- Department of Chemistry, Center for Synthetic Microbiology (SYNMIKRO), Philipps-University Marburg, Marburg, Germany
When bacteria experience growth-limiting environmental conditions, the synthesis of the hyperphosphorylated guanosine derivatives (p)ppGpp is induced by enzymes of the RelA/SpoT homology (RSH)-type protein family. High levels of (p)ppGpp induce a process called “stringent response”, a major cellular reprogramming during which ribosomal RNA (rRNA) and transfer RNA (tRNA) synthesis is downregulated, stress-related genes upregulated, messenger RNA (mRNA) stability and translation altered, and allocation of scarce resources optimized. The (p)ppGpp-mediated stringent response is thus often regarded as an all-or-nothing paradigm induced by stress. Over the past decades, several binding partners of (p)ppGpp have been uncovered displaying dissociation constants from below one micromolar to more than one millimolar and thus coincide with the accepted intracellular concentrations of (p)ppGpp under non-stringent (basal levels) and stringent conditions. This suggests that the ability of (p)ppGpp to modulate target proteins or processes would be better characterized as an unceasing continuum over a concentration range instead of being an abrupt switch of biochemical processes under specific conditions. We analyzed the reported binding affinities of (p)ppGpp targets and depicted a scheme for prioritization of modulation by (p)ppGpp. In this ranking, many enzymes of e.g., nucleotide metabolism are among the first targets to be affected by rising (p)ppGpp while more fundamental processes such as DNA replication are among the last. This preference should be part of (p)ppGpp’s “magic” in the adaptation of microorganisms while still maintaining their potential for outgrowth once a stressful condition is overcome.
The Metabolism of (p)ppGpp
Bacteria must adapt to fluctuations in their ever-changing surroundings to survive. In order to accomplish optimal resource allocation upon facing environmental shifts, the pathways through which microorganisms rewire their metabolism need to be finely and promptly tuned. An efficient control of metabolism senses stress, adjusts growth accordingly, mandates which genes – and to which extent – are expressed, and ultimately provides a fitness advantage over poorly-adapted microbial competitors. The second messenger molecules (p)ppGpp are compounds that can accomplish all this.
More than 50 years ago, the “magic spots”, which were identified as guanosine 5'-diphosphate 3'-diphosphate (ppGpp) and guanosine 5'-triphosphate 3'-diphosphate (pppGpp) and collectively referred to as (p)ppGpp or “alarmones”, were discovered by Cashel and Gallant (1969). Since then, the synthesis and degradation of (p)ppGpp as well as the (p)ppGpp-mediated response to nutrient limitations, a phenomenon known as the “stringent response”, have been subject of extensive studies. Central in the metabolism of (p)ppGpp are members of the RelA/SpoT homology (RSH)-type protein family (Atkinson et al., 2011). Briefly, when bacteria encounter nutrient-limiting conditions, RSH proteins utilize ATP as a donor substrate and, through transfer of its β‐ and γ-phosphates onto the 3'-hydroxy group of the acceptor substrate guanosine 5'-diphosphate (GDP) or guanosine 5'-triphosphate (GTP), generate ppGpp or pppGpp, respectively (Sy and Lipmann, 1973). RSH proteins also degrade (p)ppGpp through removal of the 3'-pyrophosphate moiety of (p)ppGpp, thereby regenerating GDP/GTP (Hogg et al., 2004). “Long” RSH proteins harbor (p)ppGpp hydrolase and synthetase domains whose reciprocal activities are controlled by further regulatory domains (Atkinson et al., 2011). “Short” RSH proteins consisting only of a (p)ppGpp synthetase or hydrolase domain constitute the class of small alarmone synthetases (SAS) and hydrolases (SAH), respectively (Atkinson et al., 2011). Enzymes of the GppA/PPX family are able to convert pppGpp to ppGpp opening the avenue for more intricate differential regulation by the two alarmone species (Keasling et al., 1993; Kuroda et al., 1997; Kristensen et al., 2008).
The activity of RSH proteins is subject to regulation by various mechanisms, and (p)ppGpp-inducing conditions include, e.g., the presence of stalled ribosomes (Rel/RelA; Haseltine and Block, 1973; Wendrich et al., 2002), fatty acid starvation, and carbon downshifts (SpoT; Battesti and Bouveret, 2006, 2009), enhanced transcription of (p)ppGpp synthetases through cell wall stress stimuli (SAS2/RelP; Zweers et al., 2012; Geiger et al., 2014) or allosteric stimulation through the alarmone pppGpp itself (SAS1/RelQ; Gaca et al., 2015; Steinchen et al., 2015). The intracellular concentrations of (p)ppGpp during growth of Escherichia coli amount approximately 10–40 μM during logarithmic growth – unfortunately those basal levels are still not robustly determined because they typically fall beneath or close to the limit of quantification as in two recent studies (Varik et al., 2017; Zbornikova et al., 2019) – and peak at 800 μM at the onset of stationary phase. Full induction of the stringent response during acute amino acid starvation [e.g., induced by a transfer RNA (tRNA) synthetase inhibitor] gives rise to an increase of the intracellular (p)ppGpp concentrations to approximately 1 mM (Haseltine and Block, 1973; Kuroda et al., 1997; Kriel et al., 2012; Varik et al., 2017), ultimately causing growth arrest (Schreiber et al., 1991; Svitil et al., 1993; Potrykus and Cashel, 2008).
A Priority Program of (p)ppGpp Shutdown Is Advised by Its Binding Affinities
Substantial progress has been made in the identification and characterization of (p)ppGpp binding targets (reviewed in: Kanjee et al., 2012; Steinchen and Bange, 2016; recent original works: Corrigan et al., 2016; Zhang et al., 2018; Wang et al., 2019), which fall into different cellular processes such as DNA replication, transcription, translation, ribosome biogenesis, or nucleotide metabolism (Srivatsan et al., 2008; Liu et al., 2015a; Corrigan et al., 2016; Bennison et al., 2019). Firstly, these studies provide insights as to how (p)ppGpp affect virulence, pathogenicity, persister cell and biofilm formation, heat shock response, and cell growth (Dalebroux and Swanson, 2012; He et al., 2012; Bager et al., 2016; Strugeon et al., 2016; Schafer et al., 2020). Secondly, they supply a wealth of biochemical data that quantitatively describe (p)ppGpp/protein interactions.
We were wondering whether any prioritization in the order of regulation between those targets/processes would exist. We collected and analyzed binding affinities (exemplified by the dissociation constants, Kd, of the (p)ppGpp/protein complexes), inhibitory constants (Ki) and EC50/IC50 values for the targets of (p)ppGpp and depicted a scheme of prioritization in (p)ppGpp modulation (Figure 1 and Supplementary Table S1). Proteins involved in the metabolism of amines and amino acids exhibit very high-affine (p)ppGpp binding with Kd’s as low as 0.01 μM (E. coli LdcI, Kanjee et al., 2011a,b). They are followed by a multitude of enzymes involved in the metabolism of nucleotides featuring Kd values below 10 μM (Zhang et al., 2018; Wang et al., 2019), the tRNA modification GTPase TrmE and riboswitches of the YkkC type 2a (Peselis and Serganov, 2018; Sherlock et al., 2018). Notably, their Kd values below the assumed basal levels of (p)ppGpp imply a certain degree of regulation by (p)ppGpp even under non-stringent conditions (see below). (p)ppGpp binding to targets involved in transcription [i.e., E. coli RNA polymerase (RNAP); Bhardwaj et al., 2018] and Francisella tularensis MglA-SspA (Cuthbert et al., 2017) proceeds between 2 and 25 μM. It shall be noted that (p)ppGpp elicit control over E. coli RNAP through modulation of relative transcription rates from different promoters instead of a general reduction or induction of RNAP activity (Gourse et al., 2018). These targets are succeeded by further enzymes of nucleotide metabolism with Kd values ranging between 30 and 80 μM. Except for GpmA exhibiting a Kd of 52 μM, proteins associated with carbon metabolism feature dissociation constants/IC50 values of 132–800 μM (Dietzler and Leckie, 1977; Taguchi et al., 1977; Fujita and Freese, 1979; Wang et al., 2019). In a similar range falls the inhibition of DNA replication via (p)ppGpp binding to DnaG (Wang et al., 2007; Maciag et al., 2010; Rymer et al., 2012). Enzymes of fatty acid metabolism exhibit weak binding affinities (Polakis et al., 1973; Stein and Bloch, 1976), suggesting regulation by (p)ppGpp only to take place under full stringent control (i.e., 1,000 μM (p)ppGpp). Some enzymes partaking in (p)ppGpp metabolism, i.e., the (p)ppGpp synthetases RelQ (Gaca et al., 2015; Steinchen et al., 2015) and Rel/RelA (Shyp et al., 2012; Kudrin et al., 2018; Takada et al., 2020a), are stimulated in their activity by pppGpp and to a lesser extent by ppGpp. Furthermore, the enzymes GdpP, Pde2, and PgpH involved in the degradation pathway of the second messenger cyclic-diadenosine-monophosphate (c-di-AMP) display Ki values for inhibition by ppGpp between 35 and 400 μM.
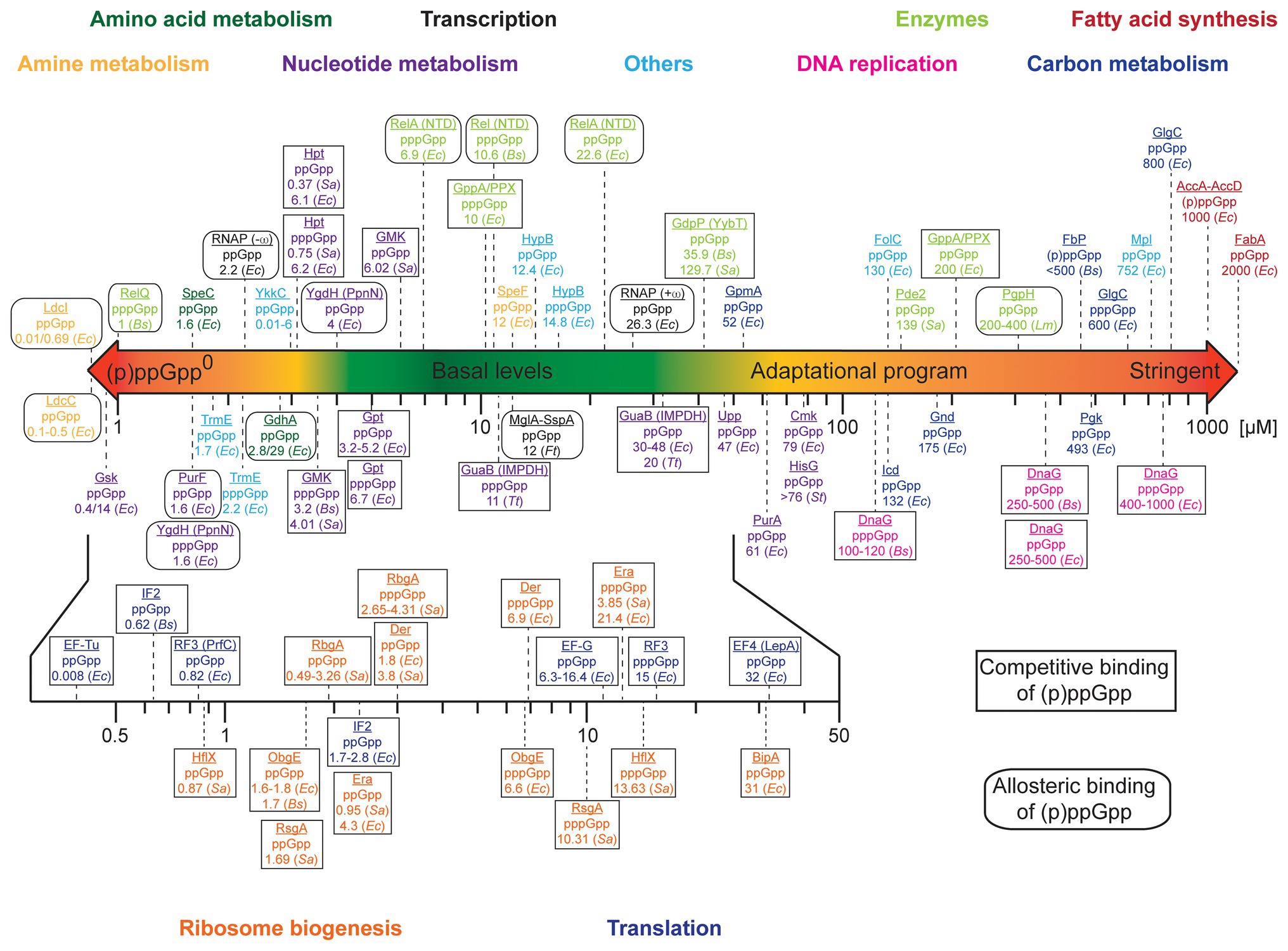
Figure 1. Scheme of prioritization of (p)ppGpp-mediated adaptation. The colored bar denotes the approximate intracellular concentrations (in μM) of (p)ppGpp in bacteria. Binding targets of (p)ppGpp were sorted according to their dissociation constants (Kd). For DnaG, “enzymes” and proteins of carbon metabolism and fatty acid synthesis, Ki or EC50/IC50 values are depicted. Proteins are color-coded according to their belonging to different groups/biochemical processes. Rectangles and rounded rectangles indicate whether (p)ppGpp binding to the target molecule is competitive or allosteric. Bacterial species are abbreviated as follows: Escherichia coli (Ec), Bacillus subtilis (Bs), Staphylococcus aureus (Sa), Listeria monocytogenes (Lm), Francisella tularensis (Ft), Thermus thermophilus (Tt), and Salmonella typhimurium (St). Further details can be found in Supplementary Table S1.
Importantly, in proteins where (p)ppGpp bind competitively to another compound (Figure 1 and Supplementary Table S1), the intracellular concentration of this compound also affects the fraction of the (p)ppGpp-bound protein. The influence of this other compound rises with its intracellular concentration. For example, the concentration of GTP under non-stringent conditions is assumed to fall between approximately 1 (Varik et al., 2017; Zbornikova et al., 2019) and 5 mM (Bennett et al., 2009) but may drop heavily due to (p)ppGpp-dependent inhibition of GTP anabolism (Kriel et al., 2012; Liu et al., 2015b). Thereby, inhibition of GTPases is governed by the (p)ppGpp to GTP ratio that is indicative for the metabolic state of the cell. Assuming, for example, similar Kd values for (p)ppGpp and GTP binding, only approximately 1% of a protein would be inhibited at 10 μM (p)ppGpp/1 mM GTP while this fraction rises to 50% at equal concentrations of both nucleotides. In fact, the Ki for the inhibition of the GTPase RbgA of 300 and 800 μM (ppGpp and pppGpp in presence of 50S ribosomal subunits determined at 1 mM GTP; Pausch et al., 2018; Supplementary Table S1) is a better estimate than the Kd value. Similar considerations apply to some enzymes of nucleotide metabolism, e.g., GMK (Nomura et al., 2014) with its substrate GMP or Gpt (Hochstadt-Ozer and Cashel, 1972) with the substrate guanine. In contrast to GTP, however, the intracellular concentrations of those nucleotides are lower, i.e., 24 μM GMP and 19 μM guanine (Bennett et al., 2009), suggesting 90% inhibition at (p)ppGpp concentrations of approximately 200 μM. In contrast, the enzymes YgdH (PpnN; Zhang et al., 2019), PurF (Wang et al., 2019), and LdcI are allosterically inhibited by (p)ppGpp, rendering inhibition by (p)ppGpp independent of the substrate concentration. Summarized, the kinetic parameters of (p)ppGpp interaction with target proteins indicate the following hierarchy of adaptation: amine and amino acid metabolism, nucleotide metabolism, translational and ribosome biogenesis GTPases, DNA replication, carbon metabolism, and fatty acid synthesis (Figure 1).
Importance of Basal (p)ppGpp Levels for Bacterial Physiology
Increasing evidence recommends that bacteria require basal (p)ppGpp levels to maintain homeostasis (Sokawa et al., 1975; Sarubbi et al., 1988; Silva and Benitez, 2006; Lemos et al., 2008; Gaca et al., 2013; Stott et al., 2015; Li et al., 2020) and strains that are completely devoid of any (p)ppGpp (known as (p)ppGpp0) reveal multiple disabilities in cell division, transcription, translation, GTP homeostasis, and antibiotic tolerance. In Bacillus subtilis, the lack of (p)ppGpp causes elevated GTP levels, which through dysregulation of the GTP-dependent transcriptional repressor CodY result in auxotrophies for branched-chain amino acids (Kriel et al., 2014). Similar CodY-mediated detrimental effects of ppGpp0-strains were observed for Enterococcus faecalis (Gaca et al., 2013) and Staphylococcus aureus (Pohl et al., 2009). In a rel-deletion strain of Synechococcus elongatus, the transcript levels of 52–67% of all genes were upregulated at least 3-fold and the levels of rRNA elevated indicating a “transcriptionally relaxed” state (Puszynska and O’Shea, 2017). Furthermore, cell size and volume increased but could be restored by synthetic compensation of (p)ppGpp (Puszynska and O’Shea, 2017). In Vibrio cholera basal (p)ppGpp has been linked to the expression of virulence factors and cell motility (Silva and Benitez, 2006). Basal (p)ppGpp was also required for the tolerance of E. faecalis against vancomycin (Abranches et al., 2009). These reports emphasize the importance of basal (p)ppGpp levels, which through high-affine binding of the alarmones (Kd < 10 μM; Figure 1) exert a regulatory function in the absence of a (p)ppGpp-stimulating trigger.
Toxic Over-Accumulation of (p)ppGpp
Intuitively, a prolonged mismatch of (p)ppGpp synthesis and degradation resulting in over-accumulation of (p)ppGpp, is detrimental for the cell. In E. coli, RelA and SpoT are responsible for the synthesis of (p)ppGpp, with only SpoT being able to hydrolyze the alarmones. As such, the role of SpoT is pivotal for preventing toxic (p)ppGpp accumulation as exemplified by SpoT-deletion strains being characterized by reduced growth rates and distorted amino acid requirements (Xiao et al., 1991). Bacteria of the Firmicutes phylum, instead of possessing RelA and SpoT, harbor the bifunctional enzyme Rel (Atkinson et al., 2011). The hydrolase activity of Rel is required to prevent toxic concentrations of (p)ppGpp, which results in severe growth defects (Gratani et al., 2018; Takada et al., 2020b). This effect appears more notorious in the presence of the two small alarmone synthetases, RelP and RelQ, the (p)ppGpp synthetase activity of which is not properly contained in absence of the (p)ppGpp-degrading Rel (Geiger et al., 2014). In Mycobacterium tuberculosis, (p)ppGpp degradation is equally essential since deletion of (p)ppGpp hydrolase activity, while retaining (p)ppGpp synthesis, lead to lethal toxicity evidenced by irregularities in colony formation irrespective of the nutritional environment (Weiss and Stallings, 2013). Notably, these M. tuberculosis strains were also impaired in their virulence in a mouse model during both acute and chronic infection, highlighting the interference with (p)ppGpp metabolism as a potential antimicrobial therapeutic strategy. Taken together, a tight regulation of the antagonistic (p)ppGpp producing and degrading activities is indispensable for many, if not all bacteria for (i) correctly adjusting (p)ppGpp levels in response to environmental cues, (ii) coordinating various cellular processes in an orchestrated manner, and (iii) avoiding lethal consequences due to (p)ppGpp over-accumulation.
Crosstalk Between (p)ppGpp and c-di-AMP
Bacteria possess a thorough toolbox of nucleotide-based second messengers to efficiently respond to diverse external cues (Pesavento and Hengge, 2009; Kalia et al., 2013; Hengge et al., 2016). Among those, c-di-AMP is a signaling molecule mainly synthetized by Gram-positive bacteria (Romling, 2008; da Aline Dias et al., 2020; He et al., 2020; Yin et al., 2020), which fulfills a pivotal role in the osmotic homeostasis (Stulke and Kruger, 2020). The dynamic range of intracellular c-di-AMP is much lower than for (p)ppGpp and lies in the one-digit μM range (in B. subtilis 1.7 μM during vegetative growth to 5.1 μM 2 h after sporulation; Oppenheimer-Shaanan et al., 2011). The importance of c-di-AMP for bacterial physiology is also highlighted by the observation that both the complete absence and over-accumulation of c-di-AMP impede growth of B. subtilis (Mehne et al., 2013; Gundlach et al., 2015). Comparison of the transcriptional profiles of S. aureus cells at high c-di-AMP or (p)ppGpp concentrations revealed an overlap of 27.9% between the two regulons (Corrigan et al., 2015). Intriguingly, (p)ppGpp inhibit the activity of the S. aureus c-di-AMP phosphodiesterase GdpP with a Ki of 129.7 ± 42.8 μM in vitro correlating with higher levels of c-di-AMP observed in vivo (Corrigan et al., 2015). Inhibition by ppGpp with an IC50 of 139 ± 5.6 μM was also reported for the second S. aureus c-di-AMP phosphodiesterase Pde2 (Bowman et al., 2016). Similar observations made for the c-di-AMP hydrolases YybT (GdpP) from B. subtilis (Rao et al., 2010) and PgpH from Listeria monocytogenes (Huynh et al., 2015). These Ki/IC50 values suggest that (p)ppGpp concentrations must rise above their basal levels to inhibit c-di-AMP degradation. Deletion of S. aureus GdpP with concomitant increase of c-di-AMP evokes elevated (p)ppGpp (Corrigan et al., 2015), thus implying that induction of one second messenger, (p)ppGpp or c-di-AMP, augments the other. Conversely, the depletion of the only c-di-AMP synthetase, dacA, in L. monocytogenes also induced a toxic accumulation of (p)ppGpp (Whiteley et al., 2015). This apparent twist might be caused in the narrow dynamic range of c-di-AMP whereby deflection in either direction triggers (p)ppGpp synthesis or indicate species-specific differences in the regulatory circuits. Nevertheless, the functional connection of c-di-AMP and (p)ppGpp is further substantiated by the observation that the essentiality of dacA in L. monocytogenes during growth in rich medium was abrogated in a (p)ppGpp0-strain or in the wild-type strain grown in minimal medium (Whiteley et al., 2015). Hereby, the activity of the GTP-dependent transcriptional regulator CodY appears critical to dacA essentiality in rich media in that mutation of codY in the (p)ppGpp0-strain renders dacA again essential. This implies that elements of the CodY regulon may be toxic in the absence of c-di-AMP-producing DacA (Whiteley et al., 2015). The GTP-loading state and activity of CodY (Sonenshein, 2007) are in turn linked to (p)ppGpp through the inhibition of GTP anabolic enzymes involved in nucleotide metabolism by the alarmones (see above). Thus, as a whole, the interdependencies between c-di-AMP and (p)ppGpp conceivably require a high degree of coordination and involve a number of different cellular processes.
The Emerging Hyperphosphorylated Nucleotides (p)ppApp
In the 1970s, the accumulation of two further magic spots designated as (p)ppApp was observed in B. subtilis during the early stages of sporulation (Rhaese and Groscurth, 1976; Rhaese et al., 1977; Nishino et al., 1979) and the (p)ppApp synthetic activity retrieved from the ribosomal fraction, which indicates the contribution of an RSH. The first RSH to catalyze (p)ppApp formation was recently identified in Methylobacterium extorquens (Sobala et al., 2019), however, (p)ppApp-producing activity was not evidenced for the E. coli RelA enzyme (Jimmy et al., 2020). Another source of (p)ppApp is the nucleoside 5'-diphosphate kinase from Streptomyces morookaensis although the enzyme being secreted raises the question of its physiological relevance for this organism (Oki et al., 1975). The high similarity of (p)ppGpp and (p)ppApp – they only differ in their nucleobase – advises a putative overlap of their target spectra. Recent studies evidence that E. coli PurF and RNAP, both of which are validated targets of (p)ppGpp, also bind (p)ppApp (Bruhn-Olszewska et al., 2018; Ahmad et al., 2019). However, while in PurF both second messengers bind in similar fashion at the same site and inhibit the enzyme with equal potency (Ahmad et al., 2019; Wang et al., 2019), the binding sites on RNAP are discrete and (p)ppApp enhance transcription from, e.g., the rrnB P1 promoter as opposed to (p)ppGpp (Bruhn-Olszewska et al., 2018). In one study, ppApp also inhibited the (p)ppGpp synthetic activity of E. coli RelA with an IC50 of 24.5 ± 3.5 μM (Beljantseva et al., 2017) raising the possibility that, in fact, (p)ppApp and (p)ppGpp are antagonists. Besides potentially fulfilling regulatory functions in their host cell, (p)ppApp also serve as toxins during interbacterial warfare (Ahmad et al., 2019). Hereby, the type VI secretion system (T6SS) effector protein Tas1 of Pseudomonas aeruginosa PA14, a (p)ppApp synthetase, is injected into the target cell and depletes the cellular ATP pools, resulting in dysregulation of the metabolome and, ultimately, cell death. Endogenous ppApp, the metabolism of which is embedded in toxin-antitoxin systems, might represent an intricate pathway to control growth (Jimmy et al., 2020). Whether the (p)ppApp molecules exert growth control independent from (p)ppGpp via a separated protein target spectrum or in a competitive manner to (p)ppGpp remains to be investigated.
Concluding Remarks
The functions of (p)ppGpp are often simplistically portrayed as a biphasic switch between relaxed and stringent environmental conditions, the latter triggering a major metabolic rearrangement through induction of (p)ppGpp synthesis. The presence of gradation in (p)ppGpp-dependent regulation of transcription is an approved mechanism (Traxler et al., 2008; Balsalobre, 2011; Traxler et al., 2011). However, the wide range of binding affinities among the (p)ppGpp-affected protein targets covering four orders of magnitude furthermore suggests a post-translational adaptational program activated hierarchically by increasing (p)ppGpp. The depicted scheme illustrates the importance of high-affine (p)ppGpp targets that explicitly require regulation, thus suggesting a protagonism of the alarmones on cell homeostasis. We believe that the post-translational sequential “dimming” of protein activities by (p)ppGpp, a conception potentially also true for related second messengers like (p)ppApp and c-di-AMP, plays a major role in successful adaptation of microorganisms.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
WS and GB conceived the study. WS and VZ performed the analysis. WS, VZ, and GB wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
GB receives support through the SPP1879 priority program of the Deutsche Forschungsgemeinschaft (DFG).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
GB thanks the SPP1879 priority program of the Deutsche Forschungsgemeinschaft (DFG) for financial support.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.02072/full#supplementary-material
References
Abranches, J., Martinez, A. R., Kajfasz, J. K., Chavez, V., Garsin, D. A., and Lemos, J. A. (2009). The molecular alarmone (p)ppGpp mediates stress responses, vancomycin tolerance, and virulence in Enterococcus faecalis. J. Bacteriol. 191, 2248–2256. doi: 10.1128/JB.01726-08
Ahmad, S., Wang, B., Walker, M. D., Tran, H. R., Stogios, P. J., Savchenko, A., et al. (2019). An interbacterial toxin inhibits target cell growth by synthesizing (p)ppApp. Nature 575, 674–678. doi: 10.1038/s41586-019-1735-9
Atkinson, G. C., Tenson, T., and Hauryliuk, V. (2011). The RelA/SpoT homolog (RSH) superfamily: distribution and functional evolution of ppGpp synthetases and hydrolases across the tree of life. PLoS One 6:e23479. doi: 10.1371/journal.pone.0023479
Bager, R., Roghanian, M., Gerdes, K., and Clarke, D. J. (2016). Alarmone (p)ppGpp regulates the transition from pathogenicity to mutualism in Photorhabdus luminescens. Mol. Microbiol. 100, 735–747. doi: 10.1111/mmi.13345
Balsalobre, C. (2011). Concentration matters!! ppGpp, from a whispering to a strident alarmone. Mol. Microbiol. 79, 827–829. doi: 10.1111/j.1365-2958.2010.07521.x
Battesti, A., and Bouveret, E. (2006). Acyl carrier protein/SpoT interaction, the switch linking SpoT-dependent stress response to fatty acid metabolism. Mol. Microbiol. 62, 1048–1063. doi: 10.1111/j.1365-2958.2006.05442.x
Battesti, A., and Bouveret, E. (2009). Bacteria possessing two RelA/SpoT-like proteins have evolved a specific stringent response involving the acyl carrier protein-SpoT interaction. J. Bacteriol. 191, 616–624. doi: 10.1128/JB.01195-08
Beljantseva, J., Kudrin, P., Jimmy, S., Ehn, M., Pohl, R., Varik, V., et al. (2017). Molecular mutagenesis of ppGpp: turning a RelA activator into an inhibitor. Sci. Rep. 7:41839. doi: 10.1038/srep41839
Bennett, B. D., Kimball, E. H., Gao, M., Osterhout, R., Van Dien, S. J., and Rabinowitz, J. D. (2009). Absolute metabolite concentrations and implied enzyme active site occupancy in Escherichia coli. Nat. Chem. Biol. 5, 593–599. doi: 10.1038/nchembio.186
Bennison, D. J., Irving, S. E., and Corrigan, R. M. (2019). The impact of the stringent response on TRAFAC GTPases and prokaryotic ribosome assembly. Cell 8:1313. doi: 10.3390/cells8111313
Bhardwaj, N., Syal, K., and Chatterji, D. (2018). The role of omega-subunit of Escherichia coli RNA polymerase in stress response. Genes Cells 23, 357–369. doi: 10.1111/gtc.12577
Bowman, L., Zeden, M. S., Schuster, C. F., Kaever, V., and Grundling, A. (2016). New insights into the cyclic di-adenosine monophosphate (c-di-AMP) degradation pathway and the requirement of the cyclic dinucleotide for acid stress resistance in Staphylococcus aureus. J. Biol. Chem. 291, 26970–26986. doi: 10.1074/jbc.M116.747709
Bruhn-Olszewska, B., Molodtsov, V., Sobala, M., Dylewski, M., Murakami, K. S., Cashel, M., et al. (2018). Structure-function comparisons of (p)ppApp vs (p)ppGpp for Escherichia coli RNA polymerase binding sites and for rrnB P1 promoter regulatory responses in vitro. Biochim. Biophys. Acta Gene Regul. Mech. 1861, 731–742. doi: 10.1016/j.bbagrm.2018.07.005
Cashel, M., and Gallant, J. (1969). Two compounds implicated in the function of the RC gene of Escherichia coli. Nature 221, 838–841. doi: 10.1038/221838a0
Corrigan, R. M., Bellows, L. E., Wood, A., and Grundling, A. (2016). ppGpp negatively impacts ribosome assembly affecting growth and antimicrobial tolerance in gram-positive bacteria. Proc. Natl. Acad. Sci. U. S. A. 113, E1710–E1719. doi: 10.1073/pnas.1522179113
Corrigan, R. M., Bowman, L., Willis, A. R., Kaever, V., and Grundling, A. (2015). Cross-talk between two nucleotide-signaling pathways in Staphylococcus aureus. J. Biol. Chem. 290, 5826–5839. doi: 10.1074/jbc.M114.598300
Cuthbert, B. J., Ross, W., Rohlfing, A. E., Dove, S. L., Gourse, R. L., Brennan, R. G., et al. (2017). Dissection of the molecular circuitry controlling virulence in Francisella tularensis. Genes Dev. 31, 1549–1560. doi: 10.1101/gad.303701.117
da Aline Dias, P., de Nathalia Marins, A., de Gabriel Guarany, A., de Robson Francisco, S., and Cristiane Rodrigues, G. (2020). The world of cyclic dinucleotides in bacterial behavior. Molecules 25:2462. doi: 10.3390/molecules25102462
Dalebroux, Z. D., and Swanson, M. S. (2012). ppGpp: magic beyond RNA polymerase. Nat. Rev. Microbiol. 10, 203–212. doi: 10.1038/nrmicro2720
Dietzler, D. N., and Leckie, M. P. (1977). Regulation of ADP-glucose synthetase, the rate-limiting enzyme of bacterial glycogen synthesis, by the pleiotropic nucleotides ppGpp and pppGpp. Biochem. Biophys. Res. Commun. 77, 1459–1467. doi: 10.1016/S0006-291X(77)80143-5
Fujita, Y., and Freese, E. (1979). Purification and properties of fructose-1,6-bisphosphatase of Bacillus subtilis. J. Biol. Chem. 254, 5340–5349.
Gaca, A. O., Kajfasz, J. K., Miller, J. H., Liu, K., Wang, J. D., Abranches, J., et al. (2013). Basal levels of (p)ppGpp in Enterococcus faecalis: the magic beyond the stringent response. mBio 4, e00646–e00613. doi: 10.1128/mBio.00646-13
Gaca, A. O., Kudrin, P., Colomer-Winter, C., Beljantseva, J., Liu, K., Anderson, B., et al. (2015). From (p)ppGpp to (pp)pGpp: characterization of regulatory effects of pGpp synthesized by the small alarmone synthetase of Enterococcus faecalis. J. Bacteriol. 197, 2908–2919. doi: 10.1128/JB.00324-15
Geiger, T., Kastle, B., Gratani, F. L., Goerke, C., and Wolz, C. (2014). Two small (p)ppGpp synthases in Staphylococcus aureus mediate tolerance against cell envelope stress conditions. J. Bacteriol. 196, 894–902. doi: 10.1128/JB.01201-13
Gourse, R. L., Chen, A. Y., Gopalkrishnan, S., Sanchez-Vazquez, P., Myers, A., and Ross, W. (2018). Transcriptional responses to ppGpp and DksA. Annu. Rev. Microbiol. 72, 163–184. doi: 10.1146/annurev-micro-090817-062444
Gratani, F. L., Horvatek, P., Geiger, T., Borisova, M., Mayer, C., Grin, I., et al. (2018). Regulation of the opposing (p)ppGpp synthetase and hydrolase activities in a bifunctional RelA/SpoT homologue from Staphylococcus aureus. PLoS Genet. 14:e1007514. doi: 10.1371/journal.pgen.1007514
Gundlach, J., Mehne, F. M., Herzberg, C., Kampf, J., Valerius, O., Kaever, V., et al. (2015). An essential poison: synthesis and degradation of cyclic di-AMP in Bacillus subtilis. J. Bacteriol. 197, 3265–3274. doi: 10.1128/JB.00564-15
Haseltine, W. A., and Block, R. (1973). Synthesis of guanosine tetra‐ and pentaphosphate requires the presence of a codon-specific, uncharged transfer ribonucleic acid in the acceptor site of ribosomes. Proc. Natl. Acad. Sci. U. S. A. 70, 1564–1568. doi: 10.1073/pnas.70.5.1564
He, H., Cooper, J. N., Mishra, A., and Raskin, D. M. (2012). Stringent response regulation of biofilm formation in Vibrio cholerae. J. Bacteriol. 194, 2962–2972. doi: 10.1128/JB.00014-12
He, J., Yin, W., Galperin, M. Y., and Chou, S. H. (2020). Cyclic di-AMP, a second messenger of primary importance: tertiary structures and binding mechanisms. Nucleic Acids Res. 48, 2807–2829. doi: 10.1093/nar/gkaa112
Hengge, R., Grundling, A., Jenal, U., Ryan, R., and Yildiz, F. (2016). Bacterial signal transduction by cyclic di-GMP and other nucleotide second messengers. J. Bacteriol. 198, 15–26. doi: 10.1128/JB.00331-15
Hochstadt-Ozer, J., and Cashel, M. (1972). The regulation of purine utilization in bacteria. V. Inhibition of purine phosphoribosyltransferase activities and purine uptake in isolated membrane vesicles by guanosine tetraphosphate. J. Biol. Chem. 247, 7067–7072.
Hogg, T., Mechold, U., Malke, H., Cashel, M., and Hilgenfeld, R. (2004). Conformational antagonism between opposing active sites in a bifunctional RelA/SpoT homolog modulates (p)ppGpp metabolism during the stringent response. Cell 117, 57–68. doi: 10.1016/S0092-8674(04)00260-0
Huynh, T. N., Luo, S., Pensinger, D., Sauer, J. D., Tong, L., and Woodward, J. J. (2015). An HD-domain phosphodiesterase mediates cooperative hydrolysis of c-di-AMP to affect bacterial growth and virulence. Proc. Natl. Acad. Sci. U. S. A. 112, E747–E756. doi: 10.1073/pnas.1416485112
Jimmy, S., Saha, C. K., Kurata, T., Stavropoulos, C., Oliveira, S. R. A., Koh, A., et al. (2020). A widespread toxin-antitoxin system exploiting growth control via alarmone signaling. Proc. Natl. Acad. Sci. U. S. A. 117, 10500–10510. doi: 10.1073/pnas.1916617117
Kalia, D., Merey, G., Nakayama, S., Zheng, Y., Zhou, J., Luo, Y., et al. (2013). Nucleotide, c-di-GMP, c-di-AMP, cGMP, cAMP, (p)ppGpp signaling in bacteria and implications in pathogenesis. Chem. Soc. Rev. 42, 305–341. doi: 10.1039/C2CS35206K
Kanjee, U., Gutsche, I., Alexopoulos, E., Zhao, B., El Bakkouri, M., Thibault, G., et al. (2011a). Linkage between the bacterial acid stress and stringent responses: the structure of the inducible lysine decarboxylase. EMBO J. 30, 931–944. doi: 10.1038/emboj.2011.5
Kanjee, U., Gutsche, I., Ramachandran, S., and Houry, W. A. (2011b). The enzymatic activities of the Escherichia coli basic aliphatic amino acid decarboxylases exhibit a pH zone of inhibition. Biochemistry 50, 9388–9398. doi: 10.1021/bi201161k
Kanjee, U., Ogata, K., and Houry, W. A. (2012). Direct binding targets of the stringent response alarmone (p)ppGpp. Mol. Microbiol. 85, 1029–1043. doi: 10.1111/j.1365-2958.2012.08177.x
Keasling, J. D., Bertsch, L., and Kornberg, A. (1993). Guanosine pentaphosphate phosphohydrolase of Escherichia coli is a long-chain exopolyphosphatase. Proc. Natl. Acad. Sci. U. S. A. 90, 7029–7033. doi: 10.1073/pnas.90.15.7029
Kriel, A., Bittner, A. N., Kim, S. H., Liu, K., Tehranchi, A. K., Zou, W. Y., et al. (2012). Direct regulation of GTP homeostasis by (p)ppGpp: a critical component of viability and stress resistance. Mol. Cell 48, 231–241. doi: 10.1016/j.molcel.2012.08.009
Kriel, A., Brinsmade, S. R., Tse, J. L., Tehranchi, A. K., Bittner, A. N., Sonenshein, A. L., et al. (2014). GTP dysregulation in Bacillus subtilis cells lacking (p)ppGpp results in phenotypic amino acid auxotrophy and failure to adapt to nutrient downshift and regulate biosynthesis genes. J. Bacteriol. 196, 189–201. doi: 10.1128/JB.00918-13
Kristensen, O., Ross, B., and Gajhede, M. (2008). Structure of the PPX/GPPA phosphatase from Aquifex aeolicus in complex with the alarmone ppGpp. J. Mol. Biol. 375, 1469–1476. doi: 10.1016/j.jmb.2007.11.073
Kudrin, P., Dzhygyr, I., Ishiguro, K., Beljantseva, J., Maksimova, E., Oliveira, S. R. A., et al. (2018). The ribosomal A-site finger is crucial for binding and activation of the stringent factor RelA. Nucleic Acids Res. 46, 1973–1983. doi: 10.1093/nar/gky023
Kuroda, A., Murphy, H., Cashel, M., and Kornberg, A. (1997). Guanosine tetra‐ and pentaphosphate promote accumulation of inorganic polyphosphate in Escherichia coli. J. Biol. Chem. 272, 21240–21243. doi: 10.1074/jbc.272.34.21240
Lemos, J. A., Nascimento, M. M., Lin, V. K., Abranches, J., and Burne, R. A. (2008). Global regulation by (p)ppGpp and CodY in Streptococcus mutans. J. Bacteriol. 190, 5291–5299. doi: 10.1128/JB.00288-08
Li, G., Zhao, Q., Luan, T., Hu, Y., Zhang, Y., Li, T., et al. (2020). Basal-level effects of (p)ppGpp in the absence of branched-chain amino acids in Actinobacillus pleuropneumoniae. J. Bacteriol. 202, e00640–e00719. doi: 10.1128/JB.00640-19
Liu, K., Bittner, A. N., and Wang, J. D. (2015a). Diversity in (p)ppGpp metabolism and effectors. Curr. Opin. Microbiol. 24, 72–79. doi: 10.1016/j.mib.2015.01.012
Liu, K., Myers, A. R., Pisithkul, T., Claas, K. R., Satyshur, K. A., Amador-Noguez, D., et al. (2015b). Molecular mechanism and evolution of guanylate kinase regulation by (p)ppGpp. Mol. Cell 57, 735–749. doi: 10.1016/j.molcel.2014.12.037
Maciag, M., Kochanowska, M., Lyzen, R., Wegrzyn, G., and Szalewska-Palasz, A. (2010). ppGpp inhibits the activity of Escherichia coli DnaG primase. Plasmid 63, 61–67. doi: 10.1016/j.plasmid.2009.11.002
Mehne, F. M., Gunka, K., Eilers, H., Herzberg, C., Kaever, V., and Stulke, J. (2013). Cyclic di-AMP homeostasis in Bacillus subtilis: both lack and high level accumulation of the nucleotide are detrimental for cell growth. J. Biol. Chem. 288, 2004–2017. doi: 10.1074/jbc.M112.395491
Nishino, T., Gallant, J., Shalit, P., Palmer, L., and Wehr, T. (1979). Regulatory nucleotides involved in the Rel function of Bacillus subtilis. J. Bacteriol. 140, 671–679. doi: 10.1128/JB.140.2.671-679.1979
Nomura, Y., Izumi, A., Fukunaga, Y., Kusumi, K., Iba, K., Watanabe, S., et al. (2014). Diversity in guanosine 3',5'-bisdiphosphate (ppGpp) sensitivity among guanylate kinases of bacteria and plants. J. Biol. Chem. 289, 15631–15641. doi: 10.1074/jbc.M113.534768
Oki, T., Yoshimoto, A., Sato, S., and Takamatsu, A. (1975). Purine nucleotide pyrophosphotransferase from Streptomyces morookaensis, capable of synthesizing pppApp and pppGpp. Biochim. Biophys. Acta 410, 262–272. doi: 10.1016/0005-2744(75)90228-4
Oppenheimer-Shaanan, Y., Wexselblatt, E., Katzhendler, J., Yavin, E., and Ben-Yehuda, S. (2011). c-di-AMP reports DNA integrity during sporulation in Bacillus subtilis. EMBO Rep. 12, 594–601. doi: 10.1038/embor.2011.77
Pausch, P., Steinchen, W., Wieland, M., Klaus, T., Freibert, S. A., Altegoer, F., et al. (2018). Structural basis for (p)ppGpp-mediated inhibition of the GTPase RbgA. J. Biol. Chem. 293, 19699–19709. doi: 10.1074/jbc.RA118.003070
Pesavento, C., and Hengge, R. (2009). Bacterial nucleotide-based second messengers. Curr. Opin. Microbiol. 12, 170–176. doi: 10.1016/j.mib.2009.01.007
Peselis, A., and Serganov, A. (2018). ykkC riboswitches employ an add-on helix to adjust specificity for polyanionic ligands. Nat. Chem. Biol. 14, 887–894. doi: 10.1038/s41589-018-0114-4
Pohl, K., Francois, P., Stenz, L., Schlink, F., Geiger, T., Herbert, S., et al. (2009). CodY in Staphylococcus aureus: a regulatory link between metabolism and virulence gene expression. J. Bacteriol. 191, 2953–2963. doi: 10.1128/JB.01492-08
Polakis, S. E., Guchhait, R. B., and Lane, M. D. (1973). Stringent control of fatty acid synthesis in Escherichia coli. Possible regulation of acetyl coenzyme a carboxylase by ppGpp. J. Biol. Chem. 248, 7957–7966.
Potrykus, K., and Cashel, M. (2008). (p)ppGpp: still magical? Annu. Rev. Microbiol. 62, 35–51. doi: 10.1146/annurev.micro.62.081307.162903
Puszynska, A. M., and O’Shea, E. K. (2017). ppGpp controls global gene expression in light and in darkness in S. elongatus. Cell Rep. 21, 3155–3165. doi: 10.1016/j.celrep.2017.11.067
Rao, F., See, R. Y., Zhang, D., Toh, D. C., Ji, Q., and Liang, Z. X. (2010). YybT is a signaling protein that contains a cyclic dinucleotide phosphodiesterase domain and a GGDEF domain with ATPase activity. J. Biol. Chem. 285, 473–482. doi: 10.1074/jbc.M109.040238
Rhaese, H. J., and Groscurth, R. (1976). Control of development: role of regulatory nucleotides synthesized by membranes of Bacillus subtilis in initiation of sporulation. Proc. Natl. Acad. Sci. U. S. A. 73, 331–335. doi: 10.1073/pnas.73.2.331
Rhaese, H. J., Hoch, J. A., and Groscurth, R. (1977). Studies on the control of development: isolation of Bacillus subtilis mutants blocked early in sporulation and defective in synthesis of highly phosphorylated nucleotides. Proc. Natl. Acad. Sci. U. S. A. 74, 1125–1129. doi: 10.1073/pnas.74.3.1125
Romling, U. (2008). Great times for small molecules: c-di-AMP, a second messenger candidate in bacteria and archaea. Sci. Signal. 1:pe39. doi: 10.1126/scisignal.133pe39
Rymer, R. U., Solorio, F. A., Tehranchi, A. K., Chu, C., Corn, J. E., Keck, J. L., et al. (2012). Binding mechanism of metalNTP substrates and stringent-response alarmones to bacterial DnaG-type primases. Structure 20, 1478–1489. doi: 10.1016/j.str.2012.05.017
Sarubbi, E., Rudd, K. E., and Cashel, M. (1988). Basal ppGpp level adjustment shown by new spoT mutants affect steady state growth rates and rrnA ribosomal promoter regulation in Escherichia coli. Mol. Gen. Genet. 213, 214–222. doi: 10.1007/BF00339584
Schafer, H., Beckert, B., Frese, C. K., Steinchen, W., Nuss, A. M., Beckstette, M., et al. (2020). The alarmones (p)ppGpp are part of the heat shock response of Bacillus subtilis. PLoS Genet. 16:e1008275. doi: 10.1371/journal.pgen.1008275
Schreiber, G., Metzger, S., Aizenman, E., Roza, S., Cashel, M., and Glaser, G. (1991). Overexpression of the relA gene in Escherichia coli. J. Biol. Chem. 266, 3760–3767.
Sherlock, M. E., Sudarsan, N., and Breaker, R. R. (2018). Riboswitches for the alarmone ppGpp expand the collection of RNA-based signaling systems. Proc. Natl. Acad. Sci. U. S. A. 115, 6052–6057. doi: 10.1073/pnas.1720406115
Shyp, V., Tankov, S., Ermakov, A., Kudrin, P., English, B. P., Ehrenberg, M., et al. (2012). Positive allosteric feedback regulation of the stringent response enzyme RelA by its product. EMBO Rep. 13, 835–839. doi: 10.1038/embor.2012.106
Silva, A. J., and Benitez, J. A. (2006). A Vibrio cholerae relaxed (relA) mutant expresses major virulence factors, exhibits biofilm formation and motility, and colonizes the suckling mouse intestine. J. Bacteriol. 188, 794–800. doi: 10.1128/JB.188.2.794-800.2006
Sobala, M., Bruhn-Olszewska, B., Cashel, M., and Potrykus, K. (2019). Methylobacterium extorquens RSH enzyme synthesizes (p)ppGpp and pppApp in vitro and in vivo, and leads to discovery of pppApp synthesis in Escherichia coli. Front. Microbiol. 10:859. doi: 10.3389/fmicb.2019.00859
Sokawa, Y., Sokawa, J., and Kaziro, Y. (1975). Regulation of stable RNA synthesis and ppGpp levels in growing cells of Escherichia coli. Cell 5, 69–74. doi: 10.1016/0092-8674(75)90093-8
Sonenshein, A. L. (2007). Control of key metabolic intersections in Bacillus subtilis. Nat. Rev. Microbiol. 5, 917–927. doi: 10.1038/nrmicro1772
Srivatsan, A., Han, Y., Peng, J., Tehranchi, A. K., Gibbs, R., Wang, J. D., et al. (2008). High-precision, whole-genome sequencing of laboratory strains facilitates genetic studies. PLoS Genet. 4:e1000139. doi: 10.1371/journal.pgen.1000139
Stein, J. P. Jr., and Bloch, K. E. (1976). Inhibition of E. coli beta-hydroxydecanoyl thioester dehydrase by ppGpp. Biochem. Biophys. Res. Commun. 73, 881–884. doi: 10.1016/0006-291X(76)90204-7
Steinchen, W., and Bange, G. (2016). The magic dance of the alarmones (p)ppGpp. Mol. Microbiol. 101, 531–544. doi: 10.1111/mmi.13412
Steinchen, W., Schuhmacher, J. S., Altegoer, F., Fage, C. D., Srinivasan, V., Linne, U., et al. (2015). Catalytic mechanism and allosteric regulation of an oligomeric (p)ppGpp synthetase by an alarmone. Proc. Natl. Acad. Sci. U. S. A. 112, 13348–13353. doi: 10.1073/pnas.1505271112
Stott, K. V., Wood, S. M., Blair, J. A., Nguyen, B. T., Herrera, A., Mora, Y. G., et al. (2015). (p)ppGpp modulates cell size and the initiation of DNA replication in Caulobacter crescentus in response to a block in lipid biosynthesis. Microbiology 161, 553–564. doi: 10.1099/mic.0.000032
Strugeon, E., Tilloy, V., Ploy, M. C., and Da Re, S. (2016). The stringent response promotes antibiotic resistance dissemination by regulating integron integrase expression in biofilms. mBio 7, e00868–e00916. doi: 10.1128/mBio.00868-16
Stulke, J., and Kruger, L. (2020). Cyclic di-AMP signaling in bacteria. Annu. Rev. Microbiol. doi: 10.1146/annurev-micro-020518-115943 [Epub ahead of print]
Svitil, A. L., Cashel, M., and Zyskind, J. W. (1993). Guanosine tetraphosphate inhibits protein synthesis in vivo. A possible protective mechanism for starvation stress in Escherichia coli. J. Biol. Chem. 268, 2307–2311.
Sy, J., and Lipmann, F. (1973). Identification of the synthesis of guanosine tetraphosphate (MS I) as insertion of a pyrophosphoryl group into the 3'-position in guanosine 5'-diphosphate. Proc. Natl. Acad. Sci. U. S. A. 70, 306–309. doi: 10.1073/pnas.70.2.306
Taguchi, M., Izui, K., and Katsuki, H. (1977). Activation of Escherichia coli phosphoenolpyruvate carboxylase by guanosine-5'-diphosphate-3'-diphosphate. FEBS Lett. 77, 270–272. doi: 10.1016/0014-5793(77)80249-4
Takada, H., Roghanian, M., Caballero-Montes, J., Van Nerom, K., Jimmy, S., Kudrin, P., et al. (2020a). Ribosome association primes the stringent factor Rel for recruitment of deacylated tRNA to ribosomal A-site. bioRxiv [Preprint]. doi: 10.1101/2020.01.17.910273
Takada, H., Roghanian, M., Murina, V., Dzhygyr, I., Murayama, R., Akanuma, G., et al. (2020b). The C-terminal RRM/ACT domain is crucial for fine-tuning the activation of ‘Long’ RelA-SpoT homolog enzymes by ribosomal complexes. Front. Microbiol. 11:277. doi: 10.3389/fmicb.2020.00277
Traxler, M. F., Summers, S. M., Nguyen, H. T., Zacharia, V. M., Hightower, G. A., Smith, J. T., et al. (2008). The global, ppGpp-mediated stringent response to amino acid starvation in Escherichia coli. Mol. Microbiol. 68, 1128–1148. doi: 10.1111/j.1365-2958.2008.06229.x
Traxler, M. F., Zacharia, V. M., Marquardt, S., Summers, S. M., Nguyen, H. T., Stark, S. E., et al. (2011). Discretely calibrated regulatory loops controlled by ppGpp partition gene induction across the ‘feast to famine’ gradient in Escherichia coli. Mol. Microbiol. 79, 830–845. doi: 10.1111/j.1365-2958.2010.07498.x
Varik, V., Oliveira, S. R. A., Hauryliuk, V., and Tenson, T. (2017). HPLC-based quantification of bacterial housekeeping nucleotides and alarmone messengers ppGpp and pppGpp. Sci. Rep. 7:11022. doi: 10.1038/s41598-017-10988-6
Wang, B., Dai, P., Ding, D., Del Rosario, A., Grant, R. A., Pentelute, B. L., et al. (2019). Affinity-based capture and identification of protein effectors of the growth regulator ppGpp. Nat. Chem. Biol. 15, 141–150. doi: 10.1038/s41589-018-0183-4
Wang, J. D., Sanders, G. M., and Grossman, A. D. (2007). Nutritional control of elongation of DNA replication by (p)ppGpp. Cell 128, 865–875. doi: 10.1016/j.cell.2006.12.043
Weiss, L. A., and Stallings, C. L. (2013). Essential roles for Mycobacterium tuberculosis Rel beyond the production of (p)ppGpp. J. Bacteriol. 195, 5629–5638. doi: 10.1128/JB.00759-13
Wendrich, T. M., Blaha, G., Wilson, D. N., Marahiel, M. A., and Nierhaus, K. H. (2002). Dissection of the mechanism for the stringent factor RelA. Mol. Cell 10, 779–788. doi: 10.1016/S1097-2765(02)00656-1
Whiteley, A. T., Pollock, A. J., and Portnoy, D. A. (2015). The PAMP c-di-AMP is essential for Listeria monocytogenes growth in rich but not minimal media due to a toxic increase in (p)ppGpp. Cell Host Microbe 17, 788–798. doi: 10.1016/j.chom.2015.05.006
Xiao, H., Kalman, M., Ikehara, K., Zemel, S., Glaser, G., and Cashel, M. (1991). Residual guanosine 3',5'-bispyrophosphate synthetic activity of relA null mutants can be eliminated by spoT null mutations. J. Biol. Chem. 266, 5980–5990.
Yin, W., Cai, X., Ma, H., Zhu, L., Zhang, Y., Chou, S. H., et al. (2020). A decade of research on the second messenger c-di-AMP. FEMS Microbiol. Rev. doi: 10.1093/femsre/fuaa019 [Epub ahead of print]
Zbornikova, E., Knejzlik, Z., Hauryliuk, V., Krasny, L., and Rejman, D. (2019). Analysis of nucleotide pools in bacteria using HPLC-MS in HILIC mode. Talanta 205:120161. doi: 10.1016/j.talanta.2019.120161
Zhang, Y. E., Baerentsen, R. L., Fuhrer, T., Sauer, U., Gerdes, K., and Brodersen, D. E. (2019). (p)ppGpp regulates a bacterial nucleosidase by an allosteric two-domain switch. Mol. Cell 74, 1239.e4–1249.e4. doi: 10.1016/j.molcel.2019.03.035
Zhang, Y., Zbornikova, E., Rejman, D., and Gerdes, K. (2018). Novel (p)ppGpp binding and metabolizing proteins of Escherichia coli. mBio 9, e02188–e02217. doi: 10.1128/mBio.02188-17
Keywords: stringent response, (p)ppGpp, alarmones, metabolism, physiology
Citation: Steinchen W, Zegarra V and Bange G (2020) (p)ppGpp: Magic Modulators of Bacterial Physiology and Metabolism. Front. Microbiol. 11:2072. doi: 10.3389/fmicb.2020.02072
Edited by:
Haike Antelmann, Freie Universität Berlin, GermanyReviewed by:
Vasili Hauryliuk, Umeå University, SwedenFabian M. Commichau, Brandenburg University of Technology Cottbus-Senftenberg, Germany
Copyright © 2020 Steinchen, Zegarra and Bange. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wieland Steinchen, d2llbGFuZC5zdGVpbmNoZW5Ac3lubWlrcm8udW5pLW1hcmJ1cmcuZGU=; Gert Bange, Z2VydC5iYW5nZUBzeW5taWtyby51bmktbWFyYnVyZy5kZQ==
 Wieland Steinchen
Wieland Steinchen Victor Zegarra
Victor Zegarra Gert Bange
Gert Bange