Abstract
Idiomarinaceae is a family of Gram-stain negative, mesophilic euryhalophiles. To provide a robust framework for the evolutionary and taxonomic relationships of bacteria of this family, we compared herein the genomes of 36 type strains and 43 non-type strains using 16S rRNA gene sequences, core genome based 78 single-copy orthologous proteins, digital DNA-DNA hybridization and average nucleotide identity (ANI) estimation. The 79 bacteria of this family were consistently divided into taxon I, taxon II, and taxon III corresponding to the three genera Idiomarina, Pseudidiomarina, and Aliidiomarina, which contained 13 putative new genospecies in addition to 35 well-defined species represented by each type strain. Furthermore, genetic diversity of this family was evident at the genus- and species levels, and exceeded that which is defined currently by the named species. In view of multiple genotypic characteristics clearly distinct from the other two genera, we propose reinstating the genus Pseudidiomarina as a monophyletic taxon. Taken together, this is the first genome-based study of the taxonomy and diversity of bacteria within the family Idiomarinaceae, and will contribute to further insights into microbial evolution and adaptation to saline environments.
Introduction
The family Idiomarinaceae, belonging to the order Alteromonadales within the class Gammaproteobacteria, was first introduced by Ivanova et al. (2004) and initially contained only the genus Idiomarina (Ivanova et al., 2000). Soon after, the second genus Pseudidiomarina was described on the basis of multiple features clearly distinct from bacteria of the genus Idiomarina (Jean et al., 2006). However, as more species were identified, the members of the genus Pseudidiomarina were reclassified in the genus Idiomarina in consideration of reduced discrepancies between the two genera (Taborda et al., 2009). More recently, a novel genus Aliidiomarina of this family was proposed by Huang et al. (2012). As a result, the family Idiomarinaceae currently embraces the two genera of Idiomarina and Aliidiomarina; the former contains 27 species with validly published names and the latter contains eight validly named species (List of Prokaryotic names with Standing in Nomenclature1), along with the effectively but not yet validly named species Aliidiomarina haloalkalitolerans (Srinivas and Anil Kumar, 2012). The members of this family are Gram-stain negative, mesophilic, non-sporulating rods with slightly curved morphology, and require NaCl for growth with the exception of Idiomarina xiamenensis (Wang et al., 2011; Albuquerque and da Costa, 2014). Almost all organisms of this family thrive in various habitats with a wide range of salinities, such as coastal and oceanic waters and sediments, solar salt-making works, saline-alkaline soil, inland hypersaline wetlands, and even submarine hydrothermal fluids, some with salinities higher than the surroundings, clearly indicating that they are typical euryhalophiles (Albuquerque and da Costa, 2014). Some bacteria of this family are well-known for distinctive features such as a primary dependence on amino acid catabolism for carbon and energy rather than on sugar fermentation (Hou et al., 2004); the production of diverse enzymes including lipase (Li et al., 2014), serine hydroxymethyl transferase (Kumar et al., 2018), N-acetylneuraminate synthase (Garcia Garcia et al., 2015), etc.; the synthesis of many kinds of nanoparticles based on selenium (Srivastava and Kowshik, 2016) and lead sulfide (Srivastava and Kowshik, 2017); and the production of Ca–Mg kutnahorite and struvite (Gonzalez-Munoz et al., 2008).
The small subunit ribosomal RNA gene (16S rRNA) has been successfully used for taxonomic classification and in analyses of community structure of Bacteria and Archaea at different taxonomic levels for over 30 years (Yarza et al., 2014). Inevitably, 16S rRNA gene analyses have also played an important role in determining the taxonomy of the family Idiomarinaceae (Poddar et al., 2014). On one hand, the establishment of this family relied mainly on the signature nucleotides of the 16S rRNA gene, including the position 143 (C), 662 (A), 682 (A), 830 (T), 856 (A) (Ivanova et al., 2004). On the other hand, based on the analysis of 16S rRNA gene sequences, the type strains of this family were classified into the four groups, labeled Group I, Group II, Group III, and Group IV. The first three of these correspond to the genus Idiomarina, while the latter corresponds to the genus Aliidiomarina (Poddar et al., 2014). However, we found, unexpectedly, that both Group I and Group II within the genus Idiomarina were actually monophyletic in multiple 16S rRNA gene trees, suggesting they represented well-defined lineages. As is well-accepted, from a viewpoint of classical taxonomy, a genus can be defined by uniting the strains from one or more species into a monophyletic cluster of a phylogenetic tree based on single or multiple molecular markers (Thompson et al., 2013). Based on the viewpoint above, the genus Idiomarina does not seem to meet the definition of a single genus, and thus need to be further investigated. Meanwhile, we further found that with the increase in number of isolated strains, the 16S rRNA gene trees of the family Idiomarinaceae presented inconsistent branching patterns and unstable topological structures, with low bootstrap values. Consequently, reliable and high resolution methods are needed for providing a robust taxonomic classification for bacteria of this family.
Presently, the integration of genomic information into microbial systematics, in addition to physiological and chemotaxonomic parameters as taxonomic criteria, is strongly recommended in the post-genomic era (Chun and Rainey, 2014). Numerous genome-based approaches have been developed and applied for species delineation, for example pan-genome analysis (Medini et al., 2005; Beukes et al., 2017), average nucleotide identity (ANI) analysis (Konstantinidis and Tiedje, 2005; Riesco et al., 2018), and digital DNA–DNA hybridization (dDDH) analysis (Meier-Kolthoff et al., 2013; Montero-Calasanz et al., 2017). The concept of the pan-genome was first introduced in 2005, and includes the core genome composed of genes shared by all strains, a dispensable genome made of genes present in a subset of the strains, and finally strain-specific genes (Medini et al., 2005; Vernikos et al., 2015). As there is a high reliability of using the core genome for bacterial taxonomy at the strain, species and genus levels, and even at higher taxonomic levels, the core genome has been used extensively in comparative genomics of multiple taxa (Colston et al., 2014; Sun et al., 2015; Hesse et al., 2018; Lugli et al., 2018). As the name implies, ANI represents the average nucleotide identity of all orthologous genes shared between two genomes (Konstantinidis and Tiedje, 2005). Owing to the high resolution of ANI, it has been enormously useful in defining species of the Bacteria and Archaea with a proposed species delineation threshold of 95–96% (Richter and Rosselló-Móra, 2009; Salvetti et al., 2018). As one of overall genome relatedness indices (OGRI), dDDH has been recognized for its reproducible, fast and easy-to-implement approaches to replace the traditional DDH (Auch et al., 2010), which for the last few decades has been used as the gold standard for prokaryotic species delineation using a 70% threshold (Wayne et al., 1987). However, the taxonomy and diversity of bacteria of the family Idiomarinaceae have not been studied yet using genome-based approaches above up to now.
With the rapid development of next-generation high throughput sequencing technologies, dozens of genomes of bacteria in the family Idiomarinaceae are currently available in public databases, and thus provide an ideal opportunity for inferring their evolutionary and taxonomic relationships in depth. In the current study, we first sequenced genomes of 27 type strains of the family Idiomarinaceae. Then, we applied four different approaches, including 16S rRNA gene sequence analysis, core genome analysis based on multiple single-copy orthologous protein sequences, dDDH analysis and Orthologous ANI (OrthoANI) analysis using USEARCH, to delineate the taxonomic classification and identification of strains within this family using a dataset of 79 genome sequences. The study revealed the feasibility and advisability of classifying the family Idiomarinaceae into three genera, and provides a robust and straightforward genome-based framework for bacteria of this family.
Materials and Methods
Bacterial Culture and Genome Sequencing
The 27 type strains studied were obtained from multiple culture collections or personal gifts, as listed in Table 1 and Supplementary Table S1. These strains were cultured aerobically on marine agar 2216 to the late exponential phase. Genomic DNA was extracted from each culture by using the SBS extraction kit (SBS Genetech, Co., Ltd., Shanghai, China) following the manufacturer’s instructions. The genomes of these strains were determined on an Illumina MiSeq platform by Shanghai Majorbio Bio-pharm Technology, Co., Ltd. (Shanghai, China). More than 1 Gbp of clean data for each strain were generated, reaching approximately 250-fold coverage depth. The high quality reads were assembled by using the software SPAdes version 3.7.0 with default parameters (Bankevich et al., 2012). The assembled contigs were submitted to the GenBank database and the assigned accession numbers are listed in Table 1.
Table 1
| Genome original name | Taxon | No. of contigs | Genome sizes (Mbp) | G + C contents (mol%) | Accession |
|---|---|---|---|---|---|
| I. abyssalis UBA4213 | I-1 | 5 | 2.68 | 46.98 | DFTL00000000– |
| Idiomarina sp. CNP8 | I-1 | 71 | 2.61 | 46.95 | PBST00000000∗ |
| I. abyssalis UBA3602 | I-1 | 47 | 2.69 | 46.97 | DFMD00000000∗ |
| I. abyssalis UBA5050 | I-1 | 68 | 2.70 | 46.95 | DIBO00000000∗ |
| I. abyssalis KMM 227T | I-1 | 12 | 2.68 | 47.15 | FPBE00000000∗ |
| Idiomarina sp. SAT134 | I-1 | 70 | 2.30 | 47.10 | PAWS00000000∗ |
| I. abyssalis UBA3100 | I-1 | 45 | 2.66 | 47.05 | DFAS00000000∗ |
| Idiomarina sp. SP350 | I-1 | 9 | 2.65 | 47.06 | PBDK00000000∗ |
| I. abyssalis UBA2690 | I-1 | 7 | 2.64 | 47.07 | DELS00000000∗ |
| Idiomarina sp. CPC33 | I-1 | 153 | 2.06 | 46.76 | NYUW00000000∗ |
| Idiomarina sp. UBA4206 | I-1 | 78 | 2.77 | 47.07 | DFTS00000000∗ |
| I. abyssalis UBA5042 | I-1 | 15 | 2.76 | 46.94 | DIBW00000000∗ |
| I. abyssalis UBA3616 | I-1 | 65 | 2.72 | 46.91 | DFLP00000000∗ |
| I. abyssalis UBA3366 | I-1 | 29 | 2.77 | 47.03 | DEQM00000000∗ |
| Idiomarina sp. EAC55 | I-1 | 34 | 2.89 | 46.78 | NZQS00000000∗ |
| Idiomarina sp. SP93 | I-1 | 20 | 2.74 | 46.95 | PAYT00000000∗ |
| I. abyssalis UBA1461 | I-1 | 25 | 2.80 | 46.99 | DCSR00000000∗ |
| I. ramblicola R22T | I-2 | 16 | 2.71 | 46.93 | PIQC00000000§ |
| Idiomarina sp. UBA1919 | I-3 | 57 | 2.96 | 46.79 | DDFV00000000∗ |
| Idiomarina sp. UBA3992 | I-3 | 62 | 2.97 | 46.78 | DGBY00000000∗ |
| Idiomarina sp. UBA3176 | I-3 | 27 | 2.93 | 46.77 | DEXU00000000∗ |
| Idiomarina sp. UBA4211 | I-3 | 163 | 2.86 | 46.72 | DFTN00000000∗ |
| Idiomarina sp. CPC80 | I-3 | 101 | 2.15 | 47.23 | NYSZ00000000∗ |
| I. loihiensis L2TRT | I-3 | 1 | 2.84 | 47.04 | AE017340∗ |
| I. loihiensis GSL 199 | I-3 | 1 | 2.84 | 47.04 | CP005964∗ |
| Idiomarina sp. NORP114 | I-3 | 61 | 2.15 | 47.11 | NVTN00000000∗ |
| I. loihiensis H2.2 | I-4 | 28 | 3.00 | 46.94 | QGUA00000000∗ |
| Idiomarina sp. 28-8 | I-5 | 110 | 2.97 | 46.91 | BANL00000000∗ |
| Idiomarina sp. UBA4520 | I-6 | 22 | 2.47 | 47.16 | DGMK00000000∗ |
| Idiomarina sp. UBA4519 | I-6 | 121 | 2.51 | 47.22 | DGML00000000∗ |
| Idiomarina sp. REDSEA-S27_B4 | I-6 | 60 | 2.26 | 47.20 | LUNZ00000000∗ |
| Idiomarina sp. REDSEA-S21_B4 | I-6 | 60 | 2.26 | 47.20 | LUNZ00000000∗ |
| I. zobellii KMM 231T | I-7 | 30 | 2.58 | 47.08 | FNCB00000000∗ |
| Idiomarina sp. UBA5185 | I-8 | 202 | 2.37 | 47.15 | DHWJ00000000∗ |
| I. piscisalsi TPS4-2T | I-9 | 24 | 2.58 | 47.03 | PIQA00000000§ |
| Idiomarina sp. X4 | I-10 | 1 | 2.62 | 47.27 | CP025000∗ |
| I. piscisalsi 10PY1A | I-11 | 1 | 2.59 | 47.36 | CP022133∗ |
| Idiomarina sp. T82-3 | I-12 | 38 | 2.72 | 47.28 | LSBQ00000000∗ |
| I. baltica UBA859 | I-12 | 137 | 2.40 | 47.48 | DBGH00000000∗ |
| I. baltica OS145T | I-12 | 70 | 2.72 | 47.30 | AAMX00000000∗ |
| Idiomarina sp. UBA3162 | I-13 | 108 | 2.60 | 47.60 | DEYI00000000∗ |
| Idiomarina sp. MD25a | I-14 | 7 | 2.68 | 47.47 | MDGT00000000∗ |
| I. fontislapidosi F23T | I-15 | 47 | 2.88 | 47.75 | PIPV00000000§ |
| Idiomarina sp. OT37-5b | I-16 | 1 | 2.89 | 51.06 | CP027188∗ |
| I. aquatica SN-14T | I-16 | 9 | 2.97 | 50.99 | PIPS00000000§ |
| Idiomarina sp. NP27 | I-17 | 87 | 3.10 | 48.07 | PBYP00000000∗ |
| I. seosinensis CL-SP19T | I-18 | 7 | 2.69 | 47.27 | PIQF00000000§ |
| I. tyrosinivorans CC-PW-9T | I-19 | 22 | 2.43 | 49.25 | PIQH00000000§ |
| I. xiamenensis 10-D-4T | I-20 | 77 | 2.90 | 49.48 | AMRG00000000 |
| Idiomarina sp. NORP21 | II-1 | 41 | 2.34 | 47.27 | NVXC00000000∗ |
| I. marina PIM1T | II-1 | 11 | 2.42 | 47.17 | PIPZ00000000§ |
| Idiomarina sp. 34-48-12 | II-2 | 283 | 2.10 | 47.52 | NCJS00000000∗ |
| I. tainanensis PIN1T | II-3 | 6 | 2.38 | 47.35 | PIQJ00000000§ |
| I. maritima CGMCC 1.7285T | II-3 | 8 | 2.44 | 47.38 | FOYU00000000∗ |
| I. woesei DSM 27808T | II-4 | 16 | 2.44 | 47.80 | LIPW00000000∗ |
| I. donghaiensis 908033T | II-5 | 16 | 2.58 | 48.07 | PIPU00000000§ |
| I. indica CGMCC 1.10824T | II-6 | 31 | 2.21 | 49.30 | FMXN00000000∗ |
| I. maritima 125B1 | II-7 | 51 | 2.78 | 47.85 | QGTT00000000∗ |
| Idiomarina sp. UBA6142 | II-8 | 129 | 1.82 | 50.42 | DIVC00000000∗ |
| I. salinarum ISL-52T | II-9 | 6 | 2.49 | 52.94 | PIQD00000000§ |
| I. homiensis PO-M2T | II-10 | 6 | 2.61 | 49.98 | PIPX00000000§ |
| I. aquimaris SW15T | II-11 | 23 | 2.80 | 50.24 | PIPT00000000§ |
| I. insulisalsae CVS-6T | II-12 | 20 | 2.57 | 52.33 | PIPY00000000§ |
| I. sediminum c121T | II-13 | 8 | 2.63 | 50.35 | PIQE00000000§ |
| I. halophila BH195T | II-14 | 11 | 2.62 | 50.58 | PIPW00000000§ |
| I. atlantica G5_TVMV8_7T | II-15 | 49 | 2.70 | 50.23 | JPIN00000000∗ |
| I. taiwanensis PIT1T | II-16 | 8 | 2.20 | 49.47 | PIQG00000000§ |
| I. aestuarii KYW314T | II-17 | 13 | 2.65 | 49.10 | PIPR00000000§ |
| I. planktonica TS-T11T | II-18 | 6 | 2.58 | 48.97 | PIQB00000000§ |
| Idiomarina sp. A28L | III-1 | 28 | 2.59 | 45.50 | AFPO00000000∗ |
| A. shirensis AIST | III-2 | 15 | 2.71 | 46.34 | PIPP00000000§ |
| A. iranensis GBPy7T | III-3 | 36 | 2.65 | 46.80 | PIPJ00000000§ |
| A. haloalkalitolerans AK5T | III-4 | 29 | 2.68 | 49.29 | PIPI00000000§ |
| A. sanyensis GYP-17T | III-5 | 48 | 2.60 | 50.82 | PIPM00000000§ |
| A. taiwanensis AIT1T | III-6 | 30 | 2.53 | 48.66 | PIPQ00000000§ |
| A. soli Y4G10-17T | III-7 | 12 | 3.10 | 51.61 | PIPO00000000§ |
| A. sedimenti GBSy1T | III-8 | 10 | 2.85 | 52.09 | PIPN00000000§ |
| A. maris CF12-14T | III-9 | 45 | 3.02 | 50.10 | PIPK00000000§ |
| A. minuta MLST1T | III-10 | 7 | 2.96 | 48.69 | PIPL00000000§ |
General feature of 79 Idiomarinaceae genomes used in this study.
I, Idiomarina; II, Pseudidiomarina; III, Aliidiomarina. The putative new genospecies are shown in bold. All type strains are marked by a superscripted capital T. The genomes from the GenBank database and this study are marked by ∗ and § after the accession, respectively.
Genome Collection and Analyses
Seventy nine Idiomarinaceae genomes, comprising the 27 obtained by this study and 52 available in the GenBank database, were collected (Table 1 and Supplementary Table S1). The strains used in this study included 36 type strains and 43 non-type strains, many of which have not been classified at the species level. Since most of the genomes analyzed in this study were in draft status, assessment of the genomes was conducted by using the software CheckM version 1.0.9 (Parks et al., 2015), which inspected the existence of gene markers specific to the Gammaproteobacteria (UID4761) lineage. The genome annotations and core genome analysis were performed by using the Pathosystems Resource Integration Center (PATRIC) resource2 (Wattam et al., 2014). The 16S rRNA gene sequences and single-copy orthologous protein sequences were obtained from each genome of these strains based on the PATRIC analysis. Pairwise similarities of 16S rRNA genes were calculated by using the software DNAMAN version 7.0 (Lynnon Biosoft, Vaudreuil, QC, Canada) with the distance method of Jukes and Cantor. Statistical analysis of 16S rRNA gene similarities, the G + C contents and the genome sizes was performed using Tukey’s post hoc test. In these analyses, the null hypothesis was rejected at the 0.05 level.
Phylogeny of 16S rRNA Gene and Core Genes
The phylogenetic analyses of 16S rRNA genes and the core genomes were performed as follows. All sequences were aligned by using the software MAFFT version 7.311 with default settings (Katoh and Standley, 2013), and then trimmed to match the length of the shortest. The core genome identified consisted of 78 concatenated protein sequences (Supplementary Table S2). The phylogenetic trees of 16S rRNA genes and core genomes were built by using the software FastTree version 2.1.10 using JTT + CAT parameters and 1000 bootstrap replicates (Price et al., 2010). Escherichia coli str. K-12 substr. MG1655 (GenBank Accession No. U00096) was used as an outgroup in all phylogenetic analyses. The visualization, annotation and management of the phylogenetic trees were performed by using the web-based tool Interactive Tree Of Life (iTOL)3 (Letunic and Bork, 2016). In addition, the analysis of signature nucleotides of the 16S rRNA genes for bacteria within this family was performed by using the software MEGA version 5.05 (Tamura et al., 2011). The locations of signature nucleotides were determined by referring to 16S rRNA gene sequence of E. coli str. K-12 substr. MG1655.
Overall Genome Relatedness Indices Calculations
The Genome-to-Genome Distance Calculator (GGDC) version 2.1 online service was employed to calculate the dDDH values between 79 genomes, applying the recommended formula 24. The OrthoANIu tool was used to calculate the ANI values between these genomes based on the improved ANI algorithm with USEARCH5 (Yoon et al., 2017). The visualization of the numerical matrix for dDDH and ANI values was carried out by using the pheatmap R package version 0.7.7.
Results
Bacterial Genomes Within the Family Idiomarinaceae
A total of 79 bacterial genomes within the family Idiomarinaceae were analyzed in this study, including five complete and 74 draft genome sequences (Table 1 and Supplementary Table S1). Based on the estimations from the CheckM, most genomes satisfied the criteria required to be considered a near-complete genome with low contamination (≥90% of completeness value and ≤5% of contamination value) (Parks et al., 2015). The general features of the 79 genomes are summarized in Table 1 and Supplementary Table S1. The genomic G + C contents of bacteria of the family Idiomarinaceae ranged from 45.50 mol% (Idiomarina sp. A28L) to 52.94 mol% (Idiomarina salinarum ISL-52T), with a mean of 48.04 ± 1.60 mol%. As shown in Supplementary Figure S1, the genomic G + C contents among the eight families of the order Alteromonadales present significant differences from each other (indicated by different letters on the boxplots). The estimated genomic sizes ranged from 2.17 Mbp (Idiomarina sp. UBA6142) to 3.67 Mbp (Idiomarina sp. NP27), with an average of 2.73 ± 0.25 Mbp (Supplementary Table S1). Intriguingly, the genomic sizes of the family Idiomarinaceae were notably smaller than those of members of the other seven families (Supplementary Figure S1), implying that the cosmopolitan bacteria of this family are examples of newly streamlined chemoheterotrophic organisms (Giovannoni et al., 2014).
16S rRNA Gene Sequences Analysis
From 48 bacterial genomes, 63 16S rRNA gene sequences were determined, including 58 complete sequences from 42 bacteria and 5 partial sequences; the 16S rRNA gene was missing from the remaining 31 genomes analyzed (Supplementary Tables S3, S4). Interestingly, each complete genome possessed four copies of the 16S rRNA gene, all of which were exhibited intragenomic heterogeneity (Figure 1). The 58 complete 16S rRNA gene sequences were used for phylogenetic and similarity analyses, and all 63 sequences for the signature nucleotides analysis. In the 16S rRNA gene tree (Figure 1), 42 strains of the family Idiomarinaceae were divided into three groups, corresponding to the genera Idiomarina, Pseudidiomarina (as originally defined) and Aliidiomarina, respectively. On the whole, the phylogenetic relationships of the members of the family Idiomarinaceae in the 16S rRNA gene tree (Figure 1) are consistent with those in the core genes tree below (Figure 2). However, two notable distinctions between the two trees were evident: (1) the two type strains, Idiomarina tyrosinivorans CC-PW-9T and I. xiamenensis 10-D-4T, formed branches separate from the three groups in 16S rRNA gene tree; (2) the two genera Aliidiomarina and Pseudidiomarina were sister groups in 16S rRNA gene tree, but the genus Aliidiomarina was more distant to the genus Pseudidiomarina relative to the genus Idiomarina in the core genome tree below.
FIGURE 1
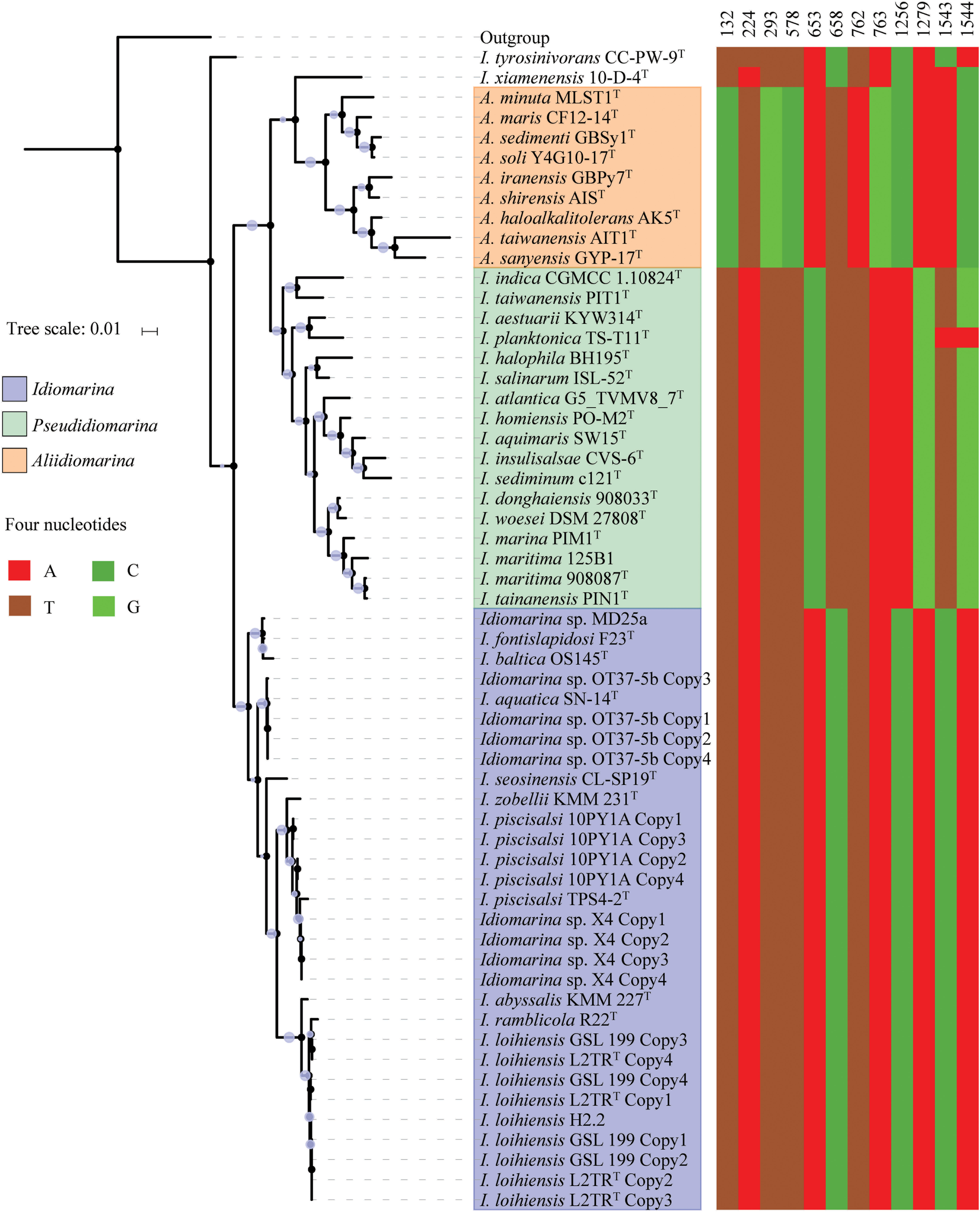
Phylogenetic tree based on 58 of 16S rRNA gene sequences (left panel) and signature nucleotides comparison (right panel) of 42 bacteria within the family Idiomarinaceae. The phylogeny was inferred by using FastTree 2.1.10 with JTT + CAT parameters and 1000 bootstrap replicates and rooted by using Escherichia coli str. K-12 substr. MG1655. Bootstrap values are indicated on the nodes with different sizes of solid circle filled by light blue. The locations of signature sites were determined by referring to 16S rRNA gene sequence of E. coli str. K-12 substr. MG1655. Each of type strains is marked by a superscripted capital T.
FIGURE 2
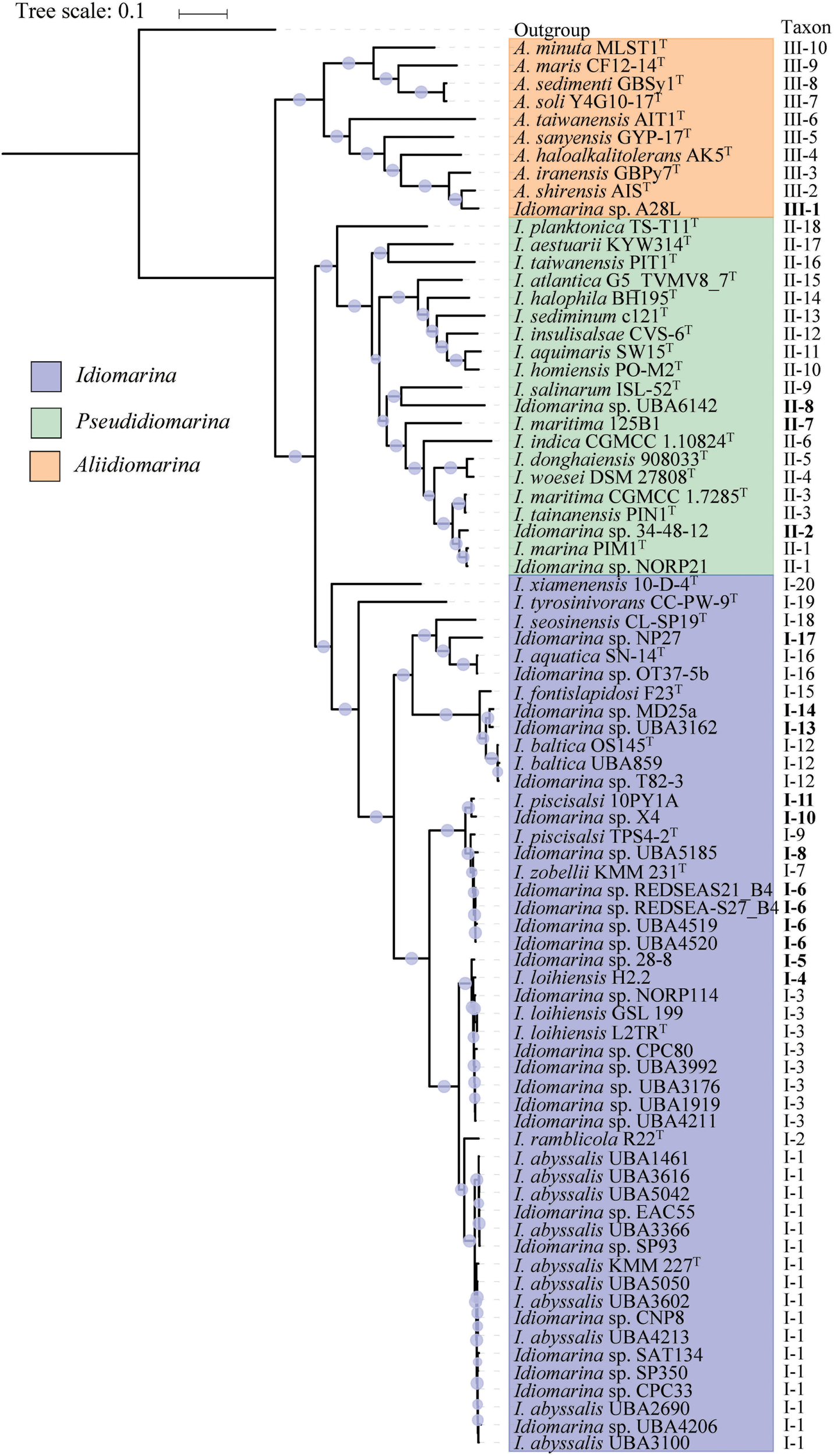
Phylogenetic tree based on core genome of 79 bacteria within the family Idiomarinaceae. The phylogeny was inferred by using FastTree 2.1.10 with JTT + CAT parameters and 1000 bootstrap replicates and rooted by using E. coli str. K-12 substr. MG1655. Bootstrap values are indicated on the nodes with different sizes of solid circle filled by light blue. The putative novel genospecies are highlighted in bold. Each of type strains is marked by a superscripted capital T.
Based on the phylogenomic analysis below, similarities and signature nucleotide analyses of the 16S rRNA genes were also conducted in this study. The intra-genus and inter-genus means of 16S rRNA gene sequences similarities for the members of the genera Idiomarina, Pseudidiomarina, and Aliidiomarina were 97.27 ± 1.66 and 93.93 ± 0.95%, 96.28 ± 0.97 and 94.10 ± 0.83%, 95.46 ± 1.59 and 93.06 ± 0.78%, respectively (Figure 3 and Supplementary Table S3). All inter-genus means among the three genera were significantly lower than the threshold value of 94.5% that has been used to define the genus (Yarza et al., 2014), indicating that the genus Pseudidiomarina should have been retained as an independent genus, rather than integrating it into the genus Idiomarina. As shown in Supplementary Table S4, the previously reported family-signature nucleotides and most group-signature nucleotides of the 16S rRNA gene were unsupported and thus should not be used in future studies. As shown in Figure 1, 12 newly specific signature nucleotides were useful for differentiating the three genera.
FIGURE 3
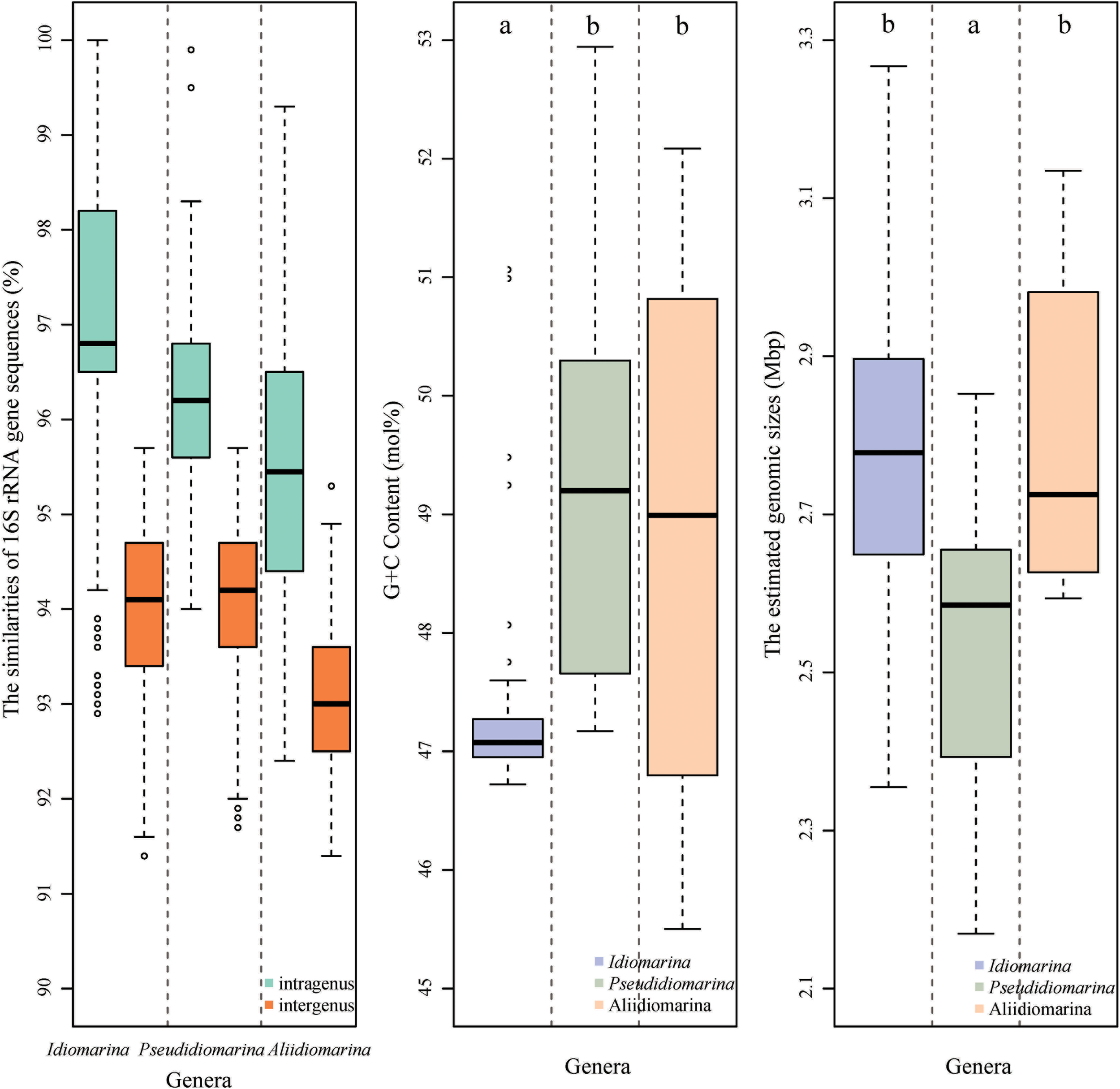
Boxplot of the similarities of 16S rRNA gene sequences (left), genomic G + C contents (middle), and genomic sizes (right) of bacteria within the family Idiomarinaceae. In the boxplot, the central rectangle spans the first quartile to the third quartile, a segment inside the rectangle shows the median, “whiskers” above and below the box show the locations of the minimum and maximum, and the unfilled circles show suspected outliers. Means with different letters at the top of figure denote significant differences (Tukey’s hsd post hoc test, α = 0.05; a < b). The estimated genomic size of a bacterium (bp) = sequencing genomic size (bp)/completeness (%).
Phylogenomic Analysis of the Family Idiomarinaceae
Prior to the phylogenomic analysis, the G + C contents and estimated sizes of the genomes of members of the three genera were analyzed by using Tukey’s post hoc test. As illustrated in Figure 3, the genomic G + C contents of members of the genera Idiomarina, Pseudidiomarina, and Aliidiomarina bacteria were 47.36 ± 0.92, 49.04 ± 1.83, and 49.38 ± 2.00 mol%, respectively, demonstrating significant differences (indicated by different letters on the boxplots, Figure 3, the same below) between the genus Idiomarina and each of the latter two genera. The estimated genome sizes of members of the three genera above were 2.80 ± 0.23, 2.52 ± 0.19, and 2.80 ± 0.20 Mbp, respectively, revealing obvious contrasts (the same as above) between the genus Pseudidiomarina and each of the other two genera.
A total of 78 orthologous protein sequences were used to infer the phylogeny of 79 bacteria of this family (Supplementary Table S2). As shown in Figure 2, the phylogenetic tree was characterized by high bootstrap values, indicating that the proteins selected were reflective of a robust evolutionary relatedness between bacteria within the family. The trees showed that bacteria of this family were grouped into three monophyletic taxa, matching exactly to the three genera, Idiomarina, Pseudidiomarina, and Aliidiomarina (defined by the three colors). Taxon I incorporated 49 strains, including 11 type strains and 38 non-type strains. Taxon II contained 20 strains, comprised of 16 type strains and 4 non-type strains, which should be restored or reclassified to the genus Pseudidiomarina rather than the genus Idiomarina. Taxon III included 10 independent branches corresponding to the nine type strains of the genus Aliidiomarina and Idiomarina sp. A28L. Taxon I and taxon II exhibited a closer relationship rather than taxon III, and were found to be sister groups. The comparison between the phylogenetic reconstructions based on 16S rRNA genes and the core genome suggested that the core genome provides a more robust phylogenetic framework for this family, and thus can be considered as the preferential approach for genus- and species-wide investigations.
The dDDH and ANI Analyses for Species Delineation
The dDDH and ANI are based on the complete genomic information (OGRI), thereby enabling accurate delineation of species and assessment of their genetic relatedness. Therefore, pairwise dDDH and ANI values of the 79 genomes for bacteria of the family Idiomarinaceae were calculated and are shown in Figure 4 and Supplementary Tables S5, S6. According to the 70% dDDH (Wayne et al., 1987) and 95% ANI thresholds (Richter and Rosselló-Móra, 2009) for species delineation, the 79 strains fell into 48 genospecies labeled by I-1 to I-20 in taxon I, II-1 to II-18 in taxon II, and III-1 to III-10 in the taxon III, each containing one to 17 strains. Taxon I included nine putative new genospecies (I-4, I-5, I-6, I-8, I-10, I-11, I-13, I-14, and I-17, shown in bold, Table 1, the same below) and 11 already described species represented by a type strain. Taxon II consisted of three putative new genospecies (II-4, II-7, and I-8) and 15 validly named species. It was notable that two type strains, Idiomarina maritima CGMCC 1.7285T and Idiomarina tainanensis PIN1T, share a dDDH of 75.8% and ANI of 97.29%, values which are higher than the two thresholds noted above, and therefore they should be considered conspecific. We propose returning these strains to the reinstated genus Pseudidiomarina and in this context it is noted that the name Pseudidiomarina tainanensisJean et al. (2009) has nomenclatural priority over Pseudidiomarina maritimaWu et al. (2009). Taxon III contained only one putative new genospecies (III-1) and nine well-defined species. Most interestingly, most of new genospecies are distributed in the taxon I corresponding to the genus Idiomarina, implying that bacteria of this taxon may possess more novel genotypes relative to those of the two other taxa. In other words, the genus Idiomarina possessed higher species diversity, with more new genospecies relative to the other two. Similarly, some species such as I-1 (I. abyssalis) and I-3 (I. loihiensis) possessed higher strain diversity compared with others. This analysis indicated that the genetic diversity of this family varies to some extent at the genus and species levels.
FIGURE 4
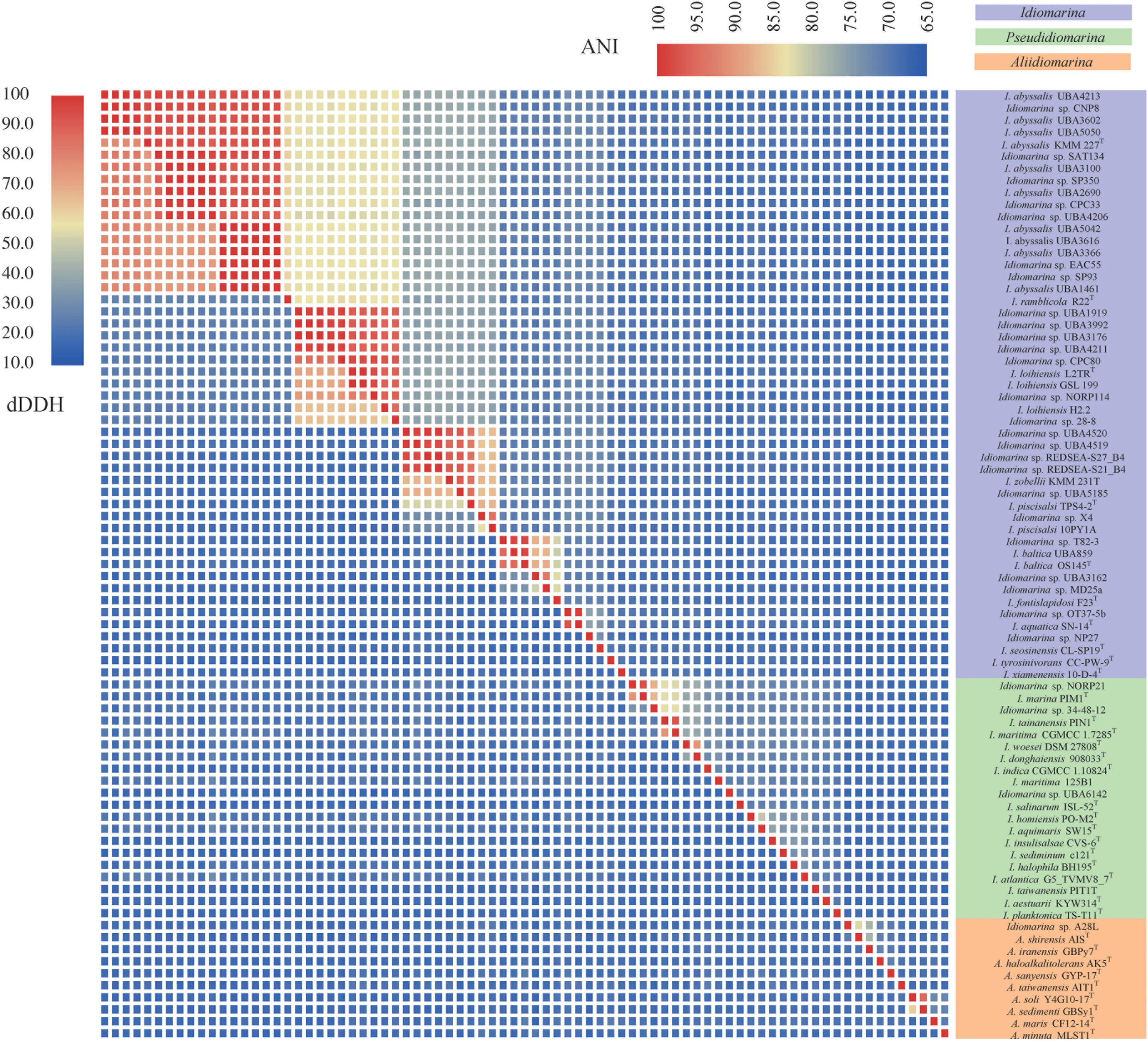
Heatmap of digital DNA-DNA hybridization values in lower-left and average nucleotide identity values in upper right for 79 genomes of bacteria within the family Idiomarinaceae. The bacteria of the three genera are shown in the three colors.
In addition, the species-level phylogenetic classification of 13 other strains of this family was determined by comparing the dDDH values with the 79 strains mentioned above (Supplementary Tables S1, S7), although they were not used in the above analysis in view of the low quality of their genomes as assessed by CheckM. As listed in Supplementary Table S1, among these strains, six appear to belong to a new genospecies named II-19, three to I-1 and the remaining four to I-8, I-12, I-17, and II-11, respectively. Therefore, based on the OGRI analysis, the proposed species designations of bacteria of this family is shown in Supplementary Table S1.
Discussion
The members of the family Idiomarinaceae are ubiquitous in various saline environments, especially in the marine environments (Albuquerque and da Costa, 2014). The widespread distribution of bacteria of this family implies that they may have as yet unexplored diversity. In this report, we performed a genome-based systematic analysis to delineate the taxonomic relationships of 79 strains within the family Idiomarinaceae. The results of 16S rRNA gene sequence analysis, the phylogenomic analysis using 78 single-copy core genes, and analysis of two overall genome relatedness indices (dDDH and ANI) were almost completely consistent, and determined the presence of three genus level taxa including 48 genospecies. These results demonstrated that the latter three genome-based methods relative to 16S rRNA gene analysis can provide a more robust taxonomic framework for bacteria of the family Idiomarinaceae at the genus and species level.
As mentioned above, the genus Pseudidiomarina was introduced to accommodate Idiomarina-like organisms without motility, DNAse activity and growth at pHs below 6, and the presence of a unique nucleotide substitution of A instead of C at position 143 in their 16S rRNA gene sequences (Jean et al., 2006). However, due to few distinguishing phenotypic and chemotaxonomic features, except for differences in 16S rRNA gene sequence signature nucleotides, the members of the genus Pseudidiomarina were later transferred to the genus Idiomarina (Taborda et al., 2009). However, the current study provides an improved taxonomic understanding of this family using genome-based methods. On one hand, the genus Pseudidiomarina was found to be an independent monophyletic group in the two phylogenetic trees, meeting with the classical definition of genus (Medini et al., 2005). On the other hand, 16S rRNA gene sequences similarities between the members of the genus Pseudidiomarina and the other strains analyzed were lower than the threshold value for delineating a genus (Yarza et al., 2014). In addition, the genomic G + C contents and genome sizes of the members of the genus Pseudidiomarina were statistically significant different compared to members of the two other genera in the family. Based on the analyses above, we propose to reinstate the taxonomic status of the genus Pseudidiomarina.
Combined with the three genome-based analyses, a considerable diversity of bacteria of the family Idiomarinaceae was demonstrated in this study. Among these genospecies clusters of this family, 35 were within the radius of an existing type strain and could thus be successfully linked to a described species. In contrast, the remaining 13 did not contain a described type strain and therefore await formal description and naming in accordance with the rules of nomenclature. In this study, 52 genomes were obtained from the GenBank database, most of which were metagenome-assembled genomes from global ocean samples collected during the Tara Oceans circumnavigation expedition (Parks et al., 2017; Tully et al., 2018). It is notable that all the isolates from the Tara Oceans project fell into taxon I (the genus Idiomarina) with the exception of Idiomarina sp. NORP21; moreover, they were mainly concentrated in three genospecies clusters (I-1, I-3, and I-6). These results suggested that the strain diversity of the above three taxa is higher than the others, and that the species diversity of the genus Idiomarina may be higher than that of the other two genera. When increasing the number of bacteria used to produce a 16S rRNA gene supertree of 717 bacteria, a similar trend of diversity was also observed (Supplementary Figure S2 and Supplementary Table S8). The genetic characteristics and underlying mechanisms of this phenomenon need to be further investigated by comparative genomic analyses in future studies. Moreover, on the basis of this study, a relation between the geographic origin and the genomic relatedness of the strains within this family warrant further investigation.
Over the past few decades, the 16S rRNA gene has been the only widely used molecular marker for prokaryotic taxonomy and is the cornerstone for the current systematic classification of the Bacteria and Archaea (Tindall et al., 2010). However, this marker is incapable of accurately distinguishing closely related strains and species, and of providing the robust phylogenetic relationships of some strains because of its low resolution (Rajendhran and Gunasekaran, 2011; Liu et al., 2013, 2015). In this study, the limitation of 16S rRNA gene analysis was also shown by comparison of the 16S rRNA gene tree and with that derived from the core genome. Moreover, based on the five complete genomes, we found that bacteria of the family Idiomarinaceae present heterogeneity in their 16S rRNA gene sequences. For example, Copy 3 of the 16S rRNA gene sequence for I. loihiensis L2TRT shares 99.8% similarity with Copy 4. Similar situations have been reported for other strains, species and genera, for example the Bacillus cereus group (Liu et al., 2015) and the genera Pseudomonas (Bodilis et al., 2012) and Aeromonas (Morandi et al., 2005). In contrast, in this study the genome-based phylogenetic analysis and relatedness indices analysis present an improved picture of the phylogeny of the family Idiomarinaceae relative to 16S rRNA gene. Thus, based on the current study and previous reports, we strongly recommend that the taxonomy and systematics of Bacteria and Archaea should take full advantage the increasing availability of genomic information (Yarza et al., 2013) and the inherent strength of genome-based approaches (Yarza et al., 2013; Chun et al., 2018; Rodriguez-R et al., 2018).
Conclusion
The family Idiomarinaceae is more diverse at the genus and species level than was previously known and comprises of three genera, in which we have identified 13 potentially novel genospecies in addition to the 35 already described species. Furthermore, the taxonomic status and correct name of strains within this family can be determined based on the genomic analyses. The study also reveals that the genus Pseudidiomarina should be reinstated in view of its distinctive phylogenetic divergence and other differences, as originally discussed (Jean et al., 2006). This study structures a robust taxonomic framework of currently available Idiomarinaceae strains, and provides an avenue to a better understanding of the functional adaptation to saline environments.
Taxonomic Consequences
Based on phenotypic and genotypic data, we propose the reclassification of Idiomarina insulisalsae as Pseudidiomarina insulisalsae comb. nov., Idiomarina aquimaris as Pseudidiomarina aquimaris comb. nov., Idiomarina indica as Pseudidiomarina indica comb. nov., Idiomarina planktonica as Pseudidiomarina planktonica comb. nov., Idiomarina woesei as Pseudidiomarina woesei comb. nov., Idiomarina halophila as Pseudidiomarina halophila comb. nov. and Idiomarina atlantica as Pseudidiomarina atlantica comb. nov.
Emended Description of the Genus Pseudidiomarina (Jean et al., 2006)
Pseudidiomarina (Pseud.i.di.o.ma.ri’na. Gr. adj. pseudes false; N.L. fem. n. Idiomarina a name of a bacterial genus; N.L. fem. n. Pseudidiomarina false Idiomarina).
The original description is based on that given by Jean et al. (2006). Members are Gram- stain negative rods belonging to the family Idiomarinaceae within the class Gammaproteobacteria. Some strains are motile by means of a single polar flagellum or peritrichous flagella, while some are non-motile. All strains are chemoheterotrophs, positive for oxidase and catalase, and strictly aerobic with the exception of the catalase negative and facultatively anaerobic Pseudidiomarina sediminum. Molecular oxygen is a universal electron acceptor while in some cases nitrate can be used as an alternative electron acceptor under anaerobic conditions. The major fatty acids are iso-C15:0, iso-C17:0 and Summed feature 9 (iso-C17:1ω9c and/or C16:0 10-methyl). The major polar lipids are phosphatidylethanolamine (PE), phosphatidylglycerol (PG), and diphosphatidylglycerol (DPG). The major respiratory quinone is ubiquinone 8 (Q-8). The DNA G + C content varies between 47.17 and 52.94 mol% (Table 1). The type species of the genus is Pseudidiomarina taiwanensis.
Emended Description of Pseudidiomarina taiwanensis (Jean et al., 2006)
Pseudidiomarina taiwanensis (tai.wan.en’sis. N.L. fem. adj. taiwanensis pertaining to Taiwan, where the type strain was isolated).
The description is identical to that given for Pseudidiomarina taiwanensis by Jean et al. (2006). Additional characteristics are as follows: the polar lipids comprise PE, PG, DPG and seven uncharacterized phospholipids (PL1-PL6 and PL10); the respiratory quinone is Q-8 (Zhong et al., 2014). The type strain, PIT1T ( = BCRC 17465T= JCM 13360T), was isolated from shallow coastal water of An-Ping Harbour, Taiwan. The DNA G + C content of the type strain is 47.35 mol%.
Emended Description of Pseudidiomarina homiensis (Kwon et al., 2006; Jean et al., 2009)
Pseudidiomarina homiensis (ho.mi.en’sis. N.L. fem. adj. homiensis referring to the Homi Cape in Korea, where the type strain was isolated).
The description is identical to that given for Idiomarina homiensis by Kwon et al. (2006). Additional characteristics are as follows: the polar lipids comprise PE, PG, DPG, phosphatidylserine (PS), an uncharacterized aminolipid (AL1) and eight uncharacterized PLs (PL1-PL8) (Chen et al., 2012). The type strain is PO-M2T ( = KACC 11514T= DSM 17923T), which was isolated from seashore sand in Pohang, Korea. The DNA G + C content of the type strain is 49.98 mol%.
Emended Description of Pseudidiomarina salinarum (Yoon et al., 2007; Jean et al., 2009)
Pseudidiomarina salinarum (sa.li.na’rum. L. gen. pl. n. salinarum of salt-works).
The description is identical to that given for Idiomarina salinarum by Yoon et al. (2007). Additional characteristics are as follows: the polar lipids comprise PE, PG, DPG and six uncharacterized PLs (PL1-PL5 and PL11); the enzyme activities of α-mannosidase in API ZYM tests and D-glucose fermentation in API 20NE are variable (Zhong et al., 2014). The type strain, ISL-52T ( = KCTC 12971T= CCUG 54359T), was isolated from a marine solar saltern of the Yellow Sea, Korea. The DNA G + C content of the type strain is 52.94 mol%.
Emended Description of Pseudidiomarina sediminum (Hu and Li, 2007)
Pseudidiomarina sediminum (se.di’mi.num. L. gen. pl. n. sediminum of sediments, pertaining to source of isolation of the type strain).
The description is identical to that given for Pseudidiomarina sediminum by Hu and Li (2007). Additional characteristics are as follows: the polar lipids comprise PE, PG, DPG, PS, three uncharacterized ALs (AL1-AL3) and three uncharacterized PLs (PL4-PL6) (Chen et al., 2012). The type strain is c121T ( = CICC 10319T= LMG 24046T), isolated from coastal sediment. The DNA G + C content of the type strain is 50.35 mol%.
Emended Description of Pseudidiomarina marina (Jean et al., 2009)
Pseudidiomarina marina (ma.ri’na. L. fem. adj. marina of the sea, marine).
The description is identical to that given for Pseudidiomarina marina by Jean et al. (2009). Additional characteristics are as follows: the polar lipids comprise PE, PG, DPG, two uncharacterized ALs (AL1–AL2), an uncharacterized PL, and three lipids (L1–L3) (Poddar et al., 2014). The type strain, PIM1T ( = BCRC 17749T= JCM 15083T), was isolated from shallow coastal seawater of An-Ping Harbour, Tainan, Taiwan. The DNA G + C content of the type strain is 47.17 mol%.
Emended Description of Pseudidiomarina tainanensis (Jean et al., 2009)
Pseudidiomarina tainanensis (tai.nan.en’sis. N.L. fem. adj. tainanensis pertaining to Tainan, Taiwan, where the type strain was isolated).
The description is identical to that given for Pseudidiomarina tainanensis by Jean et al. (2009). Additional characteristics are as follows: the polar lipids comprise PE, PG, DPG, two uncharacterized ALs (AL1–AL2), an uncharacterized PL, and L1 (Poddar et al., 2014). The type strain, PIN1T ( = BCRC 17750T= JCM 15084T), was isolated from shallow coastal seawater of An-Ping Harbour, Tainan, Taiwan. The DNA G + C content of the type strain is 47.35 mol%.
Emended Description of Pseudidiomarina donghaiensis (Wu et al., 2009)
Pseudidiomarina donghaiensis (dong.hai.en’sis. N.L. fem. adj. donghaiensis pertaining to Donghai, the Chinese name for the East China Sea).
The description is identical to that given for Pseudidiomarina donghaiensis by Wu et al. (2009). Additional characteristics are as follows: the polar lipids comprise PE, PG, DPG, an uncharacterized AL1, an uncharacterized PL, and L1 (Poddar et al., 2014). The type strain, 908033T ( = CGMCC 1.7284T= JCM 15533T), was isolated from a coastal seawater sample collected from the East China Sea, China. The DNA G + C content of the type strain is 48.07 mol%.
Emended Description of Pseudidiomarina maritima (Wu et al., 2009)
Pseudidiomarina maritima (ma.ri’ti.ma. L. fem. adj. maritima inhabiting marine environments).
The description is identical to that given for Pseudidiomarina maritima by Wu et al. (2009). Additional characteristics are as follows: the polar lipids comprise PE, PG, DPG, an uncharacterized AL, an uncharacterized PL and two Ls (L1-L2) (Poddar et al., 2014). The type strain, 908087T ( = CGMCC 1.7285T= JCM 15534T), was isolated from a coastal seawater sample collected from the East China Sea, China. The DNA G + C content of the type strain is 47.38 mol%.
Description of Pseudidiomarina insulisalsae comb. nov.
Pseudidiomarina insulisalsae (in.su.li.sal’sae. L. fem. n. insula, island; L. adj. salsus -a -um, salted; N.L. gen. n. insulisalsae, of a salted island, isolated from the Island of Sal [the salt island] in the Cape Verde Archipelago).
Basonym: Idiomarina insulisalsae (Taborda et al., 2009)
The description is identical to that given for Idiomarina insulisalsae by Taborda et al. (2009). The type strain, CVS-6T ( = CIP108836T= LMG 23123T), was isolated from the soil of a solar saltern. The DNA G + C content of the type strain is 52.33 mol%.
Emended Description of Pseudidiomarina aestuarii (Park et al., 2010)
Pseudidiomarina aestuarii (aes.tu.a’ri.i. L. gen. n. aestuarii of a tract overflowed at high tide, a salt marsh, a bay, referring to the isolation of the type strain from shallow coastal seawater).
The description is identical to that given for Pseudidiomarina aestuarii by Park et al. (2010). Additional characteristics are as follows: the polar lipids comprise PE, PG, DPG, and six uncharacterized PLs (PL1-PL5, PL12); hydrolysis of urea, gelatin, L-arginine, and the enzyme activities of leucine arylamidase and trypsin in API ZYM tests are variable (Zhong et al., 2014). The type strain is KYW314T ( = KCTC 22740T= JCM 16344T), isolated from seawater collected from the South Sea, South Korea. The DNA G + C content of the type strain is 49.10 mol%.
Description of Pseudidiomarina aquimaris comb. nov.
Pseudidiomarina aquimaris (a.qui.ma’ris. L. fem. n. aqua water; L. gen. n. maris of the sea; N.L. gen. n. aquimaris of the water of the sea).
Basonym: Idiomarina aquimaris (Chen et al., 2012).
The description is identical to that given for Idiomarina aquimaris by Chen et al. (2012). The type strain, SW15T ( = LMG 25374T = BCRC 80083T), was isolated from a sample of a reef-building coral (Isopora palifera) collected off the coast of southern Taiwan. The DNA G + C content of the type strain is 50.24 mol%.
Description of Pseudidiomarina indica comb. nov.
Pseudidiomarina indica (in’di.ca. L. fem. adj. indica Indian, referring to the Indian Ocean, where the strain was isolated).
Basonym: Idiomarina indica (Song et al., 2013).
The description is identical to that given for Idiomarina indica by Song et al. (2013). The type strain, SW104T ( = CGMCC 1.10824T = JCM 18138T), was isolated from a seawater sample collected from the India Ocean. The DNA G + C content of the type strain is 49.30 mol%.
Description of Pseudidiomarina planktonica comb. nov.
Pseudidiomarina planktonica (plank.to’ni.ca. N.L. fem. adj. planktonica (from Gr. adj. planktos wandering) living in the plankton, planktonic.)
Basonym: Idiomarina planktonica (Zhong et al., 2014).
The description is identical to that given for Idiomarina planktonica by Zhong et al. (2014). The type strain, TS-T11T ( = CGMCC 1.12458T = JCM 19263T), was isolated from a water sample of Tuosu lake in the Qaidam basin, Qinghai province, China. The DNA G + C content of the type strain is 48.97 mol%.
Description of Pseudidiomarina woesei comb. nov.
Pseudidiomarina woesei (woe’se.i. N.L. gen. n. woesei, of Woese, named after Professor Carl R. Woese, a pioneer in evolutionary microbiology).
Basonym: Idiomarina woesei (Poddar et al., 2014)
The description is identical to that given for Idiomarina woesei by Poddar et al. (2014). The type strain, W11T ( = DSM 27808T= JCM 19499T= LMG 27903T), was isolated from the Andaman Sea. The DNA G + C content of the type strain is 47.80 mol%.
Description of Pseudidiomarina halophila comb. nov.
Pseudidiomarina halophila (ha.lo’phi.la Gr. n. hals, halos salt; N.L. fem. adj. phila (from Gr. fem. adj. philê), friend, loving; N.L. fem. adj. halophila salt-loving.)
Basonym: Idiomarina halophila (Lee et al., 2015).
The description is identical to that given for Idiomarina halophila by Lee et al. (2015). The type strain, BH195T ( = KACC 17610T = NCAIM B 02544T), was isolated from the sediment of a solar saltern in Gomso, Buan-gun, Chunbuk, South Korea. The DNA G + C content of the type strain is 50.58 mol%.
Description of Pseudidiomarina atlantica comb. nov.
Pseudidiomarina atlantica (at.lan’ti.ca. L. fem. adj. atlantica, referring to the Atlantic Ocean, where the strain was isolated).
Basonym: Idiomarina atlantica (Du et al., 2015).
The description is identical to that given for Idiomarina atlantica by Du et al. (2015). The type strain, G5_TVMV8_7T ( = MCCC 1A10513T= KCTC 42141T), was isolated from the deep sea sediment of the North Atlantic Ocean.
Statements
Author contributions
YL and ZS conceived and designed the experiments. YL performed the experiments, analyzed the data, and wrote the paper. YL, QL, and ZS revised the paper.
Funding
This work was financially supported by COMRA program (Grant No. DY135-B2-01) and National Infrastructure of Natural Resources for Science and Technology Program of China (Grant No. NIMR-2018-9).
Acknowledgments
We gratefully acknowledge Prof. Aharon Oren at Hebrew University of Jerusalem (Jerusalem, Israel) and Prof. Iain Sutcliffe at Northumbria University (Newcastle upon Tyne, United Kingdom) for the kind help in through reading and comprehensive corrections on the manuscript, Dr. Li Anzhang at Guangdong Institute of Microbiology (Guangzhou, China) for the generous help in use of the software CheckM and OrthoANIu tool, Dr. Xiaoteng Fu at Third Institute of Oceanography (Xiamen, China) for the kind help in improving the manuscript, and Dr. Somboon Tanasupawat at Chulalongkorn University (Bangkok, Thailand), Dr. Cristina Sánchez-Porro at University of Tehran (Tehran, Iran), Prof. Wu Xiaolei at Peking University (Beijing, China) for the generous type strains.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.02453/full#supplementary-material
FIGURE S1Boxplot of genomic G + C contents (left panel) and genomic sizes (right panel) of bacteria from the eight families within the order Alteromonadales. In the boxplot, the central rectangle spans the first quartile to the third quartile, a segment inside the rectangle shows the median, “whiskers” above and below the box shows the locations of the minimum and maximum, and the unfilled circles show suspected outliers. Means with different letters at the top of figure denote significant differences (Tukey’s hsd post hoc test, α = 0.05; a < b < c < d).
FIGURE S2The supertree based on 16S rRNA gene sequences of 717 bacteria within the family Idiomarinaceae. The phylogeny was inferred by using FastTree 2.1.10 with JTT + CAT parameters and 1000 bootstrap replicates and rooted by using E. coli str. K-12 substr. MG1655. Bootstrap values are indicated on the nodes with different sizes of solid circle filled by light blue. Each of type strains were marked by a superscripted capital T.
TABLE S1The detailed information of 92 Idiomarinaceae genomes used in this study.
TABLE S2The 78 single-copy orthologous proteins used for tree inference.
TABLE S3The pairwise similarites (%) of 16S rRNA gene sequences of 43 strains within the family Idiomarinaceae.
TABLE S4The signature sequences of 16S rRNA gene for 48 strains within the family Idiomarinaceae.
TABLE S5The pairwise dDDH values (%) between the 79 strains used in this study.
TABLE S6The pairwise ANI values (%) between the 79 strains used in this study.
TABLE S7The pairwise dDDH values (%) between the representative strains and the other thirteen strains marked by asterisks.
TABLE S8The accession numbers of 16S rRNA gene sequences for 674 non-type strains within the family Idiomarinaceae from the GenBank database.
Footnotes
References
1
Albuquerque L. da Costa M. S. (2014). “The family idiomarinaceae,” inThe Prokaryotes: Gammaproteobacteria, edsRosenbergE.DeLongE. F.LoryS.StackebrandtE.ThompsonF. (Berlin: Springer), 361–385.
2
Auch A. F. von Jan M. Klenk H. P. Göker M. (2010). Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison.Stand. Genomic Sci.2117–134. 10.4056/sigs.531120
3
Bankevich A. Nurk S. Antipov D. Gurevich A. A. Dvorkin M. Kulikov A. S. et al (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing.J. Comput. Biol.19455–477. 10.1089/cmb.2012.0021
4
Beukes C. W. Palmer M. Manyaka P. Chan W. Y. Avontuur J. R. van Zyl E. et al (2017). Genome data provides high support for generic boundaries in Burkholderia sensu lato.Front. Microbiol.8:1154. 10.3389/fmicb.2017.01154
5
Bodilis J. Nsigue-Meilo S. Besaury L. Quillet L. (2012). Variable copy number, intra-genomic heterogeneities and lateral transfers of the 16S rRNA gene in Pseudomonas.PLoS One7:e35647. 10.1371/journal.pone.0035647
6
Chen M. H. Sheu S. Y. Chen C. A. Wang J. T. Chen W. M. (2012). Idiomarina aquimaris sp. nov., isolated from the reef-building coral Isopora palifera.Int. J. Syst. Evol. Microbiol.621536–1542. 10.1099/ijs.0.035592-0
7
Chun J. Oren A. Ventosa A. Christensen H. Arahal D. R. da Costa M. S. et al (2018). Proposed minimal standards for the use of genome data for the taxonomy of prokaryotes.Int. J. Syst. Evol. Microbiol.68461–466. 10.1099/ijsem.0.002516
8
Chun J. Rainey F. A. (2014). Integrating genomics into the taxonomy and systematics of the bacteria and Archaea.Int. J. Syst. Evol. Microbiol.64316–324. 10.1099/ijs.0.054171-0
9
Colston S. M. Fullmer M. S. Beka L. Lamy B. Gogarten J. P. Graf J. (2014). Bioinformatic genome comparisons for taxonomic and phylogenetic assignments using Aeromonas as a test case.mBio5:e02136. 10.1128/mBio.02136-14
10
Du J. Lai Q. Liu Y. Du Y. Liu X. Sun F. et al (2015). Idiomarina atlantica sp. nov., a marine bacterium isolated from the deep sea sediment of the North Atlantic Ocean.Antonie Van Leeuwenhoek107393–401. 10.1007/s10482-014-0337-7
11
Garcia Garcia M. I. Lau K. von Itzstein M. Garcia Carmona F. Sanchez Ferrer A. (2015). Molecular characterization of a new N-acetylneuraminate synthase (NeuB1) from Idiomarina loihiensis.Glycobiology25115–123. 10.1093/glycob/cwu096
12
Giovannoni S. J. Cameron Thrash J. Temperton B. (2014). Implications of streamlining theory for microbial ecology.ISME J.81553–1565. 10.1038/ismej.2014.60
13
Gonzalez-Munoz M. T. De Linares C. Martinez-Ruiz F. Morcillo F. Martin-Ramos D. Arias J. M. (2008). Ca-Mg kutnahorite and struvite production by Idiomarina strains at modern seawater salinities.Chemosphere72465–472. 10.1016/j.chemosphere.2008.02.010
14
Hesse C. Schulz F. Bull C. T. Shaffer B. T. Yan Q. Shapiro N. et al (2018). Genome-based evolutionary history of Pseudomonas spp.Environ. Microbiol.10.1111/1462-2920.14130 [Epub ahead of print],
15
Hou S. Saw J. H. Lee K. S. Freitas T. A. Belisle C. Kawarabayasi Y. et al (2004). Genome sequence of the deep-sea gamma-proteobacterium Idiomarina loihiensis reveals amino acid fermentation as a source of carbon and energy.Proc. Natl. Acad. Sci. U.S.A.10118036–18041. 10.1073/pnas.0407638102
16
Hu Z. Y. Li Y. (2007). Pseudidiomarina sediminum sp. nov., a marine bacterium isolated from coastal sediments of luoyuan bay in China.Int. J. Syst. Evol. Microbiol.572572–2577. 10.1099/ijs.0.65199-0
17
Huang S. P. Chang H. Y. Chen J. S. Jean W. D. Shieh W. Y. (2012). Aliidiomarina taiwanensis gen. nov., sp. nov., isolated from shallow coastal water.Int. J. Syst. Evol. Microbiol.62155–161. 10.1099/ijs.0.030064-0
18
Ivanova E. P. Flavier S. Christen R. (2004). Phylogenetic relationships among marine Alteromonas-like proteobacteria: emended description of the family Alteromonadaceae and proposal of Pseudoalteromonadaceae fam. nov., Colwelliaceae fam. nov., Shewanellaceae fam. nov., Moritellaceae fam. nov., Ferrimonadaceae fam. nov., Idiomarinaceae fam. nov. and Psychromonadaceae fam. nov.Int. J. Syst. Evol. Microbiol.541773–1788. 10.1099/ijs.0.02997-0
19
Ivanova E. P. Romanenko L. A. Chun J. Matte M. H. Matte G. R. Mikhailov V. V. et al (2000). Idiomarina gen. nov., comprising novel indigenous deep-sea bacteria from the Pacific Ocean, including descriptions of two species, Idiomarina abyssalis sp. nov. and Idiomarina zobellii sp. nov.Int. J. Syst. Evol. Microbiol.50901–907. 10.1099/00207713-50-2-901
20
Jean W. D. Leu T. Y. Lee C. Y. Chu T. J. Lin S. Y. Shieh W. Y. (2009). Pseudidiomarina marina sp. nov. and Pseudidiomarina tainanensis sp. nov. and reclassification of Idiomarina homiensis and Idiomarina salinarum as Pseudidiomarina homiensis comb. nov. and Pseudidiomarina salinarum comb. nov., respectively.Int. J. Syst. Evol. Microbiol.59(Pt 1), 53–59. 10.1099/ijs.0.001180-0
21
Jean W. D. Shieh W. Y. Chiu H. H. (2006). Pseudidiomarina taiwanensis gen. nov., sp. nov., a marine bacterium isolated from shallow coastal water of An-Ping Harbour, Taiwan, and emended description of the family Idiomarinaceae.Int. J. Syst. Evol. Microbiol.56899–905. 10.1099/ijs.0.64048-0
22
Katoh K. Standley D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability.Mol. Biol. Evol.30772–780. 10.1093/molbev/mst010
23
Konstantinidis K. T. Tiedje J. M. (2005). Genomic insights that advance the species definition for prokaryotes.Proc. Natl. Acad. Sci. U.S.A.1022567–2572. 10.1073/pnas.0409727102
24
Kumar A. Wu G. Wu Z. Kumar N. Liu Z. (2018). Improved catalytic properties of a serine hydroxymethyl transferase from Idiomarina loihiensis by site directed mutagenesis.Int. J. Biol. Macromol.1171216–1223. 10.1016/j.ijbiomac.2018.05.003
25
Kwon S. W. Kim B. Y. Weon H. Y. Baek Y. K. Koo B. S. Go S. J. (2006). Idiomarina homiensis sp. nov., isolated from seashore sand in Korea.Int. J. Syst. Evol. Microbiol.562229–2233. 10.1099/ijs.0.64283-0
26
Lee J. C. Kim Y. S. Yun B. S. Whang K. S. (2015). Idiomarina halophila sp. nov., isolated from a solar saltern sediment.Int. J. Syst. Evol. Microbiol.651268–1273. 10.1099/ijs.0.000094
27
Letunic I. Bork P. (2016). Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees.Nucleic Acids Res.44242–245. 10.1093/nar/gkw290
28
Li X. Qian P. Wu S. G. Yu H. Y. (2014). Characterization of an organic solvent-tolerant lipase from Idiomarina sp. W33 and its application for biodiesel production using Jatropha oil.Extremophiles18171–178. 10.1007/s00792-013-0610-0
29
Liu Y. Lai Q. Dong C. Sun F. Wang L. Li G. et al (2013). Phylogenetic diversity of the Bacillus pumilus group and the marine ecotype revealed by multilocus sequence analysis.PLoS One8:e80097. 10.1371/journal.pone.0080097
30
Liu Y. Lai Q. Göker M. Meier-Kolthoff J. P. Wang M. Sun Y. et al (2015). Genomic insights into the taxonomic status of the Bacillus cereus group.Sci. Rep.5:14082. 10.1038/srep14082
31
Lugli G. A. Milani C. Duranti S. Mancabelli L. Mangifesta M. Turroni F. et al (2018). Tracking the taxonomy of the genus Bifidobacterium based on a phylogenomic approach.Appl. Environ. Microbiol.84:e02249-17. 10.1128/AEM.02249-17
32
Medini D. Donati C. Tettelin H. Masignani V. Rappuoli R. (2005). The microbial pan-genome.Curr. Opin. Genet. Dev.15589–594. 10.1016/j.gde.2005.09.006
33
Meier-Kolthoff J. P. Auch A. F. Klenk H. P. Göker M. (2013). Genome sequence-based species delimitation with confidence intervals and improved distance functions.BMC Bioinformatics14:60. 10.1186/1471-2105-14-60
34
Montero-Calasanz M. D. C. Meier-Kolthoff J. P. Zhang D. F. Yaramis A. Rohde M. Woyke T. et al (2017). Genome-scale data call for a taxonomic rearrangement of geodermatophilaceae.Front. Microbiol.8:2501. 10.3389/fmicb.2017.02501
35
Morandi A. Zhaxybayeva O. Gogarten J. P. Graf J. (2005). Evolutionary and diagnostic implications of intragenomic heterogeneity in the 16S rRNA gene in Aeromonas strains.J. Bacteriol.1876561–6564. 10.1128/JB.187.18.6561-6564.2005
36
Park S. C. Lim C. H. Baik K. S. Lee K. H. Lee J. S. Seong C. N. (2010). Pseudidiomarina aestuarii sp. nov., a marine bacterium isolated from shallow coastal seawater.Int. J. Syst. Evol. Microbiol.602071–2075. 10.1099/ijs.0.018051-0
37
Parks D. H. Imelfort M. Skennerton C. T. Hugenholtz P. Tyson G. W. (2015). CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes.Genome Res.251043–1055. 10.1101/gr.186072.114
38
Parks D. H. Rinke C. Chuvochina M. Chaumeil P.-A. Woodcroft B. J. Evans P. N. et al (2017). Recovery of nearly 8,000 metagenome-assembled genomes substantially expands the tree of life.Nat. Microbiol.21533–1542. 10.1038/s41564-017-0012-7
39
Poddar A. Lepcha R. T. Mukherjee D. Bhattacharyya D. Das S. K. (2014). Comparative analysis of 16S rRNA signature sequences of the genus Idiomarina and Idiomarina woesei sp. nov., a novel marine bacterium isolated from the Andaman Sea.Res. Microbiol.165501–507. 10.1016/j.resmic.2014.07.008
40
Price M. N. Dehal P. S. Arkin A. P. (2010). FastTree 2–approximately maximum-likelihood trees for large alignments.PLoS One5:e9490. 10.1371/journal.pone.0009490
41
Rajendhran J. Gunasekaran P. (2011). Microbial phylogeny and diversity: small subunit ribosomal RNA sequence analysis and beyond.Microbiol. Res.16699–110. 10.1016/j.micres.2010.02.003
42
Richter M. Rosselló-Móra R. (2009). Shifting the genomic gold standard for the prokaryotic species definition.Proc. Natl. Acad. Sci. U.S.A.10619126–19131. 10.1073/pnas.0906412106
43
Riesco R. Carro L. Roman-Ponce B. Prieto C. Blom J. Klenk H. P. et al (2018). Defining the species Micromonospora saelicesensis and Micromonospora noduli under the framework of genomics.Front. Microbiol.9:1360. 10.3389/fmicb.2018.01360
44
Rodriguez-R L. M. Gunturu S. Harvey W. T. Rosselló-Mora R. Tiedje J. M. Cole J. R. et al (2018). The microbial genomes atlas (MiGA) webserver: taxonomic and gene diversity analysis of archaea and bacteria at the whole genome level.Nucleic Acids Res.46282–288. 10.1093/nar/gky467
45
Salvetti E. Harris H. M. B. Felis G. E. O’Toole P. W. (2018). Comparative genomics reveals robust phylogroups in the genus Lactobacillus as the basis for reclassification.Appl. Environ. Microbiol.84:AEM.00993-18. 10.1128/AEM.00993-18
46
Song L. Ren F. Huang Y. Dai X. Zhou Y. (2013). Idiomarina indica sp. nov., isolated from seawater.Int. J. Syst. Evol. Microbiol.632497–2500. 10.1099/ijs.0.046789-0
47
Srinivas T. N. Anil Kumar P. (2012). Aliidiomarina haloalkalitolerans sp. nov., a marine bacterium isolated from coastal surface seawater.Antonie Van Leeuwenhoek101761–768. 10.1007/s10482-011-9690-y
48
Srivastava P. Kowshik M. (2016). Anti-neoplastic selenium nanoparticles from Idiomarina sp. PR58-8.Enzyme Microb. Technol.95192–200. 10.1016/j.enzmictec.2016.08.002
49
Srivastava P. Kowshik M. (2017). Fluorescent lead(IV) sulfide nanoparticles synthesized by Idiomarina sp. strain PR58-8 for bioimaging applications.Appl. Environ. Microbiol.83:e03091-16. 10.1128/AEM.03091-16
50
Sun Z. Harris H. M. McCann A. Guo C. Argimon S. Zhang W. et al (2015). Expanding the biotechnology potential of lactobacilli through comparative genomics of 213 strains and associated genera.Nat. Commun.6:8322. 10.1038/ncomms9322
51
Taborda M. Antunes A. Tiago I. Verissimo A. Nobre M. F. da Costa M. S. (2009). Description of Idiomarina insulisalsae sp. nov., isolated from the soil of a sea salt evaporation pond, proposal to transfer the species of the genus Pseudidiomarina to the genus Idiomarina and emended description of the genus Idiomarina.Syst. Appl. Microbiol.32371–378. 10.1016/j.syapm.2009.06.005
52
Tamura K. Peterson D. Peterson N. Stecher G. Nei M. Kumar S. (2011). MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods.Mol. Biol. Evol.282731–2739. 10.1093/molbev/msr121
53
Thompson C. C. Chimetto L. Edwards R. A. Swings J. Stackebrandt E. Thompson F. L. (2013). Microbial genomic taxonomy.BMC Genomics14:913. 10.1186/1471-2164-14-913
54
Tindall B. J. Rosselló-Móra R. Busse H. J. Ludwig W. Kämpfer P. (2010). Notes on the characterization of prokaryote strains for taxonomic purposes.Int. J. Syst. Evol. Microbiol.60249–266. 10.1099/ijs.0.016949-0
55
Tully B. J. Graham E. D. Heidelberg J. F. (2018). The reconstruction of 2,631 draft metagenome-assembled genomes from the global oceans.Sci. Data5:170203. 10.1038/sdata.2017.203
56
Vernikos G. Medini D. Riley D. R. Tettelin H. (2015). Ten years of pan-genome analyses.Curr. Opin. Microbiol.23148–154. 10.1016/j.mib.2014.11.016
57
Wang L. Lai Q. Fu Y. Chen H. Wang W. Wang J. et al (2011). Idiomarina xiamenensis sp. nov., isolated from surface seawater, and proposal to transfer Pseudidiomarina aestuarii to the genus Idiomarina as Idiomarina aestuarii comb. nov.Int. J. Syst. Evol. Microbiol.61969–973. 10.1099/ijs.0.022970-0
58
Wattam A. R. Abraham D. Dalay O. Disz T. L. Driscoll T. Gabbard J. L. et al (2014). PATRIC, the bacterial bioinformatics database and analysis resource.Nucleic Acids Res.42D581–D591. 10.1093/nar/gkt1099
59
Wayne L. Brenner D. Colwell R. Grimont P. Kandler O. Krichevsky M. et al (1987). Report of the ad hoc committee on reconciliation of approaches to bacterial systematics.Int. J. Syst. Evol. Microbiol.37463–464. 10.1099/00207713-37-4-463
60
Wu Y. H. Shen Y. Q. Xu X. W. Wang C. S. Oren A. Wu M. (2009). Pseudidiomarina donghaiensis sp. nov. and Pseudidiomarina maritima sp. nov., isolated from the East China Sea.Int. J. Syst. Evol. Microbiol.591321–1325. 10.1099/ijs.0.005702-0
61
Yarza P. Spröer C. Swiderski J. Mrotzek N. Spring S. Tindall B. J. et al (2013). Sequencing orphan species initiative (SOS): filling the gaps in the 16S rRNA gene sequence database for all species with validly published names.Syst. Appl. Microbiol.3669–73. 10.1016/j.syapm.2012.12.006
62
Yarza P. Yilmaz P. Pruesse E. Glöckner F. O. Ludwig W. Schleifer K.-H. et al (2014). Uniting the classification of cultured and uncultured bacteria and archaea using 16S rRNA gene sequences.Nat. Rev. Microbiol.12:635. 10.1038/nrmicro3330
63
Yoon J. H. Jung S. Y. Jung Y. T. Oh T. K. (2007). Idiomarina salinarum sp. nov., isolated from a marine solar saltern in Korea.Int. J. Syst. Evol. Microbiol.572503–2506. 10.1099/ijs.0.65189-0
64
Yoon S. H. Ha S. M. Lim J. Kwon S. Chun J. (2017). A large-scale evaluation of algorithms to calculate average nucleotide identity.Antonie Van Leeuwenhoek1101281–1286. 10.1007/s10482-017-0844-4
65
Zhong Z. P. Liu Y. Liu H. C. Wang F. Song L. Liu Z. P. (2014). Idiomarina planktonica sp. nov., isolated from a saline lake.Int. J. Syst. Evol. Microbiol.643411–3416. 10.1099/ijs.0.065938-0
Summary
Keywords
Idiomarinaceae , genome, taxonomy, diversity, dDDH, ANI
Citation
Liu Y, Lai Q and Shao Z (2018) Genome-Based Analysis Reveals the Taxonomy and Diversity of the Family Idiomarinaceae. Front. Microbiol. 9:2453. doi: 10.3389/fmicb.2018.02453
Received
31 July 2018
Accepted
25 September 2018
Published
11 October 2018
Volume
9 - 2018
Edited by
Iain Sutcliffe, Northumbria University, United Kingdom
Reviewed by
Aharon Oren, Hebrew University of Jerusalem, Israel; Elena P. Ivanova, RMIT University, Australia; Mohammad Ali Amoozegar, University of Tehran, Iran
Updates
Copyright
© 2018 Liu, Lai and Shao.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zongze Shao, shaozz@163.com
This article was submitted to Evolutionary and Genomic Microbiology, a section of the journal Frontiers in Microbiology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.