- 1School of Life Sciences, Fujian Normal University, Fuzhou, Fujian, China
- 2Department of Chemistry, University of Nebraska-Lincoln, Lincoln, NE, United States
Siderophores are produced by bacteria in iron-restricted conditions. However, we found maltose could induce the biosynthesis of the siderophore lysochelin in Lysobacter sp. 3655 in rich media that are not compatible with siderophore production. Maltose markedly promoted cell growth, with over 300% increase in cell density (OD600) when LB medium was added with maltose (LBM). While lysochelin was not detectable when OD600 in LBM was below 5.0, the siderophore was clearly produced when OD600 reached 7.5 and dramatically increased when OD600 was 15.0. Coincidently, the transcription of lysochelin biosynthesis genes was remarkably enhanced following the increase of OD600. Conversely, the iron concentration in the cell culture dropped to 1.2 μM when OD600 reached 15.0, which was 6-fold lower than that in the starting medium. Moreover, mutants of the maltose-utilizing genes (orf2677 and orf2678) or quorum-sensing related gene orf644 significantly lowered the lysochelin yield. Transcriptomics analysis showed that the iron-utilizing/up-taking genes were up-regulated under high cell density. Accordingly, the transcription of lysochelin biosynthetic genes and the yield of lysochelin were stimulated when the iron-utilizing/up-taking genes were deleted. Finally, lysochelin biosynthesis was positively regulated by a TetR regulator (ORF3043). The lysochelin yield in orf3043 mutant decreased to 50% of that in the wild type and then restored in the complementary strain. Together, this study revealed a previously unrecognized mechanism for lysochelin biosynthetic regulation, by which the siderophore could still be massively produced in Lysobacter even grown in a rich culture medium. This finding could find new applications in large-scale production of siderophores in bacteria.
Introduction
Iron is an essential nutrient for the growth of microorganisms, since it is needed for several fundamental cellular processes and important proteins containing heme group or iron-sulfur cluster (Frausto da Silva and Williams, 2001; Miethke and Marahiel, 2007). However, microorganisms could not easily utilize iron due to its extremely low solubility, 10−18 M at neutral pH in natural habitat (Payne, 1988). To overcome iron limitation, microbes have evolved to produce siderophores that bind iron with high affinity from surroundings and then migrate iron to inside of cells through specific receptor and transport system (Wandersman and Delepelaire, 2004). Bacteria and fungi, as well as some plants, secret various types of siderophores under iron-deficiency environment (Timofeeva et al., 2022). More than 500 siderophores have been found and 300 of them are structurally characterized (Hider and Kong, 2010). Depending on their chemical nature, siderophores are classified into four general types, catecholate, hydroxamate, carboxylate, and mixed types (Barry and Challis, 2009; Ustiatik et al., 2021; Gao and Bian, 2022).
Due to the high affinity for iron, siderophores exhibit great potential for applications in agriculture, pharmaceutics, and environmental protection. The siderophore-antibiotic conjugation is an attractive strategy to enhance uptake and antibacterial potency against multi-drug resistant pathogens (Caradec et al., 2023). Siderophore-producing microbes have the potential to be developed into eco-friendly and sustainable agents to mitigate plant stresses in degraded lands (Singh et al., 2022). For practical applications, siderophores need to be generated easily with a high yield. Chemical synthesis is an option but usually unfeasible due to the cost of multi-step reactions and low yield of siderophores (Cezard et al., 2015). Therefore, using microbial fermentation for siderophore production is a promising alternative approach. Siderophores are produced only under iron-restricted conditions, which means their biosynthesis is tightly regulated. The culture media are vital for siderophores to be produced on a large scale. Usually, synthetic media are selected to cultivate the microbes for siderophores production due to the ease of controlling the iron concentration in the media (Chincholkar et al., 2007). Several synthetic media have been reported for the production of siderophores, such as minimal medium 1 (MM1) for Bacillus megaterium (Ferreira et al., 2019), minimal medium 2 (MM2) for Streptomyces olivaceus (Meiwes et al., 1990), minimal medium 9 (MM9) with casamino acids for marine bacteria (Hao et al., 2012), minimal succinate-containing medium (MMS) for Pseudomonas aeruginosa (Sasirekha and Srividya, 2016), and Burk's medium for Azotobacter vinelandii (Baars et al., 2016). Iron concentration is the key factor for siderophores biosynthesis, and the concentration that inhibits the generation of siderophores depends on the microbe and culture medium. In Bacillus sp. PZ-1, the inhibition of siderophore production was observed with the iron concentration above 5 μM, and completely inhibited with the concentration of 100 μM (Yu et al., 2017), but in Azotobacter vinelandii, the concentration of initial inhibition and complete inhibition was 1 and 10 μM, respectively (Cornish and Page, 1995). As iron is essential for microbial growth, a low iron concentration decreases cell growth and thus impairs the yield of siderophores. Therefore, it is important to find a balanced iron concentration that is low enough to stimulate siderophore production and, at the same time, not to limit the cell growth to reach a proper biomass, which directly impacts the total yield of siderophores. For Pseudomonas aeruginosa, the iron concentration of 1 μM in the culture medium supplemented with suitable carbon source could ensure the cell growth and siderophores production (Sexton and Schuster, 2017). Various carbon sources affect the production of siderophores, for example, succinate causes huge production of pyoverdines from Pseudomonas genus (Meyer and Abdallah, 1978; Hoegy et al., 2014; Vindeirinho et al., 2021), and glycerol is the optimal carbon source for the siderophores production in Bacillus megaterium, Bacillus sp. PZ-1, and Escherichia coli (Valdebenito et al., 2006; Santos et al., 2014; Yu et al., 2017).
In many bacteria, the ferric uptake regulator (Fur) is the main regulator for the biosynthesis and transportation of siderophores. When the intracellular iron reaches a certain concentration, Fur binds to ferric iron and then represses the expression of iron uptake genes directly or indirectly; when the iron concentration becomes limiting, Fur is free from ferric iron binding and the repression by Fur is abolished, allowing for iron uptake into cells (Seo et al., 2014; Beauchene et al., 2015). Other regulators have been identified as well, such as PchR for pyochelin activation and PvdS and FpvI for pyoverdines production in Pseudomonas aeruginosa (Schalk et al., 2020).
Lysobacter is a genus of rod-shaped, gliding Gram-negative bacteria widely distributed in soil and fresh water (Christensen and Cook, 1978). The genus is recognized as a new source of lead compounds for antibiotics due to their prolific production of various bioactive natural products, including polycyclic tetramate macrolactams, cyclic lipodespsipeptides, cephem-type β-lactams, and phenazines (Yue et al., 2022). Lysobacter species also exhibit potential in biocontrol of crop diseases for their potent activity to kill a variety of plant pathogenic bacteria, oomycetes, and fungi (Puopolo et al., 2018). In previous work, our lab reported the siderophore lysochelin isolated from L. enzymogenes, which was the first siderophore identified from the Lysobacter genus (Miller et al., 2023). Lysochelin is biosynthesized by the lec gene cluster through an NRPS (non-ribosomal peptide synthetase)-independent biosynthetic pathway. Its structure contains two units of 2,3-dihydroxybenzoic acid (2,3-DHB) and one unit of spermidine (Miller et al., 2023). Despite the progress, the regulatory mechanism underlying lysochelin production has not been investigated. Here, we found that maltose is a strong inducer of lysochelin production in Lysobacter sp. 3655. Maltose promoted Lysobacter cell growth to a markedly high density in rich culture media without prior removal of iron. While rich media are not a favorable condition for siderophore production, we found that both the transcription of lysochelin biosynthetic genes and the lysochelin yield were dramatically enhanced when Lysobacter cells reached a high density. Further results showed that the maltose-promoted growth significantly reduced the iron concentration in the rich media, which created an iron-restricted environment that stimulated the siderophore biosynthesis in rich media. Moreover, we found that lysochelin production in rich media was related to maltose metabolism and quorum sensing. We also found that a TetR regulator was involved in the regulation of maltose metabolism and lysochelin production. Together, the study revealed a new way for lysochelin biosynthesis in rich media, which could be applied for scale-up production of siderophores.
Methods and materials
Bacterial strains, plasmids, and growth conditions
Bacterial strains and plasmids used in this study are shown in Supplementary Table 1. Luria-Bertani (LB) broth medium was used for the growth of Lysobacter sp. 3655 and L. enzymogenes OH11. LBM (1% tryptone, 0.5% yeast extract, 1% NaCl, and 5% maltose) was used for the growth, siderophore production and RNA extraction of strain 3655 and its mutants. LB supplemented with various carbohydrates [5% sucrose (LBS), 5% glucose (LBG), 1% mannose (LBMA), 1% galactose (LBGA), 1% rhamnose (LBR), and 1% fucose (LBF)] were used to explore the siderophore-producing potential of strain 3655. MM813 (0.4% glucose, 0.3% K2HPO4, 0.138% NaH2PO4•H2O, 0.1% NH4Cl, 0.0144% MgSO4, 0.015% KCl, and 0.00111% CaCl2) was used for the lysochelin production of L. enzymogenes OH11 (Miller et al., 2023). LBM supplemented with various concentrations of FeSO4 (final concentration of 0, 1, 5, 10, 20, and 40 μM) was used to evaluate the effect of iron on lysochelin production in strain 3655. LB with different amounts of maltose (0, 0.5, 1.0, 2.5, and 5%, w/v) was also used to assess the effect of maltose on the yield of lysochelin in strain 3655. Escherichia coli strain DH 5α was cultured at 37°C in LB medium supplemented with gentamicin (Gm, 50 μg/ml) to propagate plasmids. E. coli strain S17 was used for intergeneric conjugation.
DNA manipulation and Lysobacter transformation
Chromosomal DNA and plasmids were isolated from Lysobacter sp. 3655 or E. coli according to the standard techniques (Sambrook et al., 1989). Database searching and sequence analysis were performed using the online program PSI-BLAST (Altschul et al., 1997). For Lysobacter transformation, plasmids were first introduced into the E. coli S17 and then transferred to Lysobacter sp. 3655 by intergeneric conjugation. The transformants were spread on LB plates with kanamycin (Km, 100 μg/ml) and Gentamicin (Gm, 150 μg/ml). After growing at 30°C for 72 h, the single-crossover colonies were selected and plated on LB plates containing 10% (w/v) sucrose and Km (100 μg/ml), 30°C for 72 h. Then the colonies were transferred to LB plates supplemented with Km (100 μg/ml) or Km (100 μg/ml) + Gm (150 μg/ml). The Km resistant and Gm sensitive colonies were the putative double-crossover mutants, which were selected for PCR verification.
Primers and PCR
All primers used in this study are listed in Supplementary Table 2. PCRs were carried out using Phanta® Max Super-Fidelity DNA polymerase (Vazyme) or rTaq DNA polymerase (Takara). For Phanta DNA polymerase, an initial denaturation at 95°C for 3 min was followed by 30 cycles of amplification (95°C for 15 s, 60°C for 15 s, and 72°C for 1 min), and additional 5 min at 72°C. For rTaq DNA polymerase, an initial denaturation at 95°C for 5 min was followed by 30 cycles of amplification (95°C for 30 s, 60°C for 30 s, and 72°C for 1 min), and additional 10 min at 72°C. Considering different DNA templates and primers, the annealing temperature and the elongation time were changed in some cases.
Construction of deletion mutants through homologous recombination
To construct the deletion mutant of siderophore biosynthetic genes (orf2903, orf2904, orf2905, and orf2906), maltose catabolic genes (orf2677, orf2678, and orf2742), and other relevant genes (orf644, orf4279, orf4990-orf4993, and orf3043), the DNA fragments corresponding to the upstream and downstream region of above genes were amplified by using respective primers. Then the upstream region was treated either with XhoI and HindIII (for orf2903, orf2904, orf2905, orf2906, orf644, orf4279, orf4990, orf4992, and orf4993) or with ApaI and HindIII (for orf2677, orf2678, orf2742, orf3043, and orf4991), and the downstream region was treated with HindIII and SpeI. Each pairs of the digested upstream and downstream DNA fragments were ligated into the XhoI/SpeI sites or ApaI/SpeI sites of plasmid pJQ200SK to generate the recombination plasmids. The plasmids were introduced into strain 3655 by intergeneric conjugation. After antibiotic and sucrose screening, the transformants were used for PCR verification by corresponding primers.
For mutant complementation, the DNA fragment containing the upstream region, downstream region and the deletion part of orf3043 was amplified using primers ORF3043UF/DR. Then the fragment was treated with ApaI/SpeI and ligated into the same sites of plasmid pJQ200SK to generate the recombination plasmid pJQ200SK::ORF3043C. The recombination plasmid was transferred into ΔORF3043 strain by intergeneric conjugation. After antibiotic and sucrose screening, the transformants were used for PCR verification by primers ORF3043VFO/VRO.
Growth assay of strain 3655 and its mutants
Strain 3655 and its mutants were cultured in 3 ml of LB for 24 h, then transferred to 25 ml LBM medium, 30°C for 72 h. For the growth assay of strain 3655 in different culture volumes, 1% of the 3 ml culture was transferred to 500 ml flasks containing 25, 50, 100, or 200 ml LBM medium, respectively. The OD600 values were determined every 12 h using a spectrophotometer (UV-8000, METASH).
Lysochelin extraction, HPLC and LC-MS analysis, and quantification
Strain 3655 and its mutants were grown in 3 ml of LB for 24 h, then inoculated to 25 ml LBM medium. The cultures were incubated at 30°C for 72 h. To extract lysochelin, the cultures were extracted with an equal volume of ethyl acetate (containing 0.1% TFA). The ethyl acetate extract was dried using a rotavapor (Buchi, Rotavapor R-200) to afford the crude extract, which was dissolved in 1 ml methanol. A 20 μl aliquot of each of the extracts was analyzed by HPLC (UltiMate 3000, Thermo) using a reversed phase C18 column (AcclaimTM 120 C18, 250 × 4.6 mm), equipped with a UV detector set at 318 nm, with a flow rate of 1 ml/min. The mobile phase comprised solvents A (water containing 0.05% formic acid) and B (acetonitrile containing 0.05% formic acid) using the following gradient elution program: 5–25% B in A for 0–5 min, increased to 80% B at 20 min and maintained for 1 min, then increased to 100% B for 26–28 min, and returned to 5% B in the final 2 min. Lysochelin was confirmed using LC-MS (Thermo, LCQ FLEET). The quantification of lysochelin was based on the HPLC peak aera divided by the OD600 value of the same sample. Unless otherwise specified, the yield of lysochelin was presented as fold change in comparison with the yield of the relative control strain such as the wild-type strain cultured in LBM medium.
RNA isolation and qRT-PCR
RNA was isolated from the wild-type strain 3655 and its mutants cultured in LBM medium for 24 h using SPARKeasy Improved Bacteria RNA Kit (SparkJade). RNA was reverse transcribed to complementary DNA by using EasyScript one-step gDNA removal and cDNA synthesis Supermix (Transgen). qRT-PCR was carried out in LightCycler® 96 Instrument (Roche, Inc) using PerfectStart Green qPCR Supermix (Transgen) with primers listed in Supplementary Table 2. The conditions are used as follows: 94°C for 30 s, followed by 40 cycles of 94°C for 5 s, 60°C for 15 s, and 72°C for 10 s. The 16S rRNA was used as an internal control. The relative transcriptional levels of the genes of interest were normalized to 16S rRNA and determined by using the 2−ΔΔCT method (Livak and Schmittgen, 2001). The values were presented as fold change in comparison with the relative expression levels for each gene at the first test time point in the wild-type strain. Data are presented as the averages of three independent experiments conducted in triplicate.
Determination of iron concentration
Strain 3655 was cultured in LBM medium at 30°C to the OD600 value of 5.0 and 15.0, respectively. The supernatants of two samples were saved by centrifugation (10,000 rpm, 1 min). Then the iron concentrations in LBM medium (used as control) and supernatants were determined using NexION 300D ICP-MS (Perkin Elmer, USA).
RNA sequencing and transcriptomic analysis
Strain 3655 was grown in LBM medium at 30°C to the OD600 value of 5.0 and 15.0, respectively. The cell pellets of two samples were collected by centrifugation (10,000 rpm, 1 min). RNA was extracted via Total RNA Extractor (Trizol) Kit (Sangon, China), and the purify was determined using Qubit 2.0 RNA Detection Kit (Life, USA). To reduce sequencing interference, rRNA was removed by Ribo-off rRNA Depletion Kit (Vazyme, China), and DNA was digested using DNase I (Vazyme, China). The fragmentation, reverse-transcription, 3' ends dA-Tailing, adapter ligation and DNA library construction of purified RNA were performed using VAHTSTM Stranded mRNA-seq Library Prep Kit for Illumina® (Vazyme, China).
The DNA library was sequenced by Illumina NovaSeq 6000, and the raw data were cleaned and evaluated whether the sequencing data was suitable for subsequent analysis. Then the clean reads were spliced through Rockhopper, the CDS prediction and annotation were performed using TransDecoder and NCBI Blast. The expression levels were analyzed using Salmon and WGCNA, and the differentially expressed genes were obtained by DESeq2 with a selection threshold of q-value < 0.05 and log2 fold change ≥ 1.0. Genes and Genomes Ontology (GO) and Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis were performed using topGO and clusterProfiler software, respectively.
Statistical analysis
All experiments were conducted three times, and all data were expressed as mean ± standard deviation. The SPSS (version 25.0, SPSS Inc., Chicago, ILL, USA) was used for statistical analysis that was performed using one-way analysis of variance (ANOVA) and Tukey's test at p < 0.05 or p < 0.01.
Results
Maltose induces the siderophore lysochelin production in a rich medium
During the study of bioactive natural products in Lysobacter sp. 3655, we serendipitously found that adding maltose (5%, w/v) into LB medium (LBM medium) could significantly induce the production of a compound (1) with the retention time around 9.3–9.4 min, which was the main product under UV absorption at 318 nm (Figure 1A). This compound was not detectable in the same culture grown in LB medium. Mass spectrometry gave an m/z 418.28 of [M+H]+ (Figure 1B). The retention time and mass of this compound were identical to that of lysochelin, a siderophore recently identified from L. enzymogenes OH11 (Supplementary Figure 1; Miller et al., 2023). Lysochelin is biosynthesized through the lec gene cluster in L. enzymogenes (Miller et al., 2023). We searched for homologous genes of the lec cluster in strain 3655 and found Cluster-7, which contains six genes, orf2903, orf2904, orf2905, orf2906, orf2907, and orf2908 that are homologs of lecA, lecB, lecC, lecD, lecE, and lecF, respectively. The deduced amino acid sequences of the six genes in strain 3655 share an identity of 86, 82, 92, 84, 79, and 84%, respectively, with that of the six lec genes in L. enzymogenes OH11 (Supplementary Figure 2, Supplementary Table 3). The presence of an entire lec homologous cluster in strain 3655 supports that this Lysobacter is also a lysochelin producer. To obtain evidence, we generated gene deletion mutants of orf2903, orf2904, orf2905, and orf2906 (Supplementary Figure 3). Indeed, the deletion of these genes abolished the production of compound 1 in LBM medium (Figure 1C), consistent with the compound 1 being lysochelin. Moreover, the 2,3-Dihydroxybenzoate (2,3-DHB), which is the precursor of lysochelin, was massively accumulated in the mutants of orf2904 and orf2906 (Figure 1C). The lecB homolog orf2904 encodes (2,3-dihydroxybenzoyl) adenylate synthase, which converts 2,3-DHB to its adenosylation form (2,3-DHB-AMP), and the lecD homolog orf2906 encodes the condensation enzyme that catalyzes two amide bonds between two units of 2,3-DHB-AMP and spermidine, resulting in lysochelin formation (Miller et al., 2023). The results indicated that the biosynthetic pathway of lysochelin in Lysobacter sp. 3655 is the same as that in L. enzymogenes OH11. Interestingly, the transcriptional level of the lysochelin biosynthetic genes (orf2903 and orf2905) was dramatically increased in LBM medium, whereas the transcription level of the genes in LB medium was barely detectable (Figure 1D). The results show that maltose is a potent inducer of the siderophore lysochelin biosynthesis in Lysobacter sp. 3655.
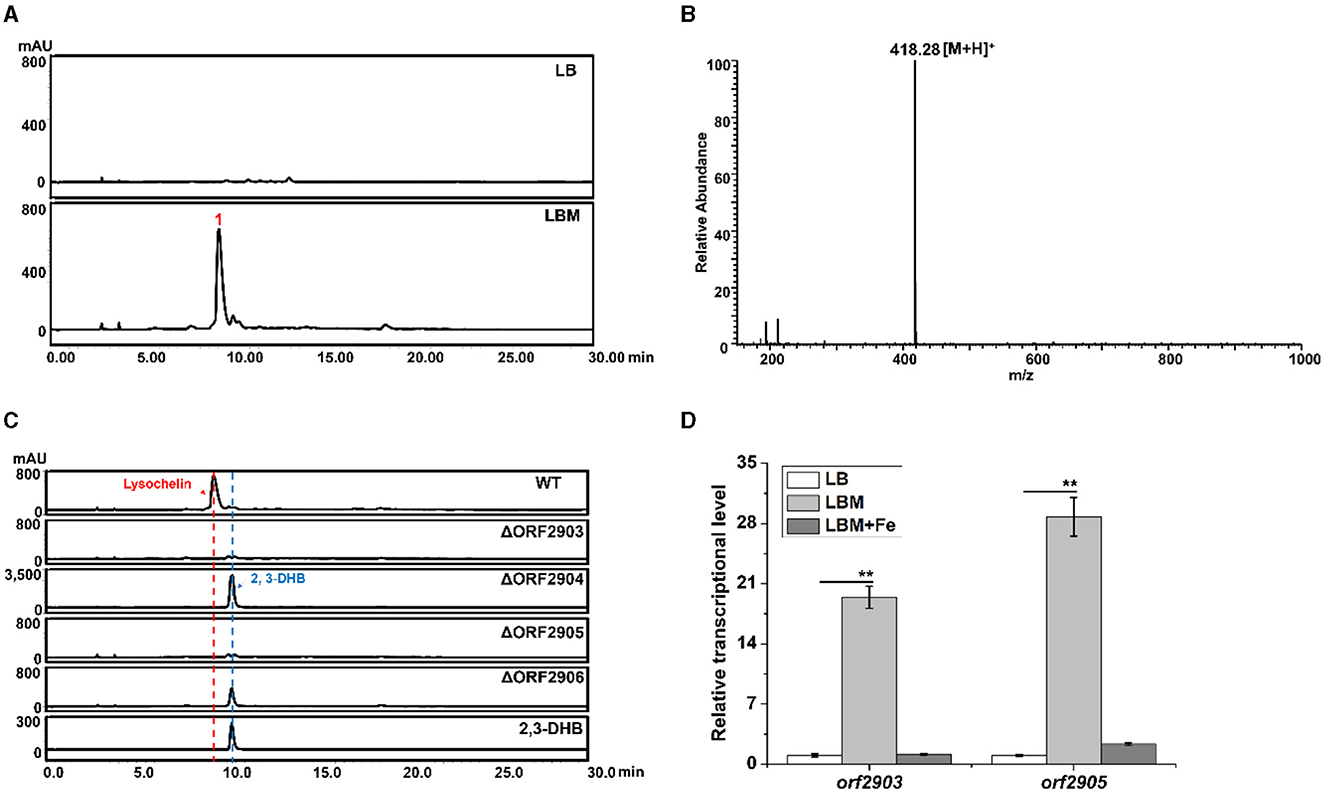
Figure 1. Production of the siderophore lysochelin in rich media. (A) HPLC analysis of the culture extracts from Lysobacter sp. 3655 wild type (WT) grown in LB medium or LBM medium for 72 h. (B) LC-MS analysis of compound 1. (C) HPLC analysis of the culture extracts from WT, deletion mutants ΔORF2903, ΔORF2904, ΔORF2905, and ΔORF2906 grown in LBM medium for 72 h. Standard 2, 3-DHB was included as a reference compound. (D) Transcriptional analysis of the lysochelin biosynthetic genes (orf2903 and orf2905) of WT grown in LB medium (white columns), LBM medium (light-gray columns), and LBM with FeSO4 (final concentration 20 μM, gray columns) for 24 h, **p < 0.01.
Maltose promotes vigorous growth of Lysobacter sp. 3655 leading to an iron-depleted condition
Siderophores are commonly produced under an iron-limited environment. For example, lysochelin was produced by L. enzymogenes grown in a modified minimal medium (MM813) without iron ion (Miller et al., 2023). LB is a eutrophic complex medium, and strain 3655 could not yield lysochelin in this medium (Figure 1A), indicating that LB is not an iron-deficient medium. However, the addition of maltose into LB could dramatically induce lysochelin production (Figure 1A). We proposed two hypotheses, one being that maltose might inhibit iron utilization in strain 3655 that would trigger the strain to synthesize lysochelin to compensate the iron deficiency in cells; the other being that maltose might cause the iron depletion in LB that would induce lysochelin biosynthesis in strain 3655. To obtain evidence, we added iron ion exogenously into LBM medium and evaluated lysochelin production. The results showed that the lysochelin level quickly decreased with the increase of iron concentration. Lysochelin was not detectable when the iron concentration reached 20 μM (Figure 2A). Furthermore, the transcriptional level of orf2903 and orf2905 in LBM returned to the level in LB when 20 μM iron was added to LBM (Figure 1D). This suggested that maltose probably might have caused a depletion of iron in LB medium which resulted in lysochelin biosynthesis in this rich medium. We compared the growth of strain 3655 cultured in LB and LBM media. Surprisingly, strain 3655 was flourishing in LBM medium with the maximum OD600 value of 18.5 at 72 h, while in LB medium strain 3655 reached its maximum growth at 36 h with the OD600 value of 6.0 (Figure 2B). This showed that maltose could dramatically promote the growth of strain 3655, which might have caused the lysochelin induction.
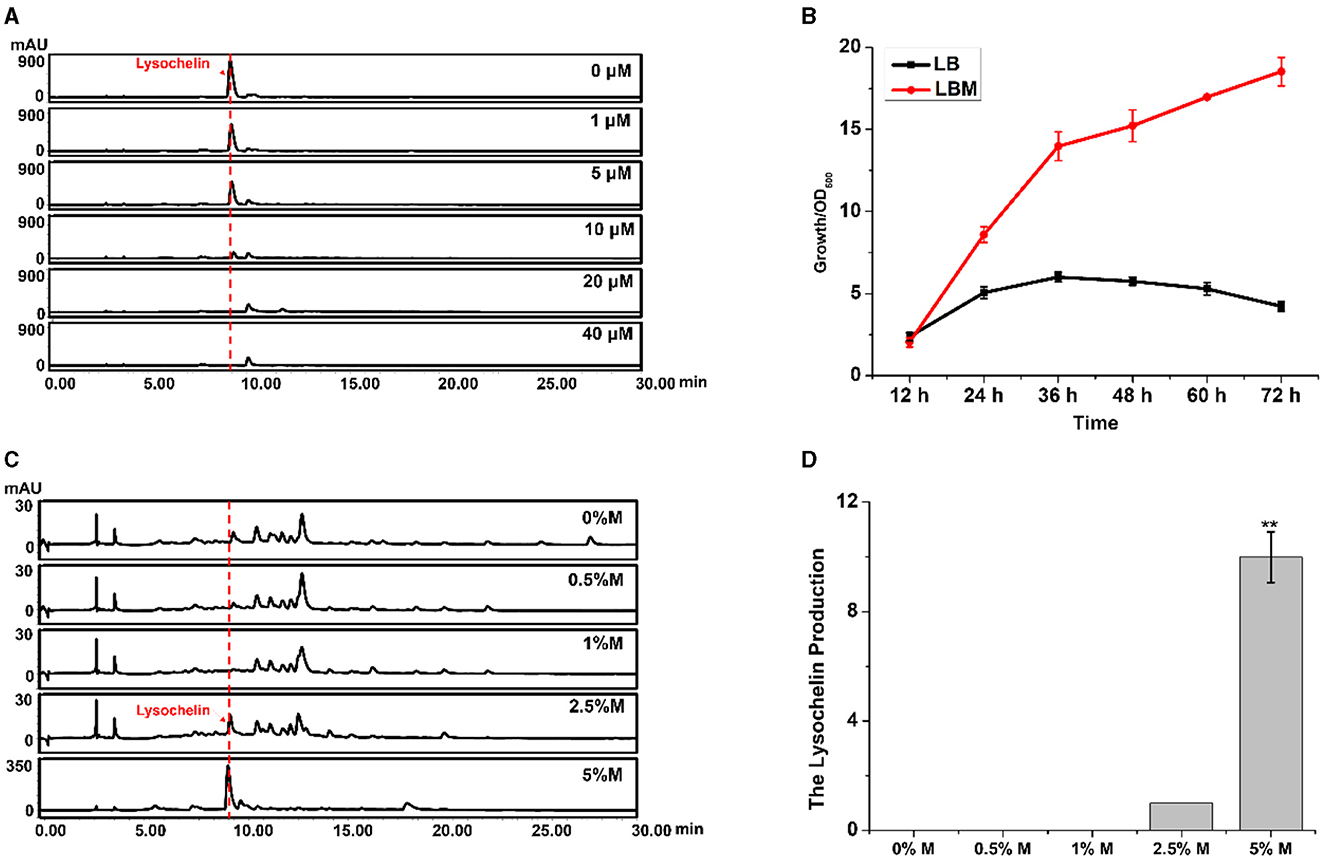
Figure 2. Effects of iron and maltose concentrations on lysochelin production. (A) HPLC analysis of lysochelin production in WT grown in LBM medium containing different concentrations of FeSO4 (final concentration 0, 1, 5, 10, 20, and 40 μM) for 72 h. (B) The growth curve of WT grown in LB medium (black line) and LBM medium (red line). (C) HPLC analysis of lysochelin production in WT grown in LB medium containing different concentrations of maltose (0, 0.5, 1, 2.5, and 5%) for 72 h. M, maltose. (D) Quantification of lysochelin in the cultures from (C). **p < 0.01.
Thus, we evaluated the effect of maltose concentration on lysochelin production. Strain 3655 could not generate lysochelin when the maltose concentration was below 1.0%, while lysochelin was clearly detected when the media contained a higher maltose concentration such as 2.5 and 5.0% (Figures 2C, D). Also, the cell density of strain 3655 was significantly enhanced at maltose concentration of 2.5 and 5.0% (Supplementary Figure 4). Moreover, strain 3655 produced a variable amount of lysochelin when cultured in different volumes of LBM medium. The yield of lysochelin was significantly decreased with the enlargement of cultural volume; the lysochelin production in 200 ml LBM medium decreased 5-fold when compared to that in 25 ml medium (Supplementary Figures 5A, B). Accordingly, the cell density of strain 3655 dramatically reduced when cultured in larger volumes (Supplementary Figure 5C). Together, the results showed that lysochelin biosynthesis was induced at high cell density and abolished at low cell density.
We investigated the point of growth at which lysochelin could be detected in strain 3655. There was not a detectable amount of lysochelin when the OD600 value was 1.0 or 5.0, a detectable amount when the OD600 value reached 7.5, and a significant amount when the OD600 value increased to 12 and then 15 (Figures 3A, B). Coincidently, the transcriptional level of orf2903 and orf2905 at the OD600 value of 12.0 was about 20–70-fold higher than that at the OD600 value of 1.0 (Figure 3C). We speculated that there was a sufficient amount of iron for the growth of strain 3655 at low cell density and, as a result, the biosynthesis of lysochelin was not turned on. However, as maltose could markedly promote strain 3655 to high-density growth, iron was used up by the “old” cells and the “new” cells would sense the iron-restriction condition, which resulted in lysochelin biosynthesis. To verify the speculation, we determined the iron concentration in LBM medium and the supernatant of cell cultures at OD600 5.0 and 15.0. The results showed that the iron concentration was 7.5 μM in LBM medium, decreased to 4.5 μM in the supernatant of low cell density (OD600 value of 5.0), and further reduced to 1.2 μM in the supernatant of high cell density (OD600 value of 15.0; Figure 3D). The data strongly supported that maltose induced lysochelin biosynthesis via promoting vigorous growth of strain 3655 to create iron-deficiency for “new” cells.
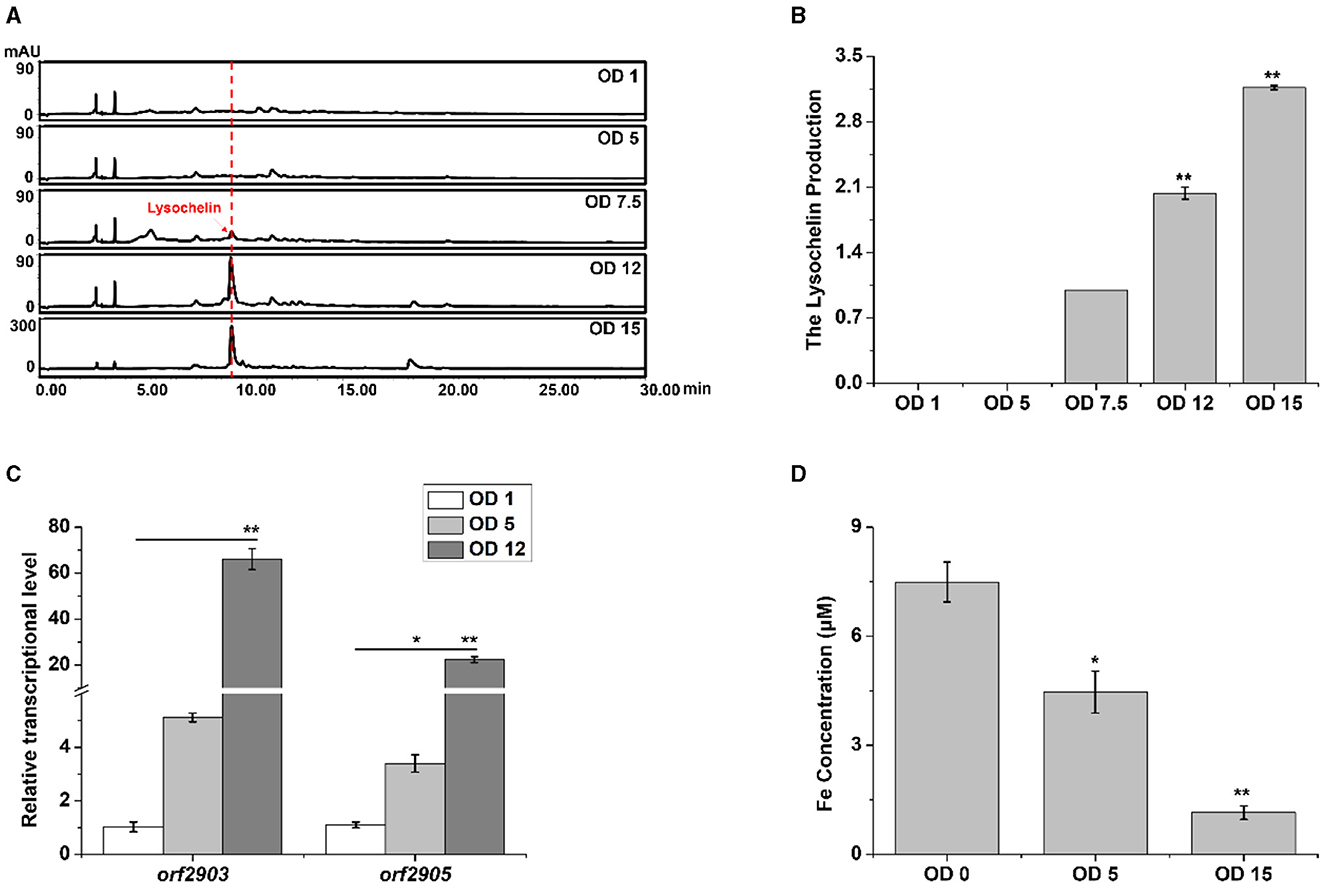
Figure 3. Induction of lysochelin biosynthesis by high cell density. (A) HPLC analysis of lysochelin production in WT grown in LBM medium at different OD600 values. (B) Quantification of lysochelin in the cultures from (A). (C) Transcriptional analysis of the lysochelin biosynthetic genes (orf2903 and orf2905) of WT grown in LBM at OD600 1 (white columns), 5 (light-gray columns), and 12 (gray columns). (D) The iron concentration in the supernatant of cell cultures at OD600 0, 5, and 15. The blank LBM medium was used as control (OD600 0). *p < 0.05, **p < 0.01.
Next, we examined the effect of other carbohydrates on induction of lysochelin biosynthesis. Glucose could facilitate the high-density growth of strain 3655 (OD600 value of 15.0) to produce lysochelin, while sucrose barely promoted the growth (OD600 value of 6.0) or lysochelin production (Supplementary Figures 6A, B). Interestingly, adding other carbohydrates, such as mannose, galactose, rhamnose, and fucose into LB medium inhibited the growth of strain 3655 (Supplementary Figure 6C). The results support the notion that lysochelin biosynthesis in strain 3655 might be triggered by the high cell density.
Lysochelin biosynthesis relates to maltose utilization and quorum sensing
Since maltose could induce lysochelin biosynthesis by promoting strain 3655 to high cell density, we investigated the effect of maltose utilization on lysochelin production. Three maltose-utilizing genes orf2677, orf2678, and orf2742 were found through homology search (Supplementary Figures 7–9). The gene orf2677 encodes a malE-like protein which is a periplasmic maltose binding protein functioning as the primary receptor for the active transport of maltose (Boos and Shuman, 1998). The gene orf2678 encodes a malF-like protein which is an intrinsic membrane protein of the transport system for maltose (Boos and Shuman, 1998). The gene orf2742 encodes maltose hydrolase responsible for cleaving maltose into two glucose molecules (Breton et al., 2005). We created the deletion mutants of orf2677, orf2678, and orf2742 (Supplementary Figure 10), and the results showed that the mutants of orf2677 or orf2678 significantly reduced lysochelin production, indicating the negative effect of an impaired maltose uptake system on siderophore biosynthesis. The yield of lysochelin in strain ΔORF2677 and ΔORF2678 decreased to 50% of that in the wild-type (WT), and accordingly the cell density of ΔORF2677 and ΔORF2678 was also lowered when compared to that of WT (Figure 4A and Supplementary Figures 11A, B). As expected, the transcriptional level of lysochelin biosynthetic genes orf2903 and orf2905 was significantly decreased in strains ΔORF2677 and ΔORF2678 (Figure 4B). However, lysochelin production and cell density of strain ΔORF2742 were similar to that of WT (Figure 4A and Supplementary Figures 11A, B), suggesting that the mutation of this maltose hydrolase alone did not cause a significant impact.
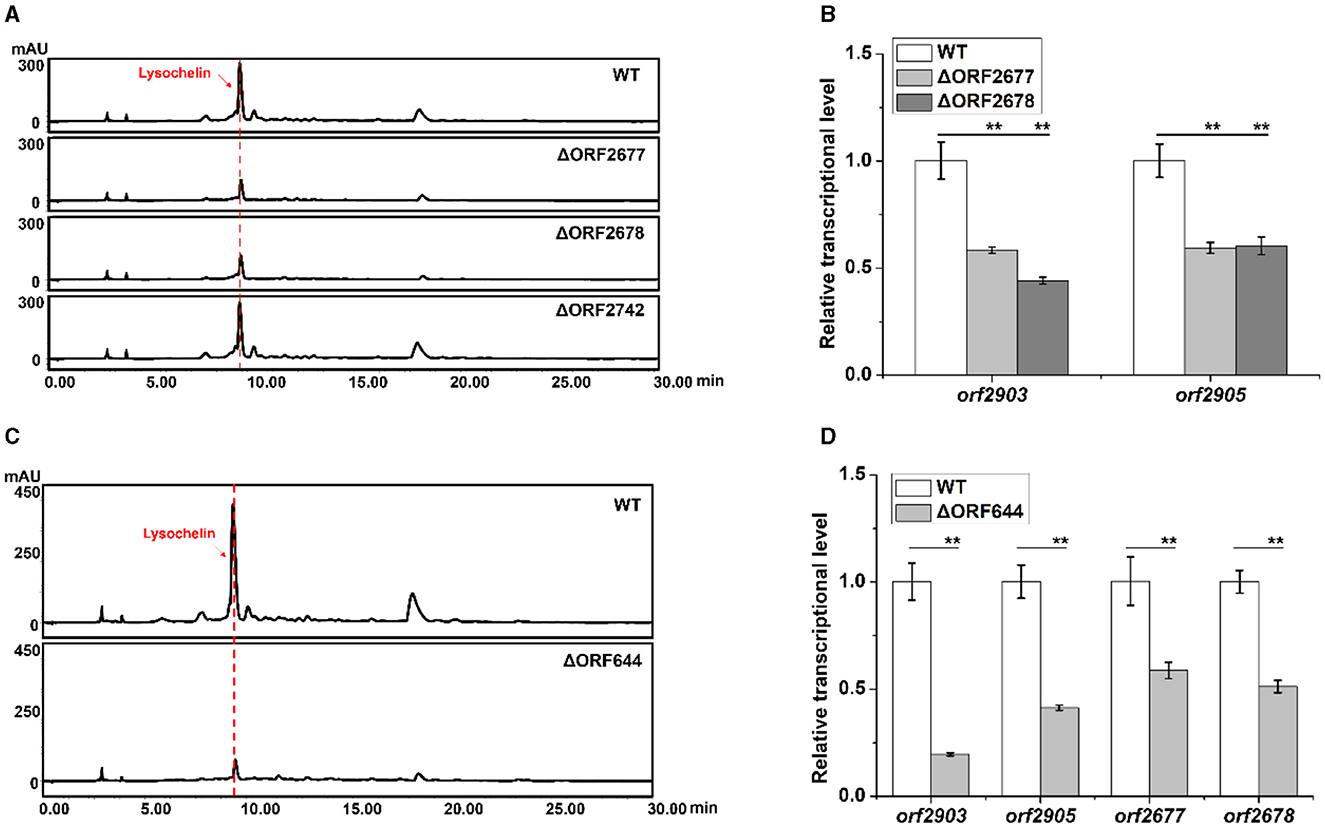
Figure 4. Effects of the maltose metabolism and quorum sensing on lysochelin production. (A) HPLC analysis of lysochelin production in WT and the deletion mutants of maltose utilization genes (ΔORF2677, ΔORF2678, and ΔORF2742) grown in LBM medium for 72 h. (B) Transcriptional analysis of lysochelin biosynthetic genes (orf2903 and orf2905) of WT (white columns), ΔORF2677 (light-gray columns), and ΔORF2678 (gray columns) grown in LBM for 24 h. (C) HPLC analysis of lysochelin production in WT and the deletion mutant of the DF signal biosynthetic gene (ΔORF644) grown in LBM medium for 72 h. (D) Transcriptional analysis of orf2903, orf2905, orf2677, and orf2678 of WT (white columns) and ΔORF644 (light-gray columns) grown in LBM for 24 h. **p < 0.01.
Quorum sensing (QS) is a cell-density-dependent regulatory mechanism mediated by diffusible chemical signal molecules in bacteria (Fuqua et al., 1994). Since lysochelin biosynthesis was connected to cell density, we figured that QS might be involved in the production of lysochelin. Diffusible factors (DF) including 3-HBA (3-hydroxybenzoic acid) and 4-HBA (4-hydroxybenzoic acid) are two QS signals that were previously identified from L. enzymogenes, and LenB2, a pteridine-dependent dioxygenase-like protein, is responsible for DF biosynthesis (Qian et al., 2013). In strain 3655, the gene orf644 encodes a homologous protein of LenB2 with 90% identity in protein sequence (Supplementary Figure 12). To test if the QS signaling is involved in lysochelin production, we constructed a deletion mutant ΔORF644 (Supplementary Figure 13). The cell density and lysochelin production in the mutant decreased to 80% and 22% of that in WT (Figure 4C and Supplementary Figures 11C, D). In accordance with the data, the transcriptional level of orf2903 and orf2905 was significantly reduced in mutant ΔORF644 (Figure 4D). Moreover, the deletion of orf644 decreased the transcriptional level of the maltose uptake genes orf2677 and orf2678, indicating that the DF signaling system might be related to maltose transportation (Figure 4D). Besides, the deletion of orf644 abolished the production of 3-HBA and 4-HBA, confirming its pivotal role in DF biosynthesis (Supplementary Figure 14). The results showed that the DF signaling system could lower lysochelin production through impairing maltose utilization and high-density growth of strain 3655.
Transcriptomics analysis identified iron-related genes
With the finding that strain 3655 massively produced lysochelin only when reached a high cell density (Figure 3A), we wanted to explore potential regulatory mechanisms underlying the cell-density-dependent lysochelin biosynthesis. We performed the transcriptomics analysis of stain 3655 cultured to different densities, including OD600 5.0 (DS group, lysochelin-deficiency) and OD600 15.0 (S group, lysochelin-producing). In total, we obtained 23.07 and 16.84 million clean reads from DS group and S group, respectively. Among them, 1,476 differentially expressed genes (DEGs) were identified, including 347 up- and 1,476 downregulated genes, 79 DEGs specifically expressed in S group and 33 DEGs specifically expressed in DS group (Supplementary Figure 15). Expectedly, some interesting genes were upregulated accompany with lysochelin biosynthesis. The transcriptional level of orf4279, encoding SUF system Fe-S cluster assembly regulator, increased 4-fold in S group (Supplementary Tables 4, 5). qRT-PCR result showed the transcriptional level of orf4279 was enhanced 24-fold at the OD600 value of 12.0 when compared to that of the OD600 value of 1.0, implying an important role in high cell density (Supplementary Figure 16A). SUF system is responsible for iron-sulfur cluster (Fe-S) biogenesis through assembly free iron and sulfides, which is crucial in bacteria for cell survival under stress conditions such as oxidation and iron starvation (Bai et al., 2018). Subsequently, we constructed the deletion mutant of orf4279 (Supplementary Figure 13) and found that the mutant ΔORF4279 could produce more lysochelin, with the yield increased by 2.5-fold when compared to that of WT, and the transcriptional level of orf2903 and orf2905 in strain ΔORF4279 increased as well (Figures 5A, B and Supplementary Figure 17A). However, the cell growth of strain ΔORF4279 slightly decreased when compared to that of WT (Supplementary Figure 17B). This seems to suggest that the inhibition of SUF system might “trick” strain ΔORF4279 to “believe” the presence of a low iron condition and, therefore, more lysochelin biosynthesis was needed to grab iron from surroundings.
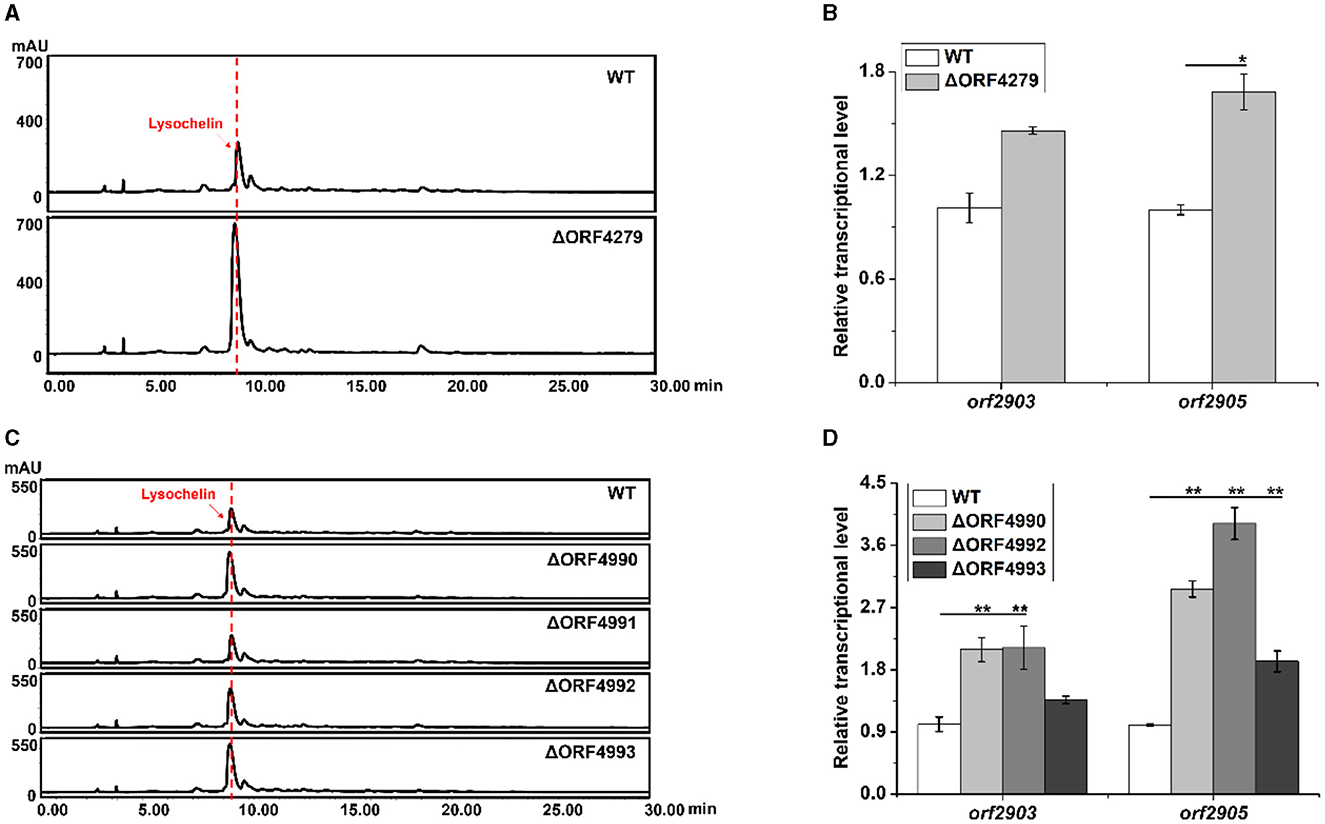
Figure 5. Effects of iron-utilizing genes on lysochelin biosynthesis. (A) HPLC analysis of lysochelin production in WT and the deletion mutant of SUF system regulator (ΔORF4279) grown in LBM medium for 72 h. (B) Transcriptional analysis of lysochelin biosynthetic genes (orf2903 and orf2905) in WT (white columns) and ΔORF4279 (light-gray columns) grown in LBM for 24 h. (C) HPLC analysis of lysochelin production in WT and the deletion mutants of RpoE family RNA polymerase sigma factor (ΔORF4990), FecR family regulator (ΔORF4991), and bacterial transferrin receptors (ΔORF4992 and ΔORF4993) grown in LBM for 72 h. (D) Transcriptional analysis of orf2903 and orf2905 in WT (white columns), ΔORF4990 (light-gray columns), ΔORF4992 (gray columns), and ΔORF4993 (dark-gray columns) grown in LBM for 24 h. *p < 0.05; **p < 0.01.
Moreover, the transcription of five clustered genes (orf4990-orf4994) was significantly upregulated in S group (Supplementary Table 4). The gene orf4990 encodes RpoE family RNA polymerase sigma factor which may function in iron-restricted condition (Karash et al., 2022). The product of orf4991 belongs to FecR family regulator that is related to regulation of ferric citrate uptake (Yokoyama et al., 2021). The gene orf4992 encodes TonB-dependent transporter, and orf4993 encodes Slam-dependent surface lipoprotein (SLP) which is homologous to transferrin-binding B (TbpB). These two proteins consist of bipartite transferrin receptor to acquire iron from transferrin (Pogoutse and Moraes, 2020). The product of orf4994 belongs to the family of surface lipoprotein assembly modulators that are essential for surface display of SLPs (Hooda et al., 2017). To investigate the effect of these genes on lysochelin production, we generated the deletion mutants for each of these genes (Supplementary Figures 10, 13). Except orf4991, the deletion of orf4990, orf4992, or orf4993 improved the yield of lysochelin, as well as the transcriptional levels of orf2903 and orf2905 (Figures 5C, D and Supplementary Figure 17C). However, the cell density of these mutants was similar to that of WT, with slightly increased in mutant ΔORF4993 (Supplementary Figure 17D). Besides, the transcriptional level of orf4990, orf4992, and orf4993 was enhanced by 11-fold, 215-fold, and 13,005-fold at the OD600 12.0 when compared to that of the OD600 1.0 (Supplementary Figure 16). The data are consistent with the notion that the block of other iron acquisition pathways can “force” the mutants to produce more lysochelin to compete for iron.
A TetR regulator regulated the biosynthesis of lysochelin
In search for potential regulators, we found that the gene orf3043 affected the biosynthesis of lysochelin. The product of this gene belongs to the TetR family of transcriptional regulators. We constructed the deletion mutant of orf3043 (Supplementary Figure 18) and found that the transcriptional level of orf2903 and orf2905 in mutant ΔORF3043 was significantly decreased when compared to that of WT (Figure 6C). Accordingly, the yield of lysochelin reduced to 50% of that in WT (Figures 6A, B). Moreover, the deletion of orf3043 reduced the transcriptional level of the maltose uptake genes orf2677 and orf2678, indicating that the TetR might regulate maltose transportation (Figure 6C). The transcriptional level of orf3043 was enhanced by 5-fold at the OD600 12.0 when compared to that of the OD600 1.0 (Supplementary Figure 16A). Next, we generated a complementary strain (ORF3043C) by reintroducing orf3043 into the mutant (Supplementary Figure 18). As expected, the production of lysochelin in the complementary strain was restored to the WT level (Figures 6A, B). While the cell density of the mutant ΔORF3043 decreased to 70% of that in WT, the density was restored to the WT level in the complementary strain ORF3043C (Figure 6D). The results indicated that the TetR regulator ORF3043 contributes to the regulation of lysochelin biosynthesis through affecting the transcription of maltose transportation genes and then lysochelin biosynthetic genes.
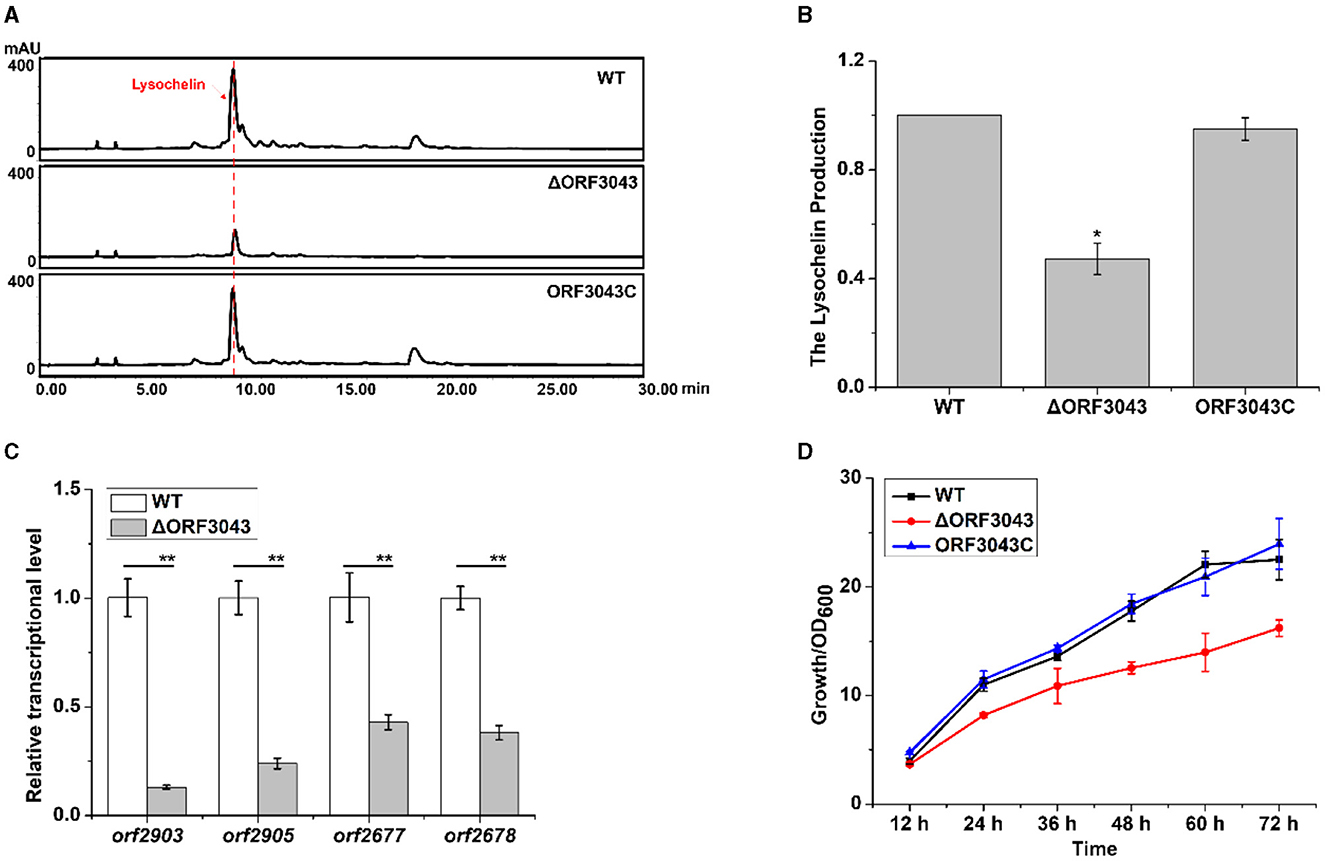
Figure 6. Regulation of lysochelin production by the TetR regulator ORF3043. (A) HPLC analysis of lysochelin production in WT, ΔORF3043 and its complementary strain ORF3043C grown in LBM medium for 72 h. (B) Quantification of lysochelin in the cultures from (A). (C) Transcriptional analysis of orf2903, orf2905, orf2677, and orf2678 in WT (white columns) and ΔORF3043 (light-gray columns) grown in LBM for 24 h. (D) The growth curve of WT (black line), ΔORF3043 (red line), and ORF3043C (blue line) grown in LBM medium. *p < 0.05; **p < 0.01.
Discussion
In this work, we found maltose dramatically induced the production of a compound in Lysobacter strain 3655 cultured in the rich medium LB. The results from gene deletion of the lec-like cluster and LC-MS analysis proved this compound to be lysochelin, the first siderophore isolated from Lysobacter (Miller et al., 2023). The production of lysochelin depended on the growth status of strain 3655 cultured in LBM medium. At low cell density (represented as the OD600 value of 5.0), the siderophore biosynthesis was completely inhibited in the rich medium, and lysochelin production was observed when the OD600 reached 7.5, and then significantly increased at high cell density (OD600 15.0). Strain 3655 grew much better in LBM than that in LB medium; in LBM the OD600 value could reach nearly 20.0. The high cell growth led to gradual depletion of iron in LBM medium. The data indicated that, when the OD600 value reached 7.5, the iron concentration was lowered to the threshold to trigger lysochelin production. As the cell density continued to increase, lysochelin was massively produced. Conversely, in the absence of maltose, strain 3655 grew in LB medium to a maximum OD600 value of 6.0, and iron was not depleted to the threshold concentration, thus no lysochelin was observed. Maltose could also promote the high-density growth and induce lysochelin biosynthesis in L. enzymogenes OH11 (Supplementary Figure 1), indicating that this might be a general phenomenon in Lysobacter, in which siderophore production can be induced through promoting the high-density growth.
The transportation of maltose into cells is the initiation of maltose metabolism. In E. coli, the periplasmic binding protein-dependent ATP binding cassette (ABC) transporter is responsible for maltose uptake, which is encoded by malEFGK genes (Boos and Shuman, 1998). We found strain 3655 contains the mal-like genes, orf2677, orf2678, orf2679, and orf2685 that are homologous to malE, malF, malG, and malK, respectively (Supplementary Figures 7, 8, 19, 20). The deletion of orf2677 or orf2678 impaired, but not completely inhibited, cell growth and lysochelin production, implying that other transport systems might be present in strain 3655. For example, the phosphotransferase system (PTS) is responsible for maltose transportation in Enterococcus faecalis (Breton et al., 2005). In E. coli, maltose is degraded into glucose and glucose-1-phosphate by three enzymes MalQ (amylomaltase), MalP (maltodextrin phosphorylase), and MalZ (maltodextrin glucosidase). Glucose and glucose-1-phosphate are then converted to glucose-6-phosphate, which enter in the glycolysis pathway with glucokinase and phosphoglucomutase, respectively (Boos and Shuman, 1998). However, the homologous genes of malQ, malP, and malZ were not found in strain 3655. In Streptococcus bovis, maltose is cleaved by a maltose hydrolase into two glucose molecules (Martin and Russell, 1987; Andersson and Rådström, 2002). Moreover, the maltose phosphorylase could convert maltose to glucose and glucose-1-phosphate in Lactococcus lactis (Levander et al., 2001; Nilsson and Rådström, 2001). In strain 3655, we found orf2742 was homologous to genes encoding maltose hydrolase (possible phosphorylase). Surprisingly, the deletion of orf2742 did not affect the cell growth and lysochelin production. Further studies are needed to explore if strain 3655 could contain other uncharacterized maltose metabolism enzymes.
The lysochelin biosynthesis was induced at high cell density in strain 3655, and the deletion of orf644, which encodes a dioxygenase for the biosynthesis of the quorum sensing (QS) signal DF, significantly decreased the cell growth and lysochelin production. The transcription of the lysochelin biosynthetic genes (orf2903 and orf2905) and the maltose transportation genes (orf2677 and orf2678) was clearly reduced in mutant ΔORF644. The results suggested that the DF signal system might regulate maltose transportation and then control lysochelin biosynthesis. The quorum sensing might also be related to the interesting observation, in which the culture volumes could make a big difference in cell density and lysochelin production even if the same concentration of maltose was used for cultures. More studies are needed to unveil the molecular details behind this phenomenon. The QS regulation of siderophores is present in many microbes. The yield of siderophore pyoverdine was decreased by 2-fold in the mutant of lasR encoding QS regulator of Pseudomonas aeruginosa (Stintzi et al., 1998). The QS antagonist furanone could stimulate pyoverdine biosynthesis in P. aeruginosa, while inhibiting pyoverdine formation in P. putida (Ren et al., 2005). However, the lack of the QS regulator CviR led to an increase of siderophore production in Chromobacterium violaceum (Batista et al., 2024). The mechanism underlying the QS-regulated siderophore production is not fully understood, but could probably be due to the divergent survival strategies adopted by various species (McRose et al., 2018). Recently, it was found that the siderophore yersiniabactin of uropathogenic Escherichia coli also functions as a QS signal, which represents a convergence of the quorum-sensing and the siderophore system (Heffernan et al., 2024). In Vibrio harveyi, QS regulates two types of siderophores in different environmental and growth contexts, producing cell-associated siderophores at low cell density and accumulating soluble siderophores at high cell density (McRose et al., 2018).
The transcriptomics analysis and qRT-PCR results showed the transcriptional level of bacterial transferrin receptor genes (orf4992 and orf4993) was greatly enhanced at high cell density under iron starvation conditions. This receptor has been found in some members of the Betaproteobacteria and Gammaproteobacteria and is vital for survival of several important human and animal pathogens (Pogoutse and Moraes, 2020). The receptor consists of two proteins including TonB-dependent transporter (TbpA) and surface lipoprotein (TbpB), which binds transferrin and then removes iron from the C-terminal lobe of transferrin and transports it across the bacterial outer membrane (Pogoutse and Moraes, 2017). Therefore, this receptor is responsible for iron acquisition from transferrin, which is an iron-carrying glycoprotein found in the mammalian circulatory system (Hughes and Friedman, 2014). This receptor is essential for colonizing and invading pathogens, such as the families of Neisseriaceae, Pasteurellaceae, and Moraxellaceae, to obtain iron from their hosts (Pogoutse and Moraes, 2017). The Lysobacter genus belongs to Gammaproteobacteria. Although they are not known to be pathogenic bacteria, they could prey on other bacteria, fungi, algae, and nematodes (Christensen and Cook, 1978; Yue et al., 2021). The presence of bacterial transferrin receptor might confer the survival advantage on Lysobacter, since the transferrin homologs are also found in invertebrates and algae (Lambert, 2012). Apart from lysochelin, strain 3655 activated bacterial transferrin receptor to enhance its ability for iron acquisition under iron restriction environment, thus improved the survivability.
Additionally, we found that the TetR family regulator ORF3043 controlled the cell growth and lysochelin production through the transcriptional regulation of maltose transportation genes (orf2677 and orf2678) and lysochelin biosynthetic genes (orf2903 and orf2905). The transcriptional level of itself was significantly enhanced at high cell density, which showed ORF3043 might function under high-density growth of strain 3655. TetR regulators have been proved to regulate siderophore production in various bacteria. A global regulator LuxT activates siderophore production via inhibiting the expression of a repressor SwrZ at low cell density of Vibrio harveyi (Eickhoff et al., 2022). In the rhizobacterium Pseudomonas fluorescens, the TetR regulator PhlH represses the expression of proteins involved in pyoverdine biosynthesis based on comparative proteomic analysis (Zhang et al., 2022). The pathway-specific regulator Orf12 from the biosynthetic cluster of 7-hydroxytropolone (7-HT) positively regulates 7-HT production through stimulating the expression of orf6-orf9 in the 7-HT cluster of Pseudomonas donghuensis HYS (Chen et al., 2018).
In summary, we found that maltose induced lysochelin production through promoting the high-density growth of Lysobacter sp. 3655, which resulted in an iron-restricted condition that activated the expression of the siderophore biosynthetic genes even when the bacterium was grown in rich media. Through generating a series of gene deletion mutants and analyzing the gene transcription and siderophore production, we showed that maltose metabolism and quorum sensing were involved in the siderophore biosynthesis in rich media. Furthermore, strain 3655 employed both the siderophore and the transferrin receptor to grab the trace amount of extracellular iron. The data also showed that a TetR regulator positively regulated lysochelin production through affecting the maltose metabolism. The studies shed new light on the regulation of siderophore biosynthesis, which could provide a new way to siderophore production through high-density scale-up fermentation.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
FZ: Writing – review & editing, Investigation, Methodology. JL: Investigation, Methodology, Writing – review & editing. LJ: Investigation, Methodology, Writing – review & editing. YZ: Investigation, Methodology, Writing – review & editing. LY: Conceptualization, Data curation, Funding acquisition, Supervision, Writing – original draft, Writing – review & editing. LD: Conceptualization, Supervision, Writing – original draft, Formal analysis, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the National Natural Science Foundation of China (32100061), the Natural Science Foundation of Fujian Province (2021J05036), and the Education Scientific Research Project of Young and Middle-aged Teachers in Fujian Province (JAT200085) to LY.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1433983/full#supplementary-material
References
Altschul, S. F., Madden, T. L., Schäffer, A. A., Zhang, J., Zhang, Z., Miller, W., et al. (1997). Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucl. Acids Res. 25, 3389–3402. doi: 10.1093/nar/25.17.3389
Andersson, U., and Rådström, P. (2002). Beta-glucose 1-phosphate interconverting enzymes in maltose- and trehalose-fermenting lactic acid bacteria. Environ. Microbiol. 4, 81–88. doi: 10.1046/j.1462-2920.2002.00268.x
Baars, O., Zhang, X., Morel, F. M. M., and Seyedsayamdost, M. R. (2016). The siderophore metabolome of Azotobacter vinelandii. Appl. Environ. Microbiol. 82, 27–39. doi: 10.1128/AEM.03160-15
Bai, Y., Chen, T., Happe, T., Lu, Y., and Sawyer, A. (2018). Iron-sulphur cluster biogenesis via the SUF pathway. Metallomics 10, 1038–1052. doi: 10.1039/C8MT00150B
Barry, S. M., and Challis, G. L. (2009). Recent advances in siderophore biosynthesis. Curr. Opin. Chem. Biol. 13, 205–215. doi: 10.1016/j.cbpa.2009.03.008
Batista, B. B., de Lima, V. M., Picinato, B. A., Koide, T., and Neto, J. F. S. (2024). A quorum-sensing regulatory cascade for siderophore-mediated iron homeostasis in Chromobacterium violaceum. mSystems 19:e0139723. doi: 10.1128/msystems.01397-23
Beauchene, N. A., Myers, K. S., Chung, D., Park, D. M., Weisnicht, A. M., Keleş, S., et al. (2015). Impact of anaerobiosis on expression of the iron responsive Fur and RyhB regulons. mBio 6, e01947–e01915. doi: 10.1128/mBio.01947-15
Boos, W., and Shuman, H. (1998). Maltose/Maltodextrin system of Escherichia coli: transport, metabolism, and regulation. Microbiol. Mol. Biol. Rev. 62, 204–229. doi: 10.1128/MMBR.62.1.204-229.1998
Breton, Y. L., Pichereau, V., Sauvageot, N., Auffray, Y., and Rince, A. (2005). Maltose utilization in Enterococcus faecalis. J. Appl. Microbiol. 98, 806–813. doi: 10.1111/j.1365-2672.2004.02468.x
Caradec, C., Anoz-Carbonell, E., Petrov, R., Billamboz, M., Antraygues, K., Cantrelle, F. X., et al. (2023). A novel natural siderophore antibiotic conjugate reveals a chemical approach to macromolecule coupling. ACS. Cent. Sci. 9, 2138–2149. doi: 10.1021/acscentsci.3c00965
Cezard, C., Farvacques, N., and Sonnet, P. (2015). Chemistry and biology of pyoverdines, Pseudomonas primary siderophores. Curr. Med. Chem. 22, 165–186. doi: 10.2174/0929867321666141011194624
Chen, M., Wang, P., and Xie, Z. (2018). A complex mechanism involving LysR and TetR/AcrR that regulates iron scavenger biosynthesis in Pseudomonas donghuensis HYS. J. Bacteriol. 200, e00087–e00018. doi: 10.1128/JB.00087-18
Chincholkar, B. L., Chaudhari, B. L., and Rane, M. R. (2007). “Microbial siderophore: a state of art,” in Microbial Siderophores, eds. A. Varma and S. B. Chincholkar (Berlin; Heidelberg: Springer-Verlag), 12.
Christensen, P., and Cook, F. D. (1978). Lysobacter, a new genus of nonfruiting, gliding bacteria with a high base ratio. Int. J. Syst. Bateriol. 28, 367–393. doi: 10.1099/00207713-28-3-367
Cornish, A. S., and Page, W. J. (1995). Production of the triacetecholate siderophore protochelin by Azotobacter vinelandii. Biometals 8, 332–338. doi: 10.1007/BF00141607
Eickhoff, M. J., Fei, C., Cong, J., and Bassler, B. L. (2022). LuxT is a global regulator of low-cell-density behaviors, including type III secretion, siderophore production, and aerolysin production, in Vibrio harveyi. mBio 13:e0362121. doi: 10.1128/mbio.03621-21
Ferreira, C. M. H., Vilas-Boas, A., Sousa, C. A., Soares, H. M. V. M., and Soares, E. V. (2019). Comparison of five bacterial strains producing siderophores with ability to chelate iron under alkaline conditions. AMB Expr. 9, 78–89. doi: 10.1186/s13568-019-0796-3
Frausto da Silva, J. J. R., and Williams, R. J. P. (2001). The Biological Chemistry of the Elements-the Inorganic Chemistry of Life, 2nd Edn. Oxford: Oxford University Press.
Fuqua, W. C., Winans, S. C., and Greenberg, E. P. (1994). Quorum sensing in bacteria: the LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 176, 269–275. doi: 10.1128/jb.176.2.269-275.1994
Gao, H., and Bian, X. (2022). Editorial: Microbial siderophores: biosynthesis, regulation, and physiological and ecological impacts. Front. Microbiol. 13:892485. doi: 10.3389/fmicb.2022.892485
Hao, L., Willis, D. K., Andrews-Polymenis, H., McClelland, M., and Barak, J. D. (2012). Requirement of siderophore biosynthesis for plant colonization by Salmonella enterica. Appl. Environ. Microbiol. 78, 4561–4570. doi: 10.1128/AEM.07867-11
Heffernan, J. R., Wildenthal, J. A., Tran, H., Katumba, G. L., McCoy, W. H., and Henderson, J. P. (2024). Yersiniabactin is a quorum-sensing autoinducer and siderophore in uropathogenic Escherichia coli. mBio 15:e0027723. doi: 10.1128/mbio.00277-23
Hider, R. C., and Kong, X. (2010). Chemistry and biology of siderophores. Nat. Prod. Rep. 27, 637–657. doi: 10.1039/b906679a
Hoegy, F., Mislin, G. L. A., and Schalk, I. J. (2014). Pyoverdine and pyochelin measurements. Methods Mol. Biol. 1149, 293–301. doi: 10.1007/978-1-4939-0473-0_24
Hooda, Y., Lai, C. C. L., and Moraes, T. F. (2017). Identification of a large family of slam-dependent surface lipoproteins in Gram-negative bacteria. Front. Cell Infect. Microbiol. 7, 207–218. doi: 10.3389/fcimb.2017.00207
Hughes, A. L., and Friedman, R. (2014). Evolutionary diversification of the vertebrate transferrin multi-gene family. Immunogenetics 66, 651–661. doi: 10.1007/s00251-014-0798-x
Karash, S., Jiang, T., and Kwon, Y. M. (2022). Genome-wide characterization of Salmonella Typhimurium genes required for the fitness under iron restriction. BMC Genomic Data 23, 55–68. doi: 10.1186/s12863-022-01069-3
Lambert, L. A. (2012). Molecular evolution of the transferrin family and associated receptors. Biochim. Biophys. Acta 1820, 244–255. doi: 10.1016/j.bbagen.2011.06.002
Levander, F., Andersson, U., and Rådström, P. (2001). Physiological role of beta-phosphoglucomutase in Lactococcus lactis. Appl. Environ. Microbiol. 67, 4546–4553. doi: 10.1128/AEM.67.10.4546-4553.2001
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta DeltaC(T)) Method. Methods 25, 402–408. doi: 10.1006/meth.2001.1262
Martin, S. A., and Russell, J. B. (1987). Transport and phosphorylation of disaccharides by the ruminal bacterium Streptococcus bovis. Appl. Environ. Microbiol. 53, 2388–2393. doi: 10.1128/aem.53.10.2388-2393.1987
McRose, D. L., Baars, O., Seyedsayamdost, M. R., and Morel, F. M. M. (2018). Quorum sensing and iron regulate a two-for-one siderophore gene cluster in Vibrio harveyi. Proc. Natl. Acad. Sci. U. S. A. 115, 7581–7586. doi: 10.1073/pnas.1805791115
Meiwes, J., Fiedler, H. P., Zahner, H., Konetschny-Rapp, S., and Jung, G. (1990). Production of desferrioxamine E and new analogues by directed fermentation and feeding fermentation. Appl. Microbiol. Biotechnol. 32, 505–510. doi: 10.1007/BF00173718
Meyer, J. M., and Abdallah, M. A. (1978). The fluorescent pigment of Pseudomonas fluorescens: biosynthesis, purification and physicochemical properties. J. Gen. Microbiol. 107, 319–328. doi: 10.1099/00221287-107-2-319
Miethke, M., and Marahiel, M. A. (2007). Siderophore-based iron acquisition and pathogen control. Microbiol. Mol. Biol. Rev. 71, 413–451. doi: 10.1128/MMBR.00012-07
Miller, A. L., Li, S., Eichhorn, C. D., Zheng, Y., and Du, L. (2023). Identification and biosynthetic study of the siderophore lysochelin in the biocontrol agent Lysobacter enzymogenes. J. Agric. Food Chem. 71, 7418–7426. doi: 10.1021/acs.jafc.3c01250
Nilsson, U., and Rådström, P. (2001). Genetic localization and regulation of the maltose phosphorylase gene, malP, in Lactococcus lactis. Microbiology 147, 1565–1573. doi: 10.1099/00221287-147-6-1565
Payne, S. M. (1988). Iron and virulence in the family Enterobacteriaceae. Crit. Rev. Microbiol. 16, 81–111. doi: 10.3109/10408418809104468
Pogoutse, A. K., and Moraes, T. F. (2017). Iron acquisition through the bacterial transferrin receptor. Crit. Rev. Biochem. Mol. Biol. 52, 314–326. doi: 10.1080/10409238.2017.1293606
Pogoutse, A. K., and Moraes, T. F. (2020). Transferrin binding protein B and transferrin binding protein A2 expand the transferrin recognition range of Histophilus somni. J. Bacteriol. 202, e00177–e00120. doi: 10.1128/JB.00177-20
Puopolo, G., Tomada, S., and Pertot, I. (2018). The impact of the omics era on the knowledge and use of Lysobacter species to control phytopathogenic micro-organisms. J. Appl. Microbiol. 124, 15–27. doi: 10.1111/jam.13607
Qian, G., Wang, Y., Liu, Y., Xu, F., He, Y., Du, L., et al. (2013). Lysobacter enzymogenes uses two distinct cell-cell signaling systems for differential regulation of secondary-metabolite biosynthesis and colony morphology. Appl. Environ. Microbiol. 79, 6604–6616. doi: 10.1128/AEM.01841-13
Ren, D., Sims, J. J., and Wood, T. K. (2005). Quorum-sensing antagonist (5Z)-4-bromo-5-(bromomethylene)-3-butyl-2(5H)-furanone influences siderophore biosynthesis in Pseudomonas putida and Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 66, 689–695. doi: 10.1007/s00253-004-1691-6
Sambrook, J., Fritsch, E. F., and Maniatis, T. (1989). Molecular Cloning: a Laboratory Manual. New York, NY: Cold Spring Harbor Laboratory Press.
Santos, S., Neto, I. F. F., Machado, M. D., Soares, H. M. V. M., and Soares, E. V. (2014). Siderophore production by Bacillus megaterium: effect of growth phase and cultural conditions. Appl. Biochem. Biotechnol. 172, 549–560. doi: 10.1007/s12010-013-0562-y
Sasirekha, B., and Srividya, S. (2016). Siderophore production by Pseudomonas aeruginosa FP6, a biocontrol strain for Rhizoctonia solani and Colletotrichum gloeosporioides causing diseases in chilli. Agric. Nat. Resour. 50, 250–256. doi: 10.1016/j.anres.2016.02.003
Schalk, I. J., Rigouin, C., and Godet, J. (2020). An overview of siderophore biosynthesis among fluorescent Pseudomonads and new insights into their complex cellular organization. Environ. Microbiol. 22, 1447–1466. doi: 10.1111/1462-2920.14937
Seo, S. W., Kim, D., Latif, H., O'Brien, E. J., Szubin, R., and Palsson, B. O. (2014). Deciphering Fur transcriptional regulatory network highlights its complex role beyond iron metabolism in Escherichia coli. Nat. Commun. 5, 4910–4930. doi: 10.1038/ncomms5910
Sexton, D. J., and Schuster, M. (2017). Nutrient limitation determines the fitness of cheaters in bacterial siderophore cooperation. Nat. Commun. 8, 230–237. doi: 10.1038/s41467-017-00222-2
Singh, P., Chauhan, P. K., Upadhyay, S. K., Singh, R. K., Dwivedi, P., Wang, J., et al. (2022). Mechanistic insights and potential use of siderophores producing microbes in rhizosphere for mitigation of stress in plants grown in degraded land. Front. Microbiol. 13:898979. doi: 10.3389/fmicb.2022.898979
Stintzi, A., Evans, K., Meyer, J. M., and Poole, K. (1998). Quorum-sensing and siderophore biosynthesis in Pseudomonas aeruginosa: lasR/lasI mutants exhibit reduced pyoverdine biosynthesis. FEMS Microbiol. Lett. 166, 341–345. doi: 10.1111/j.1574-6968.1998.tb13910.x
Timofeeva, A. M., Galyamova, M. R., and Sedykh, S. E. (2022). Bacterial siderophores: classification, biosynthesis, perspectives of use in agriculture. Plants 11, 3065–3086. doi: 10.3390/plants11223065
Ustiatik, R., Nuraini, Y., Suharjono, S., and Handayanto, E. (2021). Siderophore production of the Hg-resistant endophytic bacteria isolated from local grass in the Hg-contaminated soil. J. Ecol. Eng. 22, 129–138. doi: 10.12911/22998993/135861
Valdebenito, M., Crumbliss, A. L., Winkelmann, G., and Hantke, K. (2006). Environmental factors influence the production of enterobactin, salmochelin, aerobactin, and yersiniabactin in Escherichia coli strain Nissle 1917. Int. J. Med. Microbiol. 296, 513–520. doi: 10.1016/j.ijmm.2006.06.003
Vindeirinho, J. M., Soares, H. M. V. M., and Soares, E. V. (2021). Modulation of siderophore production by Pseudomonas fluorescens through the manipulation of the culture medium composition. Appl. Biochem. Biotechnol. 193, 607–618. doi: 10.1007/s12010-020-03349-z
Wandersman, C., and Delepelaire, P. (2004). Bacterial iron sources: from siderophores to hemophores. Ann. Rev. Microbiol. 58, 611–647. doi: 10.1146/annurev.micro.58.030603.123811
Yokoyama, T., Niinae, T., Tsumagari, K., Imami, K., Ishihama, Y., Hizukuri, Y., et al. (2021). The Escherichia coli S2P intramembrane protease RseP regulates ferric citrate uptake by cleaving the sigma factor regulator FecR. J. Biol. Chem. 296, 100673–100690. doi: 10.1016/j.jbc.2021.100673
Yu, S., Teng, C., Bai, X., Liang, J., Song, T., Dong, L., et al. (2017). Optimization of siderophore production by Bacillus sp. PZ-1 and its potential enhancement of phytoextration of Pb from soil. J. Microbiol. Biotechnol. 27, 1500–1512. doi: 10.4014/jmb.1705.05021
Yue, H., Jiang, J., Taylor, A., Leite, A., Dodds, E., and Du, L. (2021). Outer membrane vesicles-mediated co-delivery of the antifungal HSAF metabolites and lytic polysaccharide monooxygenase in the predatory Lysobacter enzymogenes. ACS Chem. Biol. 16, 1079–1089. doi: 10.1021/acschembio.1c00260
Yue, H., Miller, A. L., Khetrapal, V., Jayaseker, V., Wright, S., and Du, L. (2022). Biosynthesis, regulation, and engineering of natural products from Lysobacter. Nat. Prod. Rep. 39, 842–874. doi: 10.1039/D1NP00063B
Keywords: Lysobacter, siderophore, lysochelin, regulation, biosynthesis
Citation: Zhang F, Liu J, Jiang L, Zheng Y, Yu L and Du L (2024) Production of the siderophore lysochelin in rich media through maltose-promoted high-density growth of Lysobacter sp. 3655. Front. Microbiol. 15:1433983. doi: 10.3389/fmicb.2024.1433983
Received: 17 May 2024; Accepted: 10 June 2024;
Published: 26 June 2024.
Edited by:
Phillip E. Klebba, Kansas State University, United StatesReviewed by:
Xiaoxu Jiang, California State University, San Bernardino, United StatesKashif Shamim, University of Mississippi, United States
Copyright © 2024 Zhang, Liu, Jiang, Zheng, Yu and Du. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Lingjun Yu, eXVsajExM0Bmam51LmVkdS5jbg==; Liangcheng Du, bGR1M0B1bmwuZWR1
†These authors have contributed equally to this work
 Fang Zhang
Fang Zhang Jia Liu1†
Jia Liu1† Liangcheng Du
Liangcheng Du