Abstract
Hydrogen (H2) metabolism has attracted considerable interest because the activities of H2-producing and consuming microbes shape the global H2 cycle and may have vital relationships with the global cycling of other elements. There are many pathways of microbial H2 emission and consumption which may affect the structure and function of microbial communities. A wide range of microbial groups employ H2 as an electron donor to catalyze the reduction of pollutants such as organohalides, azo compounds, and trace metals. Syntrophy coupled mutualistic interaction between H2-producing and H2-consuming microorganisms can transfer H2 and be accompanied by the removal of toxic compounds. Moreover, hydrogenases have been gradually recognized to have a key role in the progress of pollutant degradation. This paper reviews recent advances in elucidating role of H2 metabolism involved in syntrophy and hydrogenases in environmental bioremediation. Further investigations should focus on the application of bioenergy in bioremediation to make microbiological H2 metabolism a promising remediation strategy.
Introduction
It is well established that the main sources of molecular hydrogen (H2) are geochemical and anthropogenic activities and the main sink is the biological consumption of H2 in soil ecosystems. The H2 cycle can influence air quality and climate indirectly via effects on the oxidative capacity of the atmosphere (Ehhalt and Rohrer, 2009). In addition, the H2 cycle plays an important role in microbial metabolism due to numerous microbial processes that depend on H2 production and consumption (Vignais and Billoud, 2007; Greening et al., 2015b). For example, most of the tropospheric H2 is consumed by soils due to the capacity of the majority of H2-oxidizing bacteria displaying high affinity for H2 in soils to recycle it (Constant et al., 2010). H2 is also a key metabolic compound in many anoxic ecosystems and its oxidation may support deep subsurface lithoautotrophic microbial ecosystems (Chivian et al., 2008; Nyyssonen et al., 2014; Wu et al., 2015; Bagnoud et al., 2016). The activities of H2-producing and consuming microbes therefore shape the global H2 cycle and may have vital relationships with the global cycling of other elements including carbon, sulfur, and nitrogen.
The first H2-oxidizing microorganisms were discovered in the 1900s (Kaserer, 1905; Stephenson and Stickland, 1931). The physical properties of H2 (e.g., its diffusion coefficient, 4 × 10-9 m2 s-1, and redox potential, E0’ = -0.42 V, make it relatively active in biological processes (Greening et al., 2016). H2 has dual physiological functions in microorganisms. Firstly, microbial fermentation of H2 produced by facultative or obligate fermenters can disperse excess reductant from fermentative metabolism, for example in Escherichia coli and Clostridium spp. (Trchounian et al., 2012, 2017a,b). Secondly, prokaryotic microorganisms with different metabolic processes such as hydrogen-oxidizing bacteria, methanogens and anoxygenic phototrophic bacteria can exploit H2 as an energy source and reductant (Schwartz et al., 2013). There are also a wide range of microorganisms with the ability to metabolize H2 such as aerobes and anaerobes and lithotrophs and phototrophs (Vignais and Billoud, 2007; Schwartz et al., 2013; Peters et al., 2015; Greening et al., 2016). Furthermore, recent studies show that some aerobic soil acidobacteria and actinobacteria can exploit low levels of H2 for survival in addition to growth, which challenges the traditional belief that H2 metabolism is restricted to high-H2 and low-O2 environments (Constant et al., 2010; Osborne et al., 2010; Greening et al., 2014, 2015a,b; Liot and Constant, 2016).
Hydrogenases catalyze microbial H2 production and consumption and are reversible enzymes responsible for reversible or partial catalytic reactions as follows (Equation 1).
On the basis of the metal cofactors of their H2-binding sites, these hydrogenases can be divided into three categories, namely the [NiFe]-, [FeFe]-, and [Fe]-hydrogenases (Vignais and Billoud, 2007; Schilter et al., 2016). [NiFe]-hydrogenases are closely related to both H2 production and consumption, while [FeFe]-hydrogenases are responsible mainly for the production of H2 owing to their higher turnover rate and activity compared with [NiFe]-hydrogenases (Marshall et al., 2012). However, [Fe]-hydrogenases have so far only been found in methanogenic archaea without cytochromes (Thauer et al., 2010). A recent study shows that the three different types of hydrogenase contain many subgroups based on the properties of metalloenzymes (such as metal-binding motifs, amino acid sequence phylogeny, reported biochemical characteristics and predicted genetic organization), and hydrogenase-encoding genes have also been identified in many microorganisms indicating a broad ecological distribution (Trchounian et al., 2011; Greening et al., 2016). Although the contribution of H2 metabolism to the entire ecosystem function is recognized in several environments such as hydrothermal vents, anoxic sediments and animal guts (Vignais and Billoud, 2007; Schwartz et al., 2013; Greening et al., 2016), the functions of hydrogenases in ecosystems in general remain largely unknown.
H2 metabolism plays a vital role in stability and performance in many microbial biotopes at ecosystem level (Marshall et al., 2012). It has been gradually recognized that hydrogenases may be used in bioremediation (Vignais and Billoud, 2007; Jugder et al., 2013). Numerous studies have shown that H2 can be utilized as an electron donor for reductive dehalogenation by many microorganisms and the occurrence of hydrogenases involved has been reported in dehalogenated bacteria (Seshadri et al., 2005; Rahm et al., 2006; Vignais and Billoud, 2007). In addition, microbial hydrogenases have been used in the remediation of metal-containing industrial wastes for the reduction of potentially toxic metals (Li et al., 2018). Under the impact of hydrogenases, microbial metabolic activities can influence the cycling of belowground minerals and organic matter and play a positive role in the bioremediation of both organic and inorganic pollutants (Lovley, 1993, 2008; Lovley and Coates, 2000; Vignais and Billoud, 2007). Thus, the use of hydrogenases for the remediation of polluted soils might be a promising strategy. In this review we attempt to integrate our understanding of the role of H2 metabolism in environment and environmental bioremediation processes and summarize the knowledge of H2 metabolism and hydrogenases involved in bioremediation.
Microbial H2-Producing Processes and Their Impact on the Environment
Fermentative Hydrogen Production From Organic Compounds
H2 is a key compound in the metabolism of many anaerobes, as well as a few aerobes, which owed the capacity to use this energy-rich molecule when it is available in the environment and derive electrons from its oxidation to drive energy generation. In the absence of external electron acceptors, many anaerobic bacteria can exploit carbohydrate rich substrates to produce H2 by reducing protons continuously. As described previously (Das and Veziroǧlu, 2001; Das and Veziroglu, 2008; Hallenbeck, 2009), the fermentative process generating H2 comprises two major pathways. In the first, butane 2,3 diol fermentation or mixed acid fermentation produces H2 via formate decomposition where glucose is transformed to pyruvate and then releases electrons to produce H2 under hydrogenase through a series of oxidation and reduction reactions (Figure 1A). The second is an NADH pathway in which H2 is produced by the re-oxidation of NADH (Figure 1B). In the various pyruvate metabolic pathways, H2 is usually produced by butyric acid fermentation, mixed acid fermentation, and bacterial ethanol fermentation (Ren et al., 2005). Fermentative microorganisms such as Clostridium spp. (e.g., C. butyricum and C. acetobutylicum) (Fang et al., 2006; Zhang et al., 2006), rumen flora (e.g., Butyrivibrio fibrisolvens, Eubacterium limosum, Megasphaera elsdenii, Ruminococcus flavefaciens, and Ruminococcus albus) (Miller and Wolin, 1973; Joyner et al., 1977; Miller and Wolin, 1979; Chaucheyras-Durand et al., 2010), Enterobacter spp. (e.g., E. cloacae and E. aerogenes) (Kumar and Das, 2000; Fabiano and Perego, 2002), Pyrococcus furiosus and Thermococcus litoralis (Malik et al., 1989; Rákhely et al., 1999; Schwartz et al., 2013) have been found to effectively produce H2 via different pyruvate metabolic pathways. Hydrogenase enzymes also play an important role in fermentative H2 production (Woodward et al., 2000; Trchounian et al., 2012). In general, H2 production could be catalyzed by a soluble [FeFe]-hydrogenase or a special class of membrane bound [NiFe]-hydrogenase (Ech). For example, Escherichia coli can transform intermediary fermentation products to the gaseous products H2 and CO2 by formate hydrogenlyase reaction (Figure 1A) (Sawers, 1994). Soboh et al. (2004) report that a ferredoxin-dependent [NiFe]-hydrogenase and a NADH-dependent [Fe]-hydrogenase may catalyze H2 evolution from NADH in Thermoanaerobacter tengcongensis. Production of H2 by fermentation in Thermotoga maritima is catalyzed by a heterotrimeric [FeFe]-hydrogenase and two cytoplasmic [NiFe]-hydrogenases have been identified in Thiocapsa roseopersicina (Figure 1B) (Jenney and Adams, 2008; Maróti et al., 2010).
FIGURE 1
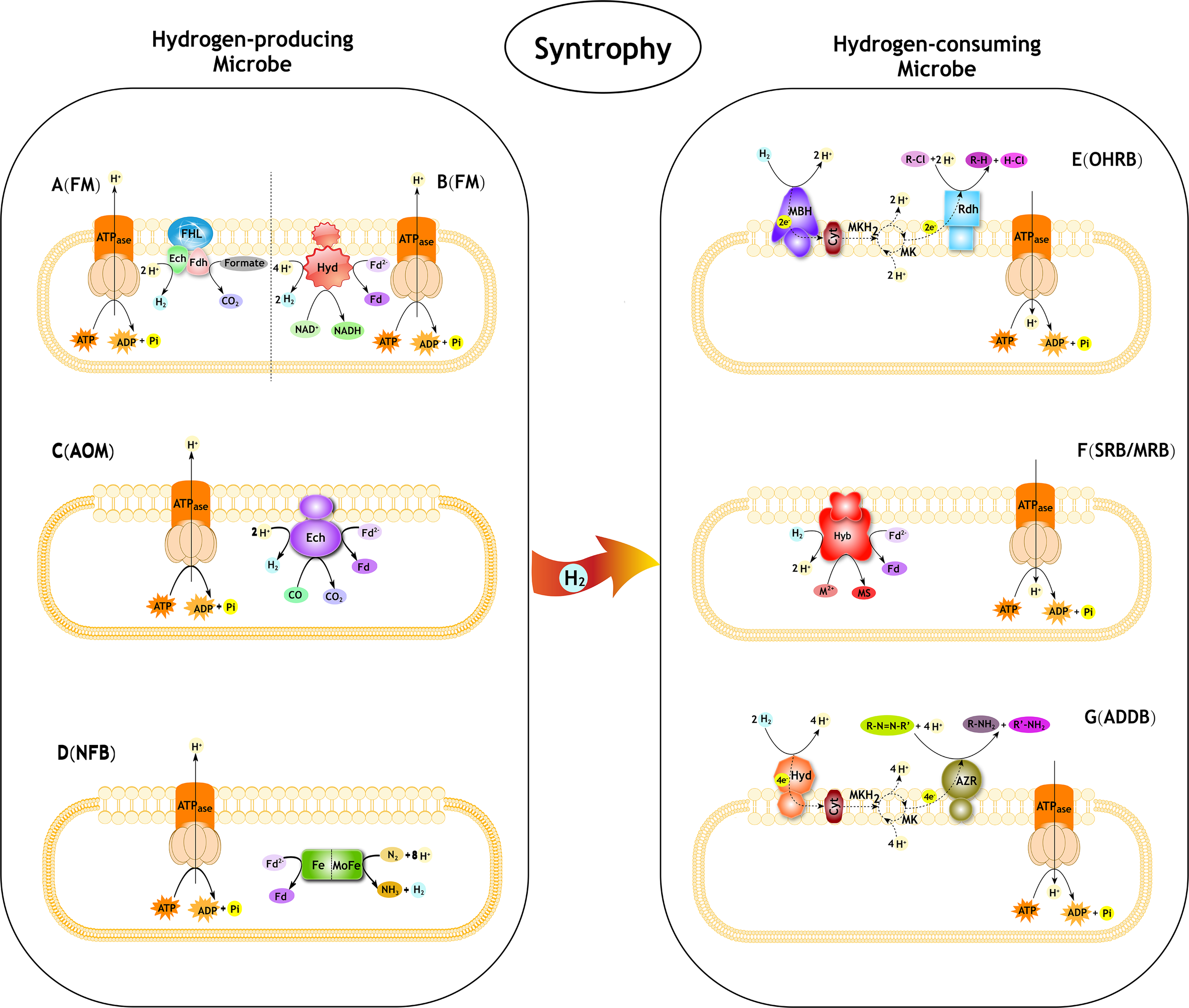
Syntrophic interactions between hydrogen-producing and hydrogen-consuming microbes in pollutant degradation. FM, Fermentative microbe; AOM, Anaerobic CO oxidation microbe; NFB, Nitrogen-fixing bacteria; OHRB, Organohalide-respiring bacteria; SRB, Sulfate-reducing bacteria; MRB, Metal-reducing bacteria; ADDB, Azo dyes decolorization bacteria. (A) The progress of formate oxidation coupled to H2 formation in E. coli (derived from Sawers, 1994; Hallenbeck, 2009; Trchounian et al., 2012). The FHL complex consist of a Ech type membrane-bound H2-evolving [NiFe] hydrogenase coupled to a formate dehydrogenase (Fdh) and membrane integral subunits. (B) The progress of re-oxidation of NADH coupled to H2 formation in T. roseopersicina (derived from Jenney and Adams, 2008; Maróti et al., 2010). The Hyd is a membrane-bound H2-evolving [FeFe] hydrogenase. Fd, ferredoxin. (C) The progress of anaerobic CO oxidation coupled to H2 formation in C. hydrogenoformans (derived from Svetlichny et al., 1991; Soboh et al., 2002). The Ech is a membrane-bound H2-evolving [NiFe] hydrogenase. Fd, ferredoxin. (D) The progress of producing H2 as a byproduct of N2 fixation. The nitrogenase complex consist of a Fe protein and MoFe protein. (E) The role of H2 in reductive dechlorination in Dehalococcoides spp. (derived from Jugder et al., 2016). MBH, membrane-bound uptake hydrogenase. Cyt, cytochrome. MK, menaquinone; MKH2, dihydromenaquinone. Rdh, reductive dehalogenase. R-Cl, organohalide. (F) The role of H2 in reductive PTEs in Desulfovibrio fructosovorans (derived from Chardin et al., 2003; Cao et al., 2014). Hyb is a membrane-bound H2-uptake [NiFe] hydrogenase. Fd, ferredoxin. M, PTEs. MS, metal sulfides. (G) The role of H2 in reductive azo compounds in Shewanella decolorationis (derived from Hong et al., 2007, 2008). Hya, membrane-bound uptake [NiFe]-hydrogenase. Cyt, cytochrome. MK, menaquinone; MKH2, dihydromenaquinone. AZR, azo reductase.R-N = N-R’, azo compounds.
Anaerobic Carbonic Monoxide (CO) Oxidation
There are several microbes owing different types of hydrogenogens that grow anaerobically in the dark and can unitize CO as the sole energy source to produce H2 (Figure 1C). Uffen (1976) and Fox et al. (1996a,b) showed that Rhodospirillum rubrum can produce H2 by oxidation of CO with the reduction of protons under the catalysis of a complex enzyme consisting of a CO-insensitive [NiFe]-hydrogenase and carbon monoxide dehydrogenase. A typical example of this group is the strictly anaerobic Carboxydothermus hydrogenoformans which contains a multienzyme membrane-bound [NiFe]-hydrogenase (Ech) complex (Figure 1C) (Svetlichny et al., 1991; Soboh et al., 2002). These enzymes together can oxidize CO and subsequently reduce the protons derived from H2O to form molecular H2. Carboxydocella thermautotrophica (Sokolova et al., 2002), Thermosinus carboxydivorans (Sokolova et al., 2004), Thermincola carboxydiphila (Sokolova et al., 2005), and Thermolithobacter carboxydivorans (Sokolova et al., 2007) are also thermophilic hydrogenogens.
Production of H2 as a Byproduct of N2 Fixation
Nitrogen fixation is one of the main processes of biogenic H2 production and is catalyzed by nitrogenase (Figure 1D). Approximately 30–50% of the entire reduction power consumed by nitrogenase is laterally tracked to H2 evolution (Brewin, 1984; Evans et al., 1987). However, H2 is not both a competitive inhibitor of N2 fixation and also represents a net loss of energy unless the H2 can be reprocessed by means of the uptake hydrogenase (Kosourov et al., 2014). Many H2-utilizing microorganisms such as the aerobic H2-oxidizing bacteria in soils reduce the loss of energy (Stein et al., 2005; Maimaiti et al., 2007; Constant et al., 2008; Osborne et al., 2010; Annan et al., 2012; Greening et al., 2015b). Many rhizobia can symbiotically fix dinitrogen in the root nodules of legumes and produce H2 concomitantly. The most-studied symbiotic nitrogen-fixing bacteria in legumes include Bradyrhizobium japonicum, Mesorhizobium mediterraneum, Sinorhizobium meliloti, and Rhizobium leguminosarum (Nour et al., 1995; Spaink, 2000; Laranjo et al., 2014). In addition, strains of Azotobacter (Cocking, 2003) and various cyanobacteria (e.g., Anabaena cylindrica, Nostoc muscorum, and Westiellopsis prolica) (Bulen et al., 1965; Fay, 1992; Nandi and Sengupta, 1998; Das and Veziroǧlu, 2001; Das and Veziroglu, 2008) can produce hydrogen through the nitrogen fixation process. Gest and Kamen (1949) report that Rhodospirillum rubrum can evolve significant amounts of H2 in the light and this is termed the photoproduction of H2 caused by nitrogenase-catalyzed reduction of protons (Bulen et al., 1965). Photoproduction of H2 was subsequently discovered in other phototrophic bacteria such as Rhodobater capsulatus, Rhodobater sphaeroides, Rhodobater palustris, Thiocapsa roseopersicina, and Halobacterium halobium (Vincenzini et al., 1982; Gogotov et al., 1991; Khan and Bhatt, 1991; Krahn et al., 1996; Fascetti et al., 1998).
Effects of Microbial Hydrogen Production on Environment
Atmospheric H2 is derived mainly from anthropogenic activities and oxidation of atmospheric methane (CH4) and non-methane hydrocarbons. An H2 mixing ratio of 0.53 ppmv is typically found in the global atmosphere (Novelli et al., 1999) and participates in atmospheric chemical cycles of H2O and greenhouse gasses as well as various pollutants (Schlegel et al., 1976; Crutzen and Fishman, 1977; Salvi and Subramanian, 2015; Talibi et al., 2017). In addition, H2 is a potential future energy carrier that may significantly affect the atmospheric H2 budget when used on a large scale (Brenninkmeijer et al., 2003; Petersen et al., 2011). It has been estimated that the total amount of H2 emissions into the troposphere each year is approximately 107 Tg (Rhee et al., 2006). Tromp et al. (2003) reported that the concentrations of stratospheric H2O and ozone and stratospheric temperatures would be affected by these H2 emissions. Moreover, the potential impacts of an increase in anthropogenic H2 emissions on the concentration of other trace gasses such as CH4 and CO) are also proposed.
About 7–11% of the global H2 pool is contributed by all oceanic, lake, and soil organisms (Schwartz et al., 2013). ‘Hot spots’ can be found in hypersaline cyanobacterial mats, with the release of H2 concentrations between 16,000 and 90,000 ppmv (Nielsen et al., 2015), which might be the main source of H2 emission from lake surface waters to the atmosphere. Numerous studies show that both cell counts of cyanobacteria and their N2 fixation rates are correlated with the H2 concentration of lake water (Conrad et al., 1983; Schütz et al., 1988; Schmidt and Conrad, 1993). Furthermore, the production of fermentation H2 and organic acids is a key component in the biogeochemistry of microbial mats, which promotes close interactions between anoxygenic phototrophs, cyanobacteria and heterotrophic bacteria (Otaki et al., 2012; Lee et al., 2014; Nielsen et al., 2015). However, almost all of the H2 produced from hypoxic sediments is also consumed by the sediments (Schwartz et al., 2013). The effects of hydrogen consumption on microbial communities in sediments therefore deserve further study.
The contribution of soils to the atmospheric H2 reservoir is more complex because soils are the main sink of the global H2 cycle, accounting for about 75 to 80% of atmospheric absorption (Constant et al., 2009; Ehhalt and Rohrer, 2009). However, nitrogen-fixing bacteria that form symbioses with legumes or free-living N2 fixing bacteria can generate large amounts of H2 as a by-product during N2 fixation (Orr et al., 2011; Mus et al., 2016). It has been estimated that H2 concentrations inside N2-fixing legume nodules range from 9,000 to 27,000 ppmv (Hunt et al., 1988; Witty, 1991; Witty and Minchin, 1998), so that diffusion losses during the growing season might reach 240,000 L H2 (Dong et al., 2003). Thus, the intensity of these H2 emissions to soils is determined by the hydrogen-metabolic capabilities of rhizobacterial symbionts (Hup+ or Hup- genotypes) in nodules through an uptake [NiFe]-hydrogenase (Evans et al., 1988; Annan et al., 2012). In the Hup+ legume rhizosphere the energy of H2 can be recycled by the [NiFe]-hydrogenase, while H2 is released into the surrounding soil in the Hup- legume rhizosphere. There is thus increasing evidence that H2 released into surrounding soils plays a key role in increasing plant biomass via the enrichment of aerobic H2-oxidizing bacteria (HOB), or plant growth-promoting rhizobacteria (PGPR) in both legumes and non-legumes (Dong et al., 2003; Maimaiti et al., 2007). Different H2 mixing ratios found in natural ecosystems may indeed lead to changes in soil microbial community structure and coordinated feedback of community functions. Constant et al. (2008) found that soil actinomycetes (such as Streptomyces sp. PCB7) are the main users of trace level of H2 in soils and might be key contributors to the function of soils as a sink in the global H2 cycle. Subsequently, Khdhiri et al. (2017) validated their own hypothesis by showing that the taxonomic response of the soil microbial community composition to H2 exposure is inconsistent across land use types. Piché-Choquette et al. (2018) revealed that H2 supports metabolic and energetic flexibility in microorganisms supplying a variety of ecosystem services via dose-response relationships between environmentally relevant H2 concentrations and the biological sinks of H2, CH4, and CO in soils.
Role of H2 in Environmental Bioremediation
The H2 produced both biogenically and abiogenically can be released and provided to support for the growth and metabolism of hydrogenotrophic prokaryotes (Karyakin et al., 2007). H2 metabolism fulfills a critical role in the ecosystems of many microbial biotopes (Vignais and Billoud, 2007; Schwartz et al., 2013; Greening et al., 2016). It is currently considered that a wide range of microbial groups employ H2 as an electron donor to catalyze the reduction of pollutants such as organohalides, azo compounds and potentially toxic elements.
Organohalides
Organohalides are recalcitrant, toxic, highly persistent, globally prevalent, and carcinogenic environmental contaminants. Organohalide-respiring bacteria (OHRB) have been isolated from polluted soils, sludges, sediments, aquifers, freshwaters, and marine habitats, and they are of considerable importance in bioremediation processes and natural halogen cycles (Zanaroli et al., 2015). Most OHRB discovered to date belong to Desulfomonile, Dehalococcoides, Dehalobacter, Desulfitobacterium, Desulfuromonas, and Sulfurospirillum (formerly Dehalospirillum) as reviewed by Jugder et al. (2015). Reductive dechlorination is an anaerobic respiration process that utilizes H2 as electron donor to dehalorespire these halogenated organics (Figure 1E) (Zanaroli et al., 2015; Agarwal et al., 2017). The process of electron via electron transport phosphorylation from the oxidation of the H2 to reductive dechlorination of organohalides involving membrane associated oxidoreductases (Figure 1E) (Jugder et al., 2016). Membrane-bound hydrogenases (MBH) are the initial oxidizers to take up the electrons released from molecular H2, which play a vital role in organohalide respiration (Jugder et al., 2013, 2015, 2016). The reductive dehalogenation of organohalides is typically catalyzed by dehalogenating enzyme systems coupled to ATP synthesis, reductive dehalogenases (Rdases) replace the halogen substituent with a hydrogen atom, reducing the toxicity and recalcitrance to biodegradation (Figure 1E) (Adrian and Loeffler, 2016; Gevorgyan et al., 2018). Sequencing data of genomes reveal that OHRB possess as many as 36 putative Rdases. After transformation to lower halogenated organics under anaerobic conditions, these toxic compounds can subsequently be mineralized by aerobic bacteria (Jugder et al., 2015).Desulfomonile tiedjei strain DCB-1 is one of the best-described dechlorinating anaerobes. The strain, first discovered by Suflita et al. (1982), reductively dechlorinates 3-chlorobenzoate while replacing the chlorine atom with hydrogen from H2, whereby providing energy for bacterial growth (Shelton and Tiedje, 1984; Dolfing and Tiedje, 1986; Dolfing and Tiedje, 1987). The strain was then noted to consume H2 with 3-chloro-, 3-bromo-, 3-iodobenzoate, tetrachloroethene (PCE), trichloroethene (TCE) (Cole et al., 1995), and chlorophenols (Mohn and Kennedy, 1992) as electron acceptors (DeWeerd et al., 1991). During the dehalogenation of 3-chlorobenzoate, formate was the most effective electron donor, followed by H2, pyruvate, and acetate.
Dehalococcoides strains are also some of the best known species capable of reductively dechlorinating a wide range of haloorganics including chlorinated benzenes, biphenyls, dioxins, ethenes, naphthalenes, and brominated diphenyl ethers. For example, tetrachloroethene is a commonly used solvent that possesses high toxicity and is a suspect carcinogen. The complete reductive dechlorination of tetrachloroethylene (PCE) and trichloroethylene (TCE) to non-toxic ethylene was first observed under methanogenic conditions by mixed cultures (Freedman and Gossett, 1989). Although H2 also served as the electron donor, methanol was more effective in sustaining the reductive dechlorination process. Holliger et al. (1993) isolated an anaerobic bacterial culture, previously named as PER-K23, from an anaerobic packed-bed column. By using H2 and formate as the only electron donors, PCE or TCE was reductively transformed to ethane via cis-1,2-dichloroethene (cis-1,2-DCE), chloroethene, and ethene, which was coupled to bacterial growth. The key role of hydrogenases in metabolizing these pollutants is underscored by the fact that both uptake (Hup type) and energy-conserving hydrogenases (Hyc or Ech type) were found in the genome of Dehalobacter restrictus PER-K23 (Rupakula et al., 2013). Maymó-Gatell et al. (1997) then isolated a dehalogenator, strain 195, and characterized it as Dehalococcoides ethenogenes. To date, Dehalococcoides species are the only bacteria known to be capable of completely dechlorinating chloroethylene. Genomic analysis of Dehalococcoides ethenogenes 195 showed that several hydrogenase genes including the membrane-bound periplasmic Hup, cytoplasmic Vhu, and membrane-bound Ech and Hyc [NiFe]-hydrogenases (Groups 1, 3, 4, and 4, respectively), and a membrane-bound Hym [Fe]-hydrogenases has potential roles in electron transport, which are capable of completing anaerobic dechlorination of the solvents PCE and TCE to vinyl chloride (VC) and ethane (Vignais et al., 2001; Morris et al., 2006).
Unlike other halorespiring bacteria, Dehalococcoides spp. use only H2 as an obligate electron donor for the dechlorination reaction, and no other electron acceptors support growth. For example, D. ethenogenes strain 195 grew only on H2 as electron donor for both bacterial growth and PCE reduction rather than formate, lactate, methanol, ethanol, glucose, pyruvate, or yeast extract (Maymó-Gatell et al., 1997). In addition, Dehalococcoides sp. CBDB1 was the first purified isolate of a bacterium relying on the energy obtained from stoichiometrical dehalorespiration of chlorobenzenes (CB) such as 1,2,3-trichlorobenzene (TCB) and 1,2,3,4-tetrachlorobenzene (TeCB) (Adrian et al., 2000). Both Dehalococcoides sp. 195 and CBDB1 exhibit reductive dehalogenation of chlorophenols (Adrian et al., 2007). Kube et al. (2005) compared the genome sequence of Dehalococcoides sp. CBDB1 with Dehalococcoides ethenogenes strain 195 and revealed that the hydrogenases previously described for strain 195 are also present in strain CBDB1. Chloroform (CF, CHCl3) is a non-polar solvent that is ubiquitous and is toxic to humans. The biodegradation of CF involves two processes, (1) dehalorespiration in which CF is dechlorinated to dichloromethane (DCM, CH2Cl2) by employing H2 as electron donor under the action of uptake hydrogenase, and (2) a fermentative process in which DCM is transformed to H2, acetate and carbon dioxide. Lee et al. (2012) report the involvement of Dehalobacter in dehalorespiration of CF [Equation (2)].
Despite these findings in respiration of organohalides, there is no real consensus on the involvement of various membrane associated components.
Potentially Toxic Elements (PTEs)
Potentially toxic elements display environmental durability, biological accumulation, and potential biological toxicity. The remediation of PTEs can be achieved by sulfate-reducing bacteria (SRB) or metal-reducing bacteria that can utilize H2 or other organic compounds as terminal electron donors to reduce the PTEs. Tebo and Obraztsova (1998) isolated the first sulfate-reducing bacterium from PTE-polluted sediments named Desulfotomaculum reducens sp. nov. strain MI-1, which can utilize H2 as terminal electron donor and metals [such as Cr(VI), Mn(IV), Fe(III), and U(VI)] as electron acceptors accompanied by bacterial growth. Thus far, more than 40 SRB species have been identified, including Desulfobacter, Desulfovibrio, Desulfotomaculum and Desulfomicrobium, and others (Leloup et al., 2009; Mizuno et al., 2012; Hussain et al., 2016; Li et al., 2018). Subsequently, due to the advantages of SRB (no secondary pollution and strong adaptability), they have been used in the bioremediation of PTEs (Li et al., 2018). Generally speaking, there are two steps involved in the mechanism of SRB removal of PTEs from wastewaters: (i) SRB utilize sulfate as electron acceptor to oxidize simple organic compounds to generate bicarbonate ion and hydrogen sulfide under anaerobic conditions [Equation (3)], and (ii) the hydrogen sulfide generated reacts with dissolved PTE to form insoluble metal sulfide precipitates [Equation (4)] (Kieu et al., 2011; Singh et al., 2011; Li et al., 2017).
Where CH2O represents simple organic compounds (such as acetate and lactate), M represents PTEs, and MS represents metal sulfides. Because of their special characteristics with the corresponding metal sulfides readily forming precipitates, SRBs have been used to treat PTE-polluted wastewaters (e.g., uranium-containing, chromium-containing and antimony-containing wastewaters, organochlorines, and other pollutants) (Li et al., 2018). Lovley and Phillips (1994) showed that the bioremediation effect of Desulfovibrio vulgaris which utilizes H2 as the electron donor catalyzed by the c3 cytochrome functions as a Cr(VI) reductase in Cr(VI)-contaminated waters was superior to the previously described Cr(VI) reductive microorganisms. Kieu et al. (2011) reported that the PTE removal efficiencies of Cu2+, Ni2+, Zn2+, and Cr6+ in anaerobic semi-continuous stirred tank reactors containing a consortium of SRB reached 94–100% after 4 weeks under experimental conditions. In addition, several microbial genera reduced uranium to form easily precipitated reduced U(IV) species, and this has been used successfully in soil remediation (Phillips et al., 1995; Fredrickson et al., 2000; Valls and De Lorenzo, 2002).
Several uptake hydrogenases were considered to have potential application in the bioremediation of PTEs. The [NiFe] uptake hydrogenases in group 1 including membrane-bound respiratory uptake hydrogenases that couple H2 oxidation to catalyze metal reduction (Figure 1F). For example, [NiFe]-uptake hydrogenase from SRB can reduce toxic chromate VI to form a less toxic product (Chardin et al., 2003). In addition, technetium VII is reduced by Desulfovibrio fructosovorans through this mechanism (Tabak et al., 2005), and hydrogenases involving in metal reduction have also been observed in other metals including ferrum (Fe) (Coppi et al., 2004), platinum (Riddin et al., 2009), and lead (Deplanche et al., 2010). A comprehensive analysis of the genome sequence of the metal-reducing bacterium (Shewanella oneidensis) has been conducted, and has predicted that an [Fe]-hydrogenase and several cytochromes are involved in the electron transport and metal reduction processes (Heidelberg et al., 2002). However, the potential application of microbes with different subgroup hydrogenases for PTE respiration is not enough, requiring further study including the biochemical investigations of these different subgroup hydrogenases.
Other Pollutants
Azo compounds undergo dissimilatory azoreduction by Shewanella decolorationis S12 under anaerobic conditions. This strain utilized azo compounds as carbon source for growth by azo reductase which is sustained by the H2 supply. The strain also catalyzed H2-dependent reduction of Fe(III) and humic substances (Coppi et al., 2004; Hong et al., 2008). Brigé et al. (2008) show that Shewanella decolorationis MR-1 utilized azo dye amaranth as electron acceptor for microbial energy conservation. Mutambanengwe et al. (2007) show the decolorization of a wide range of azo dyes with sulfate-reducing microbes (SRM) and hydrogenases might be involved in the degradation process. A multicomponent electron transfer chain has been proposed to be involved in the extracellular reduction of azo compounds. The electron transfer components consist of the cytoplasm/outer membrane, periplasm, c-type cytochromes, and menaquinone (Hong et al., 2007; Brigé et al., 2008). Hya type [NiFe]-hydrogenase or Hyd type [Fe]- hydrogenase act as a critical hub mediating the oxidization of H2 to provide electrons for azoreduction metabolism (Figure 1G) (Hong et al., 2008).
H2-dependent reduction has been reported in nitroaromatic compounds (Watrous et al., 2003). In a strict anaerobe, Clostridium acetobotulinicum, an [Fe]-hydrogenase is responsible for the reduction of nitro substituents of 2,4,6-trinitrotoluene (TNT) to the corresponding hydroxylamine in an acidogenic environment.
Factors Affecting the Utilization of Hydrogen by Degrading Bacteria in the Environment
There are many factors affecting the utilization of H2 by degrading bacteria in the environment such as H2 source, H2 transfer process and other environmental factors (including trophic hierarchies, external pH, osmotic coditions, concentration of carbon sources and their mixtures and microbial community and other physicochemical factors).
Methanogens were found to affect the interspecies H2 transfer of dehalorespiring bacteria, which might promote or inhibit the dechlorination process (Smatlak et al., 1996; Fennell et al., 1997; Yang and McCarty, 1998). Johnson et al. (2008) demonstrated the dechlorination of stress-related net cell growth by Dehalococcoides ethenogenes strain 195 (DE195) which was isolated and then transited to a smooth phase. Although Methanobacterium congolense (MC) can compete with DE195 for hydrogen, adverse effects of the dechlorination rate were not observed (Men et al., 2012). This is mainly because the H2 threshold required for dechlorination is very low, so that even though methane production consumes a large amount of H2, it does not compete for dechlorination (Yang and McCarty, 1998; Men et al., 2012). In syntrophic communities, H2-producing bacteria and H2-consuming methanogens perceive the redox conditions and affect each other’s metabolism (Stams and Plugge, 2009). Several studies have shown that the reduction dechlorination can be promoted in some communities in the presence of methanogens (Vogel and McCARTY, 1985; Heimann et al., 2006; Kong et al., 2014). In addition, a recent study found that Methylobacter seemed to be tolerant to TCE and may play a vital role in TCE degradation (Kong et al., 2014). Although many studies have assessed the association between methanogens and dechlorination bacteria, the mechanism by which methanogens affect dechlorinating communities remains unclear.
The process of forming compact aggregates involves both physicochemical and biological interactions (Stams and Plugge, 2009). When the compact aggregates are formed in anaerobic bacteria and methanogenic archaea, the rate of H2 transfer between two species increases significantly (Lettinga et al., 1988; Stams and Plugge, 2009). Several studies have also shown that the inter-microbial distances affect both their specific growth rates and biodegradation rates (Ishii et al., 2005; Stams et al., 2006; Stams and Plugge, 2009). Thus, forming compact aggregates might be an important factor influencing the biodegradation rates of degrading bacteria.
It is well known that trophic hierarchies occur because different functional members of the community provide each other with a matrix and basic cofactors, and eliminate inhibitory metabolites (Schink, 1997; Rittmann and McCarty, 2012). DeWeerd et al. (1991) reported that acetylene, molybdate, selenate, and metronidazole can inhibit dehalogenation, sulfite reduction and H2 metabolism, indicating that the reduction of sulfite and dehalogenation may share part of the same electron transport chain. However, some environmental factors might accelerate the degradation of pollutants by promoting H2 utilization. For example, cobalamin has a positive effect on the dechlorination process as a co-factor of the reductive dehalogenases (Yan et al., 2012). Desulfovibrio vulgaris Hildenborough (DVH) possesses the full set of genes required for the biosynthesis of adenosylcobalamin, a derivative of vitamin B12 which might result in an increased concentration of the corrinoid co-factor (vitamin B12) in co-cultures, taken up and utilized immediately by Dehalococcoides species (Rodionov et al., 2004). In addition, the main factors influencing H2 utilization such as external pH, osmotic conditions, concentration of carbon sources and their mixtures, microbial community and other physicochemical factors mainly affected growth and the physiological activity including uptake hydrogenase and pollutant degrading enzymes of the degrading bacteria (Richter and Gescher, 2014; Trchounian and Trchounian, 2014, 2015; Trchounian et al., 2017a).
Interspecies Hydrogen Transfer During Syntrophic Growth
Syntrophy coupling mutualistic interactions between hydrogen-/formate-producing and hydrogen-/formate-consuming microorganisms is essential for biofuel production, pollutant degradation, and global carbon cycling (Kleinsteuber et al., 2012; Sieber et al., 2012; Morris et al., 2013). When sulfate is limited or unavailable, SRBs can also mediate the transfer of H2 between species, which provides the bacterial species with a very versatile metabolism adapted to complex ecological environments. Odom and Peck (1981) first documented the transfer of the redundant H2 evolved from substrate fermentation by SRBs to other H2 consuming bacteria. Using a defined two-member continuous culture, Drzyzga et al. (2001) demonstrated that the sulfate reducer Desulfovibrio sp. strain SULF1 can use the dehalorespiring Desulfitobacterium frappieri TCE1 as a ‘biological electron acceptor’ to sustain growth. They also noted that dehalogenation of tetrachloroethene (PCE) was inhibited at sulfate concentrations above 2.5 mM, while PCE was completely dehalogenated to cis-dichloroethene (cis-DCE) with 1 mM sulfate or without sulfate addition (Drzyzga and Gottschal, 2002). In this community, Desulfovibrio vulgaris Hildenborough (DVH) can grow syntrophically with Dehalococcoides ethenogenes strain 195 (DE195), thus enhancing the robustness of bacterial growth and the dechlorination activity of trichloroethene (Men et al., 2012). The syntrophical interaction with sulfate reducers has been shown to result in more effective transfer of H2, thereby facilitating faster dechlorination and more rubust growth of dehalogenating strains compared with gaseous H2 (Men et al., 2012). The syntrophic relationship between methanogens and archaea also involves interspecies H2 transfer in the process of converting long-chain fatty acids (Stams and Plugge, 2009). Subsequently, Ziels et al. (2017) found several formate hydrogenases and dehydrogenases in the enriched genome bins (GBs) of both their codigesters. In the process of CF dechlorination, interspecies H2 transfer was observed in the form of acetogenesis and methanogenesis by Lee et al. (2012), which required syntrophic partners to maintain low H2 partial pressures.
The possible processes of syntrophic interactions between H2-producing and H2-consuming microbes in pollutant degradation are shown in Figure 1. Previous studies have shown that H2-forming bacteria and H2-utilizing bacteria sense redox conditions, influencing each other’s metabolism in syntrophic communities (Stams and Plugge, 2009). Interspecies electron transfer mechanisms underlie thermodynamically favorable syntrophic processes (Gieg et al., 2014). In anoxic environments, butyrate oxidations involving energy-dependent reactions were possible to be applied in syntrophic degradation of organohalides. For example, the standard free reaction enthalpies (ΔGo′) of butyrate oxidations and organohalide degradations were as follows [Equation (5) Müller et al., 2010; Equation (6) Jugder et al., 2016]:
Based on energy balance toward H2 production and consumption analysis, we propose that the energy-transforming reactions between H2 production and organohalide degradations might be involved in syntrophic H2 production and consumption microorganisms. Dehalogenating microorganisms (such as Dehalococcoides sp. strain BAV1 and Dehalococcoides ethenogenes strain 195) can utilize acetate as carbon source and H2 as electron donor when grown in isolation, exhibiting limited dechlorination activity and low growth rates (He et al., 2003a,b). Thus, a promising method might be to develop improved bioremediation strategies by enhancing the strong growth and dechlorination activity of dehalogenating microorganisms (Men et al., 2012). However, many interspecies H2 transfer interactions are syntrophic, and thus only present in complex microbial communities but not in pure cultures. In complex microbial consortia, H2 indirectly mediates electron shuttle between electron donors and acceptors. Hydrogenotrophic bacteria can profit from the H2 produced from their syntrophic partners, thereby transforming pollutants. Thus, both H2-producing and H2-consuming microorganisms are essential for their own growth and might also promote the degradation of pollutants (Stams and Plugge, 2009).
Conclusion and Perspectives
Metabolism of H2 including H2 production and H2 consumption have been recognized as a potential driving force affecting the structure of microbial communities and may even change community functions. Although the contribution of H2 metabolism to entire ecosystem processes is recognized in hydrothermal vents, anoxic sediments and animal guts (Vignais and Billoud, 2007; Schwartz et al., 2013), the role of H2 metabolism and hydrogenases in ecosystems are not fully elucidated. Further advances in exploiting the function of biohydrogen metabolism and related microbial communities in environmental bioremediation are expected to result from (i) using metagenome sequencing, single-gene fluorescence in situ hybridization, the functional gene arrays (GeoChip) and in situ mass spectrometry to track the dynamics of pollutant-degrading bacteria involving in H2 metabolism and the interplay between pollutant-degrading bacteria and H2-metabolism bacteria in degradation process; (ii) effects of soil conditions on H2-consuming microorganisms degrading pollutants; (iii) structural studies of hydrogenases or the synergistic action of other enzymes (such as ATPase and Rdase) involving in the process of environmental bioremediation and enhancing these enzymes activity through protein engineering; (iv) integrative analyses of genomic, transcriptomic, and epigenomic data in these environmental bioremediation process.
To date, environmentally friendly management techniques named “3B” techniques (biological carbon sequestration, bioenergy, and bioremediation) have been proposed to further enhance biodiversity and mitigate environmental stressors (Teng et al., 2012). Environmental H2 is an energy source for aerobic H2 oxidizers, sulfate reducers, acetogens and methanogens and is also a source of reducing power for anaerobic bacteria and anoxygenic phototrophs (Schwartz et al., 2013). Syntrophy coupling mutualistic interactions between H2-producing and H2-consuming microorganisms is not restricted to the transfer of reducing agents such as H2 or formate, but can also involve the exchange of organic, sulfurous and nitrogenous compounds or the removal of toxic compounds. Nevertheless, there is still a considerable need for appropriate research initiatives to apply those microbial groups to the bioremediation of contaminated soils. However, soil is a complex and dynamic biological system. From the soil to the microorganism, bioavailability of pollutants involves a full process of adsorption and desorption, transport, and uptake by microorganisms which are also affected by the soil conditions such as soil organic matter, soil minerals, soil moisture, soil aggregates and so on (Ren et al., 2018; Teng and Chen, 2019).
Proton ATPase or other membrane bound secondary transporters affect hydrogenase activity and thus H2 metabolism (Trchounian et al., 2011; Gevorgyan et al., 2018). So, structural studies of hydrogenases or other synergistic enzymes (such as ATPase and Rdase) involving in the process of environmental bioremediation are vital important in directing protein engineering, for example, in rendering these enzymes activity to promote the degradation efficiency of pollutants via identification of factors linked to the protein environment of the active site. Studies of H2 metabolism and regulation will also be important in engineering microorganisms at the cellular level to maximize the degradation efficiency of pollutants. Since hydrogenases and other synergistic enzymes have been shown to play an important role in the degradation of pollutants, it is also tempting to consider that analysis of genomic, transcriptomic, and epigenomic data of these enzymes in environmental bioremediation process will likely provide vital insights into the hydrogenase participates in degradation mechanism of pollutants.
In conclusion, this review provides a comprehensive framework for H2 production and H2 consumption in environmental bioremediation processes. The syntrophy coupling mutualistic interaction between H2-producing and H2-consuming microorganisms could be applied to the removal of toxic compounds. In addition, several uptake hydrogenases are also considered to have potential application in the bioremediation of those toxic compounds. The use of this bioenergy may provide a low-input and ecologically friendly bioremediation strategy for the future.
Statements
Author contributions
YT, YX, and XW collected the data. YT and YX drafted the article. YT, XW, and PC critically revised the article.
Funding
This study was funded by the Outstanding Youth Fund of Jiangsu Province (No. BK20150049), the National Natural Science Foundation of China (Grant Nos. 41671327 and 41571308), and the Special Project on the Basis of the National Science and Technology of China (2015FY110700).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
1
AdrianL.HansenS. K.FungJ. M.GörischH.ZinderS. H. (2007). Growth of Dehalococcoides strains with chlorophenols as electron acceptors.Environ. Sci. Technol.412318–2323. 10.1021/es062076m
2
AdrianL.LoefflerF. E. (2016). Organohalide-Respiring Bacteria.Berlin: Springer. 10.1007/978-3-662-49875-0
3
AdrianL.SzewzykU.WeckeJ.GörischH. (2000). Bacterial dehalorespiration with chlorinated benzenes.Nature408580–583. 10.1038/35046063
4
AgarwalV.MilesZ. D.WinterJ. M.EustáquioA. S.El GamalA. A.MooreB. S. (2017). Enzymatic halogenation and dehalogenation reactions: pervasive and mechanistically diverse.Chem. Rev.1175619–5674. 10.1021/acs.chemrev.6b00571
5
AnnanH.GoldingA. L.ZhaoY.DongZ. (2012). Choice of hydrogen uptake (Hup) status in legume-rhizobia symbioses.Ecol. Evol.22285–2290. 10.1002/ece3.325
6
BagnoudA.ChoureyK.HettichR. L.de BruijnI.AnderssonA. F.LeupinO. X.et al (2016). Reconstructing a hydrogen-driven microbial metabolic network in Opalinus Clay rock.Nat. Commun.7:12770. 10.1038/ncomms12770
7
BrenninkmeijerC. A.JanssenC.KaiserJ.RöckmannT.RheeT.AssonovS. (2003). Isotope effects in the chemistry of atmospheric trace compounds.Chem. Rev.1035125–5162. 10.1021/cr020644k
8
BrewinN. (1984). “Hydrogenase and energy efficiency in nitrogen fixing symbionts,” in Genes Involved in Microbe-Plant Interactions, edsVermaD. P. S.HohnT. (Vienna: Springer), 179–203.
9
BrigéA.MotteB.BorlooJ.BuysschaertG.DevreeseB.Van BeeumenJ. J. (2008). Bacterial decolorization of textile dyes is an extracellular process requiring a multicomponent electron transfer pathway.Microb. Biotechnol.140–52. 10.1111/j.1751-7915.2007.00005.x
10
BulenW.BurnsR.LeComteJ. (1965). Nitrogen fixation: hydrosulfite as electron donor with cell-free preparations of Azotobacter vinelandii and Rhodospirillum rubrum.Proc. Natl. Acad. Sci. U.S.A.53532–593. 10.1073/pnas.53.3.532
11
CaoH.WangY.LeeO. O.ZengX.ShaoZ.QianP. Y. (2014). Microbial sulfur cycle in two hydrothermal chimneys on the Southwest Indian Ridge.mBio5:e00980-13. 10.1128/mBio.00980-13
12
ChardinB.Giudici-OrticoniM. T.De LucaG.GuigliarelliB.BruschiM. (2003). Hydrogenases in sulfate-reducing bacteria function as chromium reductase.Appl. Microbiol. Biotechnol.63315–321. 10.1007/s00253-003-1390-8
13
Chaucheyras-DurandF.MassegliaS.FontyG.ForanoE. (2010). Influence of the composition of the cellulolytic flora on the development of hydrogenotrophic microorganisms, hydrogen utilization, and methane production in the rumens of gnotobiotically reared lambs.Appl. Environ. Microbiol.767931–7937.10.1128/aem.01784-10
14
ChivianD.BrodieE. L.AlmE. J.CulleyD. E.DehalP. S.DeSantisT. Z.et al (2008). Environmental genomics reveals a single-species ecosystem deep within earth.Science322275–278. 10.1126/science.1155495
15
CockingE. C. (2003). Endophytic colonization of plant roots by nitrogen-fixing bacteria.Plant Soil252169–175. 10.1023/A:1024106605806
16
ColeJ. R.FathepureB. Z.TiedjeJ. M. (1995). Tetrachloroethene and 3-chlorobenzoate dechlorination activities are co-induced in Desulfomonile tiedjei DCB-1.Biodegradation6167–172. 10.1007/BF00695347
17
ConradR.AragnoM.SeilerW. (1983). Production and consumption of hydrogen in a eutrophic lake.Appl. Environ. Microbiol.45502–510.
18
ConstantP.ChowdhuryS. P.PratscherJ.ConradR. (2010). Streptomycetes contributing to atmospheric molecular hydrogen soil uptake are widespread and encode a putative high-affinity [NiFe]-hydrogenase.Environ. Microbiol.12821–829. 10.1111/j.1462-2920.2009.02130.x
19
ConstantP.PoissantL.VillemurR. (2008). Isolation of Streptomyces sp. PCB7, the first microorganism demonstrating high-affinity uptake of tropospheric H2.ISME J.21066–1076. 10.1038/ismej.2008.59
20
ConstantP.PoissantL.VillemurR. (2009). Tropospheric H2 budget and the response of its soil uptake under the changing environment.Sci. Total Environ.4071809–1823. 10.1016/j.scitotenv.2008.10.064
21
CoppiM. V.O’NeilR. A.LovleyD. R. (2004). Identification of an uptake hydrogenase required for hydrogen-dependent reduction of Fe (III) and other electron acceptors by Geobacter sulfurreducens.J. Bacteriol.1863022–3028. 10.1128/JB.186.10.3022-3028.2004
22
CrutzenP. J.FishmanJ. (1977). Average concentrations of OH in the troposphere, and the budgets of CH4, CO, H2 and CH3CCl3.Geophys. Res. Lett.4321–324. 10.1029/GL004i008p00321
23
DasD.VezirogluT. N. (2008). Advances in biological hydrogen production processes.Int. J. Hydrogen Energ.336046–6057. 10.1016/j.ijhydene.2008.07.098
24
DasD.VeziroǧluT. N. (2001). Hydrogen production by biological processes: a survey of literature.Int. J. Hydrogen Energ.2613–28. 10.1016/S0360-3199(00)00058-6
25
DeplancheK.CaldelariI.MikheenkoI. P.SargentF.MacaskieL. E. (2010). Involvement of hydrogenases in the formation of highly catalytic Pd (0) nanoparticles by bioreduction of Pd (II) using Escherichia coli mutant strains.Microbiology1562630–2640. 10.1099/mic.0.036681-0
26
DeWeerdK.ConcannonF.SuflitaJ. M. (1991). Relationship between hydrogen consumption, dehalogenation, and the reduction of sulfur oxyanions by Desulfomonile tiedjei.Appl. Environl. Microbiol.571929–1934.
27
DolfingJ.TiedjeJ. M. (1986). Hydrogen cycling in a three-tiered food web growing on the methanogenic conversion of 3-chlorobenzoate.FEMS Microbiol. Ecol.2293–298. 10.1111/j.1574-6968.1986.tb01740.x
28
DolfingJ.TiedjeJ. M. (1987). Growth yield increase linked to reductive dechlorination in a defined 3-chlorobenzoate degrading methanogenic coculture.Arch. Microbiol.149102–105. 10.1007/BF00425073
29
DongZ.WuL.KettlewellB.CaldwellC.LayzellD. (2003). Hydrogen fertilization of soils–is this a benefit of legumes in rotation?Plant Cell Environ.261875–1879. 10.1046/j.1365-3040.2003.01103.x
30
DrzyzgaO.GerritseJ.DijkJ. A.ElissenH.GottschalJ. C. (2001). Coexistence of a sulphate-reducing Desulfovibrio species and the dehalorespiring Desulfitobacterium frappieri TCE1 in defined chemostat cultures grown with various combinations of sulphate and tetrachloroethene.Environ. Microbiol.392–99. 10.1046/j.1462-2920.2001.00157.x
31
DrzyzgaO.GottschalJ. C. (2002). Tetrachloroethene dehalorespiration and growth of Desulfitobacterium frappieri TCE1 in strict dependence on the activity of Desulfovibrio fructosivorans.Appl. Environ. Microbiol.68642–649. 10.1128/AEM.68.2.642-649.2002
32
EhhaltD. H.RohrerF. (2009). The tropospheric cycle of H2: a critical review.Tellus B61500–535. 10.1111/j.1600-0889.2009.00416.x
33
EvansH.RussellS.HanusF.Ruiz-ArguesoT. (1988). “The importance of hydrogen recycling in nitrogen fixation by legumes,” in World Crops: Cool Season Food Legumes, ed.SummerfieldR. J. (Dordrecht: Springer), 777–791. 10.1007/978-94-009-2764-3_62
34
EvansH. J.HarkerA.PapenH.RussellS. A.HanusF.ZuberM. (1987). Physiology, biochemistry, and genetics of the uptake hydrogenase in rhizobia.Annu. Rev. Microbiol.41335–361. 10.1146/annurev.mi.41.100187.002003
35
FabianoB.PeregoP. (2002). Thermodynamic study and optimization of hydrogen production by Enterobacter aerogenes.Int. J. Hydrogen Energ.27149–156. 10.1016/S0360-3199(01)00102-1
36
FangH. H.ZhuH.ZhangT. (2006). Phototrophic hydrogen production from glucose by pure and co-cultures of Clostridium butyricum and Rhodobacter sphaeroides.Int. J. Hydrogen Energ.312223–2230. 10.1016/j.ijhydene.2006.03.005
37
FascettiE.D’addarioE.TodiniO.RobertielloA. (1998). Photosynthetic hydrogen evolution with volatile organic acids derived from the fermentation of source selected municipal solid wastes.Int. J. Hydrogen Energ.23753–760. 10.1016/S0360-3199(97)00123-7
38
FayP. (1992). Oxygen relations of nitrogen fixation in cyanobacteria.Microbiol. Rev.56340–373.
39
FennellD. E.GossettJ. M.ZinderS. H. (1997). Comparison of butyric acid, ethanol, lactic acid, and propionic acid as hydrogen donors for the reductive dechlorination of tetrachloroethene.Environ. Sci. Technol.31918–926. 10.1021/es960756r
40
FoxJ. D.HeY.ShelverD.RobertsG. P.LuddenP. W. (1996a). Characterization of the region encoding the CO-induced hydrogenase of Rhodospirillum rubrum.J. Bacteriol.1786200–6208.
41
FoxJ. D.KerbyR. L.RobertsG. P.LuddenP. W. (1996b). Characterization of the CO-induced, CO-tolerant hydrogenase from Rhodospirillum rubrum and the gene encoding the large subunit of the enzyme.J. Bacteriol.1781515–1524.
42
FredricksonJ. K.ZacharaJ. M.KennedyD. W.DuffM. C.GorbyY. A.Shu-meiW. L.et al (2000). Reduction of U (VI) in goethite (α-FeOOH) suspensions by a dissimilatory metal-reducing bacterium.Geochim. Cosmochim. Acta643085–3098. 10.1016/S0016-7037(00)00397-5
43
FreedmanD. L.GossettJ. M. (1989). Biological reductive dechlorination of tetrachloroethylene and trichloroethylene to ethylene under methanogenic conditions.Appl. Environ. Microbiol.552144–2151.
44
GestH.KamenM. D. (1949). Photoproduction of molecular hydrogen by Rhodospirillum rubrum.Science109558–559. 10.1126/science.109.2840.558
45
GevorgyanH.TrchounianA.TrchounianK. (2018). Understanding the role of Escherichia coli hydrogenases and formate dehydrogenases in the F0F1-ATPase activity during the mixed acid fermentation of mixture of carbon sources.IUBMB Life701040–1047. 10.1002/iub.1915
46
GiegL. M.FowlerS. J.Berdugo-ClavijoC. (2014). Syntrophic biodegradation of hydrocarbon contaminants.Curr. Opin. Biotechnol.2721–29. 10.1016/j.copbio.2013.09.002
47
GogotovI.ZorinN.SerebriakovaL. (1991). Hydrogen production by model systems including hydrogenases from phototrophic bacteria.Int. J. Hydrogen Energ.16393–396. 10.1016/0360-3199(91)90137-8
48
GreeningC.BerneyM.HardsK.CookG. M.ConradR. (2014). A soil actinobacterium scavenges atmospheric H2 using two membrane-associated, oxygen-dependent [NiFe] hydrogenases.Proc. Natl. Acad. Sci. U.S.A.1114257–4261. 10.1073/pnas.1320586111
49
GreeningC.BiswasA.CarereC. R.JacksonC. J.TaylorM. C.StottM. B.et al (2016). Genomic and metagenomic surveys of hydrogenase distribution indicate H2 is a widely utilised energy source for microbial growth and survival.ISME J.10761–777. 10.1038/ismej.2015.153
50
GreeningC.CarereC. R.Rushton-GreenR.HaroldL. K.HardsK.TaylorM. C.et al (2015a). Persistence of the dominant soil phylum Acidobacteria by trace gas scavenging.Proc. Natl. Acad. Sci. U.S.A.11210497–10502. 10.1073/pnas.1508385112
51
GreeningC.ConstantP.HardsK.MoralesS. E.OakeshottJ. G.RussellR. J.et al (2015b). Atmospheric hydrogen scavenging: from enzymes to ecosystems.Appl. Environ. Microbiol.811190–1199. 10.1128/aem.03364-14
52
HallenbeckP. C. (2009). Fermentative hydrogen production: principles, progress, and prognosis.Int. J. Hydrogen Energ.347379–7389. 10.1016/j.ijhydene.2008.12.080
53
HeJ.RitalahtiK. M.AielloM. R.LöfflerF. E. (2003a). Complete detoxification of vinyl chloride by an anaerobic enrichment culture and identification of the reductively dechlorinating population as a Dehalococcoides species.Appl. Environ. Microbiol.69996–1003.
54
HeJ.RitalahtiK. M.YangK.-L.KoenigsbergS. S.LöfflerF. E. (2003b). Detoxification of vinyl chloride to ethene coupled to growth of an anaerobic bacterium.Nature42462–65.
55
HeidelbergJ. F.PaulsenI. T.NelsonK. E.GaidosE. J.NelsonW. C.ReadT. D.et al (2002). Genome sequence of the dissimilatory metal ion-reducing bacterium Shewanella oneidensis.Nat. Biotechnol.201118–1123. 10.1038/nbt749
56
HeimannA. C.BatstoneD. J.JakobsenR. (2006). Methanosarcina spp. Drive vinyl chloride dechlorination via interspecies hydrogen transfer.Appl. Environ. Microbiol.722942–2949. 10.1128/AEM.72.4.2942-2949.2006
57
HolligerC.SchraaG.StamsA.ZehnderA. (1993). A highly purified enrichment culture couples the reductive dechlorination of tetrachloroethene to growth.Appl. Environ. Microbiol.592991–2997.
58
HongY.GuoJ.SunG. (2008). Identification of an uptake hydrogenase for hydrogen-dependent dissimilatory azoreduction by Shewanella decolorationis S12.Appl. Microbiol. Biotechnol.80517–524. 10.1007/s00253-008-1597-9
59
HongY. G.XuM. Y.GuoJ.XuZ. C.ChenX. J.SunG. P. (2007). Respiration and growth of Shewanella decolorationis S12 with an azo compound as the sole electron acceptor.Appl. Environ. Microbiol.7364–72. 10.1128/AEM.01415-06
60
HuntS.GaitoS. T.LayzellD. B. (1988). Model of gas exchange and diffusion in legume nodules.Planta173128–141. 10.1007/BF00394497
61
HussainA.HasanA.JavidA.QaziJ. I. (2016). Exploited application of sulfate-reducing bacteria for concomitant treatment of metallic and non-metallic wastes: a mini review.3 Biotech6:119. 10.1007/s13205-016-0437-3
62
IshiiS.KosakaT.HoriK.HottaY.WatanabeK. (2005). Coaggregation facilitates interspecies hydrogen transfer between Pelotomaculum thermopropionicum and Methanothermobacter thermautotrophicus.Appl. Environ. Microbiol.717838–7845. 10.1128/AEM.71.12.7838-7845.2005
63
JenneyF. E.AdamsM. W. (2008). Hydrogenases of the model hyperthermophiles.Ann. N. Y. Acad. Sci.1125252–266. 10.1196/annals.1419.013
64
JohnsonD. R.BrodieE. L.HubbardA. E.AndersenG. L.ZinderS. H.Alvarez-CohenL. (2008). Temporal transcriptomic microarray analysis of “Dehalococcoides ethenogenes” strain 195 during the transition into stationary phase.Appl. Environ. Microbiol.742864–2872. 10.1128/AEM.02208-07
65
JoynerA.WinterW.GodboutD. (1977). Studies on some characteristics of hydrogen production by cell-free extracts of rumen anaerobic bacteria.Can. J. Microbiol.23346–353. 10.1139/m77-051
66
JugderB.ErtanH.LeeM.ManefieldM.MarquisC. P. (2015). Reductive dehalogenases come of age in biological destruction of organohalides.Trends Biotechnol.33595–610. 10.1016/j.tibtech.2015.07.004
67
JugderB.WelchJ.Aguey-ZinsouK.MarquisC. P. (2013). Fundamentals and electrochemical applications of [Ni-Fe]-uptake hydrogenases.RSC Adv.38142–8159. 10.1039/c3ra22668a
68
JugderB. E.ErtanH.BohlS.LeeM.MarquisC. P.ManefieldM. (2016). Organohalide respiring bacteria and reductive dehalogenases: key tools in organohalide bioremediation.Front. Microbiol.7:249. 10.3389/fmicb.2016.00249
69
KaryakinA. A.MorozovS. V.VoroninO. G.ZorinN. A.KaryakinaE. E.FateyevV. N.et al (2007). The limiting performance characteristics in bioelectrocatalysis of hydrogenase enzymes.Angew. Chem. Int. Ed. Engl.467244–7246. 10.1002/anie.200701096
70
KasererH. (1905). Über die Oxydation des Wasserstoffes und des Methans durch Mikroorganismen.Berlin: Hartleben.
71
KhanM. T.BhattJ. (1991). Polyethylene glycol mediated fusion of Halobacterium halobium MMT22 and Escherichia coli for enhancement of hydrogen production.Int. J. Hydrogen Energ.16683–685. 10.1016/0360-3199(91)90191-K
72
KhdhiriM.Piché-ChoquetteS.TremblayJ.TringeS. G.ConstantP. (2017). The tale of a neglected energy source: elevated hydrogen exposure affects both microbial diversity and function in soil.Appl. Environ. Microbiol.83:e00275-17. 10.1128/AEM.00275-17
73
KieuH. T.MüllerE.HornH. (2011). Heavy metal removal in anaerobic semi-continuous stirred tank reactors by a consortium of sulfate-reducing bacteria.Water Res.453863–3870. 10.1016/j.watres.2011.04.043
74
KleinsteuberS.SchleinitzK. M.VogtC. (2012). Key players and team play: anaerobic microbial communities in hydrocarbon-contaminated aquifers.Appl. Microbiol. Biotechnol.94851–873. 10.1007/s00253-012-4025-0
75
KongJ. Y.BaiY.SuY.YaoY.HeR. (2014). Effects of trichloroethylene on community structure and activity of methanotrophs in landfill cover soils.Soil Biol. Biochem.78118–127. 10.1016/j.soilbio.2014.07.018
76
KosourovS.LeinoH.MurukesanG.LynchF.SivonenK.TsygankovA. A.et al (2014). Hydrogen photoproduction by immobilized N2-fixing cyanobacteria: understanding the role of uptake hydrogenase in the long-term process.Appl. Environ. Microbiol.805807–5817. 10.1128/AEM.01776-14
77
KrahnE.SchneiderK.MüllerA. (1996). Comparative characterization of H2 production by the conventional Mo nitrogenase and the alternative “iron-only” nitrogenase of Rhodobacter capsulatus hup- mutants.Appl. Microbiol. Biotechnol.46285–290. 10.1007/s002530050818
78
KubeM.BeckA.ZinderS. H.KuhlH.ReinhardtR.AdrianL. (2005). Genome sequence of the chlorinated compound–respiring bacterium Dehalococcoides species strain CBDB1.Nat. Biotechnol.231269–1273. 10.1038/nbt1131
79
KumarN.DasD. (2000). Enhancement of hydrogen production by Enterobacter cloacae IIT-BT 08.Proc. Biochem.35589–593. 10.1016/S0032-9592(99)00109-0
80
LaranjoM.AlexandreA.OliveiraS. (2014). Legume growth-promoting rhizobia: an overview on the Mesorhizobium genus.Microbiol. Res.1692–17. 10.1016/j.micres.2013.09.012
81
LeeJ. Z.BurowL. C.WoebkenD.EverroadR. C.KuboM. D.SpormannA. M.et al (2014). Fermentation couples Chloroflexi and sulfate-reducing bacteria to Cyanobacteria in hypersaline microbial mats.Front. Microbiol.5:61. 10.3389/fmicb.2014.00061
82
LeeM.LowA.ZembO.KoenigJ.MichaelsenA.ManefieldM. (2012). Complete chloroform dechlorination by organochlorine respiration and fermentation.Environ. Microbiol.14883–894. 10.1111/j.1462-2920.2011.02656.x
83
LeloupJ.FossingH.KohlsK.HolmkvistL.BorowskiC.JørgensenB. B. (2009). Sulfate-reducing bacteria in marine sediment (Aarhus Bay, Denmark): abundance and diversity related to geochemical zonation.Environ. Microbiol.111278–1291. 10.1111/j.1462-2920.2008.01855.x
84
LettingaG.ZehnderA.GrotenhuisJ.Hulshoff PolL. (1988). “Granular anaerobic sludge; microbiology and technology,” in Proceedings of the GASMAT-Workshop, Lunteren.
85
LiX.DaiL.ZhangC.ZengG.LiuY.ZhouC.et al (2017). Enhanced biological stabilization of heavy metals in sediment using immobilized sulfate reducing bacteria beads with inner cohesive nutrient.J. Hazard. Mater.324340–347. 10.1016/j.jhazmat.2016.10.067
86
LiX.LanS. M.ZhuZ. P.ZhangC.ZengG. M.LiuY. G.et al (2018). The bioenergetics mechanisms and applications of sulfate-reducing bacteria in remediation of pollutants in drainage: a review.Ecotoxicol. Environ. Saf.158162–170. 10.1016/j.ecoenv.2018.04.025
87
LiotQ.ConstantP. (2016). Breathing air to save energy–new insights into the ecophysiological role of high-affinity [NiFe]-hydrogenase in Streptomyces avermitilis.Microbiol. Open547–59. 10.1002/mbo3.310
88
LovleyD. R. (1993). Dissimilatory metal reduction.Annu. Rev. Microbiol.47263–290. 10.1146/annurev.mi.47.100193.001403
89
LovleyD. R. (2008). The microbe electric: conversion of organic matter to electricity.Curr. Opin. Biotechnol.19564–571. 10.1016/j.copbio.2008.10.005
90
LovleyD. R.CoatesJ. D. (2000). Novel forms of anaerobic respiration of environmental relevance.Curr. Opin. Microbiol.3252–256. 10.1016/S1369-5274(00)00085-0
91
LovleyD. R.PhillipsE. J. (1994). Reduction of chromate by Desulfovibrio vulgaris and its c3 cytochrome.Appl. Environ. Microbiol.60726–728.
92
MaimaitiJ.ZhangY.YangJ.CenY. P.LayzellD. B.PeoplesM.et al (2007). Isolation and characterization of hydrogen-oxidizing bacteria induced following exposure of soil to hydrogen gas and their impact on plant growth.Environ. Microbiol.9435–444. 10.1111/j.1462-2920.2006.01155.x
93
MalikB.SuW. W.WaldH.BlumentalsI.KellyR. (1989). Growth and gas production for hyperthermophilic archaebacterium, Pyrococcus furiosus.Biotechnol. Bioeng.341050–1057. 10.1002/bit.260340805
94
MarótiJ.FarkasA.NagyI. K.MarótiG.KondorosiÉ.RákhelyG.et al (2010). A second soluble Hox-type NiFe enzyme completes the hydrogenase set in Thiocapsa roseopersicina BBS.Appl. Environ. Microbiol.765113–5123. 10.1128/AEM.00351-10
95
MarshallI. P.BerggrenD. R.AzizianM. F.BurowL. C.SempriniL.SpormannA. M. (2012). The Hydrogenase Chip: a tiling oligonucleotide DNA microarray technique for characterizing hydrogen-producing and-consuming microbes in microbial communities.ISME J.6814–826. 10.1038/ismej.2011.136
96
Maymó-GatellX.ChienY.-T.GossettJ. M.ZinderS. H. (1997). Isolation of a bacterium that reductively dechlorinates tetrachloroethene to ethene.Science2761568–1571. 10.1126/science.276.5318.1568
97
MenY.FeilH.VerBerkmoesN. C.ShahM. B.JohnsonD. R.LeeP. K.et al (2012). Sustainable syntrophic growth of Dehalococcoides ethenogenes strain 195 with Desulfovibrio vulgaris Hildenborough and Methanobacterium congolense: global transcriptomic and proteomic analyses.ISME J.6410–421. 10.1038/ismej.2011.111
98
MillerT. L.WolinM. (1973). Formation of hydrogen and formate by Ruminococcus albus.J. Bacteriol.116836–846.
99
MillerT. L.WolinM. (1979). Fermentations by saccharolytic intestinal bacteria.Am. J. Clin. Nutr.32164–172. 10.1093/ajcn/32.1.164
100
MizunoK.MorishitaY.AndoA.TsuchiyaN.HirataM.TanakaK. (2012). Genus-specific and phase-dependent effects of nitrate on a sulfate-reducing bacterial community as revealed by dsrB-based DGGE analyses of wastewater reactors.World J. Microbiol. Biotechnol.28677–686. 10.1007/s11274-011-0862-8
101
MohnW. W.KennedyK. J. (1992). Reductive dehalogenation of chlorophenols by Desulfomonile tiedjei DCB-1.Appl. Environ. Microbiol.581367–1370.
102
MorrisB. E.HennebergerR.HuberH.Moissl-EichingerC. (2013). Microbial syntrophy: interaction for the common good.FEMS Microbiol. Rev.37384–406. 10.1111/1574-6976.12019
103
MorrisR. M.SowellS.BarofskyD.ZinderS.RichardsonR. (2006). Transcription and mass-spectroscopic proteomic studies of electron transport oxidoreductases in Dehalococcoides ethenogenes.Environ. Microbiol.81499–1509. 10.1111/j.1462-2920.2006.01090.x
104
MüllerN.WormP.SchinkB.StamsA. J.PluggeC. M. (2010). Syntrophic butyrate and propionate oxidation processes: from genomes to reaction mechanisms.Environ. Microbiol. Rep.2489–499. 10.1111/j.1758-2229.2010.00147.x
105
MusF.CrookM. B.GarciaK.CostasA. G.GeddesB. A.KouriE. D.et al (2016). Symbiotic nitrogen fixation and the challenges to its extending to nonlegumes.Appl. Environ. Microbiol.823698–3710. 10.1128/AEM.01055-16
106
MutambanengweC.TogoC.WhiteleyC. (2007). Decolorization and degradation of textile dyes with biosulfidogenic hydrogenases.Biotechnol. Prog.231095–1100. 10.1021/bp070147v
107
NandiR.SenguptaS. (1998). Microbial production of hydrogen: an overview.Crit. Rev. Microbiol.2461–84. 10.1080/10408419891294181
108
NielsenM.RevsbechN. P.KühlM. (2015). Microsensor measurements of hydrogen gas dynamics in cyanobacterial microbial mats.Front. Microbiol.6:726. 10.3389/fmicb.2015.00726
109
NourS. M.Cleyet-MarelJ.-C.NormandP.FernandezM. P. (1995). Genomic heterogeneity of strains nodulating chickpeas (Cicer arietinum L.) and description of Rhizobium mediterraneum sp. nov.Int. J. Syst. Bacteriol.45640–648. 10.1099/00207713-45-4-640
110
NovelliP. C.LangP. M.MasarieK. A.HurstD. F.MyersR.ElkinsJ. W. (1999). Molecular hydrogen in the troposphere: global distribution and budget.J. Geophys. Res.10430427–30444. 10.1029/1999JD900788
111
NyyssonenM.HultmanJ.AhonenL.KukkonenI.PaulinL.LaineP.et al (2014). Taxonomically and functionally diverse microbial communities in deep crystalline rocks of the Fennoscandian shield.ISME J.8126–138. 10.1038/ismej.2013.125
112
OdomJ.PeckH.Jr. (1981). Hydrogen cycling as a general mechanism for energy coupling in the sulfate-reducing bacteria, Desulfovibrio sp.FEMS Microbiol. Lett.1247–50. 10.1111/j.1574-6968.1981.tb07609.x
113
OrrC. H.JamesA.LeifertC.CooperJ. M.CummingsS. P. (2011). Diversity and activity of free-living nitrogen-fixing bacteria and total bacteria in organic and conventionally managed soils.Appl. Environ. Microbiol.77911–919. 10.1128/AEM.01250-10
114
OsborneC. A.PeoplesM. B.JanssenP. H. (2010). Detection of a reproducible, single-member shift in soil bacterial communities exposed to low levels of hydrogen.Appl. Environ. Microbiol.761471–1479. 10.1128/AEM.02072-09
115
OtakiH.EverroadR. C.MatsuuraK.HarutaS. (2012). Production and consumption of hydrogen in hot spring microbial mats dominated by a filamentous anoxygenic photosynthetic bacterium.Microbes Environ.27293–299. 10.1264/jsme2.ME11348
116
PetersJ. W.SchutG. J.BoydE. S.MulderD. W.ShepardE. M.BroderickJ. B.et al (2015). [FeFe]-and [NiFe]-hydrogenase diversity, mechanism, and maturation.Biochim. Biophys. Acta18531350–1369. 10.1016/j.bbamcr.2014.11.021
117
PetersenJ. M.ZielinskiF. U.PapeT.SeifertR.MoraruC.AmannR.et al (2011). Hydrogen is an energy source for hydrothermal vent symbioses.Nature476176–180. 10.1038/nature10325
118
PhillipsE. J.LandaE. R.LovleyD. R. (1995). Remediation of uranium contaminated soils with bicarbonate extraction and microbial U (VI) reduction.J. Ind. Microbiol.14203–207. 10.1007/BF01569928
119
Piché-ChoquetteS.KhdhiriM.ConstantP. (2018). Dose-response relationships between environmentally-relevant H2 concentrations and the biological sinks of H2, CH4 and CO in soil.Soil Biol. Biochem.123190–199. 10.1016/j.soilbio.2018.05.008
120
RahmB. G.MorrisR. M.RichardsonR. E. (2006). Temporal expression of respiratory genes in an enrichment culture containing Dehalococcoides ethenogenes.Appl. Environ. Microbiol.725486–5491. 10.1128/AEM.00855-06
121
RákhelyG.ZhouZ. H.AdamsM. W.KovácsK. L. (1999). Biochemical and molecular characterization of the [NiFe] hydrogenase from the hyperthermophilic archaeon, Thermococcus litoralis.Eur. J. Biochem.2661158–1165. 10.1046/j.1432-1327.1999.00969.x
122
RenN.WangA.MaF. (2005). Physiological Ecology of Acid-Producing Fermentative Microbiology.Beijing: Science Press.
123
RenX. Y.ZengG. M.TangL.WangJ. J.WanJ.LiuY.et al (2018). Sorption, transport and biodegradation-an insight into bioavailability of persistent organic pollutants in soil.Sci. Total Environ.6101154–1163. 10.1016/j.scitotenv.2017.08.089
124
RheeT.BrenninkmeijerC.RöckmannT. (2006). The overwhelming role of soils in the global atmospheric hydrogen cycle.Atmos. Chem. Phys.61611–1625. 10.5194/acp-6-1611-2006
125
RichterK.GescherJ. (2014). Accelerated glycerol fermentation in Escherichia coli using methanogenic formate consumption.Bioresour. Technol.162389–391. 10.1016/j.biortech.2014.04.011
126
RiddinT.GovenderY.GerickeM.WhiteleyC. (2009). Two different hydrogenase enzymes from sulphate-reducing bacteria are responsible for the bioreductive mechanism of platinum into nanoparticles.Enzyme Microb. Technol.45267–273. 10.1016/j.enzmictec.2009.06.006
127
RittmannB. E.McCartyP. L. (2012). Environmental Biotechnology: Principles and Applications.New York, NY: Tata McGraw-Hill Education.
128
RodionovD. A.DubchakI.ArkinA.AlmE.GelfandM. S. (2004). Reconstruction of regulatory and metabolic pathways in metal-reducing δ-proteobacteria.Genome Biol.5:R90. 10.1186/gb-2004-5-11-r90
129
RupakulaA.KruseT.BoerenS.HolligerC.SmidtH.MaillardJ. (2013). The restricted metabolism of the obligate organohalide respiring bacterium Dehalobacter restrictus: lessons from tiered functional genomics.Philos. Trans. R. Soc. Lond. B Biol. Sci.368:20120325. 10.1098/rstb.2012.0325
130
SalviB.SubramanianK. (2015). Sustainable development of road transportation sector using hydrogen energy system.Renew. Sustain. Energy Rev.511132–1155. 10.1016/j.rser.2015.07.030
131
SawersG. (1994). The hydrogenases and formate dehydrogenases of Escherichia coli.Antonie Van Leeuwenhoek6657–88. 10.1007/BF00871633
132
SchilterD.CamaraJ. M.HuynhM. T.Hammes-SchifferS.RauchfussT. B. (2016). Hydrogenase enzymes and their synthetic models: the role of metal hydrides.Chem. Rev.1168693–8749. 10.1021/acs.chemrev.6b00180
133
SchinkB. (1997). Energetics of syntrophic cooperation in methanogenic degradation.Microbiol. Mol. Biol. Rev.61262–280.
134
SchlegelH. G.GottschalkG.PfenningN. (1976). “Symposium on Microbial production and utilization of gases (H2 CH4 CO),” in Proceedings of the Symposium on Microbial Production and Utilization of Gases (H2 CH4 CO), (Göttingen: Akademie der Wissenschaften).
135
SchmidtU.ConradR. (1993). Hydrogen, carbon monoxide, and methane dynamics in Lake Constance.Limnol. Oceanogr.381214–1226. 10.4319/lo.1993.38.6.1214
136
SchützH.ConradR.GoodwinS.SeilerW. (1988). Emission of hydrogen from deep and shallow freshwater environments.Biogeochemistry5295–311. 10.1007/BF02180069
137
SchwartzE.FritschJ.FriedrichB. (2013). “H2-metabolizing prokaryotes,” in The Prokaryotes, edsDworkinM.FalkowS.RosenbergE.SchleiferK. H.StackebrandtE. (New York, NY: Springer), 119–199.
138
SeshadriR.AdrianL.FoutsD. E.EisenJ. A.PhillippyA. M.MetheB. A.et al (2005). Genome sequence of the PCE-dechlorinating bacterium Dehalococcoides ethenogenes.Science307105–108. 10.1126/science.1102226
139
SheltonD. R.TiedjeJ. M. (1984). Isolation and partial characterization of bacteria in an anaerobic consortium that mineralizes 3-chlorobenzoic acid.Appl. Environ. Microbiol.48840–848.
140
SieberJ. R.McInerneyM. J.GunsalusR. P. (2012). Genomic insights into syntrophy: the paradigm for anaerobic metabolic cooperation.Annu. Rev. Microbiol.66429–452. 10.1146/annurev-micro-090110-102844
141
SinghR.KumarA.KirroliaA.KumarR.YadavN.BishnoiN. R.et al (2011). Removal of sulphate, COD and Cr (VI) in simulated and real wastewater by sulphate reducing bacteria enrichment in small bioreactor and FTIR study.Bioresour. Technol.102677–682. 10.1016/j.biortech.2010.08.041
142
SmatlakC. R.GossettJ. M.ZinderS. H. (1996). Comparative kinetics of hydrogen utilization for reductive dechlorination of tetrachloroethene and methanogenesis in an anaerobic enrichment culture.Environ. Sci. Technol.302850–2858. 10.1021/es9602455
143
SobohB.LinderD.HedderichR. (2002). Purification and catalytic properties of a CO-oxidizing: H2-evolving enzyme complex from Carboxydothermus hydrogenoformans.Eur. J. Biochem.2695712–5721. 10.1046/j.1432-1033.2002.03282.x
144
SobohB.LinderD.HedderichR. (2004). A multisubunit membrane-bound [NiFe] hydrogenase and an NADH-dependent Fe-only hydrogenase in the fermenting bacterium Thermoanaerobacter tengcongensis.Microbiol.1502451–2463. 10.1099/mic.0.27159-0
145
SokolovaT.HanelJ.OnyenwokeR.ReysenbachA.-L.BantaA.GeyerR.et al (2007). Novel chemolithotrophic, thermophilic, anaerobic bacteria Thermolithobacter ferrireducens gen. nov., sp. nov. and Thermolithobacter carboxydivorans sp. nov.Extremophiles11145–157. 10.1007/s00792-006-0022-5
146
SokolovaT.KostrikinaN.ChernyhN.TourovaT.KolganovaT.Bonch-OsmolovskayaE. (2002). Carboxydocella thermautotrophica gen. nov., sp. nov., a novel anaerobic, CO-utilizing thermophile from a Kamchatkan hot spring.Int. J. Syst. Evol. Microbiol.521961–1967.
147
SokolovaT. G.GonzalezJ. M.KostrikinaN. A.ChernyhN. A.SlepovaT. V.Bonch-OsmolovskayaE. A.et al (2004). Thermosinus carboxydivorans gen. nov., sp. nov., a new anaerobic, thermophilic, carbon-monoxide-oxidizing, hydrogenogenic bacterium from a hot pool of Yellowstone National Park.Int. J. Syst. Evol. Microbiol.542353–2359. 10.1099/ijs.0.63186-0
148
SokolovaT. G.KostrikinaN. A.ChernyhN. A.KolganovaT. V.TourovaT. P.Bonch-OsmolovskayaE. A. (2005). Thermincola carboxydiphila gen. nov., sp. nov., a novel anaerobic, carboxydotrophic, hydrogenogenic bacterium from a hot spring of the Lake Baikal area.Int. J. Syst. Evol. Microbiol.552069–2073. 10.1099/ijs.0.63299-0
149
SpainkH. P. (2000). Root nodulation and infection factors produced by rhizobial bacteria.Annu. Rev. Microbiol.54257–288. 10.1146/annurev.micro.54.1.257
150
StamsA. J.De BokF. A.PluggeC. M.Van EekertM. H.DolfingJ.SchraaG. (2006). Exocellular electron transfer in anaerobic microbial communities.Environ. Microbiol.8371–382. 10.1111/j.1462-2920.2006.00989.x
151
StamsA. J.PluggeC. M. (2009). Electron transfer in syntrophic communities of anaerobic bacteria and archaea.Nat. Rev. Microbiol.7568–577. 10.1038/nrmicro2166
152
SteinS.SelesiD.SchillingR.PattisI.SchmidM.HartmannA. (2005). Microbial activity and bacterial composition of H2-treated soils with net CO2 fixation.Soil Biol. Biochem.371938–1945. 10.1016/j.soilbio.2005.02.035
153
StephensonM.SticklandL. H. (1931). Hydrogenase: a bacterial enzyme activating molecular hydrogen: the properties of the enzyme.Biochem. J.25205–214. 10.1042/bj0250205
154
SuflitaJ. M.HorowitzA.SheltonD. R.TiedjeJ. M. (1982). Dehalogenation: a novel pathway for the anaerobic biodegradation of haloaromatic compounds.Science2181115–1117. 10.1126/science.218.4577.1115
155
SvetlichnyV.SokolovaT.GerhardtM.RingpfeilM.KostrikinaN.ZavarzinG. (1991). Carboxydothermus hydrogenoformans gen. nov., sp. nov., a CO-utilizing thermophilic anaerobic bacterium from hydrothermal environments of Kunashir Island.Syst. Appl. Microbiol.14254–260. 10.1016/S0723-2020(11)80377-2
156
TabakH. H.LensP.van HullebuschE. D.DejongheW. (2005). Developments in bioremediation of soils and sediments polluted with metals and radionuclides–1. Microbial processes and mechanisms affecting bioremediation of metal contamination and influencing metal toxicity and transport.Rev. Environ. Sci. Biotechnol.4115–156. 10.1007/s11157-005-2169-4
157
TalibiM.HellierP.LadommatosN. (2017). The effect of varying EGR and intake air boost on hydrogen-diesel co-combustion in CI engines.Int. J. Hydrogen Energy426369–6383. 10.1016/j.ijhydene.2016.11.207
158
TeboB. M.ObraztsovaA. Y. (1998). Sulfate-reducing bacterium grows with Cr (VI), U (VI), Mn (IV), and Fe (III) as electron acceptors.FEMS Microbiol. Lett.162193–199. 10.1111/j.1574-6968.1998.tb12998.x
159
TengY.ChenW. (2019). Soil microbiome: a promising strategy for contaminated soil remediation.Pedosphere.10.1016/S1002-0160(18)60061-X
160
TengY.XuZ.LuoY.ReverchonF. (2012). How do persistent organic pollutants be coupled with biogeochemical cycles of carbon and nutrients in terrestrial ecosystems under global climate change?J. Soil Sediments12411–419. 10.1007/s11368-011-0462-0
161
ThauerR. K.KasterA. K.GoenrichM.SchickM.HiromotoT.ShimaS. (2010). Hydrogenases from methanogenic archaea, nickel, a novel cofactor, and H2 storage.Annu. Rev. Biochem.79507–536. 10.1146/annurev.biochem.030508.152103
162
TrchounianK.MüllerN.SchinkB.TrchounianA. (2017a). Glycerol and mixture of carbon sources conversion to hydrogen by Clostridium beijerinckii DSM791 and effects of various heavy metals on hydrogenase activity.Int. J. Hydrogen Energy427875–7882. 10.1016/j.ijhydene.2017.01.011
163
TrchounianK.SawersR. G.TrchounianA. (2017b). Improving biohydrogen productivity by microbial dark-and photo-fermentations: novel data and future approaches.Renew. Sustain. Energy Rev.801201–1216. 10.1016/j.rser.2017.05.149
164
TrchounianK.PinskeC.SawersR. G.TrchounianA. (2011). Dependence on the F0F1-ATP synthase for the activities of the hydrogen-oxidizing hydrogenases 1 and 2 during glucose and glycerol fermentation at high and low pH in Escherichia coli.J. Bioenerg. Biomembr.43645–650. 10.1007/s10863-011-9397-9
165
TrchounianK.PoladyanA.VassilianA.TrchounianA. (2012). Multiple and reversible hydrogenases for hydrogen production by Escherichia coli: dependence on fermentation substrate, pH and the F0F1-ATPase.Crit. Rev. Biochem. Mol.47236–249. 10.3109/10409238.2012.655375
166
TrchounianK.TrchounianA. (2014). Hydrogen producing activity by Escherichia coli hydrogenase 4 (hyf) depends on glucose concentration.Int. J. Hydrogen Energy3916914–16918. 10.1016/j.ijhydene.2014.08.059
167
TrchounianK.TrchounianA. (2015). Escherichia coli hydrogen gas production from glycerol: effects of external formate.Renew. Energy83345–351. 10.1016/j.renene.2015.04.052
168
TrompT. K.ShiaR.-L.AllenM.EilerJ. M.YungY. L. (2003). Potential environmental impact of a hydrogen economy on the stratosphere.Science3001740–1742. 10.1126/science.1085169
169
UffenR. L. (1976). Anaerobic growth of a Rhodopseudomonas species in the dark with carbon monoxide as sole carbon and energy substrate.Proc. Natl. Acad. Sci. U.S.A.733298–3302. 10.1073/pnas.73.9.3298
170
VallsM.De LorenzoV. (2002). Exploiting the genetic and biochemical capacities of bacteria for the remediation of heavy metal pollution.FEMS Microbiol. Rev.26327–338. 10.1111/j.1574-6976.2002.tb00618.x
171
VignaisP. M.BilloudB. (2007). Occurrence, classification, and biological function of hydrogenases: an overview.Chem. Rev.1074206–4272. 10.1021/cr050196r
172
VignaisP. M.BilloudB.MeyerJ. (2001). Classification and phylogeny of hydrogenases.FEMS Microbiol. Rev.25455–501. 10.1111/j.1574-6976.2001.tb00587.x
173
VincenziniM.MaterassiR.TrediciM.FlorenzanoG. (1982). Hydrogen production by immobilized cells—I. light dependent dissimilation of organic substances by Rhodopseudomonas palustris.Int. J. Hydrogen Energy7231–236. 10.1016/0360-3199(82)90086-6
174
VogelT. M.McCARTYP. L. (1985). Biotransformation of tetrachloroethylene to trichloroethylene, dichloroethylene, vinyl chloride, and carbon dioxide under methanogenic conditions.Appl. Environ. Microbiol.491080–1083.
175
WatrousM. M.ClarkS.KuttyR.HuangS.RudolphF. B.HughesJ. B.et al (2003). 2, 4, 6-Trinitrotoluene reduction by an Fe-only hydrogenase in Clostridium acetobutylicum.Appl. Environ. Microbiol.691542–1547. 10.1128/AEM.69.3.1542-1547.2003
176
WittyJ. F. (1991). Microelectrode measurements of hydrogen concentrations and gradients in legume nodules.J. Exp. Bot.42765–771. 10.1093/jxb/42.6.765
177
WittyJ. F.MinchinF. R. (1998). Hydrogen measurements provide direct evidence for a variable physical barrier to gas diffusion in legume nodules.J. Exp. Bot.491015–1020. 10.1093/jxb/49.323.1015
178
WoodwardJ.OrrM.CordrayK.GreenbaumE. (2000). Biotechnology: enzymatic production of biohydrogen.Nature4051014–1015. 10.1038/35016633
179
WuX.HolmfeldtK.HubalekV.LundinD.ÅströmM.BertilssonS.et al (2015). Microbial metagenomes from three aquifers in the Fennoscandian shield terrestrial deep biosphere reveal metabolic partitioning among populations.ISME J.101192–1203. 10.1038/ismej.2015.185
180
YanJ.RitalahtiK. M.WagnerD. D.LöfflerF. E. (2012). Unexpected specificity of interspecies cobamide transfer from Geobacter spp. to organohalide-respiring Dehalococcoides mccartyi strains.Appl. Environ. Microbiol.786630–6636. 10.1128/AEM.01535-12
181
YangY.McCartyP. L. (1998). Competition for hydrogen within a chlorinated solvent dehalogenating anaerobic mixed culture.Environ. Sci. Technol.323591–3597. 10.1021/es980363n
182
ZanaroliG.NegroniA.HäggblomM. M.FavaF. (2015). Microbial dehalogenation of organohalides in marine and estuarine environments.Curr. Opin. Biotechnol.33287–295. 10.1016/j.copbio.2015.03.013
183
ZhangH.BrunsM. A.LoganB. E. (2006). Biological hydrogen production by Clostridium acetobutylicum in an unsaturated flow reactor.Water Res.40728–734. 10.1016/j.watres.2005.11.041
184
ZielsR. M.SousaD. Z.StenselH. D.BeckD. A. (2017). DNA-SIP based genome-centric metagenomics identifies key long-chain fatty acid-degrading populations in anaerobic digesters with different feeding frequencies.ISME J.12112–123. 10.1038/ismej.2017.143
Summary
Keywords
bioremediation, hydrogenase, H2 consumption, H2 metabolism, H2 production
Citation
Teng Y, Xu Y, Wang X and Christie P (2019) Function of Biohydrogen Metabolism and Related Microbial Communities in Environmental Bioremediation. Front. Microbiol. 10:106. doi: 10.3389/fmicb.2019.00106
Received
31 October 2018
Accepted
17 January 2019
Published
14 February 2019
Volume
10 - 2019
Edited by
Eric Boyd, Montana State University, United States
Reviewed by
Karen Trchounian, Yerevan State University, Armenia; Philippe Constant, Institut National de la Recherche Scientifique (INRS), Canada
Updates
Copyright
© 2019 Teng, Xu, Wang and Christie.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying Teng, yteng@issas.ac.cn
This article was submitted to Microbial Physiology and Metabolism, a section of the journal Frontiers in Microbiology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.