- 1Integrated Food Animal Management Systems, Department of Veterinary Clinical Medicine, College of Veterinary Medicine, University of Illinois at Urbana-Champaign, Champaign, IL, United States
- 2Department of Animal Medicine, College of Veterinary Medicine, Benha University, Benha, Egypt
Antimicrobials are the most commonly prescribed drugs in the swine industry. While antimicrobials are an effective treatment for serious bacterial infections, their use has been associated with major adverse effects on health. It has been shown that antimicrobials have substantial direct and indirect impacts on the swine gastrointestinal (GI) microbiota and their accompanying antimicrobial resistome. Antimicrobials have also been associated with a significant public health concern through selection of resistant opportunistic pathogens and increased emergence of antimicrobial resistance genes (ARGs). Since the mutualistic microbiota play a crucial role in host immune regulation and in providing colonization resistance against potential pathogens, the detrimental impacts of antimicrobial treatment on the microbiota structure and its metabolic activity may lead to further health complications later in life. In this review, we present an overview of antimicrobial use in the swine industry and their role in the emergence of antimicrobial resistance. Additionally, we review our current understanding of GI microbiota and their role in swine health. Finally, we investigate the effects of antimicrobial administration on the swine GI microbiota and their accompanying antibiotic resistome. The presented data is crucial for the development of robust non-antibiotic alternative strategies to restore the GI microbiota functionality and guarantee effective continued use of antimicrobials in the livestock production system.
Introduction
Recently, the swine industry has focused on sustainable pork production which maximizes value over production costs and represents a shift away from antimicrobial usage. There is an urgent need not only for higher production efficiency to meet consumer expectations, but also for the development of new phenotypes related to host vitality and robustness (Merks et al., 2012). Phenotypic development in swine is a complex multistage process, starting from conception stage and continuing throughout the entire production cycle (Pluske, 2016). There are four major criteria that drive the phenotypic development and ultimately impact swine health, including host factors, management inputs, stable microbial ecosystem, and surrounding physical environment (Figure 1). Some human data and animal experiments have revealed that the crosstalk and interaction between microbial environment and other phenotypic drivers are the key distinguishers of host health (Blaut and Clavel, 2007; Metzler and Mosenthin, 2008). The swine microbial ecosystem is composed of rich and diverse populations that harbor thousands of different microbial species (aerobic, facultative anaerobic, and strictly anaerobic), dwelling in different anatomical biogeographic locations (Metzler and Mosenthin, 2008; Holman et al., 2017). These mutualist populations have a wide range of functions, including providing colonization resistance against potential pathogens, absorbing different kind of nutrients, modulation of the host's immune system, metabolizing indigestible polysaccharides, and regulating the host's metabolism (Bischoff, 2011; Venable et al., 2016). Therefore, alteration of the swine microbial environment may detrimentally influence the host's health status and inhibit the pathogens colonization (Marchesi et al., 2016). Understanding the mechanistic pathways and abundance of these alternations are required to discover new and different management practices to promote growth rate, increase efficiency of feed utilization, and improve overall swine health.
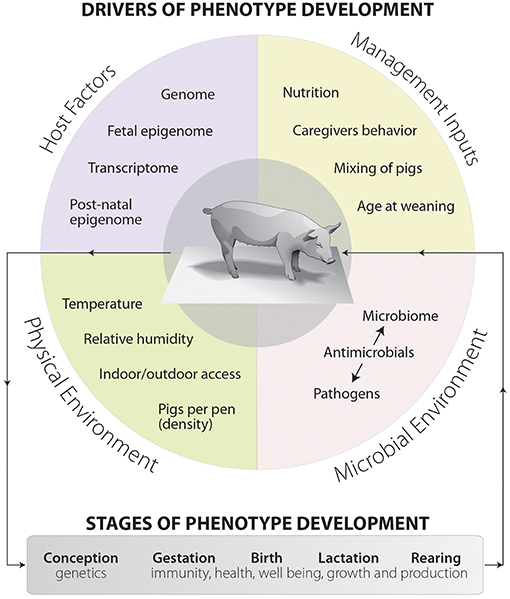
Figure 1. The drivers and main components of phenotype development in swine production systems. Phenotype development is made up of four separate but often blended sets of factors and overlapping components. These components are host factors, management inputs, microbial environment, and physical environment. The dashed arrows represent interactions between drivers and can occur between any drivers. This diagram also shows the stages of phenotype development, starting from conception and moving toward rearing.
With recent advances in our understanding of swine microbial ecosystem structures and functions, we are becoming increasingly aware of the impacts of antimicrobial on mucosal microbiota and how its use negatively impacts the host's health (Zeineldin et al., 2018b). Equally important is the potential enrichment of antimicrobial resistome between the commensal microbiota as a result of antimicrobial use, which is one of the most vital public health issues that we currently face (Wright, 2007). The detrimental impacts of antimicrobial on the GI microbiota and host health are summarized in Figure 2. Traditionally, the impacts of antimicrobial administration on GI microbiota structures and development of antimicrobial resistance were largely characterized by culture-based techniques and/or a PCR-based approach, both of which underestimate the presence of novel ARGs (Zhu et al., 2013). Consequently, culture independent platforms (real-time PCR quantification, next generation sequencing, and functional metagenomics) have been used to efficiently quantify and assess the resistant opportunistic pathogens and emergence of antimicrobial resistome (Gerzova et al., 2015). While antimicrobial intervention disrupts GI microbiota structures and function, we are just beginning to estimate the relative contribution of its use on emergence of the antimicrobial resistome.
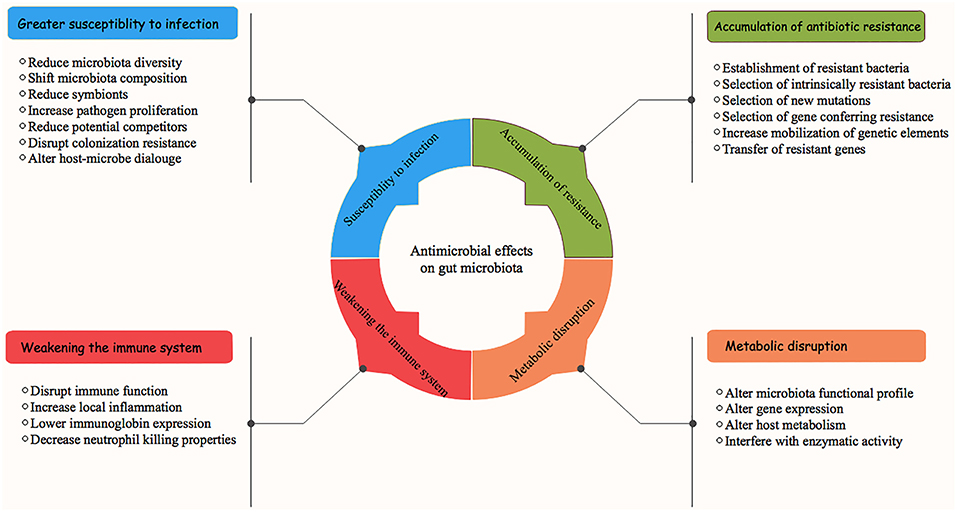
Figure 2. Antimicrobial effects on swine gastrointestinal microbiota and associated health consequences.
In this review, we present an overview of antimicrobial use in the swine industry and its association with the emergence of antibiotic resistance genes (ARGs). Additionally, we review our current understanding of GI microbiota and its role in swine health. Finally, we explore the effects of antimicrobial use on the swine GI microbiota and their accompanying antimicrobial resistome. The presented data is vital for the development of robust non-antibiotic alternative strategies to restore the GI microbiota functionality and guarantee effective continued use of antimicrobial in the livestock production system.
Antimicrobial Use in Swine Management System
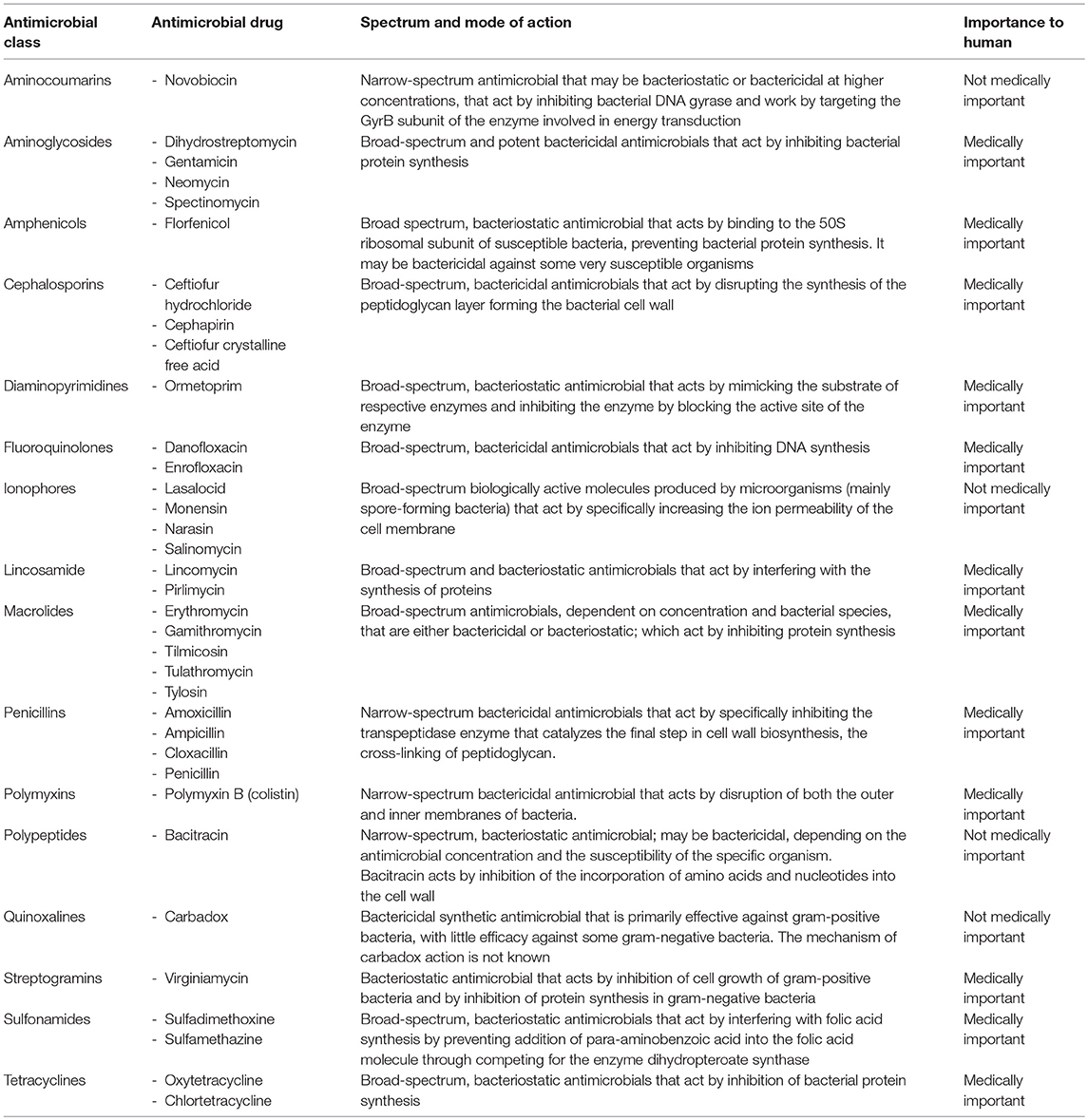
In the swine industry, antimicrobial has four potential uses: (1) disease treatment, (2) disease control, (3) disease prevention, and (4) increased the growth performance (O'Neill, 2014). It is therefore unsurprising that antimicrobial is the most commonly prescribed drug in the swine industry (Dumas et al., 2016). It is estimated that all food-producing animals consume more than 70% of antimicrobial produced worldwide. The pigs are usually raised in groups, in close proximity to one another. Many production systems use all-in, all-out management to control and prevent infectious disease outbreaks (Dewey et al., 1999). However, high contact rates provide optimal conditions for the spread of infectious diseases, many of which require the use of antimicrobials to minimize economic losses and welfare concerns. Estimates range from 62% of nursery production units and 44% of grower/finisher units (McEwen and Fedorka-Cray, 2002) to 33% of nursery units and 30% of grower/finisher units use of antimicrobial for growth promotion (Holman and Chénier, 2015). Data collected in 2001 by the USDA for US herds found that 70% used antimicrobials in starter feeds, 59% used them in grower/finisher feeds, and 46% used them in sow feeds (Cromwell, 2002), which were higher than the estimates of McEwen and Fedorka-Cray in similar populations (McEwen and Fedorka-Cray, 2002). According to the Food and Drug Administration (FDA), the available antimicrobial classes and chemotherapeutic agents (chemically synthesized agents with antimicrobial activity) for use in swine are listed in Table 1 (FDA, 2017). Certain classes of these antimicrobial are approved and validated for their ability to be successfully combined with other antimicrobials (e.g., chlortetracycline, penicillin, and sulfamethazine), whereas others cannot be combined with other antimicrobials.
The antimicrobial spectrum, administration dosage, pharmacokinetics and pharmacodynamics vary greatly according to different antimicrobial classes and their chemical structures (Cromwell, 2002). Some antimicrobials are easily absorbed after both therapeutic and subtherapeutic administration (e.g., oxytetracyclines and sulfonamides), whereas other antimicrobials are poorly absorbed after administration (e.g., bacitracin). In swine industry, the duration of antimicrobial administration typically ranges from 20 to 40 days for disease prevention and control (Stone et al., 2009). Alternatively, for growth promotion, antimicrobials are generally used for a long period of time at relatively low concentrations. While the mode of action of antimicrobial growth promotion remains poorly characterized, several potential mechanisms have been proposed. These mechanisms include decreased production of harmful metabolites (metabolic effect), increased absorption of available dietary nutrients (nutritional effect), and reduction of endemic subclinical diseases (disease control effect; Dibner and Richards, 2005). It is remarkable that antimicrobial use as a growth promoter in younger pigs is consistently efficacious while little to no response is seen in older animals (Cromwell, 2002; Skinner et al., 2014). In growing piglets, the average duration of antimicrobial use for growth promotion ranges from 22.7 to 76.8 days (Dewey et al., 1997). This prolonged exposure to sub-therapeutic antimicrobial concentrations provides ample opportunity for antimicrobial resistance to develop, particularly when compared to therapeutic use (Aarestrup et al., 2008). Consequently, there is increasing consumer desire to make sub-therapeutic antimicrobial use less frequent in livestock production (Sommer et al., 2017).
Several published studies have addressed the safety of antimicrobials, all of which could not identify a direct link between antimicrobial use in livestock and human health (Phillips et al., 2004; Chang et al., 2015). In contrast, a systematic review about restricting antibiotic use in animals and its association with antibiotic resistance in human beings concluded that antimicrobial use in food-producing animals is recognized as one of the major contributors to development of resistant organisms that result in life-threatening human infections (Landers et al., 2013). But, in general, it seems inevitable that antimicrobial administration in animals and its relationship to human health remain unquantified.
Association Between Antimicrobial Use and Antimicrobial Resistance
Since the discovery of antimicrobials, the main goal of its use in the swine industry has been to eliminate pathogenic microbes, thereby facilitating growth and restoration of beneficial microbial communities (Holman and Chénier, 2015). However, these goals are routinely complicated by presence and dissemination of ARGs among microbes (McEwen and Fedorka-Cray, 2002). Resistance to antimicrobials is a natural occurrence, developed by the microbes to help in their survival against other antibiotic-producing microorganisms in the surrounding environment (Phillips et al., 2004). In many cases, detection of clinical signs for a disease in an individual animal provokes prophylactic treatment for the whole herd (Founou et al., 2016). This approach can increase abundance of resistant bacterial strains and elevate the expression of ARGs (Langdon et al., 2016).
Traditionally, the impacts of antimicrobial treatment on emergence of antimicrobial-resistant bacteria have focused only on pathogenic bacteria (e.g., Salmonella, E. coli, Shigella, and Enterobacter; Founou et al., 2016). Many researches have investigated the association between antimicrobial use in livestock and development of antimicrobial resistance across the resident microbiota (Everaert et al., 2017; Johnson et al., 2017). When an antimicrobial is administered, it eliminates the susceptible microbial populations, leaving behind resistant strains that continue to evolve and multiply in its number (Founou et al., 2016). Selective pressure from antimicrobial exposure is exploited by antimicrobial-resistant microbes, providing them with an evolutionary advantage (Brandl et al., 2008). The resistant microbes, in presence of antimicrobials, also have a competitive advantage which facilitates its spread among other microbial populations in the surrounding ecosystem (Holmes et al., 2016). The dissemination of ARGs requires acquisition or transfer of genetic elements encoding antimicrobial resistance between the bacterial strains. The resistant bacterial populations transmit their genetic resistance pools to their progeny through vertical evolution or to other adherent bacterial species through horizontal transmission (D'Costa et al., 2007). Vertical gene transfer occurs during cell division, where resistant genes either on chromosomes or plasmids transfer to the progeny cells, leading to bacterial resistance (Lawrence, 2004). Alternately, horizontal gene transfer involves genetic pool exchange within and between the microbial populations, where genetic density and complexity of the commensal microbial community stimulate the spread of ARGs among microbes (Founou et al., 2016). The resistant genetic material is usually acquired by microbes either through conjugation, transformation, and/or transduction (Holmes et al., 2016). It is then possible for new mobile genetic element-associated transmission of antimicrobial resistance determinants to be incorporated into the bacterial chromosome or replicate independently (Sommer and Dantas, 2011). The presence of mobile genetic elements (plasmids, integrative conjugated elements, transposons, and integrons) are therefore important in transmission of antimicrobial resistance among microbes (D'Costa et al., 2007). The reservoirs of antimicrobial-resistant bacteria are ubiquitous and can merge with the GI resident microbiota through two different mechanisms (Holman and Chénier, 2015). First, the resistant bacteria can be acquired directly by the host and colonize the GI mucosal epithelium; secondly, a previously susceptible bacterial species can become resistant through induction of antibiotic-resistant mutants or through resistant gene transfer events (Crofts et al., 2017). While there is a clear association between the use of antimicrobial and emergence of antimicrobial resistance, this relationship is complex and influenced by multiple confounding factors (e.g., pathogen-host interactions, pathogen–drug interactions, rate of mutation, rate of transmission, cross-resistance, and co-selection of resistance to unrelated drugs; Holmes et al., 2016).
Effect of Antimicrobial Intervention on Swine Gastrointestinal Microbiota
The term microbiome is widely used to describe the resident populations of different organisms (bacteria, viruses, fungi, archaea, and protists) that live and/or colonize the body of multicellular host and their genetic material (Turnbaugh et al., 2007). Swine GI microbiota is not uniform and differs drastically between individuals, even individuals raised in the same management system. Additionally, the relative abundance of specific bacteria differ according to different GI biogeographic locations (Leser et al., 2002; Maradiaga et al., 2018; Yeoman et al., 2018), with richer and more diverse communities in the colon compared to the ileum and stomach (Holman and Chénier, 2015). Understanding how GI microbiome composition affects swine health is an emerging area of research (Isaacson and Kim, 2012; Zeineldin et al., 2017a). However, the exact mechanisms of how GI microbiota contributes to swine health are still unclear. There are new studies endeavoring to increase our understanding about this mechanism (Pluske et al., 2018). GI mutualistic microbiota play an important function in bile salt recycling, volatile fatty acid production, cellulose digestion, metabolism of undigested carbohydrates, and nutrient recovery (Bischoff, 2011). Additionally, GI microbiota contribute to resistance against colonization of pathogenic microbes through competition for binding sites, nutrient utilization at mucosal epithelium, and modification of local environment (Mach et al., 2015). Therefore, understanding different factors that shape swine GI microbiota and their composition, particularly in early life, are required to discover new targets and/or develop novel management practices to promote optimal GI microbiota development.
With the advancement of methodologies to assess microbiota composition (Zeineldin et al., 2017b), several considerations have been raised regarding the impact of antimicrobial administration on the resident microbial populations in swine (Bokulich et al., 2016). There are several reports and longitudinal studies that attempt to understand the impacts of antimicrobial intervention on swine GI microbiota (Gerzova et al., 2015; Holman and Chénier, 2015; Oultram et al., 2015; Bokulich et al., 2016; Founou et al., 2016; Holman et al., 2018; Zeineldin et al., 2018a). Table 2 lists a summary of the existing metagenomic studies on the impacts of antimicrobial administration on swine GI microbial communities. Commonly, antimicrobial is given to wipe out pathogenic microbes during acute infection (Dewey et al., 1999). However, several antimicrobial classes are not specific, and consequently wipe out a wide range of resident GI microbiota that are beneficial and pivotal for health (Neuman et al., 2018). Recently, a comprehensive review by Langdon et al. revealed that short and long term antimicrobial intervention in humans drastically changes both adult and neonatal microbiota structure (Leibovitz et al., 2003; Langdon et al., 2016). This shift has been associated with an increased chance of subsequent GI disease (Pettigrew et al., 2012). Although the shifts in microbiota composition occurred after antimicrobial administration, some populations have returned to a pretreatment state within 4 weeks following a single-dose treatment. Other taxa, meanwhile, failed to return to pretreatment levels even after 6 months following treatment (Jernberg et al., 2010). Similarly, shifts in the GI microbiota in other animals after antimicrobial administration (a combination of metronidazole, amoxicillin and bismuth) dissipated after cessation of treatment (Schmidt et al., 2009). The precise components responsible for GI microbiota recovery after antimicrobial administration are still undefined. Recognition of different factors that promote microbiota recovery after antimicrobial administration open up new opportunities for development of novel therapies that promote the GI health.
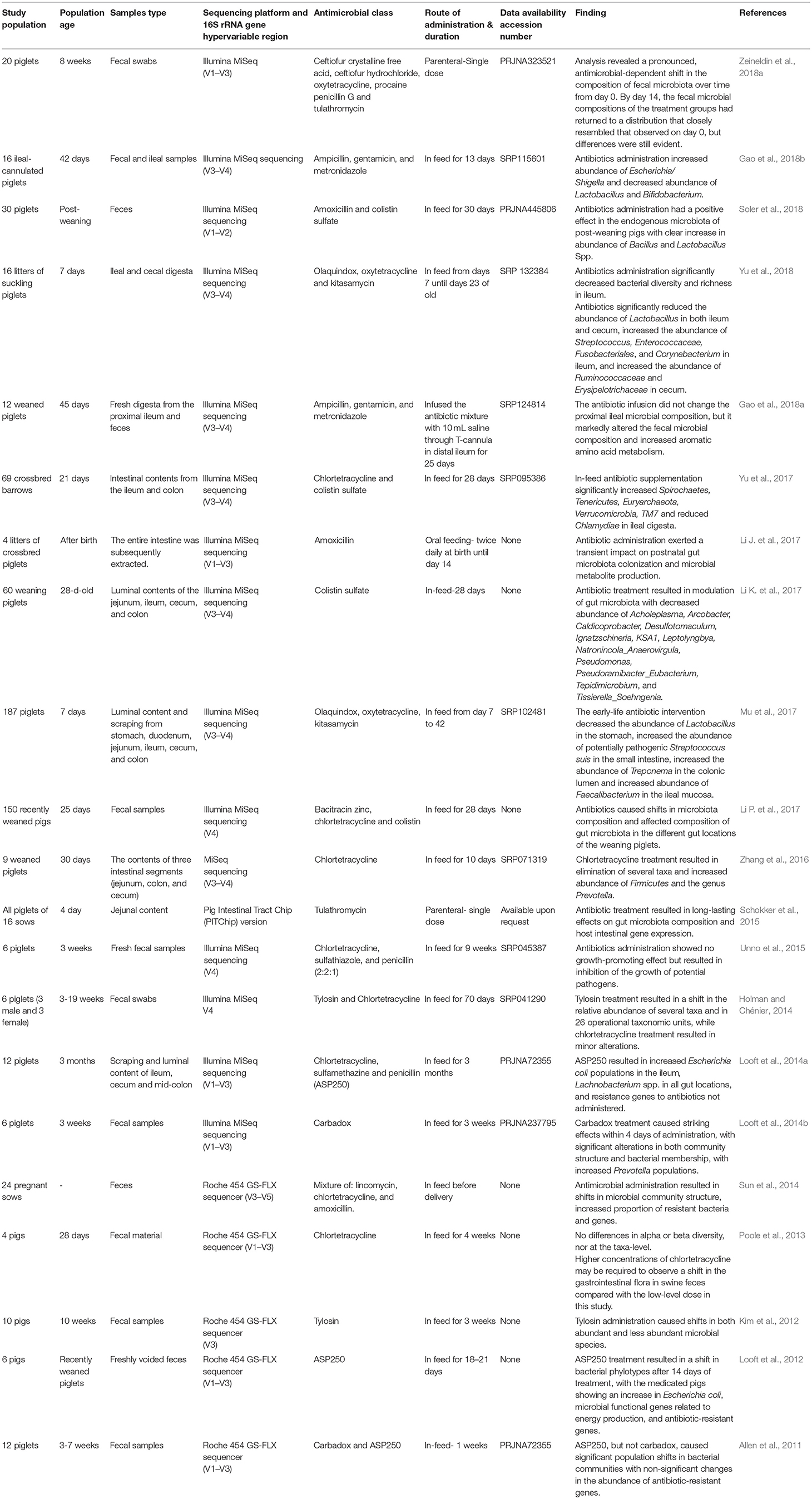
Table 2. Summary of the existing metagenomic studies on the impacts of antimicrobial intervention on the swine gastrointestinal microbiota.
It is important, when quantifying the impacts of antimicrobial intervention on swine GI microbiota structure, to consider the ages of the studied populations, route of administration and the class of the administered antimicrobial (Neuman et al., 2018). While there are some similarities between the effects of antimicrobial administration on GI microbiota structure in growing and neonatal piglets, there are also significant dissimilarities due to distinct characteristics of the neonatal microbial composition. A recent study of 16 42-day-old ileal-cannulated pigs demonstrated that oral administration of ampicillin, gentamicin, and metronidazole treatment modified GI microbial population structure and function (Gao et al., 2018b). More precisely, use of ampicillin, gentamicin, and metronidazole decreased the Lactobacillus and Bifidobacterium abundance and increased the abundance of Shigella species by 256-fold compared to the control pigs (Gao et al., 2018b). Similarly, early life amoxicillin administration in neonatal piglets during the first 14 days of life exerted transient impacts on developing gut microbiota and decreased the genes involved in short-chain fatty acid signaling and pancreatic development (Li J. et al., 2017). In neonatal piglets, early life antimicrobial administration also resulted in differential dysbiosis of GI microbiota, with major alteration between different geographical locations. For instance, a mixture of olaquindox, kitasamycin, and oxytetracycline calcium administration decreased the relative abundance of beneficial Lactobacillus species and increased the relative abundance of potentially pathogenic Streptococcus suis in both the small intestine and stomach lumen (Mu et al., 2017). In growing piglets, antimicrobial administration also induced microbiota compositional changes in both abundant and less abundant GI microbiota. For example, tylosin-treated piglets showed higher relative abundance of Lactobacillus, Eggerthella, Acetanaerobacterium, and Sporacetigenium genera compared to control piglets (Kim et al., 2012). A mixture of amoxicillin and colistin sulfate treatment in post-weaning piglets also resulted in different digestive microbiota profiles along the entire gastrointestinal tract (Soler et al., 2018). Similarly, in-feed administration of colistin sulfate and bacitracin zinc in weaned piglets caused a significant shift in GI microbiota composition along different biogeographic gut locations (Li K. et al., 2017).
Published data also suggested that different classes of antimicrobial disrupt GI microbiota in different ways. This should be included in the decision-making process for antimicrobial prescription in livestock management systems. When assessing the impacts of in-feed sub-therapeutic concentrations of two common antimicrobials (tylosin and chlortetracycline) on swine GI microbiota composition, tylosin administration resulted in a major shift in the relative abundance of several taxa, while chlortetracycline administration only resulted in minor alterations (Holman and Chénier, 2014). Similarly, oral vancomycin and metronidazole have different effects on Clostridium difficile, where only vancomycin had an obvious impact on microbial community structure (Lewis et al., 2015). The simplest mechanistic explanation for variation in the swine GI microbiota response to antimicrobial intervention is due to differences in antimicrobial spectrum, route of administration, and degree of antimicrobial resistance (Kim et al., 2012; Looft et al., 2014a,b; Schokker et al., 2015; Mu et al., 2017; Soler et al., 2018).
Gastrointestinal Microbiota as a Reservoir of Antimicrobial Resistome
The concept of the antimicrobial resistome was proposed by Gerard Wright in 2007 as a means of describing the collection of all known ARGs in the microbial ecosystem and their precursors at multiple levels (e.g., environment, pathogenic, and non-pathogenic microbes; Wright, 2007). Historically, determination of ARGs have primarily relied on conventional culture-based methods, with a focus on major pathogens that are readily cultured (Isaacson and Kim, 2012). While beneficial, these protocols do not provide information on the total amount of ARGs in the bacterial community as most species in that community cannot be cultivated, likely underestimating the complexity of the antimicrobial resistome (Henriksson et al., 1995). Although the antimicrobial resistome is theoretically accessible to all bacteria, the GI microbiota harbor a distinct antimicrobial resistome (Sundin and Wang, 2018). The known ARGs are likely to represent just a small portion of actual antimicrobial resistome populations. It is reasonable to assume that with the explosion of bacterial genome sequencing and functional metagenomics, many novel ARGs that were previously of unknown function and unrecognizable by sequence alone will be identified (D'Costa et al., 2007). The generation of more information about ARGs will be helpful in understanding the relationship between the resident microbial communities and their accompanying resistome (Boolchandani et al., 2019).
In parallel with the consecutive development of GI microbiota, the antimicrobial resistome is established during first few days of life or perhaps during prenatal phase even without prior exposure to antimicrobial treatment (Wright, 2007; Zeineldin et al., 2019). This concept endorses the theory that resistant bacteria and their antimicrobial resistome are established shortly after birth and are acquired either directly from the mother or through direct contact with resistant bacteria in surrounding environment (Gonzales-Marin et al., 2012). The GI microbiota has a large and diverse genetic pool that facilitates transmission of resistance between and within the resident commensal species (Sengupta et al., 2013). The effects of different antimicrobial intervention on emergence of the antimicrobial resistome has been extensively demonstrated (Wright, 2007; Enwemeka, 2013). In people, when the infants received antimicrobial treatment in the first 3 years of life, the GI microbiota expressed high levels of antimicrobial resistance compared to the control (Yassour et al., 2016). Similarly, the abundance of 149 ARGs conferring resistance to different classes of antimicrobials were detected in the swine feces from production units that used different antimicrobials either orally or via intramuscular injection (Zhu et al., 2013). Emergence of antimicrobial resistance determinants in pigs without prior antimicrobial administration has been also demonstrated previously (Pakpour et al., 2012; Agga et al., 2015), with the largest resistance category being against tetracycline antibiotic (Chambers et al., 2015). For instance, several tetracycline resistance genes (e.g., tetO, tetW, tetM, tetX, and tetQ), and macrolide resistance genes (e.g., ermG, ermF, and ermB) were frequently identified in the swine facilities in the absence of antimicrobial exposure (Looft et al., 2012). Similarly, our recent study showed that the neonatal piglets displayed a high frequency of ARGs without prior exposure of antibiotics (Zeineldin et al., 2019). Emergence of these ARGs without direct exposure to a known antibiotic also reveals that the swine GI antimicrobial resistome may not be affected by a reduction in antimicrobial administration in the swine industry (Holman and Chénier, 2015).
Antimicrobial Alternatives in Swine Industry
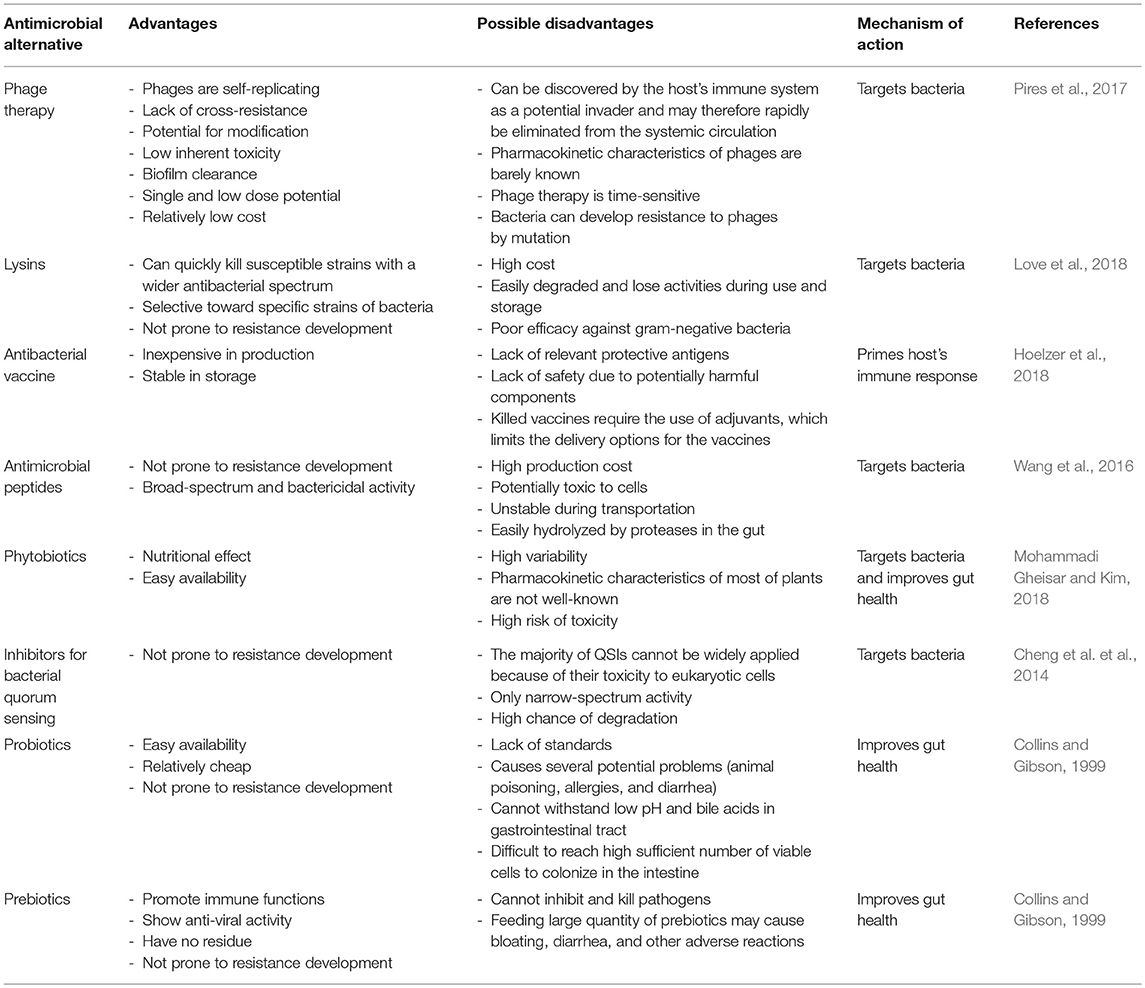
The current efforts to define the complex composition of GI microbiota and how that community responds to antimicrobial intervention would improve our ability to develop novel non-antibiotic strategies to prevent GI infection in food-producing animals, subsequently increasing animal productivity (Marchesi et al., 2016). Considering this information, different management strategies are required to reduce the deleterious consequences of antimicrobials, particularly when its administration is needed to control bacterial infections. Broad discussions of possible antimicrobial alternatives have been summarized in Table 3 and were mentioned elsewhere (Potter et al., 2008; Allen et al., 2013, 2014; Papatsiros, 2013; Czaplewski et al., 2016). In this section, we will only focus on bacteriophage therapy as an important and promising example of available antimicrobial alternatives in the swine industry.
Bacteriophage (phage) therapy involves the use of bacterial viruses (phages) to attack specific bacterial species, or a narrow group of microbes, without harming the resident autochthonous microbial communities (Kutateladze and Adamia, 2010). Because of their ubiquity in all natural environments and commercial swine facilities, as well as their specific action against pathogens, phages have been suggested as a promising antimicrobial alternative for use in swine (Zhang et al., 2015). Recent studies based on high throughput next-generation sequencing approaches highlighted the importance of phages in microbial evolution and bacterial community control (Pratama and van Elsas, 2018). In addition to GI microbiota inhabitants, the GI tract harbors diverse phage communities that have a synergistic effect along with the resident microbial communities to maintain GI health (Allen et al., 2013). Subsequent research studies demonstrated that bacteriophages attacks bacteria by attaching to the cell wall and injecting their genetic material into bacterial cytoplasm with subsequent integration into the bacterial genome. Phage populations are extensively diverse and generally grouped according to their morphological properties and life cycle into temperate (lysogenic) or virulent (lytic) phages. Virulent bacteriophages are natural predators of their bacterial hosts, they replicate using the host machinery, and complete their lifecycle by lysis of the host cell (Calero-Cáceres et al., 2019). In contrast, temperate bacteriophages integrate into the host's chromosome and produce a stable genetic relationship with the host during the process of lysogeny without creating new phage particles (Zhang et al., 2015). Despite the growing evidence that supports the medical importance of virulent bacteriophages, their functional potential in swine is not yet well-defined.
In the swine industry, bacteriophage intervention strategies have been extensively used to control various Salmonella serovars, E. coli O157:H7, enterotoxigenic E. coli-induced diarrhea and Campylobacter species (Lee and Harris, 2001; Nisbet et al., 2010; Harvey et al., 2011; Hooton et al., 2011; Cha et al., 2012). These studies have shown that phages can be effectively utilized against these pathogens. Most recently, a phage cocktail was used to reduce Salmonella typhimurium in artificially-infected market-weight swine (Wall et al., 2010; Hooton et al., 2011). Similarly, phage treatment in weaned piglets challenged with S. typhimurium via oral gavage reduced fecal and cecal Salmonella populations in phage-treated piglets compared to control piglets (Nisbet et al., 2010). Several other experiments have evaluated the antimicrobial ability of phages against E. coli infections. Oral administration of a phage cocktail was capable of reducing morbidity and mortality in enterotoxigenic E. coli-challenged pigs, even when used at the onset of clinical signs (Atterbury, 2009). Smith and Huggins also investigated the efficacy of a mixture of two phages against an enteropathogenic strain of E. coli in neonatal pigs. The results of this work indicated that phages which targeted adherence pili were more effective in controlling porcine E. coli than phages that target other pili (Smith and Huggins, 2009). Phage therapy was also associated with increased prevalence of beneficial microbes (e.g., Bifidobacterium and Lactobacillus) and decreased relative abundance of coliforms and Clostridium species in post-weaning piglets (Hosseindoust et al., 2017).
Since their discovery in 1915, phages have been proven to be harmless to humans, animals and plants. Compared to antimicrobial, phages are highly effective in killing their target bacteria without harming the rest of the microbiota in the ecosystem. Additionally, phages are relatively cheap, self-replicating, easy to isolate, and have low inherent toxicity (Sillankorva et al., 2012). Despite these advantages, there are many technical limitations in the implementation of phage therapy for treatment of infectious diseases in human and animals (Allen et al., 2014). Commercially available phages have a limited microbial range, are unstable, sensitive to temperature, have a narrow range of hosts, require rapid administration after infection, and could be neutralized by the host's immune system (Papatsiros, 2013; Zhang et al., 2015). Similarly to antimicrobial resistance, recent studies suggest that bacteriophages play a crucial role in the acquisition and emergence of the antimicrobial resistome (Calero-Cáceres et al., 2019). Phage genomes can harbor several antimicrobial resistomes belonging to different antimicrobial classes. Phage-resistant strains are believed to be generally less virulent than the phage susceptible wild types, but the use of a number of different phages in combination (phage cocktails) against many serotypes will likely alleviate this problem (Kutateladze and Adamia, 2010; Harvey et al., 2011). Therefore, high-throughput next-generation sequencing and genetic engineering will be necessary to create a more reasonable phage to optimize impact and create the best alternative to antimicrobial treatment.
Conclusion
The application of both high-throughput next-generation sequencing and functional metagenomics have clarified the effects of antimicrobial administration on commensal populations as well as on emergence of ARGs. There is, therefore, a great interest in understanding the origins, evolution and totality of antimicrobial resistance, not just in pathogenic microbes but also in whole resident microbial environment. The evidence that the commensal population harbors a previously underappreciated antimicrobial resistome should shift the paradigm of what judicious use of antimicrobials in livestock means. In addition, it raises exciting questions about the acquisition and transfer of antimicrobial resistance cross GI microbiota. A better understanding of the impacts of specific antimicrobial intervention strategies on GI microbiota and their accompanying antimicrobial resistome could open the door to the development of a novel therapeutic approach in swine production systems.
Author Contributions
MZ wrote the manuscript. BA and JL revised it. All authors have approved the manuscript submission.
Funding
The work was funded through the Integrated Food Animal Management System research program at the Department of Veterinary Clinical Medicine, University of Illinois at Urbana-Champaign.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Aarestrup, F. M., Oliver Duran, C., and Burch, D. G. S. (2008). Antimicrobial resistance in swine production. Anim. Health Res. Rev. 9, 135–148. doi: 10.1017/S1466252308001503
Agga, G. E., Arthur, T. M., Durso, L. M., Harhay, D. M., and Schmidt, J. W. (2015). Antimicrobial-resistant bacterial populations and antimicrobial resistance genes obtained from environments impacted by livestock and municipal waste. PLoS ONE 10:e132586. doi: 10.1371/journal.pone.0132586
Allen, H. K., Levine, U. Y., Looft, T., Bandrick, M., and Casey, T. A. (2013). Treatment, promotion, commotion: antibiotic alternatives in food-producing animals. Trends Microbiol. 21, 114–119. doi: 10.1016/j.tim.2012.11.001
Allen, H. K., Looft, T., Bayles, D. O., Humphrey, S., Levine, U. Y., Alt, D., et al. (2011). Antibiotics in feed induce prophages in swine fecal microbiomes. MBio 2, 1–9. doi: 10.1128/mBio.00260-11
Allen, H. K., Trachsel, J., Looft, T., and Casey, T. A. (2014). Finding alternatives to antibiotics. Ann. N.Y. Acad. Sci. 1323, 91–100. doi: 10.1111/nyas.12468
Atterbury, R. J. (2009). Bacteriophage biocontrol in animals and meat products. Microb. Biotechnol. 2, 601–612. doi: 10.1111/j.1751-7915.2009.00089.x
Bischoff, S. (2011). ‘Gut health': a new objective in medicine? BMC Med. 9:24. doi: 10.1186/1741-7015-9-24
Blaut, M., and Clavel, T. (2007). Metabolic diversity of the intestinal microbiota : implications for health and disease. J. Nutr. 137(3 Suppl. 2):751S−755S. doi: 10.1093/jn/137.3.751S
Bokulich, N. A., Chung, J., Battaglia, T., Henderson, N., Jay, M., Li, H., et al. (2016). Antibiotics, birth mode, and diet shape microbiome maturation during early life. Sci. Transl. Med. 8:343ra82. doi: 10.1126/scitranslmed.aad7121
Boolchandani, M., D'Souza, A. W., and Dantas, G. (2019). Sequencing-based methods and resources to study antimicrobial resistance. Nat. Rev. Genet. 18:1. doi: 10.1038/s41576-019-0108-4
Brandl, K., Plitas, G., Mihu, C. N., Ubeda, C., Jia, T., Fleisher, M., et al. (2008). Vancomycin-resistant enterococci exploit antibiotic-induced innate immune deficits. Nature 455, 804–807. doi: 10.1038/nature07250
Calero-Cáceres, W., Ye, M., and Balcázar, J. L. (2019). Bacteriophages as environmental reservoirs of antibiotic resistance. Trends Microbiol. doi: 10.1016/j.tim.2019.02.008. [Epub ahead of print].
Cha, S. B., Yoo, A. N., Lee, W. J., Shin, M. K., Jung, M. H., Shin, S. W., et al. (2012). Effect of bacteriophage in enterotoxigenic Escherichia coli (ETEC) infected pigs. J. Vet. Med. Sci. 74, 1037–1039. doi: 10.1292/jvms.11-0556
Chambers, L., Yang, Y., Littier, H., Ray, P., Zhang, T., Pruden, A., et al. (2015). Metagenomic analysis of antibiotic resistance genes in dairy cow feces following therapeutic administration of third generation cephalosporin. PLoS ONE 10:e0133764. doi: 10.1371/journal.pone.0133764
Chang, Q., Wang, W., Regev-Yochay, G., Lipsitch, M., and Hanage, W. P. (2015). Antibiotics in agriculture and the risk to human health: how worried should we be? Evol. Appl. 8, 240–247. doi: 10.1111/eva.12185
Cheng, G., Hao, H., Xie, S., Wang, X., Dai, M., Huang, L., et al. (2014). Antibiotic alternatives: the substitution of antibiotics in animal husbandry? Front. Microbiol. 5:217. doi: 10.3389/fmicb.2014.00217
Collins, M. D., and Gibson, G. R. (1999). Prebiotic, probiotic, and symbiotic: approaches for modulating the microbial ecology of the gut. Am. J. Clin. Nutr. 69, 1052–1057. doi: 10.1093/ajcn/69.5.1052s
Crofts, T. S., Gasparrini, A. J., and Dantas, G. (2017). Next-generation approaches to understand and combat the antibiotic resistome. Nat. Rev. Microbiol. 15, 422–434. doi: 10.1038/nrmicro.2017.28
Cromwell, G. L. (2002). Why and how antibiotics are used in swine production. Anim. Biotechnol. 13, 7–27. doi: 10.1081/ABIO-120005767
Czaplewski, L., Bax, R., Clokie, M., Dawson, M., Fairhead, H., Fischetti, V. A., et al. (2016). Alternatives to antibiotics-a pipeline portfolio review. Lancet Infect. Dis. 16, 239–251. doi: 10.1016/S1473-3099(15)00466-1
D'Costa, V. M., Griffiths, E., and Wright, G. D. (2007). Expanding the soil antibiotic resistome: exploring environmental diversity. Curr. Opin. Microbiol. 10, 481–489. doi: 10.1016/j.mib.2007.08.009
Dewey, C. E., Cox, B. D., Straw, B. E., Bush, E. J., and Hurd, H. S. (1997). Associations between off-label feed additives and farm size, veterinary consultant use, and animal age. Prev. Vet. Med. 31, 133–146. doi: 10.1016/S0167-5877(96)01077-X
Dewey, C. E., Cox, B. D., Straw, B. E., Bush, E. J., and Hurd, S. (1999). Use of antimicrobials in swine feeds in the United States. Swine Heal. Prod. 7, 19–25.
Dibner, J. J., and Richards, J. D. (2005). Antibiotic growth promoters in agriculture : history and mode of action actual usage of antimicrobials in denmark voluntary and legislated bans. Poult. Sci. 84, 634–643. doi: 10.1093/ps/84.4.634
Dumas, S. E., French, H. M., Lavergne, S. N., Ramirez, C. R., Brown, L. J., Bromfield, C. R., et al. (2016). Judicious use of prophylactic antimicrobials to reduce abdominal surgical site infections in periparturient cows: part 1-a risk factor review. Vet. Rec. 178, 654–660. doi: 10.1136/vr.i103677
Enwemeka, C. S. (2013). Antimicrobial blue light: an emerging alternative to antibiotics. Photomed. Laser Surg. 31, 509–511. doi: 10.1089/pho.2013.9871
Everaert, N., Van Cruchten, S., Weström, B., Bailey, M., Van Ginneken, C., Thymann, T., et al. (2017). A review on early gut maturation and colonization in pigs, including biological and dietary factors affecting gut homeostasis. Anim. Feed Sci. Technol. 233, 89–103. doi: 10.1016/j.anifeedsci.2017.06.011
FDA (2017). Antimicrobials Sold or Distributed for Use in Food-Producing Animals. US FOOD DRUG ADMINSTRATION.
Founou, L. L., Founou, R. C., and Essack, S. Y. (2016). Antibiotic resistance in the food chain: a developing country-perspective. Front. Microbiol. 7:1881. doi: 10.3389/fmicb.2016.01881
Gao, K., Pi, Y., Mu, C.-L., Peng, Y., Huang, Z., and Zhu, W.-Y. (2018a). Antibiotics-induced modulation of large intestinal microbiota altered aromatic amino acid profile and expression of neurotransmitters in the hypothalamus of piglets. J. Neurochem. 146, 219–234. doi: 10.1111/jnc.14333
Gao, K., Pi, Y., Peng, Y., Mu, C. L., and Zhu, W. Y. (2018b). Time-course responses of ileal and fecal microbiota and metabolite profiles to antibiotics in cannulated pigs. Appl. Microbiol. Biotechnol. 102, 2289–2299. doi: 10.1007/s00253-018-8774-2
Gerzova, L., Babak, V., Sedlar, K., Faldynova, M., Videnska, P., Cejkova, D., et al. (2015). Characterization of antibiotic resistance gene abundance and microbiota composition in feces of organic and conventional pigs from four EU countries. PLoS ONE 10:e0132892. doi: 10.1371/journal.pone.0132892
Gonzales-Marin, C., Spratt, D. A., Millar, M. R., Simmonds, M., Kempley, S. T., and Allaker, R. P. (2012). Identification of bacteria and potential sources in neonates at risk of infection delivered by Caesarean and vaginal birth. J. Med. Microbiol. 61, 31–41. doi: 10.1099/jmm.0.034926-0
Harvey, R. B., Nisbet, D. J., Anderson, R. C., Callaway, T. R., Byrd, J. A., Kogut, M. H., et al. (2011). Using Antimicrobial Cultures, Bacteriocins and Bacteriophages to Reduce Carriage of Foodborne Pathogens in Cattle and Swine (Sawston, Cambridge, UK: Woodhead Publishing Limited), 204–224.
Henriksson, A., André, L., and Conway, P. L. (1995). Distribution of lactobacilli in the porcine gastrointestinal tract. FEMS Microbiol. Ecol. 16, 55–60. doi: 10.1111/j.1574-6941.1995.tb00268.x
Hoelzer, E., Bielke, L., Raicek, M., Gay, C., Hoelzer, K., Van Immerseel, F., et al. (2018). Vaccines as alternatives to antibiotics for food producing animals. Part 1: challenges and needs. Vet. Res. 49, 1–10. doi: 10.1186/s13567-018-0560-8
Holman, D. B., Brunelle, B. W., Trachsel, J., and Allen, H. K. (2017). Meta-analysis to define a core microbiota in the swine gut. mSystems 2, e00004–17. doi: 10.1128/mSystems.00004-17
Holman, D. B., and Chénier, M. R. (2014). Temporal changes and the effect of subtherapeutic concentrations of antibiotics in the gut microbiota of swine. FEMS Microbiol. Ecol. 90, 599–608. doi: 10.1111/1574-6941.12419
Holman, D. B., and Chénier, M. R. (2015). Antimicrobial use in swine production and its effect on the swine gut microbiota and antimicrobial resistance. Can. J. Microbiol. 61, 785–798. doi: 10.1139/cjm-2015-0239
Holman, D. B., Timsit, E., Booker, C. W., and Alexander, T. W. (2018). Injectable antimicrobials in commercial feedlot cattle and their effect on the nasopharyngeal microbiota and antimicrobial resistance. Vet. Microbiol. 214, 140–147. doi: 10.1016/j.vetmic.2017.12.015
Holmes, A. H., Moore, L. S., Sundsfjord, A., Steinbakk, M., Regmi, S., Karkey, A., et al. (2016). Understanding the mechanisms and drivers of antimicrobial resistance. Lancet 387, 176–187. doi: 10.1016/S0140-6736(15)00473-0
Hooton, S. P. T., Atterbury, R. J., and Connerton, I. F. (2011). Application of a bacteriophage cocktail to reduce Salmonella Typhimurium U288 contamination on pig skin. Int. J. Food Microbiol. 151, 157–163. doi: 10.1016/j.ijfoodmicro.2011.08.015
Hosseindoust, A. R., Lee, S. H., Kim, J. S., Choi, Y. H., Noh, H. S., Lee, J. H., et al. (2017). Dietary bacteriophages as an alternative for zinc oxide or organic acids to control diarrhoea and improve the performance of weanling piglets. Vet. Med. (Praha). 62, 53–61. doi: 10.17221/7/2016-VETMED
Isaacson, R., and Kim, H. B. (2012). The intestinal microbiome of the pig. Anim. Heal. Res. Rev. 13, 100–109. doi: 10.1017/S1466252312000084
Jernberg, C., Löfmark, S., Edlund, C., and Jansson, J. K. (2010). Long-term impacts of antibiotic exposure on the human intestinal microbiota. Microbiology 156, 3216–3223. doi: 10.1099/mic.0.040618-0
Johnson, T. A., Looft, T., Severin, A. J., Bayles, D. O., Nasko, D. J., Wommack, K. E., et al. (2017). The in-feed antibiotic carbadox induces phage gene transcription in the swine gut microbiome. MBio 8, 1–14. doi: 10.1128/mBio.00709-17
Kim, H. B., Borewicz, K., White, B. A., Singer, R. S., Sreevatsan, S., Tu, Z. J., et al. (2012). Microbial shifts in the swine distal gut in response to the treatment with antimicrobial growth promoter, tylosin. Proc. Natl. Acad. Sci. U.S.A. 109, 15485–15490. doi: 10.1073/pnas.1205147109
Kutateladze, M., and Adamia, R. (2010). Bacteriophages as potential new therapeutics to replace or supplement antibiotics. Trends Biotechnol. 28, 591–595. doi: 10.1016/j.tibtech.2010.08.001
Landers, T. F., Cohen, B., Wittum, T. E., and Larson, E. L. (2013). A review of antibiotic use in food animals: perspective, policy, and potential. Public Health Rep. 127, 4–22. doi: 10.1177/003335491212700103
Langdon, A., Crook, N., and Dantas, G. (2016). The effects of antibiotics on the microbiome throughout development and alternative approaches for therapeutic modulation. Genome Med. 8, 39. doi: 10.1186/s13073-016-0294-z
Lawrence, J. G. (2004). Horizontal and vertical gene transfer: the life history of pathogens. Concepts Bact. Virulence 12, 255–271. doi: 10.1159/000081699
Lee, N., and Harris, D. (2001). The effect of bacteriophage treatment to reduce the rapid dissemination of Salmonella typhimurium in pigs. Proc. Am. Assoc. Swine Vet. (Perry, IA: American Association of Swine Veterinarians), 555–557.
Leibovitz, E., Greenberg, D., Piglansky, L., Raiz, S., Porat, N., Press, J., et al. (2003). Recurrent acute otitis media occurring within one month from completion of antibiotic therapy: relationship to the original pathogen. Pediatr. Infect. Dis. J. 22, 209–216. doi: 10.1097/01.inf.0000066798.69778.07
Leser, T. D., Amenuvor, J. Z., Jensen, T. K., Lindecrona, R. H., Boye, M., Moller, K., et al. (2002). Culture-independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl. Environ. Microbiol. 68, 673–690. doi: 10.1128/AEM.68.2.673-690.2002
Lewis, B. B., Buffie, C. G., Carter, R. A., Leiner, I., Toussaint, N. C., Miller, L. C., et al. (2015). Loss of microbiota-mediated colonization resistance to Clostridium difficile infection with oral vancomycin compared with metronidazole. J. Infect. Dis. 212, 1656–1665. doi: 10.1093/infdis/jiv256
Li, J., Yang, K., Ju, T., Ho, T., McKay, C. A., Gao, Y., et al. (2017). Early life antibiotic exposure affects pancreatic islet development and metabolic regulation. Sci. Rep. 7, 1–12. doi: 10.1038/srep41778
Li, K., Xiao, Y., Chen, J., Chen, J., He, X., and Yang, H. (2017). Microbial composition in different gut locations of weaning piglets receiving antibiotics. Asian Austral. J. Anim. Sci. 30, 78–84. doi: 10.5713/ajas.16.0285
Li, P., Niu, Q., Wei, Q., Zhang, Y., Ma, X., Kim, S. W., et al. (2017). Microbial shifts in the porcine distal gut in response to diets supplemented with Enterococcus Faecalis as alternatives to antibiotics. Sci. Rep. 7:41395. doi: 10.1038/srep41395
Looft, T., Allen, H. K., Cantarel, B. L., Levine, U. Y., Bayles, D. O., Alt, D. P., et al. (2014a). Bacteria, phages and pigs: the effects of in-feed antibiotics on the microbiome at different gut locations. ISME J. 8, 1566–1576. doi: 10.1038/ismej.2014.12
Looft, T., Allen, H. K., Casey, T. A., Alt, D. P., and Stanton, T. B. (2014b). Carbadox has both temporary and lasting effects on the swine gut microbiota. Front. Microbiol. 5:276. doi: 10.3389/fmicb.2014.00276
Looft, T., Johnson, T. A., Allen, H. K., Bayles, D. O., Alt, D. P., Stedtfeld, R. D., et al. (2012). In-feed antibiotic effects on the swine intestinal microbiome. Proc. Natl. Acad. Sci. U.S.A. 109, 1691–1696. doi: 10.1073/pnas.1120238109
Love, M. J., Bhandari, D., Dobson, R., and Billington, C. (2018). Potential for bacteriophage endolysins to supplement or replace antibiotics in food production and clinical care. Antibiotics 7:17. doi: 10.3390/antibiotics7010017
Mach, N., Berri, M., Estellé, J., Levenez, F., Lemonnier, G., Denis, C., et al. (2015). Early-life establishment of the swine gut microbiome and impact on host phenotypes. Environ. Microbiol. Rep. 7, 554–569. doi: 10.1111/1758-2229.12285
Maradiaga, N., Aldridge, B., Zeineldin, M., and Lowe, J. (2018). Gastrointestinal microbiota and mucosal immune gene expression in neonatal pigs reared in a cross-fostering model. Microb. Pathog. 121, 27–39. doi: 10.1016/j.micpath.2018.05.007
Marchesi, J. R., Adams, D. H., Fava, F., Hermes, G. D. A., Hirschfield, G. M., Hold, G., et al. (2016). The gut microbiota and host health: a new clinical frontier. Gut 65, 330–339. doi: 10.1136/gutjnl-2015-309990
McEwen, S. A., and Fedorka-Cray, P. J. (2002). Antimicrobial use and resistance in animals. Clin. Infect. Dis. 34(Suppl. 3), S93–S106. doi: 10.1086/340246
Merks, J. W., Mathur, P. K., and Knol, E. F. (2012). New phenotypes for new breeding goals in pigs. Animal 6, 535–543. doi: 10.1017/S1751731111002266
Metzler, B. U., and Mosenthin, R. (2008). A review of interactions between dietary fiber and the gastrointestinal microbiota and their consequences on intestinal phosphorus metabolism in growing pigs. Asian Austral. J. Anim. Sci. 21, 603–615. doi: 10.5713/ajas.2008.r.03
Mohammadi Gheisar, M., and Kim, I. H. (2018). Phytobiotics in poultry and swine nutrition–a review. Ital. J. Anim. Sci. 17, 92–99. doi: 10.1080/1828051X.2017.1350120
Mu, C., Yang, Y., Su, Y., Zoetendal, E. G., and Zhu, W. (2017). Differences in microbiota membership along the gastrointestinal tract of piglets and their differential alterations following an early-life antibiotic intervention. Front. Microbiol. 8:797. doi: 10.3389/fmicb.2017.00797
Neuman, H., Forsythe, P., Uzan, A., Avni, O., and Koren, O. (2018). Antibiotics in early life: dysbiosis and the damage done. FEMS Microbiol. Rev. 42, 489–499. doi: 10.1093/femsre/fuy018
Nisbet, D. J., Brabban, A., Anderson, R., Karriker, L., Poole, T. L., Krueger, N., et al. (2010). Evaluation of phage treatment as a strategy to reduce salmonella populations in growing swine. Foodborne Pathog. Dis. 8, 261–266. doi: 10.1089/fpd.2010.0671
O'Neill, J. (2014). AMR review paper-tackling a crisis for the health and wealth of nations. Rev. Antimicrob. Resist. Available online at: https://amr-review.org/sites/default/files/AMR%20Review%20Paper%20-%20Tackling%20a%20crisis%20for%20the%20health%20and%20wealth%20of%20nations_1.pdf
Oultram, J., Phipps, E., Teixeira, A. G. V., Foditsch, C., Bicalho, M. L., Machado, V. S., et al. (2015). Effects of antibiotics (oxytetracycline, florfenicol or tulathromycin) on neonatal calves' faecal microbial diversity. Vet. Rec. 177:598. doi: 10.1136/vr.103320
Pakpour, S., Jabaji, S., and Chénier, M. R. (2012). Frequency of antibiotic resistance in a swine facility 2.5 years after a ban on antibiotics. Microb. Ecol. 63, 41–50. doi: 10.1007/s00248-011-9954-0
Papatsiros, V. (2013). Alternatives to antibiotics for farm animals. CAB Rev. Perspect. Agric. Vet. Sci. Nutr. Nat. Resour. 8:32. doi: 10.1079/PAVSNNR20138032
Pettigrew, M. M., Laufer, A. S., Gent, J. F., Kong, Y., Fennie, K. P., and Metlay, J. P. (2012). Upper respiratory tract microbial communities, acute otitis media pathogens, and antibiotic use in healthy and sick children. Appl. Environ. Microbiol. 78, 6262–6270. doi: 10.1128/AEM.01051-12
Phillips, I., Casewell, M., Cox, T., De Groot, B., Friis, C., Jones, R., et al. (2004). Does the use of antibiotics in food animals pose a risk to human health? A critical review of published data. J. Antimicrob. Chemother. 53, 28–52. doi: 10.1093/jac/dkg483
Pires, D. P., Melo, L. D., Vilas Boas, D., Sillankorva, S., and Azeredo, J. (2017). Phage therapy as an alternative or complementary strategy to prevent and control biofilm-related infections. Curr. Opin. Microbiol. 39, 48–56. doi: 10.1016/j.mib.2017.09.004
Pluske, J. R. (2016). Invited review: aspects of gastrointestinal tract growth and maturation in the pre- and postweaning period of pigs. J. Anim. Sci. 94, 399–411. doi: 10.2527/jas.2015-9767
Pluske, J. R., Turpin, D. L., and Kim, J. C. (2018). Gastrointestinal tract (gut) health in the young pig. Anim. Nutr. 4, 187–196. doi: 10.1016/j.aninu.2017.12.004
Poole, T. L., Suchodolski, J. S., Callaway, T. R., Farrow, R. L., Loneragan, G. H., and Nisbet, D. J. (2013). The effect of chlortetracycline on faecal microbial populations in growing swine. J. Glob. Antimicrob. Resist. 1, 171–174. doi: 10.1016/j.jgar.2013.04.004
Potter, A., Gerdts, V., and Littel-van den Hurk, S. (2008). Veterinary vaccines: alternatives to antibiotics? Anim. Health Res. Rev. 9, 187–199. doi: 10.1017/S1466252308001606
Pratama, A. A., and van Elsas, J. D. (2018). The ‘neglected’ soil virome – potential role and impact. Trends Microbiol. 26, 649–662. doi: 10.1016/j.tim.2017.12.004
Schmidt, T. M., Young, V. B., Sogin, M. L., Morrison, H. G., Antonopoulos, D. A., and Huse, S. M. (2009). Reproducible community dynamics of the gastrointestinal microbiota following antibiotic perturbation. Infect. Immun. 77, 2367–2375. doi: 10.1128/IAI.01520-08
Schokker, D., Zhang, J., Vastenhouw, S. A., Heilig, H. G. H. J., Smidt, H., Rebel, J. M. J., et al. (2015). Long-lasting effects of early-life antibiotic treatment and routine animal handling on gut microbiota composition and immune system in pigs. PLoS ONE 10:e116523. doi: 10.1371/journal.pone.0116523
Sengupta, S., Chattopadhyay, M. K., and Grossart, H. P. (2013). The multifaceted roles of antibiotics and antibiotic resistance in nature. Front. Microbiol. 4:47. doi: 10.3389/fmicb.2013.00047
Sillankorva, S. M., Oliveira, H., and Azeredo, J. (2012). Bacteriophages and their role in food safety. Int. J. Microbiol. 2012:863945. doi: 10.1155/2012/863945
Skinner, L. D., Levesque, C. L., Wey, D., Rudar, M., Zhu, J., Hooda, S., et al. (2014). Impact of nursery feeding program on subsequent growth performance, carcass quality, meat quality, and physical and chemical body composition of growing-finishing pigs 1. J. Anim. Sci 92, 1044–1054. doi: 10.2527/jas.2013-6743
Smith, H. W., and Huggins, M. B. (2009). Effectiveness of phages in treating experimental escherichia coli diarrhoea in calves, piglets and lambs. Microbiology 129, 2659–2675. doi: 10.1099/00221287-129-8-2659
Soler, C., Goossens, T., Bermejo, A., Migura-García, L., Cusco, A., Francino, O., et al. (2018). Digestive microbiota is different in pigs receiving antimicrobials or a feed additive during the nursery period. PLoS ONE 13:e197353. doi: 10.1371/journal.pone.0197353
Sommer, M. O., and Dantas, G. (2011). Antibiotics and the resistant microbiome. Curr. Opin. Microbiol. 14, 556–563. doi: 10.1016/j.mib.2011.07.005
Sommer, M. O. A., Munck, C., Toft-Kehler, R. V., and Andersson, D. I. (2017). Prediction of antibiotic resistance: time for a new preclinical paradigm? Nat. Rev. Microbiol. 15, 689–696. doi: 10.1038/nrmicro.2017.75
Stone, J. J., Clay, S. A., Zhu, Z., Wong, K. L., Porath, L. R., and Spellman, G. M. (2009). Effect of antimicrobial compounds tylosin and chlortetracycline during batch anaerobic swine manure digestion. Water Res. 43, 4740–4750. doi: 10.1016/j.watres.2009.08.005
Sun, J., Li, L., Liu, B., Xia, J., Liao, X., and Liu, Y. (2014). Development of aminoglycoside and β-lactamase resistance among intestinal microbiota of swine treated with lincomycin, chlortetracycline, and amoxicillin. Front. Microbiol. 5:580. doi: 10.3389/fmicb.2014.00580
Sundin, G. W., and Wang, N. (2018). Antibiotic resistance in plant-pathogenic bacteria. Annu. Rev. Phytopathol. 56, 161–180. doi: 10.1146/annurev-phyto-080417-045946
Turnbaugh, P. J., Ley, R. E., Hamady, M., Fraser-Liggett, C. M., Knight, R., and Gordon, J. I. (2007). The human microbiome project. Nature 449, 804–810. doi: 10.1038/nature06244
Unno, T., Kim, J., Guevarra, R. B., and Nguyen, S. G. (2015). Effects of antibiotic growth promoter and characterization of ecological succession in swine gut microbiota. J. Microbiol. Biotechnol. 25, 431–438. doi: 10.4014/jmb.1408.08063
Venable, E. B., Bland, S. D., McPherson, J. L., and Francis, J. (2016). Role of the gut microbiota in equine health and disease. Anim. Front. 6:43. doi: 10.2527/af.2016-0033
Wall, S. K., Zhang, J., Rostagno, M. H., and Ebner, P. D. (2010). Phage therapy to reduce preprocessing Salmonella infections in market-weight swine. Appl. Environ. Microbiol. 76, 48–53. doi: 10.1128/AEM.00785-09
Wang, S., Zeng, X., Yang, Q., and Qiao, S. (2016). Antimicrobial peptides as potential alternatives to antibiotics in food animal industry. Int. J. Mol. Sci. 17:E603. doi: 10.3390/ijms17050603
Wright, G. D. (2007). The antibiotic resistome: the nexus of chemical and genetic diversity. Nat. Rev. Microbiol. 5, 175–186. doi: 10.1038/nrmicro1614
Yassour, M., Vatanen, T., Siljander, H., Hämäläinen, A. M., Härkönen, T., Ryhänen, S. J., et al. (2016). Natural history of the infant gut microbiome and impact of antibiotic treatment on bacterial strain diversity and stability. Sci. Transl. Med. 8:343ra81. doi: 10.1126/scitranslmed.aad0917
Yeoman, C. J., Ishaq, S. L., Bichi, E., Olivo, S. K., Lowe, J., and Aldridge, B. M. (2018). Biogeographical differences in the influence of maternal microbial sources on the early successional development of the bovine neonatal gastrointestinal tract. Sci. Rep. 8:3197. doi: 10.1038/s41598-018-21440-8
Yu, M., Mu, C., Zhang, C., Yang, Y., and Su, Y. (2018). Marked response in microbial community and metabolism in the ileum and cecum of suckling piglets after early antibiotics Exposure 9, 1–13. doi: 10.3389/fmicb.2018.01166
Yu, T., Zhu, C., Chen, S., Gao, L., Lv, H., Feng, R., et al. (2017). Dietary high zinc oxide modulates the microbiome of ileum and colon in weaned piglets. Front. Microbiol. 8:825. doi: 10.3389/fmicb.2017.00825
Zeineldin, M., Aldridge, B., Blair, B., Kancer, K., and Lowe, J. (2018a). Impact of parenteral antimicrobial administration on the structure and diversity of the fecal microbiota of growing pigs. Microb. Pathog. 118, 220–229. doi: 10.1016/j.micpath.2018.03.035
Zeineldin, M., Aldridge, B., Blair, B., Kancer, K., and Lowe, J. (2018b). Microbial shifts in the swine nasal microbiota in response to parenteral antimicrobial administration. Microb. Pathog. 121, 210–217. doi: 10.1016/j.micpath.2018.05.028
Zeineldin, M., Aldridge, B., and Lowe, J. (2017a). Dysbiosis of the fecal microbiota in feedlot cattle with hemorrhagic diarrhea. Microb. Pathog. 115, 123–130. doi: 10.1016/j.micpath.2017.12.059
Zeineldin, M., Lowe, J., de Godoy, M., Maradiaga, N., Ramirez, C., Ghanem, M., et al. (2017b). Disparity in the nasopharyngeal microbiota between healthy cattle on feed, at entry processing and with respiratory disease. Vet. Microbiol. 208, 30–37. doi: 10.1016/j.vetmic.2017.07.006
Zeineldin, M. M., Megahed, A., Blair, B., Burton, B., Aldridge, B., and Lowe, J. (2019). Negligible impact of perinatal tulathromycin metaphylaxis on the developmental dynamics of fecal microbiota and their accompanying antimicrobial resistome in piglets. Front. Microbiol. 10:726. doi: 10.3389/fmicb.2019.00726
Zhang, D., Ji, H., Liu, H., Wang, S., Wang, J., and Wang, Y. (2016). Changes in the diversity and composition of gut microbiota of weaned piglets after oral administration of Lactobacillus or an antibiotic. Appl. Microbiol. Biotechnol. 100, 10081–10093. doi: 10.1007/s00253-016-7845-5
Zhang, J., Li, Z., Cao, Z., Wang, L., Li, X., Li, S., et al. (2015). Bacteriophages as antimicrobial agents against major pathogens in swine : a review. J. Anim. Sci. Biotechnol. 6:39. doi: 10.1186/s40104-015-0039-7
Keywords: antimicrobial, gastrointestinal, microbiota, swine, resistome
Citation: Zeineldin M, Aldridge B and Lowe J (2019) Antimicrobial Effects on Swine Gastrointestinal Microbiota and Their Accompanying Antibiotic Resistome. Front. Microbiol. 10:1035. doi: 10.3389/fmicb.2019.01035
Received: 19 November 2018; Accepted: 24 April 2019;
Published: 15 May 2019.
Edited by:
Gilberto Igrejas, University of Trás-os-Montes and Alto Douro, PortugalReviewed by:
Jian-Qiang Su, Institute of Urban Environment (CAS), ChinaTerence Spencer Crofts, Northwestern University, United States
Copyright © 2019 Zeineldin, Aldridge and Lowe. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: James Lowe, amxvd2VAaWxsaW5vaXMuZWR1
 Mohamed Zeineldin
Mohamed Zeineldin Brian Aldridge
Brian Aldridge James Lowe
James Lowe