- 1Dipartimento di Biologia, Università di Pisa, Pisa, Italy
- 2Mikrobiologie, Institut für Biologie II, Albert-Ludwigs-Universität Freiburg, Freiburg, Germany
- 3Institut für Hydrobiologie, Technische Universität Dresden, Dresden, Germany
- 4Dipartimento di Biologia, CoNISMa, Università di Pisa, Pisa, Italy
“Candidatus Megaira polyxenophila” is a recently described member of Rickettsiaceae which comprises exclusively obligate intracellular bacteria. Interestingly, these bacteria can be found in a huge diversity of eukaryotic hosts (protist, green algae, metazoa) living in marine, brackish or freshwater habitats. Screening of amplicon datasets revealed a high frequency of these bacteria especially in freshwater environments, most likely associated to eukaryotic hosts. The relationship of “Ca. Megaira polyxenophila” with their hosts and their impact on host fitness have not been studied so far. Even less is known regarding the responses of these intracellular bacteria to potential stressors. In this study, we used two phylogenetically close species of the freshwater ciliate Paramecium, Paramecium primaurelia and Paramecium pentaurelia (Ciliophora, Oligohymenophorea) naturally infected by “Ca. Megaira polyxenophila”. In order to analyze the effect of the symbiont on the fitness of these two species, we compared the growth performance of both infected and aposymbiotic paramecia at different salinity levels in the range of freshwater and oligohaline brackish water i.e., at 0, 2, and 4.5 ppt. For the elimination of “Ca. Megaira polyxenophila” we established an antibiotic treatment to obtain symbiont-free lines and confirmed its success by fluorescence in situ hybridization (FISH). The population and infection dynamics during the growth experiment were observed by cell density counts and FISH. Paramecia fitness was compared applying generalized additive mixed models. Surprisingly, both infected Paramecium species showed higher densities under all salinity concentrations. The tested salinity concentrations did not significantly affect the growth of any of the two species directly, but we observed the loss of the endosymbiont after prolonged exposure to higher salinity levels. This experimental data might explain the higher frequency of “Ca. M. polyxenophila” in freshwater habitats as observed from amplicon data.
Introduction
Paramecium (Ciliophora, Oligohymenophorea) is a unicellular protist with a broad, nearly global distribution in fresh and brackish water bodies. This ciliate is studied, among other things (Karunanithi et al., 2019; Kelz and Mashour, 2019; Mayne et al., 2019; Soares et al., 2019; Arnaiz et al., 2020) for the abundance and diversity of its endosymbionts (Floriano et al., 2018; Garushyants et al., 2018; Grosser et al., 2018; Potekhin et al., 2018; Sabaneyeva et al., 2018; Schrallhammer et al., 2018; Castelli et al., 2019a, b; Fokin et al., 2019; Koehler et al., 2019; Lanzoni et al., 2019; Plotnikov et al., 2019). Host-symbiont interactions and their outcome have been studied for example using Holospora (Lohse et al., 2006; Hori et al., 2008; Fokin and Görtz, 2009; Nidelet et al., 2009; Duncan et al., 2013, 2018; Banerji et al., 2015; Castelli et al., 2015; Dusi et al., 2015; Garushyants et al., 2018; Grosser et al., 2018) Caedibacter (Kusch et al., 2002; Dusi et al., 2014; Grosser et al., 2018; Schu and Schrallhammer, 2018; Koehler et al., 2019) and Preeria (Bella et al., 2016; Potekhin et al., 2018). Despite an increasing number of studies, our knowledge about the impact of symbionts on Paramecium is limited, especially when considering that this ciliate is among the best studied protists in regard to host-symbiont interactions. Less is known regarding the response of these symbiotic systems exposed to additional environmental stressors. Even in a balanced system, the introduction of a stressor can have severe consequences. The impact of a symbiont could either shift towards virulent behavior (Dusi et al., 2014; Bella et al., 2016; Schu and Schrallhammer, 2018) and hence, represents an additional biotic stressor, or alternatively has a positive effect on the host’s stress response and thus on survival, e.g., for salinity stress (Smurov and Fokin, 1998; Duncan et al., 2010) or heat shock (Hori and Fujishima, 2003; Fujishima et al., 2005; Hori et al., 2008; Duncan et al., 2010). Some authors (Fokin and Sabaneyeva, 1990; Smurov and Fokin, 1998) reported a higher bacterial infection frequency in protists, i.e., Paramecium, living in brackish environments compared to those living in freshwater environments. This might imply an evolutionary advantage for symbiont-bearing microorganisms in habitats exposed to salinity stress. Members of the genus Paramecium are well known to be highly sensitive not only to temperature stress but also to increased salinity concentrations (Duncan et al., 2010). Contrary, it could indicate that stressed protists are more susceptible to infection.
In 2013, a bacterial species of the order Rickettsiales has been described as endosymbiont of different ciliates (Schrallhammer et al., 2013). All members of this order are obligate intracellular bacteria hosted by eukaryotic organisms and strictly depend on their hosts for multiplication (Dumler et al., 2001) with the notable exception of the epibiont “Candidatus Deianiraea vastatrix” (Castelli et al., 2019a). “Candidatus Megaira polyxenophila” is remarkable as it has been found associated to potential hosts spanning a huge diversity, e.g., various ciliates (Sun et al., 2009; Vannini et al., 2005; Schrallhammer et al., 2013; Zaila et al., 2017), amoebae (Hess, 2017) chlorophytes and streptophytes (Hollants et al., 2013; Kawafune et al., 2015; Yang et al., 2016) and even cnidarians (Fraune and Bosch, 2007; Sunagawa et al., 2009). Interestingly, the Megaira-infected organisms cover a surprising ecological range from freshwater lakes and ponds to brackish waters and even marine systems. Recently, this bacterium has been found associated with multiple Paramecium species, including several members of the Paramecium aurelia complex representing a group of phylogenetically very closely related species (Lanzoni et al., 2019). In this work, we found “Ca. Megaira polyxenophila” (from here referred as Ca. M. polyxenophila) naturally occurring in two species of the P. aurelia complex, i.e., Paramecium pentaurelia and Paramecium primaurelia.
The aim of this study is to analyze the role of Ca. M. polyxenophila, determining the symbiont’s effect on the growth of two phylogenetically close species of Paramecium after establishment of the corresponding symbiont-free cell lines via antibiotic treatment. The obtained infected and aposymbiotic lines were then used to compare their performance under standard laboratory conditions as well as at different salinity levels corresponding to oligohaline brackish water. Difference in growth performance depending on presence or absence of Ca. M. polyxenophila will shed light on the function of this endosymbiont and the role of this symbiosis under different environmental conditions such as increasing osmolality.
Materials and Methods
Experimental Organisms and Their Cultivation
Two different Paramecium species naturally harboring Ca. M. polyxenophila were used as endosymbiont-infected lines and to establish symbiont-free cells. The host species, as indicated in Table 1, were P. pentaurelia US_YE9 (from here on referred to as YE9) derived from Bloomington, IN, United States, whereas P. primaurelia Rio Lg_Jac 2III (from here on referred to as LgJac), was sampled in Rio de Janeiro, Brazil. Both species are freshwater protists. After antibiotic (AB) treatment (see below), we established aposymbiotic cell lines viz. YE9AB and LgJacAB. All cultures were maintained at 19°C in Cerophyll medium (CM) inoculated with Raoultella planticola DSM3069 (Enterobacteriales, Enterobacteriaceae) (Chiellini et al., 2019).
Prior to the experiment, a single cell was isolated from each cell line and was washed several times in spring water from a freshly opened bottle (San Benedetto S. p. A. Italy) in order to minimize the presence of potentially contaminating microorganisms. The cells were then fed daily for 5 days with CM inoculated with R. planticola to induce their exponential growth. These monoclonal cultures (both naturally infected and treated) are maintained since more than four years under laboratory conditions.
Identification of Paramecium Species
Paramecium species were identified using morphological characteristics (Fokin, 2010) and the cytochrome c oxidase 1 gene (COX1, Barth et al., 2006). Total DNA was extracted using a modified Chelex-based protocol as follows: approximately 100 paramecia cells of each culture were washed three times in sterile spring water and transferred in 100 μl spring water into 0.5 ml Eppendorf tubes. The tubes were stored for at least 20 min at −20°C to freeze completely. Later, 100 μl Chelex solution (Bio-Rad Laboratories, Inc., Hercules, CA, United States) was added and the samples were incubated for 20 min at 99°C. Immediately after incubation the tubes were put on ice. PCR products were obtained by using the forward primer M13 (5′-GTA AAA CGA CGG CCA G-3′, Strüder-Kypke and Lynn, 2010) for LgJac and degenerated forward primer F199dT-B (5′-TGT AAA ACG ACG GCC AGT TCA GGW GCT GCM TTA GCH ACY ATG-3′, Strüder-Kypke and Lynn, 2010) for YE9 as well as the degenerated reverse primer R1143dT (5′-CAG GAA ACA GCT ATG ACT ART ATA GGA TGM CCW CCA TAA GC-3′, Strüder-Kypke and Lynn, 2010) for both strains. Purification was performed with the NucleoSpin Gel and PCR Clean-up Kit (Macherey-Nagel GmbH & Co., KG, Düren NRW, Germany), and products were sequenced directly in both directions with the same primers used for amplification at Eurofins Genomics GmbH (Ebersberg, Germany).
In order to confirm their affiliation to the respective Paramecium species, the obtained sequences were compared to available Paramecium COX1 sequences. A Maximum likelihood (ML) tree was calculated with IQ−TREE (Nguyen et al., 2015) based on 34 sequences from 10 different Paramecium species comprising 620 characters. The alignment was trimmed to the length of the shortest sequence. The best-fit evolutionary model (Kalyaanamoorthy et al., 2017) according to Bayesian information criterion (BIC) is TPM2u+F+I+G4. Ultrafast Bootstrap support (BS) with 1000 pseudoreplicates (Hoang et al., 2018) was calculated by IQ-TREE.
Identification of “Ca. Megaira polyxenophila”
Prokaryotic SSU rRNA gene sequences were amplified by a touchdown PCR (Don et al., 1991) applying the following annealing temperatures: 58°C (30 s, 5 cycles), 54°C (30 s, 10 cycles), and 50°C (30 s, 25 cycles). For YE9, the bacterial primer combination Bac16SFor (5′-AAG AGT TTG ATC CTG GCT C-3′; modified from Neilan et al., 1997) and Bac16SRev (5′-TAC GGC TAC CTT GTT ACG AC-3′; Neilan et al., 1997) were used for both, PCR and direct sequencing from both sides. PCR on LgJac was performed with the Alphaproteobacteria specific forward primer 16S_F19b 5′-CCT GGC TCA GAA CGA ACG-3′ (Serra et al., 2016) and the Bacteria specific reverse primer 16S_R1522a 5′-GGA GGT GAT CCA GCC GCA-3′ (Serra et al., 2016) and sequenced using the internal primers 16S F343 ND 5′-TAC GGG AGG CAG CAG-3′, 16S R515 ND 5′-ACC GCG GCT GCT GGC AC-3′ and 16S F785 ND 5′-GGA TTA GAT ACC CTG GTA-3′ (Serra et al., 2016) at GATC Biotech AG (Konstanz, Germany).
A comparison with other 16S rRNA gene sequences of Ca. M. polyxenophila and Ca. Megaira venefica was performed. Therefore, we run a ML analysis with IQ−TREE based on an alignment (ARB program; Ludwig et al., 2004) of 43 sequences comprising 1343 characters, trimmed at both sides to the length of the shortest sequence. The applied best-fit evolutionary model is TIM3+F+G4 (according to BIC) and 1000 pseudoreplicates were performed for Ultrafast BS.
Fluorescence in situ Hybridization
The presence or absence of the endosymbionts was verified by performing fluorescence in situ hybridization (FISH) experiments with the universal probe EUB338 (5′-ACT CCT ACG GGA GGC AGC AG-3′) (Amann et al., 1990) in combination with the genus-specific probe Megenus_487 (5′-GCCGGGGCTTTTTCTGTTGGT-3′) detecting “Ca. Megaira” (Lanzoni et al., 2019). About 20 ciliate cells were collected, washed three times in water and fixed with 2% paraformaldehyde (PFA, final concentration) on slides as described by Szokoli et al. (2016). Hybridization and washing were carried out as described by Lanzoni et al. (2019). Images were obtained with a Leica DMR microscope, equipped with an HBO 50W/AC-L2 fluorescent lamp, a Leica DFC490 video camera, and Leica IM1000 Software (v.1.0).
Elimination of Endosymbionts via Antibiotic Treatment
The antibiotic treatment was performed in order to obtain genetically identical symbiont-free lines from the infected ones. Symbiont-free cells were obtained from both, P. pentaurelia YE9 and P. primaurelia LgJac, through antibiotic treatment (YE9AB and LgJacAB, respectively). Therefore, approximately 30 cells of the stock cultures were transferred to 500 μl of tetracycline (Carl Roth, Karlsruhe, Germany) solution (130 μg ml–1) and incubated for 24 h at 20°C. Later, the cells were washed four times. Individual cells were incubated again in tetracycline solution at 20°C for 24–48 h and then transferred to CM inoculated with R. planticola as food organism. After some rounds of cell division, single cells were treated again with tetracycline for 24 h and subsequently transferred into bacterized CM. The success of the antibiotic treatment was confirmed by FISH.
Fitness Experiment to Determine the Impact of the Endosymbiont on Host Growth at Different Salinity Conditions
In order to prime the cultures for the fitness experiment at different salinity conditions, Paramecium cells were adapted to the different salinity concentrations and fed daily for three days with bacterized CM to induce exponential growth (Supplementary Figure S1).
The cell number for each line was adjusted to approximately the same density (ca. 100 cells ml–1) with sterile CM. To set-up this experiment, a mix containing 8 ml of CM (in case of 0 ppt) or 8 ml of salinity stock solution (8 ppt for a final concentration of 2 ppt; 18 ppt for 4.5 ppt), 12 ml bacterized CM and 20 ml Paramecium culture (approximately 100 cells ml–1) were added, the total volume was set to 40 ml per line. The salinities were chosen to represent two different oligohaline brackish water conditions after a preliminary experiment to test the salinity tolerance of the two freshwater paramecia species (data not shown). This mix was then split into three experimental units for each line per salinity combination with 10 ml each (approximately 50 cells ml–1) (Supplementary Figure S1).
Each experimental unit was fed once a week with 3 ml of a 1:4 dilution of the respective salinity stock solution (sterile water, 8 or 18 ppt) and bacterized CM, to sustain the paramecia and to maintain a constant salinity concentration throughout the experiment (either 0, 2, or 4.5 ppt) for a period of 28 days.
Cell density was determined by counting under a stereomicroscope. The counting days were established as reported in Figure 1 in which T0 is the starting day of the salinity experiment. For each sample, three technical replicates each with a volume of 50–100 μl, depending on the cell number, were counted and the mean of cells per ml was calculated. Samples for cell density estimation were counted and then fixed to perform FISH as described above. These samples were collected twice per week for the duration of the experiment (28 days). Salt stock solutions were prepared by dissolving the corresponding amount of Tropic Marine Salts (Red Sea Salt Meersalz, Red Sea, Düsseldorf, Germany) in deionized water and pasteurizing for 5 min at 95°C.
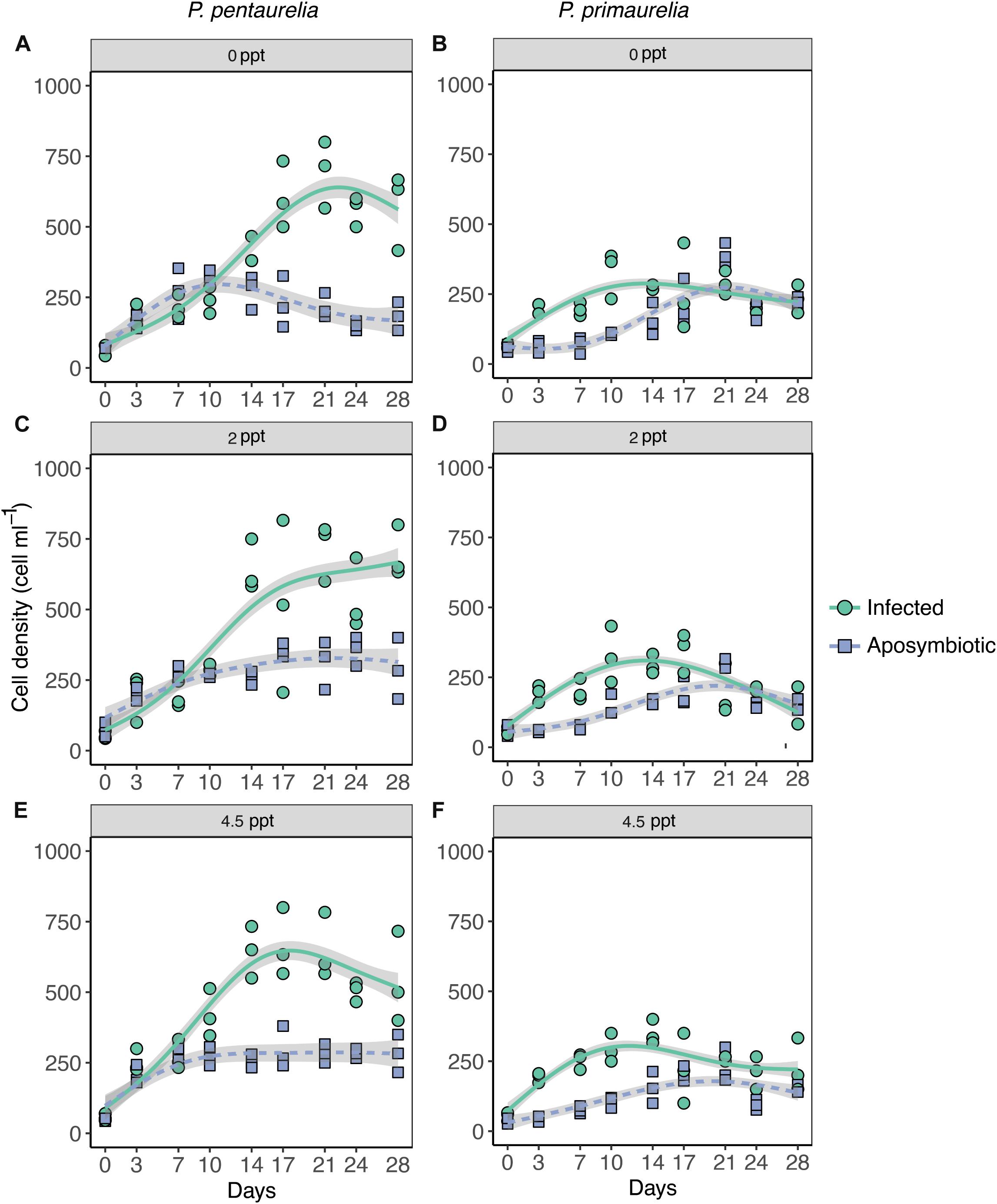
Figure 1. Growth of Paramecium lines either infected with Ca. Megaira polyxenophila or aposymbiotic at different salinity concentrations over a period of 28 days. Paramecium pentaurelia YE9 infected (green line) with Ca. Megaira polyxenophila in comparison with the respectively symbiont-free line YE9AB (purple line) at 0 ppt salinity (A), 2 ppt salinity (C), and 4.5 ppt salinity (E). Paramecium primaurelia LgJac infected (green line) with Ca. M. polyxenophila in comparison with the respectively symbiont-free (purple line) LgJacAB at 0 ppt salinity (B), 2 ppt salinity (D), and 4.5 ppt salinity (F). Data points represent three experimental units and the growth curve is represented by the mean of these three units.
Statistical Analysis
The influence of host species with different infectious status and in different salinity settings on Paramecium cell density was assessed by using a generalized additive mixed model (GAMM) with gaussian error (R, package: mgcv, version 1.7-28 (Wood, 2017). GAMMs allow assessing the influence of covariates on a response variable without specifying a priori fixed-function while accounting for non-independence of data due to successive measures in time. For this reason, time has been included both, in the fixed part, as smoothed term, and in the random part to model dependence. In particular, time has been smoothed independently for different combinations of host species, initial infection status and salinity. Fixed effect terms (infection status, salinity and host species) were modeled using a basis dimension of k = 4. The significance of the fixed terms and interactions has been obtained by using the anova.gam function of the mgcv package. The script and dataset are available as Supplementary Table S1, Supplementary Data Sheet S1. Model accuracy was estimated by the root-mean-square error (RMSE) performing a 10-fold cross-validation on the fixed part of the model using the CVgam function of the gamclass R package. The analysis was performed using R, version 3.6.0.
Results
Elimination of Ca. M. polyxenophila
Aposymbiotic cells were obtained from both, YE9 and LgJac, through antibiotic treatment producing respectively YE9AB and LgJacAB. A cell was considered successfully treated when no signal with the genus-specific probe Megenus_487 (Lanzoni et al., 2019) was observed from food vacuoles, cytoplasm, or nuclei. Successful elimination of Ca. M. polyxenophila was verified repeatedly by FISH (Figure 2). In none of the examined cells, Ca. M. polyxenophila was detected after the described tetracycline treatment. Other effects of the antibiotic treatment than symbiont elimination, e.g. delayed or abnormal cell division or population growth, were not observed.
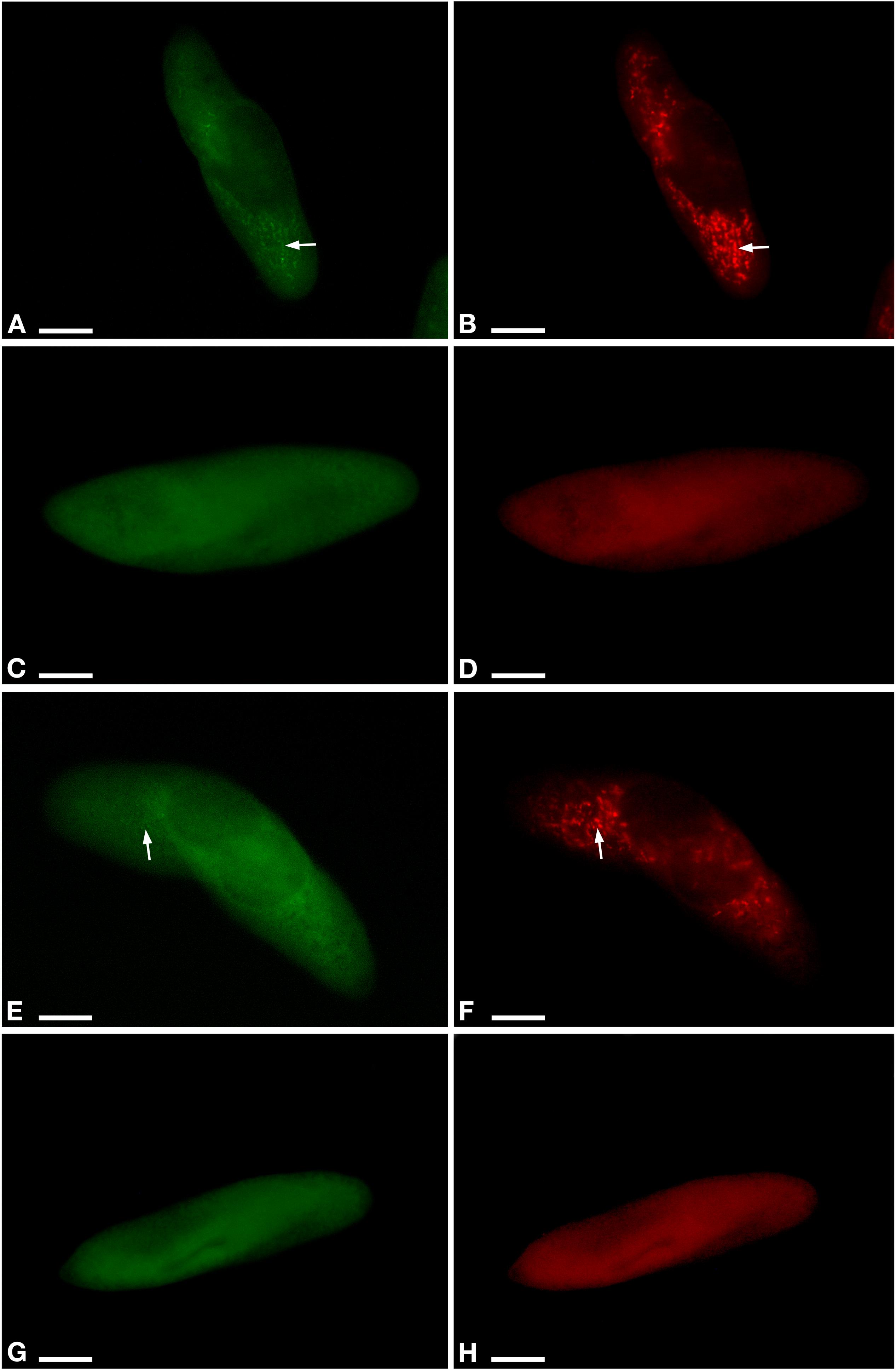
Figure 2. Fluorescent in situ hybridization for the visualization of Ca. M. polyxenophila in Paramecium pentaurelia YE9 (A,B) and Paramecium primaurelia LgJac (E,F). Fluorescent in situ hybridization for the two symbiont-free cell YE9AB (C,D) and LgJacAB (G,H) after antibiotic treatment using the universal bacterial probe EUB338 (A,C,E,G) and Megenus_487 specific for Ca. Megaira (B,D,F,H). Arrows indicate Ca. M. polyxenophila in the host’s cytoplasm. Scale bar = 10 μm.
Confirmation of Host and Endosymbiont Identities
Morphological analyses of the Paramecium strains from Bloomington and Rio de Janeiro revealed the strains belonging to the Paramecium aurelia-complex. This observation was confirmed by COX1 sequence analysis (Supplementary Figure S2). More precisely, LgJac affiliated with other P. primaurelia sequences whereas YE9 belonged to P. pentaurelia. Both affiliations received high BS support (98 and 99%, Supplementary Figure S2).
The endosymbiont’s 16S rRNA gene sequences affiliated with other sequences of Ca. M. polyxenophila with high BS support (97%, Supplementary Figure S3). The sequence similarity (data not shown) between the type strain of this species (AJ630204) and the here studied intracellular bacteria was 99.93% (LgJac) and 99.55% (YE9).
The obtained sequences are available from NCBI GenBank with following accession numbers: COX1 sequences MT362542 (LgJac) and MT362543 (YE9), 16S rRNA gene sequences MT351038 (LgJac) and MT351039 (YE9).
Fitness Impact of Ca. M. polyxenophila and Salinity on Paramecia
Time included in the model as the smoothed effect revealed a strongly significant effect on estimated degrees of freedom as expected for a nonlinear density growth (Supplementary Data Sheet S1). Among the three fixed factors, host species and infection status showed a highly significant effect (Table 2). Two interaction terms also showed a significant effect, the interaction between infection status and Paramecium species and the interaction between salinity and species (Table 2).
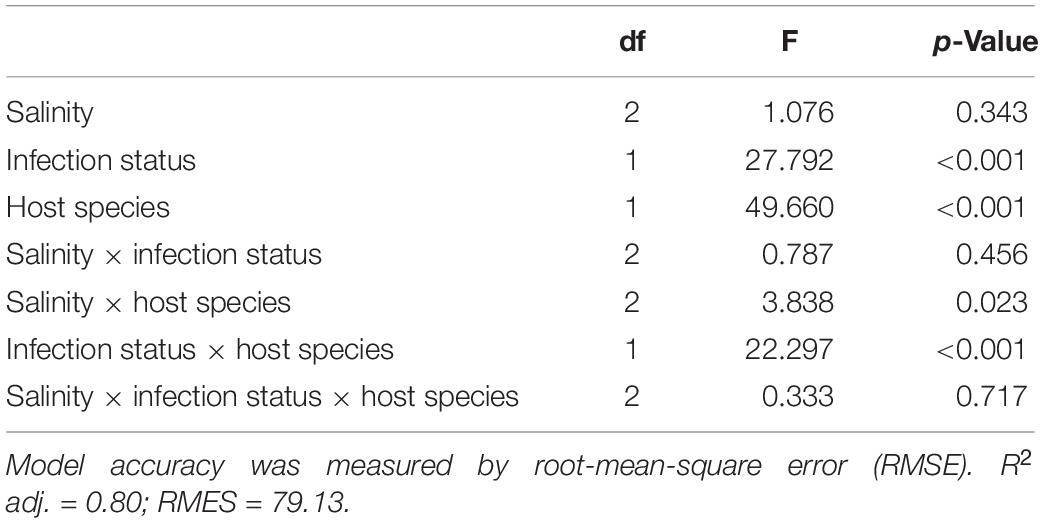
Table 2. ANOVA style-results of the generalized additive mixed models (GAMM) analysis on the salinity, infection, and species on cell density (cell ml–1).
According to the GAMM results which revealed a significant effect for host species, a substantial difference in cell densities between the two Paramecium species were observed as YE9 grew better in comparison with LgJac, reaching 800 and 400 cell ml–1, respectively (Figure 1). At the beginning of the experiment, the starting amount of cells was the same for each line (approximately 50 cells ml–1).
The inspection of growth curves and GAMM results indicated that both infected strains showed a generally higher cell density compared to their aposymbiotic counterparts. A strong significant interaction between Paramecium species and infection status was caused by the very strong difference in cell density of YE9 (between 600 and 700 cell ml–1 at the end of the experiment) compared to YE9AB (200–300 cell ml–1) which was not observed in LgJac (Figure 1). The weak significant interaction between salinity and species was due to the density decrease of LgJac and LgJacAB at the higher salinity levels, revealing its stronger salinity stress susceptibility compared to YE9 (Figure 1).
Both Paramecium species experienced growth advantages in presence of Ca. M. polyxenophila at every salinity condition (Figure 1). There was no obvious general effect of cultivation at oligohaline brackish conditions, neither on infected nor aposymbiotic lines.
Impact of Salinity on Ca. M. polyxenophila Prevalence
Infected and aposymbiotic paramecia cells of both species were observed by FISH at all sampling time points for all salinity concentrations. The aposymbiotic cells maintained their symbiont-free condition for the complete experiment duration at all tested salinities. FISH results showed that Ca. M. polyxenophila was present in YE9 and LgJac at 0 ppt with a prevalence of 100% and in a constant amount at all time points (Figure 3). For the initially Megaira-positive cells at 2 and 4.5 ppt, the 100% infection status was maintained until day 17 for both species and declined subsequently. Both species reached the lowest bacterial prevalence at the end of the experiment at 4.5 ppt (Figure 3).
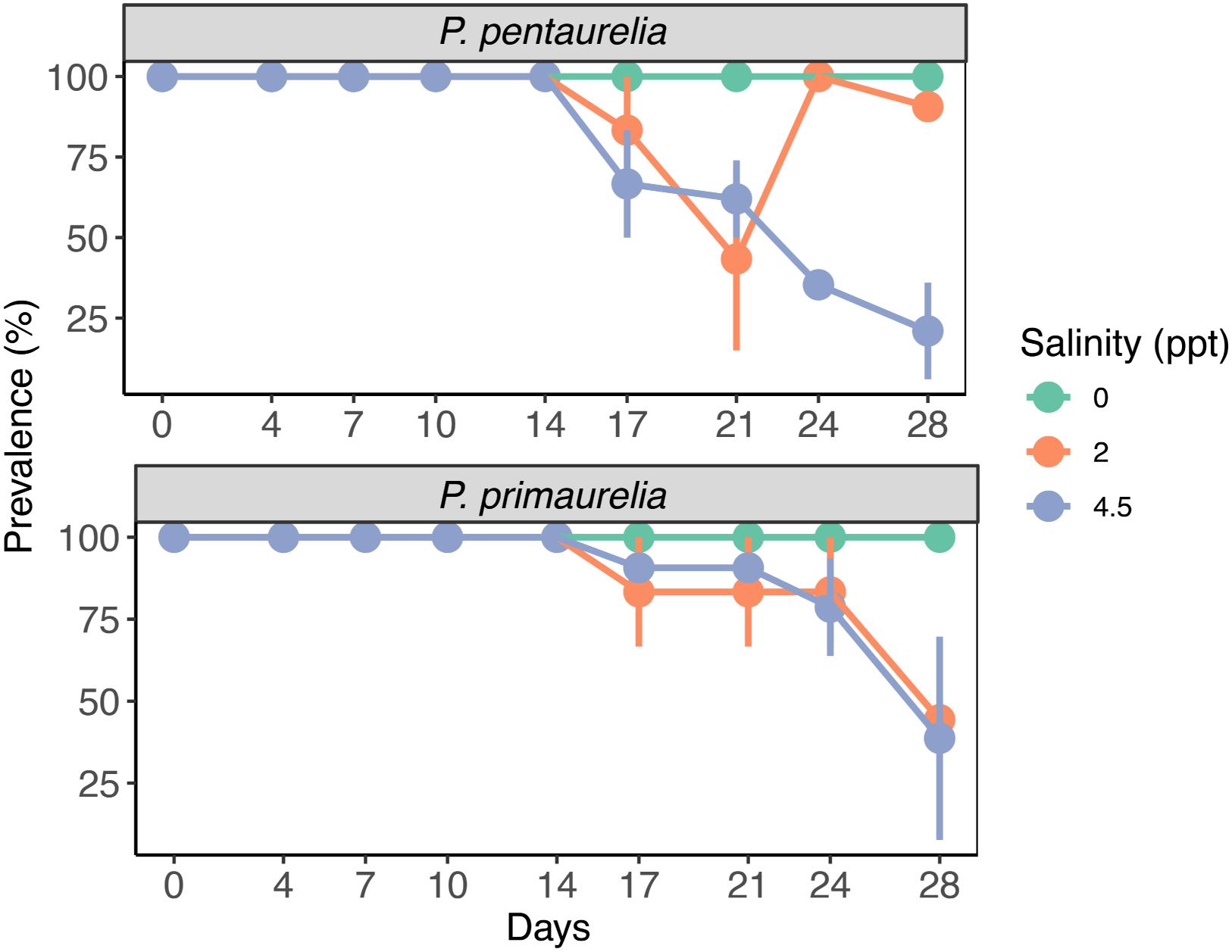
Figure 3. Bacterial prevalence (%) of infected species P. pentaurelia and P. primaurelia at three different salinity concentrations (0, 2, and 4.5 ppt) over 28 days. Error bars represent the standard error (n = 3).
While the percentage of P. pentaurelia YE9 cells with detected bacterial symbionts continuously declined after day 17 at 4.5 ppt (Figures 3, 4), at 2 ppt the strongest drop in prevalence was observed at day 21. The number of infected cells suddenly recovered and then decreased again. As our approach was to examine approximately 20 cells out of a 1 ml sample at each time point, this strong fluctuation might by chance represent an outlier with such a high number of cells which had spontaneously lost their symbionts.
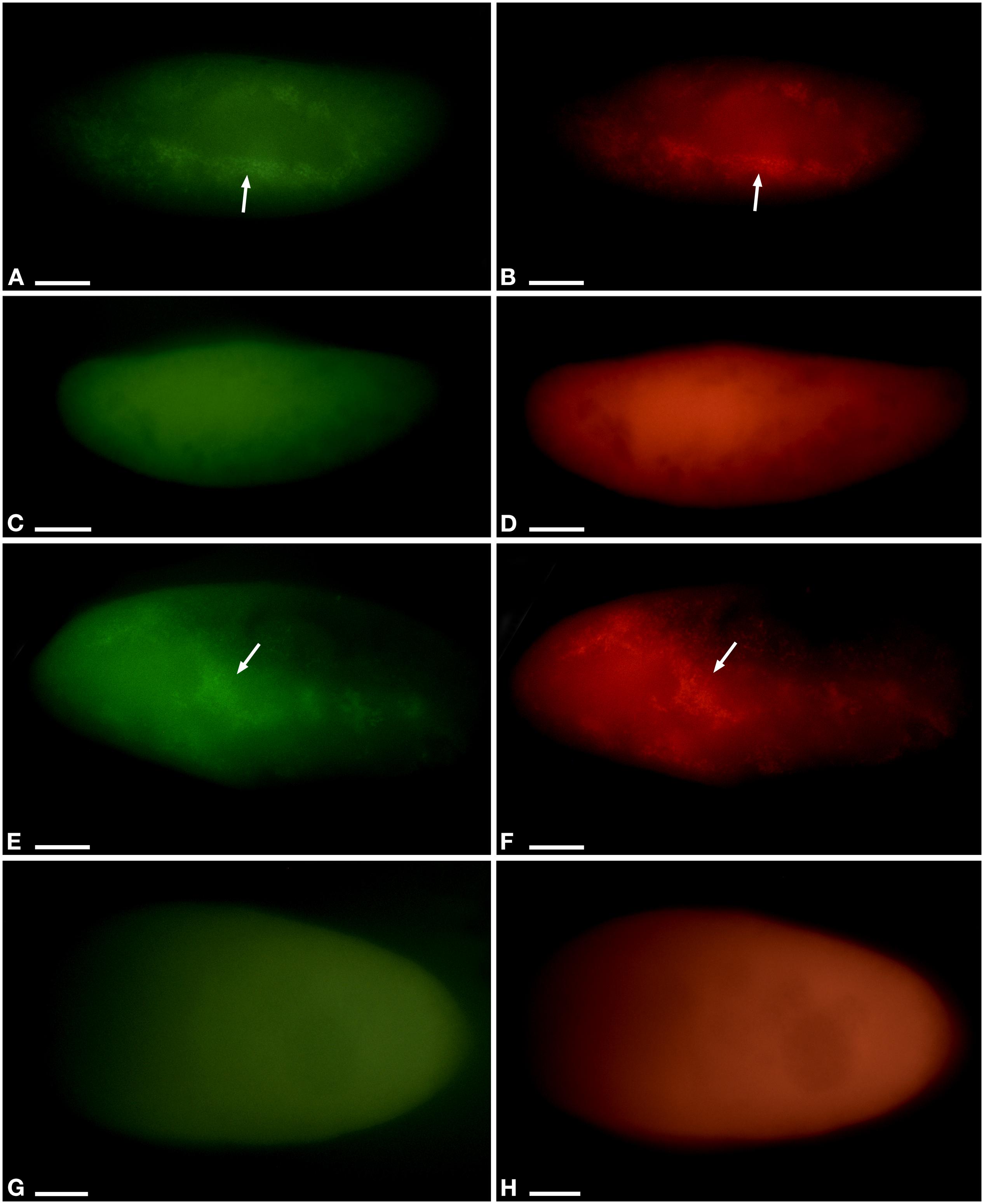
Figure 4. Fluorescent in situ hybridization of Paramecium pentaurelia YE9 (A–D) and Paramecium primaurelia LgJac (E–H). Examples for an infected cell (A,B,E,F) and one that had spontaneously lost Ca. Megaira polyxenophila (C,D,G,H) at 4.5 ppt 17 days after the beginning of the growth experiment, using the universal bacterial probe EUB338 specific for 90% of known bacteria (A,C,E,G) and Megenus_487 specific for Ca. Megaira (B,D,F,H). Arrows indicate Ca. M. polyxenophila in the host’s cytoplasm. Scale bar = 10 μm.
In case of P. primaurelia LgJac, the decrease of Megaira-positive cells was very similar at 2 and 4.5 ppt. At the end of the experiment, the infection prevalence dropped to approximately 40% (Figure 3).
Discussion
Our data suggest that the major effect of Ca. M. polyxenophila on Paramecium hosts is not harmful. While Rickettsiaceae usually are considered as obligate intracellular parasites (DeLong et al., 2014) at least in the case of Wolbachia a wide continuum of symbiotic interactions, ranging from mutualistic and essential for host development, fertility, and survival in filarial nematodes to facultative and parasitic in many arthropod species, has been observed (Landmann, 2019). Likely, the fitness effects of Ca. M. polyxenophila vary similarly between different hosts and different environmental conditions. Nevertheless, these intracellular bacteria are at least conditionally mutualistic in the two examined Paramecium species without providing essential nutritional benefits.
Both P. pentaurelia and P. primaurelia exhibit improved growth under all tested conditions in presence of the symbiont. Contrary to our expectations, the tested species showed no obvious fitness reduction by increasing salinity levels. This trend indicates that at least the here applied rather mild salt concentrations corresponding to the lower border of oligohaline brackish conditions might be below the physiological stress level of these paramecia. Like many unicellular organisms, Paramecium constantly encounter changing environmental conditions e.g., differences in water temperature between seasons, day and night, or even during a day according to the varying solar irradiation. Some fluctuations of environmental conditions remain within the physiological tolerance breadth (Krenek and Berendonk, 2009) whereas others reach stress levels. When paramecia are gradually adapted to stress conditions instead of being suddenly exposed, they can increase their resistance to this particular stressor (Smurov and Fokin, 1998; Tsukuda, 1989). As the cells examined in the fitness assay underwent an acclimatization phase prior to the experiment the applied salinity conditions remained within the organisms’ tolerance range.
Fokin and Sabaneyeva (1990) observed that ciliates living in brackish water apparently harbor more frequently bacterial infections than those in freshwater. We observed a decreasing prevalence of Ca. M. polyxenophila starting 2 weeks after the cells were exposed to increased salinity concentrations (2 and 4.5 ppt) while those at freshwater-like conditions remained to 100% infected until the end of the experiment. This is neither a confirmation nor a rejection of the hypothesis that brackish environments favor bacterial infections, but it demonstrates that elevated salt concentrations disrupt the symbiont maintenance in the Ca. M. polyxenophila-Paramecium aurelia system. Thus, we speculate that long-term exposure to mild environmental stress causes an accumulation of stress signals, for example, an accumulation of misfolded proteins or compatible solutes. While the cellular response of the host will have not necessarily immediate effects on growth, it might either impair bacterial cell division or accidentally cause their expulsion. Alternatively, the increased energy demand in order to adapt to prolonged stress exposure might result in a more directed elimination of the symbionts by lysosomal attack.
As P. pentaurelia YE9 and P. primaurelia LgJac both benefit from the presence of their symbionts, long-term exposure to brackish salinity conditions will have indirect negative effects on these paramecia.
The loss of Ca. M. polyxenophila in brackish conditions might indicate, despite the above mentioned description of this symbiont from marine and brackish host organisms, that they have an adaptation towards freshwater habitats and/or freshwater hosts. This speculation is supported by amplicon sequencing of environmental samples. More than 25% of the amplicon datasets associated with freshwater habitats screened by Lanzoni et al. (2019) revealed positive hits for Ca. M. polyxenophila, while the number of marine datasets with signatures of this symbiont was below 5%.
This study provides the first functional analysis of the frequent and promiscuous endosymbiont Ca. M. polyxenophila. So far, speculations about a possible opportunistic or parasitic lifestyle of these bacteria (Schrallhammer et al., 2013) are based only on the fact that strains of Ca. M. polyxenophila from phylogenetically and geographically distant hosts exhibit very high (99–100%) 16S rRNA gene sequence similarities. This has been considered as strong indication for horizontal transmission capabilities. So far, infection experiments with these bacteria have not been successful under laboratory conditions (Lanzoni et al., 2019). Functional studies regarding the interaction of this symbiont with its host are impaired by (i) its obligate intracellular life style, a trait Ca. M. polyxenophila has in common with other members of Rickettsiales (Castelli et al., 2016) (ii) the lack of successful protocols for experimental infection experiments, and so far (iii) for symbiont elimination to obtain genetically identical but aposymbiotic (host) organisms. A successful treatment has been established in this study. Similar approaches for generation of the aposymbiotic lines have been used in case of other endosymbionts of Paramecium (Kusch et al., 2002; Dusi et al., 2014; Bella et al., 2016; Grosser et al., 2018). The fact that members of Rickettsiaceae are naturally resistant to a variety of antibiotics (Rolain et al., 1998) did complicate the establishment of the present protocol. The aposymbiotic lines LgJacAB and YE9AB have been maintained under routine laboratory conditions since at least 5 years. While we cannot completely exclude the small chance that the antibiotic treatment affects Paramecium fitness or eliminated free-living bacteria contributing to the observed fitness effects and we would have preferred additional experiments such as infection of naïve cells, those are presently not feasible. Thus, we provide the very first insights into possible effects of Ca. M. polyxenophila on its host. As we observed differences in the impact of the endosymbiont in two very closely related host species, it is possible that in more divergent hosts an even broader spectrum of host-symbiont interactions will be observed. As Ca. M. polyxenophila infected hosts have been isolated from rather different environments (Lanzoni et al., 2019) the diversity of relevant abiotic factors might further entangle the analysis of the ecological role of this endosymbiont.
Data Availability Statement
All datasets generated for this study are included in the article/Supplementary Material.
Author Contributions
CP, FS, and MS designed the research. MS performed the molecular analysis. CP and FS performed the experiments. CP and LR performed the statistical analysis. CP, MS, GP, and LR interpreted the results. CP, FS, MS, and LR wrote the manuscript. All authors critically read and approved the manuscript.
Funding
This research was supported by the European Cooperation in Science and Technology BMBS COST Action BM1102, the European Community’s H2020 Programme H2020-MSCA-RISE 2019 under grant agreement 872767, and by the ERASMUS placement program. The article processing charge was funded by the German Research Foundation (DFG) and the University of Freiburg in the funding program Open Access Publishing.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors gratefully acknowledge Yana Eglit and Sascha Krenek for isolating and providing the strains used in this work, Fabrizio Erra for his assistance with fluorescence microscopy, Simone Gabrielli for help with the preparation of figures, and Gabriele Iannattoni for his support in the lab.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.01425/full#supplementary-material
FIGURE S1 | Schematic representation of the procedure followed for the fitness experiment.
FIGURE S2 | Species affiliation of LgJac and YE9 based on COX1 sequence analysis. Maximum likelihood tree of the genus Paramecium calculated with IQ−TREE based on an alignment of 34 sequences from 10 different Paramecium species comprising 620 characters. Here obtained sequences are reported in bold. Numbers near nodes indicate Ultrafast Bootstrap support values (below 75% not shown), numbers in brackets indicate sequences in collapsed groups. Scale bar corresponds to 0.1 sequence divergence.
FIGURE S3 | Molecular characterization of the endosymbionts of YE9 and LgJac based on 16S rRNA gene sequences. Maximum likelihood tree calculated with IQ−TREE based on 43 sequences belonging to members of the genera “Ca. Megaira” and Rickettsia comprising 1343 characters. Here obtained sequences are reported in bold. Numbers near nodes indicate Ultrafast Bootstrap support values (below 70% not shown). Scale bar corresponds to 0.01 sequence divergence. Ca. stands for Candidatus.
TABLE S1 | Data sheet for fitness analysis.
DATA SHEET S1 | R script.
References
Amann, R. I., Binder, B. J., Olson, R. J., Chisholm, S. W., Devereux, R., and Stahl, D. A. (1990). Combination of 16S rRNA-targeted oligonucleotide probes with flow cytometry for analyzing mixed microbial populations. Appl. Environ. Microbiol. 56, 1919–1925. doi: 10.1111/j.1469-8137.2004.01066.x
Arnaiz, O., Meyer, E., and Sperling, L. (2020). ParameciumDB 2019: integrating genomic data across the genus for functional and evolutionary biology. Nucleic Acids Res. 48, D599–D605.
Banerji, A., Duncan, A. B., Griffin, J. S., Humphries, S., Petchey, O. L., and Kaltz, O. (2015). Density-and trait-mediated effects of a parasite and a predator in a tri-trophic food web. J. Anim. Ecol. 84, 723–733. doi: 10.1111/1365-2656.12317
Barth, D., Krenek, S., Fokin, S. I., and Berendonk, T. U. (2006). Intraspecific genetic variation in Paramecium revealed by mitochondrial cytochrome c oxidase I sequences. J. Eukaryot. Microbiol. 53, 20–25. doi: 10.1111/j.1550-7408.2005.00068.x
Bella, C., Koehler, L., Grosser, K., Berendonk, T. U., Petroni, G., and Schrallhammer, M. (2016). Fitness impact of obligate intranuclear bacterial symbionts depends on host growth phase. Front. Microbiol. 7:2084. doi: 10.3389/fmicb.2016.02084
Castelli, M., Lanzoni, O., Fokin, S. I., Schrallhammer, M., and Petroni, G. (2015). Response of the bacterial symbiont Holospora caryophila to different growth conditions of its host. Eur. J. Protistol. 51, 98–108. doi: 10.1016/j.ejop.2014.11.006
Castelli, M., Sabaneyeva, E., Lanzoni, O., Lebedeva, N., Floriano, A. M., Gaiarsa, S., et al. (2019a). Deianiraea, an extracellular bacterium associated with the ciliate Paramecium, suggests an alternative scenario for the evolution of Rickettsiales. ISME J. 13, 2280–2294. doi: 10.1038/s41396-019-0433-9
Castelli, M., Serra, V., Senra, M. V., Basuri, C. K., Soares, C. A., Fokin, S. I., et al. (2019b). The hidden world of Rickettsiales symbionts:“Candidatus Spectririckettsia obscura,” a novel bacterium found in Brazilian and Indian Paramecium caudatum. Microb. Ecol. 77, 748–758. doi: 10.1007/s00248-018-1243-8
Castelli, M., Sassera, D., and Petroni, G. (2016). “Biodiversity of “non-model” Rickettsiales and their association with aquatic organisms,” in Rickettsiales, ed. S. Thomas (Cham: Springer), 59–91. doi: 10.1007/978-3-319-46859-4_3
Chiellini, C., Pasqualetti, C., Lanzoni, O., Fagorzi, C., Fani, R., Petroni, G., et al. (2019). Harmful effect of Rheinheimera sp. EpRS3 (Gammaproteobacteria) against the protist Euplotes aediculatus (Ciliophora, Spirotrichea): insights into the ecological role of antimicrobial compounds from environmental bacterial strains. Front. Microbiol. 10:510. doi: 10.3389/fmicb.2016.0510
DeLong, E. F., Lory, S., Stackebrandt, E., and Thompson, F. (2014). The Prokaryotes: Alphaproteobacteria And Betaproteobacteria. Berlin: Springer.
Don, R., Cox, P., Wainwright, B., Baker, K., and Mattick, J. (1991). ‘Touchdown’PCR to circumvent spurious priming during gene amplification. Nucleic Acids Res. 19:4008. doi: 10.1093/nar/19.14.4008
Dumler, J. S., Barbet, A. F., Bekker, C., Dasch, G. A., Palmer, G. H., Ray, S. C., et al. (2001). Reorganization of genera in the families Rickettsiaceae and Anaplasmataceae in the order Rickettsiales: unification of some species of Ehrlichia with Anaplasma, Cowdria with Ehrlichia and Ehrlichia with Neorickettsia, descriptions of six new species combinations and designation of Ehrlichia equi and ‘HGE agent’ as subjective synonyms of Ehrlichia phagocytophila. Int. J. Syst. Evol. Microbiol. 51, 2145–2165. doi: 10.1099/00207713-51-6-2145
Duncan, A. B., Dusi, E., Schrallhammer, M., Berendonk, T., and Kaltz, O. (2018). Population-level dynamics in experimental mixed infections: evidence for competitive exclusion among bacterial parasites of Paramecium caudatum. Oikos 127, 1380–1389. doi: 10.1111/oik.05280
Duncan, A. B., Fellous, S., Accot, R., Alart, M., Sobandi, K. C., Cosiaux, A., et al. (2010). Parasite-mediated protection against osmotic stress for Paramecium caudatum infected by Holospora undulata is host genotype specific. FEMS Microbiol. Ecol. 74, 353–360. doi: 10.1111/j.1574-6941.2010.00952.x
Duncan, A. B., Gonzalez, A., and Kaltz, O. (2013). Stochastic environmental fluctuations drive epidemiology in experimental host-parasite metapopulations. Proc. R. Soc. B Biol. Sci. 280, 20131747. doi: 10.1098/rspb.2013.1747
Dusi, E., Gougat-Barbera, C., Berendonk, T. U., and Kaltz, O. (2015). Long-term selection experiment produces breakdown of horizontal transmissibility in parasite with mixed transmission mode. Evolution 69, 1069–1076. doi: 10.1111/evo.12638
Dusi, E., Krenek, S., Schrallhammer, M., Sachse, R., Rauch, G., Kaltz, O., et al. (2014). Vertically transmitted symbiont reduces host fitness along temperature gradient. J. Evol. Biol. 27, 796–800. doi: 10.1111/jeb.12336
Floriano, A. M., Castelli, M., Krenek, S., Berendonk, T. U., Bazzocchi, C., Petroni, G., et al. (2018). The genome sequence of “Candidatus Fokinia solitaria”: insights on reductive evolution in Rickettsiales. Genome Biol. Evol. 10, 1120–1126. doi: 10.1093/gbe/evy072
Fokin, S., and Sabaneyeva, E. (1990). Paramecium (Ciliophora, Peniculina) from water with low salinity of the Barents Sea and White Sea coasts and their endobionts. Presented at the Ecology, Reproduction And Guarding Of The Bioresources Of the North Europe seas. Abstr. of the III Conference, Murmansk.
Fokin, S. I. (2010). Paramecium genus: biodiversity, some morphological features and the key to the main morphospecies discrimination. Protistology 6, 227–235.
Fokin, S. I., and Görtz, H.-D. (2009). “Diversity of Holospora bacteria in Paramecium and their characterization,” in Endosymbionts in Paramecium, Springer, 161–199.
Fokin, S. I., Serra, V., Ferrantini, F., Modeo, L., and Petroni, G. (2019). Candidatus Hafkinia simulans” gen. nov., sp. nov., a novel holospora-like bacterium from the macronucleus of the rare brackish water ciliate Frontonia salmastra (Oligohymenophorea, Ciliophora): multidisciplinary characterization of the new endosymbiont and its host. Microb. Ecol. 77, 1092–1106. doi: 10.1007/s00248-018-1311-0
Fraune, S., and Bosch, T. C. (2007). Long-term maintenance of species-specific bacterial microbiota in the basal metazoan Hydra. Proc. Natl. Acad. Sci. U.S.A. 104, 13146–13151. doi: 10.1073/pnas.0703375104
Fujishima, M., Kawai, M., and Yamamoto, R. (2005). Paramecium caudatum acquires heat-shock resistance in ciliary movement by infection with the endonuclear symbiotic bacterium Holospora obtusa. FEMS Microbiol. Lett. 243, 101–105. doi: 10.1016/j.femsle.2004.11.053
Garushyants, S. K., Beliavskaia, A. Y., Malko, D. B., Logacheva, M. D., Rautian, M. S., and Gelfand, M. S. (2018). Comparative genomic analysis of Holospora spp., intranuclear symbionts of paramecia. Front. Microbiol. 9:738. doi: 10.3389/fmicb.2016.0738
Grosser, K., Ramasamy, P., Amirabad, A. D., Schulz, M. H., Gasparoni, G., Simon, M., et al. (2018). More than the “killer trait”: infection with the bacterial endosymbiont Caedibacter taeniospiralis causes transcriptomic modulation in Paramecium host. Genome Biol. Evol. 10, 646–656. doi: 10.1093/gbe/evy024
Hess, S. (2017). Description of Hyalodiscus flabellus sp. nov. (Vampyrellida, Rhizaria) and identification of its bacterial endosymbiont,“Candidatus Megaira polyxenophila”(Rickettsiales, Alphaproteobacteria). Protist 168, 109–133. doi: 10.1016/j.protis.2016.11.003
Hoang, D. T., Chernomor, O., Von Haeseler, A., Minh, B. Q., and Vinh, L. S. (2018). UFBoot2: improving the ultrafast bootstrap approximation. Mol. Biol. Evol. 35, 518–522. doi: 10.1093/molbev/msx281
Hollants, J., Leliaert, F., Verbruggen, H., Willems, A., and De Clerck, O. (2013). Permanent residents or temporary lodgers: characterizing intracellular bacterial communities in the siphonous green alga Bryopsis. Proc. R. Soc. B Biol. Sci. 280:20122659. doi: 10.1098/rspb.2012.2659
Hori, M., Fujii, K., and Fujishima, M. (2008). Micronucleus-specific bacterium Holospora elegans irreversibly enhances stress gene expression of the host Paramecium caudatum. J. Eukaryot. Microbiol. 55, 515–521. doi: 10.1111/j.1550-7408.2008.00352.x
Hori, M., and Fujishima, M. (2003). The endosymbiotie bacterium Holospora obtusa enhances heat-shock gene expression of the host Paramecium caudatum. J. Eukaryot. Microbiol. 50, 293–298. doi: 10.1111/j.1550-7408.2003.tb00137.x
Kalyaanamoorthy, S., Minh, B. Q., Wong, T. K., von Haeseler, A., and Jermiin, L. S. (2017). ModelFinder: fast model selection for accurate phylogenetic estimates. Nat. Methods 14:587. doi: 10.1038/nmeth.4285
Karunanithi, S., Oruganti, V., Marker, S., Rodriguez-Viana, A. M., Drews, F., Pirritano, M., et al. (2019). Exogenous RNAi mechanisms contribute to transcriptome adaptation by phased siRNA clusters in Paramecium. Nucleic Acids Res. 47, 8036–8049. doi: 10.1093/nar/gkz553
Kawafune, K., Hongoh, Y., Hamaji, T., Sakamoto, T., Kurata, T., Hirooka, S., et al. (2015). Two different rickettsial bacteria invading Volvox carteri. PLoS One 10:e0116192. doi: 10.1371/journal.pone.0116192
Kelz, M. B., and Mashour, G. A. (2019). The biology of general anesthesia from Paramecium to primate. Curr. Biol. 29, R1199–R1210.
Koehler, L., Flemming, F. E., and Schrallhammer, M. (2019). Towards an ecological understanding of the killer trait-A reproducible protocol for testing its impact on freshwater ciliates. Eur. J. Protistol. 68, 108–120. doi: 10.1016/j.ejop.2019.02.002
Krenek, S., and Berendonk, T. U. (2009). A Long-term conservation tool for cell characteristics: cryopreservation of Paramecium caudatum. Protist 160, 355–363. doi: 10.1016/j.protis.2009.03.002
Kusch, J., Czubatinski, L., Nachname, S., Hübner, M., Alter, M., and Albrecht, P. (2002). Competitive advantages of Caedibacter-infected paramecia. Protist 153, 47–58. doi: 10.1078/1434-4610-00082
Landmann, F. (2019). The Wolbachia endosymbionts. Bact. Intracell. 72:139. doi: 10.1128/9781683670261.ch10
Lanzoni, O., Sabaneyeva, E., Modeo, L., Castelli, M., Lebedeva, N., Verni, F., et al. (2019). Diversity and environmental distribution of the cosmopolitan endosymbiont Candidatus Megaira. Sci. Rep. 9:1179.
Lohse, K., Gutierrez, A., and Kaltz, O. (2006). Experimental evolution of resistance in Paramecium caudatum against the bacterial parasite Holospora undulata. Evolution 60, 1177–1186. doi: 10.1111/j.0014-3820.2006.tb01196.x
Ludwig, W., Strunk, O., Westram, R., Richter, L., Meier, H., Yadhukumar, B., et al. (2004). ARB: a software environment for sequence data. Nucleic Acids Res. 32, 1363–1371. doi: 10.1093/nar/gkh293
Mayne, R., Morgan, J., Whiting, J. G., Phillips, N., and Adamatzky, A. (2019). On measuring nanoparticle toxicity and clearance with Paramecium caudatum. Sci. Rep. 9, 1–9.
Neilan, B. A., Jacobs, D., Blackall, L. L., Hawkins, P. R., Cox, P. T., and Goodman, A. E. (1997). rRNA sequences and evolutionary relationships among toxic and nontoxic cyanobacteria of the genus Microcystis. Int. J. Syst. Evol. Microbiol. 47, 693–697. doi: 10.1099/00207713-47-3-693
Nguyen, L.-T., Schmidt, H. A., Von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: a fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nidelet, T., Koella, J. C., and Kaltz, O. (2009). Effects of shortened host life span on the evolution of parasite life history and virulence in a microbial host-parasite system. BMC Evol. Biol. 9:65. doi: 10.1186/1471-2148-9-65
Plotnikov, A. O., Balkin, A. S., Gogoleva, N. E., Lanzoni, O., Khlopko, Y. A., Cherkasov, S. V., et al. (2019). High-throughput sequencing of the 16S rRNA gene as a survey to analyze the microbiomes of free-living ciliates Paramecium. Microb. Ecol. 78, 286–298. doi: 10.1007/s00248-019-01321-x
Potekhin, A., Schweikert, M., Nekrasova, I., Vitali, V., Schwarzer, S., Anikina, A., et al. (2018). Complex life cycle, broad host range and adaptation strategy of the intranuclear Paramecium symbiont Preeria caryophila comb. nov. FEMS Microbiol. Ecol. 94:fiy076.
Rolain, J., Maurin, M., Vestris, G., and Raoult, D. (1998). In vitro susceptibilities of 27 rickettsiae to 13 antimicrobials. Antimicrob. Agents Chemother. 42, 1537–1541. doi: 10.1128/aac.42.7.1537
Sabaneyeva, E., Castelli, M., Szokoli, F., Benken, K., Lebedeva, N., Salvetti, A., et al. (2018). Host and symbiont intraspecific variability: the case of Paramecium calkinsi and Candidatus Trichorickettsia mobilis. Eur. J. Protistol. 62, 79–94. doi: 10.1016/j.ejop.2017.12.002
Schrallhammer, M., Castelli, M., and Petroni, G. (2018). Phylogenetic relationships among endosymbiotic R-body producer: bacteria providing their host the killer trait. Syst. Appl. Microbiol. 41, 213–220. doi: 10.1016/j.syapm.2018.01.005
Schrallhammer, M., Ferrantini, F., Vannini, C., Galati, S., Schweikert, M., Görtz, H.-D., et al. (2013). ‘Candidatus Megaira polyxenophila’gen. nov., sp. nov.: considerations on evolutionary history, host range and shift of early divergent rickettsiae. PLoS One 8:e72581. doi: 10.1371/journal.pone.072581
Schu, M. G., and Schrallhammer, M. (2018). Cultivation conditions can cause a shift from mutualistic to parasitic behavior in the symbiosis between Paramecium and its bacterial symbiont Caedibacter taeniospiralis. Curr. Microbiol. 75, 1099–1102. doi: 10.1007/s00284-018-1493-1
Serra, V., Fokin, S. I., Castelli, M., Basuri, C. K., Nitla, V., Verni, F., et al. (2016). “Candidatus Gortzia shahrazadis”, a novel endosymbiont of Paramecium multimicronucleatum and a revision of the biogeographical distribution of Holospora-like bacteria. Front. Microbiol. 7:1704. doi: 10.3389/fmicb.2016.01704
Smurov, A., and Fokin, S. (1998). Resistance of Paramecium caudatum infected with endonuclear bacteria Holospora against salinity impact. Proc. Zool. RAS 276, 175–178.
Soares, H., Carmona, B., Nolasco, S., and Melo, L. V. (2019). Polarity in ciliate models: from cilia to cell architecture. Front. Cell Dev. Biol. 7:240. doi: 10.3389/fmicb.2016.240
Strüder-Kypke, M. C., and Lynn, D. H. (2010). Comparative analysis of the mitochondrial cytochrome c oxidase subunit I (COI) gene in ciliates (Alveolata, Ciliophora) and evaluation of its suitability as a biodiversity marker. Syst. Biodivers. 8, 131–148. doi: 10.1080/14772000903507744
Sun, H., Noe, J., Barber, J., Coyne, R., Cassidy-Hanley, D., Clark, T., et al. (2009). Endosymbiotic bacteria in the parasitic ciliate Ichthyophthirius multifiliis. Appl. Environ. Microbiol. 75, 7445–7452. doi: 10.1128/aem.00850-09
Sunagawa, S., DeSantis, T. Z., Piceno, Y. M., Brodie, E. L., DeSalvo, M. K., Voolstra, C. R., et al. (2009). Bacterial diversity and white plague disease-associated community changes in the Caribbean coral Montastraea faveolata. ISME J. 3:512. doi: 10.1038/ismej.2008.131
Szokoli, F., Castelli, M., Sabaneyeva, E., Schrallhammer, M., Krenek, S., Doak, T. G., et al. (2016). Disentangling the taxonomy of Rickettsiales and description of two novel symbionts (“Candidatus Bealeia paramacronuclearis” and “Candidatus Fokinia cryptica”) sharing the cytoplasm of the ciliate protist Paramecium biaurelia. Appl. Environ. Microbiol. 82, 7236–7247. doi: 10.1128/aem.02284-16
Tsukuda, H. (1989). Heat and osmotic tolerance in Paramecium caudatum, acclimated to different temperatures and salinities. Compar. Biochem. Physiol. Part A Physiol. 94, 333–337. doi: 10.1016/0300-9629(89)90555-0
Vannini, C., Petroni, G., Verni, F., and Rosati, G. (2005). A bacterium belonging to the Rickettsiaceae family inhabits the cytoplasm of the marine ciliate Diophrys appendiculata (Ciliophora, Hypotrichia). Microb. Ecol. 49, 434–442. doi: 10.1007/s00248-004-0055-1
Yang, A., Narechania, A., and Kim, E. (2016). Rickettsial endosymbiont in the “early-diverging” streptophyte green alga Mesostigma viride. J. Phycol. 52, 219–229. doi: 10.1111/jpy.12385
Keywords: brackish, Paramecium, stress response, symbiosis, osmolarity, GAMMs
Citation: Pasqualetti C, Szokoli F, Rindi L, Petroni G and Schrallhammer M (2020) The Obligate Symbiont “Candidatus Megaira polyxenophila” Has Variable Effects on the Growth of Different Host Species. Front. Microbiol. 11:1425. doi: 10.3389/fmicb.2020.01425
Received: 15 January 2020; Accepted: 02 June 2020;
Published: 08 July 2020.
Edited by:
Martin Kaltenpoth, Johannes Gutenberg University Mainz, GermanyReviewed by:
Harald Ronald Gruber-Vodicka, Max Planck Institute for Marine Microbiology (MPG), GermanySimon M. Dittami, UMR 8227 Laboratoire de Biologie Intégrative des Modèles Marins, France
Copyright © 2020 Pasqualetti, Szokoli, Rindi, Petroni and Schrallhammer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chiara Pasqualetti, Y2hpYXJhcGFzcXVhbGV0dGlAbGliZXJvLml0
 Chiara Pasqualetti
Chiara Pasqualetti Franziska Szokoli1,3
Franziska Szokoli1,3 Giulio Petroni
Giulio Petroni Martina Schrallhammer
Martina Schrallhammer