- 1Crop Bioprotection Research Unit, National Center for Agricultural Utilization Research, Agricultural Research Service, United States Department of Agriculture, Peoria, IL, United States
- 2Bioenergy Research Unit, National Center for Agricultural Utilization Research, Agricultural Research Service, United States Department of Agriculture, Peoria, IL, United States
Iturins and closely related lipopeptides constitute a family of antifungal compounds known as iturinic lipopeptides that are produced by species in the Bacillus subtilis group. The compounds that comprise the family are: iturin, bacillomycin D, bacillomycin F, bacillomycin L, mycosubtilin, and mojavensin. These lipopeptides are prominent in many Bacillus strains that have been commercialized as biological control agents against fungal plant pathogens and as plant growth promoters. The compounds are cyclic heptapeptides with a variable length alkyl sidechain, which confers surface activity properties resulting in an affinity for fungal membranes. Above a certain concentration, enough molecules enter the fungal cell membrane to create a pore in the cell wall, which leads to loss of cell contents and cell death. This study identified 330 iturinic lipopeptide clusters in publicly available genomes from the B. subtilis species group. The clusters were subsequently assigned into distinguishable types on the basis of their unique amino acid sequences and then verified by HPLC MS/MS analysis. The results show some lipopeptides are only produced by one species, whereas certain others can produce up to three. In addition, four species previously not known to produce iturinic lipopeptides were identified. The distribution of these compounds among the B. subtilis group species suggests that they play an important role in their speciation and evolution.
Introduction
Iturins are an important class of lipopeptides that have been widely studied for their antibiotic activities and are produced by members of the Bacillus subtilis group (Ongena and Jacques, 2008). Iturins were first reported as an antibiotic produced by B. subtilis in 1950 and named after the Ituri region in the Congo where the strain was isolated (Delcambe, 1950). The chemical structure of iturin A was later reported to be a cyclic heptapeptide with an alkyl chain (Peypoux et al., 1978). The mode of action of these lipopeptides was shown to be pore formation in cell membranes (Besson et al., 1984; Maget-Dana et al., 1985). All of them have been shown to have strong antifungal activity, and they are known active ingredients in many biological control products that target fungal plant pathogens (Ongena and Jacques, 2008). In addition to their antifungal activity, they have been shown to induce defense responses in plants (Farace et al., 2015; Park et al., 2016; Wu et al., 2018).
Several other lipopeptides have been identified as being closely related to iturins: mycosubtilin (Peypoux et al., 1976, 1986), bacillomycin L (Besson et al., 1977), bacillomycin D (Peypoux et al., 1980, 1981), bacillomycin F (Mhammedi et al., 1982), and mojavensin A (Ma et al., 2012). Along with iturin, these compounds form the iturinic lipopeptide multigene family. A defining feature of this family is that the first three amino acids of the cyclic heptapeptide are shared among members, whereas the remaining four amino acids are variable (Figure 1). In addition, family members exist as different alkyl isomers and are often produced as a mixture of closely related alkyl tails (e.g., C14, iso-C15, anteiso-C15) (Hourdou et al., 1989; Iwase et al., 2009). In this study, we only considered the nature of the cyclic peptide and disregarded the alkyl chain isomers.
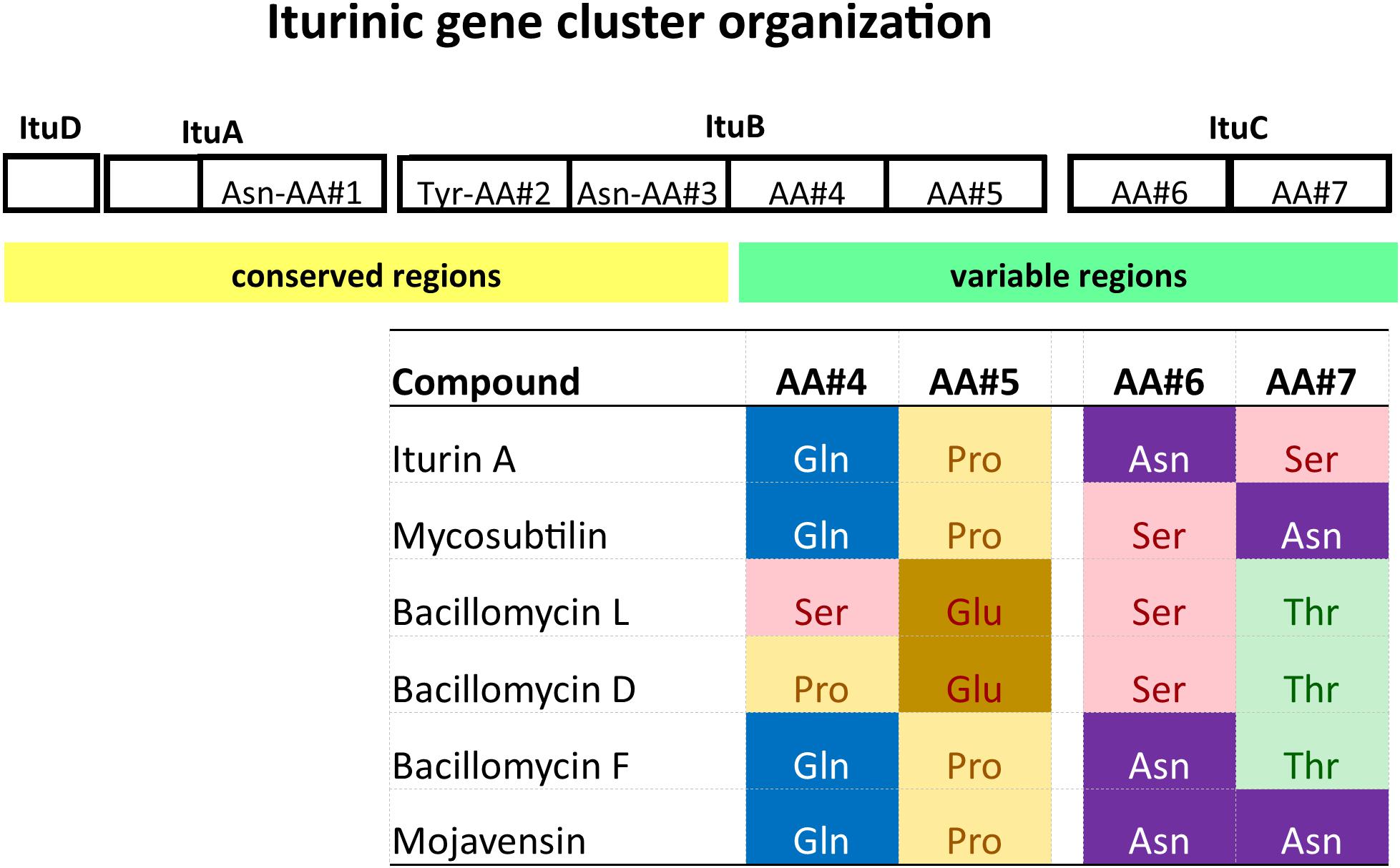
Figure 1. Gene organization and structure of a generic iturinic lipopeptide biosynthesis cluster and sequence of the cyclic heptapeptide of iturinic lipopeptides. The first three positions are Asn-Tyr-Asn for all compounds.
Iturinic lipopeptides are encoded by a cluster of four genes for biosynthesis, which is carried out by non-ribosomal peptide synthetases (NRPS). Iturinic lipopeptide genes are also characterized by modular gene organization, which was first described by Donadio et al. (1991) who defined a module as a “repeated unit” consisting of a specific sequence of nucleotides (or deduced amino acids at the protein level) within one or more open reading frames (i.e., genes) (Donadio et al., 1991). In the case of iturinic lipopeptides, there are 7 modules, each consisting of about 1,000 amino acid sites, that are spread across three genes: 1 in the second gene of the cluster, 4 in the third gene, and 2 in the fourth gene (Duitman et al., 1999; Tsuge et al., 2001; Moyne et al., 2004). Each module contributes a single amino acid that the NRPS enzymatic complex forms into a cyclic peptide consisting of seven amino acids. If an amino acid sequence comparison of all cyclic heptapeptides produced by members of the iturinic lipopeptide family is conducted, it becomes apparent that the first three amino acids of the cyclic heptapeptide are conserved among all family members, whereas the fourth through seventh amino acids may vary (Figure 1). This variation in amino acid sequence is used to assign a lipopeptide clusters to different “types.” The goal of this study is to characterize the diversity of iturin lipopeptide family members produced by the B. subtilis species group and identify evolutionary mechanisms associated with their diversification.
In recent years, the taxonomy of the B. subtilis species group has been updated and clarified as a result of whole genome sequencing (Dunlap, 2015; Dunlap et al., 2015a, b, 2016b). Prior to the availability of genome sequences, the group was plagued with taxonomic inconsistencies and misidentifications. This was primarily due to the low resolution of 16S rRNA sequencing for this group and the general lack of distinguishing phenotypes (Rooney et al., 2009). A second aspect that impacted research in this area was single dimension, low resolution mass spectroscopy (e.g., MALDI of liquid culture supernatant), which is useful in identifying parent ion masses but not useful in distinguishing between structural isomers. This problem is compounded by a number of common structural variants, such as differences in the alkyl chain length (e.g., C14, C15, C16). These all result in parent ion differences of 1 methylene unit (14.02 Da), which is the same for some common amino acid changes in iturinic peptides (e.g., serine to threonine). The nature of these parent ion molecular weight changes makes it impossible to determine their correct assignment without advanced mass spectroscopy approaches (such as, LC-MS/MS). The combination of inconsistent identification of the producing strains and the ambiguous identification of the compounds has clouded some of the past research literature in this area.
Materials and Methods
Bacterial Strains
All publicly available genomes in the B. subtilis group were downloaded from GenBank on December 30, 2018 and comprised the data set. Genomes with an unusually high number of contigs or pseudogenes were excluded from the analysis. The taxonomy of the genomes was determined using a 6 gene MLSA approach (Rooney et al., 2009) using BIGSdb software (Jolley and Maiden, 2010). Strains confirmed to belong within the B. subtilis group were included in the data set.
Iturinic Lipopeptide Cluster Mining
The genomes in the data set were BLASTed using genes from known iturinic lipopeptide gene clusters with BIGSdb software (Jolley and Maiden, 2010) in order to identify homologous iturinic lipopeptide gene clusters. Putative gene cluster homologs were sorted into groups based on species assignment and high nucleotide similarity (>94%) across the full alignment length. For a given species, if the gene clusters did not share the full alignment length (e.g., a change in amino acid module results in a 3–4 Kbp nucleotide change), a new cluster group was defined to include these homologs. A cluster groups represent a different lipopeptide within a given species. This resulted in 11 species containing one to three gene cluster types for a total of 15 gene cluster groups. A representative from each species and cluster type was selected to confirm the product of the cluster using MS/MS. A representative strain for each cluster group was obtained, except for two, Bacillus genomospecies #1 gene cluster group and Bacillus siamensis bacillomycin D group, as strains from these groups were not available in public collections and attempts to obtain them from private laboratories were unsuccessful. However, we are confident in their assignments based on high nucleotide sequence similarity (>90%) to the same verified gene cluster in closely related species. A list of links to representative ituB and ituC genes by gene cluster type and species is provided in Supplementary Table S1. In addition, the 4′-phosphopantetheinyl transferase (sfp) (GenBank#WP_003246659.1) required for NRPS activation (Quadri et al., 1998) was confirmed to be present in each genome with an identified iturinic cluster by protein BLAST.
Phylogenetic Analysis
A core genome phylogeny was used to show the taxonomic relationships that comprise the B. subtilis species group. The core genome determination and subsequent alignments were produced for all the type strains in the group with BIGSdb software (Jolley and Maiden, 2010) and consists of 991 genes. The phylogenetic tree was constructed using MEGA X software (Kumar et al., 2018). The neighbor-joining tree was determined using the Tamura-Nei model (0.40, gamma distributed with invariant sites) based on model testing under MEGA X (Kumar et al., 2018). Measures of bootstrap support for internal branches were obtain from 1,000 pseudoreplicates. A second phylogeny was determined using a 5,395 bp fragment of the ItuA gene (6963.12358 of GenBank#AB050629.1) and extracted from all the genomes containing an iturinic cluster. It was processed and analyzed as described above. The final tree was limited to one strain per species to facilitate analyses. Syntenic relationships were determined by manually examining the eight genes located before and after the iturinic lipopeptide gene cluster for at least five random references strains from each species. The average nucleotide identity (ANI) of the genomes was determined using OrthoANI software (Lee et al., 2016) and the nucleotide identity percentages of the partial ituA gene were determined by BLAST under BIGSdb software (Jolley and Maiden, 2010).
Mass Spectroscopy
Strains listed in Supplementary Table S2 as “confirmed with MS/MS” were grown in 5 ml Tryptone-Yeast-Glucose media at 37°C until the late stationary phase (∼96 h). The culture media was centrifuged at 13,000 × g for 10 min and the supernatant removed. Mass spectrometry of the supernatant samples (25 μL injections) were collected by LC-MS (Thermo Acela HPLC) through a narrow-bore (2.1 μmm × 150 mm, 3 μm particle size) C18 column (Inertsil, GL Sciences, Inc., Torrance, CA, United States) running a gradient elution of 95% A:5% B (eluent A 18 MΩ water/0.1% formic acid, eluent B 100% methanol/0.1% formic acid) to 5% A:95% B over 65 min at a flow rate of 250 μL/min, followed by a 5 min B washout and 10 min re-equilibration, while maintaining a constant column temperature of 30°C. Electrospray positive mode ionization data were collected with a linear ion trap-Orbitrap mass spectrometer (Thermo LTQ-Orbitrap Discovery) under Xcalibur 2.1 control. Prior to LC-MSn experiments the instrument was tuned and calibrated using the LTQ tune mix. Masses corresponding to iturinic lipopeptides were used to limit the collection of MS2 data to only the lipopeptides of interest (Supplementary Figure S1A). Tandem mass spectral data of iturinic peptides was collected using collision-induced dissociation [CID, collision energy (CE) = 25 and 35] in the LTQ and higher-energy collision dissociation (HCD, CE = 35 and 45) in the Orbitrap analyzer, as described in Knight et al. (2018). All tandem mass spectra were manually interpreted for assignment of the lipopeptides.
Comparison to NRPS Substrate Predictor Software
Proteins annotated as ituA, ituB, and ituC based the genes identified in our initial BLASTing and listed in Supplementary Table S1 were subjected to substrate prediction using NRPSsp software (Prieto et al., 2012).
Development of Primers for Bacillus velezensis Strains
Sequences representing ituB gene homologs were extracted from all publicly available B. velezensis genomes. These sequences were aligned using CLCbio Genomics Workbench 11.0 (Qiagen Inc., Cambridge, MA, United States) and this alignment was used to identify conserved regions of nucleotides, which were used to design primers. Primers were identified for each gene cluster type (e.g., iturin, bacillomycin D, and bacillomycin L). The forward primer (ituB-fwd, 5′-CACGAACAGACAAAACA-3′) is the same for all three sets and is located on the conserved amino acid (AA) #3 module (Figure 1), while the reverse primers are located on variable AA#4 modules (Figure 1). The reverse primers are ituB-iturin-rev, 5′-TGCGCAAAGCATCGT-3′, ituB-bacD-rev5′-CTTGCGGCGTTTGTG-3′, and ituB-R-bacL5′-GGTCGCTCCTGAATCT-3′. The primers were designed to have an annealing temperature of 55 ± 2°C. PCR testing – DNA was extracted from 1 day old cultures grown on TGY for each isolate using the QIAamp DNA Mini QIAcube Kit (Qiagen Inc., Germantown, MD, United States) according to manufacturer’s protocol instructions. PCR reactions were carried out in 25 μl volumes using 2× Amplitaq gold master mix, 50 ng of DNA and 1 μM primers (Bvel-f and Bvel-R) under the manufacturer’s instructions (AmpliTaq Gold, Invitrogen) with an annealing temperature of 55°C.
Results
Iturinic Lipopeptide Diversity at the Species Level
Genome mining identified iturinic lipopeptide gene clusters in 11 species or subspecies from the B. subtilis species group (Figure 2). It was determined that one of the clades in the reconstructed species phylogeny contained an iturinic lipopeptide gene cluster, and this clade corresponds to a hitherto unknown species based on ANI calculations (<96% with known species; Richter and Rosselló-Móra, 2009). This clade will be referred to as “Bacillus genomospecies #1.” The results show that 8 of the 11 species in the B. subtilis species group only make one iturinic compound (Figure 2), although the analyses of the species that seemingly only make one compound are limited by the number of genomes available (i.e., Bacillus tequilensis, Bacillus nakamurai, Bacillus swezeyi, and “Bacillus genomospecies #1”). In the case of the remaining single compound producing strains (Bacillus halotolerans, Bacillus subtilis subsp. spizizenii, Bacillus subtilis subsp. inaquosorum, and Bacillus atrophaeus) we feel confident that they likely produce only one compound, as we analyzed at least 9 genomes per species (Supplementary Table S2). In nearly all instances, either all of the genomes examined for a given species had an iturinic lipopeptide gene cluster or all genomes of the species lacked the cluster entirely. The two exceptions were. B. halotolerans and B. amyloliquefaciens, for which 72% (13/18) of genomes and 11% (2/19) of genomes, respectively, lacked the cluster. Supplementary Table S2 shows the assignment of the iturinic lipopeptide for all genomes in which a gene cluster was present. No strain was found to contain more than one iturinic cluster. A protein BLAST search only identified three instances in which the cluster was found in a species other than a member of the B. subtilis species group; all of these were found in strains of Paenibacillus larvae (GenBank accession numbers: CP019655, CP020557, CP003355).
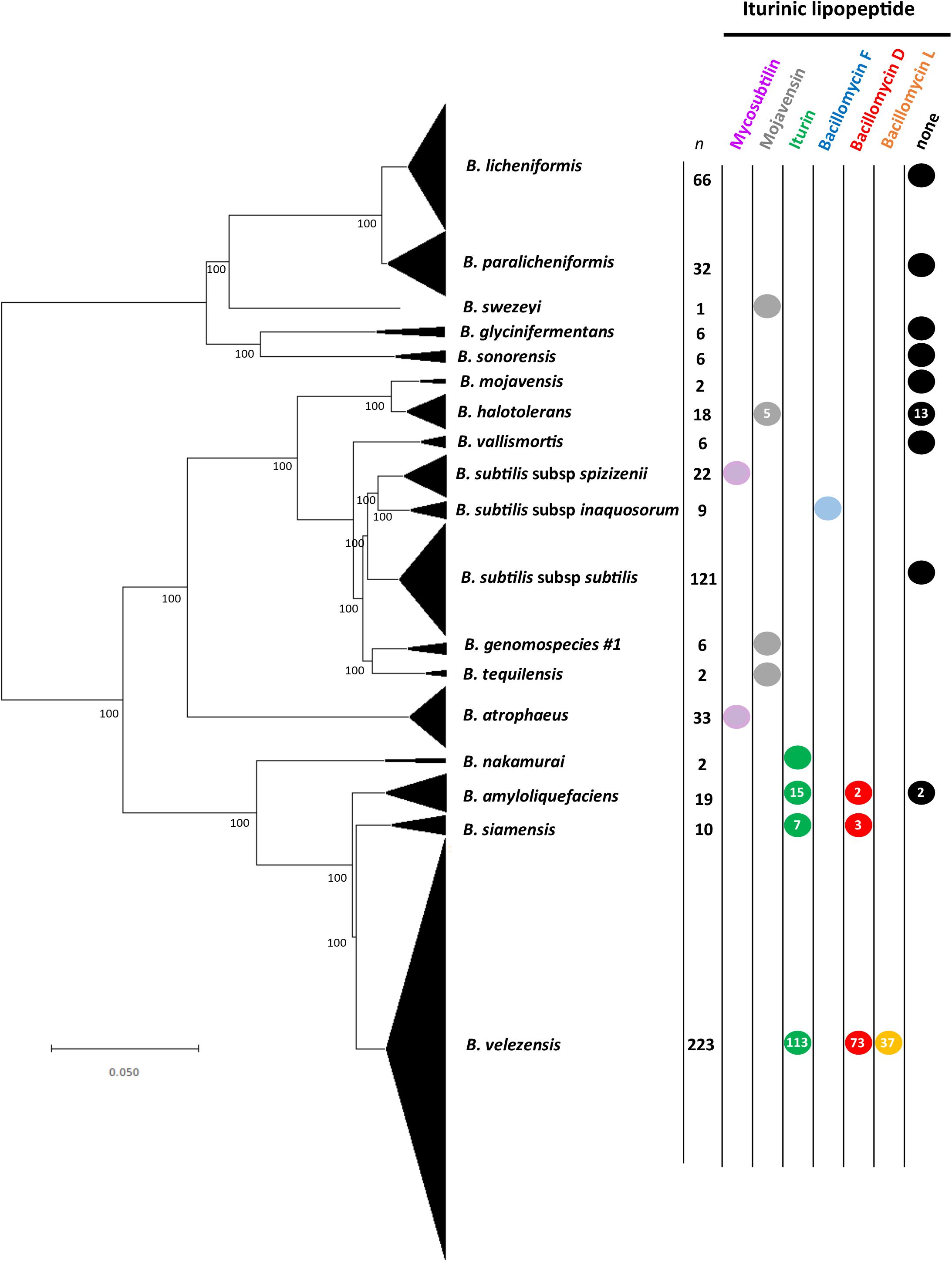
Figure 2. Neighbor-joining phylogeny showing iturinic lipopeptide occurrence across the B. subtilis species group. The tree was reconstructed from the core genomes of 584 strains. Bootstrap values >50%, based on 1,000 pseudoreplicates are indicated on branch points. The tree was rooted based on previous core genome phylogenies of the B. subtilis group (Dunlap et al., 2015b). Species in the “Bacillus pumilus” clade did not contain the cluster and were omitted from the figure to save space. The scale bar corresponds to 0.05 nucleotide substitutions per site.
Two species were identified that make their own unique lipopeptide: B. subtilis subsp. inaquosorum is the only species to produce bacillomycin F, and B. velezensis is the only species known to produce bacillomycin L. In addition, B. velezensis produces the greatest variety of lipopeptides, as some strains produce iturin or bacillomycin D instead of bacillomycin L. It is interesting to note that all iturin producers fall within the same clade, which encompasses B. amyloliquefaciens, B. nakamuari, B. siamensis, and B. velezensis. This clade also contains the only producers of bacillomycin D and bacillomycin L. It was previously believed that B. mojavensis produced only mojavensin. However, the strains apparently belong to another closely related species, B. halotolerans (Dunlap et al., 2016a). In addition, all genomes that contained a cluster also contained the sfp gene required for NRPS activation, except for two strains; B. atrophaeus ATCC 9372-1 and 1013-2. These two strains are domesticated laboratory strains that have been reported to have large genome deletions from this portion of the genome (Gibbons et al., 2011).
Iturinic Lipopeptide Diversity at the Strain Level
Because B. amyloliquefaciens, B. siamensis, and B. velezensis each produce more than one lipopeptide, we determined the distribution of the lipopeptides within the species based on core genome phylogeny (Figure 3). The results show that the different lipopeptide gene clusters appear in the core-genome phylogeny in a clade-dependent manner, which suggests unique introductions of the biosynthetic clusters. For example, it appears that, on the basis of the core genome phylogeny, the bacillomycin L cluster was likely introduced into B. velezensis 3 or 4 times. In addition, the phylogeny in Figure 3 also indicates that B. amyloliquefaciens and B. siamensis encompass distinct strain lineages that possess the iturin and bacillomycin D clusters.
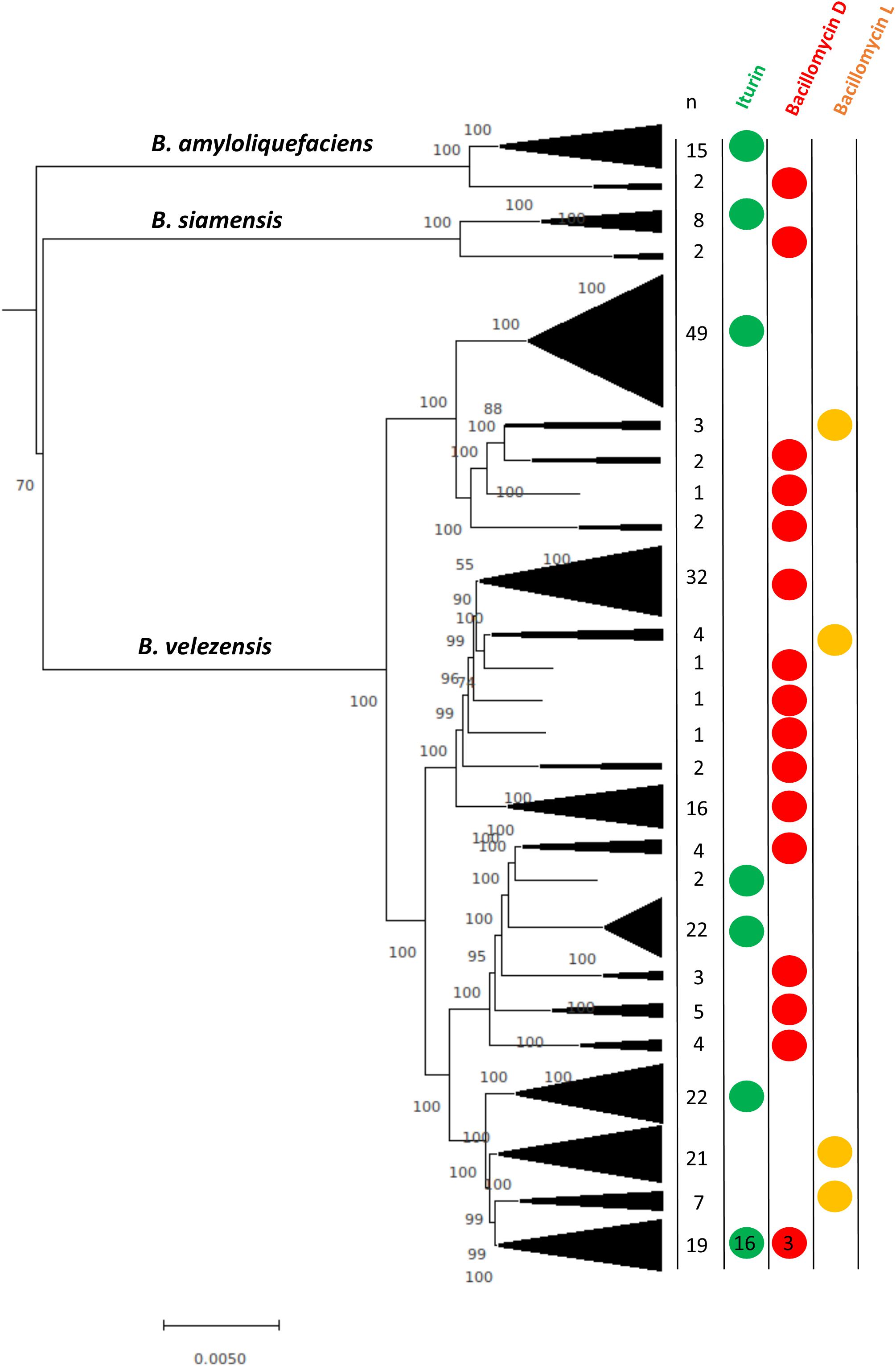
Figure 3. Neighbor-joining phylogeny showing iturinic lipopeptide occurrence among strains within the clade that encompasses B. amyloliquefaciens, B. siamensis, and B. velezensis. The tree was reconstructed from the core genomes of 250 strains. Bootstrap values >50%, based on 1,000 pseudoreplicates are indicated on branch points. The tree was rooted based on previous core genome phylogenies of the B. subtilis group (Dunlap et al., 2015b). The scale bar corresponds to 0.005 nucleotide substitutions per site.
Phylogeny Based on Conserved Cluster Gene, ituA and Synteny
To better understand the evolutionary history of the iturin cluster in the B. subtilis species group, we conducted a phylogenetic analysis using a conserved region of the cluster corresponding to a segment of the ituA gene that is found in all genomes. The results show considerable incongruence between the ituA gene phylogeny (Figure 4) and core genome phylogeny (Figure 2). We also examined the syntenic relationships of the iturinic clusters across species and discovered that many are found in the same location in the genome (Figure 4). The few notable exceptions are the two mycosubtilin producers B. subtilis subsp. spizizenii and B. atrophaeus. The cluster in these two species also do not share synteny which suggests it likely came from a horizontal gene transfer event, rather than a shared ancestor. B. swezeyi also does not share synteny with any other species, which is not necessarily surprising because it is also the most distantly related strain possessing the cluster.
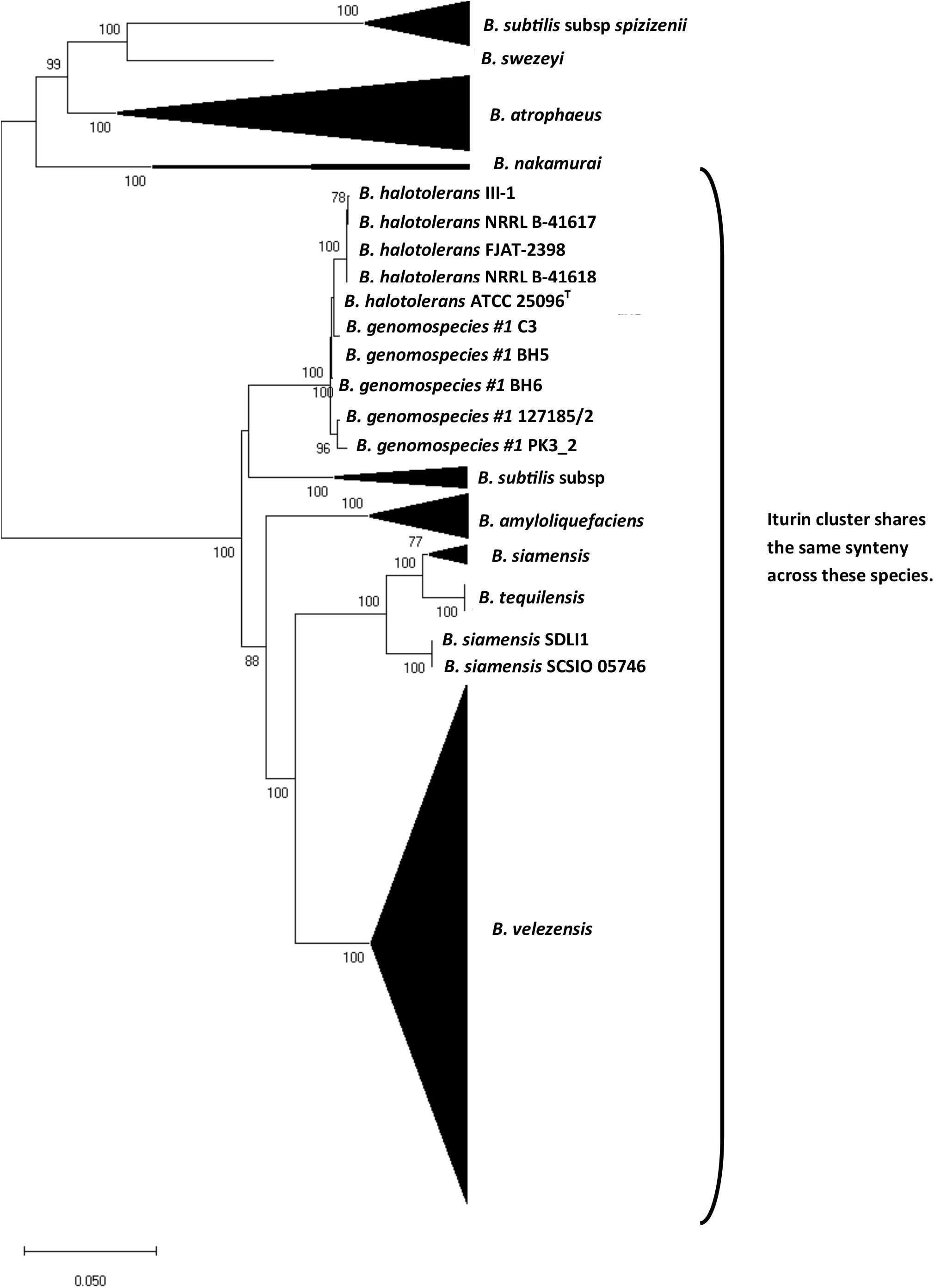
Figure 4. Neighbor-joining phylogenetic tree reconstructed from the partial ituA gene of iturinic lipopeptide producing strains from the B. subtilis group. Bootstrap values >50%, based on 1,000 pseudoreplicates are indicated on branch points. The scale bar corresponds to 0.05 nucleotide substitutions per site.
Given the incongruence between the partial ituA gene and the core genome phylogeny, we calculated the homology of the partial ituA gene and genomic ANI for representatives of each species. The results are provided in Table 1 and highlight several cases in which the partial ituA gene sequence and genomic ANI differ by more than 10% in comparisons involving the same species. For instance, the genomic ANI between B. halotolerans and Bacillus genomospecies #1 is 87.6%, whereas the partial ituA gene sequence identity is 99.1%. On the other hand, the opposite is also found in which the genomic ANI is much higher than the partial ituA gene sequence identity. For example, the genomic ANI is 94.5% whereas the partial ituA gene sequence identity is 81.9% between B. subtilis subsp. spizizenii and B. subtilis subsp. inaquosorum.
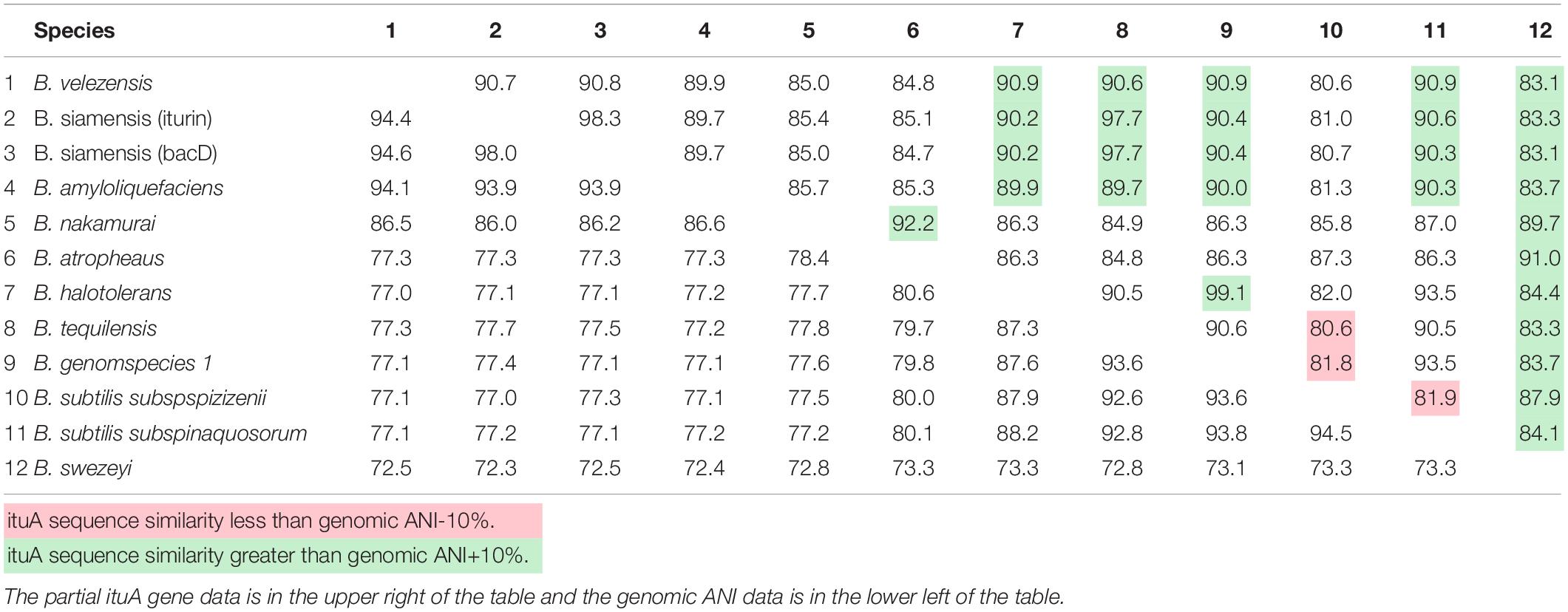
Table 1. Sequence similarity of the partial ituA gene between representative strains of iturinic lipopeptide producing species and genomic ANI values from comparisons of those same species.
Selective Primers to Determine Iturinic Cluster in B. velezensis
Because B. velezensis strains are among the most well studied iturinic lipopeptide producers and the most commonly used for commercial purposes (Dunlap, 2019), we developed a rapid, diagnostic PCR assay that uses selective primers to amplify iturinic lipopeptide gene types that can be differentiated subsequently on the basis of amplicon size through agarose gel electrophoresis. The primer sets share the same forward primer, which targets the conserved third amino acid module (Figure 1). The reverse primers target the variable fourth amino acid module in the three different clusters (iturin A, bacillomycin D, and bacillomycin L). The primer sets yield a ∼1,113 bp band for iturin, ∼1,016 bp band for bacillomycin L and a ∼732 bp band for bacillomycin D. The end result is a simple diagnostic PCR assay that provides a rapid method to determine which specific lipopeptide compound is produced by a given strain (Figure 5).
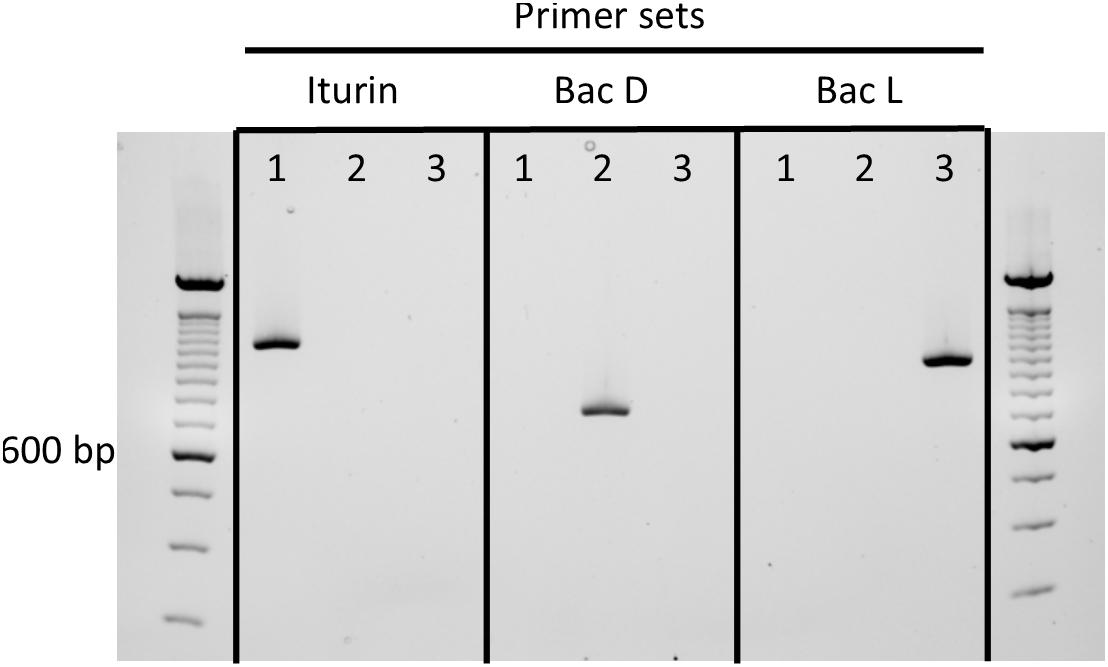
Figure 5. Agarose gel containing amplicons from selective primers sets to determine iturinic lipopeptide production in B. velezensis strains: (1) iturin, B. velezensis QST713, (2) bacillomycin D, B. velezensis FZB42, (3) bacillomycin L, B. velezensis KACC 18228. The iturin set is ituB-fwd and ituB-iturin-rev; the BacD set is ituB-fwd and ituB-bacD-rev; and the BacL set is ituB-fwd and ituB-R-bacL. A size ladder standard in 100 bp increments is included in the first and last unlabeled lanes, with a bold band corresponding to 600 bp.
Mass Spectrometric Analysis of Iturinic Lipopeptides
Iturinic lipopeptides fragment through two dominant pathways (Supplementary Figure S1A), which differ based upon where the peptide ring breaks and creates a linear peptide. Stepwise fragmentation of the terminal ends of these linear peptides can then be used to assign amino acid constituents (Supplementary Figure S1A). When the opening of the cyclic lipopeptide occurs between the β-amino acid tail and the conserved Asn at AA#1, diagnostic ions are generated from the conserved AA#1-AA#2 fragments Asn-Tyr (m/z 278.11) and the fragment for Asn-Tyr-Asn (m/z 392.15); since these fragments do not contain the lipid tail, they are invariant across all six lipopeptides and alkyl chain variants (Supplementary Figures S1B–G). Additionally, an ion of 136.076 m/z is present in all iturinic lipopeptides and appears to be generated by a double fragmentation of the tyrosine substituent. The fragmentation of the parent ions also yields fragment ions that are dependent upon the chain length of the β-amino acid; a useful ion results from the loss of 42.01 Da from the β-amino acid providing the information required to determine the length of the chain (e.g., C14 fragments to 184.20; C15 fragments to 198.22, etc. Supplementary Figures S1B–G). These fragments facilitate the assignment of 14.02 Da changes to either the β-amino acid or to the cyclic peptides (i.e., Ser to Thr). When the opening of the cyclic peptide occurs at the proline residue, short peptide fragments can be used to assign the variable residues at AA#5-AA#6-AA#7 for Iturin A (Supplementary Figure S1B), mycosubtilin (Supplementary Figure S1C), bacillomycin F (Supplementary Figure S1F), or mojavensin A (Supplementary Figure S1G) where proline occupies the AA#5 position or AA-#4-AA#5-AA#6-AA#7 for bacillomycin D where the proline occupies the AA#4 position (Supplementary Figures S1A–E). Bacillomycin L lacks a proline; thus, it has a more uniform fragmentation pattern across the β-amino acid (Supplementary Figure S1D).
Comparison to NRPS Substrate Predictor Software
Proteins representing the ituA, ituB, and ituC genes from the 15 cluster groups were subjected to substrate prediction. The software successfully predicted substrate specificity of amino acid modules in 101 out 105 modules (Supplementary Table S3). The only missed predictions were for Bacillomycin D and Bacillomycin L ituB genes, where the observed glutamate was predicted to be glutamine.
Discussion
It is well known that iturinic lipopeptides possess strong antifungal activity (Mhammedi et al., 1982; Peypoux et al., 1986; Ongena and Jacques, 2008; Ma et al., 2012; Guardado-Valdivia et al., 2018) and induce defense responses in plants (Farace et al., 2015; Park et al., 2016; Wu et al., 2018) which, in turn, has led to the commercialization of many Bacillus strains that manufacture these compounds (Dunlap, 2019). Interestingly, no known resistance mechanisms to iturinic lipopeptides have been reported in fungi. It is not clear if changes in the gene cluster encoding iturinic lipopeptides are the result of interactions with other fungi or plants, or due to an intrinsic factor such as differential genome dynamics (Rooney and Ward, 2005). Based on gene synteny, it can be inferred that the cluster was either introduced into, or evolved de novo in, the B. subtilis species group after Bacillus licheniformis and Bacillus pumilus clades arose (Figure 2). However, the sporadic distribution among the remaining members further suggests that either more than one introduction occurred or that the gene cluster evolves under a birth-and-death process (Nei and Rooney, 2005), although additional studies are needed to confirm the evolutionary processes. The fact that two mycosubtilin producers, B. atrophaeus and B. subtilis subsp. spizizenii, possess the cluster but do not share synteny with other members in the clade or with each other is consistent with the multiple introduction hypothesis. This is further supported by the incongruence of the partial ituA gene phylogeny (Figure 4) and the core genome phylogeny (Figure 2). On the other hand, the only other species that has been identified to possess an iturinic lipopeptide gene cluster is P. larvae (Sood et al., 2014), in which the gene cluster is in three genomes. The product of these P. larvae clusters (paenilarvin) shares the same cyclic heptapeptide sequence as mojavensin and only differs in the alkyl chain length (Sood et al., 2014). Although it is currently not possible to conclude with certainty the direction in which the horizontal transfer event occurred based on available data (i.e., from B. subtilis species group into P. larvae, or vice-versa), the fact that the gene cluster is found among 330 genomes and conserved across multiple species in the B. subtilis species group in contrast to being found in only three genomes in only one species of Paenibacillus tilts the evidence in favor of the transfer occurring from a Bacillus species into Paenibacillus, as the amount breadth of diversity seen in the former would have taken a substantial amount of time to have been generated. That being the case, it further suggests that the gene cluster arose in Bacillus (most likely only once) and subsequently diversified according to a birth-and-death process in light of its sporadic phylogenetic distribution across the B. subtilis species group (Figure 2). Birth-and-death evolution is one possible explanation how a compound such as mojavensin (Ma et al., 2012) can be made by various species that are not sister taxa, and it also explains why some species possess an iturinic lipopeptide gene cluster whereas others do not. This study is also agreement with a recent study that looked at the presence or absence of iturinic cluster without chemotype assignment for five of these species (Torres Manno et al., 2019). One exception between the studies we noted is Torres Manno et al. (2019) reports one Bacillus velezensis strain (TF28) that does not contain an iturinic cluster, while our study excluded the strain because it contained too many frameshifted gene, including ituB. The genome for strain TF28 appears to suffer from some assembly issues with frameshifted genes in both the iturinic cluster and in the cluster that produces the antifungal, fengycin. The strain has been reported to make both iturin (Zhang et al., 2012) and fengycin (Liu et al., 2018).
Several studies have recently reported antifungal lipopeptides in B. atrophaeus, but none have identified the active compound (Zhang et al., 2013, 2016; Chen et al., 2017; Guardado-Valdivia et al., 2018). This study was first to identify B. atrophaeus as a producer of mycosubtilin. Furthermore, it is the first to report that B. swezeyi (Dunlap et al., 2017) and B. nakamurai (Dunlap et al., 2016c) are capable of producing iturinic lipopeptides that correspond to mojavensin and iturin, respectively. In addition, this study identified that B. subtilis subsp. spizizenii and B. subtilis subsp inaquosorum only make one lipopeptide each, mycosubtilin and bacillomycin F, respectively. Recent studies based on whole genome comparisons and ANI suggest that B. subtilis subsp. spizizenii and B. subtilis subsp. inaquosorum should be promoted to species status based on established ANI guidelines of species delineation (Yi et al., 2014; Brito et al., 2018; Knight et al., 2018). The primary reason these strains were relegated to subspecies status was a lack of distinguishing phenotypes, as well as a lack of complete genome information and guidelines on how to use it for taxonomy (Rooney et al., 2009). The findings of this study support their promotion to species-level status.
Although several iturinic peptides can be putatively identified by their molecular weight (bacillomycin L, bacillomycin D, and mojavensin A), others cannot due to the isomeric nature of the amino acid substitutions and variation of alkyl tail lengths of the lipid portion of the molecule. Despite the potential for differences between molecular masses, assignments can be confounded by the presence of molecules with masses close to the iturinic peptides. This effect is pronounced when low-resolution mass analyzers are used in the absence of chromatographic separations. Therefore, liquid chromatography coupled to tandem mass spectrometry is a powerful tool to confirm the presence and identity of iturinic lipopeptides. The use of quadropole-time-of-flight or HCD cells with orbitrap mass spectrometers are particularly useful for tandem mass spectrometry due to their high mass accuracy and the ability to collect fragment data from the lower mass ranges to identify the characteristics of the amino acid substitutions. In this study, the use of HCD with orbitrap detection led to confirmation of the assignments of all iturinic peptide types. The comprehensive fragmentation reference table provided (Supplementary Figure S1A) should allow easy assignment of these compounds in future studies. In addition, this study should provide additional reference material to enhance the accuracy of NRPS substrate prediction software.
Lastly, we developed selective primers to identify the iturinic lipopeptides produced by B. velezensis. Part of the motivation for the development of these primers was the misuse or over interpretation of other commonly reported primers. Some of the most commonly used primers to detect iturin producers target the part of the cluster conserved among all iturinic lipopeptides, such as ituA (Tsuge et al., 2005) or ituD (Hsieh et al., 2008), so they are not selective for iturin specifically. Primers that are commonly used to determine bacillomycin D producers target the ituC gene (Ramarathnam et al., 2007), which shares high homology with bacillomycin L. This often leads to incorrect reporting of compound production capabilities of certain strains. Therefore, it is important to note that the primers developed in this study have only been validated for B. velezensis strains; their application to other Bacillus species must be validated before they can be reliably applied.
Data Availability
Publicly available datasets were analyzed in this study. This data can be found here: Refseq Bacillus genomes on GenBank (see Supplementary Material).
Author Contributions
CD conceived the study and conducted the bioinformatics analysis. MB conducted the mass spectroscopy experimental work and analysis. AR interpreted the evolutionary relationships. All authors wrote, read, and reviewed the final draft of the manuscript.
Funding
This study was supported by the U.S. Department of Agriculture, Agricultural Research Service in house project# 5010- 22410-019-00-D.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank Heather Walker for her technical expertise. Any opinions, findings, conclusions, or recommendations expressed in this publication are those of the author(s) and do not necessarily reflect the view of the U.S. Department of Agriculture. The mention of firm names or trade products does not imply that they are endorsed or recommended by the USDA over other firms or similar products not mentioned. The USDA is an equal opportunity provider and employer.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2019.01794/full#supplementary-material
References
Besson, F., Peypoux, F., Michel, G., and Delcambe, L. (1977). Structure de la bacillomycine L, antibiotique de Bacillus subtilis. Eur. J. Biochem. 77, 61–67. doi: 10.1111/j.1432-1033.1977.tb11641.x
Besson, F., Peypoux, F., Quentin, M. J., and Michel, G. (1984). Action of antifungal peptidolipids from Bacillus subtilis on the cell membrane of Saccharomyces cerevisiae. J. Antibiot. 37, 172–177. doi: 10.7164/antibiotics.37.172
Brito, P. H., Chevreux, B., Serra, C. R., Schyns, G., Henriques, A. O., and Pereira-Leal, J. B. (2018). Genetic competence drives genome diversity in Bacillus subtilis. Genome Biol. Evol. 10, 108–124. doi: 10.1093/gbe/evx270
Chen, Q., Liu, B., Wang, J., Che, J., Liu, G., and Guan, X. (2017). Antifungal lipopeptides produced by Bacillus sp. FJAT-14262 isolated from rhizosphere soil of the medicinal plant Anoectochilus roxburghii. Appl. Biochem. Biotechnol. 182, 155–167. doi: 10.1007/s12010-016-2317-z
Delcambe, L. (1950). Iturine, new antibiotic produced by Bacillus subtilis. C. R. Seances Soc. Biol. Fil. 144, 1431–1434.
Donadio, S., Staver, M. J., McAlpine, J. B., Swanson, S. J., and Katz, L. (1991). Modular organization of genes required for complex polyketide biosynthesis. Science 252, 675–679. doi: 10.1126/science.2024119
Duitman, E. H., Hamoen, L. W., Rembold, M., Venema, G., Seitz, H., Saenger, W., et al. (1999). The mycosubtilin synthetase of Bacillus subtilis ATCC6633: a multifunctional hybrid between a peptide synthetase, an amino transferase, and a fatty acid synthase. Proc. Natl. Acad. Sci. U.S.A. 96, 13294–13299. doi: 10.1073/pnas.96.23.13294
Dunlap, C. A. (2015). Phylogenomic analysis shows that ‘Bacillus vanillea’ is a later heterotypic synonym of Bacillus siamensis. Int. J. Syst. Evol. Microbiol. 65, 3507–3510. doi: 10.1099/ijsem.0.000444
Dunlap, C. A. (2019). Taxonomy of registered Bacillus spp. strains used as plant pathogen antagonists. Biol. Control 134, 82–86. doi: 10.1016/j.biocontrol.2019.04.011
Dunlap, C. A., Bowman, M. J., Schisler, D. A., and Rooney, A. P. (2016a). Genome analysis shows Bacillus axarquiensis is not a later heterotypic synonym of Bacillus mojavensis; reclassification of Bacillus malacitensis and Brevibacterium halotolerans as heterotypic synonyms of Bacillus axarquiensis. Int. J. Syst. Evol. Microbiol. 66, 2438–2443. doi: 10.1099/ijsem.0.001048
Dunlap, C. A., Kim, S.-J., Kwon, S.-W., and Rooney, A. P. (2016b). Bacillus velezensis is not a later heterotypic synonym of Bacillus amyloliquefaciens; Bacillus methylotrophicus, Bacillus amyloliquefaciens subsp plantarum and ‘Bacillus oryzicola’ are later heterotypic synonyms of Bacillus velezensis based on phylogenomics. Int. J. Syst. Evol. Microbiol. 66, 1212–1217. doi: 10.1099/ijsem.0.000858
Dunlap, C. A., Saunders, L. P., Schisler, D. A., Leathers, T. D., Naeem, N., Cohan, F. M., et al. (2016c). Bacillus nakamurai sp. nov., a black-pigment-producing strain. Int. J. Syst. Evol. Microbiol. 66, 2987–2991. doi: 10.1099/ijsem.0.001135
Dunlap, C. A., Kim, S.-J., Kwon, S.-W., and Rooney, A. P. (2015a). Phylogenomic analysis shows that Bacillus amyloliquefaciens subsp plantarum is a later heterotypic synonym of Bacillus methylotrophicus. Int. J. Syst. Evol. Microbiol. 65, 2104–2109. doi: 10.1099/ijs.0.000226
Dunlap, C. A., Kwon, S.-W., Rooney, A. P., and Kim, S.-J. (2015b). Bacillus paralicheniformis sp. nov., isolated from fermented soybean paste. Int. J. Syst. Evol. Microbiol. 65, 3487–3492. doi: 10.1099/ijsem.0.000441
Dunlap, C. A., Schisler, D. A., Perry, E. B., Connor, N., Cohan, F. M., and Rooney, A. P. (2017). Bacillus swezeyi sp. nov. and Bacillus haynesii sp. nov., isolated from desert soil. Int. J. Syst. Evol. Microbiol. 67, 2720–2725. doi: 10.1099/ijsem.0.002007
Farace, G., Fernandez, O., Jacquens, L., Coutte, F., Krier, F., Jacques, P., et al. (2015). Cyclic lipopeptides from Bacillus subtilis activate distinct patterns of defence responses in grapevine. Mol. Plant Pathol. 16, 177–187. doi: 10.1111/mpp.12170
Gibbons, H. S., Broomall, S. M., McNew, L. A., Daligault, H., Chapman, C., Bruce, D., et al. (2011). Genomic signatures of strain selection and enhancement in Bacillus atrophaeus var. globigii, a historical biowarfare simulant. PLoS One 6:e17836. doi: 10.1371/journal.pone.0017836
Guardado-Valdivia, L., Tovar-Pérez, E., Chacón-López, A., López-García, U., Gutiérrez-Martínez, P., Stoll, A., et al. (2018). Identification and characterization of a new Bacillus atrophaeus strain B5 as biocontrol agent of postharvest anthracnose disease in soursop (Annona muricata) and avocado (Persea americana). Microbiol. Res. 210, 26–32. doi: 10.1016/j.micres.2018.01.007
Hourdou, M. L., Besson, F., Tenoux, I., and Michel, G. (1989). Fatty acid and β-amino acid syntheses in strains of Bacillus subtilis producing iturinic antibiotics. Lipids 24, 940–944. doi: 10.1007/BF02544538
Hsieh, F. C., Lin, T. C., Meng, M., and Kao, S. S. (2008). Comparing methods for identifying Bacillus strains capable of producing the antifungal lipopeptide iturin A. Curr. Microbiol. 56, 1–5. doi: 10.1007/s00284-007-9003-x
Iwase, N., Rahman, M. S., and Ano, T. (2009). Production of iturin A homologues under different culture conditions. J. Environ. Sci. 21(Suppl. 1), S28–S32. doi: 10.1016/S1001-0742(09)60031-0
Jolley, K. A., and Maiden, M. C. (2010). BIGSdb: scalable analysis of bacterial genome variation at the population level. BMC Bioinform. 11:595. doi: 10.1186/1471-2105-11-595
Knight, C. A., Bowman, M. J., Frederick, L., Day, A., Lee, C., and Dunlap, C. A. (2018). The first report of antifungal lipopeptide production by a Bacillus subtilis subsp. inaquosorum strain. Microbiol. Res. 216, 40–46. doi: 10.1016/j.micres.2018.08.001
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Lee, I., Kim, Y. O., Park, S. C., and Chun, J. (2016). OrthoANI: an improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 66, 1100–1103. doi: 10.1099/ijsem.0.000760
Liu, Y., Zhang, J., Zhong, J., Li, J., Meng, L., and Zhang, S. (2018). Isolation and identification of antibacterial lipopeptides fengycin produced by Bacillus amyloliquefaciens TF28 and its anti-fungal mechanism studies. China Biotechnol. 38, 20–29. doi: 10.13523/j.cb.20181003
Ma, Z., Wang, N., Hu, J., and Wang, S. (2012). Isolation and characterization of a new iturinic lipopeptide, mojavensin A produced by a marine-derived bacterium Bacillus mojavensis B0621A. J. Antibiot. 65, 317–322. doi: 10.1038/ja.2012.19
Maget-Dana, R., Ptak, M., Peypoux, F., and Michel, G. (1985). Pore-forming properties of iturin A, a lipopeptide antibiotic. BBA Biomembranes 815, 405–409. doi: 10.1016/0005-2736(85)90367-0
Mhammedi, A., Peypoux, F., Besson, F., and Michel, G. (1982). Bacillomycin f, a new antibiotic of iturin group: isolation and characterization. J. Antibiot. 35, 306–311. doi: 10.7164/antibiotics.35.306
Moyne, A. L., Cleveland, T. E., and Tuzun, S. (2004). Molecular characterization and analysis of the operon encoding the antifungal lipopeptide bacillomycin D. FEMS Microbiol. Lett. 234, 43–49. doi: 10.1016/j.femsle.2004.03.011
Nei, M., and Rooney, A. P. (2005). Concerted and birth-and-death evolution of multigene families. Annu. Rev. Genet. 39, 121–152. doi: 10.1146/annurev.genet.39.073003.112240
Ongena, M., and Jacques, P. (2008). Bacillus lipopeptides: versatile weapons for plant disease biocontrol. Trends Microbiol. 16, 115–125. doi: 10.1016/j.tim.2007.12.009
Park, K., Park, Y. S., Ahamed, J., Dutta, S., Ryu, H., Lee, S. H., et al. (2016). Elicitation of induced systemic resistance of chili pepper by iturin a analogs derived from Bacillus vallismortis EXTN-1. Can. J. Plant Sci. 96, 564–570. doi: 10.1139/cjps-2015-0199
Peypoux, F., Besson, F., Michel, G., and Delcambe, L. (1981). Structure of Bacillomycin D, a new antibiotic of the iturin group. Eur. J. Biochem. 118, 323–327. doi: 10.1111/j.1432-1033.1981.tb06405.x
Peypoux, F., Besson, F., Michel, G., Lenzen, C., Dierickx, L., and Delcambe, L. (1980). Characterization of a new antibiotic of iturin group: Bacillomycin D. J. Antibiot. 33, 1146–1149. doi: 10.7164/antibiotics.33.1146
Peypoux, F., Guinand, M., Michel, G., Delcambe, L., Das, B. C., and Lederer, E. (1978). Structure of iturine A, a peptidolipid antibiotic from Bacillus subtilis. Biochemistry 17, 3992–3996. doi: 10.1021/bi00612a018
Peypoux, F., Michel, G., and Delcambe, L. (1976). Structure de la mycosubtiline, antibiotique isolé de Bacillus subtilis. Eur. J. Biochem. 63, 391–398. doi: 10.1111/j.1432-1033.1976.tb10240.x
Peypoux, F., Pommier, M. T., Michel, G., Marion, D., Ptak, M., and Das, B. C. (1986). Revised structure of mycosubtilin, a peptidolipid antibiotic from Bacillus subtilis. J. Antibiot. 39, 636–641. doi: 10.7164/antibiotics.39.636
Prieto, C., García-estrada, C., Lorenzana, D., and Martín, J. F. (2012). NRPSSP: non-ribosomal peptide synthase substrate predictor. Bioinformatics 28, 426–427. doi: 10.1093/bioinformatics/btr659
Quadri, L. E. N., Weinreb, P. H., Lei, M., Nakano, M. M., Zuber, P., and Walsh, C. T. (1998). Characterization of Sfp, a Bacillus subtilis phosphopantetheinyl transferase for peptidyl carder protein domains in peptide synthetases. Biochemistry 37, 1585–1595. doi: 10.1021/bi9719861
Ramarathnam, R., Bo, S., Chen, Y., Fernando, W. G. D., Xuewen, G., and De Kievit, T. (2007). Molecular and biochemical detection of fengycin- and bacillomycin D-producing Bacillus spp., antagonistic to fungal pathogens of canola and wheat. Can. J. Microbiol. 53, 901–911. doi: 10.1139/W07-049
Richter, M., and Rosselló-Móra, R. (2009). Shifting the genomic gold standard for the prokaryotic species definition. Proc. Natl. Acad. Sci. U.S.A. 106, 19126–19131. doi: 10.1073/pnas.0906412106
Rooney, A. P., Price, N. P. J., Ehrhardt, C., Sewzey, J. L., and Bannan, J. D. (2009). Phylogeny and molecular taxonomy of the Bacillus subtilis species complex and description of Bacillus subtilis subsp. inaquosorum subsp. nov. Int. J. Syst. Evol. Microbiol. 59, 2429–2436. doi: 10.1099/ijs.0.009126-0
Rooney, A. P., and Ward, T. J. (2005). Evolution of a large ribosomal RNA multigene family in filamentous fungi: birth and death of a concerted evolution paradigm. Proc. Natl. Acad. Sci. U.S.A. 102, 5084–5089. doi: 10.1073/pnas.0409689102
Sood, S., Steinmetz, H., Beims, H., Mohr, K. I., Stadler, M., Djukic, M., et al. (2014). Paenilarvins: iturin family lipopeptides from the honey bee pathogen Paenibacillus larvae. Chembiochem 15, 1947–1955. doi: 10.1002/cbic.201402139
Torres Manno, M. A., Pizarro, M. D., Prunello, M., Magni, C., Daurelio, L. D., Espariz, M. J. A. M., et al. (2019). GeM-Pro: a tool for genome functional mining and microbial profiling. Appl. Microbiol. Biotechnol. 103, 3123–3134. doi: 10.1007/s00253-019-09648-8
Tsuge, K., Akiyama, T., and Shoda, M. (2001). Cloning, sequencing, and characterization of the iturin A operon. J. Bacteriol. 183, 6265–6273. doi: 10.1128/JB.183.21.6265-6273.2001
Tsuge, K., Inoue, S., Ano, T., Itaya, M., and Shoda, M. (2005). Horizontal transfer of iturin a operon, itu, to Bacillus subtilis 168 and conversion into an iturin A producer. Antimicrob. Agents Chemother. 49, 4641–4648. doi: 10.1128/AAC.49.11.4641-4648.2005
Wu, G., Liu, Y., Xu, Y., Zhang, G., Shen, Q., and Zhang, R. (2018). Exploring elicitors of the beneficial rhizobacterium Bacillus amyloliquefaciens SQR9 to induce plant systemic resistance and their interactions with plant signaling pathways. Mol. Plant Microbe Interact. 31, 560–567. doi: 10.1094/MPMI-11-17-0273-R
Yi, H., Chun, J., and Cha, C.-J. (2014). Genomic insights into the taxonomic status of the three subspecies of Bacillus subtilis. Syst. Appl. Microbiol. 37, 95–99. doi: 10.1016/j.syapm.2013.09.006
Zhang, J., Xue, Q., Gao, H., Lai, H., and Wang, P. (2016). Production of lipopeptide biosurfactants by Bacillus atrophaeus 5-2a and their potential use in microbial enhanced oil recovery. Microb. Cell Fact. 15:168. doi: 10.1186/s12934-016-0574-8
Zhang, S. M., Wang, Y. X., Meng, L. Q., Li, J., Zhao, X. Y., Cao, X., et al. (2012). Isolation and characterization of antifungal lipopeptides produced by endophytic Bacillus amyloliquefaciens TF28. Afr. J. Microbiol. Res. 6, 1747–1755.
Keywords: biocontrol, probiotic, FZB42, QST713, amyloliquefaciens, fengycin, PGPR, biostimulant
Citation: Dunlap CA, Bowman MJ and Rooney AP (2019) Iturinic Lipopeptide Diversity in the Bacillus subtilis Species Group – Important Antifungals for Plant Disease Biocontrol Applications. Front. Microbiol. 10:1794. doi: 10.3389/fmicb.2019.01794
Received: 09 May 2019; Accepted: 22 July 2019;
Published: 07 August 2019.
Edited by:
Ludmila Chistoserdova, University of Washington, United StatesCopyright © 2019 Dunlap, Bowman and Rooney. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Christopher A. Dunlap, Q2hyaXN0b3BoZXIuZHVubGFwQGFycy51c2RhLmdvdg==
 Christopher A. Dunlap
Christopher A. Dunlap Michael J. Bowman
Michael J. Bowman Alejandro P. Rooney
Alejandro P. Rooney