- 1Guangdong Provincial Key Laboratory of Aquatic Product Processing and Safety, Guangdong Province Engineering Laboratory for Marine Biological Products, Guangdong Provincial Engineering Technology Research Center of Seafood, Guangdong Provincial Science and Technology Innovation Center for Subtropical Fruit and Vegetable Processing, College of Food Science and Technology, Guangdong Ocean University, Zhanjiang, China
- 2Department of Biology, College of Science, Shantou University, Shantou, China
- 3School of Life and Health Sciences, Hainan University, Haikou, China
Introduction: Research on the mechanism of marine polysaccharide utilization by Bacteroides thetaiotaomicron has drawn substantial attention in recent years. Derived from marine algae, the marine algae polysaccharides could serve as prebiotics to facilitate intestinal microecological balance and alleviate colonic diseases. Bacteroides thetaiotaomicron, considered the most efficient degrader of polysaccharides, relates to its capacity to degrade an extensive spectrum of complex polysaccharides. Polysaccharide utilization loci (PULs), a specialized organization of a collection of genes-encoded enzymes engaged in the breakdown and utilization of polysaccharides, make it possible for Bacteroides thetaiotaomicron to metabolize various polysaccharides. However, there is still a paucity of comprehensive studies on the procedure of polysaccharide degradation by Bacteroides thetaiotaomicron.
Methods: In the current study, the degradation of four kinds of marine algae polysaccharides, including sodium alginate, fucoidan, laminarin, and Pyropia haitanensis polysaccharides, and the underlying mechanism by Bacteroides thetaiotaomicron G4 were investigated. Pure culture of Bacteroides thetaiotaomicron G4 in a substrate supplemented with these polysaccharides were performed. The change of OD600, total carbohydrate contents, and molecular weight during this fermentation were determined. Genomic sequencing and bioinformatic analysis were further performed to elucidate the mechanisms involved. Specifically, Gene Ontology (GO) annotation, Clusters of Orthologous Groups (COG) annotation, and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment were utilized to identify potential target genes and pathways.
Results: Underlying target genes and pathways were recognized by employing bioinformatic analysis. Several PULs were found that are anticipated to participate in the breakdown of these four polysaccharides. These findings may help to understand the interactions between these marine seaweed polysaccharides and gut microorganisms.
Discussion: The elucidation of polysaccharide degradation mechanisms by Bacteroides thetaiotaomicron provides valuable insights into the utilization of marine polysaccharides as prebiotics and their potential impact on gut health. Further studies are warranted to explore the specific roles of individual PULs and their contributions to polysaccharide metabolism in the gut microbiota.
Introduction
Marine algae polysaccharides are complex carbohydrates extracted from a variety of marine seaweed. And they can be classified into several categories regarding their resources, structure, chemical components, and so on (Patel et al., 2023). A broad range of polysaccharides can be identified in seaweed, including sodium alginate, fucoidan, laminarin, and so on. Each class possesses particular characteristics and applications (Grundy et al., 2016; Zheng et al., 2020). Because of the special living condition of the algae, the synthesizing process of marine algae polysaccharides distantly differs from that of the terrestrial plant, for which the algae generate many active polysaccharides with novel structures and unique characteristics (Zheng et al., 2020; Xie and Cheong, 2022). Marine algae polysaccharides have garnered significant interest these years because of their biological functions in food manufacturing and pharmaceutical applications (de Jesus Raposo et al., 2015; Ebrahimzadeh et al., 2023). They might produce various positive impacts, such as anti-inflammatory, anti-virus, anti-cancer, immunomodulatory, food packaging, drug delivery, and hypoglycemic properties (Shannon and Abu-Ghannam, 2016; Zhao et al., 2018; Cheong et al., 2022; Jayakody et al., 2022; Obluchinskaya et al., 2022; Yao et al., 2022; Zhang et al., 2022).
The human gut microbiota (HGM), often referred to as the gut microbiome, comprises a diverse assembly of microorganisms that populates the digestive tract (Eckburg et al., 2005; Gomaa, 2020). These microorganisms include bacteria, fungi, and other archaea. In previous studies, it has been revealed that HGM is associated with the progression of various health disorders, including inflammatory bowel disease, anti-aging, obesity, diabetes, Parkinson’s disease, Alzheimer’s disease, asthma, and even alters how the host responds to drugs in the treatment of diseases (Ley et al., 2005; Arrieta et al., 2015; Hu et al., 2016; Leiva Gea et al., 2018; Bárcena et al., 2019; Khan et al., 2019; Sarkar and Banerjee, 2019; Contarino et al., 2023). The HGM contains the largest collection of microbes, which are embedded with millions of genes that render these hundreds of kinds of bacterial species with genes missing in our human genome. The encoded genes in the microbiota provide them with the capacity to metabolize the indigestible polysaccharides (Holscher, 2017). Still, due to insolubility or a lack of hydrolytic enzymes encoded by humans, many complex polysaccharides consumed by humans are resistant to host-mediated breakdown (Xu et al., 2020; Li et al., 2023). These carbohydrates undergo minimal degradation and assimilation in the upper gastrointestinal tract, while they function as an essential resource of nutrients and energy for the microbiota in the distal gut (Sonnenburg et al., 2010; Ye et al., 2021). Simultaneously, the absorption of carbohydrates can modulate the abundance and diversity of intestinal microbiota, making them a potential tool for treating disorders by regulating HGM (Adak and Khan, 2019; Vaughn et al., 2019).
Bacteroides predominate the gut microbial community and comprise roughly 48% of the HGM. Besides, the Bacteroides possess far more carbohydrate-active enzymes (CAZymes) than any other sequenced bacterium, making them efficient polysaccharide degraders and excellent intestine settlers (El Kaoutari et al., 2013; Lapébie et al., 2019). Bacteroides are able to quickly adapt their strategies for consuming carbohydrates to fit our various omnivorous diets. The complex nutrition-microbiota-host interaction is significantly shaped by this crucial adaptability (Lin and Zhang, 2017; Cheong et al., 2022). Additionally, Bacteroides’ considerable capacity for using polysaccharides can provide appropriate glycans for other bacterial species, promoting harmonious coexistence within intricate intestinal communities (Lapébie et al., 2019). Moreover, the Bacteroidetes show a variety of beneficial functions, including boosting host immunity, maintaining intestinal microecological balance, promoting the formation of intestinal mucosal blood vessels, and reducing intestinal inflammatory diseases (Martens et al., 2008; Romani et al., 2020). Bacteroides thetaiotaomicron, Bacteroides ovatus, and Bacteroides uniformis prove to be the dominant species of the Bacteroidetes. Among the Bacteroidetes, B. thetaiotaomicron colonizes the intestine widely and produces more CAZymes that degrade the polysaccharides into monosaccharides than other strains (Xu et al., 2003; Chaudet and Rose, 2016; Luis et al., 2017).
The CAZy1 is a specialized database designed for presenting and analyzing genomic, structural, and biochemical data related to carbohydrate-active enzymes (CAZymes). In the CAZy database, the CAZymes are divided into five categories: glycoside hydrolases (GHs), glycosyltransferases (GTs), polysaccharide lyases (PLs), carbohydrate esterases (CEs), auxiliary activities (AAs), and recognition (carbohydrate-binding module, CBM) (Park et al., 2010; Wardman et al., 2022). The degradation of the polysaccharide skeleton involves GHs and PLs in an essential way. Polysaccharide lyases break glycosidic bonds of polysaccharide in the manner of non-hydrolytic cleavage, while glycoside hydrolases break glycosidic bonds by hydrolysis and/or rearrangement (Henrissat, 1991; Davies and Henrissat, 1995; Lombard et al., 2010). Such multi-modular enzymes might or might not be made as a component of the CAZyme consortia contained within the known polysaccharide utilization loci (PULs). These PULs represent collections of genes that encode proteins with interconnected functions utilized for detecting, adhering, breaking down, and transporting specific polysaccharide (Grondin et al., 2017; Terrapon et al., 2018).
The diverse variety of GHs and PLs signify the enormous polysaccharide substrate of B. thetaiotaomicron. Thus, B. thetaiotaomicron was selected as model strain of human gut bacteria that could undergo genetic mutations for the examination of polysaccharide metabolism (Sonnenburg et al., 2006; Brown and Koropatkin, 2021). The process by which B. thetaiotaomicron utilizes complex carbohydrates has been well explored, relying on the abundance of sequencing data. B. thetaiotaomicron’s in-depth research on polysaccharide degradation provides an approach to understanding how other gut bacteria, particularly those also belonging to the genus Bacteroides, break down carbohydrates.
The impact of the interaction between polysaccharide and B. thetaiotaomicron on the host community has been gradually unrevealed these years. A wide range of metabolites produced by B. thetaiotaomicron, including short-chain fatty acids (SCFAs), vitamins, and other nutrients, have been associated with numerous health-benefiting functions. These metabolites exert their effects by influencing the gut microbiota, modulating immune responses, and contributing to the gut-brain axis (Xiao et al., 2021; Contarino et al., 2023).
In the current research, we will focus on the utilization of several marine algae polysaccharides, that is, sodium alginate, fucoidan, laminarin, and Pyropia haitanensis (Rhodophyta) polysaccharides, and their relationship with B. thetaiotaomicron G4. Besides, the genome analysis was employed to discover the genes with varying expression patterns in B. thetaiotaomicron G4. Genomic sequencing and bioinformatic analysis revealed potential target genes and pathways involved. Moreover, predicted PULs and CAZymes engaged in the breakdown of these four polysaccharides were depicted. These discoveries could contribute to a better comprehension of the interactions between marine algae polysaccharides and the microorganisms residing in the gut.
Materials and methods
Materials and chemicals
Laminarin (Eisenia bicylis source) (Phaeophyceae) was purchased from Tokyo Chemical Industry Co., Ltd. (Tokyo, Japan). Sodium alginate and fucoidan (Undaria pinnatifida source) (Phaeophyceae) were brought from Sigma-Aldrich (Saint Louis, United States). P. haitanensis polysaccharides (PHP) was prepared using a method previously reported by our research team (Qiu et al., 2020; Yu et al., 2023). In brief, the decolorized and deproteinized P. haitanensis powder was boiled in a water bath at 90°C for 2 h after adding 10 times volume distilled water (w/v). Centrifugation was used to extract the mixture’s supernatant, which was then combined with three liters of 95% ethanol. Following centrifugation for isolation, the precipitate was dissolved in water, frozen in a −20°C freezer, and subjected to freeze-drying to obtain the PHP. Standard dextrans with molecular weight from 4.66 kDa to 496 kDa were purchased from Aladdin (Shanghai, China). D-glucose, L-cysteine hydrochloride, hemin, vitamin K1 were procured from Solarbio Science & Technology (Beijing, China). Brain heart infusion (BHI) was brought from Hopebio (Qingdao, China). All other chemicals of analytical grade were acquired from XiLong Scientific Co., Ltd. (Shantou, China).
Microbial strains and medium
The microbial strain-B. thetaiotaomicron GDMCC 1.1104 (B. thetaiotaomicron G4) was bought from Guangdong Microbial Culture Collection Center (Guangzhou, China). Brain heart infusion broth (BHI) suppled with L-cysteine hydrochloride (0.5 g/L) and resazurin (0.001 g/L) was used as an enrichment medium. Basal medium containing yeast extract (2.0 g/L), peptone water (2.0 g/L), sodium chloride (0.1 g/L), potassium phosphate dibasic (0.04 g/L), potassium phosphate monobasic (0.04 g/L), magnesium sulfate heptahydrate (0.01 g/L), calcium chloride hexahydrate (0.01 g/L), sodium bicarbonate (2.0 g/L), hemin (0.05 g/L), L-cysteine hydrochloride (0.5 g/L), resazurin (0.001), bile salt (0.5 g/L), tween 80 (2 mL/L), and vitamin K1 (10 μL/L) was prepared as the previous research reported (Zeybek et al., 2020; Li S. et al., 2021). For batch cultures, the fermentation medium was created based on the basal medium supplemented with 1% (w/v) sodium alginate, fucoidan, laminarin, and PHP added as a carbohydrate source. Prior to inoculation, the pH value of medium was set within the 6.8–7.0 range using a pH meter. After being boiled, the medium was transferred to a 50 mL anaerobic culture bottle and purged with N2. Using butyl rubber stoppers and screw caps, the bottles were tightly sealed and subsequently sterilized at 121°C for 15 min. After undergoing filter sterilization with a 0.22 μm filter, the autoclaved medium was supplemented with heat-sensitive hemin and vitamin K1, after which it was cooled to 60°C. The strain was cultured in an anaerobic incubator (10% H2, 10% CO2, 80% N2) at 37°C.
Population structure and genetic diversity analysis
Identification of bacterial strain was achieved through 16S rRNA sequencing, and MEGA6.0 was used to create a systematic phylogenetic tree through the Neighbor-Joining (NJ) method (Tamura et al., 2013). The B. thetaiotaomicron G4 strain was subsequently employed for further cultivation, structural genomics analysis, functional annotation, CAZys prediction, and description of PULs.
Evaluation of bacterial growth on four polysaccharides
The freeze-dried B. thetaiotaomicron G4 was rehydrated and spread in the enrichment medium. After 48 h cultivation and passage to fresh enrichment medium twice, the bacteria in optimal growth vitality conditions were obtained and stored for further research. After the third activation, the obtained supernatant was subjected to centrifugation at 12,000 rpm at 4°C for 15 min. The bacterial pellet was rinsed and then resuspended in phosphate buffered saline (PBS; pH = 7.2) twice. After discarding the supernatant, the pellet was collected for the following processing steps.
The organisms after the last enrichment culture were used as inocula (1% v/v) and passaged to fermentation medium supplemented with 1% polysaccharides. During the fermentation, the fermentation broths were collected and taken out of the anaerobic culture bottle for further analysis at several time points (0, 12, 24, 48, and 72 h). The turbidity at OD600 was measured by a spectrophotometer (Shimadzu, Kyoto, Japan) to characterize the growth condition of B. thetaiotaomicron G4. Every fermentation operation was carried out independently three times. The fermentation medium without added organisms was used as a parallel sample for optical measurement.
Characterization of polysaccharide degradation
Total sugar contents were evaluated by the phenol-sulfuric acid method (DuBois et al., 1956), and its procedure is as follows: in short, centrifugation was employed on the fermented broths, and the supernatant was taken. After diluting the supernatant with appropriate volumes of distilled water, 0.2 mL of the distilled supernatant was added to 2 mL tubes containing 1 mL of sulfuric acid and 0.2 mL of 5% phenol (w/v). The solutions were thoroughly stirred, and the tubes were sealed with parafilm, followed by a 100°C water bath for 20 min. Then, the absorption at a wavelength of 490 nm was measured.
The standard glucose solutions were formulated through the dissolution of 100 mg of glucose in 1 L of distilled water. And the glucose solution was further diluted to the following concentrations: 0, 0.02, 0.04, 0.06, 0.08, and 0.1 mg/mL. And the sugar contents of glucose standards were also analyzed as per the procedure above. The standard curve was calculated from the absorption of glucose standards, with which the sugar contents of the fermentation samples were obtained. The measurement processes above were repeated three times.
Molecular weight analysis of the polysaccharide of fermentation broths
The supernatant of fermentation broth collected above was frozen for 48 h in a control-rate freeze dryer at −45°C while the vacuum was decreased to 11.3 Pa. After drying, the residual sugar was rehydrated in distilled water to the same volume of 1 mL. The samples’ molecular weight was assessed through high performance liquid chromatography (HPLC) as our previous study described (Yu et al., 2023). The evaporative light scattering detector (ELSD) and TSK G5000 PWXL column (30 cm × 7.8 mm, ID, 10 μL, Tosoh Co., Ltd., Tokyo, Japan) were employed in the chromatographic process. The injection of 10 μL samples into the column was complemented by a mobile phase of ammonium acetate, flowing at a rate of 0.8 mL/min. Throughout the procedure, the detector and column was maintained at 35 ± 0.1°C. By assuming that the retention time was directly proportional to the logarithm of the molecular weight of standards (4.66, 12.6, 50.0, 63.3, 126, 496 kDa), the molecular weight of the samples was determined.
Genomic DNA extraction and sequencing
The microbial DNA was obtained from the bacterial pellet by applying the TIANamp Bacteria DNA Kit (Tiangen Biotech, China) in accordance with the manufacturer’s instructions. Prior to constructing DNA library, the concentration and quality of extracted DNA were measured at 260/280 nm by NanoDrop2000 (Thermo Fisher Scientific, United States). The FEA Enzyme Mix (Tiangen Biotech, China) was utilized to break DNA samples that met the quality criteria into fragments ranging from 200 to 500 bp. Subsequently, these fragments were ligated with sequencing adaptors by DNA ligase (Sheng et al., 2021). Following adaptor ligation, the DNA fragments with attached adaptors are subjected to polymerase chain reaction (PCR) amplification. Sequencing libraries were generated with the TIANSeq Fast DNA Library Kit (Tiangen Biotech, China). The paired-end sequencing with a read length of 150 base pairs was conducted according to the manufacturer’s guidelines for whole genome sequencing, utilizing the Illumina HiSeq 2500 sequencer on the Illumina HiSeq 2500 platform (San Diego, CA, United States), achieving a 100-fold sequencing depth. After the completion of Illumina sequencing, the raw data undergo a series of data analysis operations, including quality control, statistical analysis, genome assessment, and de novo assembly (Akbar et al., 2021; Dang et al., 2022).
CGView map of the Bacteroides thetaiotaomicron G4 genome
CGView (Circular Genome Viewer, http://wishart.biology.ualberta.ca/cgview/) is a tool designed for creating interactive circular genome maps (Stothard and Wishart, 2005; Barakat et al., 2011). The genome circular map was applied to obtain a complete depiction of genomic properties, such as the distribution of genes on the forward and reverse strands, gene functional categorization based on COG categories, GC content, and genomic islands (Stothard et al., 2019).
Gene Ontology enrichment analysis
GO annotations are particularly valuable in enhancing our comprehension of the biological significance represented by the genes. Within the GO analysis, the proposed genes were examined in the GO database2 (Ashburner et al., 2000; Blake et al., 2013). The biological functions of the putative genes were established by substantial GO enrichment analysis. Protein function annotations containing three subtypes, including biological process (BP), molecular function (MF), and cellular component (CC) were assigned according to the GO database.
Clusters of Orthologous Groups annotation
The Clusters of Orthologous Group (COG) database is available at http://www.ncbi.nlm.nih.gov/COG. The website includes the following key characteristics: comprehensive list of all COGs with links to individual COG websites, COGs arranged by functional category, and COGs grouped according to functional complexes and pathways (Tatusov, 2001). Besides, COG database comprises species from various major systematic evolutionary lineages, and it is built upon integrated protein sequence comparisons of previously sequenced species (Galperin et al., 2015). Conducting EggNOG database (evolutionary genealogy of genes: non-supervised Orthologous Groups, http://eggnog.embl.de/) comparisons enables functional annotation and categorization of predicted proteins (Jensen et al., 2008; Powell et al., 2012).
Kyoto Encyclopedia of Genes and Genomes pathway enrichment analysis
KEGG is an open-access database3 of signal transduction pathways (Kanehisa et al., 2004). The candidate genes’ links to relevant biochemical metabolic and signal transduction pathways were determined through enrichment analysis (Kanehisa et al., 2006).
Carbohydrate-active enzyme annotation and PUL prediction
Relying on sequence similarity, the genomic data of B. thetaiotaomicron (BT) was queried against the combined CAZymes database (see text footnote 1) by applying dbCAN2 software with default settings4 (Huang et al., 2018). The extensive CAZymes of BT were assigned and annotated into six major module/domain families and subfamilies, including GHs, GTs, PLs, etc. The SusC-like and SusD-like genes are commonly employed as signatures for the identification and assignment of PUL loci. TonB-dependent transporters, known as SusC-like proteins, exhibit a distinct linear domain sequence, whereas SusD-like proteins are defined by either a single or a pair of domains (Cantarel et al., 2009). Utilizing the annotation information from the Pfam database,5 we sought SusC/D gene pairs and their homologous genes among all genes.
Our predictions involved classifying tandem SusC/D-like pairs based on the existence of adjacent SusC-like genes (upstream) and SusD-like genes (downstream) located on the same DNA strand. The occurrence of duplications involving SusC-like or SusD-like genes was also taken into account. Some PUL regulatory genes, such as the hybrid two-component system (HTCS) and extra-cytoplasmic functioning family of α (ECF-α), were also added to the PUL display.
Additionally, CAZymes gene loci were identified to investigate potential PULs in the B. thetaiotaomicron G4 strain. We integrated this information with available data from the PULDB database6 and literature references pertaining to PULs specific to particular polysaccharide structures (Terrapon et al., 2015). This enabled us to compile a comprehensive prediction of PULs in the genome of this particular B. thetaiotaomicron strain targeting specific polysaccharides.
Statistical analysis
The statistics were processed using GraphPad Prism 9. (GraphPad Software Inc., San Diego, CA). Data were analyzed by IBM SPSS Statistics ver. 25 (IBM Corporation, Armonk, NY, United States) using One-way ANOVA, and the significance level was determined at a p < 0.05 were considered significant.
Results
Comparative phylogenetic analysis
The 16S rRNA of B. thetaiotaomicron G4 was extracted and sequenced. Using 16S rRNA sequences, a phylogenetic tree for B. thetaiotaomicron G4 was established. Comparison of the acquired sequences with the sequences in the GenBank database using the NCBI BLAST software package revealed the greatest similarity of the genes of B. thetaiotaomicron G4 to those of members of the genus Becteroides.
In the phylogenetic analysis (Figure 1), B. thetaiotaomicron G4 and B. thetaiotaomicron VPI-5482 were clustered together closely with a bootstrap value of 99%, which indicated that they may possess the same origin. The comparative analysis of 16S rRNA between B. thetaiotaomicron G4 and B. thetaiotaomicron showed a similarity of 99.869%, which indicated that the BT strain used in the present study had a relatively high probability of belonging to B. thetaiotaomicron.
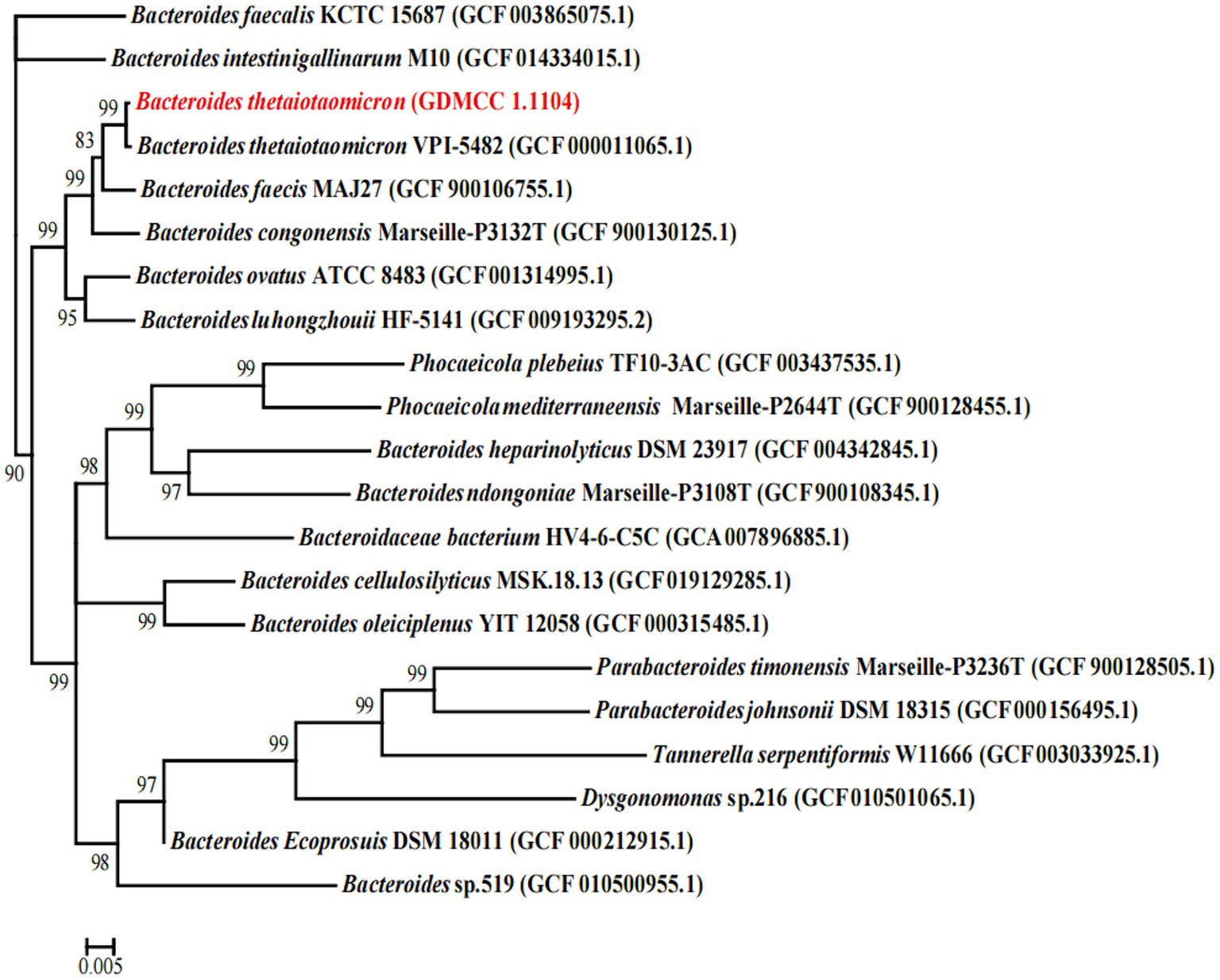
Figure 1. Phylogenetic tree showing the relatedness between the Bacteroides thetaiotaomicron G4, and the type strains of the genus Becteroides. The GenBank accession numbers of the sequences are given in parenthesis (www.ncbi.nlm.nih.gov). Bootstrap values (1,000 replicates) greater than 80% were shown. Numbers in parentheses are GenBank accession numbers. Scale bar, 0.005 substitution per nucleotide position.
The growth curve of Bacteroides thetaiotaomicron G4 on four typical marine algae polysaccharides
Bacteroides spp. allocate approximately 20% of their genome to metabolizing carbohydrates, whereas BT is capable of utilizing practically most polysaccharides. In turn, polysaccharides exert a beneficial impact on Bacteroides spp. Cultures of B. thetaiotaomicron G4 were cultivated in four different 0.5% algal polysaccharide mediums for 3 days, with fermentation samples obtained at 0, 6, 12, 24, 48, and 72 h to quantify the OD600. The growth curve (Figure 2A) showed the B. thetaiotaomicron could utilize these polysaccharides and substantially grow (OD600 > 0.5) in the basic medium containing these substates.
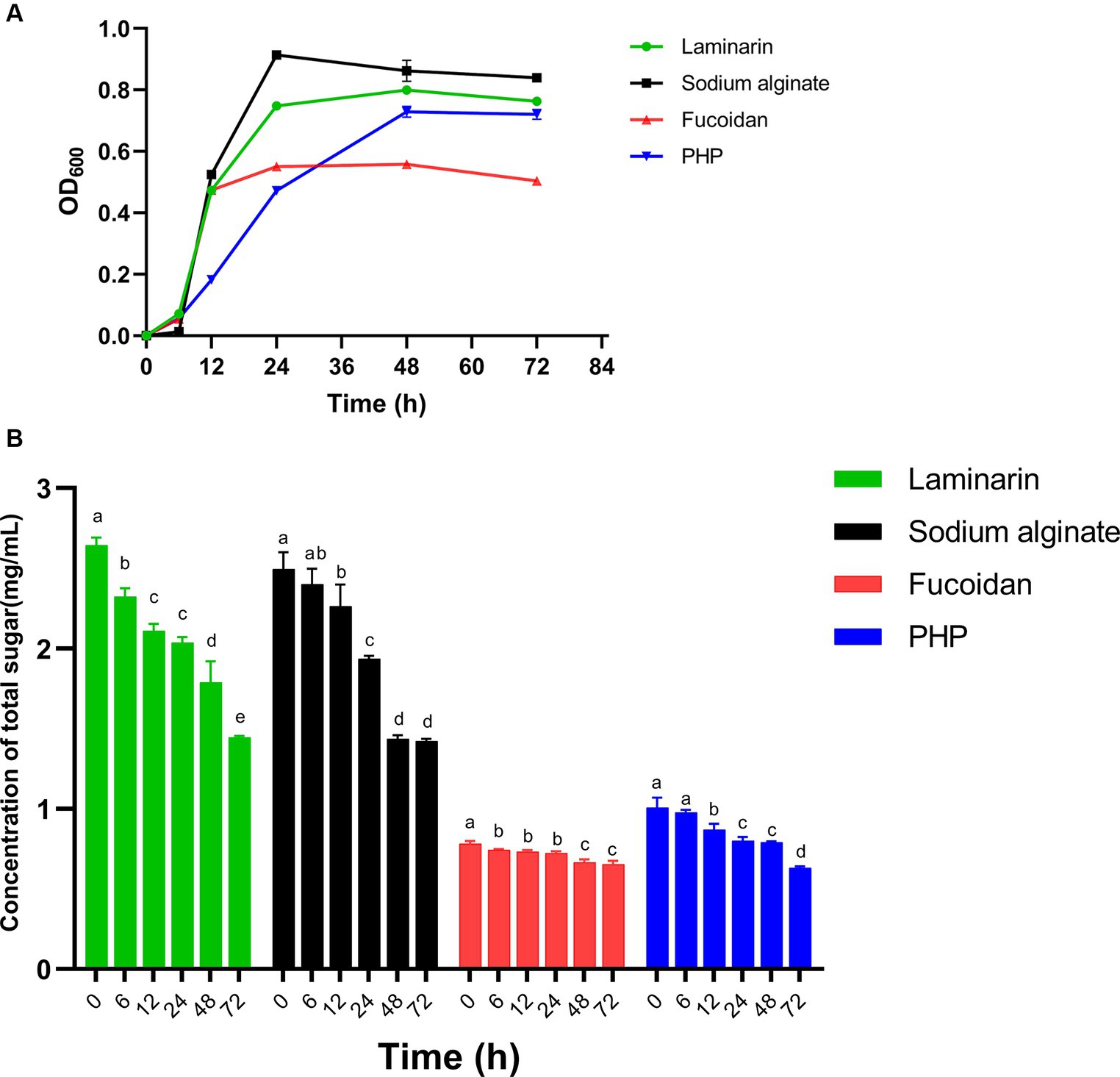
Figure 2. The utilization of four polysaccharides by Bacteroides thetaiotaomicron G4. OD600 (A) and sugar contents (B). Different lowercase alphabet letters (a–e) are significantly different at the level of p < 0.05.
The strain B. thetaiotaomicron grew in all four polysaccharide medias, with an initial lag phase within the first 6 h, and B. thetaiotaomicron entered the log phase after overcoming the lag period. After roughly 24 h of cultivation, the growth of B. thetaiotaomicron reached its stationary phase in media containing sodium alginate, fucoidan, and laminarin. In P. haitanensis polysaccharide-containing medium, the maximum cell density was reached after 48 h. The cultivation in fucoidan media revealed the relatively lowest cell density, with a maximum OD600 value of roughly 0.5, whereas the other three cultures attained OD600 values surpassing 0.7.
The experimental results indicated that BT demonstrated a greater capacity for degrading sodium alginate, laminarin, and PHP, while displaying a relatively lower capability in the degradation of fucoidan. These observations were closely tied to BT’s CAZymes production capabilities and the intrinsic properties of these enzymes, which should be further investigated in the future research.
Sugar concentration of polysaccharides residuals
The total sugar contents of bacterial culture samples, collected at different incubation times, were determined using the phenol-sulfuric acid method. Absorbance was measured at OD490 for the analysis of concentration changes (Figure 2B). Notably, there was a noteworthy decline in total sugar concentration in the sodium alginate, laminarin, and PHP, indicating putatively higher enzyme activity in the degradation of these polysaccharides. This suggests that B. thetaiotaomicron can effectively utilize these three polysaccharides, resulting in improved growth, consistent with the growth curve analysis above.
Notably, the relatively slow decline of sugar content during PHP degradation confirmed our prediction that only two PULs are involved in this process, likely attributed to lower gene expression levels associated with PHP utilization.
In the case of algal fucoidan, despite the presence of multiple polysaccharide utilization loci, there is a relatively slow decrease in carbohydrate concentration during the initial 24 h, which corresponds to the gradual increase in OD600. The enzyme activity may be hindered by suboptimal environmental factors, including pH and temperature, which resulted in the relatively low utilization of fucoidan and a slow strain growth rate.
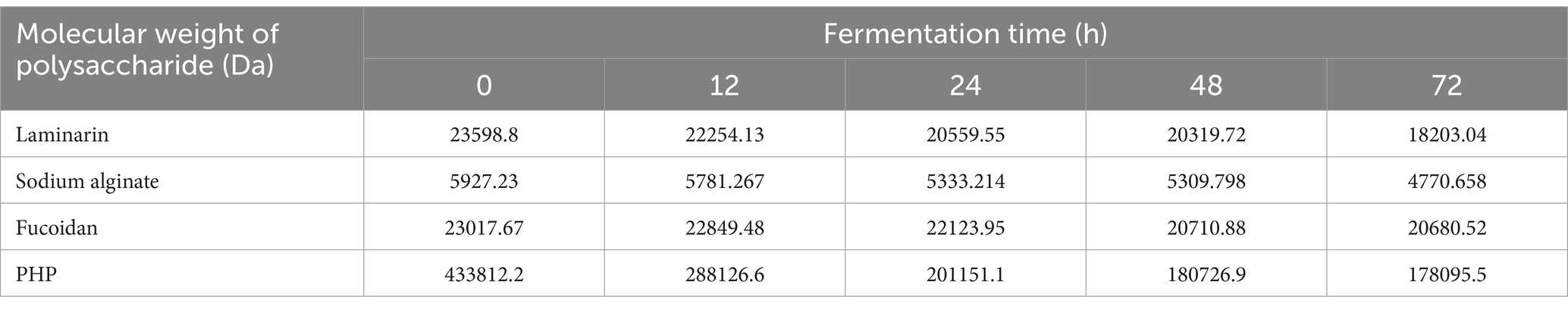
Molecular weight of polysaccharides residuals
Samples of bacterial cultures at different time points were analyzed for polysaccharide molecular weight using the HPLC-ELSD equipment. It was evident that the molecular weights of various algal polysaccharides in different carbohydrate groups had decreased (Table 1). This observation indicates that B. thetaiotaomicron can utilize the respective algal polysaccharides and break them down into smaller molecular substances.
It is worth noting that the most substantial decrease in polysaccharide molecular weight occurs during 0–24 h, corresponding with the log phase of B. thetaiotaomicron growth. In the log phase, the bacteria were characterized by rapid and high metabolic activity while extensively producing and secreting CAZymes degrading polysaccharides, which was further validated by this result.
CGView representation of genomic features of the strain
Integrating numerous forms of information into a single genome circular map helps us to acquire a more thorough and intuitive picture of the genomic properties of the bacterial strain. From the map (Figure 3), we could observe that the genes attached to carbohydrate transport and metabolism, amino acid transport, and metabolism exhibited significant expression levels. This phenomenon led to the inference that these heightened gene expressions consist of robust polysaccharide metabolism processes within B. thetaiotaomicron G4.

Figure 3. CGView map shows a full view of the genome of Bacteroides thetaiotaomicron G4. In the circular map, moving from the outermost ring to the innermost ring, the first and fourth rings represent coding sequences (CDS) on the forward and reverse DNA strands, with different colors denoting distinct COG (Cluster of Orthologous Groups) functional categories. The second and third rings depict CDS, tRNA, and rRNA on both the forward and reverse strands. The fifth ring displays the GC (Guanine-Cytosine) content, with outward extensions indicating regions where the GC content exceeds the genome-wide average GC content. Higher peaks in this ring correspond to greater deviations from the mean GC content, while inward extensions represent regions with GC content lower than the average, with higher peaks signifying larger differences from the mean GC content. The sixth ring represents GC-skew values, calculated using the formula (G − C)/(G + C). GC skew assists in distinguishing the leading and lagging DNA strands, with a general rule that the GC skew is greater than 0 for the leading strand and less than 0 for the lagging strand. Additionally, it can aid in identifying the replication origin (minimum cumulative offset) and termination site (maximum cumulative offset), which is particularly crucial for circular genomes. The innermost ring serves as a size indicator for the genome.
Gene functional enrichment analysis
To comprehend the biological activity of proposed genes, the genes of B. thetaiotaomicron G4 were annotated into three types for functional classification. The sequences in the strain’s genome were aligned with the GO (Gene Ontology) database based on sequence similarity. The results were then clustered, categorizing the genes into BP, MF, and CC categories based on their distinct functions. The top 10 subcategories of each of these three categories were further visualized in the chart (Figure 4A).
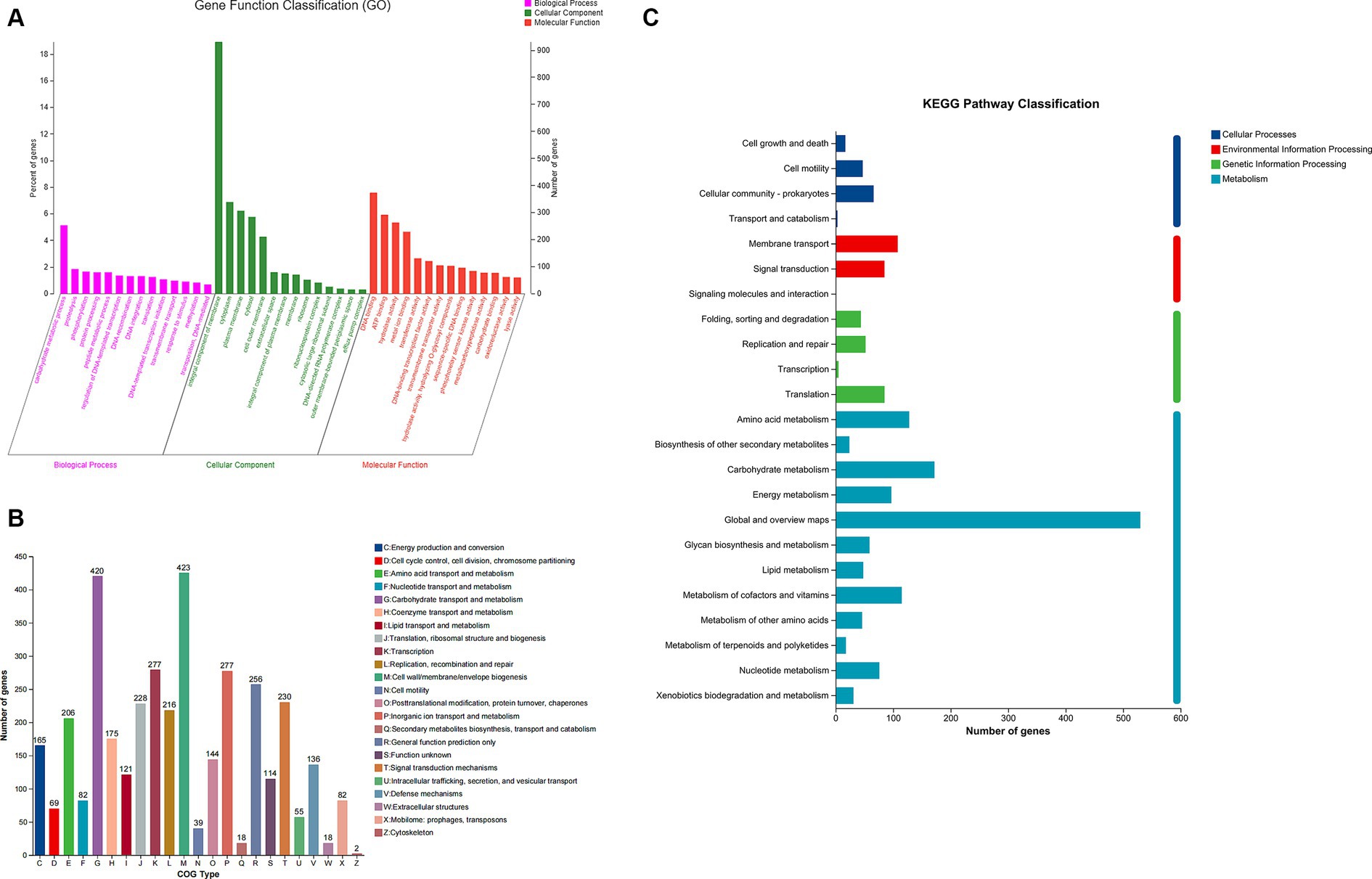
Figure 4. Gene ontology (GO) (A), Clusters of Orthologous Groups (COG) (B), and Kyoto Encyclopedia of Genes and Genomes (KEGG) (C) analysis of Bacteroides thetaiotaomicron G4.
In this study, 72.54% of genes were annotated in the databases, while the rest were not. Specifically, according to the GO classification, molecular function (5,818 genes, 49.72%), cellular component (2,747 genes, 23.47%), and biological process (3,137 genes, 26.81%) were classified. Of note, the most dominant genes were enriched (253 genes, 5.14%) in the genes related to carbohydrate metabolic processes within the biological process. Besides, among the molecular functions, hydrolase activity and hydrolyzing O-glycosyl compounds were significantly enriched (103 genes, 1.77%). These results may indicate the superior potential of carbohydrate metabolism and hydrolytic activity.
Distribution of Bacteroides thetaiotaomicron G4 by Cluster of Orthologous Groups annotation
By utilizing the Cluster of Orthologous Groups (COG) functional class designations (NCBI), essential genes were categorized into 23 types (Figure 4B). Besides, we recognized significant enrichment of essential gene groups, such as groups “G” (carbohydrate transport and metabolism), group “E” (amino acid transport and metabolism), and group “H” (coenzyme transport and metabolism), which were tightly linked to the degradation of polysaccharide. Specifically, there were 420 genes associated with groups “G,” 206 genes involved in groups “E,” and 175 genes participating in groups “H.” These COG groups included a variety of protein families that possess carbohydrate degradation, and facilitated the adaption of B. thetaiotaomicron G4 to cultural environment (Satti et al., 2018).
KEGG pathways classification
Most of the significantly enriched GO terms in our analysis were carbohydrate metabolic processes, hydrolase activity, and hydrolyzing O-glycosyl compounds. So, the KEGG was conducted to further confirm the gene functions. The KEGG classification here (Figure 4C) was mainly enriched in the following four categories: “cellular processes” (133 genes), “metabolism” (1,344 genes), “environmental information processing” (194 genes), and “genetic information processing” (186 genes). Genes assigned to “metabolism” were distributed among 12 subgroups. The pathways within these subgroups were firmly associated with carbohydrate metabolism, energy metabolism, and glycan biosynthesis and metabolism, accounting for 12.8, 7.21, and 4.39% of all genes, respectively. These results were the same as the findings in GO and COG terms, suggesting the potential importance of B. thetaiotaomicron G4 in polysaccharide degradation.
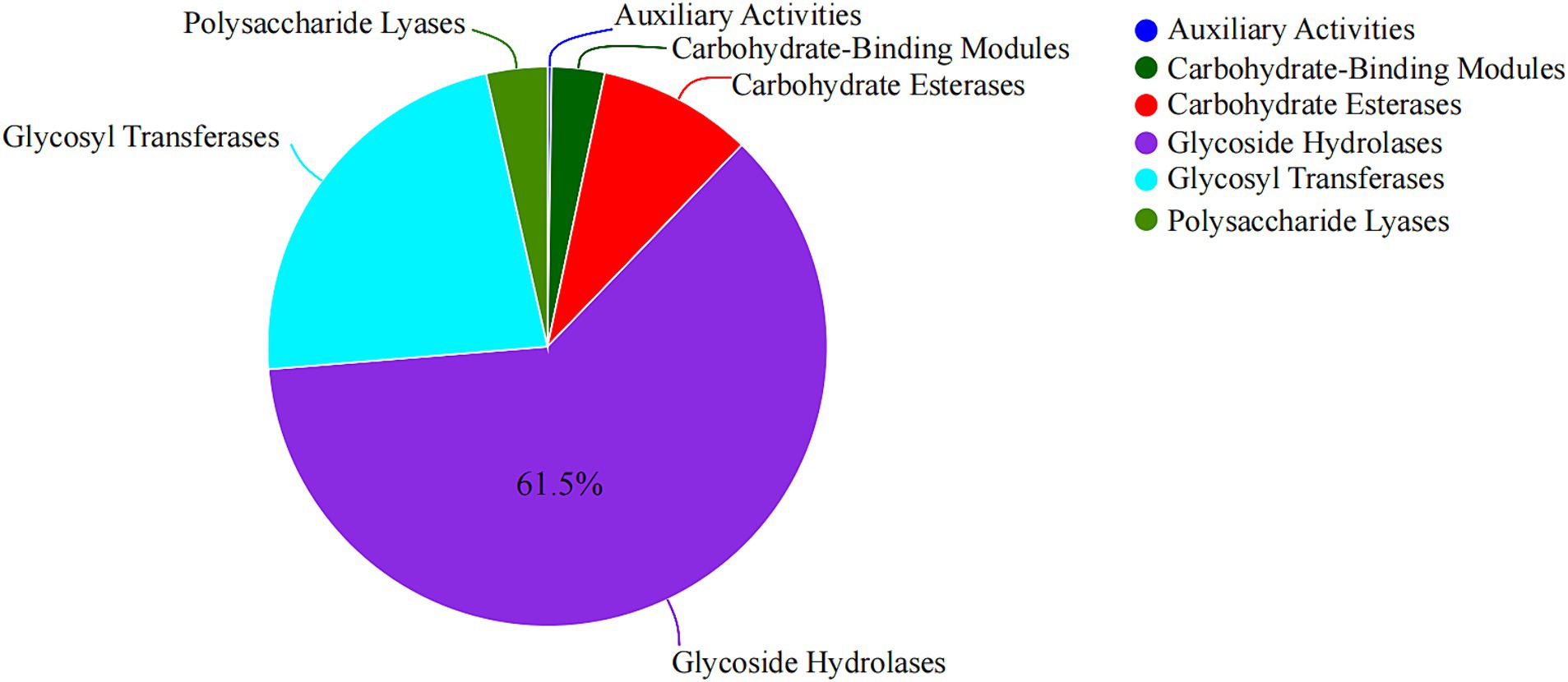
CAZymes annotations and description of the PULs dedicated to marine algae polysaccharide degradation
By sequencing, we annotated the composition and proportion of CAZymes in the B. thetaiotaomicron G4 strain (Figure 5). One hundred twenty PULs were predicted through genomic annotation of the B. thetaiotaomicron G4 strain. The whole content of these loci was presented in Supplementary Table S1. Because of the complex arrangement of PULs, we accurately completed the division of adjacent PULs based on the previous assembly strategy (Figure 6). The cluster encoded the SusC/SusD homolog pairs and various regulatory genes, such as regulator ECF-σ and HTCS.
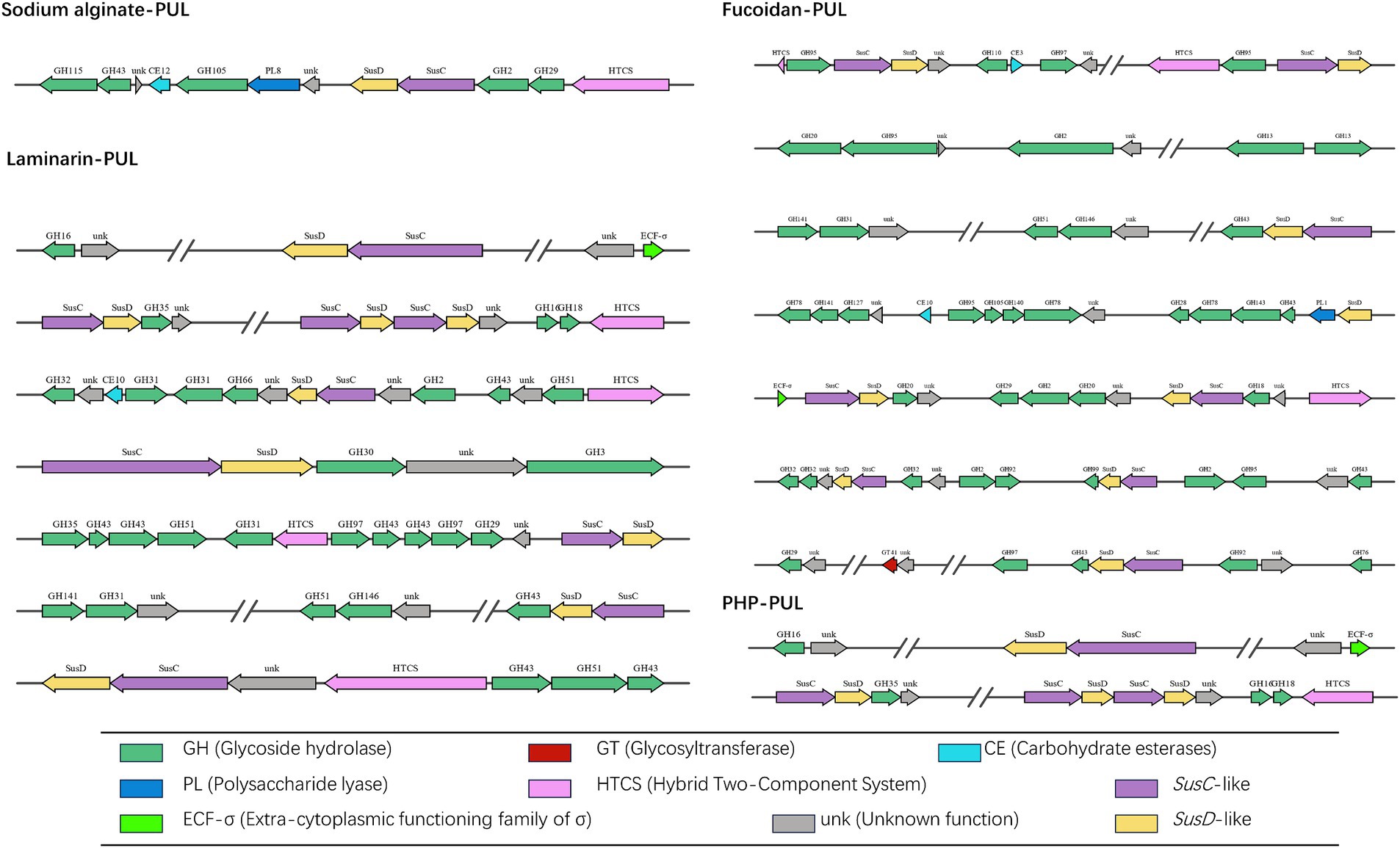
Figure 6. Schematic of the putative ten PULs involved in the degradation of four typical marine algae polysaccharide. The main functional modules involved in the degradation of sodium alginate were marked in bold red. SusC, SusC homolog; SusD, SusD homolog; ECF-σ, extra-cytoplasmic functioning family of α; HTCS, hybrid two-component system; GH, glycoside hydrolases; GTs, glycosyltransferases; CBM, carbohydrate-binding module; CE, carbohydrate esterases; PL, polysaccharide lyases; AAs, auxiliary activities; unk, unknown function.
Sodium alginate is a water-soluble linear polymer consisting of two residues of β-(1 → 4)-D-mannuronic acid (M) and α-(1 → 4)-L-guluronic acid (G) in alternating blocks, which are irregularly arranged into homopolymers (MM blocks, GG blocks) and heteropolymers (MG blocks) (Aarstad et al., 2012). The proportions of MGs in SAs originating from different species of seaweeds are also different. The enzymes reported to hydrolyze alginate predominantly belonged to PL families, such as PL 6, PL 7, and PL 8 (Stender et al., 2019). These CAZymes functionalized through the eliminative cleavage of M/G alginate. The B. thetaiotaomicron G4 strain mainly contained the PL8 family M-specific alginate lyase (EC 4.2.2.3), which catalyze the degradation of alginate by a β-elimination reaction (Davidson et al., 1977). PUL 19 were predicted to be involve in the degradation of sodium alginate, and their genomic organization was depicted (Figure 6). However, the alginate lyases face much challenges in the industrial production and utilization. To address the challenges stemming from high production costs and reduced productivity, Ramya’s research team conducted screenings on samples obtained from sponges and seaweed to identify bacteria capable of producing alginate lyases. Additionally, they optimized several parameters, including the composition of the growth medium and production processes, to maximize the yield of alginate lyase (Ramya et al., 2023). Furthermore, investigations into exo-type lyases sourced from Vibrio splendidus have unveiled their collaborative interactions and synergistic actions in the degradation of alginate (Jagtap et al., 2014). Noteworthy was the alginate lyase, AlyC6, derived from Vibrio sp. NC2, which illustrated the potential for synergy between two catalytic domains (Wang et al., 2022).
Fucoidan (U. pinnatifida source) mainly consists of the Type I fucoidan structure, which is characterized by a linear chain of α-1,3-linked fucose residues with sulfate groups attached. The minority of the U. pinnatifida source fucoidan were represented by fucose and galactose residues linked by α-1,3 or alternating α-1,3/4 glycosidic bonds (Lee et al., 2004). α-L-fucosidases can hydrolytically cleave terminal α-L-fucose residues from diverse fucose-containing glycans and glycoconjugates (EC 3.2.1.51, EC 3.2.1.63, EC 3.2.1.111, and EC 3.2.1.127). Primarily found in glycoside hydrolase (GH) families 29, 95, and 141, these fucosidases act in their specific ways in different enzymatic reactions. GH29 family enzymes demonstrate versatility by cleaving α-1,2-, α-1,3-, α-1,4-, and α-1,6-linked L-fucopyranose residues from diverse glycoconjugates. In contrast, GH95 family enzymes exhibit specificity for α-1,2-linked L-fucopyranose in oligosaccharides (see text footnote 1). The enzymes from the GH141 family demonstrated specificity for L-fucose residues, specifically those replaced at 3O- with a 2-Me-α-D-Xyl residue in pectin (Ndeh et al., 2017). To further understand the feature of fucosidases, many research have been explored. For example, a comprehensive investigation was conducted, involving both bioinformatic and biochemical analyses, to explore six alpha-L-fucosidases sourced from the marine bacterium Wenyingzhuangia fucanilytica CZ1127(T). This study marked the first in-depth exploration of fucosidase specificity, utilizing a variety of sulfated oligosaccharides linked to fucoidan (Silchenko et al., 2022). Notably, Lentimonas sp. CC4 (Verrucomicrobia) fucoidanases have been revealed to collaborate in the degradation of fucoidan obtained from diverse brown algae sources, employing substrate-specific pathways (Sichert et al., 2020). Additionally, an investigation into the effective degradation and potential industrial applications of fucosidase involved the cloning, expression, and characterization of BcFucA, a novel fucosidase originating from Bacillus cereus 2–8, in E. coli BL21 (Li Q. et al., 2021).
Laminarin is mainly β-glucans composed of β-1,3-glycosidic linkages of glucose and a small amount of β-1,6-glycosidic linkages of glucose as side chains. These endo-cleaving enzymes, known as endo-β-1,3-glucanases, can progressively cleave internal bonds within β-glucan chains (Kadam et al., 2015). Consequently, the glucan chains were internally broken into small molecular fragments. The clusters-encoded glycoside hydrolase families were predicted to be involved in the degradation of laminarin, such as GH3 (endo-1,3(4)-β-glucanase, EC 3.2.1.6), GH16 (glucan endo-1,3-β-D-glucosidase EC 3.2.1.39), and GH51 (endo-1,3(4)-β-glucanase, EC 3.2.1.6). These CZAymes could effectively catalyze the endo-hydrolysis of (1 → 3)-β-D-glucans in laminarin (Chesters and Bull, 1963; Barras and Stone, 1969). The PULs containing these families were depicted in Figure 6. Likewise, ZgLamC, a secreted laminarinase, is likely involved in the initial step of branched laminarin degradation, distinguishing it from the previously characterized laminarinase (Labourel et al., 2015). The expression of laminarinase activity in Vibrio sp. was dependent on the presence of the substrate and was repressed by the presence of glucose (Alderkamp et al., 2007). Research findings have indicated that the consumption of laminarin by Formosa spp. coincides with an increased uptake of diatom-derived peptides (Unfried et al., 2018).
The typical structure of P. haitanensis polysaccharides consists of a backbone composed of β-D-galactopyranose units linked by (1 → 3) glycosidic bonds. Additionally, the polysaccharides have two branching structures, each connected to the main chain via (1 → 4) glycosidic bonds. One branch is linked to 3,6-anhydro-α-L-galactopyranose units, and the other is linked to α-L-galactose-6-sulfate units (Zhang et al., 2004). These structural features give the polysaccharides their unique properties and functionality. During the hydrolysis process of P. haitanensis polysaccharides, the endo-β-1,3-galactanases (EC 3.2.1.181) and β-porphyranases (EC 3.2.1.178) assumed a pivotal role in the enzymatic degradation of PHP. Specifically, endo-β-1,3-galactanase primarily acts on the hydrolysis of β-D-galactopyranose-(1 → 4)-α-L-galactopyranose-6-sulfate linkages in porphyran, while porphyranase mainly catalyzes the hydrolysis of β-D-galactopyranose-(1 → 4)-α-L-galactopyranose-6-sulfate linkages in porphyrin (Correc et al., 2011; Kotake et al., 2011). In the present study, the data revealed that GH16 family was required to cleave the major structure of PHP, in which more than 20 kinds of enzymes could be produced. Similarly, an examination of the porphyranases’ sequences, responsible for the hydrolysis of consecutive methyl-porphyranobiose units, shed light on the diversity in the subsite specificity of porphyranases (Zhang et al., 2020).
B. thetaiotaomicron, a key bacterium in the human gut, shows promise in food and biotechnology. Its ability to break down complex carbohydrates offers opportunities for creating gut-friendly prebiotic foods. In biotechnology, its metabolic flexibility makes it attractive for producing bioactive compounds, biofuels, and biodegradable plastics through genetic manipulation (Liu et al., 2021; Béchon et al., 2022; Meslé et al., 2023). Additionally, it can modulate the immune system and reduce colonic inflammation, suggesting potential for treating conditions like inflammatory bowel disease (Kim et al., 2021; Li K. et al., 2021). Research is active in enhancing its probiotic traits and immune-regulating abilities, which could lead to innovative therapies leveraging the gut microbiome for better health outcomes. Overall, B. thetaiotaomicron holds vast potential for improving human health and sustainable biotechnology.
Conclusion
Marine algal polysaccharides, such as sodium alginate, fucoidan, laminarin, and Pyropia haitanensis polysaccharide, manifest remarkable attributes that underscore their essential role across diverse applications, ranging from biotechnology to healthcare. Furthermore, the pivotal role of CAZymes in polysaccharide degradation is of utmost importance for comprehending the intricate relationship between CAZymes and marine algal polysaccharides. In this study, we delved into the underlying mechanisms of polysaccharide degradation by B. thetaiotaomicron G4, focusing on four typical marine algae polysaccharides: sodium alginate, fucoidan, laminarin, and Pyropia haitanensis polysaccharides. Pure culture experiments with B. thetaiotaomicron G4 in a medium where these polysaccharides served as the carbon source were conducted. The measurement of growth curve of B. thetaiotaomicron G4, total carbohydrate content, and molecular weight of polysaccharides residuals confirmed the utilization and degradation of polysaccharides. To further validate our analyses, we employed several annotations, such as GO annotation, COG annotation, and exploration of the KEGG pathway enrichment to identify potential target genes and pathways. Our findings revealed several PULs that were likely involved in the degradation of these four polysaccharides. This comprehensive exploration of the mechanisms behind polysaccharide degradation by B. thetaiotaomicron G4 sheds light on the intricate interplay between marine seaweed polysaccharides and gut microbes. In forthcoming research, the anticipated PULs identified within this investigation will undergo further examination to elucidate the intricate process of polysaccharide degradation by B. thetaiotaomicron. These investigations may not only expand our knowledge of enzyme specificity but also hold promise for innovative applications in biotechnology, environmental sustainability, and pharmaceutical development.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, SRR26628481.
Author contributions
BY: Data curation, Investigation, Methodology, Writing – original draft. ZL: Methodology, Validation, Writing – review & editing. SZ: Conceptualization, Funding acquisition, Methodology, Supervision, Writing – review & editing. K-LC: Conceptualization, Methodology, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This work was financially supported by Key-Area Research and Development Program of Guangdong Province (2020B1111030004), The Innovative Team Program of High Education of Guangdong Province (2021KCXTD021), The Program for Scientific Research Start-up Funds of Guangdong Ocean University (060302042302), and National Natural Science Foundation of China (No. 31970101).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1332105/full#supplementary-material
Footnotes
References
Aarstad, O. A., Tøndervik, A., Sletta, H., and Skjåk-Bræk, G. (2012). Alginate sequencing: an analysis of block distribution in alginates using specific alginate degrading enzymes. Biomacromolecules 13, 106–116. doi: 10.1021/bm2013026
Adak, A., and Khan, M. R. (2019). An insight into gut microbiota and its functionalities. Cell. Mol. Life Sci. 76, 473–493. doi: 10.1007/s00018-018-2943-4
Akbar, M. A., Yusof, N. Y. M., Sahrani, F. K., Usup, G., Ahmad, A., Baharum, S. N., et al. (2021). Insights into Alexandrium minutum nutrient acquisition, metabolism and saxitoxin biosynthesis through comprehensive transcriptome survey. Biology 10:826. doi: 10.3390/biology10090826
Alderkamp, A. C., Van Rijssel, M., and Bolhuis, H. (2007). Characterization of marine bacteria and the activity of their enzyme systems involved in degradation of the algal storage glucan laminarin. FEMS Microbiol. Ecol. 59, 108–117. doi: 10.1111/j.1574-6941.2006.00219.x
Arrieta, M. C., Stiemsma, L. T., Dimitriu, P. A., Thorson, L., Russell, S., Yurist-Doutsch, S., et al. (2015). Early infancy microbial and metabolic alterations affect risk of childhood asthma. Sci. Transl. Med. 7:307ra152. doi: 10.1126/scitranslmed.aab2271
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Barakat, M., Ortet, P., and Whitworth, D. E. (2011). P2CS: a database of prokaryotic two-component systems. Nucleic Acids Res. 39, D771–D776. doi: 10.1093/nar/gkq1023
Bárcena, C., Valdés Mas, R., Mayoral, P., Garabaya, C., Durand, S., Rodríguez, F., et al. (2019). Healthspan and lifespan extension by fecal microbiota transplantation into progeroid mice. Nat. Med. 25, 1234–1242. doi: 10.1038/s41591-019-0504-5
Barras, D. R., and Stone, B. A. (1969). β1,3-glucan hydrolases from Euglena gracilis: I. The nature of the hydrolases. Biochim. Biophys. Acta 191, 329–341. doi: 10.1016/0005-2744(69)90252-6
Béchon, N., Mihajlovic, J., Lopes, A.-A., Vendrell-Fernández, S., Deschamps, J., Briandet, R., et al. (2022). Bacteroides thetaiotaomicron uses a widespread extracellular DNase to promote bile-dependent biofilm formation. Proc. Natl. Acad. Sci. U.S.A. 119:e2111228119. doi: 10.1073/pnas.2111228119
Blake, J. A., Dolan, M., Drabkin, H., Hill, D. P., Ni, L., Sitnikov, D., et al. (2013). Gene Ontology annotations and resources. Nucleic Acids Res. 41, D530–D535. doi: 10.1093/nar/gks1050
Brown, H. A., and Koropatkin, N. M. (2021). Host glycan utilization within the Bacteroidetes Sus-like paradigm. Glycobiology 31, 697–706. doi: 10.1093/glycob/cwaa054
Cantarel, B. L., Coutinho, P. M., Rancurel, C., Bernard, T., Lombard, V., and Henrissat, B. (2009). The carbohydrate-active EnZymes database (CAZy): an expert resource for glycogenomics. Nucleic Acids Res. 37, D233–D238. doi: 10.1093/nar/gkn663
Chaudet, M. M., and Rose, D. R. (2016). Suggested alternative starch utilization system from the human gut bacterium Bacteroides thetaiotaomicron. Biochem. Cell Biol. 94, 241–246. doi: 10.1139/bcb-2016-0002
Cheong, K. L., Yu, B., Chen, J., and Zhong, S. (2022). A comprehensive review of the cardioprotective effect of marine algae polysaccharide on the gut microbiota. Foods 11:3550. doi: 10.3390/foods11223550
Chesters, C. G., and Bull, A. T. (1963). The enzymic degradation of laminarin. 2. The multicomponent nature of fungal laminarinases. Biochem. J. 86, 31–38. doi: 10.1042/bj0860031
Contarino, M. F., Van Hilten, J. J., and Kuijper, E. J. (2023). Targeting the gut–brain axis with fecal microbiota transplantation: considerations on a potential novel treatment for Parkinson’s disease. Mov. Disord. Clin. Pract. 10, S21–S25. doi: 10.1002/mdc3.13621
Correc, G., Hehemann, J. H., Czjzek, M., and Helbert, W. (2011). Structural analysis of the degradation products of porphyran digested by Zobellia galactanivorans β-porphyranase A. Carbohydr. Polym. 83, 277–283. doi: 10.1016/j.carbpol.2010.07.060
Dang, H., Zhang, T., Li, Y., Li, G., Zhuang, L., and Pu, X. (2022). Population evolution, genetic diversity and structure of the medicinal legume, Glycyrrhiza uralensis and the effects of geographical distribution on leaves nutrient elements and photosynthesis. Front. Plant Sci. 12:708709. doi: 10.3389/fpls.2021.708709
Davidson, I. W., Lawson, C. J., and Sutherland, I. W. (1977). An alginate lyase from Azotobacter vinelandii phage. Microbiology 98, 223–229. doi: 10.1099/00221287-98-1-223
Davies, G., and Henrissat, B. (1995). Structures and mechanisms of glycosyl hydrolases. Structure 3, 853–859. doi: 10.1016/S0969-2126(01)00220-9
de Jesus Raposo, M. F., Bernardo de Morais, A. M., Costa, S., and de Morais, R. M. (2015). Marine polysaccharides from algae with potential biomedical applications. Mar. Drugs 13, 2967–3028. doi: 10.3390/md13052967
DuBois, M., Gilles, K. A., Hamilton, J. K., Rebers, P. A., and Smith, F. (1956). Colorimetric method for determination of sugars and related substances. Anal. Chem. 28, 350–356. doi: 10.1021/ac60111a017
Ebrahimzadeh, S., Biswas, D., Roy, S., and McClements, D. J. (2023). Incorporation of essential oils in edible seaweed-based films: a comprehensive review. Trends Food Sci. Technol. 135, 43–56. doi: 10.1016/j.tifs.2023.03.015
Eckburg, P. B., Bik, E. M., Bernstein, C. N., Purdom, E., Dethlefsen, L., Sargent, M., et al. (2005). Diversity of the human intestinal microbial flora. Science 308, 1635–1638. doi: 10.1126/science.1110591
El Kaoutari, A., Armougom, F., Gordon, J. I., Raoult, D., and Henrissat, B. (2013). The abundance and variety of carbohydrate-active enzymes in the human gut microbiota. Nat. Rev. Microbiol. 11, 497–504. doi: 10.1038/nrmicro3050
Galperin, M. Y., Makarova, K. S., Wolf, Y. I., and Koonin, E. V. (2015). Expanded microbial genome coverage and improved protein family annotation in the COG database. Nucleic Acids Res. 43, D261–D269. doi: 10.1093/nar/gku1223
Gomaa, E. Z. (2020). Human gut microbiota/microbiome in health and diseases: a review. Antonie Van Leeuwenhoek 113, 2019–2040. doi: 10.1007/s10482-020-01474-7
Grondin, J. M., Tamura, K., Dejean, G., Abbott, D. W., and Brumer, H. (2017). Polysaccharide utilization loci: fueling microbial communities. J. Bacteriol. 199:e00860. doi: 10.1128/JB.00860-16
Grundy, M. M. L., Edwards, C. H., Mackie, A. R., Gidley, M. J., Butterworth, P. J., and Ellis, P. R. (2016). Re-evaluation of the mechanisms of dietary fibre and implications for macronutrient bioaccessibility, digestion and postprandial metabolism. Br. J. Nutr. 116, 816–833. doi: 10.1017/S0007114516002610
Henrissat, B. (1991). A classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem. J. 280, 309–316. doi: 10.1042/bj2800309
Holscher, H. D. (2017). Dietary fiber and prebiotics and the gastrointestinal microbiota. Gut Microbes 8, 172–184. doi: 10.1080/19490976.2017.1290756
Hu, X., Wang, T., and Jin, F. (2016). Alzheimer’s disease and gut microbiota. Sci. China Life Sci. 59, 1006–1023. doi: 10.1007/s11427-016-5083-9
Huang, L., Zhang, H., Wu, P., Entwistle, S., Li, X., Yohe, T., et al. (2018). dbCAN-seq: a database of carbohydrate-active enzyme (CAZyme) sequence and annotation. Nucleic Acids Res. 46, D516–D521. doi: 10.1093/nar/gkx894
Jagtap, S. S., Hehemann, J. H., Polz, M. F., Lee, J. K., and Zhao, H. (2014). Comparative biochemical characterization of three exolytic oligoalginate lyases from Vibrio splendidus reveals complementary substrate scope, temperature, and pH adaptations. Appl. Environ. Microbiol. 80, 4207–4214. doi: 10.1128/AEM.01285-14
Jayakody, M. M., Vanniarachchy, M. P. G., and Wijesekara, I. (2022). Seaweed derived alginate, agar, and carrageenan based edible coatings and films for the food industry: a review. J. Food Meas. Charact. 16, 1195–1227. doi: 10.1007/s11694-021-01277-y
Jensen, L. J., Julien, P., Kuhn, M., von Mering, C., Muller, J., Doerks, T., et al. (2008). eggNOG: automated construction and annotation of orthologous groups of genes. Nucleic Acids Res. 36, D250–D254. doi: 10.1093/nar/gkm796
Kadam, S. U., Tiwari, B. K., and O’Donnell, C. P. (2015). Extraction, structure and biofunctional activities of laminarin from brown algae. Int. J. Food Sci. Technol. 50, 24–31. doi: 10.1111/ijfs.12692
Kanehisa, M., Goto, S., Hattori, M., Aoki-Kinoshita, K. F., Itoh, M., Kawashima, S., et al. (2006). From genomics to chemical genomics: new developments in KEGG. Nucleic Acids Res. 34, D354–D357. doi: 10.1093/nar/gkj102
Kanehisa, M., Goto, S., Kawashima, S., Okuno, Y., and Hattori, M. (2004). The KEGG resource for deciphering the genome. Nucleic Acids Res. 32, 277D–2280D. doi: 10.1093/nar/gkh063
Khan, I., Ullah, N., Zha, L., Bai, Y., Khan, A., Zhao, T., et al. (2019). Alteration of gut microbiota in inflammatory bowel disease (IBD): cause or consequence? IBD treatment targeting the gut microbiome. Pathogens 8:126. doi: 10.3390/pathogens8030126
Kim, K., Choe, D., Song, Y., Kang, M., Lee, S.-G., Lee, D.-H., et al. (2021). Engineering Bacteroides thetaiotaomicron to produce non-native butyrate based on a genome-scale metabolic model-guided design. Metab. Eng. 68, 174–186. doi: 10.1016/j.ymben.2021.10.005
Kotake, T., Hirata, N., Degi, Y., Ishiguro, M., Kitazawa, K., Takata, R., et al. (2011). Endo-β-1,3-galactanase from winter mushroom Flammulina velutipes. J. Biol. Chem. 286, 27848–27854. doi: 10.1074/jbc.M111.251736
Labourel, A., Jam, M., Legentil, L., Sylla, B., Hehemann, J. H., Ferrières, V., et al. (2015). Structural and biochemical characterization of the laminarinase Zg LamC GH16 from Zobellia galactanivorans suggests preferred recognition of branched laminarin. Acta Crystallogr. D 71, 173–184. doi: 10.1107/S139900471402450X
Lapébie, P., Lombard, V., Drula, E., Terrapon, N., and Henrissat, B. (2019). Bacteroidetes use thousands of enzyme combinations to break down glycans. Nat. Commun. 10:2043. doi: 10.1038/s41467-019-10068-5
Lee, J. B., Hayashi, K., Hashimoto, M., Nakano, T., and Hayashi, T. (2004). Novel antiviral fucoidan from sporophyll of Undaria pinnatifida (Mekabu). Chem. Pharm. Bull. 52, 1091–1094. doi: 10.1248/cpb.52.1091
Leiva Gea, I., Sanchez Alcoholado, L., Martin Tejedor, B., Castellano Castillo, D., Moreno Indias, I., Urda Cardona, A., et al. (2018). Gut microbiota differs in composition and functionality between children with type 1 diabetes and MODY2 and healthy control subjects: a case-control study. Diabetes Care 41, 2385–2395. doi: 10.2337/dc18-0253
Ley, R. E., Bäckhed, F., Turnbaugh, P., Lozupone, C. A., Knight, R. D., and Gordon, J. I. (2005). Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U.S.A. 102, 11070–11075. doi: 10.1073/pnas.0504978102
Li, K., Hao, Z., Du, J., Gao, Y., Yang, S., and Zhou, Y. (2021). Bacteroides thetaiotaomicron relieves colon inflammation by activating aryl hydrocarbon receptor and modulating CD4+ T cell homeostasis. Int. Immunopharmacol. 90:107183. doi: 10.1016/j.intimp.2020.107183
Li, Q., Jiang, C., Tan, H., Zhao, X., Li, K., and Yin, H. (2021). Characterization of recombinant E. coli expressing a novel fucosidase from Bacillus cereus 2–8 belonging to GH95 family. Protein Expr. Purif. 186:105897. doi: 10.1016/j.pep.2021.105897
Li, Y. F., Wu, B., Chen, J., Veeraperumal, S., Wei, J. C., Tan, K., et al. (2023). Prebiotic characteristics of added-value polysaccharides from jackfruit peel waste during in vitro digestion and fecal fermentation. LWT 187:115330. doi: 10.1016/j.lwt.2023.115330
Li, S., Zhang, B., Hu, J., Zhong, Y., Sun, Y., and Nie, S. (2021). Utilization of four galactans by Bacteroides thetaiotaomicron A4 based on transcriptome. Food Front. 2, 218–231. doi: 10.1002/fft2.82
Lin, L., and Zhang, J. (2017). Role of intestinal microbiota and metabolites on gut homeostasis and human diseases. BMC Immunol. 18:2. doi: 10.1186/s12865-016-0187-3
Liu, H., Shiver, A. L., Price, M. N., Carlson, H. K., Trotter, V. V., Chen, Y., et al. (2021). Functional genetics of human gut commensal Bacteroides thetaiotaomicron reveals metabolic requirements for growth across environments. Cell Rep. 34:108789. doi: 10.1016/j.celrep.2021.108789
Lombard, V., Bernard, T., Rancurel, C., Brumer, H., Coutinho, P. M., and Henrissat, B. (2010). A hierarchical classification of polysaccharide lyases for glycogenomics. Biochem. J. 432, 437–444. doi: 10.1042/BJ20101185
Luis, A. S., Briggs, J., Zhang, X., Farnell, B., Ndeh, D., Labourel, A., et al. (2017). Dietary pectic glycans are degraded by coordinated enzyme pathways in human colonic Bacteroides. Nat. Microbiol. 3, 210–219. doi: 10.1038/s41564-017-0079-1
Martens, E. C., Chiang, H. C., and Gordon, J. I. (2008). Mucosal glycan foraging enhances fitness and transmission of a saccharolytic human gut bacterial symbiont. Cell Host Microbe 4, 447–457. doi: 10.1016/j.chom.2008.09.007
Meslé, M. M., Gray, C. R., Dlakić, M., and DuBois, J. L. (2023). Bacteroides thetaiotaomicron, a model gastrointestinal tract species, prefers heme as an iron source, yields protoporphyrin IX as a product, and acts as a heme reservoir. Microbiol. Spectr. 11, e04815–e04822. doi: 10.1128/spectrum.04815-22
Ndeh, D., Rogowski, A., Cartmell, A., Luis, A. S., Baslé, A., Gray, J., et al. (2017). Complex pectin metabolism by gut bacteria reveals novel catalytic functions. Nature 544, 65–70. doi: 10.1038/nature21725
Obluchinskaya, E. D., Pozharitskaya, O. N., and Shikov, A. N. (2022). In vitro anti-inflammatory activities of fucoidans from five species of brown seaweeds. Mar. Drugs 20:606. doi: 10.3390/md20100606
Park, B. H., Karpinets, T. V., Syed, M. H., Leuze, M. R., and Uberbacher, E. C. (2010). CAZymes Analysis Toolkit (CAT): web service for searching and analyzing carbohydrate-active enzymes in a newly sequenced organism using CAZy database. Glycobiology 20, 1574–1584. doi: 10.1093/glycob/cwq106
Patel, A. K., Vadrale, A. P., Singhania, R. R., Michaud, P., Pandey, A., Chen, S. J., et al. (2023). Algal polysaccharides: current status and future prospects. Phytochem. Rev. 22, 1167–1196. doi: 10.1007/s11101-021-09799-5
Powell, S., Szklarczyk, D., Trachana, K., Roth, A., Kuhn, M., Muller, J., et al. (2012). eggNOG v3.0: orthologous groups covering 1133 organisms at 41 different taxonomic ranges. Nucleic Acids Res. 40, D284–D289. doi: 10.1093/nar/gkr1060
Qiu, H. M., Veeraperumal, S., Lv, J. H., Wu, T. C., Zhang, Z. P., Zeng, Q. K., et al. (2020). Physicochemical properties and potential beneficial effects of porphyran from Porphyra haitanensis on intestinal epithelial cells. Carbohydr. Polym. 246:116626. doi: 10.1016/j.carbpol.2020.116626
Ramya, P., Selvaraj, K., Suthendran, K., Sundar, K., and Vanavil, B. (2023). Optimization of alginate lyase production using Enterobacter tabaci RAU2C isolated from marine environment by RSM and ANFIS modelling. Aquac. Int. 31, 3207–3237. doi: 10.1007/s10499-023-01302-5
Romani, L., Del Chierico, F., Chiriaco, M., Foligno, S., Reddel, S., Salvatori, G., et al. (2020). Gut mucosal and fecal microbiota profiling combined to intestinal immune system in neonates affected by intestinal ischemic injuries. Front. Cell. Infect. Microbiol. 10:59. doi: 10.3389/fcimb.2020.00059
Sarkar, S. R., and Banerjee, S. (2019). Gut microbiota in neurodegenerative disorders. J. Neuroimmunol. 328, 98–104. doi: 10.1016/j.jneuroim.2019.01.004
Satti, M., Tanizawa, Y., Endo, A., and Arita, M. (2018). Comparative analysis of probiotic bacteria based on a new definition of core genome. J. Bioinforma. Comput. Biol. 16:1840012. doi: 10.1142/S0219720018400127
Shannon, E., and Abu-Ghannam, N. (2016). Antibacterial derivatives of marine algae: an overview of pharmacological mechanisms and applications. Mar. Drugs 14:81. doi: 10.3390/md14040081
Sheng, Q., Liu, C., Song, M., Xu, J., and Zhu, Z. (2021). Comprehensive transcriptome analysis of rare Carpinus putoensis plants under NO2 stress. Genes 12:754. doi: 10.3390/genes12050754
Sichert, A., Corzett, C. H., Schechter, M. S., Unfried, F., Markert, S., Becher, D., et al. (2020). Verrucomicrobia use hundreds of enzymes to digest the algal polysaccharide fucoidan. Nat. Microbiol. 5, 1026–1039. doi: 10.1038/s41564-020-0720-2
Silchenko, A. S., Rubtsov, N. K., Zueva, A. O., Kusaykin, M. I., Rasin, A. B., and Ermakova, S. P. (2022). Fucoidan-active α-L-fucosidases of the GH29 and GH95 families from a fucoidan degrading cluster of the marine bacterium Wenyingzhuangia fucanilytica. Arch. Biochem. Biophys. 728:109373. doi: 10.1016/j.abb.2022.109373
Sonnenburg, E. D., Sonnenburg, J. L., Manchester, J. K., Hansen, E. E., Chiang, H. C., and Gordon, J. I. (2006). A hybrid two-component system protein of a prominent human gut symbiont couples glycan sensing in vivo to carbohydrate metabolism. Proc. Natl. Acad. Sci. U.S.A. 103, 8834–8839. doi: 10.1073/pnas.0603249103
Sonnenburg, E. D., Zheng, H., Joglekar, P., Higginbottom, S. K., Firbank, S. J., Bolam, D. N., et al. (2010). Specificity of polysaccharide use in intestinal bacteroides species determines diet-induced microbiota alterations. Cell 141, 1241–1252. doi: 10.1016/j.cell.2010.05.005
Stender, E. G. P., Andersen, C. D., Fredslund, F., Holck, J., Solberg, A., Teze, D., et al. (2019). Structural and functional aspects of mannuronic acid-specific PL6 alginate lyase from the human gut microbe Bacteroides cellulosilyticus. J. Biol. Chem. 294, 17915–17930. doi: 10.1074/jbc.RA119.010206
Stothard, P., Grant, J. R., and Van Domselaar, G. (2019). Visualizing and comparing circular genomes using the CGView family of tools. Brief. Bioinform. 20, 1576–1582. doi: 10.1093/bib/bbx081
Stothard, P., and Wishart, D. S. (2005). Circular genome visualization and exploration using CGView. Bioinformatics 21, 537–539. doi: 10.1093/bioinformatics/bti054
Tamura, K., Stecher, G., Peterson, D., Filipski, A., and Kumar, S. (2013). MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 30, 2725–2729. doi: 10.1093/molbev/mst197
Tatusov, R. L. (2001). The COG database: new developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 29, 22–28. doi: 10.1093/nar/29.1.22
Terrapon, N., Lombard, V., Drula, E., Lapebie, P., Al-Masaudi, S., Gilbert, H. J., et al. (2018). PULDB: the expanded database of polysaccharide utilization loci. Nucleic Acids Res. 46, D677–D683. doi: 10.1093/nar/gkx1022
Terrapon, N., Lombard, V., Gilbert, H. J., and Henrissat, B. (2015). Automatic prediction of polysaccharide utilization loci in Bacteroidetes species. Bioinformatics 31, 647–655. doi: 10.1093/bioinformatics/btu716
Unfried, F., Becker, S., Robb, C. S., Hehemann, J.-H., Markert, S., Heiden, S. E., et al. (2018). Adaptive mechanisms that provide competitive advantages to marine bacteroidetes during microalgal blooms. ISME J. 12, 2894–2906. doi: 10.1038/s41396-018-0243-5
Vaughn, B. P., Rank, K. M., and Khoruts, A. (2019). Fecal microbiota transplantation: current status in treatment of GI and liver disease. Clin. Gastroenterol. Hepatol. 17, 353–361. doi: 10.1016/j.cgh.2018.07.026
Wang, X. H., Sun, X. H., Chen, X. L., Li, P. Y., Qin, Q. L., Zhang, Y. Q., et al. (2022). Synergy of the two alginate lyase domains of a novel alginate lyase from Vibrio sp. NC2 in alginate degradation. Appl. Environ. Microbiol. 88:e0155922. doi: 10.1128/aem.01559-22
Wardman, J. F., Bains, R. K., Rahfeld, P., and Withers, S. G. (2022). Carbohydrate-active enzymes (CAZymes) in the gut microbiome. Nat. Rev. Microbiol. 20, 542–556. doi: 10.1038/s41579-022-00712-1
Xiao, S., Zhang, G., Jiang, C., Liu, X., Wang, X., Li, Y., et al. (2021). Deciphering gut microbiota dysbiosis and corresponding genetic and metabolic dysregulation in psoriasis patients using metagenomics sequencing. Front. Cell. Infect. Microbiol. 11:605825. doi: 10.3389/fcimb.2021.605825
Xie, X. T., and Cheong, K. L. (2022). Recent advances in marine algae oligosaccharides: structure, analysis, and potential prebiotic activities. Crit. Rev. Food Sci. Nutr. 62, 7703–7717. doi: 10.1080/10408398.2021.1916736
Xu, J., Bjursell, M. K., Himrod, J., Deng, S., Carmichael, L. K., Chiang, H. C., et al. (2003). A genomic view of the human- Bacteroides thetaiotaomicron symbiosis. Science 299, 2074–2076. doi: 10.1126/science.1080029
Xu, S. Y., Chen, X. Q., Liu, Y., and Cheong, K. L. (2020). Ultrasonic/microwave-assisted extraction, simulated digestion, and fermentation in vitro by human intestinal flora of polysaccharides from Porphyra haitanensis. Int. J. Biol. Macromol. 152, 748–756. doi: 10.1016/j.ijbiomac.2020.02.305
Yao, W., Qiu, H. M., Cheong, K. L., and Zhong, S. (2022). Advances in anti-cancer effects and underlying mechanisms of marine algae polysaccharides. Int. J. Biol. Macromol. 221, 472–485. doi: 10.1016/j.ijbiomac.2022.09.055
Ye, M., Yu, J., Shi, X., Zhu, J., Gao, X., and Liu, W. (2021). Polysaccharides catabolism by the human gut bacterium—Bacteroides thetaiotaomicron: advances and perspectives. Crit. Rev. Food Sci. Nutr. 61, 3569–3588. doi: 10.1080/10408398.2020.1803198
Yu, B., Wang, M., Teng, B., Veeraperumal, S., Cheung, P. C. K., Zhong, S., et al. (2023). Partially acid-hydrolyzed porphyran improved dextran sulfate sodium-induced acute colitis by modulation of gut microbiota and enhancing the mucosal barrier. J. Agric. Food Chem. 71, 7299–7311. doi: 10.1021/acs.jafc.2c08564
Zeybek, N., Rastall, R. A., and Buyukkileci, A. O. (2020). Utilization of xylan-type polysaccharides in co-culture fermentations of Bifidobacterium and Bacteroides species. Carbohydr. Polym. 236:116076. doi: 10.1016/j.carbpol.2020.116076
Zhang, Y., Chang, Y., Shen, J., Mei, X., and Xue, C. (2020). Characterization of a novel porphyranase accommodating methyl-galactoses at its subsites. J. Agric. Food Chem. 68, 7032–7039. doi: 10.1021/acs.jafc.0c02404
Zhang, H., Jiang, F., Zhang, J., Wang, W., Li, L., and Yan, J. (2022). Modulatory effects of polysaccharides from plants, marine algae and edible mushrooms on gut microbiota and related health benefits: a review. Int. J. Biol. Macromol. 204, 169–192. doi: 10.1016/j.ijbiomac.2022.01.166
Zhang, Q., Li, N., Liu, X., Zhao, Z., Li, Z., and Xu, Z. (2004). The structure of a sulfated galactan from Porphyra haitanensis and its in vivo antioxidant activity. Carbohydr. Res. 339, 105–111. doi: 10.1016/j.carres.2003.09.015
Zhao, C., Yang, C., Liu, B., Lin, L., Sarker, S. D., Nahar, L., et al. (2018). Bioactive compounds from marine macroalgae and their hypoglycemic benefits. Trends Food Sci. Technol. 72, 1–12. doi: 10.1016/j.tifs.2017.12.001
Keywords: Bacteroides thetaiotaomicron , marine algae polysaccharides, CAZymes, polysaccharide utilization loci, polysaccharide metabolism
Citation: Yu B, Lu Z, Zhong S and Cheong K-L (2024) Exploring potential polysaccharide utilization loci involved in the degradation of typical marine seaweed polysaccharides by Bacteroides thetaiotaomicron. Front. Microbiol. 15:1332105. doi: 10.3389/fmicb.2024.1332105
Edited by:
Christian Sohlenkamp, National Autonomous University of Mexico, MexicoCopyright © 2024 Yu, Lu, Zhong and Cheong. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Saiyi Zhong, emhvbmdzeUBnZG91LmVkdS5jbg==; Kit-Leong Cheong, a2xjaGVvbmdAZ2RvdS5lZHUuY24=
 Biao Yu
Biao Yu Zheng Lu3
Zheng Lu3 Saiyi Zhong
Saiyi Zhong Kit-Leong Cheong
Kit-Leong Cheong