- 1School of Biology and Biological Engineering, South China University of Technology, Guangzhou, China
- 2Key Laboratory of Systems Microbial Biotechnology, Tianjin Institute of Industrial Biotechnology, Chinese Academy of Sciences, Tianjin, China
- 3College of Biotechnology, Tianjin University of Science and Technology, Tianjin, China
Corynebacterium glutamicum is an important workhorse for industrial production of diversiform bioproducts. Precise regulation of gene expression is crucial for metabolic balance and enhancing production of target molecules. Auto-inducible promoters, which can be activated without expensive inducers, are ideal regulatory tools for industrial-scale application. However, few auto-inducible promoters have been identified and applied in C. glutamicum. Here, a hyperosmotic stress inducible gene expression system was developed and used for metabolic engineering of C. glutamicum. The promoter of NCgl1418 (PNCgl1418) that was activated by the two-component signal transduction system MtrA/MtrB was found to exhibit a high inducibility under hyperosmotic stress conditions. A synthetic promoter library was then constructed by randomizing the flanking and space regions of PNCgl1418, and mutant promoters exhibiting high strength were isolated via fluorescence activated cell sorting (FACS)-based high-throughput screening. The hyperosmotic stress inducible gene expression system was applied to regulate the expression of lysE encoding a lysine exporter and repress four genes involved in lysine biosynthesis (gltA, pck, pgi, and hom) by CRISPR interference, which increased the lysine titer by 64.7% (from 17.0 to 28.0 g/L) in bioreactors. The hyperosmotic stress inducible gene expression system developed here is a simple and effective tool for gene auto-regulation in C. glutamicum and holds promise for metabolic engineering of C. glutamicum to produce valuable chemicals and fuels.
Introduction
The nonpathogenic Gram-positive Corynebacterium glutamicum is a biosafe strain recognized by FDA and widely used in biomanufacturing of amino acids, organic acids, proteins, and other chemicals (Woo and Park, 2014; Lee et al., 2016; Tsuge and Matsuzawa, 2021). Regulation and optimization of target gene expression is crucial for balance of metabolic pathway and improvement of product biosynthesis. Promoters with different properties are one of the most effective tools to control gene expression. At present, a series of promoters have been identified or developed to regulate gene expression in C. glutamicum (Becker et al., 2005; Kind et al., 2010; Patek and Nesvera, 2011; Dostalova et al., 2017; Zhang et al., 2018; Sun et al., 2020; Wang et al., 2020; Wiechert et al., 2020a,b; Henke et al., 2021). In general, there are two types of promoters: constitutive promoters and inducible promoters. The strong constitutive promoters, such as the tuf and sod promoters, are widely used to enhance the expression of target genes (Becker et al., 2005; Kind et al., 2010). However, in some cases, gene expression controlled by strong constitutive promoters may cause cellular burden and metabolic unbalance, which hinders bioproduction (Giacalone et al., 2006; Terpe, 2006). Compared to constitutive promoters, inducible promoters can initiate the gene expression at any time when required, so they are preferred for the regulation and redistribution of metabolic flux and the expression of proteins with cytotoxicity (Lipscomb et al., 2004). At the moment, tac, trc, and other inducible promoters are extensive applied in the metabolic regulation of C. glutamicum (Ben Samoun et al., 1999; Okibe et al., 2010; Lausberg et al., 2012; Sun et al., 2020; Wiechert et al., 2020b; Henke et al., 2021), but there are also limitations. The requirement of inducers that are toxic [such as isopropyl-β-D-thiogalactopyranoside (IPTG)] or exhaustible (such as gluconate) limits the use in a large scale. Besides, inducers are usually added during the cultivation, so monitoring the cell growth state and optimizing the timing of inducer addition are required (Nocadello and Swennen, 2012). Therefore, there is a demand in development of auto-inducible systems without the use of external inducers.
Recently, auto-inducible promoters that can automatically turn on gene expression in response to environmental factors, metabolites, and cell growth state have been identified and applied in several bacteria. For example, a dynamic turn-off switch (dTFS) and a dynamic turn-on switch (dTNS) in Escherichia coli were constructed using the growth phase-dependent promoter and degron (Lemke et al., 2011). This bifunctional molecular switch was used to uncouple cell growth from the biosynthesis of shikimic acid and glucaric acid (Hou et al., 2020). Pyruvate-responsive genetic circuits were constructed in Bacillus subtilis by the hybrid promoter that contained the PdhR-binding site and the pyruvate-responsive transcription factor PdhR from E. coli, which were used to balance and optimize the metabolic flux toward the production of glucaric acid (Ogasawara et al., 2007; Xu et al., 2020a). An endogenous quorum-sensing (QS) based CRISPRi circuit in Streptomyces was constructed, in which the dcas9 gene was regulated by a native QS signal-responsive promoter. The system was used for downregulating three key nodes in essential pathways to divert metabolic flux toward rapamycin biosynthesis (Tian et al., 2020). In C. glutamicum, some auto-inducible promoters have also been reported and applied. For example, based on the LysR-type transcriptional regulator (LTTR) LysG and the lysE promoter with the LysG-binding site, a sensor suitable for intracellular lysine detection was developed (Binder et al., 2012). Two growth-regulated promoters (Pcg3141 and PCP 2836) had been identified (Kim et al., 2016; Ma et al., 2018). The Pcg3141 promoter was verified with the production of glutathione S-transferase as a model protein, and the PCP_2836 promoter was used for improving the production of valine and 5-aminolevulinic acid (Kim et al., 2016; Ma et al., 2018; Zhang et al., 2020).
With strict control of fermentation conditions such as pH and dissolved oxygen, the titer of target metabolites in the bioreactors can reach 100 g/L or even 200 g/L, Lys production by C. glutamicum, for example (Xu et al., 2020b). Therefore, in the mid- and late-stage of fermentation, the accumulation of high concentrations of products or the feeding of substrates will inevitably lead to a high-salt or hyperosmotic stress, which can be used as a natural inducer that exits in almost all fermentation processes (Varela et al., 2003; Chung et al., 2017). However, hyperosmotic stress inducible promoters have not been identified so far. MtrA/MtrB, one of the two-component systems that are highly conserved in corynebacteria and mycobacteria, regulates the expression of genes involved in osmoprotection (Moker et al., 2004; Bott and Brocker, 2012). Some target genes and the consensus binding sites of MtrA have also been identified in C. glutamicum (Brocker et al., 2011). However, the MtrA/MtrB-dependent promoters responding to the hyperosmotic stress have not been identified and applied in gene regulation.
In this study, a hyperosmotic stress inducible gene expression system based on the hyperosmotic stress inducible promoter was developed in C. glutamicum. Among eight promoters regulated by the two-component system MtrA/MtrB, the promoter of NCgl1418 (PNCgl1418) showed high expression intensity and induction activity under a hyperosmotic stress. The core sequence of PNCgl1418 was identified and the activity and inducibility was further enhanced by mutagenesis and fluorescence activated cell sorting (FACS)-based high-throughput screening. To demonstrate an application of this inducible system in metabolic engineering of C. glutamicum, it was used for overexpression of lysine exporter and CRISPR-dCpf1-mediated multiplex gene repression of four endogenous genes (Li et al., 2020) to maximize lysine production. This MtrA/MtrB-dependent hyperosmotic stress inducible system may also be useful for gene regulation in microorganisms beyond C. glutamicum.
Materials and Methods
Bacterial Strains and Cultivation Conditions
The bacterial strains used in this study are listed in Supplementary Table S1. E. coli Trans1-T1 and Trans DB 3.1 were used as the host for cloning and plasmid maintenance. Wild-type C. glutamicum ATCC 13032 was used as the host for gene expression and promoter screening. C. glutamicum SCgL30, the derivative of C. glutamicum ATCC 13032 with a feedback deregulated aspartokinase (T311I; Becker and Wittmann, 2012), was used for lysine production. E. coli was cultivated at 37°C in Luria-Bertani (LB) medium (5 g/L yeast extract, 10 g/L tryptone, and 10 g/L NaCl). Kanamycin (50 μg/ml) or chloramphenicol (20 μg/ml) was added to LB medium as required. C. glutamicum was cultivated at 30°C in BHI medium containing 37 g/L bovine brain heart extract, 10 g/L (NH4)2SO4, 0.2 g/L K2HPO4, 0.3 g/L NaH2PO4, and 0.5 g/L MgSO4·7H2O. For fluorescence intensity determination, C. glutamicum was cultivated in a defined medium (CGXIIY medium) containing 50 g/L glucose, 2 g/L yeast extract, 16.5 g/L NH4Cl, 5 g/L urea, 1 g/L KH2PO4, 1 g/L K2HPO4, 42 g/L MOPS, 0.25 g/L MgSO4, 0.01 g/L FeSO4·2H2O, 0.01 g/L MnSO4·H2O, 0.001 g/L ZnSO4·7H2O, 0.2 mg/L CuSO4, 0.02 mg/L NiCl·6H2O, 0.01 g/L CaCl2, 0.03 g/L protocatechuic acid, 0.2 mg/L biotin, and 0.1 mg/L vitamin B1. Kanamycin (25 μg/ml), chloramphenicol (5 μg/ml) or IPTG (1 mM) was added when necessary.
Plasmid Manipulation
The plasmids and primers used in this study are listed in Supplementary Tables S1 and S2, respectively. The promoter regions [about 300 bp upstream of the coding sequence (CDS)] of abgT, csbD, betP, NCgl1418, NCgl1756, NCgl1838, NCgl2841, and proP were amplified from the genomic DNA of C. glutamicum ATCC 13032 by PCR with eight primer pairs abgT-F/R, csbD-F/R, betP-F/R, 1418-F/R, 1756-F/R, 1838-F/R, 2841-F/R, and proP-F/R, respectively. E. coli-C. glutamicum shuttle expression vector pXM-gfp (Sun et al., 2019) was used to characterize the promoter strength. The backbone of pXM-gfp was amplified by PCR with the primer pair pGFP-F/R and self-cyclized to construct the control plasmid pXM-con-gfp harboring no promoter for gfp. The promoters PabgT, PcsbD, PbetP, PNCgl1418, PNCgl1756, PNCgl1838, PNCgl2841, and PproP were individually inserted to pXM-con-gfp for gfp expression. The tuf promoter (the 349 bp upstream of the tuf gene) was amplified from the genomic DNA of C. glutamicum ATCC 13032 by PCR with the primer pair tuf-F/R. The backbone of pXM-PNCgl1418-gfp was amplified by PCR with the primer pair pGFP-tuf-F/pGFP-R. Then these two PCR fragments were purified and ligated using the CloneExpress® MultiS One Step Cloning Kit (Vazyme Biotech, Nanjing, China) to generate the plasmid pXM-Ptuf-gfp. The backbone of pXM-gfp was amplified by PCR with the primer pair tac-F/R and then self-cyclized to generate the plasmid pXM-Ptac-gfp. To test truncated PNCgl1418 promoters, the PNCgl1418 in pXM-PNCgl1418-gfp was replaced with truncated promoters, PNCgl1418-203, PNCgl1418-145, and PNCgl1418-94, by PCR using the forward primer 1418-203-F, 1418-145-F, and 1418-94-F, respectively, with the reverse primer pGFP-R.
E. coli-C. glutamicum shuttle expression vector pEC-XK99E was used to express lysE. The PNCgl1418 promoter and the lysE gene were amplified by PCR using the primer pairs 1418-E-F/R and lysE-F/R from the genomic DNA of C. glutamicum ATCC 13032, respectively. Then the backbone of pEC-XK99E was amplified with the primer pair pEC-F/R. These three PCR fragments were purified and ligated to generate the plasmid pEC-PNCgl1418-lysE. The PNCgl1418 on pEC-PNCgl1418-lysE was replaced with the PNCgl1418 variant (PNCgl1418-A10) by PCR using the primer pair A10-E-F/R, generating the plasmid pEC-PNCgl1418-A10-lysE.
A previously developed CRISPR-dCpf1 system (Li et al., 2020) was modified to repress the expression of target genes in response to the hyperosmotic stress. To construct an all-in-one CRISPRi tool plasmid, the backbone of pXM-07 expressing dCpf1 was divided into three parts, which were amplified with the primer pairs pXM-07-F1/R1, pXM-07-F2/R2, and pXM-07-F3/R3, respectively. A gRNA cassette consisting of a constitutive promoter (P11F), two direct repeats (DRs), a ccdB flanked by two BbsI sites, and a terminator was amplified from pEC-02 (Li et al., 2020) with the primer pair ccdB-F/R. These four PCR fragments were purified and ligated to generate the plasmid pXM-dCpf1. Two oligonucleotides (RFP-F/R) were annealed and assembled into BbsI-digested pXM-dCpf1 backbone using a Golden Gate assembly method, resulting in the plasmid pXM-dCpf1-RFP. Similarly, array-F1/R1, array-F2/R2, array-F3/R3, and array-F4/R4 were annealed, respectively, and assembled into BbsI-digested pXM-dCpf1 backbone, resulting in the plasmid pXM-dCpf1-4crRNA (the crRNA array targeting gltA, pgi, hom, and pck). All crRNAs used in this study are listed in Supplementary Table S3. To construct a hyperosmotic stress inducible CRISPRi system, the PNCgl1418 promoter was amplified from the genomic DNA of C. glutamicum ATCC 13032 by PCR using the primer pair 1418-D-F/R. The backbone of pXM-dCpf1 or pXM-dCpf1-4crRNA was amplified with the primer pairs pXM-D-F1/R1 and pXM-D-F2/R2, respectively. Then these three PCR fragments were purified and ligated to generate the plasmids pXM-PNCgl1418-con and pXM-PNCgl1418-dCpf1-4crRNA, respectively. The PNCgl1418 on pXM-PNCgl1418-con and pXM-PNCgl1418-dCpf1-4crRNA was replaced with the PNCgl1418-A10 variant by PCR using the primer pairs A10-D-F1/R1 and A10-D-F2/R2, generating the plasmid pXM-PNCgl1418-A10-con and pXM-PNCgl1418-A10-dCpf1-4crRNA, respectively.
Construction of the MtrA-Deleted Strain
Deletion of the mtrA gene in C. glutamicum ATCC 13032 chromosome was performed by the CRISPR-Cas9 system (Liu et al., 2017). Plasmid pgRNA-ΔmtrA was construct by inserting the flanking regions of mtrA into pgRNA2 and replacing the base-pairing region of gRNA targeting ldhA with a base-pairing region targeting mtrA (N20, 5'-GTGGTGCTCGGTTTGGAATC-3'). A part of pgRNA2 backbone was amplified by PCR using primer pair gRNA-mtrA/pgRNA-1. N20 for mtrA was added to the backbone by primer gRNA-mtrA. A second part of pgRNA2 backbone was amplified by PCR using primer pair pgRNA-2/pgRNA-3. The 979 and 1,008 bp flanking regions of mtrA were amplified from the genomic DNA of C. glutamicum ATCC 13032 using the primer pairs mtrA-del-up-F/R and mtrA-del-down-F/R, respectively. The four fragments were ligated using the CloneExpress® MultiS One Step Cloning Kit (Vazyme Biotech, Nanjing, China) to yield pgRNA-ΔmtrA. The two plasmids (pgRNA-ΔmtrA and pCas9) were transformed into C. glutamicum ATCC 13032 by electroporation, and the screening of ΔmtrA mutants was performed in BHI plate with 0.05 mM IPTG. Gene deletion was verified by checking the sizes of the colony PCR fragments. Tool plasmids were cured from the edited mutant before it was used for subsequent experiments.
Construction of Synthetic Promoter Libraries
The synthetic promoter libraries of PNCgl1418 were constructed using the primer pairs P-Library-F1/R1 and P-Library-F2/R2 containing degenerated oligonucleotides (Ns) by PCR using pXM-PNCgl1418-gfp as a template. The PCR product was self-ligated with T4 ligase. The ligation products were transformed into strain E. coli Trans1-T1. The quality of randomization was checked by sequencing 24 randomly picked transformants. The synthetic promoter libraries were extracted and then transformed into C. glutamicum ATCC 13032 by electroporation.
FACS Screening
The C. glutamicum library was inoculated into BHI medium and cultivated overnight at 30°C and 220 rpm. The overnight cultures were transferred into 24-well plates with 1 ml CGXIIY medium containing 0.6 M Na2SO4 to an optical density at 600 nm (OD600) of 0.5. Then, cells were cultivated for 6 h at 30°C and with shaking at 800 rpm in INFORS Microtron (INFORS HT Multitron Pro, Switzerland). The cell suspension was diluted by proper times with distilled water and sorted on FACS (MoFlo XDP, Beckman Coulter, Inc., Miami, FL, United States) using green fluorescent protein (GFP) as the reporter protein. The cells were excited by 488-nm laser and detected through a 520 band-pass filter. In the first-round screening, cells showing high fluorescence intensity (the top 0.01%) were sorted (positive sorting). The sorted cells were poured directly into 5 ml BHI medium and cultivated overnight, and then the cells were transferred into 24-well plates with 1 ml CGXIIY medium for the next round of screening. In the second-round screening, cells showing low fluorescence intensity (the bottom 1%) were sorted (negative sorting). Similarly, the third-round screening (positive sorting) was performed. After the three-round sorting, the sorted cells were spread on BHI agar plate and incubated at 30°C.
Fluorescence Intensity Determination
The C. glutamicum was inoculated into BHI medium and cultivated overnight at 30°C and 220 rpm. The overnight cultures were transferred into 24-well plates with 1 ml CGXIIY medium with or without 0.6 M Na2SO4 to an OD600 of 0.5. After cultivated at 30°C and with shaking at 800 rpm for 18 h, cells were harvested and diluted by proper times with distilled water. GFP fluorescence intensities were determined using a microplate reader (SpectraMax M5, Molecular Devices, λ excitation = 488 nm, λ emission = 520 nm). The fluorescence intensities were normalized with OD600.
Lysine Production in 24-Well Plates
C. glutamicum SCgL30 with a feedback deregulated aspartokinase (T311I; Becker and Wittmann, 2012) was used for lysine production. Strain SCgL30 and its derivatives were cultivated in BHI medium at 30°C and with shaking at 220 rpm. The overnight cultures were transferred into 24-well plates containing 1 ml fermentation medium each well to an initial OD600 of 0.5. The fermentation medium contains 80 g/L glucose, 8 g/L yeast extract, 9 g/L urea, 1.5 g/L K2HPO4·3H2O, 0.01 g/L MnSO4, 0.6 g/L MgSO4·7H2O, 0.01 g/L FeSO4·7H2O, and 42 g/L MOPS (Li et al., 2020). Then, cells were cultivated for 24 or 36 h at 30°C and with shaking at 800 rpm in INFORS Microtron (INFORS HT Multitron Pro, Switzerland).
Lysine Production in 1-L Bioreactors
Colonies grown on a BHI plate at 30°C for 24 h were picked and used to inoculate 50 ml LSS1 medium (Ohnishi et al., 2002) in 500 ml shake flasks. The cultures were then cultivated overnight at 30°C and with shaking at 220 rpm. The cultures were transferred into a 1-L jar bioreactor [T&J-MiniBox, T&J Bio-engineering (Shanghai) Co., Ltd., China] containing 500 ml LPG1 medium (Ohnishi et al., 2002) with an inoculation volume of 10%. After the sugar initially added to the bioreactor was exhausted, glucose solution (50%, w/v) containing 0.5 mg/L biotin was continuously fed to maintain the glucose concentration at about 1.5% (NH4)2SO4 solution (5.56%, w/v) was also fed to supply nitrogen source for lysine biosynthesis and cell growth. The fermentation was performed at 34°C with an agitation speed of 900 rpm and an aeration at 2.5 L/min. The pH was maintained at 7.3 with NH4OH. LSS1 medium (Ohnishi et al., 2002) contains 50 g/L sucrose, 40 g/L corn steep liquor, 8.3 g/L (NH4)2SO4, 1 g/L urea, 2 g/L KH2PO4, 0.83 g/L MgSO4·7H2O, 10 mg/L FeSO4·7H2O, 1 mg/L CuSO4·5H2O, 10 mg/L ZnSO4·7H2O, 10 mg/L β-alanine, 5 mg/L vitamin B3, 1.5 mg/L vitamin B1, 0.5 mg/L biotin, and 30 g/L CaCO3. LPG1 medium (Ohnishi et al., 2002) contains 50 g/L glucose, 20 g/L corn steep liquor, 15 g/L (NH4)2SO4, 1 g/L urea, 2.5 g/L KH2PO4, 0.75 g/L MgSO4·7H2O, 50 mg/L FeSO4·7H2O, 13 mg/L MnSO4·5H2O, 50 mg/L CaCl2·2H2O, 6.3 mg/L CuSO4·5H2O, 1.3 mg of ZnSO4·7H2O, 5 mg/L NiCl2·6H2O, 1.3 mg/L CoCl2·6H2O, 1.3 mg/L (NH4)6Mo7O24·4H2O, 23 mg/L β-alanine, 14 mg/L vitamin B3, 7 mg/L vitamin B1, and 0.5 mg/L biotin. Samples were taken periodically and glucose and lysine concentrations were quantified using an SBA-40D biosensor analyzer (Institute of Biology of Shandong Province Academy of Sciences, Shandong, China). Cell biomass was determined as OD600 with a UV-1800 spectrophotometer (Shimadzu, Kyoto, Japan) after proper dilution with distilled water.
Results
Identification of Hyperosmotic Stress Inducible Promoters
To identify the hyperosmotic stress inducible promoters, we tested the promoters of the genes showing MtrA-dependent behaviors. Eight promoters (PabgT, PcsbD, PbetP, PNCgl1418, PNCgl1756, PNCgl1838, PNCgl2841, and PproP), which control the expression of abgT (secondary transporter of the AbgT family), csbD, betP (glycine betaine transporter), NCgl1418, NCgl1756, NCgl1838, NCgl2841, and proP (proline/ectoine carrier), respectively, were selected based on a previous study (Brocker et al., 2011). These genes showed significantly decreased transcription levels (<10% of the control) upon the deletion of mtrA in C. glutamicum. Besides, the MtrA binding sites of these promoters have also been identified (Brocker et al., 2011). Since most of the exact promoters and regulatory regions of those genes have not been fully defined yet, the ~300 bp upstream of each CDS was investigated. In order to assess these MtrA-dependent promoters, GFP was used as a reporter to study the transcriptional strength of these promoters with or without 0.6 M NaCl. PabgT, PNCgl1418, PNCgl1838, and PproP showed more than 2.0-fold NaCl-induced expression activity. Among them, PNCgl1418 showed the highest induced expression intensity (Figure 1). Therefore, we chose PNCgl1418 for further analysis.
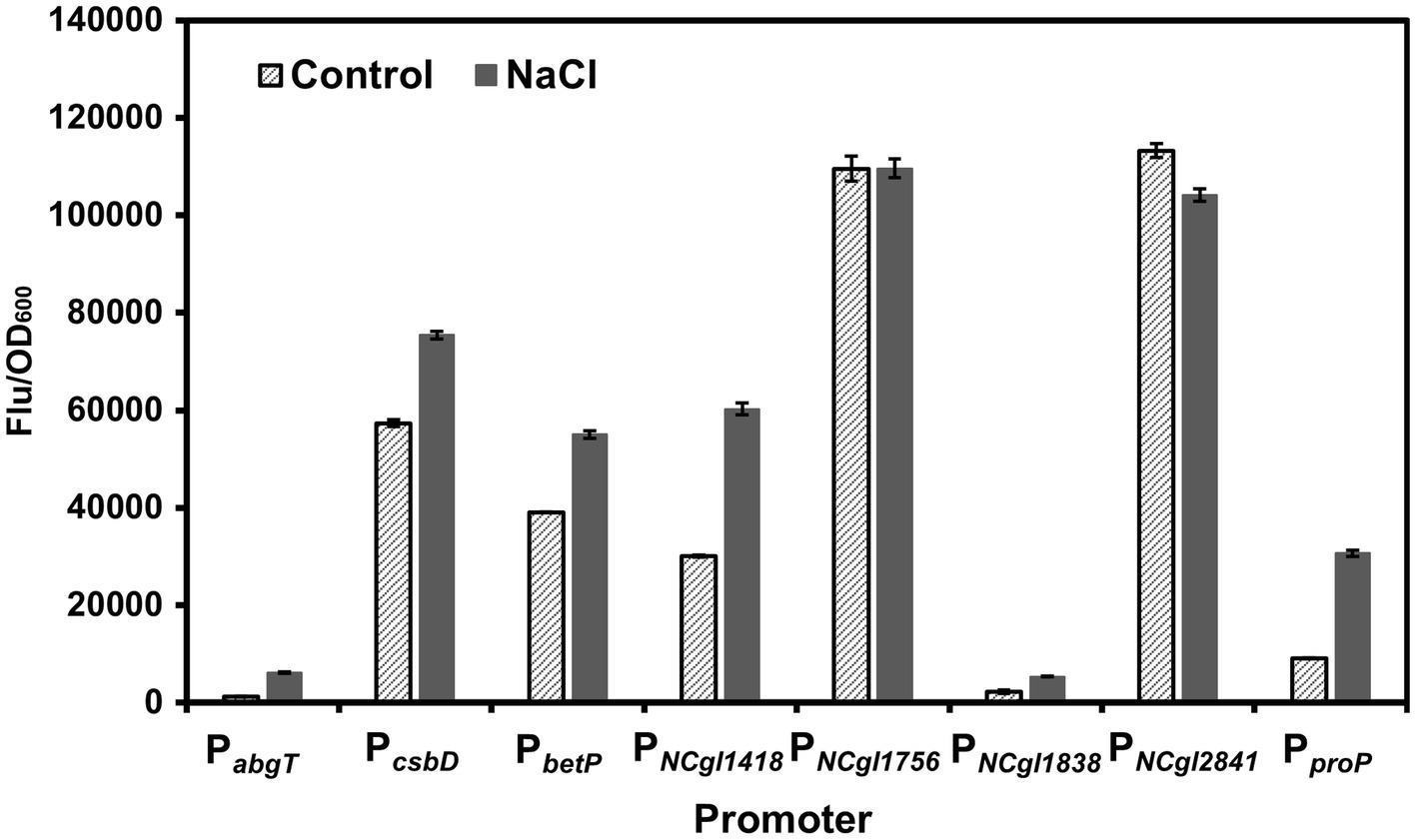
Figure 1. Screening of hyperosmotic stress inducible promoters. Expression of gfp was controlled by different MtrA/MtrB-dependent promoters (PabgT, PcsbD, PbetP, PNCgl1418, PNCgl1756, PNCgl1838, PNCgl2841, and PproP) in C. glutamicum with or without high salt stress induction. Cell fluorescence intensity was normalized to cell density and the background value of the strain containing the control vector pXM-con-gfp without a promoter was deducted. All data represent mean values from three biological replicates including standard deviations (SDs).
Characterization of the NCgl1418 Promoter
Next, the effects of different types or concentrations of inducers on induction of PNCgl1418 were investigated. Using GFP as a reporter, it was found that sulfate was the best inducer, which induced the expression activity of PNCgl1418 up to 4.0-fold (Figure 2A). PNCgl1418 showed obvious gradient induction activity in the range of 0~0.6 M Na2SO4 (Figure 2B). Compared with the endogenous strong constitutive promoter Ptuf and the widely used IPTG-inducible promoter Ptac (Becker et al., 2005; Kind et al., 2010; Rytter et al., 2014), PNCgl1418 showed about 62% expression strength upon hyperosmotic stress induction (Figure 2C). These results demonstrate that PNCgl1418 is a relative strong hyperosmotic stress inducible promoter, which has application potential for initiating the expression of target genes during the fermentation process.
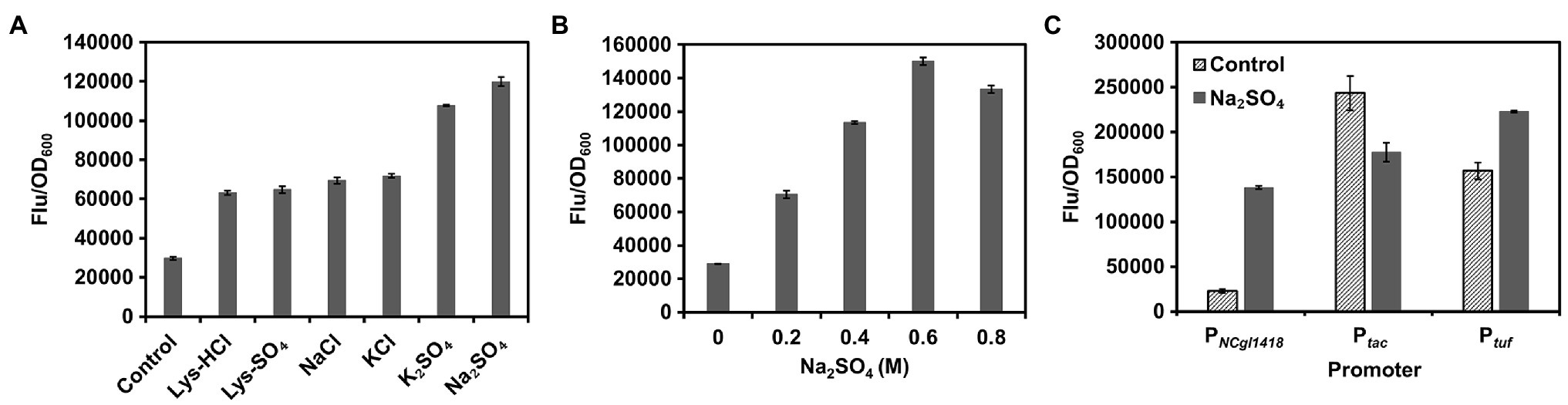
Figure 2. Characterization of PNCgl1418 promoter. (A) Expression of gfp under the control of PNCgl1418 in C. glutamicum induced by different salts. (B) Expression of gfp under the control of PNCgl1418 in C. glutamicum induced by different concentrations of sodium sulfate. (C) Comparison of PNCgl1418 with Ptuf and Ptac promoters. Expression of gfp was assayed with or without 0.6 M sodium sulfate. Cell fluorescence intensity was normalized to cell density and the background value of the strain containing the control vector pXM-con-gfp without a promoter was deducted. All data represent mean values from three biological replicates including SDs.
To determine the key functional region of PNCgl1418, three truncated variants (94, 145, and 203 bp) were constructed and used for GFP expression (Figure 3A). Although the 94 bp PNCgl1418 promoter contained the core −35 and −10 regions (Pfeifer Sancar et al., 2013), the function of the promoter completely lost. The 145 bp promoter contained the MtrA binding site but showed a decreased expression activity (74%) under hyperosmotic stress. In contrast, the 203 bp promoter maintained almost the same activity (94%) to the whole-length promoter (243 bp) under hyperosmotic stress (Figure 3B). To confirm the necessity of MtrA for PNCgl1418 functioning, a mtrA-deleted C. glutamicum strain was constructed and the plasmid harboring the PNCgl1418-controlled gfp cassette was transformed into the mutant. As expect, no GFP expression could be detected with mtrA deletion (Supplementary Figure S1). These results suggest that MtrA and its specific binding site are essential to the inducible activity of PNCgl1418. However, considering the partial activity loss of the 145 bp truncation, unknown mechanisms may also have an impact on the promoter.
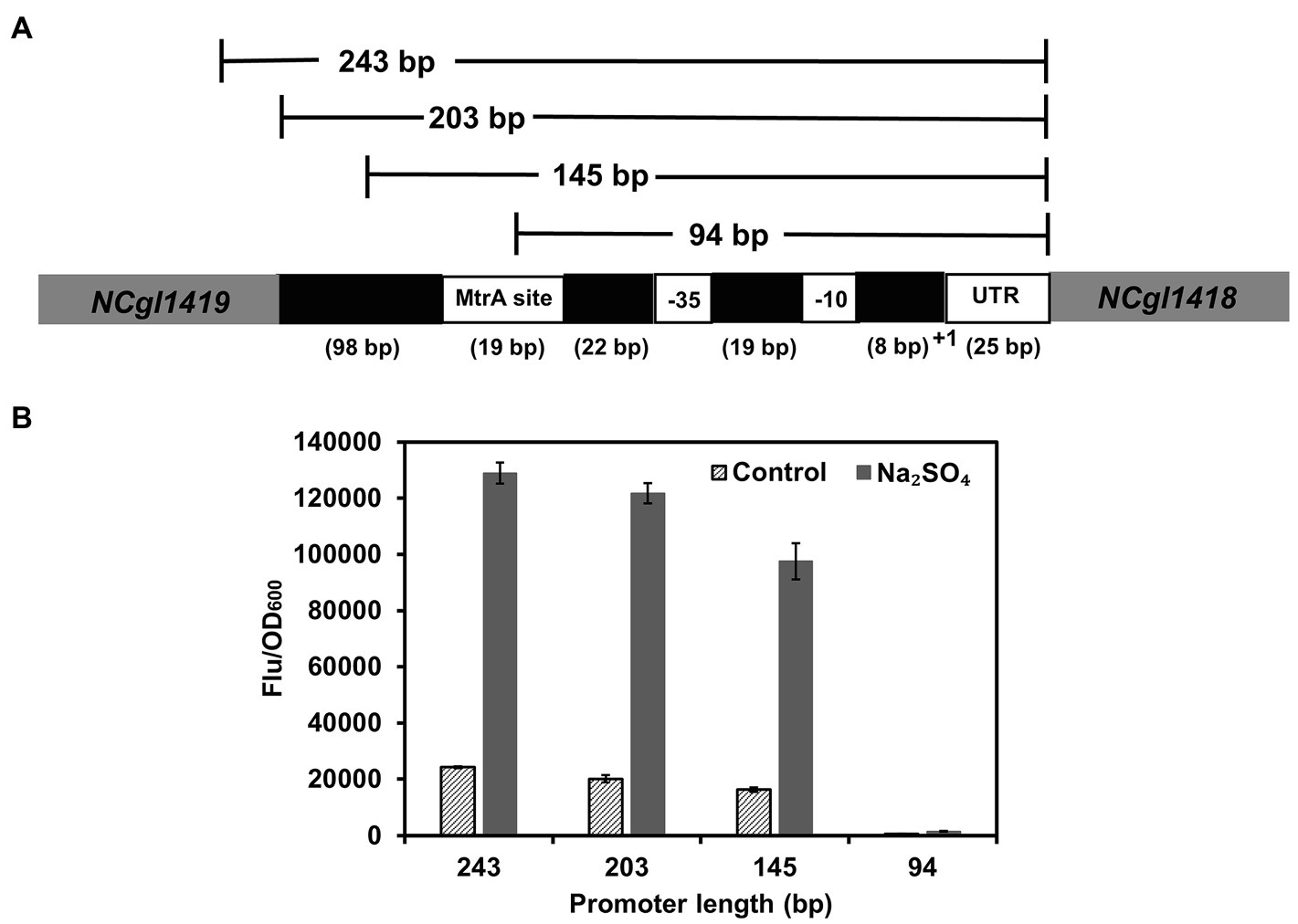
Figure 3. Identification of the core functional region of PNCgl1418 promoter. (A) Schematic diagram of the PNCgl1418 promoter truncation strategy. (B) Expression of gfp under the control of PNCgl1418 promoter variants with different length (243, 203, 145, and 94 bp) with or without 0.6 M sodium sulfate. Cell fluorescence intensity was normalized to cell density and the background value of the strain containing the control vector pXM-con-gfp without a promoter was deducted. All data represent mean values from three biological replicates including SDs.
Construction of PNCgl1418 Libraries and Isolation of Variants With Enhanced Inducible Activity
Although PNCgl1418 exhibited a relatively high inducible activity, its expression intensity was still lower than that of Ptuf and Ptac, which may hinder the application in high-level gene expression. Therefore, further engineering of this promoter is needed to increase its strength. To engineer the PNCgl1418, random promoter libraries were constructed based on the 243 bp wild-type promoter. The deduced −10 and −35 regions of PNCgl1418 were fixed to allow sigma factor binding, and the 29 bp spacer and left flanking regions were designed to have fully randomized nucleotides (Figure 4A). The randomized promoters were cloned to the upstream of the GFP reporter gene and subjected to a three-step positive-negative-positive screening to identify promoter variants with improved induction activity and expression strength. During this screening process, positive screening in the presence of 0.6 M Na2SO4 was conducted to identify high-fluorescence variants (isolation of the top 0.01% fluorescing cells) and the negative screening in the absence of 0.6 M Na2SO4 was conducted to exclude the variants with too high fluorescence intensity without induction (isolation of the bottom 1% non-fluorescing cells; Figure 4B). After three rounds of screening, the sorted cells were cultivated on agar plates, and 90 clones were randomly picked and characterized. After a secondary screening in microtiter plates, a total of 10 promoter variants with different sequences (Supplementary Figure S2) and inducible activities were obtained (Figure 4C). One promoter variant A10 (PNCgl1418-A10) showed the highest inducible activity and was chosen for further analysis. PNCgl1418-A10 was then compared with the strong constitutive promoter Ptuf and IPTG-inducible promoter Ptac with or without 0.6 M Na2SO4. The expression of GFP under the control of PNCgl1418-A10 showed an 8.5-fold induction by 0.6 M Na2SO4. The induced GFP expression level was approximately 1.8-fold higher than those of Ptuf and Ptac (Figure 4C).
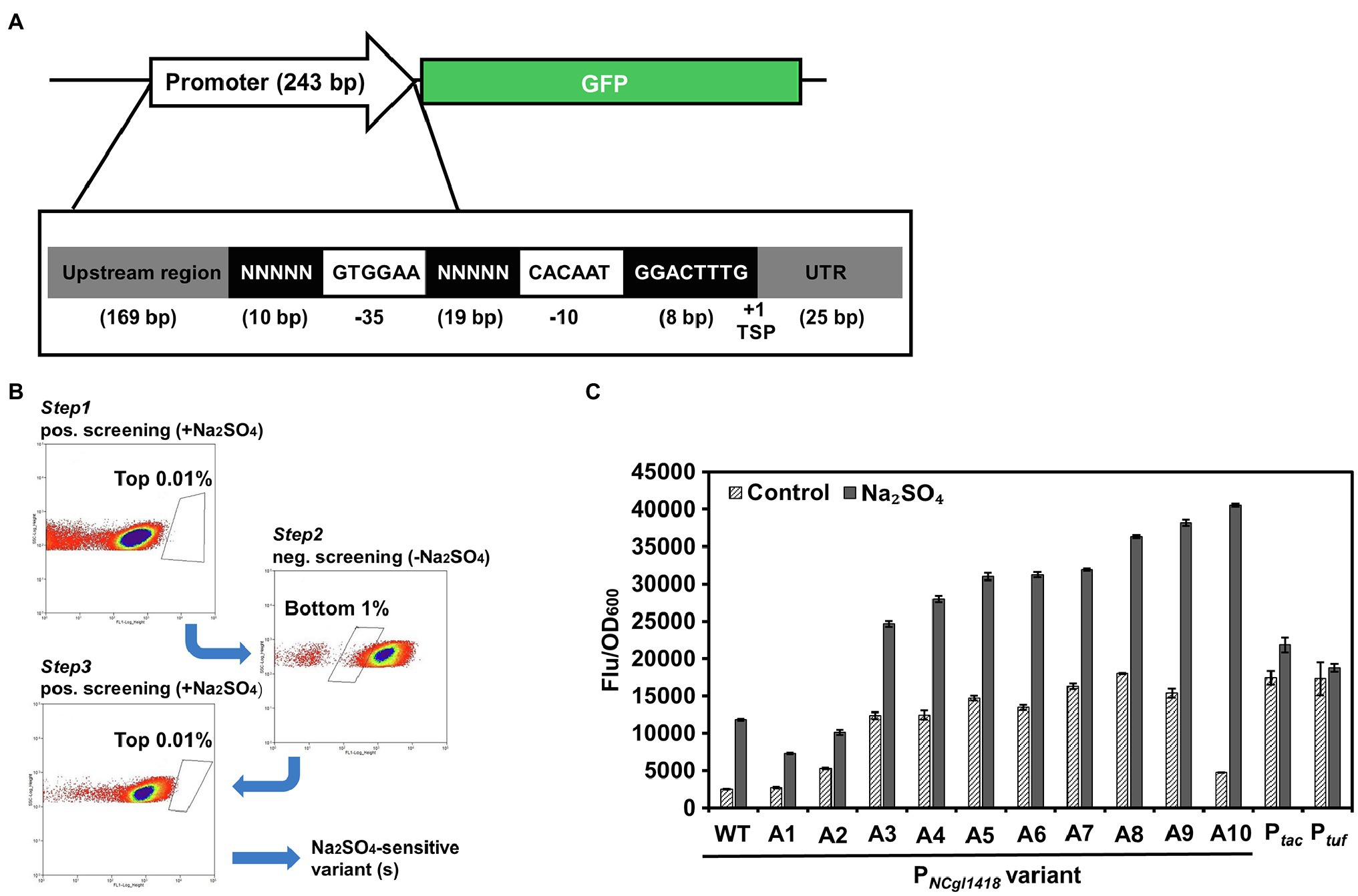
Figure 4. FACS-based positive/negative screening of the random promoter library to identify PNCgl1418 promoter variants with enhanced inducible activity. (A) Design of the random promoter library. (B) FACS plots of the three rounds of positive/negative screening. (C) Characterization of the isolated promoter variants. Cell fluorescence intensity was normalized to cell density and the background value of the strain containing the control vector pXM-con-gfp without a promoter was deducted. All data represent mean values from three biological replicates including SDs.
Application of the Hyperosmotic Stress Inducible Promoter to Optimize Lysine Biosynthesis
To demonstrate the usability of the hyperosmotic stress inducible promoters in metabolic engineering, the wild-type PNCgl1418 and PNCgl1418-A10 variant were used to regulate five genes related to lysine biosynthesis and excretion in C. glutamicum (Figure 5A). One target gene lysE encodes a lysine efflux protein and overexpression of lysE can increase lysine production by enhancing excretion of lysine (Zhou and Zeng, 2015). The other four genes, gltA, pck, pgi, and hom, are involved in the metabolic flux competition with lysine biosynthesis and their repression by CRISPRi has been proven to enhance the lysine production (Li et al., 2020).
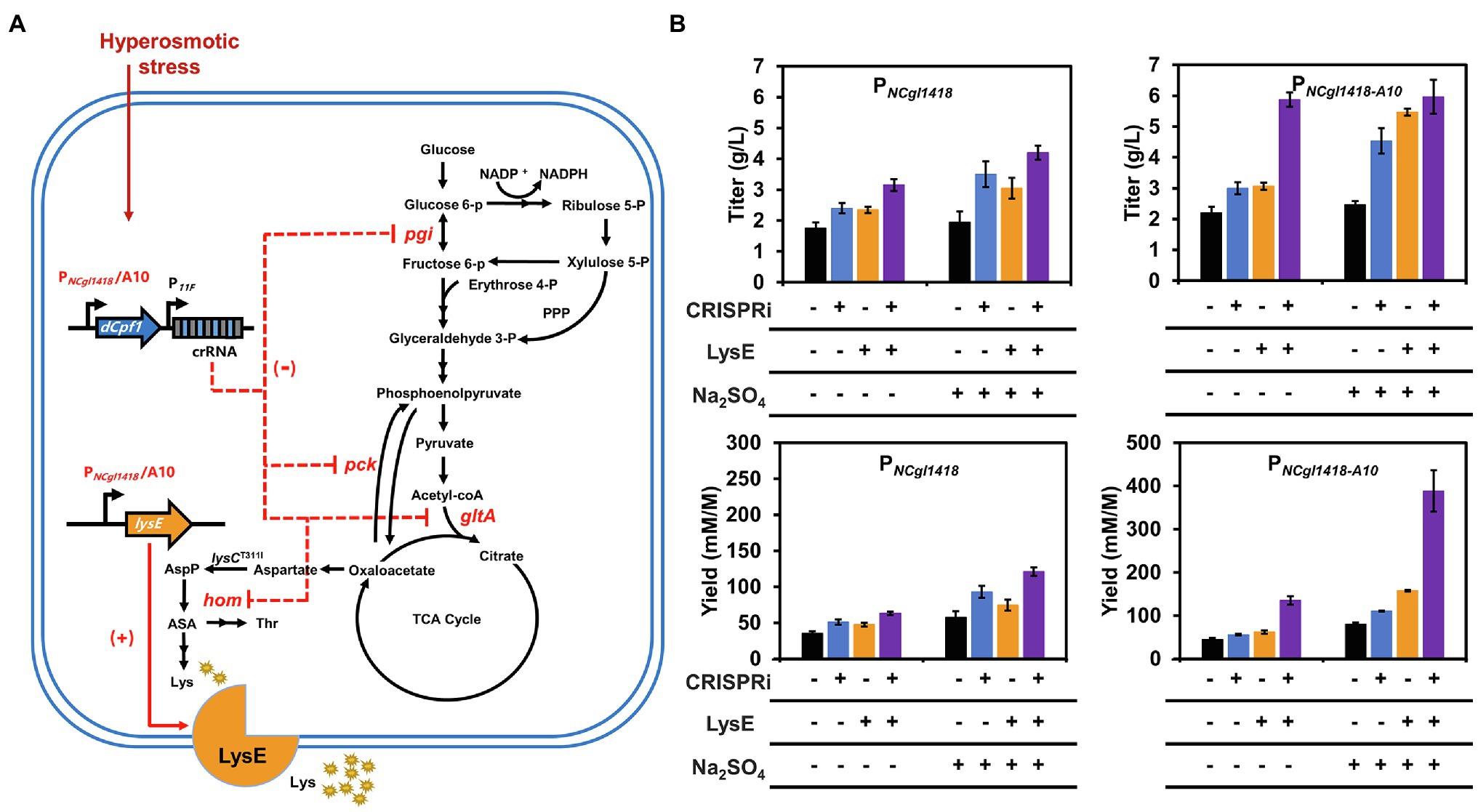
Figure 5. Enhancing lysine production by regulating the expression of target genes using the hyperosmotic stress inducible promoters. (A) Lysine biosynthesis pathway of C. glutamicum. (+) Represents gene overexpression and (−) represents gene repression. AspP, aspartyl phosphate; ASA, aspartate semialdehyde; Thr, threonine; Lys, lysine. (B) Effects of PNCgl1418 and PNCgl1418-A10 promoter-regulated lysE overexpression and/or multiple genes (gltA, pgi, hom, and pck) repression on lysine production. SCgL30 strains expressing LysE and/or dCpf1 with a crRNA array were cultivated in fermentation medium for 24 h (for PNCgl1418 promoter) or 36 h (for PNCgl1418-A10 promoter). SCgL30 strains containing the empty vector pEC-XK99E and/or the plasmid pXM-PNCgl1418-con without the crRNA array were used as controls. All data represent mean values from three biological replicates including SDs.
Firstly, we constructed an all-in-one CRIPSRi tool plasmid pXM-dCpf1 based on the previously developed two plasmids system expressing dCpf1 under the control of IPTG-inducible Ptac and crRNA array under the control of constitutive P11F (Li et al., 2020). To determine the gene repression efficiency of the all-in-one CRIPSRi system, a crRNA targeting the template strand of red fluorescent protein (RFP) encoding gene was designed and assembled to the plasmid pXM-dCpf1. A repression efficiency of 77% was achieved (Supplementary Figure S3), which was similar with the previous system using two plasmids. For combinational repression of the four target genes (gltA, pck, pgi, and hom), a crRNA array harboring four spacers was assembled to pXM-dCpf1. To achieve hyperosmotic stress inducible gene repression, the IPTG-inducible Ptac was replaced with PNCgl1418 and PNCgl1418-A10, respectively, for controlling dCpf1 expression. The two CRISPRi plasmids and a control plasmid without targeting crRNA array were individually transformed into the lysine-producing strain SCgL30 with a feedback deregulated aspartokinase (T311I; Becker and Wittmann, 2012). Fermentation in 24-well plates was performed to evaluate lysine production. Without addition of 0.6 M Na2SO4 to provide a hyperosmotic stress, repression of four target genes using PNCgl1418 enhanced lysine titer and yield by 37 and 45%, respectively (Figure 5B). When the hyperosmotic stress was exerted to cells by adding 0.6 M Na2SO4, repression of four target genes enhanced lysine titer and yield by 79 and 62%, respectively (Figure 5B). The lysine titer was improved by 92% (reached 4.5 g/L) when the stronger promoter variant PNCgl1418-A10 was used in the presence of hyperosmotic stress. The results suggest that the hyperosmotic stress inducible promoters can be used for efficiently regulating gene expression under high salt conditions for metabolic engineering purposes.
For further testing the application of the hyperosmotic stress inducible promoters in gene upregulation, lysE was selected and overexpressed in SCgL30. Without the addition of 0.6 M Na2SO4, overexpression of lysE slightly increased the lysine titer and yield. However, under the hyperosmotic stress, lysE overexpression controlled by PNCgl1418-A10 exhibited much higher lysine titer (2.3-fold) and yield (about 2.0-fold) compared with the control strain (Figure 5B). Finally, overexpression of lysE and repression of gltA, pck, pgi, and hom by CRISPRi were simultaneously performed for lysine production using the strain SLDEA10. Under the hyperosmotic stress, this combinational strategy improved lysine titer and the yield by 2.5- and 4.8-fold compared to the control, respectively (Figure 5B). It was noticed that the lysine titers of strain SLDEA10 cultivated with or without 0.6 M Na2SO4 were almost the same. This is may be due to the slower growth rate of strain SLDEA10 under hyperosmotic stress (Supplementary Figure S4). The growth retardation might be caused by the higher expression of dCpf1 and LysE or the stronger repression of the endogenous genes under hyperosmotic stress. However, the lysine yield reached 388.7 mM/M, which improved by 4.8-fold when 0.6 M Na2SO4 was added (Figure 5B). The results indicate that the promoter variant PNCgl1418-A10 has strong inductivity and the rebalance between growth and production is necessary to increase the performance of engineered strains.
Feed Batch Fermentation of the Lysine Producer With the Auto-Inducible System
Because of the limited substrate supplement and the lack of fermentation condition control, fermentation in 24-well plates cannot result in high-titer lysine production. Therefore, 0.6 M Na2SO4 was added to provide a hyperosmotic stress to induce gene expression controlled by PNCgl1418 and PNCgl1418-A10. Fed-batch fermentations are usually used to obtain high concentrations of products, which can serve as inducers for hyperosmotic stress inducible promoters such as PNCgl1418-A10. To demonstrate the auto-inducible system based on PNCgl1418-A10, fed-batch fermentations without exogenous Na2SO4 addition were conducted in 1-L bioreactors using the best lysine producer strain SLDEA10 and a control strain SLDEcon2 without the auto-inducible CRISPRi and lysE overexpression system. Overall, SLDEA10 had a better balance between growth and production than the control strain (Figure 6). Strain SLDEcon2 entered the stationary phage at 15 h with a relatively high OD600nm peak value, while the highest OD600nm was significantly reduced by 32.3% for strain SLDEA10 (Figure 6). On the contrary, the lysine biosynthesis rate of SLDEA10 was significantly higher than that of strain SLDEcon2 in the mid- and late-stage. After 33 h fermentation, 28.0 g/L of lysine was produced by strain SLDEA10, representing a 64.7% improvement compared to strain SLDEcon2 (17.0 g/L; Figure 6). These results suggest that the metabolic flux is directed from biomass synthesis to product synthesis by using the auto-inducible system based on the hyperosmotic stress inducible promoter.
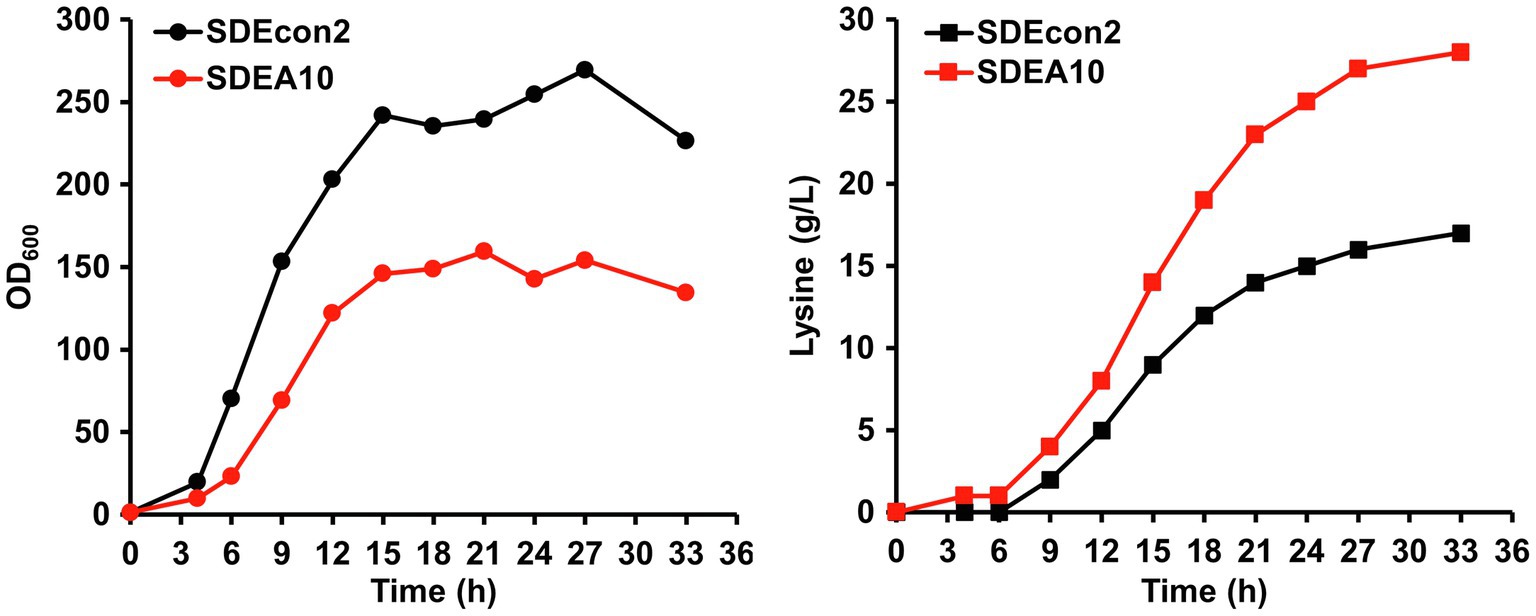
Figure 6. Fed-batch fermentation for lysine production using strains SDEcon2 (black lines) and SDEA10 (red lines). Strain SDEcon2 is strain SCgL30 harboring the empty vector pEC-XK99E and the plasmid pXM-PNCgl1418-con without the crRNA array. Strain SDEA10 is strain SCgL30 with overexpression of lysE and repression of gltA, pgi, hom, and pck controlled by PNCgl1418-A10 promoter. Cultivation was performed in 1-L bioreactors with fermentation medium.
Discussion
In the last few decades, C. glutamicum has been engineered for the production of a variety of value-added products, including amino acids, organic acids, polymer precursors, and proteins (Baritugo et al., 2018; Lee and Kim, 2018). Gene expression regulation technologies based on CRISPR and synthetic small RNAs for C. glutamicum have been developed (Liu et al., 2017; Wang et al., 2018; Gauttam et al., 2019; Sun et al., 2019; Li et al., 2020). By combining these technologies with metabolic- or QS-responding elements, dynamic regulation strategies have been developed for precise regulation of gene expression to maximize the biosynthesis of target products (Gupta et al., 2017; Chen et al., 2018; Shen et al., 2019; Liang et al., 2020; Tian et al., 2020; Hartline et al., 2021). However, the metabolic-responding elements are specific for certain metabolites and thus they are not universal for various fermentation processes. Although QS systems have good universality, their application in large-scale fermentation usually suffers from the low threshold. As an alternative, the environmental parameter-responding elements have the advantages of high universality and good controllability, which can be used to develop universal dynamic regulation systems. At present, dynamic regulation strategies based on external environmental conditions such as temperature, dissolved oxygen, pH, and illumination have been successfully developed and applied in metabolic engineering (Jong Uk et al., 2008; Binder et al., 2016; Hwang et al., 2017; Harder et al., 2018; Zhao et al., 2018; Banares et al., 2020). However, in large-scale fermentation, these environmental factors including temperature, dissolved oxygen, and pH mostly need to be controlled at a steady level to maintain cellular activity, and optogenetics tools are limited by the nonhomogeneity in large fermenters. Hyperosmotic stress, which commonly presents in the mid- and late-phase of almost all the fermentation processes, can serve as a naturally occurring inducer. Compared with the promoters based on metabolic- or QS-responding elements and those responding to external environmental conditions such as temperature, dissolved oxygen, pH, and illumination, the hyperosmotic stress inducible promoters have good universality and economy in the large-scale industrialization applications. In this study, a hyperosmotic stress inducible gene expression system was developed in C. glutamicum. The promoter of NCgl1418 identified here exhibits high expression intensity and induction activity under hyperosmotic stress caused by different salt inducers. More importantly, the induction activity was closely related to product concentration, which can be conveniently used to regulate the expression of key genes according to the product concentration. Considering the different expression intensities and induction activities by different types or concentrations of salts, optimization tests for different media may be required in practical applications.
Promoters are the core elements of the dynamic regulation systems and synthetic biology researches. Identification and modification of novel promoters with special functions can provide necessary tools for the development of new gene circuits and hyperproducing strains. At present, omics analysis under different culture conditions and bioinformatics analysis of specific recognition sequences of transcriptional regulators are the main strategies for the identification of new promoters (Dahl et al., 2013; Liu et al., 2015; Kim et al., 2016; Ma et al., 2018). However, the strength and induction activity of natural promoters are often not sufficient for practical applications. To engineer more suitable promoters, random mutation followed by high-throughput screening and redesign of the natural promoters with components from other different promoters are usually performed (Kim et al., 2016; Zhang et al., 2018; Henke et al., 2021). In this study, for isolation of engineered promoters with enhanced inducibility by hyperosmotic stress, we used a FACS screening strategy that combined three rounds of positive and negative screening. The isolated promoters showed up to a 3.4-fold increased GFP expression level compared to the original NCgl1418 promoter with much higher inducibility (up to 8.5-fold). As a proof-of-concept, lysine production was improved by regulating key genes expression with the hyperosmotic stress inducible promoters, demonstrating the great potential in metabolic engineering of C. glutamicum. Also, we observed leaky expression of the NCgl1418 promoter, like all the other kinds of inducible promoters. For a stricter control of the NCgl1418 promoter, conditional overexpression of its regulator MtrA/MtrB may be a useful strategy. In addition, promoter variants with low-level leaky expression could also be further screened by FACS. In conclusion, we identified the MtrA/MtrB-dependent NCgl1418 promoter as a hyperosmotic stress inducible promoter and developed a hyperosmotic stress inducible gene expression system in C. glutamicum. To the best of our knowledge, this is the first report on the development of a hyperosmotic stress inducible promoter in C. glutamicum. We believe this hyperosmotic stress inducible gene expression system will contribute to extend the usefulness of C. glutamicum in the industrial-scale production of various value-added biochemicals.
Data Availability Statement
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding authors.
Author Contributions
JH, JC, YW, PZ, and JS conceptualized the project, designed the study, and composed the manuscript. JH, JC, TS, YZ, and NC conducted the experiments and collected data. JH, JC, YW, XN, WP, and JL performed the data analysis. PZ, JS, and SH provided the critical feedback on the manuscript and resources. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Key R&D Program of China (2018YFA0901400 and 2019YFA0904900), Tianjin Synthetic Biotechnology Innovation Capacity Improvement Project (TSBICIP-KJGG-005), and the National Natural Science Foundation of China (32000023).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.718511/full#supplementary-material
References
Banares, A. B., Valdehuesa, K. N. G., Ramos, K. R. M., Nisola, G. M., Lee, W. K., and Chung, W. J. (2020). A pH-responsive genetic sensor for the dynamic regulation of D-xylonic acid accumulation in Escherichia coli. Appl. Microbiol. Biotechnol. 104, 2097–2108. doi: 10.1007/s00253-019-10297-0
Baritugo, K. A. G., Kim, H. T., David, Y. C., Choi, J. H., Choi, J. i., Kim, T. W., et al. (2018). Recent advances in metabolic engineering of Corynebacterium glutamicum as a potential platform microorganism for biorefinery. Biofuels Bioprod. Biorefin. 12, 899–925. doi: 10.1002/bbb.1895
Becker, J., Klopprogge, C., Zelder, O., Heinzle, E., and Wittmann, C. (2005). Amplified expression of fructose 1,6-bisphosphatase in Corynebacterium glutamicum increases in vivo flux through the pentose phosphate pathway and lysine production on different carbon sources. Appl. Environ. Microbiol. 71, 8587–8596. doi: 10.1128/AEM.71.12.8587-8596.2005
Becker, J., and Wittmann, C. (2012). Bio-based production of chemicals, materials and fuels Corynebacterium glutamicum as versatile cell factory. Curr. Opin. Biotechnol. 23, 631–640. doi: 10.1016/j.copbio.2011.11.012
Ben Samoun, K., Leblon, G., and Reyes, O. (1999). Positively regulated expression of the Escherichia coli araBAD promoter in Corynebacterium glutamicum. FEMS Microbiol. Lett. 174, 125–130. doi: 10.1111/j.1574-6968.1999.tb13558.x
Binder, D., Frohwitter, J., Mahr, R., Bier, C., Grünberger, A., Loeschcke, A., et al. (2016). Light-controlled cell factories: employing photocaged isopropyl-β-D-thiogalactopyranoside for light-mediated optimization of lac promoter-based gene expression and (+)-valencene biosynthesis in Corynebacterium glutamicum. Appl. Environ. Microbiol. 82, 6141–6149. doi: 10.1128/AEM.01457-16
Binder, S., Schendzielorz, G., Staebler, N., Krumbach, K., Hoffmann, K., Bott, M., et al. (2012). A high-throughput approach to identify genomic variants of bacterial metabolite producers at the single-cell level. Genome Biol. 13:R40. doi: 10.1186/gb-2012-13-5-r40
Bott, M., and Brocker, M. (2012). Two-component signal transduction in Corynebacterium glutamicum and other corynebacteria: on the way towards stimuli and targets. Appl. Microbiol. Biotechnol. 94, 1131–1150. doi: 10.1007/s00253-012-4060-x
Brocker, M., Mack, C., and Bott, M. (2011). Target genes, consensus binding site, and role of phosphorylation for the response regulator MtrA of Corynebacterium glutamicum. J. Bacteriol. 193, 1237–1249. doi: 10.1128/JB.01032-10
Chen, Y., Ho, J. M. L., Shis, D. L., Gupta, C., Long, J., Wagner, D. S., et al. (2018). Tuning the dynamic range of bacterial promoters regulated by ligand-inducible transcription factors. Nat. Commun. 9:64. doi: 10.1038/s41467-017-02473-5
Chung, S. C., Park, J. S., Yun, J., and Park, J. H. (2017). Improvement of succinate production by release of end-product inhibition in Corynebacterium glutamicum. Metab. Eng. 40, 157–164. doi: 10.1016/j.ymben.2017.02.004
Dahl, R. H., Zhang, F., Alonso Gutierrez, J., Baidoo, E., Batth, T. S., Redding Johanson, A. M., et al. (2013). Engineering dynamic pathway regulation using stress-response promoters. Nat. Biotechnol. 31, 1039–1046. doi: 10.1038/nbt.2689
Dostalova, H., Holatko, J., Busche, T., Rucka, L., Rapoport, A., Halada, P., et al. (2017). Assignment of sigma factors of RNA polymerase to promoters in Corynebacterium glutamicum. AMB Express 7:133. doi: 10.1186/s13568-017-0436-8
Gauttam, R., Seibold, G. M., Mueller, P., Weil, T., Weiss, T., Handrick, R., et al. (2019). A simple dual-inducible CRISPR interference system for multiple gene targeting in Corynebacterium glutamicum. Plasmid 103, 25–35. doi: 10.1016/j.plasmid.2019.04.001
Giacalone, M. J., Gentile, A. M., Lovitt, B. T., Berkley, N. L., Gunderson, C. W., and Surber, M. W. (2006). Toxic protein expression in Escherichia coli using a rhamnose-based tightly regulated and tunable promoter system. BioTechniques 40, 355–364. doi: 10.2144/000112112
Gupta, A., Reizman, I. M. B., Reisch, C. R., and Prather, K. L. J. (2017). Dynamic regulation of metabolic flux in engineered bacteria using a pathway-independent quorum-sensing circuit. Nat. Biotechnol. 35:273. doi: 10.1038/nbt.3796
Harder, B. J., Bettenbrock, K., and Klamt, S. (2018). Temperature-dependent dynamic control of the TCA cycle increases volumetric productivity of itaconic acid production by Escherichia coli. Biotechnol. Bioeng. 115, 156–164. doi: 10.1002/bit.26446
Hartline, C. J., Schmitz, A. C., Han, Y., and Zhang, F. (2021). Dynamic control in metabolic engineering: theories, tools, and applications. Metab. Eng. 63, 126–140. doi: 10.1016/j.ymben.2020.08.015
Henke, N. A., Krahn, I., and Wendisch, V. F. (2021). Improved plasmid-based inducible and constitutive gene expression in Corynebacterium glutamicum. Microorganisms 9:204. doi: 10.3390/microorganisms9010204
Hou, J., Gao, C., Guo, L., Nielsen, J., Ding, Q., Tang, W., et al. (2020). Rewiring carbon flux in Escherichia coli using a bifunctional molecular switch. Metab. Eng. 61, 47–57. doi: 10.1016/j.ymben.2020.05.004
Hwang, H. J., Kim, J. W., Ju, S. Y., Park, J. H., and Lee, P. C. (2017). Application of an oxygen-inducible nar promoter system in metabolic engineering for production of biochemicals in Escherichia coli. Biotechnol. Bioeng. 114, 468–473. doi: 10.1002/bit.26082
Jong Uk, P., Jo, J. H., Kim, Y. J., Chung, S. S., Lee, J. H., and Lee, H. H. (2008). Construction of heat-inducible expression vector of Corynebacterium-glutamicum and C-ammoniagenes: fusion of iambda operator with promoters isolated from C-ammoniagenes. J. Microbiol. Biotechnol. 18, 639–647. doi: 10.1016/S1369-8001(00)00115-3
Kim, M. J., Yim, S. S., Choi, J. W., and Jeong, K. J. (2016). Development of a potential stationary-phase specific gene expression system by engineering of SigB-dependent cg3141 promoter in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 100, 4473–4483. doi: 10.1007/s00253-016-7297-y
Kind, S., Jeong, W. K., Schroeder, H., and Wittmann, C. (2010). Systems-wide metabolic pathway engineering in Corynebacterium glutamicum for bio-based production of diaminopentane. Metab. Eng. 12, 341–351. doi: 10.1016/j.ymben.2010.03.005
Lausberg, F., Chattopadhyay, A. R., Heyer, A., Eggeling, L., and Freudl, R. (2012). A tetracycline inducible expression vector for Corynebacterium glutamicum allowing tightly regulable gene expression. Plasmid 68, 142–147. doi: 10.1016/j.plasmid.2012.05.001
Lee, M. J., and Kim, P. (2018). Recombinant protein expression system in Corynebacterium glutamicum and its application. Front. Microbiol. 9:2523. doi: 10.3389/fmicb.2018.02523
Lee, J. Y., Na, Y. A., Kim, E., Lee, H. S., and Kim, P. (2016). The Actinobacterium Corynebacterium glutamicum, an industrial workhorse. J. Microbiol. Biotechnol. 26, 807–822. doi: 10.4014/jmb.1601.01053
Lemke, J. J., Sanchez Vazquez, P., Burgos, H. L., Hedberg, G., Ross, W., and Gourse, R. L. (2011). Direct regulation of Escherichia coli ribosomal protein promoters by the transcription factors ppGpp and DksA. Proc. Natl. Acad. Sci. U. S. A. 108, 5712–5717. doi: 10.1073/pnas.1019383108
Li, M., Chen, J., Wang, Y., Liu, J., Huang, J., Chen, N., et al. (2020). Efficient multiplex gene repression by CRISPR-dCpf1 in Corynebacterium glutamicum. Front. Bioeng. Biotechnol. 8:357. doi: 10.3389/fbioe.2020.00357
Liang, C., Zhang, X., Wu, J., Mu, S., Wu, Z., Jin, J. M., et al. (2020). Dynamic control of toxic natural product biosynthesis by an artificial regulatory circuit. Metab. Eng. 57, 239–246. doi: 10.1016/j.ymben.2019.12.002
Lipscomb, M. L., Mowry, M. C., and Kompala, D. S. (2004). Production of a secreted glycoprotein from an inducible promoter system in a perfusion bioreactor. Biotechnol. Prog. 20, 1402–1407. doi: 10.1021/bp049973j
Liu, Y., Li, Q., Zheng, P., Zhang, Z., Liu, Y., Sun, C., et al. (2015). Developing a high-throughput screening method for threonine overproduction based on an artificial promoter. Microb. Cell Factories 14:121. doi: 10.1186/s12934-015-0311-8
Liu, J., Wang, Y., Lu, Y., Zheng, P., Sun, J., and Ma, Y. (2017). Development of a CRISPR/Cas9 genome editing toolbox for Corynebacterium glutamicum. Microb. Cell Factories 16:205. doi: 10.1186/s12934-017-0815-5
Ma, Y., Cui, Y., Du, L., Liu, X., Xie, X., and Chen, N. (2018). Identification and application of a growth-regulated promoter for improving L-valine production in Corynebacterium glutamicum. Microb. Cell Factories 17:185. doi: 10.1186/s12934-018-1031-7
Moker, N., Brocker, M., Schaffer, S., Kramer, R., Morbach, S., and Bott, M. (2004). Deletion of the genes encoding the MtrA-MtrB two-component system of Corynebacterium glutamicum has a strong influence on cell morphology, antibiotics susceptibility and expression of genes involved in osmoprotection. Mol. Microbiol. 54, 420–438. doi: 10.1111/j.1365-2958.2004.04249.x
Nocadello, S., and Swennen, E. F. (2012). The new pLAI (lux regulon based auto-inducible) expression system for recombinant protein production in Escherichia coli. Microb. Cell Factories 11:3. doi: 10.1186/1475-2859-11-3
Ogasawara, H., Ishida, Y., Yamada, K., Yamamoto, K., and Ishihama, A. (2007). PdhR (pyruvate dehydrogenase complex regulator) controls the respiratory electron transport system in Escherichia coli. J. Bacteriol. 189, 5534–5541. doi: 10.1128/JB.00229-07
Ohnishi, J., Mitsuhashi, S., Hayashi, M., Ando, S., Yokoi, H., Ochiai, K., et al. (2002). A novel methodology employing Corynebacterium glutamicum genome information to generate a new L-lysine-producing mutant. Appl. Microbiol. Biotechnol. 58, 217–223. doi: 10.1007/s00253-001-0883-6
Okibe, N., Suzuki, N., Inui, M., and Yukawa, H. (2010). Isolation, evaluation and use of two strong, carbon source-inducible promoters from Corynebacterium glutamicum. Lett. Appl. Microbiol. 50, 173–180. doi: 10.1111/j.1472-765X.2009.02776.x
Patek, M., and Nesvera, J. (2011). Sigma factors and promoters in Corynebacterium glutamicum. J. Biotechnol. 154, 101–113. doi: 10.1016/j.jbiotec.2011.01.017
Pfeifer Sancar, K., Mentz, A., Rueckert, C., and Kalinowski, J. (2013). Comprehensive analysis of the Corynebacterium glutamicum transcriptome using an improved RNA-seq technique. BMC Genomics 14:888. doi: 10.1186/1471-2164-14-888
Rytter, J. V., Helmark, S., Chen, J., Lezyk, M. J., Solem, C., and Jensen, P. R. (2014). Synthetic promoter libraries for Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 98, 2617–2623. doi: 10.1007/s00253-013-5481-x
Shen, X., Wang, J., Li, C., Yuan, Q., and Yan, Y. (2019). Dynamic gene expression engineering as a tool in pathway engineering. Curr. Opin. Biotechnol. 59, 122–129. doi: 10.1016/j.copbio.2019.03.019
Sun, D., Chen, J., Wang, Y., Li, M., Rao, D., Guo, Y., et al. (2019). Metabolic engineering of Corynebacterium glutamicum by synthetic small regulatory RNAs. J. Ind. Microbiol. Biotechnol. 46, 203–208. doi: 10.1007/s10295-018-02128-4
Sun, M., Gao, X., Zhao, Z., Li, A., Wang, Y., Yang, Y., et al. (2020). Enhanced production of recombinant proteins in Corynebacterium glutamicum by constructing a bicistronic gene expression system. Microb. Cell Factories 19:113. doi: 10.1186/s12934-020-01370-9
Terpe, K. (2006). Overview of bacterial expression systems for heterologous protein production: from molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Biotechnol. 72, 211–222. doi: 10.1007/s00253-006-0465-8
Tian, J., Yang, G., Gu, Y., Sun, X., Lu, Y., and Jiang, W. (2020). Developing an endogenous quorum-sensing based CRISPRi circuit for autonomous and tunable dynamic regulation of multiple targets in Streptomyces. Nucleic Acids Res. 48, 8188–8202. doi: 10.1093/nar/gkaa602
Tsuge, Y., and Matsuzawa, H. (2021). Recent progress in production of amino acid-derived chemicals using Corynebacterium glutamicum. World J. Microbiol. Biotechnol. 37:49. doi: 10.1007/s11274-021-03007-4
Varela, C., Agosin, E., Baez, M., Klapa, M., and Stephanopoulos, G. (2003). Metabolic flux redistribution in Corynebacterium glutamicum in response to osmotic stress. Appl. Microbiol. Biotechnol. 60, 547–555. doi: 10.1007/s00253-002-1120-7
Wang, Y., Gao, X., Liu, X., Li, Y., Sun, M., Yang, Y., et al. (2020). Construction of a 3A system from BioBrick parts for expression of recombinant hirudin variants III in Corynebacterium glutamicum. Appl. Microbiol. Biotechnol. 104, 8257–8266. doi: 10.1007/s00253-020-10835-1
Wang, Y., Liu, Y., Liu, J., Guo, Y., Fan, L., Ni, X., et al. (2018). MACBETH: Multiplex automated Corynebacterium glutamicum base editing method. Metab. Eng. 47, 200–210. doi: 10.1016/j.ymben.2018.02.016
Wiechert, J., Filipchyk, A., Huennefeld, M., Gaetgens, C., Brehm, J., Heermann, R., et al. (2020a). Deciphering the rules underlying xenogeneic silencing and counter-silencing of Lsr2-like proteins using CgpS of Corynebacterium glutamicum as a model. mBio 11, e02273–e02219. doi: 10.1128/mBio.02273-19
Wiechert, J., Gaetgens, C., Wirtz, A., and Frunzke, J. (2020b). Inducible expression systems based on xenogeneic silencing and counter-silencing and design of a metabolic toggle switch. ACS Synth. Biol. 9, 2023–2038. doi: 10.1021/acssynbio.0c00111
Woo, H. M., and Park, J. B. (2014). Recent progress in development of synthetic biology platforms and metabolic engineering of Corynebacterium glutamicum. J. Biotechnol. 180, 43–51. doi: 10.1016/j.jbiotec.2014.03.003
Xu, X., Li, X., Liu, Y., Zhu, Y., Li, J., Du, G., et al. (2020a). Pyruvate-responsive genetic circuits for dynamic control of central metabolism. Nat. Chem. Biol. 16, 1261–1268. doi: 10.1038/s41589-020-0637-3
Xu, J. Z., Ruan, H. Z., Yu, H. B., Liu, L. M., and Zhang, W. (2020b). Metabolic engineering of carbohydrate metabolism systems in Corynebacterium glutamicum for improving the efficiency of L-lysine production from mixed sugar. Microb. Cell Factories 19:39. doi: 10.1186/s12934-020-1294-7
Zhang, C., Li, Y., Zhu, F., Li, Z., Lu, N., Li, Y., et al. (2020). Metabolic engineering of an auto-regulated Corynebacterium glutamicum chassis for biosynthesis of 5-aminolevulinic acid. Bioresour. Technol. 318:124064. doi: 10.1016/j.biortech.2020.124064
Zhang, S., Liu, D., Mao, Z., Mao, Y., Ma, H., Chen, T., et al. (2018). Model-based reconstruction of synthetic promoter library in Corynebacterium glutamicum. Biotechnol. Lett. 40, 819–827. doi: 10.1007/s10529-018-2539-y
Zhao, E. M., Zhang, Y., Mehl, J., Park, H., Lalwani, M. A., Toettcher, J. E., et al. (2018). Optogenetic regulation of engineered cellular metabolism for microbial chemical production. Nature 555, 683–687. doi: 10.1038/nature26141
Keywords: Corynebacterium glutamicum, hyperosmotic stress, inducible promoter, MtrA, fluorescence activated cell sorting, lysine
Citation: Huang J, Chen J, Wang Y, Shi T, Ni X, Pu W, Liu J, Zhou Y, Cai N, Han S, Zheng P and Sun J (2021) Development of a Hyperosmotic Stress Inducible Gene Expression System by Engineering the MtrA/MtrB-Dependent NCgl1418 Promoter in Corynebacterium glutamicum. Front. Microbiol. 12:718511. doi: 10.3389/fmicb.2021.718511
Edited by:
Xiao-Jun Ji, Nanjing Tech University, ChinaCopyright © 2021 Huang, Chen, Wang, Shi, Ni, Pu, Liu, Zhou, Cai, Han, Zheng and Sun. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ping Zheng, emhlbmdfcEB0aWIuY2FzLmNu; Jibin Sun, c3VuX2piQHRpYi5jYXMuY24=
†These authors have contributed equally to this work
 Jingwen Huang1,2†
Jingwen Huang1,2† Jiuzhou Chen
Jiuzhou Chen Yu Wang
Yu Wang Wei Pu
Wei Pu Shuangyan Han
Shuangyan Han Ping Zheng
Ping Zheng Jibin Sun
Jibin Sun