- 1Computational and Functional Genomics Group, Centre for DNA Fingerprinting and Diagnostics, Hyderabad, India
- 2Graduate Studies, Manipal Academy of Higher Education, Manipal, India
- 3Regional Centre for Biotechnology, Faridabad, India
Tuberculosis (TB) caused by Mycobacterium tuberculosis (Mtb) accounts for nearly 1.2 million deaths per annum worldwide. Due to the emergence of multidrug-resistant (MDR) Mtb strains, TB, a curable and avertable disease, remains one of the leading causes of morbidity and mortality. Isoniazid (INH) is a first-line anti-TB drug while ethionamide (ETH) is used as a second-line anti-TB drug. INH and ETH resistance develop through a network of genes involved in various biosynthetic pathways. In this study, we identified Rv0023, an Mtb protein belonging to the xenobiotic response element (XRE) family of transcription regulators, which has a role in generating higher tolerance toward INH and ETH in Mycobacterium smegmatis (Msmeg). Overexpression of Rv0023 in Msmeg leads to the development of INH- and ETH-tolerant strains. The strains expressing Rv0023 have a higher ratio of NADH/NAD+, and this physiological event is known to play a crucial role in the development of INH/ETH co-resistance in Msmeg. Gene expression analysis of some target genes revealed reduction in the expression of the ndh gene, but no direct interaction was observed between Rv0023 and the ndh promoter region. Rv0023 is divergently expressed to Rv0022c (whiB5) and we observed a direct interaction between the recombinant Rv0023 protein with the upstream region of Rv0022c, confirmed using reporter constructs of Msmeg. However, we found no indication that this interaction might play a role in the development of INH/ETH drug tolerance.
Introduction
Tuberculosis (TB) remains a major cause of death worldwide and the leading cause by a single infectious agent (World Health Organisation, 2018). Even though the disease can be cured and managed by several multidrug regimens, the emergence of multidrug-resistant (MDR) TB is proving to be a major challenge for complete eradication of the disease. Worldwide, MDR TB constitutes 3.5% of new TB cases and 18% of previously treated cases (World Health Organisation, 2018). To overcome these challenges and to better counter resistance in Mycobacterium tuberculosis (Mtb), understanding the mechanisms and deciphering the pathways majorly responsible for generating resistance are greatly required.
Isoniazid (INH), in combination with rifampicin (RIF), ethambutol (EMB), and pyrazinamide (PZA), forms the first-line therapy for TB. Cycloserine, ethionamide (ETH), and amikacin/capreomycin are used as second-line drugs1. INH was first used as an anti-TB drug in 1952 and shortly the first INH-resistant Mtb (INHr) clinical isolates were reported (Bernstein et al., 1952; Fox, 1952; Middlebrook and Cohn, 1953). INH and ETH are prodrugs that are converted into their active forms by proteins encoded by katG (catalase peroxidase KatG) and ethA (monooxygenase EthA), respectively (Johnsson and Schultz, 1994; Johnsson et al., 1997; Baulard et al., 2000; DeBarber et al., 2000; Vannelli et al., 2002; Fraaije et al., 2004). Although katG and inhA (encoding an NADPH-dependent enoyl-ACP reductase) are the main genes involved in INH resistance, clinical isolates with mutations in the ndh gene have been identified in INH-resistant Mtb strains (Lee et al., 2001). While in the study by Lee et al. (2001), INH resistance mutations were observed only in the ndh gene, in other studies, ndh mutations occur simultaneously with mutations in other genes (Hazbón et al., 2006; Cardoso et al., 2007). The role of the ndh gene mutations in INH and ETH co-resistance in Mycobacterium smegmatis (Msmeg) and Mycobacterium bovis (Mbovis) has been shown (Vilcheze et al., 2005), while the role of ndh in conferring resistance in Mtb is yet to be determined. The gene ndh encodes the type II NADH dehydrogenase and its ortholog in Escherichia coli (E. coli) has been characterized. In E. coli, it exists in a monomeric state bound to the membrane, where it oxidizes NADH, reduces quinone, and catalyzes the transfer of electrons from reduced flavin to quinone (Jaworowski et al., 1981; Matsushita et al., 1987; Yagi, 1993; Gennis and Stewart, 1996; Kerscher, 2000). In Msmeg, mutations in ndh lead to an increase in NADH cellular concentration and inhibition of INH-NAD and ETH-NAD adducts formation (Vilcheze et al., 2005).
Xenobiotic response element (XRE) family of transcription factors are one of the most frequently occurring families of regulators in bacteria. Among the well-studied members of the XRE family are the lambda and Cro repressors, from lambda bacteriophage, and the prophage repressor Xre from Bacillus subtilis. The XRE family of regulators share a conserved N-terminal helix-turn-helix (HTH) DNA binding domain, while the C-terminal regulatory region is highly variable. The XRE family of regulators control diverse metabolic functions; e.g., SinR regulates developmental process in B. subtilis (Gaur et al., 1991), ClgR regulates Streptomyces growth and controls Clp proteolytic complex (Bellier and Mazodier, 2004), PuuR regulates putrescine utilization pathway in E. coli K-12 (Nemoto et al., 2012), and BzdR is involved in the anaerobic catabolism of benzoate in the denitrifying Azoarcus sp. strain CIB (Barragán et al., 2005). There are seven members of the XRE family of transcription regulators in Mtb: Rv0023, Rv0465c, Rv0474, Rv1129, Rv2017, Rv2021, and EspR (Rv3849). Except for EspR, which positively regulates the ESX-1 protein secretion system, the principal virulence determinant of Mtb (Raghavan et al., 2008), the remaining XRE transcription regulators in Mtb are uncharacterized.
Rv0023 is a regulator from the XRE family of transcriptional regulators, known to induce 488 genes and repress 404 genes (Rustad et al., 2014). Rv0023 regulon is enriched for the regulation of NAD reductases (Rustad et al., 2014). Rv0023 is transcribed in an operon together with Rv0024, a gene that codes for an NLPC/p60 family protein and is transcribed divergently from whiB5, belonging to the WhiB family of transcriptional regulators. The whiB5 gene product is a positive regulator of transcription and contributes to Mtb virulence and reactivation (Casonato et al., 2012). At present, very little is known about Rv0023 functions and its effect on Mtb physiology.
Here, we studied the effects of Rv0023 overexpression in Msmeg. The results show that ectopic expression of Rv0023 confers enhanced INH and ETH tolerance in Msmeg. Rv0023 ectopic expression downregulates the expression of the ndh gene and increases NADH/NAD+ levels, which are known to be mediators of INH and ETH resistance in Msmeg (Miesel et al., 1998; Vilcheze et al., 2005). We further studied the regulation of the whiB5-Rv0023 locus and identified Rv0023 as a negative regulator of whiB5. We have characterized its binding site and identified the promoters of Rv0023 and whiB5.
Materials and Methods
Bacterial Strains and Growth Condition
A complete list of strains used in this study is mentioned in Table 1. Cloning and plasmid propagation were done using E. coli strain DH5α. For protein expression, E. coli BL21 (DE3) was used. Both strains were grown in Luria Bertani (LB) medium at 37°C. Mbovis BCG Pasteur 1173P2, Msmeg mc2155, and the recombinant strains were grown in Middlebrook 7H9 (Himedia) broth supplemented with 10% OADC (oleic albumin dextrose catalase) (Himedia), 0.2% glycerol, and 0.05% Tween80 (20% stock) or on 7H10 agar without Tween80 at 37°C. Kanamycin (50 μg/ml) (Sigma-Aldrich), ampicillin (100 μg/ml) (Sigma-Aldrich), and hygromycin (50 μg/ml) (Invitrogen) were used as and when required.
Plasmids and DNA Manipulation
Cloning, genomic, and plasmid DNA isolations were done as per standard molecular biology procedures (Sambrook et al., 1989). The plasmids and primers used in this study are listed in Tables 2, 3, respectively. Overlapping extension PCR was performed to generate site-directed mutations for promoter and critical residues studies. Sequences of all clones generated were confirmed by Sanger sequencing.
Protein Expression and Purification
pET23a-0023 plasmid containing Rv0023 ORF was used to transform E. coli BL21 (DE3) strain. Cells were grown in LB broth at 37°C till mid-log phase, and then 1 mM IPTG (isopropyl 1-thio-β-D-galactopyranoside) was added to induce the expression of protein. Cells were grown for another 4 h at 37°C, after which cells were harvested and purified using Ni-NTA affinity chromatography as described earlier (Yousuf et al., 2018). The purity of recombinant protein was analyzed by 12% SDS-PAGE and protein concentration was measured by Bradford assay.
β-Galactosidase Assay
Mycobacterium smegmatis mc2155 strain was transformed with various constructs as required and grown in 7H9 media. The cultures were grown to mid-log phase and β-galactosidase activity was measured in Miller Units (MU) and all experiments were done in triplicate (Miller, 1972).
Electrophoretic Mobility Shift Assay
To verify the interaction between Rv0023 and the upstream region of whiB5, electrophoretic mobility shift assay (EMSA) was performed. The 500-bp upstream region and the 50-bp downstream region of whiB5 were PCR amplified and labeled with γ-32P ATP (3000 Ci mmol–1) using T4 polynucleotide kinase as per manufacturer’s instructions (New England Biolabs). Labeled DNA was purified using nucleotide purification kit (Qiagen). Purified labeled DNA was then incubated with increasing concentrations of recombinant Rv0023 protein in an EMSA reaction buffer (25 mM HEPES, pH 7.9, 0.1 mM EDTA, 10 mM MgCl2, 20 mM KCl, and 5% glycerol) with 50 ng/μl poly(dI–dC) as a non-specific DNA competitor. The protein–DNA complex was incubated for 45 min and was resolved on a 6% non-denaturing polyacrylamide gel in 0.5 × tris-borate (TBE) buffer. The gel was run for 3–4 h to allow sufficient resolution of protein–DNA complex.
Electrophoretic mobility shift assay with commercially synthesized overlapping oligonucleotide and motifs was performed to identify the exact binding region of Rv0023.
Primer Extension
To map the (+1) transcription start site (TSS), primer extension was performed as described earlier and in Cold Spring Harbor protocols (Carey et al., 2013; Angara et al., 2018). As the ORFs and intergenic region of whiB5-Rv0023 locus are 100% conserved between Mtb and Mbovis, we used Mbovis RNA for primer extension studies. RNA was isolated with the Qiagen RNeasy kit as per manufacturer’s instructions. SuperScript III reverse transcriptase (Invitrogen) was used to transcribe cDNA from total RNA (10–15 μg) using 5′-end labeled γ-32P ATP primers (Table 4). Primer extension products were resolved on 6% polyacrylamide/8 M urea gel in TBE buffer. The size of primer extension product was determined by generating a dideoxy sequencing ladder using pUC19 plasmid as template and M13 universal primer (USB; 70140 KT).
Stress Assay
To check the sensitivity of Msmeg toward various stress conditions, MsmegWT, MsmegpVV16, and MsmegpVV0023 cells were grown in complete 7H9 media (supplemented with 10% OADC, 0.2% glycerol, and 0.05% Tween80) at 37°C overnight. Then, the cells were centrifuged and washed with PBS. The cells were then suspended in 7H9 media and the cell concentration was adjusted to 0.02 OD at 600 nm. The sensitivity of bacterial cultures toward 0.1% SDS (Sigma Aldrich) and 5 mM hydrogen peroxide (Merck) was measured at 37°C for 6 h. Sensitivity to 250 μg/ml lysozyme (Sigma Aldrich) was measured at 37°C for 24 h. Susceptibility of Msmeg toward the antibiotics (Sigma Aldrich) INH (10 μg/ml, MIC of 5 μg/ml), RIF (10 μg/ml, MIC of 1 μg/ml), and ETH (2.5 μg/ml, MIC of 10 μg/ml) was determined at 37°C for 24 h. For determining CFU numbers for each stress condition and antibiotics, the cells were serially diluted (10-fold) and plated onto 7H10 agar plates.
Further, the strains MsmegWT, MsmegpVV16, and MsmegpVV0023 were serially diluted (10-fold) and spotted on the 7H10 agar plates containing increasing concentrations of INH (0, 5, 10, and 15 μg/ml) and ETH (0, 2.5, and 5 μg/ml). The plates were incubated at 37°C for 48 h.
qRT-PCR
Total RNA from MsmegpVV16 (vector control) and MsmegpVV0023 (Rv0023 protein overexpressed) cultures were isolated using Qiagen RNeasy kit. SuperScript III reverse transcriptase (Invitrogen) was used to generate cDNA using random hexamers. Real-time PCR was carried on the BioRad CFX96 system using gene-specific primers (Table 5) and EvaGreen qPCR mastermix (Applied Biological Materials Inc.) as per standard protocol. The fold change in expression relative to MsmegpVV16 (vector control) was calculated after normalizing to sigA. The 2−ΔΔCT method was used to calculate relative changes in gene expression (Livak and Schmittgen, 2001).
NADH/NAD+ Cellular Concentration
To measure the cellular concentration of NADH and NAD+, MsmegpVV16 and MsmegpVV0023 cells were grown to an OD600 of 0.8–1.2. Cells were collected (1 ml) by centrifugation (11,000 rpm for 2 min). The supernatant was removed and 300 μl of 0.2 M HCl (for NAD+ extraction) or 0.2 M NaOH (for NADH extraction) was added to the cells. The cells were resuspended and incubated at 50°C for 10 min after which extracts were cooled to 0°C. The bacterial suspensions were neutralized by adding 0.1 M HCl (for NADH extraction) or 0.1 M NaOH (for NAD+ extraction) dropwise while vortexing at high speed. Centrifugation was done to remove the cell debris and supernatant was transferred to a new tube and used immediately. NADH and NAD+ concentrations were obtained by spectrophotometrically measuring the rate of MTT [3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide, 4.2 mM] (Amresco), reduction by yeast alcohol dehydrogenase II (Sigma Aldrich) in the presence of PES (phenazine ethosulfate, 16.6 mM) (Sigma Aldrich) at 570 nm (Leonardo et al., 1996; San et al., 2002; Vilcheze et al., 2005). The concentration of nucleotide (NADH/NAD+) is proportional to the rate of MTT reduction.
Bioinformatics
Rv0023 protein sequence (UniProt P9WMI3) was used as a query in the SynTax web server2 for synteny analysis within the Mycobacteriaceae family (Oberto, 2013); a conserved domain database search was performed to identify HTH_3 and XRE domains (Marchler-Bauer et al., 2015). Orthologous sequences of the whiB5–Rv0023 intergenic region from different mycobacterial species were retrieved from KEGG genome database and aligned using CLUSTAL OMEGA (Sievers et al., 2011).
Statistical Analysis
Data were presented as mean ± SD. Student’s t-test and one-way ANOVA were used to determine the statistical significance between groups and values with p < 0.05 were considered to be significant. GraphPad Prism software version 5.02 was used for the statistical analysis.
Results
Overexpression of Rv0023 Confers Increased INH and ETH Tolerance in Msmeg
Rv0023 is a non-essential gene in Mtb, yet it regulates a high number of genes in the genome (Sassetti et al., 2003; Rustad et al., 2014). Msmeg genome does not harbor the ortholog of Rv0023 and hence serves as a good model to study the function of Rv0023. To ascertain the role of Rv0023, we constructed an overexpressing strain of Rv0023 in Msmeg, MsmegpVV0023. The wild-type MsmegWT, vector control MsmegpVV16, and MsmegpVV0023 strains were subjected to acid fast staining to rule out the effect of vector insertion on cellular integrity. The cells were found to be intact and acid fast (Supplementary Figure 1). To further study the role of Rv0023 in cellular physiology, MsmegWT, MsmegpVV16, and MsmegpVV0023 were subjected to different stress conditions as depicted in Figure 1A, and tolerance was measured in terms of CFU. The CFU was counted for all conditions and we observed that overexpression of Rv0023 leads to increased tolerance with regard to INH and ETH, and no effect was observed with any other stress conditions. Tolerance to INH and ETH was not seen either on the wild type or in vector control strains (Figure 1A).
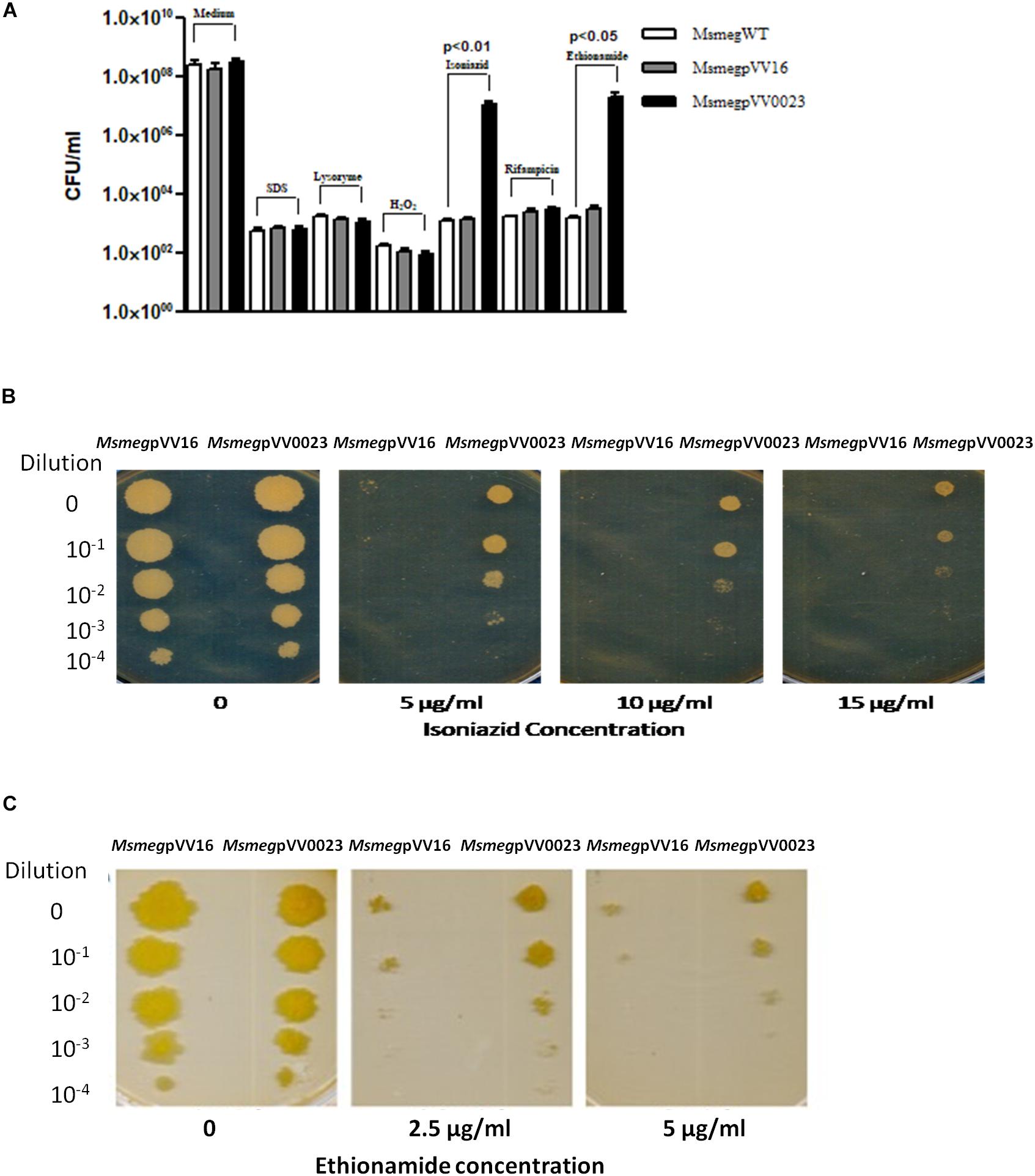
Figure 1. Survivability of Rv0023 expressing Msmeg in stress conditions. (A) The MsmegWT, MsmegpVV16, and MsmegpVV0023 were subjected to 0.1% SDS for 6 h, 250 μg of lysozyme for 24 h, 5 mM hydrogen peroxide for 6 h, INH (10 μg/ml), RIF (10 μg/ml), and ETH (5 μg/ml) for 24 h. Cells were plated on 7H10 agar plates and bacterial CFU were counted. Data are presented as the mean ± SD from three biological replicates. (B) MsmegpVV16 and MsmegpVV0023 cells were serially diluted and spotted on plates containing increasing concentrations of INH. (C) MsmegpVV16 and MsmegpVV0023 cells were serially diluted and spotted on plates containing increasing concentrations of ETH.
The role of Rv0023 in generating tolerant strains was further confirmed via spotting the wild type, vector control, and Rv0023 overexpressed strains in the presence of increasing concentrations of INH and ETH. MsmegWT, MsmegpVV16, and MsmegpVV0023 cells were serially diluted and spotted on plates containing 5, 10, and 15 μg/ml of INH and 2.5 and 5 μg/ml of ETH. Even at higher concentrations, MsmegpVV0023 showed tolerance toward INH and ETH (Figures 1B,C and Supplementary Figure 2). The results indicate that Rv0023 expression specifically contributes toward the higher tolerance of INH and ETH in Msmeg.
Rv0023 Expression in Msmeg Alters the NADH/NAD+ Levels
Isoniazid and ethionamide are both prodrugs and they are activated by the protein catalase–peroxidase KatG and the NADPH-specific flavin adenine dinucleotide-containing monooxygenase, EthA, respectively. After activation, they react with NAD+ to form INH-NAD and ETH-NAD adduct. This species then inhibits the enoyl ACP reductase InhA, which leads to the inhibition of mycolic acid biosynthesis and eventual mycobacterial cell death (Figure 2A). The major common genes to both pathways are the ndh (maintains NADH/NAD+ ratio) and inhA. Msmeg orthologs of these and other important genes involved in INH and ETH resistance were subjected to qRT-PCR quantification. Total RNA was isolated from MsmegpVV16 and MsmegpVV0023 and the expression levels compared between the two strains. We found that the expression level of ndh (MSMEG_3621) in MsmegpVV0023 was approximately twofold lower than that of MsmegpVV16. No changes in the expression levels were observed in inhA (MSMEG_3151), katG (MSMEG_6384), and ethA (MSMEG_6440) between both strains (Figure 2B). As ndh encodes NdhII, which oxidizes NADH to NAD+, we reasoned that the ratio of NADH/NAD+ might be altered in MsmegpVV0023 strain. So, we measured the cellular ratio of NADH and NAD+ in MsmegpVV16 and MsmegpVV0023 strains and found that the ratio of NADH/NAD+ is increased in MsmegpVV0023
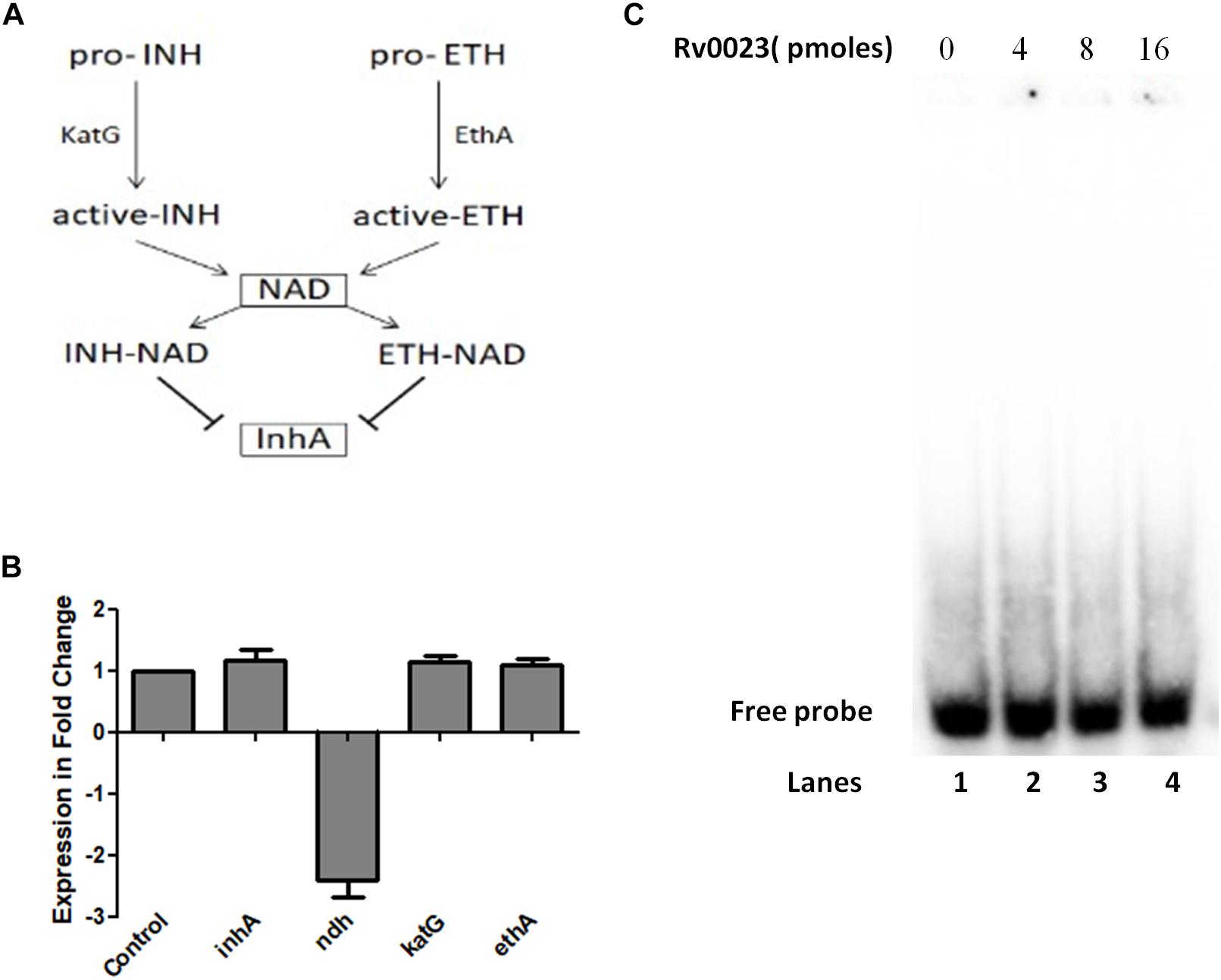
Figure 2. ndh gene regulation by Rv0023. (A) Schematic representation of mechanisms of action of INH and ETH. (B) Expression levels of genes involved in INH and ETH resistance pathway in MsmegpVV16 and MsmegpVV0023 cells. Fold change was calculated relative to the vector control pVV16. Data are presented as the mean ± SD from three biological replicates. One-way ANOVA was applied and the p-value obtained was p < 0.0001. (C) EMSA studies with radiolabeled 300 bp of upstream region of ndh. Lane 1: probe without any protein. Lanes 2–4: probe with increasing concentration of purified Rv0023 protein.
(Table 6). These results indicate that Rv0023 expression alters both the transcript levels of ndh gene and NADH/NAD+ levels. To find out whether these effects are directly mediated by Rv0023 or through some indirect means, we studied the interaction of recombinant Rv0023 with the upstream region of ndh (MSMEG_3261) gene. However, under the experimental conditions mentioned, we did not observe any such positive interaction (Figure 2C).
Rv0023 Negatively Regulates whiB5
Rv0023 is in operon with Rv0024 and is transcribed divergently from whiB5 (Figure 3A). Rv0023 and whiB5 are absent from the non-pathogenic, fast-growing Msmeg but are present in Mtb. So, we tried to find the co-occurrence of these two genes in other mycobacterial species. Synteny analysis of Rv0023 and whiB5 was performed for important species of the Mycobacterium genus using the “SynTax” web server (Figure 3B). It was observed that whiB5 is in synteny with Rv0023 and that both are present only in pathogenic species of mycobacteria (Supplementary Table 1).
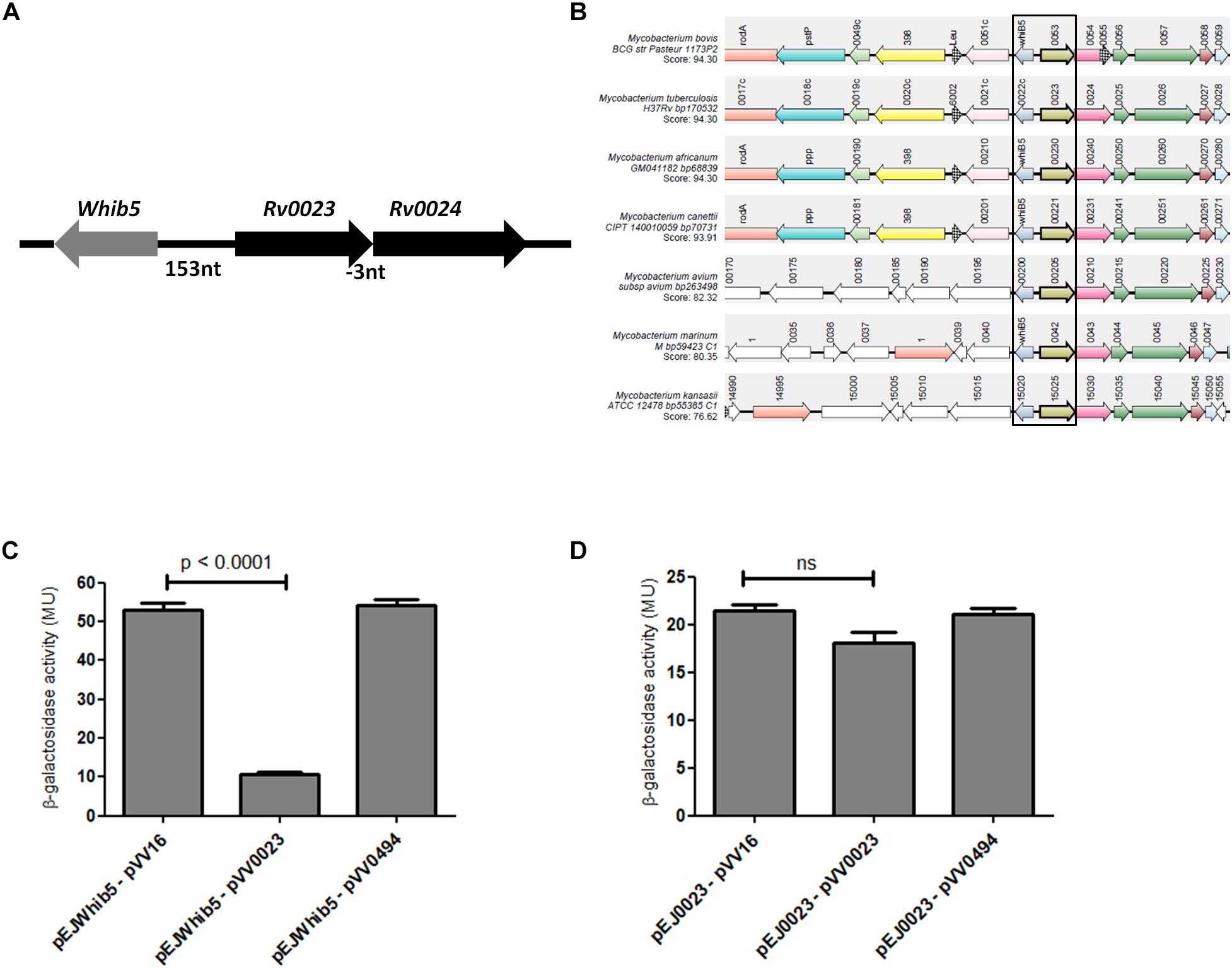
Figure 3. Rv0023 exists in a syntenic relationship with whiB5 and negatively regulates whiB5 expression. (A) Genomic organization of whiB5 and Rv0023 locus. (B) Schematic representation showing synteny of Rv0023 and whiB5 (within rectangular boxes) from seven mycobacterial species strains. (C) β-Galactosidase activity of whiB5 promoter with ectopic expression of Rv0023 protein and Rv0494 (non-specific protein for negative control). (D) β-Galactosidase activity of Rv0023 promoter in the presence of Rv0023 and Rv0494 overexpression.
The syntenic relationship of the two regulators prompted us to probe the regulation of the whiB5-Rv0023 locus. To identify the regulatory elements at this locus, upstream regions of whiB5 and Rv0023 were cloned in the promoter-less vector pEJ414 and named pEJwhiB5 and pEJ0023, respectively. The coding region of Rv0023 was cloned in the pVV16 vector to obtain pVV0023. Msmeg was transformed with pEJwhiB5 or pEJ0023. The strains were again transformed with pVV023 to obtain MsmegpEJwhiB5-pVV0023 and MsmegpEJ0023-pVV0023 double transformants. Promoter activities were measured in double transformants (MsmegpEJwhiB5-pVV0023, MsmegpEJwhiB5-pVV16, MsmegpEJ0023-pVV0023, and MsmegpEJ0023-pVV16). Overexpression of Rv0023 caused approximately fivefold decrease in whiB5 promoter activity (Figure 3C), whereas no significant change in the promoter activity of Rv0023 was noted (Figure 3D). Rv0494, a FadR transcriptional regulator, was used as a negative control, which did not affect the promoter activities of any of these genes. These data suggest that Rv0023 negatively regulates the expression of whiB5, but it is not auto-regulatory.
Rv0023 Binds to whiB5 in vitro
As we have observed that Rv0023 expression represses the promoter activity of whiB5 in Msmeg, it prompted us to further test whether Rv0023 interacts with whiB5 promoter or not. Five hundred base pairs upstream and 50 bp downstream of whiB5 gene containing the promoter were radiolabeled and EMSA was performed with the recombinant Rv0023. The radiolabeled upstream region of whiB5 interacted with the purified recombinant Rv0023 in a dose-dependent manner. No interaction of Rv0023 was observed with non-specific DNA (Figure 4A).

Figure 4. Rv0023 interaction with whiB5 upstream. (A) Rv0023 binding to whiB5 upstream in EMSA. Lane 1: radiolabeled whiB5 upstream with no protein. Lanes 2–7: whiB5 upstream with increasing concentration of Rv0023 protein. Lane 8: upstream region of Rv0494, a non-specific DNA, with no protein. Lanes 9–10: upstream region of Rv0494, a non-specific DNA, with increasing concentration of Rv0023 protein. (B) Six overlapping fragments (F1–F6) covering whiB5 and Rv0023 locus. (C) EMSA for overlapping fragments of whiB5 upstream with purified Rv0023 protein. Lanes F1–F6: fragments with (+) and without (–) Rv0023 (8 pmol) protein.
Both EMSA and β-galactosidase assays confirmed that Rv0023 interacts with whiB5 upstream and negatively regulates whiB5 expression. In order to find the exact binding site of Rv0023, the 550 bp upstream of whiB5 was divided into two fragments: UF1 of 340 bp and UF2 of 250 bp (Figure 4B). We observed that only the UF2 fragment binds to Rv0023 (Supplementary Figure 3). The UF2 fragment was further divided into six overlapping fragments, F1–F6 (Figure 4B and Table 7). Purified Rv0023 binds to fragments F1 and F2, whereas no binding was observed with other fragments (Figure 4C). So, we looked for a binding site at the overlapping region of F1 and F2. We generated new oligonucleotides (OL1–OL5) covering the F1–F2 overlapping region (Figure 5A and Table 8). EMSA with these radiolabeled fragments showed that Rv0023 binds to OL1 and OL3 fragments whereas no binding was observed with other fragments (Figure 5B). Close examination of these fragments revealed the presence of an imperfect palindrome present in both sequences, TATAcgtTTTT in OL1 and TATAcgcTTTT in OL3 (Figure 5C). To confirm the binding sites identified, we mutated TATAcgtTTTT to TGTGcgtTGTG in OL1 and TATAcgcTTTT to TGTGcgcTGTG in OL3 to obtain OL1M and OL3M, respectively (Table 8). Binding studies have shown that Rv0023 binds to the native OL1 and OL3 fragments, but not to the mutant fragments, OL1M and OL3M (Figure 5C). We conclude that there are two binding sites (TATAcgtTTTT and TATAcgcTTTT) of Rv0023 in the upstream region of whiB5 comprising an 11-bp imperfect palindromic sequence with a 3-bp spacer region.

Table 7. List of overlapping fragments used in Figure 4.

Table 8. List of overlapping fragments used in Figure 5.
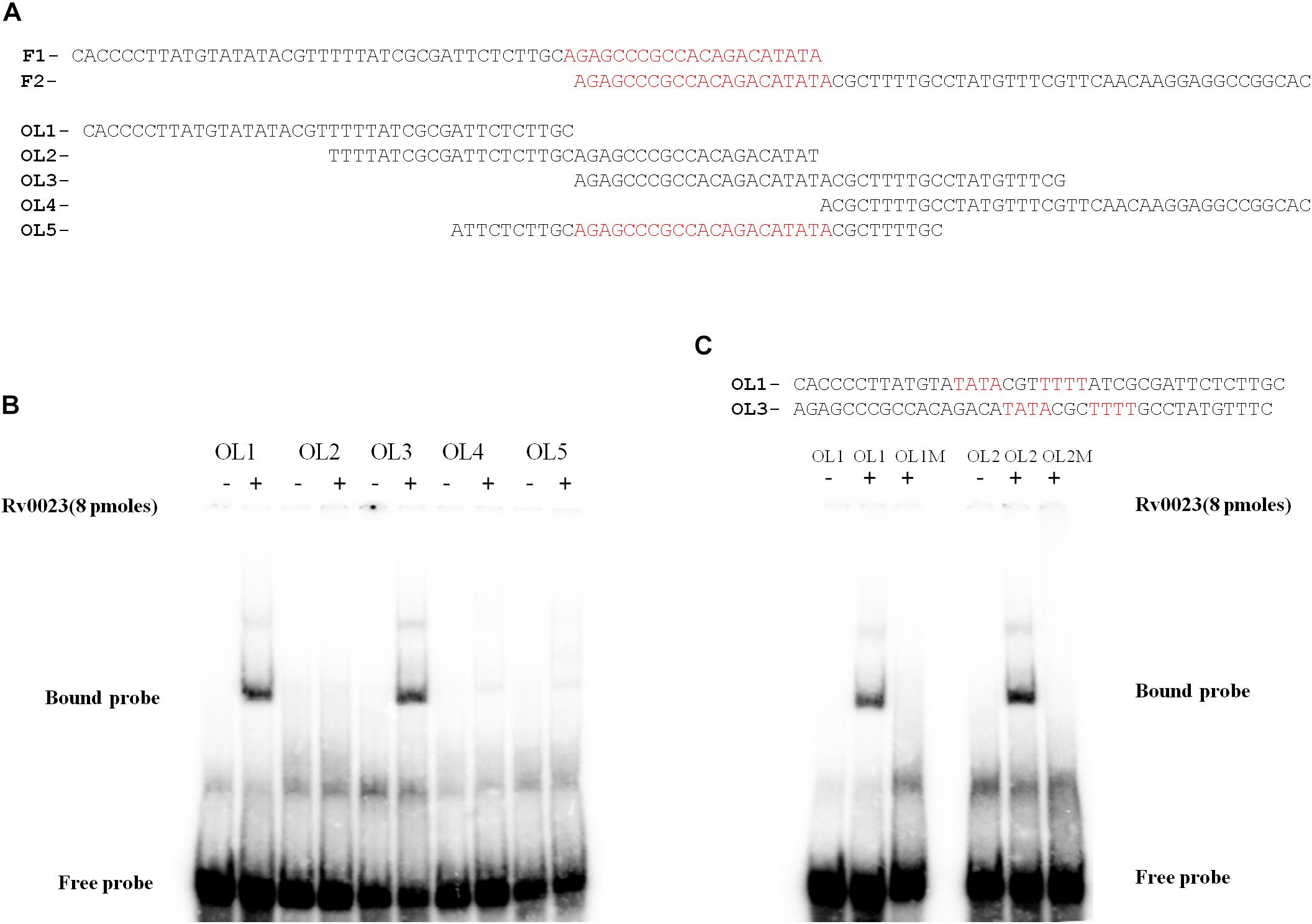
Figure 5. Identification of Rv0023 binding site. (A) OL1–OL4 spanning the F1–F2 fragments and OL5 comprising the overlapping region of F1–F2. (B) EMSA with fragments OL1–OL5 with (+) and without (–) Rv0023 (8 pmol) protein. (C) OL1 and OL3 fragments containing the imperfect palindrome, indicated in red. EMSA with mutated imperfect palindromic DNA region. Fragments with (+) and without (–) Rv0023 (8 pmol) protein.
Analysis of Rv0023 Binding Site
The importance of each residue within the Rv0023 binding site was determined by generating 39 bp probes (M0–M12) containing specific point mutations on each half of the imperfect palindrome and in the spacer region (Figure 6A). The probes were radiolabeled and were used in binding studies with recombinant Rv0023. The binding was observed with probes M0 (unaltered), M1, M4, and M5, and significant loss of binding was observed with M2, M3, M6, M7, and M8 fragments, suggesting the importance of these residues in Rv0023 binding. Changing the residues in the spacer region (M9–M10) did not affect Rv0023 binding; however, changing the length of spacer region by one base lead to significant loss of Rv0023 binding (M11–M12) (Figure 6B). As mentioned earlier, the organization of Rv0023 and whiB5 is conserved across pathogenic mycobacterial species. Therefore, we looked into the conservation of identified Rv0023 binding sites across the conserved genomes. For this, we aligned the upstream region of whiB5 orthologs from different pathogenic mycobacterial species strains and found that the two binding sites of Rv0023 are conserved across the analyzed mycobacterial species (Figure 6C). These results highlight the important residues for Rv0023 binding and its conservation across mycobacterial species.
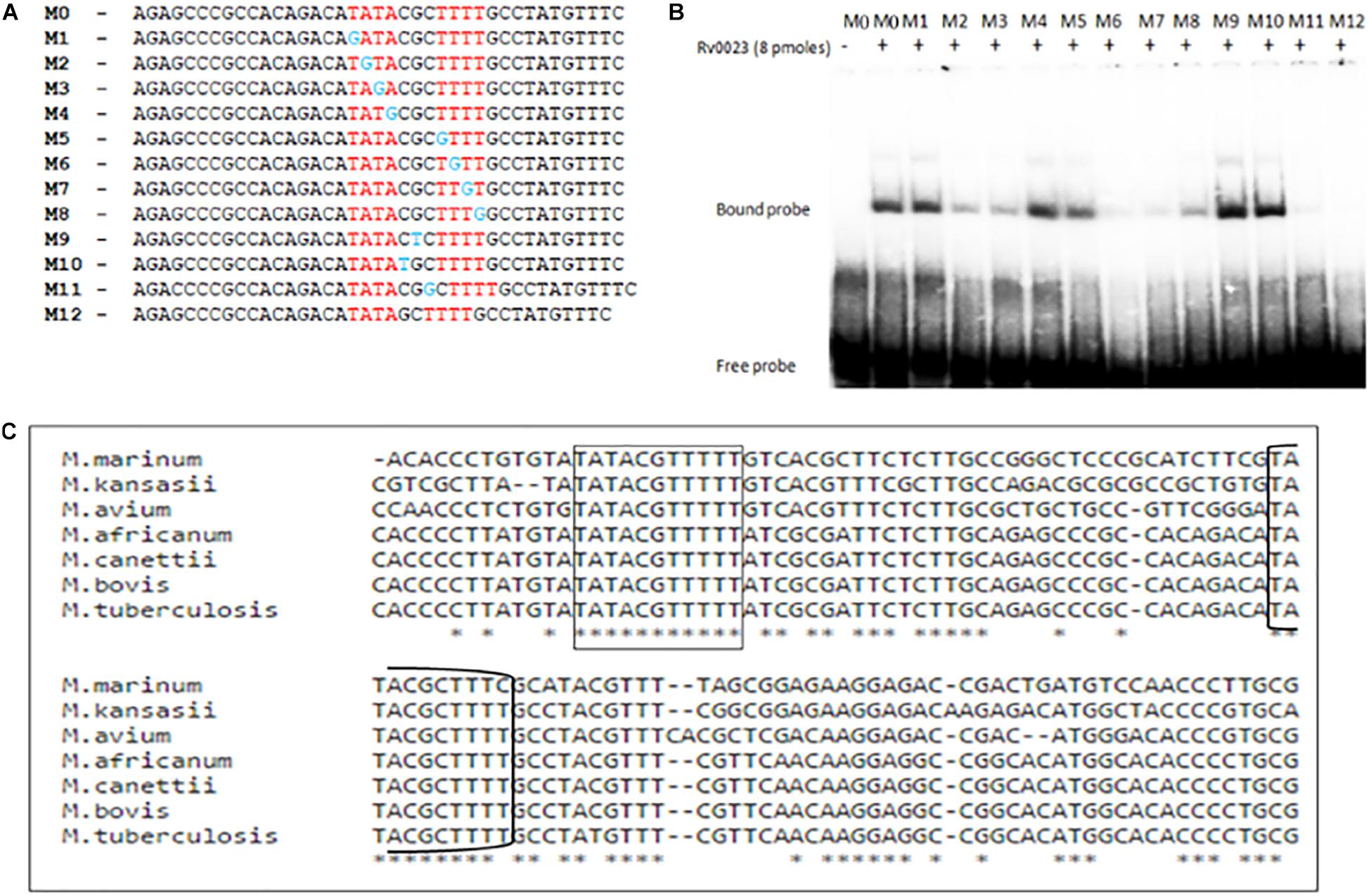
Figure 6. Identification of important residues in the imperfect palindrome for Rv0023 binding. (A) Representation of probes (∼40 bp each) containing native binding site M0 and probe with mutation at specific positions in the binding site M1–M8 and probe with mutation in spacer regions M9–M12. (B) EMSA with radiolabeled probes [native (M0), mutated (M1–M9), and altered (M9–M12)] with constant concentration of purified Rv0023 protein. Fragments with (+) and without (–) Rv0023 (8 pmol) protein. (C) Multiple sequence alignment of whiB5 upstream from selected members of mycobacterial species. The two binding sites of Rv0023 are highlighted in rectangular boxes.
Rv0023 Binding Site Overlaps With the whiB5 Promoter Region
In order to understand the mechanism of Rv0023-mediated repression of whiB5, the TSSs of whiB5 and Rv0023 were mapped. The putative −10 region of whiB5 was identified earlier by RACE (Casonato et al., 2012) (Figure 7A). To test the functionality of the putative −10 region, we cloned the 250 bp upstream of whiB5 into lacZ reporter vector (pEJwhiB5WT). The promoter activity was measured using the β-galactosidase assay in Msmeg. Mutations in the putative −10 region of whiB5 (atacgctt to gcacgcgg) abolished the promoter activity, confirming the −10 region of whiB5 (Figure 7B). To identify the promoter region of Rv0023, primer extension was performed. We identified the TSS at 228 bp upstream of Rv0023 start codon (Figure 7C). A −10-like hexamer sequence TCATAG was identified upstream of TSS. To validate the functionality of the −10 region of Rv0023, 400 bp upstream of Rv0023 was cloned into a lacZ reporter vector (pEJ0023WT). β-Galactosidase assay with WT promoter and mutated promoter (pEJ0023MUT) indicated significant decrease in the promoter activity of the mutant strain compared to WT (Figure 7D). Mapping the promoters of whiB5 and Rv0023 with Rv0023 binding site revealed that Rv0023 binding site overlaps with the promoter of whiB5 but not of the Rv0023 promoter region (Figure 7E).
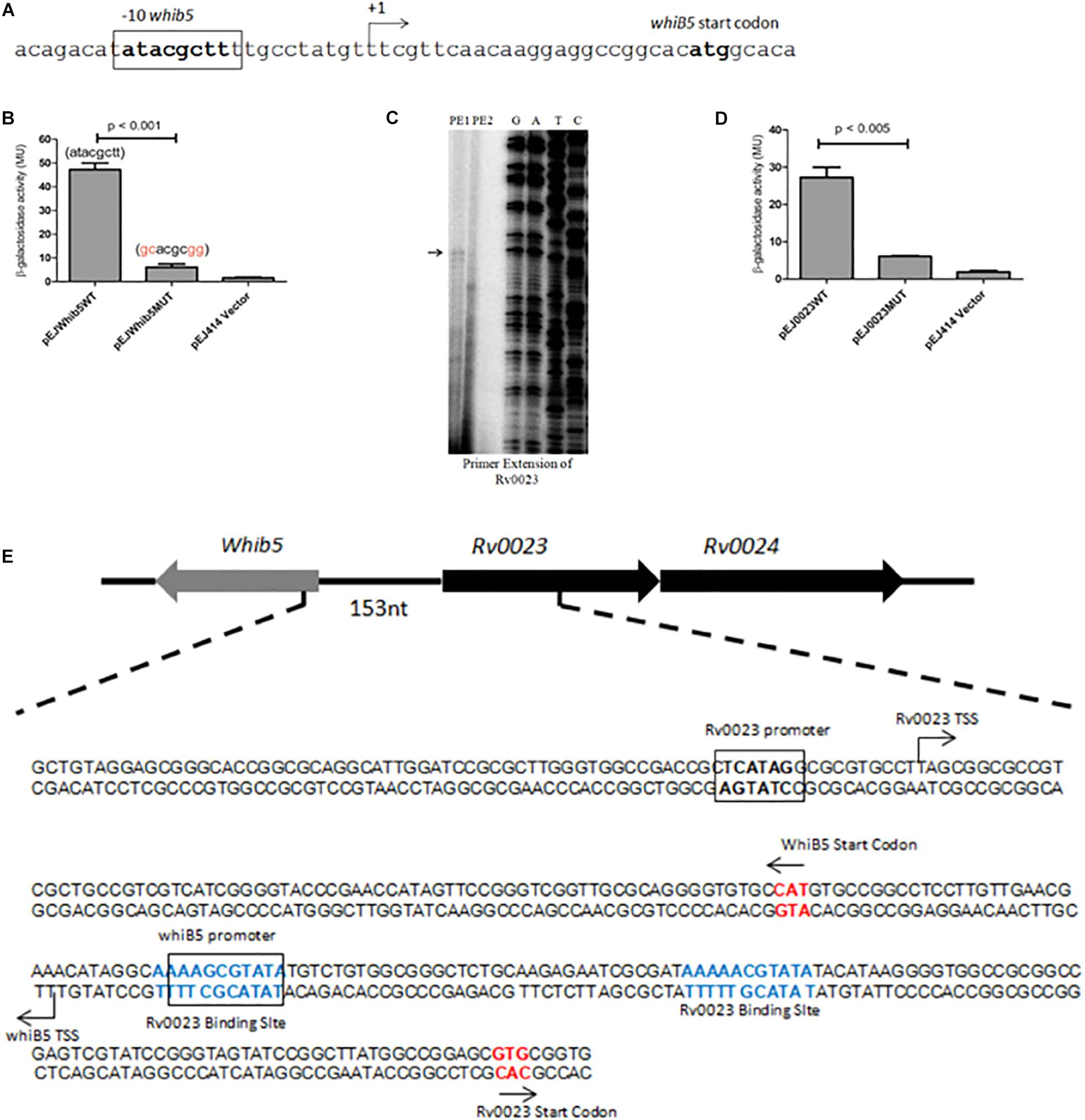
Figure 7. Identification of promoter regions of whiB5 and Rv0023. (A) Nucleotide sequence of whiB5 upstream containing whiB5 start codon and –10 region. The –10 region is highlighted in bold. (B) β-Galactosidase activity of Msmeg transformed with whiB5 wild-type promoter (pEJwhiB5WT), whiB5 mutated (pEJwhiB5MUT) promoter, and vector control. (C) Primer extension analysis of Rv0023. PE1 and PE2 indicate two primers used for primer extension assay. The primer extension product is indicated by an arrow. G, A, T, and C indicate the sequencing ladder generated by using dideoxy nucleotides. (D) β-Galactosidase activity of predicted Rv0023 wild type (pEJ0023WT) and Rv0023 mutated (pEJ0023MUT) promoter showing significantly reduced activity of mutated promoter. (E) Genomic organization of Rv0023 binding sites (shown in bold and colored) and promoter regions (bold and boxed) of whiB5 and Rv0023. Start codons are represented by an arrow.
Discussion
In this study, Rv0023, a member of the XRE family of transcriptional regulators, has been characterized, and its role in INH and ETH drug tolerance has been explored. The first part of our study concerns the role of Rv0023 in the physiology of mycobacteria. For our study, we used Msmeg as a surrogate model to study the effects of various stress conditions on wild type and Rv0023-expressing strains. It was observed that overexpression of Rv0023 confers higher tolerance toward INH and ETH in Msmeg. It was seen that the ectopic expression of Rv0023 alters the NADH/NAD+ ratio, thereby increasing drug tolerance. In the second part of the manuscript, we identified Rv0023 as a transcriptional regulator and studied the regulatory effect at whiB5-Rv0023 locus and observed that Rv0023 negatively regulates whiB5 expression but is not auto-regulatory. Rv0023 regulates whiB5 expression by binding to specific sequences in the upstream region of whiB5. Our data indicate that there are two binding sites for Rv0023 upstream of whiB5 and one of the sites overlaps with the whiB5 promoter, thereby possibly occluding the binding of RNA polymerase. The binding site of Rv0023 is conserved across mycobacterial species as confirmed by multiple sequence alignment. Apart from this, we have also characterized the binding site of Rv0023 and found important bases for binding. While analyzing the spacer region, it was found that the length of the spacer region is important for proper binding.
Isoniazid, isonicotinic acid hydrazide, is a synthetic drug and the occurrence of INH-resistant strains is significantly more frequent than other drug-resistant Mtb clinical strains (Nachega and Chaisson, 2003). ETH, 2-ethylthioisonicotinamide, is a structural analog of INH and was first synthesized in 1956 (Grumbach et al., 1956). Many genes have been found to be associated with INH and ETH resistance in clinical isolates, but the role of transcription factors regulating INH and ETH co-resistance is poorly understood. Rv0023, a transcriptional regulator, conferring higher tolerance toward INH and ETH in Msmeg, provided us a chance to explore the role of transcription factors in INH and ETH co-resistance in Mtb. To elucidate the mechanism by which Rv0023 confers drug tolerance, we investigated two main genes known to be involved in INH and ETH co-resistance, inhA (enzyme involved in the synthesis of cell wall mycolic acid) (Banerjee et al., 1994) and ndh (NADH dehydrogenase maintains the NADH/NAD + ratio) (Miesel et al., 1998). We checked their expression levels in the vector control and Rv0023-expressing Msmeg strains and differential expression was only observed in the ndh gene. The ndh gene oxidizes NADH to NAD+, which maintains the ratio of NADH/NAD+ in mycobacterial cells (Jaworowski et al., 1981). Loss of ndh function leads to higher levels of NADH in the cytosol. Previous studies have shown that the higher NADH/NAD+ levels interfere with activation of INH/ETH drugs by competitive inhibition with the formation of INH-NAD and ETH-NAD and thus confer resistance to INH/ETH in Msmeg and BCG (Miesel et al., 1998; Vilcheze et al., 2005). So, we further measured the NADH/NAD+ ratio in vector control and Rv0023-expressing Msmeg strains. The NADH/NAD+ ratio was found to be increased in the Rv0023-overexpressed Msmeg strain, suggesting that negative regulation of ndh gene by Rv0023 is probably the mechanism by which Rv0023 confers INH and ETH tolerance in Msmeg. Our results were supported by previous studies where it was shown that Rv0023 regulon is enriched for NAD reductases (Rustad et al., 2014). Rv0023 did not bind to the upstream region of ndh, indicating that the regulation occurs possibly through indirect means, which may involve multiple intermediate gene products. Further studies are required to completely decipher this mechanism.
Synteny analysis has shown that whiB5-Rv0023 locus is present mainly in pathogenic species of mycobacteria. The gene whiB5 belongs to the WhiB family of transcriptional regulators, which are exclusive to actinomycetes, such as Mycobacterium and Streptomyces spp. (Soliveri et al., 2000). Earlier studies have shown that whiB5 is a global transcriptional regulator, regulating 58 genes of diverse functions, including sigM and genes encoding for Type VII secretion systems such as esx-2 and esx-4. It was shown that whiB5 has a role in Mtb virulence and reactivation (Casonato et al., 2012). As the whole locus of whiB5-Rv0023-Rv0024 is absent in Msmeg, it is difficult to gauge the true effect of Rv0023 in the physiology of Mtb. Further investigation in Mtb may reveal important aspects of Rv0023 regulation and its effects. Hence, taking into account the earlier studies and our current data, we surmise that Rv0023 plays a significant role in modulating genes associated with NADH/NAD+ levels, which further influences mycobacterial physiology.
In conclusion, our work shows, for the first time, that Rv0023 has a role in conferring INH and ETH tolerance in Msmeg. Understanding the role of transcriptional factors in the development of drug resistance will open new avenues in the field of drug discovery and may provide important insights into Mtb physiology.
Data Availability Statement
The datasets generated for this study are available on request to the corresponding author.
Author Contributions
AR, SG, RA, and SY conceived the hypothesis and rationale of the study, analyzed the results, and wrote the manuscript. SG performed all the experiments. CR contributed in cloning of constructs and recombinant protein purification.
Funding
SG was supported by a Centre for DNA Fingerprinting and Diagnostics fellowship. RA was supported earlier by a Department of Biotechnology fellowship. SY was supported earlier by a Council of Scientific and Industrial Research fellowship at the Computational and Functional Genomics Group. CR was supported by a Centre for DNA Fingerprinting and Diagnostics fellowship.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Dr. Roger Buxton (MRC, London, United Kingdom) for pEJ414 plasmid. Research in the AR laboratory was supported by the Centre for DNA Fingerprinting and Diagnostics and Department of Biotechnology, Government of India.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.00003/full#supplementary-material
Footnotes
References
Angara, R. K., Yousuf, S., Gupta, S. K., and Ranjan, A. (2018). An IclR like protein from mycobacteria regulates leuCD operon and induces dormancy-like growth arrest in Mycobacterium smegmatis. Tuberculosis 108, 83–92. doi: 10.1016/j.tube.2017.10.009
Banerjee, A., Dubnau, E., Quemard, A., Balasubramanian, V., Um, K. S., Wilson, T., et al. (1994). inhA, a gene encoding a target for isoniazid and ethionamide in Mycobacterium tuberculosis. Science 263, 227–230. doi: 10.1126/science.8284673
Barragán, M. J., Blázquez, B., Zamarro, M. T., Mancheño, J. M., García, J. L., Díaz, E., et al. (2005). BzdR, a repressor that controls the anaerobic catabolism of benzoate in Azoarcus sp. CIB, is the first member of a new subfamily of transcriptional regulators. J. Biol. Chem. 280, 10683–10694. doi: 10.1074/jbc.m412259200
Baulard, A. R., Betts, J. C., Engohang-Ndong, J., Quan, S., McAdam, R. A., Brennan, P. J., et al. (2000). Activation of the pro-drug ethionamide is regulated in mycobacteria. J. Biol. Chem. 275, 28326–28331.
Bellier, A., and Mazodier, P. (2004). ClgR, a novel regulator of clp and lon expression in Streptomyces. J. Bacteriol. 186, 3238–3248. doi: 10.1128/jb.186.10.3238-3248.2004
Bernstein, J. W., Lott, A., Steinberg, B. A., and Yale, H. L. (1952). Chemotherapy of experimental tuberculosis. Am. Rev. Tuberc. 65, 357–374.
Cardoso, R. F., Cardoso, M. A., Leite, C. Q. F., Sato, D. N., Mamizuka, E. M., Hirata, R. D. C., et al. (2007). Characterization of ndh gene of isoniazid resistant and susceptible Mycobacterium tuberculosis isolates from Brazil. Mem. Instit. Oswaldo Cruz 102, 59–61. doi: 10.1590/s0074-02762007000100009
Carey, M. F., Peterson, C. L., and Smale, S. T. (2013). The primer extension assay. Cold Spring Harb. Protoc. 8, 164–173.
Casonato, S., Sánchez, A. C., Haruki, H., González, M. R., Provvedi, R., Dainese, E., et al. (2012). WhiB5, a transcriptional regulator that contributes to Mycobacterium tuberculosis virulence and reactivation. Infect. Immun. 80, 3132–3144. doi: 10.1128/IAI.06328-11
DeBarber, A. E., Mdluli, K., Bosman, M., Bekker, L. G., and Barry, C. E. III (2000). Ethionamide activation and sensitivity in multidrug-resistant Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. U.S.A. 97, 9677–9682. doi: 10.1073/pnas.97.17.9677
Fox, H. H. (1952). The chemical approach to the control tuberculosis. Science 116, 129–134. doi: 10.1126/science.116.3006.129
Fraaije, M. W., Kamerbeek, N. M., Heidekamp, A. J., Fortin, R., and Janssen, D. B. (2004). The prodrug activator EtaA from Mycobacterium tuberculosis is a Baeyer-Villiger monooxygenase. J. Biol. Chem. 279, 3354–3360. doi: 10.1074/jbc.m307770200
Gaur, N. K., Oppenheim, J., and Smith, I. (1991). The Bacillus subtilis sin gene, a regulator of alternate developmental processes, codes for a DNA-binding protein. J. Bacteriol. 173, 678–686. doi: 10.1128/jb.173.2.678-686.1991
Gennis, R. B., and Stewart, V. (1996). “Respiration,” in Escherichia coli and Salmonella: Cellular and Molecular Biology, Vol. 1, eds F. C. Neidhardt, R. Curtiss, III J. L. Ingraham, E. C. C. Lin, K. B. Low, B. Magasanik, et al. (Washington, DC: ASM Press), 217–261.
Grumbach, F., Rist, N., Libermann, D., Moyeux, M., Cals, S., and Clavel, S. (1956). Experimental antituberculous activity of certain isonicotinic thioamides substituted on the nucleus. CR Hebd. Seances Acad. Sci. 242, 2187–2189.
Hazbón, M. H., Brimacombe, M., Del Valle, M. B., Cavatore, M., Guerrero, M. I., Varma-Basil, M., et al. (2006). Population genetics study of isoniazid resistance mutations and evolution of multidrug-resistant Mycobacterium tuberculosis. Antimicrob. Agents Chemother. 50, 2640–2649. doi: 10.1128/aac.00112-06
Jaworowski, A., Mayo, G., Shaw, D. C., Campbell, H. D., and Young, I. G. (1981). Characterization of the respiratory NADH dehydrogenase of Escherichia coli and reconstitution of NADH oxidase in ndh mutant membrane vesicles. Biochemistry 20, 3621–3628. doi: 10.1021/bi00515a049
Johnsson, K., Froland, W. A., and Schultz, P. G. (1997). Overexpression, purification, and characterization of the catalase-peroxidase KatG from Mycobacterium tuberculosis. J. Biol. Chem. 272, 2834–2840. doi: 10.1074/jbc.272.5.2834
Johnsson, K., and Schultz, P. G. (1994). Mechanistic studies of the oxidation of isoniazid by the catalase peroxidase from Mycobacterium tuberculosis. J. Am. Chem. Soc. 116, 7425–7426. doi: 10.1021/ja00095a063
Kerscher, S. J. (2000). Diversity and origin of alternative NADH:ubiquinone oxidoreductases. Biochim. Biophys. Acta 1459, 274–283. doi: 10.1016/s0005-2728(00)00162-6
Korduláková, J., Gilleron, M., Mikuová, K., Puzo, G., Brennan, P. J., Gicquel, B., et al. (2002). Definition of the first mannosylation step in phosphatidylinositol mannoside synthesis: PimA is essential for growth of mycobacteria. J. Biol. Chem. 277, 31335–31344. doi: 10.1074/jbc.m204060200
Lee, A. S., Teo, A. S., and Wong, S. Y. (2001). Novel mutations in ndh in isoniazid-resistant Mycobacterium tuberculosis isolates. Antimicrob. Agents Chemother. 45, 2157–2159. doi: 10.1128/aac.45.7.2157-2159.2001
Leonardo, M. R., Dailly, Y., and Clark, D. P. (1996). Role of NAD in regulating the adhE gene of Escherichia coli. J. Bacteriol. 178, 6013–6018. doi: 10.1128/jb.178.20.6013-6018.1996
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2- ΔΔCT method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Marchler-Bauer, A., Derbyshire, M. K., Gonzales, N. R., Lu, S., Chitsaz, F., Geer, L. Y., et al. (2015). CDD: NCBI’s conserved domain database. Nucleic Acids Res. 43, D222–D226. doi: 10.1093/nar/gku1221
Matsushita, K., Ohnishi, T., and Kaback, H. R. (1987). NADH-ubiquinone oxidoreductases of the Escherichia coli aerobic respiratory chain. Biochemistry 26, 7732–7737. doi: 10.1021/bi00398a029
Middlebrook, G., and Cohn, M. L. (1953). Some observations on the pathogenicity of isoniazid-resistant variants of tubercle bacilli. Science 118, 297–299. doi: 10.1126/science.118.3063.297
Miesel, L., Weisbrod, T. R., Marcinkeviciene, J. A., Bittman, R., and Jacobs, W. R. (1998). NADH dehydrogenase defects confer isoniazid resistance and conditional lethality in Mycobacterium smegmatis. J. Bacteriol. 180, 2459–2467. doi: 10.1128/jb.180.9.2459-2467.1998
Miller, J. H. (1972). Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Press.
Nachega, J. B., and Chaisson, R. E. (2003). Tuberculosis drug resistance: a global threat. Clin. Infect. Dis. 36(Suppl. 1), S24–S30.
Nemoto, N., Kurihara, S., Kitahara, Y., Asada, K., Kato, K., and Suzuki, H. (2012). Mechanism for regulation of the putrescine utilization pathway by the transcription factor PuuR in Escherichia coli K-12. J. Bacteriol. 194, 3437–3447. doi: 10.1128/JB.00097-12
Oberto, J. (2013). SyntTax: a web server linking synteny to prokaryotic taxonomy. BMC Bioinf. 14:4. doi: 10.1186/1471-2105-14-4
Papavinasasundaram, K. G., Anderson, C., Brooks, P. C., Thomas, N. A., Movahedzadeh, F., Jenner, P. J., et al. (2001). Slow induction of RecA by DNA damage in Mycobacterium tuberculosis. Microbiology 147, 3271–3279. doi: 10.1099/00221287-147-12-3271
Raghavan, S., Manzanillo, P., Chan, K., Dovey, C., and Cox, J. S. (2008). Secreted transcription factor controls Mycobacterium tuberculosis virulence. Nature 454:717. doi: 10.1038/nature07219
Rustad, T. R., Minch, K. J., Ma, S., Winkler, J. K., Hobbs, S., Hickey, M., et al. (2014). Mapping and manipulating the Mycobacterium tuberculosis transcriptome using a transcription factor overexpression-derived regulatory network. Genome Biol. 15:502.
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: a Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harb Lab press.
San, K. Y., Bennett, G. N., Berrios-Rivera, S. J., Vadali, R. V., Yang, Y. T., Horton, E., et al. (2002). Metabolic engineering through cofactor manipulation and its effects on metabolic flux redistribution in Escherichia coli. Metab. Eng. 4, 182–192. doi: 10.1006/mben.2001.0220
Sassetti, C. M., Boyd, D. H., and Rubin, E. J. (2003). Genes required for mycobacterial growth defined by high density mutagenesis. Mol. Microbiol. 48, 77–84. doi: 10.1046/j.1365-2958.2003.03425.x
Sievers, F., Wilm, A., Dineen, D., Gibson, T. J., Karplus, K., Li, W., et al. (2011). Fast, scalable generation of high-quality protein multiple sequence alignments using Clustal Omega. Mol. Syst. Biol. 7:539. doi: 10.1038/msb.2011.75
Soliveri, J. A., Gomez, J., Bishai, W. R., and Chater, K. F. (2000). Multiple paralogous genes related to the Streptomyces coelicolor developmental regulatory gene whiB are present in Streptomyces and other actinomycetes. Microbiology 146, 333–343. doi: 10.1099/00221287-146-2-333
Vannelli, T. A., Dykman, A., and Ortiz de Montellano, P. R. (2002). The antituberculosis drug ethionamide is activated by a flavoprotein monooxygenase. J. Biol. Chem. 277, 12824–12829. doi: 10.1074/jbc.m110751200
Vilcheze, C., Weisbrod, T. R., Chen, B., Kremer, L., Hazbón, M. H., Wang, F., et al. (2005). Altered NADH/NAD+ ratio mediates coresistance to isoniazid and ethionamide in mycobacteria. Antimicrob. Agents Chemother. 49, 708–720. doi: 10.1128/aac.49.2.708-720.2005
Yagi, T. (1993). The bacterial energy-transducing NADH-quinone oxidoreductases. Biochim. Biophys. Acta 1141, 1–17. doi: 10.1016/0005-2728(93)90182-f
Yousuf, S., Angara, R., Vindal, V., and Ranjan, A. (2015). Rv0494 is a starvation-inducible, autoregulatory FadR-like regulator from Mycobacterium tuberculosis. Microbiology 161, 463–476. doi: 10.1099/mic.0.000017
Keywords: XRE family of protein, whiB5, isoniazid resistance, ethionamide resistance, transcription regulation
Citation: Gupta SK, Angara RK, Yousuf S, Reddy CG and Ranjan A (2020) Ectopic Expression of Rv0023 Mediates Isoniazid/Ethionamide Tolerance via Altering NADH/NAD+ Levels in Mycobacterium smegmatis. Front. Microbiol. 11:3. doi: 10.3389/fmicb.2020.00003
Received: 19 July 2019; Accepted: 03 January 2020;
Published: 07 February 2020.
Edited by:
Mattias Collin, Lund University, SwedenReviewed by:
Yu-Min Chuang, Yale University, United StatesDiana Machado, NOVA University Lisbon, Portugal
Copyright © 2020 Gupta, Angara, Yousuf, Reddy and Ranjan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Akash Ranjan, YWthc2hAY2RmZC5vcmcuaW4=; ZHIuYWthc2gucmFuamFuQGdtYWlsLmNvbQ==
†These authors have contributed equally to this work
 Shailesh Kumar Gupta
Shailesh Kumar Gupta Rajendra Kumar Angara
Rajendra Kumar Angara Suhail Yousuf1†
Suhail Yousuf1† Akash Ranjan
Akash Ranjan




