- 1Country National Laboratory of Agricultural Microbiology, Wuhan, Hubei, China
- 2Country College of Veterinary Medicine, Wuhan, Hubei, China
- 3Hubei Hongshan Laboratory, Huazhong Agricultural University, Wuhan, Hubei, China
- 4College of Biological and Food Engineering, Huanghuai University, Zhumadian, Henan, China
- 5Institute of Animal Science, Ningxia Academy of Agriculture and Forestry Sciences, Yinchuan, Ningxia, China
Ammonia is the primary component of malodorous substances in chicken farms. Currently, the microbial ammonia reduction is considered a potential method due to its low cost, high safety, and environmental friendliness. Sphingomonas sp. Z392 can significantly reduce the ammonia level in broiler coops. However, the mechanisms of ammonia nitrogen reduction by Sphingomonas sp. Z392 remain unclear. To explore the mechanisms of ammonia reduction by Sphingomonas sp. Z392, the transcriptome and metabolome analysis of Sphingomonas sp. Z392 under high ammonium sulfate level were conducted. It was found that the transcription levels of genes related to purine metabolism (RS01720, RS07605, purM, purC, purO) and arginine metabolism (glsA, argB, argD, aguA, aguB) were decreased under high ammonium sulfate environment, and the levels of intermediate products such as ornithine, arginine, IMP, and GMP also were also decreased. In addition, the ncd2 gene in nitrogen metabolism was upregulated, and intracellular nitrite content increased by 2.27 times than that without ammonium sulfate. These results suggested that under high ammonium sulfate level, the flux of purine and arginine metabolism pathways in Sphingomonas sp. Z392 might decrease, while the flux of nitrogen metabolism pathway might increase, resulting in increased nitrite content and NH3 release. To further verify the effect of the ncd2 gene on ammonia removal, ncd2 was successfully overexpressed and knocked out in Sphingomonas sp. Z392. ncd2 Overexpression exhibited the most ammonia reduction capability, the ammonia concentration of ncd2 overexpression group decreased by 43.33% than that of without Sphingomonas sp. group, and decreased by 14.17% than that of Sphingomonas sp. Z392 group. In conclusion, Sphingomonas sp. Z392 might reduce the release of NH3 by reducing the flux of purine and arginine metabolisms, while enhancing ammonia assimilation to form nitrite. In this context, ncd2 might be one of the key genes to reduce ammonia.
1 Introduction
The current environmental pollution in the livestock industry mainly comes from to feces, urine, sewage, noxious gases, noise, smoke, and dust (Ji et al., 2022), among them, the emission of malodor from livestock farms is also a major component of pollution in the animal husbandry industry. Studies have shown that large amounts of malodorous substances are generated during livestock farming and manure storage, posing a serious threat to both human and livestock health. These substances primarily include organic compounds, in addition to inorganic compounds, such as ammonia (NH3) and hydrogen sulfide (H2S), of which NH3 is particularly common and directly harmful. High ammonia concentrations in chicken coops jeopardize both human and animal health.
Currently, ammonia removal measures in chicken coops mainly include physical, chemical, feed regulation, and biological methods (Zhou and Wang, 2023). Physical methods include enhanced mechanical ventilation in the coop and the addition of physical deodorisers to the diet, which are often costly (Lubensky et al., 2019). Chemical methods include the use of inorganic metal ions, compounds, and acidifying agents, which tend to generate secondary pollution (Haq et al., 2021; Nancharaiah et al., 2016). The use of probiotics in poultry farming offers a promising approach to reducing ammonia emissions, improving animal health, and potentially promoting more sustainable and environmentally friendly agricultural practices (Chen and Ni, 2012). Microbial treatment techniques are not only characterized by strong selectivity, low cost, simple operation, high safety, non-toxic by-products, and lack of secondary pollution but can also lead to reductions in various environmental pollutants, thus positioning them as the optimal choice for bioremediation (Pang and Wang, 2021; Zhou and Wang, 2023). Different microorganisms have been reported to improve feed conversion efficiency, reduce fecal nitrogen levels in animals, and contribute to ammonia emission reduction, such as Bacillus subtilis, Clostridium butyricum, Acinetobacter (Huang et al., 2013), Pseudomonas (Jin et al., 2015), and Agrobacterium (Chen and Ni, 2012). However, few studies characterizing ammonia nitrogen-removing bacterial strains and their mechanisms have been conducted. In addition, previous studies have shown that adding Sphingomonas to broiler chicken feed significantly reduces ammonia concentrations in chicken coops during breeding (Wang et al., 2021). However, the mechanism of ammonia nitrogen reduction by Sphingomonas remains unclear. Therefore, to investigate the mechanism of ammonia removal in Sphingomonas, the transcriptome and metabolome of Sphingomonas were analyzed under different ammonium sulfate conditions, and the key genes in Sphingomonas associated with the reduction of ammonia release were screened. Subsequently, the effect of these key genes on the ability of Sphingomonas to reduce ammonia release was further investigated.
2 Materials and methods
2.1 Strains, media, and culture conditions
Sphingomonas sp. Z392 (GenBank accession no: MN108136) were isolated and stored at the Institute of Biology and Food Engineering of Huanghuai University. Escherichia coli β2155 was used for plasmid amplification.
Sphingomonas sp. Z392 and E. coli β2155 were cultured in Luriae–Bertani (LB) medium at 37°C. The heterotrophic nitrification medium (medium B) contained 0.5 g (NH4)2SO4, 5.62 g sodium succinate, and 50 mL Vicker’s salt solution (5 g K2HPO4, 2.5 g MgSO4⋅7H2O, 2.5 g NaCl, 0.05 g FeSO4⋅7H2O, 0.05 g MnSO4⋅4H2O, and 1 L distilled), distilled water 1.0 L. Moreover, media C and D were prepared by increasing the concentration of (NH4)2SO4 in medium B to 0.66 g/L and 1.06 g/L, respectively.
2.2 Transcriptome and metabolomics analysis
Sphingomonas sp. Z392 was cultured in LB medium at 37°C for 24 h. The cells were harvested by centrifugation (3,000 × g, 10 min) and washed with sterile saline, and the cell pellet was inoculated into medium B, incubated at 37°C. Then, the cells collected from medium B were added to medium C, incubated at 37°C. Finally, the cells from medium C were collected, transferred to medium D, incubated at 37°C. The cells from medium D were used for transcriptome and metabolome analysis.
Total RNA of the cells from medium D was extracted, transcriptome sequencing was performed by Gene DeNovo (Guangzhou, China) (Love et al., 2014). To obtain significant differentially expressed genes (DEGs), DESeq2 software was used with FC ≥ 2 or ≤ 0.5 (| log2(FC)| > 1) and p < 0.05 as screening conditions. Intracellular metabolites of the cells from medium D were detected by LC-MS, and metabolome were analyzed by Gene DeNovo (Guangzhou, China).
2.3 Strain construction
To screen antibiotics sensitive to Sphingomonas sp. Z392 as selection markers for genetic modification, the minimum inhibitory concentration (MIC) of gentamicin (Gm), streptomycin, and tetracycline against Sphingomonas sp. Z392 were determined. The experimental design for MIC using microplates were shown in Supplementary Tables 1, 2. For construction of ncd2 gene overexpression plasmid, ncd2 was amplified from the genomic DNA of Z392 strain. Then the PCR products were inserted into the expression plasmid p1600-gent-Flag with restriction sites NcoI and EcorI through the Gibson assembly method, generating p1600-ncd2-Flag plasmid. The Z392 strain were transformed with 5 μg p1600-ncd2-Flag plasmid by electroporation (2.5kV/cm, 200 Ω, 5 ms).
To construct ncd2 knockout strain, the targeting fragments (Δncd2:Gm) included the upstream and downstream homologous recombination arms of the ncd2 gene, and were constructed by overlap PCR. Subsequently, The targeting fragment was cloned into the suicide plasmid pCVD442, resulting in the targeting plasmid pCVD442-Δncd2:Gm. The targeting vector pCVD442-Δncd2:Gm was then electrotransformed into the E. coli β2155 strain to generate the donor strain β2155/pCVD442-Δncd2:Gm. Finally, the ncd2 gene knockout was achieved by conjugating the donor strain with the recipient strain (Sphingomonas sp. Z392), and the knockout of ncd2 was verified by PCR.
2.4 Feeding and management of the experimental broiler chickens
A total of 1, 200 one- day-old Cobb broiler chickens with similar body weights (42 ± 2 g) were randomly divided into 4 groups: a control group (CK) without Sphingomonas sp. Z392, fed with Sphingomonas sp. Z392 group (WT), fed with Z392 OVncd2 group (OV), and fed with Z392 Δncd2 (KO). The method refers to the research of Wang et al. (2021). At 7, 14, 21, 28, 35, and 42 days of age, fresh feces were collected from the broilers. The concentrations of total nitrogen, uric acid, ammonium nitrogen, and nitrate nitrogen in chicken manure were measured separately.
2.5 Nitrogen concentrations in chicken coops
The ammonia concentrations in the chicken coops were measured using an ammonia sensor (Bist et al., 2023). And total nitrogen, ammonia nitrogen, and nitrate nitrogen contents in chicken manure were measured (Such et al., 2023). Then, the determination of intracellular nitrite content was carried out using ion chromatography (Varão Moura et al., 2022).
2.6 Data and statistical analysis
All experiments were conducted in three biological replicates. The data were analyzed for statistical significance using SPSS 19.0 software. One-way analysis of variance (ANOVA) was used to express the significance of differences (P ≤ 0.05) between the groups. Figures were drawn using the GraphPad Prism 8 (GraphPad Software, USA).
3 Results
3.1 Transcriptomics analysis of Sphingomonas sp. Z392 under high level of ammonium sulfate
Previous studies have demonstrated that Sphingomonas sp. Z392 can significantly reduce ammonia nitrogen level by converting ammonium nitrogen into nitrate nitrogen in chicken manure, decreasing the emission of ammonia from chicken feces. To explore the mechanisms of Sphingomonas sp. Z392 in reducing ammonia release, in this paper, transcriptomics analysis of Sphingomonas sp. Z392 cultured under high ammonium sulfate condition was performed. As shown in Figure 1, a total of 310 differentially expressed genes (DEGs) were identified between the high ammonium sulfate group and the control group (CK_vs_T), including 95 upregulated genes and 215 downregulated genes. Subsequently, the metabolic pathways enriched by the DEGs were further analyzed. As shown in Supplementary Figure 1, the DEGs were involved in the following pathways: ribosome, photosynthesis, drug metabolism, biosynthesis of some antibiotics, purine metabolism, oxidative phosphorylation, biosynthesis of secondary metabolites, monobactam biosynthesis, VB6 metabolism, pentose phosphate pathway, glutathione metabolism, cyanoamino acid metabolism, arginine biosynthesis, arginine and proline metabolism, amino acid biosynthesis, and nitrogen metabolism.
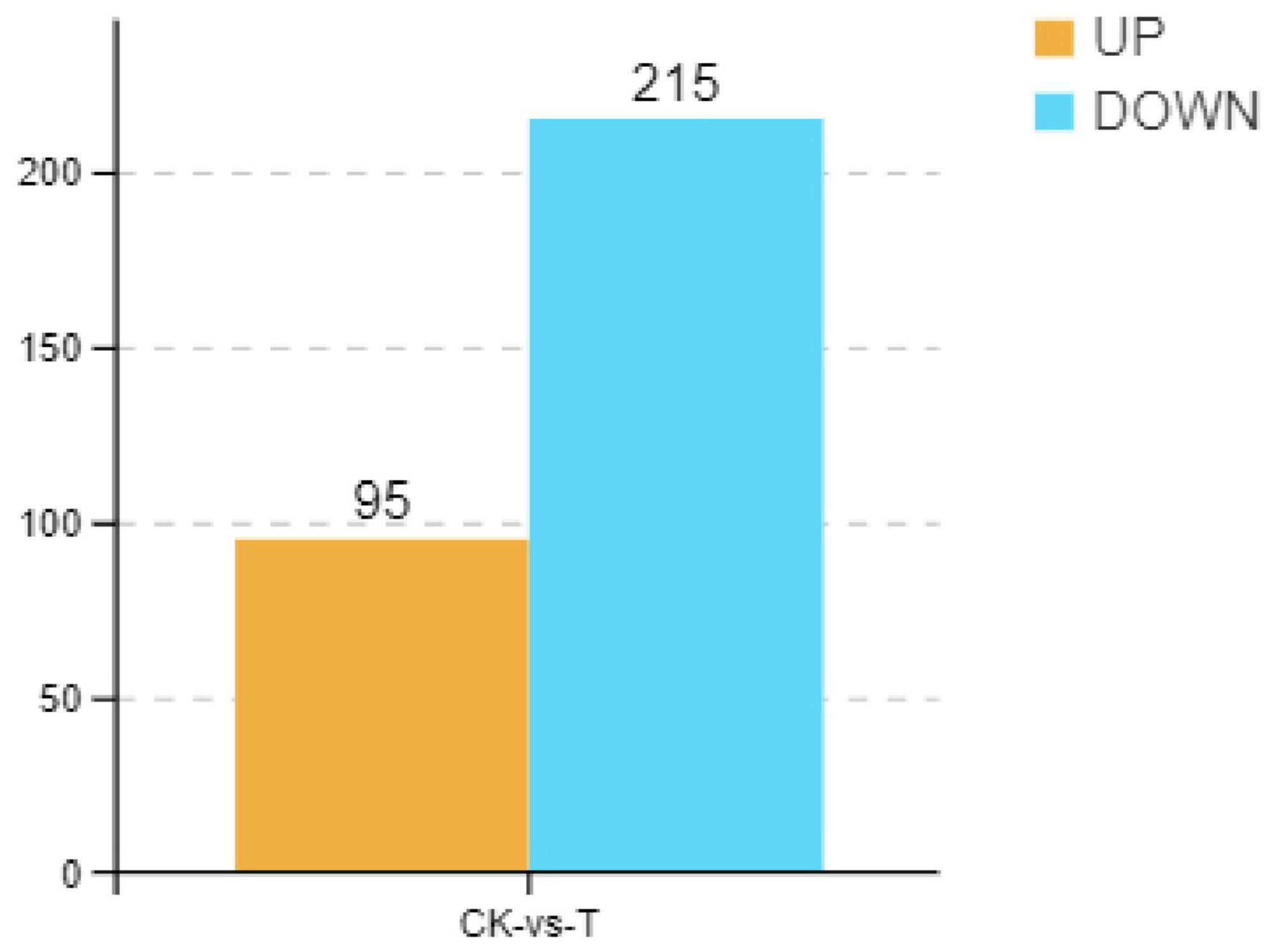
Figure 1. Differential expression genes of Sphingomonas sp. Z392 under high levels of ammonia sulfate. The Y-axis represents the number of up- and downregulated genes, orange represents upregulation, and blue represents downregulation.
Among the metabolic pathways, amino acid metabolism, purine metabolism, and nitrogen metabolism are closely associated with nitrogen. Some amino acids can act as carriers of ammonia, such as arginine and glutamine. In addition, ammonia also exists in purines, which can release ammonia during purine metabolism. When Sphingomonas sp. Z392 was cultured under high concentration of ammonium sulfate, the purine metabolism, arginine biosynthesis, arginine and proline metabolism, and nitrogen metabolism pathways were significantly enriched. These pathways were closely associated with the reduce of ammonia (Figure 2).
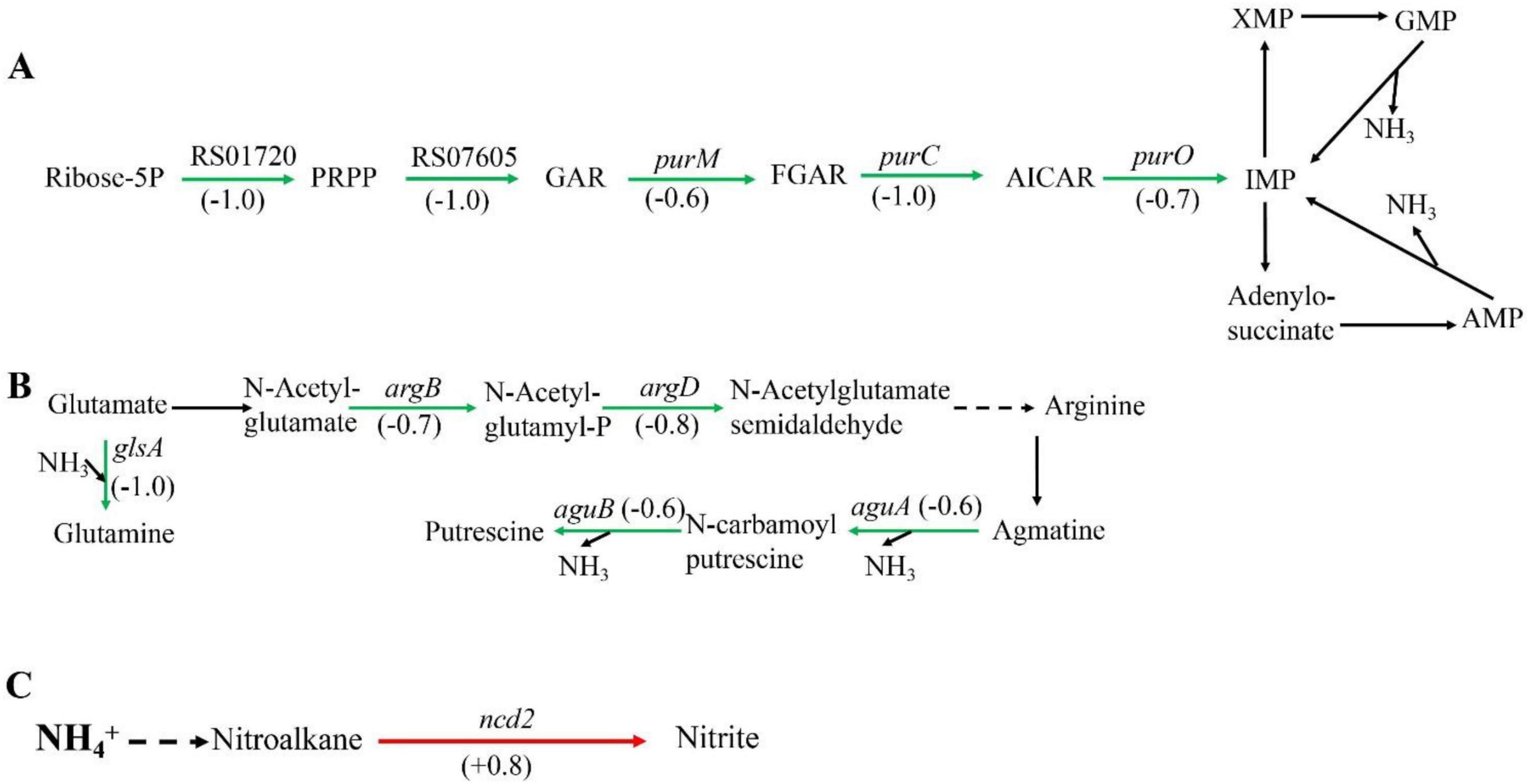
Figure 2. Significantly enriched metabolic pathways associated with ammonia degradation. (A) purine metabolism pathway. (B) Arginine metabolism pathway. (C) Nitrogen metabolism pathway. The green arrows indicate downregulation, and the red arrows indicate upregulation.
In purine metabolism, ribose 5-phosphate accepts amino groups from glutamine, aspartate, and glycine to form inosine monophosphate (IMP), which serves as a carrier for the amino groups (Pedley and Benkovic, 2017). And, IMP can interconvert with adenylic acid (AMP) and guanosine monophosphate (GMP), accompanied by the release of NH3 (Gooding et al., 2015). As shown in Figure 2A, under high concentration of ammonium sulfate, the transcription levels of RS01720, RS07605, purM, purC, and purO genes in the purine metabolism of Sphingomonas sp. Z392 were significantly downregulated. As a result, the synthesis of IMP, GMP, and AMP might be reduced, which would lead to a decrease in the production of NH3 from the purine metabolism.
In addition, the biosynthesis and metabolism of arginine are also closely related to the intracellular ammonia concentration. Glutamate is converted to arginine via catalysis by a series of enzymes. Arginine, serving as a carrier of ammonia, undergo the arginine metabolism pathway to produce putrescine, and the ammonia is released (Landete et al., 2010). As shown in Figure 2B, under high concentration of ammonium sulfate, the transcription levels of glsA, argB, and argD genes of Sphingomonas sp. Z392 in arginine biosynthesis pathway were significantly downregulated, and the transcription levels of aguB, aguA genes in arginine metabolism pathway were also significantly downregulated.
In addition to purine and arginine metabolism, intracellular ammonia in Sphingomonas sp. Z392 might form nitromethane through a series of enzymes, then nitromethane is converted to nitrite via catalysis by a nitronate monooxygenase (encoded by the ncd2 gene) (Figure 2C). When Sphingomonas sp. Z392 was cultured under high ammonium sulfate concentration, the transcription levels of ncd2 in nitrogen metabolism pathway was significantly upregulated, which would facilitate the conversion of ammonia into nitrite, enhancing the elimination for intracellular ammonia.
3.2 Metabolomics analysis of Sphingomonas sp. Z392 under high level of ammonium sulfate
The metabolites of Sphingomonas sp. Z392 from the high ammonium sulfate and control groups were identified by LC-MS/MS. To analyze the changes in differential metabolites of Sphingomonas sp. Z392 under the high ammonium sulfate, the differential expression of metabolites (DEMs) between the CK and T groups were analyzed using orthogonal partial least squares-discriminant analysis (OPLS-DA). As shown in Figure 3, the clear separation of the profiles between the T and CK groups indicated that the model had a good predictive power, suggesting the existence of DEMs between the T and CK groups.

Figure 3. OPLS-DA score map (A) and displacement test (B) of metabolomics. OPLS-DA score plot and permutation test for the metabolome of Sphingomonas sp. Z392 under high levels of ammonia sulfate.
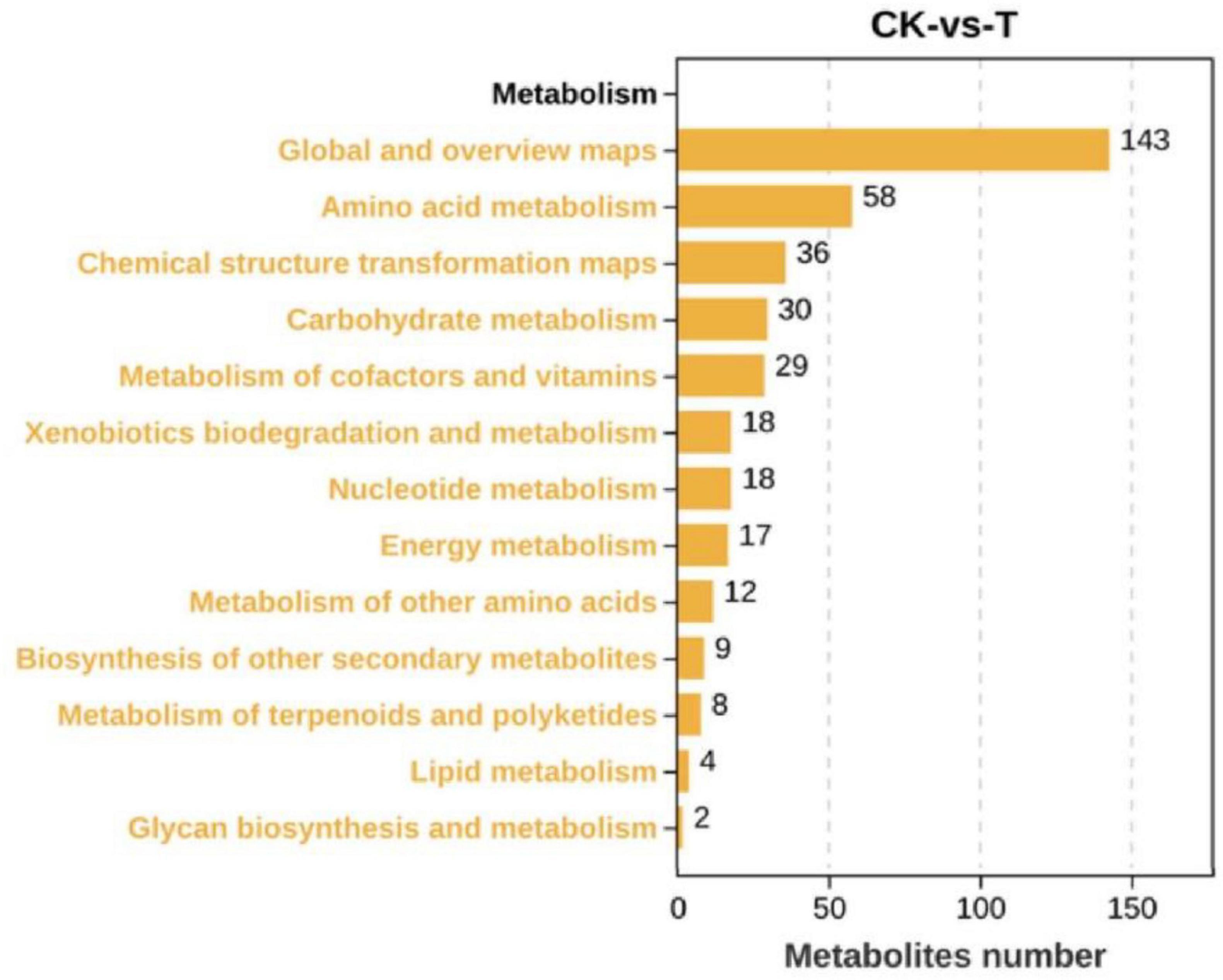
As shown in Figure 4, there were 442 DEMs, of which 223 were significantly upregulated, and 219 were significantly downregulated when the Sphingomonas sp. Z392 was cultured under the high ammonium sulfate. Then, the KEGG metabolic pathways were enriched in DEMs. As shown in Figure 5, the DEMs were involved in the following pathways: amino acid metabolism, metabolism of other amino acids, nucleotide metabolism, carbohydrate metabolism, metabolism of cofactors and vitamins, biosynthesis of secondary metabolites, lipid metabolism, energy metabolism, xenobiotics biodegradation and metabolism, and metabolism of terpenoids and polyketides. Among these pathways, amino acid and nucleotide metabolisms are closely associated with nitrogen metabolism.
Figure 4. Differential expression metabolites of Sphingomonas sp. Z392 under high levels of ammonia sulfate., the Y-axis represents the number of up- and downregulated metabolites, orange represents upregulation, and blue represents downregulation.
As shown in Figure 6, under high ammonium sulfate condition, the levels of carboxyaminoimidazole ribonucleotide (CAIR), 5-formamidoimidazole-4-carboxamide ribotide (FAICAR), IMP, and xanthosine monophosphate (XMP) of Sphingomonas sp. Z392 in purine metabolism were downregulated. In addition, under high ammonium sulfate condition, the levels of ornithine and arginine in the arginine metabolism pathway were downregulated. Therefore, under high concentration of ammonium sulfate, the concentration of arginine in Sphingomonas sp. Z392 might decrease, potentially leading to a reduced flux of arginine deamination to produce putrescine, which in turn decreases the production of NH3 from arginine decomposition.
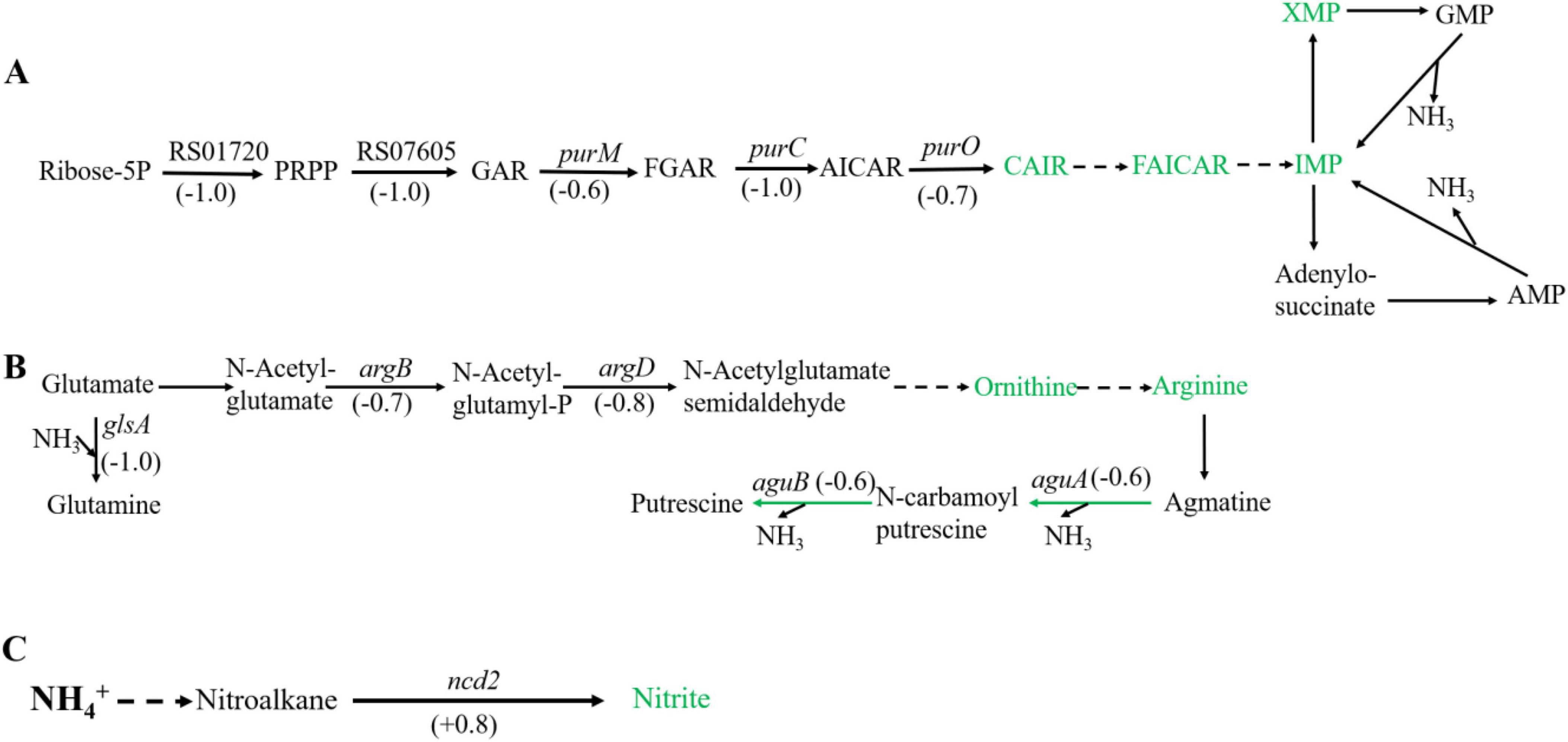
Figure 6. Differential expression metabolites in the purine metabolism (A) and arginine metabolism (B) pathways, and nitrogen metabolism pathway (C). The green metabolites indicate downregulation, and the red metabolites indicate upregulation.
Intercellular nitrite and NH3 are closely related (Figure 2C), this study used ion chromatography to detect the intercellular nitrite content of Sphingomonas sp. Z392. Under high concentration of ammonium sulfate, the intercellular nitrite content of Sphingomonas sp. Z392 was 17.67 mg/kg, which was 2.27 times that of the Sphingomonas sp. Z392 without ammonium sulfate (7.77 mg/kg) (Figure 7).The above results showed that under high ammonium sulfate concentration, the transcription levels of RS01720, RS07605, purM, purC, and purO genes in the purine metabolism were significantly downregulated, and the corresponding levels of CAIR, FAICAR, IMP, and XMP of purine metabolism were also downregulated; In arginine biosynthesis and metabolism pathways, the transcription levels of glsA, argB, argD, aguB, and aguA genes were significantly downregulated, and the levels of ornithine and arginine were also downregulated. In addition, under high ammonium sulfate concentration, the transcription level of ncd2 gene in nitrogen metabolism was significantly upregulated, and the intracellular nitrate content increased markedly.
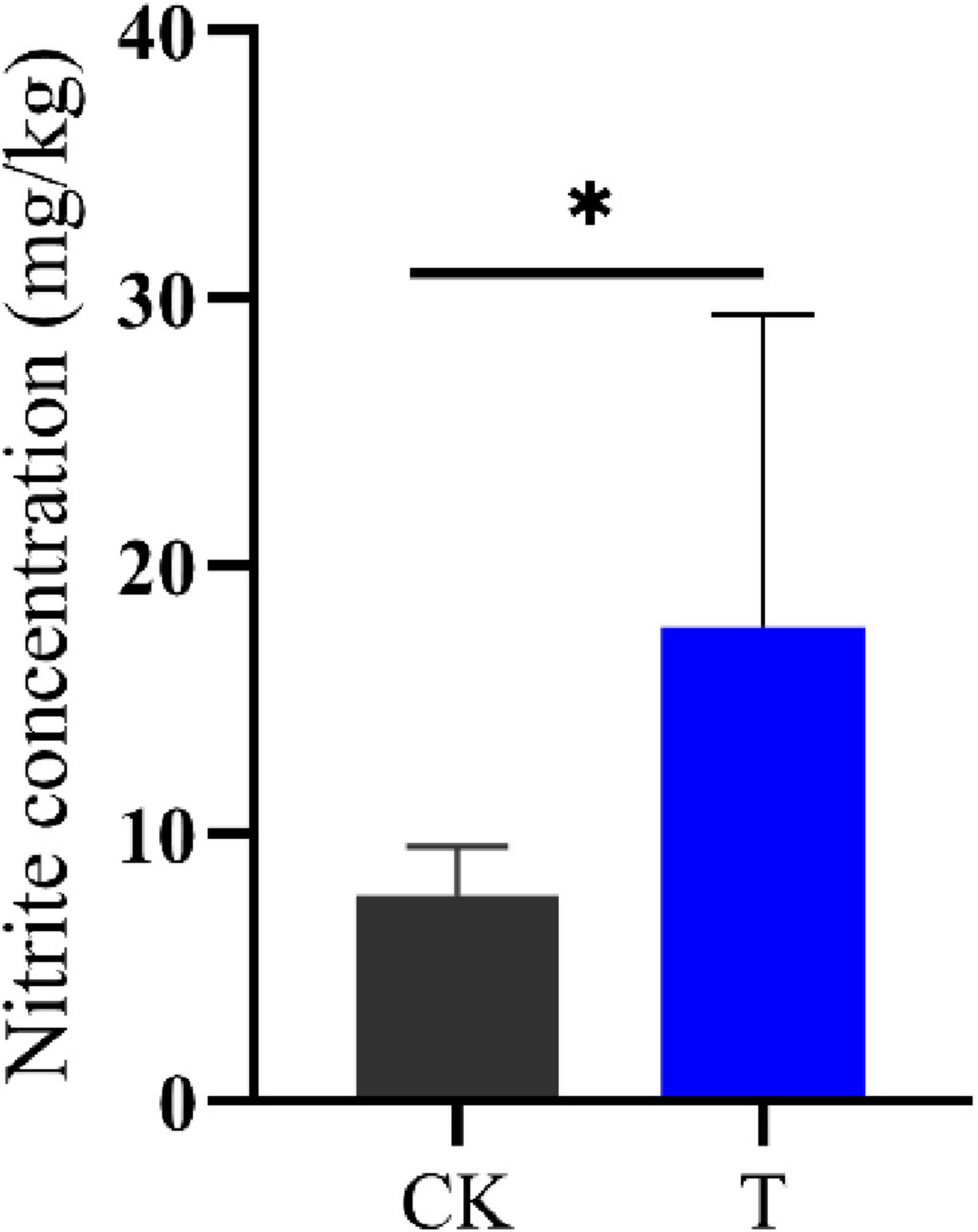
Figure 7. The nitrite concentrations of Sphingomonas sp. Z392 under conditions with (T) and without (CK) high levels of ammonia sulfate. * represents p < 0.05.
3.3 Knockout and overexpression of the ncd2 gene in Sphingomonas sp. Z392
The above results demonstrated that the ncd2 gene may contribute to ammonia reduction in Sphingomonas sp. Z392. To further explore the effect of the ncd2 gene on the ammonia degradation ability of Sphingomonas sp. Z392, this study overexpressed and knocked out the ncd2 gene in Sphingomonas sp. Z392, respectively, and named them Z392 OVncd2 and Z392 Δncd2. To validate the overexpression of ncd2, we performed quantitative RT-qPCR. As shown in Figure 8, the expression level of the ncd2 in Z392 OVncd2 was significantly increased than that of the Sphingomonas sp. Z392. Furthermore, the genomic DNA of Z392 Δncd2 was used as template, we amplified the target band with primers flanking ncd2 and confirmed the successful knockout of the ncd2 gene through sequencing.
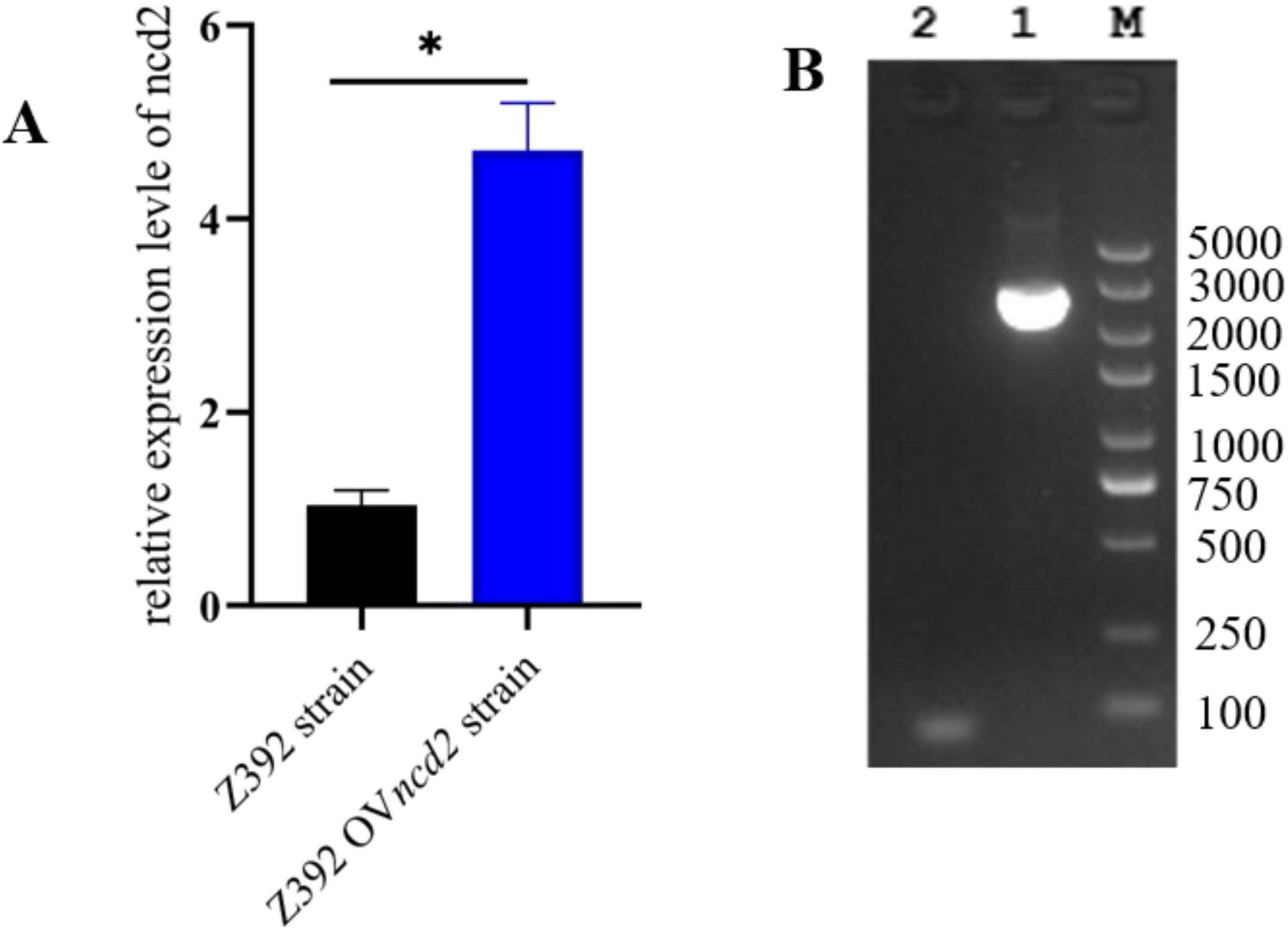
Figure 8. The verification of ncd2 overexpression and knockout in Sphingomonas sp. Z392 by RT-qPCR (A) and PCR amplification (B), respectively. Lane 1 represents band by the external primers, Lane 2 represents the negative band. M represents DL 5000 DNA Marker. * represents p < 0.05.
3.4 Effect of ncd2 on nitrogen content in chicken feces
To confirm the effect of ncd2 on ammonia reduction, the broiler chickens were fed with Sphingomonas sp. Z392, Z392 OVncd2, and Z392 Δncd2 strains, respectively. Chicken manure was collected and tested for its contents of total nitrogen, ammonia nitrogen, and nitrate nitrogen (Figure 9).
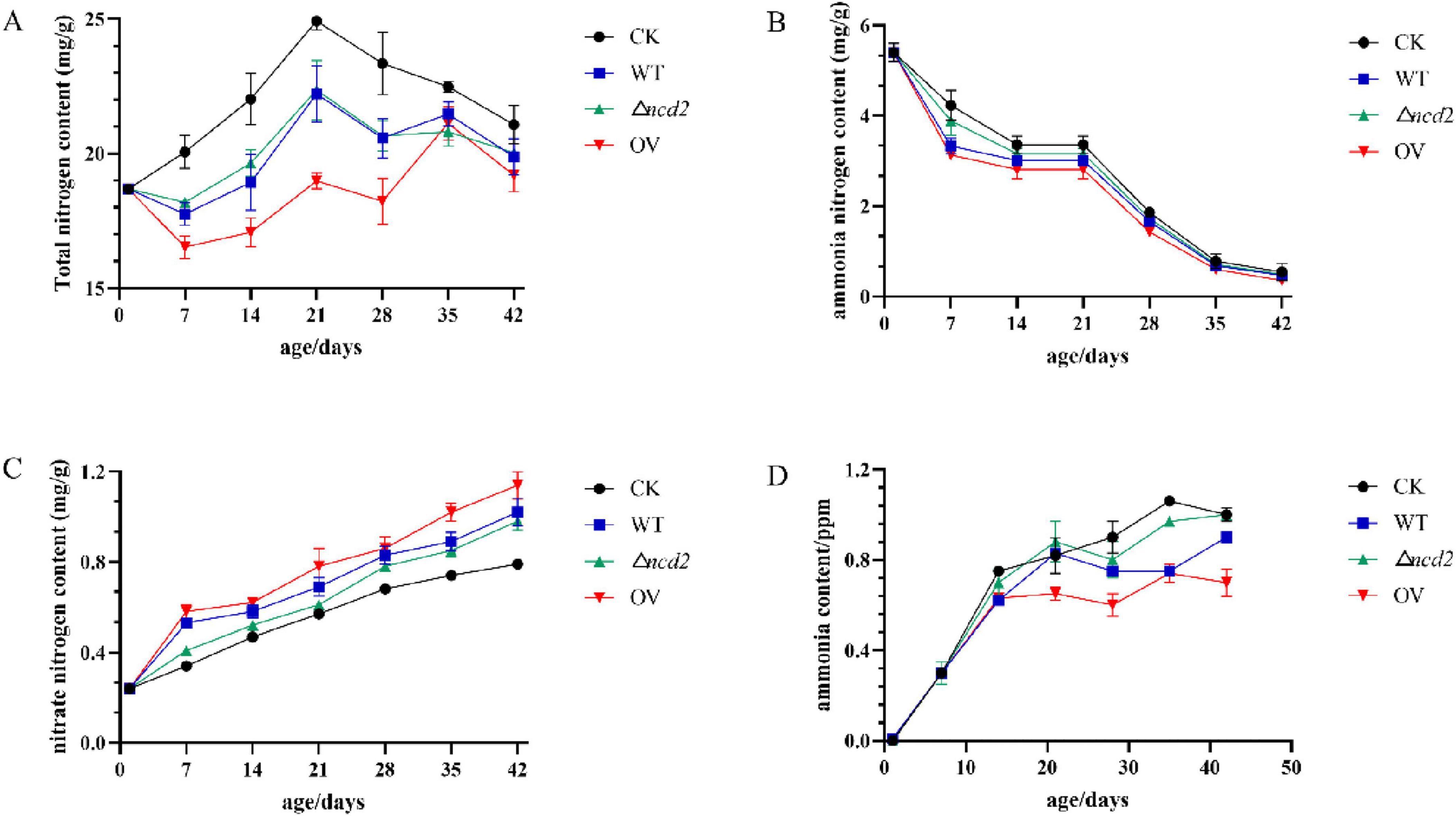
Figure 9. The changes of total nitrogen (A), ammonium nitrogen (B), nitrate nitrogen (C), and ammonia (D) contents in chicken manure fed with Spingomonas sp. Z392, Z392 OVncd2, Z392 Z392 Δncd2, and without Spingomonas sp.
When the broiler chickens were fed with Sphingomonas sp. Z392, Z392 OVncd2, and Z392 Δncd2 strains, the contents of total nitrogen and ammonium nitrogen in chicken manure were significantly decreased than that of without Sphingomonas sp., while nitrate nitrogen content was significantly increased than that of without Sphingomonas sp. Furthermore, Z392 OVncd2 strain exhibited the most ammonia reduction capability. The ammonia concentration of the group fed with Z392 OVncd2 decreased by 43.33% than that of without Sphingomonas sp. group, and decreased by 14.17% than that of group fed with Sphingomonas sp. Z392. In summary, overexpressing the ncd2 gene in Sphingomonas sp. Z392 resulted in reduced ammonia emission and ammonium nitrogen content, while increasing nitrate nitrogen content in chicken coop. Overexpression of ncd2 facilitated the conversion of ammonium nitrogen in chicken manure into nitrate nitrogen, thereby reducing ammonia emissions from chicken manure.
4 Discussion
Currently, biodegradation has received an increasing attention due to its advantages of low cost, high efficiency, simple operation, and environmental friendliness (Mi et al., 2019). The microorganisms reported to reduce ammonia mainly include Alcaligenes faecalis, Pseudomonas, Rhodococcus, Acinetobacter, and Bacillus methylotrophicus (Kim et al., 2005; Mi et al., 2019; Robertson and Kuenen, 1990; Zhao et al., 2010). However, the specific degradation mechanisms of these microorganisms toward ammonia are still unclear. Sphingomonas exhibits potential ammonia degradation ability due to its numerous pollutant degradation genes, but its systematic degradation pathway remains to be further studied.
In this study, Sphingomonas sp. Z392 was cultured under high ammonium sulfate environment, and transcriptome and metabolome analysis were performed jointly. It was found that the expression levels of genes related to purine metabolism and arginine metabolism in Sphingomonas sp. Z392 were decreased, and the levels of ornithine, arginine, IMP, and XMP were also downregulated. Purine metabolism and arginine metabolism are closely related to the intracellular ammonia concentration, under high ammonium sulfate environment, the flux of purine and arginine metabolisms might decrease, which might contribute to reduce NH3 release. Under high ammonium sulfate conditions, Sphingomonas sp. Z392 reduced the flux of purine metabolism, which not only helps to reduce ammonia release but also saves cellular energy, enhancing its survival ability in adverse environments. However, purine metabolism is closely related to the growth and reproduction of Sphingomonas sp. Z392, so the reduction of this metabolic flux can only be within a certain range, as excessive reduction may have adverse effects on cell growth and reproduction (Zhao et al., 2010). Arginine metabolism is an essential biochemical process for microorganisms to maintain normal growth and metabolism. This study found that under high ammonium sulfate conditions, the transcription levels of genes in arginine metabolism of Sphingomonas sp. Z392 decreased, resulting in reduced the levels of arginine and ornithine. Similarly, although the decrease in arginine metabolism flux could decrease NH3 release, this flux could only be reduced to a certain extent, otherwise it would affect the growth of Sphingomonas sp. Z392. Therefore, the reduction of flux in purine and arginine metabolism pathways might decrease ammonia release to some extent, but it cannot serve as the primary strategy. It is necessary to comprehensively consider the metabolic characteristics and growth requirements of Sphingomonas sp. to seek more effective and sustainable ammonia reduction solutions.
In addition, this study also found that under high ammonium sulfate environment, the transcriptional level of the ncd2 in nitrogen metabolism pathway was upregulated, accompanied by a 2.27-fold increase in intracellular nitrite content. This pathway potentially converts NH3 into nitrite through a series of complex reactions, leading to a reduction in NH3 level. The ncd2 encodes nitroalkane monooxygenase, which has been studied for the degradation of nitroalkanes (Gadda and Francis, 2010; Rolfes, 2006). After knocking out the ncd2 gene, the ammonia concentrations in the chicken coop significantly increased, while the ncd2-overexpressing strain significantly reduced the ammonia concentration in the chicken coop, which was consistent with the previous studies (Vodovoz and Gadda, 2020; Nishino et al., 2010). Similar to the case of Penicillium and Pseudomonas aeruginosa, this enzyme is also involved in the metabolism of nitroalkanes, generating malondialdehyde, nitrite, and nitrate (Torres-Guzman et al., 2021). In this study, Sphingomonas sp. Z392 could increase the removal of ammonia by enhancing ammonia assimilation to form nitrite, which would become an effective pathway for ammonia degradation. Although, this study has discovered that Sphingomonas sp. Z392 might utilize the nitroalkane monooxygenase encoded by ncd2 to convert nitromethane into nitrite, the specific pathway for ammonia to transform into nitromethane remains unknown. Current research suggested that completely ammonia-oxidizing bacteria have the potential for safe ammonia oxidation, which can directly oxidize ammonia to nitrate. The specific pathway for Sphingomonas sp. Z392 to convert ammonia into nitromethane still needs further exploration.
5 Conclusion
In this study, under high concentration of ammonium sulfate, the transcription levels of the genes related to purine metabolism and arginine metabolism pathways were significantly downregulated, and the corresponding levels of ornithine, arginine, IMP, and XMP were also downregulated in Sphingomonas sp. Z392. In addition, under high ammonium sulfate concentration, the transcription level of ncd2 gene in nitrogen metabolism was significantly upregulated, and the intracellular nitrate content increased markedly. Under high concentration of ammonium sulfate, Sphingomonas sp. Z392 might reduce the release of NH3 by decreasing the flux of purine metabolism and arginine metabolism pathway, and increasing the assimilation flux of ammonia. Furthermore, ncd2 was a key gene for ammonia reduction in Sphingomonas sp. Z392. In a word, this study not only systematically clarifies the metabolic pathway of ammonia reduction in Sphingomonas, but also points out future research directions.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: BioProject: Processed PRJNA1112986: the transcriptome of Sphingomonas sp. Z392 under high ammonium sulfate.
Ethics statement
The animal studies were approved by the Ethics Committee of Experimental Animals of Huanghuai University and strictly follows the provisions of the “Ethical Review of Experimental Animals of Huanghuai University” (Acceptance Number: 202009220005). The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
WM: Formal Analysis, Methodology, Writing – original draft, Writing – review and editing. LD: Investigation, Methodology, Writing – original draft. XH: Investigation, Methodology, Writing – original draft. WG: Data curation, Writing – original draft. LC: Data curation, Writing – original draft. GY: Data curation, Writing – original draft. GA: Writing – original draft, Writing – review and editing.
Funding
The authors declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by the Key R&D projects of Ningxia Hui Autonomous Region (2021BEE02032), the Science and Technology Development Plan Project of Henan province of China (172102110201 and 212102110005), and Natural Science Foundation of Henan Province (232300421270).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2024.1437056/full#supplementary-material
References
Bist, R. B., Subedi, S., Chai, L., and Yang, X. (2023). Ammonia emissions, impacts, and mitigation strategies for poultry production: a critical review. J. Environ. Manage. 328:116919. doi: 10.1016/j.jenvman.2022.116919
Chen, Q., and Ni, J. (2012). Ammonium removal by Agrobacterium sp. LAD9 capable of heterotrophic nitrification-aerobic denitrification. J. Biosci. Bioeng. 113, 619–623. doi: 10.1016/j.jbiosc.2011.12.012
Gadda, G., and Francis, K. (2010). Nitronate monooxygenase, a model for anionic flavin semiquinone intermediates in oxidative catalysis. Arch. Biochem. Biophys. 493, 53–61. doi: 10.1016/j.abb.2009.06.018
Gooding, J. R., Jensen, M. V., Dai, X., Wenner, B. R., Lu, D., Arumugam, R., et al. (2015). Adenylosuccinate is an insulin secretagogue derived from glucose-induced purine metabolism. Cell Rep. 13, 157–167. doi: 10.1016/j.celrep.2015.08.072
Haq, S. U., Aghajamali, M., and Hassanzadeh, H. (2021). Cost-effective and sensitive anthocyanin-based paper sensors for rapid ammonia detection in aqueous solutions. RSC Adv. 11, 24387–24397. doi: 10.1039/d1ra04069c
Huang, X., Li, W., Zhang, D., and Qin, W. (2013). Ammonium removal by a novel oligotrophic Acinetobacter sp. Y16 capable of heterotrophic nitrification-aerobic denitrification at low temperature. Bioresour. Technol. 146, 44–50. doi: 10.1016/j.biortech.2013.07.046
Ji, J., Zhao, Y., Wang, H., Jiang, L., Yuan, X., and Wang, H. (2022). Resource utilization of chicken manure to produce biochar for effective removal of levofloxacin hydrochloride through peroxymonosulfate activation: the synergetic function of graphitization and nitrogen functionality. Chemosphere 309(Pt. 1):136419. doi: 10.1016/j.chemosphere.2022.136419
Jin, R., Liu, T., Liu, G., Zhou, J., Huang, J., and Wang, A. (2015). Simultaneous heterotrophic nitrification and aerobic denitrification by the marine origin bacterium Pseudomonas sp. ADN-42. Appl. Biochem. Biotechnol. 175, 2000–2011. doi: 10.1007/s12010-014-1406-0
Kim, J. K., Park, K. J., Cho, K. S., Nam, S. W., Park, T. J., and Bajpai, R. (2005). Aerobic nitrification-denitrification by heterotrophic Bacillus strains. Bioresour. Technol. 96, 1897–1906. doi: 10.1016/j.biortech.2005.01.040
Landete, J. M., Arena, M. E., Pardo, I., Manca de Nadra, M. C., and Ferrer, S. (2010). The role of two families of bacterial enzymes in putrescine synthesis from agmatine via agmatine deiminase. Int. Microbiol. 13, 169–177. doi: 10.2436/20.1501.01.123
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Lubensky, J., Ellersdorfer, M., and Stocker, K. (2019). Ammonium recovery from model solutions and sludge liquor with a combined ion exchange and air stripping process. J. Water Process Eng. 32:100909.
Mi, J., Chen, X., and Liao, X. (2019). Screening of single or combined administration of 9 probiotics to reduce ammonia emissions from laying hens. Poult. Sci. 98, 3977–3988. doi: 10.3382/ps/pez138
Nancharaiah, Y. V., Venkata Mohan, S., and Lens, P. N. L. (2016). Recent advances in nutrient removal and recovery in biological and bioelectrochemical systems. Bioresour. Technol. 215, 173–185. doi: 10.1016/j.biortech.2016.03.129
Nishino, S. F., Shin, K. A., Payne, R. B., and Spain, J. C. (2010). Growth of bacteria on 3-nitropropionic acid as a sole source of carbon, nitrogen, and energy. Appl. Environ. Microbiol. 76, 3590–3598. doi: 10.1128/aem.00267-10
Pang, Y., and Wang, J. (2021). Various electron donors for biological nitrate removal: a review. Sci. Total Environ. 794:148699. doi: 10.1016/j.scitotenv.2021.148699
Pedley, A. M., and Benkovic, S. J. (2017). A new view into the regulation of purine metabolism: the purinosome. Trends Biochem. Sci. 42, 141–154. doi: 10.1016/j.tibs.2016.09.009
Robertson, L. A., and Kuenen, J. G. (1990). Combined heterotrophic nitrification and aerobic denitrification in Thiosphaera pantotropha and other bacteria. Antonie Van Leeuwenhoek 57, 139–152. doi: 10.1007/bf00403948
Rolfes, R. J. (2006). Regulation of purine nucleotide biosynthesis: in yeast and beyond. Biochem. Soc. Trans. 34(Pt. 5), 786–790. doi: 10.1042/bst0340786
Such, N., Mezõlaki, Á, Rawash, M. A., Tewelde, K. G., Pál, L., Wágner, L., et al. (2023). Diet composition and using probiotics or symbiotics can modify the urinary and faecal nitrogen ratio of broiler chicken’s excreta and also the dynamics of in vitro ammonia emission. Animals (Basel) 13:332. doi: 10.3390/ani13030332
Torres-Guzman, J. C., Padilla-Guerrero, I. E., Cervantes-Quintero, K. Y., Martinez-Vazquez, A., Ibarra-Guzman, M., and Gonzalez-Hernandez, G. A. (2021). Peculiarities of nitronate monooxygenases and perspectives for in vivo and in vitro applications. Appl. Microbiol. Biotechnol. 105, 8019–8032. doi: 10.1007/s00253-021-11623-1
Varão Moura, A., Aparecido Rosini Silva, A., Domingos Santo da Silva, J., Aleixo Leal Pedroza, L., Bornhorst, J., Stiboller, M., et al. (2022). Determination of ions in Caenorhabditis elegans by ion chromatography. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 1204:123312. doi: 10.1016/j.jchromb.2022.123312
Vodovoz, M., and Gadda, G. (2020). Kinetic solvent viscosity effects reveal a protein isomerization in the reductive half-reaction of Neurospora crassa class II nitronate monooxygenase. Arch. Biochem. Biophys. 695:108625. doi: 10.1016/j.abb.2020.108625
Wang, M. C., Wang, Y., Wang, G. L., Xu, L. L., and Li, E. Z. (2021). Isolation and identification of Sphingomonas sp. from chicken cecum and its ammonia-degrading activity. J. Biotech Res. 12, 65–73.
Zhao, B., He, Y. L., Hughes, J., and Zhang, X. F. (2010). Heterotrophic nitrogen removal by a newly isolated Acinetobacter calcoaceticus HNR. Bioresour. Technol. 101, 5194–5200. doi: 10.1016/j.biortech.2010.02.043
Keywords: ammonia reduction, ncd2 gene, transcriptomics, metabolomics, Sphingomonas ammonia reduction by Sphingomonas
Citation: Mingcheng W, Daoqi L, Huili X, Gailing W, Chaoying L, Yanan G and Aizhen G (2025) Multiomics-based analysis of the mechanism of ammonia reduction in Sphingomonas. Front. Microbiol. 15:1437056. doi: 10.3389/fmicb.2024.1437056
Received: 23 May 2024; Accepted: 06 December 2024;
Published: 01 May 2025.
Edited by:
Shuhao Huo, Jiangsu University, ChinaCopyright © 2025 Mingcheng, Daoqi, Huili, Gailing, Chaoying, Yanan and Aizhen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guo Aizhen, YWl6aGVuQG1haWwuaHphdS5lZHUuY24=
 Wang Mingcheng
Wang Mingcheng Liu Daoqi4
Liu Daoqi4 Guo Aizhen
Guo Aizhen