- 1Agriculture Victoria Research, AgriBio, Centre for AgriBioscience, Bundoora, VIC, Australia
- 2School of Applied Systems Biology, La Trobe University, Bundoora, VIC, Australia
- 3Agriculture Victoria Research, Australian Grains Genebank, Horsham, VIC, Australia
Global seed vaults are important, as they conserve plant genetic resources for future breeding to improve crop yield and quality and to overcome biotic and abiotic stresses. However, little is known about the impact of standard storage procedures, such as seed drying and cold storage on the seed bacterial community, and the ability to recover seed-associated bacteria after storage. In this study, soybean [Glycine max (L.) Merr.] seeds were analyzed to characterize changes in the bacterial community composition and culturability under varying storage conditions. The G. max bacterial microbiome was analyzed from undried seed, dried seed, and seed stored for 0, 3, 6, and 14months. Storage temperatures consisted of −20°C, 4°C, and room temperature (RT), with −20°C being commonly used in seed storage vaults globally. The seed microbiome of G. max was dominated by Gammaproteobacteria under all conditions. Undried seed was dominated by Pantoea (33.9%) and Pseudomonas (51.1%); however, following drying, the abundance of Pseudomonas declined significantly (0.9%), Pantoea increased significantly (73.6%), and four genera previously identified including Pajaroellobacter, Nesterenkonia, env.OPS_17, and Acidibacter were undetectable. Subsequent storage at RT, 4, or −20°C maintained high-abundance Genera at the majority of time points, although RT caused greater fluctuations in abundances. For many of the low-abundance Genera, storage at −20°C resulted in their gradual disappearance, whereas storage at 4°C or RT resulted in their more rapid disappearance. The changes in seed bacterial composition were reflected by cultured bacterial taxa obtained from the stored G. max seed. The main taxa were largely culturable and had similar relative abundance, while many, but not all, of the low-abundance taxa were also culturable. Overall, these results indicate that the initial seed drying affects the seed bacterial composition, suggesting that microbial isolation prior to seed drying is recommended to conserve these microbes. The standard seed storage condition of −20°C is most suitable for conservation of the bacterial seed microbiome, as this storage temperature slows down the loss of seed bacterial diversity over longer time periods, particularly low-abundance taxa.
Introduction
Seed vaults play a significant role in facilitating the ex situ conservation of germplasm of a range of crop species, their closely associated crop wild relatives (CWRs), and other wild species (Hay and Probert, 2013). Globally, seed vaults preserve plant genetic diversity for research and plant-breeding activities for improving food and nutritional security (Asdal and Guarino, 2018). International standards are adapted by seed vaults for long-term seed storage. According to the standard method, seeds are first kept under drying conditions at 10–15% relative humidity and 10–15°C to achieve a seed moisture content of 3–7% followed by their storage at or below −18°C. This method has been identified to work well for seeds of many plant species known as orthodox seeds (Cochrane et al., 2007).
Crop seeds are known to transmit a plant-specific core microbiota (Berg and Raaijmakers, 2018). The seed-associated microbes are reported to have plant genotype specificity and can vertically transmit from one generation to the next plant generation. Horizontal transfer of the microbes can occur via their uptake from the surrounding environment (Johnston-Monje et al., 2016; Shade et al., 2017; Adam et al., 2018; Nelson, 2018). The seed microbiome is known to harbor a wide range of microbial species (Johnston-Monje et al., 2016; Shade et al., 2017; Adam et al., 2018). The seed-borne microbes can colonize the emerging seedlings before the intake of microbes from the surrounding environment and can promote germination and early plant vigor and survival (Truyens et al., 2015). However, how seed-associated microbes influence the different growth stages during seed germination and later plant growth and provide biotic–abiotic stress resistance still need to be investigated (Adam et al., 2018; Berg and Raaijmakers, 2018).
Different seed-associated microbes were identified to protect crops against various biotic–abiotic stresses and enhance plant growth (Links et al., 2014; Mousa et al., 2016; Gdanetz and Trail, 2017; Shahzad et al., 2018; Li et al., 2020, 2021; Hone et al., 2021). In last two decades, the use of growth-promoting bacteria in agriculture has increased significantly to reduce the use of chemical fertilizers and enhance plant nutrient uptake (Rascovan et al., 2016). It is suggested that embracing greater use of beneficial microorganisms can improve crop yield and encourage biology-based agriculture (Schmidt et al., 2015).
Soybean [Glycine max (L.) Merr.] is one of the most important crops and a major source of animal feed and vegetable oil worldwide (Sugiyama et al., 2015). Due to a high protein (40–42%) and oil content (18–22%), soybean is also used for aquaculture feed and production of biofuel and as a source of protein for the human diet (Pagano and Miransari, 2016). Soybeans can play an important role in matching the food demands of the growing population by 2050, although the estimated yield increase of only 1.3% per year is not satisfactory (Pagano and Miransari, 2016). Some bacterial genera including Pseudomonas, Bacillus, Bradyrhizobium, Streptomyces, Rahnella, and Azospirillum and fungi, e.g., Piriformospora and Trichoderma, have proven to be promising in plant growth promotion in soybean production (Tsavkelova et al., 2006; Ramírez and Kloepper, 2010; Schmidt et al., 2015; Bakhshandeh et al., 2020). Notably, many crop, plant, and vegetable seeds were also reported to be inhabited by some of these bacterial genera (Johnston-Monje et al., 2016; Liu et al., 2017; Adam et al., 2018; Berg and Raaijmakers, 2018; Khalaf and Raizada, 2018; López et al., 2018; Wassermann et al., 2019a; Abdullaeva et al., 2021; Hone et al., 2021). However, despite the enormous potential of seed microbiomes to promote plant growth and sustainable agricultural practices, the impact of current international seed storage strategies on the seed microbial diversity and composition has not yet been evaluated (Berg and Raaijmakers, 2018).
Therefore, in the present study, amplicon sequencing of the V4 region of 16S rRNA was used to examine the impact of standard storage methods on the diversity and composition of the G. max seed bacterial microbiome and bacteria isolated from these seeds. The aim was to determine the effectiveness of current storage methods in maintaining the original bacterial composition and viability of seed-associated bacteria over time, thus providing an experimental basis for our understanding of the implication of seed vault storage strategies in conservation of seed bacterial microbiome.
Materials and Methods
Soybean Seed Samples
Soybean seeds (G. max-Burrinjuck) used in this study were sourced from Australian Grains Genebank, Horsham, Australia. About 1kg of seed was placed in a cloth bag and stored in a drying chamber at 15°C and 15% relative humidity for about 1month. This is the standard drying protocol used prior to storage of seed in the seed vault (FAO, 2014). At the end of the drying phase, about 100g of seed were weighed and transferred into heat-sealed aluminum bags. One bag of undried seed was also prepared to use it as control for the drying process at 0 time point. The seed bags were then transferred after 2days to the laboratory in AgriBio, Bundoora, Victoria, Australia. Seed bags containing dried seed were then stored at −20°C (±2°C), at 4°C (±2°C), and at room temperature (RT), 22°C (±2°C), for 3, 6, and 14months. Seed bags for RT were kept in an airtight plastic container at RT for 3, 6, and 14months. For further study, germinated seedlings were selected for profiling the G. max microbiome to focus on the seed-borne microbes that can colonize seedlings during germination, with the hypothesis that these microbes have a function in this process, thus focusing only on the viable microbial communities that remain after storage.
Seed Germination
For germination, G. max seeds for each time point and storage temperature were washed 10 times with an excess amount of sterile distilled water. The seeds were transferred into sterile petri dishes (12-cm diameter) by placing them between pre-water soaked Whatman™ filter paper (two sheets underneath and one on top) and then sealed with Parafilm™ and kept in darkness for 24–32h at RT. Then, the top layer of filter paper was removed, and the plates were resealed with Parafilm™ followed by a further 8–10days of incubation on a lab-benchtop under ambient light conditions. Non-germinated seeds were discarded immediately to avoid any antagonistic fungal outgrowth from these seeds. If needed, water was sprayed on seedlings during the incubation under sterile conditions. The average germination rate for the G. max seeds remained between 55 and 60% during the study. Seedlings were harvested for microbiome profiling and microbial isolation once the cotyledons reached an unfolded growth stage (Supplementary Figure S1).
DNA Extraction, 16S Amplicon Library Construction, and Sequencing
For seed microbiome profiling, 15–20 seedlings were selected for DNA extraction. Whole seedlings (root, shoot, and cotyledon) were cut into pieces of approximately 0.5–1cm using a sterile scalpel blade, collected in 1.2-ml QIAGEN collection tubes, and snap-frozen in liquid nitrogen and stored at −80°C until processed for DNA extraction. DNA extraction was performed using the MagAttract® 96 DNA plant kit using a Biomek FXᴾ Lab Automation Workstation coupled to a Synergy 2 multi-mode reader controlled by Biomek software version 4.1 and Gen 5 (2.08) software (Biotek Instruments, United States) with slight changes in manufacturer’s guidelines.
Amplicon libraries for Illumina sequencing were prepared using barcoded primer 5151f-806r, specific to V3–V4 regions of the bacterial 16s rRNA gene. Amplification of the host chloroplast and mitochondrial 16s DNA was blocked by adding peptide nucleic acids, pPNA and mPNA, respectively, to the PCR mix (Lundberg et al., 2013). PCR for 16s rRNA gene amplification was performed in a total volume of 25μl [Kapa HiFi Hotstart 2× ReadyMix DNA polymerase (Kapa Biosystems Ltd., London, United Kingdom), 50μM of pPNA and mPNA mix, 5μM of each primer, PCR-grade water, and 5μl of template DNA] under the following cycling conditions: 94°C for 3min, 30cycles of 94°C for 15s, 75°C for 10s, 55°C for 10s, 72°C for 45s, and a final elongation at 72°C for 10min using a thermal cycler (Agilent SureCycler 8,800, Agilent Technologies, United States). Libraries were further purified using AMPure XP beads (LABPLAN, Naas, Ireland). Dual indices and Illumina sequencing adapters from the Illumina Nextera XT index kits v2 B and C (Illumina, San Diego, United States) were added to the target amplicons in a second PCR step using Kapa HotStart HiFi 2× ReadyMix DNA polymerase (Kapa Biosystems Ltd., London, United Kingdom). Cycle conditions were 95°C (3min), then 10cycles of 95°C for 30s, 55°C for 30s, 72°C for 30s, then a final extension of 72°C for 5min followed by library cleanup using AMPure XP beads.
The barcoded libraries were quantified on a Nanodrop™ 1000 spectrophotometer and pooled together in an equimolar concentration. Library pools were further quantified for concentration and size using QuantiFluor® dsDNA assay (Promega Corporation, United States) and Tape station 2,200 High Sensitivity D1000 kit (Agilent Technologies, United States), respectively. Paired-end sequencing was performed on Miseq v3 (2×300bp v3 chemistry cartridge). All Illumina sequences have been submitted to the NCBI Sequence Read Archive (SRA accession PRJNA766782).
Data Analysis
The raw Illumina® paired-end reads were quality filtered and merged into a single read using PEAR with default parameters (Zhang et al., 2014). Afterward, sequencing data analysis was performed using QIIME 22020.11.1 (Bolyen et al., 2019). The primers from single-end reads were then removed using cutadapt plugin with the following parameters; error rate-0.2, flags; adapter-wildcards, read-wildcards, and discard-untrimmed (Martin, 2011). The single-end reads were then trimmed to a read length of 253bp and then dereplicated and filtered to remove chimeras. A feature table was then constructed containing the amplicon sequencing variants (ASVs) and representative sequences using the default algorithm in DADA2 (Callahan et al., 2016). The ASVs were then aligned with mafft (Katoh et al., 2002; via q2-alignment) and used to construct a phylogeny tree with fasttree2 (Price et al., 2010; via q2-phylogeny). The taxonomic classification of ASVs was performed using a naive Bayes taxonomy classifier (Bokulich et al., 2018) trained on the silva-138 release (V4 region-16s rRNA gene; Quast et al., 2013). Host-associated mitochondria and chloroplast reads and low-abundance features (<10 counts and present in at least two samples) were discarded from the data using the filter-features plugin. Alpha diversity (Observed OTUs) and beta diversity (Jaccard distance) were explored by running the core-metrics script in QIIME2 by rarefying feature tables to a read count of 6,000 sequences. The taxa classified up to the genus level were then exported and used to perform the presence/Absence test in Genedata Expressionist® Analyst™ v.10.0 using the default parameters to identify the shared features between undried seed and dried seed stored for 0, 3, 6, and 14months at RT, 4, and −20°C (Genedata, Basel, Switzerland). Statistical analyses of the 16s rRNA gene data were performed using scripts in QIIME2 2020.11.1. Alpha diversity was tested for significant differences using the Kruskal–Wallis pairwise test and Beta diversity using Analysis of Similarity (ANOSIM) test. To determine the changes in bacterial abundance after seed drying, the QIIME2 feature table was exported in biom format and one-way ANOVA test was performed on individual ASVs (>0.1% in planta) using OriginPro 2019 (v9.6.0.172).
Microbial Isolation, DNA Extraction, 16S Amplicon Library Construction, and Sequencing
For all storage temperatures, seedlings in triplicates were harvested by collecting the shoot and root tissues and discarding the seed coat. The plant tissues were cut into small pieces (0.5–1cm) and homogenized using a sterile pestle or two cycles of a Qiagen TissueLyser II for 1min at 30 Hertz in 400–500μl of 1× PBS buffer followed by centrifugation at 4,000rpm for 1min. Serial dilutions were prepared (10−1–10−4), and 20-μl aliquots, including undiluted macerate, were plated onto Reasoner’s 2A agar (R2A; Oxoid, United Kingdom), and the plates were incubated at RT for up to 10days. The bacterial growth was observed both in undiluted and diluted plates, though only undiluted plates were selected for further study to capture a snapshot of the original viable bacterial community composition that was culturable, as it is likely to contain the most diversity. The DNA was extracted by scraping all microbial colonies off the plate using a sterile plastic loop into an Eppendorf tube®. The colonies were then resuspended in 1× PBS buffer and spun at 10,000rpm for 5min. According to the manufacturer’s guidelines, the supernatant was discarded, followed by DNA extraction using Promega™ Wizard™ Genomic DNA Purification Kit (United States). The gDNA’s optical density measurements were performed in a Quantus™ Fluorometer (Promega Corporation, Madison, WI, United States), 16S rRNA bacterial genes were amplified, and libraries were prepared and sequenced using steps mentioned in Section DNA Extraction, 16S Amplicon Library Construction, and Sequencing.
Data Analysis of Cultured Microbial Libraries
The raw paired-end reads were processed and analyzed as mentioned in Section Data Analysis, except the features were only filtered based on frequency (<10 counts), and the sequences were rarefied to the read counts of 16,529 sequences.
Results
16S Amplicon Sequencing
After aligning paired-end reads, removal of low-frequency features, singletons, and chimeric and plant sequences, a total of 17,038,933 sequences were assigned to 361 Amplicon Sequence Variants (ASVs) for microbiome profiling (in planta) and 4,020,424 sequences were assigned to 155 ASVs for microbial isolation (on plates). After rarefaction and collapsing biological replicates, the ASV table was assigned to 89 genera for microbiome profiling (Supplementary Table S1) and 26 genera for microbial isolation (Supplementary Table S2).
G. max Seed Microbiome Profiling (In planta)
At the class level, the bacterial communities were mainly dominated by the presence of Gammaproteobacteria (20.1–97.0%), Bacilli (1.8–21.9%), Alphaproteobacteria (0.1–22.1%), Actinobacteria (0.2–15.2%), and Bacteroidia (0.01–20.6%). Notably, after 3months at RT, the relative abundance was almost equally dominated by Alphaproteobacteria (22.1%), Gammaproteobacteria (20.1%), Bacilli (21.9%), Bacteroidia (20.6%), and Actinobacteria (15.2%). Additionally, low-abundance bacterial classes (<0.1%) such as Polyangia (0.0038%) and Acidobacteriae (0.00083%) designated as “Others” were observed in undried seed and seed stored for 3months at −20°C, respectively (Figure 1A, Supplementary Table S1).
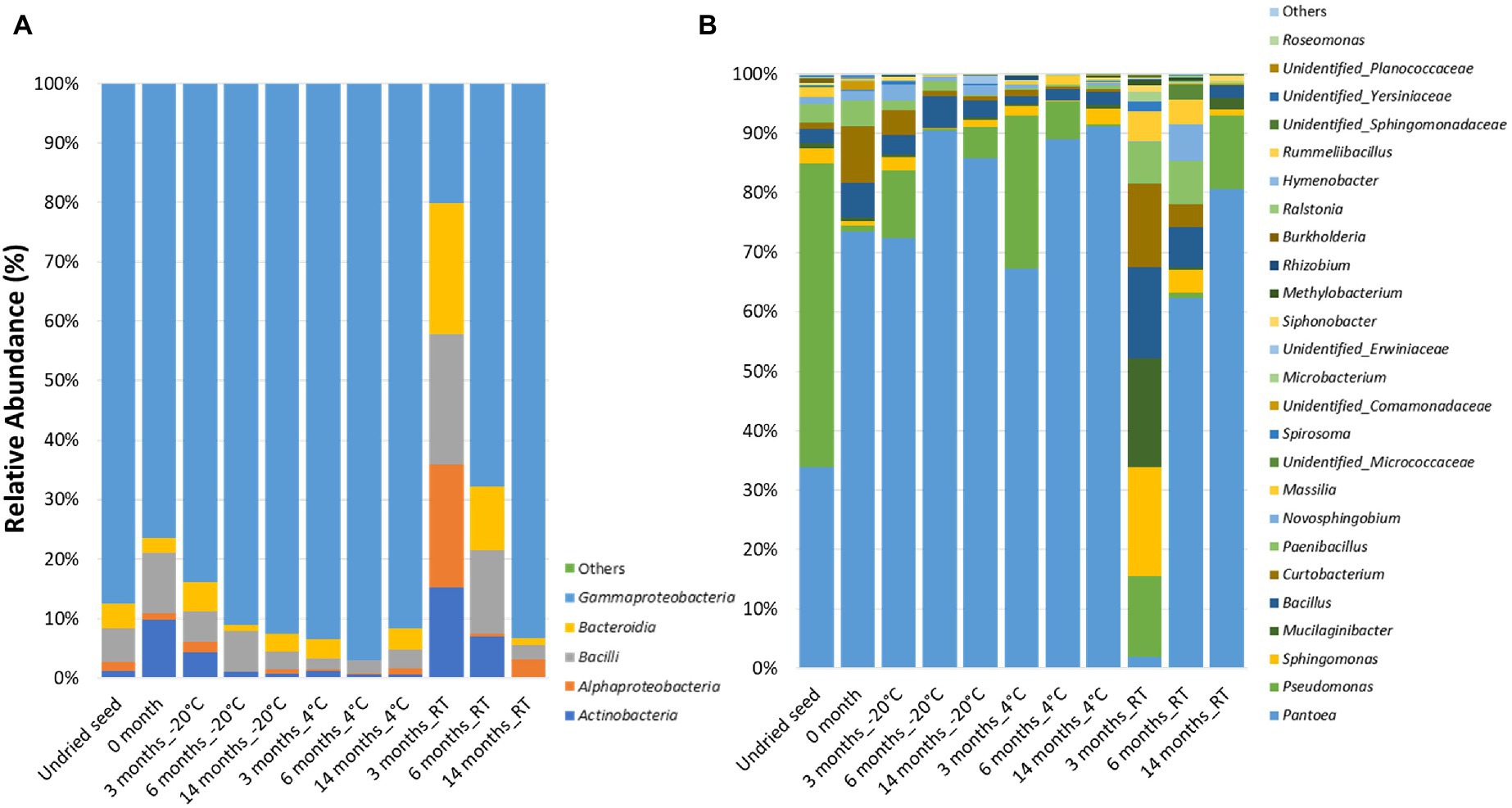
Figure 1. Relative abundance of the bacterial taxa in planta (A) at class level and (B) at genus level in undried seed and dried seed stored at different time points (0, 3, 6, and 14months) at −20°C, 4°C, and room temperature (RT). The bacterial taxa occurring with <0.1% are shown as “Others.”
At the genus level, 25 bacterial genera were identified with >0.1% relative abundance. The bacterial communities of the stored G. max seed mainly consisted of genera including Pantoea (1.8–91.2%), Pseudomonas (0.31–51.1%), Bacillus (1.4–14.9%), Sphingomonas (0.04–17.8%), Curtobacterium (0.17–13.7%), Paenibacillus (0.15–7.2%), Mucilaginibacter (0.0–17.8%), Novosphingobium (0.001–6.2%), and Massilia (0.01–4.9%) (Figure 1B). In addition to the community structure changes, the relative abundance of bacterial genera varied across all time points and storage temperatures. Interestingly, high variations in bacterial abundance were observed after 3months in seed stored at RT. In particular, majority of the ASVs mainly belonged to the genera Sphingomonas (20.7%), Mucilaginibacter (17.8%), Bacillus (14.9%), and Curtobacterium (13.7%), which were represented at much higher levels than at other time points (Figure 1B). Notably, the relative abundance of Pantoea (1.9%) was the lowest after 3months at RT compared to all other time points. The bacterial genera with >10% relative abundance in planta such as Pantoea, Pseudomonas, Mucilaginibacter, Bacillus, and Curtobacterium responded differently to drying treatment and storage temperature. It was observed that the relative abundance of Pantoea increased under cold storage, while Pseudomonas oscillates in abundance across all temperatures (Figure 1B, Supplementary Figure S2A). The bacterial genera with <10 and>1% relative abundance in planta such as Paenibacillus, Novosphingobium, Massilia, Microbacteriaceae, Spirosoma, Microbacterium, Siphonobacter, Unidentified group of Comamonadaceae, and Erwiniaceae were increased in abundance at RT especially after 3 and 6months of storage (Figure 1B, Supplementary Figure S2B). On the other hand, the bacterial genera with <1% such as Methylobacterium, Rhizobium, Burkholderia, Ralstonia, Hymenobacter, Rummeliibacillus, Roseomonas, Unidentified group of Sphingomonadaceae, Yersiniaceae, and Planococcaceae showed more variations in their abundance at 4°C and RT (Figure 1B, Supplementary Figure S2C).
Composition Differences in the Seed Microbial Communities After Different Lengths of Storage
Alpha diversity and beta diversity were used to assess the changes in bacterial diversity and composition in undried seed, dried seed, and seed stored for 0, 3, 6, and 14months at −20°C, 4°C, and RT. Based on alpha diversity, when compared to the undried seed, the number of observed features significantly (p<0.05) declined in dried seed stored at −20°C and 4°C in all time points (3, 6, and 14months) and in dried seed stored at RT for 6 and 14months. In contrast, no significant differences were observed between undried seed and dried seed (0month) and between undried seed and dried seed stored at RT for 3months. Additionally, when differences were compared for each storage temperature, the number of observed features significantly (p<0.05) declined after 3months and 6, 3, and 14months, in all time points at −20°C, 4°C, and RT, respectively (Figure 2A, Supplementary Table S3). No significant differences were observed for seed stored for 3 and 14months at −20 and 4°C, respectively.
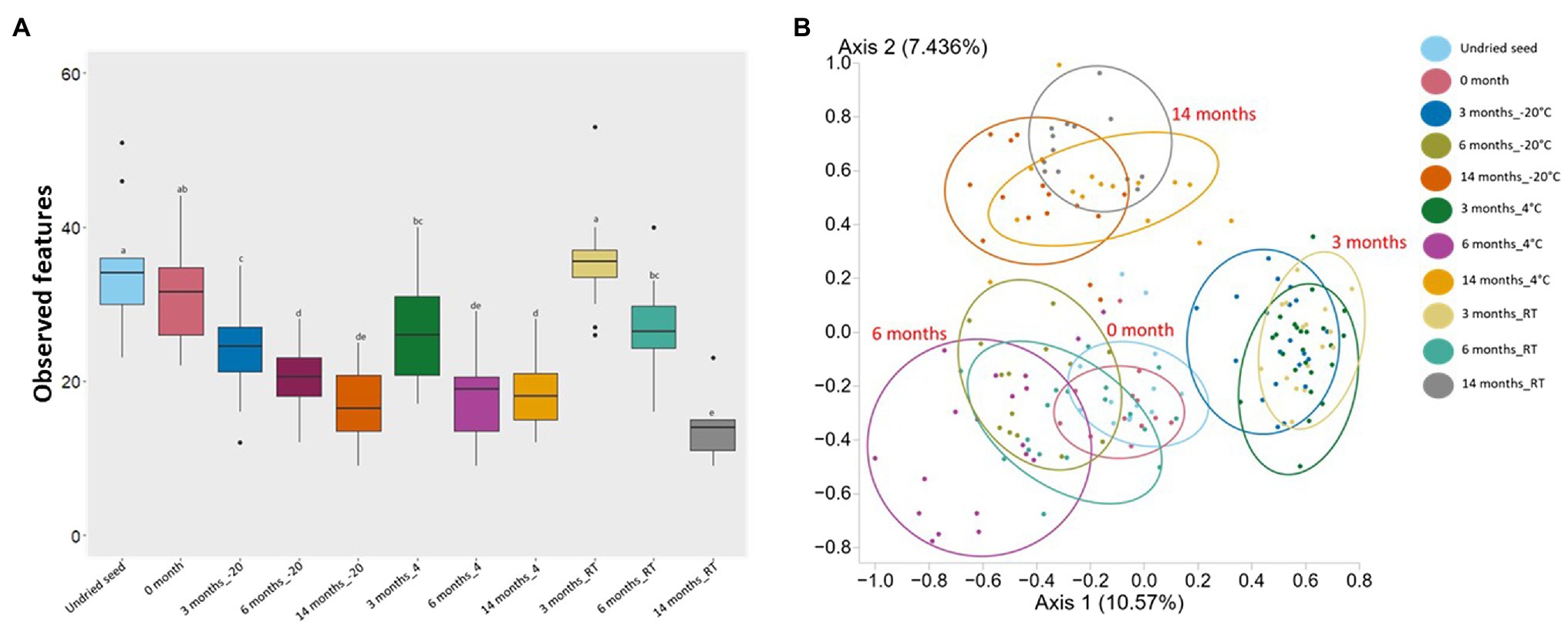
Figure 2. Alpha diversity (Observed features) and Beta diversity analysis (Jaccard dissimilarity) of the G. max seed (in planta). (A) Box-and-whisker plots showing the number of features observed under all conditions. Significant differences (p≤0.05) were assessed by ANOSIM pairwise test and are indicated by different lowercase letters (Supplementary Table S3). (B) Principal Coordinates Analysis (PCoA) plot showing the distances in the bacterial communities between undried seed and dried seed (0, 3, 6, and 14months) stored at −20°C (±2°C), 4°C (±2°C), and room temperature (RT; 22±2°C). Significant differences in bacterial composition were tested using the ANOSIM pairwise test (Supplementary Table S3).
Based on the Jaccard dissimilarity metrics, the bacterial composition significantly (p<0.05) varied under all conditions (Supplementary Table S3). These differences were also evident in the Principal Coordinates Analysis (PCoA) of the bacterial communities, where undried seed, dried seed (0month), and stored seed (3, 6, and 14months) formed a separate cluster (Figure 2B). Interestingly, the undried seed, dried seed (0month), and seed stored for 6months formed a close cluster compared to seed stored for 3 and 14months. The statistical analysis based on the presence/absence test was performed in Genedata Expressionist® Analyst™, and the number of shared genera was calculated between undried seed and dried seed (0month) and between undried seed and stored seed (3, 6, and 14months). There were 68.8% genera (33) shared between undried seed and dried seed (0month). While after 3months of storage, there were 56.3% genera (27) shared between undried seed and seed stored at RT, followed by 54.2% genera (26) with seed stored at 4°C and 47.9% genera (23) with seed stored at −20°C. Remarkably, after 6months of storage, there were 72.9% genera (35 genera) shared between undried seed and seed stored at RT followed by 64.6% (31) with seed stored at 4°C and 54.2% genera (26) with seed stored at −20°C. Next, after 14months of storage, there were 45.8% genera (22) shared between undried seed and seed stored at 4°C followed by 43.8% genera (21) seed stored at −20°C and 35.4% genera (17) seed stored at RT (Supplementary Figure S3).
Culturability of the G. max Seed Bacterial Microbiome (On Plates)
At the class level, the bacterial communities were mainly dominated by the presence of Gammaproteobacteria (0.7–91.1%), Alphaproteobacteria (0.3–42.4%), Bacilli (1.2–18.1%), Actinobacteria (0.3–31%), and Bacteroidia (0.0–7.8%). In contrast to the microbiome profiling (in planta), after 3months at RT (on plates), the relative abundance of bacterial classes was dominated by Alphaproteobacteria (42.4%), followed by Actinobacteria (31%), Bacilli (18.1%), Bacteroidia (7.8%), and Gammaproteobacteria (0.7%) (Figure 3A).
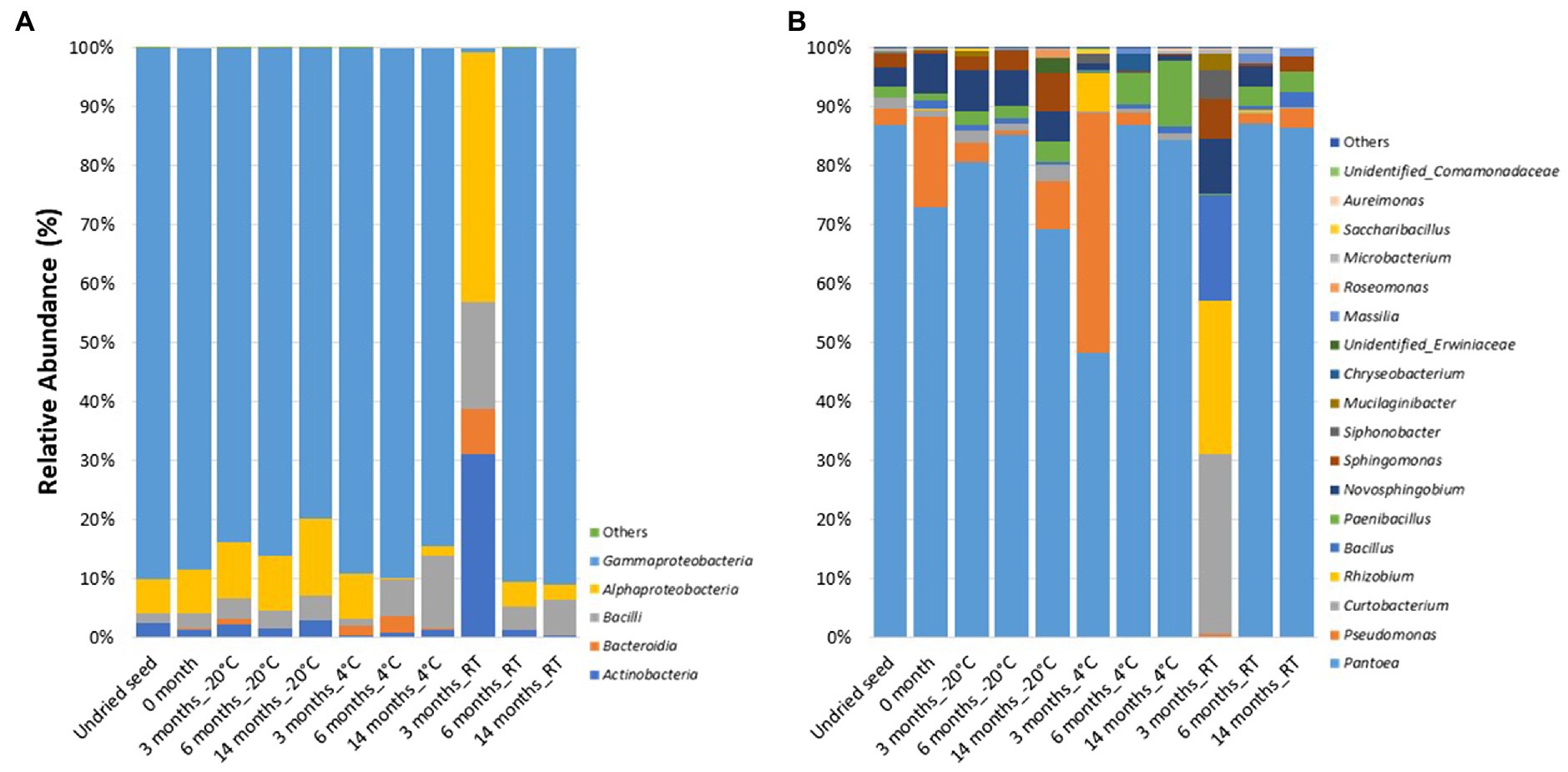
Figure 3. Relative abundance of bacterial taxa on plates (A) at class level and (B) at genus level in undried seed and dried seed stored at different time points (0, 3, 6, and 14months) at −20°C, 4°C, and room temperature (RT). The bacterial taxa occurring with less than 0.1% are shown as “Others.”
At the genus level, 18 bacterial genera were identified with >0.1% relative abundance. The bacterial communities on plates mainly consisted of genera Pantoea (0.2–87.3%), followed by Pseudomonas (0.0–40.7%), Curtobacterium (0.3–30.4%), Rhizobium (0.0–26.1%), Bacillus (0.01–17.9%), Paenibacillus (0.2–11.2%), Novosphingobium (0.0–9.3%), Sphingomonas (0.004–6.7%), Siphonobacter (0.0–4.8%), and Mucilaginibacter (0.0–3.0%). The relative abundance of these bacterial genera varied across all the time points and storage temperatures (Figure 3B). Similar to the microbiome profiling (in planta), a dramatic change in the relative abundance of bacterial genera was observed after 3months at RT (on plates). The distribution of abundance of these was different from that of the “in planta” data, with the most abundant bacteria belonging to Curtobacterium (30.4%), Rhizobium (26.1%), Bacillus (17.9%), Novosphingobium (9.3%), Sphingomonas (6.7%), Siphonobacter (4.8%), and Mucilaginibacter (3.0%). However, the relative abundance of Pantoea (0.2%) was lesser than the other time points, similar to in planta data (Figure 3B).
When compared to the 16S rRNA gene sequencing data (in planta), not all the bacterial taxa (at genus level) were culturable. The overall culturability of the bacterial genera was more consistent at −20°C storage compared to the seed stored at 4°C and RT (Figure 4A). Many of the bacterial genera present with >0.1% relative abundance in undried seed (in planta) were culturable, with −20°C providing a more stable recovery compared to 4°C and RT after 14months of storage (Figure 4B).
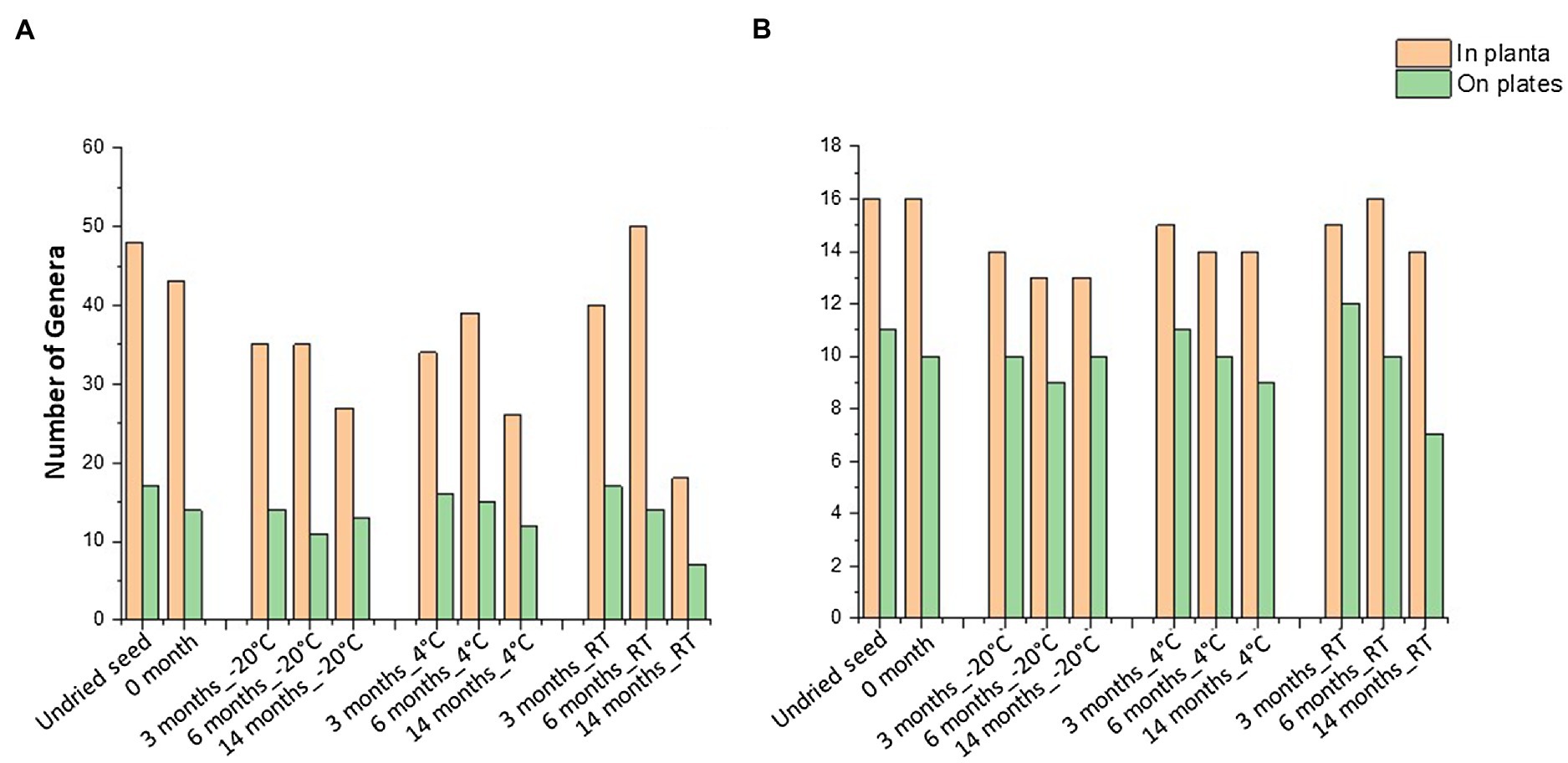
Figure 4. Culturable seed bacterial microbiome. (A) Number of genera observed in planta and on plates. (B) Number of genera with >0.1% abundance in undried seed and their culturability over time at different storage temperatures. All the seed samples belong to the undried seed, dried seed (0month), and seed stored for 3, 6, and 14months at −20°C, 4°C, and room temperature (RT).
In total, 48 genera were associated with undried seed (in planta), of which 27 genera were culturable under the conditions of this experiment. There were 16 genera present with >0.1% relative abundance in undried seed (Supplementary Table S1), of which 13 genera were culturable with the exceptions being Burkholderia (0.6%), Hymenobacter (0.2%), and Unidentified_Yersiniaceae (0.1%) (Figure 5A). There were only five bacterial genera including Pantoea, Sphingomonas, Bacillus, Curtobacterium, and Paenibacillus detected on plates under all conditions. Notably, these were also some of the abundant genera in planta. While the presence of other bacterial genera on plates varied across all time points and storage temperatures (Figure 5A).
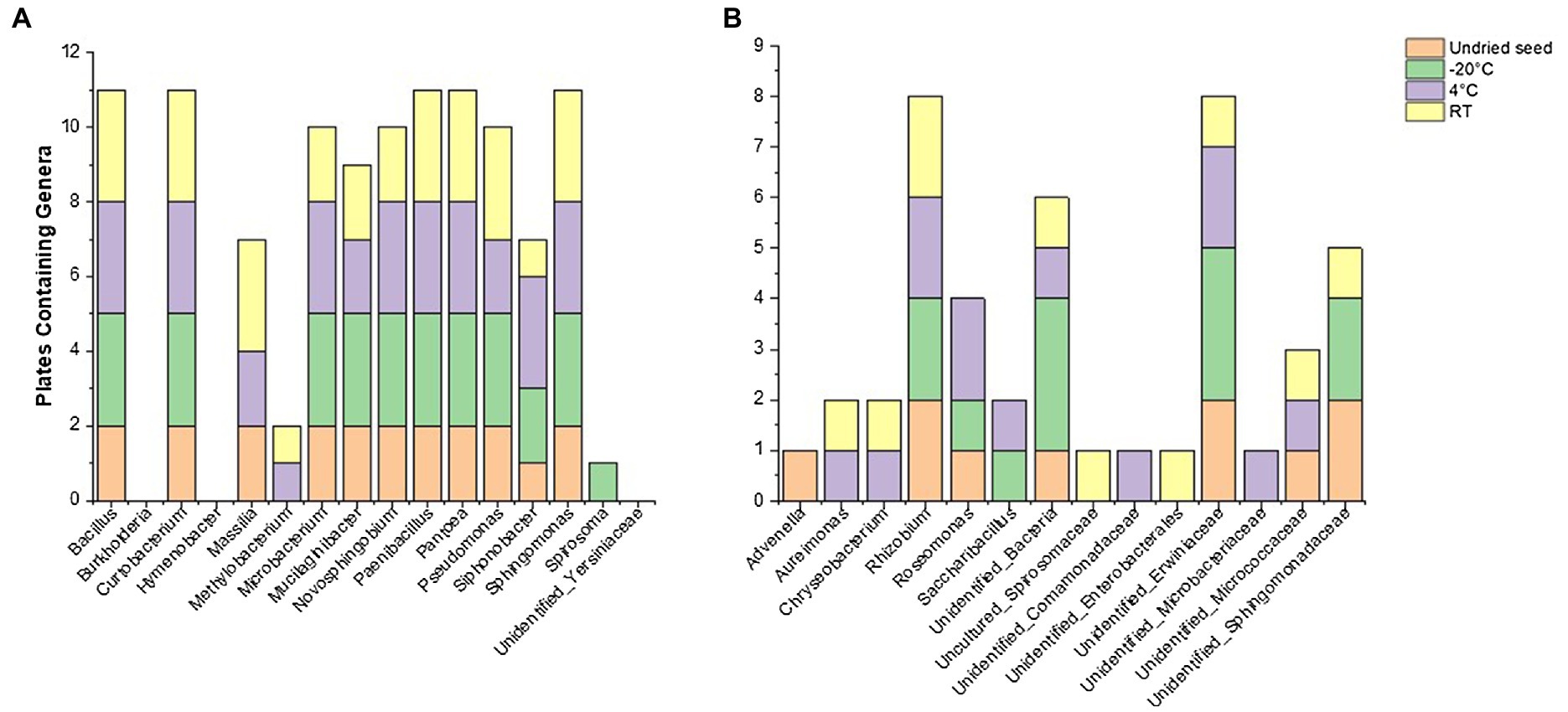
Figure 5. Culturability of bacterial genera that were associated with undried seed (in planta) across different time points (0, 3, 6, and 14months) when stored at −20°C, 4°C, and room temperature (RT). (A) Genera with >0.1% relative abundance and (B) <0.1% relative abundance in undried seed (in planta).
Of note, the abundance of Massilia declined when stored at −20°C in planta and was also not detected on plates from these seeds, showing its sensitivity to cold storage conditions (Figure 5A, Supplementary Figure S4). It was observed that some of the low-abundance bacterial genera (<0.1%) were also culturable including some unidentified bacteria (Figure 5B, Supplementary Table S2). On the other hand, some genera that were below the level of detection in undried seed and dried seed (0month) in planta including Advenella, Aureimonas, Chryseobacterium, Uncultured_Spirosomaceae and Saccharibacillus were detected on plates from varying storage temperatures (Figure 5B).
Effect of Drying on the G. max Seed Microbiota
The drying treatment of G. max seed at 15°C and 15% relative humidity for 1month was found to alter the relative abundance of bacterial genera. The seed microbiome profiling (in planta) showed a significant change in the abundance of bacterial genera after drying including Pseudomonas (51.1 to 0.9%), Pantoea (33.9 to 73.6%), Curtobacterium (1.0 to 9.5%), Sphingomonas (2.5 to 0.7%), Massilia (1.6 to 0.005%), Methylobacterium (0.3 to 0.09%), and Unidentified_ Erwiniaceae (0 to 0.03%) (Figure 6A, Supplementary Table S5). There were four low-abundance genera including Pajaroellobacter, Nesterenkonia, env.OPS_17, and Acidibacter that completely disappeared after seed drying (Supplementary Table S1).
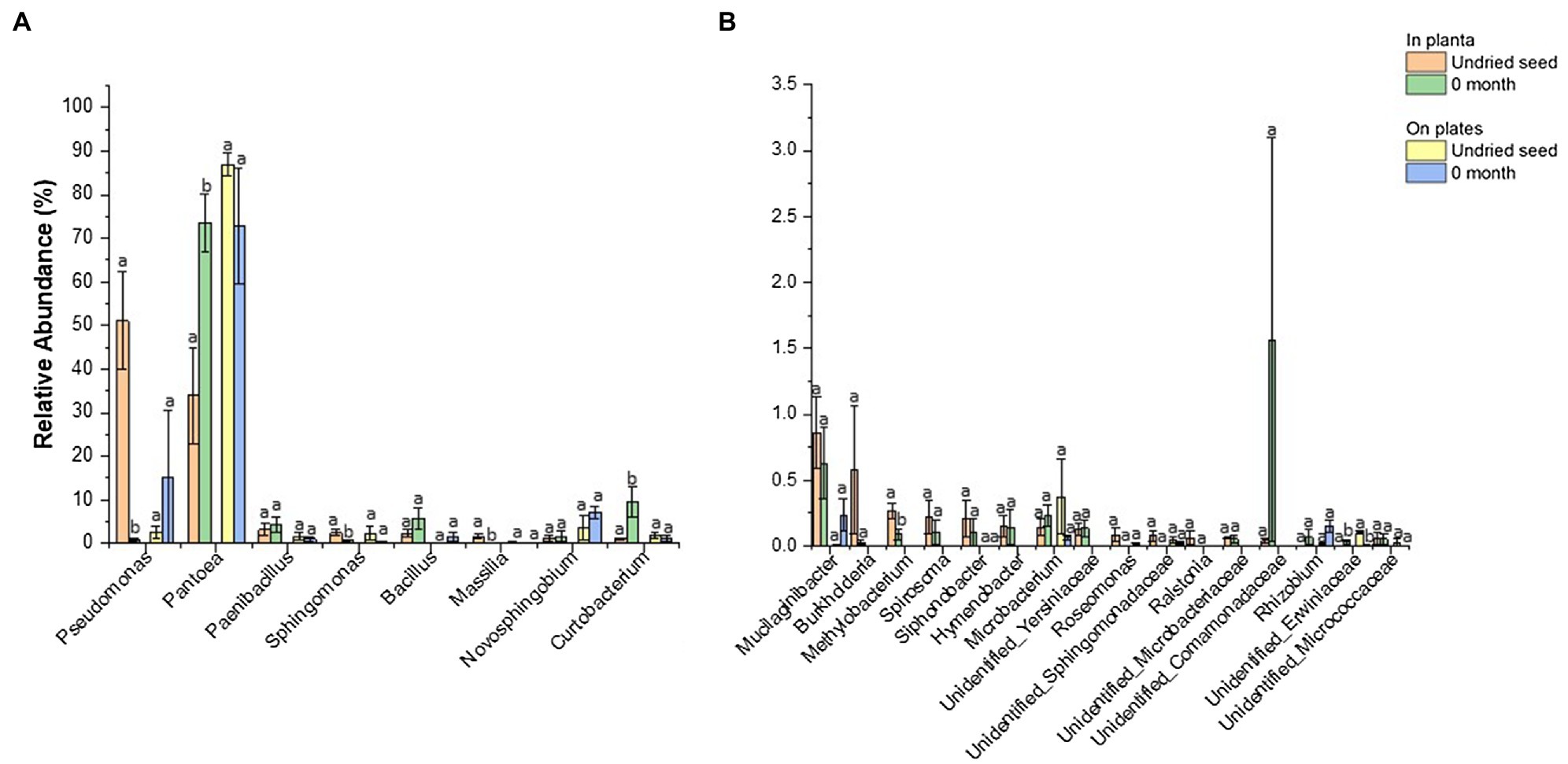
Figure 6. Comparison of relative abundance of bacterial genera detected in undried seed and dried seed (0month) in planta and on plates dataset. The abundance threshold was set based on in planta dataset (A) >1% and (B) <1% and >0.1% for statistical analysis. Significant differences (p≤0.05) were determined by one-way ANOVA test and are indicated by different lowercase letters. Error bars show SE of mean (SEM).
In contrast, no significant differences were observed for bacterial abundance on plates except for Unidentified_ Erwiniaceae (0.1 to 0.004%) (Figure 6B, Supplementary Table S5). The culturing assays, however, showed a different pattern for Pseudomonas (2 to 15%) and Pantoea (86.9 to 73.0%). It was observed that one replicate plate for dried seed (0month) was equally dominated by Pantoea (47.0%) and Pseudomonas (45.7%), contributing to an increased abundance of Pseudomonas (15.3%) and decreased abundance of Pantoea (73.0%) on plates post drying, indicating that the microbial diversity could vary from seed to seed to some extent (Figure 6B, Supplementary Table S4).
Discussion
Seed banks have been created to preserve plant and crop genetic diversity for future use. However, the value of current seed storage techniques in conserving seed-borne microbial diversity has not been investigated (Berg and Raaijmakers, 2018). In this study, we demonstrated that drying G. max seed before storage changes the abundance and composition of the seed microbiota. Moreover, we found that the different bacterial communities respond differently to the seed drying and storage temperature. Additionally, the seed bacterial composition changed more dramatically under RT storage than the cold storage (−20 and 4°C). The culturability of seed-associated microbes was found to be largely driven by abundance. Seed storage at −20°C provided a long-term stable recovery of the culturable microbes under the conditions of this experiment.
G. max Seed Microbiome Composition
Soybean seeds are known to lose viability and vigor under high temperature and relative humidity conditions (Shelar et al., 2008). Nevertheless, in this study, the impact of such environmental factors was reduced due to the optimal and stable conditions during a 14-month storage period. In general, the G. max seed microbiome consisted of bacterial classes such as Gammaproteobacteria, Alphaproteobacteria, Bacilli, Bacteroidia, and Actinobacteria. Previous studies also observed these bacterial classes for the seed of red sage (Chen et al., 2018), bean (Klaedtke et al., 2016), ryegrass (Tannenbaum et al., 2020), rice (Nakaew and Sungthong, 2018), native alpine plants (Wassermann et al., 2019a), and Brassicaceae family plants (Barret et al., 2015). It has been demonstrated that the microbes colonizing seedling during germination can confer important functional traits to the plant such as nutrient availability (Torres-Cortés et al., 2018). Representatives of the majority of the detected bacterial genera including Pantoea, Pseudomonas, Bacillus, Sphingomonas, Curtobacterium, Paenibacillus, Mucilaginibacter, Novosphingobium, and Massilia are known to have a beneficial impact on plants. For instance, Bacillus strains isolated from soybean root nodules have been observed to promote soybean plant growth when co-inoculated with Bradyrhizobium japonicum under nitrogen-free conditions (Bai et al., 2003). Also, some Pseudomonas species isolated from the soybean rhizosphere inhibited the growth of soilborne pathogenic fungi (Susilowati et al., 2011). However, other species of Pseudomonas are well known for their pathogenicity (Xin et al., 2018; Solanki et al., 2019). The endophytic bacteria from the soybean root nodules were also identified to contain plant growth-promoting traits and antagonistic properties against pathogenic fungi (Phytophthora sojae; Zhao et al., 2018).
Effect of Seed Drying on G. max Seed Microbiome Composition
Seed banks globally follow seed drying treatment prior to low-temperature storage to increase seed longevity (Hay and Probert, 2013). While this treatment is valuable for increasing seed longevity in storage, there is little evidence about the effect of seed drying on seed microbiome conservation. In this study, both the seed drying and storage temperature were identified to affect the seed bacterial composition. Some of the abundant bacteria, including Pseudomonas, Sphingomonas, Massilia, Curtobacterium, and Methylobacterium declined significantly after seed drying treatment. This change in composition corresponded to a significant increase in abundance of Pantoea from an average of 33.9% to more than 73%, with four bacterial genera remaining undetected. It was recognized that temperature, humidity, water activity, and grain moisture could affect the seed microbial community (Schmidt et al., 2018). It must be stated that the bacterial communities can respond differently to the water stress caused during the seed drying process (Esbelin et al., 2018). The exclusive increase in the abundance of Pantoea after seed drying may indicate their ability to tolerate the stress caused due to water loss. Pantoea spp. along with E. sakazakii, E. vulneris, and K. oxytoca were reported to persist over 2years when individual bacterial strains were subjected to desiccated storage. This ability was attributed to the formation of an extracellular polysaccharide that can facilitate the survival of bacterial strains during an extended desiccation period (Lehner et al., 2005; Barron and Forsythe, 2007). Notably, the results obtained with culturing assays of microbes isolated from undried seed and dried seed (0month) were in general agreement with results obtained by culture-independent 16S rRNA gene sequencing data obtained in planta. However, an opposite pattern was observed for the relative abundance pattern of Pantoea and Pseudomonas between the culturing assay and the in planta assay. Interestingly, it was identified that the differences in bacterial abundance among seed samples were responsible for this variation. This was also in line with previous studies that have identified that relative abundance of bacterial inhabitants of seed can vary significantly between seed samples of the same plant species (Barret et al., 2015; Klaedtke et al., 2016; Rybakova et al., 2017; Rezki et al., 2018; Torres-Cortés et al., 2018). It has been reported that other than plant genotype, abiotic factors such as storage, harvesting methods, and field management practices were identified as possible drivers of such variations in bacterial abundance among seed samples (Barret et al., 2015).
Effect of Storage Temperature on G. max Seed Microbiome Composition
In the present study, Gammaproteobacteria dominated the G. max seed bacterial microbiome under all conditions. Notably, this was due to an increased abundance of Pantoea after seed drying compared to other abundant genera including Pseudomonas, Sphingomonas, Mucilaginibacter, Bacillus, and Curtobacterium. Endophytic strains of Pantoea isolated from the surface-sterilized leaves of Alhagi sparsifolia Shap. and wheat roots have been shown to improve plant growth under drying conditions (Chen et al., 2017; Cherif-Silini et al., 2019). A strain of Pantoea dispersa (Selvakumar et al., 2008) isolated from a sub-alpine soil was able to grow under different temperature conditions ranging from 4 to 42°C. Notably, more variations in bacterial abundance were observed in seed stored at RT. For instance, after 3months of storage at RT, the abundance of genera including Sphingomonas, Mucilaginibacter, Bacillus, and Curtobacterium collectively accounted for about 67% of the total bacterial abundance, which was higher than that at other time points. Interestingly, this change corresponded with a significant decrease in Pantoea from an average of 62.6% in undried seed and dried seed (0month) and seed stored for 6 and 14months to only 1.8% after 3months at RT. The bacterial strains belonging to the genera Sphingomonas, Mucilaginibacter, Curtobacterium, and Bacillus were identified to be more tolerant to drying conditions (Mannisto et al., 2010; Vardharajula et al., 2011; Chimwamurombe et al., 2016; Molina-Romero et al., 2017). Thus, it is highly plausible that these bacterial strains might be benefited by the sudden environmental changes created by seed drying and packaging. It must be stated that the majority of these genera are either aerobic or facultative anaerobes. It is highly possible that a reduction in available oxygen level due to an airtight heat-sealed packaging along with the drying conditions promoted favorable conditions for these genera. Moreover, this study showed that such changes in abundance were only for a limited period and the bacterial communities were more similar to the pre-drying conditions after 6months in all storage temperatures. Šťovíček et al. (2017) in their study showed that certain anaerobic taxa in soil dominated after a rainfall, but after 5–7days, the microbial composition returned to the pre-rainfall conditions. A similar trend was also reported by Supramaniam et al. (2016) who reported a shift in soil bacterial composition initially after a short variation in temperature and water content, though the bacterial composition was more similar to the control after 4weeks. A similar trend was also observed for the cultured bacterial taxa isolated from the stored G. max seed after 3months at RT, though the dominating bacterial genera varied compared to the in planta data. The differences in the bacterial abundance among individual seed samples were identified to contributing to these variations as observed for the seed drying in the above section (Effect of Seed Drying on G. max Seed Microbiome Composition). Notably, such variations in the abundance of bacterial genera at RT were not reflected in seed under cold storage.
We have shown that the seed bacterial diversity and composition co-vary with time and storage temperature. Results indicated that the seed can be stored at RT for 6months without losing much diversity and original bacterial composition. While the bacterial diversity and composition reduced rapidly after 6months. The disappearance of the lower-abundance bacterial genera was observed as the major reason for these diversity losses during storage. Seed storage at −20°C was identified as showing a gradual disappearance of the lower-abundance genera compared to more rapid losses at 4°C and RT. Soybean seed is known to go through various biochemical changes, such as decreases in fat, water-soluble nitrogen, sugars, nitrogen solubility index, trypsin inhibitor activity, available lysin, pigment, and lipoxygenase activity of seed and increases in seed browning, free fatty acid content, and peroxidase value when stored under ambient conditions (Narayan et al., 1988; Sharma et al., 2013). For instance, the increased level of free fatty acid in soybean seed invaded by Aspergillus ruber resulted in loss of seed viability (Dhingra et al., 2001). In our study, the significant decline in bacterial diversity and composition in G. max seed, specifically the loss of lower-abundance genera at RT and 4°C, might be linked to an increased level of free fatty acid. It has been identified that free fatty acid can kill bacteria by inhibiting enzyme activity, disrupting nutrient uptake, and lysing bacterial cells directly or indirectly (e.g., toxic peroxidation and autoxidation products; Desbois and Smith, 2010).
Effect of Storage Temperature on the Culturability of G. max Seed Microbiome
Seed-associated microbes have the potential to promote plant growth and to provide sustainable ways to protect crops against various biotic and abiotic stresses in the form of seed treatments. Seed banks can play an important role in conservation of the beneficial seed-associated microbes. However, there is a need to design international conservation strategies for seed banks to protect seed microbes so that their untapped benefits for plant, human, and environment can be further explored in the future (Berg and Raaijmakers, 2018). Culturing of the seed microbes independently of stored seed has been suggested as a necessary step to ensure that the beneficial microbes remain available to use them for enhancing crop productivity (Rascovan et al., 2016; Sarhan et al., 2019). To understand the importance of independent microbe culturing, we decided to examine the impact of seed storage temperature on the culturability of microbes. Overall, the results obtained with culturing assay of the stored G. max seed agreed with results obtained by in planta 16S rRNA gene sequencing data. In the current study, the isolated bacteria mainly belonged to the Gammaproteobacteria and Alphaproteobacteria, with Pantoea, Sphingomonas, Bacillus, Curtobacterium, Rhizobium, and Paenibacillus being some of the dominant bacterial genera isolated from G. max seed throughout the study. Notably, most of these genera have been characterized with a range of beneficial features such as the ability to fix nitrogen, indole-3-acetic acid production, and antagonistic abilities against various bacteria and fungi (Silini-Cherif et al., 2012; Hansen et al., 2017; Goyal et al., 2019; Liu et al., 2019). Data indicated that the genera present with more than 0.1% relative abundance in undried seed remained culturable after 14months of storage under all conditions. Many of the bacterial genera identified by 16S rRNA gene sequencing data in planta were culturable; however, not all of them were identified in culturing assays. These results were consistent with previous findings, where majority of the microbes identified by sequencing were not detected using classical culturing approach (Sylla et al., 2013; Schmidt et al., 2014; Qaisrani et al., 2019; Solanki et al., 2019). Notably, the cold storage temperatures, especially −20°C storage provided a stable recovery for the bacterial genera present with greater than 0.1%, while the bacterial viability was adversely affected in seed stored at RT. Cabello-Olmo et al. (2020) investigated the effect of storage temperature and packaging on bacteria and yeast viability in a plant-based fermented food and indicated that the microbial content seemed to be better preserved at −20 and 4°C compared to storage at 37°C. We also observed that the genera Massilia was sensitive to cold storage. Especially at −20°C storage, their relative abundance gradually declined in planta and was poorly represented in the culturing assay. A significant reduction in the abundance of Massilia was also observed in apples after they were stored for 6months in a commercial cold storage. The cold sensitivity of Massilia was suspected as the main reason for the significant decline in abundance (Wassermann et al., 2019b).
In conclusion, our study demonstrated that standard storage methods can be used for conservation of seed-associated bacterial microbiome, especially for high-abundance genera. Given that seed drying significantly impacts the composition of G. max seed microbiome, we suggest that a fresh bacterial isolation can help to conserve the original bacterial composition. Moreover, −20°C storage has been identified as a better alternative to RT and 4°C, as the overall bacterial diversity losses including lower-abundance genera were reduced, and culturability rate was high in −20°C storage. A better understanding about the effect of the standard storage methods on the seed microbiome composition of different plant species including their wild relatives can assist in designing new international conservation strategies for seed microbiomes.
Data Availability Statement
The raw sequence files supporting the findings of this article are available in the NCBI Sequence Read Archive (SRA) under the BioProject ID PRJNA766782.
Author Contributions
TS conceptualized the study. AC prepared the article. AC, TS, JE, and RM designed the experiment. SN provided the soybean seed and helped with the seed drying. AC and JK contributed to the laboratory work. TS and RM reviewed and edited the article. TS, JE, and RM supervised the study. GS contributed to the funding acquisition. All authors have read and agreed to the submitted version of the article.
Funding
This research was supported by the Agriculture Victoria Research.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
AC received La Trobe University Full-Fee Research Scholarship, La Trobe University Postgraduate Research Scholarship, and AgriBio Scholarship. We thank Dr. Katherine Whitehouse for her assistance during seed drying and packaging. We thank Desmond Auer for his thorough reading and editing of the article.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2021.784796/full#supplementary-material
References
Abdullaeva, Y., Ambika Manirajan, B., Honermeier, B., Schnell, S., and Cardinale, M. (2021). Domestication affects the composition, diversity, and co-occurrence of the cereal seed microbiota. J. Adv. Res. 31, 75–86. doi: 10.1016/j.jare.2020.12.008
Adam, E., Bernhart, M., Müller, H., Winkler, J., and Berg, G. (2018). The Cucurbita pepo seed microbiome: genotype-specific composition and implications for breeding. Plant Soil 422, 35–49. doi: 10.1007/s11104-016-3113-9
Asdal, Å., and Guarino, L. (2018). The Svalbard global seed vault: 10 years—1 million samples. Biopreserv. Biobank. 16, 391–392. doi: 10.1089/bio.2018.0025
Bai, Y., Zhou, X., and Smith, D. L. (2003). Enhanced soybean plant growth resulting from coinoculation of bacillus strains with Bradyrhizobium japonicum. Crop Sci. 43, 1774–1781. doi: 10.2135/cropsci2003.1774
Bakhshandeh, E., Gholamhosseini, M., Yaghoubian, Y., and Pirdashti, H. (2020). Plant growth promoting microorganisms can improve germination, seedling growth and potassium uptake of soybean under drought and salt stress. Plant Growth Regul. 90, 123–136. doi: 10.1007/s10725-019-00556-5
Barret, M., Briand, M., Bonneau, S., Préveaux, A., Valière, S., Bouchez, O., et al. (2015). Emergence shapes the structure of the seed microbiota. Appl. Environ. Microbiol. 81, 1257–1266. doi: 10.1128/aem.03722-14
Barron, J. C., and Forsythe, S. J. (2007). Dry stress and survival time of Enterobacter sakazakii and other Enterobacteriaceae in dehydrated powdered infant formula. J. Food Prot. 70, 2111–2117. doi: 10.4315/0362-028x-70.9.2111
Berg, G., and Raaijmakers, J. M. (2018). Saving seed microbiomes. ISME J. 12, 1167–1170. doi: 10.1038/s41396-017-0028-2
Bokulich, N. A., Kaehler, B. D., Rideout, J. R., Dillon, M., Bolyen, E., Knight, R., et al. (2018). Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6:90. doi: 10.1186/s40168-018-0470-z
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C. C., Al-Ghalith, G. A., et al. (2019). Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 37, 852–857. doi: 10.1038/s41587-019-0209-9
Cabello-Olmo, M., Oneca, M., Torre, P., Díaz, J. V., Encio, I. J., Barajas, M., et al. (2020). Influence of storage temperature and packaging on bacteria and yeast viability in a plant-based fermented food. Foods 9:302. doi: 10.3390/foods9030302
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nMeth.3869
Chen, H., Wu, H., Yan, B., Zhao, H., Liu, F., Zhang, H., et al. (2018). Core microbiome of medicinal plant Salvia miltiorrhiza seed: a rich reservoir of beneficial microbes for secondary metabolism? Int. J. Mol. Sci. 19:672. doi: 10.3390/ijms19030672
Chen, C., Xin, K., Liu, H., Cheng, J., Shen, X., Wang, Y., et al. (2017). Pantoea alhagi, a novel endophytic bacterium with ability to improve growth and drought tolerance in wheat. Sci. Rep. 7:41564. doi: 10.1038/srep41564
Cherif-Silini, H., Thissera, B., Bouket, A. C., Saadaoui, N., Silini, A., Eshelli, M., et al. (2019). Durum wheat stress tolerance induced by endophyte pantoea agglomerans with genes contributing to plant functions and secondary metabolite arsenal. Int. J. Mol. Sci. 20:3989. doi: 10.3390/ijms20163989
Chimwamurombe, P. M., Grönemeyer, J. L., and Reinhold-Hurek, B. (2016). Isolation and characterization of culturable seed-associated bacterial endophytes from gnotobiotically grown Marama bean seedlings. FEMS Microbiol. Ecol. 92:fiw083. doi: 10.1093/femsec/fiw083
Cochrane, J., Crawford, A., and Monks, L. (2007). The significance of ex situ seed conservation to reintroduction of threatened plants. Aust. J. Bot. 55, 356–361. doi: 10.1071/BT06173
Desbois, A. P., and Smith, V. J. (2010). Antibacterial free fatty acids: activities, mechanisms of action and biotechnological potential. Appl. Microbiol. Biotechnol. 85, 1629–1642. doi: 10.1007/s00253-009-2355-3
Dhingra, O., Mizubuti, E., Napoleao, I., and Jham, G. (2001). Free fatty acid accumulation and quality loss of stored soybean seeds invaded by Aspergillus ruber.
Esbelin, J., Santos, T., and Hébraud, M. (2018). Desiccation: an environmental and food industry stress that bacteria commonly face. Food Microbiol. 69, 82–88. doi: 10.1016/j.fm.2017.07.017
FAO (2014). “Genebank standards for plant genetic resources for food and agriculture”. Food and Agriculture Organization of the United nations Rome.
Gdanetz, K., and Trail, F. (2017). The wheat microbiome under four management strategies, and potential for endophytes in disease protection. Phytobiomes 1, 158–168. doi: 10.1094/PBIOMES-05-17-0023-R
Goyal, K., Kumar, T., Sharma, P., Rao, M., Ahmed, V., and Chauhan, N. S. (2019). “Crop Improvement Through Microbial Biotechnology: A Cross Talk,” in Salt Stress, Microbes, and Plant Interactions: Mechanisms and Molecular Approaches: Volume 2. ed. M. S. Akhtar (Singapore: Springer Singapore), 69–90.
Hansen, A. P., Choudhary, D. K., Agrawal, P. K., and Varma, A. (2017). Rhizobium Biology and Biotechnology. New York:Springer.
Hay, F. R., and Probert, R. J. (2013). Advances in seed conservation of wild plant species: a review of recent research. Conserv. Physiol. 1:cot030. doi: 10.1093/conphys/cot030
Hone, H., Mann, R., Yang, G., Kaur, J., Tannenbaum, I., Li, T., et al. (2021). Profiling, isolation and characterisation of beneficial microbes from the seed microbiomes of drought tolerant wheat. Sci. Rep. 11:11916. doi: 10.1038/s41598-021-91351-8
Johnston-Monje, D., Lundberg, D. S., Lazarovits, G., Reis, V. M., and Raizada, M. N. (2016). Bacterial populations in juvenile maize rhizospheres originate from both seed and soil. Plant Soil 405, 337–355. doi: 10.1007/s11104-016-2826-0
Katoh, K., and Misawa, K., Kuma, K.-i., and Miyata, T. (2002). MAFFT: a novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Khalaf, E. M., and Raizada, M. N. (2018). Bacterial seed endophytes of domesticated cucurbits antagonize fungal and oomycete pathogens including powdery mildew. Front. Microbiol. 9:42. doi: 10.3389/fmicb.2018.00042
Klaedtke, S., Jacques, M. A., Raggi, L., Preveaux, A., Bonneau, S., Negri, V., et al. (2016). Terroir is a key driver of seed-associated microbial assemblages. Environ. Microbiol. 18, 1792–1804. doi: 10.1111/1462-2920.12977
Lehner, A., Riedel, K., Eberl, L., Breeuwer, P., Diep, B., and Stephan, R. (2005). Biofilm formation, extracellular polysaccharide production, and cell-to-cell signaling in various Enterobacter sakazakii strains: aspects promoting environmental persistence. J. Food Prot. 68, 2287–2294. doi: 10.4315/0362-028X-68.11.2287
Li, T., Mann, R., Kaur, J., Spangenberg, G., and Sawbridge, T. (2021). Transcriptomics differentiate two novel bioactive strains of Paenibacillus sp. isolated from the perennial ryegrass seed microbiome. Sci. Rep. 11:15545. doi: 10.1038/s41598-021-94820-2
Li, T., Mann, R., Sawbridge, T., Kaur, J., Auer, D., and Spangenberg, G. (2020). Novel Xanthomonas Species From the Perennial Ryegrass Seed Microbiome – Assessing the Bioprotection Activity of Non-pathogenic Relatives of Pathogens. Front. Microbiol. 11:1991. doi: 10.3389/fmicb.2020.01991
Links, M. G., Demeke, T., Gräfenhan, T., Hill, J. E., Hemmingsen, S. M., and Dumonceaux, T. J. (2014). Simultaneous profiling of seed-associated bacteria and fungi reveals antagonistic interactions between microorganisms within a shared epiphytic microbiome on Triticum and brassica seeds. New Phytol. 202, 542–553. doi: 10.1111/nph.12693
Liu, X., Li, Q., Li, Y., Guan, G., and Chen, S. (2019). Paenibacillus strains with nitrogen fixation and multiple beneficial properties for promoting plant growth. PeerJ 7:e7445. doi: 10.7717/peerj.7445
Liu, H., Zhang, L., Meng, A., Zhang, J., Xie, M., Qin, Y., et al. (2017). Isolation and molecular identification of endophytic diazotrophs from seeds and stems of three cereal crops. PLoS One 12:e0187383. doi: 10.1371/journal.pone.0187383
López, S. M. Y., Pastorino, G. N., Franco, M. E. E., Medina, R., Lucentini, C. G., Saparrat, M. C. N., et al. (2018). Microbial endophytes that live within the seeds of two tomato hybrids cultivated in Argentina. Agronomy 8:136. doi: 10.3390/agronomy8080136
Lundberg, D. S., Yourstone, S., Mieczkowski, P., Jones, C. D., and Dangl, J. L. (2013). Practical innovations for high-throughput amplicon sequencing. Nat. Methods 10, 999–1002. doi: 10.1038/nmeth.2634
Mannisto, M. K., Tiirola, M., McConnell, J., and Haggblom, M. M. (2010). Mucilaginibacter frigoritolerans sp. nov., Mucilaginibacter lappiensis sp. nov. and Mucilaginibacter mallensis sp. nov., isolated from soil and lichen samples. Int. J. Syst. Evol. Microbiol. 60, 2849–2856. doi: 10.1099/ijs.0.019364-0
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet.J 17, 10–12. doi: 10.14806/ej.17.1.200
Molina-Romero, D., Baez, A., Quintero-Hernández, V., Castañeda-Lucio, M., Fuentes-Ramírez, L. E., Bustillos-Cristales, M. D. R., et al. (2017). Compatible bacterial mixture, tolerant to desiccation, improves maize plant growth. PLoS One 12:e0187913. doi: 10.1371/journal.pone.0187913
Mousa, W. K., Shearer, C., Limay-Rios, V., Ettinger, C. L., Eisen, J. A., and Raizada, M. N. (2016). Root-hair endophyte stacking in finger millet creates a physicochemical barrier to trap the fungal pathogen Fusarium graminearum. Nat. Microbiol. 1:16167. doi: 10.1038/nmicrobiol.2016.167
Nakaew, N., and Sungthong, R. (2018). Seed phytochemicals shape the community structures of cultivable actinobacteria-inhabiting plant interiors of Thai pigmented rice. MicrobiologyOpen 7, e00591–e00591. doi: 10.1002/mbo3.591
Narayan, R., Chauhan, G. S., and Verma, N. S. (1988). Changes in the quality of soybean during storage. Part 1—effect of storage on some physico-chemical properties of soybean. Food Chem. 27, 13–23. doi: 10.1016/0308-8146(88)90032-5
Nelson, E. B. (2018). The seed microbiome: origins, interactions, and impacts. Plant Soil 422, 7–34. doi: 10.1007/s11104-017-3289-7
Pagano, M. C., and Miransari, M. (2016). “The importance of soybean production worldwide,” in in Abiotic and Biotic Stresses in Soybean Production (ed. M. Miransari (San Diego:Academic Press), 1–26.
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2 – approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490
Qaisrani, M. M., Zaheer, A., Mirza, M. S., Naqqash, T., Qaisrani, T. B., Hanif, M. K., et al. (2019). A comparative study of bacterial diversity based on culturable and culture-independent techniques in the rhizosphere of maize (Zea mays L.). Saudi J. Biol. Sci. 26, 1344–1351. doi: 10.1016/j.sjbs.2019.03.010
Quast, C., Pruesse, E., Yilmaz, P., Gerken, J., Schweer, T., Yarza, P., et al. (2013). The SILVA ribosomal RNA gene database project: improved data processing and web-based tools. Nucleic Acids Res. 41, D590–D596. doi: 10.1093/nar/gks1219
Ramírez, C. A., and Kloepper, J. W. (2010). Plant growth promotion by Bacillus amyloliquefaciens FZB45 depends on inoculum rate and P-related soil properties. Biol. Fertil. Soils 46, 835–844. doi: 10.1007/s00374-010-0488-2
Rascovan, N., Carbonetto, B., Perrig, D., Diaz, M., Canciani, W., Abalo, M., et al. (2016). Integrated analysis of root microbiomes of soybean and wheat from agricultural fields. Sci. Rep. 6, 28084. doi: 10.1038/srep28084
Rezki, S., Campion, C., Simoneau, P., Jacques, M.-A., Shade, A., and Barret, M. (2018). Assembly of seed-associated microbial communities within and across successive plant generations. Plant Soil 422, 67–79. doi: 10.1007/s11104-017-3451-2
Rybakova, D., Mancinelli, R., Wikstrom, M., Birch-Jensen, A. S., Postma, J., Ehlers, R. U., et al. (2017). The structure of the Brassica napus seed microbiome is cultivar-dependent and affects the interactions of symbionts and pathogens. Microbiome 5:104. doi: 10.1186/s40168-017-0310-6
Sarhan, M. S., Hamza, M. A., Youssef, H. H., Patz, S., Becker, M., ElSawey, H., et al. (2019). Culturomics of the plant prokaryotic microbiome and the dawn of plant-based culture media–A review. J. Adv. Res. 19, 15–27. doi: 10.1016/j.jare.2019.04.002
Schmidt, R., Köberl, M., Mostafa, A., Ramadan, E. M., Monschein, M., Jensen, K. B., et al. (2014). Effects of bacterial inoculants on the indigenous microbiome and secondary metabolites of chamomile plants. Front. Microbiol. 5:64. doi: 10.3389/fmicb.2014.00064
Schmidt, J., Messmer, M., and Wilbois, K.-P. (2015). Beneficial microorganisms for soybean (Glycine max (L.) Merr), with a focus on low root-zone temperatures. Plant Soil 397, 411–445. doi: 10.1007/s11104-015-2546-x
Schmidt, M., Zannini, E., and Arendt, E. K. (2018). Recent advances in physical post-harvest treatments for shelf-life extension of cereal crops. Foods 7:45. doi: 10.3390/foods7040045
Selvakumar, G., Kundu, S., Joshi, P., Nazim, S., Gupta, A. D., Mishra, P. K., et al. (2008). Characterization of a cold-tolerant plant growth-promoting bacterium Pantoea dispersa 1A isolated from a sub-alpine soil in the North Western Indian Himalayas. World J. Microbiol. Biotechnol. 24, 955–960. doi: 10.1007/s11274-007-9558-5
Shade, A., Jacques, M.-A., and Barret, M. (2017). Ecological patterns of seed microbiome diversity, transmission, and assembly. Curr. Opin. Microbiol. 37, 15–22. doi: 10.1016/j.mib.2017.03.010
Shahzad, R., Khan, A. L., Bilal, S., Asaf, S., and Lee, I.-J. (2018). What is there in seeds? Vertically transmitted endophytic resources for sustainable improvement in plant growth. Front. Plant Sci. 9:24. doi: 10.3389/fpls.2018.00024
Sharma, S., Kaur, A., Bansal, A., and Gill, B. S. (2013). Positional effects on soybean seed composition during storage. J. Food Sci. Technol. 50, 353–359. doi: 10.1007/s13197-011-0341-0
Shelar, V., Shaikh, R., and Nikam, A. (2008). Soybean seed quality during storage: a review. Agric. Rev. 29, 125–131.
Silini-Cherif, H., Silini, A., Ghoul, M., and Yadav, S. (2012). Isolation and characterization of plant growth promoting traits of a rhizobacteria: Pantoea agglomerans lma2. Pak. J. Biol. Sci. 15, 267–276. doi: 10.3923/pjbs.2012.267.276
Solanki, M. K., Abdelfattah, A., Britzi, M., Zakin, V., Wisniewski, M., Droby, S., et al. (2019). Shifts in the composition of the microbiota of stored wheat grains in response to fumigation. Front. Microbiol. 10:1098. doi: 10.3389/fmicb.2019.01098
Šťovíček, A., Kim, M., Or, D., and Gillor, O. (2017). Microbial community response to hydration-desiccation cycles in desert soil. Sci. Rep. 7:45735. doi: 10.1038/srep45735
Sugiyama, A., Ueda, Y., Takase, H., and Yazaki, K. (2015). Do soybeans select specific species of Bradyrhizobium during growth? Commun. Integr. Biol. 8:e992734. doi: 10.4161/19420889.2014.992734
Supramaniam, Y., Chong, C.-W., Silvaraj, S., and Tan, I. K.-P. (2016). Effect of short term variation in temperature and water content on the bacterial community in a tropical soil. Appl. Soil Ecol. 107, 279–289. doi: 10.1016/j.apsoil.2016.07.003
Susilowati, A. R. I., Wahyudi, A. T., Lestari, Y., Suwanto, A., and Wiyono, S. (2011). Potential pseudomonas isolated from soybean rhizosphere as biocontrol against soilborne phytopathogenic fungi. HAYATI J. Biosci. 18, 51–56. doi: 10.4308/hjb.18.2.51
Sylla, J., Alsanius, B. W., Krüger, E., Reineke, A., Strohmeier, S., and Wohanka, W. (2013). Leaf microbiota of strawberries as affected by biological control agents. Phytopathology 103, 1001–1011. doi: 10.1094/PHYTO-01-13-0014-R
Tannenbaum, I., Kaur, J., Mann, R., Sawbridge, T., Rodoni, B., and Spangenberg, G. (2020). Profiling the Lolium perenne microbiome: from seed to seed. Phytobiomes J. 4, 281–289. doi: 10.1094/PBIOMES-03-20-0026-R
Torres-Cortés, G., Bonneau, S., Bouchez, O., Genthon, C., Briand, M., Jacques, M.-A., et al. (2018). Functional microbial features driving community assembly during seed germination and emergence. Front. Plant Sci. 9:902. doi: 10.3389/fpls.2018.00902
Truyens, S., Weyens, N., Cuypers, A., and Vangronsveld, J. (2015). Bacterial seed endophytes: genera, vertical transmission and interaction with plants. Environ. Microbiol. Rep. 7, 40–50. doi: 10.1111/1758-2229.12181
Tsavkelova, E., Klimova, S. Y., Cherdyntseva, T., and Netrusov, A. (2006). Microbial producers of plant growth stimulators and their practical use: a review. Appl. Biochem. Microbiol. 42, 117–126. doi: 10.1134/S0003683806020013
Vardharajula, S., Zulfikar Ali, S., Grover, M., Reddy, G., and Bandi, V. (2011). Drought-tolerant plant growth promoting Bacillus spp.: effect on growth, osmolytes, and antioxidant status of maize under drought stress. J. Plant Interact. 6, 1–14. doi: 10.1080/17429145.2010.535178
Wassermann, B., Cernava, T., Müller, H., Berg, C., and Berg, G. (2019a). Seeds of native alpine plants host unique microbial communities embedded in cross-kingdom networks. Microbiome 7:108. doi: 10.1186/s40168-019-0723-5
Wassermann, B., Kusstatscher, P., and Berg, G. (2019b). Microbiome response to hot water treatment and potential synergy with biological control on stored apples. Front. Microbiol. 10:2502. doi: 10.3389/fmicb.2019.02502
Xin, X.-F., Kvitko, B., and He, S. Y. (2018). Pseudomonas syringae: what it takes to be a pathogen. Nat. Rev. Microbiol. 16, 316–328. doi: 10.1038/nrmicro.2018.17
Zhang, J., Kobert, K., Flouri, T., and Stamatakis, A. (2014). PEAR: a fast and accurate Illumina paired-end re Ad mergeR. Bioinformatics 30, 614–620. doi: 10.1093/bioinformatics/btt593
Keywords: seed vault, storage strategies, seed bacterial microbiomes, conservation, culturability
Citation: Chandel A, Mann R, Kaur J, Norton S, Edwards J, Spangenberg G and Sawbridge T (2021) Implications of Seed Vault Storage Strategies for Conservation of Seed Bacterial Microbiomes. Front. Microbiol. 12:784796. doi: 10.3389/fmicb.2021.784796
Edited by:
Satish Kumar Verma, Banaras Hindu University, IndiaReviewed by:
Dheeraj Kumar Singh, Banaras Hindu University, IndiaBliss Ursula Furtado, Nicolaus Copernicus University in Toruń, Poland
Amit Kishore Singh, Tilka Manjhi Bhagalpur University, India
Copyright © 2021 Chandel, Mann, Kaur, Norton, Edwards, Spangenberg and Sawbridge. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ankush Chandel, YW5rdXNoLmNoYW5kZWxAYWdyaWN1bHR1cmUudmljLmdvdi5hdQ==
 Ankush Chandel
Ankush Chandel Ross Mann
Ross Mann Jatinder Kaur
Jatinder Kaur Sally Norton
Sally Norton Jacqueline Edwards
Jacqueline Edwards German Spangenberg
German Spangenberg Timothy Sawbridge
Timothy Sawbridge