Merlin, J. C. (1985). Resonance Raman spectroscopy of carotenoids and carotenoid-containing systems. Pure Appl. Chem. 57, 785–792. doi: 10.1351/pac198557050785
Mohammadi, M., Burbank, L., and Roper, M. C. (2012). Biological role of pigment production for the bacterial phytopathogen Pantoea stewartii subsp. stewartii. Appl. Environ. Microbiol. 78, 6859–6865. doi: 10.1128/AEM.01574-1512
Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue cultures. Physiol. Plant. 15, 473–497. doi: 10.1111/j.1399-3054.1962.tb08052.x
Nanda, A. K., Andrio, E., Marino, D., Pauly, N., and Dunand, C. (2010). Reactive oxygen species during plant-microorganism early interactions. J. Integr. Plant Biol. 52, 195–204. doi: 10.1111/j.1744-7909.2010.00933.x
O’Toole, G. A., and Kolter, R. (1998). Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28, 449–461. doi: 10.1046/j.1365-2958.1998.00797.x
Patten, C. L., and Glick, B. R. (1996). Bacterial biosynthesis of indole-3-acetic acid. Can. J. Microbiol. 42, 207–220. doi: 10.1139/m96-032
Pelletier, D. A., Hurst, G. B., Foote, L. J., Lankford, P. K., McKeown, C. K., Lu, T. Y., et al. (2008). A general system for studying protein-protein interactions in Gram-negative bacteria. J. Proteome Res. 7, 3319–3328. doi: 10.1021/pr8001832
Perrig, D., Boiero, M. L., Masciarelli, O. A., Penna, C., Ruiz, O. A., Cassan, F. D., et al. (2007). Plant-growth-promoting compounds produced by two agronomically important strains of Azospirillum brasilense, and implications for inoculant formulation. Appl. Microbiol. Biotechnol. 75, 1143–1150. doi: 10.1007/s00253-007-0909–909
Raaijmakers, J. M., Paulitz, T. C., Steinberg, C., Alabouvette, C., and Moenne-Loccoz, Y. (2009). The rhizosphere: a playground and battlefield for soilborne pathogens and beneficial microorganisms. Plant Soil 321, 341–361. doi: 10.1007/s11104-008-9568-6
Robert, B. (1990). Resonance Raman studies of bacterial reaction centers. Biochim. Biophys. Acta 1017, 99–111. doi: 10.1016/0005-2728(90)90140-y
Rodriguez, H., and Fraga, R. (1999). Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol. Adv. 17, 319–339. doi: 10.1016/S0734-9750(99)00014-2
Romling, U. (2002). Molecular biology of cellulose production in bacteria. Res. Microbiol. 153, 205–212. doi: 10.1016/S0923-2508(02)01316-5
Ruppel, S., Hecht-Buchholz, C., Remus, R., Ortmann, U., and Schmelzer, R. (1992). Settlement of the diazotrophic, phytoeffective bacterial strain Pantoea agglomerans on and within winter wheat: an investigation using ELISA and transmission electron microscopy. Plant Soil 145, 261–273. doi: 10.1007/BF00010355
Sandmann, G., and Misawa, N. (1992). New functional assignment of the carotenogenic genes crtB and crtE with constructs of these genes from Erwinia species. FEMS Microbiol. Lett. 69, 253–257. doi: 10.1111/j.1574-6968.1992.tb05162.x
Santos, R., Herouart, D., Puppo, A., and Touati, D. (2000). Critical protective role of bacterial superoxide dismutase in rhizobium-legume symbiosis. Mol. Microbiol. 38, 750–759. doi: 10.1046/j.1365-2958.2000.02178.x
Schafer, A., Tauch, A., Jager, W., Kalinowski, J., Thierbach, G., and Puhler, A. (1994). Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 145, 69–73. doi: 10.1016/0378-1119(94)90324-7
Schulz, H., Baranska, M., and Baranski, R. (2005). Potential of NIR-FT-Raman spectroscopy in natural carotenoid analysis. Biopolymers 77, 212–221. doi: 10.1002/bip.20215
Sedkova, N., Tao, L., Rouviere, P. E., and Cheng, Q. (2005). Diversity of carotenoid synthesis gene clusters from environmental Enterobactetiaceae strains. Appl. Environ. Microbiol. 71, 8141–8146. doi: 10.1128/Aem.71.12.8141-8146.2005
Sergeeva, E., Hirkala, D. L. M., and Nelson, L. M. (2007). Production of indole-3-acetic acid, aromatic amino acid aminotransferase activities and plant growth promotion by Pantoea agglomerans rhizosphere isolates. Plant Soil. 297, 1–13. doi: 10.1007/s11104-007-9314-5
Shakya, M., Gottel, N., Castro, H., Yang, Z. K., Gunter, L., Labbe, J., et al. (2013). A multifactor analysis of fungal and bacterial community structure in the root microbiome of mature Populus deltoides trees. PLoS ONE 8:e76382. doi: 10.1371/journal.pone.0076382
Somers, E., Vanderleyden, J., and Srinivasan, M. (2004). Rhizosphere bacterial signalling: a love parade beneath our feet. Crit. Rev. Microbiol. 30, 205–240. doi: 10.1080/10408410490468786
Spaepen, S., Vanderleyden, J., and Remans, R. (2007). Indole-3-acetic acid in microbial and microorganism-plant signaling. FEMS Microbiol. Rev. 31, 425–448. doi: 10.1111/j.1574-6976.2007.00072.x
Stockwell, V. O., Johnson, K. B., Sugar, D., and Loper, J. E. (2010). Control of fire blight by Pseudomonas fluorescens A506 and Pantoea vagans C9-1 applied as single strains and mixed inocula. Phytopathology 100, 1330–1339. doi: 10.1094/Phyto.03.10.0097
Tang, Y. W., and Bonner, J. (1947). The enzymatic inactivation of indoleacetic acid; some characteristics of the enzyme contained in pea seedlings. Arch. Biochem. 13, 11–25.
To, K. Y., Lai, E. M., Lee, L. Y., Lin, T. P., Hung, C. H., Chen, C. L., et al. (1994). Analysis of the gene cluster encoding carotenoid biosynthesis in Erwinia herbicola Eho13. Microbiology 140(Pt 2), 331–339. doi: 10.1099/13500872-140-2–331
Torres, M. A., Jones, J. D., and Dangl, J. L. (2006). Reactive oxygen species signaling in response to pathogens. Plant Physiol. 141, 373–378. doi: 10.1104/pp.106.079467
Tram, G., Korolik, V., and Day, C. J. (2013). MBDS Solvent: an improved method for assessment of biofilms. Adv. Microbiol. 3, 200–204. doi: 10.4236/aim.2013.32030
Verma, S. C., Ladha, J. K., and Tripathi, A. K. (2001). Evaluation of plant growth promoting and colonization ability of endophytic diazotrophs from deep water rice. J. Biotechnol. 91, 127–141. doi: 10.1016/S0168-1656(01)00333-9
Walcott, R. R., Gitaitis, R. D., Castro, A. C., Sanders, F. H., and Diaz-Perez, J. C. (2002). Natural infestation of onion seed by Pantoea ananatis, causal agent of center rot. Plant Dis. 86, 106–111. doi: 10.1094/Pdis.2002.86.2.106
Walterson, A. M., and Stavrinides, J. (2015). Pantoea: insights into a highly versatile and diverse genus within the Enterobacteriaceae. FEMS Microbiol. Rev. 39, 968–984. doi: 10.1093/femsre/fuv027
Wang, Y., Ohara, Y., Nakayashiki, H., Tosa, Y., and Mayama, S. (2005). Microarray analysis of the gene expression profile induced by the endophytic plant growth-promoting rhizobacteria, Pseudomonas fluorescens FPT9601-T5 in Arabidopsis. Mol. Plant Microbe Interact. 18, 385–396. doi: 10.1094/MPMI-18–0385
Wasim, M., Bible, A. N., Xie, Z., and Alexandre, G. (2009). Alkyl hydroperoxide reductase has a role in oxidative stress resistance and in modulating changes in cell-surface properties in Azospirillum brasilense Sp245. Microbiology 155, 1192–1202. doi: 10.1099/mic.0.022541-22540
Weston, D. J., Pelletier, D. A., Morrell-Falvey, J. L., Tschaplinski, T. J., Jawdy, S. S., Lu, T. Y., et al. (2012). Pseudomonas fluorescens induces strain-dependent and strain-independent host plant responses in defense networks, primary metabolism, photosynthesis, and fitness. Mol. Plant Microbe Interact. 25, 765–778. doi: 10.1094/MPMI-09-11-0253
Withnall, R., Chowdhry, B. Z., Silver, J., Edwards, H. G. M., and de Oliveira, L. F. C. (2003). Raman spectra of carotenoids in natural products. Spectrochim. Acta Part A 59, 2207–2212. doi: 10.1016/s1386-1425(03)00064–67
Wojtaszek, P. (1997). Oxidative burst: an early plant response to pathogen infection. Biochem. J. 322(Pt 3), 681–692. doi: 10.1042/bj3220681
Wong, H. C., Fear, A. L., Calhoon, R. D., Eichinger, G. H., Mayer, R., Amikam, D., et al. (1990). Genetic organization of the cellulose synthase operon in Acetobacter xylinum. Proc. Natl. Acad. Sci. U.S.A. 87, 8130–8134. doi: 10.1073/pnas.87.20.8130
Zamioudis, C., and Pieterse, C. M. (2012). Modulation of host immunity by beneficial microbes. Mol Plant Microbe Interact 25, 139–150. doi: 10.1094/mpmi-06-11-0179
Keywords: Pantoea, carotenoids, crtB, indole-3-acetic acid, poplar, rhizosphere, zeaxanthin
Citation: Bible AN, Fletcher SJ, Pelletier DA, Schadt CW, Jawdy SS, Weston DJ, Engle LN, Tschaplinski T, Masyuko R, Polisetti S, Bohn PW, Coutinho TA, Doktycz MJ and Morrell-Falvey JL (2016) A Carotenoid-Deficient Mutant in Pantoea sp. YR343, a Bacteria Isolated from the Rhizosphere of Populus deltoides, Is Defective in Root Colonization. Front. Microbiol. 7:491. doi: 10.3389/fmicb.2016.00491
Received: 19 January 2016; Accepted: 24 March 2016;
Published: 18 April 2016.
Edited by:
Stéphane Hacquard, Max Planck Institute for Plant Breeding Research, GermanyReviewed by:
Birgit Mitter, AIT Austrian Institute of Technology, AustriaAnton Hartmann, Helmholtz Zentrum München– German Research Center for Environmental Health, Germany
Ann M. Stevens, Virginia Tech, USA
Copyright © 2016 Bible, Fletcher, Pelletier, Schadt, Jawdy, Weston, Engle, Tschaplinski, Masyuko, Polisetti, Bohn, Coutinho, Doktycz and Morrell-Falvey. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jennifer L. Morrell-Falvey, bW9ycmVsbGpsMUBvcm5sLmdvdg==
 Amber N. Bible
Amber N. Bible Sarah J. Fletcher
Sarah J. Fletcher Dale A. Pelletier
Dale A. Pelletier Christopher W. Schadt
Christopher W. Schadt Sara S. Jawdy
Sara S. Jawdy David J. Weston
David J. Weston Nancy L. Engle1
Nancy L. Engle1 Teresa A. Coutinho
Teresa A. Coutinho Mitchel J. Doktycz
Mitchel J. Doktycz Jennifer L. Morrell-Falvey
Jennifer L. Morrell-Falvey