- 1School of Agriculture, Ningxia University, Yinchuan, China
- 2Department of Preventive Veterinary Medicine, College of Veterinary Medicine, Northwest A&F University, Yangling, China
- 3Scientific Observing and Experimental Station of Veterinary Pharmacology and Diagnostic Technology, Ministry of Agriculture, Yangling, China
- 4Division of Immunology, Virginia-Maryland Regional College of Veterinary Medicine, University of Maryland, College Park, College Park, MD, United States
Avian hepatitis E virus (HEV) is a single-stranded, positive-sense RNA virus with a complete genome of approximately 6.6 kb in size. To date, four major genotypes of avian HEV have been identified and classified into the Orthohepevirus B genus of the family Hepeviridae. The avian HEV associated with hepatitis-splenomegaly syndrome, big liver and spleen disease or hepatic rupture hemorrhage syndrome in chickens is genetically and antigenically related to mammalian HEV. With the increased genotypes of avian HEV identified, a broader host tropism is also notable in the epidemiological studies. Due to the lack of an efficient cell culture system, the mechanisms of avian HEV replication and pathogenesis are still poorly understood. The recent identification and characterization of animal strains of avian HEV has demonstrated the virus’ ability of cross-species infection. Although it has not yet been detected in humans, the potential threat of a zoonotic HEV capable of transmission to humans needs to be taken into consideration. This review article focuses on the current knowledge regarding avian HEV in virology, epidemiology, pathogenesis, clinical presentation, transmission, diagnosis and prevention.
HIGHLIGHTS
- The mechanisms of avian HEV replication and pathogenesis are still poorly understood due to the lack of an efficient cell culture system.
- A broader host tropism is also notable in the epidemiological studies with the increased genotypes of avian HEV identified.
- The recent identification and characterization of animal strains of avian HEV has demonstrated the virus’ ability of cross-species infection.
- The potential threat of a zoonotic HEV capable of transmission to humans needs to be taken into consideration.
Introduction
History
In the early 1980s, big liver and spleen disease (BLS) was firstly characterized in chickens in Australia, and was considered to be the most significant disease affecting commercial broiler breeder flocks (Handlinger and Williams, 1988). It was clinically described as a sudden reduction in egg production, splenomegaly, hepatomegaly, and increased mortality (Handlinger and Williams, 1988). The causative agent of this disease, big liver and spleen virus (BLSV), was isolated in the late 1990s, and based on sequence analysis of a 523 bp nucleic acid fragment, it was found that this sequence of BLSV had a 62% homology with the helicase gene of human hepatitis E virus (HEV) (Payne et al., 1999; Zhao et al., 2017). Not long after, another disease affecting chickens, hepatitis-splenomegaly syndrome (HSS) was first described in British Columbia, Canada in 1991 and in eastern Canada and the United States in 1997 (Ritchie and Riddell, 1991; Riddell, 1997). It is characterized by pale to white combs with red edges, abdomens with red ascites, poor to moderate body condition, the regression of ovaries and oviducts, enlarged livers and spleens and increased mortality in 32-week-old broiler breeders and commercial egg laying hens (Riddell, 1997). In 2001, the HEV-related virus, named avian HEV, was characterized from HSS afflicted chickens in the United States. Based on genomic sequencing, this novel virus displayed approximately 50 and 80% nucleotide (nt) sequence identity with mammalian HEV and BLSV, respectively, suggesting that BLS and HSS are caused by variants of the same avian HEV (Haqshenas et al., 2001; Huang et al., 2002). Phylogenetic analyses indicated that avian HEV belongs to a separate floating genus within the family Hepeviridae (Meng, 2008; Yugo and Meng, 2013). In addition, in 2004, avian HEV isolated from presumed healthy flocks was shown to cluster with BLSV and form a distinct branch, which indicated these two are variant strains of the same virus (Sun et al., 2004).
To date, avian HEV is considered as a major causative agent of both BLS and HSS in chickens based on these findings. Avian HEV has now been detected in chickens in a number of countries, including the United States, Hungary, Canada, Australia, Spain, China, Korea, Poland, Israel and the cross-border region Austria and Czechia (Haqshenas et al., 2001; Agunos et al., 2006; Morrow et al., 2008; Bilic et al., 2009; Peralta et al., 2009; Marek et al., 2010; Zhao et al., 2010; Kwon et al., 2012; Konicek et al., 2016).
Classification
Avian HEV belongs to the Orthohepevirus B species of the family Hepeviridae (Emerson and Purcell, 2003). The family contains two genera: Orthohepevirus (all mammalian and avian HEV isolates) and Piscihepevirus (cutthroat trout virus) (Smith et al., 2014; Spahr et al., 2018). There are four species within the genus Orthohepevirus: A to D. Orthohepevirus A consists of the strains isolated from human, pig, wild boar, deer, mongoose, rabbit, moose, and camel (Li et al., 2005; Kim et al., 2011; Izopet et al., 2012; Lee et al., 2016; Meng, 2016; Spahr et al., 2018). Within Orthohepevirus A, there are eight genotypes (1 to 8). Genotypes 1 and 2 are obligate human pathogens (Meng, 2013), whereas genotypes 3 and 4 can infect both human and animals (swine, rabbits, deer and mongooses) (Sridhar et al., 2017). Genotypes 5 and 6 are isolated from wild boars (Takahashi et al., 2011), and genotypes 7 and 8 are found in dromedary and Bactrian camels (Woo et al., 2014; Sridhar et al., 2017). Orthohepevirus B includes the viruses isolated from chickens and wild birds and named avian HEV (Meng, 2016; Reuter et al., 2016). Orthohepevirus C is composed of the strains isolated from rat, greater bandicoot, Asian musk shrew, ferret and mink (Meng, 2016), and Orthohepevirus D is isolated from bat (Meng, 2016). The zoonotic characteristic of HEV has raised great public health concerns regarding cross-species transmission and new genotype discovery in recent years (Mushahwar, 2008; Meng, 2010, 2011).
Within Orthohepevirus B, different avian HEV strains share approximately 73 to 100% identity to one another, and approximately 30 to 50% identity with mammalian HEVs (Huang et al., 2002). Although avian HEV only shares ∼50% identity to human and swine HEV strains, the genomic organization is relatively conserved (Haqshenas et al., 2001; Huang et al., 2004). Now, four major genotypes (1 to 4) were distinguished by genome-wide sequencing (Figure 1) and share approximately 82% identity at the nucleotide level (Syed et al., 2017). Genotype 1, was primarily isolated from BLS chickens in Australia and was also reported to cause the decrease in egg production in chickens during an outbreak in Korea (Payne et al., 1999; Kwon et al., 2012). Genotype 2, was primarily isolated from HSS chickens in the United States and was detected in BLS flocks in Central Europe and Poland, and also isolated from chickens with hepatitis-splenomegaly syndrome in Korea (Haqshenas et al., 2001; Moon et al., 2016; Matczuk et al., 2019). Genotype 3, was isolated from HSS chickens in Europe and from hens with decreased egg laying in China (Morrow et al., 2008; Zhao et al., 2010). Genotype 4, was isolated from affected flocks in Hungary and from bile samples of commercial egg-layer flocks in Taiwan region, which were initially presumed to be healthy (Bányai et al., 2012; Hsu and Tsai, 2014). Recently, a new isolate from chickens suffering from hepatic rupture hemorrhage syndrome (HRHS) in China was shown to be associated with avian HEV and named avian HEV-Hebei, phylogenetic analyses constructed according to the partial helicase and partial capsid gene showed in a single branch in avian HEV groups but distinctly separate from other avian HEV strains, which may further extend the genotypes of avian HEV (Figure 1) (Su et al., 2018a, b). This isolate belongs to a single branch, distinctly separated from other reference avian HEV strains (Figure 1). While all of these genotypes tend to be geographically different, each genotype of avian HEV belongs to a single serotype (Marek et al., 2010; Meng, 2010; Zhao et al., 2015).
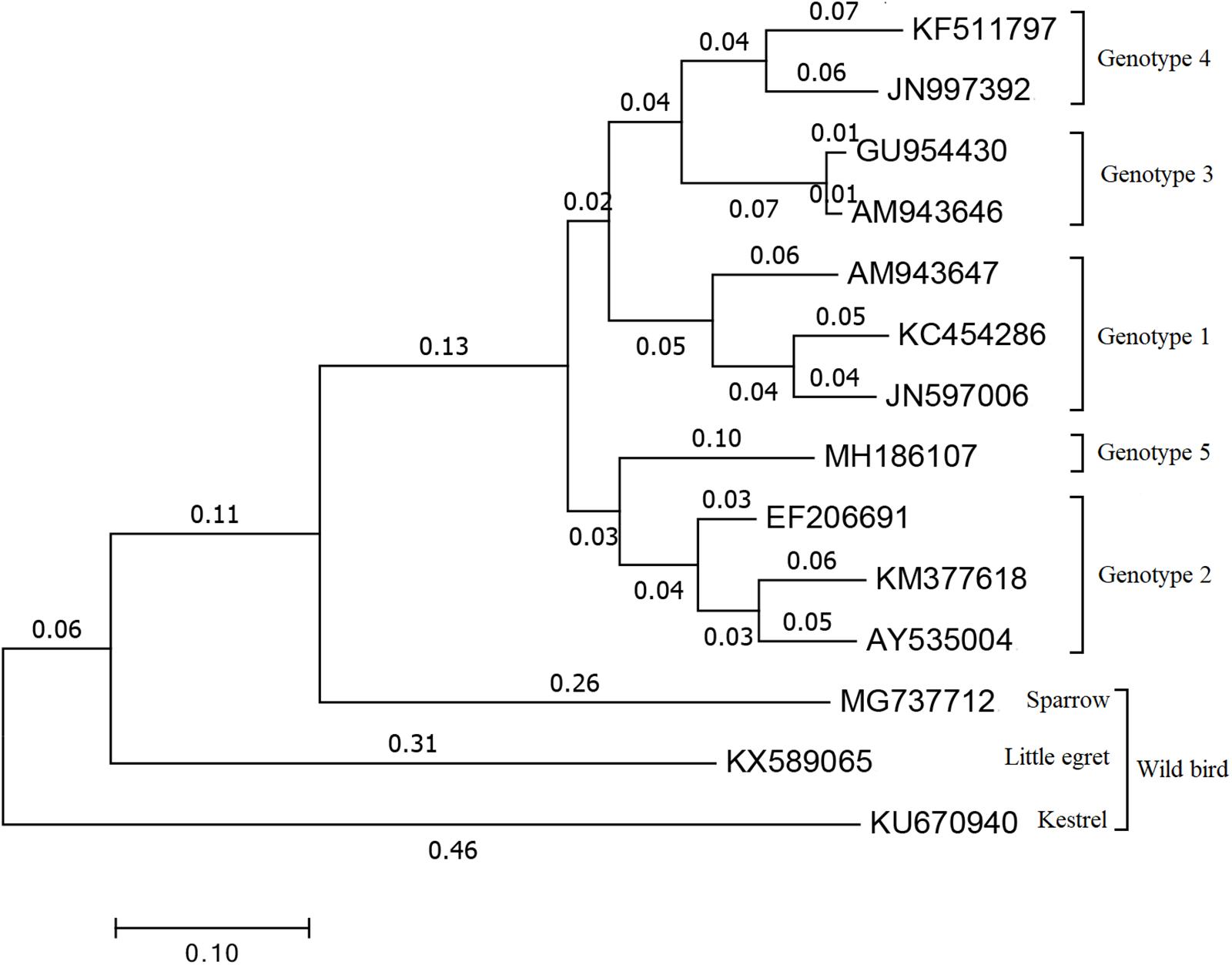
Figure 1. Phylogenetical tree of the current genotypes of different avian HEV strains based on the complete or near complete genomes. The tree was built by MEGA 7.0 using the neighbor-joining (NJ) method with 1000 bootstrap replicates. GenBank accession numbers represents different HEV strains. Genotype 1: AM943647 is for avian HEV strain in Australia, JN597006 and KC454286 are for avian HEV strain in Korea. Genotype 2: AY535004 and EF206691 are two U.S. avian HEV strain, KM377618 is avian HEV strain in Korea. Genotype 3: AM943646 represents European avian HEV strain and GU954430 represents Chinese avian HEV strain. Genotype 4: JN997392 is for avian HEV strain in Hungary and KF511797 is for avian HEV strain in Taiwan region. Genotype 5: MH186107 stands for the recent new type avian HEV strain which isolated in Hebei, China. MG737712 is for sparrow, KX589065 is for little egret and KU670940 is for kestrel which are all representing avian HEV strain isolated from wild birds.
Virology
Virion Properties
Avian HEV is a single-stranded, positive-sense RNA virus which is sensitive to high temperature and iodinated disinfectants, but with certain resistance to acidic and mild alkaline conditions (Meng, 2010). The virions are icosahedral and symmetrical with a diameter of approximately 30–35 nm in size (Meng, 2010). The buoyant density of avian HEV is 1.39–1.40 g/cm3 in cesium chloride (CsCL) and 1.29 g/cm3 in potassium tartrate and glycerol according to a report by Meng (2010).
Genome Organization
The full-length genome of avian HEV is ∼6.6 kb in length, which is 600 bp shorter than mammalian HEV (Figure 2). It contains a short 5′non-coding region (NCR), followed by three open reading frames (ORFs) and a short 3′NCR terminated by a poly(A) tract (Kabrane-Lazizi et al., 1999). The ORF2 and ORF3 are partially overlapped, but neither overlaps with ORF1 (Meng, 2010). The length of the 5′NCR of avian HEV is 24 bp, which is 2–4 bp shorter than that of most human and swine HEV (Huang et al., 2004). The ORF1 (nt 25 to 4620) of avian HEV encodes a viral non-structural protein 1531 amino acids (aa) in length with multiple functional domains based on computer predictions, including a methyltransferase (Methyl, aa 56–241), papain-like cysteine protease (PLP, aa 433–593), hypervariable region (HVR, aa 673–802), helicase (Hel, aa 984–1216) and RNA-dependent RNA polymerase (RdRp, aa 1231–1720) (Huang et al., 2004, 2007; Bilic et al., 2009). The typical motifs I-VI of the helicase superfamily I and the motifs I-III of viral methyltransferase are found conserved throughout the alpha-like virus super-group (Huang et al., 2004).
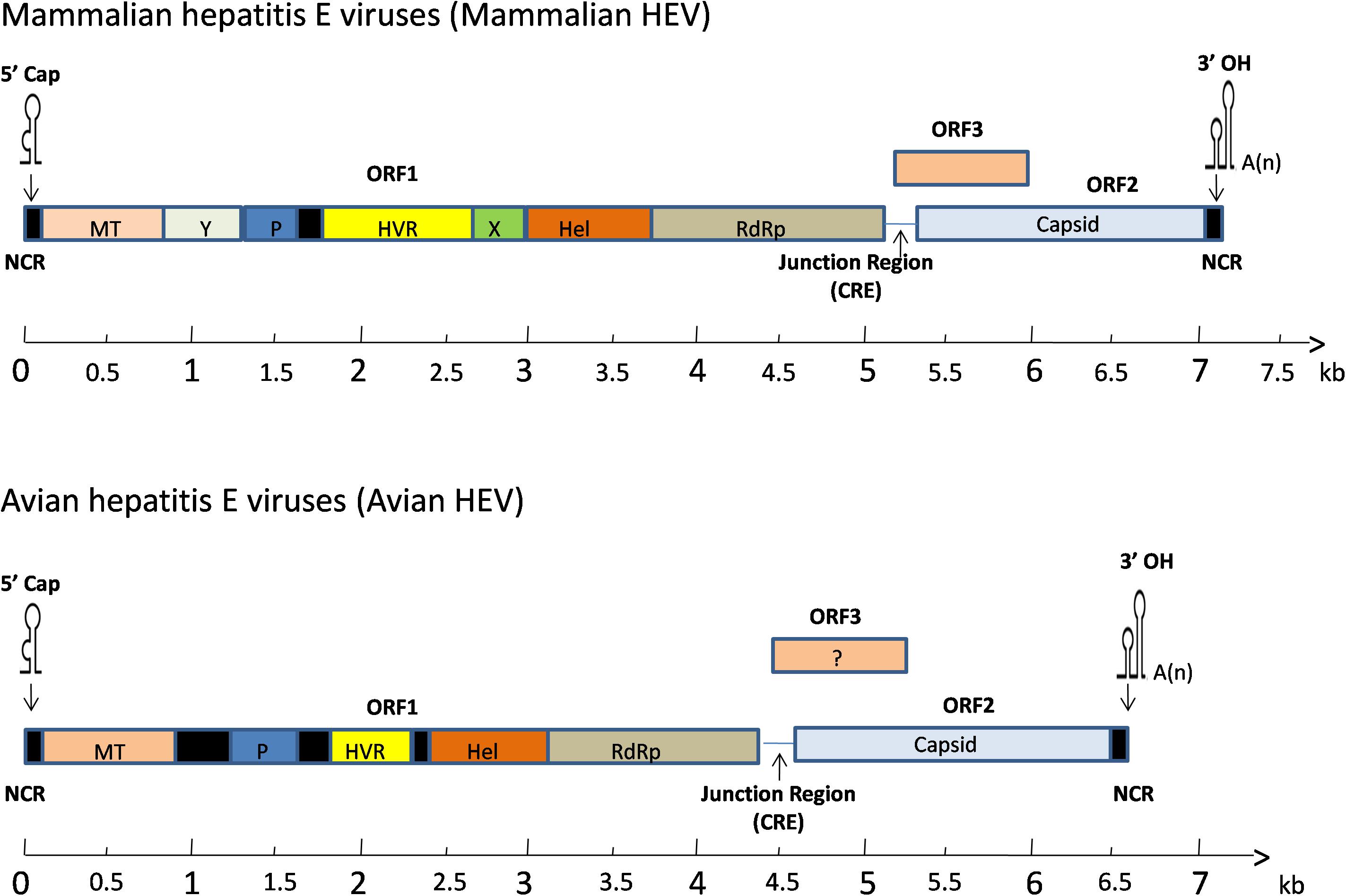
Figure 2. A schematic diagram of comparative genomic organization of mammalian and avian HEV. The three open reading frames (ORFs) are labeled with different colors and shown as above. The avian HEV genome is approximately 600 bp smaller than the mammalian HEV. ORF1 encodes non-structural proteins with the putative functional domains. ORF2 encodes the capsid protein, and ORF3 encodes a phosphoprotein. The ORF2 and ORF3 are partially overlapped, but neither overlaps with ORF1. The genome is capped at the 5′ end and contains a poly(A) tail at the 3′ end. The open reading frame is flanked by non-coding regions (NCR) at the 5′ and 3′ ends of the genome. A junction region exists between ORF1 and ORF3 for mammalian and avian HEV, which is characterized with a stem-loop structure and a cis-reactive element (CRE). MT, methyltransferase; P, a papain-like cysteine protease; HVR, hypervariable region; Hel, helicase; RdRp, RNA-dependent RNA polymerase.
ORF1 accounts for the largest portion in the genome and its products play a very important role in the proper assembly and release of virus particles (Ahmad et al., 2011). The ORF2 (nt 4707 to 6527) encodes the capsid protein of 606 aa in length, sharing 48–49% aa identity with swine and human HEVs (Haqshenas et al., 2001). Six linear antigenic domains (I–VI) have been characterized in avian HEV ORF2 protein, which are located in aa 389–410, 461–492, 556–566, 583–600, 339–389, and 23–85, respectively (Dong et al., 2011; Wang et al., 2014). Domains I and V contains epitopes, which are common among avian, human, and swine HEVs (Wang et al., 2015). The domains II and VI are unique to avian HEV and can induce immune responses in chickens, while domains III and IV only elicit a weak immune response (Zhao et al., 2015). Collectively, the capsid protein contains the most immunogenic epitopes capable of inducing neutralizing antibodies, which are a potential target for vaccine development (Meng, 2008). The smallest one, ORF3 (nt 4654 to 4917) encodes a protein of 87 aa (Huang et al., 2004). Three antigenic domains on ORF3 at amino acids (aa) 1–28, 55–74 and 75–88 were identified in the Chinese isolate of avian HEV (CaHEV) (Zhao et al., 2013b). The aa 75–88 region was shown to be a dominant antigenic domain which contains at least two epitopes, though the neutralizing characteristics of these three antigenic domains are still unknown (Zhao et al., 2013b). The downstream of ORF2 is the 3′NCR of avian HEV, excluding the poly(A) tail, consists of 123 nt (Bilic et al., 2009).
Epidemiology
Avian HEV is considered to be the main causative agent of BLS and HSS (Haqshenas et al., 2001; Huang et al., 2002), which has been reported in Australia (Handlinger and Williams, 1988), the United States (Haqshenas et al., 2001), Canada (Ritchie and Riddell, 1991; Agunos et al., 2006) and Europe (Morrow et al., 2008). In addition, molecular epidemiologic investigations show high HEV-positive rates in chicken flocks in the United States, and that the virus can cause subclinical infections in these chickens (Sun et al., 2004). Serologic investigations show that the positive rate of chicken flocks and chickens are 71 and 30%, 57 and 28%, 95.08 and 40.57% in the United States (2002), Korea (2012) and throughout Taiwan (2013), respectively (Huang et al., 2002; Kwon et al., 2012; Hsu and Tsai, 2014). There is a 20% positive rate for anti-avian HEV antibodies in chickens from Brazil (Vitral et al., 2005), and in Spain, positive rates for farms span a wide range from 20 to 80% (Peralta et al., 2009). In China, the avian HEV seropositive rate in chicken flocks is 35.9% (Zhao et al., 2013a), while in Poland, there is a 56.1% positive rate for avian HEV genotype 2 among breeder broiler and egg laying hen flocks (Matczuk et al., 2019). In the United States between the years 2012 and 2014, 141 chickens from 10 organic layer flocks showed a 40% decrease in the rate of egg production and slightly increased mortality (up to 1% per week), avian HEV RNA was detected in 10 livers out of 141 chickens from two different affected flocks (Crespo et al., 2015). Recently, a disease known as HRHS was proven to be associated with avian HEV, and has emerged in many large-scale layers and breeding farms in China since 2016 (Su et al., 2018a). During this time, over 270,000 chickens from four farms in the Guangdong, Anhui, Hebei and Jilin provinces of China were positive for avian HEV. Those affected flocks showed several death peaks at ages 1 to 5 weeks, 17 to 20 weeks and 27 to 40 weeks. The cumulative mortality was approximately 15%, and the egg laying peak was significantly delayed with a laying rate decreased by 20% (Su et al., 2018b).
Whether infections with avian HEV are age-associated or not, it’s still known of ambiguity as there have been a number of conflicting results. One study showed that 17% of chickens less than 18 weeks old and 36% of adult chickens in the United States were positive for avian HEV antibodies using a truncated recombinant ORF2 antigen (Huang et al., 2002). On the other hand, another report on commercial egg-type chicken flocks in the United States, identified avian HEV in 17-week-old chickens but no infections in 30 to 80-week-old chickens (Gerber et al., 2014). A third study showed that high antibody titers in infected chickens can be expected within the ages of 20–29 weeks (Zhao et al., 2013a). Meanwhile, 10% of chickens at 60 weeks old were also found to be avian HEV seropositive (Zhao et al., 2013a). The result is variable, the different region and flocks may contribute to this issue. Thus, a systematic artificially infection experiment should carry out to confirm age-associate result.
In addition to chickens, avian HEV has also been identified in wild aquatic birds, such as the little egret in Hungary, which may represent a potential novel Orthohepevirus species (Reuter et al., 2016). An avian-like HEV from sparrow feces in the Midwestern United States has also been detected, which may also represent a novel HEV which is related to chicken and little egret HEVs (Yang et al., 2018). In both affected and presumed healthy poultry flocks across Russia and Kazakhstan, a high genetic divergence of avian HEV has also been detected (Sprygin et al., 2012). However, the avian HEV isolates from wild bird to cause chicken infection or the ones from chicken to cause wild bird infection are still uncertain (Reuter et al., 2016). Based on the phylogenetic tree, we speculate that the avian HEV strains from chicken and wild bird have an independent evolution from the same HEV ancestor. In addition, based on the comparisons between the different avian HEV isolates, they have the possible ability for cross-transmission (Meng, 2010).
Co-infection is common in clinical case, avian HEV co-infected with other pathogens has also been reported (Sun et al., 2016; Yang et al., 2016). For example, an epidemiology study showed that both avian HEV and avian leucosis virus subgroup J infection is endemic in those flocks with HSS in China (Sun et al., 2016). Additionally, avian HEV and Marek’s disease virus (MDV) was found to be co-prevalent in a flock of egg-type chickens in China with a double virus positive rate of ∼30% (Yang et al., 2016).
Clinical Presentation
In chickens, subclinical infections of avian HEV are very common in flocks in the United States and other countries, but the morbidity and mortality associated with HSS (or BLS) and other avian HEV infections in the field are relatively low (Huang et al., 2002; Sun et al., 2004; Peralta et al., 2009). The mortality rate is between 0.3 and 1% for avian HEV genotypes 1–3 infections (Meng et al., 2008; Nikonova and Zinyakov, 2012; Yugo et al., 2016). The clinical presentation caused by the virus can be variable. Broiler breeders which have been inoculated with avian HEV genotype 1, typically show more gross ovarian and microscopic lesions than younger chickens inoculated with genotype 2, while avian HEV genotype 3 infection of laying hens can exhibit BLS or HSS, but shows decreased egg production without gross ovary and microscopic lesions (Billam et al., 2005; Park et al., 2015; Zhao et al., 2017). There are several other diseases that could be caused by avian HEV, which are listed as follows (Table 1).
Pathogenesis
Considering the economic losses and the increased clinical infection rates, avian HEV pathogenesis research has become a necessity. The current method to study avian HEV pathogenicity is limited since the virus cannot be efficiently propagated in cell culture, and can only proliferate within living organisms or animals. As such, the pathogenicity of avian HEV remains largely uncharacterized (Meng, 2010; Syed et al., 2017).
Billam et al. (2005) infected 60-week-old specific-pathogen-free (SPF) adult chickens by a natural route, to systematically study HEV pathogenesis and replication in a homologous animal model. They found avian HEV RNA in both the bile and liver of infected chickens, indicating that the virus replicates in the liver. Billam et al. (2008) showed that viral replication initially takes place in gastrointestinal tissues including the colorectal, cecal, jejunal, ileal, duodenal, and cecal tonsil tissues prior to reaching the liver (Billam et al., 2008; Yugo et al., 2016). Based on these results, it can be inferred that the entry of avian HEV first occurs from an oral-route, and then replicates and accumulates in the digestive system. In addition to viral replication, virus attachment to host cells is considered to be another crucial step in viral infections (Schneider-Schaulies, 2000; Yugo et al., 2016). It is reported that the capsid protein region from amino acid 471 to 507 is critical for avian HEV attachment to host cells (Zhang et al., 2016) and the C-terminal region of the capsid ORF2 protein is recognized as putative binding sites for both cellular receptors and neutralizing antibodies (Ahmad et al., 2011). Wang et al. (2018) utilized Lactococcuslactis as a delivery vector expressing higher levels of ΔORF2 (avian HEV truncated ORF2 protein spanning amino acids 249–606 protein) specific IgG antibodies, in order to neutralize or bind to avian HEV and induce effective mucosal immunity, prevented the virus from invading into host cells. These results also showed effective control of avian HEV infection in chickens. Better understanding of viral pathogenesis and replication both in vivo and in vitro is crucial for controlling and diminishing avian HEV infections.
Transmission
Avian HEV is believed to be transmitted mainly by the fecal-oral route through contaminated feed and water in different flocks (Haqshenas et al., 2002; Huang et al., 2002). Furthermore, another new route of transmission has been successfully identified by Billam et al. (2005) experiment in which avian HEV infection was successfully reproduced via nasal and oral route inoculation in SPF chickens. To determine whether avian HEV can be transmitted vertically, Guo et al. (2007) collected and tested embryonated eggs starting from 1 week prior to virus inoculation to 5 weeks post-infection (p.i.), but all of the hatched chicks were negative for avian HEV infection. Conversely, Troxler et al. (2014) research, demonstrated that avian HEV RNA could be detected at week 25 in chicks hatched from laid eggs, indicating the possibility of vertical transmission routes.
Within the eight species of Orthohepevirus A, genotypes 1 and 2 exclusively infect humans, whereas genotypes 3, 4, 7 and 8 can cross species barriers and are denoted as zoonotic genotypes (Purcell and Emerson, 2008; Doceul et al., 2016; Lee et al., 2016; Spahr et al., 2018), and according to the very recent study, Orthohepevirus C was identified to be zoonotic (Murphy et al., 2019). However, Orthohepevirus A (genotypes 5, 6) and D have not been labeled as zoonotic diseases (Liu et al., 2018). Surprisingly, the avian HEV (a member of Orthohepevirus B) also can be transmitted across species barriers, though they do not infect rhesus monkeys or pigs (Sun et al., 2004), but has been reported to infect turkeys under experimental conditions (Sun et al., 2004) and as well as other birds under natural conditions (wild aquatic bird, little egret, song thrush, little owl, feral pigeon and common buzzard), suggesting that avian HEV may have a limited host range and may not infect humans, and that chickens are likely not a reservoir for avian HEV (Huang et al., 2004). Meanwhile, genotypes 1 and 3 are prevalent in wild birds of 4 different species (song thrush, little owl, feral pigeon and common buzzard), which indicates avian HEV transmission between chickens and wild birds (Meng, 2010; Zhang et al., 2017). The presence of HEV in wild birds indicates that wild birds might play a role as a reservoir for avian HEV (Zhang et al., 2017). In addition to wild birds and turkeys, a mixed animal group comprised of chickens, ducks, geese and rabbits also can be infected with avian HEV genotype 3, demonstrating avian HEV could be a cross-species transmission pathogen (Liu et al., 2018). Though avian HEV has not been reported to cause zoonotic infections, its expanded host range and its ability to infect across species raises additional concerns (Meng, 2010).
For the mammalian HEV, it was reported that the ORF1 is considered to play an indispensable role in determining host tropism (Chatterjee et al., 2016). However, there were also some reports which HEV ORF2 and ORF3 protein, host factors, especially host immune status, may also minorly contribute to cross-species transmission of HEV (Ding et al., 2017). For avian HEV, it was also documented that more mutated amino acids in ORF1 of the avian HEV isolated from the rabbit, suggesting that ORF1 maybe also play a role in the cross-transmission of avian HEV (Liu et al., 2018). However, the underlying molecular mechanism that ORF1 plays to alter host cell tropism during HEV cross-species infection is not yet clear.
Diagnosis
The diagnosis for clinical manifestation of avian HEV is based on the detection of specific viral RNA, as the clinical signs and pathologic lesions are atypical (Crespo et al., 2015). All presumptive diagnoses must rely on laboratory tests, primarily based on RT-PCR for virus detection, enzyme-linked immunosorbent assay (ELISA) for avian HEV ORF2 and ORF3 specific IgG antibody detection and electron microscope (EM) for virus particles detection (Meng, 2010). Other lesion detection markers such as serum lactate dehydrogenase (LDH) and serum alanine aminotransferase (ALT) could possibly be used as a supplementary diagnostic method since both are shown to be increased after intravenous (i.v.) and oronasal inoculation.
Avian HEV-specific RT-PCR assays have been developed for the detection of avian HEV infections in chickens (Sun et al., 2004). The nested RT-PCR assays were universally used to detect avian HEV RNA from the fecal, serum, bile, liver and spleen samples. To determine avian HEV infection in the chickens, two truncated genes (ORF1 and ORF2) were amplified by nested RT-PCR at the same time. Recently, the pair of primers designed by Sun et al. (2004) were universally detected different genotypes of avian HEV RNA. Compared with conventional RT-PCR, duplex TaqMan real-time RT-PCR assay was shown to be highly specific (Troxler et al., 2011). A SYBR Green real-time RT-PCR assay was developed for the rapid, specific, and reproducible diagnosis of avian HEV infection in chickens by Zhao et al. (2015), which can accurately detect avian HEV RNA in serum, liver, spleen, and fecal samples with more sensitivity than conventional RT-PCR.
The indirect ELISA (iELISA) methods were also improved, using a newly developed truncated recombinant ORF2 antigen of avian HEV genotype 2 strain to determine avian HEV seroprevalence (Huang et al., 2002). Zhao et al. (2013b) used a truncated ORF2 protein from an avian HEV genotype 3 strain isolated in China (CaHEV) as the coating antigen, and got a sensitivity of 96.1% and a specificity of 95.8%, with 97% corresponding with Western blot results, which can be used for the detection of chicken immunoglobulin G antibodies (IgG) against avian HEV for Chinese poultry farms. The blocking enzyme-linked immunosorbent assay (bELISA) had a sensitivity of 98.3% and a specificity of 93.3% with CaHEV and had no cross-reaction with other anti-avian virus antibodies which can be used for detection of antibodies against avian HEV and showed high reproducibility compared with indirect ELISA and Western blot methods (Liu et al., 2014). Furthermore, two indirect ELISAs, using the truncated capsid proteins separately from the prototype avian HEV genotype 2 and genotype 3 as coated antigens, can be used effectively for the sero-epidemiological detection (Huang et al., 2002). Generally, the C-terminal region of capsid protein (268 amino acids) expressed by bacterial system were used as the coated antigen in ELISA assay (Haqshenas et al., 2002). Four antigenic domains were identified from the region (Zhao et al., 2015). Out of which, the domain I (aa 389–410) and domain V (aa 339–389) can induce a strong immune response in chickens, so this is the optimal selected domain as an antigen for detection of antibodies against avian HEV (Zhao et al., 2014). In addition, the ORF3 protein with lower sensitivity and specificity comparing with ORF2 protein was also used as the antigen to detect antibodies against avian HEV (Syed et al., 2017).
However, the specificity and consistency of the ELISA assays using the different genotypes avian HEV as coating antigen is unknown, since avian HEV isolates from different geographic regions are genetically heterogenic (Huang et al., 2002; Zhao et al., 2013b). Most of these assays are based on antigens expressed by a single avian HEV genotype; these might be a limitation for detecting all other avian HEV genotypes (Zhao et al., 2013b). To efficiently detect HEV infection from different sources, the identification of conserved epitopes may facilitate the development of diagnosis kits for region-specific avian HEV detection.
Prevention and Control
Avian HEV infection is common in chicken flocks, however, there is no effective measure to prevent the disease from spreading (Reuter et al., 2016). Due to the lack of effective commercial vaccines and drugs for preventing disease in chickens, blocking fecal-oral transmission should prevent the spread of virus infection, while the implementation of strict biosecurity regulations at chicken farms may limit the spread of the virus (Molaei et al., 2006; Meng, 2010; Zhao et al., 2017). From Liu et al. (2017) study, cage-free compared to caged animals showed higher positive rates for both antibodies and RNA of avian HEV. The control of chicken excrement pollution could reduce avian HEV infection while implementing a caged living/raising method can help to better prevent HEV transmission. Conversely, Sun et al. (2016) believed epidemiological investigation and elimination of infected chickens to be the only efficient methods for avian HEV prevention and control due to the subclinical and persistent infections in chickens and the lack of availability of vaccines or treatment options (Sun et al., 2016; Nimgaonkar et al., 2017). The most effective measures for avian HEV prevention and control should combine two methods in clinical work since it’s the core of biosecurity-based process.
Truncated avian HEV ORF2 contains immunodominant epitopes, which can induce a protective humoral immune response (Schofield et al., 2000; Meng et al., 2001). Guo et al. (2007) showed that recombinant ORF2 epitope identification can promote the development of efficient vaccines. While the complete ORF3 proteins were evaluated for immunoprotection of chickens against CaHEV infection, Syed et al. (2017) proved the expression of ORF3 protein only provides partial immune protection.
Most of these recombinant ORF2/ORF3 products depend on an Escherichia coli-bacterial expression system (Dong et al., 2011; Zhao et al., 2013b), while Wang et al. (2018) utilized Lactococcuslactis as a delivery vector for truncated avian HEV ORF2 protein, and demonstrated that avian HEV ORF2 protein can induce ideal protection against hepatitis and liver injury caused by avian HEV, indicating that vaccine production using different vectors may differentially alter host immune responses.
Conclusion and Future Perspectives
HEV is an important pathogen affecting both humans and animals, yet many questions remain unanswered, and more research needs to be done to better understand pathogenesis and to aid in the development of vaccines and treatments (Meng, 2010; Yugo et al., 2016). A vaccine for human use, based solely on the capsid protein of a genotype 1 HEV isolate, named Hecolin® , has been approved in China (Nan et al., 2018), but commercial vaccine for avian HEV is unavailable. Though avian HEV is not currently considered a zoonotic disease, it possesses cross-species infection ability with wide prevalence in most countries, which has aroused more attention for avian HEV prophylaxis and treatment. Vaccine development using synthetic peptides against avian HEV may help to minimize the risks of cross-species transmission and limit potential food borne transmission routes (Meng, 2010). The detection of avian HEV is based on the capsid protein (contains immunodominant epitopes and induces a protective humoral immune response) and antigenic domains for laboratory avian HEV serum antibody, but not all these antigens are commercially available (Syed et al., 2017; Nan et al., 2018). Further epitope identification can improve the accuracy of diagnosis kit and the efficacy of subunit vaccines (Nan et al., 2018). Additional pathogenesis studies on the virus could also provide new insights to combat HEV infection. With the accumulation of these fundamental works, it is promising that upcoming avian HEV vaccines will successfully protect against avian HEV, a broad cross-species virus.
Author Contributions
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
Funding
This study was supported by grants from the National Natural Science Foundation China to QZ (31672583). QZ is a Tang Scholar of Northwest A&F University recipient.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors acknowledge Ashley B. Strickland (Division of Immunology, Virginia-Maryland Regional College of Veterinary Medicine, University of Maryland, College Park, MD, United States) and Dr. Mohamed Yosri (Regional Center for Mycology and Biotechnology, Al-Azhar University, Cairo, Egypt) for providing useful suggestions for this review.
References
Agunos, A., Yoo, D., Youssef, S., Ran, D., Binnington, B., and Hunter, D. (2006). Avian hepatitis E virus in an outbreak of hepatitis–splenomegaly syndrome and fatty liver haemorrhage syndrome in two flaxseed-fed layer flocks in Ontario. Avian Pathol. 35, 404–412. doi: 10.1080/03079450600920976
Ahmad, I., Holla, R. P., and Jameel, S. (2011). Molecular virology of hepatitis E virus. Virus Res. 161, 47–58. doi: 10.1016/j.virusres.2011.02.011
Bányai, K., Tóth, G., Ivanics, E., Glávits, R., Szentpáli-Gavallér, K., and Dán, A. (2012). Putative novel genotype of avian hepatitis E virus, Hungary, 2010. Emerg. Infect. Dis. 18, 1365–1368. doi: 10.3201/eid1808.111669
Bilic, I., Jaskulska, B., Basic, A., Morrow, C. J., and Hess, M. (2009). Sequence analysis and comparison of avian hepatitis E viruses from Australia and Europe indicate the existence of different genotypes. J. Gen. Virol. 90, 863–873. doi: 10.1099/vir.0.007179-0
Billam, P., Huang, F., Sun, Z., Pierson, F., Duncan, R., Elvinger, F., et al. (2005). Systematic pathogenesis and replication of avian hepatitis E virus in specific-pathogen-free adult chickens. J. Virol. 79, 3429–3437. doi: 10.1128/jvi.79.6.3429-3437.2005
Billam, P., Pierson, F. W., Li, W., Leroith, T., Duncan, R. B., and Meng, X. J. (2008). Development and validation of a negative-strand-specific reverse transcription-PCR assay for detection of a chicken strain of hepatitis E Virus: identification of nonliver replication sites. J. Clin. Microbiol. 46, 2630–2634. doi: 10.1128/JCM.00536-08
Chatterjee, S. N., Devhare, P. B., Pingle, S. Y., Paingankar, M. S., Arankalle, V. A., and Lole, K. S. (2016). Hepatitis E virus (HEV)-1 harbouring HEV-4 non-structural protein (ORF1) replicates in transfected porcine kidney cells. J. Gen. Virol. 97, 1829–1840. doi: 10.1099/jgv.0.000478
Crespo, R., Opriessnig, T., Uzal, F., and Gerber, P. F. (2015). Avian hepatitis E virus infection in organic layers. Avian Dis. 59, 388–393. doi: 10.1637/11070-032215-reg.1
Ding, Q., Heller, B., Capuccino, J. M., Song, B., Nimgaonkar, I., Hrebikova, G., et al. (2017). Hepatitis E virus ORF3 is a functional ion channel required for release of infectious particles. Proc. Natl. Acad. Sci.U.S.A. 114, 1147–1152. doi: 10.1073/pnas.1614955114
Doceul, V., Bagdassarian, E., Demange, A., and Pavio, N. (2016). Zoonotic hepatitis E virus: classification, animal reservoirs and transmission routes. Viruses 8:270. doi: 10.3390/v8100270
Dong, S., Zhao, Q., Lu, M., Sun, P., Qiu, H., Zhang, L., et al. (2011). Analysis of epitopes in the capsid protein of avian hepatitis E virus by using monoclonal antibodies. J. Virol. Methods 171, 374–380. doi: 10.1016/j.jviromet.2010.11.025
Gerber, P. F., Trampel, D. W., and Opriessnig, T. (2014). Identification and characterization of avian hepatitis E virus in 2013 outbreaks of hepatitis-splenomegaly syndrome in two US layer operations. Avian Pathol. 43, 357–363. doi: 10.1080/03079457.2014.935755
Guo, H., Zhou, E., Sun, Z., and Meng, X.-J. (2007). Egg whites from eggs of chickens infected experimentally with avian hepatitis E virus contain infectious virus, but evidence of complete vertical transmission is lacking. J. Gen. Virol. 88, 1532–1537. doi: 10.1099/vir.0.82689-0
Handlinger, J., and Williams, W. (1988). An egg drop associated with splenomegaly in broiler breeders. Avian Dis. 32, 773–778.
Haqshenas, G., Huang, F., Fenaux, M., Guenette, D., Pierson, F., Larsen, C., et al. (2002). The putative capsid protein of the newly identified avian hepatitis E virus shares antigenic epitopes with that of swine and human hepatitis E viruses and chicken big liver and spleen disease virus. J. Gen. Virol. 83, 2201–2209. doi: 10.1099/0022-1317-83-9-2201
Haqshenas, G., Shivaprasad, H., Woolcock, P., Read, D., and Meng, X. (2001). Genetic identification and characterization of a novel virus related to human hepatitis E virus from chickens with hepatitis–splenomegaly syndrome in the United States. J. Gen. Virol. 82, 2449–2462. doi: 10.1099/0022-1317-82-10-2449
Hsu, I. W. Y., and Tsai, H.-J. (2014). Avian hepatitis E virus in chickens, Taiwan, 2013. Emerg. Infect. Dis. 20, 149–151. doi: 10.3201/eid2001.131224
Huang, F., Haqshenas, G., Shivaprasad, H., Guenette, D., Woolcock, P., Larsen, C., et al. (2002). Heterogeneity and seroprevalence of a newly identified avian hepatitis E virus from chickens in the United States. J. Clin. Microbiol. 40, 4197–4202. doi: 10.1128/jcm.40.11.4197-4202.2002
Huang, F., Sun, Z., Emerson, S., Purcell, R., Shivaprasad, H., Pierson, F., et al. (2004). Determination and analysis of the complete genomic sequence of avian hepatitis E virus (avian HEV) and attempts to infect rhesus monkeys with avian HEV. J. Gen. Virol. 85, 1609–1618. doi: 10.1099/vir.0.79841-0
Huang, Y., Opriessnig, T., Halbur, P., and Meng, X. (2007). Initiation at the third in-frame AUG codon of open reading frame 3 of the hepatitis E virus is essential for viral infectivity in vivo. J. Virol. 81, 3018–3026. doi: 10.1128/jvi.02259-06
Izopet, J., Dubois, M., Bertagnoli, S., Lhomme, S., Marchandeau, S., Boucher, S., et al. (2012). Hepatitis E virus strains in rabbits and evidence of a closely related strain in humans, France. Emerg. Infect. Dis. 18, 1274–1281. doi: 10.3201/eid1808.120057
Kabrane-Lazizi, Y., Meng, X.-J., Purcell, R.H., and Emerson, S.U. (1999). Evidence that the genomic RNA of hepatitis E virus is capped. J. Virol. 73, 8848–8850.
Kim, Y. M., Jeong, S.-H., Kim, J. Y., Song, J. C., Lee, J. H., Kim, J.-W., et al. (2011). The first case of genotype 4 hepatitis E related to wild boar in South Korea. J. Clin. Virol. 50, 253–256. doi: 10.1016/j.jcv.2010.11.005
Konicek, C., Vodržka, P., Bartčk, P., Knotek, Z., Hess, C., Račka, K., et al. (2016). Detection of zoonotic pathogens in wild birds in the cross-border region Austria–Czech Republic. J. Wildlife Dis. 52, 850–861. doi: 10.7589/2016-02-038
Kwon, H. M., Sung, H. W., and Meng, X.-J. (2012). Serological prevalence, genetic identification, and characterization of the first strains of avian hepatitis E virus from chickens in Korea. Virus Genes 45, 237–245. doi: 10.1007/s11262-012-0761-6
Lee, G.-H., Tan, B.-H., Teo, E. C.-Y., Lim, S.-G., Dan, Y.-Y., Wee, A., et al. (2016). Chronic infection with camelid hepatitis E virus in a liver transplant recipient who regularly consumes camel meat and milk. Gastroenterology 150, 355.–357.. doi: 10.1053/j.gastro.2015.10.048
Li, T.-C., Chijiwa, K., Sera, N., Ishibashi, T., Etoh, Y., Shinohara, Y., et al. (2005). Hepatitis E virus transmission from wild boar meat. Emerg. Infect. Dis. 11, 1958–1960. doi: 10.3201/eid1112.051041
Liu, B., Fan, M., Zhang, B., Chen, Y., Sun, Y., Du, T., et al. (2018). Avian hepatitis E virus infection of duck, goose, and rabbit in northwest China. Emerg. Microbes Infect. 7:76.
Liu, B., Sun, Y., Chen, Y., Du, T., Nan, Y., Wang, X., et al. (2017). Effect of housing arrangement on fecal-oral transmission of avian hepatitis E virus in chicken flocks. BMC Vet. Res. 13:282. doi: 10.1186/s12917-017-1203-4
Liu, B., Zhao, Q., Sun, Y., Wang, X., Zhao, J., Du, T., et al. (2014). Development of a blocking ELISA for detection of antibodies against avian hepatitis E virus. J. Virol. Methods 204, 1–5. doi: 10.1016/j.jviromet.2014.03.023
Marek, A., Bilic, I., Prokofieva, I., and Hess, M. (2010). Phylogenetic analysis of avian hepatitis E virus samples from European and Australian chicken flocks supports the existence of a different genus within the Hepeviridae comprising at least three different genotypes. Vet. Microbiol. 145, 54–61. doi: 10.1016/j.vetmic.2010.03.014
Matczuk, A. K., ćwiek, K., and Wieliczko, A. (2019). Avian hepatitis E virus is widespread among chickens in Poland and belongs to genotype 2. Arch. Virol. 164, 595–599. doi: 10.1007/s00705-018-4089-y
Meng, J., Dai, X., Chang, J. C., Lopareva, E., Pillot, J., Fields, H. A., et al. (2001). Identification and characterization of the neutralization epitope (s) of the hepatitis E virus. Virology 288, 203–211. doi: 10.1006/viro.2001.1093
Meng, X.-J. (2011). From barnyard to food table: the omnipresence of hepatitis E virus and risk for zoonotic infection and food safety. Virus Res. 161, 23–30. doi: 10.1016/j.virusres.2011.01.016
Meng, X.-J. (2013). Zoonotic and foodborne transmission of hepatitis E virus. Semin. Liver Dis. 33, 041–049. doi: 10.1055/s-0033-1338113
Meng, X.-J. (2016). Expanding host range and cross-species infection of hepatitis E virus. PLoS Pathog. 12:e1005695. doi: 10.1371/journal.ppat.1005695
Meng, X. (2008). Hepatitis E virus (hepevirus). Encyclopedia of Virology, 3rd Edn: Oxford: Elsevier, 377–383.
Meng, X. (2010). Hepatitis E virus: animal reservoirs and zoonotic risk. Vet. Microbiol. 140, 256–265. doi: 10.1016/j.vetmic.2009.03.017
Meng, X., Shivaprasad, H., and Payne, C. (2008). Hepatitis E virus infections. Dis. Poul. 12, 443–452.
Molaei, G., Andreadis, T. G., Armstrong, P. M., Anderson, J. F., and Vossbrinck, C. R. (2006). Host feeding patterns of Culex mosquitoes and West Nile virus transmission, Northeastern United States. Emerg. Infect. Dis. 12, 468–474. doi: 10.3201/eid1203.051004
Moon, H.-W., Lee, B.-W., Sung, H. W., Yoon, B.-I., and Kwon, H. M. (2016). Identification and characterization of avian hepatitis E virus genotype 2 from chickens with hepatitis-splenomegaly syndrome in Korea. Virus Genes 52, 738–742. doi: 10.1007/s11262-016-1351-9
Morrow, C. J., Samu, G., Mátrai, E., Klausz, A., Wood, A. M., Richter, S., Jaskulska, B., et al. (2008). Avian hepatitis E virus infection and possible associated clinical disease in broiler breeder flocks in Hungary. Avian Pathol. 37, 527–535. doi: 10.1080/03079450802356946
Murphy, E. G., Williams, N. J., Jennings, D., Chantrey, J., Verin, R., Grierson, S., and Bennett, M. (2019). First detection of Hepatitis E virus (Orthohepevirus C) in wild brown rats (Rattus norvegicus) from Great Britain. Zoonoses and Public Health. doi: 10.1111/zph.12581 [Epub ahead of print].
Mushahwar, I. K. (2008). Hepatitis E virus: molecular virology, clinical features, diagnosis, transmission, epidemiology, and prevention. J. Med. Virol. 80, 646–658. doi: 10.1002/jmv.21116
Nan, Y., Wu, C., Zhao, Q., Sun, Y., Zhang, Y.-J., and Zhou, E.-M. (2018). Vaccine development against zoonotic hepatitis E virus: open questions and remaining challenges. Front. Microbiol. 9:266. doi: 10.3389/fmicb.2018.00266
Nikonova, Z. B., and Zinyakov, N. G. (2012). Avian hepatitis E virus identified in Russian chicken flocks exhibits high genetic divergence based on the ORF2 capsid gene AU - Sprygin, A. V. Avian Pathol. 41, 459–463. doi: 10.1080/03079457.2012.711464
Nimgaonkar, I., Ding, Q., Schwartz, R. E., and Ploss, A. (2017). Hepatitis E virus: advances and challenges. Nat. Rev. Gastroenterol. Hepatol. 15, 96–110. doi: 10.1038/nrgastro.2017.150
Park, S.-J., Lee, B.-W., Moon, H.-W., Sung, H. W., Yoon, B.-I., Meng, X.-J., et al. (2015). Construction of an infectious cDNA clone of genotype 1 avian hepatitis E virus: characterization of its pathogenicity in broiler breeders and demonstration of its utility in studying the role of the hypervariable region in virus replication. J. Gen. Virol. 96, 1015–1026. doi: 10.1099/vir.0.000045
Payne, C., Ellis, T., Plant, S., Gregory, A., and Wilcox, G. (1999). Sequence data suggests big liver and spleen disease virus (BLSV) is genetically related to hepatitis E virus. Vet. Microbiol. 68, 119–125. doi: 10.1016/s0378-1135(99)00067-x
Peralta, B., Biarnés, M., Ordóñez, G., Porta, R., Martín, M., Mateu, E., et al. (2009). Evidence of widespread infection of avian hepatitis E virus (avian HEV) in chickens from Spain. Vet. Microbiol. 137, 31–36. doi: 10.1016/j.vetmic.2008.12.010
Purcell, R., and Emerson, S. (2008). Hepatitis E: an emerging awareness of an old disease. J. Hepatol. 48, 494–503. doi: 10.1016/j.jhep.2007.12.008
Reuter, G., Boros, A., Mátics, R., Kapusinszky, B., Delwart, E., and Pankovics, P. (2016). A novel avian-like hepatitis E virus in wild aquatic bird, little egret (Egretta garzetta), in Hungary. Infect. Genet. Evol. 46, 74–77. doi: 10.1016/j.meegid.2016.10.026
Riddell, C. (1997). Hepatitis-splenomegaly syndrome. Diseases of poultry. Iowa State University Press, Ames, 1041.
Ritchie, S. J., and Riddell, C. (1991). British Columbia.“Hepatitis-splenomegaly” syndrome in commercial egg laying hens. Can. Vet. J. 32, 500–501.
Schneider-Schaulies, J. (2000). Cellular receptors for viruses: links to tropism and pathogenesis. J. Gen. Virol. 81, 1413–1429. doi: 10.1099/0022-1317-81-6-1413
Schofield, D., Glamann, J., Emerson, S., and Purcell, R. (2000). Identification by phage display and characterization of two neutralizing chimpanzee monoclonal antibodies to the hepatitis E virus capsid protein. J. Virol. 74, 5548–5555. doi: 10.1128/jvi.74.12.5548-5555.2000
Smith, D. B., Simmonds, P., Jameel, S., Emerson, S. U., Harrison, T. J., Meng, X.-J., et al. (2014). Consensus proposals for classification of the family Hepeviridae. J. Gen. Virol. 95, 2223–2232. doi: 10.1099/vir.0.068429-0
Spahr, C., Knauf-Witzens, T., Vahlenkamp, T., Ulrich, R. G., and Johne, R. (2018). Hepatitis E virus and related viruses in wild, domestic and zoo animals: a review. Zoonoses Public Health 65, 11–29. doi: 10.1111/zph.12405
Sprygin, A., Nikonova, Z., and Zinyakov, N. (2012). Avian hepatitis E virus identified in Russian chicken flocks exhibits high genetic divergence based on the ORF2 capsid gene. Avian Pathol. 41, 459–463. doi: 10.1080/03079457.2012.711464
Sridhar, S., Teng, J. L. L., Chiu, T.-H., Lau, S. K. P., and Woo, P. C. Y. (2017). Hepatitis E virus genotypes and evolution: emergence of camel hepatitis E variants. Int. J. Mol. Sci. 18:869. doi: 10.3390/ijms18040869
Su, Q., Li, Y., Meng, F., Cui, Z., Chang, S., and Zhao, P. (2018a). Hepatic rupture hemorrhage syndrome in chickens caused by a novel genotype avian hepatitis E virus. Vet. Microbiol. 222, 91–97. doi: 10.1016/j.vetmic.2018.06.019
Su, Q., Li, Y., Zhang, Y., Zhang, Z., Meng, F., Cui, Z., et al. (2018b). Characterization of the novel genotype avian hepatitis E viruses from outbreaks of hepatic rupture haemorrhage syndrome in different geographical regions of China. Transbound. Emerg. Dis. 65, 2017–2026. doi: 10.1111/tbed.12987
Sun, Y., Du, T., Liu, B., Syed, S. F., Chen, Y., Li, H., et al. (2016). Seroprevalence of avian hepatitis E virus and avian leucosis virus subgroup J in chicken flocks with hepatitis syndrome, China. BMC Vet. Res. 12:261. doi: 10.1186/s12917-016-0892-4
Sun, Z., Larsen, C., Huang, F., Billam, P., Pierson, F., Toth, T., et al. (2004). Generation and infectivity titration of an infectious stock of avian hepatitis E virus (HEV) in chickens and cross-species infection of turkeys with avian HEV. J. Clin. Microbiol. 42, 2658–2662. doi: 10.1128/jcm.42.6.2658-2662.2004
Syed, S. F., Sun, Y., Du, T., Chen, Y., Liu, B., Wang, X., et al. (2017). Evaluation of recombinant Chinese avian hepatitis E virus (CaHEV) ORF2 and ORF3 proteins for protection of chickens against CaHEV infection. Vaccine 35, 3482–3489. doi: 10.1016/j.vaccine.2017.05.030
Tablante, N., Vaillancourt, J. P., and Julian, R. (1994). Necrotic, haemorrhagic, hepatomegalic hepatitis associated with vasculitis and amyloidosis in commercial laying hens. Avian Pathol. 23, 725–732. doi: 10.1080/03079459408419041
Takahashi, M., Nishizawa, T., Sato, H., Sato, Y., Nagashima, S., and Okamoto, H. (2011). Analysis of the full-length genome of a hepatitis E virus isolate obtained from a wild boar in Japan that is classifiable into a novel genotype. J. Gen. Virol. 92, 902–908. doi: 10.1099/vir.0.029470-0
Troxler, S., Marek, A., Prokofieva, I., Bilic, I., and Hess, M. (2011). TaqMan real-time reverse transcription-PCR assay for universal detection and quantification of avian hepatitis E virus from clinical samples in the presence of a heterologous internal control RNA. J. Clin. Microbiol. 49, 1339–1346. doi: 10.1128/JCM.01626-10
Troxler, S., Pać, K., Prokofieva, I., Liebhart, D., Chodakowska, B., Furmanek, D., et al. (2014). Subclinical circulation of avian hepatitis E virus within a multiple-age rearing and broiler breeder farm indicates persistence and vertical transmission of the virus. Avian Pathol. 43, 310–318. doi: 10.1080/03079457.2014.924616
Vitral, C. L., Pinto, M. A., Lewis-Ximenez, L. L., Khudyakov, Y. E., Dos Santos, D. R., and Gaspar, A. M. C. (2005). Serological evidence of hepatitis E virus infection in different animal species from the Southeast of Brazil. Mem. Inst. Oswaldo Cruz 100, 117–122. doi: 10.1590/s0074-02762005000200003
Wang, D., Zhang, Y., Ma, C., Ma, D., Zhao, Q., Wang, F., et al. (2018). Live recombinant Lactococcuslactis expressing avian hepatitis virus ORF2 protein: immunoprotection against homologous virus challenge in chickens. Vaccine 36, 1108–1115. doi: 10.1016/j.vaccine.2018.01.003
Wang, L., Sun, Y., Du, T., Wang, C., Xiao, S., Mu, Y., et al. (2014). Identification of an antigenic domain in the N-terminal region of avian hepatitis E virus (HEV) capsid protein that is not common to swine and human HEVs. J. Gen. Virol. 95, 2710–2715. doi: 10.1099/vir.0.069021-0
Wang, X., Zhao, Q., Dang, L., Sun, Y., Gao, J., Liu, B., et al. (2015). Characterization of two novel linear B-cell epitopes in the capsid protein of avian hepatitis E virus (HEV) that are common to avian, swine, and human HEVs. J. Virol. 89, 5491–5501. doi: 10.1128/JVI.00107-15
Woo, P. C., Lau, S. K., Teng, J. L., Tsang, A. K., Joseph, M., Wong, E. Y., et al. (2014). New hepatitis E virus genotype in camels, the Middle East. Emerg. Infect. Dis. 20, 1044–1048. doi: 10.3201/eid2006.140140
Yang, C., Wang, L., Shen, H., Zheng, Y., Gauger, P. C., Chen, Q., et al. (2018). Detection and genomic characterization of new avian-like hepatitis E virus in a sparrow in the United States. Arch. Virol. 163, 2861–2864. doi: 10.1007/s00705-018-3925-4
Yang, S., Wang, L., and Sun, S. (2016). Natural infection with avian hepatitis E virus and marek’s disease virus in brown layer chickens in China. Avian Dis. 60, 698–704. doi: 10.1637/11386-013016-Reg.1
Yugo, D. M., Hauck, R., Shivaprasad, H. L., and Meng, X.-J. (2016). Hepatitis virus infections in poultry. Avian Dis. 60, 576–588.
Yugo, D. M., and Meng, X.-J. (2013). Hepatitis E virus: foodborne, waterborne and zoonotic transmission. Int. J. Environ. Res. Public Health 10, 4507–4533. doi: 10.3390/ijerph10104507
Zhang, X., Bilic, I., Marek, A., Glösmann, M., and Hess, M. (2016). C-Terminal amino acids 471-507 of avian hepatitis E virus capsid protein are crucial for binding to avian and human cells. PLoS One 11:e0153723. doi: 10.1371/journal.pone.0153723
Zhang, X., Bilic, I., Troxler, S., and Hess, M. (2017). Evidence of genotypes 1 and 3 of avian hepatitis E virus in wild birds. Virus Res. 228, 75–78. doi: 10.1016/j.virusres.2016.11.028
Zhao, Q., Liu, B., Sun, Y., Du, T., Chen, Y., Wang, X., et al. (2017). Decreased egg production in laying hens associated with infection with genotype 3 avian hepatitis E virus strain from China. Vet. Microbiol. 203, 174–180. doi: 10.1016/j.vetmic.2017.03.005
Zhao, Q., Sun, Y., Zhao, J., Hu, S., Zhao, F., Chen, F., et al. (2013a). Development and application of an indirect ELISA for detection of antibodies against avian hepatitis E virus. J. Virol. Methods 187, 32–36. doi: 10.1016/j.jviromet.2012.08.026
Zhao, Q., Sun, Y.-N., Hu, S.-B., Wang, X.-J., Xiao, Y.-H., Hsu, W. H., et al. (2013b). Characterization of antigenic domains and epitopes in the ORF3 protein of a Chinese isolate of avian hepatitis E virus. Vet. Microbiol. 167, 242–249. doi: 10.1016/j.vetmic.2013.08.006
Zhao, Q., Sun, Y., Zhang, G., Widen, F., and En-Min, Z. (2014). Antigenic domains of avian HEV: bases for serological diagnosis of HEV in avian, swine and human. Br. J. Virol. 1, 103–105.
Zhao, Q., Syed, S. F., and Zhou, E.-M. (2015). Antigenic properties of avian hepatitis E virus capsid protein. Vet. Microbiol. 180, 10–14. doi: 10.1016/j.vetmic.2015.08.016
Keywords: avian HEV, big liver and spleen virus, hepatitis-splenomegaly syndrome, hepatic rupture hemorrhage syndrome, genotypes, virology
Citation: Sun P, Lin S, He S, Zhou E-M and Zhao Q (2019) Avian Hepatitis E Virus: With the Trend of Genotypes and Host Expansion. Front. Microbiol. 10:1696. doi: 10.3389/fmicb.2019.01696
Received: 08 May 2019; Accepted: 09 July 2019;
Published: 24 July 2019.
Edited by:
Jianrong Li, The Ohio State University, United StatesReviewed by:
Yao-Wei Huang, Zhejiang University, ChinaMijia Lu, The Ohio State University, United States
Copyright © 2019 Sun, Lin, He, Zhou and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Shenghu He, aGVzaGVuZ2h1MzA4QDE2My5jb20=; En-Min Zhou, emhvdWVtQG53c3VhZi5lZHUuY24=; Qin Zhao, cWluemhhb18yMDA0QG53c3VhZi5lZHUuY24=
†These authors have contributed equally to this work
 Peng Sun
Peng Sun Shaoli Lin4†
Shaoli Lin4† En-Min Zhou
En-Min Zhou Qin Zhao
Qin Zhao