- 1Department of Laboratory Medicine, Zhujiang Hospital, Southern Medical University, Guangzhou, China
- 2Department of Clinical Laboratory, The First People’s Hospital of Foshan, Foshan, China
- 3State Key Laboratory of Pathogen and Biosecurity, Beijing Institute of Microbiology and Epidemiology, Beijing, China
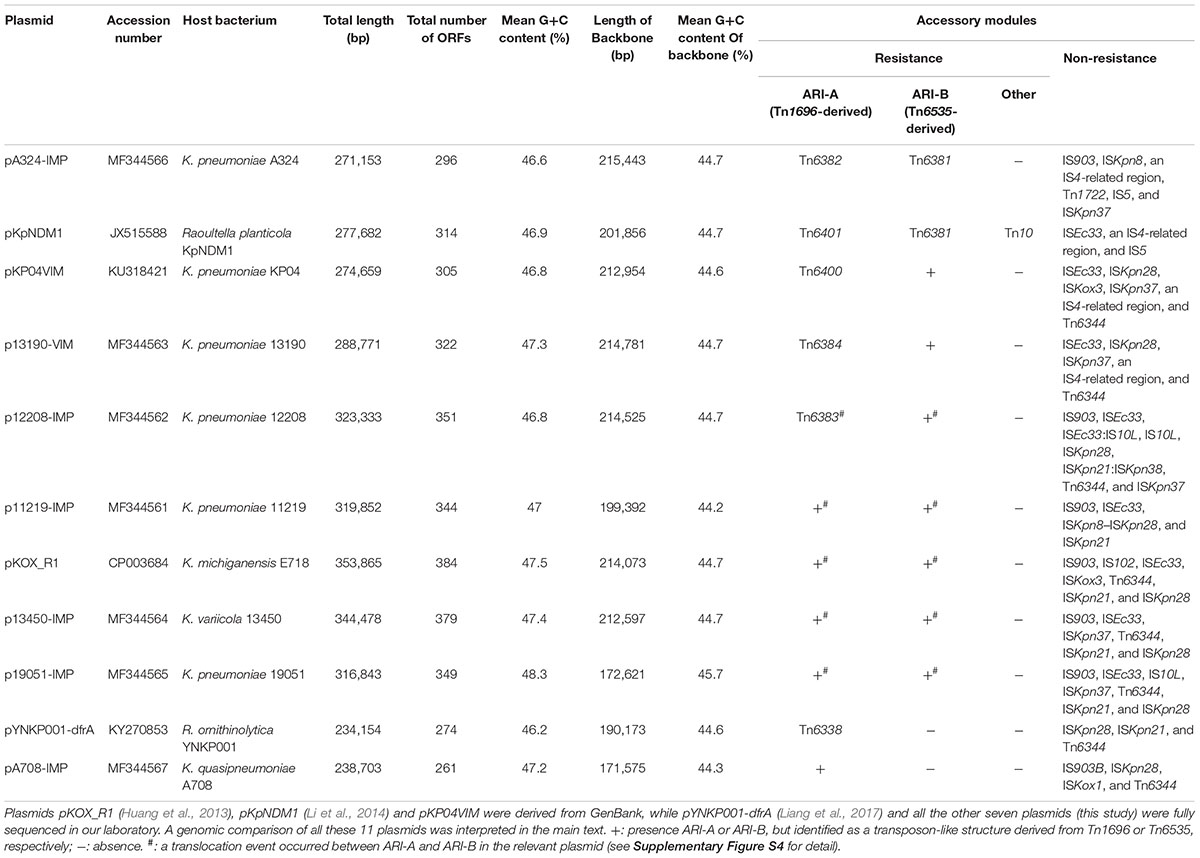
IncHI plasmids could be divided into five different subgroups IncHI1–5. In this study, the complete nucleotide sequences of seven blaIMP- or blaVIM-carrying IncHI5 plasmids from Klebsiella pneumoniae, K. quasipneumoniae, and K. variicola were determined and compared in detail with all the other four available sequenced IncHI5 plasmids. These plasmids carried conserved IncHI5 backbones composed of repHI5B and a repFIB-like gene (replication), parABC (partition), and tra1 (conjugal transfer). Integration of a number of accessory modules, through horizontal gene transfer, at various sites of IncHI5 backbones resulted in various deletions of surrounding backbone regions and thus considerable diversification of IncHI5 backbones. Among the accessory modules were three kinds of resistance accessory modules, namely Tn10 and two antibiotic resistance islands designated ARI-A and ARI-B. These two islands, inserted at two different fixed sites (one island was at one site and the other was at a different site) of IncHI5 backbones, were derived from the prototype Tn3-family transposons Tn1696 and Tn6535, respectively, and could be further discriminated as various intact transposons and transposon-like structures. The ARI-A or ARI-B islands from different IncHI5 plasmids carried distinct profiles of antimicrobial resistance markers and associated mobile elements, and complex events of transposition and homologous recombination accounted for assembly of these islands. The carbapenemase genes blaIMP-4, blaIMP-38 and blaVIM-1 were identified within various class 1 integrons from ARI-A or ARI-B of the seven plasmids sequenced in this study. Data presented here would provide a deeper insight into diversification and evolution history of IncHI5 plasmids.
Introduction
Plasmids of the H incompatibility (IncH) group show two types of surface exclusion and incompatibility interactions, namely IncHI and IncHII (Taylor and Grant, 1977). The IncHI group can be further divided into five subgroups IncHI1 to IncHI5 based on their nucleotide sequence homology (Liang et al., 2017), but the incompatibility interactions between these subgroups are still unclear because no one has actually done real incompatibility tests. IncHI plasmids, often >200 kb in size, have a wide host range including Enterobacteriaceae species and several other Gram-negative organisms (Maher and Taylor, 1993). IncHI1–5 have different replication gene profiles, namely repHI1A+repHI1B+repFIA-like, repHI2A+repHI2C, repHI3B+repFIB-like, repHI4A+repHI4B, and repHI5B+repFIB-like, respectively (Liang et al., 2017). IncHI plasmids generally possess two conjugal transfer regions tra1 and tra2, and the ability of conjugative transfer is thermosensitive and the transfer efficiency is optimal between 22 and 30°C, but inhibited at 37°C (Sherburne et al., 2000). IncHI plasmids are important vectors of genes encoding for resistance not only to heavy metals (such as mercuric ions, copper, silver ions, tellurite, arsenate, and arsenite) but to antibiotics (such as β-lactams including carbapenems, quinolones, aminoglycosides, tetracyclines, amphenicols, and fosfomycin) (Cain and Hall, 2012).
Currently only four fully sequenced IncHI5 plasmids are available (last accessed June 28th, 2017), including pKOX_R1 (Accession No. CP003684) (Huang et al., 2013), pKpNDM1 (Accession No. JX515588) (Li et al., 2014), pKP04VIM (Accession No. KU318421), and pYNKP001-dfrA (Accession No. KY270853) (Liang et al., 2017). This study presents the complete nucleotide sequences of six blaIMP-carrying IncHI5 plasmids and a blaVIM-carrying one, and further comprehensive genomic comparison of all the 11 available sequenced IncHI5 plasmids enable to gain a deeper insight into genomic variation and evolution of IncHI5 plasmids.
Materials and Methods
Bacterial Strains
Klebsiella quasipneumoniae A708 and K. pneumoniae A324 were isolated in 2014 from the blood specimens of two different patients from a teaching hospital in Guangzhou City, China. K. pneumoniae 13190 and 12208, and K. variicola 13450 were recovered in 2013 from the sputum, sputum and blood specimens of three different patients from a teaching hospital in Hangzhou City, China, respectively. K. pneumoniae 11219 was isolated in 2013 from a sputum specimen of a patient from a teaching hospital in Hefei City, China. K. pneumoniae 19051 was recovered in 2011 from a urine specimen of a patient from a public hospital in Ningbo, China.
Phenotypic Assays
Activity of Ambler class A/B/D carbapenemases in bacterial cell extracts was determined by a modified CarbaNP test (Wei et al., 2016). Bacterial antimicrobial susceptibility was tested by BioMérieux VITEK 2 and interpreted as per the 2017 CLSI guidelines (CLSI, 2017).
Conjugal Transfer
Conjugal transfer experiments were carried out with the rifampin-resistant Escherichia coli EC600 used as a recipient and the blaIMP-positive A324 isolate as a donor. Three milliliters of overnight cultures of each of donor and recipient bacteria were mixed together, harvested and resuspended in 80 μL of Brain Heart Infusion (BHI) broth (BD Biosciences). The mixture was spotted on a 1 cm2 hydrophilic nylon membrane filter with a 0.45 μm pore size (Millipore) that was placed on BHI agar (BD Biosciences) plate and then incubated for mating at 22°C for 24 h. Bacteria were washed from filter membrane and spotted on Muller-Hinton (MH) agar (BD Biosciences) plates containing 2500 μg/mL rifampin together with 4 μg/mL meropenem for selecting an E. coli transconjugant carrying blaIMP (pA324-IMP).
Electroporation
To prepare competent cells for electroporation, 200 mL of overnight culture of E. coli TOP10 in Super Optimal Broth (SOB) at an optical density (OD600) of 0.4 to 0.6 was washed three times with electroporation buffer (0.5 M mannitol and 10% glycerol) and concentrated into a final volume of 2 mL. One microgram of DNA were mixed with 100 μL of competent cells for electroporation at 25 μF, 200 Ω and 2.5 Kv. The resulting cells were suspended in 500 μL of SOB and an appropriate aliquot was spotted on SOB agar plates containing 4 μg/mL meropenem for selecting of an electroporant carrying blaIMP (pA324-IMP).
Sequencing and Sequence Assembly
Genomic DNA was isolated from each of the A708, 13190, 11219, 12208, 13450, and 19051 isolates using a Qiagen blood & cell culture DNA maxi kit. Genome sequencing was performed with a sheared DNA library with average size of 15 kb (ranged from 10 to 20 kb) on a PacBio RSII sequencer (Pacific Biosciences, Menlo Park, CA, United States), as well as a paired-end library with an average insert size of 400 bp (ranged from 150 to 600 bp) on a HiSeq sequencer (Illumina, San Diego, CA, United States). The paired-end short Illumina reads were used to correct the long PacBio reads utilizing proovread (Hackl et al., 2014), and then the corrected PacBio reads were assembled de novo utilizing SMARTdenovo1.
Plasmid DNA was isolated from the A324-IMP-TOP10 electroporant using a Large Construct Kit (Qiagen, Germany) and then sequenced from a mate-pair library with average insert size of 5 kb (ranged from 2 to 10 kb) using a MiSeq sequencer (Illumina, San Diego, CA, United States). Quality control, removing adapters and low quality reads, were performed using Trimmomatic 0.36 (Bolger et al., 2014). The filtered clean reads were then assembled using Newbler 2.6 (Nederbragt, 2014), followed by extraction of the consensus sequence with CLC Genomics Workbench 3.0 (Qiagen Bioinformatics). Gapfiller V1.11 (Boetzer and Pirovano, 2012) was used for gap closure.
Sequence Annotation and Comparison
Open reading frames and pseudogenes were predicted using RAST 2.0 (Brettin et al., 2015) combined with BLASTP/BLASTN searches (Boratyn et al., 2013) against the UniProtKB/Swiss-Prot database (Boutet et al., 2016) and the RefSeq database (O’Leary et al., 2016). Annotation of resistance genes, mobile elements, and other features was carried out using the online databases including CARD (Jia et al., 2017), ResFinder (Zankari et al., 2012), ISfinder (Siguier et al., 2006), INTEGRALL (Moura et al., 2009), and Tn Number Registry (Roberts et al., 2008). Multiple and pairwise sequence comparisons were performed using MUSCLE 3.8.31 (Edgar, 2004) and BLASTN, respectively. Gene organization diagrams were drawn in Inkscape 0.48.12.
Phylogenetic Analysis
The backbone regions of indicative plasmids were aligned using MUMmer 3.0 (Kurtz et al., 2004). Inference of homologous recombination was performed using ClonalFrameML (Didelot and Wilson, 2015) to remove recombination-associated single-nucleotide polymorphisms (SNPs). A maximum-likelihood tree was constructed from recombination-free SNPs using MEGA7 (Kumar et al., 2016) with a bootstrap iteration of 1000.
Nucleotide Sequence Accession Numbers
The complete nucleotide sequences of plasmids p11219-IMP, p12208-IMP, p13190-VIM, p13450-IMP, p19051-IMP, pA324-IMP, and pA708-IMP, and those of the A708, 12208, 13450, 11219, 13190, and 19051 chromosomes were submitted to GenBank under Accession Nos. MF344561 to MF344567, CP030171 to CP030174, CP026017, and CP022023, respectively.
Results and Discussion
Overview of Sequenced IncHI5 Plasmids
The seven plasmids sequenced in the present work varied in size from about 238 kb to nearly 345 kb with variation in the number of predicted ORFs from 261 to 379 (Table 1 and Supplementary Figure S1). All these plasmids belonged to the IncHI5 group, because each contained a conserved IncHI5 backbone especially including the IncHI5-type replication gene repHI5B and an additional repFIB-like gene (Liang et al., 2017). Table 1 also lists the features of the previously four sequenced IncHI5 plasmids. Further comparative genomics of all these 11 plasmids revealed that the IncHI5 backbones were interrupted by various accessory modules (defined as acquired DNA regions associated and bordered with mobile elements) inserted at different sites. In addition, while pKOX_R1 (Huang et al., 2013), the first sequenced IncHI5 plasmid, was used previously as the IncHI5 reference (Liang et al., 2017), the newly sequenced pA324-IMP seemed a more appropriate reference in this analysis because it contained the most complete IncHI5 backbone (Supplementary Figure S2).
This large collection of IncHI5 plasmids allowed us to accurately distinguish backbone and accessory modules. This has allowed us to gain a deeper understanding of the evolution and diversification of IncHI5 plasmids.
General Comparison of Backbone Sequences
Pairwise sequence comparison using BLASTN showed that these 11 plasmids had >99% nucleotide identity across >73% of their backbone sequences (Supplementary Table S1). The major IncHI5 backbone genes or gene loci (including repHI5B together with its iterons and repFIB-like for replication, parABC for partition, and tra1 for conjugal transfer) were conserved among all these 11 plasmids. Two conjugal transfer regions tra1 and tra2 were found in IncHI5 plasmids, but some of these plasmids lost tra2, which would impair their self-transferability (Supplementary Table S3). A total of 574 core SNPs (among them 115 were recombination-free) were identified from the backbone regions of these 11 plasmids. A maximum likelihood phylogenetic tree was constructed using these 115 recombination-free SNPs, and accordingly these 11 plasmids could be assigned into three clades I, II, and III (Figure 1).
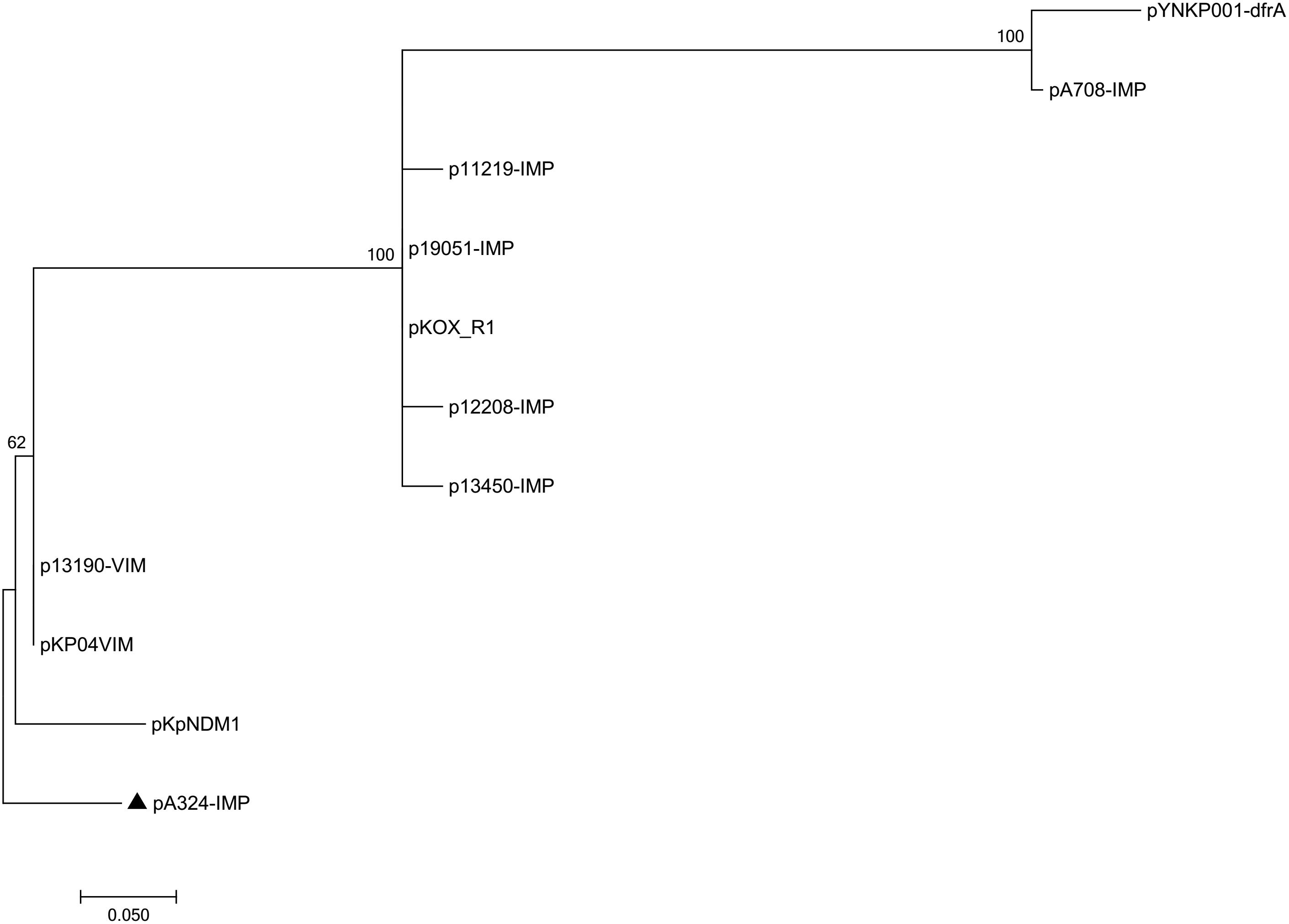
Figure 1. Maximum-likelihood tree. The degree of support (percentage) for each cluster of associated taxa, as determined by bootstrap analysis, is shown next to each branch. The bar corresponds to the scale of sequence divergence. The triangle indicates IncHI5 reference plasmid pA324-IMP.
Classification of Accessory Modules
These 11 plasmids harbored different profiles of accessory modules (Table 1) and different collections of resistance genes (Supplementary Table S2). The accessory modules were further divided into resistance (containing resistance genes) and non-resistance (containing no resistance genes) ones (Table 1). The resistance accessory modules included Tn10 (an IS10-composite transposon carrying class B tetracycline-resistance genes) and two antibiotic resistance islands designated ARI-A and ARI-B. The presence and modular organization of ARI-A and ARI-B islands in the seven newly sequence plasmids and pYNKP001-dfrA (Liang et al., 2017) were validated by a set of PCR amplifications (see Figures 3, 4 for location of PCR primers and expected amplicons) that targeted various key jointing fragments of these islands and their surrounding backbone regions, using the genomic DNA of each corresponding wild-type isolate as template. The non-resistance accessory modules were composed of 12 different insertion sequences (ISs), four distinct IS-related regions, and two cryptic transposons Tn6344 (Supplementary Figure S3) and Tn1722.
Massive Gene Acquisition and Loss in IncHI5 Plasmids
At least 10 major events of gene acquisition/loss accounted for modular diversity of these 11 plasmids across their genomes (Figure 2). First, IS903 was inserted at a site upstream of orf444 in five plasmids, additionally resulting in a 42.5-kb deletion (containing the whole tra2 region) in p19051-IMP. Second, ISEc33, ISEc33 and IS102 were inserted at different sites within the tra2 regions of pKpNDM1, p12208-IMP and pKOX_R1, respectively. Third, IS903B in pA708-IMP, ISEc33 in six plasmids, and ISEc33:IS10L in p12208-IMP were inserted within hnhc (HNH endonuclease), splitting it into two separate parts Δhnhc-5′ and Δhnhc-3′. Fourth, in pKpNDM1, Tn10 was inserted at a site within orf648, splitting it into two separate parts Δorf648-5′ and Δorf648-3′. Fifth, compared to the backbone region from orf633 to hokG in pA324-IMP as a prototype structure, various insertions occurred in all the other plasmids: (i) insertion of ISKpn28 upstream of hokG resulted in deletion of a 34.3-kb region in pA708-IMP and that of a 24.8-kb region in pYNKP001-dfrA, respectively; however, upstream-of-hokG insertion of ISKpn8 in pA324-IMP, that of ISKpn28 in six plasmids, and that of ISKpn8–ISKpn28 in p11219-IMP did not cause deletions; and (ii) insertion of ARI-B at a site within xerC2 occurred in the following nine plasmids, which led to a 3.2-kb deletion in seven plasmids, a 14.0-kb deletion in pKpNDM1, and no deletion in pA324-IMP. Sixth, six plasmids had complete tra1 regions; by contrast, four additional plasmids had undergone insertion of an IS4-related region at a site downstream of tivF3 (resulting in a 3.1-kb deletion), and ISkox1 was inserted at a site between tivF3 and orf171 in pA708-IMP (no further deletion occurred). All the above insertions and/or deletions within tra1 and tra2 might impair self-transferability of corresponding plasmids. Seventh, ISKpn21 in five plasmids, ISKox3 in pKP04VIM, and ISKpn21:ISKpn38 in p12208-IMP were inserted at a site downstream of orf294. Eighth, Tn1722 was inserted at a site between orf333 and orf261 in pA324-IMP. Ninth, IS5 or Tn6344 was inserted at a site between luxR and orf183 in all the night plasmids except for p11219-IMP and p13450-IMP, and additionally the Tn6344 insertion resulted in a 1.8-kb deletion in p13450-IMP. Tenth, the ARI-A islands were inserted at a site downstream of orf342 in all the 11 plasmids, which resulted in the 12.6-kb deletion (covering terE-5′ and terABCDZW) in pA708-IMP and the 15.1-kb deletion (covering the complete ter gene cluster) in p11219-IMP; additionally, ISKpn37 was inserted at a site within terF in six plasmids. The above insertions would impair tellurium resistance gene expression.

Figure 2. Sites of insertion of accessory modules. Genes are denoted by arrows. Genes, mobile elements and other features are colored based on function classification. Shading denotes regions of homology (>95% nucleotide identity).
In conclusion, massive gene acquisition and loss were found in IncHI5 plasmids: a wealth of accessory modules were integrated at various sites of IncHI5 backbones, which resulted in various deletions of surrounding backbone regions and thus considerable diversification of IncHI5 backbones.
Tn1696-Related ARI-A Islands
The ARI-A islands (Figure 3) were found in all the 11 plasmids analyzed and identified as Tn1696 derivatives. Tn1696, a unit transposon belonging to the Tn21 subgroup of Tn3 family, was generated from insertion of a class 1 integron In4 into the resolution (res) site of a primary backbone structure: IRL (inverted repeat left)–tnpA (transposase)–tnpR (resolvase)–res–mer (mercury resistance locus)–IRR (inverted repeat right) (Partridge et al., 2001).
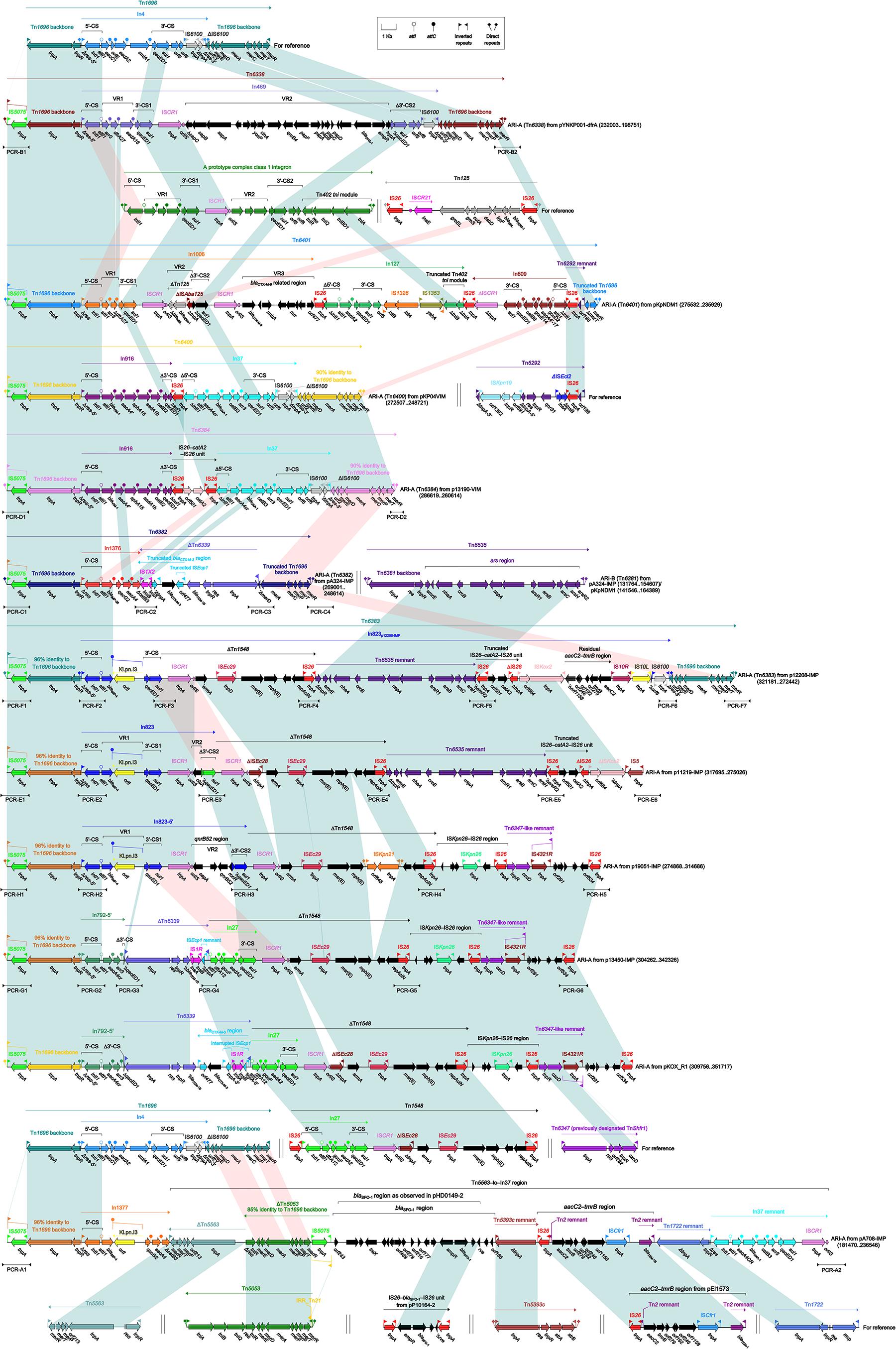
Figure 3. Organization of ARI-A islands and comparison to related regions. Genes are denoted by arrows. Genes, mobile elements and other features are colored based on their functional classification. Shading denotes regions of homology (nucleotide identity > 95%). Numbers in brackets indicate nucleotide positions within corresponding plasmids. The accession numbers of Tn1696 (Partridge et al., 2001), Tn125 (Poirel et al., 2012), Tn6292 (Wei et al., 2016), Tn1548 (Galimand et al., 2005), Tn6347, Tn5563 (Yeo et al., 1998), Tn5053 (Kholodii et al., 1993), IS26–blaSFO–1–IS26 unit (Sun et al., 2016), Tn5393c (L’Abée-Lund and Sørum, 2000), aacC2–tmrB region (Partridge et al., 2012), and Tn1722 (Allmeier et al., 1992) for reference are U12338, JN872328, KU886034, AF550415, CP000447, U88088, L40585, KX710093, AF262622, JX101693, and X61367, respectively. Arrowheads indicated location of PCR primers and expected amplicons.
Being similar to Tn1696, the ARI-A islands from six plasmids had paired terminal 38-bp IRL/IRR and were further bracketed by 5-bp direct repeats (DRs; target site duplication signals for transposition), and thus they were identified as unit transposons designated Tn6338, Tn6401, Tn6400, Tn6384, Tn6382, and Tn6383, respectively. The remaining five ARI-A islands carried only IRLs (interrupted by IS5075 that was a hunter of terminal IRL/IRR of Tn21 subgroup transposons (Partridge and Hall, 2003)) but did not harbor IRRs (due to truncation at 3′-terminal regions of these islands), and thus they were identified as transposon-like structures rather than intact transposons.
In conclusion, the ARI-A islands was inserted at a site downstream of orf342 in all 11 plasmids, and further discriminated as six intact transposons (among them Tn6384, Tn6382, and Tn6383 were novel) and five transposon-like structures. These 11 islands were derived from Tn1696 but differed from it mainly by insertion of distinct integrons or integron-related regions instead of In4 in Tn1696. These integrons could be divided into concise integrons each containing a single gene cassette (GC) array, and complex integrons each harboring one or more variable regions (VRs) in addition to the GC array. These GCs and VRs commonly carried antibiotic resistance genes.
Tn6535-Related ARI-B Islands
The ARI-B islands (Figure 4) as found in nine plasmids were identified as the derivatives of a prototype arsenic-resistance (ars) unit transposon Tn6535. As observed in the chromosome (Accession No. CP009706) of Hafnia alvei FB1 (Tan et al., 2014), Tn6535 was assembled from integration of an ars region with a Tn3-family core transposition module tnpA–res–tnpR.
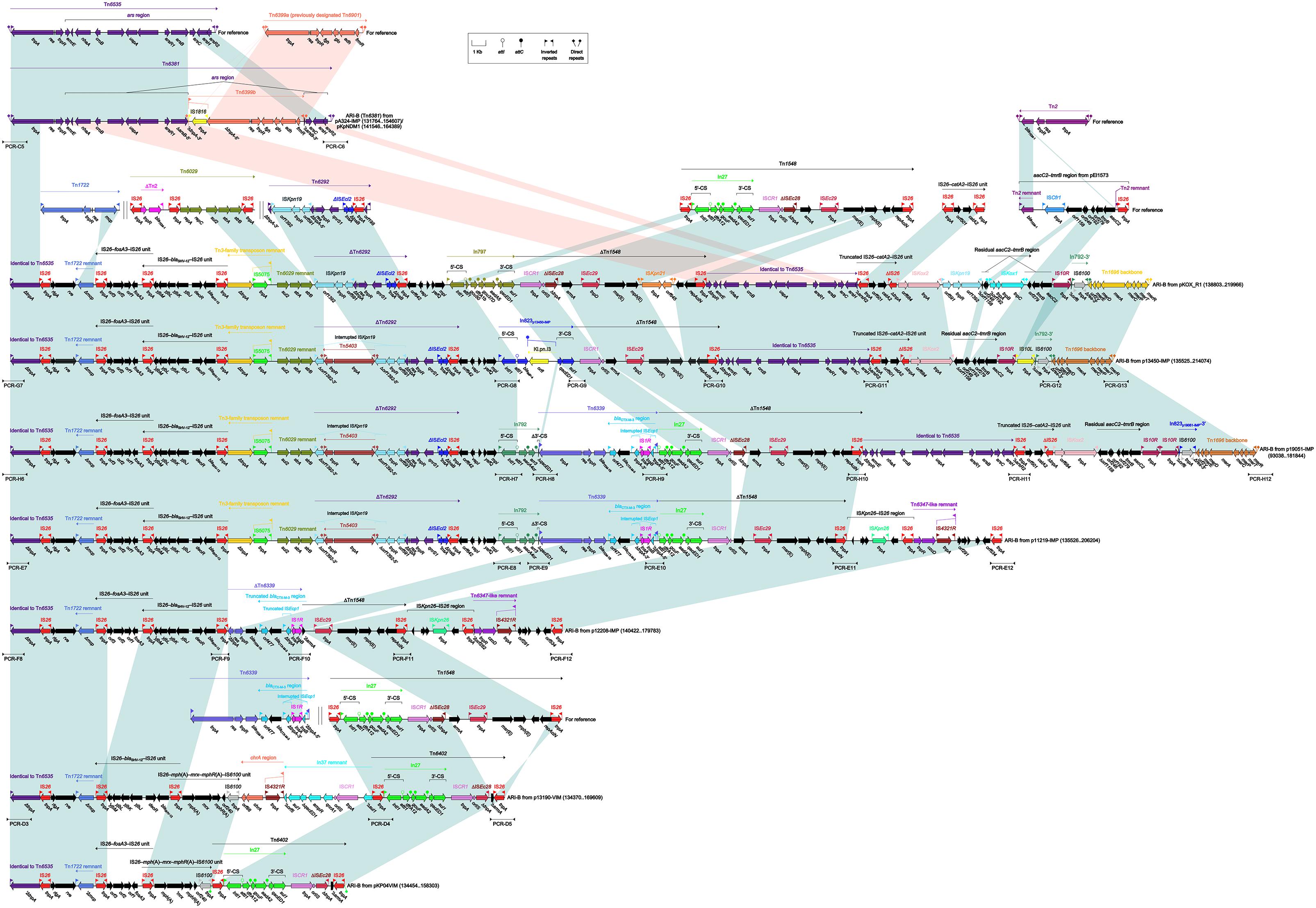
Figure 4. Organization of ARI-B islands and comparison to related regions. Genes are denoted by arrows. Genes, mobile elements and other features are colored based on their functional classification. Shading denotes regions of homology (nucleotide identity > 95%). Numbers in brackets indicate nucleotide positions within corresponding plasmids. The accession numbers of Tn6535 (Tan et al., 2014), Tn6399a (Murata et al., 2002), Tn1722 (Allmeier et al., 1992), Tn6029 (Cain et al., 2010), Tn6292 (Wei et al., 2016), Tn1548 (Galimand et al., 2005), IS26–catA2–IS26 unit (Xiang et al., 2015), Tn2 (Bailey et al., 2011), aacC2–tmrB region (Partridge et al., 2012), and Tn6339 (Liang et al., 2017) for reference are CP009706, AP004237, X61367, HQ840942, KU886034, AF550415, KY270851, HM749967, JX101693, and CP003684, respectively. Arrowheads indicated location of PCR primers and expected amplicons.
The ARI-B island from pA324-IMP or pKpNDM1 was identified as an unit transposon designated Tn6381, with presence of paired terminal 38-bp IRL/IRR and 5-bp DRs. Tn6381 differed from Tn6535 by insertion of a Tn3-family unit transposon Tn6399b at a site within arsB. Tn6399a (it was previously designated Tn6901 because it was 6901 bp in length) was found in plasmid Rts1 from Proteus vulgaris (Murata et al., 2002) and carried several alcohol-metabolism genes (Chen et al., 2013), while interruption of tnpA by IS1618 insertion turned Tn6399a into Tn6399b. Compared to Tn6535 and Tn6381, all the other ARI-B islands contained only IRLs but not IRRs and identified as transposon-like structures. Various types of insertion events occurred within these ARI-B islands, leading to truncation of prototype regions as found in Tn6535 and Tn6381 as well as integration of foreign resistance markers and associated mobile elements.
ARI-BpKOX_R1 contained at least 10 resistance loci, including IS26–fosA3–IS26 unit, IS26–blaSHV -12–IS26 unit, a 3-kb Tn6029 remnant, ΔTn6292, In797, ΔTn1548, ΔTn6535, a truncated IS26–catA2–IS26 unit, a residual aacC2–tmrB region, and a mer region (Liang et al., 2017). The resistance markers from ARI-Bp13450-IMP and ARI-Bp19051-IMP differed from pKOX_R1 by replacement of In797 with In823 and In792, respectively; moreover, Tn6339 (containing blaTEM-1B and blaCTX-M-3) was inserted at a site between In792 and ΔTn1548 in p19051-IMP. The resistance markers from ARI-Bp11219-IMP were composed of IS26–fosA3–IS26, IS26–blaSHV -12–IS26, a Tn6029 remnant, ΔTn6292, In792, Tn6339, and ΔTn1548. Similarly, IS26–fosA3–IS26, IS26–blaSHV -12–IS26, ΔTn6339, and ΔTn1548 were present in ARI-Bp12208-IMP. The resistance markers from ARI-Bp13190-VIM consisted of IS26–blaSHV -12–IS26, IS26–mph(A)–mrx–mphR(A)–IS6100, chrA region, a In37 remnant and Tn6402, while those from ARI-BpKP04VIM were composed of IS26–fosA3–IS26, IS26–mph(A)–mrx–mphR(A)–IS6100 and Tn6402. Tn6402 was an IS26-composite transposon (delimited by 4-bp DRs at both ends) derived from Tn1548. Tn6402 differed from Tn1548 by deletion of arsB-3′–ISEc29–msr(E)–mph(E)–repAciN and inversion of the 3′-end copy of IS26.
In conclusion, the ARI-B islands, integrated at a site within xerC2 in nine plasmids, could be further identified as Tn6381 and eight transposon-like structures. These nine ARI-B islands were derived from Tn6535 but differed from it by insertion of various collections of mobile elements and associated resistance genes into the original Tn6381 backbone.
All the transposon-like structures of ARI-A or ARI-B could not be annotated as intact transposons because they lacked paired terminal inverted repeats, and complex transposition and homologous recombination events accounted for assembly and diversification of these transposons and transposon-like structures. The ARI-A or ARI-B islands from different IncHI5 plasmids carried distinct profiles of resistance markers and associated mobile elements, promoting accumulation and spread of antimicrobial resistance among bacterial species.
Translocation of Large Regions Across ARI-A and ARI-B
Compared to the intact transposons Tn6338, Tn6401, Tn6400, Tn6384, Tn6382, and Tn6383 (corresponding to ARI-A), and Tn6381 (corresponding to ARI-B), two kinds of translocation events across ARI-A and ARI-B occurred in each of the following five plasmids (Supplementary Figure S4): (i) exchange of the ΔTn1548–to–IS26 region (finally observed in ARI-B) and the ΔTn1548–to–Tn6535 region (finally observed in ARI-A) in each of p12208-IMP and p11219-IMP; and (ii) movement of the Δorf6–mer region from ARI-A to ARI-B in each of pKOX_R1, p13450-IMP, and p19051-IMP. These two kinds of translocation might be mediated by the common regions ΔTn1548 and IS6100, respectively.
Carbapenemase Genes and Related Integrons
As for the seven plasmids sequenced in this study, the carbapenemase genes blaIMP-4, blaIMP-38, and blaVIM-1 were identified within In823p11219-IMP/p12208-IMP/p19051-IMP, or In1377pA708-IMP, In1376pA324-IMP, and In916p13190-VIM respectively, from the ARI-A islands, while a blaIMP-4 gene was found within In823p13450-IMP from the ARI-B island. Of all the integrons identified in these seven plasmids, In1376 and In1377 were novel.
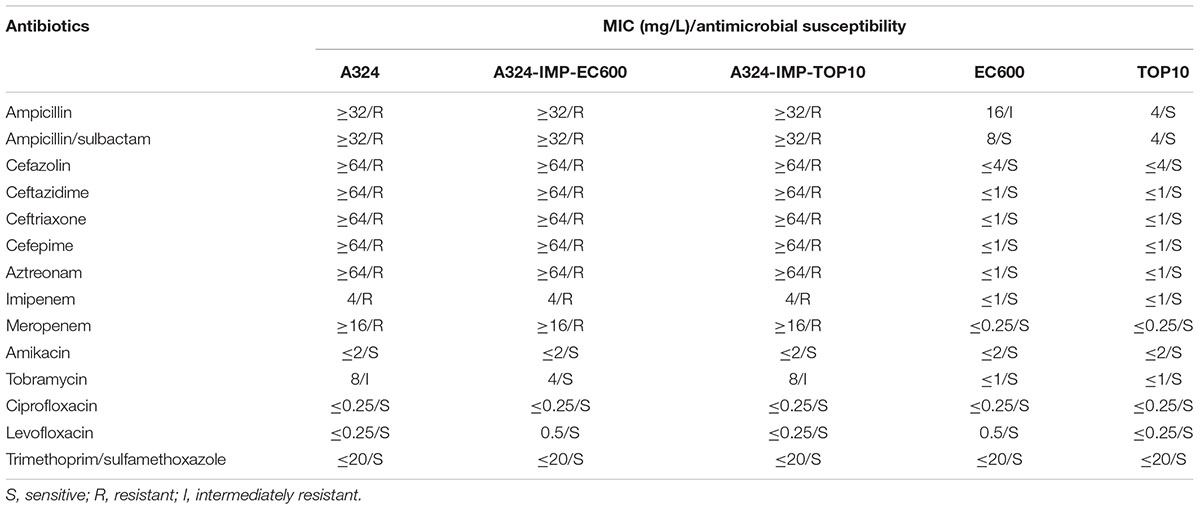
Transferability and Antimicrobial Susceptibility
As a representative IncHI5 plasmid, pA324-IMP could be transferred from the A324 isolate into E. coli EC600 and TOP10 through conjugation and electroporation, respectively, generating the A324-IMP-EC600 transconjugant and the A324-IMP-TOP10 electroporant, respectively. This was consistent with the presence of two complete sets of tra1 and tra2 genes in pA324-IMP, making it self-transferable. All the above three strains had class B carbapenemase activity (data not shown), and were resistant to all the cephalosporins and carbapenems tested (Table 2), which were resulted from production of IMP or VIM enzymes in these strains.
Author Contributions
DZ and LJ conceived the study and designed experimental procedures. QL, XJ, LH, ZY, and WY performed the experiments. QL, XJ, BG, YZ, and HY analyzed the data. QL, XJ, YT, and WL contributed reagents and materials. DZ, QL, XJ, and LJ wrote the manuscript.
Funding
This work was supported by the National Key R&D Program (2017YFC1200800) of China.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2018.03318/full#supplementary-material
FIGURE S1 | Schematic maps of sequenced plasmids. Genes are denoted by arrows, and the backbone and accessory module regions are highlighted in black and color, respectively. The innermost circle presents GC-skew [(G-C)/(G+C)], with a window size of 500 bp and a step size of 20 bp. The next-to-innermost circle presents GC content. The accession numbers of pYNKP001-dfrA (Liang et al., 2017), pKP04VIM, pKpNDM1 (Li et al., 2014), and pKOX_R1 (Huang et al., 2013) for reference are KY270853, KU318421, JX515588, and CP003684, respectively.
FIGURE S2 | Linear comparison of plasmid genome sequences. Genes are denoted by arrows. Genes, mobile elements and other features are colored based on function classification. Shading denotes regions of homology (>95% nucleotide identity). The accession numbers of pYNKP001-dfrA (Liang et al., 2017), pKP04VIM, pKpNDM1 (Li et al., 2014), and pKOX_R1 (Huang et al., 2013) for reference are KY270853, KU318421, JX515588, and CP003684, respectively.
FIGURE S3 | Organization of Tn6344 and comparison to related region. Genes are denoted by arrows. Genes, mobile elements and other features are colored based on their functional classification. Shading denotes regions of homology (nucleotide identity > 95%).
FIGURE S4 | Translocation between ARI-A and ARI-B islands. Genes are denoted by arrows. Genes, mobile elements and other features are colored based on their functional classification. Arrowheads indicated location of PCR primers and expected amplicons.
TABLE S1 | (a) Pairwise comparison of IncHI5 sequences using BLASTN. (b) Pairwise comparison of plamid backbone sequences using BLASTN.
TABLE S2 | Drug resistance genes in sequenced IncHI5 plasmids.
TABLE S3 | Conjugation transfer features of IncHI5 plasmids analyzed.
Footnotes
References
Allmeier, H., Cresnar, B., Greck, M., and Schmitt, R. (1992). Complete nucleotide sequence of Tn 1721 : gene organization and a novel gene product with features of a chemotaxis protein. Gene 111, 11–20. doi: 10.1016/0378-1119(92)90597-I
Bailey, J. K., Pinyon, J. L., Anantham, S., and Hall, R. M. (2011). Distribution of the blaTEM gene and blaTEM-containing transposons in commensal Escherichia coli. J. Antimicrob. Chemother. 66, 745–751. doi: 10.1093/jac/dkq529
Boetzer, M., and Pirovano, W. (2012). Toward almost closed genomes with GapFiller. Genome Biol. 13:R56. doi: 10.1186/gb-2012-13-6-r56
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Boratyn, G. M., Camacho, C., Cooper, P. S., Coulouris, G., Fong, A., Ma, N., et al. (2013). BLAST: a more efficient report with usability improvements. Nucleic Acids Res. 41, W29–W33. doi: 10.1093/nar/gkt282
Boutet, E., Lieberherr, D., Tognolli, M., Schneider, M., Bansal, P., Bridge, A. J., et al. (2016). UniProtKB/Swiss-Prot, the manually annotated section of the UniProt KnowledgeBase: how to use the entry view. Methods Mol. Biol. 1374, 23–54. doi: 10.1007/978-1-4939-3167-5_2
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5:8365. doi: 10.1038/srep08365
Cain, A. K., and Hall, R. M. (2012). Evolution of IncHI2 plasmids via acquisition of transposons carrying antibiotic resistance determinants. J. Antimicrob. Chemother. 67, 1121–1127. doi: 10.1093/jac/dks004
Cain, A. K., Liu, X., Djordjevic, S. P., and Hall, R. M. (2010). Transposons related to Tn1696 in IncHI2 plasmids in multiply antibiotic resistant Salmonella enterica serovar Typhimurium from Australian animals. Microb. Drug Resist. 16, 197–202. doi: 10.1089/mdr.2010.0042
Chen, L., Chavda, K. D., Fraimow, H. S., Mediavilla, J. R., Melano, R. G., Jacobs, M. R., et al. (2013). Complete nucleotide sequences of blaKPC-4- and blaKPC-5-harboring IncN and IncX plasmids from Klebsiella pneumoniae strains isolated in New Jersey. Antimicrob. Agents Chemother. 57, 269–276. doi: 10.1128/AAC.01648-12
CLSI (2017). Performance Standards for Antimicrobial Susceptibility Testing: Twenty-Seventh Informational Supplement M100-S27. Wayne, PA: CLSI.
Didelot, X., and Wilson, D. J. (2015). ClonalFrameML: efficient inference of recombination in whole bacterial genomes. PLoS Comput. Biol. 11:e1004041. doi: 10.1371/journal.pcbi.1004041
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Galimand, M., Sabtcheva, S., Courvalin, P., and Lambert, T. (2005). Worldwide disseminated armA aminoglycoside resistance methylase gene is borne by composite transposon Tn1548. Antimicrob. Agents Chemother. 49, 2949–2953. doi: 10.1128/AAC.49.7.2949-2953.2005
Hackl, T., Hedrich, R., Schultz, J., and Forster, F. (2014). proovread: large-scale high-accuracy PacBio correction through iterative short read consensus. Bioinformatics 30, 3004–3011. doi: 10.1093/bioinformatics/btu392
Huang, T. W., Wang, J. T., Lauderdale, T. L., Liao, T. L., Lai, J. F., Tan, M. C., et al. (2013). Complete sequences of two plasmids in a blaNDM-1-positive Klebsiella oxytoca isolate from Taiwan. Antimicrob. Agents Chemother. 57, 4072–4076. doi: 10.1128/AAC.02266-12
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., et al. (2017). CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573. doi: 10.1093/nar/gkw1004
Kholodii, G. Y., Yurieva, O. V., Lomovskaya, O. L., Gorlenko, Z., Mindlin, S. Z., and Nikiforov, V. G. (1993). Tn5053, a mercury resistance transposon with integron’s ends. J. Mol. Biol. 230, 1103–1107. doi: 10.1006/jmbi.1993.1228
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kurtz, S., Phillippy, A., Delcher, A. L., Smoot, M., Shumway, M., Antonescu, C., et al. (2004). Versatile and open software for comparing large genomes. Genome Biol. 5:R12. doi: 10.1186/gb-2004-5-2-r12
L’Abée-Lund, T. M., and Sørum, H. (2000). Functional Tn5393-like transposon in the R plasmid pRAS2 from the fish pathogen Aeromonas salmonicida subspecies salmonicida isolated in Norway. Appl. Environ. Microbiol. 66, 5533–5535. doi: 10.1128/AEM.66.12.5533-5535.2000
Li, J., Lan, R., Xiong, Y., Ye, C., Yuan, M., Liu, X., et al. (2014). Sequential isolation in a patient of Raoultella planticola and Escherichia coli bearing a novel ISCR1 element carrying blaNDM-1. PLoS One 9:e89893. doi: 10.1371/journal.pone.0089893
Liang, Q., Yin, Z., Zhao, Y., Liang, L., Feng, J., Zhan, Z., et al. (2017). Sequencing and comparative genomics analysis of the IncHI2 plasmids pT5282-mphA and p112298-catA and the IncHI5 plasmid pYNKP001-dfrA. Int. J. Antimicrob. Agents 49, 709–718. doi: 10.1016/j.ijantimicag.2017.01.021
Maher, D., and Taylor, D. E. (1993). Host range and transfer efficiency of incompatibility group HI plasmids. Can. J. Microbiol. 39, 581–587. doi: 10.1139/m93-084
Moura, A., Soares, M., Pereira, C., Leitao, N., Henriques, I., and Correia, A. (2009). INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25, 1096–1098. doi: 10.1093/bioinformatics/btp105
Murata, T., Ohnishi, M., Ara, T., Kaneko, J., Han, C. G., Li, Y. F., et al. (2002). Complete nucleotide sequence of plasmid Rts1: implications for evolution of large plasmid genomes. J. Bacteriol. 184, 3194–3202. doi: 10.1128/JB.184.12.3194-3202.2002
Nederbragt, A. J. (2014). On the middle ground between open source and commercial software - the case of the Newbler program. Genome Biol. 15:113. doi: 10.1186/gb4173
O’Leary, N. A., Wright, M. W., Brister, J. R., Ciufo, S., Haddad, D., McVeigh, R., et al. (2016). Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res. 44, D733–D745. doi: 10.1093/nar/gkv1189
Partridge, S. R., Brown, H., Stokes, H., and Hall, R. (2001). Transposons Tn1696 and Tn21 and their integrons In4 and In2 have independent origins. Antimicrob. Agents Chemother. 45, 1263–1270. doi: 10.1128/AAC.45.4.1263-1270.2001
Partridge, S. R., Ginn, A. N., Paulsen, I. T., and Iredell, J. R. (2012). pEl1573 Carrying blaIMP-4, from Sydney, Australia, is closely related to other IncL/M plasmids. Antimicrob. Agents Chemother. 56, 6029–6032. doi: 10.1128/AAC.01189-12
Partridge, S. R., and Hall, R. M. (2003). The IS1111 family members IS4321 and IS5075 have subterminal inverted repeats and target the terminal inverted repeats of Tn21 family transposons. J. Bacteriol. 185, 6371–6384. doi: 10.1128/JB.185.21.6371-6384.2003
Poirel, L., Bonnin, R. A., Boulanger, A., Schrenzel, J., Kaase, M., and Nordmann, P. (2012). Tn125-related acquisition of blaNDM-like genes in Acinetobacter baumannii. Antimicrob. Agents Chemother. 56, 1087–1089. doi: 10.1128/AAC.05620-11
Roberts, A. P., Chandler, M., Courvalin, P., Guedon, G., Mullany, P., Pembroke, T., et al. (2008). Revised nomenclature for transposable genetic elements. Plasmid 60, 167–173. doi: 10.1016/j.plasmid.2008.08.001
Sherburne, C. K., Lawley, T. D., Gilmour, M. W., Blattner, F. R., Burland, V., Grotbeck, E., et al. (2000). The complete DNA sequence and analysis of R27, a large IncHI plasmid from Salmonella typhi that is temperature sensitive for transfer. Nucleic Acids Res. 28, 2177–2186. doi: 10.1093/nar/28.10.2177
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Sun, F., Zhou, D., Sun, Q., Luo, W., Tong, Y., Zhang, D., et al. (2016). Genetic characterization of two fully sequenced multi-drug resistant plasmids pP10164-2 and pP10164-3 from Leclercia adecarboxylata. Sci. Rep. 6:33982. doi: 10.1038/srep33982
Tan, J. Y., Yin, W. F., and Chan, K. G. (2014). Gene clusters of Hafnia alvei strain FB1 important in survival and pathogenesis: a draft genome perspective. Gut Pathog. 6:29. doi: 10.1186/1757-4749-6-29
Taylor, D. E., and Grant, R. B. (1977). Incompatibility and surface exclusion properties of H1 and H2 plasmids. J. Bacteriol. 131, 174–178.
Wei, F., Zhou, D., Qian, W., Luo, W., Zhang, D., Qiang, S., et al. (2016). Dissemination of IMP-4-encoding pIMP-HZ1-related plasmids among Klebsiella pneumoniae and Pseudomonas aeruginosa in a Chinese teaching hospital. Sci. Rep. 6:33419. doi: 10.1038/srep33419
Xiang, D. R., Li, J. J., Sheng, Z. K., Yu, H. Y., Deng, M., Bi, S., et al. (2015). Complete sequence of a novel IncR-F33:A-:B- plasmid pKP1034 harboring fosA3, blaKPC-2, blaCTX-M-65, blaSHV-12, and rmtB from an epidemic Klebsiella pneumoniae sequence Type 11 strain in China. Antimicrob. Agents Chemother. 60, 1343–1348. doi: 10.1128/AAC.01488-15
Yeo, C. C., Tham, J. M., Kwong, S. M., Yiin, S., and Poh, C. L. (1998). Tn5563, a transposon encoding putative mercuric ion transport proteins located on plasmid pRA2 of Pseudomonas alcaligenes. FEMS Microbiol. Lett. 165, 253–260. doi: 10.1111/j.1574-6968.1998.tb13154.x
Keywords: IncHI5 plasmids, IMP, VIM, mobile elements, multidrug resistance
Citation: Liang Q, Jiang X, Hu L, Yin Z, Gao B, Zhao Y, Yang W, Yang H, Tong Y, Li W, Jiang L and Zhou D (2019) Sequencing and Genomic Diversity Analysis of IncHI5 Plasmids. Front. Microbiol. 9:3318. doi: 10.3389/fmicb.2018.03318
Received: 20 August 2018; Accepted: 20 December 2018;
Published: 14 January 2019.
Edited by:
Katy Jeannot, UMR 6249 Chrono Environnement, FranceReviewed by:
Antonio Juárez, University of Barcelona, SpainChristopher Morton Thomas, University of Birmingham, United Kingdom
Copyright © 2019 Liang, Jiang, Hu, Yin, Gao, Zhao, Yang, Yang, Tong, Li, Jiang and Zhou. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lingxiao Jiang, amlhbmctbGluZ3hpYW9AMTYzLmNvbQ== Dongsheng Zhou, ZG9uZ3NoZW5nemhvdTE5NzdAZ21haWwuY29t
†These authors have contributed equally to this work
 Quanhui Liang
Quanhui Liang Xiaoyuan Jiang3†
Xiaoyuan Jiang3† Zhe Yin
Zhe Yin Wenhui Yang
Wenhui Yang Yigang Tong
Yigang Tong Dongsheng Zhou
Dongsheng Zhou