- 1Zhejiang Provincial Key Laboratory of Biometrology and Inspection & Quarantine, College of Life Sciences, China Jiliang University, Hangzhou, China
- 2Institute for Pharmaceutical Sciences, Pharmaceutical Biology and Biotechnology, University of Freiburg, Freiburg, Germany
The nucleoside antibiotic toyocamycin (TM) is a potential fungicide that can control plant diseases, and it has become an attractive target for research. Streptomyces diastatochromogenes 1628, a TM-producing strain, was isolated by our laboratory and was considered to be a potent industrial producer of TM. Recently, the putative TM biosynthetic gene cluster (toy cluster) in S. diastatochromogenes 1628 was found by genome sequencing. In this study, the role of toy cluster for TM biosynthesis in S. diastatochromogenes 1628 was investigated by heterologous expression, deletion, and complementation. The extract of the recombinant strain S. albusJ1074-TC harboring a copy of toy cluster produced TM as shown by HPLC analysis. The Δcluster mutant completely lost its ability to produce TM. TM production in the complemented strain was restored to a level comparable to that of the wild-type strain. These results confirmed that the toy cluster is responsible for TM biosynthesis. Moreover, the introduction of an extra copy of the toy cluster into S. diastatochromogenes 1628 led to onefold increase in TM production (312.9 mg/l vs. 152.1 mg/l) as well as the transcription of all toy genes. The toy gene cluster was engineered in which the native promoter of toyA gene, toyM gene, toyBD operon, and toyEI operon was, respectively, replaced by permE∗ or SPL57. To further improve TM production, the engineered toy gene cluster was, respectively, introduced and overexpressed in S. diastatochromogenes 1628 to generate recombinant strains S. diastatochromogenes 1628-EC and 1628-SC. After 84 h, S. diastatochromogenes 1628-EC and 1628-SC produced 456.5 mg/l and 638.9 mg/l TM, respectively, which is an increase of 2- and 3.2-fold compared with the wild-type strain.
Introduction
Streptomyces spp. are Gram-positive bacteria with a complex morphological differentiation and a highly developed secondary metabolism (Li and Tan, 2017; Jones and Elliot, 2018). They are well-known to produce a wide variety of bioactive natural products including antibiotics (Baltz, 2016; Kemung et al., 2018; Liu et al., 2018). Strain improvement is main goal of fermentation-based antibiotics production (Wang et al., 2010; Yamada et al., 2015). With the development of genetic engineering and metabolic engineering in Streptomyces spp. and an increasing knowledge of biosynthetic pathways, enhancement of antibiotic production has been addressed by using a rational approach (Chen et al., 2010; Tan et al., 2015; Palazzotto et al., 2019). Genetic manipulations of regulators, controlling the production of antibiotics or key structural genes governing the biosynthesis of antibiotics, are common and generally applicable strategies to achieve high-level production of antibiotics (Vicente et al., 2014; Martínez-Burgo et al., 2015; Zhu et al., 2017).
Because of its broad range of biological activities against pathogenic fungi, the nucleoside antibiotic toyocamycin (TM) has become an interesting target for research. TM is produced by Streptomyces diastatochromogenes 1628, but the production level is always very low (Ma et al., 2014a). In this context, efforts to improve TM production have been made by the overexpression of structural genes (Tao et al., 2016) or favorable genes (Ma et al., 2014a; Ma et al., 2014b) and the pathway-specific regulatory gene toyA (Xu et al., 2019) or ribosome engineering technology (Ma et al., 2016). Expression of an extra copy of toyG and frr led to a 65.5% increase in TM production (Tao et al., 2016). In another study, expression of an extra copy of pathway-specific regulator toyA driven by different promoters led to up to twofold increase in TM production (Xu et al., 2019).
Despite this, it cannot be ignored that more than a single rate-limiting step is often in the biosynthetic pathway of secondary metabolites that leads to the complexity of genetic manipulations. As almost all genes responsible for the biosynthesis of secondary metabolites are clustered together, the introduction of an extra copy of the entire biosynthetic gene cluster into the parent strain is considered to be an effective strategy to increase the yield of antibiotics, in which regulatory genes, structural genes and antibiotic-resistant genes are expressed simultaneously to maintain their compatibility. Successful experiments have confirmed this strategy (Liao et al., 2010; Murakami et al., 2011; Du et al., 2013; Jiang et al., 2013; Zhou et al., 2014; Li et al., 2017). For example, Liao et al. (2010) introduced an extra copy of the nikkomycin biosynthetic gene cluster and this led to a 4-fold and 1.8-fold increase in nikkomycin Z and nikkomycin X, respectively. In the work described by Zhou et al. (2014), three to five copies of validamycin (VAL-A) biosynthetic gene cluster were integrated into S. hygroscopicus 5008 and this led to a 34% enhancement of VAL-A production in shake-flask fermentation. In addition, the work of Li et al. (2017) led to an improved pristinamycin II production which was achieved by the introduction of five copies of the pristinamycin cluster.
Recently, the draft genome sequence of S. diastatochromogenes 1628 was obtained (unpublished). After analysis of the genome sequence by antiSMASH, preliminary data indicated the presence of 44 putative gene clusters including the TM biosynthetic gene cluster (toy cluster), which ranges from toyA to toyI (GenBank accession No. KY022432). Ten genes were controlled by four units: toyA, toyM, toyEFGHI operon (toyEI), and toyBCD operon (toyBD) (Figure 1). Surprisingly, each gene in the toy cluster showed the highest similarity to a corresponding gene from S. ahygroscopicus (JX576291) (Supplementary Table S1). However, thus far, the relationship of this published toy gene cluster with TM biosynthesis in S. ahygroscopicus has yet to be reported. Moreover, the overall gene organization and the sequence of the toy gene cluster differs from the gene organization and sequence of the toy gene cluster (GenBank accession No. EU573979) (Supplementary Table S1) of Streptomyces rimosus (McCarty and Bandarian, 2008).
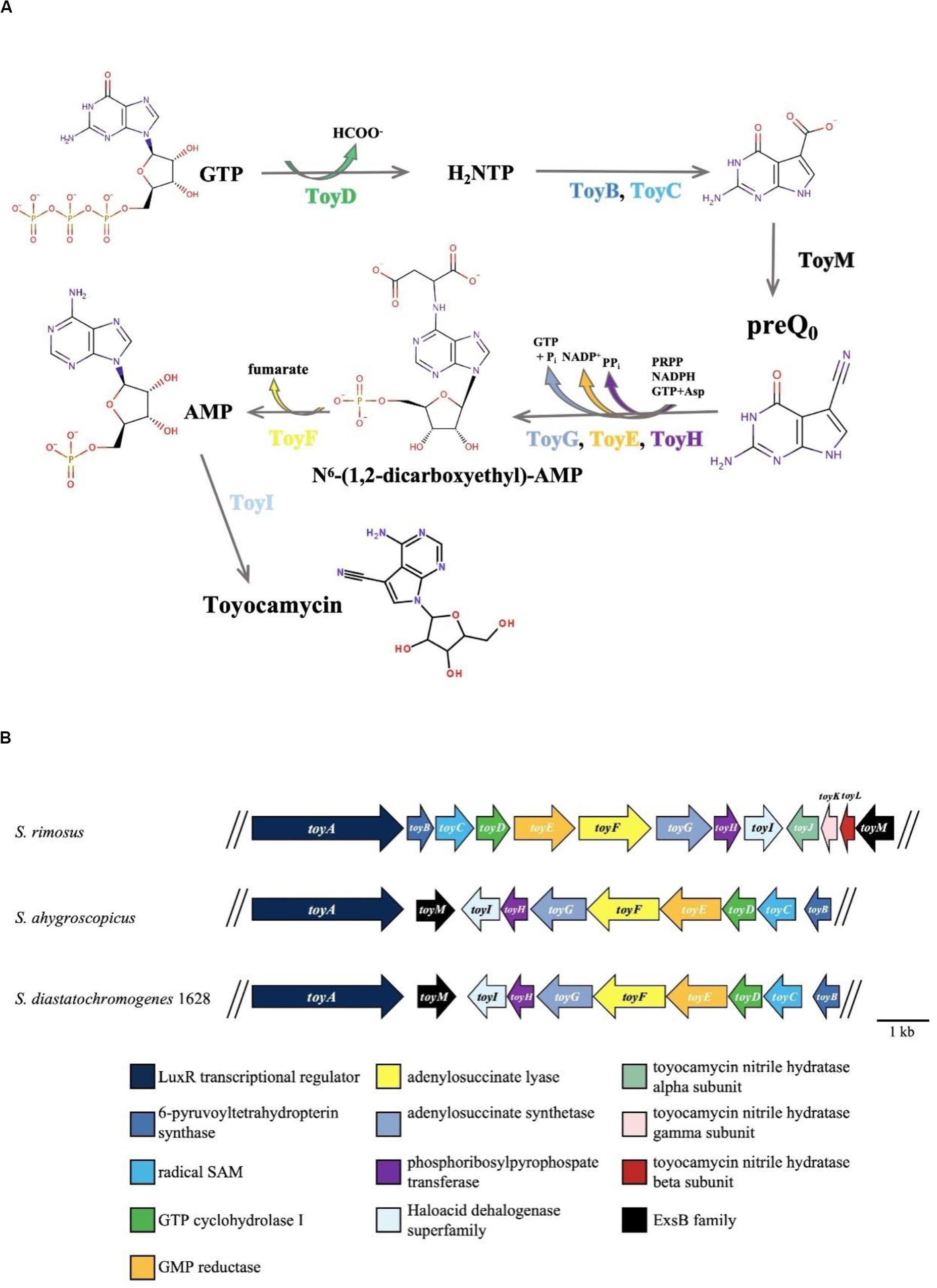
Figure 1. TM biosynthesis pathway and involved genes. (A) Putative TM biosynthesis pathway in S. rimosus (McCarty and Bandarian, 2008). (B) Organization of the toyocamycin gene clusters from S. rimosus (upper), S. ahygroscopicus (middle), and S. diastatochromogenes 1628 (lower).
In this context, two obvious questions are: whether putative toy cluster is responsible for TM biosynthesis in S. diastatochromogenes 1628? Whether the amplification of toy cluster could enhance the TM production in S. diastatochromogenes 1628?
To address the first question, in this study, the relationship between toy cluster and TM biosynthesis in S. diastatochromogenes 1628 was revealed by gene deletion and complementation experiment. The role of toy cluster was also confirmed by its heterologous expression in S. albus J1074.
To address the second question, the native toy cluster was cloned and overexpressed in parent strain S. diastatochromogenes 1628, in order to improve TM production. Furthermore, to overexpress the toy cluster, gain higher transcription levels of all toy genes, and further enhance TM production, the native toy cluster was engineered by replacing the native promoters of toyA, toyM, and toyEI, and of the toyBD operon by the strong constitutive promoter permE∗ and the synthetic promoter SPL57, respectively. The higher TM production was achieved by introduction of an extra copy of engineered cluster comparing to that by native cluster.
Materials and Methods
Materials
Q5 High-Fidelity Master Mix with GC-buffer was purchased from NEB. PCR reagents, restriction endonucleases, Miniprep, and Gel Extraction kits were purchased from TaKaRa Biotechnology Co. Ltd. Oligonucleotide primer synthesis and DNA sequencing of PCR products were performed by Shanghai Sunny Biotechnology Co. Ltd. China.
Bacterial Strains, Plasmids, and Primers
The plasmids and strains used in this study are listed in Table 1 and Supplementary Table S2. TM producer S. diastatochromogenes 1628 has been deposited in the China General Microbiological Culture Collection Center (CGMCC No. 2060) (Ma et al., 2014a). E. coli JM109 was used as general host for gene cloning. The methylation-deficient strain E. coli ET12567/pUZ8002 was used as the donor for plasmid transfer to Streptomyces.
Plasmids pSET152, pGUS-SPL-57, and pGUS-ermE∗ were described previously (Siegl et al., 2013).
Media and Growth Conditions
Escherichia coli strains were cultured using liquid or solid Luria-Bertani medium containing appropriate antibiotics at 37°C. Antibiotics were used in the following concentrations: apramycin (100 μg/ml), chloramphenicol (25 μg/ml), ampicillin (100 μg/ml), and kanamycin (50 μg/ml). S. diastatochromogenes 1628 was incubated at 28 °C and grown in solid mannitol soya flour (MS) medium (Zhao et al., 2019) for sporulation and conjugation. To generate spores, S. diastatochromogenes 1628 and its derivates were spread on MS medium and incubated for 4–5 days at 28°C. Collected spores were washed with water and preserved in water/glycerol (1:1, v/v) at −80°C.
DNA Manipulations
General procedures for DNA manipulation were conducted according to the method described by Sambrook and Russel (2001). Intergeneric conjugation of Streptomyces and E. coli was performed as described by Kieser et al. (2000).
Amplification and Sequencing of Toy Cluster
The toy cassette1 (harboring toyA gene with its 300-bp promoter regions, toyM gene, toyI gene and toyH gene) and toy cassette2 (harboring toyB gene with its 300-bp promoter regions, toyC, toyD, toyE, toyF, and toyG genes) were obtained through PCR amplification by using genome DNA of S. diastatochromogenes 1628 as the template. The toy cassette1 and cassette2 were amplified using primers Pcass1-F/R and Pcass2-F/R, respectively (Supplementary Table S3). High-fidelity PCR was performed using Q5 DNA polymerase (NEB) to obtain DNA fragments used for plasmid construction. PCR amplification was started at 98°C for 30 s, followed by 30 cycles of denaturation at 98°C for 20 s, annealing at 64°C for 20 s, and extension at 72°C for 6 min. After 30 cycles, a 5-min extension at 72°C was performed. Plasmid pSET152 was digested with XbaI and EcoRV and was fused to toy cassette1 and cassette2, using the Gibson assembly method to generate the recombinant plasmid pSET152::ncluster. Sequencing of the inserted 11410-bp native toy gene cluster confirmed that the gene did not contain any mutations.
Construction of Toy Cluster Disruption (ΔCluster) Mutant and Its Complementation
Disruption of the toy cluster was performed by gene replacement via homologous recombination as described by Xu et al. (2019). For construction of toy cluster-disruption (Δcluster) mutant, a 3.0-kb fragment upstream of the toyA start codon was amplified by PCR using Pup-F/R (Supplementary Table S3) and a 3.0-kb fragment downstream of the toyB start codon was amplified by PCR using Pdown-F/R (Supplementary Table S3). The kanamycin-resistance gene (neo) was amplified from plasmid pET28a (Novagen) by PCR using primers Pkan-F/R (Supplementary Table S3). The three resulting DNA fragments were ligated into the XbaI and EcoRV sites of pKC1132 to yield pKC1132-Δcluster by using Gibson DNA assembly methods as described by Gibson et al. (2008). Subsequently, introduction of the constructed pKC1132-Δcluster into the wild-type strain S. diastatochromogenes 1628 was conducted by intergeneric conjugation. The Δcluster mutants were named S. diastatochromogenes 1628-Δcluster. Mutants were selected by both apramycin sensitivity (Aprs) and kanamycin-resistance (Kanr). The genotype of mutant S. diastatochromogenes 1628-Δcluster was confirmed by PCR.
For the complementation of the toy cluster in S. diastatochromogenes 1628-Δcluster, the plasmid pSET152::ncluster and the empty vector pSET152 as a control were introduced into mutant S. diastatochromogenes 1628-Δcluster by conjugation, resulting in the complemented strain 1628-Δcluster/pSET152::ncluster and the control strain 1628-Δcluster/pSET152, respectively.
Heterologous Expression/Overexpression in S. albus J1074/S. diastatochromogenes 1628
Introduction of the constructed pSET152::ncluster into S. albus J1074 and S. diastatochromogenes 1628 was conducted by conjugation to yield recombinant strains S. albus J1074-TC and S. diastatochromogenes 1628-TC, respectively. Recombinant strains were confirmed using apramycin resistance and PCR.
Engineered Toy Cluster by Insertion of Promoter permE∗ and SPL57
The construction of plasmid pSET152::ecluster was conducted by the following steps. First, three DNA fragments (717 bp of toyM gene, 4893 bp of toyEI cassette, and 1768 bp of toyBD cassette) were amplified from S. diastatochromogenes 1628 genomic DNA by using primers P1/P2, P3/P4, and P5/P6 (Supplementary Table S4), respectively. These fragments were, respectively, digested with SpeI/EcoRV and ligated into corresponding sites of pGUS-ermE∗ to generate plasmids pM, pEI, and pBD, respectively, in each of which the gusA gene in pGUS-ermE∗ was removed and replaced by the toyM gene, toyEI operon and toyBD operon. Second, three DNA fragments permE∗-toyM, permE∗-toyEI, and permE∗-toyBD were amplified from pM, pEI, and pBD by using primers P7/P8, P9/P10, and P11/P12 (Supplementary Table S4), respectively. The obtained three DNA fragments were fused into the plasmid permE∗-toyA linearized by EcoRV using the Gibson assembly method to generate the recombinant plasmid pSET152::ecluster, in which the toyA gene, toyM gene, toyBD operon and toyEI operon were all driven by promoter permE∗. The inserted genes or cassettes were sequenced using sequencing primers. Sequencing results confirmed that no mutations were introduced.
A similar procedure described in the construction of plasmid pSET152::ecluster was performed to construct plasmid pSET152::scluster. The gusA gene in plasmid pGUS-SPL57 was replaced by toyM gene, toyEI operon, and toyBD operon to yield plasmids p57M, p57EI, and p57BD, respectively, in each of which the toyM gene, toyEI operon, and toyBD operon were placed under the control of synthetic promoter SPL57. Three DNA fragments SPL57-toyM, SPL57-toyEI, and SPL57-toyBD were then amplified from p57M, p57EI, and p57BD by using primers P13/P14, P15/P16, and P17/P18 (Supplementary Table S4), respectively. These were fused into the plasmid pSPL57-toyA linearized by EcoRV using the Gibson assembly method to generate the recombinant plasmid pSET152::scluster, in which toyA gene, toyM gene, toyBD operon, and toyEI operon all driven by promoterSPL57. The inserted genes or cassettes were sequenced using sequencing primers. Sequencing results confirmed that no mutations were introduced.
Overexpression of Engineered Toy Cluster in S. diastatochromogenes 1628 Strain
The constructed plasmids pSET152::ecluster and pSET152::scluster were introduced into E. coli ET12567/pUZ8002 and then transferred into S. diastatochromogenes 1628 by conjugation to generate recombinant strains S. diastatochromogenes 1628-EC and S. diastatochromogenes 1628-SC, respectively. The recombinant strains were selected in the presence of apramycin at concentration of 50 μg/ml. The genotype of recombinant strains was verified by PCR using specific primers, binding to the apramycin resistance gene as part of the plasmid, which integrated into the chromosome.
Analysis of Gene Transcriptional Levels by qRT-PCR
The extraction of RNA, design of primers, and analysis of transcriptional level of toy genes were performed as described by Wang et al. (2018). The qRT-PCR experiments were carried out in triplicate using RNA samples from three independent experiments.
Production and Analysis of TM
Production and HPLC analysis of TM were conducted according to the method described by Ma et al. (2014b). Liquid chromatography-mass spectrometry (LC-MS) was performed on an Agilent 6120 Quadrupole MSD mass spectrometer (Agilent Technologies, United States) equipped with an Agilent 1290 Series Quaternary LC system and an Eclipse Plus C18 column (100 × 2.1 mm, 1.8 μm).
Statistical Analysis
All experiments were conducted at least three times, and the results were expressed as mean ± standard deviations. Statistical analysis was performed with Student’s t-test. Samples with p values < 0.05 were considered statistically significant.
Results
Toy Cluster Is Responsible for TM Biosynthesis by Heterologous Expression and Deletion
To investigate whether the putative toy cluster is involved in TM biosynthesis, a DNA fragment containing the putative toy cluster was cloned as described in the section “Materials and Methods.” To ensure the fidelity of the toy cluster sequence, amplification of the toy cluster was divided into cassette1 and cassette2, in which the original toy cluster sequences and direction from toyA to toyB of S. diastatochromogenes 1628 was retained. Then cassette1 and cassette2 were fused into plasmid pSET152 by Gibson assembly to generate recombinant pSET152::ncluster (Figure 2A). The plasmid pSET152::ncluster was confirmed by digesting with XbaI and EcoRV (Figure 2B) and was then passed through E.coli ET12567 (pUZ8002) and subsequently introduced into S. albusJ1074 by intergeneric conjugation, to generate the recombinant strain S. albus J1074-TC resistant to 50 μg/ml apramycin. The integration of plasmid pSET152::ncluster into the chromosome of S. albus J1074 was verified by PCR assay (Supplementary Figure S1). In addition, the heterologous expression of toy gene cluster in S. albus J1074 was confirmed by quantitative RT-PCR analysis (Supplementary Figure S2). The recombinant strain S. albus J1074-TC and wild-type strain S. albus J1074 were cultured in shake-flask fermentation. As shown in Figure 3, culture filtrate from the recombinant strain S. albus J1074-TC was subjected to HPLC analysis, and a distinct peak appeared at a position corresponding to that of the standard sample of TM, whereas the culture filtrates from the negative control strains (S. albus J1074 and S. albus J1074-pSET152) lacked this peak. This peak was further identified as TM by means of LC-MS, whose negative molecular ion was at m/z290.2 [M-H]– (Figure 3). Thus S. albus J1074-TC could produce TM, albeit in a small amount.
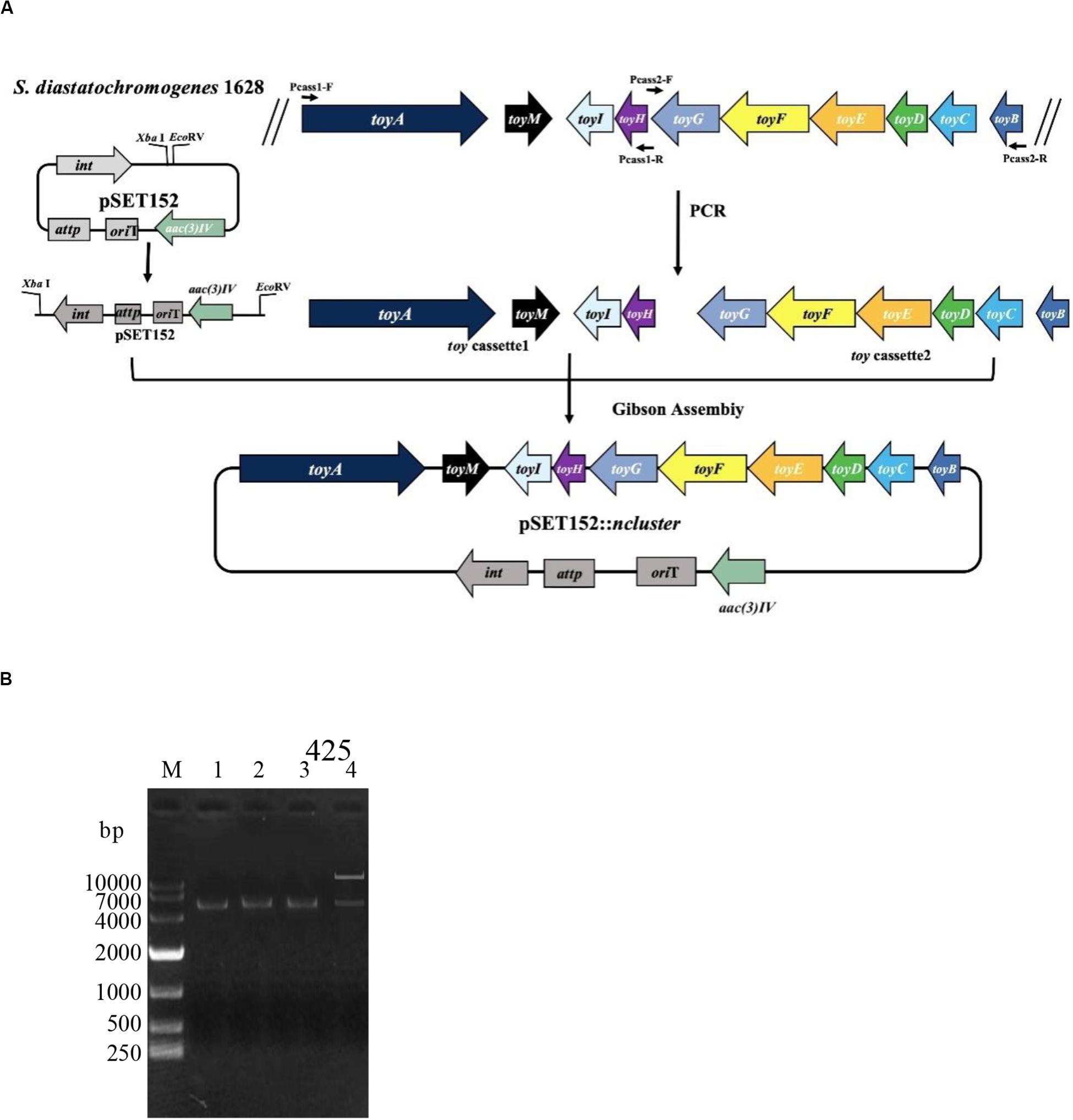
Figure 2. Schematic representation of the construction of pSET152::ncluster. (A) Cassette1 harbors toyA gene with its 300-bp promoter regions, toyM gene, toyI gene and toyH gene; Cassette2 harbors toyB gene with its 300-bp promoter regions, toyC gene, toyD gene, toyE gene, toyF gene, and toyG gene. Plasmid pSET152 was digested with XbaI and EcoRV and was fused to cassette1 and cassette2, using the Gibson assembly method to generate the recombinant plasmid pSET152::ncluster. (B) Identification of plasmid pSET152::ncluster. M: DL10000 DNA Marker. Lane 1, The PCR product of the 5.4-kb cassette1 were amplified by using the primers Pcass1-F/R; Lane 2, The PCR product of the 5.6-kb cassette2 was amplified by using the primers Pcass2-F/R; Lane 3, Plasmid pSET152 was digested and linearized by XbaI and EcoRV; Lane 4, The pSET152::ncluster was digested by XbaI and EcoRV.
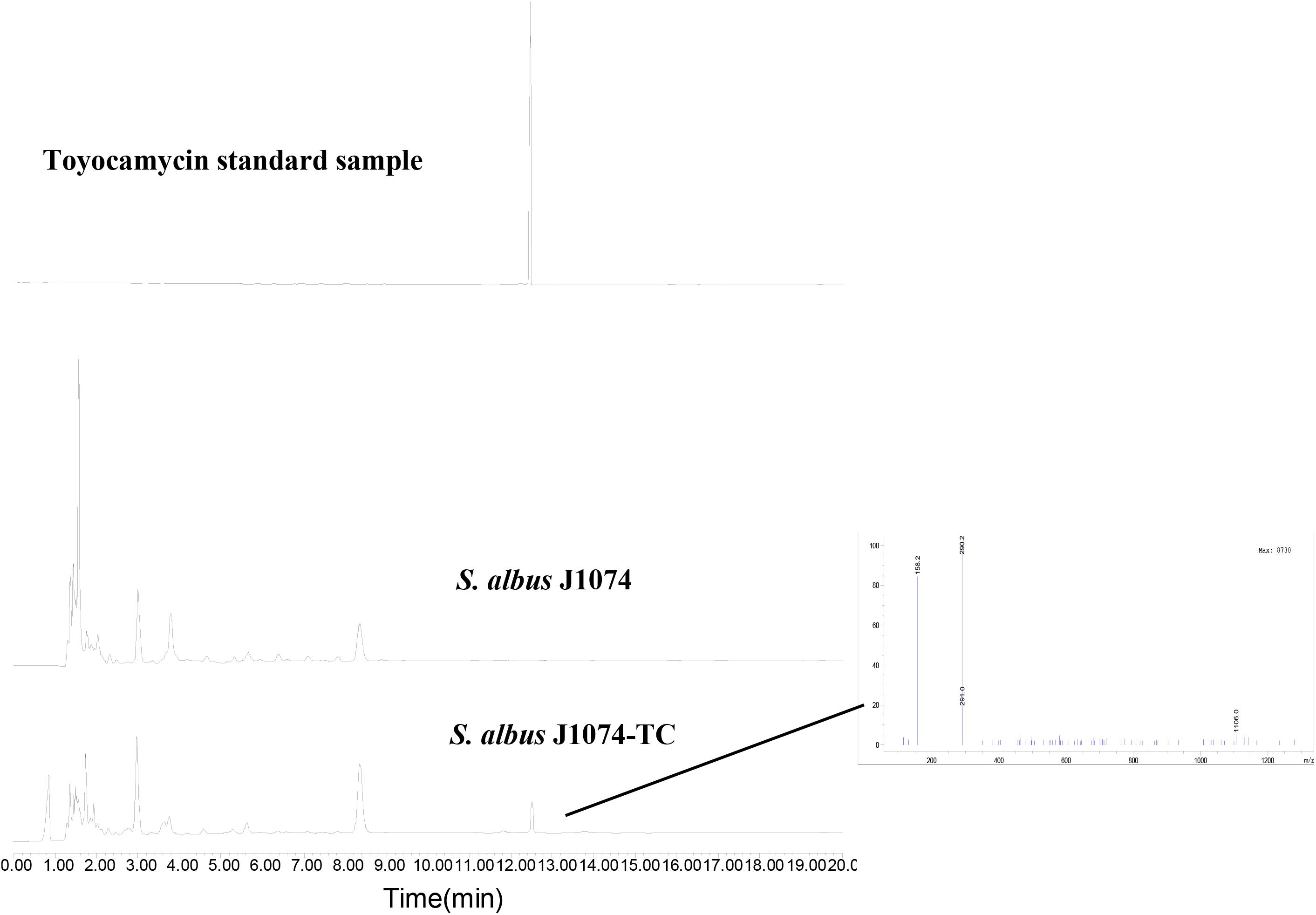
Figure 3. Heterologous expression of toy cluster in S. albus J1074. HPLC analysis of TM isolated from fermentation extracts of S. albus J1074 and recombinant strain S. albus J1074-TC; LC-MS (negative mode) of the distinct peak in the recombinant strain S. albus J1074-TC. LC-MS was performed on an Agilent 6120 Quadrupole MSD mass spectrometer (Agilent Technologies, United States) equipped with an Agilent 1290 Series Quaternary LC system and an Eclipse Plus C18 column (100 × 2.1 mm, 1.8 μm).
The toy cluster was completely replaced by a neo gene in the S. diastatochromogenes 1628 chromosome via double-crossover homologous recombination (Supplementary Figure S3A), to generate mutant S. diastatochromogenes 1628-Δcluster. The genotype of mutant 1628-Δcluster was verified via PCR analysis by using the primers PkanF/R, PkanF/PdownR, and PupF/PkanR, respectively. In mutant 1628-Δcluster, a single band 1.5-kb neo gene can be detected (Supplementary Figure S3B), suggesting that a double crossover event had occurred. Mutant 1628-Δcluster and wild-type strain S. diastatochromogenes 1628 were cultured in fermentation medium and their TM production was determined. HPLC analysis indicated that mutant S. diastatochromogenes 1628-Δcluster completely lost its ability to produce TM (Supplementary Figure S3C). The Δcluster mutant was then complemented by introducing pSET152::ncluster, to generate the complemented strain S. diastatochromogenes 1628-Δcluster/pSET152::ncluster. TM production in complemented strain S. diastatochromogenes 1628-Δcluster/pSET152::ncluster was restored to a level comparable to that in the wild-type strain S. diastatochromogenes 1628 under the normal fermentation condition (Supplementary Figure S3C). These results suggest that the cloned toy cluster is responsible for TM biosynthesis.
Enhancement of TM Production by Addition of an Extra Copy of Native Toy Cluster in S. diastatochromogenes 1628
We investigated whether an extra copy number of the native toy cluster increases TM production in S. diastatochromogenes 1628. The plasmid pSET152::ncluster was introduced into S. diastatochromogenes 1628 by intergeneric conjugation, to generate the recombinant strain 1628-TC (Figure 4A), which is resistant to 50 μg/ml apramycin. To verify the integration of plasmid pSET152::ncluster into the chromosome of S. diastatochromogenes 1628, genomic DNA of S. diastatochromogenes 1628-TC was isolated and used as template to perform PCR (Supplementary Figure S1). A large increase in TM production was observed for the recombinant strain 1628-TC harboring an extra copy of toy cluster (Figure 4B). After 84 h, the amount of TM produced by 1628-TC reached the highest level of 312.9 mg/l, a 105.7% increase in the yield of TM produced by S. diastatochromogenes 1628 (152.1 mg/l). In addition, introduction of either plasmid pSET152 or pSET152::ncluster had no effect on the cell growth of S. diastatochromogenes 1628 (Supplementary Figure S4).
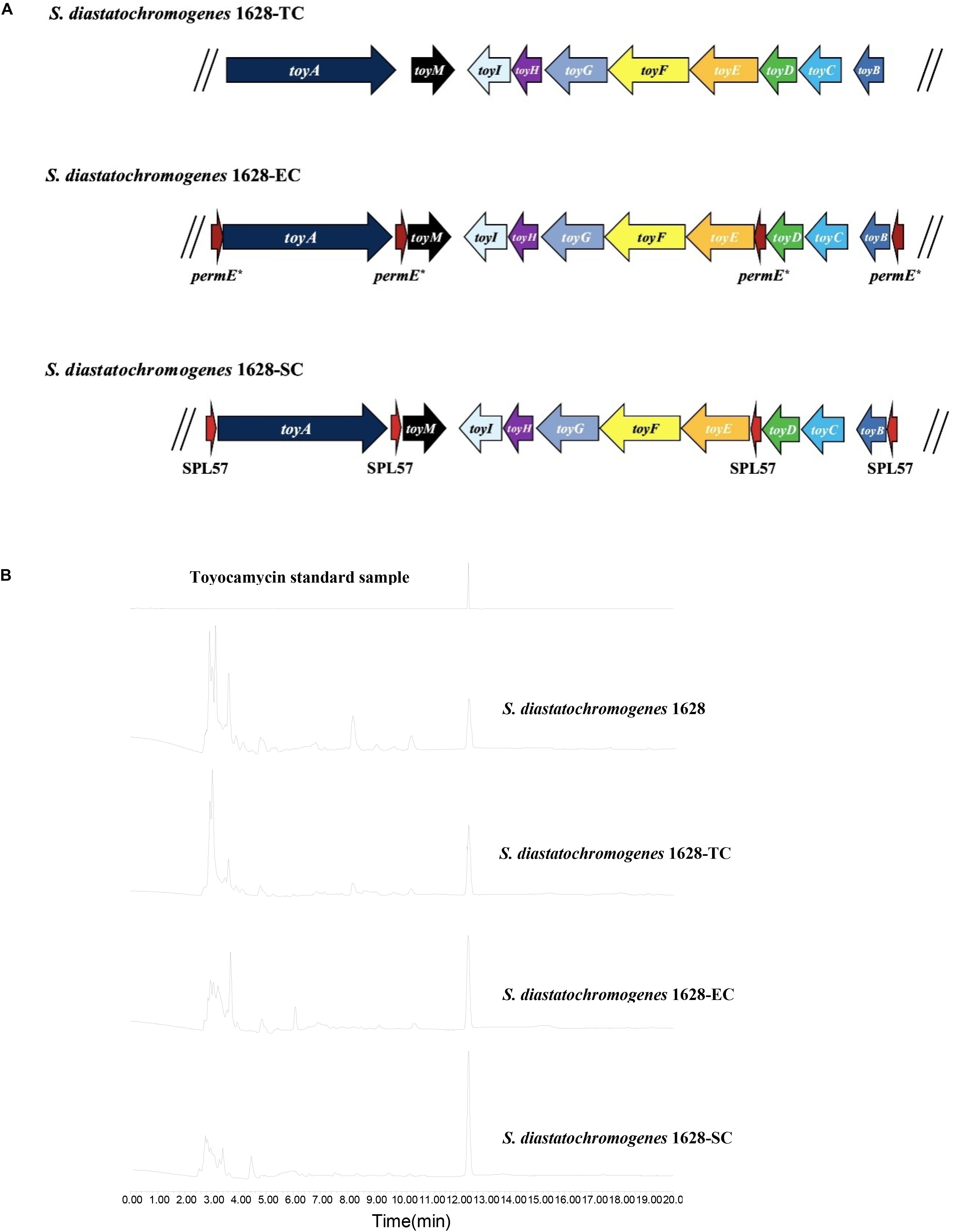
Figure 4. Characteristics of recombinant strains harboring an extra copy of toy cluster and their TM productions. (A) S. diastatochromogenes 1628-TC: S. diastatochromogenes 1628 harboring an extra copy of native toy gene cluster; S. diastatochromogenes 1628-EC: S. diastatochromogenes 1628 harboring an extra copy of engineered toy gene cluster in which four permE* promoters were inserted, the native promoter of toyA gene, toyM gene, toyBD operon and toyEI operon was, respectively, replaced by permE*; S. diastatochromogenes 1628-SC: S. diastatochromogenes 1628 harboring an extra copy of engineered toy gene cluster in which four SPL57 promoters were inserted, the native promoter of toyA gene, toyM gene, toyBD operon and toyEI operon was, respectively, replaced by SPL57. (B) HPLC analysis of TM isolated from fermentation extracts of the recombinant strain S. diastatochromogenes 1628-TC, S. diastatochromogenes 1628-EC, and S. diastatochromogenes 1628-SC.
Quantitative real-time RT-PCR analysis was performed to assess the transcriptional levels of ten toy genes located in the toy cluster in recombinant strain 1628-TC and wild-type strain 1628, after 36, and 72 h of fermentation. Compared with wild-type, the transcriptional levels of all toy genes were greatly increased in the recombinant strain 1628-TC (Figure 5). We speculated that overexpression of native toy cluster thus enhances gene expression at the transcriptional level of all toy genes and further increases TM production in the recombinant strain 1628-TC.
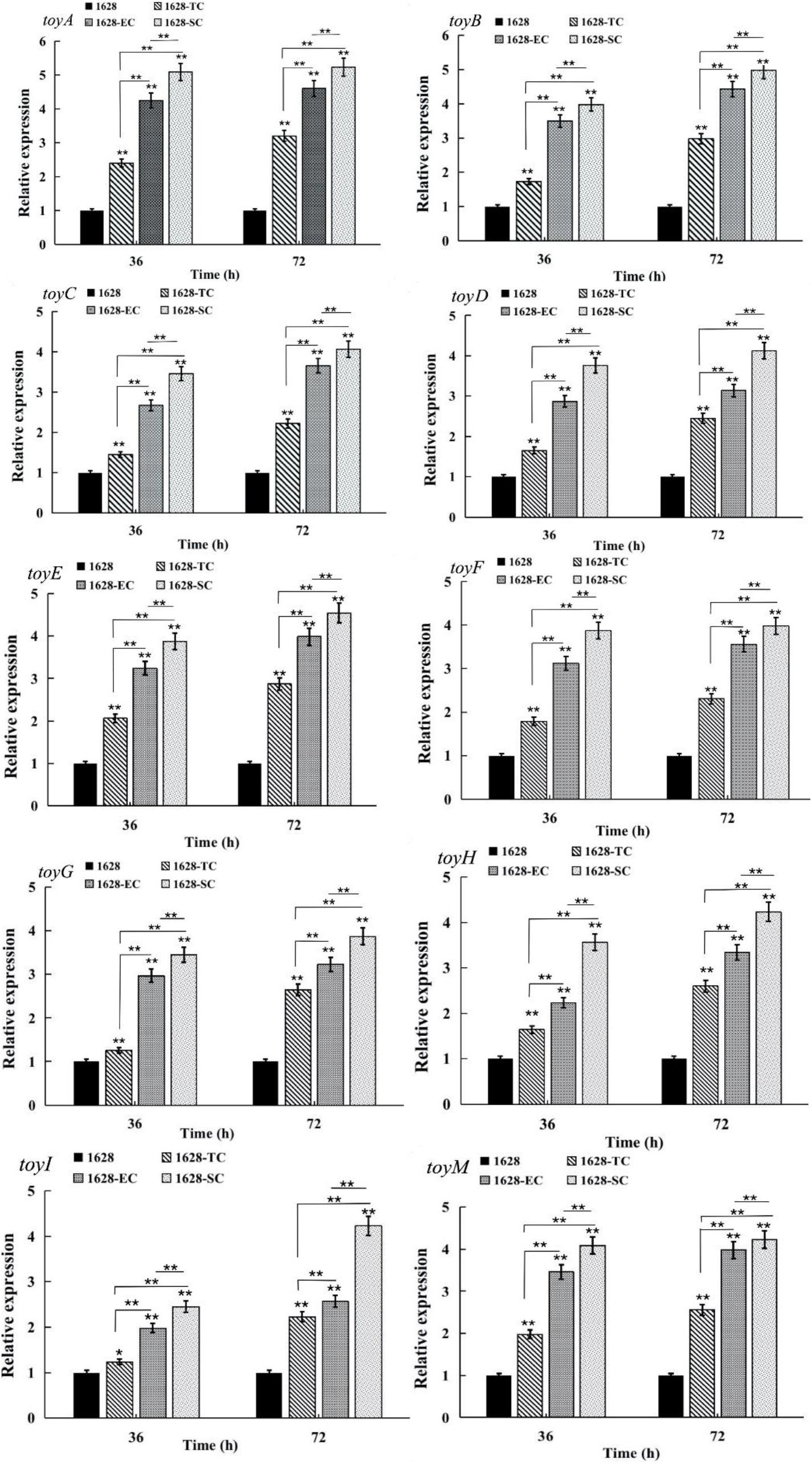
Figure 5. Comparison of the transcription levels of genes involved in TM production in different strains obtained by quantitative reverse transcription-PCR (qRT-PCR). 1628: S. diastatochromogenes 1628; 1628-TC: S. diastatochromogenes 1628-TC; 1628-EC: S. diastatochromogenes 1628-EC; 1628-SC: S. diastatochromogenes 1628-SC. The cells were harvested from the fermentation broth after 36 and 72 h. Error bars were calculated by measuring the standard deviations of the data from three replicates of each sample.(**) indicates highly statistically significant results (P-value < 0.01).
Engineered Toy Cluster and Its Overexpression to Improve TM Production
To overexpress the toy cluster, gain higher transcription levels of all toy genes, and further enhance TM production, we set out to engineer the native toy cluster as presented in Supplementary Figure S5. Briefly, toyA gene, toyM gene, toyBD operon and toyEI operon were placed under the control of constitutive promoter permE∗ or synthetic promoter SPL57. The introduction of the 4 permE∗ or SPL57 promoters and the assembly of four DNA fragments were conducted as described in the section “Materials and Methods,” yielding recombinant plasmids pSET152::ecluster and pSET152::scluster, respectively. Plasmids pSET152::ecluster and pSET152::scluster were introduced into S. diastatochromogenes 1628 by conjugation to generate recombinant strains 1628-EC and 1628-SC (Figure 4A), respectively.
Quantitative RT-PCR analysis revealed that the transcriptional levels of all the toy genes in S. diastatochromogenes 1628-EC and S. diastatochromogenes 1628-SC were markedly increased compared with those in S. diastatochromogenes 1628-TC, harboring an extra copy of native toy cluster (Figure 5). This result suggested that engineering toy cluster through insertion of promoter permE∗ or SPL57 successfully increased transcription of all the toy genes located in the toy cluster. We asked next whether the increased transcription of toy genes could improve the TM production.
Recombinant strains S. diastatochromogenes 1628-EC and S. diastatochromogenes 1628-SC, as well as S. diastatochromogenes 1628 as a control, were studied in shake-flask fermentation. As shown in Figure 6A, overexpression of engineered toy cluster had no significant effect on cell growth compared to that of the wild-type strain S. diastatochromogenes 1628. In contrast, it exhibited great promotional effect on TM production (Figure 4B). After 84 h, S. diastatochromogenes 1628-EC and 1628-SC increased TM levels to 456.5 mg/l and 638.9 mg/l, respectively (Figure 6B).
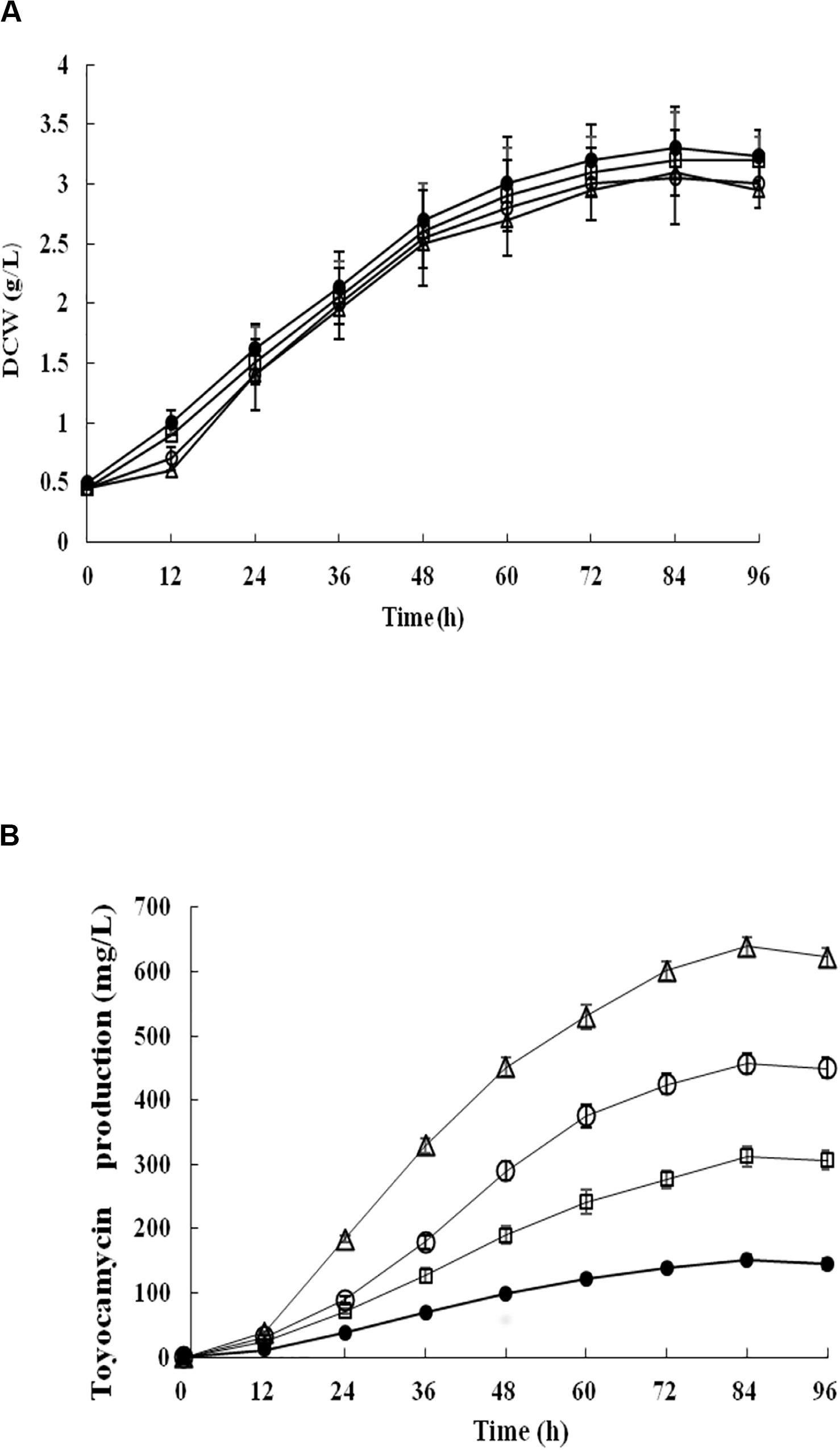
Figure 6. Detection and comparison of cell growth (A) and TM concentration (B) of the wild-type strain S. diastatochromogenes 1628 (filled circle), the recombinant strains S. diastatochromogenes 1628-TC (open square), S. diastatochromogenes 1628-EC (open circle) and S. diastatochromogenes 1628-SC (open triangle) in shake-flask culture experiment. All shake-flask fermentations were carried out in 250 ml flasks with a working volume of 40 ml at 200 rpm and 28°C. The medium was inoculated at 5% (v/v). Error bars were calculated from three different batches of fermentation.
To determine the stability of engineered toy cluster in the chromosome of S. diastatochromogenes 1628, samples, taken from several continuous shake-flask fermentations of two recombinant strains S. diastatochromogenes 1628-EC and 1628-SC in the absence of apramycin, were streaked on the MS agar medium with apramycin (50 μg/ml), and the TM concentration was measured. The results showed that the cells still conferred apramycin resistance and exhibited a similar high level of TM production (data not shown). These results suggest that the integration of an extra copy of an engineered toy cluster in the chromosome of S. diastatochromogenes 1628 is genetically stable.
Discussion
The nucleoside antibiotic TM is highly efficient against a broad range of plant pathogenic fungi. McCarty and Bandarian (2008) were the first to report the involvement of toy genes (toy A to toyM) in TM biosynthesis in Streptomyces rimosus (ATCC 14673). Putative functions of toy genes were analyzed. Four genes (toyD, toyB, toyC, and toyM) are responsible for the biosynthesis of 7-cyano-7-deazaguanine (preQ0). PreQ0 is converted to TM by ToyH, ToyE, ToyG, ToyF, and ToyI (McCarty and Bandarian, 2008; Mccarty et al., 2009; Battaglia et al., 2011). S. diastatochromogenes 1628 was shown to produce TM (Ma et al., 2014a). To support our research on TM biosynthesis, the genome of S. diastatochromogenes 1628 recently was sequenced, and the TM biosynthetic gene cluster (toy cluster) was predicted. Heterologous expression is an effective method to identify relationship between gene cluster and secondary metabolite. Therefore, to check whether the putative toy cluster was required for TM biosynthesis in S. diastatochromogenes 1628, we attempted to express the toy cluster in other Streptomyces spp. host cells as well.
Because of its fast growth and efficient genetic system, the S. albus J1074 strain is one of the most widely used for the heterologous production of bioactive natural products (Kallifidas et al., 2018; Myronovskyi et al., 2018). Therefore, in this study, S. albus J1074 was employed as a host for heterologous expression of the toy cluster. A DNA fragment harboring the entire toy cluster with the natural promoter was cloned and integrated into the chromosome of S. albus J1074 to generate S. albus J1074-TC. The strain was able to produce TM (16.9 mg/l) as shown by HPLC analysis. The lower production rate could be explained either by a lack of positive regulatory network or by an inadequate supply of important precursors. But our result clearly indicates that the toy cluster (GenBank accession No. KY022432) is involved in TM biosynthesis in S. diastatochromogenes 1628. It also suggests that differences in the location and orientation of genes in the known TM cluster do not affect their biological functions. Results were confirmed by gene deletion- and complementation-experiments.
The long-term goal of our research is to improve TM production of S. diastatochromogenes 1628 by rational design. This will eventually lead to an increased TM production and opens the way for more potent and useful TM analogs for industrial application.
Amplification of the entire biosynthetic gene cluster led to an improvement of the production of secondary metabolites in other strains (Du et al., 2015; Liu et al., 2017). Therefore, we constructed a strain (S. diastatochromogenes 1628-TC) harboring an extra copy of the toy cluster. TM production as well as the transcription of all toy genes was higher in this strain compared to that of wild-type strain S. diastatochromogenes 1628. To improve TM production further, we subsequently engineered the toy cluster to increase transcription of all toy genes. In our earlier study, the constitutive promoter permE∗, which is commonly used for gene expression in streptomycetes, was selected to increase the expression of favorable genes to improve TM production in S. diastatochromogenes 1628 (Ma et al., 2014b; Tao et al., 2016). Siegl et al. (2013) developed a synthetic promoter library for actinomycetes based on the −10 and −35 consensus sequences of the constitutive and widely used ermEp1 promoter. The synthetic promoters exhibited a wide range of promoter strengths (2% to 319%) compared with the ermEp1 promoter. Interestingly, promoter SPL-57 was shown to be much stronger than permE∗. Moreover, the synthetic promoter SPL57 showed high activity comparable with that of permE∗ in S. diastatochromogenes 1628 by using GUS as reporter gene (Xu et al., 2017). Recently, overexpression of the pathway-specific regulator toyA separately driven by the promoters permE∗ and SPL57 in S. diastatochromogenes 1628 led to a great increase in TM production compared to the 1628 strain (Xu et al., 2019). Replacement of native promoters with strong constitutive or synthetic promoters became a key strategy, resulting in an increase of gene transcription most probably by overcoming the regulatory barriers of the cell (Hammer et al., 2006; Myronovskyi and Luzhetskyy, 2016; Zhou et al., 2017; Horbal et al., 2018). In this study, the promoters permE∗ and SPL57 were introduced into the cluster resulting inpSET152::ecluster and pSET152::scluster. Notably, in pSET152::ecluster and pSET152::scluster, toy genes are arranged in tandem in the same direction as in the native toy cluster. Only the native promoters of the toyA gene, toyM gene, toyBD operon and toyEI operon were replaced by permE∗or SPL57. Recombinant strains S. diastatochromogenes 1628-EC and 1628-SC increased TM levels to 456.5 mg/l and 638.9 mg/l, respectively, resulting in a 2- and 3.2-fold increase compared to that of wild-type strain S. diastatochromogenes 1628, respectively. Meanwhile, the highest TM level produced by S. diastatochromogenes 1628-EC and 1628-SC was 45.9% and 104.2% higher than that produced by the S. diastatochromogenes 1628-TC (312.9 mg/l), respectively. In addition, we were able to show that toy genes were stronger expressed in these strains than in the wt strain. Based on our results we can conclude that the expression of clusters containing “artificial” promoters is more efficient than the expression of cluster containing the native promoters.
The largest improvement in TM production was detected in recombinant S. diastatochromogenes 1628-SC. This result shows that the promoter SPL57 is more suitable for the expression of the toy gene to improve TM production than the promoter permE∗. A similar phenomenon was also found in our previous study (Xu et al., 2019), in which overexpression of toyA driven by SPL57 exhibited a greater increase in TM production compared to that of toyA driven by permE∗. It is important to mention that co-expression of the whole cluster containing the “artificial” promoters led to a higher TM production than co-expression of only toyA behind an artificial promoter (using permE∗(456.5 mg/l vs. 309.3 mg/l; or SPL57 (638.9 mg/l vs. 456.3 mg/l). The stable integration of the engineered toy cluster in the chromosome of S. diastatochromogenes 1628 did not show any effect on cell growth.
In order to further optimize TM production, the engineered toy cluster was introduced into the resistant (Rifr) mutant S. diastatochromogenes 1628-T15 (Ma et al., 2016). Preliminary results indicate an explosive increase in TM production in this strain (data not shown). We are now planning to further investigate the strain, perform genome engineering (Huang et al., 2015; Li et al., 2017) and a precursor feeding strategy (Zabala et al., 2013; Gubbens et al., 2017; Zheng et al., 2019) to further improve TM production.
Conclusion
This study confirmed that the putative TM biosynthetic gene cluster is responsible for TM biosynthesis in S. diastatochromogenes 1628. Amplification of specially engineered toy cluster greatly improved the TM production in S. diastatochromogenes 1628. This work importantly provides an effective strategy to improve TM production in S. diastatochromogenes 1628, and it can potentially be extended to other antibiotic overproduction strains.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
Author Contributions
ZM designed the research and wrote the manuscript. YH, ZL, JX, and XX conducted the experiments. AB revised the manuscript. XY checked the final version of the manuscript. All authors read and approved the manuscript.
Funding
This work was supported by the Excellent Youth Fund of Zhejiang Province, China (LR17C140002) and the National Natural Science Foundation of China (31772213 and 31972320).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.02074/full#supplementary-material
References
Baltz, R. H. (2016). Genetic manipulation of secondary metabolite biosynthesis for improved production in Streptomyces and other actinomycetes. J. Ind. Microbiol. Biotechnol. 43, 343–370. doi: 10.1007/s10295-015-1682-x
Battaglia, U., Long, J. E., Searle, M. S., and Moody, C. J. (2011). 7-Deazapurine biosynthesis: NMR study of toyocamycin biosynthesis in Streptomyces rimosus using 2-13C-7-15N-adenine. Org. Biomol. Chem. 9, 2227–2232. doi: 10.1039/c0ob01054e
Chen, Y., Smanski, M. J., and Shen, B. (2010). Improvement of secondary metabolite production in Streptomyces by manipulating pathway regulation. Appl. Microbiol. Biotechnol. 86, 19–25. doi: 10.1007/s00253-009-2428-3
Du, D., Wang, L., Tian, Y., Liu, H., Tan, H., and Niu, G. (2015). Genome engineering and direct cloning of antibiotic gene clusters via phage phi BT1 integrase-mediated site-specific recombination in Streptomyces. Sci. Rep. 5:8740. doi: 10.1038/srep08740
Du, D., Zhu, Y., Wei, J., Tian, Y., Niu, G., and Tan, H. (2013). Improvement of gougerotin and nikkomycin production by engineering their biosynthetic gene clusters. Appl. Microbiol. Biotechnol. 97, 6383–6396. doi: 10.1007/s00253-013-4836-7
Gibson, D. G., Benders, G. A., Axelrod, K. C., Zaveri, J., Algire, M. A., Moodie, M., et al. (2008). One-step assembly in yeast of 25 overlapping DNA fragments to form a complete synthetic Mycoplasma genitalium genome. Proc. Natl. Acad. Sci. U.S.A. 105, 20404–20409. doi: 10.1073/pnas.0811011106
Gubbens, J., Wu, C., Zhu, H., Filippov, D. V., Florea, B. I., Rigali, S., et al. (2017). Intertwined precursor supply during biosynthesis of the catecholate-hydroxamate siderophores qinichelins in Streptomyces sp. MBT76. ACS Chem. Biol. 12, 2756–2766. doi: 10.1021/acschembio.7b00597
Hammer, K., Mijakovic, I., and Jensen, P. R. (2006). Synthetic promoter libraries–tuning of gene expression. Trends Biotechnol. 24 (2), 53–55. doi: 10.1016/j.tibtech.2005.12.003
Horbal, L., Marques, F., Nadmid, S., Mendes, M. V., and Luzhetskyy, A. (2018). Secondary metabolites overproduction through transcriptional gene cluster refactoring. Metab. Eng. 49, 299–315. doi: 10.1016/j.ymben.2018.09.010
Huang, H., Zheng, G., Jiang, W., Hu, H., and Lu, Y. (2015). One-step high-efficiency CRISPR/Cas9-mediated genome editing in Streptomyces. Acta Biochim. Biophys. Sin. 47 (4), 231–243. doi: 10.1093/abbs/gmv007
Jiang, L., Wei, J., Li, L., Niu, G., and Tan, H. (2013). Combined gene cluster engineering and precursor feeding to improve gougerotin production in Streptomyces graminearus. Appl. Microbiol. Biotechnol. 97, 10469–10477. doi: 10.1007/s00253-013-5270-6
Jones, S. E., and Elliot, M. A. (2018). ‘Exploring’ the regulation of Streptomyces growth and development. Curr. Opin. Microbiol. 42, 25–30. doi: 10.1016/j.mib.2017.09.009
Kallifidas, D., Jiang, G., Ding, Y., and Luesch, H. (2018). Rational engineering of Streptomyces albus J1074 for the overexpression of secondary metabolite gene clusters. Microb. Cell Fact. 17:25. doi: 10.1186/s12934-018-0874-2
Kemung, H. M., Tan, L. T., Khan, T. M., Chan, K. G., Pusparajah, P., Goh, B. H., et al. (2018). Streptomyces as a prominent resource of future anti-MRSA drugs. Front. Microbiol. 9:2221. doi: 10.3389/fmicb.2018.02221
Kieser, T., Bibb, M. J., Buttner, M. J., Chater, K. F., and Hopwood, D. A. (2000). Practical Streptomyces Genetics. Norwich: John Innes Foundation.
Li, L., Zheng, G., Chen, J., Ge, M., Jiang, W., and Lu, Y. (2017). Multiplexed site-specific genome engineering for overproducing bioactive secondary metabolites in actinomycetes. Metab. Eng. 40, 80–92. doi: 10.1016/j.ymben.2017.01.004
Li, Y., and Tan, H. (2017). Biosynthesis and molecular regulation of secondary metabolites in microorganisms. Sci. China Life Sci. 60, 935–938. doi: 10.1007/s11427-017-9115-x
Liao, G., Li, J., Li, L., Yang, H., Tian, Y., and Tan, H. (2010). Cloning, reassembling and integration of the entire nikkomycin biosynthetic gene cluster into Streptomyces ansochromogenes lead to an improved nikkomycin production. Microb. Cell Fact. 9:6. doi: 10.1186/1475-2859-9-6
Liu, P., Zhu, H., Zheng, G., Jiang, W., and Lu, Y. (2017). Metabolic engineering of Streptomyces coelicolor for enhanced prodigiosins (RED) production. Sci. China Life Sci. 60, 948–957. doi: 10.1007/s11427-017-9117-x
Liu, R., Deng, Z., and Liu, T. (2018). Streptomyces species: ideal chassis for natural product discovery and overproduction. Metab. Eng. 50, 74–84. doi: 10.1016/j.ymben.2018.05.015
Ma, Z., Liu, J., Bechthold, A., Tao, L., Shentu, X., Bian, Y., et al. (2014a). Development of intergeneric conjugal gene transfer system in Streptomyces diastatochromogenes 1628 and its application for improvement of toyocamycin production. Curr. Microbiol. 68, 180–185. doi: 10.1007/s00284-013-0461-z
Ma, Z., Tao, L., Bechthold, A., Shentu, X., Bian, Y., and Yu, X. (2014b). Overexpression of ribosome recycling factor is responsible for improvement of nucleotide antibiotic-toyocamycin in Streptomyces diastatochromogenes 1628. Appl. Microbiol. Biotechnol. 98, 5051–5058. doi: 10.1007/s00253-014-5573-2
Ma, Z., Luo, S., Xu, X., Bechthold, A., and Yu, X. (2016). Characterization of representative rpoB gene mutations leading to a significant change in toyocamycin production of Sreptomyces diastatochromogenes 1628. J. Ind. Microbiol. Biotechnol. 43, 463–471. doi: 10.1007/s10295-015-1732-4
Makitrynskyy, R., Tsypik, O., Nuzzo, D., Paululat, T., Zechel, D. L., and Bechthold, A. (2020). Secondary nucleotide messenger c-di-GMP exerts a global control on natural product biosynthesis in streptomycetes. Nucleic Acids Res. 48, 1583–1598. doi: 10.1093/nar/gkz1220
Martínez-Burgo, Y., Álvarez-Álvarez, R., Rodríguez-García, A., and Liras, P. (2015). The pathway-specific regulator claR of Streptomyces clavuligerus has a global effect on the expression of genes for secondary metabolism and differentiation. Appl. Environ. Microb. 81, 6637–6648. doi: 10.1128/AEM.00916-15
McCarty, R. M., and Bandarian, V. (2008). Deciphering deazapurine biosynthesis: pathway for pyrrolopyrimidine nucleosides toyocamycin and sangivamycin. Chem. Biol. 15, 790–798. doi: 10.1016/j.chembiol.2008.07.012
Mccarty, R. M., Somogyi, A., Lin, G., Jacobsen, N. E., and Bandarian, V. (2009). The deazapurine biosynthetic pathway revealed: in vitro enzymatic synthesis of preQ0 from guanosine-5’-triphosphate in four steps. Biochemistry 48, 3847–3852. doi: 10.1021/bi900400e
Murakami, T., Burian, J., Yanai, K., Bibb, M. J., and Thompson, C. J. (2011). A system for the targeted amplification of bacterial gene clusters multiplies antibiotic yield in Streptomyces coelicolor. Proc. Natl. Acad. Sci. U.S.A. 108 (38), 16020–16025. doi: 10.1073/pnas.1108124108
Myronovskyi, M., and Luzhetskyy, A. (2016). Native and engineered promoters in natural product discovery. Nat. Prod. Rep. 33 (8), 1006–1019. doi: 10.1039/c6np00002a
Myronovskyi, M., Rosenkränzer, B., Nadmid, S., Pujic, P., Normand, P., and Luzhetskyy, A. (2018). Generation of a cluster-free Streptomyces albus chassis strains for improved heterologous expression of secondary metabolite clusters. Metab. Eng. 49, 316–324. doi: 10.1016/j.ymben.2018.09.004
Palazzotto, E., Tong, Y., Lee, S. Y., and Weber, T. (2019). Synthetic biology and metabolic engineering of actinomycetes for natural product discovery. Biotechnol. Adv. 37:107366. doi: 10.1016/j.biotechadv.2019.03.005
Sambrook, J., and Russel, D. W. (2001). Molecular Cloning: A Laboratory Manual, 3rd Edn. New York, NY: Cold Spring Harbor Laboratory.
Siegl, T., Tokovenko, B., Myronovskyi, M., and Luzhetskyy, A. (2013). Design, construction and characterisation of a synthetic promoter library for fine-tuned gene expression in actinomycetes. Metab. Eng. 19, 98–106. doi: 10.1016/j.ymben.2013.07.006
Tan, G., Peng, Y., Lu, C., Bai, L., and Zhong, J. (2015). Engineering validamycin production by tandem deletion of gamma-butyrolactone receptor genes in Streptomyces hygroscopicus 5008. Metab. Eng. 28, 74–81. doi: 10.1016/j.ymben.2014.12.003
Tao, L., Ma, Z., Xu, X., Bechthold, A., Bian, Y., Shentu, X., et al. (2016). Engineering Streptomyces diastatochromogenes 1628 to increase the production of toyocamycin. Eng. Life Sci. 15, 779–787. doi: 10.1007/s00253-019-09959-w
Vicente, C. M., Santos-Aberturas, J., Payero, T. D., Barreales, E. G., De Pedro, A., and Aparicio, J. F. (2014). PAS-LuxR transcriptional control of filipin biosynthesis in S. avermitilis. Appl. Microbiol. Biotechnol. 98, 9311–9324. doi: 10.1007/s00253-014-5998-7
Wang, J., Xu, J., Luo, S., Ma, Z., Bechthold, A., and Yu, X. (2018). AdpAsd, a positive regulator for morphological development and toyocamycin biosynthesis in Streptomyces diastatochromogenes 1628. Curr. Microbiol. 75, 1345–1351. doi: 10.1007/s00284-018-1529-6
Wang, L. Y., Huang, Z. L., Li, G., Zhao, H. X., Xing, X. H., Sun, W. T., et al. (2010). Novel mutation breeding method for Streptomyces avermitilis using an atmospheric pressure glow discharge plasma. J. Appl. Microbiol. 108, 851–858. doi: 10.1111/j.1365-2672.2009.04483.x
Xu, J., Song, Z., Xu, X., Ma, Z., Bechthold, A., and Yu, X. (2019). ToyA, a positive pathway-specific regulator for toyocamycin biosynthesis in Streptomyces diastatochromogenes 1628. Appl. Microbiol. Biotechnol. 103, 7071–7084. doi: 10.1007/s00253-019-09959-w
Xu, X., Wang, J., Bechthold, A., Ma, Z., and Yu, X. (2017). Selection of an efficient promoter and its application in toyocamycin production improvement in Streptomyces diastatochromogenes 1628. World J. Microbiol. Biotechnol. 33:30. doi: 10.1007/s11274-016-2194-1
Yamada, R., Higo, T., Yoshikawa, C., China, H., Yasuda, M., and Ogino, H. (2015). Random mutagenesis and selection of organic solvent-stable haloperoxidase from Streptomyces aureofaciens. Biotechnol. Prog. 31, 917–924. doi: 10.1002/btpr.2117
Zabala, D., Braña, A. F., Flórez, A. B., Salas, J. A., and Méndez, C. (2013). Engineering precursor metabolite pools for increasing production of antitumor mithramycins in Streptomyces argillaceus. Metab. Eng. 20, 187–197. doi: 10.1016/j.ymben.2013.10.002
Zhao, Y., Song, Z., Ma, Z., Bechthold, A., and Yu, X. (2019). Sequential improvement of rimocidin production in Streptomyces rimosus M527 by introduction of cumulative drug-resistance mutations. J. Ind. Microbiol. Biotechnol. 46, 697–708. doi: 10.1007/s10295-019-02146-w
Zheng, J., Li, Y., Guan, H., Zhang, J., and Tan, H. (2019). Enhancement of neomycin production by engineering the entire biosynthetic gene cluster and feeding key precursors in Streptomyces fradiae CGMCC 4.576. Appl. Microbiol. Biotechnol. 103, 2263–2275. doi: 10.1007/s00253-018-09597-8
Zhou, S., Du, G., Kang, Z., Li, J., Chen, J., Li, H., et al. (2017). The application of powerful promoters to enhance gene expression in industrial microorganisms. World J. Microbiol. Biotechnol. 33:23. doi: 10.1007/s11274-016-2184-3
Zhou, T. C., Kim, B. G., and Zhong, J. J. (2014). Enhanced production of validamycin A in Streptomyces hygroscopicus 5008 by engineering validamycin biosynthetic gene cluster. Appl. Microbiol. Biotechnol. 98, 7911–7922. doi: 10.1007/s00253-014-5943-9
Keywords: toyocamycin, Streptomyces diastatochromogenes 1628, cluster, heterologous expression, overproduction
Citation: Ma Z, Hu Y, Liao Z, Xu J, Xu X, Bechthold A and Yu X (2020) Cloning and Overexpression of the Toy Cluster for Titer Improvement of Toyocamycin in Streptomyces diastatochromogenes. Front. Microbiol. 11:2074. doi: 10.3389/fmicb.2020.02074
Received: 10 May 2020; Accepted: 06 August 2020;
Published: 02 September 2020.
Edited by:
Ning-Yi Zhou, Shanghai Jiao Tong University, ChinaCopyright © 2020 Ma, Hu, Liao, Xu, Xu, Bechthold and Yu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiaoping Yu, eXV4aWFvcGluZzE5NjMwMzA2QDE2My5jb20=
 Zheng Ma
Zheng Ma Yefeng Hu
Yefeng Hu Zhijun Liao
Zhijun Liao Jie Xu
Jie Xu Xianhao Xu
Xianhao Xu Andreas Bechthold
Andreas Bechthold Xiaoping Yu
Xiaoping Yu