- 1Key Laboratory of Pathogenic Fungi and Mycotoxins of Fujian Province, Key Laboratory of Biopesticide and Chemical Biology of Education Ministry, and School of Life Sciences, Fujian Agriculture and Forestry University, Fuzhou, China
- 2College of Life Science, Jiangsu Normal University, Xuzhou, China
- 3Longyan City Corporation of Fujian Tobacco Corporation, Longyan, China
Aspergillus flavus is an opportunistic pathogenic fungus for both plant and animal that produces carcinogenic toxins termed aflatoxins (AFs). To identify possible genetic targets to reduce AF contamination, in this study, we have characterized a novel A. flavus Set3, and it shares sequence homology with the yeast protein Set3. The set3 deletion mutants present no difference in growth rate but alterations in asexual development and secondary metabolite production when compared to the A. flavus wild type. Specifically, deletion of set3 gene decreases conidiophore formation and conidial production through downregulating expression of brlA and abaA genes. In addition, normal levels of set3 are required for sclerotial development and expression of sclerotia-related genes nsdC and sclR. Further analyses demonstrated that Set3 negatively regulates AF production as well as the concomitant expression of genes in the AF gene cluster. Importantly, our results also display that A. flavus Set3 is involved in crop kernel colonization. Taking together, these results reveal that a novel Set3 plays crucial roles in morphological development, secondary metabolism, and fungal virulence in A. flavus.
Introduction
As both plant and animal opportunistic pathogenic fungus, Aspergillus flavus is responsible for serious health and economic impacts worldwide by producing carcinogenic mycotoxins termed aflatoxins (AFs). Many agriculturally important oilseed crops, such as peanuts, maize, and tree nuts, can be contaminated by A. flavus and AFs (Amaike and Keller, 2011). AFs are also responsible for numerous health problems, including acute aflatoxicosis, immunosuppression, liver cancer, and even death in many animal species and human. These diseases are highly linked to the consumption of large amounts of AFs due to ingestion of contaminated crops (Hedayati et al., 2007; Klich, 2007). Economically, AF contamination leads to substantial monetary losses yearly, due in large part to rejection or reduced value of contaminated crops as well as costs associated with monitoring and detection in developed countries (Wu et al., 2014). AFs and other mycotoxins are estimated to contaminate one quarter of the world’s crops. Specifically, the health risks are a major concern in developing countries because of the lack of strict regulations or monitoring AF levels in commodities prior to consumption (Groopman et al., 2008).
Nowadays, approaches such as chemical or physical methods are insufficient to control A. flavus colonization and AF contamination, since A. flavus caused extensive infestations by generating asexual spores called conidia (Adams et al., 1998; Hedayati et al., 2007; Amaike and Keller, 2011). New strategies, such as those depending on genetic approaches, could contribute to the development of new methodologies to decrease dissemination and survival of this organism, as well as AF biosynthesis. Therefore, it is quite important to explore genetic regulatory pathways that control A. flavus morphogenesis and AF biosynthesis. Previous studies have revealed that AF biosynthesis is controlled by regulatory cluster pathways (Payne and Brown, 1998; Yu et al., 2004), and increasing literatures showed that AFs are regulated not only by cluster genes (Amare and Keller, 2014; Nie et al., 2018) but also by other signal pathways (Roze et al., 2004), transcriptional regulators (Affeldt et al., 2014; Cary et al., 2017; Chang et al., 2017), and epigenetic regulators (Lan et al., 2016; Zhi et al., 2017; Pfannenstiel et al., 2018).
Set3 is a signature of chromatin-associated protein, which was first characterized in yeast by its feature of containing plant homeodomain (PHD) finger and Su(var)3-9, Enhancer-of-zeste, Trithorax (SET) domains (Pijnappel et al., 2001). Nowadays, Set3 protein had been identified in various eukaryotic cells, and these proteins encompass several roles, such as histone methyltransferase activity, and protein-protein interactions with other factors involved in chromatin regulation. Current data demonstrated that Set3 participates in multiple cellular functions, including meiosis-specific repression of sporulation (Pijnappel et al., 2001), promotion of Ty1 retrotransposon integration at tRNA genes (Mou et al., 2006), signaling secretory stress upon the PKC cell integrity pathway (Cohen et al., 2008), the white-opaque transition and pathogenicity in Candida albicans (Hnisz et al., 2010), as well as the environmental stress response (Torres-Machorro et al., 2015; Yu et al., 2016). In budding yeast, Set3, Hos2, Sif2, and Snt1 form the functional core of a histone deacetylase complex named Set3/Hos2 complex (Set3C) (Pijnappel et al., 2001). Recently, Set3C is found to play both repressive and activating roles in transcription, depending on the context of the region to which it is recruited (Kim and Buratowski, 2009). Set3C is predominantly recruited to the 5′ transcribed region of genes to reduce the histone acetylation level (Hnisz et al., 2012). A recent study also showed that Set3 can regulate transcription independent of Set3C (Yu et al., 2016).
Although the roles of Set3 in many organisms have been studied, the function of Set3 in A. flavus has not been characterized. Herein, by using gene knockout strategy, we identified a novel Set3 in A. flavus, encoding a putative SET and a PHD domain protein. Our results reveal that Set3 is involved in morphological development, secondary metabolism, and virulence of the agriculturally and medically important fungus A. flavus.
Materials and Methods
Strains and Growth Conditions
The uracil auxotrophic strain A. flavus PTSΔku70ΔpyrG (SRRC collection number 1709) (Chang et al., 2010) was used as recipient strain for gene knockout, and PTSΔku70ΔpyrG:: AfpyrG was used as wild-type strain (WT). For phenotype assays, all utilized strains were cultured on potato dextrose agar (PDA, BD Difco™, USA) media for growth assays at 37°C, on yeast extract sucrose (YES, 20 g/l yeast extract, 150 g/l sucrose, 1g/l MgSO4•7H2O) media at 29°C for aflatoxin analysis, and on sclerotia-inducing Wickerham media (WKM, 2 g/l yeast extract, 3 g/l peptone, 5 g/l cornsteep solids, 2 g/l dextrose, 30 g/l sucrose, 2 g/l NaNO3, 1 g/l K2HPO4•3H2O, 0.5 g/l MgSO4•7H2O, 0.2 g/l KCl, 0.1 g/l FeSO4•7H2O) (Lan et al., 2016) for sclerotia analysis. Each strain was cultured on three plates at least for technical replicates, and each experiment was repeated for three times.
Phylogenetic Tree and Domain Analysis
Amino acid sequences of Saccharomyces cerevisiae Set3 (GenBank accession number: NP_012954.3) were used as a query, and basic local alignment search tool algorithm was used to download sequences of Set3 protein (Aspergillus spp. Candida albicans, Fusarium graminearum, Magnaporthe oryzae, Neurospora crassa, Arabidopsis thaliana, Drosophila melanogaster, Danio rerio, Mus musculus, Homo sapiens) from National Center for Biotechnology Information resources (NCBI, http://www.ncbi.nlm.nih.gov/). A neighbor-joining phylogenetic tree was constructed by the MEGA 6.0 software. The visualized Set3 domain was generated by DOG 2.0 software (downloaded from http://dog.biocuckoo.org/).
Construction of Knockout and Complemented Mutant Strains
To construct set3 knockout mutant (Δset3) strain, previous approach was used (Yang et al., 2016a) Primers utilized in this study were listed in Table 1. The entire gene deletion cassettes were amplified with specific primers. Overlap polymerase chain reaction (PCR) method was performed as described earlier (Szewczyk et al., 2006), and then, fusion PCR products were transformed into the PTSΔku70ΔpyrG protoplasts of A. flavus. For constructing set3 complemented (Δset3-com) strain, PCR products of native promoter and open reading frame for Set3, combined with plasmid pPTR1 (Takara, Japan) containing the marker gene ptrA, were re-introduced into the protoplasts of the gene deletion strains. Fungal transformants were preliminary analyzed by PCR and reverse transcription PCR (RT-PCR) and further verified by southern blot as reported earlier in our group (Yang et al., 2016a).
Microscopic Examination of Set3-mCherry Subcellular Localization
A. flavus Set3-mCherry strains were prepared using a published method (Yang et al., 2016b), and the primers were listed in Table 1. To assess Set3-mCherry localization, fresh mycelia were analyzed using the Leica confocal SP8 microscope (Leica, Heidelberg, Germany). The nuclei of mycelia were observed after samples were stained with 1 μg/ml 4′,6-diamidino-2-phenylindole (DAPI, Sigma, USA).
Analysis of Fungal Conidia and Sclerotia
Spores (106 conidia/ml) from each strain were top-agar inoculated on PDA media for conidia assays and on WKM media for sclerotia analysis. For conidia analysis, cultures were incubated at 37°C in darkness for 5 days, and conidia were collected in triplicate from 10-mm cores that taken from equivalent zones of the fungal surface of PDA, and the collected samples were homogenized and diluted in 3 ml of 0.05% Tween-20 and counted by a hemocytometer (Qiujing, Shanghai, China). For sclerotia analysis, after 7 days grown on WKM media, each plate was sprayed with 75% ethanol to wash away the mycelia mat to allow the enumeration of the sclerotia. Sclerotia were collected and counted with the light microscope (Leica, Heidelberg, Germany). Each strain was assessed on five plates, and each experiment was repeated three times.
Stress Response Assays
The WT, the ∆set3-1, the ∆set3-2, and the ∆set3-com strains were inoculated onto PDA agar with oxidative stress agent H2O2 (2.5 and 5 mM) and cell wall stress agent Congo red (CR, 200 and 500 μg/ml), at 37°C in darkness for 3 days, respectively. To analyze the role of Set3 in stress response of A. flavus, the relative inhibition rates were calculated, according to the formula listed in the brackets {(diameter of colony without inhibitor − diameter of colony with inhibitor)/diameter of colony without inhibitor}. The experiments were performed in three repetitions.
Determination of Aflatoxin Production
To analyze aflatoxins (AFs), each strain was cultured in YES liquid media at 29°C for 3 days (180 r/min). Extracted AF samples were assessed by thin layer chromatography (TLC) and high performance liquid chromatography (HPLC) methods as previously described (Lan et al., 2016).
Briefly, 106 conidia of the WT, the ∆set3-1, the ∆set3-2, and the ∆set3-com strains were inoculated in 50 ml YES liquid medium, and cultures were incubated at 29°C. After 72 h, the cultures were combined with 25 ml chloroform in 250 ml flask, which were shaken for 30 min. The mycelia were then collected, dried completely, and weighed. Next, the organic layer of each sample was taken to a new plate, completely dried, and resuspended in chloroform solvent (1 ml/mg of mycelia). Then, the extracts (10 ml/sample) were loaded onto silica TLC plates (Haiyang Chemical, Qingdao, China) and separated in developing solvent (chloroform: acetone = 9:1). The TLC plates were exposed to UV radiation (365 nm) and photographed using a Quantum ST5 imaging system (Vilber Lourmat Deutschl and GmbH, Eberhardzell, Germany).
For HPLC experiment, the aflatoxin extracts were dissolved in methanol, filtered (0.22 μm), and performed by a Mycotox™ column (Waters, Milford, USA) at 42°C. The column was equilibrated in running solvent (water: methanol: acetonitrile = 56: 22: 22), and 10 μl samples were injected, and isocratic runs were conducted for 15 min in 100% running solvent at a flow rate of 1.0 ml/min. Aflatoxins were analyzed using a fluorescent detector (Waters, Milford, USA) with excitation and emission wave lengths of 365 and 455 nm, respectively. Aflatoxin production for each strain was analyzed using three flasks, and each experiment was repeated three times.
Crops Infection Experiments
Peanuts and maize seed colonization assays were performed using a published procedure (Lan et al., 2016). The peanut cotyledons and maize seeds infected with utilized strains were incubated at 28°C. After 5 days incubation, host seeds were harvested in 50 ml Falcon tubes and then vortexed for 2 min to release conidia into 20 ml sterile water supplemented with 0.05% Tween-80. The aflatoxin from the infected host seeds was extracted and analyzed as previously described (Lan et al., 2016).
Quantitative Real-Time PCR Analysis
For qRT-PCR analysis, mycelia of all tested strains were collected from PDA, WKM, and YES cultures for total RNA isolation with TRIzol reagent (Biomarker Technologies, Beijing, China). qRT-PCR was performed with Piko real-time PCR system (Thermo Fisher Scientific, Finland) by using the qPCR SuperMix (TransGen Biotech, Beijing, China). All utilized qRT-PCR primers were listed in Table 2. The relative quantification of expression level for each gene was calculated following the 2−ΔΔCt method, and the expression of actin was used as internal control. Each sample for qRT-PCR assays was conducted with technical triplicates, and the experiment was repeated three times.
Statistical Analysis
All data were presented with the means ± SD (standard deviation). The significant differences (statistical significances) among groups were calculated with ANOVA and least significant difference (LSD) tests. The statistical analysis and significance were performed with the software GraphPad Prism5 (La Jolla, CA, USA), and the difference is regarded to be statistically significant when p < 0.05.
Results
Identification and Analysis of Set3 in A. flavus
There were no previous reports of Set3 in Aspergillus species, so the Set3 amino acid sequence from model fungus Saccharomyces cerevisiae (GenBank accession number: NP_012954.3) was used with a basic local alignment search tool algorithm, then a putative protein that contains a PHD finger and a SET domain protein was identified in A. flavus designated Set3 (AFLA_134050). A. flavus Set3 presents 24% identity and 52% similarity with S. cerevisiae Set3, while it showed 61% similarity with the model filamentous fungus Aspergillus nidulans (AN5891.2, a putative protein). Analysis of Set3 proteins indicated that all of those Set3 proteins share conserved structures consisting of SET and PHD domains among fungi, plants, and animals (Figure 1A). A phylogenetic tree of evolutionary relationship of these Set3 proteins was constructed, revealing that the Set3 protein is conserved among Aspergillus species (Figure 1B).

Figure 1. Characterization of Set3 protein of A. flavus. (A) Domains from Set3 proteins were characterized by SMART, and DOG2.0 software was used to visualize protein domains. (B) Phylogenetic relationship of Set3 from different species was analyzed.
Subcellular Localization of A. flavus Set3
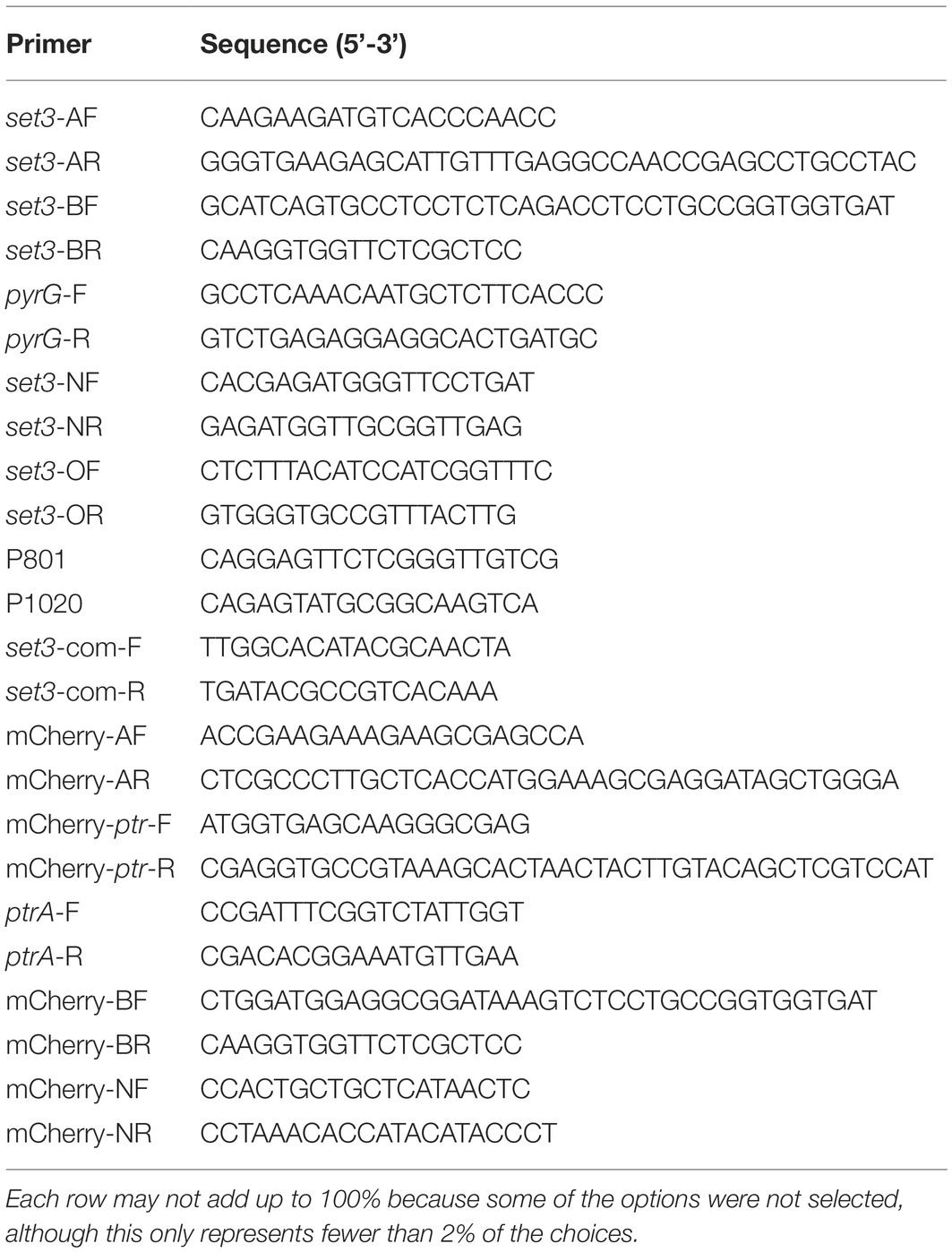
For subcellular localization analysis, a Set3-mCherry fusion generated with its native promoter was constructed and transformed into A. flavus auxotrophic strain PTSΔku70ΔpyrG. The construction strategy was shown in Figure 2A, and the resulting transformed strains exhibited a similar phenotype with WT strain, suggesting that the mCherry-tag did not affect the function of Set3 of A. flavus (data not shown). The results in Figure 2B showed that the mCherry fluorescence was dispersed in whole cytoplasm. By staining with 4,6-diamidino-2-phenylindole (DAPI), we also found that A. flavus Set3 is localized not only in cytoplasm but also in nucleus (Figure 2B).
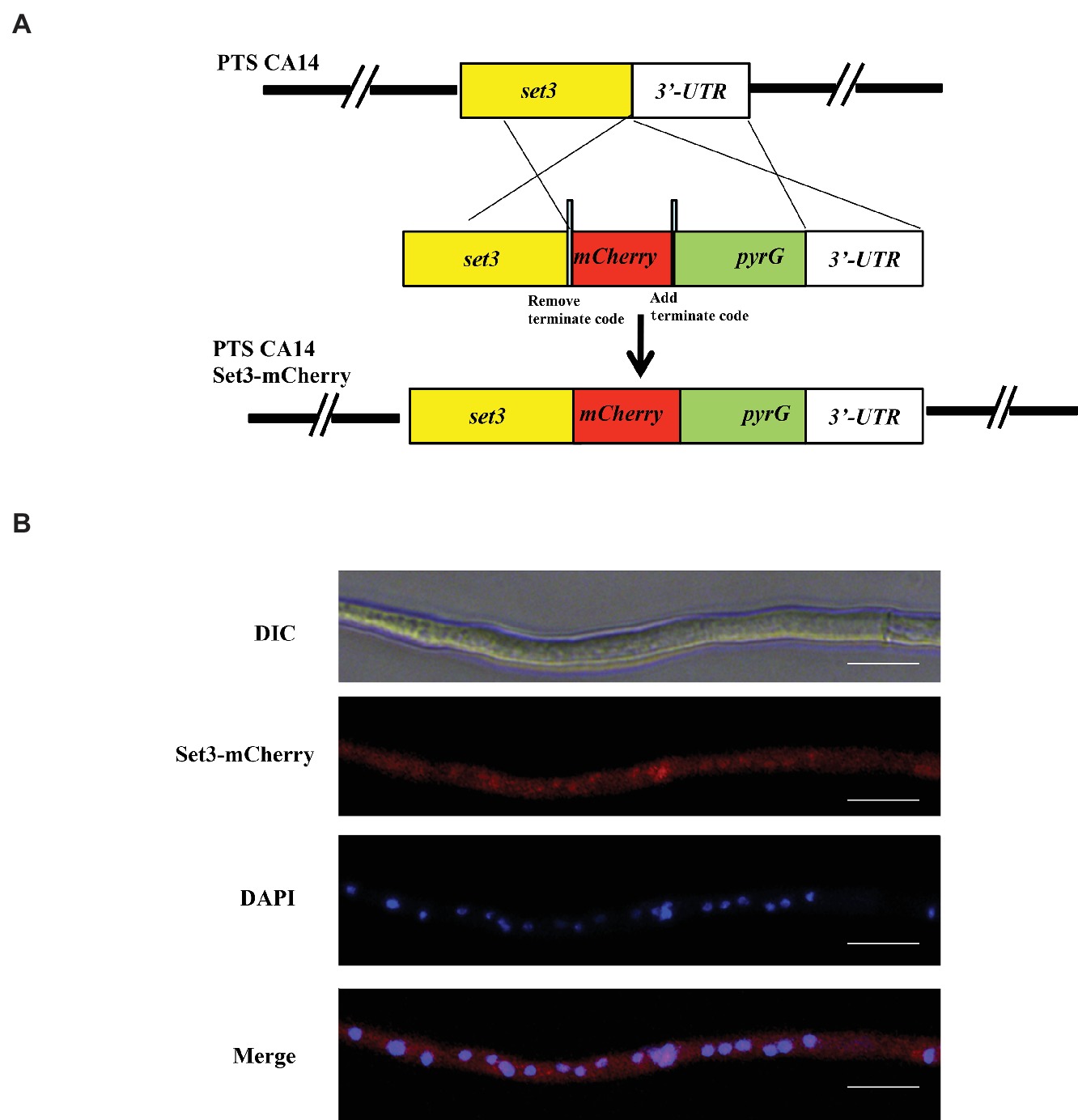
Figure 2. Subcellular localization of A. flavus Set3. (A) Construction strategy of Set3-mCherry strains. (B) Fluorescent image of Set3-mCherry during the hyphae growth period, and the nucleus was stained with DAPI. Bars = 20 μm.
Set3 Does Not Affect Growth Rate, but Involves in Hyphal Development
To gain an insight into the function of Set3 in morphogenesis of A. flavus, we generated set3 gene deletion mutants (Δset3-1 and Δset3-2) and complementation strain (Δset3-com), which are illustrated in Figure 3A. Transformants were confirmed by diagnostic PCR (Figure 3B). Expression levels of set3 in WT, Δset3, and Δset3-com strains were analyzed by RT-PCR, and the results showed that set3 gene transcript level was not expressed in those deletion strains, whereas set3 was detected in both the WT and Δset3-com strains (Figure 3C). The deletion strains were further verified by Southern blot (Figure 3D). In this study, we selected two deletion strains Δset3-1 and Δset3-2 for further analysis. In the morphological study, our results showed that colony growth was not significantly altered in Δset3 strains in comparison to the WT and Δset3-com strains (Figure 3E). However, the Δset3-1 and Δset3-2 strains presented more fluffy phenotype when compared to WT and Δset3-com strains (Figure 3F), suggesting that Set3 involves in hyphal growth in A. flavus.
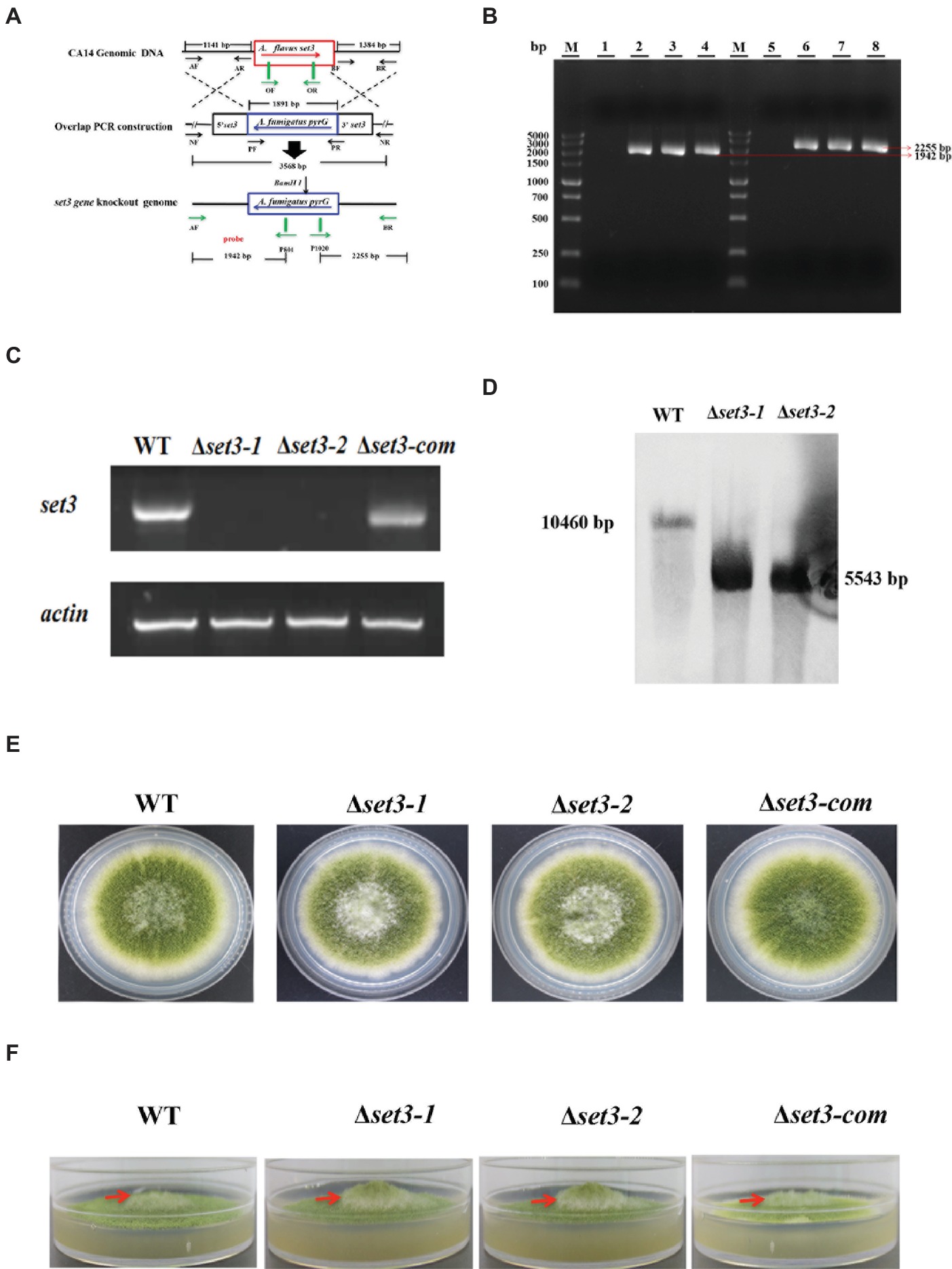
Figure 3. Construction of the set3 deleted (Δset3-1 and Δset3-2), complemented strains (Δset3-com), and growth analysis. (A) Construction strategy for Δset3 strain using homolog recombination. (B) The deleted and complemented strains were verified by PCR analysis with genomic DNA as template, lane 1~4: PCR examination on upstream of WT, Δset3-1, Δset3-2, and Δset3-com strains, lane 5~8: PCR examination on downstream of WT, Δset3-1, Δset3-2, and Δset3-com strains. (C) RT-PCR was used to confirm the transcript levels of set3 gene in deleted and complemented strains. (D) Southern blot was conducted to confirm the deletion mutants. (E) Colony morphology of WT, Δset3, and Δset3-com strains, grown on PDA media at 37°C for 5 days. (F) The Δset3-1 and Δset3-2 strain showed fluffier phenotype when compared to WT and Δset3-com strains.
Set3 Regulates Conidia Formation
In addition to fungal growth, Δset3-1 and Δset3-2 strains were found to decrease severely in conidiation when compared to WT strains (Figure 4B). For analysis of defect in conidiation, we further examined formation of conidiophores, and the result showed the Δset3-1 and Δset3-2 strains generate less normal conidiophores than WT strains (Figure 4A). Next, we checked the expression levels of genes brlA and abaA, which encode transcript factors related to conidiation. The results indicated that the transcript levels of both brlA (p < 0.05) and abaA (p < 0.05) were significantly reduced in the Δset3-1 and Δset3-2 strains, when compared to the WT andΔset3-com strains (Figures 4C,D). All these results indicated that set3 regulates conidia formation in A. flavus.
Figure 4. Deletion of set3 caused defects of conidiation in A. flavus. (A) Conidiophores of WT, Δset3-1, Δset3-2, and Δset3-com strains were observed by microscope after 12 h incubation, and bars = 200 μm. (B) Conidia production of WT, Δset3-1, Δset3-2, and Δset3-com strains. (C) Transcript levels of conidia-related gene brlA among WT, Δset3-1, Δset3-2, and Δset3-com strains. (D) Transcript levels of conidia-related gene abaA among WT, Δset3-1, Δset3-2, and Δset3-com strains. Different letters represent p < 0.05.
Set3 Positively Affects Sclerotia Production
A. flavus produces sclerotia to adapt unsuitable environment (Horn et al., 2009). To determine involvement of Set3 in sclerotia formation, all the strains were cultured on the sclerotia-inducing Wickerham media (WKM) at 37°C for 7 days. The results indicated that sclerotia production in the Δset3-1 and Δset3-2 strains was significantly decreased and less matured than that of the WT and complemented strains (p < 0.05) (Figures 5A,B). To confirm these findings, we performed qRT-PCR to check transcript levels of the sclerotia-related genes, nsdC and sclR. The results revealed that gene expression levels of nsdC (p < 0.05) and sclR (p < 0.05) were significantly lower in the Δset3-1 and Δset3-2 strains than WT and Δset3-com strains (Figures 5C,D). These above results showed that set3 plays an important role in sclerotia production in A. flavus.
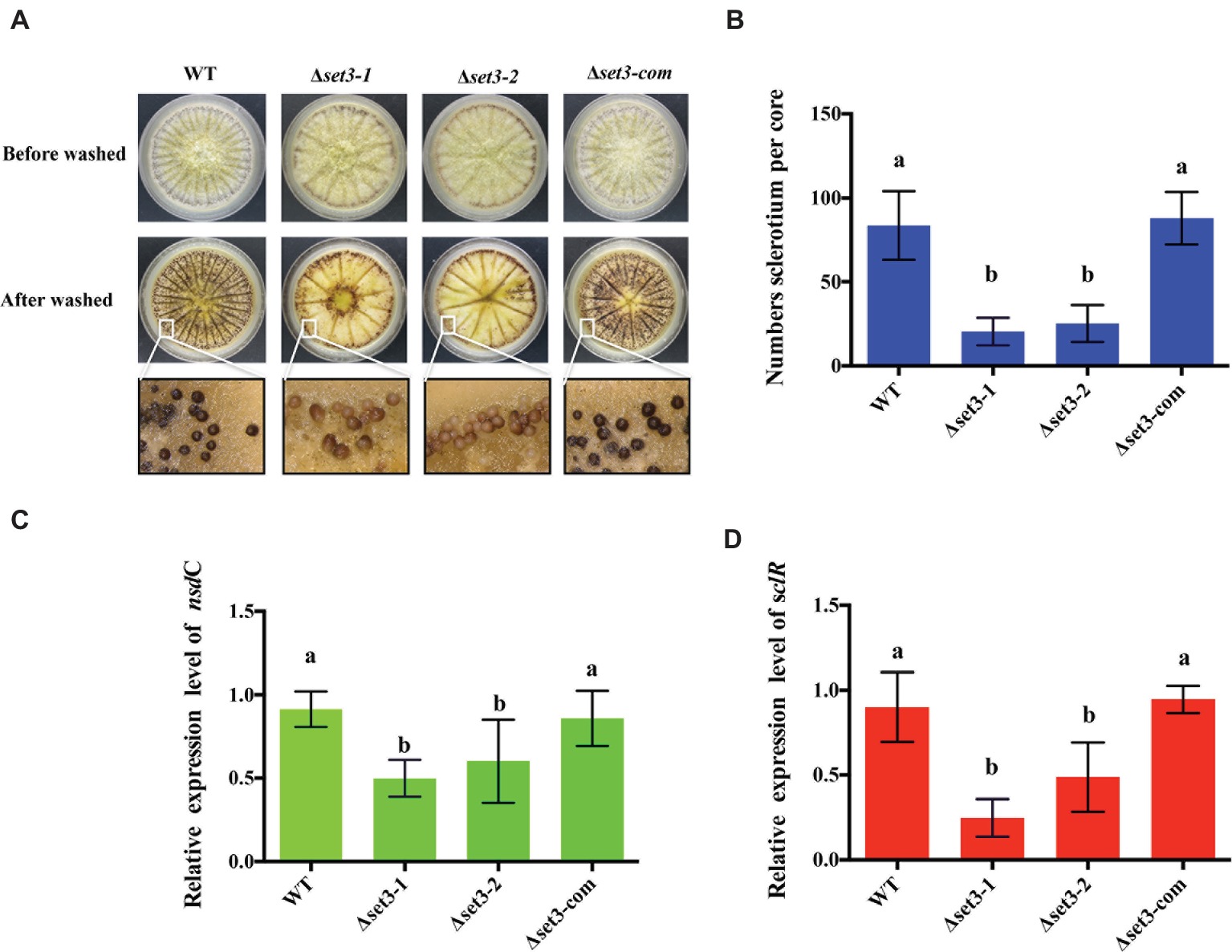
Figure 5. Deletion of set3 caused defects of sclerotia production in A. flavus. (A) Phenotypic analyses of WT, Δset3-1, Δset3-2, and Δset3-com strains grown on WKM media at 37°C for 7 days. (B) Sclerotia production of WT, Δset3-1, Δset3-2, and Δset3-com strains. (C,D) Gene transcript level of sclerotia-related genes nsdC and sclR among WT, Δset3-1, Δset3-2, and Δset3-com strains, respectively. Different letters represent p < 0.05.
Set3 Plays Important Roles in Responses to Oxidative and Cell Wall Stresses
To verify whether A. flavus Set3 was involved in stress responses, we measured several environmental stress responses by adding various stress agents into the tested media. As shown in Figures 6A,B, the Δset3-1 and Δset3-2 strains showed more endurance (p < 0.05) than WT and Δset3-com strains when induced by oxidative stress agents (2.5 mM and 5 mM H2O2), suggesting that the Δset3-1 and Δset3-2 strains were less sensitive to the oxidative stress. Additionally, our results displayed that the relative growth inhibition of the deletion strains was significantly higher (p < 0.05) than that of WT and Δset3-com strains when induced by cell wall integrity stress agent Congo Red (CR, 200 and 500 μg/ml) (Figures 6A,C). Whereas there was no inhibition growth difference among the WT, Δset3-1, Δset3-2, and Δset3-com strains with the addition of osmotic stress (sodium chloride, NaCl) and genotoxic stress (methyl methanesulfonate, MMS) agents (p > 0.05) (data not shown). All these results suggested that Set3 participates in oxidative and cell wall stress responses in A. flavus.
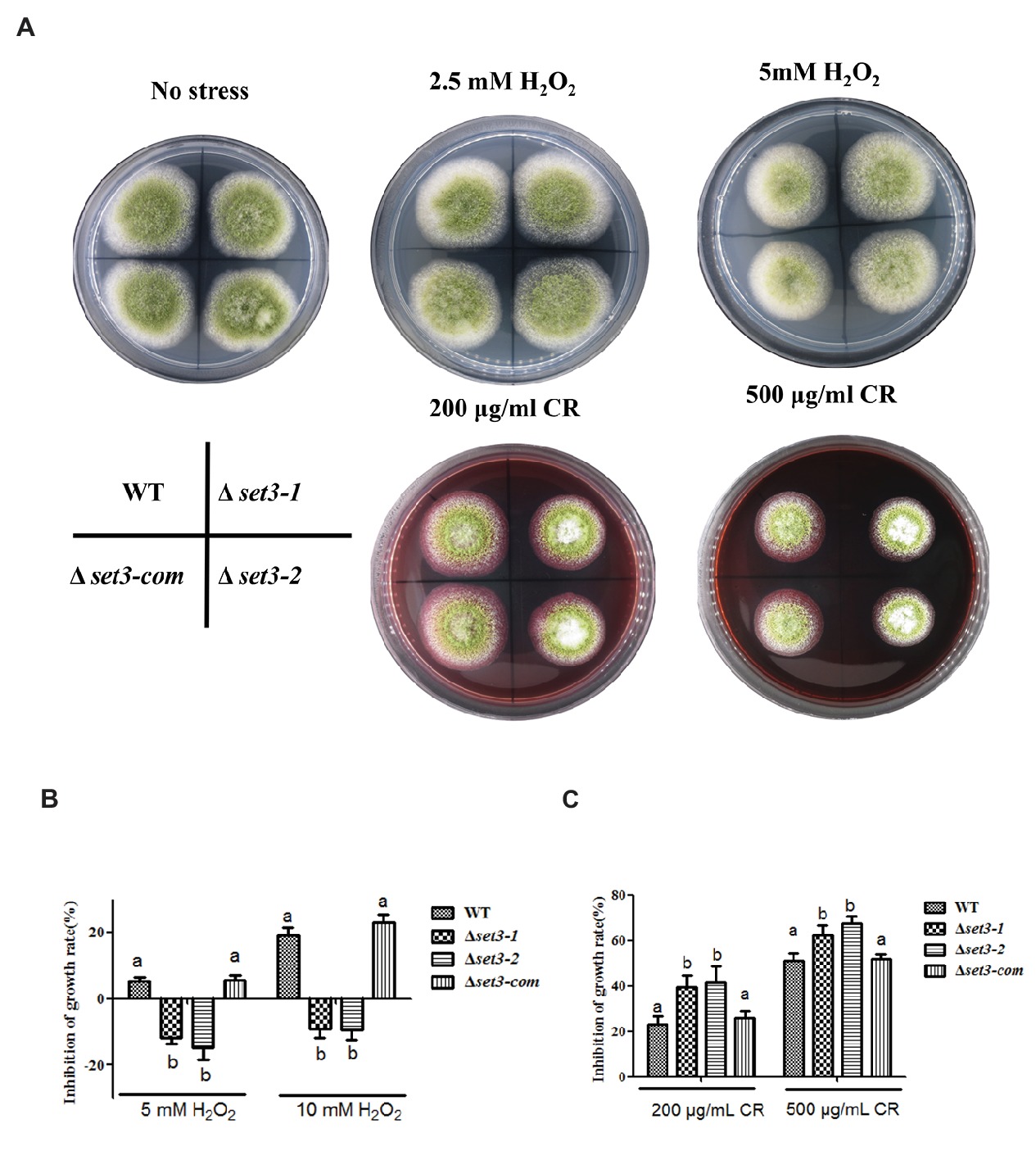
Figure 6. Deletion of set3 affects oxidative and cell wall stress responses in A. flavus. (A) Colony morphology of WT, Δset3-1, Δset3-2, and Δset3-com strains cultured on PDA media with oxidative stress agents (2.5 mM and 5 mM H2O2) and cell wall integrity stresses agent (200 μg/ml and 500 μg/ml CR) at 37°C for 7 days. (B) Inhibition growth rate induced by oxidative stress agents of WT, Δset3-1, Δset3-2, and Δset3-com strains. (C) Inhibition growth rate induced by cell wall integrity stress agent of WT, Δset3-1, Δset3-2, and Δset3-com strains. Different letters represent p < 0.05.
Set3 Negatively Regulates Aflatoxin Production
To examine if Set3 plays a role in aflatoxin (AFs) production, content of AFs in Δset3-1 and Δset3-2 cultures as well as in WT and complemented strains were assayed. The results showed that deletion of set3 gene resulted in a significant increase (>100%) (p < 0.05) in aflatoxin B1 (AFB1) levels in comparison with those in WT and Δset3-com strains (Figures 7A,B). These findings were further confirmed by high performance layer chromatography (HPLC) analysis, showing both AFB1 and aflatoxin B2 (AFB2) production were upregulated in Δset3-1 and Δset3-2 strains (Figure 7C). In addition, we detected transcript levels of genes relevant to aflatoxin biosynthesis. The qRT-PCR results indicated that both Δset3-1 and Δset3-2 strains increased the transcript levels of the candidate genes for AFs biosynthesis, including aflR, aflS, aflC, aflO, aflP, and aflQ, when compared to that of WT and Δset3-com strains (Figure 7D). These above results implied that set3 negatively regulates AF production in A. flavus.
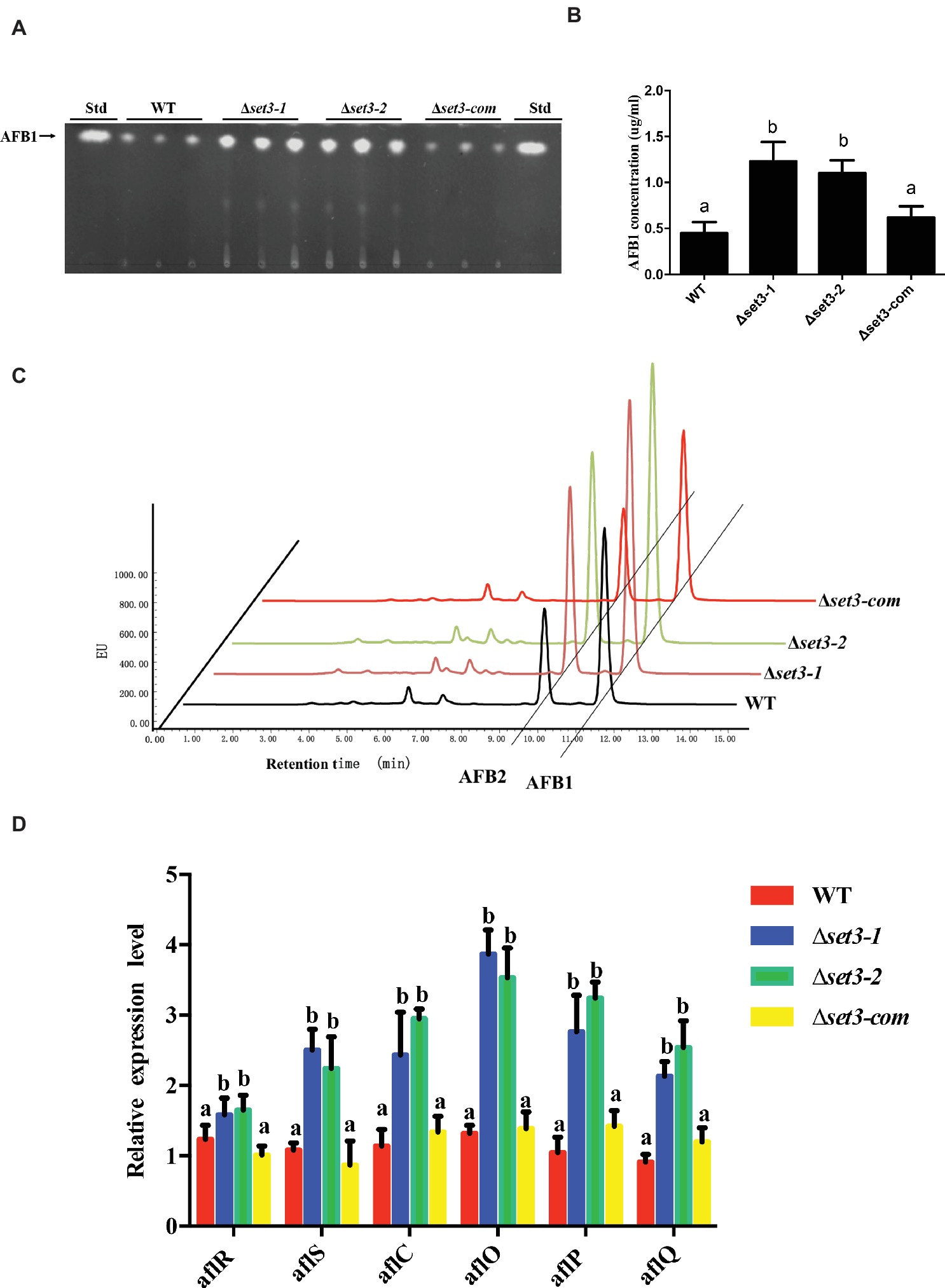
Figure 7. Aflatoxin production of WT, Δset3-1, Δset3-2, and Δset3-com strains. (A) Aflatoxins were detected by thin-layer chromatography (TLC) after grown on YES media for 3 days at 28°C in the dark. (B) Relative aflatoxin production in (A) was qualified. (C) HPLC analysis of aflatoxin production in WT, Δset3-1, Δset3-2, and Δset3-com strains after grown on YES media for 3 days at 28°C in the dark. (D) Transcript level of aflatoxin-related genes aflR, aflS, aflC, aflO, aflP, aflQ from WT, Δset3-1, Δset3-2, and Δset3-com strains. Different letters represent p < 0.05.
Set3 Is Involved in Crop Kernel Colonization
To determine the roles of Set3 in kernel virulence, peanuts and maize kernel seeds were inoculated with WT strain, the Δset3-1, Δset3-2, and Δset3-com strains. Visually, both the Δset3-1 and Δset3-2 strains showed less able to infect and sporulate on host seeds (Figures 8A,D). After 5 days inoculation at 28°C, we assayed conidia amount from the host seeds, and the results showed that Δset3-1 and Δset3-2 strains were impaired to generate the conidia in comparison with the WT and complemented strains (p < 0.05) (Figures 8B,E). The aflatoxin from the infected seeds was subsequently assessed, and the results in Figures 8C,F showed that the Δset3-1 and Δset3-2 strains produced more AF contents (p < 0.05) in both peanut and maize seeds. All these results indicated that set3 in A. flavus is involved in colonization to crops.
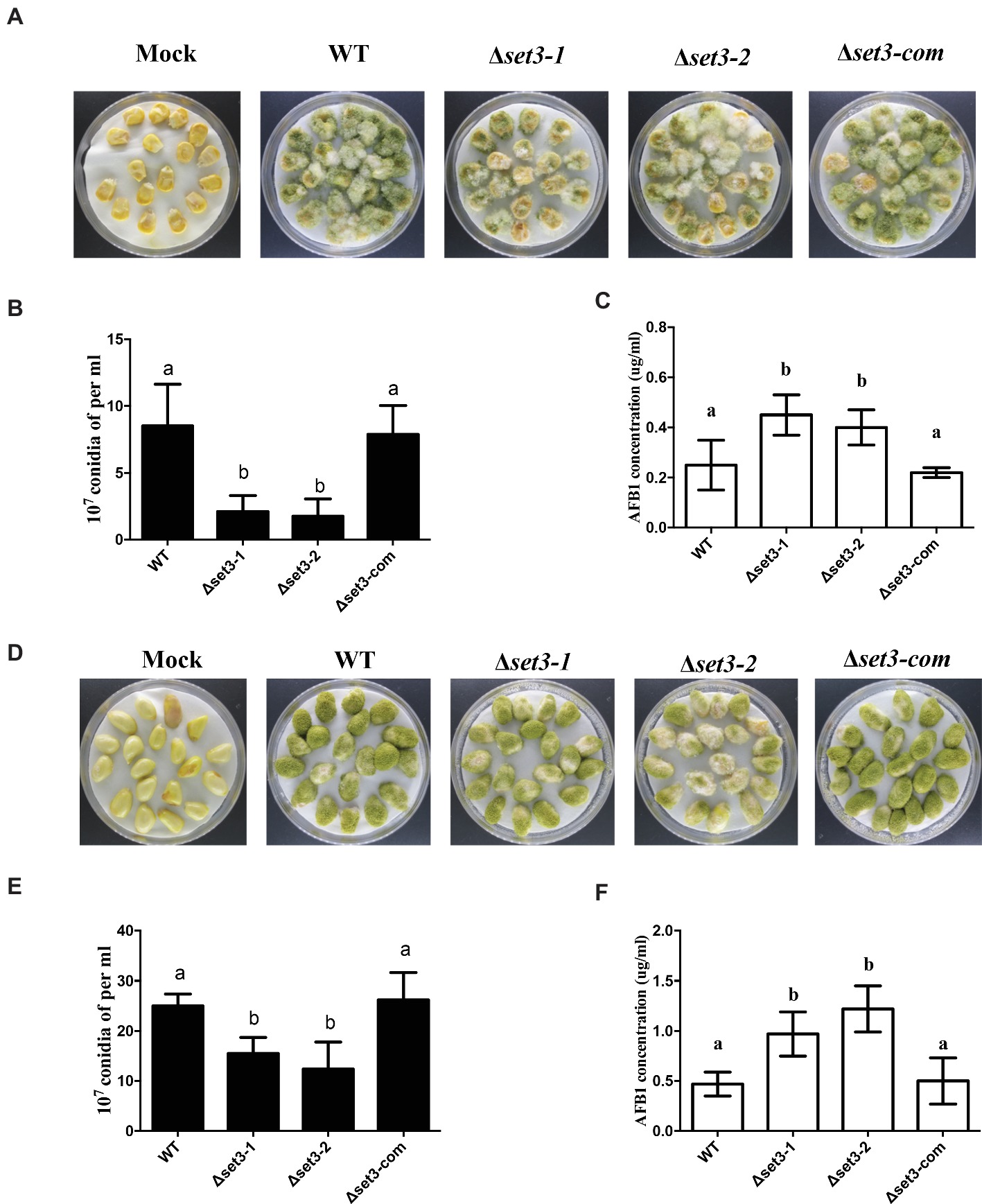
Figure 8. Crops infection of WT, Δset3-1, Δset3-2, and Δset3-com strains. (A) Phenotype of all strains grown on living maize seeds after grown in darkness for 5 days. (B) Amount of conidia was measured from the infected maize seeds. (C) Aflatoxin production was detected from the infected maize seeds. (D) Phenotype of all strains grown on peanut seeds after 5 days in darkness. (E) Amount of conidia was measured from the infected peanut seeds. (F) Aflatoxin production was detected from the infected peanut seeds. Mock represents that crop kernels were inoculated with sterile water as a control group. Different letters represent p < 0.05.
Discussion
During recent years, SET and PHD domain orthologs have been documented to play crucial roles in increasingly organisms from fungi to animals (Pijnappel et al., 2001; Hnisz et al., 2010; Nobile et al., 2014; Yun et al., 2014; McElroy et al., 2017). Our in silico analysis indicated that the predicted Set3 protein sequences were conserved within its corresponding homologs (Figure 1). A. flavus Set3 shows 100% identity to its homolog in important industrial fungus Aspergillus oryzae, and 61% identity to its homolog in the model Aspergillus species A. nidulans. Though it only shares 45% similarity with the model plant species Arabidopsis thaliana and 38% similarity with Drosophila elegans, the whole analyzed organisms harbor the conserved PHD and SET domain, implying that Set3 is important for survival. In yeast, Set3 is a non-essential gene, for survival, while with a mutant phenotype of defective transcription kinetics (Wang et al., 2002; Hnisz et al., 2012). Deletion of upset gene, the Drosophila homolog of SET3, was found to be lethal in both sexes in flies (McElroy et al., 2017). What’s more, MLL5 (SET3 homolog in mammals) has been linked to several different cellular processes, including cell cycle progression (Deng et al., 2007), hematopoiesis (Heuser et al., 2009), oncogenesis (Emerling et al., 2002), and DNA methylation (Yun et al., 2014). In this study, our results indicated that Set3 protein positively regulates conidiation, sclerotial development, and cell wall stress response, whereas it negatively controls AF biosynthesis and oxidative stress response in A. flavus.
Here, we investigated the effects of Set3 on the fungal biology in A. flavus. Deletion of set3 gene produces more hyphal in A. flavus (Figure 3), which means that Set3 functions as a repressor for the hyphal development in filamentous fungi. These findings are consistent with the report on the pleiomorphic fungal pathogen, Candida albicans, which showed that the Set3/Hos2 histone deacetylase complex (Set3C) acts as a crucial repressor of the yeast-to-filament transition (Hnisz et al., 2010), and inactivation of set3 gene resulted in biofilm perturbation in this fungus (Nobile et al., 2014). Our results also revealed that Set3 is a positive regulator of A. flavus asexual development, as a significant reduction in the conidial production of the Δset3 colonies was observed when compared to that of WT (Figure 4). These were accompanied by a reduction in expression of brlA and abaA, essential genes in the central regulatory pathway that controls asexual development (Adams et al., 1988, 1998). Unlike its positive roles in conidiation in A. flavus, Set3C represses genes in early/middle of the yeast sporulation program, including key meiotic regulators Ime2 and Ndt80 (Pijnappel et al., 2001). Besides conidiation, Set3 also engages in sclerotia formation (Figure 5A), sexual development structures that allow this fungus to survive extreme environmental conditions (Wicklow, 1987; Cary et al., 2012), and it was well supported by the obvious downregulation of the sclerotia-related transcription factors nsdC (Figure 5C) and sclR (Figure 5D). All these observations indicated that Set3 plays diverse roles in cellular functions of filamentous fungi.
In natural environments, cells can experience rapidly changing conditions and must correspondingly change their gene expression patterns to adapt (Feil and Fraga, 2012). Set3 binding was enriched for stress-related genes, and it plays both positive and negative roles in cell defense (Hnisz et al., 2012; Kim et al., 2012). Set3C was important in regulating gene induction during the stress response, including changes in the carbon sources (Kim et al., 2012), nitrogen starvation (Pijnappel et al., 2001), and DNA damage (Sharma et al., 2007). Here, deletion of A. flavus set3 caused less sensitive to oxidative stress (Figure 6). Previous study showed that a paralog to Set3 known as Set4 also contains a PHD finger and a divergent SET domain, and it can interact with chromatin, which directly localizes to stress response genes upon regulating ROS (Tran et al., 2018). Oxidative stress response is highly related to reactive oxygen species (ROS) (Schieber and Chandel, 2014). Therefore, it is reasonable to infer that A. flavus Set3 regulates oxidative stress response in the same pathway. On the contrary, Δset3 mutants showed more sensitive to cell wall stress than WT (Figure 6). Exposure to Congo red (CR) lowers the content of cell wall chitin, and the effects of Set3 on A. flavus cell wall integrity may be due to its regulation of the cell wall chitin accumulation factor Smp1 or the oligosaccharyltransferase Stt3 (Hagiwara et al., 2011). All these results suggest the diverse roles of Set3 in environmental stress responses. Therefore, we postulate that Set3 is likely to contribute to each cell defense through distinct molecular mechanisms in A. flavus; however, further investigation will be required to reveal the mechanisms driving the stress-responsive regulation by Set3.
Although the biosynthesis pathway of AFs has been well characterized, the regulatory mechanism is complicated and has not been fully understood. Specially, the involvement of both SET and PHD domain protein was not reported yet in control of secondary metabolism. Our results found that inactivation of Set3 promoted AF production and its related genes’ expression (Figure 7), suggesting that Set3 acted as a repressor in AF biosynthesis. Set3 and HosA, as the core subunit of Set3C histone deacetylase complex, had been shown similar biological functions in most studies (Pijnappel et al., 2001; Cohen et al., 2008; Hnisz et al., 2010; Torres-Machorro et al., 2015). In another study, we identified a key Set3C histone deacetylase component HosA (homolog to Hos2) of A. flavus, unexpectedly, deletion of hosA seriously reduced the AF production. This might be due to that HosA was required for bounding directly to AF biosynthesis cluster genes (data unpublished). We speculated that Set3 and HosA were independently involved in regulation of AF biosynthesis, not only restricted to function as the Set3C complex, but also might play roles in other pathways or functional complexes to control AF biosynthesis. Functional data on SET domain proteins have related to chromatin regulation, and in certain cases, epigenetic mechanisms. Specifically, Set3 proteins have been identified as histone methyltransferase (Kim and Buratowski, 2009), and they participated in Hst1-Sum1 complex (Pijnappel et al., 2001). From the upregulation of AF biosynthesis regulatory genes aflR and aflS in Δset3 strains (Figure 7D), it is possible that inactivation of Set3 may cause alteration of regulatory genes for post-translation modification. Taking together, these results further revealed that regulatory mechanism for AFs biosynthesis is highly complicated.
Previous study had been shown that C. albicans Δset3 displayed strongly attenuated virulence in a mouse model of systemic infection (Hnisz et al., 2010), but the role of Set3 in virulence is still unknown in filamentous fungus. A. flavus has potential to infect oilseed crops by sporulation on injured seeds, therefore, to contaminate the hosts with AFs. Although the physiological significance of these SET domains remains unknown, Set3 may be relevant to fungal virulence of A. flavus, on the basis of the reduction of conidiation and increase of aflatoxin biosynthesis as a result of the inactivation of set3. This idea is further supported by the colonization phenotypes of the set3 mutants on both peanut and maize seeds (Figure 8).
In conclusion, we identified a novel Set3 consisting of a functional SET and a PHD domain in A. flavus. Our results suggested that A. flavus Set3 plays important roles in reproduction, AFs biosynthesis, and fungal virulence and provides a novel sight for developing new fungal control strategies. Whereas further studies are required to discover the SET and PHD protein machinery and the molecular mechanism of Set3 cross-talk with the other crucial signal pathways in A. flavus.
Author Contributions
HL, LW, and SW conceived and designed the experiment. HL, RS, KF, XL, YC, and LW performed the experiments. HL, LW, KY, and SW analyzed the data. HL, JT, FZ, KY, GY, and SW wrote the manuscript.
Funding
The research was supported by the National Natural Science Foundation of China (No. 31772105) and the Natural Science Foundation of Fujian Province, China (No. 2018J07002).
Conflict of Interest Statement
XL was employed by the company Longyan City Corporation of Fujian Tobacco Corporation.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Adams, T. H., Boylan, M. T., and Timberlake, W. E. (1988). brlA is necessary and sufficient to direct conidiophore development in Aspergillus nidulans. Cell 54, 353–362. doi: 10.1016/0092-8674(88)90198-5
Adams, T. H., Wieser, J. K., and Yu, J.-H. (1998). Asexual sporulation in Aspergillus nidulans. Microbiol. Mol. Biol. Rev. 62, 35–54.
Affeldt, K. J., Carrig, J., Amare, M., and Keller, N. P. (2014). Global survey of canonical Aspergillus flavus G protein-coupled receptors. MBio 5, e01501–e01514. doi: 10.1128/mBio.01501-14
Amaike, S., and Keller, N. P. (2011). Aspergillus flavus. Annu. Rev. Phytopathol. 49, 107–133. doi: 10.1146/annurev-phyto-072910-095221
Amare, M. G., and Keller, N. P. (2014). Molecular mechanisms of Aspergillus flavus secondary metabolism and development. Fungal Genet. Biol. 66, 11–18. doi: 10.1016/j.fgb.2014.02.008
Cary, J. W., Harris-Coward, P., Scharfenstein, L., Mack, B. M., Chang, P.-K., Wei, Q., et al. (2017). The Aspergillus flavus homeobox gene, hbx1, is required for development and aflatoxin production. Toxins 9, 315. doi: 10.3390/toxins9100315
Cary, J. W., Harris-Coward, P. Y., Ehrlich, K. C., Mack, B. M., Kale, S. P., Larey, C., et al. (2012). NsdC and NsdD affect Aspergillus flavus morphogenesis and aflatoxin production. Eukaryot. Cell 12, 00069–00012. doi: 10.1128/ec.00069-12
Chang, P.-K., Scharfenstein, L. L., Li, R. W., Arroyo-Manzanares, N., De Saeger, S., and Di Mavungu, J. D. (2017). Aspergillus flavus aswA, a gene homolog of Aspergillus nidulans oefC, regulates sclerotial development and biosynthesis of sclerotium-associated secondary metabolites. Fungal Genet. Biol. 104, 29–37. doi: 10.1016/j.fgb.2017.04.006
Chang, P.-K., Scharfenstein, L. L., Wei, Q., and Bhatnagar, D. (2010). Development and refinement of a high-efficiency gene-targeting system for Aspergillus flavus. J. Microbiol. Methods 81, 240–246. doi: 10.1016/j.mimet.2010.03.010
Cohen, T., Mallory, M., Strich, R., and Yao, T.-P. (2008). Hos2p/Set3p deacetylase complex signals secretory stress through the Mpk1p cell integrity pathway. Eukaryot. Cell 7, 1191–1199. doi: 10.1128/EC.00059-08
Deng, H., Bao, X., Zhang, W., Girton, J., Johansen, J., and Johansen, K. M. (2007). Reduced levels of Su (var) 3-9 but not Su (var) 2-5 (HP1) counteract the effects on chromatin structure and viability in loss-of-function mutants of the JIL-1 histone H3S10 kinase. Genetics 177, 79–87. doi: 10.1534/genetics.107.075143
Emerling, B. M., Bonifas, J., Kratz, C. P., Donovan, S., Taylor, B. R., Green, E. D., et al. (2002). MLL5, a homolog of Drosophila trithorax located within a segment of chromosome band 7q22 implicated in myeloid leukemia. Oncogene 21, 4849–4854. doi: 10.1038/sj.onc.1205615
Feil, R., and Fraga, M. F. (2012). Epigenetics and the environment: emerging patterns and implications. Nat. Rev. Genet. 13, 97–109. doi: 10.1038/nrg3142
Groopman, J. D., Kensler, T. W., and Wild, C. P. (2008). Protective interventions to prevent aflatoxin-induced carcinogenesis in developing countries. Annu. Rev. Public Health 29, 187–203. doi: 10.1146/annurev.publhealth.29.020907.090859
Hagiwara, D., Mizuno, T., and Abe, K. (2011). Characterization of the conserved phosphorylation site in the Aspergillus nidulans response regulator SrrA. Curr. Genet. 57, 103–114. doi: 10.1007/s00294-010-0330-2
Hedayati, M., Pasqualotto, A., Warn, P., Bowyer, P., and Denning, D. (2007). Aspergillus flavus: human pathogen, allergen and mycotoxin producer. Microbiology 153, 1677–1692. doi: 10.1099/mic.0.2007/007641-0
Heuser, M., Yap, D. B., Leung, M., De Algara, T. R., Tafech, A., Mckinney, S., et al. (2009). Loss of MLL5 results in pleiotropic hematopoietic defects, reduced neutrophil immune function, and extreme sensitivity to DNA demethylation. Blood 113, 1432–1443. doi: 10.1182/blood-2008-06-162263
Hnisz, D., Bardet, A. F., Nobile, C. J., Petryshyn, A., Glaser, W., Schöck, U., et al. (2012). A histone deacetylase adjusts transcription kinetics at coding sequences during Candida albicans morphogenesis. PLoS Genet. 8:e1003118. doi: 10.1371/journal.pgen.1003118
Hnisz, D., Majer, O., Frohner, I. E., Komnenovic, V., and Kuchler, K. (2010). The Set3/Hos2 histone deacetylase complex attenuates cAMP/PKA signaling to regulate morphogenesis and virulence of Candida albicans. PLoS Pathog. 6:e1000889. doi: 10.1371/journal.ppat.1000889
Horn, B. W., Moore, G. G., and Carbone, I. (2009). Sexual reproduction in Aspergillus flavus. Mycologia 101, 423–429. doi: 10.3852/09-011
Kim, T., and Buratowski, S. (2009). Dimethylation of H3K4 by Set1 recruits the Set3 histone deacetylase complex to 5′ transcribed regions. Cell 137, 259–272. doi: 10.1016/j.cell.2009.02.045
Kim, T., Xu, Z., Clauder-Münster, S., Steinmetz, L. M., and Buratowski, S. (2012). Set3 HDAC mediates effects of overlapping noncoding transcription on gene induction kinetics. Cell 150, 1158–1169. doi: 10.1016/j.cell.2012.08.016
Klich, M. A. (2007). Aspergillus flavus: the major producer of aflatoxin. Mol. Plant Pathol. 8, 713–722. doi: 10.1111/j.1364-3703.2007.00436.x
Lan, H., Sun, R., Fan, K., Yang, K., Zhang, F., Nie, X. Y., et al. (2016). The Aspergillus flavus histone acetyltransferase AflGcnE regulates morphogenesis, aflatoxin biosynthesis, and pathogenicity. Front. Microbiol. 7:1324. doi: 10.3389/fmicb.2016.01324
Mcelroy, K. A., Jung, Y. L., Zee, B. M., Wang, C. I., Park, P. J., and Kuroda, M. I. (2017). upSET, the Drosophila homologue of SET3, is required for viability and the proper balance of active and repressive chromatin marks. G3 116, 625–635. doi: 10.1534/g3.116.037788
Mou, Z., Kenny, A. E., and Curcio, M. J. (2006). Hos2 and Set3 promote integration of Ty1 retrotransposons at tRNA genes in Saccharomyces cerevisiae. Genetics 172, 2157–2167. doi: 10.1534/genetics.105.054072
Nie, X., Li, B., and Wang, S. (2018). Epigenetic and posttranslational modifications in regulating the biology of Aspergillus species. Adv. Appl. Microbiol. 105, 191–226. doi: 10.1016/bs.aambs.2018.05.004
Nobile, C. J., Fox, E. P., Hartooni, N., Mitchell, K. F., Hnisz, D., Andes, D. R., et al. (2014). A histone deacetylase complex mediates biofilm dispersal and drug resistance in Candida albicans. MBio 5, e01201–e01214. doi: 10.1128/mBio.01201-14
Payne, G., and Brown, M. (1998). Genetics and physiology of aflatoxin biosynthesis. Annu. Rev. Phytopathol. 36, 329–362. doi: 10.1146/annurev.phyto.36.1.329
Pfannenstiel, B. T., Greco, C., Sukowaty, A. T., and Keller, N. P. (2018). The epigenetic reader SntB regulates secondary metabolism, development and global histone modifications in Aspergillus flavus. Fungal Genet. Biol. 120, 9–18. doi: 10.1016/j.fgb.2018.08.004
Pijnappel, W. P., Schaft, D., Roguev, A., Shevchenko, A., Tekotte, H., Wilm, M., et al. (2001). The S. cerevisiae SET3 complex includes two histone deacetylases, Hos2 and Hst1, and is a meiotic-specific repressor of the sporulation gene program. Genes Dev. 15, 2991–3004. doi: 10.1101/gad.207401
Roze, L. V., Beaudry, R. M., Keller, N. P., and Linz, J. E. (2004). Regulation of aflatoxin synthesis by FadA/cAMP/protein kinase A signaling in Aspergillus parasiticus. Mycopathologia 158, 219–232. doi: 10.1023/B:MYCO.0000041841.71648.6e
Schieber, M., and Chandel, N. S. (2014). ROS function in redox signaling and oxidative stress. Curr. Biol. 24, R453–R462. doi: 10.1016/j.cub.2014.03.034
Sharma, V. M., Tomar, R. S., Dempsey, A. E., and Reese, J. C. (2007). Histone deacetylases RPD3 and HOS2 regulate the transcriptional activation of DNA damage-inducible genes. Curr. Biol. 24, R453–R462. doi: 10.1128/MCB.02311-06
Szewczyk, E., Nayak, T., Oakley, C. E., Edgerton, H., Xiong, Y., Taheri-Talesh, N., et al. (2006). Fusion PCR and gene targeting in Aspergillus nidulans. Nat. Protoc. 1, 3111–3120. doi: 10.1038/nprot.2006.405
Torres-Machorro, A. L., Clark, L. G., Chang, C. S., and Pillus, L. (2015). The set3 complex antagonizes the MYST acetyltransferase Esa1 in the DNA damage response. Mol. Cell. Biol. 35, 3714–3725. doi: 10.1128/mcb.00298-15
Tran, K., Jethmalani, Y., Jaiswal, D., and Green, E. M. (2018). Set4 is a chromatin-associated protein, promotes survival during oxidative stress, and regulates stress response genes in yeast. J. Biol. Chem. 293, 14429–14443. doi: 10.1074/jbc.RA118.003078
Wang, A., Kurdistani, S. K., and Grunstein, M. (2002). Requirement of Hos2 histone deacetylase for gene activity in yeast. Science 298, 1412–1414. doi: 10.1126/science.1077790
Wicklow, D. T. (1987). Survival of Aspergillus flavus sclerotia in soil. Br. Mycol. Soc. 89, 131–134.
Wu, F., Groopman, J. D., and Pestka, J. J. (2014). Public health impacts of foodborne mycotoxins. Annu. Rev. Food Sci. Technol. 5, 351–372. doi: 10.1146/annurev-food-030713-092431
Yang, K., Liang, L., Ran, F., Liu, Y., Li, Z., Lan, H., et al. (2016a). The DmtA methyltransferase contributes to Aspergillus flavus conidiation, sclerotial production, aflatoxin biosynthesis and virulence. Sci. Rep. 6:23259. doi: 10.1038/srep23259
Yang, K., Qin, Q., Liu, Y., Zhang, L., Liang, L., and Lan, H. (2016b). Adenylate cyclase AcyA regulates development, aflatoxin biosynthesis and fungal virulence in Aspergillus flavus. Front. Cell. Infect. Microbiol. 6, 190. doi: 10.3389/fcimb.2016.00190
Yu, J., Chang, P.-K., Ehrlich, K. C., Cary, J. W., Bhatnagar, D., Cleveland, T. E., et al. (2004). Clustered pathway genes in aflatoxin biosynthesis. Appl. Environ. Microbiol. 70, 1253–1262. doi: 10.1128/AEM.70.3.1253-1262.2004
Yu, Y., Zhou, H., Deng, X., Wang, W., and Lu, H. (2016). Set3 contributes to heterochromatin integrity by promoting transcription of subunits of Clr4-Rik1-Cul4 histone methyltransferase complex in fission yeast. Sci. Rep. 6:31752. doi: 10.1038/s41598-016-0011-6
Yun, H., Damm, F., Yap, D., Schwarzer, A., Chaturvedi, A., Jyotsana, N., et al. (2014). Impact of MLL5 expression on decitabine efficacy and DNA methylation in acute myeloid leukemia. Haematologica 99, 1456–1464. doi: 10.3324/haematol.2013.101386
Keywords: Set3, regulate, reproduction, aflatoxin biosynthesis, Aspergillus flavus
Citation: Lan H, Wu L, Fan K, Sun R, Yang G, Zhang F, Yang K, Lin X, Chen Y, Tian J and Wang S (2019) Set3 Is Required for Asexual Development, Aflatoxin Biosynthesis, and Fungal Virulence in Aspergillus flavus. Front. Microbiol. 10:530. doi: 10.3389/fmicb.2019.00530
Edited by:
Weiguo Fang, Zhejiang University, ChinaReviewed by:
Massimo Reverberi, Sapienza University of Rome, ItalyMaureen Wright, Agricultural Research Service, United States Department of Agriculture, United States
Copyright © 2019 Lan, Wu, Fan, Sun, Yang, Zhang, Yang, Lin, Chen, Tian and Wang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shihua Wang, d3NoeXlsQHNpbmEuY29t
†These authors have contributed equally to this work
 Huahui Lan
Huahui Lan Lianghuan Wu1†
Lianghuan Wu1† Kunlong Yang
Kunlong Yang Jun Tian
Jun Tian Shihua Wang
Shihua Wang