- Laboratory of Medical and Molecular Mycology, Department of Clinical and Toxicological Analyses, Federal University of Rio Grande do Norte, Natal, Brazil
Candida tropicalis has emerged as one of the most important Candida species. It has been widely considered the second most virulent Candida species, only preceded by C. albicans. Besides, this species has been recognized as a very strong biofilm producer, surpassing C. albicans in most of the studies. In addition, it produces a wide range of other virulence factors, including: adhesion to buccal epithelial and endothelial cells; the secretion of lytic enzymes, such as proteinases, phospholipases, and hemolysins, bud-to-hyphae transition (also called morphogenesis) and the phenomenon called phenotypic switching. This is a species very closely related to C. albicans and has been easily identified with both phenotypic and molecular methods. In addition, no cryptic sibling species were yet described in the literature, what is contradictory to some other medically important Candida species. C. tropicalis is a clinically relevant species and may be the second or third etiological agent of candidemia, specifically in Latin American countries and Asia. Antifungal resistance to the azoles, polyenes, and echinocandins has already been described. Apart from all these characteristics, C. tropicalis has been considered an osmotolerant microorganism and this ability to survive to high salt concentration may be important for fungal persistence in saline environments. This physiological characteristic makes this species suitable for use in biotechnology processes. Here we describe an update of C. tropicalis, focusing on all these previously mentioned subjects.
Introduction
In the last decades, medicine advances related to the discovery of several medical devices which seek for a longer survival of patients with several infirmities, such as AIDS, hematological malignancies, cancer, and other immunosuppressive diseases promoted a longer lifespan. On the other hand, the number of opportunistic fungal infections increased, mainly the ones caused by the Candida genus (Pincus et al., 2007; Araújo et al., 2017). In this context, Candida tropicalis emerges as one of the most important Candida species in terms of epidemiology and virulence. It is able to produce true hyphae, an exclusive property of Candida albicans and its sibling species Candida dubliniensis. C. tropicalis has also been considered a strong biofilm producer species and is highly adherent to epithelial and endothelial cells (Marcos-Zambrano et al., 2014). In addition, several recent investigations have reported the recovery of C. tropicalis resistant to the antifungal drugs currently available, such as the azoles derivatives, amphotericin B, and echinocandins (Choi et al., 2016; Seneviratne et al., 2016). In addition, C. tropicalis has been considered an osmotolerant microorganism and this ability to survive to high salt concentration may be important for fungal persistence in saline environments, contributing to the expression of virulence factors in vitro and resistance to antifungal drugs (Zuza-Alves et al., 2016). This property explains C. tropicalis potential use in biotechnological processes such as the production of xylitol from corn fiber and the ethanol from marine algae (Rao et al., 2006; Ra et al., 2015).
Biology and Taxonomy
C. tropicalis was originally isolated from a patient with fungal bronchitis in 1910 and named Oidium tropicale (Castellani, 1912). It is a yeast belonging to the filo Ascomycota, from the Hemiascomycetes class (Blandin et al., 2000), which has a single Order created in 1960 by Kudrjavzev, called Saccharomycetales (Kirk et al., 2001). This monophyletic lineage comprises about 1,000 known species, including several yeasts of medical importance such as C. tropicalis (Diezmann et al., 2004).
According to Kurtzman et al. (2011) C. tropicais colonies on Sabouraud Dextrose Agar (SDA) are white to cream, with a creamy texture and smooth appearance and may have slightly wrinkled edges. Therefore, it is indistinguishable from other Candida species. After 7 days of microculture on cornmeal agar containing Tween 80, incubated at 25°C, spherical or ovoid blastoconidia, which may be grouped in pairs or alone, measuring ~4–8 × 5–11 μm, pseudohyphae in branched chains, and even true hyphae may be observed (Silva et al., 2012; Figure 1). With respect to the biochemical characteristics, it is known that C. tropicalis is capable of fermenting galactose, sucrose, maltose, and trehalose, besides assimilating these and others carbohydrates through the oxidative pathway (Kurtzman et al., 2011).
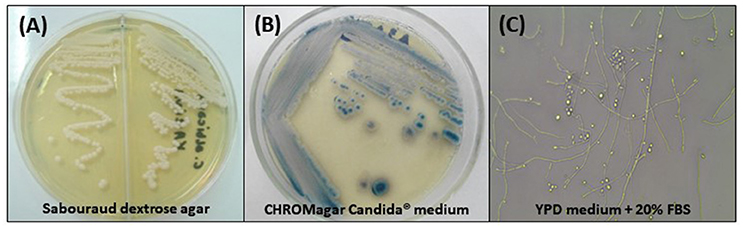
Figure 1. Phenotypic characteristics of Candida tropicalis. (A) Cream-colored, dull, smooth colonies, after 48 h of incubation at 30°C on Sabouraud dextrose agar; (B) Colonies with typical dark blue color on CHROMagar Candida® medium after 96 h of incubation at 35°C; (C) Micromorphological aspects after incubation in YPD medium containing 20% fetal bovine serum (FBS) for 7 days at 30°C, 400x: blastoconidia in single or branched chains, true hyphae and abundant pseudohyphae.
Genetic Characteristic
C. tropicalis is a diploid yeast, whose genome was sequenced in 2009 (strain MYA-3404) in a study conducted by Butler et al. (2009). It has a genomic size of 14.5 Mb, containing 6,258 genes encoding proteins and a guanine-cytosine content of 33.1%. The number of chromosomes is not known with precision, but Doi et al. reported 12 chromosomes per cell for C. tropicalis (Doi et al., 1992).
It has been widely believed that C. tropicalis is an asexual yeast. However, some studies performed recently have reported that mating between diploid cells a and α, generating a/α tetraploid cells may occur (Porman et al., 2011; Xie et al., 2012; Seervai et al., 2013). Such mating is regulated by colony phenotypic switching, where cells change from a white to an opaque state. Seervai et al. (2013) demonstrated that tetraploid strains of C. tropicalis can be induced to undergo parasexual cycle without meiotic reduction. This process results in a or α diploid cells competent for mating, being able to form tetraploid cells, which show chromosomal instability after incubation and return to the diploid state after ~240 generations (Seervai et al., 2013). Genetic recombination has also been demonstrated, besides ploidy changes (aneuploidies and polyploidy), affecting cells gene expression and protein production (Morrow and Fraser, 2013). This reduction in ploidy is considered a mechanism of adaptation and may be associated with cell stress (Berman and Hadany, 2012). This adaptive mechanism may also generate karyotype variation within the host, and may be induced by various stressors, such as thermal shock, exposure to UV light, and growth in l-sorbose or d-arabinose as the only carbon source (Legrand et al., 2008; Arbour et al., 2009; Bouchonville et al., 2009; Morrow and Fraser, 2013). It is important to emphasize again that meiosis occurrence has never been described in C. tropicalis.
C. tropicalis has greater genetic similarity with C. albicans than the other Candida species of medical interest (Butler et al., 2009), as may be observed in Figure 2. This intimate evolutionary relationship is also evident in phenotypic and biochemical characteristics of both species. Phylogenetically, this pattern of evolution can be explained due to predominant clonal reproduction. However, with recombination events frequent enough to generate a population with similar characteristics (Wu et al., 2014).
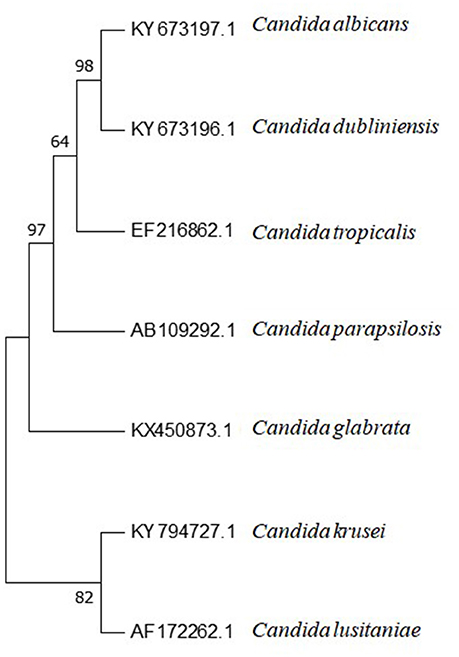
Figure 2. Phylogenetic tree of Candida spp. internal transcribed spacer 1 (ITS1)-5.8S ribosomal RNA gene and internal transcribed spacer 2 (ITS2) complete sequences and their accession numbers, obtained from Genbank database at https://www.ncbi.nlm.nih.gov. Sequences were aligned using BioEdit software (v7.2.61). Aligned sequences were used for phylogenetic analysis conducted with Mega 7.0.26 Software. The method used for tree constructions was maximum parsimony. Phylogram stability was accessed by bootstrapping with 1,000 pseudoreplications.
Identification
Conventional Methods for Candida tropicalis Identification
C. tropicalis has been quite reasonably well-identified with phenotypic methods until the present moment (Table 1). This is contradictory to some other Candida spp., where molecular identification is mandatory due to the existence of cryptic species.
Although the classical methodology is of easy execution, it is very laborious and time-consuming making it difficult to be used in microbiology routine laboratories (Table 1; Pincus et al., 2007; Sariguzel et al., 2015).
The use of chromogenic media, with different substrates that react with specific enzymes of the main Candida species induce the formation of colonies with different colors and has been used for the presumptive identification of C. tropicalis. They have all been used for the screening of distinct species, besides being used to check the purity of Candida colonies and may be helpful to detect mixed infections. Quite a few number of different chromogenic culture media are currently commercially available for yeasts identification, and they have been successfully used for the initial screening of C. tropicalis colonies (Table 1).
Several commercially available kits used for yeasts identification based on carbohydrates used by oxidative pathways have been in the market in order to facilitate the process used for yeasts identification (Table 1). C. tropicalis identification with commercial methods have been performed since 1975; since then, several papers have been published in the literature evaluating the efficiency of this method. In a recent study by Stefaniuk and colaborators), the API ID32C system (bioMérieux) was used for the identification of 124 Candida clinical isolates, where 21 C. tropicalis isolates (100% of cases) were accurately identified (Stefaniuk et al., 2016). In a study performed by Alfonso et al., with 240 isolates of different Candida species, the authors found the accurate identification of 34 isolates of C. tropicalis with the API ID 32C system (Alfonso et al., 2010). Gundes et al. compared the efficiency of different commercial methods used in the identification 116 yeasts of medical interest, demonstrating the accuracy of 87% (101 out of 116) for API 20C® against 82.7% (96 out of 116) with Candifast® system. However, C. tropicalis was accurately identified in 100% of cases with both methods (Gündeş et al., 2001). The AuxaColor™ Kit (Bio-Rad) identification has been shown to be accurate in 63.8–95.2% of cases (Pincus et al., 2007). Recently, in a meta-analysis performed by Posteraro et al. (2015), including a total of 26 studies that evaluated yeasts identification methods, they observed that C. tropicalis was accurately identified in 168 out of 184 cases tested with AuxaColor™ and 55 out of 66 cases by using API ID32C® (Posteraro et al., 2015).
Besides semi-automated methods currently available, there are other methods completely automated for yeasts identification (Table 1). Won and collaborators performed a study that compared the efficiency of several medically important yeast species identification with the automated systems Vitek2® and BD Phoenix™. This study included a total of 341 isolates, from 49 species and C. tropicalis (36 isolates) was accurately identified in 34 cases with BD Phoenix™ System and in 32 occasions with Vitek2® (Won et al., 2014).
The conventional methods of identification including the classical methods, semi-automated and automated systems may not be completely accurate on some cases and may lead to an incomplete identification, needing supplementary tests or even give a wrong identification for some species (Marcos and Pincus, 2013; Chao et al., 2014). Therefore, molecular biology advances are of extreme importance for microorganism's identification because of the fact they are more accurate, and may reduce costs involving identification during the whole process, resulting in a decreased time for the release of results (Chao et al., 2014; Posteraro et al., 2015).
Molecular Methods and Proteomics for the Identification of Candida tropicalis
Recently, the evaluation of the protein profile of each species has been used as the basis for yeasts identification and has been proven as more efficient than the conventional methods (Santos et al., 2011; Chao et al., 2014; Stefaniuk et al., 2016). The protein profile by mass spectrophotometry is a simple methodology of easy sample preparation and short time for analysis (Table 2; Keceli et al., 2016).
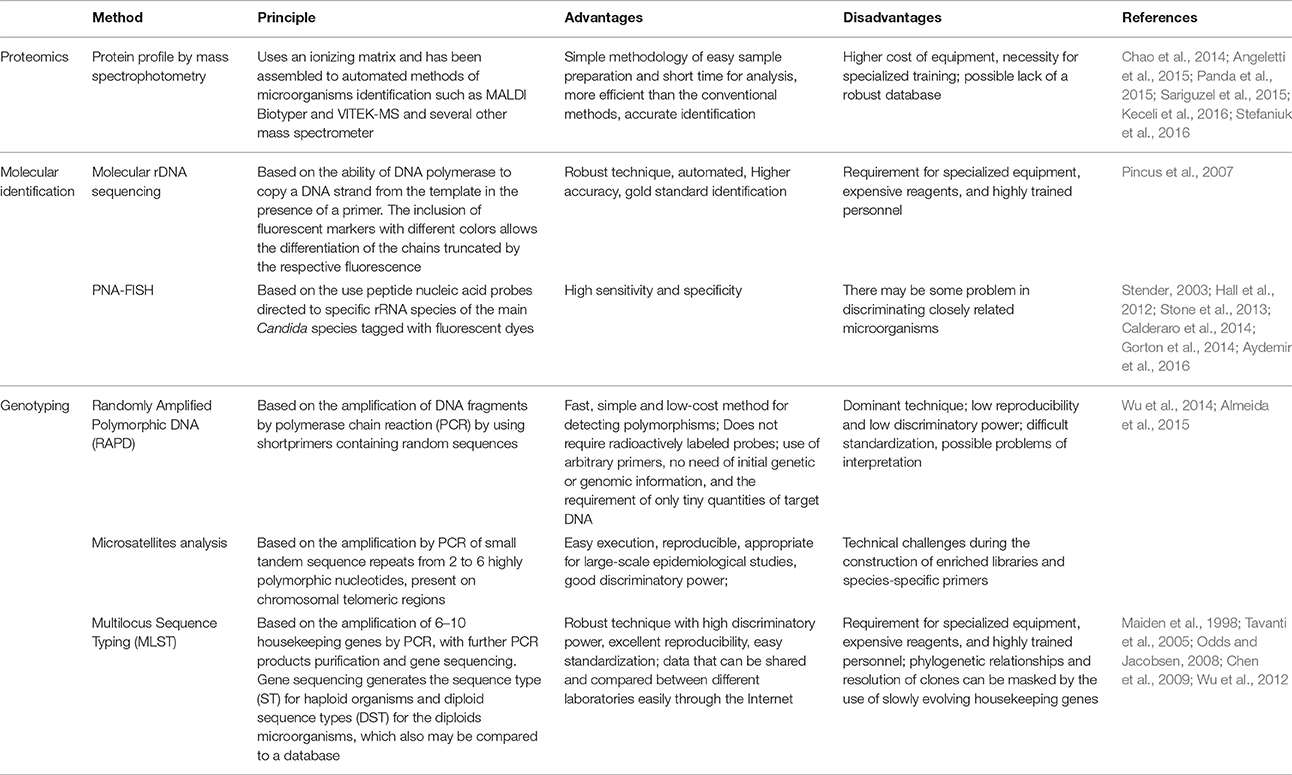
Table 2. Molecular methods and proteomics for the identification and genotyping of Candida tropicalis.
The accurate identification of C. tropicalis by proteomics analysis has been demonstrated in several studies which compared identification methods (Chao et al., 2014; Angeletti et al., 2015; Panda et al., 2015; Sariguzel et al., 2015; Keceli et al., 2016; Stefaniuk et al., 2016). C. tropicalis was accurately identified in 22/22 (100%) (Sow et al., 2015), in 21/21 (100%) (Stefaniuk et al., 2016), in 18/18 (100%) (Angeletti et al., 2015), in 17/17 (100%) (Chao et al., 2014), in 13/13 (100%) (Keceli et al., 2016), and in 2/2 (100%) by VITEK-MS (Sariguzel et al., 2015). The system performance of the MALDI Biotyper system also showed satisfactory results for the identification of C. tropicalis, where the accurate identification was found for 21/21 (100%) (Stefaniuk et al., 2016), 17/17 (100%), and in 18/18 (100%) (Angeletti et al., 2015) of cases.
Several studies have also been performed to evaluate PNA-FISH performance for different Candida species isolated from different anatomic sites, where conclusive results for C. tropicalis ranged from 96 to 100% of cases (Table 2; Hall et al., 2012; Stone et al., 2013; Calderaro et al., 2014; Gorton et al., 2014).
Although the methods used for microorganism's identification by using PNA-FISH and protein profile analysis using mass spectrophotometry techniques are accurate and have high sensitivity and specificity, molecular sequencing has been considered the gold standard technique for microorganisms identification recently (Keceli et al., 2016). rDNA ITS region sequencing has been quite satisfactorily used for C. tropicalis identification elsewhere. The main target for yeasts DNA molecular sequencing is the ribosomal (rDNA) region (Pincus et al., 2007). This region contains conserved domains separated by variable regions (the small sub unities 18S and 5.8S, besides the large subunit 26S, while these sub unities are separated by the interespacer regions ITS1 and ITS2) which contain species-specific sequences used as the preferential target for universal primers used of identification (Table 2; Merseguel et al., 2015; Shi et al., 2015; Benedetti et al., 2016).
Candida tropicalis Genotyping
Genotyping methods have largely been used recently to investigate a genetic correlation of different strains of the same species or even among different species (Table 2). These methods may be applied to the investigation of infections caused by similar or identical strains, besides the observation of possible micro-evolution or strains substitution during colonization and infection (da Costa et al., 2012; Almeida et al., 2015).
Recently, Almeida and collaborators employed RAPD technique with three different random primers (OPA-18, OPE-18, and P4) to evaluate the genetic variability of 15 clinical isolates of C. tropicalis obtained from patients with candiduria (Almeida et al., 2015). The analyses of the dendrogram constructed with DNA bands with the best discriminatory power primer (OPA-18) showed four well-defined clusters (I, II, III, and IV), where cluster I and II showed above 90% similarity among them, while clusters III and IV had 70% similarity.
Da Costa et al. (2012) genotyped by RAPD 15 strains of C. tropicalis oral isolates with primers OPA-01, OPA-09, OPB-11, OPE-18, and SEQ-06 (da Costa et al., 2012). OPA-01 showed the best discriminatory power, presenting ten distinct patterns for C. tropicalis isolates, with 80% similarity (da Costa et al., 2012). Another study using primers OPE-03, RP4-2, OPE-18, and AP50-with 12 catheter tip and urine isolates, obtained 9 different clusters with similarities coefficients (SABs) ranging from 0.8 to 1.0, where different strains were considered unrelated (if SAB was bellow 0.8), moderately related (SAB 0.8–0.89), highly related (SAB 0.90–0.99), and identical (SAB 1.0) (Marol and Yücesoy, 2008).
Almeida et al. (2015) typed 15 isolates of C. tropicalis with microsatellites and obtained the presence of five different alleles with the marker URA3 and eight different allelic combinations with the CT14 locus, being this marker considered to have a better discriminatory power than the URA3 locus (Almeida et al., 2015).
By evaluating 65 clinical isolates of C. tropicalis obtained from different anatomic sites, Wu et al. used different markers of sequence tandem repeats, as follows: Ctrm1, Ctrm7, Ctrm10, Ctrm12, Ctrm15N, Ctrm21, Ctrm24, and Ctrm28 and selected six loci for population genetic analyses (Ctrm1, Ctrm10, Ctrm12, Ctrm21, Ctrm24 and Ctrm28), obtaining a total of 7 (Ctrm24 e Ctrm28) to 27 (Ctrm1) distinct genotypes (Wu et al., 2014).
The methodology known as MLST (Multilocus Sequence Typing) was originally described by Maiden et al. (1998). Therefore, by using MLST, strains from different geographic regions and various anatomic sources may be analyzed and compared. Strains maintenance, substitution, and multiple colonization may be investigated (Maiden et al., 1998; Chen et al., 2009; Wu et al., 2012, 2014).
The first MLST studies on C. tropicalis were performed in 2005, by Tavanti et al. with DNA sequencing of 6 housekeeping genes (ICL1, MDR1, SAPT2, SAPT4, XYR1, and ZWF1α). In this study, 106 isolates of C. tropicalis (104 human clinical isolates and 2 from animal origin) were evaluated, where 87 DSTs where obtained, grouped within three different highly related clades (Tavanti et al., 2005). In the study performed by Wu et al., with 58 strains of C. tropicalis from different anatomic sites by MLST, 52 different DSTs grouped within 6 different clades where obtained (Wu et al., 2012). Therefore, MLST is considered a very robust molecular technique used for typing with high discriminatory power, being widely used to evaluate intra-specific variability for different microorganisms including C. tropicalis (Tavanti et al., 2005; Odds and Jacobsen, 2008; Chen et al., 2009; Wu et al., 2012).
Virulence Factors
The ability of yeasts to adhere, infect, and cause diseases altogether is defined as a potential of virulence or pathogenicity. According to Cauchie et al. (in press), it was previously believed that species of the Candida genus were passively involved in the process of establishment of infection. However, it is now established that these yeasts play an active role in the infectious process through the action of several virulence factors (Cauchie et al., in press).
Adhesion to Epithelial and Endothelial Cells
Adhesion of blastoconidia to host cells is considered the first step for both colonization and the establishment of Candida infections and involves interactions between fungal cells and host surfaces (Cannon and Chaffin, 2001). It is a complex and multiphase process, including different factors, such as the microorganism involved, the composition of adhesion surfaces and several environmental factors (Silva-Dias et al., 2012).
Galan-Ladero et al. (2013) performed a study with 29 C. tropicalis isolates with hydrophobicity potential (Galan-Ladero et al., 2013). The cell wall structure is composed by hydrophobic proteins embedded in a cellular matrix which may favor the initial interaction, because hydrophobic particles tend to attach to a high variety of plastic materials and host proteins such as laminin, fibrinogen, and fibronectin (Tronchin et al., 2008).
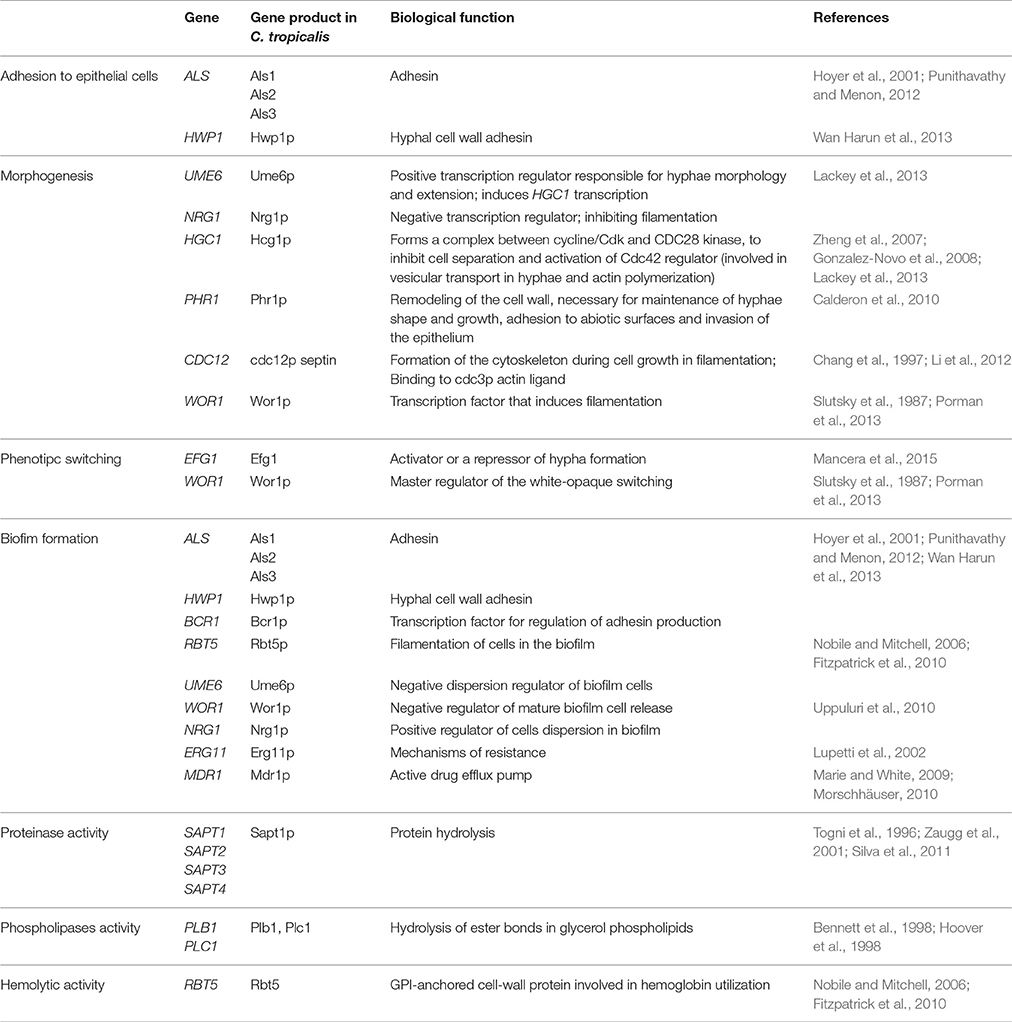
Genes which codify proteins related to adhesion processes are differentially expressed, accordingly to a variety of hosts and environmental conditions (Sohn et al., 2006; Verstrepen and Klis, 2006). Despite the fact that ALS genes (Table 3) are highly involved with adhesion in C. albicans, it has been reported that several Candida species also have the ability to adhere to human buccal and vaginal epithelial cells, besides to the gastrointestinal epithelia of mice and several different plastic materials, motivating studies on adhesion in Non-C. albicans Candida (NCAC) species (Klotz et al., 1983).
Punithavathy and Menon (2012) evaluated the presence of ALS genes in 48 isolates of C. tropicalis obtained from HIV-negative and positive patients. The authors found that 12 isolates (25%) expressed the ALS1 gene, 24 isolates (50%) expressed ALS2, and 23 of them (48%) showed ALS3 expression (Punithavathy and Menon, 2012).
HWP1 (“Hyphal wall protein”) gene codify another important adhesin present on the hyphal cell wall (Table 3). In vitro studies demonstrated the presence of high amounts of Hwp1p at hyphal cell walls, while low amounts are present in blastoconidia (Naglik et al., 2006) and pseudo-hyphae (Snide and Sundstrom, 2006). The HWP1 gene is involved in adhesion to human buccal epithelial cells (HBEC), codifying the first protein needed for biofilm formation (Sundstrom et al., 2002; Nobile et al., 2008).
The expression of this adhesin was recently reported for C. tropicalis in a study performed in Malaysia (Wan Harun et al., 2013) which investigated the presence of HWP1 in NCAC species by using mRNA expression. HWP1 mRNA transcription was positively regulated in C. tropicalis, indicating the ability of this species to express this adhesin. This study suggests that HWP1 in C. tropicalis shares an identical sequence with C. albicans. Therefore, this is contradictory with the description of the presence of HWP1 only in C. albicans (Ten Cate et al., 2009).
In fact, most of the studies report C. albicans as more adherent than other NCAC species, but C. tropicalis is considered the second most adherent species of the Candida genus (Calderone and Gow, 2002; Lyon and de Resende, 2006; Biasoli et al., 2010). For instance, Costa et al. evaluated the ability of adherence of Candida isolates obtained from the oral cavity of HIV individuals, patients with candidemia and catheter tips and found C. albicans as the most adherent species (average of 227.5 cells/100 HBEC) while C. tropicalis showed in average 123.5 cells/100 HBEC (Costa et al., 2010). Conversely, another study investigating adhesion by oral isolates of C. albicans and C. tropicalis to laminin and fibronectin detected by ELISA, reported C. tropicalis adhesion significantly higher than what was found for C. albicans (da Costa et al., 2012).
More recently, Menezes et al. (2013) evaluated the ability Candida spp. clinical isolates to adhere to glass cover slips (Menezes et al., 2013). They found higher adherence of C. tropicalis than C. albicans and yeasts belonging to the C. parapsilosis complex. A recent study performed in Brazil, with isolates from the oral cavity of kidney transplant recipients also demonstrated high ability of adherence to HBEC by C. tropicalis (Chaves et al., 2013). In this study, while C. albicans isolates showed about 237 cells/150 HBECs in average, an isolate of C. tropicalis had 335 cells/150 HBECs, reinforcing the remarkable role of adhesion as an important virulence factor in C. tropicalis.
Morphogenesis and Phenotypic Switching
Subsequently to the adhesion step to host cells, bud-to-hyphae transition (also called morphogenesis) is highly relevant to some pathogenic yeasts, including Candida spp. (Calderone and Gow, 2002). It is one of the most important steps for the establishment of candidiasis and is considered a necessary step for several virulence processes, including invasion of host epithelial layers, endothelial rupture, survival to phagocytic cells attack, biofilm formation, and thigmotropism (Lackey et al., 2013).
Studies concerning morphogenesis are very well-established for C. albicans, with very well-established environmental signals, transcription regulators, and target genes involved in filamention (Gustin et al., 1998; Kumamoto and Vinces, 2005; Wapinski et al., 2007; Lackey et al., 2013). However, there are considerably less studies concerning morphogenesis in other NCAC species. Several Candida species may develop pseudo-hyphae, but quite a few are able to form true hypahe, including C. albicans, C. dubliniensis, and C. tropicalis. The latter do not show the same degree of filamentation than C. albicans; however, because of the fact they are frequently associated with infectious processes, they certainly have mechanisms of adaptation that may favor filamentation in specific environmental conditions (Lackey et al., 2013).
Galan-Ladero et al. (2013) evaluated the filamentation among C. tropicalis isolates obtained from different anatomic sites of patients admitted in a Spanish tertiary hospital (Galan-Ladero et al., 2013). The authors described high levels of filamentation for 76.6% of the isolates at the specific environmental conditions. Wapinski et al. (2007) reported that at least 55 out of the 105 genes involved in C. albicans filamentation are conserved in C. tropicalis (Wapinski et al., 2007).
Lackey et al. (2013) induced C. tropicalis cells filamentation and analyzed gene expression at the conditions provided (Lackey et al., 2013). They found significant filamentation in serum and glucose medium at 37°C. Optical microscopy showed the presence of elongated yeast-like cells, pseudohyphae, and true hyphae that were shorter than the ones found in C. albicans. They also verified that the negatively regulated gene NRG1 has an important role in inhibiting filamentation in other NCAC species, suggesting that this gene may be related with poorer filamentation found among these species (Table 3). The UME6 gene is transcriptionally induced during filamentation in C. tropicalis, similarly to what happens in C. albicans (Table 3; Banerjee et al., 2013).
Porman et al. (2013) reported the elevated expression of the transcriptional regulator WOR1 (Table 3) in C. tropicalis cells cultivated on Spider medium (Porman et al., 2013). The micromorphological analysis of isolates with wrinkled phenotype showed that the most filamentous strains had WOR1 overexpression. Wor1p homologs were also found in Saccharomyces cerevisiae (Cain et al., 2012) and Histoplasma capsulatum (Nguyen and Sil, 2008), controlling morphological transition within these species. This finding may suggest the existence of a common ancestor gene found in the C. tropicalis genome (Porman et al., 2013).
In addition, Wor1 which is the master regulator of the white-opaque switching, a phenomenon which is related to the reversible transition of cells from a white phase to an opaque phase, where cells are larger and elongated, while colonies have wrinkled appearance (Slutsky et al., 1987). Besides morphology, these two different cell types exhibit dramatic differences regarding to the preferred anatomic sites they colonize and infect, in addition to specific responses to environmental and nutritional signals and mating behavior (Mancera et al., 2015). In C. tropicalis, WOR1 overexpression direct cells to the opaque phase which is involved in biofilm formation and morphogenesis (Porman et al., 2013).
It was described in the C. tropicalis genome an ortholog of the transcription factor Efg1 (enhanced filamentous growth), commonly found in C. albicans (Table 3; Mancera et al., 2015). The deletion of both alleles of the EFG1 gene revealed that Efg1p is essential for filamentation, biofilm formation, and white-opaque switching in C. tropicalis, similarly to C. albicans, indicating conservation in the function of this ortholog gene.
Zhang Y. et al. (2016) reported a gray phenotype in C. tropicalis recently, whose cells are small and elongated, show intermediate mating competence and virulence in rats' animal models (Zhang Y. et al., 2016).
Biofilm Formation
The ability of yeast cells to form biofilms is an important determinant of virulence in Candida spp. and has been considered the main form of microbial growth recently (Donlan and Costerton, 2002; Fanning and Mitchell, 2012). Biofilms are complex structures formed by a community of microorganisms adhered to solid surfaces of either biotic or abiotic nature. Therefore, in vitro biofilm formation may be organized by three important steps, as follows: adhesion and colonization of yeast cells on a surface; cellular growth and proliferation, forming a basal layer; and pseudohyphal and/or true hyphal formation (for the species that are able to form filaments), with the subsequent secretion of an exopolymeric extracellular matrix which embeds microorganisms with low growth rates and altered phenotypes (Hawser and Douglas, 1995; Baillie and Douglas, 1999; Chandra et al., 2001; Ramage et al., 2001; Douglas, 2003). The exopolymeric matrix (EPS) may be secreted by different populations of either unique or multiple microbial species (Adam et al., 2002). Some advantages of biofilm formation include: the protection of microorganisms against environmental damage, nutrients availability, metabolic cooperation, and the acquisition of genetic modification (Douglas, 2002).
The formation of the microbial community involves a cascade of molecular mechanisms and fine alterations in gene expression (Nobile and Mitchell, 2006; Araújo et al., 2017). Signaling molecules which naturally occur in fungal cells as a response to environmental stimuli are part of this process present in the Candida genus (Ramage et al., 2006). This regulation is called “quorum sensing” (QS) mechanism and is the main communication form among several microorganisms correlated to population density (Albuquerque and Casadevall, 2012).
Farnesol is kind of self-regulator, a sesquiterpene with the ability to inhibit biofilm formation and altering the expression of 274 genes in C. albicans, specifically involved in filamentation. Weber et al. (2010) investigated the role of farnesol in biofilm formation of C. tropicalis. They found that besides inhibiting cellular aggregates, cells of the C. tropicalis mature biofilm were also influenced by farnesol, which may be related to their dispersion to other body sites (Nickerson et al., 2006; Ramage et al., 2006).
The initial step for biofilm formation is dependent of cellular adhesion cells to substrates and further formation of a basal layer (Nobile and Mitchell, 2006). C. tropicalis adhesins are also involved in biofilm formation (Table 3; Punithavathy and Menon, 2012; Wan Harun et al., 2013), and are regulated by the BCR1 gene (also considered a cell wall regulator). In addition, the RBT5 gene was also found in the C. tropicalis genome (Fitzpatrick et al., 2010).
Other genes involved in C. tropicalis biofilm formation are WOR1, UME6, NRG1, ERG11, and MDR1 (Table 3). Besides being involved with morphogenesis and phenotypic switching, WOR1 is one of the main transcriptional factors involved in biofilm formation (Xie et al., 2012; Porman et al., 2013).
UME6 and NRG1 are key transcription regulators directly involved in morphogenesis in C. tropicalis (Finkel and Mitchell, 2011). The overexpession of UME6 reduces the liberation of mature sessile cells, while the decreased expression of NRG1 promotes cells dispersion (Uppuluri et al., 2010).
With respect to the expression of resistance genes to antifungal drugs, ERG11 (ergosterol biosynthesis), and MDR1 (multidrug resistance) genes (Table 3) are related with resistance to fluconazole. Bizerra et al. (2008) reported the increased expression of these genes in sessile cells of C. tropicalis isolated from vulvovaginal candidiasis (VVC) and uroculture resistant to both fluconazole and amphotericin B. Punithavathy and Menon (2012) also demonstrated higher resistance to fluconazole of sessile cells liberated from mature biofilms of C. tropicalis.
There are evidences that biofilm cells formed on medical devices constantly released in the bloodstream guarantee the successful establishment of disseminated candidiasis (Fanning and Mitchell, 2012). Marcos-Zambrano et al. (2014) investigated biofilm formation in different Candida species obtained from episodes of fungemia and found C. tropicalis isolates were the strongest biofilm producers. In fact, another study reported that the high thickness of the EPS matrix of C. tropicalis biofilm cells may impair oxygen and nutrients diffusion to cells, and may be responsible for the lower metabolic activity (Alnuaimi et al., 2013).
Pannanusorn et al. (2013) also described C. tropicalis as the most efficient biofilm producers among bloodstream isolates as compared to other NCAC species. Paiva et al. (2012) evaluated the in vitro biofilm formation by C. tropicalis isolates obtained from VVC. This species was also considered the strongest biofilm producer compared to C. albicans, yeasts belonging to the C. parapsilosis complex, C. glabrata, and C. guilliermondii. A similar trend was also described by Udayalaxmi et al. (2014) with strains isolated from the urogenital tract (samples from vaginal fluid and urine) of patients from a tertiary hospital in the South of India. Therefore, C. tropicalis has been considered an important biofilm producer species of the Candida genus.
Lytic Enzymes
In order to facilitate host tissues invasions, several pathogenic microbes secrete lytic enzymes such as proteinases, phospholipases, and hemolysins to destroy, alter, or damage the integrity of host membranes, leading to the dysfunction, or rupture of host cells (Sanita et al., 2014).
Pathogenic Candida species produce a great variety of hydrolases, including secreted aspartic proteinases (Saps). These proteins have been intensely investigated, and possess a wide range of substrates, including collagen, queratin, and mucin. They have the ability to degrade epithelial barriers, antibodies, complement, and cytokines (Hube and Naglik, 2001), and are encoded by a great gene family. The SAP gene family is composed by 10 genes and was initially described in C. albicans (Ruchel et al., 1983). These genes are differentially regulated and expressed under several laboratory conditions and are activated during different stages of infections in vivo. In addition, some of the SAP genes are more important to superficial rather than systemic infections, and are also involved in other pathogenic process in C. albicans, such as adhesion, host tissue invasion, and immunological system cells evasion (Hube and Naglik, 2001).
It is well-known since 1983 that C. tropicalis is able to secrete proteinases as one of the most important determinants of virulence of this species (Macdonald and Odds, 1983; Ruchel et al., 1983). In 1991, Togni et al. reported the nucleotide sequence of a gene involved with the extracellular secretion of proteinases by this yeast, while in 1996 the same authors reported the secretion of Sapt1p by C. tropicalis (Togni et al., 1991, 1996). Subsequently, the crystallographic structure of this protein was published, and was considered very similar to the Sap2p of C. albicans (Symersky et al., 1997).
A study performed by Zaugg et al. (2001) suggested the existence of a SAPT gene family in the C. tropicalis genome, leading to four genes cloning: SAPT (1–4; Table 3). However, only Sapt1p was purified from culture supernatant and biochemically characterized.
Silva et al. (2011) investigated epithelial invasion by C. tropicalis using a reconstituted human buccal epithelia model. All the isolates tested were able to colonize this tissue and cause a great damage after 24 h. Real time PCR showed that SAPT2-4 transcripts were detected, while SAPT1 expression was rarely observed. In addition, the authors showed that there was no increase in SAPT1 expression, suggesting that the high invasive capacity of C. tropicalis may not be related with the specific expression of this gene. Following the same trend, Togni et al. (1996) reported that SAPT1 gene disruption in C. tropicalis seemed to have low effect in attenuation of virulence in mice, in a model of systemic infection.
Costa et al. (2010) evaluated proteinase activity of 15 isolates of C. albicans and 15 of C. tropicalis obtained from the saliva of dental patients in Brazil. All C. tropicalis isolates showed higher enzymatic production than C. albicans. These results are contradictory to most of the studies which suggest higher proteinase activity in C. albicans than in C. tropicalis (Zaugg et al., 2001; Sachin et al., 2012).
In addition to the secretion of proteinases, the secretion of phospholipases constitutes important determinants of virulence in Candida spp. This heterogeneous group of enzymes catalyzes the hydrolysis of ester bonds in glycerol phospholipids, with each enzyme participating in a specific reaction (Ghannoum, 2000). Secretion of phospholipases is therefore considered a key attribute for invasion of host epithelia, since phospholipids are major components of all cell membranes. In addition, the breakdown of these molecules promotes great instability in host cells, resulting in cellular lysis (Schaller et al., 2005).
One of the first studies that analyzed the production of phospholipases in Candida spp. was published in 1984 by Samaranayake et al. which demonstrated the secretion of these enzymes only in C. albicans isolates, without any detection in C. tropicalis (Samaranayake et al., 1984). However, other authors later reported phospholipase activity in isolates of this species. A recent study conducted by Jiang et al. (2016) with 52 strains of C. tropicalis found phospholipase activity in 31 isolates from different clinical sources. However, strains showed low enzyme production. Another study with 29 strains of several anatomic sites obtained from hospitalized patients, described low or no phospholipase activity in C. tropicalis (Galan-Ladero et al., 2010).
Conversely, a study conducted by Deorukhkar et al. (2014) investigating the expression of several virulence factors in 125 clinical isolates of this species concluded that the secretion of phospholipases was the main determinant of virulence expressed by these strains. The authors suggest that the variability of results between different authors may be a result of biological differences among the isolates tested.
Related to the expression of these enzymes in the presence of antifungal drugs, Anil and Samaranayake (2003) analyzed the effect of previous exposure of C. albicans and C. tropicalis to antifungal drugs on extracellular phospholipase activity. They concluded that the enzymatic activity of both species reduced significantly after previous exposure to nystatin and amphotericin B. In fact, they showed that C. albicans had greater phospholipase expression than C. tropicalis.
Phospholipases are classified into four major groups, named from A to D, all already well-described for C. albicans (Schaller et al., 2005). However, a few studies address this gene regulation in C. tropicalis. Phospholipase B (PLB; Table 3) is known to catalyze the hydrolytic cleavage of sn-1 and acyclic glycerophospholipid sn-2 esters (Ghannoum, 2000) and is primarily responsible for phospholipase activity in C. albicans (Schaller et al., 2005).
In 1998, Hoover et al. published an investigation with degenerate oligonucleotides (derived from conserved regions of the PLB1 gene of S. cerevisiae and other fungi) to amplify homologous fragments of PLB1 in C. albicans and C. tropicalis by PCR. The main PCR product obtained was a 540 bp fragment with a high probability of PLB1-correspondence of other fungi, and significant homology was found between the deduced amino acid sequence of the PCR product of C. albicans and C. tropicalis and the corresponding regions of PLB1 sequence of S. cerevisiae, Torulaspora delbrueckii, and Penicillium notatum (~70–75% resemblance, ~55–65% identity). In that same year, Bennett et al. (1998) evaluated the presence of homologous sequences to C. albicans PLC in NCAC species, including five isolates of C. tropicalis. A DNA sequence homologous to CAPLC1 was detected in only three of these isolates. Thus, the need for further studies addressing the molecular mechanisms related to phospholipase activity in C. tropicalis is evident.
The hemolysins are another group of proteins that significantly contribute for the dissemination of Candida infections, specifically in facilitating hyphal penetration in host tissues (Luo et al., 2004; Tsang et al., 2007). Hemolytic factors secreted by fungi cause hemoglobin liberation from red blood cells for further utilization by yeasts as an iron source (Giolo and Svidzinski, 2010). This chemical element is an essential cofactor to a great number of metabolic processes, such as oxygen transport, gene expression regulation, and DNA synthesis. Therefore, the ability of iron acquisition is of fundamental importance for microorganisms survival and establishment of infectious processes (Giolo and Svidzinski, 2010).
Manns et al. (1994) reported iron acquisition from erythrocytes by C. albicans as a consequence of a protein factor that promoted host cells lysis. In 1997, Tanaka et al. reported that this factor is liberated from the culture medium supernatant, and concluded that it was a cell wall manoprotein. The same phenomenon was observed in C. tropicalis (Favero et al., 2014), and although this factor is known as directly involved with yeasts pathogenicity, it is still poorly understood (Favero et al., 2011).
The study conducted by Luo et al. (2001) was the first one to show differences in hemolysin production by different Candida species on SDA plates containing sheep blood. The authors also observed that the hemolysis induced by this method could be divided into categories according to the standard microbiological nomenclature, including: total hemolysis (beta), partial hemolysis (alpha), or hemolysis absence (gama). In this study with 80 isolates of 14 different Candida species, all the five isolates of C. tropicalis showed a large clear halo around colonies, proving the ability of C. tropicalis in producing beta hemolysis. Similarly, Favero et al. (2011) detected hemolysin production in C. tropicalis strains after incubation in both solid and liquid SDA containing either human or sheep blood.
A study produced by Rossoni et al. (2013) evaluated the hemolytic activity in different Candida species obtained from the oral cavity of HIV positive patients. Strong hemolytic activity was observed in 75% of C. tropicalis isolates evaluated, only after C. albicans. Similar results were found for Candida isolates obtained from different anatomical sites (blood, synovial, and peritoneal liquid) where, again, C. albicans proceeded C. tropicalis in hemolysins production (de Melo Riceto et al., 2015).
Contradictory to these results, Favero et al. (2014) analyzing clinical Candida spp. isolates from bloodstream infection, reported low hemolytic activity in C. albicans, while C. tropicalis was the species tested with greater hemolysins production.
The genetic regulation of hemolysins production in the Candida genus was not still largely investigated (Anil et al., 2014). It is known that in C. glabrata, the HLP gene (hemolysin-like protein) encodes a protein associated with hemolytic activity (Luo et al., 2004). In C. albicans the Csap is involved with iron acquisition from host erythrocytes during hyphal development (Okamoto-Shibayama et al., 2014). This enzyme is a member of the Rbt5 protein (Table 3), also described in C. tropicalis, as previously mentioned. However, there are currently no studies in the literature concerning the genetic elucidation of hemolytic activity in C. tropicalis.
In vivo Models of Infection by Candida tropicalis
The characterization of the expression of most variable virulence factors by Candida spp. and other fungi are necessary for the understanding each particular pathway involved in microorganisms pathogenicity (Takakura et al., 2003; Solis and Filler, 2012; de Campos Rasteiro et al., 2014). However, experiments performed in vivo involve different variables which cannot be controlled like what happens in experimental conditions in vitro, including the presence of body fluids, pH variation, commensal microorganisms, and their metabolites and host response during infection. Therefore, in vivo experimental models are needed for the global understanding of infectious disease pathogenicity, interactions with host cells and immune response as well as it is a more appropriate approach to evaluate new therapeutic strategies (Takakura et al., 2003; Solis and Filler, 2012; de Campos Rasteiro et al., 2014).
Several studies have been described in the literature with animal models of infections by C. tropicalis using mice (Bayegan et al., 2010; Mariné et al., 2010; Koga-Ito et al., 2011; Chen et al., 2014; Nash et al., 2016a; Wang et al., 2016; Zhang Y. et al., 2016). Nash et al. (2016a) evaluated the co-infection of six different Candida species (C. albicans, C. tropicalis, C. parapsilosis, C. krusei, C. dubliniensis, and C. glabrata) with Staphylococcus aureus, intraperitoneally inoculated. They evaluated mortality rates and attributed a score of 1–4 to evaluate characteristics of morbidity (creepy hair, absence of mobility, arched posture, and ocular secretion). C. tropicalis associated with S. aureus showed the second highest mortality rate (behind C. albicans) and a mortality index of 3 (Nash et al., 2016a).
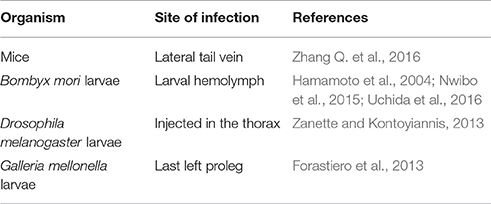
Animal models of systemic infections may be induced by the inoculation of C. tropicalis via the lateral tail vein (Table 4; Zhang Y. et al., 2016). This via of infection was established by Zhang Y. et al. (2016) to evaluate virulence of a new phenotype described by C. tropicalis, the “Grey phenotype,” besides the other phenotypes already described (White–Opaque). After systemic infection through the tail vein with strains of each phenotype (Gray, White, and Opaque), animal organs have been removed and macerated and fungal load was evaluated. The authors found that cells of the Gray phenotype showed intermediate distribution, but greater than cells with the White phenotype for all the organs evaluated (kidney, lungs, spleen, liver, and brain), 24 h and 7 days after infection.
Other mice models of Candida infections have been described in the literature such as the VVC model described by Fidel et al. (1997), where doses of estradiol valerate are subcutaneously administered (0.1 mg/100 μl of sesame oil) in the vagina of animals infected with 5 × 104 cells/20 μl PBS in order to successfully establish the vaginal infection (Fidel et al., 1996, 1997; Garvey et al., 2015; Nash et al., 2016b). Nevertheless, to the best of our knowledge, they were still not employed for the experimental investigation of VVC caused by C. tropicalis.
Alternative models of experimental infections have been broadly used for virulence and interactions with the host studies (Table 4; Hamamoto et al., 2004; Forastiero et al., 2013; Mesa-Arango et al., 2013; Zanette and Kontoyiannis, 2013; de Souza et al., 2015; Ishii et al., 2015; Shu et al., 2016). Several factors are considered as an advantage for the utilization of a model of infection using larvae, including an easier manipulation and lower maintenance cost (de Souza et al., 2015; Ishii et al., 2015). The Silkworm—Bombyx mori, (Lepidoptera: Bombycidae) produces a large enough larvae for antifungal drugs distribution studies (Nwibo et al., 2015; Uchida et al., 2016). B. mori larvae were used as a C. tropicalis model of infection in order to evaluate the effective dose of both fluconazole and amphotericin B (Hamamoto et al., 2004). When C. tropicalis was inoculated into the larval hemolymph followed by antifungal drugs administration, it was obtained an effective dose for 50% of them (ED 50%) of 1.8 μg/g of larvae for amphotericin B and fluconazole, being in agreement with previous animal models using mice previously performed (Hamamoto et al., 2004). Therefore, it confirms its possible use for C. tropicalis virulence studies.
Drosophila melanogaster larvae (Diptera: Drosophilidae), known as fruit flies also have been used as an animal model to study microbial interactions with innate immune response (Alarco et al., 2004). Zanette and Kontoyiannis satisfactorily used this model to investigate C. tropicalis strains with or without paradoxical growth (Zanette and Kontoyiannis, 2013).
Galleria mellonella larvae (Lepidoptera: Pyralidae) have been used as another invertebrate model to investigate fungal and host interactions (Champion et al., 2016), in systemic studies of antimicrobial efficiency (Wei et al., 2016), evaluation of virulence in immunosuppressive models (Torres et al., 2016), immunomodulatory response (Fuchs et al., 2016), and antifungal resistance (Souza et al., 2015).
G. mellonella infection by C. tropicalis was used to investigate cross resistance to azoles or multidrug resistance among them and amphotericin B (Forastiero et al., 2013). Two hours after infection with C. tropicalis, different antifungal drugs were applied (fluconazole, voriconazole, amphotericin B, and anidulafungin). In this study, 80% of the untreated infected larvae died between day 3 and 4 of infection, while better survival rates were observed for animals inoculated with susceptible strains (10 mg/kg/day of voriconazole and 9 mg/kg/day of fluconazol). When the larvae were infected with strains resistant to the azoles with the same therapeutic doses, survival rates were equivalent to the group that was untreated. This study demonstrates the reliable application of the use of the G. mellonella model for the study of infection by Candida as well as for the evaluation of antifungal action (Forastiero et al., 2013).
Superficial and Systemic Infections
C. tropicalis belongs to the normal human microbiota and is present on the skin, gastrointestinal, genitourinary, and respiratory tracts of humans (Basu et al., 2003; Oksuz et al., 2007; Negri et al., 2010). This yeast has been associated with superficial and systemic infections all over the world, specifically in neutropenic patients, or in individuals with a reduction of the microbiota by antimicrobial use or presenting damage in gastrintestinal mucosa (Colombo et al., 2006).
C. tropicalis is classified as the third or fourth NCAC species more commonly isolated in the clinical practice (Pfaller et al., 2010; Peman et al., 2012), but it is considered the most prevalent yeast in Asia (Chakrabarti et al., 2009; Kothavade et al., 2010; Adhikary and Joshi, 2011) and the second or the third more isolated species in Brazil and other Latin America countries (20.9 and 13.2%, respectively) (Pfaller et al., 2010). The expressive increase in isolation of this yeast in cases of both superficial and systemic infections in different casuistic all over the world emphasizes its emergent character.
The clinical aspects of Candida infections may vary according with the body site affected. Oral candidiasis, VVC, and onychomycosis are superficial mycoses caused by this genus, while systemic candidiasis involves blood and deep-seated organs such as the lungs and gastrintestinal tract (Jacobs and Nall, 1990).
Oral candidiasis is an opportunistic infection caused by Candida commonly found in the eldery (due to low immunity caused by age), HIV patients, malnourished individuals and those submitted to systemic steroid therapy, denture wearers, and people with xerostomia (Muadcheingka and Tantivitayakul, 2015). Clinical manifestations are divided into white and erythematous forms. The white form is characterized by whitish lesions and includes pseudomembranous candidiasis and hyperplastic candidiasis. The erythematous form presents with red lesions, including acute atrophic candidiasis, chronic atrophic candidiasis, median rhomboid glossitis, angular cheilitis, and linear gingival erythema. There are also three forms which are not classified into these two clinical categories, which are chronic mucocutaneous candidiasis, cheilocandidiasis, and chronic multifocal candidiasis (Millsop and Fazel, 2016).
In Brazil, a study performed by da Silva-Rocha et al. (2014) investigated Candida species distribution of isolates obtained from the oral cavity of kidney transplant recipients from two geographic regions of Brazil (Northeast and South). The authors found that C. tropicalis was the second most prevalent species, corresponding to 4.5% of the isolates.
A prevalence study of Candida species obtained from oral candidiasis was carried out in Thailand with 250 strains isolated from 207 patients and C. tropicalis was the third most isolated species (10.4%) (Muadcheingka and Tantivitayakul, 2015). Similarly, in a study conducted in the northwest of Ethiopia with 215 oral cavity isolates from HIV positive patients, this yeast was also the third most prevalent species, with a percentage of isolation equal to 14.1%. More interestingly, 8% of them were resistant to fluconazole and 4% to ketoconazole, itraconazole, and fluocytosine (Mulu et al., 2013). Another Indian study concluded that there was a significant increase in Candida infections in oral cancer patients who underwent chemotherapy or radiotherapy, where NCAC species predominated, mainly C. tropicalis, occurring in 42.8% of cases (Jain et al., 2016).
VVC is an infection of the vulva and vagina caused by different Candida spp. (de Medeiros et al., 2014; Sobel, 2016). C. tropicalis is generally described as the third most prevalent Candida species in VVC, preceded by C. albicans and C. glabrata in most of the studies (Dias et al., 2011; Kanagal et al., 2014; Ragunathan et al., 2014).
A study developed in India by Vijaya et al. (2014) with 300 women of reproductive age with clinical signs of VVC reported C. tropicalis as the second most prevalent Candida species, corresponding to 26.4% of the isolates. Of these, 42.9% were resistant to fluconazole and 14.3% to voriconazole.
An investigation in Iran with 67 Candida isolates obtained from vaginal secretion samples from patients with VVC found that C. tropicalis was present in 5.9% of cases, with 100% resistance to fluconazole, 50% resistance to clotrimazole, 25% to ketoconazole, and 75% against terbinafine. In addition, all isolates showed dose-dependent susceptibility (DDS) to itraconazole (Salehei et al., 2012).
Nevertheless, C. tropicalis is reported to a lesser degree in cases of onychomycosis in relation to other species such as C. albicans and C. parapsilosis species complex, promoting paronychial infection mainly in immunosuppressed patients and individuals in extreme age (elderly and children) (Aghamirian and Ghiasian, 2010; Cambuim et al., 2011).
A study developed in South Korea reports the prevalence of the Candida genus in 59% of cases of onychomycosis in pediatric patients. The authors obtained 39 isolates, where only 2.6% of them belonged to C. tropicalis (Kim et al., 2013). Another study developed in Mexico analyzing 166 samples of dystrophic nails reports C. tropicalis as one of the less prevalent Candida species in onychomycosis, corresponding to 4.2% of the isolates. However, 14.2% were resistant to fluconazole, itraconazole, and ketoconazole (Manzano-Gayosso et al., 2011), reinforcing its clinical importance.
However, contradictory results were found in a Brazilian study with 200 Candida isolates obtained from nail infections that reported a prevalence of 26% for C. tropicalis. These authors also observed high antifungal drugs resistance in these isolates, including 30.6% resistance to fluconazole, 25% to itraconazole, 9.6% to ketoconazole, and 96.2% resistance to terbinafine (Figueiredo et al., 2007).
According to McCarty and Pappas (2016), invasive infection by Candida species is commonly associated with medical care, where they may be the third or fourth cause of bloodstream infection (BSI). Risk factors for systemic candidiasis are well-known and include the presence of central venous catheter (CVC), the exposure to broad spectrum antibacterial agents, prolonged staying in the ICU with or without mechanic ventilation (more than 3 days), complex surgery, the presence of necrotizing pancreatitis, hemodialysis, and immunosuppressive conditions (McCarty and Pappas, 2016). Candidemia caused by C. tropicalis infection has a greater association with skin petechia than other Candida species (Manzano-Gayosso et al., 2011), and this species was described as the most common etiological agent of invasive infection associated with the hospital environment in India (Giri and Kindo, 2012).
According to Kontoyiannis et al. (2001), C. tropicalis produces more persistent systemic infections than C. albicans, leading to a longer stay in the hospital environment. Other studies have associated C. tropicalis infections with a higher mortality rate when compared to other NCAC species, even when compared to C. albicans (Krcmery et al., 1999; Kontoyiannis et al., 2001; Eggimann et al., 2003). This factor may be related to the known higher virulence of both species as well as to a higher antifungal resistance by C. tropicalis.
Recently, an important multicenter study was carried out in 29 Spanish hospitals, where C. tropicalis was isolated in 7.6% of a total of 781 cases of candidemia and 20% of them were resistant to azoles (Guinea et al., 2014). Another multicenter study in China with 389 isolates from patients with candidemia admitted to intensive care units found C. tropicalis as the third most isolated species (17.2%), while resistance to fluconazole was observed in 37.3% of isolates of this species, as well as 10% of them were resistant to voriconazole (Liu et al., 2014).
A research carried out in Malaysia with 82 bloodstream isolates and peritoneal fluid reports C. tropicalis as responsible for 18.3% of the isolates obtained. Resistance to ketoconazole was observed in 20.9% of the clinical strains, in addition to 13.4% resistance to itraconazole (Santhanam et al., 2013). A study performed by Chang et al. (2013) with isolates from 152 cases of candidemia in Taiwan reported a prevalence of 19.7% for C. tropicalis.
In Brazil, a study conducted by Oliveira (2011) investigated candidemia in a pediatric hospital in Sao Paulo from 2007 to 2010. C. tropicalis was the second most isolated Candida species (24%), only preceded by C. albicans. More recently, a multicenter surveillance study involving 16 public and private hospitals in the five Brazilian regions (North, Northeast, Center-West, Southeast and South) was conducted, which investigated 137 episodes of systemic infections. NCAC species were responsible for 65.7% of the total infections and C. tropicalis was the third most isolated Candida species (15.3%) (Doi et al., 2016).
It is known that candidemia is the most common form of invasive candidiasis, but there are other less frequent clinical manifestations with C. tropicalis as an etiological agent (McCarty and Pappas, 2016). For example, a case of acute disseminated candidiasis in a pediatric patient with aplastic anemia (Fong et al., 1988); the formation of fungal vegetation in a mitral valve prosthesis, causing endocarditis (Nagaraja et al., 2005); and the development of septic arthritis in a cancer patient on chemotherapy with diabetes secondary to corticosteroid therapy that had a negative outcome (Vicari et al., 2003).
Another unusual clinical form of candidiasis is endophthalmitis, which is considered an important indicator of systemic infection in hospitalized patients (Donahue et al., 1994). C. tropicalis seems to be an important etiological agent of this infirmity, being classified as the fourth species of the genus to promote ocular infection in adult and pediatric patients attended at two medical centers in the USA (Dozier et al., 2011).
Disseminated chronic candidiasis is another condition of low occurrence characterized by the presence of histopathological evidence of candidiasis in the spleen, liver, and kidneys, or radiological evidence of hepatosplenic or renal candidiasis (Al-Anazi and Al-Jasser, 2006). Xu et al. (2010) described the isolation of C. tropicalis in a patient with acute leukemia whose computerized tomography showed multiple hypodense lesions in the liver and spleen. This yeast was also isolated from the kidneys of a patient diagnosed with acute lymphocytic leukemia (Sun et al., 2006) and was associated with a higher mortality rate than other Candida species involved in this disease (Al-Anazi and Al-Jasser, 2006).
Finally, C. tropicalis is more rarely found as an etiological agent of respiratory tract infections (Garczewska et al., 2016). This was the second most common yeast species in patients with cystic fibrosis, preceded only by C. albicans. Similarly, another study reports C. tropicalis as the second most common Candida species in cases of pulmonary co-infection with Mycobacterium tuberculosis (Kali et al., 2013).
Antifungal Susceptibility
The high incidence of severe infections caused by C. tropicalis has attracted attention, especially considering the evident increase in the reports of resistance of this yeast to antifungal drugs, which is a serious therapeutic problem.
Resistance to azoles in this species has been extensively reported, especially to fluconazole. In this respect, Anil and Samaranayake (2003) argue that the increasing global use of this drug is one of the main causes for the dominant tendency of infections caused by NCAC species to the detriment of C. albicans. It is known that there are several factors involved in the development of Candida spp. antifungal resistance in clinical settings, including indiscriminate antifungal therapy use in nosocomial infections (Joseph-Horne and Hollomon, 1997). However, studies on the molecular mechanisms underlying this phenomenon are still necessary.
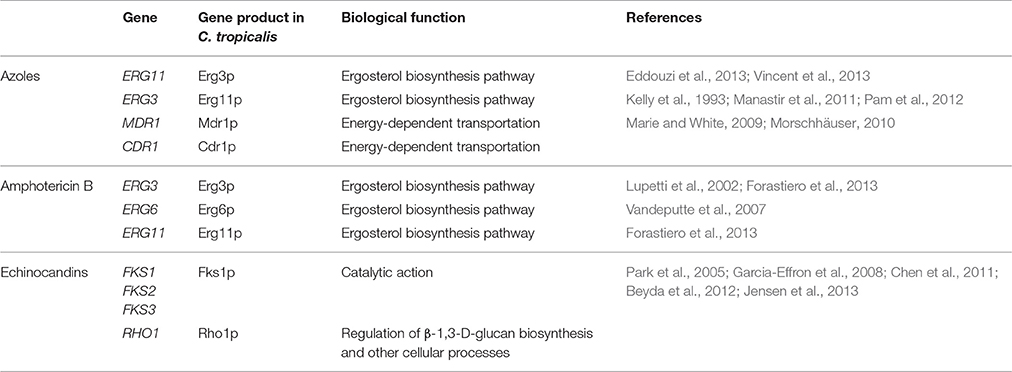
With regard to azoles, the action target of these compounds is the enzyme 14 α-lanosterol demetilase (Erg11p), a product of the ERG11 gene (Table 5), which is part of the ergosterol biosynthesis pathway (Lupetti et al., 2002). Ergosterol is the predominant component of the cell membrane of fungi, and influences various cellular functions such as membrane fluidity and integrity, as well as the adequate activity of various enzymes anchored to it, such as proteins related to nutrient transport and chitin synthesis. Therefore, the azoles cause depletion of ergosterol and accumulation of 14 α-methyl steroids harmful to cells, inhibiting growth of fungal cells (Lupetti et al., 2002).
Sanglard and Odds (2002) report different mechanisms that may lead to resistance to azoles. The first is the action of multidrug transporters or efflux pumps, which leads to a decrease in drug concentration within the fungal cell (Pfaller, 2012). The positive regulation of MDR1 (“multidrug resistance gene”) and CDR1 (“Candida drug resistance”) genes (Table 5), are related to the active efflux of azoles in several Candida species, including C. tropicalis (Marie and White, 2009; Morschhäuser, 2010). The induction of efflux caused by CDR genes tends to affect all azoles. In contrast, efflux pumps encoded by MDR genes in Candida are normally selective for fluconazole (Pfaller, 2012).
Another pathway leading to azole resistance is the occurrence of amino acid substitutions in Erg11p, which is the target of these drugs, generating changes in protein conformation Forastiero et al., 2013). Increased ERG11 gene expression results in the production of a large amount of 14 α-lanosterol demethylase, favoring the continuous synthesis of ergosterol, and the maintenance of cell integrity, which allows the fungus to resist the action of the drugs (Manastir et al., 2011). This factor may occur as a function of a point mutation in ERG11 (Kelly et al., 1993). Pam et al. (2012) detected this point mutation in a C. tropicalis isolate with DDS to fluconazole and demonstrated increased ERG11 expression.
Eddouzi et al. (2013) studied the molecular mechanisms of drug resistance in a clinical isolate of C. tropicalis with multidrug resistance to fluconazole, voriconazole, and amphotericin B, obtained from a hospital in Tunisia. Analysis of sterol production by mass spectrometry and gas chromatography revealed accumulation of 14α-methylfecosterol, 4,14α-dimethylzimosterol, and 14α-methyl-3β, 6α-diol, indicating change in Erg3p (Table 5). Another study reported the occurrence of ERG3 mutation in a C. tropicalis isolate, with substitution of a phenylalanine for serine in portion 258, a residue that is absolutely conserved in this protein (Vincent et al., 2013).
A study conducted in five Chinese hospitals investigated resistance to these drugs in 52 clinical isolates of C. tropicalis (Jiang et al., 2013). Resistance to fluconazole was observed in 34.6% of the isolates, while 40.4% were resistant to itraconazole and only 7.7% to voriconazole. The authors suggest that voriconazole has a more potent activity against the clinical isolates of C. tropicalis than the other drugs tested.
Despite the number of studies involving strains of C. tropicalis resistant to azoles, there are still relatively less studies regarding the resistance of this species to other drugs, such as amphotericin B. This compound is the third most commonly used antifungal in clinical practice (Seneviratne et al., 2016) and is part of the class of polyenes. Its fungicidal activity comes from the ability to selectively bind to the ergosterol of the fungal cell, inducing the formation of pores in the plasma membrane, resulting in intense osmotic imbalance and rapid collapse of the cell (Brajtburg et al., 1990). A recent study reported that the production of reactive oxygen species is also part of the fungicidal mechanism of action of amphotericin B (Forastiero et al., 2013).
Amphotericin B resistance seems to be a rare phenomenon in yeasts, but Woods and Bard in 1974 demonstrated the development of resistance to this drug in two isolates of C. tropicalis obtained from the urine of a patient with pyelonephritis (Woods et al., 1974). A subsequent study of these strains revealed the existence of a mutation in the ergosterol of the cell membrane, exactly at the binding site of amphotericin B (Drutz and Lehrer, 1978). Also in the 1970s, Merz and Sandford reported the isolation of eight strains of C. tropicalis resistant to amphotericin B, obtained from urine of transplanted patients, with the same mutation in ergosterol (Merz and Sandford, 1979).
Reports of isolation of C. tropicalis resistant to this drug have been progressively increasing over the years. In 1988, Powderly et al., reported that the development of resistance to amphotericin B is most observed in patients with some kind of immunosuppression and who frequently use this drug (Powderly et al., 1988). Resistance to this polyene is believed to result from changes in ergosterol, or changes in the plasma membrane itself (Seneviratne et al., 2016).
Lupetti et al. (2002) postulated that resistance to amphotericin B in Candida species generally occurs due to defects in ergosterol biosynthesis and most likely results from mutations in the ERG3 gene. In addition to the ERG3 gene, mutations in ERG6 can generate resistance to polyenes, a phenomenom already described in C. tropicalis (Vandeputte et al., 2007). A study conducted by Forastiero et al. (2013) showed that concomitant mutations in the ERG11 and ERG3 genes lead to multidrug resistance between amphotericin B and azoles (fluconazole and itraconazole) in C. tropicalis.
In addition to amphotericin B, echinocandins have been increasingly used in the treatment of invasive infections, being the first new class of echinocandins that target the fungal cell wall, blocking β-1,3-D-glucan synthase (Perlin, 2007). It has been described that these drugs have an excellent range of action against the main Candida species, including C. tropicalis (Pfaller et al., 2008).
A surveillance study carried out in 2008 by Pfaller et al. (2008) with 5,346 Candida spp. isolates obtained from candidemia infection showed a 12% prevalence for C. tropicalis, and all of them were 100% susceptible for the three echinocandins tested (caspofungin, anidulafungin, and micafungin).
Despite the extensive use of these drugs for more than a decade, the incidence of resistance in the Candida genus remains very low (Beyda et al., 2012). More recent surveillance studies have indicated an incidence of 2.9–3.1% Candida spp. resistance to echinocandins (Arendrup et al., 2010; Castanheira et al., 2010). However, recently Garcia-Effron et al. (2010) reported the isolation of three strains of C. tropicalis resistant to caspofungin obtained from patients with hematological malignancies. A study by Eschenauer et al. (2014) with 185 isolates of C. tropicalis reports 1.4% resistance to caspofungin, anidulafungin, and micafungin.
Another study described the presence of the paradoxical effect (or paradoxical growth) in 15 isolates of C. tropicalis in the presence of high concentrations of echinocandins (Soczo et al., 2007). This phenomenon was first documented by Stevens et al. (2004) for caspofungin in C. albicans, and is defined as fungal growth in the presence of echinocandin concentrations above MIC in broth microdilution susceptibility tests performed according to the guidelines of the Clinical Laboratory Standards Institute (CLSI, previously NCCLS) (Melo et al., 2007).
The target enzyme of echinocandins, called glucan synthetase, possesses at least two subunits: Fks1p (encoded by the FKS1, FKS2 and FKS3 genes) and Rho1p (Table 5; Beyda et al., 2012). Fks1p has a catalytic action and Rho1p is a regulatory protein of several cellular processes, including the biosynthesis of β-1,3-D-glucan (Chen et al., 2011). In general, the reduction of C. tropicalis susceptibility to echinocandins occurs by response to adaptive stress or mutations in the FKS genes.
With regard to adaptive responses, it is known that the fungal cell wall is a dynamic structure that presents a compensatory mechanism to increase the production of one or more components that are eventually inhibited, such as that produced by the action of echinocandins. A study by Chen et al. (2014) investigated the role of calcineurin in C. tropicalis, which is one of the main signaling pathways for the compensatory increase of chitin synthesis in C. albicans. Calcineurin is a phosphatase that regulates numerous stress response processes in fungi, including stress promoted on the cell wall (Cowen, 2009). The study demonstrated that, in fact, calcineurin is responsible for this effect on C. tropicalis against micafungin, since it promotes the thickening of the chitin layer of the cell wall as a function of β-1,3-D-glucan depletion.
In the case of mutations in the FKS1 gene, it is already well-established that substitutions in specific gene regions cause reduced susceptibility to echinocandins, being quite associated with therapeutic failure (Perlin, 2007). Mutations in the FKS1 gene in C. tropicalis were already described (Park et al., 2005). Garcia-Effron et al. (2008) demonstrated that 7.5% (3/40) of clinical isolates of C. tropicalis showed resistance to caspofungin because of amino acid substitutions in Fks1p. Jensen et al. (2013) also performed an investigation alterations in FKS1, with isolates of C. tropicalis from patients with acute lymphoblastic leukemia and found that after 8–8.5 weeks of treatment with caspofungin, two isolates showed resistance to the three echinocandins. Multilocus sequencing of FKS1 revealed progressive development of heterozygosis, and finally the presence of homozygous mutation, leading to substitutions of amino acids S80P and S80S.
The low level of resistance of C. tropicalis to echinocandins and lower side effects, since they target the wall of the fungal cell, make them vital in cases of resistance to fluconazole and amphotericin B, with a broad spectrum of action against C. tropicalis (Pfaller et al., 2008).
Natural Products with Antifungal Properties Against Candida Tropicalis
Several groups have been dedicated to the study of products of natural origin with antifungal action, in order to identify and isolate compounds with effective activity, safety in use and low toxicity against pathogenic fungi (Correia et al., 2016).
There are several parts of the plants used to search for biological activity, with emphasis on antifungal action, such as leaves (Morais-Braga et al., 2016), stem bark (Mendes de Toledo et al., 2015), roots, seeds, and essential oils (Asdadi et al., 2015), that may be isolated used or in synergism with synthetic antifungal drugs, such as fluconazole (Mendes de Toledo et al., 2015).
The use of essential oils of Vitex agnus was used in a study by Asdadi et al. (2015) in clinical strains of Candida isolated from hospital infection. The extraction product was tested against Candida isolates using the principle of disc diffusion and broth macrodilution, according to the standardization of CLSI (Salari et al., 2016). It was observed that for the isolates of C. tropicalis, 10 μl of essential oils produced halos of inhibition of growth of 58 mm, superior to the halos of control drugs such as amphotericin B (8 mm), and fluconazole (21 mm) (Salari et al., 2016).
Salvia rhytidoa Benth., A plant belonging to the family Lamiaceae, typical in Iran was used to evaluate the antifungal activity in several Candida isolates by Salari et al. (2016). A total of 96 clinical isolates of Candida, including 11 C. tropicalis strains were tested using broth microdilution with the methanolic extract, according to CLSI protocols (Salari et al., 2016). It was observed that for C. tropicalis the MIC range had a variation of 100–6.26 μg/ml. Similarly, Siqueira et al. found biological activity using a red propolis alcoholic extract, with an MIC range of 64–32 μg/ml (Siqueira et al., 2015) for this species.
In relation to plants found mainly in Brazilian territory, Correia et al. (2016) carried out an important study with different plants found in the Brazilian Cerrado, a region with an important number of species used in popular medicine, mainly in studies of essential oils with anti-Candida activity (Nordin et al., 2013; Correia et al., 2016).
In a study conducted by Morais-Braga et al. (2016) the interaction of aqueous and hydroethanolic extracts of Psidium brownianum was observed in association with fluconazole. The IC 50 values for fluconazole were 68.10 μg/ml for C. tropicalis CTINCQS 40042 and 41.11 μg/ml for C. tropicalis CTLM 23, obtained by broth microdilution. When in combination with fluconazole, the aqueous and hydroethanolic extracts of P. brownianum showed a significant reduction in IC 50 values, ranging from 37.2–3.10 μg/ml for CTINCM 40042 and 13.66–6.94 μg/ml for CTLM 23.
All these studies involving the evaluation of vegetal products with biological activity, especially against C tropicalis, has reinforced the great importance and necessity of the emergence of alternative and less toxic sources of treatment, alone or in combinations with different antifungal drugs in commercially available.
C. Tropicalis Osmotic Stress Response and Biotechnological Applications
Several virulence attributes are expressed by fungi in response to stress conditions induced by the environment (Brown et al., 2014), and some yeasts can tolerate high salt concentrations, developing physical and genetic mechanisms to neutralize the two mains deleterious effects of osmotic stress, which are toxicity and loss of water and cellular turgidity (Garcia et al., 1997; Beales, 2004).
A study conducted by Rodriguez et al. (1996) reported the gene isolation involved with osmotic adaptation in C. tropicalis, a true homolog of HAL3, called CtHAL3. In fact, C. tropicalis is able to grow in culture medium with more than 10–15% sodium chloride and has been isolated from the hypersaline environment for the first time from Dead Sea samples (Butinar et al., 2005). Bastos et al. (2000) reported the isolation of this yeast from a sample of Amazonian forest enriched with high salt concentration.
Garcia et al. (1997) carried out one of the few studies investigating the mechanisms of osmotic adaptation of C. tropicalis, analyzing ion extrusion. The results showed that Na+/K+-ATPase transporters are activated immediately after exposure to hypersaline environment, promoting rapid efflux of ions and restoring intracellular osmotic equilibrium.
Exposure to sodium chloride (NaCl) leads to high osmotic stress in fungal cells, promoting rapid loss of water that leads to reduced size and loss of cellular turgidity (Kühn and Klipp, 2012).
With regard to C. tropicalis, Garcia et al. (1997) reported that the accumulation of glycerol necessary for the restoration of a normal cellular physiology occurred only after the stationary phase. In addition, they found that there is a preponderant role of efflux pumps in the osmotic adaptation of C. tropicalis to the detriment of the nonionic compensatory mechanisms of water loss and turgor. Besides, the accumulation of intracellular glycerol seems to be less efficient than the activation of the Ion efflux pumps (Garcia et al., 1997).
Therefore, C. tropicalis is considered an osmotolerant yeast, since it can grow well in environments with high osmotic pressure, but this condition is not essential to its survival (Tokuoka, 1993). Such property is often associated with its use in industrial and biotechnological practices.
In the food industry, osmolytic strains of C. tropicalis improve xylitol production (Kwon et al., 2006; Misra et al., 2012). Rao et al. (2006) used C. tropicalis strains in hypersaline solution to produce xylitol from corn fiber and sugarcane bagasse. Another example of industrial application of this species is the production of ethanol from algae (Ra et al., 2015).
C. tropicalis is still widely used in bioremediation processes. Al-Araji et al. (2007) reported the use of this yeast in the commercial recovery of petroleum spillage. In 2011, Farag and Soliman reported the high degradability of crude oil and hydrocarbons by C. tropicalis (Farag and Soliman, 2011). Benard and Tuah (2016) also evaluated this property under conditions simulating sea water. In addition, Yan et al. (2005) demonstrated the high potential for degradation of phenol by C. tropicalis in saline medium. Microorganisms with this capacity are called biosorbents, found to correct pollution processes without causing damage to ecosystems (Leitão et al., 2007).
Halotolerance also provides a longer permanence of C. tropicalis in the coastal environment, allowing greater opportunity for contamination of bathers. Prolonged persistence in the marine environment may also lead to adaptation to high concentrations of other ions and UV light. This whole process can be reflected in genetic alterations that results in selection pressure (Krauke and Sychrova, 2008).
Recently, our group was involved in the investigation of osmotolerance and its relation to virulence expression in vitro with C. tropicalis isolated from the coastal environment. We found that these strains can fully express virulence attributes and may show a high persistence capacity on the coastal environment, because they all tolerated high salt concentration. In addition, they showed high MICs to several antifungal drugs used in current clinical practice, demonstrating that environmental isolates may have pathogenic potential and suggesting that the persistence of yeasts in the sand environment may have leaded to the overexpression of efflux pumps, that may partially explain the reason why C. tropicalis isolates not previously exposed to antifungal drugs had high levels of resistance to azoles and amphotericin B (Zuza-Alves et al., 2016).
Concluding Remarks
In conclusion, this review highlights important aspects of C. tropicalis biology and clinical relevance. This species may be easily identified by classical taxonomy, commercial, proteomics, and molecular methods and no cryptic sibling species has been discovered. This asexual yeast closely related to C. albicans may be considered of high virulence, which can be verified in animal models of superficial and systemics infections, plus its ability to form true hyphae and complex biofilm in vitro, besides the ability to secret proteinases, phospholipases and hemolisins. C. tropicalis is classified as the third or fourth NCAC species more commonly isolated in the clinical practice, while may be the second more frequently isolated Candida species in Latin America and Asia. Several mechanisms of antifungal resistance have been elucidated, including ERG and FKS gene families' mutations and efflux pumps. Some natural products have also been investigated as new potential use for future development of antifungal compounds active against C. tropicalis. This species is considered osmotolerant and this characteristic has been recently demonstrated to influence the expression of virulence factors and primary antifungal resistance. This ability to survive to high salt concentrations is a property that explains C. tropicalis potential use for biotechnological processes, including ethanol production through the fermentation of sea algae. Therefore, for all the factors previously described, C. tropicalis may be indubitably considered one of the most important Candida species.
Author Contributions
DZ and WS prepared the manuscript. GC designed all topics and revised the manuscript. All authors approved the final manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
C. albicans, Candida albicans; C. tropicalis, Candida tropicalis; CLSI, Clinical and Laboratory Standards Institute; MALDI-TOF/MS, Matrix-assisted laser desorption time-of-flight mass; ELISA, Enzyme-linked immunosorbent assay.
References
Adam, B., Baillie, G. S., and Douglas, L. J. (2002). Mixed species biofilms of Candida albicans and Staphylococcus epidermidis. J. Med. Microbiol. 51, 344–349. doi: 10.1099/0022-1317-51-4-344
Adhikary, R., and Joshi, S. (2011). Species distribution and anti-fungal susceptibility of Candidaemia at a multi super-specialty center in Southern India. Indian J. Med. Microbiol. 29, 309–311. doi: 10.4103/0255-0857.83920
Aghamirian, M. R., and Ghiasian, S. A. (2010). Onychomycosis in Iran: epidemiology, causative agents and clinical features. Nihon Ishinkin Gakkai Zasshi 51, 23–29. doi: 10.3314/jjmm.51.23
Al-Anazi, K., and Al-Jasser, A. (2006). Candidaemia in patients with haematological disorders and stem cell transplant. Libyan J. Med. 1, 140–155. doi: 10.3402/ljm.v1i2.4673
Al-Araji, L., Rahman, R. N. Z. R. A., Basri, M., and Salleh, A. B. (2007). Microbial surfactant. Asia Pac. J. Mol. Biol. Biotechnol. 15, 99–105.
Alarco, A.-M., Marcil, A., Chen, J., Suter, B., Thomas, D., and Whiteway, M. (2004). Immune-deficient Drosophila melanogaster: a model for the innate immune response to human fungal pathogens. J. Immunol. 172, 5622–5628. doi: 10.4049/jimmunol.172.9.5622
Albuquerque, P., and Casadevall, A. (2012). Quorum sensing in fungi–a review. Med. Mycol. 50, 337–345. doi: 10.3109/13693786.2011.652201
Alfonso, C., Lopez, M., Arechavala, A., del Perrone, M. C., Guelfand, L., and Bianchi, M. (2010). Presumptive identification of Candida spp. and other clinically important yeasts: usefulness of Brilliance Candida Agar. Rev. Iberoam. Micol. 27, 90–93. doi: 10.1016/j.riam.2010.01.008
Almeida, A. A., Nakamura, S. S., Fiorini, A., Grisolia, A. B., Svidzinski, T. I., and Oliveira, K. M. (2015). Genotypic variability and antifungal susceptibility of Candida tropicalis isolated from patients with candiduria. Rev. Iberoam. Micol. 32, 153–158. doi: 10.1016/j.riam.2014.06.003
Alnuaimi, A. D., O'Brien-Simpson, N. M., Reynolds, E. C., and McCullough, M. J. (2013). Clinical isolates and laboratory reference Candida species and strains have varying abilities to form biofilms. FEMS Yeast Res. 13, 689–699. doi: 10.1111/1567-1364.12068
Angeletti, S., Lo Presti, A., Cella, E., Dicuonzo, G., Crea, F., Palazzotti, B., et al. (2015). Matrix-assisted laser desorption/ionization time of flight mass spectrometry (MALDI-TOF MS) and Bayesian phylogenetic analysis to characterize Candida clinical isolates. J. Microbiol. Methods 119, 214–222. doi: 10.1016/j.mimet.2015.11.003
Anil, S., and Samaranayake, L. (2003). Brief exposure to antimycotics reduces the extracellular phospholipase activity of Candida albicans and Candida tropicalis. Chemotherapy 49, 243–247. doi: 10.1159/000072448
Anil, S., Hashem, M., Vellappally, S., Patil, S., Bandara, H. M., and Samaranayake, L. P. (2014). Sub-inhibitory concentrations of antifungals suppress hemolysin activity of oral Candida albicans and Candida tropicalis isolates from HIV-infected individuals. Mycopathologia 178, 207–215. doi: 10.1007/s11046-014-9802-0
Araújo, D., Henriques, M., and Silva, S. (2017). Portrait of Candida species biofilm regulatory network genes. Trends Microbiol. 25, 62–75. doi: 10.1016/j.tim.2016.09.004
Arbour, M., Epp, E., Hogues, H., Sellam, A., Lacroix, C., Rauceo, et al. (2009). Widespread occurrence of chromosomal aneuploidy following the routine production of Candida albicans mutants. FEMS Yeast Res. 9, 1070–1077. doi: 10.1111/j.1567-1364.2009.00563.x
Arendrup, M. C., Garcia-Effron, G., Lass-Florl, C., Lopez, A. G., Rodriguez-Tudela, J. L., Cuenca-Estrella, M., et al. (2010). Echinocandin susceptibility testing of Candida species: comparison of EUCAST EDef 7.1, CLSI M27-A3, Etest, disk diffusion, and agar dilution methods with RPMI and isosensitest media. Antimicrob. Agents Chemother. 54, 426–439. doi: 10.1128/AAC.01256-09
Asdadi, A., Hamdouch, A., Oukacha, A., Moutaj, R., Gharby, S., Harhar, H., et al. (2015). Study on chemical analysis, antioxidant and in vitro antifungal activities of essential oil from wild Vitex agnus-castus L. seeds growing in area of Argan Tree of Morocco against clinical strains of Candida responsible for nosocomial infections. J. Mycol. Med. 25, e118–e27. doi: 10.1016/j.mycmed.2015.10.005
Aydemir, G., Koc, A. N., and Atalay, M. A. (2016). Evaluation of peptide nucleic acid fluorescent in situ hybridization (PNA FISH) method in the identifi cation of Candida species isolated from blood cultures. Mikrobiyol. Bull. 50, 293–299. doi: 10.5578/mb.22092
Baillie, G. S., and Douglas, L. J. (1999). Role of dimorphism in the development of Candida albicans biofilms. J. Med. Microbiol. 48, 671–679. doi: 10.1099/00222615-48-7-671
Banerjee, M., Uppuluri, P., Zhao, X. R., Carlisle, P. L., Vipulanandan, G., Villar, C. C., et al. (2013). Expression of UME6, a key regulator of Candida albicans hyphal development, enhances biofilm formation via Hgc1- and Sun41-dependent mechanisms. Eukaryot. Cell 12, 224–232. doi: 10.1128/EC.00163-12
Bastos, A. E. R., Moon, D. H., Rossi, A., Trevors, J. T., and Tsai, S. M. (2000). Salt-tolerant phenol-degrading microorganisms isolated from Amazonian soil samples. Arch. Microbiol. 174, 346–352. doi: 10.1007/s002030000216
Basu, S., Gugnani, H. C., Joshi, S., and Gupta, N. (2003). Distribution of Candida species in different clinical sources in Delhi, India, and proteinase and phospholipase activity of Candida albicans isolates. Rev. Iberoam. Micol. 20, 137–140.
Bayegan, S., Majoros, L., Kardos, G., Kemény-Beke, A., Miszti, C., Kovacs, R., et al. (2010). In vivo studies with a Candida tropicalis isolate exhibiting paradoxical growth in vitro in the presence of high concentration of caspofungin. J. Microbiol. 48, 170–173. doi: 10.1007/s12275-010-9221-y
Beales, N. (2004). Adaptation of microorganisms to cold temperatures, weak acid preservatives, low pH, and osmotic stress: a review. Compr. Rev. Food Sci. Food Saf. 3, 1–20. doi: 10.1111/j.1541-4337.2004.tb00057.x
Benard, L. D., and Tuah, P. M. (2016). Biodegradation of sabah light crude oil by locally isolated Candida tropicalis RETL-Cr1 and Pseudomonas aeruginosa BAS-Cr1. Trans. Sci. Technol. 3, 101–106.
Benedetti, V. P., Savi, D. C., Aluizio, R., Adamoski, D., Kava-Cordeiro, V., Galli-Terasawa, L. V., et al. (2016). Analysis of the genetic diversity of Candida isolates obtained from diabetic patients and kidney transplant recipients. Mem. Inst. Oswaldo Cruz 111, 417–422. doi: 10.1590/0074-02760160042
Bennett, D. E., McCreary, C. E., and Coleman, D. C. (1998). Genetic characterization of a phospholipase C gene from Candida albicans: presence of homologous sequences in Candida species other than Candida albicans. Microbiology 144, 55–72. doi: 10.1099/00221287-144-1-55
Berman, J., and Hadany, L. (2012). Does stress induce (para) sex? Implications for Candida albicans evolution. Trends Genet. 28, 197–203. doi: 10.1016/j.tig.2012.01.004
Beyda, N. D., Lewis, R. E., and Garey, K. W. (2012). Echinocandin resistance in Candida species: mechanisms of reduced susceptibility and therapeutic approaches. Ann. Pharmacother. 46, 1086–1096. doi: 10.1345/aph.1R020
Biasoli, M. S., Tosello, M. E., Luque, A. G., and Magaro, H. M. (2010). Adherence, colonization and dissemination of Candida dubliniensis and other Candida species. Med. Mycol.. 48, 291–297. doi: 10.1080/13693780903114942
Bizerra, F. C., Nakamura, C. V., de Poersch, C., Estivalet Svidzinski, T. I., Borsato Quesada, R. M., Goldenberg, S., et al. (2008). Characteristics of biofilm formation by Candida tropicalis and antifungal resistance. FEMS Yeast Res. 8, 442–450. doi: 10.1111/j.1567-1364.2007.00347.x
Blandin, G., Ozier-Kalogeropoulos, O., Wincker, P., Artiguenave, F., and Dujon, B. (2000). Genomic exploration of the hemiascomycetous yeasts: 16. Candida tropicalis. FEBS Lett. 487, 91–94. doi: 10.1016/S0014-5793(00)02287-0
Bouchonville, K., Forche, A., Tang, K. E., Selmecki, A., and Berman, J. (2009). Aneuploid chromosomes are highly unstable during DNA transformation of Candida albicans. Eukaryot. Cell 8, 1554–1566. doi: 10.1128/EC.00209-09
Bowman, P. I., and Ahearn, D. G. (1976). Evaluation of commercial systems for the identification of clinical yeast isolates. J. Clin. Microbiol. 4, 49–53.
Brajtburg, J., Powderly, W. G., Kobayashi, G. S., and Medoff, G. (1990). Amphotericin B: current understanding of mechanisms of action. Antimicrob. Agents Chemother. 34, 183–188.
Brown, A. J., Budge, S., Kaloriti, D., Tillmann, A., Jacobsen, M. D., Yin, Z., et al. (2014). Stress adaptation in a pathogenic fungus. J. Exp. Biol. 217, 144–155. doi: 10.1242/jeb.088930
Butinar, L., Santos, S., Spencer-Martins, I., Oren, A., and Gunde-Cimerman, N. (2005). Yeast diversity in hypersaline habitats. FEMS Microbiol. Lett. 244, 229–234. doi: 10.1016/j.femsle.2005.01.043
Butler, G., Rasmussen, M. D., Lin, M. F., Santos, M. A., Sakthikumar, S., Munro, C. A., et al. (2009). Evolution of pathogenicity and sexual reproduction in eight Candida genomes. Nature 459, 657–662. doi: 10.1038/nature08064
Cain, C. W., Lohse, M. B., Homann, O. R., Sil, A., and Johnson, A. D. (2012). A conserved transcriptional regulator governs fungal morphology in widely diverged species. Genetics 190, 511–521. doi: 10.1534/genetics.111.134080
Calderaro, A., Martinelli, M., Motta, F., Larini, S., Arcangeletti, M. C., Medici, M. C., et al. (2014). Comparison of peptide nucleic acid fluorescence in situ hybridization assays with culture-based matrix-assisted laser desorption/ionization-time of flight mass spectrometry for the identification of bacteria and yeasts from blood cultures and cerebrospinal fluid cultures. Clin. Microbiol. Infect. 20, 468–475. doi: 10.1111/1469-0691.12490
Calderone, R. A., and Gow, N. A. (2002). “Host recognition by Candida species,” in Candida and Candidiasis, ed R. A. Calderone (Washington, DC: ASM Press), 67–86.
Calderon, J., Zavrel, M., Ragni, E., Fonzi, W. A., Rupp, S., and Popolo, L. (2010). PHR1, a pH-regulated gene of Candida albicans encoding a glucan-remodelling enzyme, is required for adhesion and invasion. Microbiology 156, 2484–2494. doi: 10.1099/mic.0.038000-0
Cambuim, I. I., Macedo, D. P., Delgado, M., de Lima, K. M., Mendes, G. P., Souza-Motta, C. M., et al. (2011). Clinical and mycological evaluation of onychomycosis among Brazilian HIV/AIDS patients. Rev. Soc. Bras. Med. Trop. 44, 40–42. doi: 10.1590/S0037-86822011000100010
Cannon, R. D., and Chaffin, W. L. (2001). Colonization is a crucial factor in oral candidiasis. J. Dent. Educ. 65, 785–778.
Castanheira, M., Woosley, L. N., Diekema, D. J., Messer, S. A., Jones, R. N., and Pfaller, M. A. (2010). Low prevalence of fks1 hot spot 1 mutations in a worldwide collection of Candida strains. Antimicrob. Agents Chemother. 54, 2655–2659. doi: 10.1128/AAC.01711-09
Castellani, A. (1912). Observations on the fungi found in tropical bronchomycosis. Lancet 179, 13–15. doi: 10.1016/S0140-6736(00)51698-5
Cauchie, M., Desmet, S., and Lagrou, K. (in press). Candida and its dual lifestyle as a commensal a pathogen. Res. Microbiol. doi: 10.1016/j.resmic.2017.02.005
Chakrabarti, A., Chatterjee, S. S., Rao, K. L., Zameer, M. M., Shivaprakash, M. R., Singhi, S., et al. (2009). Recent experience with fungaemia: change in species distribution and azole resistance. Scand. J. Infect. Dis. 41, 275–284. doi: 10.1080/00365540902777105
Champion, O. L., Wagley, S., and Titball, R. W. (2016). Galleria mellonella as a model host for microbiological and toxin research. Virulence 7, 840–845. doi: 10.1080/21505594.2016.1203486
Chandra, J., Kuhn, D. M., Mukherjee, P. K., Hoyer, L. L., McCormick, T., and Ghannoum, M. A. (2001). Biofilm formation by the fungal pathogen Candida albicans: development, architecture, and drug resistance. J. Bacteriol. 183, 5385–5394. doi: 10.1128/JB.183.18.5385-5394.2001
Chang, F., Drubin, D., and Nurse, P. (1997). cdc12p, a protein required for cytokinesis in fission yeast, is a component of the cell division ring and interacts with profilin. J. Cell Biol. 137, 169–182. doi: 10.1083/jcb.137.1.169
Chang, T. P., Ho, M. W., Yang, Y. L., Lo, P. C., Lin, P. S., Wang, A. H., et al. (2013). Distribution and drug susceptibilities of Candida species causing candidemia from a medical center in central Taiwan. J. Infect. Chemother. 19, 1065–1071. doi: 10.1007/s10156-013-0623-8
Chao, Q. T., Lee, T. F., Teng, S. H., Peng, L. Y., Chen, P. H., Teng, L. J., et al. (2014). Comparison of the accuracy of two conventional phenotypic methods and two MALDI-TOF MS systems with that of DNA sequencing analysis for correctly identifying clinically encountered yeasts. PLoS ONE 9:e109376. doi: 10.1371/journal.pone.0109376
Chaves, G. M., Diniz, M. G., da Silva-Rocha, W. P., de Souza, L. B., Gondim, L. A., Ferreira, M. A., et al. (2013). Species distribution and virulence factors of Candida spp. isolated from the oral cavity of kidney transplant recipients in Brazil. Mycopathologia 175, 255–263. doi: 10.1007/s11046-013-9640-5
Chen, K.-W., Chen, Y.-C., Lin, Y.-H., Chou, H.-H., and Li, S.-Y. (2009). The molecular epidemiology of serial Candida tropicalis isolates from ICU patients as revealed by multilocus sequence typing and pulsed-field gel electrophoresis. Infect. Genet. Evol. 9, 912–920. doi: 10.1016/j.meegid.2009.06.011
Chen, S. C., Slavin, M. A., and Sorrell, T. C. (2011). Echinocandin antifungal drugs in fungal infections: a comparison. Drugs 71, 11–41. doi: 10.2165/11585270-000000000-00000
Chen, Y. L., Yu, S. J., Huang, H. Y., Chang, Y. L., Lehman, V. N., Silao, F. G., et al. (2014). Calcineurin controls hyphal growth, virulence, and drug tolerance of Candida tropicalis. Eukaryot. Cell 13, 844–854. doi: 10.1128/EC.00302-13
Choi, M. J., Won, E. J., Shin, J. H., Kim, S. H., Lee, W. G., Kim, M. N., et al. (2016). Resistance mechanisms and clinical features of fluconazole-nonsusceptible Candida tropicalis isolates compared with fluconazole-less-susceptible isolates. Antimicrob. Agents Chemother. 60, 3653–3661. doi: 10.1128/AAC.02652-15
Colombo, A. L., Nucci, M., Park, B. J., Nouer, S. A., Arthington-Skaggs, B., da Matta, D. A., et al. (2006). Epidemiology of candidemia in Brazil: a nationwide sentinel surveillance of candidemia in eleven medical centers. J. Clin. Microbiol. 44, 2816–28123. doi: 10.1128/JCM.00773-06
Correia, A. F., Silveira, D., Fonseca-Bazzo, Y. M., Magalhães, P. O., Fagg, C. W., da Silva, E. C., et al. (2016). Activity of crude extracts from Brazilian cerrado plants against clinically relevant Candida species. BMC Complement. Altern. Med. 16:203. doi: 10.1186/s12906-016-1164-3
Costa, C. R., Passos, X. S., de Souza, L. K., de Lucena, P. A., de Fernandes, O. F., and da Silva, M. R. (2010). Differences in exoenzyme production and adherence ability of Candida spp. isolates from catheter, blood and oral cavity. Rev. Inst. Med. Trop. Sao Paulo 52, 139–143. doi: 10.1590/S0036-46652010000300005
Cowen, L. E. (2009). Hsp90 orchestrates stress response signaling governing fungal drug resistance. PLoS Pathog. 5:e1000471. doi: 10.1371/journal.ppat.1000471
da Costa, K. R., Ferreira, J. C., Lavrador, M. A., Baruffi, M. D., and Candido, R. C. (2012). Virulence attributes and genetic variability of oral Candida albicans and Candida tropicalis isolates. Mycoses 55, e97–e105. doi: 10.1111/j.1439-0507.2011.02125.x
da Silva-Rocha, W. P., Lemos, V. L., Svidizisnki, T. I., Milan, E. P., and Chaves, G. M. (2014). Candida species distribution, genotyping and virulence factors of Candida albicans isolated from the oral cavity of kidney transplant recipients of two geographic regions of Brazil. BMC Oral Health 14:20. doi: 10.1186/1472-6831-14-20
de Campos Rasteiro, V. M., da Costa, A. C. B. P., Araújo, C. F., De Barros, P. P., Rossoni, R. D., Anbinder, A. L., et al. (2014). Essential oil of Melaleuca alternifolia for the treatment of oral candidiasis induced in an immunosuppressed mouse model. BMC Complement. Altern. Med. 14:1. doi: 10.1186/1472-6882-14-489
de Medeiros, M. A. P., de Melo, A. P. V., Gonçalves, S. S., Milan, E. P., and Chaves, G. M. (2014). Genetic relatedness among vaginal and anal isolates of Candida albicans from women with vulvovaginal candidiasis in north-east Brazil. J. Med. Microbiol. 63, 1436–1445. doi: 10.1099/jmm.0.076604-0
de Melo Riceto, É. B., de Paula Menezes, R., Penatti, M. P. A., and dos Santos Pedroso, R. (2015). Enzymatic and hemolytic activity in different Candida species. Rev. Iberoam. Micol. 32, 79–82. doi: 10.1016/j.riam.2013.11.003
de Souza, P. C., Morey, A. T., Castanheira, G. M., Bocate, K. P., Panagio, L. A., Ito, F. A., et al. (2015). Tenebrio molitor (Coleoptera: Tenebrionidae) as an alternative host to study fungal infections. J. Microbiol. Methods 118, 182–186. doi: 10.1016/j.mimet.2015.10.004
Deorukhkar, S. C., Saini, S., and Mathew, S. (2014). Virulence factors contributing to pathogenicity of Candida tropicalis and its antifungal susceptibility profile. Int. J. Microbiol. 2014:456878. doi: 10.1155/2014/456878
Dias, L. B., de Souza Carvalho Melhem, M., Szeszs, M. W., Filho, J. M., and Hahn, R. C. (2011). Vulvovaginal candidiasis in Mato Grosso, Brazil: pregnancy status, causative species and drugs tests. Braz. J. Microbiol. 42, 1300–1307. doi: 10.1590/S1517-83822011000400009
Diezmann, S., Cox, C. J., Schonian, G., Vilgalys, R. J., and Mitchell, T. G. (2004). Phylogeny and evolution of medical species of Candida and related taxa: a multigenic analysis. J. Clin. Microbiol. 42, 5624–5635. doi: 10.1128/JCM.42.12.5624-5635.2004
Doi, A. M., Pignatari, A. C., Edmond, M. B., Marra, A. R., Camargo, L. F., Siqueira, R. A., et al. (2016). Epidemiology and microbiologic characterization of nosocomial candidemia from a Brazilian national surveillance program. PLoS ONE 11:e0146909. doi: 10.1371/journal.pone.0146909
Doi, M., Homma, M., Chindamporn, A., and Tanaka, K. (1992). Estimation of chromosome number and size by pulsed-field gel electrophoresis (PFGE) in medically important Candida species. J. Gen. Microbiol. 138, 2243–2251. doi: 10.1099/00221287-138-10-2243
Donahue, S. P., Greven, C. M., Zuravleff, J. J., Eller, A. W., Nguyen, M. H., Peacock, J. E., et al. (1994). Intraocular candidiasis in patients with candidemia: clinical implications derived from a prospective multicenter study. Ophthalmology 101, 1302–1309. doi: 10.1016/S0161-6420(94)31175-4
Donlan, R. M., and Costerton, J. W. (2002). Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15, 167–193. doi: 10.1128/CMR.15.2.167-193.2002
Douglas, L. J. (2002). Medical importance of biofilms in Candida infections. Rev. Iberoam. Micol. 19, 139–143.
Douglas, L. J. (2003). Candida biofilms and their role in infection. Trends Microbiol. 11, 30–36. doi: 10.1016/S0966-842X(02)00002-1
Dozier, C. C., Tarantola, R. M., Jiramongkolchai, K., and Donahue, S. P. (2011). Fungal eye disease at a tertiary care center: the utility of routine inpatient consultation. Ophthalmology 118, 1671–1676. doi: 10.1016/j.ophtha.2011.01.038
Drutz, D. J., and Lehrer, R. I. (1978). Development of amphotericin B-resistant Candida tropicalis in a patient with defective leukocyte function. Am. J. Med. Sci. 276, 77–92.
Eddouzi, J., Parker, J. E., Vale-Silva, L. A., Coste, A., Ischer, F., Kelly, S., et al. (2013). Molecular mechanisms of drug resistance in clinical Candida species isolated from Tunisian hospitals. Antimicrob. Agents Chemother. 57, 3182–3193. doi: 10.1128/AAC.00555-13
Eggimann, P., Garbino, J., and Pittet, D. (2003). Epidemiology of Candida species infections in critically ill non-immunosuppressed patients. Lancet Infect. Dis. 3, 685–702. doi: 10.1016/S1473-3099(03)00801-6
Eschenauer, G. A., Nguyen, M. H., Shoham, S., Vazquez, J. A., Morris, A. J., Pasculle, W. A., et al. (2014). Real-world experience with echinocandin MICs against Candida species in a multicenter study of hospitals that routinely perform susceptibility testing of bloodstream isolates. Antimicrob. Agents Chemother. 58, 1897–1906. doi: 10.1128/AAC.02163-13
Fanning, S., and Mitchell, A. P. (2012). Fungal biofilms. PLoS Pathog. 8:e1002585. doi: 10.1371/journal.ppat.1002585
Farag, S., and Soliman, N. A. (2011). Biodegradation of crude petroleum oil and environmental pollutants by Candida tropicalis strain. Braz. Arch. Biol. Technol. 54, 821–830. doi: 10.1590/S1516-89132011000400023
Favero, D., Franca, E. J., Furlaneto-Maia, L., Quesada, R. M., and Furlaneto, M. C. (2011). Production of haemolytic factor by clinical isolates of Candida tropicalis. Mycoses 54, e816–e820. doi: 10.1111/j.1439-0507.2011.02035.x
Favero, D., Furlaneto-Maia, L., Franca, E. J., Goes, H. P., and Furlaneto, M. C. (2014). Hemolytic factor production by clinical isolates of Candida species. Curr. Microbiol. 68, 161–166. doi: 10.1007/s00284-013-0459-6
Fidel, P. L., Cutright, J. L., Tait, L., and Sobel, J. D. (1996). A murine model of Candida glabrata vaginitis. J. Infect. Dis. 173, 425–431. doi: 10.1093/infdis/173.2.425
Fidel, P., Cutright, J. L., and Sobel, J. D. (1997). Efficacy of D0870 treatment of experimental Candida vaginitis. Antimicrob. Agents Chemother. 41, 1455–1459.
Figueiredo, V. T., de Assis Santos, D., Resende, M. A., and Hamdan, J. S. (2007). Identification and in vitro antifungal susceptibility testing of 200 clinical isolates of Candida spp. responsible for fingernail infections. Mycopathologia 164, 27–33. doi: 10.1007/s11046-007-9027-6
Finkel, J. S., and Mitchell, A. P. (2011). Genetic control of Candida albicans biofilm development. Nat. Rev. Microbiol. 9, 109–118. doi: 10.1038/nrmicro2475
Fitzpatrick, D. A., O'Gaora, P., Byrne, K. P., and Butler, G. (2010). Analysis of gene evolution and metabolic pathways using the Candida Gene Order Browser. BMC Genomics 11:290. doi: 10.1186/1471-2164-11-290
Fong, P. H., Chan, H. L., Lee, Y. S., and Wong, H. B. (1988). Acute disseminated cutaneous candidiasis. Ann. Acad. Med. Singap. 17, 551–553.
Forastiero, A., Mesa-Arango, A. C., Alastruey-Izquierdo, A., Alcazar-Fuoli, L., Bernal-Martinez, L., Pelaez, T., et al. (2013). Candida tropicalis antifungal cross-resistance is related to different azole target (Erg11p) modifications. Antimicrob. Agents Chemother. 57, 4769–4781. doi: 10.1128/AAC.00477-13
Fuchs, B. B., Li, Y., Li, D., Johnston, T., Hendricks, G., Li, G., et al. (2016). Micafungin elicits an immunomodulatory effect in Galleria mellonella and mice. Mycopathologia 181, 17–25. doi: 10.1007/s11046-015-9940-z
Galan-Ladero, M. A., Blanco-Blanco, M. T., Hurtado, C., Perez-Giraldo, C., Blanco, M. T., and Gomez-Garcia, A. C. (2013). Determination of biofilm production by Candida tropicalis isolated from hospitalized patients and its relation to cellular surface hydrophobicity, plastic adherence and filamentation ability. Yeast 30, 331–339. doi: 10.1002/yea.2965
Galan-Ladero, M., Blanco, M., Sacristán, B., Fernández-Calderón, M., Pérez-Giraldo, C., and Gomez-Garcia, A. (2010). Enzymatic activities of Candida tropicalis isolated from hospitalized patients. Med. Mycol. 48, 207–210. doi: 10.3109/13693780902801242
Garcia, M. J., Rios, G., Ali, R., Bellés, J. M., and Serrano, R. (1997). Comparative physiology of salt tolerance in Candida tropicalis and Saccharomyces cerevisiae. Microbiology 143, 1125–1131. doi: 10.1099/00221287-143-4-1125
Garcia-Effron, G., Chua, D. J., Tomada, J. R., DiPersio, J., Perlin, D. S., Ghannoum, M., et al. (2010). Novel FKS mutations associated with echinocandin resistance in Candida species. Antimicrob. Agents Chemother. 54, 2225–2227. doi: 10.1128/AAC.00998-09
Garcia-Effron, G., Kontoyiannis, D. P., Lewis, R. E., and Perlin, D. S. (2008). Caspofungin-resistant Candida tropicalis strains causing breakthrough fungemia in patients at high risk for hematologic malignancies. Antimicrob. Agents Chemother. 52, 4181–4183. doi: 10.1128/AAC.00802-08
Garczewska, B., Jarzynka, S., Kus, J., Skorupa, W., and Augustynowicz-Kopec, E. (2016). Fungal infection of cystic fibrosis patients—single center experience. Pneumonol. Alergol. Pol. 84, 151–159. doi: 10.5603/PiAP.2016.0017
Garvey, E., Hoekstra, W., Schotzinger, R., Sobel, J., Lilly, E., and Fidel, P. (2015). Efficacy of the clinical agent VT-1161 against fluconazole-sensitive and-resistant Candida albicans in a murine model of vaginal candidiasis. Antimicrob. Agents Chemother. 59, 5567–5573. doi: 10.1128/AAC.00185-15
Ghannoum, M. A. (2000). Potential role of phospholipases in virulence and fungal pathogenesis. Clin. Microbiol. Rev. 13, 122–143. doi: 10.1128/CMR.13.1.122-143.2000
Giolo, M. P., and Svidzinski, T. I. E. (2010). Fisiopatogenia, epidemiologia e diagnóstico laboratorial da candidemia. J. Bras. Patol. Med. Lab. 46, 225–234. doi: 10.1590/S1676-24442010000300009
Giri, S., and Kindo, A. J. (2012). A review of Candida species causing blood stream infection. Indian J. Med. Microbiol. 30, 270–278. doi: 10.4103/0255-0857.99484
Gonzalez-Novo, A., Correa-Bordes, J., Labrador, L., Sanchez, M., Vazquez de Aldana, C. R., and Jimenez, J. (2008). Sep7 is essential to modify septin ring dynamics and inhibit cell separation during Candida albicans hyphal growth. Mol. Biol. Cell. 19, 1509–1518. doi: 10.1091/mbc.E07-09-0876
Gorton, R. L., Ramnarain, P., Barker, K., Stone, N., Rattenbury, S., McHugh, T. D., et al. (2014). Comparative analysis of Gram's stain, PNA-FISH and Sepsityper with MALDI-TOF MS for the identification of yeast direct from positive blood cultures. Mycoses 57, 592–601. doi: 10.1111/myc.12205
Grant, M. L., Parajuli, S., Deleon-Gonsalves, R., Potula, R., and Truant, A. L. (2016). Comparative evaluation of the BD phoenix yeast ID panel and remel RapID yeast plus system for yeast identification. Can. J. Infect. Dis. Med. Microbiol. 2016:4094932. doi: 10.1155/2016/4094932
Guinea, J., Zaragoza, O., Escribano, P., Martin-Mazuelos, E., Peman, J., Sanchez-Reus, F., et al. (2014). Molecular identification and antifungal susceptibility of yeast isolates causing fungemia collected in a population-based study in Spain in 2010 and 2011. Antimicrob. Agents Chemother. 58, 1529–1537. doi: 10.1128/AAC.02155-13
Gündeş, S., Gulenc, S., and Bingol, R. (2001). Comparative performance of Fungichrom I, Candifast and API 20C Aux systems in the identification of clinically significant yeasts. J. Med. Microbiol. 50, 1105–1110. doi: 10.1099/0022-1317-50-12-1105
Gustin, M. C., Albertyn, J., Alexander, M., and Davenport, K. (1998). MAP kinase pathways in the yeast Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 62, 1264–1300.
Hall, L., Le Febre, K. M., Deml, S. M., Wohlfiel, S. L., and Wengenack, N. L. (2012). Evaluation of the yeast traffic light PNA FISH probes for identification of Candida species from positive blood cultures. J. Clin. Microbiol. 50, 1446–1448. doi: 10.1128/JCM.06148-11
Hamamoto, H., Kurokawa, K., Kaito, C., Kamura, K., Razanajatovo, I. M., Kusuhara, H., et al. (2004). Quantitative evaluation of the therapeutic effects of antibiotics using silkworms infected with human pathogenic microorganisms. Antimicrob. Agents Chemother. 48, 774–779. doi: 10.1128/AAC.48.3.774-779.2004
Hawser, S. P., and Douglas, L. J. (1995). Resistance of Candida albicans biofilms to antifungal agents in vitro. Antimicrob. Agents Chemother. 39, 2128–2131. doi: 10.1128/AAC.39.9.2128
Hoover, C. I., Jantapour, M. J., Newport, G., Agabian, N., and Fisher, S. J. (1998). Cloning and regulated expression of the Candida albicans phospholipase B (PLB1) gene. FEMS Microbiol. Lett. 167, 163–169. doi: 10.1111/j.1574-6968.1998.tb13223.x
Hoyer, L. L., Fundyga, R., Hecht, J. E., Kapteyn, J. C., Klis, F. M., and Arnold, J. (2001). Characterization of agglutinin-like sequence genes from non-albicans Candida and phylogenetic analysis of the ALS family. Genetics 157, 1555–1567.
Hube, B., and Naglik, J. (2001). Candida albicans proteinases: resolving the mystery of a gene family. Microbiology 147(Pt 8), 1997–2005. doi: 10.1099/00221287-147-8-1997
Ishii, M., Matsumoto, Y., and Sekimizu, K. (2015). Usefulness of silkworm as a model animal for understanding the molecular mechanisms of fungal pathogenicity. Drug Discov. Ther. 9, 234–237. doi: 10.5582/ddt.2015.01052
Jacobs, P. H., and Nall, L. (1990). Antifungal Drug Therapy: A Complete Guide for the Practitioner. New York, NY: CRC Press.
Jain, M., Shah, R., Chandolia, B., Mathur, A., Chauhan, Y., Chawda, J., et al. (2016). The oral carriage of Candida in oral cancer patients of indian origin undergoing radiotherapy and/or chemotherapy. J. Clin. Diagn. Res. 10, ZC17–ZC20. doi: 10.7860/JCDR/2016/15702.7180
Jensen, R. H., Johansen, H. K., and Arendrup, M. C. (2013). Stepwise development of a homozygous S80P substitution in Fks1p, conferring echinocandin resistance in Candida tropicalis. Antimicrob. Agents Chemother. 57, 614–617. doi: 10.1128/AAC.01193-12
Jiang, C., Dong, D., Yu, B., Cai, G., Wang, X., Ji, Y., et al. (2013). Mechanisms of azole resistance in 52 clinical isolates of Candida tropicalis in China. J. Antimicrob. Chemother. 68, 778–785. doi: 10.1093/jac/dks481
Jiang, C., Li, Z., Zhang, L., Tian, Y., Dong, D., and Peng, Y. (2016). Significance of hyphae formation in virulence of Candida tropicalis and transcriptomic analysis of hyphal cells. Microbiol. Res. 192, 65–72. doi: 10.1016/j.micres.2016.06.003
Joseph-Horne, T., and Hollomon, D. W. (1997). Molecular mechanisms of azole resistance in fungi. FEMS Microbiol. Lett. 149, 141–149. doi: 10.1111/j.1574-6968.1997.tb10321.x
Kali, A., Charles, M. P., Noyal, M. J., Sivaraman, U., Kumar, S., and Easow, J. M. (2013). Prevalence of Candida co-infection in patients with pulmonary tuberculosis. Australas. Med. J. 6, 387–391. doi: 10.4066/AMJ.2013.1709
Kanagal, D., Vineeth, V., Kundapur, R., Shetty, H., and Rajesh, A. (2014). Prevalence of vaginal candidiasis in pregnancy among coastal south Indian women. J. Womens Health. Issues Care 3:6. doi: 10.4172/2325-9795.1000168
Keceli, S. A., Dundar, D., and Tamer, G. S. (2016). Comparison of vitek matrix-assisted laser desorption/ionization time-of-flight mass spectrometry versus conventional methods in Candida identification. Mycopathologia 181, 67–73. doi: 10.1007/s11046-015-9944-8
Kelly, S. L., Arnoldi, A., and Kelly, D. E. (1993). Molecular genetic analysis of azole antifungal mode of action. Biochem. Soc. Trans. 21, 1034–1038. doi: 10.1042/bst0211034
Kim, D. M., Suh, M. K., and Ha, G. Y. (2013). Onychomycosis in children: an experience of 59 cases. Ann. Dermatol. 25, 327–334. doi: 10.5021/ad.2013.25.3.327
Kirk, P., Cannon, P., David, J., and Stalpers, J. (2001). Ainsworth & Bisby's Dicitionary of the Fungi. Wallingford: Ed CAB International.
Klotz, S. A., Drutz, D. J., Harrison, J. L., and Huppert, M. (1983). Adherence and penetration of vascular endothelium by Candida yeasts. Infect. Immun. 42, 374–384.
Koga-Ito, C. Y., Komiyama, E. Y., de Paiva Martins, C. A., Vasconcellos, T. C., Cardoso Jorge, A. O., Carvalho, Y. R., et al. (2011). Experimental systemic virulence of oral Candida dubliniensis isolates in comparison with Candida albicans, Candida tropicalis and Candida krusei. Mycoses 54, e278–e85. doi: 10.1111/j.1439-0507.2010.01899.x
Kontoyiannis, D. P., Vaziri, I., Hanna, H. A., Boktour, M., Thornby, J., Hachem, R., et al. (2001). Risk factors for Candida tropicalis fungemia in patients with cancer. Clin. Infect. Dis. 33, 1676–1681. doi: 10.1086/323812
Kothavade, R. J., Kura, M. M., Valand, A. G., and Panthaki, M. H. (2010). Candida tropicalis: its prevalence, pathogenicity and increasing resistance to fluconazole. J. Med. Microbiol. 59(Pt 8), 873–880. doi: 10.1099/jmm.0.013227-0
Krauke, Y., and Sychrova, H. (2008). Functional comparison of plasma-membrane Na+/H+ antiporters from two pathogenic Candida species. BMC Microbiol. 8:80. doi: 10.1186/1471-2180-8-80
Krcmery, V. Jr., Mrazova, M., Kunova, A., Grey, E., Mardiak, J., Jurga, L., et al. (1999). Nosocomial Candidaemias due to species other than Candida albicans in cancer patients. Support. Care Cancer 7, 428–431. doi: 10.1007/s005200050304
Kühn, C., and Klipp, E. (2012). Zooming in on yeast osmoadaptation. Adv. Sys. Biol. 739, 293–310. doi: 10.1007/978-1-4419-7210-1_17
Kumamoto, C. A., and Vinces, M. D. (2005). Contributions of hyphae and hypha-co-regulated genes to Candida albicans virulence. Cell. Microbiol. 7, 1546–1554. doi: 10.1111/j.1462-5822.2005.00616.x
Kurtzman, C., Fell, J. W., and Boekhout, T. (2011). The Yeasts: A Taxonomic Study. Amsterdam: Elsevier.
Kwon, S. G., Park, S. W., and Oh, D. K. (2006). Increase of xylitol productivity by cell-recycle fermentation of Candida tropicalis using submerged membrane bioreactor. J. Biosci. Bioeng. 101, 13–18. doi: 10.1263/jbb.101.13
Lackey, E., Vipulanandan, G., Childers, D. S., and Kadosh, D. (2013). Comparative evolution of morphological regulatory functions in Candida species. Eukaryot. Cell 12, 1356–1368. doi: 10.1128/EC.00164-13
Legrand, M., Forche, A., Selmecki, A., Chan, C., Kirkpatrick, D. T., and Berman, J. (2008). Haplotype mapping of a diploid non-meiotic organism using existing and induced aneuploidies. PLoS Genet. 4:e1. doi: 10.1371/journal.pgen.0040001
Leitão, A. L., Duarte, M. P., and Oliveira, J. S. (2007). Degradation of phenol by a halotolerant strain of Penicillium chrysogenum. Int. Biodeterior. Biodegrad. 59, 220–225. doi: 10.1016/j.ibiod.2006.09.009
Li, L., Zhang, C., and Konopka, J. B. (2012). A Candida albicans temperature-sensitive cdc12-6 mutant identifies roles for septins in selection of sites of germ tube formation and hyphal morphogenesis. Eukaryot. Cell 11, 1210–1218. doi: 10.1128/EC.00216-12
Liu, W., Tan, J., Sun, J., Xu, Z., Li, M., Yang, Q., et al. (2014). Invasive candidiasis in intensive care units in China: in vitro antifungal susceptibility in the China-SCAN study. J. Antimicrob. Chemother. 69, 162–167. doi: 10.1093/jac/dkt330
Luo, G., Samaranayake, L. P., and Yau, J. Y. (2001). Candida species exhibit differential in vitro hemolytic activities. J. Clin. Microbiol. 39, 2971–2974. doi: 10.1128/JCM.39.8.2971-2974.2001
Luo, G., Samaranayake, L. P., Cheung, B. P., and Tang, G. (2004). Reverse transcriptase polymerase chain reaction (RT-PCR) detection of HLP gene expression in Candida glabrata and its possible role in in vitro haemolysin production. APMIS 112, 283–290. doi: 10.1111/j.1600-0463.2004.apm11204-0509.x
Lupetti, A., Danesi, R., Campa, M., Del Tacca, M., and Kelly, S. (2002). Molecular basis of resistance to azole antifungals. Trends Mol. Med. 8, 76–81. doi: 10.1016/S1471-4914(02)02280-3
Lyon, J. P., and de Resende, M. A. (2006). Correlation between adhesion, enzyme production, and susceptibility to fluconazole in Candida albicans obtained from denture wearers. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. Endod. 102, 632–638. doi: 10.1016/j.tripleo.2005.12.015
Macdonald, F., and Odds, F. C. (1983). Virulence for mice of a proteinase-secreting strain of Candida albicans and a proteinase-deficient mutant. J. Gen. Microbiol. 129, 431–438. doi: 10.1099/00221287-129-2-431
Maiden, M. C., Bygraves, J. A., Feil, E., Morelli, G., Russell, J. E., Urwin, R., et al. (1998). Multilocus sequence typing: a portable approach to the identification of clones within populations of pathogenic microorganisms. Proc. Natl. Acad. Sci. U.S.A. 95, 3140–3145. doi: 10.1073/pnas.95.6.3140
Manastir, L., Ergon, M. C., and Yucesoy, M. (2011). Investigation of mutations in Erg11 gene of fluconazole resistant Candida albicans isolates from Turkish hospitals. Mycoses 54, 99–104. doi: 10.1111/j.1439-0507.2009.01766.x
Mancera, E., Porman, A. M., Cuomo, C. A., Bennett, R. J., and Johnson, A. D. (2015). Finding a missing gene: EFG1 regulates morphogenesis in Candida tropicalis. G3 (Bethesda). 5, 849–856. doi: 10.1534/g3.115.017566
Manns, J. M., Mosser, D. M., and Buckley, H. R. (1994). Production of a hemolytic factor by Candida albicans. Infect. Immun. 62, 5154–5156.
Manzano-Gayosso, P., Mendez-Tovar, L. J., Arenas, R., Hernandez-Hernandez, F., Millan-Chiu, B., Torres-Rodriguez, J. M., et al. (2011). Onychomycosis-causing yeasts in four Mexican dermatology centers and their antifungal susceptibility to azolic compounds. Rev. Iberoam. Micol. 28, 32–35. doi: 10.1016/j.riam.2010.11.002
Marcos, J. Y., and Pincus, D. H. (2013). Fungal diagnostics: review of commercially available methods. Methods Mol. Biol. 968, 25–54. doi: 10.1007/978-1-62703-257-5_2
Marcos-Zambrano, L. J., Escribano, P., Bouza, E., and Guinea, J. (2014). Production of biofilm by Candida and non-Candida spp. isolates causing fungemia: comparison of biomass production and metabolic activity and development of cut-off points. Int. J. Med. Microbiol. 304, 1192–1198. doi: 10.1016/j.ijmm.2014.08.012
Marie, C., and White, T. C. (2009). Genetic basis of antifungal drug resistance. Curr. Fungal Infect. Rep. 3, 163–169. doi: 10.1007/s12281-009-0021-y
Mariné, M., Pastor, F. J., and Guarro, J. (2010). Efficacy of posaconazole in a murine disseminated infection by Candida tropicalis. Antimicrob. Agents Chemother. 54, 530–532. doi: 10.1128/AAC.01136-09
Marol, S., and Yücesoy, M. (2008). Molecular epidemiology of Candida species isolated from clinical specimens of intensive care unit patients. Mycoses 51, 40–49. doi: 10.1111/j.1439-0507.2007.01435.x
McCarty, T. P., and Pappas, P. G. (2016). Invasive candidiasis. Infect. Dis. Clin. North Am. 30, 103–124. doi: 10.1016/j.idc.2015.10.013
Melo, A. S., Colombo, A. L., and Arthington-Skaggs, B. A. (2007). Paradoxical growth effect of caspofungin observed on biofilms and planktonic cells of five different Candida species. Antimicrob. Agents Chemother. 51, 3081–3088. doi: 10.1128/AAC.00676-07
Mendes de Toledo, C. E. M., Santos, P. R., de Mello, J. C. P., Filho, B. P. D., Nakamura, C. V., and Ueda-Nakamura, T. (2015). Antifungal properties of crude extracts, fractions, and purified compounds from bark of Curatella americana L. (Dilleniaceae) against Candida species. Evid. Based Complement. Alternat. Med. 2015:673962. doi: 10.1155/2015/673962
Menezes, T. O., Gillet, L. C., Menezes, S. A., Feitosa, R. N., Ishak, M. O., Ishak, R., et al. (2013). Virulence factors of Candida albicans isolates from the oral cavities of HIV-1-positive patients. Curr. HIV Res. 11, 304–308. doi: 10.2174/1570162X113119990042
Merseguel, K. B., Nishikaku, A. S., Rodrigues, A. M., Padovan, A. C., Ferreira, R. C., de Azevedo Melo, A. S., et al. (2015). Genetic diversity of medically important and emerging Candida species causing invasive infection. BMC Infect. Dis. 15:57. doi: 10.1186/s12879-015-0793-3
Merz, W. G., and Sandford, G. R. (1979). Isolation and characterization of a polyene-resistant variant of Candida tropicalis. J. Clin. Microbiol. 9, 677–680.
Mesa-Arango, A. C., Forastiero, A., Bernal-Martínez, L., Cuenca-Estrella, M., Mellado, E., and Zaragoza, O. (2013). The non-mammalian host Galleria mellonella can be used to study the virulence of the fungal pathogen Candida tropicalis and the efficacy of antifungal drugs during infection by this pathogenic yeast. Med. Mycol. 51, 461–472. doi: 10.3109/13693786.2012.737031
Millsop, J. W., and Fazel, N. (2016). Oral candidiasis. Clin. Dermatol. 34, 487–494. doi: 10.1016/j.clindermatol.2016.02.022
Misra, S., Raghuwanshi, S., Gupta, P., Dutt, K., and Saxena, R. K. (2012). Fermentation behavior of osmophilic yeast Candida tropicalis isolated from the nectar of Hibiscus rosa sinensis flowers for xylitol production. Antonie Van Leeuwenhoek 101, 393–402. doi: 10.1007/s10482-011-9646-2
Morais-Braga, M. F. B., Sales, D. L., Carneiro, J. N. P., Machado, A. J. T., dos Santos, A. T. L., de Freitas, M. A., et al. (2016). Psidium guajava L. and Psidium brownianum Mart ex DC.: Chemical composition and anti–Candida effect in association with fluconazole. Microb. Pathog. 95, 200–207. doi: 10.1016/j.micpath.2016.04.013
Morrow, C. A., and Fraser, J. A. (2013). Ploidy variation as an adaptive mechanism in human pathogenic fungi. Semin. Cell Dev. Biol. 4, 339–346. doi: 10.1016/j.semcdb.2013.01.008
Morschhäuser, J. (2010). Regulation of multidrug resistance in pathogenic fungi. Fungal Genet. Biol. 47, 94–106. doi: 10.1016/j.fgb.2009.08.002
Muadcheingka, T., and Tantivitayakul, P. (2015). Distribution of Candida albicans and non-albicans Candida species in oral candidiasis patients: correlation between cell surface hydrophobicity and biofilm forming activities. Arch. Oral Biol. 60, 894–901. doi: 10.1016/j.archoralbio.2015.03.002
Mulu, A., Kassu, A., Anagaw, B., Moges, B., Gelaw, A., Alemayehu, M., et al. (2013). Frequent detection of ‘azole’ resistant Candida species among late presenting AIDS patients in northwest Ethiopia. BMC Infect. Dis. 13:82. doi: 10.1186/1471-2334-13-82
Nagaraja, P., Mathew, T., and Shetty, D. (2005). Candida tropicalis causing prosthetic valve endocarditis. Indian J. Med. Microbiol. 23:139. doi: 10.4103/0255-0857.16059
Naglik, J. R., Fostira, F., Ruprai, J., Staab, J. F., Challacombe, S. J., and Sundstrom, P. (2006). Candida albicans HWP1 gene expression and host antibody responses in colonization and disease. J. Med. Microbiol. 55(Pt 10), 1323–1327. doi: 10.1099/jmm.0.46737-0
Nash, E. E., Peters, B. M., Fidel, P. L., and Noverr, M. C. (2016a). Morphology-independent virulence of Candida species during polymicrobial intra-abdominal infections with Staphylococcus aureus. Infect. Immun. 84, 90–98. doi: 10.1128/IAI.01059-15
Nash, E. E., Peters, B. M., Lilly, E. A., Noverr, M. C., and Fidel, P. L. Jr. (2016b). A murine model of Candida glabrata Vaginitis shows no evidence of an inflammatory immunopathogenic response. PLoS ONE 11:e0147969. doi: 10.1371/journal.pone.0147969
Negri, M., Martins, M., Henriques, M., Svidzinski, T. I., Azeredo, J., and Oliveira, R. (2010). Examination of potential virulence factors of Candida tropicalis clinical isolates from hospitalized patients. Mycopathologia 169, 175–182. doi: 10.1007/s11046-009-9246-0
Nguyen, V. Q., and Sil, A. (2008). Temperature-induced switch to the pathogenic yeast form of Histoplasma capsulatum requires Ryp1, a conserved transcriptional regulator. Proc. Natl. Acad. Sci. U.S.A. 105, 4880–4885. doi: 10.1073/pnas.0710448105
Nickerson, K. W., Atkin, A. L., and Hornby, J. M. (2006). Quorum sensing in dimorphic fungi: farnesol and beyond. Appl. Environ. Microbiol. 72, 3805–3813. doi: 10.1128/AEM.02765-05
Nobile, C. J., and Mitchell, A. P. (2006). Genetics and genomics of Candida albicans biofilm formation. Cell. Microbiol. 8, 1382–1391. doi: 10.1111/j.1462-5822.2006.00761.x
Nobile, C. J., Schneider, H. A., Nett, J. E., Sheppard, D. C., Filler, S. G., Andes, D. R., et al. (2008). Complementary adhesin function in C. albicans biofilm formation. Curr. Biol. 18, 1017–1024. doi: 10.1016/j.cub.2008.06.034
Nordin, M. A., Wan Harun, W. H., and Abdul Razak, F. (2013). Antifungal susceptibility and growth inhibitory response of oral Candida species to Brucea javanica Linn. extract. BMC Complement. Altern. Med. 13:342. doi: 10.1186/1472-6882-13-342
Nwibo, D. D., Hamamoto, H., Matsumoto, Y., Kaito, C., and Sekimizu, K. (2015). Current use of silkworm larvae (Bombyx mori) as an animal model in pharmaco-medical research. Drug Discov. Ther. 9, 133–135. doi: 10.5582/ddt.2015.01026
Odds, F. C., and Jacobsen, M. D. (2008). Multilocus sequence typing of pathogenic Candida species. Eukaryot. Cell 7, 1075–1084. doi: 10.1128/EC.00062-08
Okamoto-Shibayama, K., Kikuchi, Y., Kokubu, E., Sato, Y., and Ishihara, K. (2014). Csa2, a member of the Rbt5 protein family, is involved in the utilization of iron from human hemoglobin during Candida albicans hyphal growth. FEMS Yeast Res. 14, 674–677. doi: 10.1111/1567-1364.12160
Oksuz, S., Sahin, I., Yildirim, M., Gulcan, A., Yavuz, T., Kaya, D., et al. (2007). Phospholipase and proteinase activities in different Candida species isolated from anatomically distinct sites of healthy adults. Jpn. J. Infect. Dis. 60, 280–283.
Oliveira, V. K. P. D. (2011). Ocorrência das Espécies de Leveduras Isoladas de Sangue e Cateter de Pacientes Internados em Hospital Público Infantil de São Paulo (Período 2007 a 2010). Master's thesis, Universidade de São Paulo, São Paulo.
Paiva, L. C., Vidigal, P. G., Donatti, L., Svidzinski, T. I., and Consolaro, M. E. (2012). Assessment of in vitro biofilm formation by Candida species isolates from vulvovaginal candidiasis and ultrastructural characteristics. Micron 43, 497–502. doi: 10.1016/j.micron.2011.09.013
Pam, V. K., Akpan, J. U., Oduyebo, O. O., Nwaokorie, F. O., Fowora, M. A., Oladele, R. O., et al. (2012). Fluconazole susceptibility and ERG11 gene expression in vaginal Candida species isolated from Lagos Nigeria. Int. J. Mol. Epidemiol. Genet. 3, 84–90.
Panda, A., Ghosh, A. K., Mirdha, B. R., Xess, I., Paul, S., Samantaray, J. C., et al. (2015). MALDI-TOF mass spectrometry for rapid identification of clinical fungal isolates based on ribosomal protein biomarkers. J. Microbiol. Methods 109, 93–105. doi: 10.1016/j.mimet.2014.12.014
Pannanusorn, S., Fernandez, V., and Romling, U. (2013). Prevalence of biofilm formation in clinical isolates of Candida species causing bloodstream infection. Mycoses 56, 264–272. doi: 10.1111/myc.12014
Park, S., Kelly, R., Kahn, J. N., Robles, J., Hsu, M. J., Register, E., et al. (2005). Specific substitutions in the echinocandin target Fks1p account for reduced susceptibility of rare laboratory and clinical Candida sp. isolates. Antimicrob Agents Chemother. 49, 3264–3273. doi: 10.1128/AAC.49.8.3264-3273.2005
Peman, J., Canton, E., Quindos, G., Eraso, E., Alcoba, J., Guinea, J., et al. (2012). Epidemiology, species distribution and in vitro antifungal susceptibility of fungaemia in a Spanish multicentre prospective survey. J. Antimicrob. Chemother. 67, 1181–1187. doi: 10.1093/jac/dks019
Perlin, D. S. (2007). Resistance to echinocandin-class antifungal drugs. Drug Resist. Updat. 10, 121–130. doi: 10.1016/j.drup.2007.04.002
Pfaller, M. A. (2012). Antifungal drug resistance: mechanisms, epidemiology, and consequences for treatment. Am. J. Med. 125(1 Suppl.), S3–S13. doi: 10.1016/j.amjmed.2011.11.001
Pfaller, M. A., Boyken, L., Hollis, R. J., Kroeger, J., Messer, S. A., Tendolkar, S., et al. (2008). In vitro susceptibility of invasive isolates of Candida spp. to anidulafungin, caspofungin, and micafungin: six years of global surveillance. J Clin Microbiol. 46, 150–156. doi: 10.1128/JCM.01901-07
Pfaller, M. A., Castanheira, M., Diekema, D. J., Messer, S. A., Moet, G. J., and Jones, R. N. (2010). Comparison of European Committee on Antimicrobial Susceptibility Testing (EUCAST) and Etest methods with the CLSI broth microdilution method for echinocandin susceptibility testing of Candida species. J. Clin. Microbiol. 48, 1592–1599. doi: 10.1128/JCM.02445-09
Pincus, D. H., Orenga, S., and Chatellier, S. (2007). Yeast identification–past, present, and future methods. Med. Mycol. 45, 97–121. doi: 10.1080/13693780601059936
Porman, A. M., Alby, K., Hirakawa, M. P., and Bennett, R. J. (2011). Discovery of a phenotypic switch regulating sexual mating in the opportunistic fungal pathogen Candida tropicalis. Proc. Natl. Acad. Sci. U.S.A. 108, 21158–21163. doi: 10.1073/pnas.1112076109
Porman, A. M., Hirakawa, M. P., Jones, S. K., Wang, N., and Bennett, R. J. (2013). MTL-independent phenotypic switching in Candida tropicalis and a dual role for Wor1 in regulating switching and filamentation. PLoS Genet. 9:e1003369. doi: 10.1371/journal.pgen.1003369
Posteraro, B., Efremov, L., Leoncini, E., Amore, R., Posteraro, P., Ricciardi, W., et al. (2015). Are the conventional commercial yeast identification methods still helpful in the era of new clinical microbiology diagnostics? A meta-analysis of their accuracy. J. Clin. Microbiol. 53, 2439–2450. doi: 10.1128/JCM.00802-15
Powderly, W. G., Kobayashi, G. S., Herzig, G. P., and Medoff, G. (1988). Amphotericin B-resistant yeast infection in severely immunocompromised patients. Am. J. Med. 84, 826–832. doi: 10.1016/0002-9343(88)90059-9
Punithavathy, P., and Menon, T. (2012). Characterization of gene family that mediates the adhesion of biofilms formed by Candida tropicalis isolated from HIV and non-HIV patients. BMC Infect. Dis. 12:1. doi: 10.1186/1471-2334-12-S1-O8
Ra, C. H., Jung, J. H., Sunwoo, I. Y., Kang, C. H., Jeong, G. T., and Kim, S. K. (2015). Detoxification of Eucheuma spinosum hydrolysates with activated carbon for ethanol production by the salt-tolerant yeast Candida tropicalis. J. Microbiol. Biotechnol. 25, 856–862. doi: 10.4014/jmb.1409.09038
Ragunathan, L., Poongothai, G. K., Sinazer, A. R., Kannaiyan, K., Gurumurthy, H., Jaget, N., et al. (2014). Phenotypic characterization and antifungal susceptibility pattern to fluconazole in Candida species isolated from vulvovaginal candidiasis in a tertiary care hospital. J. Clin. Diagn. Res. 8, DC01–DC04. doi: 10.7860/JCDR/2014/7434.4311
Ramage, G., Martinez, J. P., and Lopez-Ribot, J. L. (2006). Candida biofilms on implanted biomaterials: a clinically significant problem. FEMS Yeast Res. 6, 979–986. doi: 10.1111/j.1567-1364.2006.00117.x
Ramage, G., Vande Walle, K., Wickes, B. L., and Lopez-Ribot, J. L. (2001). Standardized method for in vitro antifungal susceptibility testing of Candida albicans biofilms. Antimicrob. Agents Chemother. 45, 2475–2479. doi: 10.1128/AAC.45.9.2475-2479.2001
Rao, R. S., Jyothi, C. P., Prakasham, R. S., Sarma, P. N., and Rao, L. V. (2006). Xylitol production from corn fiber and sugarcane bagasse hydrolysates by Candida tropicalis. Bioresour. Technol. 97, 1974–1978. doi: 10.1016/j.biortech.2005.08.015
Rodriguez, P. L., Ali, R., and Serrano, R. (1996). CtCdc55p and CtHal3p: Two putative regulatory proteins from Candida tropicalis with long acidic domains. Yeast 12, 1321–1329. doi: 10.1002/(SICI)1097-0061(199610)12:13<1321::AID-YEA27>3.0.CO;2-6
Rossoni, R. D., Barbosa, J. O., Vilela, S. F., Jorge, A. O., and Junqueira, J. C. (2013). Comparison of the hemolytic activity between C. albicans and non-albicans Candida species. Braz. Oral Res. 27, 484–489. doi: 10.1590/S1806-83242013000600007
Ruchel, R., Uhlemann, K., and Boning, B. (1983). Secretion of acid proteinases by different species of the genus Candida. Zentralbl. Bakteriol. Mikrobiol. Hyg. A. 255, 537–548. doi: 10.1016/S0174-3031(83)80013-4
Sachin, C., Ruchi, K., and Santosh, S. (2012). In vitro evaluation of proteinase, phospholipase and haemolysin activities of Candida species isolated from clinical specimens. IJMBR 1, 153–157. doi: 10.14194/ijmbr.1211
Salari, S., Bakhshi, T., Sharififar, F., Naseri, A., and Ghasemi, N. A. P. (2016). Evaluation of antifungal activity of standardized extract of Salvia rhytidea Benth. (Lamiaceae) against various Candida isolates. J. Mycol. Med. 26, 323–330. doi: 10.1016/j.mycmed.2016.06.003
Salehei, Z., Seifi, Z., and Mahmoudabadi, A. (2012). Sensitivity of vaginal isolates of Candida to eight antifungal drugs isolated from Ahvaz, Iran. Jundishapur J. Microbiol. 5, 574–577. doi: 10.5812/jjm.4556
Samaranayake, L. P., Raeside, J. M., and MacFarlane, T. W. (1984). Factors affecting the phospholipase activity of Candida species in vitro. Sabouraudia 22, 201–207.
Sanglard, D., and Odds, F. C. (2002). Resistance of Candida species to antifungal agents: molecular mechanisms and clinical consequences. Lancet Infect. Dis. 2, 73–85. doi: 10.1016/S1473-3099(02)00181-0
Sanita, P. V., Zago, C. E., Mima, E. G., Pavarina, A. C., Jorge, J. H., Machado, A. L., et al. (2014). In vitro evaluation of the enzymatic activity profile of non-albicans Candida species isolated from patients with oral candidiasis with or without diabetes. Oral Surg. Oral Med. Oral Pathol. Oral Radiol. 118, 84–91. doi: 10.1016/j.oooo.2014.03.020
Santhanam, J., Nazmiah, N., and Aziz, M. N. (2013). Species distribution and antifungal susceptibility patterns of Candida species: is low susceptibility to itraconazole a trend in Malaysia? Med. J. Malaysia 68, 343–347.
Santos, C., Lima, N., Sampaio, P., and Pais, C. (2011). Matrix-assisted laser desorption/ionization time-of-flight intact cell mass spectrometry to detect emerging pathogenic Candida species. Diagn. Microbiol. Infect. Dis. 71, 304–308. doi: 10.1016/j.diagmicrobio.2011.07.002
Sariguzel, F., Berk, E., Koc, A., Sav, H., and Aydemir, G. (2015). Evaluation of CHROMagar Candida, VITEK2 YST and VITEK® MS for identification of Candida strains isolated from blood cultures. Infez. Med. 23, 318–322.
Schaller, M., Borelli, C., Korting, H. C., and Hube, B. (2005). Hydrolytic enzymes as virulence factors of Candida albicans. Mycoses 48, 365–377. doi: 10.1111/j.1439-0507.2005.01165.x
Seervai, R. N. H., Jones, S. K. Jr., Hirakawa, M. P., Porman, A. M., and Bennett, R. J. (2013). Parasexuality and ploidy change in Candida tropicalis. Eukaryot. Cell 12, 1629–1640. doi: 10.1128/EC.00128-13
Seneviratne, C. J., Rajan, S., Wong, S. S., Tsang, D. N., Lai, C. K., Samaranayake, L. P., et al. (2016). Antifungal susceptibility in serum and virulence determinants of Candida bloodstream isolates from Hong Kong. Front. Microbiol. 7:216. doi: 10.3389/fmicb.2016.00216
Shi, X. Y., Yang, Y. P., Zhang, Y., Li, W., Wang, J. D., Huang, W. M., et al. (2015). Molecular identification and antifungal susceptibility of 186 Candida isolates from vulvovaginal candidiasis in southern China. J. Med. Microbiol. 64, 390–393. doi: 10.1099/jmm.0.000024
Shu, C., Sun, L., and Zhang, W. (2016). Thymol has antifungal activity against Candida albicans during infection and maintains the innate immune response required for function of the p38 MAPK signaling pathway in Caenorhabditis elegans. Immunol. Res. 64, 1013–1024. doi: 10.1007/s12026-016-8785-y
Silva, S., Hooper, S. J., Henriques, M., Oliveira, R., Azeredo, J., and Williams, D. W. (2011). The role of secreted aspartyl proteinases in Candida tropicalis invasion and damage of oral mucosa. Clin. Microbiol. Infect. 17, 264–722. doi: 10.1111/j.1469-0691.2010.03248.x
Silva, S., Negri, M., Henriques, M., Oliveira, R., Williams, D. W., and Azeredo, J. (2012). Candida glabrata, Candida parapsilosis and Candida tropicalis: biology, epidemiology, pathogenicity and antifungal resistance. FEMS Microbiol. Rev. 36, 288–305. doi: 10.1111/j.1574-6976.2011.00278.x
Silva-Dias, A., Miranda, I. M., Rocha, R., Monteiro-Soares, M., Salvador, A., Rodrigues, A. G., et al. (2012). A novel flow cytometric protocol for assessment of yeast cell adhesion. Cytometry A 81, 265–270. doi: 10.1002/cyto.a.21170
Siqueira, A. B. S., Rodriguez, L. R. N. D. A., Santos, R. K. B., Marinho, R. R. B., Abreu, S., Peixoto, R. F., et al. (2015). Antifungal activity of propolis against Candida species isolated from cases of chronic periodontitis. Braz. Oral Res. 29, 1–6. doi: 10.1590/1807-3107BOR-2015.vol29.0083
Slutsky, B., Staebell, M., Anderson, J., Risen, L., Pfaller, M., and Soll, D. (1987). “White-opaque transition”: a second high-frequency switching system in Candida albicans. J. Bacteriol. 169, 189–197. doi: 10.1128/jb.169.1.189-197.1987
Snide, J., and Sundstrom, P. (2006). “A characterization of HWP1 promoter activation in pseudohyphal cells in Candida albicans,” in Proceedings of the 8th ASM Conference on Candida and Candidiasis (Denver, CO: ASM Press).
Sobel, J. D. (2016). Recurrent vulvovaginal candidiasis. Am. J. Obstet. Gynecol. 214, 15–21. doi: 10.1016/j.ajog.2015.06.067
Soczo, G., Kardos, G., Varga, I., Kelentey, B., Gesztelyi, R., and Majoros, L. (2007). In vitro study of Candida tropicalis isolates exhibiting paradoxical growth in the presence of high concentrations of caspofungin. Antimicrob. Agents Chemother. 51, 4474–4476. doi: 10.1128/AAC.00880-07
Sohn, K., Senyurek, I., Fertey, J., Konigsdorfer, A., Joffroy, C., Hauser, N., et al. (2006). An in vitro assay to study the transcriptional response during adherence of Candida albicans to different human epithelia. FEMS Yeast Res. 6, 1085–1093. doi: 10.1111/j.1567-1364.2006.00130.x
Solis, N. V., and Filler, S. G. (2012). Mouse model of oropharyngeal candidiasis. Nat. Protoc. 7, 637–642. doi: 10.1038/nprot.2012.011
Souza, A. C. R., Fuchs, B. B., Pinhati, H. M., Siqueira, R. A., Hagen, F., Meis, J. F., et al. (2015). Candida parapsilosis resistance to fluconazole: molecular mechanisms and in vivo impact in infected Galleria mellonella larvae. Antimicrob. Agents Chemother. 59, 6581–6587. doi: 10.1128/AAC.01177-15
Sow, D., Fall, B., Ndiaye, M., Ba, B. S., Sylla, K., Tine, R., et al. (2015). Usefulness of MALDI-TOF mass spectrometry for routine identification of Candida species in a resource-poor setting. Mycopathologia 180, 173–179. doi: 10.1007/s11046-015-9905-2
Stefaniuk, E., Baraniak, A., Fortuna, M., and Hryniewicz, W. (2016). Usefulness of CHROMagar Candida medium, biochemical methods–API ID32C and VITEK 2 compact and two MALDI-TOF MS systems for Candida spp. identification. Pol. J. Microbiol. 65, 111–114. doi: 10.5604/17331331.1197283
Stender, H. (2003). PNA FISH: an intelligent stain for rapid diagnosis of infectious diseases. Expert Rev. Mol. Diagn. 3, 649–655. doi: 10.1586/14737159.3.5.649
Stevens, D. A., Espiritu, M., and Parmar, R. (2004). Paradoxical effect of caspofungin: reduced activity against Candida albicans at high drug concentrations. Antimicrob. Agents Chemother. 48, 3407–3411. doi: 10.1128/AAC.48.9.3407-3411.2004
Stone, N. R., Gorton, R. L., Barker, K., Ramnarain, P., and Kibbler, C. C. (2013). Evaluation of PNA-FISH yeast traffic light for rapid identification of yeast directly from positive blood cultures and assessment of clinical impact. J. Clin. Microbiol. 51, 1301–1302. doi: 10.1128/JCM.00028-13
Sun, H. Y., Chiu, Y. S., Tang, J. L., Wang, J. L., Chang, S. C., and Chen, Y. C. (2006). The usefulness of the Platelia Candida antigen in a patient with acute lymphocytic leukemia and chronic disseminated candidiasis. Med. Mycol. 44, 647–650. doi: 10.1080/13693780600735445
Sundstrom, P., Balish, E., and Allen, C. M. (2002). Essential role of the Candida albicans transglutaminase substrate, hyphal wall protein 1, in lethal oroesophageal candidiasis in immunodeficient mice. J. Infect. Dis. 185, 521–530. doi: 10.1086/338836
Symersky, J., Monod, M., and Foundling, S. I. (1997). High-resolution structure of the extracellular aspartic proteinase from Candida tropicalis yeast. Biochemistry 36, 12700–12710. doi: 10.1021/bi970613x
Takakura, N., Sato, Y., Ishibashi, H., Oshima, H., Uchida, K., Yamaguchi, H., et al. (2003). A novel murine model of oral candidiasis with local symptoms characteristic of oral thrush. Microbiol. Immun. 47, 321–326. doi: 10.1111/j.1348-0421.2003.tb03403.x
Tavanti, A., Davidson, A. D., Johnson, E. M., Maiden, M. C., Shaw, D. J., Gow, N. A., et al. (2005). Multilocus sequence typing for differentiation of strains of Candida tropicalis. J. Clin. Microbiol. 43, 5593–5600. doi: 10.1128/JCM.43.11.5593-5600.2005
Ten Cate, J., Klis, F., Pereira-Cenci, T., Crielaard, W., and De Groot, P. (2009). Molecular and cellular mechanisms that lead to Candida biofilm formation. J. Dent. Res. 88, 105–115. doi: 10.1177/0022034508329273
Togni, G., Sanglard, D., Falchetto, R., and Monod, M. (1991). Isolation and nucleotide sequence of the extracellular acid protease gene (ACP) from the yeast Candida tropicalis. FEBS Lett. 286, 181–185. doi: 10.1016/0014-5793(91)80969-A
Togni, G., Sanglard, D., Quadroni, M., Foundling, S. I., and Monod, M. (1996). Acid proteinase secreted by Candida tropicalis: functional analysis of preproregion cleavages in C. tropicalis and Saccharomyces cerevisiae. Microbiology 142 (Pt 3), 493–503. doi: 10.1099/13500872-142-3-493
Tokuoka, K. (1993). Sugar-and salt-tolerant yeasts. J. Appl. Microbiol. 74, 101–110. doi: 10.1111/j.1365-2672.1993.tb03002.x
Torres, M. P., Entwistle, F., and Coote, P. J. (2016). Effective immunosuppression with dexamethasone phosphate in the Galleria mellonella larva infection model resulting in enhanced virulence of Escherichia coli and Klebsiella pneumoniae. Med. Microbiol. Immunol. 205, 333–343. doi: 10.1007/s00430-016-0450-5
Tronchin, G., Pihet, M., Lopes-Bezerra, L. M., and Bouchara, J. P. (2008). Adherence mechanisms in human pathogenic fungi. Med. Mycol. 46, 749–772. doi: 10.1080/13693780802206435
Tsang, C. S., Chu, F. C., Leung, W. K., Jin, L. J., Samaranayake, L. P., and Siu, S. C. (2007). Phospholipase, proteinase and haemolytic activities of Candida albicans isolated from oral cavities of patients with type 2 diabetes mellitus. J. Med. Microbiol. 56(Pt 10), 1393–1398. doi: 10.1099/jmm.0.47303-0
Uchida, R., Namiguchi, S., Ishijima, H., and Tomoda, H. (2016). Therapeutic effects of three trichothecenes in the silkworm infection assay with Candida albicans. Drug Discov. Ther. 10, 44–48. doi: 10.5582/ddt.2016.01013
Udayalaxmi Jacob, S., and D'Souza, D. (2014). Comparison between virulence factors of Candida albicans and Non-Albicans species of Candida isolated from genitourinary tract. J Clin Diagn Res. 8, DC15–DC17. doi: 10.7860/JCDR/2014/10121.5137
Uppuluri, P., Pierce, C. G., Thomas, D. P., Bubeck, S. S., Saville, S. P., and Lopez-Ribot, J. L. (2010). The transcriptional regulator Nrg1p controls Candida albicans biofilm formation and dispersion. Eukaryot. Cell 9, 1531–1537. doi: 10.1128/EC.00111-10
Vandeputte, P., Tronchin, G., Berges, T., Hennequin, C., Chabasse, D., and Bouchara, J. P. (2007). Reduced susceptibility to polyenes associated with a missense mutation in the ERG6 gene in a clinical isolate of Candida glabrata with pseudohyphal growth. Antimicrob. Agents Chemother. 51, 982–9890. doi: 10.1128/AAC.01510-06
Verstrepen, K. J., and Klis, F. M. (2006). Flocculation, adhesion and biofilm formation in yeasts. Mol. Microbiol. 60, 5–15. doi: 10.1111/j.1365-2958.2006.05072.x
Vicari, P., Feitosa Pinheiro, R., de Chauffaille, M. L., Yamamoto, M., and Figueiredo, M. S. (2003). Septic arthritis as the first sign of Candida tropicalis fungaemia in an acute lymphoid leukemia patient. Braz. J. Infect. Dis. 7, 426–428. doi: 10.1590/S1413-86702003000600012
Vijaya, D., Dhanalakshmi, T. A., and Kulkarni, S. (2014). Changing trends of vulvovaginal candidiasis. J. Lab. Phys. 6, 28–30. doi: 10.4103/0974-2727.129087
Vincent, B. M., Lancaster, A. K., Scherz-Shouval, R., Whitesell, L., and Lindquist, S. (2013). Fitness trade-offs restrict the evolution of resistance to amphotericin B. PLoS Biol. 11:e1001692. doi: 10.1371/journal.pbio.1001692
Wan Harun, W. H., Jamil, N. A., Jamaludin, N. H., and Nordin, M. A. (2013). Effect of Piper betle and Brucea javanica on the differential expression of Hyphal Wall Protein (HWP1) in Non-Candida albicans Candida (NCAC) Species. Evid. Based Complement. Alternat. Med. 2013:397268. doi: 10.1155/2013/397268
Wang, T., Pan, D., Zhou, Z., You, Y., Jiang, C., Zhao, X., et al. (2016). Dectin-3 deficiency promotes colitis development due to impaired antifungal innate immune responses in the gut. PLoS Pathog. 12:e1005662. doi: 10.1371/journal.ppat.1005662
Wapinski, I., Pfeffer, A., Friedman, N., and Regev, A. (2007). Natural history and evolutionary principles of gene duplication in fungi. Nature 449, 54–61. doi: 10.1038/nature06107
Weber, K., Schulz, B., and Ruhnke, M. (2010). The quorum-sensing molecule E, E-farnesol—its variable secretion and its impact on the growth and metabolism of Candida species. Yeast 27, 727–739. doi: 10.1002/yea.1769
Wei, W.-j., Yang, H.-f., Ye, Y., and Li, J.-b. (2016). Galleria mellonella as a model system to assess the efficacy of antimicrobial agents against Klebsiella pneumoniae infection. J. Chemother. 29, 252–256. doi: 10.1080/1120009X.2016.1156892
Won, E. J., Shin, J. H., Kim, M. N., Choi, M. J., Joo, M. Y., Kee, S. J., et al. (2014). Evaluation of the BD Phoenix system for identification of a wide spectrum of clinically important yeast species: a comparison with Vitek 2-YST. Diagn. Microbiol. Infect. Dis. 79, 477–480. doi: 10.1016/j.diagmicrobio.2014.05.011
Woods, R. A., Bard, M., Jackson, I. E., and Drutz, D. J. (1974). Resistance to polyene antibiotics and correlated sterol changes in two isolates of Candida tropicalis from a patient with an amphotericin B-resistant funguria. J. Infect. Dis. 129, 53–58. doi: 10.1093/infdis/129.1.53
Wu, Y., Zhou, H. J., Che, J., Li, W. G., Bian, F. N., Yu, S. B., et al. (2014). Multilocus microsatellite markers for molecular typing of Candida tropicalis isolates. BMC Microbiol. 14:245. doi: 10.1186/s12866-014-0245-z
Wu, Y., Zhou, H., Wang, J., Li, L., Li, W., Cui, Z., et al. (2012). Analysis of the clonality of Candida tropicalis strains from a general hospital in Beijing using multilocus sequence typing. PLoS ONE 7:e47767. doi: 10.1371/journal.pone.0047767
Xie, J., Du, H., Guan, G., Tong, Y., Kourkoumpetis, T. K., Zhang, L., et al. (2012). N-acetylglucosamine induces white-to-opaque switching and mating in Candida tropicalis, providing new insights into adaptation and fungal sexual evolution. Eukaryot. Cell 11, 773–782. doi: 10.1128/EC.00047-12
Xu, B., Shi, P., Wu, H., Guo, X., Wang, Q., and Zhou, S. (2010). Utility of FDG PET/CT in guiding antifungal therapy in acute leukemia patients with chronic disseminated candidiasis. Clin. Nucl. Med. 35, 567–750. doi: 10.1097/RLU.0b013e3181e4db84
Yan, J., Jianping, W., Hongmei, L., Suliang, Y., and Zongding, H. (2005). The biodegradation of phenol at high initial concentration by the yeast Candida tropicalis. Biochem. Eng. J. 24, 243–247. doi: 10.1016/j.bej.2005.02.016
Zanette, R. A., and Kontoyiannis, D. P. (2013). Paradoxical effect to caspofungin in Candida species does not confer survival advantage in a Drosophila model of candidiasis. Virulence 4, 497–498. doi: 10.4161/viru.25523
Zaugg, C., Borg-Von Zepelin, M., Reichard, U., Sanglard, D., and Monod, M. (2001). Secreted aspartic proteinase family of Candida tropicalis. Infect. Immun. 69, 405–412. doi: 10.1128/IAI.69.1.405-412.2001
Zhang, Q., Tao, L., Guan, G., Yue, H., Liang, W., Cao, C., et al. (2016). Regulation of filamentation in the human fungal pathogen Candida tropicalis. Mol. Microbiol. 99, 528–545. doi: 10.1111/mmi.13247
Zhang, Y., Tao, L., Zhang, Q., Guan, G., Nobile, C. J., Zheng, Q., et al. (2016). The gray phenotype and tristable phenotypic transitions in the human fungal pathogen Candida tropicalis. Fungal Genet. Biol. 93, 10–16. doi: 10.1016/j.fgb.2016.05.006
Zhao, L., de Hoog, G. S., Cornelissen, A., Lyu, Q., Mou, L., Liu, T., et al. (2016). Prospective evaluation of the chromogenic medium CandiSelect 4 for differentiation and presumptive identification of non-Candida albicans Candida species. Fungal Biol. 120, 173–178. doi: 10.1016/j.funbio.2015.09.006
Zheng, X. D., Lee, R. T., Wang, Y. M., Lin, Q. S., and Wang, Y. (2007). Phosphorylation of Rga2, a Cdc42 GAP, by CDK/Hgc1 is crucial for Candida albicans hyphal growth. EMBO J. 26, 3760–3769. doi: 10.1038/sj.emboj.7601814
Zuza-Alves, D. L., de Medeiros, S. S., de Souza, L. B., Silva-Rocha, W. P., Francisco, E. C., de Araújo, M. C., et al. (2016). Evaluation of virulence factors in vitro, resistance to osmotic stress and antifungal susceptibility of Candida tropicalis isolated from the coastal environment of Northeast Brazil. Front. Microbiol. 7:1783. doi: 10.3389/fmicb.2016.01783
Keywords: Candida tropicalis, virulence factors, antifungal resistance, phenotypic and molecular identification, update
Citation: Zuza-Alves DL, Silva-Rocha WP and Chaves GM (2017) An Update on Candida tropicalis Based on Basic and Clinical Approaches. Front. Microbiol. 8:1927. doi: 10.3389/fmicb.2017.01927
Received: 23 May 2017; Accepted: 21 September 2017;
Published: 13 October 2017.
Edited by:
Hector Mora Montes, Universidad de Guanajuato, MexicoReviewed by:
Nicolas Papon, University of Angers, FranceIrene Castano, Institute for Scientific and Technological Research, Mexico
Copyright © 2017 Zuza-Alves, Silva-Rocha and Chaves. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guilherme M. Chaves, Z3VpbGhlcm1lLmNoYXZlc0B1ZnJuZXQuYnI=
 Diana L. Zuza-Alves
Diana L. Zuza-Alves Walicyranison P. Silva-Rocha
Walicyranison P. Silva-Rocha Guilherme M. Chaves
Guilherme M. Chaves