- 1College of Life Sciences, Nanjing Normal University, Nanjing, China
- 2Zhejiang Provincial Key Laboratory for Water Environment and Marine Biological Resources Protection, College of Life and Environmental Sciences, Wenzhou University, Wenzhou, China
- 3Hainan Key Laboratory of Herpetological Research, College of Fisheries and Life Sciences, Hainan Tropical Ocean University, Sanya, China
- 4Herpetological Research Center, College of Life and Environmental Sciences, Hangzhou Normal University, Hangzhou, China
Environmental temperature affects the composition, structure, and function of the gut microbial communities in host animals. To elucidate the role of gut microbiota in thermal adaptation, we designed a 2 species × 3 temperatures experiment, whereby we acclimated adult males of two agamid lizard species (warm-climate Leiolepis reevesii and cold-climate Phrynocephalus przewalskii) to 20, 28, and 36°C for 2 weeks and then collected their fecal and small-intestinal samples to analyze and compare the microbiota using 16S rRNA gene amplicon sequencing technology. The fecal microbiota displayed more pronounced interspecific differences in microbial community than the small-intestinal microbiota in the two species occurring in thermally different regions. The response of fecal and small-intestinal microbiota to temperature increase or decrease differed between the two species, with more bacterial taxa affected by acclimation temperature in L. reevesii than in P. przewalskii. Both species, the warm-climate species in particular, could cope with temperature change by adjusting the relative abundance of functional categories associated with metabolism and environmental information processing. Functional genes associated with carbohydrate metabolism were enhanced in P. przewalskii, suggesting the contribution of the fecal microbiota to cold-climate adaptation in P. przewalskii. Taken together, our results validate the two hypotheses tested, of which one suggests that the gut microbiota should help lizards adapt to thermal environments in which they live, and the other suggests that microbial communities should be thermally more sensitive in warm-climate lizards than in cold-climate lizards.
Introduction
The co-evolution of a host and its microbes colonizing different parts of the body including the body surface, oral cavity, gastrointestinal tract, and genitourinary tract is determined by complex symbiotic relationships (Ley et al., 2008; Chevalier et al., 2015). Numerous internal and external factors potentially affect the microbial community composition and structure, but the effects of temperature are more important and pervasive compared to many other external factors (Kohl and Yahn, 2016; Fontaine et al., 2018; Moeller et al., 2020; Zhu et al., 2021; Du et al., 2022; Huang et al., 2022). For example, warming decreases the relative abundance of Firmicutes in the gut microbiome and increases the ratio of putative pathogens in the western fence lizard Sceloporus occidentalis (Moeller et al., 2020) and alters the composition of gut microbial communities in the red-backed salamander Plethodon cinereus (Fontaine et al., 2018). The reduced relative abundance of bacteria of the genera Cellvibrio and Stenotrophomonas may adversely influence digestive performance in P. cinereus (Fontaine et al., 2018). Other studies on the brown bear Ursus arctos (Sommer et al., 2016), the Mongolian gerbil Meriones unguiculatus (Khakisahneh et al., 2020), and the thirteen-lined ground squirrel Ictidomys tridecemlineatus (Regan et al., 2022) provide indirect and direct evidence for the role of the microbiota in host physiology and survival. For example, compared with germ-free mice colonized with the gut microbiota from hibernating brown bears, germ-free mice colonized with the gut microbiota from summer brown bears trigger obesity, suggesting that seasonal changes in the gut microbiota can affect host energy metabolism and thereby result in mass gains of the body and fat (Sommer et al., 2016). Another example from I. tridecemlineatus shows that gut ureolytic microbes convert urea nitrogen into metabolites that can be absorbed by the host to promote protein balance during hibernation (Regan et al., 2022).
Host animals used to examine the effect of temperature on gut microbiota are either from the field, or maintained under controlled conditions in the laboratory. Field studies have examined the effects of external factors including seasonal temperature fluctuations on gut microbiota in a diverse array of vertebrate taxa hibernating [e.g., the brown frog Rana dybowskii (Tong et al., 2020), the Chinese alligator Alligator sinensis (Tang K.-Y. et al., 2019), I. tridecemlineatus (Chiang et al., 2022), U. arctos (Sommer et al., 2016), the greater horseshoe bat Rhinolophus ferrumequinum (Xiao et al., 2019), and the least horseshoe bat Rhinolophus pusillus (Liu et al., 2023)], or not hibernating [e.g., the plateau pika Ochotona curzoniae and the yak Bos mutus (Fu et al., 2020)] in winter. These field studies have yielded many interesting findings. In A. sinensis, for example, the increased relative abundance of bacteria of the genus Bacteroides in winter promotes the degradation of host mucin glycans, which is beneficial for hibernating Chinese alligators to pass the winter fasting period (Tang K.-Y. et al., 2019). Studies conducted under controlled conditions in the laboratory have paid more attention to the effect of temperature on gut microbiota. These laboratory studies consistently show that temperature increase or decrease affects the microbial community composition and structure, but conclusions are often drawn based on single species from a single sampling locality (e.g., Khakisahneh et al., 2020; Moeller et al., 2020; Arango et al., 2021; Hassenrück et al., 2021; Zhu et al., 2021; see also Chen et al., 2022). Whether and how the responses of gut microbiota to temperature change differ between host species distributed in thermally different regions or using thermally different habitats remain a sparsely studied area, but data on this topic are of great value in elucidating the role of gut microbiota in thermal adaptation. For example, gut microbial communities in tadpoles are more plastic in the invasive American bullfrog Lithobates catesbeianus than in the non-invasive green frog Lithobates clamitans, making the former species better able to adapt to a changing thermal environment (Fontaine and Kohl, 2020). Further work could usefully conduct comparative studies on multiple species from thermally contrasting environments to determine the role of gut microbiota in thermal adaptation.
Here, we focused on two oviparous lizard species of the family Agamidae, the Reeves’ butterfly lizard Leiolepis reevesii and the Przewalski’s toad-headed lizard Phrynocephalus przewalskii, living in thermally contrasting environments. L. reevesii is a medium-sized (up to 166 mm snout vent length, SVL), warm-climate lizard occurring in South China (Guangdong, Guangxi, and Hainan) and Vietnam (Figure 1A; Du et al., 2011), where air temperatures vary from 7.0 to 35.5°C.1 P. przewalskii is a small (up to 66 mm SVL), cold-climate lizard occurring in North and Northwest China (Gansu, Inner Mongolia, Ningxia, and Qinghai) (Figure 1A; Wang, 2011), where air temperatures vary from −24.8 to 38.9°C (see text footnote 1). We designed a 2 species × 3 temperatures experiment, whereby we acclimated adult males of the two lizard species to 20, 28, and 36°C for 2 weeks and then collected their fecal and small-intestinal samples to analyze and compare the microbiota using 16S rRNA gene amplicon sequencing technology. We tested the following two hypotheses. First, the gut microbiota should help lizards adapt to thermal environments in which they live and, as such, gut microbial genes related to metabolic function should differ between the cold- and warm-climate species. Second, gut microbial communities should be more sensitive to temperature change in the warm-climate species living in thermally more stable environments.
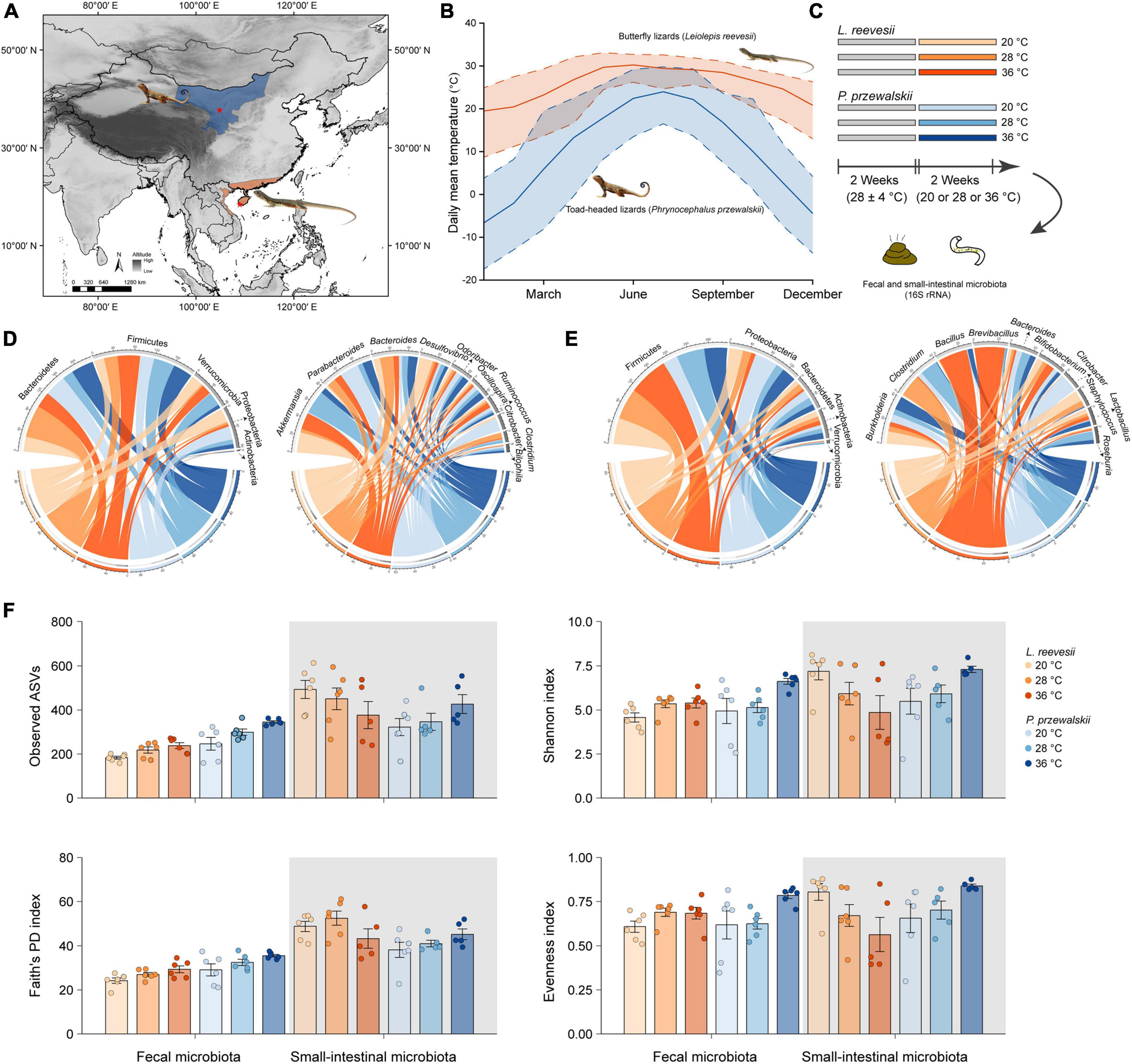
Figure 1. (A) Sampling localities (asterisks) of two agamid lizard species, the Reeves’ butterfly lizard Leiolepis reevesii having a warm-climate distribution (orange) and the Przewalski’s toad-headed lizard Phrynocephalus przewalskii having a cold-climate distribution (blue). (B) Daily mean (solid line), maximum (upper dashed line) and minimum (lower dashed line) temperatures of the two sampling localities during 2010–2019 (http://www.cma.gov.cn). (C) A 2 species × 3 acclimation temperatures experiment testing the hypotheses associated with the role of gut microbiota in thermal adaptation. (D) The relative abundance of the fecal microbiota at the phylum and genus levels in L. reevesii and P. przewalskii acclimated to three temperatures (20, 28, and 36°C) for 2 weeks. (E) The relative abundance of the small-intestinal microbiota at the phylum and genus levels in L. reevesii and P. przewalskii acclimated to three temperatures (20, 28, and 36°C) for 2 weeks. (F) Alpha diversity matrices of microbial communities in L. reevesii and P. przewalskii.
Materials and methods
Animal collection, experimental design, and sample collection
In mid-July 2020, we collected adult males of L. reevesii (103–152 mm SVL; 25.4–37.5 g body mass) from Ledong (18°29′N, 108°57′E), Hainan, South China, and adult males of P. przewalskii (52–61 mm SVL; 4.7–7.6 g body mass) from Zhongwei (37°27′N, 104°52′E), Ningxia, Northwest China (Figure 1A). Daily mean, maximum and minimum temperatures are higher in Ledong than in Zhongwei throughout the year (Figure 1B). All lizards were transferred to our laboratory in Sanya, Hainan, where four individuals of the same species were housed together in each 1.2 × 0.6 × 0.5 m (length × width × height) glass cage with a soil substrate (c. 120 mm depth). All cages were placed in a room for 2 weeks; temperatures in the room varied from 24 to 32°C with a mean of 28°C, and lights were on a 12: 12 light-dark cycle. Lizards were provided with mealworms (larvae of Tenebrio molitor) and water enriched with vitamins and minerals ad libitum (Figure 1C).
We then conducted a 2 species (L. reevesii and P. przewalskii) × 3 temperatures (20, 28, and 36°C) experiment in early August 2020. We chose the three temperatures for the following reasons: 20°C is the lowest nest temperature ever recorded in L. reevesii (Lin, 2008), temperatures around 28°C are optimal for egg incubation (embryonic development) in both L. reevesii (Lin et al., 2007) and P. przewalskii (Wang et al., 2013b), and 36°C is within the range of body temperatures selected by these two species in a thermal gradient (Lin, 2008; Wang et al., 2013a). We equally assigned lizards of each species into one of three temperature treatments. Specifically, we acclimated six individuals of each species to one of three temperatures (20, 28, and 36°C) for 2 weeks, and then collected fecal and small-intestinal samples (Figure 1C). Prior to the experiment, three AAPS (artificial atmospheric phenomena simulator) rooms and all experimental items were disinfected with alcohol wipes and ultraviolet lamps, and lizards were individually weighed to the nearest 0.01 on a Mettler balance and measured for SVL to the nearest 0.1 mm using Mitutoyo digital calipers, and then housed in 1.2 × 0.6 × 0.5 m glass cages. There were 12 lizards (six of each species) in each AAPS room, in which temperature was set at 20 ± 1, 28 ± 1, or 36 ± 1°C and fluorescent lights were switched on at 07:00 h and off at 19:00 h throughout the experiment. Neither in L. reevesii nor in P. przewalskii did mean values for SVL and body mass differ among the three temperature treatments (one-way ANOVA: F2,15 < 1.808 and P > 0.198 in both species). Mealworms sterilized with an ultraviolet lamp 1 h before feeding and distilled water were provided ad libitum for 12 days. Feces of each lizard were collected in sterile centrifuge tubes under sterile conditions in the last 2 days of the experiment. We collected small-intestinal contents immediately after collection of fecal samples. All fecal and small-intestinal samples were individually labeled and stored at −80°C for later DNA extraction (Figure 1C).
DNA extraction and sequencing
Total genomic DNA was extracted from the fecal and small-intestinal samples using the CTAB/SDS method, and concentration and purity were assessed with Nanodrop (Thermo Fisher Scientific, Waltham, MA, USA) and 1.2% agarose gels. The V3–V4 region of the bacterial 16S rRNA was amplified by PCR using the forward primer 338F (5′-ACTCCTACGGGAGGCAGCA-3′) and the reverse primer 806R (5′-GGACTACHVGGGTWTCTAAT-3′) with 7–12 bp unique barcodes. PCR reaction mixture contained 2 μl of DNA template, 5 μl of Q5 reaction buffer (5×), 5 μl of GC buffer (5×), 2 μl of dNTPs (2.5 mM), 1 μl of each primer (10 μM), 0.25 μl of Q5 DNA Polymerase, and 8.75 μl of ddH2O. The PCR thermal cycling conditions were set as follows: initial denaturation at 98°C for 2 min, followed by 25 cycles of denaturation at 98°C for 15 s, annealing at 55°C for 30 s, and extension at 72°C for 30 s; and final elongation at 72°C for 5 min. All samples were mixed with an equal molar amount from the purified PCR product of each sample, and library was prepared using the TruSeq Nano DNA LT Library Prep Kit (Illumina, Sangon Biotech Co., Ltd., Shanghai, China). Purified amplicons DNA libraries were sequenced using a MiSeq Reagent Kit V3 (Illumina, Sangon Biotech Co., Ltd., Shanghai, China) on an Illumina MiSeq platform (San Diego, CA, USA) in accordance with Frasergen Bioinformatics (Wuhan, Hubei, China).
Bioinformatics
We used the Quantitative Insights Into Microbial Ecology 2 (QIIME2 2020.8)2 (Caporaso et al., 2010) to process and analyze raw reads, the Cutadapt plugin to trim primers, and the DADA2 plugin to filter reads based on quality, merge paired reads, remove chimeras, and assign reads to amplicon sequence variants (ASVs) (Callahan et al., 2016). We then used the Greengenes 13_8_99% OTUs reference database classifier (DeSantis et al., 2006) to assign the sequence from each ASV, and removed reads identified as the sequences from archaea, chloroplast, and mitochondria. We retained ASVs with more than 100 reads and found in more than two samples, thereby avoiding large partial sample deviations. A phylogenetic tree was constructed using the Mafft alignment algorithm and the Fasttree maximum likelihood estimation (Katoh et al., 2002; Price et al., 2010). To standardize the ASVs abundance information, the sequencing depth was rarefied at 25,000 according to the 95% of the lowest number of sequences of one sample. Alpha and beta diversity metric values were calculated using the “core-metrics-phylogenetic” pipeline through the QIIME2 q2-diversity plugin.
Data analyses
Four indexes of alpha diversity were chosen, including observed ASVs, Faith’s phylogenetic diversity (Faith’s PD), Shannon index, and Pielou’s evenness index. We used Wilcoxon signed-rank test to examine the differences in these four indexes between the fecal and small-intestinal microbiota, Mann–Whitney test to examine the differences between the two species, and Kruskal–Wallis test to examine the differences among the three temperature treatments.
We calculated three beta diversity distance matrixes, including Bray–Curtis dissimilarity, unweighted UniFrac (which takes the presence and absence of microbial lineages into account, showing the community membership), and weighted UniFrac (which takes the relative abundances of microbial lineages into account, showing the community structure) distances. We used principal coordinates analysis (PCoA) ordinations to visualize sample clustering patterns based on three matrixes. Furthermore, we used the adonis function in vegan package (Oksanen et al., 2017) to perform the permutational multivariate analysis of variance (PERMANOVA, permutation = 9,999) and thereby examine the effects of microbiota source (fecal versus small-intestinal microbiota), host species, temperature treatment, and the interaction between microbiota source and host species on these three distance matrixes. We used the betadisper function in vegan package to perform a beta dispersion test for each matrix and thereby test the homogeneity of dispersion between the fecal and small-intestinal microbiota.
We respectively used Mann–Whitney test and Similarity Percentages (SIMPER) test to identify the phyla and ASVs contributing to the differences in microbial community between the two species. We used a multinomial species classification method (clamtest, clamtest function in vegan) to categorize ASVs into four classes, rare, generalist, warm-climate (L. reevesii) specialist, and cold-climate (P. przewalskii) specialist. We used linear discriminant analysis (LDA) effect size (LEfSe; Segata et al., 2011) to compare the taxa with total relative abundances >1% (from the phylum to genus levels) among the three temperature treatments. Biomarker species in each treatment were detected by Kruskal–Wallis test and linear discriminant analysis.
We compared the differences in bacterial phenotype and gene function among the three temperature treatments and thereby explored the effect of temperature on microbial function. Bacterial phenotypes were inferred through the BugBase web server, including aerobic, anaerobic and facultatively anaerobic bacteria, bacteria containing mobile elements, and biofilm-forming, Gram-negative, Gram-positive, potentially pathogenic, and stress-tolerant bacteria (Ward et al., 2017). Predictive functional profiles of microbial communities were identified by PICRUSt2 based on the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (Douglas et al., 2020). The LEfSe analysis was performed with a threshold criterion of LDA > 3 and P < 0.05 to compare the relative abundances of function genes from KEGG pathways Level 2 to Level 3 (KEGG orthology, KO) between the two species, and among the three temperature treatments in each species.
Descriptive statistics were expressed as mean ± standard error (SE). All P-values were adjusted for false discovery rate using the Benjamini–Hochberg procedure based on the p.adjust function in stats package (Bolar, 2019), and the significance level was set at P = 0.05.
Results
Composition of the fecal and small-intestinal microbiota
We obtained 2,960,930 fecal (36 samples) and 2,963,949 small-intestinal (33 samples) raw reads, from which we further obtained 1,655,845 fecal (mean ± SE: 45,996 ± 2,342) and 1,709,938 (51,816 ± 944) small-intestinal high-quality reads. Of the 33 small-intestinal samples, 17 were collected from L. reevesii (6 acclimated to 20°C treatment, 6 acclimated to 28°C, and 5 acclimated to 36°C), and 16 from P. przewalskii (6 acclimated to 20°C, 5 acclimated to 28°C, and 5 acclimated to 36°C). The rarefaction curves of estimated ASVs and Shannon index indicated that sequence depths were sufficient to describe of the bacterial community, and that species richness was higher in the small-intestinal microbiota than in the fecal microbiota (Supplementary Figure 1). After normalization, we identified 1,728 ASVs of 14 phyla, 31 classes, 41 orders, 79 families, and 117 genera in the fecal microbiota, and 2,716 ASVs of 20 phyla, 52 classes, 92 orders, 137 families, and 216 genera in the small-intestinal microbiota.
The top four dominant bacterial phyla in the fecal microbiota were Bacteroidetes (41.5% ± 5.5% in L. reevesii, and 36.2% ± 4.6% in P. przewalskii), Firmicutes (30.3% ± 4.0% in L. reevesii, and 36.0% ± 3.9% in P. przewalskii), Verrucomicrobia (15.5% ± 4.3% in L. reevesii, and 11.7% ± 4.0% in P. przewalskii), and Proteobacteria (12.1% ± 3.6% in L. reevesii, and 14.8% ± 4.8% in P. przewalskii) in both species (Figure 1D and Supplementary Figure 2). The top four dominant fecal bacterial genera were Akkermansia (15.4% ± 4.3%), Bacteroides (9.8% ± 1.7%), Desulfovibrio (7.5% ± 2.5%), and Parabacteroides (6.9% ± 1.9%) in L. reevesii, and were Parabacteroides (13.0% ± 3.9%), Akkermansia (11.7% ± 4.0%), Bacteroides (7.2% ± 1.3%), and Oscillospira (5.8% ± 0.7%) in P. przewalskii (Figure 1D and Supplementary Figure 2).
The top four dominant small-intestinal bacterial phyla were Firmicutes (61.7% ± 4.9% in L. reevesii, and 41.6% ± 5.1% in P. przewalskii), Proteobacteria (38.9% ± 3.6% in L. reevesii, and 38.9% ± 3.6% in P. przewalskii), Bacteroidetes (17.9% ± 3.2% in L. reevesii, and 30.6% ± 5.4% in P. przewalskii), and Actinobacteria (9.3% ± 2.0% in L. reevesii, and 8.9% ± 2.0% in P. przewalskii) in both species (Figure 1E and Supplementary Figure 2). The top four abundant small-intestinal bacterial genera were Brevibacillus (7.6% ± 5.4%), Bacillus (7.5% ± 4.9%), Clostridium (7.1% ± 3.9%), and Burkholderia (6.3% ± 1.1%) in L. reevesii, and were Burkholderia (7.5% ± 1.6%), Clostridium (6.5% ± 4.6%), Bacteroides (5.7% ± 1.4%), and Citrobacter (5.7% ± 5.7%) in P. przewalskii (Figure 1E and Supplementary Figure 2).
Influence of host species on microbial community
Observed ASVs, Faith’s PD, and Shannon indexes revealed that alpha diversity was higher in the small-intestinal microbiota as compared to the fecal microbiota (Wilcoxon signed-rank test, all P < 0.03), but evenness index did not show a significant difference in alpha diversity between the small-intestinal and fecal microbiota (Figure 1F; Wilcoxon signed-rank test, W = 201, P = 0.16). For the fecal microbiota, observed ASVs and Faith’s PD index were greater in P. przewalskii than in L. reevesii (Mann–Whitney test, both P < 0.01), whereas Shannon and evenness indexes did not differ significantly between the two species (Figure 1F; Mann–Whitney test, both P > 0.07). For the small-intestinal microbiota, none of the four alpha diversity indexes differed significantly between the two species (Figure 1F; Mann–Whitney test, all P > 0.06). Bray–Curtis dissimilarity, unweighted UniFrac, and weighted UniFrac distances revealed that microbiota source (fecal versus small-intestinal microbiota), host species, and their interaction affected beta diversity of microbial communities (Figure 2A and Table 1). Bray–Curtis dissimilarity (P < 0.01) and weighted UniFrac (P < 0.01) both revealed that distance to group centroid was significantly greater in the fecal than in small-intestinal microbiota (Figure 2A). Host species explained about 16.2% (Bray–Curtis dissimilarity), 29.9% (unweighted UniFrac), or 11.7% (weighted UniFrac) of variation in fecal microbial communities, and about 6.4% (Bray–Curtis dissimilarity), 11.3% (unweighted UniFrac), or 8.2% (weighted UniFrac) of variation in small-intestinal microbial communities (Table 1).
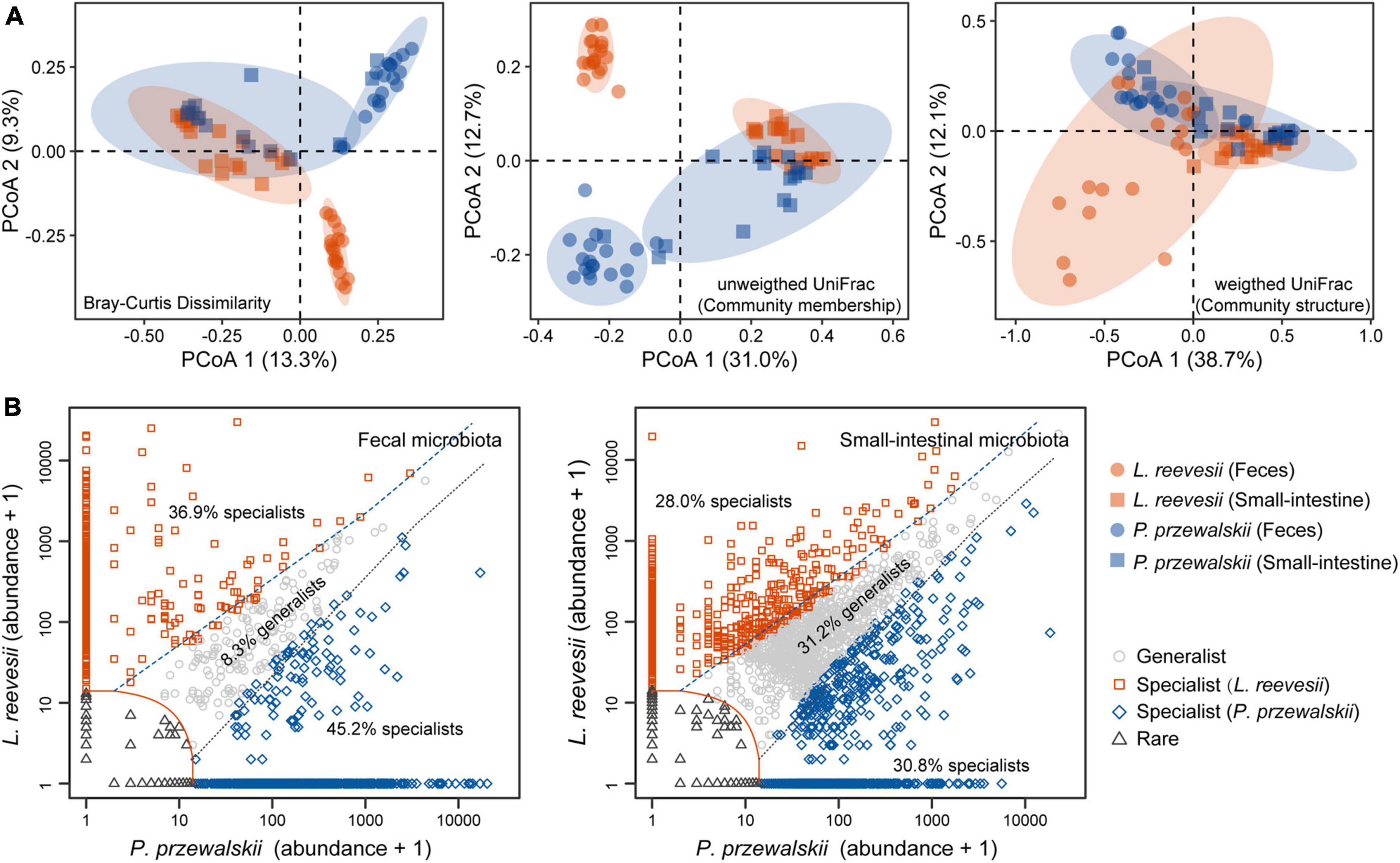
Figure 2. Beta diversity of the fecal and small-intestinal microbiota and distinct microbial taxa classified in L. reevesii and P. przewalskii. (A) Results of principal coordinates analysis (PCoA) based on Bray–Curtis dissimilarity, unweighted UniFrac (community membership) and weighted UniFrac (community structure) distances. (B) Four classes categorized by multinomial species classification method (clamtest): for the fecal microbiota, 9.6% were classified as rare, 8.3% as generalist, 36.9% as L. reevesii specialist, and 45.2% as P. przewalskii specialist; for the small-intestinal microbiota, 10.0% were classed as rare, 31.2% as generalist, 28.0% as L. reevesii specialist, and 30.8% as P. przewalskii specialist.
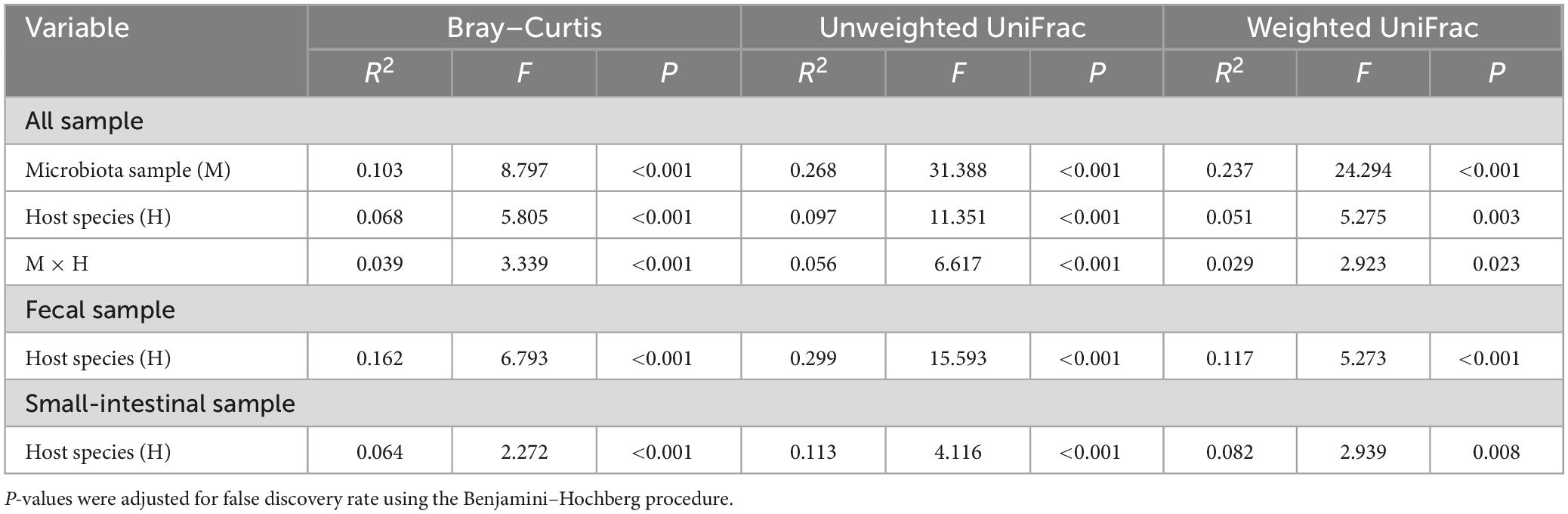
Table 1. The permutational multivariate analysis of variance (PERMANOVA) of microbiota samples (the fecal and small-intestinal microbiota) based on Bray–Curtis dissimilarity, unweighted UniFrac, and weighted UniFrac distances in two host lizard species (L. reevesii and P. przewalskii) from thermally different regions.
Host-specific microbial taxa resulted in beta diversity divergence between L. reevesii and P. przewalskii. The relative abundances of the top four dominant phyla were similar between the two species. In both species, Bacteroidetes, Firmicutes, Proteobacteria, and Verrucomicrobia were the top four dominant phyla (with a relative abundance >1%) in the fecal microbiota, and Firmicutes, Proteobacteria, Bacteroidetes, and Actinobacteria were the top four dominant phyla in the small-intestinal microbiota. The relative abundance of Firmicutes was significantly higher in L. reevesii than in P. przewalskii (Mann–Whitney test, U = 63, P < 0.01). SIMPER analysis revealed that five dominant fecal ASVs of the phyla Bacteroidetes, Proteobacteria, and Verrucomicrobia explained 13.1% of variation in Bray–Curtis dissimilarity, and that five small-intestinal dominant ASVs of the phyla Firmicutes and Proteobacteria explained 14.1% of variation in Bray–Curtis dissimilarity (Supplementary Table 1). Of the ASVs in the fecal microbiota, 9.6% were classed as rare, 8.3% as generalist, 36.9% as L. reevesii specialist, and 45.2% as P. przewalskii specialist (Figure 2B). Of the ASVs in the small-intestinal microbiota, 10.0% were classed as rare, 31.2% as generalist, 28.0% as L. reevesii specialist, and 30.8% as P. przewalskii specialist (Figure 2B). In the fecal microbiota, functional categories associated with carbohydrate metabolism were enriched in P. przewalskii (LDA = 3.01, P < 0.01).
Influence of acclimation temperature on alpha and beta diversity
Neither in the fecal microbiota nor in the small-intestinal microbiota did the four alpha diversity indexes (observed ASVs, Faith’s PD, Shannon index, and evenness index) differ significantly among L. reevesii acclimated to three temperatures (Figure 1F and Supplementary Table 2; Kruskal–Wallis test, all P > 0.07). The four alpha diversity indexes in the small-intestinal microbiota also did not differ significantly among P. przewalskii acclimated to three temperatures (Figure 1F and Supplementary Table 2; Kruskal–Wallis test, all P > 0.06). In the fecal microbiota of P. przewalskii we found the following. First, species richness inferred from observed ASVs was highest in the 36°C treatment and lowest in the 20°C treatment, with the 28°C treatment in between (Figure 1F and Supplementary Table 2; Kruskal–Wallis test, H2,18 = 9.07, P = 0.02). Second, both Shannon index and evenness index were apparently greater in P. przewalskii acclimated to 36°C than in their conspecifics acclimated to 20 and 28°C (Figure 1F and Supplementary Table 2; Kruskal–Wallis test, both P < 0.03).
Bray–Curtis dissimilarity (feces: R2 = 0.15, P = 0.04; small intestine: R2 = 0.23, P < 0.01), unweighted UniFrac (feces: R2 = 0.21, P < 0.01; small-intestine: R2 = 0.19, P < 0.01), and weighted UniFrac (feces: R2 = 0.27, P = 0.02; small-intestine: R2 = 0.22, P = 0.02) distances revealed that microbial communities differed among L. reevesii acclimated to three temperatures (Figure 3A). Weighted UniFrac distance differed among P. przewalskii acclimated to three temperatures (Figure 3A; feces: R2 = 0.21, P = 0.02).
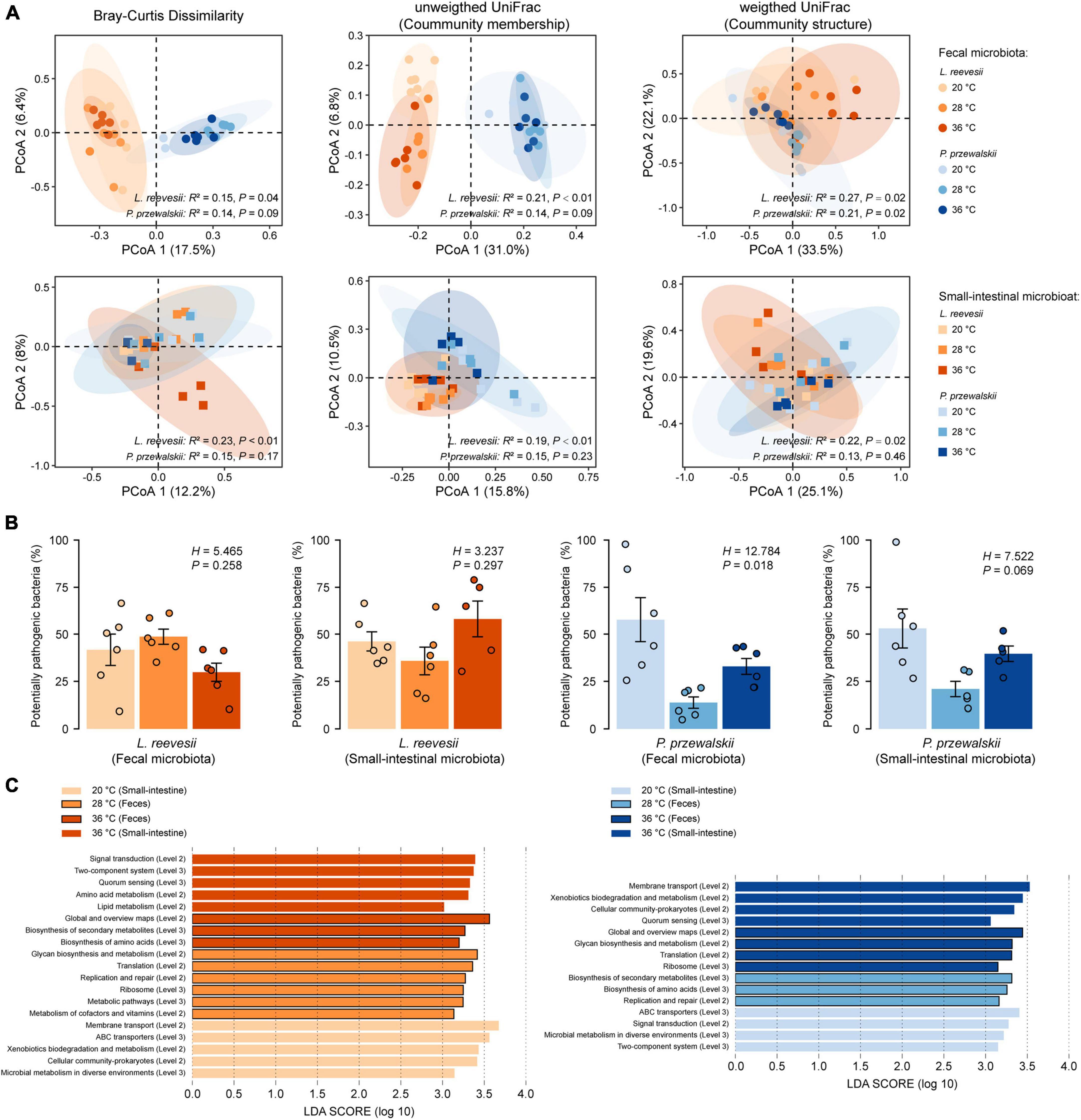
Figure 3. Beta diversity of the fecal and small-intestinal microbiota and the differences in bacterial phenotype and gene function among lizards acclimated to three temperatures (20, 28, and 36°C) for 2 weeks. (A) Results of principal coordinates analysis (PCoA) based on Bray–Curtis dissimilarity, unweighted UniFrac and weighted UniFrac distances. (B) The relative abundance of potentially pathogenic bacterial (inferred from BugBase) within the fecal and small-intestinal microbiota in L. reevesii and P. przewalskii. (C) KEGG pathway (Level 2 and Level 3) enrichment in lizards acclimated under three thermal conditions (LDA > 3 and P < 0.05).
More bacterial taxa are affected by acclimation temperature in L. reevesii
The LEfSe results (LDA > 3, P < 0.05) showed that bacterial taxa were significantly associated with temperature in L. reevesii, with two phyla (Bacteroidetes and Firmicutes) and seven genera of the two phyla in the fecal microbiota and one phylum (Proteobacteria) and six genera of the phyla Actinobacteria, Firmicutes, and Proteobacteria in the small-intestinal microbiota varying among the three temperature treatments (Supplementary Figure 3 and Table 2). In P. przewalskii, two phyla (Bacteroidetes and Verrucomicrobia) and four genera in the fecal microbiota varied among the three temperature treatments (Supplementary Figure 3 and Table 2). Specifically, the relative abundances of one phylum (Firmicutes) and four genera of the phylum were highest in the 36°C treatment in L. reevesii, and the relative abundances of two genera of the phylum Bacteroidetes were highest in the 36°C treatment in P. przewalskii. The relative abundance of one phylum (Proteobacteria) and three genera of the phylum were highest in the 20°C treatment and lowest in the 36°C treatment in L. reevesii (Table 2). Bacteria of the genera Clostridium, Parabacteroides, and Propionibacterium were enriched in L. reevesii acclimated to 28°C, and bacteria of the genera Akkermansia and Clostridium were enriched in P. przewalskii acclimated to the same temperature (Table 2). The relative abundance of the phylum Bacteroidetes increased in L. reevesii acclimated to 28°C but decreased in P. przewalskii acclimated to the same temperature (Supplementary Figures 3, 4).
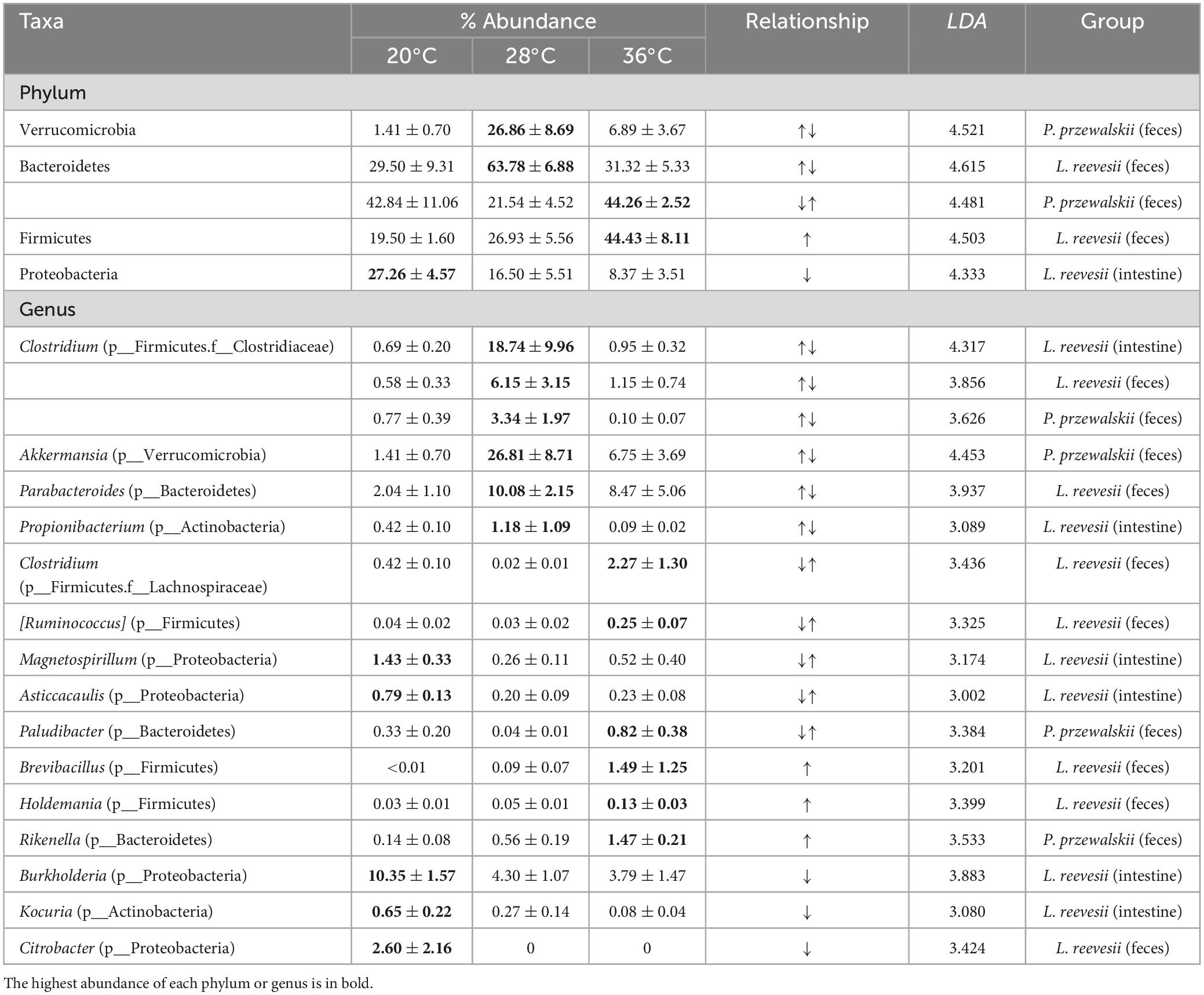
Table 2. Descriptive statistics, expressed as mean ± SE, for the relative abundances of the phyla and genera differing significantly among the three temperature treatments in two host lizard species.
Microbial functions are similar between L. reevesii and P. przewalskii
The relative abundance of potential pathogenic bacteria in the fecal microbiota increased dramatically in P. przewalskii acclimated to 20°C (Figure 3B; H2,18 = 12.784, P = 0.018). All other functional categories (aerobic, anaerobic and facultatively anaerobic bacteria, bacteria containing mobile elements, and biofilm-forming, Gram-negative, Gram-positive, and stress-tolerant bacteria) did not differ significantly among the three temperature treatments in both species (Supplementary Figure 5; Kruskal–Wallis test, all P > 0.05).
The relative abundances of the taxa increased at a given acclimation temperature differed between L. reevesii and P. przewalskii, but microbial functions enriched at the three acclimation temperatures were similar between the two species. The LEfSe results (LDA > 3, P < 0.05) showed that the two lizards were similar in the following two aspects. First, microbial pathways involved in global and overview maps, glycan biosynthesis and metabolism, and xenobiotics biodegradation and metabolism differed significantly among the three temperature treatments. Second, five functional categories (KEGG Level 2) associated with cellular processes, environmental information processing, and genetic information processing differed significantly among the three temperature treatments (Figure 3C). More specifically, two functional categories (Global and overview maps, replication and repair) significantly decreased in lizards acclimated to 20°C, and three functional categories [KEGG Level 3, including in metabolism in diverse environments (ko01120), ABC transporters (ko02010) and quorum sensing (ko02024)] significantly increased in lizards acclimated to 20 and 36°C (Figure 3C).
Some differences in the microbial function existed between L. reevesii and P. przewalskii. Specifically, three functional categories associated with cellular community-prokaryotes, membrane transport, and xenobiotics biodegradation and metabolism were enriched in L. reevesii acclimated to 20°C and P. przewalskii acclimated to 36°C (Figure 3C). On the contrary, signal transduction was enriched in L. reevesii acclimated to 36°C and P. przewalskii acclimated to 20°C (Figure 3C). Amino acid metabolism (LDA = 3.33, P < 0.01) and lipid metabolism (LDA = 3.01, P < 0.01) was enriched in L. reevesii acclimated to 36°C; metabolism of cofactors and vitamins (LDA = 3.14, P < 0.01) was enriched in L. reevesii acclimated to 28°C, but they did not differ among P. przewalskii acclimated to the three temperatures (Figure 3C).
Discussion
Interspecific differences in microbial community were more pronounced in the fecal microbiota than in the small-intestinal microbiota. Bacteroidetes and Firmicutes were the top two dominant phyla in the fecal microbiota and Firmicutes and Proteobacteria were the top two dominant phyla in the small-intestinal microbiota in both species. These findings are consistent with earlier studies on amphibians (Zhou et al., 2020; Zhu et al., 2021), lizards (Ren et al., 2016; Kohl et al., 2017; Zhang et al., 2018; Montoya-Ciriaco et al., 2020; Chen et al., 2022), snakes (Tang W.-J. et al., 2019; Smith et al., 2021; Zhong et al., 2022), and turtles (Qu et al., 2020) where Bacteroidetes, Firmicutes, and Proteobacteria are the top three dominant phyla in the fecal and/or gut microbiota. An earlier study on three sympatric lizard species (Liolaemus parvus, Liolaemus ruibali, and Phymaturus williamsi) shows the stomach, hindgut and fecal microbiota display more pronounced interspecific differences in microbial community than the small-intestinal microbiota (Kohl et al., 2017). We also found that the fecal microbial community was more dissimilar than the small-intestinal microbial community. For example, ASVs of the genera Akkermansia, Butyricicoccus, and Cronobacter were classed as species-specific taxa in the fecal microbial community, but as generalists in the small-intestinal microbial community. More species-specific (L. reevesii or P. przewalskii) taxa and fewer generalists led to more pronounced interspecific differences in the fecal microbial community than in the small-intestinal microbial community.
The fecal and small-intestinal microbiota could help lizards adapt to thermal environments in which they live, as revealed by the following two pieces of evidence. First, alpha diversity was significantly higher in the cold-climate species (P. przewalskii) than in the warm-climate species (L. reevesii). High species diversity and functional redundancy are indicators of microbial resilience to return to ecosystem stability after disruption (Sommer et al., 2017). For example, to satisfy the physiological or energetic demands of the host in high-altitude and cold habitats, alpha diversity of the gut microbiota increases with altitude in the plateau pika O. curzoniae (Li et al., 2019) and the domestic yak Bos grunniens (Fan et al., 2020). Second, functional genes associated with carbohydrate metabolism were enhanced in P. przewalskii, conforming to the metabolic cold adaption hypothesis which predicts an increase in the metabolic rate of ectotherms from cold environments compared with their more temperate counterparts (Clarke, 2003; Schaefer and Walters, 2010). In a recent study on two Phrynocephalus lizards occurring in thermally different regions, Chen et al. (2022) found that the gut microbiota had a higher abundance of functional categories associated with metabolism in a high-altitude species (Phrynocephalus erythrurus) occurring from 4,500 to 5,300 m than in a low-altitude species (P. przewalskii) occurring from 500 to 1,700 m. Here, we found that, as in cattle where Firmicutes assimilate nutrients more efficiently in warm tropical environments (Zhang et al., 2022), the relative abundance of Firmicutes in the small-intestinal microbiota was higher in the warm-climate species than in the cold-climate species. Firmicutes are more efficient in dietary caloric intake than Bacteroidetes (Turnbaugh et al., 2006; Krajmalnik-Brown et al., 2012; Semova et al., 2012), and short-chain fatty acids produced by Firmicutes can be directly absorbed by the host as an energy source to promote mass gain (den Besten et al., 2013). That Firmicutes play a more important role in the absorption of nutrients in the warm-climate species further proves the contribution of microbial community to thermal adaptation in lizards.
Environmental temperature alters alpha and beta diversities of the gut microbiota in a diverse array of animal taxa (e.g., Bestion et al., 2017; Fontaine et al., 2018; Zhu et al., 2019, 2021; Cao et al., 2022). Our results showed that the cold-climate species responded to temperature increase or decrease by adjusting alpha diversity of microbial community, whereas the warm-climate species did so by adjusting beta diversity of microbial community. Reduced alpha diversity has been reported for P. cinereus (Fontaine et al., 2018), the common lizard Zootoca vivipara (Bestion et al., 2017), and the domestic chicken Gallus domesticus (Zhu et al., 2019) exposed to warm thermal conditions. Reduced alpha diversity may result in a decrease in genetic diversity and thereby inhibit the host’s ability to respond to environmental stress and resist infection (Pollock et al., 2019). In this study, we found that, as in mice exposed to a high temperature of 36°C (Cao et al., 2022), alpha diversity (observed ASVs and Shannon indices) of the fecal microbiota increased with increasing temperatures in P. przewalskii. Such a temperature-dependent change in alpha diversity may be due to the relatively high proportions of Parabacteroides and Citrobacter in P. przewalskii acclimated to 20°C (Figure 1D). The increased relative abundance of some bacterial genera such as Citrobacter and Pseudomonas of the phylum Proteobacteria reduces microbial diversity in the gut of the brown tree frog Polypedates megacephalus artificially hibernating in the laboratory (Weng et al., 2016). In L. reevesii, however, we found that neither in the fecal microbiota nor in the small-intestinal microbiota did alpha diversity differ significantly among the three temperature treatments. This finding is consistent with previous studies on the northern leopard frog Rana pipiens (Kohl and Yahn, 2016) and the tropical clawed frog Xenopus tropicalis (Li et al., 2020). Temperature increase or decrease led to higher microbial heterogeneity in L. reevesii than in P. przewalskii (Figure 3), indicating that microbial communities were more changeable in the warm-climate species. Plasticity in the gut microbial community potentially helps host animals adapt to harsh environments with extreme climate events, limited food resources, or toxic exposure (Alberdi et al., 2016). The responses of microbial community to temperature increase or decrease differed between the two agamid lizards, and this difference presumably results from the fact that they use thermally different habitats and differ in thermal tolerance (Lin, 2008; Qu et al., 2011; Wang, 2013).
The microbial composition was thermally more sensitive in L. reevesii than in P. przewalskii. Fecal or small-intestinal bacteria of the phyla Bacteroidetes, Firmicutes, and Proteobacteria and 12 genera belonging to the phyla Actinobacteria, Bacteroidetes, Firmicutes, and Proteobacteria were affected by acclimation temperature in L. reevesii. However, in P. przewalskii, only fecal bacteria of the phyla Bacteroidetes and Verrucomicrobia and four genera belonging to the phyla Bacteroidetes, Firmicutes, and Verrucomicrobia were affected by acclimation temperature. The trends of variation in the relative abundance of Bacteroidetes and Verrucomicrobia differed between the two lizard species. More specifically, bacteria of the phylum Bacteroidetes enriched in the 20 and 36°C treatments in P. przewalskii, bacteria of the phylum Bacteroidetes enriched in the 28°C treatment in L. reevesii, and bacteria of the phylum Verrucomicrobia enriched in the 28°C treatment in P. przewalskii. Bacteria of the phyla Bacteroidetes and Verrucomicrobia can use host-derived mucin glycans in the absence of dietary substrates (Sonnenburg et al., 2005; Carey et al., 2013). It has been reported for R. dybowskii (Tong et al., 2020), I. tridecemlineatus (Dill-McFarland et al., 2014; Stevenson et al., 2014), and U. arctos (Sommer et al., 2016) that the increased relative abundance of Bacteroidetes or Verrucomicrobia and the decreased relative abundance of Firmicutes during hibernation help hosts adapt to long-term fasting in winter. The increased relative abundance of Verrucomicrobia was observed in L. reevesii acclimated to 20°C, though statistically not significant. Additionally, as in Andrias davidianus (Zhu et al., 2021), the relative abundance of Firmicutes and Proteobacteria increased at low or high acclimation temperatures in L. reevesii. In Drosophila subobscura, decisive effects of heat stress on the gut microbiota composition differ between the sexes, with warming reducing the relative abundances of Firmicutes in females, increasing the relative abundances of Firmicutes, and reducing the relative abundances of Proteobacteria in males (Jaramillo and Castañeda, 2021). The result for male D. subobscura is consistent with this study based on male adults of L. reevesii. High Firmicutes abundance (or Firmicutes/Bacteroidetes ratio) is related to obesity in humans (Clemente et al., 2012), and to weight gain and immune function in birds (Zhang et al., 2015) and mammals (Ley et al., 2005). The Firmicutes/Bacteroidetes ratio increased with elevated temperature in L. reevesii, indicating an increased capacity for energy harvest. However, in cold-climate vertebrates such as the Brandt’s vole Lasiopodomys brandtii (Bo et al., 2019) and the Tibetan sheep Ovis aries (Fan et al., 2021), cold stress increases the Firmicutes/Bacteroidetes ratio to maintain the host’s metabolic balance. Compared with active counterparts, hibernating animals often have a higher relative abundance of Proteobacteria in their gut microbiota (Weng et al., 2016; Song et al., 2020; Tong et al., 2020; Cao et al., 2023). Such a difference is also evident in animals not hibernating in winter (Sun et al., 2016; Ferguson et al., 2018), indicating that the bacterial phylum Proteobacteria may have an important role in helping hosts adapt to fasting (starvation) or cold conditions. It is worth noting that most taxa of the phylum Proteobacteria have pathogenic or antipathogenic functions (Wiggins et al., 2011; Ramsey et al., 2015). The relative abundances of putative pathogens did not differ significantly among three temperature treatments in L. reevesii, but were enriched in P. przewalskii acclimated to 20°C. The accelerated colonization of pathogenic bacteria in the gut at low temperatures has been observed in other cold-climate vertebrates (Liu et al., 2022; Tong et al., 2023). Bacteria of the genus Akkermansia can enhance metabolic homeostasis and anti-inflammatory properties (Everard et al., 2013; Shin et al., 2014). The relative abundance of Akkermansia bacteria decreased in P. przewalskii acclimated to 20°C, suggesting that, as in X. tropicalis (Li et al., 2020), low-temperatures exposure may lead to a decrease in immunity in this cold-climate lizard. Temperature-dependent changes in the microbial composition differed between L. reevesii and P. przewalskii, and this difference can be explained by the following two reasons. First, host genetic background is a key determinant of the microbial composition and structure (Kokou et al., 2018; Weinstein et al., 2021). Second, host thermal tolerance modulates the microbial response to temperature change (Kokou et al., 2018). Overall, our data showed that the microbial community was thermally more sensitive in the warm-climate lizard than in the cold-climate lizard, and that lizards can adapt to temperature increase or decrease by adjusting the gut microbiota.
Although the influence of acclimation temperature on the microbial composition and structure differed between L. reevesii and P. przewalskii, the two species exhibited similar microbial functions enriched at different acclimation temperatures. Pathways related to environmental information processing (i.e., ABC transporters and two-component system) were enriched in lizards acclimated to 20 or 36°C, confirming the existence of a functional response of the gut microbial community to temperature (Nachin et al., 2005). ABC transporters pathway uses ATP hydrolysis to transport a wide variety of substrates (i.e., sugars, amino acids, glycans, and phospholipids) (Gottesman and Ambudkar, 2001). This pathway might promote food absorption to help the host survive in the season or environment that is cold and/or short of food (Xia et al., 2021). Two-component system pathway is involved in the information communication between microbes and their environment, allowing bacteria to sense and respond to environmental change. Consistent with a study on goitred gazelles (Gazella subgutturosa) exposed to low temperatures in winter (Qin et al., 2022), our study showed that the relative abundant of two-component system was significantly higher in P. przewalskii acclimated to the lowest temperature (20°C). This finding suggests the role of two-component system in cold adaptation. Additionally, the temperature-dependence of pathways associated with amino acid metabolism, lipid metabolism, and metabolism of cofactors and vitamins seen in L. reevesii suggests that metabolic functions of bacteria are thermally more sensitive in warm-climate lizards. Consistent with studies on the Wistar rat Rattus norvegicus (Qu et al., 2021) and the rice frog Fejervarya limnocharis (Huang and Liao, 2021), amino acid metabolism and lipid metabolism were enriched in L. reevesii acclimated to 36°C. Overall, both species of lizards studied herein, L. reevesii in particular, could cope with temperature changes by adjusting the relative abundance of functional categories associated with metabolism and environmental information processing.
Conclusion
With the findings of this study, we can draw the following conclusions. First, the fecal microbiota displays more pronounced interspecific differences in microbial community than the small-intestinal microbiota in both species of lizards occurring in thermally different regions. Second, the response of fecal and small-intestinal microbiota to temperature increase or decrease differs between the warm-climate (L. reevesii) and cold-climate (P. przewalskii) species, with more bacterial taxa affected by acclimation temperature in L. reevesii than in P. przewalskii. Third, both species, the warm-climate species in particular, can cope with temperature changes by adjusting the relative abundance of functional categories associated with metabolism and environmental information processing. Fourth, the fecal microbiota contributes to cold-climate adaptation in P. przewalskii, as revealed by the fact that functional genes associated with carbohydrate metabolism were enhanced in this species. Taken together, our results validate the hypotheses tested, of which one suggests that the gut microbiota should help lizards adapt to thermal environments in which they live and one suggests that microbial communities should be thermally more sensitive in warm-climate lizards than in cold-climate lizards.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA901369.
Ethics statement
The animal study was approved by the Laboratory Animal Care and Animal Ethics Committee of Nanjing Normal University and Hainan Tropical Ocean University. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
X-MZ: Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. J-QC: Investigation, Writing – review & editing. YD: Funding acquisition, Investigation, Writing – review & editing. C-XL: Investigation, Resources, Writing – review & editing. Y-FQ: Data curation, Formal analysis, Writing – review & editing. L-HL: Conceptualization, Funding acquisition, Investigation, Supervision, Writing – original draft, Writing – review & editing. XJ: Conceptualization, Funding acquisition, Investigation, Project administration, Resources, Supervision, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by grants from the National Natural Science Foundation of China (32370519, 31870390, and 31971414), Natural Science Foundation of Zhejiang Province (LY23C030003), and Finance Science and Technology Project of Hainan Province (ZDYF2018219).
Acknowledgments
We thank Shan Su, Xia-Qiu Tao, and Lin Zhu who contributed to the field sampling. We also thank Yun Shang and Yan-Lin Xue for assistance in the laboratory.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1374209/full#supplementary-material
Supplementary Figure 1 | Rarefaction curves based on observed ASVs (A) and Shannon index (B) for each sample.
Supplementary Figure 2 | The relative abundance of the fecal (A) and small-intestinal (B) microbiota at the phylum and genus levels in L. reevesii and P. przewalskii. LC, LM, and LH represent L. reevesii acclimated to 20, 28, and 36°C, respectively; PC, PM, and PH represent P. przewalskii acclimated to 20, 28, and 36°C, respectively.
Supplementary Figure 3 | Biomarker taxa of the fecal and small-intestinal microbiota identified by LEfSe in lizards acclimated under three thermal conditions. (A) Fecal microbiota of L. reevesii; (B) small-intestinal microbiota of L. reevesii; (C) fecal microbiota of P. przewalskii; (D) small-intestinal microbiota of P. przewalskii. Letters p, c, o, f, and g represent phylum, class, order, family, and genus, respectively.
Supplementary Figure 4 | The relative abundance of key bacteria in lizards acclimated under three thermal conditions.
Supplementary Figure 5 | The relative abundance of potential microbial phenotypes (inferred from BugBase, except potentially pathogenic bacteria) within the fecal (A) and small-intestinal (B) microbiota in L. reevesii and P. przewalskii.
Footnotes
References
Alberdi, A., Aizpurua, O., Bohmann, K., Zepeda-Mendoza, M. L., and Gilbert, M. T. P. (2016). Do vertebrate gut metagenomes confer rapid ecological adaption? Trends Ecol. Evol. 31, 689–699. doi: 10.1016/j.tree.2016.06.008
Arango, R. A., Schoville, S. D., Currie, C. R., and Carlos-Shanley, C. (2021). Experimental warming reduces survival, cold tolerance, and gut prokaryotic diversity of the eastern subterranean termite, Reticulitermes flavipes (Kollar). Front. Microbiol. 12:632715. doi: 10.3389/fmicb.2021.632715
Bestion, E., Jacob, S., Zinger, L., Di Gesu, L., Richard, M., White, J., et al. (2017). Climate warming reduces gut microbiota diversity in a vertebrate ectotherm. Nat. Ecol. Evol. 1:0161. doi: 10.1038/s41559-017-0161
Bo, T.-B., Zhang, X.-Y., Wen, J., Deng, K., Qin, X.-W., and Wang, D.-H. (2019). The microbiota-gut-brain interaction in regulating host metabolic adaptation to cold in male Brandt’s voles (Lasiopodomys brandtii). ISME J. 13, 3037–3053. doi: 10.1038/s41396-019-0492-y
Bolar, K. (2019). STAT: Interactive Document for Working with Basic Statistical Analysis. Vienna: R Foundation for Statistical Computing.
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13:581. doi: 10.1038/nmeth.3869
Cao, H.-W., Shi, Y.-P., Wang, J., Niu, Z.-Y., Wei, L., Tian, H.-B., et al. (2023). The intestinal microbiota and metabolic profiles of Strauchbufo raddei underwent adaptive changes during hibernation. Integr. Zool. [Online ahead of print]. doi: 10.1111/1749-4877.12749
Cao, Y., Liu, Y., Dong, Q.-Y., Wang, T., and Niu, C. (2022). Alterations in the gut microbiome and metabolic profile in rats acclimated to high environmental temperature. Microb. Biotechnol. 15, 276–288. doi: 10.1111/1751-7915.13772
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Carey, H. V., Walters, W. A., and Knight, R. (2013). Seasonal restructuring of the ground squirrel gut microbiota over the annual hibernation cycle. Am. J. Physiol. Reg. I 304, R33–R42. doi: 10.1152/ajpregu.00387.2012
Chen, J.-Q., Zhang, L.-W., Zhao, R.-M., Wu, H.-X., Lin, L.-H., Li, P., et al. (2022). Gut microbiota differs between two cold-climate lizards distributed in thermally different regions. BMC Ecol. Evol. 22:120. doi: 10.1186/s12862-022-02077-8
Chevalier, C., Stojanović, O., Colin, D. J., Suarez-Zamorano, N., Tarallo, V., Veyrat-Durebex, C., et al. (2015). Gut microbiota orchestrates energy homeostasis during cold. Cell 163, 1360–1374. doi: 10.1016/j.cell.2015.11.004
Chiang, E., Deblois, C. L., Carey, H. V., and Suen, G. (2022). Characterization of captive and wild 13-lined ground squirrel cecal microbiotas using Illumina-based sequencing. Anim. Microbiome 4:1. doi: 10.1186/s42523-021-00154-9
Clarke, A. (2003). Costs and consequences of evolutionary temperature adaptation. Trends Ecol. Evol. 18, 573–581. doi: 10.1016/j.tree.2003.08.00
Clemente, J. C., Ursell, L. K., Parfrey, L. W., and Knight, R. (2012). The impact of the gut microbiota on human health: An integrative view. Cell 148, 1258–1270. doi: 10.1016/j.cell.2012.01.035
den Besten, G., van Eunen, K., Groen, A. K., Venema, K., Reijngoud, D. J., and Bakker, B. M. (2013). The role of short-chain fatty acids in the interplay between diet, gut microbiota, and host energy metabolism. J. Lipid Res. 54, 2325–2340. doi: 10.1194/jlr.R036012
DeSantis, T. Z., Hugenholtz, P., Larsen, N., Rojas, M., Brodie, E. L., Keller, K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microb. 72, 5069–5072. doi: 10.1128/AEM.03006-05
Dill-McFarland, K. A., Neil, K. L., Zeng, A., Sprenger, R. J., Kurtz, C. C., Suen, G., et al. (2014). Hibernation alters the diversity and composition of mucosa-associated bacteria while enhancing antimicrobial defence in the gut of 13-lined ground squirrels. Mol. Ecol. 23, 4658–4669. doi: 10.1111/mec.12884
Douglas, G. M., Maffei, V. J., Zaneveld, J. R., Yurgel, S. N., Brown, J. R., Taylor, C. M., et al. (2020). PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 38, 685–688. doi: 10.1038/s41587-020-0548-6
Du, Y., Chen, J.-Q., Liu, Q., Fu, J.-C., Lin, C.-X., Lin, L.-H., et al. (2022). Dietary correlates of oral and gut microbiota in the water monitor lizard, Varanus salvator (Laurenti, 1768). Front. Microbiol. 12:771527. doi: 10.3389/fmicb.2021.771527
Du, Y., Lin, C.-X., Lin, L.-H., Qiu, Q.-B., and Ji, X. (2011). Ontogenetic shifts in sexual dimorphism and female reproduction in the Reeves’s Butterfly Lizard Leiolepis reevesii from Hainan, China. J. Herpetol. 45, 399–405. doi: 10.1670/10-100.1
Everard, A., Belzer, C., Geurts, L., Ouwerkerk, J. P., Druart, C., Bindels, L. B., et al. (2013). Cross-talk between Akkermansia muciniphila and intestinal epithelium controls diet-induced obesity. Proc. Natl. Acad. Sci. U. S. A. 110, 9066–9071. doi: 10.1073/pnas.1219451110
Fan, Q.-S., Cui, X.-X., Wang, Z.-F., Chang, S.-H., Wanapat, M., Yan, T.-H., et al. (2021). Rumen microbiota of Tibetan sheep (Ovis aries) adaptation to extremely cold season on the Qinghai-Tibetan Plateau. Front. Vet. Sci. 8:673822. doi: 10.3389/fvets.2021.673822
Fan, Q.-S., Wanapat, M., Yan, T.-H., and Hou, F. J. (2020). Altitude influences microbial diversity and herbage fermentation in the rumen of yaks. BMC Microbiol. 20:370. doi: 10.1186/s12866-020-02054-5
Ferguson, L. V., Dhakal, P., Lebenzon, J. E., Heinrichs, D. E., Bucking, C., and Sinclair, B. J. (2018). Seasonal shifts in the insect gut microbiome are concurrent with changes in cold tolerance and immunity. Funct. Ecol. 32, 2357–2368. doi: 10.1111/1365-2435.13153
Fontaine, S. S., and Kohl, K. D. (2020). Gut microbiota of invasive bullfrog tadpoles responds more rapidly to temperature than a noninvasive congener. Mol. Ecol. 29, 2449–2462. doi: 10.1111/mec.15487
Fontaine, S. S., Novarro, A. J., and Kohl, K. D. (2018). Environmental temperature alters the digestive performance and gut microbiota of a terrestrial amphibian. J. Exp. Biol. 221:jeb187559. doi: 10.1242/jeb.187559
Fu, H.-B., Zhang, L.-Z., Fan, C., Liu, C.-F., Li, W.-J., Cheng, Q., et al. (2020). Environment and host species identity shape gut microbiota diversity in sympatric herbivorous mammals. Microb. Biotechnol. 14, 1300–1315. doi: 10.1111/1751-7915.13687
Gottesman, M. M., and Ambudkar, S. V. (2001). Overview: ABC transporters and human disease. J. Bioenerg. Biomembr. 33, 453–458. doi: 10.1023/a:1012866803188
Hassenrück, C., Reinwald, H., Kunzmann, A., Tiedemann, I., and Gärdes, A. (2021). Effects of thermal stress on the gut microbiome of juvenile milkfish (Chanos chanos). Microorganisms 9:5. doi: 10.3390/microorganisms9010005
Huang, C.-H., and Liao, W.-B. (2021). Seasonal variation in gut microbiota related to diet in Fejervarya limnocharis. Animals 11:1393. doi: 10.3390/ani11051393
Huang, G.-P., Wang, L., Li, J., Hou, R., Wang, M., Wang, Z.-L., et al. (2022). Seasonal shift of the gut microbiome synchronizes host peripheral circadian rhythm for physiological adaptation to a low-fat diet in the giant panda. Cell Rep. 38:110203. doi: 10.1016/j.celrep.2021.110203
Jaramillo, A., and Castañeda, L. E. (2021). Gut microbiota of Drosophila subobscura contributes to its heat tolerance and is sensitive to transient thermal stress. Front. Microbiol. 12:654108. doi: 10.3389/fmicb.2021.654108
Katoh, K., Misawa, K., Kuma, K., and Miyata, T. (2002). MAFFT: A novel method for rapid multiple sequence alignment based on fast Fourier transform. Nucleic Acids Res. 30, 3059–3066. doi: 10.1093/nar/gkf436
Khakisahneh, S., Zhang, X.-Y., Nouri, Z., and Wang, D.-H. (2020). Gut microbiota and host thermoregulation in response to ambient temperature fluctuations. mSystems 5, e514–e520. doi: 10.1128/mSystems.00514-20
Kohl, K. D., Brun, A., Magallanes, M., Brinkerhoff, J., Laspiur, A., Acosta, J. C., et al. (2017). Gut microbial ecology of lizards: Insights into diversity in the wild, effects of captivity, variation across gut regions and transmission. Mol. Ecol. 26, 1175–1189. doi: 10.1111/mec.13921
Kohl, K. D., and Yahn, J. (2016). Effects of environmental temperature on the gut microbial communities of tadpoles. Environ. Microbiol. 18, 1561–1565. doi: 10.1111/1462-2920.13255
Kokou, F., Sasson, G., Nitzan, T., Doron-Faigenboim, A., Harpaz, S., Cnaani, A., et al. (2018). Host genetic selection for cold tolerance shapes microbiome composition and modulates its response to temperature. eLife 7:e36398. doi: 10.7554/eLife.36398
Krajmalnik-Brown, R., Ilhan, Z. E., Kang, D. W., and Dibaise, J. K. (2012). Effects of gut microbes on nutrient absorption and energy regulation. Nutr. Clin. Pract. 27, 201–214. doi: 10.1177/0884533611436116
Ley, R. E., Bäckhed, F., Turnbaugh, P., Lozupone, C. A., Knight, R. D., and Gordon, J. I. (2005). Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. U. S. A. 102, 11070–11075. doi: 10.1073/pnas.0504978102
Ley, R. E., Hamady, M., Lozupone, C., Turnbaugh, P. J., Ramey, R. R., Bircher, J. S., et al. (2008). Evolution of mammals and their gut microbes. Science 320, 1647–1651. doi: 10.1126/science.1155725
Li, H., Zhou, R., Zhu, J.-X., Huang, X.-D., and Qu, J.-P. (2019). Environmental filtering increases with elevation for the assembly of gut microbiota in wild pikas. Microb. Biotechnol. 12, 976–992. doi: 10.1111/1751-7915.13450
Li, J.-Y., Rui, J.-P., Li, Y.-L., Tang, N., Zhan, S.-P., Jiang, J.-P., et al. (2020). Ambient temperature alters body size and gut microbiota of Xenopus tropicalis. Sci. China Life Sci. 63, 915–925. doi: 10.1007/s11427-019-9540-y
Lin, C.-X., Du, Y., Qiu, Q.-B., and Ji, X. (2007). Relatively high but narrow incubation temperatures in lizards depositing eggs in warm and thermally stable nests. Acta Zool. Sin. 53, 437–445.
Lin, L.-H. (2008). Population Density, Thermal Requirement and Geographic Pattern of Genetic Structure in the Reeves’ Butterfly Lizard (Leiolepis reevesii) from Hainan, Southern China. Nanjing: Nanjing Normal University.
Liu, S., Xiao, Y.-H., Wang, X.-F., Guo, D.-G., Wang, Y.-M., and Wang, Y. (2023). Effects of microhabitat temperature variations on the gut microbiotas of free-living hibernating animals. Microbiol. Spectr. 11:e00433-23. doi: 10.1128/spectrum.00433-23
Liu, W.-L., Yang, J., Meng, Y., Wu, D.-Y., Cui, L.-X., Li, T., et al. (2022). The divergent effects of moderate climate warming on the gut microbiota and energetic state of cold-climate lizards from open and semi-closed microhabitats. Front. Microbiol. 13:1050750. doi: 10.3389/fmicb.2022.1050750
Moeller, A. H., Lvey, K., Cornwall, M. B., Herr, K., Rede, J., Taylor, E. N., et al. (2020). The lizard gut microbiome changes with temperature and is associated with heat tolerance. Appl. Environ. Microb. 86:e01181-20. doi: 10.1128/AEM.01181-20
Montoya-Ciriaco, N., Gómez-Acata, S., Muñoz-Arenas, L. C., Dendooven, L., Estrada-Torres, A., de la Vaga-Pérez, A. H. D., et al. (2020). Dietary effects on gut microbiota of the mesquite lizard Sceloporus grammicus (Wiegmann, 1828) across different altitudes. Microbiome 8:6. doi: 10.1186/s40168-020-0783-6
Nachin, L., Nannmark, U., and Nyström, T. (2005). Differential roles of the universal stress proteins of Escherichia coli in oxidative stress resistance, adhesion, and motility. J. Bacteriol. 187, 6265–6272. doi: 10.1128/JB.187.18.6265-6272.2005
Oksanen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2017). Vegan: Community Ecology Package. Vienna: R Foundation for Statistical Computing.
Pollock, F. J., Lamb, J. B., van de Water, J. A. J. M., Smith, H. A., Schaffelke, B., Willis, B. L., et al. (2019). Reduced diversity and stability of coral-associated bacterial communities and suppressed immune function precedes disease onset in corals. R. Soc. Open Sci. 6:190355. doi: 10.1098/rsos.190355
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2-approximately maximum likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490
Qin, W., Song, P.-F., and Zhang, S.-D. (2022). Seasonal and soil microbiota effects on the adaptive strategies of wild goitered gazelles based on the gut microbiota. Front. Microbiol. 13:918090. doi: 10.3389/fmicb.2022.918090
Qu, Q., Li, H., Bai, L., Zhang, S.-W., Sun, J.-Q., Lv, W. J., et al. (2021). Effects of heat stress on gut microbiome in rats. Indian J. Microbiol. 61, 338–347. doi: 10.1007/s12088-021-00948-0
Qu, Y.-F., Li, H., Gao, J.-F., Xu, X.-F., and Ji, X. (2011). Thermal preference, thermal tolerance and the thermal dependence of digestive performance in two Phrynocephalus lizards (Agamidae), with a review of species studied. Curr. Zool. 57, 684–700. doi: 10.1093/czoolo/57.6.684
Qu, Y.-F., Wu, Y.-Q., Zhao, Y.-T., Lin, L.-H., Du, Y., Li, P., et al. (2020). The invasive red-eared slider turtle is more successful than the native Chinese three-keeled pond turtle: Evidence from the gut microbiota. PeerJ 8:e10271. doi: 10.7717/peerj.10271
Ramsey, J. P., Mercurio, A., Holland, J. A., Harris, R. N., and Minbiole, K. P. C. (2015). The cutaneous bacterium Janthinobacterium lividum inhibits the growth of Trichophyton rubrum in vitro. Int. J. Dermatol. 54, 156–159. doi: 10.1111/ijd.12217
Regan, M. D., Chiang, E., Liu, Y.-X., Tonelli, M., Verdoorn, K. M., Gugel, S. R., et al. (2022). Nitrogen recycling via gut symbionts increases in ground squirrels over the hibernation season. Science 375, 460–463. doi: 10.1126/science.abh2950
Ren, T.-T., Kahrl, A. F., Wu, M., and Cox, R. M. (2016). Does adaptive radiation of a host lineage promote ecological diversity of its bacterial communities? A test using gut microbiota of Anolis lizards. Mol. Ecol. 25, 4793–4804. doi: 10.1111/mec.13796
Schaefer, J., and Walters, A. (2010). Metabolic cold adaptation and developmental plasticity in metabolic rates among species in the Fundulus notatus species complex. Funct. Ecol. 24, 1087–1094. doi: 10.1111/j.1365-2435.2010.01726.x
Segata, N., Izard, J., Waldron, L., Gevers, D., Miropolsky, L., Garrett, W. S., et al. (2011). Metagenomic biomarker discovery and explanation. Genome Biol. 12:R60. doi: 10.1186/gb-2011-12-6-r60
Semova, I., Carten, J. D., Stombaugh, J., Mackey, L. C., Knight, R., Farber, S. A., et al. (2012). Microbiota regulate intestinal absorption and metabolism of fatty acids in the zebrafish. Cell Host Microbe 12, 277–288. doi: 10.1016/j.chom.2012.08.003
Shin, N.-R., Lee, J.-C., Lee, H.-Y., Kim, M.-S., Whon, T.-W., Lee, M.-S., et al. (2014). An increase in the Akkermansia spp. population induced by metformin treatment improves glucose homeostasis in diet-induced obese mice. Gut 63, 727–735. doi: 10.1136/gutjnl-2012-303839
Smith, S. N., Colston, T. J., and Siler, C. D. (2021). Venomous snakes reveal ecological and phylogenetic factors influencing variation in gut and oral microbiomes. Front. Microbiol. 12:657754. doi: 10.3389/fmicb.2021.657754
Sommer, F., Anderson, J. M., Bharti, R., Raes, J., and Rosenstiel, P. (2017). The resilience of the intestinal microbiota influences health and disease. Nat. Rev. Microbiol. 15, 630–638. doi: 10.1038/nrmicro.2017.58
Sommer, F., Ståhlman, M., Ilkayeva, O., Arnemo, J. M., Kindberg, J., Josefsson, J., et al. (2016). The gut microbiota modulates energy metabolism in the hibernating brown bear Ursus arctos. Cell Rep. 14, 1655–1661. doi: 10.1016/j.celrep.2016.01.026
Song, X.-W., Zhang, J.-W., Song, J.-H., and Zhai, Y.-Y. (2020). Decisive effects of life stage on the gut microbiota discrepancy between two wild populations of hibernating Asiatic toads (Bufo gargarizans). Front. Microbiol. 12:665849. doi: 10.3389/fmicb.2021.665849
Sonnenburg, J. L., Xu, J., Leip, D. D., Chen, C.-H., Westover, B. P., Weatherford, J., et al. (2005). Glycan foraging in vivo by an intestine-adapted bacterial symbiont. Science 307, 1955–1959. doi: 10.1126/science.1109051
Stevenson, T. J., Duddleston, K. N., and Buck, C. L. (2014). Effects of season and host physiological state on the diversity, density, and activity of the arctic ground squirrel cecal microbiota. Appl. Environ. Microb. 80, 5611–5622. doi: 10.1128/AEM.01537-14
Sun, B.-H., Wang, X., Bernstein, S., Huffman, M. A., Xia, D.-P., Gu, Z.-Y., et al. (2016). Marked variation between winter and spring gut microbiota in free-ranging Tibetan macaques (Macaca thibetana). Sci. Rep. 6:26035. doi: 10.1038/srep26035
Tang, K.-Y., Wang, Z.-W., Wan, Q.-H., and Fang, S.-G. (2019). Metagenomics reveals seasonal functional adaptation of the gut microbiome to host feeding and fasting in the Chinese alligator. Front. Microbiol. 10:2409. doi: 10.3389/fmicb.2019.02409
Tang, W.-J., Zhu, G.-X., Shi, Q., Yang, S.-J., Ma, T.-Y., Mishra, S. K., et al. (2019). Characterizing the microbiota in gastrointestinal tract segments of Rhabdophis subminiatus: Dynamic changes and functional predictions. MicrobiologyOpen 8:e00789. doi: 10.1002/mbo3.789
Tong, Q., Diong, W.-J., Xu, M.-D., Hu, Z.-F., Guo, P., Han, X. Y., et al. (2023). Characteristics and a comparison of the gut microbiota in two frog species at the beginning and end of hibernation. Front. Microbiol. 14:1057398. doi: 10.3389/fmicb.2023.1057398
Tong, Q., Hu, Z.-F., Du, X.-P., Bie, J., and Wang, H.-B. (2020). Effects of seasonal hibernation on the similarities between the skin microbiota and gut microbiota of an amphibian (Rana dybowskii). Microb. Ecol. 79, 898–909. doi: 10.1007/s00248-019-01466-9
Turnbaugh, P. J., Ley, R. E., Mahowald, M. A., Magrini, V., Mardis, E. R., and Gordon, J. I. (2006). An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 444, 1027–1031. doi: 10.1038/nature05414
Wang, N.-B. (2013). The Comparison of Thermoregulation between Phrynocephalus erythrurus and Phrynocephalus przewalskii. Gansu: Lanzhou University.
Wang, Z. (2011). Adapting to Extreme Climate: The Evolution of Viviparity in Phrynocephalus lizards. Nanjing: Nanjing Normal University.
Wang, Z., Lu, H.-L., Ma, L., and Ji, X. (2013a). Differences in thermal preference and tolerance among three Phrynocephalus lizards (Agamidae) with different body sizes and habitat use. Asian Herpetol. Res. 4:214–220. doi: 10.3724/SP.J.1245.2013.00214
Wang, Z., Ma, L., Shao, M., and Ji, X. (2013b). Differences in incubation length and hatchling morphology among five oviparous Phrynocephalus lizards (Agamidae) from China. Asian Herpetol. Res. 4, 225–232. doi: 10.3724/SP.J.1245.2013.00225
Ward, T., Larson, J., Meulemans, J., Hillmann, B., Lynch, J., Sidiropoulos, D., et al. (2017). BugBase predicts organism level microbiome phenotypes. bioRxiv [Preprint]. doi: 10.1101/133462
Weinstein, S. B., Martínez-Mota, R., Stapleton, T. E., Klure, D. M., Greenhalgh, R., Orr, T. J., et al. (2021). Microbiome stability and structure is governed by host phylogeny over diet and geography in woodrats (Neotoma spp.). Proc. Natl. Acad. Sci. U. S. A. 118:e2108787118. doi: 10.1073/pnas.2108787118
Weng, F. C.-H., Yang, Y.-J., and Wang, D. (2016). Functional analysis for gut microbes of the brown tree frog (Polypedates megacephalus) in artificial hibernation. BMC Genom. 17:1024. doi: 10.1186/s12864-016-3318-6
Wiggins, P. J., Smith, J. M., Harris, R. N., and Minbiole, K. P. C. (2011). Gut of red-backed salamanders (Plethodon cinereus) may serve as a reservoir for an antifungal cutaneous bacterium. J. Herpetol. 45, 329–332. doi: 10.1670/10-231.1
Xia, T.-R., Yao, Y.-F., Wang, C., Dong, M.-M., Wu, Y.-H., Li, D.-Y., et al. (2021). Seasonal dynamics of gut microbiota in a cohort of wild Tibetan macaques (Macaca thibetana) in western China. Glob. Ecol. Conserv. 25:e01409. doi: 10.1016/j.gecco.2020.e01409
Xiao, G.-H., Liu, S., Xiao, Y.-H., Zhu, Y., Zhao, H.-B., Li, A.-Q., et al. (2019). Seasonal changes in gut microbiota diversity and composition in the greater horseshoe bat. Front. Microbiol. 10:2247. doi: 10.3389/fmicb.2019.02247
Zhang, L., Li, J., Yun, T.-T., Qi, W.-T., Liang, X.-X., Wang, Y.-W., et al. (2015). Effects of pre-encapsulated and pro-encapsulated Enterococcus faecalis on growth performance, blood characteristics, and cecal microflora in broiler chickens. Poult. Sci. 94, 2821–2830. doi: 10.3382/ps/pev262
Zhang, W.-Y., Li, N., Tang, X.-L., Liu, N.-F., and Zhao, W. (2018). Changes in intestinal microbiota across an altitudinal gradient in the lizard Phrynocephalus vlangalii. Ecol. Evol. 8, 4695–4703. doi: 10.1002/ece3.4029
Zhang, X.-H., Cui, K., Wen, X.-B., Li, L.-B., Yu, X.-C., Li, B.-L., et al. (2022). The association between gut microbiome diversity and composition and heat tolerance in cattle. Microorganisms 10:1672. doi: 10.3390/microorganisms10081672
Zhong, J., Guo, K., Liao, Z.-L., Hu, S.-C., Du, Y., and Ji, X. (2022). Comparative analysis of the gut microbiota composition between two sea snakes, Hydrophis curtus, and Hydrophis cyanocinctus. Coral Reefs 41, 53–62. doi: 10.1007/s00338-021-02200-z
Zhou, J., Nelson, T. M., Lopez, C. R., Sarma, R. R., Zhou, S.-J., and Rollins, L. A. (2020). A comparison of nonlethal sampling methods for amphibian gut microbiome analyses. Mol. Ecol. Resour. 20, 844–855. doi: 10.1111/1755-0998.13139
Zhu, L.-F., Zhu, W., Zhao, T., Chen, H., Zhao, C.-L., Xu, L.-L., et al. (2021). Environmental temperatures affect the gastrointestinal microbes of the Chinese giant salamander. Front. Microbiol. 12:543767. doi: 10.3389/fmicb.2021.543767
Keywords: 16S rRNA gene sequencing, cold-climate lizard, fecal and small-intestinal microbiota, thermal adaptation, warm-climate lizard
Citation: Zhu X-M, Chen J-Q, Du Y, Lin C-X, Qu Y-F, Lin L-H and Ji X (2024) Microbial communities are thermally more sensitive in warm-climate lizards compared with their cold-climate counterparts. Front. Microbiol. 15:1374209. doi: 10.3389/fmicb.2024.1374209
Received: 21 January 2024; Accepted: 01 April 2024;
Published: 15 April 2024.
Edited by:
Ekaterina Avershina, Oslo University Hospital, NorwayReviewed by:
Sunil Banskar, University of Arizona, United StatesTingbei Bo, Institute of Zoology (CAS), China
Copyright © 2024 Zhu, Chen, Du, Lin, Qu, Lin and Ji. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiang Ji, eGppQHd6dS5lZHUuY24=; Long-Hui Lin, bGlubGhAaHpudS5lZHUuY24=
 Xia-Ming Zhu
Xia-Ming Zhu Jun-Qiong Chen
Jun-Qiong Chen Yu Du
Yu Du Chi-Xian Lin
Chi-Xian Lin Yan-Fu Qu
Yan-Fu Qu Long-Hui Lin
Long-Hui Lin Xiang Ji
Xiang Ji