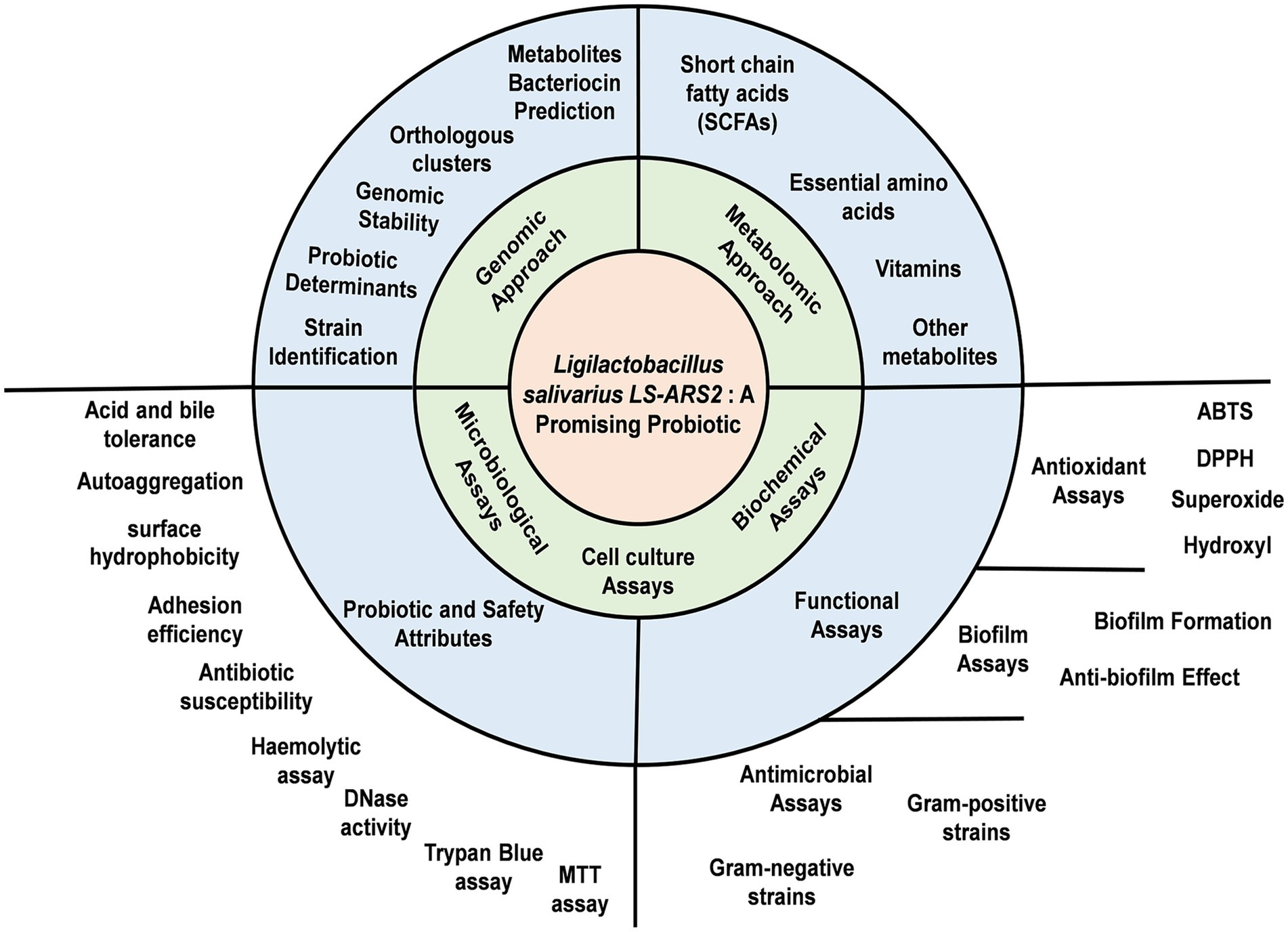
- 1Biochemistry and Cell Biology Laboratory, School of Basic Sciences, Indian Institute of Technology Bhubaneswar, Odisha, India
- 2S. K. Dash Center of Excellence of Biosciences and Engineering & Technology (SKBET), Indian Institute of Technology Bhubaneswar, Odisha, India
Background: Probiotics restore microbial balance and prevent gut-inflammation. Therefore, finding out novel probiotic strains is a demand. As gut-microbe, benefits of Ligilactobacillus salivarius (LS) are established. However, strain-specific detailed studies are limited. Here, we illustrate probiotic attributes of novel LS-ARS2 for its potential application as food-supplement and/or therapeutic to improve gut-health.
Methods: Whole genome sequencing (WGS) and phylogenetic analysis confirm the strain as LS. To establish probiotic properties, acid-bile tolerance, auto-aggregation, cell-surface-hydrophobicity, biofilm-formation, and adhesion-assays are performed. To ensure safety attributes, antibiotic-susceptibility, hemolytic, DNase, trypan-blue, and MTT assays are done. ABTS, DPPH, superoxide, hydroxyl free radical scavenging assays are used to determine anti-oxidant potential. Antibacterial assays, including co-culture assay with pathogen and pathogenic biofilm-inhibition assays, are performed to explore antibacterial efficacy. To characterize metabolic-profile of LS-ARS2-derived cell-free-supernatant (CFS), HRMS analysis are carried out. Consequently, WGS-analyses predict potential molecular associations related to functional outcomes.
Results: We find LS-ARS2 a remarkable fast-growing strain that shows acid and bile tolerance (>60% survival rate), indicating promising gut-sustainability. High auto-aggregation capacity (>80%), robust cell-surface hydrophobicity (>85%), and adhesion efficacy to Caco-2 cells illustrate significant potential of LS-ARS2 for gut colonization. Fascinatingly, LS-ARS2 is able to form biofilm within 24 h (p < 0.0001), rare among LS strains, indicating the potential of the strain for efficient stay in the gut. The strain ensures safety attributes. LS-ARS2-WGS analysis recognizes probiotic-specific determinants, predicts genomic stability, identifies orthologous-clusters for diverse functions, and predicts metabolites and bacteriocins. HRMS-studies with LS-ARS2-CFS further validate the presence of diverse beneficial metabolites with antimicrobial and immunomodulatory potential. LS-ARS2 shows significant antioxidant properties in ABTS (>60%), DPPH (>10 U/mL), superoxide (>70%), and hydroxyl free radical scavenging assays (>70%). Further, LS-ARS2 shows antimicrobial activities against Gram-positive Methicillin-resistant Staphylococcus aureus (MRSA) and Gram-negative multidrug-resistant clinical strains enterotoxigenic Escherichia coli, Vibrio cholerae, and Shigella flexneri. Anti-Salmonella effect of LS-ARS2 is prominent (p < 0.0001). Most interestingly, LS-ARS2-CFS inhibits MRSA-biofilm (p < 0.0001), again rare among LS strains.
Conclusion: LS-ARS2 is a novel, fast-growing, biofilm-forming probiotic with significant antioxidant, antibacterial, and anti-biofilm potentials, suggesting the promising applications of LS-ARS2 for combating pathogenic biofilms and improving gut-health. However, further in vivo studies would facilitate their potential applications.
1 Introduction
The human gut is colonized by 100 trillion bacteria along with fungi, viruses, protozoans, and archaea, which is collectively called gut-microbiota (Saxami et al., 2023). The gut microbiome is a dynamic ecosystem that manifests a symbiotic relationship with the host. It regulates host metabolism and immunity and preserves intestinal barrier function by restoring a balance between beneficial and pathogenic bacteria in the gut. However, several factors, like diets rich in processed food and refined carbohydrates, unhealthy lifestyles, and uncontrolled medications, jeopardize the equilibrium of gut-microbial composition, which is termed dysbiosis (Pedroza Matute and Iyavoo, 2023). As a result, barrier integrity is disrupted, and permeability is increased, leading to the entry of metabolites and endotoxins from pathogens. This triggers the activation of the immune cells and intestinal inflammation. Constant elevation of inflammatory mediators further promotes chronic inflammatory diseases like cancer, chronic respiratory diseases, atherosclerosis, stroke, and type 2 diabetes mellitus (Hakansson and Molin, 2011). As reported by the World Health Organization (WHO), inflammation-associated diseases are emerging as the most severe health hazard, with around 60% of all deaths across the world (Furman et al., 2019). Interestingly, probiotics can restore microbial balance in the intestine and reduce the severity of gut-inflammation (Patra et al., 2022; Patra et al., 2021). Therefore, probiotic intake as a diet or food supplement could be a preventive strategy for these inflammatory diseases.
Lactic acid bacteria (LAB) are the major source of probiotics. However, not all LAB are probiotics and should be evaluated for their probiotic attributes and safety profile (Ayed et al., 2024). According to the International Scientific Association for Probiotics and Prebiotics (ISAPP), for probiotic application, the strain must be tolerant to harsh conditions (acidic and alkaline) to survive during passage through the gastrointestinal tract. It should have auto-aggregation and adhesion capacity to adhere and colonize the intestinal mucosa. After colonization, the bacteria must have beneficial effects, like the secretion of bioactive metabolites, and/or should depict antimicrobial effects for pathogens. Most importantly, it should be safe for further application (Navarré et al., 2024). Although many potential probiotic strains have been identified, their strain-specific behaviors, safety issues, and lack of reproducibility in animal studies hold their future application (Dash et al., 2023). Therefore, the search for novel probiotics is in constant demand. Moreover, studies on gut-inflammatory diseases have shown that the general physiological benefits of probiotics are strain-specific (McFarland et al., 2018). Therefore, the selection of novel probiotic strains requires well-rounded sequential evaluations for crucial probiotic properties.
Probiotics originating from the intestinal microbiota have a more preventive effect on host health than probiotics isolated from other sources (Timmerman et al., 2006). Lactobacillus salivarius is such a LAB that is majorly found in the oropharyngeal-gastrointestinal tract (OGT), milk, vagina, and oral cavities of humans (Yang et al., 2024; Guerrero Sanchez et al., 2022). Lactobacillus salivarius was recently reclassified from the Lactobacillus genus to a new genus called Ligilactobacillus (here onwards, we will follow the same), which refers to its vertebrate host (Guerrero Sanchez et al., 2022). Being a native of the intestinal microbiota, L. salivarius helps to maintain a healthy and balanced gut-microbiome with multiple health-benefits (Yang et al., 2024; Zhou et al., 2020; Quilodrán-Vega et al., 2020; Yang et al., 2023; Sevillano et al., 2023; Alba et al., 2025). However, despite their promising potential, compared to other LAB, in-depth studies on Ligilactobacillus salivarius strains are scarce (Jiang et al., 2023) (Supplementary Figure S1).
In this above-mentioned background, this study aims to comprehensively characterize the probiotic potential of the strain LS-ARS2 through whole genome sequencing (WGS) and functional validation. We systematically elucidate the physiological and functional prospects of the strain. For this, we perform genomic, and metabolomic analyses with functional studies to project LS-ARS2 as a multifaceted probiotic. WGS and phylogenetic analysis confirm the strain as Ligilactobacillus salivarius. Genomic predictions and functional assays validate key probiotic traits, including acid and bile tolerance, autoaggregation, cell-surface hydrophobicity, strong adhesion, and robust biofilm-forming capabilities of remarkably fast-growing LS-ARS2. The strain also exhibits significant antioxidant properties and potent antimicrobial activity against multi-drug resistant Gram-positive and Gram-negative pathogens, with its cell-free supernatant (CFS) effectively inhibiting pathogenic biofilms. Metabolic profiling through high-resolution mass spectrometry (HRMS) highlights the secretion of diverse health-promoting metabolites. Collectively, this study establishes LS-ARS2 as a promising probiotic candidate with potential applications as a functional food supplement and biotherapeutic for the improvement of gut-health.
2 Materials and methods
2.1 Bacterial culture
Pure culture of LS-ARS2 was procured from Microbial Type Culture Collection (MTCC), Institute of Microbial Technology (IMTECH), Chandigarh, India. The strain was maintained on MRS agar and MRS broth (HiMedia) in anaerobic conditions.
2.2 Reaffirmation of the novel strain using whole genome sequencing
2.2.1 Isolation of DNA and preparation of library
LS-ARS2 was grown in MRS broth anaerobically for 12 h at 37°C. The genomic DNA was isolated using a DNA mini kit [QiaAmp DNA Mini Kit (Cat# 51306)] and quantified by Qubit Fluorometer 3 using the Qubit dsDNA High Sensitivity Assay Kit (Invitrogen, Cat# Q32854). Then, it was further processed for library preparation with the 5,300 Fragment Analyzer (3.1.0.12) using ProSize data analysis software 4.0.0.3. DNA libraries were subjected to pair-end sequencing using the NovaSeq6000 platform (MedGenome, Bangalore, India) with a read length of 151 bp. The generated sequence data was assessed for quality control and processed to generate FASTQ files. The 16S rRNA gene sequencing was performed, and the obtained consensus sequence was analyzed with the NCBI-BLAST tool in comparison with similar sequences present in the repository database. Neighbor-joining (NJ) phylogenetic tree was constructed using MEGA12 to identify the strain (Kumar et al., 2024).
2.2.2 Determination of the strain identity by whole genome sequencing and genome assembly
The whole genome of the strain was sequenced [NovaSeq6000 (MedGenome, Bangalore, India)]. Initially, the reads were refined for contamination with human DNA. According to the alignment to the human genome (around 10.06–27.65%), the reads were filtered for further alignment with a reference genome. The fastq file generated was checked for parameters like distribution of sequence quality score, base quality score, GC content in the reads, average base content per read, over-represented sequences, PCR amplification issue, and adapters. These parameters assisted to ensure the precision of the sequencing results. The sequences of fastq files were trimmed according to the quality report to retain only high-quality sequences for further analysis. Fastq mcf (1.04.803) was used for adapter trimming, a process in which adapter sequences are removed from the 3′ end of the reads. Adapter trimming eliminates the chances of inference (due to adapters) in the alignment of the reads to a reference. For reference-guided assembly, the adapter trimmed reads were mapped to the suggested reference species Ligilactobacillus salivarius strain 609_LSAL to get the coverage and assembled fasta. The aligned consensus fasta files were generated [Samtools tools (version 1.2)]. Further, the depth statistics and coverage were generated using bedtools (version 2.0) and an in-house Perl script. The reference-aligned reads were used to predict variants using GATK, and the variants were annotated using the snpEff. The annotation of the primarily assembled genome was performed by applying the Prokaryotic Genome Annotation System (Prokka version 1.14.6) to predict genes, CDS, etc. (Seemann, 2014; Li and Durbin, 2009). The whole genome data can be retrieved from NCBI with the Bioproject Number PRJNA1024881.
2.3 Evaluation of probiotic attributes of LS-ARS2
2.3.1 Tolerance to acidic pH
An acid tolerance assay was performed, followed by modifications to the previous publication (Abouloifa et al., 2020). LS-ARS2 overnight culture was inoculated (1% v/v) in MRS broth adjusted to pH 4 and pH 3 with HCl (1 N, Merck), followed by incubation for 0, 1, 3, and 5 h at 37°C (Lab Companion, Korea). MRS broth (pH 6.5) was a control. After each time point, the culture was serially diluted in PBS (pH 7.4), 100 μL culture of appropriate dilution was plated on MRS agar, and incubated under anaerobic conditions at 37°C for 24 h. Colonies were counted, and the viable cells, or the biomass (Log10 CFU/mL), were calculated. Survival rate (%) = [biomass in pH 4 or 3/biomass in control pH 6.5] × 100. The experiment was performed in triplicate.
2.3.2 Tolerance to bile salt
A bile tolerance assay was investigated according to the previous publication with modifications (Abouloifa et al., 2020). 1% of overnight culture was inoculated in MRS broth containing 0.3 and 1% (w/v) bile salts (HiMedia). MRS without bile salt was a control. After incubation for 0, 1, 3, and 5 h at 37°C, each culture was serially diluted in PBS (pH 7.4), and 100 μL of the appropriate diluted culture was spread on MRS agar plate, thereafter incubated for 24 h at 37°C under anaerobic conditions. After 24 h, the biomass (Log10 CFU/mL) was calculated by counting the colonies. Survival rate (%) = [biomass in bile salt 0.3% or 1% / biomass in control] x 100. The experiment was performed in triplicate.
2.3.3 Determination of self-aggregation property of LS-ARS2
The aggregation property was evaluated according to the previous publications with modifications (Fonseca et al., 2021; Campana et al., 2017). 1% overnight culture of LS-ARS2 was sub-cultured in MRS broth under anaerobic conditions till OD600 = 0.5–0.6. The culture was then harvested at 5000 rpm, washed using PBS (pH 7.4) twice, and dissolved in the same (PBS, pH 7.4). The absorbance of the culture was adjusted to approximately 108 CFU/mL (OD600 0.25–0.26 ± 0.1, A0) (UV-1800 UV Spectrophotometer, Shimadzu, Japan), vortexed for 10 s, and incubated for 1, 2, 3, 4, 5, 6, 12, and 24 h at 37°C. After each time interval, the absorbance of the upper suspension was measured at 600 nm (Atime). The auto-aggregation percentage was determined by: Auto-aggregation (%) = [1 – (ATime/A0) × 100]. L. acidophilus DDS1, a widely used probiotic in clinical studies, was used as a positive control. The experiment was performed in triplicate.
2.3.4 Evaluation of cell surface hydrophobicity of LS-ARS2
The surface hydrophobicity of LS-ARS2 was evaluated using the Bacterial Attachment to Hydrocarbons (BATH) method, followed by the previous publication with modifications (Rosenberg et al., 1980). The overnight culture was washed with phosphate urea magnesium sulphate (PUM) buffer and dissolved in 10 mL PUM to reach OD600 (A0) of 0.8–0.9. The adjusted cell suspension (4.8 mL) and n-hexadecane (Sigma)/xylene (Merck) (0.8 mL) were mixed, followed by incubation for 10 min. The cell suspension was vortexed for 2 min and kept at 37°C for 2 h for the separation of phases. The absorbance of the lower aqueous phase was measured at 600 nm (A). Cell surface hydrophobicity (H%) was calculated as: Hydrophobicity (%) = [(1 – A/A0) × 100]. Probiotic strain L. acidophilus DDS1 was used as a positive control. The experiment was performed in triplicate.
2.3.5 Determination of adhesion property of LS-ARS2 using human colon adenocarcinoma cells (Caco-2)
Adhesion ability was studied using Caco-2 cell lines (ATCC), maintained in complete DMEM (Himedia), supplemented with 10% heat-inactivated FBS (Gibco) in 5% CO2 (Galaxy 48R, New Brunswick, Germany). 2×104 Caco-2 cells/well were seeded in six-well tissue culture plates (Dash et al., 2023; Ayala et al., 2019). After 80% confluent, the monolayer was washed using PBS (pH 7.4) twice, followed by incubation in serum-free DMEM overnight. Overnight bacteria at MOI (100:1) were co-cultured with Caco-2 cells and incubated for 2 h in a 5% CO2 atmosphere at 37°C. Next, cells were washed using PBS (pH 7.4) five times, trypsinized, and gently aspirated. After serial dilution, 100 μL of appropriate dilution was plated on MRS agar followed by incubation for 24 h at 37°C under anaerobic conditions. The attachment efficiency was determined by dividing the average number of bacteria (colonies after 24 h) attached per Caco-2 cell in each well (CFU/cell). The experiment was performed in triplicate.
2.3.6 Elucidation of adhesion properties of LS-ARS2 using phase-contrast confocal microscopy
To investigate the adhesion property, Caco-2 cells were seeded on poly-L lysine-coated (Sigma) coverslips (HiMedia) in 35 mm plates using complete DMEM and incubated up to 80% confluency (Dash et al., 2023). The cell monolayer was then washed using PBS (pH 7.4), followed by overnight incubation in serum-free DMEM. The bacteria at MOI (100:1) were co-cultured along with Caco-2 cells for 2 h, and unattached bacteria were washed by PBS (pH 7.4). The bacteria attached to Caco-2 cells were fixed with 4% paraformaldehyde for 30 min at 37°C, followed by washing with water. 15 μL of the mounting solution was placed on a glass slide, and the coverslip was placed on the mounting solution upside down. Then, the cells were dried at room temperature, and finally, images were taken in a confocal microscope (FV3000, Olympus, Japan).
2.3.7 Evaluation of biofilm-forming ability of LS-ARS2
The biofilm-forming ability of LS-ARS2 was investigated as described earlier with minor changes (Rocchetti et al., 2024; Baldassarri et al., 2006). Briefly, the LS-ARS2 strain was grown in MRS broth for 18 h under anaerobic conditions. Then, the culture was diluted in MRS broth till OD600 = 0.1 and distributed in 24-well plates. The plates were incubated anaerobically for 24 h at 37°C in a moist chamber. After the incubation period, the media was discarded, and the wells were washed with PBS (pH 7.4), and dried for 1 h at 60°C. The remaining bacterial cells were stained for 45 min with 0.1% (w/v) crystal violet (SRL) solution (dissolved in 95% ethanol), washed with PBS (pH 7.4), and air-dried. The stain was dissolved in 33% acetic acid (Merck), and absorbance was measured at 570 nm. Probiotic strains L. acidophilus DDS1 and L. rhamnosus GG were used as positive controls. The assay was performed in triplicate.
2.4 Determination of safety attributes of LS-ARS2
2.4.1 Prediction of prophage, CRISPR sequences, and antibiotic-resistant genes using genome analysis
PHAge Search Tool Enhanced Release (PHASTER) was implemented for the characterization of prophage sequences in the genome of LS-ARS2 (Arndt et al., 2016). CRISPRCasFinder 1.1.2 (Grissa et al., 2007) was applied to screen CRISPR, Cas sequences, and truncated Cas sequences. The Comprehensive Antibiotic Resistance Database (CARD 3.3.0) (Jia et al., 2016) and the Resistance Gene Identifier Tool (RGI 6.0.3) were employed to find out the genes responsible for antibiotic resistance in the strain sequence using the criteria of perfect and strict hit and high-quality coverage (Zankari et al., 2012). The ResFinder 4.4.2 server was adopted to recognize the genes associated with acquired antimicrobial resistance with a threshold for %ID selected as 90.00% and minimum length selected as 60% and/or chromosomal mutations (Bortolaia et al., 2020). The identification of genes encoding virulence factors and toxins in the LS-ARS2 genome was performed using a BLAST search against the virulence factor database (VFDB) available at http://www.mgc.ac.cn/cgi-bin/VFs/v5/main.cgi (Liu et al., 2019). The presence of any pathogenic elements in the genome of LS-ARS2 was examined using the PathogenFinder tool (Cosentino et al., 2013).
2.4.2 Evaluation of antibiotic susceptibility of LS-ARS2
The susceptibility of LS-ARS2 to antibiotics was checked by the disc diffusion pattern following the previous publication (Fonseca et al., 2021). Overnight culture was inoculated on MRS agar plates, and discs containing antibiotics (HiMedia) were placed. Inhibition zone diameters (mm) were measured after incubation for 24 h at 37°C under anaerobic conditions. The susceptibility of the strain to the antibiotics was categorized as resistant (R), intermediate susceptible (I), or susceptible (S) according to CLSI guidelines (CLSI, 2020). The experiment was performed in triplicate.
2.4.3 Assessment of haemolytic activity of LS-ARS2
Overnight culture was spot-inoculated on a sheep blood agar plate (defibrinated, 5% w/v, HiMedia) and incubated for 24 h at 37°C (Fonseca et al., 2021). The hydrolysis of blood cells resulted in a clear zone surrounding the colonies, which was considered β-hemolysis, whereas green-hued zones surrounding colonies designated partial hydrolysis, and were considered α-hemolysis, finally, no zone surrounding colonies was considered γ-hemolysis or non-haemolytic. Staphylococcus aureus ATCC 25923 was employed as a β-hemolysis positive control, Escherichia coli ATCC 25922 as a positive control for α-hemolysis, and reference probiotic strain L. acidophilus DDS1 for γ-hemolysis. The experiment was performed in triplicate.
2.4.4 Estimation of DNase activity of LS-ARS2
Overnight culture was spot-inoculated on a DNase agar plate (HiMedia) and incubated at 37°C for 24 h (Fonseca et al., 2021). Staphylococcus aureus ATCC 25923 was considered a positive control, and the reference probiotic strain L. acidophilus DDS1 was a negative control for DNase activity. After incubation, the plates were flooded with 1 N HCl to develop a clear zone around the DNase producer colony. No clear zone around colonies was considered as negative DNase activity. The experiment was performed in triplicate.
2.4.5 Evaluation of safety properties of LS-ARS2 for human colon adenocarcinoma cells HCT116
The viability of LS-ARS2-treated colon cancer cells HCT116 (ATCC) was checked by the trypan blue exclusion assay based on the method described earlier, with modifications (Pradhan et al., 2016). Briefly, 1×105 HCT116 cells/well were seeded in complete DMEM (Himedia) supplemented with 10% heat-inactivated FBS (Gibco), and after attachment, the cells were treated with LS-ARS2 at 100 MOI and incubated for 6, 12, and 24 h. The media was replenished every 3–4 h to avoid deprivation of the nutrients. After each time point, the cells were harvested and mixed with the same volume of 0.4% trypan blue (HiMedia). HCT116 without LS-ARS2 treatment was considered a negative control. The living and the dead cells were counted. Cell viability was calculated as the number of viable cells divided by the total number of cells. As a reference, probiotic strain L. acidophilus DDS1 was used. The experiment was performed in triplicate.
2.4.6 Assessment of safety properties of LS-ARS2-derived CFS in the stationary phase of Caco-2 cells
Cytotoxicity of LS-ARS2-derived CFS was assessed on Caco-2 cells according to the previously narrated method with modifications (Pereira et al., 2022). LS-ARS2 was grown in MRS broth, followed by transfer in complete DMEM for 18 h in a static condition at 37°C. The culture was then centrifuged at 4000 rpm for 20 min at 4°C, followed by filtration through a 0.22 μm filter to collect the CFS. Briefly, 2 × 104 Caco-2 cells/well were seeded in 96-well plates in complete DMEM and allowed to grow for 7 days, with renewal of the media every 48 h. The confluent monolayer of Caco-2 in the stationary phase mimics the normal intestine cells. Hence, the viability of the cells after treatment assessed the safety of LS-ARS2-CFS for the host cells (Pereira et al., 2022). On day 7, cells were treated with different concentrations of CFS and pH-neutralized CFS (10, 20, 30, 40, 50, and 60% CFS, diluted with a complete medium) and incubated for 24 h. Caco-2 cells without treatment with CFS were considered a negative control. After the incubation period, 10 μL of MTT reagent (Sigma) was added and incubated for 4 h. Then, 100 μL DMSO (SRL) was added, and MTT absorbance was measured at 570 nm with background correction at 670 nm on the Spectramax iD3. The experiment was performed in duplicate with two technical repeats.
2.5 Evaluation of antioxidant properties of LS-ARS2
2.5.1 Preparation of the intact and heat-lysed cells
Overnight cultures were washed by PBS (pH 7.4) twice. The cell pellet was re-suspended in distilled water to a final OD600 = 1.0. Heat-lysed cells were prepared by heating samples at 95°C (water bath) for 30 min (Ramalho et al., 2019).
2.5.2 Assessment of ABTS cation radical scavenging capacity of LS-ARS2
The ABTS cation radical scavenging assay was implemented as stated earlier with minor changes (Kim et al., 2022). A working solution of ABTS (Sigma) was prepared by combining 7 mM ABTS and 2.45 mM potassium persulphate (HiMedia) in equal volume (1:1 v/v). The working solution was diluted with methanol (HiMedia, HPLC) up to OD734 = 0.7. 0.6 mL of samples (intact cells or heat-lysed cells) were mixed with 1.2 mL of ABTS solution and incubated for 30 min (dark, room temperature). Distilled water was used as a control. ABTS scavenging rate (%) = (Ac – As)/Ac x 100, where AC is the absorbance of the control and AS is the absorbance of the test sample measured at 734 nm. Probiotic strain L. acidophilus DDS1 was considered a positive control. The experiment was performed in triplicate.
2.5.3 Determination of DPPH free radical scavenging capacity of LS-ARS2
The DPPH free radical scavenging assay was carried out following the previously described method with minor modifications (Ramalho et al., 2019). 1 mL of sample (intact cells or heat-lysed cells) was mixed with 1 mL 0.2 mM DPPH (CDH) solution in methanol (HiMedia). Mixed vigorously and incubated for 30 min (dark, room temperature). Distilled water was used as a control. DPPH radical scavenging activity (U/mL) = ABSC – ABSS/S x 100, where ABSC and ABSS stand for the absorbance of the control and the test samples measured at 517 nm, respectively, and S is the sample volume (mL). Reference probiotic strain L. acidophilus DDS1 was considered a positive control. The experiment was performed in triplicate.
2.5.4 Evaluation of superoxide anion scavenging activity of LS-ARS2
The superoxide anion scavenging activity was evaluated according to the method described earlier with minor modifications (Gao et al., 2013). 0.8 mL of the sample (intact cells or heat-lysed cells) was added to 0.2 mL of Tris–HCl (Sigma) solution (0.1 M, pH 8). Subsequently, 0.1 mL of pyrogallol (3 mM, Sigma) was added, mixed, and incubated in the dark at room temperature (25°C) for 30 min. The control group was taken as an equal volume of deionized water. Superoxide anion radical scavenging ability (%) = [1 – (As – A1)/A0] x 100, where As = the absorbance (320 nm) of the sample with pyrogallol, A1 = the absorbance of the sample solution lacking pyrogallol, and A0 = the absorbance of the blank solution with pyrogallol. Reference probiotic strain L. acidophilus DDS1 was considered a positive control. The experiment was performed in triplicate.
2.5.5 Estimation of hydroxyl radical scavenging ability of LS-ARS2
The hydroxyl radical scavenging assay was evaluated using the earlier method with minor modifications (Xiong et al., 2019). Equal volumes of 2.5 mM 1,10-phenanthroline (Sigma), 0.2 M sodium phosphate buffer (Sigma), and FeSO4 (2.5 mM, Merck) were vortexed and incubated for 5–7 min. Then, an equal volume of H2O2 (0.12% v/v, CDH) and a sample (intact cells or heat-lysed cells) were added, vortexed, and incubated in a water bath at 37°C for 60 min. The control group was taken as an equal volume of deionized water instead of the sample. Hydroxyl radical scavenging activity (%) = [(As – A1)/(A0 – A1)] × 100, where As = the absorbance (536 nm) of the sample containing H2O2, A1 = the absorbance of the sample deprived of H2O2, and A0 = the absorbance of the solution with H2O2 and without a sample. Reference probiotic strain L. acidophilus DDS1 was considered a positive control. The experiment was performed in triplicate.
2.6 Determination of antimicrobial properties of LS-ARS2
2.6.1 Preparation of the cell-free supernatant (CFS)
LS-ARS2 was grown anaerobically in MRS broth at 37°C for 24 h. The culture was centrifuged at 4000 rpm for 20 min at 4°C. The supernatant was sterilized by filtration through a 0.22 μm PVDF filter (HiMedia) and used for the experiments (Chen et al., 2019).
2.6.2 Evaluation of the antimicrobial activity of LS-ARS2 using Gram-positive and Gram-negative pathogens, including muti-drug resistant hospital strains
The antimicrobial effect was assessed for enteric pathogens by the agar well diffusion assay using the published method with modifications (Abouloifa et al., 2020). The studied pathogens include enteric Gram-negative strain Salmonella typhimurium ATCC 14028, Gram-positive Methicillin-resistant Staphylococcus aureus ATCC 700699 (MRSA), Staphylococcus aureus ATCC 25923, as well as clinically isolated Gram-negative multi-drug-resistant strains like Escherichia coli (ETEC) BCH 04067, Vibrio cholerae BCH 09616, and Shigella Flexneri BCH 06745. The pathogenic strains were swabbed at a concentration of 0.5 McFarland (OD600 = 0.5) on Mueller Hinton agar (HiMedia), and 6 mm wells were made on agar medium. The wells were loaded with 100 μL CFS of LS-ARS2. The antimicrobial activity was determined by measuring the clear zone of inhibition around the wells in MHA plates after overnight incubation at 37°C. For the negative control, MRS broth (uninoculated) was used. L. rhamnosus GG (LR-GG), an established probiotic, was used as a positive control. The experiment was performed in triplicate.
2.6.3 Co-culture experiments of LS-ARS2 with Salmonella typhimurium
The antibacterial effect of LS-ARS2 was further validated by co-culture assay with Salmonella typhimurium (ATCC 14028) as described earlier with slight modifications (Dash et al., 2023). Overnight cultures of LS-ARS2 and S. typhimurium were centrifuged and washed with PBS (pH 7.4). First, different doses of LS-ARS2 (106, 107, and 108 CFU/mL, respectively) were co-cultured with the pathogen (106 CFU/mL) till 24 h, and the viable cells were counted on SS agar plates (HiMedia). To perform the time-kill assay, S. typhimurium was co-cultured together with LS-ARS2 in a 1:1 ratio (106: 106 CFU/mL) for multiple time points (0, 3, 6, 12, 24 h), and respective colonies of S. typhimurium (ST) were counted. ST without LS-ARS2 was considered a control. The experiment was performed in triplicate.
2.6.4 Calculation of minimum inhibitory percentage (MIP) of LS-ARS2 CFS using broth microdilution method
For the determination of MIP, a microdilution assay was implemented according to the method published earlier (Chen et al., 2019). The overnight culture of S. typhimurium was centrifuged and washed by PBS (pH 7.4) twice. CFS was used in three sets: set 1: used as it is; set 2: heated at 95°C for 15 min; set 3: pH of the CFS was neutralized using NaOH. 100 μL of Mueller Hinton broth (MHB, HiMedia) comprising 105 CFU/mL S. typhimurium was mixed with different percentages (i.e., 1, 5, 10, 20, 30, 40, 50%) of the CFS diluted in MRS in a microtiter plate. Un-inoculated MHB and MRS were applied as negative controls (blank). The plates were incubated for more than 18 h, and the absorbance was measured at 600 nm using a microplate reader. From the same microtiter plate, cultures were spot-inoculated on MHA plates to evaluate bacteriostatic or bactericidal effects on the pathogen. The standard probiotic LR-GG was used as a positive control. The experiment was performed in triplicate.
2.6.5 Evaluation of anti-biofilm properties of LS-ARS2 CFS
The antibiofilm property of cell-free supernatant of LS-ARS2 was assessed by the crystal violet staining method according to the previously published method with modifications (Lee et al., 2021). Overnight-grown MRSA strain was adjusted to OD600 = 0.1 in Tryptone Soya Broth (TSB, HiMedia) and dispensed into each well of 24-well plates with 100 mM glucose (Merck) and NaCl (Merck). CFS of LS-ARS2 was added to the pathogen at a range of concentrations of 1.25–40% v/v (1/8 MIP to 4 MIP). After incubation at 37°C for 24 h, the medium was removed, and staining was performed as mentioned above (section 2.3.7.). Un-inoculated MRS with TSB broth was used as a control. The biofilm formation rate (%) was calculated according to the following formula: Biofilm formation rate (%) = (ODSample/ODControl) x 100. The experiment was performed in triplicate.
2.6.6 Prediction of bacteriocins using the whole genome sequence of LS-ARS2
Biosynthetic gene clusters (BGCs) are closely associated groups of genes on a genome that together work to synthesize specialized metabolites. The BGCs of antimicrobial compounds (bacteriocins) of the LS-ARS2 genome were explored by the web server, BAGEL4 (de Jong et al., 2006).
2.7 Determination of functional annotations of the LS-ARS2 genome
The prediction of genes in the LS-ARS2 genome was performed using Prokka (1.14.6), and the functional annotation was done using Emapper. The cluster of orthologous groups (COG) intended for the protein-coding genes was determined by Egg-NOG mapper (version 2.1.12) (Dietzsch et al., 2021) using the online Egg-NOG database (version 5.0). The presence of carbohydrate-active enzyme (CAZymes) encoding genes in the LS-ARS2 genome was explored with the meta server, dbCAN3 (Zheng et al., 2023). Additionally, the genomic determinants linked to various probiotic traits were identified through comprehensive annotation of the LS-ARS2 genome, employing multiple bioinformatic tools, including KEGG mapper and RAST (Kanehisa et al., 2016; Sethi et al., 2024).
2.8 Prediction of primary and secondary metabolites using the whole genome sequence of LS-ARS2
The LS-ARS2 genome sequence was analyzed in gutSMASH (Specialized Primary Metabolite Analysis from Anaerobic Bacteria) for the prediction of potential primary metabolites (Pascal Andreu et al., 2021). Similarly, the secondary metabolites of the strain were predicted using antiSMASH version 6.0.1 (Antibiotic and Secondary Metabolites Shell) using strictness “strict” (Blin et al., 2021).
2.9 Elucidation of metabolite profiling of LS-ARS2 by high-resolution mass spectroscopy (HRMS)
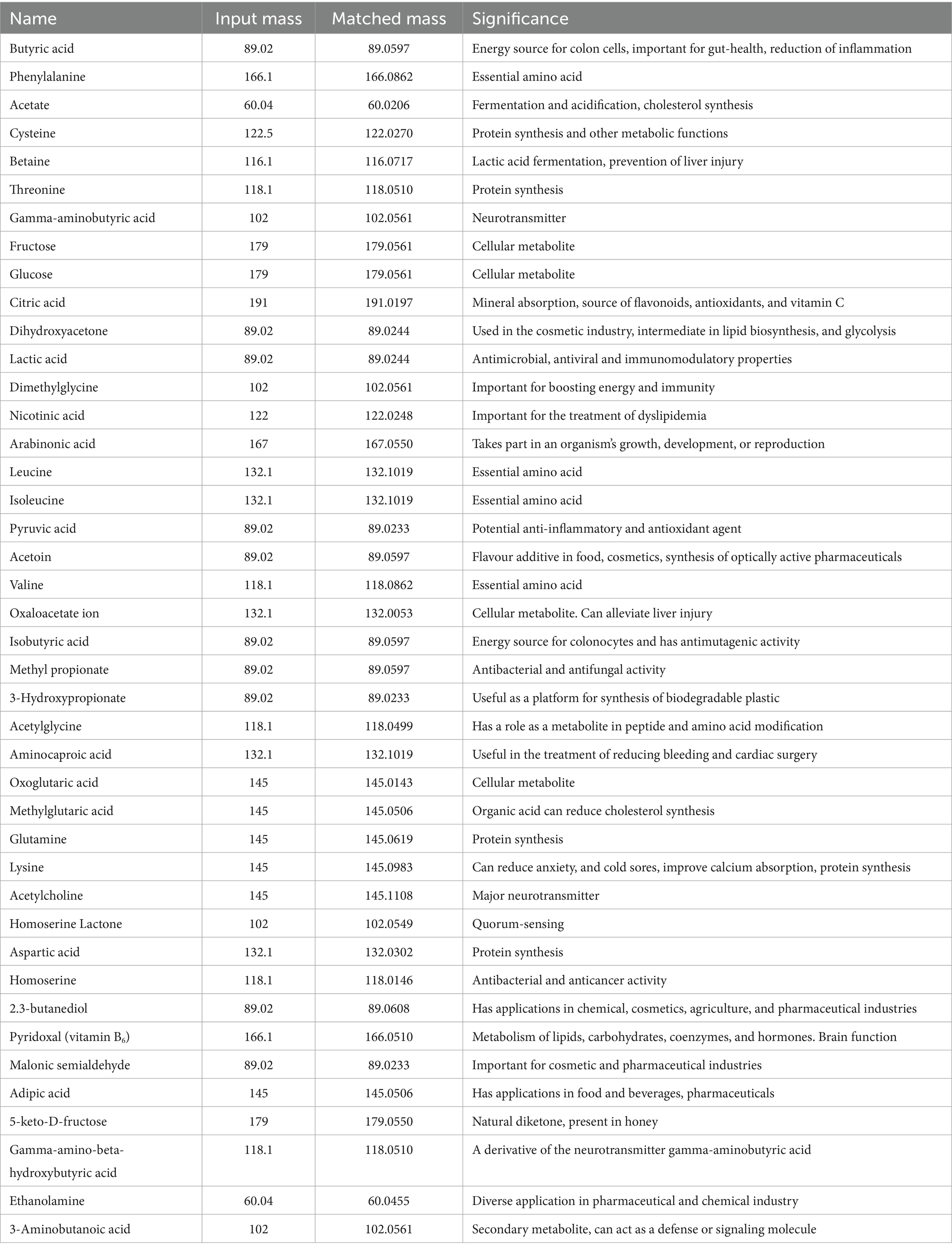
The metabolite composition of the LS-ARS2-derived CFS was analyzed using an Exactive™ Plus Orbitrap high-resolution mass spectrometer linked in tandem to an Ultimate 3,000 high-performance liquid chromatography column (Thermo Scientific, United States) (Dash et al., 2023). Liquid chromatography-mass spectrometry (LC–MS)-grade water, methanol, acetonitrile, and formic acid were used (Fisher Scientific, USA). Equal volumes of CFS and methanol (1:1 ratio) were methodically combined and sterilized by syringe filtration using a 0.22 μm filter. For mass spectrometry, around 0.5 mL of the filtered solution was added to the DP ID vial (Cat# C4000-1 W, Thermo Scientific, USA). The components in the sample were sorted out by a C18 column (Hypersil BDS, 250 mm × 2.1 mm, 5 μm; ThermoScientific, USA). The mobile phase contained acetonitrile and water (1:1) containing formic acid (0.1%). The flow rate of the sample was adjusted to 3 μL/min, the temperature of the column was set at 30°C, and the pressure was 700 bar. Electrospray ionization at a potential of 3 eV was used to ionize the molecules. The sample run time was 5 min, and both ions (positive and negative) were detected within the 50–750 m/z scan range. Metabolite annotation of the mass peaks was performed through the National Metabolomics Data Repository (NMDR). The peaks were then annotated with the Metabolomics Workbench database1 using public sources such as LIPID MAPS, ChEBI, HMDB, BMRB, PubChem, NP Atlas, and KEGG. For compound consolidation, mass tolerance (the acceptable deviation from the obtained m/z) was set at 0.2 ppm. The results of the analysis were exported with the following filtration criteria: delta PPM ranges from −0.2 ppm to +0.2 ppm and the complete annotated name of the compound. Both positive and negative mode analyses were performed simultaneously, which were combined at a later stage, and duplicates were removed. The analyzed molecules were clarified on the basis of the published literature and recorded in the table.
2.10 Statistical analysis
Statistical analysis was performed by two-way ANOVA (for comparison between more than two groups and two parameters) and one-way ANOVA (for comparison between more than two groups and one parameter) to determine significant differences between the treatments using GraphPad Prism (Version 8). The differences were considered statistically significant between all the treatment groups when p ≤ 0.05.
3 Results
3.1 Whole genome sequencing features of LS-ARS2
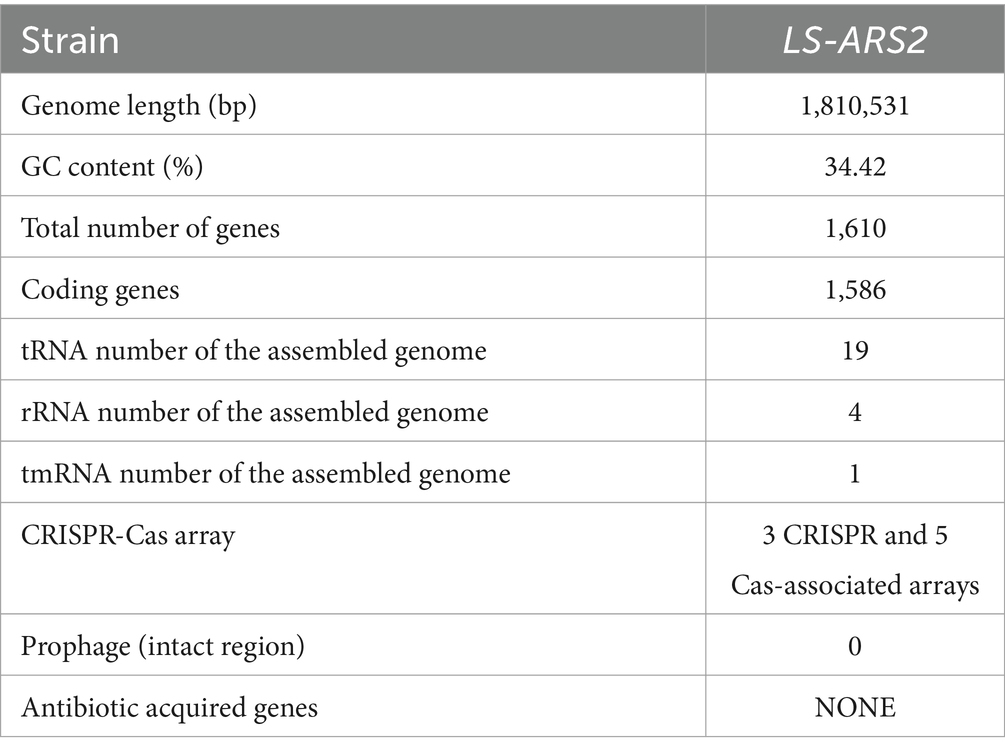
Our whole genome sequence study revealed that the genome of LS-ARS2 was a single circular chromosome. The complete genome sequence contained 1,810,531 bp. Average GC content was found to be 34.4% (Figure 1A). The coding DNA sequences (CDSs) were 1,586, and the identified genes were 1,610 (Table 1). The chromosome contained 4 rRNAs, 19 tRNAs, and 1 tmRNA encoding sequence. The overall summary of the genome assembly was included in Supplementary Table S1.
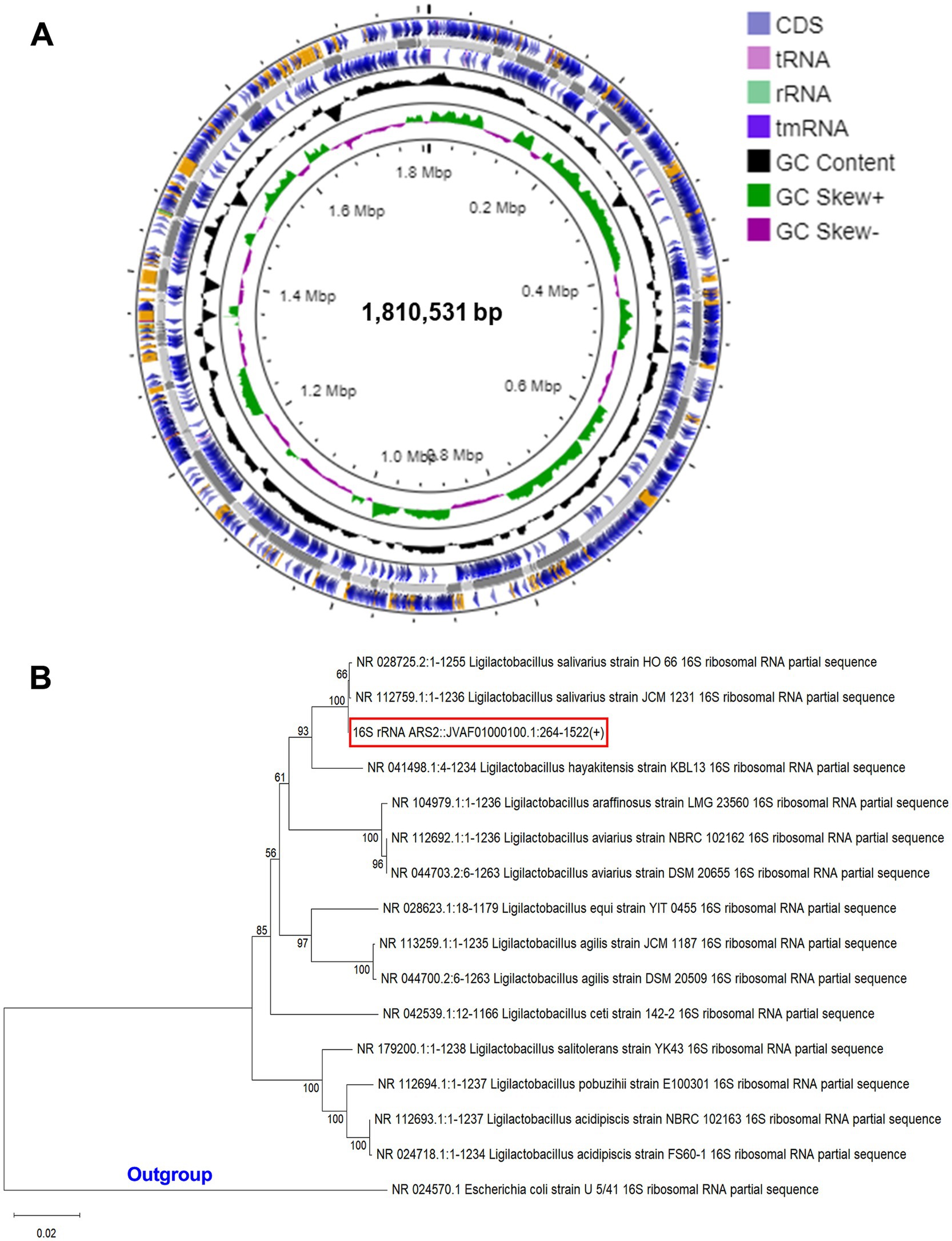
Figure 1. (A) Circular genome diagram of LS-ARS2. From the inner to the outer: the first ring represented the genome size (1,810,531 bp); the second depicted the GC skew (G + C/G – C); the third and fourth showed forward and reverse CDS (coding sequences) annotated with Prokka (the sites of CDSs/rRNA/tRNA/tmRNA on the genome are marked). (B) Phylogenetic analysis based on NCBI blast and MEGA 12 reaffirmed that LS-ARS2 belongs to Lactobacillus salivarius. The phylogenetic tree is constructed based on the neighbor-joining method and indicates that the closest clad for LS-ARS2 is Ligilactobacillus salivarius strain HO 66. Escherichia coli U 5/41 is used as an outgroup strain.
3.2 Phylogenetic analysis and taxonomic position reaffirmed the strain as Ligilactobacillus salivarius
A BLAST search was performed with all the Ligilactobacillus complete genomes to identify the species closest to our strain. Phylogenetic analysis showed the close relations of our strain with other species of the genus Ligilactobacillus and grouped in a single clade with Ligilactobacillus salivarius strain HO66 (Figure 1B). This phylogenetic analysis result corroborated with the average nucleotide identity (ANI) analysis, where 98% of ANI identity was found for LS-ARS2 with L. salivarius DSM 20555 (Tenea and Ortega, 2021). Moreover, as per 16S rRNA extracted using Barrnapp, Ligilactobacillus salivarius strain HO 66 showed the highest identity with our strain. Based on this, our strain was inferred as Ligilactobacillus salivarius.
3.3 Probiotic attributes of LS-ARS2: genomic analysis coupled with in vitro investigations
3.3.1 Survival potential of LS-ARS2: acid and bile tolerance
Probiotics should be able to survive the harsh environment (temperature, low pH, high bile salts, osmotic pressure, and oxidative stress) of the GI tract to reach the small intestine, where they show beneficial effects.
Interestingly, LS-ARS2 appears as a fast-growing probiotic. A fast growth rate is not so common among LAB and could facilitate the strain for faster colonization in the gut. Our WGS analysis and functional annotation identified several stress response genes in the LS-ARS2 genome, like universal stress proteins, yugI, usp6, and uspA, which are reported to be induced under several stress conditions. LS-ARS2 genome also harbors numerous heat shock proteins like heat-shock related regulators (hrcA, ctsR), molecular chaperons (dnaK, dnaJ, grpE, groL, groS, hslO, hslU), and protease encoding genes (hslU, hslV, clpX, clpL, clpP, clpC, clpE, clpB) which might protect LS-ARS2 under heat-shock conditions by preventing intracellular protein aggregation and maintaining membrane stabilization. Additionally, the cpsC gene encoding cold-shock proteins was identified in the LS-ARS2 genome, which might enable the strain to sustain cold stress conditions (Kandasamy et al., 2024).
Being a food supplement, probiotics encounter acidic pH in the stomach after ingestion. Bacteria are reported to evolve direct and specific strategies like ATPases and amino acid decarboxylation-antiporter reactions to remove different stress factors actively. In the present study, functional annotation of the LS-ARS2 genome identified numerous ATPase-associated genes, which are directly involved in the translocation of protons across the membrane. ATPases are one of the major proton pumps used by Gram-positive bacteria for the extrusion of protons from the cytoplasm by proton motive force. Among them, the most remarkable is F0F1 ATP synthase, which is an operon consisting of eight genes, atpA-atpH, which maintains a stable pH in the bacterial cytoplasm to sustain the acidic stress (Kandasamy et al., 2024). Moreover, several ABC transporters, acyltransferase (plsC), pyruvate kinase (pyk), alkaline shock proteins (asp23), sodium proton antiporter (nhaC, which maintains pH and Na+ homeostasis), ABC-type Na + efflux pump, and permease component was detected in the LS-ARS2 genome, which constitutes a stress tolerance machinery helping the strain to survive under acid stress condition (Kandasamy et al., 2024) (Supplementary Table S2).
Further, the acid tolerance assay showed LS-ARS2 exhibited 80 and 61% survival in pH 3 after 3 and 5 h of incubation, respectively (Figure 2A). Interestingly, there was no significant difference in the biomass with control MRS up to 1 h of incubation for both pH conditions (p > 0.05). Moreover, the viable count increased significantly (****p < 0.0001) after 3 and 5 h of incubation at pH 4. However, in pH 3, the strain survived well up to 3 h (p > 0.05), but the viable cells decreased significantly after 5 h of incubation (****p < 0.0001). Therefore, LS-ARS2 was able to survive in the acidic pH 3 and pH 4.
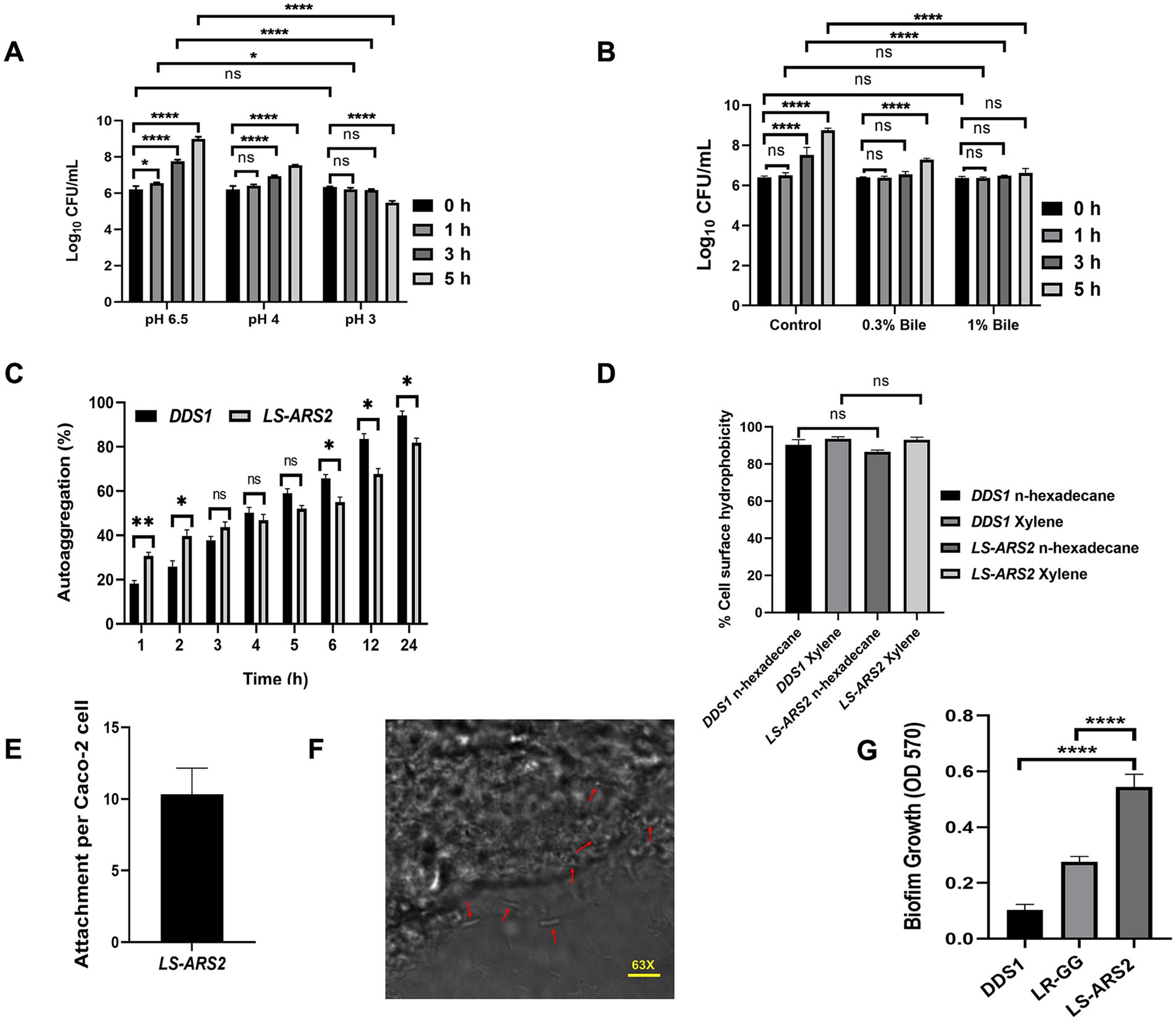
Figure 2. Probiotic properties of LS-ARS2. (A) Acid tolerance of LS-ARS2. Survival of LS-ARS2 at pH 3 and pH 4 at different time intervals indicated the viability of the strain at acidic pH. Data was presented as mean ± SD from three independent biological replicates. Statistical analysis was done using a two-way ANOVA with a Bonferroni post hoc test; *p < 0.05; ****p < 0.0001. (B) Bile tolerance of LS-ARS2. Viability of LS-ARS2 in the presence of 0.3 and 1% bile salt for 3 and 5 h of incubation time indicated tolerance of the strain at high bile concentrations. Data was represented as mean ± SD based on three independent biological replicates. Statistical analysis was done using a two-way ANOVA with a Bonferroni post hoc test; *p < 0.05; ****p < 0.0001. (C) Autoaggregation attributes of LS-ARS2. The aggregation property of LS-ARS2 was assessed side by side and compared with the reference probiotic strain L. acidophilus DDS1 at different time points up to 24 h. The results were depicted as mean ± SD based on three independent biological replicates. Statistical analysis was done using two-way ANOVA with Bonferroni post hoc test; p > 0.05. (D) Cell surface hydrophobicity property of LS-ARS2. The hydrophobicity percentage of LS-ARS2 was evaluated with n-hexadecane and xylene (2 h of incubation) in comparison to the reference strain L. acidophilus DDS1. All the results were shown as mean ± SD based on three independent biological replicates. Statistical analysis was done using one-way ANOVA with Bonferroni post hoc test; *p < 0.05; ****p < 0.0001. (E) Adhesion efficacy of LS-ARS2. The attachment efficiency of LS-ARS2 was shown with intestinal epithelial Caco-2 cells. (F) Visualization of Adhesion property of LS-ARS2. Phase contrast confocal microscopy images of the Caco-2 cells after co-culturing with LS-ARS2 showed efficient adhesion to Caco-2 cells. (G) Biofilm-forming ability of LS-ARS2. The assay was performed compared with two standard probiotic strains, L. acidophilus DDS1 and L. rhamnosus GG (LR-GG). Results were represented as mean ± SD based on three independent biological replicates. Statistical analysis was done using one-way ANOVA with Bonferroni post hoc test; ****p < 0.0001.
Next, the probiotic faces the alkaline conditions of the small intestine due to bile acids. WGS analysis identified inorganic pyrophosphatase (ppaC), which maintains membrane integrity and surface tension of bacteria under bile stress conditions. Additionally, several other genes were detected contributing to bile tolerance mechanism of LS-ARS2, like cyclopropane-fatty-acyl-phospholipid [CFP] synthase (cfa), which enhance lipid synthesis, glutamine synthetase (glnA), oligopeptide transporting proteins (oppA-F), ABC transporters (glnR, glnP, glnQ, glnH, glnPH2), and sodium-bile acid symporter family (Kandasamy et al., 2024) (Supplementary Table S2). Amino acid decarboxylation-antiporter reactions increase the intracellular pH by decarboxylation of amino acid transported into the cell. In the LS-ARS2 genome, genes encoding Orn/Lys/Arg decarboxylase, C-terminal domain, and possible lysine decarboxylase are annotated, which might be responsible for such mechanism in the removal of stress factors (Dash et al., 2023). Apart from these, LS-ARS2 contains an abundance of genes involved in carbohydrate metabolism, amino acid metabolism, replication and transcription, and major proteins involved in cell-wall biosynthesis, which reflects the adaptive ability of the strains in various stress conditions (Heunis et al., 2014).
To validate the bile tolerance ability of LS-ARS2, we performed a bile tolerance assay. The bile salt concentration varies from 0.2–0.3% depending on the type and amount of food ingested (Hsiung et al., 2021). The strain exhibited 87 and 83% survival in 0.3% (w/v) bile salt after 3 and 5 h of incubation, respectively. The strain also showed a survival of 86 and 76% in 1% (w/v) bile salt after the above-mentioned incubation time (Figure 2B). Interestingly, the survivability of the strain was similar to the control versus 0.3% or 1% bile salt up to 1 h (p > 0.05). Moreover, no significant changes were observed in viable count between 0 and 3 h for both 0.3 and 1% bile salt (p > 0.05). In addition, the biomass was found to increase significantly (****p < 0.0001) after 5 h in the presence of 0.3% bile. Therefore, the strain could potentially be able to survive at high bile salt.
In summary, our functional annotation and genomic analysis together with the results from biochemical assays strongly supported the functional capabilities and survival of the strain LS-ARS2 in harsh environments (heat, acid, bile, cold, osmotic) in the GI tract.
3.3.2 Colonization potential of LS-ARS2: self-aggregation, cell surface hydrophobicity property, and adhesion efficacy in Caco-2 cells
Probiotics are expected to colonize in the gut and form a strong barrier on the epithelial mucosa that restricts the entry of pathogens.
Functional annotation of the genes by COG analysis exhibited the abundant presence of glycosyltransferases in the LS-ARS2 genome that might provide a specific benefit to adhere and colonize in the GI tract (Dash et al., 2023). The genomic analysis of the LS-ARS2 genome identified putative adhesion-related genes such as maltose phosphorylase (mapA), lipoprotein signal peptidase II (lspA), a sortase family protein (srtA), and enolase (eno), which are cell-surface proteins responsible for the high adhesion potential of the strain (Kandasamy et al., 2024). The envelope of Lactobacilli contains many cell-surface proteins involved in Lactobacillus-host interactions. Among them, sortase-dependent proteins (SDPs) and S-layer proteins (SLPs) are best characterized for providing adhesion properties (Lebeer et al., 2008). In the present study, srtA (a sortase family protein) and lspA (involved in adhesion through prolipoproteins) were found by genome COG analysis supporting the adhesion potential of LS-ARS2. The genome of LS-ARS2 also contained several cell wall biosynthetic proteins such as dltD, dltC, dltA, dltB, and dltX, which might influence adhesion capacity and immune response in the host cells (Dash et al., 2023; Lebeer et al., 2008) (Supplementary Table S2).
Since aggregation of microbes is important for gut-adhesion and colonization, evaluation of self-aggregation is considered an important selection criterion for potential probiotics. Our strain showed a significant increase in autoaggregation capacity from 30.6 to 67.6% within 1–12 h, while it showed the highest autoaggregation ability (81.75%) after 24 h (Figure 2C). The auto-aggregation ability shown by the reference probiotic strain L. acidophilus DDS1 was also comparable to LS-ARS2 (nsp > 0.05, *p < 0.05, **p < 0.05).
Surface hydrophobicity of the bacterial cells is also pivotal for gut-colonization (Hsiung et al., 2021). LS-ARS2 showed significant surface hydrophobicity with n-hexadecane (86.59%) and xylene (93%) (Figure 2D). The hydrophobicity of LS-ARS2 was similar to the reference strain L. acidophilus DDS1 (p > 0.05). This result indicated the potential adhesion capacity of the strain with complex hydrophobic substratum.
One of the most important properties of probiotics is their capacity to adhere to the gut epithelial cells. Therefore, next, we wanted to study directly the adhesion potential of the strain for colon epithelial cells (Caco-2). Significant adhesion capacity (10–12 of LS-ARS2/ well-differentiated Caco-2 cells) was observed after 2 h of incubation (Figure 2E). The representative phase-contrast confocal microscopy images also reaffirmed the substantial attachment of LS-ARS2 to Caco-2 cells. Therefore, the strain indeed showed promising adhesion ability for the human gut lining (Figure 2F).
Therefore, genomic analysis together with in vitro assays indicate the potential of the strain LS-ARS2 to colonize in the gut.
3.3.3 Biofilm forming ability of LS-ARS2
Biofilm formation is reported to be a beneficial property of Lactobacillus sp. since the formation of biofilm showed better colonization, survival, and prolonged persistence in the gastrointestinal mucosa in the host (Salas-Jara et al., 2016). Probiotic-derived biofilm was also reported to secrete exopolysaccharides, vitamins, enzymes, and many metabolites to enhance the growth of gut-microbiota and inhibit pathogenic adhesion (Gao et al., 2022).
Biofilm-forming ability among L. salivarius strains is rare and strain-specific. However, our WGS analysis of LS-ARS2 revealed the presence of veg and luxS genes in the LS-ARS2 genome. It is worth mentioning that veg is a widely reported stimulator for biofilm formation in Gram-positive bacteria (Lei et al., 2013). Further, through quorum sensing, luxS-derived autoinducer-2 (AI-2) plays a crucial role in the adhesion and formation of biofilm in Lactobacillus spp. (Mgomi et al., 2023). Therefore, the presence of the veg and luxS genes in the LS-ARS2 genome encouraged us to explore the biofilm-forming ability of LS-ARS2 experimentally.
It was extremely exciting to find that LS-ARS2 was able to form robust biofilm within 24 h without any additional carbon source or supplements; such efficacy is rare among Ligilactobacillus salivarius strains, indicating the possibility of the strain for efficient and longer sustainability in the gut. Moreover, biofilm formation by LS-ARS2 was found to be more robust than the reference probiotic strains DDS1 and LR-GG (****p < 0.0001) (Figure 2G). LS-ARS2-biofilm could, therefore, offer a promising application potential of the strain for improving gut-health.
3.4 Safety attributes of LS-ARS2: WGS analysis and in vitro validation
3.4.1 Genome stability of LS-ARS2
Numerous Lactobacillus species contain hidden prophages with the potential of cross-contamination or modulation of the intestinal microecology when released (Pei et al., 2021). Therefore, it is essential to perform a comprehensive scanning of prophages in probiotic candidates for their safety evaluation. While analyzing the genome sequence of LS-ARS2, we found four prophage regions (PHASTER). However, all of them were incomplete prophage regions (Supplementary Table S3). Moreover, bacteria harbour adaptive antiphage defense mechanisms like the CRISPR/Cas system for their protection from phage attacks (Pei et al., 2021; Roberts et al., 2024). In our strain, three CRISPR arrays and five Cas-associated sequences (CRISPRFinder) were predicted (Supplementary Table S4). Next, a previous study indicated that Lactobacillus genomes carrying type I or type III CRISPR-Cas systems harbored fewer intact prophages (Pei et al., 2021). Indeed, TypeIE Cas-associated systems were the most prevalent in the genome of our strain as well. Hence, the presence of a TypeI Cas-associated system in LS-ARS2 might act as an antiphage defense system and could cause the absence of intact prophage regions.
For the identification of virulence factors (VFs), the genomic sequence of LS-ARS2 was analyzed against two VF databases: a core dataset comprising experimentally validated VFs and a comprehensive dataset encompassing all known and predicted VF-related genes. While there was no significant hit in the experimentally validated core dataset, analysis against the full dataset revealed the presence of VFG015903 (argK) phaseolotoxin-insensitive ornithine carbamoyltransferase (bits score 56, identities 25%). According to VFDB, this ornithine carbamoyltransferase enzyme is encoded by Pseudomonas syringae pv. phaseolicola 1448A, which is insensitive to phaseolotoxin-mediated inhibition (Staskawicz et al., 1980). Additionally, the analysis of the pathogenic potential by the PathogenFinder tool classified LS-ARS2 as a non-human pathogen, with no matches to known pathogenic families and a very low probability (0.159) of being a human pathogen.
Microorganisms may acquire antibiotic-resistant genes by horizontal gene transfer (Jia et al., 2016). Several Lactobacillus strains carry d-Ala-d-lactate in their peptidoglycan rather than the d-Ala-d-Ala dipeptide, which makes them resistant to vancomycin (Rychen et al., 2018). For example, L. plantarum strains were found with intrinsic vancomycin resistance genes (Deghorain et al., 2007). The genome of LS-ARS2 was also predicted to have one strict antibiotic-resistant gene (CARD database and RGI analysis, Supplementary Table S5). However, the same was not found with Resfinder analysis.
3.4.2 Antibiotic susceptibility of LS-ARS2
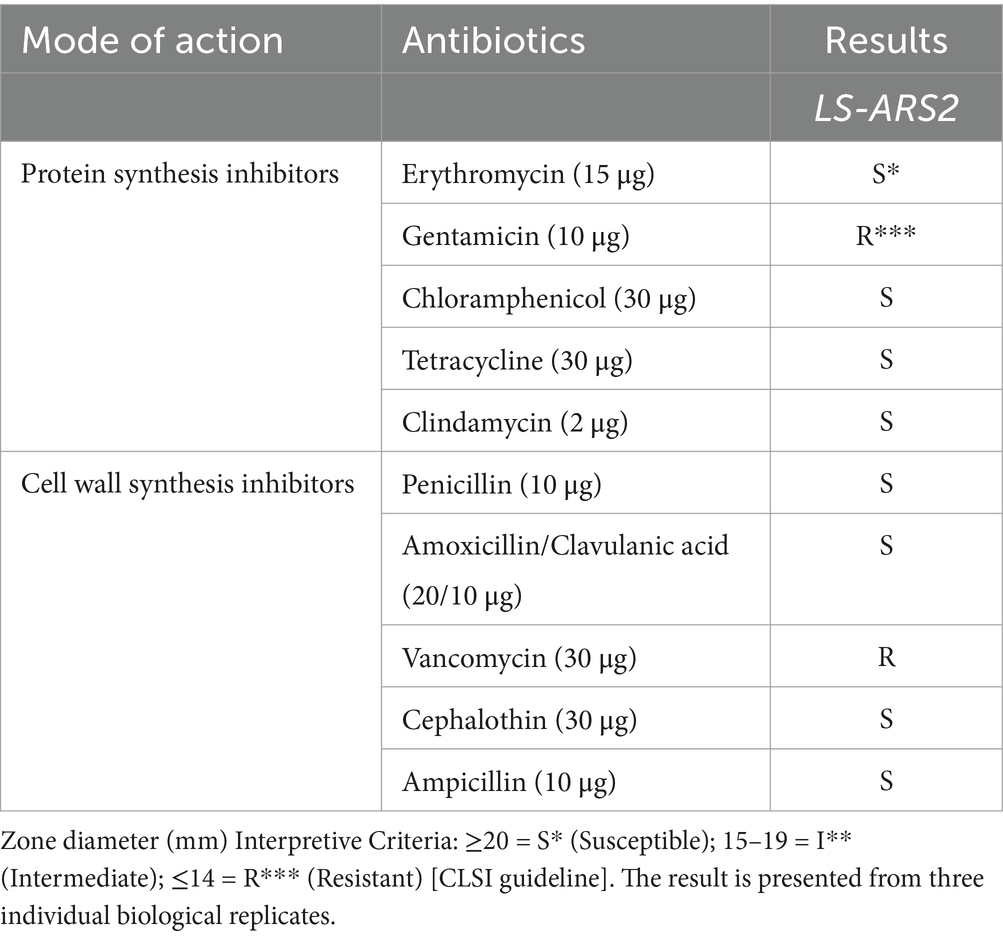
Extensive use of antibiotics with food supplements can accumulate genes responsible for antibiotic-resistance in gut-bacteria. These antibiotic-resistant genes can be acquired by the pathogens sharing the corresponding intestinal niche, which appears as a clinical threat (Wong et al., 2015). Therefore, to project any probiotic as a food supplement, the assessment of the antibiotic susceptibility profile is crucial to ensure its safe application. In our study, LS-ARS2 was sensitive to penicillin, erythromycin, clindamycin, tetracycline, ampicillin, chloramphenicol, and amoxicillin/clavulanic acid. However, it was found to be resistant to vancomycin and gentamicin, similar to many probiotic strains (Table 2). Therefore, LS-ARS2 was sensitive to the majority of the conventional antibiotics.
3.4.3 Non-haemolytic and DNase negative properties of LS-ARS2
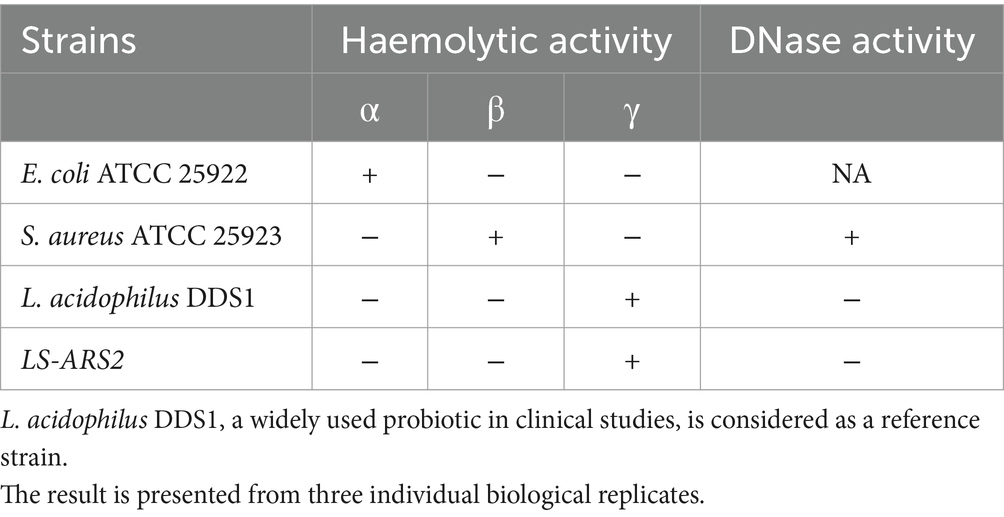
The non-haemolytic nature of LS-ARS2 was signified by the nonappearance of any transparent zone at the blood agar plate. Further, our strain did not show any halo zone around the colonies at the DNase agar plate, which suggested the absence of DNase activity (Table 3). Therefore, in silico studies and in vitro assays assured the non-pathogenic nature of LS-ARS2.
3.4.4 Safety attributes of LS-ARS2 in human cell lines
To further secure the safety attributes of LS-ARS2, both the whole cell and cell-free supernatant of the strain were tested on human colon cancer cells. The trypan blue exclusion assay ensured that the application of LS-ARS2 did not show any notable toxicity to HCT116. A similar result was also observed for the standard probiotic strain L. acidophilus DDS1 (Figures 3A,B). In the MTT assay, the CFS of LS-ARS2 also did not show any notable toxicity as compared to the control (complete DMEM without CFS) to the stationary-phase Caco-2 cells, which mimic the normal intestinal cells (Figures 3C,D). MTT and trypan blue exclusion assays, therefore, ensured the safe application of the strain on human cells.
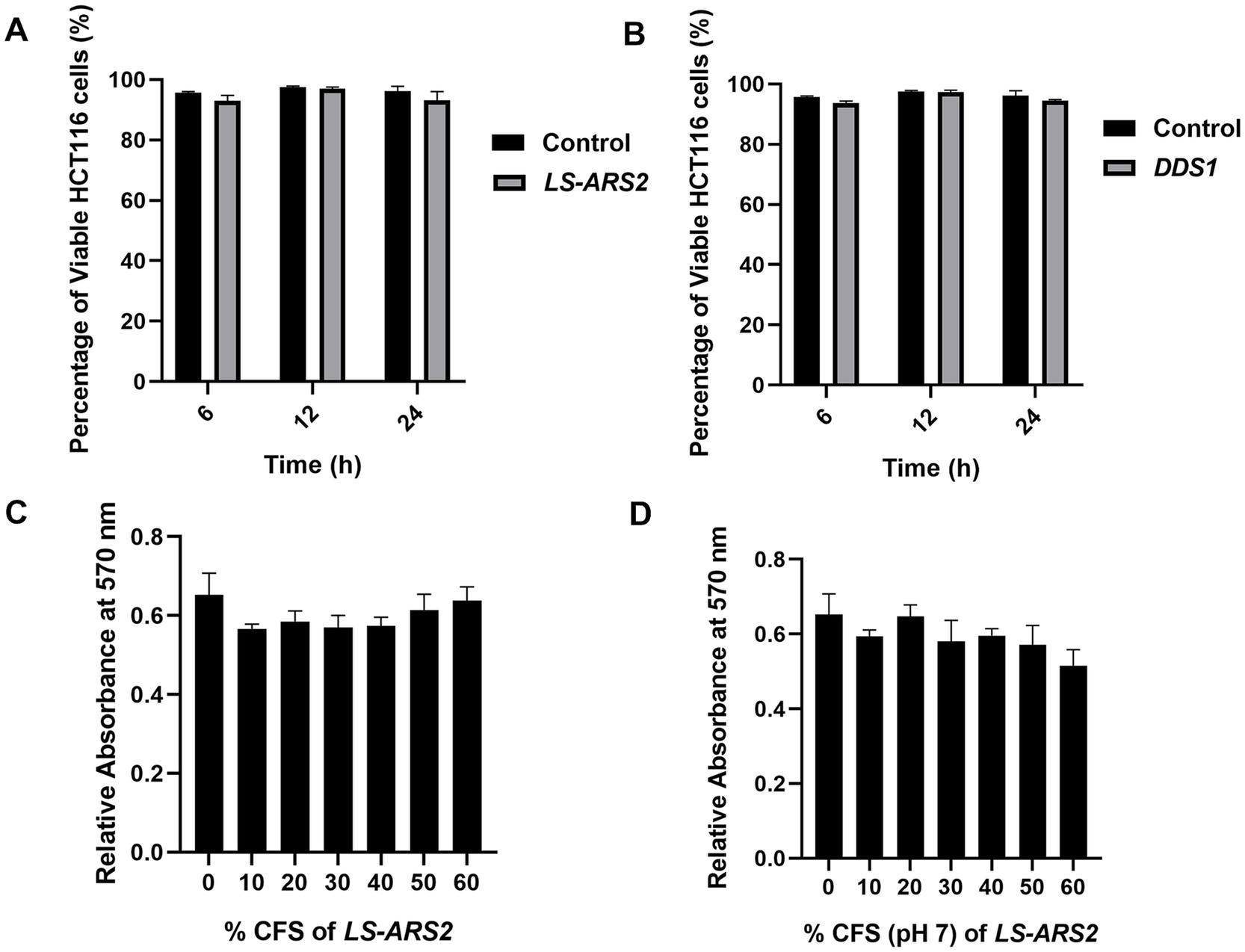
Figure 3. (A,B) Trypan blue exclusion assay. The result showed the percentage viability of human colorectal cancer cell HCT116 after treatment with (A) LS-ARS2 and (B) L. acidophilus DDS1. All the data were represented as the mean ± SD based on three independent biological replicates. (C,D) MTT assay. (C) LS-ARS2 CFS and (D) pH-neutralized CFS (pH 7) of LS-ARS2 on Caco-2 cells showed non-toxic nature of LS-ARS2-derived CFS.
3.5 Functional attributes of LS-ARS2: antioxidant potential of LS-ARS2
Probiotics with antioxidant potential could alleviate the oxidative stress in the host and, hence, could serve as a natural antioxidant. Functional classification of the LS-ARS2 genes indicated the presence of three oxidative stress mechanisms: Glutathione: Redox cycle, Glutathione: Biosynthesis and gamma-glutamyl cycle, and Glutaredoxins. Further digging into the LS-ARS2 genome annotated multiple genes responsible for antioxidant activities (Supplementary Table S2). The genes encoding whole thioredoxin (trxa, trxb, tpx) and NADH (ndh, npr) antioxidant systems were detected, which are reported to be associated with scavenging of reactive oxygen species (ROS), detoxifying peroxidases, and protecting cells from oxidative stress (Kandasamy et al., 2024; Zhang et al., 2018). Additionally, the genes encoding for glutathione synthetase (gshF) and glutaredoxin (nrdH) were identified, which are important for the regulation of cellular redox homeostasis (Xiong et al., 2018). Further, genes associated with the methionine sulfoxide reductase system (msrA and msrB) were recognized in the genome of LS-ARS2, which are reported to protect cellular proteins from damage by ROS-mediated oxidation (Oien and Moskovitz, 2019). These genomic signatures encouraged us to find out the potential antioxidant properties of LS-ARS2 experimentally.
3.5.1 Cation scavenging ability of LS-ARS2 by ABTS assay
In the ABTS cation radical scavenging assay, the intact cells of LS-ARS2 showed a 62.14% ABTS scavenging rate, and the heat-lysed cells showed a 71% scavenging rate. Meanwhile, the ABTS scavenging rate of reference strain L. acidophilus DDS1 was 47.59% for intact cells and 57.10% for heat-lysed cells (Figure 4A). Therefore, the ABTS scavenging rate of LS-ARS2 was found to be significant, even higher (*p < 0.05) than the reference probiotic strain L. acidophilus DDS1 for both intact and heat-lysed cells.
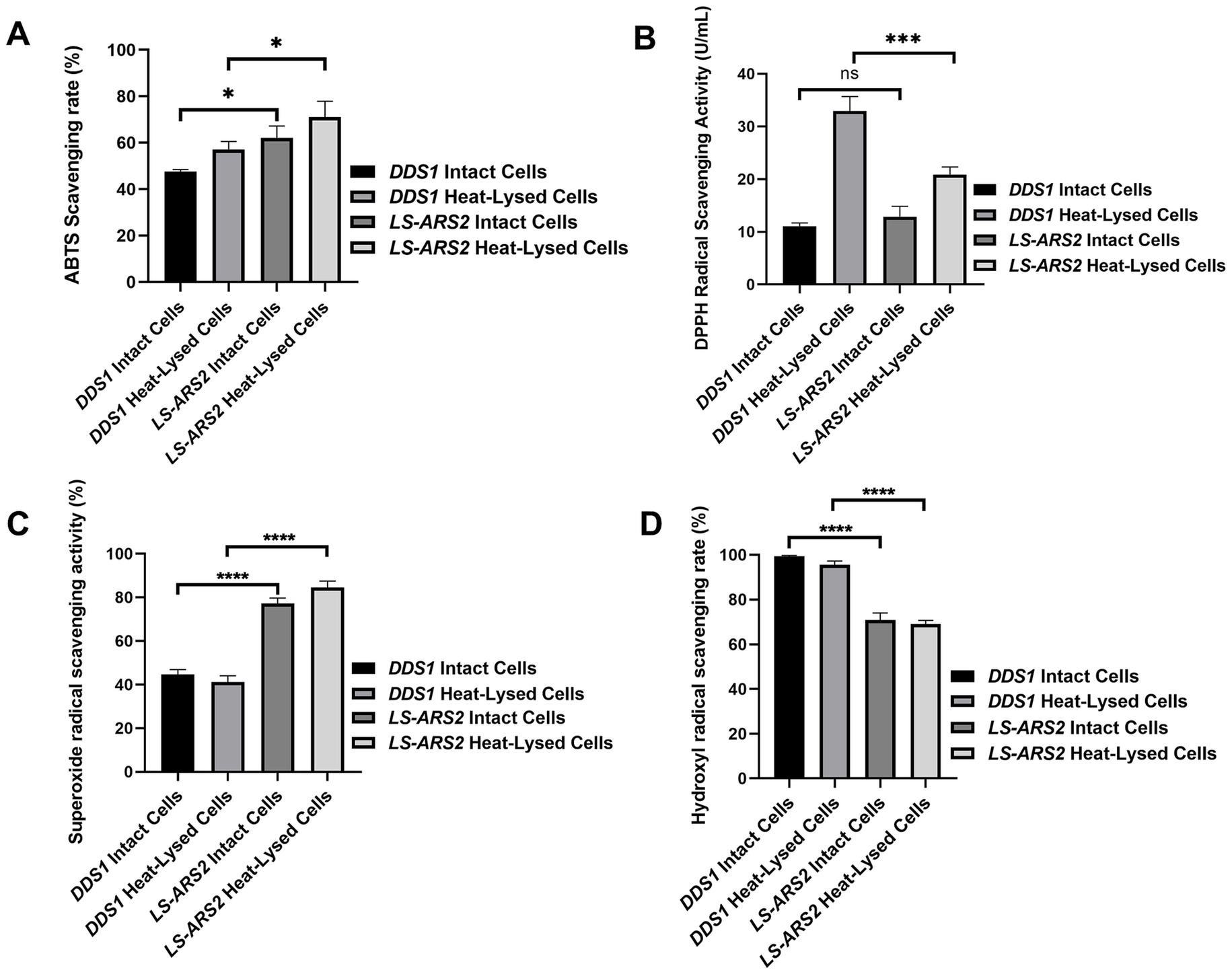
Figure 4. Anti-oxidant properties of LS-ARS2. (A) ABTS cation radical scavenging assay. The result depicted a significant cation radical scavenging rate of intact and heat-lysed cells of LS-ARS2. (B) DPPH free radical scavenging assay. The result demonstrated the free radical scavenging activity of LS-ARS2 intact and heat-lysed cells. (C) Superoxide free radical scavenging assay. The data showed the superoxide anion scavenging capacity of LS-ARS2 intact and heat-lysed cells. (D) Hydroxyl free radical scavenging assay. The result indicated hydroxyl radical scavenging ability of LS-ARS2 intact and heat-lysed cells. All the antioxidant activities were compared with the clinically used probiotic L. acidophilus DDS1. The results were shown as mean ± SD from three independent biological replicates. Statistical analysis was done using one-way ANOVA with a Bonferroni post hoc test. *p < 0.05; ****p < 0.0001.
3.5.2 Free radical scavenging capacity of LS-ARS2 by DPPH assay
DPPH is used extensively to estimate the free radical scavenging efficacy of antioxidant substances. The transfer of an electron or hydrogen atom to DPPH radical causes a decrease in the absorbance at 517 nm proportionally to the increase of non-radical DPPH (yellow) (Gao et al., 2013). DPPH free radical scavenging activity of LS-ARS2 whole cell was found to be 12.8 U/mL, which was comparable (p > 0.05) to reference probiotic strain L. acidophilus DDS1 whole cell (11.08 U/mL). However, the antioxidant activity of LS-ARS2 heat-lysed cells was found to be significant (20.8 U/mL) but less than the reference probiotic strain L. acidophilus DDS1 (32.9 U/mL) (***p < 0.0001) (Figure 4B). It is to be noted that intact bacteria cells show the functional properties of whole cells, whereas heat-lysed form reveals the functionality of intracellular components (Piqué et al., 2019). The intracellular components are strain-specific properties of the probiotics. Therefore, less free-radical scavenging activity of LS-ARS2 heat-lysed cells than that of DDS1 indicated more significant antioxidant activity of intracellular components of DDS1 than LS-ARS2. However, the free radical scavenging capacity of LS-ARS2 whole cells appeared to be comparable with that of the reference strain, indicating the potential use of LS-ARS2 as a promising antioxidant agent.
3.5.3 Superoxide radical scavenging ability of LS-ARS2
Superoxide anions serve as precursors of singlet oxygen, which indirectly initiates lipid oxidation. The superoxide anion scavenging rate of intact cells of LS-ARS2 was 77.24%, and heat-lysed cells were 84.61%. Meanwhile, the scavenging rate of reference probiotic strain L. acidophilus DDS1 intact cells was 44.68%, and heat-lysed cells were 41.15% (Figure 4C). Hence, LS-ARS2 depicted remarkable superoxide anion scavenging potency, even higher (****p < 0.0001) than the reference probiotic strain L. acidophilus DDS1 for both intact and heat-lysed cells.
3.5.4 Hydroxyl radical scavenging capacity of LS-ARS2
Hydrogen peroxide generates hydroxyl radicals in the body as a by-product of various biological processes. This toxic radical initiates indirect oxidation of lipids or proteins and subsequent damage to tissues (Gao et al., 2013). Probiotics may scavenge the radical and reduce such tissue damage. The hydroxyl radical scavenging rate of LS-ARS2 intact cells was 70.83%, and heat-lysed cells were 69.09%. Reference probiotic strain L. acidophilus DDS1 exhibited a 99.41% hydroxyl radical scavenging rate for intact cells and 95.58% for heat-lysed cells. Therefore, the hydroxyl radical scavenging rate of LS-ARS2 was moderate for both intact cells and heat-lysed cells, although less than the reference probiotic strain L. acidophilus DDS1 (****p < 0.0001) (Figure 4D). However, functional properties like hydroxyl radical scavenging capacity are distinct and independent attributes of probiotics. Less activity of LS-ARS2 than the reference strain could indicate the limitation of the use of LS-ARS2 as a hydroxyl radical scavenging agent.
3.6 Functional attributes of LS-ARS2: promising antimicrobial properties
3.6.1 Co-culture assay indicated the significant anti-Salmonella effect of the strain LS-ARS2
Salmonella typhimurium (ST) is one of the major food pathogens that trigger gut inflammation and life-threatening diarrheal diseases. Therefore, we wanted to evaluate the antimicrobial activity of LS-ARS2 against ST. While characterizing the CFS of LS-ARS2, the heat-denatured CFS indicated a zone of inhibition (13.6 mm ± 0.5) similar to the untreated CFS (13.6 mm ± 0.5) in the agar well diffusion assay, whereas the inhibitory effect decreased (0 mm) significantly in the pH-neutralized CFS. This result indicated that the acidic nature of CFS could be the crucial factor for the antibacterial effects of LS-ARS2 (Figure 5A). This result was similar to the reference probiotic strain L. rhamnosus GG (LR-GG) (p > 0.05). The broth microdilution method showed that 10% of the CFS was enough to inhibit the visible growth of the pathogen and was considered the MIP (Figure 5B). Agar spot assay also suggested that 10% CFS depicted a bacteriostatic effect, whereas 20% CFS was required for a bactericidal effect (Figure 5C). The result shown by LR-GG was comparable to LS-ARS2 (Figures 5B–G). When LS-ARS2 was co-cultured with ST, it effectively hindered the viability of the pathogen with time. A significant (****p < 0.0001) reduction in the viable count of the pathogen was observed after 12 h of co-culture, and complete growth inhibition was observed after 24 h (Figure 6A).
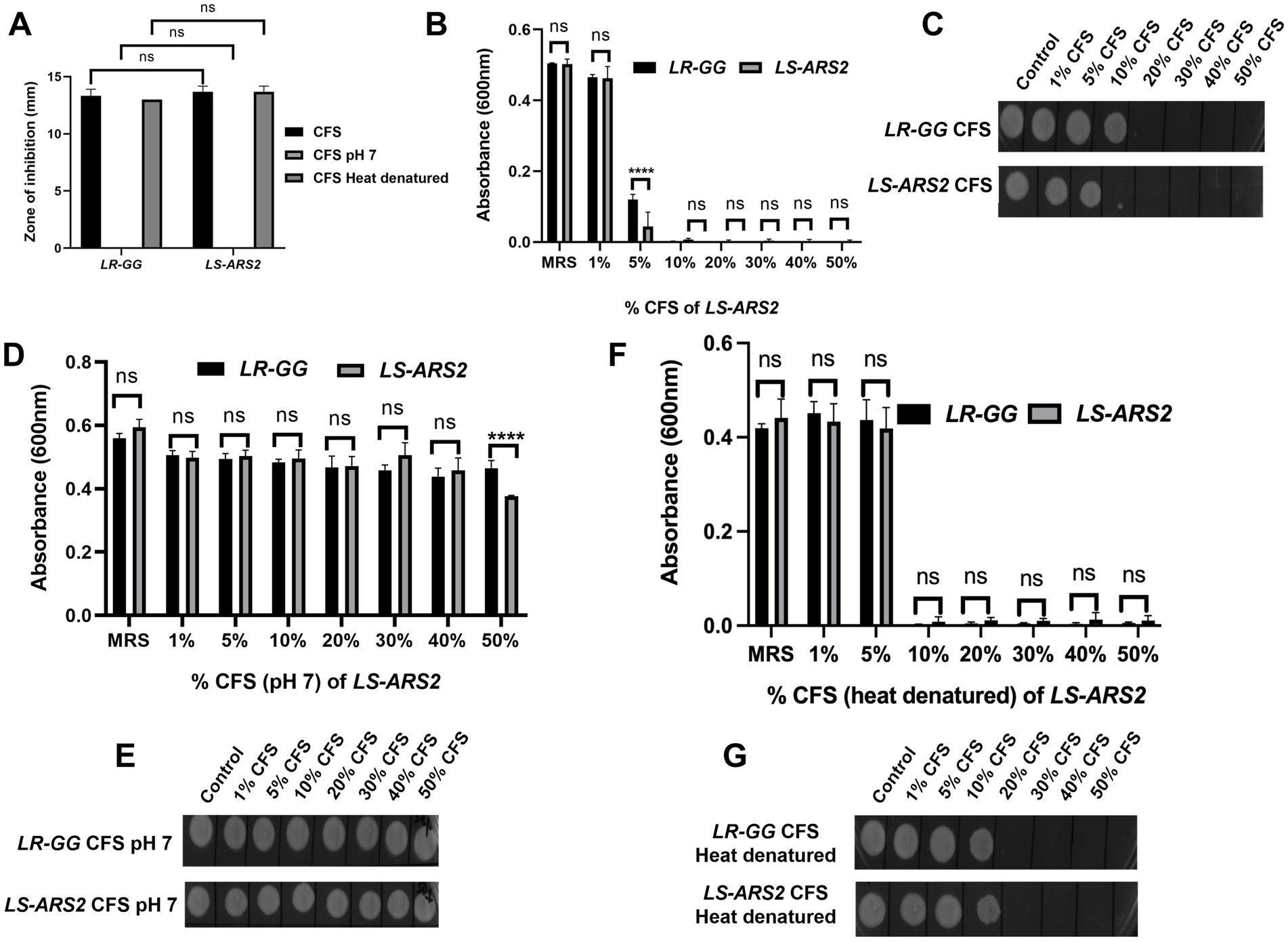
Figure 5. Antimicrobial properties of LS-ARS2. (A) Agar well diffusion assay. The results showed the effect of normal, pH-neutralized, and heat-denatured cell-free supernatant (CFS) of LS-ARS2 and LR-GG (positive control) on the growth of S. typhimurium (ST). The results were shown as mean ± SD based on three independent biological replicates. Statistical analysis was done using two-way ANOVA with Bonferroni post hoc test *p < 0.05; ****p < 0.0001. (B,D,F) Determination of the minimum inhibitory percentage (MIP) of LS-ARS2 CFS for ST. ST was incubated in various percentages of (B) normal, (D) pH-neutralized, and (F) heat-denatured CFS of LS-ARS2 and LR-GG to estimate the MIP of the CFS against the pathogen. The data presented from three biological replicates; *p < 0.05; ****p < 0.0001 using two-way ANOVA with Bonferroni post hoc test. (C,E,G) Estimation of the viability of ST in the presence of LS-ARS2 CFS. From every well of ST grown in LS-ARS2 CFS and the probiotic LR-GG CFS, culture was spotted to find out the bactericidal or bacteriostatic effect (C,E,G). All the data are represented as the mean ± SD from three independent biological replicates.
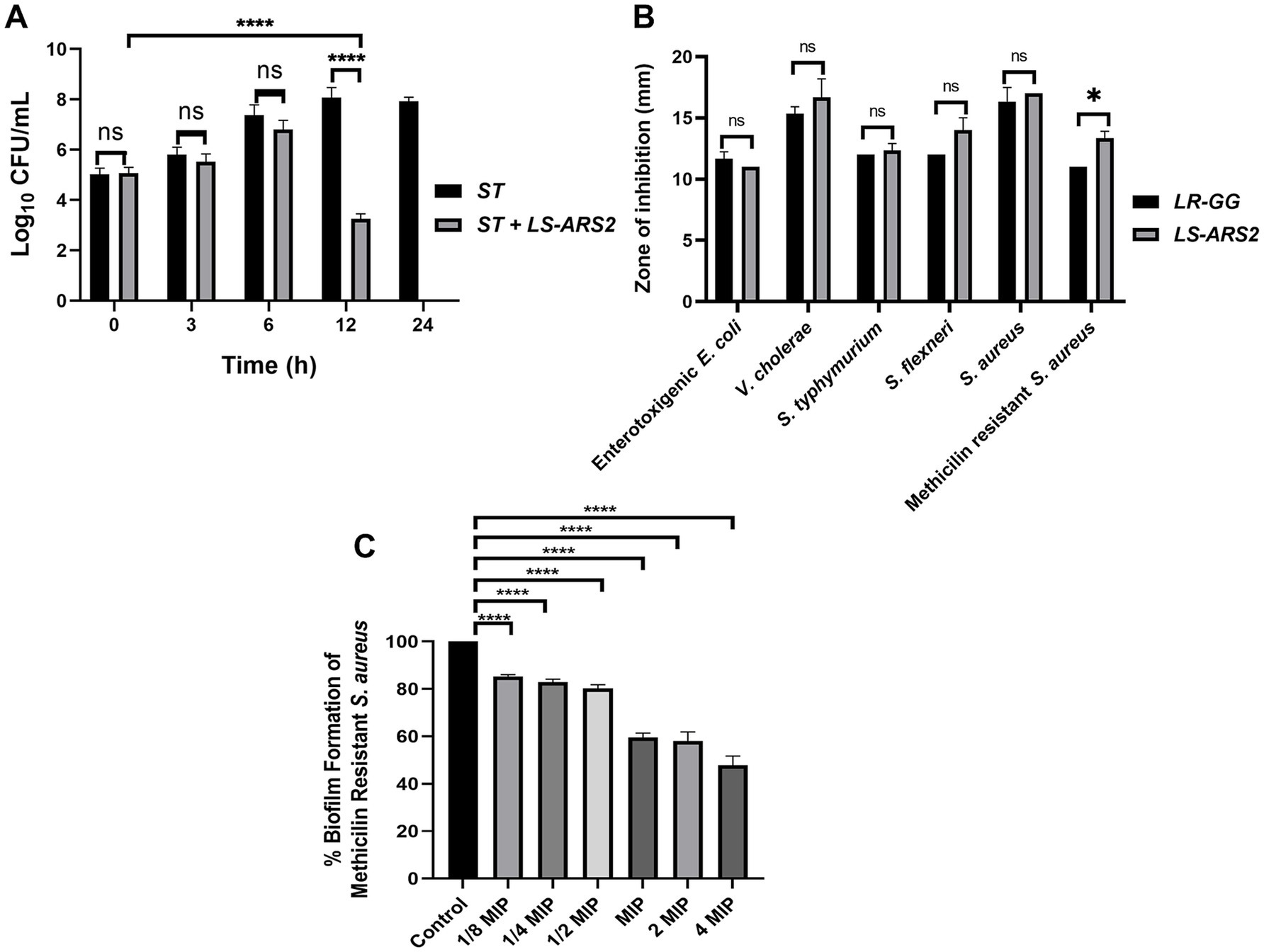
Figure 6. Antimicrobial properties of LS-ARS2. (A) Co-culture assay. Co-culture of S. typhimurium (ST) with LS-ARS2 at multiple time periods illustrated the LS-ARS2 cytotoxic effect to the pathogen. After 12 h of co-culture, a significant reduction (****p < 0.0001) in the viable cell number of S. typhimurium was observed. Whereas, after 24 h of culture, the viable cell number of ST was decreased to zero (data presented from three biological replicates with multiple technical repeats; *p < 0.05; ****p < 0.0001 using a two-way ANOVA with a Bonferroni post hoc test). (B) Agar well diffusion assay. The result showed diverse inhibition zones, signifying the antimicrobial activity of LS-ARS2 and LR-GG (positive control) against multiple Gram-positive and Gram-negative enteric pathogens. The data were represented as mean ± SD based on three independent biological replicates. Statistical analysis was done using two-way ANOVA with Bonferroni post hoc test; p > 0.05, *p < 0.05. (C) Anti-biofilm assay. Cell-free supernatant (CFS) of LS-ARS2 showed a significant reduction in the biofilm formation of Methicillin-resistant Staphylococcus aureus ATCC 700699. The results were represented as the mean ± SD based on three independent biological replicates. Statistical analysis was done using one-way ANOVA with a Bonferroni post hoc test; ****p < 0.0001.
3.6.2 Robust antimicrobial potential of LS-ARS2 across the diverse pathogen
It is desirable for a potential probiotic to possess robust antimicrobial potential for a wide range of pathogens. Indeed, LS-ARS2 demonstrated remarkable inhibitory activity against all studied Gram-positive and Gram-negative enteric pathogens (Figure 6B). Among them, the strain showed the highest antimicrobial activity for S. aureus, V. cholerae, and S. flexneri. It should be emphasized that V. cholerae and S. flexneri were multi-drug-resistant clinical isolates. Moreover, the strain also showed a significant inhibitory effect against Gram-positive methicillin-resistant S. aureus. The antimicrobial activity of LR-GG was comparable to LS-ARS2 (Figure 6B) (p > 0.05).
3.6.3 Anti-biofilm effect of LS-ARS2 for methicillin-resistant Staphylococcus aureus (MRSA)
The pathogenic biofilms pose serious hazards in the food and clinical industries, affecting public health. Therefore, safe applications like probiotics are encouraged to reduce the prevalence of the growth of pathogenic biofilms. MRSA is known to be a strong biofilm-former, and MRSA-biofilm is a part of their virulence factor (Kaushik et al., 2024).
To estimate the anti-biofilm effect of LS-ARS2, first, the MIP of CFS for MRSA was determined (10% of LS-ARS2-CFS, Supplementary Figure S2). Further, 1/8 MIP to 4 MIP (1.25–40%) of LS-ARS2-derived CFS were used to study the formation of MRSA-biofilm. Our result indicated a significant reduction (****p < 0.0001) in the formation of MRSA-biofilm at all studied concentrations. Moreover, LS-ARS2-derived CFS was able to reduce biofilm formation even at 1.25%. These results illustrated the potential of the strain to mitigate the formation of pathogenic biofilm (Figure 6C).
3.6.4 Prediction of bacteriocin in the LS-ARS2 genome
The anti-Salmonella effect of LS-ARS2 was eliminated in the CFS, in which the pH was neutralized, indicating the presence of pH-sensitive bacteriocins. This result motivated us to dig further into the LS-ARS2 genome in search of bacteriocin-encoding gene signatures. It was extremely exciting to find that one area of interest (AOI) region responsible for BCG was identified in the genome of LS-ARS2 (Figure 7). The BCG was located within contig 32.1 of the salivaricin_P_chain_b class. The AOI of contig 32.1 resides with salivaricin_P_chain_b (bit score = 134.806) and salivaricin_P_chain_a (bit score = 122.479) (Barrett et al., 2007; Elnar et al., 2025). Besides, LS-ARS2 contains various ORFs encoding miscellaneous bacteriocins (Tenea and Ascanta, 2022), accessory transport, and sensor proteins, as indicated in Figure 7 (Tenea and Ascanta, 2022).
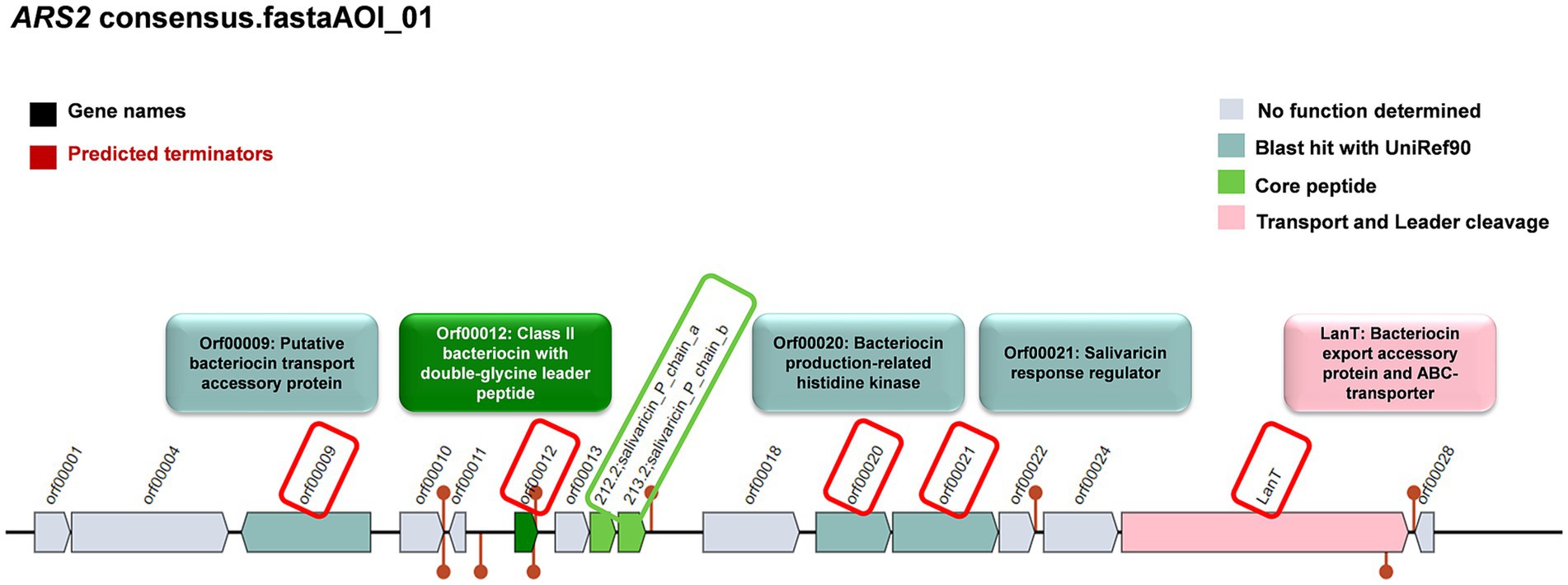
Figure 7. Prediction of genes for bacteriocin and accessory proteins in biosynthetic gene clusters (BCGs) organization in the genome of LS-ARS2 (BAGEL4 analysis).
3.7 Functional attributes of LS-ARS2: prediction from genomic analyses
3.7.1 Association of beneficial metabolite-producing pathways in the LS-ARS2 genome
The functional potential of LS-ARS2 was evaluated by annotating its coding sequences using the clusters of orthologous groups (COG) and KEGG databases. The KEGG annotation classified 1,016 CDS (64.1%) into 37 functional classes, 174 pathways, and 30 complete modules (Supplementary Tables S6–S8). Within the annotated KEGG functional categories, the LS-ARS2 genome showed 30 enriched KEGG pathways. Among them, metabolic pathways contained the highest gene count (approximately 40), followed by the pathway responsible for the biosynthesis of secondary metabolites (approximately 20). Further, the KEGG pathway related to microbial metabolism in diverse environments with an approximate gene count of 10 indicated the adaptation capability of LS-ARS2 in various niches. Moreover, the strain contained numerous genes involved in major metabolic pathways like amino acid biosynthesis, carbon and nucleotide metabolism, and biosynthesis of cofactors. Interestingly, abundant genes associated with essential amino acid metabolism (methionine, threonine, phenylalanine, and tryptophan) were detected in the LS-ARS2 genome, which indicated the potential health-benefits of the strain (Figure 8A).
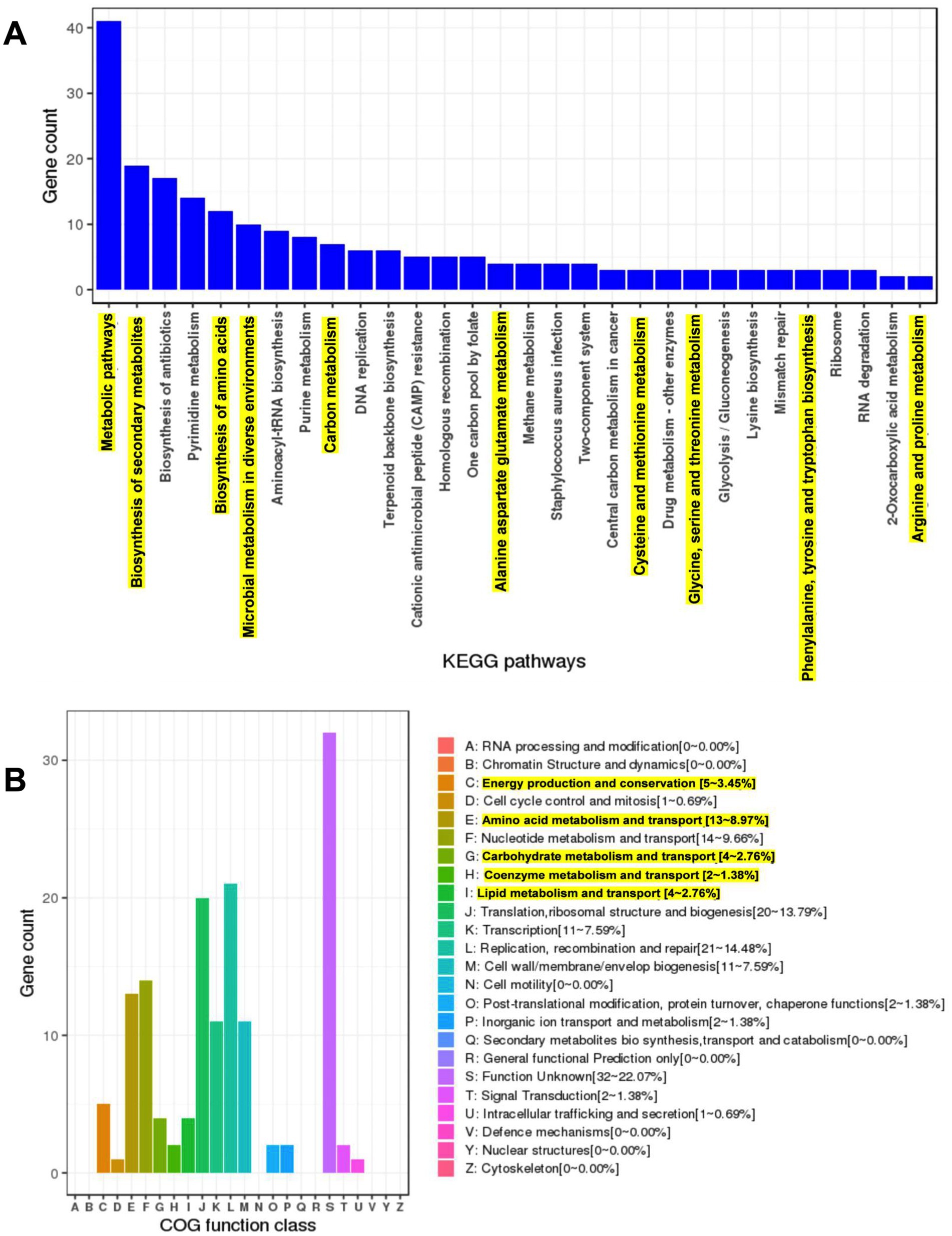
Figure 8. Genome mining of the LS-ARS2 genome and prediction of major pathways and COG categories. (A) The KEGG enriched pathways. Top 30 KEGG enriched pathways were analysed from the genome of LS-ARS2. Metabolic pathways, biosynthesis of secondary metabolites, and biosynthesis of antibiotics were three major pathways. Highlighted KEGG pathways showed the gene count of significant metabolite-producing pathways. (B) WGS analysis identified major COG categories in the LS-ARS2 genome. Highlighted COG categories showed the abundant gene count responsible for the production of beneficial metabolites.
Next, the protein-coding genes of the LS-ARS2 genome were found to be 1,586, among which 1,527 (96.28%) genes were classified into diverse COG functions. The annotated genes were found to be involved in several housekeeping physiological processes such as replication, recombination and repair (21–14.48%), translation, ribosomal structure and biogenesis (20–13.79%), nucleotide metabolism and transport (14–9.66%), amino acid metabolism and transport (13–8.97%), transcription (11–7.59%), cell wall/ membrane/ envelope biogenesis (11–7.59%), lipid metabolism and transport (4–2.76%), post-translational modification, protein turnover and chaperone functions (2–1.38%), inorganic ion transport and metabolism (2–1.38%), signal transduction (2–1.38%), cell cycle control and mitosis (1–0.69%), intracellular trafficking and secretion (1–0.69%). Functional annotation revealed the association of significant COG categories like energy production and conversion (5–3.45%), amino acid metabolism and transport (13–8.97%), carbohydrate metabolism and transport (4–2.76%), coenzyme metabolism and transport (2–1.38%), and lipid metabolism and transport (4–2.76%) (Figure 8B). Corroborating with the KEGG pathways, the COG analysis also showed the abundance of the genes that could be responsible for the production of beneficial metabolites (amino acids, vitamins) and amino acid or carbohydrate metabolism. The enrichment of metabolic genes in the LS-ARS2 genome further strengthened the potential of the strain to survive in diverse environments.
In-depth functional classification of the LS-ARS2 genome indicated the presence of genes involved in the metabolism of key sugars like glucose (n = 21), fructose and mannose (n = 18), galactose (n = 13), starch and sucrose (n = 18), amino sugar and nucleotide sugar (n = 23), and various other metabolic pathways (Supplementary Table S7). Apart from that, 94 coding sequences were identified for the metabolism of amino acids and derivatives. Among them, 58 were detected to be associated with the metabolism of essential amino acids like valine, isoleucine, leucine (n = 6), lysine (n = 13), methionine (n = 21), phenylalanine (n = 3), threonine (n = 13), tryptophan (n = 2) (Supplementary Table S7). Moreover, the KEGG functional annotation of the LS-ARS2 genome pointed out 55 coding sequences associated with the metabolism of cofactors and vitamins, including thiamine, riboflavin, nicotinate, vitamin B6, and many more. Such genomic and functional richness strengthens the potential application of LS-ARS2 as an enriched food supplement. Additionally, comprehensive functional annotation of the LS-ARS2 genome enabled to detection of the coding sequences associated with carbohydrate fermentation, resulting in the generation of short-chain fatty acids (SCFAs) such as pyruvate (n = 22), propanoate (n = 10), butanoate (n = 7). These carbohydrate metabolic pathways ultimately lead to the synthesis of key SCFAs (acetate, propionate, butyrate, pyruvate, lactate, succinate), characteristic of probiotic activity, underscoring the functional potential of LS-ARS2. It is noteworthy to mention here that the presence of genes associated with the metabolism pathways of diverse secondary molecules like terpenoids (n = 10) and polyketides (n = 4) in the LS-ARS2 genome emphasized the application of the strain in nutraceuticals and pharmaceutical industries (Supplementary Table S7).
3.7.2 Detection of carbohydrate-active enzymes (CAZymes)
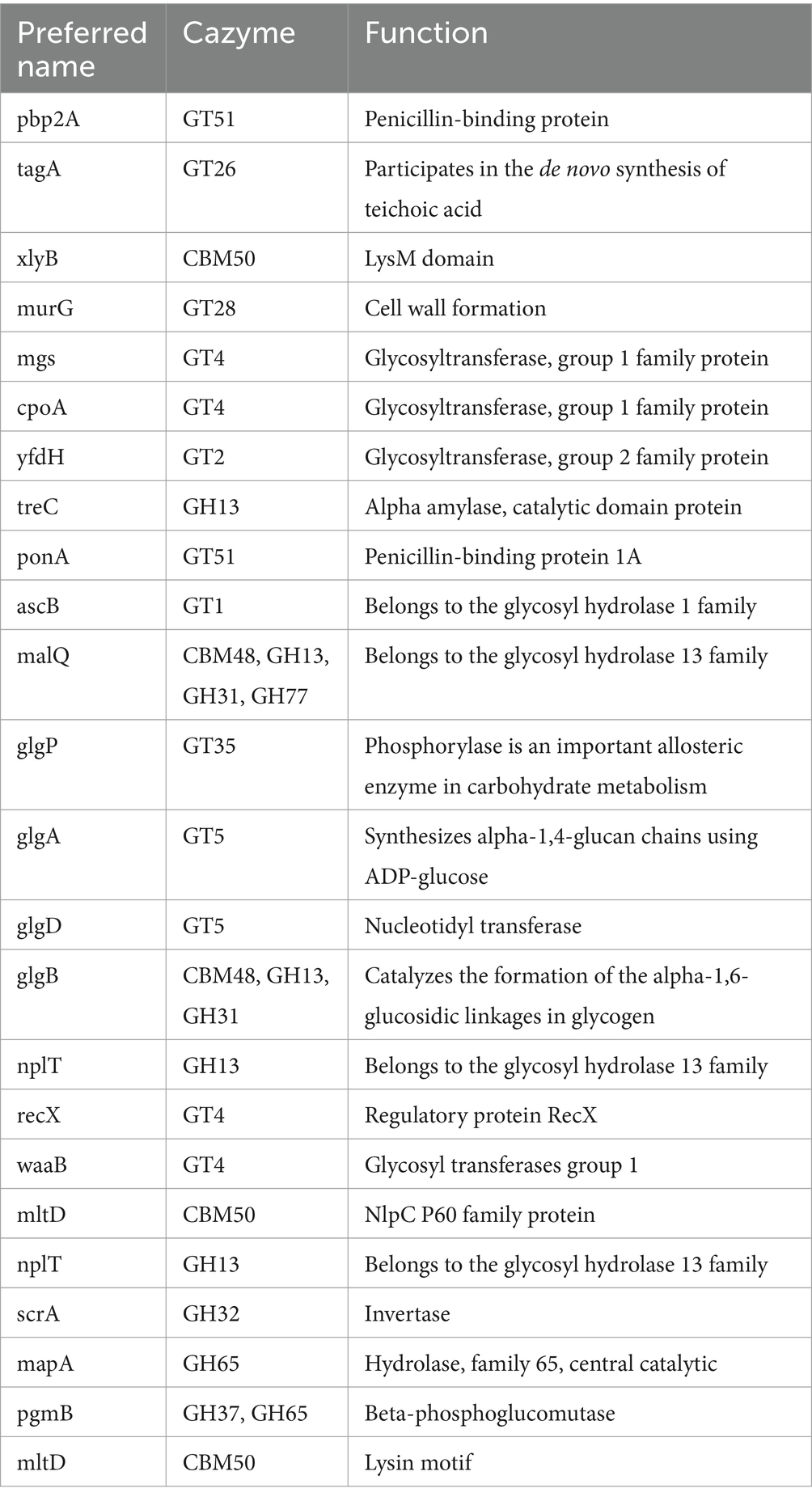
Carbohydrate active enzymes (CAZymes) enable bacteria to adapt to harsh environments by utilizing complex carbohydrate molecules (Liang et al., 2024). KEGG pathways and COG analysis revealed the abundance of genes associated with carbohydrate metabolism and transport, which could enable microbial metabolism in diverse environments. Functional classification of the LS-ARS2 genome indicated the presence of three pathways involved in carbohydrate fermentation: acetoin, butanediol metabolism, mixed acid fermentation, and lactate fermentation. Further mining into the LS-ARS2 genome revealed the presence of CAZymes that belonged to the glycoside hydrolase family (GH), glycosyltransferases family (GTs), and carbohydrate-binding modules (CBM) (Table 4). CAZymes of the GH family are responsible for the hydrolysis of complex carbohydrates. In the LS-ARS2 genome, GH13, GH31, GH77, GH32, GH65, and GH37 were detected, which represents 38.89% of the total CAZyme encoding genes (Supplementary Figure S3). Among these, GH31, GH32, and GH65 are reported to participate in the metabolism of monosaccharides and oligosaccharides such as glucose, galactose, fructose, xylose, or arabinose (Kavanova et al., 2024). Whereas CAZyme families GH13 and GH77 are involved in the hydrolysis of polysaccharides such as α-glucans, β-glucans, or arabinoxylans. The GH37 family is known to be associated with the degradation of trehalose (Shrestha et al., 2024). In addition, CAZymes involved in the transfer of sugar moieties are termed glycosyltransferases (GTs), which play a key role in forming surface structures sensed by the host immune system (Kandasamy et al., 2024). In the LS-ARS2 genome, GTs were the most abundantly annotated CAZymes (47.22%), which included GT1, GT2, GT4, GT5, GT26, GT28, GT35, and GT51. Moreover, two types of carbohydrate binding modules (CBM), CBM48 and CBM50, were detected in the LS-ARS2 genome, representing 13.89% of the CAZymes encoding genes. CBM helps in the binding of some carbohydrate hydrolases to augment the hydrolysis of polysaccharides. The functional annotation also revealed the presence of a few sugar transporter genes (pts13C, ptsH, ptsI) and the PTS system of mannose sorbose and fructose family sugars (manA, manL, manY, manN) in the LS-ARS2 genome. Additionally, PTS system genes of fructose family sugars (dhaK, dhaL, dhaM), and glucitol or sorbitol group of sugars (srlA, srlB, srlE, srlM) were also identified in the LS-ARS2 genome. Moreover, lactose and cellobiose-specific PTS systems like mtlA, mtlD, mtlF, and mtlR were identified in the genome of LS-ARS2. The presence of PTS system encoding genes indicated the strong carbon transporting ability of LS-ARS2, which is crucial to adapting multiple habitats (Kandasamy et al., 2024). Fascinatingly, genes encoding alcohol dehydrogenase, phosphoketolase, phosphate acetyltransferase, and pyruvate formate-lyase were identified in the LS-ARS2 genome, which is important for homo- or hetero-fermentative pathways making LS-ARS2 an attractive choice in the food industry (Kandasamy et al., 2024) (Supplementary Table S2).
3.8 Metabolomics of LS-ARS2: genomic prediction coupled with experimental validation by HRMS analysis
3.8.1 Prediction of primary and secondary metabolites in the LS-ARS2 genome
Microorganisms produce various substances (primary and secondary metabolites) that modulate host metabolism and immunity. Metabolic gene cluster (MGC) is defined as a cluster of genes that encodes different enzymes of the same metabolic pathway. The gutSMASH tool predicts genes involved in bioenergetics and metabolism in anaerobic bacteria (Pascal Andreu et al., 2021). In our study, gutSMASH found one MGC region, pyruvate to acetate-formate type (region 58.1), in the LS-ARS2 genome (Figure 9A). This region encodes for short-chain fatty acids (SCFAs), primarily acetate, butyrate, and propionate, which could have a positive impact on gut-health (Donia and Fischbach, 2015). The KnownClusterBlast output indicated high sequence similarity between the predicted LS-ARS2 MGC and other Lactobacillus strains (Figure 9B). Interestingly, among all the Lactobacillus strains, the highest gene similarity of the Pyruvate2acetate-formate region was found with other L. salivarius strains with the same region and known function. The KnownClusterBlast output thus strongly supports the potential production of primary metabolites (SCFAs) from LS-ARS2. The gutSMASH results nicely corroborate with the previous KEGG pathways and COG analysis, where abundant gene count was detected for metabolic pathways (specifically carbohydrate metabolism and transport) and energy production. The output of the gutSMASH further correlated with the functional prediction where several genes, like nagA, pdh, fabI, acc, poxB, gabD, and fadB4, associated with the SCFA biosynthesis were identified in the genome of LS-ARS2.
Figure 9. Prediction of genes for primary metabolites in Metabolic Gene Clusters (MGCs) of the LS-ARS2 genome. (A) GutSMASH run of the LS-ARS2 genome. GutSMASH predicted one MGC region, type pyruvate 2 acetate-formate, which is responsible for the production of SCFAs. (B) Gene Cluster Comparative Analysis. Comparative analysis between different bacterial reference genes (pre-computed in gutSMASH) for the most similar metabolic gene clusters (MGCs) based on a gutSMASH run. Genes with the same colour indicated putative homologs based on significant Blast hits between LS-ARS2 and the reference bacterial genes. The KnownClusterBlast analysis showed the gene similarity of the predicted region (Pyruvate2acetate-formate) with MGCs associated with known functions of other Lactobacillus reference genomes. The ClusterBlast analysis output showed that the predicted metabolic gene cluster (MGC) (Pyruvate2acetate-formate) does not have homologous MGCs among other Lactobacillus. White genes have no relationship. (C) Detection of beneficial metabolites in the LS-ARS2 CFS using HRMS analysis (also see Table 5 for details).
The KEGG pathway analysis indicated the richness of genes responsible for the biosynthesis of secondary metabolites, which are vital for the antibacterial, antifungal, and antitumor activities of the microorganisms (Tenea and Ascanta, 2022). The web server, antiSMASH, predicts NRPs, PKs, terpenes, and RiPP-like peptides with various known or putative secondary metabolites in the genome of bacteria. Fascinatingly, further genome mining of the LS-ARS2 genome by antiSMASH predicted one secondary metabolite-producing region (region 39.1) named T3PKS region (T3PKS: Chal_sti_synt_N, Chalcone and Stilbene synthases, N-terminal domain) within the LS-ARS2 genome (Supplementary Table S9). Polyketide synthases (PKSs) are related to the synthesis of antibiotics and pharmaceutical products and have various roles in food processing (Naughton et al., 2017). Microbial Type III PKSs (T3PKS) are involved in the biosynthesis of some lipidic compounds and various secondary metabolites with significant biological functions (Katsuyama and Ohnishi, 2012). Notably, the results from antiSMASH analysis aligned with genomic predictions from the KEGG pathways analysis, particularly those associated with the metabolism of terpenoids and polyketides (Supplementary Table S6).
3.8.2 Beneficial metabolites in LS-ARS2 CFS: HRMS analyses
The detailed genome mining and functional annotation of the LS-ARS2 genome showed the abundance of the genes responsible for synthesizing SCFAs, vitamins, essential amino acids, and cellular metabolites (Figures 8A,B, 9A,B; Supplementary Tables S2, S7). Next, we explored metabolic profiling of the strain to integrate WGS-based functional predictions with the HRMS-generated metabolites. The whole genome mining by gutSMASH tool identified the major SCFA-producing MGC, pyruvate2acetate-formate, in the LS-ARS2 genome. Further, the thorough investigation of the KEGG pathways and functional annotations of the LS-ARS2 genome predicted the gene signatures like acyP, pdhC, accC, accD, accB, fabD, pykfadB4, and argE, associated with the SCFAs biosynthesis (Supplementary Table S2). Notably, the HRMS analysis indicated the presence of a wide range of organic acids (for example, lactic acid, acetic acid, butyric acid, and homoserine) in the CFS of LS-ARS2, correlating metabolic pathways and gene signatures predicted in the genome. Several SCFAs, like butyric acid, isobutyric acid, lactic acid, acetic acid, and hydroxypropionic acid, were also prominent in CFS, aligning with the genomic identification of genes involved in SCFA biosynthesis (Figure 9C). These compounds have been shown to function as major antimicrobial components and immunomodulators (Dash et al., 2023). The detection of acetic acid, hydroxypropionate, butyric acid, and lactic acid in the metabolic profile strongly correlated with the genes encoding enzymes linked to glycolysis, pyruvate, propanoate, butanoate metabolism, and fermentation pathways. Additionally, the presence of many essential amino acids like leucine, isoleucine, valine, phenylalanine, threonine, and lysine in the CFS of LS-ARS2 was consistent with the genomic prediction of KEGG pathways involved in amino acid metabolism (Figure 8A; Supplementary Table S7) (see discussion). The WGS-genome mining identified KEGG pathways associated with the metabolism of vitamin B6 and nicotinate and nicotinamide (Supplementary Table S7). The functional annotation further underscored the gene signatures, including pdxK, nadD, birA, folA, thiT, adk, which are involved in vitamin metabolism. The identification of vitamins such as pyridoxal (vitamin B6) and nicotinic acid (vitamin B3) by metabolomic analysis in the LS-ARS2-derived CFS provided a robust correlation between the predicted metabolic pathways and gene signatures with the HRMS metabolite profiling (Table 5; Figure 9C). Therefore, our integrated genome mining and HRMS analysis beautifully validated the findings from the WGS analysis and confirmed the presence of beneficial metabolites in the CFS of LS-ARS2, reinforcing the functional capabilities of the strain.
4 Discussion
Despite their promising potential as probiotics, very few L. salivarius strains are available at an industrial scale as commercial products (Guerrero Sanchez et al., 2022). Our results illustrate that LS-ARS2 is a biofilm-former LAB, that exhibits significant antioxidant, antibacterial, as well as antibiofilm potential. Metabolic profiling of LS-ARS2-CFS indicates the presence of diverse health-promoting metabolites as well as other compounds with various applications as food, poultry, and cosmetic products. Therefore, LS-ARS2 appears as a promising candidate for diverse lucrative applications, particularly in food and therapeutic industries (see below for details).
First, the WGS analysis, together with the in vitro assays, establishes the probiotic attributes of LS-ARS2. The strain could potentially withstand the acidic and alkaline environments of the stomach and small intestine, respectively. Biofilm formation ability, aggregation efficacy, as well as adhesion and colonization potential, could facilitate LS-ARS2 for an efficient and longer stay in the gut. Moreover, the safety attributes of the strain are also assured, although at the in vitro level.
The dysbiotic gut often disrupts the intricate oxidative balance (McCord, 1985). Supraphysiological levels of all the reactive oxygen species (ROS), like hydrogen peroxide and superoxide anions, cause severe tissue damage and organ dysfunction (Barry, 1991). The excess level of ROS triggers chronic inflammation that results in gut-inflammatory diseases (ulcerative colitis, intestinal bowel syndrome, colorectal cancer) as well as diseases related to gut-organ axes (Ren et al., 2023). The significant antioxidant activities of LS-ARS2 reveal the potential of the strain to reduce oxidative stress in the gut. Interestingly, the functional annotation of the LS-ARS2 genome beautifully corroborates our in vitro investigations (Figure 4), indicating that supplementation of LS-ARS2 could contribute to the oxidative balance in the gut.
Chronic infections by pathogenic biofilms are clinically challenging. MRSA, particularly in biofilm form, is a leading cause of hospital-acquired infections that cause significant morbidity and mortality. MRSA-biofilm facilitates the pathogen to invade, spread, and resist antimicrobial treatments (Ciofu et al., 2022). In this background, it is extremely exciting to find that the CFS of LS-ARS2 inhibits MRSA-biofilm. Application of LS-ARS2 could therefore provide a green therapeutic window for combating pathogenic-biofilm.
WGS analysis predicts the presence of the MGC region (encodes SCFAs) in the genome of LS-ARS2. The HRMS study indicates the presence of diverse SCFAs (acetate, propionate, butyrate, lactic acid, etc.) in the CFS of LS-ARS2. Thus, genomic characterization coupled with metabolomic studies (Figure 9) confirms that, like other probiotic bacteria, SCFAs in the CFS of LS-ARS2 could offer beneficial roles to the host-gut. These SCFAs eliminate pathogens by entering through their cell membrane and acidification of their alkaline cytoplasm. Further, SCFAs have numerous health-promoting properties, including antibacterial, antiviral, immunomodulatory, and anticancer effects (Dash et al., 2023). Specifically, butyrate is an energy source for colonocytes (Pedroza Matute and Iyavoo, 2023). Therefore, intake of LS-ARS2 could facilitate maintaining gut-health.
Bacteriocins, produced by LAB, are ribosomally synthesized antimicrobial peptides. BAGEL4 analysis of the LS-ARS2 genome shows the presence of one class II bacteriocin-producing BCG (salivaricin_P_chain_b) and bacteriocin accessory as well as export proteins, which supports the probable presence of bacteriocin in the LS-ARS2 genome. Bacteriocins and associated proteins retain their activity preferably at an acidic pH rather than at an alkaline or neutral pH (Magnusson and Schnürer, 2001). In the present study, the antimicrobial effect of the LS-ARS2 CFS against Salmonella was not found for pH-neutralized CFS, indicating the possible existence of pH-sensitive bacteriocin-like peptides in the CFS.
HRMS study indicates LS-ARS2 CFS consists of metabolite betaine, which is an essential osmolyte and neuroprotectant with significant health-improving roles (Arumugam et al., 2021). LS-ARS2 CFS also consists of gamma-aminobutyric acid (GABA), one of the principal neurotransmitters. GABA significantly regulates nerve cell hyperactivity associated with anxiety, stress, convulsions, epilepsy, Parkinson’s, and Alzheimer’s diseases. Several studies have also reported the immunological and antimicrobial functions of GABA (Bs et al., 2021). Further, genomic analysis reveals the presence of CAZymes in the LS-ARS2 genome. Bacterial strains use CAZymes for carbohydrate degradation. Therefore, CAZymes in the LS-ARS2 not only could participate in utilizing complex carbohydrates and thrive in the host intestinal ecosystem; they also could further stimulate the growth of other probiotics by providing the carbon source for them. Therefore, as a nutritional supplement, LS-ARS2 with CAZymes could facilitate the overall good bacterial distribution and improve gut-health.
Food supplements are added to the diet to maintain the nutritional balance of the body. HRMS analysis indicates LS-ARS2 CFS is enriched with several beneficial metabolites that highlight the potential of the strain to be used as a dietary supplement. For instance, essential amino acids like threonine, lysine, phenylalanine, leucine, isoleucine, and valine are detected in the CFS of LS-ARS2. Next, vitamins nicotinic acid (vitamin B3) and pyridoxal (vitamin B6) are also found in LS-ARS2 CFS, indicating that application of the strain could reduce the severity of clinical vitamin deficiencies (Gasperi et al., 2019; Stach et al., 2021). Moreover, LS-ARS2 CFS is enriched with metabolites like pyruvic acid, oxaloacetate, and oxoglutaric acid, which are reported to have health-promoting effects (Dash et al., 2023; Wernerman et al., 1990).
Besides, LS-ARS2 CFS is also enriched with metabolites important for food processing and cosmetic industries. Metabolites like citric acid, dihydroxyacetone, and acetoin have extensive applications in the poultry, food, and cosmetic industries (Dash et al., 2023). In our genomic study, antiSMASH predicts the presence of the T3PKS region, which is known to encode substances with enormous functions in food processing industries. Other detected metabolites in LS-ARS2 CFS, such as adipic acid, can be used as food additives (Horn et al., 1957), whereas 3-Hydroxypropeonate, 2.3-butanediol, and malonic semialdehyde have diverse applications in the pharmaceutical, cosmetics, and agriculture industries (Zhao and Tian, 2021; Gu et al., 2022; Dash et al., 2023).
5 Conclusion
We present a full-fledged study on the probiotic attributes of LS-ARS2. (a) The remarkable anti-oxidant and anti-bacterial potential of LS-ARS2 encourages further studies of the strain for health-promoting applications. (b) The strong biofilm-forming ability of LS-ARS2 ensures efficient and longer stay in the GI tract. The significant anti-biofilm activity of LS-ARS2 CFS for MRSA again suggests the use of LS-ARS2 as a promising antimicrobial agent to inhibit pathogenic biofilm formation. (c) As a gut microbe, LS-ARS2 could naturally re-establish the gut-microbial composition and thereby improve the metabolic status of the gut and prevent dysbiosis. Further, the identification of beneficial metabolites in LS-ARS2-CFS with antimicrobial and immunomodulatory properties indicates the therapeutic application of LS-ARS2 in different diseases. However, in vivo studies with gut-inflammatory disease models would reaffirm that.
The potential ability of LS-ARS2 to stably colonize in the GI tract and other probiotic attributes, including beneficial metabolic profiling of LS-ARS2-CFS, indicate the promising application of the strain as a food supplement. Further, myriads of secondary metabolites with industrial applications were detected in LS-ARS2 CFS, which supports the potential use of the strain in the food as well as poultry and cosmetic industries. Together, the study indicates that LS-ARS2 demands further studies to determine its potential application in the future.
In summary, the study offers a novel and promising probiotic strain, LS-ARS2, that demands in vivo research, particularly focusing on gut-disease models for their potential applications in the near future.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article.
Author contributions
SP: Conceptualization, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing, Data curation, Software. BP: Data curation, Methodology, Software, Writing – review & editing. AR: Conceptualization, Supervision, Methodology, Formal analysis, Investigation, Validation, Visualization, Writing – original draft, Writing – review & editing, Funding acquisition, Resources, Project administration.
Funding
The author(s) declare financial support was received for the research and/or publication of this article. This study was supported by the intramural funding provided by IIT Bhubaneswar. SP receives an institutional fellowship from IIT Bhubaneswar. BP is supported by the endowment grant to IIT Bhubaneswar from the Dr. Dash Foundation, United States.
Acknowledgments
We are grateful to Dr. Nrisingha Dey, ILS Bhubaneswar, for providing us Gram-positive pathogens [Methicillin-resistant Staphylococcus aureus ATCC 700699 (MRSA) and Staphylococcus aureus ATCC 25923] used in this study. All the Gram-negative clinical isolates [Escherichia coli (ETEC) BCH 04067, Vibrio cholerae BCH 09616, Shigella Flexneri BCH 06745] were generous gifts from Dr. Asish Kumar Mukhopadhyay, NICED Kolkata. We are thankful to Dr. Tushar Kant Beuria and his group, ILS Bhubaneswar, for providing technical guidance for the biofilm assays. We would like to thank Prof. Surajit Mandal, West Bengal University of Animal and Fishery Sciences, Kolkata, for his suggestions on the surface hydrophobicity assay. We would like to convey our thanks to Prof. Chandan Goswami, NISER, Bhubaneswar, for providing the confocal microscopy facility.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The author(s) declare that no Gen AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fmicb.2025.1535388/full#supplementary-material
Footnotes
References
Abouloifa, H., Rokni, Y., Bellaouchi, R., Ghabbour, N., Karboune, S., Brasca, M., et al. (2020). Characterization of probiotic properties of antifungal Lactobacillus strains isolated from traditional fermenting green olives. Probiotics Antimicrob. Proteins 12, 683–696. doi: 10.1007/s12602-019-09543-8
Alba, C., Arroyo, R., Fernandez, L., Narbad, A., and Rodriguez, J. M. (2025). Characterization of a Ligilactobacillus salivarius strain isolated from a cheese seal which was last used in 1936. Food Secur. 13. doi: 10.3390/foods13132005
Arndt, D., Grant, J. R., Marcu, A., Sajed, T., Pon, A., Liang, Y., et al. (2016). PHASTER: a better, faster version of the PHAST phage search tool. Nucleic Acids Res. 44, W16–W21. doi: 10.1093/nar/gkw387
Arumugam, M. K., Paal, M. C., Donohue, T. M. Jr., Ganesan, M., Osna, N. A., and Kharbanda, K. K. (2021). Beneficial effects of betaine: a comprehensive review. Biology 10:456. doi: 10.3390/biology10060456
Ayala, D. I., Cook, P. W., Franco, J. G., Bugarel, M., Kottapalli, K. R., Loneragan, G. H., et al. (2019). A systematic approach to identify and characterize the effectiveness and safety of novel probiotic strains to control foodborne pathogens. Front. Microbiol. 10:1108. doi: 10.3389/fmicb.2019.01108
Ayed, L., M’hir, S., Nuzzolese, D., Di Cagno, R., and Filannino, P. (2024). Harnessing the health and techno-functional potential of lactic acid Bacteria: a comprehensive review. Food Secur. 13:1538. doi: 10.3390/foods13101538
Baldassarri, L., Creti, R., Recchia, S., Imperi, M., Facinelli, B., Giovanetti, E., et al. (2006). Therapeutic failures of antibiotics used to treat macrolide-susceptible Streptococcus pyogenes infections may be due to biofilm formation. J. Clin. Microbiol. 44, 2721–2727. doi: 10.1128/jcm.00512-06
Barrett, E., Hayes, M., O'Connor, P., Gardiner, G., Fitzgerald, G. F., Stanton, C., et al. (2007). Salivaricin P, one of a family of two-component antilisterial bacteriocins produced by intestinal isolates of Lactobacillus salivarius. Appl. Environ. Microbiol. 73, 3719–3723. doi: 10.1128/AEM.00666-06
Barry, H. (1991). Reactive oxygen species in living systems. Am. J. Med. 91, S14–S22. doi: 10.1016/0002-9343(91)90279-7
Blin, K., Shaw, S., Kloosterman, A. M., Charlop Powers, Z., Van Wezel, G. P., Medema, M. H., et al. (2021). antiSMASH 6.0: improving cluster detection and comparison capabilities. Nucleic Acids Res. 49, W29–W35. doi: 10.1093/nar/gkab335
Bortolaia, V., Kaas, R. S., Ruppe, E., Roberts, M. C., Schwarz, S., Cattoir, V., et al. (2020). ResFinder 4.0 for predictions of phenotypes from genotypes. J. Antimicrob. Chemother. 75, 3491–3500. doi: 10.1093/jac/dkaa345
Bs, S., Thankappan, B., Mahendran, R., Muthusamy, G., Femil Selta, D. R., and Angayarkanni, J. (2021). Evaluation of GABA production and probiotic activities of Enterococcus faecium BS5. Probiotics Antimicrob. Proteins 13, 993–1004. doi: 10.1007/s12602-021-09759-7
Campana, R., van Hemert, S., and Baffone, W. (2017). Strain-specific probiotic properties of lactic acid bacteria and their interference with human intestinal pathogens invasion. Gut Pathog. 9:12. doi: 10.1186/s13099-017-0162-4
Chen, C. C., Lai, C. C., Huang, H. L., Huang, W. Y., Toh, H. S., Weng, T. C., et al. (2019). Antimicrobial activity of Lactobacillus species against carbapenem-resistant Enterobacteriaceae. Front. Microbiol. 10:789. doi: 10.3389/fmicb.2019.00789
Ciofu, O., Moser, C., Jensen, P. Ø., and Høiby, N. (2022). Tolerance and resistance of microbial biofilms. Nat. Rev. Microbiol. 20, 621–635. doi: 10.1038/s41579-022-00682-4
CLSI (2020). Clinical and laboratory standards institute. Performance standards for antimicrobial susceptibility testing. 30th Edn.
Cosentino, S., Voldby Larsen, M., Moller Aarestrup, F., and Lund, O. (2013). PathogenFinder--distinguishing friend from foe using bacterial whole genome sequence data. PLoS One 8:e77302. doi: 10.1371/journal.pone.0077302
Dash, J., Sethi, M., Deb, S., Parida, D., Kar, S., Mahapatra, S., et al. (2023). Biochemical, functional and genomic characterization of a new probiotic Ligilactobacillus salivarius F14 from the gut of tribes of Odisha. World J. Microbiol. Biotechnol. 39:171. doi: 10.1007/s11274-023-03626-z
de Jong, A., van Hijum, S. A., Bijlsma, J. J., Kok, J., and Kuipers, O. P. (2006). BAGEL: a web-based bacteriocin genome mining tool. Nucleic Acids Res. 34, W273–W279. doi: 10.1093/nar/gkl237
Deghorain, M., Goffin, P., Fontaine, L., Mainardi, J. L., Daniel, R., Errington, J., et al. (2007). Selectivity for D-lactate incorporation into the peptidoglycan precursors of Lactobacillus plantarum: role of Aad, a VanX-like D-alanyl-D-alanine dipeptidase. J. Bacteriol. 189, 4332–4337. doi: 10.1128/jb.01829-06
Dietzsch, S., Braesigk, A., Seidel, C., Remmele, J., Kitzing, R., Schlender, T., et al. (2021). Types of deviation and review criteria in pretreatment central quality control of tumor bed boost in medulloblastoma—an analysis of the German radiotherapy quality control panel in the SIOP PNET5 MB trial. Strahlenther. Onkol. 198, 282–290. doi: 10.1007/s00066-021-01822-0
Donia, M. S., and Fischbach, M. A. (2015). Small molecules from the human microbiota. Science 349:1254766. doi: 10.1126/science.1254766
Elnar, A., Jang, Y., Eum, B., Kang, M., Hwang, G., Kil, D., et al. (2025). Distinct phenotypes of salivaricin-producing Ligilactobacillus salivarius isolated from the gastrointestinal tract of broiler chickens and laying hens. Poult. Sci. 104:104537. doi: 10.1016/j.psj.2024.104537
Fonseca, H. C., de Sousa Melo, D., Ramos, C. L., Dias, D. R., and Schwan, R. F. (2021). Probiotic properties of lactobacilli and their ability to inhibit the adhesion of enteropathogenic bacteria to Caco-2 and HT-29 cells. Probiotics Antimicrob. Proteins 13, 102–112. doi: 10.1007/s12602-020-09659-2
Furman, D., Campisi, J., Verdin, E., Carrera Bastos, P., Targ, S., Franceschi, C., et al. (2019). Chronic inflammation in the etiology of disease across the life span. Nat. Med. 25, 1822–1832. doi: 10.1038/s41591-019-0675-0
Gao, D., Gao, Z., and Zhu, G. (2013). Antioxidant effects of Lactobacillus plantarum via activation of transcription factor Nrf2. Food Funct. 4, 982–989. doi: 10.1039/C3FO30316K
Gao, J., Sadiq, F. A., Zheng, Y., Zhao, J., He, G., and Sang, Y. (2022). Biofilm-based delivery approaches and specific enrichment strategies of probiotics in the human gut. Gut Microbes 14:2126274. doi: 10.1080/19490976.2022.2126274
Gasperi, V., Sibilano, M., Savini, I., and Catani, M. V. (2019). Niacin in the central nervous system: an update of biological aspects and clinical applications. Int. J. Mol. Sci. 20:974. doi: 10.3390/ijms20040974
Grissa, I., Vergnaud, G., and Pourcel, C. (2007). CRISPRFinder: a web tool to identify clustered regularly interspaced short palindromic repeats. Nucleic Acids Res. 35, W52–W57. doi: 10.1093/nar/gkm360
Gu, S., Zhao, Z., Yao, Y., Li, J., and Tian, C. (2022). Designing and constructing a novel artificial pathway for malonic acid production biologically. Front. Bioeng. Biotechnol. 9:820507. doi: 10.3389/fbioe.2021.820507
Guerrero Sanchez, M., Passot, S., Campoy, S., Olivares, M., and Fonseca, F. (2022). Ligilactobacillus salivarius functionalities, applications, and manufacturing challenges. Appl. Microbiol. Biotechnol. 106, 57–80. doi: 10.1007/s00253-021-11694-0
Hakansson, A., and Molin, G. (2011). Gut microbiota and inflammation. Nutrients 3, 637–682. doi: 10.3390/nu3060637
Heunis, T., Deane, S., Smit, S., and Dicks, L. M. (2014). Proteomic profiling of the acid stress response in Lactobacillus plantarum 423. J. Proteome Res. 13, 4028–4039. doi: 10.1021/pr500353x
Horn, H. J., Holland, E., and Hazleton, L. (1957). Food additives, safety of Adipic acid as compared with citric and tartaric acid. J. Agric. Food Chem. 5, 759–762. doi: 10.1021/jf60080a007
Hsiung, R. T., Fang, W. T., LePage, B. A., Hsu, S. A., Hsu, C. H., and Chou, J. Y. (2021). In vitro properties of potential probiotic indigenous yeasts originating from fermented food and beverages in Taiwan. Probiotics Antimicrob. Proteins 13, 113–124. doi: 10.1007/s12602-020-09661-8
Jia, B., Raphenya, A. R., Alcock, B., Waglechner, N., Guo, P., Tsang, K. K., et al. (2016). CARD 2017: expansion and model-centric curation of the comprehensive antibiotic resistance database. Nucleic Acids Res. 45, D566–D573. doi: 10.1093/nar/gkw1004
Jiang, Y. H., Yang, R. S., Lin, Y. C., Xin, W. G., Zhou, H. Y., Wang, F., et al. (2023). Assessment of the safety and probiotic characteristics of Lactobacillus salivarius CGMCC20700 based on whole-genome sequencing and phenotypic analysis. Front. Microbiol. 14:1120263. doi: 10.3389/fmicb.2023.1120263
Kandasamy, S., Lee, K. H., Yoo, J., Yun, J., Kang, H. B., Kim, J. E., et al. (2024). Whole genome sequencing of Lacticaseibacillus casei KACC92338 strain with strong antioxidant activity, reveals genes and gene clusters of probiotic and antimicrobial potential. Front. Microbiol. 15:1458221. doi: 10.3389/fmicb.2024.1458221
Kanehisa, M., Sato, Y., and Morishima, K. (2016). BlastKOALA and GhostKOALA: KEGG tools for functional characterization of genome and metagenome sequences. J. Mol. Biol. 428, 726–731. doi: 10.1016/j.jmb.2015.11.006
Katsuyama, Y., and Ohnishi, Y. (2012). “Type III polyketide synthases in microorganisms” in Methods Enzymology, vol. 515 (Elsevier), 359–377.
Kaushik, A., Kest, H., Sood, M., Steussy, B. W., Thieman, C., and Gupta, S. (2024). Biofilm producing methicillin-resistant Staphylococcus aureus (MRSA) infections in humans: clinical implications and management. Pathogens 13:76. doi: 10.3390/pathogens13010076
Kavanova, K., Kostovova, I., Moravkova, M., Kubasova, T., Babak, V., and Crhanova, M. (2024). Comparative genome analysis and characterization of the probiotic properties of lactic acid Bacteria isolated from the gastrointestinal tract of wild boars in the Czech Republic. Probiotics Antimicrob. Proteins 1-19. doi: 10.1007/s12602-024-10259-7
Kim, S., Lee, J. Y., Jeong, Y., and Kang, C. H. (2022). Antioxidant activity and probiotic properties of lactic acid bacteria. Fermentation 8:29. doi: 10.3390/fermentation8010029
Kumar, S., Stecher, G., Suleski, M., Sanderford, M., Sharma, S., and Tamura, K. (2024). MEGA12: molecular evolutionary genetic analysis version 12 for adaptive and green computing. Mol. Biol. Evol. 41:msae263. doi: 10.1093/molbev/msae263
Lebeer, S., Vanderleyden, J., and De Keersmaecker, S. C. (2008). Genes and molecules of lactobacilli supporting probiotic action. Microbiol. Mol. Biol. Rev. 72, 728–764. doi: 10.1128/mmbr.00017-08
Lee, J. E., Lee, N. K., and Paik, H. D. (2021). Antimicrobial and anti-biofilm effects of probiotic Lactobacillus plantarum KU200656 isolated from kimchi. Food Sci. Biotechnol. 30, 97–106. doi: 10.1007/s10068-020-00837-0
Lei, Y., Oshima, T., Ogasawara, N., and Ishikawa, S. (2013). Functional analysis of the protein veg, which stimulates biofilm formation in Bacillus subtilis. J. Bacteriol. 195, 1697–1705. doi: 10.1128/jb.02201-12
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with burrows–wheeler transform. Bioinformatics 25, 1754–1760. doi: 10.1093/bioinformatics/btp324
Liang, X., Dai, N., Yang, F., Zhu, H., Zhang, G., and Wang, Y. (2024). Molecular identification and safety assessment of the potential probiotic strain Bacillus paralicheniformis HMPM220325 isolated from artisanal fruit dairy products. Food Funct. 15, 747–765. doi: 10.1039/D3FO04625G
Liu, B., Zheng, D., Jin, Q., Chen, L., and Yang, J. (2019). VFDB 2019: a comparative pathogenomic platform with an interactive web interface. Nucleic Acids Res. 47, D687–D692. doi: 10.1093/nar/gky1080
Magnusson, J., and Schnürer, J. (2001). Lactobacillus coryniformis subsp. coryniformis strain Si3 produces a broad-spectrum proteinaceous antifungal compound. Appl. Environ. Microbiol. 67, 1–5. doi: 10.1128/AEM.67.1.1-5.2001
McCord, J. M. (1985). Oxygen-derived free radicals in postischemic tissue injury. N. Engl. J. Med. 312, 159–163. doi: 10.1056/NEJM198501173120305
McFarland, L. V., Evans, C. T., and Goldstein, E. J. (2018). Strain-specificity and disease-specificity of probiotic efficacy: a systematic review and meta-analysis. Front. Med. 5:124. doi: 10.3389/fmed.2018.00124
Mgomi, F. C., Yang, Y. R., Cheng, G., and Yang, Z. Q. (2023). Lactic acid bacteria biofilms and their antimicrobial potential against pathogenic microorganisms. Biofilms 5:100118. doi: 10.1016/j.bioflm.2023.100118
Naughton, L. M., Romano, S., O’Gara, F., and Dobson, A. D. (2017). Identification of secondary metabolite gene clusters in the Pseudovibrio genus reveals encouraging biosynthetic potential toward the production of novel bioactive compounds. Front. Microbiol. 8:1494. doi: 10.3389/fmicb.2017.01494
Navarré, A., Nazareth, T., Luz, C., Meca, G., and Escrivá, L. (2024). Characterization of lactic acid bacteria isolated from human breast milk and their bioactive metabolites with potential application as a probiotic food supplement. Food Funct. 15, 8087–8103. doi: 10.1039/D4FO02171A
Oien, D. B., and Moskovitz, J. (2019). Genetic regulation of longevity and age-associated diseases through the methionine sulfoxide reductase system. Biochim. Biophys. Acta Mol. basis Dis. 1865, 1756–1762. doi: 10.1016/j.bbadis.2018.11.016
Pascal Andreu, V., Roel Touris, J., Dodd, D., Fischbach, M. A., and Medema, M. H. (2021). The gutSMASH web server: automated identification of primary metabolic gene clusters from the gut microbiota. Nucleic Acids Res. 49, W263–W270. doi: 10.1093/nar/gkab353
Patra, S., Sahu, N., Saxena, S., Pradhan, B., Nayak, S. K., and Roychowdhury, A. (2022). Effects of probiotics at the interface of metabolism and immunity to prevent colorectal cancer-associated gut inflammation: a systematic network and meta-analysis with molecular docking studies. Front. Microbiol. 13:878297. doi: 10.3389/fmicb.2022.878297
Patra, S., Saxena, S., Sahu, N., Pradhan, B., and Roychowdhury, A. (2021). Systematic network and meta-analysis on the antiviral mechanisms of probiotics: a preventive and treatment strategy to mitigate SARS-CoV-2 infection. Probiotics Antimicrob. Proteins 13, 1138–1156. doi: 10.1007/s12602-021-09748-w
Pedroza Matute, S., and Iyavoo, S. (2023). Exploring the gut microbiota: lifestyle choices, disease associations, and personal genomics. Front. Nutr. 10:1225120. doi: 10.3389/fnut.2023.1225120
Pei, Z., Sadiq, F. A., Han, X., Zhao, J., Zhang, H., Ross, R. P., et al. (2021). Comprehensive scanning of prophages in Lactobacillus: distribution, diversity, antibiotic resistance genes, and linkages with CRISPR-Cas systems. Msystems 6, e01211–e01220. doi: 10.1128/msystems.01211-20
Pereira, J., Castro, M. M., Santos, F., Jesus, A. R., Paiva, A., Oliveira, F., et al. (2022). Selective terpene based therapeutic deep eutectic systems against colorectal cancer. Eur. J. Pharm. Biopharm. 175, 13–26. doi: 10.1016/j.ejpb.2022.04.008
Piqué, N., Berlanga, M., and Miñana Galbis, D. (2019). Health benefits of heat-killed (Tyndallized) probiotics: an overview. Int. J. Mol. Sci. 20:2534. doi: 10.3390/ijms20102534
Pradhan, B., Guha, D., Ray, P., Das, D., and Aich, P. (2016). Comparative analysis of the effects of two probiotic bacterial strains on metabolism and innate immunity in the RAW 264.7 murine macrophage cell line. Probiotics Antimicrob. Proteins 8, 73–84. doi: 10.1007/s12602-016-9211-4
Quilodrán-Vega, S., Albarracin, L., Mansilla, F., Arce, L., Zhou, B., Islam, M. A., et al. (2020). Functional and genomic characterization of Ligilactobacillus salivarius TUCO-L2 isolated from Lama glama milk: a promising immunobiotic strain to combat infections. Front. Microbiol. 11:608752. doi: 10.3389/fmicb.2020.608752
Ramalho, J. B., Soares, M. B., Spiazzi, C. C., Bicca, D. F., Soares, V. M., Pereira, J. G., et al. (2019). In vitro probiotic and antioxidant potential of Lactococcus lactis subsp. cremoris LL95 and its effect in mice behaviour. Nutrients 11:901. doi: 10.3390/nu11040901
Ren, J., Li, H., Zeng, G., Pang, B., Wang, Q., and Wei, J. (2023). Gut microbiome-mediated mechanisms in aging-related diseases: are probiotics ready for prime time? Front. Pharmacol. 14:1178596. doi: 10.3389/fphar.2023.1178596
Roberts, A., Spang, D., Sanozky-Dawes, R., Nethery, M. A., and Barrangou, R. (2024). Characterization of Ligilactobacillus salivarius CRISPR-Cas systems. Msphere 9, e00171–e00124. doi: 10.1128/msphere.00171-24
Rocchetti, M. T., Russo, P., De Simone, N., Capozzi, V., Spano, G., and Fiocco, D. (2024). Immunomodulatory activity on human macrophages by cell-free supernatants to explore the probiotic and postbiotic potential of Lactiplantibacillus plantarum strains of plant origin. Probiotics Antimicrob. Proteins 16, 911–926. doi: 10.1007/s12602-023-10084-4
Rosenberg, M., Gutnick, D., and Rosenberg, E. (1980). Adherence of bacteria to hydrocarbons: a simple method for measuring cell-surface hydrophobicity. FEMS Microbiol. Lett. 9, 29–33. doi: 10.1111/j.1574-6968.1980.tb05599.x
Rychen, G., Aquilina, G., Azimonti, G., Bampidis, V., de Lourdes Bastos, M., Bories, G., et al. (2018). Guidance on the characterisation of microorganisms used as feed additives or as production organisms. EFSA J. 16:e05206. doi: 10.2903/j.efsa.2018.5206
Salas-Jara, M. J., Ilabaca, A., Vega, M., and García, A. (2016). Biofilm forming Lactobacillus: new challenges for the development of probiotics. Microorganisms 4:35. doi: 10.3390/microorganisms4030035
Saxami, G., Kerezoudi, E. N., Eliopoulos, C., Arapoglou, D., and Kyriacou, A. (2023). The gut–organ axis within the human body: gut dysbiosis and the role of prebiotics. Life 13:2023. doi: 10.3390/life13102023
Seemann, T. (2014). Prokka: rapid prokaryotic genome annotation. Bioinformatics 30, 2068–2069. doi: 10.1093/bioinformatics/btu153
Sethi, M., Ok, A., Dash, J., Parida, D., Kar, S., Mishra, S., et al. (2024). Whole genome mining and characterization of a new probiotic strain Levilactobacillus brevis ILSH3 from Handia: an ethnic fermented beverage of Odisha, India. Probiotics Antimicrob Proteins 12, 1–9. doi: 10.1007/s12602-024-10217-3
Sevillano, E., Lafuente, I., Peña, N., Cintas, L. M., Muñoz-Atienza, E., Hernández, P. E., et al. (2023). Evaluation of safety and probiotic traits from a comprehensive genome-based in silico analysis of Ligilactobacillus salivarius P1CEA3, isolated from pigs and producer of Nisin S. Food Secur. 13:107. doi: 10.3390/foods13010107
Shrestha, P., Karmacharya, J., Han, S. R., Lee, J. H., and Oh, T. J. (2024). Elucidation of bacterial trehalose-degrading trehalase and trehalose phosphorylase: physiological significance and its potential applications. Glycobiology 34:cwad084. doi: 10.1093/glycob/cwad084
Stach, K., Stach, W., and Augoff, K. (2021). Vitamin B6 in health and disease. Nutrients 13:3229. doi: 10.3390/nu13093229
Staskawicz, B., Panopoulos, N., and Hoogenraad, N. (1980). Phaseolotoxin-insensitive ornithine carbamoyltransferase of Pseudomonas syringae pv. Phaseolicola: basis for immunity to phaseolotoxin. J. Bacteriol. 142, 720–723. doi: 10.1128/jb.142.2.720-723.1980
Tenea, G. N., and Ascanta, P. (2022). Bioprospecting of ribosomally synthesized and post-translationally modified peptides through genome characterization of a novel probiotic Lactiplantibacillus plantarum UTNGt21A strain: a promising natural antimicrobials factory. Front. Microbiol. 13:868025. doi: 10.3389/fmicb.2022.868025
Tenea, G. N., and Ortega, C. (2021). Genome characterization of Lactiplantibacillus plantarum strain UTNGt2 originated from Theobroma grandiflorum (white cacao) of Ecuadorian Amazon: antimicrobial peptides from safety to potential applications. Antibiotics 10:383. doi: 10.3390/antibiotics10040383
Timmerman, H., Veldman, A., Van den Elsen, E., Rombouts, F., and Beynen, A. (2006). Mortality and growth performance of broilers given drinking water supplemented with chicken-specific probiotics. Poult. Sci. 85, 1383–1388. doi: 10.1093/ps/85.8.1383
Wernerman, J., Hammarqvist, F., and Vinnars, E. (1990). α-Ketoglutarate and postoperative muscle catabolism. Lancet 335, 701–703. doi: 10.1016/0140-6736(90)90811-I
Wong, A., Ngu, D. Y. S., Dan, L. A., Ooi, A., and Lim, R. L. H. (2015). Detection of antibiotic resistance in probiotics of dietary supplements. Nutr. J. 14, 95–96. doi: 10.1186/s12937-015-0084-2
Xiong, Z. Q., Kong, L. H., Wang, G. Q., Xia, Y. J., Zhang, H., Yin, B. X., et al. (2018). Functional analysis and heterologous expression of bifunctional glutathione synthetase from Lactobacillus. J. Dairy Sci. 101, 6937–6945. doi: 10.3168/jds.2017-14142
Xiong, L., Ni, X., Niu, L., Zhou, Y., Wang, Q., Khalique, A., et al. (2019). Isolation and preliminary screening of a Weissella confusa strain from giant panda (Ailuropoda melanoleuca). Probiotics Antimicrob. Proteins 11, 535–544. doi: 10.1007/s12602-018-9402-2
Yang, J., Shang, P., Zhang, B., Wang, J., Du, Z., Wang, S., et al. (2023). Genomic and metabonomic methods reveal the probiotic functions of swine-derived Ligilactobacillus salivarius. BMC Microbiol. 23:242. doi: 10.1186/s12866-023-02993-9
Yang, Y., Song, X., Wang, G., Xia, Y., Xiong, Z., and Ai, L. (2024). Understanding Ligilactobacillus salivarius from probiotic properties to omics technology: a review. Food Secur. 13:895. doi: 10.3390/foods13060895
Zankari, E., Hasman, H., Cosentino, S., Vestergaard, M., Rasmussen, S., Lund, O., et al. (2012). Identification of acquired antimicrobial resistance genes. J. Antimicrob. Chemother. 67, 2640–2644. doi: 10.1093/jac/dks261
Zhang, W., Ji, H., Zhang, D., Liu, H., Wang, S., Wang, J., et al. (2018). Complete genome sequencing of Lactobacillus plantarum ZLP001, a potential probiotic that enhances intestinal epithelial barrier function and defense against pathogens in pigs. Front. Physiol. 9:1689. doi: 10.3389/fphys.2018.01689
Zhao, P., and Tian, P. (2021). Biosynthesis pathways and strategies for improving 3-hydroxypropionic acid production in bacteria. World J. Microbiol. Biotechnol. 37:117. doi: 10.1007/s11274-021-03091-6
Zheng, J., Ge, Q., Yan, Y., Zhang, X., Huang, L., and Yin, Y. (2023). dbCAN3: automated carbohydrate-active enzyme and substrate annotation. Nucleic Acids Res. 51, W115–W121. doi: 10.1093/nar/gkad328
Zhou, B., Albarracin, L., Indo, Y., Arce, L., Masumizu, Y., Tomokiyo, M., et al. (2020). Selection of immunobiotic Ligilactobacillus salivarius strains from the intestinal tract of wakame-fed pigs: functional and genomic studies. Microorganisms 8:1659. doi: 10.3390/microorganisms8111659
Glossary
GI – gastrointestinal
WHO – World Health Organization
WGS – Whole Genome Sequencing
ATCC – American Type Culture Collection
MRS – De Man Rogosa Sharpe agar
DMEM – Dulbecco’s Modified Eagle Medium
FBS – Fetal Bovine Serum
MTCC – Microbial Type Culture Collection
PBS – Phosphate Buffered Saline
MOI – Multiplicity Of Infection
SS – Salmonella Shigella
DPPH – 1,1-diphenyl-2-picrylhydrazyl
ABTS – 2,2′-azino-bis (3-ethylbenzothiazoline-6-sulfonic acid)
PCR – polymerase chain reaction
CFU – colony forming unit
ANI – Average Nucleotide Identity
CRISPR – Clustered Regularly Interspaced Short Palindromic Repeats
SCFA – short-chain fatty acid
PKs – Polyketide Synthases
NRPs – Nonribosomal peptides
RiPP – Ribosomally synthesized and post-translationally modified peptides
Keywords: probiotics, microbial dysbiosis, oxidative stress, whole genome sequencing analysis, biofilm formation and anti-biofilm potential, metabolic profile analysis, food supplement, biotherapeutic
Citation: Patra S, Pradhan B and Roychowdhury A (2025) Complete genome sequence, metabolic profiling and functional studies reveal Ligilactobacillus salivarius LS-ARS2 is a promising biofilm-forming probiotic with significant antioxidant, antibacterial, and antibiofilm potential. Front. Microbiol. 16:1535388. doi: 10.3389/fmicb.2025.1535388
Edited by:
K. B. Arun, Christ University, IndiaReviewed by:
Kumaraswamy Jeyaram, Institute of Bioresources and Sustainable Development (IBSD), IndiaChand Ram, National Dairy Research Institute (ICAR), India
Copyright © 2025 Patra, Pradhan and Roychowdhury. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Anasuya Roychowdhury, YXJveWNob3dkaHVyeUBpaXRiYnMuYWMuaW4=
 Sinjini Patra
Sinjini Patra Biswaranjan Pradhan
Biswaranjan Pradhan Anasuya Roychowdhury
Anasuya Roychowdhury