- 1Department of Forest Mycology and Plant Pathology, Swedish University of Agricultural Sciences, Uppsala, Sweden
- 2Department of Agricultural, Forest and Food Sciences (DISAFA), University of Torino, Grugliasco, Italy
Introduction: The fungal secretome comprise diverse proteins that are involved in various aspects of fungal lifestyles, including adaptation to ecological niches and environmental interactions. The aim of this study was to investigate the composition and activity of fungal secretomes in mycoparasitic and beneficial fungal-plant interactions.
Methods: We used six Clonostachys spp. that exhibit saprotrophic, mycotrophic and plant endophytic lifestyles. Genome-wide analyses was performed to investigate the composition, diversity, evolution and gene expression of Clonostachys secretomes in relation to their potential role in mycoparasitic and endophytic lifestyles.
Results and discussion: Our analyses showed that the predicted secretomes of the analyzed species comprised between 7 and 8% of the respective proteomes. Mining of transcriptome data collected during previous studies showed that 18% of the genes encoding predicted secreted proteins were upregulated during the interactions with the mycohosts Fusarium graminearum and Helminthosporium solani. Functional annotation of the predicted secretomes revealed that the most represented protease family was subclass S8A (11–14% of the total), which include members that are shown to be involved in the response to nematodes and mycohosts. Conversely, the most numerous lipases and carbohydrate-active enzyme (CAZyme) groups appeared to be potentially involved in eliciting defense responses in the plants. For example, analysis of gene family evolution identified nine CAZyme orthogroups evolving for gene gains (p ≤ 0.05), predicted to be involved in hemicellulose degradation, potentially producing plant defense-inducing oligomers. Moreover, 8–10% of the secretomes was composed of cysteine-enriched proteins, including hydrophobins, important for root colonization. Effectors were more numerous, comprising 35–37% of the secretomes, where certain members belonged to seven orthogroups evolving for gene gains and were induced during the C. rosea response to F. graminearum or H. solani. Furthermore, the considered Clonostachys spp. possessed high numbers of proteins containing Common in Fungal Extracellular Membranes (CFEM) modules, known for their role in fungal virulence. Overall, this study improves our understanding of Clonostachys spp. adaptation to diverse ecological niches and establishes a basis for future investigation aiming at sustainable biocontrol of plant diseases.
1. Introduction
Fungal secreted proteins (secretome) play an important role in fungal biology and adaption to various ecological niches, and environmental interactions ranging from mutualism to parasitism and interference competition (Druzhinina et al., 2012; Pellegrin et al., 2015; Lu and Edwards, 2016). Genes encoding secreted proteins typically encompass 4–15% of the total gene numbers in fungal genomes (Girard et al., 2013; Pellegrin et al., 2015). These proteins are classified into various functional groups such as carbohydrate-active enzymes (CAZymes), proteases, lipases and oxidoreductases needed for nutrient acquisition, self-protection and biotic interactions with microbes, plants and animals (Kim et al., 2016; Guzmán-Guzmán et al., 2017; Feldman et al., 2020). Moreover, 40–60% of predicted fungal secretome proteins are typically shorter than 300 amino acids (aa) and are referred to as small secreted proteins (SSPs; Pellegrin et al., 2015; Kim et al., 2016). SSPs are often cysteine-rich, lack known protein modules or catalytic domains and certain members act as effectors mediating communication between organisms ranging from beneficial to detrimental interactions (Kim et al., 2016; Selin et al., 2016).
SSPs are mainly studied in the context of pathogenic fungal-plant interactions, and their composition and role in parasitic fungal-fungal and beneficial fungal-plant interactions is still poorly investigated. In fungi used for biological control of plant diseases, for example members of the Trichoderma and Clonostachys genera, the function of certain SSPs including hydrophobins, cerato-platanins and LysM module-containing proteins in regulating interactions with fungal hosts (mycohosts) and plant hosts are shown (Dubey et al., 2012, 2014; Guzmán-Guzmán et al., 2017; Ramírez-Valdespino et al., 2019; Jensen et al., 2021). In addition, a SSP family with Common in Fungal Extracellular Membranes (CFEM) modules has been identified in T. atroviride (Kulkarni et al., 2003; Druzhinina et al., 2012; Zhang et al., 2015; Guzmán-Guzmán et al., 2017). Certain members of this family have been shown to be induced during interactions with host plants indicating their roles as putative effector proteins (Guzmán-Guzmán et al., 2017). A role of CFEM-containing proteins in fungal pathogenesis has been demonstrated in the plant pathogenic fungi Botrytis cinerea, Magnaporthe oryzae, and Colletotrichum graminicola (Kou et al., 2017; Zhu et al., 2017; Gong et al., 2020).
Clonostachys spp. including C. rosea, C. byssicola, C. chloroleuca, C. rhizophaga and C. solani, are filamentous ascomycetes fungi with a multi-trophic mode of lifestyle. These fungi can be found as saprotrophs in various ecological niches including soil and dead organic matter (Moreira et al., 2016; Jensen et al., 2021). Certain species can thrive in rhizospheres where they can colonize the plant root surfaces and establish a beneficial relationship with the plant host as endophytes (Sutton et al., 2002; Karlsson et al., 2015; Saraiva et al., 2015; Maillard et al., 2020; Jensen et al., 2021). In addition, these species can live as necrotrophic mycoparasites by killing and feeding on their mycohosts (Alvindia and Natsuaki, 2008; Dugan et al., 2012; Sun et al., 2018; Jensen et al., 2021). The antagonistic ability of certain strains of C. rosea against plant parasitic nematodes has also been reported (Iqbal et al., 2018a,b, 2020). To succeed in these distinct ecological niches, Clonostachys spp. have evolved capabilities for decomposition of organic materials, competition with other microorganisms for nutrients and space in soil and rhizosphere, and interference competition through antibiosis and mycoparasitism (Morandi et al., 2000; Li et al., 2002; Saraiva et al., 2015; Sun et al., 2017; Fatema et al., 2018). Due to these properties, certain Clonostachys strains are used as efficient biological control agents against fungal plant diseases in agricultural and horticultural production systems (Jensen et al., 2000; Sutton et al., 2002; Xue et al., 2008; Cota et al., 2009; Salamone et al., 2018).
The ability of Clonostachys spp. to inhabit broad ecological niches is reflected by their genomic characteristics (Karlsson et al., 2015; Broberg et al., 2021). For example, copy number of genes coding for enzymes associated with biosynthesis of specialized metabolites such as polyketide synthases, non-ribosomal peptide synthetase and cytochrome P450 monooxygenases are expanded in C. rosea compared to plant pathogenic Fusarium spp. and mycoparasitic Trichoderma spp. (Karlsson et al., 2015; Broberg et al., 2021). Similarly, the ATP-binding cassette (ABC) and major facilitator superfamily (MFS) membrane transporter gene families, associated with efflux of endogenous and exogenous specialized metabolites, are also expanded (Karlsson et al., 2015; Nygren et al., 2018; Broberg et al., 2021). Among the CAZyme gene families, auxiliary activity (AA) family 9 lytic polysaccharide monooxygenases, AA7 gluco-and chitooligosaccharide oxidases, AA3 glucose-methanol-choline oxidoreductases, polysaccharide lyase family 1 (PL1) pectin/pectate lyases and certain proteases are also evolving under selection for increased gene copy numbers (Karlsson et al., 2015; Atanasova et al., 2018; Iqbal et al., 2018a,b; Broberg et al., 2021). However, analysis of gene family evolution of genes specifically coding for secreted proteins in Clonostachys is yet to be comprehensively investigated.
In this study, we performed prediction and in-depth analysis of the secretomes of six Clonostachys spp., including C. byssicola, C. chloroleuca, C. rhizophaga, C. rosea, C. solani and Clonostachys sp. CBS 192.96 with the hypothesis that the composition of Clonostachys spp. secreteome is shaped to accomplish their saprotrophic, mycotrophic and plant endophytic lifestyles. Our analysis was focused on predicted lipases, proteases, oxidoreductases, CAZymes, cysteine-rich SSPs, putative effectors and CFEM proteins and their possible roles in mycoparasitic and beneficial fungal-plant interactions. This revealed the presence of many proteins with a known role in antagonism against mycohosts and nematodes, including chitinases, endopolygalacturonases, subtilisin-like peptidases and phospholipases A. Moreover, several enzyme classes including hemicellulose and cellulose degradation enzymes, SSPs and effectors with putative role in fungus-plant interactions were identified.
2. Materials and methods
2.1. Prediction of fungal secretomes
Based on the genome sequences of C. rosea IK726 (Genbank: GCA_902827195.2), C. byssicola CBS 245.78 (GCA_902006505.2), C. chloroleuca CBS 570.77 (GCA_902074915.2), C. rhizophaga CBS 906.72A (GCA_902077795.2), C. solani 1703 (GCA_902141235.2) and Clonostachys sp. CBS 192.96 (Karlsson et al., 2015; Broberg et al., 2018, 2021; Supplementary file 1), the predicted secretomes were generated using procedures described previously (Gogleva et al., 2018). In short, SignalP ver. 4.0 (Petersen et al., 2011) was used to predict secretion signal peptides, while proteins with transmembrane domains were identified with TMHMM ver. 2 (Krogh et al., 2001). TargetP ver. 2 (Armenteros et al., 2019) was used to identify proteins putatively targeted to mitochondria, while PredGPI (Pierleoni et al., 2008) was used to predict proteins with a glycosylphosphatidylinositol (GPI) anchor. The complete bash script is available as Supplementary file 2. Proteins with predicted signal peptides but lacking transmembrane domains, GPI anchors or mitochondrial targeting signatures, were considered to be secreted. OrthoFinder ver. 2.5.2 (Emms and Kelly, 2019) was used to identify orthogroups of the secreted proteins. For comparative purposes, secretome prediction and InterProScan annotation was performed on the genomes of Neurospora crassa OR74A (GCA_000182925.2), T. atroviride IMI 206040 (GCA_000171015.2), T. reesei QM6a (GCA_000167675.2 v2.0), T. virens Gv29-8 (GCA_000170995.2), F. graminearum PH-1 (GCA_000240135.3), F. verticillioides 7600 (GCA_000149555.1) and F. vanetteni 77–13-4 (Coleman et al., 2009).
2.2. Functional annotation and gene ontology enrichment analyses
Gene ontology (GO) enrichment analyses were performed using the agriGO toolkit (Tian et al., 2017) with the Fisher statistical test and maximum adjusted p-value of 0.05. The FDR adjustment method was used to adjust the p-value. The GO annotation for the proteome of C. rosea was obtained from a previous study (Piombo et al., 2021).
The composition of the Clonostachys secretomes was analyzed with focus on predicted proteases, lipases, CAZymes, oxidoreductases, cysteine rich proteins and effectors, due to their potential role in environmental interactions. InterProScan v. 5.46–81 (Jones et al., 2014) was used to predict modules in the proteomes, and lipases, oxidoreductases and proteases were specifically identified based on the presence of InterProScan modules. Lipases and oxidoreductases were further classified in classes depending on the type of detected module, while proteases were classified according to the Merops database (Rawlings et al., 2010), using the BLAST algorithm (Altschul et al., 1990). EffectorP 3.0 was used to predict putative effectors (Sperschneider and Dodds, 2021), and the dbCAN meta server was used to predict CAZymes using both the HMMER and DIAMOND tools (Buchfink et al., 2015; Zhang et al., 2018).
2.3. CFEM identification and analysis
Proteins predicted to contain CFEM modules using InterProScan v. 5.46–81 (Jones et al., 2014) were considered to be CFEM proteins. For phylogenetic analysis, CFEM proteins were aligned with mafft v.7.453 in E-INS-I mode, suggested for sequences containing large unalignable regions (Katoh and Standley, 2013), and the phylogenetic trees were generated with iqtree v.2.1.3 (Nguyen et al., 2015) with 500 bootstrap replicates and the option “MFP” (ModelFinder) to find the best substitution model. Visualization was carried out with Figtree v.1.4.4 (Rambaut, 2018). The same programs were used for the phylogenetic analysis of the concatenated CFEM modules of each species, but mafft was used in L-INS-I mode, recommended when working with less than 200 sequences.
2.4. Study of gene family evolution
Computational analysis of gene family evolution (CAFE) v.5 (Mendes et al., 2020) was used to estimate accelerated rates of gene gain or loss, associated with lineages. The significance threshold was set at 0.05. The phylogenetic tree necessary for CAFE analysis was obtained as described in Broberg et al. (2021), using concatenated gene sequences of ATP citrate lyase (acl1), RNA polymerase II large subunit (rpb1), translation elongation factor 1-α (tef1) and β-tubulin (tub) for the considered species.
2.5. Mining of gene expression data
To investigate transcriptional regulation of genes coding for secreted proteins, differentially expressed genes of C. rosea interacting with the mycohosts Botrytis cinerea, F. graminearum and Helminthoisporum solani were retrieved from four previously published studies (Lysøe et al., 2017; Demissie et al., 2018, 2020; Nygren et al., 2018). When the studies used different version of the C. rosea genome, proteins from different versions were considered the same if they had a match in a BLAST analysis with 90% minimum identity and query coverage. Each proteome was used as both query and database for the BLAST analyses, and only proteins with a match in both database-query combinations were accepted.
3. Results
3.1. Prediction of Clonostachys secretomes and mining of gene expression data
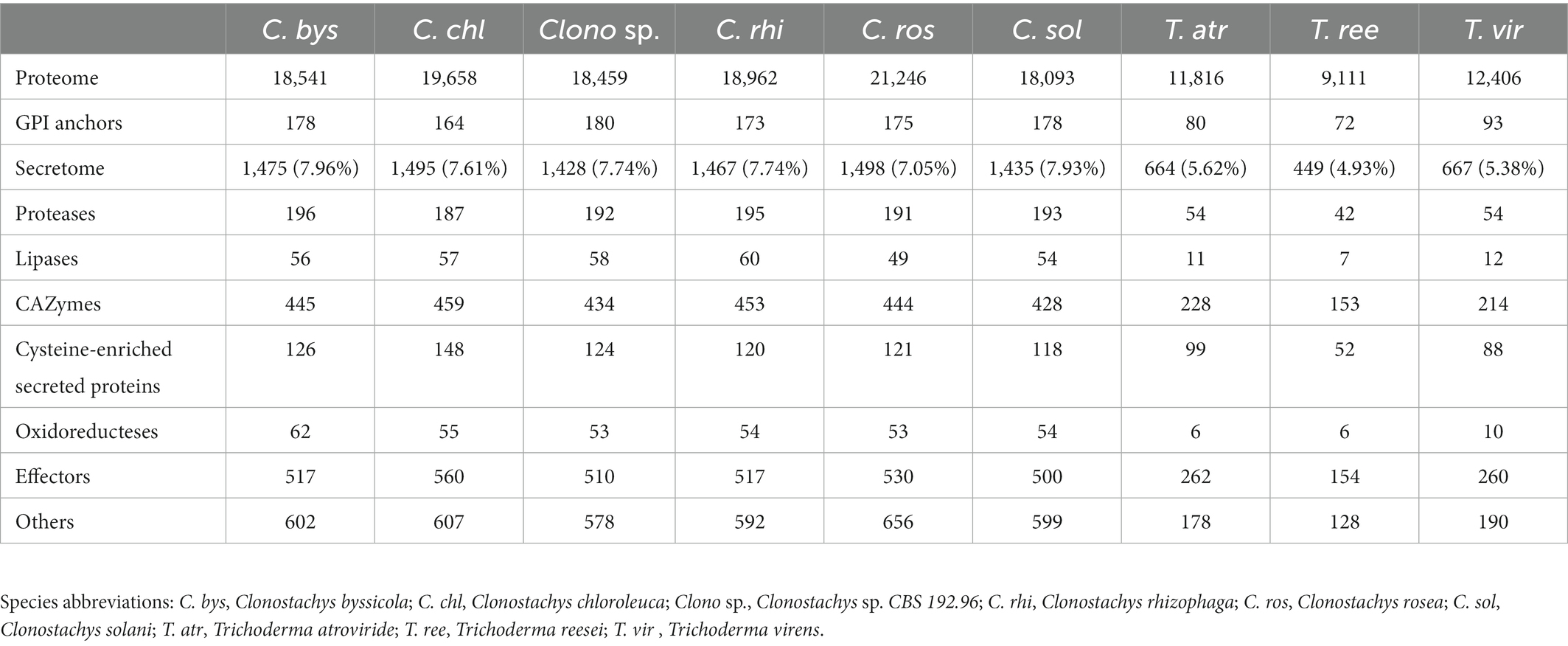
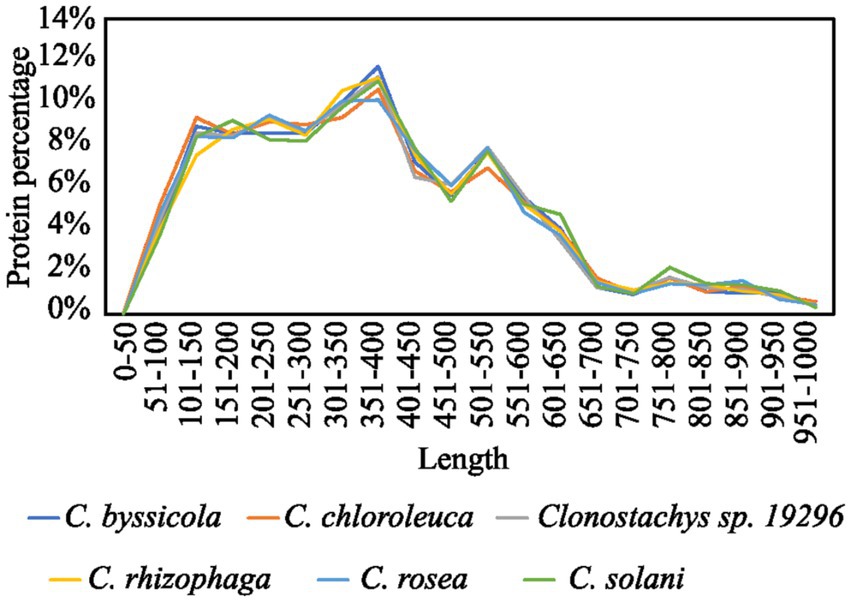
The secretomes of six Clonostachys spp. was predicted to contain 1,428 to 1,498 proteins, amounting to between 7.1% (C. rosea) and 8.0% (C. byssicola) of their proteomes (Table 1; Supplementary file 3). In the considered Trichoderma spp., the proportion of secreted proteins in their predicted secretomes accounted for 4.9% in T. reesei to 5.6% in T. atroviride. More than 85% of the Clonostachys spp. secreted proteins were less than 600 aa in length (Figure 1), among which a majority of proteins (56%) were 100–400 aa in length with the highest proportion (around 12%) at 350–400 aa. No differences were found in proportion and length distribution of secreted proteins between the analyzed Clonostachys species. Mining of available RNA-seq data (Lysøe et al., 2017; Demissie et al., 2018, 2020; Nygren et al., 2018) identified 274 genes upregulated in C. rosea during interaction with the mycohosts F. graminearum or H. solani (Figure 2; Supplementary Table S1).
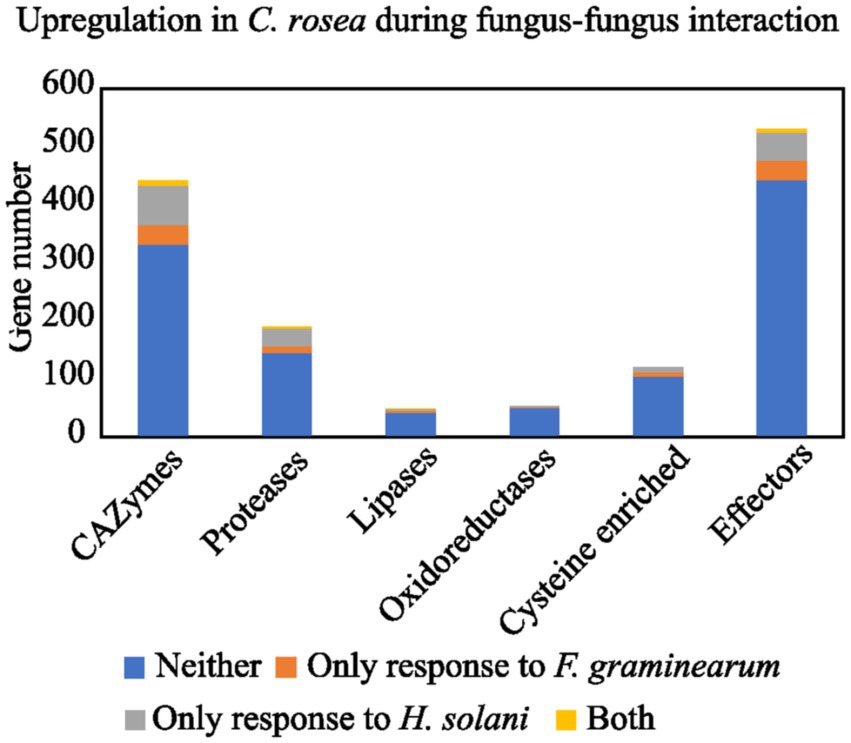
Figure 1. Number of C. rosea transcripts, coding for secreted proteins, upregulated during contact with H. solani and/or F. graminearum. The numbers are shown for CAZymes, proteases, lipases, oxidoreductases, cysteine enriched proteins and effectors. The gene expression data was retrieved from previously performed transcriptome analysis of C. rosea during interactions with H. solani or F. graminearum (Lysøe et al., 2017; Demissie et al., 2018, 2020; Nygren et al., 2018).
3.2. Gene ontology enrichment analysis of predicted Clonostachys secretomes
GO enrichment analysis based on the annotated C. rosea proteome (Piombo et al., 2021) revealed that 57 biological processes were enriched (p ≤ 0.05) in the C. rosea secretome in respect to the rest of the proteome (Figure 3). The analysis was run on the proteome of C. rosea as it had the best available functional annotation among the considered Clonostachys spp., and had underwent multiple functional analyses in previous studies (Broberg et al., 2018, 2021; Piombo et al., 2021). The majority of the enriched biological processes were related to metabolic and catabolic activity on several compounds including carbohydrates, proteins and lipids. The terms response to fungus (GO: 0009620), defense response to fungus (GO: 0050832), cell wall organization (GO: 0071555), cell wall organization or biogenesis (GO: 0071554) were also enriched (Figure 3).
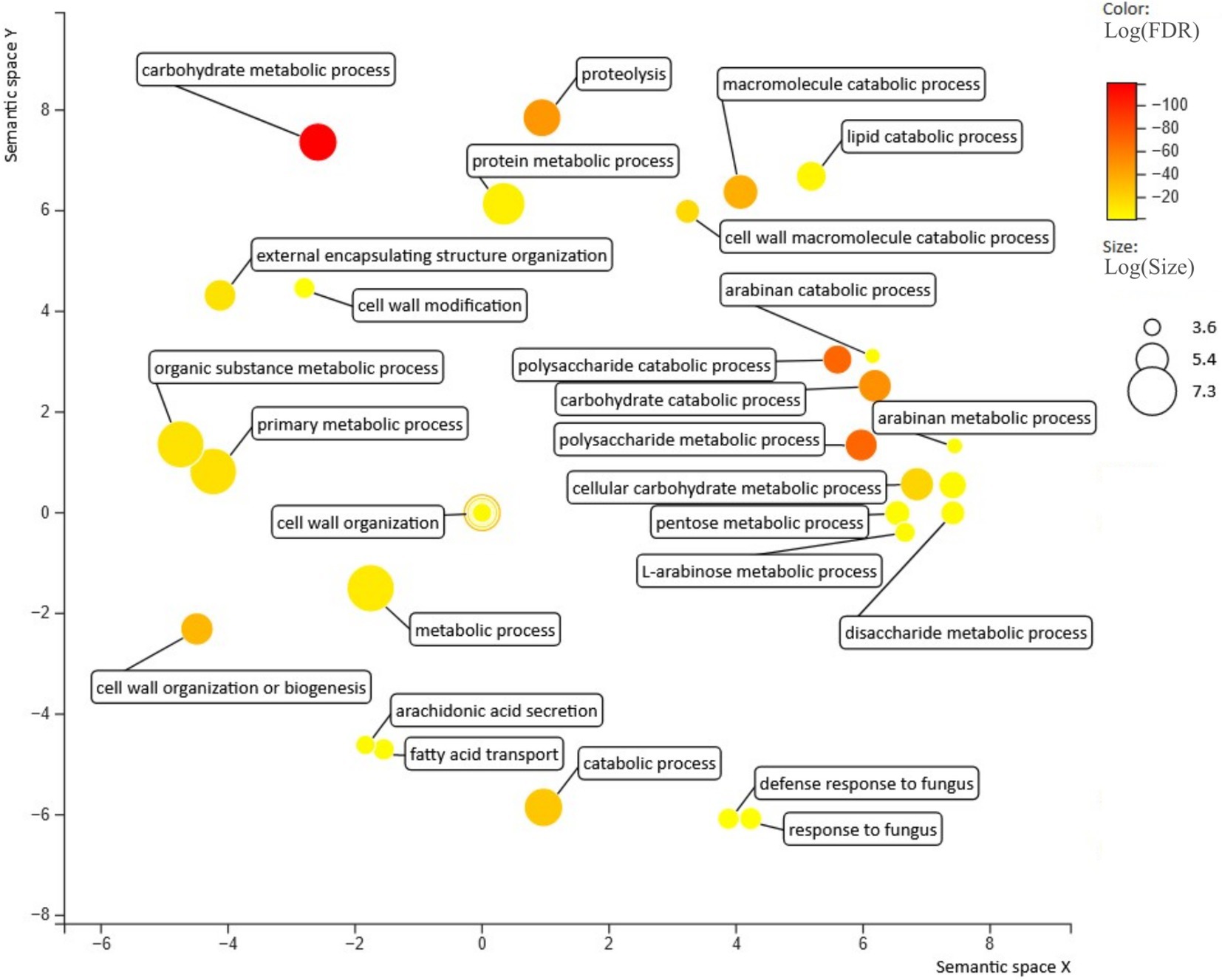
Figure 3. Biological processes enriched in the secretome predicted for C. rosea. Figure generated with revigo (Supek et al., 2011).
3.3. Composition of predicted Clonostachys secretomes
3.3.1. Carbohydrate-active enzymes
A large number of CAZymes (428–459 genes) comprising 30% of the secretomes on average, were detected in the Clonostachys species. The percentage of CAZymes in Clonostachys spp. was lower compared to the predicted proportion of 30 to 35% in the considered Trichoderma spp. (Table 1). The highest number of predicted CAZymes in Clonostachys spp. were classified as GH43 (arabinofuranosidases, arabinases and xylosidases), followed by AA7 (glucooligosaccharide oxidases), AA9 (lytic polysaccharide monooxygenases), AA3 (glucose-methanol-choline oxidoreductases), GH5 (hydrolases with many substrates) and GH28 (polygalacturonases) (Supplementary Table S2). The number of detected GH18 chitinases was between six and nine, while three or four GH20 N-acetylglucosaminidases were identified (Supplementary Table S2).
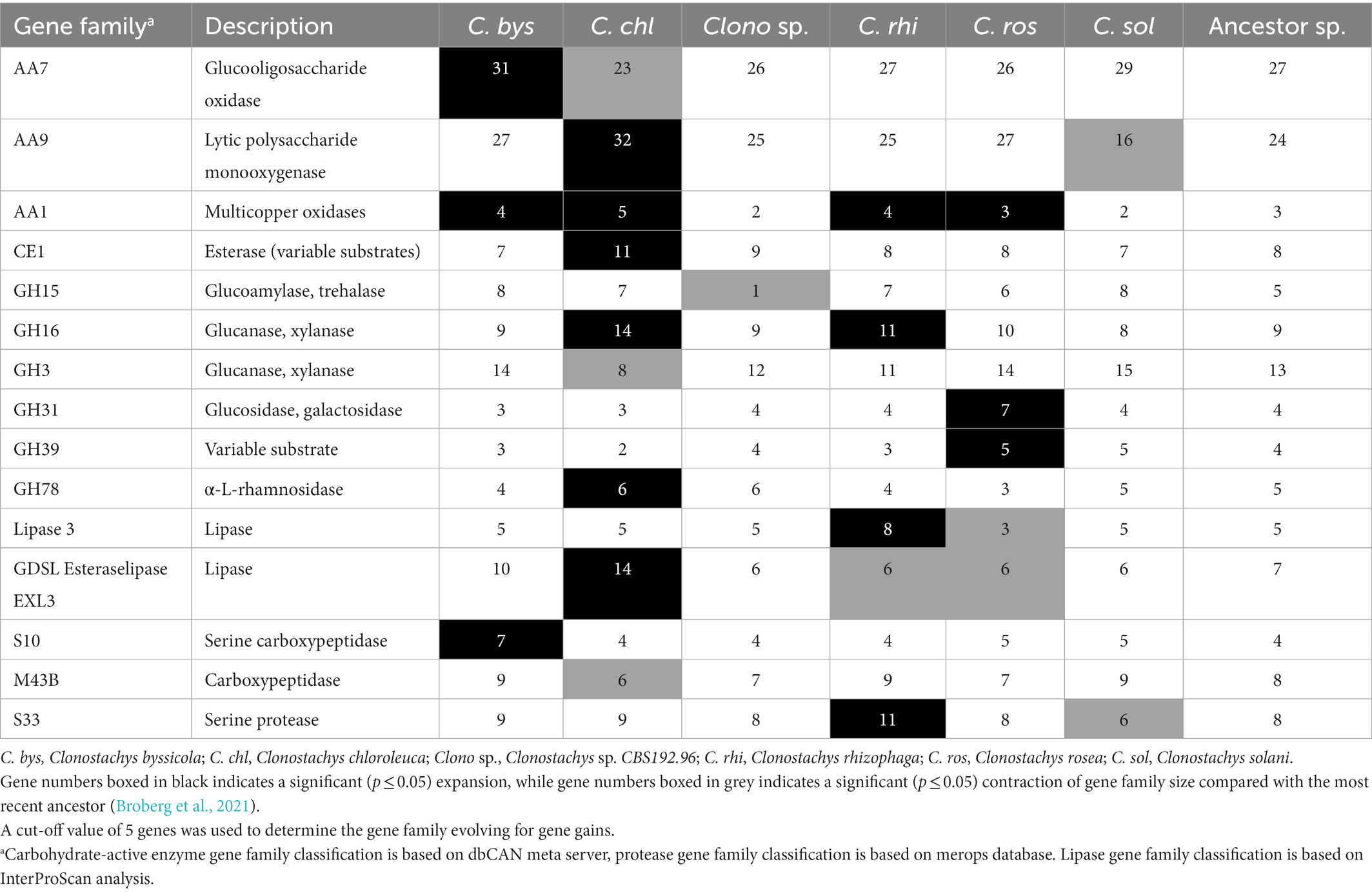
CAFE analysis identified ten CAZyme families evolving for gene gains or losses (p ≤ 0.05) in the Clonostachys spp. (Table 2). These included different classes of enzymes degrading cellulose, glucan, xylan, trehalose and other components of the fungal and plant cell wall, including AA9, GH3, GH15, GH16, GH31 and GH78 (p ≤ 0.037). The highest number of secreted CAZyme families (five families) evolving for gene gains were identified in C. chloroleuca followed by three families in C. rosea (Table 2). Intriguingly, only GH15 was evolving for gene gains or losses in Clonostachys sp. CBS 192.96, with a significant (p = 0.001) decrease from five genes in the ancestral species to one gene in Clonostachys. sp. CBS 192.96. Conversely, only family AA9 was evolving for gene gains or losses in C. solani, with a decrease from 24 to 16 genes compared with the ancestral lineage (p < 0.001). Mining of gene expression data also revealed that family AA9 was the most represented class by far among the CAZymes significantly upregulated in C. rosea during the interaction with F. graminearum or H. solani (Supplementary Table S1), with 12 out of 111 upregulated CAZymes belonging to this class, followed by GH7, GH12 and GH28 with 5 members each.
3.3.2. Proteases, lipases, and oxidoreductases
On average 12% of the Clonostachys secretomes (187–196 genes) were classified as proteases of several Merops classifications (Rawlings et al., 2012; Table 1), while only around 8.5% of the secretome (41–54 genes) was comprised of proteases in Trichoderma spp. (Table 1). The dominant groups in Clonostachys spp. were serine proteases (S8A serine endopeptidase subtilisins, S33 serine proteasesand S1A chymotrypsins), carboxypeptidases (M14A) and metallo-endopeptidases (M43B), all families with many more members in Clonostachys spp. than in Trichoderma spp. (Supplementary Table S2). Gene family evolution analysis identified three protease families (S10, S33 and M43B) as evolving for gene gains or losses in the considered Clonostachys spp. (Table 2). The S10 serine carboxypeptidase gene family was significantly (p = 0.004) expanded from four to seven genes in C. byssicola, while subfamily M43B was significantly contracted in C. chloroleuca (p = 0.004) compared to the number in the ancestral species, while family S33 was expanded in C. rhizophaga and contracted in C. solani (Table 2). Among the 191 secreted proteases identified in C. rosea, 46 were upregulated during the interactions with F. graminearum or H. solani (Figure 2; Supplementary Table S1). The most frequent classes in this subgroup were serine endopeptidases of family S8A and S1A, with six genes each.
Predicted lipases amounted to 3.8% of the total secretomes on average in Clonostachys spp., but only 1.7% in Trichoderma spp. (Table 1). The most represented groups in Clonostachys spp. were phospholipases A2, lipases 5, lysophospholipases L1 and the GDSL-like Lipase/Acylhydrolase family (Supplementary Table S2). Lipases 3 and GDSL esteraselipase exl3 were predicted to be evolving for gene gains. The gene copy number of the lipases 3 gene family was expanded in C. rhizophaga (p = 0.001) and contracted (p = 0.013) in C. rosea, while the gene copy number of the GDSL esteraselipase exl3 gene family was expanded (p = 0.001) in C. chloroleuca but contracted (p ≤ 0.031) in C. rhizophaga and C. rosea (Table 2). Seven lipase encoding genes were significantly upregulated in C. rosea during the interactions with F. graminearum or H. solani (Figure 1; Supplementary Table S1).
Numerous predicted oxidoreductases were detected in the Clonostachys secretomes, ranging from 62 in C. byssicola to 53 in C. rosea and Clonostachys sp. CBS 192.96. This amounted to 3.8% of the secretome on average, against the 1.3% in Trichoderma spp. (Table 1). The most represented class was the AA3 glucose-methanol-choline oxidoreductases, which amounted to around one third of the total in all the considered Clonostachys species. The second most frequent family in Clonostachys spp. was the flavin-containing amine oxidoreductases, which varied from 10 genes in C. chloroleuca to six genes in C. rosea (Supplementary Table S2). Three oxidoreductase encoding genes were upregulated in C. rosea during the interactions with F. graminearum or H. solani (Figure 1; Supplementary Table S1).
3.3.3. Cysteine-enriched proteins and effectors
On average, 8.5% of the secretomes (118 out of 1478 proteins) consisted of proteins shorter than 300 aa and with more than 4% of cysteine residues and were considered cysteine-enriched. Conversely, at least 11.5% of secreted proteins were classified as cysteine-enriched in all Trichoderma spp. (Table 1). The GO enrichment analysis of cysteine-enriched proteins showed enrichment (p ≤ 0.05) in molecular functions related to lytic activity, specifically carbon–oxygen lyase activity (GO:0016835), carbon–oxygen lyase activity acting on polysaccharides (GO:0016837), pectate lyase activity (GO:0030570) and lyase activity (GO:0016829).
More than one third of the secretomes (500 of 563 proteins, 36% on average) consisted of putative effectors, while this amount was on average 40% in Trichoderma spp. (Table 1). However, less than half of the detected effectors contained a known InterProScan amino acidic motif, while the rest was uncharacterized. Several known effector classes already identified in Trichoderma spp. (Guzmán-Guzmán et al., 2017) such as serine proteases, metalloproteases, LysM proteins, cerato-platanins, thioredoxins and CFEM proteins, were detected among the predicted effectors in Clonostachys spp. (Supplementary Table S2). Gene expression analysis identified 88 C. rosea effector genes to be induced in response to F. graminearum or H. solani (Figure 2; Supplementary Table S1). Predicted effector proteins were enriched (p ≤ 0.05) in GO terms related to cell wall degradation and penetration in plant tissues, including cellulase activity (GO:0008810), pectate lyase activity (GO:0030570) and polysaccharide catabolic process (GO:0000272), but also in GO terms referring to transport and localization of lipids and acids, such as acid secretion (GO:0046717) and lipid transport (GO:0006869) (Figure 4).
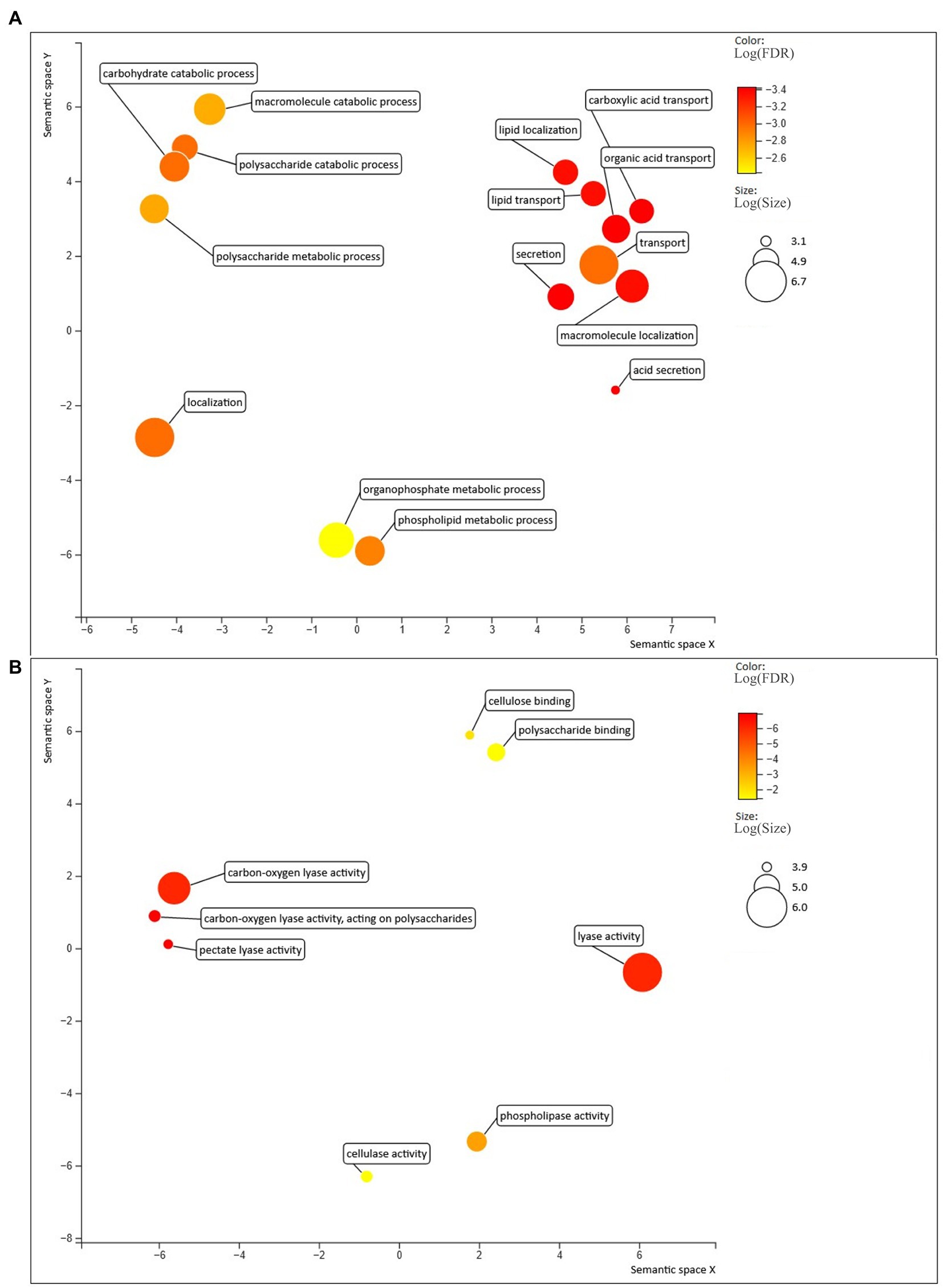
Figure 4. Biological processes (A) and molecular functions (B) enriched in the predicted effectors, compared with the rest of the C. rosea secretome. Figure generated with Revigo (Supek et al., 2011).
Between 93% (Clonostachys sp. CBS 192.96) and 98% (C. rhizophaga) of the cysteine-enriched secreted proteins were also predicted to be effectors (Supplementary Figure S1). One particular class of cysteine-enriched proteins was hydrophobins, where C. chloroleuca had the highest number of secreted proteins (11 proteins) among the analyzed species, followed by C. rosea with six proteins and C. solani with three predicted secreted hydrophobins (Supplementary Table S2).
3.4. Analysis of evolution of gene family composition
An orthofinder analysis grouped the genes encoding predicted secreted proteins of the Clonostachys spp. into 1,547 orthogroups, of which 816 contained at least one gene from each included species (Supplementary Table S3). CAFE analysis identified a total of 38 orthogroups (involving 74 genes) evolving for gene gains (p ≤ 0.05) (Table 3). Twenty-one orthogroups were found to consist of 37 C. rosea genes coding for various families of glycosyl hydrolases. Among those, eight predicted proteins contained additional carbohydrate-binding modules (CBMs) and 13 were found to be differentially expressed during interspecific interaction with F. gramineaum or H. solani (Table 3). Seven orthogroups containing 15 C. rosea genes were predicted to encode various families of proteases including cuticle-degrading proteases, serine-type endopeptidases, subtilisin-like proteases and metallocarboxypeptidases. Orthogroup OG0000107 contained aspartyl proteases, a class involved in the response to both fungi and plants in Trichoderma spp. (Viterbo et al., 2004; Kredics et al., 2005; Table 3). Furthermore, nine orthogroups composed of 16 putative C. rosea effectors were identified, of which seven were found to be induced in response to F. graminearum or H. solani (Table 3).
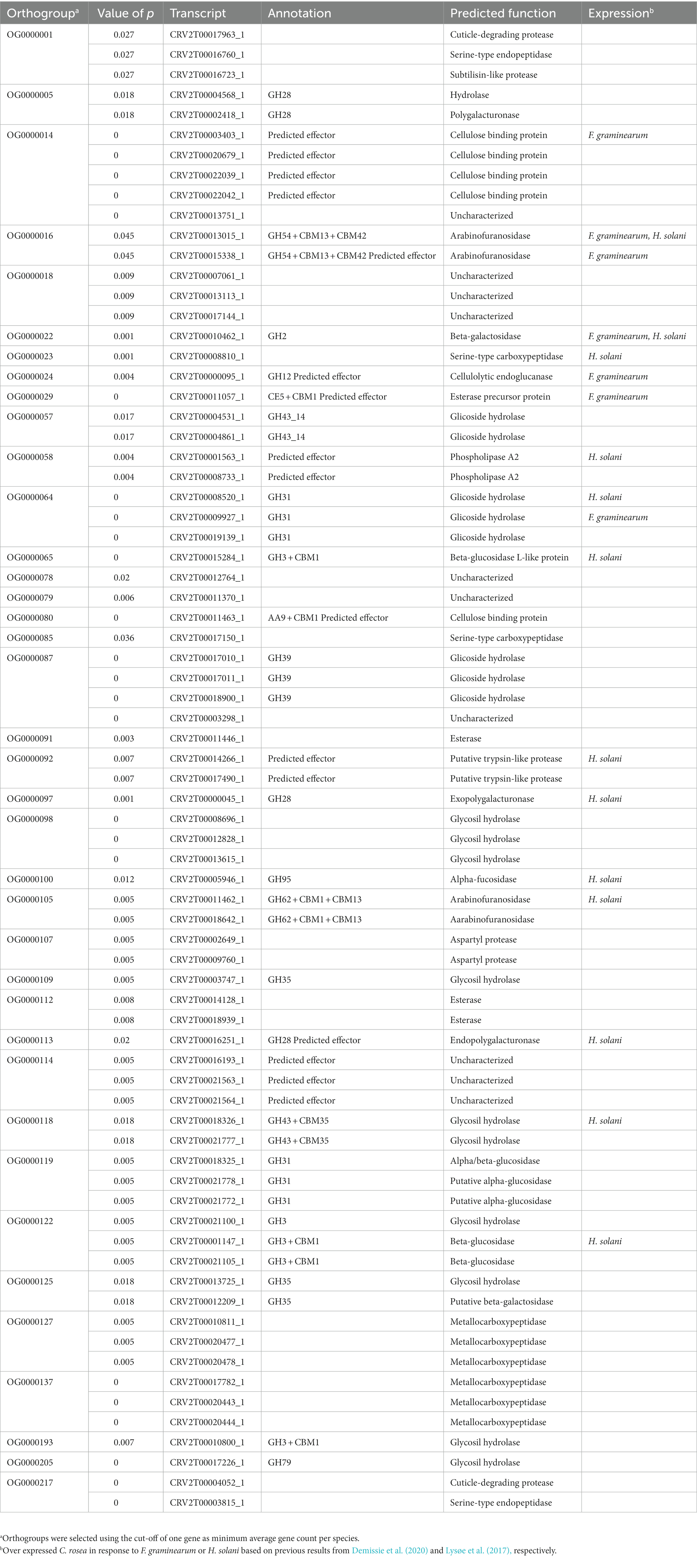
Table 3. Orthogroups evolving for gene gains predicted by Orthofinder v. 2.5.2 in Clonostachys secretomes.
3.5. Identification and sequence analysis of CFEM proteins
Proteins with CFEM modules are considered to play an important role in fungi during interactions with other organisms (Srivastava et al., 2014; Zhang et al., 2015; Sabnam and Barman, 2017; Arya et al., 2020). The number of predicted CFEM proteins in Clonostachys varied from 21 in Clonostachys sp. CBS 192.96 to 32 in C. chloroleuca. However, the number of predicted secreted CFEM proteins ranged from only one in Clonostachys sp. CBS 192.96 to four in C. chloroleuca (Table 4). The total number of predicted CFEM proteins in Clonostachys spp. (21–32 genes) was higher compared with mycoparasitic T. atroviride, T. virens and T. reesei (14–17 genes). However, the CFEM gene copy number in the plant pathogenic F. graminearum, F. verticillioides and F. vanetteni (18–23 CFEM genes) was comparable to the number in Clonostachys spp. (Table 4). The predicted subcellular localization of CFEM proteins was found to be similar between Clonostachys and Fusarium species, with around half of them being transmembrane proteins, 40% GPI-anchored and only 8% secreted. In contrast, Trichoderma spp. CFEMs were predicted to have a different subcellular localization pattern, with 32% transmembrane proteins, 38% GPI-anchored and 17% secreted (Table 4). CFEM proteins may contain one or more copies of the CFEM module (Kulkarni et al., 2003). A conserved domain analysis identified a single CFEM module present in each predicted protein in the considered Clonostachys spp., except for one protein in each species that had two modules (Supplementary Table S4). Gene expression analysis identified seven genes coding for CFEM proteins in C. rosea that were upregulated in response to F. graminearum or H. solani (Supplementary Table S4).
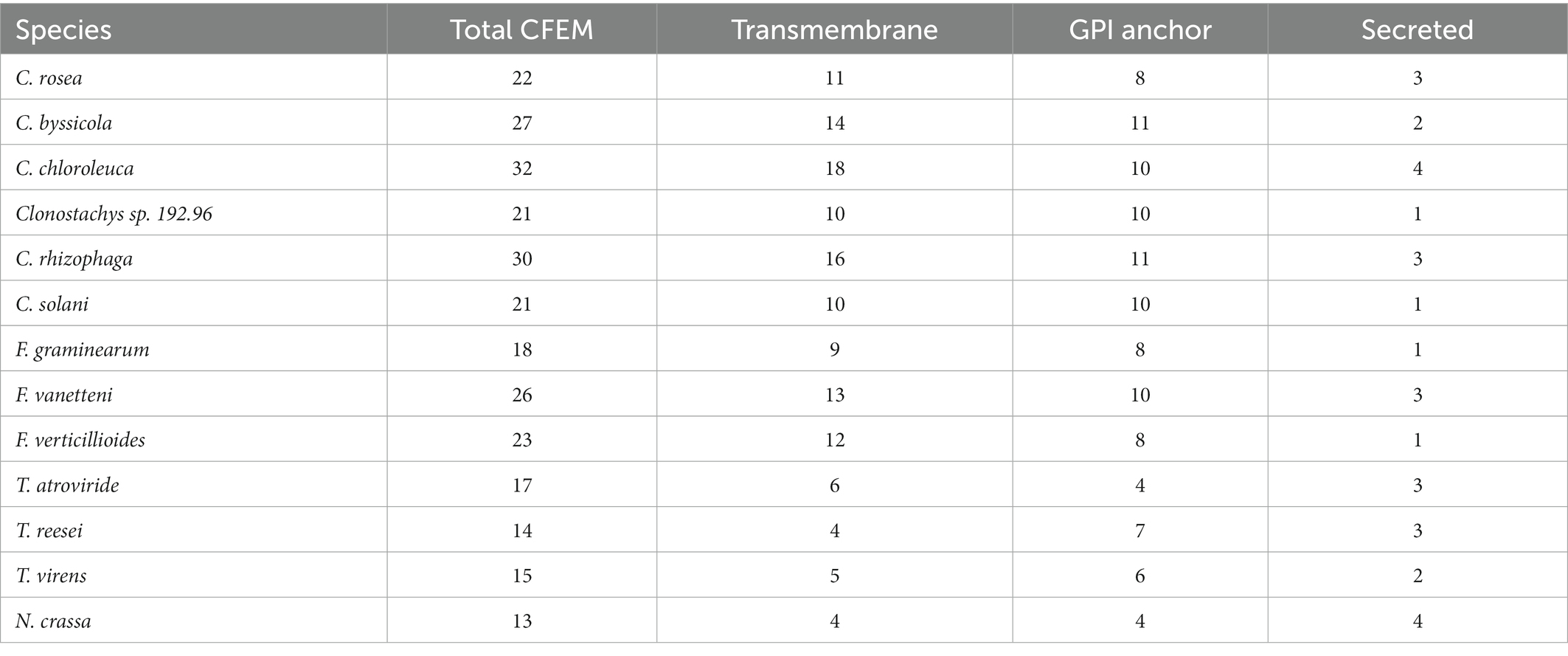
Table 4. Number of proteins with CFEM domains found in the transmemebrane, GDP-anchored and secreted portion of the proteomes for each species of interest.
An analysis with CAFE identified gene gains (p ≤ 0.05) in the ancestral lineage leading to C. byssicola, C. chloroleuca, C. rhizophaga and C. rosea, followed by additional gains in C. chloroleuca and losses in C. rosea (Figure 5). A phylogenetic analysis of predicted CFEM proteins from Clonostachys spp. together with above-mentioned species of Trichoderma, Fusarium and N. crassa showed that the CFEM proteins of Clonostachys spp. typically clustered in monophyletic groups, indicating recent diversification, even though some of them were orthologous to CFEM proteins in Fusarium and Trichoderma species (Figure 6). The phylogenetic tree further displayed low resolution among the deeper branches, sometimes in combination with incongruence with the species phylogeny, which may suggest a birth-and-death evolutionary process in combination with sequence divergence. Among the predicted CFEM proteins, the branches containing C. rosea proteins CRV2T00010850_1, CRV2T00012038_1, CRV2T00008709_1, CRV2T00021845_1, CRV2T00018221_1, CRV2T00019286_1, CRV2T00016013_1 and CRV2T00014542_1 were expanded in the Clonostachys genus (Figure 6), and they either had transmembrane domains or a GPI-anchor.
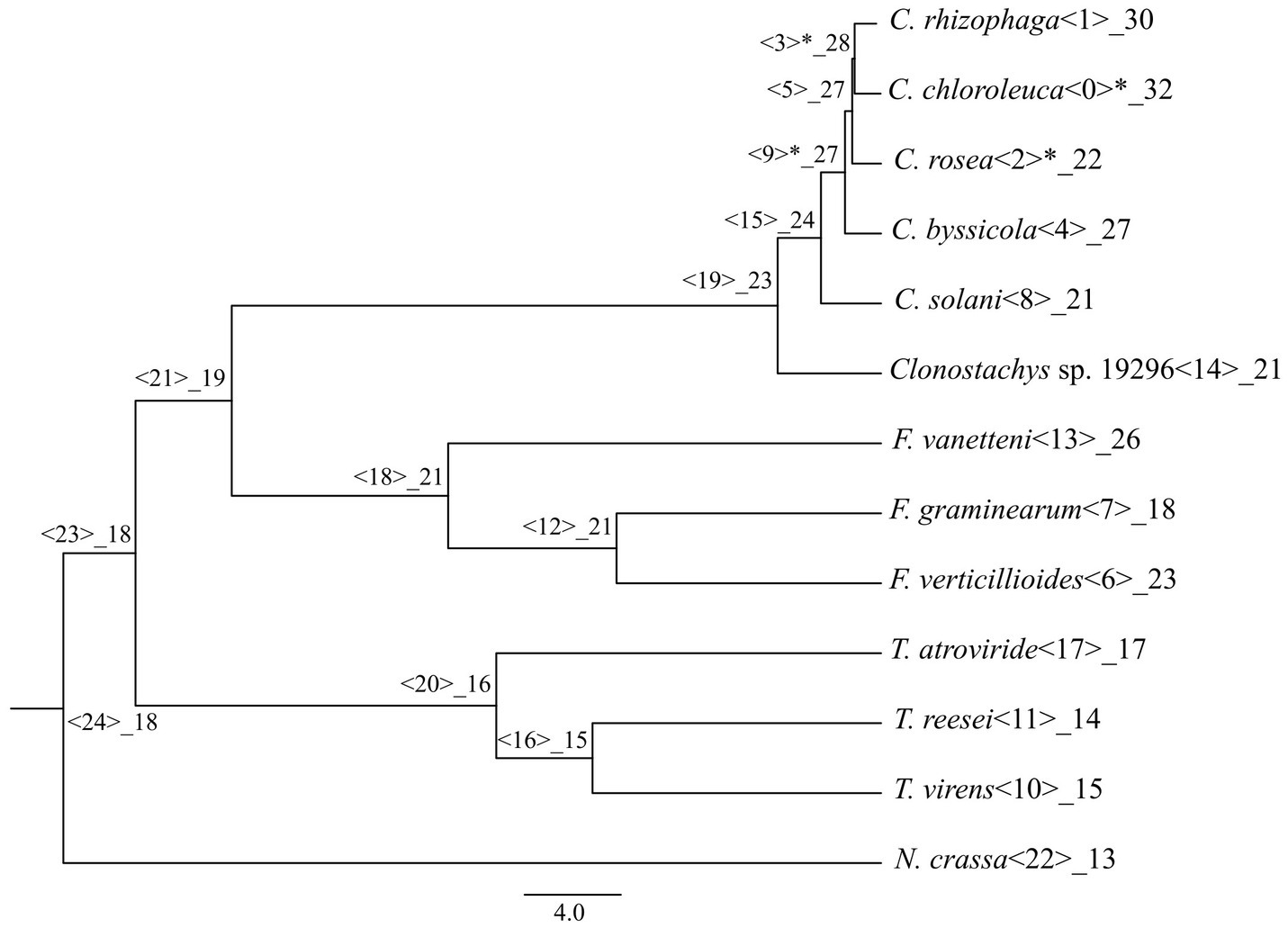
Figure 5. Number of CFEM proteins gained or lost during the evolution of the considered species, mapped on a phylogenetic tree obtained in Broberg et al. (2021). Significant changes are marked with the asterisk.
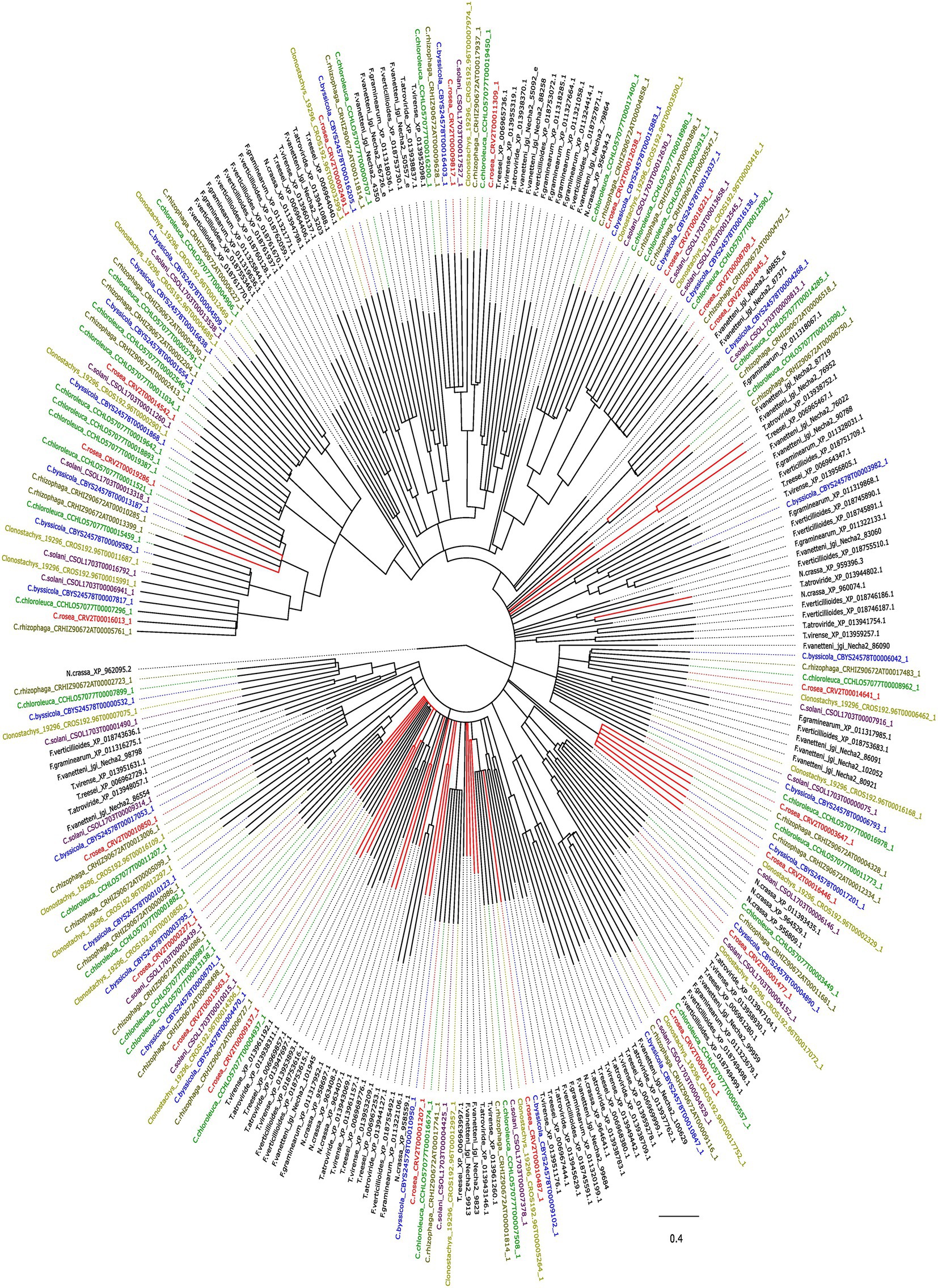
Figure 6. Phylogenetic tree showing the evolutionary relationship between the CFEM proteins in the species of interest. Red lines indicate secreted proteins. Bootstraps value lesser than 70% were condensed. The tree was generated with iqtree v.1.6.12 and visualized with figtree v.1.4.4.
4. Discussion
The predicted secretomes of the considered Clonostachys spp. amount to 7.7% of their proteomes on average, more than what was predicted for Trichoderma spp. The secretomes comprise a similar number of predicted proteins in all the considered Clonostachys spp., with enriched GO terms relating to proteolysis, catabolism of carbohydrates and response to fungus. We therefore hypothesize a role of secreted proteins in nutrient acquisition and fungal antagonism, which correlates well with the fact that 18% of the C. rosea secreted proteins are encoded by genes that are differentially expressed during the response to plant pathogenic fungi (Lysøe et al., 2017; Demissie et al., 2018, 2020; Nygren et al., 2018). The proportionally higher number of proteases, lipases and oxidoreductases in Clonostachys spp., compared with Trichoderma spp., together with the proportionally lower number of CAZymes, suggests different evolutionary trajectories in the two genera, driven by differences in their ecological strategies. The Clonostachys secretome included many proteins involved in fungal cell wall catabolism, such as C. rosea chitinase ChiC2 (CRV2T00000260_1), whose gene deletion cause a reduction in the growth inhibitory activity of culture filtrates against B. cinerea and Rhizoctonia solani (Tzelepis et al., 2015). An ortholog of this gene is present in all Clonostachys spp. considered in this study. However, the total number of identified GH18 chitinases range between six and nine genes in Clonostachys spp., which is lower compared to the gene copy number (12–21 genes) predicted in Trichoderma spp. More specifically, the major difference between Clonostachys and Trichoderma relates to the number of subgroup C killer toxin-like chitinases, hypothesized to be involved in permeabilization of mycohost cell walls for toxin entry (Tzelepis and Karlsson, 2019), suggesting intrinsic differences in the mode of action of these mycoparasites. Another class of secreted CAZymes operating on fungal cell walls are GH16 endo-β-(1,3)-glucanases, with potential roles in cell wall morphogenesis and catabolism (Mouyna et al., 2016). This class is evolving for increased gene copy numbers in the analyzed Clonostachys spp. with on average nine genes, while only five are found in Trichoderma spp. It is possible that the abundance of GH16 enzymes in their secretomes allow Clonostachys spp. to better modify and adapt their cell wall for the interaction with their hosts, or simply indicate that GH16 endo-β-(1,3)-glucanases are involved in cell wall degradation of the fungal prey.
Likewise, several CAZyme families with a putative role in deconstruction of plant cell walls evolve for increased paralog numbers in Clonostachys, possibly mediating nutrient uptake or plant host colonization. Family AA9 is involved in the degradation of cellulose and the high number of AA9 genes has already been observed in both C. rosea and C. byssicola (Karlsson et al., 2015; Gomes et al., 2020). In the present study, we identified a large difference, between 16 and 32 predicted AA9 enzymes, in the secretomes of different Clonostachys spp. indicating an involvement in ecological niche adaptation. Moreover, AA9 was the most frequent CAZyme class in C. rosea secreted proteins involved in the response to plant pathogens. AA9 enzymes need exogenous electron donors to function correctly, and it has been hypothesized that these could come from AA3 glucose-methanol-choline oxidoreductases (Vaaje-Kolstad et al., 2010), which is also the most numerous class of secreted oxidoreductases in the considered Clonostachys species. Additional gene families evolving for gene gene gains or losses include GH31 and GH3 where many members are putatively involved in hemicellulose degradation, including glucosidases, xylosidases and alpha-L-arabinofuranosidases. Enzymes of this class are also present in CAZyme family GH43, which is the most abundant in Clonostachys spp. with up to 39 members, while another abundant class, GH28 (13 to 16 members), include enzymes predicted to degrade pectin (Markovič and Janeček, 2001). Cell-wall degradation results in oligomers, such as xyloglucan, which can serve as damage-associated molecular patterns (DAMPs) and activate plant immunity reactions, including pattern-triggered immunity (PTI) and induced systemic resistance (ISR), resulting in the defense-inducing activity of Clonostachys spp. (Beliën et al., 2006; Souza et al., 2017; Claverie et al., 2018). Interestingly, Trichoderma spp. had on average a lower number of GH28, GH3, GH31, GH43, AA9 and AA3 enzymes, again emphasizing the different mechanistic strategies that underlie the ecological opportunism of these two genera. It is possible that Clonostachys spp. perform their biocontrol action through a greater induction of defense on host plants through partial plant cell wall degradation, while Trichoderma spp. have a greater capacity for the direct degradation of the fungal cell wall. Additional studies are needed to confirm this. Recent proteomic studies have shown an increased production of secreted plant cell wall degradation enzymes in Trichoderma spp. upon contact with the plant hosts Phaseolus vulgaris and Arabidopsis thaliana, and Clonostachys spp. might behave similarly (González-López et al., 2021; da Silva et al., 2022). Alternatively, these genomic adaptations may indicate a greater capacity for saprophytic growth of Clonostachys spp., which may also influence its usefulness in biocontrol applications.
Another process facilitated by secreted glycoside hydrolases is plant root colonization. In particular, all the considered Clonostachys spp. have a homolog of the PG1 protein, a class GH28 CAZyme involved in tomato root colonization in T. harzianum (Morán-Diez et al., 2009), and the C. rosea homolog of this gene (CRV2T00004567_1) is involved in the response to F. graminearum (Demissie et al., 2020). Another gene family with members involved in the interaction with plant hosts is the hydrophobins, necessary for plant colonization and pathogenicity in the pathogen M. oryzae (Talbot et al., 1996; Kim et al., 2005). Among the secreted C. rosea hydrophobins, we identified hyd3 (CRV2T00012494_1), a F. graminearum responsive gene (Demissie et al., 2020) whose deletion causes a reduction in root colonization (Dubey et al., 2014). This protein has a homolog in all considered Clonostachys spp., and it is similar (45% aa identity) to hydrophobin HFB2-6 of T. asperelleum, which has a function in root colonization and promotes jasmonic acid and salicylic acid signal transduction pathways in poplars (Huang et al., 2015). C. chloroleuca has almost twice as many hydrophobins, both secreted and otherwise, compared with other Clonostachys spp., suggesting that hydrophobins have evolved specific functions in the preferred ecological niche of this species.
Protease subfamily S8A is highly represented in the Clonostachys secretomes and it has previously been shown to evolve for gene gains in C. rosea (Iqbal et al., 2018a,b) and the same is true for the whole S8 family in Trichoderma spp. (Druzhinina et al., 2012). This family contains the serine endopeptidase subtilisin and its homologs, which have proven roles in biocontrol of fungi (Fan et al., 2014; Zhang et al., 2017) and nematodes (Ahman et al., 2002; Fekete et al., 2008). Numerous proteases of this class are proven to be involved in the response to mycohosts and nematodes, and this class was the most abundant one among the secreted C. rosea proteases found to be responsive to plant pathogens, together with class S1A. For example, the protease genes prs6 and prs16 are induced in C. rosea during the response to F. graminearum (Iqbal et al., 2018a,b; Demissie et al., 2020), while prs11, prs14 and prs16 are induced during parasitism of the potato pathogen H. solani (Lysøe et al., 2017). The serine protease prC gene is expressed in C. rosea when the fungus is degrading nematode cuticle material and is also involved in resistance to oxidative stress (Zou C. et al., 2010; Zou C.-G. et al., 2010). Many more members of these classes were detected in the secretomes of Clonostachys spp. than Trichoderma spp., indicating that Clonostachys spp. rely more on this type of proteases for their proteolytic action against mycohosts. However, serine proteases were also observed to be secreted in greater quantities upon T. harzianum interaction with P. vulgaris, suggesting a role in the interaction with the plant (da Silva et al., 2022).
The most numerous lipases in Clonostachys spp. proved to be GDSL-like lipases, which can potentially contribute to ethylene-based resistance in plants (Kwon et al., 2009; Gottwald et al., 2012). C. rosea and other species are known for inducing defense responses in plant hosts (Kamou et al., 2020; Sun et al., 2020), and this class of lipases could bolster that action. A subfamily of this class, GDSL esteraselipases exl3, are among the lipase families predicted to evolve for gene gains in Clonostachys species. Notably, we detected more than six Clonostachys GDSL-like lipases for each Trichoderma sp. in the secretomes, giving another indication that Clonostachys spp. have a greater part of their secretome dedicated to influencing plant hosts defense reactions. Among the most represented lipase families are also phospholipases A2, normally involved in nutrient acquisition but also in the modulation of host’s immune response (Köhler et al., 2006). One such gene (JK757061.1) is induced during T. harzianum colonization of tomatoes (Mehrabi-Koushki et al., 2012), and phospholipase A activity is a key mechanism by which Trichoderma spp. rupture the biological membranes of other fungi (Minchiotti et al., 2021).
C. rosea LysM protein LYSM2 (CRV2T00011102_1) is also predicted to be secreted. Deletion of the lysm2 gene resulted in C. rosea mutants with impaired biocontrol capabilities towards B. cinerea and F. graminearum, and also altered the suppression of wheat defense genes PR1 and PR4 (Dubey et al., 2020). An ortholog of this gene is present in all considered Clonostachys spp. except for C. rhizophaga. Other secreted proteins of interest include homologs of the effector cerato-platanin protein EPL1, involved in induction of defense reaction in maize, cotton, beans and Nicothiana bentamiana (Djonović et al., 2006; Djonovic et al., 2007; Crutcher et al., 2015; Gomes et al., 2015; Cheng et al., 2018). The thioredoxin-like effector class is also present in the secretomes of all considered species, with three proteins present in all Clonostachys spp. and one (CRV2T00013356_1) involved in the response to H. solani in C. rosea (Lysøe et al., 2017). This class is normally involved in apoplastic reactive oxygen species scavenging to protect plant pathogens from oxidative stress during the interaction with the plant, and it could play a similar role in Clonostachys spp., which are known to withstand high amounts of oxidative stress (Viefhues et al., 2014; Li et al., 2016).
Among the orthogroups detected with Orthofinder, 38 are evolving for gene gains and 16 of them contain genes involved in the C. rosea response to either F. graminearum or H. solani. Among these, nine contain CAZymes putatively involved in hemicellulose degradation and seven include putative effectors. Effector proteins in biocontrol fungi are typically necessary to resist and suppress the defense responses of plant hosts in order to allow plant colonization (Mendoza-Mendoza et al., 2018; Nogueira-Lopez et al., 2018; Romero-Contreras et al., 2019). Additionally, these orthogroups include proteases involved in the degradation of the plant cuticle, which is fundamental to initiate defense responses (Xia et al., 2009; Aragón et al., 2017). Orthogroup OG0000113 in particular included homologs of TvPG2, an endopolygalacturonase regulating the induction of plant defense in T. virens (Sarrocco et al., 2017). Yet other orthogroups consisted of trypsin proteases, which are a part of the biological control action of T. atroviride (Grinyer et al., 2005). C. rosea genes from these two orthogroups (CRV2T00016251_1 and CRV2T00014266_1) are both induced during the response to H. solani (Lysøe et al., 2017).
Several proteins with CFEM domains are predicted in the considered Clonostachys species. Such proteins are particularly numerous in fungal pathogens and they often act as cell-surface receptors, signal transducers, adhesion molecules or proteins involved in appressorium formation (Choi and Dean, 1997; Kulkarni et al., 2003; Zhang et al., 2015; Sabnam and Barman 2017). Interestingly, the considered Clonostachys spp. have higher numbers of CFEM proteins compared with Trichoderma species. In non-pathogenic fungi, CFEM can have a role in interactions with plants, and one member is upregulated in T. atroviride during plant host interaction (Guzmán-Guzmán et al., 2017), suggesting that their high number in Clonostachys spp. may be tied to a role in plant host perception and colonization. The high number of CFEM proteins could therefore help Clonostachys spp. to interact with a high variety of plant hosts, possibly by facilitating adhesion. Some of them, however, could also play a part in the interaction with mycohosts. In particular, the transmembrane CFEM protein CRV2T00016013_1 is part of a phylogenetic group evolving for gene gains in Clonostachys and is induced in C. rosea in response to F. graminearum (Demissie et al., 2020). Furthermore, the GPI-anchored protein CRV2T00009137_1 is the only CFEM protein encoded by a gene induced in response to both H. solani and F. graminearum (Lysøe et al., 2017; Demissie et al., 2020), indicating a general function in interspecific fungal interactions. This could be related to the known role of CFEM proteins as signal transducers (Kulkarni et al., 2003; Sabnam and Barman 2017).
5. Conclusion
This work investigates the composition of the predicted secretome of Clonostachys spp. and highlights its potential role in the mycoparasitic lifestyle and ecological opportunism of these commercially important fungi. Presence of proteins with a known role in fungal antagonism, including the chitinase CHIC2, the LysM protein LYSM2 and the endopolygalacturonase PG2 homolog, as well as several subtilisin-like peptidases and phospholipases A, emphasize the potential contribution of antibiosis to the biocontrol property of Clonostachys. Clonostachys spp. secretomes contained more CAZymes with a predicted function to degrade hemicellulose compared with Trichoderma spp., which however contained more chitinases. This may suggest that Trichoderma spp. are more adapted to degrade the cell wall of their mycohosts but less suited to induce defense reactions on their plant hosts. Moreover, we detected an unexpectedly high number of CFEM proteins in Clonostachys spp., typically more frequently found in plant pathogens, which also highlight the intimate interaction between Clonostachys spp. and plants, with potential consequences for their biocontrol activity. In summary, Clonostachys and Trichoderma species superficially share the same ecological lifestyle as rhizosphere-competent mycoparasites and opportunistic plant mutualists. Together with previous studies (Karlsson et al., 2015; Nygren et al., 2018; Broberg et al., 2021), the current work emphasizes several differences in the genomic characteristics of these two genera that show that convergent evolution resulted in adaptation of different underlying mechanisms for these apparent ecological similarities. This may have important consequences for the commercial exploitation of these fungi for biocontrol applications.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
MD, MK, and DJ conceived and designed the analysis. EP and MG performed the analyses with inputs from MK and MD. EP, MG, DJ, MK, and MD discussed and interpreted the results. EP wrote the first draft of the manuscript. MD and MK revised the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This work was financially supported by the Swedish Research Council for Environment, Agricultural Sciences and Spatial Planning (FORMAS; grant numbers 2018-01420, 2021-01461), and Carl Tryggers Stiftelse för Vetenskaplig Forskning (CTS 19: 82).
Acknowledgments
MK acknowledges the SLU Centre for Biological Control (CBC) at the Swedish University of Agricultural Sciences.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1112673/full#supplementary-material
References
Ahman, J., Johansson, T., Olsson, M., Punt, P. J., van den Hondel, C. A., and Tunlid, A. (2002). Improving the pathogenicity of a nematode-trapping fungus by genetic engineering of a subtilisin with nematotoxic activity. Appl. Environ. Microbiol. 68, 3408–3415. doi: 10.1128/AEM.68.7.3408-3415.2002
Altschul, S. F., Gish, W., Miller, W., Myers, E. W., and Lipman, D. J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. doi: 10.1016/S0022-2836(05)80360-2
Alvindia, D. G., and Natsuaki, K. T. (2008). Evaluation of fungal epiphytes isolated from banana fruit surfaces for biocontrol of banana crown rot disease. Crop Prot. 27, 1200–1207. doi: 10.1016/j.cropro.2008.02.007
Aragón, W., Reina-Pinto, J. J., and Serrano, M. (2017). The intimate talk between plants and microorganisms at the leaf surface. J. Exp. Bot. 68, 5339–5350. doi: 10.1093/jxb/erx327
Armenteros, J. J. A., Salvatore, M., Emanuelsson, O., Winther, O., Von Heijne, G., Elofsson, A., et al. (2019). Detecting sequence signals in targeting peptides using deep learning. Life Sci. 2:alliance. 2. doi: 10.26508/lsa.201900429
Arya, G. C., Srivastava, D. A., Pandaranayaka, E. P. J., Manasherova, E., Prusky, D. B., Elad, Y., et al. (2020). Characterization of the role of a non-GPCR membrane-bound CFEM protein in the pathogenicity and germination of Botrytis cinerea. Microorganisms. 8:1043. doi: 10.3390/microorganisms8071043
Atanasova, L., Dubey, M., Grujić, M., Gudmundsson, M., Lorenz, C., Sandgren, M., et al. (2018). Evolution and functional characterization of pectate lyase PEL12, a member of a highly expanded Clonostachys rosea polysaccharide lyase 1 family. BMC Microbiol. 18, 1–19. doi: 10.1186/s12866-018-1310-9
Beliën, T., Van Campenhout, S., Robben, J., and Volckaert, G. (2006). Microbial endoxylanases: effective weapons to breach the plant cell-wall barrier or, rather, triggers of plant defense systems? Mol. Plant-Microbe Interact. 19, 1072–1081. doi: 10.1094/MPMI-19-1072
Broberg, M., Dubey, M., Iqbal, M., Gudmundssson, M., Ihrmark, K., Schroers, H. J., et al. (2021). Comparative genomics highlights the importance of drug efflux transporters during evolution of mycoparasitism in Clonostachys subgenus Bionectria (fungi, Ascomycota, Hypocreales). Evol. Appl. 14, 476–497. doi: 10.1111/eva.13134
Broberg, M., Dubey, M., Sun, M. H., Ihrmark, K., Schroers, H. J., Li, S. D., et al. (2018). Out in the cold: identification of genomic regions associated with cold tolerance in the biocontrol fungus Clonostachys rosea through genome-wide association mapping. Front. Microbiol. 9:2844. doi: 10.3389/fmicb.2018.02844
Buchfink, B., Xie, C., and Huson, D. H. (2015). Fast and sensitive protein alignment using DIAMOND. Nat. Methods 12, 59–60. doi: 10.1038/nmeth.3176
Cheng, C.-H., Shen, B.-N., Shang, Q.-W., Liu, L.-Y. D., Peng, K.-C., Chen, Y.-H., et al. (2018). Gene-to-gene network analysis of the mediation of plant innate immunity by the eliciting plant response-like 1 (Epl1) elicitor of Trichoderma formosa. Mol. Plant-Microbe Interact. 31, 683–691. doi: 10.1094/MPMI-01-18-0002-TA
Choi, W., and Dean, R. A. (1997). The adenylate cyclase gene MAC1 of Magnaporthe grisea controls appressorium formation and other aspects of growth and development. Plant Cell 9, 1973–1983.
Claverie, J., Balacey, S., Lemaître-Guillier, C., Brulé, D., Chiltz, A., Granet, L., et al. (2018). The cell wall-derived xyloglucan is a new DAMP triggering plant immunity in Vitis vinifera and Arabidopsis thaliana. Front. Plant Sci. 9:1725. doi: 10.3389/fpls.2018.01725
Coleman, J. J., Rounsley, S. D., Rodriguez-Carres, M., Kuo, A., Wasmann, C. C., Grimwood, J., et al. (2009). The genome of Nectria haematococca: contribution of supernumerary chromosomes to gene expansion. PLoS Genet. 5:e1000618. doi: 10.1371/journal.pgen.1000618
Cota, L. V., Maffia, L. A., Mizubuti, E. S. G., and Macedo, P. E. F. (2009). Biological control by Clonostachys rosea as a key component in the integrated management of strawberry gray mold. Biol. Control 50, 222–230. doi: 10.1016/j.biocontrol.2009.04.017
Crutcher, F. K., Moran-Diez, M. E., Ding, S., Liu, J., Horwitz, B. A., Mukherjee, P. K., et al. (2015). A paralog of the proteinaceous elicitor SM1 is involved in colonization of maize roots by Trichoderma virens. Fungal Biol. 119, 476–486. doi: 10.1016/j.funbio.2015.01.004
da Silva, F. L., Aquino, E. N., da Cunha, D. C., Hamann, P. R. V., Magalhães, T. B., Steindorff, A. S., et al. (2022). Analysis of Trichoderma harzianum TR 274 secretome to assign candidate proteins involved in symbiotic interactions with Phaseolus vulgaris. Biocatal. Agric. Biotechnol. 43:102380. doi: 10.1016/j.bcab.2022.102380
Demissie, Z. A., Foote, S. J., Tan, Y., and Loewen, M. C. (2018). Profiling of the transcriptomic responses of Clonostachys rosea upon treatment with fusarium graminearum secretome. Front. Microbiol. 9:1061. doi: 10.3389/fmicb.2018.01061
Demissie, Z. A., Witte, T., Robinson, K. A., Sproule, A., Foote, S. J., Johnston, A., et al. (2020). Transcriptomic and exometabolomic profiling reveals antagonistic and defensive modes of Clonostachys rosea action against fusarium graminearum. Mol. Plant-Microbe Interact. 33, 842–858. doi: 10.1094/MPMI-11-19-0310-R
Djonović, S., Pozo, M. J., Dangott, L. J., Howell, C. R., and Kenerley, C. M. (2006). Sm1, a proteinaceous elicitor secreted by the biocontrol fungus Trichoderma virens induces plant defense responses and systemic resistance. Mol. Plant-Microbe Interact. 19, 838–853. doi: 10.1094/MPMI-19-0838
Djonovic, S., Vargas, W. A., Kolomiets, M. V., Horndeski, M., Wiest, A., and Kenerley, C. M. (2007). A proteinaceous elicitor Sm1 from the beneficial fungus Trichoderma virens is required for induced systemic resistance in maize. Plant Physiol. 145, 875–889. doi: 10.1104/pp.107.103689
Druzhinina, I. S., Shelest, E., and Kubicek, C. P. (2012). Novel traits of Trichoderma predicted through the analysis of its secretome. FEMS Microbiol. Lett. 337, 1–9. doi: 10.1111/j.1574-6968.2012.02665.x
Dubey, M. K., Jensen, D. F., and Karlsson, M. (2014). Hydrophobins are required for conidial hydrophobicity and plant root colonization in the fungal biocontrol agent Clonostachys rosea. BMC Microbiol. 14, 18–14. doi: 10.1186/1471-2180-14-18
Dubey, M. K., Ubhayasekera, W., Sandgren, M., Jensen, D. F., and Karlsson, M. (2012). Disruption of the Eng18b ENGase gene in the fungal biocontrol agent Trichoderma atroviride affects growth, conidiation and antagonistic ability. PLoS One 7:e36152. doi: 10.1371/journal.pone.0036152
Dubey, M., Vélëz, H., Broberg, M., Jensen, D. F., and Karlsson, M. (2020). LysM proteins regulate fungal development and contribute to hyphal protection and biocontrol traits in Clonostachys rosea. Front. Microbiol. 11:679. doi: 10.3389/fmicb.2020.00679
Dugan, F. M., Lupien, S. L., and Chen, W. (2012). Clonostachys rhizophaga and other fungi from chickpea debris in the Palouse region of the Pacific northwest, USA. North Am. Fungi. 7, 1–11. doi: 10.2509/naf2012.007.006
Emms, D. M., and Kelly, S. (2019). OrthoFinder: phylogenetic orthology inference for comparative genomics. Genome Biol. 20, 1–14. doi: 10.1186/s13059-019-1832-y
Fan, H., Liu, Z., Zhang, R., Wang, N., Dou, K., Mijiti, G., et al. (2014). Functional analysis of a subtilisin-like serine protease gene from biocontrol fungus Trichoderma harzianum. J. Microbiol. 52, 129–138. doi: 10.1007/s12275-014-3308-9
Fatema, U., Broberg, A., Jensen, D. F., Karlsson, M., and Dubey, M. (2018). Functional analysis of polyketide synthase genes in the biocontrol fungus Clonostachys rosea. Sci. Rep. 8, 1–17. doi: 10.1038/s41598-018-33391-1
Fekete, C., Tholander, M., Rajashekar, B., Ahrén, D., Friman, E., Johansson, T., et al. (2008). Paralysis of nematodes: shifts in the transcriptome of the nematode-trapping fungus Monacrosporium haptotylum during infection of Caenorhabditis elegans. Environ. Microbiol. 10, 364–375. doi: 10.1111/j.1462-2920.2007.01457.x
Feldman, D., Yarden, O., and Hadar, Y. (2020). Seeking the roles for fungal small-secreted proteins in affecting saprophytic lifestyles. Front. Microbiol. 11:455. doi: 10.3389/fmicb.2020.00455
Girard, V., Dieryckx, C., Job, C., and Job, D. (2013). Secretomes: the fungal strike force. Proteomics 13, 597–608. doi: 10.1002/pmic.201200282
Gogleva, A., Drost, H.-G., and Schornack, S. (2018). SecretSanta: flexible pipelines for functional secretome prediction. Bioinformatics 34, 2295–2296. doi: 10.1093/bioinformatics/bty088
Gomes, E. V., Do Nascimento Costa, M., De Paula, R. G., De Azevedo, R. R., Da Silva, F. L., Noronha, E. F., et al. (2015). The Cerato-Platanin protein Epl-1 from Trichoderma harzianum is involved in mycoparasitism, plant resistance induction and self cell wall protection. Sci. Rep. 5, 1–13. doi: 10.1038/srep17998
Gomes, H. A. R., Moreira, L. R. S., Júnior, A. C. S., Fontes, W., Santana, R. H., Kruger, R. H., et al. (2020). Evaluation of different secretomes produced by Clonostachys byssicola as tools to holocellulose breakdown. Int. Biodeterior. Biodegradation 148:104880. doi: 10.1016/j.ibiod.2019.104880
Gong, A., Jing, Z., Zhang, K., Tan, Q., Wang, G., and Liu, W. (2020). Bioinformatic analysis and functional characterization of the CFEM proteins in maize anthracnose fungus Colletotrichum graminicola. J. Integr. Agric. 19, 541–550. doi: 10.1016/S2095-3119(19)62675-4
González-López, M. D. C., Jijón-Moreno, S., Dautt-Castro, M., Ovando-Vázquez, C., Ziv, T., Horwitz, B. A., et al. (2021). Secretome analysis of Arabidopsis–Trichoderma atroviride interaction unveils new roles for the plant glutamate: glyoxylate aminotransferase GGAT1 in plant growth induced by the fungus and resistance against Botrytis cinerea. Int. J. Mol. Sci. 22:6804. doi: 10.3390/ijms22136804
Gottwald, S., Samans, B., Lück, S., and Friedt, W. (2012). Jasmonate and ethylene dependent defence gene expression and suppression of fungal virulence factors: two essential mechanisms of fusarium head blight resistance in wheat? BMC Genomics 13, 1–22. doi: 10.1186/1471-2164-13-369
Grinyer, J., Hunt, S., McKay, M., Herbert, B. R., and Nevalainen, H. (2005). Proteomic response of the biological control fungus Trichoderma atroviride to growth on the cell walls of Rhizoctonia solani. Curr. Genet. 47, 381–388. doi: 10.1007/s00294-005-0575-3
Guzmán-Guzmán, P., Alemán-Duarte, M. I., Delaye, L., Herrera-Estrella, A., and Olmedo-Monfil, V. (2017). Identification of effector-like proteins in Trichoderma spp. and role of a hydrophobin in the plant-fungus interaction and mycoparasitism. BMC Genet. 18, 1–20. doi: 10.1186/s12863-017-0481-y
Huang, Y., Mijiti, G., Wang, Z., Yu, W., Fan, H., Zhang, R., et al. (2015). Functional analysis of the class II hydrophobin gene HFB2-6 from the biocontrol agent Trichoderma asperellum ACCC30536. Microbiol. Res. 171, 8–20. doi: 10.1016/j.micres.2014.12.004
Iqbal, M., Broberg, M., Haarith, D., Broberg, A., Bushley, K. E., Brandström Durling, M., et al. (2020). Natural variation of root lesion nematode antagonism in the biocontrol fungus Clonostachys rosea and identification of biocontrol factors through genome-wide association mapping. Evol. Appl. 13, 2264–2283. doi: 10.1111/eva.13001
Iqbal, M., Dubey, M., Gudmundsson, M., Viketoft, M., Jensen, D. F., and Karlsson, M. (2018a). Comparative evolutionary histories of fungal proteases reveal gene gains in the mycoparasitic and nematode-parasitic fungus Clonostachys rosea. BMC Evol. Biol. 18, 1–17. doi: 10.1186/s12862-018-1291-1
Iqbal, M., Dubey, M., Mcewan, K., Menzel, U., Franko, M. A., Viketoft, M., et al. (2018b). Evaluation of clonostachys rosea for control of plant-parasitic nematodes in soil and in roots of carrot and wheat. Phytopathology 108, 52–59. doi: 10.1094/PHYTO-03-17-0091-R
Jensen, D. F., Dubey, M., Jensen, B., and Karlsson, M. (2021). “Clonostachys rosea for the control of plant diseases” in Microbial bioprotectants for plant disease management, ed. Jürgen Köhl and Willem Ravensberg (Location: London BDS Publishing), 429–471.
Jensen, B., Knudsen, I. M. B., and Jensen, D. F. (2000). Biological seed treatment of cereals with fresh and long-term stored formulations of Clonostachys rosea: biocontrol efficacy against fusarium culmorum. Eur. J. Plant Pathol. 106, 233–242. doi: 10.1023/A:1008794626600
Jones, P., Binns, D., Chang, H. Y., Fraser, M., Li, W., McAnulla, C., et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240. doi: 10.1093/bioinformatics/btu031
Kamou, N. N., Cazorla, F., Kandylas, G., and Lagopodi, A. L. (2020). Induction of defense-related genes in tomato plants after treatments with the biocontrol agents pseudomonas chlororaphis ToZa7 and Clonostachys rosea IK726. Arch. Microbiol. 202, 257–267. doi: 10.1007/s00203-019-01739-4
Karlsson, M., Durling, M. B., Choi, J., Kosawang, C., Lackner, G., Tzelepis, G. D., et al. (2015). Insights on the evolution of mycoparasitism from the genome of Clonostachys rosea. Genome Biol. Evol. 7, 465–480. doi: 10.1093/gbe/evu292
Katoh, K., and Standley, D. M. (2013). MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780. doi: 10.1093/molbev/mst010
Kim, S., Ahn, I., Rho, H., and Lee, Y. (2005). MHP1, a Magnaporthe grisea hydrophobin gene, is required for fungal development and plant colonization. Mol. Microbiol. 57, 1224–1237. doi: 10.1111/j.1365-2958.2005.04750.x
Kim, K.-T., Jeon, J., Choi, J., Cheong, K., Song, H., Choi, G., et al. (2016). Kingdom-wide analysis of fungal small secreted proteins (SSPs) reveals their potential role in host association. Front. Plant Sci. 7:186. doi: 10.3389/fpls.2016.00186
Köhler, G. A., Brenot, A., Haas-Stapleton, E., Agabian, N., Deva, R., and Nigam, S. (2006). Phospholipase A2 and phospholipase B activities in fungi. Biochim. Biophys. Acta (BBA)-molecular cell biol. Lipids 1761, 1391–1399. doi: 10.1016/j.bbalip.2006.09.011
Kou, Y., Tan, Y. H., Ramanujam, R., and Naqvi, N. I. (2017). Structure–function analyses of the Pth11 receptor reveal an important role for CFEM motif and redox regulation in rice blast. New Phytol. 214, 330–342. doi: 10.1111/nph.14347
Kredics, L., Antal, Z., Szekeres, A., Hatvani, L., Manczinger, L., Vágvölgyi, C. S., et al. (2005). Extracellular proteases of Trichoderma species. Acta Microbiol. Immunol. Hung. 52, 169–184. doi: 10.1556/AMicr.52.2005.2.3
Krogh, A., Larsson, B., Von Heijne, G., and Sonnhammer, E. L. L. (2001). Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 305, 567–580. doi: 10.1006/jmbi.2000.4315
Kulkarni, R. D., Kelkar, H. S., and Dean, R. A. (2003). An eight-cysteine-containing CFEM domain unique to a group of fungal membrane proteins. Trends Biochem. Sci. 28, 118–121. doi: 10.1016/S0968-0004(03)00025-2
Kwon, S. J., Jin, H. C., Lee, S., Nam, M. H., Chung, J. H., Kwon, S. I., et al. (2009). GDSL lipase-like 1 regulates systemic resistance associated with ethylene signaling in Arabidopsis. Plant J. 58, 235–245. doi: 10.1111/j.1365-313X.2008.03772.x
Li, Y.-B., Han, L.-B., Wang, H.-Y., Zhang, J., Sun, S.-T., Feng, D.-Q., et al. (2016). The thioredoxin GbNRX1 plays a crucial role in homeostasis of apoplastic reactive oxygen species in response to Verticillium dahliae infection in cotton. Plant Physiol. 170, 2392–2406. doi: 10.1104/pp.15.01930
Li, G. Q., Huang, H. C., Kokko, E. G., and Acharya, S. N. (2002). Ultrastructural study of mycoparasitism of Gliocladium roseum on Botrytis cinerea. Bot. Bull. Acad. Sin. 43, 211–218. doi: 10.7016/BBAS.200207.0211
Lu, S., and Edwards, M. C. (2016). Genome-wide analysis of small secreted cysteine-rich proteins identifies candidate effector proteins potentially involved in fusarium graminearum− wheat interactions. Phytopathology 106, 166–176. doi: 10.1094/PHYTO-09-15-0215-R
Lysøe, E., Dees, M. W., and Brurberg, M. B. (2017). A three-way transcriptomic interaction study of a biocontrol agent (Clonostachys rosea), a fungal pathogen (Helminthosporium solani), and a potato host (Solanum tuberosum). Mol. Plant-Microbe Interact. 30, 646–655. doi: 10.1094/MPMI-03-17-0062-R
Maillard, F., Andrews, E., Moran, M., Kennedy, P. G., Van Bloem, S. J., and Schilling, J. S. (2020). Stem-inhabiting fungal communities differ between intact and snapped trees after hurricane Maria in a Puerto Rican tropical dry forest. For. Ecol. Manag. 475:118350. doi: 10.1016/j.foreco.2020.118350
Markovič, O., and Janeček, Š. (2001). Pectin degrading glycoside hydrolases of family 28: sequence-structural features, specificities and evolution. Protein Eng. 14, 615–631. doi: 10.1093/protein/14.9.615
Mehrabi-Koushki, M., Rouhani, H., and Mahdikhani-Moghaddam, E. (2012). Differential display of abundantly expressed genes of Trichoderma harzianum during colonization of tomato-germinating seeds and roots. Curr. Microbiol. 65, 524–533. doi: 10.1007/s00284-012-0189-1
Mendes, F. K., Vanderpool, D., Fulton, B., and Hahn, M. W. (2020). CAFE 5 models variation in evolutionary rates among gene families. Bioinformatics 36, 5516–5518. doi: 10.1093/bioinformatics/btaa1022
Mendoza-Mendoza, A., Zaid, R., Lawry, R., Hermosa, R., Monte, E., Horwitz, B. A., et al. (2018). Molecular dialogues between Trichoderma and roots: role of the fungal secretome. Fungal Biol. Rev. 32, 62–85. doi: 10.1016/j.fbr.2017.12.001
Minchiotti, M. C., Vargas, L. I., and Madoery, R. R. (2021). Phospholipase a activity and the biocontrol potential of Trichoderma harzianum and Trichoderma atroviride. Biocontr. Sci. Technol. 31, 1231–1247. doi: 10.1080/09583157.2021.1940864
Morandi, M. A. B., Sutton, J. C., and Maffia, L. A. (2000). Effects of host and microbial factors on development of Clonostachys rosea and control of Botrytis cinerea in rose. Eur. J. Plant Pathol. 106, 439–448. doi: 10.1023/A:1008738513748
Morán-Diez, E., Hermosa, R., Ambrosino, P., Cardoza, R. E., Gutiérrez, S., Lorito, M., et al. (2009). The ThPG1 endopolygalacturonase is required for the Trichoderma harzianum–plant beneficial interaction. Mol. Plant-Microbe Interact. 22, 1021–1031. doi: 10.1094/MPMI-22-8-1021
Moreira, G. M., Abreu, L. M., Carvalho, V. G., Schroers, H.-J., and Pfenning, L. H. (2016). Multilocus phylogeny of Clonostachys subgenus Bionectria from Brazil and description of Clonostachys chloroleuca sp. nov. Mycol. Prog. 15, 1031–1039. doi: 10.1007/s11557-016-1224-6
Mouyna, I., Aimanianda, V., Hartl, L., Prevost, M., Sismeiro, O., Dillies, M., et al. (2016). GH16 and GH81 family β-(1, 3)-glucanases in aspergillus fumigatus are essential for conidial cell wall morphogenesis. Cell. Microbiol. 18, 1285–1293. doi: 10.1111/cmi.12630
Nguyen, L. T., Schmidt, H. A., Von Haeseler, A., and Minh, B. Q. (2015). IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274. doi: 10.1093/molbev/msu300
Nogueira-Lopez, G., Greenwood, D. R., Middleditch, M., Winefield, C., Eaton, C., Steyaert, J. M., et al. (2018). The apoplastic secretome of Trichoderma virens during interaction with maize roots shows an inhibition of plant defence and scavenging oxidative stress secreted proteins. Front. Plant Sci. 9:409. doi: 10.3389/fpls.2018.00409
Nygren, K., Dubey, M., Zapparata, A., Iqbal, M., Tzelepis, G. D., Durling, M. B., et al. (2018). The mycoparasitic fungus Clonostachys rosea responds with both common and specific gene expression during interspecific interactions with fungal prey. Evol. Appl. 11, 931–949. doi: 10.1111/eva.12609
Pellegrin, C., Morin, E., Martin, F. M., and Veneault-Fourrey, C. (2015). Comparative analysis of secretomes from ectomycorrhizal fungi with an emphasis on small-secreted proteins. Front. Microbiol. 6:1278. doi: 10.3389/fmicb.2015.01278
Petersen, T. N., Brunak, S., Von Heijne, G., and Nielsen, H. (2011). SignalP 4.0: discriminating signal peptides from transmembrane regions. Nat. Methods 8, 785–786. doi: 10.1038/nmeth.1701
Pierleoni, A., Martelli, P. L., and Casadio, R. (2008). PredGPI: a GPI-anchor predictor. BMC Bioinformatics. 9, 1–11. doi: 10.1186/1471-2105-9-392
Piombo, E., Vetukuri, R. R., Broberg, A., Kalyandurg, P. B., Kushwaha, S., Funck Jensen, D., et al. (2021). Role of dicer-dependent RNA interference in regulating mycoparasitic interactions. Microbiol. Spectr. 9:e0109921. doi: 10.1128/Spectrum.01099-21
Rambaut, A. (2018). FigTree v. 1.4.4. Available at: http://tree.bio.ed.ac.uk/software/figtree/
Ramírez-Valdespino, C. A., Casas-Flores, S., and Olmedo-Monfil, V. (2019). Trichoderma as a model to study effector-like molecules. Front. Microbiol. 10:1030. doi: 10.3389/fmicb.2019.01030
Rawlings, N. D., Barrett, A. J., and Bateman, A. (2010). MEROPS: the peptidase database. Nucleic Acids Res. 38, D227–D233. doi: 10.1093/nar/gkp971
Rawlings, N. D., Barrett, A. J., and Bateman, A. (2012). MEROPS: The database of proteolytic enzymes, their substrates and inhibitors. Nucleic Acids Res. 40, D343–D350.
Romero-Contreras, Y. J., Ramírez-Valdespino, C. A., Guzmán-Guzmán, P., Macías-Segoviano, J. I., Villagómez-Castro, J. C., and Olmedo-Monfil, V. (2019). Tal6 from Trichoderma atroviride is a LysM effector involved in mycoparasitism and plant association. Front. Microbiol. 10:2231. doi: 10.3389/fmicb.2019.02231
Sabnam, N., and Barman, S. R. (2017). WISH, a novel CFEM GPCR is indispensable for surface sensing, asexual and pathogenic differentiation in rice blast fungus. Fungal Genet. Biol. 105, 37–51. doi: 10.1016/j.fgb.2017.05.006
Salamone, A. L., Gundersen, B., and Inglis, D. A. (2018). Clonostachys rosea, a potential biological control agent for Rhizoctonia solani AG-3 causing black scurf on potato. Biocontrol Sci. Tech. 28, 895–900. doi: 10.1080/09583157.2018.1498063
Saraiva, R. M., Czymmek, K. J., Borges, Á. V., Caires, N. P., and Maffia, L. A. (2015). Confocal microscopy study to understand Clonostachys rosea and Botrytis cinerea interactions in tomato plants. Biocontrol Sci. Tech. 25, 56–71. doi: 10.1080/09583157.2014.948382
Sarrocco, S., Matarese, F., Baroncelli, R., Vannacci, G., Seidl-Seiboth, V., Kubicek, C. P., et al. (2017). The constitutive endopolygalacturonase TvPG2 regulates the induction of plant systemic resistance by Trichoderma virens. Phytopathology 107, 537–544. doi: 10.1094/PHYTO-03-16-0139-R
Selin, C., De Kievit, T. R., Belmonte, M. F., and Fernando, W. (2016). Elucidating the role of effectors in plant-fungal interactions: progress and challenges. Front. Microbiol. 7:600. doi: 10.3389/fmicb.2016.00600
Souza, C. D. A., Li, S., Lin, A. Z., Boutrot, F., Grossmann, G., Zipfel, C., et al. (2017). Cellulose-derived oligomers act as damage-associated molecular patterns and trigger defense-like responses. Plant Physiol. 173, 2383–2398. doi: 10.1104/pp.16.01680
Sperschneider, J., and Dodds, P. (2021). EffectorP 3.0: prediction of apoplastic and cytoplasmic effectors in fungi and oomycetes. Mol. Plant-Microbe Interact. 35, 146–156. doi: 10.1094/MPMI-08-21-0201-R
Srivastava, V. K., Suneetha, K. J., and Kaur, R. (2014). A systematic analysis reveals an essential role for high-affinity iron uptake system, haemolysin and CFEM domain-containing protein in iron homoeostasis and virulence in Candida glabrata. Biochem. J. 463, 103–114. doi: 10.1042/BJ20140598
Sun, Z. B., Li, S. D., Ren, Q., Xu, J. L., Lu, X., and Sun, M. H. (2020). Biology and applications of Clonostachys rosea. J. Appl. Microbiol. 129, 486–495. doi: 10.1111/jam.14625
Sun, Z.-B., Sun, M.-H., Zhou, M., and Li, S.-D. (2017). Transformation of the endochitinase gene Chi67-1 in Clonostachys rosea 67-1 increases its biocontrol activity against Sclerotinia sclerotiorum. AMB Express 7, 1–9. doi: 10.1186/s13568-016-0313-x
Sun, Z.-B., Wang, Q., Zhang, J., Jiang, W.-Z., Li, S.-D., Ma, G.-Z., et al. (2018). The transcription factor-encoding gene crtf is involved in Clonostachys chloroleuca mycoparasitism on Sclerotinia sclerotiorum. Microbiol. Res. 210, 6–11. doi: 10.1016/j.micres.2018.03.002
Supek, F., Bošnjak, M., Škunca, N., and Šmuc, T. (2011). REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS One 6: e21800.
Sutton, J. C., Liu, W., Huang, R., and Owen-Going, N. (2002). Ability of Clonostachys rosea to establish and suppress sporulation potential of Botrytis cinerea in deleafed stems of hydroponic greenhouse tomatoes. Biocontrol Sci. Tech. 12, 413–425. doi: 10.1080/09583150220146004
Talbot, N. J., Kershaw, M. J., Wakley, G. E., De Vries, O. M. H., Wessels, J. G. H., and Hamer, J. E. (1996). MPG1 encodes a fungal hydrophobin involved in surface interactions during infection-related development of Magnaporthe grisea. Plant Cell 8, 985–999. doi: 10.2307/3870210
Tian, T., Liu, Y., Yan, H., You, Q., Yi, X., Du, Z., et al. (2017). agriGO v2. 0: a GO analysis toolkit for the agricultural community, 2017 update. Nucleic Acids Res. 45, W122–W129. doi: 10.1093/nar/gkx382
Tzelepis, G., Dubey, M., Jensen, D. F., and Karlsson, M. (2015). Identifying glycoside hydrolase family 18 genes in the mycoparasitic fungal species Clonostachys rosea. Microbiol. (United Kingdom). 161, 1407–1419. doi: 10.1099/mic.0.000096
Tzelepis, G., and Karlsson, M. (2019). Killer toxin-like chitinases in filamentous fungi: structure, regulation and potential roles in fungal biology. Fungal Biol. Rev. 33, 123–132. doi: 10.1016/j.fbr.2018.11.001
Vaaje-Kolstad, G., Westereng, B., Horn, S. J., Liu, Z., Zhai, H., Sørlie, M., et al. (2010). An oxidative enzyme boosting the enzymatic conversion of recalcitrant polysaccharides. Science 330, 219–222. doi: 10.1126/science.1192231
Viefhues, A., Heller, J., Temme, N., and Tudzynski, P. (2014). Redox systems in Botrytis cinerea: impact on development and virulence. Mol. Plant-Microbe Interact. 27, 858–874. doi: 10.1094/MPMI-01-14-0012-R
Viterbo, A., Harel, M., and Chet, I. (2004). Isolation of two aspartyl proteases from Trichoderma asperellum expressed during colonization of cucumber roots. FEMS Microbiol. Lett. 238, 151–158. doi: 10.1016/j.femsle.2004.07.028
Xia, Y., Gao, Q.-M., Yu, K., Lapchyk, L., Navarre, D., Hildebrand, D., et al. (2009). An intact cuticle in distal tissues is essential for the induction of systemic acquired resistance in plants. Cell Host Microbe 5, 151–165. doi: 10.1016/j.chom.2009.01.001
Xue, A., Chen, Y., Voldeng, H., Savard, M., and Tian, X. (2008). Biological control of fusarium head blight of wheat with Clonostachys rosea strain ACM941. Cereal Res. Commun. 36, 695–699. doi: 10.1556/CRC.36.2008.Suppl.B.62
Zhang, H., Wang, N. A., Wang, Y., Wang, J., Zheng, H., and Liu, Z. (2017). Subtilisin-like serine protease gene TghSS42 from Trichoderma ghanense ACCC 30153 was successfully expressed in Escherichia coli and recombinant protease rTghSS42 exhibited antifungal ability to five phytopathogens. Biocontrol Sci. 22, 145–152. doi: 10.4265/bio.22.145
Zhang, Z.-N., Wu, Q.-Y., Zhang, G.-Z., Zhu, Y.-Y., Murphy, R. W., Liu, Z., et al. (2015). Systematic analyses reveal uniqueness and origin of the CFEM domain in fungi. Sci. Rep. 5, 1–7. doi: 10.1038/srep13032
Zhang, H., Yohe, T., Huang, L., Entwistle, S., Wu, P., Yang, Z., et al. (2018). DbCAN2: a meta server for automated carbohydrate-active enzyme annotation. Nucleic Acids Res. 46, W95–W101. doi: 10.1093/nar/gky418
Zhu, W., Wei, W., Wu, Y., Zhou, Y., Peng, F., Zhang, S., et al. (2017). BcCFEM1, a CFEM domain-containing protein with putative GPI-anchored site, is involved in pathogenicity, conidial production, and stress tolerance in Botrytis cinerea. Front. Microbiol. 8:1807. doi: 10.3389/fmicb.2017.01807
Zou, C., Tao, N., Liu, W., Yang, J., Huang, X., Liu, X., et al. (2010). Regulation of subtilisin-like protease prC expression by nematode cuticle in the nematophagous fungus Clonostachys rosea. Environ. Microbiol. 12, 3243–3252. doi: 10.1111/j.1462-2920.2010.02296.x
Keywords: biocontrol, Clonostachys, mycoparasitism, small secreted cysteine-rich proteins, effector, CFEM proteins, antagonism, secretome
Citation: Piombo E, Guaschino M, Jensen DF, Karlsson M and Dubey M (2023) Insights into the ecological generalist lifestyle of Clonostachys fungi through analysis of their predicted secretomes. Front. Microbiol. 14:1112673. doi: 10.3389/fmicb.2023.1112673
Edited by:
Sabrina Sarrocco, University of Pisa, ItalyReviewed by:
Roberta Marra, University of Naples Federico II, ItalyEliane Ferreira Noronha, University of Brasilia, Brazil
Prasun K. Mukherjee, Bhabha Atomic Research Centre (BARC), India
Copyright © 2023 Piombo, Guaschino, Jensen, Karlsson and Dubey. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mukesh Dubey, ✉ TXVrZXNoLkR1YmV5QHNsdS5zZQ==
 Edoardo Piombo
Edoardo Piombo Micol Guaschino
Micol Guaschino Dan Funck Jensen
Dan Funck Jensen Magnus Karlsson
Magnus Karlsson Mukesh Dubey
Mukesh Dubey