- 1Key Lab of Plant Pathology of Hubei Province, Huazhong Agricultural University, Wuhan, China
- 2Laboratory of Plant Pathology, Department of Agronomy, Jilin Agricultural University, Changchun, China
- 3School of Environmental Sciences, University of Guelph, Guelph, ON, Canada
- 4Department of Biology, University of Saskatchewan, Saskatoon, SK, Canada
Colletotrichum higginsianum is a widely distributed fungus attacking many cruciferous species. To investigate pathogenic mechanisms of the pathogen on the host Arabidopsis thaliana, we screened and obtained a virulence-deficient mutant Ch-1-T513 in a T-DNA insertion mutant library of C. higginsianum. The mutant Ch-1-T513 produced yellow colony centers with distorted multi-branching hyphal tips as well as producing few conidia. Heavily swollen hyphae in the mutant could be observed, and intra-hyphal hyphae were found to be formed in the balloon-shaped hyphae. The mutant failed to produce lesions on 12-day-old Arabidopsis seedlings, and invasive hyphae did not differentiate into large primary and thin secondary hyphae after appressorial formation on Arabidopsis leaves, but formed abundant bulbous hyphae in epidermal cells. Southern blot analysis showed Ch-1-T513 had double-site T-DNA integrations. The mutant had insertions upstream of genes for a major facilitator superfamily (MFS) transporter, ChMfs1 and an aldo/keto reductase, ChAkr. Complementation experiments by transforming genomic sequences from a wild-type strain into the insertion mutant demonstrated that ChMfs1 is involved in the Ch-1-T513 phenotype. The complementation strain C-ChMfs1-1 exhibited normal hyphal morphology, conidiation, and pathogenicity identical to the wild-type. The results demonstrate that ChMfs1 is involved in intra-hyphal hyphae production, conidiation, and pathogenicity in C. higginsianum. To our knowledge, this is the first report of a MFS transporter gene in a phytopathogenic fungus associated with intra-hyphal hyphae formation, playing a key role in infection of its plant host.
Introduction
Colletotrichum higginsianum as a hemibiotrophic fungal pathogen causes anthracnose disease on many cruciferous plants, including economically important species, Brassica spp. as well as Arabidopsis thaliana (O’Connell et al., 2004). To infect plants, anthracnose conidia typically produce germination hyphae that differentiate into melanized appressoria, which in turn produce penetration pegs and enter into plant cell. Once inside the plant cell, the fungus forms swollen biotrophic primary hyphae which proliferate by invading living epidermal cells. These necrotrophic secondary hyphae spread more quickly across host cells causing chlorosis and necrosis. A. thaliana is an important model host with a completely sequenced genome available and ease of genetic analysis as well as an extensive mutant collection (O’Connell et al., 2004). Thus, the C. higginsianum–Arabidopsis pathosystem is an attractive model system for studying molecular mechanisms of plant–microbe interactions (O’Connell et al., 2004; Huser et al., 2009).
In natural environments, microorganisms always produce toxic chemicals to compete with other organisms and are capable of tolerating toxicity of these natural chemicals. Microorganisms have active transport systems which actively secrete synthetic and natural toxic compounds into the external environment. The ATP-binding cassette (ABC) and the major facilitator superfamily (MFS) of transporters are the two families which play an important role in these transport processes. ABC transporters are known well as primary active transporter systems. They hydrolyze nucleotide triphosphates and utilize the ATP energy to mediate membrane transport (Higgins, 1992). MFS transporters are regarded as secondary active transport systems and are not able to hydrolyze ATP. The proton-motive force involving membrane potential and electrochemical proton gradient drives transport of compounds though membranes (Lewis, 1994).
The MFS is one of the two largest superfamilies of membrane transporters present in eukaryotes and prokaryotes, and has members which function as symporters, antiporters, or uniporters. In plant pathogens, the function of several MFS transporters has been characterized and identified. The MFS transporters, Bcmfs1 from Botrytis cinerea and MgMfs1 from Mycosphaerella graminicola, are required for protection against fungicides and natural toxic compounds (Hayashi et al., 2002; Roohparvar et al., 2007). In several species of plant pathogens, MFS transporters are involved in secretion of phytotoxins including the host-specific toxin HC-toxin in Cochliobolus carbonum (Pitkin et al., 1996), non-host-specific toxins cercosporin in Cercospora kikuchii (Callahan et al., 1999), and trichothecenes in Fusarium sporotrichioides (Alexander et al., 1999). There are functional differences of the MFS transporters in several fungal species. We previously obtained a virulence-deficient mutant Ch-1-T513 from a T-DNA insertion mutant library containing over 5000 mutants in C. higginsianum using a high-throughout pathogenicity assay on Arabidopsis leaves. In this study, we found that the mutant producing intra-hyphal hyphae during infection phases had insertions upstream of genes for a MFS transporter and an aldo/keto reductase. In separate complementation studies for these two genes corresponding to the insertions, we demonstrated that the MFS transporter named ChMfs1 is responsible for the mutant Ch-1-T513 phenotype. This study demonstrates that ChMfs1 is involved in pathogenicity and formation of intra-hyphal hyphae during infection phases of C. higginsianum and complements a novel function of MFS transporters.
Materials and Methods
Strains, Vectors, and Plants
The C. higginsianum strain, IMI349061 (Table 1), originating from diseased plants of Brassica campestris, is maintained at CABI Bioscience (Egham, Surrey, United Kingdom). The Agrobacterium tumefaciens strain EHA105 was used in fungal transformation as the T-DNA donor (Qin et al., 2011). Agrobacterium was grown at 28°C on Luria Bertani (LB) agar supplemented with 50 μg each of kanamycin, rifampicin, and streptomycin per milliliter. Escherichia coli competent cell DH5α was used for plasmid transformation.
Bacteria harboring binary vector pTFCM including the hygromycin B phosphotransferase (hph) cassette were used for C. higginsianum transformation (Li et al., 2005). Plasmid pNeo3300III with the neomycin resistance cassette were used for gene complementation vector construction (Liu et al., 2013).
The host plant A. thaliana ecotype Col-0 was used for all inoculation experiments. Arabidopsis seeds were placed on a peat-based compost, and incubated in a growth chamber (65–80% relative humidity) with a 16-h in light (40 μmol m-2 s-1, 400–700 nm) at 23°C, and 8-h dark at 18°C. The soil was composed of a perlite:vermiculite:sphagnum (1:1:1) mixture (Peilei, Qingzhou, China) and irrigated with a mineral nutrient solution as needed (Liu et al., 2013).
Molecular Analysis of Mutant Ch-1-T513
In previous work, A. tumefaciens-mediated transformation system was used to generate an insertional mutant library of C. higginsianum wild-type strain Ch-1. Among 5012 transformants, we obtained mutant Ch-1-T513 which produced an intra-hyphal hyphae. Thermal asymmetric interlaced-polymerase chain reaction (TAIL-PCR) and inverse PCR were used for amplifying sequences flanking T-DNA insertions (Qin et al., 2011; Liu et al., 2013). The PCR products were cloned into the pMD18-T vector (TaKaRa, Dalian, China) and sequenced. And then the flanking sequences were used in BLAST against the C. higginsianum genome database1.
Functional Complementation of Mutant Ch-1-T513
To verify that the two T-DNA integration events detected in the mutant Ch-1-T513 were responsible for the observed auxotrophy and pathogenicity phenotypes, ATMT was used to complement the mutant separately with a vector harboring a wild-type copy of the corresponding gene, ChMfs1 or ChAkr. The complementation vectors designated pNeo3300IIIChMfs1-C and pNeo3300IIIChAkr-C were constructed. The 3.8-kb PCR product contained a 2000-bp upstream sequence, a full-length ChMfs1 gene, and a 900-bp downstream sequence, and was amplified from genomic DNA of wild-type strain Ch-1 using primers COMPsp-BamHI-513-MFS/COMPap-BamHI-513-MFS (Table 2) and enzyme TaKaRa LA Taq (5 U/μl) (TaKaRa, Dalian, China). The BamHI-digested PCR fragment was then ligated to the BamHI site in pCAMBIA3300III to obtain pNeo3300IIICh-Mfs1-C. The 1.28-kb PCR product, containing a full-length ChAkr gene coding region, was amplified from the Ch-1 genomic DNA using primers COMPsp-BamHI-513-Akr/COMPap-BamHI-513-Akr (Table 2), and cloned into the pMD18-T vector. The BamHI-digested PCR fragment was then ligated to the BamHI site in pCAMBIA3300III to obtain pNeo3300IIIChAkr-C. The two complementation vectors were separately transformed into A. tumefaciens EHA105 and then integrated into corresponding mutant Ch-1-T513. The ATMT protocol was modified from that of Liu et al. (2013). Since the mutant Ch-1-T513 did not grow on potato dextrose agar (PDA) amended with antibiotic G418 (Amresco, Solon, OH, United States), the neomycin resistance cassette was chosen as the selectable marker for the complementation transformation. Complementation strains were screened on PDA containing 80 μg/ml G418 (Liu et al., 2013). Target gene and hph fragments of ChMfs1 or ChAkr complementation transformants were primarily investigated by PCR amplification using primers MFSSP/MFSAP (for ChMfs1) and AkrSP/AkrAP (for ChAkr) (Table 2), and then Southern blot and gene expression were used for further confirmation. The complemented strains were then analyzed for phenotype and pathogenicity.
Phenotypic Analysis
To analyze hyphal morphology, 7-day-old mycelial plugs (5 mm in diameter) were transferred onto fresh PDA plates, and then incubated at 25°C in dark conditions. Hyphae picked from edge and center of the colony on PDA were examined by light microscopy (Nikon, Tokyo, Japan). The extent of radial hyphal growth was measured after 7 days.
For conidiation, strains were cultured on PDA plates at 25°C in dark conditions for 7 days. Conidia were harvested from 9 cm diameter Petri dishes by suspending in 5 ml sterile distilled water per disc, and centrifuging at 3000 × g for 2 min. The solution was decanted, and then were counted with a hemocytometer under a light microscope.
Pathogenicity Tests
Conidial suspensions (106 spores/ml) were prepared from the wild-type and mutants, and 10 ml was sprayed onto foliage of 12-day-old Arabidopsis plants for each pot (7 cm in diameter). After enclosing plants inside plastic translucent bags lined with wet tissue paper to provide high humidity, inoculated plants were incubated at 25°C with 16-h photoperiod at 40 μmol m-2 s-1. Symptoms were evaluated and leave tissues were sampled for light microscopy at 6 dpi.
Light and Electron Microscopy
To evaluate fungal infection, inoculated leaf tissues were cleared in methanol/chloroform/glacial acetic acid (6:3:1) solution. To calculate the number of acervulus formation on leaves, infected tissues were then stained with lactophenol-trypan blue and cleared in chloral hydrate (Takahara et al., 2012). All leaf samples were viewed by differential interference contrast microscopy.
For scanning electron microscopy (SEM, following Cao et al., 2011), strains were cultured on PDA and sterilized glass slides were inserted in the media at an oblique angle by sterilized tweezers. After 7 days, the glass slides were coated with mycelia and conidia. The fungal samples were fixed in phosphate-buffered 2.5% glutaricdialdehyde in a series of increasing ethanol concentrations (30, 50, 70, 85, 95, and 100%), and dried using isoamyl acetate as the intermediate fluid in Critical Point Dryer (Model 13200-AB, SPI SUPPLIES, West Chester, PA, United States). Samples were sputter-coated with gold palladium using Auto Fine Coater (Model JFC-1600, NTC, Japan), and the mycelia of strains were observed with a JSM-6390/LV SEM (NTC, Japan).
For transmission electron microscopy (TEM, following Cao et al., 2011), hyphae were grown on PDA medium overlaid with cellophane membranes at 25°C for 7 days, collected, and prefixed with phosphate-buffered 2.5% glutaraldehyde (pH 7.2) at 4°C for 6 h. After rinsing several times with phosphate buffer, leaf tissues were fixed with phosphate-buffered 1.5% osmium tetroxide (pH 7.2) at 4°C for 2 h. The sections were then dehydrated with ethanol and embedded in SPI-812. Ultrathin sections were stained with 2% uranyl acetate and lead citrate, and observed with a Hitachi H-7650 electron microscopy (Hitachi, Tokyo, Japan).
Nucleic Acid Manipulation, Southern Blotting, and RT-PCR
Hyphae were grown on PDA overlaid with cellophane membranes at 25°C, and genomic DNA was extracted using the CTAB method. For Southern blot analysis, 15 μg of DNA was completely digested for 24 h at 37°C with SacI harboring only one recognition site in pTFCM. And then the digest was fractionated in a 0.8% agarose gel and mounted onto positively charged nylon membranes. The hygromycin resistance gene (hph) was excised from the pTFCM vector (Gong et al., 2007), and labeled with digoxigenin (DIG)-dUTP, using the PCR DIG Probe Synthesis Kit (Roche, Mannheim, Germany) following manufacturer’s instructions. Hybridization was detected using the DIG Luminescence Detection Kit (Roche, Mannheim, Germany). The nylon membrane was hybridized with probe P (Figure 1A). RNA isolation from 4-day-old mycelia was conducted using TRIzol Plus RNA Purification Kit (Invitrogen, Carlsbad, CA, United States), and then treated with DNase I (RNase Free) (Takara, Dalian, China). The first-strand cDNA was synthesized by RevertAid First-strand cDNA Synthesis Kit (Thermo Fisher Scientific Inc., Waltham, MA, United States). Expressions of ChMfs1 gene in mutant strains and wild-type were examined by RT-PCR, and a 270-bp fragment was amplified with primers MFSF/MFSR (Table 2). The actin gene of C. higginsianum amplified with primers ActinF/ActinR was used as a reference gene (Table 2). PCR conditions were 34 cycles of 94°C for 30 s, 55°C for 30 s, and 72°C for 20 s and with a final extension at 72°C for 5 min. PCR reactions were run on a T-100 Thermal Cycler (Bio-Rad, United States).
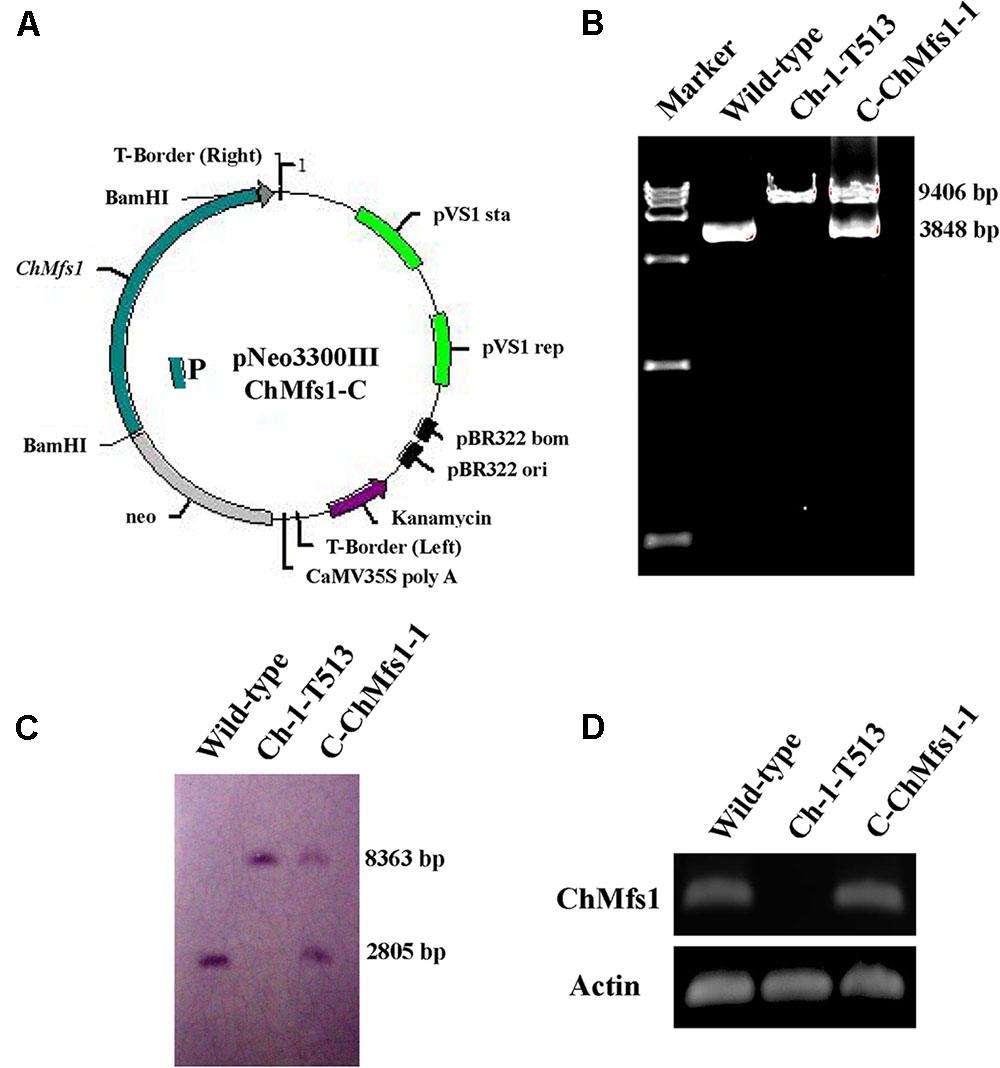
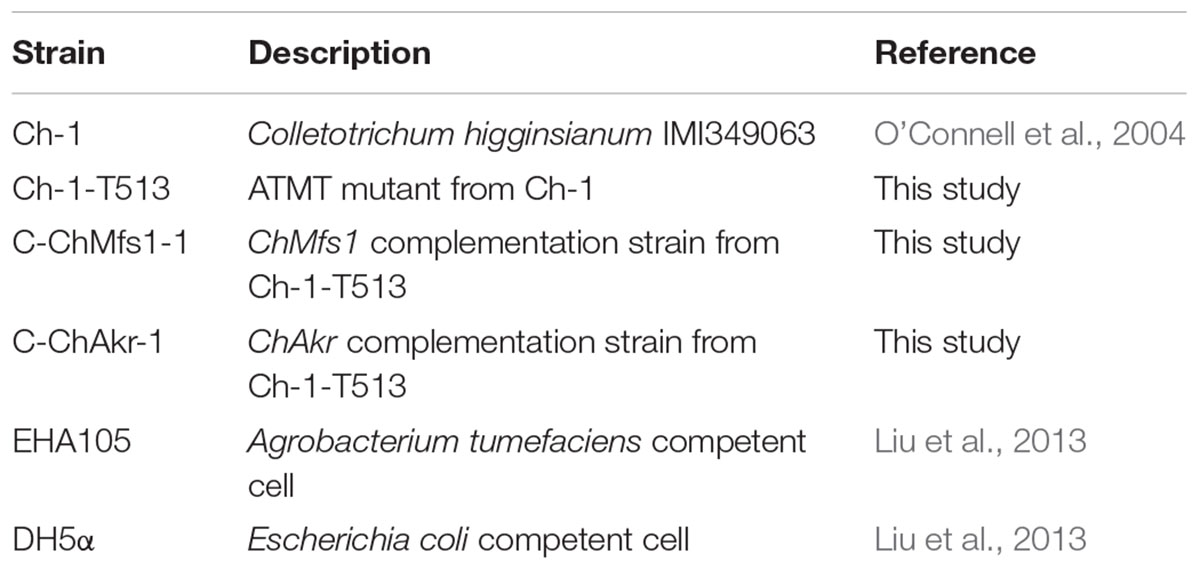
FIGURE 1. Complementation of ChMfs1 gene in C. higginsianum. (A) Strategic map of gene complementation vector construction and sites for restriction enzymes at the ChMfs1 genomic region. A bold line below the construct represents the sequence used as a probe (probe 1) in Southern blot analysis. (B) PCR analysis of wild-type Ch-1, T-DNA insertion mutant Ch-1-T513, and complementation strain C-ChMfs1-1. Marker in the image is DL15000. The 9406-bp band containing the sequences of ChMfs1 gene and inserted hph cassette from T-DNA insertion vector pTFCM was amplified in the mutant Ch-1-T513 and complementation strain C-ChMfs1-1 while a 3848-bp fragment containing ChMfs1 gene was obtained in the wild-type and C-ChMfs1-1. (C) Southern hybridization analysis of wild-type, T-DNA insertion mutant Ch-1-T513, and complementation strain C-ChMfs1-1. Genomic DNAs were digested with SacI and separated in 0.8% agarose gel. Blot was hybridized with the probe 1 amplified from genomic DNA of wild-type. (D) RT-PCR analysis of the transcription of wild-type Ch-1, T-DNA insertion mutant Ch-1-T513, and complementation strain C-ChMfs1-1. The 4-day old mycelia from PDA plates were collected and used for RNA isolation. Actin was used as the reference gene.
Bioinformatics
Primers were designed with Primer Premier (version 5.02). Open reading frames were analyzed using FGENESH (Softberry Inc., Mount Kisco, NY, United States). Protein domain and motif predictions were performed with SMART software3. Top matching protein sequences from different organisms were downloaded from the GenBank database. Sequences were aligned using ClustalX (version 2.04), and a phylogenetic tree was generated using the MEGA software (version 5.05) with the Neighbor-joining algorithm. Bootstrap percentage values for branching support based on 1000 replicates are shown on the branches.
Results
Identification of ATMT Mutant Ch-1-T513
In previous work, for genome wide screening and identification of genes involved in pathogenicity in C. higginsianum, 5012 transformants were generated using the ATMT method and screened for the phenotypes and the ability to cause disease on Arabidopsis plants (Liu et al., 2013). Among transformants, one mutant, Ch-1-T513, showing abnormal hyphal morphology and conidial production was identified as a nonpathogenic strain, defective in pathogenesis on Arabidopsis leaves.
The integration pattern of the vector pTFCM in the mutant Ch-1-T513 was determined by Southern blotting, and two hybridizing bands were detected in genomic DNA from Ch-1-T513, indicating that Ch-1-T513 had double-site T-DNA integrations in the genome. To identify the location of the T-DNA insertion in the C. higginsianum genome, 2.1 and 1.8 kb genomic DNA flanking sequences were obtained using TAIL-PCR and inverse PCR, respectively. The flanking sequence of the ATMT mutant Ch-1-T513 was used to search the C. higginsianum genome database using BLASTx, pinpointing CH063_12120 and CH063_09290 as the targeted sequences of the T-DNA insertions. One T-DNA integration occurred 82 bp upstream of the CH063_12120 with significant similarity to a MFS transporter gene of Coccidioides posadasii (GenBank EFR28508.1; e-value = 7e-95) as well as other similarly annotated genes with low e-value matching, and the other T-DNA insertion was located 411 bp upstream of the CH063_09290 with significant similarity to an aldo/keto reductase gene of Glomerella graminicola (GenBank EFQ31134.1; e-value = 0) as well as other highly matching genes of similar annotation. Hence, these two T-DNA insertion genes were designated as ChMfs1 and ChAkr.
Insertion of T-DNA at the 82 bp Upstream of a MFS Transporter Gene Locus Is Responsible for the Ch-1-T513 Mutant Phenotype
To determine whether altered phenotypes and attenuated virulence in mutant Ch-1-T513 could be restored, we reintroduced a copy of the wild-type ChMfs1, by transforming mutant Ch-1-T513 with vector pNeo3300IIIChMfs1-C (Figure 1A). Potential complementation strains were confirmed by PCR, Southern blot, and gene expression analysis. In PCR analysis, the high molecular weight band of 9406 bp containing the sequences of ChMfs1 gene and inserted hph cassette from T-DNA insertion vector pTFCM was amplified by ChMfs1 gene amplification primers MFSSP/MFSAP in the mutant Ch-1-T513 and complementation strain C-ChMfs1-1, while a 3848-bp fragment containing the ChMfs1 gene was obtained in the wild-type and C-ChMfs1-1 (Figure 1B). This revealed that inserted hph cassette was reliably detected in both transformants Ch-1-T513 and C-ChMfs1-1, meanwhile the ChMfs1 gene was identified in wild-type and C-ChMfs1-1. In Southern blotting, a fragment from the ChMfs1 gene was used as the probe (P) (Figure 1A), and a band of approximately 2805 bp was detected in wild-type and C-ChMfs1-1 (Figure 1C), similarly indicating that the sequence of ChMfs1 was detected in the complementation strain C-ChMfs1-1. To examine the expression levels of ChMfs1, RT-PCR analysis of the mutant Ch-1-T513, complementation strain C-ChMfs1-1, and wild-type was conducted. In contrast with wild-type and C-ChMfs1-1, the ChMfs1 gene expression was not detected in the mutant Ch-1-T513 (Figure 1D). The complementation transformant C-ChMfs1-1 exhibited a normal phenotype identical to the wild-type and recovered virulence on Arabidopsis leaves. The other corresponding gene (aldo/keto reductase gene) designated ChAkr was also reintroduced into the mutant Ch-1-T513 with vector pNeo3300IIIChAkr-C. We use the same methods to obtain the ChAkr complementation strain C-ChAkr-1. Complementation of the insertion at the locus of the aldo/keto reductase gene alone did not restore pathogenicity (data not shown).
ChMfs1 Is Highly Conserved in Phytopathogenic Fungi
Sequence analysis revealed that ChMfs1 contains a MFS_1 domain at the N-terminus and transmembrane region toward the C-terminus (Figure 2A). A phylogenetic tree of MFS homolog proteins identified among various fungal genomes is shown in Figure 2B. The phylogenetic relationship of ChMfs1 to other MFS proteins revealed that MFS proteins in filamentous fungi are separated from those of unicellular yeasts, with ChMfs1 being most similar to that of Colletotrichum gloeosporioides and most distant from that of the basidiomycete Ustilago maydis and other yeasts (Figure 2B). The most distantly related sequences are still within 28% identity, so this indicates that ChMfs1 proteins are well conserved in fungi.
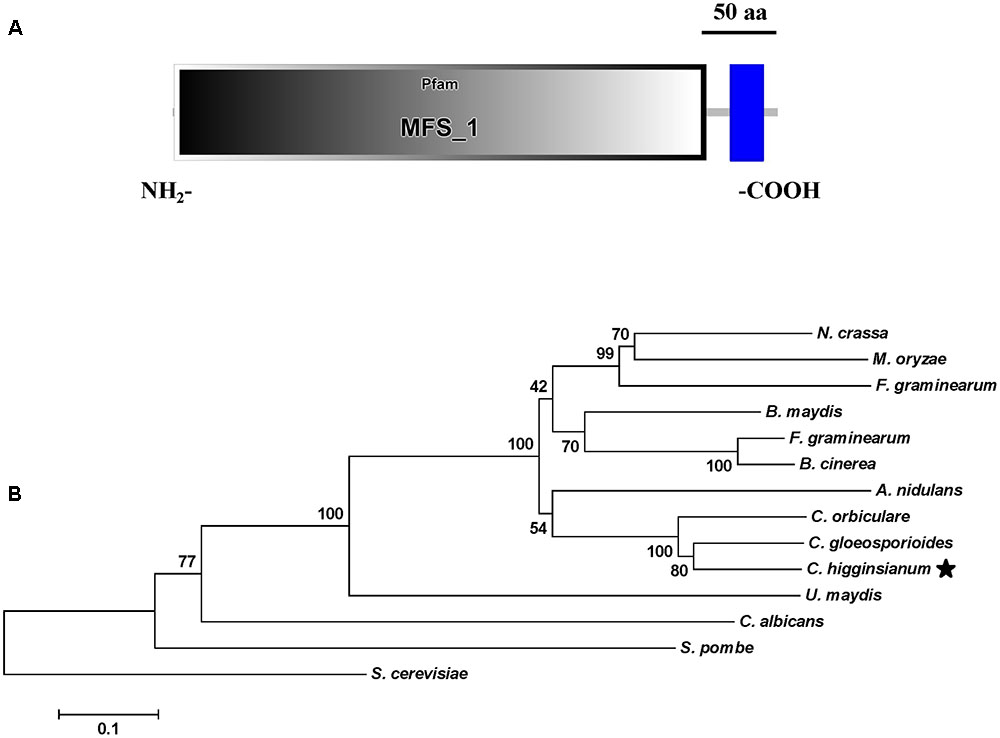
FIGURE 2. Functional domain identification and phylogenetic tree. (A) A conserved MFS_1 domain in ChMfs1 was predicted using the SMART website. (B) Phylogenetic tree of putative ChMfs1 identified in 14 fungal genomes. All of the ChMfs1 proteins were downloaded from the NCBI database and their accession numbers are listed as follows: Aspergillus nidulans, XP_662973.1; Bipolaris maydis, XP_014083759.1; B. cinerea, XP_001560975.1; Candida albicans, KGU20518.1; C. gloeosporioides, XP_007272693.1; C. higginsianum, XP_018155340.1; Colletotrichum orbiculare, ENH82883.1; Fusarium graminearum, XP_011327285.1; Magnaporthe oryzae, XP_003712384.1; Neurospora crassa, XP_958596.1; Saccharomyces cerevisiae, AJU18388.1; Schizosaccharomyces pombe, NP_592802.1; Sclerotinia sclerotiorum, APA06707.1; Ustilaginoidea virens, KDB12461.1; and Ustilago maydis, XP_011392086.1. The numbers at branch nodes are bootstrap percentages out of 1000 replications.
ChMfs1 Is Required for Colony Morphology and Conidiation
Growth and morphology of the wild-type, the T-DNA insertion mutant Ch-1-T513, and the complementation strain C-ChMfs1-1 were monitored. After growth on PDA for 7 days, the mutant Ch-1-T513 produced few aerial mycelium and became yellow in the colony centers, whereas the wild-type and C-ChMfs1-1 produced dark brown colonies with abundant white aerial mycelium (Figure 3A). However, the growth rate of the complementation strain C-ChMfs1-1 and the mutant was still significantly lower than that of the wild-type (Table 3). Examination under the microscope revealed that hyphal tips of the mutant Ch-1-T513 were twisted in contrast to a polar, linear hyphal growth of wild-type, and C-ChMfs1-1 which regained the ability to form normal hyphal morphology (Figure 3B). The conidiation of all tested strains was further examined on PDA plates post 7-day incubation. Although there were no morphological defects observed in conidia of the wild-type, Ch-1-T513, and C-ChMfs1-1, conidial abundance of C-ChMfs1-1 was recovered from the mutant Ch-1-T513 which was reduced by 84% of the wild-type (Table 3 and Figure 3C). The results indicate that ChMfs1 plays an important role in hyphal morphology and conidiation.
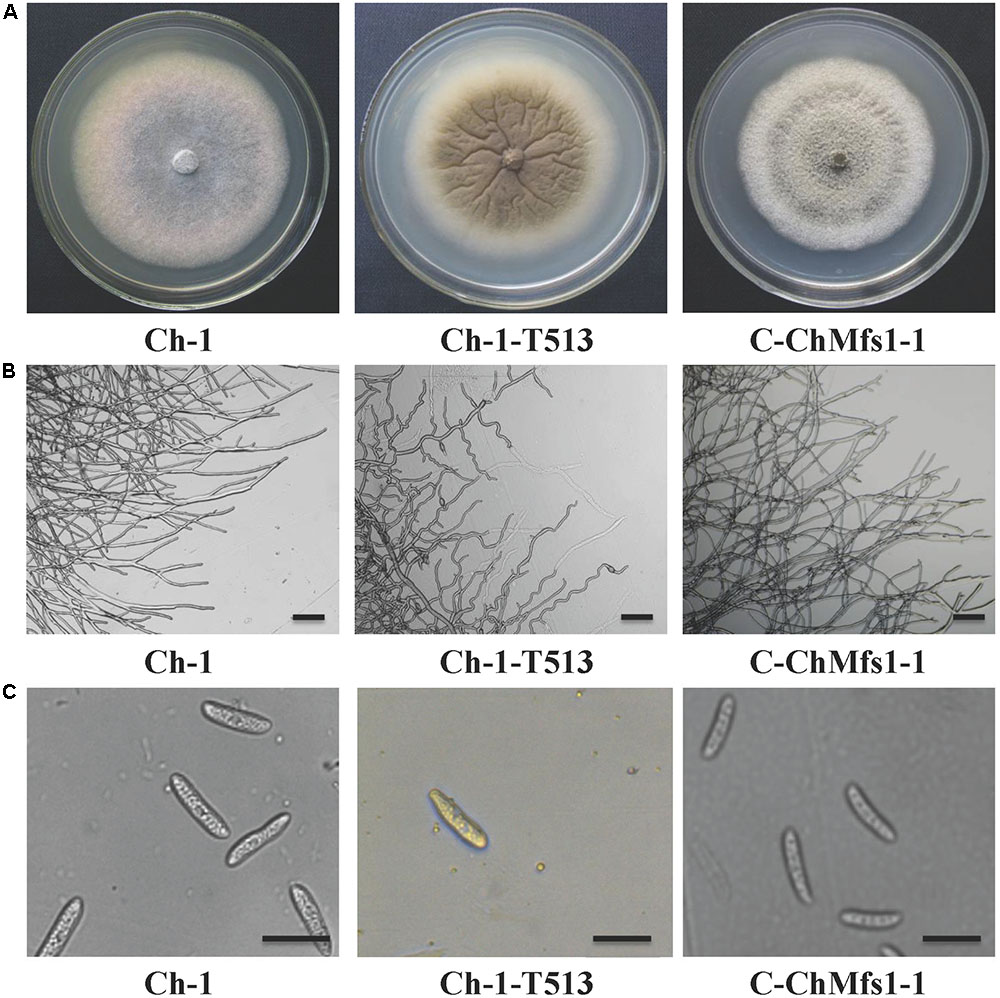
FIGURE 3. Hyphal and conidial morphology of ChMfs1 mutant and complementation strain. (A) Colony morphology of wild-type Ch-1, T-DNA insertion mutant Ch-1-T513, and complementation strain C-ChMfs1-1 on PDA plates for 7 days. The mutant Ch-1-T513 produced yellow color in the center of colonies while the wild-type and C-ChMfs1-1 produced dark brown colonies. (B) Hyphae tips of the strains. Twisted hyphae were observed for the mutant Ch-1-T513 while normal hyphae were found in wild-type and C-ChMfs1-1. Hyphae tips picked from the edge of the colonies were examined by light microscopy. Scale bar = 10 μm. (C) Conidial morphology was not altered in the mutant Ch-1-T513. Conidia of wild-type, Ch-1-T513, and C-ChMfs1-1 were washed from PDA medium grown at 25°C in dark conditions for 7 days. Scale bar = 10 μm.
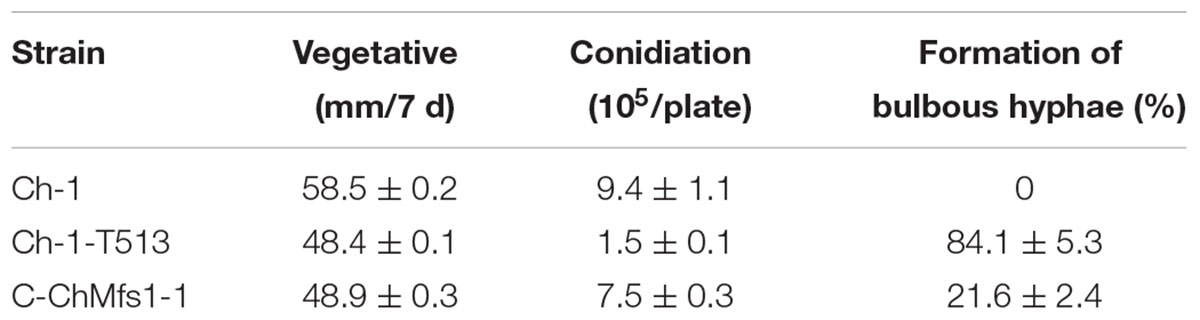
TABLE 3. Growth, conidiation, and formation of bulbous hyphae of Colletotrichum higginsianum mutants.
ChMfs1 Is Involved in Producing Intra-hyphal Hyphae of C. higginsianum
Heavily swollen hyphae visible under SEM in the mutant Ch-1-T513 almost fully reverted to wild-type in the complementation strain C-ChMfs1-1 (Figure 4A). Under TEM, intra-hyphal hyphae were seen to form in the balloon-shaped hyphae of the mutant Ch-1-T513. Moreover, single hyphal cells often contained more than one intra-hyphal hypha strand, where the cell walls were clearly delimited from that of the enclosing hyphae in the mutant Ch-1-T513 (Figure 4B). Intra-hyphal hyphae (84.1%) were frequently observed in the mutant Ch-1-T513 but were not found in the wild-type. Lower incidence of intra-hyphal hyphae (21.6%) was observed in the complementation strain C-ChMfs1-1 (Table 3 and Figure 4A). This revealed that ChMfs1 in the complementation strain could partially restore hyphal morphology. All of these observations suggest that ChMfs1 is involved in producing intra-hyphal hyphae of C. higginsianum.
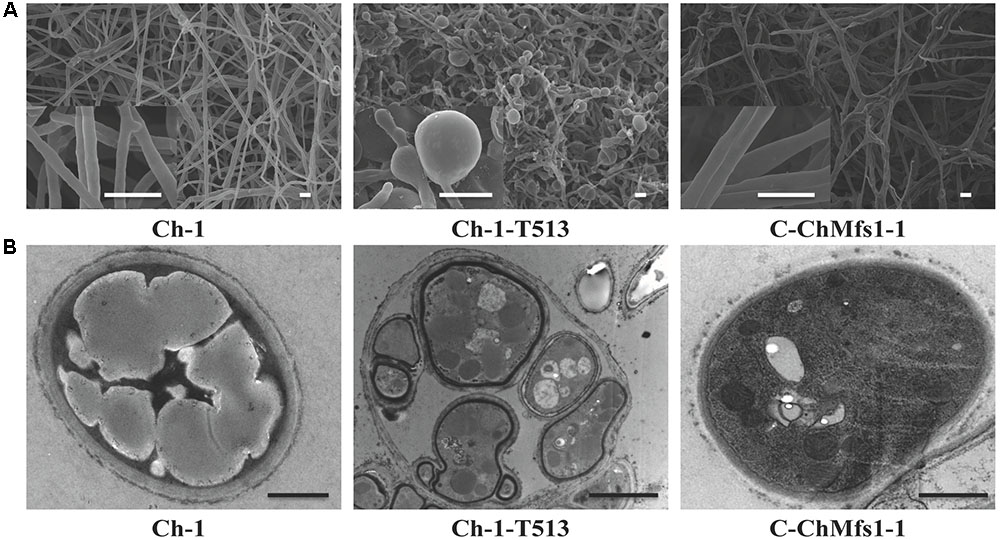
FIGURE 4. Scanning and transmission electron micrographs of hyphae in ChMfs1 mutants. (A) Scanning electron micrographs of hyphae in wild-type Ch-1, T-DNA insertion mutant Ch-1-T513, and complementation strain C-ChMfs1-1. Swollen balloon-shaped hyphae were observed in the mutant Ch-1-T513 while complementation strain C-ChMfs1-1 recovered the hyphal morphology. Scale bar = 10 μm. (B) Transmission electron micrographs of hyphae in wild-type, Ch-1-T513, and C-ChMfs1-1. One hypha contained more than one intra-hyphal hypha in the mutant Ch-1-T513 in comparison to the wild-type and the complementation strain. The intra-hyphal strand of hypha has cell walls which clearly distinguish it from being a vacuole. Scale bar = 1 μm.
ChMfs1 Is Associated with Pathogenicity of C. higginsianum
After inoculation with conidial suspensions on 12-day-old Arabidopsis leaves, the mutant Ch-1-T513 failed to produce lesions, while the complementation strain C-ChMfs1-1 could cause necrotic lesions similar to the wild-type at 6 dpi (Figure 5A). Under microscopy, dark appressoria were observed to form on Arabidopsis leaves, but invasive hyphae of the mutant Ch-1-T513 did not differentiate into large primary and thin secondary hyphae, but formed abundant bulbous hyphae in epidermal cells and only three acervuli per leaf were produced (Figures 5B,C). However, conidia of C-ChMfs1-1 and the wild-type formed darkly melanized appressoria which produced penetration pegs into the epidermal cells and resulted in many acervuli (156 acervuli/leaf for C-ChMfs1-1 and 218 for the wild-type) (Figures 5B,C). The mutant Ch-1-T513 produced balloon-shaped hyphae in acervuli with few setae in lesions, whereas the wild-type and C-ChMfs1-1 produced normal acervuli with black setae in lesions (Figure 5D). The abnormal acervuli could not produce enough conidia to let the mutant Ch-1-T513 enter into infection cycle again. Therefore, we concluded that ChMfs1 was involved in the formation of invasive hyphae after penetration and pathogenicity of C. higginsianum.
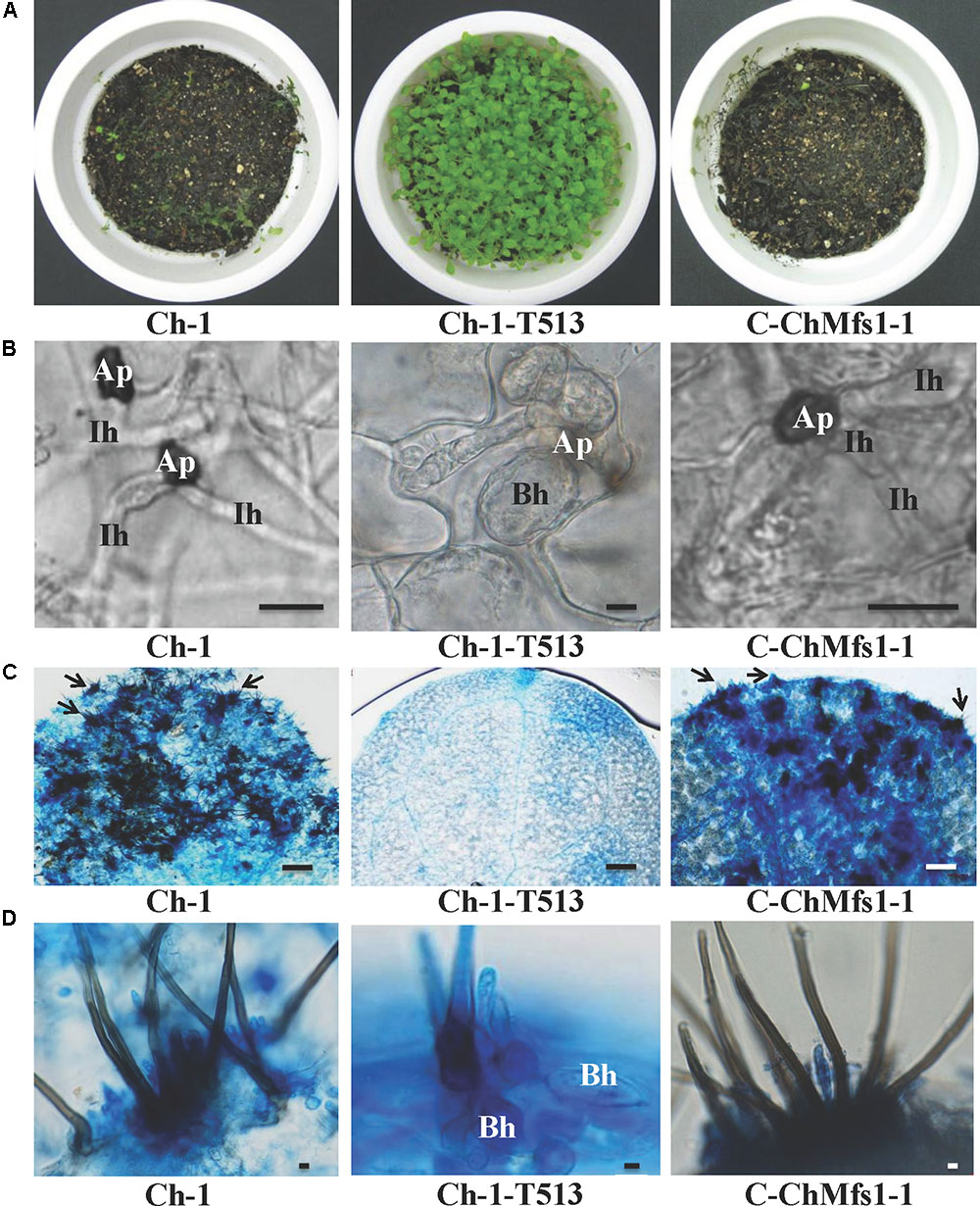
FIGURE 5. Deficiency in pathogenicity and acervulus formation of ChMfs1 mutants on Arabidopsis. (A) Symptoms on 12-day-old seedlings at 6 dpi sprayed with conidial suspensions of wild-type Ch-1, T-DNA insertion mutant Ch-1-T513, and complementation strain C-ChMfs1-1. The wild-type and C-ChMfs1-1 caused completely collapsed and macerated lesions, whereas the mutant Ch-1-T513 caused no symptom. (B) Development of infection structures on Arabidopsis leaves at 4 dpi. Production of dark appressoria and infection hyphae on the surface of Arabidopsis leaves were found in wild-type and C-ChMfs1-1, but infection hyphae were not differentiated from appressoria in the mutant Ch-1-T513. Ap, appressoria; Ih, infection hyphae; Bh, balloon-shaped hyphae. Scale bar = 10 μm. (C) Acervulus formation on Arabidopsis leaves at 4 dpi. Few acervulus were found by mutant Ch-1-T513, but numerous acervulus were produced by the wild-type and complementation strain C-ChMfs1-1. Each arrow indicates an acervulus consisting of several melanized seta. Scale bar = 30 μm. (D) Balloon-shaped hyphae (Bh) formation on acervulus. Few balloon-shaped hyphae were found in the acervulus of the complementation strain C-ChMfs1-1. But numerous balloon-shaped hyphae were produced in acervulus by mutant Ch-1-T513. Scale bar = 5 μm.
Discussion
In this study, we obtained an avirulent mutant Ch-1-T513 by ATMT in C. higginsianum which showed intra-hyphal hyphae. The mutant had double-site T-DNA integrations with insertions upstream of genes for a MFS transporter (ChMfs1) and an aldo/keto reductase (ChAkr). Complementation experiments using the wild-type sequences corresponding to the two T-DNA insertion genes showed that ChMfs1 alone is responsible for the phenotype of mutant Ch-1-T513. In human or animal pathogenic yeasts of Candida genus, a number of MFS transporters are well reported to be involved in host infection and virulence (Costa et al., 2014; Shah et al., 2014; Santos et al., 2017). However, intra-hyphal hyphae were not found in MFS transporter deletion mutants. A few MFS transporters from filamentous fungi also can act as virulence factors. This was reported for the HC toxin transporter from C. carbonum, the cercosporin transporter from C. kikuchii and the trichothecene transporter from F. sporotrichioides (Alexander et al., 1999; Callahan et al., 1999; Pitkin and Mullon, 1999). Among them, MFS transporters for secreting HC toxin or trichothecenes are located in a gene cluster that also encode enzymes required for biosynthesis of these toxins. However, these MFS proteins mentioned above have low level of homology to ChMfs1 in C. higginsianum (with identities up to 31%), suggesting that there are significantly different domains between ChMfs1 and these reported MFS proteins from C. kikuchii, C. carbonum, and F. sporotrichioides. Hence, ChMfs1 in C. higginsianum is the first reported to be involved in pathogenicity and producing intra-hyphal hyphae. We believe that abnormal hyphae in the mutant Ch-1-T513 might be a key factor affecting virulence of C. higginsianum.
Intra-hyphal strands of hyphae refer to an interesting phenomenon where viable hyphae grow within dead or declining hypha cells, and this enables fungi to adapt to adverse conditions (Lane and Garrison, 1970). The formation of intra-hyphal hyphae is widely observed in many plant pathogenic fungi such as Ceratocystis dryocoetidis (Kendrick and Molnar, 1965), Monilinia fructigena (Calonge, 1968), Verticillium albo-atrum (Brown and Wyllie, 1970), V. dahliae (Wright, 1970), Alternaria kikuchiana (Ishizaki et al., 1976), Botryosphaeria dothidea (Kim et al., 2001), Ustilaginoidea virens (Kim and Park, 2007), and Elsinoë fawcettii (Kim and Hyun, 2007). Several impact factors had been reported to have the potential to induce or be associated with the formation of intra-hyphal hyphae in fungi. These include anaerobic cultural conditions (Miller and Anderson, 1961), stress from toxins and antibiotics (Lowry and Sussman, 1966; Ishizaki et al., 1976), injury and aging of mycelia (Chan and Stephen, 1967; Carbonell, 1969), fluidity of cultural media (Kim and Hyun, 2007), and mutation or deletion of genes associated with hyphal growth (Horiuchi et al., 1999; Kim et al., 2004; Takeshita et al., 2006; Martín-Urdíroz et al., 2008; Kim et al., 2009). In this study, formation of intra-hyphal hyphae was observed in the ChMfs1 responding mutant Ch-1-T513 of C. higginsianum. We hypothesized that destruction of this key MFS transporter might result in deficiency of transport of toxic compounds, and abundant toxic compounds accumulated in the fungal cells. And then intra-hyphal hyphae were generated to resist adverse environment. This abnormal mycelial structure also could affect formation of infection structures. This is the first report of a MFS transporter gene associated with such structure formation, and also involved in reduced sporulation and inhibited formation of normal acervuli. These abnormal characteristics are similar to those of found with PDA cultures of the ΔCgChsV mutant strain of C. graminicola (Werner et al., 2007).
Formation of intra-hyphal hyphae in different fungi has been reported to have different origins. In M. fructigena, intra-hyphal hyphae originated from a zone between the cell membrane and the cell wall within a normal hyphal cell next to a degenerated hyphal cell (Calonge, 1968). These were found to originate from septa of existing hyphae in C. dryocoetidis and V. albo-atrum (Kendrick and Molnar, 1965; Brown and Wyllie, 1970). In A. kikuchiana, intra-hyphal hyphae formation was observed to be involved in the cell wall of parental hyphae, which expanded inward to divide the cytoplasm of the parental hyphal cells into several parts (Ishizaki et al., 1976). In this study, we found that formation of intra-hyphal hyphae is involved in pathogenicity and indentation of the hyphal cell wall at the penetration site of infected hyphae, indicating that mechanical force might be associated with the infection process.
Fungicides play an important role in modern agriculture by protecting crops against yield loss. Our results indicate the MFS transporter gene ChMfs1 is a significant virulence factor affecting the infection of C. higginsianum on cruciferous plants. In future work, this virulence factor as a molecular target of C. higginsianum or even Colletotrichum spp. could be used for fungicide or inhibitor design in anthracnose disease management.
Conclusion
In summary, the MFS transporter gene ChMfs1 is important for hyphal morphology, conidation, and pathogenicity in C. higginsianum. Future studies to elucidate molecular mechanisms responsible for the formation of intra-hyphal hyphae in Ch-1-T513 are needed, and such information may lead to development of novel strategies to manage diseases caused by C. higginsianum and other Colletotrichum species.
Author Contributions
Designed the experiments: LL, LZ, JH, and YW. Performed the experiments: LL and YY. Analyzed the experiment data: LL, LZ, YL, and JG. Contributed reagents/materials/analysis tools: LL and LZ. Wrote the paper: LL, LZ, and TH. All authors have read and approved the final manuscript.
Funding
The work was supported by grants funded by the National Natural Science Foundation of China (No. 31101399), the Specialized Research Fund for the Doctoral Program of Higher Education of China (No. 20110146120033), and the Programme of Introducing Talents of Discipline to University (No. D17014).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are grateful to Dr. Daohong Jiang for providing the vectors and Dr. Guoqing Li for reviewing this manuscript.
Footnotes
- ^http://genome.jgi.doe.gov/Colhi1/Colhi1.home.html
- ^http://www.premierbiosoft.com/primerdesign/
- ^http://smart.embl-heidelberg.de/
- ^http://www.clustal.org/clustal2/
- ^http://www.megasoftware.net/index.php
References
Alexander, N. J., McCormick, S. P., and Hohn, T. M. (1999). TRI12, a trichothecene efflux pump from Fusarium sporotrichioides: gene isolation and expression in yeast. Mol. Gen. Genet. 261, 977–984. doi: 10.1007/s004380051046
Brown, M. F., and Wyllie, T. D. (1970). Ultrastructure of microsclerotia of Verticillium albo-atrum. Phytopathology 60, 538–542. doi: 10.1094/Phyto-60-538
Callahan, T. M., Rose, M. S., Meade, M. J., Ehrenshaft, M., and Upchurch, R. G. (1999). CFP, the putative cercosporin transporter of Cercospora kikuchii, is required for wild type cercosporin production, resistance, and virulence on soybean. Mol. Plant Microbe Interact. 12, 901–910. doi: 10.1094/MPMI.1999.12.10.901
Calonge, F. D. (1968). Origin and development of intrahyphal hyphae in Sclerotinia fructigena. Mycologia 60, 933–942. doi: 10.2307/3757395
Cao, J. B., Zhou, Y., Zhang, L., Zhang, J., Yang, L., Qin, L. H., et al. (2011). DsRNA-free transmissible hypovirulence associated with formation of intra-hyphal hyphae in Botrytis cinerea. Fungal Biol. 115, 660–671. doi: 10.1016/j.funbio.2011.04.008
Carbonell, L. M. (1969). Ultrastructure of dimorphic transformation in Paracoccidioides brasiliensis. J. Bacteriol. 100, 1076–1082.
Chan, C., and Stephen, R. C. (1967). Intrahyphal hyphae in the genus Linderina. Can. J. Bot. 45, 1995–1998. doi: 10.1139/b67-217
Costa, C., Dias, P. J., Sá-Correia, I., and Teixeira, M. C. (2014). MFS multidrug transporters in pathogenic fungi: do they have real clinical impact? Front. Physiol. 5:197. doi: 10.3389/fphys.2014.00197
Gong, X., Fu, Y., Jiang, D., Li, G., Yi, X., and Peng, Y. (2007). L-Arginine is essential for conidiation in the filamentous fungus Coniothyrium minitans. Fungal Genet. Biol. 44, 1368–1379. doi: 10.1016/j.fgb.2007.07.007
Hayashi, K., Schoonbeek, H. J., and De Waard, M. A. (2002). Bcmfs1, a novel major facilitator superfamily transporter from Botrytis cinerea, provides tolerance towards the natural toxic compounds camptothecin and cercosporin and towards fungicides. Appl. Environ. Microbiol. 68, 4996–5004. doi: 10.1128/AEM.68.10.4996-5004.2002
Higgins, C. F. (1992). ABC transporters: from microorganisms to man. Annu. Rev. Cell Biol. 8, 67–113. doi: 10.1146/annurev.cb.08.110192.000435
Horiuchi, H., Fujiwara, M., Yamashita, S., Ohta, A., and Takagi, M. (1999). Proliferation of intra-hyphal hyphae caused by disruption of csmA, which encodes a class V chitin synthase with a myosin motor-like domain in Aspergillus nidulans. J. Bacteriol. 181, 3721–3729.
Huser, A., Takahara, H., Schmalenbach, W., and O’Connell, R. J. (2009). Discovery of pathogenicity genes in the crucifer anthracnose fungus Colletotrichum higginsianum, using random insertional mutagenesis. Mol. Plant Microbe Interact. 22, 143–156. doi: 10.1094/MPMI-22-2-0143
Ishizaki, H., Mitsuoka, K., Kohno, M., and Hunoh, H. (1976). Effect of poly-oxin on fungi III: electron microscopic observation of mycelia of Alternaria kikuchiana Tanaka. Ann. Phytopathol. Soc. Japan 42, 35–41.
Kendrick, W. B., and Molnar, A. C. (1965). A new Ceratocystis and its Verticicladiella imperfect stage associated with the bark beetle Dryocoetes confusus on Abies lasiocarpa. Can. J. Bot. 43, 39–43. doi: 10.1139/b65-005
Kim, J. E., Lee, H. J., Lee, J., Kim, K. W., Yun, S. H., Shim, W. B., et al. (2009). Gibberella zeae chitin synthase genes, GzCHS5 and GzCHS7, are required for hyphal growth, perithecia formation, and pathogenicity. Curr. Genet. 55, 449–459. doi: 10.1007/s00294-009-0258-6
Kim, K. W., and Hyun, J. W. (2007). Nonhost-associated proliferation of intra-hyphal hyphae of citrus scab fungus Elsinoe fawcettii: refining the perception of cell-within-a-cell organization. Micron 38, 565–571. doi: 10.1016/j.micron.2006.10.007
Kim, K. W., and Park, E. W. (2007). Ultrastructure of spined conidia and hyphae of the rice false smut fungus Ustilaginoidea virens. Micron 38, 626e631. doi: 10.1016/j.micron.2006.09.006
Kim, K. W., Park, E. W., Kim, Y. H., Ahn, K. K., Kim, P. G., and Kim, K. S. (2001). Latency- and defense-related ultrastructural characteristics of apple fruit tissues infected with Botryosphaeria dothidea. Phytopathology 91, 165–172. doi: 10.1094/PHYTO.2001.91.2.165
Kim, M. J., Park, S. M., Kim, Y. H., Cha, B. J., Yang, M. S., and Kim, D. H. (2004). Deletion of a hypoviral-regulated cppk1 gene in a chestnut blight fungus, Cryphonectria parasitica, results in microcolonies. Fungal Genet. Biol. 41, 482–492. doi: 10.1016/j.fgb.2003.12.006
Lane, J. W., and Garrison, R. G. (1970). Electronmicroscopy of self-parasitism by Histoplasma capsulatum and Blastomyces dermatitidis. Mycopathologia 40, 271–276. doi: 10.1007/BF02051780
Lewis, K. (1994). Multidrug resistance pumps in bacteria: variations on a theme. Trends Biochem. Sci. 19, 119–123. doi: 10.1016/0968-0004(94)90204-6
Li, M., Gong, X., Zheng, J., Jiang, D., Fu, Y., and Hou, M. (2005). Transformation of Coniothyrium minitans, a parasite of Sclerotinia sclerotiorum, with Agrobacterium tumefaciens. FEMS Microbiol. Lett. 243, 323–329. doi: 10.1016/j.femsle.2004.12.033
Liu, L., Zhao, D., Zheng, L., Hsiang, T., Wei, Y., Fu, Y., et al. (2013). Identification of virulence genes in the crucifer anthracnose fungus Colletotrichum higginsianum by insertional mutagenesis. Microb. Pathog. 64, 6–17. doi: 10.1016/j.micpath.2013.06.001
Lowry, R. J., and Sussman, A. S. (1966). Intra-hyphal hyphae in “clock” mutants of Neurospora. Mycologia 58, 541–548. doi: 10.2307/3757032
Martín-Urdíroz, M., Roncero, M. I. G., González-Reyes, J. A., and Ruiz-Roldán, C. (2008). ChsVb, a class VII chitin synthase involved in septation, is critical for pathogenicity in Fusarium oxysporum. Eukaryot. Cell 7, 112–121. doi: 10.1128/EC.00347-07
Miller, C. W., and Anderson, N. A. (1961). Proliferation of conidiophores and intra-hyphal hyphae in Aspergillus niger. Mycologia 53, 433–436.
O’Connell, R. J., Herbert, C., Sreenivasaprasad, S., Khatib, M., EsquerreTugaye, M. T., and Dumas, B. (2004). A novel Arabidopsis-Colletotrichum pathosystem for the molecular dissection of plant-fungal interactions. Mol. Plant Microbe Interact. 17, 272–282. doi: 10.1094/MPMI.2004.17.3.272
Pitkin, J. W., Panaccione, D. G., and Walton, J. D. (1996). A putative cyclic peptide efflux pump encoded by the TOXA gene of the plant pathogenic fungus Cochliobolus carbonum. Microbiology 142, 1557–1565. doi: 10.1099/13500872-142-6-1557
Pitkin, Z., and Mullon, C. (1999). Evidence of absence of porcine endogenous retrovirus (PERV) infection in patients treated with a bioartificial liver support system. Artif. Organs 23, 829–833. doi: 10.1046/j.1525-1594.1999.06444.x
Qin, L., Gong, X., Xie, J., Jiang, D., Cheng, J., Li, G., et al. (2011). Phosphoribosylamidotransferase, the first enzyme for purine de novo synthesis, is required for conidiation in the sclerotial mycoparasite Coniothyrium minitans. Fungal Genet. Biol. 48, 956–965. doi: 10.1016/j.fgb.2011.06.007
Roohparvar, R., De Waard, M. A., Kema, G. H., and Zwiers, L. H. (2007). MgMfs1, a major facilitator superfamily transporter from the fungal wheat pathogen Mycosphaerella graminicola, is a strong protectant against natural toxic compounds and fungicides. Fungal Genet. Biol. 44, 378–388. doi: 10.1016/j.fgb.2006.09.007
Santos, R., Costa, C., Mil-Homens, D., Romão, D., de Carvalho, C. C., Pais, P., et al. (2017). The multidrug resistance transporters CgTpo1_1 and CgTpo1_2 play a role in virulence and biofilm formation in the human pathogen Candida glabrata. Cell. Microbiol. 19:e12686. doi: 10.1111/cmi.12686
Shah, A. H., Singh, A., Dhamgaye, S., Chauhan, N., Vandeputte, P., Suneetha, K. J., et al. (2014). Novel role of a family of major facilitator transporters in biofilm development and virulence of Candida albicans. Biochem. J. 460, 223–235. doi: 10.1042/BJ20140010
Takahara, H., Huser, A., and O’Connell, R. J. (2012). Two arginine biosynthesis genes are essential for pathogenicity of Colletotrichum higginsianum on Arabidopsis. Mycology 3, 54–64. doi: 10.1080/21501203.2011.654353
Takeshita, N., Yamashita, S., Ohta, A., and Horiuchi, H. (2006). Aspergillus nidulans class V and VI chitin synthases CsmA and CsmB, each with a myosin motor-like domain, perform compensatory functions that are essential for hyphal tip growth. Mol. Microbiol. 59, 1380–1394. doi: 10.1111/j.1365-2958.2006.05030.x
Werner, S., Sugui, J. A., Steinberg, G., and Deising, H. B. (2007). A Chitin synthase with a myosin-like motor domain is essential for hyphal growth, appressorium differentiation, and pathogenicity of the maize anthracnose fungus Colletotrichum graminicola. Mol. Plant Microbe Interact. 12, 1555–1567. doi: 10.1094/MPMI-20-12-1555
Keywords: Colletotrichum higginsianum, infection, intra-hyphal hyphae, major facilitator superfamily transporter, pathogenicity
Citation: Liu L, Yan Y, Huang J, Hsiang T, Wei Y, Li Y, Gao J and Zheng L (2017) A Novel MFS Transporter Gene ChMfs1 Is Important for Hyphal Morphology, Conidiation, and Pathogenicity in Colletotrichum higginsianum. Front. Microbiol. 8:1953. doi: 10.3389/fmicb.2017.01953
Received: 22 June 2017; Accepted: 22 September 2017;
Published: 10 October 2017.
Edited by:
Dominique Sanglard, University of Lausanne, SwitzerlandReviewed by:
Priya Uppuluri, University of California, Los Angeles, United StatesMiguel Cacho Teixeira, Universidade de Lisboa, Portugal
Copyright © 2017 Liu, Yan, Huang, Hsiang, Wei, Li, Gao and Zheng. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) or licensor are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lu Zheng, bHV6aGVuZ0BtYWlsLmh6YXUuZWR1LmNu
†These authors have contributed equally to this work.
 Liping Liu
Liping Liu Yaqin Yan
Yaqin Yan Junbin Huang1
Junbin Huang1 Yangdou Wei
Yangdou Wei Lu Zheng
Lu Zheng