- Department of Virology, College of Medicine, University of Ibadan, Ibadan, Nigeria
In 2019, 38 million people lived with HIV-1 infection resulting in 690,000 deaths. Over 50% of this infection and its associated deaths occurred in Sub-Saharan Africa. The West African region is a known hotspot of the HIV-1 epidemic. There is a need to develop an HIV-1 vaccine if the HIV epidemic would be effectively controlled. Few protective cytotoxic T Lymphocytes (CTL) epitopes within the HIV-1 GAG (HIV_gagconsv) have been previously identified to be functionally conserved among the HIV-1 M group. These epitopes are currently the focus of universal HIV-1 T cell-based vaccine studies. However, these epitopes’ phenotypic and genetic properties have not been observed in natural settings for HIV-1 strains circulating in the West African region. This information is critical as the usefulness of universal HIV-1 vaccines in the West African region depends on these epitopes’ occurrence in strains circulating in the area. This study describes non-synonymous substitutions within and without HIV_gagconsv genes isolated from 10 infected Nigerians at the early stages of HIV-1 infection. Furthermore, we analyzed these substitutions longitudinally in five infected individuals from the early stages of infection till after seroconversion. We identified three non-synonymous substitutions within HIV_gagconsv genes isolated from early HIV infected individuals. Fourteen and nineteen mutations outside the HIV_gagconsv were observed before and after seroconversion, respectively, while we found four mutations within the HIV_gagconsv. These substitutions include previously mapped CTL epitope immune escape mutants. CTL immune pressure likely leaves different footprints on HIV-1 GAG epitopes within and outside the HIV_gagconsv. This information is crucial for universal HIV-1 vaccine designs for use in the West African region.
Introduction
Thirty-eight million people lived with HIV-1 infection in 2019, with 690,000 AIDS-related deaths. Over 50% of HIV infection and about 63% of its associated deaths occurred in Sub-Saharan Africa (UNAIDS, 2020). The rate of new infections also increased in the region despite a 40% global reduction in new infections since 1998 (UNAIDS, 2020; WHO, 2020). About 69% of these new HIV infections occurred in Western and Central Africa. Nigeria accounts for over 60% of new HIV infections in West and Central Africa. The country also has one of the massive HIV epidemic globally (UNAIDS, 2020; WHO, 2020). Combination antiretroviral therapies (cART) have effectively suppressed viremia to undetectable levels, increasing survival and quality of life. It has also decreased infectiousness in infected individuals (Herout et al., 2016; Hanke, 2019).
However, just about half of infected individuals are on antiretroviral therapy. Sub-Sahara Africa has the lowest access to treatment despite being the highest hit region with the virus (UNAIDS, 2019). Access to treatment is unlikely to increase to optimal levels because of economic, social, and pharmacologic challenges associated with cART use (Hanke, 2019; Ndung’u et al., 2019). Moreover, cART is not curative as persons on treatment have to use drugs to control the emergence of latent reservoir HIV (immune escape and drug-resistant strains) for the rest of their lives (Papuchon et al., 2013; Hanke, 2019). Therefore, to eliminate the HIV epidemic, especially in African countries, there is the need to develop a safe, cost-effective, durable, and accessible HIV-1 vaccine(Ondondo et al., 2016; Ndung’u et al., 2019; Bekker et al., 2020; Lé tourneau et al., 2020).
Induction of broadly neutralizing antibodies is the mainstay of all protective anti-viral vaccines. However, this has been very difficult to generate for HIV-1 because of the virus genes’ plasticity (Ondondo et al., 2016; Hanke, 2019; Ndung’u et al., 2019). Studies have shown that broadly neutralizing antibodies develop very late in infection after the latent reservoir landscape is already established (Burton et al., 2004; Muema et al., 2017). Therefore, at present, two arms of universal HIV-1 vaccine research is ongoing, with a group focusing on generating robust neutralizing antibodies and the other on effective CTL immune responses. Findings from these fields will hopefully be merged into one universal HIV vaccine (Ondondo et al., 2016). Cytotoxic T Lymphocytes (CTL), commonly referred to as CD8 + T cells have been extensively shown to control HIV-1 replication, especially during the early stages of infection (Ndhlovu et al., 2015; Leitman et al., 2016; Ndung’u et al., 2019). Previous studies have associated expansion of CTL with the control of acute infections (Borrow et al., 1994; Koup et al., 1994; Ogg et al., 1998; Goulder and Watkins, 2004; Goonetilleke et al., 2009). Initially, several studies associated CTL control of acute HIV infections with HLA protection (Gillespie et al., 2002; Brumme et al., 2008; Honeyborne et al., 2010; Mori et al., 2014); however, recent studies have shown HIV/AIDS outcomes are better predicted by the (i) magnitude and breadth of CTL responses as well as (ii) presentation of functionally conserved CTL epitopes during the early stages of infection (Balla-Jhagjhoorsingh et al., 1999; Lazaro et al., 2011; Matthews et al., 2012; Kløverpris et al., 2014; Ndhlovu et al., 2015; Radebe et al., 2015; Koofhethile et al., 2016).
Recently, the conserved region T cell-based vaccine strategy was developed (Létourneau et al., 2007; Hanke, 2019). This strategy is aimed at eliciting effective CTL responses by (a) using functionally conserved HIV-1 proteins for vaccine constructs, (b) blocking common HIV immune escape paths, and (c) including epitopes associated with low viral load in untreated people (Ondondo et al., 2016; Hanke, 2019). Using proteomic and bioinformatics methods, which included the Shannon entropy algorithm, 14 highly conserved consensus HIV-1 proteins were assembled into a chimeric vaccine construct (Barouch et al., 2010; Santra et al., 2010; Borthwick et al., 2014; Abdul-Jawad et al., 2016). This construct induced high frequencies of HIV-1 specific T cells capable of inhibiting HIV-1 replication in vitro and rhesus monkeys (Barouch et al., 2010; Abdul-Jawad et al., 2016). The immune coverage of the construct against diverse HIV strains was also noted (Santra et al., 2010). The construct was further developed into just six functionally conserved regions, which spanned six areas, namely, the whole of Gag p24, one part in Gag p15, and four regions of Pol overlapping with protease, polymerase, and integrase (Ondondo et al., 2016). This second-generation vaccine construct was showed to possess strong immunogenicity in mice (Mothe et al., 2015). The construct also elicited CD8 T cells, which correlated with high CD4 T-cell count in untreated patients (Ondondo et al., 2016).
Three conserved epitopes out of the six, namely Gag 240–249, TSTLQEQIGT; Gag 162–172, KAFSPEVIPMF and Gag 203–212, ETINEEAAEW, were shown to be functionally conserved and possessed high immune coverage among diverse HIV-1 strains. One of these epitopes- Gag 162–172, KAFSPEVIPMF is found within the major homology region (MHR) of the HIV-1 Gag gene. CTL epitopes within the MHR are promiscuously presented by similar HLA alleles (Carlson et al., 2012). The MHR, found within the capsid gene, is a conserved motif among retroviruses (Honeyborne et al., 2010; Tanaka et al., 2016). This region is essential in particle assembly and viral infectivity (Reicin et al., 1995). Furthermore, CTL epitopes within the MHR are also hydrophobic, highly immunogenic, and immuno-dominant (Tenzer et al., 2009; Bennett et al., 2010; Kløverpris et al., 2015; Yang et al., 2015).
It is also hypothesized that escape mutations within the MHR are likely to be deleterious to HIV as they seem to be associated with fitness costs (Martinez-Picado et al., 2006; Troyer et al., 2009; Liu et al., 2014). Ondondo et al. and others identified Gag 240–249, TSTLQEQIGT, and Gag 203–212, ETINEEAAEW as functionally conserved epitopes with high coverage within HIV-1 diverse strains (Fischer et al., 2007; Abdul-Jawad et al., 2016; Ondondo et al., 2016). These epitopes also showed robust T cell immune responses when assembled in vaccine constructs and tested in macaques (Mothe et al., 2015; Ondondo et al., 2016). However, despite ongoing research in this field, insufficient data exists on these CTL epitopes’ functionally conserved abilities in early HIV-1 infected individuals’ in real-world scenarios. There is also a dearth of information on the rate of non-synonymous substitutions in these epitopes compared to other epitopes of the HIV-1 GAG region in natural settings. This knowledge gap is prominent in Sub-Saharan African countries where diverse HIV strains circulate.
These countries are also of very high priority in the development of HIV-1 vaccines (Ndung’u et al., 2019). When developed, HIV-1 vaccines must be effective against strains circulating in African countries (Ndung’u et al., 2019). Despite the West African epicenter having one of the highest numbers of diverse circulating HIV-1 strains, very few longitudinal studies on HIV-1 have been reported from the region. Escape due to CTL epitopes, which is driven by frequencies of non-synonymous and synonymous substitutions (Kosakovsky Pond et al., 2008; Garcia-knight et al., 2016), outside the functionally conserved region may be a crucial factor to consider in the design of therapeutic and universal HIV vaccines (Murakoshi et al., 2019). They may also provide opportunities for compensatory mutations on replicative fitness (Crawford et al., 2011).
This study describes non-synonymous substitutions within and without the three functionally conserved epitopes (HIV_gagconsv) of HIV-1 GAG genes isolated from 10 infected Nigerians at the early stage of HIV-1 infection. These individuals were part of a previously described study (Olusola et al., 2017). Furthermore, using phylogenetic tools, programs and databases in the Los Alamos National Laboratory HIV Sequence Database1, we analyze these substitutions longitudinally in five infected individuals from the early stages of infection until after seroconversion.
Methods
Study Sites and Patient Population
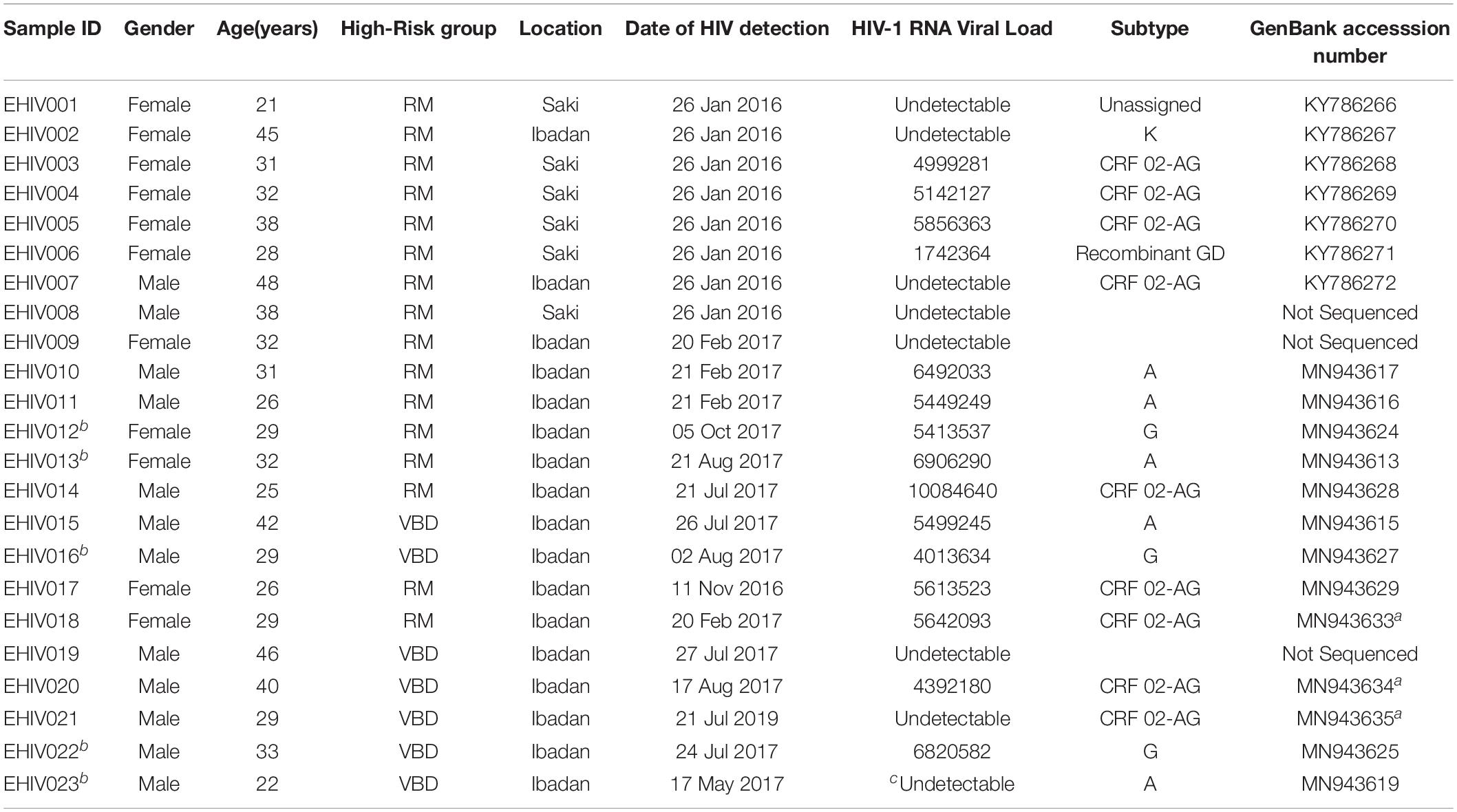
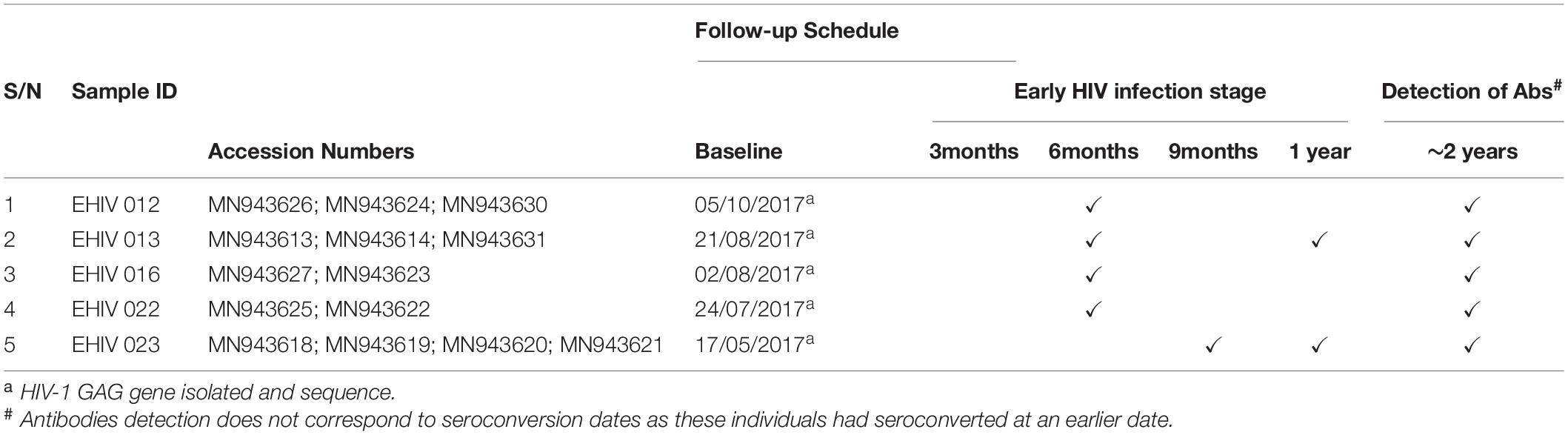
Twenty-three individuals at the early stages of HIV-1 infection identified and described previously were recruited for this study (see Table 1). Ten out of these individuals were studied for early HIV-1 infection. Another five individuals identified in 2017 were followed up until after seroconversion. The profile of the follow-up schedule is shown in Table 2. These individuals were screened for HIV-1 infection at every visit using the earlier described protocol (Olusola et al., 2017, 2020b).
Recruitment of Participants, Sample Collection, and Processing
Participants were recruited for this study after obtaining informed consent. Experiments were conducted with the understanding and the consent of participants. Socio-demographic data of participants were collected using a structured questionnaire. Feedback on results was provided within a week of sample collection. Individuals were counseled and encouraged to continue the presentation for testing at the scheduled intervals. Five milliliters of whole blood were collected in EDTA bottles from participants at every visit. Plasma was separated from the samples immediately after collection, stored at −20°C, and transported in a cold chain to a central laboratory for analysis. The samples were then stored at −80°C until analyzed. Blood samples were analyzed for HIV antigen/antibody, serum creatinine, HIV-1 RNA viral load (at baseline), and HIV-1 GAG DNA.
Identification of Early HIV-1 Infection and Detection of Antibodies
The updated CDC algorithm of laboratory testing for the diagnosis of early and chronic HIV-1 infection was used for this study, as previously described (Olusola et al., 2017).
Clinical Chemistry Assay for Serum Creatinine
Plasma samples were analyzed on a Roche cobas® C11 blood chemistry analyzer (Roche Diagnostics, Indianapolis, United States). Each sample was analyzed to determine the level of serum creatinine, according to the manufacturer’s instruction. Normal reference ranges for plasma creatinine is 62–133 μmol/L.
HIV-1 RNA Viral Load Testing
Serum samples collected at baseline were tested for Plasma HIV-1 viral load (copies/ml) using the COBAS® Ampliprep/COBAS TaqMan96®HIV-1 Test, v2:0 (Roche Molecular Diagnostics, Branchburg, NJ, United States) according to manufacturer’s instruction or by an in house real-time PCR protocol. Briefly, the in-house real-time PCR protocol entails a two-step reaction. First, reverse transcription PCR for cDNA which is a 25 μL reaction utilizing 5 μL of extracted RNA, 12.5 μL of 2X superscript III-RT polymerase reaction mix, and 1 μL superscript III RT/Platinum Taq high fidelity mix (Jena Bioscience, Jena, Germany) as well as random hexamers (1 μL) and RNase free water (5.5 μL). Thermal cycling was performed at 20°C for 10 min, 45°C for 30 min, 70°C for 15 min and an RNase H step of 37°C for 20 min using Applied Biosystem 7500 Fast Real-Time PCR system (Thermo Fisher Scientific, MA, United States). The second stage involved a quantitative real-time PCR targeting a 140bp Nef- Env region of HIV-1. The 20 μL reaction utilized 5 μL of cDNA, 10 μL of qPCR SYBR Master UNG (Jena Bioscience, Jena, Germany) as well as 0.6 μL each of Nef8343 (ATGGGTGGCAAGTGGTCAAAAG) (Tcherepanova et al., 2008) and Env3out (TTGCTACTTGTGATTGCTCCATGT) primers (Keele et al., 2008). Inqaba Biotechnology, South Africa synthesized the primers, and thermal cycling was performed at 50°C for 2 min, 95°C for 2 min and then 35cycles of 95°C for 15 s, 55°C for 20 s using Applied Biosystem 7500 Fast Real-Time PCR system (Thermo Fisher Scientific, MA, United States). Quantitation standards were used to interpolate the quantitative values of the HIV-1 RNA viral load for samples.
PCR Amplification and Sequencing of the GAG Gene
Total DNA was extracted from whole blood samples collected at each visit using guanidium thiocyanate in house protocol. A fragment of the gag-pol region (900 base pairs) of the virus was amplified using previously published primers and cycling conditions by Gall 2012 (Gall et al., 2012) with slight modifications. Briefly, PCR was performed using platinum TaqDNA High fidelity polymerase (Jena Bioscience). Each 25 μl reaction mixture contained 12.5 μl reaction mix (2x), 4.5 μl RNase-free water, 1 μl each of each primer (20 pmol/μl), 1 μl Platinum Taq DNA High Fidelity mix, and 5 μl of template DNA. Pan-HIV-1_1R (CCT CCA ATT CCY CCT ATC ATT TT) and Pan-HIV-1_2F (GGG AAG TGA YAT AGC WGG AAC) were used. Cycling conditions were 94°C for 5 min; 35 cycles of 94°C for 15 s, 58°C for 30 s, and 68°C for 1 min 30 s; and finally, 68°C for 10 min. Positive HIV samples that were undetectable using the above-stated primers were retested using another set of GAG primers for nested PCR as described previously (Vidal et al., 2000). Positive PCR reactions were shipped on ice to Macrogen, South Korea, for Big Dye sequencing using the same amplification primers (Pan-HIV-1_1R and Pan-HIV-1_2F; or G60 andG25).
Detection of HIV-1 Subtypes and Phylogenetic Analysis
The sequences were cleaned and edited using Chromas and Bioedit software. Subtyping was performed using a combination of four subtyping tools: The Rega HIV-1 Subtyping Tool, version 3.02, Comet, version 2.23, National Center for Biotechnology Information, Bethesda, MD4 and jpHMMM: Improving the reliability of recombination prediction in HIV-15. The first three tools were used simultaneously, while jpHMMM was used to resolve discordant subtypes. Phylogenetic analyses were performed using MEGA software version 10. Alignment of sequences was performed using MAFFTS online software. Genetic distances were inferred using the Tamura-Nei model, and a phylogenetic tree was generated using the maximum likelihood method. The robustness of the tree was evaluated with 1000 bootstrap replicates. All consensus nucleotide sequences obtained in this study were submitted to GenBank database and assigned accession numbers MN943617-635.
Non-synonymous Substitutions in Cytotoxic T Lymphocytes (CTL) Epitopes Within HIV_gagconsv of HIV-1 GAG Gene Isolated From 10 Early Infected Individuals
Reference GAG sequences for subtypes G, A, and CRF02-AG were downloaded from the Los Alamos National Laboratory HIV Sequence Database6. Deduced amino acid (aa) sequences were translated for both reference and sample sequences with the standard genetic code using Bioedit software. CTL epitope corresponding to the three highly conserved sites (HIV_gagconsv), namely KAFSPEVIPMFSALSEGATPQD, DTINEEAAEWDR, and TSTLQEQIR (Yang et al., 2015; Ondondo et al., 2016; Hanke, 2019; Lé tourneau et al., 2020), were used for comparison and identification of amino acid substitution. HIV-1 GAG sequences identified as subtypes A, G, and CRF02_AG in this study were aligned with Reference A(GenBank accession numbers DQ676872; AB253421 and AB253429), G(GenBank accession numbers AF084936; AF061641; U88826 and AY612637), and CRF02_AG(GenBank accession numbers L39106 and DQ168578) sequences respectively.
Non-synonymous Substitutions in Cytotoxic T Lymphocytes (CTL) Epitopes Outside the HIV_gagconsv of HIV-1 GAG Gene
The Virus Epidemiology Signature Patterns Analysis7 program was used to identify variations in other sites HIV-1 GAG sequence outside the HIV_gagconsv corresponding to CTL epitope regions (Korber and Myers, 1992). Already defined CTL epitope in HIV-1 database can be found in https://www.hiv.lanl.gov/content/immunology/ctl_search. The VESPA program is a user-friendly amino and nucleic acids signature pattern analysis tool. The program can calculate numbers of variations in an amino acid sequence relative to background sequence(s) using bioinformatics algorithms. By selecting positions where the most common character in a query set differs from that in the background set, differences between groups of sequences can quickly be detected. This analysis invariably can also identify conserved sequence signature patterns. The frequencies of distinguishing amino acids in each set can also be determined. The program has previously been used for HIV-1 and Chikungunya sequence analysis (Ou et al., 1992; Salvatierra and Florez, 2017). This program can also distinguish non-synonymous substitutions from synonymous substitutions based on the threshold settings. Similarity scores or thresholds represent Hamming distances (Nowak et al., 1991) or the number of point mutations between two aligned sequences, calculated using the score: (1 — D) X 100% where D is the hamming distance. This algorithm has previously been described in Nowak et al. (1991), Korber and Myers (1992).
With a threshold setting of 100%, only non-synonymous substitutions that are not due to chance were reported in this study. Sequences were aligned using CLC Main Workbench version 6.5, after which VESPA analysis was performed.
Deduced amino acid (aa) sequences were translated for both reference and sample sequences with the standard genetic code using Bioedit. HIV-1 GAG sequences identified as subtypes A, G, and CRF02_AG in this study were aligned with Reference A (GenBank accession numbers DQ676872; AB253421 and AB253429), G (GenBank accession numbers AF084936; AF061641; U88826 and AY612637), and CRF02_AG (GenBank accession numbers L39106 and DQ168578) sequences respectively. Only amino acid replacements with 100% non-synonymous substitution between reference sequences and sample sequences were considered. Percentage substitution rates were calculated by finding the ratio of the number of substitutions to the total possible substitution sites. These mutations were compared with the Los Alamos National Laboratory HIV Immunology Database for CTL/CD8 + Epitope Variants and Escape Mutants8.
Ethical Approval
This research was conducted following the declaration of Helsinki. Experiments were conducted with the understanding and the consent of each participant. Ethical approvals for this research were obtained from the University of Ibadan/University College Hospital (UI/UCH) Research and Ethics Committee (UI/EC/15/0076) and the Oyo State Ministry of Health Committee on Human Research (AD13/479/951). All results were delinked from patient identifiers and anonymized.
Eligibility/Exclusion Criteria
Only individuals between 18 and 65 years of age were included in the study. Individuals who already knew their HIV status were excluded from the study.
Data Management and Statistical Analysis
Statistical analyses were performed using SPSS version 20. Data are expressed as means ± standard deviations. Statistical significance was estimated using the Kruskal-Wallis test, with SPSS package version 12.0, while Statistical significance was defined as P-values = 0.05.
Results
Participants’ Characteristics
Twenty-three individuals were identified to be at the early stages of HIV-1 infection. Figure 1 shows the phylogeny of HIV-1 subtypes. Out of the ten early infected persons studied, five were infected with Subtype A, three with subtype G, and the rest CRF02-AG. Five individuals at the early stages of HIV-1 infection were followed up until after seroconversion. However, samples in which antibodies were detected were collected after these individuals had seroconverted. The participants were identified to be at the early stages of infection at different periods in 2017. One individual in October, July, and May each and two in August. Three of the individuals were males and were voluntary blood donors. The remaining two females were identified when referred for malaria antigen test. Three individuals were infected with HIV-1 subtype G, while the other two were infected with subtype A (Table 1).
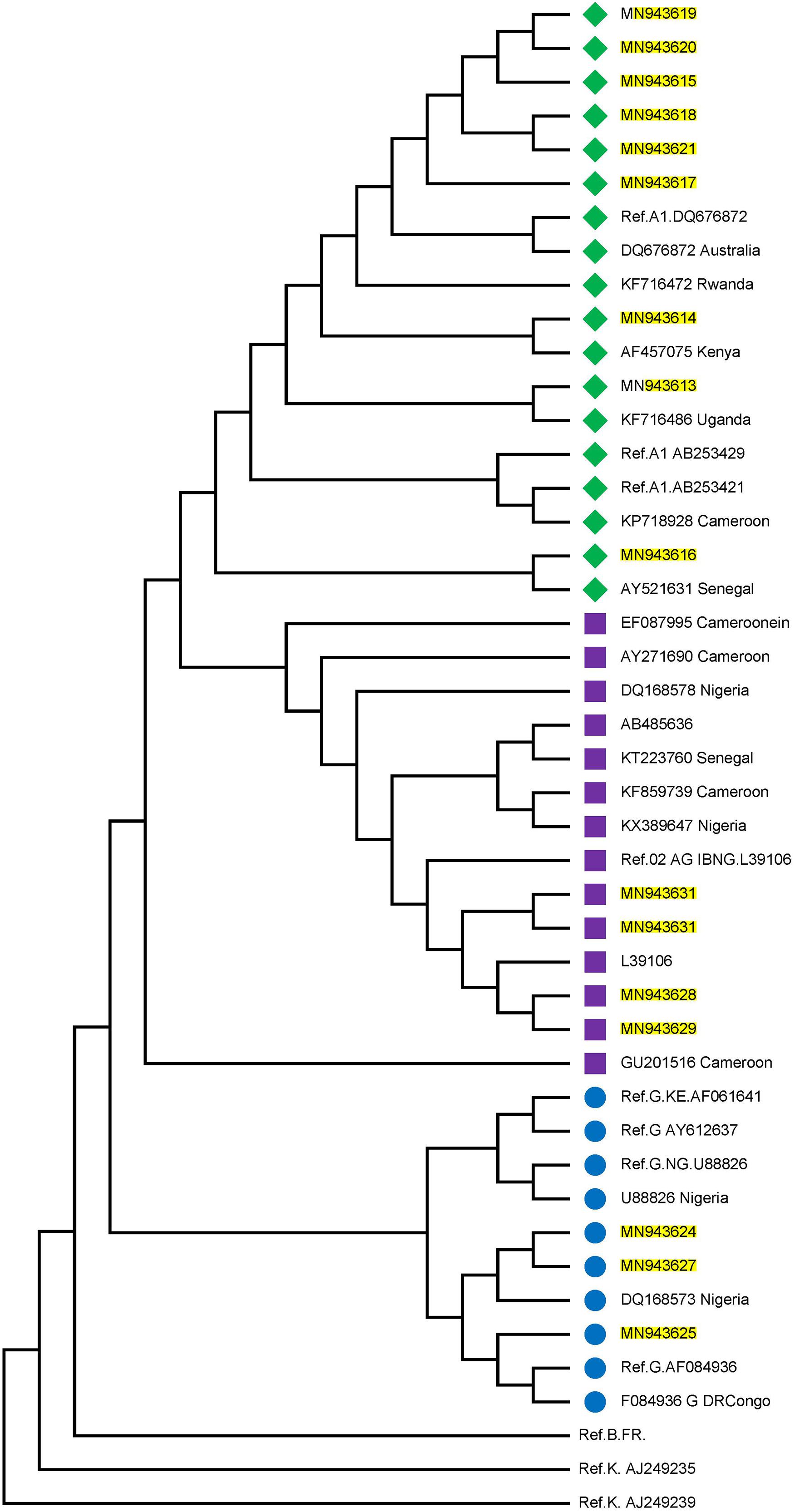
Figure 1. Phylogenetic tree of the P17/P24 regions of the GAG gene of HIV-1. Reference subtypes are indicated with Ref. before their accession numbers. Other sequences are shown with their accession numbers and country of isolation. Subtypes obtained from samples in this study are shown with their accession numbers only. Subtypes A, G, and CRF02_AG were identified with green diamond, blue circle, and pink square symbols respectively. Multiple sequence alignment and phylogenetic tree were constructed using MAFFTS and Maximum Parsimony algorithm in MEGA 6 software. Statistical significance of the tree topology was tested by 1000 bootstrap replication.
Table 2 shows the analysis of the samples collected from these individuals from baseline until after seroconversion. In four individuals, blood samples were collected at three time points, twice during the early stages of infection and once after seroconversion. Samples were collected four times in the fifth individual, thrice during the early stages of infection and once after seroconversion. HIV-1 GAG sequences of these infected individuals were determined at every time point of blood collection.
Phylogenetic Analysis
Figure 1 shows the estimated phylogeny of HIV-1 subtypes with respect to reference sequences available in the HIV Los Alamos National HIV Sequence database. As shown in the Figure, Subtypes A, G, and CRF02_AG were identified with green, blue, and pink symbols, respectively. HIV-1 subtypes A identified in this study were closely related to Ref A1 DQ676872 (from Nigeria) and subtypes AF457075, KF716486 and AY521631 from Kenya, Uganda and Senegal, respectively. Those identified as subtypes G and CRF02-AG were closely related to the Nigerian subtypes DQ168573 and Ref.02 AG IBNG. L39106 respectively.
Substitutions in HIV_gagconsv of CTL Epitope Regions of Subtype A HIV-1 GAG Gene During Early Infection
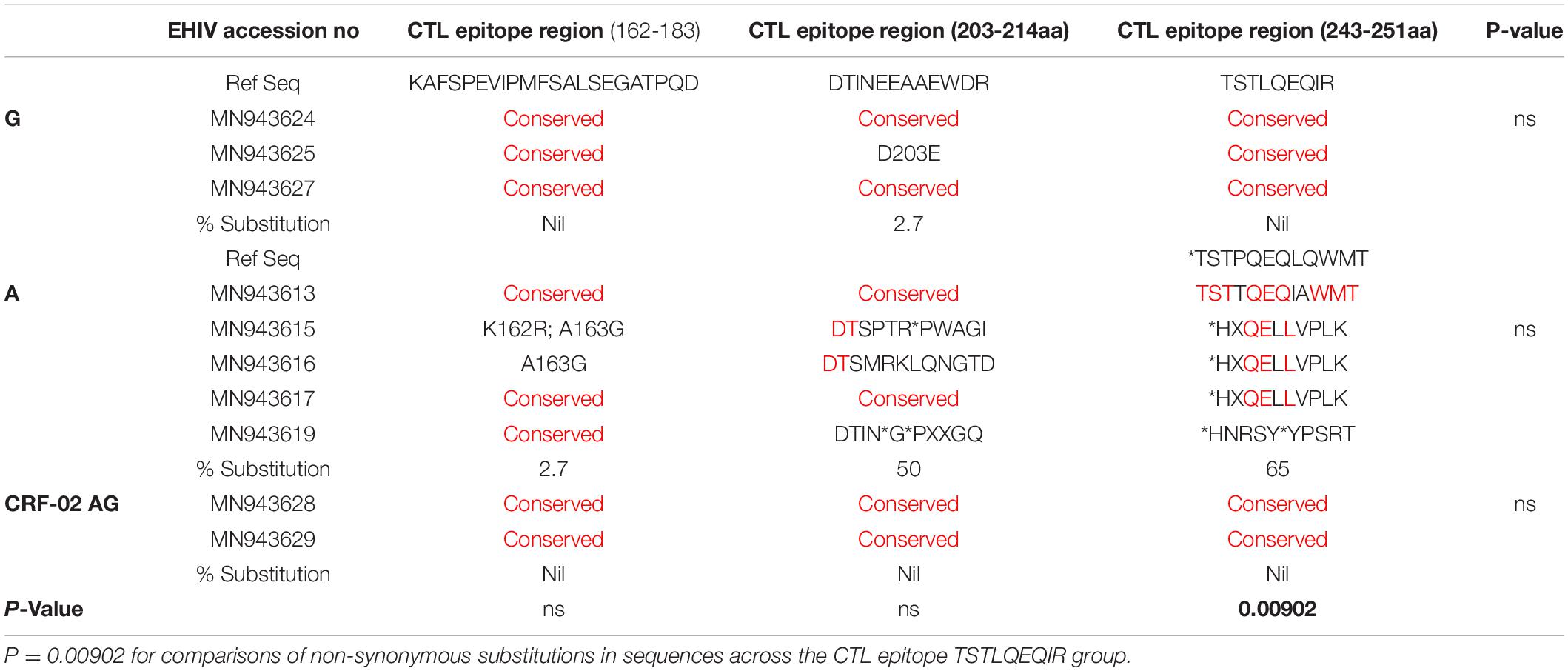
We compared intra and inter variations among 10 HIV-1 GAG sequences isolated from persons at the early stage of infection. These sequences were grouped by subtypes and analyzed alongside reference sequences. As shown in Table 3, variations occurred mostly in HIV-1 Subtype A at the CTL epitope region of 243-251aa. The conserved epitope of TSTLQEQIR was not found in both the reference subtypes and those from early infected individuals. HIV-1 Subtype A also had the highest variations (50%) for HIV_gagconsv corresponding to the CTL epitope region (203-214aa). Although the HIV_gagconsv for CTL epitope region 162-183aa was the most conserved among the subtypes, a substitution rate of 2.7% was found for HIV-1 subtype A isolates. The CTL escape region (162-183) KAFSPEVIPMFSALSEGATPQD had the lowest frequency of mutations. However, sample MN943615 had two mutations-K162R and A163G, while MN943616 had mutation A163G only.
Sequences Isolated After Seroconversion Are Associated With a Higher Rate of Substitutions in HIV_gagconsv of CTL Epitope Regions
Substitutions in the HIV_gagconsv of subtype B HIV-1 GAG gene have been shown to come with a fitness cost. These substitutions have shown to be deleterious to HIV-1 strains’ eventual survival and transmission. However, minimal information exists on substitutions associated with immune epitopes during early non-subtype B HIV-1 infection. We followed five individuals from the early stage of infection till after seroconversion. Three of these individuals were infected with subtype G, while the remaining two were subtypes A (see Table 4). There were no substitutions in CTL epitope regions for EHIV016, while EHIV022 had single aa substitutions before and after seroconversion.
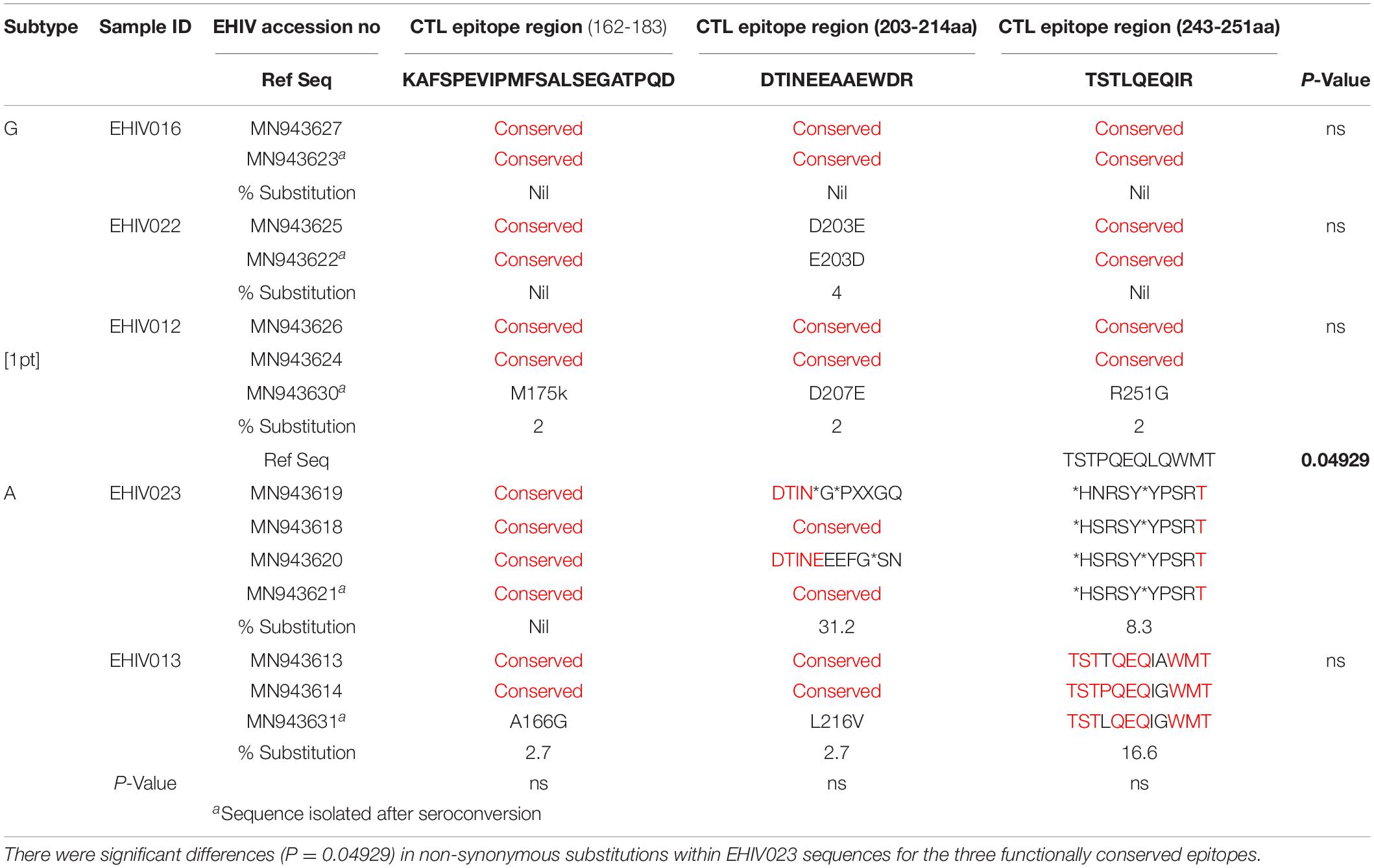
Table 4. Substitutions associated with HIV_gagconsv in HIV-1 GAG gene isolated from 5 individuals followed from early HIV infection till after seroconversion.
EHIV012 had single aa substitutions each after seroconversion in the three CTL epitope regions studied. These substitutions have not been reported before, to the best of our knowledge. For HIV-1 subtype A samples, EHIV023 had very high substitution rates in the CTL epitope region of 203-214aa (31.2%), although there were reversions after seroconversion. Amino acid substitutions were also observed in the CTL epitope region spanning 243-251aa (8.3%). Significant differences (P = 0.04929) in substitution rates before and after seroconversion across two of the three CTL epitope regions were observed for the EHIV023 sample (see Table 4). Substitutions were more associated with the HIV-1 GAG gene sequenced after seroconversion for EHIV013 across the three CTL epitope regions studied. The two amino acid substitutions, A166G and L216V, observed in EHIV013, occurred after seroconversion and were not previously reported.
Amino Acid Signature Patterns in Variable Sites of HIV-1 GAG Genes
In this study, only sequences with non-synonymous substitutions compared to the reference sequence were analyzed. In Table 5, these substitutions were compared between the reference sequence and sequences isolated from individuals in the early stages of infection. In contrast, in Tables 6, 7, the substitutions were compared within sequences isolated per sample spanning early HIV infection till after seroconversion. As shown in Table 5, non-synonymous substitutions were mostly observed in subtype G. However, a substitution, E105K, observed in CRF02_AG had been previously identified as a variant not recognized by the HXB2 epitope (Li et al., 2007). Subtype A had a frameshift mutation at aa85-88. There were no substitutions in sequences isolated from sample EHIV016, while EHIV 012 had the highest substitution within Subtype G sequences (see Table 6). EHIV022 and EHIV 012 had substitutions within sequences at aa positions of 106 and 110-113.

Table 5. Frequency of non-synonymous substitutions in sites outside the HIV_gagconsv of HIV-1 GAG gene amino acids.
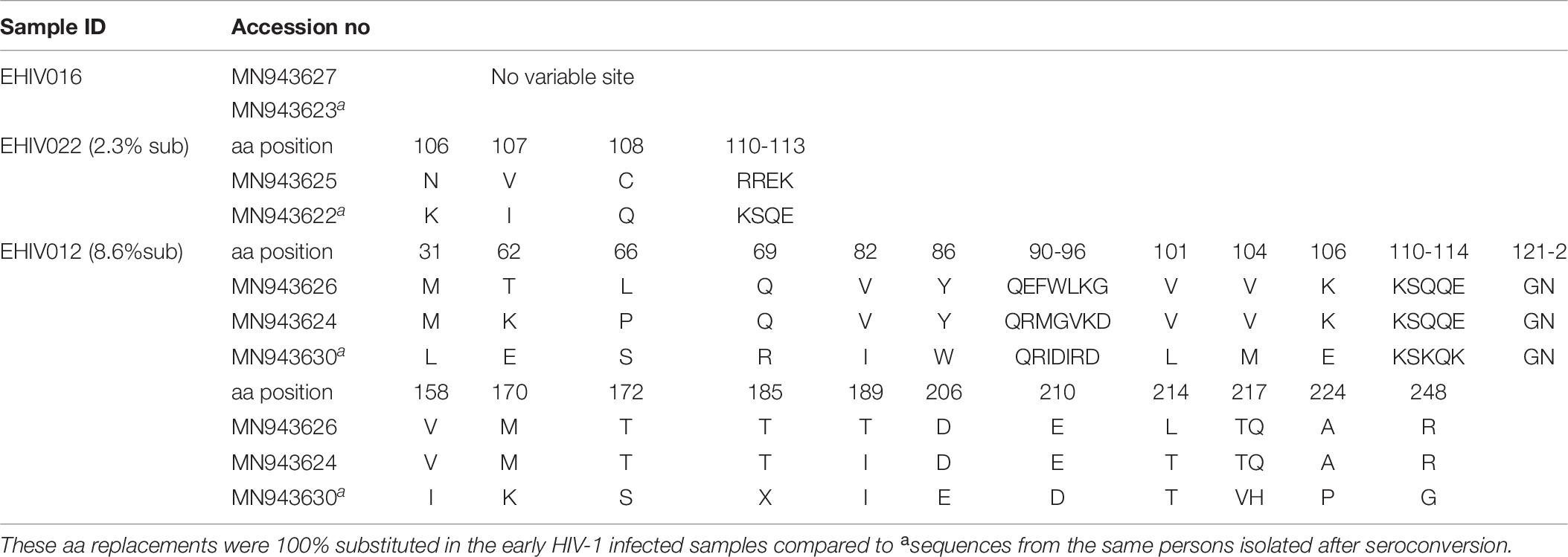
Table 6. Frequency of non-synonymous substitutions in variable sites outside the HIV_gagconsv of HIV-1 Subtype G GAG gene isolated from 3 individuals followed up from early infection till after seroconversion.
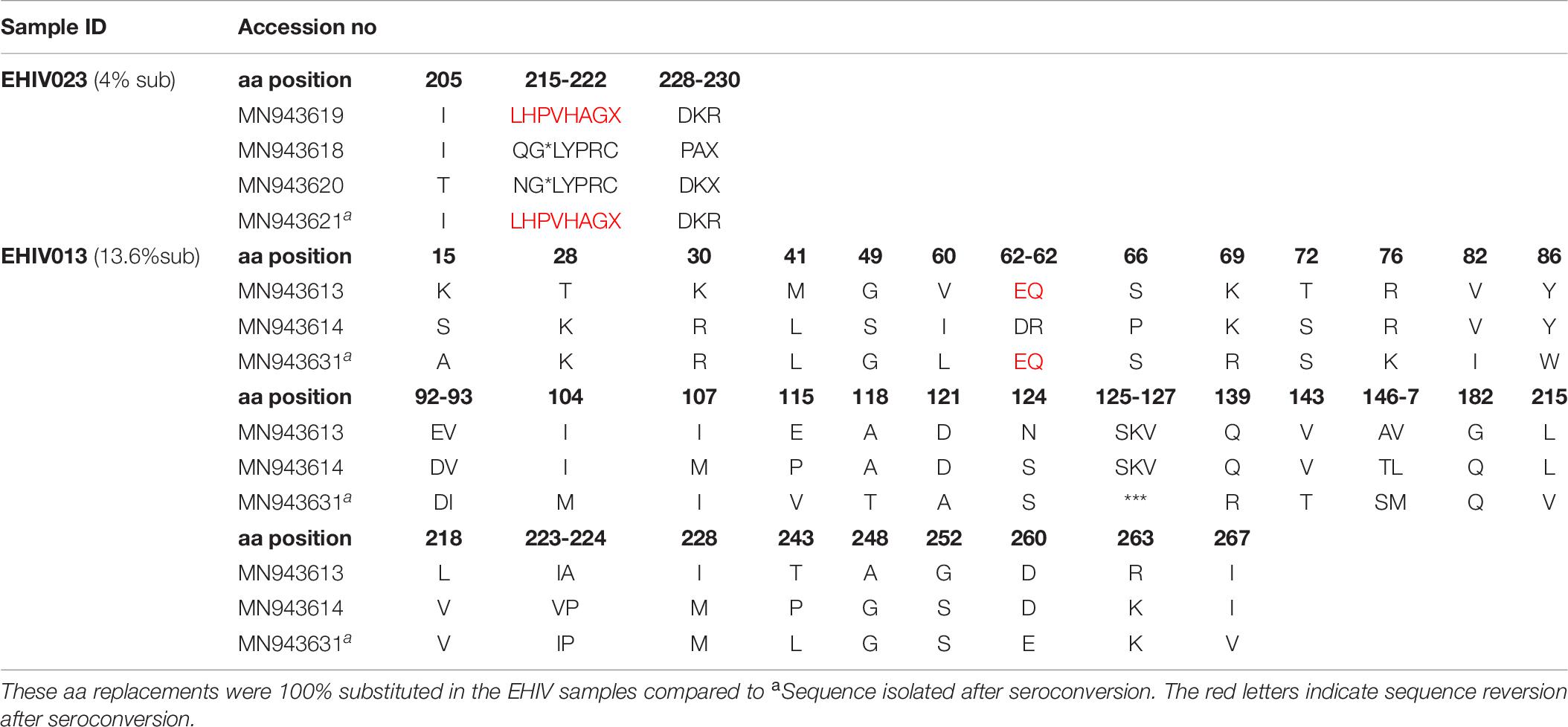
Table 7. Frequency of non-synonymous substitutions in variable sites outside HIV_gagconsv of HIV-1 Subtype A GAG gene isolated from 2 individuals followed up from early infection till after seroconversion.
Lysine was the commonest aa used at position 106 (3/5), while a single substitution was observed for glutamate and asparagine. KSQ was the commonest aa usage in positions 110-113 (3/5); other aa in these positions were RRE (1/5) and KSK (1/5). In Table 7, non-synonymous substitutions associated with HIV-1 subtype A are described. EHIV013 had the highest number of substitutions spanning aa region 15 to 267 in this study. Samples EHIV023 and EHIV013 had substitutions within sequences at aa positions 215, 218, and 228. In position 215, Leucine was the highest aa used (4/7), while glutamine, asparagine, and valine were present in one sequence each. Valine (4/7) and Leucine (3/7) were the only aa used in position 218. Aspartate (3/7) was the highest aa used in position 228, while methionine (2/7), proline (1/7), and isoleucine (1/7) were also present in some isolates.
Non-synonymous Substitutions Associated With Immune Escape Variants Are More Within Epitopes Outside the HIV_gagconsv
Although four substitutions (E105K/CRF02AG; E203D/subtype G; K162R and A163G/subtype A) previously associated with immune escape were observed in HIV_gagconsv, more substitutions were found in regions outside the HIV_gagconsv. Out of the five individuals followed up, two had substitutions previously associated with immune escape strains in GAG gene sites outside the HIV_gagconsv (see Figures 2, 3). Most mutations associated with previously described immune escape strains were identified after seroconversion in this study. Fourteen mutations in 16 HIV-1 GAG sites outside the HIV_gagconsv were identified before seroconversion, while 21 mutations in 23 HIV-1 GAG sites outside the HIV_gagconsv were identified after seroconversion. As shown in Figures 2, 3, EHIV012 had three mutations before (L31M, L101V, and S172T) and after (V82I, Y86W, and F172S) seroconversion. H28K, M30R, A224P, and A248G mutations were identified pre and post seroconversion, while V82I and Y86W mutations were identified only after seroconversion.
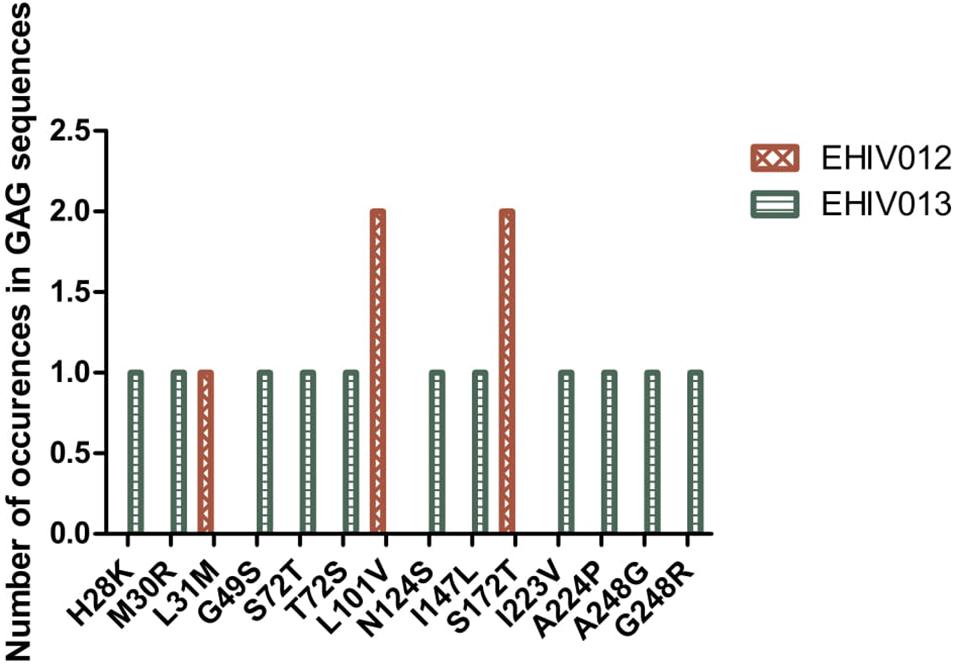
Figure 2. Distribution of mutations associated with escape in EHIV012 and EHIV013 before seroconversion. Three individuals had mutations previously associated with escape, diminished responses, non-susceptible forms, etc. These mutations were compiled from the Los Alamos National Laboratory HIV Immunology Database for CTL/CD8 + Epitope Variants and Escape Mutations8. The list of all the identified mutations is presented in Supplementary Table 1. Two individuals (EHIV012 and EHIV013) had mutations outside the GAG HIV_GAGCONSV, while EHIV022 had a mutation corresponding to escape (Murakoshi) – E203D. Figure 3 shows the distribution of mutations associated with escape in EHIV 012(Red Bars) and EHIV013 (Green Bars). The number of occurrences of the mutations in GAG sequences is shown in the Y-axis while the X-axis shows aa mutations.
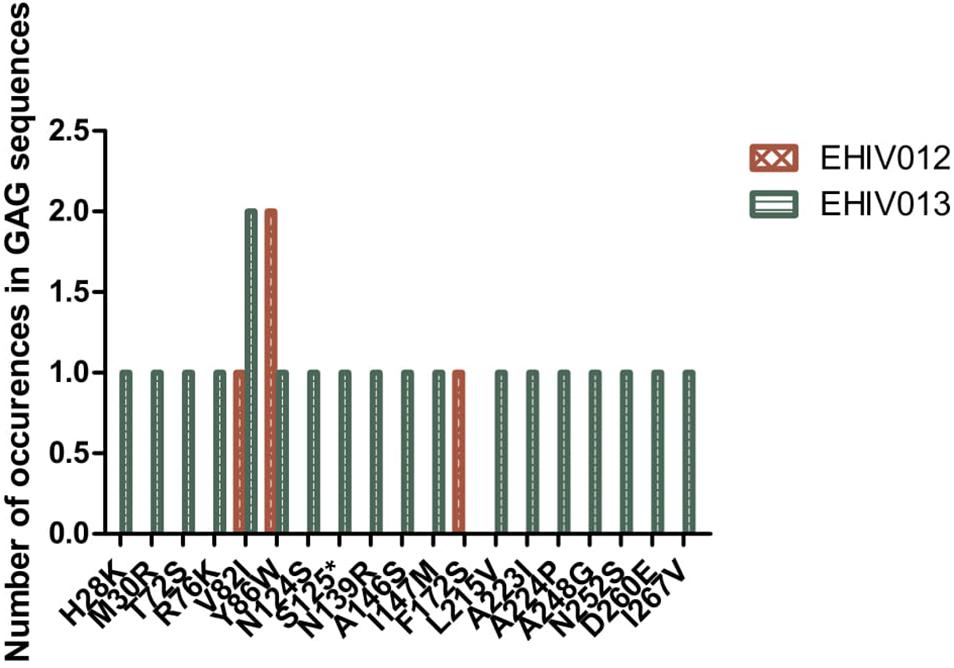
Figure 3. Distribution of mutations associated with escape in EHIV012 and EHIV013 after seroconversion. Three individuals had mutations previously associated with escape, diminished responses, non-susceptible forms, etc. These mutations were compiled from the Los Alamos National Laboratory HIV Immunology Database for CTL/CD8 + Epitope Variants and Escape Mutations8. The list of all the identified mutations is presented in Supplementary Table 1. Two individuals (EHIV012 and EHIV013) had mutations outside the GAG HIV_GAGCONSV, while EHIV022 had a mutation corresponding to escape (Murakoshi) – E203D. Figure 4 shows the distribution of mutations associated with escape in EHIV 012(Red Bars) and EHIV013 (Green Bars). The number of occurrences of the mutations in GAG sequences is shown in the Y-axis while the X-axis shows aa mutations.
Serum Creatinine Concentration During Longitudinal Follow Up
We had earlier reported high levels of creatinine among HIV-infected Africans and African-Americans during the early stages of infection (Olusola et al., 2017, 2020b). In one of these studies (Olusola et al., 2020b), we also showed that immune activation of CTL was ongoing and correlated positively with high creatinine levels in early HIV-1 infected individuals. However, the effect of antibodies on creatinine levels were not observed in the previous study (Olusola et al., 2020b). As shown in Figure 4, there were differences in serum creatinine concentrations between early HIV-1 infection and seroconversion for the three individuals studied among the five persons followed up. EHIV 023 had lowest serum creatinine concentration at baseline (0.9 mg/dl) and after seroconversion (0.8 mg/dl) while EHIV 022 had the highest serum concentration of creatinine at baseline (1.1 mg/dl) and after seroconversion (1.0 mg/dl). Sample EHIV016 had no serum creatinine concentration changes from baseline until after seroconversion (1.0 mg/dl).
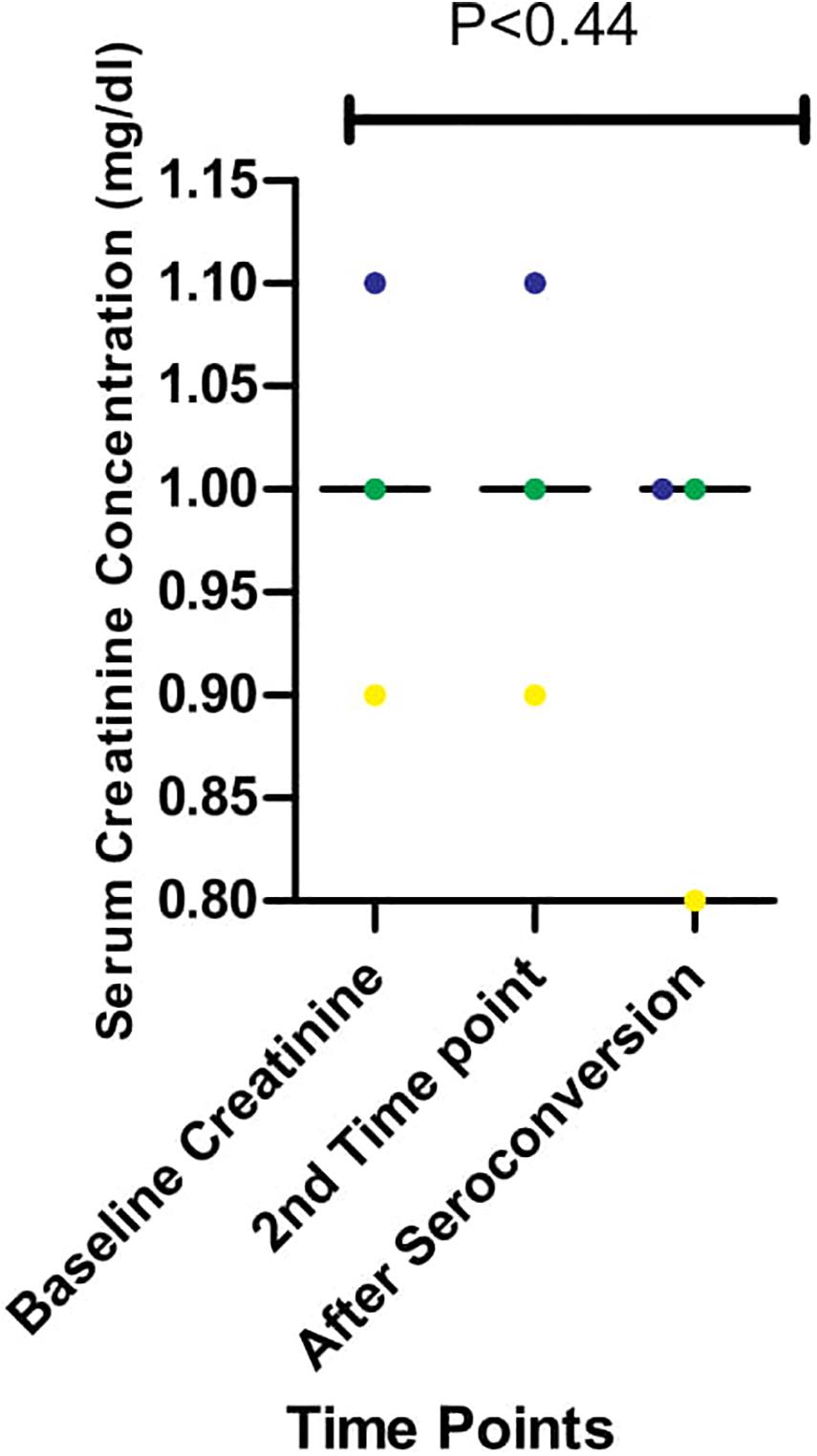
Figure 4. Serum creatinine concentration during longitudinal follow up of three individuals from early infection till after seroconversion. The three individuals followed up are represented in blue, yellow, and green. Serum creatinine levels for these individuals were measured in three-time, Baseline, second-time, and after seroconversion. Significant differences in the levels of serum creatinine (measured in mg/dl) observed across the three-time points (P < 0.44) were calculated using 1-way ANOVA.
Discussion
This study shows that diverse HIV-1 subtypes circulate in Nigeria, as subtypes A, G, and CRF02_AG were identified. Our study identified three non-synonymous substitutions within the HIV_gagconsv of HIV-1 GAG genes isolated from 10 early infected Nigerians. One substitution was, however, observed outside the HIV_gagconsv epitopes. Three (E105K, K162R, and A163G) of these substitutions have been previously related to immune escape (Currier et al., 2005; Li et al., 2007). These substitutions were associated with subtypes A and CRF02_AG. Most subtype G substitutions within the HIV_gagconsv were related to periods after seroconversion, while subtype A with early HIV infection.
Although few, substitutions within the HIV_gagconsv is a significant call for concern. Recent T cell-based vaccine studies have reported the likelihood of a single substitution within HIV_gagconsv affecting the immunogenicity vaccine constructs (Ondondo et al., 2016; Hanke, 2019). Immunogenicity of functionally conserved epitopes is a foundational requirement for an effective universal T cell-based HIV-1 vaccine (Létourneau et al., 2007). However, studies have shown that D203E mutations in the ETINEEAAEW epitope do not impact the epitope’s function and strain diversity coverage (Ondondo et al., 2016). Mutations after seroconversion may be due to the pressure of viral evasion mechanisms such as NEF mediated evasion of antibodies and HIV-1 subtype differences (Buckheit et al., 2012; Omondi et al., 2019). Studies have shown that these factors may account for replication and increased viremia during HIV-1 infection (Buckheit et al., 2012; Omondi et al., 2019).
We observed fourteen and nineteen mutations previously associated with immune escape outside the HIV_gagconsv before and after seroconversion, respectively, in this study. This finding is a far cry from the four observed within the HIV_gagconsv. It is also in accordance with previous reports that observed minimal non-synonymous substitutions in CTL epitopes within the HIV_gagconsv (Amicosante et al., 2002; Lazaro et al., 2011; Ondondo et al., 2016). APOBEC-3G has previously been noted as a major cause of hypermutation in the HIV-1 proviral landscape during the early stages of infection (Lee et al., 2019). Identifying these functionally conserved epitopes in our study participants lends credence to their immunodominance and high strain diversity coverage (Ondondo et al., 2016; Shu et al., 2020). However, the high rate of non-synonymous substitutions outside the HIV_gagconsv epitopes implies that escape mutants outside the HIV_gagconsv are more likely to be integrated into the HIV-1 latent cellular reservoir landscape. This is because latent reservoir strains are established during the early stages of infection and are known to compose majorly of CTL immune escape strains (Deng et al., 2014; Gounder et al., 2015; Roberts et al., 2015; Leitman et al., 2017). Therefore, these latent reservoirs may require a broad CTL response for clearance, as previously alluded to Deng et al. (2014). If this is the case, then using functionally conserved HIV-1 DNA vaccines for therapeutic clearance of latent reservoirs (hybrid HIV-1 cure) may be very difficult.
Therapeutic HIV-1 vaccines are intended to be used after cART stoppage, particularly against latent reservoir strains. However, as shown in this study, immune escape strains generated during the early stages of infection, which are likely significant constituents of the latent reservoir, may lead to therapeutic vaccine failures (Daucher et al., 2008; Ondondo et al., 2016; Hanke, 2019; Lé tourneau et al., 2020). Since immune escape occurs during the early stages of infection, cART must commence early to reduce the reservoir size and the incorporation of immune escape variants into the reservoir landscape (Brockman et al., 2015; Takata et al., 2017). Other studies have alluded that post-treatment control may be possible if treatment commences at the early stages of HIV infection since blips observed after cART stoppage are mainly due to immune escape variants incorporated into reservoir cells (Conway and Perelson, 2014). However, rapid and high magnitude CTL responses observed during the early stages of infection (Ndhlovu et al., 2015) may be affected by early treatment (Takata et al., 2017; Lee et al., 2019; Ndhlovu et al., 2019). This treatment can impair subsequent CTL responses during cART stoppage in post-treatment control trials. This study has shown that immune escape variants may be from those arising from CTL epitopes outside the HIV_gagconsv. However, functional and molecular studies on the nature and characteristics of HIV-1 strains in latent reservoirs need to be carried out to ascertain our claims. Furthermore, we cannot fully corroborate this theory in our study because of the relatively low sample size.
Also, universal HIV-1 vaccines are supposed to be broadly effective against all HIV-1 clades. However, immune escape strains encoding CTL epitopes outside the HIV_gagconsv may reduce the sensitivities of these vaccines. Observations of CTL escape mutants after seroconversion in this study suggest that immune pressures by other cells other than CTL may aid the generation of CTL immune escape mutants. Mapping HIV immune epitopes in different regions of the genome will further clarify this hypothesis (Matthews et al., 2012; Adland et al., 2013). As observed previously in other studies, CTL epitope KAFSPEVIPMF was the most conserved (Garcia-knight et al., 2016; Gama Caetano et al., 2018). This epitope has been associated with very low frequencies of CTL response selective pressures and has been a choice for many T cell-based HIV vaccines (Hanke, 2019). However, two substitutions previously associated with immune escape, K162R, and A163G were observed in this epitope for Subtype A during the early stages of infection.
On the other hand, CTL epitope DTINEEAAEWDR was associated with more substitutions, although most of these mutations reverted to wild type after seroconversion. This phenomenon was observable in both subtypes A and G. It seems that although more mutations were observed during the early stages of HIV infection, reversions of these mutations occurred later on in infection. Previous studies have also associated CTL epitopes’ reversions with the early stages of HIV infection (Li et al., 2007). This epitope was included in the second generation of functionally conserved HIV DNA vaccines because of its high conservation and coverage of strains diversity (Abdul-Jawad et al., 2016; Hanke, 2019). The immunogenicity of this epitope in the vaccine construct was also observed in macaques and untreated HIV infected individuals (Ondondo et al., 2016). These unique properties of this epitope were also observed in this study. However, a non-synonymous substitution, E203D, was observed in CTL epitope DTINEEAAEWDR. The selection of this epitope for immune escape strains has been previously described (Murakoshi et al., 2019). However, the substitution has also been shown not to impact the epitope’s immunogenicity as a vaccine construct (Ondondo et al., 2016). While the substitution may not affect function, it may be integrated into the latent reservoir landscape. Hence, the amino acid position should be excluded in the epitope’s design as a DNA vaccine construct.
CTL epitope TSTLQEQIR was conserved for subtype G. However, this epitope does not possess high coverage for subtype A sequences. Previous studies have documented that TSTLQEQIR may have lower HIV-1 strain diversity coverage, may be presented early, and probably associated with elite controllers (O’Connell et al., 2011; Balamurugan et al., 2013; Ondondo et al., 2016). This epitope in T cell-based HIV-1 DNA vaccines has shown moderate coverage but a strong HIV-1 specific CTL (Ondondo et al., 2016). The epitope may not be functional against subtype A and may probably be expressed by rare HLAs since it was identified in only six individuals in this study. This study does not favor using the epitope in a vaccine construct for the West African region where subtype A predominantly circulates. However, the epitope’s association with HIV-1 RNA viral load control and NEF gene downregulation (O’Connell et al., 2011; Balamurugan et al., 2013) is a plus for its use in a therapeutic HIV-1 DNA vaccine.
Several previously recognized immune escape substitutions were observed in this study. Majority of these substitutions emanated from epitopes outside the HIV_gagconsv. This is the first longitudinal study from West Africa on the kinetics of previously recognized functionally conserved epitopes of the HIV-1 GAG gene to the best of our knowledge. It is worthy of note that these epitopes have already been used in second-generation T cell HIV-1 DNA vaccines as a proof of concept. Our study provides real-life evidence of the immunodominance, conservative, and highly diverse strain coverage of these epitopes. These properties form the basis of the strategies employed in the design of conserved region vaccines.
We have also shown that numerous non-synonymous substitutions associated with CTL epitopes outside the HIV_gagconsv occur during the early stages of HIV-1 infection among HIV-1 subtypes and recombinant forms circulating in West Africa. It is essential to state that these substitutions were identified from HIV-1 DNA sequences against plasma RNA used in a similar study (Gounder et al., 2015). Proviral sequences have previously been associated with rare mutations on CTL epitopes (Gama Caetano et al., 2018; Lee et al., 2019). These substitutions may have to be considered in designing universal and therapeutic vaccines for HIV-1 strains circulating in West African countries (Ndung’u et al., 2019; Shu et al., 2020). Recent studies have shown the significant role of poorly recognized CTL epitopes in viral escape (Grossman et al., 2019).
While H28K, M30R, A224P, and A248G non-synonymous substitutions were observed before and after seroconversion, others, namely V82I and Y86W, were consistently identified after seroconversion. V82I has been previously identified with the emergence of higher viral loads in studies among HIV-infected individuals (Arcia et al., 2018; Karlsson et al., 2020). On the other hand, Y86W was associated with HIV-1 clade B and E (Fukada et al., 2002). Besides previous studies, we have also reported the high levels of creatinine among HIV-infected Africans during the early stages of infection(Bruggeman et al., 2000; Marras et al., 2002; Olusola et al., 2017, 2020b). In our study, we also showed that immune activation of CTL was ongoing and correlated positively with high creatinine levels in these early HIV-1 infected individuals (Olusola et al., 2020b). In this present study, we showed that a reduction in creatinine concentrations occurred after seroconversion. CTL immune pressures may be associated with high creatinine levels in Africans. However, this needs further investigation.
In summary, we have shown that there is a high genetic diversity of HIV-1 strains in Nigeria. Also, very high frequencies of non-synonymous substitutions occur in the HIV-1 GAG gene during the early stages of infection up until seroconversion. These substitutions include previously mapped CTL epitope immune escape mutants that are frequent in epitopes outside the HIV_gagconsv. Observation of the immunodominance of functionally conserved epitopes used in current T cell-based HIV-1 DNA vaccines in this study emphasizes the usefulness of these vaccines in a region where it is highly needed (Ndung’u et al., 2019). However, future directions for slight modifications to the use of the epitopes in the West African region are also noted. CTL immune pressure likely leaves different footprints and signature patterns on HIV-1 GAG epitopes within and outside the HIV_gagconsv.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary Material.
Ethics Statement
The studies involving human participants were reviewed and approved. This research was conducted in accordance with the declaration of Helsinki. Experiments were conducted with the understanding and the consent of each participant. Ethical approvals for this research were obtained from the University of Ibadan/University College Hospital (UI/UCH) Research and Ethics Committee (UI/EC/15/0076) and the Oyo State Ministry of Health Committee on Human Research (AD13/479/951). All results were delinked from patient identifiers and anonymized. The patients/participants provided their written informed consent to participate in this study.
Author Contributions
BO, DO, and GO conceptualized and designed the study. BO preformed the experiments, analysed and interpreted the data as well as wrote the first draft of manuscript. DO and GO supervised the work and reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by the University of Ibadan MEPI Junior Faculty Research Training Program (UI-MEPI-J) mentored research award through National Institute of Health (NIH) USA grant funded by Fogarty International Centre, the office of AIDS Research and National Human Genome Research Institute of NIH, the Health Resources and Services Administration (HRSA) and the Office of the U.S. Global AIDS Coordinator under award number D43TW010140 to DO. The funders had no role in study design, data collection, analysis, decision to publish, or manuscript preparation.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
This manuscript has been released as a pre-print at Medrxiv, doi: https://doi.org/10.1101/2020.04.09.20057703 (Olusola et al., 2020a).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2020.615721/full#supplementary-material
Footnotes
- ^ https://www.hiv.lanl.gov
- ^ http://dbpartners.stanford.edu/RegaSubtyping/
- ^ http://comet.retrovirology.lu
- ^ http://www.ncbi.nlm.nih.gov/Blast.cgi
- ^ http://jphmm.gobics.de/submission_hiv
- ^ https://www.hiv.lanl.gov/content/sequence/NEWALIGN/align.html
- ^ http://www.hiv.lanl.gov/content/sequence/VESPA/vespa.html
- ^ https://www.hiv.lanl.gov/content/immunology/variants/ctl_variant.html
References
Abdul-Jawad, S., Ondondo, B., Van Hateren, A., Gardner, A., Elliott, T., Korber, B., et al. (2016). Increased valency of conserved-mosaic vaccines enhances the breadth and depth of Epitope recognition. Mol. Ther. 24, 375–384. doi: 10.1038/mt.2015.210
Adland, E., Carlson, J. M., Paioni, P., Kløverpris, H., Shapiro, R., Ogwu, A., et al. (2013). Nef-specific CD8+ T cell responses contribute to HIV-1 immune control. PLoS One 8:e73117. doi: 10.1371/journal.pone.0073117
Amicosante, M., Gioia, C., Montesano, C., Casetti, R., Topino, S., D’Offizi, G., et al. (2002). Computer-based design of an HLA-haplotype and HIV-clade independent cytotoxic T-lymphocyte assay for monitoring HIV-specific immunity. Mol. Med. 8, 798–807. doi: 10.1007/bf03402084
Arcia, D., Ochoa, R., Hernández, J. C., Cristiam, M., Díaz, F. J., Velilla, P. A., et al. (2018). Potential immune escape mutations under inferred selection pressure in HIV-1 strains circulating in Medellin, Colombia. Infect. Genet. Evol. 69, 267–278.
Balamurugan, A., Ali, A., Boucau, J., Le Gall, S., Ng, H. L., and Yang, O. O. (2013). HIV-1 Gag Cytotoxic T lymphocyte epitopes vary in presentation kinetics relative to HLA class I downregulation. J. Virol. 87, 8726–8734.
Balla-Jhagjhoorsingh, S. S., Koopman, G., Mooij, P., Haaksma, T. G., Teeuwsen, V. J., Bontrop, R. E., et al. (1999). Conserved CTL epitopes shared between HIV-infected human long-term survivors and chimpanzees. J. Immunol. 162, 2308–2314.
Barouch, D. H., O’Brien, K. L., Simmons, N. L., King, S. L., Abbink, P., Maxfield, L. F., et al. (2010). Mosaic HIV-1 vaccines expand the breadth and depth of cellular immune responses in rhesus monkeys. Nat. Med. 16, 319–323. doi: 10.1038/nm.2089
Bekker, L. G., Tatoud, R., Dabis, F., Feinberg, M., Kaleebu, P., Marovich, M., et al. (2020). The complex challenges of HIV vaccine development require renewed and expanded global commitment. Lancet 395, 384–388. doi: 10.1016/s0140-6736(19)32682-0
Bennett, M. S., Joseph, A., Ng, H. L., Goldstein, H., and Yang, O. O. (2010). Fine-tuning of T-cell receptor avidity to increase HIV epitope variant recognition by cytotoxic T lymphocytes. AIDS 24, 2619–2628. doi: 10.1097/qad.0b013e32833f7b22
Borrow, P., Lewicki, H., Hahn, B. H., Shaw, G. M., and Oldstone, M. B. (1994). Virus-specific CD8+ cytotoxic T-lymphocyte activity associated with control of viremia in primary human immunodeficiency virus type 1 infection. J. Virol. 68, 6103–6110. doi: 10.1128/jvi.68.9.6103-6110.1994
Borthwick, N., Ahmed, T., Ondondo, B., Hayes, P., Rose, A., Ebrahimsa, U., et al. (2014). Vaccine-elicited human T cells recognizing conserved protein regions inhibit HIV-1. Mol. Ther. 22, 464–475. doi: 10.1038/mt.2013.248
Brockman, M. A., Jones, R. B., and Brumme, Z. L. (2015). Challenges and opportunities for T-cell-mediated strategies to eliminate HIV reservoirs. Front. Immunol. 6:506. doi: 10.3389/fimmu.2015.00506
Bruggeman, L. A., Ross, M. D., Tanji, N., Cara, A., Dikman, S., Gordon, R. E., et al. (2000). Renal epithelium is a previously unrecognized site of HIV-1 infection. J. Am. Soc. Nephrol. 11, 2079–2087.
Brumme, Z. L., Brumme, C. J., Carlson, J., Streeck, H., John, M., Eichbaum, Q., et al. (2008). Marked epitope-and allele-specific differences in rates of mutation in human immunodeficiency type 1 (HIV-1) Gag, Pol, and Nef Cytotoxic T-Lymphocyte Epitopes in Acute/Early HIV-1 Infection. J. Virol. 82, 9216–9227. doi: 10.1128/jvi.01041-08
Buckheit, R. W., Allen, T. G., Alme, A., Salgado, M., O’Connell, K. A., Huculak, S., et al. (2012). Host factors dictate control of viral replication in two HIV-1 controller/chronic progressor transmission pairs. Nat. Commun. 3:716.
Burton, D. R., Desrosiers, R. C., Doms, R. W., Koff, W. C., Kwong, P. D., Moore, J. P., et al. (2004). HIV vaccine design and the neutralizing antibody problem. Nat. Immunol. 5, 233–236.
Carlson, J. M., Listgarten, J., Pfeifer, N., Tan, V., Kadie, C., Walker, B. D., et al. (2012). Widespread impact of HLA restriction on immune control and escape pathways of HIV-1. J. Virol. 86, 5230–5243. doi: 10.1128/jvi.06728-11
Conway, J. M., and Perelson, A. S. (2014). Post-treatment control of HIV infection. Proc Natl Acad Sci U.S.A. 112, 5467–5472. doi: 10.1073/pnas.1419162112
Crawford, H., Matthews, P. C., Schaefer, M., Carlson, J. M., Leslie, A., Kilembe, W., et al. (2011). The Hypervariable HIV-1 Capsid Protein Residues Comprise HLA-Driven CD8+ T-Cell Escape Mutations, and Covarying HLA-Independent Polymorphisms. J. Virol. 85, 1384–1390. doi: 10.1128/jvi.01879-10
Currier, J. R., Harris, M. E., Cox, J. H., McCutchan, F. E., Birx, D. L., Maayan, S., et al. (2005). Immunodominance and cross-reactivity of B5703-restricted CD8 T lymphocytes from HIV type 1 subtype C-infected Ethiopians. AIDS Res. Hum. Retroviruses. 21, 239–245. doi: 10.1089/aid.2005.21.239
Daucher, M., Price, D. A., Brenchley, J. M., Lamoreaux, L., Metcalf, J. A., Rehm, C., et al. (2008). Virological outcome after structured interruption of antiretroviral therapy for human immunodeficiency virus infection is associated with the functional profile of virus-specific CD8+ T cells. J. Virol. 82, 4102–4114. doi: 10.1128/jvi.02212-07
Deng, K., Pertea, M., Rongvaux, A., Wang, L., Durand, C. M., Ghiaur, G., et al. (2014). Broad CTL response is required to clear latent HIV-1 due to dominance of escape mutations. Nature 517, 381–385. doi: 10.1038/nature14053
Fischer, W., Perkins, S., Theiler, J., Bhattacharya, T., Yusim, K., Funkhouser, R., et al. (2007). Polyvalent vaccines for optimal coverage of potential T-cell epitopes in global HIV-1 variants. Nat. Med. 13, 100–106. doi: 10.1038/nm1461
Fukada, K., Tomiyama, H., Wasi, C., Matsuda, T., Kusagawa, S., Sato, H., et al. (2002). Cytotoxic T-cell recognition of HIV-1 cross-clade and clade-specific epitopes in HIV-1-infected Thai and Japanese patients. AIDS 16, 701–711. doi: 10.1097/00002030-200203290-00005
Gall, A., Ferns, B., Morris, C., Watson, S., Cotten, M., Robinson, M., et al. (2012). Universal Amplification, Next-Generation Sequencing, and Assembly of HIV-1 Genomes. J. Clin. Microbiol. 50, 3838–3844.
Gama Caetano, D., Heloise Côrtes, F., Bello, G., Lopes Maia Teixeira, S., Hoagland, B., Grinsztejn, B., et al. (2018). Next-generation sequencing analyses of the emergence and maintenance of mutations in CTL epitopes in HIV controllers with differential viremia control. Retrovirology 15:62.
Garcia-knight, M. A., Slyker, J., Payne, B. L., Pond, S. L. K., De Silva, T. I., Chohan, B., et al. (2016). Viral Evolution and Cytotoxic T Cell Restricted Selection in Acute Infant HIV-1 Infection. Nat. Publ. Gr. 6:29536.
Gillespie, G. M. A., Kaul, R., Dong, T., Yang, H. B., Rostron, T., Bwayo, J., et al. (2002). Cross-reactive cytotoxic T lymphocytes against a HIV-1 p24 epitope in slow progressors with B∗57. AIDS 16, 961–972. doi: 10.1097/00002030-200205030-00002
Goonetilleke, N., Liu, M. K. P., Salazar-Gonzalez, J. F., Ferrari, G., Giorgi, E., Ganusov, V. V., et al. (2009). The first T cell response to transmitted/founder virus contributes to the control of acute viremia in HIV-1 infection. J. Exp. Med. 206, 1253–1272. doi: 10.1084/jem.20090365
Goulder, P. J. R., and Watkins, D. I. (2004). HIV and SIV CTL escape: Implications for vaccine design. Nat. Rev. Immunol. Nat. Publ. Group 4, 630–640. doi: 10.1038/nri1417
Gounder, K., Padayachi, N., Mann, J. K., Radebe, M., Mokgoro, M., Van Der Stok, M., et al. (2015). High frequency of transmitted HIV-1 Gag HLA class I-driven immune escape variants but minimal immune selection over the first year of clade C infection. PLoS One 10:e0119886. doi: 10.1371/journal.pone.0119886
Grossman, M. A., Hofmann, C., Ng, H. L., and Yang, O. O. (2019). Suboptimal stimulation by weak agonist epitope variants does not drive dysfunction of HIV-1-specific cytotoxic T lymphocyte clones. AIDS 33, 1565–1574. doi: 10.1097/qad.0000000000002259
Hanke, T. (2019). Aiming for protective T-cell responses: a focus on the first generation conserved-region HIVconsv vaccines in preventive and therapeutic clinical trials. Exp. Rev. Vaccines 18, 1029–1041. doi: 10.1080/14760584.2019.1675518
Herout, S., Mandorfer, M., Breitenecker, F., Reiberger, T., Grabmeier-Pfistershammer, K., Rieger, A., et al. (2016). Impact of early initiation of antiretroviral therapy in patients with acute HIV infection in vienna. Austria. Sluis-Cremer N, editor. PLoS One 11:e0152910. doi: 10.1371/journal.pone.0152910
Honeyborne, I., Codoñer, F. M., Leslie, A., Tudor-Williams, G., Luzzi, G., Ndung’u, T., et al. (2010). HLA-Cw∗03-restricted CD8+ T-cell responses targeting the HIV-1 gag major homology region drive virus immune escape and fitness constraints compensated for by intracodon variation. J. Virol. 84, 11279–11288. doi: 10.1128/jvi.01144-10
Karlsson, A. C., Iversen, A. K. N., Chapman, J. M., De Oliveira, T., Spotts, G., Mcmichael, A. J., et al. (2020). Sequential broadening of CTL responses in early HIV-1 infection is associated with viral escape. PLoS One 21:e225. doi: 10.1371/journal.pone.0000225
Keele, B. F., Giorgi, E. E., Salazar-gonzalez, J. F., Decker, J. M., Pham, K. T., Salazar, M. G., et al. (2008). Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U.S.A. 105, 7552–7557.
Kløverpris, H. N., Adland, E., Koyanagi, M., Stryhn, A., Harndahl, M., Matthews, P. C., et al. (2014). HIV subtype influences HLA-B∗07:02-associated HIV disease outcome. AIDS Res. Hum. Retrovirus. 30, 468–475. doi: 10.1089/aid.2013.0197
Kløverpris, H. N., McGregor, R., McLaren, J. E., Ladell, K., Harndahl, M., Stryhn, A., et al. (2015). CD8 + TCR Bias and Immunodominance in HIV-1 Infection. J. Immunol. 194, 5329–5345. doi: 10.4049/jimmunol.1400854
Koofhethile, C. K., Ndhlovu, Z. M., Thobakgale-Tshabalala, C., Prado, J. G., Ismail, N., Mncube, Z., et al. (2016). CD8+ T cell breadth and ex vivo virus inhibition capacity distinguish between Viremic Controllers with and without Protective HLA Class I Alleles. J. Virol. 90, 6818–6831. doi: 10.1128/jvi.00276-16
Korber, B., and Myers, G. (1992). Signature pattern analysis: a method for assessing viral sequence relatedness. AIDS Res. Hum. Retroviruses 8, 1549–1560. doi: 10.1089/aid.1992.8.1549
Kosakovsky Pond, S. L., Poon, A. F. Y., Zárate, S., Smith, D. M., Little, S. J., Pillai, S. K., et al. (2008). Estimating selection pressures on HIV-1 using phylogenetic likelihood models. Stat. Med. 27, 4779–4789. doi: 10.1002/sim.3192
Koup, R. A., Safrit, J. T., Cao, Y., Andrews, C. A., McLeod, G., Borkowsky, W., et al. (1994). Temporal association of cellular immune responses with the initial control of viremia in primary human immunodeficiency virus type 1 syndrome. J. Virol. 68, 4650–4655. doi: 10.1128/jvi.68.7.4650-4655.1994
Lazaro, E., Kadie, C., Stamegna, P., Zhang, S. C., Gourdain, P., Lai, N. Y., et al. (2011). Variable HIV peptide stability in human cytosol is critical to epitope presentation and immune escape. J. Clin. Invest. 121, 2480–2492. doi: 10.1172/jci44932
Lé tourneau, S., Im, E.-J., Mashishi, T., Brereton, C., Bridgeman, A., Yang, H., et al. (2020). Design and Pre-Clinical Evaluation of a Universal HIV-1 Vaccine. PLoS One 2:e984. doi: 10.1371/journal.pone.0000984
Lee, G. Q., Reddy, K., Einkauf, K. B., Gounder, K., Chevalier, J. M., Dong, K. L., et al. (2019). HIV-1 DNA sequence diversity and evolution during acute subtype C infection. Nat. Commun. 110.
Leitman, E. M., Thobakgale, C. F., Adland, E., Ansari, M. A., Raghwani, J., Prendergast, A. J., et al. (2016). Role of HIV-specific CD8 + T cells in pediatric HIV cure strategies after widespread early viral escape. J. Exp. Med. 214, 3239–3262. doi: 10.1084/jem.20162123
Leitman, E. M., Thobakgale, C. F., Adland, E., Ansari, M. A., Raghwani, J., Prendergast, A. J., et al. (2017). Role of HIV-specific CD8+ T cells in pediatric HIV cure strategies after widespread early viral escape. J. Exp. Med. 214, 3239–3261. doi: 10.1084/jem.20162123
Létourneau, S., Im, E. J., Mashishi, T., Brereton, C., Bridgeman, A., Yang, H., et al. (2007). Design and pre-clinical evaluation of a universal HIV-1 vaccine. PLoS One 2:e984. doi: 10.1371/journal.pone.0000984
Li, B., Gladden, A. D., Altfeld, M., Kaldor, J. M., Cooper, D. A., Kelleher, A. D., et al. (2007). Rapid reversion of sequence polymorphisms dominates early human immunodeficiency virus type 1 evolution. J. Virol. 81, 193–201. doi: 10.1128/jvi.01231-06
Liu, Y., Rao, U., McClure, J., Konopa, P., Manocheewa, S., Kim, M., et al. (2014). Impact of mutations in highly conserved amino acids of the HIV-1 Gag-p24 and Env-gp120 proteins on viral replication in different genetic backgrounds. Menéndez-Arias L, editor. PLoS One 9:e94240. doi: 10.1371/journal.pone.0094240
Marras, D., Bruggeman, L. A., Gao, F., Tanji, N., Mansukhani, M. M., Cara, A., et al. (2002). Replication and compartmentalization of HIV-1 in kidney epithelium of patients with HIV-associated nephropathy. Nat. Med. 8, 522–526. doi: 10.1038/nm0502-522
Martinez-Picado, J., Prado, J. G., Fry, E. E., Pfafferott, K., Leslie, A., Chetty, S., et al. (2006). Fitness cost of escape mutations in p24 Gag in association with control of human immunodeficiency virus type 1. J. Virol. 80, 3617–3623. doi: 10.1128/jvi.80.7.3617-3623.2006
Matthews, P. C., Koyanagi, M., Kløverpris, H. N., Harndahl, M., Stryhn, A., Akahoshi, T., et al. (2012). Differential clade-specific HLA-B∗3501 association with HIV-1 disease outcome is linked to immunogenicity of a single Gag epitope. J. Virol. 86, 12643–12654. doi: 10.1128/jvi.01381-12
Mori, M., Wichukchinda, N., Miyahara, R., Rojanawiwat, A., Pathipvanich, P., Maekawa, T., et al. (2014). HLA-B∗35: 05 is a protective allele with a unique structure among HIV-1 CRF01-AE-infected Thais, in whom the B∗57 frequency is low. AIDS 28, 959–967. doi: 10.1097/qad.0000000000000206
Mothe, B., Hu, X., Llano, A., Rosati, M., Olvera, A., Kulkarni, V., et al. (2015). A human immune data-informed vaccine concept elicits strong and broad T-cell specificities associated with HIV-1 control in mice and macaques. J. Transl. Med. 13:60. doi: 10.1186/s12967-015-0392-5
Muema, D. M., Macharia, G. N., Olusola, B. A., Hassan, A. S., Fegan, G. W., Berkley, J. A., et al. (2017). Proportions of circulating follicular helper T cells are reduced and correlate with memory B cells in HIV-infected children. PLoS One 12:e0175570. doi: 10.1371/journal.pone.0175570
Murakoshi, H., Kuse, N., Akahoshi, T., Zhang, Y., Chikata, T., Borghan, M. A., et al. (2019). Broad Recognition of Circulating HIV-1 by HIV-1-Specific Cytotoxic T-Lymphocytes with Strong Ability to Suppress HIV-1 Replication. J. Virol. 93, e001480–18.
Ndhlovu, Z. M., Kamya, P., Mewalal, N., Kløverpris, H. N., Nkosi, T., Pretorius, K., et al. (2015). Magnitude and kinetics of CD8+ T cell activation during hyperacute HIV infection impact viral set point. Immunity 43, 591–604. doi: 10.1016/j.immuni.2015.08.012
Ndhlovu, Z. M., Kazer, S. W., Nkosi, T., Ogunshola, F., Muema, D. M., Anmole, G., et al. (2019). Augmentation of HIV-specific T cell function by immediate treatment of hyperacute HIV-1 infection. Sci. Transl. Med. 11:eaau0528. doi: 10.1126/scitranslmed.aau0528
Ndung’u, T., McCune, J. M., and Deeks, S. G. (2019). Why and where an HIV cure is needed and how it might be achieved. Nature 576, 397–405. doi: 10.1038/s41586-019-1841-8
Nowak, M. A., Anderson, R. M., McLean, A. R., Wolfs, T. F. W., Goudsmit, J., and May, R. M. (1991). Antigenic diversity thresholds and the development of AIDS. Science 254, 963–969. doi: 10.1126/science.1683006
O’Connell, K. A., Hegarty, R. W., Siliciano, R. F., and Blankson, J. N. (2011). Viral suppression of multiple escape mutants by de novo CD8+T cell responses in a human immunodeficiency virus-1 Infected elite suppressor. Retrovirology 8:63. doi: 10.1186/1742-4690-8-63
Ogg, G. S., Jin, X., Bonhoeffer, S., Dunbar, P. R., Nowak, M. A., Monard, S., et al. (1998). Quantitation of HIV-1-specific cytotoxic T lymphocytes, and plasma load of viral RNA. Science 279, 2103–2106. doi: 10.1126/science.279.5359.2103
Olusola, B., Olaleye, D. O., and Odaibo, G. N. (2020a). Non-synonymous substitutions in Hiv-1 gag are frequent in epitopes outside the major hydrophobic region and associated with subtype differences. medRxiv[Preprint]. doi: 10.1101/2020.04.09.20057703
Olusola, B. A., Kabelitz, D., Olaleye, D. O., and Odaibo, G. N. (2020b). Early HIV infection is associated with reduced proportions of gamma delta T subsets as well as high creatinine, and urea levels. Scand. J. Immunol. 91:e12868.
Olusola, B. A., Olaleye, D. O., and Odaibo, G. N. (2017). Early HIV infection among persons referred for malaria parasite testing in Nigeria. Arch. Virol. 163, 439–445. doi: 10.1007/s00705-017-3599-3
Omondi, F. H., Chandrarathna, S., Mujib, S., Brumme, C. J., Jin, S. W., Sudderuddin, H., et al. (2019). HIV Subtype and Nef-mediated immune evasion function correlate with viral reservoir size in early-treated individuals downloaded from. jvi.asm.org 1. J. Virol. 93, 1832–1850.
Ondondo, B., Murakoshi, H., Clutton, G., Abdul-Jawad, S., Wee, E. G. T., Gatanaga, H., et al. (2016). Novel conserved-region T-cell mosaic vaccine with high global HIV-1 coverage is recognized by protective responses in untreated infection. Mol. Ther. 24, 832–842. doi: 10.1038/mt.2016.3
Ou, C. Y., Ciesielski, C. A., Myers, G., Bandea, C. I., Luo, C. C., Korber, B. T. M., et al. (1992). Molecular epidemiology of HIV transmission in a dental practice. Science 256, 1165–1171.
Papuchon, J., Pinson, P., Lazaro, E., Reigadas, S., Guidicelli, G., Taupin, J. L., et al. (2013). Resistance mutations and CTL epitopes in archived HIV-1 DNA of patients on antiviral treatment: toward a new concept of vaccine. PLoS One 8:e69029. doi: 10.1371/journal.pone.0069029
Radebe, M., Gounder, K., Mokgoro, M., Ndhlovu, Z. M., Mncube, Z., Mkhize, L., et al. (2015). Broad and persistent Gag-specific CD8+ T-cell responses are associated with viral control but rarely drive viral escape during primary HIV-1 infection. AIDS 29, 23–33. doi: 10.1097/qad.0000000000000508
Reicin, A. S., Paik, S., Berkowitz, R. D., Luban, J., Lowy, I., and Goff, S. P. (1995). Linker insertion mutations in the human immunodeficiency virus type 1 gag gene: effects on virion particle assembly, release, and infectivity. J. Virol. 69, 642–650. doi: 10.1128/jvi.69.2.642-650.1995
Roberts, H. E., Hurst, J., Robinson, N., Brown, H., Flanagan, P., Vass, L., et al. (2015). Structured observations reveal slow HIV-1 CTL escape. PLoS Genet. 11:e1004914. doi: 10.1371/journal.pgen.1004914.g002
Salvatierra, K., and Florez, H. (2017). Pathogen sequence signature analysis (PSSA): a software tool for analyzing sequences to identify microorganism genotypes. F1000Research 6:21. doi: 10.12688/f1000research.10393.1
Santra, S., Liao, H. X., Zhang, R., Muldoon, M., Watson, S., Fischer, W., et al. (2010). Mosaic vaccines elicit CD8+ T lymphocyte responses that confer enhanced immune coverage of diverse HIV strains in monkeys. Nat. Med. 16, 324–328. doi: 10.1038/nm.2108
Shu, J., Shen, W., Liu, H., Zhou, Y., Li, J., Zhuang, Y., et al. (2020). The immunologic dominance of an epitope within a rationally designed poly-epitope vaccine is influenced by multiple factors. Vaccine 38, 2913–2924.
Takata, H., Buranapraditkun, S., Kessing, C., Fletcher, J. L. K., Muir, R., Tardif, V., et al. (2017). Delayed differentiation of potent effector CD8+ T cells reducing viremia and reservoir seeding in acute HIV infection. Sci. Transl. Med. 9:eaag1809. doi: 10.1126/scitranslmed.aag1809
Tanaka, M., Robinson, B. A., Chutiraka, K., Geary, C. D., Reed, J. C., and Lingappa, J. R. (2016). Mutations of conserved residues in the major homology region arrest assembling HIV-1 gag as a membrane-targeted intermediate containing genomic RNA and cellular proteins. J. Virol. 15, 1944–1963. doi: 10.1128/jvi.02698-15
Tcherepanova, I., Harris, J., Starr, A., Cleveland, J., Ketteringham, H., Calderhead, D., et al. (2008). Multiplex RT-PCR amplification of HIV genes to create a completely autologous DC-based immunotherapy for the treatment of HIV infection. PLoS One 3:e1489. doi: 10.1371/journal.pone.0001489
Tenzer, S., Wee, E., Burgevin, A., Stewart-Jones, G., Friis, L., Lamberth, K., et al. (2009). Antigen processing influences HIV-specific cytotoxic T lymphocyte immunodominance. Nat. Immunol. 10, 636–646. doi: 10.1038/ni.1728
Troyer, R. M., McNevin, J., Liu, Y., Zhang, S. C., Krizan, R. W., Abraha, A., et al. (2009). Variable fitness impact of HIV-1 escape mutations to cytotoxic T lymphocyte (CTL) response. PLoS Pathog. 5:e1000365. doi: 10.1371/journal.ppat.1000365
Vidal, N., Peeters, M., Mulanga-Kabeya, C., Nzilambi, N., Robertson, D., Ilunga, W., et al. (2000). Unprecedented degree of human immunodeficiency virus type 1 (HIV-1) Group M genetic diversity in the democratic republic of congo suggests that the HIV-1 pandemic originated in central Africa. J. Virol. 74, 10498–10507. doi: 10.1128/jvi.74.22.10498-10507.2000
Keywords: non-synonymous substitutions, HIV-1 GAG, MHR, HIV-1 subtypes, HIV cure
Citation: Olusola BA, Olaleye DO and Odaibo GN (2021) Non-synonymous Substitutions in HIV-1 GAG Are Frequent in Epitopes Outside the Functionally Conserved Regions and Associated With Subtype Differences. Front. Microbiol. 11:615721. doi: 10.3389/fmicb.2020.615721
Received: 09 October 2020; Accepted: 15 December 2020;
Published: 11 January 2021.
Edited by:
Taisuke Izumi, Henry M Jackson Foundation for the Advancement of Military Medicine (HJF), United StatesReviewed by:
George William Carnell, University of Cambridge, United KingdomViraj Kulkarni, Texas Biomedical Research Institute, United States
Copyright © 2021 Olusola, Olaleye and Odaibo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Georgina N. Odaibo, Z2VvcmdpbmFvZGFpYm9AZ21haWwuY29t
 Babatunde A. Olusola
Babatunde A. Olusola David O. Olaleye
David O. Olaleye