- 1The Second Affiliated Hospital and Yuying Children’s Hospital, Wenzhou Medical University, Wenzhou, China
- 2Key Laboratory of Medical Genetics of Zhejiang Province, Key Laboratory of Laboratory Medicine, Ministry of Education, School of Laboratory Medicine and Life Sciences, Wenzhou Medical University, Wenzhou, China
- 3Institute of Biomedical Informatics, School of Laboratory Medicine and Life Sciences, Wenzhou Medical University, Wenzhou, China
- 4Institute of Translational Medicine, Baotou Central Hospital, Baotou, China
- 5Department of Clinical Laboratory, Zhejiang Hospital, Hangzhou, China
A novel plasmid-encoded aminoglycoside 3''-nucleotidyltransferase ANT(3")-IId, was discovered in Acinetobacter lwoffi strain H7 isolated from a chick on an animal farm in Wenzhou, China. The whole-genome of A. lwoffii H7 consisted of one chromosome and five plasmids (pH7-250, pH7-108, pH7-68, pH7-48, and pH7-11). ant(3")-IId was identified as being encoded on pH7-250, sharing the highest amino acid identity of 50.64% with a function-known resistance gene, ant(3")-IIb (KB849358.1). Susceptibility testing and enzyme kinetic parameter analysis were conducted to determine the function of the aminoglycoside 3"-nucleotidyltransferase. The ant(3")-IId gene conferred resistance to spectinomycin and streptomycin [the minimum inhibitory concentration (MIC) levels of both increased 16-fold compared with the control strain]. Consistent with the MIC data, kinetic analysis revealed a narrow substrate profile including spectinomycin and streptomycin, with Kcat/Km ratios of 4.99 and 4.45×103M−1 S−1, respectively. Sequencing analysis revealed that the ant(3")-IId gene was associated with insertion sequences (IS) element [ΔISAba14-ΔISAba14-hp-orf-orf-orf1-ant(3")-IId], and ant(3")-IId were identified in plasmids from various Acinetobacter species. This study of the novel aminoglycoside 3"-nucleotidyltranferase ANT(3")-IId helps us further understand the functional and sequence characteristics of aminoglycoside 3"-nucleotidyltranferases, highlights the risk of resistance gene transfer among Acinetobacter species and suggests that attention should be given to the emergence of new aminoglycoside 3"-nucleotidyltranferase genes.
Introduction
Acinetobacter lwoffii (formerly Mima polymorpha, or Acinetobacter calcoaceticus var. lwoffii) is a Gram-negative aerobic bacillus that inhabits the oropharynx, skin, and perineum (Rathinavelu et al., 2003). As an opportunistic pathogen, A. lwoffii can cause infections in patients with impaired or compromised immune systems (Ku et al., 2000; Regalado et al., 2009). Aminoglycosides are highly potent, broad-spectrum antibiotics that act through inhibition of bacterial protein synthesis. Their potent bactericidal activity relies upon binding specifically to the 16S rRNA of the 30S ribosomal subunit, thus interfering with protein synthesis (Mehta and Champney, 2003). There are a large number of aminoglycoside antibiotics, and with the long-term overuse of antimicrobials, multidrug-resistant bacteria, including A. lwoffii, have become prevalent worldwide (Mittal et al., 2015). Aminoglycoside resistance mechanisms mainly include methylation of 16S rRNA (Doi et al., 2016), active efflux pumps (Aires et al., 1999), modification of outer membrane permeability or diminished inner membrane transport (Over et al., 2001), and aminoglycoside-modifying enzymes (AMEs; Shaw et al., 1993).
In the clinical setting, resistance to aminoglycosides is most commonly mediated by the presence of various AMEs, including acetyltransferases (AACs), nucleotidyltransferases (ANTs), and phosphotransferases (APHs; Garneau-Tsodikova and Labby, 2016). Among them, ANTs mediate the inactivation of aminoglycosides by catalyzing the transfer of an AMP group from the donor substrate ATP to a hydroxyl group in the aminoglycoside molecule (Ramirez and Tolmasky, 2010). Based on their position specificities for aminoglycoside modification, these enzymes are further divided into several subtypes. There are five classes of ANTs that catalyze adenylation at the 6, 9, 4', 2'' and 3" positions, namely, ANT(6), ANT(9), ANT(4'), ANT(2''), and ANT(3"), respectively (Ramirez and Tolmasky, 2010). The ANT(3") enzymes are the most commonly found ANT enzymes and include two main subclasses (I-II) that specify resistance to spectinomycin and streptomycin. The ant(3")-I genes (also known as aadA genes) exist as gene cassettes and are part of a large number of integrons, plasmids, and transposons (Parent and Roy, 1992; Naas et al., 1999; Peters et al., 2001). There are more than 20 genes encoding the ANT(3")-I-type enzymes, identified as aadA1 through aadA31 (some numbers are missing). To date, a novel subclass of aminoglycoside 3"-nucleotidyltransferase, ANT(3")-II, has been identified in Acinetobacter spp., which comprises numerous variants distributed among three main clades [ANT(3")-IIa, ANT(3")-IIb, and ANT(3")-IIc; Zhang et al., 2017]. The ant(3")-II gene not only conferred phenotypic resistance in a given species but was also frequently horizontally transferred between different Acinetobacter species (Zhang et al., 2017).
In this work, we identified a novel aminoglycoside 3"-nucleotidyltransferase gene encoding an ANT(3")-II-type enzyme, designated ANT(3")-IId, encoded on a plasmid in A. lwoffii isolated from the environment. The function, kinetic parameters and genetic context of the ant(3")-IId gene were characterized.
Materials and Methods
Bacterial Strains
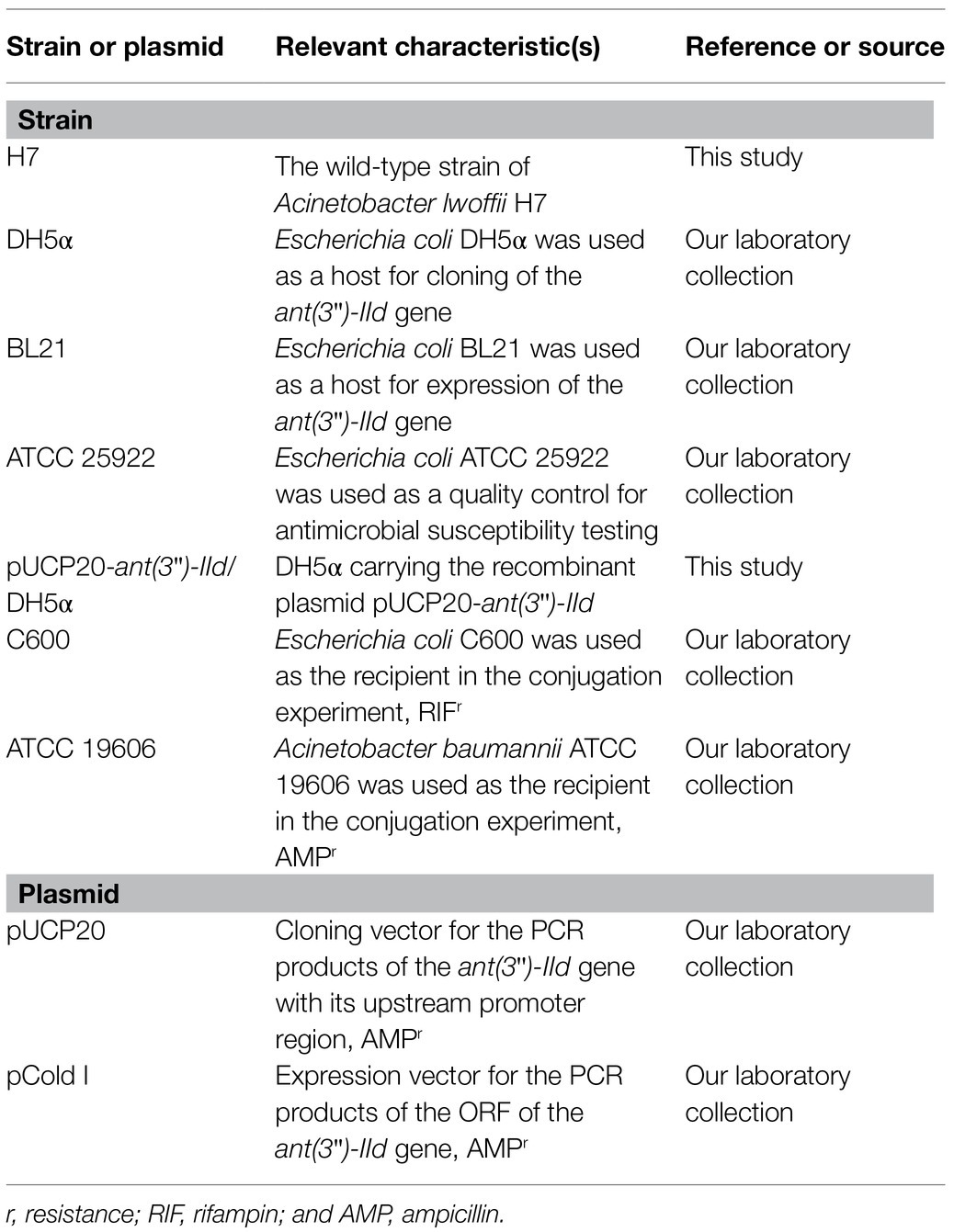
Acinetobacter lwoffii H7 was isolated from an anal swab of a chick from an animal farm in Wenzhou, China. Species identification was initially conducted by the Vitek-60 microorganism auto-analysis system (BioMerieux Corporate, Craponne, France). Further species identification was performed by 16S rRNA gene homology comparison and average nucleotide identity (ANI) analysis using FastANI (Jain et al., 2018). The strains and plasmids used in this work are listed in Table 1.
Antibiotic Susceptibility Testing
The minimum inhibitory concentrations (MICs) were determined using the agar dilution method following the guidelines of the Clinical and Laboratory Standards Institute (CLSI), and the susceptibility patterns were interpreted according to the CLSI breakpoint criteria (CLSI, 2019). The antibiotics tested in this work included 10 aminoglycoside antibiotics (kanamycin, neomycin, ribostamycin, tobramycin, sisomicin, netilmicin, spectinomycin, amikacin, micronomicin, and streptomycin), six β-lactam antibiotics [aztreonam, ceftazidime, cefepime, cefoxitin, meropenem, and ampicillin (AMP)], two quinolone antibiotics (nalidixic acid and ciprofloxacin), two chloramphenicol antibiotics (chloramphenicol and florfenicol), fosfomycin, tetracycline, and polymyxin B. Additionally, as no CLSI breakpoints existed for streptomycin and spectinomycin, so the MIC results for the two antibiotics were interpreted according to the publications by Hu et al. (2017) and Jouybari et al. (2021), respectively. Escherichia coli ATCC 25922 was used as a reference strain for quality control.
Molecular Cloning of the ant(3")-IId Gene
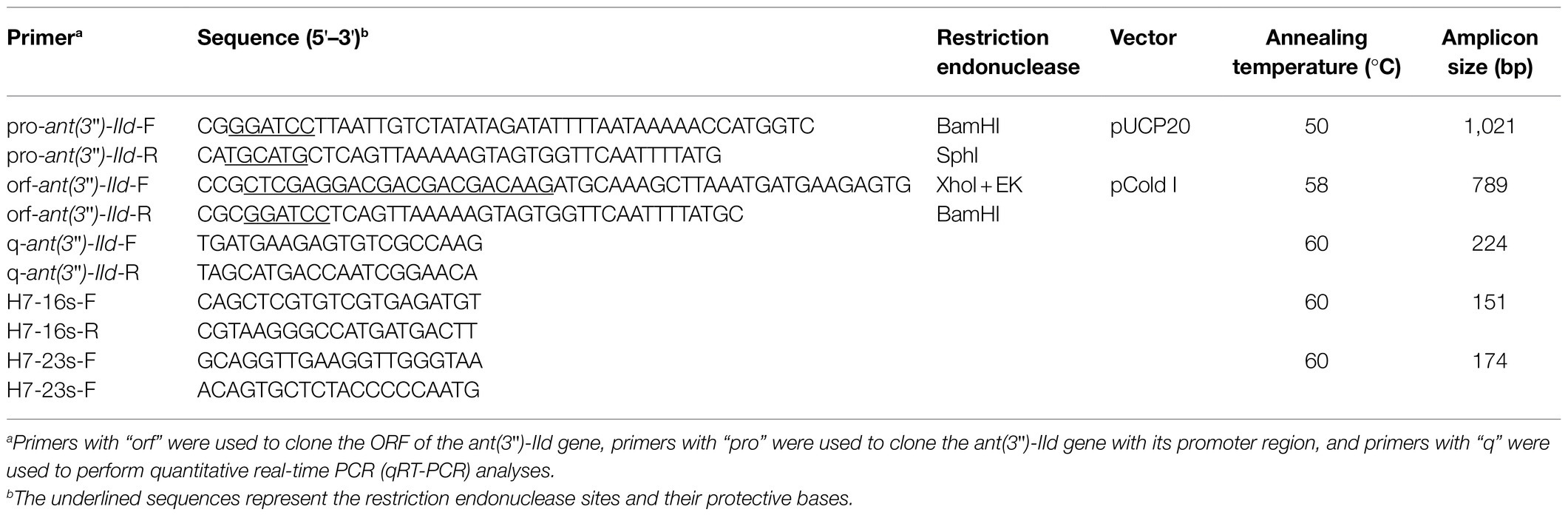
The gene encoding ANT(3")-IId was amplified along with its promoter region by PCR with the primers listed in Table 2. The PCR product was digested with BamHI and SphI and ligated into the pUCP20 vector with a T4 DNA ligase cloning kit (Takara Bio, Inc., Dalian, China). The recombinant plasmid was transformed into competent E. coli DH5α cells by the calcium chloride method, and the transformant was cultured on Luria-Bertani (LB) agar plates supplemented with 100μg/ml AMP. The size and sequence of the cloned insert was confirmed by restriction enzyme digestion and DNA sequencing.
Quantitative RT-PCR Analyses
To analyze the expression level of ANT(3")-IId, overnight cultures of A. lwoffii H7 were diluted in fresh LB with or without supplementation with 1/4 MIC of streptomycin or spectinomycin and grown to mid-log phase. Total RNA was extracted using TRIzol reagent according to the manufacturer’s instructions. RNA purity and concentration were determined spectrophotometrically. DNA-free RNA was confirmed by PCR amplification of the A. lwoffii 16S rRNA and 23S rRNA genes. cDNA was synthesized using the PrimeScript RT-PCR Kit (Takara, Dalian, China), and real-time PCR (RT-PCR) was performed using ChamQ Universal SYBR qPCR Master Mix (Vazyme) according to the manufacturer’s protocol. The primers used for quantitative RT-PCR (qRT-PCR) are listed in Table 2. Relative gene expression was calculated using the 2−ΔΔCT method with 16S rRNA and 23S rRNA as the reference genes.
Plasmid Conjugation Experiment
To detect the transferability of the plasmid pH7-250, E. coli C600 (susceptible to rifampicin at >2,048μg/ml) and Acinetobacter baumannii ATCC 19606 (susceptible to ampicillin at >128μg/ml) were used as the recipients in conjugation experiments using the filter mating method. The transconjugant was selected on LB plate supplemented with rifampin (RIF; 512μg/ml) or ampicillin (128μg/ml) plus spectinomycin (16mg/ml) or ribostamycin (16mg/ml) and incubated overnight at 37°C. The candidate transconjugant was further analyzed by PCR and sequencing for the presence of resistance genes.
Expression and Purification of the ANT(3")-IId Enzyme
ANT(3")-IId was overexpressed from E. coli BL21(DE3)/pCold I-ANT(3")-IId and purified as described previously with some modifications (Zhang et al., 2017). In detail, the ant(3")-IId gene was cloned with an N-terminal His6 tag and enterokinase cleavage site into the pCold I vector under the control of the cspA promoter using the cold-shock system (Qing et al., 2004). Protein expression was induced with 1mM isopropyl-β-D-thiogalactoside (IPTG) when the culture reached an OD600 of 0.6–0.8 at 37°C, and incubation was continued for an additional 16–20h at 16°C. Cells were harvested by centrifugation (5,000×g, 10min) at 4°C, resuspended in lysis buffer (20mM Tris-HCl, 150mM NaCl, 3mM β-mercaptoethanol, 0.5% Nonidet-P-40; pH 8.0; Shi et al., 2015), and disrupted by sonication. Cellular debris was removed by centrifugation (10,000×g, 30min) at 4°C. The lysates were incubated with pre-equilibrated nickel-nitrilotriacetic acid (Ni-NTA) agarose resin (Beyotime Biotechnology, Shanghai, China) for 8h at 4°C with gentle agitation. The mixture containing the recombinant protein was then loaded onto a column and purified using standard Ni-NTA affinity chromatography. The His6 tag was removed by incubation with enterokinase for 3h at 25°C. The digested ANT(3")-IId was purified further using a Ni-NTA column to remove the free His6 tag. The purity of ANT(3")-IId was checked by SDS-PAGE, and the protein concentration was determined spectrophotometrically by using a BCA protein assay kit (Thermo Fisher Scientific, Rockford, IL, United States).
Enzyme Kinetics
The kinetic assay used to monitor activity was performed as reported previously with slight modifications (Kim et al., 2006). The ANT(3")-IId activity was measured by coupling the enzymatic reaction to the reactions of UDP-glucose pyrophosphorylase, phosphoglucomutase, and glucose-6-phosphate dehydrogenase. The catalytic activity of aminoglycoside 3"-nucleotidyltransferase was assayed by monitoring the accumulation of NADPH at 340nm with a Synergy™ Neo2 Multi-Mode Microplate Reader (Biotek, United States). The reaction mixtures contained 50mM HEPES (pH 7.5), 10mM MgCl2, 0.2mM UDP-glucose, 0.2mM glucose 1,6-bisphosphate, 0.2mM NADP, 0.2mM dithiothreitol, 2units/ml UDP-glucose pyrophosphorylase, 20units/ml phosphoglucomutase, 20units/ml glucose-6-phosphate dehydrogenase, 1mM ATP, 2μl of ANT(3")-IId, and variable concentrations of aminoglycoside (1–125μM) in a total volume of 0.2ml. Reactions were initiated by addition of the enzyme.
Genome Sequencing, Assembly, Annotation, and Bioinformatic Analysis
The whole-genomic DNA of A. lwoffii H7 was extracted using an AxyPrep Bacterial Genomic DNA Miniprep kit (Axygen Biosciences, Union City, CA, United States). Whole-genome sequencing was achieved using the Illumina HiSeq-2500 and PacBio RS II platforms by Shanghai Personal Biotechnology Co., Ltd. (Shanghai, China). The PacBio long reads were initially assembled by SPAdes v3.14.1 (Bankevich et al., 2012) and Canu v2.1 (Koren et al., 2017). Further correction was conducted by using Pilon (Walker et al., 2014) to improve assembly quality through mapping short reads aligned to the draft of the whole-genome assembly. The ORFs were predicted and annotated using Prokka v1.14.0 (Seemann, 2014) and further annotated by DIAMOND (Buchfink et al., 2015) against the UniProtKB/Swiss-Prot and NCBI nonredundant protein databases with an e-value threshold of 1e-5. Annotation of resistance genes was performed using Resistance Gene Identifier (RGI) v4.0.3 in the Comprehensive Antibiotic Resistance Database (CARD; Mcarthur et al., 2013). The molecular weight and pI value of ANT(3")-IId were predicted using ProtParam (Bohm et al., 2020). GView was used to construct basic genomic features (Petkau et al., 2010). Annotation of MGEs was performed using Isfinder (Siguier et al., 2006) and INTEGRALL (Moura et al., 2009). GenoPlotR was used to generate the figure showing structural comparisons and the nucleotide identities between several segments in a linear fashion (Guy et al., 2010). Comparisons of the nucleotide sequences were performed using BLASTN. Multiple sequence alignment and neighbor-joining phylogenetic tree construction were performed using the MAFFT program and MEGAX with a bootstrap value of 1,000 replicates, respectively (Katoh and Standley, 2013; Kumar et al., 2018). Protein sequence motifs of ANT(3")-IId were determined using the MEME Suite.1 A predicted amino acid (aa) sequence pairwise identity matrix was generated using MatGAT (Campanella et al., 2003).
Nucleotide Sequence Accession Numbers
The nucleotide sequences of the chromosome, five plasmids (pH7-250, pH7-108, pH7-68, pH7-48, and pH7-11) of A. lwoffii H7 and the ant(3")-IId gene reported in this study have been deposited in GenBank under accession numbers CP072549, CP072550, CP072551, CP072552, CP072553, CP072554, and MW984426, respectively.
Results and Discussion
Characteristics and the Resistance Profile of A. lwoffii H7
The 16S rRNA gene homology analysis suggested that the 16S rRNA gene of H7 had the closest relationship with that of A. lwoffii JCM6840 (NR_113346.1, with 95.00% coverage and 99.52% identity), and the ANI analysis revealed that the chromosome sequences of A. lwoffii 12CE1 (NZ_CP059081.1) shared the highest identity of 95.71% with H7. Finally, we grouped the strain into the species A. lwoffii and named it A. lwoffii H7.
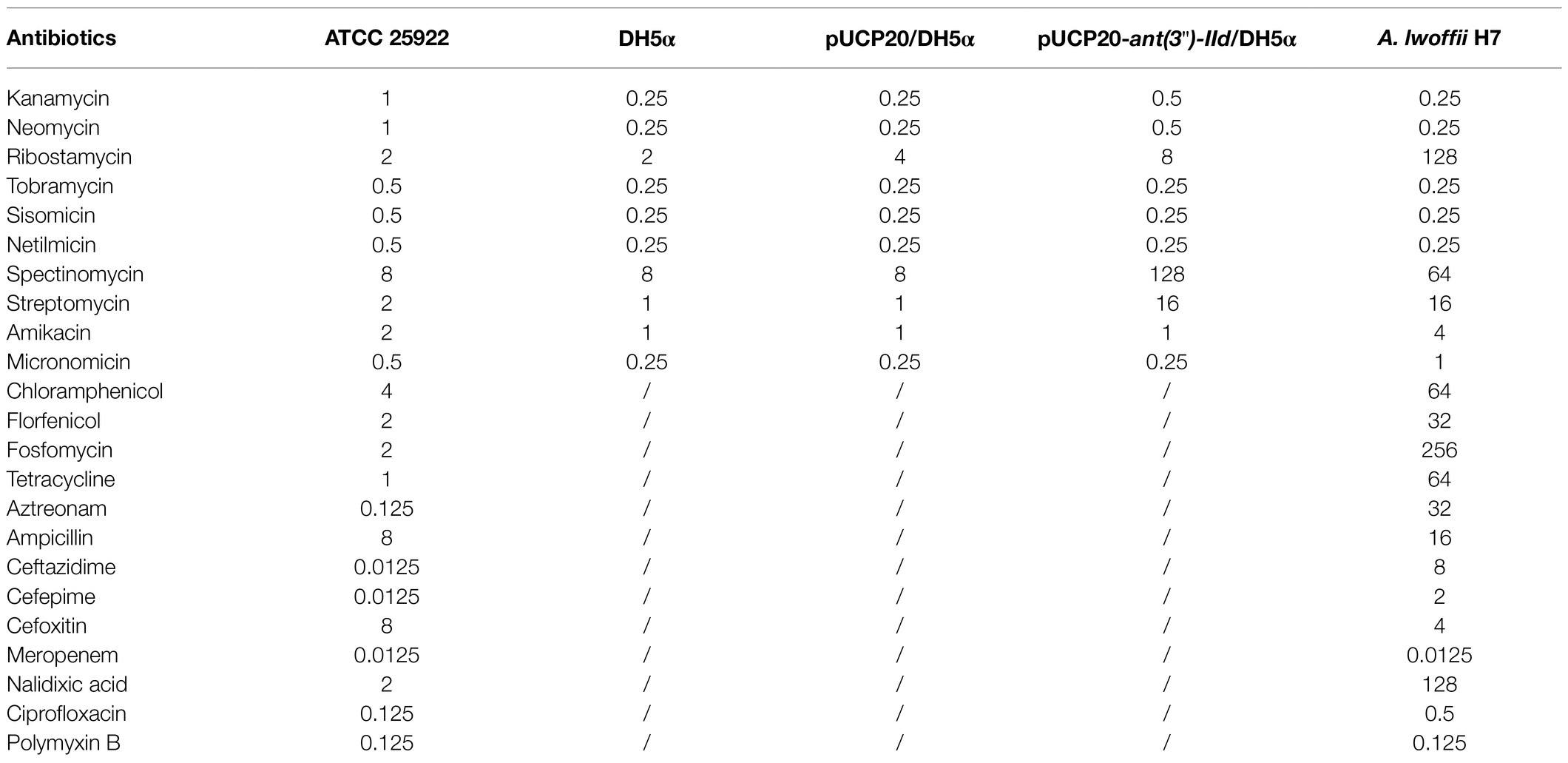
The in vitro susceptibility test showed that A. lwoffii H7 exhibited resistance to eight of the 23 antibiotics tested. It showed higher MIC levels (≥64μg/ml) for ribostamycin, spectinomycin, tetracycline, chloramphenicol, fosfomycin, and nalidixic acid. It was susceptible to many other antibiotics, including aminoglycosides (such as kanamycin, neomycin, and tobramycin) and all the β-lactams tested (except aztreonam; Table 3).
General Features of A. lwoffii H7
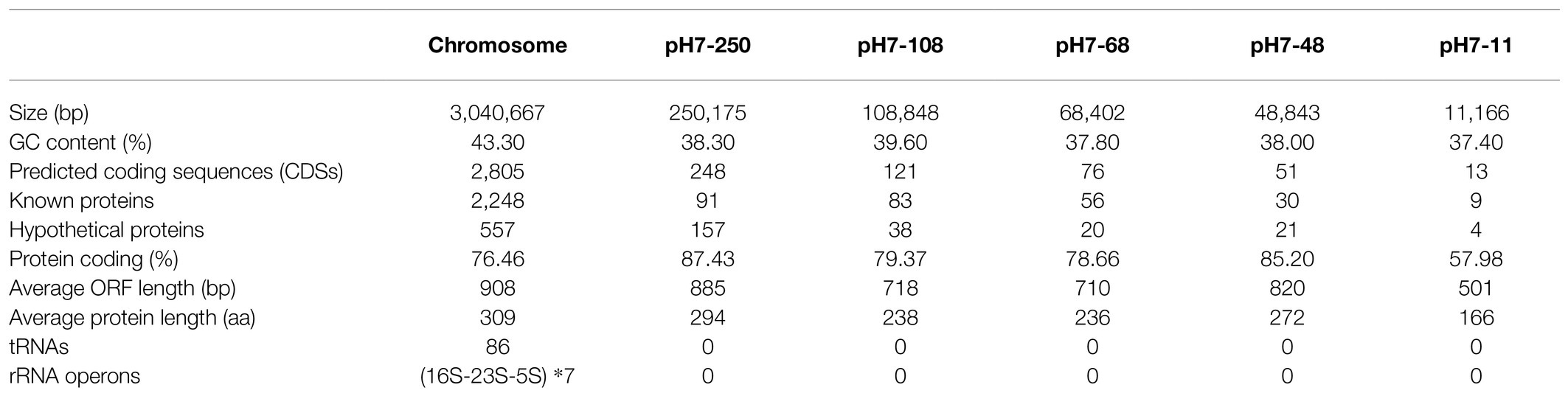
To better understand the molecular mechanism of drug resistance, the complete genome sequence of H7 was determined. The whole-genome consisted of a chromosome and five plasmids, designated pH7-250, pH7-108, pH7-68, pH7-48, and pH7-11 (Table 4). The chromosome of H7 was 3,040,667 in length, with 43.3% GC content, and encoded 2,805 coding sequences (CDSs). A total of 14 genes with ≥80% similarity with the known antibiotic resistance genes (ARGs) were identified in the whole genome, of which three ARGs were located on the chromosome, 10 and 1 were located on the plasmids pH7-250 and pH7-48, respectively (Supplementary Table S3). When analyzing the resistance mechanism of the isolate against the aminoglycoside antibiotic (spectinomycin), we found an insert sequence (IS) element-related putative ant(3")-II gene (finally named ant(3")-IId in this work) encoded on plasmid pH7-250 and then cloned it to confirm whether it was functional.
ANT(3")-IId Confers Resistance to Streptomycin and Spectinomycin
Among the resistance genes of known function, ant(3")-IId showed the highest aa sequence identity (50.64%) with the aminoglycoside 3"-nucleotidyltransferase ANT(3")-IId (ENU91137.1). Of the 10 aminoglycoside antibiotics tested and compared with the control (E. coli DH5α harboring the vector pUCP20 only), the recombinant clone with ant(3")-IId [pUCP20-ant(3")-IId/E. coli DH5α] increased the MIC levels of both streptomycin and spectinomycin by 16-fold, while no significant increase in MIC level was identified for the other aminoglycosides (Table 3).
The qRT-PCR results revealed that ant(3")-IId was transcriptionally active under growth in LB free of antibiotics, and treatment with streptomycin or spectinomycin did not result in any significant fluctuation in the mRNA abundance of ant(3")-IId (data not shown).
The 3"-nucleotidyltransferase activity and kinetic parameters of ANT(3")-IId against aminoglycosides were in accordance with the MIC results of the cloned ant(3")-IId. This enzyme adenylated streptomycin and spectinomycin with Kcat/Km ratios of 4.99 and 4.45×103M−1 S−1, respectively.
Comparative Analysis of ANT(3")-IId
The ant(3")-IId gene is 789bp in length and encodes a 262-amino-acid protein with a molecular mass of 30.38kDa and a pI value of 5.01. It showed an overall identity of 63.7–66.0% with the currently known ANT(3")-II family enzymes. A phylogenetic tree containing ANT(3")-IId and other identified ANTs, including ANT(2'), ANT(4), ANT(6), ANT(9), and ANT(3"), the sequences of which were collected from the GenBank database and previous publications (Shaw et al., 1993; Ramirez and Tolmasky, 2010), revealed that this protein clustered closest to a branch composed of ANT(3")-IIa, ANT(3")-IIb, ANT(3")-IIc, and AadA27 (Figure 1). Sequence comparison assays revealed that this protein shared 50.64, 48.99, 48.40, and 48.00% identity with the four previously reported ANT(3")-II enzymes ANT(3")-IIb, ANT(3")-IIa, AadA27, and ANT(3")-IIc, respectively (Figure 2). Therefore, ANT(3")-IId was assigned as a novel lineage of the ANT(3")-II family.
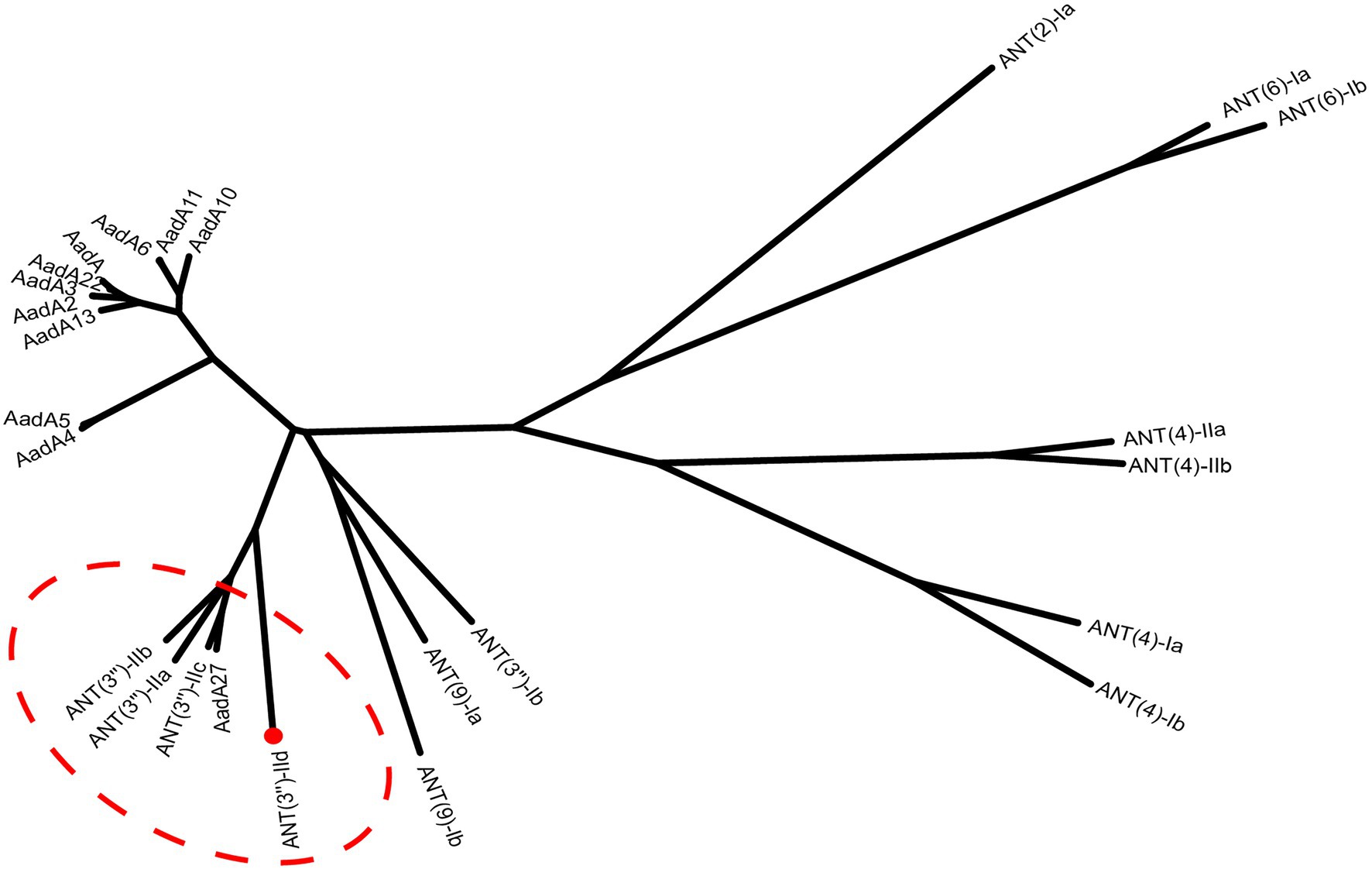
Figure 1. A phylogenetic tree showing the relationship of ANT(3")-IId with other functionally characterized ANTs. ANT(3")-II enzymes are highlighted with a red dashed ellipse. ANT(3")-IId from our study is highlighted with a red filled circle and shown with a red dot.
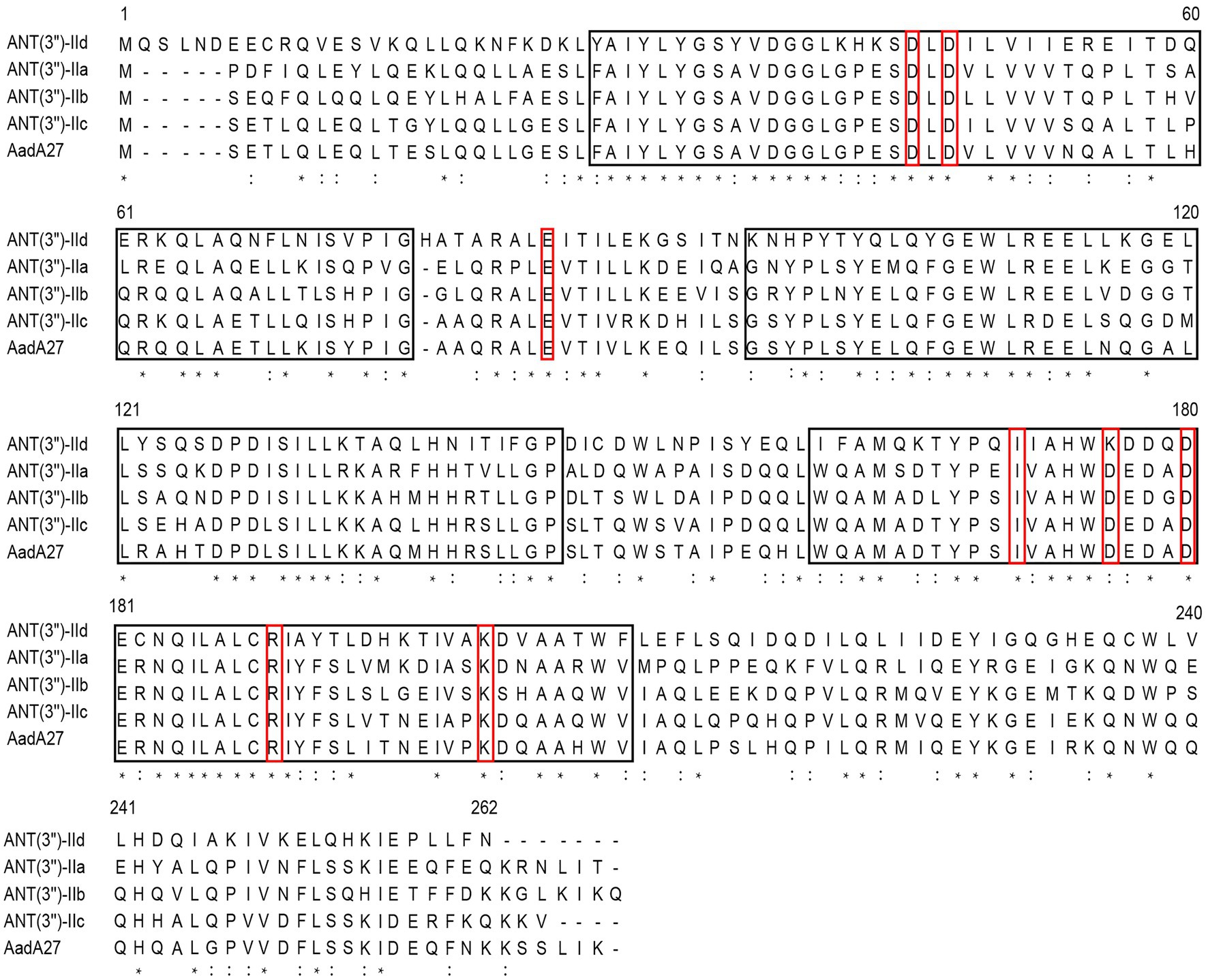
Figure 2. Sequence comparison assay of ANT(3")-II subgroup proteins. The ANT(3")-II proteins and their accession numbers: ANT(3")-IId (this work), ANT(3")-IIa (EEX02086.1), ANT(3")-IIb (ENU91137.1), ANT(3")-IIc (ENU37733.1), and AadA27 (CTQ57092.1). The asterisks indicate fully conserved residues; exclamation marks indicate strongly similar residues. The black frames are the conserved motif sites predicted by the MEME program, and the red frames are some functional positions that have been studied. The numbers correspond to the amino acid residues in each full-length protein.
By searching for homologous genes of ant(3")-IId in the NCBI nucleotide database, a total of 15 sequences were retrieved. Notably, these 15 gene sequences were identical to ant(3")-IId (100% identity and 100% coverage). Similar to ant(3")-IId of A. lwoffii from Zhejiang Province, China, these 15 sequences were all from the genus Acinetobacter, i.e., Acinetobacter indicus (4), Acinetobacter schindleri (3), Acinetobacter towneri (4), A. baumannii (1), Acinetobacter pisocicola (1), and (2) from two Acinetobacter spp. strains, in addition to A. lwoffii H7 from this work. All of them were isolated from four different provinces (Guangdong, Henan, Shanxi, and Jiangsu), China (Supplementary Table S1).
Encoded on the plasmids, the ant(3")-IId genes were related to the IS elements. Different insert sequences (such as ISAba21 and ISAba14) were identified next to or in the proximal region of the ant(3")-IId gene. In the upstream region of ant(3")-IId of this work were two truncated insert sequences (ΔISAba14-ΔISAba14), and the gene array of ΔISAba14-ΔISAba14-hp-orf-orf-orf1-ant(3")-IId was flanked by a pair of 7-bp perfect direct repeats (DRs), which was characteristic of a typical transposon structure. The different MGEs related to ant(3")-IId were identified in the plasmids from various Acinetobacter species (Figure 3).
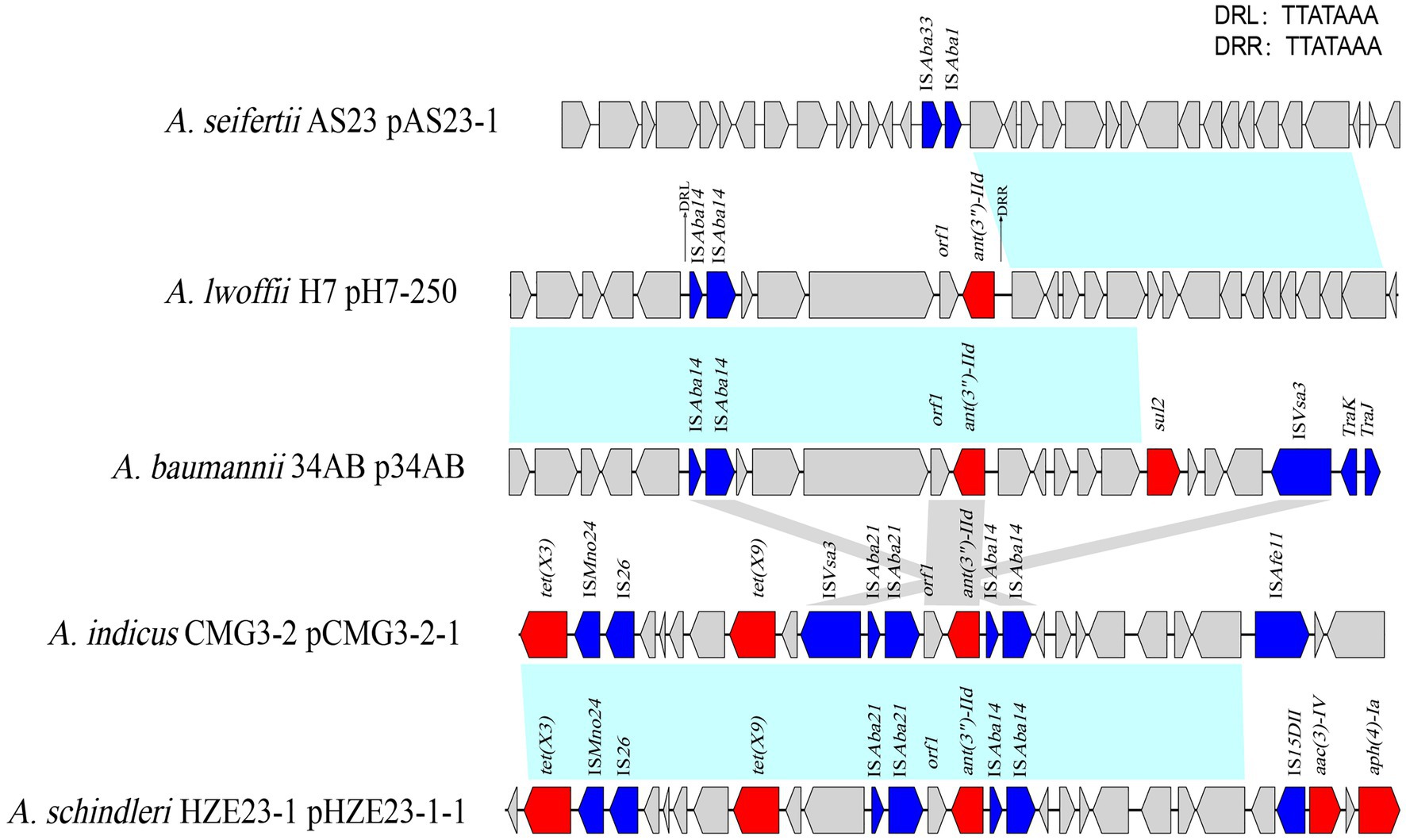
Figure 3. Comparative analysis of the genomic context of the ant(3")-IId gene-related region. The ant(3")-IId genes in different species were compared. Genes are denoted by arrows and colored according to gene function classification. Blue shading denotes regions of homology (>80% nucleotide sequence identity). The detected direct repeat sequence is marked in the upper right corner. The accession numbers of the sequences are as follows: Acinetobacter seifertii plasmid pAS23-1 (CP061673.1), Acinetobacter baumannii plasmid p34AB (NZ_MK134375.1), Acinetobacter indicus plasmid pCMG3-2-1 (CP044446.1), and Acinetobacter schindleri plasmid pHZE23-1-1 (CP044464.1).
Comparative Genomic Analysis of the Plasmid pH7-250
In addition to ant(3")-IId, 10 other known resistance genes (similarity ≥80%) were identified as being encoded on the plasmid pH7-250, including two aminoglycoside resistance genes [aph(3')-VIa and aac(6')-Ib9], one β-lactam resistance gene (blaPER-1), one chloramphenicol/florfenicol resistance gene (floR), one truncated quaternary ammonium compound resistance gene (qacEΔ1), two sulfanilamide resistance genes (sul1), one rifampicin resistance gene (arr-3), and two macrolide resistance genes (mphE and msrE). These resistance genes were clustered in two regions (designated resistance regions A and B), with floR, mphE, and msrE in one region (7.1kb in size) and the other eight in the other region (39.9kb in size). All the resistance genes were related to MGEs (Figure 4). In resistance region A, mphE and msrE were surrounded by two inversely oriented pdif (XerC-XerD) sites (Blackwell and Hall, 2017), and floR was near ISAlw4. In resistance region B, as mentioned above, ant(3")-IId was related to a transposon, and aph(3')-VIa was flanked by a part of the intact insert sequence ISAba125, while the other four resistance genes were carried by a typical class 1 integron with a complete 5'-CS (intI1), 3'-CS region (qacEΔ1/sul1), and variable region [arr-3/aac(6')-Ib9]. Fifteen plasmid genome sequences with the highest similarities with the pH7-250 genome (>80.0% coverage and >90.0% identity), including p34AB of A. baumannii (NZ_MK134375.1, 89% coverage and 99.69% identity), pOXA58_010055 of Acinetobacter sp. (NZ_CP032285.1, 93% coverage and 99.81% identity) and pAHTJR1 of Acinetobacter haemolyticus (CP038010.1, 91% coverage and 99.73% identity), were retrieved from the NCBI nucleotide database (Figure 4; Supplementary Table S2). Of these plasmid sequences, only p34AB of A. baumannii also had the ant(3")-IId gene. The complete class 1 integron structure with the variable region [arr-3/aac(6')-Ib9] on pH7-250 and the MGEs carrying aph(3')-VIa were identical to those on pOXA58_010055 and pAHTJR1. The msrE-mphE dif module was also found in these three plasmids. Additionally, some type IV secretion system (T4SS) proteins, including DotG/IcmE/VirB10, DotI-like, DotD/TraH, T4SS_TraI, DotA/TraY and TadA, and plasmid-partitioning proteins, including ParM/StbA and ParB/RepB/Spo0J, were predicted in pH7-250. Unfortunately, the plasmid failed to be transferred into recipient cells through conjugation.
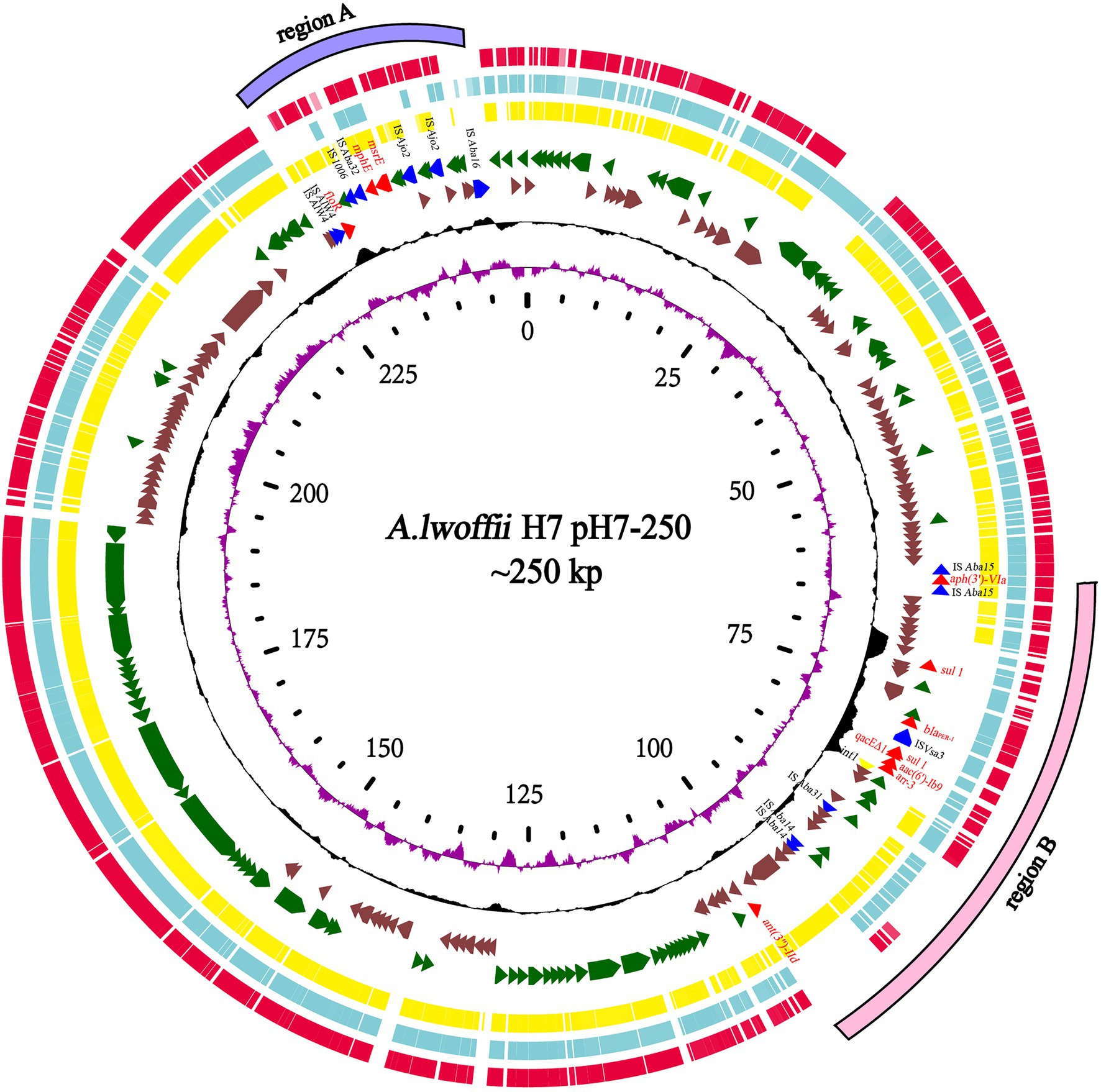
Figure 4. Genomic comparison of the Acinetobacter lwoffii H7 plasmid pH7-250 with other plasmids. From outside to inside: circle 1 shows two regions of resistance genes (region A in purple and region B in pink); circles 2, 3, and 4 are homologous regions of the Acinetobacter haemolyticus TJR01 plasmid pAHTJR1 (CP038010.1), Acinetobacter sp. WCHA55 plasmid pOXA58_010055(NZ_CP032285.1), and A. baumannii 34AB plasmid p34AB (NZ_MK134375.1) compared to those of A. lwoffii H7, while unmatched regions are left blank; circles 5 and 6 display predicted ORFs encoded in the forward strand and reverse strand, respectively; circles 7 and 8 represent the GC content and GC skew, respectively; and circle 9 shows the scale in kb.
Discussion
In this work, a novel aminoglycoside 3"-nucleotidyltransferase gene designated ant(3")-IId, showing the ability to confer resistance to streptomycin and spectinomycin, was identified as being encoded in a plasmid from A. lwoffii H7 isolated from chick. ANT(3") enzymes include two main subclasses (I-II) that confer specific resistance to streptomycin and spectinomycin by adenylating the 3'-OH position of the streptomycin glucosamine ring and the 9'-OH position of the spectinomycin actinamine ring (Ramirez and Tolmasky, 2010). In addition to ANT(3")-IId, two other aminoglycoside 3"-nucleotidyltransferases with the highest aa sequence identity with ANT(3")-IId, ANT(3")-IIb (ENU91137.1, 50.64%), and ANT(3")-IIa (EEX02086.1, 48.99%), also showed resistance to the two antibiotics. Compared with the controls, the recombinant carrying ant(3")-IId showed 16-fold increased MIC levels for both streptomycin and spectinomycin (from 1 to 16μg/ml and 8 to 128μg/ml, respectively), while both ANT(3")-IIb and ANT(3")-IIa increased the MIC levels (≥64-fold) of both streptomycin and spectinomycin [ANT(3")-IIb, from 4 to 256μg/ml and 32 to 2048μg/ml; ANT(3")-IIa, from 4 to 512μg/ml and 32 to 2048μg/ml, respectively; Zhang et al., 2017].
It has been reported that the four aa residues E87, W112, D182, and 185H/N and the other two residues W173 and D178 of AadA (CAA48215.1, an aminoglycoside adenylyl transferase with a known structural mechanism) were verified to be determinants of spectinomycin and streptomycin resistance, respectively, and this protein shared 34.30% amino acid sequence identity with ANT(3")-IId. These two proteins had the first four residues (E87, W112, D182, and 185H/N) in common but varied in the last two residues (W173I and D178K; Stern et al., 2018). The publication described the construction of the chromosomal mutants W173A and D178A in the aadA gene, and the generated strains were subjected to in vivo MIC tests with streptomycin and spectinomycin. The MIC values for the two mutants were reduced 10- and 5-fold for streptomycin (from 128 to 12μg/ml and 24μg/ml, respectively) but remained close to the WT MIC values for spectinomycin (from 192 to 192μg/ml and 128μg/ml, respectively; Stern et al., 2018). Therefore, the low resistance levels to streptomycin and spectinomycin and the corresponding low kcat/Km ratio of ANT(3")-IId might be the result of aa residues variations (most likely W173I and D178K) in commonly conserved loci of the functional domains, and this remains to be clarified in future studies.
The ant(3")-IId genes identified in this work or found in the other 15 Acinetobacter strains were all encoded on the plasmids. None were found to be encoded on the chromosome. When analyzing the plasmid group of these plasmids, to our surprise, no rep gene was predicted in any of them. These ant(3")-IId genes were all related to the MGEs of the different structures. Except for the one on the plasmid p34AB of A. baumannii, which shared the same sequence structure ΔISAba14-ΔISAba14-hp-orf-orf-orf1-ant(3″)-IId with pH7-250 of A. lwoffii H7 in this work, the others all showed different sequence structures from each other. lSAba14 and lSAba21 both belong to the same IS3 family, and this structure of the left-end extremity of ISAba21 and the right-end extremity of ISAba14 was first reported in A. baumannii, which formed a composite transposon named Tn2114 at the origin of acquisition of blaRTG-5 (Bonnin et al., 2012). Because plasmid-carried resistance genes generally originate from bacterial chromosomes, the origin of this plasmid-encoded resistance gene remained unknown, even though it appeared in different Acinetobacter species through horizontal gene transfer.
Conclusion
In this work, we reported the complete sequence and function of a novel aminoglycoside 3"-nucleotidyltransferase gene, ant(3")-IId, present on pH7-250 in A. lwoffii H7. The plasmid pH7-250, with an unknown maintenance mechanism, harbors 11 ARGs. The ant(3")-IId gene is located inside a composite transposon structure, which has been found in other Acinetobacter strains, suggesting a potential threat in the future because of its high horizontal transfer capability.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: The datasets presented in this study can be found in the GenBank database as GenBank: CP072549, CP072550, CP072551, CP072552, CP072553, and CP072554 for the chromosome and five plasmids (pH7-250, pH7-108, pH7-68, pH7-48, and pH7-11) of A. lwoffii H7 genome sequence, GenBank: MW984426 for the ant(3")-IId gene.
Author Contributions
JLL, KZ, QL, PZ, HML, XZ, and HZ collected the strains and performed the experiments. KZ, HLL, JWL, XL, and KL analyzed the experimental results. JLL, XD, and TX performed the bioinformatics analysis. JLL, KZ, and QB wrote the manuscript. MZ, YH, and PR designed the work. All authors contributed to the article and approved the submitted version.
Funding
This study was supported by Zhejiang Provincial Natural Science Foundation of China (LY19C060002 and LQ17H190001), the National Natural Science Foundation of China (81960381 and 81973382), and the Science and Technology Project of Inner Mongolia Autonomous Region China (201802125).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors would like to acknowledge all study participants and individuals who contributed to this study.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.728216/full#supplementary-material
Footnotes
References
Aires, J. R., Kohler, T., Nikaido, H., and Plesiat, P. (1999). Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 43, 2624–2628. doi: 10.1128/AAC.43.11.2624
Bankevich, A., Nurk, S., Antipov, D., Gurevich, A. A., Dvorkin, M., Kulikov, A. S., et al. (2012). SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J. Comput. Biol. 19, 455–477. doi: 10.1089/cmb.2012.0021
Blackwell, G. A., and Hall, R. M. (2017). The tet39 determinant and the msrE-mphE genes in acinetobacter plasmids are Each part of discrete modules flanked by inversely oriented pdif (XerC-XerD) sites. Antimicrob. Agents Chemother. 61, e00780–e00717. doi: 10.1128/AAC.00780-17
Bohm, M. E., Razavi, M., Marathe, N. P., Flach, C. F., and Larsson, D. G. J. (2020). Discovery of a novel integron-borne aminoglycoside resistance gene present in clinical pathogens by screening environmental bacterial communities. Microbiome 8:41. doi: 10.1186/s40168-020-00814-z
Bonnin, R. A., Poirel, L., and Nordmann, P. (2012). A novel and hybrid composite transposon at the origin of acquisition of Bla(RTG-5) in Acinetobacter baumannii. Int. J. Antimicrob. Agents 40, 257–259. doi: 10.1016/j.ijantimicag.2012.04.002
Buchfink, B., Xie, C., and Huson, D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Campanella, J. J., Bitincka, L., and Smalley, J. (2003). MatGAT: an application that generates similarity/identity matrices using protein or DNA sequences. BMC Bioinform. 4:29. doi: 10.1186/1471-2105-4-29
CLSI (2019). Performance Standards for Antimicrobial Susceptibility Testing, CLSI Supplement M100. 29th Edn. Wayne, PA: Clinical and Laboratory Standards Institute.
Doi, Y., Wachino, J. I., and Arakawa, Y. (2016). Aminoglycoside resistance: the emergence of acquired 16S ribosomal RNA methyltransferases. Infect. Dis. Clin. N. Am. 30, 523–537. doi: 10.1016/j.idc.2016.02.011
Garneau-Tsodikova, S., and Labby, K. J. (2016). Mechanisms of resistance to aminoglycoside antibiotics: overview and perspectives. MedChemComm 7, 11–27. doi: 10.1039/C5MD00344J
Guy, L., Kultima, J. R., and Andersson, S. G. (2010). genoPlotR: comparative gene and genome visualization in R. Bioinformatics 26, 2334–2335. doi: 10.1093/bioinformatics/btq413
Hu, Y., Liu, L., Zhang, X., Feng, Y., and Zong, Z. (2017). In vitro activity of neomycin, streptomycin, paromomycin and apramycin against carbapenem-resistant enterobacteriaceae clinical strains. Front. Microbiol. 8:2275. doi: 10.3389/fmicb.2017.02275
Jain, C., Rodriguez, R. L., Phillippy, A. M., Konstantinidis, K. T., and Aluru, S. (2018). High throughput ANI analysis of 90K prokaryotic genomes reveals clear species boundaries. Nat. Commun. 9:5114. doi: 10.1038/s41467-018-07641-9
Jouybari, M. A., Ahanjan, M., Mirzaei, B., and Goli, H. R. (2021). Role of aminoglycoside-modifying enzymes and 16S rRNA methylase (ArmA) in resistance of Acinetobacter baumannii clinical isolates against aminoglycosides. Rev. Soc. Bras. Med. Trop. 54:e05992020. doi: 10.1590/0037-8682-0599-2020
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kim, C., Hesek, D., Zajicek, J., Vakulenko, S. B., and Mobashery, S. (2006). Characterization of the bifunctional aminoglycoside-modifying enzyme ANT(3″)-ii/AAC(6′)-IId from Serratia marcescens. Biochemistry 45, 8368–8377. doi: 10.1021/bi060723g
Koren, S., Walenz, B. P., Berlin, K., Miller, J. R., Bergman, N. H., and Phillippy, A. M. (2017). Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 27, 722–736. doi: 10.1101/gr.215087.116
Ku, S. C., Hsueh, P. R., Yang, P. C., and Luh, K. T. (2000). Clinical and microbiological characteristics of bacteremia caused by Acinetobacter lwoffii. Eur. J. Clin. Microbiol. Infect. Dis. 19, 501–505. doi: 10.1007/s100960000315
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi: 10.1093/molbev/msy096
Mcarthur, A. G., Waglechner, N., Nizam, F., Yan, A., Azad, M. A., Baylay, A. J., et al. (2013). The comprehensive antibiotic resistance database. Antimicrob. Agents Chemother. 57, 3348–3357. doi: 10.1128/AAC.00419-13
Mehta, R., and Champney, W. S. (2003). Neomycin and paromomycin inhibit 30S ribosomal subunit assembly in Staphylococcus aureus. Curr. Microbiol. 47, 237–243. doi: 10.1007/s00284-002-3945-9
Mittal, S., Sharma, M., Yadav, A., Bala, K., and Chaudhary, U. (2015). Acinetobacter lwoffii an emerging pathogen in neonatal ICU. Infect. Disord. Drug Targets 15, 184–188. doi: 10.2174/1871526515666150826114745
Moura, A., Soares, M., Pereira, C., Leitao, N., Henriques, I., and Correia, A. (2009). INTEGRALL: a database and search engine for integrons, integrases and gene cassettes. Bioinformatics 25, 1096–1098. doi: 10.1093/bioinformatics/btp105
Naas, T., Poirel, L., and Nordmann, P. (1999). Molecular characterisation of In51, a class 1 integron containing a novel aminoglycoside adenylyltransferase gene cassette, aadA6, in Pseudomonas aeruginosa. Biochim. Biophys. Acta 1489, 445–451.
Over, U., Gur, D., Unal, S., and Miller, G. H., Aminoglycoside Resistance Study Group (2001). The changing nature of aminoglycoside resistance mechanisms and prevalence of newly recognized resistance mechanisms in Turkey. Clin. Microbiol. Infect. 7, 470–478. doi: 10.1046/j.1198-743x.2001.00284.x
Parent, R., and Roy, P. H. (1992). The chloramphenicol acetyltransferase gene of Tn2424: a new breed of cat. J. Bacteriol. 174, 2891–2897. doi: 10.1128/jb.174.9.2891-2897.1992
Peters, E. D., Leverstein-Van Hall, M. A., Box, A. T., Verhoef, J., and Fluit, A. C. (2001). Novel gene cassettes and integrons. Antimicrob. Agents Chemother. 45, 2961–2964. doi: 10.1128/AAC.45.10.2961-2964.2001
Petkau, A., Stuart-Edwards, M., Stothard, P., and Van Domselaar, G. (2010). Interactive microbial genome visualization with GView. Bioinformatics 26, 3125–3126. doi: 10.1093/bioinformatics/btq588
Qing, G., Ma, L. C., Khorchid, A., Swapna, G. V., Mal, T. K., Takayama, M. M., et al. (2004). Cold-shock induced high-yield protein production in Escherichia coli. Nat. Biotechnol. 22, 877–882. doi: 10.1038/nbt984
Ramirez, M. S., and Tolmasky, M. E. (2010). Aminoglycoside modifying enzymes. Drug Resist. Updat. 13, 151–171. doi: 10.1016/j.drup.2010.08.003
Rathinavelu, S., Zavros, Y., and Merchant, J. L. (2003). Acinetobacter lwoffii infection and gastritis. Microbes Infect. 5, 651–657. doi: 10.1016/S1286-4579(03)00099-6
Regalado, N. G., Martin, G., and Antony, S. J. (2009). Acinetobacter lwoffii: bacteremia associated with acute gastroenteritis. Travel Med. Infect. Dis. 7, 316–317. doi: 10.1016/j.tmaid.2009.06.001
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Shaw, K. J., Rather, P. N., Hare, R. S., and Miller, G. H. (1993). Molecular genetics of aminoglycoside resistance genes and familial relationships of the aminoglycoside-modifying enzymes. Microbiol. Rev. 57, 138–163. doi: 10.1128/mr.57.1.138-163.1993
Shi, J., Jin, Y., Bian, T., Li, K., Sun, Z., Cheng, Z., et al. (2015). SuhB is a novel ribosome associated protein that regulates expression of MexXY by modulating ribosome stalling in Pseudomonas aeruginosa. Mol. Microbiol. 98, 370–383. doi: 10.1111/mmi.13126
Siguier, P., Perochon, J., Lestrade, L., Mahillon, J., and Chandler, M. (2006). ISfinder: the reference centre for bacterial insertion sequences. Nucleic Acids Res. 34, D32–D36. doi: 10.1093/nar/gkj014
Stern, A. L., Van Der Verren, S. E., Kanchugal, P. S., Nasvall, J., Gutierrez-De-Teran, H., and Selmer, M. (2018). Structural mechanism of AadA, a dual-specificity aminoglycoside adenylyltransferase from Salmonella enterica. J. Biol. Chem. 293, 11481–11490. doi: 10.1074/jbc.RA118.003989
Walker, B. J., Abeel, T., Shea, T., Priest, M., Abouelliel, A., Sakthikumar, S., et al. (2014). Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One 9:e112963. doi: 10.1371/journal.pone.0112963
Zhang, G., Leclercq, S. O., Tian, J., Wang, C., Yahara, K., Ai, G., et al. (2017). A new subclass of intrinsic aminoglycoside nucleotidyltransferases, ANT(3")-II, is horizontally transferred among Acinetobacter spp. by homologous recombination. PLoS Genet. 13:e1006602. doi: 10.1371/journal.pgen.1006602
Keywords: ANT(3")-IId, aminoglycoside-modifying enzyme, aminoglycoside 3"-nucleotidyltranferase, Acinetobacter lwoffii, resistance
Citation: Liang J, Zhou K, Li Q, Dong X, Zhang P, Liu H, Lin H, Zhang X, Lu J, Lin X, Li K, Xu T, Zhang H, Bao Q, Zhu M, Hu Y and Ren P (2021) Identification and Characterization of a Novel Aminoglycoside 3''-Nucleotidyltransferase, ANT(3'')-IId, From Acinetobacter lwoffii . Front. Microbiol. 12:728216. doi: 10.3389/fmicb.2021.728216
Edited by:
Benjamin Andrew Evans, University of East Anglia, United KingdomReviewed by:
Dana Marshall, Meharry Medical College, United StatesZhongming Ge, Massachusetts Institute of Technology, United States
Copyright © 2021 Liang, Zhou, Li, Dong, Zhang, Liu, Lin, Zhang, Lu, Lin, Li, Xu, Zhang, Bao, Zhu, Hu and Ren. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mei Zhu, emh1bWVpX2RAMTYzLmNvbQ==; Yunliang Hu, aHV5dW5saWFuZzY2QDE2My5jb20=; Ping Ren, NDQzMjgwNjhAcXEuY29t
†These authors have contributed equally to this work
 Jialei Liang
Jialei Liang Kexin Zhou
Kexin Zhou Qiaoling Li1,2†
Qiaoling Li1,2† Xu Dong
Xu Dong Peiyao Zhang
Peiyao Zhang Hongmao Liu
Hongmao Liu Hailong Lin
Hailong Lin Junwan Lu
Junwan Lu Xi Lin
Xi Lin Kewei Li
Kewei Li Teng Xu
Teng Xu Hailin Zhang
Hailin Zhang Qiyu Bao
Qiyu Bao Mei Zhu
Mei Zhu Ping Ren
Ping Ren