- Department of Plant and Microbial Biology, University of Zurich, Zurich, Switzerland
Members of the genus Burkholderia (β-proteobacteria) have only recently been shown to be able to establish a nitrogen-fixing symbiosis with several legumes, which is why they are also referred to as β-rhizobia. Therefore, very little is known about the competitiveness of these species to nodulate different legume host plants. In this study, we tested the competitiveness of several Burkholderia type strains (B. diazotrophica, B. mimosarum, B. phymatum, B. sabiae, B. symbiotica and B. tuberum) to nodulate four legumes (Phaseolus vulgaris, Macroptilium atropurpureum, Vigna unguiculata and Mimosa pudica) under our closely defined growth conditions. The assessment of nodule occupancy of these species on different legume host plants revealed that B. phymatum was the most competitive strain in the three papilionoid legumes (bean, cowpea and siratro), while B. mimosarum outcompeted the other strains in mimosa. The analysis of phenotypes known to play a role in nodulation competitiveness (motility, exopolysaccharide production) and additional in vitro competition assays among β-rhizobial strains suggested that B. phymatum has the potential to be a very competitive legume symbiont.
Introduction
The rhizobia-legume symbiosis is estimated to contribute nearly half of all current biological nitrogen fixation and is an essential element of agricultural sustainability (Vitousek et al., 2002). This symbiosis is manifested by the formation of root nodules on legume plants, in which rhizobia fix atmospheric nitrogen to the benefit of the plant and in exchange for nutrient supply and protection against environmental stresses (Downie, 2010; Oldroyd et al., 2011). Rhizobia are taxonomically diverse and polyphyletic (Masson-Boivin et al., 2009; Sprent et al., 2017). Until recently, all known examples of legume-rhizobial symbiosis were confined to the α-proteobacteria group (Young and Haukka, 1996). This changed with the discovery of nodulating Burkholderia and Cupriavidus strains, both of which belong to the β-proteobacteria (Chen et al., 2001; Moulin et al., 2001; Bontemps et al., 2010). The number of legume nodulating strains belonging to the environmental clade of the genus Burkholderia, for which the new genus Paraburkholderia has been proposed recently (Sawana et al., 2014; Beukes et al., 2017), has increased rapidly over the last years (Gyaneshwar et al., 2011; Howieson et al., 2013; Lemaire et al., 2015). These strains of the Paraburkholderia clade were shown to nodulate a wide range of legumes, among them several Mimosa species (sub-family Mimosoideae) and Papilionoid legumes from different tribes and from several regions of the world (Chen et al., 2003; Chen et al., 2005a; Elliott et al., 2007b; dos Reis et al., 2010; Liu et al., 2012; Mishra et al., 2012; Bournaud et al., 2013). All nodulating and diazotrophic Burkholderia species described so far, with the exception of the South African strains B. tuberum (Elliott et al., 2007a) and B. dipogonis (Liu et al., 2014), were either isolated from or are able to nodulate Mimosa plants (Bournaud et al., 2013). The diversity of mimosoid-nodulating Burkholderia is high and includes species such as B. phymatum, which is able to nodulate several important legumes including the common bean (Phaseolus vulgaris) (Talbi et al., 2010; Moulin et al., 2014). Previous studies have shown that an individual legume plant can be infected by more than one rhizobial strain (Barrett and Parker, 2006; Klonowska et al., 2012; Mishra et al., 2012; Melkonian et al., 2014). For example, Mimosa spp. was found to host Rhizobium spp., Burkholderia and Cupriavidus strains (Barrett and Parker, 2006; dos Reis et al., 2010; Gyaneshwar et al., 2011; Bontemps et al., 2016). Two previous studies showed that β-rhizobia are very dominant in the genus Mimosa (Elliott et al., 2009; Melkonian et al., 2014). In particular, Elliott et al. (2009) showed that B. mimosarum was more competitive than Cupriavidus taiwanensis and other tested α-rhizobia on several Mimosa species, while Melkonian et al. (2014) reported that B. phymatum and B. tuberum were more competitive than α-rhizobia on M. pudica. In that same study, they also tested the effects of environmental conditions on the competitiveness of Burkholderia against other rhizobial species (i.e., Cupriavidus) and showed that nitrogen-limited conditions favor the dominance of Burkholderia species (Elliott et al., 2009). Other studies showed that β-rhizobial competitiveness was greatly affected by soil pH (Garau et al., 2009; Angus et al., 2013). Beside environmental conditions, competition has been shown to depend on the geographical position and on symbiont and host plant genetic diversity (Dowling and Broughton, 1986; Mishra et al., 2012; Melkonian et al., 2014; Bontemps et al., 2016). For the well-studied rhizobia from the α-subclass of proteobacteria, several phenotypic traits were shown to be important for their nodulation competitiveness (Triplett and Sadowsky, 1992). These included motility (Mellor et al., 1987; Liu et al., 1989), exopolysaccharide (EPS) and lipopolysaccharide (LPS) production (Bhagwat et al., 1991; Zdor and Pueppke, 1991; Geddes et al., 2014), antibiotic production (Schwinghamer and Belkengren, 1968; Robleto et al., 1998) and the catabolism of certain compounds such as proline, myo-inositol, and glycerol (Jiménez-Zurdo et al., 1995; Fry et al., 2001; Kohler et al., 2010; Ding et al., 2012).
In this work, we aimed to study the competition among β-rhizobial strains of the genus Burkholderia using – in addition to the well-studied β-rhizobial plant host Mimosa pudica – three papilionoid legumes of major economic importance in agriculture: P. vulgaris (bean), Macroptilium atropurpureum (siratro) and Vigna unguiculata (cowpea). We used sterile vermiculite and constant growth conditions to investigate the competitiveness of several different Burkholderia type strains (B. diazotrophica LMG26031, B. mimosarum LMG23256, B. phymatum LMG21445, B. sabiae LMG24235, B. symbiotica LMG26032, B. tuberum LMG21444) to nodulate a single legume. Seeds from four different host plants (M. pudica, bean, cowpea and siratro) were inoculated with a bacterial mixture and after 3 or 4 weeks, the nodule occupancy rates of each species on different legumes were assessed by employing specific PCRs and recA sequencing. B. phymatum revealed to be the most competitive strain in nodulating the roots of three of the four tested legumes under the closely defined growth conditions that we used. B. mimosarum is shown to outcompete the other strains in M. pudica. The assessment of traits important for competitiveness provided further support that B. phymatum is very competitive, as the strain (i) was among the most motile strains, (ii) produced more EPSs compared to the other strains, and (iii) outcompeted the other strains in vitro.
Materials and Methods
Bacterial Strains, Plasmids and Growth Conditions
The bacterial strains, plasmids and primers employed in this work are listed in Supplementary Table S1. Burkholderia strains were cultivated under aerobic conditions at 30°C in LB medium without salt (10 g of tryptone and 5 g yeast extract per liter). Bacterial cultures for competition plant tests were washed in defined buffered AB-minimal medium (Clark and Maaloe, 1967) without nitrogen and with 10 mM sodium citrate as the carbon source, while for phenotypical tests they were washed in LB medium without salt. Growth of the Burkholderia strains on plates was assessed in AB-minimal medium (Clark and Maaloe, 1967) containing 10 mM glucose, 15 mM succinic acid or 10 mM sodium citrate as carbon source; PIA (Pseudomonas Isolation Agar, Difco) and in LB medium without salt. To assess liquid growth in LB without salt, cultures were grown at 30°C with agitation (220 rpm) in 250 ml Erlenmeyer flasks containing 100 ml of medium. Independent duplicates for all the bacterial strains were tested.
Plant Growth Conditions
Common bean seedlings (Phaseolus vulgaris, cv. Negro jamapa; kindly provided by Professor Eulogio Bedmar, Granada, Spain) were surface sterilized as previously described (Talbi et al., 2010). Macroptilium atropurpureum (siratro), Vigna unguiculata (cowpea) and Mimosa pudica (mimosa) were surface sterilized as described previously (Klonowska et al., 2012). Seeds were subsequently deposited on 0.8% agar plates and incubated in the dark at 30°C. After one and a half days, germinated seedlings were planted into 250 ml autoclaved yogurt-jars containing vermiculite (VTT-Group, Muttenz, Switzerland) and 170 ml diluted Jensen medium (Hahn and Hennecke, 1984). The pH in the pots was 6.5. Direct seed inoculation with the individual bacterial strains or with the specific bacterial mixtures was performed. The plants were grown with the following parameters: temperature, 22°C at night and 25°C during the day; light, approximately 16 h (200 μMol intensity); humidity 60%. The plants were harvested 21 days post infection (dpi) for bean and cowpea or 28 dpi for siratro and mimosa.
Determination of Symbiotic Effectiveness
Nodule number and nodule dry weight were determined as described previously (Göttfert et al., 1990). For the determination of the shoot dry weight and N content, six plants for each of the bacteria–plant combinations tested were dried at 60°C for 72 h. Shoots were first weighed, crushed in a mortar and placed in one or more 2 ml Eppendorf. Each sample was homogenized using a Tissuelyser (Qiagen, Hilden, Germany) and a glass bead of 7 mm diameter (Assistent, Rhön, Germany) for 2 min at 30 Hertz. After homogenization, 1.5–2 mg of the samples were taken for the determination of the total N content using LECO TruSpec CHM micro instrument (LECO Corporation, St. Joseph, MI, United States), which is based on the Dumas or combustion method.
Competition Assays In Planta
The strains were grown in 4 ml LB medium without salt until an optical density (OD600) of 2. At this optical density, all strains were in the same growth phase (end of exponential phase). After washing the cultures twice in minimal medium without nitrogen, the OD600 of each culture was adjusted to 0.01 and subsequently diluted 1000-fold. This dilution was plated to ensure that the same amount of cells was used for each strain in the mix. The bacterial mixture containing around 102 colony forming units (CFU) each of B. diazotrophica, B. mimosarum, B. phymatum, B. sabiae and B. tuberum was immediately applied on bean and siratro germinated seeds (Mix 1). The same mix excluding B. tuberum was used for cowpea (Mix 2). Mimosa seeds were inoculated with 102 CFU each of B. diazotrophica, B. mimosarum, B. phymatum and B. symbiotica (Mix 3). The bacterial mixtures were prepared in equal volumes of cell suspension (1 ml/seedling). The plants in sterilized vermiculite were grown as described above and regularly watered with deionized water. Two independent experiments were performed and per experiment 4–6 plants for bean, cowpea, mimosa and siratro were analyzed. After 21 dpi (for bean and cowpea) or 28 dpi (for siratro and mimosa), roots were surface sterilized by soaking for 10 s in 100% ethanol, followed by 3 min in 2.5% sodium hypochlorite and then washed 5 times with sterile deionized water (Vincent, 1970). All the nodules present on the root of each plant were individually collected in an Eppendorf containing 100 μl LB without salt and 100 μl 50% glycerol. Afterward, each nodule of one plant was crushed and the extract was plated onto solid 0.06% YEM medium. Single colonies and 2 μl from the nodule extract were tested by PCR using specific primers (Supplementary Table S1): for B. phymatum (Bphy_comp_F, TGCGCTGCTTTCCATTTCAC and Bphy_comp_R, AGTAGTCGCTGCTATCGTGC), for B. mimosarum (Bmim_comp_F, GCACTTTACGTCCAGACACG and Bmim_comp_R, CGTCGTGAGTCAGGTAACCA) and for B. tuberum (Btub_comp_F, GCCGAACTAGGATTGTACGC and Btub_comp_R, CGCGAACTCCAGACACAATA). If no product was obtained, PCR were performed with Burkholderia degenerate recA primers (recABurk1_F, GATCGARAAGCAGTTCGGCAA and recABurk1_R, TTGTCCTTGCCCTGRCCGAT), which amplified a product with all the six Burkholderia strains used (Mishra et al., 2012). To identify the strains specifically, the obtained PCR product was purified and sequenced (at Microsynth AG, Switzerland). Further proof for all the nodules infected by B. tuberum was obtained by plating the nodule extract on PIA plates, a medium that allows selective growth of B. tuberum. Nodules obtained from the second plant competition test were directly tested by PCR from the crushed material. The nodule occupancy rate for each legume was calculated by dividing the number of nodules occupied by one strain by the total number of nodules analyzed and present on the roots.
Phenotypical Analysis Including Competition Assays In Vitro
Exopolysaccharide production was visualized on modified YEM medium plates (1% mannitol, 0.06% yeast extract) (Zlosnik et al., 2008). Plates were incubated for 4 days at 30°C. Swimming motility was tested by inoculating cells onto plates containing LB without salt supplemented with 0.1% casamino acids that were solidified with 0.2% agar (Eberl et al., 1996; Lardi et al., 2015). Plates were incubated for 30 h at 30°C. Competition experiments on plate (in vitro) were performed in 50:50 ratio for B. phymatum against B. tuberum, B. mimosarum against B. tuberum and B. mimosarum against B. phymatum GFP-tagged (Elliott et al., 2007b). Cultures were grown o/n in LB medium without salt until an OD600 of 3 and washed once with LB without salt. The inoculum was normalized at OD600 = 0.1 (corresponding to a CFU of 108). The bacteria of interest were equally mixed in a 50:50 volume and 5 μl were spotted onto an LB plate without salt and incubated for 24 h at 30°C. The cells were recollected in 1 ml of LB without salt and CFU were counted on LB without salt plates and onto selective plates, i.e., PIA for B. tuberum and LB medium without salt containing tetracycline for B. phymatum GFP.
Statistical Analyses
Swimming motility was statistically analyzed using unpaired t-test (∗∗∗∗p < 0.0001, ∗∗∗p < 0.001, ∗∗p < 0.01 and ∗p < 0.05). The analyses were performed using GraphPad Prism software version 7.0c. For the competition assay in vitro standard deviation (SD) was calculated and inserted into the histograms. For determination of dry weight, N content and nodule dry weight ANOVA and Tukey’s test were performed. The correlation between N content, nodule dry weight and nodule number was analyzed with GraphPad Prism software version 7.0c.
Results
Growth and Nodulation Behavior of the Burkholderia Strains Used in Competition
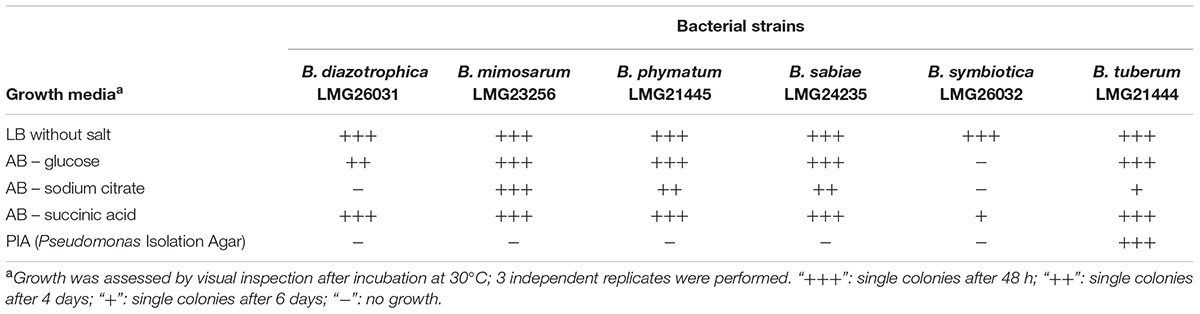
Before setting up the competition experiments, the growth behavior of the different Burkholderia strains, i.e., B. diazotrophica [originally isolated from Mimosa candollei; (Sheu et al., 2013)], B. mimosarum [originally isolated from Mimosa pigra, (Chen et al., 2006)], B. phymatum [isolated from Mimosa species (Elliott et al., 2007b)], B. sabiae [originally isolated from Mimosa caesalpiniifolia, (Chen et al., 2008)], B. symbiotica [originally isolated from Mimosa cordistipula, (Sheu et al., 2012)] and B. tuberum [originally isolated from Aspalathus carnosa, (Moulin et al., 2001; Vandamme et al., 2002)], was tested in five different media (Table 1). All six strains were able to grow within 48 h in LB without salt and in AB minimal medium with succinic acid as carbon source, with the exception of B. symbiotica that needed 6 days to grow on AB minimal medium with succinic acid. When AB minimal medium was supplemented with sodium citrate as carbon source, B. diazotrophica and B. symbiotica were not able to grow. Additionally, B. symbiotica could not use glucose as carbon source. B. tuberum was the only strain able to grow on PIA, which contains a broad-spectrum antimicrobial (Irgasan) that is commonly used to select for Burkholderia strains (Kost et al., 2014).
A growth kinetic analysis in LB liquid medium without salt over 25 h showed that all strains displayed similar growth rates and reached the stationary phase at an OD600 of approximately 3 (Supplementary Figure S1).
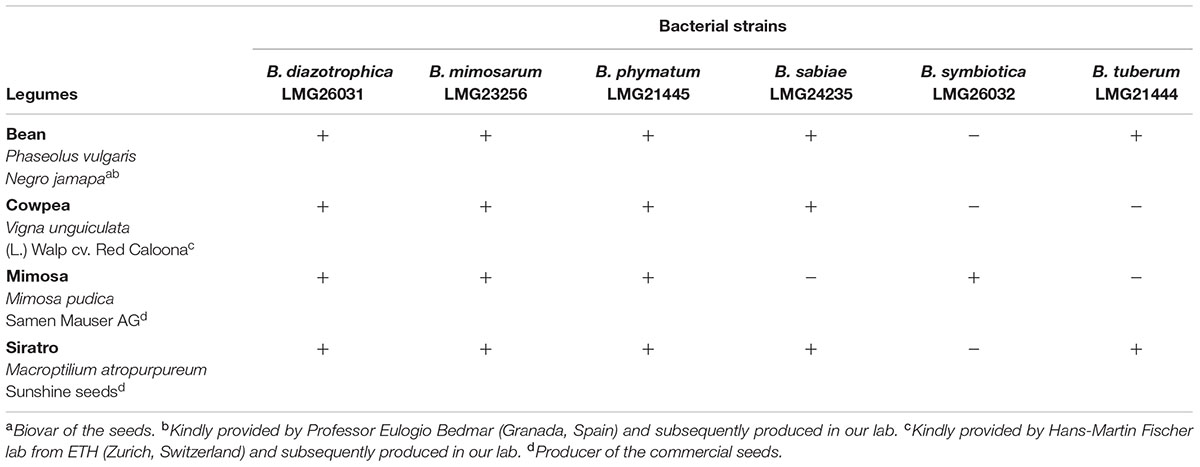
Next, we tested nodulation for each of the overall 24 different Burkholderia-legume combinations (Table 2). B. diazotrophica, B. mimosarum and B. phymatum were able to nodulate all the four tested legumes, i.e., M. pudica (tribe Mimoseae), P. vulgaris (bean; tribe Phaseoleae), M. atropurpureum (siratro; tribe Phaseoleae) and V. unguiculata (cowpea; tribe Phaseoleae). B. sabiae formed nodules on all legumes except on mimosa. While B. tuberum induced nodules in bean and siratro, B. symbiotica was the most host-specific strain tested and only able to nodulate mimosa.
Capacity of Different Burkholderia Strains to Compete for Nodulation of Different Legumes
Nodulation competitiveness of B. diazotrophica, B. mimosarum, B. phymatum, B. sabiae, B. symbiotica and B. tuberum was determined by competition experiments using different host plants. Importantly, we performed all experiments by growing the plants in sterile vermiculite and under our own closely defined growth conditions (see Materials and Methods and Figure 1). Commercial seeds from the mimosoid legume M. pudica were inoculated with a 1 ml mixture containing around 100 cells (Supplementary Figure S2) of the following strains, which we had shown before (s. above) to be able to nodulate M. pudica: B. diazotrophica, B. mimosarum, B. phymatum and B. symbiotica (mix 3) (Figure 2 and Table 2). V. unguiculata seeds were infected with a mixture containing B. diazotrophica, B. mimosarum, B. phymatum and B. sabiae (mix 2) (Figure 2 and Table 2). The same mix with B. diazotrophica, B. mimosarum, B. phymatum, B. sabiae and in addition B. tuberum (mix 1) was used to inoculate the commercial seeds of M. atropurpureum and the seeds of P. vulgaris (Figure 2 and Table 2).
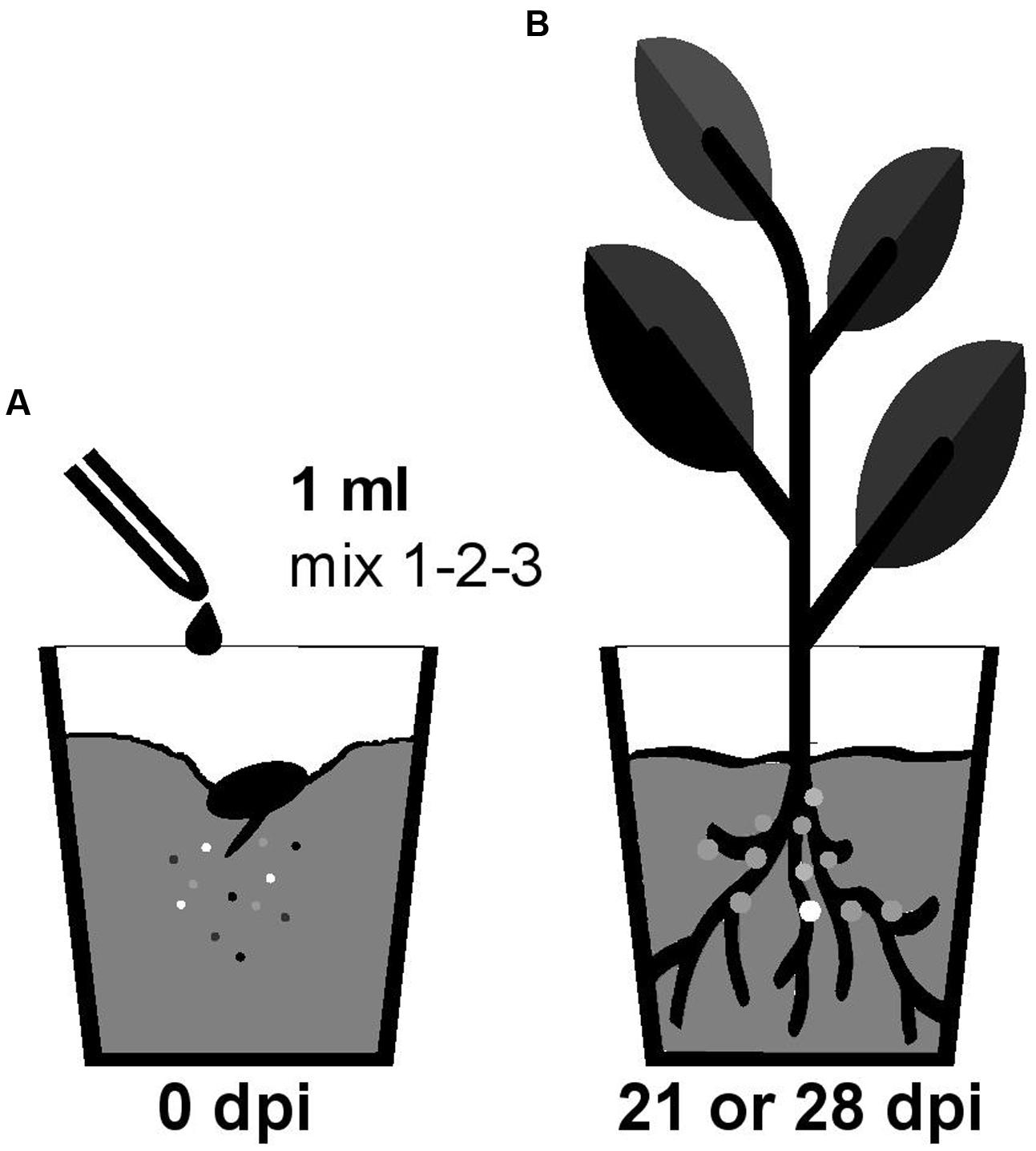
FIGURE 1. Workflow of the competition experiment in planta. Germinated seedlings were inoculated with 1 ml of bacterial mix (mix 1 for siratro and bean; mix 2 for cowpea and mix 3 for mimosa) (A). After 21 (bean and cowpea) or 28 (mimosa and siratro) days all the bacteria present within nodules of the root apparatus were identified by PCR with degenerate primers and sequencing (B). The different colors represent the different Burkholderia type strains present in the mix (A) and inside the nodules (B).
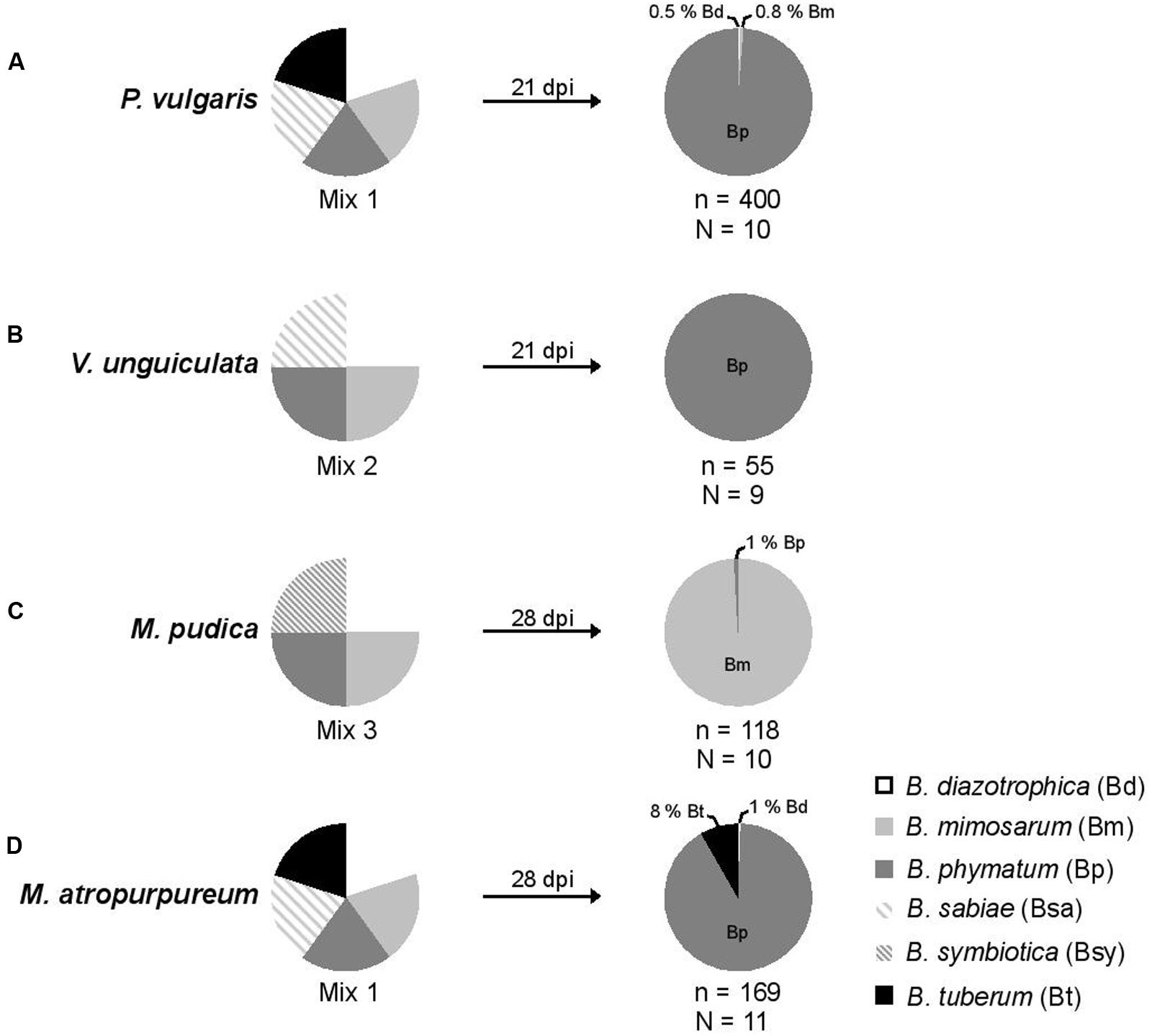
FIGURE 2. Relative nodule occupancy of several nodulating and nitrogen-fixing Burkholderia strains in four legumes. For each tested legume, the ratio of each bacteria in the initial inoculum is shown followed by the ratio obtained 21 (bean and cowpea) or 28 (mimosa and siratro) days post infection (dpi). A mixture of five Burkholderia strains – B. diazotrophica, B. mimosarum, B. phymatum, B. sabiae, and B. tuberum (mix 1) – was inoculated on bean (P. vulgaris) (A) and siratro (M. atropurpureum) (D). The same bacterial mixture excluding B. tuberum (mix 2) was applied onto cowpea (V. unguiculata) (B). For mimosa (M. pudica) (C) four Burkholderia strains were inoculated: B. diazotrophica, B. mimosarum, B. phymatum and B. symbiotica (mix 3). “N” indicates the number of analyzed plants and “n” the number of analyzed nodules.
In order to provide the best possible analysis of the nodule occupancy, we decided to harvest and individually assess each and every single root nodule (typically ranging from a minimum of 2 in V. unguiculata to a maximum of 47 in P. vulgaris) per plant. After 3 or 4 weeks all nodules from one plant were collected and subsequently analyzed for occupancy using strain-specific PCRs (see Materials and Methods section for details). The identity of the strains was verified by amplifying and sequencing the recA gene (Mishra et al., 2012). The results showed that bean was preferentially nodulated by B. phymatum (99% of the nodules), and only 2 and 3 nodules out of 400 analyzed were occupied by B. diazotrophica and B. mimosarum, respectively (Figure 2A). While B. phymatum was the most competitive strain on siratro (91% nodule occupancy), 8% of the siratro nodules (14) contained B. tuberum, and one nodule was occupied by B. diazotrophica (Figure 2D). The nodule occupancy of B. phymatum in cowpea was 100% in all the 9 analyzed plants (55 nodules overall) (Figure 2B). In mimosa B. mimosarum occupied 99% of the nodules. Only one nodule out of 118 analyzed nodules was occupied by B. phymatum (Figure 2C). Using this strategy, we did not identify any nodules colonized by two species (mixed nodules).
Competitiveness for Nodulation versus Symbiotic Performance
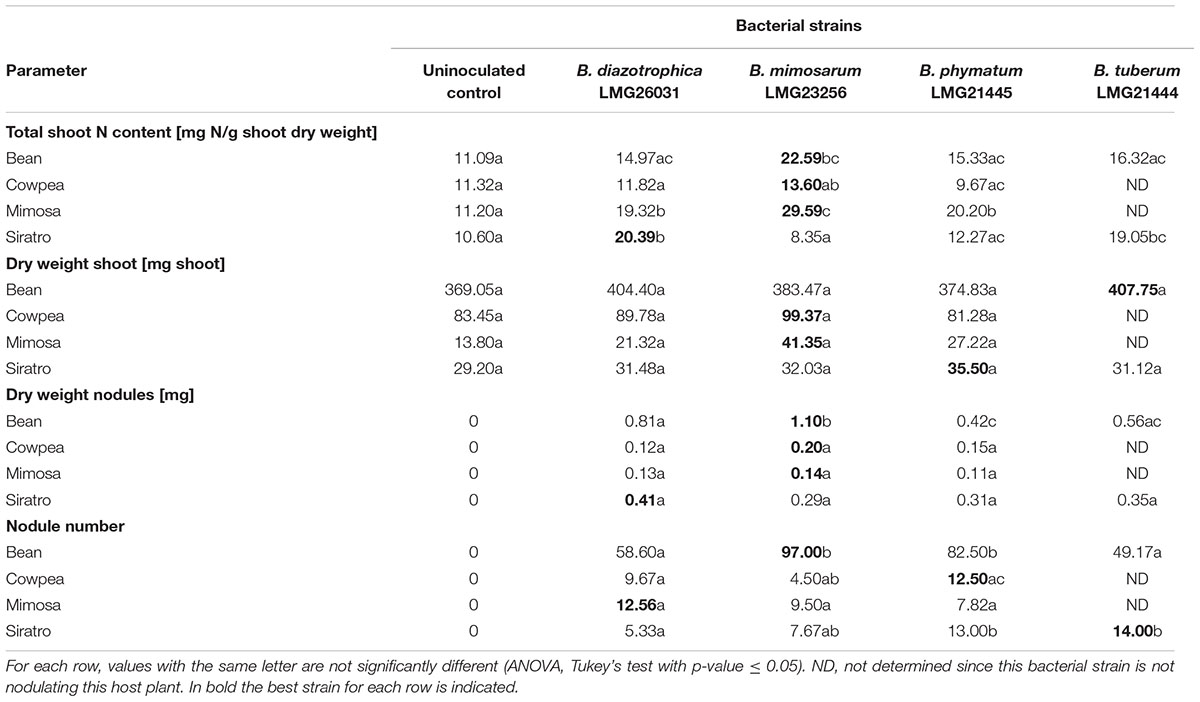
The performance of a single strain inoculated in a single legume was assessed by recording shoot dry mass, shoot N content, nodule number per plant and dry weight per nodule (Table 3). We tried all legumes in combinations with the four most competitive strains – B. diazotrophica, B. mimosarum, B. phymatum and B. tuberum – (Table 3) by using the same plant growth conditions we applied for competition experiments (see Materials and Methods). Competition experiments on bean revealed that B. phymatum is the most competitive strain and only few nodules were colonized by B. mimosarum and B. diazotrophica (s. above). The symbiotic efficiency on bean differed depending on the inoculated strain. Plants inoculated with B. mimosarum showed a higher shoot N content when compared with the other three strains tested (Table 3). Bean plants infected with B. tuberum also displayed a higher shoot dry weight and N content compared to the most competitive strain B. phymatum. Inoculation of single strains on cowpea revealed that the otherwise best competitor B. phymatum was not performing well in this host plant. In fact, the shoot dry weight and the N content of plants inoculated with B. phymatum was similar to that measured in uninoculated control plants (Table 3). In M. pudica, B. mimosarum was not only outcompeting the other strains but also performed best in terms of shoot dry weight and provided N content (Table 3). Based on the determination of N content, the most efficient strains in providing nitrogen to siratro plants were B. diazotrophica and B. tuberum (Table 3). Although to a lesser extent than B. phymatum, B. tuberum was able to colonize few nodules in competition with the other bacterial strains (Figure 2). In contrast, the best competitor in siratro, B. phymatum, was less efficient in providing nitrogen to host plants compared to B. diazotrophica and B. tuberum when inoculated alone.
For some Burkholderia – legume combinations we observed a positive correlation between shoot N contents and nodule dry weight and a negative correlation between shoot N contents and number of nodules. For instance, in siratro B. diazotrophica produced the highest shoot N content and nodule dry weight, but showed the lowest number of nodules. In cowpea, B. mimosarum had the same effect. Indeed, the legume usually regulates the number of nodules according to their activity, which is proportional to their size (Ferguson et al., 2010). In the case of common bean, that was shown to have a relaxed autoregulation of nodulation (AON) (George and Robert, 1991) no negative correlation between N content and nodule number was observed.
Phenotypic Analysis of the Burkholderia Strains Used in Competition
Two phenotypes known to be relevant for nodulation competitiveness in α-Rhizobia were assessed (Triplett and Sadowsky, 1992): EPS production and motility. Figure 3 shows that on an agar plate containing mannitol the most competitive strain on bean, cowpea and siratro, B. phymatum, was more mucoid compared to the other five strains, suggesting an increased EPS production. This difference among the tested strains is visible even after a longer incubation of the plates (6 days). In addition, B. phymatum was also able to swim; in fact, it was the second-best swimmer after B. sabiae (Figure 4). While B. mimosarum’s swimming ability was about one third of that of B. sabiae, the other three strains – B. diazotrophica, B. symbiotica and B. tuberum – were drastically impaired in their swimming capabilities compared to B. sabiae.
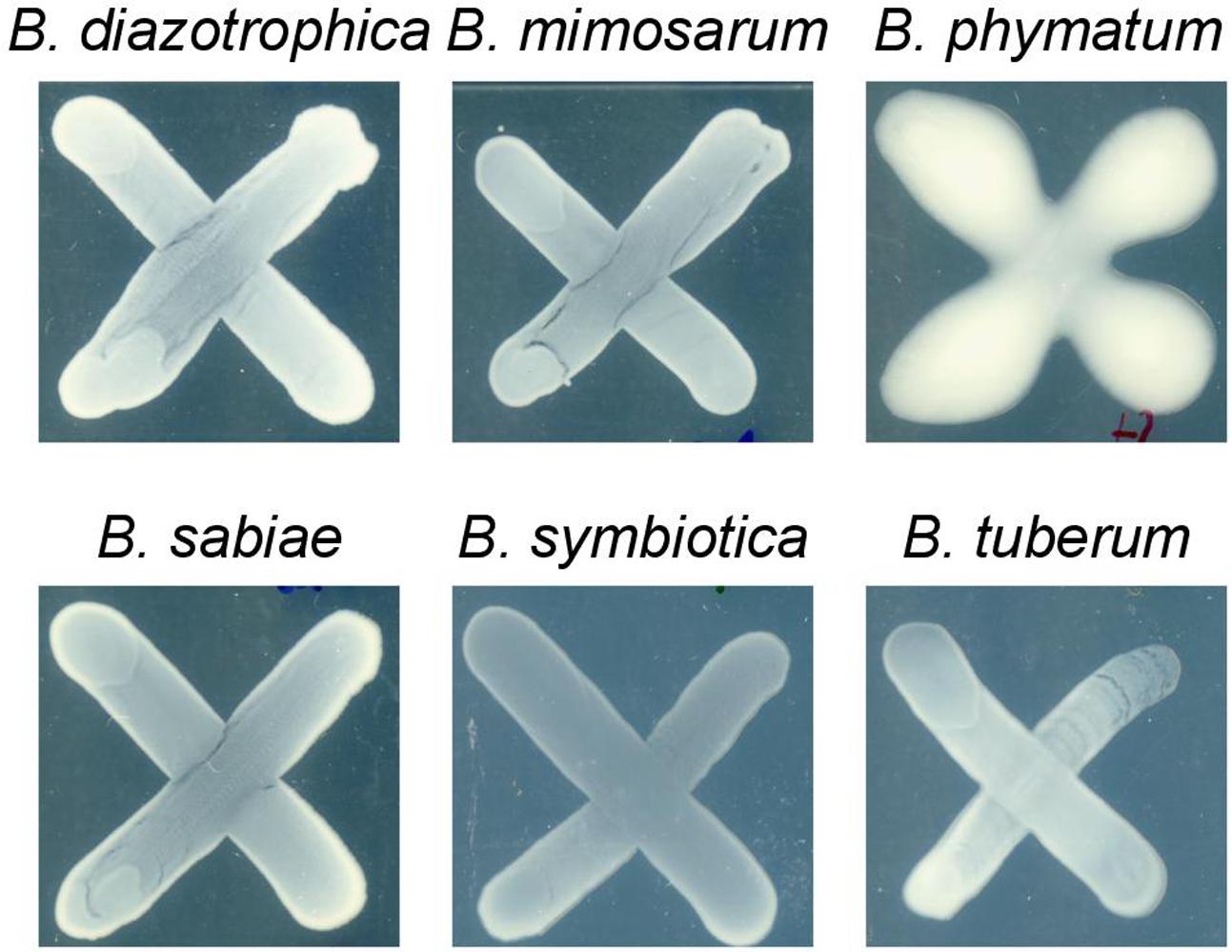
FIGURE 3. Exopolysaccharides (EPS) production in six tested Burkholderia strains. The plates were supplemented with 0.06% of yeast extract and incubated for 4 days. At least 3 independent replicates were performed per each strain.
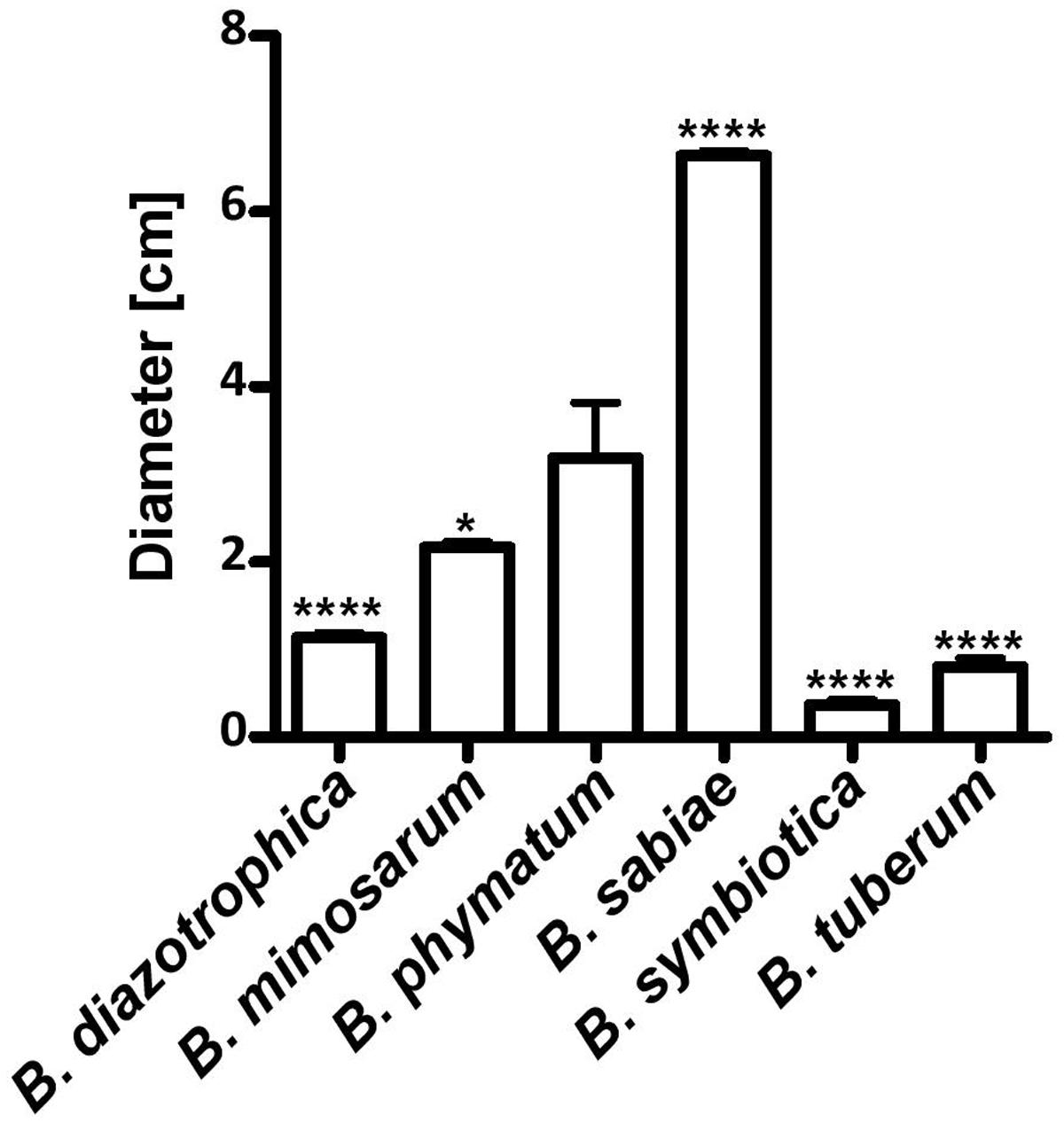
FIGURE 4. Motility of six nodulating and nitrogen-fixing Burkholderia type strains. Swimming motility on LB without salt plates was measured after 30 h incubation. Error bars indicate SD. At least 3 independent replicates were performed. Asterisks indicate a significant difference between B. phymatum and the indicated strain (∗∗∗∗p-value < 0.0001; ∗p-value < 0.05; unpaired two-tailed t-test).
We next evaluated if the competition occurs already under free-living conditions. For this we mixed two strains in equal amount and incubated them on an agar plate for 24 h. As a control, each strain was spotted alone on the plate and was incubated for the same time. The following combinations were tested: B. phymatum – B. mimosarum, B. phymatum – B. tuberum and B. mimosarum – B. tuberum (Figure 5). In the first combination, B. phymatum was outcompeting B. mimosarum and after 24 h only 8% of the cells were B. mimosarum (Figure 5A and Supplementary Figure S3A). When competing B. phymatum against B. tuberum, we recovered about 88% B. phymatum and 12% B. tuberum (Figure 5B and Supplementary Figure S3B). The last combination tested revealed that B. mimosarum outcompeted B. tuberum (94% against 6% of recovered cells) (Figure 5C and Supplementary Figure S3C). Interestingly, while the amount of B. phymatum cells growing alone or as a mixture remained constant after 24 h, the number of cells of the outcompeted strain (B. mimosarum or B. tuberum) was always around 10-fold lower after competition (Supplementary Figure S3).
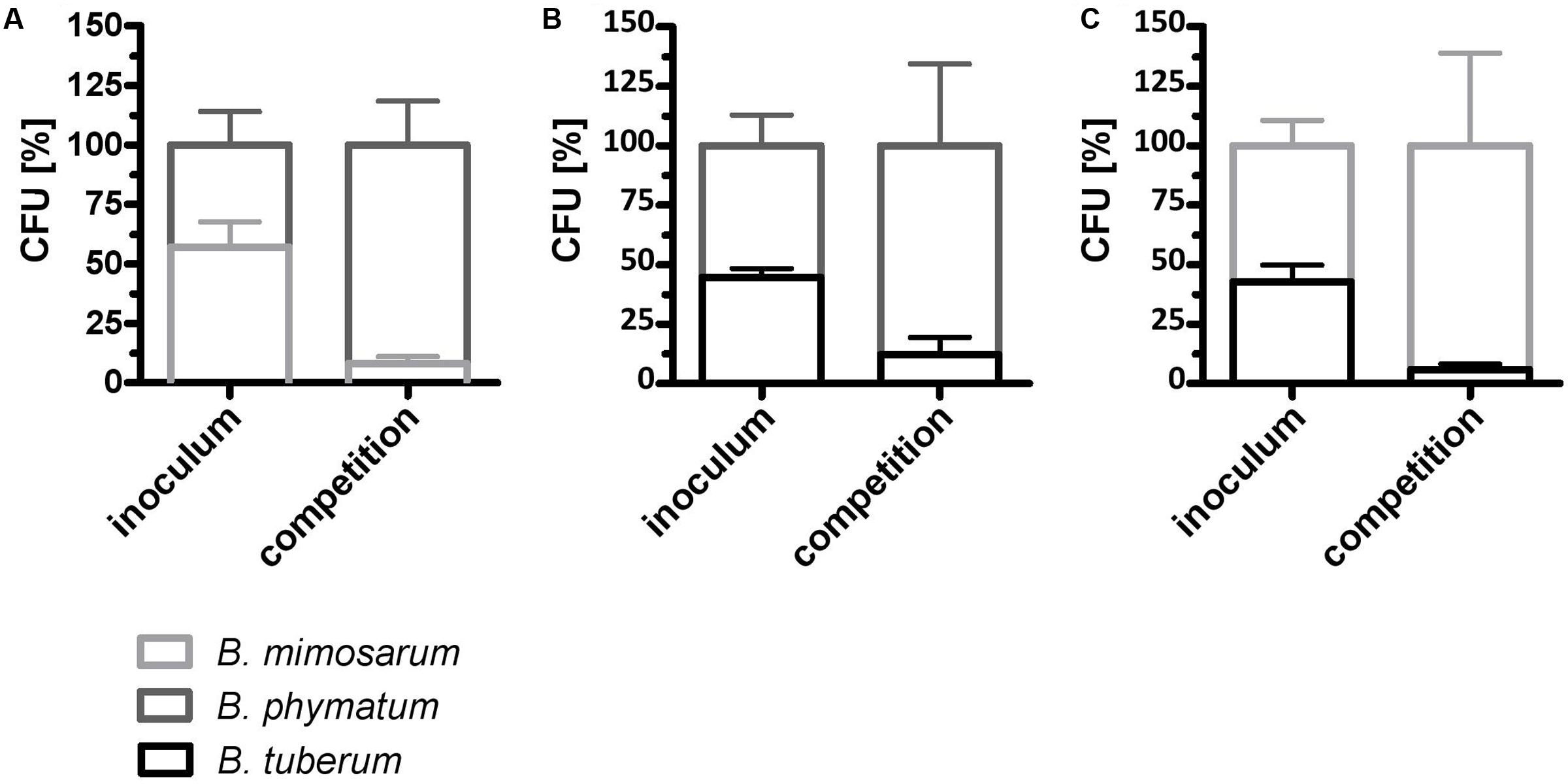
FIGURE 5. Rhizobial competition on plate. A mixture of B. phymatum GFP-tagged against B. mimosarum (A), B. phymatum against B. tuberum (B) and of B. mimosarum against B. tuberum (C) was tested for competition for 24 h on plate. The percentage of colony forming units (CFU) of the two competing strains in the inoculum (inoculum) and recovered after 24 h of contact (competition) is shown. Error bars indicate standard deviation (SD). At least 3 independent replicates were analyzed.
Discussion
Several Burkholderia strains (β-rhizobia) have recently been isolated from root nodules and were found to efficiently nodulate a range of legumes belonging to the Mimosoideae [Mimosa spp., (Elliott et al., 2007b)] and to the Papilionoideae (Elliott et al., 2007a) subfamilies. B. phymatum has been shown to be highly promiscuous with many mimosoid legumes (Moulin et al., 2014) and in fynbos species (Lemaire et al., 2016). Several legumes have been shown to associate with both α- and β-rhizobia (Barrett and Parker, 2006; Elliott et al., 2007a; Gyaneshwar et al., 2011; Klonowska et al., 2012) and often display a higher affinity for α- or β-rhizobia depending on their geographical origin (Melkonian et al., 2014; Bontemps et al., 2016). Trapping experiments with different Mimosa species recovered mainly strains belonging to the Burkholderia genus (Barrett and Parker, 2005; Chen et al., 2005a,b; Parker et al., 2007; Bontemps et al., 2010; dos Reis et al., 2010; Liu et al., 2012; Mishra et al., 2012) and competition experiments showed that Burkholderia strains outcompete C. taiwanensis and α-rhizobia on the three major invasive Mimosa species M. diplotricha, M. pigra and M. pudica (Elliott et al., 2009). Among the Burkholderia strains, B. phymatum was shown to be very competitive on Mimosa independently of the geographical origin of the seeds (Melkonian et al., 2014). Nodulation competitiveness in the Rhizobium-legume symbiosis is a complex process. In the field, it depends at least on three factors, namely the rhizobial genome, the legume genome and the environment. To investigate the role of genomic factors of both the host plant and its rhizobial partner in the symbiosis, we have established and used a particular and defined environment throughout all our experiments (inoculation of a germ-free seed with a defined mixture of β-rhizobial strains, plant grown in sterile vermiculite under constant growth conditions). We competed several nodulating and nitrogen fixing Burkholderia type strains using four different legumes belonging to two sub-families: M. pudica from the Mimosoideae and P. vulgaris (bean), M. atropurpureum (siratro) and V. unguiculata (cowpea) from the Papilionoideae. P. vulgaris and V. unguiculata are major grain legumes relevant for agricultural sustainability worldwide. V. unguiculata can also be used as forage plant and M. atropurpureum is a perennial tropical forage legume1. We show here for the first time that B. phymatum was dominating over the other five strains in the nodules of the three papilionoid legumes bean, cowpea and siratro. B. mimosarum revealed to be the most competitive strain on M. pudica. The dominance of B. mimosarum over B. phymatum on M. pudica confirms the data reported by Liu et al. (2012), which were sampling M. pudica nodules from various locations in China. However, these B. mimosarum strains were shown to harbor nodA from B. phymatum. In contrast, two other studies performed on M. pudica grown in sterile conditions (Elliott et al., 2009; Melkonian et al., 2014) showed that B. phymatum was more competitive than B. mimosarum. The presence of more strains in our rhizobial mixture used to inoculate the plants, the origin of the seeds and the different growth conditions could possibly explain the different outcomes of the experiments.
While B. phymatum has so far only been isolated from one location in its native South America (Mishra et al., 2012), it is quite widespread in association with invasive Mimosa species in Asia Elliott et al., 2007b; Liu et al., 2011; Gehlot et al., 2013) and is also found in common beans cultivated in Morocco (Talbi et al., 2010). B. mimosarum on the other hand is very widespread in South America (Chen et al., 2005a; Bontemps et al., 2010; Mishra et al., 2012) and Asia (Chen et al., 2005b; Elliott et al., 2009; Liu et al., 2012; Gehlot et al., 2013).
Our results suggested that the competitiveness of B. phymatum is not always proportional to its symbiotic performance. In fact, while B. phymatum was the most competitive strain on the three papilionoid legumes, B. mimosarum was the most efficient strain on bean and cowpea in terms of N content and nodule dry weight. Siratro plants were performing better when inoculated with B. diazotrophica and B. tuberum. This suggests that all three papilionoid legumes (bean, cowpea and siratro) do not favor those rhizobia that would supply them with a higher amount of nitrogen. This is not the case with M. pudica, which seems to promote association with the strain B. mimosarum, which is providing the highest shoot N content and nodule dry weight (Figure 2 and Table 3).
The mechanisms that confer a competitive advantage to Burkholderia strain for nodulation are not known. From the rhizobial side, it has been shown for α-rhizobia that cell-surface structures such as LPS (Bhagwat et al., 1991; Lagares et al., 1992) and EPS (Zdor and Pueppke, 1991; Geddes et al., 2014) are important for competition. Moreover, in Bradyrhizobium diazoefficiens it has been reported that a flagella mutant was less competitive than the wild type for legume nodulation suggesting that motility of the strain is an important phenotype for a successful competition (Liu et al., 1989; Althabegoiti et al., 2011). Interestingly, B. phymatum was the strain producing more EPS on mannitol plates (Figure 4), suggesting that this phenotype may contribute to the nodulation competitiveness of the strain. Inspection of B. phymatum genome revealed the presence of a potential EPS cluster, which shares homologies with the cepacian cluster identified in B. cenocepacia strain H111 (Ferreira et al., 2010; Moulin et al., 2014). Preliminary tests suggested that, similar to the situation in B. cenocepacia (Lardi et al., 2015), the expression of the B. phymatum EPS cluster is induced in nitrogen limiting conditions, which are known to be important to establish a successful infection (Lardi et al., unpublished data). Additionally, the assessment of motility behavior showed that B. phymatum was the second-best motile strain (Figure 5). As described above, this trait may confer an additional competitive advantage to B. phymatum. However, since direct seed inoculation was performed, motility may not be an essential trait in our setup.
Competition test performed in vitro suggest that B. phymatum is able to inhibit other Burkholderia strains such as B. mimosarum and B. tuberum also on plates. Although a much higher amount of cells as well as a lower volume have been used for this in vitro competition assay (performed with two bacterial species) compared to the in planta competition test, the results suggest that rhizobia-rhizobia inhibition may also influence the outcome of the competition for nodulation. Several mechanisms are known to underlie growth inhibition including the contact-dependent growth inhibition (CDI) system, which is based on two-partner secretion proteins that suppress the growth of neighboring cells unless the recipient bacterium produces a corresponding immunity protein (Aoki et al., 2010). This system is widely distributed in the genus Burkholderia (Nikolakakis et al., 2012; Garcia et al., 2016). Growth inhibition can also be mediated via a type VI secretion system (T6SS), which is similar to a phage tail spike and can penetrate adjacent eukaryotic or prokaryotic target cells and deliver toxic effector proteins that can cause contact-dependent killing. A T6SS was first identified in the α-rhizobial strain Rhizobium leguminosarum (Bladergroen et al., 2003) and shown to be important for nodulation of Pisum sativum (pea). Many Gram-negative bacteria use this secretion system for killing competitors (Mougous et al., 2006; Pukatzki et al., 2006; Schwarz et al., 2010; Bernal et al., 2017). Both the CDI system and the type VI secretion system are present in B. phymatum genome (Moulin et al., 2014). Another possibility to explain this growth inhibition phenotype could be that B. phymatum produces a compound, which is inhibitory for the growth of other Burkholderia strains present in the mixture. Additional experiments will be required to investigate the possible contribution of these traits to the exceptional competitiveness of B. phymatum.
In the future, it would be interesting to change our standard experimental settings and analyze the shift in competitiveness between β-rhizobial symbionts as well as perform competition experiments between selected α- and β-rhizobia to identify a competitive Rhizobium strain, which could be used as inoculant providing these agriculturally important legumes with high levels of nitrogen.
Author Contributions
Conceived and designed the experiments: ML and GP. Performed the experiments: ML, GPu, and SdC. Analyzed the data: ML, SdC, LE, and GP. Wrote the paper: ML and GP.
Funding
This work was supported by the Swiss National Science Foundation (grant 31003A_153374 to GP).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We acknowledge Lionel Moulin and Agnieszka Klonowska for providing the strain B. phymatum-GFP. We thank Christian Ahrens for his feedback on the manuscript, Filip Kocovski for helping with graphics and René Husi, Sebastian Hug and Yilei Liu for precious help in processing the samples for determination of the total plant nitrogen content. We are also grateful to the two referees for useful comments.
Supplementary Material
The Supplementary Material for this article can be found online at: http://journal.frontiersin.org/article/10.3389/fmicb.2017.01527/full#supplementary-material
Footnotes
References
Althabegoiti, M. J., Covelli, J. M., Perez-Gimenez, J., Quelas, J. I., Mongiardini, E. J., Lopez, M. F., et al. (2011). Analysis of the role of the two flagella of Bradyrhizobium japonicum in competition for nodulation of soybean. FEMS Microbiol. Lett. 319, 133–139. doi: 10.1111/j.1574-6968.2011.02280.x
Angus, A. A., Lee, A., Lum, M. R., Shehayeb, M., Hessabi, R., Fujishige, N. A., et al. (2013). Nodulation and effective nitrogen fixation of Macroptilium atropurpureum (siratro) by Burkholderia tuberum, a nodulating and plant growth promoting beta-proteobacterium, are influenced by environmental factors. Plant Soil 369, 543–562. doi: 10.1007/s11104-013-1590-7
Aoki, S. K., Diner, E. J., de Roodenbeke, C. T., Burgess, B. R., Poole, S. J., Braaten, B. A., et al. (2010). A widespread family of polymorphic contact-dependent toxin delivery systems in bacteria. Nature 468, 439–442. doi: 10.1038/nature09490
Barrett, C. F., and Parker, M. A. (2005). Prevalence of Burkholderia sp. nodule symbionts on four mimosoid legumes from Barro Colorado Island, Panama. Syst. Appl. Microbiol. 28, 57–65. doi: 10.1016/j.syapm.2004.09.002
Barrett, C. F., and Parker, M. A. (2006). Coexistence of Burkholderia, Cupriavidus, and Rhizobium sp. nodule bacteria on two Mimosa spp. in Costa Rica. Appl. Environ. Microbiol. 72, 1198–1206. doi: 10.1128/AEM.72.2.1198-1206.2006
Bernal, P., Allsopp, L. P., Filloux, A., and Llamas, M. A. (2017). The Pseudomonas putida T6SS is a plant warden against phytopathogens. ISME J. 11, 972–987. doi: 10.1038/ismej.2016.169
Beukes, C. W., Palmer, M., Manyaka, P., Chan, W. Y., Avontuur, J. R., van Zyl, E., et al. (2017). Genome data provides high support for generic boundaries in Burkholderia sensu lato. Front. Microbiol. 8:1154. doi: 10.3389/fmicb.2017.01154
Bhagwat, A. A., Tully, R. E., and Keister, D. L. (1991). Isolation and characterization of a competition defective Bradyrhizobium japonicum mutant. Appl. Environ. Microbiol. 57, 3496–3501.
Bladergroen, M. R., Badelt, K., and Spaink, H. P. (2003). Infection-blocking genes of a symbiotic Rhizobium leguminosarum strain that are involved in temperature-dependent protein secretion. Mol. Plant Microbe Interact. 16, 53–64. doi: 10.1094/Mpmi.2003.16.1.53
Bontemps, C., Elliott, G. N., Simon, M. F., Dos Reis, F. B. D., Gross, E., Lawton, R. C., et al. (2010). Burkholderia species are ancient symbionts of legumes. Mol. Ecol. 19, 44–52. doi: 10.1111/j.1365-294X.2009.04458.x
Bontemps, C., Rogel, M. A., Wiechmann, A., Mussabekova, A., Moody, S., Simon, M. F., et al. (2016). Endemic Mimosa species from Mexico prefer alphaproteobacterial rhizobial symbionts. New Phytol. 209, 319–333. doi: 10.1111/nph.13573
Bournaud, C., de Faria, S. M., dos Santos, J. M., Tisseyre, P., Silva, M., Chaintreuil, C., et al. (2013). Burkholderia species are the most common and preferred nodulating symbionts of the Piptadenia group (tribe Mimoseae). PLoS ONE 8:e63478. doi: 10.1371/journal.pone.0063478
Chen, W. M., de Faria, S. M., Chou, J. H., James, E. K., Elliott, G. N., Sprent, J. I., et al. (2008). Burkholderia sabiae sp. nov., isolated from root nodules of Mimosa caesalpiniifolia. Int. J. Syst. Evol. Microbiol. 58(Pt 9), 2174–2179. doi: 10.1099/ijs.0.65816-0
Chen, W. M., de Faria, S. M., Straliotto, R., Pitard, R. M., Simoes-Araujo, J. L., Chou, J. H., et al. (2005a). Proof that Burkholderia strains form effective symbioses with legumes: a study of novel Mimosa-nodulating strains from South America. Appl. Environ. Microbiol. 71, 7461–7471. doi: 10.1128/AEM.71.11.7461-7471.2005
Chen, W. M., James, E. K., Chou, J. H., Sheu, S. Y., Yang, S. Z., and Sprent, J. I. (2005b). β-Rhizobia from Mimosa pigra, a newly discovered invasive plant in Taiwan. New Phytol. 168, 661–675. doi: 10.1111/j.1469-8137.2005.01533.x
Chen, W. M., James, E. K., Coenye, T., Chou, J. H., Barrios, E., de Faria, S. M., et al. (2006). Burkholderia mimosarum sp. nov., isolated from root nodules of Mimosa spp. from Taiwan and South America. Int. J. Syst. Evol. Microbiol. 56(Pt 8), 1847–1851. doi: 10.1099/ijs.0.64325-0
Chen, W. M., Laevens, S., Lee, T. M., Coenye, T., De Vos, P., Mergeay, M., et al. (2001). Ralstonia taiwanensis sp. nov., isolated from root nodules of Mimosa species and sputum of a cystic fibrosis patient. Int. J. Syst. Evol. Microbiol. 51(Pt 5), 1729–1735. doi: 10.1099/00207713-51-5-1729
Chen, W. M., Moulin, L., Bontemps, C., Vandamme, P., Bena, G., and Boivin-Masson, C. (2003). Legume symbiotic nitrogen fixation by beta-proteobacteria is widespread in nature. J. Bacteriol. 185, 7266–7272. doi: 10.1128/JB.185.24.7266-7272.2003
Clark, D. J., and Maaloe, O. (1967). DNA replication and division cycle in Escherichia coli. J. Mol. Biol. 23, 99–112. doi: 10.1016/S0022-2836(67)80070-6
Ding, H., Yip, C. B., Geddes, B. A., Oresnik, I. J., and Hynes, M. F. (2012). Glycerol utilization by Rhizobium leguminosarum requires an ABC transporter and affects competition for nodulation. Microbiology 158(Pt 5), 1369–1378. doi: 10.1099/mic.0.057281-0
dos Reis, F. B. Jr., Simon, M. F., Gross, E., Boddey, R. M., Elliott, G. N., and Neto, N. E. (2010). Nodulation and nitrogen fixation by Mimosa spp. in the Cerrado and Caatinga biomes of Brazil. New Phytol. 186, 934–946. doi: 10.1111/j.1469-8137.2010.03267.x
Dowling, D. N., and Broughton, W. J. (1986). Competition for nodulation of legumes. Annu. Rev. Microbiol. 40, 131–157. doi: 10.1146/annurev.mi.40.100186.001023
Downie, J. A. (2010). The roles of extracellular proteins, polysaccharides and signals in the interactions of rhizobia with legume roots. FEMS Microbiol. Rev. 34, 150–170. doi: 10.1111/j.1574-6976.2009.00205.x
Eberl, L., Christiansen, G., Molin, S., and Givskov, M. (1996). Differentiation of Serratia liquefaciens into swarm cells is controlled by the expression of the flhD master operon. J. Bacteriol. 178, 554–559. doi: 10.1128/jb.178.2.554-559.1996
Elliott, G. N., Chen, W. M., Bontemps, C., Chou, J. H., Young, J. P. W., Sprent, J. I., et al. (2007a). Nodulation of Cyclopia spp. (Leguminosae, Papilionoideae) by Burkholderia tuberum. Ann. Bot. 100, 1403–1411. doi: 10.1093/aob/mcm227
Elliott, G. N., Chen, W. M., Chou, J. H., Wang, H. C., Sheu, S. Y., Perin, L., et al. (2007b). Burkholderia phymatum is a highly effective nitrogen-fixing symbiont of Mimosa spp. and fixes nitrogen ex planta. New Phytol. 173, 168–180. doi: 10.1111/j.1469-8137.2006.01894.x
Elliott, G. N., Chou, J. H., Chen, W. M., Bloemberg, G. V., Bontemps, C., Martinez-Romero, E., et al. (2009). Burkholderia spp. are the most competitive symbionts of Mimosa, particularly under N-limited conditions. Environ. Microbiol. 11, 762–778. doi: 10.1111/j.1462-2920.2008.01799.x
Ferguson, B. J., Indrasumunar, A., Hayashi, S., Lin, M. H., Lin, Y. H., Reid, D. E., et al. (2010). Molecular analysis of legume nodule development and autoregulation. J. Integr. Plant Biol. 52, 61–76. doi: 10.1111/j.1744-7909.2010.00899.x
Ferreira, A. S., Leitão, J. H., Silva, I. N., Pinheiro, P. F., Sousa, S. A., Ramos, C. G., et al. (2010). Distribution of cepacian biosynthesis genes among environmental and clinical Burkholderia strains and role of cepacian exopolysaccharide in resistance to stress conditions. Appl. Environ. Microbiol. 76, 441–450. doi: 10.1128/AEM.01828-09
Fry, J., Wood, M., and Poole, P. S. (2001). Investigation of myo-inositol catabolism in Rhizobium leguminosarum bv. viciae and its effect on nodulation competitiveness. Mol. Plant Microbe Interact. 14, 1016–1025. doi: 10.1094/Mpmi.2001.14.8.1016
Garau, G., Yates, R. J., Deiana, P., and Howieson, J. G. (2009). Novel strains of nodulating Burkholderia have a role in nitrogen fixation with papilionoid herbaceous legumes adapted to acid, infertile soils. Soil Biol. Biochem. 41, 125–134. doi: 10.1016/j.soilbio.2008.10.011
Garcia, E. C., Perault, A. I., Marlatt, S. A., and Cotter, P. A. (2016). Interbacterial signaling via Burkholderia contact-dependent growth inhibition system proteins. Proc. Natl. Acad. Sci. U.S.A. 113, 8296–8301. doi: 10.1073/pnas.1606323113
Geddes, B. A., Gonzalez, J. E., and Oresnik, I. J. (2014). Exopolysaccharide production in response to medium acidification is correlated with an increase in competition for nodule occupancy. Mol. Plant Microbe Interact. 27, 1307–1317. doi: 10.1094/MPMI-06-14-0168-R
Gehlot, H. S., Tak, N., Kaushik, M., Mitra, S., Chen, W. M., Poweleit, N., et al. (2013). An invasive Mimosa in India does not adopt the symbionts of its native relatives. Ann. Bot. 112, 179–196. doi: 10.1093/aob/mct112
George, M. L., and Robert, F. M. (1991). Autoregulatory response of Phaseolus vulgaris L. to symbiotic mutants of Rhizobium leguminosarum bv. phaseoli. Appl. Environ. Microbiol. 57, 2687–2692.
Göttfert, M., Hitz, S., and Hennecke, H. (1990). Identification of nodS and nodU, two inducible genes inserted between the Bradyrhizobium japonicum nodYABC and nodIJ genes. Mol. Plant Microbe Interact. 3, 308–316. doi: 10.1094/MPMI-3-308
Gyaneshwar, P., Hirsch, A. M., Moulin, L., Chen, W. M., Elliott, G. N., Bontemps, C., et al. (2011). Legume-nodulating betaproteobacteria: diversity, host range, and future prospects. Mol. Plant Microbe Interact. 24, 1276–1288. doi: 10.1094/MPMI-06-11-0172
Hahn, M., and Hennecke, H. (1984). Localized mutagenesis in Rhizobium japonicum. Mol. Gen. Genet. 193, 46–52. doi: 10.1007/Bf00327412
Howieson, J. G., De Meyer, S. E., Vivas-Marfisi, A., Ratnayake, S., Ardley, J. K., and Yates, R. J. (2013). Novel Burkholderia bacteria isolated from Lebeckia ambigua - A perennial suffrutescent legume of the fynbos. Soil Biol. Biochem. 60, 55–64. doi: 10.1016/j.soilbio.2013.01.009
Jiménez-Zurdo, J. I., van Dillewijn, P., Soto, M. J., de Felipe, M. R., Olivares, J., and Toro, N. (1995). Characterization of a Rhizobium meliloti proline dehydrogenase mutant altered in nodulation efficiency and competitiveness on alfalfa roots. Mol. Plant Microbe Interact. 8, 492–498. doi: 10.1094/MPMI-8-0492
Klonowska, A., Chaintreuil, C., Tisseyre, P., Miche, L., Melkonian, R., Ducousso, M., et al. (2012). Biodiversity of Mimosa pudica rhizobial symbionts (Cupriavidus taiwanensis, Rhizobium mesoamericanum) in New Caledonia and their adaptation to heavy metal-rich soils. FEMS Microbiol. Ecol. 81, 618–635. doi: 10.1111/j.1574-6941.2012.01393.x
Kohler, P. R. A., Zheng, J. Y., Schoffers, E., and Rossbach, S. (2010). Inositol catabolism, a key pathway in Sinorhizobium meliloti for competitive host nodulation. Appl. Environ. Microbiol. 76, 7972–7980. doi: 10.1128/Aem.01972-10
Kost, T., Stopnisek, N., Agnoli, K., Eberl, L., and Weisskopf, L. (2014). Oxalotrophy, a widespread trait of plant-associated Burkholderia species, is involved in successful root colonization of lupin and maize by Burkholderia phytofirmans. Front. Microbiol. 4:421. doi: 10.3389/fmicb.2013.00421
Lagares, A., Caetanoanolles, G., Niehaus, K., Lorenzen, J., Ljunggren, H. D., Puhler, A., et al. (1992). A Rhizobium meliloti lipopolysaccharide mutant altered in competitiveness for nodulation of alfalfa. J. Bacteriol. 174, 5941–5952. doi: 10.1128/jb.174.18.5941-5952.1992
Lardi, M., Aguilar, C., Pedrioli, A., Omasits, U., Suppiger, A., Carcamo-Oyarce, G., et al. (2015). σ54-dependent response to nitrogen limitation and virulence in Burkholderia cenocepacia strain H111. Appl. Environ. Microbiol. 81, 4077–4089. doi: 10.1128/AEM.00694-15
Lemaire, B., Chimphango, S. B., Stirton, C., Rafudeen, S., Honnay, O., Smets, E., et al. (2016). Biogeographical patterns of legume-nodulating Burkholderia spp.: from African fynbos to continental scales. Appl. Environ. Microbiol. 82, 5099–5115. doi: 10.1128/AEM.00591-16
Lemaire, B., Dlodlo, O., Chimphango, S., Stirton, C., Schrire, B., Boatwright, J. S., et al. (2015). Symbiotic diversity, specificity and distribution of rhizobia in native legumes of the Core Cape Subregion (South Africa). FEMS Microbiol. Ecol. 91, 1–17. doi: 10.1093/femsec/fiu024
Liu, R. L., Tran, V. M., and Schmidt, E. L. (1989). Nodulating competitiveness of a nonmotile Tn7-mutant of Bradyrhizobium japonicum in nonsterile soil. Appl. Environ. Microbiol. 55, 1895–1900.
Liu, W. Y., Ridgway, H. J., James, T. K., James, E. K., Chen, W. M., Sprent, J. I., et al. (2014). Burkholderia sp. induces functional nodules on the South African invasive legume Dipogon lignosus (Phaseoleae) in New Zealand soils. Microb. Ecol. 68, 542–555. doi: 10.1007/s00248-014-0427-0
Liu, X., Wei, S., Wang, F., James, E. K., Guo, X., Zagar, C., et al. (2012). Burkholderia and Cupriavidus spp. are the preferred symbionts of Mimosa spp. in southern China. FEMS Microbiol. Ecol. 80, 417–426. doi: 10.1111/j.1574-6941.2012.01310.x
Liu, X. Y., Wu, W., Wang, E. T., Zhang, B., Macdermott, J., and Chen, W. X. (2011). Phylogenetic relationships and diversity of β-rhizobia associated with Mimosa species grown in Sishuangbanna, China. Int. J. Syst. Evol. Microbiol. 61, 334–342. doi: 10.1099/ijs.0.020560-0
Masson-Boivin, C., Giraud, E., Perret, X., and Batut, J. (2009). Establishing nitrogen-fixing symbiosis with legumes: how many rhizobium recipes? Trends Microbiol. 17, 458–466. doi: 10.1016/j.tim.2009.07.004
Melkonian, R., Moulin, L., Bena, G., Tisseyre, P., Chaintreuil, C., Heulin, K., et al. (2014). The geographical patterns of symbiont diversity in the invasive legume Mimosa pudica can be explained by the competitiveness of its symbionts and by the host genotype. Environ. Microbiol. 16, 2099–2111. doi: 10.1111/1462-2920.12286
Mellor, H. Y., Glenn, A. R., Arwas, R., and Dilworth, M. J. (1987). Symbiotic and competitive properties of motility mutants of Rhizobium trifolii Ta1. Arch. Microbiol. 148, 34–39. doi: 10.1007/Bf00429644
Mishra, R. P., Tisseyre, P., Melkonian, R., Chaintreuil, C., Miche, L., Klonowska, A., et al. (2012). Genetic diversity of Mimosa pudica rhizobial symbionts in soils of French Guiana: investigating the origin and diversity of Burkholderia phymatum and other beta-rhizobia. FEMS Microbiol. Ecol. 79, 487–503. doi: 10.1111/j.1574-6941.2011.01235.x
Mougous, J. D., Cuff, M. E., Raunser, S., Shen, A., Zhou, M., Gifford, C. A., et al. (2006). A virulence locus of Pseudomonas aeruginosa encodes a protein secretion apparatus. Science 312, 1526–1530. doi: 10.1126/science.1128393
Moulin, L., Klonowska, A., Caroline, B., Booth, K., Vriezen, J. A., Melkonian, R., et al. (2014). Complete genome sequence of Burkholderia phymatum STM815(T), a broad host range and efficient nitrogen-fixing symbiont of Mimosa species. Stand. Genomic Sci. 9, 763–774. doi: 10.4056/sigs.4861021
Moulin, L., Munive, A., Dreyfus, B., and Boivin-Masson, C. (2001). Nodulation of legumes by members of the beta-subclass of Proteobacteria. Nature 411, 948–950. doi: 10.1038/35082070
Nikolakakis, K., Amber, S., Wilbur, J. S., Diner, E. J., Aoki, S. K., Poole, S. J., et al. (2012). The toxin/immunity network of Burkholderia pseudomallei contact-dependent growth inhibition (CDI) systems. Mol. Microbiol. 84, 516–529. doi: 10.1111/j.1365-2958.2012.08039.x
Oldroyd, G. E., Murray, J. D., Poole, P. S., and Downie, J. A. (2011). The rules of engagement in the legume-rhizobial symbiosis. Annu. Rev. Genet. 45, 119–144. doi: 10.1146/annurev-genet-110410-132549
Parker, M. A., Wurtz, A. K., and Paynter, Q. (2007). Nodule symbiosis of invasive Mimosa pigra in Australia and in ancestral habitats: a comparative analysis. Biol. Invasions 9, 127–138. doi: 10.1007/s10530-006-0009-2
Pukatzki, S., Ma, A. T., Sturtevant, D., Krastins, B., Sarracino, D., Nelson, W. C., et al. (2006). Identification of a conserved bacterial protein secretion system in Vibrio cholerae using the Dictyostelium host model system. Proc. Natl. Acad. Sci. U.S.A. 103, 1528–1533. doi: 10.1073/pnas.0510322103
Robleto, E. A., Kmiecik, K., Oplinger, E. S., Nienhuis, J., and Triplett, E. W. (1998). Trifolitoxin production increases nodulation competitiveness of Rhizobium etli CE3 under agricultural conditions. Appl. Environ. Microbiol. 64, 2630–2633.
Sawana, A., Adeolu, M., and Gupta, R. S. (2014). Molecular signatures and phylogenomic analysis of the genus Burkholderia: proposal for division of this genus into the emended genus Burkholderia containing pathogenic organisms and a new genus Paraburkholderia gen. nov harboring environmental species. Front. Genet. 5:429. doi: 10.3389/Fgene.2014.00429
Schwarz, S., Hood, R. D., and Mougous, J. D. (2010). What is type VI secretion doing in all those bugs? Trends Microbiol. 18, 531–537. doi: 10.1016/j.tim.2010.09.001
Schwinghamer, E. A., and Belkengren, R. P. (1968). Inhibition of rhizobia by a strain of Rhizobium trifolii - some properties of antibiotic and of strain. Arch. Mikrobiol. 64, 130–145. doi: 10.1007/Bf00406972
Sheu, S. Y., Chou, J. H., Bontemps, C., Elliott, G. N., Gross, E., dos Reis Junior, F. B., et al. (2013). Burkholderia diazotrophica sp. nov., isolated from root nodules of Mimosa spp. Int. J. Syst. Evol. Microbiol. 63(Pt 2), 435–441. doi: 10.1099/ijs.0.039859-0
Sheu, S. Y., Chou, J. H., Bontemps, C., Elliott, G. N., Gross, E., James, E. K., et al. (2012). Burkholderia symbiotica sp. nov., isolated from root nodules of Mimosa spp. native to north-east Brazil. Int. J. Syst. Evol. Microbiol. 62, 2272–2278. doi: 10.1099/ijs.0.037408-0
Sprent, J. I., Ardley, J., and James, E. K. (2017). Biogeography of nodulated legumes and their nitrogen-fixing symbionts. New Phytol. 215, 40–56. doi: 10.1111/nph.14474
Talbi, C., Delgado, M. J., Girard, L., Ramirez-Trujillo, A., Caballero-Mellado, J., and Bedmar, E. J. (2010). Burkholderia phymatum strains capable of nodulating Phaseolus vulgaris are present in Moroccan soils. Appl. Environ. Microbiol. 76, 4587–4591. doi: 10.1128/AEM.02886-09
Triplett, E. W., and Sadowsky, M. J. (1992). Genetics of competition for nodulation of legumes. Ann. Rev. Microbiol. 46, 399–428. doi: 10.1146/annurev.micro.46.1.399
Vandamme, P., Goris, J., Chen, W. M., de Vos, P., and Willems, A. (2002). Burkholderia tuberum sp. nov. and Burkholderia phymatum sp. nov., nodulate the roots of tropical legumes. Syst. Appl. Microbiol. 25, 507–512. doi: 10.1078/07232020260517634
Vincent, J. M. (1970). A Manual for the Practical Study of Root Nodule Bacteria. Oxford: Blackwell Scientific Publications.
Vitousek, P. M., Hattenschwiler, S., Olander, L., and Allison, S. (2002). Nitrogen and nature. Ambio 31, 97–101. doi: 10.1639/0044-74472002031
Young, J. P. W., and Haukka, K. E. (1996). Diversity and phylogeny of rhizobia. New Phytol. 133, 87–94. doi: 10.1111/j.1469-8137.1996.tb04344.x
Zdor, R. E., and Pueppke, S. G. (1991). Nodulation competitiveness of Tn5-induced mutants of Rhizobium fredii Usda208 that are altered in motility and extracellular polysaccharide production. Can. J. Microbiol. 37, 52–58. doi: 10.1139/m91-008
Keywords: Rhizobium, Burkholderia, legume, symbiosis, nodulation, competitiveness
Citation: Lardi M, de Campos SB, Purtschert G, Eberl L and Pessi G (2017) Competition Experiments for Legume Infection Identify Burkholderia phymatum as a Highly Competitive β-Rhizobium. Front. Microbiol. 8:1527. doi: 10.3389/fmicb.2017.01527
Received: 09 May 2017; Accepted: 28 July 2017;
Published: 15 August 2017.
Edited by:
Benjamin Gourion, UMR2594 Laboratoire des Interactions Plantes Microorganismes (LIPM), FranceReviewed by:
Anibal Roberto Lodeiro, CONICET, ArgentinaEuan James, James Hutton Institute, United Kingdom
Copyright © 2017 Lardi, de Campos, Purtschert, Eberl and Pessi. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gabriella Pessi, Z2FicmllbGxhLnBlc3NpQGJvdGluc3QudXpoLmNo
 Martina Lardi
Martina Lardi Samanta Bolzan de Campos
Samanta Bolzan de Campos Gabriela Purtschert
Gabriela Purtschert Gabriella Pessi
Gabriella Pessi