- School of Biotechnology, University of Jammu, Jammu, India
Dynamic consortium of microbial communities (bacteria, fungi, protists, viruses, and nematodes) colonizing multiple tissue types and coevolving conclusively with the host plant is designated as a plant microbiome. The interplay between plant and its microbial mutualists supports several agronomic functions, establishing its crucial role in plant beneficial activities. Deeper functional and mechanistic understanding of plant-microbial ecosystems will render many “ecosystem services” by emulating symbiotic interactions between plants, soil, and microbes for enhanced productivity and sustainability. Therefore, microbiome engineering represents an emerging biotechnological tool to directly add, remove, or modify properties of microbial communities for higher specificity and efficacy. The main goal of microbiome engineering is enhancement of plant functions such as biotic/abiotic stresses, plant fitness and productivities, etc. Various ecological-, biochemical-, and molecular-based approaches have come up as a new paradigm for disentangling many microbiome-based agromanagement hurdles. Furthermore, multidisciplinary approaches provide a predictive framework in achieving a reliable and sustainably engineered plant-microbiome for stress physiology, nutrient recycling, and high-yielding disease-resistant genotypes.
Introduction
Scientific research advances over the eons of time have propelled microbial coevolution and diversification as important forces in sculpturing and carving every accessible part of nature (Saleem et al., 2017). The taxonomically diverse microbial communities interacting with different components of ecosystem are acknowledged to be a major trait in terrestrialization of plants. Therefore, this multiorganismal assemblage and its synergistic relationships with the host shape the “holobiont” framework (Cooke et al., 2019). The holistic and interactive colonization of plants by ecologically diverse microbial communities is designated as plant microbiome (Foo et al., 2017).
The microbiota can exist persistently in, on, and around different tissues during plant life cycle (Nelson, 2018). Bacteria, fungi, protozoa, archaea, and viruses comprise diverse microbiota teaming with the plant. The study of multitrophic interactions between the two has greatly elaborated ecoevolutionary and functional understanding of host-microbe interactions. Rhizosphere, endosphere, and phyllosphere are the major microecosystems where bidirectional chemical dialog directly contributes to plant development, physiology, and systemic defenses and indirectly produce root exudates and other metabolites acting as nutrient sources and signals for modulating microbial composition (Mueller and Sachs, 2015). Host genotypic traits, developmental stage, soil properties, and environmental conditions harmonize the structural and functional dynamics of microbiome (Rossmann et al., 2017). This symbiotic interactome confers many adaptive advantages to plant growth and development viz. nutrient acquisition, stress resilience, modulation of hormone levels, disease resistance, enable toxin production, and increased root exudation (Singh et al., 2020).
Opening up of new high-throughput community analyses methods, next-generation sequencing techniques and meta-“omics” tools have greatly unraveled the multitrophic interactions present in the black box of plant microbiome (Ahmad et al., 2019). With the advent of these high-throughput technologies, plant beneficial microbes can be manipulated. In this scenario, microbiome engineering may be an alternative way to understand, manipulate, and develop corresponding technologies for developing microbial communities crucial to plant health and productivity (Prasad et al., 2018). The present review fosters various practical ways by which plant-microbial mutualism can be manipulated to enhance plant performance and agricultural productivity.
Interactive Dialog Within Plant Microbiome
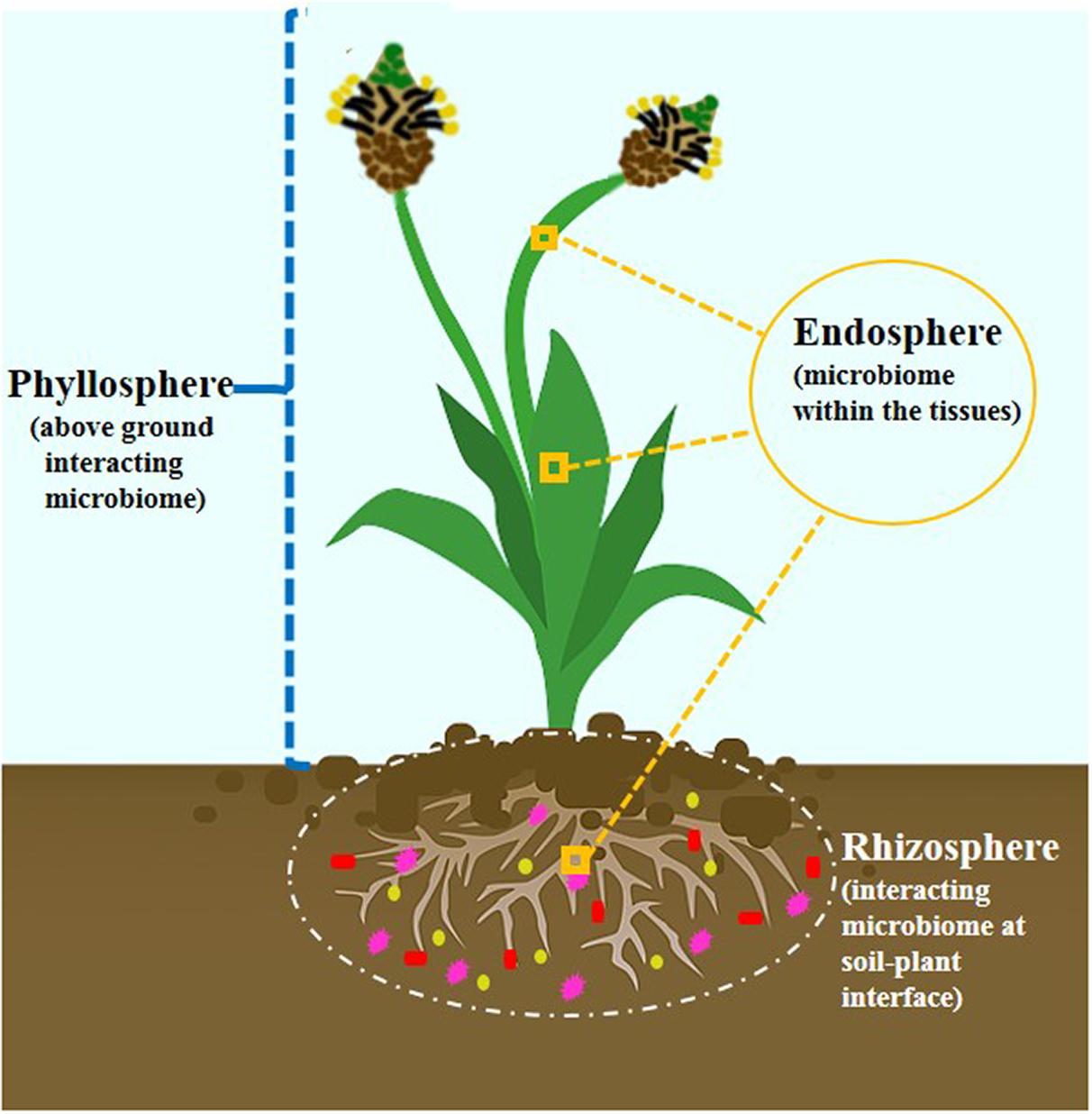
Decades of research have predicted that plants are naturally colonized by a congregation of microbes. The recognition was converted from single entities to a community conception. These microbes are aggregated in micropockets thriving either outside (rhizosphere), inside (endosphere), or on (phyllosphere) the plant. These three main microecosystems (Figure 1) are regions of chemical communications and are interconnected (Vandenkoornhuyse et al., 2015). The critical interface where plant roots interact intimately with the physicochemical and biotic components of the soil thereby harboring tremendous diversity of microbial communities is referred to as rhizosphere (Wang et al., 2020). Plant-root exudation and microbial substrate utilization traits are the main driving forces for promoting the abundance and metabolic synchronization of microbial communities (Brunel et al., 2020). Santoyo et al. (2016) have reviewed rhizosphere as a nutrient-rich reservoir for facilitating establishment and development of endophytic community. The plant endosphere is defined by complex microbial communities inhabiting and enduring within plant tissues with nonharming nature. Unlike rhizosphere, it receives additional advantage as one habitat without being affected by microenvironment variation and dynamics (Compant et al., 2019). In addition, phyllosphere is another microhabitat which comprises aboveground portions of plant forming an oligotrophic environment for hosting diverse microbial communities.
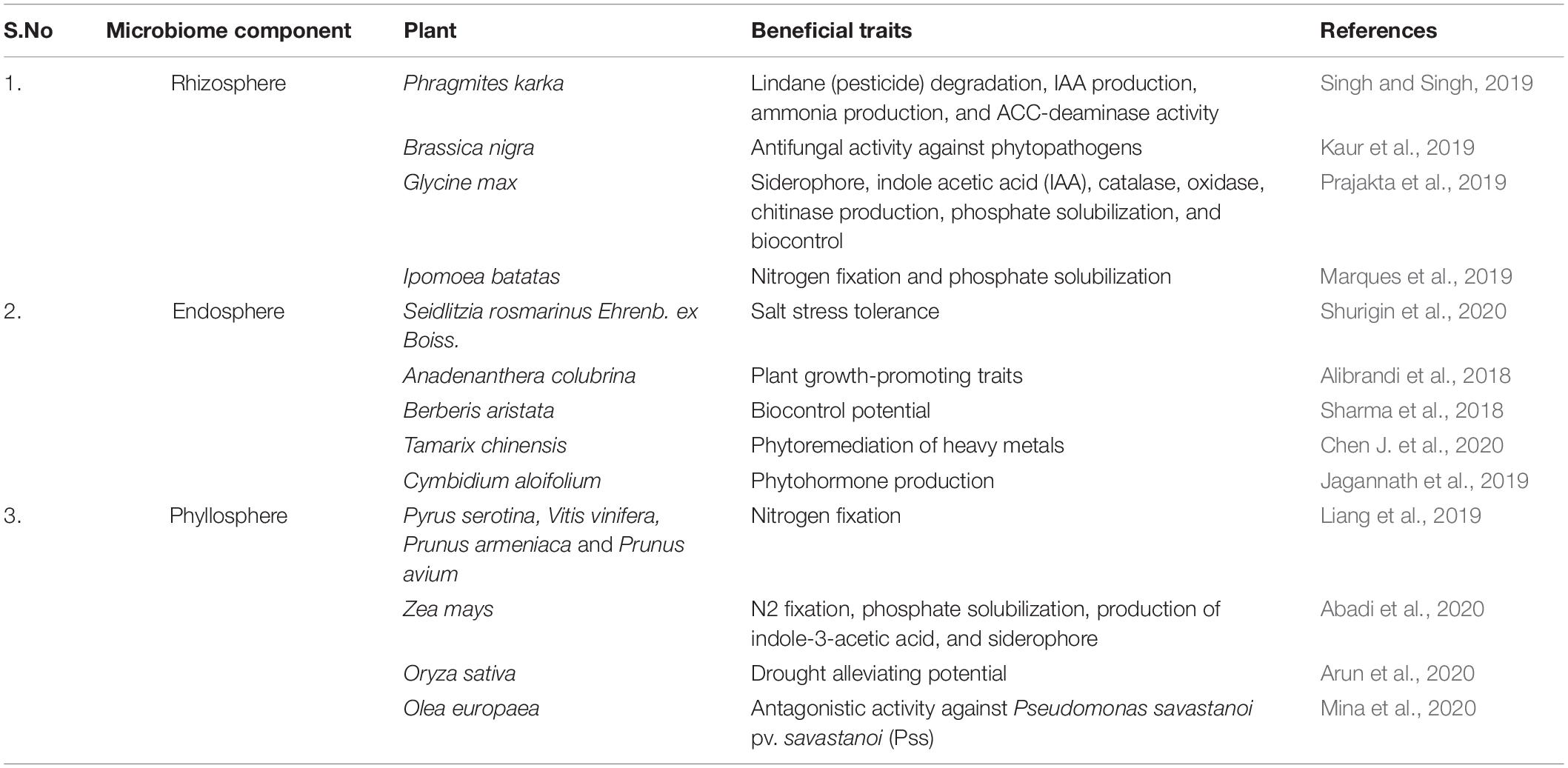
Evolutionary experiments have underpinned the crucial role of microbial phylotypes in ecosystem functions (Liu et al., 2020). These dynamic microenvironments represent a fundamental way to connect bottom-up and top-down microbial diversity with each other. Collective microbial environments pose additional advantage by coupling Green Revolution and agricultural sustainability. The beneficial microbes in the microhabitat offer multifarious prospects to agroecological systems viz. nutrient solubilization, phytohormone production, nitrogen fixation, bioremediation, improved tolerance to biotic and abiotic stresses, alter plant phenology, modify morphological and size-related traits, and ultimately play a major role in mediating plant responses to climate change and variation (Han et al., 2016; Carrion et al., 2019; Liang et al., 2019). Molecular signaling and trophic interactions between the plant-shaped microhabitat mold the composition and activities of associated microbial populations (Chen X. et al., 2020). Furthermore, mounting evidence have highlighted the potential of beneficial microbes in enhancing plant beneficial traits (Table 1).
Knowledge of the chemical communication between plant and plant, microbe and microbe, and plant and microbe is essential to unveil the complexity of these interactions. Reductionist approaches reveal that plants produce an impressive cocktail of insoluble and soluble low molecular weight organic compounds, known as root exudates/semiochemicals (Quiza et al., 2015; Zhang et al., 2015). This arsenal of secondary metabolites causes inducible structural and physiological modifications in the rhizosphere for the establishment of the microbial communities (Senthil-Kumar and Mysore, 2013). Most of these plant metabolites are synthesized through complex isoprenoid, phenylpropanoid, alkaloid, or fatty acid/polyketide biosynthetic trackways. Flavonoids, coumarins (phytoalexins), benzoxazinoids, strigolactones, terpenoids, malic acid, camalexin, and ethylene are important chemical mediators in the beneficial plant-microbe interaction (Stringlis et al., 2019). The plant exudates are either actively produced or elicited when exposed to certain threats and environmental conditions (Morel and Castro-Sowinski, 2013). These rhizodeposits generate quorum sensing response in microbial communities and also secrete various signaling compounds such as antibiotics, volatile molecules, phytohormones, organic acids, amino acids, sugars, and surface receptors (pattern-recognition receptors) (Zhang et al., 2015; Arif et al., 2020).
Literature reports have addressed the importance of exudates in aboveground and belowground “signalomics,” encouraging symbiosis, minimizing diseases, combating herbivores and pathogen attack, and upregulating growth-promoting traits (Velmourougane et al., 2017; Chagas et al., 2018). The importance of root exudates in modifying the structure and activity of rhizosphere microbiome is well investigated by Lombardi et al. (2018). Their study demonstrated that the roots of stressed plants release peroxidases and oxylipins which act as a chemoattractant for establishing the growth of biocontrol fungus Trichoderma harzianum. Shaposhnikov et al. (2020) also demonstrated the role of root exudates in creating a favorable environment for promoting the growth of beneficial rhizobacteria over phytopathogens. They assessed that barley root releases a repertoire of aromatic carboxylic acids as antimicrobial compounds for suppressing the growth of phytopathogenic fungus Fusarium culmorum. In contrast, fewer antimicrobial root exudates were released in the presence of antagonistic bacterium Pseudomonas fluorescens, therefore, enhancing the assembly of root-beneficial rhizobacteria. Another study conducted by Jin et al. (2019) reported the potential of maize root exudates in inducing chemotactic response, swarming motility, and biofilm formation. Their study establishes that the various components of maize root exudates such as organic acids, amino acids, and sugars significantly enhanced the chemotaxis, cluster movement, and biofilm formation of Bacillus velezensis in rhizospheric soil and root tissue. These observations gained significant focus in understanding plant-microbe interaction via plant root exudates/semiochemicals. It is worthwhile to understand and manipulate the functions of this “second genome” for future diagnostics and therapy in plants.
Microbiome Engineering
Amidst climate change and human population growth, the global demand for increasing crop production is expected (Hamilton et al., 2016). For sustainable agriculture, plant-microbial mutualism will help by imparting resistance against biotic and abiotic stresses by enhancing plant health and productivity.
The diversity and composition of microbiota culminates in imparting unique characteristics to the host plant and exert a selection pressure in both natural and managed environments. Lumibao et al. (2020) reported elementary insights on how rhizospheric microbial variation in an engineered ecosystem can greatly influence plant provenance. Their study established the crucial role of feedback mechanisms at plant-soil-microbe interface in shaping the plant intraspecific variation in a depauperate ecosystem. However, the reverse hypothesis recognizing the role of plant genotype in leaving phylogenetic signatures on the rhizosphere microbiomes has been proved experimentally for the first time by Perez-Izquierdo et al. (2019). Their finding establishes a good communication between host-genotypic effects and the environmental effects leading to better performance.
Similarly, engineering the banana endospheric bacterial cell walls (Enterobacteriaceae) with 1-aminocyclopropane-1-carboxylate (ACC) deaminase serves as a promising method in imparting resistance against Fusarium wilt progression in banana. Synthetic microbial community has been implicated in studying the priority effects and keystone species in the phyllosphere of gnotobiotic Arabidopsis model system (Carlstrom et al., 2019). Such a valuable approach helps in testing the fundamental principles responsible for shaping community structure in the phyllosphere. Therefore, engineering the incredible diversity of microbes associated with the host plant serves as a stepping stone in the elucidation of microbiota structure and gene functions. Herein, undermentioned techniques have been enlisted to modify/engineer beneficial microbiome function and services that ensure plant productivity through several generations.
Soil Amendments
The interconnection of climatic, edaphic, and biotic conditions collectively regulate the physicochemical properties of the soil. Soil structure, organic matter, pH, texture, temperature, quality, and quantity of carbon inputs (root exudates) and nutrient status create conducive environment for the distribution and functional activities of microbial species (Gomez-Sagasti et al., 2018 and Igiehon and Babalola, 2018). Plant metabolites in the root region exert feedback effects on the selection of the microorganisms for colonization. This elicits several responses leading to healthier ecosystem functionality. Such characteristics pay dividend to both plant health and soil-type–specific microbial assembly (Luo et al., 2017).
Soil amendments can include organic additions which may optimize productivity leading to stable agroecosystem function. Organic amendments initiate a cascade of events between different ecosystem trophic levels by manipulating soil biophysical properties, heterogenous distribution of microbial species, and plant traits. The application of organic amendments influences the composition, distribution, and structure of microbial community (Cesarano et al., 2017). They possess multitude advantages propagating with significant changes in soil fertility, plant and microbial biomass, root traits, tissue elemental composition, substrate utilization, and decomposability (Ganesh et al., 2015).
The essential role of organically managed soil systems over conventional systems have been potentially studied by Lupatini et al. (2017). They assessed the composition and diversification of microbial taxa by employing next-generation sequence techniques based on the 16S rRNA gene. Their study provides a comprehensive insight into the phylogenetic richness, diversity, and heterogeneity of soil microbiota being effected by various soil health treatments. This notion that organic amendments positively influence the soil chemistry, microbiota composition, and crop productivity (Eruca sativa) was further supported by the study of Bonanomi et al. (2020). Frequent addition of organic substrates like biochar, alfalfa hay, and glucose offers a reliable and effective approach for the sustainable management of soil fertility. Similarly, Obermeier et al. (2020) also studied the effects of pelletized spent mushroom substrate (organic substrate) in combination with mineral fertilizer on agricultural soil health and performance of Hordeum vulgare L. Their results highlighted the effect of organic additions in enhancing plant performance and soil bacterial diversity.
Bonanomi et al. (2020) focused on the conducive effects of organic manipulation strategies in inducing plant growth and suppression of plant pathogens. The substantial organic additions work by enhancing the decomposition process which triggers symbiotic microbial colonization and indirectly results into pathogen suppression either by direct parasitism or by production of secondary metabolites with biocontrol potential. The idea of using microbial amendments to enhance crop productivity as investigated by their studies reap potential benefits to the agroecosystem by alleviating the toxic effects of oligotrophication and reduced external inputs. A comprehensive understanding on marked functional implications of soil amendments in manipulating plant-microbe links has been overviewed by Tosi et al. (2020). They reviewed latest trends in optimizing the unique scenario of interactive microbiome at plant-soil interface. Besides organic additions, agricultural practices like rotation, cover crops, intercropping, tillage practices, and multicropping leverage benefit in shaping the diversification of rhizospheric and endospheric microbiota.
In general, application of organic amendments regulates soil nitrogen cycle, improves soil water retention, and improves carbon sequestration and mitigation of climate change effects which ultimately influences plant health and productivity.
Microbe-Based Manipulations
Host-associated microbial community forms synergistic relationships and displays population-based behavior having immense implications for human food security, biodiversity, and agricultural productivity. These interactions influence the physiology of the host by addressing urgent environmental challenges. Hence, it represents an appealing target for in situ microbial engineering.
The frequent application of microbial communities as bioinoculants for increased nutrient mobilization, stress resilience, and plant growth-promoting traits has been well documented (ALKahtani et al., 2020; Alok et al., 2020). Magallon-Servin et al. (2020) designed a consortia of phosphate rock-solubilizing bacteria for testing their capacity in phosphate solubilization, biofilm formation, and root colonization. Their study underscores the promotion of maize seedling growth in low-phosphorus soil amendments under greenhouse conditions by using selected biocompatible multispecies consortia. Similarly, Veeramachaneni and Ramachandrudu (2020) conducted a study for assessing the influence of bioinoculants on oil palm seedlings (Elaeis guineensis Jacq.). They reported the efficacy of microbial-based crop amendment in modulating the dynamics and enzymatic potential of rhizospheric microbes thereby promoting the establishment of healthy oil seed crop. Integration of reductionist approaches by vertical transmission of microbial species is of paramount importance in agricultural settings for upregulating plant growth and development, some of the examples of which have been tabulated (Table 2). This is often associated with navigating the plant system for manipulating genetic, biochemical, physical, and metabolic parameters of the plant (Agler et al., 2016; Qiu et al., 2019; Vannier et al., 2019; Khan et al., 2020).
Recently, there is a surge of interest in accelerating the progress of microbiome research with profound effects on environmental fluctuations. In this context, a series of reductive experiments were carried out for evaluating the capability of endophytic Salicaceae consortia in mitigating abiotic stresses (Aghai et al., 2019). The results of the study affirmed the employment of multispecies consortia as seed or field amendment in garnering a positive conifer seedling performance. Similar to this, Syed et al. (2020) also developed a consortium of plant growth-promoting rhizobacteria (PGPR) in introducing resistance mechanisms against early-stage diseases in Arachis hypogaea. Therefore, such an addition of PGPR consortia is not only significant as biocontrol agent but also imparts multitude growth characteristics to the plant.
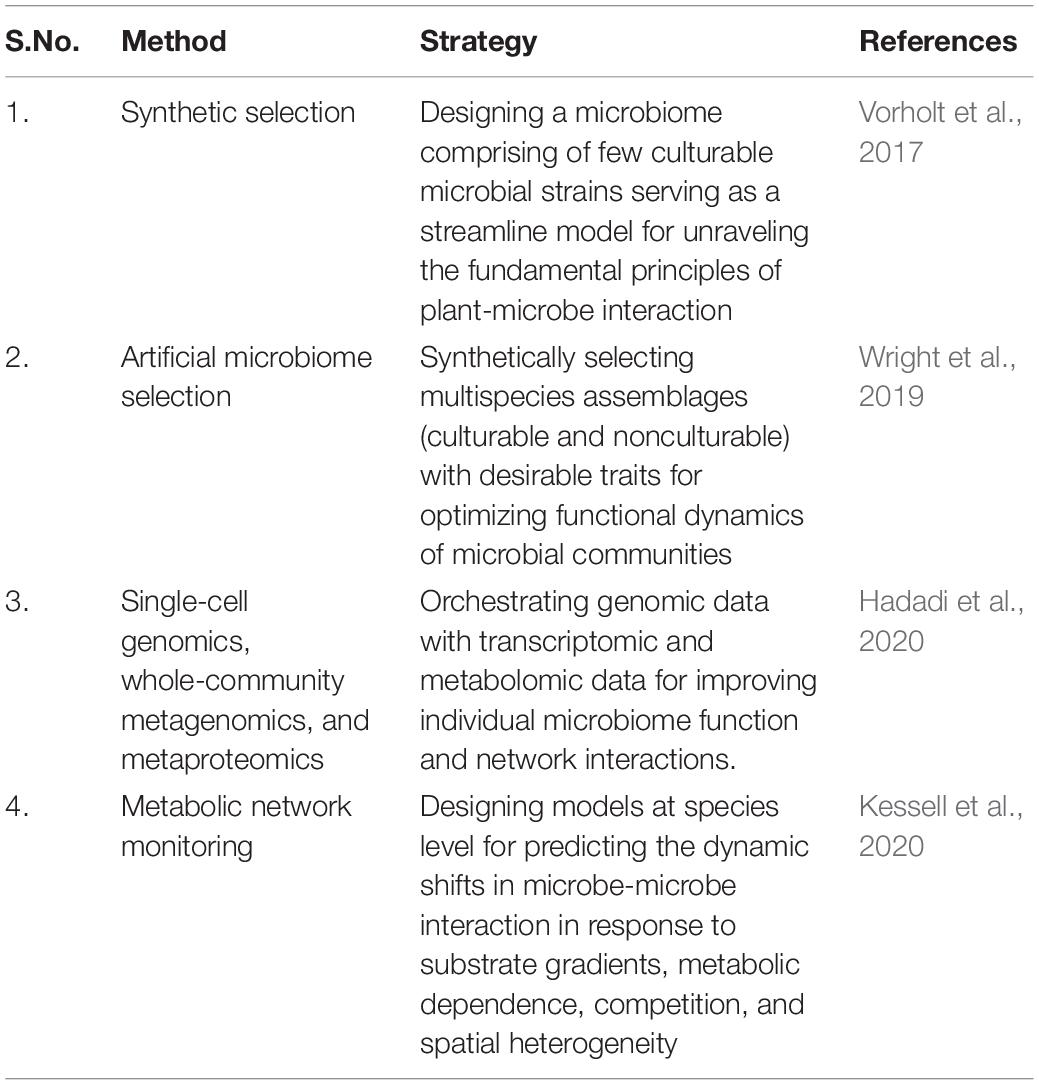
For addressing the knowledge gaps between microbe and microbe interaction dynamics, synthetic biology has emerged as an effective probe. It is a field for designing, predicting, and manipulating the behavior of natural microbial populations with focus on specific applications. Recent advancements in next-generation sequencing (NGS) platform, gene editing technologies, culture independent analysis, meta-“omics” and bioinformatics tools have provided a top–down approach for creatively synthesizing synthetic microbial consortia. Synthetic biologists are honing their ability for revolutionizing agricultural productivity by introducing genes from other photosynthetic microorganisms or by in vivo construction of synthetic metabolic pathways (Ogawa et al., 2015; Long et al., 2016; Schwander et al., 2016).
Recently, a novel strategy was introduced known as synthetic microbial communities (SynComs) based on tailoring plant-microbe interactions by recruiting and manipulating microbial consortia with enhanced colonization, prevalence, and stress-resilient traits. Such synthetically designed microbial inoculants bridge ecology and genetics of microbial communities in sustainable agricultural outcomes (de Souza et al., 2020). Such microbial-based manipulations hold great promise in challenging the global stagnation in terms of crop production, biomass requirement, and climate uncertainties.
Plant-Based Manipulations
To address the global and environmental challenges including climatic variations, desertification, salinization, emergence of new diseases, and resistance to biotic and abiotic stresses, there is a need to look for holistic and sustainable approaches that ensure the global conservation of food resources (Garcia et al., 2020 and Ulian et al., 2020). In this context, plant-based manipulation strategies focused on advanced molecular biology techniques, biochemical approaches, mutagenesis, selection, and breeding of crop varieties have significantly unraveled the plant system against various environmental changes (Table 3). Furthermore, the continued success of renowned Green Revolution can be achieved by high-throughput transcription profiling, identification of specific protein network on large scale, molecular modeling and their association with environmental changes for improved agricultural traits and increased yields (Mahfouz et al., 2016; Jangra et al., 2017).
With the emergence of site-specific nucleases (TALENs), targeted mutagenesis, genome-editing tools like CRISPR/Cas systems, the discovery of novel traits, trait development, site-specific genome modifications have been enabled across the plant kingdom. The use of CRISPR/Cas systems in the targeted engineering of crop traits has been well studied. Butt et al. (2018) studied the CRISPR/Cas9-mediated targeted disruption of carotenoid cleavage dioxygenase (ccd7) in rice (Oryza sativa), controlling a key step in strigolactone biosynthesis and resulting in increased tillering ccd7 mutants. Such an unprecedented engineering of plant architecture traits underpins the single-base level control of CRISPR/Cas systems. In concomitant with the previous study, IR64 rice lines resistant to rice tungro spherical virus (RTSV) were developed by targeted mutagenesis of eIF4G. The CRISPR/Cas9-induced mutations resulted in the generation of novel eIF4G alleles for the production of RTSV resistance varieties (Macovei et al., 2018). The application of CRISPR/Cas9 system has also been reported in preferable endogenous gene modifications (Okuzaki et al., 2018). Gene editing of an agronomically targeted fatty acid desaturase 2 gene (FAD2) in Brassica napus using the CRISPR/Cas9 system resulted in desirable mutant alleles with increased oleic content. Such a cutting edge plant-genome engineering techniques stand poised in shaping and sustainably conserving the architecture of “Green Revolution” (Sedeek et al., 2019).
Simkin et al. (2017) generated transgenic Arabidopsis plants with altered combinations of sedoheptulose 1,7−bisphosphatase (SBPase), fructose 1,6−bisphophate aldolase (FBPA), and the glycine decarboxylase−H protein (GDC−H) gene coding for three different enzymes of the Calvin–Benson cycle and photorespiratory pathway. This multigene stacking results in cumulative increase in photosynthesis, biomass, and seed yield. Designing synthetic carbon-conserving photorespiration bypass routes by metabolic engineering can also be used as a strategy in this direction. Trudeau et al. (2018) used this strategy for two enzymes namely, acetyl-CoA synthetase and propionyl-CoA reductase. These enzymes enable the formation of glycolate reduction module, i.e., recycling of glycolate to ribulose 1,5-bisphosphate without the loss of CO2. Lopez-Calcagno et al. (2019) engineered tobacco plants for increased biomass by overexpressing photorespiration-limiting H-protein of the glycine cleavage system using the leaf−specific promoter ST−LS1. CRISPR/Cas9−mediated editing of SlJAZ2 has been used to prevent stomatal reopening and thus providing resistance. Ortigosa et al. (2019), designed tomato resistant varieties against Pseudomonas syringae pv. tomato (Pto) DC3000 pathogen (causing bacterial speck disease) using this strategy.
The prerequisite for targeted improvement of traits is the use of molecular scissors capable of introducing site-specific double-stranded breaks (DSBs). Such nuclease-induced breaks stimulate the cell’s DNA repair machinery for repairing these DSBs either by nonhomologous end-joining (NHEJ) or by homology-directed repair (HDR). NHEJ results in frameshift mutations creating functional knockouts of a gene. Unlike NHEJ, HDR introduces specific point mutations or a new gene sequence by precisely modifying/repairing genome with oligonucleotide template with homologous ends or arms to the DSB flanking sequence. Such programmable site-specific techniques are used for targeted improvement of crop traits (Butt et al., 2018; Sedeek et al., 2019). Therefore, harnessing engineered variants of eukaryotic genes will pave way for significant improvement in plant yield and food security. Also, key advances in functional genomics will anticipate forward synergies in genomics and plant-breeding techniques for improved crop genetic resources, new cultivar development, and targeted crop improvement.
From an agronomic perspective, key gene players of biosynthetic and metabolic pathway can be targeted at the transcriptional, posttranscriptional, and posttranslational levels for the manipulation and better understanding of pathways. Gene clusters like Phenylalanine ammonia lyase (PALs), Flowering Locus T-Like (FTL), Calcium–dependent protein kinases (CDKs), and other regulatory networks has undergone substantial editing with better performance-tailored plants (Nagegowda and Gupta, 2020; Yadav et al., 2020 and Schulz et al., 2021).
Furthermore, the introduction of microbial genes in plant system for improving biomass yield and photosynthetic efficiency has been well presented in the study of Hay et al. (2017). They transformed the soybean (Glycine max cv. Thorne) with cyanobacterial inorganic carbon transporter B (ictB) gene and found that transgenic plant showed increased photosynthetic rates and biomass production. In accordance with this finding, constitutive expression of cyanobacterial fructose-1,6/sedoheptulose-1,7-bisphosphatase in Glycine max leads to significantly higher carbon assimilation and seed yield under elevated CO2 and temperature conditions as reported by Kohler et al. (2017). Not only this, diverse strategies focusing on improving nutrient uptake, use of broad-host range plasmids for engineering microbes, and improved CO2 fixation efficiency were also targeted. The genetic transformation of cytochrome c6 (UfCyt c6) gene from Ulva fasciata Delile in tobacco was reported for the first time by Yadav et al. (2018). The study reported higher quantum yields of PSII, stomatal conductance, and transpiration rate in transgenic tobacco plants, hence, providing deeper insights into in vivo realization of photosynthetic enhancement by improving photosynthetic electron transfer and water use efficiency. Another feasible approach focused on improving carbon fixation efficiency was studied by Yu et al. (2018). They reported the in vitro designing and investigated the in vivo functional demonstration of synthetic malyl-CoA-glycerate (MCG) pathway in Escherichia coli and photosynthetic organism Synechococcus elongates PCC7942o for optimal synthesis of acetyl-CoA. Nonetheless, the elucidation of data from omics−based system biology has come up as an innovative and powerful tool for heterologous engineering of plant traits.
Therefore, deep understanding of various techniques at soil, microbe, and plant level will surely open a new window in harmonizing the complex assemblages of phytomicrobiome. Furthermore, combined analysis will act as selective force in sculpting intrinsically complex, heterogenous, and dynamic microbiome assembly. However, these manipulation strategies are hindered by current technical gaps, thereby hampering future commercialization and adoption strategies. These include root exudate composition, root architecture, induced systemic response, competition by indigenous soil microflora (at root-soil interface), poor microbial colonization, environmental stress conditions, limited persistence, inability of mutual recognition (at plant-root interface), host genotype, lack of in situ manipulation tools and standardization protocols (at the host level). These knowledge gaps can further be addressed by QTL mapping and integration of various meta-omics approaches (metagenomics, metatranscriptomics, and metabolomics). Implementation of synthetic biology techniques for the discovery of novel phages, plasmids, and transposable elements for in situ microbiome engineering will possibly help in the establishment and success of healthier microbiomes.
Concluding Remarks
In view of the complexities of microbial interactions, “microbe-friendly” plants or genetically engineered/edited plant genome will lead to field success by strengthening plant health and preparedness against environmental fluctuations. This microbial fortification will enhance gene expression, enzymatic parameters, nutrient uptake, and biocontrol response. Thus, strategic manipulation and inoculation of microbial multispecies will have more impactful response on plant growth performance by minimizing chemical farm inputs. In this scenario, microbiome engineering offers exciting opportunities for understanding and engineering individual organisms to the entire ecosystems. This envisioned technology will reveal a vast diversity and elegance underlying natural microbial ecosystems at the frontier of nutritional and ecological demands. The transcendental role of microbes in the field of basic sciences, human health, and agriculture is worth mentioning at the frontier of synthetic biology strategies. However, many of the associated technologies of bioengineering are still in its infancy and require a regulated framework for examining its future in crop improvement programs.
Author Contributions
All authors listed have made substantial, direct and intellectual contribution to the work, and approved it for publication.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank the Department of Biotechnology, Govt. of India, Bioinformatics Centre at School of Biotechnology and Instrumentation facility created under the PURSE, FIST (DST, Govt. of India), SAP (UGC, New Delhi), and RUSA (MHRD) programs.
References
Abadi, V. A. J. M., Sepehri, M., Rahmani, H. A., Zarei, M., Ronaghi, A., Taghavi, S. M., et al. (2020). Role of dominant phyllosphere bacteria with plant growth promoting characteristics on growth and nutrition of maize (Zea mays L.). J. Soil Sci. Plant Nutr. 20, 2348–2363. doi: 10.1007/s42729-020-00302-1
Aghai, M. M., Khan, Z., Joseph, M. R., Stoda, A. M., Sher, A. W., Ettl, G. J., et al. (2019). The effect of microbial endophyte consortia on Pseudotsuga menziesii and Thuja plicata survival, growth, and physiology across edaphic gradients. Front. Microbiol. 10:1353. doi: 10.3389/fmicb.2019.01353
Agler, M. T., Ruhe, J., Kroll, S., Morhenn, C., Kim, S.-T., Weigel, D., et al. (2016). Microbial hub taxa link host and abiotic factors to plant microbiome variation. PLoS Biol. 14:e1002352. doi: 10.1371/journal.pbio.1002352
Ahmad, M., Nadeem, S. M., and Zahir, Z. A. (2019). “Plant-microbiome interactions in agroecosystem: an application,” in Microbiome in Plant Health and Disease, eds V. Kumar, R. Prasad, M. Kumar, and D. Choudhary (Singapore: Springer), 251–291. doi: 10.1007/978-981-13-8495-0_12
Alibrandi, P., Cardinale, M., Rahman, M. M., Strati, F., Cina, P., de Viana, M. L., et al. (2018). The seed endosphere of Anadenanthera colubrina is inhabited by a complex microbiota, including Methylobacterium spp. and Staphylococcus spp. with potential plant-growth promoting activities. Plant Soil 422, 81–99. doi: 10.1007/s11104-017-3182-4
ALKahtani, M. D. F., Fouda, A., Attia, K. A., Al-Otaibi, F., Eid, A. M., Ewais, E. D., et al. (2020). isolation and characterization of plant growth promoting endophytic bacteria from desert plants and their application as bioinoculants for sustainable agriculture. Agronomy 10:1325. doi: 10.3390/agronomy10091325
Alok, D., Annapragada, H., Singh, S., Ghosh, P., Sengupta, A., Basu, D., et al. (2020). Symbiotic nitrogen fixation and endophytic bacterial community structure in Bt-transgenic chickpea (Cicer arietinum L). Sci. Rep. 10:5453. doi: 10.1038/s41598-020-62199-1
Arif, I., Batool, M., and Schenk, P. M. (2020). Plant microbiome engineering: expected benefits for improved crop growth and resilience. Trends Biotechnol. 38, P1385–P1396. doi: 10.1016/j.tibtech.2020.04.015
Arun, K. D., Sabarinathan, K. G., Gomathy, M., Kannan, R., and Balachandar, D. (2020). Mitigation of drought stress in rice crop with plant growth-promoting abiotic stress-tolerant rice phyllosphere bacteria. J. Basic Microbiol. 60, 768–786. doi: 10.1002/jobm.202000011
Bonanomi, G., De Filippis, F., Zotti, M., Idbella, M., Cesarano, G., Al-Rowaily, S., et al. (2020). Repeated applications of organic amendments promote beneficial microbiota, improve soil fertility and increase crop yield. Appl. Soil Ecol. 156:103714. doi: 10.1016/j.apsoil.2020.103714
Brunel, C., Pouteau, R., Dawson, W., Pester, M., Ramirez, K. S., and Van Kleunen, M. (2020). Towards unraveling macroecological patterns in rhizosphere microbiomes. Trends Plant Sci. 25, 1017–1029. doi: 10.1016/j.tplants.2020.04.015
Butt, H., Jamil, M., Wang, J. Y., Al-Babili, S., and Mahfouz, M. (2018). Engineering plant architecture via CRISPR/Cas9-mediated alteration of strigolactone biosynthesis. BMC Plant Biol. 18:174. doi: 10.1186/s12870-018-1387-1
Carlstrom, C. I., Field, C. M., Bortfeld-Miller, M., Muller, B., Sunagawa, S., and Vorholt, J. A. (2019). Synthetic microbiota reveal priority effects and keystone strains in the Arabidopsis phyllosphere. Nat. Ecol. Evol. 3, 1445–1454. doi: 10.1038/s41559-019-0994-z
Carrion, V. J., Perez-Jaramillo, J., Cordovez, V., Tracanna, V., De Hollander, M., Ruiz-Buck, D., et al. (2019). Pathogen-induced activation of disease-suppressive functions in the endophytic root microbiome. Science 366, 606–612. doi: 10.1126/science.aaw9285
Cesarano, G., De Filippis, F., La Storia, A., Scala, F., and Bonanomi, G. (2017). Organic amendment type and application frequency affect crop yields, soil fertility and microbiome composition. Appl. Soil Ecol. 120, 254–264. doi: 10.1016/j.apsoil.2017.08.017
Chagas, F. O., de Cassia Pessotti, R., Caraballo-Rodriguez, A. M., and Pupo, M. T. (2018). Chemical signaling involved in plant–microbe interactions. Chem. Soc. Rev. 47, 1652–1704.
Chen, J., Li, N., Han, S., Sun, Y., Wang, L., Qu, Z., et al. (2020). Characterization and bioremediation potential of nickel-resistant endophytic bacteria isolated from the wetland plant Tamarix chinensis. FEMS Microbiol. Lett. 367:fnaa098. doi: 10.1093/femsle/fnaa098
Chen, X., Wicaksono, W. A., Berg, G., and Cernava, T. (2020). Bacterial communities in the plant phyllosphere harbour distinct responders to a broad-spectrum pesticide. Sci. Total Environ. 751:141799. doi: 10.1016/j.scitotenv.2020.141799
Compant, S., Samad, A., Faist, H., and Sessitsch, A. (2019). A review on the plant microbiome: Ecology, functions, and emerging trends in microbial application. J. Adv. Res. 19, 29–37. doi: 10.1016/j.jare.2019.03.004
Cooke, I., Mead, O., Whalen, C., Boote, C., Moya, A., Ying, H., et al. (2019). Molecular techniques and their limitations shape our view of the holobiont. Zoology. 137:125695. doi: 10.1016/j.zool.2019.125695
de Souza, R. S. C., Armanhi, J. S. L., and Arruda, P. (2020). From microbiome to traits: designing synthetic microbial communities for improved crop resiliency. Front. Plant Sci. 11:1179. doi: 10.3389/fpls.2020.01179
Foo, J. L., Ling, H., Lee, Y. S., and Chang, M. W. (2017). Microbiome engineering: current applications and its future. Biotechnol J. 12:1600099. doi: 10.1002/biot.201600099
Ganesh, K. S., Sundaramoorthy, P., and Nagarajan, M. (2015). Organic soil amendments: potential sourcefor heavy metal accumulation. World Sci. News 16, 28–39.
Garcia, S. N., Osburn, B. I., and Jay Russell, M. T. (2020). One health for food safety, food security, and sustainable food production. Front. Sustain. Food Syst. 4:1. doi: 10.3389/fsufs.2020.00001
Gomez-Sagasti, M. T., Hernandez, A., Artetxe, U., Garbisu, C., and Becerril, J. M. (2018). How valuable are organic amendments as tools for the phytomanagement of degraded soils? The knowns, known, unknowns, and unknowns. Front. Sustain. Food Syst. 2:68. doi: 10.3389/fsufs.2018.00068
Gupta, S., and Pandey, S. (2020). Enhanced salinity tolerance in the common bean (Phaseolus vulgaris) plants using twin ACC deaminase producing rhizobacterial inoculation. Rhizosphere 16:100241. doi: 10.1016/j.rhisph.2020.100241
Hadadi, N., Pandey, V., Chiappino-Pepe, A., Morales, M., Gallart-Ayala, H., Mehl, F., et al. (2020). Mechanistic insights into bacterial metabolic reprogramming from omics-integrated genome-scale models. NPJ Syst. Biol. Appl. 6, 1–11.
Hamilton, C. E., Bever, J. D., Labb, J., Yang, X., and Yin, H. (2016). Mitigating climate change through managing constructed-microbial communities in agriculture. Agr. Ecosyst. Environ. 216, 304–308. doi: 10.1016/j.agee.2015.10.006
Han, T., You, C., Zhang, L., Feng, C., Zhang, C., Wang, J., et al. (2016). Biocontrol potential of antagonist Bacillus subtilis Tpb55 against tobacco black shank. Biocontrol 61, 195–205. doi: 10.1007/s10526-015-9705-0
Hay, W. T., Bihmidinem, S., Mutlu, N., Le Hoang, K., Awada, T., Weeks, D. P., et al. (2017). Enhancing soybean photosynthetic CO2 assimilation using a cyanobacterial membrane protein, ictB. J. Plant Physiol. 212, 58–68. doi: 10.1016/j.jplph.2017.02.003
Igiehon, N. O., and Babalola, O. O. (2018). Rhizosphere microbiome modulators: contributions of nitrogen fixing bacteria towards sustainable agriculture. Int. J. Environ. Res. Public Health 15:574. doi: 10.3390/ijerph15040574
Jagannath, S., Konappa, N. M., Alurappa, R., and Chowdappa, S. (2019). production, characterization of indole acetic acid and its bioactive potential from endophytic fungi of Cymbidium aloifolium L. J. Biol. Active Prod. Nat. 9, 387–409. doi: 10.1080/22311866.2019.1688684
Jangra, S., Mishra, A., Kamboj, D., Yadav, N. R., and Yadav, R. C. (2017). “Engineering abiotic stress tolerance traits for mitigating climate change”, in Plant Biotechnology: Recent Advancements and Developments, eds S. Gahlawat, R. Salar, P. Siwach, J. Duhan, S. Kumar, and P. Kaur (Singapore: Springer), 59–73. doi: 10.1007/978-981-10-4732-9_3
Jin, Y., Zhu, H., Luo, S., Yang, W., Zhang, L., Li, S., et al. (2019). Role of maize root exudates in promotion of colonization of Bacillus velezensis strain S3-1 in rhizosphere soil and root tissue. Curr. Microbiol. 76, 855–862.
Kaur, T., Rani, R., and Manhas, R. K. (2019). Biocontrol and plant growth promoting potential of phylogenetically new Streptomyces sp. MR14 of rhizospheric origin. AMB Expr. 9:125. doi: 10.1186/s13568-019-0849-7
Kessell, A. K., McCullough, H. C., Auchtung, J. M., Bernstein, H. C., and Song, H. S. (2020). Predictive interactome modeling for precision microbiome engineering. Curr. Opin. Chem. Eng. 30, 77–85. doi: 10.1016/j.coche.2020.08.003
Khan, S., Hauptman, R., and Kelly, L. (2020). Engineering the microbiome to prevent adverse events: challenges and opportunities. Ann. Rev. Pharmacol. Toxicol. 61, 159–179. doi: 10.1146/annurev-pharmtox-031620-031509
Kohler, I. H., Ruiz-Vera, U. M., VanLoocke, A., Thomey, M. L., Clemente, T., and Long, S. P. (2017). Expression of cyanobacterial FBP/SBPase in soybean prevents yield depression under future climate conditions. J. Exp. Bot. 68, 715–726. doi: 10.1093/jxb/erw435
Kumar, A., Singh, S., Mukherjee, A., Rastogi, R. P., and Verma, J. P. (2020). Salt-tolerant plant growth-promoting Bacillus pumilus strain JPVS11 for enhancing plant growth attributes of rice and soil health management under salinity stress. Microbiol. Res. 9:126616. doi: 10.1016/j.micres.2020.126616
Lau, E. T., Tani, A., Khew, C. Y., Chua, Y. Q., and San Hwang, S. (2020). Plant growth-promoting bacteria as potential bio-inoculants and biocontrol agents to promote black pepper plant cultivation. Microbiol. Res. 240:126549. doi: 10.1016/j.micres.2020.126549
Liang, H., Liu, S., Wu, S., Xu, D., Jin, F., Faiola, X., et al. (2019). BaiGenetic diversity of diazotrophs and total bacteria in the phyllosphere of Pyrus serotina, Prunus armeniaca, Prunus avium, and Vitis vinifera. Can. J. Microbiol. 65, 1–11. doi: 10.1139/cjm-2018-0588
Liu, H., Brettell, L. E., and Singh, B. (2020). Linking the phyllosphere microbiome to plant health. Trends Plant Sci. 25, 841–844. doi: 10.1016/j.tplants.2020.06.003
Lombardi, N., Vitale, S., Turrà, D., Reverberi, M., Fanelli, C., Vinale, F., et al. (2018). Root exudates of stressed plants stimulate and attract Trichoderma soil fungi. Mol Plant Microbe Interact. 31, 982–994.
Long, B. M., Rae, B. D., Rolland, V., Forster, B., and Price, G. D. (2016). Cyanobacterial CO2-concentrating mechanism components: function and prospects for plant metabolic engineering. Curr. Opin. Plant Biol. 31, 1–8. doi: 10.1016/j.pbi.2016.03.002
Lopez-Calcagno, P. E., Fisk, S., Brown, K. L., Bull, S. E., South, P. F., and Raines, C. A. (2019). Overexpressing the H-protein of the glycine cleavage system increases biomass yield in glass house and field-grown transgenic tobacco plants. Plant Biotechnol. J. 17, 141–151. doi: 10.1111/pbi.12953
Lumibao, C. Y., Bernik, B. M., Formel, S. K., Kandalepas, D., Mighell, K. L., Pardue, J., et al. (2020). Rhizosphere microbial communities reflect genotypic and trait variation in a salt marsh ecosystem engineer. Am. J. Bot. 107, 941–949. doi: 10.1002/ajb2.1497
Luo, Z., Feng, W., Luo, Y., Baldock, J., and Wang, E. (2017). Soil organic carbon dynamics jointly controlled by climate, carbon inputs, soil properties and soil carbon fractions. Glob. Change Biol. 23, 4430–4439. doi: 10.1111/gcb.13767
Lupatini, M., Korthals, G. W., de Hollander, M., Janssens, T. K., and Kuramae, E. E. (2017). Soil microbiome is more heterogeneous in organic than in conventional farming system. Front. Microbiol. 7:2064. doi: 10.3389/fmicb.2016.02064
Macovei, A., Sevilla, N. R., Cantos, C., Jonson, G. B., Slamet-Loedin, I., Cermak, T., et al. (2018). Novel alleles of rice eIF4G generated by CRISPR/Cas9-targeted mutagenesis confer resistance to Rice tungro spherical virus. Plant Biotechnol. J. 16, 1918–1927. doi: 10.1111/pbi.12927
Magallon-Servin, P., Antoun, H., Taktek, S., Taktek, S., and de-Bashan, L. E. (2020). Designing a multi-species inoculant of phosphate rock-solubilizing bacteria compatible with arbuscular mycorrhizae for plant growth promotion in low-P soil amended with PR. Biol. Fertil. Soils 56, 521–536. doi: 10.1007/s00374-020-01452-1
Mahfouz, M. M., Cardi, T., and Neal Stewart, C. (2016). Next-generation precision genome engineering and plant biotechnology. Plant Cell Rep. 35, 1397–1399. doi: 10.1007/s00299-016-2009-8
Marques, J. M., Mateus, J. R., da Silva, T. F., Couto, C. R., Blank, A. F., and Seldin, L. (2019). nitrogen fixing and phosphate mineralizing bacterial communities in sweet potato rhizosphere show a genotype-dependent distribution. Diversity. 11:231. doi: 10.3390/d11120231
Mina, D., Pereira, J. A., Lino-Neto, T., and Baptista, P. (2020). Screening the olive tree phyllosphere: search and find potential antagonists against Pseudomonas savastanoi pv. savastanoi. Front. Microbiol. 11:2051. doi: 10.3389/fmicb.2020.02051
Morel, M., and Castro-Sowinski, S. (2013). “The complex molecular signaling network in microbe–plant interaction,” in Plant Microbe Symbiosis: Fundamentals and Advances, ed. N. K. Arora (New Delhi: Springer), 169–199. doi: 10.1007/978-81-322-1287-4_6
Mueller, U. G., and Sachs, J. L. (2015). Engineering microbiomes to improve plant and animal health. Trends Microbiol. 23, 606–617. doi: 10.1016/j.tim.2015.07.009
Mukherjee, P., Mitra, A., and Roy, M. (2019). Halomonas rhizobacteria of Avicennia marina of Indian Sundarbans promote rice growth under saline and heavy metal stresses through exopolysaccharide production. Front. Microbiol. 10:1207. doi: 10.3389/fmicb.2019.01207
Nagegowda, D. A., and Gupta, P. (2020). Advances in the biosynthesis, regulation, and metabolic engineering of plant specialized terpenoids. Plant Sci. 294:110457. doi: 10.1016/j.plantsci.2020.110457
Nelson, E. B. (2018). The seed microbiome: origins, interactions, and impacts. Plant Soil 422, 7–34. doi: 10.1007/s11104-017-3289-7
Obermeier, M. M., Minarsch, E. L., Durai Raj, A. C., Rineau, F., and Schröder, P. (2020). Changes of soil-rhizosphere microbiota after organic amendment application in a Hordeum vulgare L. short-term greenhouse experiment. Plant Soil 455, 489–506. doi: 10.1007/s11104-020-04637-7
Ogawa, T., Tamoi, M., Kimura, A., Mine, A., Sakuyama, H., Yoshida, E., et al. (2015). Enhancement of photosynthetic capacity in Euglena gracilis by expression of cyanobacterial fructose-1,6-/sedoheptulose-1,7-bisphosphatase leads to increases in biomass and wax ester production. Biotechnol. Biofuels 8:80. doi: 10.1186/s13068-015-0264-5
Okuzaki, A., Ogawa, T., Koizuka, C., Kaneko, K., Inaba, M., Imamura, J., et al. (2018). CRISPR/Cas9-mediated genome editing of the fatty acid desaturase 2 gene in Brassica napus. Plant Physiol. Biochem. 131, 63–69. doi: 10.1016/j.plaphy.2018.04.025
Ortigosa, A., Gimenez-Ibanez, S., Leonhardt, N., and Solano, R. (2019). Design of a bacterial speck resistant tomato by CRISPR/Cas9-mediated editing of Sl JAZ 2. Plant Biotechnol. J. 17, 665–673. doi: 10.1111/pbi.13006
Perez-Izquierdo, L., Zabal-Aguirre, M., Gonzalez-Martinez, S. C., Buee, M., Verdu, M., Rincon, A., et al. (2019). Plant intraspecific variation modulates nutrient cycling through its below ground rhizospheric microbiome. J. Ecol. 107, 1594–1605.
Prajakta, B. M., Suvarna, P. P., Raghvendra, S. P., and Alok, R. R. (2019). Potential biocontrol and superlative plant growth promoting activity of indigenous Bacillus mojavensis PB-35(R11) of soybean (Glycine max) rhizosphere. SN Appl. Sci. 1:1143. doi: 10.1007/s42452-019-1149-1
Prasad, R., Gill, S. S., and Tuteja, N. (2018). New and Future Developments in Microbial Biotechnology and Bioengineering: Crop Improvement through Microbial Biotechnology. Amsterdam: Elsevier.
Qiu, Z., Egidi, E., Liu, H., Kaur, S., and Singh, B. K. (2019). New frontiers in agriculture productivity: optimised microbial inoculants and in situ microbiome engineering. Biotechnol. Adv. 37:107371. doi: 10.1016/j.biotechadv.2019.03.010
Quiza, L., St-Arnaud, M., and Yergeau, E. (2015). Harnessing phytomicrobiome signaling for rhizosphere microbiome engineering. Front. Plant Sci. 6:107. doi: 10.3389/fpls.2015.00507
Rossmann, M., Sarango-Flores, S. W., Chiaramonte, J. B., Kmit, M. C. P., and Mendes, R. (2017). “Plant microbiome: composition and functions in plant compartments,” in The Brazilian Microbiome, eds V. Pylro and L. Roesch (Cham: Springer), 7–20. doi: 10.1007/978-3-319-59997-7_2
Saleem, M., Meckes, N., Pervaiz, Z. H., and Traw, M. B. (2017). Microbial interactions in the phyllosphere increase plant performance under herbivore biotic stress. Front. Microbiol. 20:41. doi: 10.3389/fmicb.2017.00041
Santoyo, G., Moreno-Hagelsieb, G., del Carmen Orozco-Mosqueda, M., and Glick, B. R. (2016). Plant growth-promoting bacterial endophytes. Microbiol. Res. 183, 92–99. doi: 10.1016/j.micres.2015.11.008
Schulz, P., Piepenburg, K., Lintermann, R., Herde, M., Schottler, M. A., Schmidt, L. K., et al. (2021). Improving plant drought tolerance and growth under water limitation through combinatorial engineering of signalling networks. Plant Biotechnol. J. 19:74.
Schwander, T., Von Borzyskowski, L. S., Burgener, S., Cortina, N. S., and Erb, T. J. (2016). A synthetic pathway for the fixation of carbon dioxide in vitro. Science 354, 900–904. doi: 10.1126/science.aah5237
Sedeek, K. E., Mahas, A., and Mahfouz, M. (2019). Plant genome engineering for targeted improvement of crop traits. Front. Plant Sci. 10:114. doi: 10.3389/fpls.2019.00114
Senthil-Kumar, M., and Mysore, K. S. (2013). Nonhost resistance against bacterial pathogens: retrospectives and prospects. Ann. Rev. Phytopathol. 51, 407–427. doi: 10.1146/annurev-phyto-082712-102319
Sexton, W. K., Fidero, M., Spain, J. C., Jiang, L., Bucalo, K., Cruse-Sanders, J. M., et al. (2019). Characterization of endophytic bacterial communities within greenhouse and field-grown rhizomes of three rare pitcher plant species (Sarracenia oreophila, S. leucophylla, and S. purpurea spp. venosa) with an emphasis on nitrogen-fixing bacteria. Plant Soil 447, 257–279. doi: 10.1007/s11104-019-04372-8
Shaposhnikov, A. I., Shakhnazarova, V. Y., Vishnevskaya, N. A., Borodina, E. V., and Strunnikova, O. K. (2020). Aromatic carboxylic acids in barley-root exudates and their influence on the growth of Fusarium culmorum and Pseudomonas fluorescens. Appl. Biochem. Microbiol. 56, 344–351.
Sharma, R., Gal, L., Garmyn, D., Bisaria, V. S., Sharma, S., and Piveteau, P. (2020). Evidence of biocontrol activity of bioinoculants against a human pathogen, Listeria monocytogenes. Front. Microbiol. 11:350. doi: 10.3389/fmicb.2020.00350
Sharma, S., Gupta, S., Dhar, M. K., and Kaul, S. (2018). Diversity and bioactive potential of culturable fungal endophytes of medicinal shrub Berberis aristata DC.: a first report. Mycobiology 46, 370–381. doi: 10.1080/12298093.2018.1538068
Shurigin, V., Egamberdieva, D., Li, L., Davranov, K., Panosyan, H., Birkeland, N. K., et al. (2020). Endophytic bacteria associated with halophyte Seidlitzia rosmarinus Ehrenb. ex Boiss. from saline soil of Uzbekistan and their plant beneficial traits. J. Arid. Land 12, 730–740. doi: 10.1007/s40333-020-0019-4
Simkin, A. J., Lopez-Calcagno, P. E., Davey, P. A., Headland, L. R., Lawson, T., Timm, S., et al. (2017). Simultaneous stimulation of sedoheptulose 1, 7-bisphosphatase, fructose 1, 6-bisphophate aldolase and the photorespiratory glycine decarboxylase-H protein increases CO2 assimilation, vegetative biomass and seed yield in Arabidopsis. Plant Biotechnol. J. 15, 805–816. doi: 10.1111/pbi.12676
Singh, A., Kumar, M., Verma, S., Choudhary, P., and Chakdar, H. (2020). “Plant microbiome: trends and prospects for sustainable agriculture,” in Plant Microbe Symbiosis, eds A. Varma, S. Tripathi, and R. Prasad (Cham: Springer), 129–151. doi: 10.1007/978-3-030-36248-5_8
Singh, A. V., Prasad, B., and Goel, R. (2018). Plant growth promoting efficiency of phosphate solubilizing Chryseobacterium sp. PSR 10 with different doses of N and P fertilizers on lentil (Lens culinaris var. PL-5) growth and yield. Int. J. Curr. Microbiol. Appl. Sci. 7, 2280–2289. doi: 10.20546/ijcmas.2018.705.265
Singh, T., and Singh, D. K. (2019). Rhizospheric Microbacterium sp. P27 showing potential of lindane degradation and plant growth promoting traits. Curr. Microbiol. 76, 888–895. doi: 10.1007/s00284-019-01703-x
Stringlis, I. A., Jonge, R. D., and Pieterse, C. M. J. (2019). The age of coumarins in plant–microbe interactions. Plant Cell Physiol. 60, 1405–1419. doi: 10.1093/pcp/pcz076
Syed, S., Tollamadugu, N. P., and Lian, B. (2020). Aspergillus and Fusarium control in the early stages of Arachis hypogaea (groundnut crop) by plant growth-promoting rhizobacteria (PGPR) consortium. Microbiol. Res. 240:126562. doi: 10.1016/j.micres.2020.126562
Tosi, M., Mitter, E. K., Gaiero, J., and Dunfield, K. (2020). It takes three to tango: the importance of microbes, host plant, and soil management to elucidate manipulation strategies for the plant microbiome. Can J. Microbiol. 66, 413–433. doi: 10.1139/cjm-2020-0085
Trudeau, D. L., Edlich-Muth, C., Zarzycki, J., Scheffen, M., Goldsmith, M., Khersonsky, O., et al. (2018). Design and in vitro realization of carbon-conserving photorespiration. Proc. Natl. Acad. Sci. U.S.A. 115, e11455–e11464. doi: 10.1073/pnas.1812605115
Ulian, T., Diazgranados, M., Pironon, S., Padulosi, S., Liu, U., Davies, L., et al. (2020). Unlocking plant resources to support food securityand promote sustainable agriculture. Plants, People, Planet. 2, 421–445. doi: 10.1002/ppp3.10145
Vandenkoornhuyse, P., Quaiser, A., Duhamel, M., Le Van, A., and Dufresne, A. (2015). The importance of the microbiome of the plant holobiont. New Phytol. 206, 1196–1206. doi: 10.1111/nph.13312
Vannier, N., Agler, M., and Hacquard, S. (2019). Microbiota-mediated disease resistance in plants. PLoS Pathog. 15:e1007740. doi: 10.1371/journal.ppat.1007740
Veeramachaneni, S., and Ramachandrudu, K. (2020). Changes in growth, microbial and enzyme activities in oil palm nursery in response to bioinoculants and chemical fertilizers. Arch. Agron. Soil Sci. 66, 545–558.
Velmourougane, K., Saxena, G., and Prasanna, R. (2017). “Plant-microbe interactions in the rhizosphere: mechanisms and their ecological benefits,” in Plant-Microbe Interactions in Agro-Ecological Perspectives, eds D. Singh, H. Singh, and R. Prabha (Singapore: Springer), 193–219. doi: 10.1007/978-981-10-6593-4_7
Vorholt, J. A., Vogel, C., Carlstrom, C. I., and Muller, D. B. (2017). Establishing causality: opportunities of synthetic communities for plant microbiome research. Cell Host Microbe 22, 142–155. doi: 10.1016/j.chom.2017.07.004
Wang, X., Whalley, W. R., Miller, A. J., White, P. J., Zhang, F., and Shen, J. (2020). Sustainable cropping requires adaptation to a heterogeneous rhizosphere. Trends Plant Sci. 25, 1194–1202. doi: 10.1016/j.tplants.2020.07.006
Wright, R. J., Gibson, M. I., and Christie-Oleza, J. A. (2019). Understanding microbial community dynamics to improve optimal microbiome selection. Microbiome 7:85. doi: 10.1186/s40168-019-0702-x
Yadav, S. K., Khatri, K., Rathore, M. S., and Jha, B. (2018). Introgression of UfCyt c 6, a thylakoid lumen protein from a green seaweed Ulva fasciata Delile enhanced photosynthesis and growth in tobacco. Mol. Biol. Rep. 45, 1745–1758. doi: 10.1007/s11033-018-4318-1
Yadav, V., Wang, Z., Wei, C., Amo, A., Ahmed, B., Yang, X., et al. (2020). Phenylpropanoid pathway engineering: an emerging approach towards plant defense. Pathogens 9:312.
Yu, H., Li, X., Duchoud, F., Chuang, D. S., and Liao, J. C. (2018). Augmenting the Calvin–Benson–Bassham cycle by a synthetic malyl-CoA-glycerate carbon fixation pathway. Nat. Commun. 9:2008. doi: 10.1038/s41467-018-04417-z
Keywords: biocontrol agents, biogeochemical processes, microbiome, microbiome engineering, omics tools, plant efficacy, mutualism
Citation: Kaul S, Choudhary M, Gupta S and Dhar MK (2021) Engineering Host Microbiome for Crop Improvement and Sustainable Agriculture. Front. Microbiol. 12:635917. doi: 10.3389/fmicb.2021.635917
Received: 30 November 2020; Accepted: 16 April 2021;
Published: 28 May 2021.
Edited by:
Alok Kumar Srivastava, National Bureau of Agriculturally Important Microorganisms (ICAR), IndiaReviewed by:
Ajar Nath Yadav, Eternal University, IndiaYangchun Xu, Nanjing Agricultural University, China
Zhiguang Qiu, Western Sydney University, Australia
Copyright © 2021 Kaul, Choudhary, Gupta and Dhar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sanjana Kaul, c2Fucm96aWVAcmVkaWZmbWFpbC5jb20=
†ORCID: Sanjana Kaul, orcid.org/0000-0002-6256-0586; Malvi Choudhary, orcid.org/0000-0001-8874-1820; Suruchi Gupta, orcid.org/0000-0002-2994-0640; Manoj K. Dhar, orcid.org/0000-0002-8777-6244
 Sanjana Kaul
Sanjana Kaul Malvi Choudhary
Malvi Choudhary Suruchi Gupta
Suruchi Gupta Manoj K. Dhar
Manoj K. Dhar