- 1Department of Biological Sciences, Virginia Institute of Marine Science, College of William & Mary, Williamsburg, VA, United States
- 2Department of Biology, Romberg Tiburon Center for Environmental Studies, San Francisco State University, Tiburon, CA, United States
- 3Ocean Sciences, University of California, Santa Cruz, Santa Cruz, CA, United States
- 4Department of Ecology, Environment and Plant Sciences, Stockholm University, Stockholm, Sweden
Diazotrophic cyanobacteria, those capable of fixing di-nitrogen (N2), are considered one of the major sources of new nitrogen (N) in the oligotrophic tropical ocean, but direct incorporation of diazotrophic N into food webs has not been fully examined. In the Amazon River-influenced western tropical North Atlantic (WTNA), diatom diazotroph associations (DDAs) and the filamentous colonial diazotrophs Trichodesmium have seasonally high abundances. We sampled epipelagic mesozooplankton in the Amazon River plume and WTNA in May–June 2010 to investigate direct grazing by mesozooplankton on two DDA populations: Richelia associated with Rhizosolenia diatoms (het-1) and Hemiaulus diatoms (het-2), and on Trichodesmium using highly specific qPCR assays targeting nitrogenase genes (nifH). Both DDAs and Trichodesmium occurred in zooplankton gut contents, with higher detection of het-2 predominantly in calanoid copepods (2.33–16.76 nifH copies organism-1). Abundance of Trichodesmium was low (2.21–4.03 nifH copies organism-1), but they were consistently detected at high salinity stations (>35) in calanoid copepods. This suggests direct grazing on DDAs, Trichodesmium filaments and colonies, or consumption as part of sinking aggregates, is common. In parallel with the qPCR approach, a next generation sequencing analysis of 16S rRNA genes identified that cyanobacterial assemblage associated with zooplankton guts was dominated by the non-diazotrophic unicellular phylotypes Synechococcus (56%) and Prochlorococcus (26%). However, in two separate calanoid copepod samples, two unicellular diazotrophs Candidatus Atelocyanobacterium thalassa (UCYN-A) and Crocosphaera watsonii (UCYN-B) were present, respectively, as a small component of cyanobacterial assemblages (<2%). This study represents the first evidence of consumption of DDAs, Trichodesmium, and unicellular cyanobacteria by calanoid copepods in an area of the WTNA known for high carbon export. These diazotroph populations are quantitatively important in the global N budget, widespread and hence, the next step is to accurately quantify grazing. Nonetheless, these results highlight a direct pathway of diazotrophic N into the food web and have important implications for biogeochemical cycles, particularly oligotrophic regions where N2 fixation is the main source of new nitrogen.
Introduction
Primary production in the marine environment is mostly limited by nitrogen availability (Gruber, 2008). In the open ocean dissolved inorganic nitrogen (DIN) is rare, diazotrophic organisms, those able to utilize N2 through the process of biological N2 fixation, play a significant role as drivers of primary production by provision of new nitrogen (N) (Dugdale and Goering, 1967). Often the most abundant and best investigated of open ocean diazotrophs are cyanobacteria. The non-heterocyst forming, filamentous cyanobacterium Trichodesmium has been a major focus and is especially dominant throughout the tropical and subtropical oceans. Estimations of N2 fixation by Trichodesmium vary, but globally it is a significant source of new N to the open ocean (Capone et al., 1997; Capone, 2001; Sohm et al., 2011b) through varied pathways including exudation (Glibert and Bronk, 1994; Bronk and Steinberg, 2008), programmed cell death (Berman-Frank et al., 2004) and viral lysis (Hewson et al., 2004). Diatom diazotroph associations (DDAs) are also of considerable interest because they are capable of expansive blooms and high rates of N2 fixation (Carpenter et al., 1999; Foster et al., 2007; Subramaniam et al., 2008; Villareal et al., 2012). High densities, including blooms, have been observed in many tropical river plumes including the Amazon (Foster et al., 2007; Subramaniam et al., 2008), Congo (Foster et al., 2009), and Mekong (Grosse et al., 2010; Bombar et al., 2011). Blooms of DDAs are also important for enhancing carbon export from surface waters (Cooley and Yager, 2006; Subramaniam et al., 2008; Karl et al., 2012; Yeung et al., 2012).
More recently other diazotrophs including Archaea, heterotrophic bacteria, and several lineages of unicellular cyanobacteria have been identified as equally important N2 fixers in the open ocean (Zehr et al., 1998, 2003; Leigh, 2000; Goebel et al., 2010; Halm et al., 2012; Thompson et al., 2012). Work with the prior two is still limited (Sohm et al., 2011b) but unicellular cyanobacteria have broad distributions, and thus the range of N2 fixation now includes areas of the world’s ocean outside the tropical and subtropical latitudes (e.g., cooler temperate regions) (Needoba et al., 2007; Goebel et al., 2010; Moisander et al., 2010; Sohm et al., 2011b; Farnelid et al., 2016). High N2-fixation rates have been directly measured (Martínez-Pérez et al., 2016) for unicellular diazotrophs that are comparable to previous estimates for Trichodesmium (Falcon et al., 2004; Montoya et al., 2004) indicating an additional source of diazotrophically derived nitrogen available in the food web.
However, less is known about the impacts of N2-fixers on secondary production. For example, compared to other dominant primary producers, grazing by zooplankton is not considered a major pathway for new N to enter the food web from Trichodesmium. Toxicity and unpalatability of Trichodesmium are thought to be the major deterrents for grazing by most zooplankton (O’Neil and Roman, 1994; O’Neil, 1998; Carpenter and Capone, 2008), although a few genera of harpacticoid copepods are known to feed on Trichodesmium (O’Neil and Roman, 1994; O’Neil et al., 1996; O’Neil, 1998). Moreover, the harpacticoid, Macrosetella gracilis relies on Trichodesmium, using the colonies as a habitat and a substrate for juvenile development (Björnberg, 1965; Bottger-Schnack and Schnack, 1989; O’Neil and Roman, 1994; Sheridan et al., 2002). The calanoid copepod Acartia tonsa was also observed to graze on Trichodesmium during a bloom along the coast of North Carolina (Guo and Tester, 1994). A recent review by Turner (2014) highlights the varied responses of zooplankton to toxic algae (including a number of cyanobacteria) with grazing impact varying considerably according to predator and prey species and their environment.
Low δ15N ratios of both suspended particles and zooplankton in tropical and subtropical waters (Montoya et al., 2002; Landrum et al., 2011; Karl et al., 2012; Hunt et al., 2016) indicate diazotrophic nitrogen (ND) incorporation into zooplankton and the food web. However, this methodology does not distinguish which zooplankton actively consumes diazotrophs or the source of ND incorporated into the zooplankton (e.g., DDAs, Trichodesmium, unicellular cyanobacteria). As mentioned previously Trichodesmium is not considered a major prey item for oceanic zooplankton and, to our knowledge, no prior studies have observed direct consumption of DDAs by zooplankton. Furthermore, a modeling study by Stukel et al. (2014) suggests that release from grazing pressure due to decreased zooplankton populations stimulates DDA blooms in the WTNA. A number of studies (e.g., Pfannkuche and Lochte, 1993; Gorsky et al., 1999; Wilson and Steinberg, 2010; Stukel et al., 2013) have found the non-diazotrophic unicellular cyanobacterium, Synechococcus, is grazed by zooplankton either individually or as a component of aggregates. Comparatively only two studies show direct consumption of the diazotrophic unicellular groups. In the coastal North Atlantic, Scavotto et al. (2015) showed the symbiotic diazotrophic cyanobacterium Atelocyanobacterium thalassa (UCYN-A) occurred in copepod guts and suggested copepod grazers targeted the larger (4–5 μm) prymnesiophyte host rather than individual cells of the cyanobacteria (<1 μm). More recently, Hunt et al. (2016) showed in a mesocosm experiment performed in an oligotrophic lagoon in the southwest Pacific that one of the diazotrophic unicellular cyanobacteria groups (UCYN-C) was grazed by zooplankton and potentially contributed 28–73% ND to the food web. We are not aware of any studies to date indicating grazing on the diazotroph Crocosphaera watsonii (UCYN-B), which can be found in singular, aggregate, or symbiotic (with diatoms) forms (Carpenter and Janson, 2000; Foster et al., 2013; Thompson and Zehr, 2013), although UCYN-B does produce extracellular polysaccharide (EPS) which may act as a grazing deterrent (Liu and Buskey, 2000; Sohm et al., 2011a).
In contrast to the paucity of studies on zooplankton-diazotroph grazing in the marine environment, freshwater literature provides many examples of these interactions. The number of toxic cyanobacteria blooms in freshwater and estuarine systems have increased due to eutrophication and climate change (Paerl and Otten, 2013). Subsequently, considerable effort has focused on cyanobacteria bloom successional patterns and fate (Ger et al., 2014; Gerphagnon et al., 2015). Similar to the results of O’Neil and Roman (1994) and O’Neil (1998) in the marine environment, many freshwater studies have suggested morphology, toxicity, and unpalatability of cyanobacteria as deterrents to zooplankton grazing (von Elert et al., 2003; Ger et al., 2014). Yet size-selective herbivorous copepods target cyanobacteria (Bouvy et al., 2001), and Kâ et al. (2011) observed that copepods, cladocerans, and rotifers actively graze and fragment larger filamentous cyanobacteria. While there are differences between freshwater and marine planktonic food webs, it seems improbable that dominant freshwater grazers (e.g., copepods and cladocerans) are able to adapt and consume large-scale, often toxic, cyanobacterial blooms as a food source (Kâ et al., 2011; Gerphagnon et al., 2015) while their marine counterparts entirely avoid feeding on cyanobacteria.
The goal of this study was to investigate if mesozooplankton grazers in the Amazon River plume-influenced WTNA directly grazed upon diazotrophic organisms. In concert with this study, we recently reported elevated mesozooplankton grazing in the Amazon River plume relative to non-plume influenced waters (Conroy et al., 2016), although the community-based pigment approach used did not distinguish the exact prey items. Here, we used a molecular quantification method targeting the nifH genes, encoding nitrogenase enzymes, to detect two DDA populations and Trichodesmium in the gut contents of zooplankton in the WTNA. We also used a next generation sequencing (NGS) analysis of 16S rRNA genes to investigate the cyanobacterial composition in zooplankton.
Materials and Methods
Study Area
Samples were collected from 9 stations in the Amazon River plume-influenced region of the WTNA (between 0 and 13°N and 44–57°W) as part of the Amazon INfluence on the Atlantic: CarbOn export from Nitrogen fixation by DiAtom Symbioses (ANACONDAS) project. Data presented here are from a cruise aboard the R/V Knorr May 22–June 24, 2010, during the period of peak plume discharge (Figure 1) and a large scale DDA bloom of Hemiaulus-Richelia (Goes et al., 2014). Underway monitoring of sea surface salinity (SSS) and photosynthetic pigments as well as satellite monitoring of plume indicators such as chromophoric dissolved organic matter (CDOM) were the measurements used to determine station location. Station categorizations were determined by SSS at time of the respective tow for each sample. We sampled from 1 plume station (23) with SSS < 30; 4 mesohaline stations (2, 3, 19, and 21) with SSS between 30 and 35; and 4 oceanic stations (5, 6, 20, and 27) with SSS > 35.
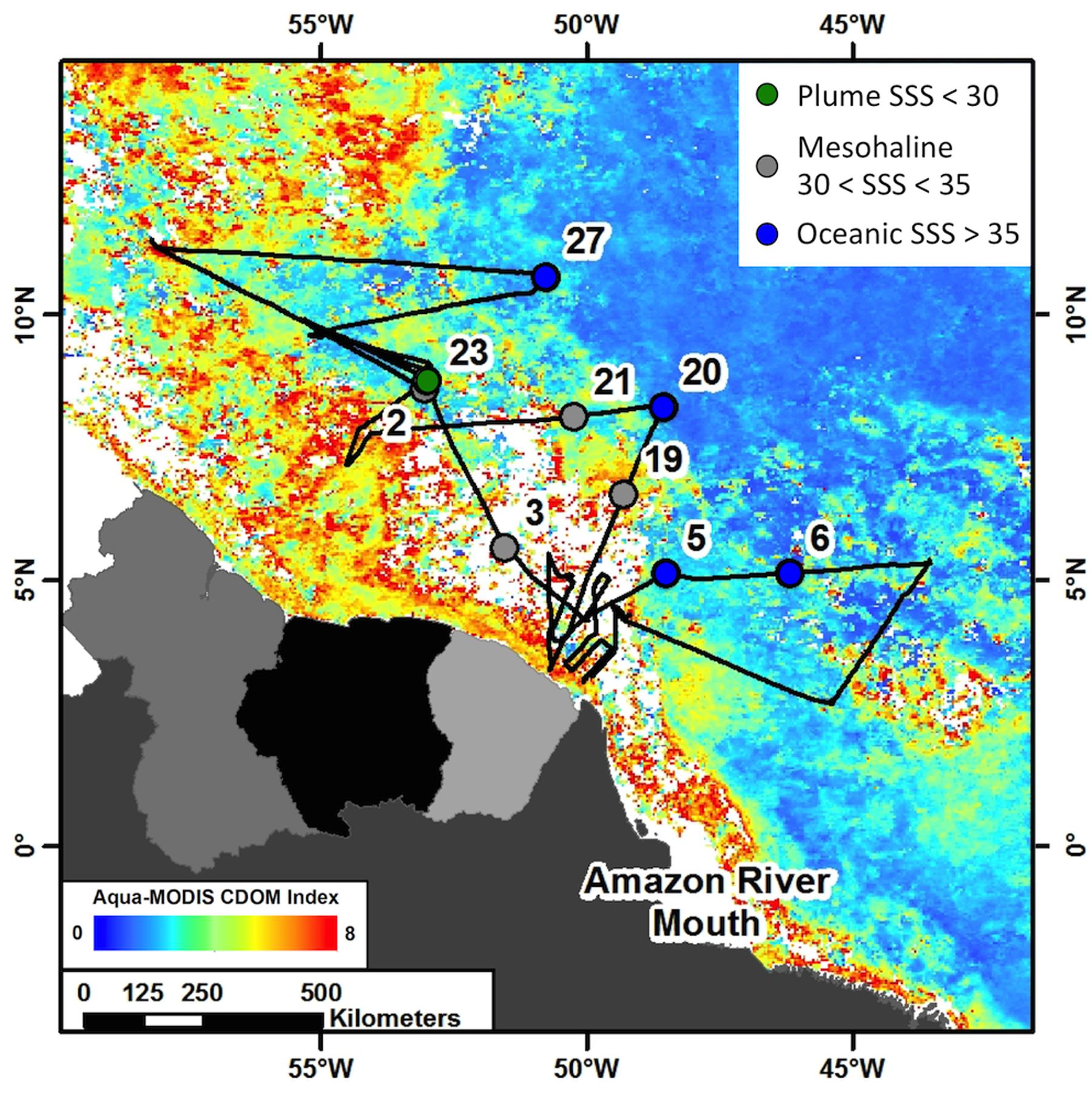
FIGURE 1. Cruise track from ANACONDAS cruise in May–June, 2010 with stations sampled for analyses used in this study labeled. Cruise track is overlaid on monthly averaged chromophoric dissolved organic matter (CDOM) concentration from Aqua-MODIS satellite data (oceancolor.gsfc.nasa.gov). Colors represent station categories based on sea surface salinity (SSS). Stations with SSS < 30 are considered “plume” and shown in green, stations with 30 < SSS < 35 are considered “mesohaline” and shown in gray and stations with SSS > 35 are considered “oceanic” and shown in blue.
Mesozooplankton Collection
Mesozooplankton were collected with a 1-m Multiple Opening and Closing Net and Environmental Sensing System (MOCNESS; Wiebe et al., 1976) fitted with ten 202 μm mesh nets. Daytime tows were performed between 1000 and 1400 h while nighttime tows were collected between 2200 and 0200 h (local time). Four depth intervals within the top 150 m were sampled with the MOCNESS, although molecular analysis was performed on animals collected from the two shallowest depth intervals (0–25 and 25–50 m) since shipboard observations and cell abundances detected the highest number of microscopically identified diazotrophs (e.g., Trichodesmium, DDAs) in the upper 50 m. Once the nets were onboard, zooplankton were immediately anesthetized with carbonated water to prevent gut evacuation (Gannon and Gannon, 1975). Samples were subsequently split into either ¼ or ½ of the total sample volume using a Folsom plankton splitter, then size fractionated using nested sieves into the following size fractions: 0.2–0.5, 0.5–1.0, 1.0–2.0, 2.0–5.0, and >5.0 mm. Each size fraction was then concentrated onto a pre-weighed, 0.2 mm Nitex mesh filter, rinsed with Milli-Q to remove salt, and frozen at -80°C until processed in the laboratory.
Phytoplankton Cell Abundances
To compare grazing at stations with different diazotroph assemblages, mesozooplankton selection for molecular analysis was based on phytoplankton distribution from Goes et al. (2014) utilizing pigment analysis, physical and chemical properties described in Loick-Wilde et al. (2016), and phytoplankton cell counts presented here. Cells for microscopy counts were collected directly from the CTD at distinct depths (5–6) in the upper 150 m and approximately at midday (local noon). Stations 2, 23, and 27 were sampled once per day for 2 days. The entire contents of the Niskin bottles were gravity filtered onto a 47 mm diameter Poretics (Millipore) membrane filter with a pore size of 10 μm. The filter was removed and mounted onto an oversize microscope slide (75 mm × 50 mm × 1 mm) and examined under 400× magnification using a Zeiss Axioskop Epifluorescence microscope (Zeiss, Berlin, Germany). Colonial and single trichomes of Trichodesmium, as well as the symbiotic Richelia, were identified by phycoerythrin and chlorophyll a (Chl a) excitation with green (510–560 nm) and blue (450–490 nm) excitation wavelengths, respectively. The diatoms associated with Richelia were identified as Hemiaulus or Rhizosolenia spp. based on cell ultrastructure. In many instances the entire filter was scanned and symbiotic diatom cells enumerated as number of Richelia heterocysts L-1 for each respective diatom and for Trichodesmium as colonies L-1 (col L-1) or single trichomes L-1. In some instances when symbiotic cells were at high densities, several smaller grid areas (62.5 μm2) of the filter were scanned, and at least 1000 cells were enumerated and corrections to cell abundance were made by the area scanned (Supplementary Table S1).
DNA Extraction and Quantitative PCR (qPCR) Assays
Zooplankton from the 0.5–1.0 and 1.0–2.0 mm size fractions were selected after visual inspection under a stereomicroscope. Harpacticoid copepods (Macrosetella gracilis and Miracia spp.) and the decapod shrimp Lucifer faxoni were identified to species or genus level, while calanoid copepods, fish larvae, and decapod larvae from the family Thalassinidae were not (Supplementary Table S2). These targets were chosen based on mesozooplankton community composition of each station, onboard microscopic counts from the cruise, pigment concentrations (Goes et al., 2014), and stable isotope analysis (Loick-Wilde et al., 2015) for the same cruise. Calanoid copepods were selected, as they were present across all salinities and with only a few exceptions were the most abundant taxa in all samples, while the harpacticoid copepods were selected because of known associations with Trichodesmium. The L. faxoni, fish, and decapod larvae were included given their periodic high abundance. The taxa analyzed were mostly calanoid copepods (and proportionately representative of the zooplankton community composition; Conroy and Steinberg, in preparation), with the relative taxonomic representation as follows: calanoid copepods (69.4%), harpacticoid copepods (19.4%), decapod larvae including crab megalopae and Thalassinidae larvae (4.2%), fish larvae (4.2%), and L. faxoni (2.8%) (Supplementary Table S2).
Animals were sorted and placed in autoclaved artificial seawater and inspected for exterior contamination with phytodetritus in appendages and mouthparts. Animals were picked clean of any obvious large particles using a needle and forceps, and following the procedure of Boling et al. (2012) subsequently rinsed five times with autoclaved artificial seawater. Animals were inspected again for contaminating phytoplankton and cyanobacteria using blue (450–490 nm) and green (510–560 nm) excitation filters on an epifluorescent compound microscope at 200–450× magnifications. This procedure ensured animals chosen for molecular analysis were phytoplankton-free on their exterior. Between 25 and 50 animals were pooled per DNA extraction (Supplementary Table S1); the number of individuals varied depending on size and availability of target and the results from preliminary PCR assays which determined the lowest number of pooled individuals needed for consistent amplification (see below). Samples were extracted using a modification to the Qiagen DNeasy® Blood and Tissue Kit Animal Tissue (Spin-Column) protocol. Briefly, a 12-h lysis step was performed and all recommended reagent volumes were halved during the extraction. The final elution volume was 35 μl in the provided AE buffer.
We performed qPCR assays for three of the major diazotrophs in the WTNA, two DDAs (het-1, Richelia associated with Rhizosolenia diatoms and het-2, Richelia associated with Hemiaulus diatoms), and Trichodesmium spp. using the previously described oligonucleotides (Church et al., 2005a,b; Foster et al., 2007) and a modified TaqMAn assay (see below). A total of 72 samples were analyzed for all three targets (het-1, het-2, Trichodesmium) with the exception of two calanoid copepod samples collected from St. 2 that were not run with the het-1 assay, and six samples from St. 3 were not run with the Trichodesmium due to low template (Supplementary Table S2). In preliminary attempts (data not shown) to optimize the extraction and detection by qPCR, we identified a minimum number of individuals for replicable amplification. From those results we used a minimum of 25 pooled individuals per extraction but, unless limited by abundance of the taxa in a sample, we pooled 50 individuals per extraction (Supplementary Table S2).
For all TaqMan PCR, the 12 μL reactions contained 6.25 μL TaqMan 2X Master Mix (Applied Biosystems), 0.5 μL forward and reverse 0.5 μM primers, 0.25 μL fluorogenic 0.5 μM probe, 2.5 μL of nuclease free water, and 2 μL of DNA template. All reactions were run in triplicate, and for the no-template controls, 2 mL of 5-kD filtered nuclease free water was added to each reaction. All qPCR assays were performed on an ABI 7500 Fast machine (Applied Biosystems) with the following thermal cycling conditions: 50°C for 2 min, 95°C for 10 min, and 45 cycles of 95°C for 15 s, followed by 60°C for 1 min. Gene copy abundances were calculated from the mean cycle threshold (Ct) value of three replicates and the standard curve for the appropriate primer and probe set. For each primer and probe set, triplicate standard curves were made from 10-fold dilution series ranging from 108 to 1 gene copies per reaction. The standard curves were made from linearized plasmids of the target nifH. Regression analyses of the results (number of cycles = Ct) of the duplicate standard curves were analyzed in Excel. In some samples only one of the three replicates produced an amplification signal; these were noted as detectable, but not quantifiable (dnq). For samples where two or three of the replicates amplified the values were averaged and reported as nifH-gene copies per organism. We note that while we report gene copies per organism, as is convention with qPCR, we do not scale our numbers to an estimation of feeding rate. Instead we consider amplification of our targets as confirmation of direct grazing on either DDAs or Trichodesmium (see detailed explanation in Discussion).
16S rRNA Gene Sequencing of Zooplankton
Sequencing of 16S rRNA genes in zooplankton (containing their gut contents) followed a modified protocol (Arfken et al., 2015) using the Ion Torrent PGM sequencer (Life Technologies). DNA concentrations from the extractions were measured on a NanoDrop 2000 (Thermo Scientific) and PCR was performed on samples normalized to 5 ng μl-1 per reaction, except for fish larvae, which were normalized to 20 ng μl-1 (see Supplementary Table S3 for samples included in sequencing). The V4 hypervariable region of 16S rRNA genes was targeted with 515F and 805R primers for PCR reactions using GoTaq Green Master Mix (Promega) (Caporaso et al., 2011). The 805R primers have barcodes with fusion sequences while the 515F contains fusion sequences only. PCR reactions were performed as follows: an initial denaturing step for 3 min at 95°C, then 30 cycles of 30 s at 95°C, 1 min at 55°C, 1 min at 72°C, and finally at 72°C for 5 min. PCR products for each sample were then combined and sequencing was conducted. Barcoded samples were sequenced using the Ion Torrent 400 base pair (bp) sequencing protocol with samples pooled onto a 316 chip. We note that for the 16S rRNA analysis we consider our results representative of the zooplankton “microbiome,” similar to other studies (Scavotto et al., 2015; Shoemaker and Moisander, 2015), however, we focus our results on the cyanobacteria and are confident that the cleaning methods adapted from Boling et al. (2012) were adequate enough and our results represent the cyanobacteria consumed by the various mesozooplankton.
Statistical Analyses and Bioinformatics Pipeline
Sequencing output was downloaded from the Torrent Server using Torrent Suite v3.0 to obtain the FastQ file. A total of 33 libraries were created using the barcoded sequences. Denoising was performed with Acacia (Bragg et al., 2012) and the open-source bioinformatics program Mothur (Schloss et al., 2009) was used to trim and align the sequences. Chimera sequences were removed using UCHIME and remaining sequences for each library were classified using the Greengenes database1 at confidence of greater than or equal to 0.80. Relative abundance of each taxa was calculated by dividing the number of classified sequences by total number of sequences in each library. All cyanobacteria sequences identified through Greengenes database were subsampled and subsequently reclassified through the SILVAngs pipeline2 with the SILVA 128 reference library (Quast et al., 2013). Additionally, principal coordinate analysis (PCoA) was performed on square-root transformed relative abundance data in PRIMER 7 to identify patterns in microbial diversity between libraries. The raw sequence reads have been submitted to National Center for Biotechnology Information3 and are available under Bioproject number PRJNA387277.
Results
Plume and Oceanic Diazotroph Phytoplankton Assemblages
At all stations, except Station 6, the Hemiaulus-Richelia DDA was more abundant than the Rhizosolenia-Richelia DDA. As such, below we only report the former, although all cell counts are available in Supplementary Table S1 for comparison.
The diazotroph assemblage within the low salinity plume (SSS < 30) at station 23 was dominated by Hemiaulus-Richelia DDAs (39.4–7.29 × 103 heterocysts L-1; Supplementary Table S1) primarily in the upper 75 m (Figure 2). At several stations in the mesohaline plume (30 > SSS > 35; stations 2, 3, 19, and 21), cell counts for stations 2 and 19 showed the DDA assemblage described by Goes et al. (2014) was dominated by Hemiaulus-Richelia, and the highest cell density of the cruise (7.66 × 105 heterocysts L-1; Supplementary Table S1) occurred at 20 m at Station 2 (Figure 2). At station 19 in the upper 100 m, Hemiaulus-Richelia and Trichodesmium were similar in abundance, ranging from 34.0–2.39 × 103 heterocysts L-1 and 415–1.59 × 103 trichomes L-1, respectively (Supplementary Table S1). In comparison, stations 3 and 21 had low DDA abundances. At station 3, Hemiaulus-Richelia abundance (8.4 heterocysts L-1) was low in surface waters (∼2 m), free-living Richelia cells in the surface 20 m were the highest observed on the cruise (34.9–338 heterocysts L-1; Supplementary Table S2). Goes et al. (2014) characterized the phytoplankton community of Station 21 as dominated by Trichodesmium and Synechococcus spp. based on pigments, and microscopy confirmed Trichodesmium as the most dominant cyanobacterial diazotroph in the phytoplankton assemblage, with 4.6–141 trichomes L-1 in the surface 50 m (Figure 2).
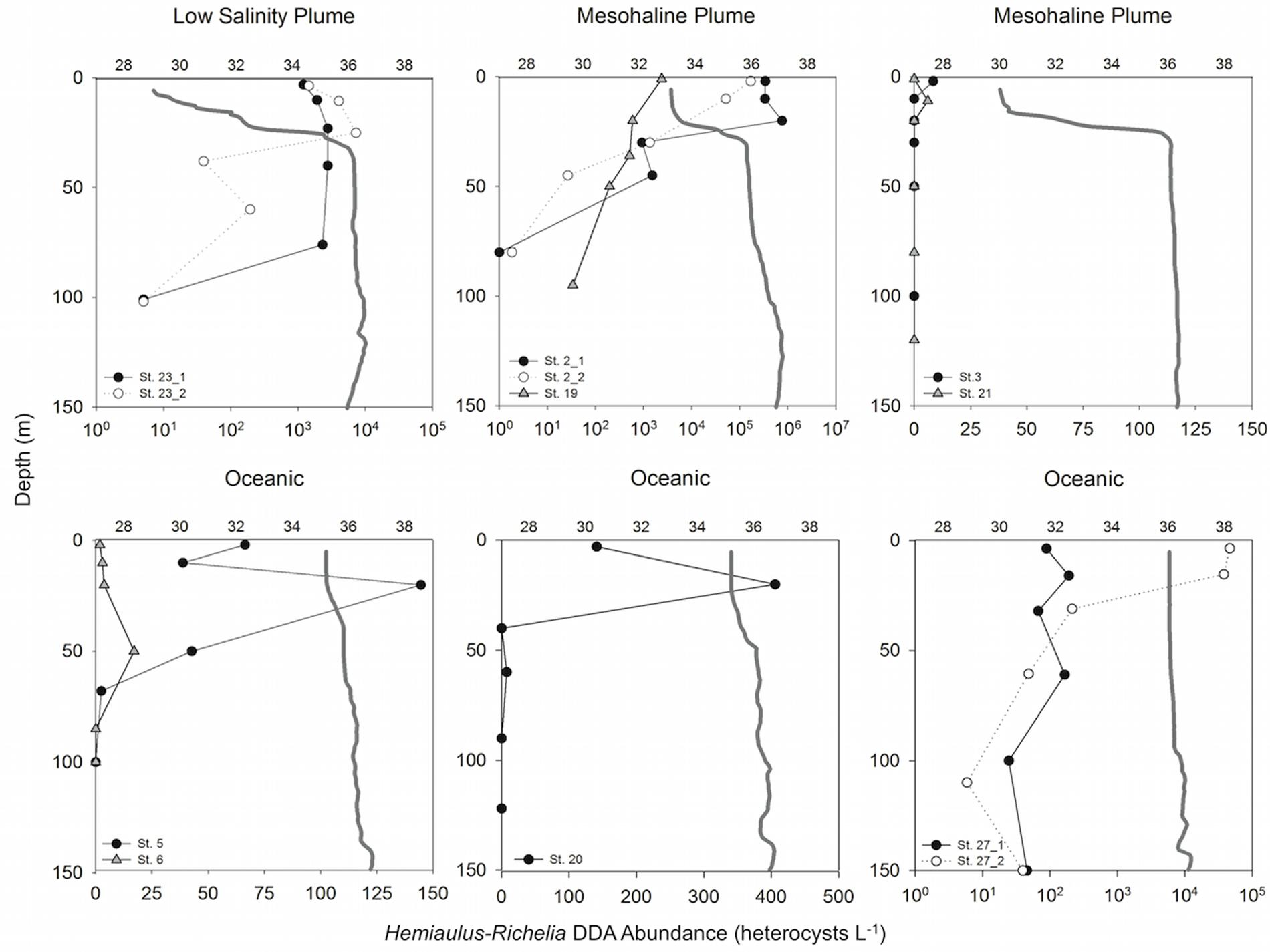
FIGURE 2. Depth profiles of Hemiaulus-Richelia DDA abundance (heterocysts L-1) for each station sampled with representative salinity profiles (gray solid lines). For Stations 23, 2, and 27, open circles are abundances for the second day sampled. Upper x-axes indicate salinity; note different scale bars on lower x-axes for abundance.
Oceanic stations (SSS > 35; stations 5, 6, 20, and 27) were dominated by Trichodesmium and Synechococcus spp. Trichodesmium was present at all four oceanic stations but was not always the most abundant diazotroph in the phytoplankton assemblage (Supplementary Table S1) as previously described in Goes et al. (2014) based on pigments. At Stations 6 and 27 Hemiaulus-Richelia were more abundant than Trichodesmium, particularly at Station 27 with a 1–2 order of magnitude difference (5.9–4.66 × 104 heterocysts L-1 vs. 25.6–1.25 × 103 trichomes L-1 in the surface 150 m, respectively; Supplementary Table S1).
Quantitative PCR of Gut Contents
DDAs– Both the het-1 (Richelia associated with Rhizosolenia diatoms) and het-2 (Richelia associated with Hemiaulus diatoms), targets were successfully amplified from the gut content extractions (Table 1 and Supplementary Table S2). Het-2 was the most common target amplified across all extractions, with a total of 25 samples amplified–24 from calanoid copepods and one from Macrosetella gracilis. Of those, 11 (10 calanoids and 1 M. gracilis), were detectable not quantifiable (dnq). The remaining samples detected het-2 with a range of 1.6–16.8 nifH-copies/organism. All stations except for station 3, the furthest inshore mesohaline station, had samples that amplified the het-2 target.
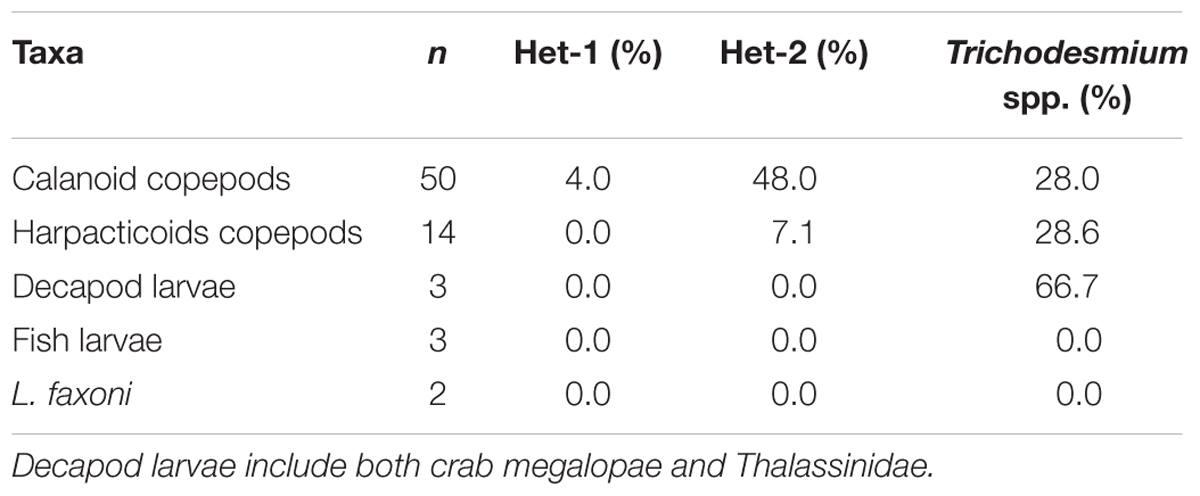
TABLE 1. Percentage of samples (n), for each taxa, exhibiting positive qPCR amplification of Het-1, Het-2, and Trichodesmium spp. in molecular assays.
In comparison to het-2, the other DDA, het-1, was detected only in calanoid copepods collected at night at Station 19 (Table 1 and Supplementary Table S2). Each of the two size fractions at this station had very low detection with the smaller calanoid copepods dnq and larger calanoid copepods 0.10 nifH copies/organism. No pattern was observed between night (n = 12) and day (n = 13) samples for het-2, whereas the only het-1 detection occurred at night (Supplementary Table S2).
Trichodesmium– The nifH of Trichodesmium spp. was also successfully detected in the gut contents of the mesozooplankton samples (Table 1 and Supplementary Table S2). Twenty samples out of 72 (14%) showed amplification: 14 in calanoid copepods (7 dnq), 4 in M. gracilis (3 dnq), and 2 in crab megalopae (1 dnq) (Figure 2). All oceanic stations (St. 5, 6, 20, and 2; Figure 1), as well as one mesohaline station (St. 19), included in this analysis detected Trichodesmium. Values ranged from 1.1 to 4.0 Trichodesmium nifH copies/organism with the highest value found in crab megalopae. No pattern was observed between day (n = 11) and night (n = 9) samples for detecting Trichodesmium.
Characterization of Zooplankton Gut Contents
Sample selection for 16S rRNA NGS analysis was guided by our qPCR assays and resulted in the 33 samples selected for analysis. Not all samples with amplification from the qPCR were included due to template limitation, but a representative number of samples provided over 1.7 million sequences for analysis (see Supplementary Table S3 for NGS samples).
PCoA of 16S rRNA sequences showed a tight clustering of two groups (Figure 3). Calanoid copepods (blue squares in Figure 3) and all harpacticoid copepods (green triangles in Figure 3) clustered by taxa. Other variables analyzed (salinity, size fraction, depth interval, time of day) resulted in no significant grouping.
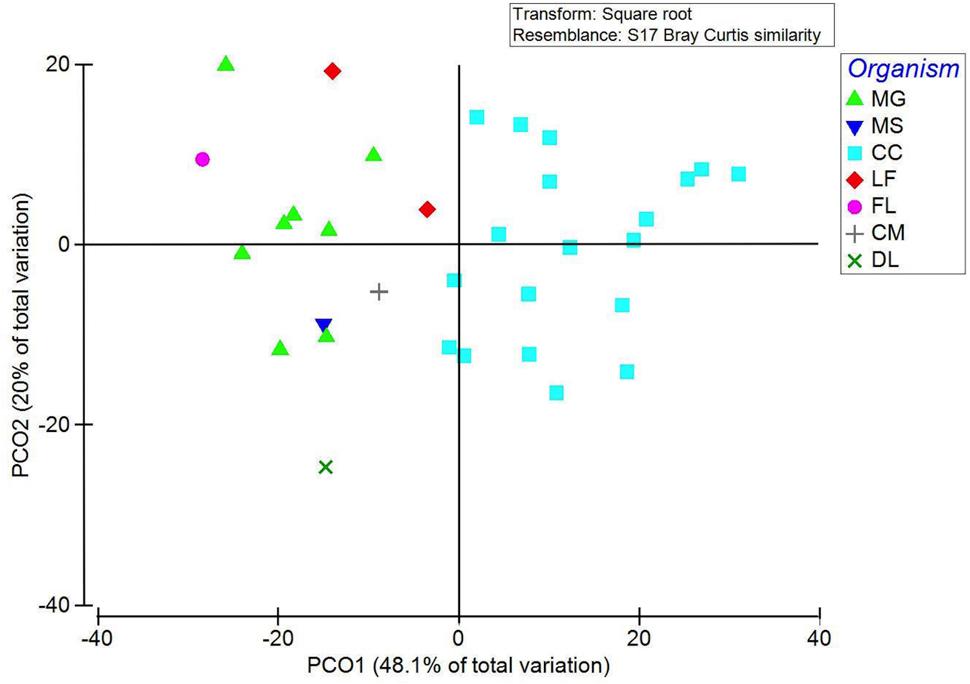
FIGURE 3. PCoA of 16S rRNA NGS of gut microbial community shown by zooplankton taxonomic group (organism). PCO1 and PCO2 explain 48.1 and 20% of the variation, respectively. Samples primarily grouped by taxonomy with the major clusters being calanoid copepods (blue squares) and harpacticoid copepods including both Macrosetella gracilis and Miracia spp. (green triangles). Zooplankton taxonomic groups are abbreviated as follows: MG, Macrosetella gracilis; MS, Miracia spp.; CC, calanoid copepod; LF, Lucifer faxoni; FL, fish larvae; CM, crab megalopae, and DL, decapod larvae.
Proteobacteria were the most abundant phyla represented in our samples, with 16 of the 33 samples having over 50% of sequences associated with proteobacteria (Figure 4). Cyanobacteria represented between 0 and 45% of bacterial composition across all samples and represented 11.3% of total sequences (Supplementary Table S3). In the non-calanoid copepod samples (n = 14), the decapod shrimp L. faxoni, had cyanobacterial sequences associated with its gut which represented >1% of total sequences. Harpactacoid copepods M. gracilis and Miracia spp. had cyanobacterial sequences present but were all <1% of total sequences, and the fish larvae sample analyzed was devoid of cyanobacterial sequences (Figure 4 and Supplementary Table S3).
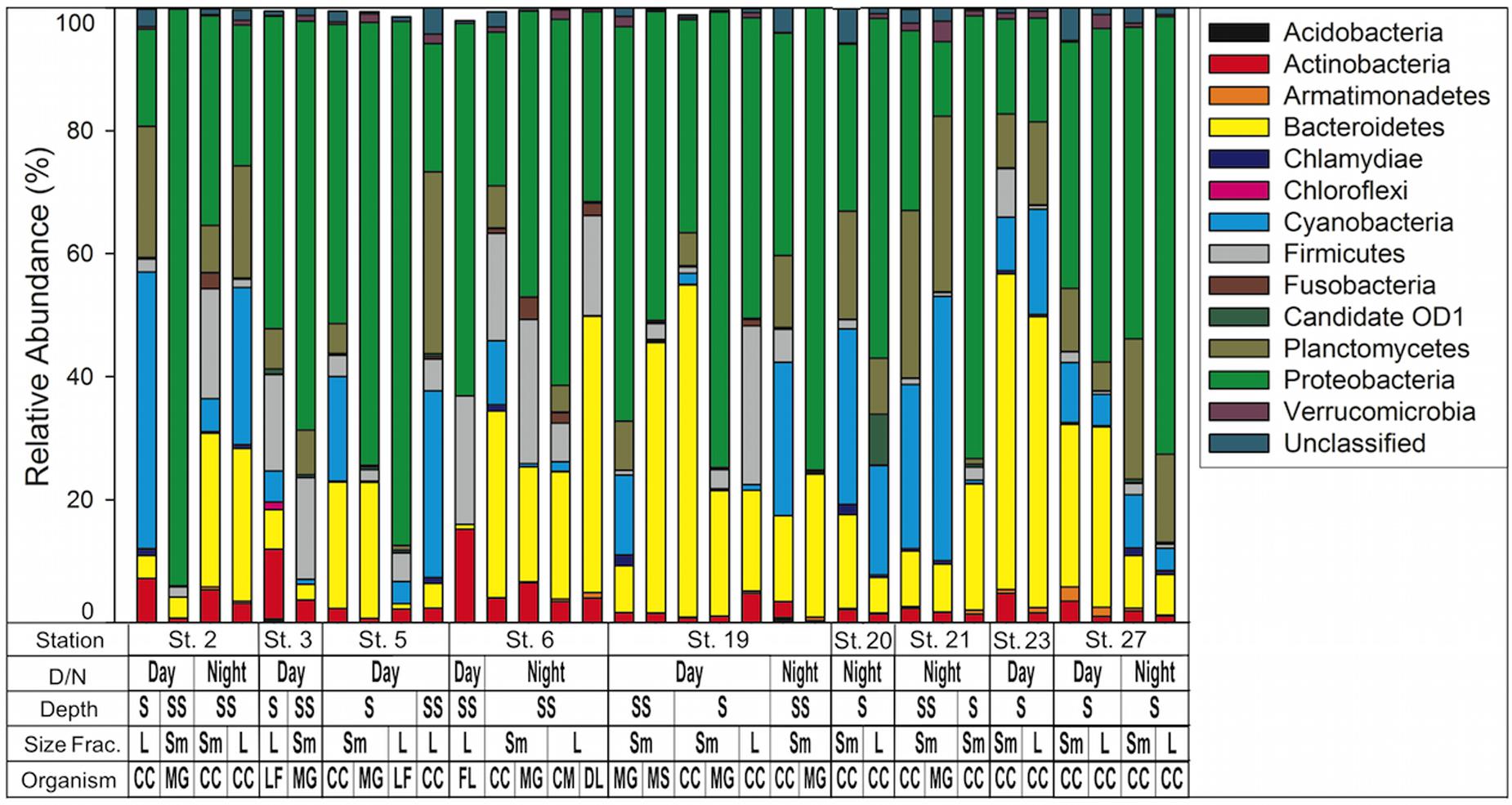
FIGURE 4. Bacterial community composition by phylum as determined by 16S rRNA NGS analysis. Phyla are listed in key to right. Key below graph identifies each sample with categories as follows: D/N, day or night; Depth S = Surface 0–25 m, SS = Sub Surface 25–50 m; Size fraction L = 1.0–2.0 mm, Sm = 0.5–1.0 mm; Organism CC, calanoid copepod; MG, Macrosetella gracilis; MS, Miracia spp; LF, Lucifer faxoni; FL, fish larvae; CM, crab megalopae, and DL, decapod larvae.
When composition of only cyanobacterial sequences (n = 197,298) was analyzed with the GreenGenes database, the most abundant class was Synechococcophycideae (Figure 5). When investigated to genus level, the most abundant within this class was Synechococcus (n = 83,745, representing 42.4% of all cyanobacterial sequences). Prochlorococcus was the next largest contributor at the genus level (n = 22,734 representing 11.4% of all cyanobacterial sequences) although 21.2% of cyanobacterial sequences in Synechococcophycideae were unidentifiable to genus classification. Additionally, in a several samples chloroplast sequences composed up to 100% of the cyanobacterial sequences (Figure 5).
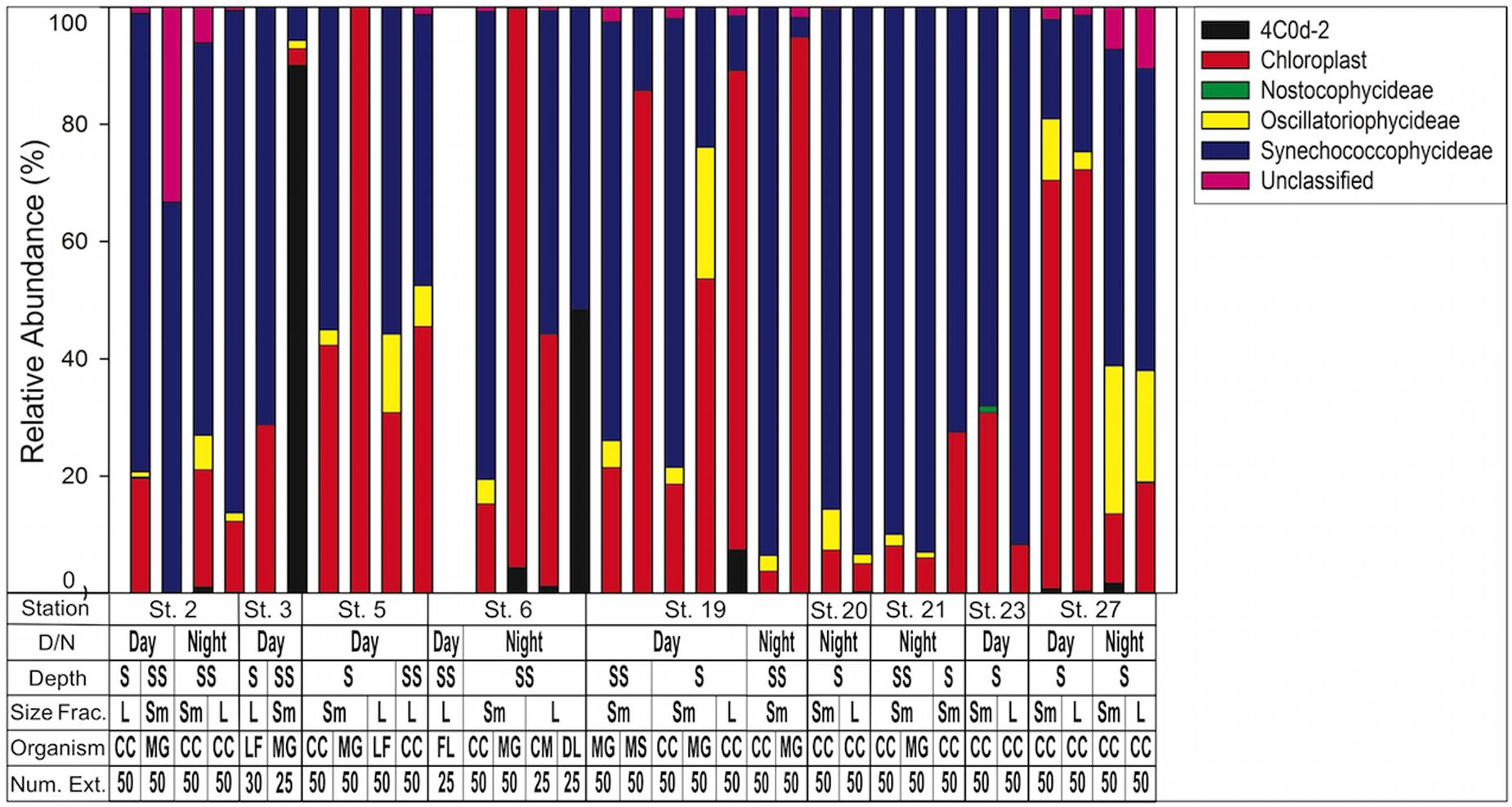
FIGURE 5. Cyanobacteria assemblage composition determined by 16S rRNA NGS analysis. Classes are listed in key to right. Note chloroplasts are included in the Green Genes database although not technically cyanobacteria. 4C0d-2 are closely related to cyanobacteria but recently proposed candidates for the new phylum melainabacteria. Key below graph identifies each sample with categories as follows: D/N, day or night; Depth S = Surface 0–25 m, SS = Sub Surface 25–50 m; Size fraction L = 1.0–2.0 mm, Sm = 0.5–1.0 mm; Organism CC, calanoid copepod; MG, Macrosetella gracilis; MS, Miracia spp.; LF, Lucifer faxoni; FL, Fish Larvae; DL, Decapod Larvae; CM, Crab Megalopae; Num Ext., animals per extraction.
Reclassification of cyanobacterial sequences based on SILVA 128 database agreed that Synechococcus and Prochlorococcus were still the most abundant representatives (56 and 26%, respectively). However, at station 5 and station 21 UCYN-A and UCYN-B sequences were found within the smallest size fraction calanoid copepod samples, representing 1–2% of sequences.
Discussion
Studying the pathway of diazotrophically derived production (ND) in the marine food web is challenging; however, with several of the molecular-based approaches, new insights can be achieved. For example, using nifH qPCR assays with highly specific oligonucleotides allowed detection of DDAs and Trichodesmium in the gut contents of mesozooplankton. Moreover, our parallel 16S rRNA NGS libraries further elucidated the greater cyanobacterial diversity in zooplankton guts. Templates derived from the harpacticoid copepod M. gracilis, commonly considered the major grazer of Trichodesmium, were often below detection for the Trichodesmium qPCR assays. Although our sample size for harpacticoid copepods was low. We also found evidence that crab megalopae consume Trichodesmium. Finally, our 16S rRNA NGS analysis concluded that mesozooplankton taxa frequently consume the non-diazotrophic picoplanktonic cyanobacteria Synechococcus and Prochlorococcus spp., and are capable of consumption of other diazotrophic unicellular cyanobacteria such as UCYN-A and UCYN-B. However, these results provide insights on cyanobacteria-zooplankton interactions with important implications for the pathways of ND entering marine food webs and for N cycling in the WTNA.
Diazotroph Consumption by Mesozooplankton
DDAs–Stable isotope studies in the subtropical and tropical Atlantic Ocean (Montoya et al., 2002; Landrum et al., 2011; Loick-Wilde et al., 2015), including the region of our study, established that new N attributed to N2-fixation from diazotrophs is incorporated in the planktonic food web, but the pathways were not elucidated (i.e., through the microbial loop, exudation, or grazing). The qPCR results of the gut content show ND can enter the food web via consumption of DDAs, as both DDA targets (het-1 and het-2) were detected in the zooplankton gut contents. This is the first evidence for consumption of DDAs. These results are consistent with DDA distributions observed in our microscopy, where het-2 was the most dominant of the two DDAs present during our study, as well as in a prior studies of the Amazon-influenced WTNA (Carpenter et al., 1999; Foster et al., 2007; Luo et al., 2012). Consumption of DDAs is also consistent with the decreased δ15N content of zooplankton reported by Loick-Wilde et al. (2016) in the mesohaline plume, particularly at Stations 2 and 19 (their Figure 9). At St. 19, the only station where het-1 was detected in the guts of mesozooplankton, microscope counts observed both Hemiaulus and Rhizosolenia symbioses were present. However, Hemiaulus-Richelia symbioses (het-2) were 1–2 orders of magnitude more abundant and found deeper in the water column than the Rhizosolenia-Richelia symbiosis (het-1).
The pattern of DDA distribution in the WTNA largely follows the nutrient and surface salinity gradients outlined in Subramaniam et al. (2008), with DDAs occurring in the mesohaline region of the plume (Goes et al., 2014; Loick-Wilde et al., 2015). However, the diazotrophic cyanobacterial counts and molecular results reported here indicate there is not a strict boundary between the salinity regimes for DDA presence, particularly along the transition from mesohaline to oceanic waters. All four of the oceanic stations included in this study had low abundances of het-2 symbioses along with amplification of this target in zooplankton gut contents. These stations were characterized as an oceanic assemblage with abundant Trichodesmium and Synechococcus by Goes et al. (2014) (note Station 6 was not included in their study). Therefore, het-2 consumption at all four oceanic stations sampled is unexpected and can be explained by plume meanders or development of frontal zones between mesohaline and oceanic waters. These physical features are common in river plumes and represent areas of zooplankton aggregation and increased grazing (Woodson et al., 2005; True et al., 2015; Woodson and Litvin, 2015) and could promote episodic, opportunistic feeding in oceanic mesozooplankton.
Our results, while novel, are not unexpected. Conroy et al. (2016) characterized the mesozooplankton food web from grazing rate estimations at low salinity plume and mesohaline waters with SSS < 33 as an export style food web. Export food webs are characterized by shorter diatom and mesozooplankton food chains compared to retention food webs dominated by microzooplankton and the microbial loop (Michaels and Silver, 1988; Legendre and Michaud, 1998; Pomeroy et al., 2007). Both DDAs investigated form chains, often suggested as a grazing deterrent, yet copepods actively graze diatoms of similar size (Bergkvist et al., 2012). Therefore in the plume and mesohaline stations DDA consumption supports a shorter export food web; DDA consumption we observed at oceanic stations is likely more occasional, but when it occurs could increase export.
Trichodesmium were consumed by zooplankton at all four oceanic stations, as well as at two mesohaline stations. Calanoid copepods were again the primary consumers, although the highest nifH gene copies organism-1 were amplified from crab megalopae. Despite the higher abundance of M. gracilis at oceanic stations relative to plume stations (Conroy et al., in preparation) and its known association with, and grazing on, Trichodesmium (O’Neil and Roman, 1994; O’Neil et al., 1996; O’Neil, 1998), this species only showed one amplification for Trichodesmium that was not dnq (Figure 2 and Supplementary Table S2). This is not to say M. gracilis does not graze on Trichodesmium, which was reported to be a pathway of atmospheric nitrogen incorporation into zooplankton in the eastern tropical Atlantic (Sandel et al., 2015), but that this result is an artifact of our methodology, discussed in detail below.
Prior to our study there was little evidence of calanoid copepod grazing on Trichodesmium, and a general consensus emerged that it did not occur or was severely limited (Capone et al., 1997; Carpenter and Capone, 2008). The detection of Trichodesmium in the guts of calanoid copepods in the WTNA is novel and builds on other limited evidence for calanoid grazing on Trichodesmium globally (Hawser et al., 1992; Guo and Tester, 1994). Calanoid copepods were typically the most abundant organism across all stations in the three smallest size fractions (0.2–0.5, 0.5–1.0, and 1.0–2.0 mm) (Conroy et al., in preparation), as also found in zooplankton community composition studies in the subtropical North Atlantic (Steinberg et al., 2008; Eden et al., 2009). Thus, grazing by copepods on Trichodesmium in the oligotrophic tropical and subtropical ocean where Trichodesmium is abundant (Capone et al., 1997, 2005) has potential for a significant input of ND into the food web. Furthermore, these results support the observed low δ15N of mesozooplankton at oceanic stations (Loick-Wilde et al., 2015), was due, at least in part, to direct grazing on Trichodesmium. Similar evidence was recently reported in the Eastern tropical North Atlantic where δ15N of zooplankton was correlated with Trichodesmium spp. colony abundance (Sandel et al., 2015).
Other work from stable isotope analysis suggests that decreases in δ15N in Trichodesmium-dominated waters is due to ND exudation and incorporation into the food web via the microbial loop (Montoya et al., 2002; Mulholland, 2007) rather than direct grazing, predominately due their potentially allelopathic toxins (Hawser et al., 1992). In the eastern tropical North Atlantic Ocean, Sandel et al. (2015) observed decreased δ15N in carnivorous, omnivorous, and Trichodesmium-grazing copepods. Aside from the known Trichodesmium grazers included in that study (M. gracilis and M. efferata) they did not suggest direct consumption of Trichodesmium by other copepod species. However, Hawser et al. (1992) showed that toxicity was species dependent and that not all Trichodesmium spp. are toxic to zooplankton. Furthermore, Guo and Tester (1994) investigated the effect of Trichodesmium sp. on the calanoid copepod Acartia tonsa after a meander from the Gulf Stream transported Trichodesmium inshore toward Albemarle Sound, North Carolina. They found no toxic effects of Trichodesmium on A. tonsa when fed healthy cells, but observed toxic effects, including distended guts and mortality, when fed aging or senescing cells or when treated with a filtered cell homogenate (Guo and Tester, 1994). Given the ephemeral exposure A. tonsa has to Trichodesmium as a predominantly coastal copepod, it is reasonable that calanoid copepods exposed to Trichodesmium during development in the open ocean would have a similar ability to consume Trichodesmium. Furthermore, a study from the tropical North Atlantic between the Cape Verde Islands and Barbados reports that δ15N values for zooplankton and Trichodesmium are similar, so that direct consumption is the likeliest explanation (McClelland et al., 2003, see their Figure 6). Zooplankton grazing on Trichodesmium is further supported by zooplankton-cyanobacteria interactions from freshwater habitats. Kâ et al. (2011) performed feeding experiments with calanoid copepods, cladocerans, and rotifers and found grazing could be a significant factor in controlling filamentous cyanobacteria. While we are unable to scale our numbers to estimate the grazing impact on Trichodesmium (see below), our results support that Trichodesmium is directly consumed by mesozooplankton.
Microbial and Cyanobacterial Diversity Associated with Zooplankton
Pairing the highly specific qPCR approach with a 16S rRNA NGS approach provided insight into the broader zooplankton-cyanobacterial dynamics in the WTNA. Our 16S rRNA sequences show abundant phyla that varied between samples, with proteobacteria and cyanobacteria consistently a large percentage of all sample sequences. Similar to findings of Shoemaker and Moisander (2015) from the subtropical North Atlantic, our results indicate a distinct partitioning of microbes based on taxonomic groups rather than environmental factors (Figure 3). While an understanding of the complete microbiome of zooplankton is important (Scavotto et al., 2015; Shoemaker and Moisander, 2015), particularly in the oligotrophic ocean where microenvironments (e.g., zooplankton) support unique bacterial assemblages (Azam and Malfatti, 2007), we limit the scope of our discussion to the cyanobacteria given their potential role in primary production, N2 fixation, and carbon export.
Predominance of Synechococcus and to a lesser extent Prochlorococcus sequences within the most abundant cyanobacteria class (Figure 5) is similar to results from the same cruise investigating diversity of free-living and particle-associated cells. Metatranscriptomic studies conducted by Satinsky et al. (2014) showed that the prokaryotic assemblage was dominated by the Synechococcaceae family of cyanobacteria (Satinsky et al., 2014, See Supplementary Figure S1 of that study). Those results are limited to one station, which we did not sample; however, it does provide insight into why our sequences were dominated by Synechococcus and Prochlorococcus. Synechococcus and Prochlorococcus are globally abundant (Flombaum et al., 2013) but generally considered too small to be directly consumed by most mesozooplankton. On the other hand, consumption of the latter are often considered indirectly through feeding on marine snow aggregates or fecal pellets (Wilson and Steinberg, 2010), or by ingesting microzooplankton which previously consumed small cells. Satinsky et al. (2014) showed a higher percentage of Synechococcaceae cyanobacteria in “particle-associated” sequences compared to “free-living,” while HPLC pigment data showed abundant Synechococcus throughout the plume and WTNA (Goes et al., 2014). We conclude that predominance of Synechococcus and Prochlorococcus sequences in the crustacean zooplankton are likely from consumption of cyanobacteria-containing aggregates and/or consumption of microzooplankton, which had consumed picocyanobacteria previously.
The presence of UCYN-A and UCYN-B sequences in two different calanoid copepod samples indicates a pathway of ND directly into the food web (Zehr, 2011; Thompson et al., 2012, 2014). Scavotto et al. (2015) is the only other study to show consumption of UCYN-A by zooplankton with their results showing the calanoid copepod Acartia spp. regularly had UCYN-A in full gut contents. To our knowledge there are no studies reporting UCYN-B consumption by zooplankton. UCYN-A is comparable in size to Synechococcus and Prochlorococcus, but UCYN-A is suggested to be an obligate, or at least mutualistic, symbiont with a larger haptophyte (Thompson et al., 2012; Farnelid et al., 2016) and UCYN-B can be found in free-living, colonial and symbioses with diatoms. Therefore, UCYN-A and UCYN-B may be directly targeted by zooplankton or consumed as components of larger sinking aggregates in the water column.
We did not analyze water column UCYN-A or UCYN-B abundance; however, results from prior studies in the tropical Atlantic suggest both are widely distributed, particularly in the eastern Atlantic and outer most regions of the plume-influenced WTNA (Foster et al., 2007; Goebel et al., 2010; Moisander et al., 2010). Globally, UCYN-A and UCYN-B are observed throughout the tropics and thus our results suggest a broad potential pathway for ND from unicellular diazotrophs into planktonic food webs, particularly in areas such as the south Pacific where high concentrations for both are observed (Moisander et al., 2010; Farnelid et al., 2016).
Lastly, we offer explanations for the chloroplast sequences identified in the cyanobacterial sequences (Figure 5). Our NGS protocol required a PCR amplification step and it is possible that these chloroplasts could represent a bias introduced during amplification (Acinas et al., 2005). More likely is that the sequences represent phytoplankton, likely cyanobacteria, that were ingested but are not included in the most recent GreenGenes reference taxonomy (McDonald et al., 2012).
Perspectives for Scaling up to Grazing and Method Improvements
While molecular methods to quantify zooplankton grazing rates are becoming more common (Nejstgaard et al., 2008; Troedsson et al., 2009; Cleary et al., 2012, 2016; Shoemaker and Moisander, 2015), we hesitate to extend our qPCR results beyond a qualitative assessment of grazing. The nifH Taqman probes are highly specific, thus other non-targeted organisms consumed, which our NGS analysis confirms, would not be detected by the qPCR assays but could interfere with the amplification (Kanagawa, 2003; Nejstgaard et al., 2008). On the other hand, given the evidence of polyploidy in heterocystous cyanobacteria (Griese et al., 2011; Sukenik et al., 2012) and recently observed in Trichodesmium (Sargent et al., 2016) there is potential to overestimate the nifH copy number by qPCR. However, target DNA degradation and inefficient DNA extraction in the gut is a significant factor in underestimating zooplankton grazing, particularly in copepods, with molecular methods (Simonelli et al., 2009; Troedsson et al., 2009). Conroy et al. (2016) observed elevated grazing in both the low salinity plume stations and in the intermediate mesohaline stations utilizing the gut pigment method, yet our qPCR assays all yielded low gene copies per organism regardless of station category. Similarly, Nejstgaard et al. (2008) found a pattern of underrepresentation of gut contents when comparing qPCR estimates to gut pigment estimates. Regardless of the low gene copies, we are confident our results are indicative of grazing on DDAs and Trichodesmium given the high specificity of our qPCR assays and our sanitation techniques. In order to account for low gene copies due to gut degradation, we suggest future work include controlled grazing experiments using a culture of the targeted diazotroph for analyses so that a differential length amplification qPCR (dla-qPCR) method, similar to that utilized in Troedsson et al. (2009), could account for DNA degradation, and be used to estimate grazing rate on diazotrophs. Furthermore, the molecular methods utilized in this study could be paired with fluorescent in situ hybridization as well as analysis of gut contents through traditional electron microscopy. Lastly, there is the potential for contamination from environmental DNA (eDNA) that at present is not accounted for in this or other studies. While likely a low percentage of the genetic material analyzed, methodologies such as the one used here, that will likely have low amplification, need to consider a method for determining contamination from eDNA.
Conclusion
Diazotrophic nitrogen incorporation into the planktonic food web has long been observed through the use of nitrogen stable isotope analysis. While an extremely robust method, stable isotope analysis lacks the nuance to determine the exact pathways ND enters the planktonic food web. We provide direct evidence that two DDAs, Hemiaulus-Richelia and Rhizosolenia-Richelia, are consumed by mesozooplankton. We further show that Trichodesmium is consumed by calanoid and harpacticoid copepods, as well as some decapod larvae. Lastly, we show that unicellular cyanobacteria, particularly non-diazotrophic Synechococcus and Prochlorococcus, as well as diazotrophic UCYN-A and UCYN-B, are consumed by zooplankton, likely as components of aggregates or in symbioses with microalgae (e.g., UCYN-A). Grazing on UCYN-A and UCYN-B provides an additional and previously undocumented pathway for ND incorporation into the food web in the WTNA. This study has important implications for our understanding of cyanobacterial-zooplankton dynamics in a changing ocean. Increased stratification, due to warming surface waters, is expected to elevate the importance of N2-fixation in the oligotrophic open ocean (Doney et al., 2012), and our results suggest that mesozooplankton that consume diazotrophs would likely benefit. Our results suggest the need for investigation beyond the WTNA, as in other areas with DDA blooms such as the Congo, Niger, and Mekong River plumes (Foster et al., 2009; Grosse et al., 2010; Bombar et al., 2011), the South Pacific Ocean (Turk-Kubo et al., 2015), as well as globally where Trichodesmium (Capone et al., 1997, 2005) and unicellular cyanobacteria are important diazotrophs. Questions do remain, however, concerning what other mesozooplankton taxa target diazotrophs, and what are their grazing rates and impacts on the bloom-forming DDAs and Trichodesmium. Further work in these areas is needed to extend our results to multiple taxa and other regions, and to quantify specific pathways of diazotrophic nitrogen incorporation into the food web.
Author Contributions
BC, DS, BS, and RF conceived the zooplankton sampling and experimental design. BC, DS, BS, AK, EC, and RF acquired and analyzed the data. BC, DS, and RF wrote the paper, and BC, DS, BS, AK, EC, and RF edited the manuscript. All authors approved the final submitted manuscript.
Funding
This research was funded by National Science Foundation grants OCE-0934036 to DS, DGE-0840804 to BC, OCE-1321373 to BS, OCE-0934035 to EC, and OCE-0929015 to RF. Knut and Alice Wallenberg Foundation (Stockholm Sweden) also support the contribution by RF.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank the captain and crew of the R/V Knorr for their support and hard work during the research cruise. Special thanks to Joe Cope, Miram Gleiber, and Jason Landrum for their assistance with zooplankton collection and processing at sea. Additional thanks to Chief Scientist and Lead PI Patricia Yager as well as all other members of the ANACONDAS team. Lastly, thanks to the Jon Zehr’s lab at UC Santa Cruz for providing lab space and input during the initial training and method development for this manuscript. This research was part of BC’s Ph.D. dissertation which is available in its entirety through the College of William & Mary (Conroy, 2016). This paper is Contribution No. 3651 of the Virginia Institute of Marine Science, College of William & Mary.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01436/full#supplementary-material
Footnotes
References
Acinas, S. G., Sarma-Rupavtarm, R., Klepac-Ceraj, V., and Polz, M. F. (2005). PCR-induced sequence artifacts and bias: insights from comparison of two 16S rRNA clone libraries constructed from the same sample. Appl. Environ. Microbiol. 71, 8966–8969. doi: 10.1128/AEM.71.12.8966-8969.2005
Arfken, A. M., Song, B., and Mallin, M. A. (2015). Assessing hog lagoon waste contamination in the Cape Fear Watershed using Bacteroidetes 16S rRNA gene pyrosequencing. Appl. Microbiol. Biotechnol. 99, 7283–7293. doi: 10.1007/s00253-015-6784-x
Azam, F., and Malfatti, F. (2007). Microbial structuring of marine ecosystems. Nat. Rev. Microbiol. 5, 782–791. doi: 10.1038/nrmicro1747
Bergkvist, J., Thor, P., Henrik Jakobsen, H., Wängberg, S.-Å., and Selander, E. (2012). Grazer-induced chain length plasticity reduces grazing risk in a marine diatom. Limnol. Oceanogr. 57, 318–324. doi: 10.4319/lo.2012.57.1.0318
Berman-Frank, I., Bidle, K. D., Haramaty, L., and Falkowski, P. G. (2004). The demise of the marine cyanobacterium, Trichodesmium spp., via an autocatalyzed cell death pathway. Limnol. Oceanogr. 49, 997–1005. doi: 10.1038/ismej.2013.121
Björnberg, T. K. S. (1965). Observations on the development and the biology of the Miracidae Dana (Copepoda: Crustacea). Bull. Mar. Sci. 15, 512–520.
Boling, W. B., Sinclair, G. A., and Wawrik, B. (2012). Identification of calanoid copepod prey species via molecular detection of carbon fixation genes. Mar. Biol. 159, 1165–1171. doi: 10.1007/s00227-011-1877-2
Bombar, D., Moisander, P., Dippner, J., Foster, R., Voss, M., Karfeld, B., et al. (2011). Distribution of diazotrophic microorganisms and nifH gene expression in the Mekong River plume during intermonsoon. Mar. Ecol. Prog. Ser. 424, 39–52. doi: 10.3354/meps08976
Bottger-Schnack, R., and Schnack, D. (1989). Vertical distribution and population structure of Macrosetella gracilis (Copepoda: Harpacticoida) in the Red Sea in relation to the occurrence of Oscillatoria (Trichodesmium) spp. (Cyanobacteria). Mar. Ecol. Prog. Ser. 52, 17–31. doi: 10.3354/meps052017
Bouvy, M., Pagano, M., and Troussellier, M. (2001). Effects of a cyanobacterial bloom (Cylindrospermopsis raciborskii) on bacteria and zooplankton communities in Ingazeira reservoir (northeast Brazil). Aquat. Microb. Ecol. 25, 215–227. doi: 10.3354/ame025215
Bragg, L., Stone, G., Imelfort, M., Hugenholtz, P., and Tyson, G. W. (2012). Fast, accurate error-correction of amplicon pyrosequences using Acacia. Nat. Methods 9, 425–426. doi: 10.1038/nmeth.1990
Bronk, D. A., and Steinberg, D. K. (2008). “Chapter 8 - Nitrogen Regeneration,” in Nitrogen in the Marine Environment, 2nd Edn, eds D. G. Capone, D. A. Bronk, M. R. Mulholland, and E. J. Carpenter (San Diego, CA: Academic Press), 385–467.
Capone, D. G. (2001). Marine nitrogen fixation: What’s the fuss? Curr. Opin. Microbiol. 4, 341–348. doi: 10.1016/S1369-5274(00)00215-0
Capone, D. G., Burns, J. A., Montoya, J. P., Subramaniam, A., Mahaffey, C., Gunderson, T., et al. (2005). Nitrogen fixation by Trichodesmium spp.: an important source of new nitrogen to the tropical and subtropical North Atlantic Ocean. Glob. Biogeochem. Cycles 19:GB2024. doi: 10.1029/2004GB002331
Capone, D. G., Zehr, J. P., Paerl, H. W., Bergman, B., and Carpenter, E. J. (1997). Trichodesmium, a globally significant marine cyanobacterium. Science 276, 1221–1229. doi: 10.1126/science.276.5316.1221
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. U.S.A. 108, 4516–4522. doi: 10.1073/pnas.1000080107
Carpenter, E. J., and Capone, D. G. (2008). “Chapter 4 - nitrogen fixation in the marine environment,” in Nitrogen in the Marine Environment, 2nd Edn, eds D. G. Capone, D. A. Bronk, M. R. Mulholland, and E. J. Carpenter (San Diego, CA: Academic Press), 141–198.
Carpenter, E. J., and Janson, S. (2000). Intracellular cyanobacterial symbionts in the marine diatom Climacodium frauenfeldianum (bacillariophyceae). J. Phycol. 36, 540–544. doi: 10.1046/j.1529-8817.2000.99163.x
Carpenter, E. J., Montoya, J. P., Burns, J., Mulholland, M. R., Subramaniam, A., and Capone, D. G. (1999). Extensive bloom of a N2-fixing diatom/cyanobacterial association in the tropical Atlantic Ocean. Mar. Ecol. Prog. Ser. 185, 273–283. doi: 10.3354/meps185273
Church, M. J., Jenkins, B. D., Karl, D. M., and Zehr, J. P. (2005a). Vertical distributions of nitrogen-fixing phylotypes at Station ALOHA in the oligotrophic North Pacific Ocean. Aquat. Microb. Ecol. 38, 3–14. doi: 10.3354/ame038003
Church, M. J., Short, C. M., Jenkins, B. D., Karl, D. M., and Zehr, J. P. (2005b). Temporal patterns of nitrogenase gene (nifH) expression in the oligotrophic North Pacific Ocean. Appl. Environ. Microbiol. 71, 5362–5370. doi: 10.1128/AEM.71.9.5362-5370.2005
Cleary, A., Durbin, E., and Rynearson, T. (2012). Krill feeding on sediment in the Gulf of Maine (North Atlantic). Mar. Ecol. Prog. Ser. 455, 157–172. doi: 10.3354/meps09632
Cleary, A. C., Durbin, E. G., Rynearson, T. A., and Bailey, J. (2016). Feeding by Pseudocalanus copepods in the Bering Sea: trophic linkages and a potential mechanism of niche partitioning. Deep Sea Res. II 134, 181–189. doi: 10.1016/j.dsr2.2015.04.001
Conroy, B. (2016). Zooplankton Community Composition and Grazing in the Amazon River Plume and Western Tropical North Atlantic Ocean. Ph.D. thesis, College of William and Mary, Williamsburg, VA. doi: 10.21220/V53K52
Conroy, B. J., Steinberg, D. K., Stukel, M. R., Goes, J. I., and Coles, V. J. (2016). Meso- and microzooplankton grazing in the Amazon River plume and western tropical North Atlantic. Limnol. Oceanogr. 61, 825–840. doi: 10.1002/lno.10261
Cooley, S. R., and Yager, P. L. (2006). Physical and biological contributions to the western tropical North Atlantic Ocean carbon sink formed by the Amazon River plume. J. Geophys. Res. 111:C08018. doi: 10.1029/2005JC002954
Doney, S. C., Ruckelshaus, M., Emmett Duffy, J., Barry, J. P., Chan, F., English, C. A., et al. (2012). Climate change impacts on marine ecosystems. Annu. Rev. Mar. Sci. 4, 11–37. doi: 10.1146/annurev-marine-041911-111611
Dugdale, R. C., and Goering, J. J. (1967). Uptake of new and regenerated forms of nitrogen in primary productivity. Limnol. Oceanogr. 12, 196–206. doi: 10.4319/lo.1967.12.2.0196
Eden, B. R., Steinberg, D. K., Goldthwait, S. A., and McGillicuddy, D. J. (2009). Zooplankton community structure in a cyclonic and mode-water eddy in the Sargasso Sea. Deep Sea Res. I 56, 1757–1776. doi: 10.1016/j.dsr.2009.05.005
Falcon, L. I., Carpenter, E. J., Cipriano, F., Bergman, B., and Capone, D. G. (2004). N2 fixation by unicellular bacterioplankton from the Atlantic and Pacific Oceans: phylogeny and in situ rates. Appl. Environ. Microbiol. 70, 765–770. doi: 10.1128/AEM.70.2.765-770.2004
Farnelid, H., Turk-Kubo, K., Muñoz-Marín, M., and Zehr, J. (2016). New insights into the ecology of the globally significant uncultured nitrogen-fixing symbiont UCYN-A. Aquat. Microb. Ecol. 77, 125–138. doi: 10.3354/ame01794
Flombaum, P., Gallegos, J. L., Gordillo, R. A., Rincón, J., Zabala, L. L., Jiao, N., et al. (2013). Present and future global distributions of the marine Cyanobacteria Prochlorococcus and Synechococcus. Proc. Natl. Acad. Sci. U.S.A. 110, 9824–9829. doi: 10.1073/pnas.1307701110
Foster, R. A., Subramaniam, A., Mahaffey, C., Carpenter, E. J., Capone, D. G., and Zehr, J. P. (2007). Influence of the Amazon River plume on distributions of free-living and symbiotic cyanobacteria in the western tropical North Atlantic Ocean. Limnol. Oceanogr. 52, 517–532. doi: 10.4319/lo.2007.52.2.0517
Foster, R. A., Subramaniam, A., and Zehr, J. P. (2009). Distribution and activity of diazotrophs in the Eastern Equatorial Atlantic. Environ. Microbiol. 11, 741–750. doi: 10.1111/j.1462-2920.2008.01796.x
Foster, R. A., Sztejrenszus, S., and Kuypers, M. M. M. (2013). Measuring carbon and N2 fixation in field populations of colonial and free-living unicellular cyanobacteria using nanometer-scale secondary ion mass spectrometry1. J. Phycol. 49, 502–516. doi: 10.1111/jpy.12057
Gannon, J. E., and Gannon, S. A. (1975). Observations on the narcotization of crustacean zooplankton. Crustaceana 28, 220–224. doi: 10.1163/156854075X01099
Ger, K. A., Hansson, L.-A., and Lürling, M. (2014). Understanding cyanobacteria-zooplankton interactions in a more eutrophic world. Freshw. Biol. 59, 1783–1798. doi: 10.1111/fwb.12393
Gerphagnon, M., MacArthur, D. J., Latour, D., Gachon, C. M. M., Van Ogtrop, F., Gleason, F. H., et al. (2015). Microbial players involved in the decline of filamentous and colonial cyanobacterial blooms with a focus on fungal parasitism. Environ. Microbiol. 17, 2573–2587. doi: 10.1111/1462-2920.12860
Glibert, P. M., and Bronk, D. A. (1994). Release of dissolved organic nitrogen by marine diazotrophic cyanobacteria, Trichodesmium spp. Appl. Environ. Microbiol. 60, 3996–4000.
Goebel, N. L., Turk, K. A., Achilles, K. M., Paerl, R., Hewson, I., Morrison, A. E., et al. (2010). Abundance and distribution of major groups of diazotrophic cyanobacteria and their potential contribution to N2 fixation in the tropical Atlantic Ocean: diazotrophic cyanobacteria in the tropical North Atlantic. Environ. Microbiol. 12, 3272–3289. doi: 10.1111/j.1462-2920.2010.02303.x
Goes, J. I., Gomes, H., do, R., Chekalyuk, A. M., Carpenter, E. J., Montoya, J. P., et al. (2014). Influence of the Amazon River discharge on the biogeography of phytoplankton communities in the western tropical north Atlantic. Prog. Oceanogr. 120, 29–40. doi: 10.1016/j.pocean.2013.07.010
Gorsky, G., Chrétiennot-Dinet, M. J., Blanchot, J., and Palazzoli, I. (1999). Picoplankton and nanoplankton aggregation by appendicularians: fecal pellet contents of Megalocercus huxleyi in the equatorial Pacific. J. Geophys. Res. 104, 3381–3390. doi: 10.1029/98JC01850
Griese, M., Lange, C., and Soppa, J. (2011). Ploidy in cyanobacteria. FEMS Microbiol. Lett. 323, 124–131. doi: 10.1111/j.1574-6968.2011.02368.x
Grosse, J., Bombar, D., Doan, H. N., Nguyen, L. N., and Voss, M. (2010). The Mekong River plume fuels nitrogen fixation and determines phytoplankton species distribution in the South China Sea during low and high discharge season. Limnol. Oceanogr. 55, 1668–1680. doi: 10.4319/lo.2010.55.4.1668
Gruber, N. (2008). “Chapter 1 – The marine nitrogen cycle: overview and challenges,” in Nitrogen in the Marine Environment, 2nd Edn, eds D. G. Capone, D. A. Bronk, M. R. Mulholland, and E. J. Carpenter (San Diego, CA: Academic Press), 1–50.
Guo, C., and Tester, P. A. (1994). Toxic effect of the bloom-forming Trichodesmium sp. (cyanophyta) to the copepod Acartia tonsa. Nat. Toxins 2, 222–227. doi: 10.1002/nt.2620020411
Halm, H., Lam, P., Ferdelman, T. G., Lavik, G., Dittmar, T., LaRoche, J., et al. (2012). Heterotrophic organisms dominate nitrogen fixation in the South Pacific Gyre. ISME J. 6, 1238–1249. doi: 10.1038/ismej.2011.182
Hawser, S. P., O’neil, J. M., Roman, M. R., and Codd, G. A. (1992). Toxicity of blooms of the cyanobacterium Trichodesmium to zooplankton. J. Appl. Phycol. 4, 79–86. doi: 10.1007/BF00003963
Hewson, I., Govil, S. R., Capone, D. G., Carpenter, E. J., and Fuhrman, J. A. (2004). Evidence of Trichodesmium viral lysis and potential significance for biogeochemical cycling in the oligotrophic ocean. Aquat. Microb. Ecol. 36, 1–8. doi: 10.3354/ame036001
Hunt, B. P. V., Bonnet, S., Berthelot, H., Conroy, B. J., Foster, R. A., and Pagano, M. (2016). Contribution and pathways of diazotroph-derived nitrogen to zooplankton during the VAHINE mesocosm experiment in the oligotrophic New Caledonia lagoon. Biogeosci. Discuss. 13, 3131–3145. doi: 10.5194/bg-2015-614
Kâ, S., Mendoza-Vera, J. M., Bouvy, M., Champalbert, G., N’Gom-Kâ, R., and Pagano, M. (2011). Can tropical freshwater zooplankton graze efficiently on cyanobacteria? Hydrobiologia 679, 119–138. doi: 10.1007/s10750-011-0860-8
Kanagawa, T. (2003). Bias and artifacts in multitemplate polymerase chain reactions (PCR). J. Biosci. Bioeng. 96, 317–323. doi: 10.1016/S1389-1723(03)90130-7
Karl, D. M., Church, M. J., Dore, J. E., Letelier, R. M., and Mahaffey, C. (2012). Predictable and efficient carbon sequestration in the North Pacific Ocean supported by symbiotic nitrogen fixation. Proc. Natl. Acad. Sci. U.S.A. 109, 1842–1849. doi: 10.1073/pnas.1120312109
Landrum, J. P., Altabet, M. A., and Montoya, J. P. (2011). Basin-scale distributions of stable nitrogen isotopes in the subtropical North Atlantic Ocean: contribution of diazotroph nitrogen to particulate organic matter and mesozooplankton. Deep Sea Res. I 58, 615–625. doi: 10.1016/j.dsr.2011.01.012
Legendre, L., and Michaud, J. (1998). Flux of biogenic carbon in oceans: size-dependent regulation by pelagic food webs. Mar. Ecol. Prog. Ser. 164, 1–11. doi: 10.3354/meps164001
Leigh, J. A. (2000). Nitrogen fixation in methanogens: the archaeal perspective. Curr. Issues Mol. Biol. 2, 125–131.
Liu, H., and Buskey, E. J. (2000). The exopolymer secretions (EPS) layer surrounding Aureoumbra lagunensis cells affects growth, grazing, and behavior of protozoa. Limnol. Oceanogr. 45, 1187–1191. doi: 10.4319/lo.2000.45.5.1187
Loick-Wilde, N., Weber, S. C., Conroy, B. J., Capone, D. G., Coles, V. J., Medeiros, P. M., et al. (2015). Nitrogen sources and net growth efficiency of zooplankton in three Amazon River plume food webs. Limnol. Oceanogr. 61, 460–481. doi: 10.1002/lno.10227
Loick-Wilde, N., Weber, S. C., Conroy, B. J., Capone, D. G., Coles, V. J., Medeiros, P. M., et al. (2016). Nitrogen sources and net growth efficiency of zooplankton in three Amazon River plume food webs. Limnol. Oceanogr. 61, 460–481. doi: 10.1002/lno.10227
Luo, Y.-W., Doney, S. C., Anderson, L. A., Benavides, M., Berman-Frank, I., Bode, A., et al. (2012). Database of diazotrophs in global ocean: abundance, biomass and nitrogen fixation rates. Earth Syst. Sci. Data 4, 47–73. doi: 10.5194/essd-4-47-2012
Martínez-Pérez, C., Mohr, W., Löscher, C. R., Dekaezemacker, J., Littmann, S., Yilmaz, P., et al. (2016). The small unicellular diazotrophic symbiont, UCYN-A, is a key player in the marine nitrogen cycle. Nat. Microbiol. 1:16163. doi: 10.1038/nmicrobiol.2016.163
McClelland, J. W., Holl, C. M., and Montoya, J. P. (2003). Relating low δ15N values of zooplankton to N2-fixation in the tropical North Atlantic: insights provided by stable isotope ratios of amino acids. Deep Sea Res. I 50, 849–861. doi: 10.1016/S0967-0637(03)00073-6
McDonald, D., Price, M. N., Goodrich, J., Nawrocki, E. P., DeSantis, T. Z., Probst, A., et al. (2012). An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6, 610–618. doi: 10.1038/ismej.2011.139
Michaels, A. F., and Silver, M. W. (1988). Primary production, sinking fluxes and the microbial food web. Deep Sea Res. A 35, 473–490. doi: 10.1016/0198-0149(88)90126-4
Moisander, P. H., Beinart, R. A., Hewson, I., White, A. E., Johnson, K. S., Carlson, C. A., et al. (2010). Unicellular cyanobacterial distributions broaden the oceanic N2 fixation domain. Science 327, 1512–1514. doi: 10.1126/science.1185468
Montoya, J. P., Carpenter, E. J., and Capone, D. G. (2002). Nitrogen fixation and nitrogen isotope abundances in zooplankton of the oligotrophic North Atlantic. Limnol. Oceanogr. 47, 1617–1628. doi: 10.1371/journal.pone.0131258
Montoya, J. P., Holl, C. M., Zehr, J. P., Hansen, A., Villareal, T. A., and Capone, D. G. (2004). High rates of N2 fixation by unicellular diazotrophs in the oligotrophic Pacific Ocean. Nature 430, 1027–1031. doi: 10.1038/nature02824
Mulholland, M. R. (2007). The fate of nitrogen fixed by diazotrophs in the ocean. Biogeosciences 4, 37–51. doi: 10.5194/bg-4-37-2007
Needoba, J. A., Foster, R. A., Sakamoto, C., Zehr, J. P., and Johnson, K. S. (2007). Nitrogen fixation by unicellular diazotrophic cyanobacteria in the temperate oligotrophic North Pacific Ocean. Limnol. Oceanogr. 52, 1317–1327. doi: 10.4319/lo.2007.52.4.1317
Nejstgaard, J. C., Frischer, M. E., Simonelli, P., Troedsson, C., Brakel, M., Adiyaman, F., et al. (2008). Quantitative PCR to estimate copepod feeding. Mar. Biol. 153, 565–577. doi: 10.1007/s00227-007-0830-x
O’Neil, J. M. (1998). The colonial cyanobacterium Trichodesmium as a physical and nutritional substrate for the harpacticoid copepod Macrosetella gracilis. J. Plankton Res. 20, 43–59. doi: 10.1093/plankt/20.1.43
O’Neil, J. M., Metzler, P. M., and Glibert, P. M. (1996). Ingestion of 15N2-labelled Trichodesmium spp. and ammonium regeneration by the harpacticoid copepod Macrosetella gracilis. Mar. Biol. 125, 89–96. doi: 10.1007/BF00350763
O’Neil, J. M., and Roman, M. R. (1994). Ingestion of the cyanobacterium Trichodesmium spp. by pelagic harpacticoid copepods Macrosetella, Miracia and Oculosetella. Hydrobiologia 292, 235–240. doi: 10.1007/BF00229946
Paerl, H. W., and Otten, T. G. (2013). Harmful cyanobacterial blooms: causes, consequences, and controls. Microb. Ecol. 65, 995–1010. doi: 10.1007/s00248-012-0159-y
Pfannkuche, O., and Lochte, K. (1993). Open ocean pelago-benthic coupling: cyanobacteria as tracers of sedimenting salp faeces. Deep Sea Res. I 40, 727–737. doi: 10.1016/0967-0637(93)90068-E
Pomeroy, L. R., Williams, P. J., Azam, F., and Hobbie, J. E. (2007). The microbial loop. Oceanography 20, 28–33. doi: 10.5670/oceanog.2007.45
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Sandel, V., Kiko, R., Brandt, P., Dengler, M., Stemmann, L., Vandromme, P., et al. (2015). Nitrogen fuelling of the pelagic food web of the tropical Atlantic. PLoS ONE 10:e0131258. doi: 10.1371/journal.pone.0131258
Sargent, E. C., Hitchcock, A., Johansson, S. A., Langlois, R., Moore, C. M., LaRoche, J., et al. (2016). Evidence for polyploidy in the globally important diazotroph Trichodesmium. FEMS Microbiol. Lett. 363:fnw244. doi: 10.1093/femsle/fnw244
Satinsky, B. M., Crump, B. C., Smith, C. B., Sharma, S., Zielinski, B. L., Doherty, M., et al. (2014). Microspatial gene expression patterns in the Amazon River Plume. Proc. Natl. Acad. Sci. U.S.A. 111, 11085–11090. doi: 10.1073/pnas.1402782111
Scavotto, R. E., Dziallas, C., Bentzon-Tilia, M., Riemann, L., and Moisander, P. H. (2015). Nitrogen-fixing bacteria associated with copepods in coastal waters of the North Atlantic Ocean. Environ. Microbiol. 17, 3754–3765. doi: 10.1111/1462-2920.12777
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Sheridan, C. C., Steinberg, D. K., and Kling, G. W. (2002). The microbial and metazoan community associated with colonies of Trichodesmium spp.: a quantitative survey. J. Plankton Res. 24, 913–922. doi: 10.1093/plankt/24.9.913
Shoemaker, K. M., and Moisander, P. H. (2015). Microbial diversity associated with copepods in the North Atlantic subtropical gyre. FEMS Microbiol. Ecol. 91:fiv064. doi: 10.1093/femsec/fiv064
Simonelli, P., Troedsson, C., Nejstgaard, J. C., Zech, K., Larsen, J. B., and Frischer, M. E. (2009). Evaluation of DNA extraction and handling procedures for PCR-based copepod feeding studies. J. Plankton Res. 31, 1465–1474. doi: 10.1093/plankt/fbp087
Sohm, J. A., Edwards, B. R., Wilson, B. G., and Webb, E. A. (2011a). Constitutive extracellular polysaccharide (EPS) production by specific isolates of Crocosphaera watsonii. Front. Microbiol. 2:229. doi: 10.3389/fmicb.2011.00229
Sohm, J. A., Webb, E. A., and Capone, D. G. (2011b). Emerging patterns of marine nitrogen fixation. Nat. Rev. Microbiol. 9, 499–508. doi: 10.1038/nrmicro2594
Steinberg, D. K., Cope, J. S., Wilson, S. E., and Kobari, T. (2008). A comparison of mesopelagic mesozooplankton community structure in the subtropical and subarctic North Pacific Ocean. Deep Sea Res. II 55, 1615–1635. doi: 10.1016/j.dsr2.2008.04.025
Stukel, M. R., Coles, V. J., Brooks, M. T., and Hood, R. R. (2014). Top-down, bottom-up and physical controls on diatom-diazotroph assemblage growth in the Amazon River plume. Biogeosciences 11, 3259–3278. doi: 10.5194/bg-11-3259-2014
Stukel, M. R., Décima, M., Selph, K. E., Taniguchi, D. A. A., and Landry, M. R. (2013). The role of Synechococcus in vertical flux in the Costa Rica upwelling dome. Prog. Oceanogr. 11, 49–59. doi: 10.1016/j.pocean.2013.04.003
Subramaniam, A., Yager, P. L., Carpenter, E. J., Mahaffey, C., Björkman, K., Cooley, S., et al. (2008). Amazon River enhances diazotrophy and carbon sequestration in the tropical North Atlantic Ocean. Proc. Natl. Acad. Sci. U.S.A. 105, 10460–10465. doi: 10.1073/pnas.0710279105
Sukenik, A., Kaplan-Levy, R. N., Welch, J. M., and Post, A. F. (2012). Massive multiplication of genome and ribosomes in dormant cells (akinetes) of Aphanizomenon ovalisporum (Cyanobacteria). ISME J. 6, 670–679. doi: 10.1038/ismej.2011.128
Thompson, A., Carter, B. J., Turk-Kubo, K., Malfatti, F., Azam, F., and Zehr, J. P. (2014). Genetic diversity of the unicellular nitrogen-fixing cyanobacteria UCYN-A and its prymnesiophyte host. Environ. Microbiol. 16, 3238–3249. doi: 10.1111/1462-2920.12490
Thompson, A. W., Foster, R. A., Krupke, A., Carter, B. J., Musat, N., Vaulot, D., et al. (2012). Unicellular cyanobacterium symbiotic with a single-celled eukaryotic alga. Science 337, 1546–1550. doi: 10.1126/science.1222700
Thompson, A. W., and Zehr, J. P. (2013). Cellular interactions: lessons from the nitrogen-fixing cyanobacteria. J. Phycol. 49, 1024–1035. doi: 10.1111/jpy.12117
Troedsson, C., Simonelli, P., Nägele, V., Nejstgaard, J. C., and Frischer, M. E. (2009). Quantification of copepod gut content by differential length amplification quantitative PCR (dla-qPCR). Mar. Biol. 156, 253–259. doi: 10.1007/s00227-008-1079-8
True, A. C., Webster, D. R., Weissburg, M. J., Yen, J., and Genin, A. (2015). Patchiness and depth-keeping of copepods in response to simulated frontal flows. Mar. Ecol. Prog. Ser. 539, 65–76. doi: 10.3354/meps11472
Turk-Kubo, K. A., Frank, I. E., Hogan, M. E., Desnues, A., Bonnet, S., and Zehr, J. P. (2015). Diazotroph community succession during the VAHINE mesocosm experiment (New Caledonia lagoon). Biogeosciences 12, 7435–7452. doi: 10.5194/bg-12-7435-2015
Turner, J. T. (2014). Planktonic marine copepods and harmful algae. Harmful Algae 32, 81–93. doi: 10.1016/j.hal.2013.12.001
Villareal, T. A., Brown, C. G., Brzezinski, M. A., Krause, J. W., and Wilson, C. (2012). Summer diatom blooms in the North Pacific subtropical gyre: 2008–2009. PLoS ONE 7:e33109. doi: 10.1371/journal.pone.0033109
von Elert, E., Martin-Creuzburg, D., and Le Coz, J. R. (2003). Absence of sterols constrains carbon transfer between cyanobacteria and a freshwater herbivore (Daphnia galeata). Proc. Biol. Sci. 270, 1209–1214. doi: 10.1098/rspb.2003.2357
Wiebe, P. H., Burt, K. H., Boyd, S. H., and Morton, A. W. (1976). A multiple opening/closing net and environmental sensing system for sampling zooplankton. J. Mar. Res. 34, 313–326.
Wilson, S., and Steinberg, D. (2010). Autotrophic picoplankton in mesozooplankton guts: evidence of aggregate feeding in the mesopelagic zone and export of small phytoplankton. Mar. Ecol. Prog. Ser. 412, 11–27. doi: 10.3354/meps08648
Woodson, C. B., and Litvin, S. Y. (2015). Ocean fronts drive marine fishery production and biogeochemical cycling. Proc. Natl. Acad. Sci. U.S.A. 112, 1710–1715. doi: 10.1073/pnas.1417143112
Woodson, C. B., Webster, D. R., Weissburg, M. J., and Yen, J. (2005). Response of copepods to physical gradients associated with structure in the ocean. Limnol. Oceanogr. 50, 1552–1564. doi: 10.4319/lo.2005.50.5.1552
Yeung, L. Y., Berelson, W. M., Young, E. D., Prokopenko, M. G., Rollins, N., Coles, V. J., et al. (2012). Impact of diatom-diazotroph associations on carbon export in the Amazon River plume. Geophys. Res. Lett. 39:L18609. doi: 10.1029/2012GL053356
Zehr, J. P. (2011). Nitrogen fixation by marine cyanobacteria. Trends Microbiol. 19, 162–173. doi: 10.1016/j.tim.2010.12.004
Zehr, J. P., Jenkins, B. D., Short, S. M., and Steward, G. F. (2003). Nitrogenase gene diversity and microbial community structure: a cross-system comparison. Environ. Microbiol. 5, 539–554. doi: 10.1046/j.1462-2920.2003.00451.x
Keywords: mesozooplankton, cyanobacteria, diazotroph, grazing, Amazon River plume, North Atlantic Ocean, nitrogen incorporation
Citation: Conroy BJ, Steinberg DK, Song B, Kalmbach A, Carpenter EJ and Foster RA (2017) Mesozooplankton Graze on Cyanobacteria in the Amazon River Plume and Western Tropical North Atlantic. Front. Microbiol. 8:1436. doi: 10.3389/fmicb.2017.01436
Received: 02 March 2017; Accepted: 14 July 2017;
Published: 03 August 2017.
Edited by:
Ajit Subramaniam, Lamont Doherty Earth Observatory (LDEO), United StatesReviewed by:
Rainer Kiko, GEOMAR Helmholtz Centre for Ocean Research Kiel (HZ), GermanyAssaf Sukenik, Israel Oceanographic & Limnological Research, Israel
Copyright © 2017 Conroy, Steinberg, Song, Kalmbach, Carpenter and Foster. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Brandon J. Conroy, YnJhbmRvbi5qLmNvbnJveS5wdWJzQGdtYWlsLmNvbQ==
 Brandon J. Conroy
Brandon J. Conroy Deborah K. Steinberg
Deborah K. Steinberg Bongkuen Song
Bongkuen Song Andrew Kalmbach
Andrew Kalmbach Edward J. Carpenter
Edward J. Carpenter Rachel A. Foster
Rachel A. Foster