- 1Food Microbiology, Wageningen University and Research, Wageningen, Netherlands
- 2Laboratory of Biochemistry, Wageningen University and Research, Wageningen, Netherlands
Bacterial microcompartments (BMCs) are proteinaceous prokaryotic organelles that enable the utilization of substrates such as 1,2-propanediol and ethanolamine. BMCs are mostly linked to the survival of particular pathogenic bacteria by providing a growth advantage through utilization of 1,2-propanediol and ethanolamine which are abundantly present in the human gut. Although a 1,2-propanediol utilization cluster was found in the probiotic bacterium Propionibacterium freudenreichii, BMC-mediated metabolism of 1,2-propanediol has not been demonstrated experimentally in P. freudenreichii. In this study we show that P. freudenreichii DSM 20271 metabolizes 1,2-propanediol in anaerobic conditions to propionate and 1-propanol. Furthermore, 1,2-propanediol induced the formation of BMCs, which were visualized by transmission electron microscopy and resembled BMCs found in other bacteria. Proteomic analysis of 1,2-propanediol grown cells compared to L-lactate grown cells showed significant upregulation of proteins involved in propanediol-utilization (pdu-cluster), DNA repair mechanisms and BMC shell proteins while proteins involved in oxidative phosphorylation were down-regulated. 1,2-Propanediol utilizing cells actively produced vitamin B12 (cobalamin) in similar amounts as cells growing on L-lactate. The ability to metabolize 1,2-propanediol may have implications for human gut colonization and modulation, and can potentially aid in delivering propionate and vitamin B12 in situ.
Introduction
Propionibacterium freudenreichii is a Gram-positive, non-spore forming bacterium which has been linked to several potential health promoting effects, such as reducing intestinal inflammation, immunomodulation, modulation of intestinal motility and absorption, reduction of pathogen adhesion and enhancement of bifidobacteria [reviewed by Cousin et al. (2011)]. P. freudenreichii is able to cope with a large variety of stresses (oxidative, bile salt, and temperature; Falentin et al., 2010) and consequently has good survival capabilities in the upper intestinal tract (Huang and Adams, 2004). Survival of P. freudenreichii in the intestinal environment is supported by expression of pathways involved in the metabolism of substrates present in the intestinal environment, such as propanediol (Saraoui et al., 2013).
1,2-propanediol is a major end product from anaerobic degradation of rhamnose or fucose by the human intestinal microbiota and serves as an important carbon source for 1,2-propanediol utilizing bacteria, which can metabolize 1,2-propanediol into propionate, generating ATP, and into 1-propanol for maintaining redox balance. The metabolism of 1,2-propanediol produces the toxic intermediate propionaldehyde (Sampson and Bobik, 2008). Some bacteria can protect themselves from toxic intermediates by encapsulating the enzymatic processes in self-assembling proteinaceous organelles called bacterial microcompartments (BMCs; Axen et al., 2014). BMCs are typically about 40–200 nm in diameter and are made of three types of shell proteins: hexamers, pseudohexamers, and pentamers (Kerfeld et al., 2018). Based on the highly conserved domain of shell proteins, Axen et al. (2014) predicted the presence of these organelles in 23 different bacterial phyla, including Actinobacteria which includes the species P. freudenreichii. However, no experimental evidence of BMC mediated utilization of 1,2-propanediol has been shown in P. freudenreichii and its role as carbon source is yet to be elucidated.
Bacterial microcompartment-mediated catabolism of substrates involving a toxic aldehyde intermediate is driven by three core enzymes: aldehyde dehydrogenase, alcohol dehydrogenase, and phosphotransacylase (Axen et al., 2014). The signature enzyme in the propanediol utilization (pdu) pathway is propanediol dehydratase, a vitamin B12-dependent enzyme catalyzing the reaction of 1,2-propanediol to propionaldehyde (Cheng et al., 2011; Axen et al., 2014). Vitamin B12 is produced in high quantities by P. freudenreichii (Burgess et al., 2009) and it also requires vitamin B12 as a cofactor for a key enzyme in its characteristic Wood–Werkman cycle. It is suggested that Actinobacteria such as P. freudenreichii obtained the pdu cluster through horizontal gene transfer from Clostridiales (Ravcheev et al., 2019), in which acquisition may have been supported by the ability of P. freudenreichii to produce vitamin B12 de-novo (Roessner et al., 2002). BMCs are also found in some pathogenic bacteria, such as Salmonella enterica, Enterococcus faecalis, Listeria monocytogenes, pathogenic Escherichia coli, and Clostridium perfringens (Kerfeld et al., 2018). BMC-mediated utilization of 1,2-propanediol increases competitive fitness of pathogens in the gut and consequently has been linked to virulence (Jakobson and Tullman-Ercek, 2016). However, symbiotic relationships depending on 1,2-propanediol metabolism have also been shown for beneficial Lactobacillus reuteri and Bifidobacterium breve (Cheng et al., 2020). The ability to degrade 1,2-propanediol may have similar implications for the bifidogenic capacity reported for P. freudenreichii (Kaneko et al., 1994) and consequences for gut modulation and competition with pathogenic bacteria. Furthermore, the active production of vitamin B12 during metabolism of 1,2-propanediol has not been studied yet in P. freudenreichii.
In this study we present evidence for BMC-mediated anaerobic growth of P. freudenreichii on 1,2-propanediol, evidenced by substrate utilization, propionate and 1-propanol production, and vitamin B12 synthesis. Using transmission electron microscopy (TEM) and proteomics, we confirmed the presence of BMCs, Pdu BMC shell proteins and enzymes in pdu-induced P. freudenreichii.
Materials and Methods
Strains, Culture Conditions, and Growth Measurement
Propionibacterium freudenreichii DSM 20271 was grown anaerobically (Anoxomat modified atmosphere, MART; 10% CO2, 5% H2, and 85% N2) at 30°C in 50 mL tubes containing 40 mL complex media containing per liter: 10 g tryptone, 5 g yeast extract, and 5 g KH2PO4 supplemented with 100 mM L-lactate or 1,2-propanediol. All media was set at pH 7.0 by addition of 5 M NaOH and was filter sterilized through 0.2 μm filters into sterile flasks. OD600 measurements and extracellular metabolite samples were taken daily for a time period of 7 days.
Analysis of Extracellular Metabolites Using High Performance Liquid Chromatography
Culture samples were taken at various time intervals and were analyzed for extracellular metabolites by high performance liquid chromatography. 1 mL culture was centrifuged at 17,000 × g for 1 min and the supernatant was collected. 0.5 mL supernatant was treated with 0.25 mL Carrez A and 0.25 mL Carrez B, vortexed and centrifuged at 17,000 × g for 2 min. 200 μL supernatant was stored in HPLC vials at −20°C upon analysis. HPLC was performed as described by Zeng et al. (2019). Quantification was performed by addition of a standard curve containing L-lactate, acetate, propionate, 1,2-propanediol, and 1-propanol.
Transmission Electron Microscopy
Propionibacterium freudenreichii cultures were grown anaerobically at 30°C in 100 mM L-lactate or 1,2-propanediol containing media. Samples were collected after 6 days of incubation (early stationary phase for 1,2-propanediol-grown cells). About 10 μg of dry cell biomass was fixed for 2 h in 2.5% (v/v) glutaraldehyde in 0.1 M sodium cacodylate buffer (pH 7.2). After rinsing in the same buffer, a post-fixation was done in 1% (w/v) OsO4 for 1 h at room temperature. The samples were dehydrated using ethanol. The dehydrated cell pellets were then embedded in resin (Spurr HM20) for 10 h at 70°C. Thin sections (<100 nm) of polymerized resin samples were obtained with microtomes. After staining with 2% (w/v) aqueous uranyl acetate, the samples were analyzed with a Jeol 1400 plus TEM with 120 kV setting as described by Zeng et al. (2019).
Vitamin B12 Quantification
After 7 days of incubation the vitamin B12 (cobalamin) concentration was determined using a microbiological assay (Vitafast vitamin B12 kit, R-biopharm) for P. freudenreichii grown in 66 mM L-lactate and 49 mM 1,2-propanediol medium. Briefly, 1 mL of culture was disrupted by bead beating (lysing matrix B, mp-bio) 2× 1 min at 4.5 m/s with 1 min on ice in between. Samples were centrifuged and diluted with water to appropriate concentrations for the test kit and were heat-extracted for 30 min at 95°C. The samples were cooled to room temperature and vitamin B12 detection was performed according to the manufacturer protocol.
Proteomics
Propionibacterium freudenreichii cells were cultured in media supplemented with 100 mM L-lactate or 100 mM 1,2-propanediol for 7 days. Cell pellets were harvested by centrifugation of 1 mL of sample at 17,000 × g for 1 min in table top centrifuges and cell pellets were frozen at -80 degrees. Samples were washed twice with 100 mM Tris (pH 8) and resuspended in 100 μl 100 mM Tris. Samples were lysed by sonication for 45 s twice while cooling 1 min on ice. Protein content was determined using Pierce Coomassie protein assay and samples were diluted to 1 μg/μl using Tris–HCl pH 8. Samples were prepared according to the filter assisted sample preparation protocol (FASP; Wiśniewski et al., 2009) with the following steps: reduction with 15 mM dithiothreitol, alkylation with 20 mM acrylamide, and digestion with sequencing grade trypsin overnight. Each prepared peptide sample was analyzed by injecting (18 μl) into a nanoLC-MS/MS (Thermo nLC1000 connected to a LTQ-Orbitrap XL) as described previously (Lu et al., 2011). LCMS data with all MS/MS spectra were analyzed with the MaxQuant quantitative proteomics software package (Cox et al., 2014) as described before (Smaczniak et al., 2012; Wendrich et al., 2017).
A protein database with the protein sequence of P. freudenreichii DSM 20271 (ID:UP000032238) was downloaded from UniProt. Filtering and further bioinformatics and statistical analysis of the MaxQuant ProteinGroups file were performed with Perseus (Tyanova et al., 2016). Reverse hits and contaminants were filtered out. Protein groups were filtered to contain minimally two peptides for protein identification of which at least one is unique and at least one is unmodified. Also, each group required three valid values in at least one of the two experimental groups. A volcano plot was prepared based on the Student’s t-test difference between samples. Volcano plots were produced in Rstudio using EnhancedVolcano (Blighe, 2018). Proteins were considered to be significantly different amongst sample if p < 0.05 and 4-fold change difference was detected. KEGG gene set enrichment analysis was performed using Clusterprofiler (Yu et al., 2012) and 2-fold change difference amongst proteins. The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium via the PRIDE (Vizcaíno et al., 2016) partner repository with the dataset identifier PXD024700.
Predicting BMCs Shell Proteins
The Hidden Markov Models (HMMs) of two BMC shell protein domains listed as Pf00936 and Pf03319 were retrieved from the Pfam database to predict BMC shell proteins as described in Axen et al., 2014; Zeng et al., 2019. Shell proteins were predicted by a HMM search using the HMMER package and a local protein database of P. freudenreichii DSM 20271 genome [CP010341.1 (Deptula et al., 2017)]. All hits with an e-value less than or equal to 1e-05 that correspond to a genomic record from Genbank, RefSeq, EMBL, or DDBJ databases were accepted as BMC shell protein homologs (Supplementary File 1).
Results
Growth Performance and Metabolite Production on 1,2-Propanediol and L-Lactate
Anaerobic growth of P. freudenreichii on 100 mM 1,2-propanediol was monitored for 6 days with daily sampling in triplicate. As a control, P. freudenreichii was cultured on 100 mM L-lactate. P. freudenreichii was able to completely metabolize 100 mM 1,2-propanediol during a period of 6 days (see Figure 1). This resulted in the production of 37.5 ± 0.1 mM 1-propanol and 43.2 ± 0.5 mM propionate. No production of acetate was observed, and with the current HPLC method used, approximately 28 mM of expected C3 compounds is missing, conceivably volatile propionaldehyde. In the control cultures, 100 mM L-lactate was consumed within 2 days, resulting in 65.1 + 0.1 mM propionate and 30.5 ± 0.1 mM acetate, close the expected molar ratio of 2:1 (Seeliger et al., 2002). Growth on 1,2-propanediol resulted in slower growth, with a stationary phase after 120 h vs 48 h in L-lactate. Our results clearly demonstrate that P. freudenreichii can grow on 1,2-propanediol, metabolizing it to propionate and 1-propanol.
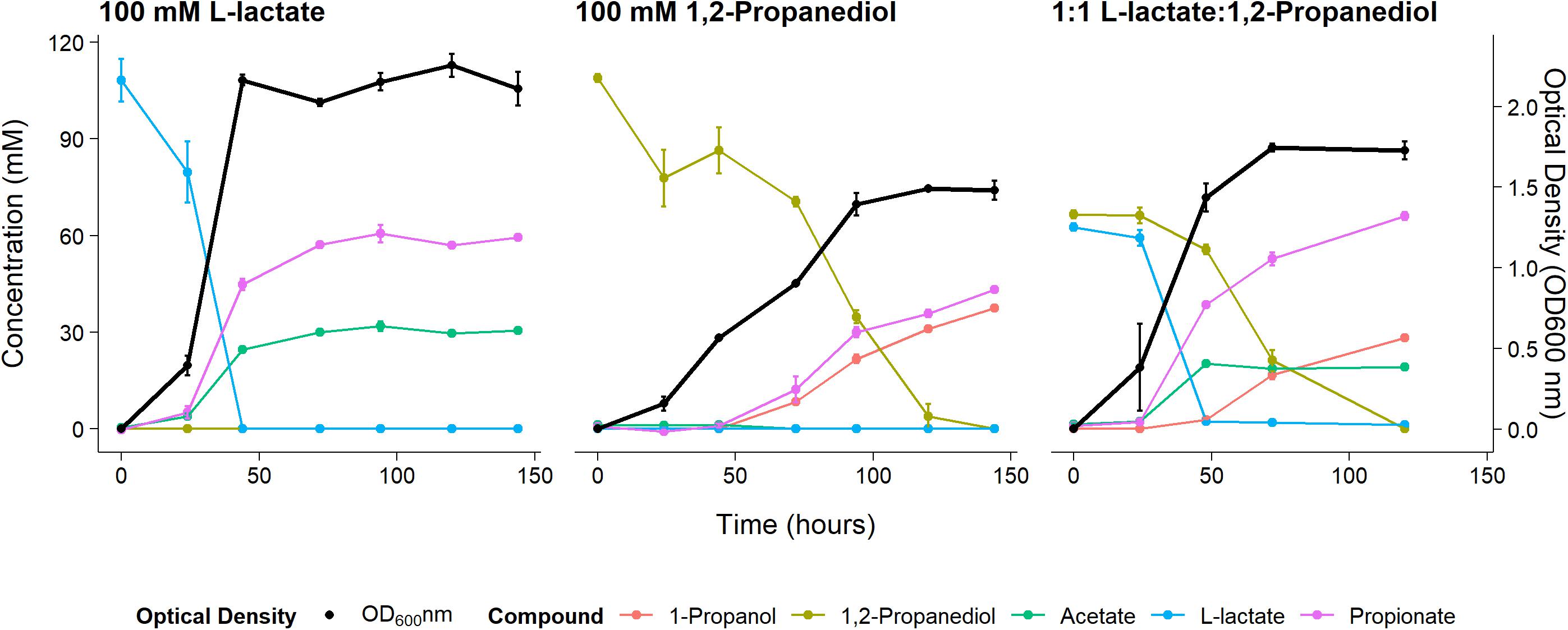
Figure 1. Substrate consumption, metabolite production and biomass formation of P. freudenreichii grown on 100 mM L-lactate, 100 mM 1,2-Propanediol, and 1:1 1,2-Propanediol:L-lactate for 6 days. Error bars = standard error (n = 3 biological replicates).
To monitor metabolism of 1,2-propanediol in the presence of other carbon sources P. freudenreichii was grown in media containing both L-lactate and 1,2-propanediol. When NAD+/NADH pools are shared between the cytosol and the BMC as discussed by Ferlez et al. (2019), the highest ATP yield from 1,2-propanediol in the presence of L-lactate can be obtained by co-fermenting 1,2-propanediol and L-lactate solely to propionate (see Supplementary Text 1). The expected ATP yield with L-lactate and 1,2-propanediol as mixed substrates is higher compared to cells growing solely on L-lactate (see Supplementary Text 1). We found production of 67.1 ± 1.5 mM propionate, 19.0 ± 1.0 mM of acetate, and 28.3 ± 2.8 mM 1-propanol in cells growing on the mixed substrates. Assuming 1-propanol:propionate is produced in a 1:1 ratio from 1,2-propanediol and acetate:propionate is produced in a 1:2 ratio from L-lactate, 28 mM of the propionate originates from the 1,2-propanediol metabolism and 38 mM propionate originates from L-lactate metabolism. This matches the total found propionate of 67.1 ± 1.5 mM propionate in the samples. In mixed substrate conditions the amounts of total products formed indicate no apparent loss of C3 compounds, which may indicate reduced loss of volatile propionaldehyde. Biomass formation with 1:1 L-lactate:1,2-propanediol-grown cells was found to be lower compared to 100 mM L-lactate-grown cells. These results indicate no apparent energetic benefit from mixed substrate conditions compared to mono substrate conditions, pointing toward independent pathways. Our results show mixed-substrate metabolism influences the total amount of short-chain fatty acid production, but pathway interactions are not apparent.
Vitamin B12 Production
To demonstrate active vitamin B12 production under 1,2-propanediol utilizing conditions, P. freudenreichii was grown for 7 days in either yeast extract medium supplemented with 66 mM L-lactate or 49 mM 1,2-propanediol. Biomass formation, substrate utilization, and vitamin B12 production by P. freudenreichii was monitored after 7 days of incubation. Incubation vessels were not opened in-between to prevent side effects by oxygen-dependent stimulation of vitamin B12 production, as has been shown for P. freudenreichii (Quesada-Chanto et al., 1998). Again, complete utilization of L-lactate and 1,2-propanediol was found (Supplementary File 2).
Vitamin B12 was produced both when grown on L-lactate as on 1,2-propanediol (1.84 ± 0.11 μg/mmol L-lactate vs 2.19 ± 0.03 μg/mmol 1,2-propanediol). The specific vitamin B12 production when grown on L-lactate and on 1,2-propanediol was 270 ± 33 μg/g cells and 288 ± 27 μg/g cells, respectively, (Figure 2). No significant difference was thus found for the production of vitamin B12 per g cells when growing on either L-lactate supplemented media or 1,2-propanediol supplemented media. In complex media, 1,2-propanediol as carbon source thus supports vitamin B12 production by P. freudenreichii.
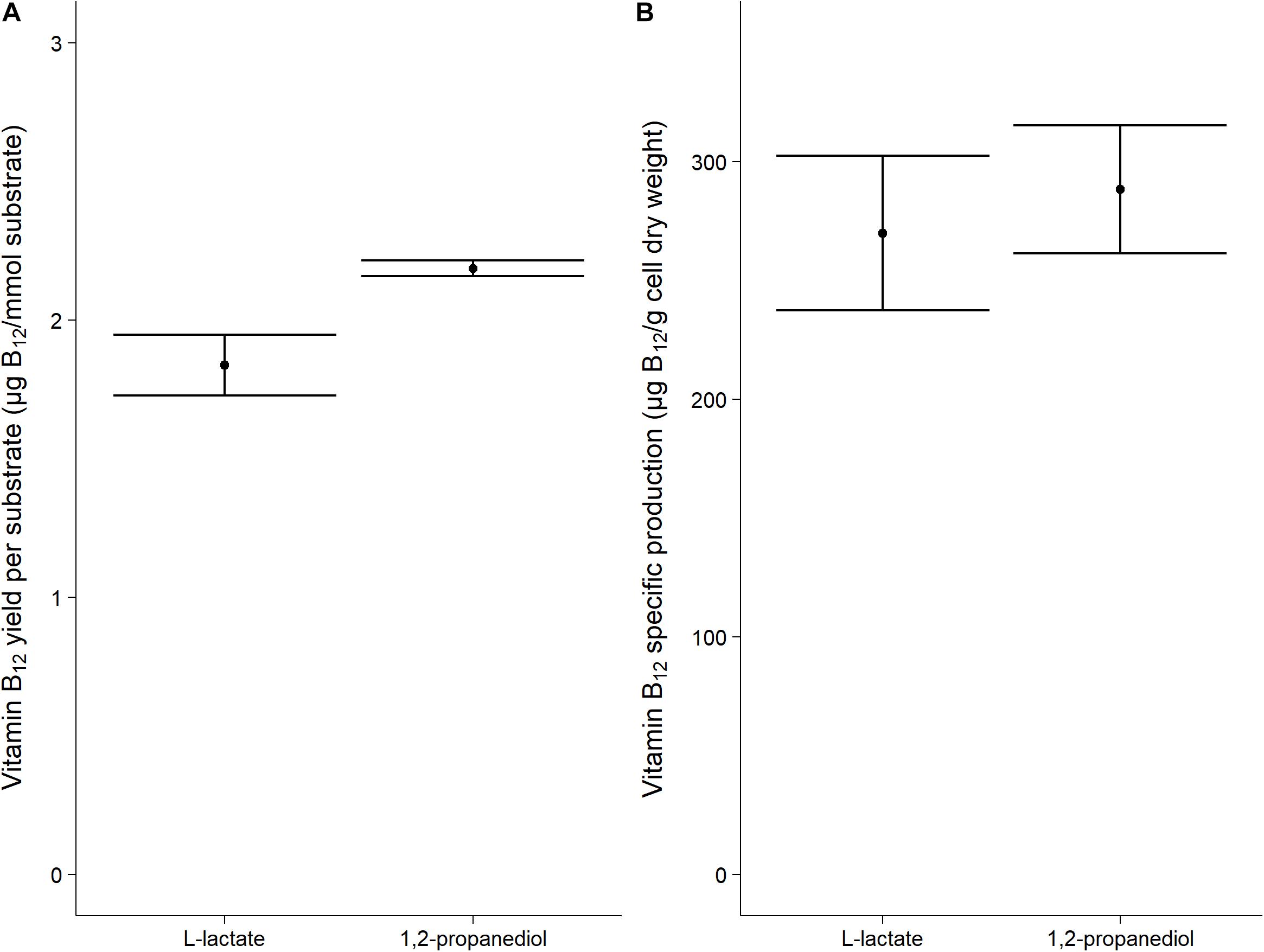
Figure 2. Vitamin B12 formation in cells grown on L-lactate or 1,2-propanediol. (A) vitamin B12 yield in μg/mmol substrate (B) Biomass specific vitamin B12 production in μg/g cell dry weight. Error bars = standard error (n = 3 biological replicates).
Proteomics and Electron Microscopy of L-Lactate and 1,2-Propanediol Grown Cells
Cells growing in media supplemented with L-lactate (control) and with 1,2-propanediol were visualized using TEM. Thin sections of cells grown on 1,2-propanediol supplemented media clearly display cellular structures which were not found in the cells grown on L-lactate supplemented media (see Figure 3), and those structures resemble BMC structures found in other bacteria including L. monocytogenes (Zeng et al., 2019), S. enterica (Crowley et al., 2008), and E. coli (Toraya et al., 1979).
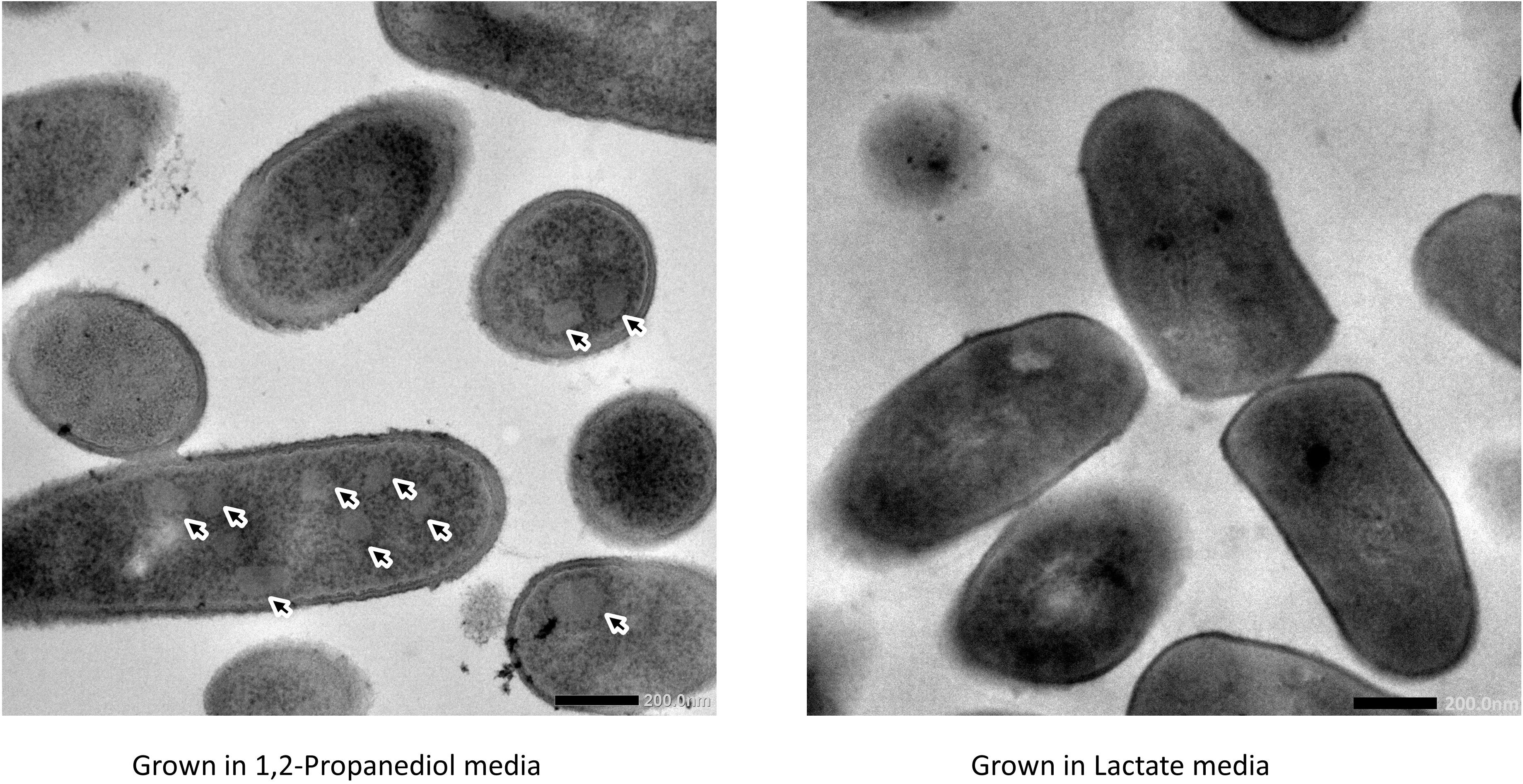
Figure 3. Visualization of bacterial microcompartments in propanediol media (left) and absence in lactate media (right).
Proteome analysis revealed that BMC structural shell proteins (PduA, PduB, PduK, PduJ, PduN, and PduM) and enzymes involved in 1,2-propanediol utilization (PduL, PduC, PduD, PduE, PduP, PduO, and PduQ) were significantly more abundant in cells grown with 1,2-propanediol as a substrate compared to cells grown on L-lactate (Figure 4). Lactate permease, succinate dehydrogenase subunits A, C1 (SdhA, SdhC1) were found to be significantly more abundant in the L-lactate-grown cells compared to 1,2-propanediol-grown cells, indicating a decrease of proteins in lactate-degradation pathways such as the Wood-Werkman cycle. Gene set enrichment analysis of KEGG pathways (see Supplementary File 3) revealed upregulation of the propanediol degradation pathway in 1,2-propanediol grown cells compared to L-lactate grown cells (adjusted p-value < 0.10). DNA repair mechanisms were also found to be activated in 1,2-propanediol-grown cells (adjusted p-value < 0.10). In 1,2-propanediol-grown cells significant suppression of oxidative phosphorylation was found compared to L-lactate grown cells (adjusted p-value < 0.01). The measured consumption of 1,2-propanediol, production of 1-propanol and propionate, visualized cellular structures, and strong upregulation of the pdu loci proteins provide evidence for activation of BMC-dependent 1,2-pdu in P. freudenreichii DSM 20271.
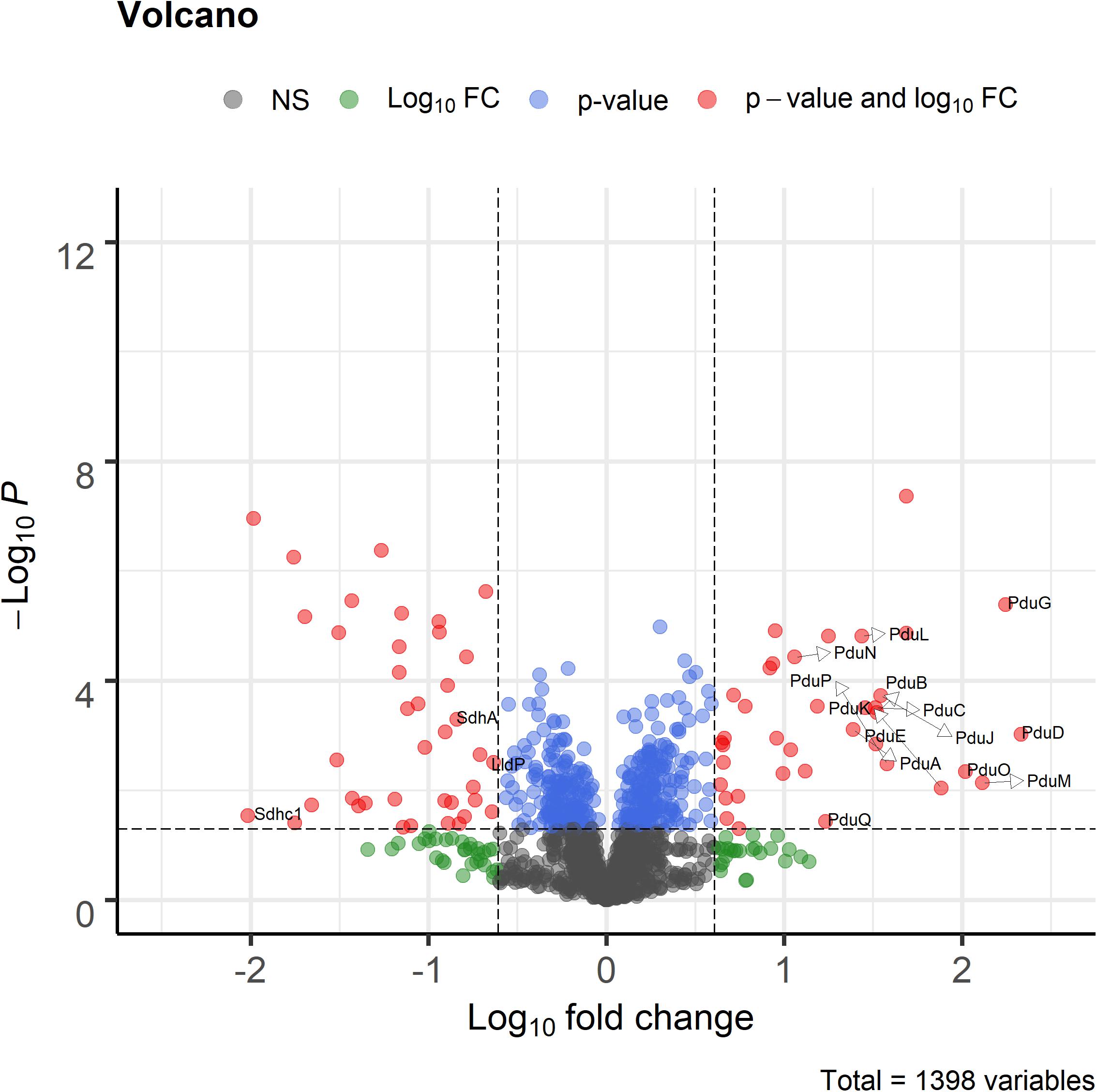
Figure 4. Volcano plot of proteomic analysis of cells grown in 1,2-propanediol media and L-lactate media. Positive log10 fold change indicate upregulation in 1,2-propanediol-grown cells, negative log10 fold change indicate down regulation in 1,2-propanediol-grown cells compared to lactate-grown cells. Red dots indicate proteins with p < 0.05 and 4-fold difference expression. Blue dots indicate only p < 0.05, green dots only 4-fold difference. Black dots indicate non-significant, non-differentially expressed proteins (n = 3 biological replicates).
Overview of BMC-Dependent 1,2-Propanediol Metabolism Model in P. freudenreichii
Subsequent analysis of the pdu cluster in P. freudenreichii DSM 20271 identified two distant loci, with locus 1 starting from RM25_0852 to RM25_0857 and locus 2 starting from RM25_1258 to RM25_1273 (Figure 5A). Locus 1 contains four genes which are pocR encoding a transcriptional regulator, pduQ encoding 1-propanol dehydrogenase, pduV with unknown function and pduU encoding BMC shell protein. Locus 2 carries 14 genes including 6 genes encoding BMC shell proteins and 8 genes encoding enzymes for the 1,2-propanediol degradation pathway and is not preceded by any known transcriptional regulators.
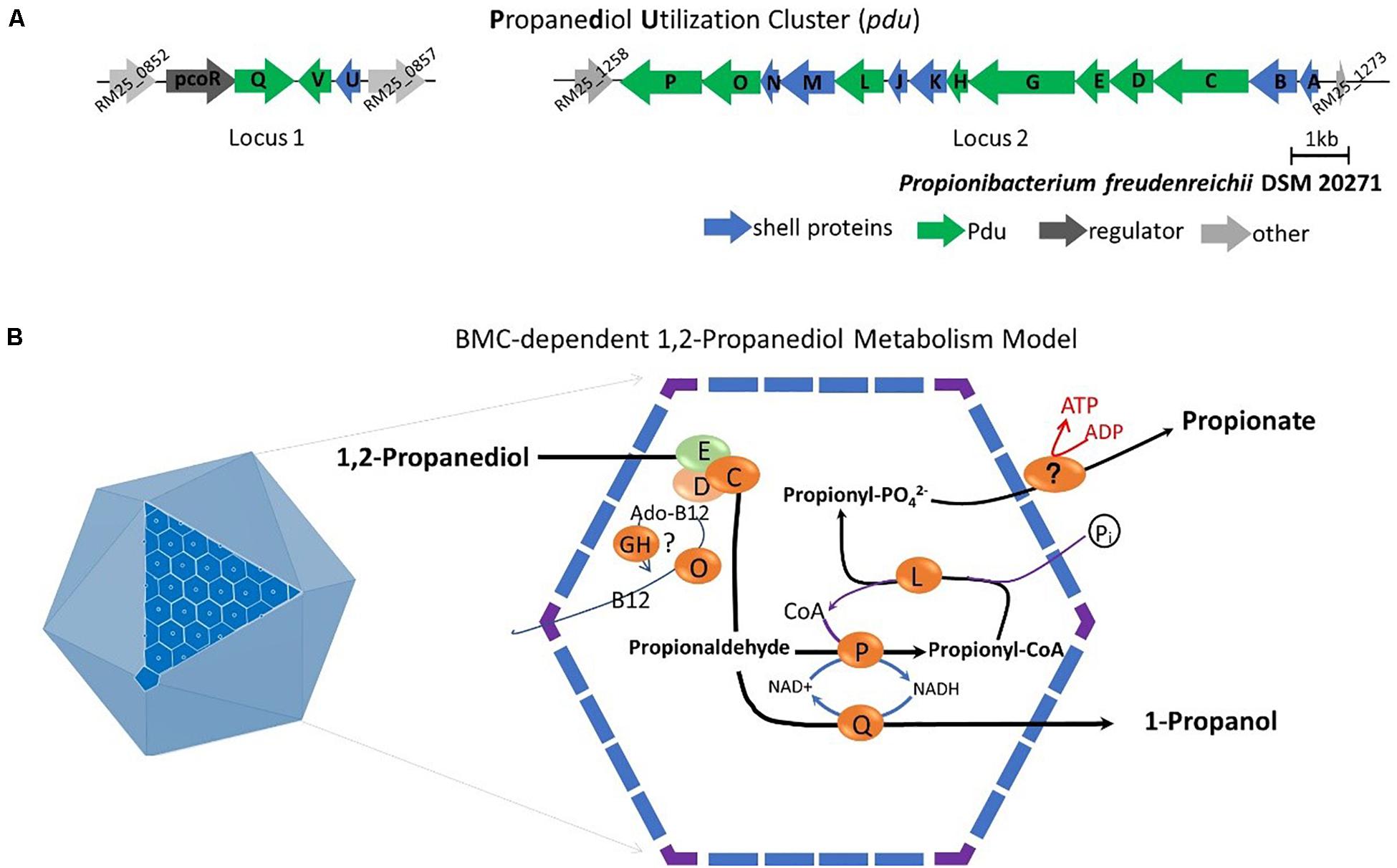
Figure 5. (A) Analysis of the pdu gene cluster (Details in Supplementary File 1). Characters in orange represent Pdu enzymes, in blue represent BMC shell proteins, in dark represent regulator and in gray represent unannotated proteins with gene ID. (B) Model of BMC-dependent 1,2-propanediol metabolism. In the left, the icosahedral diagram represent BMC with one surface showing the assembly of shell proteins. In the right, the metabolic pathway of 1,2-propanediol metabolism. PduCDE, B12-dependent diol dehydratase; PduP, CoA-dependent propionaldehyde dehydrogenase; PduGH, diol dehydratase reactivase; PduO, corrinoid adenosyltransferase; PduS, Vitamin B12 reductase; PduL, phosphate propanoyltransferase; PduW, propionate kinase; and PduQ, propanol dehydrogenase. See text for details.
Based on the structural studies of BMCs (Sutter et al., 2017; Greening and Lithgow, 2020) and our understanding of 1,2-propanediol metabolism (Sampson and Bobik, 2008; Kerfeld et al., 2018; Zeng et al., 2019), we propose the model of BMC-dependent 1,2-propanediol metabolism in P. freudenreichii, with the predicted BMC shell proteins PduA, PduB, PduK, PduJ, PduM, PduN, and PduU constituting the self-organized icosahedral organelle (Kerfeld et al., 2018; Greening and Lithgow, 2020). As illustrated in Figure 5B, the catabolism of 1,2-propanediol starts with the conversion of 1,2-propanediol to propionaldehyde by vitamin B12-dependent diol dehydratase PduCDE. The toxic propionaldehyde is then converted to propionate by the enzyme CoA-dependent propionaldehyde dehydrogenase PduP, followed by action of phosphate propanoyltransferase PduL, and potential propionate kinase located in the cytoplasm, resulting in the end product propionate and the production of ATP. The other end product is produced following conversion of propionaldehyde by propanol dehydrogenase PduQ into 1-propanol. The diol dehydratase reactivase PduGH and corrinoid adenosyltransferase PduO are linked to the supply and recycling of vitamin B12.
Discussion
Propionibacterium freudenreichii is commonly found in the rumen and colon of animals and in the human intestine (Bryant, 1959). In these environments fucose and rhamnose are degraded to 1,2-propanediol by the present microbiota (Xue et al., 2008). We showed 1,2-propanediol can be further metabolized into propionate and 1-propanol by P. freudenreichii, thereby supporting anaerobic growth. Previously, a locus containing 15 genes involved in pdu was detected in P. freudenreichii (Falentin et al., 2010). In vivo gene expression analysis showed the pdu operon to be expressed in P. freudenreichii cells contained in the colon environment of a pig (Saraoui et al., 2013), pointing toward pdu in intestinal environments. Interestingly, in this study we identified the pdu cluster distributed in two different loci in P. freudenreichii DSM 20271. The presence of the pdu cluster seems to be species specific in propionic acid bacteria, as blasting the pdu cluster of P. freudenreichii against Cutibacterium acnes, Acidipropionibacterium acidipropionici, A. thoenii, and A. jensenii did not result significant hits for key components in the cluster (see Supplementary File 4). Presence of 1,2-propanediol in the medium induced expression of the two loci in P. freudenreichii DSM 20271 and resulted in BMC formation, 1,2-propanediol metabolism and consequently propionate and 1-propanol production. Our results show that expression of the pdu cluster from two different loci results in effective BMC formation and 1,2-propanediol metabolism. Indeed, BMC genes are split into two or more loci in 40% of the prokaryotic genomes containing BMCs (Abdul-Rahman et al., 2013). In P. freudenreichii DSM 20271, locus 1 is preceded by transcriptional activator pocR, which has been linked to activation of the pdu cluster in Salmonella (Chen et al., 1994). We did not find any annotated transcriptional regulator in the vicinity of locus 2. The presence of two different loci in many prokaryotic genomes suggests expression is controlled by additional regulators next to PocR. Indeed heterologous expression of BMCs uncoupled from their cognate transcriptional regulators has been reported previously (Wilson, 2021). The transcriptional regulation and activation of the two pdu loci and the role of pocR in P. freudenreichii requires further attention.
Upregulation of the pdu cluster and DNA repair mechanisms clearly indicated the crucial role of BMCs to protect P. freudenreichii from the toxic intermediate propionaldehyde produced in the degradation pathway. The ability to utilize substrates producing toxic intermediates upon degradation results in a competitive advantage to other gut microbiota (Jakobson and Tullman-Ercek, 2016), as shown for ethanolamine utilization by S. enterica during intestinal inflammation (Thiennimitr et al., 2011). Interestingly, the presence of genes encoding metabolosomes for the utilization of ethanolamine and propanediol has been linked to pathogenicity and aids in anaerobic growth and colonization of foodborne pathogens L. monocytogenes, C. perfringens, and S. typhimurium (Korbel et al., 2005; De Weirdt et al., 2012). It has been suggested for beneficial bacterium L. reuteri that competition for 1,2-propanediol could result in decreased proliferation of pathogens (Cheng et al., 2020). P. freudenreichii is considered to be non-pathogenic and has the generally recognized as safe (GRAS) status (Meile et al., 2008). Substrate competition for 1,2-propanediol by P. freudenreichii may thus exert similar effects as suggested for L. reuteri on growth of pathogenic bacteria. This is in line with reports that P. freudenreichii can decrease adhesion of pathogens to human intestinal mucus cells (Collado et al., 2007). Furthermore, P. freudenreichii stimulates the growth of beneficial bifidobacteria (Satomi et al., 1999; Hojo et al., 2002) thereby promoting a healthy gut microbiota. The role of BMC-mediated 1,2-propanediol utilization by P. freudenreichii and its importance for modulating gut microbiota composition, both by substrate competition and promoting other beneficial microbiota, requires further investigation.
Our study shows the potential of P. freudenreichii to substantially contribute to the production of propionate in the human gut. Next to 1,2-propanediol, also lactic acid (Macfarlane and Gibson, 1997) is a major end-product of microbial fermentation, which consequently can also be fermented by P. freudenreichii and additionally contributes to the production of propionate. In mixed substrate conditions both pathways remained active, albeit without apparent interaction based on metabolite formation. However, mixed substrate conditions decreased the loss of C3, suggesting less loss of volatile propionaldehyde, thus potentially more efficient BMC assembly. The assembly of BMCs in mixed substrate condition requires further attention. Propionate is linked to many putative health effects [reviewed by (Hosseini et al., 2011)] and can further stimulate bifidobacteria (Kaneko et al., 1994). The production of propionate from 1,2-propanediol further supports the potential of P. freudenreichii as a probiotic.
The metabolism of 1,2-propanediol requires propanediol dehydratase, which is vitamin B12-dependant (Axen et al., 2014). Vitamin B12 is actively produced by P. freudenreichii as it is essential cofactor in a key enzyme in the Wood-Werkman cycle, methylmalonyl-CoA mutase. Synthesis of vitamin B12 in P. freudenreichii follows the anaerobic pathway (Roessner et al., 2002), enabling production of vitamin B12 for anaerobic metabolism of lactate. Here we show vitamin B12 is produced in similar amounts in complex medium when cells metabolize 1,2-propanediol or L-lactate as carbon source. In S. typhimurium pocR mediated expression of vitamin B12 is induced in the presence of propanediol (Richter-Dahlfors et al., 1994). We also identified pocR upstream of pdu loci 1 in P. freudenreichii DSM 20271, but as discussed before its exact regulatory role in P. freudenreichii remains to be elucidated. Vitamin B12 production is also regulated by a vitamin B12 regulated riboswitch in P. freudenreichii. In Propionibacterium strain UT1 expression of vitamin B12 biosynthesis occurred at vitamin B12 concentrations of 750 μM, much higher compared to the vitamin B12 concentrations found in this study (∼90 nM). The role of this riboswitch for vitamin B12 production during metabolism of 1,2-propanediol also remains to be elucidated. Based on our findings, we hypothesize that P. freudenreichii occupies a lactate and propanediol-rich niche in the gut environment. Symbiotic relationships have been shown for the production of vitamin B12 (Belzer et al., 2017; Sokolovskaya et al., 2020), which may be hard-wired in the vitamin B12 production of P. freudenreichii. As next to vitamin B12-dependent lactate metabolism, BMC-mediated 1,2-propanediol metabolism supports anaerobic growth of P. freudenreichii, thereby contributing to in situ vitamin B12 production in the gut.
This study presents evidence for BMC-mediated vitamin B12-dependent utilization of 1,2-propanediol in P. freudenreichii. We have shown that 1,2-propanediol supports anaerobic growth of P. freudenreichii. It is conceivable that utilization of 1,2-propanediol could aid colonization of Propionibacterium freudenreichii in the human gut to exert beneficial effects, such as delivering vitamin B12 and propionate in situ, but these aspects require further study.
Data Availability Statement
The original contributions presented in the study are publicly available. This data can be found here: https://www.ebi.ac.uk/pride/archive/projects/PXD024700.
Author Contributions
AD and ZZ designed and performed the experiments. SB, AD, and ZZ performed proteomics and analyzed data. ZZ, AD, ES, RN, and TA analyzed data. ZZ, AD, ES, and TA wrote the manuscript. All authors read, edited and approved the manuscript.
Funding
The authors declare that this study received funding from Arla Foods (Denmark). The funder was not involved in the study design, collection, analysis, interpretation of data, the writing of this article or the decision to submit it for publication. ZZ was supported by a grant from the China Scholarship Council.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Wageningen Electron Microscopy Centre (WEMC) for the use of their transmission electron microscope and for their help in sample preparation.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.679827/full#supplementary-material
References
Abdul-Rahman, F., Petit, E., and Blanchard, J. L. (2013). The distribution of polyhedral bacterial microcompartments suggests frequent horizontal transfer and operon reassembly. J. Phylog. Evol. Biol. 1:118.
Axen, S. D., Erbilgin, O., and Kerfeld, C. A. (2014). A taxonomy of bacterial microcompartment loci constructed by a novel scoring method. PLoS Comput. Biol. 10:e1003898. doi: 10.1371/journal.pcbi.1003898
Belzer, C., Chia, L. W., Aalvink, S., Chamlagain, B., Piironen, V., Knol, J., et al. (2017). Microbial metabolic networks at the mucus layer lead to diet-independent butyrate and Vitamin B12 production by intestinal symbionts. mBio 8:e00770–17. doi: 10.1128/mBio.00770-17
Blighe, K. (2018). Enhancedvolcano: Publication-Ready Volcano Plots With Enhanced Colouring and Labeling. Vienna: R Foundation for Statistical Computing.
Bryant, M. P. (1959). Bacterial species of the rumen. Bacteriol. Rev. 23, 125–153. doi: 10.1128/br.23.3.125-153.1959
Burgess, C. M., Smid, E. J., and van Sinderen, D. (2009). Bacterial vitamin B2, B11 and B12 overproduction: an overview. Int. J. Food Microbiol. 133, 1–7. doi: 10.1016/j.ijfoodmicro.2009.04.012
Chen, P., Andersson, D. I., and Roth, J. R. (1994). The control region of the pdu/cob regulon in Salmonella typhimurium. J. Bacteriol. 176, 5474–5482. doi: 10.1128/jb.176.17.5474-5482.1994
Cheng, C. C., Duar, R. M., Lin, X., Perez-Munoz, M. E., Tollenaar, S., Oh, J. H., et al. (2020). Ecological importance of cross-feeding of the intermediate metabolite 1,2-propanediol between bacterial gut symbionts. Appl. Environ. Microbiol. 86, e190–e120. doi: 10.1128/AEM.00190-20
Cheng, S., Sinha, S., Fan, C., Liu, Y., and Bobik, T. A. (2011). Genetic analysis of the protein shell of the microcompartments involved in coenzyme B12-dependent 1, 2-propanediol degradation by Salmonella. J. Bacteriol. 193, 1385–1392. doi: 10.1128/jb.01473-10
Collado, M. C., Meriluoto, J., and Salminen, S. (2007). In vitro analysis of probiotic strain combinations to inhibit pathogen adhesion to human intestinal mucus. Food Res. Int. 40, 629–636. doi: 10.1016/j.foodres.2006.11.007
Cousin, F. J., Mater, D. D., Foligné, B., and Jan, G. (2011). Dairy propionibacteria as human probiotics: a review of recent evidence. Dairy Sci. Technol. 91, 1–26.
Cox, J., Hein, M. Y., Luber, C. A., Paron, I., Nagaraj, N., and Mann, M. (2014). Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Prot. 13, 2513–2526. doi: 10.1074/mcp.m113.031591
Crowley, C. S., Sawaya, M. R., Bobik, T. A., and Yeates, T. O. (2008). Structure of the PduU shell protein from the Pdu microcompartment of Salmonella. Structure 16, 1324–1332. doi: 10.1016/j.str.2008.05.013
De Weirdt, R., Crabbe, A., Roos, S., Vollenweider, S., Lacroix, C., van Pijkeren, J. P., et al. (2012). Glycerol supplementation enhances L. reuteri’s protective effect against S. Typhimurium colonization in a 3-D model of colonic epithelium. PLoS One 7:e37116. doi: 10.1371/journal.pone.0037116
Deptula, P., Laine, P. K., Roberts, R. J., Smolander, O.-P., Vihinen, H., Piironen, V., et al. (2017). De novo assembly of genomes from long sequence reads reveals uncharted territories of Propionibacterium freudenreichii. BMC Genomics 18:790. doi: 10.1186/s12864-017-4165-9
Falentin, H., Deutsch, S.-M., Jan, G., Loux, V., Thierry, A., Parayre, S., et al. (2010). The complete genome of Propionibacterium freudenreichii CIRM-BIA1T, a hardy actinobacterium with food and probiotic applications. PLoS One 5:e11748. doi: 10.1371/journal.pone.0011748
Ferlez, B., Sutter, M., and Kerfeld, C. A. (2019). Glycyl radical enzyme-associated microcompartments: redox-replete bacterial organelles. mBio 10, e2327–18.
Greening, C., and Lithgow, T. (2020). Formation and function of bacterial organelles. Nat. Rev. Microbiol. 18, 677–689.
Hojo, K., Yoda, N., Tsuchita, H., Ohtsu, T., Seki, K., Taketomo, N., et al. (2002). Effect of ingested culture of Propionibacterium freudenreichii ET-3 on fecal microflora and stool frequency in healthy females. Biosci. Microflora 21, 115–120. doi: 10.12938/bifidus1996.21.115
Hosseini, E., Grootaert, C., Verstraete, W., and Van de Wiele, T. (2011). Propionate as a health-promoting microbial metabolite in the human gut. Nutr. Rev. 69, 245–258. doi: 10.1111/j.1753-4887.2011.00388.x
Huang, Y., and Adams, M. C. (2004). In vitro assessment of the upper gastrointestinal tolerance of potential probiotic dairy propionibacteria. Int. J. Food Microbiol. 91, 253–260. doi: 10.1016/j.ijfoodmicro.2003.07.001
Jakobson, C. M., and Tullman-Ercek, D. (2016). Dumpster diving in the gut: bacterial microcompartments as part of a host-associated lifestyle. PLoS Pathogens 12:e1005558. doi: 10.1371/journal.ppat.1005558
Kaneko, T., Mori, H., Iwata, M., and Meguro, S. (1994). Growth stimulator for bifidobacteria produced by Propionibacterium freudenreichii and several intestinal bacteria. J. Dairy Sci. 77, 393–404. doi: 10.3168/jds.s0022-0302(94)76965-4
Kerfeld, C. A., Aussignargues, C., Zarzycki, J., Cai, F., and Sutter, M. (2018). Bacterial microcompartments. Nat. Rev. Microbiol. 16, 277–290.
Korbel, J. O., Doerks, T., Jensen, L. J., Perez-Iratxeta, C., Kaczanowski, S., Hooper, S. D., et al. (2005). Systematic association of genes to phenotypes by genome and literature mining. PLoS Biol. 3:e134. doi: 10.1371/journal.pbio.0030134
Lu, J., Boeren, S., De Vries, S., Van Valenberg, H., Vervoort, J., and Hettinga, K. (2011). Filter-aided sample preparation with dimethyl labeling to identify and quantify milk fat globule membrane proteins. J. Prot. 75, 34–43. doi: 10.1016/j.jprot.2011.07.031
Macfarlane, G. T., and Gibson, G. R. (1997). “Carbohydrate fermentation, energy transduction and gas metabolism in the human large intestine,” in Gastrointestinal Microbiology: Gastrointestinal Ecosystems and Fermentations, Vol. 1, eds R. I. Mackie and B. A. White (Boston, MA: Springer US), 269–318. doi: 10.1007/978-1-4615-4111-0_9
Meile, L., Le Blay, G., and Thierry, A. (2008). Safety assessment of dairy microorganisms: propionibacterium and Bifidobacterium. Int. J. Food Microbiol. 126, 316–320. doi: 10.1016/j.ijfoodmicro.2007.08.019
Quesada-Chanto, A., Schmid-Meyer, A., Schroeder, A., Carvalho-Jonas, M., Blanco, I., and Jonas, R. (1998). Effect of oxygen supply on biomass, organic acids and vitamin B12 production by Propionibacterium shermanii. World J. Microbiol. Biotechnol. 14, 843–846.
Ravcheev, D. A., Moussu, L., Smajic, S., and Thiele, I. (2019). Comparative genomic analysis reveals novel microcompartment-associated metabolic pathways in the human gut microbiome. Front. Genet. 10:636. doi: 10.3389/fgene.2019.00636
Richter-Dahlfors, A., Ravnum, S., and Andersson, D. (1994). Vitamin B12 repression of the cob operon in Salmonella typhimurium: translational control of the cbiA gene. Mol. Microbiol. 13, 541–553. doi: 10.1111/j.1365-2958.1994.tb00449.x
Roessner, C. A., Huang, K.-x, Warren, M. J., Raux, E., and Scott, A. I. (2002). Isolation and characterization of 14 additional genes specifying the anaerobic biosynthesis of cobalamin (vitamin B12) in Propionibacterium freudenreichii (P. shermanii) The GenBank accession numbers for the sequences reported in this paper are AY033235, AY033236, U13043 and U51164. Microbiology 148, 1845–1853. doi: 10.1099/00221287-148-6-1845
Sampson, E. M., and Bobik, T. A. (2008). Microcompartments for B12-dependent 1, 2-propanediol degradation provide protection from DNA and cellular damage by a reactive metabolic intermediate. J. Bacteriol. 190, 2966–2971. doi: 10.1128/jb.01925-07
Saraoui, T., Parayre, S., Guernec, G., Loux, V., Montfort, J., Cam, A. L., et al. (2013). A unique in vivo experimental approach reveals metabolic adaptation of the probiotic Propionibacterium freudenreichii to the colon environment. BMC Genomics 14:911. doi: 10.1186/1471-2164-14-911
Satomi, K., Kurihara, H., Isawa, K., Mori, H., and Kaneko, T. (1999). Effects of culture-powder of Propionibacterium freudenreichii ET-3 on fecal microflora of normal adults. Biosci. Microflora 18, 27–30. doi: 10.12938/bifidus1996.18.27
Seeliger, S., Janssen, P. H., and Schink, B. (2002). Energetics and kinetics of lactate fermentation to acetate and propionate via methylmalonyl-CoA or acrylyl-CoA. FEMS Microbiol. Lett. 211, 65–70. doi: 10.1111/j.1574-6968.2002.tb11204.x
Smaczniak, C., Li, N., Boeren, S., America, T., Van Dongen, W., Goerdayal, S. S., et al. (2012). Proteomics-based identification of low-abundance signaling and regulatory protein complexes in native plant tissues. Nat. Protocols 7:2144. doi: 10.1038/nprot.2012.129
Sokolovskaya, O. M., Shelton, A. N., and Taga, M. E. (2020). Sharing vitamins: cobamides unveil microbial interactions. Science 369:eaba0165. doi: 10.1126/science.aba0165
Sutter, M., Greber, B., Aussignargues, C., and Kerfeld, C. A. (2017). Assembly principles and structure of a 6.5-MDa bacterial microcompartment shell. Science 356, 1293–1297. doi: 10.1126/science.aan3289
Thiennimitr, P., Winter, S. E., Winter, M. G., Xavier, M. N., Tolstikov, V., Huseby, D. L., et al. (2011). Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc. Natl. Acad. Sci. U.S.A. 108, 17480–17485. doi: 10.1073/pnas.1107857108
Toraya, T., Honda, S., and Fukui, S. (1979). Fermentation of 1, 2-propanediol and 1, 2-ethanediol by some genera of Enterobacteriaceae, involving coenzyme B12-dependent diol dehydratase. J. Bacteriol. 139, 39–47. doi: 10.1128/jb.139.1.39-47.1979
Tyanova, S., Temu, T., Sinitcyn, P., Carlson, A., Hein, M. Y., Geiger, T., et al. (2016). The Perseus computational platform for comprehensive analysis of (prote)omics data. Nat. Methods 13, 731–740. doi: 10.1038/nmeth.3901
Vizcaíno, J. A., Csordas, A., del-Toro, N., Dianes, J. A., Griss, J., Lavidas, I., et al. (2016). 2016 update of the PRIDE database and related tools. Nucleic Acids Res. 44(D1), D447–D456.
Wendrich, J. R., Boeren, S., Möller, B. K., Weijers, D., and De Rybel, B. (2017). In Vivo Identification of Plant Protein Complexes Using IP-MS/MS Plant Hormones. Berlin: Springer, 147–158.
Wilson, J. W. (2021). Manipulating microcompartment operons to study mechanism and function. Curr. Opin. Microbiol. 60, 66–72. doi: 10.1016/j.mib.2021.01.014
Wiśniewski, J. R., Zougman, A., Nagaraj, N., and Mann, M. (2009). Universal sample preparation method for proteome analysis. Nat. Methods 6:359. doi: 10.1038/nmeth.1322
Xue, J., Murrieta, C. M., Rule, D. C., and Miller, K. W. (2008). Exogenous or L-rhamnose-derived 1, 2-propanediol is metabolized via a pduD-dependent pathway in Listeria innocua. Appl. Environ. Microbiol. 74, 7073–7079. doi: 10.1128/aem.01074-08
Yu, G., Wang, L.-G., Han, Y., and He, Q.-Y. (2012). clusterProfiler: an R package for comparing biological themes among gene clusters. Omics J. Integr. Biol. 16, 284–287. doi: 10.1089/omi.2011.0118
Keywords: bacterial microcompartments, Propionibacterium freudenreichii, vitamin B12, metabolosome, propanediol utilization cluster, probiotic, intestine
Citation: Dank A, Zeng Z, Boeren S, Notebaart RA, Smid EJ and Abee T (2021) Bacterial Microcompartment-Dependent 1,2-Propanediol Utilization of Propionibacterium freudenreichii. Front. Microbiol. 12:679827. doi: 10.3389/fmicb.2021.679827
Received: 12 March 2021; Accepted: 16 April 2021;
Published: 12 May 2021.
Edited by:
Ulrike Kappler, The University of Queensland, AustraliaReviewed by:
Chiranjit Chowdhury, National Chemical Laboratory (CSIR), IndiaBhawani Chamlagain, University of Helsinki, Finland
Copyright © 2021 Dank, Zeng, Boeren, Notebaart, Smid and Abee. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Tjakko Abee, dGpha2tvLmFiZWVAd3VyLm5s; Eddy J. Smid, ZWRkeS5zbWlkQHd1ci5ubA==
†These authors have contributed equally to this work and share first authorship
 Alexander Dank
Alexander Dank Zhe Zeng
Zhe Zeng Sjef Boeren
Sjef Boeren Richard A. Notebaart
Richard A. Notebaart Eddy J. Smid
Eddy J. Smid Tjakko Abee
Tjakko Abee