Abstract
Phytopathogenic Verticillia cause Verticillium wilt on numerous economically important crops. Plant infection begins at the roots, where the fungus is confronted with rhizosphere inhabiting bacteria. The effects of different fluorescent pseudomonads, including some known biocontrol agents of other plant pathogens, on fungal growth of the haploid Verticillium dahliae and/or the amphidiploid Verticillium longisporum were compared on pectin-rich medium, in microfluidic interaction channels, allowing visualization of single hyphae, or on Arabidopsis thaliana roots. We found that the potential for formation of bacterial lipopeptide syringomycin resulted in stronger growth reduction effects on saprophytic Aspergillus nidulans compared to Verticillium spp. A more detailed analyses on bacterial-fungal co-cultivation in narrow interaction channels of microfluidic devices revealed that the strongest inhibitory potential was found for Pseudomonas protegens CHA0, with its inhibitory potential depending on the presence of the GacS/GacA system controlling several bacterial metabolites. Hyphal tip polarity was altered when V. longisporum was confronted with pseudomonads in narrow interaction channels, resulting in a curly morphology instead of straight hyphal tip growth. These results support the hypothesis that the fungus attempts to evade the bacterial confrontation. Alterations due to co-cultivation with bacteria could not only be observed in fungal morphology but also in fungal transcriptome. P. protegens CHA0 alters transcriptional profiles of V. longisporum during 2 h liquid media co-cultivation in pectin-rich medium. Genes required for degradation of and growth on the carbon source pectin were down-regulated, whereas transcripts involved in redox processes were up-regulated. Thus, the secondary metabolite mediated effect of Pseudomonas isolates on Verticillium species results in a complex transcriptional response, leading to decreased growth with precautions for self-protection combined with the initiation of a change in fungal growth direction. This interplay of bacterial effects on the pathogen can be beneficial to protect plants from infection, as shown with A. thaliana root experiments. Treatment of the roots with bacteria prior to infection with V. dahliae resulted in a significant reduction of fungal root colonization. Taken together we demonstrate how pseudomonads interfere with the growth of Verticillium spp. and show that these bacteria could serve in plant protection.
Introduction
The genus Verticillium comprises soil-borne plant pathogens causing vascular wilt disease in numerous crops. The amphidiploid species Verticillium longisporum induces a stem striping disease in rapeseed. In field trials it was found that symptoms occur close to the harvest and might only in some cases result in yield losses (Depotter et al., 2019). Still a recent survey demonstrates that Verticillium stem striping caused by V. longisporum belongs to the top ten biotic threats of oilseed rape in Europe (Zheng et al., 2020). In addition, temperature increase caused by global warming might exacerbate the effects of fungal plant disease (Siebold and von Tiedemann, 2013). V. longisporum is the result of several independent hybridization events between Verticillium dahliae or V. dahliae-like species (named D1–D3) as well as a so far unknown species named A1 (Depotter et al., 2016). V. longisporum isolates can differ in their pathogenicity toward hosts. Even pathogenic isolates have mechanisms to tame their virulence and limit the damage to the host plant (Harting et al., 2021).
The haploid V. dahliae is the economically most important fungus for causing Verticillium wilt, which infects up to several hundred different plants and is further broadening its host range (EFSA Panel on Plant Health [PLH], 2014). Host plants include crops such as strawberry, tomato, olive, lettuce, hops or sunflowers. Verticillium wilt can lead to severe yield losses and control of the fungus with fungicides remains difficult. The impact on agricultural productivity of Verticillium species highlights the need for novel efficient but environmentally friendly strategies for effective control of Verticillium-induced disease.
Verticillia infect their host plants through the roots, mainly at positions where the endodermis is not fully developed. They first colonize the vascular system and later other tissues of the plant. Once the fungus has entered the plant and reached the xylem vessels, it produces conidia, which are distributed in the plant with the sap stream. At suitable sites, conidia get trapped, germinate and colonize adjacent cells (Fradin and Thomma, 2006). Transport processes in the cell are hindered by increased fungal colonization and potentially also by plant defense mechanisms, as for example the accumulation of phenolic compounds, leading to the development of disease symptoms (Fradin and Thomma, 2006; Kunej et al., 2020). In addition, the fungal infection can also induce cellular changes inside the plant, which allow compensation of the transport processes (Reusche et al., 2014). Verticillium spp. produce microsclerotia as long-term resting structures in the dying host, which reach the soil with degrading plant tissue. The melanized cell wall of the resting structures enables the fungus to survive extreme environmental conditions such as radiation, temperature changes and reactive oxygen species but also shelters the fungus against other microorganisms (Belozerskaya et al., 2017; Casadevall et al., 2017; Fan et al., 2020). In one of the next growing seasons, microsclerotia re-establish infections once a susceptible host is present (Wilhelm, 1955; Fradin and Thomma, 2006).
The fungus recognizes appropriate hosts by their root exudates, forms a germ tube and grows toward the root surface (Acharya et al., 2020; Zhang et al., 2020). Verticillium spp. have to cope with organisms inhabiting the plant rhizosphere in this hyphal growth phase after microsclerotia germination and prior to root penetration (Deketelaere et al., 2017). Verticillium spp. can form hyphopodia, which are swollen hyphal tips, to enter the root. The plant rhizosphere is colonized by a variety of different organisms, including plant-beneficial bacteria. The use of bacteria against plant pathogens is a useful alternative instead of chemical toxins, which improve crop quality and output but can lead to environmental damages (Deketelaere et al., 2017; Syed Ab Rahman et al., 2018). Plants can shape their own root microbiome by attracting certain bacteria (Rudrappa et al., 2008; Berendsen et al., 2018; Huang et al., 2019a), which can not only improve nutrient availability for the plant, but also may act in pest control. V. dahliae in turn can shape the soil microbiome in order to enhance infection (Snelders et al., 2020).
Rhizosphere bacteria including the genera Bacillus or Pseudomonas reduce Verticillium spp. growth in co-culture with potential plant growth promoting or biocontrol functions (Hayat et al., 2010; Hollensteiner et al., 2017; Nesemann et al., 2018). In general, bacteria can prevent pathogen infection by a variety of mechanisms. These include the competition for space and/or nutrients, parasitism (mycophagy), enhancement of plant immunity or the production of antimicrobial compounds (Leveau and Preston, 2008; Köhl et al., 2019). Fluorescent pseudomonads secrete various compounds with bioactive properties as potential biocontrol agents of Verticillium wilt (Deketelaere et al., 2017). These include 2,4-diacetylphloroglucinol (DAPG), phenazines, hydrogen cyanide (HCN), pyoluteorin, and cyclic lipodepsipeptides such as syringomycin and syringopeptin (Maurhofer et al., 1994; Haas and Défago, 2005; Berry et al., 2010).
The polyketide DAPG acts on mitochondria, where it eliminates the proton gradient and thereby leads to increased respiration without production of ATP resulting in growth inhibition (Troppens et al., 2013). DAPG and another polyketide, pyoluteorin, enhance their own production through autoregulation and mutually inhibit the synthesis of each other (Schnider-Keel et al., 2000; Brodhagen et al., 2004). Hydrogen cyanide interferes with the mitochondrial respiratory chain through blockage of oxygen reduction (Hamel, 2011). Phenazines are small pigmented metabolites, which lead to formation of reactive oxygen species (ROS), reactive nitrogen species or iron starvation in Aspergillus fumigatus (Briard et al., 2015). Closely related Pseudomonas strains with beneficial or detrimental effects for Arabidopsis thaliana can differ in the genes encoding enzymes required to produce lipopeptides, which are regulated via quorum sensing (Melnyk et al., 2019). Lipopeptides can also have antifungal properties (Sinden et al., 1971; Berry et al., 2010; Meena and Kanwar, 2015; Toral et al., 2018; Desmyttere et al., 2019). Cyclic lipodepsipeptides as for instance syringomycin and syringopeptin can be inserted into plasma membranes to form ion channels resulting in ion leakage and cell death (Hutchison and Gross, 1997). The inhibitory potential of bacteria in fungal co-cultures varies considerably in the presence of different nutrients affecting fungal spore germination or growth (Michelsen and Stougaard, 2012; Nesemann et al., 2018). We could show that Pseudomonas isolates from soil are not only able to inhibit plant pathogenic Verticillium strains but also moderately affect growth of soil saprophytes of the genus Aspergillus (Nesemann et al., 2018). Aspergillus nidulans is an important and well-studied model organism and is widely distributed in the soil. Depending on the environmental conditions, it can form mitotic spores, which are distributed by the wind or fruiting bodies as overwintering structures with meiotic spores (Krijgsheld et al., 2013). Another ubiquitous saprophyte is Aspergillus fumigatus. Humans are exposed to conidia of this fungus on a regular basis without the development of disease but in immunocompromised patients, the fungus can cause invasive aspergillosis (Kwon-Chung and Sugui, 2013; Shlezinger et al., 2017).
A broader understanding on how Pseudomonas spp. act on or interact with fungal pathogens in different environments is needed to explore potential applications in plant protection. The aim of our study was to compare the inhibitory effects of various fluorescent pseudomonads in different artificial co-culture settings to elucidate the antagonistic effects in a controlled environment with a focus on the rapeseed pathogen V. longisporum. For individual experiments we also included V. dahliae, which is a relative of the V. longisporum parental strain as well as the saprophytic soil fungi A. nidulans and A. fumigatus. Pseudomonas isolates with different genomic potentials including the genes to produce lipopeptides and DAPG were examined. Co-cultivation in small interaction channels of microfluidic devices filled with liquid medium, which enabled visualization of growth and polarity effects on single hyphae level was compared to co-cultures on plates. Fungal transcriptome changes were determined in the absence or presence of the strong inhibitor P. protegens CHA0 (P_DAPG). The ability of different Pseudomonas spp. to interfere with V. dahliae plant root colonization as early and important step for host protection was quantified. Our data revealed that different Pseudomonas isolates induce morphological and transcriptional alterations when controlling growth of phytopathogenic Verticillium spp. and reduce fungal root colonization.
Materials and Methods
Fungal and bacterial strains used in this study are listed in Supplementary Table 1.
Cultivation of Fungal Strains
For spore production, V. dahliae and V. longisporum were grown in liquid simulated xylem medium (SXM), modified according to Neumann and Dobinson (2003) as previously described by Hollensteiner et al. (2017). A. nidulans and A. fumigatus were grown on solid minimal medium [1% (w/v) glucose, 1 x AspA solution (70 mM NaNO3, 7 mM KCl, 11.2 mM KH2PO4, pH 5.5 with KOH), 1 x (v/v) trace element solution (18 μM FeSO4, 174 μM EDTA, 76 μM ZnSO4, 178 μM H3BO3, 25 μM MnCl2, 7.1 μM CoCl2, 6.4 μM CuSO4, 6.2 μM (NH4)6Mo7O24; pH 6.5 with KOH) (Hill and Käfer, 2001), 2 mM MgSO4, 2% agar] at 37°C in light or darkness, respectively.
Bacterial Fungal Co-cultivation on Solid Medium
The co-cultivation experiments on solid SXM were performed as previously described in Hollensteiner et al. (2017) and Nesemann et al. (2018). Spores of Verticillium spp. were harvested after 7 days of incubation, washed and resuspended in sterile water. After two to three days of incubation Aspergillus spp. spores were harvested into a physiological solution (0.96% NaCl, 0.05% Tween 80). The spore concentration was determined with a particle counter (Beckman Coulter, Brea, CA, United States) or in case of A. fumigatus with a Thoma cell counting chamber (hemocytometer; Paul Marienfeld GMBH and CO. KG, Lauda-Königshofen, Germany). 1 × 105 spores of the respective fungus were spread on SXM plates. The concentration of bacterial cultures was determined by photometry (Eppendorf BioPhotometer® D30, Hamburg, Germany). 60 μl bacterial culture grown in liquid SXM (OD600nm = 1; 7 × 107 colony-forming bacteria) were added to a hole in the plate center.
The bacteria tested included different strains, which contained or lacked genes involved in lipopeptide biosynthesis. Pseudomonas brassicacearum DF41 and Pseudomonas sp. N2C3 possess the complete cluster for the production of lipopeptides with homologous genes to P. syringae encoding syringomycin and syringopeptin synthetases (Melnyk et al., 2019). The respective genes are absent in the closely related strains N2E2 or WCS365. Different N2C3 deletion strains were examined, including ΔSYR, impaired in syringomycin synthesis and ΔSYP, missing genes for syringopeptin production. The ΔSYR/ΔSYP double deletion strain and a strain missing the transcriptional regulator LUXR (ΔLUXR) are impaired in the synthesis of both lipopeptides.
After 4 days of incubation in the light at 25°C the inhibition radius was measured. Inhibition was defined as area, where no fungal growth was observed. The experiment was conducted in three biological replicates, each consisting of two to four technical replicates.
Determination of Bacterial Doubling Time
For comparison of doubling times, bacterial strains were inoculated in 50 ml SXM to an OD600 of 0.2 and cultures were incubated at 25°C shaking for 12 h. OD600 was measured directly after inoculation and then every 2 h. Growth curves were plotted and the respective doubling time (Td) was calculated based on the exponential growth phase of the individual bacterial strains (Allen and Waclaw, 2019). The experiment was conducted in three biological replicates.
Bacterial Fungal Co-cultivation in Microfluidic Devices
Verticillium longisporum was inoculated with a piece of agar from a fungal colony at one side of the device (Stanley et al., 2014), which was filled with liquid pectin-rich SXM. Then, the device was incubated at 25°C, which allowed individual hyphae of the fungus to cross a small connecting bridge (10 μm width) and enter the interaction channels (110 μm width, 6650 μm length). Once the fungus entered these interaction channels, bacterial cultures (40 μl, OD600nm = 1; 5 × 107 CFU) were added at the opposite end and the device was further incubated at 25°C. The number of devices used for each experiment is indicated in the respective figure legend. Fungal growth was evaluated once the fungus of the non-treated control reached the end of the channels using a fluorescence microscope (Zeiss Axio Observer Z.1 system (Carl Zeiss AG, Germany) with Laser Lunch System (Model 3iL32, Intelligent Imaging Innovations Inc., Colorado, United States), Zeiss Plan-Apochromat 40×/1,4 oil objective, QuantEM:512SC camera (Photometrics, AZ, United States) and the Slide Book 5.0 imaging software (Intelligent Imaging Innovations Inc.) with an exposure time of 150 ms for GFP).
Sample Preparation and Evaluation of Transcriptomic Data
1350 ml liquid SXM were inoculated with 9 × 106 spores of V. longisporum Vl43 and incubated for five days at 25°C under constant agitation at 150 rpm. After slight sedimentation, 350 ml supernatant were discarded. The remaining culture was equally separated to eight cultures of 125 ml each. P. protegens P_DAPG (CHA0) cultures were also grown in SXM at 25°C and 150 rpm to an OD600nm = 1. After centrifugation at 5000 g bacteria were resuspended in fresh SXM to an OD600nm = 3. To six of the fungal cultures, 25 ml bacterial culture were added resulting in a co-culture with an OD600nm = 0.5. Two fungal cultures mixed with 25 ml SXM served as control. All cultures were further incubated under agitation at 25°C. After 60, 120, and 180 min, respectively two co-cultures were filtered through Miracloth (EMD Millipore Corp., Billerica, MA, United States) and the mycelium remaining in the filter was frozen in liquid nitrogen. The control cultures without bacteria were harvested after 120 min, thus further analysis was conducted for data obtained from the 120 min time point. RNA extraction was performed using the RNeasy Plant Mini Kit (Qiagen, Hilden, Germany). RNA sequencing was conducted by GATC Biotech AG (Konstanz, Germany). By selecting the poly-A+-part of the eukaryotic mRNA, only fungal RNA was sequenced. The sequences were released at NCBI under the Accession number SRP0683481.
The produced RNAseq data were analyzed using the GALAXY platform (Afgan et al., 2018) maintained by the GWDG (Gesellschaft für wissenschaftliche Datenverarbeitung mbH Göttingen). In short, the raw reads from the sequencing were mapped against the Verticillium longisporum genome (GCA_001268145.1, (Fogelqvist et al., 2018)) using Bowtie 2 (Langmead and Salzberg, 2012). The produced alignment files were then used together with the GFF file for V. longisporum (GCA_001268145.1, (Fogelqvist et al., 2018) in the tool HTSeq (Anders et al., 2015) to calculate the number of mapped reads per feature and generate the corresponding matrices, required for the subsequent analysis. The output files from HTSeq were then used for the identification of statistically significant differentially expressed genes with the tool DESeq2 (Love et al., 2014). For the produced data cut off criteria were applied regarding the p-value (p < 0.0001) and Log2-fold-change. Transcripts with a Log2-fold-change > 2 were regarded as most up-regulated and those with a Log2-fold-change < −2 were regarded as most down-regulated (Supplementary Tables 2, 3). The GO enrichment analysis was performed separately for the down-regulated and up-regulated genes using the g:Profiler (Raudvere et al., 2019). Genes within significantly enriched GO categories (Supplementary Tables 4, 5) were further investigated using EnsemblFungi (Yates et al., 2020) with embedded InterPro (Mitchell et al., 2019). Some predicted proteins were additionally compared by BLAST with NCBI (Agarwala et al., 2018) and AspGD (Cerqueira et al., 2014). Cellular localization of selected candidates was predicted using DeepLoc 1.0 (Almagro Armenteros et al., 2017).
Arabidopsis thaliana Root Infection With V. dahliae in Presence of Bacteria
Arabidopsis thaliana (Col-0, N1902; Nottingham Arabidopsis Stock Centre) seeds were surface sterilized and grown as described by Bui et al. (2019) and Harting et al. (2020) with following modifications.
Escherichia coli and Pseudomonas strains were inoculated in Lysogeny broth (LB; Bertani, 1951) and grown over the day at 30°C, under constant agitation. From this culture, 50 ml LB were inoculated to an OD600nm = 0.00001 or in case of the P. synxantha or P. brassicacearum isolates to an OD600nm = 0.001. After overnight incubation at 30°C or 37°C (E. coli) the bacterial cultures were centrifuged at room temperature, with 2500 rpm for 5 min. The supernatant was discarded and the cells were washed in dH2O twice. Eventually the cells were resuspended in dH2O and adjusted to an OD600nm = 0.001 in dH2O.
20-day-old A. thaliana seedlings were transferred to H2O agarose plates (1% agarose (Biozym LE Agarose) in dH2O) and roots were treated with the respective bacterial suspension (OD600nm = 0.001) or dH2O as a control. For that, 50 μl were applied to the hypocotyl of an individual A. thaliana Col-0 plant. Bacteria were spread as the 50 μl drop ran down the root. The plants were further incubated under long day conditions. On the next day all plants were shifted to fresh H2O agarose plates to only keep bacteria on the root. The following day, the plants were infected with GFP-expressing V. dahliae JR2 (Tran et al., 2014) via root dipping in 50 ml of a 1 × 105 spores per ml suspension as described by Bui et al. (2019). Five days after infection, roots were stained using 0.0025% propidium iodide/0.0005% silwet staining solution and colonization of the roots was analyzed using fluorescence microscopy (see section “Bacterial Fungal Co-cultivation in Microfluidic Devices”; EC Plan-Neofluar 20×/0.50 objective and Slide Book 6.0 imaging software were used). For one biological replicate two independent roots were analyzed per treatment. Per root, 10–15 stacks consisting of several individual pictures were acquired to image the different layers of the complete root. Positions were chosen randomly along the root. From each stack, a projection was created. The experiment was performed in three or four replicates (without bacteria, with the N2C3 wild type and respective mutants). Fluorescence quantification was performed using mask statistics feature of Slidebook 6.0 software package (Intelligent Imaging Innovations). Mean GFP intensity was normalized to mean root area. Mean values of biological replicates were normalized to the control of only fungus without bacteria.
Results
Pseudomonas spp. Specifically Inhibit Different Fungi in Growth on Plate
We investigated whether soil inhabiting pseudomonads with or without the ability to form the lipopetides syringomycin and/or syringopeptin have effects on growth of the plant pathogenic fungus Verticillium longisporum. For comparison, the haploid parental relative V. dahliae was included. Cultivations of only fungi or co-cultivation with E. coli were used as controls. As culture medium simulated xylem medium (SXM) was used, which was initially developed to mimic the conditions inside the plant. SXM mainly contains pectin, which is found in plant debris in the soil and is significantly different from plant xylem sap, which is a nutrient-poor unbalanced medium (Singh et al., 2010). V. longisporum is able to distinguish between these two environments, which is supported by two different secretomes, which are expressed by the fungus when grown in these two media (Leonard et al., 2020).
Both bacterial strains with lipopeptide clusters (DF41 and N2C3) were able to inhibit growth of Verticillium spp. (Figures 1A,B). This effect depends on the presence of the gene for the transcriptional regulator LuxR or genes involved in synthesis of syringomycin (ΔLUXR, ΔSYR, ΔSYR/ΔSYP). Absence of only the gene for syringopeptin synthesis (ΔSYP) decreased the antagonistic effect toward Verticillium spp. compared to the respective wild type N2C3. The closely related strains WCS365 and N2E2, which do not encode genes for lipopeptide synthesis, showed a dissimilar effect on Verticillium fungi. Whereas the strain WCS365 was not able to inhibit growth of both isolates, N2E2 showed a strong antagonistic effect against V. dahliae, but no significant growth inhibition of V. longisporum. This suggests that the bacterial isolate N2E2 produces other bioactive substances that are specifically harmful to V. dahliae.
FIGURE 1
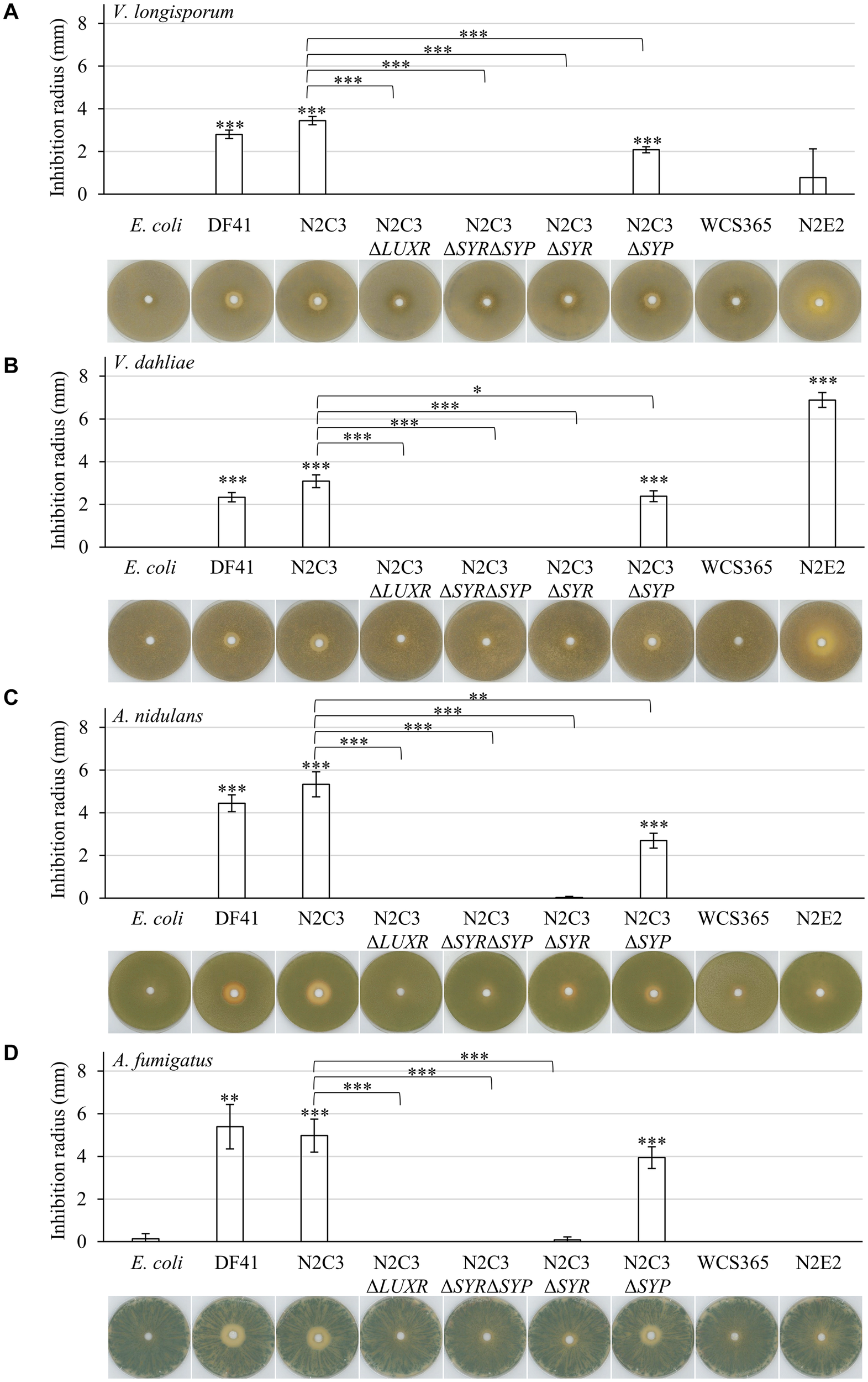
Lipopeptide producing fluorescent pseudomonads inhibit plant pathogenic Verticillium spp. as well as saprophytic Aspergillus spp. Co-cultivation of 1 × 105 fungal spores with 7 × 107 colony-forming bacteria on pectin-rich simulated xylem medium. Plates were incubated in the light for four days at 25°C. The following bacterial isolates were used: Pseudomonas brassicacearum DF41 and Pseudomonas sp. N2C3 (cluster for the production of lipopeptides present), N2C3 deletion strains ΔLUXR (neither syringomycin nor syringopeptin synthesis), ΔSYRΔSYP (neither syringomycin nor syringopeptin synthesis), ΔSYR (no syringomycin synthesis), ΔSYP (no syringopeptin synthesis), WCS365 and N2E2 (both no cluster for lipopeptide synthesis), E. coli as control. Each cultivation was performed in three biological replicates with two to four technical replicates each. Bars represent the mean of biological replicates with standard deviation. Statistical differences were calculated with two-tailed Student’s T-test (∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001). Differences to E. coli control are indicated directly on top of the bars, differences between bacterial wild types and respective mutant strains are indicated with connecting lines. Representative plates are shown beneath the diagrams. Only zones without any fungal growth were measured. Co-cultivation was performed with the rapeseed pathogen V. longisporum Vl43 (A), the tomato pathogen V. dahliae JR2 (B), the saprophyte A. nidulans A4 (C) and the opportunistic human pathogen A. fumigatus AfS35 (D). Fungal growth inhibition by the tested Pseudomonas isolates was increased toward Aspergillus spp. compared to Verticillium spp. The antagonistic effect of N2C3 was more dependent on syringomycin than on syringopeptin.
Both Verticillium species are hemibiotrophic pathogens with only a short saprophytic phase. We further examined bacterial antagonism on specialized saprophytes, as the soil fungus Aspergillus nidulans or A. fumigatus, which can occasionally be an opportunistic human pathogen in immunocompromised individuals. The overall effect of lipopeptide producing strains on growth of Aspergillus spp. is more pronounced than the antagonistic activity toward Verticillium spp. (Figures 1C,D; Supplementary Table 6). Syringomycin contributes more to fungal growth inhibition than syringopeptin. The two strains lacking essential genes for lipopeptide synthesis, WCS365 and N2E2, were not able to affect hyphal growth of the two Aspergillus isolates.
As control we compared doubling times of the different bacterial wild type isolates as well as respective mutant strains to exclude a fitness effect. Growth of the isolates DF41, N2C3 and N2E2 was similar, whereas the isolate WCS365 even grew significantly better under the tested conditions (Supplementary Figure 1). Comparison of the isolate N2C3 and the mutant strains impaired in lipopeptide production shows similar doubling times without significant differences (Supplementary Figure 1). This suggests that the observed changes in fungal inhibition are not the result of differences in bacterial growth.
Taken together, presence of the genes required for syringomycin production is a prerequisite to inhibit growth of plant pathogenic Verticillium species and to a greater extend also the saprophyte A. nidulans and the opportunistic human pathogen A. fumigatus.
V. longisporum Polar Hyphal Growth Is Reduced by Fluorescent Pseudomonads
Co-cultivation of Verticillium hyphae with Pseudomonas spp. might affect growth differently when agar plates are compared to spatially restricted device conditions. Small interaction channels of microfluidic devices (Stanley et al., 2014) allowed confrontation assays and microscopic analysis of single hyphae. Bacterial isolates from the rhizosphere were used for these experiments. The analyzed strains exhibited a stronger antagonistic potential toward V. longisporum compared to the lipopeptide producing bacteria (Nesemann et al., 2018). The growth inhibition potential of the rhizosphere bacteria on plate was dependent on the medium and the genomic potential to produce different metabolites Pseudomonas fluorescens DSM8569 (P_rhizo) was isolated from the rhizosphere of rapeseed (Berg and Ballin, 1994). P_rhizo lacks the entire cluster for the production of phenazines and also part of the cluster for 2-4-diacetylphloroglucinol (DAPG) formation (Nesemann et al., 2015a, 2018). Pseudomonas synxantha 2-79 (P_phen), which was isolated from wheat rhizosphere (Weller and Cook, 1983), has the potential to produce phenazines (Nesemann et al., 2015b, 2018). Pseudomonas protegens CHA0 (P_DAPG) was isolated from the rhizosphere of tobacco (Stutz et al., 1986). P_DAPG lacks the genes for phenazine synthesis, but has genes for DAPG, pyoluteorin and HCN synthesis and also the GacS/GacA control system for the production of several metabolites (Jousset et al., 2014). The production of DAPG, HCN and pyoluteorin in the P_DAPG strain was also experimentally verified (Laville et al., 1992; Schnider-Keel et al., 2000).
Growth of single hyphae of the rapeseed pathogen V. longisporum Vl43 in presence of Pseudomonas isolates P_rhizo, P_phen, P_DAPG and mutant strains were monitored in microfluidic interaction devices depicted in Supplementary Figure 2 (Stanley et al., 2014). Mycelium of the fungal strain ectopically overexpressing GFP was inoculated at one side of the interaction device. Hyphae were grown into the device filled with pectin-rich simulated xylem medium (SXM). As the hyphae entered the interaction channels of the device, bacterial culture was added at the opposite side for co-cultivation. Fungal growth was quantified by measuring how far hyphae grew through the device relative to the control (100%) using the scale of the device.
Specific associations of bacteria with fungal hyphae were never observed during these experiments. Bacteria were instead distributed throughout the whole medium. Co-cultivation with the bacteria resulted in measurable reductions of fungal growth compared to the non-treated control (Figures 2A,B) or co-cultivation with E. coli (Supplementary Figures 3A,B). Incubation with P_rhizo decreased fungal growth to approximately 42% (Supplementary Figures 3A,B). V. longisporum hyphal growth was reduced from 100% to approximately 6% when co-cultivated with P_DAPG (Figures 2A,B). This was the strongest bacterial effect observed. The doubling time of P_DAPG is decreased compared to the other rhizosphere isolates. However, the overall difference in doubling times is rather small, therefore the impact of these differences might be neglectable (Supplementary Figures 1A,B). Incubation with P_phen decreased fungal growth to approximately 30%. A mutant bacterial strain lacking parts of the phenazine metabolite gene cluster improved fungal growth in comparison to P_phen (decreased to only approximately 57% compared to non-treated control; Supplementary Figure 3), although no significant differences in the doubling time were observed (Supplementary Figure 3C). This suggests that the effect of P_phen on fungal growth is partially mediated by phenazines.
FIGURE 2
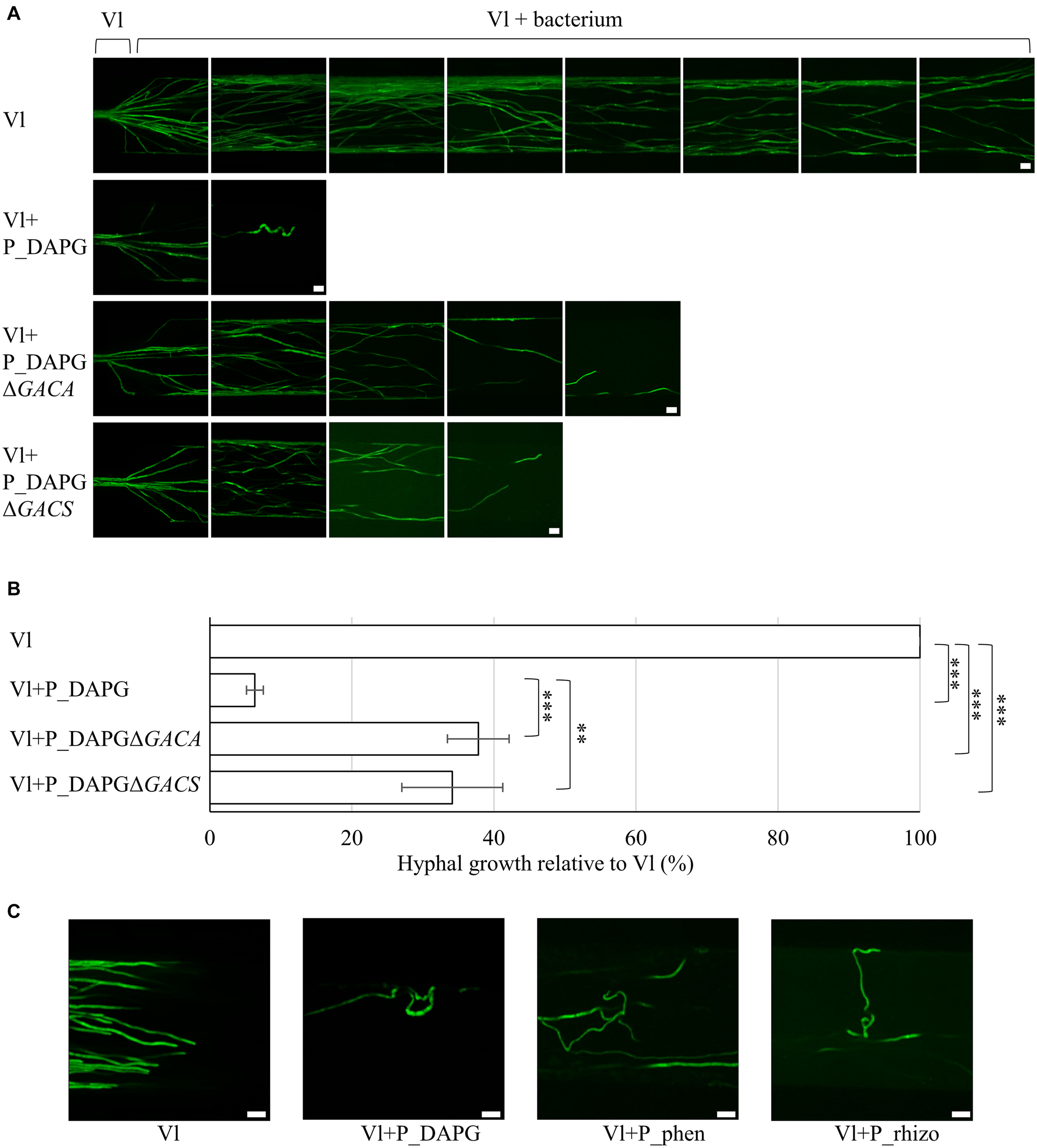
Fluorescent pseudomonads inhibit fungal growth and induce polarity changes in microfluidic devices. Co-cultivation was performed in liquid pectin-rich simulated xylem medium. V. longisporum Vl43 producing high amounts of GFP (Vl) was inoculated at one side of the device with an agar block containing hyphae. The device was incubated at 25°C until hyphae entered the microchannels. As soon as the fungal hyphae reached the beginning of the interaction channels fluorescent pseudomonads were inoculated at the opposite end of the channel. Bacteria spread throughout the device. (A) Representative micrographs of a V. longisporum strain producing high amounts of GFP without (Vl) and with fluorescent bacteria with genes for DAPG synthesis (Vl + P_DAPG) and respective mutant strains, which lack the GacS/GacA two-component control system for the synthesis of various metabolites (Vl + P_DAPGΔGACS, Vl + P_DAPGΔGACA). Images were acquired from three devices with 28 interaction channels each. Fungal growth inhibition partially depends on the presence of GacS/GacA. Scale bars: 20 μm. (B) Quantification of fungal growth in devices relative to V. longisporum growth without bacteria (Vl), which was set to 100% for each device. Bars represent the mean of three devices and error bars the respective standard deviation. Statistical significance was calculated with two-tailed Student’s T-test (∗∗p < 0.01, ∗∗∗p < 0.001). (C) Representative micrographs of a V. longisporum strain producing high amounts of GFP without (Vl) and with fluorescent bacteria with genes for DAPG (Vl + P_DAPG) or phenazines (Vl + P_phen) synthesis and a bacterium isolated from the rhizosphere of rapeseed (Vl + P_rhizo). Polar growth of V. longisporum is altered when cultivated together with different bacterial isolates resulting in a curly phenotype. Two (Vl + P_phen and Vl + P_rhizo) or three (Vl + P_DAPG) devices were evaluated. Scale bars: 20 μm.
Further analysis focused on Pseudomonas P_DAPG, which caused the strongest inhibitory effect of the three bacterial wild type strains on the fungus. Several bacterial P_DAPG mutant derivatives were analyzed, which are impaired in the production of different metabolites. The focus was on strains, which did not show any statistically significant difference in the doubling times compared to wild type (Supplementary Figure 4). Deletion of a structural gene for a DAPG biosynthetic enzyme (P_DAPGΔPHLA) resulted in a strain without DAPG but increased pyoluteorin production (Schnider-Keel et al., 2000). Lack of the gene for the repressor of the DAPG operon (P_DAPGΔPHLF) resulted in increased DAPG production (Schnider-Keel et al., 2000). P_DAPGΔPHLA and P_DAPGΔPHLF did not lead to strong changes in fungal growth inhibition compared to P_DAPG. Deletion of genes for HCN (P_DAPGΔHCN and P_DAPGΔANR) resulted in not detectable or strongly reduced HCN production (Laville et al., 1998). In the strain P_DAPGΔPLT no pyoluteorin was produced (Maurhofer et al., 1994). Both, HCN and pyoluteorin, are dispensable for the inhibitory potential of the bacteria (Supplementary Figure 5). Deletion strains of genes encoding GacS or GacA (P_DAPGΔGACS and P_DAPGΔGACA), which function together in a two-component system to control production of multiple metabolites, did not produce DAPG, HCN or pyoluteorin (Laville et al., 1992; Zuber et al., 2003). Co-cultivation with both strains caused significantly less fungal repression (38% and 34% relative to 100% fungal growth without bacteria or reduction to 6% in presence of P_DAPG; Figures 2A,B). This suggests an important role of the GacS/GacA two-component signal transduction pathway in the bacterial-fungal interaction. As there was still an inhibitory effect observable these results further indicate that there are also GacS/GacA independent inhibitory effects.
An interesting observation of the microscopy studies with microfluidic interaction devices was that polarity of single hyphal growth was disturbed when the fungus was confronted with either of the bacterial isolates P_DAPG, P_phen and P_rhizo, respectively (Figure 2C). The untreated V. longisporum control (Vl) grew in a polar manner, whereas many hyphal tips started to redirect growth and showed an overall curly phenotype when the bacterial antagonist was added to the device. This curly phenotype was less pronounced in co-cultivation with bacterial mutant strains deficient in GacS/GacA system (P_DAPGΔGACS, P_DAPGΔGACA, Figure 2A) or phenazine production (P_phenΔPHZ;Supplementary Figure 3A), which is in line with our observations regarding fungal growth inhibition.
In summary, these data suggest that fluorescent Pseudomonas strains induce polarity changes of the fungus when space is limited. Bacteria are able to significantly decrease growth of single fungal hyphae. This effect on growth is most severe when a bacterial strain is used with a broad potential for the synthesis of different metabolites, which is dependent on the GacS/GacA system that controls the production of several metabolites. Such strong morphological growth defects correlate most likely with significant transcriptomic changes in fungal cells.
P. protegens P_DAPG Induces Transcriptional Changes in V. longisporum for Substrate Utilization and Oxidation-Reduction Processes
RNA sequencing was applied to analyze how co-cultivation induces changes in the fungal transcriptome. Pre-grown fungal cultures were co-cultivated with the bacterium P_DAPG (CHA0), which showed the strongest inhibition potential in previous experiments, in liquid SXM in submerged cultures for 120 min. RNAs were extracted and sequenced. Fungal culture without bacteria served as control. Reads were mapped onto the genome of V. longisporum VL1 (Fogelqvist et al., 2018) and data were further processed as described in the Materials and Methods section.
We found a total of 2151 transcripts which were significantly differentially expressed (p-value < 0.0001). 1407 genes were found to be up-regulated in the presence of P. protegens P_DAPG (Log2-fold-change > 2; Supplementary Table 2). 744 transcripts were down-regulated when the fungus was confronted with bacteria (Log2-fold-change < −2; Supplementary Table 3). The number of proteins encoded by significantly regulated genes, which could be assigned to a GO term was limited, but several candidates could be assigned to one or more significantly enriched categories (Tables 1, 2). The conserved domains of these candidates were further analyzed using the EnsemblFungi webpage (Yates et al., 2020) (Supplementary Tables 4, 5). 11 significantly down-regulated transcripts encode proteins, which are likely involved in the degradation of pectin. They were sorted to the biological process categories ‘cell wall organization’, ‘cell wall organization or biogenesis’, ‘external encapsulating structure organization’ and ‘cellular component organization’. In addition, eight of them were also assigned to the molecular function ‘polygalacturonase activity’. Other identified down-regulated candidates are connected to cellular transport processes. These proteins are represented in the categories ‘SNARE complex disassembly’, ‘cellular component organization’, ‘vesicle-mediated transport’ and ‘structural constituent of cytoskeleton’. For 16 candidates the GO terms ‘tRNA aminoacylation for protein translation’, ‘tRNA aminoacylation’, ‘catalytic activity, acting on RNA’, ‘ligase activity, forming carbon-oxygen bonds’, ‘aminoacyl-tRNA ligase activity’ and ‘catalytic activity, acting on a tRNA’ were identified. Further 11 down-regulated transcripts encode proteins that are predicted to act on RNA (GO term ‘catalytic activity, acting on RNA’). The three candidates from the group ‘N-glycan processing’ are predicted to be involved in protein glycosylation. Additional candidates were grouped into the categories ‘monosaccharide catabolic process’ and ‘monosaccharide metabolic process’. One GO term of the group cellular component with eight proteins was enriched, namely ‘proteasome core complex’.
TABLE 1
| GO category | GO term | Adjusted p-value | Number of candidates |
| Biological process | tRNA aminoacylation for protein translation (GO:0006418) | 0.005049635 | 16 |
| Biological process | Monosaccharide catabolic process (GO:0046365) | 0.006376761 | 5 |
| Biological process | Cell wall organization (GO:0071555) | 0.009380084 | 12 |
| Biological process | External encapsulating structure organization (GO:0045229) | 0.009380084 | 12 |
| Biological process | Monosaccharide metabolic process (GO:0005996) | 0.013036605 | 11 |
| Biological process | SNARE complex disassembly (GO:0035494) | 0.014166236 | 4 |
| Biological process | tRNA aminoacylation (GO:0043039) | 0.01478278 | 16 |
| Biological process | Cellular component organization (GO:0016043) | 0.020421763 | 34 |
| Biological process | Cell wall organization or biogenesis (GO:0071554) | 0.024867501 | 14 |
| Biological process | Vesicle-mediated transport (GO:0016192) | 0.028088849 | 28 |
| Biological process | N-glycan processing (GO:0006491) | 0.032130268 | 3 |
| Cellular component | Proteasome core complex (GO:0005839) | 0.047398555 | 8 |
| Molecular function | Structural constituent of cytoskeleton (GO:0005200) | 5.69437E-05 | 7 |
| Molecular function | Catalytic activity, acting on RNA (GO:0140098) | 0.000109369 | 27 |
| Molecular function | Polygalacturonase activity (GO:0004650) | 0.002643383 | 8 |
| Molecular function | Ligase activity, forming carbon-oxygen bonds (GO:0016875) | 0.00650204 | 16 |
| Molecular function | Aminoacyl-tRNA ligase activity (GO:0004812) | 0.00650204 | 16 |
| Molecular function | Catalytic activity, acting on a tRNA (GO:0140101) | 0.014137349 | 19 |
| Molecular function | Phosphoglycerate mutase activity (GO:0004619) | 0.049844805 | 3 |
GO term enrichment of proteins encoded by genes down-regulated after fungal co-cultivation with P_DAPG for 120 min (p-value < 0.0001 and Log2-fold-change < −2).
TABLE 2
| GO category | GO term | Adjusted p-value | Number of candidates |
| Biological process | Cellular aldehyde metabolic process (GO:0006081) | 0.002079647 | 8 |
| Biological process | Aldehyde biosynthetic process (GO:0046184) | 0.002655749 | 6 |
| Biological process | Vitamin B6 biosynthetic process (GO:0042819) | 0.002655749 | 6 |
| Biological process | Vitamin B6 metabolic process (GO:0042816) | 0.002655749 | 6 |
| Biological process | Pyridoxal phosphate metabolic process (GO:0042822) | 0.002655749 | 6 |
| Biological process | Protoporphyrinogen IX metabolic process (GO:0046501) | 0.003121445 | 8 |
| Biological process | Protoporphyrinogen IX biosynthetic process (GO:0006782) | 0.003121445 | 8 |
| Biological process | Heme biosynthetic process (GO:0006783) | 0.004950441 | 11 |
| Biological process | Porphyrin-containing compound biosynthetic process (GO:0006779) | 0.011773111 | 11 |
| Biological process | Porphyrin-containing compound metabolic process (GO:0006778) | 0.01751914 | 11 |
| Biological process | Tetrapyrrole biosynthetic process (GO:0033014) | 0.01751914 | 11 |
| Biological process | Pyridoxal phosphate biosynthetic process (GO:0042823) | 0.024094575 | 6 |
| Biological process | Tetrapyrrole metabolic process (GO:0033013) | 0.025526976 | 11 |
| Biological process | Proline metabolic process (GO:0006560) | 0.030412694 | 8 |
| Molecular function | Oxygen binding (GO:0019825) | 0.001584484 | 6 |
| Molecular function | Oxidoreductase activity (GO:0016491) | 0.002147285 | 176 |
| Molecular function | Proline dehydrogenase activity (GO:0004657) | 0.031266391 | 5 |
| Molecular function | Amine-lyase activity (GO:0016843) | 0.036201599 | 3 |
| Molecular function | Pyridoxal 5’-phosphate synthase (glutamine hydrolyzing) activity (GO:0036381) | 0.036201599 | 3 |
GO term enrichment of proteins encoded by genes up-regulated after fungal co-cultivation with P_DAPG for 120 min (p-value < 0.0001 and Log2-fold-change > 2).
The largest group of transcripts (176), which were found to be up-regulated in response to bacterial co-cultivation, could be assigned to the molecular function GO term ‘oxidoreductase activity’. This group contains a great variety of different enzymes, which are linked to different cellular processes. One example of this category is a potential manganese/iron superoxide dismutase (BN1708_001526), which might be involved in stress response. Genes encoding proteins, which might be involved in ER redox homeostasis were also identified in this group (BN1708_006230 and BN1708_014150). Five encoded proteins of the oxidoreductases also belong to the group ‘proline dehydrogenase activity’ and are also associated with the biological process ‘proline metabolic process’, to which three additional candidates were also assigned. Six candidates were sorted to the GO term ‘oxygen binding’, including putative protoglobins/globins. Three proteins were categorized to the GO terms ‘amine-lyase activity’ and ‘pyridoxal 5’-phosphate synthase (glutamine hydrolyzing) activity’. Several biological process associated GO terms were significantly enriched in the group of up-regulated transcripts (Table 2). This includes proteins involved in the biosynthesis of aldehydes, vitamin B6 and heme.
These co-cultivation data suggest that the presence of the bacterium induces a V. longisporum response of reduced transcription of genes, which encode enzymes for the degradation of pectin, the carbon source in the surrounding medium. Additionally, cellular transport and protein biosynthesis as well as degradation are presumably down-regulated. In parallel, P. protegens P_DAPG causes a significantly higher fungal expression of genes with oxidoreductase activities. The bacterial induction of changes in fungal growth, morphology and transcription might be sufficient to protect plants from fungal infection.
Pseudomonas spp. Protect Roots From V. dahliae Colonization
Verticillium fungi colonize their host plants starting at the roots. This is also the place where they might encounter bacteria, which inhabit the plant rhizosphere, and where mutually antagonistic interactions take place. Root colonization of V. dahliae in presence of respective bacteria was monitored to examine whether Pseudomonas strains can inhibit Verticillium spp. at the entry point of their plant hosts. Roots of the model plant Arabidopsis thaliana were used for these experiments. Two days prior to infection, roots of the seedlings were treated with a fresh bacterial suspension. Plants were treated with sterile water as control. V. dahliae wild type ectopically expressing high amounts of GFP was used for fungal infection. Roots were inspected by fluorescence microscopy five days after infection.
Roots of plants, which were not treated with bacteria but only water, showed a strong colonization with fungal hyphae on the root surface (Figure 3A). Similarly, roots of plants, which were exposed to E. coli cells, were successfully colonized by the fungus. In contrast, less fungal hyphae could be detected on roots, which have been treated with a suspension containing cells of different Pseudomonas strains. Large parts of the root were free of fungal hyphae, other parts were only slightly colonized (Figure 3A; only pictures with fungal hyphae are shown). Green fluorescence of hyphae was quantified relative to the root surface inspected and normalized to the control without bacteria (Figure 3B). Colonization was decreased to approximately 8% when P_DAPG was applied to the plants before infection. Even the GACS or GACA deletion could reduce the detected fungus on the root surface significantly (27% and 12% compared to the untreated control), which could be due to Gac-independent compounds.
FIGURE 3
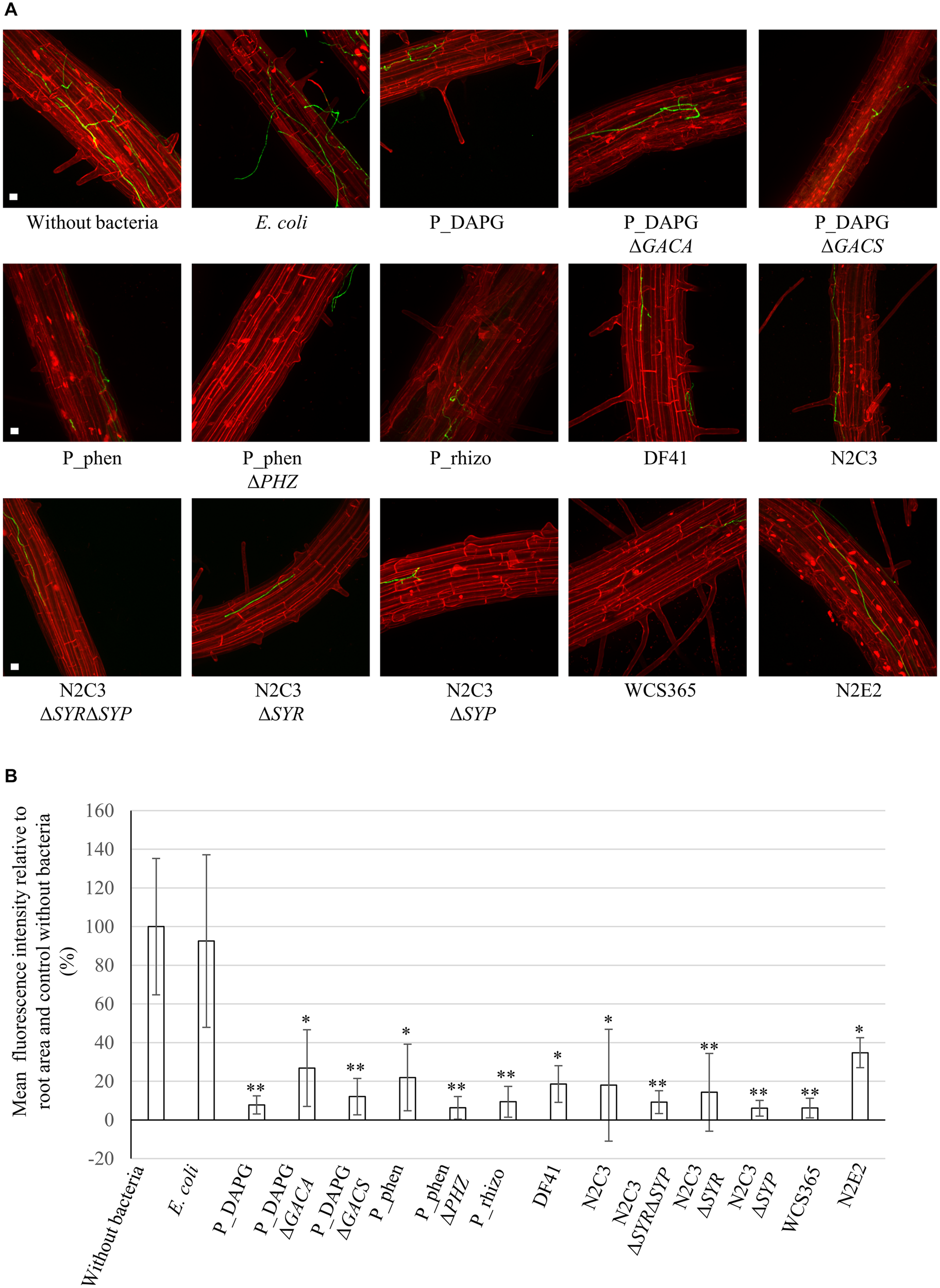
Verticillium dahliae avoids efficient colonization of Arabidopsis thaliana roots in the presence of Pseudomonas strains. A. thaliana seedlings were pre-grown on plate and roots were coated with water (without bacteria) or a bacterial suspension of the indicated strains (OD600nm = 0.001). Two days later, plants were infected by root dipping in spore suspension of V. dahliae JR2 expressing GFP. After five days, fungal colonization of the root surface was investigated by fluorescence microscopy. Roots were stained with propidium iodide. Data are derived from three to four independent experiments. Per experiment, fungal colonization of two roots was analyzed for each individual treatment. For each root 10-15 stacks of single micrographs from randomly chosen root sections were pictured and projections created. (A) Single projection pictures generated from stacks of micrographs for each treatment. Roots of the control without bacteria or with E. coli were colonized by fungal hyphae. When Pseudomonas strains were applied to the roots prior to infection, large parts of the roots remained free of fungus or showed only mild colonization. The pictures show examples of fungal hyphae, which were detected on the root surface. Scale bars: 20 μm. (B) Quantification of green fluorescence of fungal hyphae relative to the root area and the respective control without bacteria. The mean value of treatment without bacteria was set as 100%. Depicted is the mean of three to four biological replicates conducted as described above. Error bars indicate the standard deviation between biological replicates. Significant differences to the control without bacteria were calculated with two-tailed Student’s T-test (∗p < 0.05, ∗∗p < 0.01). Differences to control are indicated on top of the bars. There was no significant difference between wild type strains and their respective mutants. All Pseudomonas isolates are able to reduce fungal colonization of the root surface.
The bacterial isolates P_phen and P_rhizo also efficiently inhibited fungal growth, as only 22% or 9% of fungal hyphae were detected on the root surface compared to the control. The function of P_phen in root protection was independent of the cluster for phenazine production, as the phenazine deficient mutant showed similar protection capacities (P_phenΔPHZ) as the wild type. The lipopeptide producing strains DF41 and N2C3 were also able to significantly reduce the number of fungal hyphae on the root (19% and 18% of hyphae compared to the control). This effect was independent from the production of syringomycin and/or syringopeptin as respective deletion strains (N2C3ΔSYP, N2C3ΔSYR, N2C3ΔSYRΔSYP) or the non-producing isolate WCS365 also prevented the fungus to efficiently colonize the root surface. The isolate N2E2, which showed a strong inhibitory potential toward V. dahliae on plate, was able to significantly inhibit fungal colonization of the root but overall showed the weakest effect of all tested bacteria with 35% of fungal hyphae on the root compared to the control.
Taken together, these data suggest that treatment of roots prior to Verticillium infection significantly reduced the number of fungal hyphae on the root surface. This effect was not significantly dependent on the genomic potential to produce phenazines, lipopeptides or several metabolites controlled by the GacS/GacA system. This suggests a general protective effect of different Pseudomonas isolates on the root surface against the plant pathogen V. dahliae. By reducing the fungal burden on the root, the bacteria might decrease the intensity of Verticillium wilt, accordingly.
Discussion
Treatment of the devastating Verticillium wilt disease is difficult as the most efficient agent for soil fumigation, methyl bromide is no longer allowed, because it can harm the environment (Carroll et al., 2018). Other fungicides are ineffective once the fungus has entered the plant (Sande et al., 2011; Deketelaere et al., 2017; Huang et al., 2019b). The use of antagonistic organisms is an alternative to protect plants from fungal pathogen infection (Uppal et al., 2008; Angelopoulou et al., 2014; Deketelaere et al., 2017; Mulero-Aparicio et al., 2020). Verticillium wilt symptoms of cotton plants can be attenuated by the addition of P. protegens or P. donghuensis isolates with an inhibitory effect of the metabolite 7-hydroxytropolone, which can be monitored during co-cultivation on plates (Tao et al., 2020). Here, the impacts of different bacterial Pseudomonas spp. with various genetic potentials for secondary metabolite formation were dissected at different interaction levels with amphidiploid Verticillium longisporum or haploid V. dahliae. This includes plant root colonization, polarity changes at the fungal hyphal tips, pectin substrate degradation, detoxification of bacterial compounds and the fungal growth rate in general. The complex bacterial influence on the fungus, which was analyzed, is summarized in Figure 4.
FIGURE 4
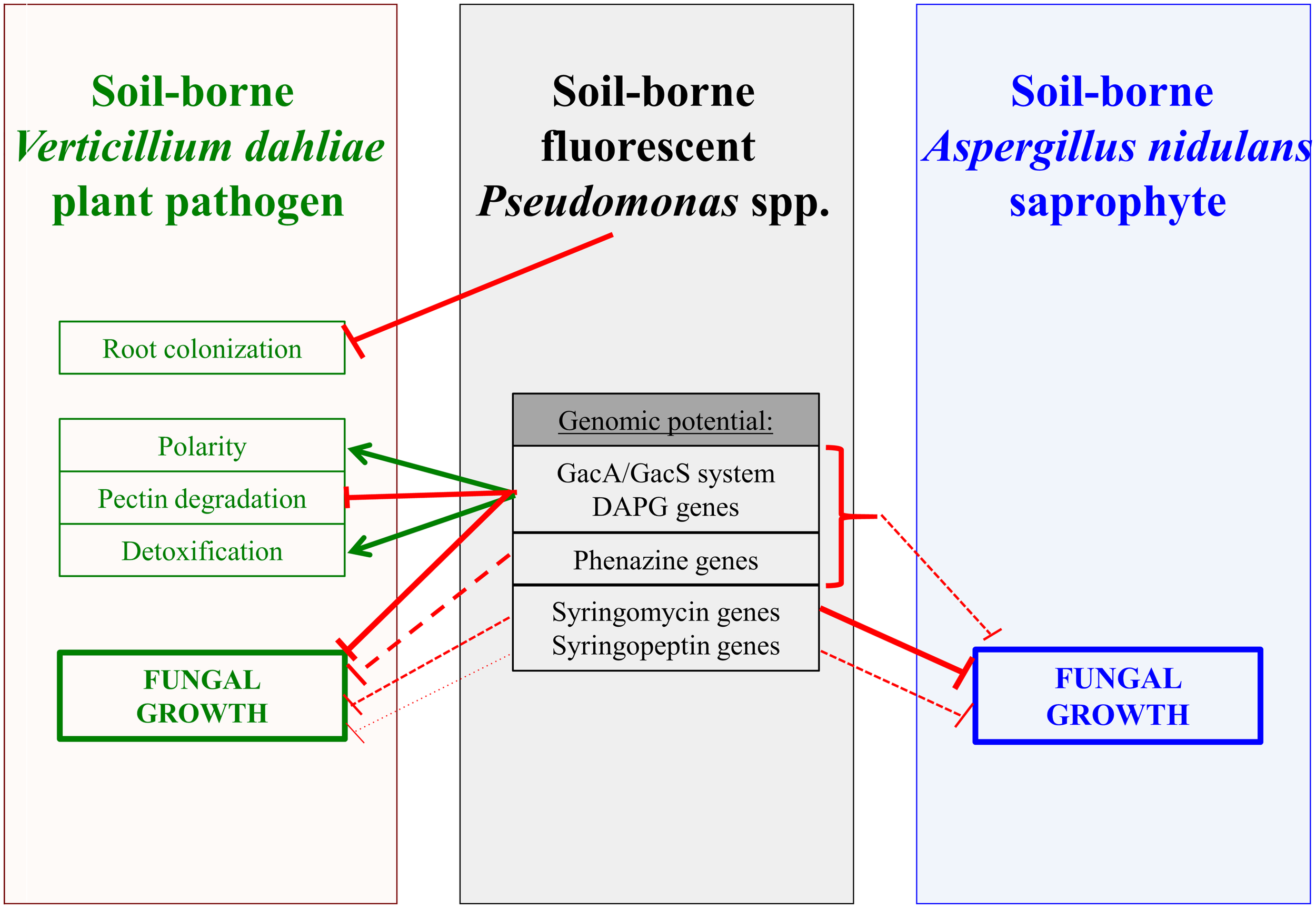
Interplay of Pseudomonas strains with fungal phytopathogen Verticillium dahliae or saprophyte Aspergillus nidulans. Fluorescent pseudomonads isolated from different rhizospheres or groundwater affect Verticillium dahliae and its amphidiploid hybrid V. longisporum as well as growth of A. nidulans. Inhibitory and promoting activities of the bacteria are indicated by red connectors and green arrows, respectively. Dashed lines and smaller line width indicate weaker effects. The bacterial GacS/GacA two component system controls the formation of several bacterial metabolites. For details see text. DAPG: 2,4-diacetylphloroglucinol (DAPG).
The root is the fungal entry point into the plant. Treatment of A. thaliana roots with a bacterial solution of low amounts of different Pseudomonas isolates two days prior to V. dahliae spore infection is sufficient to decrease the number of fungal hyphae on the root surface significantly (Figure 3). Competition for space has been suggested as one potential mechanism (Deketelaere et al., 2017). Plants can also manipulate their root microbiome to attract beneficial bacteria that can help to reduce pathogen attack (Pascale et al., 2020). In our experiments, we applied the bacteria prior to fungal infection, but did not examine if simultaneous or later application of bacteria would still allow reduction of pathogen colonization. An important contribution to the decrease in fungal colonization presumably is bacterial inhibition of fungal spore germination, which can even not be re-established after co-cultivation when bacteria were removed (Nesemann et al., 2018). In addition, the fungus might sense and avoid roots where bacteria are present. In nature, colonization of the host usually starts with fungal hyphae instead of conidia, which derive from stress-resistant microsclerotia, which can survive in the soil for several years (Wilhelm, 1955). Fungal hyphal growth also suggests an avoidance behavior toward Pseudomonas isolates and might not necessarily depend on the production of specific bacterial metabolites. Confrontation of single fungal hyphae with various Pseudomonas strains in microfluidic devices resulted not only in a slowdown of fungal growth, but also in polarity changes at the tip of the hyphae (Figure 2). The fungus is not able to evade the bacterium in these interaction devices filled with liquid medium. The tips of single hyphae start to redirect growth in the channels, leading to a curly phenotype. This suggests that the fungus senses its environment, including the presence of bacteria and/or secretion of bacterial metabolites. Co-cultures of the plant pathogenic fungus Rhizoctonia solani with Serratia spp. bacteria resulted in a different type of morphology change, with swollen fungal hyphae linked to increased branch and septa formation. The R. solani fungal cell wall is modified in co-culture with the bacterium, which is reflected in the transcriptome where genes involved in chitin metabolism as well as genes for ergosterol biosynthesis are down-regulated (Gkarmiri et al., 2015). Growth direction changes of V. longisporum in co-culture with Pseudomonas spp. might be a strategy to avoid contact with the bacteria and potentially harmful metabolites.
Confrontation of the fungus with the bacterium does not only lead to a slowdown but might even stop fungal growth. Analysis of different mutant strains of the Pseudomonas P_DAPG with Verticillium in microfluidic devices revealed that the contribution of bacterial genes for single metabolites as DAPG, pyoluteorin or HCN to fungal growth inhibition is significantly lower than the impairment of the bacterial GacS/GacA two-component regulatory system, which controls the formation of a cocktail of metabolites. This is consistent with earlier findings of bacterial-fungal surface co-cultures on plates (Nesemann et al., 2018). V. longisporum is able to distinguish between pectin-rich medium, plant xylem sap, and other culture media and adapts its secretion response accordingly (Leonard et al., 2020). The perception of external signals and an adequate fungal response requires intracellular signaling pathways, which have important functions in development and virulence of Verticillium spp. (Rauyaree et al., 2005; Qi et al., 2016; Li et al., 2019; Yu et al., 2019; Starke et al., 2021). Transcriptional changes of the fungus caused during bacterial co-cultivation with P_DAPG in liquid medium revealed that transcripts encoding proteins required for the degradation of pectin as main carbon source of the co-culture medium were among the most down-regulated transcripts within 120 min (Supplementary Table 5). This slowdown in metabolism presumably correlates with reduced fungal growth. It is an open question, whether bacteria cause nutrient starvation, and the decreased fungal expression of pectinolytic genes is a result of this depletion. Cellular transport processes as well as transcripts potentially involved in protein synthesis and turnover are also down-regulated (Table 1). Similar responses have been observed for other fungi during growth in unfavorable conditions. When the human opportunistic pathogen A. fumigatus is cultured in human blood, it also senses the environment and then down-regulates energy-consuming processes while turning into a kind of resting mycelium (Irmer et al., 2015).
Botrytis cinerea induces the expression of ABC transporters in the presence of phenazines or DAPG (Schoonbeek et al., 2002). Some of the V. longisporum transporter encoding genes with significantly increased expression (e.g., the potential ABC transporters BN1708_002864 and BN1708_002867, Supplementary Table 2) might be required for the export of toxic bacterial substances from the fungal cell. Significantly induced V. longisporum transcripts that encode the carbon-nitrogen hydrolase (BN1708_019975 and BN1708_004875, Supplementary Table 2) potentially protect against bacterial HCN formation as part of the fungal detoxification and stress response. HCN acts on the electron transport chain (Hamel, 2011). This suggests that an increased expression of genes encoding alternative oxidases (BN1708_000782 and BN1708_017690; Supplementary Tables 2, 4) is a possible reaction of the fungus to cope with bacterial metabolites. Similarly, increased expression of the manganese/iron superoxide dismutase might be a direct stress response of the fungus.
Fungal secondary metabolite clusters can be induced in combination with genes for detoxification during fungal-bacterial interactions (Gkarmiri et al., 2015). The presence of Pseudomonas P_DAPG hardly affects the expression of V. longisporum secondary metabolite genes. BLAST searches for genes encoding several polyketide synthases, non-ribosomal peptide synthases, a hybrid polyketide synthase-non-ribosomal peptide synthase as well as genes for the production of terpenes and other secondary metabolites (Shi-Kunne et al., 2019) revealed that only the genes potentially coding for a non-ribosomal peptide synthase (BN1708_006789 and BN1708_014277) were up-regulated in our RNAseq analysis. Regulation of secondary metabolism and development is regulated by the velvet family of transcription factors. The V. dahliae velvet protein Vel1 is required for initial root colonization, transport within the plant by conidiation, control of secondary metabolism as well as resting structure formation for long time survival in the soil (Höfer et al., 2021).
Transcription of three glycoside hydrolase family 24 member genes was induced in Coprinopsis cinerea co-cultivated with bacteria and bacterial lysis was confirmed with purified proteins (Kombrink et al., 2019). One hint for antibacterial Verticillium activity is the increased transcript level (BN1708_003720, Supplementary Table 2) of the gene encoding a glycoside hydrolase for potential degradation of bacterial cell wall peptidoglycan. The predicted extracellular localization supports that the fungus secretes the enzyme against bacteria in the liquid medium.
The potential of Pseudomonas isolates to control growth of different fungi, which they might encounter in soil, varies considerably. Pseudomonas strains P_DAPG, P_phen and P_rhizo, isolated from the rhizosphere of different plants, had a greater inhibitory potential toward the plant pathogen V. dahliae than toward the saprophyte A. nidulans or the opportunistic human pathogen A. fumigatus when cultivated on solid medium (Nesemann et al., 2018). In contrast, the inhibitory potential of lipopetide producing Pseudomonas brassicacearum DF41, which inhibits the growth of Sclerotinia sclerotiorum (Berry et al., 2010, 2012), is significantly higher toward Aspergillus strains than to Verticillium spp. The overall effect of syringomycin is stronger than of syringopeptin (Figure 1). Lipopeptides act on the plasma membrane of plants and fungi and increase their permeability (Hutchison et al., 1995; Hutchison and Gross, 1997). Nonapeptide lactones including syringomycin have more antimicrobial activity, whereas syringopeptins are rather phytotoxic (Iacobellis et al., 1992; Lavermicocca et al., 1997; Dalla Serra et al., 1999). The addition of cell wall degrading enzymes in the soil, which permeabilize the cell wall, can increase the antifungal activity of syringopeptins, which are rather big molecules (Fogliano et al., 2002). Syringomycin is lethal for Aspergillus spp. (De Lucca et al., 1999). Germinating conidia from different fungal species from diseased grapes are killed by a combination of syringomycin and rhamnolipids (Takemoto et al., 2010).
In summary, fluorescent Pseudomonas strains with their metabolic diversity have a great and not yet fully explored potential to protect plants from fungal infections, such as Verticillium wilt. It is important to investigate the potential of respective bacteria to protect natural host plants as rapeseed from V. longisporum and tomato, lettuce, strawberry or olive trees from V. dahliae, respectively. Plant protection happens on multiple levels including control of hyphal growth and root colonization. The interactions need to be further explored to fully understand the mechanism by which inhibition, colonization reduction or ultimately plant protection occur.
Statements
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, SRP068348.
Author contributions
The study was designed and conceived by RH, AN, KN, CS, MK, CH, SB-S, and GB. RH, AN, KN, AH, EB, MStö, and KH performed and analyzed the experiments. Formal analysis was undertaken by all authors. The whole project was supervised by RH, SB-S, GB. RH, AN, KN, SB-S, and GB wrote the manuscript. All authors interpreted the results, revised, and approved the final manuscript.
Funding
This research has been funded by the Deutsche Forschungsgemeinschaft DFG BR1502-1 (https://www.dfg.de) to GB, the IRTG 2172 ‘PRoTECT’ program of the Goettingen Graduate Center of Neurosciences, Biophysics, and Molecular Biosciences to AN and GB and by ETH Zürich (ETH Research Grants ETH-34 11-2 and ETH-45 16-1) to MK. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Acknowledgments
We thank N. Scheiter for technical assistance and M. Leonard, I. Maurus, J. Starke, C. Sasse, B. Popova, M. Landesfeind, M. Nowrousian and M. Aebi for support and help with experimental work. We acknowledge support by the Open Access Publication Funds of the Göttingen University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.652468/full#supplementary-material
Footnotes
References
1
Acharya B. Ingram T. W. Oh Y. Adhikari T. B. Dean R. A. Louws F. J. (2020). Opportunities and challenges in studies of host-pathogen interactions and management of Verticillium dahliae in tomatoes.Plants91–31. 10.3390/plants9111622
2
Afgan E. Baker D. Batut B. van den Beek M. Bouvier D. Cech M. et al (2018). The galaxy platform for accessible, reproducible and collaborative biomedical analyses: 2018 update.Nucleic Acids Res.46W537–W544. 10.1093/nar/gky379
3
Agarwala R. Barrett T. Beck J. Benson D. A. Bollin C. Bolton E. et al (2018). Database resources of the national center for biotechnology information.Nucleic Acids Res.46D8–D13. 10.1093/nar/gkx1095
4
Allen R. J. Waclaw B. (2019). Bacterial growth: a statistical physicist’s guide.Rep. Prog. Phys.82:016601. 10.1088/1361-6633/aae546.Bacterial
5
Almagro Armenteros J. J. Sønderby C. K. Sønderby S. K. Nielsen H. Winther O. (2017). DeepLoc: prediction of protein subcellular localization using deep learning.Bioinformatics333387–3395. 10.1093/bioinformatics/btx431
6
Anders S. Pyl P. T. Huber W. (2015). HTSeq - a Python framework to work with high-throughput sequencing data.Bioinformatics31166–169. 10.1093/bioinformatics/btu638
7
Angelopoulou D. J. Naska E. J. Paplomatas E. J. Tjamos S. E. (2014). Biological control agents (BCAs) of Verticillium wilt: influence of application rates and delivery method on plant protection, triggering of host defence mechanisms and rhizosphere populations of BCAs.Plant Pathol.631062–1069. 10.1111/ppa.12198
8
Belozerskaya T. A. Gessler N. N. Aver‘yanov A. A. (2017). “Melanin pigments of fungi,” in Fungal Metabolites, edsMerillonJ. M.RamawatK. (Berlin: Springer International Publishing), 263–291. 10.1007/978-3-319-25001-4
9
Berendsen R. L. Vismans G. Yu K. Song Y. de Jonge R. Burgman W. P. et al (2018). Disease-induced assemblage of a plant-beneficial bacterial consortium.ISME J.121496–1507. 10.1038/s41396-018-0093-91
10
Berg G. Ballin G. (1994). Bacterial antagonists to Verticillium dahliae kleb.J. Phytopathol.14199–110. 10.1111/j.1439-0434.1994.tb01449.x
11
Berry C. Fernando W. G. Dilantha Loewen P. C. de Kievit T. R. (2010). Lipopeptides are essential for Pseudomonas sp. DF41 biocontrol of sclerotinia sclerotiorum.Biol. Control55211–218. 10.1016/j.biocontrol.2010.09.011
12
Berry C. L. Brassinga A. K. C. Donald L. J. Fernando W. G. D. Loewen P. C. de Kievit T. R. (2012). Chemical and biological characterization of sclerosin, an antifungal lipopeptide.Can. J. Microbiol.581027–1034. 10.1139/w2012-079
13
Bertani G. (1951). Studies on lysogenesis. I. the mode of phage liberation by lysogenic Escherichia coli.J. Bacteriol.62293–300. 10.1128/JB.62.3.293-300.1951
14
Briard B. Bomme P. Lechner B. E. Mislin G. L. A. Lair V. Prévost M.-C. et al (2015). Pseudomonas aeruginosa manipulates redox and iron homeostasis of its microbiota partner Aspergillus fumigatus via phenazines.Sci. Rep.5:8220. 10.1038/srep08220
15
Brodhagen M. Henkels M. D. Loper J. E. (2004). Positive autoregulation and signaling properties of pyoluteorin, an antibiotic produced by the biological control organism Pseudomonas fluorescens Pf-5.Appl. Environ. Microbiol.701758–1766. 10.1128/AEM.70.3.1758-1766
16
Bui T. Harting R. Braus-Stromeyer S. A. Tran V. Leonard M. Höfer A. et al (2019). Verticillium dahliae transcription factors Som1 and Vta3 control microsclerotia formation and sequential steps of plant root penetration and colonisation to induce disease.New Phytol.2212138–2159. 10.1111/nph.15514
17
Carroll C. L. Carter C. A. Goodhue R. E. Lawell C.-Y. C. L. Subbarao K. V. (2018). A review of control options and externalities for Verticillium wilts.Phytopathology108160–171. 10.1094/PHYTO-03-17-0083-RVW
18
Casadevall A. Cordero R. J. B. Bryan R. Nosanchuk J. Dadachova E. (2017). Melanin, radiation, and energy transduction in fungi.Microbiol. Spectr.5:FUNK-0037-2016.
19
Cerqueira G. C. Arnaud M. B. Inglis D. O. Skrzypek M. S. Binkley G. Simison M. et al (2014). The Aspergillus genome database: multispecies curation and incorporation of RNA-Seq data to improve structural gene annotations.Nucleic Acids Res.42D705–D710. 10.1093/nar/gkt1029
20
Dalla Serra M. Fagiuoli G. Nordera P. Bernhart I. Della Volpe C. Di Giorgio D. et al (1999). The interaction of lipodepsipeptide toxins from Pseudomonas syringae pv. syringae with biological and model membranes: a comparison of syringotoxin, syringomycin, and two syringopeptins.Mol. Plant-Microbe Interact.12391–400. 10.1094/mpmi.1999.12.5.391
21
De Lucca A. J. Jacks T. J. Takemoto J. Vinyard B. Peter J. Navarro E. et al (1999). Fungal lethality, binding, and cytotoxicity of syringomycin-E.Antimicrob. Agents Chemother.43371–373. 10.1128/aac.43.2.371
22
Deketelaere S. Tyvaert L. França S. C. Höfte M. (2017). Desirable traits of a good biocontrol agent against Verticillium wilt.Front. Microbiol.8:1186. 10.3389/fmicb.2017.01186
23
Depotter J. R. L. Deketelaere S. Inderbitzin P. von Tiedemann A. Höfte M. Subbarao K. V. et al (2016). Verticillium longisporum, the invisible threat to oilseed rape and other brassicaceous plant hosts.Mol. Plant Pathol.171004–1016. 10.1111/mpp.12350
24
Depotter J. R. L. Thomma B. P. H. J. Wood T. A. (2019). Measuring the impact of Verticillium longisporum on oilseed rape (Brassica napus) yield in field trials in the United Kingdom.Eur. J. Plant Pathol.153321–326. 10.1007/s10658-018-1537-1531
25
Desmyttere H. Deweer C. Muchembled J. Sahmer K. Jacquin J. Coutte F. et al (2019). Antifungal activities of Bacillus subtilis lipopeptides to two Venturia inaequalis strains possessing different tebuconazole sensitivity.Front. Microbiol.10:2327. 10.3389/fmicb.2019.02327
26
EFSA Panel on Plant Health [PLH] (2014). Scientific opinion on the pest categorisation of Verticillium dahliae Kleb.EFSA J.12:3928. 10.2903/j.efsa.2014.3928
27
Fan R. Gong X. Gao L. Shang W. Hu X. Xu X. (2020). Temporal dynamics of the survival of Verticillium dahliae microsclerotia with or without melanin in soils amended with biocontrol agents.Eur. J. Plant Pathol.157521–531. 10.1007/s10658-020-02014-2019
28
Fogelqvist J. Tzelepis G. Bejai S. Ilbäck J. Schwelm A. Dixelius C. (2018). Analysis of the hybrid genomes of two field isolates of the soil-borne fungal species Verticillium longisporum.BMC Genomics19:14. 10.1186/s12864-017-4407-x
29
Fogliano V. Ballio A. Gallo M. Woo S. Scala F. Lorito M. (2002). Pseudomonas lipodepsipeptides and fungal cell wall-degrading enzymes act synergistically in biological control.Mol. Plant-Microbe Interact.15323–333. 10.1094/MPMI.2002.15.4.323
30
Fradin E. F. Thomma B. P. H. J. (2006). Physiology and molecular aspects of Verticillium wilt diseases caused by V. dahliae and V. albo-atrum.Mol. Plant Pathol.771–86. 10.1111/j.1364-3703.2006.00323.x
31
Gkarmiri K. Finlay R. D. Alström S. Thomas E. Cubeta M. A. Högberg N. (2015). Transcriptomic changes in the plant pathogenic fungus Rhizoctonia solani AG-3 in response to the antagonistic bacteria Serratia proteamaculans and Serratia plymuthica.BMC Genomics16:630. 10.1186/s12864-015-1758-z
32
Haas D. Défago G. (2005). Biological control of soil-borne pathogens by fluorescent pseudomonads.Nat. Rev. Microbiol.3307–319. 10.1038/nrmicro1129
33
Hamel J. (2011). A review of acute cyanide poisoning with a treatment update.Crit. Care Nurse3172–81. 10.4037/ccn2011799
34
Harting R. Höfer A. Tran V.-T. Weinhold L.-M. Barghahn S. Schlüter R. et al (2020). The Vta1 transcriptional regulator is required for microsclerotia melanization in Verticillium dahliae.Fungal Biol.124490–500. 10.1016/j.funbio.2020.01.007
35
Harting R. Starke J. Kusch H. Pöggeler S. Maurus I. Schlüter R. et al (2021). A 20 kb lineage-Specific genomic region tames virulence in pathogenic amphidiploid Verticillium longisporum.Mol. Plant Pathol.10.1111/mpp.13071
36
Hayat R. Ali S. Amara U. Khalid R. Ahmed I. (2010). Soil beneficial bacteria and their role in plant growth promotion: a review.Ann. Microbiol.60579–598. 10.1007/s13213-010-0117-111
37
Hill T. W. Käfer E. (2001). Improved protocols for Aspergillus minimal medium: trace element and minimal medium salt stock solutions.Fungal Genet. Rep.4820–21. 10.4148/1941-4765.1173
38
Höfer A. M. Harting R. Aßmann N. F. Gerke J. Schmitt K. Starke J. et al (2021). The velvet protein Vel1 controls initial plant root colonization and conidia formation for xylem distribution in Verticillium wilt.PLoS Genet.17:e1009434. 10.1371/journal.pgen.1009434
39
Hollensteiner J. Wemheuer F. Harting R. Kolarzyk A. M. Diaz Valerio S. M. Poehlein A. et al (2017). Bacillus thuringiensis and Bacillus weihenstephanensis inhibit the growth of phytopathogenic Verticillium species.Front. Microbiol.7:2171. 10.3389/fmicb.2016.02171
40
Huang A. C. Jiang T. Liu Y.-X. Bai Y.-C. Reed J. Qu B. et al (2019a). A specialized metabolic network selectively modulates Arabidopsis root microbiota.Science364:eaau6389. 10.1126/science.aau6389
41
Huang B. Yan D. Wang X. Wang X. Fang W. Zhang D. et al (2019b). Soil fumigation alters adsorption and degradation behavior of pesticides in soil.Environ. Pollut.246264–273. 10.1016/j.envpol.2018.12.003
42
Hutchison M. L. Gross D. C. (1997). Lipopeptide phytotoxins produced by Pseudomonas syringae pv. syringae: comparison of the biosurfactant and ion channel-forming activities of syringopeptin and syringomycin.Mol. Plant-Microbe Interact.10347–354. 10.1094/MPMI.1997.10.3.347
43
Hutchison M. L. Tester M. A. Gross D. C. (1995). Role of biosurfactant and ion channel-forming activities of syringomycin in transmembrane ion flux: a model for the mechanism of action in the plant-pathogen interaction.Mol. Plant-Microbe Interact.8610–620. 10.1094/MPMI-8-0610
44
Iacobellis N. S. Lavermicocca P. Grgurina I. Simmaco M. Ballio A. (1992). Phytotoxic properties of Pseudomonas syringae pv. syringae toxins.Physiol. Mol. Plant Pathol.40107–116.
45
Irmer H. Tarazona S. Sasse C. Olbermann P. Loeffler J. Krappmann S. et al (2015). RNAseq analysis of Aspergillus fumigatus in blood reveals a just wait and see resting stage behavior.BMC Genomics16:640. 10.1186/s12864-015-1853-1851
46
Jousset A. Schuldes J. Keel C. Maurhofer M. Daniel R. Scheu S. et al (2014). Full-genome sequence of the plant growth-promoting bacterium Pseudomonas protegens CHA0.Genome Announc.2:e00322-14. 10.1128/genomeA.00322-314
47
Köhl J. Kolnaar R. Ravensberg W. J. (2019). Mode of action of microbial biological control agents against plant diseases: relevance beyond efficacy.Front. Plant Sci.10:845. 10.3389/fpls.2019.00845
48
Kombrink A. Tayyrov A. Essig A. Stöckli M. Micheller S. Hintze J. et al (2019). Induction of antibacterial proteins and peptides in the coprophilous mushroom Coprinopsis cinerea in response to bacteria.ISME J.13588–602. 10.1038/s41396-018-0293-298
49
Krijgsheld P. Bleichrodt R. van Veluw G. J. Wang F. Müller W. H. Dijksterhuis J. et al (2013). Development in Aspergillus.Stud. Mycol.741–29. 10.3114/sim0006
50
Kunej U. Mikulič-Petkovšek M. Radišek S. Štajner N. (2020). Changes in the phenolic compounds of hop (Humulus lupulus L.) induced by infection with Verticillium nonalfalfae, the causal agent of hop Verticillium wilt.Plants91–16. 10.3390/plants9070841
51
Kwon-Chung K. J. Sugui J. A. (2013). Aspergillus fumigatus-What makes the species a ubiquitous human fungal pathogen?PLoS Pathog.9:e1003743. 10.1371/journal.ppat.1003743
52
Langmead B. Salzberg S. L. (2012). Fast gapped-read alignment with Bowtie 2.Nat. Methods9357–359. 10.1038/nmeth.1923
53
Lavermicocca P. Iacobellis N. S. Simmaco M. Graniti A. (1997). Biological properties and spectrum of activity of Pseudomonas syringae pv. syringae toxins.Physiol. Mol. Plant Pathol.50129–140. 10.1006/pmpp.1996.0078
54
Laville J. Blumer C. Von Schroetter C. Gaia V. Défago G. Keel C. et al (1998). Characterization of the hcnABC gene cluster encoding hydrogen cyanide synthase and anaerobic regulation by ANR in the strictly aerobic biocontrol agent Pseudomonas fluorescens CHA0.J. Bacteriol.1803187–3196. 10.1128/jb.180.12.3187-3196.1998
55
Laville J. Voisard C. Keel C. Maurhofer M. Défago G. Haas D. (1992). Global control in Pseudomonas fluorescens mediating antibiotic synthesis and suppression of black root rot of tobacco.Proc. Natl. Acad. Sci. U S A.891562–1566. 10.1073/pnas.89.5.1562
56
Leonard M. Kühn A. Harting R. Maurus I. Nagel A. Starke J. et al (2020). Verticillium longisporum elicits media-dependent secretome responses with capacity to distinguish between plant-related environments.Front. Microbiol.11:1876. 10.3389/fmicb.2020.01876
57
Leveau J. H. J. Preston G. M. (2008). Bacterial mycophagy: definition and diagnosis of a unique bacterial-fungal interaction.New Phytol.177859–876. 10.1111/j.1469-8137.2007.02325.x
58
Li J.-J. Zhou L. Yin C. M. Zhang D.-D. Klosterman S. J. Wang B.-L. et al (2019). The Verticillium dahliae Sho1-MAPK pathway regulates melanin biosynthesis and is required for cotton infection.Environ. Microbiol.214852–4874. 10.1111/1462-2920.14846
59
Love M. I. Huber W. Anders S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2.Genome Biol.15:550. 10.1186/s13059-014-0550-558
60
Maurhofer M. Keel C. Haas D. Défago G. (1994). Pyoluteorin production by Pseudomonas fluorescens strain CHA0 is involved in the suppression of Pythium damping-off of cress but not of cucumber.Eur. J. Plant Pathol.100221–232. 10.1007/BF01876237
61
Meena K. R. Kanwar S. S. (2015). Lipopeptides as the antifungal and antibacterial agents: applications in food safety and therapeutics.Biomed Res. Int.2015:473050. 10.1155/2015/473050
62
Melnyk R. A. Hossain S. S. Haney C. H. (2019). Convergent gain and loss of genomic islands drive lifestyle changes in plant-associated Pseudomonas.ISME J.131575–1588. 10.1038/s41396-019-0372-375
63
Michelsen C. F. Stougaard P. (2012). Hydrogen cyanide synthesis and antifungal activity of the biocontrol strain Pseudomonas fluorescens In5 from greenland is highly dependent on growth medium.Can. J. Microbiol.58381–390. 10.1139/w2012-004
64
Mitchell A. L. Attwood T. K. Babbitt P. C. Blum M. Bork P. Bridge A. et al (2019). InterPro in 2019: improving coverage, classification and access to protein sequence annotations.Nucleic Acids Res.47D351–D360. 10.1093/nar/gky1100
65
Mulero-Aparicio A. Varo A. Agustí-Brisach C. López-Escudero F. J. Trapero A. (2020). Biological control of Verticillium wilt of olive in the field.Crop Prot.128:104993. 10.1016/j.cropro.2019.104993
66
Nesemann K. Braus-Stromeyer S. A. Harting R. Höfer A. Kusch H. Ambrosio A. B. et al (2018). Fluorescent pseudomonads pursue media-dependent strategies to inhibit growth of pathogenic Verticillium fungi.Appl. Microbiol. Biotechnol.102817–831. 10.1007/s00253-017-8618-8615
67
Nesemann K. Braus-Stromeyer S. A. Thuermer A. Daniel R. Braus G. H. (2015a). Draft genome sequence of the beneficial rhizobacterium Pseudomonas fluorescens DSM 8569, a natural isolate of oilseed rape (Brassica napus).Genome Announc.3:e10037-15. 10.1128/genomeA.00137-115
68
Nesemann K. Braus-Stromeyer S. A. Thuermer A. Daniel R. Mavrodi D. V. Thomashow L. S. et al (2015b). Draft genome sequence of the phenazine-producing Pseudomonas fluorescens strain 2-79.Genome Announc3:e00130-15. 10.1128/genomeA.00130-115
69
Neumann M. J. Dobinson K. F. (2003). Sequence tag analysis of gene expression during pathogenic growth and microsclerotia development in the vascular wilt pathogen Verticillium dahliae.Fungal Genet. Biol.3854–62. 10.1016/S1087-1845(02)00507-508
70
Pascale A. Proietti S. Pantelides I. S. Stringlis I. A. (2020). Modulation of the root microbiome by plant molecules: the basis for targeted disease suppression and plant growth promotion.Front. Plant Sci.10:1741. 10.3389/fpls.2019.01741
71
Qi X. Zhou S. Shang X. Wang X. (2016). VdSho1 regulates growth, oxidant adaptation and virulence in Verticillium dahliae.J. Phytopathol.1641064–1074. 10.1111/jph.12527
72
Raudvere U. Kolberg L. Kuzmin I. Arak T. Adler P. Peterson H. et al (2019). g:Profiler: a web server for functional enrichment analysis and conversions of gene lists (2019 update).Nucleic Acids Res.47W191–W198. 10.1093/nar/gkz369
73
Rauyaree P. Ospina-Giraldo M. D. Kang S. Bhat R. G. Subbarao K. V. Grant S. J. et al (2005). Mutations in VMK1, a mitogen-activated protein kinase gene, affect microsclerotia formation and pathogenicity in Verticillium dahliae.Curr. Genet.48109–116. 10.1007/s00294-005-0586-580
74
Reusche M. Truskina J. Thole K. Nagel L. Rindfleisch S. Tran V. T. et al (2014). Infections with the vascular pathogens Verticillium longisporum and Verticillium dahliae induce distinct disease symptoms and differentially affect drought stress tolerance of Arabidopsis thaliana.Environ. Exp. Bot.10823–37. 10.1016/j.envexpbot.2013.12.009
75
Rudrappa T. Czymmek K. J. Paré P. W. Bais H. P. (2008). Root-secreted malic acid recruits beneficial soil bacteria.Plant Physiol.1481547–1556. 10.1104/pp.108.127613
76
Sande D. Mullen J. Wetzstein M. Houston J. (2011). Environmental impacts from pesticide use: a case study of soil fumigation in Florida tomato production.Int. J. Environ. Res. Public Health84649–4661. 10.3390/ijerph8124649
77
Schnider-Keel U. Seematter A. Maurhofer M. Blumer C. Duffy B. Gigot-Bonnefoy C. et al (2000). Autoinduction of 2,4-diacetylphloroglucinol biosynthesis in the biocontrol agent Pseudomonas fluorescens CHA0 and repression by the bacterial metabolites salicylate and pyoluteorin.J. Bacteriol.1821215–1225. 10.1128/JB.182.5.1215-1225.2000
78
Schoonbeek H. J. Raaijmakers J. M. De Waard M. A. (2002). Fungal ABC transporters and microbial interactions in natural environments.Mol. Plant-Microbe Interact.151165–1172. 10.1094/MPMI.2002.15.11.1165
79
Shi-Kunne X. Jové R. D. P. Depotter J. R. L. Ebert M. K. Seidl M. F. Thomma B. P. H. J. (2019). In silico prediction and characterisation of secondary metabolite clusters in the plant pathogenic fungus Verticillium dahliae.FEMS Microbiol. Lett.366:fnz081. 10.1093/femsle/fnz081
80
Shlezinger N. Irmer H. Dhingra S. Beattie S. R. Cramer R. A. Braus G. H. et al (2017). Sterilizing immunity in the lung relies on targeting fungal apoptosis-like programmed cell death.Science3571037–1041. 10.1126/science.aan0365
81
Siebold M. von Tiedemann A. (2013). Effects of experimental warming on fungal disease progress in oilseed rape.Glob. Chang. Biol.191736–1747. 10.1111/gcb.12180
82
Sinden S. L. DeVay J. E. Backman P. A. (1971). Properties of syringomycin, a wide spectrum antibiotic and phytotoxin produced by Pseudomonas syringae, and its role in the bacterial canker disease of peach trees.Physiol. Plant Pathol.1199–200. 10.1016/0048-4059(71)90029-90024
83
Singh S. Braus-Stromeyer S. A. Timpner C. Tran V. T. Lohaus G. Reusche M. et al (2010). Silencing of Vlaro2 for chorismate synthase revealed that the phytopathogen Verticillium longisporum induces the cross-pathway control in the xylem.Appl. Microbiol. Biotechnol.851961–1976. 10.1007/s00253-009-2269-2260
84
Snelders N. C. Rovenich H. Petti G. C. Rocafort M. van den Berg G. C. M. Vorholt J. A. et al (2020). Microbiome manipulation by a soil-borne fungal plant pathogen using effector proteins.Nat. Plants61365–1374. 10.1038/s41477-020-00799-5
85
Stanley C. E. Stöckli M. Van Swaay D. Sabotič J. Kallio P. T. Künzler M. et al (2014). Probing bacterial-fungal interactions at the single cell level.Integr. Biol.6935–945. 10.1039/c4ib00154k
86
Starke J. Harting R. Maurus I. Leonard M. Bremenkamp R. Heimel K. et al (2021). Unfolded protein response and scaffold independent pheromone MAP kinase signaling control Verticillium dahliae growth, development, and plant pathogenesis.J. Fungi7:305. 10.3390/jof7040305
87
Stutz E. W. Defago G. Kern H. (1986). Naturally occuring fluorescent pseudomonads involved in suppression of black root rot of tobacco.Phytopathology76181–185. 10.1094/Phyto-76-181
88
Syed, Ab Rahman S. F. Singh E. Pieterse C. M. J. Schenk P. M. (2018). Emerging microbial biocontrol strategies for plant pathogens.Plant Sci.267102–111. 10.1016/j.plantsci.2017.11.012
89
Takemoto J. Y. Bensaci M. De Lucca A. J. Cleveland T. E. Gandhi N. R. Palmer Skebba V. (2010). Inhibition of fungi from diseased grape by syringomycin E-rhamnolipid mixture.Am. J. Enol. Vitic.61120–124.
90
Tao X. Zhang H. Gao M. Li M. Zhao T. Guan X. (2020). Pseudomonas species isolated via high-throughput screening significantly protect cotton plants against Verticillium wilt.AMB Express10:193. 10.1186/s13568-020-01132-1131
91
Toral L. Rodríguez M. Béjar V. Sampedro I. (2018). Antifungal activity of lipopeptides from Bacillus XT1 CECT 8661 against Botrytis cinerea.Front. Microbiol.9:1315. 10.3389/fmicb.2018.01315
92
Tran V.-T. Braus-Stromeyer S. A. Kusch H. Reusche M. Kaever A. Kühn A. et al (2014). Verticillium transcription activator of adhesion Vta2 suppresses microsclerotia formation and is required for systemic infection of plant roots.New Phytol.202565–581. 10.1111/nph.12671
93
Troppens D. M. Dmitriev R. I. Papkovsky D. B. O’Gara F. Morrissey J. P. (2013). Genome-wide investigation of cellular targets and mode of action of the antifungal bacterial metabolite 2,4-diacetylphloroglucinol in Saccharomyces cerevisiae.FEMS Yeast Res.13322–334. 10.1111/1567-1364.12037
94
Uppal A. K. El Hadrami A. Adam L. R. Tenuta M. Daayf F. (2008). Biological control of potato Verticillium wilt under controlled and field conditions using selected bacterial antagonists and plant extracts.Biol. Control4490–100. 10.1016/j.biocontrol.2007.10.020
95
Weller D. M. Cook R. J. (1983). Suppression of take-all of wheat by seed treatments with fluorescent pseudomonads.Phytopathology73463–469. 10.1094/Phyto-73-463
96
Wilhelm S. (1955). Longevity of the Verticillium wilt fungus in the laboratory and field.Phytopathology45180–181.
97
Yates A. D. Achuthan P. Akanni W. Allen J. Allen J. Alvarez-Jarreta J. et al (2020). Ensembl 2020.Nucleic Acids Res.48D682–D688. 10.1093/nar/gkz966
98
Yu J. Li T. Tian L. Tang C. Klosterman S. J. Tian C. et al (2019). Two Verticillium dahliae MAPKKKs, VdSsk2 and VdSte11, have distinct roles in pathogenicity, microsclerotial formation, and stress adaptation.mSphere4:e00426-19. 10.1128/mSphere.00426-419
99
Zhang X. Cheng W. Feng Z. Zhu Q. Sun Y. Li Y. et al (2020). Transcriptomic analysis of gene expression of Verticillium dahliae upon treatment of the cotton root exudates.BMC Genomics21:155. 10.1186/s12864-020-6448-6449
100
Zheng X. Koopmann B. Ulber B. von Tiedemann A. (2020). A global survey on diseases and pests in oilseed rape—current challenges and innovative strategies of control.Front. Agron.2:590908. 10.3389/fagro.2020.590908
101
Zuber S. Carruthers F. Keel C. Mattart A. Blumer C. Pessi G. et al (2003). GacS sensor domains pertinent to the regulation of exoproduct formation and to the biocontrol potential of Pseudomonas fluorescens CHA0.Mol. Plant-Microbe Interact.16634–644. 10.1094/MPMI.2003.16.7.634
Summary
Keywords
Verticillium dahliae , Verticillium longisporum , fluorescent pseudomonads, plant pathogen, fungal growth inhibition, bacterial-fungal interaction, microfluidic device
Citation
Harting R, Nagel A, Nesemann K, Höfer AM, Bastakis E, Kusch H, Stanley CE, Stöckli M, Kaever A, Hoff KJ, Stanke M, deMello AJ, Künzler M, Haney CH, Braus-Stromeyer SA and Braus GH (2021) Pseudomonas Strains Induce Transcriptional and Morphological Changes and Reduce Root Colonization of Verticillium spp.. Front. Microbiol. 12:652468. doi: 10.3389/fmicb.2021.652468
Received
12 January 2021
Accepted
26 April 2021
Published
24 May 2021
Volume
12 - 2021
Edited by
Georgios Tzelepis, Swedish University of Agricultural Sciences, Sweden
Reviewed by
Sotiris Tjamos, Agricultural University of Athens, Greece; Claudio Valverde, National University of Quilmes, Argentina
Updates
Copyright
© 2021 Harting, Nagel, Nesemann, Höfer, Bastakis, Kusch, Stanley, Stöckli, Kaever, Hoff, Stanke, deMello, Künzler, Haney, Braus-Stromeyer and Braus.
This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gerhard H. Braus, gbraus@gwdg.de
†These authors have contributed equally to this work
This article was submitted to Microbe and Virus Interactions with Plants, a section of the journal Frontiers in Microbiology
Disclaimer
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.