- 1Department of Laboratory Medicine, Nanjing Drum Tower Hospital, The Affiliated Hospital of Nanjing University Medical School, Nanjing, Jiangsu, China
- 2Department of Acute Infectious Disease Control and Prevention, Jiangsu Provincial Center for Disease Control and Prevention, Nanjing, China
Pseudomonas aeruginosa is an opportunistic human pathogen notorious for its remarkable capacity of multi-drug resistance, and has become one of the most important model bacteria in clinical bacteriology research. Quantitative real-time PCR is a reliable method widely used in gene expression analysis, for which the selection of a set of appropriate housekeeping genes is a key prerequisite for the accuracy of the results. However, it is easy to overlook that the expression level of housekeeping gene may vary in different conditions, especially in the condition of molecular microbiology assays, where tested strains are generally cultured under the pre-set antibiotic selection pressures, and how this affects the stability of commonly used housekeeping genes remains unclear. In this study, the expression stability of ten classic housekeeping genes (algD, gyrA, anr, nadB, recA, fabD, proC, ampC, rpoS, and rpsL) under the pressure of eight laboratory commonly used antibiotics (kanamycin, gentamycin, tetracycline, chloramphenicol, hygromycin B, apramycin, tellurite, and zeocin) were tested. Results showed that the stability of housekeeping gene expression was indeed affected by the types of antibiotics added, and of course the best reference gene set varied for different antibiotics. This study provides a comprehensive summary of the effects of laboratory antibiotics on the stability of housekeeping genes in P. aeruginosa, highlighting the necessity to select housekeeping genes according to the type of antibiotics used in the initial stage of experiment.
Introduction
Pseudomonas aeruginosa is a gram-negative, rod-shaped bacterium belonging to the Pseudomonadaceae family. It is a ubiquitous organism found in many environments including soil, water, animals and humans (Moradali et al., 2017). Pseudomonas aeruginosa is seldom a member of normal microbial flora in most healthy adults, but a severe menace to immunocompromised patients, associated with an array of life-threatening infections, including cystic fibrosis, ventilator-associated pneumonia, urinary tract infections, burn injury infections, and bloodstream infections (Malhotra et al., 2019).
In bacteriology research, P. aeruginosa serves as a model organism for studying biofilm formation, type VI secretion system (T6SS) and quorum sensing signaling, etc. (Lee and Zhang, 2015; Pena et al., 2019; Thi et al., 2020). Clinically, P. aeruginosa is notorious for its complicated antimicrobial resistance mechanisms, which can develop antibacterial resistance either exogenously by obtaining various resistance gene cassettes, or endogenously by altering the expression level of chromosomally encoded enzymes (Breidenstein et al., 2011). Both strategies would severely limit therapeutic options and make the pathogen difficult to eradicate in clinical anti-infection treatments (Botelho et al., 2019). For these reasons, numerous studies that aim to reveal the deep patterns of P. aeruginosa have been performed using expressed sequences tags (ESTs), luciferase reporter system, and quantitative real-time PCR (Bustin and Mueller, 2005; Parkinson and Blaxter, 2009; England et al., 2016).
The quantitative real-time PCR is a reliable technique that is used extensively for gene expression analysis due to its high sensitivity and repeatability (Bustin and Mueller, 2005). This method could be performed as either an absolute measurement using a pre-prepared external standard curve, or as a relative measurement by comparing the expression level of a target gene with that of a presumed constantly expressed “housekeeping gene.” The expression level of housekeeping genes may be affected by a variety of factors, and therefore a selection process with caution is required (Kozera and Rapacz, 2013). For instance, in most molecular microbiology assays, tested strains which carry pre-introduced antimicrobial resistance cassette are firstly plated on the selective agar medium for colonies formation, and cultured under the same antimicrobial pressure during subsequent experimental process. Because of the great differences in pharmacological properties among various types of antibiotics, the expression level of pre-set housekeeping gene might be affected with varying degrees by different antibiotics. This means that the choice of housekeeping gene would be compromised according to the type of antibiotic selected, but there is still no literature available for researchers to refer to.
In this study, the reference strain P. aeruginosa PAO1 was set as the test strain, and the growth environment under the pressure of 8 antibiotics that could be used in molecular microbiology assays was simulated by adding 1/2 MIC of each antibiotic (Ducas-Mowchun et al., 2019). Then, a total of 10 candidate housekeeping genes were selected, and their expression levels were validated one by one by using quantitative real-time PCR. The aim of this study is to give some help to the initial step of reference gene selection in the studies of P. aeroginosa, and the experimental results show that it is necessary to select the housekeeping genes according to the different types of antibiotics used.
Materials and methods
Strains, resistance tests and culture conditions
Pseudomonas aeruginosa PAO1 strain was inoculated from a Lysogeny-Broth (LB) agar plates, and cultured overnight in LB liquid medium at 37°C in a rotary incubator shaker (200 rpm). Mid-exponential phase cultures were obtained after 1:100 dilution of overnight culture, and incubation at 37°C with shaking until OD600 ~ 0.5. Resistance profile of kanamycin, gentamycin, tetracycline, chloramphenicol, hygromycin B, apramycin, tellurite, and zeocin (Sangon) was determined by broth micro dilution assays as described by Clinical and Laboratory Standards Institute (2015). Experimental conditions by which strains were cultured under the pressure of antibiotics were simulated by adding 1/2 minimum inhibitory concentration (MIC) of corresponding antibiotics and incubation at 37°C with shaking overnight.
Total RNA isolation and quantitative real-time PCR
Total RNA of antibiotic-induced strains was extracted from 5 ml mid-exponential phase cultures by using the Spin Column Bacteria Total RNA Purification Kit (Sangon) as described by the manufacturer. Genomic DNA was eliminated by RNase-free DNase Set (Sangon) treatment during the extraction procedure. Finally, total RNA was eluted from the column in a volume of 40 μl DEPC-treated water, and all RNA samples were uniformly diluted to ~25 μg/ml. This step is critical for performing successful calculations. The step of reverse transcription and real-time PCR were continuously carried out in the same reaction tube to effectively prevent contamination. Total RNA samples were transcribed to cDNA and amplified by using One Step TB Green Primescript Plus RT-PCR Kit (TaKaRa) according to the manufacturer’s protocol. Real-time PCR was carried out in the CFX96 ThermalCycler (Bio-Rad) under the reaction condition as 42°C at 5 min, 95°C at 10 s for cDNA synthesis, then followed by 40 cycles of 95°C for 5 s, 60°C for 20 s with data acquisition. At last, a melt curve analysis was performed using the temperature range 60°C to 95°C at 0.5°C intervals to confirm the specific amplification of a single PCR product. Primers were designed by using NCBI primer-Blast (Table 1). Cycle threshold (CT) values were determined with the Bio-Rad CFX Manager software and were used to calculate the gene expression level. All reactions were performed in triplicate, and the mean value was calculated.
Results
MIC values of several antibiotics against Pseudomonas aeruginosa PAO1
The MIC values of kanamycin, tetracycline, chloramphenicol, gentamycin, apramycin, hygromycin B, zeocin, and tellurite were tested (Table 2), and the sub-MIC antibiotics were added during subsequent experiments to simulate the circumstances of molecular microbiology assays in which bacteria are cultured under the pressure of the above antibiotics.

Table 2. The minimum inhibitory concentrations (MICs) of the tested antibiotics against Pseudomonas aeruginosa PAO1.
The MICs were determined using the broth microdilution assay. Assays were performed in triplicates, and the mean value was calculated.
Expression profiling of selected housekeeping genes
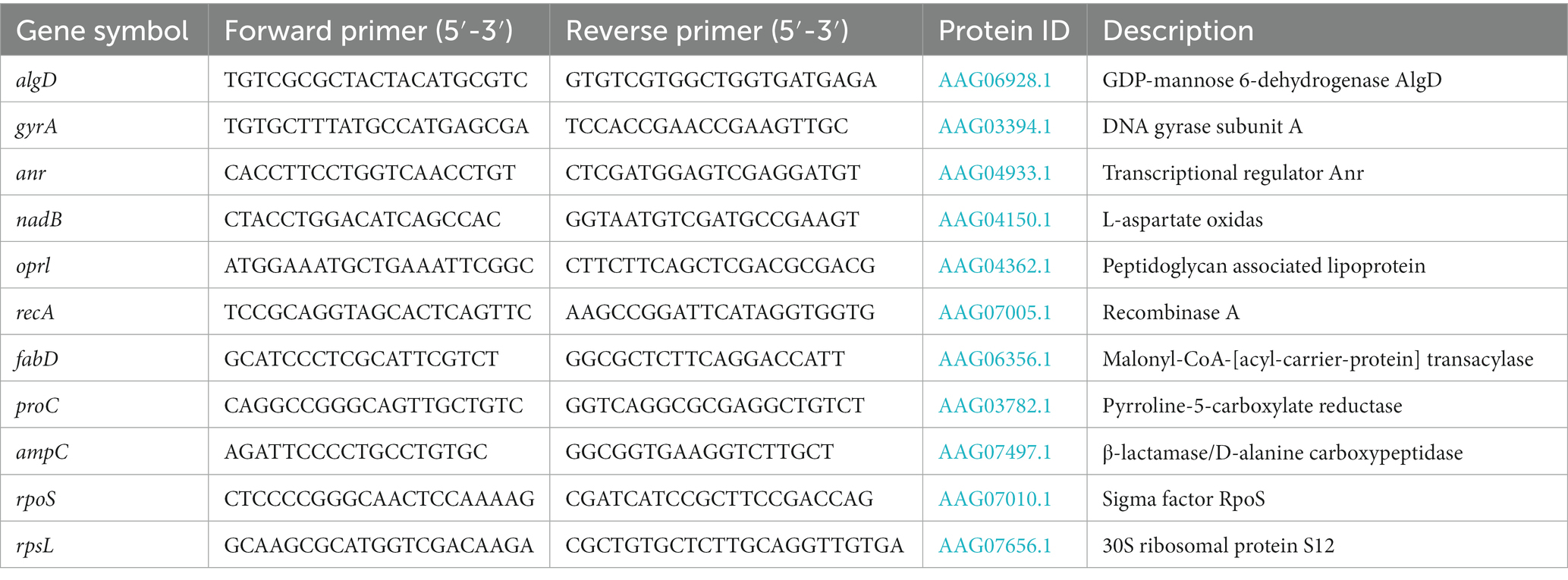
In this study, a total of ten housekeeping genes with different functions that have been used in other studies as internal controls of P. aeruginosa were selected (Table 1; Alqarni et al., 2016). The quantitative real-time PCR was then performed to validate the expression profile of candidate housekeeping genes by comparing the raw CT values of each gene under the pressure of different antibiotics. Briefly, CT values directly indicate the mRNA abundance of target genes, lower CT value indicates higher abundance and vice versa. The CT ranges of each tested gene under different antibiotic pressures are shown in Figure 1. CT values of most candidate housekeeping genes such as algD, gyrA, anr, nadB, recA, rpoS, and rpsL basically appeared around 21 cycles, while fabD and ampC were slightly higher above average. The proC showed the highest CT value around 28 cycles, indicating that its expression level is the lowest among all tested genes.
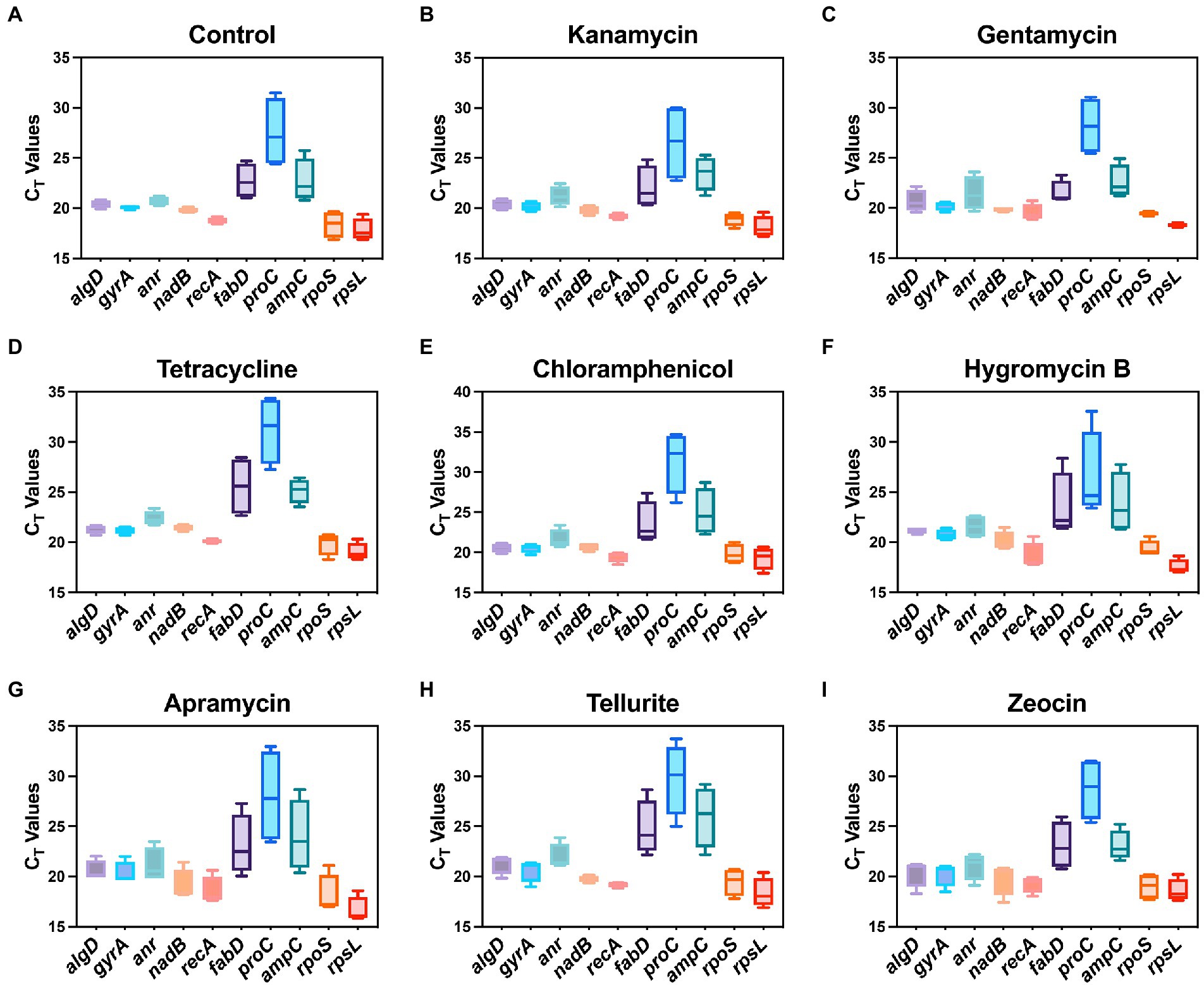
Figure 1. Range of real-time PCR cycle threshold (CT) values of 10 housekeeping genes in P. aeruginosa under 8 antibiotic pressures. (A) Control; (B) kanamycin; (C) gentamycin; (D) tetracycline; (E) chloramphenicol; (F) hygromycin B; (G) apramycin; (H) tellurite; (I) zeocin.
Stability analysis of Pseudomonas aeruginosa housekeeping genes
To calculate the stability value of selected P. aeruginosa housekeeping gene in this study, dedicated algorithms including geNorm, NormFinder, BestKeeper and RefFinder were used, and an overall ranking is then carried out according to the comprehensive results.
geNorm algorithm
geNorm is a classic algorithm used to determine the stability of candidate reference (housekeeping) genes by calculating a gene expression normalization factor (M value) based on the geometric mean of a user-defined number of reference genes. Genes with the lowest M value reflect the highest stability in terms of gene expression and vice versa, and each calculation provides two recommended housekeeping genes. Following this criterion, recA and nadB were found as suitable housekeeping genes in the control group which had no antibiotic pressure (Figure 2A). Under the pressure of kanamycin and chloramphenicol, gyrA and algD were found to be the most stable housekeeping genes (Figures 2B,E), while rpoS and rpsL registered the lowest M values under the pressure of gentamycin and hygromycin B (Figures 2C,F). In the group of tetracycline, nadB and gyrA were found to be the most stable housekeeping genes (Figure 2D), as well as the circumstance of recA and rpsL under the pressure of apramycin (Figure 2G). In the tellurite-treated samples, recA and nadB showed excellent stability (Figure 2H), while anr and gyA were found to be the most stable housekeeping genes under the zeocin pressure (Figure 2I).
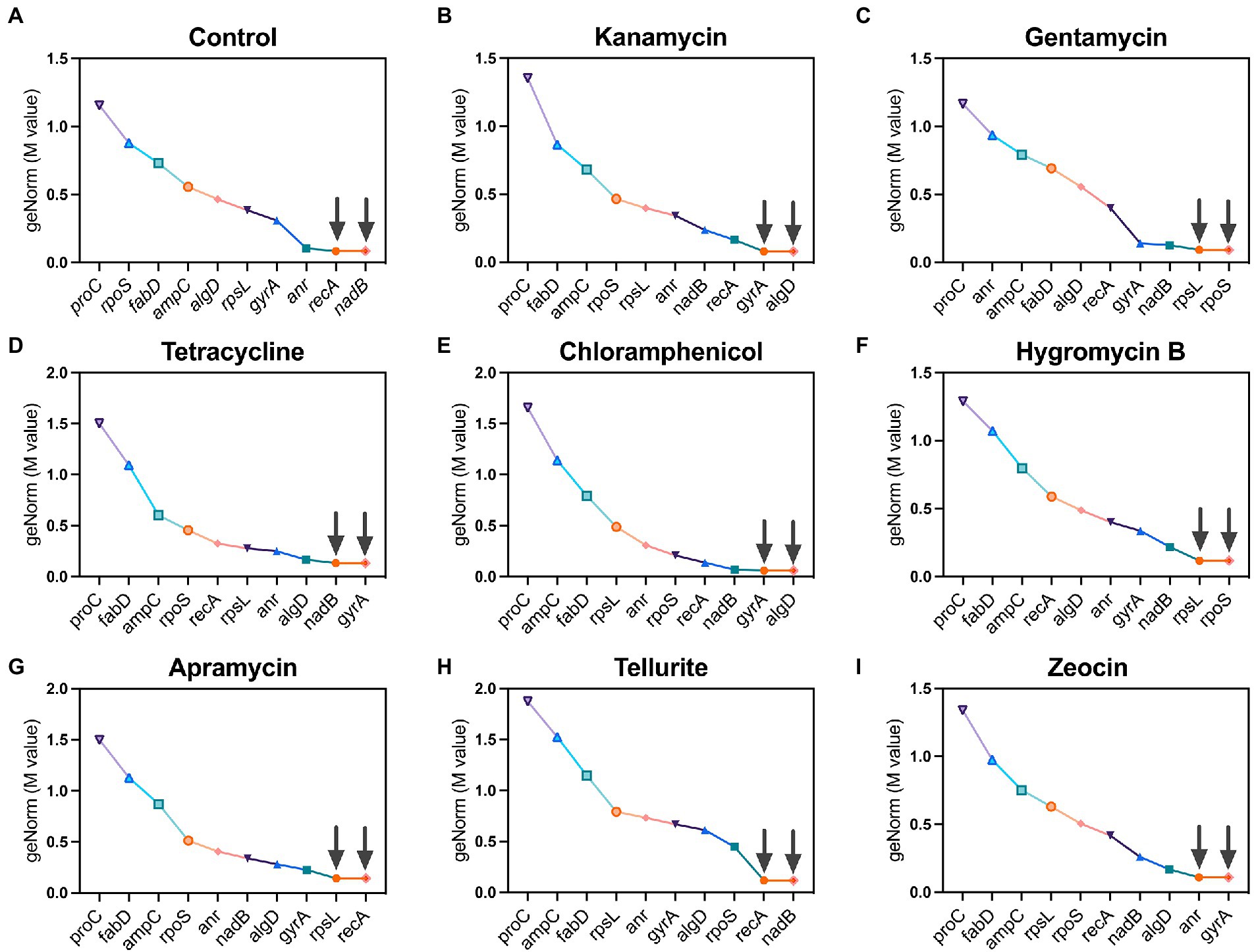
Figure 2. Ranking of 10 candidate housekeeping genes based on stability values (M values) calculated by geNorm algorithm. (A) Control; (B) kanamycin; (C) gentamycin; (D) tetracycline; (E) chloramphenicol; (F) hygromycin B; (G) apramycin; (H) tellurite; (I) zeocin.
NormFinder ranking
NormFinder was also used to screen the suitable candidate reference genes in this study. Similar to geNorm, this algorithm could rank the most competent housekeeping gene by calculating a stability value, with lower values representing more stability and vice versa. Under the rule of NormFinder algorithm, anr and rpsL exhibited excellent performance, and anr was the best housekeeping gene under the pressure of tetracycline, chloramphenicol, hygromycin B, apramycin and control group, while rpsL was the most stable gene in kanamycin, tellurite and zeocin group. Otherwise, recA was considered the best housekeeping gene only under the pressure of gentamycin (Table 3).
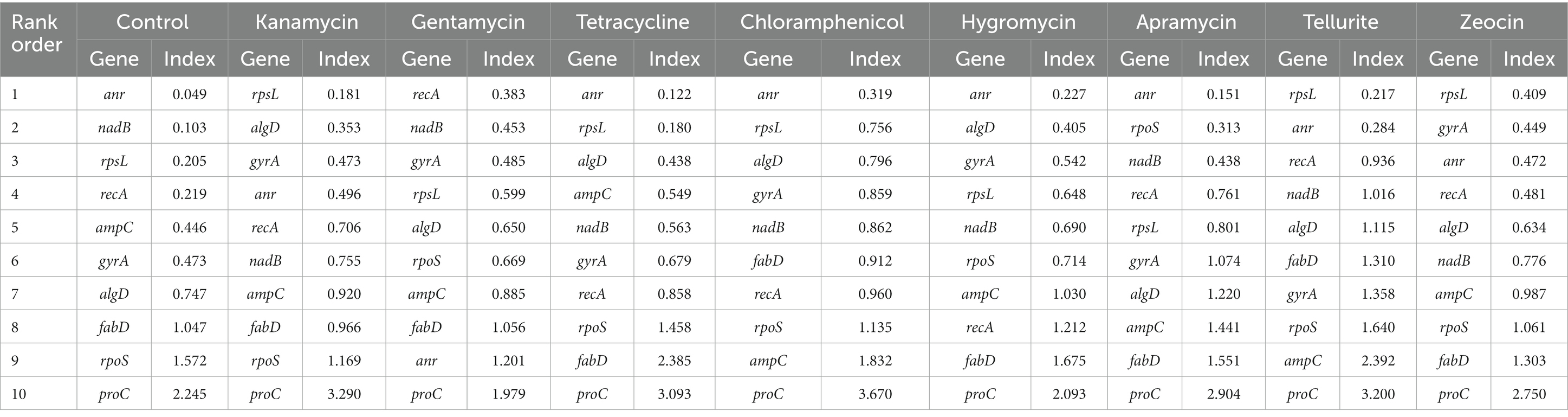
Table 3. Expression stability of seleceted Pseudomonas aeruginosa housekeeping genes calculated by NormFinder under different antibiotic stress.
BestKeeper ranking
BestKeeper is also a housekeeping gene ranking tool base on Microsoft Excel (Pfaffl et al., 2004). It could export a series of indices, of which standard deviation (SD) and coefficient of variance (CV) are two main parameters. Similarly, lower values represent higher level of stability, and only data with an SD < 1 is considered acceptable. The ranking of the housekeeping gene tested in our study by this algorithm was slightly different from the two methods mentioned above: rpoS was considered to be the most stable gene under the influence of kanamycin and gentamycin (Figures 3B,C), recA outperformed in tetracycline and tellurite (Figures 3D,H), algD was the most stable gene in apramycin and hygromycin B (Figures 3F,G), and gyrA and anr were considered the most stable gene in the control and chloramphenicol groups, respectively, (Figures 3A,E).
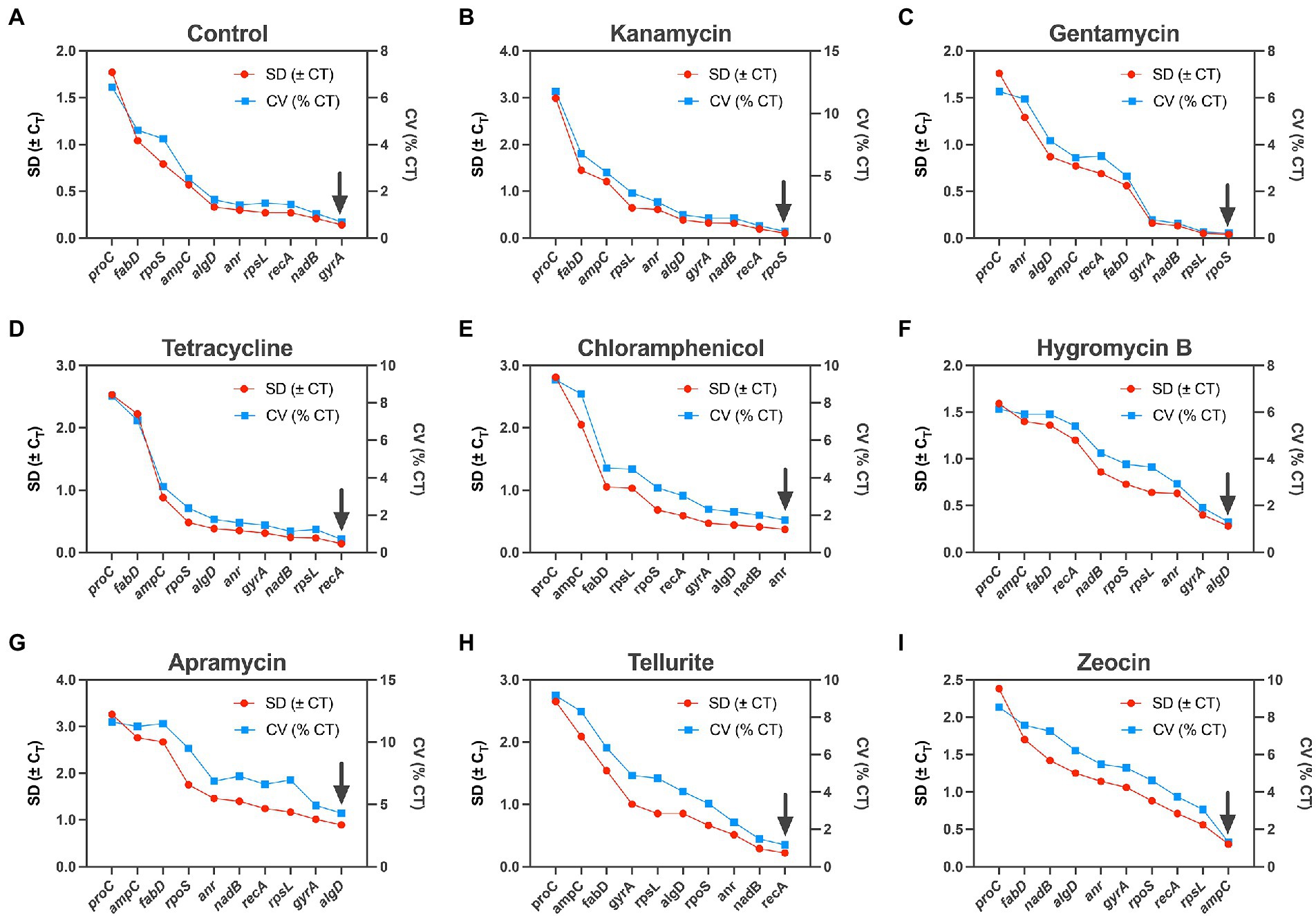
Figure 3. Expression analysis of 10 candidate housekeeping genes by BestKeeper algorithm. (A) Control; (B) kanamycin; (C) gentamycin; (D) tetracycline; (E) chloramphenicol; (F) hygromycin B; (G) apramycin; (H) tellurite; (I) zeocin.
Comprehensive ranking by RefFinder
RefFinder was used to generate a ranking order in light of the apparent ranking generated by geNorm, NormFinder, and BestKeeper (Xie et al., 2012). This algorithm assesses gene stability based on the above three algorithms plus ∆CT, and then generates a comprehensive ranking by ordering the geometric mean of multiple results. Similarly, the gene with the most stable expression level receives the lowest score. In general, as shown in Figure 4, nadB was the best housekeeping gene in the control group, gentamycin and tetracycline, algD performed best in kanamycin and chloramphenicol, recA got the lowest score in apramycin and tellurite group, and anr and gyrA were the best in hygromycin B and zeocin.
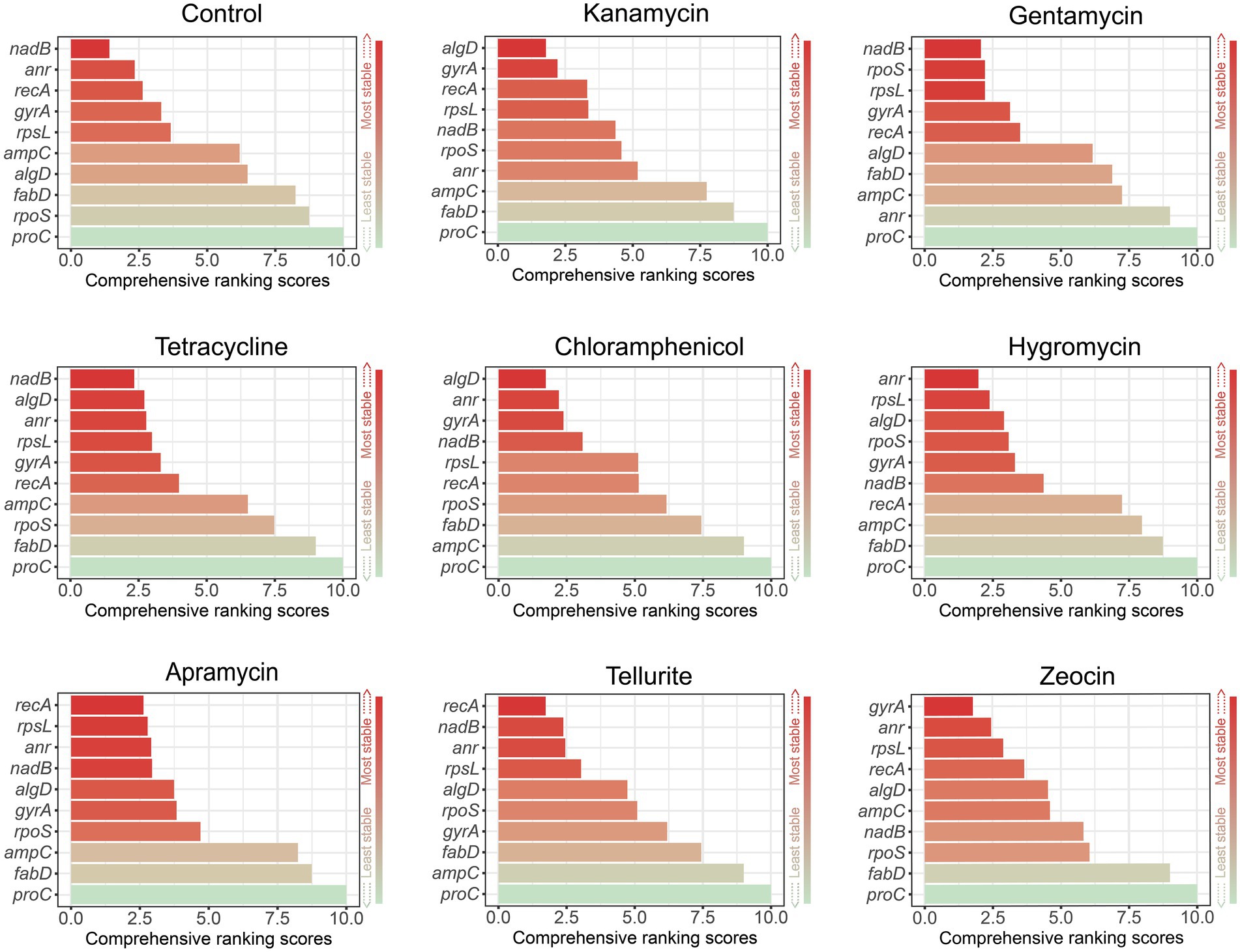
Figure 4. Comprehensive ranking by RefFinder according to the results of geNorm, NormFinder, and BestKeeper. Lower scores (red) indicate the higher expression stability.
Discussion
Over the past two decades, researchers have made great efforts to understand the natural characteristics of the human pathogen P. aeruginosa, and quantitative real-time PCR has proven indispensable in the study of this infamous, hard-to-eradicate pathogen. This technique has many advantages over other RNA quantification methods, such as high sensitivity, high reproducibility, and wide quantification range. At the same time, however, several variables must be carefully controlled to ensure the quality of the experimental results, such as the quality of the RNA sample extraction and reverse transcription process, a moderate amount of starting cDNA mass, an appropriate number of technical and biological replicates, and, of course, an appropriate selection of “housekeeping genes” as reference internal controls.
Ideally, this internal control gene should be highly conserved in the target species and held steady in ever-changing experimental conditions. However, it is not wise to attempt to chase a single all-around housekeeping gene as internal control, as no reference gene has been reported yet with verified stable expression under all circumstances, even for conserved genes involved in basic cellular processes throughout the life of bacteria. Several studies have shown that the expression levels of some commonly used housekeeping genes can fluctuate due to changes in the external environment, rendering them unsuitable as internal controls for experiments. Costaglioli et al. (2014) reported that the RNA expression levels of P. aeruginosa in 13 sputum specimens from nine cystic fibrosis patients showed that the commonly used housekeeping genes clpX and oprL were not as stable as PA2875 and PA3340 found by transcriptome sequencing, which are more suitable as housekeeping genes for clinical sample studies (Costaglioli et al., 2014). In another study, to evaluate the stability of P. aeruginosa housekeeping genes, Savli et al. (2003) collected 23 strains of P. aeruginosa with different resistance phenotypes and evaluated the stability of the six commonly used housekeeping genes and found that only proC and rpoD met the requirements for housekeeping genes, while ampC, fabD, pbp-2 and rpoS were not recommended due to their poor stability (Savli et al., 2003). In addition, a study on Pseudomonas brassicacearum G20 showed that growth stage and growth environment should also be considered when selecting housekeeping genes. Bai et al. (2020) selected nine candidate housekeeping genes based on previous studies and tested the RNA expression level of the tested strain at different growth phases, temperatures, and environmental iron levels (Bai et al., 2020). After RNA extraction, quantitative real-time PCR and data analysis using BestKeeper, NormFinder and GeNorm algorithms, it was found that among the nine candidate housekeeping genes, only rho, rpoD and gyrA were suitable reference genes for P. brassicacearum GS20 (Bai et al., 2020).
The above study illustrates that the instability of housekeeping genes due to external environmental influences should be fully considered at the beginning of the experimental design, but this does not seem to have received enough attention, as many studies still select housekeeping genes solely based on the references they refer to. Therefore, it is very important to select one or several of suitable housekeeping gene in the initial stage of research, and it is for this reason that algorithms such as geNorm, NormFinder, BestKeeper, and RefFinder can be utilized, which can score and rank the stability of multiple candidate genes through programs to help us select the most suitable gene as internal control.
In most molecular microbiology assays, e.g., protein expression, gene deletion, bacterial two-hybrid system, transposon mutagenesis, etc., it is a common practice to introduce antibiotic resistance cassette carried by plasmid into the wild-type strains to be used as screening markers for subsequent experiments, resulting in the tested strains actually being cultured under the pressure of antibiotic throughout the experimental process (Sanchez-Romero et al., 1998; Ducas-Mowchun et al., 2019). Furthermore, since different types of antibiotics have different antimicrobial mechanisms, it is reasonable to assume that their effects on housekeeping gene stability are also heterogeneous. Therefore, it is valuable to clarify this issue so that we can select the appropriate housekeeping gene according to the type of antibiotics in the early stage of the experiment.
In this study, we selected 10 candidate housekeeping genes for P. aeruginosa PAO1 based on previous studies and determined their stability under the pressure of 8 antibiotics, among which kanamycin, gentamycin, tetracycline, and chloramphenicol are regular antibiotics widely used in most laboratories, while hygromycin B, apramycin, tellurite, and zeocin have rarely been used in the study of P. aeruginosa. These last four unconventional antibiotics are mainly used for cell culture or other purposes, but in the age of bacterial multidrug resistance, they have been already increasingly being used as select markers in the molecular microbiology studies (Blondelet-Rouault et al., 1997; Sanchez-Romero et al., 1998; Ducas-Mowchun et al., 2019). In general, the experimental results showed that different algorithms ranked the candidate housekeeping genes in different order, and no single reference gene was found to be competent for housekeeping gene under all antibiotic pressures (Figures 2, 3; Table 3). To draw a clear conclusion, RefFinder software was used to normalize the results of several algorithms, which showed that in the molecular microbiology assays, algD should be the first choice of housekeeping gene when we use kanamycin or chloramphenicol as screening antibiotics, and recA when we use apramycin or tellurite. On the other hand, when gentamycin or tetracycline are used as select agents, nadB is the best internal control gene, and anr and gyrA should be the first choice for hygromycin B and zeocin repectively (Figure 4). Actually, there is no need for us to pay too much attention to the actual ranking order of the best housekeeping genes, because several recent studies have reached a consensus that in the quantitative real-time PCR experiment, the selection of multiple housekeeping genes as internal control instead of one is required. Therefore, our work can be used in the initial stage of the experiment to guide the selection of internal controls based on the type of antibiotic used, and two or more genes can be easily selected as internal controls according to the comprehensive ranking of housekeeping genes shown in Figure 4. It is worth noting that there may be other excellent housekeeping genes that have not yet been discovered. In future work, transcriptome sequencing can be used to screen for genes that are stably expressed under different conditions and make them “candidate housekeeping genes,” and further validate their stability using qRT-PCR. This study provides a comprehensive summary of the effects of commonly used laboratory antibiotics on the stability of housekeeping genes in P. aeruginosa to provide guidance and suggestions for housekeeping gene selection in future studies.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Author contributions
ChaL, HS, and XC designed the project and drafted the manuscript. LM and ChuL conducted the experiments. JL performed the statistical analysis. HX, JS, and MH helped with the writing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the National Natural Science Foundation of China (81902124 and 82002205).
Acknowledgments
We would like to thank LIZhen (Sichuan Academy of Botanical Engineering) for contributing the analysis tool: RefFinder (http://blooge.cn/RefFinder/), and Li Yang for his help in writing this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Alqarni, B., Colley, B., Klebensberger, J., McDougald, D., and Rice, S. A. (2016). Expression stability of 13 housekeeping genes during carbon starvation of Pseudomonas aeruginosa. J. Microbiol. Methods 127, 182–187. doi: 10.1016/j.mimet.2016.06.008
Bai, B., Ren, J., Bai, F., and Hao, L. (2020). Selection and validation of reference genes for gene expression studies in pseudomonas brassicacearum GS20 using real-time quantitative reverse transcription PCR. PLoS One 15:e0227927. doi: 10.1371/journal.pone.0227927
Blondelet-Rouault, M. H., Weiser, J., Lebrihi, A., Branny, P., and Pernodet, J. L. (1997). Antibiotic resistance gene cassettes derived from the omega interposon for use in E. coli and Streptomyces. Gene 190, 315–317. doi: 10.1016/S0378-1119(97)00014-0
Botelho, J., Grosso, F., and Peixe, L. (2019). Antibiotic resistance in Pseudomonas aeruginosa - mechanisms, epidemiology and evolution. Drug Resist. Updat. 44:100640. doi: 10.1016/j.drup.2019.07.002
Breidenstein, E. B., de la Fuente-Núñez, C., and Hancock, R. E. (2011). Pseudomonas aeruginosa: all roads lead to resistance. Trends Microbiol. 19, 419–426. doi: 10.1016/j.tim.2011.04.005
Bustin, S. A., and Mueller, R. (2005). Real-time reverse transcription PCR (qRT-PCR) and its potential use in clinical diagnosis. Clin. Sci. (Lond.) 109, 365–379. doi: 10.1042/CS20050086
Clinical and Laboratory Standards Institute. Methods for Dilution Antimicrobial Susceptibility Tests for Bacteria that Grow Aerobically—Tenth Edition: Approved Standard M07-A10. CLSI, Wayne, PA, USA, (2015).
Costaglioli, P., Barthe, C., Fayon, M., Christoflour, N., Bui, S., Derlich, L., et al. (2014). Selection of Pseudomonas aeruginosa reference genes for RT-qPCR analysis from sputum of cystic fibrosis patients. Mol. Cell. Probes 28, 10–12. doi: 10.1016/j.mcp.2013.09.003
Ducas-Mowchun, K., De Silva, P. M., Crisostomo, L., Fernando, D. M., Chao, T. C., Pelka, P., et al. (2019). Next generation of Tn7-based single-copy insertion elements for use in multi-and pan-drug-resistant strains of Acinetobacter baumannii. Appl. Environ. Microbiol. 85:e00066–19. doi: 10.1128/AEM.00066-19
England, C. G., Ehlerding, E. B., and Cai, W. (2016). Nano Luc: a small luciferase is brightening up the field of bioluminescence. Bioconjug. Chem. 27, 1175–1187. doi: 10.1021/acs.bioconjchem.6b00112
Kozera, B., and Rapacz, M. (2013). Reference genes in real-time PCR. J. Appl. Genet. 54, 391–406. doi: 10.1007/s13353-013-0173-x
Lee, J., and Zhang, L. (2015). The hierarchy quorum sensing network in Pseudomonas aeruginosa. Protein Cell 6, 26–41. doi: 10.1007/s13238-014-0100-x
Malhotra, S., Hayes, D. Jr., and Wozniak, D. J. (2019). Cystic fibrosis and Pseudomonas aeruginosa: the host-microbe interface. Clin. Microbiol. Rev. 32:e00138-18. doi: 10.1128/CMR.00138-18
Moradali, M. F., Ghods, S., and Rehm, B. H. (2017). Pseudomonas aeruginosa lifestyle: a paradigm for adaptation, survival, and persistence. Front. Cell. Infect. Microbiol. 7:39. doi: 10.3389/fcimb.2017.00039
Parkinson, J., and Blaxter, M. (2009). Expressed sequence tags: an overview. Methods Mol. Biol. 533, 1–12. doi: 10.1007/978-1-60327-136-3_1
Pena, R. T., Blasco, L., Ambroa, A., González-Pedrajo, B., Fernández-García, L., López, M., et al. (2019). Relationship between quorum sensing and secretion systems. Front. Microbiol. 10:1100. doi: 10.3389/fmicb.2019.01100
Pfaffl, M. W., Tichopad, A., Prgomet, C., and Neuvians, T. P. (2004). Determination of stable housekeeping genes, differentially regulated target genes and sample integrity: best keeper--excel-based tool using pair-wise correlations. Biotechnol. Lett. 26, 509–515. doi: 10.1023/B:BILE.0000019559.84305.47
Sanchez-Romero, J. M., Diaz-Orejas, R., and De Lorenzo, V. (1998). Resistance to tellurite as a selection marker for genetic manipulations of pseudomonas strains. Appl. Environ. Microbiol. 64, 4040–4046. doi: 10.1128/AEM.64.10.4040-4046.1998
Savli, H., Karadenizli, A., Kolayli, F., Gundes, S., Ozbek, U., and Vahaboglu, H. (2003). Expression stability of six housekeeping genes: a proposal for resistance gene quantification studies of Pseudomonas aeruginosa by real-time quantitative RT-PCR. J. Med. Microbiol. 52, 403–408. doi: 10.1099/jmm.0.05132-0
Thi, M. T. T., Wibowo, D., and Rehm, B. H. A. (2020). Pseudomonas aeruginosa biofilms. Int. J. Mol. Sci. 21:8671. doi: 10.3390/ijms21228671
Keywords: housekeeping gene, Pseudomonas aeruginosa, qRT-PCR, BestKeeper, geNorm, NormFinder, RefFinder
Citation: Meng L, Cao X, Li C, Li J, Xie H, Shi J, Han M, Shen H and Liu C (2023) Housekeeping gene stability in Pesudomonas aeruginosa PAO1 under the pressure of commonly used antibiotics in molecular microbiology assays. Front. Microbiol. 14:1140515. doi: 10.3389/fmicb.2023.1140515
Edited by:
Abdelali Daddaoua, University of Granada, SpainReviewed by:
Riti Mann, University of Technology Sydney, AustraliaAhmed S. Khairalla, University of Regina, Canada
Copyright © 2023 Meng, Cao, Li, Li, Xie, Shi, Han, Shen and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chang Liu, bGl1Y2hhbmdiaW9AMTYzLmNvbQ==; Han Shen, c2hlbmhhbjEwMzY2QHNpbmEuY29t
†These authors have contributed equally to this work
 Lingning Meng1†
Lingning Meng1† Xiaoli Cao
Xiaoli Cao Jiping Shi
Jiping Shi Mei Han
Mei Han Han Shen
Han Shen Chang Liu
Chang Liu