- 1State Key Laboratory for Conservation and Utilization of Subtropical Agro-bioresources, Guangdong Province Key Laboratory of Microbial Signals and Disease Control, Integrative Microbiology Research Centre, South China Agricultural University, Guangzhou, China
- 2Laboratory of Insect Toxicology, and Key Laboratory of Pesticide and Chemical Biology, Ministry of Education, South China Agricultural University, Guangzhou, China
D-phenothrin is one of the most popular pyrethroid insecticides for its broad spectrum and high insecticidal activity. However, continuous use of D-phenothrin has resulted in serious environmental contamination and raised public concern about its impact on human health. Biodegradation of D-phenothrin has never been investigated and its metabolic behaviors remain unknown. Here, a novel bacterial strain P31 was isolated from active sludge, which completely degraded (100%) D-phenothrin at 50 mg⋅L-1 in 72 h. Based on the morphology, 16S rRNA gene and Biolog tests, the strain was identified as Pseudomonas fulva. Biodegradation conditions were optimized as 29.5°C and pH 7.3 by utilizing response surface methodology. Strain P31 depicted high tolerance and strong D-phenothrin degradation ability through hydrolysis pathway. Strain P31 degraded D-phenothrin at inhibition constant (Ki) of 482.1673 mg⋅L-1 and maximum specific degradation constant (qmax) of 0.0455 h-1 whereas critical inhibitor concentration remained as 41.1189 mg⋅L-1. The 3-Phenoxybenzaldehyde and 1,2-benzenedicarboxylic butyl dacyl ester were identified as the major intermediate metabolites of D-phenothrin degradation pathway through high-performance liquid chromatography and gas chromatography-mass spectrometry. Bioaugmentation of D-phenothrin-contaminated soils with strain P31 dramatically enhanced its degradation, and over 75% of D-phenothrin was removed from soils within 10 days. Moreover, the strain illustrated a remarkable capacity to degrade other synthetic pyrethroids, including permethrin, cyhalothrin, β-cypermethrin, deltamethrin, fenpropathrin, and bifenthrin, exhibiting great potential in bioremediation of pyrethroid-contaminated environment.
Introduction
D-phenothrin [(1R-trans)-2,2-dimethyl-3-(2-methyl-1-propenyl)-(3-phenoxyphenyl) methyl ester] is a type I synthetic pyrethroid (SP) insecticide which contains a cyclopropanecarboxylic acid group linked to an aromatic alcohol through a central ester bond (Supplementary Figure 1). D-phenothrin is widely used to control insects related to public health including mosquitoes, ticks, mites, human head lice and scabies, as well as insect pests of animals and agricultural crops (Scott et al., 2015). SP insecticides are analogs of pyrethrins found in Chrysanthemum cinerariaefolium flowers (Cycoń and Piotrowska-Seget, 2016). Applications of SPs have significantly increased over the past two decades because of their ability to paralyze the target insects and low mammalian toxicity as compared to organochlorines and organophosphates (Scott et al., 2015). Half-life of pyrethroids is usually less than 600 days and are generally considered safe for humans and animals (Laskowski, 2002), but pyrethroid residues are frequently found in groundwater, sediments, foods, and mammals (Delgado-Moreno et al., 2011; Kuivila et al., 2012; Markle et al., 2014). Pyrethroid bioaccumulation was first reported in wild river fish in Iberian river basins of Spain (Corcellas et al., 2015), and in 100% honeybee wax samples (García et al., 2017). Pyrethroid intoxication to endocrine activity causing estrogenic or antiestrogenic effects and immune responses in fish and mammals was also reported (Ansari et al., 2011; Saillenfait et al., 2015; Brander et al., 2016). In addition, D-phenothrin exposure to humans and mammals can lead to DNA damage in vitro (Nagy et al., 2014; Atmaca and Aksoy, 2015). Such serious environmental contaminations of SPs are not only effecting the wild life but human population as well. This situation demands for the advanced remediating strategies to tackle pyrethroid-polluted environments.
To remove pesticide residues from the environment, several remediation technologies such as photodecomposition, fenton degradation, ozonation, adsorption, incineration, and biodegradation have been developed (Arora et al., 2017; Cycoń et al., 2017; Morillo and Villaverde, 2017; Zhan et al., 2018a). Recently, bioremediation through bioaugmentation and/or biostimulation, has emerged as the most cost-effective and eco-friendly approach to breakdown pesticides in soils (Gao et al., 2012; Cycoń and Piotrowska-Seget, 2016). Different pyrethroid-degrading strains, such as Sphingobium sp. JZ-1 (Wang et al., 2009), Stenotrophomonas sp. ZS-S-01 (Chen et al., 2011d), Ochrobactrum anthropic YZ-1 (Zhai et al., 2012), Brevibacterium aureum DG-12 (Chen et al., 2013), Serratia marcescens (Cycoń et al., 2014), Bacillus thuringiensis ZS-19 (Chen et al., 2015), Pseudomonas aeruginosa GF31 (Tang et al., 2017), and Acinetobacter baumannii ZH-14 (Zhan et al., 2018b) have already been isolated from contaminated soils. However, microbial degradation of D-phenothrin and its pathway has never been investigated.
The objectives of this study were: (1) to investigate the microbial degradation of D-phenothrin and its biochemical degradation pathway; (2) to determine the biodegradation kinetics of D-phenothrin; and (3) to evaluate the potentials of isolate for bioremediation of D-phenothrin-contaminated environment.
Materials and Methods
Chemicals and Media
D-phenothrin (98% purity) was obtained from Jiangsu Yangnong Chemical Group Co., Ltd., China. Technical-grade permethrin, cyhalothrin, β-cypermethrin, deltamethrin, fenpropathrin, and bifenthrin were purchased from Sigma–Aldrich, United States and high-performance liquid chromatography (HPLC)-grade acetonitrile was purchased from Fisher Scientific, United States. All other chemicals and solvents were of analytical grade. SPs were dissolved in dimethyl sulfoxide (DMSO) or acetone at a stock concentration of 10 g⋅L-1, and stored in dark bottles at 4°C.
Mineral salt medium (MSM) containing (g⋅L-1) (NH4)2SO4, 2; MgSO4⋅7H2O, 0.2; CaCl2⋅2H2O, 0.01; FeSO4⋅7H2O, 0.001; Na2HPO4⋅12H2O, 1.5; and KH2PO4, 1.5; and Luria-Bertani (LB) medium containing (g⋅L-1) tryptone, 10; yeast extract, 5; and NaCl, 10 were used in this study. Both culture media were adjusted to pH 7.3 and sterilized at 121°C for 20 min.
Isolation of D-Phenothrin-Degrading Bacterial Strains
Active sludge from an aerobic pyrethroid-manufacturing wastewater treatment system in Zhongshan, China, was used to achieve D-phenothrin-degrading bacterial isolates. Isolation and enrichment of degrading bacterial isolates was carried out according to Chen et al. (2011b,c). Initial culture enrichment was performed in a 250-mL Erlenmeyer flask containing 50 mL of sterilized MSM supplemented with 5 g of activated sludge and 50 mg⋅L-1 of D-phenothrin. Enrichment culture was incubated in an incubator shaker at 180 rpm and 30°C for 7 days. After 7 days, 0.5 mL of enrichment culture was transferred to new Erlenmeyer flasks containing 50 mL of fresh MSM and 100, 200, 400, and 800 mg⋅L-1 D-phenothrin, respectively. After five rounds of transferring, the enrichment medium was serially diluted and spread on MSM plates along with 50 mg⋅L-1 D-phenothrin to isolate individual colonies. After purifying the obtained isolates, their D-phenothrin degradation potential was investigated. Concentrations of D-phenothrin residues in culture fluids were detected by HPLC (Waters 2690, United States).
Identification and Characterization of Strain P31
Colony and cellular morphologies of strain P31 were studied under electron microscope (BH-2 Olympus, Japan) and scanning electron microscope (XL-30 ESEM, Philips Optoelectronics Co., Ltd., Holland) on LB plates and slides, respectively.
Strain P31 was identified by sequencing 16S rRNA, which is the most conserved DNA region in prokaryotes (Festa et al., 2013). Genomic DNA was prepared by using MasterPureTM DNA Purification Kit (Epicentre Biotechnologies, United States). 16S rRNA gene was amplified with reference (Escherichia coli) primers 27F (5V-AGA GTT TGA TCC TGG CTC AG-3V) and 1492R (5V-TAC GGY TAC CTT GTT ACG ACT T-3V) and NCBI BLAST tool was used to compare 16S rRNA sequences. Bacterium was further identified by Biolog Microbial Identification System (GP2 MicroPlateTM, BIOLOG, Hayward, CA, United States).
Growth and Degradation Characteristics of Strain P31
Strain P31 was thawed and inoculated in LB medium for 12 h. Bacterial cells were harvested by centrifugation (5 min, 1950 × g) and washed twice with sterile saline (0.9% NaCl) to prepare a concentrated inoculum solution (Chen et al., 2011a). A total of 0.3 g⋅L-1 of inoculum was transferred to 50 mL of MSM medium containing 50 mg⋅L-1 of D-phenothrin, and kept at 29.5°C and 200 rpm in a rotary shaker. Initial OD600 of the culture was about 0.05. All set-ups were prepared in triplicates, and non-inoculated cultures served as controls. Samples were collected at 0, 12, 24, 36, 48, 60, and 72 h, respectively. Residual concentrations of D-phenothrin were detected by HPLC.
Optimization of D-Phenothrin Degradation Conditions
Based on Box–Behnken design, optimized conditions of D-phenothrin biodegradation were determined according to the response surface methodology (RSM) (Ghevariya et al., 2011; Zhang et al., 2011; Chen et al., 2013). For statistical calculations, the variables Xi (the uncoded value of the ith independent variable) were coded as xi (the coded value of the ith independent variable) according to the following equation:
where xi is the value of Xi at the center point, and ΔXi is the step change value.
Based on the preliminary results of one-factor-at-a-time experiments (data not shown), critical factors such as temperature, pH and inoculum size were selected as independent variables. Levels of the variables and experimental Box–Behnken design are shown in Supplementary Table 1. Accordingly, based on Box–Behnken design 15 experiments were carried out to build quadratic models, with three replications of the center points to estimate experimental error. Experimental data of the model experiments was used to predict optimal conditions according to the following equation:
where Xi are the input variables, which influence the response variable Yi, ϸ0 is the offset term, ϸi, ϸii, and ϸij are the first-order, quadratic, and interaction coefficients, respectively, i and j are the index numbers for the factors, and ei is the residual error.
SAS 9.0 was used for statistical analysis and building regression model to predict optimal processing parameters.
Dynamics and Substrate Range Characteristics of Strain P31
Degradation dynamics parameters were determined by measuring specific degradation constant at various D-phenothrin concentrations (25, 50, 100, 200, 400, and 800 mg⋅L-1) under optimal reaction conditions. D-phenothrin residual concentrations were detected and specific degradation constant (q) at different initial concentrations was assessed according to substrate inhibition model (Luong, 1987), as follows.
where qmax is the maximum specific degradation constant, Ks is the half-saturation constant, Ki is the substrate inhibition constant, and S is the inhibitor concentration. Kinetic parameters were calculated by non-linear regression method in MATLAB 7.8.
To assess the degradation ability of strain P31 against various SPs, MSM medium was supplemented with D-phenothrin, permethrin, cyhalothrin, β-cypermethrin, deltamethrin, fenpropathrin, or bifenthrin at 50 mg⋅L-1 along with the optimal inoculum size. Each treatment was performed in triplicate and incubated at optimum conditions whereas non-inoculated samples served as control. Samples of different treatments were collected at regular intervals and degradation was measured as described above.
Identification of D-Phenothrin Metabolic Products
To identify D-phenothrin and its metabolic products of biodegradation, the strain P31 was grown in MSM media containing 50 mg⋅L-1 of D-phenothrin. Non-inoculated samples containing the same amount of D-phenothrin served as control. Samples of different treatments were collected at regular intervals as described above, and extracted after acidification to pH 2 (2 M HCl) according to Tallur et al. (2008). Metabolites were detected by gas chromatography-mass spectrometry (GC-MS) (Agilent 6890N/5975, United States). To identify the metabolites, mass spectrometry analyses were matched with authentic standard compounds from the National Institute of Standards and Technology (NIST, United States) library database.
Biodegradation of D-Phenothrin in Soils
Soil samples were collected at the depth of 5–20 cm from a field, which had never been treated with D-phenothrin or fertilizers, in South China Agricultural University, Guangzhou, China. Physicochemical properties of the soils were as (g/kg of dry weight): organic matter, 10.5; total N, 0.5; total P, 0.4; total K, 18.2; and pH, 6.9 and has sandy loam texture (sand, 65.0%; silt, 28.0%; clay, 7.0%). Soil samples were air-dried in the laboratory to the suitable moisture level, sieved (2 mm), and used for bioremediation studies (Chen et al., 2012, 2014).
Sterile (at 121°C for 1 h) and nonsterile soils (nSSs) were used to investigate bioremediation ability of strain P31. A total of 600 g of sterile or nSSs were placed in 500-mL Erlenmeyer flasks, and D-phenothrin solution was added to a final concentration of 50 mg⋅kg-1 of soil in acetone solution. Erlenmeyer flasks were kept opened to allow aerobic conditions. Water contents were adjusted to 40% of water-holding capacity with deionized water (Cycoń et al., 2014). After mixing, the strain P31 suspension was introduced into the soils by drip irrigation to a final concentration of 1.0 × 107 CFU⋅g-1 of soil and incubated in a sterile incubator at 30°C. Sterile or nSS samples having insecticides but without strain P31 suspension were used as controls. Triplicate soil samples were kept in a dark thermostatic chamber for 10 days at 30°C.
Soil samples (20 g) for D-phenothrin residue analysis were collected on day 0, 2, 4, 6, 8, and 10. Samples were ultrasonically extracted with 40 mL of ethyl acetate for 15 min, and then centrifuged at 4000 × g for 6 min to obtain supernatants. Supernatants were pooled and loaded onto a chromatography column (35 cm length × 1 cm i.d., pre-eluted with 40 mL of ethyl acetate), packed with anhydrous sodium sulfate, silica gel, and neutral alumina. Column was eluted three times, with 60 mL of ethyl acetate. Eluates were evaporated and residue was re-dissolved in 1.5 mL methanol for HPLC analysis.
Degradation constant (k) was determined according to the first-order kinetic model (Eq. 4) (Cycoń et al., 2009):
where C0 is the initial concentration of D-phenothrin at zero time, Ct is the concentration of D-phenothrin at time t, k, and t are the degradation constant (day-1) and degradation period in days, respectively.
D-phenothrin half-life (t1/2) in different soils was calculated by using Eq. 5:
where k is the degradation constant (day-1).
Chemical Analysis
Analysis of D-phenothrin residues in filtrates was carried on a Phenomenex C18 reversed phase column (250 × 4.60 mm, 5 μm) with a Water 2690 HPLC system. A mixture of acetonitrile and water (90:10) was used as the mobile phase at the flow rate of 1.0 mL⋅min-1. Based on retention time (RT) and peak area of pure standard, array detection was carried out from 190 to 400 nm (total scan) at detection limit of 0.01 mg⋅L-1. Detection wavelengths of D-phenothrin, permethrin, cyhalothrin, β-cypermethrin, deltamethrin, fenpropathrin, and bifenthrin were as 276.8, 276.8, 276.8, 276.8, 250.9, 276.8, and 354 nm, respectively.
Analysis of D-phenothrin metabolic products, was performed on Agilent 6890N/5975 GC-MS system equipped with auto-sampler, an on-column, split/splitless capillary injection system with array detection from 30 to 500 nm (total scan). Chromatographic column was a HP-5MS capillary column (30.0 m × 250 μm × 0.25 μm), and column temperature was initially held at 90°C for 2 min, then increased to 150°C at the rate of 6°C⋅min-1 for 1 min, raised to 180°C at the rate of 10°C⋅min-1 for 4 min and finally increased to 260°C at the rate of 20°C⋅min-1 for 10 min. Carrier gas (helium) flow rate was 1.5 mL⋅min-1 and the injection volume was 1.0 μL with splitless sampling at 250°C. Temperatures corresponding to transfer line and the ion trap were 280°C and 230°C, respectively, and the ionization energy was 70 eV (Chen et al., 2014, 2015).
Nucleotide Sequence Accession Number
The GenBank accession number of strain P31 16S rRNA gene is KY229868.
Results
Isolation of D-Phenothrin-Degrading Strain
Based on the isolation and enrichment results, 34 morphologically different strains (grown on MSM containing 800 mg⋅L-1 D-phenothrin) were attained from activated sludge and named as P01∼34, respectively. Results showed that strain P31 possessed the highest degradation capacity among 34 strains and degraded 100% of D-phenothrin within 72 h of incubation. Degradation of D-phenothrin by each isolated strain within 72 h of incubation is shown in Supplementary Table 2. Moreover, strain P31 was also found to efficiently utilize permethrin, cyhalothrin, β-cypermethrin, deltamethrin, fenpropathrin, and bifenthrin as the sole source of carbon. Thus, strain P31 was selected for further studies.
Identification and Characterization of Strain P31
Physio-biochemical properties of the strain P31 are shown in Supplementary Table 3. Strain P31 is a Gram-negative, obligatory aerobic and rod-shaped bacterium of 1.5∼4.0 μm length and 0.5∼1.0 μm width. Colonies grown on LB agar plates were primrose yellow, opaque, and circular with smooth margin. This bacterial isolate showed positive reactions in catalase, oxidase, citrate utilization, nitrate reduction, hemolysis, and arginine dihydrolase tests, but remained negative in anaerobic growth, gelatin liquefaction, urease production, indole production, and esculin hydrolysis tests. Growth was observed over a temperature range of 20∼40°C, a salinity range of 2∼5% NaCl, and a pH range of 5∼10. It was positive in the utilization of glycerol, α-D-glucose, D-fructose, L-fucose, L-alanine, dextrin, L-serine, inosine, D-galactose, D-fucose, methyl pyruvate, sodium bromate; and negative in the utilization of sucrose, pectin, gelatin, D-maltose, D-trehalose, D-cellobiose, stachyose, D-turanose, gentiobiose, D-raffinose, D-melibiose, and β-methyl-D-glucoside.
According to BLAST analysis, partial sequence showed high similarity to 16S rRNA gene sequence of Pseudomonas group and closely clustered with strain f2 (2012) (GenBank accession No. JQ717288) and strain SRZ19 (GenBank accession No. KU877341), having sequence identities over 99%. Similarity and distance between strain P31 and the model strain (P. fulva) were 0.617 and 4.355, respectively, according to Biolog microbial identification system. Therefore, the strain P31 was confirmed as P. fulva based on the morphology, physio-biochemical properties, 16S rRNA gene analysis, and Biolog microbial identification system.
Growth and Degradation Characteristics of Strain P31
Growth of strain P31 in MSM media with D-phenothrin as the sole carbon resource and its degradation characteristics are shown in Figure 1. Strain P31 showed rapid growth without lag phase during initial period (0∼12 h) indicating that the strain utilized D-phenothrin as growth substance. Cell growth hiked after 12 h of incubation, and significant degradation of D-phenothrin was noted during this period. Cell growth increased rapidly at logarithmic phase (12∼24 h), and approximately 57.0% of the D-phenothrin was degraded by strain P31 in 24 h. During this time, the concentration of D-phenothrin decreased with a concomitant increase of cell number. Subsequently, cell growth slowed down at stationary phase (36∼60 h), and about 77.0–86.4% of the initial dose was eliminated within 36–48 h, respectively. Strain P31 reached to maximum level of growth at 60 h post inoculation and caused the maximum D-phenothrin biodegradation of 96.8%. Finally, the strain did not show growth at decline phase (> 72 h) and the residual amount of D-phenothrin was not detectable by HPLC at 72 h post incubation. Approximately 9.0% degradation was observed in the non-inoculated control.
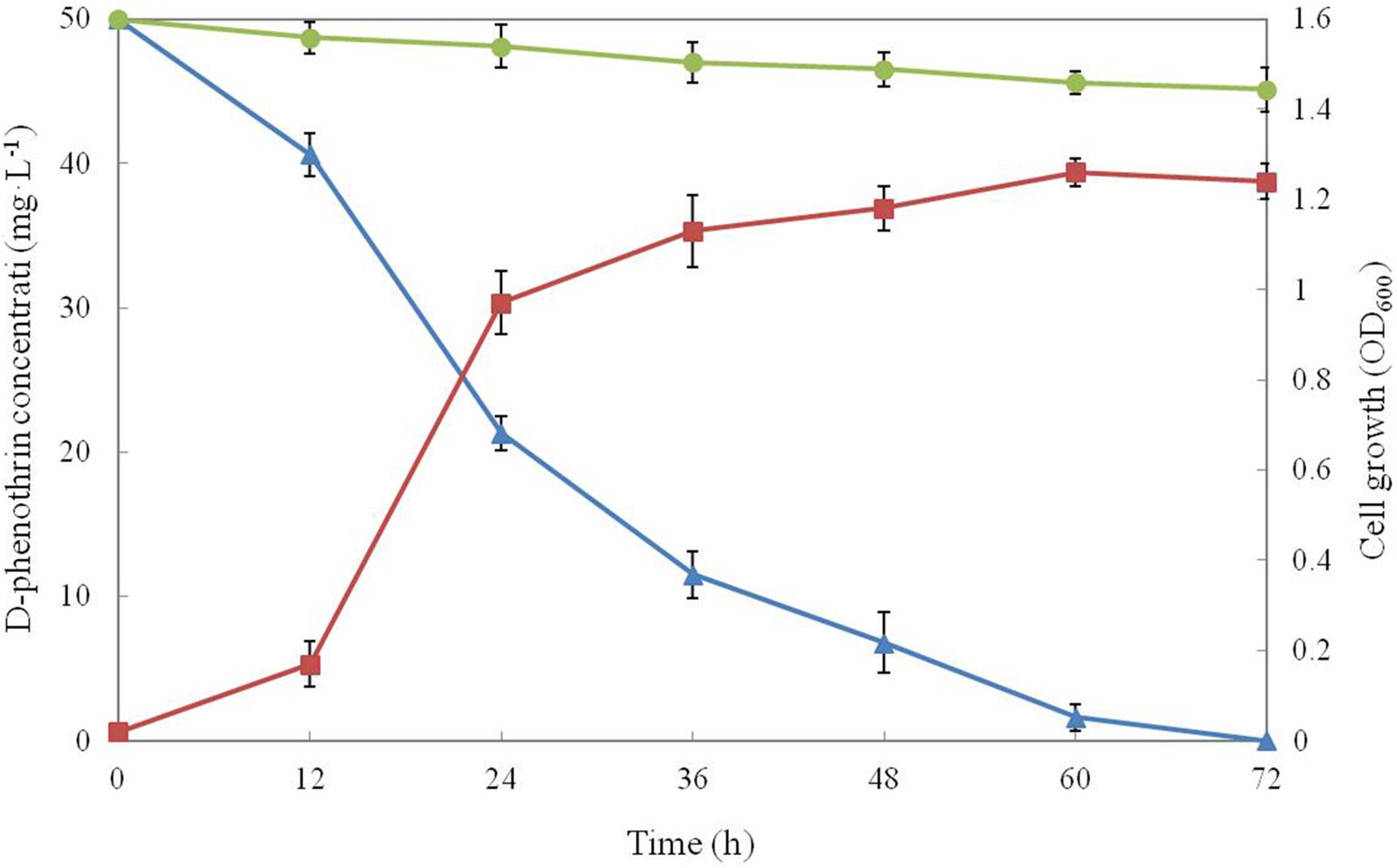
FIGURE 1. Biodegradation of D-phenothrin (50 mg⋅L-1) during growth of strain P31. Symbol: •, D-phenothrin control;  , D-phenothrin degradation by strain P31;
, D-phenothrin degradation by strain P31;  , cell growth. Data represent mean values of three replicatesov with standard deviation.
, cell growth. Data represent mean values of three replicatesov with standard deviation.
Optimization of D-Phenothrin Degradation Conditions
Supplementary Table 1 demonstrates the level and range of three independent variables, including temperature (X1), pH (X2), and inoculum size (X3). The three parameters were taken at a central coded value considered as zero and investigated at three different levels (-1, 0, and +1). In this case, the result of a three-level factorial design, a total number of 15 experiments were employed to match the second-order polynomial model in accordance with Box–Behnken design. Statistical combinations of critical parameters along with the maximum observed and predicted degradation are displayed in Supplementary Table 1. These predicted values were in close proximity to the observed ones in all sets of experiments. Highest degradation of 100% and the lowest of 80.4% were observed. Two regression equations, Eq. 6a for coded values and Eq. 6b for actual experimental values, which are analogous to Eq. 2, showed the degradation rate (Y) as a function of the test variables temperature (X1), pH (X2), and inoculum size (X3).
Analysis of variance (ANOVA) is essential to test the significance and adequacy of the model (Ghevariya et al., 2011) and ANOVA of the quadratic regression model is tabulated in Supplementary Table 4. As shown in the table, a very low probability value (P-value < 0.05), a large value of the regression coefficient (R2 = 0.9905) and a low coefficient of variation (CV = 1.10) demonstrated that the established quadratic polynomial model for D-phenothrin degradation by strain P31 presented the actual relationship of response and variables. Regression analysis reveals that the linear terms of temperature (X1), pH (X2), and inoculum size (X3) are important factors in the D-phenothrin degradation by strain P31. Square terms of X12 and pH X22, interaction terms of X1X3 and X2X3 also showed significant effects (P < 0.05) on D-phenothrin degradation by strain P31, however, square terms of X32 and interaction term of X1X2 played insignificant role (P > 0.05) in degradation (Supplementary Table 4).
For better understanding of the results, a 3D response surface plot is given in Supplementary Figure 2, which illustrates the effects of temperature (X1) and pH (X2) on D-phenothrin degradation with inoculum size (X3) as a constant. Supplementary Figure 2 has clear stationary point, indicating that maximum degradation could be achieved inside the design boundaries. Model depicted maximum degradation of 100% at temperature 29.5°C, pH 7.3 and an inoculum size of 0.3 g⋅L-1. Thus, the optimum culturing conditions for D-phenothrin degradation by strain P31 were concluded to be temperature 29.5°C, pH 7.3 and an inoculum size of 0.3 g⋅L-1.
Biodegradation Kinetics of D-Phenothrin by Strain P31
D-phenothrin biodegradation at different initial D-phenothrin concentrations is presented in Figure 2A. Strain P31 utilized and degraded D-phenothrin up to a concentration of 800 mg⋅L-1, and no lag period was observed. D-phenothrin was degraded completely within 72 h at initial concentrations below 50 mg⋅L-1. During 72 h incubation, strain P31 displayed 91.2, 88.3, 82.6, and 80.4% of degradation at D-phenothrin initial concentrations of 100, 200, 400, and 800 mg⋅L-1, respectively. Increased concentration of D-phenothrin had a slight effect on biodegradation performance of strain P31, indicating that the D-phenothrin degradation by this strain is concentration dependent. Relationship between initial D-phenothrin concentration and specific degradation constant by strain P31 is shown in Figure 2B. According to the substrate inhibition model (Eq. 3), Ks and Ki values were determined by non-linear regression method as 3.5066 and 482.1673 mg⋅L-1, respectively. Critical inhibitor concentration (Sm) was calculated as 41.1189 mg⋅L-1 whereas the overall qmax value remained 0.0455 h-1. As shown in Figure 2B, at the initial concentration of D-phenothrin below 50 mg⋅L-1, q value was rapidly increased. At the initial concentrations of D-phenothrin above 100 mg⋅L-1, q value gradually declined with the increase of D-phenothrin. These results illustrated that the biodegradation of D-phenothrin by strain P31 could be partially inhibited at high concentration of D-phenothrin.
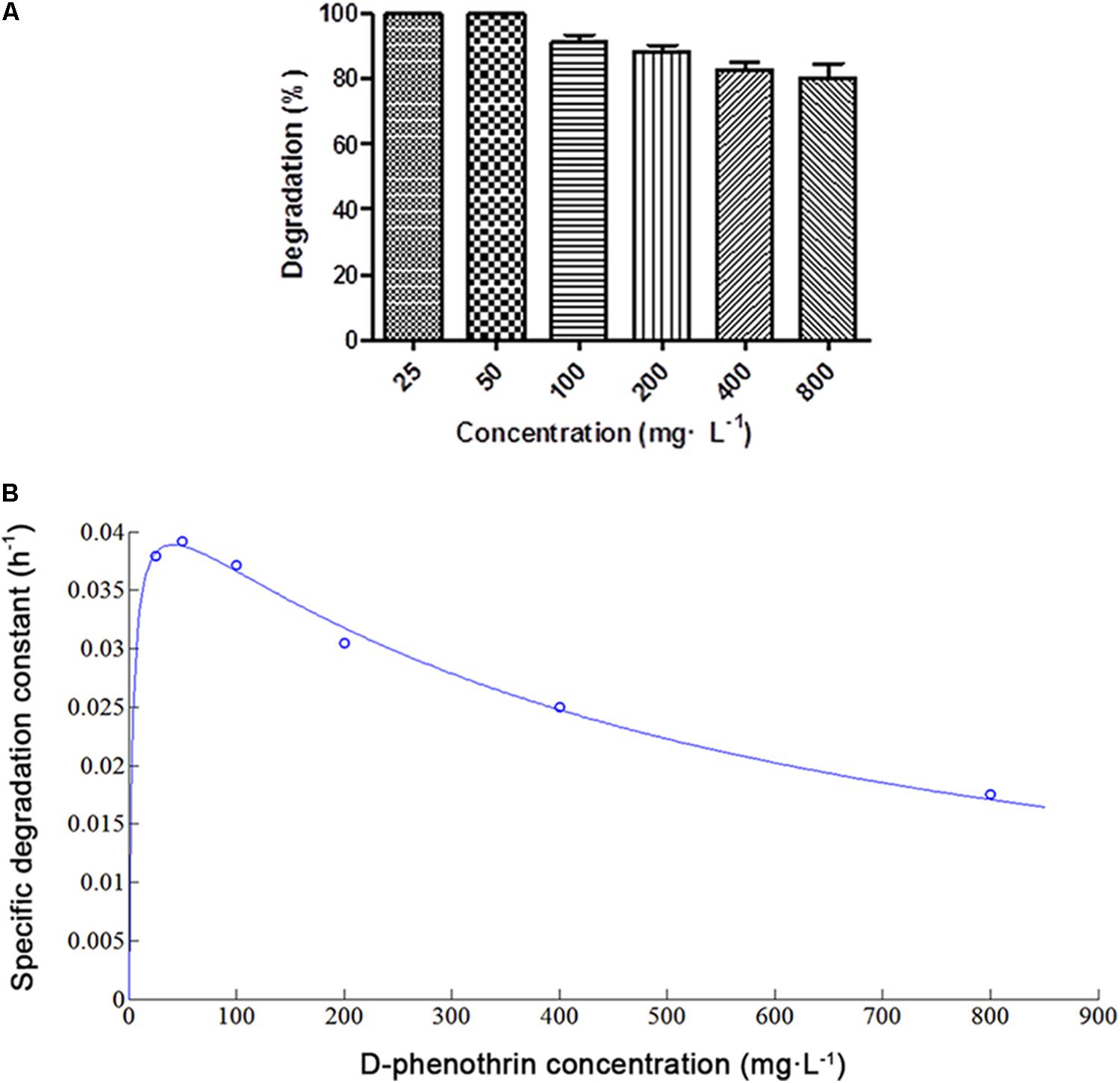
FIGURE 2. Biodegradation of D-phenothrin at different initial concentrations by strain P31 within 72 h. (A) Degradation of D-phenothrin at different concentrations; (B) Relationship between initial D-phenothrin concentration and specific degradation constant by strain P31. Data represent mean values of three replicates with standard deviation.
Biodegradation Kinetics of Various SPs
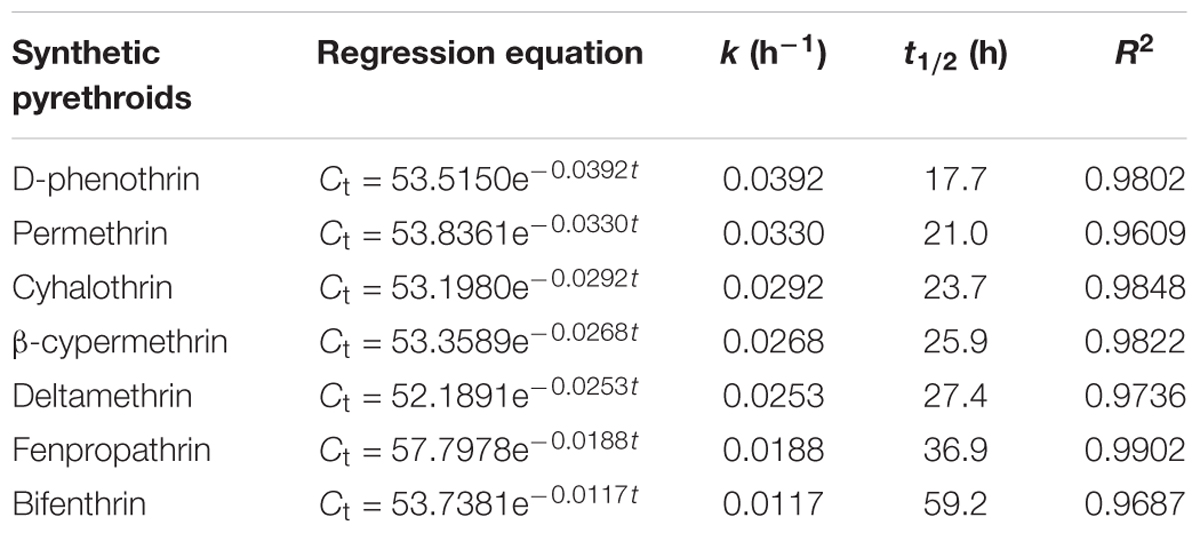
Strain P31 revealed capability to utilize various SPs as food source including D-phenothrin, permethrin, cyhalothrin, β-cypermethrin, deltamethrin, fenpropathrin, and bifenthrin, having degradation efficiency of 100, 96.7, 89.2, 86.6, 83.1, 71.4, and 57.9% within 72 h, respectively (Figure 3). To further quantify the biodegradation capacity of strain P31 against different SPs, the degradation constant (k) and half-life (t1/2) were determined by using the first-order kinetic model. Degradation kinetic parameters of various SPs by strain P31 are summarized in Table 1. Biodegradation process was characterized by a k ranging from 0.0117 to 0.0392 h-1. The t1/2 value of the above mentioned SPs with strain P31 were calculated as 17.7, 21.0, 23.7, 25.9, 27.4, 36.9, and 59.2 h, respectively, and are significantly lower as compared to those in the environment.
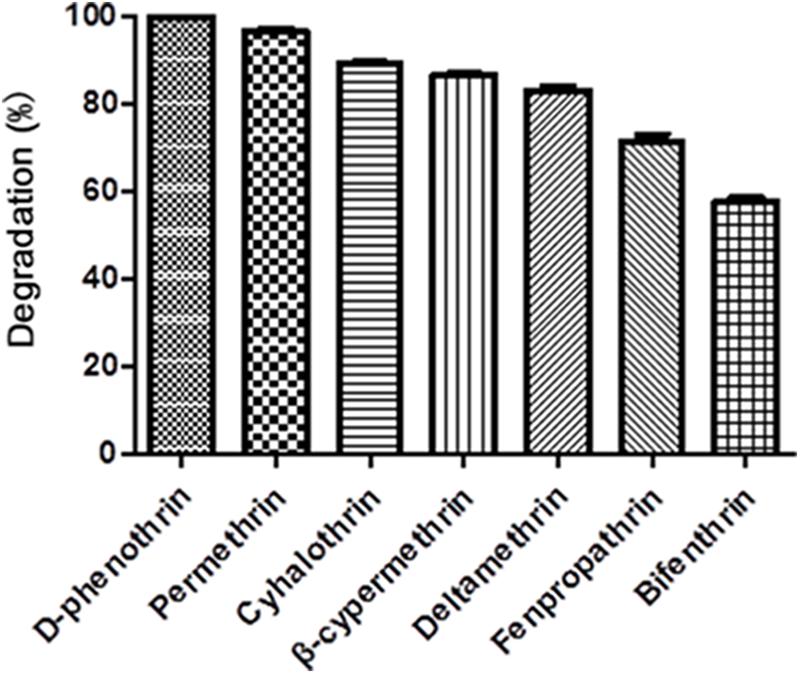
FIGURE 3. Degradation of various synthetic pyrethroids by strain P31 within 72 h. Data represent mean values of three replicates with standard deviation.
Metabolic Products and Degradation Pathway of D-Phenothrin
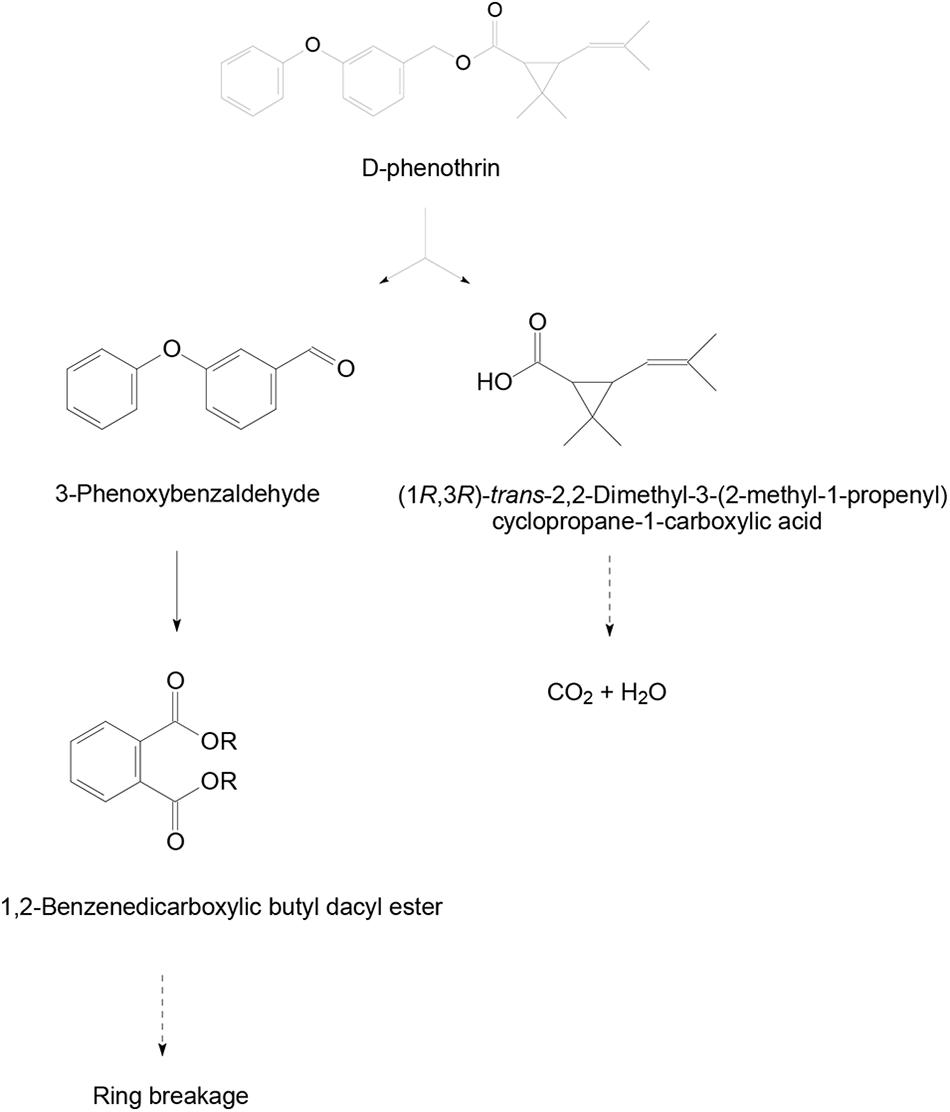
Samples obtained at different intervals were analyzed through GC-MS to identify the metabolic products during the bacterial degradation process. In the initial sample of 0 and 12 h, a significant peak was detected at RT of 26.710 min, displaying a characteristic mass fragment [M+] at m/z 350 with major fragment ions at m/z 123 and 183, which was identified as D-phenothrin and named as compound A. Subsequently, the compound A disappeared concomitantly with the formation of two new compounds. According to the similarity of their fragment RTs and molecular ions to those of corresponding authentic compounds in the NIST library database, the two compounds were characterized as 3-phenoxybenzaldehyde (compound B) and 1,2-benzenedicarboxylic butyl dacyl ester (compound C), respectively (Supplementary Figure 3). It is the first time that these metabolic products were detected in the degradation pathway of D-phenothrin. It is noteworthy that all these metabolites were transitory and faded away without any non-cleavable metabolites at the end of experiment. Chemical structures, RT, and characteristic ions of the mass spectra are shown in Figure 4.
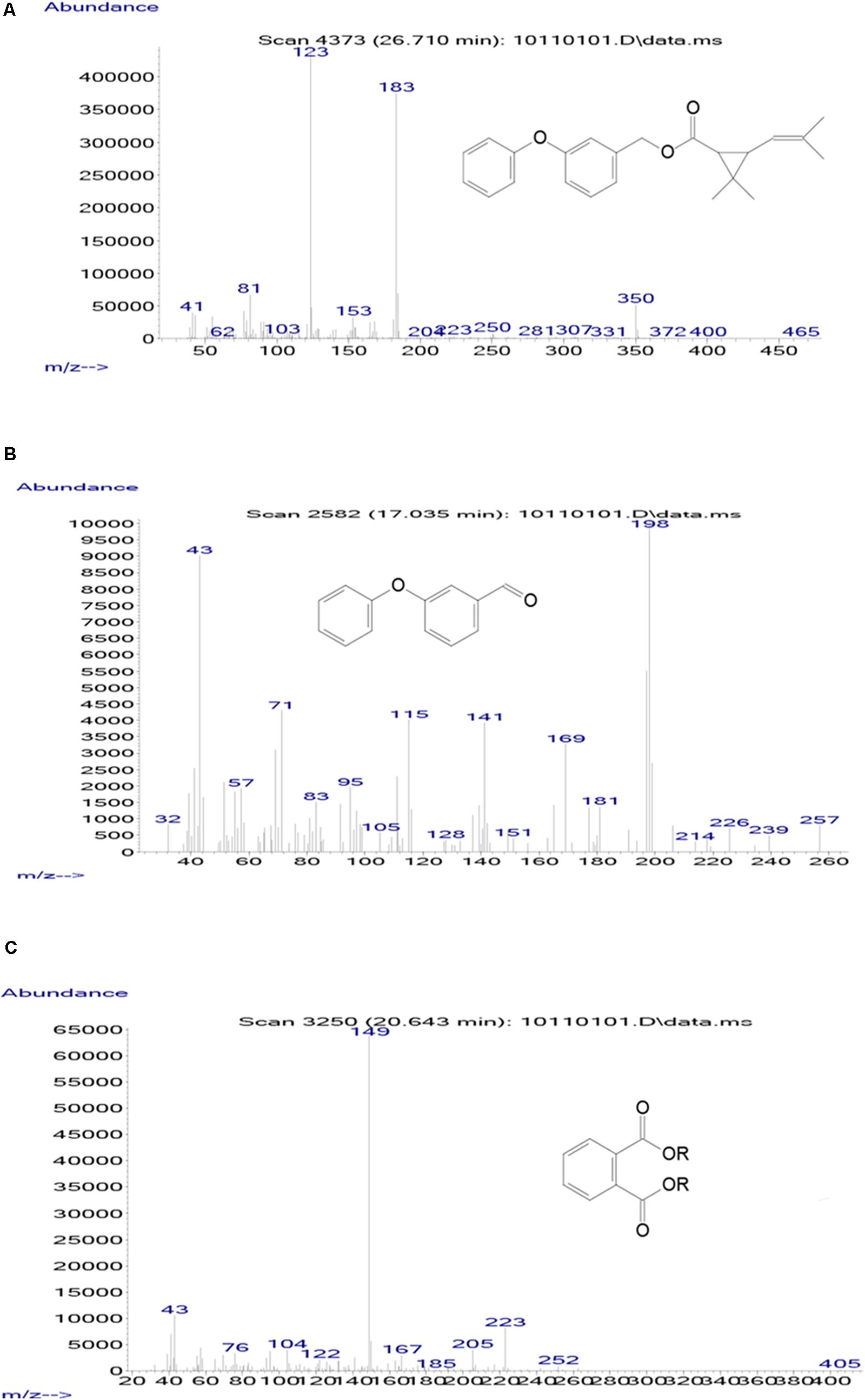
FIGURE 4. GC-MS analysis of the metabolites produced from D-phenothrin degradation by strain P31. (A–C) The characteristic ions of compounds A–C in GC-MS. Retention time of each compound was 26.710, 17.035, and 20.643 min, respectively, and identified as D-phenothrin, 3-phenoxybenzaldehyde, and 1,2-benzenedicarboxylic butyl dacyl ester, respectively.
A degradation pathway of D-phenothrin in strain P31 was proposed based on the chemical structures of D-phenothrin and metabolites formed during fermentation (Figure 5). D-phenothrin was first hydrolyzed by cleavage of the carboxylester linkage to produce 3-phenoxybenzaldehyde and (1R,3R)-trans-2,2-dimethyl-3-(2-methyl-1-propenyl) cyclopropane-1-carboxylic acid. Then, the intermediate product 3-phenoxybenzaldehyde was further degraded with diaryl cleavage to form 1,2-benzenedicarboxylic butyl dacyl ester. Eventually, D-phenothrin was degraded by strain P31 without any persistent accumulative product.
Biodegradation of D-Phenothrin in Soils
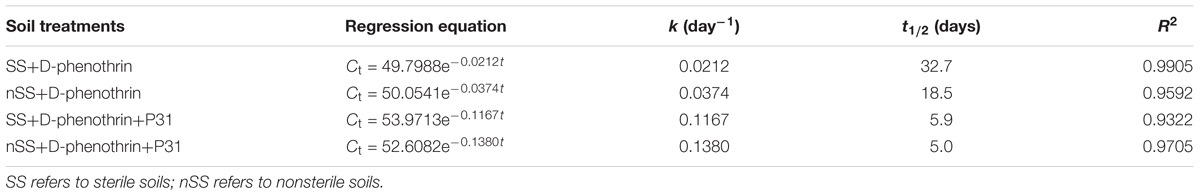
The ability of strain P31 to degrade D-phenothrin in contaminated soils was evaluated under controlled conditions (Figure 6). Degradation process was further investigated using the first-order kinetic model as shown in Table 2. D-phenothrin degraded rapidly after strain P31 inoculation (1 × 107 CFU⋅g-1 of soil) in nSS at the beginning of incubation, and approximately 77.0% of D-phenothrin at initial concentration of 50 mg⋅kg-1 was removed within 10 days without any lag phase. Degradation process was characterized by a degradation constant (k) of 0.1380 day-1 and t1/2 value of 5.0 days (Table 2). By contrast, only 34.6% of added pesticide was degraded in the control group inoculated with autochthonous microorganisms, at t1/2 value of 18.5 days. About 75.4% of initial D-phenothrin was removed from the sterile soil (SS) within 10 days at k-value of 0.1167 day-1 and t1/2 of 5.9 days. In the non-inoculated control, this activity decreased to only 19.1% in SS at t1/2 of 32.7 days. It is noteworthy that the degradation of D-phenothrin in nSS was higher as compared to SS.
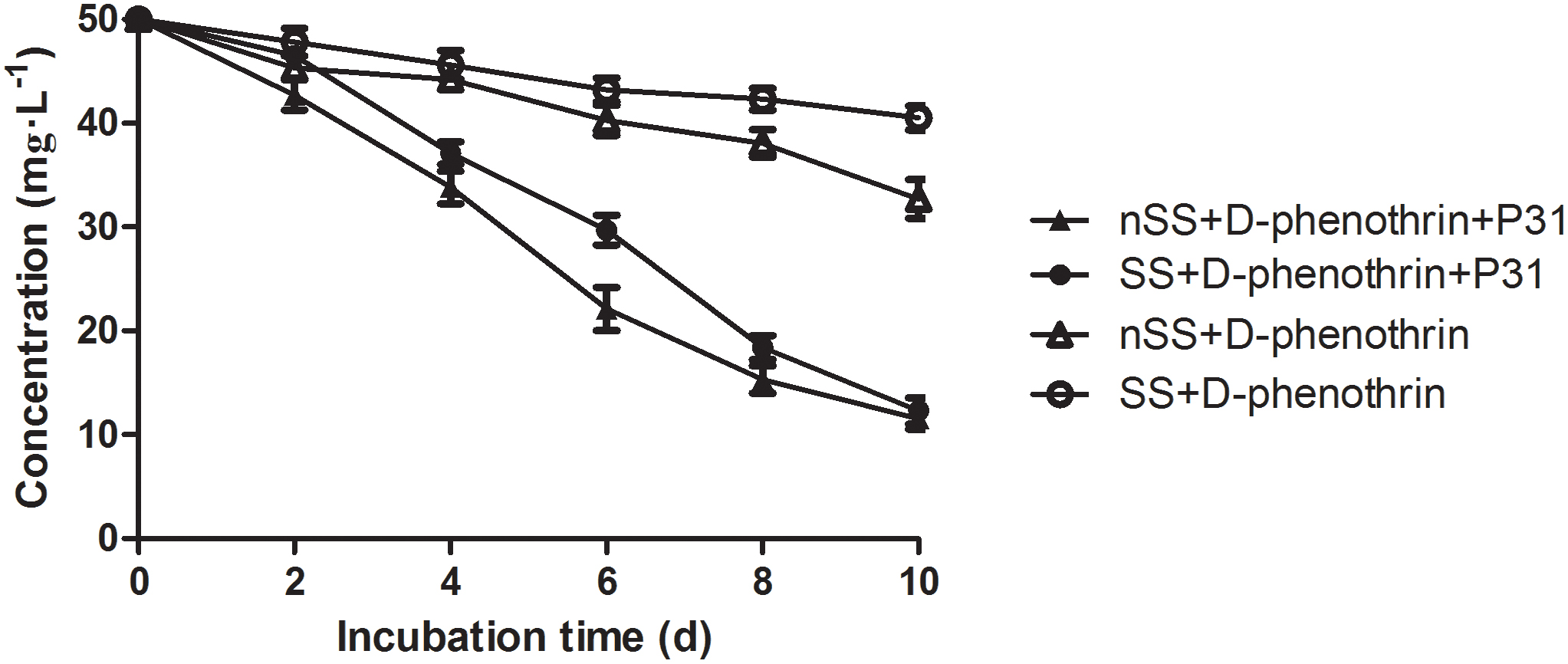
FIGURE 6. Degradation kinetics of D-phenothrin in sterile and nSSs. Data represent mean values of three replicates with standard deviation. SS refers to sterile soils; nSS refers to nonsterile soils.
Discussion
Pseudomonas bacterial strains are known for their metabolic capability, environmental versatility and ability to degrade different xenobiotic compounds including permethrin (Maloney et al., 1988), 3-phenoxybenzoate (Topp and Akhtar, 1991), β-cyfluthrin (Saikia et al., 2005), cypermethrin (Jilani and Khan, 2006), diazinon (Cycoń et al., 2009), 4-chloro-2-nitrophenol (Arora and Bae, 2014), 4-chloro-3-nitrophenol (Arora et al., 2014), fenpropathrin (Song et al., 2015), and β-cypermethrin (Zhang et al., 2011; Tang et al., 2017). Pseudomonas fulva, one of the highly potent degrading microbes of genus Pseudomonas, is a ubiquitous environmental bacterium. Nevertheless, the potential use of P. fulva in biodegradation of organic compounds has not received the attention it deserves. Our studies demonstrated that P. fulva can utilize various organic pollutants as food source and thus it can be successfully colonized in nutrient deficient niches.
Generally, it is considered that the screening and enrichment of environmental microorganisms is of great importance for selecting desirable degrading bacteria (Yang et al., 2011; Liu et al., 2015; Xiao et al., 2015). In this study, 34 morphologically different strains, which grew well in MSM with D-phenothrin as the sole carbon source, were selected from pyrethroid-contaminated active sludge through enrichment culture technique, and the most active bacterium strain P31 was chosen for further studies. This is the first time that a D-phenothrin-degrading strain P31 was isolated which completely degraded D-phenothrin in 72 h. Unlike other environmental bacteria, P. fulva P31 has remarkable capacity to degrade a variety of SPs including D-phenothrin, permethrin, cyhalothrin, β-cypermethrin, deltamethrin, fenpropathrin, and bifenthrin which are widely used insecticides and are associated with environmental contamination. Study reveals that this isolate has great potential as a degrader in cleaning-up pyrethroid residues from the environment (Delgado-Moreno et al., 2011; Kuivila et al., 2012; Markle et al., 2014).
The RSM provides reliable information for the optimization of key process parameters to enhance biodegradation by employing polynomial equation of empirical model to experimental data (Bezerra et al., 2008). Box–Behnken design has great advantages as it requires fewer experimental runs to efficiently estimate quadratic surfaces for the optimization of responses (Ghevariya et al., 2011). During this study, RSM based on Box–Behnken design was successfully explored to improve the biodegradation process in strain P31, and optimum D-phenothrin biodegradation conditions were determined as 29.5°C, pH 7.3, and an inoculum size of 0.3 g⋅L-1. In addition, two mathematical models (Eqs. 6a and 6b) could be used to predict and optimize D-phenothrin degradation conditions by strain P31 within given parameters. Models indicated maximum degradation rate of 100% under the optimum conditions. Similar enhanced degradation through RSM has been reported in a variety of degrading microbes such as Achromobacter xylosoxidans CG542 (Ghevariya et al., 2011), Pseudomonas aeruginosa CH7 (Zhang et al., 2011), Brevibacterium aureum DG-12 (Chen et al., 2013), and Bacillus subtilis BSF01 (Xiao et al., 2015).
Biodegradation process is often inhibited by high concentration of substrates, which limits their use in bioremediation of variable environments. Previous studies reported that the metabolic activity of pyrethroid-degrading microbes lead to complete catabolite repression at high substrate concentrations < 200 mg⋅L-1 (Jilani and Khan, 2006; Lin et al., 2011). Contrarily, increased concentrations of D-phenothrin did not cause much effect on the biodegradation performance of strain P31, and more importantly it did not inhibit cell growth (Figure 2B). It is noteworthy that the isolate tolerated and rapidly degraded D-phenothrin up to a concentration, as high as 800 mg⋅L-1, at Ki and qmax of 482.1673 mg⋅L-1 and 0.0455 h-1, respectively. This feature provides significant advantage to survive under various environments and utilize even the high concentrations of xenobiotic (Chen et al., 2013, 2014).
Hydrolysis plays a significant role in the biodegradation of SPs, which could be attributed to the presence of cyclopropane carboxylic acid moieties connected to aromatic alcohols with a central ester (or ether) bond in SPs. Ester bond is generally considered susceptible to degrading microbes (Stok et al., 2004; Heidari et al., 2005; Tallur et al., 2008; Wang et al., 2009; Zhang et al., 2011; Chen et al., 2015; Tang et al., 2017). In this study, D-phenothrin was initially hydrolyzed via cleavage of the ester bond to yield the main intermediate products 3-phenoxybenzaldehyde and 1,2-benzenedicarboxylic butyl dacyl ester (Figure 4). The 3-Phenoxybenzaldehyde is a common product of type II SPs, as also was detected in the biodegradation process of fenvalerate (Chen et al., 2011b), deltamethrin (Chen et al., 2011c), fenpropathrin (Chen et al., 2014), and β-cypermethrin (Xiao et al., 2015; Tang et al., 2017). However, this metabolite has been rarely observed in type I SPs. Involvement of metabolite 1,2-benzenedicarboxylic butyl dacyl ester in SPs biodegradation has never been reported. It is worth noting that the intermediate products were transient and disappeared gradually, without further accumulation of metabolites at the end of experiment, suggesting that the isolate may harbor a complete metabolic pathway for the degradation and metabolism of D-phenothrin. Based on metabolites analysis, a degradation pathway of D-phenothrin in strain P31 has been proposed during this study (Figure 5). The proposed D-phenothrin degradation pathway may provide new insights into the biogeocycle of this pesticide.
During this study, potential of the strain P31 for bioremediation of pyrethroid-contaminated soils was investigated under laboratory conditions. Bioaugmentation of D-phenothrin-contaminated soils with strain P31 significantly enhanced the degradation of D-phenothrin, and 75.4∼77.0% of the initial dose of D-phenothrin (50 mg⋅kg-1) was removed from soils within 10 days at t1/2 value of 5.0∼5.9 days; while in the control group without strain P31 it took 18.5∼32.7 days. Results indicate biodegradation as the primary mechanism of SPs dissipation in the environment whereas its abiotic breakdown is less important (Khan et al., 1988; Maloney et al., 1988). Notably, at a final bacterial count of 1 × 107 cells⋅g-1 of soil, strain P31 rapidly degraded D-phenothrin in the beginning of incubation without any lag phase. These results contradict the findings of Cycoń et al. (2014) who reported the slow degradation of deltamethrin during initial 14 days, in the soil bioaugmented with Serratia marcescens DeI-1 and DEI-2 and only 10∼14% of the initial dose was removed. Usually, the introduced bacteria need to adapt to the soil conditions, xenobiotics and to the proliferation of autochthonous microorganisms utilizing this pollutant as a carbon and energy source (Zhang et al., 2012; Cycoń and Piotrowska-Seget, 2016). Unlike other bacterial isolates, P. fulva P31 has an exceptional ability to adapt and thrive in nutrient deficient ecological niches that makes it a potent strain for various applications.
Conclusion
D-phenothrin-degrading bacterial strain P. fulva P31 was isolated and characterized in terms of its physiology, biochemistry, and biodegradation ability. Strain P31 revealed remarkable capacity to degrade a wide range of SPs that cause environmental contaminations. Based on metabolites analysis, we also proposed a D-phenothrin degradation pathway in strain P31. Moreover, this particular strain exhibited great advantages in bioremediation of pyrethroid-contaminated soils because of its adaptability in water and soil environments. These insights will facilitate to develop new strategies to curb pesticide residues related environmental pollution.
Author Contributions
SC designed the experiments; JY, YF, HZ, KZ, and SC performed the experiments; JY, FY, and SC performed the data analysis; LZ provided the scientific expertise; JY, JL, FY, and SC wrote the manuscript.
Funding
This study was partially funded by grants from the National Natural Science Foundation of China (31401763), the National Key Project for Basic Research (2015CB150600), Guangdong Natural Science Funds for Distinguished Young Scholar (2015A030306038), the Science and Technology Planning Project of Guangdong Province (2016A020210106, 2017A010105008), and Pearl River S&T Nova Program of Guangzhou (201506010006).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.01003/full#supplementary-material
References
Ansari, A. R., Rahman, S., Kaur, M., Anjum, S., and Raisuddin, S. (2011). In vivo cytogenetic and oxidative stress-inducing effects of cypermethrin in freshwater fish, Channa punctata Bloch. Ecotoxol. Environ. Saf. 74, 150–156. doi: 10.1016/j.ecoenv.2010.08.036
Arora, P. K., and Bae, H. (2014). Biotransformation and chemotaxis of 4-chloro-2-nitrophenol by Pseudomonas sp. JHN. Microb. Cell Fact. 13:110. doi: 10.1186/s12934-014-0110-7
Arora, P. K., Srivastava, A., Garg, S. K., and Singh, V. P. (2017). Recent advances in degradation of chloronitrophenols. Bioresour. Technol. 250, 902–909. doi: 10.1016/j.biortech.2017.12.007
Arora, P. K., Srivastava, A., and Singh, V. P. (2014). Degradation of 4-chloro-3-nitrophenol via a novel intermediate, 4-chlororesorcinol by Pseudomonas sp. JHN. Sci. Rep. 4:4475. doi: 10.1038/srep04475
Atmaca, E., and Aksoy, A. (2015). D-Phenothrin-induced oxidative DNA damage in rat liver and kidney determined by HPLC-ECD/DAD. Environ. Toxicol. 30, 607–613. doi: 10.1002/tox.21938
Bezerra, M. A., Santelli, R. E., Oliveir, E. P., Villar, L. S., and Escaleira, L. A. (2008). Response surface methodology (RSM) as a tool for optimization in analytical chemistry. Talanta 76, 965–977. doi: 10.1016/j.talanta.2008.05.019
Brander, S. M., Gabler, M. K., Fowler, N. L., Connon, R. E., and Schlenk, D. (2016). Pyrethroid pesticides as endocrine disruptors: molecular mechanisms in vertebrates with a focus on fishes. Environ. Sci. Technol. 50, 8977–8992. doi: 10.1021/acs.est.6b02253
Chen, S. H., Chang, C. Q., Deng, Y. Y., An, S. W., Dong, Y. H., Zhou, J. N., et al. (2014). Fenpropathrin biodegradation pathway in Bacillus sp DG-02 and its potential for bioremediation of pyrethroid-contaminated soils. J. Agric. Food Chem. 62, 2147–2157. doi: 10.1021/jf404908j
Chen, S. H., Deng, Y. Y., Chang, C. Q., Lee, J., Cheng, Y. Y., Cui, Z. N., et al. (2015). Pathway and kinetics of cyhalothrin biodegradation by Bacillus thuringiensis strain ZS-19. Sci. Rep. 5:8784. doi: 10.1038/srep08784
Chen, S. H., Dong, Y. H., Chang, C. Q., Deng, Y. Y., Zhang, X. F., Zhong, G. H., et al. (2013). Characterization of a novel cyfluthrin-degrading bacterial strain Brevibacterium aureum and its biochemical degradation pathway. Bioresour. Technol. 132, 16–23. doi: 10.1016/j.biortech.2013.01.002
Chen, S. H., Geng, P., Xiao, Y., and Hu, M. Y. (2012). Bioremediation of β-cypermethrin and 3-phenoxybenzaldehyde contaminated soils using Streptomyces aureus HP-S-01. Appl. Microbiol. Biotechnol. 94, 505–515. doi: 10.1007/s00253-011-3640-5
Chen, S. H., Hu, M. Y., Liu, J. J., Zhong, G. H., Yang, L., Rizwan-Ul-Haq, M., et al. (2011a). Biodegradation of beta-cypermethrin and 3-phenoxybenzoic acid by a novel Ochrobactrum lupini DG-S-01. J. Hazard. Mater. 187, 433–440. doi: 10.1016/j.jhazmat.2011.01.049
Chen, S. H., Hu, Q. B., Hu, M. Y., Luo, J. J., Weng, Q. F., and Lai, K. P. (2011b). Isolation and characterization of a fungus able to degrade pyrethroids and 3-phenoxybenzaldehyde. Bioresour. Technol. 102, 8110–8116. doi: 10.1016/j.biortech.2011.06.055
Chen, S. H., Lai, K. P., Li, Y. N., Hu, M. Y., Zhang, Y. B., and Zeng, Y. (2011c). Biodegradation of deltamethrin and its hydrolysis product 3-phenoxybenzaldehyde by a newly isolated Streptomyces aureus strain HP-S-01. Appl. Microbiol. Biotechnol. 90, 1471–1483. doi: 10.1007/s00253-011-3136-3
Chen, S. H., Yang, L., Hu, M. Y., and Liu, J. J. (2011d). Biodegradation of fenvalerate and 3-phenoxybenzoic acid by a novel Stenotrophomonas sp. strain ZS-S-01 and its use in bioremediation of contaminated soils. Appl. Microbiol. Biotechnol. 90, 755–767. doi: 10.1007/s00253-010-3035-z
Corcellas, C., Eljarrat, E., and Barceló, D. (2015). First report of pyrethroid bioaccumulation in wild river fish: a case study in Iberian river basins (Spain). Environ. Int. 75, 110–116. doi: 10.1016/j.envint.2014.11.007
Cycoń, M., Mrozik, A., and Piotrowska-Seget, Z. (2017). Bioaugmentation as a strategy for the remediation of pesticide-polluted soil: a review. Chemosphere 172, 52–71. doi: 10.1016/j.chemosphere.2016.12.129
Cycoń, M., and Piotrowska-Seget, Z. (2016). Pyrethroid-degrading microorganisms and their potential for the bioremediation of contaminated soils: a review. Front. Microbiol. 7:1463. doi: 10.3389/fmicb.2016.01463
Cycoń, M., Wójcik, M., and Piotrowska-Seget, Z. (2009). Biodegradation of the organophosphorus insecticide diazinon by Serratia sp. and Pseudomonas sp. and their use in bioremediation of contaminated soil. Chemosphere 76, 494–501. doi: 10.1016/j.chemosphere.2009.03.023
Cycoń, M., Żmijowska, A., and Piotrowska-Seget, Z. (2014). Enhancement of deltamethrin degradation by soil bioaugmentation with two different strains of Serratia marcescens. Int. J. Environ. Sci. Technol. 11, 1305–1316. doi: 10.1007/s13762-013-0322-0
Delgado-Moreno, L., Lin, K., Veiga-Nascimento, R., and Gan, J. (2011). Occurrence and toxicity of three classes of insecticides in water and sediment in two southern California coastal watersheds. J. Agric. Food Chem. 59, 9448–9456. doi: 10.1021/jf202049s
Festa, F., Steel, J., Bian, X., and Labaer, J. (2013). High-throughput cloning and expression library creation for functional proteomics. Proteomics 13, 1381–1399. doi: 10.1002/pmic.201200456
Gao, Y., Chen, S. H., Hu, M. Y., Hu, Q. B., Luo, J. J., and Li, Y. N. (2012). Purification and characterization of a novel chlorpyrifos hydrolase from Cladosporium cladosporioides Hu-01. PLoS One 7:e38137. doi: 10.1371/journal.pone.0038137
García, M. D., Duque, S. U., Fernández, A. B., Sosa, A., and Fernández-Alba, A. R. (2017). Multiresidue method for trace pesticide analysis in honeybee wax comb by GC-QqQ-MS. Talanta 163, 54–64. doi: 10.1016/j.talanta.2016.10.083
Ghevariya, C. M., Bhatt, J. K., and Dave, B. P. (2011). Enhanced chrysene degradation by halotolerant Achromobacter xylosoxidans using response surface methodology. Bioresour. Technol. 102, 9668–9674. doi: 10.1016/j.biortech.2011.07.069
Heidari, R., Devonshire, A. L., Campbell, B. E., Dorrian, S. J., Oakeshott, J. G., and Russell, R. J. (2005). Hydrolysis of pyrethroids by carboxylesterases from Lucilia cuprina and Drosophila melanogaster with active sites modified by in vitro mutagenesis. Insect Biochem. Mol. Biol. 35, 597–609. doi: 10.1016/j.ibmb.2005.02.018
Jilani, S., and Khan, M. A. (2006). Biodegradation of cypermethrin by Pseudomonas in a batch activated sludge process. Int. J. Environ. Sci. Technol. 3, 371–380. doi: 10.1007/BF03325946
Khan, S. U., Bekhi, R. M., Tapping, R. I., and Akhbar, M. H. (1988). Deltamethrin residues in an organic soil under laboratory conditions and its degradation by bacterial strain. J. Agric. Food Chem. 36, 636–638. doi: 10.1021/jf00081a057
Kuivila, K. M., Hladik, M. L., Ingersoll, C. G., Kemble, N. E., Moran, P. W., Calhoun, D. L., et al. (2012). Occurrence and potential sources of pyrethroid insecticides in stream sediments from seven US metropolitan areas. Environ. Sci. Technol. 46, 4297–4303. doi: 10.1021/es2044882
Laskowski, D. A. (2002). Physical and chemical properties of pyrethroids. Rev. Environ. Contam. Toxicol. 174, 49–170. doi: 10.1007/978-1-4757-4260-2_3
Lin, Q. S., Chen, S. H., Hu, M. Y., Rizwan-ul-Haq, M., Yang, L., and Li, H. (2011). Biodegradation of cypermethrin by a newly isolated actinomycetes HU-S-01 from wastewater sludge. Int. J. Environ. Sci. Technol. 8, 45–56. doi: 10.1007/BF03326194
Liu, J., Chen, S. H., Ding, J., Xiao, Y., Han, H. T., and Zhong, G. H. (2015). Sugarcane bagasse as support for immobilization of Bacillus pumilus HZ-2 and its use in bioremediation of mesotrione-contaminated soils. Appl. Microbiol. Biotechnol. 99, 10839–10851. doi: 10.1007/s00253-015-6935-0
Luong, J. H. (1987). Generalization of monod kinetics for analysis of growth data with substrate inhibition. Biotechnol. Bioeng. 29, 242–248. doi: 10.1002/bit.260290215
Maloney, S. E., Maule, A., and Smith, A. R. (1988). Microbial transformation of the pyrethroid insecticides: permethrin, deltamethrin, fastac, fenvalerate, and fluvalinate. Appl. Environ. Microbiol. 54, 2874–2876.
Markle, J. C., van Buuren, B. H., Moran, K., and Barefoot, A. C. (2014). “Pyrethroid pesticides in municipal wastewater: a baseline survey of publicly owned treatment works facilities in California in 2013,” in Describing the Behavior and Effects of Pesticides in Urban and Agricultural Settings, eds R. L. Jones and S. H. Jackson (Washington, DC: American Chemical Society), 177–194.
Morillo, E., and Villaverde, J. (2017). Advanced technologies for the remediation of pesticide-contaminated soils. Sci. Total Environ. 586, 576–597. doi: 10.1016/j.scitotenv.2017.02.020
Nagy, K., Rácz, G., Matsumoto, T., Ádány, R., and Ádám, B. (2014). Evaluation of the genotoxicity of the pyrethroid insecticide phenothrin. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 770, 1–5. doi: 10.1016/j.mrgentox.2014.05.001
Saikia, N., Das, S. K., Patel, B. K. C., Niwas, R., Singh, A., and Gopal, M. (2005). Biodegradation of beta-cyfluthrin by Pseudomonas stutzeri strain S1. Biodegradation 16, 581–589. doi: 10.1016/j.mrgentox.2014.05.001
Saillenfait, A. M., Ndiaye, D., and Sabaté, J. P. (2015). Pyrethroids: exposure and health effects-an update. Int. J. Hyg. Environ. Health 218, 281–292. doi: 10.1016/j.ijheh.2015.01.002
Scott, J. G., Yoshimizu, M. H., and Kasai, S. (2015). Pyrethroid resistance in Culex pipiens mosquitoes. Pestic. Biochem. Phys. 120, 68–76. doi: 10.1016/j.pestbp.2014.12.018
Song, H., Zhou, Z., Liu, Y., Deng, S., and Xu, H. (2015). Kinetics and mechanism of fenpropathrin biodegradation by a newly isolated Pseudomonas aeruginosa sp. strain JQ-41. Curr. Microbiol. 71, 326–332. doi: 10.1007/s00284-015-0852-4
Stok, J. E., Huang, H. Z., Jones, P. D., Wheelock, C. E., Morisseau, C., and Hammock, B. D. (2004). Identification, expression, and purification of a pyrethroid-hydrolyzing carboxylester from mouse liver microsomes. J. Biol. Chem. 279, 29863–29869. doi: 10.1074/jbc.M403673200
Tallur, P. N., Megadi, V. B., and Ninnekar, H. Z. (2008). Biodegradation of cypermethrin by Micrococcus sp. strain CPN 1. Biodegradation 28, 77–82. doi: 10.1007/s10532-007-9116-8
Tang, A. X., Liu, H., Liu, Y. Y., Li, Q. Y., and Qing, Y. M. (2017). Purification and characterization of a novel β-cypermethrin-degrading aminopeptidase from Pseudomonas aeruginosa GF31. J. Agric. Food Chem. 65, 9412–9418. doi: 10.1021/acs.jafc.7b03288
Topp, E., and Akhtar, M. H. (1991). Identification and characterization of a Pseudomonas strain capable of metabolizing phenoxybenzoates. Appl. Environ. Microbiol. 57, 1294–1300.
Wang, B. Z., Guo, P., Hang, B. J., Li, L., He, J., and Li, S. P. (2009). Cloning of a novel pyrethroid-hydrolyzing carboxylesterase gene from Sphingobium sp. strain JZ-1 and characterization of the gene product. Appl. Environ. Microbiol. 75, 5496–5500. doi: 10.1128/AEM.01298-09
Xiao, Y., Chen, S. H., Gao, Y. Q., Hu, W., Hu, M. Y., and Zhong, G. H. (2015). Isolation of a novel beta-cypermethrin degrading strain Bacillus subtilis BSF01 and its biodegradation pathway. Appl. Microbiol. Biotechnol. 99, 2849–2859. doi: 10.1007/s00253-014-6164-y
Yang, L., Chen, S., Hu, M., Hao, W., Geng, P., and Zhang, Y. B. (2011). Biodegradation of carbofuran by Pichia anomala strain HQ-C-01 and its application for bioremediation of contaminated soils. Biol. Fertil. Soils 47, 917–923. doi: 10.1007/s00374-011-0602-0
Zhai, Y., Li, K., Song, J., Shi, Y., and Yan, Y. (2012). Molecular cloning, purification and biochemical characterization of a novel pyrethroid-hydrolyzing carboxylesterase gene from Ochrobactrum anthropi YZ-1. J. Hazard. Mater. 221–222, 206–212. doi: 10.1016/j.jhazmat.2012.04.031
Zhan, H., Feng, Y. M., Fan, X. H., and Chen, S. H. (2018a). Recent advances in glyphosate biodegradation. Appl. Microbiol. Biotechnol. doi: 10.1007/s00253-018-9035-0 [Epub ahead of print].
Zhan, H., Wang, H. S., Liao, L. S., Feng, Y. M., Fan, X. H., Zhang, L. H., et al. (2018b). Kinetics and novel degradation pathway of permethrin in Acinetobacter baumannii ZH-14. Front. Microbiol. 9:98. doi: 10.3389/fmicb.2018.00098
Zhang, C., Wang, S. H., and Yan, Y. C. (2011). Isomerization and biodegradation of beta-cypermethrin by Pseudomonas aeruginosa CH7 with biosurfactant production. Bioresour. Technol. 102, 7139–7146. doi: 10.1016/j.biortech.2011.03.086
Keywords: bioremediation, D-phenothrin, Pseudomonas fulva, metabolites, degradation pathway
Citation: Yang J, Feng Y, Zhan H, Liu J, Yang F, Zhang K, Zhang L and Chen S (2018) Characterization of a Pyrethroid-Degrading Pseudomonas fulva Strain P31 and Biochemical Degradation Pathway of D-Phenothrin. Front. Microbiol. 9:1003. doi: 10.3389/fmicb.2018.01003
Received: 05 February 2018; Accepted: 30 April 2018;
Published: 16 May 2018.
Edited by:
Sabine Kleinsteuber, Helmholtz-Zentrum für Umweltforschung (UFZ), GermanyReviewed by:
Maria Fátima Carvalho, Centro Interdisciplinar de Pesquisa Marine e Ambiental (CIIMAR), PortugalFernando Govantes, Universidad Pablo de Olavide, Spain
Copyright © 2018 Yang, Feng, Zhan, Liu, Yang, Zhang, Zhang and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shaohua Chen, c2hjaGVuQHNjYXUuZWR1LmNu
†These authors have contributed equally to this work.
 Jingjing Yang1†
Jingjing Yang1† Yanmei Feng
Yanmei Feng Hui Zhan
Hui Zhan Shaohua Chen
Shaohua Chen