- 1Division of Bacterial Diseases, State Key Laboratory of Veterinary Biotechnology, Harbin Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Harbin, China
- 2Shanghai Veterinary Research Institute, Chinese Academy of Agricultural Sciences, Shanghai, China
- 3Section of Paediatrics, Department of Medicine, Imperial College London, London, UK
Antimicrobial peptides are essential to the innate immune defense of the mammal against bacterial infection. However, pathogenic bacteria have evolved multiple strategies to resist and evade antimicrobial peptides, which is vital to bacterial survival and colonization in hosts. PR-39 is a linear porcine antimicrobial peptide containing 39 amino acid residues with a high proline content. Resistance to antimicrobial peptide PR-39 has been observed in Actinobacillus pleuropneumoniae. However, little is known about the factors required for this resistance. In the present study, PR-39 exposure increased the expression of the sapA gene in A. pleuropneumoniae. The sapA gene, which encodes a putative peptide transport periplasmic protein, was deleted from this bacterium. The ΔsapA mutant showed increased sensitivity to PR-39 compared to the wild-type MD12 and complemented PΔsapA strains. However, the ΔsapA mutant did not exhibit any alterations in outer membrane integrity. Scanning electron microscopy showed that the ΔsapA mutant displayed morphological defects, as indicated by a deformed and sunken shape after PR-39 treatment. In addition, disruption of the SapA protein led to reduced colonization and attenuated virulence of A. pleuropneumoniae in the BALB/c mouse model. Collectively, these data suggest that SapA acts as one mechanism for A. pleuropneumoniae to counteract PR-39-mediated killing. To the best of our knowledge, this is the first study to show a mechanism underlying antimicrobial peptide resistance in A. pleuropneumoniae.
Introduction
Actinobacillus pleuropneumoniae is a Gram-negative bacterial pathogen responsible for porcine pleuropneumonia, which is a highly contagious respiratory disease that causes major economic losses to the swine industry worldwide (Chiers et al., 2010; Bossé et al., 2017). This pathogen mainly causes damage to respiratory tissue, leading to hemorrhagic, fibrinous and necrotic lung lesions (Bossé et al., 2002; Li et al., 2016). The ability of A. pleuropneumoniae to adhere to, colonize, and invade its host, and host factors such as innate and adaptive immune responses are crucial to the outcome of this disease (Chiers et al., 2010).
Antimicrobial peptides, also known as host defense peptides, are important components of innate immunity as a first line of defense against bacterial infection (Band and Weiss, 2015). Relative to other mammals, the pig has the most diverse set of cathelicidins (Wessely-Szponder et al., 2010). According to their primary amino acid structures, porcine cathelicidins divide into three subgroups: linear proline-rich cathelicidins (including PR-39, Prophenin 1 and 2), disulfide-rich Protegrins 1–5, and α-helix-rich porcine myeloid antimicrobial peptides (PMAP)-23, PMAP-36, and PMAP-37 (Sang and Blecha, 2009). The proline-rich antimicrobial peptide PR-39 contains 39 amino acid residues with high contents of proline (49%) and arginine (26%) (Zhang et al., 2000). PR-39 manifests antibacterial activity against a variety of Gram-negative bacteria and some Gram-positive bacteria, including multidrug-resistant clinical isolates (Linde et al., 2001). Like other proline-rich peptides, PR-39 kills bacteria without inducing lysis through pore-forming mechanisms. Instead, it translocates across the membrane and disrupts DNA and protein synthesis (Pranting et al., 2008). In addition to its antibacterial activity, PR-39 exerts other important functions, including immunomodulation, wound repair, and the prevention of inflammation during tissue injury (Shi et al., 1996; Veldhuizen et al., 2014).
PR-39 is prominent in tissue of the upper and lower respiratory tract of healthy pigs, and of pigs infected with A. pleuropneumoniae, and plays a pivotal role in the innate immune defense of the pig against A. pleuropneumoniae infections (Hennig-Pauka et al., 2012). The concentration of PR-39 has been shown to be significantly elevated in bronchoalveolar lavage fluid (BALF) of pigs chronically infected with A. pleuropneumoniae (Hennig-Pauka et al., 2006). However, the minimal inhibitory concentration (MIC) of PR-39 for A. pleuropneumoniae was 5-fold higher than that of Escherichia coli, suggesting the resistance of A. pleuropneumoniae to PR-39 (Hennig-Pauka et al., 2006). This resistance observed in vitro is consistent with the ability of the pathogen to persist in pig respiratory tissue for long periods. The cause of this resistance is not yet clear. A. pleuropneumoniae may have already evolved several strategies to control or evade killing by PR-39 in vivo, and the ability to adapt to PR-39 exposure is vital to the pathogenicity of A. pleuropneumoniae.
One of the important strategies for bacterial evasion of antimicrobial peptides involves the aid of transporter systems (Band and Weiss, 2015). The Sap transporter system is important for resistance to antimicrobial peptides in several Gram-negative pathogens, including Haemophilus ducreyi, non-typeable Haemophilus influenzae, and Salmonella enterica serovar Typhimurium (Parra-Lopez et al., 1993; Mason et al., 2005; Mount et al., 2010). In general, the Sap transporter consists of five proteins: SapA is a periplasmic solute binding protein, SapB and SapC are permease subunits of the transporter, and SapD and SapF function as ATPase proteins for providing energy to translocate the substrate(s) across the bacterial inner membrane (Parra-Lopez et al., 1993; Mount et al., 2010).
Analysis of the A. pleuropneumoniae serovar 5 strain L20 genome sequence (NC_009053.1) reveals the presence of a predicted intact sap operon. The nucleotide sequence of sapA of A. pleuropneumoniae L20 is similar to that of the sapA genes of H. ducreyi strain 35000HP and non-typeable H. influenzae strain 86-028NP, with 68.1 and 53.6% identity, respectively. The amino acid sequence of SapA exhibited 71.1 and 44.4% sequence identity with the SapA protein of H. ducreyi strain 35000HP and non-typeable H. influenzae strain 86-028NP, respectively. The SapA protein of A. pleuropneumoniae has been shown to be expressed in vivo during the chronic stage of the disease and is responsible for persistence of A. pleuropneumoniae (Baltes et al., 2007). In addition, the sapF gene has been reported to be up-regulated in BALF (Lone et al., 2009). Interestingly, PR-39 translocates across the cellular membrane of bacteria via some sort of transport system (Li et al., 2014). It is therefore here hypothesized that the Sap transporter is involved in the resistance of A. pleuropneumoniae to PR-39. To test this hypothesis, a nonpolar, unmarked deletion mutation in the sapA gene was constructed to investigate the role of SapA protein in PR-39 resistance of A. pleuropneumoniae. This is the first demonstration of the mechanism underlying antimicrobial peptide resistance in A. pleuropneumoniae and may improve comprehension of the role of the SapA protein in the persistence and pathogenicity of A. pleuropneumoniae.
Materials and Methods
Ethics Statement
Animal experiments were approved by Animal Ethics Committee of Harbin Veterinary Research Institute of the Chinese Academy of Agricultural Sciences (CAAS) and carried out in strict accordance with the recommendations of the Animal Ethics Procedures and Guidelines of the People's Republic of China. All efforts were made to minimize animal suffering.
Bacterial Strains and Growth Conditions
The bacterial strains and plasmids used for this study are described in Table 1. The A. pleuropneumoniae strains were cultured in a brain heart infusion (BHI, Difco Laboratories, Detroit, MI, USA) medium supplemented with 10 μg/ml nicotinamide adenine dinucleotide (NAD) (Sigma-Aldrich, U.S.). For culture of A. pleuropneumoniae transconjugants (single crossovers), BHI medium was supplemented with 10 μg/ml of NAD and 7 μg/ml of chloramphenicol. E. coli ATCC 25922 strain and S. enterica ATCC 51741 strain (American Type Culture Collection, ATCC) were cultured in a Luria-Bertani (LB, Difco Laboratories, Detroit, MI, USA) medium. E. coli β2155 was grown in LB medium supplemented with 1 mM diaminopimelic acid (DAP) (Sigma-Aldrich, U.S.). All strains were routinely grown at 37°C.
Minimum Inhibitory Concentration (MIC) and Minimal Bactericidal Concentration (MBC) Analysis
A microdilution broth method was performed to determine the minimal inhibitory concentration (MIC) of antimicrobial peptide PR-39 according to the broth micro dilution guideline of the Clinical and Laboratory Standards Institute (CLSI, 2013). PR-39 was purchased from AnaSpec (San Jose, CA). Standardized bacterial suspensions of log-phase cultures of E. coli ATCC 25922, S. enterica ATCC 51741, A. pleuropneumoniae strains ATCC 27090, S-8 and MD12 were prepared and diluted to a concentration of 1 × 106 CFU/ml. MIC determinations were performed using commercially sterile 96-well microtiter plates (Costar 3599, U.S.A.). The MIC value was determined as the lowest concentration of PR-39 that prevented visible growth. Then 20 μl of each bacteria-peptide suspension in the 96-well microtiter plates was plated onto LB or BHI agar plates and incubated for 20 h at 37°C. The MBC value was determined as the lowest concentration of PR-39 that showing no visible growth on the plates (Hu et al., 2016).
In vitro Growth Assays
The A. pleuropneumoniae wild-type strain MD12 was grown in 5 ml of BHI medium for 15 h, and then diluted to an optical density at 600 nm (OD600) of 0.1. Fresh cultures in 5 ml of BHI medium were supplemented with PR-39 (concentration range 0–0.2 μM) and incubated while shaking at 37°C. Growth was monitored by measuring the OD600 values at an interval of 1 h using the Eppendorf BioPhotometer (Eppendorf, Germany).
RNA Isolation and qRT-PCR
For RNA isolation, A. pleuropneumoniae MD12 strain was grown to mid-logarithmic phase in 3 ml of BHI medium supplemented with PR-39 (concentration range 0–0.2 μM). The cultures were harvested by centrifugation at 10,000 g at 4°C. Total RNA was extracted using RNeasy kit (Qiagen) and cDNA was synthesized using the PrimeScript RT reagent kit (TaKaRa, Japan) according to the manufacturer's instructions. The primers used for analysis of sapA expression are listed in Table S1. The cDNA samples were amplified using SYBR Green I (TakaRa). Quantitative real-time polymerase chain reactions (qRT-PCR) were performed in a MicroAmp Optical 96-well reaction plate using a Stratagene Mx3000P system (Agilent Technologies, Germany). Amplification efficiency was evaluated using a standard curve generated by qRT-PCR using the cDNA dilution series with three replicates. The stability of the six housekeeping genes recF, glyA, rho, tpiA, pykA (Nielsen and Boye, 2005) and syp (Lone et al., 2009) was examined using the program geNorm (Vandesompele et al., 2002). The geometric mean of the best-scoring reference genes glyA, tpiA, and syp was used to normalize the target gene expression levels. The qRT-PCR experiments were performed in triplicate with three independent biological replicates. Relative expression levels were analyzed by a threshold cycle (ΔΔCt) method to calculate the fold change in gene expression (Pfaffl, 2001).
Construction of Gene Deletion Mutant
The primers used for the construction of the deletion mutant ΔsapA are listed in Table S1. Primers AUF/AUR, and ADF/ADR were used to amplify the two segments flanking the sapA gene. Using single-overlap extension PCR (SOE PCR), the fragment with a 570 bp internal in-frame deletion in the sapA gene (from nt 24 to 593) was generated, and cloned into the conjugative vector pEMOC2 (Baltes et al., 2003) to produce the plasmid pEMΔsapA. Using E. coli β2155 and a single-step transconjugation system (Dehio and Meyer, 1997; Oswald et al., 1999), plasmid pEMΔsapA was used to introduce the sapA mutation into the wild-type strain MD12. After two homologous recombination steps, the A. pleuropneumoniae ΔsapA mutant was verified by sequencing and PCR analyses using AJDF/AJDR primers.
Complementation of the A. pleuropneumoniae ΔsapA Mutant
The 2,471 bp PCR product including the entire sapA open reading frame (ORF) and 675 bp of the upstream region containing the native promoter was amplified with the primers AHBF/AHBR (Table S1). The PCR reaction was performed under the following conditions: 95°C for 3 min, 30 cycles with 94°C for 30 s, 52 °C for 30 s and 72°C for 2 min, the final extension at 72°C for 8 min. The PCR product was digested with SalI/SacI and ligated to SalI/SacI-digested pGZRS-19 plasmid (West et al., 1995), yielding plasmid pGZRS-sapA. The recombined plasmid pGZRS-sapA was confirmed by DNA sequencing (Comate Bioscience Co., Ltd.) and electroporated into the ΔsapA mutant for trans complementation. The electroporation conditions were set to 2,500 V, 200 Ω, and 25μF. Transformants were selected on BHI agar containing 20 μg/ml of ampicillin. The complemented mutant strain, verified by colony PCR and DNA sequencing, was designated PΔsapA.
Bactericidal Assays
Bactericidal assays were performed as described previously (Mason et al., 2005). The A. pleuropneumoniae strains MD12, ΔsapA, and PΔsapA were grown in BHI medium to OD600 0.8. Cells of each strain from the broth cultures were harvested and diluted in PBS (pH 7.4) to a concentration of 106 CFU/ml. The wells of a sterile, polystyrene 96-well microtiter plate (Costar 3599, U.S.A.) were filled with 90 μl of PBS. PR-39 was serially diluted in the wells and each well retained 90 μl of the appropriate concentration (0.5–4 μM) of PR-39. Ten microliters of the bacterial suspension were added to each well, and the plate was incubated for 0.5–3 h at 37°C. Bacteria incubated with PBS served as controls. Serial dilutions of the bacteria were plated on BHI agar. The bactericidal effect was expressed as the percentage of surviving cells, using the bacterial counts obtained with bacteria incubated in PBS as 100%.
SDS-EDTA Sensitivity Assay
SDS-EDTA sensitivity assay was performed as described previously (Carpenter et al., 2014). A. pleuropneumoniae strains MD12, ΔsapA, and PΔsapA were incubated in BHI medium at 37°C with shaking at 180 rpm to OD600 1.0. Each strain was serially diluted with PBS, and 2 μl of these dilutions were spotted in triplicate onto fresh BHI agar plates containing 0.1% SDS and 0.5 mM EDTA. All the plates were incubated overnight at 37°C.
NPN Uptake Assay
The 1-N-phenylnaphthylamine (NPN) uptake assay was performed as described previously (Martinez De Tejada and Moriyon, 1993). A. pleuropneumoniae strains MD12, ΔsapA, and PΔsapA were grown to OD600 0.6 and harvested by centrifugation at 2,500 g for 15 min. Pellets were washed three times and resuspended in 5 mM HEPES buffer (pH 7.2) containing 10 μM NPN (Sigma–Aldrich, USA). NPN uptake into the A. pleuropneumoniae ΔvacJ mutant was used as a positive control, as vacJ encodes VacJ lipoprotein and the membrane permeability of this mutant was increased as described previously (Xie et al., 2016). Fluorescence was measured using the EnVision Multilabel Reader (PerkinElmer, UK), with emission at 420 nm and excitation at 350 nm.
Scanning Electron Microscopy
The A. pleuropneumoniae strains MD12, ΔsapA and PΔsapA were cultivated in BHI medium at 37°C to mid-logarithmic growth phase. Cells of each strain from the broth cultures (106 CFU/ml) were incubated with 4 μM PR-39 for 1 h, and harvested by centrifugation. The cells were washed three times with PBS, and fixed overnight using 2.5% glutaraldehyde at 4°C. Dehydration was performed in upgraded ethanol (washed once with each of 50, 70, 85, 95%, and three times with 100%). Then the samples were dried using a critical point drying method and sputter-coated with gold. The cell morphology of all samples was visualized using a scanning electron microscope (JSM-7500F, JEOL, Japan).
Mouse In vivo Experiments
The BALB/c mouse model has been acknowledged as an appropriate one to assess A. pleuropneumoniae infection (Chiang et al., 2009; Seo et al., 2013). Specific-pathogen-free, 6-week-old female BALB/c mice (Beijing Vital River Laboratory Animal Co., Ltd.) were purchased from the VitalRiver Laboratories (VRL, Beijing, China). A. pleuropneumoniae strains MD12 and ΔsapA were cultured in BHI medium at 37°C, and harvested during the mid-exponential phase and washed three times with sterile PBS. A total of 50 mice were randomly divided into 5 groups (n = 10/group). Group 1 and group 2 were respectively inoculated intraperitoneally with 100 μl of PBS containing 108 and 107 CFU of MD12. Group 3 and group 4 were respectively inoculated intraperitoneally with 100 μl of PBS containing 108 and 107 CFU of ΔsapA. Non-infected mice in the control group were inoculated with 100 μl of sterile PBS (pH 7.4). The health status and the weight of the mice were monitored twice daily for a 14-day period and humane endpoints used to determine if the mice met criteria to be euthanized (Nemzek et al., 2004). These criteria included weight loss >10–15%, lethargy, inability to stand, anorexia or flocked together for more than 6 h. Mice meeting criteria were euthanized by cervical dislocation under isoflurane anesthesia.
Enumeration of Bacterial Load in Organs
A total of 10 specific-pathogen-free, 6 week-old female BALB/c mice were randomly divided into 2 groups (n = 5), and each group was intraperitoneally administered with 5.0 × 106 CFU of the MD12 strain or the ΔsapA mutant. Three days post-infection, mice from each group were humanely euthanized and the organs of lung, liver, and kidney were removed aseptically. Samples were weighed, and homogenized using a tissue homogenizer (100 mg weight/ml of PBS). Viable counts in serial dilutions of homogenates were determined following culture on BHI agar plates for 24 h at 37°C.
Statistical Analysis
Statistical analyses were performed using GraphPad Prism version 5.01 (GraphPad Software Inc., U.S.A.). The data are expressed as the means ± standard deviation. The statistical analysis of the data was performed using one-way ANOVA, two-way ANOVA, or the Student's t-test. P-values less than 0.05 were considered statistically significant.
Results
Antibacterial Activity of PR-39
To explore the antibacterial activity of PR-39, the MICs of PR-39 for A. pleuropneumoniae strains ATCC 27090, S-8, MD12, E. coli ATCC 25922 and S. enterica ATCC 51741 were measured (Table S2). For E. coli and S. enterica, the MICs of PR-39 ranged from 0.5 to 1 μM, the MBCs of PR-39 were 1 μM. However, PR-39 had higher MICs for A. pleuropneumoniae strains than E. coli and S. enterica, ranging from 4 to 8 μM. Similarly, the MBCs of PR-39 for A. pleuropneumoniae strains were 8 μM, much higher than those of E. coli and S. enterica. These results showed that A. pleuropneumoniae exhibited a certain resistance to PR-39 compared to E. coli and S. enterica.
A. pleuropneumoniae Exposure to PR-39 Upregulated Expression of the sapA Gene
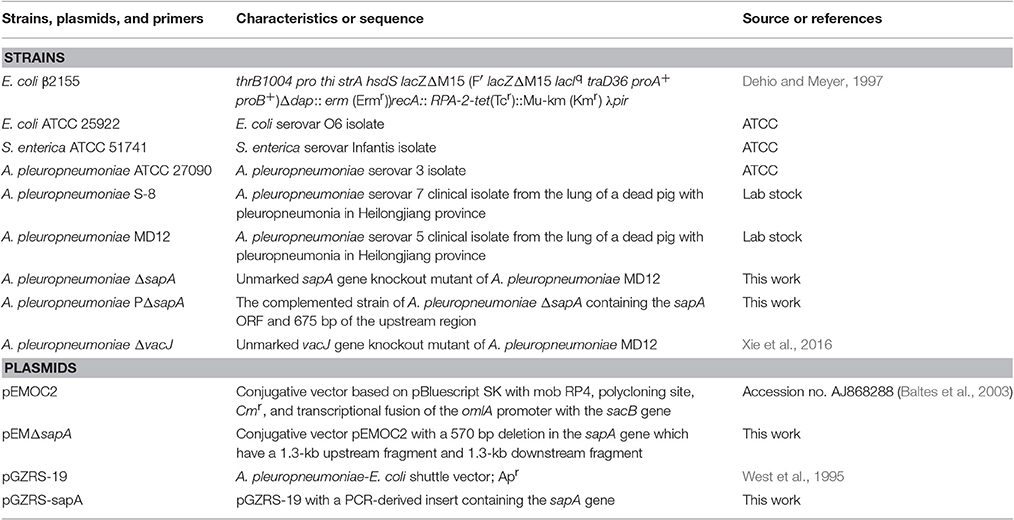
The expression of the sapA gene was analyzed using qRT-PCR in A. pleuropneumoniae MD12 when exposed to PR-39. Three housekeeping genes glyA, tpiA, and syp were selected to normalize sapA gene expression levels, and PCR efficiency for each gene was not less than 1.92. When the MD12 strain was exposed to sublethal concentrations of PR39, growth curves were similar to that of untreated bacteria (Figure 1A), but the transcription levels of sapA were higher than that of untreated bacteria (Figure 1B). In the presence of increasing concentrations of PR-39, the expression of sapA was upregulated in a dose- dependent manner, suggesting sapA may contribute to a resistance mechanism in A. pleuropneumoniae MD12 against PR-39.
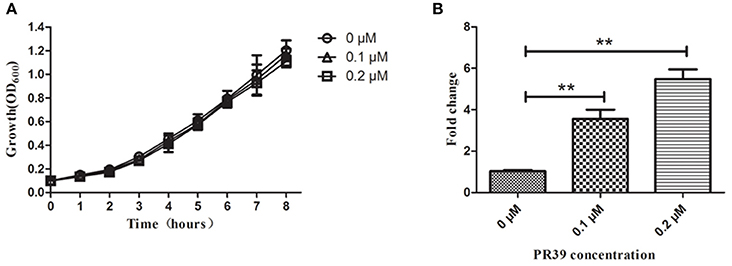
Figure 1. The expression of sapA gene of A. pleuropneumoniae upon exposure to PR-39 by qRT-PCR. (A) A. pleuropneumoniae strain MD12 was grown in the presence of increasing, yet sublethal concentrations of PR-39. (B) Transcriptional alteration of sapA was examined by quantitative analysis of mRNA expression levels in MD12 when exposed to increasing, yet sublethal concentrations of PR-39. Values represent three independent assays in triplicate ± SD, **p < 0.01.
Construction of A. pleuropneumoniae ΔsapA Mutant and Its Complemented Strain
Analysis of the A. pleuropneumoniae L20 genome sequence revealed the presence of an intact sap operon (Figure 2A). This operon consists of four genes: sapA (APL_RS04170), sapB (APL_RS04165), sapC (APL_RS04160), sapD (APL_RS04155). However, sapF (APL_RS06520) is not linked to the sapABCD locus.
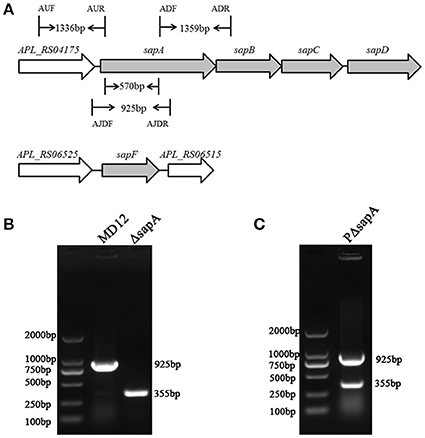
Figure 2. Chromosomal inactivation of A. pleuropneumoniae sapA gene. (A) Schematic representation of the A. pleuropneumoniae sap operon locus. The sapA gene was inactivated using double-crossover homologous recombination. Binding locations for the primers AUF/AUR and ADF/ADR used to amplify the two flanking regions (1,336 and 1,359 bp, respectively) of the sapA gene are shown in the schematic, and primers AJDF/AJDR were here used to identify the sapA-deleted mutant (355 bp) and wild-type MD12 strain (925 bp). (B) PCR identification of the ΔsapA mutant using the primers AJDF/AJDR. (C) PCR identification of the complemented strain PΔsapA using the primers AJDF/AJDR.
To investigate the function of the SapA protein, an in-frame-deletion mutant of sapA in A. pleuropneumoniae was constructed using double-crossover homologous recombination and confirmed by PCR and DNA sequencing (Figures 2A,B, Supplementary Materials). PCR with primers AJDF/AJDR was used to amplify the 925 bp amplicon from the wild-type MD12 strain, and the 355 bp amplicon from the sapA deletion mutant ΔsapA (Figure 2B). The ΔsapA mutant contains a 570 bp in-frame deletion in the sapA gene. The results of qRT-PCR showed that the transcription levels of the downstream genes sapB, sapC, and sapD were unaffected, confirming that the mutation in ΔsapA was nonpolar (Figure S1). The complemented mutant strain PΔsapA was generated using the plasmid pGZRS-sapA, with transformants selected on plates containing ampicillin, and confirmed by PCR (Figure 2C).
Mutation in sapA Enhanced Sensitivity of A. pleuropneumoniae to PR-39
To determine whether the SapA protein has a role in the survival of A. pleuropneumoniae upon exposure to the antimicrobial peptide PR-39, the MD12, ΔsapA, and PΔsapA strains were tested in a bactericidal assay. Cells of each strain were incubated with specific concentrations of PR-39 for 3 h. The results showed that the mutant devoid of sapA was significantly more sensitive to PR-39 over a concentration range of 0.5–4 μM than was the isogenic wild type strain (Figure 3A). In addition, following the elongation of treatment time, the sensitivity of ΔsapA to PR-39 was increased (Figure 3B). Trans-complementation with sapA expressed on pGZRS-19 partially restored resistance to PR-39. These findings indicate that the A. pleuropneumoniae SapA protein is required for the bacterium's resistance to the PR-39.
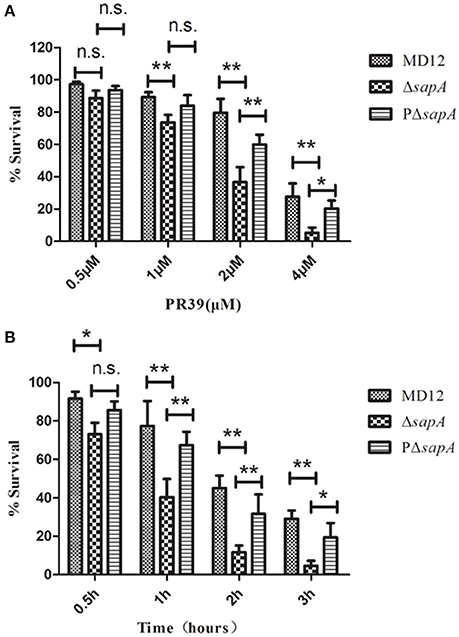
Figure 3. Sensitivity of the sapA mutant to PR-39. MD12, ΔsapA, and PΔsapA strains were incubated with (A) increasing concentrations of PR-39 for 3 h; and (B) 4 μM of PR-39 for increasing periods. Each sample was plated on BHI agar for identification of viable bacteria. Values represent three independent assays in triplicate ± SD, n.s. = not significant, *p < 0.05, **p < 0.01.
To exclude the possibility of impaired outer membrane integrity due to the deletion of the sapA gene, the sensitivity of the MD12, ΔsapA, and PΔsapA strains to SDS-EDTA was analyzed. As shown in Figure S2A, all these strains did not exhibit sensitivity to SDS-EDTA. In addition, the outer membrane integrity of each strain was further evaluated using the fluorescent probe NPN, which exhibits fluorescence weakly in aqueous but strongly in hydrophobic environments (Lee et al., 2015). In Figure S2B, no significant difference in uptake of NPN was observed between MD12, ΔsapA, and PΔsapA, while NPN fluorescence was significantly higher in the ΔvacJ mutant whose membrane permeability was increased as described previously (Xie et al., 2016). These data indicated that the sapA gene deletion did not cause alterations in the outer membrane integrity in A. pleuropneumoniae.
Morphology of A. pleuropneumoniae ΔsapA upon Exposure to PR-39
To further confirm the increased sensitivity of the ΔsapA strain to PR-39, the morphology of the MD12, ΔsapA, and PΔsapA strains treated with PR-39 was assessed using scanning electron microscopy. After PR-39 treatment, a significant morphological variation was observed among the MD12, ΔsapA, and PΔsapA strains (Figure 4). MD12 displayed a smooth surface, which is typical of this A. pleuropneumoniae strain (Figure 4). However, when exposed to PR39, cells of the ΔsapA mutant had an irregular and crinkled appearance and a sunken shape compared to that of the wild-type MD12 and complemented PΔsapA strains (Figure 4). These data indicated that the A. pleuropneumoniae SapA protein functions, at least to some extent, to protect this pathogen from the lethal effects of PR-39.
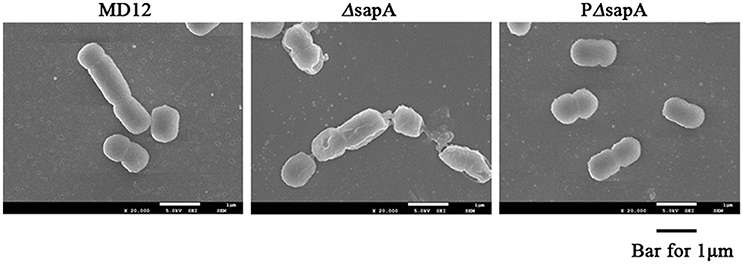
Figure 4. Scanning electron microscopy of A. pleuropneumoniae strains treated with PR-39. MD12, ΔsapA, and PΔsapA in the mid- logarithmic phase were harvested and treated with 4 μM PR-39 for 30 min. Scale bar, 100 nm. The cells of ΔsapA displayed a relatively deformed cell shape compared to that of the wild-type MD12 strain and the complemented strain PΔsapA.
Loss of sapA Attenuates the Virulence of A. pleuropneumoniae in the BALB/c Mouse Model
To address whether sapA deletion affected the virulence of A. pleuropneumoniae, BALB/c mice were inoculated intraperitoneally with wild type strain MD12 and the ΔsapA mutant at various doses. The MD12 strain gave rise to a higher mortality rate than ΔsapA (Figure 5), which suggested that the deletion of sapA attenuates the virulence of A. pleuropneumoniae.
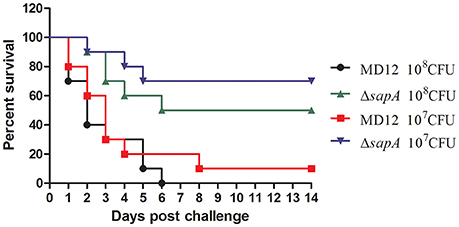
Figure 5. Survival of mice following intraperitoneally challenge with A. pleuropneumoniae strains. Percentage of surviving mice after challenged with 1 × 108 or 1 × 107 CFU of MD12 or ΔsapA strain.
The capacity of the MD12 and ΔsapA strains to colonize mice was then tested. The A. pleuropneumoniae load in tissues of systemically infected mice was determined by culturing the lungs, livers, and kidneys homogenates 3 days post-infection. As shown in Figure 6, the viable counts in lung were significantly decreased in the ΔsapA mutant-infected mice compared with the WT-infected mice (P < 0.01). Similarly, significant differences (P < 0.05) in bacterial loads were also found between the MD12-inoculated and ΔsapA-inoculated mice in livers and kidneys (Figure 6). Taken together, the results showed that the ΔsapA mutant of A. pleuropneumoniae displayed a reduced ability to colonize BALB/c mice.
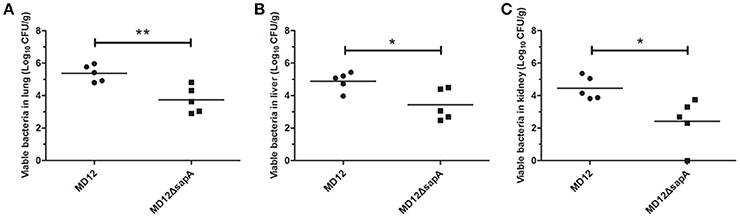
Figure 6. Bacterial loads in organs from BALB/c mice infected with A. pleuropneumoniae. Mice were infected with the WT MD12 or ΔsapA strain, and bacterial loads in (A) lung, (B) liver, (C) kidney were examined 3 days post infection. The data shown are the means of bacterial colonies from five mice, and error bars indicate standard deviations. Data presented are the mean values and standard deviations of 5 biological replicates, *p < 0.05, **p < 0.01.
Discussion
Antimicrobial peptides are an essential part of innate immune defenses that inhibit pathogen infection and contribute to clearance of bacterial colonization (Band and Weiss, 2015). Upon encountering invasive pathogens, hosts can generate the specific innate immune signaling events to induce production of specific antimicrobial peptides in response to the invasion of pathogens (Plichta et al., 2012). However, to adapt to the environments of elevated antimicrobial peptides, bacteria have evolved multiple countermeasures to resist and evade antimicrobial peptide-mediated killing (Band and Weiss, 2015). Resistance to porcine antimicrobial peptides is vital to survival and colonization of A. pleuropneumoniae in host environments (Hennig-Pauka et al., 2006). However, little is known about the factors required for this resistance. This study demonstrated that the putative peptide transport periplasmic protein SapA of A. pleuropneumoniae is involved in resistance to PR-39-mediated killing. This is the first study to show an antimicrobial peptide resistance mechanism in A. pleuropneumoniae.
PR-39 has been shown to be essential to the innate immune defense of the pig against A. pleuropneumoniae infection (Hennig-Pauka et al., 2012). In this study, the MICs of E. coli and A. pleuropneumoniae isolates were measured in the laboratory, and an MIC of 1 μM was determined for E. coli, but 4–8 μM for A. pleuropneumoniae strains. This finding is in accordance with the previous report by Hennig-Pauka et al. suggesting innate resistance of A. pleuropneumoniae to PR-39 (Hennig-Pauka et al., 2006). This resistance contributes to the promotion of A. pleuropneumoniae survival and colonization in the host for extended periods of time. Most notably, the mode of action of PR-39 killing bacteria does not involve the formation of pores, but translocation across the membrane via some sort of transport system and the targeting of intracellular molecules (Li et al., 2014). Thus, we hypothesized that certain peptide transport systems would be required for resistance of A. pleuropneumoniae to PR-39. Analysis of the A. pleuropneumoniae L20 genome sequence revealed the presence of an intact sap operon (Figure 2A). This operon consists of four genes, but does not contain the sapF gene: sapA (APL_RS04170), which encodes a putative periplasmic binding protein; sapB (APL_RS04165) and sapC (APL_RS04160), which encode putative permease components; and sapD (APL_RS04155) which encodes the ATPase components. The unlinked sapF (APL_RS06520) is predicted to encode the ATPase component of this transporter. The peptide transport periplasmic protein SapA, which has been found to be expressed in vivo during the chronic stage of A. pleuropneumoniae infection (Baltes et al., 2007), has drawn considerable attention. Additionally, the sapF gene has been reported to be up-regulated in A. pleuropneumoniae when grown in pig BALF (Lone et al., 2009). The results of the present study showed that inactivation of the sapA gene significantly enhanced sensitivity of A. pleuropneumoniae to PR-39, indicating that the SapA protein is required for A. pleuropneumoniae resistance to the PR-39 (Figures 3, 4).
Until now, several transporter systems have been shown to mediate resistance against antimicrobial peptides. Mutation in the yejF gene of S. enterica from the yejABEF operon encoding an ATP-binding cassette (ABC) peptide import system, reduced resistance to polymyxin B, human defensin (HBD)-1 and HBD-2 (Eswarappa et al., 2008). Additionally, in the pathogens Neisseria meningitidis and H. ducreyi, a periplasmic membrane fusion protein MtrC actively transports cathelicidin LL-37 out of the bacterial membrane to promote resistance to LL-37 (Tzeng et al., 2005; Rinker et al., 2011). Furthermore, the Sap transporter was also demonstrated to contribute to antimicrobial peptide resistance in other Gram-negative species. In non-typeable H. influenzae, mutation in the sapA gene led to reduced resistance to the chinchilla beta defensin 1 (cBD-1) (Mason et al., 2005). The Sap transporter in H. ducreyi confers resistance to LL-37, but not to α-or β-defensins (Mount et al., 2010). In Vibrio fischeri, SapA did not confer resistance to any of the eight tested antimicrobial peptides but was implicated in in vitro growth and in vivo colonization (Lupp et al., 2002). The present study showed that the SapA protein rendered A. pleuropneumoniae resistant to the porcine antimicrobial peptide PR-39 (Figures 3, 4). Taken together, these studies suggested that the SapA protein served multiple functions to satisfy the requirements of different bacterial species over the course of infection.
In vivo colonization by A. pleuropneumoniae is a complicated process, during which the evasion from the host innate immune plays an important role. Another goal of this study was to clarify whether SapA is essential for A. pleuropneumoniae colonization and pathogenicity in a mouse model. The data presented in this study showed that the ΔsapA mutant displayed an attenuated virulence and reduced bacterial colonization, compared with wild type strain (Figures 5, 6). These results may be explained by the decreased ability of the ΔsapA mutant to resist PR-39 mediated killing in vivo. This is highly consistent with the data presented by a previous study which showed that the SapA protein was required for colonization and virulence of non-typeable H. influenzae in a chinchilla model (Mason et al., 2005). It is clear from these data that the sapA gene product is involved in colonization and virulence in A. pleuropneumoniae infection.
The lung is the major target of A. pleuropneumoniae infection (Bossé et al., 2002; Chiers et al., 2010). PR-39 expression has been reported to be increased in BALF and epithelial lining fluid (ELF) after A. pleuropneumoniae infection of pigs (Hennig-Pauka et al., 2006). The concentration of PR-39 in BALF ranged from 0.4 to 75.9 nM and concentrations in ELF are approximately 6- to 40-fold higher than in BALF in infected pigs, which are lower than the MICs of A. pleuropneumoniae strains. At sub-inhibitory concentrations, PR-39 in combination with other antimicrobial factors in the lower respiratory tract may result in a synergistic antimicrobial effect against A. pleuropneumoniae. The sub-inhibitory concentrations of PR-39 might also let A. pleuropneumoniae adapt to the host environment and is in accordance with the in vivo observation that A. pleuropneumoniae persists in respiratory epithelium of pigs for extended periods. In addition to its antimicrobial properties, PR-39 is also involved in many other biological processes, such as chemotaxis of neutrophils, promotion of wound healing, and inhibition of apoptosis (Shi et al., 1996; Veldhuizen et al., 2014). In A. pleuropneumoniae chronic infection, the involvement of PR-39 in these biological processes might be its primary role.
In conclusion, this study demonstrates that the SapA protein in A. pleuropneumoniae promotes resistance to antimicrobial peptide PR-39, and it is the first mechanism of antimicrobial peptide resistance identified in A. pleuropneumoniae. In addition, disruption of the SapA protein led to reduced colonization and attenuated virulence of A. pleuropneumoniae in the BALB/c mouse model. This has shed light on the role of SapA protein in the pathogenicity of A. pleuropneumoniae. Of note, though the sapA deletion increased sensitivity of A. pleuropneumoniae to PR-39, the MIC of ΔsapA was still more than that of E. coli (data not shown), indicating the presence of other PR39 resistance mechanisms in A. pleuropneumoniae. Future studies should include attempts to determine whether other proteins, such as secreted proteases or two-component systems, participate in the resistance to PR39, and unravel the mechanism of detection and signal transduction that takes place when A. pleuropneumoniae encounters PR-39.
Author Contributions
FX and CW designed the experiments, FX and GL conducted experiments, YW, ShL, and NC performed the experiments, FX and YW analyzed the data and drafted the manuscript, CW, PL, and SiL finalized the manuscript. All authors read and approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This research was supported by grants from Special Fund for Agro-scientific Research in the Public Interest (201303034), Natural Science Foundation of Heilongjiang Province of China (C2016067 and QC2016044), the project of Harbin Science and Technology innovative talents (2015RQQYJ073), and the State's Key Project of Research and Development Plan (2016YFD0500700). We thank Dr. Gerald-F. Gerlach (Institute for Microbiology, Department of Infectious Diseases, University of Veterinary Medicine Hannover, Germany) for the generous donation of E. coli β2155 strain and vector pEMOC2. PL was supported by the United Kingdom Biotechnology and Biological Sciences Research Council (BB/K020765/1).
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.00811/full#supplementary-material
Figure S1. Transcriptional levels of downstream genes of sapA in MD12 and ΔsapA strains. Transcriptional levels of sapB, sapC, sapD genes were examined by qRT-PCR. Values represent two independent assays in triplicate ±SD.
Figure S2. The outer membrane integrity of A. pleuropneumoniae. (A) SDS-EDTA sensitivity assay. Cultures were grown until mid-log phase, and 2 μl of each dilution, indicated on the left, was spotted in triplicate onto BHI agar plate supplemented with 0.1% SDS and 0.5 mM EDTA. (B) NPN uptake assay. Changes in fluorescence following the addition of the hydrophobic fluorescent probe NPN for the MD12, ΔsapA, PΔsap, and ΔvacJ strains are shown. Values represent two independent assays in triplicate ±SD, n.s. = not significant, **p < 0.01.
Table S1. Primers used in this study.
Table S2. Minimum inhibitory concentrations and minimal bactericidal concentration of PR-39 for E. coli, S. enterica, and A. pleuropneumoniae strains.
References
Baltes, N., Buettner, F. F., and Gerlach, G. F. (2007). Selective capture of transcribed sequences (SCOTS) of Actinobacillus pleuropneumoniae in the chronic stage of disease reveals an HlyX-regulated autotransporter protein. Vet. Microbiol. 123, 110–121. doi: 10.1016/j.vetmic.2007.03.026
Baltes, N., Tonpitak, W., Hennig-Pauka, I., Gruber, A. D., and Gerlach, G. F. (2003). Actinobacillus pleuropneumoniae serotype 7 siderophore receptor FhuA is not required for virulence. FEMS Microbiol. Lett. 220, 41–48. doi: 10.1016/S0378-1097(03)00064-8
Band, V. I., and Weiss, D. S. (2015). Mechanisms of antimicrobial peptide resistance in Gram-negative bacteria. Antibiotics (Basel) 4, 18–41. doi: 10.3390/antibiotics4010018
Bossé, J. T., Janson, H., Sheehan, B. J., Beddek, A. J., Rycroft, A. N., Kroll, J. S., et al. (2002). Actinobacillus pleuropneumoniae: pathobiology and pathogenesis of infection. Microbes Infect. 4, 225–235. doi: 10.1016/S1286-4579(01)01534-9
Bossé, J. T., Li, Y., Rogers, J., Fernandez Crespo, R., Li, Y., Chaudhuri, R. R., et al. (2017). Whole Genome Sequencing for Surveillance of Antimicrobial Resistance in Actinobacillus pleuropneumoniae. Front. Microbiol. 8:311. doi: 10.3389/fmicb.2017.00311
Carpenter, C. D., Cooley, B. J., Needham, B. D., Fisher, C. R., Trent, M. S., Gordon, V., et al. (2014). The Vps/VacJ ABC transporter is required for intercellular spread of Shigella flexneri. Infect. Immun. 82, 660–669. doi: 10.1128/IAI.01057-13
Chiang, C. H., Huang, W. F., Huang, L. P., Lin, S. F., and Yang, W. J. (2009). Immunogenicity and protective efficacy of ApxIA and ApxIIA DNA vaccine against Actinobacillus pleuropneumoniae lethal challenge in murine model. Vaccine 27, 4565–4570. doi: 10.1016/j.vaccine.2009.05.058
Chiers, K., De Waele, T., Pasmans, F., Ducatelle, R., and Haesebrouck, F. (2010). Virulence factors of Actinobacillus pleuropneumoniae involved in colonization, persistence and induction of lesions in its porcine host. Vet. Res. 41, 65. doi: 10.1051/vetres/2010037
CLSI (2013). Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria Isolated from Animals. Wayne, PA: Clinical and Laboratory Standards Institute.
Dehio, C., and Meyer, M. (1997). Maintenance of broad-host-range incompatibility group P and group Q plasmids and transposition of Tn5 in Bartonella henselae following conjugal plasmid transfer from Escherichia coli. J. Bacteriol. 179, 538–540.
Eswarappa, S. M., Panguluri, K. K., Hensel, M., and Chakravortty, D. (2008). The yejABEF operon of Salmonella confers resistance to antimicrobial peptides and contributes to its virulence. Microbiology 154, 666–678. doi: 10.1099/mic.0.2007/011114-0
Hennig-Pauka, I., Jacobsen, I., Blecha, F., Waldmann, K. H., and Gerlach, G. F. (2006). Differential proteomic analysis reveals increased cathelicidin expression in porcine bronchoalveolar lavage fluid after an Actinobacillus pleuropneumoniae infection. Vet. Res. 37, 75–87. doi: 10.1051/vetres:2005043
Hennig-Pauka, I., Koch, R., Hoeltig, D., Gerlach, G. F., Waldmann, K. H., Blecha, F., et al. (2012). PR-39, a porcine host defence peptide, is prominent in mucosa and lymphatic tissue of the respiratory tract in healthy pigs and pigs infected with Actinobacillus pleuropneumoniae. BMC Res. Notes 5:539. doi: 10.1186/1756-0500-5-539
Hu, F., Wu, Q., Song, S., She, R., Zhao, Y., Yang, Y., et al. (2016). Antimicrobial activity and safety evaluation of peptides isolated from the hemoglobin of chickens. BMC Microbiol. 16:287. doi: 10.1186/s12866-016-0904-3
Lee, E., Kim, J. K., Jeon, D., Jeong, K. W., Shin, A., and Kim, Y. (2015). Functional roles of aromatic residues and helices of papiliocin in its antimicrobial and anti-inflammatory activities. Sci. Rep. 5:12048. doi: 10.1038/srep12048
Li, W., Tailhades, J., O'brien-Simpson, N. M., Separovic, F., Otvos, L. Jr., Hossain, M. A., et al. (2014). Proline-rich antimicrobial peptides: potential therapeutics against antibiotic-resistant bacteria. Amino Acids 46, 2287–2294. doi: 10.1007/s00726-014-1820-1
Li, Y., Cao, S., Zhang, L., Lau, G. W., Wen, Y., Wu, R., et al. (2016). A TolC-Like protein of Actinobacillus pleuropneumoniae is involved in antibiotic resistance and biofilm formation. Front. Microbiol. 7:1618. doi: 10.3389/fmicb.2016.01618
Linde, C. M., Hoffner, S. E., Refai, E., and Andersson, M. (2001). In vitro activity of PR-39, a proline-arginine-rich peptide, against susceptible and multi-drug-resistant Mycobacterium tuberculosis. J. Antimicrob. Chemother. 47, 575–580. doi: 10.1093/jac/47.5.575
Lone, A. G., Deslandes, V., Nash, J. H., Jacques, M., and Macinnes, J. I. (2009). Modulation of gene expression in Actinobacillus pleuropneumoniae exposed to bronchoalveolar fluid. PLoS ONE 4:e6139. doi: 10.1371/journal.pone.0006139
Lupp, C., Hancock, R. E., and Ruby, E. G. (2002). The Vibrio fischeri sapABCDF locus is required for normal growth, both in culture and in symbiosis. Arch. Microbiol. 179, 57–65. doi: 10.1007/s00203-002-0502-7
Martinez de Tejada, G., and Moriyon, I. (1993). The outer membranes of Brucella spp. are not barriers to hydrophobic permeants. J. Bacteriol. 175, 5273–5275.
Mason, K. M., Munson, R. S. Jr., and Bakaletz, L. O. (2005). A mutation in the sap operon attenuates survival of nontypeable Haemophilus influenzae in a chinchilla model of otitis media. Infect. Immun. 73, 599–608. doi: 10.1128/IAI.73.1.599-608.2005
Mount, K. L., Townsend, C. A., Rinker, S. D., Gu, X., Fortney, K. R., Zwickl, B. W., et al. (2010). Haemophilus ducreyi SapA contributes to cathelicidin resistance and virulence in humans. Infect. Immun. 78, 1176–1184. doi: 10.1128/IAI.01014-09
Nemzek, J. A., Xiao, H. Y., Minard, A. E., Bolgos, G. L., and Remick, D. G. (2004). Humane endpoints in shock research. Shock 21, 17–25. doi: 10.1097/01.shk.0000101667.49265.fd
Nielsen, K. K., and Boye, M. (2005). Real-time quantitative reverse transcription-PCR analysis of expression stability of Actinobacillus pleuropneumoniae housekeeping genes during in vitro growth under iron-depleted conditions. Appl. Environ. Microbiol. 71, 2949–2954. doi: 10.1128/AEM.71.6.2949-2954.2005
Oswald, W., Tonpitak, W., Ohrt, G., and Gerlach, G. (1999). A single-step transconjugation system for the introduction of unmarked deletions into Actinobacillus pleuropneumoniae serotype 7 using a sucrose sensitivity marker. FEMS Microbiol. Lett. 179, 153–160.
Parra-Lopez, C., Baer, M. T., and Groisman, E. A. (1993). Molecular genetic analysis of a locus required for resistance to antimicrobial peptides in Salmonella typhimurium. EMBO J. 12, 4053–4062.
Pfaffl, M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45. doi: 10.1093/nar/29.9.e45
Plichta, J. K., Nienhouse, V., and Radek, K. A. (2012). Integrating “omics” technologies to conceptualize dynamic antimicrobial peptide responses. Front. Immunol. 3:284. doi: 10.3389/fimmu.2012.00284
Pranting, M., Negrea, A., Rhen, M., and Andersson, D. I. (2008). Mechanism and fitness costs of PR-39 resistance in Salmonella enterica serovar Typhimurium LT2. Antimicrob. Agents. Chemother. 52, 2734–2741. doi: 10.1128/AAC.00205-08
Rinker, S. D., Trombley, M. P., Gu, X., Fortney, K. R., and Bauer, M. E. (2011). Deletion of mtrC in Haemophilus ducreyi increases sensitivity to human antimicrobial peptides and activates the CpxRA regulon. Infect. Immun. 79, 2324–2334. doi: 10.1128/IAI.01316-10
Sang, Y., and Blecha, F. (2009). Porcine host defense peptides: expanding repertoire and functions. Dev. Comp. Immunol. 33, 334–343. doi: 10.1016/j.dci.2008.05.006
Seo, K. W., Kim, S. H., Park, J., Son, Y., Yoo, H. S., Lee, K. Y., et al. (2013). Nasal immunization with major epitope-containing ApxIIA toxin fragment induces protective immunity against challenge infection with Actinobacillus pleuropneumoniae in a murine model. Vet. Immunol. Immunopathol. 151, 102–112. doi: 10.1016/j.vetimm.2012.10.011
Shi, J., Ross, C. R., Leto, T. L., and Blecha, F. (1996). PR-39, a proline-rich antibacterial peptide that inhibits phagocyte NADPH oxidase activity by binding to Src homology 3 domains of p47 phox. Proc. Natl. Acad. Sci. U.S.A. 93, 6014–6018.
Tzeng, Y. L., Ambrose, K. D., Zughaier, S., Zhou, X., Miller, Y. K., Shafer, W. M., et al. (2005). Cationic antimicrobial peptide resistance in Neisseria meningitidis. J. Bacteriol. 187, 5387–5396. doi: 10.1128/JB.187.15.5387-5396.2005
Vandesompele, J., De Preter, K., Pattyn, F., Poppe, B., Van Roy, N., De Paepe, A., et al. (2002). Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, RESEARCH0034. doi: 10.1186/gb-2002-3-7-research0034
Veldhuizen, E. J., Schneider, V. A., Agustiandari, H., Van Dijk, A., Tjeerdsma-Van Bokhoven, J. L., Bikker, F. J., et al. (2014). Antimicrobial and immunomodulatory activities of PR-39 derived peptides. PLoS ONE 9:e95939. doi: 10.1371/journal.pone.0095939
Wessely-Szponder, J., Majer-Dziedzic, B., and Smolira, A. (2010). Analysis of antimicrobial peptides from porcine neutrophils. J. Microbiol. Methods 83, 8–12. doi: 10.1016/j.mimet.2010.07.010
West, S. E., Romero, M. J., Regassa, L. B., Zielinski, N. A., and Welch, R. A. (1995). Construction of Actinobacillus pleuropneumoniae-Escherichia coli shuttle vectors: expression of antibiotic-resistance genes. Gene 160, 81–86.
Xie, F., Li, G., Zhang, W., Zhang, Y., Zhou, L., Liu, S., et al. (2016). Outer membrane lipoprotein VacJ is required for the membrane integrity, serum resistance and biofilm formation of Actinobacillus pleuropneumoniae. Vet. Microbiol. 183, 1–8. doi: 10.1016/j.vetmic.2015.11.021
Keywords: Actinobacillus pleuropneumoniae, SapA, antimicrobial peptide resistance, PR-39, Virulence
Citation: Xie F, Wang Y, Li G, Liu S, Cui N, Liu S, Langford PR and Wang C (2017) The SapA Protein Is Involved in Resistance to Antimicrobial Peptide PR-39 and Virulence of Actinobacillus pleuropneumoniae. Front. Microbiol. 8:811. doi: 10.3389/fmicb.2017.00811
Received: 09 February 2017; Accepted: 20 April 2017;
Published: 10 May 2017.
Edited by:
Yuji Morita, Aichi Gakuin University, JapanReviewed by:
Margaret E. Bauer, Indiana University School of Medicine, USAKevin Mason, Ohio State University at Columbus, USA
Isabel Hennig-Pauka, Veterinärmedizinische Universität Wien, Austria
Copyright © 2017 Xie, Wang, Li, Liu, Cui, Liu, Langford and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunlai Wang, Y2h1bmxhaS53QGh2cmkuYWMuY24=
 Fang Xie1
Fang Xie1 Paul R. Langford
Paul R. Langford Chunlai Wang
Chunlai Wang