- Department of Earth and Environmental Sciences, University of Michigan, Ann Arbor, MI, USA
Anoxygenic cyanobacteria that use sulfide as the electron donor for photosynthesis are a potentially influential but poorly constrained force on Earth’s biogeochemistry. Their versatile metabolism may have boosted primary production and nitrogen cycling in euxinic coastal margins in the Proterozoic. In addition, they represent a biological mechanism for limiting the accumulation of atmospheric oxygen, especially before the Great Oxidation Event and in the low-oxygen conditions of the Proterozoic. In this study, we describe the draft genome sequence of Geitlerinema sp. PCC 9228, formerly Oscillatoria limnetica ‘Solar Lake’, a mat-forming diazotrophic cyanobacterium that can switch between oxygenic photosynthesis and sulfide-based anoxygenic photosynthesis (AP). Geitlerinema possesses three variants of psbA, which encodes protein D1, a core component of the photosystem II reaction center. Phylogenetic analyses indicate that one variant is closely affiliated with cyanobacterial psbA genes that code for a D1 protein used for oxygen-sensitive processes. Another version is phylogenetically similar to cyanobacterial psbA genes that encode D1 proteins used under microaerobic conditions, and the third variant may be cued to high light and/or elevated oxygen concentrations. Geitlerinema has the canonical gene for sulfide quinone reductase (SQR) used in cyanobacterial AP and a putative transcriptional regulatory gene in the same operon. Another operon with a second, distinct sqr and regulatory gene is present, and is phylogenetically related to sqr genes used for high sulfide concentrations. The genome has a comprehensive nif gene suite for nitrogen fixation, supporting previous observations of nitrogenase activity. Geitlerinema possesses a bidirectional hydrogenase rather than the uptake hydrogenase typically used by cyanobacteria in diazotrophy. Overall, the genome sequence of Geitlerinema sp. PCC 9228 highlights potential cyanobacterial strategies to cope with fluctuating redox gradients and nitrogen availability that occur in benthic mats over a diel cycle. Such dynamic geochemical conditions likely also challenged Proterozoic cyanobacteria, modulating oxygen production. The genetic repertoire that underpins flexible oxygenic/anoxygenic photosynthesis in cyanobacteria provides a foundation to explore the regulation, evolutionary context, and biogeochemical implications of these co-occurring metabolisms in Earth history.
Introduction
With a long evolutionary history and wide ecological success, cyanobacteria are pivotal mediators of Earth’s geochemical cycles, most notably through oxygenic photosynthesis (OP). This metabolism emerged early in cyanobacteria (Blankenship, 2010; Farquhar et al., 2010), and oxygenic cyanobacteria that colonized newly formed continental margins and shallow seas in the Archean (Reddy and Evans, 2009; Lalonde and Konhauser, 2015) were the leading mechanism for partial oxygenation of these shallow regions (Planavsky et al., 2014; Satkoski et al., 2015). Although, cyanobacterial OP is the major biological force behind Earth’s oxygenation (Blankenship, 2010; Crowe et al., 2013; Planavsky et al., 2014), the biological and geological processes that influence cyanobacterial oxygen production and thus underpin Earth’s oxygenation are still under debate (Lyons et al., 2014).
The balance of oxygenic and anoxygenic photosynthesis (AP) has been proposed as a biological mechanism to explain the delay and variability in oxygenation (Johnston et al., 2009). This includes competition between AP bacteria and OP cyanobacteria, interactions between different cyanobacterial groups with varying degrees of AP or OP specialization, as well as cellular regulation of the photosynthetic modes within metabolically flexible cyanobacterial groups. Atmospheric oxygenation is considered key evidence for OP, but the emergence and development of AP in cyanobacteria is less clear. Assuming an early evolution of photosynthetic flexibility in cyanobacteria, AP cyanobacteria may have had an important role in sustaining ancient ecosystems from the end of the Archean through the Proterozoic, especially in times of global and local variability of O2, fixed nitrogen, and alternative electron donors for photosynthesis such as H2S and Fe(II) (Canfield, 1998; Scott et al., 2008; Lyons et al., 2014; Sperling et al., 2015).
Molecular innovations equipped cyanobacteria for OP and nitrogen fixation in a dynamic environment. The development of OP required the linkage of two light-driven reaction centers: photosystem II, which produces oxygen from the oxidation of water; and photosystem I, which transfers electrons from plastoquinone to ferredoxin (Blankenship, 2002). This coupled system capitalized on the wide availability of H2O compared to more limited supply of electron donors for AP such as Mn(II), Fe(II), and H2S (Blankenship, 2002). Homologous proteins D1 and D2, encoded by the psbA and psbD genes, form the core of PSII and anchor the water oxidizing complex (Ferreira et al., 2004; Fischer et al., 2015). Modern cyanobacteria have multiple versions of psbA to cope with different oxygen levels and light regimes (Mohamed et al., 1993). Cyanobacterial adaptations to an aerobic lifestyle are also reflected in the nif genes for nitrogen fixation, which initially emerged in methanogens in an anoxic environment (Raymond et al., 2004; Boyd et al., 2011). Reflecting increasing oxygen levels, cyanobacterial genomes lost and recruited nitrogenase (nif)-related genes, and shifted expression patterns and regulation that enabled nitrogen fixation in an oxic world (Boyd et al., 2015).
Studies of modern cyanobacteria have provided insights into how sulfide may have modulated the balance of OP and AP in ancient ecosystems. The influence of sulfide on cyanobacterial photosynthesis ranges from complete inhibition at even low levels of H2S to resilience or resistance to sulfide toxicity. In some cyanobacteria, sulfide exposure may induce AP (Cohen et al., 1986; Miller and Bebout, 2004), which does not include PSII and thus does not produce O2. Instead, the sulfide quinone reductase (SQR, coded for by the sqr gene) oxidizes sulfide to sulfur and transfers electrons to PSI (Arieli et al., 1994; Theissen et al., 2003). Such AP cyanobacteria have been documented in hypersaline lakes (Cohen et al., 1975a), sinkholes (Voorhies et al., 2012), and sulfidic springs (Chaudhary et al., 2009; Bühring et al., 2011; Klatt et al., 2016). Studied cyanobacteria have different mechanisms for the transition between OP and AP, such as protein synthesis (Oren and Padan, 1978), a dependence on light quantity and spectrum, and kinetics and affinities between enzymes and quinones (Klatt et al., 2015a). The physiology of AP cyanobacteria and their potential importance in modern and ancient ecosystems have been previously explored, yet the genomic basis for this flexible metabolism is poorly understood.
Geitlerinema sp. PCC 9228, formerly Oscillatoria limnetica ‘Solar Lake’, is a model anoxygenic photosynthetic cyanobacterium. The organism was cultured from the low-light sulfidic hypolimnion of Solar Lake, below a layer of green and purple sulfur bacteria (Cohen et al., 1975b). Filamentous cyanobacteria such as Geitlerinema are rare in oxic and well-illuminated surface waters, and most numerous in the euxinic hypolimnion at which they receive a fraction of surface irradiance (Cohen et al., 1977b). In laboratory experiments at light intensities similar to or higher than in situ levels, Geitlerinema performs OP, but transitions fully to sulfide-based AP at micromolar concentrations of sulfide (Cohen et al., 1986). Under sulfidic conditions, Geitlerinema can also fix nitrogen and produce hydrogen (Belkin and Padan, 1978; Belkin et al., 1982). Its SQR has been isolated (Arieli et al., 1994), sequenced (Bronstein et al., 2000), and phylogenetically characterized (Pham et al., 2008; Marcia et al., 2010a; Gregersen et al., 2011). Geitlerinema is a model organism for studying the physiology of flexible AP/OP, diazotrophic cyanobacteria and their influence on modern and ancient systems. In this study, we analyzed a draft genome of Geitlerinema and characterized the genes related to nitrogen fixation, AP, and OP. These results provide a genomic foundation for metabolic flexibility in response to varying sulfide, oxygen, and light levels that was observed in previous physiology studies (Belkin and Padan, 1978; Belkin et al., 1982; Cohen et al., 1986; Shahak et al., 1987).
Materials and Methods
Culturing and Sequencing
The original strain was isolated from the sulfidic water column of Solar Lake, Israel (Cohen et al., 1975b), and was kindly provided by A. Oren for culturing. A monoalgal culture was grown in modified Chu’s 11 in Turks Island Salts medium at room temperature (average 22.0°C) and ambient light in a 125 mL Erhlenmeyer flask. We extracted whole community DNA using the MPBio FastDNA SpinKit and Fastprep-24 Bead Beater (MP Biomedicals, Solon, OH, USA) following the default protocol, except that 0.3 g of beads were used for bead beating. DNA was quantified using Quant-IT PicoGreen (Invitrogen, Grand Island, NY, USA) and submitted to the University of Michigan DNA Sequencing Core for library preparation and Illumina HiSeq 2 × 100 bp paired-end sequencing.
Assembly
Using wrappers provided at https://github.com/Geo-omics/scripts, reads were dereplicated with custom perl scripts, trimmed using Sickle (version 1.33) (Joshi and Fass, 2011), and assembled using IDBA-UD (version 1.1.1) (Peng et al., 2012) with the following parameters: –mink 65 –maxk 85 –step 10 –pre_correction. Tetranucleotide frequency was used to bin scaffolds by emergent self-organizing maps (Dick et al., 2009), with a minimum contig length of 500 bp and a window size of 10,000 bp (Supplementary Figure S1). The cyanobacterial bin of scaffolds >1000 bp in length was submitted to the Integrated Microbial Genomes Expert Review (IMG-ER) automated pipeline from Joint Genomes Institute (JGI) for annotation of genes and pathways (IMG accession number: 2660238729; Supplementary Table S1). Raw reads, assembled scaffolds, and gene annotations were submitted to NCBI (project: PRJNA302164).
Phylogenetic Analysis
Phylogenetic analyses of genes of interest (Table 1) was performed with maximum likelihood and the PROTGAMMAGTR algorithm in RAxML 8.1.15 and bootstrapped 1000 times (Stamatakis, 2014). psbA. sqr, and nifHDK gene phylogenies included 44 cyanobacterial isolate genomes and genomic bins from Hot Lake and Middle Island Sinkhole cyanobacterial mat metagenomes (Cole et al., 2014; Voorhies et al., 2016; Supplementary Table S2). Each of these cyanobacterial genome has at least one sqr gene, and 30 of the 44 genomes have a nifHDK gene set. sqr and nifHDK genes from Sulfuricurvum kujiense YK-1 DSM 16994, and other bacterial sqr genes (Supplementary Table S3) were included for context. Translated genes (amino acid sequences) were aligned with clustal-omega (version 1.2.0) (Sievers et al., 2011). Alignments for nifH. nifD, and nifK were concatenated, and the concatenated alignment was used for analysis. Halothece halophytica chlLNB genes were used to root the nifHDK tree (Boyd et al., 2015). Flavocytochrome c:sulfide dehydrogenase (FCSD) genes formed an outgroup for the sqr tree (Marcia et al., 2010a). The most divergent psbA from Gloeobacter kilaueensis JS-1 was used as an outgroup for the psbA tree (Cardona et al., 2015). Phylogenetic trees were drawn with FigTree version 1.4.2 (Rambaut, 2012).
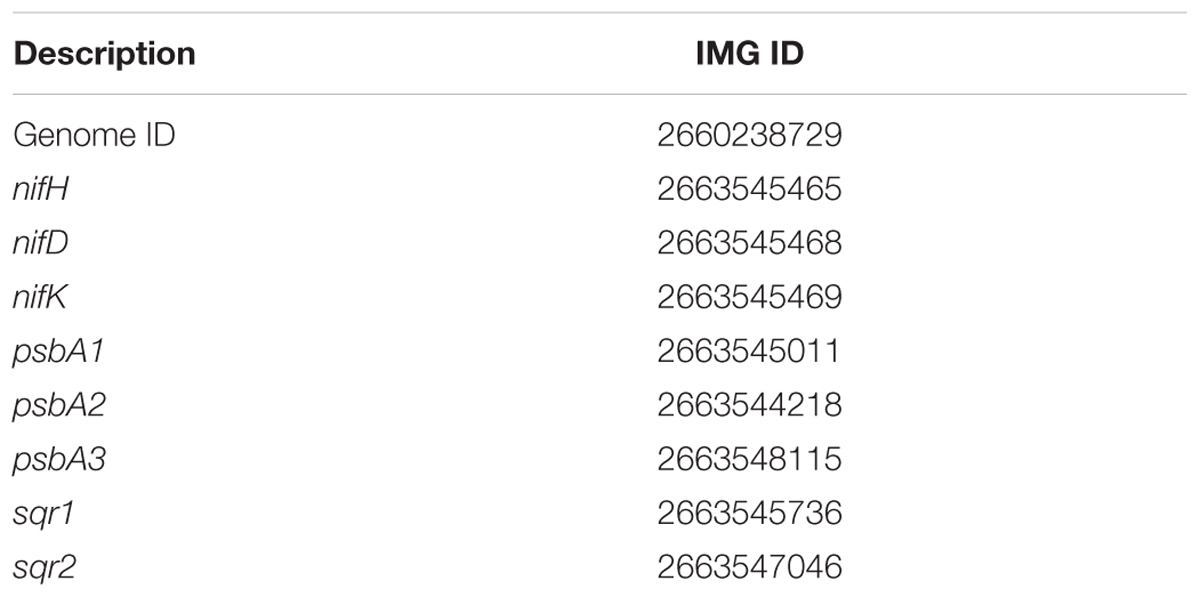
TABLE 1. Integrated Microbial Genomes (IMG) accession numbers of genome and select genes of interest of Geitlerinema sp. PCC 9228.
Results
Overview
The 4.77 Mb draft genome of Geitlerinema sp. PCC 9228 (henceforth “Geitlerinema”) contains 3,969 protein coding genes on 195 scaffolds. Coverage is on average 905x across the genome. The genome has 100% of universally conserved bacterial genes expected to be present (Raes et al., 2007; Alneberg et al., 2014).
Carbon Metabolism
Geitlerinema has key genes coding for proteins involved in photosynthetic and respiratory electron flow, including photosystem II (psbADBCEFO), succinate dehydrogenase (sdhABC), type-1 NADPH dehydrogenase (ndhA-M), cytochrome b6f (petADBCEJ), photosystem I (psaABCDEFK), and cytochrome c oxidation (coxABC) (Supplementary Table S1; Mulkidjanian et al., 2006). It has genes for the Calvin-Benson-Bassham cycle, carbon dioxide concentrating proteins (ccmK1K2MN), and Rubisco large and small subunits (rbcLS) for carbon fixation (Figure 1). Superoxide dismutase genes (sodC and sodN) to cope with superoxide formation during photosynthesis and aerobic respiration are also present in the genome. In ancestral cyanobacteria, these enzymes would have been critical for defense against increased production of reactive oxygen species alongside increasing O2 fluxes (Blank and Sánchez-Baracaldo, 2010; Fischer et al., 2016).
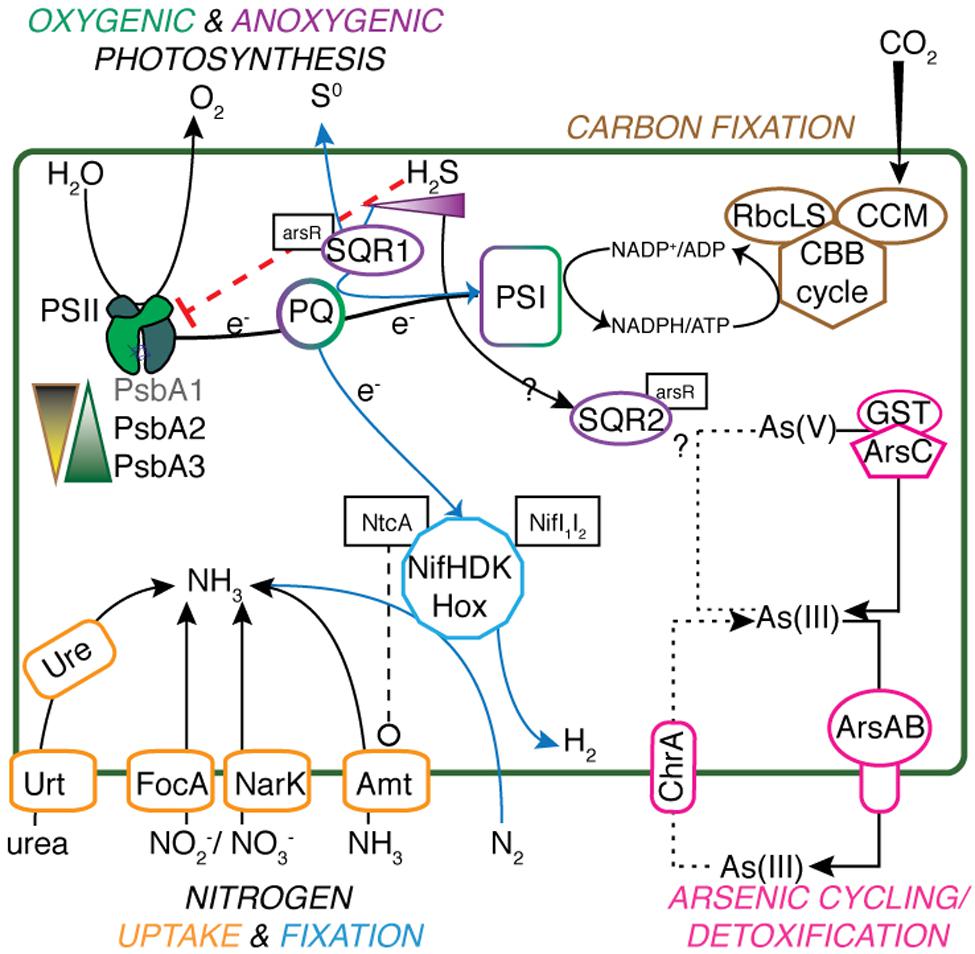
FIGURE 1. Metabolic schematic of Geitlerinema sp. PCC 9228. Genes for major processes [psbA1-3, sqr1, ccm, rbcLS, nifHDK, hox, arsABC, ure, urt, focA, narK, amt, glutathione S-transferase (GST)], and associated regulatory genes (arsR, ntcA, nifI1I2) are presented. Suppression of functions is indicated with red dashed lines ending in a flat line. Other interactions are indicated with black dashed lines ending with open circles. Transfer and production of key reactants and products are indicated with solid lines. Putative arsenite import is indicated with a dotted line, and anoxygenic photosynthesis-related processes have a solid blue line.
Geitlerinema has genes encoding a complete tricarboxylic acid (TCA) cycle, including 2-oxoglutarate dehydrogenase and succinyl CoA synthase to link synthesis of 2-oxoglutarate through succinyl-coA with succinate (Steinhauser et al., 2012). The genome also has the genes for acetolactate synthases and succinate-semialdehyde dehydrogenase to interconvert 2-OG and succinate through succinic semialdehyde in an alternative closure to the TCA cycle (Zhang and Bryant, 2011). Though it has shc, encoding for squalene-hopene cyclase, Geitlerinema lacks the hpnP gene for hopanoid methylation (Ricci et al., 2015). 2-methylhopanes, derived from 2-methylhopanoids, have been used as a bacterial biomarker in the geologic record (Summons and Lincoln, 2012). Geitlerinema has genes for acyl-ACP reductase and fatty aldehyde decarbonylase, key enzymes in an alkane biosynthesis pathway unique to cyanobacteria (Schirmer et al., 2010; Coates et al., 2014).
In the chlorophyll synthesis pathway, we identified both the aerobic oxidative ester cyclase chlE common to all cyanobacteria (Mulkidjanian et al., 2006), and the oxygen independent ester cyclase bchE. Functional bchE is common in anoxygenic phototrophic bacteria such as green sulfur bacteria and heliobacteria (Sousa et al., 2013), and is only rarely present in cyanobacteria, such as Synechocystis sp. PCC 6803 (Minamizaki et al., 2008). Geitlerienema has the light-dependent NADPH-protochlorophyllide oxidoreductase that produces chlorophyllide in the penultimate step of the pathway. This gene originated in cyanobacteria and is limited to cyanobacteria and phototrophic eukaryotes (Blankenship, 2002; Yang and Cheng, 2004). However, the genome lacks the light-independent oxidoreductase chlLNB, which allows for chlorophyll synthesis in the dark and is observed in all photosynthetic phyla (Blankenship, 2002). Due to their homology, Geitlerinema’s genes for nifHDK are the closest matches to H. halophytica’s chlLNB. Coverage estimates for nifHDK genes are consistent with the rest of the genome, indicating there was no mis-assembly of chlLNB reads into nifHDK genes.
Nitrogen Metabolism
The cyanobacterial genome has genes for a variety of pathways of nitrogen acquisition, including an ammonia transporter gene amt, cyanate lyase cynS, nitrate/nitrite transporters narK and focA, and nitrate assimilation-related genes nitrate and nitrite reductases nirA. nirC, and narB (Figure 1). The cyanobacterium also has urea transporters (urtABCD), urease genes (ureABCDFG), and genes for transport of neutral, branched, and polar amino acids.
Geitlerinema has a comprehensive operon for nitrogen fixation (nifVXSU, nifHDKEB; Raymond et al., 2004). Additional nitrogenase-related proteins are located on the same contig (iscA. nifI1I2, ferredoxin, nifN; Figure 2; Boyd et al., 2015). iscA is commonly observed in aerobic diazotrophs, and its recruitment into the genome is linked to a transition to aerobic lifestyle (Boyd et al., 2015). The cyanobacterium has glnB, a member of the PII signal transduction protein family that regulates nitrogen-related proteins, and ntcA, which controls expression of glnB (Forchhammer, 2004). The nifI1I2 gene is also a member of the PII protein family, but is characteristic of diazotrophic anaerobes that regulate their nitrogenase activity post-translation, such as Desulfovibirio and Clostridium (Boyd et al., 2015).
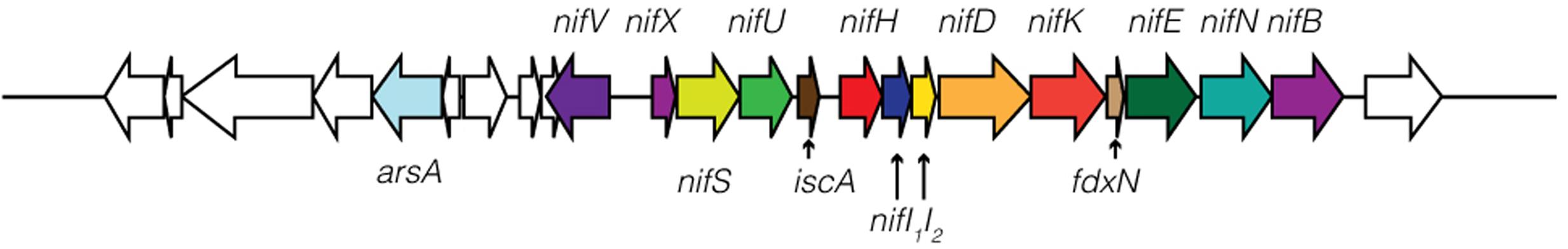
FIGURE 2. Schematic of nitrogenase genes in Geitlerinema sp. PCC 9228. nifXSU, iscA, nifHI1I2DK, fdxN, and nifENB are arranged in an apparent operon. nifV and a regulatory arsenic-related gene arsA are located upstream of the nifHDK operon.
Phylogenetic analysis shows that nifHDK genes from Geitlerinema clusters with those from two cultured cyanobacterial genomes (Pleurocapsa sp. 7327, Microcoleus chthonoplastes) and a cyanobacterial genome-from-meta genomic bin (Phormidium OSCR; bootstrap = 100; Supplementary Figure S2). Like Geitlerinema, their nitrogenase operons also hold nifI1I2. Geitlerinema has a bidirectional NiFe hydrogenase gene set (hoxEFUYH and hoxW) with its transcriptional regulator (lexA) and hydrogenase maturation proteins (hypBAEDC) (Figure 1). Unlike Geitlerinema, typical nitrogen-fixing cyanobacteria have an uptake hydrogenase (hupSL) to consume H2 produced in nitrogen fixation. hupSL is under similar transcriptional regulation as nitrogen acquisition genes like dinitrogenase, making expression of hupSL dependent on nitrogen limitation (Tamagnini et al., 2007). In contrast, the bidirectional hydrogenase can be present in both diazotrophic and non-diazotrophic cyanobacteria and is expressed under more diverse conditions. It may, for instance, be used in fermentation or to direct electrons during photosynthesis (Tamagnini et al., 2007). Geitlerinema produces hydrogen during sulfide-dependent AP in the presence of bioavailable nitrogen, as well as in the absence of CO2, suggesting the nitrogenase-independent hox gene set may be responsible for hydrogen evolution (Belkin and Padan, 1978; Belkin et al., 1982).
Photosystem II Assembly
The core proteins in photosystem II are encoded by psbA and psbD (Blankenship, 2002). Geitlerinema has one version of psbD and three versions of the psbA gene (Figure 1). The standard psbA, hereafter referred to as psbA3, is in a large clade of typical oxygenic psbA designated “group 4” after (Cardona et al., 2015) (Figure 3). All but one of the isolate genomes in this analysis have at least one copy of this form of psbA, which is used in OP in aerobic conditions (Supplementary Table S2). Coverage estimates and paired-end information suggest that Geitlerinema has two copies of psbA3. It is the sole gene on its contig, and pairs of reads that map to the ends of the psbA3 match to the ends or beginnings of four other contigs. Those portions have 100% identity to the ∼80 bp beginning and end of psbA3. The De Bruijn graph-based assembly algorithm used in this analysis frequently prematurely assembles identical copies of genes (Nagarajan and Pop, 2013). The approach accurately assembled the two copies of psbA3, but could not automatically bridge the copies.
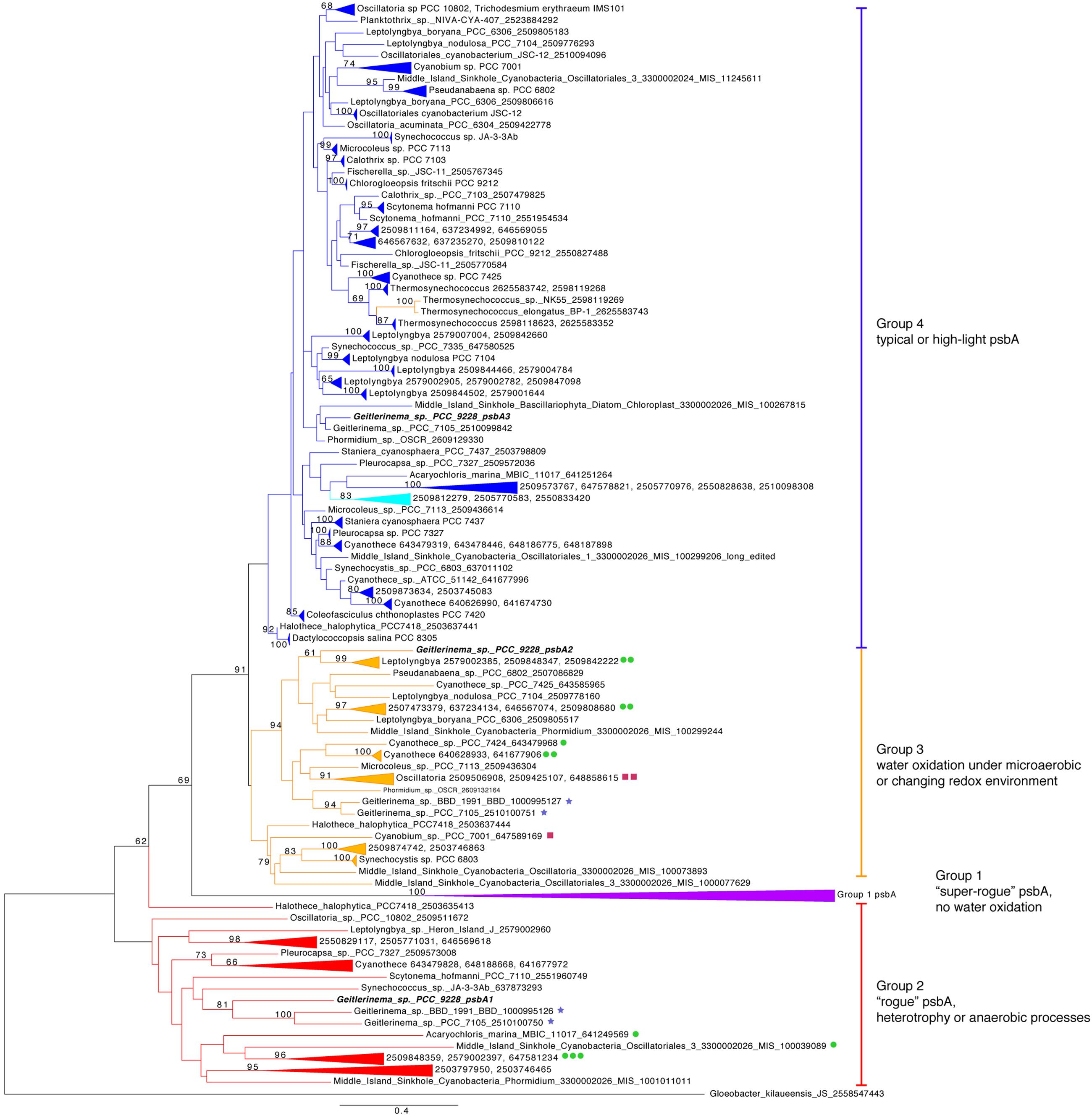
FIGURE 3. Phylogenetic tree of psbA. Genes are colored by groups 1–4 modeled after (Cardona et al., 2015). The outgroup is a Gloeobacter kilaueensis JS psbA, which is most similar to psbD and is unable to oxidize water (Cardona et al., 2015). Purple indicates group 1, red genes belong to group 2, orange genes are members of group 3, and blue standard psbAs belong to group 4. Genetic proximity to sqr types I (green circle), II (maroon squares), and VI (blue stars) sqr genes are indicated. Fourteen psbA genes are five or fewer genes from sqr, and the largest gap is 19 genes. Geitlerinema sp. PCC 9228 has members of groups 2–4.
One additional version of psbA, henceforth referred to as psbA1, has 85% sequence identity to psbA3 (blastx). psbA1 is located in a different operon but on the same scaffold near ntcA and photosynthetic subunits psbN and psbH. psbA1 is in a well-supported clade with genes from Geitlerinema sp. PCC 7105 (2510100750) and Geitlerinema sp. BBD 1991 (BBD_1000995126; bootstrap = 81; Figure 3). This gene is also in a larger “group 2” (Cardona et al., 2015) that includes psbA genes from cyanobacterial genomic bins (MIS_1001011011, MIS_100039089) sourced from a low-oxygen cyanobacterial mat in the Middle Island Sinkhole (Voorhies et al., 2016). Group 2 genes encode D1 proteins used under growth conditions that do not favor water oxidation (Cardona et al., 2015), such as heterotrophy in the dark (Park et al., 2013), photosynthetic electron transport inhibition (Kiss et al., 2012), or oxygen-sensitive processes such as nitrogen fixation (Toepel et al., 2008; Wegener et al., 2015). Finally, the third variant, psbA2, is 59–88% similar to psbA1 (discontinuously; tblastx), and 90% similar to psbA3 (over 99% of their lengths; blastx). psbA2 is in a clade with genes from a Middle Island Sinkhole cyanobacterium (MIS_100299244), Geitlerinema sp. BBD1991 (BBD_1000995127) and Phormidium sp. OSCR (2609132164; bootstrap = 94). Proteins from these “group 3” psbA genes (Cardona et al., 2015) are expressed under microaerobic conditions, with modified electron transfer to cope with the changing redox environment (Figure 1; Sicora et al., 2004, 2009; Sugiura et al., 2012).
Sulfide Oxidation
Two copies of the gene for sulfide quinone reductase, sqr, are in the genome of Geitlerinema (Supplementary Figure S4). This gene is involved in the oxidation of sulfide for detoxification or to harvest electrons for AP, such as in purple and green sulfur bacteria (Theissen et al., 2003; Marcia et al., 2009, 2010a; Gregersen et al., 2011). Each sqr is located upstream of arsenic resistance arsR-type genes, putatively involved in transcriptional regulation of sqr under conditions such as sulfide exposure (Nagy et al., 2014). One sqr, referred to as sqr1 hereafter, matches the previously cloned and sequenced sqr gene of Geitlerinema (Bronstein et al., 2000). This enzyme mediates the reduction of plastoquinone and oxidation of sulfide in sulfide-dependent AP (Arieli et al., 1994). On the sqr phylogenetic tree, Geitlerinema’s canonical sqr groups with other cyanobacterial versions that are considered type I (Marcia et al., 2010a; Gregersen et al., 2011; bootstrap = 100; Figure 4; Supplementary Figure S5). Hydrogen sulfide affinities for cyanobacterial sqr in this cluster are high, with Km in the micromolar range (Arieli et al., 1991; Bronstein et al., 2000). Geitlerinema. Coleofasciculus (formerly Microcoleus), Halothece, and proteobacterial members in this cluster have been shown to grow with this sqr on sulfide-induced AP (Oren and Padan, 1978; Cohen et al., 1986; Jørgensen et al., 1986; Schütz et al., 1999).
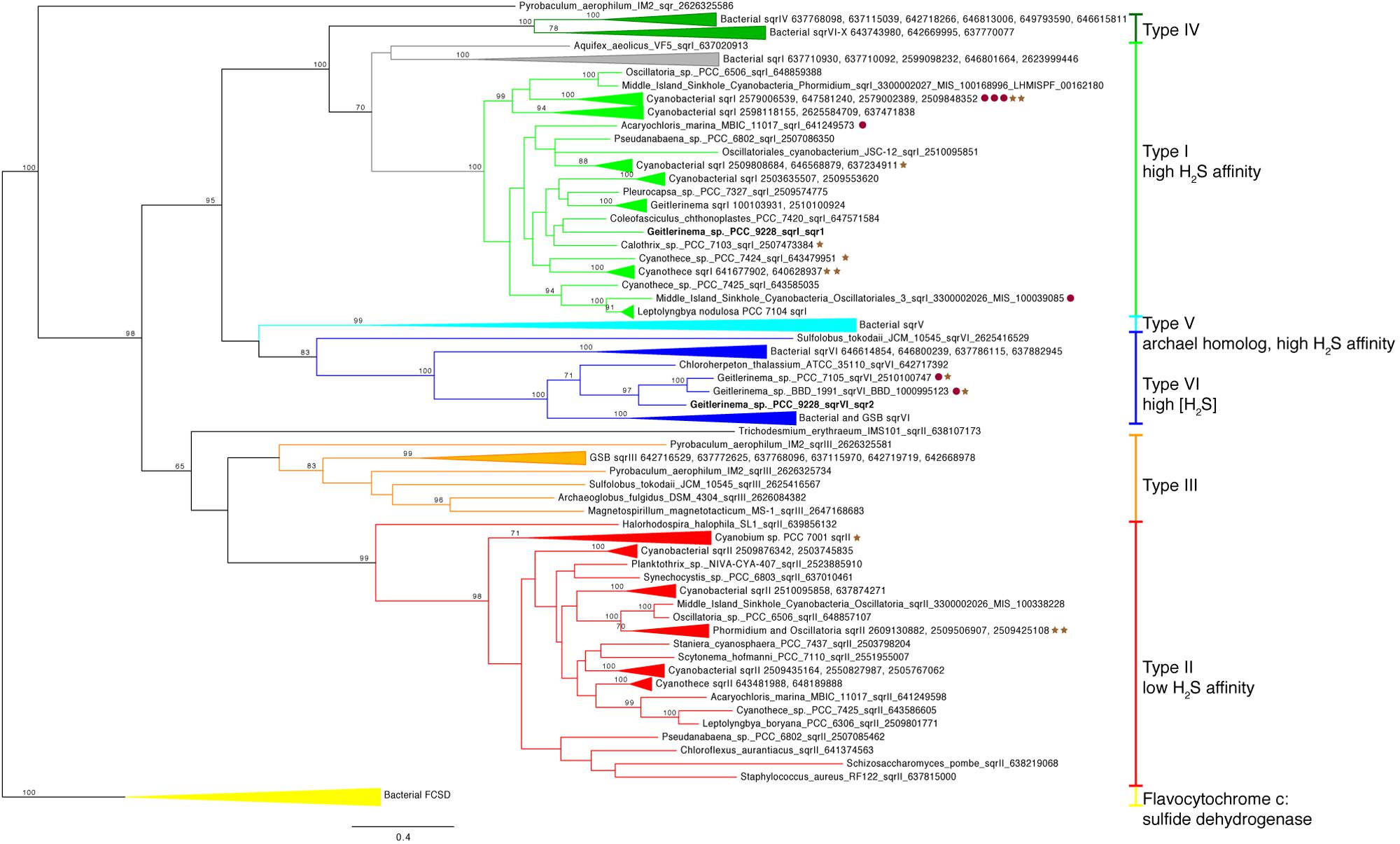
FIGURE 4. Phylogenetic tree of sulfide quinone reductase (sqr). Genes are colored by types I–VI modeled after (Marcia et al., 2010a). Bacterial FCSD (yellow) includes representatives of flavocytochrome c:sulfide dehydrogenase. Green sulfur bacteria have sqr belonging to types III (orange), V (light blue), IV (dark green), and VI (blue). Cyanobacterial sqr are limited to types I (light green), II (red), and VI. Proximity to psbA versions on the same contig (see Figure 3) are indicated with red circles (group 2) and brown stars (group 3). No sqr genes in this analysis were genetically proximal to groups 1 or 4 psbA genes. Geitlerinema sp. PCC 9228 has types VI and I sqr.
The second sqr gene, referred to as sqr2 hereafter, is 25–50% identical over discontinuous fragments to the sqr1 gene (tblastx). It is most similar (tblastx 48–63% identity to overlapping fragments that span the length of the gene) to that of Chloroherpeton thalassium ATCC35110, a green sulfur bacterium, with similar identity to the sqr genes of other sulfur oxidizing bacteria. sqr2 clusters phylogenetically with two other cyanobacterial sqr [Geitlerinema sp. PCC 7105 and BBD 1991 (bootstrap = 97)], among a group of known type VI versions (bootstrap = 100; Figure 4; Supplementary Figure S5). They include sqr from thiotrophic proteobacteria (Marcia et al., 2010a) and green sulfur bacteria (Gregersen et al., 2011), including that of Chlorobium tepidum used in sulfide oxidation when sulfide exceeds 4 mM (Chan et al., 2009).
Trace Metal Resistance
Arsenic resistance genes arsB (an arsenite efflux pump) and arsC (arsenate reductase; Slyemi and Bonnefoy, 2011; van Lis et al., 2013), and arsR, the putative regulatory protein of sqr, are in an arsRBC operon downstream of sqr2. Upstream of sqr2 are genes encoding glutathione S-transferase and a multidrug efflux pump (Supplementary Figure S4). Another arsB is located downstream of sqr1 but in a different operon. In combination with arsB, two arsenite-tranporting ATPases arsA genes are present in the genome, one located on the same scaffold as the nitrogenase gene suite and the other on the longest scaffold in the dataset. Geitlerinema does not have known arsenite oxidizing genes (aioAB. arxA) or respiratory arsenate reductase genes (arrA), based on gene annotation and BLAST searches against known genes (Hoeft et al., 2010; Slyemi and Bonnefoy, 2011). It has an annotated chromate transporter (chrA) on another scaffold that has 52% positive match (blastp) to a similarly annotated gene in Synechocystis sp. PCC6803 that functions as an arsenite uptake transporter for arsenite oxidation (Nagy et al., 2014; Figure 1). On another scaffold, a gene belonging to the DUF302 protein superfamily of unknown function is quite similar (75% positive, 56% identical blastp) to a Synechocystis gene in the sqr1 plasmid operon, as well as to a potential arsenic oxidase gene in Agrobacterium (61% positive, 44% identical, blastp; Nagy et al., 2014). Geitlerinema also has a methyltransferase similar (84% positive, blastx) to the arsM gene of Synechocystis used in arsenite methylation for detoxification (Yin et al., 2011).
Discussion
Previous investigations of cyanobacterial cultures capable of OP and sulfide-dependent AP targeted physiology, biochemistry, and limited genetic analyses (Cohen et al., 1986; Arieli et al., 1994; Bronstein et al., 2000; Miller and Bebout, 2004; Klatt et al., 2015a). However, the broader genetic characteristics of this cyanobacterial metabolic flexibility have not been thoroughly evaluated. In this study, we analyzed the genome of a model anoxygenic photosynthetic cyanobacterium, Geitlerinema sp. PCC 9228. The organism was isolated from a sulfidic, low-light environment, and numerous physiology studies have documented its ability to fix nitrogen, its high affinity for sulfide, and its oxygenic and anoxygenic photosynthetic capabilities (Cohen et al., 1975b, 1986; Belkin et al., 1982). We comprehensively evaluated the draft genome of Geitlerinema for key metabolic genes for oxygen, nitrogen, and carbon cycling, and provide the genetic evidence of its metabolic versatility.
Photosynthesis Is Optimized for Light and Redox
Our results indicate that Geitlerinema sp. PCC 9228 and other sulfide-tolerant and/or sulfide-using cyanobacteria have multiple different types of psbA genes that are inferred to meet physiological and biochemical needs under changing light and redox conditions. The psbA gene encodes the D1 protein that directly supports the water-oxidizing cluster in PSII (Ferreira et al., 2004; Fischer et al., 2015). Due to its role in water oxidation, this protein experiences high levels of oxidative damage and degradation. By impacting water oxidation, changes in redox such as sulfide inhibition of the water oxidizing complex in PSII (Miller and Bebout, 2004) and/or non-optimal light conditions lead to excessive energy (Klatt et al., 2015b) and influence the risk and magnitude of oxidative damage to D1 (Aro et al., 1993). Thus, cyanobacteria that experience such dynamic conditions may use another of the multiple psbA genes in their genomic repertoire under different light and oxygen regimes to mitigate oxidative damage (Schaefer and Golden, 1989; Campbell et al., 1999; Sicora et al., 2009; Gan et al., 2014; Cardona et al., 2015; Ho et al., 2016). Geitlerinema has two copies of psbA for OP under high oxygen and/or light levels (both psbA3; group 4), one gene for non-oxygen evolving PSII (psbA1; group 2), and one gene for microaerobic conditions (psbA2; group 3; Figures 1 and 3). We infer that these distinct photosynthetic genes reflect the adaptation of Geitlerinema to a variably lit, low-oxygen, and sulfidic lifestyle.
During high light conditions, when Geitlerinema conducts OP (Cohen et al., 1975a, 1986), the psbA3 (group 4) genes are likely used to reduce photoinhibition and oxidative stress, as observed in Synechococcus sp. PCC 7942, Synechocystis sp. PCC6803, and Thermosynechococcus elongatus (Schaefer and Golden, 1989; Sicora et al., 2006; Kós et al., 2008; Sugiura et al., 2010). Other cyanobacteria synthesize identical D1 proteins from different genes at high light versus regular light conditions (Sugiura et al., 2010). Two copies of psbA3 in its genome may equip Geitlerinema to continue OP in times of sufficient and/or high light, such as when mixed up into the epilimnion or in holomixis. Given that psbA3 clusters with standard and high-light variant genes (Figure 3), it remains to be seen if one or both of Geitlerinema’s two copies of psbA3 is cued to the different light intensities the organism may experience in its natural habitat.
The psbA1 and psbA2 genes likely provide Geitlerinema with the flexibility to conduct photosynthesis and nitrogen fixation under varying oxygen and/or sulfide concentrations. With sufficient light, the cyanobacterium may rely upon psbA2 (group 3), which is keyed for microaerobic conditions. Changing redox environment promotes synthesis of psbA2-like variants (group 3) in Thermosynechococcus elongatus (Sugiura et al., 2012) and Synechocystis sp. PCC6803 (Sicora et al., 2004). However, light levels are lower than required for OP in the hypolimnion of Solar Lake, the prime habitat of Geitlerinema. Together with continuous sulfide exposure, these conditions often favor AP in cyanobacteria (Klatt et al., 2015a, 2016) and in green sulfur bacteria (Hanson et al., 2013; Findlay et al., 2015). Groups 2 and 3 psbA genes in the current study are in close chromosomal proximity to sqr genes (Figure 3), suggesting an intriguing but currently untested linkage between modified PSII and sulfide exposure. Alternative D1 proteins have been hypothesized to be directly involved in the transfer of electrons from donors other than water (Murray, 2012), such as in sulfide oxidation during nitrogen fixation (Becraft et al., 2015; Olsen et al., 2015). However, whether alternative D1 proteins are directly participating in this unusual photochemistry or serve to inactivate PSII in AP, remains to be tested.
Very little sulfide is required to limit OP in Geitlerinema (Cohen et al., 1986), and in the transition between oxygenic to AP, continuous exposure induces synthesis of proteins such as SQR (Arieli et al., 1991). SQR has a higher affinity for plastoquinone than PSII (Klatt et al., 2015a), thus in this induction period and with continual sulfide exposure, Geitlerinema may also reformulate its photosystem II machinery to reduce oxygen production. Finally, the psbA1 (group 2) gene is also likely used to disable oxygen production during times of nitrogen fixation. Non-oxygen producing group 2 enzymes have been implicated as structural ‘placeholders’ that may allow oxygen-sensitive processes such as nitrogen fixation to occur (Sicora et al., 2009; Murray, 2012; Wegener et al., 2015). Further experiments targeting gene and protein expression will confirm the physiological roles of the variant psbA genes.
Variant psbA genes such as those observed in Geitlerinema likely permitted ancestral and potentially AP-capable cyanobacteria to meet metabolic requirements in dynamic physicochemical conditions. The importance of AP cyanobacteria and their metabolisms on Archean and Proterozoic oxygen levels is linked to the timing of OP. Mat-forming cyanobacteria in modern hypersaline and hot spring ecosystems likely face the same environmental challenges as their stromatolite-forming ancestors (Stal, 1995; Grotzinger and Knoll, 1999). As in modern systems, the metabolisms and mat-building lifestyles of ancestral cyanobacterial populations would have promoted light and redox dynamics on spatial (depth) and temporal (diel) scales (Canfield, 2005; Blank and Sánchez-Baracaldo, 2010; Lalonde and Konhauser, 2015; Sumner et al., 2015). Genomic and physiological studies on contemporary mat-forming cyanobacteria and their responses to changing environmental parameters, such as different versions of psbA keyed to light and/or oxygen levels, inform potential genetic and physiological strategies in ancient cyanobacteria. Given the long geologic history of variable and low atmospheric oxygen concentrations, the first appearance of alternative psbA in cyanobacteria is uncertain. The functional differences and potential heterotachy in these homologous genes dictate caution when evaluating their timing and evolutionary order. However, the basal arrangements of group 2 to group 3/4, and group 3 to group 4 in the phylogenetic tree hint at an ancestral development of water oxidation when atmospheric oxygen was low (Cardona et al., 2015). The perpetuation of diverse and evolutionarily old psbA genes in cyanobacterial genomes allow for efficient metabolic functioning regardless of oxygen levels/needs (Murray, 2012). These variant genes were likely retained in cyanobacterial genomes for oxygen-sensitive processes, such as nitrogen fixation, in an increasingly oxidizing environment.
Nitrogen Acquisition Strategies in Variable Redox Conditions
Geitlerinema has a suite of genes for uptake of nitrogen (nitrate, nitrite, urea) as well as nitrogen fixation through a comprehensive nitrogenase gene suite (Figures 1 and 2). Sulfide-dependent AP was measured in select strains grown in nitrogen-replete media, including Geitlerinema sp. PCC 9228, Coleofasciculus chthonoplastes, and Pseudanabaena FS39, suggesting nitrate assimilation occurs under AP conditions (Cohen et al., 1986; Klatt et al., 2015a). Geitlerinema is also capable of AP-dependent nitrogen fixation (Belkin et al., 1982), potentially using sulfide to scavenge residual oxygen or donate electrons to nitrogenase (Stal, 2012).
Clues about the cyanobacterial transition from an anaerobic to aerobic lifestyle are apparent in regulatory genes for nitrogen acquisition. Due to its evolution under anoxia and strict requirement for anaerobic conditions, cyanobacterial diazotrophy in an increasingly oxidizing environment required new adaptations (Boyd et al., 2011; Stüeken et al., 2015). In the nif operon of Geitlerinema is nifI1I2, which encodes a signal transduction protein of the PII family that is present in diazotrophic archaea and select anaerobic bacteria, but has not been studied in cyanobacteria (Forchhammer, 2004; Boyd et al., 2015). In those organisms, NifI1I2 inhibits nitrogenase activity when the cell is no longer nitrogen limited (Kessler et al., 2001). Geitlerinema’s nifHDK gene suite is phylogenetically grouped with those from three other cyanobacterial genomes that also have nifI1I2 in their nitrogenase operons (Supplementary Figure S2), and members of this cluster are adapted to low-oxygen and/or sulfidic conditions. C. chthonoplastes can continue to operate OP at low sulfide concentrations and is also capable of sulfide-dependent AP (Cohen et al., 1986). Similar to Geitlerinema and its psbA1. Pleurocapsa sp. PCC 7327 also has a psbA gene that codes for a rogue D1 protein typically used in anoxic conditions (Wegener et al., 2015).
Geitlerinema Possesses Genetic Versatility for Sulfide Oxidation
Sulfide quinone reductase (SQR, encoded by sqr) oxidizes sulfide to elemental sulfur, and this process can be coupled with lithotrophic or phototrophic growth in bacteria and archaea or detoxification of sulfide in eukaryotes (Theissen et al., 2003; Marcia et al., 2010a). The genome of Geitlerinema sp. PCC 9228 holds two sqr operons. The first operon has the well-studied high-affinity sqr1 (type I), which is translated after a few hours of exposure to micromolar levels of H2S (Oren and Padan, 1978) and enables Geitlerinema to grow by sulfide-based AP (Cohen et al., 1986). The transcriptional regulatory gene arsR, located downstream of sqr1, likely controls its expression (Nagy et al., 2014). Though lateral gene transfer has distributed various sqr types among bacteria, archaea, and eukarya (Theissen et al., 2003), type I sqr genes in cyanobacteria such as Geitlerinema sqr1 form a distinct and well-supported cyanobacterial phylogenetic subclade within a bacterial clade (Figure 4). This cyanobacterial clade includes genes from AP-capable or sulfide-tolerant members such as C. chthonoplastes. Synechocystis sp. PCC6803, H. halophytica, and Geitlerinema sp. BBD (Cohen et al., 1986; Theissen et al., 2003; Marcia et al., 2010a; Nagy et al., 2014; Den Uyl et al., 2016). SQR is an evolutionarily ancient enzyme that was widespread in organisms during the Proterozoic (Theissen et al., 2003), during which the oceans had variable redox conditions including euxinia. Although the evolutionary history of sqr in cyanobacteria appears to be complex and remains unresolved, the sqr1 gene is thought to be endogenous to cyanobacteria (Theissen et al., 2003). As a critical component of sulfide-based AP, this gene would have enabled ancestral cyanobacteria to thrive during the Proterozoic, when periods of photic zone euxinia would have favored AP and tempered oxygen production (Johnston et al., 2009). Whether the endogenous type I SQR may have permitted ancestral cyanobacteria to oxidize sulfide for energy, or cyanobacteria retooled this detoxifying enzyme for AP, is an open question.
The role of the second sqr operon in Geitlerinema is unknown. In addition to sulfide consumption through sqr1-based AP (Oren and Padan, 1978), Geitlerinema has a second sulfide donation site on the immediate donor side of PSI, which is not inducible (i.e., does not require protein synthesis) and does not significantly contribute to proton translocation (Shahak et al., 1987). However, this response operates at even higher concentrations of sulfide (Km in the mM range) without saturation (Shahak et al., 1987; Arieli et al., 1991). The enzyme mediating this response to high sulfide is unknown, thus it could be encoded by this sqr2. Only 10 of the 44 cyanobacterial genomes and genomic bins examined in this study (including Geitlerinema sp. PCC 9228) have more than one version of sqr. Seven genomes in this subset, such as Synechocystis sp. PCC6803 (Nagy et al., 2014), each have two versions sqr, one of which is phylogenetically similar to sqr1, and the other a eukaryotic homolog (type II). Only three genomes, all of them Geitlerinema species, have genes phylogenetically similar to sqr2 (type VI) as well as an sqr1-like version. The sulfide physiology of Geitlerinema sp. PCC7105 is unknown, but Geitlerinema sp. BBD is a known sulfide-resistant photosynthetic cyanobacterium (Den Uyl et al., 2016). Green sulfur bacteria, thiotrophic proteobacteria, and members of Aquificaceae have multiple versions of sqr or homologs with a range of sulfide affinities. Aquifex aeolicus expresses its types I and VI sqr, similar to Geitlerinema’s sqr1 and sqr2, even when not growing thiotrophically (Marcia et al., 2010a,b). On the other hand, Chlorobium is capable of sulfide-dependent growth at mM H2S by using its type VI sqr, and other versions are used at lower sulfide concentrations (Chan et al., 2009). Phylogenetic clustering of sqr2 with green sulfur bacterial sqrs, including one used at high sulfide levels, supports a similar role in Geitlerinema. Like sqr1. sqr2 also has a transcriptional regulatory gene arsR in the operon. These observations raise the possibility that Geitlerinema is capable of growing phototrophically on different sulfide levels through its sqr1 and sqr2. Our results also suggest that such versatility was likely achieved through a combination of evolutionary processes including vertical descent within ancestral cyanobacteria (sqr1, type I clade) as well as lateral transfer from other groups (sqr2, type VI clade). When these genes became part of the AP-cyanobacterial repertoire is uncertain.
Chromosomal examination of cyanobacterial sqr and groups 2 and 3 psbA genes underscores a potential relationship between sulfide exposure and reformulation of the water-oxidizing complex in periods of AP and/or OP. Of the evaluated cyanobacterial genomes with sqr, 15 of the genomes have sqr types I, II, or VI in close genetic proximity to low-oxygen or anaerobic psbA genes. These genomes include metagenomic bins from Middle Island Sinkhole, where sulfide-based AP has been demonstrated (Voorhies et al., 2012), two other species of Geitlerinema, four Cyanothece species, and two Leptolyngbya species, among others. Because of variable contig lengths, it is unknown if psbA1 and/or psbA2 of Geitlerinema sp. PCC 9228 are in proximity to one or both of its sqr genes. However, the small intergenetic spaces between anoxygenic/micro-oxygenic psbA varieties and sqr in the other genomes hints at potential linked transcriptional regulation of these genes. Additionally, of the cyanobacterial sqr genes, 27 were in close proximity to arsR-like transcriptional regulators. These genes and conditions of their expression are an attractive target for further studies.
Potential Trace Metal Oxidation
Different versions of sqr in Geitlerinema sp. PCC 9228 may also be linked to trace metal metabolism and resistance. In close proximity to sqr2 there are genes for arsC. arsB. arsR, and glutathione (GST) S-transferase, and on other contigs there are genes for arsenite transporting ATP-ases arsA, another arsB, a methyltransferase similar to arsenite methylator arsM, and a chromate transporter similar to a cyanobacterial arsenite importer. These genes are involved in arsenate reduction, arsenite transport, transcriptional regulation, and mediation of arsenic resistance (Oden et al., 1994; Mukhopadhyay et al., 2002; Cameron and Pakrasi, 2010; Slyemi and Bonnefoy, 2011). Synechocystis sp. PCC6803 uses an arsR gene to regulate genomic arsBHC expression during arsenic exposure (López-Maury et al., 2003), and is able to grow in mM concentrations of arsenite and arsenate (Sánchez-Riego et al., 2014). An arsR-like transcriptional regulator is also adjacent to sqr1 in Geitlerinema (Bronstein et al., 2000), and the proximity of arsR to sqr2 suggests a similar regulatory role. Arsenic resistance genes are widely distributed among bacteria and archaea, and may be found in environments that do not have measurable arsenite (Mukhopadhyay et al., 2002; Oremland and Stolz, 2005). The hydrology of the habitat of Geitlerinema, Solar Lake, is driven primarily by seawater seepage through the sand bar, with minor meteoric input (Cohen et al., 1977a). Usual sources of arsenic, such as hydrothermal hot springs in hypersaline environments or weathering of arsenic-rich clays (Oremland et al., 2009), are absent from this system. As such, these genes may have been inherited from ancestral cyanobacteria inhabiting a metal-rich environment (Oremland et al., 2009), be used to detoxify other metals such as antimony (Nagy et al., 2014), or cope with reactive oxygen species and oxidative stress (Latifi et al., 2009; Takahashi et al., 2011).
The proximity of the sqr2 to arsenic-related genes (arsRBC, glutathione S-transferase), taken together with experimental results from related organisms and genes, hints at an unexplored potential metabolism in Geitlerinema: AP using arsenite as the electron donor, as in anoxygenic bacteria (Kulp et al., 2008; Hoeft et al., 2010; Edwardson et al., 2014). Oscillatoria-like cyanobacterial biofilms in an arsenic-rich hot spring, and Synechocystis sp. PCC 6803, can perform light-dependent arsenite oxidation (Kulp et al., 2008; Nagy et al., 2014). In Synechocystis sp. PCC 6803, the same sqr and arsR genes from its plasmid sqr operon, which enable light-dependent sulfide oxidation, are expressed during arsenite (As(III)) oxidation in the light (Nagy et al., 2014). The cyanobacterium imports arsenite through a chromate transporter (suoT, on the same plasmid operon), stores mM arsenite intracellularly without detriment, and exports excess arsenite through arsB (Nagy et al., 2014). The arrangement of Geitlerinema’s arsenic-related genes on its sqr2 operon is similar to the plasmid sqr operon of Synechocystis sp. PCC 6803. Arsenite oxidation has been linked to electron transfer to quinones (Jiang et al., 2009) and energetics support the potential of cyanobacterial plastoquinone as an arsenite oxidant (Nagy et al., 2014), but this pathway has not been explored in cyanobacteria. The co-transcription of sqr1 in Synechocystis with arsenite uptake genes during arsenite exposure (Nagy et al., 2014), and the well-characterized electron-stripping mechanism of sqr1 on sulfide (Arieli et al., 1994), hints at a role for sqr in arsenite oxidation.
Arsenite-dependent cyanobacterial AP is intriguing due to the role of arsenite-based primary production on ancient Earth. Arsenic was likely more abundant on Earth’s surface during the Archean than present (Oremland et al., 2009). In anoxic marine basins that dominated the biosphere 2.7 Ga ago, arsenite-dependent microbial autotrophy putatively cycled nitrogen, carbon, and arsenic (Sforna et al., 2014). This metabolism continues in modern hypersaline hot springs and subsurface aquifers that have elevated arsenic levels (Oremland and Stolz, 2005; Kulp et al., 2008; Hoeft et al., 2010). The genome of Geitlerinema lacks proteobacterial arsenite-oxidizing genes, and instead has genes similar to those used for light-dependent arsenite oxidation in Synechocystis sp. PCC 6803 (Nagy et al., 2014). Verifying arsenite-dependent AP in cyanobacteria, and conclusively linking the sqr2 gene in Geitlerienema to that process, would clarify the potential role of anoxygenic cyanobacteria in arsenic cycling in both modern and ancient ecosystems.
In summary, analysis of the genome of Geitlerinema sp. PCC9228 complements prior physiology studies by providing the genetic foundation for its metabolisms of nitrogen fixation, facultative OP, and sulfide-based AP. We find multiple versions of psbA, encoding a key protein for water oxidation, which may enable a sensitive response to varying conditions of light, oxygen and sulfide. Nitrogen fixation is linked to oxygen level and production via the nifI1I2 regulator in the nif operon and via non-oxygen producing psbA, respectively. Multiple versions of sqr likely address a range of sulfide concentrations and may also be linked to responses to metals and oxidative stress and perhaps even arsenite oxidation. Aerobic versatility encoded in the genome of Geitlerinema, coupled with diazotrophic regulation and concentration-specific sulfide responses, permit Geitlerinema to thrive in periodic sulfidic, microoxic, and poorly lit conditions of Solar Lake (Cohen et al., 1975a). Such dynamic geochemical conditions likely also challenged cyanobacteria during variable sulfide and oxygen levels of the Archean and Proterozoic (Satkoski et al., 2015; Sperling et al., 2015). This study of Geitlerinema and its unique gene assemblage addresses both the ambiguous role of Archean cyanobacteria in oxygen production/mitigation prior to the development of OP and the Great Oxidation Event, as well as the contributions of AP cyanobacteria to Proterozoic biogeochemistry. The phylogeny and diversity of genes responsible for metabolic versatility in Geitlerinema suggest a blend of genetic strategies for the anoxic early environment—such as methanogen-like modulation of nitrogen fixation and non-water oxidizing photosynthetic proteins—with post-oxidation strategies such as specific photosynthetic proteins for micro-oxic as well as oxic conditions, and different SQRs for fluctuating sulfide concentrations. Phototrophs capable of versatile AP/OP, such as Geitlerinema, would have had the advantage over organisms metabolically limited to either oxic or sulfidic conditions. Their continuous photosynthesis likely supported other microorganisms with fixed nitrogen and carbon, sulfide removal, and intermittent oxygen production. Furthermore, conditional production of oxygen at variable concentrations would have had strong influences on the structure and metabolic needs of their associated microbial communities through development of oxygen refugia and/or oases. Further research into Geitlerinema’s growth and transcriptional regulation will uncover the fine-tuned response of AP/OP cyanobacteria to changing redox conditions. In turn, we can relate the scope of these dual metabolisms and their modern physiologies to their ancestors’ impacts on ecology and geochemistry as Earth slowly and discontinuously became oxygenated.
Author Contributions
SG designed the study and conducted phylogenetic analyses. GD supervised the analyses. SG and GD contributed to the manuscript.
Funding
This work was supported by NSF grants EAR 1035955 and EAR 1637066 to GD. SG was supported by a University of Michigan Novak Fellowship.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Tanai Cardona and Eric Boyd for providing cyanobacterial psbA and nifHDK sequences used in this study, and for helpful discussion. Prof. Aharon Oren kindly provided the culture of Geitlerinema sp. PCC 9228. We thank Paul Den Uyl for culturing the cyanobacterium and extracting DNA for sequencing, and for providing sequences from Geitlerinema sp. BBD for analyses. We thank Sunit Jain and Judith Klatt of the University of Michigan Geomicrobiology Lab for bioinformatics assistance and thoughtful feedback and discussion.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2016.01546
FIGURE S1 | Emergent self-organized map (ESOM) of Geitlerinema sp. PCC 9228 genomic contigs, clustered on the basis of tetranucleotide frequency.
FIGURE S2 | Phylogenetic tree of nifHDK. The clade with Geitlerinema sp. PCC 9228 includes three other cyanobacterial genomes that also have nifI1I2. The outgroup is the homologous chlLNB gene suite from Halothece halophytica.
FIGURE S3 | Full phylogenetic tree of psbA (with non-collapsed clades). Genes are colored by groups 1–4 modeled after (Cardona et al., 2015). The outgroup is a Gloeobacter kilaueensis JS psbA that is most like psbD and is unable to oxidize water. Purple indicates group 1, red genes belong to group 2, orange genes are members of group 3, and blue standard psbAs belong to group 4. Genetic proximity to types I (green circle), II (maroon squares), and VI (blue stars) sqr genes are indicated. Fourteen psbA genes are five or fewer genes from sqr, and the largest gap is 19 genes. Geitlerinema sp. PCC 9228 has members of groups 2–4.
FIGURE S4 | Schematic of sqr operons in Geitlerinema sp. PCC 9228. (A) Canonical sqr (type I) with regulatory gene arsR collocated downstream. (B) Novel sqr (type VI) with arsR, arsenite transport gene arsB, and arsenate reductase arsC all downstream, and glutathione S-transferase and a multidrug efflux pump are upstream.
FIGURE S5 | Full phylogenetic tree of sulfide quinone reductase (sqr). Genes are colored by types I–VI modeled after (Marcia et al., 2010a). Bacterial FCSD (yellow) includes representatives of flavocytochrome c:sulfide dehydrogenase. Green sulfur bacteria have sqr belonging to types III (orange), V (light blue), IV (dark green), and VI (blue). Cyanobacterial sqr are limited to types I (light green), II (red), and VI. Proximity to psbA versions on the same contig (see Figure 3; Supplementary Figure S3) are indicated with red circles (group 2) and brown stars (group 3). No sqr genes in this analysis were genetically proximal to groups 1 or 4 psbA genes. Geitlerinema sp. PCC 9228 has types VI and I sqr.
References
Alneberg, J., Bjarnason, B. S., de Bruijn, I., Schirmer, M., Quick, J., Ijaz, U. Z., et al. (2014). Binning metagenomic contigs by coverage and composition. Nat. Methods 11, 1144–1146. doi: 10.1038/nmeth.3103
Arieli, B., Padan, E., and Shahak, Y. (1991). Sulfide-induced sulfide-quinone reductase activity in thylakoids of Oscillatoria limnetica. J. Biol. Chem. 266, 104–111.
Arieli, B., Shahak, Y., Taglicht, D., Hauska, G., and Padan, E. (1994). Purification and characterization of sulfide-quinone reductase, a novel enzyme driving anoxygenic photosynthesis in Oscillatoria limnetica. J. Biol. Chem. 269, 5705–5711.
Aro, E.-M., Virgin, I., and Andersson, B. (1993). Photoinhibition of photosystem II. inactivation, protein damage and turnover. Biochim. Biophys. Acta 1143, 113–134. doi: 10.1016/0005-2728(93)90134-2
Becraft, E. D., Wood, J. M., Rusch, D., Kühl, B. M., Jensen, S. I., Bryant, D. A., et al. (2015). The molecular dimension of microbial species: 1. ecological distinctions among, and homogeneity within, putative ecotypes of Synechococcus inhabiting the cyanobacterial mat of mushroom spring, yellowstone national park. Front. Microbiol. 6:590. doi: 10.3389/fmicb.2015.00590
Belkin, S., Arieli, B., and Padan, E. (1982). Sulfide dependent electron transport in Oscillatoria limnetica. Isr. J. Bot. 31, 199–200. doi: 10.1080/0021213X.1982.10676943
Belkin, S., and Padan, E. (1978). Sulfide-dependent hydrogen evolution in the cyanobacterium Oscillatoria limnetica. FEBS Lett. 94, 291–294. doi: 10.1016/0014-5793(78)80959-4
Blank, C. E., and Sánchez-Baracaldo, P. (2010). Timing of morphological and ecological innovations in the cyanobacteria – a key to understanding the rise in atmospheric oxygen. Geobiology 8, 1–23. doi: 10.1111/j.1472-4669.2009.00220.x
Blankenship, R. E. (2002). Molecular Mechanisms of Photosynthesis, 1st Edn. Oxford: Blackwell Science Ltd.
Blankenship, R. E. (2010). Early evolution of photosynthesis. Plant Physiol. 154, 434–438. doi: 10.1104/pp.110.161687
Boyd, E. S., Anbar, A. D., Miller, S., Hamilton, T. L., Lavin, M., and Peters, J. W. (2011). A late methanogen origin for molybdenum-dependent nitrogenase. Geobiology 9, 221–232. doi: 10.1111/j.1472-4669.2011.00278.x
Boyd, E. S., Costas, A. M. G., Hamilton, T. L., Mus, F., and Peters, J. W. (2015). Evolution of molybdenum nitrogenase during the transition from anaerobic to aerobic metabolism. J. Bacteriol. 197, 1690–1699. doi: 10.1128/JB.02611-14
Bronstein, M., Schütz, M., Hauska, G., Padan, E., and Shahak, Y. (2000). Cyanobacterial sulfide-quinone reductase: cloning and heterologous expression. J. Bacteriol. 182, 3336–3344. doi: 10.1128/JB.182.12.3336-3344.2000
Bühring, S., Sievert, S. M., Jonkers, H. M., Ertefai, T., Elshahed, M. S., Krumholz, L. R., et al. (2011). Insights into chemotaxonomic composition and carbon cycling of phototrophic communities in an artesian sulfur-rich spring (Zodletone, Oklahoma, USA), a possible analog for ancient microbial mat systems. Geobiology 9, 166–179. doi: 10.1111/j.1472-4669.2010.00268.x
Cameron, J. C., and Pakrasi, H. B. (2010). Essential role of glutathione in acclimation to environmental and redox perturbations in the cyanobacterium synechocystis sp. PCC 6803. Plant Physiol. 154, 1672–1685. doi: 10.1104/pp.110.162990
Campbell, D. A., Clarke, A. K., Gustafsson, P., and Öquist, G. (1999). Oxygen-dependent electron flow influences photosystem II function and psbA gene expression in the cyanobacterium Synechococcus sp. PCC 7942. Physiol. Plant. 105, 746–755. doi: 10.1034/j.1399-3054.1999.105420.x
Canfield, D. E. (1998). A new model for proterozoic ocean chemistry. Nature 396, 450–453. doi: 10.1038/24839
Canfield, D. E. (2005). The early history of atmospheric oxygen: homage to Robert M. Garrels. Annu. Rev. Earth Planet. Sci. 33, 1–36. doi: 10.1146/annurev.earth.33.092203.122711
Cardona, T., Murray, J. W., and Rutherford, A. W. (2015). Origin and evolution of water oxidation before the last common ancestor of the Cyanobacteria. Mol. Biol. Evol. 32, 1310–1328. doi: 10.1093/molbev/msv024
Chan, L.-K., Morgan-Kiss, R. M., and Hanson, T. E. (2009). Functional analysis of three sulfide:quinone oxidoreductase homologs in Chlorobaculum tepidum. J. Bacteriol. 191, 1026–1034. doi: 10.1128/JB.01154-08
Chaudhary, A., Haack, S. K., Duris, J. W., and Marsh, T. L. (2009). Bacterial and archaeal phylogenetic diversity of a cold sulfur-rich spring on the shoreline of Lake Erie, Michigan. Appl. Environ. Microbiol. 75, 5025–5036. doi: 10.1128/AEM.00112-09
Coates, R. C., Podell, S., Korobeynikov, A., Lapidus, A., Pevzner, P., Sherman, D. H., et al. (2014). Characterization of cyanobacterial hydrocarbon composition and distribution of biosynthetic pathways. PloS ONE 9:e85140. doi: 10.1371/journal.pone.0085140
Cohen, Y., Jørgensen, B. B., Padan, E., and Shilo, M. (1975a). Sulphide-dependent anoxygenic photosynthesis in the cyanobacterium Oscillatoria limnetica. Nature 257, 489–492. doi: 10.1038/257489a0
Cohen, Y., Jørgensen, B. B., Revsbech, N. P., and Poplawski, R. (1986). Adaptation to hydrogen sulfide of oxygenic and anoxygenic photosynthesis among cyanobacteria. Appl. Environ. Microbiol. 51, 398–407.
Cohen, Y., Krumbein, W. E., Goldberg, M., and Shilo, M. (1977a). Solar Lake (Sinai). 1. Physical and chemical limnology. Limnol. Oceanogr. 22, 597–608. doi: 10.4319/lo.1977.22.4.0597
Cohen, Y., Krumbein, W. E., and Shilo, M. (1977b). Solar Lake (Sinai). 2. Distribution of photosynthetic microorganisms and primary production. Limnol. Oceanogr. 22, 609–620. doi: 10.4319/lo.1977.22.4.0609
Cohen, Y., Padan, E., and Shilo, M. (1975b). Facultative anoxygenic photosynthesis in the cyanobacterium Oscillatoria limnetica. J. Bacteriol. 123, 855–861.
Cole, J. K., Hutchison, J. R., Renslow, R. S., Kim, Y.-M., Chrisler, W. B., Engelmann, H. E., et al. (2014). Phototrophic biofilm assembly in microbial-mat-derived unicyanobacterial consortia: model systems for the study of autotroph-heterotroph interactions. Front. Microbiol. 5:109. doi: 10.3389/fmicb.2014.00109
Crowe, S. A., Døssing, L. N., Beukes, N. J., Bau, M., Kruger, S. J., Frei, R., et al. (2013). Atmospheric oxygenation three billion years ago. Nature 501, 535–538. doi: 10.1038/nature12426
Den Uyl, P. A., Richardson, L. L., Jain, S., and Dick, G. J. (2016). Unraveling the physiological roles of the cyanobacterium Geitlerinema sp. BBD and other black band disease community members through genomic analysis of a mixed culture. PloS ONE 11:e0157953. doi: 10.1371/journal.pone.0157953
Dick, G. J., Andersson, A. F., Baker, B. J., Simmons, S. L., Thomas, B. C., Yelton, A. P., et al. (2009). Community-wide analysis of microbial genome sequence signatures. Genome Biol. 10:R85. doi: 10.1186/gb-2009-10-8-r85
Edwardson, C. F., Planer-Friedrich, B., and Hollibaugh, J. T. (2014). Transformation of monothioarsenate by haloalkaliphilic, anoxygenic photosynthetic purple sulfur bacteria. FEMS Microbiol. Ecol. 90, 858–868. doi: 10.1111/1574-6941.12440
Farquhar, J., Zerkle, A. L., and Bekker, A. (2010). Geological constraints on the origin of oxygenic photosynthesis. Photosynth. Res. 107, 11–36. doi: 10.1007/s11120-010-9594-0
Ferreira, K. N., Iverson, T. M., Maghlaoui, K., Barber, J., and Iwata, S. (2004). Architecture of the photosynthetic oxygen-evolving center. Science 303, 1831–1838. doi: 10.1126/science.1093087
Findlay, A. J., Bennett, A. J., Hanson, T. E., and Luther, G. W. (2015). Light-dependent sulfide oxidation in the anoxic zone of the chesapeake bay can be explained by small populations of phototrophic bacteria. Appl. Environ. Microbiol. 81, 7560–7569. doi: 10.1128/AEM.02062-15
Fischer, W. W., Hemp, J., and Johnson, J. E. (2015). Manganese and the evolution of photosynthesis. Orig. Life Evol. Biosph. 45, 351–357. doi: 10.1007/s11084-015-9442-5
Fischer, W. W., Hemp, J., and Valentine, J. S. (2016). How did life survive Earth’s great oxygenation? Curr. Opin. Chem. Biol. 31, 166–178. doi: 10.1016/j.cbpa.2016.03.013
Forchhammer, K. (2004). Global carbon/nitrogen control by PII signal transduction in cyanobacteria: from signals to targets. FEMS Microbiol. Rev. 28, 319–333. doi: 10.1016/j.femsre.2003.11.001
Gan, F. S. Z., Rockwell, N. C., Martin, S. S., Lagarias, J. C., and Bryant, D. A. (2014). Extensive remodeling of a cyanobacterial photosynthetic apparatus in far-red light. Science 345, 1312–1317. doi: 10.1126/science.1256963
Gregersen, L. H., Bryant, D. A., and Frigaard, N. U. (2011). Mechanisms and evolution of oxidative sulfur metabolism in green sulfur bacteria. Front. Microbiol. 2:116. doi: 10.3389/fmicb.2011.00116
Grotzinger, J. P., and Knoll, A. H. (1999). Stromatolites in precambrian carbonates: evolutionary mileposts or environmental dipsticks? Annu. Rev. Earth Planet. Sci. 27, 313–358. doi: 10.1146/annurev.earth.27.1.313
Hanson, T. E., Luther, G. W. III., and Findlay, A. J. (2013). Phototrophic sulfide oxidation: environmental insights and a method for kinetic analysis. Front. Microbiol. 4:382. doi: 10.3389/fmicb.2013.00382
Ho, M. Y., Shen, G., Canniffe, D. P., Zhao, C., and Bryant, D. A. (2016). Light-dependent chlorophyll f synthase is a highly divergent paralog of PsbA of photosystem II. Science 353. doi: 10.1126/science.aaf9178
Hoeft, S. E., Kulp, T. R., Han, S., Lanoil, B., and Oremland, R. S. (2010). Coupled arsenotrophy in a hot spring photosynthetic biofilm at Mono Lake, California. Appl. Environ. Microbiol. 76, 4633–4639. doi: 10.1128/AEM.00545-10
Jiang, J., Bauer, I., Paul, A., and Kappler, A. (2009). Arsenic redox changes by microbially and chemically formed semiquinone radicals and hydroquinones in a humic substance model quinone. Environ. Sci. Technol. 43, 3639–3645. doi: 10.1021/es803112a
Johnston, D. T., Wolfe-Simon, F., Pearson, V., and Knoll, A. H. (2009). Anoxygenic photosynthesis modulated proterozoic oxygen and sustained Earth’s middle age. Proc. Natl. Acad. Sci. U.S.A. 106, 16925–16929. doi: 10.1073/pnas.0909248106
Jørgensen, B. B., Cohen, Y., and Revsbech, N. P. (1986). Transition from anoxygenic to oxygenic photosynthesis in a microcoleus chthonoplastes cyanobacterial mat. Appl. Environ. Microbiol. 51, 408–417.
Joshi, N. A., and Fass, J. N. (2011). Sickle: A Sliding-Window, Adaptive, Quality-Based Trimming Tool for FastQ Files. Available at: https://github.com/najoshi/sickle.
Kessler, P. S., Daniel, C., and Leigh, J. A. (2001). Ammonia switch-off of nitrogen fixation in the methanogenic archaeon Methanococcus maripaludis: mechanistic features and requirement for the novel GlnB homologues, NifI(1) and NifI(2). J. Bacteriol. 183, 882–889. doi: 10.1128/JB.183.3.882-889.2001
Kiss,É, Kós, P. B., Chen, M., and Vass, I. (2012). A unique regulation of the expression of the psbA. psbD, and psbE genes, encoding the D1, D2 and cytochrome b559 subunits of the photosystem II complex in the chlorophyll d containing cyanobacterium Acaryochloris marina. Biochim. Biophys. Acta 1817, 1083–1094. doi: 10.1016/j.bbabio.2012.04.010
Klatt, J. M., Al-Najjar, M. A. A., Yilmaz, P., Lavik, G., de Beer, D., and Polerecky, L. (2015a). Anoxygenic photosynthesis controls oxygenic photosynthesis in a cyanobacterium from a sulfidic spring. Appl. Environ. Microbiol. 81, 2025–2031. doi: 10.1128/AEM.03579-14
Klatt, J. M., Haas, S., Yilmaz, P., de Beer, D., and Polerecky, L. (2015b). Hydrogen sulfide can inhibit and enhance oxygenic photosynthesis in a cyanobacterium from sulfidic springs. Environ. Microbiol. 17, 3301–3313. doi: 10.1111/1462-2920.12791
Klatt, J. M., Meyer, S., Häusler, S., Macalady, J. L., de Beer, D., and Polerecky, L. (2016). Structure and function of natural sulphide-oxidizing microbial mats under dynamic input of light and chemical energy. ISME J. 10, 921–933. doi: 10.1038/ismej.2015.167
Kós, P. B., Deák, Z., Cheregi, O., and Vass, I. (2008). Differential regulation of psbA and psbD gene expression, and the role of the different D1 protein copies in the Cyanobacterium Thermosynechococcus elongatus BP-1. Biochim. Biophys. Acta 1777, 74–83. doi: 10.1016/j.bbabio.2007.10.015
Kulp, T. R., Hoeft, S. E., Asao, M., Madigan, M. T., Hollibaugh, J. T., Fisher, J. C., et al. (2008). Arsenic(III) fuels anoxygenic photosynthesis in hot spring biofilms from mono lake, California. Science 321, 967–970. doi: 10.1126/science.1160799
Lalonde, S. V., and Konhauser, K. O. (2015). Benthic perspective on Earth’s oldest evidence for oxygenic photosynthesis. Proc. Natl. Acad. Sci. U.S.A. 112, 995–1000. doi: 10.1073/pnas.1415718112
Latifi, A., Ruiz, M., and Zhang, C.-C. (2009). Oxidative stress in cyanobacteria. FEMS Microbiol. Rev. 3, 258–278. doi: 10.1111/j.1574-6976.2008.00134.x
López-Maury, L., Florencio, F. J., and Reyes, J. C. (2003). Arsenic sensing and resistance system in the cyanobacterium Synechocystis sp. strain PCC 6803. J. Bacteriol. 185, 5363–5371. doi: 10.1128/JB.185.18.5363-5371.2003
Lyons, T. W., Reinhard, C. T., and Planavsky, N. J. (2014). The rise of oxygen in Earth’s early ocean and atmosphere. Nature 506, 307–315. doi: 10.1038/nature13068
Marcia, M., Ermler, U., Peng, G., and Michel, H. (2009). The structure of Aquifex aeolicus sulfide:quinone oxidoreductase, a basis to understand sulfide detoxification and respiration. Proc. Natl. Acad. Sci. U.S.A. 106, 9625–9630. doi: 10.1073/pnas.0904165106
Marcia, M., Ermler, U., Peng, G., and Michel, H. (2010a). A new structure-based classification of sulfide:quinone oxidoreductases. Proteins 78, 1073–1083. doi: 10.1002/prot.22665
Marcia, M., Langer, J. D., Parcej, D., Vogel, V., Peng, G., and Michel, H. (2010b). Characterizing a monotopic membrane enzyme. biochemical, enzymatic and crystallization studies on Aquifex aeolicus sulfide:quinone oxidoreductase. Biochim. Biophys. Acta 1798, 2114–2123. doi: 10.1016/j.bbamem.2010.07.033
Miller, S. R., and Bebout, B. M. (2004). Variation in sulfide tolerance of photosystem II in phylogenetically diverse cyanobacteria from sulfidic habitats. Appl. Environ. Microbiol. 70, 736–744. doi: 10.1128/AEM.70.2.736-744.2004
Minamizaki, K., Mizoguchi, T., Goto, T., Tamiaki, H., and Fujita, Y. (2008). Identification of two homologous genes, chlAI and chlAII, that are differentially involved in isocyclic ring formation of chlorophyll a in the cyanobacterium Synechocystis sp. PCC 6803. J. Biol. Chem. 283, 2684–2692. doi: 10.1074/jbc.M708954200
Mohamed, A., Eriksson, J., Osiewacz, H. D., and Jansson, C. (1993). Differential expression of the psbA genes in the cyanobacterium Synechocystis 6803. Mol. Gen. Genet. 238, 161–168. doi: 10.1007/BF00279543
Mukhopadhyay, R., Rosen, B. P., Phung, L. T., and Silver, S. (2002). Microbial arsenic: from geocycles to genes and enzymes. FEMS Microbiol. Rev. 26, 311–325. doi: 10.1111/j.1574-6976.2002.tb00617.x
Mulkidjanian, A. Y., Koonin, E. V., Makarova, K. S., Mekhedov, S. L., Sorokin, A., Wolf, Y. I., et al. (2006). The cyanobacterial genome core and the origin of photosynthesis. Proc. Natl. Acad. Sci. U.S.A. 103, 13126–13131. doi: 10.1073/pnas.0605709103
Murray, J. W. (2012). Sequence variation at the oxygen-evolving centre of photosystem II: a new class of ‘rogue’ cyanobacterial D1 proteins. Photosynth. Res. 110, 177–184. doi: 10.1007/s11120-011-9714-5
Nagarajan, N., and Pop, M. (2013). Sequence assembly demystified. Nat. Rev. Genet. 14, 157–167. doi: 10.1038/nrg3367
Nagy, C. I., Vass, I., Rákhely, G., Vass, I. Z., Tóth, A., Duzs, A., et al. (2014). Coregulated genes link sulfide:quinone oxidoreductase and arsenic metabolism in Synechocystis sp. strain PCC6803. J. Bacteriol. 196, 3430–3440. doi: 10.1128/JB.01864-14
Oden, K. L., Gladysheva, T. B., and Rosen, B. P. (1994). Arsenate reduction mediated by the plasmid-encoded ArsC protein is coupled to glutathione. Mol. Microbiol. 12, 301–306. doi: 10.1111/j.1365-2958.1994.tb01018.x
Olsen, M. T., Nowack, S., Wood, J. M., Becraft, E. D., LaButti, K., Lipzen, A., et al. (2015). The molecular dimension of microbial species: 3. comparative genomics of Synechococcus strains with different light responses and in situ diel transcription patterns of associated putative ecotypes in the mushroom spring microbial mat. Front. Microbiol. 6:604. doi: 10.3389/fmicb.2015.00604
Oremland, R. S., Saltikov, C. W., Wolfe-Simon, F., and Stolz, J. F. (2009). Arsenic in the evolution of earth and extraterrestrial ecosystems. Geomicrobiol. J. 26, 522–536. doi: 10.1080/01490450903102525
Oremland, R. S., and Stolz, J. F. (2005). Arsenic, microbes and contaminated aquifers. Trends Microbiol. 13, 45–49. doi: 10.1016/j.tim.2004.12.002
Oren, A., and Padan, E. (1978). Induction of anaerobic, photoautotrophic growth in the cyanobacterium Oscillatoria limnetica. J. Bacteriol. 133, 558–563.
Park, J.-J., Lechno-Yossef, S., Wolk, C. P., and Vieille, C. (2013). Cell-specific gene expression in anabaena variabilis grown phototrophically, mixotrophically, and heterotrophically. BMC Genomics 14:759. doi: 10.1186/1471-2164-14-759
Peng, Y., Leung, H. C. M., Yiu, S. M., and Chin, F. Y. L. (2012). IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics 28, 1420–1428. doi: 10.1093/bioinformatics/bts174
Pham, V. H., Yong, J.-J., Park, S.-J., Yoon, D.-N., Chung, W.-H., and Rhee, S.-K. (2008). Molecular analysis of the diversity of the sulfide:quinone reductase (Sqr) gene in sediment environments. Microbiology 154, 3112–3121. doi: 10.1099/mic.0.2008/018580-0
Planavsky, N. J., Asael, D., Hofmann, A., Reinhard, C. T., Lalonde, S. V., Knudsen, A., et al. (2014). Evidence for oxygenic photosynthesis half a billion years before the great oxidation event. Nat. Geosci. 7, 283–286. doi: 10.1038/ngeo2122
Raes, J., Korbel, J. O., Lercher, M. J., von Mering, C., and Bork, P. (2007). Prediction of effective genome size in metagenomic samples. Genome Biol. 8:R10. doi: 10.1186/gb-2007-8-1-r10
Rambaut, A. (2012). FigTree Version 1.4.0. Available at: http://tree.bio.ed.ac.uk/software/figtree
Raymond, J., Siefert, J. L., Staples, C. R., and Blankenship, R. E. (2004). The natural history of nitrogen fixation. Mol. Biol. Evol. 21, 541–554. doi: 10.1093/molbev/msh047
Reddy, S. M., and Evans, D. A. D. (2009). Palaeoproterozoic supercontinents and global evolution: correlations from core to atmosphere. Geol. Soc. Spec. Publications 323, 1–26. doi: 10.1144/SP323.1
Ricci, J. N., Michel, A. J., and Newman, D. K. (2015). Phylogenetic analysis of HpnP reveals the origin of 2-methylhopanoid production in alphaproteobacteria. Geobiology 13, 267–277. doi: 10.1111/gbi.12129
Sánchez-Riego, A. M., López-Maury, L., and Florencio, F. J. (2014). Genomic responses to arsenic in the cyanobacterium Synechocystis sp. PCC 6803. PloS ONE 9:e96826. doi: 10.1371/journal.pone.0096826
Satkoski, A. M., Beukes, N. J., Li, W., Beard, B. L., and Johnson, C. M. (2015). A redox-stratified ocean 3.2 billion years ago. Earth Planet. Sci. Lett. 430, 43–53. doi: 10.1016/j.epsl.2015.08.007
Schaefer, M. R., and Golden, S. S. (1989). Differential expression of members of a cyanobacterial psbA gene family in response to light. J. Bacteriol. 171, 3973–3981.
Schirmer, A., Rude, M. A., Li, X., Popova, E., and del Cardayre, S. B. (2010). Microbial biosynthesis of alkanes. Science 329, 559–562. doi: 10.1126/science.1187936
Schütz, M., Maldener, I., Griesbeck, C., and Hauska, G. (1999). Sulfide-quinone reductase from Rhodobacter capsulatus: requirement for growth, periplasmic localization, and extension of gene sequence analysis. J. Bacteriol. 181, 6516–6523.
Scott, C., Lyons, T. W., Bekker, A., Shen, Y., Poulton, S. W., Chu, X., et al. (2008). Tracing the stepwise oxygenation of the proterozoic ocean. Nature 452, 456–459. doi: 10.1038/nature06811
Sforna, M. C., Philippot, P., Somogyi, A., van Zuilen, M. A., Medjoubi, K., Schoepp-Cothenet, B., et al. (2014). Evidence for arsenic metabolism and cycling by microorganisms 2.7 billion years ago. Nat. Geosci. 7, 811–815. doi: 10.1038/ngeo2276
Shahak, Y., Arieli, B., Binder, B., and Padan, E. (1987). Sulfide-dependent photosynthetic electron flow coupled to proton translocation in thylakoids of the cyanobacterium Oscillatoria limnetica. Arch. Biochem. Biophys. 259, 605–615. doi: 10.1016/0003-9861(87)90527-3
Sicora, C., Wiklund, R., Jansson, C., and Vass, I. (2004). Charge stabilization and recombination in photosystem II containing the D1′ protein product of the psbA1 gene in Synechocystis 6803. Phys. Chem. Chem. Phys. 6, 4832–4837. doi: 10.1039/B406695B
Sicora, C. I., Appleton, S. E., Brown, C. M., Chung, J., Chandler, J., Cockshutt, A. M., et al. (2006). Cyanobacterial psbA families in Anabaena and Synechocystis encode trace, constitutive and UVB-induced D1 isoforms. Biochim. Biophys. Acta 1757, 47–56. doi: 10.1016/j.bbabio.2005.11.002
Sicora, C. I., Ho, F. M., Salminen, T., Styring, S., and Aro, E.-M. (2009). Transcription of a ‘Silent’ cyanobacterial psbA gene is induced by microaerobic conditions. Biochim. Biophys. Acta 1787, 105–112. doi: 10.1016/j.bbabio.2008.12.002
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using clustal omega. Mol. Syst. Biol. 7, 539–539. doi: 10.1038/msb.2011.75
Slyemi, D., and Bonnefoy, V. (2011). How prokaryotes deal with arsenic. Environ. Microbiol. Rep. 4, 571–586. doi: 10.1111/j.1758-2229.2011.00300.x
Sousa, F. L., Shavit-Grievink, L., Allen, J. F., and Martin, W. F. (2013). Chlorophyll biosynthesis gene evolution indicates photosystem gene duplication, not photosystem merger, at the origin of oxygenic photosynthesis. Genome Biol. Evol. 5, 200–216. doi: 10.1093/gbe/evs127
Sperling, E. A., Wolock, C. J., Morgan, A. S., Gill, B. C., Kunzmann, M., Halverson, G. P., et al. (2015). Statistical analysis of iron geochemical data suggests limited late proterozoic oxygenation. Nature 523, 451–454. doi: 10.1038/nature14589
Stal, L. J. (1995). Physiological ecology of cyanobacteria in microbial mats and other communities. New Phytol. 131, 1–32. doi: 10.1111/j.1469-8137.1995.tb03051.x
Stal, L. J. (2012). “Cyanobacterial mats and stromatolites,” in Ecology of Cyanobacteria II, ed. B. A. Whitton (Dordrecht: Springer), 65–125. doi: 10.1007/978-94-007-3855-3_4
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313. doi: 10.1093/bioinformatics/btu033
Steinhauser, D., Fernie, A. R., and Araújo, W. L. (2012). Unusual cyanobacterial TCA cycles: not broken just different. Trends Plant Sci. 17, 503–509. doi: 10.1016/j.tplants.2012.05.005
Stüeken, E. E., Buick, R., Guy, B. M., and Koehler, M. C. (2015). Isotopic evidence for biological nitrogen fixation by molybdenum-nitrogenase from 3.2 Gyr. Nature 520, 666–669. doi: 10.1038/nature14180
Sugiura, M., Kato, Y., Takahashi, R., Suzuki, H., Watanabe, T., Noguchi, T., et al. (2010). Energetics in photosystem II from Thermosynechococcus elongatus with a D1 protein encoded by either the psbA1 or psbA3 gene. Biochim. Biophys. Acta 1797, 1491–1499. doi: 10.1016/j.bbabio.2010.03.022
Sugiura, M., Ogami, S., Kusumi, M., and Un, S. (2012). Environment of TYRZ in photosystem II from Thermosynechococcus elongatus in which PsbA2 Is the D1 protein. J. Biol. Chem. 287, 13336–13347. doi: 10.1074/jbc.M112.340323
Summons, R. E., and Lincoln, S. A. (2012). “Biomarkers: informative molecules for studies in geobiology,” in Fundamentals of Geobiology, eds A. H. Knoll, D. E. Canfield, and K. O. Konhauser (Chichester: John Wiley & Sons, Ltd) doi: 10.1002/9781118280874.ch15
Sumner, D. Y., Hawes, I., Mackey, T. J., Jungblut, A. D., and Doran, P. T. (2015). Antarctic microbial mats: a modern analog for archean lacustrine oxygen oases. Geology 43, 887–890. doi: 10.1130/G36966.1
Takahashi, H., Kopriva, S., Giordano, M., Saito, K., and Hell, R. (2011). Sulfur assimilation in photosynthetic organisms: molecular functions and regulations of transporters and assimilatory enzymes. Annu. Rev. Plant Biol. 62, 157–184. doi: 10.1146/annurev-arplant-042110-103921
Tamagnini, P., Leitão, E., Oliveira, P., Ferreira, D., Pinto, F., Harris, D. J., et al. (2007). Cyanobacterial hydrogenases: diversity, regulation and applications. FEMS Microbiol. Rev. 31, 692–720. doi: 10.1111/j.1574-6976.2007.00085.x
Theissen, U., Hoffmeister, M., Grieshaber, M., and Martin, W. (2003). Single eubacterial origin of eukaryotic sulfide:quinone oxidoreductase, a mitochondrial enzyme conserved from the early evolution of eukaryotes during anoxic and sulfidic times. Mol. Biol. Evol. 20, 1564–1574. doi: 10.1093/molbev/msg174
Toepel, J., Welsh, E., Summerfield, T. C., Pakrasi, H. B., and Sherman, L. A. (2008). Differential transcriptional analysis of the cyanobacterium Cyanothece sp. strain ATCC 51142 during light-dark and continuous-light growth. J. Bacteriol. 190, 3904–3913. doi: 10.1128/JB.00206-08
van Lis, R., Nitschke, W., Duval, S., and Schoepp-Cothenet, B. (2013). Arsenics as bioenergetic substrates. Biochim. Biophys. Acta 1827, 176–188. doi: 10.1016/j.bbabio.2012.08.007
Voorhies, A. A., Biddanda, B. A., Kendall, S. T., Jain, S., Marcus, D. N., Nold, S. C., et al. (2012). Cyanobacterial life at low O2: community genomics and function reveal metabolic versatility and extremely low diversity in a great lakes sinkhole mat. Geobiology 10, 250–267. doi: 10.1111/j.1472-4669.2012.00322.x
Voorhies, A. A., Eisenlord, S. D., Marcus, D. N., Duhaime, M. B., Biddanda, B. A., Cavalcoli, J. D., et al. (2016). Ecological and genetic interactions between cyanobacteria and viruses in a low-oxygen mat community inferred through metagenomics and metatranscriptomics. Environ. Microbiol. 18, 358–371. doi: 10.1111/1462-2920.12756
Wegener, K. M., Nagarajan, A., and Pakrasi, H. B. (2015). An atypical psbA gene encodes a sentinel D1 protein to form a physiologically relevant inactive photosystem II complex in cyanobacteria. J. Biol. Chem. 290, 3764–3774. doi: 10.1074/jbc.M114.604124
Yang, J., and Cheng, Q. (2004). Origin and evolution of the light-dependent protochlorophyllide oxidoreductase (LPOR) genes. Plant Biol. 6, 537–544. doi: 10.1055/s-2004-821270
Yin, X.-X., Chen, J., Qin, J., Sun, G.-X., Rosen, B. P., and Zhu, Y.-G. (2011). Biotransformation and volatilization of arsenic by three photosynthetic cyanobacteria. Plant Physiol. 156, 1631–1638. doi: 10.1104/pp.111.178947
Keywords: cyanobacteria, anoxygenic photosynthesis, great oxidation event, nitrogenase, photosystem II D1 protein, sulfide quinone reductase
Citation: Grim SL and Dick GJ (2016) Photosynthetic Versatility in the Genome of Geitlerinema sp. PCC 9228 (Formerly Oscillatoria limnetica ‘Solar Lake’), a Model Anoxygenic Photosynthetic Cyanobacterium. Front. Microbiol. 7:1546. doi: 10.3389/fmicb.2016.01546
Received: 17 June 2016; Accepted: 15 September 2016;
Published: 13 October 2016.
Edited by:
Robert Eugene Blankenship, Washington University in St. Louis, USAReviewed by:
James Hemp, California Institute of Technology, USALuis M. Rubio, Technical University of Madrid, Spain
Copyright © 2016 Grim and Dick. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gregory J. Dick, Z2RpY2tAdW1pY2guZWR1 Sharon L. Grim, c2dyaW1AdW1pY2guZWR1
 Sharon L. Grim
Sharon L. Grim Gregory J. Dick
Gregory J. Dick