- 1Center of Infectious Diseases, West China Hospital, Sichuan University, Chengdu, China
- 2Division of Infectious Diseases, State Key Laboratory of Biotherapy, Chengdu, China
- 3Laboratory of Clinical Microbiology, Department of Laboratory Medicine, West China Hospital, Sichuan University, Chengdu, China
- 4Department of Infection Control, West China Hospital, Sichuan University, Chengdu, China
- 5Center for Pathogen Research, West China Hospital, Sichuan University, Chengdu, China
Klebsiella grimontii is a newly identified species closely related to Klebsiella oxytoca, but carbapenem resistance was not identified in the species before. We found a carbapenem-resistant K. oxytoca-like clinical strain, WCHKG020121. The strain was subjected to whole genome sequencing using Illumina HiSeq X10. The precise species identification was established based on average nucleotide identity (ANI) and in silico DNA–DNA hybridization (isDDH) between strain WCHKG020121 and type strains of Klebsiella species. Antimicrobial resistance genes were identified from the genome sequence. The sequence of the blaKPC-2-carrying plasmid was completed using PCR and Sanger sequencing. Conjugation experiments were performed to obtain the plasmid carrying blaKPC-2. All K. grimontii genomes were retrieved from GenBank and were analyzed for antimicrobial resistance genes. Strain WCHKG020121 was resistant to imipenem and meropenem (MIC for both, 32 μg/ml) but was susceptible to colistin (1 μg/ml). Strain WCHKG020121 was initially identified as K. oxytoca using Vitek II but it actually belongs to K. grimontii as it had a 98.81% ANI and 83.4% isDDH value with K. grimontii type strain. Strain WCHKG020121 had blaKPC-2; by contrast, none of other K. grimontii genomes carry any known carbapenemase genes. blaKPC-2 was carried by a 95,734-bp plasmid, designated pKPC2_020121, which contained two different FII(Y) replicons. pKPC2_020121 was closest (93% coverage, 99% identity) to blaKPC-2-carrying plasmids from Enterobacter hormaechei recovered in 2014 at the same hospital. pKPC2_020121 was not self-transmissible, which could be explained by the absence of a conjugation essential gene, traY. In conclusion, we reported the first K. grimontii strain that produced the KPC carbapenemase. Carbapenem resistant K. grimontii may represent a new threat.
Introduction
Klebsiella grimontii is a newly identified species of the Klebsiella genus within the Enterobacteriaceae family (Passet and Brisse, 2018). K. grimontii is closely related to Klebsiella oxytoca and has been previously known as the ko6 phylogenetic group of K. oxytoca (Fevre et al., 2005; Passet and Brisse, 2018). Like K. oxytoca, a relatively common human pathogen (Herzog et al., 2014), K. grimontii is also associated with human infections such as bacteremia and soft tissue infection and has been found in France, Germany, and South Africa (Passet and Brisse, 2018). Carbapenems, such as imipenem and meropenem, are the main choice to treat severe infections caused by the Enterobacteriaceae, but carbapenem-resistant Enterobacteriaceae has emerged as a major threat for human health (Holt et al., 2015). Carbapenem-resistant Klebsiella spp., in particular Klebsiella pneumoniae, has been found worldwide and blaKPC is a major determinant conferring carbapenem resistance (Tzouvelekis et al., 2012). The international dissemination of blaKPC is largely mediated by ST258 K. pneumoniae (Munoz-Price et al., 2013), while in China ST11 is the major type of carbapenem-resistant Klebsiella (Qi et al., 2011). The plasmids carrying blaKPC remain largely unexplored but IncF plasmids may act as a major vehicle mediating the dissemination of blaKPC (Chmelnitsky et al., 2014). However, carbapenem-resistant K. grimontii has not been reported before. We have found and characterized a carbapenem-resistant K. grimontii clinical strain, which is reported here.
Materials and Methods
Strain and in vitro Susceptibility
Strain WCHKG020121 was recovered from a human sputum sample in 2017 in China. Initial species identification was performed using Vitek II (bioMérieux, Marcy-l’Étoile, France). MICs of amikacin, aztreonam, aztreonam/avibactam, ceftazidime, ceftazidime/avibactam, ciprofloxacin, colistin, imipenem, meropenem, piperacillin/tazobactam, tigecycline, and trimethoprim/sulfamethoxazole were determined using the broth microdilution method of the Clinical and Laboratory Standards Institute (Clinical and Laboratory Standards Institute [CLSI], 2017). As there are no breakpoints of colistin and tigecycline from CLSI, those defined by EUCAST1 were applied. As this study was to characterize the bacterial strain and ethical approval was not required according to the Ethical Committee of West China Hospital. No patient information was included in this study.
Whole Genome Sequencing and Analysis
Genomic DNA of strain WCHKG020121 was prepared using the QIAamp DNA mini kit (Qiagen, Hilden, Germany) and was subjected to whole genome sequencing using the HiSeq X10 platform (Illumina, San Diego, CA, United States). Reads were trimmed using Trimmomatic (Bolger et al., 2014) and were then assembled to contigs using the SPAdes program v3.12.0 (Bankevich et al., 2012) with careful mode turned on. Annotation of the genomic sequence was carried out using the Prokka program v1.12 (Seemann, 2014). The precise species identification was established based on average nucleotide identity (ANI) and in silico DNA-DNA hybridization (isDDH) between strain WCHKG020121 and type strains of Klebsiella species (Table 1) using JSpeciesWS (Richter et al., 2016) and GGDC (formula 2) (Meier-Kolthoff et al., 2013), respectively. Antimicrobial resistance genes were identified from the genome sequence using the ABRicate program2 to query the ResFinder database3. The sequence of the blaKPC-2-carrying plasmid, a carbapenemase-encoding gene, was completed using PCR and Sanger sequencing to close gaps between contigs. Plasmid replicon types were determined using the PlasmidFinder tool at https://cge.cbs.dtu.dk/services/PlasmidFinder/ and the allele types of IncF plasmids were assigned using the IncF replicon typing tool (Villa et al., 2010).
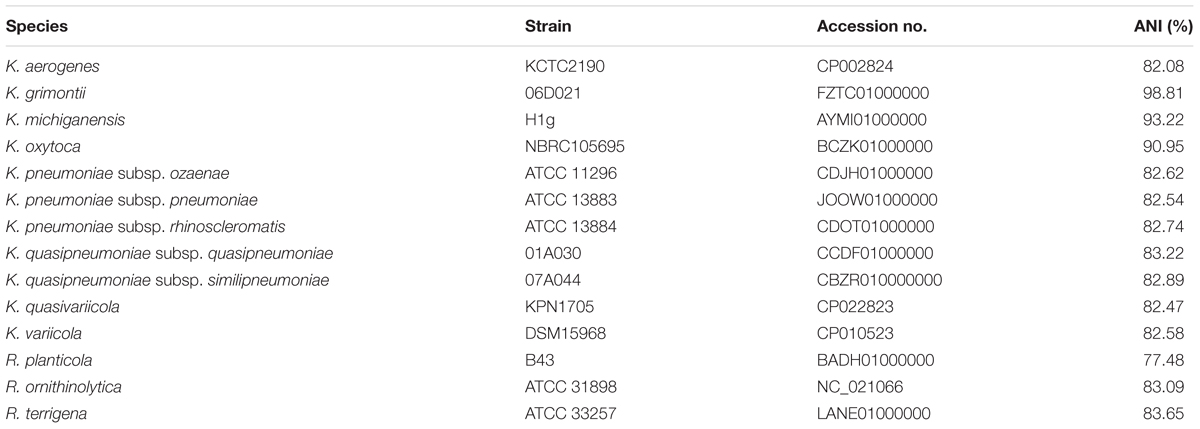
TABLE 1. Average nucleotide identity (ANI) and isDDH values between strain WCHKG020121 and the type strains of Klebsiella species.
Nucleotide Sequence Accession Number
The draft whole-genome sequence of strain WCHKG020121 and the complete sequence of pKPC2_020121 have been deposited into GenBank under the accession no. QBDY00000000 and MH192342, respectively.
Analysis on K. grimontii Genomes Available in GenBank
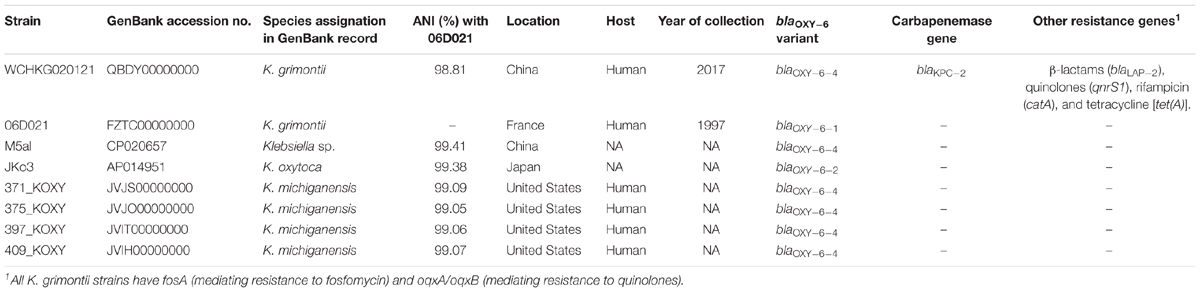
Klebsiella grimontii carries the chromosomally based blaOXY-6 β-lactamase gene, which is the marker of K. grimontii (Passet and Brisse, 2018). All genomes containing blaOXY-6 were therefore retrieved from GenBank (accessed by May 1, 2018, Table 2) and were subjected to the precise species identification using ANI with the type strain of K. grimontii as described above. Antimicrobial resistance genes were identified using ResFinder.
Mating
Conjugation experiments were carried out in blood heart infusion broth (Oxoid, Hampshire, United Kingdom) and on nitrocellulose filters (GE Life Science, Pittsburgh, PA, United States) at both 30 and 37° as described previously (Coque et al., 2002; Novais et al., 2006; Valenzuela et al., 2007). An azide-resistant Escherichia coli strain J53 was used as the recipient. For the broth method, the donor and recipient were mixed at a ratio of 1:10 and the mixture was incubated overnight. For the filter method, the donor and recipient were mixed at a ratio of 1:1 and the mixture was incubated for 4 h. Transconjugants were then selected on LB agar plates containing 4 μg/ml meropenem and 150 μg/ml azide.
Results and Discussion
Strain WCHKG020121 was resistant to aztreonam (MIC, 512 μg/ml), ceftazidime (64 μg/ml), imipenem (32 μg/ml), meropenem (32 μg/ml), piperacillin/tazobactam (512/4 μg/ml) and tigecycline (4 μg/ml), intermediate to ciprofloxacin (2 μg/ml) and susceptible to amikacin (1 μg/ml), aztreonam/avibactam (<0.125/4 μg/ml), ceftazidime/avibactam (0.25/4 μg/ml), colistin (1 μg/ml), and trimethoprim/sulfamethoxazole (<0.5/9.5 μg/ml).
Whole genome sequencing generated 6,957,050 reads and 2.09 Gb clean bases, which were de novo assembled to 127 contigs (108 > 1,000 bp; N50 107,740 bp). The draft genome of strain WCHKG020121 was 6.28 Mb with a 55.56% GC content. Strain WCHKG020121 was initially identified as K. oxytoca using Vitek II. However, strain WCHKG020121 had 98.81% ANI value with strain 06D021T, the type strain of K. grimontii (Passet and Brisse, 2018), while the ANI values between strain WCHKG020121 and types strains of other Klebsiella spp. were 82.54 to 93.22% (Table 1). A =95–96% ANI value (Richter and Rossello-Mora, 2009) is commonly used to define a bacterial species. The isDDH value between strain WCHKG020121 and the type strain of K. grimontii was 83.4%, which is above the 70% cutoff to define a bacterial species. Therefore, strain WCHKG020121 actually belongs to K. grimontii.
The strain had nine antimicrobial resistance genes mediating resistance to β-lactams (blaKPC-2, blaLAP-2, blaOXY-6-4), fosfomycin (fosA), quinolones (oqxA, oqxB, qnrS1), rifampicin (catA), and tetracycline [tet(A)]. There are seven Klebsiella genomes containing blaOXY-6 available in the GenBank(Table 2). Although these strains were commonly reported as K. oxytoca or Klebsiella michiganensis in their records in GenBank, they had >99% ANI values with K. grimontii type strain 06D021 (Table 2), which clearly suggests that the strains actually belonged to K. grimontii. None of these K. grimontii strains carried any known carbapenemase genes (Table 2), although the susceptibility data of carbapenems against these strains were not available. To our knowledge, this is the first report of a carbapenemase-producing K. grimontii, which expands the species spectrum of carbapenem-resistant Enterobacteriaceae. Curiously, all K. grimontii genomes analyzed contained fosA, oqxA, and oqxB, being needed more analyses to verify if they could be intrinsic of this species. In contrast, none of other antimicrobial resistance genes (blaLAP-2, qnrS1, catA, and [tet(A)]) seen in strain WCHKG020121 was present in other K. grimontii genomes, suggesting that these genes were acquired by strain WCHKG020121.
blaKPC-2 was carried by a 95,734-bp plasmid, which is designated pKPC2_020121 here. pKPC2_020121 contained two different FII(Y) replicons, in which the 227-bp allele used by the PlasmidFinder tool to define FII was 91.63% identical between the two replicons. pKPC2_020121 was closest (93% coverage, 99% identity) to 88,213-bp blaKPC-2-carrying plasmids pKPC2-EC14653, pKPC2_EClY2402 and pKPC2_EClY2403 (GenBank accession no. KP868646, KY399972, and KY399973; Figure 1). The three plasmids were identical except several nucleotide differences and were found in three Enterobacter hormaechei isolates, which were recovered in 2014 at our hospital and were likely of a common strain (Yang et al., 2018). Nonetheless, pKPC2-EC14653/pKPC2_EClY2402/pKPC2_EClY2403 had only one FII(Y) replicon (Figure 1). A 7-kb region containing the additional FII(Y) replicon on pKPC2_020121 were absent from the three plasmids; otherwise, pKPC2_020121 were almost identical to pKPC2-EC14653/pKPC2_EClY2402/pKPC2_EClY2403 with only a few nucleotide mutations or insertions/deletions. It is likely that pKPC2_020121 and pKPC2-EC14653/pKPC2_EClY2402/pKPC2_EClY2403 had originated from a common plasmid to mediate inter-species transfer of blaKPC-2 at our hospital and pKPC2_020121 might have acquired the additional FII(Y) replicon during its transfer. The presence of an additional replicon may facilitate the host plasmid to adapt to different strains of different species.
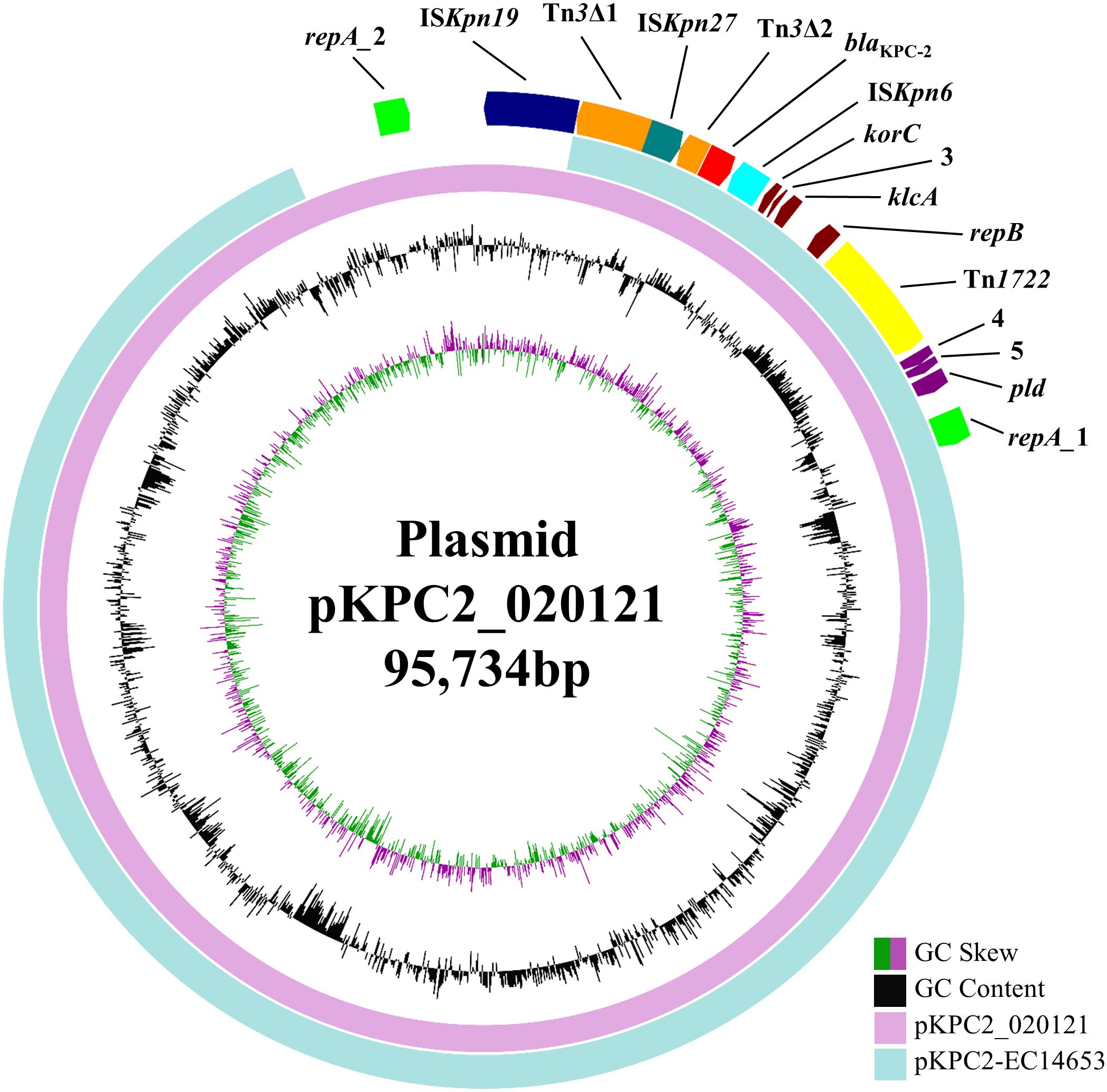
FIGURE 1. pKPC2_020121 vs. pKPC2-EC14653. pKPC2_020121 (in pink) is aligned with pKPC2-EC14653 (GenBank accession no. KP868646; in light blue), a plasmid from E. hormaechei recovered in 2014 at our hospital, using BRIG (Alikhan et al., 2011). pKPC2_020121 has two FII(Y) replicon, while pKPC2-EC14653 has one. GC content and GC skew are indicated. The genetic context of blaKPC-2 is shown in Figure 2. The annotation of the genetic components were added manually using the Microsoft PowerPoint program.
Transconjugants were not obtained despite repeated attempts. We found that the relaxosome protein-encoding gene traY, which is an essential component of the conjugation module, was absent from pKPC2_020121. This could explain that pKPC2_020121 was not self-transmissible. Nonetheless, in our previous study, we have found that pKPC2-EC14653 could be transferred in the presence of a self-transmissible FII plasmid (Wu et al., 2015) suggesting that pKPC2_020121 may also utilize this mechanism to realize its transmission. Plasmid replicon typing revealed that strain WCHKG020121 had an FII(K) and an FIB(K) replicon in addition to pKPC2_020121, suggesting that there is one more FII plasmid in the strain.
The genetic context of blaKPC-2 on pKPC2_020121 was almost identical to those on pKPC2-EC14653/pKPC2_EClY2402/pKPC2_EClY2403 (Figure 2). The only difference is that the Tn3 transposon upstream of blaKPC-2 was truncated by ISKpn19, resulting in the absence of the Tn3 inverted repeat (IR) from pKPC2_020121.
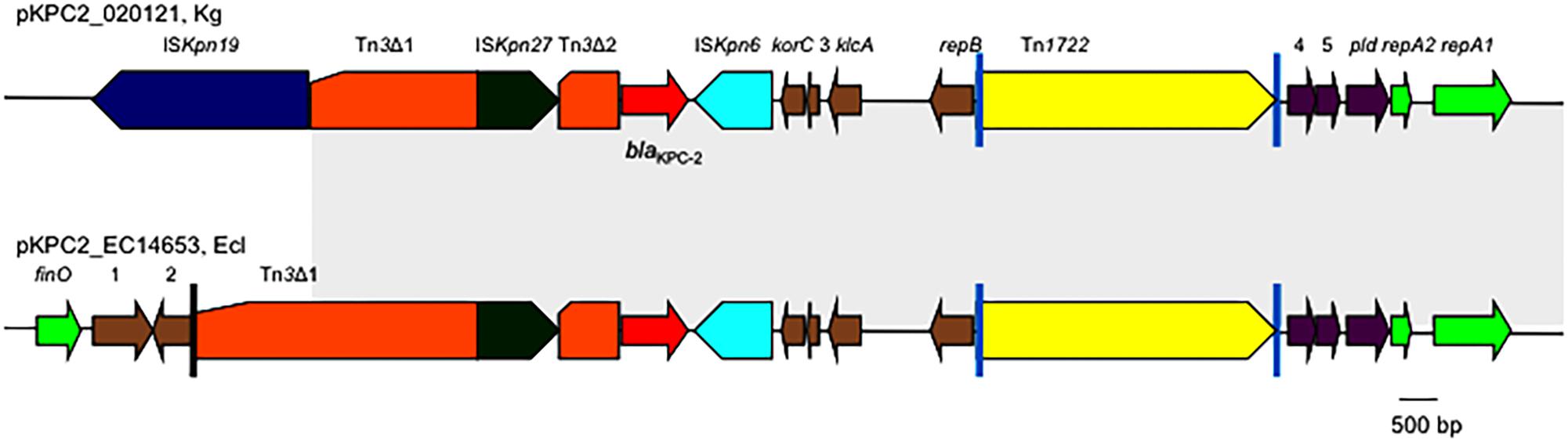
FIGURE 2. The genetic context of blaKPC-2 on pKPC2_020121 and pKPC2-EC14653. The genetic context of blaKPC-2 on pKPC2_020121 is compared with that on pKPC2-EC14653 with the identical region highlighted in gray. Poles in black are the inverted repeats (IR) of Tn3, while poles in cyan are the IR of Tn1722. orfs without known function are indicated by numbers 1–5. This figure was drawn manually using the Microsoft PowerPoint program.
Conclusion
We reported a carbapenem-resistant strain of the newly recognized species K. grimontii. Carbapenem resistance was due to blaKPC-2, which was carried by a plasmid containing two FII(Y) replicons. The blaKPC-2-carrying plasmid had circulated in different species at the hospital for several years.
Author Contributions
ZZ designed the study. LL, YH, YX, and MK performed the experiments. LL, YF, and ZZ analyzed and interpreted the data. ZZ wrote the manuscript.
Funding
This work was supported by grants from the National Natural Science Foundation of China (Project Nos. 81572030 and 81772233), a joint grant from the National Natural Science Foundation of China (Project No. 81661130159), and the Newton Advanced Fellowship, Royal Society, United Kingdom (NA150363).
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Footnotes
- ^http://www.eucast.org/
- ^https://github.com/tseemann/abricate
- ^https://cge.cbs.dtu.dk/services/ResFinder/
References
Alikhan, N. F., Petty, N. K., Ben Zakour, N. L., and Beatson, S. A. (2011). BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. doi: 10.1186/1471-2164-12-402
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Chmelnitsky, I., Shklyar, M., Leavitt, A., Sadovsky, E., Navon-Venezia, S., Ben Dalak, M., et al. (2014). Mix and match of KPC-2 encoding plasmids in Enterobacteriaceae-comparative genomics. Diagn. Microbiol. Infect. Dis. 79, 255–260. doi: 10.1016/j.diagmicrobio.2014.03.008
Clinical and Laboratory Standards Institute [CLSI] (2017). Performance Standards for Antimicrobial Susceptibility Testing; Twenty-Seventh Informational Supplement. M100-S27. Wayne, PA: Clinical and Laboratory Standards Institute.
Coque, T. M., Oliver, A., Perez-Diaz, J. C., Baquero, F., and Canton, R. (2002). Genes encoding TEM-4, SHV-2, and CTX-M-10 extended-spectrum β-lactamases are carried by multiple Klebsiella pneumoniae clones in a single hospital (Madrid, 1989 to 2000). Antimicrob. Agents Chemother. 46, 500–510. doi: 10.1128/AAC.46.2.500-510.2002
Fevre, C., Jbel, M., Passet, V., Weill, F. X., Grimont, P. A., and Brisse, S. (2005). Six groups of the OXY β-Lactamase evolved over millions of years in Klebsiella oxytoca. Antimicrob. Agents Chemother. 49, 3453–3462. doi: 10.1128/AAC.49.8.3453-3462.2005
Herzog, K. A., Schneditz, G., Leitner, E., Feierl, G., Hoffmann, K. M., Zollner-Schwetz, I., et al. (2014). Genotypes of Klebsiella oxytoca isolates from patients with nosocomial pneumonia are distinct from those of isolates from patients with antibiotic-associated hemorrhagic colitis. J. Clin. Microbiol. 52, 1607–1616. doi: 10.1128/JCM.03373-13
Holt, K. E., Wertheim, H., Zadoks, R. N., Baker, S., Whitehouse, C. A., Dance, D., et al. (2015). Genomic analysis of diversity, population structure, virulence, and antimicrobial resistance in Klebsiella pneumoniae, an urgent threat to public health. Proc. Natl. Acad. Sci. U.S.A. 112, E3574–E3581. doi: 10.1073/pnas.1501049112
Meier-Kolthoff, J. P., Auch, A. F., Klenk, H. P., and Goker, M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions. BMC Bioinformatics 14:60. doi: 10.1186/1471-2105-14-60
Munoz-Price, L. S., Poirel, L., Bonomo, R. A., Schwaber, M. J., Daikos, G. L., Cormican, M., et al. (2013). Clinical epidemiology of the global expansion of Klebsiella pneumoniae carbapenemases. Lancet Infect. Dis. 13, 785–796. doi: 10.1016/S1473-3099(13)70190-7
Novais, A., Canton, R., Valverde, A., Machado, E., Galan, J. C., Peixe, L., et al. (2006). Dissemination and persistence of blaCTX-M-9 are linked to class 1 integrons containing CR1 associated with defective transposon derivatives from Tn402 located in early antibiotic resistance plasmids of IncHI2, IncP1-α, and IncFI groups. Antimicrob. Agents Chemother. 50, 2741–2750. doi: 10.1128/AAC.00274-06
Passet, V., and Brisse, S. (2018). Description of Klebsiella grimontii sp. nov. Int. J. Syst. Evol. Microbiol. 68, 377–381. doi: 10.1099/ijsem.0.002517
Qi, Y., Wei, Z., Ji, S., Du, X., Shen, P., and Yu, Y. (2011). ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J. Antimicrob. Chemother. 66, 307–312. doi: 10.1093/jac/dkq431
Richter, M., and Rossello-Mora, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 106, 19126–19131. doi: 10.1073/pnas.0906412106
Richter, M., Rossello-Mora, R., Oliver Glockner, F., and Peplies, J. (2016). JSpeciesWS: a web server for prokaryotic species circumscription based on pairwise genome comparison. Bioinformatics 32, 929–931. doi: 10.1093/bioinformatics/btv681
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Tzouvelekis, L. S., Markogiannakis, A., Psichogiou, M., Tassios, P. T., and Daikos, G. L. (2012). Carbapenemases in Klebsiella pneumoniae and other Enterobacteriaceae: an evolving crisis of global dimensions. Clin. Microbiol. Rev. 25, 682–707. doi: 10.1128/CMR.05035-11
Valenzuela, J. K., Thomas, L., Partridge, S. R., Van Der Reijden, T., Dijkshoorn, L., and Iredell, J. (2007). Horizontal gene transfer in a polyclonal outbreak of carbapenem-resistant Acinetobacter baumannii. J. Clin. Microbiol. 45, 453–460. doi: 10.1128/JCM.01971-06
Villa, L., Garcia-Fernandez, A., Fortini, D., and Carattoli, A. (2010). Replicon sequence typing of IncF plasmids carrying virulence and resistance determinants. J. Antimicrob. Chemother. 65, 2518–2529. doi: 10.1093/jac/dkq347
Wu, W., Feng, Y., Carattoli, A., and Zong, Z. (2015). The emergence of Enterobacter cloacae producing both KPC and NDM carbapenemases: characterization by whole genome sequencing. Antimicrob. Agents Chemother. 59, 6625–6628. doi: 10.1128/AAC.01275-15
Keywords: carbapenemase, KPC-2, Klebsiella, Klebsiella grimontii, Klebsiella oxytoca
Citation: Liu L, Feng Y, Hu Y, Kang M, Xie Y and Zong Z (2018) Klebsiella grimontii, a New Species Acquired Carbapenem Resistance. Front. Microbiol. 9:2170. doi: 10.3389/fmicb.2018.02170
Received: 28 May 2018; Accepted: 23 August 2018;
Published: 11 September 2018.
Edited by:
Raffaele Zarrilli, University of Naples Federico II, ItalyReviewed by:
Jason Sahl, Northern Arizona University, United StatesFilipa Grosso, Universidade do Porto, Portugal
Copyright © 2018 Liu, Feng, Hu, Kang, Xie and Zong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhiyong Zong, em9uZ3poaXlAc2N1LmVkdS5jbg==
†These authors have contributed equally to this work
 Lu Liu1,2†
Lu Liu1,2† Yu Feng
Yu Feng Zhiyong Zong
Zhiyong Zong