- 1Center for Food Safety, Department of Food Science, University of Arkansas, Fayetteville, AR, United States
- 2Meat Science and Animal Biologics Discovery, Animal and Dairy Sciences, University of Wisconsin-Madison, Madison, WI, United States
Salmonella Reading is an ongoing public health issue in the turkey industry, leading to significant morbidity in humans in the United States. Pre-harvest intervention strategies that contribute to the reduction of foodborne pathogens in food animals, such as the yeast fermentation metabolites of Original XPCTM (XPC), may become the key to multi-hurdle farm to fork strategies. Therefore, we developed an anaerobic in vitro turkey cecal model to assess the effects of XPC on the ceca of commercial finisher tom turkeys fed diets void of XPC and antibiotics. Using the in vitro turkey cecal culture method, ceca were tested with and without XPC for their anti-Salmonella Reading and the previously defined anti-Typhimurium (ST97) effects. Ultimately, the anti-Salmonella effects were independent of serovar (P > 0.05). At 0 h post inoculation (hpi), Salmonella levels were equivalent between treatments at 7.3 Log10 CFU/mL, and at 24 hpi, counts in XPC were reduced by 5 Log10 CFU/mL, which was 2.1 Log10 lower than the control (P < 0.05). No differences in serovar prevalence existed (P > 0.05), with a 92% reduction in Salmonella positive XPC-treated ceca cultures by 48 hpi (P < 0.05). To evaluate changes to the microbiota independent of the immune response, the 16S rDNA was sequenced using the Illumina MiSeq platform. Data indicated a profound effect of time and treatment for the reduction of Salmonella irrespective of serovar. XPC sustained diversity metrics compared to the control, demonstrating a reduction in diversity over time (Q < 0.05).
Introduction
By July 19, 2018, an outbreak of Salmonella enterica serovar Reading linked to ground turkey had effected 26 states in the United States. By the end of the outbreak, 358 cases were reported across 42 states and Washington DC, resulting in 133 hospitalizations and one death directly from the consumption or cross-contamination of S. Reading contaminated ground turkey meat (Centers for Disease Control and Prevention [CDC], 2019). In parallel, between April of 2017 and January of 2020, a total of 130 cases of S. Reading [British Columbia (33), Alberta (44), Saskatchewan (8), Manitoba (25), Ontario (9), Quebec (2), New Brunswick (1), Prince Edward Island (1), Northwest Territories (1), and Nunavut (6)] were confirmed across Canada (Public Health Agency of Canada [PHAC], 2021). As a result, the U.S. Food Safety and Inspection Service (FSIS), Canadian Food Inspection Agency, and other agencies across North America and the world have reached out to the turkey industry to find solutions to end this escalating public health crisis.
Salmonella Reading is a common serovar associated with poultry meat, as well as ground beef and pork, with a prevalence of 4.1 and 4.0% among raw swine carcasses and ground pork tested (Schlosser et al., 2000). Additionally, S. Reading is consistently ranked among the top 10 most isolated serovars in ground turkey, representing 25% of the 1.7% Salmonella positive samples (Food Safety Inspection Service [FSIS], 2014). In 2009, a survey of Oklahoma retail ground poultry meat representing five brands and six supermarket chains in Tulsa, Oklahoma, indicated that 47% (66/141) of the ground turkey meat was positive for Salmonella (Gad et al., 2018). Nevertheless, the incidence of foodborne illness remains relatively low by comparison. In addition to Salmonella and more specifically, S. Reading, being isolated frequently among chicken and turkey commodities, Salmonella isolates recovered from ground meat are phenotypically diverse, exhibiting different antimicrobial resistance and pulse-field gel electrophoresis patterns (Zhao et al., 2006). Therefore, there is a critical need for further interventions at the farm and processing levels in order to reduce the incidence of S. Reading and other S. enterica serovars.
Original XPCTM is a concentrated Saccharomyces cerevisiae fermentate that is added to animal feed to maximize the nutritional benefits (Diamond V Original XPCTM (XPC), 2020). In addition to extensive nutritional characteristics, fermentation increases the presence of lactic acid bacteria, promotes basic pH, and produces high amounts of organic acids which potentially help sustain the feed from pathogen colonization prior to ingestion and promote a good gastrointestinal environment in animals (Canibe and Jenses, 2012). The fermentation metabolites of XPC have demonstrated anti-Salmonella effects against multiple Salmonella serovars (Feye et al., 2016a,b; Rubinelli et al., 2016; Park et al., 2017; Roto et al., 2017). These effects include the reduction of multimodal antibiotic resistance and attenuating the virulence of the pathogen, specifically (Feye et al., 2019). A likely mechanism associated with XPC is rooted in the demonstrated changes to the microbiota in response to XPC supplementation, shifting the metabolites present, and ultimately reducing Salmonella fitness (Park et al., 2017; Rubinelli et al., 2016; Feye et al., 2019). The same mechanism likely drives the immune-modulating response that reduces peripheral inflammation while increasing the phagocytosis of Salmonella (Feye et al., 2016b, 2019; Nelson et al., 2020).
Due to the significance of S. Reading outbreaks and the previously demonstrated anti-Salmonella effects of XPC, it is the objective of the current study to determine whether the presence of XPC could lead to the reduction of a nalidixic acid-resistant (64 μg/mL) strain of S. Reading in an in vitro turkey cecal culture model. The in vitro fermentation cecal model is a robust method to research microbial interactions while avoiding host factors, such as an immune response (Feye et al., 2019; Poeker et al., 2019). As this model was untested in turkeys, a previous strain of nalidixic acid-resistant Salmonella Typhimurium that was successfully reduced by XPC in the in vitro chicken cecal model was used as a control. Therefore, if S. Reading and Typhimurium were reduced equally or at least followed the same trend, the model should represent the cecal microbial ecology and its response to the presence of XPC.
Materials and Methods
Tom Turkey Ceca Procurement
There were two independent trials for this experiment, with five ceca being collected at each trial from a USDA inspected commercial processing plant located in Arkansas, United States, and transported to the Center for Food Safety at the University of Arkansas on ice (n = 10). The turkeys were processed as a component of standard processing production practices by a commercial collaborator and were exempt from the Institutional Animal Care and Use Committee. The ceca were aseptically collected by commercial collaborators and were at the laboratory within one hour of slaughter for use in the experiment.
In vitro Tom Turkey Cecal Culture Model
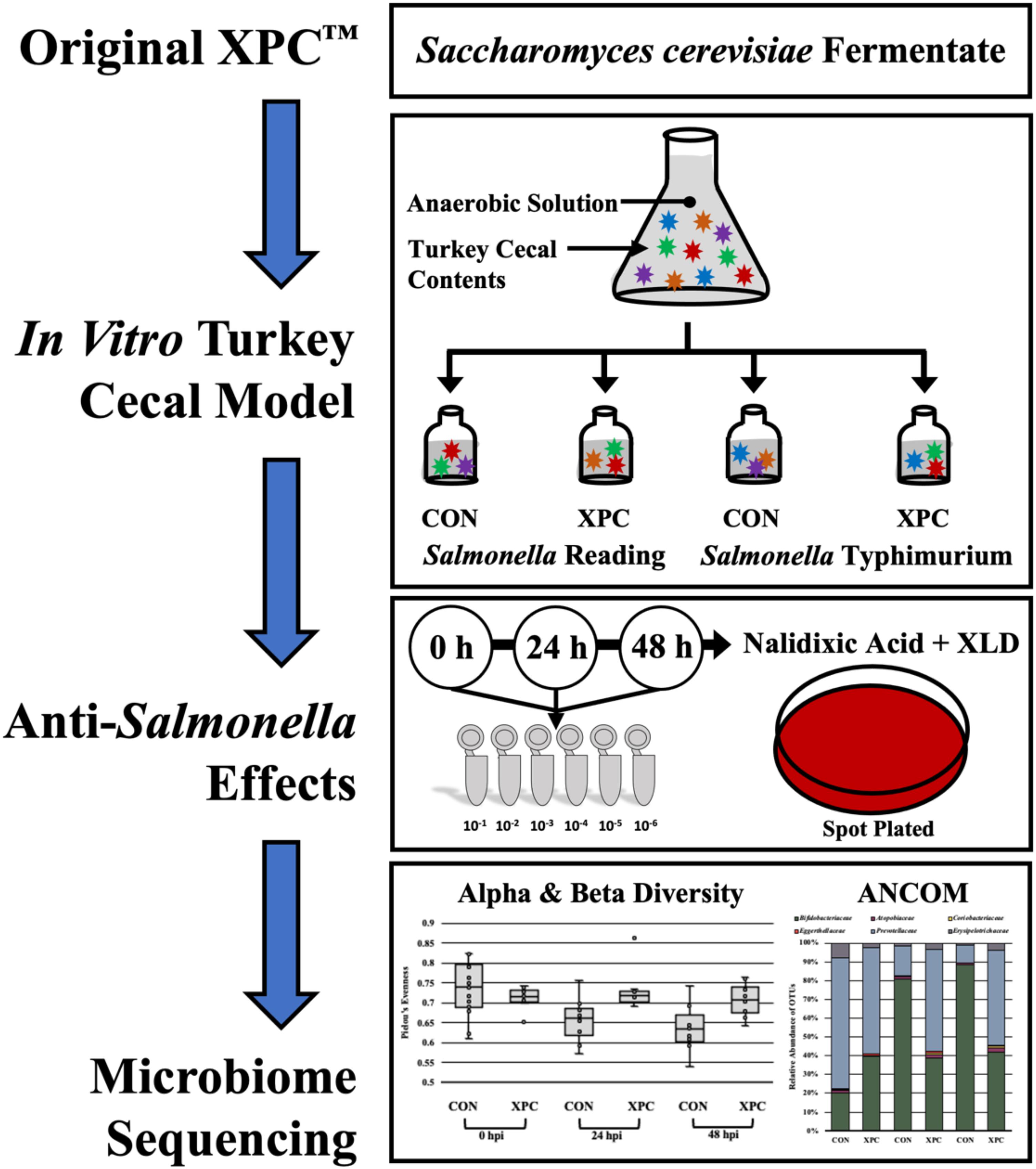
The cecal cultures were prepared in an anaerobic chamber (7% H2, 5% CO2, and 88% N2) to closely mimic the cecal environment (Rubinelli et al., 2016; Figure 1). Once introduced to the anaerobic chamber, a 0.1 g portion of ceca contents from the proximal end of the ceca inferior to the ileal–cecal junction was cut, and the contents were aliquoted into sterile 1.5 mL microcentrifuge tubes. Cecal contents were subsequently re-suspended in 1 mL of Anaerobic Dilution Solution (ADS). The ADS medium consisted of 0.45 g/L K2 HPO4, 0.45 g/L KH2PO4, (NH2)2SO4, 0.9 g/L NaCl, 0.1875 g/L MgSO4⋅7H2O, 0.12 g/L CaCl2⋅2H2O, 1 mL of 0.1% resazurin, and 0.05% v/v cysteine HCl and was autoclaved. Filter sterilized 0.4% CO2 was added post autoclaving (Rubinelli et al., 2016). Prior to the start of the experiment, the ADS was allowed to become anaerobic. As the blue resazurin dye became clear, it indicated minimal oxygen present in the system.
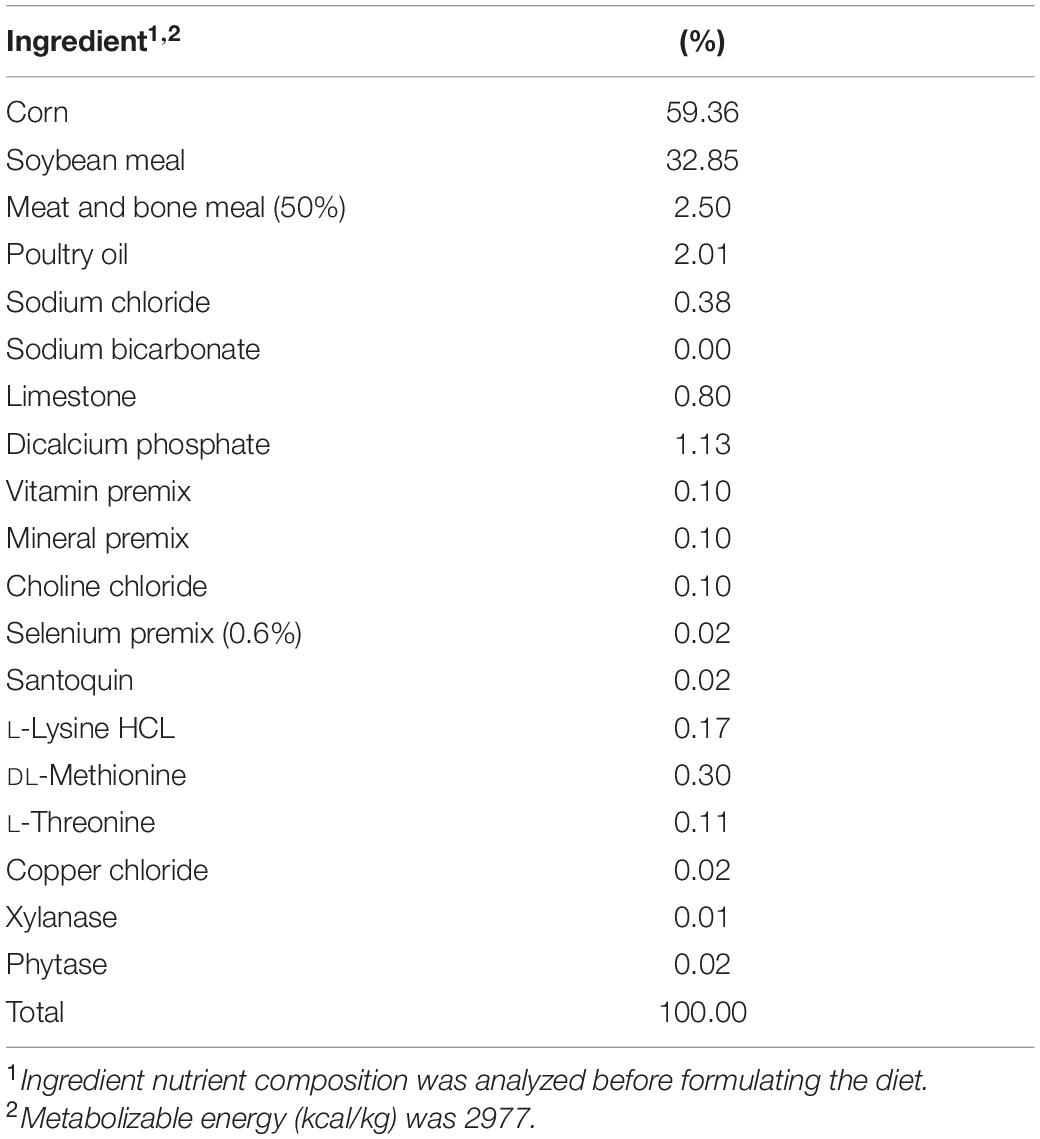
Approximately, 100 mg of cecal content was resuspended in 900 μL of ADS and vortexed until completely homogenized. Following, a 1:3000 dilution was performed by adding 1 mL of the re-suspended ceca into sterile Erlenmeyer flasks containing 299 mL ADS each, and the cecal cultures were homogenized by pipetting. A 20 mL portion from each flask was then aliquoted to each of four serum bottles containing the following treatments: (1) 0.25 g of the basal diet feed inoculated with S. Typhimurium; (2) 0.25 g of the basal diet inoculated with S. Reading; (3) 0.25 g of the basal diet + 0.2 g XPC (1.25% w/v) inoculated with S. Typhimurium; and (4) 0.25 g of the basal diet + 0.2 g XPC (1.25% w/v) inoculated with S. Reading. The basal diet used in the current study was formulated to meet the recommend nutrient requirements for broilers according to the NRC guidelines (Table 1; National Research Council [NRC], 1994). Although chicken feed was utilized in the current study, the metabolizable energy and protein composition of the basal diet was comparative to standard commercial turkey diets (National Research Council [NRC], 1994). The XPC was a fresh shipment from Diamond V and was received as a standard representation of their commercial production and was visually free of mold, insects, or other contaminants.
Once the ADS solution was clear, and therefore anaerobic, the bottles were subsequently sealed with a thick rubber stopper and crimped to retain the anaerobic headspace. The serum bottles were pre-incubated at 42°C and oscillated at 130 rpm for 24 h. Pre-incubation was chosen as previously published studies indicate that XPC is effective after it has been metabolized by the cecal contents (Rubinelli et al., 2016).
Salmonella Inocula Preparation and Inoculation
Salmonella Reading used in the current study was provided by Diamond V (Cedar Rapids, IA, United States), where the company obtained the donated isolate during the S. Reading outbreak among raw turkey products in the United States (Centers for Disease Control and Prevention [CDC], 2019). The S. Typhimurium (ST97) strain used in the study was isolated from chicken by the Poultry Health Laboratory at the University of Arkansas (Park et al., 2017). Both S. Reading and Typhimurium were resistant to nalidixic acid at 64 μg/mL prior to obtaining the cultures from Diamond V and the Poultry Health Laboratory. In order to use both S. Reading and Typhimurium as marker strains throughout the study, significant care was taken to ensure that selective pressure (64 μg/mL of nalidixic acid) was maintained throughout the culture phase before use and during stock preparations. Before initiating the experiment, frozen stocks of S. Reading and Typhimurium (ST97) from the Center for Food Safety’s culture collection were inoculated into tryptic soy broth (TSB; BD Biosciences, San Jose, CA, United States). Cultures were prepared in an oscillating incubator (125 rpm) aerobically at 37°C for 12–16 h. Immediately following, cultures were streaked onto Xylose Lysine Deoxycholate (XLD, HiMedia, West Chester, PA, United States) containing 64 μg/mL of nalidixic acid (Alfa Aesar, Haverhill, MA, United States) and incubated in an oscillating incubator (125 rpm) overnight (12–16 h) at 37°C, aerobically. One colony was used to inoculate 35 mL of TSB and incubated overnight under the aforementioned conditions. The inoculum was prepared as previously described (Rubinelli et al., 2016).
After the pre-incubation of the turkey cecal cultures with and without XPC, cultures were inoculated with the marker strains of S. Reading or Typhimurium according to Rubinelli et al. (2016). As such, cultures designated as the Salmonella treatment bottles were inoculated with one of the two serovars of Salmonella, with 106 cells/mL S. Typhimurium ST97 or 106 cells/mL S. Reading. After inoculation and the zero timepoint enumeration, serum bottles were sealed with a thick rubber stopper and crimped to retain the anaerobic headspace.
Salmonella Enumeration
As the tom turkey cecal cultures were inoculated with S. Reading and Typhimurium, a 1 mL aliquot was taken for the zero timepoint enumeration. The bottles were then resealed with the thick rubber stoppers and crimped inside of the anaerobic chamber to retain the anaerobic headspace. Cultures were then placed into the oscillating incubator at 42°C and oscillated at 130 rpm until the 24 and 48 h timepoints occurred where they were subsampled under anaerobic conditions and plated. At 0, 24, and 48 h, the aliquots were serially diluted (1:10) with 1× phosphate buffered saline (PBS). Tom turkey cecal cultures were spot plated on XLD agar containing 20 μg/mL nalidixic acid in duplicate and allowed to dry completely prior to inverting the plates (Jett et al., 1997; Herigstad et al., 2001; Naghili et al., 2013). The plates were incubated at 37°C for 24 h, and colonies were enumerated. Prevalence was identified by positive plates using the spot plate method, not by enrichment. Therefore, while the term prevalence was used throughout the study, it is not a true measure of prevalence but that the colonies were not detected. The necessary caveats should be considered when evaluating data presented herein as traditional enrichment may yield different results.
The input strain was confirmed via an antibiogram as the Salmonella strains were 64 μg/mL resistant to nalidixic acid. Briefly, Salmonella isolated from XLD media containing 20 μg/mL were individually isolated into a 96-well flat-bottom plate containing 200 μL of sterile, pre-warmed TSB. The plate was incubated statically for 24 h at 37°C and pin replicated into individual plates containing nalidixic acid at 64 μg/mL. The concentration of nalidixic acid (64 μg/mL) was the chosen resistance level provided by Diamond V Mills. All colonies enumerated throughout this study were subjected to this antibiogram. Irrespective of turbidity, turbid wells were considered positive for resistance. Additionally, non-inoculated controls were included to control for environmental contamination.
DNA Extraction and Microbiome Sequencing
All timepoints were collected for microbiome analysis and stored in 1.5 mL microcentrifuge tubes at −20°C until they could be processed. Individual aliquots of each sample (200 mg) were extracted using the Fast Mini Stool Kit (Qiagen, Hilden, Germany) using standard protocols and subsequently diluted to 10 ng/μL using a NanodropTM 1000 spectrophotometer (Thermo Scientific, Waltham, MA, United States). Following the Kozich et al. (2013) protocol, 16S rDNA gene sequencing libraries targeting the V4 region were prepared using a high-fidelity polymerase, Pfx, under the recommended PCR conditions (Invitrogen, Carlsbad, CA, United States). Libraries were then quantified with the KAPA Library quantification kit (Kapa Biosystems, Inc., Wilmington, MA, United States) specified for Illumina platforms and with a broad range dsDNA kit for a Qubit fluorometer (Invitrogen, Carlsbad, CA, United States). Library amplicon size in base pairs (bp) was determined using an Agilent bioanalyzer (Agilent Technologies, Santa Clara, CA, United States). The library was then diluted to 20 pM with HT1 buffer and 10% PhiX, loaded into a Miseq V2 cartridge, and sequenced on an Illumina MiSeq as per standard Illumina practices (Illumina, San Diego, CA, United States). Resulting sequences (fastq files) were downloaded from Illumina BaseSpace and uploaded to a GitHub depository1. Data were also uploaded to NCBI Sequence Read Archive (SRA) under the accession PRJNA716729.
Bioinformatic Analyses
Although a total of 10 cecal cultures were sampled directly for microbiome analysis at each timepoint and inocula (n = 10), eight samples per treatment and timepoint passed quality control measures prior to sequencing. Therefore, only eight replications per treatment and inocula are represented in the microbiome data.
The subsequent data were downloaded from Illumina BaseSpace, de-multiplexed, and locally uploaded into QIIME2.2019.10 (Bolyen et al., 2018). Sequences were filtered for quality and trimmed via DADA2, with chimeras filtered by consensus and quality (Callahan et al., 2016) (via q2−dada2). A contingency filter removed frequencies of less than 3 per sample to reduce noise. The phylogenetic trees were created in mafft (Katoh et al., 2002) (via q2−alignment), with the taxonomical alignments aligned to SILVA full OTU sequence with a confidence limit of 95% (Bokulich et al., 2018). Alpha and Beta diversity analyses were rarified at 8000 reads, which was when the read samples reached their maximum and sustained differences in diversity. Alpha diversity was assessed for richness with the Shannon diversity index and evenness using Pielou’s Evenness. Beta-diversity metrics were analyzed with quantitative indices, such as Jaccard dissimilarity index and weighted UniFrac distance matrix (Lozupone et al., 2007), using the Analysis of Similarity (ANOSIM) function, which takes into account dispersion and the average variation of the populations (Anderson, 2001). Significant features of time were plotted along the X-axis and visualized using the Emperor PCoA plot. The Alpha diversity analytics included the Kruskal–Wallis tests for pairwise differences in the treatment groups (Kruskal and Wallis, 1952). The differential abundance was identified using the ANCOM analysis, which was chosen due to the low sample size (Mandal et al., 2015).
Statistical Analyses
Per sample tested, the arithmetic mean colony-forming unit counts of the two technical replicates per biological replicate and treatment were taken and imported into Excel (Microsoft, Redmond, CA, United States). The colony-forming unit counts were Log10 transformed, and the document was exported into SAS statistical software (Version 9.4, SAS Institute Inc., Cary, NC, United States). All zeros were assumed zero throughout this trial as they were below the limit of detection. Salmonella CFU data were analyzed for the main effects (Treatment, Serovar, Time) and evaluated with a full factorial interaction with Time as a repeated measure. Means were separated using least squares means (LSMEANS) with the PDIFF option. Prevalence was a binomial response defined as positive (growth) or negative (no growth) and analyzed with a χ2 analysis using the Nominal Logistic function. Significance was defined at P ≤ 0.05.
Microbiome main effects were considered significant if the main effect had P ≤ 0.05, and the pairwise effect had Q ≤ 0.05 with each statistical measurement within the QIIME2 pipeline. The Q-value represents the P-value as adjusted for a strict false discovery rate, which is already incorporated into the QIIME2.2019.10 pipeline. All microbiome analyses were done in accordance with the QIIME2 pipeline and aforementioned packages defined in the previous method section.
Results
XPC Exhibits Reduced Salmonella Reading Load
Salmonella Reading was evaluated alongside S. Typhimurium to determine if the reduction of Salmonella was serovar specific. The preadaptation step was taken to ensure that the metabolites of XPC were properly liberated by the microbiota (Rubinelli et al., 2016). Samples from the turkey cecal cultures were taken immediately after inoculation 0 h (s) post-inoculation (hpi), 24 hpi, and 48 hpi (n = 10, N = 160, k = 4, 3 timepoints). There were no differences in the prevalence of the Salmonella between the two serovars utilized in this study (P = 0.7735, Supplementary Tables 1, 2). As there was no significant serovar effect, the serovars were pooled to create a protected pool (n = 20, N = 160). Using Time as a repeated measure, the effect of time and treatment, and their subsequent interaction was significant (P < 0.0001). Therefore, as time progressed, there was a decrease in Salmonella prevalence (Main Effect of Time, P < 0.0001; Table 2). There was an 11% reduction of Salmonella prevalence at 24 hpi and a 61% reduction by 48 hpi. As for the effect of treatment (Main Effect of Treatment, P < 0.001), the addition of XPC reduced Salmonella prevalence by 25% compared to the control (Table 1). There was a 92% drop in Salmonella prevalence over time (0–48 h hpi) in the XPC-treated turkey cecal cultures (Treatment × Time Interaction, P < 0.0001; Table 2). In the control turkey cecal cultures, there was a 30% decrease in the prevalence of Salmonella over time (0–48 hpi). At 48 hpi, the control turkey cecal cultures had a prevalence of 70% where XPC-treated cultures had a 7.58% Salmonella prevalence.
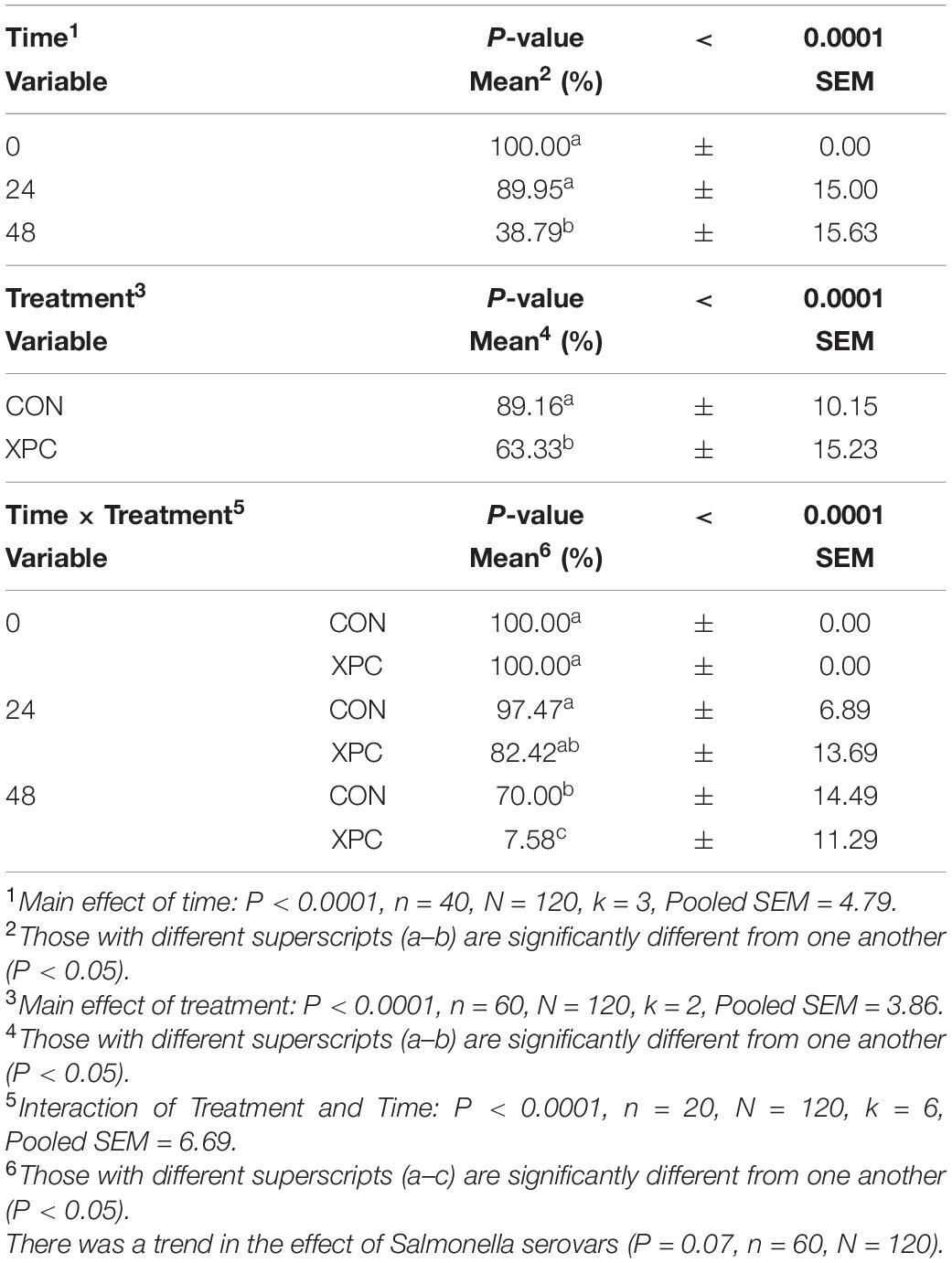
Table 2. The main effect of time, treatment, and the interaction of treatment × time on the prevalence of overall Salmonella when inoculated into an in vitro turkey cecal model with and without Original XPCTM at 0, 24, and 48 h post inoculation (hpi) over two trials.
Using time as a repeated measure, the main effect of serovar was not significant throughout this study for any analyses (P = 0.7779, Supplementary Tables 3, 4). However, the main effects of time and treatment, as well as their interaction (treatment × time), were significant (P < 0.01; Table 3). Pairwise contrasts were performed to examine the interactions at each timepoint. Over time (Main Effect of Time, P < 0.0001), Salmonella was reduced from 7.32 to 3.20 and from 3.20 to 1.19 Log10 CFU/mL from 0 to 48 hpi. With the main effect of treatment, turkey cecal cultures treated with XPC resulted in lower concentrations of Salmonella compared to those not treated, 3.26 and 4.55 Log10 CFU/mL, respectively (Main Effect of Treatment, P < 0.0001). Overall, the interaction of treatment and time (Treatment × Time Interaction, P < 0.0001, Table 3) resulted in a stepwise decrease in Salmonella load in the CON and XPC groups. However, there was no difference between the XPC or control group at the start of the trial. At 24 hpi, there was a 3 Log10 CFU/mL (7.31–4.22 Log10 CFU/mL) reduction in the control group versus a 5 Log10 reduction (7.33–2.19 Log10 CFU/mL) of Salmonella in the XPC-treated group. By 48 h, the XPC-treated group had almost reduced Salmonella completely as compared to the control, 0.25 and 2.12 Log10 CFU/mL of ceca culture, respectively. The un-inoculated controls remained negative throughout the time frame of the course of the study. All Salmonella isolated were confirmed to be resistant to 64 μg/mL nalidixic acid through an antibiogram; therefore, the plate concentration did not pick up any additional native Salmonella with the 20 μg/mL resistance profile.
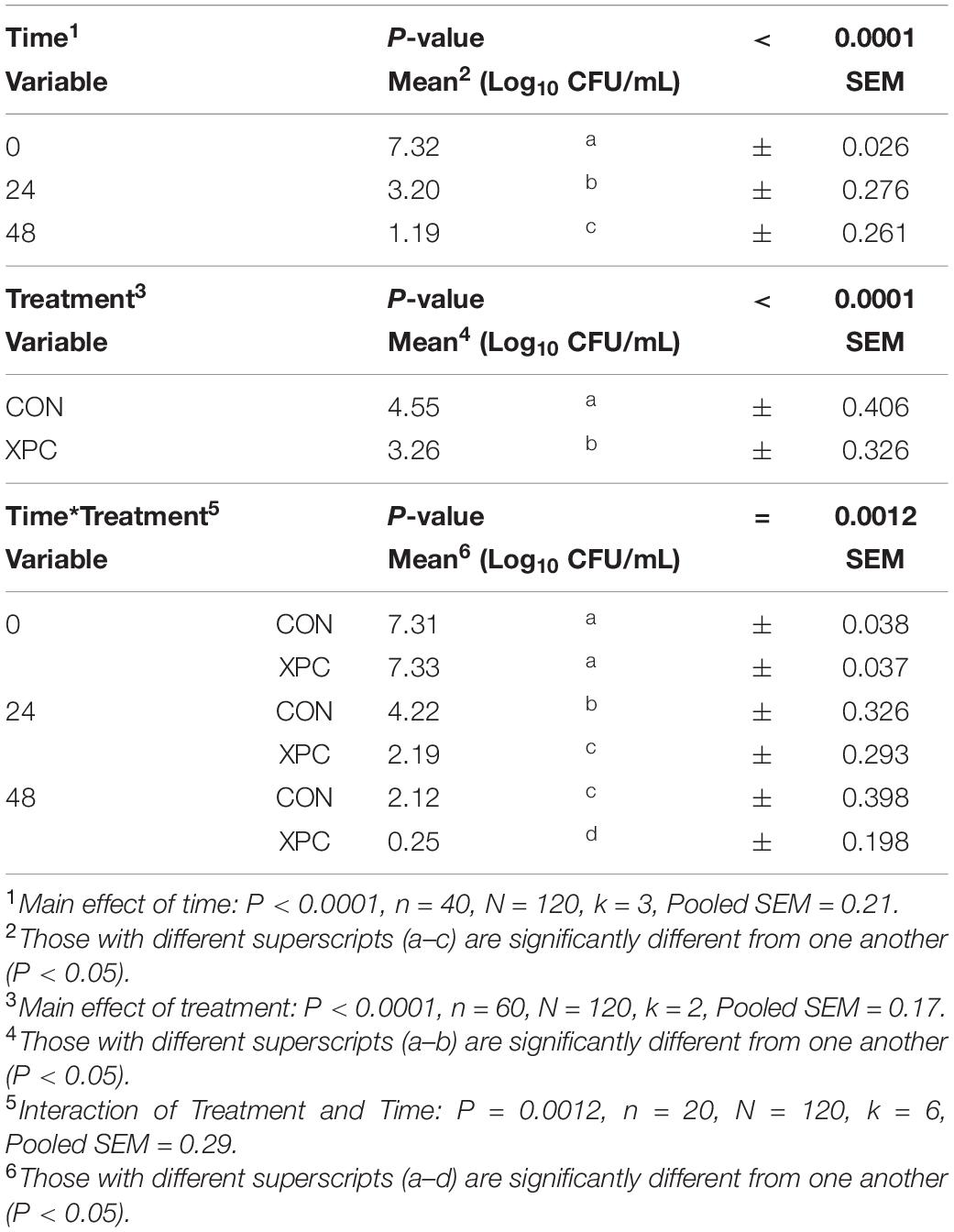
Table 3. The effect of time and treatment and subsequent interaction on the total load of Salmonella at 0, 24, and 48 h post-inoculation (hpi) over two trials where Salmonella Typhimurium and S. Reading were inoculated into an in vitro turkey cecal model with and without Original XPCTM.
Microbiota Analysis
In congruence with the microbiological results, the microbiota results demonstrated no difference between serovars and therefore were pooled to create a protected pool (n = 16, N = 96). As such, XPC-treated ceca had a higher Shannon diversity index at 0 hpi (Figure 2A and Table 4). As time progressed throughout the in vitro study, the control group demonstrated a stepwise reduction in diversity through 48 hpi. However, the XPC-treated groups exhibited an increased richness that stabilized during the trial. This effect was mirrored in Pielou’s evenness, where evenness declined with each passing timepoint in the CON group while evenness narrowed initially in the XPC group and then stabilized with greater evenness at 48 h (Figure 2B and Table 4).
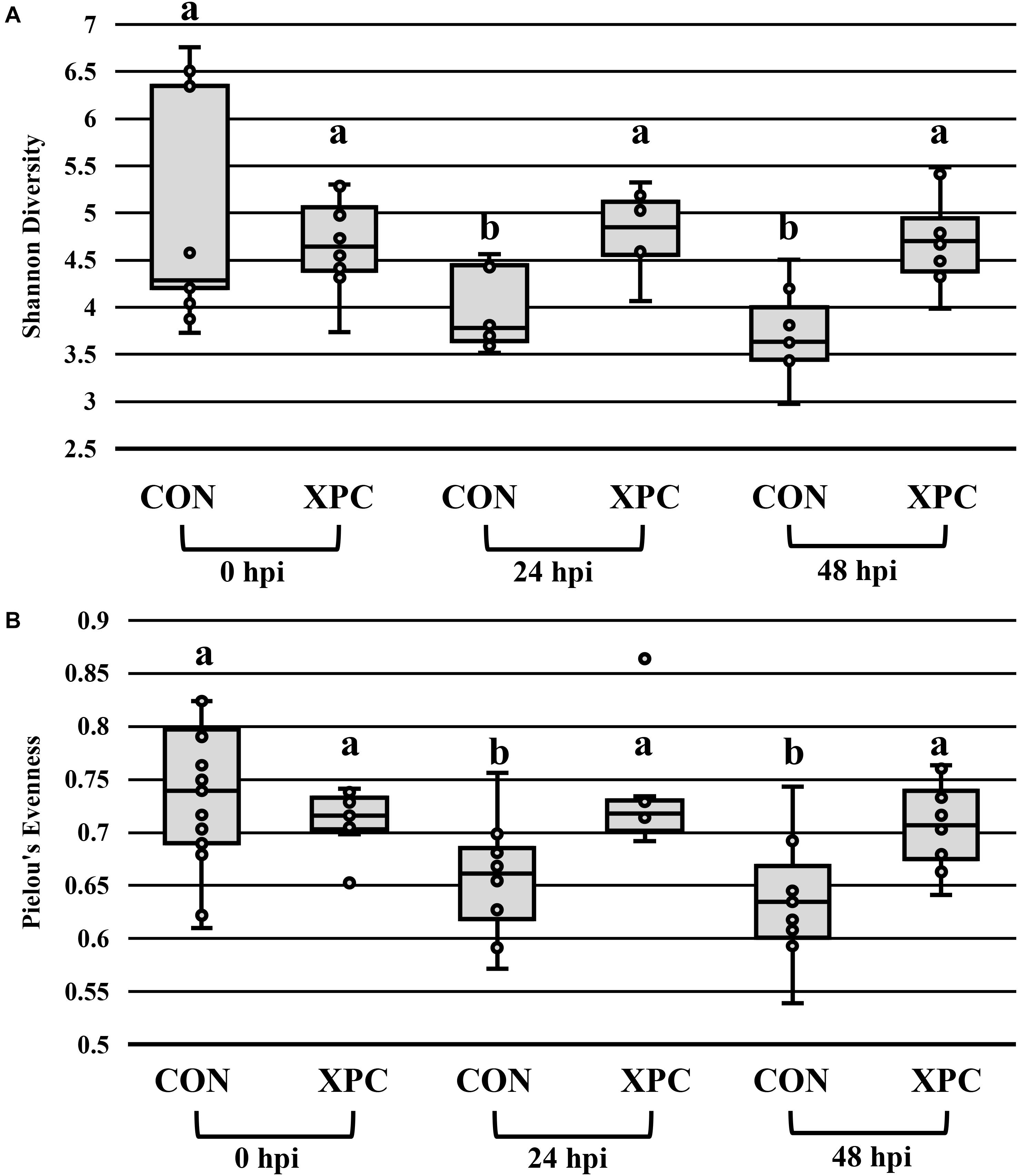
Figure 2. The effect of treatment on the richness (A) and evenness (B) of the microbiota of turkey ceca inoculated with Salmonella Typhimurium and Reading during an in vitro turkey cecal model with and without the supplementation of Original XPCTM at 0, 24, and 48 h post-inoculation (hpi) (P < 0.05, Q < 0.05, n = 16, N = 96). Those with different superscripts (a–b) are significantly different than one another.
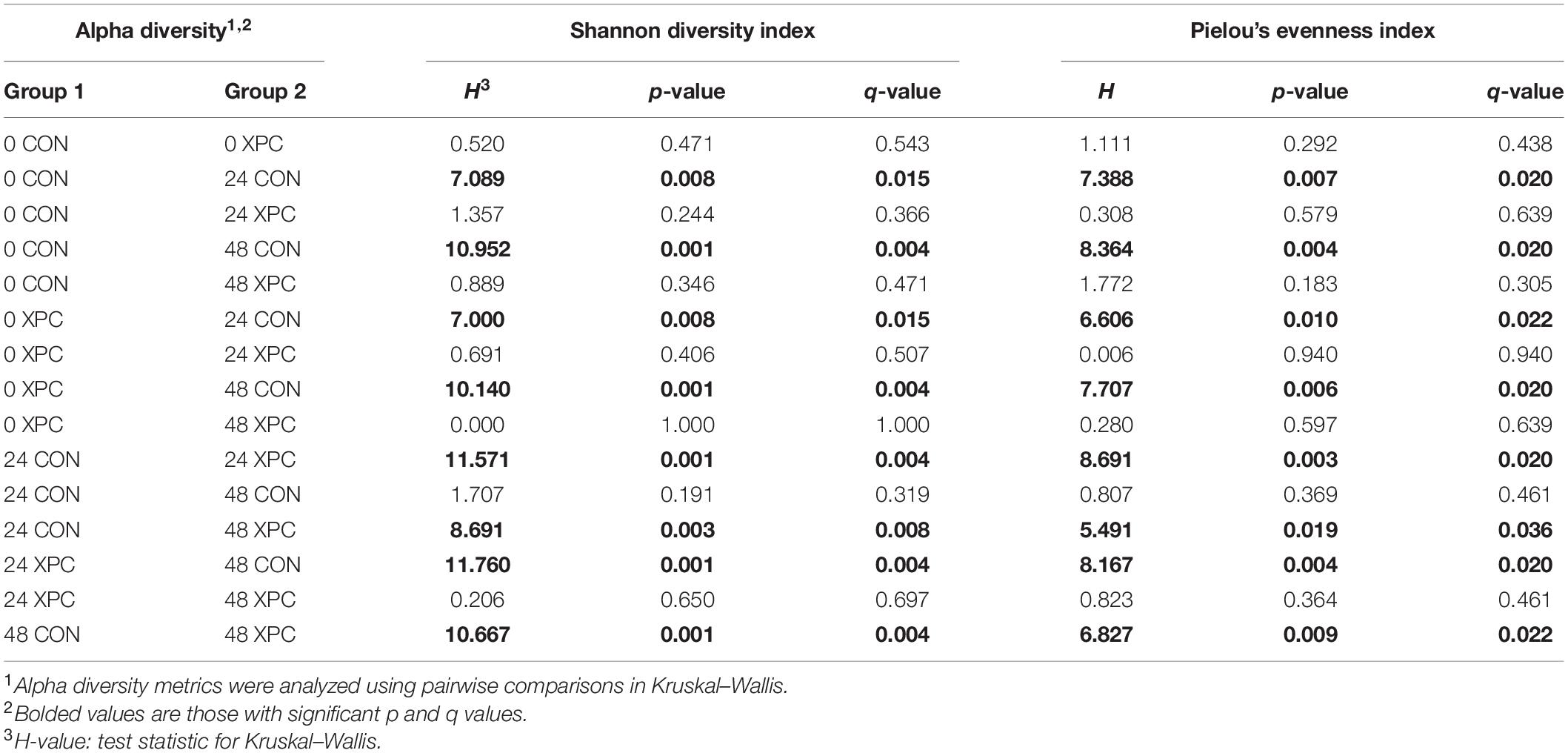
Table 4. The effect of treatment on the richness and evenness of the turkey cecal microbiota at 0, 24, and 48 h post-inoculation (hpi) of Salmonella Typhimurium and Reading during an in vitro turkey cecal model with and without the supplementation of Original XPCTM (P < 0.05, n = 16, N = 96).
Beta diversity describes the binary differences in community structure and the differences in variation each treatment has for the quantity of OTUs present. The metric does not specify how a sample is compositionally dissimilar, but it does delineate that differences exist. The ANOSIM indicated that the Jaccard dissimilarity matrix was different by time and treatment (Table 5). Therefore, the effect of time was plotted as a continuous variable on a PCoA plot, and the individual treatments were visualized (Figure 3). The effect of treatment was significant, with differences becoming more profound over time (Q < 0.05; Figure 3A and Table 5). The XPC-treated groups were more diverse by the Jaccard dissimilarity matrix. Weighted Unifrac distance matrix was used to determine how the phylogenetic weight of the abundance OTUs contributed to the analysis. Like the Jaccard dissimilarity matrix that is based on presence/absence, the weighted Unifrac distance matrix is quantitative. However, besides just binary differences in diversity weighed by abundance, the weighted Unifrac distance matrix incorporates the phylogenetic branch length-weight into the analysis (Figure 3B and Table 5). In short, the use of XPC resulted in the stability of beta diversity over time (Q < 0.05). Irrespective of Alpha or Beta diversity index, there was no difference between the two serovars.
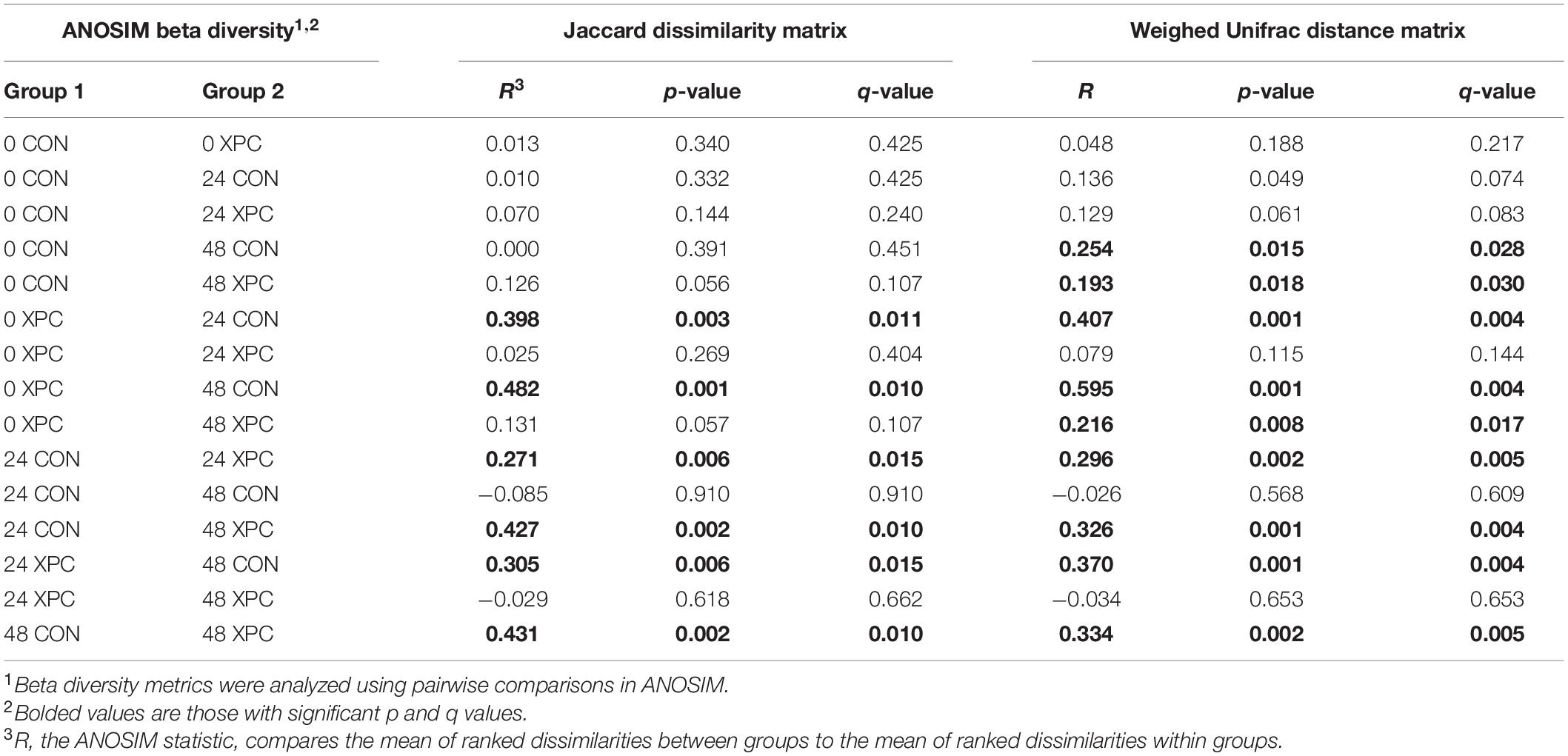
Table 5. The effect of treatment on the Jaccard dissimilarity and weighted UniFrac distance matrices of the turkey cecal microbiota at 0, 24, and 48 h post-inoculation (hpi) of Salmonella Typhimurium and Reading during an in vitro turkey cecal model with and without the supplementation of Original XPCTM (P < 0.05, n = 16, N = 96).
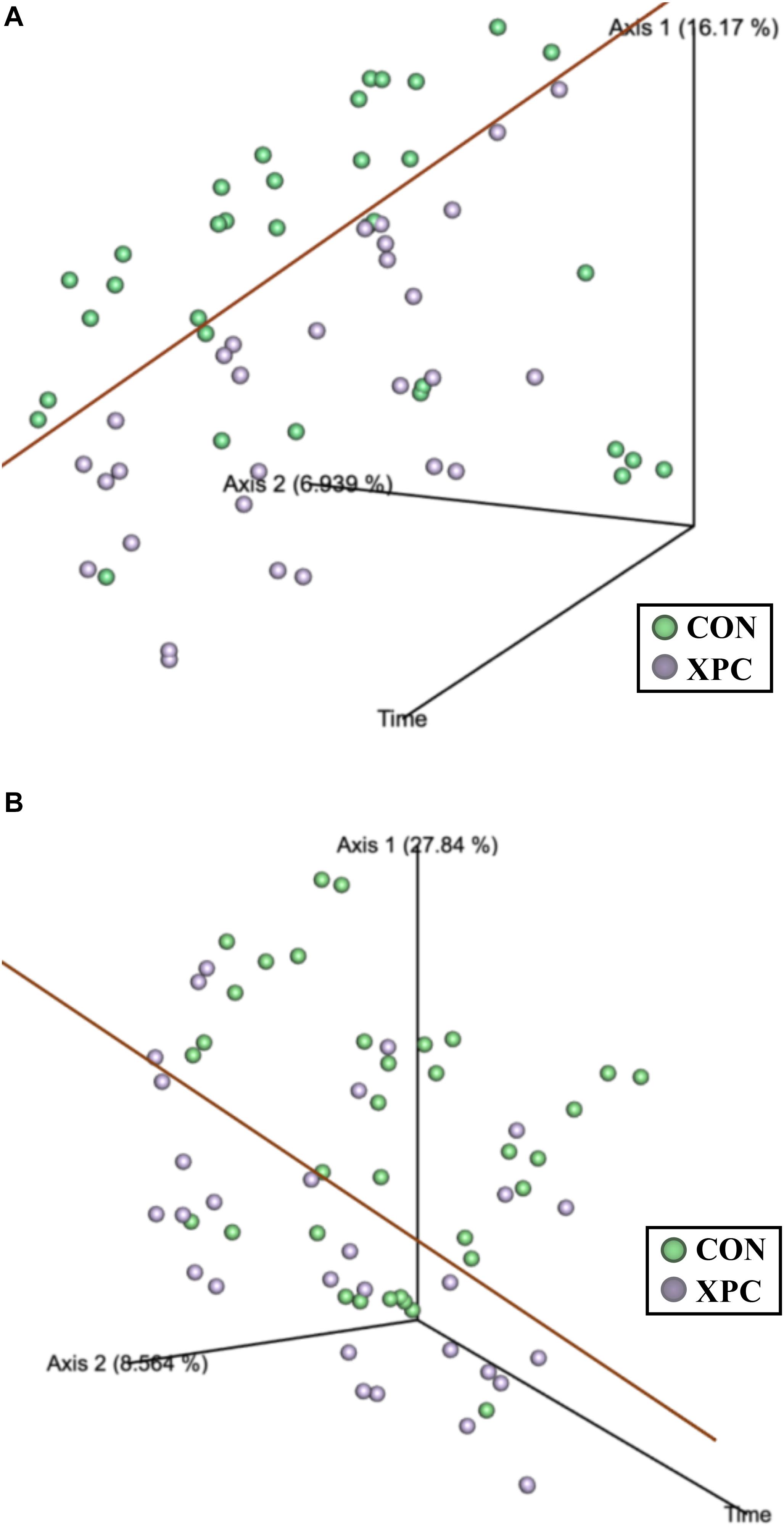
Figure 3. The effect of treatment on of the Jaccard dissimilarity (A) and weighted UniFrac distance (B) matrices of the turkey cecal microbiota at 0, 24, and 48 h post-inoculation (hpi) of Salmonella Typhimurium and Reading during an in vitro turkey cecal model with and without the supplementation of Original XPCTM (P < 0.05, Q < 0.05, n = 16, N = 96). The green data points represent the control cecal cultures, whereas the purple data points represent the XPC treated turkey cecal cultures.
In order to ascertain the compositional differences between the treatment groups, analysis of composition of microbiomes (ANCOM) was used. ANCOM incorporates a strict false discovery rate that is capable of increasing statistical power. When compared in a pairwise fashion, the differences are pairwise by treatment. However, when comparing treatments over time, the OTUs are correlated to the treatment effects. The family level analysis was chosen as the family level dynamics are better annotated and could lend themselves to general structural changes by treatment. Additionally, as no diversity indices or plate count results indicated a difference between the two Salmonella serovars, all Salmonella results were treated the same for these analyses. As such, this brought the total number of replicates per treatment per timepoint to 16 (n = 16), which enhanced the statistical power of the study.
Figure 4 documents the total effects of time by treatment and their associated OTUs (P < 0.05). At 0 hpi, XPC groups had an increase in Bifidobacteriaceae, Eggerthellaceae, and Prevotellaceae, and a contraction in the Atopobiaceae, Coriobacteriaceae, and Erysipelotrichaceae abundance of families as compared to the CON. By 24 hpi, the CON group had an increase in Bifidobacteriaceae. Meanwhile, the XPC group exhibited an increase in Eggerthellaceae, Prevotellaceae, Atopobiaceae, and Coriobacteriaceae. By 48 h, when Alpha and Beta diversities were at their lowest for CON, significant populations were dominated by Bifidobacteriaceae in the CON group. The XPC-treated group had an increase in Eggerthellaceae, Prevotellaceae, and Coriobacteriaceae.
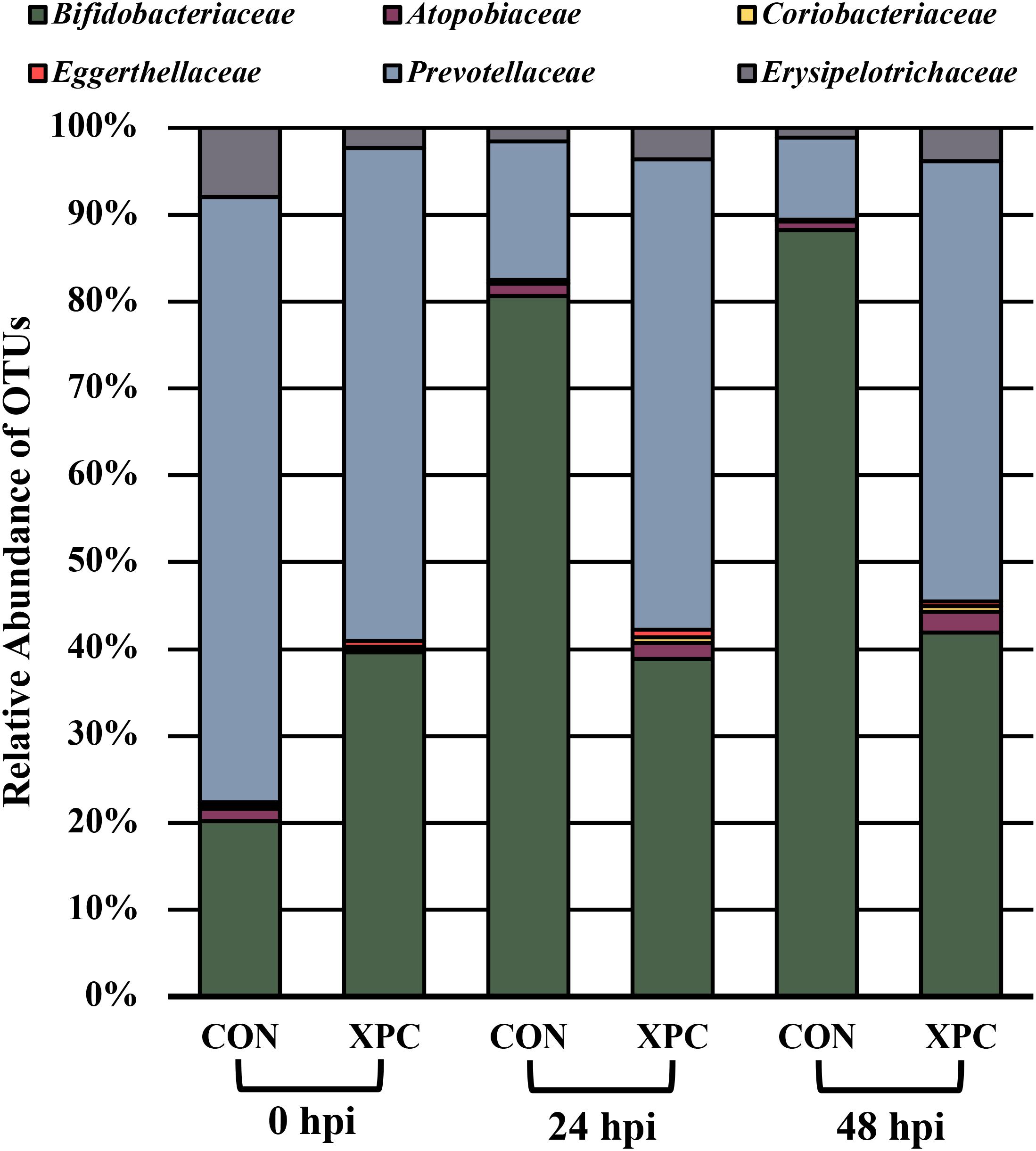
Figure 4. The effect of treatment on the composition of the turkey cecal microbiota at 0, 24, and 48 h post-inoculation (hpi) of Salmonella Typhimurium and Reading during an in vitro turkey cecal model with and without the supplementation of Original XPCTM. Sequences were compared with ANCOM at the family level (P < 0.05; n = 16; N = 96; W = 89, 76, 76, 73, 69, and 66).
In order to understand which populations had the greatest leverage at each timepoint, the sequences were filtered by hpi and compared with ANCOM. While significant fluctuations associated with treatment occurred when all comparisons were together, the only difference that emerged was Eggerthellaceae, where XPC had over three times more OTUs present than the CON (Figure 5A; Q < 0.05). By 24 hpi, Coriobacteriacea was higher in XPC than the control as well (Figure 5B; Q < 0.05). However, by 48 hpi, CON had less Eggerthellaceae and Tannerellaceae, and Bacteriodales (order), but an increase in Ruminococcaceae. Meanwhile, XPC-treated groups had an increase in Bacteriodales (Figure 5C).
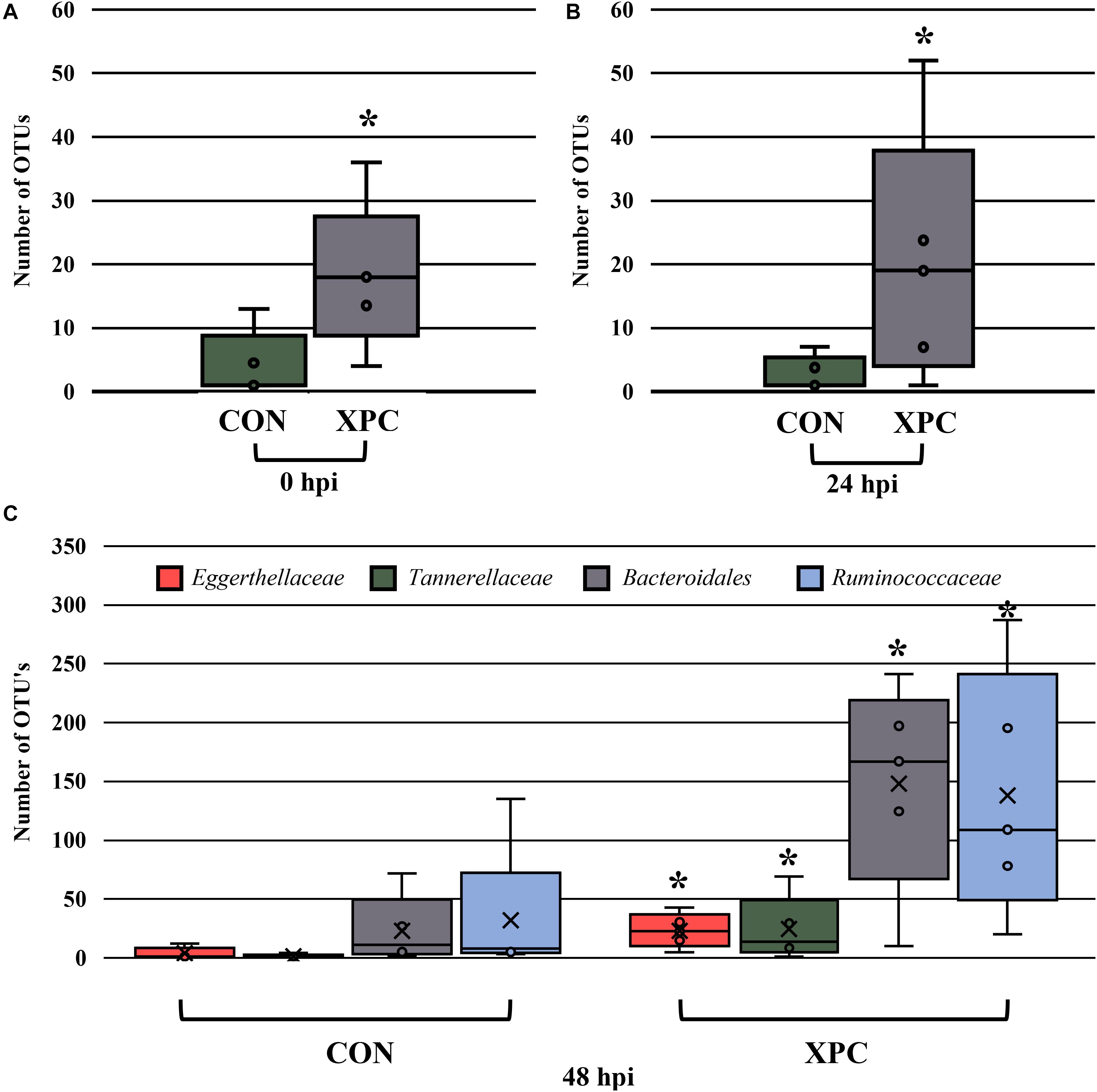
Figure 5. The effect of treatment on the composition of the turkey cecal microbiota at 0 (A), 24 (B), and 48 (C) h post-inoculation (hpi) of Salmonella Typhimurium and Reading during an in vitro turkey cecal model with and without the supplementation of Original XPCTM. Sequences were filtered by hpi and compared with ANCOM at the family level (P < 0.05, Q < 0.05, n = 16, N = 96). At 0 h, Eggerthellaceae was the only significant different family identified. At 24 hpi, Coriobacteriacea was different, and at 48 hpi, Eggerthellaceae, Tannerellaceae, Bacteriodales, and Ruminococcaceae were different. Those with an asterisk (*) have a significantly greater abundance than those without one.
Discussion
In the current study, we evaluated an in vitro cecal model using turkey ceca. To date, it is the first study to develop and utilize this model to our knowledge. With additional validation, the in vitro anaerobic culture method could be valuable as a screening tool to feed amendment producers looking to understand the biology and efficacy of a particular feed component. This is especially relevant, when producers need a rapid understanding of their product’s potential impact during a public health crisis, such as what was presented in the current study with S. Reading. This rapid screening could help evaluate feed amendment derivatives to determine optimal dosages and combinations.
An important nuance of this study was the incorporation of S. Typhimurium, which was historically used in this in vitro model in broiler chicken ceca with significant consistency and success (Rubinelli et al., 2016, 2017; Kim et al., 2018). Ultimately, both serovars were key for the early validation of this model. This is especially true since early studies indicate that the total reduction of Salmonella using turkey ceca is more rapid than traditional cecal culture with chickens. Additional comparative microbiota studies directly comparing the chicken and turkey ceca will be essential if tools to work in both species are developed and attempts at comparisons are to be developed.
The use of XPC decreased the prevalence of Salmonella significantly throughout the study and parallels what has been observed in previous research in vitro and in vivo (Feye et al., 2016a,b, 2019; Rubinelli et al., 2016; Park et al., 2017; Roto et al., 2017; McGinnis et al., 2019). Additionally, XPC effectively reduced the pathogenic load of each Salmonella strain throughout the study. Interestingly, there was no difference in XPC’s ability to reduce the prevalence nor load of the two serovars preferentially. This is contrary to previously reported studies that indicate XPC may be more effective against different serovars (Feye et al., 2016a). However, the current research is confined to S. Typhimurium versus S. Reading. It should be expanded to evaluate the anti-Salmonella effects of XPC against all major poultry serovars of public health importance in both species as well as multiple strains within serovar to determine the range of anti-Salmonella effects fully. It should not be assumed this trend holds nor the chicken trends extrapolate to the turkey model as the systems are biologically distinct. Ultimately, some evidence suggests that serovars may respond differently to unique environments (Foley et al., 2013). Therefore, if recommendations regarding XPC administration are to become broadly adopted for the reduction of multiple Salmonella serovars, further analyses are required.
The diversity metrics in this study were also telling for the model. Previous data do not demonstrate a stepwise decrease in richness and diversity using chicken models. This difference could be explained by the limitations associated with the turkey microbiota, which tends to have a lower diversity initially. In addition, both the intestinal and cecal microbiomes of turkeys have been characterized by Wei et al. (2013) as being distinct from their chicken counterparts. Wei and colleagues attributed this demarcation to several factors, including breed, physiological, and anatomical differences, along with the slower digesta passage rate in turkeys. Ultimately, a reduction in diversity may suffer more profound and stepwise losses once the energy of the system narrows due to loss of nutrient availability. However, XPC appears to sustain diversity, which may ultimately point to changes in the metabolism of the microbiota. Likely, the narrowing is artificial, but it is demonstrative of the bottlenecks associated with turkey microbial ecology (Moxon and Kussell, 2017). More studies need to evaluate the metabolomics of the system and how sustained in vitro modeling impacts data interpretation.
Finally, the compositional changes associated with this model are a bit unique. Typically, in chicken models, an increase in Ruminococcaceae and Archeae occur as time progresses (Rubinelli et al., 2017; Feye et al., 2020). However, in this study, less common microorganisms persisted, which was likely due to the microbiota of the turkey. Unfortunately, the biological function of the turkey microbiota is poorly established. Therefore, speculating on the possible contribution of this consortium to the effects of XPC is premature at this time and awaits further research.
Conclusion
XPC continues to reduce Salmonella as compared to the control using an in vitro approach. The benefits of this approach are that Salmonella reduction is purely microbiota mediated as the immune system and other host factors are not present. The current study was novel as it not only demonstrated the preliminary development of an in vitro cecal culture model using turkey ceca but demonstrated that Salmonella was reduced consistently using XPC regardless of serovar. However, it is of interest to screen XPC against multiple S. enterica serovars to determine if XPC demonstrates similar anti-Salmonella characteristics across all serovars as demonstrated in the current study.
Ultimately, the microbiota may become a better diagnostic tool when one considers the effects of the microbial ecology structure rather than the compositional biomarker approach. Not only is it more general, but it also does not assume specific biological meaning. In particular, models such as the one presented herein describe how the microbiota shifts independent of the immune system. From a mechanistic perspective, this is important as it indicates that XPC has greater global shifts to the microbiota, which in turn could be changing the immune system than previously described. Therefore, it may not be which organisms and when, but how the population shifts that leads to speaks to the global changes of the microbiota that impact host physiology. Understandably, this model should be further compared to an animal infection model to further delineate the anti-Salmonella effects of XPC.
Data Availability Statement
The data presented in the study are deposited in the NCBI Sequence Read Archive (RSA) repository under the accession number PRJNA716729.
Author Contributions
KF and SR conceived the trial. KF and PR conducted the benchwork for the trial. KF analyzed the data. KF and DD prepared the figures, wrote the draft, and handled edits. EO handled edits, formatting, and submissions. All authors were able to edit the publication prior to submission.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors would like to thank David Kenyon from Diamond V for his commitment to the acquisition of tom turkey ceca throughout the trial. In addition, the authors would like to acknowledge Hilary O. Pavlidis and Evan W. Chaney from Diamond V for their input and efforts made regarding the current study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.645301/full#supplementary-material
Footnotes
References
Anderson, M. J. (2001). A new method for non-parametric multivariate analysis of variance. Austral Ecol. 26, 32–46. doi: 10.1111/j.1442-9993.2001.01070.pp.x
Bokulich, N. A., Kaehler, B. D., Rideout, J. R., Dillon, M., Bolyen, E., Knight, R., et al. (2018). Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6:90. doi: 10.1186/s40168-018-0470-z
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C., Al-Ghalith, G. A., et al. (2018). QIIME 2: reproducible, interactive, scalable, and extensible microbiome data science. PeerJ Preprints 6:e27295v2. doi: 10.7287/peerj.preprints.27295v2
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Canibe, N., and Jenses, B. B. (2012). Fermented liquid feed-microbial and nutritional aspects and impact on enteric diseases in pigs. Anim. Feed Sci. Technol. 173, 17–40. doi: 10.1016/j.anifeedsci.2011.12.021
Centers for Disease Control and Prevention [CDC]. (2019). Multidrug-Resistant Salmonella Infections Linked to Raw Turkey Products. Available online at: https://www.cdc.gov/salmonella/reading-07-18/ (accessed August 14, 2020).
Diamond V Original XPCTM (XPC) (2020). Product Profile. Available online at: https://www.diamondv.com/wp-content/uploads/productprofile_xpc_original.pdf (accessed December 21, 2020).
Feye, K. M., Andereson, K. L., Scott, M. F., Henry, D. L., Dorton, K. L., Depenbusch, B. E., et al. (2016a). Abrogation of Salmonella and E. coli O157:H7 in feedlot cattle fed a proprietary Saccharomyces cerevisiae fermentation prototype. J. Vet. Sci. Technol. 7:350. doi: 10.4172/2157-7579.1000350
Feye, K. M., Anderson, K. L., Scott, M. F., McIntyre, D. R., and Carlson, S. A. (2016b). Inhibition of virulence, antibiotic resistance, and fecal shedding of multiple-antibiotic resistant Salmonella Typhimurium in broilers fed Original XPC. Poult. Sci. 95, 2901–2910. doi: 10.3382/ps/pew254
Feye, K. M., Carroll, J. A., Anderson, K. L., Whittaker, J., Schmidt-McCormack, G., McIntyre, D. R., et al. (2019). Saccharomyces cerevisiae fermentation products that mitigate foodborne Salmonella. Front. Vet. Sci. 6:107. doi: 10.3389/fvets.2019.00107
Feye, K. M., Rubinelli, P. M., Chaney, W. E., Pavlidis, H. O., Kogut, M. H., and Ricke, S. C. (2020). The preliminary development of an in vitro poultry cecal culture model to evaluate the effects of Original XPCTM for the reduction of Campylobacter jejuni and its potential effects on the microbiota. Front. Microbiol. 10:3062. doi: 10.3389/fmicb.2019.03062
Foley, S. L., Johnson, T. J., Ricke, S. C., Nayak, R., and Danzeisen, J. (2013). Salmonella pathogenicity and host adaptation in chicken-associated serovars. Microbiol. Mol. Biol. Rev. 77, 582–607. doi: 10.1128/MMBR.00015-13
Food Safety Inspection Service [FSIS] (2014). Serotypes of Salmonella Isolates from Meat and Poultry Products January 1998 through December 2014. Available online at: https://www.fsis.usda.gov/wps/wcm/connect/3866026a-582d-4f0e-a8ce-851b39c7390f/Salmonella-Serotype-Annual-2014.pdf?MOD=AJPERES (accessed August 20, 2020).
Gad, A. H., Abo-Shama, U. H., Harclerode, K. K., and Fakhr, M. K. (2018). Prevalence, serotyping, molecular typing, and antimicrobial resistance of Salmonella isolated from conventional and organic retail ground poultry. Front. Microbiol. 9:2653. doi: 10.3389/fmicb.2018.02653
Herigstad, B., Hamilton, M., and Heersink, J. (2001). How to optimize the drop plate method for enumerating bacteria. J. Microbiol. Methods 44, 121–129. doi: 10.1016/S0167-7012(00)00241-4
Jett, B. D., Hatter, K. L., Huycke, M. M., and Gilmore, M. S. (1997). Simplified agar plate method for quantifying viable bacteria. Biotechniques 23, 648–650. doi: 10.2144/97234bm22
Katoh, K., Misawa, K., Kuma, K., and Miyata, T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Kim, S. A., Rubinelli, P. M., Park, S. H., and Ricke, S. C. (2018). Ability of Arkansas LaKast and LaKast hybrid rice bran to reduce Salmonella typhimurium in chicken cecal incubations and effects on cecal microbiota. Front. Microbiol. 9:134. doi: 10.3389/fmicb.2018.00134
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K., and Schloss, P. D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120. doi: 10.1128/AEM.01043-13
Kruskal, W. H., and Wallis, W. A. (1952). Use of ranks in one-criterion variance analysis. J. Am. Stat. Assoc. 47, 583–621. doi: 10.1080/01621459.1952.10483441
Lozupone, C. A., Hamady, M., Kelley, S. T., and Knight, R. (2007). Quantitative and qualitative beta diversity measures lead to different insights into factors that structure microbial communities. Appl. Environ. Microbiol. 73, 1576–1585. doi: 10.1128/AEM.01996-06
Mandal, S., Treuren, W. V., White, R. A., Eggesbø, M., Knight, R., and Peddada, S. D. (2015). Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb. Ecol. Health Dis. 26:27663. doi: 10.3402/mehd.v26.27663
McGinnis, J., Byrd, J. A., Pavlidis, H. O., and Chaney, W. E. (2019). “The effects of feeding Original XPC on reducing Salmonella prevalence and numbers in ceca samples and carcass rinses taken from commercial broilers,” in Proceedings of the International Association for Food Protection Annual Meeting, July 2019, Louisville, KY, 3–222.
Moxon, R., and Kussell, E. (2017). The impact of bottlenecks on microbial survival, adaptation, and phenotypic switching in host-pathogen interactions. Evolution 71, 2803–2816. doi: 10.1111/evo.13370
Naghili, H., Tajik, H., Mardani, K., Razavi Rouhani, S. M., Ehsani, A., and Zare, P. (2013). Validation of drop plate technique for bacterial enumeration by parametric and nonparametric tests. Vet. Res. Forum 4, 179–183.
National Research Council [NRC] (1994). Nutrient Requirements of Poultry: Ninth Revised Edition. Washington, DC: The National Academies Press. doi: 10.17226/2114
Nelson, J. R., Sobotik, E. B., Athrey, G., and Archer, G. S. (2020). Effects of supplementing yeast fermentate in the feed or drinking water on stress susceptibility, plasma chemistry, cytokine levels, antioxidant status, and stress-and immune-related gene expression of broiler chickens. Poult. Sci. 99, 3312–3318. doi: 10.1016/j.psj.2020.03.037
Park, S. H., Kim, S. A., Lee, S. I., Rubinelli, P. M., Roto, S. M., Pavlidis, H. O., et al. (2017). Original XPCTM effect on Salmonella Typhimurium and cecal microbiota from three different ages of birds when incubated in an anaerobic in vitro culture system. Front. Microbiol. 8:1070. doi: 10.3389/fmicb.2017.01070
Poeker, S. A., Lacroix, C., de Wouters, T., Spalinger, M. R., Scharl, M., and Geirnaert, A. (2019). Stepwise development of an in vitro continuous fermentation model for the murine caecal microbiota. Front. Microbiol. 10:1166. doi: 10.3389/fmicb.2019.01166
Public Health Agency of Canada [PHAC] (2021). Public Health Notice — Outbreak of Salmonella Illnesses Linked to Raw Turkey and Raw Chicken. Available online at: https://www.canada.ca/en/public-health/services/public-health-notices/2018/outbreak-salmonella-illnesses-raw-turkey-raw-chicken.html (accessed March 3, 2021).
Roto, S. M., Park, S. H., Lee, S. I., Kaldhone, P., Pavlidis, H. O., Frankenbach, S. B., et al. (2017). Effects of feeding Original XPC(to broilers with a live coccidiosis-vaccine under industry conditions: Part 1. Growth performance and Salmonella inhibition. Poult. Sci. 96, 1831–1837. doi: 10.3382/ps/pew445
Rubinelli, P. M., Roto, S. M., Kim, S., Park, S., Pavlidis, H. O., McIntyre, D., et al. (2016). Reduction of Salmonella Typhimurium by fermentation metabolites of Diamond V Original XPC in an in vitro anaerobic mixed chicken cecal culture. Front. Vet. Sci. 3:83. doi: 10.3389/fvets.2016.00083
Rubinelli, P. R., Kim, S. A., Park, S. H., Roto, S. M., Nealon, N. J., Ryan, E. P., et al. (2017). Differential effects of rice bran cultivars to limit Salmonella Typhimurium in chicken cecal in vitro incubations and impact on the cecal microbiome and metabolome. PLoS ONE 12:e0185002. doi: 10.1371/journal.pone.0185002
Schlosser, W., Hogue, A., Ebel, E., Rose, B., Umholtz, R., Ferris, K., et al. (2000). Analysis of Salmonella serotypes from selected carcasses and raw ground products sampled prior to the implementation of the Pathogen Reduction: hazard analysis and critical control point final rule in the US. Int. J. Food Microbiol. 58, 107–111. doi: 10.1016/s0168-1605(00)00293-2
Wei, S., Morrison, M., and Yu, Z. (2013). Bacterial census of poultry intestinal microbiome. Poult. Sci. 92, 671–683. doi: 10.3382/ps.2012-02822
Keywords: Salmonella, turkey, XPC, cecal culture method, yeast fermentate
Citation: Feye KM, Dittoe DK, Rubinelli PM, Olson EG and Ricke SC (2021) Yeast Fermentate-Mediated Reduction of Salmonella Reading and Typhimurium in an in vitro Turkey Cecal Culture Model. Front. Microbiol. 12:645301. doi: 10.3389/fmicb.2021.645301
Received: 22 December 2020; Accepted: 11 March 2021;
Published: 15 April 2021.
Edited by:
Arun K. Bhunia, Purdue University, United StatesReviewed by:
Sunae Kim, Ewha Womans University, South KoreaYosra A. Helmy, The Ohio State University, United States
Copyright © 2021 Feye, Dittoe, Rubinelli, Olson and Ricke. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Steven C. Ricke, c3JpY2tlQHdpc2MuZWR1
 Kristina M. Feye
Kristina M. Feye Dana K. Dittoe
Dana K. Dittoe Peter M. Rubinelli
Peter M. Rubinelli Elena G. Olson2
Elena G. Olson2 Steven C. Ricke
Steven C. Ricke