- 1‘Iolani School, Honolulu, HI, United States
- 2Center for Genes, Environment and Health, National Jewish Health, Denver, CO, United States
- 3Rock Canyon High School, Highlands Ranch, CO, United States
- 4Department of Cellular and Molecular Biology, School of Medicine, The University of Texas Health Science Center at Tyler, Tyler, TX, United States
As environmental opportunistic pathogens, nontuberculous mycobacteria (NTM) can cause severe and difficult to treat pulmonary disease. In the United States, Hawai’i has the highest prevalence of infection. Rapid growing mycobacteria (RGM) such as Mycobacterium abscessus and M. porcinum and the slow growing mycobacteria (SGM) including M. intracellulare subspecies chimaera are common environmental NTM species and subspecies in Hawai’i. Although iron acquisition is an essential process of many microorganisms, iron acquisition via siderophores among the NTM is not well-characterized. In this study, we apply genomic and microbiological methodologies to better understand iron acquisition via siderophores for environmental and respiratory isolates of M. abscessus, M. porcinum, and M. intracellulare subspecies chimaera from Hawai’i. Siderophore synthesis and transport genes, including mycobactin (mbt), mmpL/S, and esx-3 were compared among 47 reference isolates, 29 respiratory isolates, and 23 environmental Hawai’i isolates. Among all reference isolates examined, respiratory isolates showed significantly more siderophore pertinent genes compared to environmental isolates. Among the Hawai’i isolates, RGM M. abscessus and M. porcinum had significantly less esx-3 and mbt genes compared to SGM M. chimaera when stratified by growth classification. However, no significant differences were observed between the species when grown on low iron culture agar or siderophore production by the chrome azurol S (CAS) assay in vitro. These results indicate the complex mechanisms involved in iron sequestration and siderophore activity among diverse NTM species.
1. Introduction
Nontuberculous mycobacteria (NTM) are found in a variety of environmental habitats and biofilms including those collected from showerheads, water faucets, air conditioning units, hospital water faucets, and soil (Honda et al., 2018). There are more than 195 different species of rapid growing mycobacteria (RGM) and slow growing mycobacteria (SGM) that have been identified to date (Parte et al., 2020), yet only a handful of these species are considered opportunistic pathogens that cause pulmonary disease (PD) particularly in susceptible individuals (Falkinham, 1996). NTM PD is becoming a mounting public health concern as the number of cases continue to increase globally (Adjemian et al., 2012).
In the United States (U.S.), Hawai’i had the highest period prevalence of NTM PD calculated at 396 cases/100,000 persons according to a Medicare Part B beneficiary analysis (Adjemian et al., 2012). In our prior research, we demonstrated frequent recovery of viable RGM including Mycobacterium abscessus and Mycobacterium porcinum from the Hawai’i environment. We also reported the preponderance of the SGM, Mycobacterium intracellulare subspecies chimaera, among both respiratory and environmental samples from this geographic NTM hot spot (Honda et al., 2016).
Iron is an essential element for microbial growth, used in various biological processes including electron transport, and iron plays a major role in metabolic and cellular pathways including energy generation, DNA replication, transcriptional regulation (Crosa, 1989; Skaar, 2010). In the human body, iron mainly exists in complex forms bound to protein (e.g., hemoprotein) as heme compounds (e.g., hemoglobin or myoglobin), heme enzymes, or nonheme compounds (e.g., flavin-iron enzymes, transferring, and ferritin) (McDowell, 1992; Abbaspour et al., 2014). Minute amounts, i.e., 10−18 M concentration of free iron are available or 0.4–0.9 mg Fe/g dry weight in blood or human lungs, respectively (Griffiths et al., 1980; Takemoto et al., 1991). Under this competitive iron-restricted environment, bacteria that have the capacity to sequester and use iron may be selectively advantaged. In contrast, iron is more readily available and in higher quantity in the exogenous environment. Hawai’i soil, for example, contains 95,550 mg/kg of iron on average but iron content varies with rock type as basaltic and mafic rocks range from 56,000–87,000 mg/kg (Hawai’i Department of Health Hazard Evaluation and Emergency Response, 2012).
To acquire exogenous iron, many bacteria exploit siderophores. For mycobacteria, intracellular and extracellular iron-chelating siderophores synergistically scavenge iron from the environment (Gobin et al., 1995) and are critical to pathogenicity (Braun and Winkelmann, 1988; De Voss et al., 2000). Mycobactin (mbt) is an intracellular siderophore produced by most mycobacteria, especially pathogenic species, under iron-limiting conditions and is restricted to the cell envelope (De Voss et al., 2000). Based on models for Mycobacterium tuberculosis (Mtb), two gene complexes are needed to synthesize mycobactin: mbt-1 and mbt-2. Mbt-1 is comprised of ten genes, ranging from mbtA to mbtJ. mbt-2 is comprised of four genes ranging from mbtK to mbtN (Sritharan, 2016). Exochelin is an extracellular peptide siderophore produced under iron-deficient conditions and exclusive to RGM. The mycobacterial membrane large/small proteins (mmpL/S) complex and the type VII secretion system, esx-3, are used to scavenge iron from the surroundings, facilitating acquisition of iron from mycobactin (Siegrist et al., 2009; Szekely and Cole, 2016). esx-3 involves a homologous gene system relating to protein production for iron acquisition. However, beyond confirmation of their existence, NTM siderophore biology is significantly understudied and most research in this area predates the current genomic era (Hall and Ratledge, 1982 1984 1985; Barclay et al., 1985).
The aim of the current study was to provide an updated assessment of mycobacterial iron acquisition via siderophores. This was achieved by leveraging genomic data from respiratory and environmental Hawai’i NTM isolates (Honda et al., 2016; Virdi et al., 2021) as well as NTM genomic data from the NCBI GenBank public database. Mycobacterial iron-acquisition gene comparisons among the RGM and SGM and between respiratory and environmental isolates from Hawai’i were assessed. To further confirm siderophore production, in vitro microbiological tests were performed using the same respiratory and environmental isolates from Hawai’i under low iron culture conditions.
2. Materials and methods
2.1. NTM isolates used in this study
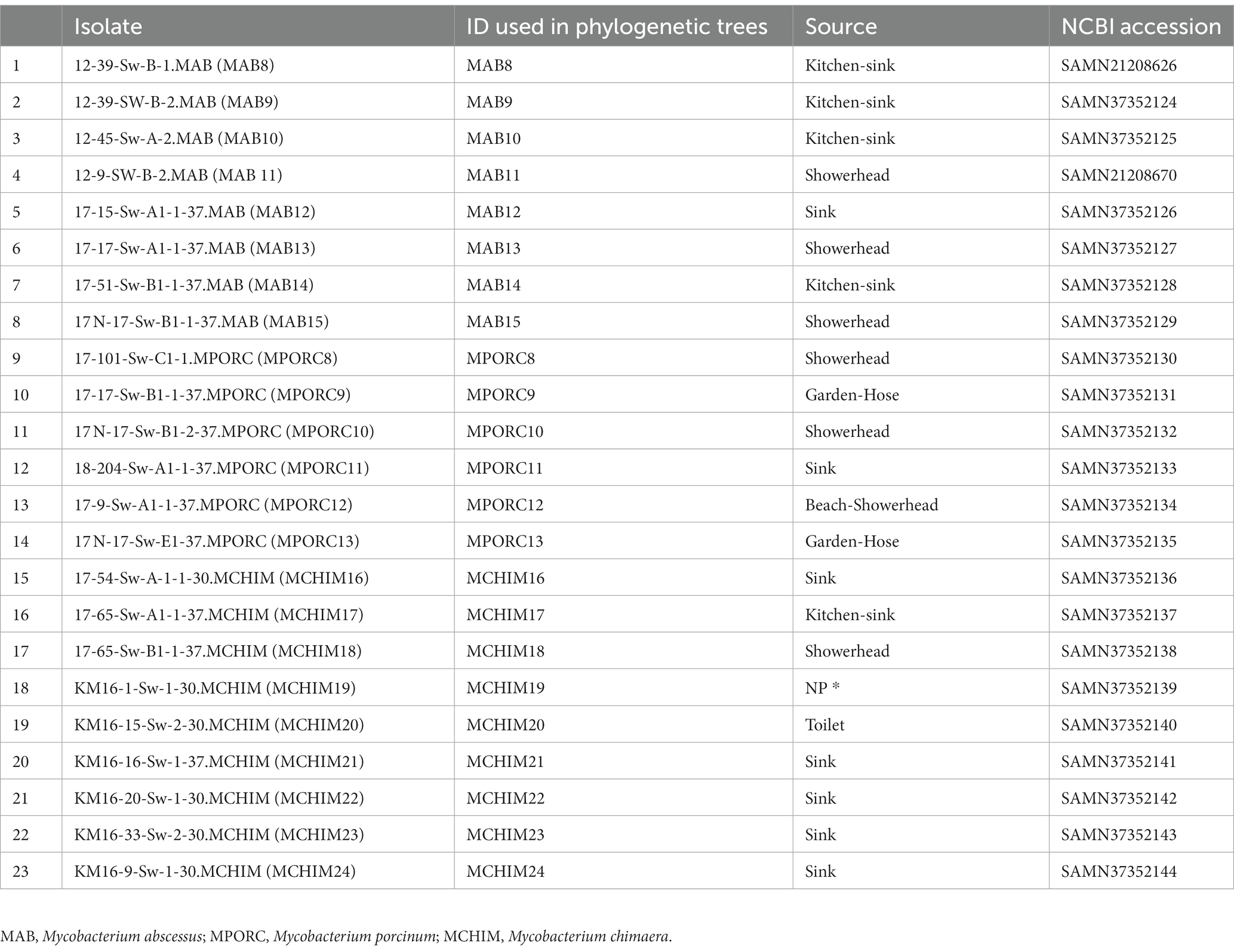
We collected 51 Hawai’i NTM isolates, including 28 respiratory and 23 environmental NTM isolates (Tables 1, 2) for analysis. Respiratory isolates were from de-identified individuals living in Hawai’i with suspected mycobacterial infections whose sputum had been submitted for mycobacterial culture and processed by the Diagnostic Laboratory Services, Inc (Aiea, HI). Environmental NTM isolates were collected by our group from indoor household water biofilms including household shower heads and sink faucets. A minority of water biofilms were collected from outdoor sources in Hawai’i including beach showerheads and garden hoses (Honda et al., 2016; Virdi et al., 2021). A description of isolate collection processes and DNA extraction methods are published (Honda et al., 2016; Virdi et al., 2021). All environmental isolates tested in this study were recovered from O’ahu, Hawai’i. Respiratory and environmental isolates were stratified as RGM or SGM. For the Hawai’i isolate panel tested, M. abscessus and M. porcinum were included as representative RGM. M. intracellulare subspecies chimaera (referred herein as M. chimaera) was used as a representative SGM. Ethical review and approval were waived because the clinical isolates used in this study were de-identified patient residual isolates and not considered human subject research.
2.2. Whole-genome sequencing
DNA isolation for Illumina whole-genome sequencing (WGS) and genome assembly were performed as previously described (Epperson and Strong, 2020; Hasan et al., 2021). To contextualize the Hawai’i isolate data, DNA sequences for 47 additional non-Hawai’i respiratory and environmental NTM were downloaded from the publicly available National Center for Biotechnology Information (NCBI) GenBank database and are referred in this study as “NTM reference” isolates (Supplementary Tables 1A,B). Reference isolates used were M. abscessus subsp. abscessus (n = 1), M. abscessus subsp. massiliense (n = 2), M. abscessus subsp. bolletii (n = 1), M. avium (n = 5), M. intracellulare (n = 4), M. paraintracellulare (n = 1), M. chimaera (n = 1), M. marseillense (n = 1), M. sinensis (n = 1), M. terrae (n = 1), and M. kansasii (n = 1). There was no reference available for M. porcinum.
2.3. Siderophore gene analyses
The Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Kanehisa et al., 2019) was used to interrogate NTM siderophore genes. Genes related to iron acquisition were identified by utilizing the KEGG pathway database to generate a list of each gene in the mbt, mmpL/S, and esx-3 gene pathways for each NTM species. All gene sequences with “mbt” in the gene name were acquired through the KEGG database.
Control metrics were performed to ensure the quality of the genome assemblies for each species: genome length (Supplementary Figure 1A), number of contigs (Supplementary Figure 1B), and the number of annotated genes (Supplementary Figure 1C). To assess the presence or absence of mycobactin (mbt), mmpL/S, and esx-3 genes, WGS were annotated using Prokka (Seemann, 2014) and compiled into a pan-genome using Roary (Page et al., 2015). The genomes of respiratory and environmental Hawai’i NTM were parsed for mbt, mmpL, and esx-3 genes, and the sequences of each gene for each isolate were aligned in a multiple sequence alignment using Geneious Prime (Version 2020.1).1 For each gene, phylogenetic trees were created to investigate genetic variation between species and between respiratory and environmental isolates within each species as well.
2.4. In vitro siderophore screening
Siderophore assays were adapted from prior work by Hall and Ratledge (1982) and Louden et al. (2011). The NTM Hawai’i panel (Tables 1, 2) was streaked for isolation onto Middlebrook 7H10 OADC (oleic acid albumin dextrose catalase) plates and incubated at 37°C for 7 days for RGM and up to 21 days for M. chimaera to generate stock cultures. From stock cultures, isolates were picked and streaked for isolation onto low iron culture media (glycerol, L-asparagine, potassium phosphate monobasic, zinc chloride, manganese (II) chloride, magnesium sulfate heptahydrate) with agar. Low iron plates were incubated at 37°C for 9–16 days for RGM and up to 28 days for M. chimaera. Growth on low iron agar plates was indicative of viable bacteria and siderophore production. No growth on low iron plates was recorded as no siderophore production. In parallel, low iron culture broth was inoculated with the same panel of NTM isolates and incubated for 1 month. The chrome azurol S (CAS) assay was used to detect siderophore activity and adapted from Louden et al. (2011). 1 mL of low iron NTM cultures were dispensed into wells of a 24 well plate with 100 μL of CAS reagent per the Louden protocol. Samples were incubated at room temperature for 15 min. Siderophore activity was noted by an eye-visible color change from blue to yellow/orange 24 h post inoculation. The results of the CAS test and the low iron media plates were used to conclude which isolates produced siderophores in low iron environments. Mycobacterium neoaurum and Mycobacterium fortuitum were, respectively, used as positive and negative controls for the CAS assay.
2.5. Statistical analyses
Statistical tests were done in R Studio and Prism. The One Sample Student’s t-test was used to compare the group with a higher or lesser number of genes between respiratory and environmental groups in the KEGG dataset. The Mann–Whitney-Wilcoxon test was used to determine whether the number of genes of respiratory and environmental groups for the Hawai’i data set were identical. The Kruskal-Wallis test was used to compare the number of genes in the comparison of respiratory/environmental and RGM/SGM from KEGG and to compare the number of genes between M. abscessus, M. chimaera, and M. porcinum from Hawai’i. The Games-Howell nonparametric post-hoc test was used to further compare the number of genes in the comparison of respiratory/environmental and RGM/SGM from different pairings of each group from KEGG. The Fisher’s exact test was used to compare siderophore growth presence in RGM/SGM and respiratory/environmental samples for each in vitro analysis due to the small sample size.
3. Results
3.1. mbt genes are more abundant among respiratory NTM isolates compared to environmental NTM isolates
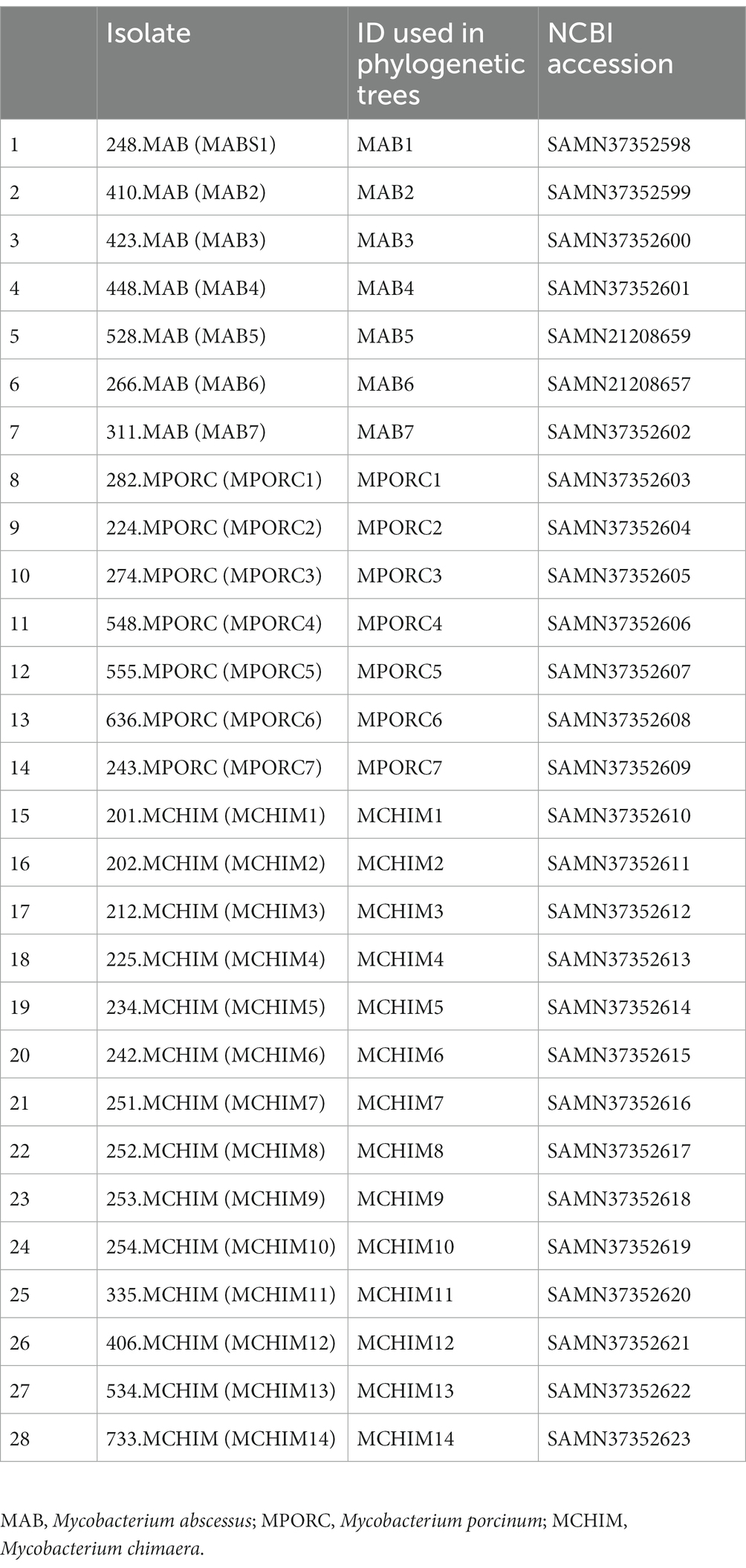
The mbt genes available through KEGG were identified from 47 NTM isolates including 20 respiratory NTM, 27 environmental NTM, and two species from the Mtb complex for comparison (Mtb H37Rv and M. canetti). Overall, respiratory NTM isolates showed significantly more mbt genes (n = 10.7 genes) compared to environmental NTM (n = 8.3 genes) (p = 0.0002) (Figure 1A). When the respiratory and environmental isolates were stratified by RGM and SGM, the mean number of mbt genes was statistically different between respiratory/RGM and environmental/RGM group (p < 0.05), the respiratory/SGM and environmental/RGM group (p < 0.005), and the environmental/RGM and environmental/SGM group (p < 0.005) (Figure 1B).
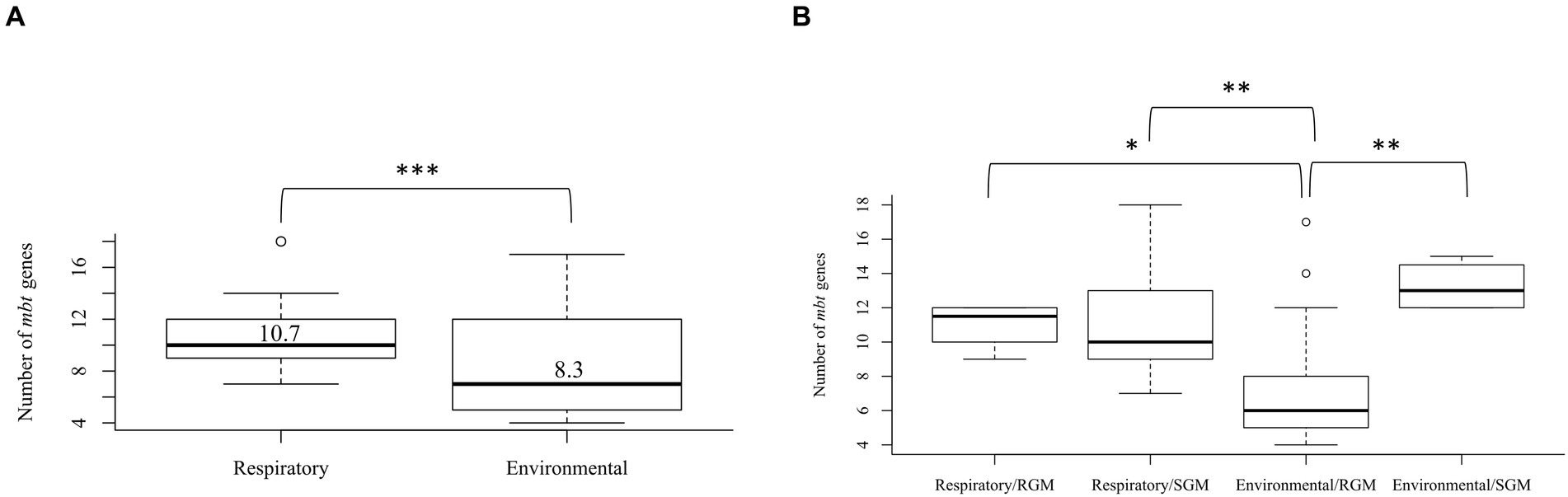
Figure 1. Varying number of mycobactin (mbt) genes between respiratory and environmental reference NTM isolates (Supplementary Tables 1A, B). Box plot showing (A) the number of mbt genes stratified by respiratory vs. environmental derived NTM; ***p = 0.0001989. (B) Number of mbt genes for respiratory vs. environmental NTM stratified by rapid growing mycobacteria (RGM) and slow growing mycobacteria (SGM). The central rectangle of the plot spans the interquartile range (IQR). The bar inside the rectangle represents the median, and the bars above and below show the location of the maximum and minimum, respectively. The small circle represents outliers. *p < 0.05; **p < 0.005.
3.2. Mycobacterium chimaera shows more mbt genes compared to M. abscessus or M. porcinum
To study the differences between the number of mbt genes among NTM species that can cause respiratory disease, 51 isolates were compared including seven respiratory and eight environmental isolates of M. abscessus, seven respiratory and six environmental isolates of M. porcinum, and 14 respiratory and nine environmental isolates of M. chimaera. Comparing both respiratory and environmental isolates, gene duplications were observed for mbtC, mbtD, mbtE, and mbtF, generally located outside of the mbt locus (average: 888,213 bp; range: [1,328:2,671,854 bp]), and varied by gene and species. Among the RGM M. abscessus and M. porcinum isolates, only one mbtD gene was identified. M. abscessus showed an average number of 14 mbt genes while M. porcinum showed an average of 17 genes, which was significantly less than the 27 mbt genes for M. chimaera (p = 0.005) (Figure 2A). Of note, the mbt-2 gene complex, mbtL, mbtM, and mbtN genes were absent from M. abscessus and mbtI and mbtL genes were missing from M. porcinum. All ten mbt-1 and four mbt-2 genes were detected in M. chimaera accounting for its significantly higher average number of mbt genes.
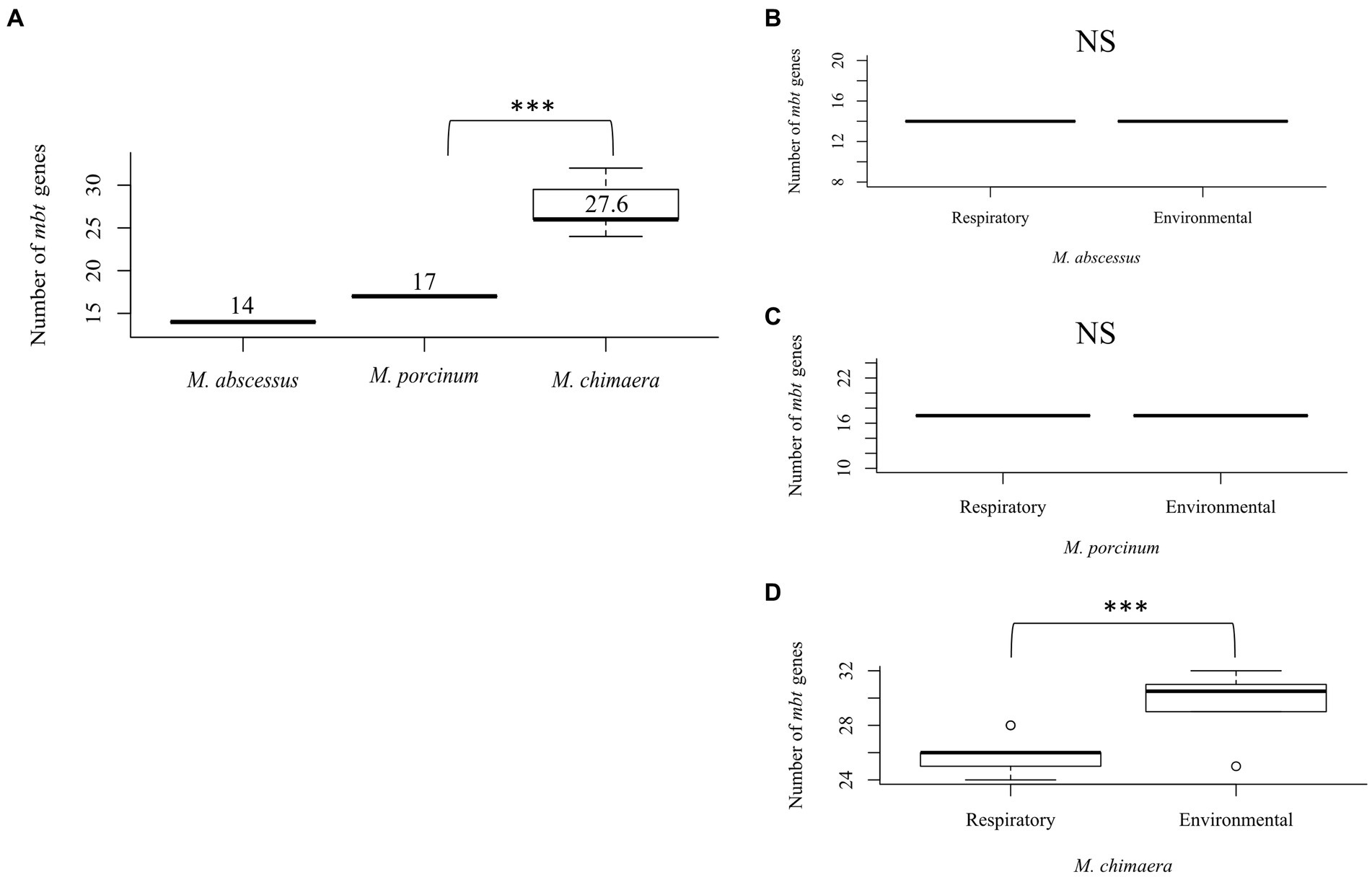
Figure 2. Mycobacterium chimaera shows more mbt genes compared to M. abscessus and M. porcinum (A) Distribution of the number of mbt genes among all M. abscessus, M. chimaera, and M. porcinum isolates from Hawai’i. ***p = 0.0005097. Number of mbt genes between respiratory or environmental (B) M. abscessus, (C) M. porcinum, (D) M. chimaera. ***p = 0.0005097. NS, not significant. Isolates studied in Figure 2 are listed in Tables 1 2.
Similar numbers of mbt genes were observed between respiratory and environmental M. abscessus (Figure 2B) and respiratory and environmental M. porcinum (Figure 2C) isolates. However, respiratory M. chimaera isolates showed significantly lower numbers of mbt genes compared to environmental M. chimaera isolates (p = 0.005) (Figure 2D).
We constructed a phylogenetic tree to examine the genetic relatedness of mbtA genes among M. abscessus, M. porcinum, and M. chimaera. Each isolate contained one copy of the mbtA gene. M. abscessus mbtA is genetically similar to M. porcinum mbtA genes, as revealed by the close proximity in location on the phylogenetic tree (Figure 3). All sequences from respiratory and environmental isolates of M. porcinum were identical. This was also observed for M. abscessus, except for one respiratory and two environmental isolates which shared a single nucleotide polymorphism (SNP). For M. chimaera, two environmental isolates showed the same 13 SNPs while the rest of the samples were identical in sequence. Similar results can be seen for the mbt synthase G, mbtG gene (Supplementary Figure 2), where respiratory and environmental isolates within M. abscessus and M. porcinum and had identical gene sequences compared to two sub-branches for the M. chimaera cluster.

Figure 3. mbtA is genetically similar among the NTM. Neighbor-joining phylogenetic tree based on the mbtA gene. Scale bar indicates number of substitutions per site.
For the mycobactin synthases mbtM gene, two copies of the gene were identified in M. porcinum and M. chimaera. All M. porcinum isolates had identical sequences for both mbtM copies. For one copy of mbtM in M. chimaera, the two environmental isolates sharing the mbtA variant also shared an mbtM copy variant with 18 SNPs. The other mbtM copy had the same relationships as within the mbtG gene of M. chimaera. This pattern of relationships was also observed in the mbtM and mbtN genes in M. porcinum and M. chimaera; however, these genes are single-copy in all isolates (Supplementary Figures 3A,B).
3.3. mmpl/S and esx-3 secretion system associated genes in NTM from Hawai’i
Because of their roles in siderophore and iron transport, we also investigated the number of mmpL/S and esx-3 secretion system associated genes in the same sample of Hawai’i NTM isolates. There were 13 mmpL proteins based on a model of Mtb, but we focused on mmpL/S3, mmpL/S4, and mmpL/S5 for their roles in either siderophore or heme export (Szekely and Cole, 2016). While the number of mmpL345 and mmpS345 genes tended to follow the trend of M. abscessus > M. porcinum > M. chimaera, no significant differences were observed (Supplementary Figures 4A,B). When the number of mmpL genes were compared between respiratory or environmental isolates among the three different species, no significant differences were found (Supplementary Figures 5A–C).
The esx-3 secretion system is required for mycobacterial iron acquisition through mbt (Siegrist et al., 2009). The esx-3 locus contains the genes for the esxG and esxH proteins, and supporting genes identified as eccA3, eccB3, eccC3, eccD3, eccE3, espG3, and mycP3 that are required for protein export and iron acquisition (Siegrist et al., 2014). While esx-3 gene numbers did not vary among the RGM isolates (Figures 4A,B), environmental M. chimaera isolates showed significantly more numbers of esx-3 genes compared to respiratory M. chimaera isolates (p = 0.013) which appear to be due to gene duplications of the mycP3 gene (Figure 4C). Among Hawai’i M. chimaera, we found 1–3 copies of espG3 or mycP3 per genome. The duplicate copies of espG3 and mycP3 were inconsistently located throughout the genome. The average distance between espG3 copies was 1,979,955 nt (range: 560,978–2,891,545 nt) and the average distance between mycP3 copies was 2,636,478 nt (range: 479,373 – 4,820,547 nt).
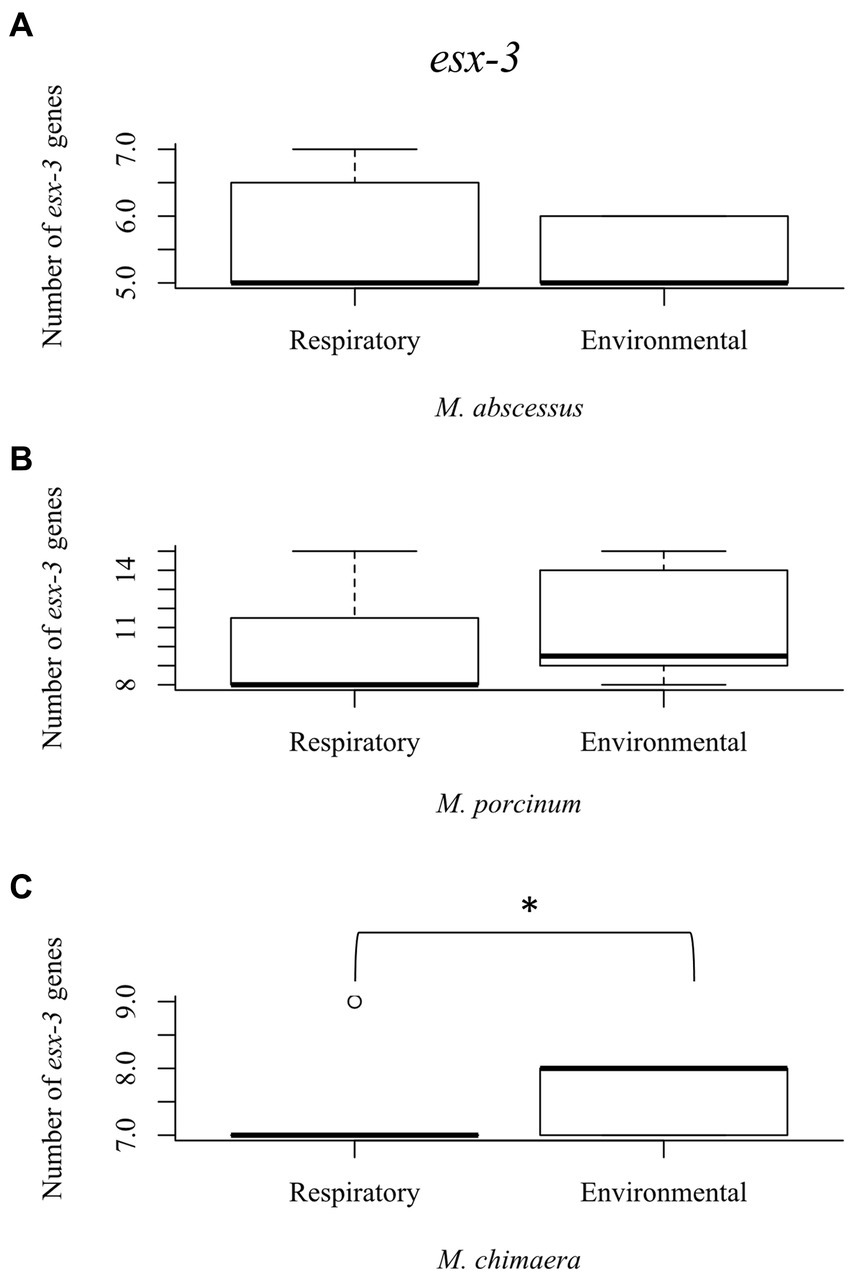
Figure 4. Mycobacterium chimaera shows significantly more esx-3 genes compared to rapid growing mycobacteria. Box plot showing the number of esx-3 genes in respiratory and environmental (A) M. abscessus; (B) M. porcinum; and (C) M. chimaera. *p = 0.01346.
3.4. In vitro siderophore production is higher in RGM than in SGM
Next, two in vitro methodologies, including growth on low iron media plates and the CAS detection assay, were used to screen the isolates for siderophore production in low iron environments and results are summarized in Supplementary Tables 3, 4. Of the environmental M. abscessus, M. porcinum, and M. chimaera isolates tested, 88% (n = 7/8), 100% (n = 6/6), and 11% (n = 1/9), respectively showed visible colonies indicative of growth on low iron plates. Of the respiratory M. abscessus, M. porcinum, and M. chimaera isolates tested, 100% (n = 7/7), 86% (n = 6/7), and 29% (n = 4/14), respectively showed visible colonies indicative of growth on low iron plates.
No differences were observed between respiratory and environmental NTM growth on low iron plates (p > 0.9999) (data not shown). When these data were stratified by RGM (M. abscessus and M. porcinum) and SGM (M. chimaera) categories, growth on low iron agar was more frequently observed for the RGM compared to the SGM (p < 0.0001) (Figure 5A, left panel). This trend was also true when the isolates were stratified as respiratory RGM and SGM isolates (p = 0.018) (Figure 5A, middle panel) and environmental RGM and SGM isolates (p = 0.0002) (Figure 5A, right panel). No statistical differences were observed between the number of environmental and respiratory isolates that grew or did not grow on low iron agar.
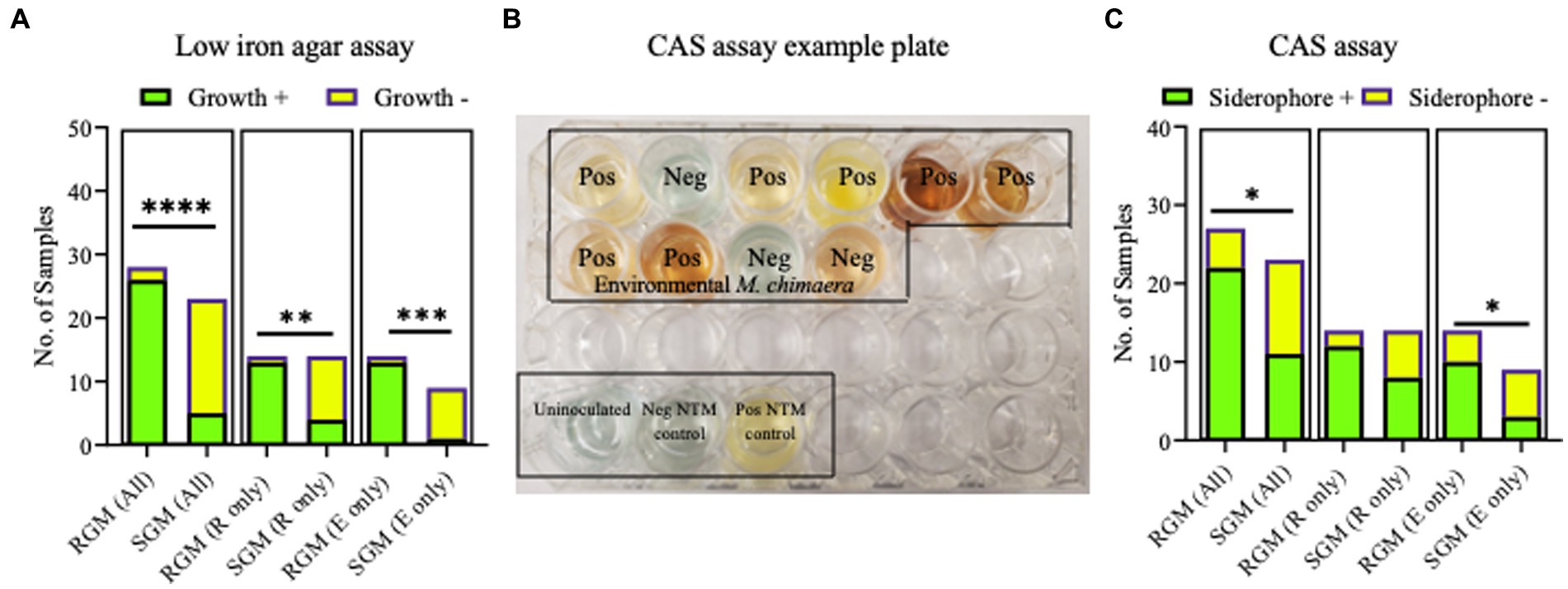
Figure 5. Growth on low iron agar and positive CAS assays are more often observed for rapid growing mycobacteria (RGM) compared to slow growing mycobacteria (SGM). (A) Stacked bar graphs showing +/− growth for RGM and SGM on low iron agar plates. R, respiratory derived isolates. E, environment derived isolates. Low iron RGM vs SGM ****p = <0.0001, ***p = 0.0002, *p = 0.0175. (B) To demonstrate CAS assay outcomes, we show an example of positive and negative media color change results for 10 environmental M. chimaera isolates, black box. M. neoaurum and M. fortuitum were used as positive and negative control isolates, respectively along with an uninoculated well (bottom of image, black box). (C) Outcomes for all CAS assays shown as stacked bar graphs. R, respiratory derived isolates. E, environment-derived isolates. *p < 0.05.
The universal siderophore assay, CAS, was used as a second assay to assess siderophore production by NTM isolates. When a strong iron chelator such as a siderophore removes iron from the media a color change from green/blue to yellow/orange occurs. In Figure 5B, we show an example of CAS assay outcomes using 10 environmental M. chimaera isolates. Eight isolates (80%) showed a yellow/orange color change (positive) and two (20%) showed a green/blue color indicative of a negative test. M. neoaurum and M. fortuitum were used as positive and negative controls, respectively along with an uninoculated well. Finally, we tested the collection of environmental and respiratory NTM by the CAS assay. Among environmental M. abscessus, M. porcinum, and M. chimaera isolates, 88% (n = 7/8), 50% (n = 3/6), and 30% (n = 3/10), respectively showed positive color change for CAS. Of the respiratory M. abscessus, M. porcinum, and M. chimaera isolates tested, 100% (n = 7/7), 71% (n = 5/7), and 57% (n = 8/14), respectively showed positive color change for CAS (Supplementary Tables 3, 4). We did not observe any significant difference in CAS outcomes between environmental and respiratory isolate groups or when the isolates were sub-categorized by species. We observed more CAS positive tests in RGM versus SGM (p < 0.05) (Figure 5C, left panel) and among environmental isolates (e.g., environmental RGM vs. environmental SGM) (p < 0.05) (Figure 5C, middle panel), but not when comparing within respiratory isolates (e.g., respiratory RGM vs. respiratory SGM) (Figure 5C, right panel).
Because all isolates studied were tested for siderophore activity via growth on low iron plates and by the CAS assay, we determined the frequency at which the outcomes of the two assays matched or mismatched. We highlight in red font the between assay matches per isolate tested in Supplementary Tables 1A,B. Overall, among all 28 RGM isolates tested, 22 (79%) showed low iron assay outcomes matched CAS assay outcomes compared to 21% (6/28) mismatches (Supplementary Figure 6). When analyzed separately, assay concurrency was more often observed for respiratory isolates compared to environmental RGM isolates (93% vs. 64%) (Supplementary Figure 7). Taking an additional step, we separated the outcomes for M. abscessus and M. porcinum. Generally, low iron assay results aligned with CAS assay outcomes for respiratory M. abscessus (7/7) and M. porcinum (6/7) isolates, agreeing 100 and 86%, respectively, of the time. Among the environmental M. abscessus and M. porcinum isolates tested, the two assay outcomes agreed 75% (6/8) and 50% (3/6), respectively.
Similarly, for the SGM, 70% (16/23) M. chimaera isolates studied showed low iron assay outcomes matched CAS assay outcomes compared to 30% (7/23) mismatches (Supplementary Figure 6). By stratifying all M. chimaera isolates into respiratory and environmental categories, concurrency between the two tests was more often observed for environmental M. chimaera isolates compared to respiratory M. chimaera isolates (89% vs. 57%) (Supplementary Figure 7).
The combined outcomes from genomic analyses and in vitro assays for the three NTM species and subspecies tested in this study are summarized in a cartoon, Figure 6.
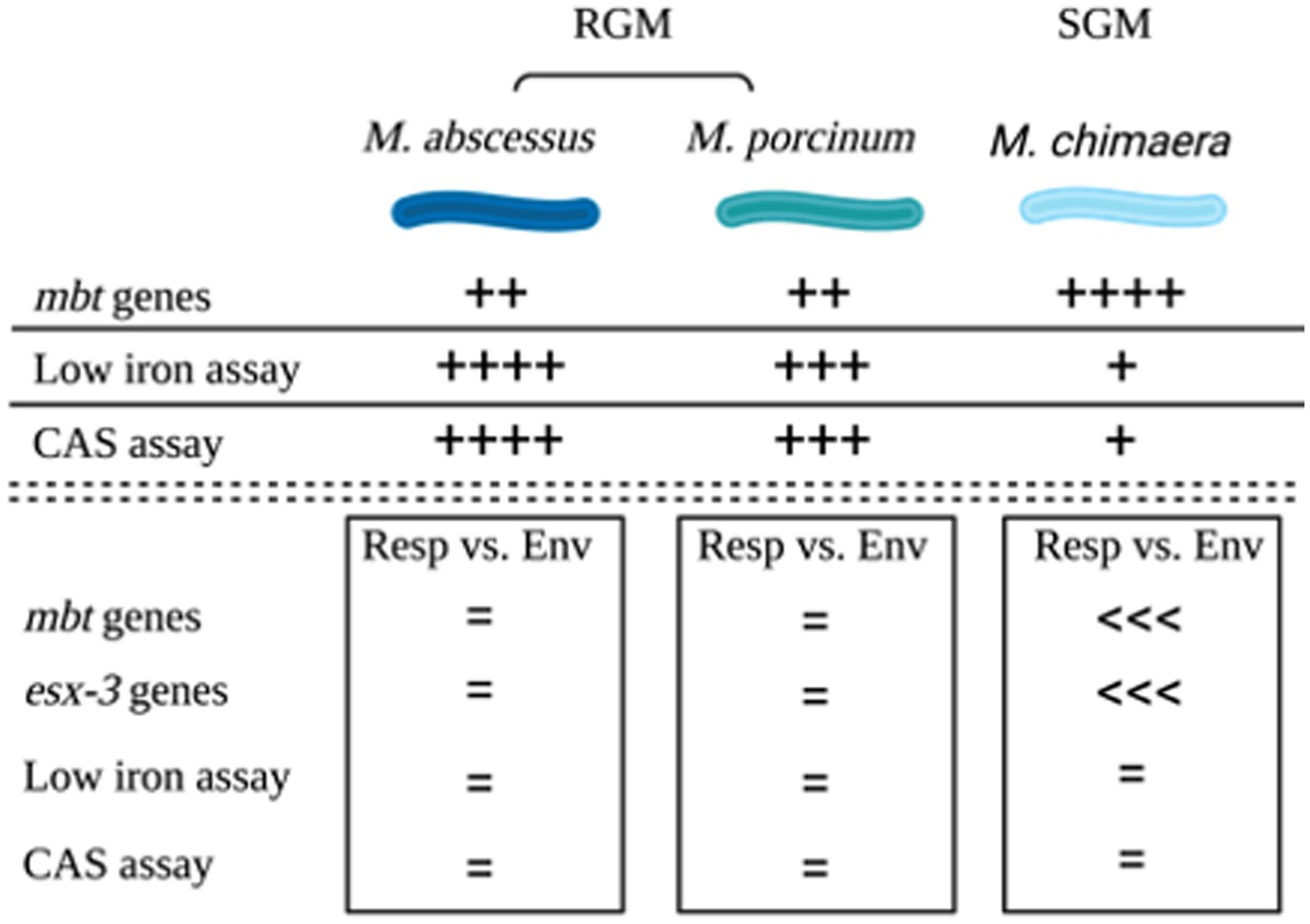
Figure 6. Summary. Cartoon summarizing the outcomes of genomic comparisons and applied microbiological assays to assess siderophores among M. abscessus, M. porcinum, and M. chimaera respiratory and environmental isolates. Image created using BioRender.
4. Discussion
NTM PD is a growing cause of concern as its prevalence has been increasing in Hawai’i, nationwide, and globally. A panel of NTM from Hawai’i comprised of environmental and respiratory isolates was used to examine differences, if any, between M. abscessus, M. porcinum, and M. chimaera isolates in terms of iron acquisition by siderophores when stratified by their (1) recovered niche and (2) growth type (i.e., RGM or SGM).
In the U.S., the mean amount of iron in surface soil (0–5 centimeters) is 214 μg/L or 0.214 mg/kg (Smith et al., 2013; Brittenham et al., 2018). For comparison, this is lower than the mean amount of iron in Hawaiian Island soils that is around 95,500 mg/kg (Hawai’i Department of Health Hazard Evaluation and Emergency Response, 2012). Drinking water in the U.S. seldom contains more than 300 μg/L (Services). The normal range for serum iron is 6 to 17 μg/L and iron is present in the cytoplasm of host cells at very low levels (Agoro and Mura, 2019). In prolonged iron starvation environments such as the granuloma, Mtb can show high expression of mbt genes and siderophore production (Agoro and Mura, 2019). Due to the limited amount of iron in the body compared to the overabundance of iron in the extracellular niches that environmental NTM inhabit, we hypothesized that respiratory NTM would be more likely to produce siderophores due to the limited access to iron in the lung, compared to environmental NTM isolates. To test this hypothesis, we performed complementary genomic and microbiological analyses to assess siderophore production.
Among the reference NTM gene sequences parsed through the KEGG database that we examined, respiratory NTM showed significantly higher mean number of mbt genes compared to environmentally isolated species (Figure 1A). On the other hand, the number of mbt genes were similar among the respiratory and environmental NTM isolates from Hawai’i based on the phylogenetic trees and multiple sequence alignments (Figure 3; Supplementary Figures 2, 3). For the Hawai’i isolates, we detected all ten mbt-1 and four mbt-2 genes in SGM M. chimaera compared to the RGM species (containing nine to ten mbt-1 and one to three mbt-2 genes) suggesting M. chimaera is selectively advantaged in iron acquisition from the environment compared to M. abscessus and M. porcinum. When we further analyzed for the presence of mbt genes where each species was stratified by their recovered niches, there was no statistical difference in mbt gene numbers between the respiratory and environmental RGM isolates of M. abscessus and M. porcinum (Figures 2B,C). However, for SGM M. chimaera from Hawai’i, environmental isolates showed more mbt genes compared to respiratory isolates (Figure 2D). However, despite differences in the detection of siderophore genes among the isolates tested, we did not observe significant phenotypic differences in the in vitro assay outcomes.
Besides mbt, we also examined two other iron acquiring systems for mycobacteria, mmpL/S and esx-3. mmpL3, plays a key role in the acquisition of iron from heme (Tullius et al., 2011). mmpL4 and mmpL5 both have similar functions in iron acquisition via siderophores (Wells et al., 2013). In contrast to mbt genes of the SGM, the total number of mmpL proteins are more abundant in RGM compared to SGM, especially in the M. abscessus species (Viljoen et al., 2017). In our study, although not statistically significant, the number of mmpL/S genes tended to be higher in RGM compared to SGM (Supplementary Figure 4). The abundance of mmpL genes could allow the RGM to persist in various environments, and M. abscessus has been observed to evolve and acquire genetic material rapidly (Sapriel et al., 2016). Although, it is unknown how esx-3 plays a role in iron acquisition, it is essential for mycobacterial growth in iron-limited conditions. Similar to mbt, the environmental M. chimaera isolates showed significantly more esx-3 genes than respiratory M. chimaera isolates (Figure 4C). These genetic results support siderophore producing advantages among Hawai’i M. chimaera environmental isolates that likely facilitate survival in exogenous niches. M. chimaera was the most common NTM recovered from the environment of Hawai’i (Honda et al., 2016); thus, this subspecies may have evolved methods to procure iron in nutrient limited environmental conditions. We have already shown that M. chimaera-derived from the Hawai’i environment grows better in the presence of iron oxide minerals such as hematite after 24 h in culture and binds to its surface (Glickman et al., 2020).
To complement our genomic analysis, we tested our NTM Hawai’i isolate panel for siderophore production by two microbiological assays, growth on low iron culture media and the CAS assay. Such assays have already been applied to study siderophore production in a variety of gram-negative and gram-positive bacteria (Hall and Ratledge, 1982; Louden et al., 2011; Ferreira et al., 2019). Based on the results from our genomic analyses, we posited that siderophore production would be more frequently observed in vitro for M. chimaera isolates compared to M. abscessus and M. porcinum isolates. However, iron scavenging from culture media containing low amounts of iron and production of siderophores was more frequently observed for the RGM M. abscessus and M. porcinum isolates compared to SGM M. chimaera categories (Figures 5A,C). We also observed discrepancies across the outcomes for the microbiological assays; that is, siderophore test outcomes did not match 100% of the time. For example, for 22 of the 28 RGM isolates tested and for 16 of 23 SGM isolates tested, low iron assay results matched CAS assay results 79 and 70% of the time, leaving mismatch outcomes for 21 and 30% of the isolates tested (Supplementary Figure 6). The reasons for the incongruency between the genomic analyses and in vitro outcomes and between the two microbiological tests used in our study are not known. Other studies have demonstrated SGM show significantly more inorganic ion transport gene clusters than RGM (Bachmann et al., 2019). Alternatively, discrepancies may be related to innate adaptation to exogenous iron or differences in siderophore secretion as our in vitro assays detect secreted siderophores. There may also be features of each culture media (e.g., dextrose, glycerol, or other components) that may influence siderophore activity which may also vary per isolate and between isolates of different growth rates (e.g., RGM and SGM). Future studies to reassess NTM siderophore production as supplemental iron is added to culture media, utilizing similar media components per assay, and repeating low iron tests using liquid culture would help to disentangle some of these outcomes.
This study has some limitations. For the gene sequences taken through the KEGG database, there was limited information on some of the genomes. For instance, there is a lack of clear information on isolation details. The data from those NTM isolates were excluded from the dataset; thus, reducing the sample size and variety of isolates analyzed. Secondly, the genomes from Hawai’i were from bacteria isolated from the island of O’ahu. While O’ahu is home to the majority of the state’s population, it would be prudent to explore iron-acquisition mechanisms by NTM recovered from other islands and other geographic areas. In addition, the in vitro CAS assay analyses of extracellular siderophores was not included in the gene analysis of intracellular siderophores and other iron-acquisition processes. A follow up transcriptomic study would be helpful to explore other genes and proteins involved with iron metabolism (e.g., transferrin, ferritin) and discern ones that are modulated with low iron exposure.
To expand upon this study, future studies may include similar experiments to analyze extracellular siderophore genes and siderophore activity using a more diversified panel of SGM and RGM species with larger numbers of environmental and respiratory isolates to further verify differences among NTM. In addition, other factors affecting iron acquisition such as competition could be analyzed through a combined analysis of other NTM and non-NTM isolates. More importantly, the roles of mbt, mmpL/S, and esx-3 in iron acquisition for M. abscessus, M. porcinum, and M. chimaera should be further evaluated for their roles in virulence in the context of lung immune cell infections, as has been performed for Mtb and Salmonella (De Voss et al., 2000; Saha et al., 2019).
In summary, our data provide new insights into iron acquisition genes and the presence and activity of NTM siderophores. Clinical studies should be performed to understand the role of dietary iron in individuals with NTM PD. Outcomes of these studies may highlight NTM iron acquisition genes or siderophores as new targets for drug development.
Data availability statement
All relevant data is contained within the article. The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author. Information for existing publicly accessible datasets is contained within the article. The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material. The data presented in this study are deposited in NCBI, accession numbers: SAMN37352598-SAMN37352623, SAMN21208657, SAMN21208659, SAMN37352124-SAMN37352144, SAMN21208626, SAMN212086270.
Author contributions
CT: Data curation, Investigation, Methodology, Visualization, Writing – review & editing, Formal analysis, Software, Validation, Writing – original draft. NO: Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – review & editing. AM: Data curation, Formal analysis, Investigation, Validation, Visualization, Writing – review & editing. SD: Formal analysis, Investigation, Validation, Visualization, Writing – review & editing. YC: Writing – review & editing, Funding acquisition, Methodology, Project administration, Resources, Software, Supervision, Writing – original draft. MS: Methodology, Software, Supervision, Writing – review & editing. NH: Methodology, Software, Supervision, Writing – review & editing, Data curation, Formal analysis, Investigation, Validation, Visualization, Writing – original draft. JH: Data curation, Formal analysis, Investigation, Methodology, Supervision, Visualization, Writing – original draft, Writing – review & editing, Conceptualization, Funding acquisition, Project administration, Resources.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was funded, in part, by the National Science Foundation grant #1743587, Division of Environmental Biology, as part of the Ecology and Evolution of Infectious Diseases program. JH also acknowledges support from the Padosi Foundation.
Acknowledgments
The authors would like to acknowledge Diagnostic Laboratory Services, Inc. (Aiea, Hawai’i) at the time clinical NTM isolates were procured for their assistance and enthusiasm for this project.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2023.1268963/full#supplementary-material
Footnotes
References
Abbaspour, N., Hurrell, R., and Kelishadi, R. (2014). Review on iron and its importance for human health. J. Res. Med. Sci. 19, 164–174. Available at: https://pubmed.ncbi.nlm.nih.gov/24778671/.
Adjemian, J., Olivier, K. N., Seitz, A. E., Holland, S. M., and Prevots, D. R. (2012). Prevalence of nontuberculous mycobacterial lung disease in U.S. Medicare beneficiaries. Am. J. Respir. Crit. Care Med. 185, 881–886. doi: 10.1164/rccm.201111-2016OC
Agoro, R., and Mura, C. (2019). Iron supplementation therapy, a friend and foe of mycobacterial infections? Pharmaceuticals (Basel) 12, 1–28. doi: 10.3390/ph12020075
Bachmann, N. L., Salamzade, R., Manson, A. L., Whittington, R., Sintchenko, V., Earl, A. M., et al. (2019). Key transitions in the evolution of rapid and slow growing Mycobacteria identified by comparative genomics. Front. Microbiol. 10:3019. doi: 10.3389/fmicb.2019.03019
Barclay, R., Ewing, D. F., and Ratledge, C. (1985). Isolation, identification, and structural analysis of the mycobactins of Mycobacterium avium, Mycobacterium intracellulare, Mycobacterium scrofulaceum, and Mycobacterium paratuberculosis. J Bacteriol. 164, 896–903. doi: 10.1128/JB.164.2.896-903.1985
Braun, V., and Winkelmann, G. (1988). "Microbial iron transport structure and function of siderophores ": Springer Berlin/Heidelberg, 67–99.
Brittenham, G., Hoffman, R., Benz, E., and Silberstein, L. (2018). Disorders of iron homeostasis: iron deficiency and overload. Philadelphia, PA: Elsevier
Crosa, J. H. (1989). Genetics and molecular biology of siderophore-mediated iron transport in bacteria. Microbiol. Rev. 53, 517–530. doi: 10.1128/mr.53.4.517-530.1989
De Voss, J. J., Rutter, K., Schroeder, B. G., Su, H., Zhu, Y., and Barry, C. E. 3rd (2000). The salicylate-derived mycobactin siderophores of Mycobacterium tuberculosis are essential for growth in macrophages. Proc. Natl. Acad. Sci. U. S. A. 97, 1252–1257. doi: 10.1073/pnas.97.3.1252
Epperson, L. E., and Strong, M. (2020). A scalable, efficient, and safe method to prepare high quality DNA from mycobacteria and other challenging cells. J. Clin. Tuberc. Other Mycobact. Dis. 19:100150. doi: 10.1016/j.jctube.2020.100150
Falkinham, J. O. 3rd (1996). Epidemiology of infection by nontuberculous mycobacteria. Clin. Microbiol. Rev. 9, 177–215. doi: 10.1128/CMR.9.2.177
Ferreira, C. M. H., Vilas-Boas, A., Sousa, C. A., Soares, H., and Soares, E. V. (2019). Comparison of five bacterial strains producing siderophores with ability to chelate iron under alkaline conditions. AMB Express 9:78. doi: 10.1186/s13568-019-0796-3
Glickman, C. M., Virdi, R., Hasan, N. A., Epperson, L. E., Brown, L., Dawrs, S. N., et al. (2020). Assessment of soil features on the growth of environmental nontuberculous mycobacterial isolates from Hawai'i. Appl. Environ. Microbiol. 86:e00121-20. doi: 10.1128/AEM.00121-20
Gobin, J., Moore, C. H., Reeve, J. R. Jr., Wong, D. K., Gibson, B. W., and Horwitz, M. A. (1995). Iron acquisition by Mycobacterium tuberculosis: isolation and characterization of a family of iron-binding exochelins. Proc. Natl. Acad. Sci. U. S. A. 92, 5189–5193. doi: 10.1073/pnas.92.11.5189
Griffiths, E., Rogers, H. J., and Bullen, J. J. (1980). Iron, plasmids and infection. Nature 284, 508–509. doi: 10.1038/284508a0
Hall, R. M., and Ratledge, C. (1982). A simple method for the production of mycobactin, the lipid-soluble siderophore, from mycobacteria. FEMS Microbiol. Lett. 15, 133–136. doi: 10.1111/j.1574-6968.1982.tb00053.x
Hall, R. M., and Ratledge, C. (1984). Mycobactins as chemotaxonomic characters for some rapidly growing mycobacteria. J Gen Microbiol. 130, 1883–92. doi: 10.1099/00221287-130-8-1883
Hall, R. M., and Ratledge, C. (1985). Equivalence of mycobactins from Mycobacterium senegalense, Mycobacterium farcinogenes and Mycobacterium fortuitum. J Gen Microbiol. 131, 1691–6. doi: 10.1099/00221287-131-7-1691
Hasan, N. A., Davidson, R. M., Epperson, L. E., Kammlade, S. M., Beagle, S., Levin, A. R., et al. (2021). Population genomics and inference of Mycobacterium avium complex clusters in cystic fibrosis care centers, United States. Emerg. Infect. Dis. 27, 2836–2846. doi: 10.3201/eid2711.210124
Hawaii Department of Health Hazard Evaluation and Emergency Response. (2012). Hawaiian Islands Soil Metal Background Evaluation Report, May 2012 [Online]. Available at: https://health.hawaii.gov/heer/files/2012/05/Hawaiian-Islands-Soil-Metal-Background-Evaluation-Report-May-2012.pdf (Accessed October 26, 2023).
Honda, J. R., Hasan, N. A., Davidson, R. M., Williams, M. D., Epperson, L. E., Reynolds, P. R., et al. (2016). Environmental nontuberculous mycobacteria in the Hawaiian islands. PLoS Negl. Trop. Dis. 10:e0005068. doi: 10.1371/journal.pntd.0005068
Honda, J. R., Virdi, R., and Chan, E. D. (2018). Global environmental nontuberculous mycobacteria and their contemporaneous man-made and natural niches. Front. Microbiol. 9:2029. doi: 10.3389/fmicb.2018.02029
Kanehisa, M., Sato, Y., Furumichi, M., Morishima, K., and Tanabe, M. (2019). New approach for understanding genome variations in KEGG. Nucleic Acids Res. 147, D590–D595. doi: 10.1093/nar/gky962
Louden, B. C., Haarmann, D., and Lynne, A. M. (2011). Use of blue agar CAS assay for siderophore detection. J. Microbiol. Biol. Educ. 12, 51–53. doi: 10.1128/jmbe.v12i1.249
Page, A. J., Cummins, C. A., Hunt, M., Wong, V. K., Reuter, S., Holden, M. T., et al. (2015). Roary: rapid large-scale prokaryote pan genome analysis. Bioinformatics 31, 3691–3693. doi: 10.1093/bioinformatics/btv421
Parte, A. C., Sarda Carbasse, J., Meier-Kolthoff, J. P., Reimer, L. C., and Goker, M. (2020). List of prokaryotic names with standing in nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 70, 5607–5612. doi: 10.1099/ijsem.0.004332
Saha, P., Xiao, X., Yeoh, B. S., Chen, Q., Katkere, B., Kirimanjeswara, G. S., et al. (2019). The bacterial siderophore enterobactin confers survival advantage to Salmonella in macrophages. Gut Microbes 10, 412–423. doi: 10.1080/19490976.2018.1546519
Sapriel, G., Konjek, J., Orgeur, M., Bouri, L., Frezal, L., Roux, A. L., et al. (2016). Genome-wide mosaicism within Mycobacterium abscessus: evolutionary and epidemiological implications. BMC Genomics 17:118. doi: 10.1186/s12864-016-2448-1
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Siegrist, M. S., Steigedal, M., Ahmad, R., Mehra, A., Dragset, M. S., Schuster, B. M., et al. (2014). Mycobacterial Esx-3 requires multiple components for iron acquisition. MBio 5, e01073–e01014. doi: 10.1128/mBio.01073-14
Siegrist, M. S., Unnikrishnan, M., Mcconnell, M. J., Borowsky, M., Cheng, T. Y., Siddiqi, N., et al. (2009). Mycobacterial Esx-3 is required for mycobactin-mediated iron acquisition. Proc. Natl. Acad. Sci. U. S. A. 106, 18792–18797. doi: 10.1073/pnas.0900589106
Skaar, E. P. (2010). The battle for iron between bacterial pathogens and their vertebrate hosts. PLoS Pathog. 6:e1000949. doi: 10.1371/journal.ppat.1000949
Smith, D.B., Cannon, W.F., Woodruff, L.G., Solano, F., Kilburn, J.E., and Fey, D.L. (2013). Geochemical and mineralogical data for soils of the conterminous United States. Data Series. (Reston, VA).
Sritharan, M. (2016). Iron Homeostasis in Mycobacterium tuberculosis: Mechanistic Insights into Siderophore-Mediated Iron Uptake. J Bacteriol. 198, 2399–409. doi: 10.1128/JB.00359-16
Szekely, R., and Cole, S. T. (2016). Mechanistic insight into mycobacterial MmpL protein function. Mol. Microbiol. 99, 831–834. doi: 10.1111/mmi.13306
Takemoto, K., Kawai, H., Kuwahara, T., Nishina, M., and Adachi, S. (1991). Metal concentrations in human lung tissue, with special reference to age, sex, cause of death, emphysema and contamination of lung tissue. Int. Arch. Occup. Environ. Health 62, 579–586. doi: 10.1007/BF00381111
Tullius, M. V., Harmston, C. A., Owens, C. P., Chim, N., Morse, R. P., Mcmath, L. M., et al. (2011). Discovery and characterization of a unique mycobacterial heme acquisition system. Proc. Natl. Acad. Sci. U. S. A. 108, 5051–5056. doi: 10.1073/pnas.1009516108
Viljoen, A., Dubois, V., Girard-Misguich, F., Blaise, M., Herrmann, J. L., and Kremer, L. (2017). The diverse family of MmpL transporters in mycobacteria: from regulation to antimicrobial developments. Mol. Microbiol. 104, 889–904. doi: 10.1111/mmi.13675
Virdi, R., Lowe, M. E., Norton, G. J., Dawrs, S. N., Hasan, N. A., Epperson, L. E., et al. (2021). Lower recovery of nontuberculous mycobacteria from outdoor Hawai’i environmental water biofilms compared to indoor samples. Microorganisms 9:224. doi: 10.3390/microorganisms9020224
Keywords: nontuberculous mycobacteria, Hawai’i, siderophores, bioinformatics, CAS assay
Citation: Tan CG, Oberlag NM, McGowan AE, Dawrs SN, Chan YL, Strong M, Hasan NA and Honda JR (2023) Genomic and microbiological analyses of iron acquisition pathways among respiratory and environmental nontuberculous mycobacteria from Hawai’i. Front. Microbiol. 14:1268963. doi: 10.3389/fmicb.2023.1268963
Edited by:
Matt Johansen, University of Technology Sydney, AustraliaReviewed by:
Avishek Mitra, Oklahoma State University, United StatesAndre Kipnis, Universidade Federal de Goiás, Brazil
Copyright © 2023 Tan, Oberlag, McGowan, Dawrs, Chan, Strong, Hasan and Honda. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jennifer R. Honda, SmVubmlmZXIuSG9uZGFAVVRUeWxlci5lZHU=
†Present addresses:
Cara G. Tan, Department of Economics, University of Hawai’i at Manoa, Honolulu, HI, United States
Nicole M. Oberlag, Department of Biology, Claremont McKenna College, Claremont, CA, United States
Acelyn E. McGowan, Krieger School of Arts and Sciences, Johns Hopkins University, Baltimore, MD, United States
Stephanie N. Dawrs, Division of Pulmonary Science and Critical Care Medicine, University of Colorado Anschutz Medical Campus, Aurora, CO, United States
 Cara G. Tan
Cara G. Tan Nicole M. Oberlag
Nicole M. Oberlag Acelyn E. McGowan
Acelyn E. McGowan Stephanie N. Dawrs2†
Stephanie N. Dawrs2† Michael Strong
Michael Strong Nabeeh A. Hasan
Nabeeh A. Hasan Jennifer R. Honda
Jennifer R. Honda