- 1Antibiotics, Biocides, Residues and Resistance Unit, Fougères Laboratory, ANSES, Fougères, France
- 2Micalis Institute, INRA, AgroParisTech, Université Paris-Saclay, Jouy-en-Josas, France
- 3MaIAGE, INRA, Université Paris-Saclay, Jouy-en-Josas, France
Biofilms are dynamic habitats which constantly evolve in response to environmental fluctuations and thereby constitute remarkable survival strategies for microorganisms. The modulation of biofilm functional properties is largely governed by the active remodeling of their three-dimensional structure and involves an arsenal of microbial self-produced components and interconnected mechanisms. The production of matrix components, the spatial reorganization of ecological interactions, the generation of physiological heterogeneity, the regulation of motility, the production of actives enzymes are for instance some of the processes enabling such spatial organization plasticity. In this contribution, we discussed the foundations of architectural plasticity as an adaptive driver of biofilms through the review of the different microbial strategies involved. Moreover, the possibility to harness such characteristics to sculpt biofilm structure as an attractive approach to control their functional properties, whether beneficial or deleterious, is also discussed.
Introduction
The traditional perception of microbes as unicellular life forms has deeply changed over the last decades with the collection of scientific evidences showing that microorganisms predominantly live in dense and complex communities known as biofilms. Biofilms are classically defined as aggregates of cells adhering to a surface or interface and often embedded in an extracellular matrix of polymeric substances. They constitute one of the most successful mode of life on Earth (Flemming et al., 2016). They are consequently found in natural, industrial, medical, household environments and, from the human point of view, they can be either beneficial or detrimental. Indeed, microbial biofilms are involved in essential nutrient cycling or biotechnological processes as well as in severe chronic infections and biodeterioration phenomenon (for instance Beech and Sunner, 2004; Bjarnsholt, 2013; Berlanga and Guerrero, 2016). Positive or negative impacts directly result from the ability of microorganisms to express specific functions in these complex communities compared to the single planktonic state. The higher resistance of biofilm cells to antimicrobials compared to that of their planktonic counterparts is a telling example of such specific functional properties and should be relied to the structural characteristics of the community (Bridier et al., 2011). Indeed, both the microbial growth and the production of matrix lead to the rise of a biological edifice offering progressively a protective structure to inhabitants able to hinder penetration and action of antimicrobials. The development of three-dimensional biofilm structure also generates physicochemical gradients and physiological heterogeneity with slow growth resistant phenotypes for instance (Stewart and Franklin, 2008). Recently, Berleman et al. (2016) demonstrated the central role of multicellular bacterial community structure in the colonization of surface by Myxococcus xanthus. Indeed, the authors showed that extracellular polymeric substances (EPS) synthesis led to the creation of microchannels which govern both bacterial motility and cell-to-cell interactions and finally organize multicellular behavior during swarm migration. In contrast, a mutant lacking EPS showed a deficiency of cell orientation and poor colony migration. As biofilms are mostly complex associations of strains and/or species in our environments, spatial arrangement of genotypes within biofilms also governs strain interactions and the evolution of social phenotypes as immediate neighbors in the structure are more affected by the social behaviors (Nadell et al., 2016). Spatial organization of genotypes and social interactions will thus govern the whole community architecture and functions (Liu et al., 2016). Functional properties of a biofilm therefore emerge from the construction and shaping of the microbial structure like many of the emergent properties of natural communities relying on the creation of biogenic structures by habitat-forming organisms (Flemming et al., 2016).
The close relationships between the architecture of a biofilm and its functional properties emphasizes the need to better describe and understand cell behavior, from single cell to multicellular scale, during biofilm structure development and maturation. Recent technological advances in methodologies including imaging and microscopy, molecular techniques, and physico-chemical assays, enabled the development of novel approaches dedicated to biofilm studies (Azeredo et al., 2017). The possibility to observe biofilm using high resolution and non-destructive methods now allows investigating the dynamics of multicellular structure development and the fate of each of its individual cellular components in parallel. For instance, the key architectural transitions and associated biophysical and genetic mechanisms supporting the developmental program of Vibrio cholerae biofilms have been recently disclosed using single-cell live imaging (Drescher et al., 2016; Yan et al., 2016). This kind of observations has clearly improved our understanding of spatio-temporal development of biofilms and has finally increasingly supported the intimate connection between structural modulations and the emergence of functional features and survival strategies. Indeed, the ability of biofilms to adapt their structure in response to internal or external stimuli, called hereafter the architectural plasticity, appears as a key factor affecting the fitness of individuals within the whole microbial community. Interestingly, the role of plasticity in bacterial survival was already demonstrated at the cellular scale. Bacteria are able to alter their morphology and to produce specific morphotypes conferring survival advantages in hostile environments. This was showed for numbers of bacterial pathogens for which filamentation is essential in the resistance to phagocytosis and overall for persistence during infection (Justice et al., 2008; Justice et al., 2014).
In this review, we will discuss the central role of architectural plasticity in the emergence of functional properties of biofilms and as a communal bacterial response to many harsh conditions and external attacks. We will also deal with the various mechanisms developed by microorganisms to build and modify the three-dimensional community and, with the existing strategies for humans to sculpt biofilm architecture in order to control their function.
Biofilm Architecture Plasticity as a Collective Response to Environmental Fluctuations
The starting point of the development of the three-dimensional biofilm structure corresponds to the transition from planktonic state to sessile mode of life which occurs in response to diverse environmental cues and cell-to-cell signaling molecules. The translation of perceived signals to specific genetic expression and finally to a series of dramatic metabolic and phenotypic changes involves complex regulatory networks and diverse molecules including the second messenger cyclic-di-GMP (c-di-GMP) in number of bacterial species (Kostakioti et al., 2013; Romling et al., 2013). A correlation between high intracellular levels of c-di-GMP and biofilm formation has indeed been shown for a variety of species and various biofilm determinants including flagella rotation, exopolysaccharide production, surface adhesin expression, secondary metabolite production, antimicrobial resistance and other stress responses (Valentini and Filloux, 2016). In addition, the quorum sensing (QS), which is a cell-to-cell signaling system making bacteria able to communicate with each other via the production and detection of signaling molecules, enable the regulation of communal behaviors (Srivastava and Waters, 2012). The interconnection between QS and c-di-GMP pathways enables bacteria to act collectively through coordinated response to cellular signals or environmental conditions. It was showed for instance in V. Cholerae that QS and c-di-GMP pathways are strongly intertwined at many levels and that their integration play a key role in the control of the expression of vpsT, a transcriptional activator that induces biofilms formation (Srivastava et al., 2011). Similarly, Ueda and Wood (2009) demonstrated in Pseudomonas aeruginosa that the transcription of the tpbA gene encoding a tyrosine phosphatase involved in synthesis of polysaccharides and biofilm formation, is under the direct control of QS while this enzyme is also involved in the regulation of intracellular c-di-GMP concentrations. Such observation clearly highlights the convergence of the two signaling processes and the connection between the environment, cell populations and finally biofilm formation.
Using this sensor system, bacteria are able to coordinate their activities during the different steps of biofilm development leading to complex three-dimensional structures. Recurrent developmental stages can be schematically defined in bacteria ranging from initial adhesion to irreversible attachment, formation of microcolonies, macrocolonies development and maturation of architecture and then dispersion (Monds and O’Toole, 2009). Nevertheless, the development program and its dynamic are actually very specific and largely depend on nutrient conditions, pH, temperature, hydrodynamics conditions, species involved, etc… Fundamentally, the shaping of specific biofilm architecture reflects the impact of local growth conditions (Toyofuku et al., 2015). Numerous studies in various bacterial species reported the impact of temperature, hydrodynamics or nutrient concentration on biofilm structure suggesting an adaptation of biofilm shaping to optimally fit to growth conditions (Stoodley et al., 1998; Yang et al., 2006; Abdallah et al., 2015). The changes of biofilm structure alter the diffusibility of substances and enables metabolic adaptation under various conditions by optimizing nutrient and waste product exchange for instance (Toyofuku et al., 2015).
This is illustrated in Figure 1 where confocal images of biofilms with various architectures were used as an input of a modeling pipeline, which simulates diffusion of a chemical molecule through biofilm and thus reflects its diffusive capabilities. The diffusion coefficient maps obtained suggested that biofilm architecture is a determinant driver of the chemical compound density map at steady state, presenting a diversity of situations, from quasi-uniform distributions to strong gradients.
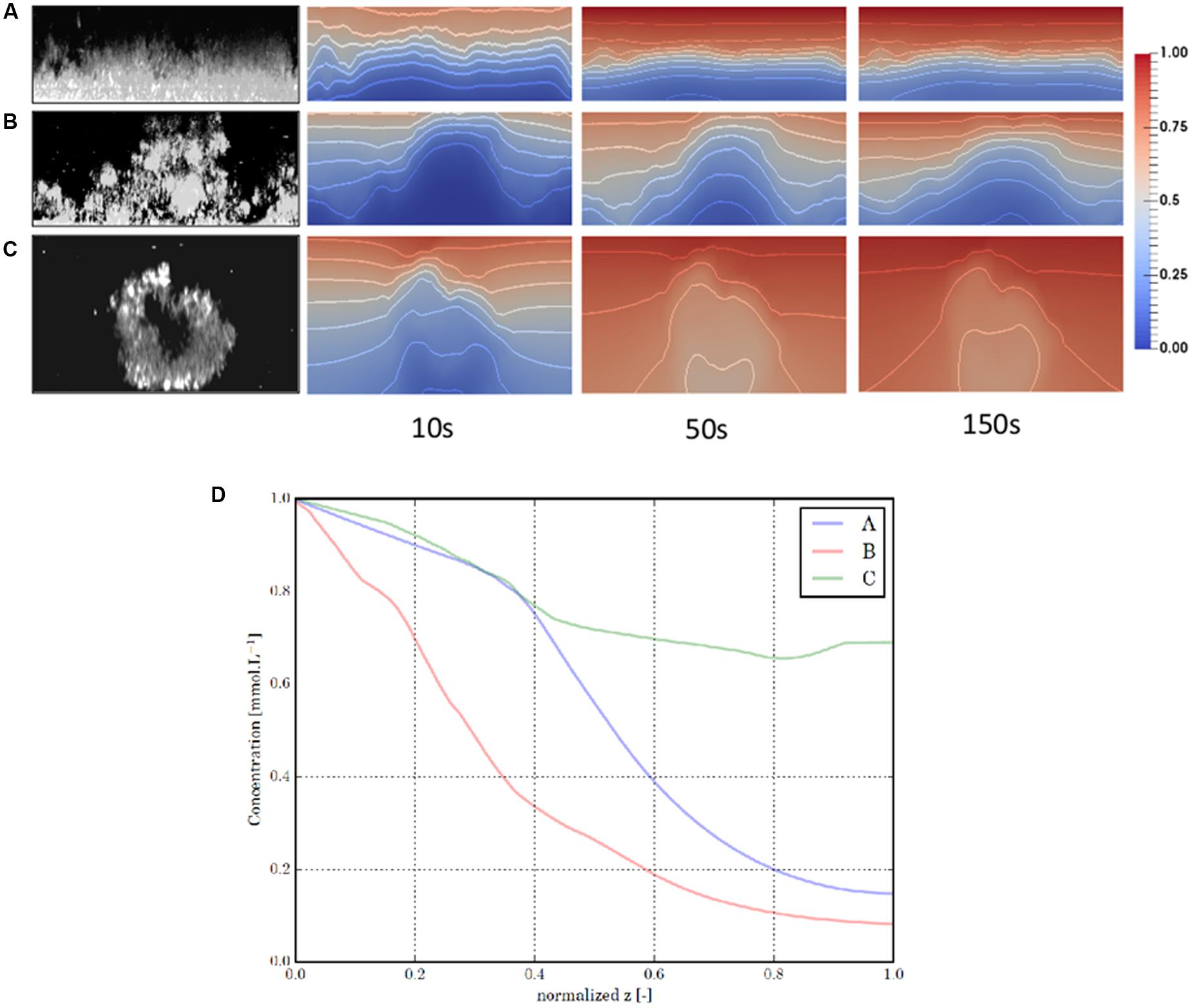
FIGURE 1. Modeling of diffusion in biofilm of various architecture. CLSM sections of three characteristic biofilm structures were displayed in the first column: (A) a flat (Escherichia coli), (B) a mushroom-like (Pseudomonas putida and Pseudomonas aeruginosa) and (C) a egg-like structure (Salmonella enterica). Those images are used as an input of a modeling pipeline which simulates the diffusion of a chemical component through the biofilm, from a bulk source located in the upper boundary of the image. Based on the biofilm images, we construct for each structure a heterogeneous diffusion coefficient map that reflects the diffusive capabilities of the biofilm: the higher the local bacterial density, the lower the local diffusion coefficient. Next, this tensor is inserted in a reaction-diffusion equation together with a reaction function that mimics the consumption of the component by the bacteria. The consumption rate also varies with the local bacterial density. We display three snapshots of the simulated component distribution, at time t = 10, 50, and 150 s when the steady-state is reached. Isolines are displayed every 0.1 to better represent the distribution gradients. We finally display a cut in z of the component distribution in each biofilm at steady-state (D). The cut plane of a given biofilm crosses its point of minimal component concentration at steady-state. To facilitate the comparison, we normalized the z-coordinates of the different graphs. We can see that the biofilm structure is a determinant driver of the component density map at steady-state, presenting a diversity of situation, from quasi-uniform distributions (structure C) to strong gradients (structures A and B).
Accordingly, it is clear that structural adjustments of biofilm clearly lead to both the modulation of phenotypic heterogeneity and the way each bacterium perceive its local microenvironment. This architectural plasticity provides thereby an efficient way to adapt to various stresses for microorganisms. Many demonstrations of this phenomenon occur in our environments as for instance, stream biofilms in rivers, which dynamically adapt and evolve in response to the streambed environment and flow intermittency through modifications of their physical structure, species composition and through spatial re-organization (Battin et al., 2016; Sabater et al., 2016). The intimate relation between architectural differentiation and community composition suggests that this micro-scale process is an important driver of the biofilm adaptation to the fluctuations of stream conditions, especially to compensate hydrodynamic perturbations and changes in quantity and quality of nutrients (Besemer et al., 2009).
Another concrete illustration of adaptation through biofilm structure modulation is the stimulation of biofilm production in different bacterial species exposed to antimicrobials, metals and a large range of other molecules (Hoffman et al., 2005; Perrin et al., 2009; Shemesh et al., 2010; Marchal et al., 2011; Chen et al., 2015). In many cases, the presence of subinhibitory concentration of such toxic molecules induces the sur-expression of genes coding for matrix components that finally lead to an increase of biofilm production and a modification of its three-dimensional structure (Shemesh et al., 2010). In line with this, it was showed in Thiomonas sp. that arsenic exposure lead to an increase of EPS production and cell death within microcolonies creating hollow voids structure that is subsequently followed by active dispersal of cells (Marchal et al., 2011). Authors suggested that the survival and persistence of Thiomonas sp. under selective pressure of arsenic exposure relied on its ability to rapidly develop biofilm followed by the dispersal of a more resistant population.
Architectural plasticity of biofilms thus gives the opportunity to bacteria to constantly reorganize their direct microenvironments to face adverse conditions and to better harness surrounding resources (Table 1). Structural adaptations can occur through various active processes which mostly involve a differential expression of genes or a genetic plasticity in response to conditions changes. The diverse mechanisms, directly or indirectly involved in the shaping of biofilm architecture, are discussed in the next section.
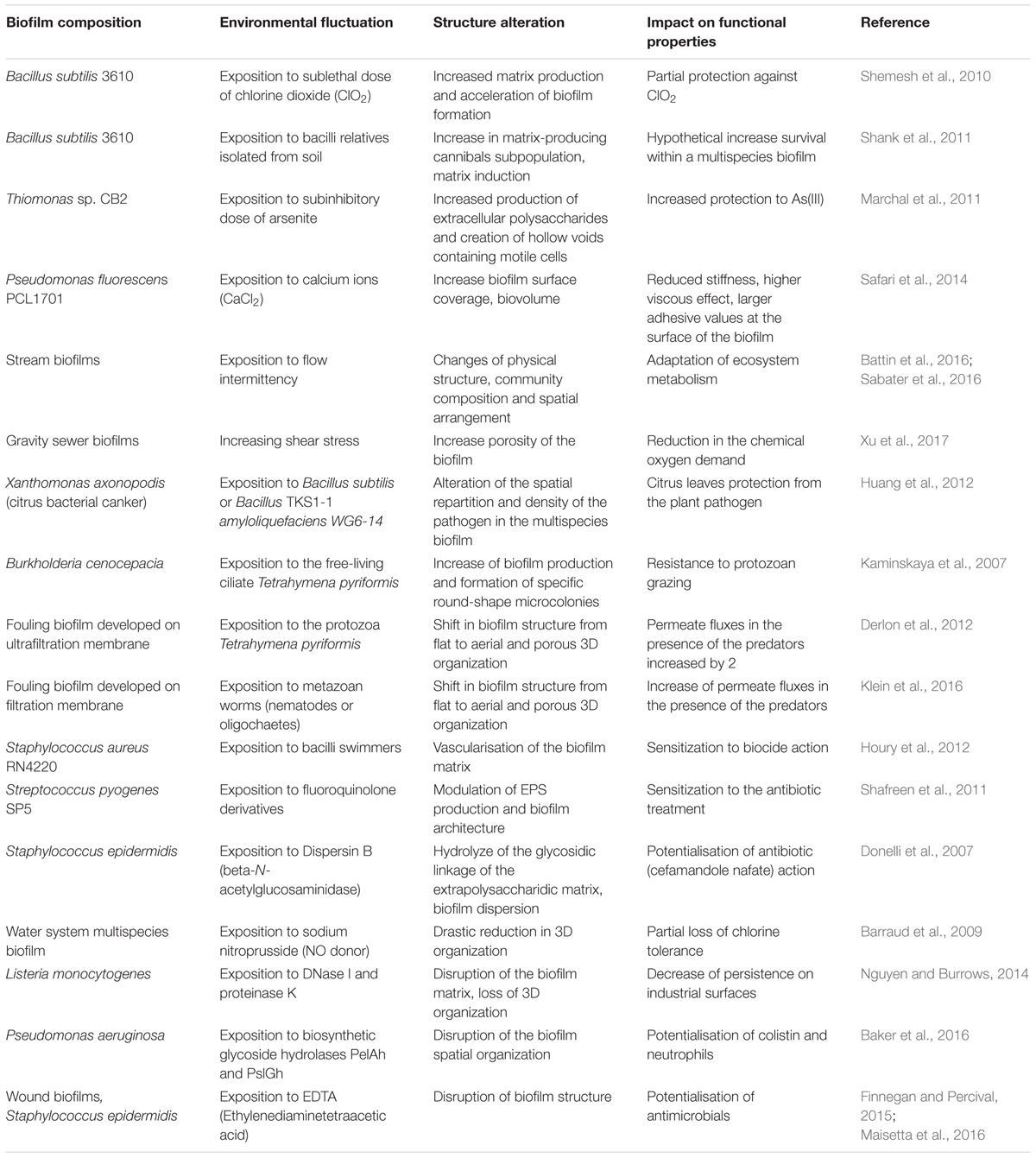
TABLE 1. Examples of biofilm structural responses to environmental fluctuations associated with the alteration of community functions.
Microbial Systems to Shape Biofilm Structure
Microorganisms harness an arsenal of complementary mechanisms to tailor biofilm architecture. They range from regulation of cell motility to modification of cellular morphology, production of matrix components, generation of genetic and physiological heterogeneity or subpopulation interactions. Examples of modulations of biofilm architecture in response of various environmental conditions and depending on bacterial composition are displayed in Figure 2.
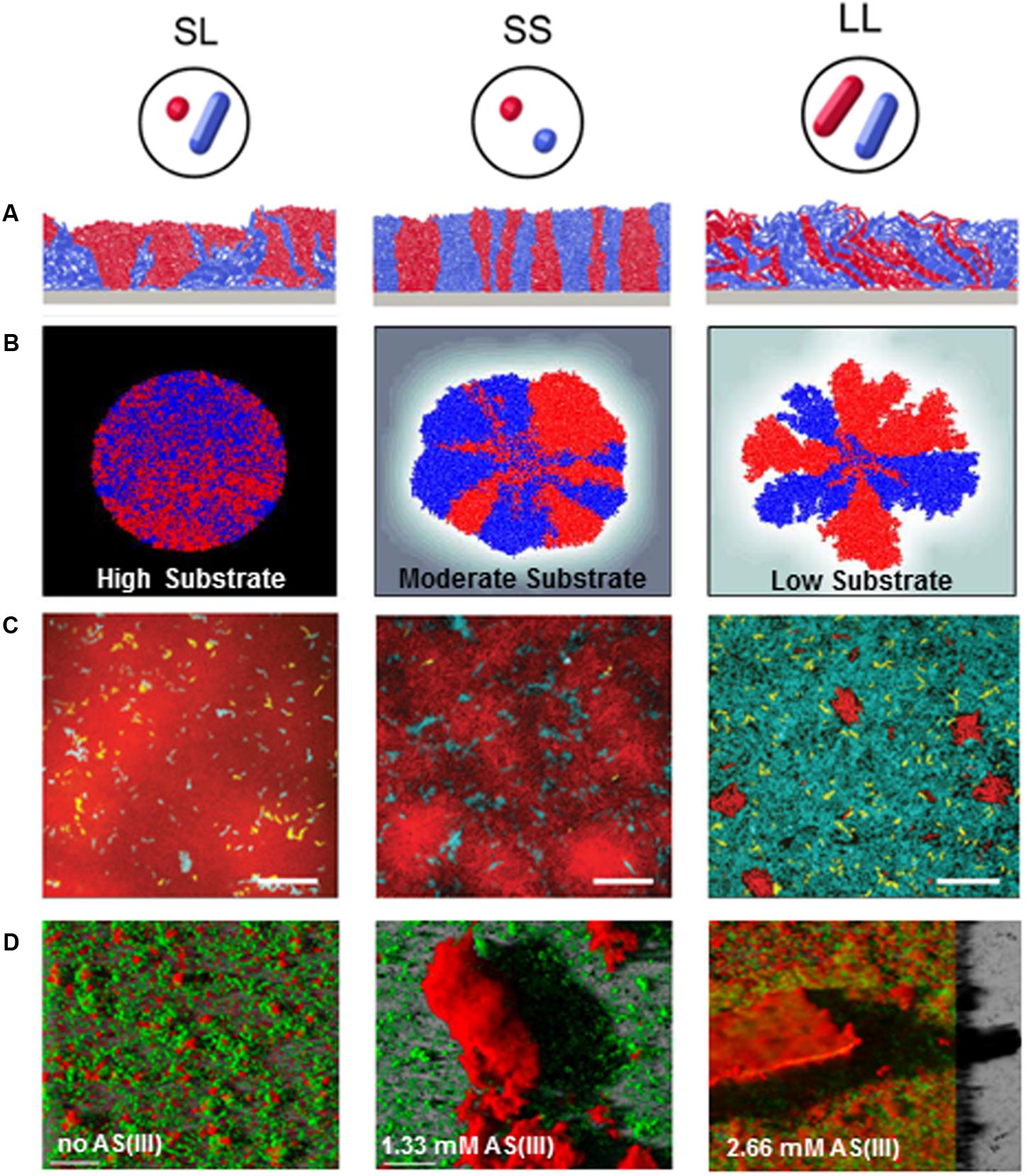
FIGURE 2. Biofilm architectural modulations in response to environmental stimuli or depending on bacterial composition. (A) Impact of cell morphology on biofilm spatial organization. Images displayed 2D sections from simulations where biofilms exponentially grown from 1:1 mixtures of red- and blue-labeled strains form distinct 3D patterns depending on the coccal (S) or rod-like (L) morphology of the strain (Adapted from Smith et al., 2017). (B) Impact of substrate availability (High, moderate and low availability) on biofilm architecture and lineage segregation. Simulations were started with a 1:1 mixture of red and blue cells, where cell color served a neutral marker for lineage segregation. Substrate concentration decrease was associated to a higher spatial segregation of cell lineages (Adapted from Nadell et al., 2010). (C) Impact of disturbance frequency on Vibrio cholerae biofilm spatial organization and strain competition. Images are optical sections taken from the bottom cell layer of biofilms initiated with a 1:1:1 mixture of wild-type strain (teal): a mutant strain hyper-secreting biofilm matrix (red): a mutant strain that is unable to produce extracellular matrix (yellow) cells in microfluidic devices (scale bar: 20 μm). Biofilms grew under continuous nutrient provision (left), or underwent disturbance events every 12 h (middle) or every 6 h (right). Each disturbance event consist in stopping the flow during 2 h to lead to nutrient limitation. Cells were then allowed to disperse to a new microfluidic chamber by pumping the dispersed cells from the initial chamber to the new chamber. After a 2 h-incubation, flow was resumed to pump fresh medium in the newly colonized chamber and enable biofilm growth (adapted from Yan et al., 2017). (D) Impact of toxic on EPS production and biofilm structure in Thiomonas sp. CB2. Images show three dimensional confocal reconstruction of 7 day-old biofilms cultivated in the absence, or in the presence of 1.33 and 2.67 mM Arsenic [As(III)]. Biofilms were stained using SYTO9 (cells, green) and ConA (exopolysaccharides, red) (Adapted from Marchal et al., 2011).
Genetic and Physiological Adaptation at Single Cell Scale
Physical and chemical microenvironments within the biofilm (e.g., varied conditions of pH, osmotic strength, nutrients or exposure to sublethal concentrations of biocide) induce heterogeneous metabolic activity and adaptive responses among biofilm cells (Bridier et al., 2011; Giaouris et al., 2015). During biofilm development, the population displays multiple phenotypes (Sauer et al., 2002). At the single cell scale, the diversity of cell properties are due to either the phenotypic adaptation driven by up- or down- regulation of gene expression, or the appearance of genetic mutants driven by an increased level of mutation in biofilm environment.
Gene regulation at different stages of biofilm formation, compared to the free-living mode of life, can be studied through the comparison of transcriptomic (Waite et al., 2006; Moreno-Paz et al., 2010; Guilhen et al., 2016), proteomic (Sauer, 2003; Resch et al., 2006; Vilain and Brozel, 2006; Giaouris et al., 2013; Qayyum et al., 2016) or metabolomic (Wong et al., 2015; Stipetic et al., 2016) profiles revealing up- or down- regulated functions. For example, in mature biofilms of P. aeruginosa, more than 50% of proteins are upregulated and more than 100 proteins are de novo synthesized in comparison to planktonic cells (Sauer et al., 2002). The multiple phenotypes described in biofilm communities do not correspond to a simple mixture of planktonic cells at different growth stages. The biofilm proteome of Bacillus cereus was for example demonstrated as unique and different from those of exponential and stationary-phase planktonic cells (Vilain and Brozel, 2006). Compared lipidomics between planktonic and biofilm cells also support the idea of specific biofilm phenotypes. Indeed, in various growth conditions, the biofilm cell membrane of different bacterial strains was shown to be more saturated than their planktonic counterparts, whatever their growth phase (Dubois-Brissonnet et al., 2016). In addition, the spatialized environments in biofilms promote the generation and fixation of a phenotypic diversity compared to selection of only one or very few clones in well mixed environment (Traverse et al., 2013; Martin et al., 2016). The spatial distribution of the biofilm multiple phenotypes can be visualized within the biofilm thickness through the observation of different patterns of physiological characteristics (growth rate, mRNA, proteins synthesis or CsgD production) using for example Gfp reporter systems (Werner et al., 2004; Lenz et al., 2008; Stewart and Franklin, 2008; Serra et al., 2013).
Regulation of genes which differentiate planktonic and biofilm protein patterns are numerous and can be partitioned in several categories: metabolism (carbon catabolism, aerobic/anaerobic metabolism, membrane and transport), stress responses and adaptation, motility and attachment (flagellin, surface proteins), EPS production and quorum-sensing signaling (Whiteley et al., 2001; Sauer et al., 2002; Khemiri et al., 2016). Both transcriptional and post-transcriptional regulation occur: the first is slow but may be important for the long-term stability of the biofilm (Guttenplan and Kearns, 2013); the second is described to be mainly controlled by the c-di-GMP intracellular level. As mentioned previously, elevated intracellular levels of c-di-GMP generally promote EPS synthesis and biofilm formation, while decreased levels reduce biofilm formation (Martinez and Vadyvaloo, 2014).
Besides, biofilms can constitute an optimal environment for both cell to cell exchanges of genetic material and genetic mutations in biofilm inhabitants. They offer a panel of ideal characteristics for horizontal gene transfer through conjugation and transformation. These include the presence of high cell density favoring physical contact between biofilm bacteria and of a matrix that is rich in communication signals and in extracellular DNA (eDNA) (Madsen et al., 2012). In addition to providing transferable genetic elements (Hannan et al., 2010), the eDNA plays a central role in triggering natural competence in biofilm bacteria (Molin and Tolker-Nielsen, 2003). Horizontal gene transfer has therefore been described in several studies revealing that conjugation levels were 700–1000-fold higher in biofilms compared to planktonic bacterial cells (Król et al., 2013; Savage et al., 2013). This “permeability” of biofilm bacteria to heterologous mobile genetic elements is likely to shape the evolution of biofilm bacteria and to enhance their relatedness (Madsen et al., 2012). Another mechanism yielding genetic evolution in biofilms is linked to a higher mutation rate of certain biofilm bacteria. Important studies down this line have been performed in P. aeruginosa. Initial observations reported that genetic diversification occurred through a recA-dependant mechanism within short-term growth in biofilms and yielded mutants with multiple novel traits including motility, nutrition requirements, morphology, biofilm phenotypes, and stress resistance (Boles et al., 2004). The study of mutations in-situ within biofilms using a gfp gene containing a +1 frameshift mutation showed that mutations occurred in microcolony structures and increased at a frequency 100 to 1800-fold higher than that observed in planktonic cultures (Conibear et al., 2009). The underlying mechanism is linked to the mismatch repair system (MRS) which monitors the fidelity of DNA replication and recombination through its two main components MutS and MutL (Oliver et al., 2002). Using mutS deficient derivatives of P. aeruginosa and a flow-cell biofilm model system, Luján et al. (2011) showed that the mutants yielded enhanced phenotypic and morphological diversities over wild type strains in structured biofilms. Interestingly, the generated morphotypic variants showed increased competitiveness over the parental strain. This is to correlate to the high prevalence (30–60%) of mutator strains due to alterations in the mutS and mutL genes in P. aeruginosa chronic infections while detection of mutators is rare in P. aeruginosa acute infections (Gutiérrez et al., 2004; Feliziani et al., 2010).
Altogether, this overall genetic plasticity of bacteria in biofilm yields a rapid development of diversity among members of biofilm communities and is likely to shape the biofilm structure because of the co-development of the different morphologies and phenotypes. This provides the biofilm with what has been termed the “insurance hypothesis” in ecology that considers that the stability of many biological communities relies on their diversity which increases the chance that some members will be able to withstand environmental variations that the community may encounter (Boles et al., 2004). This enhanced clonal diversity in biofilms is a real challenge in the control of pathogen and detrimental biofilms as they may rapidly adapt to environmental stresses such as treatments with antimicrobials (Macià et al., 2011; Koch et al., 2014; Van Meervenne et al., 2014). In contrast, this diversity is a real benefit in biotechnological issues in which biofilms can be exploited in numerous applications and under many different environmental conditions (Berlanga and Guerrero, 2016; Piard and Briandet, 2016).
Cell Adaptation with Direct Impact on Biofilm Structure
Individual adaptative responses of biofilm cells, due to heterogeneous environments within their complex living place, lead to individual phenotypic changes, such as individual cell morphology and motility or modification of matrix production.
Bacterial motility, within or outside the biofilm structure, is a major driver of the community plasticity. Once associated to a surface, most of the bacterial cells transfer from a motile to a non-motile state. P. aeruginosa for example becomes non-motile as soon as it attaches irreversibly to a surface and forms clusters with non-motile cells during the maturation of the biofilm (Sauer et al., 2002). In accordance, transcriptional profiles of P. aeruginosa biofilms showed that motility genes are downregulated compared to planktonic cells (Whiteley et al., 2001). The B. subtilis motility is also inhibited under biofilm conditions (Guttenplan et al., 2010). In the short-term, motility is inhibited at multiple levels through accumulation of intracellular c-di-GMP (Ahmad et al., 2013; Guttenplan and Kearns, 2013). In the longer term, regulation relies on transcriptional repression which is slow but may be important for the long-term stability of the biofilm (Guttenplan and Kearns, 2013). In mature biofilms, maintenance of motility for the majority of the cells can destabilize multicellular aggregates and regulation of biofilm plasticity likely shifts to other determinants including EPS production (Guttenplan and Kearns, 2013). Nevertheless, some motile minor isogenic subpopulations can coexist with sessile biofilm cells, creating transients pores within a mature biofilm structure, altering the diffusion-limitation properties of the matrix (Houry et al., 2012; Turonova et al., 2015). In Campylobacter biofilms, an unusual continued expression of the motility complex was described by proteomics in the whole population which suggests a crucial role of the measured motility in this biofilm phenotype (Kalmokoff et al., 2006). Similarly, flagellar hook protein (FlgE) was expressed in biofilm cells but not in planktonic cells of Cronobacter sakazakii (Ye et al., 2016).
Flagella synthesis and movement are highly regulated in response to environmental conditions. During biofilm maturation, starvation stress occurs in the growing biofilm structure, along with a lack of oxygen and accumulation of by-products and QS signaling molecules. All these factors are important drivers of microbial dispersion (Huynh et al., 2012; Martinez and Vadyvaloo, 2014; Solano et al., 2014). Non-coding small RNAs were also recently identified as players of this dissemination process (Chambers and Sauer, 2013). In the well described P. aeruginosa biofilm cycle, dispersion is a consequence of the return to a motile state of a subpopulation of bacterial cells in the center of a cluster. This return is possible through phage-mediated localized cell death (hollow-voids) along with the synthesis of enzymes that can degrade extracellular substances (Webb et al., 2003; Sauer et al., 2004). Dispersion is heterogeneously distributed at the surface of the biofilm and can induce modification of the whole biofilm topography. Motility up- and down- regulation is thus an important driver of the biofilm structure plasticity through its role in attachment, cluster formation and disruption.
Besides, individual cell morphology can also have a great impact on the organization of the population within the biofilm consortium (Smith et al., 2017). Growing in biofilm state, some coccoid-shaped bacteria or small rod can elongate and multiple morphotypes of isogenic cells can appear in different layers. Two different shapes of Lactococcus lactis were observed in 16 h flow-cell biofilms: coccoid cells were localized in the depth of the structure while a stratum of elongated filaments rises on the interfacial layers of the structure (Perez-Nunez et al., 2011). Similarly, different morphotypes of uropathogenic Escherichia coli were observed from coccoid form to elongated rods, through different stages of biofilm formation. Filamentous bacteria were observed on the edge of late biofilm in connection with detaching cells (Justice et al., 2004). The filamentation was shown to be a response to stressful environment and is essential for uropathogenic E. coli virulence (Justice et al., 2006, 2008).
In addition, EPS are the cement of biofilm architecture and their modulation trigger direct alteration of the spatial structure (Ziemba et al., 2016). EPS content includes water and biopolymers originating from biofilm microorganisms including polysaccharides, proteins, lipids, and eDNA (Flemming, 2011; Fong and Yildiz, 2015; Limoli et al., 2015). From an anthropomorphic biofilm perspective, the matrix has been described as the house of bacteria and as such its structure and composition are unique according to the inhabiting bacteria and the environment (Stoodley et al., 1999; Watnick and Kolter, 2000; Flemming et al., 2007; Flemming, 2011). The EPS matrix cannot be considered as a homogeneous slimy material, but rather as the sum of multiple microhabitats with different local environments (oxygen concentrations, pH-values, redox potential, shear forces, etc.). This stratification governs biofilm heterogeneity in which bacterial groups distribute themselves according to their preferred particular microenvironment and to symbiotic relationships (Watnick and Kolter, 2000; Stewart and Franklin, 2008; Flemming, 2011). This heterogeneity in space is doubled by heterogeneity in time: EPS evolves with the biofilm aging and appears as a dynamic structure due to various events including degradation of matrix elements by bacterial enzymes, dissolution of EPS components, incorporation of new material, etc… (Sutherland, 2001). It can also be noted that most of the different components of the matrix are associated by non-convalent interactions suggesting that dissociation can occur through local modifications of the EPS physicochemical properties (pH, ionic strength, hydration, etc…) (Neu and Lawrence, 2016). This poorly characterized plasticity of the EPS matrix makes it the least understood component of biofilms biology and as such has been termed the “dark matter” of biofilms (Flemming and Wingender, 2010; Flemming et al., 2016).
In an attempts to characterize the signals governing matrix formation and modification in biofilms, Shemesh et al. (2010) showed that exposure of B. subtilis and P. aeruginosa to sublethal doses of a biocide (chlorine dioxide, CIO2) stimulate biofilm formation. The transcription of two major operons involved in matrix production [epsA-epsO involved in polysaccharide (PS) production and yqxM-sipW-tasA involved in amyloid production] was shown to be increased by CIO2 via the membrane-bound kinase KinC. Interestingly, kinC mutants unable to make a matrix were hypersensitive to CIO2. Another kinase within the epsA-epsO operon, the EpsAB tyrosine kinase, is involved in regulation of PS production by a seemingly QS mechanism (Elsholz et al., 2014). The membrane sensor EpsA is able to sense the presence of PS and control kinase activity. In the absence of PS, the kinase is inactivated by autophosphorylation while the presence of PS inhibits autophosphorylation and stimulates the phosphorylation of glycosyltransferases and thereby the synthesis of PS. This positive feedback loop therefore ties PS synthesis to the external concentration of PS. This opens exciting perspectives in applications in which exogenous polysaccharides could be used either as inducers of the biofilm way of life or as modulators of the matrix structure. Also this raises the question whether PS produced by one biofilm bacteria could trigger PS production in another biofilm bacterium. A part of the answer probably relies on the yet unknown specificity of the sensor EpsA toward the different PS produced by a biofilm bacterial community. In an attempt to explore such interbacterial interactions, Shank et al. (2011) investigated whether soil bacteria were able to affect the biofilm development in B. subtilis. Using a fluorescent reporter fused to the tapA promoter, the coculture screening test showed that most strains able to induce matrix production in B. subtilis belonged to the Bacillus genus suggesting that interactions occur mostly with close relatives. Two mechanisms were dissected. One involves the activation of the sensor kinase KinD while the other is kinase independent and involves the master regulator Spo0A (Shank et al., 2011).
Species belonging to Thiomonas species are frequent in arsenic polluted sites and play key roles in arsenic natural remediation (Marchal et al., 2011). Exposure of Thiomonas sp. to sublethal arsenite concentration yielded biofilms with an up to six-fold increase in PS production concomitantly to a 83-fold increase in cell death and cell lysis. This was accompanied with a complex rearrangement of the biofilm structure into PS covered mushroom-like structures in which eDNA was a key player as treatment with a nuclease abolished such phenomenon. eDNA is indeed a crucial component of the biofilm matrix and is involved in multiple interactions with other EPS components including PS and amyloids (Hu et al., 2012; Liao et al., 2014; Schwartz et al., 2016). In Staphylococcus aureus, the cidA and lrgA genes act as holins and antiholins, respectively, and regulate cell lysis in an analogous way to that observed in bacteriophage-mediated cell lysis. While wild-type S. aureus produced biofilm with distinct mushroom-like 3D structures that are characteristic of mature biofilms, both a cidA mutant deficient in lysis and a lrg mutant deficient in the inhibition of CidA-mediated lysis produced biofilms lacking 3D mushroom-like structures (Mann et al., 2009). S. aureus is also able to produce and secrete Nuc, a thermostable nuclease. Analysis of the biofilm formed by a nuc mutant showed increased amounts of mushroom-like structures. Also, treatment of the S. aureus biofilms with DNAseI in flow cell chambers completely removed biofilms. Altogether this suggests that different bacterial factors are able to modulate the level of available eDNA that appear critical in the shaping of biofilm structure and dispersal.
Cell Adaptation with Indirect Changes on Biofilm Structure via Increased Resistance and Persistence
Physiological adaptation of individual cells within the biofilm community may lead to an increased resistance to biocides and antibiotics. Stresses such as starvation (oxygen or nutrients) in the depth of the biofilm or contact with sublethal concentrations of antimicrobials during disinfection can induce a bacterial stress response and higher tolerance to biocides (Mangalappalli-Illathu et al., 2008; Bridier et al., 2011). The higher individual cell resistance can be explained by several mechanisms. An overexpression of enzymes that are able to degrade biocides (catalase, superoxide dismutase) was described in biofilm under the control of QS (Hassett et al., 1999). P. aeruginosa membrane efflux pumps were shown to be up-regulated for cell cultivated in biofilm although their exact role in the biology of these sessile communities needs to be clarified (Zhang and Mah, 2008; Soto, 2013). Moreover, by limiting biocide intracellular penetration, the observed increase in membrane saturation in biofilm cells compared with their planktonic counterparts can be another resistance mechanism (Dubois-Brissonnet et al., 2016). After repeated antimicrobial treatments, the development of the most resistant surviving cells in the biofilm structure will modify the spatio-temporal dimension of the biofilm architecture.
Interactions between Biofilm Subpopulations
Multispecies biofilm is a result of cell–cell and cell–environment interactions such as cooperation, competition or exploitation that create heterogeneity in biofilms (Liu et al., 2016). These specific interactions between species are involved in the spatial organization of biofilms in which they are more favored than in planktonic environments. They maintain their diversity and stability by generating more physiological and functional heterogeneity (Nadell et al., 2009; Pamp et al., 2009; Rendueles and Ghigo, 2015; Kragh et al., 2016; Liu et al., 2016; Pande et al., 2016). Indeed, in specific environment, some species cannot form biofilm alone, but grow in association with others species in multispecies biofilm showing interspecific cooperation interactions between subpopulations (Palmer et al., 2001). Specific interactions and spatial organization within biofilm create fitness effect through social phenotypes. A telling example is the symbiotic two-species consortium formed by Pseudomonas putida and Acinetobacter sp. strain C6 which has evolved in a non-random spatial organization where P. putida exclusively attached and grew on pre-existent colonies formed by Acinetobacter sp. strain C6. Resulting evolved communities were characterized by an increased fitness and productivity (Hansen et al., 2007). Microscopic time-lapse observations revealed that cell clusters were arranged according to a uniform pattern and that such structure results from the moving along the surface and the fusion of early microcolonies (Haagensen et al., 2015). These observations illustrate the improvement of community fitness through the active spatial structuration of its individuals and theirs interactions, and thereby the stabilization of their symbiotic relations.
Similar observations were made by describing the evolution of communities derived from a clonal Burkholderia cenocepacia biofilms (Poltak and Cooper, 2011). The authors highlighted the emergence of three variants and their persistence in mixed communities displaying enhanced productivity than any monoculture. The authors demonstrated that such productivity gains were due to the asymmetrical cross-feeding between the different ecotypes and the expansion and restructuration of biofilm space that constructed new niches. Overall, the fitness of cooperative or competitive phenotypes largely depends on neighboring cells that finally influences the spatial arrangement of genotypes within biofilms (Nadell et al., 2016; Stubbendieck et al., 2016). Reciprocally, the spatial structuring of genotypes within biofilm greatly influences the evolution of social phenotypes (Nadell et al., 2016). Many social phenotypes are regulated by QS through the secretion of diffusible signaling peptide (Nadell et al., 2016; Perchat et al., 2016). Studies showed how interspecies QS may have a role in competition interactions. In a P. putida–P. aeruginosa mixed-species biofilm, it was demonstrated a spatial repulsion between the two isolates (Fernandez-Piñar et al., 2011; Bridier et al., 2014). Indeed, both populations secreted molecules which negatively alter the growth of each other; P. aeruginosa secreted quinolone, a QS signaling molecule which inhibits biofilm formation of P. putida, and in the same way, P. putida secreted putisolvin which is regulated by QS and inhibits P. aeruginosa biofilm formation (Diggle et al., 2003; Kuiper et al., 2004; Fernandez-Piñar et al., 2011; Bridier et al., 2014). Other systems can have an important role in interspecies interactions such as communication and transport including outer membrane vesicles (OMVs) (Wang et al., 2015). OMVs could promote bacterial interactions and thereby participate to the architectural integrity of biofilms (Schwechheimer and Kuehn, 2015). In Helicobacter pylori, Franciscella, P. aeruginosa, V. cholera and P. putida, vesicles are involved in biofilm formation by increasing hydrophobicity of cells surface and by participating to the matrix formation (Yonezawa et al., 2011; Baumgarten et al., 2012; van Hoek, 2013; Altindis et al., 2014; Murphy et al., 2014; Wang et al., 2015). OMVs can also have an interspecies interference property in biofilms when they are coupled with an antimicrobial action and alter bacteria in biofilms (Schooling and Beveridge, 2006). Species interactions contribute thus through various way to shape biofilm architecture. Actually, processes related to intra-species interactions, as for instance cell death, can also play a key role in biofilm structuring. Localized cell death is known to trigger wrinkle formation of biofilm by focusing mechanical forces and instigate vertical extending of the biofilm (Asally et al., 2012; Rendueles et al., 2014; Nadell et al., 2016). Overall, it has been showed that cell death plays an important role in the development of multicellular biofilms and the subsequent dispersal of surviving cells (Webb et al., 2003; Mai-Prochnow et al., 2004). In Bacillus subtilis biofilms, subpopulations of cells use a cannibalistic strategy involving the production and secretion of two toxins to lyse sensitive siblings which then provide nutrients for the cannibals. Interestingly, cannibal cells correspond to the subpopulation producing the extracellular matrix, the production of toxins and matrix being triggered by surfactin, a paracrine signal whose production is controlled by the QS signaling peptide ComX (Lopez et al., 2009). This process finally promotes matrix producer subpopulations and enables the development of biofilm structure through an increase of matrix production.
This extraordinary large diversity of means provides to microorganisms the ability to dynamically shape biofilm architecture and functions using complementary mechanisms. Numbers of processes involved in architecture plasticity are thus inter-related through complex regulation networks enabling the targeted adaptation through the sensing of a wide range of environmental conditions.
Tuning Biofilms Architecture to Control Their Functions?
Sculpting biofilm spatial organization represents an attractive approach to control their overall functions, whether beneficial or deleterious (Table 1). The structure of those surface-associated communities can be faceted by governing their local environmental or by exposing them to molecular and biological effectors. Illustrations of such shaping are displayed in Figure 3.
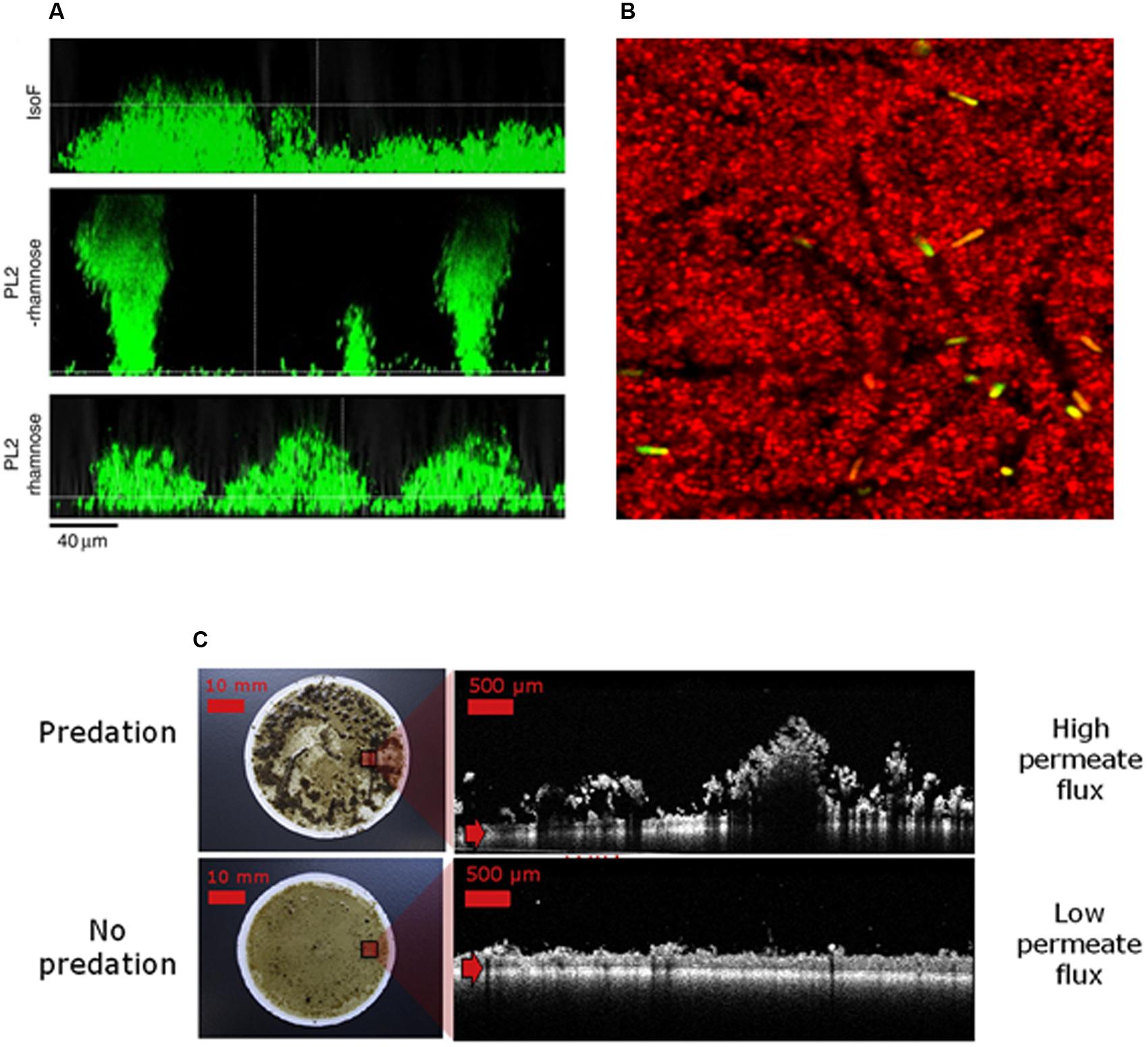
FIGURE 3. Sculpting biofilm architecture to control their function. (A) Role of biosurfactants in Pseudomonas putida biofilm architecture. Images displayed vertical sections of biofilms grew in flow chamber during 3 days in presence or absence of rhamnoses. IsoF correspond to the wild-type strain and PL2 strain to a conditional mutant in which the native promoter region of psoA (a gene coding a large non-ribosomal peptide synthethase which directs the biosynthesis of the two cyclic lipopeptide biosurfactants putisolvin I and II) has been replaced with the rhamnose-inducible PrhaB promoter (adapted from Carcamo-Oyarce et al., 2015). Addition of 0.2% rhamnose in growth medium of PL2 lead to the recovery of the flat wild-type biofilm structure suggesting that putisolvins promote the colonization of the substratum. (B) Predation by protozoa affects biofilms spatial organization during gravity-driven dead-end ultrafiltration and induces higher permeate fluxes (adapted from Derlon et al., 2012). (C) Green bacilli creates transient pores in the biofilm matrix of Staphylococcus aureus, leading to an increased sensitivity to biocide action as described in Houry et al. (2012) (courtesy of Julien Deschamps, INRA).
Manipulating Biofilm Local Environment
Within a biofilm, individual cells have the ability to monitor their direct environment (nutrients, pH, ionic strength, oxygen, surface…). The integration of these various external signals leads to specific cellular responses that can be exploited to alter the community structure/function.
In this line, Sauer et al. (2004) elegantly demonstrated that a sudden increase in carbon substrate or pH of the growing medium lead to significant change in P. aeruginosa biofilm structure. Changing the glutamate concentration of the media from 2 to 20 mM triggers a total loss of the biofilm tridimensional structure in less than 60 min. This massive loss of surface-associated biomass observed was correlated with the induction of a subpopulation of bacteria with an increased expression of flagella and a decreased expression of pilus, allowing their dispersal in the flow. Similarly, Staphylococcus epidermidis biofilm exposed to a high osmotic pressure (from 86 to 776 mM NaCl) decreased the average bacterial local number density by 10-fold (Stewart et al., 2013). Increasing the flow shear stress applied on P. aeruginosa biofilm reduced the formation of self-aggregating clusters, in particular through a significant down regulation of genes involved in extracellular polysaccharide synthesis (Crabbé et al., 2008; Dingemans et al., 2016). Exposing a gravity sewer biofilm to increasing shear stress (from 1.12 to 1.45 mPa) affected porosity of the biostructure (from 70 down to 55%) and reduced the chemical oxygen demand in the sewers from 40 to 32% (Xu et al., 2017). Growing the microaerophilic human pathogen Campylobacter jejuni under aerobic condition (20% O2) stimulates the kinetic of biofilm development (Reuter et al., 2010) and the complexity in their architecture (Turonova et al., 2015). Desiccation of the biofilm occurs periodically in various environments including soils, industrial surfaces or hypersaline ponds (Habimana et al., 2014; Decho, 2016; Lennon and Lehmkuhl, 2016). In the latter environment, the EPS attains a glass state upon extreme desiccation that presumably protects the biofilm inhabitants and allows them to resume activities upon rehydratation. When grown at the air interface, Bacillus subtilis developed a biofilm protected by a hydrophobic raincoat layer formed by the BslA surface-active protein (Arnaouteli et al., 2016). This interfacial layer of water-repellent proteins also protects the biofilm inhabitants from ethanol and biocide action (Epstein et al., 2011). When the biofilm structure limits antimicrobial penetration and prevents the contact with the microbial target, exposition to pulsating waves of energy (e.g., ultrasonic waves) can amplify the antimicrobial effect. This so called bio-acoustic effect is likely associate with a deformation of the biofilm and a better penetration in the EPS of the toxic compounds (Qian et al., 1996; Peterson et al., 2015). Another often neglected environmental parameter to shape biofilm is the nature of the substratum. Greene et al. (2016) demonstrated that it was possible to alter the biofilm structure of Acinetobacter baumannii only by changing its carrier nature. While an important structured biofilm was able to grow in 4 days on polycarbonate coupons, only sparse adhering cells were visible in the same condition on glass (biofilm biovolume decreased from more than 2.5 to below 0.1 μm3/μm2). Not only the spatial arrangement of the cell were altered by the nature of the substratum, but also the bacterial physiology as reported by the live/dead ratio that ranged from less than 2 for biofilm grown on rubber to almost 8 for cell grown on stainless steel. Muszanska et al. (2012) demonstrated that coating silicone rubber with a brush polymer alters the biofilm structure (including a strong decrease in the polysaccharidic matrix) and the susceptibility to the gentamycin antibiotic. From those observations, authors suggested that the antimicrobial treatments of biofilm-associated infections could be more effective on material protected with such active antibiofilm coatings. Similarly, Valle et al. (2006) observed that treating abiotic surfaces with group II capsular polysaccharides drastically reduces both initial adhesion and biofilm architecture by important nosocomial pathogens.
All these examples illustrate the possibility to manipulate the structure/function association of microbial biofilms by controlling one (or a combination) of parameter(s) in their local environment.
Reprogramming Biofilm Structure/Function with Specific Molecular Triggers
External cues can be put in used to act both directly on the biofilm EPS properties or reprogram individual cell physiology and transcriptional expression patterns. A large palette of exogeneous enzymes has the ability to degrade specific moieties of the complex biofilm matrix. Those EPS-degrading enzymes can act specifically on extracellular polysaccharides (dispersin B), proteins (proteinase K, trypsin) or eDNA (DNase I) (Boles and Horswill, 2011). Cocktails of such enzymes are proposed in the food-industry to target persistent deleterious biofilms (Lequette et al., 2010; Nguyen and Burrows, 2014). Dispersin B that hydrolyzes the glycosidic linkages of PNAG was found to be efficient in a range of pathogenic bacteria and is being commercially developed as a wound care gel (Kaplan et al., 2003). Enzymes from bacteriophages can dissolve extracellular polysaccharides of the matrix and reverse the biofilm tolerance to antibiotics and other antimicrobial treatments (Chan and Abedon, 2015). Bacteriophage enzymes were able to reduce the alginate EPS viscosity by up to 40% in P. aeruginosa biofilm (Hanlon et al., 2001). Using purified EPS depolymerase isolated from an Enterobacter agglomerans bacteriophage, Skillman et al. (1998) demonstrated a change in the physical properties of the EPS from a two species biofilms resulting in the effective removal of both species. Another telling example is the use of the biosynthetic glycoside hydrolases PelAh and PslGh that were able to disrupt the spatial organization of a pre-existing P. aeruginosa biofilm within 1 h, potentiating the action of colistin and neutrophils (Baker et al., 2016). By targeting the cell wall, the hydrolases LySMP was able to reduce the biofilm structure of Streptococcus suis by more than 80% and facilitate the action of several antibiotics on sessile communities (Meng et al., 2011).
Amyloids fibers are the “neglected child” of the EPS matrix (Dueholm and Nielsen, 2016) while evidence is rinsing that those proteinous assemblages are important drivers of the matrix viscoelastic properties (Lembré et al., 2014). D-amino acids (with some controversy) and parthenolide were identified as molecular inhibitors targeting the polymerisation or anchorage to the cell wall of TasA, the main Bacillus subtilis EPS amyloid (Kolodkin-Gal et al., 2010; Leiman et al., 2013; Romero et al., 2013). P. aeruginosa produces cis-2-decenoic acid, a small messenger molecule responsible for the induction of the biofilm dispersion response in a range of Gram-negative and Gram-positive bacteria. It has been shown to alter biofilm structure and to reverse tolerance to conventional antimicrobial agents (Marques et al., 2015). The matrix reprogrammation can also be triggered by biofilm cells exposition to sublethal concentration of antimicrobials. Schilcher et al. (2016) observed that subinhibitory concentrations of clindamycin upregulated the expression of major biofilm-associated genes in S. aureus biofilm and shift the composition of the biofilm matrix toward higher eDNA content.
In addition to soluble molecular effectors, microorganisms are able to respond to organic and inorganic volatiles in their local headspace, some of which influencing their ability to form biofilm (Audrain et al., 2015). Nitric oxide (NO) is a volatile messenger able to trigger biofilm dispersion. Barraud et al. (2009) demonstrated that exposing a multispecies biofilms in water system to 500 nM sodium nitroprusside (NO donor) almost totally abolished the biofilm spatial organization, increasing by 20 the efficacy of the conventional chlorine treatment. On the opposite, ammonia, a volatile produced by many bacteria, stimulates biofilm formation in Bacillus licheniformis and other relatives (Nijland and Burgess, 2010). Similarly, it was showed that self-produced acetic acid was used as volatile signals to stimulate and coordinate the timing of biofilm formation in B. subtilis (Chen et al., 2015). This behavioral biofilm response triggered by odorant molecules was compared to olfaction; it opens doors to new biofilm control strategies based on airborne volatile metabolites.
Guided Biofilm Ecology to Shape the Biofilm Structures and Functions
As mentioned previously, biofilm architecture and functions are intimately related to their microbial content and the spatial repartition of their inhabitants. In, several fields including health, agriculture, food processing and environment, new strategies emerged to manipulate biofilm functions by guided biofilm ecology. The effectors of these approaches are selected organisms that can alter population structures in the targeted community such as bacteria, bacteriophages, molds, yeasts, microalgae, amoeba, and metazoans.
A family of microbial probiotics are put in used on the market to combat human biofilm-associated infections (Vuotto et al., 2014). Specific inhabitants of the oral microbiome such as Porphyromonas gingivalis are responsible of the production of unpleasant malodorant volatile sulfur compounds (halitosis) (Lee and Baek, 2014). Different reports described a beneficial long term effect of combining conventional oral mouthwashes chemical pretreatment with probiotic therapies involving lactic acid bacteria such as the bacteriocin producing Streptococcus salivarius K12 (Masdea et al., 2012; Jamali et al., 2016). Using an agent-based spatially explicit model approach, Bucci et al. (2011) demonstrated that the competitive dynamic of bacteriocin producing strain in a multispecies biofilm strongly depends on a single critical bacteriocin-range parameter that measures the threshold distance from a focal bacteriocin-producing cell whose fitness is higher than that of sensitive cell. Similarly, the biofilm of Aggregatibacter actinomycetemcomitans involved in chronic periodontal diseases was degraded after exposition to a Lactobacillus probiotic altering the biofilm structure (Jaffar et al., 2016). Lactobacillus rhamnosus GG and Lactococcus lactis HY449 both affect the spatial organization of model oral biofilms and reduced the count of oral pathogens in the community (Jiang et al., 2016; Kim and Lee, 2016). The most widespread use of probiotic is the treatment of gastrointestinal diseases. Rieu et al. (2014) demonstrated that a Lactobacillus-induced host immunomodulation response was strongly enhanced when the potential probiotic was cultivated as a structured biofilm in contrast with free-cells. This fundamental discovery leads to the exploration of new biofilm-based formulations to increase their in vivo beneficial effects (Cheow et al., 2014).
In the medical area, an emerging research field to overcome bacterial antibioresistance (super bugs) and chronic biofilm-associated infections (BAI) is the bacteriophage therapy (Maura and Debarbieux, 2011; Chan and Abedon, 2015). In the lab, bacteriophages were efficient in mice models to treat a (biofilm associated) P. aeruginosa acute lung infection (Debarbieux et al., 2010). Exposing Clostridium difficile colony biofilms to a cocktail of selected phages lead to the emergence of lysed zones and elongated cells morphotypes in the structure, but the loss of cell viability observed in early stages decreased with biofilm age (Nale et al., 2016). While highly effective on free-cells, the architecture of biofilm, the diversity of cell types and the presence of matrix likely limit the phages efficacy to treat chronic BAI. Only few human phase II trials explored this approach to treat human patient with only mitigated success (Wright et al., 2009). There is a clear need for larger scale trials and deeper research on phage and biofilm interactions in this promising emerging field (Servick, 2016).
From farms to forks, the microbiological control of raw and processed food through the food chain is still mainly ensure by the use of chemical products including pesticides, antibiotics or disinfectants. Their massive use raised some important environmental and health concerns and stressed out the need for alternative sustainable approaches. In the crop field, a recent paper pinpointed the biofilm mode of life as an important driver of the efficacy of microbial biocontrol agents (Pandin et al., 2017). Indeed, different studies showed evidence that biocontrol agent are able to form protective biofilms on crop that develop antagonistic properties against unwanted microorganisms (Zeriouh et al., 2014). The associated mechanisms likely involved many of the biofilm traits, including spatial competition, cell-cell signaling and the production of antimicrobials (Zhou et al., 2016). Spraying antagonistic Bacillus subtilis TSK1-1 or Bacillus amyloliquefaciens WG6-14 on citrus leaf surface alter the spatial organization and the density of Xanthomonas axonopodis pv. citri, a pathogenic bacteria involved in citrus canker (Huang et al., 2012). A comparative transcriptome analysis of the biocontrol agent Bacillus amyloliquefaciens FZB42 as response to biofilm formation showed an up regulation of the lci gene encoding an antimicrobial peptide, and of operons involved in the production of the extracellular matrix (Kröber et al., 2016). It was also shown that the architecture of those protective biofilm can be stimulated by plant metabolites such as root exudates (Espinosa-Urgel et al., 2002). Similar protecting biofilms are envisioned in the feed/food environments to protect livestock building and the surface of food processing equipments from pathogen persistence (Mariani et al., 2011; Piard and Briandet, 2016). Habimana et al. (2011) demonstrated using confocal imaging and a simplified individual based model that exposing sessile cells of Listeria monocytogenes to Lactococcus lactis engaged a spatial race to interfacial nutrients resulting in a total loss of the pathogen multiplication. It was also recently shown that motile bacilli can create transient pores in Staphylococcus aureus biofilms, sensitizing the pathogenic structure to biocide action (Houry et al., 2012).
The use of organisms to shape new biofilm functions is also emerging in environmental sciences. Derlon and his collaborators nicely demonstrated that predation mediated by added protozoa (Tetrahymena pyriformis) triggers strong architectural change of an ultrafiltration membrane biofilm (from flat to heterogeneous and porous structure), increasing by 2 the permeate flux (Derlon et al., 2012). The same group also demonstrated that metazoan worms, including the nematode Plectus aequalis and the oligochaetes Aelosoma hemprichi, were also able to remodel membrane fouling biofilm structure and to increase significantly the membrane efficacy (Derlon et al., 2013; Klein et al., 2016).
The microflora of stone monuments is mainly composed of microbial biofilms and lichens. Scientists of this field implicate these complex ecosystems in stone damage while others pinpointed their bioprotective role (Pinna, 2014). Application of biofilm-induced calcium carbonate precipitation is an emerging tool for the bioremineralisation of stone and cultural heritage (Dhami et al., 2014). Dick et al. (2006) evaluated the performance of Bacillus sphaericus biofilms to restore deteriorated Euville limestone, a stone used for building and sculpturing in France. They demonstrated an important surface colonization and the presence of dense calcium carbonate crystals on biofilms formed on the treated stone. Similar biocalcifying effect was observed with Bacillus subtilis on deteriorated globigerina limestone (Micallef et al., 2016).
Environmental biofilms are largely involved in global biogeochemical cycles (Singer et al., 2010). Through human intensive activities and the resulting environmental changes, we are unintentionally affecting and remodeling those natural ecosystems. At the Paris climate conference (COP21) in 2015, 195 countries adopted a legally binding global climate deal. The agreement sets out a global action plan to put the world on track to avoid dangerous climate change by limiting global warming to well below 2°C above pre-industrial levels (Rhodes, 2016). Indeed, a 2°C warming in flowing water is already enough to drive significant changes in freshwater biofilm structure/function by inducing a complex reorganization in the network of interactions among microbial populations within the biofilm matrix (Romani et al., 2014).
Conclusion
Architectural plasticity of biofilm constitutes a central process to actively adapt to stress and to increase productivity and fitness of microbial communities in response to changing environmental conditions. Considering dynamics of biofilm structure is thus required to better understand the emergence of novel functional properties and to decipher the communal mechanisms underlying microbial behavior, from single cell to multicellular community. Although our ability to predict and manage the functional properties and adaptation strategies of these complex dynamic communities is yet limited, the increasing development of predictive modeling approaches and the improvement of integration of experiments and models should, in a near future, enable to better link composition, dynamic organization and function of microbial communities (Widder et al., 2016). Recent technological advances in single-cell analytic methods have led to the generation of quantities of novel interesting data on individual microbial behaviors which still are to be exploited through individual-based modeling approach for instance, to provide insights into self-organized spatial patterns and to construct a realistic vision of biofilm at both the individual and community levels (Hellweger et al., 2016).
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
CP is supported by a grant from Rgion Ile-de-France, DIM ASTREA. We warmly acknowledge A. Canette from the MIMA2 microscopy platform and J. Deschamps (INRA) for biofilm imaging.
References
Abdallah, M., Khelissa, O., Ibrahim, A., Benoliel, C., Heliot, L., Dhulster, P., et al. (2015). Impact of growth temperature and surface type on the resistance of Pseudomonas aeruginosa and Staphylococcus aureus biofilms to disinfectants. Int. J. Food Microbiol. 214, 38–47. doi: 10.1016/j.ijfoodmicro.2015.07.022
Ahmad, I., Wigren, E., Le Guyon, S., Vekkeli, S., Blanka, A., El Mouali, Y., et al. (2013). The EAL-like protein STM1697 regulates virulence phenotypes, motility and biofilm formation in Salmonella typhimurium. Mol. Microbiol. 90, 1216–1232. doi: 10.1111/mmi.12428
Altindis, E., Fu, Y., and Mekalanos, J. J. (2014). Proteomic analysis of Vibrio cholerae outer membrane vesicles. Proc. Natl. Acad. Sci. U.S.A. 111, E1548–E1556. doi: 10.1073/pnas.1403683111
Arnaouteli, S., MacPhee, C. E., and Stanley-Wall, N. R. (2016). Just in case it rains: building a hydrophobic biofilm the Bacillus subtilis way. Curr. Opin. Microbiol. 34, 7–12. doi: 10.1016/j.mib.2016.07.012
Asally, M., Kittisopikul, M., Rue, P., Du, Y., Hu, Z., Cagatay, T., et al. (2012). Localized cell death focuses mechanical forces during 3D patterning in a biofilm. Proc. Natl. Acad. Sci. U.S.A. 109, 18891–18896. doi: 10.1073/pnas.1212429109
Audrain, B., Farag, M. A., Ryu, C. M., and Ghigo, J. M. (2015). Role of bacterial volatile compounds in bacterial biology. FEMS Microbiol. Rev. 39, 222–233. doi: 10.1093/femsre/fuu013
Azeredo, J., Azevedo, N. F., Briandet, R., Cerca, N., Coenye, T., Costa, A. R., et al. (2017). Critical review on biofilm methods. Crit. Rev. Microbiol. 43, 313–351. doi: 10.1080/1040841X.2016.1208146
Baker, P., Hill, P. J., Snarr, B. D., Alnabelseya, N., Pestrak, M. J., Lee, M. J., et al. (2016). Exopolysaccharide biosynthetic glycoside hydrolases can be utilized to disrupt and prevent Pseudomonas aeruginosa biofilms. Sci. Adv. 2:e1501632. doi: 10.1126/sciadv.1501632
Barraud, N., Schleheck, D., Klebensberger, J., Webb, J. S., Hassett, D. J., Rice, S. A., et al. (2009). Nitric oxide signaling in Pseudomonas aeruginosa biofilms mediates phosphodiesterase activity, decreased cyclic di-GMP levels, and enhanced dispersal. J. Bacteriol. 191, 7333–7342. doi: 10.1128/JB.00975-09
Battin, T. J., Besemer, K., Bengtsson, M. M., Romani, A. M., and Packmann, A. I. (2016). The ecology and biogeochemistry of stream biofilms. Nat. Rev. Microbiol. 14, 251–263. doi: 10.1038/nrmicro.2016.15
Baumgarten, T., Sperling, S., Seifert, J., von Bergen, M., Steiniger, F., Wick, L. Y., et al. (2012). Membrane vesicle formation as a multiple-stress response mechanism enhances Pseudomonas putida DOT-T1E cell surface hydrophobicity and biofilm formation. Appl. Environ. Microbiol. 78, 6217–6224. doi: 10.1128/AEM.01525-12
Beech, I. B., and Sunner, J. (2004). Biocorrosion: towards understanding interactions between biofilms and metals. Curr. Opin. Biotechnol. 15, 181–186. doi: 10.1016/j.copbio.2004.05.001
Berlanga, M., and Guerrero, R. (2016). Living together in biofilms: the microbial cell factory and its biotechnological implications. Microb. Cell Fact. 15:165. doi: 10.1186/s12934-016-0569-5
Berleman, J. E., Zemla, M., Remis, J. P., Liu, H., Davis, A. E., Worth, A. N., et al. (2016). Exopolysaccharide microchannels direct bacterial motility and organize multicellular behavior. ISME J. 10, 2620–2632. doi: 10.1038/ismej.2016.60
Besemer, K., Hodl, I., Singer, G., and Battin, T. J. (2009). Architectural differentiation reflects bacterial community structure in stream biofilms. ISME J. 3, 1318–1324. doi: 10.1038/ismej.2009.73
Bjarnsholt, T. (2013). The role of bacterial biofilms in chronic infections. APMIS Suppl. 136, 1–51. doi: 10.1111/apm.12099
Boles, B. R., and Horswill, A. R. (2011). Staphylococcal biofilm disassembly. Trends Microbiol. 19, 449–455. doi: 10.1016/j.tim.2011.06.004
Boles, B. R., Thoendel, M., and Singh, P. K. (2004). Self-generated diversity produces ”insurance effects” in biofilm communities. Proc. Natl. Acad. Sci. U.S.A. 101, 16630–16635. doi: 10.1073/pnas.0407460101
Bridier, A., Briandet, R., Bouchez, T., and Jabot, F. (2014). A model-based approach to detect interspecific interactions during biofilm development. Biofouling 30, 761–771. doi: 10.1080/08927014.2014.923409
Bridier, A., Briandet, R., Thomas, V., and Dubois-Brissonnet, F. (2011). Resistance of bacterial biofilms to disinfectants: a review. Biofouling 27, 1017–1032. doi: 10.1080/08927014.2011.626899
Bucci, V., Nadell, C. D., and Xavier, J. B. (2011). The evolution of bacteriocin production in bacterial biofilms. Am. Nat. 178, E162–E173. doi: 10.1086/662668
Carcamo-Oyarce, G., Lumjiaktase, P., Kummerli, R., and Eberl, L. (2015). Quorum sensing triggers the stochastic escape of individual cells from Pseudomonas putida biofilms. Nat. Commun. 6:5945. doi: 10.1038/ncomms6945
Chambers, J. R., and Sauer, K. (2013). Small RNAs and their role in biofilm formation. Trends Microbiol. 21, 39–49. doi: 10.1016/j.tim.2012.10.008
Chan, B. K., and Abedon, S. T. (2015). Bacteriophages and their enzymes in biofilm control. Curr. Pharm. Des. 21, 85–99. doi: 10.2174/1381612820666140905112311
Chen, Y., Gozzi, K., Yan, F., and Chai, Y. (2015). Acetic acid acts as a volatile signal to stimulate bacterial biofilm formation. mBio 6:e00392. doi: 10.1128/mBio.00392-15
Cheow, W. S., Kiew, T. Y., and Hadinoto, K. (2014). Controlled release of Lactobacillus rhamnosus biofilm probiotics from alginate-locust bean gum microcapsules. Carbohydr. Polym. 103, 587–595. doi: 10.1016/j.carbpol.2014.01.036
Conibear, T. C., Collins, S. L., and Webb, J. S. (2009). Role of mutation in Pseudomonas aeruginosa biofilm development. PLoS ONE 4:e6289. doi: 10.1371/journal.pone.0006289
Crabbé, A., De Boever, P., Van Houdt, R., Moors, H., Mergeay, M., and Cornelis, P. (2008). Use of the rotating wall vessel technology to study the effect of shear stress on growth behaviour of Pseudomonas aeruginosa PA01. Environ. Microbiol. 10, 2098–2110. doi: 10.1111/j.1462-2920.2008.01631.x
Debarbieux, L., Leduc, D., Maura, D., Morello, E., Criscuolo, A., Grossi, O., et al. (2010). Bacteriophages can treat and prevent Pseudomonas aeruginosa lung infections. J. Infect. Dis. 201, 1096–1104. doi: 10.1086/651135
Decho, A. (2016). “Unique and baffling aspects of the matrix: EPS syneresis and glass formation during desiccation,” in The Perfect Slime, Microbial extracellular Polymeric Substances (EPS), eds H. C. Flemming, T. R. Neu, and J. Wingender (London: IWA Publishing), 207–226.
Derlon, N., Koch, N., Eugster, B., Posch, T., Pernthaler, J., Pronk, W., et al. (2013). Activity of metazoa governs biofilm structure formation and enhances permeate flux during Gravity-Driven Membrane (GDM) filtration. Water Res. 47, 2085–2095. doi: 10.1016/j.watres.2013.01.033
Derlon, N., Peter-Varbanets, M., Scheidegger, A., Pronk, W., and Morgenroth, E. (2012). Predation influences the structure of biofilm developed on ultrafiltration membranes. Water Res. 46, 3323–3333. doi: 10.1016/j.watres.2012.03.031
Dhami, N. K., Reddy, M. S., and Mukherjee, A. (2014). Application of calcifying bacteria for remediation of stones and cultural heritages. Front. Microbiol. 5:304. doi: 10.3389/fmicb.2014.00304
Dick, J., De Windt, W., De Graef, B., Saveyn, H., Van der Meeren, P., De Belie, N., et al. (2006). Bio-deposition of a calcium carbonate layer on degraded limestone by Bacillus species. Biodegradation 17, 357–367. doi: 10.1007/s10532-005-9006-x
Diggle, S. P., Winzer, K., Chhabra, S. R., Worrall, K. E., Camara, M., and Williams, P. (2003). The Pseudomonas aeruginosa quinolone signal molecule overcomes the cell density-dependency of the quorum sensing hierarchy, regulates rhl-dependent genes at the onset of stationary phase and can be produced in the absence of LasR. Mol. Microbiol. 50, 29–43. doi: 10.1046/j.1365-2958.2003.03672.x
Dingemans, J., Monsieurs, P., Yu, S. H., Crabbe, A., Forstner, K. U., Malfroot, A., et al. (2016). Effect of shear stress on Pseudomonas aeruginosa isolated from the cystic fibrosis lung. mBio 7:e00813-16. doi: 10.1128/mBio.00813-16
Donelli, G., Francolini, I., Romoli, D., Guaglianone, E., Piozzi, A., Ragunath, C., et al. (2007). Synergistic activity of dispersin B and cefamandole nafate in inhibition of staphylococcal biofilm growth on polyurethanes. Antimicrob. Agents Chemother. 51, 2733–2740. doi: 10.1128/AAC.01249-06
Drescher, K., Dunkel, J., Nadell, C. D., van Teeffelen, S., Grnja, I., Wingreen, N. S., et al. (2016). Architectural transitions in Vibrio cholerae biofilms at single-cell resolution. Proc. Natl. Acad. Sci. U.S.A. 113, E2066–E2072. doi: 10.1073/pnas.1601702113
Dubois-Brissonnet, F., Trotier, E., and Briandet, R. (2016). The biofilm lifestyle involves an increase in bacterial membrane saturated fatty acids. Front. Microbiol. 7:1673. doi: 10.3389/fmicb.2016.01673
Dueholm, M. S., and Nielsen, P. H. (2016). “Amyloids - a neglected child of the slime,” in The Perfect Slime, Microbial Extracellular Polymeric Substances (EPS), eds H. C. Flemming, T. R. Neu, and J. Wingender (London: IWA Publishing), 113–133.
Elsholz, A. K., Wacker, S. A., and Losick, R. (2014). Self-regulation of exopolysaccharide production in Bacillus subtilis by a tyrosine kinase. Genes Dev. 28, 1710–1720. doi: 10.1101/gad.246397.114
Epstein, A. K., Pokroy, B., Seminara, A., and Aizenberg, J. (2011). Bacterial biofilm shows persistent resistance to liquid wetting and gas penetration. Proc. Natl. Acad. Sci. U.S.A. 108, 995–1000. doi: 10.1073/pnas.1011033108
Espinosa-Urgel, M., Kolter, R., and Ramos, J. L. (2002). Root colonization by Pseudomonas putida: love at first sight. Microbiology 148(Pt 2), 341–343. doi: 10.1099/00221287-148-2-341
Feliziani, S., Lujan, A. M., Moyano, A. J., Sola, C., Bocco, J. L., Montanaro, P., et al. (2010). Mucoidy, quorum sensing, mismatch repair and antibiotic resistance in Pseudomonas aeruginosa from cystic fibrosis chronic airways infections. PLoS ONE 5:e12669. doi: 10.1371/journal.pone.0012669
Fernandez-Piñar, R., Camara, M., Dubern, J. F., Ramos, J. L., and Espinosa-Urgel, M. (2011). The Pseudomonas aeruginosa quinolone quorum sensing signal alters the multicellular behaviour of Pseudomonas putida KT2440. Res. Microbiol. 162, 773–781. doi: 10.1016/j.resmic.2011.06.013
Finnegan, S., and Percival, S. L. (2015). EDTA: an antimicrobial and antibiofilm agent for use in wound care. Adv. Wound Care 4, 415–421. doi: 10.1089/wound.2014.0577
Flemming, H. C. (2011). The perfect slime. Colloids Surf. B Biointerfaces 86, 251–259. doi: 10.1016/j.colsurfb.2011.04.025
Flemming, H. C., Neu, T. R., and Wozniak, D. J. (2007). The EPS matrix: the ”house of biofilm cells”. J. Bacteriol. 189, 7945–7947. doi: 10.1128/JB.00858-07
Flemming, H. C., and Wingender, J. (2010). The biofilm matrix. Nat. Rev. Microbiol. 8, 623–633. doi: 10.1038/nrmicro2415
Flemming, H. C., Wingender, J., Szewzyk, U., Steinberg, P., Rice, S. A., and Kjelleberg, S. (2016). Biofilms: an emergent form of bacterial life. Nat. Rev. Microbiol. 14, 563–575. doi: 10.1038/nrmicro.2016.94
Fong, J. N., and Yildiz, F. H. (2015). Biofilm matrix proteins. Microbiol. Spectr. 3:MB-0004-2014. doi: 10.1128/microbiolspec.MB-0004-2014
Giaouris, E., Chorianopoulos, N., Doulgeraki, A., and Nychas, G. J. (2013). Co-culture with Listeria monocytogenes within a dual-species biofilm community strongly increases resistance of Pseudomonas putida to benzalkonium chloride. PLoS ONE 8:e77276. doi: 10.1371/journal.pone.0077276
Giaouris, E., Heir, E., Desvaux, M., Hebraud, M., Moretro, T., Langsrud, S., et al. (2015). Intra- and inter-species interactions within biofilms of important foodborne bacterial pathogens. Front. Microbiol. 6:841. doi: 10.3389/fmicb.2015.00841
Greene, C., Wu, J., Rickard, A. H., and Xi, C. (2016). Evaluation of the ability of Acinetobacter baumannii to form biofilms on six different biomedical relevant surfaces. Lett. Appl. Microbiol. 63, 233–239. doi: 10.1111/lam.12627
Guilhen, C., Charbonnel, N., Parisot, N., Gueguen, N., Iltis, A., Forestier, C., et al. (2016). Transcriptional profiling of Klebsiella pneumoniae defines signatures for planktonic, sessile and biofilm-dispersed cells. BMC Genomics 17:237. doi: 10.1186/s12864-016-2557-x
Gutiérrez, O., Juan, C., Pérez, J. L., and Oliver, A. (2004). Lack of association between hypermutation and antibiotic resistance development in Pseudomonas aeruginosa isolates from intensive care unit patients. Antimicrob. Agents Chemother. 48, 3573–3575. doi: 10.1128/AAC.48.9.3573-3575.2004
Guttenplan, S. B., Blair, K. M., and Kearns, D. B. (2010). The EpsE flagellar clutch is bifunctional and synergizes with EPS biosynthesis to promote Bacillus subtilis biofilm formation. PLoS Genet. 6:e1001243. doi: 10.1371/journal.pgen.1001243
Guttenplan, S. B., and Kearns, D. B. (2013). Regulation of flagellar motility during biofilm formation. FEMS Microbiol. Rev. 37, 849–871. doi: 10.1111/1574-6976.12018
Haagensen, J. A., Hansen, S. K., Christensen, B. B., Pamp, S. J., and Molin, S. (2015). Development of spatial distribution patterns by biofilm cells. Appl. Environ. Microbiol. 81, 6120–6128. doi: 10.1128/AEM.01614-15
Habimana, O., Guillier, L., Kulakauskas, S., and Briandet, R. (2011). Spatial competition with Lactococcus lactis in mixed-species continuous-flow biofilms inhibits Listeria monocytogenes growth. Biofouling 27, 1065–1072. doi: 10.1080/08927014.2011.626124
Habimana, O., Nesse, L. L., Moretro, T., Berg, K., Heir, E., Vestby, L. K., et al. (2014). The persistence of Salmonella following desiccation under feed processing environmental conditions: a subject of relevance. Lett. Appl. Microbiol. 59, 464–470. doi: 10.1111/lam.12308
Hanlon, G. W., Denyer, S. P., Olliff, C. J., and Ibrahim, L. J. (2001). Reduction in exopolysaccharide viscosity as an aid to bacteriophage penetration through Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 67, 2746–2753. doi: 10.1128/AEM.67.6.2746-2753.2001
Hannan, S., Ready, D., Jasni, A. S., Rogers, M., Pratten, J., and Roberts, A. P. (2010). Transfer of antibiotic resistance by transformation with eDNA within oral biofilms. FEMS Immunol. Med. Microbiol. 59, 345–349. doi: 10.1111/j.1574-695X.2010.00661.x
Hansen, S. K., Rainey, P. B., Haagensen, J. A., and Molin, S. (2007). Evolution of species interactions in a biofilm community. Nature 445, 533–536. doi: 10.1038/nature05514
Hassett, D. J., Ma, J. F., Elkins, J. G., McDermott, T. R., Ochsner, U. A., West, S. E., et al. (1999). Quorum sensing in Pseudomonas aeruginosa controls expression of catalase and superoxide dismutase genes and mediates biofilm susceptibility to hydrogen peroxide. Mol. Microbiol. 34, 1082–1093. doi: 10.1046/j.1365-2958.1999.01672.x
Hellweger, F. L., Clegg, R. J., Clark, J. R., Plugge, C. M., and Kreft, J. U. (2016). Advancing microbial sciences by individual-based modelling. Nat. Rev. Microbiol. 14, 461–471. doi: 10.1038/nrmicro.2016.62
Hoffman, L. R., D’Argenio, D. A., MacCoss, M. J., Zhang, Z., Jones, R. A., and Miller, S. I. (2005). Aminoglycoside antibiotics induce bacterial biofilm formation. Nature 436, 1171–1175. doi: 10.1038/nature03912
Houry, A., Gohar, M., Deschamps, J., Tischenko, E., Aymerich, S., Gruss, A., et al. (2012). Bacterial swimmers that infiltrate and take over the biofilm matrix. Proc. Natl. Acad. Sci. U.S.A. 109, 13088–13093. doi: 10.1073/pnas.1200791109
Hu, W., Li, L., Sharma, S., Wang, J., McHardy, I., Lux, R., et al. (2012). DNA builds and strengthens the extracellular matrix in Myxococcus xanthus biofilms by interacting with exopolysaccharides. PLoS ONE 7:e51905. doi: 10.1371/journal.pone.0051905
Huang, T. P., Tzeng, D. D., Wong, A. C., Chen, C. H., Lu, K. M., Lee, Y. H., et al. (2012). DNA polymorphisms and biocontrol of Bacillus antagonistic to citrus bacterial canker with indication of the interference of phyllosphere biofilms. PLoS ONE 7:e42124. doi: 10.1371/journal.pone.0042124
Huynh, T. T., McDougald, D., Klebensberger, J., Al Qarni, B., Barraud, N., Rice, S. A., et al. (2012). Glucose starvation-induced dispersal of Pseudomonas aeruginosa biofilms is cAMP and energy dependent. PLoS ONE 7:e42874. doi: 10.1371/journal.pone.0042874
Jaffar, N., Ishikawa, Y., Mizuno, K., Okinaga, T., and Maeda, T. (2016). Mature biofilm degradation by potential probiotics: Aggregatibacter actinomycetemcomitans versus Lactobacillus spp. PLoS ONE 11:e0159466. doi: 10.1371/journal.pone.0159466
Jamali, Z., Aminabadi, N. A., Samiei, M., Sighari Deljavan, A., Shokravi, M., and Shirazi, S. (2016). Impact of chlorhexidine pretreatment followed by probiotic Streptococcus salivarius strain K12 on halitosis in children: a randomised controlled clinical trial. Oral. Health Prev. Dent. 14, 305–313. doi: 10.3290/j.ohpd.a36521
Jiang, Q., Stamatova, I., Kainulainen, V., Korpela, R., and Meurman, J. H. (2016). Interactions between Lactobacillus rhamnosus GG and oral micro-organisms in an in vitro biofilm model. BMC Microbiol. 16:149. doi: 10.1186/s12866-016-0759-7
Justice, S. S., Harrison, A., Becknell, B., and Mason, K. M. (2014). Bacterial differentiation, development, and disease: mechanisms for survival. FEMS Microbiol. Lett. 360, 1–8. doi: 10.1111/1574-6968.12602
Justice, S. S., Hung, C., Theriot, J. A., Fletcher, D. A., Anderson, G. G., Footer, M. J., et al. (2004). Differentiation and developmental pathways of uropathogenic Escherichia coli in urinary tract pathogenesis. Proc. Natl. Acad. Sci. U.S.A. 101, 1333–1338. doi: 10.1073/pnas.0308125100
Justice, S. S., Hunstad, D. A., Cegelski, L., and Hultgren, S. J. (2008). Morphological plasticity as a bacterial survival strategy. Nat. Rev. Microbiol. 6, 162–168. doi: 10.1038/nrmicro1820
Justice, S. S., Hunstad, D. A., Seed, P. C., and Hultgren, S. J. (2006). Filamentation by Escherichia coli subverts innate defenses during urinary tract infection. Proc. Natl. Acad. Sci. U.S.A. 103, 19884–19889. doi: 10.1073/pnas.0606329104
Kalmokoff, M., Lanthier, P., Tremblay, T. L., Foss, M., Lau, P. C., Sanders, G., et al. (2006). Proteomic analysis of Campylobacter jejuni 11168 biofilms reveals a role for the motility complex in biofilm formation. J. Bacteriol. 188, 4312–4320. doi: 10.1128/JB.01975-05
Kaminskaya, A., Pushkareva, V., Moisenovich, M., Stepanova, T., Volkova, N., Romanova, J., et al. (2007). Stimulation of biofilm formation by insertion of Tetrahymena pyriformis wells within Burkholderia cenocepacia biofilms. Mol. Genet. Microbiol. Virol. 22, 186–194. doi: 10.3103/s0891416807040088
Kaplan, J. B., Meyenhofer, M. F., and Fine, D. H. (2003). Biofilm growth and detachment of Actinobacillus actinomycetemcomitans. J. Bacteriol. 185, 1399–1404. doi: 10.1128/JB.185.4.1399-1404.2003
Khemiri, A., Jouenne, T., and Cosette, P. (2016). Proteomics dedicated to biofilmology: what have we learned from a decade of research? Med. Microbiol. Immunol. 205, 1–19. doi: 10.1007/s00430-015-0423-0
Kim, Y. J., and Lee, S. H. (2016). Inhibitory effect of Lactococcus lactis HY 449 on cariogenic biofilm. J. Microbiol. Biotechnol. 26, 1829–1835. doi: 10.4014/jmb.1604.04008
Klein, T., Zihlmann, D., Derlon, N., Isaacson, C., Szivak, I., Weissbrodt, D. G., et al. (2016). Biological control of biofilms on membranes by metazoans. Water Res. 88, 20–29. doi: 10.1016/j.watres.2015.09.050
Koch, G., Yepes, A., Forstner, K. U., Wermser, C., Stengel, S. T., Modamio, J., et al. (2014). Evolution of resistance to a last-resort antibiotic in Staphylococcus aureus via bacterial competition. Cell 158, 1060–1071. doi: 10.1016/j.cell.2014.06.046
Kolodkin-Gal, I., Romero, D., Cao, S., Clardy, J., Kolter, R., and Losick, R. (2010). D-amino acids trigger biofilm disassembly. Science 328, 627–629. doi: 10.1126/science.1188628
Kostakioti, M., Hadjifrangiskou, M., and Hultgren, S. J. (2013). Bacterial biofilms: development, dispersal, and therapeutic strategies in the dawn of the postantibiotic era. Cold Spring Harb. Perspect. Med. 3:a010306. doi: 10.1101/cshperspect.a010306
Kragh, K. N., Hutchison, J. B., Melaugh, G., Rodesney, C., Roberts, A. E., Irie, Y., et al. (2016). Role of multicellular aggregates in biofilm formation. mBio 7:e00237-16. doi: 10.1128/mBio.00237-16
Kröber, M., Verwaaijen, B., Wibberg, D., Winkler, A., Puhler, A., and Schluter, A. (2016). Comparative transcriptome analysis of the biocontrol strain Bacillus amyloliquefaciens FZB42 as response to biofilm formation analyzed by RNA sequencing. J. Biotechnol. 231, 212–223. doi: 10.1016/j.jbiotec.2016.06.013
Król, J. E., Wojtowicz, A. J., Rogers, L. M., Heuer, H., Smalla, K., Krone, S. M., et al. (2013). Invasion of E. coli biofilms by antibiotic resistance plasmids. Plasmid 70, 110–119. doi: 10.1016/j.plasmid.2013.03.003
Kuiper, I., Lagendijk, E. L., Pickford, R., Derrick, J. P., Lamers, G. E., Thomas-Oates, J. E., et al. (2004). Characterization of two Pseudomonas putida lipopeptide biosurfactants, putisolvin I and II, which inhibit biofilm formation and break down existing biofilms. Mol. Microbiol. 51, 97–113. doi: 10.1046/j.1365-2958.2003.03751.x
Lee, S. H., and Baek, D. H. (2014). Effects of Streptococcus thermophilus on volatile sulfur compounds produced by Porphyromonas gingivalis. Arch. Oral. Biol. 59, 1205–1210. doi: 10.1016/j.archoralbio.2014.07.006
Leiman, S. A., May, J. M., Lebar, M. D., Kahne, D., Kolter, R., and Losick, R. (2013). D-amino acids indirectly inhibit biofilm formation in Bacillus subtilis by interfering with protein synthesis. J. Bacteriol. 195, 5391–5395. doi: 10.1128/JB.00975-13
Lembré, P., Di Martino, P., and Vendrely, C. (2014). Amyloid peptides derived from CsgA and FapC modify the viscoelastic properties of biofilm model matrices. Biofouling 30, 415–426. doi: 10.1080/08927014.2014.880112
Lennon, J. T., and Lehmkuhl, B. K. (2016). A trait-based approach to bacterial biofilms in soil. Environ. Microbiol. 18, 2732–2742. doi: 10.1111/1462-2920.13331
Lenz, A. P., Williamson, K. S., Pitts, B., Stewart, P. S., and Franklin, M. J. (2008). Localized gene expression in Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 74, 4463–4471. doi: 10.1128/AEM.00710-08
Lequette, Y., Boels, G., Clarisse, M., and Faille, C. (2010). Using enzymes to remove biofilms of bacterial isolates sampled in the food-industry. Biofouling 26, 421–431. doi: 10.1080/08927011003699535
Liao, S., Klein, M. I., Heim, K. P., Fan, Y., Bitoun, J. P., Ahn, S. J., et al. (2014). Streptococcus mutans extracellular DNA is upregulated during growth in biofilms, actively released via membrane vesicles, and influenced by components of the protein secretion machinery. J. Bacteriol. 196, 2355–2366. doi: 10.1128/JB.01493-14
Limoli, D., Jones, C., and Wozniak, D. (2015). “Bacterial extracellular polysaccharides in biofilm formation and function,” in Microbial Biofilms, 2nd Edn, eds. M. Ghannoum, M. Parsek, M. Whiteley, and P. Mukherjee (Washington, DC: ASM Press), 223–247. doi: 10.1128/microbiolspec.MB-0011-2014
Liu, W., Roder, H. L., Madsen, J. S., Bjarnsholt, T., Sorensen, S. J., and Burmolle, M. (2016). Interspecific bacterial interactions are reflected in multispecies biofilm spatial organization. Front. Microbiol. 7:1366. doi: 10.3389/fmicb.2016.01366
Lopez, D., Vlamakis, H., Losick, R., and Kolter, R. (2009). Cannibalism enhances biofilm development in Bacillus subtilis. Mol. Microbiol. 74, 609–618. doi: 10.1111/j.1365-2958.2009.06882.x
Luján, A. M., Macia, M. D., Yang, L., Molin, S., Oliver, A., and Smania, A. M. (2011). Evolution and adaptation in Pseudomonas aeruginosa biofilms driven by mismatch repair system-deficient mutators. PLoS ONE 6:e27842. doi: 10.1371/journal.pone.0027842
Macià, M. D., Perez, J. L., Molin, S., and Oliver, A. (2011). Dynamics of mutator and antibiotic-resistant populations in a pharmacokinetic/pharmacodynamic model of Pseudomonas aeruginosa biofilm treatment. Antimicrob. Agents Chemother. 55, 5230–5237. doi: 10.1128/AAC.00617-11
Madsen, J. S., Burmolle, M., Hansen, L. H., and Sorensen, S. J. (2012). The interconnection between biofilm formation and horizontal gene transfer. FEMS Immunol. Med. Microbiol. 65, 183–195. doi: 10.1111/j.1574-695X.2012.00960.x
Mai-Prochnow, A., Evans, F., Dalisay-Saludes, D., Stelzer, S., Egan, S., James, S., et al. (2004). Biofilm development and cell death in the marine bacterium Pseudoalteromonas tunicata. Appl. Environ. Microbiol. 70, 3232–3238. doi: 10.1128/AEM.70.6.3232-3238.2004
Maisetta, G., Grassi, L., Di Luca, M., Bombardelli, S., Medici, C., Brancatisano, F. L., et al. (2016). Anti-biofilm properties of the antimicrobial peptide temporin 1Tb and its ability, in combination with EDTA, to eradicate Staphylococcus epidermidis biofilms on silicone catheters. Biofouling 32, 787–800. doi: 10.1080/08927014.2016.1194401
Mangalappalli-Illathu, A. K., Vidovic, S., and Korber, D. R. (2008). Differential adaptive response and survival of Salmonella enterica serovar enteritidis planktonic and biofilm cells exposed to benzalkonium chloride. Antimicrob. Agents Chemother. 52, 3669–3680. doi: 10.1128/AAC.00073-08
Mann, E. E., Rice, K. C., Boles, B. R., Endres, J. L., Ranjit, D., Chandramohan, L., et al. (2009). Modulation of eDNA release and degradation affects Staphylococcus aureus biofilm maturation. PLoS ONE 4:e5822. doi: 10.1371/journal.pone.0005822
Marchal, M., Briandet, R., Halter, D., Koechler, S., DuBow, M. S., Lett, M. C., et al. (2011). Subinhibitory arsenite concentrations lead to population dispersal in Thiomonas sp. PLoS ONE 6:e23181. doi: 10.1371/journal.pone.0023181
Mariani, C., Oulahal, N., Chamba, J. F., Dubois-Brissonnet, F., Notz, E., and Briandet, R. (2011). Inhibition of Listeria monocytogenes by resident biofilms present on wooden shelves used for cheese ripening. Food Control 22, 1357–1362. doi: 10.1016/j.foodcont.2011.02.012
Marques, C. N., Davies, D. G., and Sauer, K. (2015). Control of biofilms with the fatty acid signaling molecule cis-2-decenoic acid. Pharmaceuticals 8, 816–835. doi: 10.3390/ph8040816
Martin, M., Holscher, T., Dragos, A., Cooper, V. S., and Kovacs, A. T. (2016). Laboratory evolution of microbial interactions in bacterial biofilms. J. Bacteriol. 198, 2564–2571. doi: 10.1128/JB.01018-15
Martinez, L. C., and Vadyvaloo, V. (2014). Mechanisms of post-transcriptional gene regulation in bacterial biofilms. Front. Cell Infect. Microbiol. 4:38. doi: 10.3389/fcimb.2014.00038
Masdea, L., Kulik, E. M., Hauser-Gerspach, I., Ramseier, A. M., Filippi, A., and Waltimo, T. (2012). Antimicrobial activity of Streptococcus salivarius K12 on bacteria involved in oral malodour. Arch. Oral. Biol. 57, 1041–1047. doi: 10.1016/j.archoralbio.2012.02.011
Maura, D., and Debarbieux, L. (2011). Bacteriophages as twenty-first century antibacterial tools for food and medicine. Appl. Microbiol. Biotechnol. 90, 851–859. doi: 10.1007/s00253-011-3227-1
Meng, X., Shi, Y., Ji, W., Meng, X., Zhang, J., Wang, H., et al. (2011). Application of a bacteriophage lysin to disrupt biofilms formed by the animal pathogen Streptococcus suis. Appl. Environ. Microbiol. 77, 8272–8279. doi: 10.1128/AEM.05151-11
Micallef, R., Vella, D., Sinagra, E., and Zammit, G. (2016). Biocalcifying Bacillus subtilis cells effectively consolidate deteriorated Globigerina limestone. J. Ind. Microbiol. Biotechnol. 43, 941–952. doi: 10.1007/s10295-016-1768-0
Molin, S., and Tolker-Nielsen, T. (2003). Gene transfer occurs with enhanced efficiency in biofilms and induces enhanced stabilisation of the biofilm structure. Curr. Opin. Biotechnol. 14, 255–261. doi: 10.1016/S0958-1669(03)00036-3
Monds, R. D., and O’Toole, G. A. (2009). The developmental model of microbial biofilms: ten years of a paradigm up for review. Trends Microbiol. 17, 73–87. doi: 10.1016/j.tim.2008.11.001
Moreno-Paz, M., Gomez, M. J., Arcas, A., and Parro, V. (2010). Environmental transcriptome analysis reveals physiological differences between biofilm and planktonic modes of life of the iron oxidizing bacteria Leptospirillum spp. in their natural microbial community. BMC Genomics 11:404. doi: 10.1186/1471-2164-11-404
Murphy, K., Park, A. J., Hao, Y., Brewer, D., Lam, J. S., and Khursigara, C. M. (2014). Influence of O polysaccharides on biofilm development and outer membrane vesicle biogenesis in Pseudomonas aeruginosa PAO1. J. Bacteriol. 196, 1306–1317. doi: 10.1128/JB.01463-13
Muszanska, A. K., Nejadnik, M. R., Chen, Y., van den Heuvel, E. R., Busscher, H. J., van der Mei, H. C., et al. (2012). Bacterial adhesion forces with substratum surfaces and the susceptibility of biofilms to antibiotics. Antimicrob. Agents Chemother. 56, 4961–4964. doi: 10.1128/AAC.00431-12
Nadell, C. D., Drescher, K., and Foster, K. R. (2016). Spatial structure, cooperation and competition in biofilms. Nat. Rev. Microbiol. 14, 589–600. doi: 10.1038/nrmicro.2016.84
Nadell, C. D., Foster, K. R., and Xavier, J. B. (2010). Emergence of spatial structure in cell groups and the evolution of cooperation. PLoS Comput. Biol. 6:e1000716. doi: 10.1371/journal.pcbi.1000716
Nadell, C. D., Xavier, J. B., and Foster, K. R. (2009). The sociobiology of biofilms. FEMS Microbiol. Rev. 33, 206–224. doi: 10.1111/j.1574-6976.2008.00150.x
Nale, J. Y., Chutia, M., Carr, P., Hickenbotham, P. T., and Clokie, M. R. (2016). ‘Get in early’; biofilm and wax moth (Galleria mellonella) models reveal new insights into the therapeutic potential of Clostridium difficile Bacteriophages. Front. Microbiol. 7:1383. doi: 10.3389/fmicb.2016.01383
Neu, T. R., and Lawrence, J. R. (2016). “The extracellular matrix - an intractable part of biofilm systems,” in The Perfect Slime, Microbial Extracellular Polymeric Substances (EPS), eds H. C. Flemming, T. R. Neu, and J. Wingender (London: IWA Publishing), 25–60.
Nguyen, U. T., and Burrows, L. L. (2014). DNase I and proteinase K impair Listeria monocytogenes biofilm formation and induce dispersal of pre-existing biofilms. Int. J. Food Microbiol. 187, 26–32. doi: 10.1016/j.ijfoodmicro.2014.06.025
Nijland, R., and Burgess, J. G. (2010). Bacterial olfaction. Biotechnol. J. 5, 974–977. doi: 10.1002/biot.201000174
Oliver, A., Baquero, F., and Blazquez, J. (2002). The mismatch repair system (mutS, mutL and uvrD genes) in Pseudomonas aeruginosa: molecular characterization of naturally occurring mutants. Mol. Microbiol. 43, 1641–1650. doi: 10.1046/j.1365-2958.2002.02855.x
Palmer, R. J. Jr., Kazmerzak, K., Hansen, M. C., and Kolenbrander, P. E. (2001). Mutualism versus independence: strategies of mixed-species oral biofilms in vitro using saliva as the sole nutrient source. Infect. Immun. 69, 5794–5804. doi: 10.1128/IAI.69.9.5794-5804.2001
Pamp, S. J., Sternberg, C., and Tolker-Nielsen, T. (2009). Insight into the microbial multicellular lifestyle via flow-cell technology and confocal microscopy. Cytometry A 75, 90–103. doi: 10.1002/cyto.a.20685
Pande, S., Kaftan, F., Lang, S., Svatos, A., Germerodt, S., and Kost, C. (2016). Privatization of cooperative benefits stabilizes mutualistic cross-feeding interactions in spatially structured environments. ISME J. 10, 1413–1423. doi: 10.1038/ismej.2015.212
Pandin, C., Le Coq, D., Canette, A., Aymerich, S., and Briandet, R. (2017). Should the biofilm mode of life be taken into consideration for microbial biocontrol agents? Microb. Biotechnol. 10, 719–734. doi: 10.1111/1751-7915.12693
Perchat, S., Talagas, A., Poncet, S., Lazar, N., Li de la Sierra-Gallay, I., Gohar, M., et al. (2016). How quorum sensing connects sporulation to necrotrophism in Bacillus thuringiensis. PLoS Pathog. 12:e1005779. doi: 10.1371/journal.ppat.1005779
Perez-Nunez, D., Briandet, R., David, B., Gautier, C., Renault, P., Hallet, B., et al. (2011). A new morphogenesis pathway in bacteria: unbalanced activity of cell wall synthesis machineries leads to coccus-to-rod transition and filamentation in ovococci. Mol. Microbiol. 79, 759–771. doi: 10.1111/j.1365-2958.2010.07483.x
Perrin, C., Briandet, R., Jubelin, G., Lejeune, P., Mandrand-Berthelot, M. A., Rodrigue, A., et al. (2009). Nickel promotes biofilm formation by Escherichia coli K-12 strains that produce curli. Appl. Environ. Microbiol. 75, 1723–1733. doi: 10.1128/AEM.02171-08
Peterson, B. W., He, Y., Ren, Y., Zerdoum, A., Libera, M. R., Sharma, P. K., et al. (2015). Viscoelasticity of biofilms and their recalcitrance to mechanical and chemical challenges. FEMS Microbiol. Rev. 39, 234–245. doi: 10.1093/femsre/fuu008
Piard, J. C., and Briandet, R. (2016). Lactic Acid Bacteria Biofilms. Biotechnology of Lactic Acid Bacteria: Novel Applications, 2nd Edn. Hoboken, NJ: Wiley-Blackwell, 341–361.
Pinna, D. (2014). Biofilms and lichens on stone monuments: do they damage or protect? Front. Microbiol. 5:133. doi: 10.3389/fmicb.2014.00133
Poltak, S. R., and Cooper, V. S. (2011). Ecological succession in long-term experimentally evolved biofilms produces synergistic communities. ISME J. 5, 369–378. doi: 10.1038/ismej.2010.136
Qayyum, S., Sharma, D., Bisht, D., and Khan, A. U. (2016). Protein translation machinery holds a key for transition of planktonic cells to biofilm state in Enterococcus faecalis: A proteomic approach. Biochem. Biophys. Res. Commun. 474, 652–659. doi: 10.1016/j.bbrc.2016.04.145
Qian, Z., Stoodley, P., and Pitt, W. G. (1996). Effect of low-intensity ultrasound upon biofilm structure from confocal scanning laser microscopy observation. Biomaterials 17, 1975–1980. doi: 10.1016/0142-9612(96)00022-1
Rendueles, O., and Ghigo, J. M. (2015). Mechanisms of competition in biofilm communities. Microbiol. spectr. 3:MB-0009-2014. doi: 10.1128/microbiolspec.MB-0009-2014
Rendueles, O., Beloin, C., Latour-Lambert, P., and Ghigo, J. M. (2014). A new biofilm-associated colicin with increased efficiency against biofilm bacteria. ISME J. 8, 1275–1288. doi: 10.1038/ismej.2013.238
Resch, A., Leicht, S., Saric, M., Pasztor, L., Jakob, A., Gotz, F., et al. (2006). Comparative proteome analysis of Staphylococcus aureus biofilm and planktonic cells and correlation with transcriptome profiling. Proteomics 6, 1867–1877. doi: 10.1002/pmic.200500531
Reuter, M., Mallett, A., Pearson, B. M., and van Vliet, A. H. (2010). Biofilm formation by Campylobacter jejuni is increased under aerobic conditions. Appl. Environ. Microbiol. 76, 2122–2128. doi: 10.1128/AEM.01878-09
Rhodes, C. J. (2016). The 2015 Paris climate change conference: COP21. Sci. Prog. 99, 97–104. doi: 10.3184/003685016X14528569315192
Rieu, A., Aoudia, N., Jego, G., Chluba, J., Yousfi, N., Briandet, R., et al. (2014). The biofilm mode of life boosts the anti-inflammatory properties of Lactobacillus. Cell. Microbiol. 16, 1836–1853. doi: 10.1111/cmi.12331
Romani, A. M., Borrego, C. M., Diaz-Villanueva, V., Freixa, A., Gich, F., and Ylla, I. (2014). Shifts in microbial community structure and function in light- and dark-grown biofilms driven by warming. Environ. Microbiol. 16, 2550–2567. doi: 10.1111/1462-2920.12428
Romero, D., Sanabria-Valentin, E., Vlamakis, H., and Kolter, R. (2013). Biofilm inhibitors that target amyloid proteins. Chem. Biol. 20, 102–110. doi: 10.1016/j.chembiol.2012.10.021
Romling, U., Galperin, M. Y., and Gomelsky, M. (2013). Cyclic di-GMP: the first 25 years of a universal bacterial second messenger. Microbiol. Mol. Biol. Rev. 77, 1–52. doi: 10.1128/MMBR.00043-12
Sabater, S., Timoner, X., Borrego, C., and Acuña, V. (2016). Stream biofilm responses to flow intermittency: from cells to ecosystems. Front. Environ. Sci. 4:14. doi: 10.3389/fenvs.2016.00014
Safari, A., Habimana, O., Allen, A., and Casey, E. (2014). The significance of calcium ions on Pseudomonas fluorescens biofilms - a structural and mechanical study. Biofouling 30, 859–869. doi: 10.1080/08927014.2014.938648
Sauer, K. (2003). The genomics and proteomics of biofilm formation. Genome Biol. 4:219. doi: 10.1186/gb-2003-4-6-219
Sauer, K., Camper, A. K., Ehrlich, G. D., Costerton, J. W., and Davies, D. G. (2002). Pseudomonas aeruginosa displays multiple phenotypes during development as a biofilm. J. Bacteriol. 184, 1140–1154. doi: 10.1128/jb.184.4.1140-1154.2002
Sauer, K., Cullen, M. C., Rickard, A. H., Zeef, L. A., Davies, D. G., and Gilbert, P. (2004). Characterization of nutrient-induced dispersion in Pseudomonas aeruginosa PAO1 biofilm. J. Bacteriol. 186, 7312–7326. doi: 10.1128/JB.186.21.7312-7326.2004
Savage, V. J., Chopra, I., and O’Neill, A. J. (2013). Staphylococcus aureus biofilms promote horizontal transfer of antibiotic resistance. Antimicrob. Agents Chemother. 57, 1968–1970. doi: 10.1128/AAC.02008-12
Schilcher, K., Andreoni, F., Dengler Haunreiter, V., Seidl, K., Hasse, B., and Zinkernagel, A. S. (2016). Modulation of Staphylococcus aureus biofilm matrix by subinhibitory concentrations of clindamycin. Antimicrob. Agents Chemother. 60, 5957–5967. doi: 10.1128/AAC.00463-16
Schooling, S. R., and Beveridge, T. J. (2006). Membrane vesicles: an overlooked component of the matrices of biofilms. J. Bacteriol. 188, 5945–5957. doi: 10.1128/JB.00257-06
Schwartz, K., Ganesan, M., Payne, D. E., Solomon, M. J., and Boles, B. R. (2016). Extracellular DNA facilitates the formation of functional amyloids in Staphylococcus aureus biofilms. Mol. Microbiol. 99, 123–134. doi: 10.1111/mmi.13219
Schwechheimer, C., and Kuehn, M. J. (2015). Outer-membrane vesicles from Gram-negative bacteria: biogenesis and functions. Nat. Rev. Microbiol. 13, 605–619. doi: 10.1038/nrmicro3525
Serra, D. O., Richter, A. M., Klauck, G., Mika, F., and Hengge, R. (2013). Microanatomy at cellular resolution and spatial order of physiological differentiation in a bacterial biofilm. mBio 4:e100103-13. doi: 10.1128/mBio.00103-13
Servick, K. (2016). DRUG DEVELOPMENT. Beleaguered phage therapy trial presses on. Science 352:1506. doi: 10.1126/science.352.6293.1506
Shafreen, R. M., Srinivasan, S., Manisankar, P., and Pandian, S. K. (2011). Biofilm formation by Streptococcus pyogenes: modulation of exopolysaccharide by fluoroquinolone derivatives. J. Biosci. Bioeng. 112, 345–350. doi: 10.1016/j.jbiosc.2011.06.013
Shank, E. A., Klepac-Ceraj, V., Collado-Torres, L., Powers, G. E., Losick, R., and Kolter, R. (2011). Interspecies interactions that result in Bacillus subtilis forming biofilms are mediated mainly by members of its own genus. Proc. Natl. Acad. Sci. U.S.A. 108, E1236–E1243. doi: 10.1073/pnas.1103630108
Shemesh, M., Kolter, R., and Losick, R. (2010). The biocide chlorine dioxide stimulates biofilm formation in Bacillus subtilis by activation of the histidine kinase KinC. J. Bacteriol. 192, 6352–6356. doi: 10.1128/JB.01025-10
Singer, G., Besemer, K., Schmitt-Kopplin, P., Hodl, I., and Battin, T. J. (2010). Physical heterogeneity increases biofilm resource use and its molecular diversity in stream mesocosms. PLoS ONE 5:e9988. doi: 10.1371/journal.pone.0009988
Skillman, L. C., Sutherland, I. W., and Jones, M. V. (1998). The role of exopolysaccharides in dual species biofilm development. J. Appl. Microbiol. 85 (Suppl. 1), 13S–18S. doi: 10.1111/j.1365-2672.1998.tb05278.x
Smith, W. P., Davit, Y., Osborne, J. M., Kim, W., Foster, K. R., and Pitt-Francis, J. M. (2017). Cell morphology drives spatial patterning in microbial communities. Proc. Natl. Acad. Sci. U.S.A. 114, E280–E286. doi: 10.1073/pnas.1613007114
Solano, C., Echeverz, M., and Lasa, I. (2014). Biofilm dispersion and quorum sensing. Curr. Opin. Microbiol. 18, 96–104. doi: 10.1016/j.mib.2014.02.008
Soto, S. M. (2013). Role of efflux pumps in the antibiotic resistance of bacteria embedded in a biofilm. Virulence 4, 223–229. doi: 10.4161/viru.23724
Srivastava, D., Harris, R. C., and Waters, C. M. (2011). Integration of cyclic di-GMP and quorum sensing in the control of vpsT and aphA in Vibrio cholerae. J. Bacteriol. 193, 6331–6341. doi: 10.1128/JB.05167-11
Srivastava, D., and Waters, C. M. (2012). A tangled web: regulatory connections between quorum sensing and cyclic Di-GMP. J. Bacteriol. 194, 4485–4493. doi: 10.1128/JB.00379-12
Stewart, E. J., Satorius, A. E., Younger, J. G., and Solomon, M. J. (2013). Role of environmental and antibiotic stress on Staphylococcus epidermidis biofilm microstructure. Langmuir 29, 7017–7024. doi: 10.1021/la401322k
Stewart, P. S., and Franklin, M. J. (2008). Physiological heterogeneity in biofilms. Nat. Rev. Microbiol. 6, 199–210. doi: 10.1038/nrmicro1838
Stipetic, L. H., Dalby, M. J., Davies, R. L., Morton, F. R., Ramage, G., and Burgess, K. E. (2016). A novel metabolomic approach used for the comparison of Staphylococcus aureus planktonic cells and biofilm samples. Metabolomics 12:75. doi: 10.1007/s11306-016-1002-0
Stoodley, P., Boyle, J. D., DeBeer, D., and Lappin-Scott, H. M. (1999). Evolving perspectives of biofilm structure. Biofouling 14, 75–90. doi: 10.1080/08927019909378398
Stoodley, P., Dodds, I., Boyle, J. D., and Lappin-Scott, H. M. (1998). Influence of hydrodynamics and nutrients on biofilm structure. J. Appl. Microbiol. 85(Suppl. 1), 19S–28S. doi: 10.1111/j.1365-2672.1998.tb05279.x
Stubbendieck, R. M., Vargas-Bautista, C., and Straight, P. D. (2016). Bacterial communities: interactions to scale. Front. Microbiol. 7:1234. doi: 10.3389/fmicb.2016.01234
Sutherland, I. W. (2001). The biofilm matrix–an immobilized but dynamic microbial environment. Trends Microbiol. 9, 222–227. doi: 10.1016/S0966-842X(01)02012-1
Toyofuku, M., Inaba, T., Kiyokawa, T., Obana, N., Yawata, Y., and Nomura, N. (2015). Environmental factors that shape biofilm formation. Biosci. Biotechnol. Biochem. 80, 7–12. doi: 10.1080/09168451.2015.1058701
Traverse, C. C., Mayo-Smith, L. M., Poltak, S. R., and Cooper, V. S. (2013). Tangled bank of experimentally evolved Burkholderia biofilms reflects selection during chronic infections. Proc. Natl. Acad. Sci. U.S.A. 110, E250–E259. doi: 10.1073/pnas.1207025110
Turonova, H., Briandet, R., Rodrigues, R., Hernould, M., Hayek, N., Stintzi, A., et al. (2015). Biofilm spatial organization by the emerging pathogen Campylobacter jejuni: comparison between NCTC 11168 and 81-176 strains under microaerobic and oxygen-enriched conditions. Front. Microbiol. 6:709. doi: 10.3389/fmicb.2015.00709
Ueda, A., and Wood, T. K. (2009). Connecting quorum sensing, c-di-GMP, pel polysaccharide, and biofilm formation in Pseudomonas aeruginosa through tyrosine phosphatase TpbA (PA3885). PLoS Pathog. 5:e1000483. doi: 10.1371/journal.ppat.1000483
Valentini, M., and Filloux, A. (2016). Biofilms and cyclic di-GMP (c-di-GMP) signaling: lessons from Pseudomonas aeruginosa and other bacteria. J. Biol. Chem. 291, 12547–12555. doi: 10.1074/jbc.R115.711507
Valle, J., Da Re, S., Henry, N., Fontaine, T., Balestrino, D., Latour-Lambert, P., et al. (2006). Broad-spectrum biofilm inhibition by a secreted bacterial polysaccharide. Proc. Natl. Acad. Sci. U.S.A. 103, 12558–12563. doi: 10.1073/pnas.0605399103
van Hoek, M. L. (2013). Biofilms: an advancement in our understanding of Francisella species. Virulence 4, 833–846. doi: 10.4161/viru.27023
Van Meervenne, E., De Weirdt, R., Van Coillie, E., Devlieghere, F., Herman, L., and Boon, N. (2014). Biofilm models for the food industry: hot spots for plasmid transfer? Pathog. Dis. 70, 332–338. doi: 10.1111/2049-632X.12134
Vilain, S., and Brozel, V. S. (2006). Multivariate approach to comparing whole-cell proteomes of Bacillus cereus indicates a biofilm-specific proteome. J. Proteome Res. 5, 1924–1930. doi: 10.1021/pr050402b
Vuotto, C., Longo, F., and Donelli, G. (2014). Probiotics to counteract biofilm-associated infections: promising and conflicting data. Int. J. Oral Sci. 6, 189–194. doi: 10.1038/ijos.2014.52
Waite, R. D., Paccanaro, A., Papakonstantinopoulou, A., Hurst, J. M., Saqi, M., Littler, E., et al. (2006). Clustering of Pseudomonas aeruginosa transcriptomes from planktonic cultures, developing and mature biofilms reveals distinct expression profiles. BMC Genomics 7:162. doi: 10.1186/1471-2164-7-162
Wang, W., Chanda, W., and Zhong, M. (2015). The relationship between biofilm and outer membrane vesicles: a novel therapy overview. FEMS Microbiol. Lett. 362:fnv117. doi: 10.1093/femsle/fnv117
Watnick, P., and Kolter, R. (2000). Biofilm, city of microbes. J. Bacteriol. 182, 2675–2679. doi: 10.1128/JB.182.10.2675-2679.2000
Webb, J. S., Thompson, L. S., James, S., Charlton, T., Tolker-Nielsen, T., Koch, B., et al. (2003). Cell death in Pseudomonas aeruginosa biofilm development. J. Bacteriol. 185, 4585–4592. doi: 10.1128/JB.185.15.4585-4592.2003
Werner, E., Roe, F., Bugnicourt, A., Franklin, M. J., Heydorn, A., Molin, S., et al. (2004). Stratified growth in Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 70, 6188–6196. doi: 10.1128/AEM.70.10.6188-6196.2004
Whiteley, M., Bangera, M. G., Bumgarner, R. E., Parsek, M. R., Teitzel, G. M., Lory, S., et al. (2001). Gene expression in Pseudomonas aeruginosa biofilms. Nature 413, 860–864. doi: 10.1038/35101627
Widder, S., Allen, R. J., Pfeiffer, T., Curtis, T. P., Wiuf, C., Sloan, W. T., et al. (2016). Challenges in microbial ecology: building predictive understanding of community function and dynamics. ISME J. 10, 2557–2568. doi: 10.1038/ismej.2016.45
Wong, H. S., Maker, G. L., Trengove, R. D., and O’Handley, R. M. (2015). Gas chromatography-mass spectrometry-based metabolite profiling of Salmonella enterica serovar Typhimurium differentiates between biofilm and planktonic phenotypes. Appl. Environ. Microbiol. 81, 2660–2666. doi: 10.1128/AEM.03658-14
Wright, A., Hawkins, C. H., Anggard, E. E., and Harper, D. R. (2009). A controlled clinical trial of a therapeutic bacteriophage preparation in chronic otitis due to antibiotic-resistant Pseudomonas aeruginosa; a preliminary report of efficacy. Clin. Otolaryngol. 34, 349–357. doi: 10.1111/j.1749-4486.2009.01973.x
Xu, J., Li, M., He, Q., Sun, X., Zhou, X., Su, Z., et al. (2017). Effect of flow rate on growth and oxygen consumption of biofilm in gravity sewer. Environ. Sci. Pollut. Res. Int. 24, 427–435. doi: 10.1007/s11356-016-7710-1
Yan, J., Nadell, C. D., and Bassler, B. L. (2017). Environmental fluctuation governs selection for plasticity in biofilm production. ISME J. 11, 1569–1577. doi: 10.1038/ismej.2017.33
Yan, J., Sharo, A. G., Stone, H. A., Wingreen, N. S., and Bassler, B. L. (2016). Vibrio cholerae biofilm growth program and architecture revealed by single-cell live imaging. Proc. Natl. Acad. Sci. U.S.A. 113, E5337–E5343. doi: 10.1073/pnas.1611494113
Yang, Y., Sreenivasan, P. K., Subramanyam, R., and Cummins, D. (2006). Multiparameter assessments to determine the effects of sugars and antimicrobials on a polymicrobial oral biofilm. Appl. Environ. Microbiol. 72, 6734–6742. doi: 10.1128/AEM.01013-06
Ye, Y., Jiao, R., Gao, J., Li, H., Ling, N., Wu, Q., et al. (2016). Proteins involved in responses to biofilm and planktonic modes in Cronobacter sakazakii. LWT Food Sci. Technol. 65, 1093–1099. doi: 10.1016/j.lwt.2015.09.039
Yonezawa, H., Osaki, T., Woo, T., Kurata, S., Zaman, C., Hojo, F., et al. (2011). Analysis of outer membrane vesicle protein involved in biofilm formation of Helicobacter pylori. Anaerobe 17, 388–390. doi: 10.1016/j.anaerobe.2011.03.020
Zeriouh, H., de Vicente, A., Perez-Garcia, A., and Romero, D. (2014). Surfactin triggers biofilm formation of Bacillus subtilis in melon phylloplane and contributes to the biocontrol activity. Environ. Microbiol. 16, 2196–2211. doi: 10.1111/1462-2920.12271
Zhang, L., and Mah, T. F. (2008). Involvement of a novel efflux system in biofilm-specific resistance to antibiotics. J. Bacteriol. 190, 4447–4452. doi: 10.1128/JB.01655-07
Zhou, H., Luo, C., Fang, X., Xiang, Y., Wang, X., Zhang, R., et al. (2016). Loss of GltB inhibits biofilm formation and biocontrol efficiency of Bacillus subtilis Bs916 by altering the production of gamma-polyglutamate and three lipopeptides. PLoS ONE 11:e0156247. doi: 10.1371/journal.pone.0156247
Keywords: microbial biofilm, spatial dynamic, structure/function, adaptative response
Citation: Bridier A, Piard J-C, Pandin C, Labarthe S, Dubois-Brissonnet F and Briandet R (2017) Spatial Organization Plasticity as an Adaptive Driver of Surface Microbial Communities. Front. Microbiol. 8:1364. doi: 10.3389/fmicb.2017.01364
Received: 30 March 2017; Accepted: 05 July 2017;
Published: 20 July 2017.
Edited by:
Sara María Soto, ISGlobal, SpainReviewed by:
Akos T. Kovacs, Technical University of Denmark, DenmarkGiovanna Batoni, University of Pisa, Italy
Copyright © 2017 Bridier, Piard, Pandin, Labarthe, Dubois-Brissonnet and Briandet. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Romain Briandet, cm9tYWluLmJyaWFuZGV0QGlucmEuZnI=
 Arnaud Bridier
Arnaud Bridier Jean-Christophe Piard2
Jean-Christophe Piard2 Caroline Pandin
Caroline Pandin Simon Labarthe
Simon Labarthe Romain Briandet
Romain Briandet