- 1Key Laboratory of Experimental Marine Biology, Institute of Oceanology, Chinese Academy of Sciences, Qingdao, China
- 2Laboratory for Marine Biology and Biotechnology, Qingdao National Laboratory for Marine Science and Technology, Qingdao, China
Vibrio sp. is the most serious pathogen in marine aquaculture, and the development of anti-Vibrio agents is urgently needed. However, it is extreme lack of high-throughput screening (HTS) model for searching anti-Vibrio compounds. Here, we established a protein-based HTS screening model to identify agents targeting peptide deformylase (PDF) of Vibrio anguillarum. To find potential anti-Vibrio compounds, crude extracts derived from marine actinomycetes were applied for screening with this model. Notably, crude extract of strain Streptomyces sp. NHF165 inhibited dramatically both on V. anguillarum PDF (VaPDF) activity and V. anguillarum cell growth. And actinonin was further identified as the functional component. Anti-VaPDF and anti-V. anguillarum activities of actinonin were dose-dependent, and the IC50 values were 6.94 and 2.85 μM, respectively. To understand the resistance of V. anguillarum against actinonin, spontaneous V. anguillarum mutants with resistance against actinonin were isolated. Surprisingly, for the resistant strains, the region between 774 and 852 base pairs was found to be absent in the gene folD which produces 10-formyl-tetrahydrofolate, a donor of N-formyl to Met-tRNAfmet. When compared to the wild type strain, ΔfolD mutant showed eight times of minimum inhibition concentration on actinonin, however, the folD complementary strain could not grow on the medium supplemented with actinonin, which suggested that folD gene mutation was mainly responsible for the actinonin resistance. To our knowledge, this is the first report showing that marine derived Streptomyces sp. could produce actinonin with anti-VaPDF activity and the resistance against actinonin by V. anguillarum is mediated by mutation in folD gene.
Introduction
Sudden outbreak of diseases is a major setback in aquaculture, and it leads to high mortality and severe economic loss in all producing countries. Marine Vibrio species are associated with large-scale losses of penaeids and also cause diseases to fish (Letchumanan et al., 2015b). Vibrio anguillarum is the causative agent of vibriosis, a deadly haemorrhagic septicaemic disease affecting various marine and fresh/brackish water fish, bivalves and crustaceans. In both aquaculture and larviculture, this disease is responsible for severe economic losses worldwide (Frans et al., 2011). Vibrio species inhabit aquatic environments at temperatures ranging from 10 to 30°C and are highly susceptible to antibiotics (Shaw et al., 2014). Therefore, antibiotics is one of the main choices for controlling the proliferation of Vibrio sp. in aquaculture. Oxytetracycline, tetracycline, quinolones, sulphonamides and trimethoprim are antimicrobial agents permitted and utilized in the Asian aquaculture industry (Letchumanan et al., 2015a). However, extensive use of antibiotics has been postulated to be a major contributing factor in the rising incidence of antimicrobial resistance in pathogenic bacteria. Three fundamental mechanisms of antimicrobial resistance have been summarized: (1) prevention of access to target, (2) changes in antibiotic targets by mutation, and (3) modification (and protection) of targets (Blair et al., 2015). New resistance mechanisms are constantly being described, such as combined novel gene mph(G) coding macrolide phosphotransferase and gene mef(C) coding efflux pump were found to be responsible for high-level macrolide resistance Vibrio sp. (Nonaka et al., 2015).
To find novel anti-Vibrio sp. agents, screening models targeting Vibrio sp. whole cells or proteins involved in quorum sensing have been widely used (Zhang et al., 2016; Zhao et al., 2016). Because of serious antibiotics resistance, screening models with new targets are always needed. Peptide deformylase (PDF) is a class of metalloprotease responsible for catalyzing the removal of N-formyl group from N-terminal methionine following translation in prokaryotes. The widespread occurrence, conservation, and essential nature of deformylase in bacteria make it an attractive target for antibacterial drug discovery (Giglione et al., 2000; Sangshetti et al., 2015). PDF is widely used in human bacteria infection treatment caused by Staphylococcus aureus, Streptococcus pneumonia, Helicobacter pylori, Haemophilus influenza and Mycobacterium tuberculosis, etc (Sharma et al., 2009; Peyrusson et al., 2015). PDF inhibitors, GSK-1322322, BB-83698 and LBM-415, have entered into clinical developments (Sangshetti et al., 2015).
However, very little was investigated about PDF of aquaculture pathogen V. anguillarum. Actually, like other gram-negative organisms, V. anguillarum has one chromosomal copy of pdf gene, and no results have been published regarding PDF as an anti-Vibrio sp. target in marine aquaculture. Actinonin was reported in 1962 (Gordon et al., 1962) and was the first characterized PDF inhibitor (Chen et al., 2000). Up to now, resistance to actinonin has been reported in Staphylococcus aureus, Streptococcus pneumonia, Bacillus subtilis, Haemophilus influenza, Streptococcus pyogenes and Escherichia coli. Mechanisms causing actinonin resistance were also investigated in these strains. Genes pdf, folD, fmt, and glyA involved in translation initiation were the most frequency mutation sites (Margolis et al., 2000, 2001; Duroc et al., 2009).
Natural products are essential for the novel antibiotics screening. A lot of compounds had been developed to efficient antibiotics and applied in diseases treatment of human and aquaculture (Varoglu et al., 1997; Vinothkumar and Parameswaran, 2013). It is well known that the biodiversity of the marine environment and the associated chemical diversity constitute a practically unlimited resource of new bioactive substance, and the bioactive compounds from marine microorganisms have been exploited for decades (Varoglu et al., 1997). Marine actinomycete is one of the most efficient organisms of natural bioactive metabolite producers. The genus Streptomyces is considered as the most prolific producer of bioactive agents amongst actinomycete (Miao and Davies, 2010). Interestingly, Streptomyces sp. isolated from arctic were found to have biofilm inhibitory activity against Vibrio sp. by attenuating the signal molecules N-acylated homoserine lactones’ activity (You et al., 2007), and Streptomyces producing siderophores derived from nearshore marine sediments were found to inhibit the growth of Vibrio sp. by competition for iron in the aquatic environment (You et al., 2005).
In this study, we established an high-throughput screening (HTS) model targeting PDF of pathogenic bacterium V. anguillarum YN isolated from infected Scophthalmus maximus samples. Actinomycetes from eight different South China Sea sediments were isolated and corresponding crude extracts were prepared and subjected to anti-V. anguillarum agents screening. Actinonin produced by marine Streptomyces sp. NHF165 exhibited high inhibitory both on V. anguillarum PDF (VaPDF) activity and V. anguillarum cell growth. Furthermore, actinonin-resistant V. anguillarum mutants were obtained and the mechanism of resistance was also elucidated.
Materials and Methods
V. anguillarum PDF (VaPDF) Expression and Purification
The pdf gene was amplified from V. anguillarum YN genome DNA by PCR using the following primers: For: 5′-CGCGGATCCATGTCTGTATTACAAG-3′ (the underlined region indicates BamH I site) and Rev: 5′-CCGCTCGAGTTAGTTTTTTTCGTTATAG-3′ (the underlined region indicates Xho I site). PCR products were cloned into pMD18-T vector (TaKaRa). After sequence confirmation, PCR products were inserted in the multiple cloning site of vector pET30a(+) (Novagen) and the resulting plasmid was designated as pET30a(+)::pdf. Plasmid pET30a(+)::pdf was transformed into E. coli BL21(DE3) cells. Recombinant PDF was expressed and purified as follows. Briefly, cells harboring plasmids pET30a(+)::pdf were grown to an absorbance at 600 nm (A600) of 0.6 and induced with 0.5 mM isopropyl-D-thiogalactopyranoside at 16°C overnight. Cells were harvested by centrifugation, washed in HEPES buffer (25 mM, pH 7.4) and resuspended in HEPES (pH 7.4)-75 mM KCl-10% glycerol (buffer A). Then cells were lysed by sonication and centrifugated at 25,000 ×g. The supernatant was loaded onto a 5 ml HisTrap FF column (GE healthcare) and equilibrated in buffer A. The column was further washed and eluted with a gradient of imidazole from 0 to 300 mM using ÄKTA protein purification system (GE healthcare).
Anti-VaPDF Screening Assay
Peptide deformylase catalyzes the removal of the N-formyl group from formyl-Met-Ala-Ser. The free amino group reacts with fluorescamine to form highly fluorescent products which can be monitored with a TECAN Infinite M1000 PRO multi-mode microplate reader by exciting at 390 nm and emission at 470 nm. For screening, assays were performed in black flat-bottom 96-well microplates (Corning). First, 49.5 μl reaction solution (20 nM VaPDF, 1 mM formyl-Met-Ala-Ser and 25 mM HEPES, pH 7.4) was dispensed in each well and then 0.5 μl dimethylsulfoxide (DMSO) or samples dissolved in DMSO (4 mg/ml) was dispensed. Plates were incubated at 37°C for 30 min. Then fluorescamine was added to a final concentration of 60 μg/ml. The fluorescence intensity (FI) of each well was detected. The inhibitory values were calculated as (FIsample-FInegativecontrol)/(FIpositivecontrol-FInegativecontrol) × 100%.
Dimethylsulfoxide was chosen as negative control and heat-inactivated VaPDF as positive control during measurements. The Z′ factor and CV values were calculated as follows:
Z′ = 1–3(SDFImax-SDFImin)/(MeanFImax-MeanFImin), SD: standard deviation. The theoretical value is between 0.5 and 1. CV(%) = SDFImax/MeanFImax or CV(%) = SDFImin/MeanFImin. The acceptable value of CV for HTS assay is less than 10%.
Anti-V. anguillarum Cell Based Assay
The anti-V. anguillarum assay utilized strain V. anguillarum YN which was isolated from infected Scophthatmus maximus sample. The activities of crude extracts or compounds against V. anguillarum were determined in a clear flat-bottom 96-well plate. V. anguillarum YN was grown at 28°C to mid-log phase in Luria Bertani (LB) medium ( peptone 10 g, yeast extract 5 g, NaCl 10 g, in 1000 ml distilled water, pH 7.0). Then the culture was diluted to A600 = 0.025 with LB medium. 80 μl bacterial suspension was added to each well, followed by adding 0.8 μl of sample solution (4 mg/ml). DMSO served as the negative control and chloramphenicol as the positive control. The plate was incubated at 28°C for 15 h and the growth of V. anguillarum YN was measured by detecting A600 of each well.
Marine Actinomycetes Isolation and Crude Extracts Preparation
Sediment samples were collected using the mud sampler in the South China Sea during 26th April to 23th May 2010 (Supplementary Table S1). The samples were transported to laboratory in an insulated container at 4°C and then stored at -80°C. All samples were pretreated using dispersion and differential centrifugation (DDC) method (Hopkins et al., 1991) to enrich for spore-forming actinomycetes. Five different agar media were selected for spreading sediment samples: (1) M1 agar: raffinose 10.0 g, L-histidine 1.0 g, K2HPO4 1.0 g, MgSO4.7H2O 0.5 g, FeSO4.7H2O 0.01 g, agar 15.0 g; (2) M2 agar: trehalose 5.0 g, proline1.0 g, (NH4)2SO4 1.0 g, NaCl 1.0 g, CaCl2 2.0 g, K2HPO4 1.0 g, MgSO4.7H2O 1.0 g, agar 20.0 g; (3) M3 agar: humic acid 1.0 g, KCl 1.7 g, NaH2PO4 0.5 g, MgSO4.7H2O 0.5 g, FeSO4.7H2O 0.01 g, CaCO3 0.02 g, agar 15.0 g; (4) M4 agar: glycerol 12.5 g, arginine 1.0 g, K2PO4 1.0 g, NaCl 0.5 g, MgSO4.7H2O 0.5 g, CuSO4.5H2O 0.001 g, trace salt solution 1.0 ml, agar 15.0 g, trace salt solution contains FeSO4.7H2O 0.001 g, MgCl2.4H2O 0.001 g, ZnSO4.7H2O 0.001 g, distilled water 1000 ml; (5) M5 agar: soluble starch 10.0 g, hydrolyzed casein 0.3 g, NaCl 5.0 g, KNO3 2.0 g, K2HPO4 2.0 g, MgSO4.7H2O 0.5 g, CaCO3 0.02 g, FeSO4.7H2O 0.01 g, agar 15.0 g. All media were prepared using the artificial seawater and adjusted to pH 7.5 and were supplemented with nalidixic acid (20 μg/ml) and nystatin (100 μg/ml) or cycloheximide (100 μg/ml) to inhibit the growth of fungi and Gram-negative bacteria. Spreaded plates were incubated at 28°C for 1 month. Actinomycetes were selected and transferred to GT agar medium until pure cultures were obtained for further study (GT agar medium: soluble starch 20 g, L-asparagine 0.5 g, KNO3 1.0 g, K2HPO4.H2O 0.5 g, NaCl 0.5 g, MgSO4.7H2O 0.5 g, distilled water 1000 ml, pH 7.5). Pure actinomycetes were maintained on GT slants at 4°C and 25% (v/v) glycerol suspensions at -80°C. Morphological features of spores and mycelia were observed by light microscopy (model BH2; Olympus) and scanning electron microscopy (Quanta 200). For crude extracts preparation, all the selected strains were cultured in 250 ml flask containing 40 ml fermentation medium (MPG medium consisting of glucose 10.0 g, millet meal 20.0 g, cotton seed gluten meal 20.0 g, MOPS 20.0 g, distilled water 1000 ml, pH 7.2). The liquid cultures were grown for 7 days at 28°C with shaking at 160 rpm. An equal volume of ethyl acetate was added to the liquid cultures for extraction and evaporated to give crude extracts.
16S rRNA Gene Amplification and Phylogenetic Analysis
The 16S rRNA genes were amplified by using universal bacterial primers: 27F and 1492R (Lane, 1991). PCR products were sent to Sangon Biotech (Shanghai, China) Co. Ltd. for DNA sequencing and deposited in GenBank (accession numbers: KU500358- KU500370, KU312336- KU312339, KU529470- KU529472, KU550963, JQ911670). The 16S rRNA gene sequences were compared with available 16S rRNA gene sequences from GenBank database by using BLAST program1 to determine an approximate phylogenetic affiliation. Neighbour-joining (NJ) tree was constructed using software package Mega version 6.0 (Tamura et al., 2013). Bootstrap re-sampling method with 1000 replicates was used in evaluating the topology of the phylogenetic trees (Felsenstein, 1985).
Compound Separation and Identification
The fermentation of active strain Streptomyces sp. NHF165 was carried out in 1000 ml flask containing 250 ml MPG medium that inoculated 3 ml seed culture of strain Streptomyces sp. NHF165. The fermentation broth was cultured at 28°C for 7 days on a rotary shaker at 160 rpm. After fermentation, total broth (10 L) was fractionated by centrifugation. Supernatant was extracted with the same volume ethyl acetate thrice. The evaporated ethyl acetate phase crude extract was applied on a Sephadex LH-20 column [elution reagent, dichloromethane:methanol = 2:1 (v/v)] and separated into 10 fractions. The sixth fraction with anti-VaPDF activity was subjected to a preparative HPLC C18 column (9.4 mm × 250 mm, 5 μm, Agilent) using acetonitrile and water as mobile phase at 3 ml/min to give pure compound 1 (5.2 mg) and 2 (3.5 mg). And the compounds were identified by checking NMR data.
Resistant Mechanism Study of Actinonin against V. anguillarum
To isolate V. anguillarum resistant to actinonin, exponential-phase cells were inoculated into Mueller-Hinton (MH) broth supplemented with 25 μM of actinonin and incubated for 1 day at 28°C. Then 100 μl culture was plated onto MH agar containing 25 μM of actinonin. Resistant colonies were picked and restreaked for single-cell colonies on the same plate. Purified resistant mutants were frozen at -80°C in LB with 10% DMSO. Growth curves for wild type and mutant strains were tested using MH broth without actinonin at 28°C for 25 h. The growth was monitored at different time points by reading A600. Cells were also plated on minimal medium (MM) agar (Duroc et al., 2009) to test the growth. For MICs (minimum inhibition concentration) determination, actinonin was serially diluted twofold from 1000 to 0.49 μM in each column using a clear flat-bottom 96-well plate. The plate was incubated at 28°C for 15 h, and after incubation, the plate was read under absorbance at 600 nm. In this study, the MIC was defined as the lowest actinonin concentration which prevented V. anguillarum growth (an A600 value < 0.05).
The PCR primers used for DNA amplification of the pdf, folD, fmt, and glyA genes were designed from the appropriate sequences of the corresponding public genome sequences from NCBI website2. PCR amplification was performed with both wild type and mutants genome DNAs of V. anguillarum. PCR products were confirmed by sequencing in Sangon Biotech (Shanghai, China) Co. Ltd. Alignment of the DNA sequences of the pdf, folD, fmt, and glyA genes from wild type and mutant strains was carried out using software package Mega version 6.0. To confirm whether mutation of gene folD leads to resistance, complementary experiment was taken out. Briefly, full length of folD was amplified from wild V. anguillarum genome DNA by PCR and ligated into vector pACYC184 (Milton et al., 1992), which was transformed conjugately into mutant V. anguillarum by a donor strain E. coli 17-1. The positive clones were selected on LB agar containing tetracycline.
Expression changes in transcription level between wild type and ΔfolD strain were compared by performing RT-PCR. RNA was extracted from 2 ml culture broth of bacterial samples using an Ultrapure RNA Kit (CWBio) as described by the manufacturer. 1 μg total RNA of each sample was subjected to reverse transcription using random hexamers to prepare cDNAs. RT-PCR was optimized with a SYBR Premix Ex Taq kit (TaKaRa) for each primer pair (Table 1). Each cDNA sample was independently quantified three times, with two technical replicates of each. Relative mRNA levels were calculated.
Results
Establishment and Validation of Screening Model Targeting VaPDF
The genome sequence of V. anguillarum on NCBI web was used as a major reference to clone the pdf gene. The sequencing result showed that the length of pdf gene of V. anguillarum YN was 510 bp (including stop codon) which encodes a 19.21 kDa “Class I” PDF (Giglione et al., 2000) (Figure 1A), and the GenBank accession number of this gene is KU214433. BLAST result showed that its encoding protein VaPDF had 98.0% identity to other types of Vibrio sp. PDFs in amino acid sequence. VaPDF shared three highly conserved characteristic stretches (Baldwin et al., 2002): motif 1 (GIGLAATQ), motif 2 (EGCLS), and motif 3 (HELDH) (Supplementary Figure S1) with other types of PDFs.
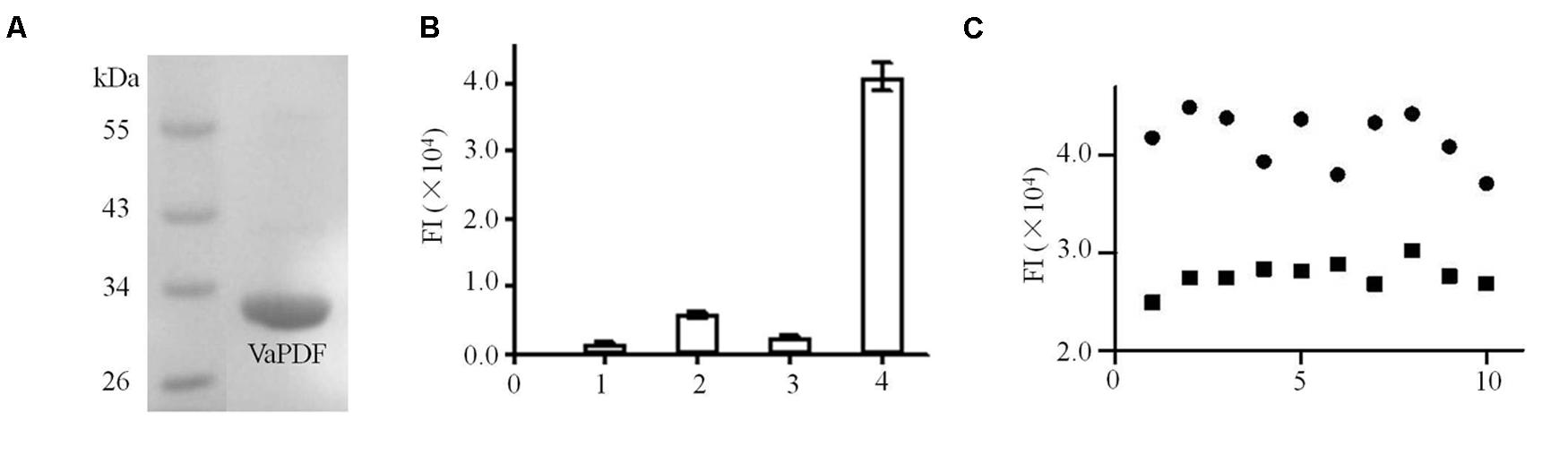
FIGURE 1. Purification and activity assays of Vibrio anguillarum PDF (VaPDF). (A) The purified VaPDF with 6 × His tag. (B) Activity assays of purified VaPDF. 1, HEPES buffer. 2, HEPES buffer + formyl-Met-Ala-Ser. 3, HEPES buffer + VaPDF. 4, HEPES buffer + formyl-Met-Ala-Ser + VaPDF. Data are representative of three independent experiments. Error bars indicate standard deviation. (C) Validation of VaPDF assay. Fluroscence intensity of positive controls and negative controls were detected (n = 10). Solid circle, negative control; solid square, positive control.
Activity of targeting protein is essential for the establishment of screening model. Based on previous data, PDFs purified from Leptospira interrogans etc. catalyzed the removal of a formyl group from the N-termini of nascent polypeptides (Li et al., 2002). Consistently, the purified VaPDF catalyzed the removal of the N-formyl group from formyl-Met-Ala-Ser (Figure 1B) and the free N-formyl group could reacted with fluorescamine to form highly fluorescent products. The optimized reaction conditions were determined as 40 nM VaPDF, 1 mM substrate in 25 mM HEPES buffer (pH 7.4) for 30 min at 37°C. The VaPDF screening model can tolerate up to 2% DMSO (Supplementary Figure S2). Moreover, the Z′ factor was calculated in order to evaluate the PDF assay for HTS. In this model, the value of Z′ factor was 0.71(≥0.5) which is considered acceptable for HTS. The CV values were CVFImax = 6.7% and CVFImin = 5.1%. Both values were less than the threshold value of 10% that is recognized as delineation of correct assays (Figure 1C).
Selective Isolation of Actinomycetes
To find potential novel compounds against V. anguillarum with our HTS model mentioned above, we sought to isolate marine actinomycetes derived natural products for the screening. Totally, 84 actinobacterial strains were isolated from eight marine sediment samples based on the characteristic colonial morphology. As expected, the predominant population of marine actinomycetes was similar to the previous report with marine sediment samples (Maldonado et al., 2005), which showed that Streptomyces was the most abundant species, then was the Micromonospora. Other rare actinomycetes were also recovered from sediment samples. Thereafter, 22 strains were selected and subjected to 16S rRNA gene sequence analysis. GenBank accession numbers were shown in Table 1. Results indicated that these 22 strains shared 99% of similarities with their closest strains. And they belonged to eight genera, which were Micromonospora, Nocardiopsis, Prauserella, Promicromonospora, Saccharopolyspora, Salinispora, Streptomycetes, and Verrucosispora. The phylogenetic affiliation was investigated and the results were presented in Figure 2.
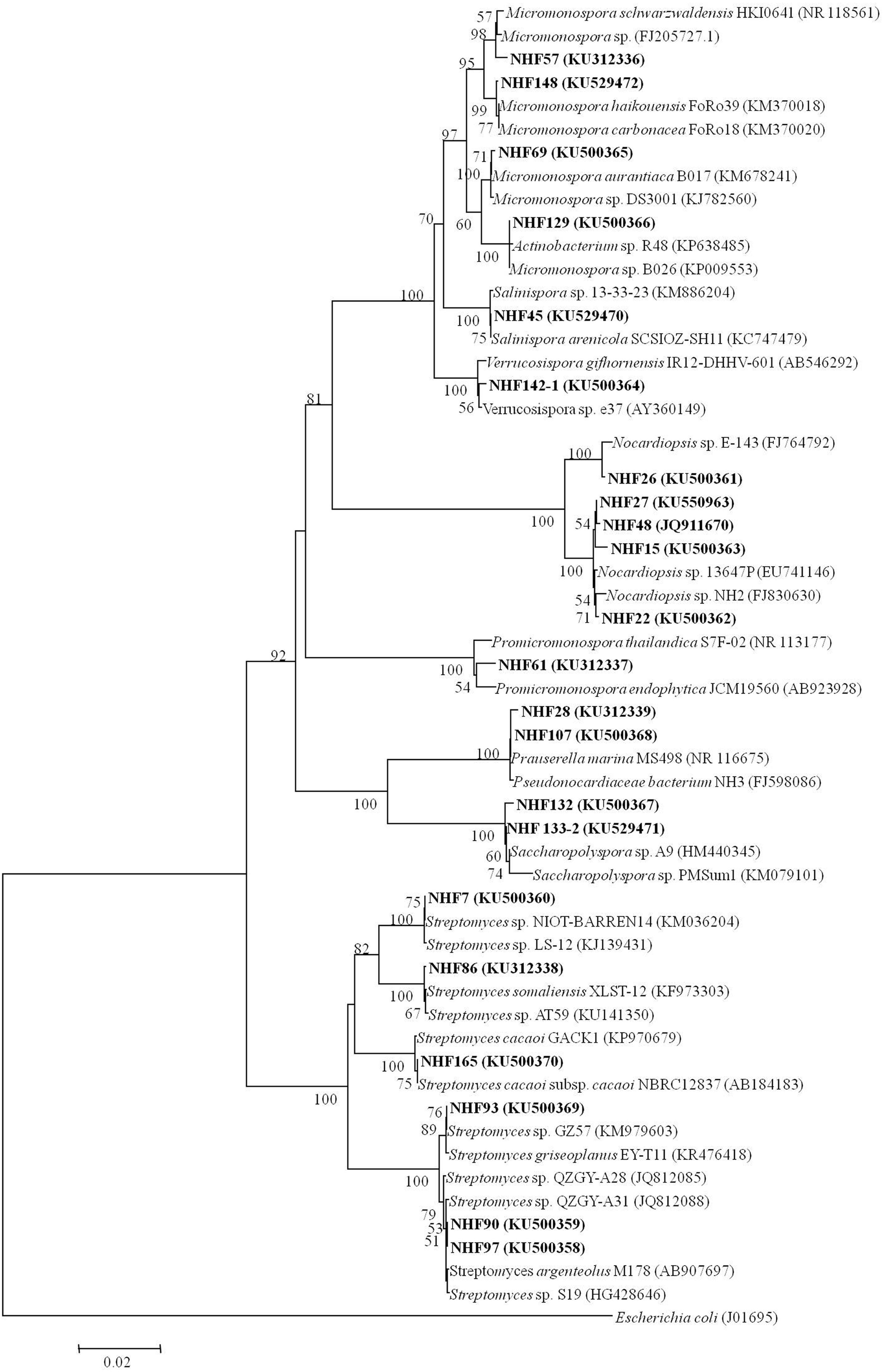
FIGURE 2. Neighbor-joining tree showing the phylogenetic relationships of actinobacterial 16S rRNA gene sequences of obtained strains from South China Sea sediments. Bar, 0.02. Bootstrap values of >50% (for 1000 replicates) are shown.
HTS for Crude Extracts of Marine Actinomycetes
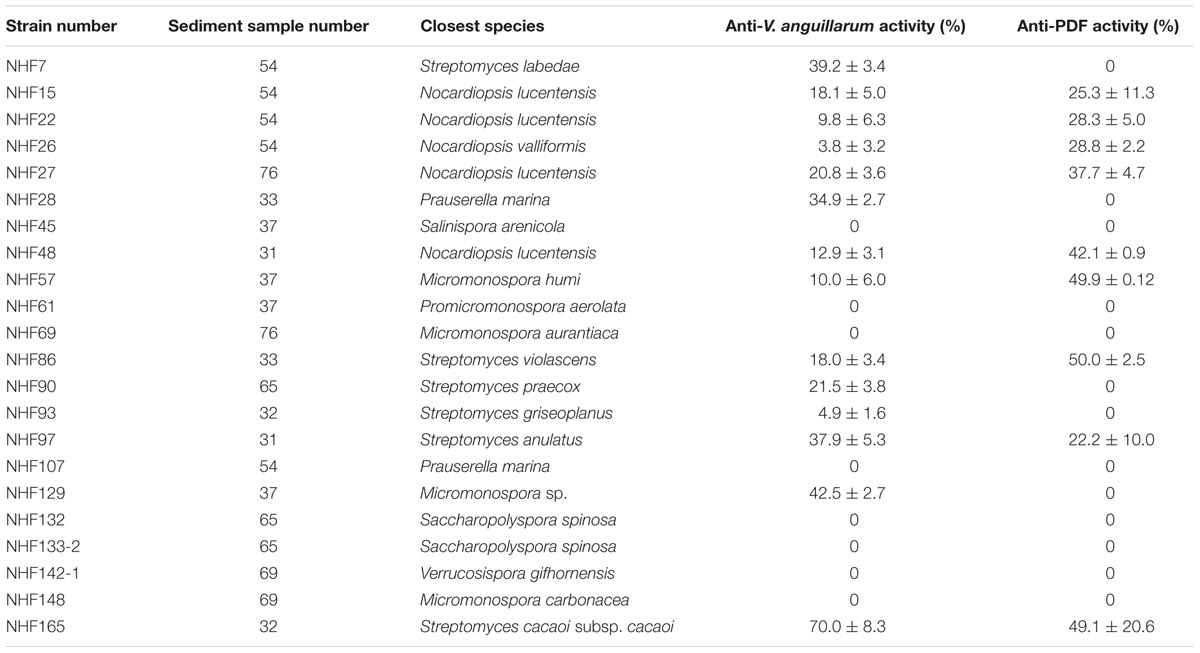
To identify the anti-VaPDF activity of different marine actinomycetes mentioned above with the present HTS model, the corresponding crude extracts were prepared with ethyl acetate extraction method. Thereafter, the crude extracts were used for screening to discover anti-VaPDF agents. For the first round screening, each crude extract was added to a final concentration of 20 μg/ml to the reaction system. Screening results showed that crude extracts isolated from strains NHF27, NHF48, NHF57, NHF69, NHF86, and NHF165 exhibited anti-VaPDF activity with minimum 30% inhibition. Active crude extracts were produced by strains affiliated to genera Micromonospora, Nocardiopsis, and Streptomyces. To confirm the anti-vibrio activities of above active crude extracts, anti-V. anguillarum YN cell activity results were also checked and shown in Table 1. Notably, crude extract isolated from strain Streptomyces sp. NHF165 exhibited the highest inhibitory both on VaPDF activity and V. anguillarum YN cell growth. Therefore, Streptomyces sp. NHF165 was chosen for further study. Strain NHF165 had a highest 16S rRNA gene similarity (>99%) with Streptomyces cacaoi subsp. Cacaoi, and colonies of this strain appeared to be yellow substrate mycelium and white aerial mycelium. Oval spores were produced along the long, straight and smooth aerial mycelium after 7 days of cultivation on medium GT (Figure 3).
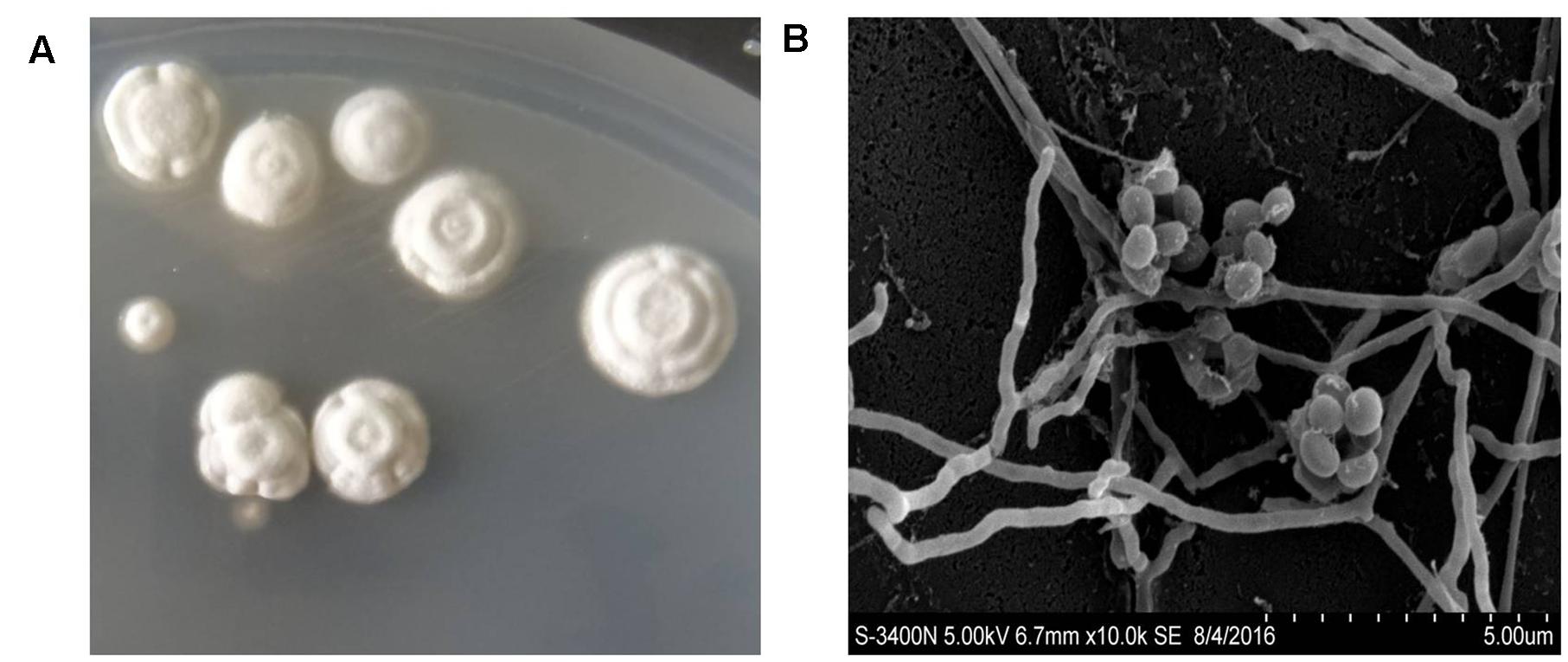
FIGURE 3. Characteristics of Streptomyces sp. NHF165. (A) Colony characteristics of Streptomyces sp. NHF165. (B) Scanning electron micrograph of Streptomyces sp. NHF165 grown on GT agar at 28°C for 7 days. Bar = 5 μm.
Structure Elucidation of Compounds Produced by Streptomyces sp. NHF165
To identify the exact structure of compound with anti-VaPDF activity isolated form Streptomyces sp. NHF165, the corresponding crude extract was separated with sephadex LH-20. The purification results showed that fraction 6 contained the main anti-VaPDF constitute. Then fraction 6 was further separated with HPLC with C18 column and two compounds were finally obtained (1 and 2). Their structures were elucidated by UV, 1D NMR, 2D NMR (1H-1H COSY, 1H-13C HSQC, 1H-13C HMBC). ESI-MS data revealed molecular ion peaks at m/z 386.2961 [M+H]+, and 408.2498 [M+Na]+ for compound 1 (Umezawa et al., 1985). The compound 1 with anti-VaPDF activity was identified by comparing the NMR data with previous published data, and it was considered to be actinonin (Figure 4A) (Umezawa et al., 1985). The total yield of actinonin was 5.3 mg per 10 L broth. Correspondingly, this marine derived-actinonin inhibited the VaPDF activity in a dose-dependent manner and the IC50 was 6.94 μM. The IC50 of this actinonin on V. anguillarum cell viability was 2.85 μM (Figures 4B,C).
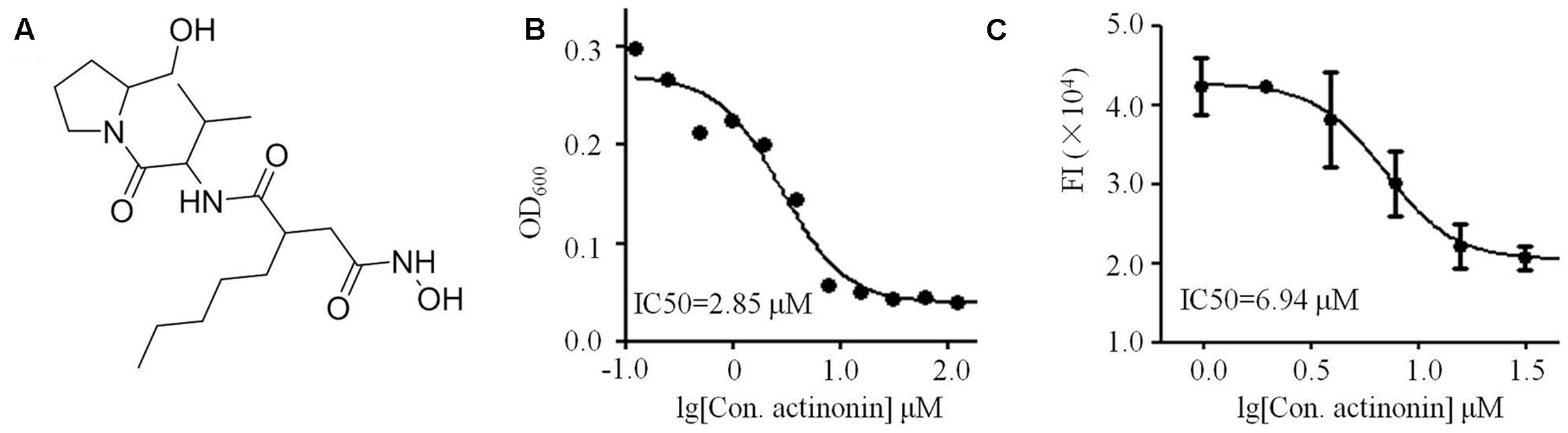
FIGURE 4. Characterization of marine derived actinonin. (A) Structure of actinonin isolated from Streptomyces sp. NHF165. (B) Anti-V. anguillarum IC50 value of actinonin. (C) Anti-VaPDF IC50 value of actinonin.
Compound 2 was obtained as light brown amorphous powder. Its HRESIMS revealed a molecular ion peak of m/z 565.2565 for C32H33N6O4 [M+H]+ (Calcd. 565.2485) and suggested 564 as the molecular weight and C32H32N6O4 as the molecular formula. UV spectrum with the maximal absorbance at 206, 228, and 288 nm.13C NMR spectrum of compound 2 revealed signals of 32 carbons, including four amide carbonyl 13C resonances were suggested by signals of δC 165.2, C-13; 165.6, C-35; 166.0, C-16; 169.1, C-32. The 1H and 13C NMR spectra in combination with 1H–1H COSY and 1H–1C HSQC NMR data indicated signals of two substituted benzene groups (1, 2- substituted benzene: δH 7.16, d, 12.0, δC 123.8, C-5; δH 6.62, t, overlap, δC 118.1, C-6; δH 6.98, t, 7.2, δC128.1, C-7; δH 6.65, d, 6.0, δC 109.5, C-8; δC 133.0, C-4; δC 149.4, C-9 and 1, 2, 4- substituted benzene: δH 7.21, d, 6.0, δC 111.5, C-20; δH 7.03, d, 12.0, δC 119.2, C-21; δH 7.62, s, δC 114.6, C-23; δC 134.1, C-22, δC 127.1, C-24; δC 134.8, C-25). 1H–13C HMBC NMR data revealed HN-26 connected with C-24, C-25, C-27 (δC 124.7), C-28 (δC 109.6), and H-27 connected with C-24 and C-25. The 1, 2, 4-substituted benzene moiety was an indole structure. Combined 13C and HMBC spectrum, C-30, 32, 33, 35, 36, 37, 38 signals showed a diketopiperazine moiety. H-29 [δH 3.23, dd (14.4, 4.2); 3.06, dd (12.0, 6.0)] connected with C-24, C-27, C-35, and HN-31 (δH 7.7) connected with C-32, C-35. These data suggested this group was a condensation product of tryptophan and proline. The HMBC signals from H-2 (δH 5.63, s) to C-4, C-9 and from HN-1 (δH 6.61) to C-2 (δC 81.1), C-3 (δC 58.7), C-4, C-8, and C-9 demonstrated that the 1, 2- substituted benzene moiety was an indoline structure. C-11, 13, 15, 16, 17, 18, 19 signals were assigned to another diketopiperazine moiety. A methylene group contributed to establish connectivity of indoline and diketopiperazine moieties. Signal from H-2 to C-16 demonstrated the connection of C-2 to N-10. Signal from H-2 to C-22 showed the connection of C-3 to C-22. ROESY data showed signals from H2 to H-11 and H-21 which suggested H-1, H-11 and indolyl diketopiperazine structure on the same side. Thus the structure of 2 was established (Supplementary Figure S3). It was apparent that compound 2 was related to asperazine derived from a marine fungi Aspergillus niger (Varoglu et al., 1997). Compound 2 was shown to be a new compound of indolyl diketopiperazine analogs, and it showed no activities against V. anguillarum or VaPDF.
Resistance Mechanism of V. anguillarum against Actinonin
The resistance of V. anguillarum YN to actinonin was challenged on MH agar with 25 μM actinonin. The frequency of resistance in V. anguillarum YN was 5 × 10-6. Notably, the mutants were stable, as re-streaking on actinonin-free MH agar did not lose resistance, and no phenotypic differences between wild type and mutant were observed for this strain. Compared with parent strains, V. anguillarum YN mutants grew at much slower rates when cultured in MH broth (Figure 5A) and showed 8 × MIC to actinonin (Figure 5B). Moreover, these mutants showed resistance to actinonin but still remained susceptibility to streptomycin, chloramphenicol, carbenicillin, kanamycin, and ampicillin as wild type strains do.
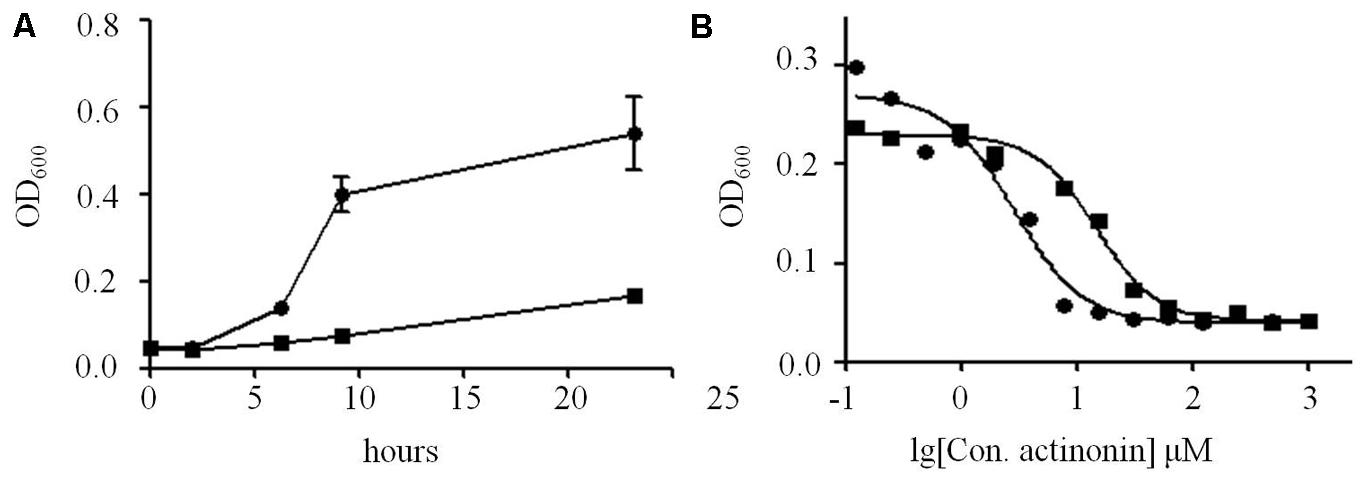
FIGURE 5. Proliferation differences between V. anguillarum mutant and wild type strains. (A) Growth of V. anguillarum mutants and wild type strains in MH broth, measured as optical density at 600 nm. (B) MIC tests of actinonin on V. anguillarum mutants and wild type strains. Solid circle, wild type strain; solid square, mutant strain.
In order to understand the mutation details, open reading frame regions of pdf, folD, fmt, and glyA DNA sequence from the mutant strains were amplified, sequenced and aligned with those from parent strains. The results showed that no mutation was retrieved in pdf, fmt, and glyA, and all five mutant strains harbored a mutation in folD gene possessing deletion of base pairs 774–852 (Supplementary Figure S4). As reported, folD catalyzes the formation of 10-formyl-tetrahydrofolate (THF), which supplies N-formyl group to Met-tRNAfMet. To our knowledge, ΔfolD mutants have been described only in species Salmonella enterica and B. subtilis (Duroc et al., 2009). None of the resistant strains could grow on MM medium, which consisted with the results described previously (Duroc et al., 2009). To determine whether mutation of gene folD is the main cause for the actinonin resistance of V. anguillarum, complementary experiment was performed. Plasmid pACYC184::folD was successfully constructed and introduced into ΔfolD mutants to get pACYC184::folD/ΔfolD strains. Complementary strains could not grow on MH agar with 25 μM actinonin in this study, which further confirmed that folD gene mutation was responsible for actinonin resistance in V. anguillarum.
To understand the expression changes between wild type and mutant strains, genes involved in translation initiation (pdf), amino acid biosynthesis (gtlB), metabolites biosynthesis (srfAC), ATP production (atpH), cell protection (ahpF), ABC transporter (fhuD), TCA cycle (pdhA) were checked with RT-PCR (Supplementary Table S2) and the expression of rplL gene was used as a reference for the determination of induction levels. Significant expression changes of pdf, atpH, and ahpF genes were observed for genes encoding functions of the intermediary metabolism (Figure 6). pdf and atpH genes were significantly down-regulated, which suggested that the translation initiation was hampered by less N-formyl group supply. However, the expression of gene ahpF corresponding for protecting cells was significantly up-regulated. Thus, in the tested condition, V. anguillarum mutants developed an adaptation mechanism to survive in high concentration of actinonin.
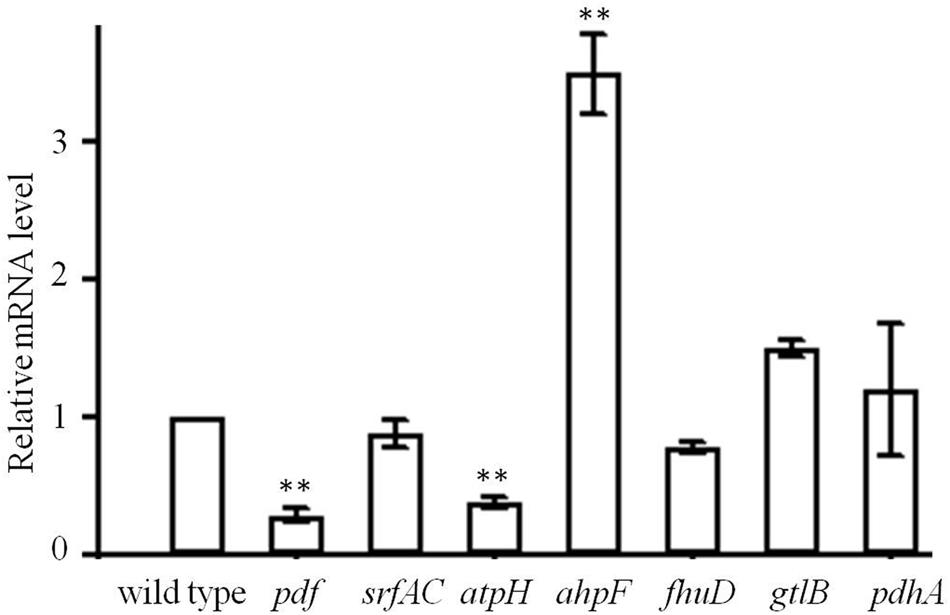
FIGURE 6. Trancription level of pdf, srfAC, atpH, ahpF, fhuD, gtlB, and pdhA in the mutant of V. anguillarum. Wild type was used as a control. Values reported are the mean of three replicates. Error bars indicate the standard deviation. ∗∗p < 0.01 was taken to indicate statistical distinct significance.
Discussion
Vibrio anguillarum is an opportunistic fish pathogen that is common to marine and estuarine environments. It has been identified as the main cause of vibriosis, a potentially fatal septicemia that affects fish and shellfish in marine aquaculture, with consequent economic losses (Frans et al., 2011). To find novel antibiotics against V. anguillarum is urgently needed.
It is now widely accepted that the traditional screening methods are unlikely to generate many promising molecules. Alternative strategies must therefore be developed to find new compounds. One possible strategy is to identify a molecular target at the outset and then to screen the available libraries of chemical compounds looking for hits with potent inhibitory capacities in vitro with HTS model. However, it is extreme lack of HTS model for searching anti-Vibrio compounds. For this approach, the identification of a good target is vital. PDF has been suggested as a possible candidate that may fulfill all those criteria for HTS and has become a promising and attractive bacterial target to explore for the discovery of new antibacterial agents (Giglione et al., 2000). We confirmed VaPDF shared the three highly conserved characteristic stretches and was essential for V. anguillarum growth. Therefore, active agents against VaPDF can be potential drugs for vibriosis treatment. Due to the lack of effective anti-Vibrio HTS methods, we first developed a protein-based assay based on VaPDF activity and screened crude extracts derived from marine actinomycetes.
In recent years, great attention has been paid to the isolation and characterization of actinomycetes from marine environment, which provides a valuable source for discovering bioactive metabolites. South China Sea located in the southeast of China with tropical oceanic climate and was poorly studied. Therefore, we chose deep-sea sediment samples collected from South China Sea to isolate anti-Vibrio actinomycetes, which might be used in marine aquaculture industry. Totally, 84 actinobacterial strains belonging to eight genera were obtained. The predominant numbers of Streptomyces and Micromonospora strains is in line with the results reported previously (Maldonado et al., 2005). Representative strains isolated in the present study showed bioactivities against VaPDF and V. anguillarum cell. Among 22 strains, 14 strains showed anti-bacteria activity against V. anguillarum and 9 strains showed anti-activity against VaPDF. These strains belonged to genera Streptomyces, Micromonospora, and Nocardiopsis.
As is well known, Streptomyces could produce diverse range of secondary metabolites with relevant anti-inflammatory, antimicrobial, antioxidant activities (Dubert et al., 2015) and are potential probiotics in aquaculture (Tan et al., 2016). Streptomyces rubrolavendulae M56 isolated from the sediments of Bay of Bengal could significantly exclude the pathogenic Vibrio spp. in co-culture experiments (Augustine et al., 2015). Addition of 1% wet cell mass of marine Streptomyces strains can reduce mortality rate of nauplii and adult Artemia caused by both V. harveyi and V. proteolyticus (Das et al., 2010). Crude extract of Streptomyces sp. LCJ94 showed good inhibitory activities against V. harveyi, V. vulnificus, V. alginolyticus with the MIC values of 250, 250, and 500 μg/ml, respectively (Mohanraj and Sekar, 2013). In this study, Streptomyces sp. NHF165 exhibited the highest activity against V. anguillarum, and the functional component was finally determined as actinonin. Actinonin was isolated from soil Streptomyces in 1962 and was reported to be an inhibitor targeting E. coli PDF and M. tuberculosis PDF (Sharma et al., 2009). Our discovery is the first report to show that marine derived actinonin possesses anti-Vibrio activity via targeting VaPDF. Considering Streptomyces sp. NHF165 with high yield (5.3 mg/10 L) and low IC50 of actinonin on V. anguillarum (2.85 μM), it might be a good candidate for the management of vibriosis in marine aquaculture industry. On the other hand, as a natural product, actinonin shows derivative of L-prolinol and hydroxamic acid of the type R-CO-NHOH and some structural relationship to other polypeptide antibiotics. Hence, it will be very interesting to dig the conserved DNA sequence of non-ribosomal peptide synthetases (NRPS) adenylation domain (Ayuso-Sacido and Genilloud, 2005) in the genomic DNA of Streptomyces sp. NHF165 in the future.
Nowadays, antibiotics have been routinely applied to water to treat and prevent bacterial disease in fish and shellfish culture industries. However, extensive use of antibiotics goes with development of resistant strains, especially resistant vibrios. Characterization of antibiotic-resistant vibrios is necessary to elucidate mechanism of resistance. Vibrio strains with resistance to chloramphenicol, tetracycline, amoxicillin, or streptomycin were successfully isolated from hatchery larval cultures, and R-plasmids harboring resistant genes (chloramphenicol acetyltransferase, tetracycline resistance markers, etc.) were elucidated (Dubert et al., 2015). In other report, about 63% of the isolated V. parahaemolyticus strains were resistant to ampicillin, cephalexin, or kanamycin (Bhattacharya et al., 2000). Hence, appearance of resistance to actinonin is a predictable consequence, and it is necessary to study the resistance mechanism of V. anguillarum against actinonin.
It was reported that mechanisms causing PDF inhibitor resistance involve (i) mutations in the target gene, (ii) bypassing of the formylation pathway, or (iii) efflux of PDF inhibitor (Duroc et al., 2009). Notably, we could amplify genes involved in translation initiation including pdf, fmt, and glyA but failed to get folD fragment from mutants, and then we confirmed a fragment deletion happened in the gene folD. Interestingly, similar mutations in the gene fold of S. enterica and B. subtilis had been described previously (Duroc et al., 2009). The loss of function of folD could inactivate translation initiation pathway that uses 10-formyl-THF, which led to a dramatic decrease of growth rate of ΔfolD mutants. It is proposed that, in addition to folD, mutations in the genes involved in efflux pump, modification of actinonin or coding enzymes that degrade actinonin might also happened. Additionally, the RT-PCR results showed the expression of genes pdf, atpH, and ahpF were significantly regulated, which suggested that V. anguillarum mutants might develop an adaptation mechanism to survive in high concentration of actinonin.
Collectively, it is evident that VaPDF can be a good target for anti-Vibrio agents screening. And actinomycetes isolated from marine could be promising candidates for treating pathogens in marine aquaculture. It will also be very interesting to find more anti-Vibrio compounds with the present HTS model and develop the corresponding anti-bacteria drugs in the future.
Author Contributions
NY and CS conceived and designed the experiments. NY performed all of the experiments. NY and CS analyzed the data, prepared the figures and wrote the paper. All authors reviewed the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This work was supported by the Scientific and Technological Innovation Project of Qingdao National Laboratory for Marine Science and Technology (No.2015ASKJ02-3), AoShan Talents Program supported by Qingdao National Laboratory for Marine Science and Technology (No.2015ASTP) and “100-Talent Project” of Chinese Academy of Sciences for Chaomin Sun.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01467
Footnotes
References
Augustine, D., Jacob, J. C., and Philip, R. (2015). Exclusion of Vibrio spp. by an antagonistic marine actinomycete Streptomyces rubrolavendulae M56. Aquac. Res. 47, 2951–2960. doi: 10.1111/are.12746
Ayuso-Sacido, A., and Genilloud, O. (2005). New PCR primers for the screening of NRPS and PKS-I systems in actinomycetes: detection and distribution of these biosynthetic gene sequences in major taxonomic groups. Microb. Ecol. 49, 10–24. doi: 10.1007/s00248-004-0249-6
Baldwin, E. T., Harris, M. S., Yem, A. W., Wolfe, C. L., Vosters, A. F., Curry, K. A., et al. (2002). Crystal structure of type II peptide deformylase from Staphylococcus aureus. J. Biol. Chem. 277, 31163–31171. doi: 10.1074/jbc.M202750200
Bhattacharya, M., Choudhury, P., and Kumar, R. (2000). Antibiotic- and metal-resistant strains of Vibrio parahaemolyticus isolated from shrimps. Microb. Drug Resist. 6, 171–172. doi: 10.1089/107662900419492
Blair, J. M., Webber, M. A., Baylay, A. J., Ogbolu, D. O., and Piddock, L. J. (2015). Molecular mechanisms of antibiotic resistance. Nat. Rev. Microbiol. 13, 42–51. doi: 10.1038/nrmicro3380
Chen, D. Z., Patel, D. V., Hackbarth, C. J., Wang, W., Dreyer, G., Young, D. C., et al. (2000). Actinonin, a naturally occurring antibacterial agent, is a potent deformylase inhibitor. Biochemistry 39, 1256–1262. doi: 10.1021/bi992245y
Das, S., Ward, L. R., and Burke, C. (2010). Screening of marine Streptomyces spp. for potential use as probiotics in aquaculture. Aquaculture 305, 32–41. doi: 10.1016/j.aquaculture.2010.04.001
Dubert, J., Osorio, C. R., Prado, S., and Barja, J. L. (2015). Persistence of antibiotic resistant Vibrio spp. in Shellfish Hatchery Environment. Microb. Ecol doi: 10.1007/s00248-015-0705-5 [Epub ahead of print].
Duroc, Y., Giglione, C., and Meinnel, T. (2009). Mutations in three distinct loci cause resistance to peptide deformylase inhibitors in Bacillus subtilis. Antimicrob. Agents Chemother. 53, 1673–1678. doi: 10.1128/AAC.01340-08
Felsenstein, J. (1985). Confidence-limits on phylogenies - an approach using the bootstrap. Evolution 39, 783–791. doi: 10.2307/2408678
Frans, I., Michiels, C. W., Bossier, P., Willems, K. A., Lievens, B., and Rediers, H. (2011). Vibrio anguillarum as a fish pathogen: virulence factors, diagnosis and prevention. J. Fish Dis. 34, 643–661. doi: 10.1111/j.1365-2761.2011.01279.x
Giglione, C., Pierre, M., and Meinnel, T. (2000). Peptide deformylase as a target for new generation, broad spectrum antimicrobial agents. Mol. Microbiol. 36, 1197–1205. doi: 10.1046/j.1365-2958.2000.01908.x
Gordon, J. J., Kelly, B. K., and Miller, G. A. (1962). Actinonin: an antibiotic substance produced by an actinomycete. Nature 195, 701–702. doi: 10.1038/195701b0
Hopkins, D. W., Macnaughton, S. J., and Odonnell, A. G. (1991). A Dispersion and differential centrifugation technique for representatively sampling microorganisms from soil. Soil Biol. Biochem. 23, 217–225. doi: 10.1016/0038-0717(91)90055-O
Lane, D. J. (1991). “16S/23S rRNA sequencing,” in Nucleic Acid Techniques in Bacterial Systematics, eds E. Stackebrandt and M. Goodfellow (New York, NY: John Wiley and Sons).
Letchumanan, V., Pusparajah, P., Tan, L. T., Yin, W. F., Lee, L. H., and Chan, K. G. (2015a). Occurrence and antibiotic resistance of Vibrio parahaemolyticus from shellfish in selangor, Malaysia. Front. Microbiol. 6:1417. doi: 10.3389/fmicb.2015.01417
Letchumanan, V., Yin, W. F., Lee, L. H., and Chan, K. G. (2015b). Prevalence and antimicrobial susceptibility of Vibrio parahaemolyticus isolated from retail shrimps in Malaysia. Front. Microbiol. 6:33. doi: 10.3389/fmicb.2015.00033
Li, Y. K., Chen, Z. F., and Gong, W. M. (2002). Enzymatic properties of a new peptide deformylase from pathogenic bacterium Leptospira interrogans. Biochem. Biophys. Res. Commun. 295, 884–889. doi: 10.1016/S0006-291x(02)00777-5
Maldonado, L. A., Stach, J. E. M., Pathom-aree, W., Ward, A. C., Bull, A. T., and Goodfellow, M. (2005). Diversity of cultivable actinobacteria in geographically widespread marine sediments. Antonie Van Leeuwenhoek 87, 11–18. doi: 10.1007/s10482-004-6525-0
Margolis, P., Hackbarth, C., Lopez, S., Maniar, M., Wang, W., Yuan, Z., et al. (2001). Resistance of Streptococcus pneumoniae to deformylase inhibitors is due to mutations in defB. Antimicrob. Agents Chemother. 45, 2432–2435. doi: 10.1128/AAC.45.9.2432-2435.2001
Margolis, P. S., Hackbarth, C. J., Young, D. C., Wang, W., Chen, D., Yuan, Z., et al. (2000). Peptide deformylase in Staphylococcus aureus: resistance to inhibition is mediated by mutations in the formyltransferase gene. Antimicrob. Agents Chemother. 44, 1825–1831. doi: 10.1128/AAC.44.7.1825-1831.2000
Miao, V., and Davies, J. (2010). Actinobacteria: the good, the bad, and the ugly. Antonie Van Leeuwenhoek 98, 143–150. doi: 10.1007/s10482-010-9440-6
Milton, D. L., Norqvist, A., and Wolfwatz, H. (1992). Cloning of a metalloprotease gene involved in the virulence mechanism of Vibrio-Anguillarum. J. Bacteriol. 174, 7235–7244.
Mohanraj, G., and Sekar, T. (2013). Antagonistic activity of marine Streptomyces sp LCJ94 against the shrimp pathogens. Ann. Biol. Res. 4, 224–227.
Nonaka, L., Maruyama, F., Suzuki, S., and Masuda, M. (2015). Novel macrolide-resistance genes, mef(C) and mph(G), carried by plasmids from Vibrio and Photobacterium isolated from sediment and seawater of a coastal aquaculture site. Lett. Appl. Microbiol. 61, 1–6. doi: 10.1111/lam.12414
Peyrusson, F., Butler, D., Tulkens, P. M., and van Bambeke, F. (2015). Cellular pharmacokinetics and intracellular activity of the novel peptide deformylase inhibitor GSK1322322 against Staphylococcus aureus laboratory and clinical strains with various resistance phenotypes: Studies with human THP-1 monocytes and J774 murine macrophages. Antimicrob. Agents Chemother. 59, 5747–5760. doi: 10.1128/Aac.00827-15
Sangshetti, J. N., Khan, F. A. K., and Shinde, D. B. (2015). Peptide deformylase: a new target in antibacterial, antimalarial and anticancer drug discovery. Curr. Med. Chem. 22, 214–236. doi: 10.2174/0929867321666140826115734
Sharma, A., Sharma, S., Khuller, G. K., and Kanwar, A. J. (2009). In vitro and ex vivo activity of peptide deformylase inhibitors against Mycobacterium tuberculosis H37Rv. Int. J. Antimicrob. Agents 34, 226–230. doi: 10.1016/j.ijantimicag.2009.04.005
Shaw, K. S., Rosenberg Goldstein, R. E., He, X., Jacobs, J. M., Crump, B. C., and Sapkota, A. R. (2014). Antimicrobial susceptibility of Vibrio vulnificus and Vibrio parahaemolyticus recovered from recreational and commercial areas of chesapeake bay and maryland coastal bays. PLoS ONE 9:e89616. doi: 10.1371/journal.pone.0089616
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tan, L. T. H., Chan, K. G., Lee, L. H., and Goh, B. H. (2016). Streptomyces bacteria as potential probiotics in aquaculture. Front. Microbiol 7:79. doi: 10.3389/Fmicb.2016.00079
Umezawa, H., Aoyagi, T., Tanaka, T., Suda, H., Okuyama, A., Naganawa, H., et al. (1985). Production of actinonin, an inhibitor of aminopeptidase-M, by actinomycetes. J. Antibiot. 38, 1629–1630. doi: 10.7164/antibiotics.38.1629
Varoglu, M., Corbett, T. H., Valeriote, F. A., and Crews, P. (1997). Asperazine, a selective cytotoxic alkaloid from a sponge-derived culture of Aspergillus niger. J. Org. Chem. 62, 7078–7079. doi: 10.1021/jo970568z
Vinothkumar, S., and Parameswaran, P. S. (2013). Recent advances in marine drug research. Biotechnol. Adv. 31, 1826–1845. doi: 10.1016/j.biotechadv.2013.02.006
You, J. L., Cao, L. X., Liu, G. F., Zhou, S. N., Tan, H. M., and Lin, Y. C. (2005). Isolation and characterization of actinomycetes antagonistic to pathogenic Vibrio spp. from nearshore marine sediments. World J. Microbiol. Biotechnol. 21, 679–682. doi: 10.1007/sl1274-004-3851-3
You, J. L., Xue, X. L., Cao, L. X., Lu, X., Wang, J., Zhang, L. X., et al. (2007). Inhibition of Vibrio biofilm formation by a marine actinomycete strain A66. Appl. Microbiol. Biot. 76, 1137–1144. doi: 10.1007/s00253-007-1074-x
Zhang, W., Liang, W., and Li, C. (2016). Inhibition of marine Vibrio sp. by pyoverdine from Pseudomonas aeruginosa PA1. J. Hazard. Mater. 302, 217–224. doi: 10.1016/j.jhazmat.2015.10.003
Keywords: peptide deformylase, high-throughput screening assay, Vibrio anguillarum, marine Streptomyces sp. NHF165, resistance mechanism, actinonin
Citation: Yang N and Sun C (2016) The Inhibition and Resistance Mechanisms of Actinonin, Isolated from Marine Streptomyces sp. NHF165, against Vibrio anguillarum. Front. Microbiol. 7:1467. doi: 10.3389/fmicb.2016.01467
Received: 22 June 2016; Accepted: 01 September 2016;
Published: 13 September 2016.
Edited by:
Learn-Han Lee, Monash University Malaysia Campus, MalaysiaReviewed by:
Carmen Wacher, National Autonomous University of Mexico, MexicoVengadesh Letchumanan, University of Malaya, Malaysia
Copyright © 2016 Yang and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chaomin Sun, c3VuY2hhb21pbkBxZGlvLmFjLmNu
 Na Yang
Na Yang Chaomin Sun
Chaomin Sun