- 1National Navel Orange Engineering Research Center, College of Life and Environmental Sciences, Gannan Normal University, Ganzhou, China
- 2Institute of Translational Medicine, Nanchang University, Nanchang, China
Huanglongbing (HLB) is the most devastating citrus disease worldwide, and suppression of the Asian citrus psyllid (Diaphorina citri) is regarded as an effective method to inhibit the spread of HLB. In this study, we isolated a strain named as Serratia marcescens KH-001 from D. citri nymphs suffering from disease, and evaluated its killing effect on D. citri via toxicity test and effect on microbial community in D. citri using high-throughput sequencing. Our results indicated that S. marcescens KH-001 could effectively kill 83% of D. citri nymphs, while the fermentation products of S. marcescens KH-001 only killed 40% of the D. citrinymphs. High-throughput sequencing results indicated that the S. marcescens KH-001 increased the OTU numbers from 62.5 (PBS buffer) to 81.5, while significantly lowered the Shannon index compared with Escherichia coli DH5α (group E) (p < 0.05). OTU analysis showed that the S. marcescens KH-001 had significantly reduced the relative abundance of endosymbionts Wolbachia, Profftella, and Carsonella in group S compared with that in other groups (p < 0.05). Therefore, the direct killing effect of the fermentation products of S. marcescens KH-001 and the indirect effect via reducing the numbers of endosymbionts (Wolbachia, Profftella, and Carsonella) of D. citri endow S. marcescens KH-001 a sound killing effect on D. citri. Further work need to do before this strain is used as a sound biological control agents.
Introduction
Huanglongbing (HLB, also known as citrus greening) is a devastating citrus disease caused by Candidatus Liberibacter spp., which is first reported in China in 1943 and has been reported in at least 40 citrus producing countries (Bové, 2006). Till now, there is no cure for HLB once the trees get infected, and traditional HLB managements rely on psyllid control, aggressive removal of infected trees and planting of disease-free nursery trees (Grafton-Cardwell et al., 2013; Alvarez et al., 2016). However, growers are reluctant to follow these traditional HLB managements because it is costly and difficult to replant trees and get them into production, therefore most growers choose not to remove diseased trees if they remain productive (Hall and Gottwald, 2011).
Three different species of Candidatus Liberibacter have been identified, e.g., Candidatus Liberibacter asiaticus (Las), Candidatus Liberibacter africanus, and Candidatus Liberibacter americanus. Among them, Las is the most widely distributed HLB pathogen and is vectored by Asian citrus psyllid (ACP) Diaphorina citri Kuwayama (Hemiptera: Liviidae) (Bové, 2006). D. citri depends heavily on young citrus flush for survival and reproduction, and the feeding and reproductive behavior of D. citri plays an important role in the spread of Las. D. citri adults prefer emerging plant tissues for oviposition (Halbert and Manjunath, 2004), which make young trees particularly prone to Las infection. Moreover, the volatile methyl salicylate compound released by the infected trees attracts D. citri adults (Mann et al., 2012). Therefore, suppression or reduction of D. citri has been the primary method to inhibit the spread of HLB (Saha et al., 2017).
Biological control is one of the most promising approaches to control pests (Peshin et al., 2009; Alvarez et al., 2016), which offers an alternative to chemical pest control for its high selectivity, neglected effect on environment (Abdel-Baky and Abdel-Salam, 2003). Here, we isolated a bacterium from D. citri nymphs suffered from disease in citrus orchard (Gannan Normal University, Ganzhou City, Jiangxi Province), which was identified as Serratia marcescens using biochemical identification and molecular biological identification. Then this stain was named as S. marcescens KH-001 and stored at China Center for Type Culture Collection (Registration No. CCTCC M 2017465).
As we known, the microbiome hosted in D. citri played an important role on host growth, development, spawning (Coletta-Filho et al., 2005). As a powerful tool, high-throughput sequencing technology can detect almost all the DNA signatures of microbes within specific environments, even the bacteria with a low numbers or in the dormant metabolic state (Kobayashi et al., 2006; Wang et al., 2011; Zhang et al., 2012), and the high distinguish ability and sensitivity of the high-throughput sequencing provided us the actual microbial composition in D. citri. Therefore, our group investigated the killing effects of fermented supernatant of S. marcescens KH-001, Escherichia coli DH5α and S. marcescens KH-001 on D. citri, and we also studied their effects on the parasitic microbial diversity in D. citri using high-throughput sequencing method, to explore the possible killing mechanism of S. marcescens KH-001 on D. citri.
Materials and Methods
Bacteria Isolation and Identification
The D. citri nymphs suffered from disease in citrus orchard (Gannan Normal University, Ganzhou City, Jiangxi Province) were sampled, then they received surface sterilization using 75% ethanol for 5 min, and washed for three times with sterile water in a clean bench. Then washed D. citri were placed onto the LB solid medium, and the D. citri were broken down using transferring loop and cultured for 24 h at 28°C. Then the red colonies were picked out and purified using agar-streak method. Forty five strains were randomly selected and identified using biochemical identification and molecular biological identification, and a strain of S. marcescens were finally selected based on its sound killing effect on D. citri nymphs, which was named as S. marcescens KH-001 and stored at China Center for Type Culture Collection (Registration No. CCTCC M 2017465).
Virulence of the S. marcescens KH-001 on D. citri
The D. citri Kuwayama (Hemiptera: Liviidae) were collected from citrus orchard (Tandong Town, Ganzhou City, Jiangxi Province 25°47′38′′ N, 114°25′4′′ E), seedling at 27 ± 1°C, 68% RH, and 14:10 h L:D photoperiod. The D. citri was maintained on Murraya exotica L. and was consecutive propagated for 20 generations before experiment.
Serratia marcescens KH-001 and E. coli DH5α were cultured in 5 ml LB medium and cultured for 6 h at 28°C. Then 1% of the above-mentioned mixture was inoculated into 5 ml LB medium and cultured for another 12 h. The cultures were centrifuged at 4500 ×g for 10 min at 4°C, and were resuspended using PBS buffer. Then D. citri (Fifth instar nymphs) were randomly divided into four groups: C, treated with PBS (N = 40); A, treated with the fermented supernatant of S. marcescens KH-001 (N = 40); E, treated with the E. coli DH5α containing in PBS buffer (N = 40); S, treated with the S. marcescens KH-001 containing in PBS buffer (N = 40). For all groups, 0.4 μl liquid were dropped onto the back of D. citri, and all D. citri were covered with cling film, and placed in the incubator (27 ± 1°C, 14 h light:10 h dark), and the survival rate of D. citri were recorded every 3 h.
Extraction of Genome DNA and High-Throughput Sequencing
As it is difficult to isolated sufficient DNAs from a single D. citri, so 10 D. citri were mixed as one sample for the DNA extraction. The genomic DNAs of each sample were extracted by the TIANamp Genomic DNA kit (TIANGEN) (Yu et al., 2015). The extracted DNAs were used as templates, and the universal primer pair 338F/806R with the respective barcode for ease of identification were used to amplify the V3–V4 region of 16S ribosomal (r)RNA genes of all samples (GenBank Accession No. SUB3652522). PCR reactions, pyrosequencing of the PCR amplicons and quality control of raw data were performed as described previously (Xu et al., 2015)
Bioinformatics and Multivariate Statistics
To eliminate the low-quality sequences, PyroNoise algorithm in Mothur (version 1.33.3) were used (Schloss et al., 2009), and the Quantitative Insights Into Microbial Ecology (QIIME) platform (version 1.8.0) was implemented for Bioinformatics analysis (Caporaso et al., 2010). Briefly, 16S rRNA operational taxonomic units (OTUs) were clustered using an open-reference OTU picking protocol based on 97% nucleotide similarity with the UCLUST algorithm (Davenport et al., 2014), and ChimeraSlayer was applied to remove chimeric sequences (Haas et al., 2011). The relative abundance of each OTU was determined as a proportion of the sum of sequences for every sample, taxonomic relative abundance profiles were generated based on OTU annotation, and the microbial community structure was evaluated by biodiversity. Shannon index, phylogenetic diversity, chao1 index and the observed number of species were used to evaluate α diversity, and the weighted and unweighted UniFrac distances were used to evaluate β diversity using QIIME pipeline.
Statistical Analysis
The statistical significance of the relationships among different groups was evaluated by Wilcoxon and Kruskal–Wallis tests and Spearman’s rank correlation. The data were presented as mean ± SD, and p < 0.05 or p < 0.01 was considered statistically significant (Lu et al., 2014).
Results
Bacterial Isolation and Its Virulence on D. citri Nymphs
To isolate the potential bacteria killing D. citri nymphs, we sampled the D. citri nymphs suffering disease and 45 bacteria were isolated from them. Finally, S. marcescens KH-001 were selected for our further evaluation.
Then we treated the artificial rearing D. citri nymphs using PBS buffer (group C), supernatant of S. marcescens KH-001 (group A), E. coli DH5α (group E), and S. marcescens KH-001 (group S). In Figure 1, our results indicated that no D. citri nymphs was dead in negative control group C, only 20% of D. citri nymphs were killed by the control strain of E. coli DH5α. Interestingly, the supernatant of S. marcescens KH-001 killed about 40% of D. citri nymphs, while S. marcescens KH-001 possessed the best killing rate of 83% in group S (p < 0.05).
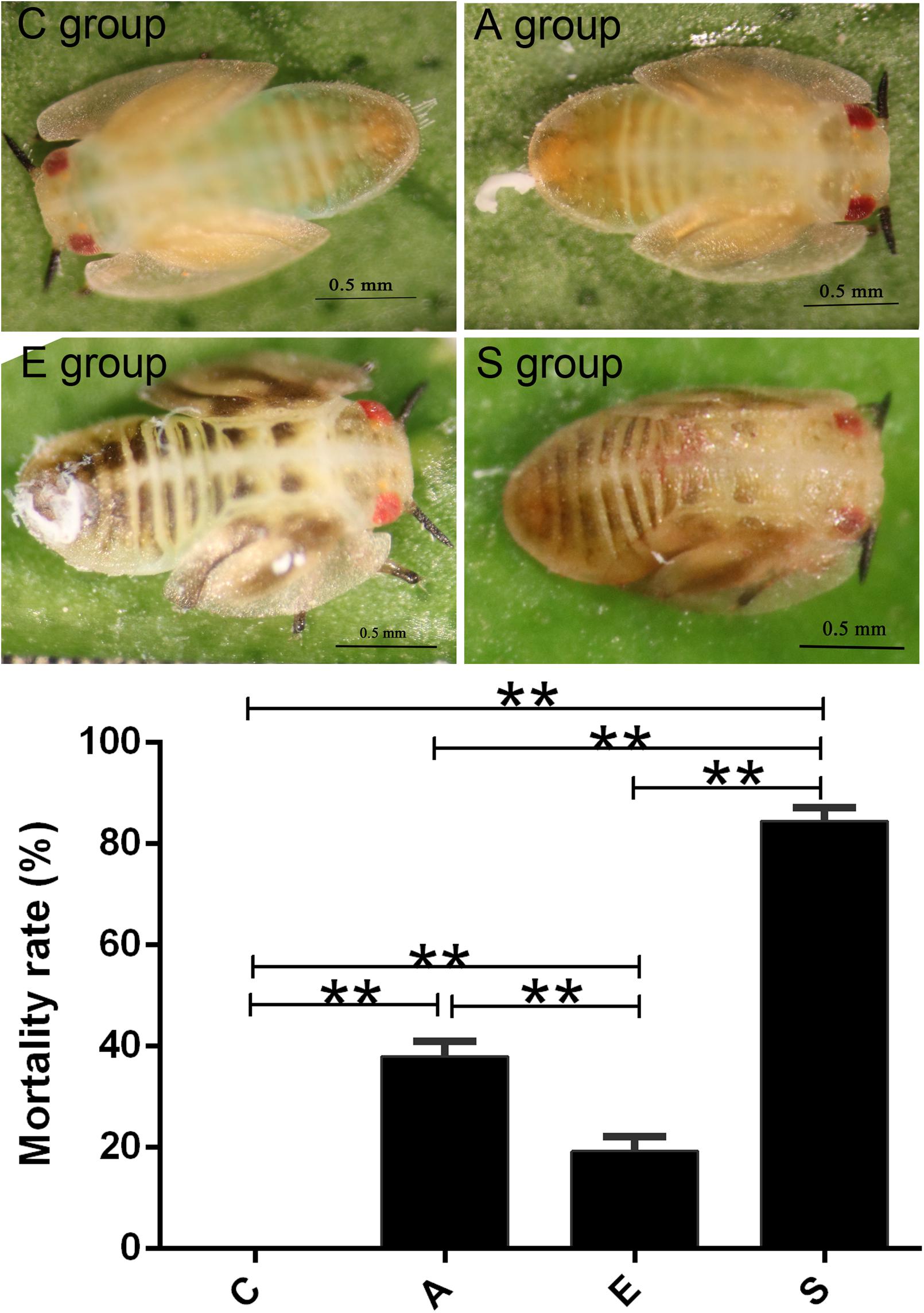
FIGURE 1. Killing effect of the PBS (C), Fermentation product of Serratia marcescens KH-001 (A), Escherichia coli DH5α (E), and S. marcescens (S) KH-001 on Diaphorina citri at 24 h. ∗p < 0.05; ∗∗p < 0.01.
The OUT Analysis of Groups C, A, E, and S
To evaluate the effects of fermented supernatant of S. marcescens KH-001, E. coli DH5α and S. marcescens KH-001 on microbial diversity in D. citri, 16S rRNA amplicon sequencing analysis was used to sequence the V3-V4 hypervariable region. All the effective tags of all samples were clustered and those sequences with over 97% similarity were considered as one OTU. In total, 1,062,236 usable raw sequences and 1,331 OTUs were obtained from all the samples with an average of 83.18 OTUs per group (Figure 2A). Moreover, the Shannon index indicated that the relative abundance of group E was higher than group A (p < 0.05), and the relative abundance of group S was significantly lower than that in group E (p < 0.01).
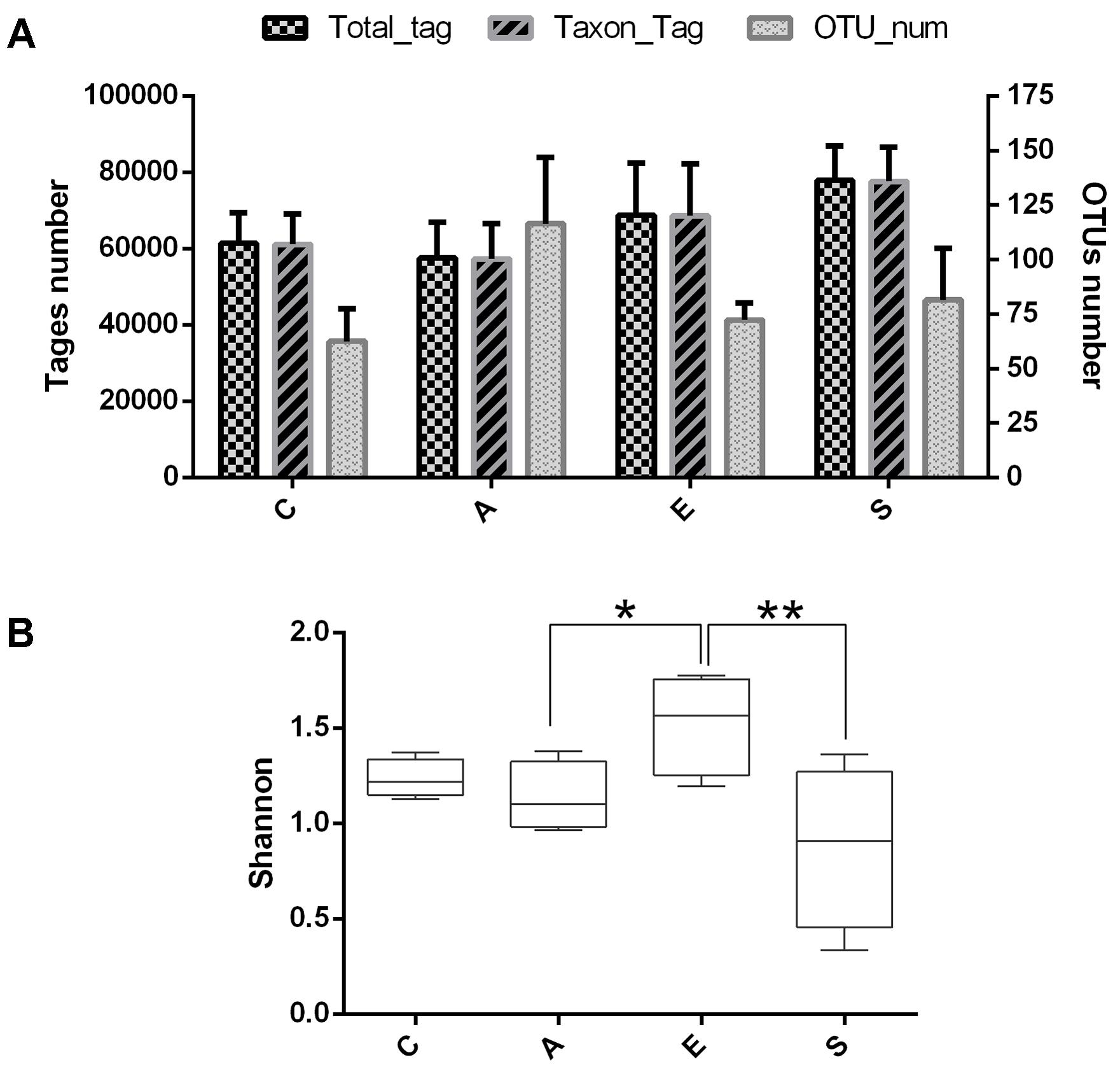
FIGURE 2. The OUT analysis of groups C, A, E, and S. (A) Number of total tags, taxon tags, and OTUs in groups C, A, E, and S. (B) The alpha-diversity distances calculated using phylotype relative abundance measurements among groups C, A, E, and S. C, PBS group; A, Fermentation product of S. marcescens KH-001 group; E, E. coli DH5α group; S, S. marcescens KH-001 group. ∗p < 0.05; ∗∗p < 0.01.
The β Diversity of the Microbial Community in Groups C, A, E, and S
As shown in Figure 3A, data of the top 10 microorganism populations was analyzed using unweighted Pair-group Method with Arithmetic Mean (UPGMA) to check the similarity between different groups (24). Proteobacteria, Firmicutes, Actinobacteria, and Bacteroidetes constituted four common dominant phyla and accounted for >90% of the total sequencing number in all groups. When treated with fermented supernatant of S. marcescens KH-001 (A), E. coli DH5α (E) and S. marcescens KH-001 (S), the relative abundance of Firmicutes and Bacteroidetes received an obvious reduction, while the abundance of Proteobacteria and Actinobacteria were greatly enhanced (Figure 3A). Moreover, the UPGMA and principal component analysis (PCA) results indicated that the microbial composition in groups C, E, and S possessed high similarity, while group A scattered away from groups C, E, and S (Figure 3B).
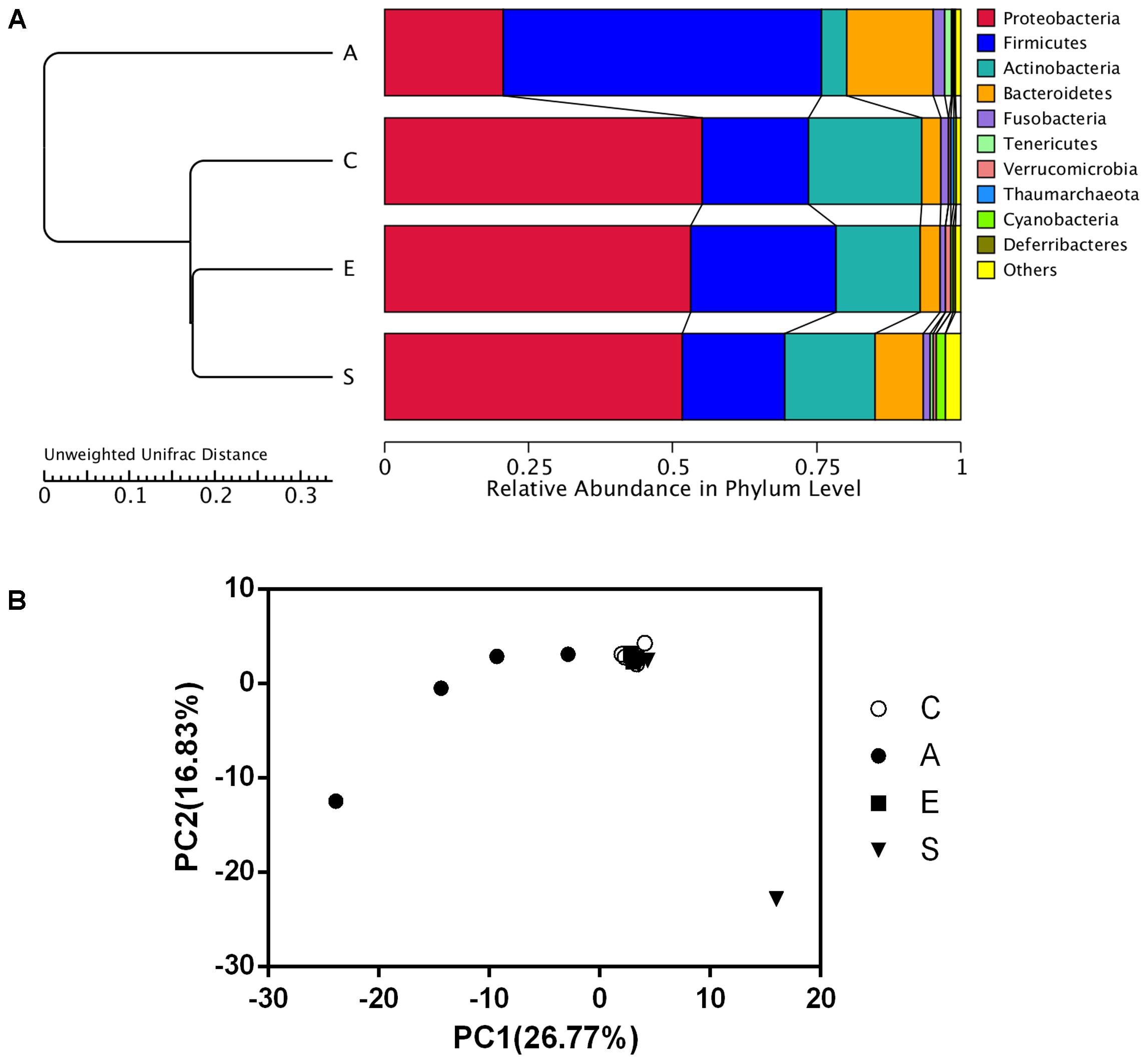
FIGURE 3. The UPGMA clustering tree Based on the Weighted UniFrac distance at phylum level (A) and the principle component analysis (PCA) of the microbial diversity (B) in groups C, A, E, and S. C, PBS group; A, Fermentation product of S. marcescens KH-001 group; E, E. coli DH5α group; S, S. marcescens KH-001 group.
The Specificity of Bacterial Communities in Groups C, A, E, and S
The Venn figure reflecting the difference among groups C, A, E and S was shown in Figure 4A, there were 96, 205, 103, and 130 OTUs in groups C, A, E, and S, respectively. The comparison among all groups was done and only 34 common OTUs were detected. In addition, the statistically significant differences among groups C, A, E, and S indicated that the relative abundance of Wolbachia, Anaplasmataceae, Alphaproteobacteria, Gammaproteobacteria, Rickettsiales, Carsonella in C group was significantly higher than that in groups A, E, and S (Figure 4B, p < 0.05).
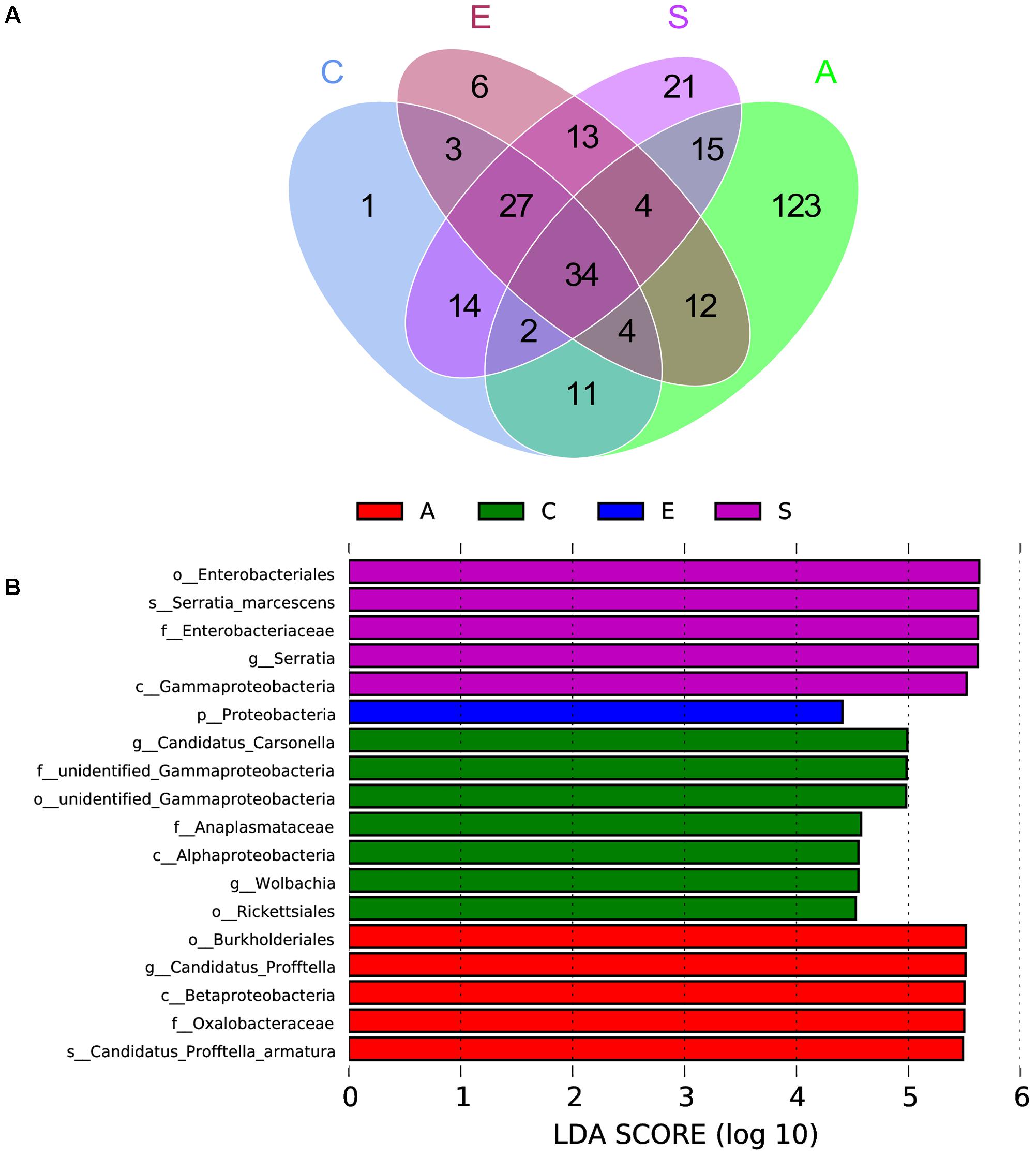
FIGURE 4. The scalar-Venn representation of shared genera among microbiome in groups C, A, E, and S (A) and the taxonomic representation of statistically significant differences among groups C, A, E, and S (B). C, PBS group; A, Fermentation product of S. marcescens KH-001 group; E, E. coli DH5α group; S, S. marcescens KH-001 group.
The Relative Abundance of Bacteria Related to D. citri
Based on the sequencing results, we compared the relative abundance of Serratia, Wolbachia, Profftella, and Carsonella in groups C, A, E, and S. The Figure 5 showed that the Serratia in group S was significantly higher than that in groups C, A, and E (p < 0.01); the Wolbachia in group C was significantly higher than that in group S (p < 0.01); the Profftella and Carsonella in group S was significantly lower than that in groups C, A, and E (p < 0.01).
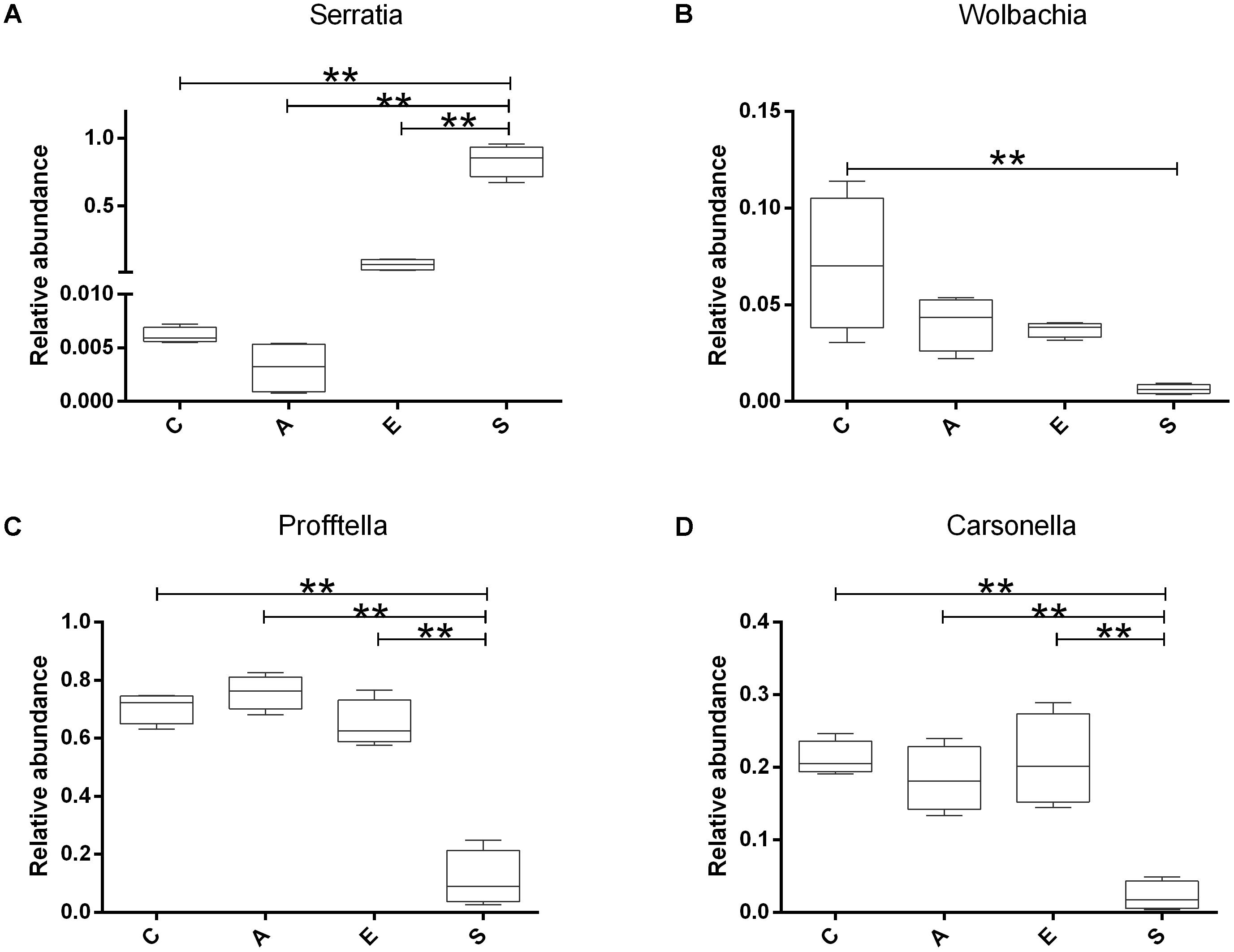
FIGURE 5. The relative abundance of Serratia (A), Wolbachia (B), Profftella (C), and Carsonella (D) in groups C, A, E, and S. C, PBS group; A, Fermentation product of S. marcescens KH-001 group; E, E. coli DH5α group; S, S. marcescens KH-001 group. ∗p < 0.05; ∗∗p < 0.01.
Discussion
HBL were caused by the Candidatus Liberibacter spp. in phloem. It is estimated that the HBL occurred in more than 40 countries, and infected about billions of citrus plants, which caused 30–100% of the Citrus production (Crafts-Brandner, 2002; Bové, 2006; Iftikhar et al., 2016). In Florida, a production loss of $1.7 billion between 2006 and 2011 were observed, and consumers and producers lost an estimated $1 billion in the 2012–2013 season (Alvarez et al., 2016).
Diaphorina citri Kuwayama (Hemiptera: Liviidae) was the most important insect vector for HBL (Hall and Hentz, 2016; Saha et al., 2017). Numerous researches indicated that insect borne pathogen could affect the growth and development of media insects, e.g., the tomato yellow leaf curl virus could decrease the Fecundity and survival of Bemisia tabaci, and the Alfalfa mosaic virus could lower the survival rate and population growth rate of Aphids (Donaldson and Gratton, 2007). Therefore, our group sampled the D. citri nymphs suffered from disease in citrus orchard, and isolated a potential strain (S. marcescens KH-001) to kill the D. citri nymphs based on the previous report (Arp et al., 2017).
First of all, we compared the killing effect of supernatant of S. marcescens KH-001 (group A) and S. marcescens KH-001 (group S) on D. citri nymphs, and the PBS (group C), and E. coli DH5α (group E) were used as control groups. The virulence experiment indicated that S. marcescens KH-001 could effectively kill 83% of D. citri nymphs, while the killing rates were only 40 and 20% in group A (supernatant of S. marcescens KH-001) and group E (E. coli DH5α), indicating that the directly killing effect of the fermentation products of S. marcescens KH-001 was not the sole reason to kill D. citri nymphs (Figure 1).
A recent study indicated that D. citri experienced significant mortality when exposed to S. marcescens because of the lacking of antimicrobial peptides and the Imd pathway, revealing a reduced innate immune system defensed against Gram negative bacteria (Arp et al., 2017). However, no study is carried out to explore the relationship tween S. marcescens and the microbial diversity in D. citri. In nature, a surprisingly high number of endosymbionts harbored in the arthropods, and these endosymbionts had greatly influenced the nutrition, reproduction, evolution, and survival of their host (Zug and Hammerstein, 2012). Therefore, the high-throughput sequencing technology was used to investigate the effect of S. marcescens KH-001 on the microbial diversity hosting in D. citri nymphs.
Our results indicated that an average of 83.18 OTUs per group was obtained, the supernatant of S. marcescens KH-001 and S. marcescens KH-001 increased the OTU numbers in groups A and S from 62.5 (group C) to 116.5 and 81.5, respectively (Figure 1). For group E, though E. coli DH5α possessed little effect on the OTU numbers compared with group C, its Shannon index was significantly higher than that in groups A and S (p < 0.05). As we known, the Shannon index is an important indicator of bacterial diversity, and the higher microbial diversity is regarded as a key point for host to defense the alien invasions (Chen et al., 2011, 2012; Zhang et al., 2015; Erdman, 2016; Kakihana et al., 2016). Therefore, the significant reduction of Shannon index caused by S. marcescens KH-001 indicated that S. marcescens KH-001 could effectively kill D. citri nymphs via reducing the microbial diversity to weak their defense to external interference (Figure 2).
In addition, data of top 10 microorganism populations was analyzed using UPGMA to check the similarity among different groups (24). Proteobacteria, Firmicutes, Actinobacteria, and Bacteroidetes constituted four common dominant phyla and accounted for >90% of the total sequencing number in all groups. When treated with fermented supernatant of S. marcescens KH-001 (A), E. coli DH5α (E) and S. marcescens KH-001, the relative abundance of Firmicutes and Bacteroidetes received an obvious reduction, while the abundance of Proteobacteria and Actinobacteria were greatly enhanced (Figure 3A). Moreover, the UPGMA and PCA results indicated that the microbial composition in groups C, E, and S possessed high similarity, scattering away from group A (Figure 3B).
Next, we compared the OTUs in groups C, A, E, and S (Figure 4). The Venn results indicated there were 96, 205, 103, and 130 OTUs in groups C, A, E and S, and their common OTUs was only 34, and the unique OTUs specifically contained in groups C, A, E, and S were 1, 123, 6, and 21 (Figure 4A), and the relative number of Wolbachia, Anaplasmataceae, Alphaproteobacteria, Gammaproteobacteria, Rickettsiales, Carsonella in C group were significantly higher than that in other groups (p < 0.05) (Figure 4B).
In previous studies, researchers found that the endosymbionts of Wolbachia, Profftella, and Carsonella possessed high infection rate on D. citri (Guidolin and Cônsoli, 2013; Sun et al., 2014; Arp et al., 2017). Of which Wolbachia was regarded as one of the world’s most common parasitic microbes infected a high proportion of insects. As many as 25 to 70% of all insect species were estimated to be potential hosts of Wolbachia, and some host species could not reproduce, or even survive, without Wolbachia infection (Werren et al., 1995; Kozek et al., 2007). Carsonella was an endosymbiont was not only parasitic in its host insect, but also supplied the host with some essential amino acids (Thao et al., 2001; Tortora et al., 2007). Profftella was a Betaproteobacterium, could produce a defensive polyketide (diaphorin) and it could potential influence the biology of D. citri (Kondo et al., 2005; Fischer and Seitz, 2012). Therefore, we compared the relative abundance of Serratia, Wolbachia, Profftella, and Carsonella in groups C, A, E, and S (Figure 5). Our results indicated that the Serratia in group S was significantly higher than other groups (p < 0.01), and the few OTU numbers of Serratia in groups C, A, and E indicated that the Serratia was a common genius in D. citri nymphs (Figure 5A). Moreover, the addition of S. marcescens KH-001 had significantly reduced the endosymbionts of Wolbachia, Profftella and Carsonella, while the supernatant produced by the S. marcescens KH-001 posted little effect on these three genuses (Figure 5). Considering the important role of Wolbachia, Profftella, and Carsonella in the reproduce, development and survive of D. citri, we safely proposed that S. marcescens KH-001 might play its sound killing effect on D. citri via reducing the numbers of endosymbionts (Wolbachia, Profftella, and Carsonella) in D. citri (Figures 1, 4, 5).
In the present study, our group isolated a S. marcescens from D. citri nymphs suffering disease, and our results indicated that this stain could effetely kill D. citri via reducing the number of endosymbiosis, together with the direct killing effect of the supernatant (Arp et al., 2017). We first explored the potential relationship between the killing effect of S. marcescens KH-001 and the microbial diversity of endosymbionts in D. citri, and provided basic data to use this strain as sound biological control agent. However, limited by the differences of host and geographical location, interactions between symbionts and HLB pathogen, much more work is need before this strain is applied in the biological control of the HBL via killing the insect vector of D. citri.
Author Contributions
TC and NZ designed the experiment. WH, FK, and ZL performed the experiments. TC analyzed the data and wrote the manuscript. All authors discussed the results and commented on the manuscript.
Funding
This study was supported by National Natural Science Foundation of China (Grant Nos. 31560602, 81503364, and 31560264), Key Program of Science and Technology Planning Project of Jiangxi Province (Grant No. 20171BBF60076), Natural Science Foundation of Jiangxi Province (Grant Nos. 20161BAB214178, 20171BAB214029, 20171BCB23028, and 20175526), and Foundation of Jiangxi Province Educational Committee (Grant Nos. GJJ 170832 and GJJ 151007).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Abdel-Baky, N. F., and Abdel-Salam, A. (2003). Natural incidence of Cladosporium spp. as a bio-control agent against whiteflies and aphids in Egypt. J. Appl. Entomol. 127, 228–235. doi: 10.1046/j.1439-0418.2003.00662.x
Alvarez, S., Rohrig, E., Solís, D., and Thomas, M. H. (2016). Citrus greening disease (Huanglongbing) in Florida: economic impact, management and the potential for biological control. Agric. Res. 5, 109–118. doi: 10.1007/s40003-016-0204-z
Arp, A. P., Martini, X., and Pelz-Stelinski, K. S. (2017). Innate immune system capabilities of the Asian citrus psyllid. Diaphorina citri. J. Invertebr. Pathol. 148, 94–101. doi: 10.1016/j.jip.2017.06.002
Bové, J. M. (2006). Huanglongbing: a destructive, newly-emerging, century-old disease of citrus. J. Plant Pathol. 88, 7–37.
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Chen, T., Jiang, S., Xiong, S., Wang, M., Zhu, D., and Wei, H. (2012). Application of denaturing gradient gel electrophoresis to microbial diversity analysis in Chinese Douchi. J. Sci. Food Agric. 92, 2171–2176. doi: 10.1002/jsfa.5604
Chen, T., Tan, Q., Wang, M., Xiong, S., Jiang, S., Wu, Q., et al. (2011). Identification of bacterial strains in viili by molecular taxonomy and their synergistic effects on milk curd and exopolysaccharides production. Afr. J. Biotechnol. 10, 16969–16975.
Coletta-Filho, H., Takita, M., Targon, M., and Machado, M. (2005). Analysis of 16S rDNA sequences from citrus huanglongbing bacteria reveal a different “Ca. Liberibacter” strain associated with citrus disease in Sao Paulo. Plant Dis. 89, 848–852. doi: 10.1094/PD-89-0848
Crafts-Brandner, S. J. (2002). Plant nitrogen status rapidly alters amino acid metabolism and excretion in Bemisia tabaci. J. Insect Physiol. 48, 33–41. doi: 10.1016/S0022-1910(01)00140-8
Davenport, M., Poles, J., Leung, J. M., Wolff, M. J., Abidi, W. M., Ullman, T., et al. (2014). Metabolic alterations to the mucosal microbiota in inflammatory bowel disease. Inflamm. Bowel Dis. 20, 723–731. doi: 10.1097/MIB.0000000000000011
Donaldson, J. R., and Gratton, C. (2007). Antagonistic effects of soybean viruses on soybean aphid performance. Environ. Entomol. 36, 918–925. doi: 10.1093/ee/36.4.918
Erdman, S. E. (2016). Gut microbiota: microbes offer engineering strategies to combat cancer. Nat. Rev. Gastroenterol. Hepatol. 13, 125–126. doi: 10.1038/nrgastro.2016.14
Fischer, H., and Seitz, F. (2012). Characterization of the relative abundance of the citrus pathogen Ca. liberibacter asiaticus in the microbiome of its insect vector, Diaphorina citri, using High Throughput 16S rRNA Sequencing. Open Microbiol. J. 6, 29–33. doi: 10.2174/1874285801206010029
Grafton-Cardwell, E. E., Stelinski, L. L., and Stansly, P. A. (2013). Biology and management of Asian citrus psyllid, vector of the huanglongbing pathogens. Annu. Rev. Entomol. 58, 413–432. doi: 10.1146/annurev-ento-120811-153542
Guidolin, A. S., and Cônsoli, F. L. (2013). Molecular characterization of Wolbachia strains associated with the invasive Asian citrus psyllid Diaphorina citri in Brazil. Microb. Ecol. 65, 475–486. doi: 10.1007/s00248-012-0150-7
Haas, B. J., Gevers, D., Earl, A. M., Feldgarden, M., Ward, D. V., Giannoukos, G., et al. (2011). Chimeric 16S rRNA sequence formation and detection in Sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21, 494–504. doi: 10.1101/gr.112730.110
Halbert, S. E., and Manjunath, K. L. (2004). Asian citrus psyllids (Sternorrhyncha: Psyllidae) and greening disease of citrus: a literature review and assessment of risk in Florida. Fla. Entomol. 87, 330–353. doi: 10.1653/0015-4040(2004)087[0330:ACPSPA]2.0.CO;2
Hall, D. G., and Gottwald, T. R. (2011). Pest management practices aimed at curtailing citrus huanglongbing disease. Outlooks Pest Manage. 22, 189–192. doi: 10.1564/22aug11
Hall, D. G., and Hentz, M. G. (2016). Asian citrus psyllid (Hemiptera: Liviidae) tolerance to heat. Ann. Entomol. Soc. Am. 107, 641–649. doi: 10.1603/AN13169
Iftikhar, Y., Rauf, S., Shahzad, U., and Zahid, M. A. (2016). Huanglongbing: pathogen detection system for integrated disease management – A review. J. Saudi Soc. Agric. Sci. 15, 1–11. doi: 10.1016/j.jssas.2014.04.006
Kakihana, K., Fujioka, Y., Suda, W., Najima, Y., Kuwata, G., Sasajima, S., et al. (2016). Fecal microbiota transplantation for patients with steroid-resistant/dependent acute graft-versus-host disease of the gut. Blood 128, 2083–2088. doi: 10.1182/blood-2016-05-717652
Kobayashi, N., Bauer, T. W., Tuohy, M. J., Lieberman, I. H., Krebs, V., Togawa, D., et al. (2006). The comparison of pyrosequencing molecular Gram stain, culture, and conventional Gram stain for diagnosing orthopaedic infections. J. Orthop. Res. 24, 1641–1649. doi: 10.1002/jor.20202
Kondo, N., Shimada, M., and Fukatsu, T. (2005). Infection density of Wolbachia endosymbiont affected by co-infection and host genotype. Biol. Lett. 1, 488–491. doi: 10.1098/rsbl.2005.0340
Kozek, W. J., Rao, R. U., and Hoerauf, A. (2007). The discovery of Wolbachia in arthropods and nematodes - a historical perspective. Issues Infect. Dis. 5, e16–e18. doi: 10.1159/000104228
Lu, K., Ryan, P. A., Schlieper, K. A., Graffam, M. E., Levine, S., Wishnok, J. S., et al. (2014). Arsenic exposure perturbs the gut microbiome and its metabolic profile in mice: an integrated metagenomics and metabolomics analysis. Environ. Health Perspect. 122, 284–291. doi: 10.1289/ehp.1307429
Mann, R. S., Ali, J. G., Hermann, S. L., Tiwari, S., Pelz-Stelinski, K. S., Alborn, H. T., et al. (2012). Induced release of a plant-defense volatile ‘deceptively’attracts insect vectors to plants infected with a bacterial pathogen. PLoS Pathog. 8:e1002610. doi: 10.1371/journal.ppat.1002610
Peshin, R., Bandral, R. S., Zhang, W., Wilson, L., and Dhawan, A. K. (2009). Integrated Pest Management: A Global Overview of History, Programs and Adoption. Berlin: Springer, 1–49.
Saha, S., Hosmani, P. S., Villalobos-Ayala, K., Miller, S., Shippy, T., Flores, M., et al. (2017). Improved annotation of the insect vector of citrus greening disease: biocuration by a diverse genomics community. Database 2017:bax032. doi: 10.1093/database/bax032
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microb. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Sun, L. Q., Yin, Y. P., Wang, F., Xiao-Fang, W. U., and Wang, Z. K. (2014). Correlation of candidatus liberibacter asiaticus and the endophytic community in diaphorina citri. Sci. Agric. Sin. 47, 2151–2161.
Thao, M. L., Clark, M. A., Burckhardt, D. H., Moran, N. A., and Baumann, P. (2001). Phylogenetic analysis of vertically transmitted psyllid endosymbionts (Candidatus Carsonella ruddii) based on atpAGD and rpoC: comparisons with 16S-23S rDNA-derived phylogeny. Curr. Microbiol. 42, 419–421. doi: 10.1007/s002840010240
Tortora, G. J., Funke, B. R., and Case, C. L. (2007). Microbiology: An Introduction, 9th Edn. San Francisco, CA: Addison-Wesley Longman.
Wang, T., Cai, G., Qiu, Y., Fei, N., Zhang, M., Pang, X., et al. (2011). Structural segregation of gut microbiota between colorectal cancer patients and healthy volunteers. ISME J. 6, 320–329. doi: 10.1038/ismej.2011.109
Werren, J. H., Windsor, D., and Guo, L. (1995). Distribution of Wolbachia among Neotropical Arthropods. R. Soc. Proc. B Biol. Sci. 262, 197–204. doi: 10.1371/journal.pone.0059619
Xu, J., Lian, F. M., Zhao, L. H., Zhao, Y., Chen, X., Zhang, X., et al. (2015). Structural modulation of gut microbiota during alleviation of type 2 diabetes with a Chinese herbal formula. ISME J. 9, 552–562. doi: 10.1038/ismej.2014.177
Yu, X., Wu, X., Qiu, L., Wang, D., Gan, M., Chen, X., et al. (2015). Analysis of the intestinal microbial community structure of healthy and long-living elderly residents in Gaotian Village of Liuyang City. Appl. Microbiol. Biotechnol. 99, 9085–9095. doi: 10.1007/s00253-015-6888-3
Zhang, C., Zhang, M., Pang, X., Zhao, Y., Wang, L., and Zhao, L. (2012). Structural resilience of the gut microbiota in adult mice under high-fat dietary perturbations. ISME J. 6, 1848–1857. doi: 10.1038/ismej.2012.27
Zhang, Q., Pan, Y., Yan, R., Zeng, B., Wang, H., Zhang, X., et al. (2015). Commensal bacteria direct selective cargo sorting to promote symbiosis. Nat. Immunol. 16, 918–926. doi: 10.1038/ni.3233
Keywords: Serratia marcescens, Diaphorina citri, OTU, high-throughput sequencing, Huanglongbing (HLB)
Citation: Hu W, Kuang F, Lu Z, Zhang N and Chen T (2018) Killing Effects of an Isolated Serratia marcescens KH-001 on Diaphorina citri via Lowering the Endosymbiont Numbers. Front. Microbiol. 9:860. doi: 10.3389/fmicb.2018.00860
Received: 08 November 2017; Accepted: 13 April 2018;
Published: 01 May 2018.
Edited by:
Brigitte Mauch-Mani, University of Neuchâtel, SwitzerlandReviewed by:
Alexander Ignatov, Research Center of Biotechnology of the Russian Academy of Sciences, RussiaAdalbert Balog, Sapientia Hungarian University of Transylvania, Romania
Copyright © 2018 Hu, Kuang, Lu, Zhang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tingtao Chen, Y2hlbnRpbmd0YW8xOTg0QDE2My5jb20= Ning Zhang, emhuaW5nODczNEAxNjMuY29t
 Wei Hu1
Wei Hu1 Zhanjun Lu
Zhanjun Lu Tingtao Chen
Tingtao Chen