- College of Life Sciences, Yan’an University, Yan’an, China
Microbial community succession during the enrichment of crude-oil-degrading bacteria was analyzed using Illumina high-throughput sequencing to guide bacterial isolation and construction of a bacterial consortium. Community change occurred in 6 days; the most abundant phylum changed from Proteobacteria to Actinobacteria; the most abundant genera were Dietzia and unspecified_Idiomarinaceae. Two crude oil-degrading strains, Rhodococcus sp. OS62-1 and Dietzia sp. OS33, and one weak-crude-oil-degrading strain, Pseudomonas sp. P35, were isolated. A consortium comprising Rhodococcus sp. OS62-1 and Pseudomonas sp. P35 showed the highest crude-oil-degrading efficiency, reaching 85.72 ± 3.21% within 7 days, over a wide pH range (5–11) and salinity (0–80 g·L−1). Consumption of saturated hydrocarbons, aromatic hydrocarbons, and resins was greater by the consortium than by a single strain, as was degradation of short-chain-alkanes (C13–C17) according to gas-chromatography. The bacterial consortium provides technical support for bioremediation of crude oil pollution.
Introduction
Crude oil is one of the essential compounds in both the energy and chemical industries (Li et al., 2016). However, crude oil leakage caused by natural or human factors results in serious environmental pollution and sustainability problems worldwide (Gu and Wang, 2015; Muthukumar et al., 2022). Most of the components in crude oil can be stored and bioaccumulated in animals and humans, causing serious health issues (Elumalai et al., 2021). Degradation of crude oil pollutants is slow under natural conditions (Varjani and Upasani, 2019). There are numerous methods to repair crude oil pollution, including physicochemical and biological methods (Prendergast and Gschwend, 2014). However, physicochemical methods possess several side effects. Microbial remediation has attracted considerable attention because it is environmentally friendly, economical, and efficient (Kuyukina et al., 2013; Govarthanan et al., 2020).
Because of the complexity of crude oil composition, the biodegradation efficiency by a single microbe is extremely low (Li et al., 2016). Owing to the synergistic effect between strains, consortia possess a wide range of substrate spectra, higher robustness, and better adaptation to complex environments (Varjani and Upasani, 2019). The bacterial consortium constructed using surfactant producer Acinetobacter sp. Y2 and crude oil-degrading Scedosporium sp. ZYY possessed higher biodegradation rate and lower surface tension than the single strain (Atakpa et al., 2022). The bacterial consortium constructed using Dietzia sp. CN-3 and Acinetobacter sp. HC8-3S achieved higher crude oil degradation efficiency and better pH and NaCl tolerance than a single strain (Chen et al., 2020). Therefore, constructing a consortium is essential for microbial remediation of crude oil pollution (Balba et al., 1998; Suja et al., 2014).
To date, due to the lack of relevant theoretical guidance, most of the constructed consortiums are compounded between several crude oil-degrading bacteria, challenging to adapt to the real polluted environment. In fact, in crude oil-contaminated environments, only a tiny proportion of microbes can degrade crude oil, most of which are non or weak oil-degrading microbes (Aislabie et al., 2006). Enrichment culture procedures are usually required before isolating crude oil-degrading bacteria (Phulpoto et al., 2021). Under the stimulation of nutrients and crude oil, microbial communities will undergo the corresponding succession, and crude oil-degrading bacteria will be further enriched. Therefore, exploring the community succession of crude oil degrading bacteria enrichment culture and revealing the microbial community structure under enrichment culture will help to provide a reference for the construction of consortium in the selection of strains. With the development of high-throughput sequencing technology, the microbial diversity in crude oil-contaminated soil (Bidja Abena et al., 2020; Wang et al., 2020; Huang et al., 2021), water bodies (Harayama et al., 2004), and sediments (Chen et al., 2017) has been continuously described. However, the changes in the microbial community during the process of enrichment cultivation is poorly understood.
In the present study, Illumina high-throughput sequencing technology was used to analyze microbial community succession during the enrichment process to guide the isolation of microbes from the enrichment culture and construction of a crude-oil-degradation consortium. Then, the degradation effects of the consortium and single strain were evaluated by first-order kinetics, gas chromatography analysis (GC), and chromatographic separation. Finally, the tolerance of the consortium and single strain to NaCl and pH was also tested. This study provides a theoretical basis for utilizing microorganisms in the remediation of crude oil pollution.
Materials and methods
Chemicals and media
All chemicals used in the study were of analytical or high-purity grade. Crude oil (density: 0.742 g·cm−3) and contaminated soil (crude oil content: 1328.451 mg·kg−1) were obtained from the YANCHANG Oilfield in northern Shaanxi, China. The crude oil-contaminated soil was collected in a sterilized polyethylene bottle, immediately transported to the laboratory, and stored at 4°C until further use.
Two different culture media were used in this study. The enrichment medium (ERM) used for the enrichment cultivation, microbial isolation, and biodegradation test in this study contained (NH4)2SO4 (1 g·L−1), NaNO3 (2 g·L−1), KH2PO4 (5 g·L−1), MgSO4·7H2O (0.3 g·L−1), NaCl (5 g·L−1), microelements (1 ml, Supplementary Table S1), and crude oil (10 g·L−1) at pH 8.5. Peptone yeast glucose (PYG) medium was used for the strain proliferation, which composition was as follows: peptone (5 g·L−1), yeast extract (0.5 g·L−1), glucose (5 g·L−1), beef extract (3 g·L−1), MgSO4·7H2O (1.5 g·L−1) and NaCl (10 g·L−1) at pH 7.5. All media were sterilized at 121°C for 15 min before use.
Methods
Enrichment cultivation
Crude oil-contaminated soil was treated to remove impurities, such as small stones, roots, and leaves, and was subsequently mixed completely. As a control, 5 g of soil sample was collected and stored at −80°C until further use. For enrichment cultivation, a 10 g soil sample was added to 1 l ERM and cultured at 28°C and 180 rpm. Every 3 days, 40 ml of the enrichment culture was collected, 30 ml of the culture was centrifuged at 12,000 rpm for 20 min at 4°C to obtain the pellet, and 10 ml was stored at 4°C for bacterial isolation. The pellets were stored at −80°C until the end of enrichment cultivation (15 days). Three replicate experiments were performed. The frozen soil sample and collected enrichment culture pellets were used for DNA extraction, and Illumina high-throughput sequencing that was performed at Wekemo Tech Co., Ltd. (Shenzhen, China).
Illumina high-throughput sequencing
The microbial communities in the crude oil-contaminated soil and pellets collected from the enrichment culture were sequenced using the Illumina Novaseq platform (Illumina, San Diego, CA, United States). Soil, ERC1, ERC2, ERC3, ERC4, and ERC5 represent soil samples and enrichment cultures on days 3, 6, 9, 12, and 15, respectively. DNA from all samples was extracted using the E.Z.N.A.® Soil DNA Kit (Omega Bio-Tek, Norcross, GA, United States) according to the manufacturer’s protocol. The V3-V4 hypervariable regions of the 16S rRNA gene were amplified using primers 341F (5′-CCTAYGGGRBGCASCAG-3′) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′; Niem et al., 2020). The DNA libraries were constructed using FastPfu Polymerase for PCR to amplify DNA, which was then purified using the AxyPrep DNA Gel Extraction Kit (Axygen Biosciences, Union City, CA, United States) and quantified using QuantiFluor™-ST (Promega, United States) according to the manufacturer’s protocol.
The raw reads were analyzed using the QIIME2 dada 2.0 plugin to obtain the feature table of amplicon sequence variants (ASVs), exact sequence variants, or sub-operational taxonomic units (sub-OTUs; Callahan et al., 2016). Different features contained different sequences at the single-nucleotide level. To minimize the effects of sequencing depth on data analysis, the number of reads in each sample was rarefied to 38,834. The generated ASVs were aligned to the GREENGENES 13_8 database classifier with 99% similarity using the QIIME2 feature-classifier plugin to generate a taxonomic table (Bokulich et al., 2018). Contaminated mitochondrial and chloroplast sequences were filtered and removed using the QIIME2 feature-table plugin.
The 16S rRNA Illumina libraries were deposited in the NCBI small read archive (SRA) dataset under the BioProject accession numbers PRJNA776725 and SRA accession numbers SRR16702634-SRR16702651.
Isolation and identification of bacteria
The 10 ml enrichment cultures were homogenized, and 1 ml of each enrichment culture was taken out and mixed thoroughly. The mixed bacterial suspension was used for bacterial isolation using the dilution-plating method on the ERM and PYG media.
Cell morphology was examined using scanning electron microscopy (JSM-7610f, JEOL, Japan) and Gram staining. For 16S rRNA gene sequencing and phylogenetic analysis, 16S rRNA gene sequences were obtained as previously described (Deng et al., 2020) and the GenBank accession numbers of strains OS62-1, OS33, and P35 were MZ149267, MZ149266, and ON115028, respectively. The 16S rRNA gene sequence of the strain was compared with those available from Ezbiocloud database (Chun et al., 2007). The phylogenetic tree was analyzed by MEGA 5.1 using the Neighbour-Joining (N-J) method (Hollingsworth and Ennos, 2004).
Crude-oil-degradation test
To evaluate the capability of the isolated microbial strains to biodegrade crude oil, the purified strains were first inoculated into fresh PYG medium and cultured at 28°C and 160 rpm for 2 days. After centrifugation at 8,000 rpm for 5 min, the cells were collected and washed 2–3 times using sterile water. The concentration of the bacterial solution was adjusted to OD600 = 1.00 for standby. The proportion of bacteria in the consortium was 1:1 throughout the entire experimental process. Then, the microbial (0.1%, v/v) was added to ERM and cultured at 160 rpm for 7 days. Finally, the residual crude oil was extracted with an equal volume of C2Cl4, dried in anhydrous sodium sulfate, and concentrated via vacuum rotary evaporation. The residual crude oil concentration was determined using an infrared spectrometric oil detector (OIL-480, China Invent, China). The crude-oil-biodegradation rate formula is as follows:
where η is the biodegradation efficiency of crude oil (%), C0 is the initial crude oil concentration in the medium (mg·L−1), and Ct is the crude oil concentration in the medium at time t (mg·L−1).
Kinetics of crude-oil-degradation in microcosm
To further evaluate the biodegradation characteristics of crude oil by the consortium and the single strain, the concentration of residual crude oil and strain growth were measured at an interval of 1 day. Strain growth was evaluated using the plate counting method on PYG agar plates. Crude-oil-degradation was determined using a first-order kinetic model (Kachienga and Momba, 2017). The formula used is as follows:
Specifically, t is the degradation time (days), Ct is the crude oil concentration at time (t), C0 is the initial crude oil concentration, k is the degradation rate constant (day−1), t1/2 is the half-life of crude-oil-degradation (days).
GC analysis
The components of residual crude oil after biodegradation by single strains and the bacterial consortium were measured using GC (7820A, Agilent Technologies, United States) with a capillary column (HP-5 model, 30 m × 0.25 mm × 0.25 μm). The operating conditions were as follows: helium was used as the carrier gas at a flow rate of 2 ml·min−1; the column temperature was 60°C for 1 min, with a ramp to 290°C t a rate of 8°C min−1, raised to 320°C at a rate of 30°C·min−1, which was maintained for 7 min. Petroleum hydrocarbon mixed standards (1,000 μg·mL−1) were used as internal standards to calibrate GC measurements (Liu et al., 2009).
Degradation of different crude oil components
The chromatographic separation of saturated, aromatic, resin, and asphaltene fractions in residual crude oil after biodegradation followed the method described by Márquez et al. (1999). Briefly, the residual crude oil after biodegradation was dissolved in 30 ml n-hexane, asphaltene was filtered by degreasing cotton and dissolved in CHCl3. The filtrate was then concentrated to 2 ml and added to a glass column containing neutral alumina (200–300 mesh, Kermel, China). The saturated fraction was extracted with 30 ml n-hexane, yielding a colorless fraction. The aromatic fraction was extracted using a 2:1 (v/v) CH2Cl2 / C6H14 mixture (20 ml), yielding a yellowish fraction. The resin fraction was first separated with 10 ml ethanol and then extracted with 10 ml chloroform. The solvents in the fractions were evaporated and weighed.
Effect of culture conditions on crude-oil-biodegradation
Crude-oil-biodegradation of the potent isolate was carried out in the presence of different pH and salinity levels using a one-variable-at-a-time approach, keeping other parameters constant. Different pH (3.0–11.0) and salinity (0–70 g·L−1) values were prepared in the ERM. The bacterial consortium and single strain were inoculated into each medium (0.1%, v/v) and then cultured at 28°C and 180 rpm for 7 days to determine the concentration of remaining crude oil.
Statistical analysis
All experiments were performed in triplicate, and the data are expressed as mean ± standard deviation (SD). ORIGIN 2020, SPSS 22.0, and R 4.1.3 software were used for data analysis and statistics, with p < 0.05, indicating a significant statistical difference.
Results and discussion
Microbial diversity during enrichment cultivation
Illumina high-throughput sequencing generated 1,304,557 high-quality reads from 6,244 features at single-nucleotide resolution. After rarefication to 38,834 reads for each sample, 5,825 features were generated from all 18 samples.
The crude oil-containing soil samples showed significantly higher observed_OTUs index and Shannon index than the enrichment samples (Supplementary Table S2). The observed_OTUs decreased sharply from 1,548 ± 185 in the soil samples to 153 ± 2 in the enrichment samples on day 3 (ERC1). However, when enrichment was continued to days 6, 9, and 12, the observed_OTUs index increased to 294 ± 118, 217 ± 70, and 630 ± 232, respectively. Then, on day 15, the observed_OTUs index decreased to 179.866 ± 57.960. Similarly, the Shannon index for the soil samples was higher than that of the enrichment cultures and was reduced from 7.902 ± 0.551 in the soil samples to 2.788 ± 0.169 in ERC1. The lowest Shannon index was observed on day 9 (1.298 ± 0.306). The other alpha diversity indices, including the Chao1, Faith_PD, and Simpson indices, showed similar trends (Supplementary Table S2). Thus, as described in previous studies (Jin and Kim, 2017; Mu et al., 2018), the microbial alpha diversity in the enrichment culture samples was considerably lower than that in the soil samples.
Microbial community structure
We examined bacterial taxa and their relative abundances in the soil and enrichment culture samples at different time points. In soil, the predominant phyla were Actinobacteria (45.20 ± 7.25%), Proteobacteria (26.48 ± 4.46%), Firmicutes (10.00 ± 1.80%), and Bacteroidetes (14.24 ± 11.02%; Figure 1A). Unlike the microbial community composition described in the investigation of historic oilfield-contaminated soil in the Loess Plateau (Gao et al., 2019), the dominant phylum in this study was Actinobacteria, rather than Proteobacteria, possibly owing to differences in contamination levels. In ERC1, Proteobacteria predominated (93.39 ± 1.29%), followed by Actinobacteria (4.28 ± 1.32%), and Firmicutes (1.95 ± 0.58%). The relative abundances of the other phyla in ERC1 were below 1%. In ERC2-5, Actinobacteria and Proteobacteria were the predominant phyla (58.86 ± 19.72% and 38.52 ± 19.40%, respectively). Compared to ERC1, the relative abundance of Actinobacteria was significantly higher in ERC2-5 (Student’s t-test, p = 0.003, 0.011, 0.037, 0.048 for ERC2-5 compared to ERC1, respectively), whereas that of Proteobacteria was significantly lower (Student’s t-test, p = 0.001, 0.012, 0.041, 0.046 for ERC2-5 compared to ERC1, respectively). Despite fluctuations in relative abundance, Actinobacteria and Proteobacteria were the predominant phyla in ERC2-5 (Figure 1A).
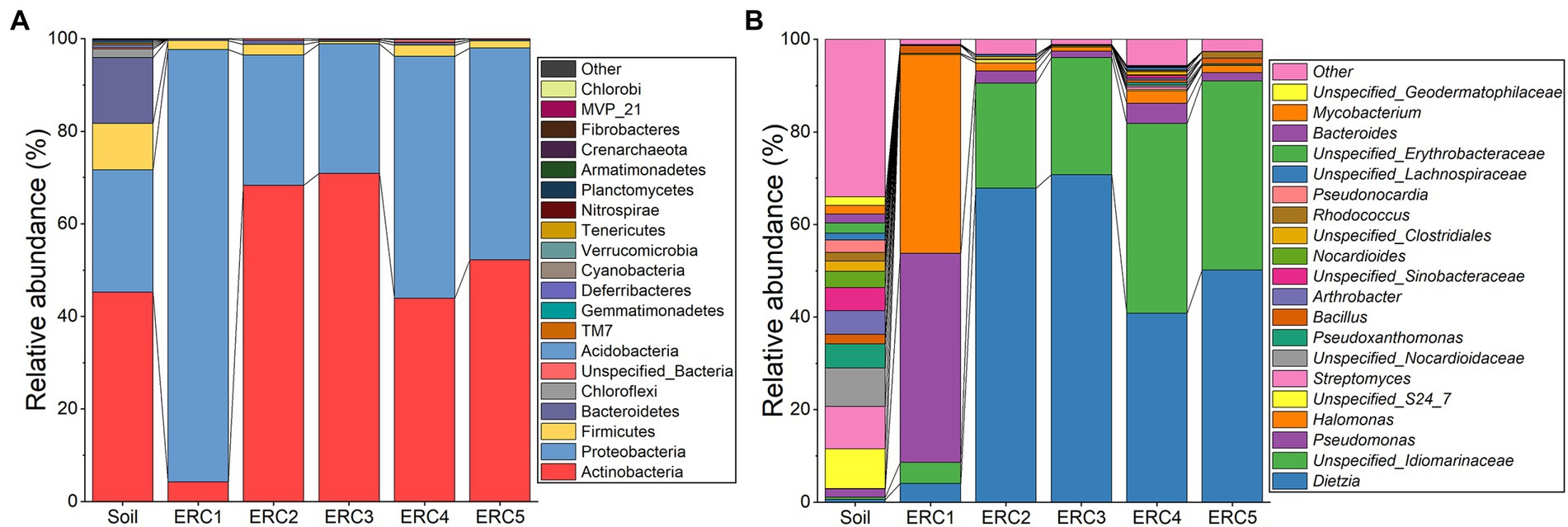
Figure 1. Microbial community composition during enrichment culture at (A) phylum and (B) genus levels (relative abundance top 20).
The top 20 identified genera are shown in Figure 1B. A noticeable difference was observed in the distribution of genera among the soil, ERC1, and ERC2-5 samples. The relative abundance of the top 20 genera in the soil samples displayed a more uniform distribution than those in the ERC samples. In ERC1, Pseudomonas and Halomonas showed high relative abundances (45.13 ± 10.90% and 42.88 ± 11.78%, respectively), followed by Dietzia and an unspecified_Idiomarinaceae genus (4.07 ± 1.27% and 4.58 ± 0.90%, respectively; Figure 1B). In ERC2, the relative abundances of Dietzia and unspecified_Idiomarinaceae were increased dramatically, to 67.91 ± 7.92% and 22.63 ± 2.34%, respectively. ERC3-5 showed a bacterial composition similar to that of ERC2.
The composition of microbial communities reached a relatively steady state on day 6 of enrichment cultivation, indicating that a duration of 1 week was sufficient for enrichment cultivation under these conditions. Dietzia sp. have been reported to be marker microorganisms of crude oil pollution, and many Dietzia sp. have an excellent ability to degrade crude oil (Wang et al., 2014; Huang et al., 2021; Venil et al., 2021). The high abundance of the genera Halomonas and Pseudomonas in ERC1 implied they played important roles in the initial stages of crude-oil-degradation, whereas the abundance of Dietzia and unspecified_Idiomarinaceae suggested that their roles were important in the later stages of crude-oil-degradation. Unfortunately, no strains belonging to unspecified_Idiomarinaceae and Halomonas were isolated in this study, and their roles could not be investigated.
Biodegradation of crude oil by defined consortium
Three kinds of bacteria, OS62-1, OS33, and P35, were obtained; OS62-1 and OS33 were isolated using ERM medium and P35 was isolated using PYG medium. After 3 days of incubation on PYG agar plates, strain OS62-1 formed circular, smooth, pink colonies (0.5–1.0 mm). The strain OS62-1 was Gram-positive. Scanning electron microscopy revealed that it was a coccus (0.5–1.0 μm) bacterium. The strain OS33 was a Gram-positive bacilli bacterium (2.0 × 0.5 μm), forming wet, orange and opaque colonies (0.5–1.0 mm). Strain P35 formed wet, light yellow, opaque, and shiny colonies (1.0–2.0 mm), which were Gram-negative bacilli (3.0 × 1.0 μm; Supplementary Figure S1).
The 16S rRNA gene sequence comparisons indicated that strain OS62-1 showed a high similarity (100%) to the previously isolated bacterium Rhodococcus qingshengii cqsV23. The strain OS33 possessed 100% sequence similarity with the strain Dietzia maris DSM 43672. The strain P35 possessed 99.51% sequence similarity with the strain Pseudomonas songnenensis MXR1709B06. Phylogenetic trees based on the 16S rRNA sequences of strains OS62-1, OS33, and P35 were constructed using neighbor-joining (Figures 2A–C). Based on the phylogenetic analysis results, strains OS62-1, OS33, and P35 were tentatively suggested to be members of the genera Rhodococcus, Dietzia and Pseudomonas, respectively.
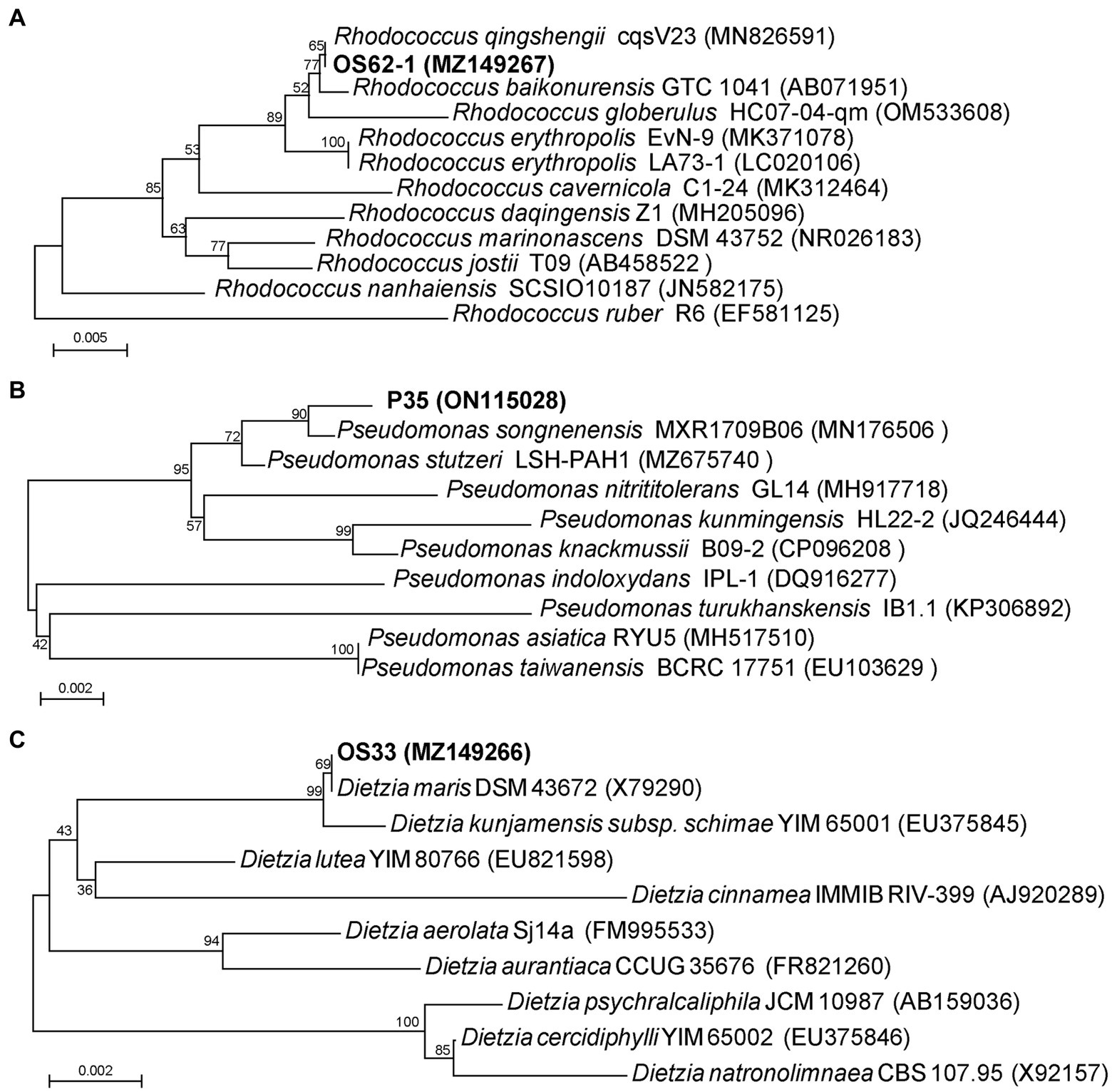
Figure 2. Phylogenetic tree based on 16S rRNA sequences of OS62-1 (A), P35 (B), OS33 (C) and related strains. Bar = 0.01 nucleotide substitution per locus.
The propagation speed and morphology of Rhodococcus sp., OS62-1, and Dietzia sp. OS33 were good during the biodegradation of crude oil. As shown in Figure 3, the biodegradation rates of Rhodococcus sp. OS62-1, Dietzia sp. OS33, and Pseudomonas sp. P35 over 7 days were 72.28 ± 3.60%, 65.46 ± 3.57%, and 13.61 ± 4.12%, respectively. It has been extensively reported that Rhodococcus is one of the groups with most potential for biodegradation and it can degrade crude oil, especially aromatic compounds (Li et al., 2020). In this study, the control group also exhibited an 8.51 ± 1.06% degradation rate, which may be due to the loss of crude oil in the extraction process. Although many Pseudomonas sp. were proved to be good crude-oil-degrading bacteria, the crude-oil-degradation efficiency of Pseudomonas sp. P35 was only slightly higher than that in the control group, suggested that the Pseudomonas sp. P35 can be considered a very weak-oil-degrading bacterium.
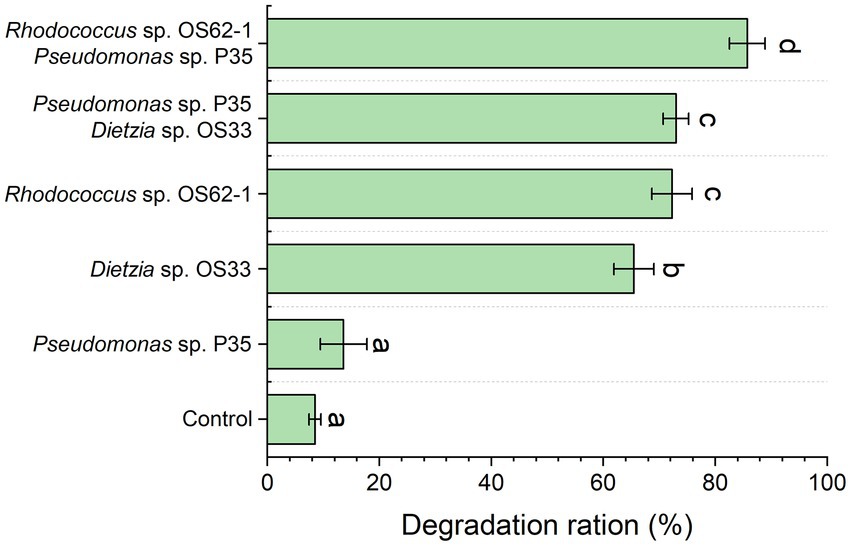
Figure 3. The crude oil degradation efficiency of the isolated strains and the constructed consortium was tested for 7 days. Groups sharing different letters (a, b, c) indicate significant differences between two treatments by one-way ANOVA test (p < 0.05).
To further study the role of the weak oil-degrading bacterium Pseudomonas sp. P35, we combined it with Rhodococcus sp. OS62-1 and Dietzia sp. OS33 in a 1:1 ratio to construct a bacterial consortium and tested the crude-oil-degradation efficiency. Pseudomonas sp. P35 formed good symbiotic relationships with Rhodococcus sp. OS62-1 and Dietzia sp. OS33 and the crude-oil-degradation efficiency over 7 days increased to 85.72 ± 3.21% and 72.97 ± 2.30%, respectively (Figure 3). Although some studies have shown that Pseudomonas can promote the degradation ability of some petroleum degrading bacteria (Isaac et al., 2015; Hu et al., 2020; Feng et al., 2021), our study showed that Pseudomonas had different promoting effects on different petroleum degrading bacteria. The Pseudomonas sp. P35 had a stronger ability to promote the crude oil degradation of Rhodococcus sp. OS62-1 than Dietzia sp. OS33. Therefore, we selected Rhodococcus sp. OS62-1 and Pseudomonas sp. P35 for further study.
Kinetics of crude-oil-degradation
Microcosms for crude-oil-biodegradation were prepared by inoculating Rhodococcus sp. OS62-1, Pseudomonas sp. P35, and the consortium into 100 ml of ERM amended with 10,000 mg·L−1 crude oil. The biodegradation rate of crude oil in the ERM inoculated with Pseudomonas sp. P35 was extremely weak (Figure 4A); its rate constant (k) was only 0.031 d−1 (R2 = 0.96), and its half-life (t1/2) was 22.36 d (Table 1). On the second day, the crude oil inoculated with Rhodococcus sp. OS62-1 dispersed in the ERM as tiny particles, the turbidity of the medium increased with obvious emulsification, and the degradation rate of crude oil accelerated (Figure 4B). The rate constant (k) was 0.201 d−1 (R2 = 0.96), and the half-life (t1/2) was 3.45 d (Table 1). Unfortunately, the biosurfactants produced by Rhodococcus sp. OS62-1 were not successfully separated, suggested that the surfactants might attached to the cells tightly. Compared to the single strain, the crude oil degradation rate constant (k) of the consortium increased to 0.315 d−1 (R2 = 0.99), and the half-life (t1/2) was shortened to 2.20 d (Table 1). The biomass of Rhodococcus sp. OS62-1 and Pseudomonas sp. P35 also increased significantly in the consortium than in the single strain tests (Figure 4C), suggested that the two strains can assist each other in crude oil degradation.
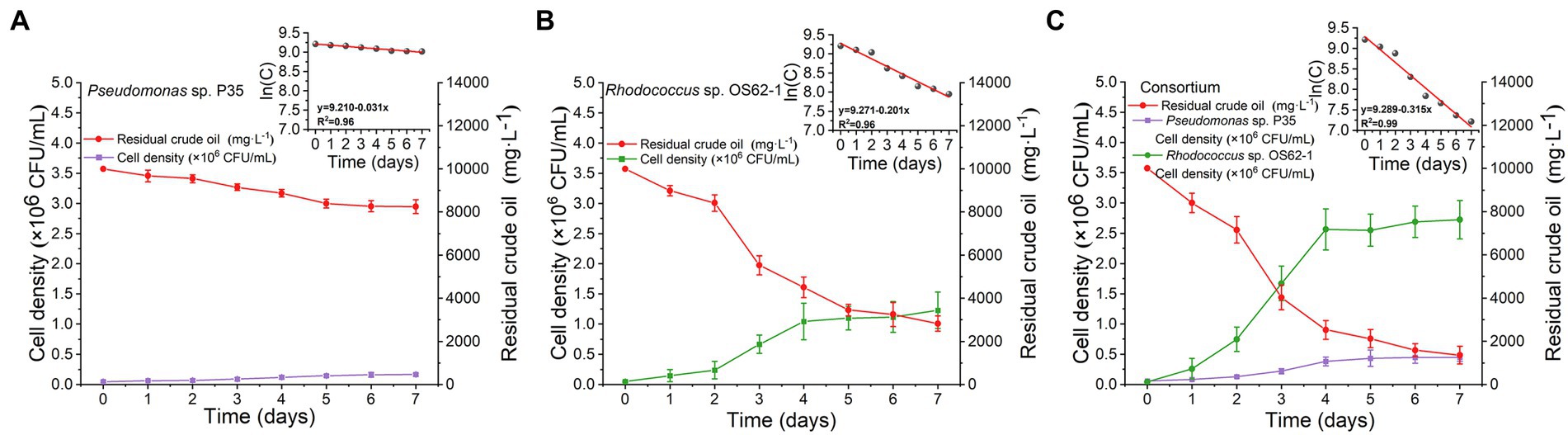
Figure 4. Time course of strain cell density and crude oil biodegradation by (A) Pseudomonas sp. P35, (B) Rhodococcus sp. OS62-1 and (C) Consortium in 10,000 mg·L−1 crude oil amended ERM. The values represent the average of two independent experiments with three replicates each.
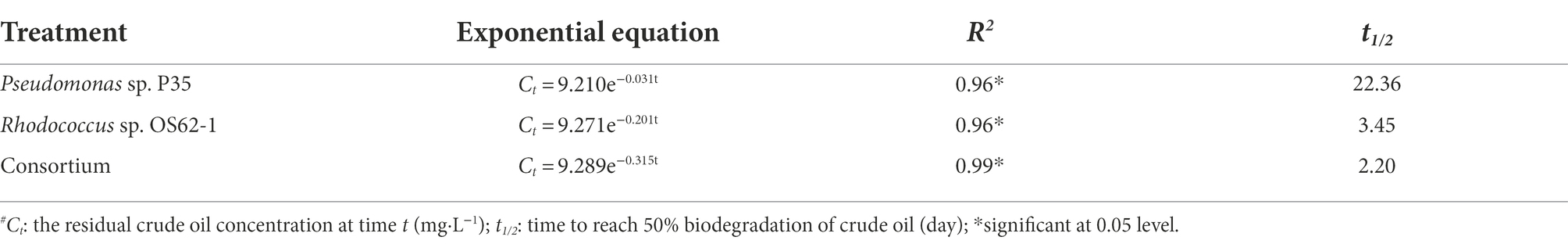
Table 1. First-order kinetics equations and the corresponding half-life times of crude oil biodegradation by Rhodococcus sp. OS62-1, Pseudomonas sp. P35, and Consortium at 28°C in 7 days.
It is important to evaluate the biodegradation kinetics of crude oil to develop effectively and predicted bioremediation models (Chettri and Singh, 2019). In this study, the crude oil concentration decreased with incubation time, and the data were in good agreement with the first-order kinetic equation (R2 > 0.95). There are several reports on crude-oil-biodegradation by either a single strain or a consortium fitted to first-order reaction kinetics at low crude oil concentrations (Chen et al., 2008; Gu et al., 2016). Moreover, the consortium and Rhodococcus sp. OS62-1 degraded crude oil at a relatively high concentration for a short period of 7 days. However, when incubated for longer than 4 days, the degradation efficiency of crude oil began to slow owing to nutrient deficiency (Singh et al., 2014) or accumulation of toxic substances (Chettri et al., 2016).
GC analysis
The biodegradation ratios of n-alkanes (C13–C37) in crude oil were calculated by semi-quantitative analysis of the GC data for residual and fresh crude oil. According to previous studies (Li et al., 2013), the carbon atom number ranges of short-chain alkanes, long-chain alkanes, and heavy long-chain alkanes are considered to be C10–C17, C18–C30 and C31–C38, respectively. The concentrations of only a few long-chain alkanes (C17–C21) were slightly decreased by Pseudomonas sp. P35 (Figure 5), providing further evidence for its weak oil-degrading bacteria status. Rhodococcus sp. OS62-1 can use C13–C33 n-alkanes in crude oil as carbon sources, similar to R. erythropolis M-25 and R. zopfii P2-29 (Pi et al., 2017). The biodegradation ratio of the total crude oil by the consortium was 85.84%, which was 10.76% more than that of the Rhodococcus sp. OS62-1 (75.08%). More specifically, 94.61% of short-chain alkanes, 77.57% of long-chain alkanes, and 36.09% of heavy long-chain alkanes were degraded by the consortium. Although Rhodococcus sp. OS62-1 performed well in the degradation of short-chain alkanes, the consortium seemed to have a higher degradation efficiency especially for C13-C17 n-alkanes, which almost completely disappeared, illustrating the powerful performance of the consortium.
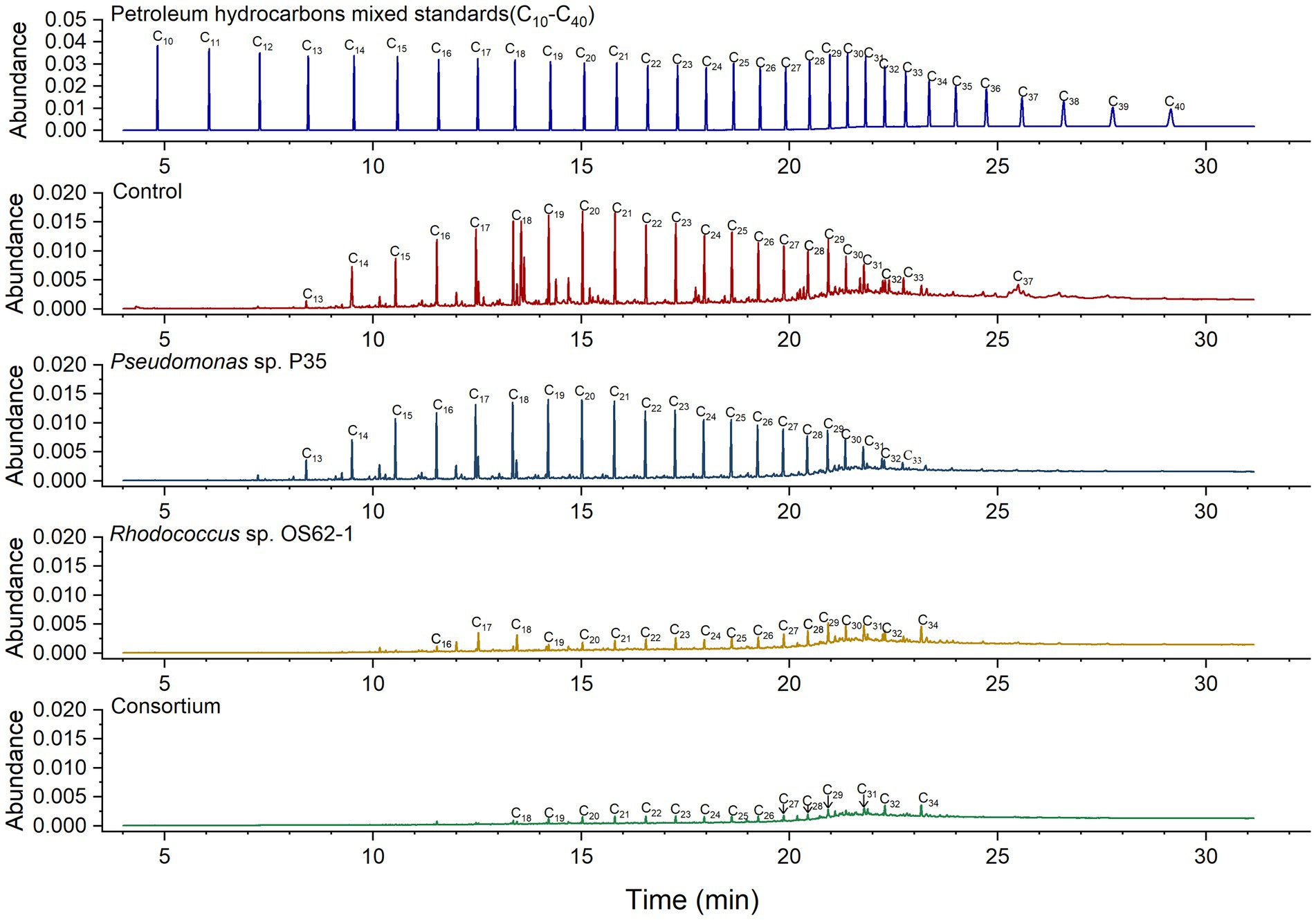
Figure 5. Gas chromatogram (GC) of biodegradation efficiency of saturated hydrocarbons in crude oil by single strain and consortium treatment in 7 days.
Measurement of crude oil components
Biodegradation of different components of crude oil by single strain and the consortium was analyzed using chromatographic separation. The contents of the diverse components are displayed in Figure 6. The biodegradation efficiency of the consortium was significantly higher (p < 0.05) than that of the single strain for the different components. The contents of saturated alkanes decreased from 0.428 ± 0.028 g to 0.055 ± 0.028 g after treatment with the consortium, which was significantly higher (p < 0.05) by more than 48.11% compared to the Rhodococcus sp. OS62-1 (0.106 ± 0.062 g). A similar situation also occurred for aromatic hydrocarbons, the content of aromatic hydrocarbons in residual crude oil after treatment with the consortium was 0.035 ± 0.008 g, which was significantly lower (p < 0.05) than the decrease with Rhodococcus sp. OS62-1 (0.065 ± 0.009).
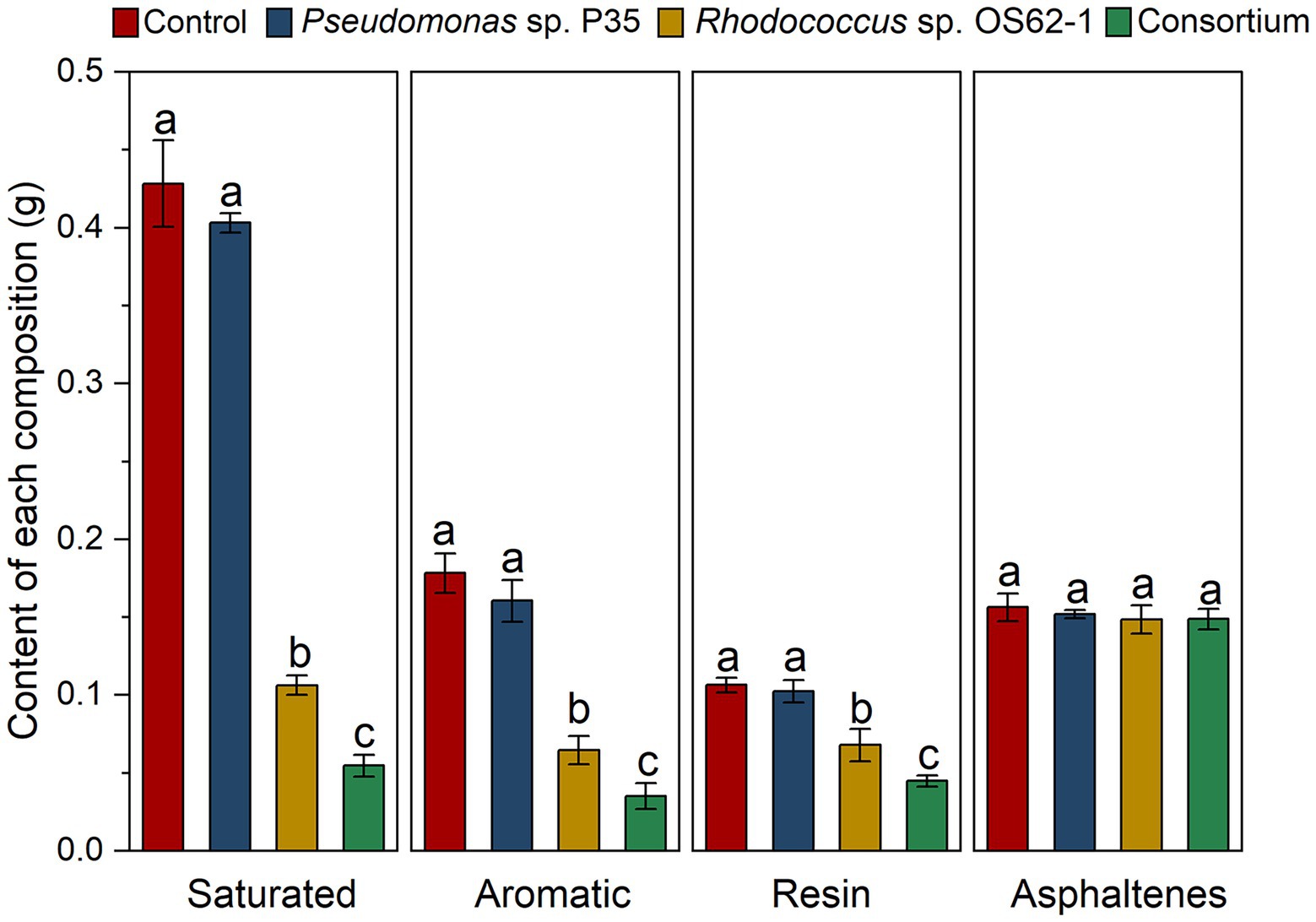
Figure 6. Changes of contents of different components in crude oil under different treatments in 7 days. Groups sharing different letters (a, b, c) indicate significant differences between two treatments by one-way ANOVA test (p < 0.05).
Owing to their complex structures, resins and asphaltenes in crude oil are extremely difficult to degrade (Ding et al., 2021). Excitingly, resin content also decreased significantly after treatment by Rhodococcus sp. OS62-1 and consortium. At present, there have been many reports on the application of microorganisms to solve the problem of resin deposition in crude oil transportation (Patel and Bhaskaran, 2018; Wang et al., 2022). The main applied bacteria are Bacillus sp., Pseudomonas sp., Ochrobactrum sp., but the reports on the degradation of resin by Rhodococcus sp. are limited. In this study, Rhodococcus sp. OS62-1 significantly decreased the resin content in crude oil from 0.107 ± 0.005 g to 0.068 ± 0.011 g (p < 0.05). Meanwhile, the consortium also performed better than Rhodococcus sp. OS62-1 in resin removal, indicating that it had great potential in resin removal. Compared with resin, the number of heteroatoms in asphaltene is higher, and its degradation is more difficult (Ding et al., 2021). The asphaltene content in this study did not change significantly, as described by (Chaineau et al., 1995), who found that asphaltenes were completely retained after saturation and aromatic hydrocarbons were completely degraded by microorganisms.
Effect of culture conditions on crude-oil-biodegradation
It is necessary to understand the effects of physical and chemical factors (e.g., pH and salinity) on biodegradation because these factors significantly affect the biological activity and biodegradation efficiency of microorganisms (Khanpour-Alikelayeh et al., 2020). The pH tolerance range of Rhodococcus sp. OS62-1 and Pseudomonas sp. P35 was 5–11, and the salinity tolerance range of Pseudomonas sp. P35 was 0–80 g·L−1 which was wider than that of Rhodococcus sp. OS62-1 (0–50 g·L−1). Because the resistance of microorganisms to environmental salinity above 30 g·L−1 expands the opportunities for their use in bioremediation of oil-contaminated ground and water ecosystems (Iminova et al., 2022), the halotolerant characteristics of these two strains will lay the foundation for their practical applications.
Based on the adaptability of Rhodococcus sp. OS62-1, the effects of pH and salinity on crude-oil-biodegradation by a single strain and a consortium were studied. As shown in Figure 7, the crude-oil-biodegradation rates of Rhodococcus sp. OS62-1 were higher than 20% at NaCl concentrations ranging from 0–40 g·L−1; the optimum NaCl concentration was 5 g·L−1 (75.83 ± 1.73%). Because of the synergistic effects of Rhodococcus sp. OS62-1 and Pseudomonas sp. P35, the crude-oil-biodegradation efficiency of the consortium was significantly higher than that of the single strain in NaCl concentrations of 0–60 g·L−1 (p < 0.05). Like Rhodococcus sp. OS62-1, the optimal NaCl concentration for biodegradation by the consortium was also 5 g·L−1 (85.89 ± 1.21%). When the NaCl concentration was 70 g·L−1, no significant difference in crude oil content was detected between the treated and untreated flasks after 7 days of incubation.
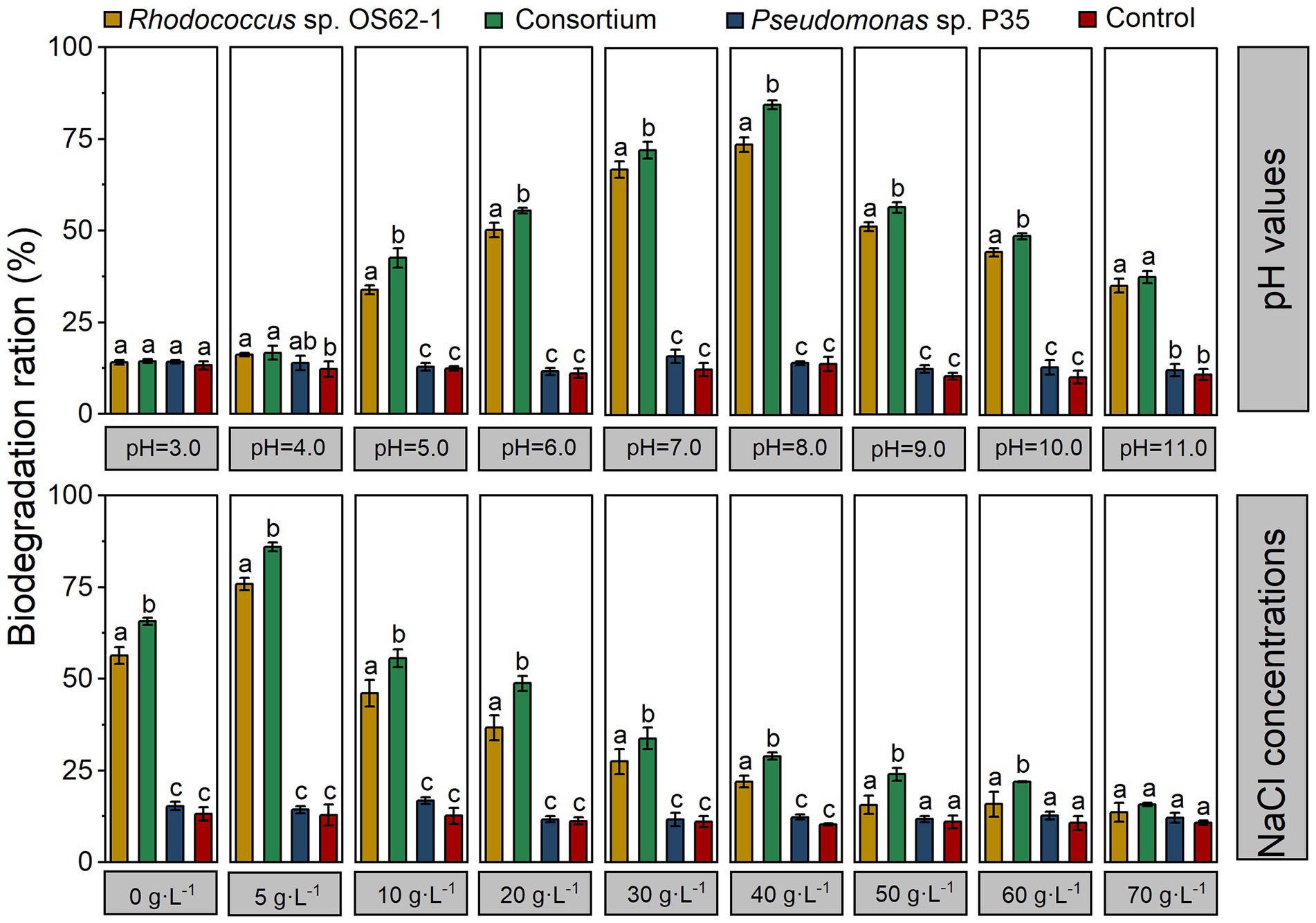
Figure 7. The degradation efficiency of crude oil by the single strain and consortium under different pH values and NaCl concentrations in 7 days. Groups sharing different letters (a, b, c) indicate significant differences between two treatments by one-way ANOVA test (p < 0.05).
To investigate effects of pH on crude-oil-degradation activity, experiments were carried out at different pH ranges (3–11). In the ranges of pH 5–10, the crude-oil-biodegradation efficiencies of the consortium were significantly higher than those of the single strain (p < 0.05), and varied from 42.54 ± 2.61% to 84.58 ± 1.21%. The biodegradation rate was higher than 55% within the pH range of 6–9, and the optimum pH was 8 (84.58 ± 1.21%). It is worth noting that the degradation efficiency of crude oil remained at 37.66% ± 1.67 in strong alkaline conditions (pH = 11).
Crude oil pollution is often accompanied by soil salinization (Huang et al., 2021), so the bacteria that are planned for use in the course of remediation must have salt and alkali resistance. According to previous studies, pH and NaCl concentrations are significant environmental factors affecting crude-oil-biodegradation (Kästner et al., 1998), and slightly saline (Zaki et al., 2015) and alkaline (Khanpour-Alikelayeh et al., 2020) conditions result in better hydrocarbon degradation performance. In this study, the constructed consortium showed the highest degradation efficiency at pH = 8.0 and NaCl concentration of 5 g·L−1; the pH tolerance range was 5–11 and the NaCl tolerance range was 0–60 g·L−1. Therefore, it can be concluded that this bacterial consortium can be used for real crude oil pollution of soil and water.
Conclusion
The Illumina high-throughput sequencing results showed that the microbial community changes occurred within 6 days during the enrichment process and the Dietzia sp. OS33 and Pseudomonas sp. P35 possessed high abundance during the enrichment cultivation, while the Rhodococcus sp. OS62-1 possessed very low relative abundance throughout the enrichment process. However, the consortium composed of Rhodococcus sp. OS62-1 and Pseudomonas sp. P35 achieved the highest crude-oil-degradation efficiency. Based on the results of the first-order kinetic model, GC analysis, chromatographic separation, and NaCl and pH tolerance test, the constructed consortium has higher crude oil degradation efficiency and better environmental tolerance than a single strain. In summary, our results demonstrated the microbial changes during the enrichment process and constructed a consortium with high crude oil degradation efficiency and environmental tolerance.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
TY: experimentation and writing—original draft. YJ: conceptualization and methodology. JA: data curation and software. JW, XinL, and YG: experimentation. XH: methodology and resources. ZD: methodology and supervision. XiaoL: project administration, and writing—review and editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by Special Scientific Research Project of Shaanxi Education Department (no. 21JK0992); Special fund for science and technology of Yan’an City (no. 2019–27; no. 203010105); and Doctor Support Project of Yan’an University (no. YDBK2019-43).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2022.1044448/full#supplementary-material
References
Aislabie, J., Saul, D. J., and Foght, J. M. (2006). Bioremediation of hydrocarbon-contaminated polar soils. Extremophiles 10, 171–179. doi: 10.1007/s00792-005-0498-4
Atakpa, E. O., Zhou, H., Jiang, L., Ma, Y., Liang, Y., Li, Y., et al. (2022). Improved degradation of petroleum hydrocarbons by co-culture of fungi and biosurfactant-producing bacteria. Chemosphere 290:133337. doi: 10.1016/j.chemosphere.2021.133337
Balba, M. T., Al-Awadhi, N., and Al-Daher, R. (1998). Bioremediation of oil-contaminated soil: microbiological methods for feasibility assessment and field evaluation. J. Microbiol. Methods 32, 155–164. doi: 10.1016/S0167-7012(98)00020-7
Bidja Abena, M. T., Chen, G., Chen, Z., Zheng, X., Li, S., Li, T., et al. (2020). Microbial diversity changes and enrichment of potential petroleum hydrocarbon degraders in crude oil-, diesel-, and gasoline-contaminated soil. 3 Biotech 10:42. doi: 10.1007/s13205-019-2027-7
Bokulich, N. A., Kaehler, B. D., Rideout, J. R., Dillon, M., Bolyen, E., Knight, R., et al. (2018). Optimizing taxonomic classification of marker-gene amplicon sequences with QIIME 2’s q2-feature-classifier plugin. Microbiome 6:90. doi: 10.1186/s40168-018-0470-z
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Chaineau, C. H., Morel, J. L., and Oudot, J. (1995). Microbial degradation in soil microcosms of fuel oil hydrocarbons from drilling cuttings. Environ. Sci. Technol. 29, 1615–1621. doi: 10.1021/es00006a027
Chen, W., Kong, Y., Li, J., Sun, Y., Min, J., and Hu, X. (2020). Enhanced biodegradation of crude oil by constructed bacterial consortium comprising salt-tolerant petroleum degraders and biosurfactant producers. Int. Biodeterior. Biodegradation 154:105047. doi: 10.1016/j.ibiod.2020.105047
Chen, C., Liu, Q., Liu, C., and Yu, J. (2017). Effect of different enrichment strategies on microbial community structure in petroleum-contaminated marine sediment in Dalian, China. Mar. Pollut. Bull. 117, 274–282. doi: 10.1016/j.marpolbul.2017.02.004
Chen, J., Wong, M. H., Wong, Y. S., and Tam, N. F. Y. (2008). Multi-factors on biodegradation kinetics of polycyclic aromatic hydrocarbons (PAHs) by Sphingomonas sp. a bacterial strain isolated from mangrove sediment. Mar. Pollut. Bull. 57, 695–702. doi: 10.1016/j.marpolbul.2008.03.013
Chettri, B., Mukherjee, A., Langpoklakpam, J. S., Chattopadhyay, D., and Singh, A. K. (2016). Kinetics of nutrient enhanced crude oil degradation by Pseudomonas aeruginosa AKS1 and Bacillus sp. AKS2 isolated from Guwahati refinery, India. Environ. Pollut. 216, 548–558. doi: 10.1016/j.envpol.2016.06.008
Chettri, B., and Singh, A. K. (2019). Kinetics of hydrocarbon degradation by a newly isolated heavy metal tolerant bacterium Novosphingobium panipatense P5: ABC. Bioresour. Technol. 294:122190. doi: 10.1016/j.biortech.2019.122190
Chun, J., Lee, J.-H., Jung, Y., Kim, M., Kim, S., Kim, B. K., et al. (2007). EzTaxon: a web-based tool for the identification of prokaryotes based on 16S ribosomal RNA gene sequences. Int. J. Syst. Evol. Microbiol. 57, 2259–2261. doi: 10.1099/ijs.0.64915-0
Deng, Z., Jiang, Y., Chen, K., Li, J., Zheng, C., Gao, F., et al. (2020). One biosurfactant-producing bacteria Achromobacter sp. A-8 and its potential use in microbial enhanced oil recovery and bioremediation. Front. Microbiol. 11:247. doi: 10.3389/fmicb.2020.00247
Ding, Y., Wyckoff, K. N., He, Q., Cao, X., and Huang, B. (2021). Biodegradation of waste asphalt shingle by white rot fungi. J. Clean. Prod. 310:127448. doi: 10.1016/j.jclepro.2021.127448
Elumalai, P., Parthipan, P., Huang, M., Muthukumar, B., Cheng, L., Govarthanan, M., et al. (2021). Enhanced biodegradation of hydrophobic organic pollutants by the bacterial consortium: impact of enzymes and biosurfactants. Environ. Pollut. 289:117956. doi: 10.1016/j.envpol.2021.117956
Feng, S., Gong, L., Zhang, Y., Tong, Y., Zhang, H., Zhu, D., et al. (2021). Bioaugmentation potential evaluation of a bacterial consortium composed of isolated Pseudomonas and Rhodococcus for degrading benzene, toluene and styrene in sludge and sewage. Bioresour. Technol. 320:124329. doi: 10.1016/j.biortech.2020.124329
Gao, S., Liang, J., Teng, T., and Zhang, M. (2019). Petroleum contamination evaluation and bacterial community distribution in a historic oilfield located in loess plateau in China. Appl. Soil Ecol. 136, 30–42. doi: 10.1016/j.apsoil.2018.12.012
Govarthanan, M., Ameen, F., Kamala-Kannan, S., Selvankumar, T., Almansob, A., Alwakeel, S. S., et al. (2020). Rapid biodegradation of chlorpyrifos by plant growth-promoting psychrophilic Shewanella sp. BT05: an eco-friendly approach to clean up pesticide-contaminated environment. Chemosphere 247:125948. doi: 10.1016/j.chemosphere.2020.125948
Gu, H., Lou, J., Wang, H., Yang, Y., Wu, L., Wu, J., et al. (2016). Biodegradation, biosorption of phenanthrene and its trans-membrane transport by Massilia sp. WF1 and phanerochaete chrysosporium. Front. Microbiol. 7:38. doi: 10.3389/fmicb.2016.00038
Gu, J. D., and Wang, Y. S. (2015). Coastal and marine pollution and ecotoxicology. Ecotoxicology 24, 1407–1410. doi: 10.1007/s10646-015-1528-3
Harayama, S., Kasai, Y., and Hara, A. (2004). Microbial communities in oil-contaminated seawater. Curr. Opin. Biotechnol. 15, 205–214. doi: 10.1016/j.copbio.2004.04.002
Hollingsworth, P. M., and Ennos, R. A. (2004). Neighbour joining trees, dominant markers and population genetic structure. Heredity 92, 490–498. doi: 10.1038/sj.hdy.6800445
Hu, B., Wang, M., Geng, S., Wen, L., Wu, M., Nie, Y., et al. (2020). Metabolic exchange with non-alkane-consuming Pseudomonas stutzeri SLG510A3-8 improves n-alkane biodegradation by the alkane degrader Dietzia sp. strain DQ12-45-1b. Appl. Environ. Microbiol. 86, e02931–e02919. doi: 10.1128/AEM.02931-19
Huang, L., Ye, J., Jiang, K., Wang, Y., and Li, Y. (2021). Oil contamination drives the transformation of soil microbial communities: co-occurrence pattern, metabolic enzymes and culturable hydrocarbon-degrading bacteria. Ecotoxicol. Environ. Saf. 225:112740. doi: 10.1016/j.ecoenv.2021.112740
Iminova, L., Delegan, Y., Frantsuzova, E., Bogun, A., Zvonarev, A., Suzina, N., et al. (2022). Physiological and biochemical characterization and genome analysis of Rhodococcus qingshengii strain 7B capable of crude oil degradation and plant stimulation. Biotechnol. Rep. 35:e00741. doi: 10.1016/j.btre.2022.e00741
Isaac, P., Martínez, F. L., Bourguignon, N., Sánchez, L. A., and Ferrero, M. A. (2015). Improved PAHs removal performance by a defined bacterial consortium of indigenous Pseudomonas and Actinobacteria from Patagonia, Argentina. Int. Biodeterior. Biodegradation 101, 23–31. doi: 10.1016/j.ibiod.2015.03.014
Jin, C. E., and Kim, M. N. (2017). Change of bacterial community in oil-polluted soil after enrichment cultivation with low-molecular-weight polyethylene. Int. Biodeterior. Biodegradation 118, 27–33. doi: 10.1016/j.ibiod.2017.01.020
Kachienga, L., and Momba, M. N. B. (2017). Kinetics of petroleum oil biodegradation by a consortium of three protozoan isolates (Aspidisca sp., Trachelophyllum sp. and Peranema sp.). Biotechnol. Rep. 15, 125–131. doi: 10.1016/j.btre.2017.07.001
Kästner, M., Breuer-Jammali, M., and Mahro, B. (1998). Impact of inoculation protocols, salinity, and pH on the degradation of polycyclic aromatic hydrocarbons (PAHs) and survival of PAH-degrading bacteria introduced into soil. Appl. Environ. Microbiol. 64, 359–362. doi: 10.1128/AEM.64.1.359-362.1998
Khanpour-Alikelayeh, E., Partovinia, A., Talebi, A., and Kermanian, H. (2020). Investigation of Bacillus licheniformis in the biodegradation of Iranian heavy crude oil: a two-stage sequential approach containing factor-screening and optimization. Ecotoxicol. Environ. Saf. 205:111103. doi: 10.1016/j.ecoenv.2020.111103
Kuyukina, M. S., Ivshina, I. B., Kamenskikh, T. N., Bulicheva, M. V., and Stukova, G. I. (2013). Survival of cryogel-immobilized Rhodococcus strains in crude oil-contaminated soil and their impact on biodegradation efficiency. Int. Biodeterior. Biodegradation 84, 118–125. doi: 10.1016/j.ibiod.2012.05.035
Li, J., Wu, C., Chen, S., Lu, Q., Shim, H., Huang, X., et al. (2020). Enriching indigenous microbial consortia as a promising strategy for xenobiotics’ cleanup. J. Clean. Prod. 261:121234. doi: 10.1016/j.jclepro.2020.121234
Li, X., Zhao, L., and Adam, M. (2016). Biodegradation of marine crude oil pollution using a salt-tolerant bacterial consortium isolated from Bohai Bay, China. Mar. Pollut. Bull. 105, 43–50. doi: 10.1016/j.marpolbul.2016.02.073
Li, C., Zhou, Z.-X., Jia, X.-Q., Chen, Y., Liu, J., and Wen, J.-P. (2013). Biodegradation of crude oil by a newly isolated strain Rhodococcus sp. JZX-01. Appl. Biochem. Biotechnol. 171, 1715–1725. doi: 10.1007/s12010-013-0451-4
Liu, Y.-C., Zhou, T.-T., Zhang, J., Xu, L., Zhang, Z.-H., Shen, Q.-R., et al. (2009). Molecular characterization of the alkB gene in the thermophilic Geobacillus sp. strain MH-1. Res. Microbiol. 160, 560–566. doi: 10.1016/j.resmic.2009.08.010
Márquez, N., Ysambertt, F., and De La Cruz, C. (1999). Three analytical methods to isolate and characterize vanadium and nickel porphyrins from heavy crude oil. Anal. Chim. Acta 395, 343–349. doi: 10.1016/S0003-2670(99)00304-9
Mu, D. S., Liang, Q. Y., Wang, X. M., Lu, D. C., Shi, M. J., Chen, G. J., et al. (2018). Metatranscriptomic and comparative genomic insights into resuscitation mechanisms during enrichment culturing. Microbiome 6:230. doi: 10.1186/s40168-018-0613-2
Muthukumar, B., Al Salhi, M. S., Narenkumar, J., Devanesan, S., Tentu Nageswara, R., Kim, W., et al. (2022). Characterization of two novel strains of Pseudomonas aeruginosa on biodegradation of crude oil and its enzyme activities. Environ. Pollut. 304:119223. doi: 10.1016/j.envpol.2022.119223
Niem, J. M., Billones-Baaijens, R., Stodart, B., and Savocchia, S. (2020). Diversity profiling of grapevine microbial endosphere and antagonistic potential of endophytic Pseudomonas against grapevine trunk diseases. Front. Microbiol. 11:477. doi: 10.3389/fmicb.2020.00477
Patel, D. D., and Bhaskaran, L. (2018). Study on paraffin wax degrading ability of Pseudomonas nitroreducens isolated from oil wells of Gujarat, India. Pet. Sci. Technol. 36, 583–590. doi: 10.1080/10916466.2018.1437634
Phulpoto, I. A., Hu, B., Wang, Y., Ndayisenga, F., Li, J., and Yu, Z. (2021). Effect of natural microbiome and culturable biosurfactants-producing bacterial consortia of freshwater lake on petroleum-hydrocarbon degradation. Sci. Total Environ. 751:141720. doi: 10.1016/j.scitotenv.2020.141720
Pi, Y., Chen, B., Bao, M., Fan, F., Cai, Q., Ze, L., et al. (2017). Microbial degradation of four crude oil by biosurfactant producing strain Rhodococcus sp. Bioresour. Technol. 232, 263–269. doi: 10.1016/j.biortech.2017.02.007
Prendergast, D. P., and Gschwend, P. M. (2014). Assessing the performance and cost of oil spill remediation technologies. J. Clean. Prod. 78, 233–242. doi: 10.1016/j.jclepro.2014.04.054
Singh, A. K., Sherry, A., Gray, N. D., Jones, D. M., Bowler, B. F. J., and Head, I. M. (2014). Kinetic parameters for nutrient enhanced crude oil biodegradation in intertidal marine sediments. Front. Microbiol. 5:160. doi: 10.3389/fmicb.2014.00160
Suja, F., Rahim, F., Taha, M. R., Hambali, N., Rizal Razali, M., Khalid, A., et al. (2014). Effects of local microbial bioaugmentation and biostimulation on the bioremediation of total petroleum hydrocarbons (TPH) in crude oil contaminated soil based on laboratory and field observations. Int. Biodeterior. Biodegradation 90, 115–122. doi: 10.1016/j.ibiod.2014.03.006
Varjani, S., and Upasani, V. N. (2019). Influence of abiotic factors, natural attenuation, bioaugmentation and nutrient supplementation on bioremediation of petroleum crude contaminated agricultural soil. J. Environ. Manag. 245, 358–366. doi: 10.1016/j.jenvman.2019.05.070
Venil, C. K., Malathi, M., and Devi, P. R. (2021). Characterization of Dietzia maris AURCCBT01 from oil-contaminated soil for biodegradation of crude oil. 3 Biotech 11:291. doi: 10.1007/s13205-021-02807-7
Wang, W., Cai, B., and Shao, Z. (2014). Oil degradation and biosurfactant production by the deep sea bacterium Dietzia maris as-13-3. Front. Microbiol. 5:711. doi: 10.3389/fmicb.2014.00711
Wang, X., Guan, X., Zhang, X., Xiang, S., Zhang, R., and Liu, M. (2020). Microbial communities in petroleum-contaminated seasonally frozen soil and their response to temperature changes. Chemosphere 258:127375. doi: 10.1016/j.chemosphere.2020.127375
Wang, W., He, Y., Wang, B., Dong, M., Zhang, H., and Shen, C. (2022). Experimental study on wax removal and viscosity reduction of waxy crude oil by Ochrobactrum intermedium. J. Pet. Sci. Eng. 213:110445. doi: 10.1016/j.petrol.2022.110445
Keywords: crude oil, biodegradation, consortium, Rhodococcus, Pseudomonas
Citation: Yu T, Liu X, Ai J, Wang J, Guo Y, Liu X, He X, Deng Z and Jiang Y (2022) Microbial community succession during crude oil-degrading bacterial enrichment cultivation and construction of a degrading consortium. Front. Microbiol. 13:1044448. doi: 10.3389/fmicb.2022.1044448
Edited by:
Xiaowu Huang, Guangdong Technion-Israel Institute of Technology (GTIIT), ChinaReviewed by:
Naga Raju Maddela, Technical University of Manabi, EcuadorSivakumar Nallusamy, Sultan Qaboos University, Oman
Copyright © 2022 Yu, Liu, Ai, Wang, Guo, Liu, He, Deng and Jiang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yingying Jiang, anl5QHlhdS5lZHUuY24=
†These authors have contributed equally to this work and share first authorship
 Tianfei Yu
Tianfei Yu Xiaodong Liu
Xiaodong Liu Jiamin Ai
Jiamin Ai Zhenshan Deng
Zhenshan Deng Yingying Jiang
Yingying Jiang